1. Introduction
Inter-fractional variation during the delivery of multi-fractional external beam radiation therapy (RT) is a major issue impacting the effectiveness of RT. The current standard of care to address the inter-fractional variations is to use image-guided RT (IGRT) to reposition the patient based on the pre-treatment daily images while delivering the same treatment plan (i.e., reference plan) for all fractions. It has been well documented that IGRT repositioning primarily addresses translational error only and cannot fully account for all the inter-fractional changes [1–6]. Adaptive radiation therapy (ART), particularly online adaptive re-planning (OLAR), is being developed to fully address the inter-fractional variations [7–12]. Major steps in OLAR include (1) acquiring a daily image, (2) delineating the targets and organs at risk (OAR) on the daily image, (3) creating a new treatment plan by adapting a reference plan to the daily anatomy, (4) performing quality assurance on the adapted plan, and (5)delivering the adaptive plan for the fraction. Recently, MRI-guided ART (MRgART) is being rapidly introduced into the clinics due to the advent of the MR-Linac [13, 14]. With the superior soft tissue contrast, biological information, and real-time imaging available from MRI, MRgART is substantially improving RT delivery for many tumor sites [15–18] . However, the current process of MRgART, particularly OLAR, is complex, labor-intensive, and time-consuming. It has been reported by several early adopters of MRgART that the OLAR process can take up to 90 minutes [19–22] .
In our clinic, MRgART is performed using a 1.5 Tesla MRI-Linac (Unity, Elekta AB) [16, 22] . The system offers two adaptive planning techniques to address the inter-fractional variations based on daily MRI: (i) Adapt-to-Position (ATP), in which an adaptive plan is generated and optimized using the original contours of the reference images with multi-leaf collimators (MLC) shifted based on the co-registration of the daily MRI and the reference image, and ii) Adapt-to-Shape (ATS), in which a new plan is generated based on the anatomy of the day from the daily MRI. While ATP is a simple workflow like the conventional IGRT repositioning (shifting MLC in ATP instead of shifting patient in IGRT), ATS is a full OLAR process that fully corrects for inter-fractional variations. Based on our experience, the times required for ATS workflow can be more than twice the ATP process [22] . Such prolonged time has been a major bottleneck issue preventing OLAR from routine practice. On the other hand, OLAR is not necessary for each treatment fraction. Omari et al. showed that approximately 1/3 of treatment fractions for pancreatic cancer did/would not require OLAR [23]. Thus, in clinical practice, it is desirable to determine when OLAR is necessary before substantial effort (e.g., re-contouring on daily image) is spent. Currently, whether to perform OLAR for a treatment fraction is determined either empirically prior to the fraction (e.g., based on the fractionation scheme or the specific tumor site) or by subjectively reviewing of anatomic change between the daily and reference images immediately after the daily image acquisition.
Several researchers proposed various ideas to determine the necessity of OLAR objectively and automatically. For instance, Brown et al. reported predictive factors that may be utilized to identify head and neck cancer patients likely requiring ART [24]. Lim et al. showed that Jacobian determinant histogram (JDH) obtained from deformable image registration (DIR) between reference (planning) and daily CTs can be used to determine the necessity of OLAR [25] . However, this method may not be applicable to MRI primarily due to the large variation in MR intensity that may mask the trends of organ deformation in JDH. Very recently, Parchur et al. proposed to use the change of structural similarity index measure (SSIM) between the reference and daily MRIs to predict OLAR necessity in MRgART. However, they reported that SSIM, a first order image feature, might be too simple to measure subtle anatomic changes, resulting in false positive prediction of OLAR necessity in at least 5% of the cases [26].
It is known that high-order image features, such as wavelet features, can reveal more subtle, deep seated imaging properties than first-order features. It has been shown that wavelet features are useful for classification in a variety of applications [27–33].Wavelet decompositions are one of the most recent additions to the multiscale signal processing techniques where a set of cascaded filter banks are used to provide a complete image representation and perform decomposition according to both scale and orientation. Wavelet transform provides multiscale representation of the image that carries both spatial position and spatial frequency information at the same time. Coarse-scale wavelet coefficients represent low-resolution image components and fine-scale wavelet coefficients denote high resolution components [28–30] . Zhou et al. showed that MRI wavelet-transformed textures outperformed volumetric textures in radiomic prediction of pathological response to neoadjuvant chemotherapy for patients with locally advanced breast cancer [31]. Çinarer et al. showed that a deep learning algorithm with wavelet radiomic features is highly effective in grading gliomas with 96.15% accuracy [32]. Chaplot et al. used Daubechies wavelet transform to extract MRI features that can correctly classify cancer patients using a self-organizing map and support vector machines [33] .
In this study we developed a model based on multiscale wavelet features extracted from reference and daily MRIs to predict the necessity of OLAR automatically and objectively in MRgART. We used the MRI data collected on the Unity system for pancreatic cancer patients and built a machine learning classifier to identify appropriate multiscale texture features that can distinguish ATP versus ATS planning. We developed the prediction model such that it can be executed immediately after the daily MRI acquisition without any segmentation, avoiding any unnecessary effort if OLAR is not needed.
2. Methods
A classifier based on multiscale wavelet features was developed to predict whether OLAR is needed for a daily fraction. The correlation (e.g., change) between the reference and daily MRIs (CRD) of a feature was calculated in a region of interest (ROI). A high CRD (strong correlation) would indicate small anatomic difference between the reference and daily images, implying that OLAR is not necessary, and that ATP would be sufficient for the fraction, while a low CRD (weak correlation) would indicate substantial anatomic difference (e.g., organ deformation) between the daily and reference images, thus, making OLAR (e.g., ATS) necessary for the fraction. Machine learning algorithms were used to identify appropriate features that can differentiate ATS and ATP groups. The major steps for the development of the classifier are shown in Figure 1 and will be described in the subsequent subsections.
Figure 1.
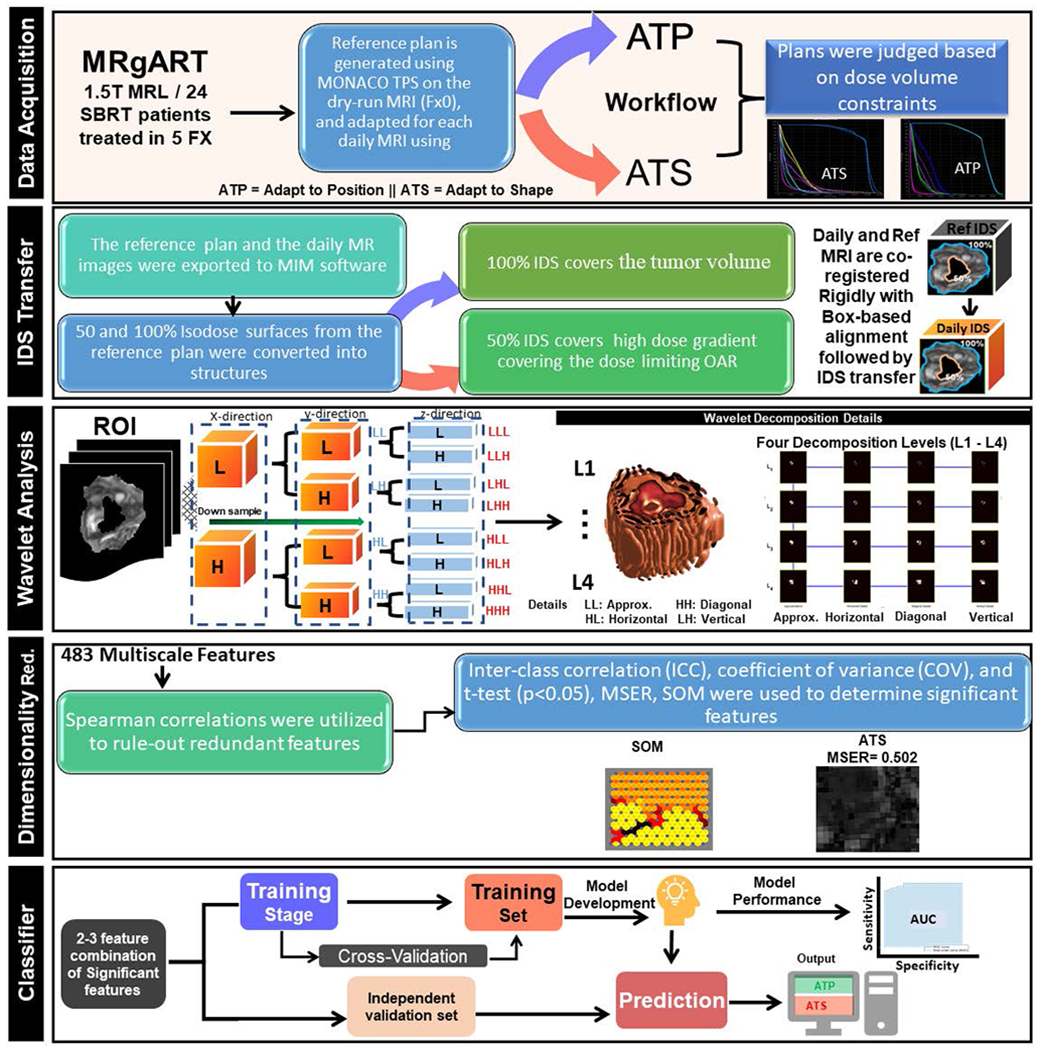
Major steps from data acquisition to classifier development and testing showing data acquisition, IDS transfer, wavelet analysis, dimensionality reduction and classifier building to automatically determine the necessity of online adaptive replanning using multiscale wavelet-based features
2.1. Image Data
In this retrospective study, approved by the institutional Internal Review Board (IRB), a total of 119 daily MRI sets along with the reference images were acquired during routine MRgART on a Unity system for 24 pancreatic cancer patients treated in 5 fractions (one daily image missing for a patient). The collected daily and reference images were the motion average images calculated from 4D MRI that included a stack of image slices. The daily and reference MRI sets were used to build and test the prediction model for OLAR necessity. Each of the patients was treated with SBRT of 33-40 Gy in 5 fractions with a reference MRI taken before the treatment (fraction 0), resulting in one reference and five daily MRI sets for each patient. The patient demographic information followed natural distribution as there was no specific inclusion or exclusion criteria in patient selection. All MRIs were the motion average images derived from 4D MRI [22] using a software of our design [34].
For each case, a reference plan was generated on the reference MRI. Both ATP and ATS plans were created for each fraction (daily MRI set) using the ATS or ATP workflows with a MRgART treatment planning system (Monaco V5.4, Elekta, Stockholm, Sweden). Fractions were categorized into ATP and ATS groups based on commonly used dose volume criteria in our clinic (presented in Table 1 Supplementary material). Daily fraction whose ATS plan superior to the ATP plan was categorized as an ATS fraction, while a fraction with its ATP plan comparable to the ATS plan was labeled as an ATP fraction. These classifications of the 119 datasets, including 33 ATP and 86 ATS sets constituted the ground truth for the model training and validation described below.
2.2. Image preprocessing and registration
All image and the plan data were analyzed using a software tool (MIM software Inc.). All MRIs were standardized (bias correction, and intensity normalization) to minimize the impact of MR intensity variations and allow robust extraction of quantitative data from the images. The bias correction and denoising were performed in MIM to correct for the radiofrequency coil inhomogeneity using an intensity-based nonparametric bias correction method followed by a Gaussian filter to smooth the noisy pixels. The intensity normalization was done by linearly normalizing the image intensities to a maximum of 10240. More details of these preprocessing steps are reported previously by our group [35]. Each daily MRI was rigidly registered to the reference MRI followed by a local registration using a box-based alignment tool to minimize registration error (Figure 1 IDS transfer section). The obtained images were used subsequently for wavelet feature extraction through wavelet decompositions as described below.
2.3. Region of interest selection
To select the optimum ROI for the wavelet feature extraction, the 100%, 80% and 50% isodose surface (IDS) from the reference plan were converted into structures in MIM and subsequently transferred to the registered daily image. Typically, the 100% IDS covers the tumor volume while the region between 100 and 50% IDSs contains the dose limiting OAR(s) with high dose gradient. Image features extracted in the ROI spheres enclosed by the 100%-80%, 80%-50%, and 100%-50% IDSs in both reference and daily images were analyzed to determine optimal ROI where the significant features can be extracted to build a machine learning classification model as shown in Figure 1, IDS transfer subsection.
2.4. Wavelet decomposition
To detect patterns that are not visible in the native images, multi-resolution analysis was applied using 3D wavelet decomposition to texturize each of the ROIs. The image was decomposed into approximation (low frequency) components and detail (high frequency) components using wavelet and scaling functions to perform the multi-resolution analysis. As shown in Figure 1, Wavelet Analysis subsection, first, the image was convolved in the x-direction by a low pass filter and then down sampling it to produce the approximation sub-band denoted by ‘L’, then convolved by a high pass filter to produce the detail sub-band denoted by ‘H’, following Equation 1 below.
| Equaion (1) |
The approximation and details sub-bands were then convolved in the same manner in the y-direction to produce the approximation detail denoted ‘LL’ and 3 detail sub-ands, the vertical detail ‘LH’, horizontal detail ‘HL’, and the diagonal detail ‘HH’. The process was repeated in the same manner in the z-direction to produce the subsequent sub-bands (‘LLL’, ‘HLL’,‘LHL’,‘HHL’,‘LLH’,‘HLH’,‘LHH’,‘HHH’). For instance, an ‘LHH’ means that the low-pass (scaling) filter with down sampling was applied to the rows of the image, followed by the high pass (wavelet) filter with down sampling applied to the columns then a high pass filter applied to the 3rd dimension. The second decomposition level was achieved by considering the approximation sub-band (LL) as the original image and convolving it again with high pass and low pass filters in the same manner and so on [32]. For this analysis, four decomposition levels were selected as the best trade-off between weakening tree structure and increased computation cost. A total of 438 wavelet features was extracted from the different decomposition levels in each ROI on the daily and the reference images. The relative net change of the wavelet feature value on the daily image (average over the entire ROI) from the corresponding value on the reference MRI were calculated to determine appropriate features that exhibit different CRD between the ATP and ATs groups.
2.5. Dimensionality reduction
To eliminate redundant features, spearman correlation was applied (r<0.9). Inter-class correlation (ICC), coefficient of variance (COV), and t-test (p<0.05) were used to determine significant features that exhibited feature change between the ATS and ATP groups. To further reduce the number of features to be used to build the classification model, the identified significant features were fed into a self-organized artificial neural network feature map to select features with least overlap between the ATS and ATP groups. The self-organized map (SOM) consisted of several nodes whose weight was the position in the input space and the classification was performed by finding the node that was the closest in distance to the input space vector. A maximally Stable Extremal Regions (MSER) algorithm was also applied to establish correspondence points between the daily and the reference images and to determine the final significant features with low redundancy and high quality MSER for ATS and ATP, respectively (Figure 1, Dimensionality red. subsection)
2.6. Model building with machine learning
The classification model was built and tested using daily MRIs of either ATS or ATP fractions. Combinations of 2-3 of the significant wavelet features found after the dimensionality reduction process were used to build multivariate Bayesian classifier models with a leave one out cross-validation method. The 119 daily MRI datasets were divided into 89 sets for training and cross validation and 30 sets (10 ATP and 20 ATS fractions) for independent testing. For a multivariate normal Bayesian classification to change from probabilities to discriminants, the strategy is to maximize the class conditional probability or to minimize the distance to the centroid of each of the training classes (i.e., minimum Mahalonobis distance). The generated model was saved, and the independent validation set was used to test the model by calculating the minimum Mahalonobis distance of the new data set to the centroid of the training class as shown in Figure 1, classifier subsection.
3. Results
Based on the Spearman correlation analysis, a total of 123 out of 438 features from different wavelet decomposition levels were found to be non-redundant (r<0.9). Comparing the data extracted from different ROI indicates that the max significant feature differences between the ATP and ATS were observed in the shell enclosed by the 100%-50% IDS. Consequently, this ROI typically representing the high dose gradient region, was selected for remaining analysis. Among the 123 features selected, 82 showed high ICCs for ATP and lower ICCs for ATS. Based on the box plots of the ICC for the ATP and ATS fractions shown in Figure 2 (right), a cutoff correlation coefficient of 0.6 for ICC was found to be appropriate to determine the candidate features for building the classifier. Sixty-seven of these features had a COV greater than 9% for ATS and lower than 5% for ATP. Figure 2 (left) shows box plots of the COV for ATP and ATS fractions. A large COV indicates a substantial image (anatomic) changes between the reference and daily images, implying the necessity of ATS for the fraction. In addition, 38 of the 67 features passed the t-test with p value <0.05.
Figure 2.
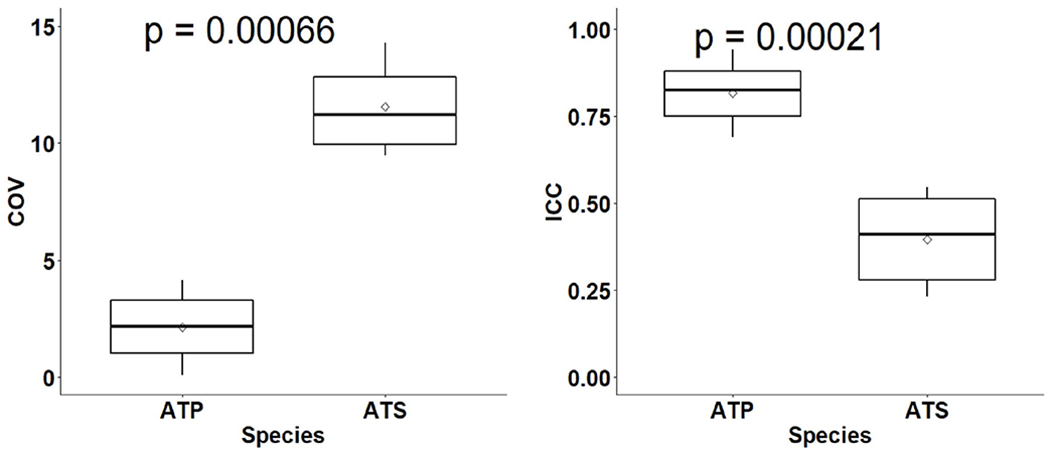
Box plots of ICC (right) and COV (left) for the ATP and ATS groups. The boxplots show the median and interquartile range for each group, and the diamond shape in the middle represents the mean of the group
Figure 3 shows example box plots of two features (HLL2 mean, and HHL2 ClusterTend) exhibiting significant t-test p-values between the ATP and ATS groups.
Figure 3.
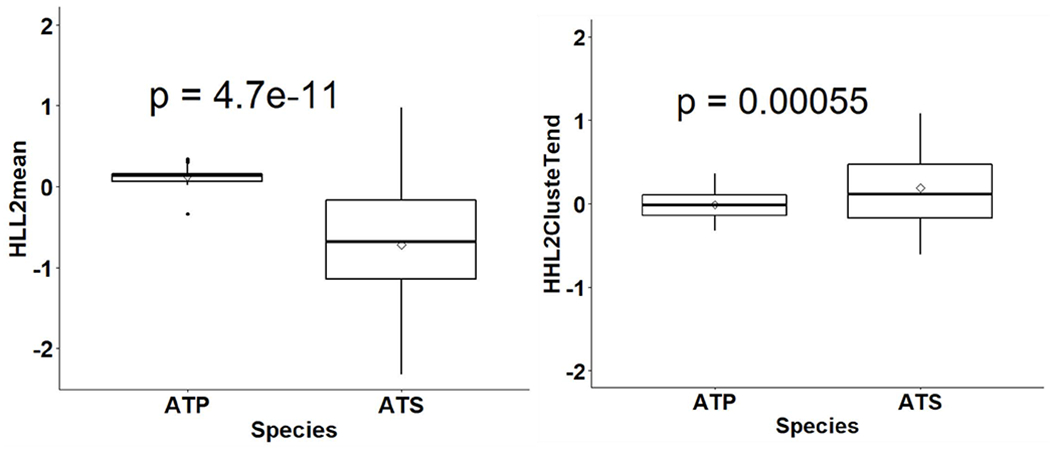
Box plots showing median and interquartile ranges for two features, HHL2 ClusteTend (right) and HLL2 mean (left), for the ATS and ATP groups with a diamond shape in the middle showing mean of each group and a t-test p-value on top showing significant difference.
The SOM analysis resulted in 25 features that showed the least overlap between the two groups. The MSER process yielded 12 final reproducible features with high MSER (> 0.8) for ATP and low MSER (<0.6) for ATS. These final features were used to build the classifier using 2-3 features combination at a time. As shown in Figure 4, the best performing model was a three-feature combination (HLL2 mean, HHL1 Homogenity, HHL2 ClusterTend), which can predict OLAR necessity with a cross validated AUC of 0.98. The model with a two-feature combination (HLL2 mean, HHL2 ClusterTend) can achieve AUC of 0.97. The execution of the model to predict OLAR necessarily took up to 38 seconds using a hardware of Intel® Core™i7-6700 CPU @3.4GHz 32GB RAM x64-based processor.
Figure 4.
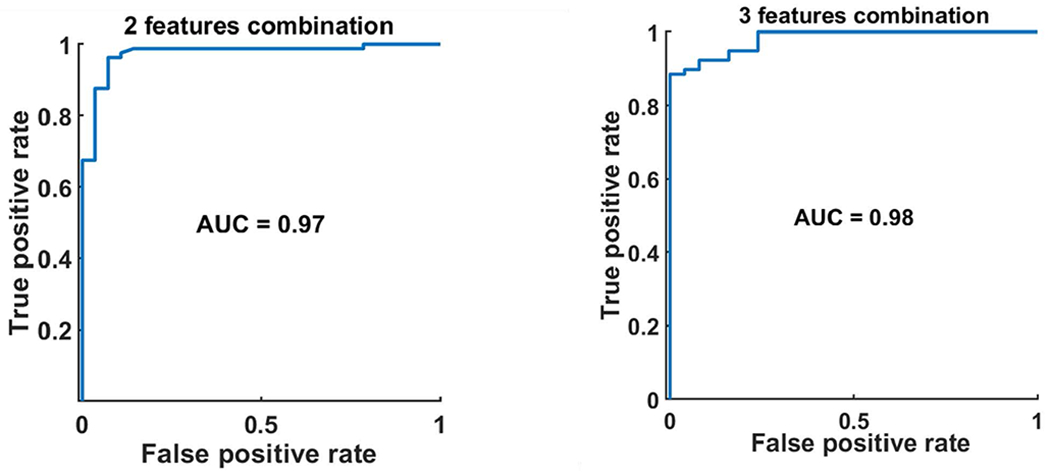
AUC of the ROC of Bayesian classifier based on combined 2 (left) and 3 (right) features to determine the necessity of OLAR.
4. Discussion
Ideally, OLAR (e.g., ATS) should be performed for every fraction during MRgART if OLAR (ATS) required no or minimal additional time and effort compared to the current IGRT repositioning (e.g., ATP). With current available technology OLAR process is labor intensive and time-consuming. Thus, being able to determine its necessity would avoid the unnecessary effort in the current practice of MRgART. To achieve this, we have developed a model using MRI wavelet features to accurately predict whether OLAR is needed for a given daily MRI set during MRgART. The obtained model can be implemented in the following process immediately after a daily MRI set is acquired: (1) rigidly registering the daily MRI with the reference MRI, (2) populating 100% and 50% IDSs from the reference to daily MRI, (3) extracting the three wavelet features (HLL2 mean, HHL1 Homogenity, HHL2 ClusterTend) from the ROI enclosed by the 100%-50% IDSs on the daily MRI, (4) feeding the extracted features to the developed model, (5) calculating the minimum Mahalonobis distance to the centroid of each trained class to determine if OLAR is needed.
As a next step, we are implementing the developed model as a workflow in MIM. In our clinical practice, the daily MRI is automatically transferred to MIM for registration and re-contouring if necessary. There is no additional image transferring time for this implementation. The model prediction time of 38 seconds includes the preprocessing, the extraction of three significant features from daily and reference images, and the execution of the pre-trained model. The model performance should not depend on dose prescription and/or adaptive dose constraints as online adaptation is justified as long as the adaptive plan is superior to the non-adaptive plan (e.g., ATP, IGRT).
Due to the large inherited MR intensity variation, the image preprocessing is essential to minimize effect of the intensity variation for the quantitative image feature exaction. In addition, the accuracy of the image registration is important for accurate and consistent model prediction. The rigid registration with the box-based alignment in MIM is similar to the common IGRT process (e.g., ATP) where the images are often rigidly registered with local adjustment to minimize registration error in high-dose region (e.g., PTV or PTV plus a few cm expansion). In this quantitative analysis, we used the relative net change of the image feature values, rather than the absolute values, further reducing the impact of registration uncertainties. We believe that the minimized registration error should not significantly affect the model prediction.
Compared to the SSIM model previously reported [24], the current model based on wavelet features is more accurate (AUC of 0.98), particularly, with improved prediction for daily MRIs with minimal anatomic difference (e.g., unobvious organ deformation) from the reference MRI. This is because the wavelet-based multiscale texture features can encode many invisible patterns that may not be possible with the first-order texture features, such as SSIM. Using the machine learning algorithm, the appropriate wavelet features can be determined to represent the underlying residual structural characteristics relate to the dosimetric properties of the daily RT plans. Because the quality of the ATS plan is generally better or equivalent to that of the ATP plan for a given daily MRI set, the current classifier was built using a conservative approach such that the ATS is indicated if the difference between ATS and ATP is minimal. In addition, the model was biased toward the ATS given the fact that the number of ATP plans used in the model training set was smaller than that of the ATS plans. With these conservative considerations, the misclassifications (2%) by the model, which occurs for the fractions with minimal differences between ATP and ATS plans, would not cause any clinically meaning impact.
To further test the reliability and applicability of the model, and to explore whether the obtained model can be translated to other clinics, we validated the model with 40 additional independent datasets. This independent validation showed that the model was able to successfully classify all the cases into the appropriate class except for one ATP case being judged as ATS. A close examination of the misclassification case indicated that the ATP and ATS plans were comparable, hence, difficult for discrimination.
Furthermore, the success of the independent test implies that the developed classifier may be used in other institutions using the same MRgRT system and the similar MRI sequences. For the institutions using different system or sequence, the method presented in this work may be applied to identify appropriate wavelet features and to build the corresponding model.
The newly developed classifier can be implemented into a robust automated tool for clinical decision making during MRgART workflow. The previously developed SSIM model can be included in the tool as a secondary, independent check of the primary prediction by the wavelet model. Such a primary-secondary two-tier prediction process would increase clinician’s confidence in the fully automated decision-making process. The automated process should be very fast as no user intervention is needed. If OLAR is determined to be necessary, the remaining OLAR steps can be immediately started. Otherwise, the simple repositioning workflow can proceed. As a next step, we will develop a graphical user interface to fully automate the process.
5. Conclusion
A machine learning classifier based on changes of wavelet features between the daily MRI of a treatment fraction and the reference image was developed to predict if OLAR is beneficial for the fraction during MRgART. The obtained wavelet-based multiscale features can reveal underlying residual anatomic changes between reference and daily MRIs, resulting in accurate model prediction with an AUC of 98%. The use of the wavelet prediction model can be fully automated and incorporated into MRgART workflow to quickly and objectively determine if OLAR (e.g., ATS) is needed immediately after the daily MRI is acquired.
Supplementary Material
Acknowledgments
The research was partially supported by the Medical College of Wisconsin Meinerz and Fotsch Foundations and the USA National Cancer Institute of the National Institutes of Health under award number R01CA247960. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- [1].Hall WA, Paulson E, Li XA, Magnetic resonance linear accelerator technology and adaptive radiation therapy: An overview for clinicians, CA Cancer J Clin (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Paulson ES, Ahunbay E, Chen X, 4D-MRI driven MR-guided online adaptive radiotherapy for abdominal stereotactic body radiation therapy on a high field MR-Linac: Implementation and initial clinical experience, Clin Transl Radiat Oncol 23 (2020) 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brown E, Owen R, Harden F, Predicting the need for adaptive radiotherapy in head and neck cancer, Radiother Oncol 116 (1) (2015) 57–63. [DOI] [PubMed] [Google Scholar]
- [4].Hilado S, Lim G, Naguib LA, Rng, Implementation of wavelets and artificial neural networks in colonic histopathological classification, J Adv Comput Intell Intell Inform 18 (2014) 792–792. [Google Scholar]
- [5].Srivastava V, Purwar K A Five-Level Wavelet Decomposition and Dimensional Reduction Approach for Feature Extraction and Classification of MR and CT Scan Images, Applied Computational Intelligence and Soft Computing, (2017). [Google Scholar]
- [6].Saravanan N, Ramachandran KI, Incipient gear box fault diagnosis using discrete wavelet transform (DWT) for feature extraction and classification using artificial neural network (ANN), Expert Systems with Applications 37 (6) (2010) 4168–4181. [Google Scholar]
- [7].Chaddad A, Daniel P, Niazi T, Radiomics Evaluation of Histological Heterogeneity Using Multiscale Textures Derived From 3D Wavelet Transformation of Multispectral Images, Frontiers in Oncology 8(96) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou J, Lu J, Gao C, Predicting the response to neoadjuvant chemotherapy for breast cancer: wavelet transforming radiomics in MRI, BMC Cancer 20 (100) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Çinarer G, Emiroğlu BG, Yurttakal AH, Prediction of Glioma Grades Using Deep Learning with, Wavelet Radiomic Features. Appl. Sci 10 (2020) 6296–6296. [Google Scholar]
- [10].Chaplot S, Patnaik L, Jagannathan N, Classification of magnetic resonance brain images using wavelets as input to support vector machine and neural network, Biomedical Signal Processing and Control 1 (1) (2006) 86–92. [Google Scholar]
- [11].Lim SN, Ahunbay EE, Nasief H, Indications of Online Adaptive Replanning Based on Organ Deformation, Pract Radiat Oncol 10 (2) (2020) 95–102. [DOI] [PubMed] [Google Scholar]
- [12].Sterzing F, Engenhart-Cabillic R, Flentje M, Image-guided radiotherapy: a new dimension in radiation oncology, Deutsches Arzteblatt international 108 (16) (2011) 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xing L, Thorndyke B, Schreibmann E, Overview of image-guided radiation therapy, Med Dosim 31 (2) (2006) 91–112. [DOI] [PubMed] [Google Scholar]
- [14].Stoiber M, Lechsel G, Giske K, Quantitative assessment of image-guided radiotherapy for paraspinal tumors, Int J Radiat Oncol Biol Phys 75 (3) (2009) 933–973. [DOI] [PubMed] [Google Scholar]
- [15].Santos JDL, Popple R, Agazaryan N, Image Guided Radiation Therapy (IGRT) Technologies for Radiation Therapy Localization and Delivery Int, J. Radiation Oncology Biol. Phys 87 (2013) 33–45. [DOI] [PubMed] [Google Scholar]
- [16].Court LE, Radiation Therapy Physics and Treatment Optimization (2014).
- [17].Xin S, Setup errors in cone-beam computed tomography and their effects on acute radiation toxicity in cervical cancer radiotherapy, Genet Mol Res 14 (3) (2015) 10937–10980. [DOI] [PubMed] [Google Scholar]
- [18].Liu F, Erickson B, Peng C, Li XA, Characterization and Management of Interfractional Anatomic Changes for Pancreatic Cancer Radiotherapy, Int J Radiation Oncol Biol. Phys 83 (2012) 423–429. [DOI] [PubMed] [Google Scholar]
- [19].Yan D, Vicini F, Wong J, Martinez A, Adaptive radiation therapy, Phys. Med. Biol 42 (1997) 123–132. [DOI] [PubMed] [Google Scholar]
- [20].Ahunbay E, Ates O, Li XA, An online replanning method using warm start optimization and aperture morphing for flattening-filter-free beams, Med. Phys 43 (2016) 4575–4584. [DOI] [PubMed] [Google Scholar]
- [21].Ahunbay E, Peng C, Chen GP, An on-line replanning scheme for interfractional variations, Med Phys 35 (2008) 3607–3615. [DOI] [PubMed] [Google Scholar]
- [22].Ahunbay E, Peng C, Holmes S, An on-line replanning scheme for interfractional variations, Int. J. Radiation Oncology Biol. Phys 77 (2010) 1561–1572. [Google Scholar]
- [23].Mutic S, Dempsey JF, The View Ray system: magnetic resonance-guided and controlled radiotherapy, Semin Radiat Oncol 24 (3) (2014) 196–199. [DOI] [PubMed] [Google Scholar]
- [24].Parchur AK, Lim S, Nasief H, Auto-Detection of Structural Similarity Index Map from Daily MRI as An Indicator for MR-Guided Online Adaptation Replanning, Med. Phys. proceeding of annual meeting (2021). [Google Scholar]
- [25].Omari E, Nasief H, Zhang Y, Should Daily Online Adaptive Replanning be Utilized for Every Treatment in MRI-Guided Radiotherapy for Pancreatic Cancer?, Int J Radiat Oncol Biol Phys 111 (3S) (2021) 66–66. [Google Scholar]
- [26].Li T, Thongphiew D, Zhu X, Lee RW, Adaptive prostate IGRT combining online re-optimization and re-positioning: a feasibility study, Phys. Med. Biol 56 (2011) 1243–1258. [DOI] [PubMed] [Google Scholar]
- [27].Lagendijk J, Raaymakers BW, Vulpen MV, The Magnetic Resonance Imaging-Linac System, Seminars in Radiation Oncology 24 (3) (2014) 207–209. [DOI] [PubMed] [Google Scholar]
- [28].Glide-Hurst CK, Adaptive Radiation Therapy (ART) Strategies and Technical Considerations: A State-of-the-art Review from NRG Oncology, Int J Radiat Oncol Biol Phys 109 (4) (2021) 1054–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Henke L, Kashani R, Robinson C, Phase I trial of stereotactic MR-guided online adaptive radiation therapy (SMART) for the treatment of oligometastatic or unresectable primary malignancies of the abdomen, Radiother Oncol 126 (3) (2018) 519–526. [DOI] [PubMed] [Google Scholar]
- [30].Henke LE, Olsen JR, Contreras JA, Stereotactic MR-guided online adaptive radiation therapy (SMART) for ultracentral thorax malignancies: Results of a phase 1 trial. Advances in Radiation Oncology.4(1):201–209, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lamb J, Cao M, Kishan A, Online Adaptive Radiation Therapy: Implementation of a New Process of Care, Cureus 9 (8) (2017) 1618–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Güngör G, Serbez İ, Temur B, Time Analysis of Online Adaptive Magnetic Resonance Guided Radiation Therapy Workflow According to Anatomical Sites, Practical Radiation Oncology 11 (1) (2021) 11–21. [DOI] [PubMed] [Google Scholar]
- [33].Lagendijk JJ, Raaymakers BW, Vulpen MV, The magnetic resonance imaging linac system, Semin Radiat Oncol 24 (3) (2014) 207–209. [DOI] [PubMed] [Google Scholar]
- [34].Mickevicius NJ, Paulson ES, On the use of low-dimensional temporal subspace constraints to reduce reconstruction time and improve image quality of accelerated 4D-MRI, Radiother Oncol 158 (2021) 215–223 [DOI] [PubMed] [Google Scholar]
- [35].Zhang Y, Paulson E, Lim S, et al. A Patient-Specific Auto segmentation Strategy Using Multi-Input Deformable Image Registration for Magnetic Resonance Imaging – Guided Online Adaptive Radiation Therapy: A Feasibility Study. Advances in Radiation Oncology. 2020;5(6): 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


