Abstract
Machine learning represents a growing subfield of artificial intelligence with much promise in the diagnosis, treatment, and tracking of complex conditions, including neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases. While no definitive methods of diagnosis or treatment exist for either disease, researchers have implemented machine learning algorithms with neuroimaging and motion-tracking technology to analyze pathologically relevant symptoms and biomarkers. Deep learning algorithms such as neural networks and complex combined architectures have proven capable of tracking disease-linked changes in brain structure and physiology as well as patient motor and cognitive symptoms and responses to treatment. However, such techniques require further development aimed at improving transparency, adaptability, and reproducibility. In this review, we provide an overview of existing neuroimaging technologies and supervised and unsupervised machine learning techniques with their current applications in the context of Alzheimer’s and Parkinson’s diseases.
Keywords: Alzheimer’s disease, clinical detection, deep learning, machine learning, neurodegenerative disorders, neuroimaging, Parkinson’s disease
Introduction
Healthcare represents one of the most prolific fields for the development and deployment of artificial intelligence (AI) technologies, medical imaging, wearable sensors, augmented and virtual reality, and more (Myszczynska et al., 2020). With today’s staggering abundance of big data, AI represents a particularly noteworthy emerging field, as it aims to automate human intelligence and emulate cognitive functions with the help of a wide range of mechanisms (Choi et al., 2020; Emmert-Streib et al., 2020). AI has already shown extraordinary promise in several areas, from the development of early diagnostic tools to the successful completion of robot-assisted surgery. However, the most recent growth and development in AI have come via advancements in machine learning (Myszczynska et al., 2020).
Machine learning (ML) is a subfield of AI that consists of algorithms targeted toward recognizing patterns and extracting noteworthy features from large datasets (Cao et al., 2018). Once these patterns are identified and learned, ML algorithms can be used to classify and predict future results. In healthcare, ML can be used on data from various sources, assisting in diagnosis as well as disease management, tracking, and outcome prediction. For instance, real-time remote monitoring by ML systems can detect disease severity, record symptoms, and register a patient’s response to treatment (Belić et al., 2019).
As with other medical disciplines, ML has been well integrated into the field of neurology, specifically concerning the computer-aided identification, monitoring, and management of symptoms associated with neurodegenerative movement disorders like Parkinson’s disease (PD). In PD, the pathogenic accumulation of alpha-synuclein in Lewy bodies and Lewy neurites drives a gradual loss of dopaminergic neurons in a dark-colored midbrain region known as the substantia nigra pars compacta (de Miranda and Greenamyre, 2017; Mahajani et al., 2019; Raina et al., 2020; Psol et al., 2021; Garg et al., 2022). This loss of dopaminergic neurons occurs long before the clinical characteristics of PD manifest and contributes to a variety of motor and non-motor symptoms (Giacomini et al., 2015; Marotta et al., 2016; Mahajani et al., 2021; Raina et al., 2021). Motor symptoms of PD range from rigidity and bradykinesia, or slow, impaired movement to resting tremors and postural instability. Other, non-motor symptoms of PD include constipation, olfactory dysfunction, disturbed sleep, cognitive and behavioral changes, and depression (de Miranda and Greenamyre, 2017; Kouli et al., 2018; MacMahon Copas et al., 2021). Yet, despite being the second most prevalent neurogenerative disorder today, currently no definitive method exists for the antemortem diagnosis of PD and no viable therapeutic strategy for disease treatment (DeMaagd and Phillip, 2015; de Miranda et al., 2017).
However, ML algorithms coupled with wearable devices have been used to address some of the challenges associated with PD. For instance, ML has been used to differentiate between PD and other disorders that present themselves similarly and to track and manage PD progression. ML-integrated tools possess great potential in clinical practice due to their increased accuracy, reliability, accessibility, and efficiency in clinical decision-making (Myszczynska et al., 2020).
As such, ML has also been used to track disease progression and to provide a source of differential diagnosis in the context of Alzheimer’s disease (AD). The most common form of dementia, AD is characterized by the abnormal accumulation of two proteins: amyloid-beta (Aβ) and tau. Misfolded amyloid precursor protein aggregates to form extracellular Aβ plaques, and hyper-phosphorylated tau forms intracellular neurofibrillary tangles. As neurofibrillary tangles and Aβ plaques form, synaptic degeneration, and neuronal death follow, driving a neurodegenerative progression through various brain regions (Blennow et al., 2006; eTure and Dickson, 2019; Soria Lopez et al., 2019; Wakhloo et al., 2022).
AD pathogenesis can be divided into stages depending on how far this neurodegeneration progresses and where neurofibrillary tangles and Aβ plaques are present. Early phase AD brains bear neurodegenerative features in the trans-entorhinal cortex. These features spread to the limbic regions in mid-phase AD brains and eventually to the iso-cortical regions in the late phases of the disease (Braak and Braak, 1991; Otero-Garcia et al., 2022). The symptoms accompanying this gradual neurodegenerative spread include the progressive impairment of cognitive functions, specifically speech, recognition, episodic memory, decision making, and orientation (Colombi et al., 2013; Cortelli et al., 2015; Mahajani et al., 2017; Wakhloo et al., 2020). As AD progresses, patients also sometimes suffer from apraxia, or the inability to perform learned movements despite possessing the desire, understanding, and physical ability required to do so (Blennow et al., 2006). Like PD, there exists neither a conclusive method for the diagnosis of AD nor an effective therapy capable of treating more than the symptoms of the disease (Briejyeh and Karaman, 2020).
Most prominent clinical symptoms develop after the brain has taken significant damage. Physicians are unable to diagnose these diseases before irreversible damage has been done, due to the lack of efficient diagnostic tools. It is, therefore, imperative to develop non-invasive methods of disease detection. Currently, neuroimaging can assist clinicians in staging and screening for specific identification of diseases. The use of AI, with ML and deep learning (DL) algorithms could assist clinicians in preclinical diagnosis of such diseases. To this extent, the combination of ML algorithms and neuroimaging techniques has granted researcher insights that may lead to a method for early AD diagnosis. For instance, recent studies have used ML to differentiate between the brains of patients with AD and those with mild cognitive impairment (MCI), the precursor to AD (Li et al., 2012; Suk et al., 2016; Saboo et al., 2022). Results of these studies allow researchers to identify biomarkers capable of predicting disease trajectory (Li et al., 2012; Suk et al., 2016) and of explaining individual vulnerability or resilience to cognitive decline (Saboo et al., 2022). Most importantly, the methods used in ML/neuroimaging studies provide a roadmap for the future identification of cognitively vulnerable individuals and the development of new therapeutic interventions (Saboo et al., 2022). In this review, we aim to outline the ML/DL modeling tools available to researchers and highlight certain use case scenarios.
Search Strategy
The references cited in this review have been obtained from the following databases: PubMed, Google Scholar, and Science Direct. We referenced full-text review articles, randomized control trials, meta-analyses, and textbooks. No limits were used.
Neuroimaging
Today’s neuroimaging technologies have proven capable of illustrating the brain’s anatomy with a resolution comparable to that achieved with high-quality images taken of thin tissue slices in vitro (Shen et al., 2017). Technological advancements in medical image processing have led to its widespread use and contributed to the development of new areas of exploration for the prediction and future diagnosis of neurodegenerative diseases (Noor et al., 2019). We discuss the neuroimaging techniques capable of identifying functional and anatomical changes related to neurodegeneration below.
Magnetic resonance imaging
Magnetic resonance imaging (MRI) uses a set of powerful magnets to generate magnetic waves capable of forming two or three-dimensional images of the brain without the need for radioactive tracers. MRI enables the imaging and evaluation of functional neural activity in the cortical regions. The images generated by MRI allow clinicians and scientists to study both functional and structural abnormalities of the brain in neurogenerative diseases (Ahmed et al., 2019).
Functional magnetic resonance imaging
Functional MRI (fMRI) determines the small changes that occur in blood flow with certain brain activity. It is used to determine the part of the brain responsible for critical functions, assess the effect of stroke, or to guide brain treatments (Dijkhuizen et al., 2012). Two most common types of fMRI are Quantitative Susceptibility Mapping and Diffusion Tensor Imaging. Quantitative Susceptibility Mapping detects the difference in magnetic susceptibility between healthy and diseased tissues, whereas Diffusion Tensor Imaging exploits the sensitivity of magnetic resonance signal into water molecules with small random motion (Liu et al., 2015; Ruetten et al., 2019). Quantitative Susceptibility Mapping is generally used to quantitatively assess tissue properties, whereas Diffusion Tensor Imaging studies the reorganization of the brain in different stroke models.
Arterial spin labeling
Arterial spin labeling (ASL) is one of the most widely used MRI techniques in clinical diagnosis. This non-invasive method can measure brain perfusion, providing a reliable technique to evaluate the cerebral blood flow of an individual suspected to have a neurodegenerative disorder (Arevalo-Rodriguez et al., 2021). ASL-MRI is also used to augment routine diagnostic procedures by providing a source of data for differential diagnosis, particularly in the differentiation between AD and frontotemporal dementia (Mas, 2018). 3D pseudo-continuous ASL-MRI data and tissue segmentation methods of the entire supratentorial cortex and ten gray matter regions are used to quantify cerebral blood flow and gray matter volume (Arevalo-Rodriguez et al., 2021). ASL-MRI is generally complemented with other cognitive examinations and/or questionnaires (De Vis et al., 2018). For instance, to distinguish patients suffering from AD from patients with frontotemporal dementia and controls, the Mini-Mental State Examination is used. Mini-Mental State Examination is a 30-question quiz developed by Folstein and McHugh in 1975 to help clinicians grade the cognitive state of patients (Upton, 2020). The quiz assesses attention, orientation, memory, registration, recall, calculation, language, and the ability to draw a complex polygon (Mas, 2018).
Positron emission tomography
Positron emission tomography (PET) measures radiation to generate two or three-dimensional images that record the circulation of bloodborne radiotracers throughout the brain (Fleisher et al., 2020). The primary benefit of PET is that it provides a visualization of brain activity and function, illustrating blood flow, oxygen level, and glucose metabolism in functional brain tissues (Bao et al., 2021). Combining PET scans with structural MRI could significantly improve the accuracy of neurological disease identification. The images obtained after using the radiotracer fluorodeoxyglucose and PET have been optimized to illustrate patterns characteristic of neurodegenerative diseases (Fleisher et al., 2020; Bao et al., 2021). Fluorodeoxyglucose-PET plays a crucial role in the early detection and monitoring of AD, illustrating pathophysiological changes in patient brains (Fleisher et al., 2020; Bao et al., 2021; Ni et al., 2021).
Single-photon emission computed tomography
Single-photon emission computed tomography (SPECT) is a functional nuclear imaging technique reliant upon radioactive tracers, or SPECT agents. SPECT is primarily used for the evaluation of regional cerebral blood flow. Regional cerebral blood flow is a measure of the rate of delivery of arterial blood to the capillary bed in brain tissues per unit time. Its output comes in the form of two- or three-dimensional images. SPECT has previously assisted in distinguishing between AD and white matter vascular dementia cases by analyzing the semi-quantitative circumferential profile (Ahmed et al., 2019).
Machine Learning Approaches
ML algorithms facilitate the clinical decision-making process by automatically classifying and predicting disease progression using computer-aided diagnosis, rather than the hands-on interpretation by medical experts (Shen et al., 2017; Myszczynska et al., 2020). ML models are trained by multiple techniques, including transfer learning using pre-trained weights, ensemble model construction, and new model development. Training data can be retrieved from multiple open source platforms such as Kaggle, IEEEDataPort, and Grand Challenge as well as from specialized neuro data repositories, such as NeuroVault, Whole Brain Atlas, Temple EEG database, SchizConnect, The Pain Repository, Open Access Series of Imaging Studies (OASIS), Glucose Imaging in Parkinsonian Syndromes Project (GLIMPS), Brain-CODE, Alzheimer’s Disease Neuroimaging Initiative (ADNI), OpenNeuro, and Collaborative Research in Computational Neuroscience (CRCNS).
Deep learning models
Deep learning (DL) is an emerging soft computing technique in ML that often relies on layered algorithmic structures known as neural networks (Ahmed et al., 2019; Myszczynska et al., 2020). A DL architecture is referred to as a “hybrid model” when combined with a traditional ML architecture, such as a support vector machine (SVM), as a classifier. DL and neural networks have been implemented in translational studies ranging in focus from sequence binding (Alipanahi et al., 2015; Trabelsi et al., 2019) to structural and image analysis (Shen et al., 2017; Saboo et al., 2022). Much ongoing research is focused on adapting neural network structures for disease detection and treatment analysis applications (Ahmed et al., 2019; Figure 1).
Figure 1.
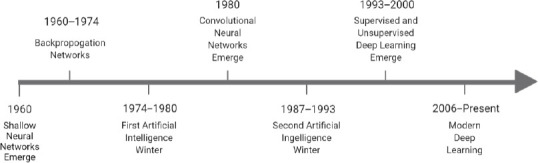
The evolution of deep learning techniques over time.
The concept of neural networks originated in the 1940s and expanded with the development of shallow neural networks in the 1960s. Several artificial intelligence winters stemmed from intellectual roadblocks in the field. Eventually, the supervised and unsupervised deep learning methods widely utilized in modern research arose, as deep neural networks became a critical component of computing (Emmert-Streib et al., 2020- for a more in-depth history, see this review as well as Schmidhuber, 2015).
The ML mechanisms involved in DL techniques are optimized via a model training process, where the computer is provided with a dataset (input) and an associated collection of outputs (Gautam and Sharma, 2020). The job of the ML model is to adapt its algorithm to fit an identified relationship between the two sets of information (Choi et al., 2020). This learning process falls into one of four categories, depending on the amount of labeling applied to a training dataset. In supervised learning models, entirely labeled training data provide an answer key that a model can use to evaluate and inform its accuracy (Segovia et al., 2018). In contrast, unsupervised models draw from entirely unlabeled training data and must rely on independently drawn conclusions and extracted features. In between these two categories are semi-supervised and reinforcement learning (Segovia et al., 2018). Semi-supervised models use both labeled and unlabeled training data, while reinforcement learning models train using an optimization-focused reward system (Kang and Jameson, 2018; Choi et al., 2020). This review will focus on supervised and unsupervised approaches, as reinforcement learning is currently not well-suited for medical analyses (Choi et al., 2020).
Supervised models
Algorithms that can identify patterns and generate hypotheses by using externally provided information to predict future outcomes are known as supervised ML algorithms (Choi et al., 2020). Supervised models are primarily used for data classification, regression, and to predict desired outcomes. Some of the common techniques proposed to solve problems using supervised ML algorithms include rule-based techniques, logic-based techniques, instance-based techniques, and stochastic techniques (Singh et al., 2016). Several commonly used supervised learning methods are described below.
Between the input and output, DL neural networks consist of multiple hidden layers responsible for data processing and computation. Hidden layers consist of a convolutional layer, a pooling layer, and a fully connected layer with an activation function. The convolutional layer is the main building block of a neural network. It consists of several filters, or kernels, whose parameters are learned throughout model training (Zhao et al., 2021). Examples of well-characterized and widely used DL models include convolutional neural networks (CNN), deep neural networks (DNN), and recurrent neural networks (RNN) (Uddin et al., 2019).
Built to imitate the alternating layers of cells present in the visual cortex of the human brain, CNN are composed of three layers: the convolutional layer, the pooling layer, and the fully connected layer. CNN implement a one-way model, in which information is transmitted from the input layer to the output layer only (Zhao et al., 2021). This feed-forward network structure can be implemented in both a supervised and an unsupervised manner (Rovini et al., 2018). CNN represent the most widely used DL approach for biomedical image analysis (Cao et al., 2018) and are designed to handle multiple array data, such as signals data two- and three-dimensional images. Some of the most widely used CNN include Alxenet, Lenet, R-CNN, Zfnet, GoogleNet, and ResNet (Zhao et al., 2021).
DNN are composed of multiple layers for transformation and use artificial neural networks as distinct processing layers. RNN are neural networks capable of forming internal memory, rendering them optimal for the analysis of sequential data (Rovini et al., 2018; Uddin et al., 2019). Bayesian networks are a statistical model used to represent the probabilistic relationship between a set of variables. Another statistical model is the logistic regression model, which deploys an online gradient descent method for improved probabilistic interpretation. This model is mainly applied when the dependent variables are dichotomous (Singh et al., 2016).
Decision trees are designed to deal with inseparable information and other data that include nominal, numeric, textual, missing, or redundant values. The random forest method utilizes an ensemble of decision trees. It is robust to noise, scalable, and does not overfit (Singh et al., 2016; Uddin et al., 2019). As described previously, neural networks are constructed to emulate the neural structures, learning abilities, and processing methods of the human brain. These models are deployed to solve non-linear problems (Uddin et al., 2019).
SVM is a complex supervised algorithm currently considered one of the best ML algorithms (Uddin et al., 2019). SVM models are deployed when data are not linearly separable to prevent overfitting. In contrast, k-nearest neighbor (k-NN) models are non-parametric classification algorithms that assign unlabeled sample points to the class of the nearest previously labeled sample point (Singh et al., 2016).
As Table 1 illustrates, all complex models underperform due to poor parameter choice. However, SVM and k-NN have outperformed all supervised ML algorithms as described in Table 1. SVM and k-NN are considered one of the best-supervised machine learning algorithms since SVM is robust in comparison to linear regression, handles multiple features, does not overfit, and performs very well in classifying semi-structured and unstructured data such as text and images. Whereas k-NN is a simple algorithm that can classify subjects quickly, is capable of handling noise and missing values, and is mainly used to solve regression and classification problems (Uddin et al., 2019). Tree-based algorithms have historically performed better than other algorithms. Figure 2 exhibits the different supervised algorithms in use and their modes of implementation.
Table 1.
The potential applications, advantages, limitations, and varied accuracies of commonly used supervised learning algorithms
| Algorithm | Potential applications | Advantages | Limitations | Accuracy | |
|---|---|---|---|---|---|
|
| |||||
| Test | Cross-validation | ||||
| Bayesian Network | Classification of documents, medical prognosis system | Takes less time to train the model and can interpret the relationship among predictors. | Cannot deal handle high dimensional data and efficiency of the model decreases with the increase of data. | 0.73 | 0.77 |
| Logistic Regression | Crash types, and injury severity, handle the nonlinearity in data. | The model is capable of handling nonlinear data and interprets the output as probability. | Suffer multicollinearity and require large data to provide stable results. | 0.75 | 0.79 |
| Random Forests | Identifies a cluster of patients, object detection, and classification of microarray data. | Scalable, fast, robust to noise, does not overfit and provides explanation and visualization of the output. | As the number of trees increases the algorithm slows down. | 0.77 | 0.82 |
| SVM | Text classification | High accuracy, does not overfit, accuracy and performance of the model is independent of features, excellent generalizability, and | Slow training speed, highly complex model, and the performance of the model highly depends upon the selected parameters | 0.76 | 0.82 |
| k-NN | Vision and computational geometry | Suitable for multi-modal classes, the model is independent of the joint distribution of sample points | Low efficiency, output depends upon the selection of the K value, the model is adversely affected by the noise and irrelevant features, and performance varies according to the size of the data set. | 0.63 | 0.81 |
| Neural Networks | Image classification | Deals with the relationship which may be either nonlinear or dynamic, independent of variables, robust to irrelevant input or noise, and used for | Takes time to train, performance is sensitive to the chosen parameters and the size of hidden layers. | 0.74 | 0.81 |
The difference between cross-validation and test accuracy demonstrates the degree of over-fitting implicit in each model (Singh et al., 2016).
Figure 2.
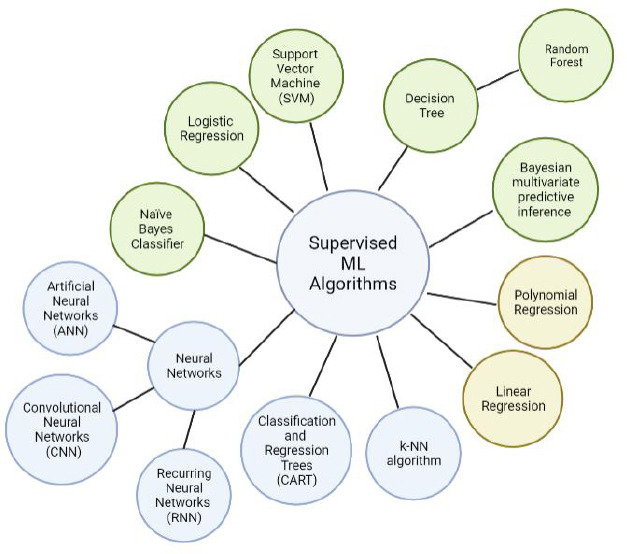
A relationship map of currently used supervised learning models.
Yellow circles denote regression algorithms, green circles denote classification algorithms, and blue circles mark algorithms that can be implemented for both purposes. The neural networks category can be grossly deconstructed into artificial neural networks (ANN), convolutional neural networks (CNN), and recurring neural networks (RNN). The random forest model represents a collection of individual decision trees. Created with BioRender.com.
Unsupervised models
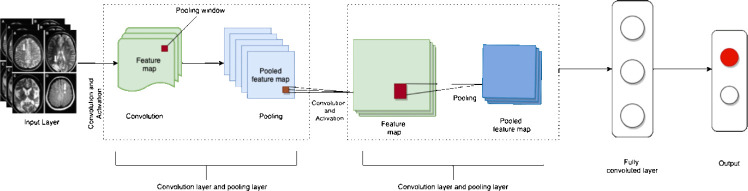
Unlike supervised algorithms, unsupervised algorithms learn without an answer key, supervisor, or source of external information to fall back on (Kang and Jameson, 2018; Choi et al., 2020). Figure 3 demonstrates the general architecture of unsupervised neural networks. Unsupervised neural networks and other algorithms are mainly used for clustering and feature reduction (Noor et al., 2020).
Figure 3.
A representation of the basic architecture of an unsupervised neural network.
First, feature maps are made from MRI, CT, or PET images. Next, pooling layers summarize the feature maps. This step is repeated through multiple neural layers. Then, the final feature maps are sent to the final fully connected layer, where predictions and masks are made, leading to the eventual model output. Adapted from Gautam and Sharma (2020).
Unsupervised models are also known as self-supervised algorithms. Autoencoders (AE), which are used for compression and other functionalities, represent a unique form of self-supervised learning (Singh et al., 2015). AE are deployed to implement a feedforward approach to generating output from an input (Singh et al., 2015). In this feedforward approach, multiple inputs enter a layer and are multiplied by their respective weights. The generated outputs are then added together to obtain a sum of all weighted input values (Lu et al., 2020).
Deep autoencoders are unsupervised models with a stacked structure composed of three layers: the input (encoder), the output (decoder), and the hidden layer (code). Sparse AE are another commonly used unsupervised ML model that relies on an axisymmetric single hidden layer for feature extraction. Stacked AE are used for the prediction of disease using time-frequency features of speech signals. Stacked autoencoders are made up of three layers: an input layer, hidden layers (which generate learned features), and an output layer in the same dimension as the input layer, demonstrating the reconstruction of the inputs (Li et al., 2019).
Beyond the scope of artificial neural networks, other generative learning methods have also been implemented to solve complex problems. Generative learning methods include models like the deep belief network (Zhao et al., 2021). Developed in 2006, the deep belief network is an unsupervised model composed of complex layers of the restricted Boltzmann machine algorithm with pre-trained weights. This unsupervised generative neural network is comprised of two layers: visible and hidden. Generative adversarial networks represent another generative learning technique structured based on image-to-image translation. As a result, this class of model is also known as pix2pix (Zhao et al., 2021). Other common unsupervised ML algorithms used in the detection of neurodegenerative disorders include K-means and self-organizing maps, described in more detail below.
K-means: One of the simplest unsupervised ML algorithms, the K-means method represents a popular clustering algorithm used to divide a given dataset between a pre-determined number of clusters (k) based on recognized similarities and dissimilarities between data points (Raval and Jani, 2016; Ahmed et al., 2020). The model operates by randomly selecting data points to represent each cluster. These data points, known as centroids, center and define the characteristics of each cluster throughout the sorting process (Ahmed et al., 2020). However, using the k-means model presents several difficulties. For instance, varying k-value selection leads to varying convergence results (Ahmed et al., 2020). Similarly, different randomized centroid placements can generate different results (Das et al., 2017). To optimize the k-center placement, researchers attempt to place them as far as possible from one another (Das et al., 2017).
Self-organizing maps (SOM): SOM are considered one of the most efficient unsupervised neural network techniques. The basic architecture of SOM is based on competitive learning. SOM are primarily used for the cluster analysis of high-dimensional data. This technique aims to reduce the dimensionality of complex data, organizing a simplified representation of each datapoint spatially within a two-dimensional map (Sarmiento et al., 2017). Datapoints on the map are known as neurons. These neurons build a two-dimensional lattice that acts as the output layer of the SOM, where high-dimensional input space is mapped within the plane. The completed map acts as a spectrum, where the discrete features of each simplified datapoint resemble its immediate neighbor (Sarmiento et al., 2017).
Deep Learning Applications
The four essential steps for image processing include acquiring an existing dataset, analyzing, and identifying new patterns in the data. This is followed by preprocessing the raw data and making predictions. Based on the accuracy of the model, it can be then trained on other new datasets (Figure 4).
Figure 4.
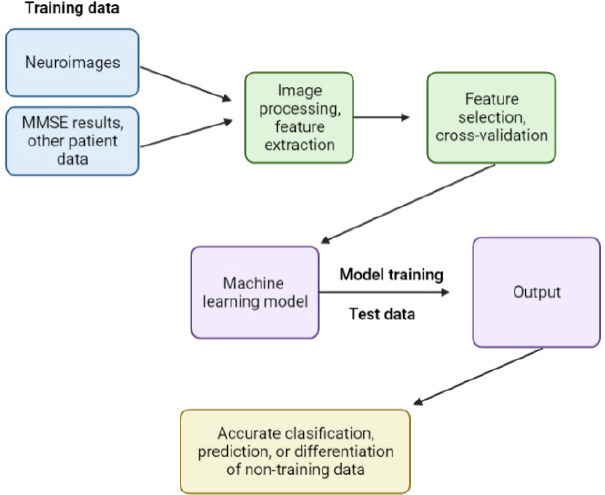
An overview of the step-by-step process by which machine learning and computer-aided diagnosis techniques process and analyze clinical and neuroimaging data to identify features associated with neurodegenerative diseases.
First, images and clinical data are processed, and features of interest are identified. Then, the identified features are extracted and cross-validated across data types. The machine learning model establishes patterns in the training dataset that can be used to classify or make predictions based on any comparable future dataset. Created with BioRender.com. MMSE: Mini-Mental State Examination.
However, the primary limitation of DL applications for the analysis of biomedical images remains the availability of only a small sample of training data in a space where additional images are not easily obtainable (Shen et al., 2017). The more complex the DL model, the more parameters it must internalize (Ying, 2019). Therefore, to construct and train a sufficiently sophisticated DL algorithm without overfitting the model, researchers must find creative ways to expand their dataset (Shen et al., 2017). To accomplish this, researchers use a range of different techniques. Data augmentation, for example, involves the artificial inflation of the existing training dataset by transforming existing images (Shorten and Khoshgoftaar, 2019). Dimensionality reduction can also be used to reduce the existing number of model parameters, or models can be pre-trained with external data (Shen et al., 2017).
Evaluation of Classifier Performance
Evaluation of classifier performance is done in terms of sensitivity or recall. Eq (1) is used to calculate the percentage of cases correctly identified as true, or specificity. Eq (2) calculates the percentage of cases correctly identified as false, or precision. Eq (3) is employed to determine the percentage of cases correctly identified as true about all diagnosed as true, otherwise labeled accuracy (Uddin et al., 2019). Eq (4) determines the percentage of cases that are correctly identified with respect to all subjects, and F-score Eq (5) calculates a weighted average of specificity and sensitivity. To obtain all the above measurement values, True Positive (TP), False Positive (FP), True Negative (TP), and False Negative (FN) are calculated (Uddin et al., 2019).
Eq (1) Recall = TP/(TP+FN)
Eq (2) Specificity = TN/(TN+FP)
Eq (3) Precision = TP/(TP+FP)
Eq (4) Accuracy = (TP+TN)/(TP+TN+FP+FN)
Eq (5) F-score = 2 × (Precision × Recall)/(Precision+Recall)
Machine Learning Classifiers for Alzheimer’s Disease
Currently, some of the most common DL techniques used for clinical AD prognosis include DNN, restricted Boltzmann machine algorithm, DBM, deep belief network, AE, Sparse AE, and Stacked AE (Altinkaya et al., 2020). Each architecture has been developed to distinguish between multi-modal neuroimaging data from cognitively normal controls and brains with MCI, commonly known as the prodromal or pre-symptomatic phase of AD. Once trained, these DL models can be used to predict the conversion of MCI to AD (Bringas et al., 2019; Altinkaya et al., 2020). AD research using DL algorithms is still evolving to achieve higher accuracy, as summarized in Table 2.
Table 2.
ML algorithms developed for the classification of AD, CN, and MCI over the past ten years and their accuracies
| Author | Study | Method | Architecture | Accuracy |
|---|---|---|---|---|
| Suk and Shen, 2013 | AD/CN classification; MCI/CN classification; AD to MCI conversion | Feature representation with a stacked autoencoder from MRI and PET data | Data processing and training: SAE; Classifier: SVM | AD/CN classification: 95.9% MCI/CN classification: 85.0% MCI to AD prediction: 75.8% |
| Ngiam et al., 2011; Liu et al., 2015 |
AD/CN classification | Extraction of complementary information from multimodal neuroimaging data. | SAE, Softmax logistic regressor, and zero-masking strategy | 91.40% |
| Liu et al., 2014 | AD/CN classification | Extraction of complementary information from multimodal neuroimaging data | Stacked SAE and Softmax regression layer | 87% |
| Li et al., 2014 | AD/CN Classification and MCI to AD Conversion Prediction | Subjects with MRI and PET scans encode non-linear relationship between MRI and PET images. Trained network used to estimate PET patterns for subjects with only MRI data. | 3D CNN | AD/CN classification: 92.87% MCI to AD prediction: 72.44% |
| Li et al., 2014; Vu et al., 2017; Choi and Jin 2018 | AD/CN classification and MCI to AD conversion | Subjects with MRI and FDG PET scans encode non-linear relationship between MRI and PET images. Trained network used to estimate PET patterns for subjects with only MRI data. | 3D CNN models, SAE and 3D CNN | 96.00% 91.10% AD/CN classification: 92.87% MCI to AD prediction: 72.44% |
| Li et al., 2014; Vu et al., 2017; Choi and Jin, 2018; Liu et al., 2018b | AD/CN classification and MCI to AD conversion prediction | Subjects with MRI and FDG PET scans encode non-linear relationship between MRI and PET images. Trained network used to estimate PET patterns for subjects with only MRI data. Used three independent data sets (Training: ADNI-1; testing: ADNI-2 and MIRIAD) | 3D CNN models SAE and 3D CNN, 3D CNN | 96.00% 91.10% AD/CN classification: 92.87% MCI to AD prediction: 72.44% ADNI-2: 91.09 MIRIAD: 92.75% |
| Li et al., 2014; Vu et al., 2017 | AD/CN classification and MCI to AD conversion prediction | Subjects with MRI and PET scans encode non-linear relationship between MRI and PET images. Trained network used to estimate PET patterns for subjects with only MRI data | SAE and 3D CNN3D CNN | 91.10% AD/CN classification: 92.87% MCI to AD prediction: 72.44% |
| Vu et al., 2017 | AD/CN Classification | MRI and FDG PET scans | SAE and 3D CNN | 91.10% |
| Cheng and Liu, 2017; Cheng et al., 2017 | AD/CN classification | Neuroimages from MRI and PET scans. Results combined in 3D CNN Feature extraction from MRI images. | Two 3D CNN | Two 3D CNN: 89.6% |
| Single 3D CNN | Single 3D CNN: 87.2% | |||
| Korolev et al., 2017; Cheng and Liu, 2017 | AD/CN classification | Manual feature extraction. Neuroimages from MRI and PET scans. Results combined in 3D CNN | Plain (VoxCNN) | 80% |
| Residual Neural Networks (ResNet) Two 3D CNNs | 89.60% | |||
| Aderghal et al., 2017; Korolev et al., 2017 | AD/CN classification | 2D slices from hippocampal region in axial, sagittal, and coronal directions. Manual feature extraction | 2D CNN plain (VoxCNN) | 85.90% |
| residual neural networks (ResNet) | 80% | |||
| Cheng et al., 2017; Lu et al., 2018 |
AD/CN classification and MCI to AD conversion prediction | Extraction of features from MRI images. Pre-training: SAE; final prediction: DNN | Single 3D CNN | Single 3D CNN: 87.2% |
| SAE and DNN | AN/CN classification: 84.6% MCI to AD prediction: 82.93% |
|||
| Aderghal et al., 2017; Liu et al., 2018a | Intra-slice and inter-slice features for AD/CN classification | Decomposed 3D PET images into 2D slices, used 2D slices from hippocampal region in axial, sagittal, and coronal directions | Combination of 2D CNN and RNN + 2D CNN | 91.20% 85.90% |
| Lu et al., 2018 | AD/CN classification and MCI to AD conversion prediction | SAE for pre-training, DNN for final step | SAE and DNN | AN/CN classification: 84.6% MCI to AD prediction: 82.93% |
| Liu et al., 2018b; Liu et al., 2018a |
Intra-slice and inter-slice features for AD/CN classification | Three independent data sets of 3D PET images decomposed into 2D slices. (Training: ADNI-1; Testing: ADNI-2, MIRIAD) | 3D CNN Combination of 2D CNN and RNNs | ADNI-2: 91.09 MIRIAD: 92.75%, 91.2% |
From the available literature, the 3D CNN model developed by Choi and Jin (2018) appears to outperform all ML algorithms with an accuracy of 96.0%.
Data mining identifies patterns and relationships, classifies complex data, and extracts useful information from the recorded data. It is a common technique used on K-means datasets for the automatic classification of normal control individuals, MCI, and AD (Nabeel et al., 2021). Extracting information from real cases helps in the early clinical detection of AD. A recent study by Uddin et al. (2019) demonstrates the comparison of supervised ML architectures for AD classification. The classification was based on various performance metrics, including accuracy, precision, recall, and F1 score, to determine the efficiency of each classifying architecture. The study’s results demonstrated that the bagging method, which combines several learning models in parallel, outperformed all other ML architectures. Other studies have identified XG-Boost as the most powerful architecture for the classification of MCI subjects (Gautam and Sharma, 2020; Noor et al., 2020).
Additional research coupled with AI-integrated wearable sensors has identified CERAD change scores as a valuable tool for the early detection of MCI (Gautam and Sharma, 2020; Noor et al., 2020). These studies suggest that these architectures and sensors can be implemented in a clinical setting as a diagnostic tool (Taeho et al., 2019). Integrating data mining and ML algorithms seems to promise more accurate disease predictions and classifications of AD and other forms of dementia (Uddin et al., 2019).
Machine Learning Classifiers for Parkinson’s Disease
ML analysis of simple drawing tasks and handwriting is widely used in the early detection of PD. Several supervised ML algorithms are also increasingly utilized for PD symptom tracking. Figure 5 illustrates a general workflow for the supervised ML algorithms implemented in clinical practice for the identification of PD using motor symptoms (Rovini et al., 2018; Hoq et al., 2021).
Figure 5.
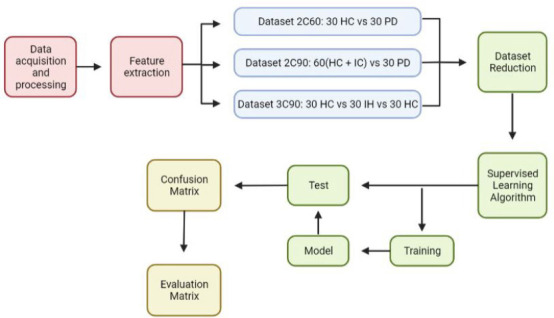
An illustration of the supervised ML algorithms used in comparative motor pre-clinical assessments for the classification of PD.
After motor data is acquired from all groups, the algorithm extracts notable distinguishing features, distributing the subjects into three different datasets. The reference dataset (2C60, 2C90, 3C90; Rovini et al., 2018). The dimensionality of the dataset is reduced before being fed into the supervised learning algorithm itself, where training takes place, then model testing. The algorithm output comes in the form of a confusion matrix, which is transformed into an evaluation matrix capable of categorizing disease types based on their distinguishing features. Created with BioRender.com.
To identify irregular motor characteristics implicated in PD and to differentiate between abnormal and normal hand movements, researchers implement classifier-based supervised ML algorithms on data derived from patients given horizontal drawing tasks (Hoq et al., 2021). Naïve Bayes classification has been combined with a set of metrics related to the velocity and spatiotemporal tracing of the subject’s pen to accurately differentiate between PD patients and normal control participants. Similarly, Archimedean spiral tracing performed by individuals with PD can undergo automatic analysis by a supervised model (Rovini et al., 2018). The results of algorithm-scored drawings display striking similarities to drawings independently scored by clinical experts (Nair et al., 2020). These tests are performed on patients’ personal computers using the relevant supporting software. Therefore, since these tests can be performed via smartphones and tablets, they represent an accessible and cost-effective approach to the analysis of motor function (Woodzinski et al., 2019).
Two common ML-based hardware systems used in clinical practice for the identification of PD are Parkinson’s KinetiGraph and the Kinesia system (Power et al., 2019). These systems are designed specifically for the identification of dyskinesia and bradykinesia in individuals suffering from PD. The KinetiGraph system is worn on the wrist to measure wrist acceleration (Cancela et al., 2016), whereas Kinesia is worn on either a finger or wrist and detects motion via a built-in accelerometer and gyroscope (Belić et al., 2019).
These system outputs are analyzed using supervised ML algorithms, such as SVM. For instance, Kinesia recordings have been used to classify PD tremor severity. Gait analysis algorithms have also shown promising results in the early identification of motor disorders (Heldman et al., 2017). For instance, RNN and long short-term memory have been used to classify gait analysis recordings, drawing from a database that includes measures of stride-to-stride footfall times. These algorithms have proven capable of differentiating between healthy controls and individuals suffering from PD, Huntington’s disease, and motor neuron disease, with accuracies ranging from 95% to 100%. However, in SVM, an ensemble of classifiers performs better than a single classifier. Existing algorithms can also be improved to optimize performance (Heldman et al., 2017; Belić et al., 2019).
Bayesian multivariate predictive inference platforms have also been applied to clinical data to study PD progression. Latourelle and colleagues published a study in which they trained a model for assessments of motor progression and of the complete molecular and genetic information obtained from a group of 117 healthy controls and 312 participants with PD over two years. To identify novel predictors of motor progression in the early stages of PD, a total of 17,499 features were included in the model. Progression modeling identified common factors for faster motor regression, including higher baseline motor scores, old age, and male sex. However, it also identified new predictors such as genetic variation and biomarkers found in patients’ cerebrospinal fluid (Latourelle et al., 2017).
As shown in Table 3, the SVM ML classifier appears to perform best for the differentiation of PD patients from control subjects (Cai et al., 2017). SVM algorithms report the highest accuracy scores in all performance measures except for precision and F-score, in which they rank second. In the existing literature, authors have also compared the accuracy of regression models for tracking the development of PD using metrics such as the measured error between paired predictions, the mean squared error, and the coefficient of correlation (Nair et al., 2020). Studies have also reported that least square SVM outperforms both multilayer perceptron neural networks and general regression neural networks in the differentiation between healthy and PD patients (Cai et al., 2017). At this point, SVM remains the most suitable method for modeling vocal features for PD prognosis and monitoring (Perumal and Sankar, 2016).
Table 3.
ML algorithms developed for the classification of PD over the past ten years and their accuracies
| Reference | Title | Method | Architecture | Accuracy |
|---|---|---|---|---|
| Mancini et al., 2011 | Trunk accelerometry reveals postural instability in untreated PD | Posture: (i) EO gazes straight ahead at art poster 6 m continuously. (ii) EC, upright standing position; (iii) EC cognitive task (ECT). | Linear mixed model; ROC | Accuracy for F95: 0.90; FD: 0.82; RMS: 0.93; for jerk (EO) for untreated PD/HC classification. |
| Sant’Anna et al., 2011 | New measure of movement symmetry in early PD patients using symbolic processing of inertial sensor data | Walking 30 m hallway at preferred speed (2 minutes) | t-test, ROC; ICC | Accuracy for PD/HC classification: 0.872; ICC: 0.949 |
| Rigas et al., 2012 | Assessment of tremor activity in PD using set of wearable sensors | Resting task (resting in bed, on a chair, standing with hand support); Postural task; kinetic tasks (finger to nose, finger to finger, walking, and picking) | HMM (Leaving one patient out) | Accuracy for posture and action detection: 81%; Accuracy for tremor severity classification: 87% |
| Scanlon et al., 2013 | Accelerometry-based study of lower and upper limb tremors in PD | Resting task, Postural task with distracting task [upper and lower limbs, both dexterity dominant and non-dominant] (each 8.2 seconds) | Mann-Whitney U and Wilcoxon signed-rank texts | IIVF50 for RT (P = 0.032), (P = 0.017) lower in the DD lower limb of PwPD compared to HC |
| Chen et al., 2014 | Postural sway in idiopathic rapid eye movement sleep behavior disorder as a potential marker of prodromal Parkinson’s disease | Upright standing position: arms crossed by chest, looking ahead (every 30 seconds) (i) eyes open (EO) (ii) feet together eyes closed (EC) (iii) feet together EO dual-task (EODT) (iv) feet together EC dual-task (ECDT) (v) tandem standing EO (TEO) | ANOVA, t-test, Pearson chi-square test | Differences in jerk between PD/HC for EODT (P = 0.030), ECDT (P = 0.015), and TEO (P = 0.023) |
| Kostikis et al., 2015 | Smartphone-based tool for assessing Parkinsonian hand tremor | Resting and Postural task (every 30 seconds) | Pearson coefficient; Bag DT | AUC for PwPD/HC classification: 0.94 |
| Perumal and Sankar, 2016 | Gait and tremor assessment for patients with PD using wearable sensors | Gait and Tremor (60 seconds) | ANOVA, LDA 5-fold cross-validation, ROC | Mean accuracy for Gait: 91.58%, ROC: 0.72 AUC for PD/HC classification: 90% Features able to differentiate PD tremor from atypical PD tremor |
| Cai et al., 2017 | New hybrid intelligent framework for predicting PD | Voice recordings of 31 subjects, including 23 PD patients (16 males, 7 females) and 8 healthy controls (3 males, 5 females). Each subject provided an average of six 36-second long phonations of vowels (95 samples total) | BFO-SVM, KELM | Acc of BFO-SVM: 96.84%, sensitivity: 98.75%, Specificity: 90.83% |
| Rumman et al., 2018 | Early detection of PD using image processing and artificial neural networks | SPECT Image dataset retrieved from PPMI database. ANN trained twice: first with ROI area of known subjects, then ROI area of unknown subjects | ANN | Accuracy: 94% Sensitivity: 100% Specificity: 88% |
| Woodzinski et al., 2019 | Deep learning approach to PD detection using voice recordings and convolutional neural networks for image classification | 100 voice recordings divided into 10 folds 90/10% (training and validation data). Included 50 HC and 50 PD patient recordings. PC-GITA database created to evaluate the model | LSTM, ResNet with 18 layers | F1-score, Precision, and recall: 0.92 Accuracy: 0.917 |
| Nair et al., 2020 | Predicting early-stage drug-induced Parkinsonism using unsupervised and supervised machine learning | Kinematic walking data | Logistic regression model | Logistic regression accuracy: 0.94, specificity: 0.96, sensitivity: 0.89 |
| Powers et al., 2021 | Longitudinal, remote smartwatch monitoring of PD motor defects to inform treatment decisions | Motor Fluctuations Monitor for Parkinson’s Disease (MM4PD) algorithm trained on smartwatch tremor data from 3 studies (343 PD, 171 controls), used to track symptom changes with activity, medication, etc. | Apple Proprietary Algorithm | Post-treatment symptom changes matching clinical expectations: 94% |
Conclusion and Outlook
To efficiently implement machine learning and data mining techniques for the clinical detection of the neurodegenerative disorders, more training data should be made available. More patient information such as postmortem data needs to be present to avoid a high error rate. However, a semi-supervised algorithm must be implemented using a clustering approach for high accuracy results. Literature also reveals that while implementing ML algorithms especially neural networks on different neuroimages such as MRI, ASL, PET, and SPECT images must be combined with SVM and k-NN for improved results since SVM is robust to linear regressions and is best at differentiating PD from control subjects whereas k-NN is a simple algorithm, classifies subjects quickly and handles noise along with unlabeled data. However, for the classification of AD patients in the clinical setting 3D CNN models should be implemented since 3D models provide promising results. Moreover, recent research also reveals that Kohonen unsupervised self-organizing map and least-squares support vector machine when performed on Structural MRI could detect structural changes of early PD. Singh and colleagues implemented this technique on a large dataset which validates the robustness and value of the technique (Singh et al., 2018). In summary, using combined neuroimaging, multi-modal, and clinical data could further enhance the diagnosis and early detection of neurodegenerative disorders. To implement machine learning in clinical settings for diagnosis and other applications, further validation and optimization are required to make it reliable and accurate. Despite the challenges in deciphering machine learning into clinical settings, it can assist clinicians in improving differential diagnosis of Parkinsonism, AD patients and early detection of neurodegenerative disorders, which can drastically reduce the error rate and help in diagnosing PD at a pre-motor stage so that early treatment is started to slow down the progression of neurodegenerative diseases like Parkinson’s and Alzheimer’s disease.
Researchers intend to develop DL classification models based on time series to learn patients’ temporal patterns. Even though great success has been achieved in the diagnosis of brain disorders using functional MRI (fMRI) images, these successes remain far from providing an effective clinical diagnosis (Yin et al., 2022). To implement fMRI in a clinical setting, researchers must first develop reliable and explainable biomarkers. Future DL models coupled with neuroimaging should be capable of classifying more than one single disorder against healthy controls with high accuracy (Yin et al., 2022). Implementing complementary parameters such as electronic medical records, EEG, and structural MRI images with fMRI could help yield better results (Yin et al., 2022).
Other fusion methods still in development include a cross-modal representation-based method for fMRI images that show enhanced performance over traditional DL models. However, there remains a need for more training samples for multimodal fusions (Yan et al., 2022).
Another important yet relatively new field in neuroimaging is neural architecture search (NAS) techniques (Yan et al., 2022). NAS automatically selects, composes, and parameterizes DL models to achieve maximum accuracy and optimal model performance on provided fMRI or neural images (Yan et al., 2022). NAS techniques show additional promise due to their optimization of search space, search strategy, and performance estimation strategy (Yan et al., 2022). The search space is defined as potential neural architectures that can be implemented using the NAS algorithm (Yan et al., 2022). The search strategy is termed as how this search space is explored. And finally, the performance estimation strategy references the performance evaluation parameters that evaluate NAS algorithm performance on various training datasets (Yan et al., 2022).
Scientists are working on developing wearable sensors with embedded digital signal readout software for the early diagnosis and symptom monitoring of neurodegenerative diseases (Asci et al., 2022). For instance, ongoing research seeks to develop electrochemical/biocatalytic sensors for the detection of L-Dopa, a dopaminergic precursor molecule, in patients at risk for PD. These wearable, minimally invasive sensors allow researchers and clinicians to correlate plasma levels of L-Dopa and the severity of motor symptoms, tailoring the treatment accordingly (Asci et al., 2022). Small inertial sensors such as tri-axial accelerometers and gyroscopes can be placed on different body parts to examine patient motor activity (Mughal et al., 2022). Capable of recording 3D kinematic and spatial-temporal data related to the body’s spatial orientation and motion (Avalle et al., 2021), these sensors can be integrated with random forest algorithms for signal pattern recognition and classification (Asci et al., 2022). Such classifications could be further integrated into clinical settings to objectively analyze motor symptoms and quantify PD severity and progression (Mughal et al., 2022).
In contrast, infrared ambient sensors can be placed in the home as well as in smartphones, tablets, or wristwatches to monitor behavioral symptoms of AD (Gillani and Arslan, 2021). These sensors monitor signals related to patients’ interest in and interaction with their environment, providing information that is processed using ML algorithms to predict the patient’s declining cognitive functionality (Perumal and Sankar, 2016). This feature provides continuous feedback helping caregivers and health professionals by providing autonomous patient support and monitoring disease progression (Gillani and Arslan, 2021).
Other wearable sensors such as Neuroglass are also being developed for the early detection of neurodegenerative disorders (Asci et al., 2022). Neuroglass seeks to integrate sensors capable of tracking head movement, velocity, acceleration, blood pressure, body temperature, blood oxygenation, electroencephalography, electro-oculography, and trans-cranium impedance (Avalle et al., 2021). However, Neuroglass’ primary technique measures eye motion and ocular tremor. Clinicians and scientists believe that eye and head movement quantification could represent an effective approach for the early detection of neurological disorders, drastically reducing diagnostic error (Avalle et al., 2021).
Whereas the integration of ML and wearable technology could soon emerge as a predictive tool for the early diagnosis of neurological diseases, important areas for further research still remain. Areas of investigation which require work include the detection of pre-symptomatic cognitive decline at the cellular level, improvements in GAIT and movement analysis for patients using wearable sensors, patient memory recollection using video modeling, and understanding the role of telemedicine in the treatment of neurological diseases.
Footnotes
Abbreviations: AD: Alzheimer’s disease; AE: auto encoders; AI: artificial intelligence; ANN: artificial neural networks; ASL: arterial spin labeling; CN: cognitively normal; CNN: convolutional neural networks; DBM: deep belief network; DL: deep learning; DNN: deep neural networks; EC: eyes closed; ECDT: EC dual-task; EO: eyes open; EODT: EO dual-task; fMRI: functional magnetic resonance imaging; HC: healthy controls; k-NN: k-nearest neighbor; MCI: mild cognitive impairment; ML: machine learning; MM4PD: motor fluctuations monitor for Parkinson’s disease; MRI: magnetic resonance imaging; NAS: neural architecture search; PD: Parkinson’s disease; PET: positron emission tomography; ResNet: residual neural networks; RNN: recurring neural networks; ROC: receiver’s operating curve; SOM: self-organizing maps; SPECT: single-photon emission computed tomography; SVM: support vector machine; TEO: tandem standing EO.
Conflicts of interest: The authors declare no conflicts of interest.
C-Editors: Zhao M, Liu WJ, Qiu Y; T-Editor: Jia Y
References
- 1.Ahmed M, Seraj R, Mohammed Shamsul Islam S. The k-means algorithm:a comprehensive survey and performance evaluation. Electronics. 2020;9:1295. [Google Scholar]
- 2.Ahmed MR, Zhang Y, Feng Z, Lo B, Inan OT, Liao H. Neuroimaging and machine learning for dementia diagnosis:recent advancements and future prospects. IEEE Rev Biomed Eng. 2019;12:19–33. doi: 10.1109/RBME.2018.2886237. [DOI] [PubMed] [Google Scholar]
- 3.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 4.Altinkaya E, Polat K, Barakli B. Detection of Alzheimer's disease and dementia states based on deep learning from MRI images:a comprehensive review. J Institute Electronics Comput. 2020;1:39–53. [Google Scholar]
- 5.Arevalo-Rodriguez I, Smailagic N, Roqué-Figuls M, Ciapponi A, Sanchez-Perez E, Giannakou A, Pedraza OL, Bonfill Cosp X, Cullum S. Mini-Mental State Examination (MMSE) for the early detection of dementia in people with mild cognitive impairment (MCI) Cochrane Database Syst Rev. 2021;7:CD010783. doi: 10.1002/14651858.CD010783.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asci F, Vivacqua G, Zampogna A, D'Onofrio V, Mazzeo A, Suppa A. Wearable electrochemical sensors in Parkinson's disease. Sensors. 2022;22:951. doi: 10.3390/s22030951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avalle M, Belotti V, Frascio M, Razzoli R. Development of a wearable device for the early diagnosis of neurodegenerative diseases. IOP Conf Ser:Mater Sci Eng. 2021;1038:012033. [Google Scholar]
- 8.Bao W, Xie F, Zuo C, Guan Y, Huang YH. PET neuroimaging of Alzheimer's disease:radiotracers and their utility in clinical research. Front Aging Neurosci. 2021;13:114. doi: 10.3389/fnagi.2021.624330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belić M, Bobić V, Badža M, Šolaja N, Đurić-Jovičić M, Kostić VS. Artificial intelligence for assisting diagnostics and assessment of Parkinson's disease—A review. Clin Neurol Neurosurg. 2019;184:105442. doi: 10.1016/j.clineuro.2019.105442. [DOI] [PubMed] [Google Scholar]
- 10.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 11.Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 12.Breijyeh Z, Karaman R. Comprehensive review on Alzheimer's disease:causes and treatment. Molecules. 2020;25:5789. doi: 10.3390/molecules25245789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bringas S, Salomón S, Duque R, Montaña JL, Lage C. A convolutional neural network-based method for human movement patterns classification in Alzheimer's disease. Proceedings. 2019;31:72. [Google Scholar]
- 14.Cai Z, Gu J, Chen H. A new hybrid intelligent framework for predicting Parkinson's disease. IEEE Access. 2017;5:17188–17200. [Google Scholar]
- 15.Cao C, Liu F, Tan H, Song D, Shu W, Li W, Zhou Y, Bo X, Xie Z. Deep learning and its applications in biomedicine. Genomics Proteomics Bioinformatics. 2017;16:17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen TZ, Xu GJ, Zhou GA, Wang JR, Chan P, Du YF. Postural sway in idiopathic rapid eye movement sleep behavior disorder:a potential marker of prodromal Parkinsons disease. Brain Res. 2014;1559:26–32. doi: 10.1016/j.brainres.2014.02.040. [DOI] [PubMed] [Google Scholar]
- 17.Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Trans Vis Sci Tech. 2020;9:14. doi: 10.1167/tvst.9.2.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombi I, Mahajani S, Frega M, Gasparini L, Chiappalone M. Effects of antiepileptic drugs on hippocampal neurons coupled to micro-electrode arrays. Front Neuroeng. 2013;6:10. doi: 10.3389/fneng.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortelli P, Brusco A, Brussino A, Giorgio E, Antonarakis SE, Pennacchio L, Spielmann M, Di Gregorio E, Capellari S, Bartoletti Stella A, Terlizzi R, Parchi P, Liguori R, Zanigni S, Tonon C, Lodi R, Vaula G, Contestabile A, Mahajani S, Giacomini C, et al. Clinical, neuroradiological and molecular investigation of Adult-onset Autosomal Dominant LeukoDystrophy (ADLD):dissection of Lamin B1-mediated pathophysiological mechanisms in cellular and mouse models. XIII Scientific Convention. 2015:39–40. [Google Scholar]
- 20.Das KD, Saji AJ, Kumar CS. Frequency analysis of gait signals for detection of neurodegenerative diseases. International Conference on Circuit, Power and Computing Technologies (ICCPCT) 2017;20:1–6. doi:10.1109/ICCPCT.2017.8074273.de Miranda BR, Greenamyre JT (2017) Etiology and pathogenesis of Parkinson's disease. In:Oxidative Stress and Redox Signaling in Parkinson's Disease (Franco R, Doorn JA, Rochet JC, eds). Chapter 1, pp1-26. doi:10.1039/9781782622888-00001. [Google Scholar]
- 21.De Vis JB, Peng SL, Chen X, Li Y, Liu P, Sur S, Rodrigue KM, Park DC, Lu H. Arterial-spin-labeling (ASL) perfusion MRI predicts cognitive function in elderly individuals:a 4-year longitudinal study. J Magn Reson Imaging. 2018;48:449–458. doi: 10.1002/jmri.25938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeMaagd G, Philip A. Parkinson's disease and its management:part 1:disease entity, risk factors, pathophysiology, clinical presentation, and diagnosis. P T. 2015;40:504–532. [PMC free article] [PubMed] [Google Scholar]
- 23.DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegeneration. 2019;14:32. doi: 10.1186/s13024-019-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijkhuizen RM, van der Marel K, Otte WM, Hoff EI, van der Zijden JP, van der Toorn A, van Meer MP. Functional MRI and diffusion tensor imaging of brain reorganization after experimental stroke. Transl Stroke Res. 2012;3:36–43. doi: 10.1007/s12975-011-0143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duc NT, Ryu S, Qureshi MNI, Choi M, Lee KH, Lee B. 3D-deep learning based automatic diagnosis of Alzheimer's disease with joint MMSE prediction using resting-state fMRI. Neuroinformatics. 2020;18:71–86. doi: 10.1007/s12021-019-09419-w. [DOI] [PubMed] [Google Scholar]
- 26.Emmert-Streib F, Yang Z, Feng H, Tripathi S, Dehmer M. An introductory review of deep learning for prediction models with big data. Front Artif Intell. 2020;3:4. doi: 10.3389/frai.2020.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleisher AS, Pontecorvo MJ, Devous MD, Sr, Lu M, Arora AK, Truocchio SP, Aldea P, Flitter M, Locascio T, Devine M, Siderowf A, Beach TG, Montine TJ, Serrano GE, Curtis C, Perrin A, Salloway S, Daniel M, Wellman C, Joshi AD, Irwin DJ, et al. Positron Emission Tomography Imaging with [18F] flortaucipir and postmortem assessment of Alzheimer disease neuropathologic changes. JAMA Neurol. 2020;77:829–839. doi: 10.1001/jamaneurol.2020.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg P, Maass F, Sundaram SM, Mollenhauer B, Mahajani S, van Riesen C, Kügler S, Bähr M. The relevance of synuclein autoantibodies as a biomarker for Parkinson's disease. Mol Cell Neurosci. 2022;121:103746. doi: 10.1016/j.mcn.2022.103746. [DOI] [PubMed] [Google Scholar]
- 29.Gautam R, Sharma M. Prevalence and diagnosis of neurological disorders using different deep learning techniques:a meta-analysis. J Med Syst. 2020;44:49. doi: 10.1007/s10916-019-1519-7. [DOI] [PubMed] [Google Scholar]
- 30.Giacomini C, Mahajani S, Ruffilli R, Marotta R, Gasparini L. Lamin B1 protein is required for dendrite development in primary mouse cortical neurons. Mol Biol Cell. 2016;27:35–47. doi: 10.1091/mbc.E15-05-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillani N, Arslan T. Intelligent sensing technologies for the diagnosis, monitoring and therapy of alzheimer's disease:a systematic review. Sensors (Basel) 2021;21:4249. doi: 10.3390/s21124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heldman DA, Harris DA, Felong T, Andrzejewski KL, Dorsey ER, Giuffrida JP, Goldberg B, Burack MA. Telehealth management of Parkinson's disease using wearable sensors:an exploratory study. Digit Biomark. 2017;1:43–51. doi: 10.1159/000475801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoq M, Uddin MN, Park SB. Vocal feature extraction-based artificial intelligent model for Parkinson's disease detection. Diagnostics (Basel) 2021;11:1076. doi: 10.3390/diagnostics11061076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo T, Nho K, Saykin AJ. Deep learning in Alzheimer's disease:diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci. 2019;11:220. doi: 10.3389/fnagi.2019.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang M, Jameson NJ. Machine Learning:Fundamentals. Pecht MG, Kang M, editors. Prognostics and Health Management of Electronics. 2018:85–109. Wiley Online Library. [Google Scholar]
- 36.Kostikis N, Hristu-Varsakelis D, Arnaoutoglou M, Kotsavasiloglou C. A smartphone-based tool for assessing parkinsonian hand tremor. IEEE J Biomed Health Inform. 2015;19:1835–1842. doi: 10.1109/JBHI.2015.2471093. [DOI] [PubMed] [Google Scholar]
- 37.Kouli A, Torsney K, Kuan W. Parkinson's disease:etiology, neuropathology, and pathogenesis. In: Stoker TB, Greenland JC, editors. Parkinson's disease:pathogenesis and clinical aspects [Internet] Brisbane (AU): Codon Publications; 2018. Chapter 1. [PubMed] [Google Scholar]
- 38.Latourelle JC, Beste MT, Hadzi TC, Miller RE, Oppenheim JN, Valko MP, Wuest DM, Church BW, Khalil IG, Hayete B, Venuto CS. Large-scale identification of clinical and genetic predictors of motor progression in patients with newly diagnosed Parkinson's disease:a longitudinal cohort study and validation. Lancet Neurol. 2017;16:908–916. doi: 10.1016/S1474-4422(17)30328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Lei H, Zhou F, Gardezi G, Lei B. Longitudinal and multi-modal data learning for Parkinson's disease diagnosis via stacked sparse auto-encoder 2019 IEEE. 16th International Symposium on Biomedical Imaging (ISBI 2019) 2019:384–387. doi:10.1109/ISBI.2019.8759385. [Google Scholar]
- 40.Liu C, Wei H, Gong N, Cronin M, Dibb R, Decker K. Quantitative susceptibility mapping:contrast mechanisms and clinical applications. Tomography. 2015;1:3–17. doi: 10.18383/j.tom.2015.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu S, Wang S, Zhang Y. A classification method for brain MRI via MobileNet and feedforward network with random weights. Pattern Recognit Lett. 2020;140:252–260. [Google Scholar]
- 42.Soria Lopez JA, González HM, Léger GC. Alzheimer's disease. Handb Clin Neurol. 2019;167:231–255. doi: 10.1016/B978-0-12-804766-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 43.MacMahon Copas AN, McComish SF, Fletcher JM, Caldwell MA. The pathogenesis of Parkinson's disease:a complex interplay between astrocytes, microglia, and T lymphocytes? Front Neurol. 2021;12:666737. doi: 10.3389/fneur.2021.666737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahajani S, Giacomini C, Marinaro F, De Pietri Tonelli D, Contestabile A, Gasparini L. Lamin B1 levels modulate differentiation into neurons during embryonic corticogenesis. Sci Rep. 2017;7:4897. doi: 10.1038/s41598-017-05078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahajani S, Raina A, Fokken C, Kügler S, Bähr M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019;10:898. doi: 10.1038/s41419-019-2133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahajani S, Bähr M, Kügler S. Patterning inconsistencies restrict the true potential of dopaminergic neurons derived from human induced pluripotent stem cells. Neural Regen Res. 2021;16:692–693. doi: 10.4103/1673-5374.295316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancini M, Horak FB, Zampieri C, Carlson-Kuhta P, Nutt JG, Chiari L. Trunk accelerometry reveals postural instability in untreated Parkinson's disease. Parkinsonism Relat Disord. 2011;17:557–562. doi: 10.1016/j.parkreldis.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marotta R, Catelani T, Pesce M, Giacomini C, Mahajani S, Gasparini L. Role of Lamin B1 in structuring the cell nucleus in eukaryotic cells. The 16th European Microscopy Congress. 2016;2016:1011–1012. [Google Scholar]
- 49.Mas J. Receiver Operating Characteristic (ROC) Analysis. In: Camacho Olmedo M, Paegelow M, Mas JF, Escobar F, editors. Geomatic Approaches for Modeling Land Change Scenarios. Lecture Notes in Geoinformation and Cartography. Cham: Springer; 2018. [Google Scholar]
- 50.Mughal H, Javed AR, Rizwan M, Almadhor AS, Kryvinska N. Parkinson's disease management via wearable sensors:a systematic review. IEEE Access. 2022;10:35219–35237. [Google Scholar]
- 51.Myszczynska MA, Ojamies PN, Lacoste AMB, Neil D, Saffari A, Mead R, Hautbergue GM, Holbrook JD, Ferraiuolo L. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat Rev Neurol. 2020;16:440–456. doi: 10.1038/s41582-020-0377-8. [DOI] [PubMed] [Google Scholar]
- 52.Nabeel M, Majeed S, Awan M, Muslih-ud-Din H, Wasique M, Nasir R. Review on effective disease prediction through data mining techniques. Int J Electr Eng Inform. 2021 doi:10.15676/ijeei.2021.13.3.13. [Google Scholar]
- 53.Nair P, Trisno R, Baghini MS, Pendharkar G, Chung H. Predicting early stage drug induced Parkinsonism using unsupervised and supervised machine learning. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:776–779. doi: 10.1109/EMBC44109.2020.9175343. [DOI] [PubMed] [Google Scholar]
- 54.Ni R, Nitsch RM. Recent developments in Positron Emission Tomography tracers for proteinopathies imaging in dementia. Front Aging Neurosci. 2021 doi: 10.3389/fnagi.2021.751897. doi:10.3389/fnagi.2021.751897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noor MBT, Zenia NZ, Kaiser MS, Mahmud M, AI Mamun S. Detecting neurodegenerative disease from MRI:a brief review on a deep learning perspective. In: Liang P, Goel V, Shan C, editors. Brain Informatics. Lecture Notes in Computer Science. Vol. 11976. Cham: Springer; 2019. [Google Scholar]
- 56.Otero-Garcia M, Mahajani S, Wakhloo D, Tang W, Xue Y, Morabito S, Pan J, Oberhauser J, Madira A, Shakouri T, Deng Y, Allison T, He Z, Lowry W, Kawaguchi R, Swarup V, Cobos I. Molecular signatures underlying neurofibrillary tangle susceptibility in Alzheimer's disease. Neuron S0896-6273(22)00600-6. 2022 doi: 10.1016/j.neuron.2022.06.021. doi:10.1016/j.neuron.2022.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perumal SV, Sankar R. Gait and tremor assessment for patients with Parkinson's disease using wearable sensors. ICT Express. 2016;2:168–174. [Google Scholar]
- 58.Powers R, Etezadi-Amoli M, Arnold EM, Kianian S, Mance I, Gibiansky M, Trietsch D, Alvarado AS, Kretlow JD, Herrington TM, Brillman S, Huang NC, Lin P, Pham HA, Ullal A. Smartwatch inertial sensors continuously monitor real-world motor fluctuations in Parkinson's disease. Sci Transl Med. 2021;13:eabd7865. doi: 10.1126/scitranslmed.abd7865. [DOI] [PubMed] [Google Scholar]
- 59.Psol M, Darvas SG, Leite K, Mahajani SU, Bähr M, Kügler S. Dementia with Lewy bodies-associated β-synuclein mutations V70M and P123H cause mutation-specific neuropathological lesions. Hum Mol Genet. 2021;30:247–264. doi: 10.1093/hmg/ddab036. [DOI] [PubMed] [Google Scholar]
- 60.Raina A, Mahajani S, Bähr M, Kügler S. Neuronal trans-differentiation by transcription factors Ascl1 and Nurr1:induction of a dopaminergic neurotransmitter phenotype in cortical GABAergic neurons. Mol Neurobiol. 2020;57:249–260. doi: 10.1007/s12035-019-01701-x. [DOI] [PubMed] [Google Scholar]
- 61.Raina A, Leite K, Guerin S, Mahajani SU, Chakrabarti KS, Voll D, Becker S, Griesinger C, Bähr M, Kügler S. Dopamine promotes the neurodegenerative potential of β-synuclein. J Neurochem. 2021;156:674–691. doi: 10.1111/jnc.15134. [DOI] [PubMed] [Google Scholar]
- 62.Raval UR, Jani C. Implementing and improvisation of K-means clustering algorithm. Int J Comput Sci Mobile Comput. 2016;5:191–203. [Google Scholar]
- 63.Rigas G, Tzallas AT, Tsipouras MG, Bougia P, Tripoliti EE, Baga D, Fotiadis DI, Tsouli SG, Konitsiotis S. Assessment of tremor activity in the Parkinson's disease using a set of wearable sensors. IEEE Trans Inf Technol Biomed. 2012;16:478–487. doi: 10.1109/TITB.2011.2182616. [DOI] [PubMed] [Google Scholar]
- 64.Rovini E, Maremmani C, Moschetti A, Esposito D, Cavallo F. Comparative motor pre-clinical assessment in Parkinson's disease using supervised machine learning approaches. Ann Biomed Eng. 2018;46:2057–2068. doi: 10.1007/s10439-018-2104-9. [DOI] [PubMed] [Google Scholar]
- 65.Ruetten PPR, Gillard JH, Graves MJ. Introduction to quantitative susceptibility mapping and susceptibility weighted imaging. Br J Radiol. 2019;92:20181016. doi: 10.1259/bjr.20181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rumman M, Tasneem AN, Farzana S, Pavel MI, Alam MA. Early detection of Parkinson's disease using image processing and artificial neural network. Joint 7th International Conference on Informatics, Electronics &Vision (ICIEV) and 2018 2nd International Conference on Imaging, Vision &Pattern Recognition (icIVPR) 2018:256–261. doi:10.1109/ICIEV.2018.8641081. [Google Scholar]
- 67.Saboo KV, Hu C, Varatharajah Y, Przybelski SA, Reid RI, Schwarz CG, Graff-Radford J, Knopman DS, Machulda MM, Mielke MM, Petersen RC, Arnold PM, Worrell GA, Jones DT, Jack CR, Jr, Iyer RK, Vemuri P. Deep learning identifies brain structures that predict cognition and explain heterogeneity in cognitive aging. Neuroimage. 2022;251:119020. doi: 10.1016/j.neuroimage.2022.119020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sant'Anna A, Salarian A, Wickström N. A new measure of movement symmetry in early Parkinson's disease patients using symbolic processing of inertial sensor data. IEEE Trans Biomed Eng. 2011;58:2127–2135. doi: 10.1109/TBME.2011.2149521. [DOI] [PubMed] [Google Scholar]
- 69.Sarmiento JR, Lao A, Solano GA. Pathway-based human disease clustering tool using self-organizing maps. 2017 8th International Conference on Information, Intelligence, Systems &Applications (IISA) 2017:1–6. doi:10.1109/IISA.2017.8316389. [Google Scholar]
- 70.Scanlon BK, Levin BE, Nation DA, Katzen HL, Guevara-Salcedo A, Singer C, Papapetropoulos S. An accelerometry-based study of lower and upper limb tremor in Parkinson's disease. J Clin Neurosci. 2013;6:827–830. doi: 10.1016/j.jocn.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 71.Schmidhuber J. Deep learning in neural networks:an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 72.Segovia F, Górriz JM, Ramírez J, Martinez-Murcia FJ, García-Pérez M. Using deep neural networks along with dimensionality reduction techniques to assist the diagnosis of neurodegenerative disorders. Log J IGPL. 2018;26:618–628. doi: 10.1093/jigpal/jzy026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6 doi: 10.1186/s40537-021-00492-0. doi:10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh A, Thakur N, Sharma A. A review of supervised machine learning algorithms. 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom) 2016:1310–1315. [Google Scholar]
- 76.Singh G, Samavedham L. Unsupervised learning-based feature extraction for differential diagnosis of neurodegenerative diseases:a case study on early-stage diagnosis of Parkinson disease. J Neurosci Methods. 2015;256:30–40. doi: 10.1016/j.jneumeth.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Singh G, Samavedham L, Lim EC;Alzheimer's Disease Neuroimaging Initiative, Parkinson Progression Marker Initiative. Determination of imaging biomarkers to decipher disease trajectories and differential diagnosis of neurodegenerative diseases (DIsease TreND) J Neurosci Methods. 2018;305:105–116. doi: 10.1016/j.jneumeth.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):583–590. doi: 10.1007/978-3-642-40763-5_72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taeho J, Nho K, Saykin AJ. Deep learning in Alzheimer's disease:diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci. 2019;11:220. doi: 10.3389/fnagi.2019.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trabelsi A, Chaabane M, Ben-Hur A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics. 2019;35:i269–i277. doi: 10.1093/bioinformatics/btz339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19:281. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Upton J. Mini-Mental State Examination. In: Gellman MD, editor. Encyclopedia of Behavioral Medicine. Cham: Springer; 2020. pp. 1402–1403. [Google Scholar]
- 83.Wakhloo D, Scharkowski F, Curto Y, Javed Butt U, Bansal V, Steixner-Kumar AA, Wüstefeld L, Rajput A, Arinrad S, Zillmann MR, Seelbach A, Hassouna I, Schneider K, Qadir Ibrahim A, Werner HB, Martens H, Miskowiak K, Wojcik SM, Bonn S, Nacher J, et al. Functional hypoxia drives neuroplasticity and neurogenesis via brain erythropoietin. Nat Commun. 2020;11:1313. doi: 10.1038/s41467-020-15041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wakhloo D, Oberhauser J, Madira A, Mahajani S. From cradle to grave:neurogenesis, neuroregeneration and neurodegeneration in Alzheimer's and Parkinson's diseases. Neural Regen Res. 2022;17:2606–2614. doi: 10.4103/1673-5374.336138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wutao Y, Li L, Wu F. Deep learning for brain disorder diagnosis based on fMRI images. Neurocomputing. 2022;469:332–345. [Google Scholar]
- 86.Yan W, Qu G, Hu W, Abrol A, Cai B, Qiao C, Plis S, Wang Y, Sui J, Calhoun V. Deep learning in neuroimaging:promises and challenges. IEEE Signal Process Mag. 2022;39:87–98. [Google Scholar]
- 87.Ying X. An overview of overfitting and its solutions. J Phys Conf Ser. 2019;1168:022022. [Google Scholar]
- 88.Zhao Y, Ma B, Jiang P, Zeng D, Wang X, Li S. Prediction of Alzheimer's disease progression with multi-information generative adversarial network. IEEE J Biomed Health Inform. 2021;25:711–719. doi: 10.1109/JBHI.2020.3006925. [DOI] [PubMed] [Google Scholar]