Abstract
During the study of lignicolous freshwater fungi from Thailand, three pleurotheciaceous species were collected from freshwater habitats in Thailand. Two were identified as Pleurotheciumaquaticum and Rhexoacrodictysfimicola, and the third is a new species Dematipyriformamuriformis sp. nov.. Rhexoacrodictys is accepted in Pleurotheciaceae based on phylogenetic analysis. Rhexoacrodictysnigrospora is transferred to Dematipyriforma based on phylogenetic analysis and morphological characters. Pleurotheciumaquaticum and Rhexoacrodictysfimicola are reported from Thailand for the first time.
Keywords: 1 new combination, 1 new taxon, freshwater fungi, phylogeny, Pleurotheciales, taxonomy
Introduction
Pleurotheciales was introduced by Réblová et al. (2016) to accommodate a single family Pleurotheciaceae. The order was originally placed in Hypocreomycetidae by Réblová et al. (2016). Hongsanan et al. (2017) showed that Pleurotheciales clustered with Conioscyphales, Fuscosporellales and Savoryellales in a monophyletic clade within Sordariomycetes. Hence, they transferred Pleurotheciales to a newly introduced subclass Savoryellomycetidae based on phylogenetic analysis and the placement has been confirmed and accepted by Dayarathne et al. (2019) and Hyde et al. (2020a).
Pleurotheciaceae was introduced by Réblová et al. (2016) with Pleurothecium Höhn. as the type genus. Currently, Adelosphaeria, Anapleurothecium, Coleodictyospora, Dematipyriforma, Helicoascotaiwania, Melanotrigonum, Neomonodictys, Phaeoisaria, Phragmocephala, Pleurotheciella, Pleurothecium, Saprodesmium, and Sterigmatobotrys are accepted in this family (Hyde et al. 2020a; Wijayawardene et al. 2020; Dong et al. 2021). The sexual morphs of Pleurotheciaceae share dark, papillate, perithecial, astromatic, immersed to superficial ascomata, unitunicate asci with a distinct non-amyloid apical annulus, and fusiform to ellipsoidal, septate, hyaline ascospores (Réblová et al. 2016; Luo et al. 2018a; Hyde et al. 2020a). The asexual morphs of Pleurotheciaceae are diverse in morphology, comprising acrodictys-like (Monotosporella), (Hyde and Yanna 2002; Sadowski et al. 2012), helicoön-like (Helicoascotaiwania, Dayarathne et al. 2019; Réblová et al. 2020), monodictys-like (Neomonodictys, Hyde et al. 2020b) and dactylaria-like taxa (Pleurotheciella, Phaeoisaria and Pleurothecium, Réblová et al. 2016; Luo et al. 2018a). Species in Pleurotheciaceae are cosmopolitan with a worldwide distribution and have been reported from both aquatic and terrestrial habitats (Réblová et al. 2016, 2020; Hernandez-Restrepo et al. 2017; Luo et al. 2018a, 2019; Hyde et al. 2020a, b).
In this study, three new collections are placed in Dematipyriforma, Rhexoacrodictys and Pleurothecium respectively. The monotypic genus Dematipyriforma was introduced to accommodate an endophytic species, D.aquilaria which was collected from wood of Aquilariacrassna (Sun et al. 2017). Dematipyriforma was originally placed in Savoryellales (Sun et al. 2017). However, Dong et al. (2021) showed that Dematipyriforma clustered within Pleurotheciales and sister to Rhexoacrodictys and Saprodesmium. In addition, the morphology of Dematipyriforma is similar to Neomonodictys in Pleurotheciales. Therefore, they transferred Dematipyriforma to Pleurotheciales based on phylogenetic analysis and morphological characteristics. Rhexoacrodictys was introduced by Baker et al. (2002) to accommodate species previously identified as Acorcdictys (i.e., A.erecta, A.fimicola, A.fuliginosa and A.queenslandica) and wherein Rhexoacrodictyserecta was designated as the type. Two additional species R.martini and R.broussonetiae were subsequently added to the genus based on morphological characteristics (Delgado 2009; Xiao et al. 2018). While R.martini and R.queenslandica were transferred to Distoseptispora and Junewangia based on phylogenetic analysis (Xia et al. 2017). Currently, four species are accepted in Rhexoacrodictys. Pleurothecium was established by Höhnel (1919) with P.recurvatum (Morgan) Höhn as type species. Pleurothecium species are characterized by macronematous, mononematous, septate, brown conidiophores, polyblastic, sympodially extended, denticulate conidiogenous cells and solitary, septate, hyaline or pigmented or bicolored conidia (Goos 1969; Matsushima 1975, 1980; Subramanian and Bhat 1989; Matsushima and Matsushima 1996; Cooper 2005; Arzanlou et al. 2007; Wu and Zhang 2009; Réblová et al. 2012; Monteiro et al. 2016; Luo et al. 2018a). Presently, 11 species are accepted in the genus. Most Pleurothecium species are reported as saprobes from freshwater or terrestrial habitats (Wu and Zhang 2009; Réblová et al. 2012; Monteiro et al. 2016; Luo et al. 2018a).
We are currently investigating the diversity of lignicolous freshwater fungi from the Greater Mekong Subregion (Hyde et al. 2016). Thailand is an area of the Greater Mekong Subregion with rich fungal biodiversity. Freshwater fungi have been studied in Thailand over several decades initiated by Tubaki et al. (1983) who found 40 Ingoldian fungi in the stream foams. Many new freshwater taxa have since been reported in Thailand, especially a large number of lignicolous freshwater ascomycetes (Sivichai et al. 1998, 2000, 2002; Jones et al. 1999; Sivichai and Boonyene 2004; Zhang et al. 2011; Luo et al. 2019; Dong et al. 2020; Calabon et al. 2021, 2022). Until 2020, more than 302 freshwater taxa had been reported from Thailand (Zhang et al. 2011; Calabon et al. 2021). In this study, we introduce three taxa of Pleurotheciaceae, collected from freshwater habitats in Thailand. With phylogenetic analysis of ITS, LSU, SSU, RPB2 and TEF1-α sequence data, they are placed in Dematipyriforma, Pleurothecium and Rhexoacrodictys within Pleurotheciaceae. Of these three species, one is identified as Pleurotheciumaquaticum, one as Rhexoacrodictysfimicola, and the third as a new species in Dematipyriforma. In addition, Rhexoacrodictysnigrospora is transferred to Dematipyriforma based on morphological and phylogenetic evidence.
Materials and methods
Collection, isolation and morphology
Submerged decaying woods were collected from the streams in Thailand. The sample incubation, examination and morphological studies were referred to the methods described by Luo et al. (2018b). Single spore isolations were followed the methods outlined by Senanayake et al. (2020). Specimens (dry wood with fungal material) were deposited in the herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand and Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS). Pure cultures were deposited in Mae Fah Luang University Culture Collection (MFLUCC) and Kunming Institute of Botany culture collection (KUNCC). Faces of Fungi and Index Fungorum numbers were registered as outlined in Jayasiri et al. (2015) and Index Fungorum (2022). The descriptions are added to it GMS database (Chaiwan et al. 2021).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fungal mycelium (Rhexoacrodictyserecta and Pleurotheciumaquaticum) or directly from the conidiamatal tissue thalli of fungi (Dematipyriformamuriformis) as outlined by Wanasinghe et al. (2018). The Ezup Column Fungi Genomic DNA Purification Kit (Sangon Biotech, China) was used to extract DNA following the manufacturer’s instructions. ITS, LSU, SSU, RPB2 and TEF1-α gene regions were amplified using the primer pairs ITS5/ITS4, LR0R/LR7, NS1/NS4, fRPB2-5F/fRPB2-7cR and 983F/2218R, respectively (Vilgalys and Hester 1990; White et al. 1990; Liu et al. 1999). The amplification was performed in a 25 μl reaction volume containing 9.5 μl ddH2O, 12.5 μl 2 × Taq PCR Master Mix with blue dye (Sangon Biotech, China), 1 μl of DNA template and 1 μl of each primer (10 μM). The amplification condition for ITS, LSU, SSU, RPB2 and TEF1-α were followed Luo et al. (2018b). DNA sequencing of PCR products were carried out using the above-mentioned PCR primers at Tsingke Biological Engineering Technology and Services Co. (Yunnan, P.R. China).
Phylogenetic analyses
The taxa used in the phylogenetic analysis were obtained from previous studies (Table 1) (Hernandez-Restrepo et al. 2017; Luo et al. 2018a, 2019; Dayarathne et al. 2019; Hyde et al. 2020b; Réblová et al. 2020; Boonmee et al. 2021; Dong et al. 2021) and downloaded from GenBank. SEQMAN v. 7.0.0 (DNASTAR, Madison, WI) was used to assemble the consensus sequences and MAFFT v.7 online program (http://mafft.cbrc.jp/alignment/server/) was used to align the sequences (Katoh et al. 2019). BioEdit was used to manually adjust the alignments and the alignment fasta file was converted to Phylip format by Alivew (Hall 2021; Larsson 2014).
Table 1.
Taxa used in this study; the ex-type strains were indicated in bold, newly generated sequences are indicated by * after the species name.
| Species | Strain number | GenBank accession number | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | SSU | RPB2 | TEF1-α | ||
| Adelosphaeriacatenata | CBS 138679 | KT278721 | KT278707 | KT278692 | KT278743 | – |
| Anapleurotheciumbotulisporum | CBS 132713 | KY853423 | KY853483 | – | – | – |
| Ascotaiwanialignicola | NIL00005 | HQ446341 | HQ446364 | HQ446284 | HQ446419 | HQ446307 |
| Ascotaiwaniasawadae | SS00051 | HQ446340 | HQ446363 | HQ446283 | HQ446418 | HQ446306 |
| Bactrodesmiastrumobovatum | FMR 6482 | FR870264 | FR870266 | – | – | – |
| Bactrodesmiastrumpyriforme | FMR 10747 | FR870263 | FR870265 | – | – | – |
| Bactrodesmiumabruptum | CBS 144404 | MN699391 | MN699408 | MN699365 | MN704288 | MN704313 |
| Bactrodesmiumleptopus | CBS 144542 | MN699388 | MN699423 | MN699374 | MN704297 | MN704321 |
| Bactrodesmiumobovatum | CBS 144077 | MN699395 | MN699424 | MN699375 | MN704298 | MN704322 |
| Canalisporiumexiguum | SS00809 | GQ390296 | GQ390281 | GQ390266 | HQ446436 | – |
| Canalisporiumgrenadoideum | SS03615 | – | GQ390267 | GQ390252 | HQ446420 | HQ446309 |
| Coleodictyosporamuriformis | MFLUCC 18–1243 | MW981642 | MW981648 | MW981704 | – | – |
| Coleodictyosporamuriformis | MFLUCC 18–1279 | MW981643 | MW981649 | MW981705 | – | – |
| Conioscyphahoehnelii | FMR 11592 | KY853437 | KY853497 | HF937348 | – | – |
| Conioscyphalignicola | CBS 335.93 | – | AY484513 | JQ437439 | JQ429260 | – |
| Conioscyphaperuviana | ILL41202 | – | KF781539 | – | – | – |
| Conioscyphapleiomorpha | FMR 13134 | KY853438 | KY853498 | – | – | – |
| Dematipyriformaaquilaria | CGMCC 3.17268 | KJ138621 | KJ138623 | KJ138622 | – | – |
| Dematipyriformamuriformis * | MFLU 21–0146 | OM654773 | OM654770 | – | – | OM672032 |
| Dematipyriformanigrospora | MFLUCC 21-0096 | MZ538524 | MZ538558 | – | – | MZ567100 |
| Dematipyriformanigrospora | MFLUCC 21-0097 | MZ538525 | MZ538559 | MZ538574 | MZ567113 | MZ567101 |
| Fuscosporellapyriformis | MFLUCC 16–0570 | – | KX550896 | KX550900 | KX576872 | – |
| Helicoascotaiwaniafarinosa | ILLS 53605 | – | AY094189 | – | – | – |
| Helicoascotaiwaniafarinosa | DAOMC 241947 | JQ429145 | JQ429230 | – | – | – |
| Helicoascotaiwanialacustris | CBS 145963 | – | MN699430 | MN699382 | MN704304 | MN704329 |
| Helicoascotaiwanialacustris | CBS 145964 | MN699400 | MN699431 | MN699383 | MN704305 | – |
| Helicoascotaiwanialacustris | CBS 146144 | MN699401 | MN699432 | MN699384 | MN704306 | – |
| Leotialubrica | AFTOL-ID1 | DQ491484 | AY544644 | AY544746 | DQ470876 | DQ028596 |
| Melanotrigonumovale | CBS 138815 | KT278722 | KT278711 | KT278698 | KT278747 | – |
| Microglossumrufum | AFTOL-ID 1292 | – | DQ470981 | DQ471033 | DQ470933 | DQ471104 |
| Monotosporellasetosa | HKUCC3713 | – | AF132334 | – | – | – |
| Mucisporaobscuriseptata | MFLUCC 15–0618 | – | KX550892 | KX550897 | – | – |
| Mucisporaphangngaensis | MFLUCC 16–0865 | – | MG388210 | MG388207 | – | – |
| Neomonodictysmuriformis | MFLUCC 16–1136 | MN644509 | MN644485 | – | – | MN646856 |
| Obliquifusoideumguttulatum | MFLUCC 18–1233 | MW981645 | MW981650 | MW981706 | – | – |
| Parafuscosporellagarethii | FF00725.01 | – | KX958430 | KX958428 | KX958432 | – |
| Parafuscosporellamoniliformis | MFLUCC 15–0626 | – | KX550895 | KX550899 | – | – |
| Parafuscosporellamucosa | MFLUCC 16–0571 | – | MG388211 | MG388208 | – | – |
| Phaeoisariaaquatica | MFLUCC 16–1298 | MF399237 | MF399254 | – | MF401406 | – |
| Phaeoisariaclematidis | MFLUCC 17–1968 | MG837022 | MG837017 | MG837027 | – | – |
| Phaeoisariafasciculata | CBS 127885 | – | KT278705 | KT278693 | KT278741 | – |
| Phaeoisariafiliformis | MFLUCC 18–0214 | MK878381 | MK835852 | MK834785 | – | MN200285 |
| Phaeoisariaguttulata | MFLUCC 17–1965 | MG837021 | MG837016 | MG837026 | – | – |
| Phaeoisariapseudoclematidis | MFLUCC 11–0393 | – | KP744501 | KP753962 | – | – |
| Phaeoisariasedimenticola | CGMCC 3.14949 | – | JQ031561 | – | – | – |
| Phaeoisariasedimenticola | S-908 | MK878380 | MK835851 | – | – | MN200284 |
| Phaeoisariasparsa | FMR11939 | – | HF677185 | – | – | – |
| Phragmocephalastemphylioides | DAOM 673211 | KT278730 | KT278717 | – | – | – |
| Pleurotheciellaaquatica | MFLUCC 17–0464 | MF399236 | MF399253 | MF399220 | MF401405 | – |
| Pleurotheciellacentenaria | DAOM 229631 | – | JQ429234 | JQ429246 | JQ429265 | – |
| Pleurotheciellafusiformis | MFLUCC 17–0115 | MF399232 | MF399249 | MF399217 | MF401402 | – |
| Pleurotheciellaguttulata | KUMCC 15–0296 | MF399240 | MF399257 | MF399223 | MF401409 | – |
| Pleurotheciellakrabiensis | MFLUCC 18–0852 | MG837018 | MG837013 | MG837023 | – | – |
| Pleurotheciellalunata | MFLUCC 17–0111 | MF399238 | MF399255 | MF399221 | MF401407 | – |
| Pleurotheciellarivularia | CBS 125238 | – | JQ429232 | JQ429244 | JQ429263 | – |
| Pleurotheciellarivularia | CBS 125237 | – | JQ429233 | JQ429245 | JQ429264 | – |
| Pleurotheciellasaprophytica | MFLUCC 16–1251 | MF399241 | MF399258 | MF399224 | MF401410 | – |
| Pleurotheciellasubmersa | MFLUCC 17–1709 | MF399243 | MF399260 | MF399226 | MF401412 | – |
| Pleurotheciellasubmersa | MFLUCC 17–0456 | MF399244 | MF399261 | MF399227 | MF401413 | – |
| Pleurotheciellatropica | MFLUCC 16–0867 | MG837020 | MG837015 | MG837025 | – | – |
| Pleurotheciellauniseptata | DAOM 673210 | KT278729 | KT278716 | – | – | – |
| Pleurotheciumaquaticum | MFLUCC 17–1331 | MF399245 | MF399263 | – | – | – |
| Pleurotheciumaquaticum * | KUMCC 21-0477 | OM654775 | OM654772 | OM654807 | OM672034 | OM672033 |
| Pleurotheciumfloriforme | MFLUCC 15–0628 | NR_156614 | NG_059791 | – | – | – |
| Pleurotheciumobovoideum | CBS 209.95 | EU041784 | EU041841 | – | – | – |
| Pleurotheciumpulneyense | MFLUCC 16–1293 | – | MF399262 | MF399228 | MF401414 | – |
| Pleurotheciumrecurvatum | CBS 138686 | – | KT278715 | KT278702 | – | – |
| Pleurotheciumsemifecundum | CBS 131271 | – | JQ429240 | JQ429254 | JQ429270 | – |
| Rhexoacrodictyserecta | HSAUPmyr4622 | KU999964 | KX033556 | KX033526 | – | – |
| Rhexoacrodictyserecta | IFRD500–016 | MT555421 | MT559123 | MT555735 | – | – |
| Rhexoacrodictyserecta | HSAUP myr6489 | KU999963 | KX033555 | KX033525 | – | – |
| Rhexoacrodictysfimicola | HMAS 47737 | KU999960 | KX033553 | KX033522 | – | – |
| Rhexoacrodictysfimicola | HMAS 42882 | KU999962 | KX033554 | KX033524 | – | – |
| Rhexoacrodictysfimicola | HMAS 43690 | KU999957 | KX033550 | KX033519 | – | – |
| Rhexoacrodictysfimicola * | MFLUCC 18–0340 | OM654774 | OM654771 | OM654806 | – | – |
| Saprodesmiumdematiosporium | KUMCC 18–0059 | MW981646 | MW981647 | MW981707 | – | – |
| Savoryellaaquatica | SS03801 | – | HQ446372 | HQ446292 | HQ446405 | HQ446326 |
| Savoryellalignicola | NF00204 | – | HQ446378 | HQ446300 | HQ446413 | HQ446334 |
| Sterigmatobotrysmacrocarpa | MR2973 | – | GU017317 | – | – | – |
| Sterigmatobotrysrudis | DAOM 229838 | JQ429152 | JQ429241 | JQ429256 | JQ429272 | – |
| Sterigmatobotrysuniseptata | MFLUCC 15–0358 | MK878379 | MK835850 | MK834784 | – | – |
Maximum likelihood (ML) analysis generated using the RAxML-HPC2 on XSEDE (v.8.2.8) in the CIPRES Science Gateway (https://www.phylo.org, Stamatakis 2006; Stamatakis et al. 2008; Miller et al. 2010) with rapid bootstrap analysis, followed by 1000 bootstrap replicates, using the GTR+I+G model of evolution.
Bayesian analysis was performed by MrBayes v. 3.2 (Ronquist et al. 2012), best-fit model of DNA evolution for the Bayesian inference analysis was estimated by MrModeltest v. 2.2 (Nylander 2004) and the GTR+I+G model was selected for LSU, ITS, RPB2 and TEF1-α, GTR+G model was selected for SSU. Posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002) was defined by Bayesian Markov Chain Monte Carlo (BMCMC) sampling method in MrBayes v. 3.0b4 (Huelsenbeck and Ronquist 2001). Six simultaneous Markov Chains were run for 50,000,000 generations and trees were sampled every 500th generation (resulting in 100,000 trees). The first 20% trees that represented the burn-in phase were discarded and the remaining 80% (post burn-in) trees used for calculating posterior probabilities (PP) for the majority rule consensus tree.
Phylogenetic trees were visualized with FigTree v. 1.4.2 (Rambaut 2014) and edited in Microsoft Office PowerPoint 2019 (Microsoft Inc., United States). Newly generated sequences in this study were deposited in GenBank.
Results
Phylogenetic analyses
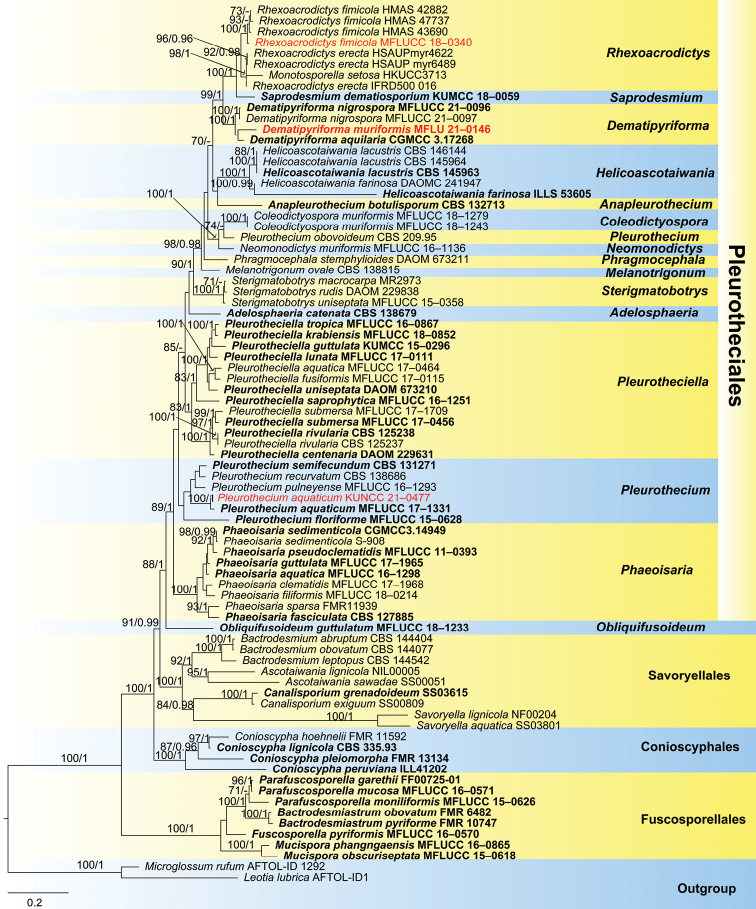
The dataset of combined ITS, LSU, SSU, RPB2 and TEF1-α sequence data comprises 81 strains with 4257 characters including gaps (ITS: 509 bp, LSU: 1006 bp, SSU: 862 bp, RPB2: 1032 bp, TEF1-α: 848 bp). Leotialubrica (AFTOL-ID1) and Microglossumrufum (AFTOL-ID 1292) were used as outgroup taxa. RAxML and Bayesian analyses were conducted and resulted in generally congruent topologies. The best RAxML tree with a final likelihood value of –45872.924927 is presented. The matrix had 2433 distinct alignment patterns, with 44.65% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.234712, C = 0.261626, G = 0.290634, T = 0.213028; substitution rates AC = 1.347806, AG = 2.754719, AT = 1.490447, CG = 1.095887, CT = 6.696475, GT = 1.000000; gamma distribution shape parameter α = 0.316898.
In the phylogenetic analysis, Dematipyriformamuriformis (MFLU 21–0146) clustered with the ex-type strain of D.aquilaria (CGMCC 3.17268) with low support (Fig. 1). The new isolate of Rhexoacrodictysfimicola (MFLUCC 18–0340) clustered with three strains of R.fimicola (HMAS 42882, HMAS 43690 and HMAS 47737) with 100% ML/1.00 PP support (Fig. 1). Pleurotheciumaquaticum (KUNCC 21–0477) clustered with the ex-type strain of P.aquaticum (MFLUCC 17–1331) with 100% ML/1.00 PP support (Fig. 1).
Figure 1.
Phylogram based on a combined ITS, LSU SSU, RPB2 and TEF1-α sequence data of selected members of four orders of the Savoryellomycetidae. Bootstrap support values for maximum likelihood (ML) greater than 70% and Bayesian posterior probabilities (PP) greater than 0.95 are given as ML/PP above the nodes. Newly obtained sequences are indicated in red and ex-type strains are in bold.
Taxonomy
. Dematipyriforma muriformis
D.F. Bao, K.D. Hyde & Z.L. Luo sp. nov.
9771DF07-5B27-5031-9B8E-090279283AA0
http://www.indexfungorum.org/names/NamesRecord.asp?RecordID=553383
https://www.facesoffungi.org/?s=FoF10414
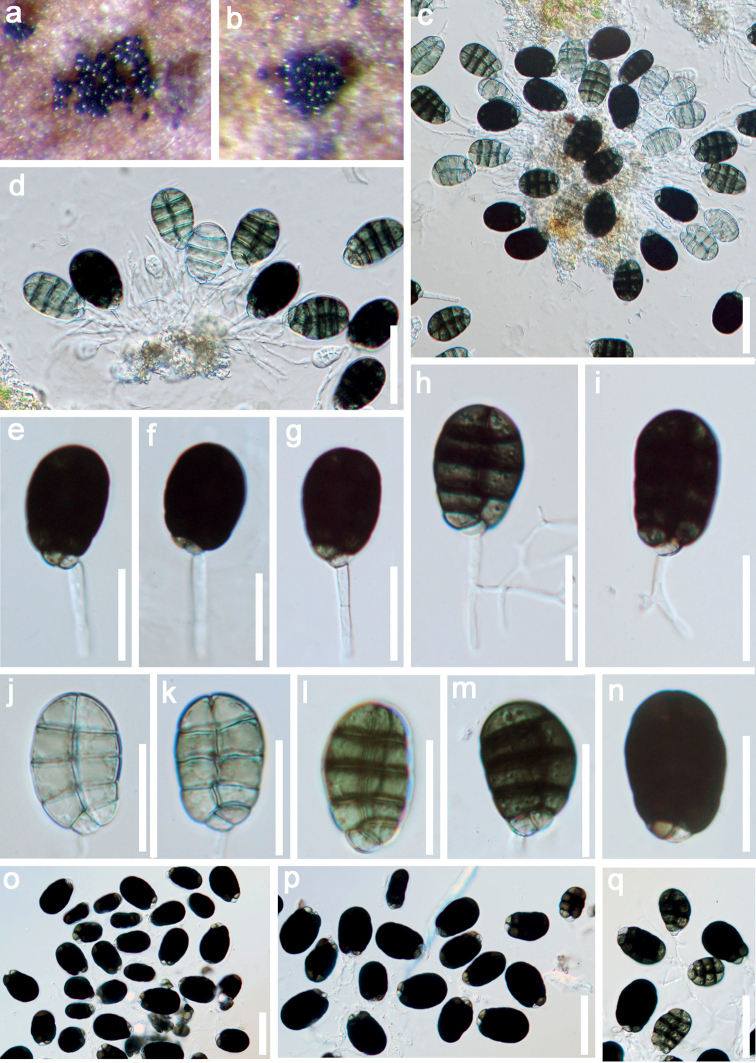
Figure 2.
Dematipyriformamuriformis (MFLU 21–0146, holotype) a, b colonies on wood c–d conidiomata e–i conidiophore with conidia j–q conidia. Scale bars: 30 μm (c–d, o–q); 20 μm (e–n).
Etymology.
Referring to the muriform conidia of this species.
Holotype.
MFLU 21–0146.
Description.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies on substratum superficial, scattered, black, shining, granulate. Mycelium immersed, composed of hyaline, branched, septate, smooth, hyphae. Conidiomata sporodochial, subhyaline. Conidiophores 10–26.5 × 2–3 μm (x‒ = 18.2 × 2.3 μm, n = 20), micronematous to semi-macronematous, mononematous, fasciculate, simple or branched, hyaline, cylindrical, smooth. Conidiogenous cells monoblastic, integrated, terminal, determinate, hyaline, smooth. Conidia 23–26 × 15.5–18 μm (x‒ = 24.6 × 16.7 μm, n = 30), acrogenous, solitary, smooth, thick-walled, ellipsoidal to obovoid, muriform, rounded at apex, pointed at base, with 3–5 transverse septa, 1-longitudinal septum in all cells and rarely in end cells, slightly constricted at septa, subhyaline to pale olivaceous when young, olive to dark brown at maturity.
Material examined.
Thailand, Bangkok Province, Bang Kapi District, on decaying wood submerged in a freshwater stream, 3 October 2017, Z.L. Luo, Bsite 4–3–1 (MFLU 21–0146, holotype; KUN-HKAS 122858, isotype).
Notes.
In the phylogenetic analysis, Dematipyriformamuriformis clustered with the ex-type strain of D.aquilaria (CGMCC 3.17268) within Pleurotheciaceae with low support (Fig. 1). The ITS blast result in NCBI GenBank showed that D.muriformis (MFLU 21–0146) is 92.36% and 91.92% similar to D.nigrospora (MFLUCC 21-0097) and D.aquilaria (CGMCC 3.17268) respectively.
Dematipyriformamuriformis resembles D.aquilaria in having micronematous, mononematous, smooth septate conidiophores, monoblastic, integrated, terminal, determinate conidiogenous cells and solitary, muriform conidia. However, D.muriformis differs from D.aquilaria in having hyaline conidiophores and slightly smaller conidia (23–26 × 15.5–18 vs. 25–37.5 × 15–22.5 μm). In addition, conidia of D.muriformis are subhyaline to pale olivaceous when young, olive to dark brown at maturity, with 3–5 transverse septa, 1-longitudinal septum in all cells and rarely in end cells. Whereas, D.aquilaria has pale grey olivaceous to pale brown conidia with 4–5 transverse septa and 0–2 longitudinal septa (Sun et al. 2017).
Dematipyriformamuriformis shares some similar characteristics with Neomonodictys taxa in Pleurotheciaceae, such as monoblastic, integrated, terminal, determinate conidiogenous cells and muriform conidia. Neomonodictys, however, lacks sporodochial conidiomata and conidia of Neomonodictys are subglobose to globose, while, Dematipyriformamuriformis has ellipsoidal to obovoid conidia (Hyde et al. 2020b).
. Dematipyriforma nigrospora
(Boonmee, D.F. Bao & K.D. Hyde) D.F. Bao, K.D. Hyde & Z.L. Luo comb. nov.
A6148170-3B36-5B96-9677-CAD862AE897E
http://www.indexfungorum.org/names/NamesRecord.asp?RecordID=553384
≡ Rhexoacrodictysnigrospora Boonmee, D.F. Bao & K.D. Hyde, in Boonmee et al., Fungal Diversity 111: 200 (2021).
Holotype.
Thailand, Phetchabun Province, on decaying bark, 25 July 2019, S. Boonmee, LSP03 (MFLU 21–0073).
Descriptions and illustrations.
Notes.
Rhexoacrodictysnigrospora was introduced by Boonmee et al. (2021) based on morphological characters and phylogenetic analysis. In our phylogenetic analysis, R.nigrospora clustered with two Dematipyriforma species (D.aquilaria and D.muriformis) in a distinct clade within Pleurotheciaceae (Fig. 1). Therefore, we transfer Rhexoacrodictysnigrospora to Dematipyriforma, as Dematipyriformanigrospora comb. nov.
Dematipyriformanigrospora resembles D.muriformis in having micronematous or semi-macronematous, mononematous conidiophores and monoblastic, polyblastic, integrated, terminal conidiogenous cells. However, D.nigrospora differs from D.muriformis in having brown to dark brown conidiophores and globose to subglobose, dark brown to black conidia (Boonmee et al. 2021). Conidiophores of D.muriformis are hyaline and conidia are ellipsoidal to obovoid, muriform, and subhyaline to pale olivaceous when young, olive to dark brown at maturity.
. Rhexoacrodictys fimicola
(M.B. Ellis & Gunnell) W.A. Baker & Morgan-Jones, in Baker, Partridge & Morgan-Jones, Mycotaxon 82: 103 (2002)
3D33BD6A-675E-542C-8B19-614ECB4162C6
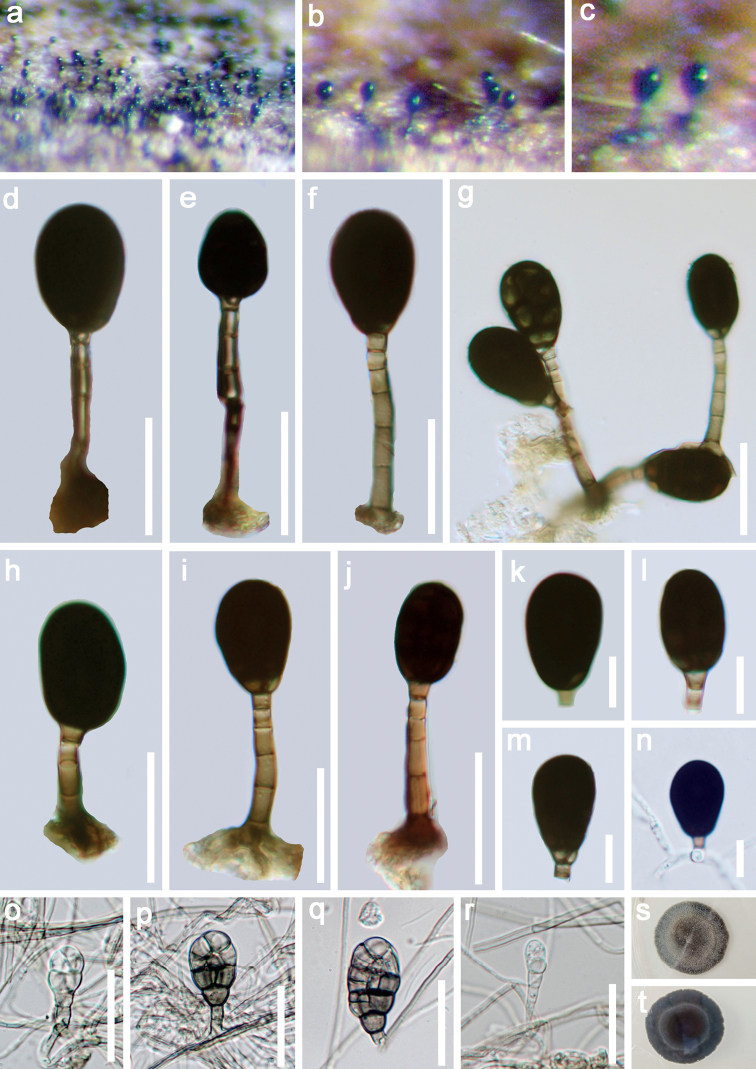
Figure 3.
Rhexoacrodictysfimicola (MFLU 21–0147, new record) a–c colonies on wood d–j conidiophores with conidia k–m conidi n germinating conidium o–r re-produced asexual morph of Rhexoacrodictysfimicolas–t culture on PDA from surface and reverse. Scale bars: 20 μm (d–j, o–r); 10 μm (k–n).
Holotype.
Maya, Perak, on elephant dung, September 1958, A.H.S, Onions, IMI 76413.
Description.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies on the substratum superficial, effuse, hairy or velvety, black. Mycelium mostly immersed, composed of branched, septate, smooth, pale brown hyphae. Conidiophores (17.5–)20–44.5 (–65.5) × 2.5–4.0 μm (x‒ = 32.2 × 3.4 μm, n = 20), macronematous, mononematous, erect, straight or slightly flexuous, thick-walled, smooth, orange-brown or brown, 3–7-septate. Conidiogenous cells monoblastic, integrated, terminal. Conidia 16.5–24 × 11–15 μm (x‒ = 20.3 × 13 μm, n = 30), solitary, dry, acrogenous, broadly oval to subglobose, muriform, transversely and longitudinally septate, with transverse septa typically spanning the whole conidial width, with longitudinal septa typically incomplete, short; dark-blackish brown to black, smooth, narrowly truncate at the base.
Cultural characteristics.
Conidia germinating on PDA within 24 h. Germ tubes produced from the basal cell. Colonies on PDA reaching 3 cm diameter in 30 days at 20–25 °C, pale brown, with dense, tight mycelia on the surface, sparse at the margin, reverse dark brown, with smooth margin. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, monoblastic, integrated, hyaline to pale brown, smooth. Conidia broad oval to subglobose, muriform, strongly constricted at all the septa, hyaline when young, brown to grayish-brown when aged, smooth-walled.
Material examined.
Thailand, Bangkok Province, Bang Kapi District, on decaying wood submerged in a freshwater stream, 3 October 2017, Z.L. Luo, Bsite 4–3–2 (MFLU 21–0147 = KUN-HKAS 122859), living culture, MFLUCC 18–0340.
Notes.
In the phylogenetic analysis, our new isolate MFLUCC 18–0340 clustered with three strains of Rhexoacrodictysfimicola (HMAS 42882, HMAS 43690 and HMAS 47737) with strong support (100% ML/ 1.00 PP). The nucleotide BLASTn search of ITS showed that our new strain (MFLUCC 18–0340) has 99.7%, 99.3% and 99.1% similarities with strain HMAS 43690, HMAS 47737 and HMAS 42882 of Rhexoacrodictysfimicola, respectively. Morphologically, our new collection is similar to R.fimicola in having macronematous, mononematous, indeterminate conidiophores, integrated, terminal, monoblastic, pale brown conidiogenous cells and broadly oval to subglobose, transversely and longitudinally septate, smooth, brown to black conidia, with the size of conidia and conidiophores are overlapping (Ellis 1961; Baker et al. 2002). Based on both phylogeny and morphology, we identified our species as R.fimicola.
Rhexoacrodictysfimicola was originally introduced by Ellis (1961) as Acrodictysfimicola. Baker et al. (2002) transferred A.fimicola to Rhexoacrodictys based on morphological characteristics. Rhexoacrodictysfimicola has been reported on Bambusavulgaris and elephant dung from Africa and Malaysia respectively. Our collection, on the other hand, was collected from freshwater habitats and represents the first time it was reported from Thailand.
. Pleurothecium aquaticum
Z.L. Luo, H.Y. Su & K.D. Hyde, in Luo, Hyde, Bhat, Jeewon, Maharachchikumbura, Bao, Li, Su, Yang & Su, Mycol. Prog. 17(5): 526 (2018)
7644B4BA-2F53-5575-A238-F9CE05C64759
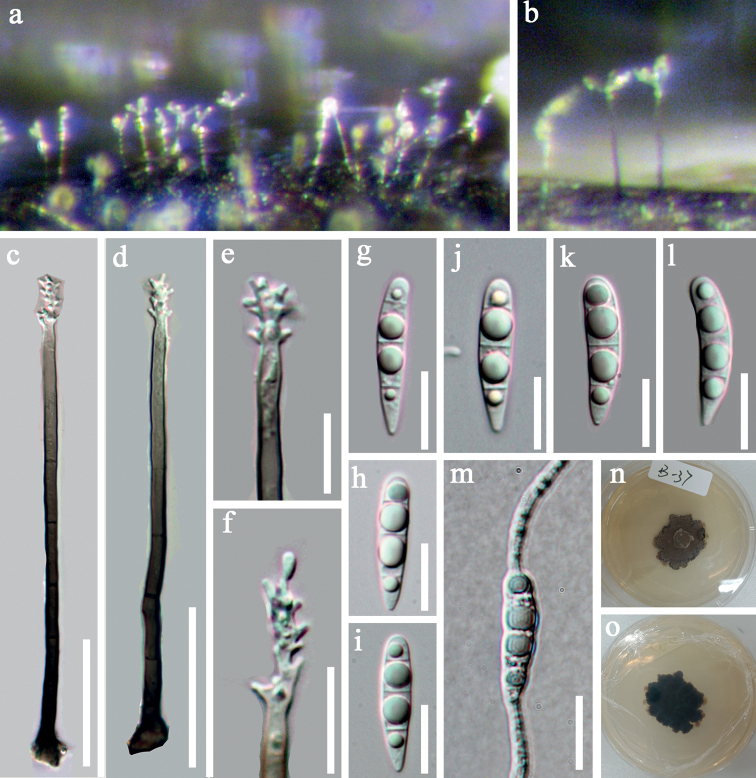
Figure 4.
Pleurotheciumaquaticum (MFLU 21–0148, new record) a, b colonies on wood c, d conidiophores e, f conidiogenous cells g–i conidia m germinating conidium n, o culture on PDA from surface and reverse. Scale bars: 30 μm (c, d); 10 μm (e–m).
Description.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: colonies on substratum, effuse, shining, dark brown to black. Mycelium partly immersed, composed of septate, branched, smooth, dark brown hyphae. Conidiophores 84–110 × 3–4 μm (x‒ = 97 × 3.4 μm, n = 10), macronematous, mononematous, erect, simple, unbranched, straight or slightly flexuous, 5–8-septate, dark brown, pale towards apex, smooth. Conidiogenous cells integrated, polyblastic, terminal, hyaline, denticulate, smooth. Conidia 18–22 × 4–5 μm (x‒ = 20 × 4.5 μm, SD = 4 n = 30), acrogenous, solitary, clavate, mostly curved, rounded at apex, tapering at base, hyaline, 3-septate, with guttulate cells, smooth.
Cultural characteristics.
Conidia germinating on PDA within 24 h. Germ tubes produced from the basal and apical cells. Colonies on PDA reaching 2.3 cm diameter in 30 days at 20–25 °C, with dense mycelia, dry, rigid, rugose, dark brown, reverse dark brown.
Material examined.
Thailand, Prachuap Khan, on submerged decaying wood, 15 August 2017, V. Kumar, site1–24–2 (MFLU 21–0148 = KUN-HKAS 122857), living culture, KUNCC 21–0477.
Notes.
In the phylogenetic analysis, our new collection KUNCC 21–0477 clustered with the ex-type strain of Pleurotheciumaquaticum (MFLUCC 17–1331) with high (100% ML/1.00 PP). In addition, the ITS and LSU BLASTn search on NCBI GenBank showed that our new strain is 99.88% and 97.45% similarities to the ex-type of P.aquaticum (MFLUCC 17–1331). The new collection is morphologically similar to P.aquaticum in having macronematous, mononematous, septate, brown, pale brown towards the apex conidiophores, integrated, terminal, polyblastic, denticulate conidiogenous cells and hyaline, cylindrical or clavate, rounded at the apex, obtuse and tapering towards base, 3-septate conidia. We therefore identified our new collection as P.aquaticum. Pleurotheciumaquaticum was introduced by Luo et al. (2018a) collected from freshwater habitats in China. Our new collection, on the other hand, was collected from Thailand and is a new record for Thailand.
Discussion
Pleurotheciaceae is a diverse family. The sexual morphs of Pleurotheciaceae are quite similar and difficult to distinguish without molecular data (Réblová et al. 2016; Hyde et al. 2020a). However, the asexual morphs in the family are morphologically diverse. Most genera have mononematous, macrounematous conidiophores (Anapleurothecium, Pleurothecium, Pleurotheciella and Rhexoacrodictys) (Réblová et al. 2016; Luo et al. 2018a, 2019; Hyde et al. 2020a), whereas some genera have synnematous conidiophores (Phaeoisaria and Phragmocephala) (Höhnel 1919; Mason and Hughes 1951; Seifert et al. 2011; Wijayawardene et al. 2012; Su et al. 2015; Réblová et al. 2016; Luo et al. 2018a), and others with micronematous or reduced conidiophores (Neomonodictys and Sterigmatobotrys). (Hyde et al. 2020b). Conidiogenous cells of Anapleurothecium, Pleurothecium, Phaeoisaria and Pleurotheciella are polyblastic and denticulate (Réblová et al. 2012, 2016; Monteiro et al. 2016; Luo et al. 2018a). Phragmocephala and Monotosporella have monoblastic conidiogenous cells (Mason and Hughes 1951; Hyde and Yanna 2002; Wijayawardene et al. 2012; Su et al. 2015). Conidia of Pleurotheciaceae are diverse in their shape, color and septation. Conidia of Sterigmatobotrys are fusiform and in persistent chains (Heuchert et al. 2018); Helicoascotaiwania has helicosporous conidia (Dayarathne et al. 2019); Anapleurothecium, Melanotrigonum, Pleurothecium, Phaeoisaria and Pleurotheciella have clavate, ellipsoidal, obovoidal, fusiform-cylindrical, hyaline or brown, aseptate or transversely septate conidia (Réblová et al. 2012, 2016; Monteiro et al. 2016; Hernandez-Restrepo et al. 2017; Luo et al. 2018a); Monotosporella, Neomonodictys and Phragmocephalahave ellipsoidal or subglobose to globose conidia (Mason and Hughes 1951; Hyde and Yanna 2002;Wijayawardene et al. 2012; Su et al. 2015; Hyde et al. 2020b). However, conidia of Neomonodictys are muriform (Hyde et al. 2020b), whereas, Phragmocephala and Monotosporella have transversely septate conidia.
In this study, we introduced a new asexual species, Dematipyriformamuriformis based on both morphology and phylogeny. Dematipyriforma was introduced by Sun et al. (2017) with a single species D.aquilaria which was reported as an endophyte from Aquilariacrassna in China. While our new species is a saprobe isolated on submerged wood from freshwater habitats in Thailand. In addition, Rhexoacrodictysnigrospora is transferred to Dematipyriforma in this study. Currently, three species are accepted in the genus. Morphologically, the muriform conidia of Dematipyriforma are similar to Neomonodictys, Saprodesmium and Coleodictyospora. However, Dematipyriforma can be distinguished from Neomonodictys by the shape of conidia (ellipsoidal to obovoid vs. subglobose to globose) and conidiophores (semi-micronematous to macronematous vs. micronematous or lacking conidiophores, Hyde et al. 2020b). Dematipyriforma differs from Coleodictyospora in the conidia lacking a semi-gelatinous sheath (Dong et al. 2021). Dematipyriforma is distinct from Saprodesmium by the micronematous to semi-macronematous, simple or branched, hyaline, cylindrical, conidiophores, whereas, conidiophores of Saprodesmium are micronematous, unbranched, consisted of 1–4 subglobose smooth, hyaline cells (Dong et al. 2021).
Rhexoacrodictys comprises six species of which four species (R.erecta, R.fimicola, R.martini and R.queenslandica) have sequence data available in the GenBank. Among them, R.martini and R.queenslandica were transferred to Distoseptispora and Junewangia based on phylogenetic analysis (Xia et al. 2017). However, sequence data of R.martini are doubted by several studies (Sun et al. 2020; Shen et al. 2021), as its morphology does not fit with the characters of Distoseptispora. Rhexoacrodictyserecta and R.fimicola clustered within Pleurotheciaceae (Luo et al. 2019; Dong et al. 2021). The placement of Rhexoacrodictys was questionable since it was established. Baker et al. (2002) established the genus; however, they did not mention the placement of the genus. Xia et al. (2017) firstly provided sequence data for Rhexoacrodictyserecta (Type species of Rhexoacrodictys) and R.fimicola based on their fresh collections, their phylogenetic analysis showed that R.erecta and R.fimicola clustered within Savoryellaceae. However, they did not include the related orders (Conioscyphales, Fuscosporellales and Pleurotheciales) in Savoryellomycetidae. Luo et al. (2019) found that R.erecta and R.fimicola grouped in Pleurotheciaceae. Recently, Dong et al. (2021) obtained the same result as Luo et al. (2019). However, Boonmee et al. (2021) and Wijayawardene et al. (2022) placed Rhexoacrodictys in Savoryellaceae (Savoryellales). Our result is consistent with Luo et al. (2019) and Dong et al. (2021), the two species clustered within Pleurotheciaceae (Fig. 1). On the other hand, the morphology of Rhexoacrodictys is similar to Dematipyriforma, Neomonodictys and Saprodesmium, in having muriform conidia, micronematous conidiophores and holoblastic, monoblastic conidiogenous cells. Therefore, we formally accepted Rhexoacrodictys in Pleurotheciaceae (Pleurotheciales) based on morphological characters and phylogenetic analysis.
In our phylogenetic analysis, Rhexoacrodictyserecta and R.fimicola clustered with Monotosporellasetosa which is the type species of Monotosporella. Morphologically, R.erecta and R.fimicola fit well within the genus concept of Monotosporella in having macronematous, mononematous, brown, septate conidiophores, monoblastic, percurrent conidiogenous cells and acrogenous, brown septate conidia (Hughes 1958; Baker et al. 2002; Hyde and Yanna 2002). However, the strain of Monotosporellasetosa (HKUCC 3713) lacks a morphological description. Therefore, further study is necessary to clarify the relationship between Rhexoacrodictys and Monotosporella, whether they should be combined into one genus or not. In addition, our phylogenetic analysis showed that three strains of R.erecta clustered with Monotosporellasetosa. However, M.erecta differs from M.setosa in having transverse and longitudinal septation, while, conidia of M.setosa only have transverse septa. Only LSU sequence data is available for M.setosa, which is not significant to distinguish in the phylogenetic tree, but morphologically they are quite distinct. Hence, we maintain them as two distinct species, however, further morphological and phylogenetic analysis is required to clarify the relationship between these two species.
In our phylogenetic analysis, Pleurotheciumobovoideum was placed distant from Pleurothecium and close to Neomonodictysmuriformis and Coleodictyosporamuriformis which is consistent with recent studies (Luo et al. 2018a, 2019; Hyde et al. 2020b). Pleurotheciumobovoideum was introduced by Arzanlou et al. (2007) based on morphological characters. However, their analysis showed that P.obovoideum clustered with Ascotaiwaniahughesii and with more sequence data now available for Pleurothecium species, P.obovoideum is shown phylogenetically distinct from Pleurothecium. Morphologically, P.obovoideum is similar to Pleurothecium in having distinct brown conidiophores, polyblastic, denticulate conidiogenous cells and pale brown, ellipsoidal to obovate conidia. However, conidia of P.obovoideum are aseptate and solitary or in short chains whereas the conidia of Pleurothecium are solitary and unicellular or septate. Thus, the placement of P.obovoideum needs revision in the future with more evidence.
Supplementary Material
Acknowledgements
We would like to thank the National Natural Science Foundation of China (Project ID: 32060005 and 31970021) for financial support. This study was also supported by the Yunnan Fundamental Research Project (grant NO. 202101AU070137, 202201AW070001) and Thailand research fund “Macrofungi diversity research from the Lancang-Mekong Watershed and Surrounding areas (Grant no. DBG6280009)”. Dan-Feng Bao would like to thank Shaun Pennycook from Landcare Research, Auckland, New Zealand, for advising on the taxon names. Wen-Li Li is acknowledged for her help with DNA extraction and PCR amplification.
Citation
Bao D-F, Bhat DJ, Boonmee S, Hyde KD, Luo Z-L, Nalumpang S (2022) Lignicolous freshwater ascomycetes from Thailand: Introducing Dematipyriforma muriformis sp. nov., one new combination and two new records in Pleurotheciaceae. MycoKeys 93: 57–79. https://doi.org/10.3897/mycokeys.93.87797
Contributor Information
Zong-Long Luo, Email: 514992672@qq.com.
Sarunya Nalumpang, Email: sarunya.v@cmu.ac.th.
References
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW. (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. 10.3114/sim.2007.58.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker WA, Partridge EC, Morgan-Jones G. (2002) Notes on hyphomycetes LXXXVII. Rhexoacrodictys, a new segregate genus to accommodate four species previously classified in Acrodictys. Mycotaxon 82: 95–113.
- Boonmee S, Wanasinghe DN, Calabon MS, Huanraluek N, Chandrasiri SKU, Jones EBG, Rossi W, Leonardi M, Singh SK, Rana S, Singh PN, Maurya DK, Lagashetti AC, Choudhary D, Dai YC, Zhao CL, Mu YH, Yuan HS, He SH, Phookamsak R, Jiang HB, Martín MP, Dueñas M, Telleria MT, Kałucka IL, Jagodziński AM, Liimatainen K, Pereira DS, Phillips AJL, Suwannarach N, Kumla J, Khuna S, Lumyong S, Potter TB, Shivas RG, Sparks AH, Vaghefi N, Abdel-Wahab MA, Abdel-Aziz FA, Li GJ, Lin WF, Singh U, Bhatt RP, Lee HB, Nguyen TTT, Kirk PM, Dutta AK, Acharya K, Sarma VV, Niranjan M, Rajeshkumar KC, Ashtekar N, Lad S, Wijayawardene NN, Bhat DJ, Xu RJ, Wijesinghe SN, Shen HW, Luo ZL, Zhang JY, Sysouphanthong P, Thongklang N, Bao DF, Aluthmuhandiram JVS, Abdollahzadeh J, Javadi A, Dovana F, Usman M, Khalid AN, Dissanayake AJ, Telagathoti A, Probst M, Peintner U, Garrido-Benavent I, Bóna L, Merényi Z, Boros L, Zoltán B, Stielow JB, Jiang N, Tian CM, Shams E, Dehghanizadeh F, Pordel A, Javan-Nikkhah M, Denchev TT, Denchev CM, Kemler M, Begerow D, Deng CY, Harrower E, Bozorov T, Kholmuradova T, Gafforov Y, Abdurazakov A, Xu JC, Mortimer PE, Ren GC, Jeewon R, Maharachchikumbura SSN, Phukhamsakda C, Mapook A, Hyde KD. (2021) Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111(1): 1–335. 10.1007/s13225-021-00489-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabon MS, Jones EBG, Boonmee S, Doilom M, Lumyong S, Hyde KD. (2021) Five novel freshwater ascomycetes indicate high undiscovered diversity in lotic habitats in Thailand. Journal of Fungi (Basel, Switzerland) 7(2): e117. 10.3390/jof7020117 [DOI] [PMC free article] [PubMed]
- Calabon MS, Hyde KD, Jones EBG, Luo ZL, Dong W, Hurdeal VG, Gentekaki E, Rossi W, Leonardi M, Thiyagaraja V, Lestari AS, Shen H-W, Bao D-F, Boonyuen N, Zeng M. (2022) Freshwater fungal numbers. Fungal Diversity 114(1): 3–235. 10.1007/s13225-022-00503-2 [DOI] [Google Scholar]
- Chaiwan N, Gomdola D, Wang S, Monkai J, Tibpromma S, Doilom M, Wanasinghe DN, Mortimer PE, Lumyong S, Hyde KD. (2021) an online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere : Journal of Fungal Biology 12(1): 1513–1526. 10.5943/mycosphere/12/1/19 [DOI] [Google Scholar]
- Cooper JA. (2005) New Zealand hyphomycetes fungi: Additional records, new species and notes on interesting collections. New Zealand Journal of Botany 43(1): 323–349. 10.1080/0028825X.2005.9512957 [DOI] [Google Scholar]
- Dayarathne MC, Maharachchikumbura SSN, Jones EBG, Wei D, Devadatha B, Yang J, Ekanayake H, De Silva W, Sarma VV, AlSadi AM, Khongphinitbunjong K, Hyde KD, Zhao RL. (2019) Phylogenetic revision of Savoryellaceae and evidence for its ranking as a subclass. Frontiers in Microbiology 10: 840. 10.3389/fmicb.2019.00840 [DOI] [PMC free article] [PubMed]
- Delgado G. (2009) South Florida microfungi: Veramycellabispora, a new palmicolous anamorphic genus and species, with some new records for the continental USA. Mycotaxon 107(1): 357–373. 10.5248/107.357 [DOI] [Google Scholar]
- Dong W, Wang B, Hyde KD, McKenzie EHC, Bhat DJ, Raja HA, Tanaka K, Abdel-Wahab MA, Abdel-Aziz FA, Doilom M, Phookamsak R, Hongsanan S, Wanasinghe DN, Yu XD, Wang GN, Yang H, Yang J, Thambugala AN, Tian Q, Luo ZL, Yang JB, Miller AN, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105(1): 319–575. 10.1007/s13225-020-00463-5 [DOI] [Google Scholar]
- Dong W, Jeewon R, Hyde KD, Yang EF, Zhang H, Yu XD, Wang GN, Suwannarach N, Doilom M, Dong Z. (2021) Five novel taxa from freshwater habitats and new taxonomic insights of Pleurotheciales and Savoryellomycetidae. Journal of Fungi (Basel, Switzerland) 7(9): 711. 10.3390/jof7090711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MB. (1961) Dematiaceous hyphomycetes. II. Mycological Papers 79: 1–23. [Google Scholar]
- Goos RD. (1969) The genus Pleurothecium. Mycologia 61(6): 1048–1053. 10.1080/00275514.1969.12018832 [DOI] [Google Scholar]
- Hall T. (2021) Bioedit Version 6.0.7. http://www.mbio.ncsu.edu/bioedit/bioedit.html [accessed on 18 May 2021]
- Hernandez-Restrepo M, Gene J, Castaneda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J. (2017) Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. 10.1016/j.simyco.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuchert B, Braun U, Diederich P, Ertz D. (2018) Taxonomic monograph of the genus Taeniolella s. lat. (Ascomycota). Fungal Systematics and Evolution 2: 69–261. 10.3114/fuse.2018.02.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhnel F. (1919) Fünfte vorlaüfige Mitteilungen mykologischer Ergebnisse (Nr. 399–500). Berichte der Deutschen Botanischen Gesellschaft 37: 153–161. [Google Scholar]
- Hongsanan S, Maharachchikumbura SSN, Hyde KD, Samarakoon MC, Jeewon R, Zhao Q, Al-Sadi AM, Bahkali AH. (2017) An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Diversity 84(1): 25–41. 10.1007/s13225-017-0384-2 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hughes SJ. (1958) Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Canadian Journal of Botany 36(6): 727–836. 10.1139/b58-067 [DOI] [Google Scholar]
- Hyde KD, Yanna HYDE. (2002) New saprobic fungi on fronds of palms from northern Queensland, Australia. Australian Systematic Botany 15(6): 755–764. 10.1071/SB01015 [DOI] [Google Scholar]
- Hyde KD, Fryar S, Tian Q, Bahkali AH, Xu JC. (2016) Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the affects of global warming on biodiversity and function? Fungal Ecology 19: 190–200. 10.1016/j.funeco.2015.07.002 [DOI]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. (2020a) Refined families of Sordariomycetes. Mycosphere : Journal of Fungal Biology 11(1): 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei D, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li J, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang SK, Thiyagaraja V, Lu YZ, Jayawardena RS, Dong W, Yang EF, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu JC, Sheng J. (2020b) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Index Fungorum (2022) Index Fungorum. http://www.indexfungorum.org/Names/Names.asp [accessed on 10 May 2022]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-Ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, Ghobad-Nejhad M, Nilsson H, Pang KL, Pereira OL, Phillips AJL, Raspe O, Rollins AW, Romero AI, Etayo J, Selcuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, De Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I. (2015) The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74(1): 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jones EBG, Wong SW, Sivichai S, Au DWT, Hywel-Jones NL. (1999) Lignicolous freshwater ascomycota from Thailand: Micropeltopsisquinquecladiopsis sp. nov. Mycological Research 103(6): 729–735. 10.1017/S0953756298007618 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) Mafft online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView:afast andlightweight alignment viewer and editor for large datasets. Bioinformatics 30(22): 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Luo ZL, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY. (2018a) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17(5): 511–530. 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Bhat DJ, Bao DF, Li WL, Su HY. (2018b) Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere : Journal of Fungal Biology 9(3): 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY. (2019) Freshwater Sordariomycetes. Fungal Diversity 99(1): 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Mason EW, Hughes SJ. (1951) Phragmocephala, gen. nov. hyphomycetarum. Naturalist 1951: 97–105. [Google Scholar]
- Matsushima T. (1975) Icones microfungorum a Matsushima lectorum. Nippon Printing Co, Osaka.
- Matsushima T. (1980) Saprophytic microfungi from Taiwan, part 1, hyphomycetes. Matsushima Mycological Memoirs no. 1. Matsushima Fungus Collection, Kobe, Japan.
- Matsushima K, Matsushima T. (1996) Fragmentamycologica – II. Matsushima mycological memoirs no. 9: 31–40. Matsushima Fungus Collection, Kobe, Japan.
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop 2010 (GCE), New Orleans, Louisiana, November 2010, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Monteiro JS, Gusmão LFP, Castañeda-Ruiz RF. (2016) Pleurotheciumbicoloratum & Sporidesmiopsispluriseptata spp. nov. from Brazil. Mycotaxon 131(1): 145–152. 10.5248/131.145 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModel test v2 Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala.
- Rambaut A. (2014) FigTree v14: tree figure drawing tool. http://treebioedacuk/software/figtree
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution 43(3): 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Réblová M, Seifert KA, Fournier J, Štěpánek V. (2012) Phylogenetic classification of Pleurothecium and Pleurotheciella gen. nov. and its dactylaria-like anamorph (Sordariomycetes) based on nuclear ribosomal and protein-coding genes. Mycologia 104(6): 1299–1314. 10.3852/12-035 [DOI] [PubMed] [Google Scholar]
- Réblová M, Seifert KA, Fournier J, Štěpánek V. (2016) Newly recognised lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia 37: 57–81. 10.3767/003158516X689819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Hernández-Restrepo M, Fournier J, Nekvindová J. (2020) New insights into the systematics of Bactrodesmium and its allies and introducing new genera, species and morphological patterns in the Pleurotheciales and Savoryellales (Sordariomycetes). Studies in Mycology 95: 415–466. 10.1016/j.simyco.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systems biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski EM, Beimforde C, Gube M, Rikkinen J, Singh H, Seyfullah LJ, Heinrichs J, Nascimbene PC, Reitner J, Schmidt AR. (2012) The anamorphic genus Monotosporella (Ascomycota) from Eocene amber and from modern Agathis resin. Fungal Biology 116(10): 1099–1110. 10.1016/j.funbio.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. (2011) The genera of Hyphomycetes. CBS Biodiversity Series 9:1–997. CBS-KNAW Fungal Biodiversity Centre, Utrecht. 10.3767/003158511X617435 [DOI]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TTT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 11(1): 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shen HW, Bao DF, Hyde KD, Su HY, Bhat DJ, Luo ZL. (2021) Two novel species and two new records of Distoseptispora from freshwater habitats in China and Thailand. MycoKeys 84: 79–101. 10.3897/mycokeys.84.71905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivichai S, Boonyene N. (2004) Freshwater fungi. In: Jones EBG, Tanticharoen M, Hyde KD. (Eds) Thai Fungal Diversity.BIOTEC, Pathum Thani, Thailand, 95–106.
- Sivichai S, Hywel-jones N, Jones EBG. (1998) Lignicolous freshwater Ascomycota from Thailand: 1. Ascotaiwaniasawada and its anamorph state Monotosporella. Mycoscience 39(3): 307–311. 10.1007/BF02464013 [DOI]
- Sivichai S, Hywel-jones N, Somrithipol S. (2000) Lignicolous freshwater Ascomycota from Thailand: Melanochaeta and Sporoschisma anamorphs. Mycological Research 104(4): 478–485. 10.1017/S0953756299001604 [DOI] [Google Scholar]
- Sivichai S, Jones EBG, Hywel-Jones N. (2002) Fungal colonisation of wood in a freshwater stream at Tad Ta Phu, Khao Yai National Park, Thailand. Fungal Diversity 10: 113–129. 10.1016/0006-3207(72)90026-2 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22(21): 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web-servers. Systems Biology 75(5): 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Su HY, Udayanga D, Luo ZL, Manamgoda DS, Zhao YC, Yang J, Liu XY, McKenzie EHC, Zhou DQ, Hyde KD. (2015) Hyphomycetes from aquatic habitats in Southern China: Species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226(3): 201–216. 10.11646/phytotaxa.226.3.1 [DOI] [Google Scholar]
- Subramanian CV, Bhat DJ. (1989) Hyphomycetes from South India I. Some new taxa. Kavaka 15: 41–74. [Google Scholar]
- Sun LY, Li HY, Sun X, Guo LD. (2017) Dematipyriformaaquilaria gen. et sp. nov., a new hyphomycetous taxon from Aquilariacrassna. Cryptogamie, Mycologie 38(3): 341–351. 10.7872/crym/v38.iss3.2017.341 [DOI] [Google Scholar]
- Sun YR, Goonasekara ID, Thambugala KM, Jayawardena RS, Wang Y, Hyde KD. (2020) Distoseptisporabambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodiversity Data Journal 8: e53678. 10.3897/BDJ.8.e53678 [DOI] [PMC free article] [PubMed]
- Tubaki K, Watanabe K, Manoch L. (1983) Aquatic hyphomycetes from Thailand. Transactions of the British Mycological Society 451–457.
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li WJ, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu J, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89(1): 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocol Guide Methods Appl 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, McKenzie EHC, Hyde KD. (2012) Towards incorporating anamorphic fungi in a natural classification – checklist and notes for 2011. Mycosphere : Journal of Fungal Biology 3(2): 157–228. 10.5943/mycosphere/3/2/5 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk P, Kolaříková Z, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castrov I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu JC, Wang Y, Fenghua T, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCM, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Kuhnert E, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Pu L, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Lateef AA, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Erdoğdu M. (2020) Outline of fungi and fungus-like taxa. Mycosphere 11(1): 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa (2021). Mycosphere : Journal of Fungal Biology 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Wu YM, Zhang TY. (2009) New species of Phialosporostilbe and Pleurothecium from soil. Mycotaxon 110(1): 1–4. 10.5248/110.1 [DOI] [Google Scholar]
- Xia JW, Ma YR, Li Z, Zhang XG. (2017) Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Scientific Reports 7(1): 7888. 10.1038/s41598-017-08318-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZJ, Li XX, Wang HD, Song PY, Tang L. (2018) Rhexoacrodictysbroussonetiae sp. nov. from Guizhou, China. Mycotaxon 133(1): 149–152. 10.5248/133.149 [DOI] [Google Scholar]
- Zhang H, Jones EBG, Zhou DQ, Bahkali AH, Hyde KD. (2011) Checklist of freshwater fungi in Thailand. Cryptogamie. Mycologie 32(2): 199–217. 10.7872/crym.v32.iss2.2011.199 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genomics 3(1): 4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.