Abstract
Objectives
To explore the correlation between tumor endothelial marker 1 (TEM1) and matrix metalloproteinase 2 (MMP-2) in uterine sarcoma and their roles in the progression of uterine sarcoma.
Methods
Uterine leiomyosarcoma (uLMS, n = 25) and uterine leiomyoma (n = 25) specimens were collected from a total of 50 patients. Immunohistochemistry assay was conducted to determine the expression of TEM1, MMP-2 and MMP-9. TEM1 over expression (hTEM1) and low expression (shRNA-TEM1) MES-SA cell lines were established as in vitro uterine sarcoma models. MMP-2 mRNA, protein expression and enzymatic activity were verified using qPCR, Western blot and gelatin zymography respectively. MMP-2 expression was downregulated using MMP-2 siRNA in hTEM1 MES-SA cells to better study the role of MMP-2. The invasive and migratory capacities of hTEM1, shRNA-TEM1, and hTEM1 treated with MMP-2 siRNA MES-SA cells were determined using transwell assays. Extracellular matrix (ECM) remodeling mediated by TEM1 was examined using cell-ECM adhesion and fluorescent gelatin-ECM degradation assays. The immunofluorescence of F-actin was examined to analyze the formation of invadopodia. Subcutaneous and intraperitoneal xenografts were established to validate the role of TEM1 in promoting uterine sarcoma metastasis.
Results
TEM1 and MMP-2 were expressed in 92% (n = 23) and 88% (n = 22) of uterine leiomyosarcoma specimens, respectively. Both TEM1 and MMP-2 were highly expressed in 100% (n = 17) of high stage (III-IV) uterine leiomyosarcoma specimens. In addition, TEM1 expression was positively correlated with MMP-2 expression in uterine leiomyosarcoma. The successful establishment of in vitro uterine sarcoma models was confirmed with qPCR and Western blotting tests. TEM1 promoted the invasion and metastasis of uterine sarcoma in vivo and in vitro. MMP-2 expression and activity were up-regulated in hTEM1 cells but down-regulated in shRNA-TEM1 cells. Importantly, MMP-2 knockdown impaired the invasive and migratory capacity of hTEM1 cells. TEM1 promoted ECM remodeling by increasing cell-ECM adhesion and ECM degradation. TEM1 overexpression also induced the formation of invadopodia.
Conclusion
TEM1 was co-expressed and positively correlated with MMP-2 in uterine leiomyosarcoma specimens. In addition, both TEM1 and MMP-2 were associated with tumor development. TEM1 promoted uterine sarcoma progression by regulating MMP-2 activity and ECM remodeling.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-023-00613-6.
Keywords: Tumor endothelial marker 1/TEM1, Uterine leiomyosarcoma, Matrix metalloproteinase 2/MMP-2, Metastasis, Invasion, Migration
Introduction
Uterine sarcomas are rare gynecological malignancies affecting 3–9% of all uterine cancers. Uterine leiomyosarcomas (uLMS) account for 60–70% of all uterine sarcomas compared with 10% for endometrial stromal sarcoma and undifferentiated uterine sarcomas [1]. The prognosis of patients with uterine sarcoma is usually poor because of its aggressiveness and early metastatic nature. A recent US Surveillance reported a 42% overall five-year survival rate for uterine leiomyosarcomas patients and death within 5 years for all patients with late stage (stage III–IV) uterine leiomyosarcomas [2]. Currently, Systemic chemotherapy is the mainstay treatment for patients after enbloc surgery [3].
TEM1 (tumor endothelial marker 1), also known as endosialin or CD248, is dynamically expressed on pericytes, fibroblasts of tumor stroma, but barely expressed in benign and normal tissues [4–6]. Recent studies have found that TEM1 is overexpressed in solid tumors and participate in tumor angiogenesis and tumor progress [7–10]. Ontuxizumab (MORAb-004), a humanized anti-TEM1 monoclonal antibody, was recently studied for its anti-solid tumor efficacy in preclinical and clinical trials [11–14]. Previously, we reported that the TEM1 protein was highly expressed in 19 subtypes of sarcomas, including tumor vessels and tumor cells, with an overall positive rate of 96% [15]. All 13 samples of uterine leiomyosarcoma were TEM1-positive, among which 9 were positive in both tumors and vessels whereas 4 were positive in vessels only. TEM1 is usually up-regulated in tumor cells independent of the TEM1 expression levels in vessels or pericytes [7, 15], indicating that other than angiogenesis, TEM1 might play multiple roles in tumorigenesis and progression of uterine sarcoma.
The extracellular matrix (ECM) provides a dynamic supportive structure for cellular components and modulates tissue homeostasis [16]. the degradation and remodeling of ECM is considered as an essential step in the metastatic dissemination of cancer cells, which facilitates cancer progression [17]. Evidence showed that TEM1 interaction with ECM proteins promoted cell adhesion and migration [6]. In benign and/or malignant tissues, TEM1 is co-expressed with ECM components and enzymes involved in ECM remodeling, including collagens I, III, V, XIII, MMP-2, MMP-14 and lysyl oxidase (LOX) [16, 18]. These ECM-related molecules also participate in angiogenesis and epithelial-mesenchymal transition (EMT). TEM1 protein was reported to bind to fibronectin (FN) and mediate cell adhesion and migration in vivo [6]. An in vivo study showed the possible association between TEM1 and MMP-9 in fibrosarcoma cells [19]. Other studies found that TEM1 and MMP-2 were colocalized in tissue areas of early angiogenic processes [18, 20], suggesting a possible functional association between TEM1 expression and MMPs activity in cell migration and invasion. However, there are limited data supporting the association between TEM1 and MMP-2 and their roles in uterine sarcoma.
In the last decade, studies on TEM1 have mostly focused on its role in promoting angiogenesis [18, 21–23] but not its robust interaction with ECM components in tumorigenesis. The expression of TEM1 and MMP-2 and their correlation with uterine leiomyosarcoma was explored. By establishing TEM1 overexpression and knockdown MES-SA cell models, we demonstrated that TEM1 regulated the expression of MMP-2 and promoted the invasion and migration of uterine sarcoma cells through ECM remodeling.
Materials and methods
Clinical specimens
Clinical sarcoma specimens were collected from patients at Tongji Hospital 2015 to 2020, which is affiliated to Tongji University. The study protocol was approved by Ethics Committee of Tongji Hospital. Patients who provided informed consent to undergo staging using FIGO staging system were enrolled [24]. Specimens were obtained during surgeries and stored in liquid nitrogen preserved of paraffin block. Standard formalin-fixed paraffin-embedded (FFPE) techniques were conducted as previously described [15]. Briefly, FFPE tissue sections were blocked in matched normal sera after dewaxing and epitope retrieval. Slides were incubated with primary and second antibodies. Subsequently, the sections were stained with Hematoxylin (Beyotime, China) for 10 min before observation. The slides were observed using the Bond Polymer Refine Detection System. Protein expression levels were independently determined by two senior pathologists on a scale of 0, 1 + , 2 + and 3 + (0 = negative, 1 + = weak, 2 + = medium, 3 + = strong) according to the staining intensity of vasculature, stromal cells and/or tumor cells. The Long H-score was calculated by multiplying overall staining intensity with the percentage of positive cells [25–28]. The staining intensity scores ranged from 0 to 3 and the positive percentage increased from 0 to 100. Theoretically, the final H-score values obtained would range from 0 to 300.
Cells and reagents
MES-SA cell line was obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China) and cultured in McCoy’s 5α medium (Invitrogen, USA) supplemented with 10% FBS (Invitrogen, USA) and 100 Unit/ml penicillin/streptomycin (Sangon Biotech, China). Cells were cultured at 37 °C and in 5% CO2 incubator following the American Type Culture Collection (ATCC) guidelines. Anti-TEM1 (ab67273), anti-MMP-2 (ab92536) and GAPDH (ab9485) antibodies were purchased from Abcam (Cambridge, MA, USA).
Establishment of TEM1 overexpression and knockdown cell lines
A customized TEM1-overexpression lentivirus vector was constructed by KeyGen Biotech (Nanjing, China). MES-SA cells were transfected with the TEM1-lentivirus (LV-hTEM1-6His-OE) or control lentivirus (LV-vector) and selected using 2 µg/ml puromycin (Sigma–Aldrich, St. Louis, MO, USA). Selected cells were cultured in conditioned medium without puromycin to generate multiple monoclonal cell lines. The cells were subjected to Western blot analysis to detect TEM1 expression.
Short hairpin RNAs (shRNAs) targeting TEM1 (shRNA-TEM1) were used to knock down TEM1. Commercially inventoried lentivirus vectors expressing shRNA (LV-Htem1-shRNA) and negative control (LV-NC-shRNA) were constructed by KeyGen Biotech (Nanjing, China) with the following sequences: shRNA-TEM1 sequence: (sense) 5’-CACCGGTGGCTTCGAGTGTTATTGTCTCGAGACAATAACACTCGAAGCCACCTTTTTTC-3’, (antisense) 5’-AAAAGGTGGCTTCGAGTGTTATTGTCTCGAGACAATAACACTCGAAGCCACCAAAAAAGAGCT-3’; shRNA-NC (scramble) sequence: (sense) 5′‐TAACTAGTAACGGCTGCTCCCTCGAGGGAGCAGCCGTTACTAGTTTTTTTTC‐3′; antisense: 5′‐ATTGATCATTGCCGACGAGGCTCGAGCCTCGTCGGCAATGATCAAAAAAAAGAGCT‐3′. And pLL3.7 vector containing shRNA-TEM1 or shRNA-NA was combined with pRSVRev vectors and pMD.G vectors (Clonetech, USA). Subsequently, packaging vectors and lentiviral vector DNAs and were transfected into 293 T cells and harvested after 72 h. Supernatants containing lentiviruses were purified, concentrated and quantified. MES-SA cells were transfected with LV-hTEM1-shRNA or LV-NC-shRNA lentivirus and selected using 2 µg/mL puromycin.
RNA extraction, cDNA synthesis and real-time quantitative PCR
Total RNA was extracted using TRIzol reagent and reverse transcribed using High-capacity cDNA Reverse Transcription Kit. The cDNA samples generated were subjected to qPCR analysis using TaqMan Universal Master Mix II and commercially inventoried primers for TEM1 and MMP-2 (Applied Biosystems, USA). cDNA samples were then quantified using an ABI ViiA 7 System (Applied Biosystems, USA). 18S probe was used as an endogenous control. The reaction program was set as follows: 95 °C for 15 min followed by denaturation (95 °C for 10 s), annealing (for 30 s), and elongation (for 30 s). In total, 40 cycles of annealing and elongation steps were performed. The qPCR reactions were repeated three times and 2−ΔΔCT method was used to quantify relative gene expression levels.
Western blot
Total protein was extracted from cell lysates or tumor specimens. Protein concentrations were determined using a bicinchoninic acid (BCA) kit (Sangon Biotech, China). Total protein (20 μg per sample) was electrophoretically separated on a 4–20% gradient sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and transferred to PVDF membranes (Invitrogen, USA). The blots were incubated with 5% non-fat milk and probed with TEM1 (1:5000 dilution) and MMP-2 (1:2500 dilution) antibodies. The secondary antibody horseradish peroxidase (HRP) conjugated with goat anti-rabbit immunoglobulin G (1:5000 dilution) was then added to the membranes before protein detection. An endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (1:5000 dilution) was used as a control. Reactive proteins were detected using TMB Substrate Solution (Beyotime, China) through GBOX chemiXR5 imaging system (Gene company, Hongkong, China).
siRNA transfection and knockdown
MMP-2 siRNA was constructed using previously published sequences [29] by GenePharma (Shanghai, China). MES-SA cells were grown in serum-free medium to 50% confluency. Cells were then transfected with 20 μmol/L MMP-2 siRNA or Scramble siRNA using lipofectamine 2000 and incubated for 6 h. Cells were washed and cultured in the conditioned medium for observation at 24, 48 and 72 h. The expression of MMP-2 protein was detected using Western blot. MMP-2 siRNA sequence: F) 5-AACACAGCCTTCTCCTCCTG, R) 5-CACCTACACCAAGAACTTCC; β-actin sequence: F) TTGCGTTACACCCTTTCTTG, R) ACTGCTGTCACCTTCACCG.
Gelatin zymography
The cell supernatant was concentrated and mixed with 4X non-reducing sample buffer (Invitrogen, USA). Equivalent amounts of total protein supernatants (8 µg each lane) were loaded on a 7.5% SDS-PAGE gel containing 1 mg/mL gelatin. After separation, the gels were washed for 30 min in 2.5% Triton X-100 (Invitrogen, USA) to remove the SDS and incubated in developing buffer (Invitrogen, USA) for 15 h at 37 °C. The gels were then rinsed with distilled water and stained with Coomassie blue to detect MMP-2 activity.
Cell migration assay
The migration ability of cells was determined using the transwell migration assay (Corning, New York, USA). Briefly, serum-free medium was added to the upper chamber containing 1 × 105 cells and 20% serum-contained medium was added in the lower chamber. After incubation overnight, cells on the upper membrane of upper chamber were removed clearly, and migrated cells on the lower membrane were fixed with 4% paraformaldehyde, stained with crystal violet and counted under optical microscope. The percentage of migrated cells was calculated as the ratio of migratory cells against the total number of seeded cells. All assays were performed three times.
Cell invasion assay
The cell invasion ability of cells was evaluated using the transwell invasion assay. Briefly, the upper chamber was precoated with 0.5 mg/ml Matrigel matrix and serum-starved MES-SA cells (1 × 105) were added. In addition, medium containing 20% serum was added to the lower chamber. The chambers were incubated 72 h, after which cells that invaded to the lower membrane of upper chamber were fixed with 4% paraformaldehyde and stained with crystal violet. The number of invaded cells was immediately counted under a microscope. All assays were performed three times.
ECM adhesion assay
96-well plates were precoated with human fibronectin, laminin, or vitronectin overnight at 4 °C and blocked with 1% BSA for 1 h at 37 °C. Cells were added to the plates at 1 × 104 cells per well and incubated at 37 °C for 2 h. The wells were washed twice with PBS. Next, 200 µL serum-free medium was added to the wells and the OD value was tested using the CCK-8 assay kit.
ECM degradation assay
The gelatin degradation assay was conducted as described by Artym et al. [30]. Briefly, 18 mm acid washed coverslips were seeded in 12-wells tissue-culture plate. 1 mL of 50 μg/mL solution of poly- L-lysine was added to each well and incubated for 20 min at room temperature. The coverslips were washed three times with PBS and then incubated with 1 mL of 0.5% glutaraldehyde for 15 min at room temperature. The coverslips were washed again with PBS and coated with 0.2 mg/ml FITC-coupled gelatin (G-13187, ThermoFisher, USA) for 15 min. 3 × 104 of MES-SA or MBA-MD-231 cells were seeded on gelatin-coated coverslips for 5 to 8 h and then immunofluorescence was measured. Ten random fields per sample were captured using a laser confocal microscope to calculate gelatin degradation. To better observe the degradation behavior of cells, three-dimensional (3D) videos were reconstructed by collecting sequential Z-sections. Image J software was used to distinguish degraded (black) areas from nondegraded (green) area by thresholding. The percentage of degradation for each field was calculated based on the ratio of degraded regions to total regions.
In vivo study
Fifteen BALB/C nude female mice (4 weeks old; 10–12 g; Viton Lever, Beijing, China) were divided into three groups (five mice per group): the hTEM1-MES-SA cell group, shRNA-TEM1-MES-SA cell group and NC-MES-SA cell group. The cell density in each group was adjusted to 5 × 106 cells/mL and 0.2 mL was injected subcutaneously into the left armpit of the nude mice. Mice were housed in a semi-barrier system with constant temperature and humidity. The animals were euthanized after 4 weeks. The tumor volumes were measured and calculated using the following formula: volume (cm3) = (width2 × length)/2.
Another fifteen BALB/C nude female mice were randomly divided into three groups with five mice in each group. 1 × 106 of MES-SA shRNA-TEM1 cells, hTEM1 cells and NC cells in 0.2 ml of PBS was injected intraperitoneally. The mice were sacrificed on day 28 post transplantation and the number of metastases were calculated.
All animal experiments were performed in accordance with the institutional guidelines and approved by the Institutional Animal Care and Use Committee of Tongji Hospital of Tongji University.
Statistical analysis and data presentation
Pearson’s correlation and linear regression analyses were conducted to determine the correlation between TEM1, MMP-2 and MMP-9 expression in clinical specimens. For qPCR tests, 18 s probe was used as the internal control. In the Western blotting test, protein expression was normalized to the expression of GAPDH. Statistical and graphing data were analyzed using GraphPad Prism 5 software. Graphical data were presented as the mean values with standard deviations. Comparisons between groups were analyzed using unpaired student's t-test or Two-way ANOVA. A P value of less than 0.05 was considered statistically significant. Each experiment was repeated at least two times in triplicates.
Results
TEM1 and MMP-2 are co-expressed in uterine leiomyosarcoma but not in leiomyoma specimens
Our previous study found that TEM1 was overexpressed in 19 sarcoma subtypes [15]. Here, we studied TEM1, MMP-2 and MMP-9 expression in 50 samples of uterine leiomyosarcoma (n = 25) and leiomyoma (n = 25) tissues (Table 1). The expression levels of TEM1, MMP-2 and MMP-9 were quantified using Long H-scoring system, as previously described [31–33]. Our analysis revealed that TEM1 and MMP-2 were expressed in 92% (23 positive) and 88% (22 positive) of the 25 uterine leiomyosarcoma tissues, respectively. In contrast, only weak (median of 20 and 35 for TEM1 and MMP-2, respectively) or negative TEM1 (10 negative) and MMP-2 (18 negative) staining were seen in 25 leiomyoma tissues. According to FIGO classification, 8 specimens were low-grade (stage I-II) and 17 were high-grade (stage III-IV). TEM1 and MMP-2 were highly expressed in all 17 of high stage (III-IV) uterine sarcoma specimens and 62.5% or 5 of 8 of low-grade specimens. Interestingly, previous studies demonstrated a possible association between TEM1 and MMP-9 in fibrosarcoma cells [19]. However, in our study, MMP-9 was 72% negative (21 of 25) or only weakly expressed (median Long-H score of 80) in uterine leiomyosarcoma which was similar to that of leiomyoma tissues (22 of 25 negative). Only weak or negative staining of TEM1 (9 positive, 45%), MMP-2 (3 positive, 15%) and MMP-9 (2 positive, 10%) were detected in normal uterine myometrium. This suggested that MMP-9 might not play an important role in tumorigenesis of uterine leiomyosarcoma (Table 1).
Table 1.
TEM1, MMP-2 and MMP-9 expression in,uterine leiomyosarcoma (uLMS), leiomyoma and normal uterine tissues
| Group | Total (n) | FIGO classification | N (n) | TEM1 expressionA | MMP-2 expressionB | Mmp-9 expressionC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg | Min | Max | Mean | Total positive | Neg | Min | Max | Mean | Total positive | Neg | Min | Max | Mean | Total positive | ||||
| uLMS | 25 | I | 4 | 2 | 0 | 144 | 43.5 | 23 (92%) | 2 | 0 | 95 | 28.8 | 22 (88%) | 4 | 0 | 0 | 0 | 4 (16%) |
| II | 4 | 0 | 80 | 235 | 146.3 | 1 | 0 | 170 | 77.5 | 4 | 0 | 0 | 0 | |||||
| III | 8 | 0 | 80 | 230 | 162.9 | 0 | 35 | 180 | 99.1 | 5 | 0 | 85 | 20 | |||||
| IV | 9 | 0 | 35 | 300 | 236.1 | 0 | 10 | 190 | 135.6 | 8 | 0 | 160 | 17.8 | |||||
| Leiomyoma | 25 | / | / | 10 | 0 | 120 | 20 | 15 (60%) | 18 | 0 | 85 | 35 | 7 (28%) | 22 | 0 | 65 | 15 | 3 (12%) |
| Normal tissues | 20 | / | / | 11 | 0 | 70 | 12.5 | 9 (45%) | 17 | 0 | 30 | 3.0 | 3 (15%) | 18 | 0 | 20 | 2.0 | 2 (10%) |
AThe mean of TEM1 Long H-Score are positively correlated with tumor stages, r = 0.9554, p < 0.01
BThe mean of MMP-2 Long H-Score are positively correlated with tumor stages, r = 0.9602, p < 0.01
CThe mean of MMP-9 Long H-Score are not correlated with tumor stages, r = 0.1205, p > 0.05
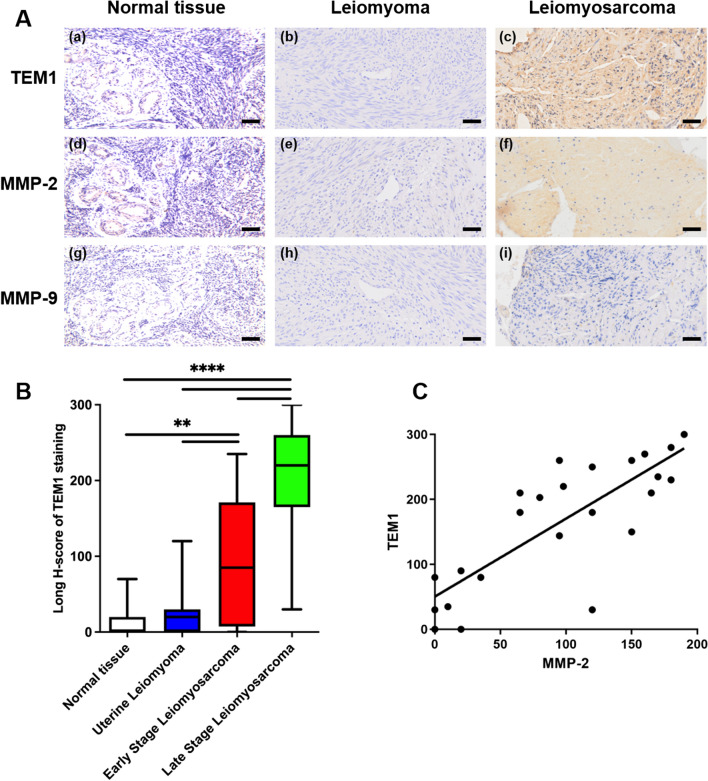
Figure 1 A shows representative images of TEM1 (a–c), MMP-2 (d–f) and MMP-9 (g–i) stained tissues from a patient with uterine myometrium (normal tissues), a patient with leiomyoma and another patient with stage III uterine leiomyosarcoma using serial tumor sections. The Long H-scores of TEM1, MMP-2 and MMP-9 were 280 (80% 3 + , 20% 2 +), 130 (70% 1 + , 30% 2 +) and 0 in this stage III leiomyosarcoma tissue. The boxplot showed that TEM1 staining was positive in 92% of leiomyosarcoma tissues with a median score of 203 (Range, 0–300), significantly higher than those of leiomyoma tissues (P < 0.01; Fig. 1B) and uterine myometrium tissues (P < 0.0001; Fig. 1B). What is more, the Long H-scores of TEM1 in late stage uterine leiomyosarcoma was significantly higher than those with early stage (P < 0.01; Fig. 1C)
Fig. 1.
The expression of TEM1, MMP-2 and MMP-9 in different uterine samples. A Representative immunohistochemical results of TEM1, MMP-2 and MMP-9 staining. TEM1 and MMP-2 were negative in uterine myometrium (normal tissue) and leiomyoma, strong positive in leiomyosarcoma; MMP-9 staining was negative or weakly expressed in all samples. Scale bar, 50 μm. B Boxplot for Long H-scores of TEM1 in normal myometrium, leiomyoma and leiomyosarcoma tissues. TEM1 Long H-scores were positively correlated with tumor stages, r = 0.9554. **, P < 0.01; ****, P < 0.0001. C TEM1 expression was positively correlated with MMP-2 expression in uterine leiomyosarcoma tissues. Correlation curves are shown with r square, and P‐values from Pearson´s correlation analysis. R square = 0.6487, Pearson r = 0.8054, P < 0.0001
Altogether, these results indicate that TEM1 and MMP-2 were co-expressed in uterine leiomyosarcoma tissues, especially high-grade lesions, suggesting poor prognosis in uterine leiomyosarcoma patients. MMP-2, but not MMP-9, might work together with TEM1 in uterine leiomyosarcoma progression.
TEM1 and MMP-2 expression in uterine leiomyosarcoma tissues are positively correlated
To determine the correlations among TEM1, MMP-2 and MMP-9 in uterine leiomyosarcoma tissues, protein expression levels were calculated using Long H-scores and Pearson’s correlation across all patients. Table 1 shows that the median values of both TEM1 and MMP-2 Long H-Score were positively correlated with tumor stages (TEM1, Pearson’s r = 0.9554, P < 0.01; MMP-2, Pearson’s r = 0.9602, P < 0.01). However, we found no correlation between median MMP-9 Long H-Score values and tumor stages (Pearson’s r = 0.1205, P > 0.05). Moreover, the correlation curve in Fig. 1C revealed a significantly positive correlation between TEM1 and MMP-2 expression (r squared = 0.6487, Pearson r = 0.8054, P < 0.0001), suggesting a functional association between TEM1 and MMP-2 in promoting tumor progression of uterine leiomyosarcomas. However, no correlation was found between TEM1 and MMP-9 expression (r square = 0.0202, Pearson r = 0.1420, P = 0.5999).
TEM1 overexpression and knockdown in vitro model are established
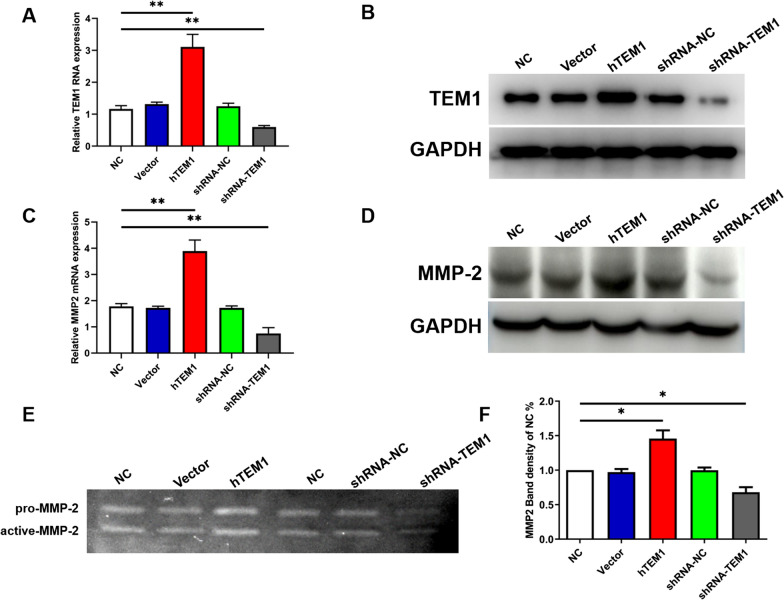
TEM1 overexpression (hTEM1) and knockdown (shRNA-TEM1) MES-SA cell lines were established using a lentivirus-mediated TEM1 vector and shRNA. Compared with control vector MES-SA (vector) cells, TEM1 mRNA and protein levels were significantly higher in hTEM1 MES-SA cells using qPCR and Western blot (Fig. 2A, B). In contrast, TEM1 mRNA and protein expression levels were significantly lower in shRNA-TEM1 MES-SA cells (shRNA-TEM1) than in shRNA-NC-TEM1 MES-SA (shRNA-NC) cells (P < 0.01). TEM1 mRNA and protein expression levels in vector and shRNA-NC group were not significantly different from those of non-treated negative control wild-type MES-SA (NC) cells (P > 0.05).
Fig. 2.
The expression level of MMP-2 regulated by TEM1. Lentivirus-mediated hTEM1 MES-SA control vector (Vector), TEM1 overexpression vector (hTEM1), shRNA NC vector (shRNA-NC) and shRNA-TEM1 vector were transfected into MES-SA cell lines (NC) for the establishment of TEM1 overexpression and low expression MES-SA cell lines. TEM1 mRNA A and protein B expression were determined using qPCR and Western blot assays, respectively. The protein expression level of MMP-2 mRNA C and protein D in all cell lines. MMP-2 expression was upregulated in hTEM1 cells and downregulated in shRNA-TEM1 cells. Gelatin zymography showing elevated expression of MMP-2 in hTEM1 cells and decreased expression of MMP-2 in shRNA-TEM1 cells E, based on band density using Image J (F). *, P < 0.05; **, P < 0.01
TEM1 regulates expression of MMP-2 in MES-SA cells
We determined MMP-2 mRNA and protein expression levels in hTEM1 MES-SA (hTEM1) cells, shRNA-TEM1 MES-SA (shRNA-TEM1) cells, shRNA-NC-TEM1 MES-SA (shRNA-NC) cell, vector MES-SA (vector) cells and wild-type MES-SA (NC) cells using qPCR and Western blot (Fig. 2C, D). Results showed that MMP-2 mRNA and protein expression levels were significantly higher in hTEM1 cells than in vector cells (P < 0.01). In addition, MMP-2 mRNA and protein expression levels were significantly down-regulated in shRNA-TEM1 cells, compared with shRNA-NC cells (P < 0.01). To determine whether the activation of MMP-2 was also up-regulated by TEM1, we conducted gelatin zymography with cell supernatants from each group (Fig. 2E). Compared with vector cells, hTEM1 cells exhibited a higher level of activated MMP-2 with a higher band density, whereas shRNA-TEM1 cells showed a lower level of activated MMP-2 with a lower band density (both P < 0.05; Fig. 2F). These results suggested that TEM1 up-regulated MMP-2 expression and activity in MES-SA cells.
Loss of MMP-2 inhibits migration and invasion ability of hTEM1 MES-SA cells
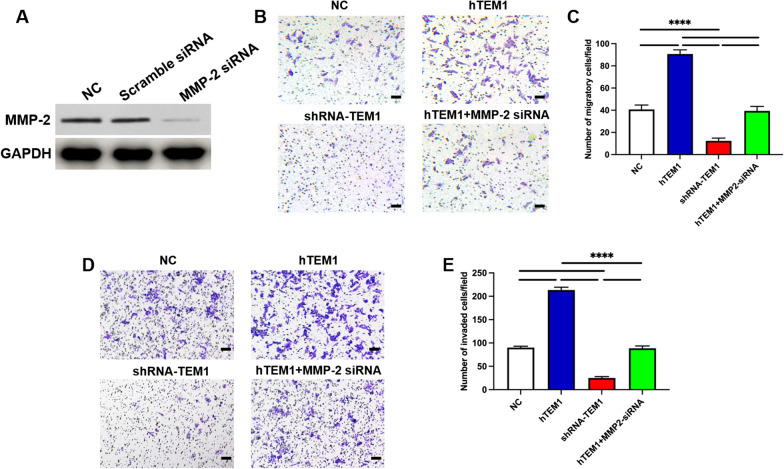
Our limited data [34] suggested that TEM1 promoted migration and invasion of MES-SA cells. However, it is unknown whether MMP-2 participates in TEM1-promoted migration and invasion of MES-SA cells. Effect of MMP-2 gene knockdown in hTEM1 MES-SA cells using an MMP-2-targeted siRNA were confirmed by Western Blot (Fig. 3A). The invasion and migration abilities of cells were subsequently determined using transwell assays. The migration and invasion abilities of hTEM1 MES-SA cells were inhibited by MMP-2 siRNA (both P < 0.05) and were not significantly different from those of control cells (P > 0.05; Fig. 3B–D).
Fig. 3.
TEM1 regulating MES-SA cell migration and invasion via MMP-2. Western blotting results confirming knock-down of MMP-2 protein by MMP-2 siRNA A. Both cell migration ability B and C and invasion ability D and E were enhanced in hTEM1 cells and inhibited in shRNA-TEM1 treated cells. And hTEM1 MES-SA cells transfected with MMP-2 siRNA (hTEM1 + MMP-2 siRNA) showed comparable invasion and migration ability to NC group. ****, P < 0.0001. All assays were replicated three times in triplicates. Magnification, 100X; bar scale, 100 μm
MMP-2 is necessary for TEM1- mediated ECM adhesion
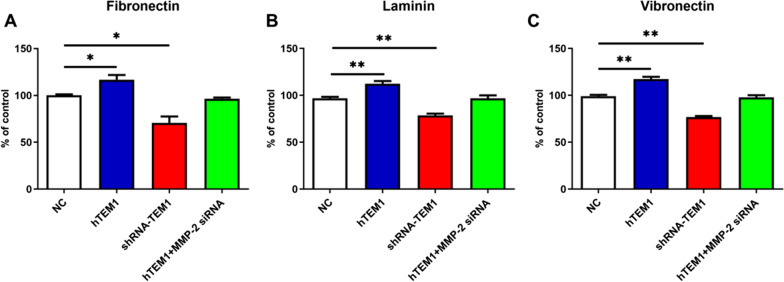
Cell-ECM adhesion is a critical step in cancer distant metastasis [35–37]. Previous studies revealed that TEM1 co-expressed with ECM components and promoted the adhesion of cells to fibronectin, increasing cell migration in vitro [18]. Our study explored changes in TEM1-mediated adhesion ability of MES-SA cells to ECM components, including fibronectin, laminin and vitronectin, using Cell-ECM adhesion assay. As shown in Fig. 4A–C, hTEM1 cells displayed higher adhesion ability to ECM components compared with negative control cells (all P < 0.05). MMP-2 siRNA significantly decreased the number of adhered hTEM1 cells in each group (P < 0.5), leading to its adherent ability not significantly different from negative control cells (P > 0.05). These results indicated that TEM1 promoted the adhesion ability of MES-SA cells to ECM components in the presence of MMP-2.
Fig. 4.
TEM1 promoted MES-SA cell adhesion to extracellular matrix (ECM) via MMP-2. NC, hTEM1, shRNA-TEM1, and hTEM1 + MMP-2 siRNA cells were selected to adhere to fibronectin (Fn) A, laminin (Laminin) B, and Vitronectin (Vn) C in the ECM-coated 12-well plate for 72 h respectively. The numbers of adhesion cells were calculated based on the CCK-8 assay. *, P < 0.05; **, P < 0.01. All assays were replicated three times in triplicates
MMP-2 is necessary for TEM1-mediated ECM degradation
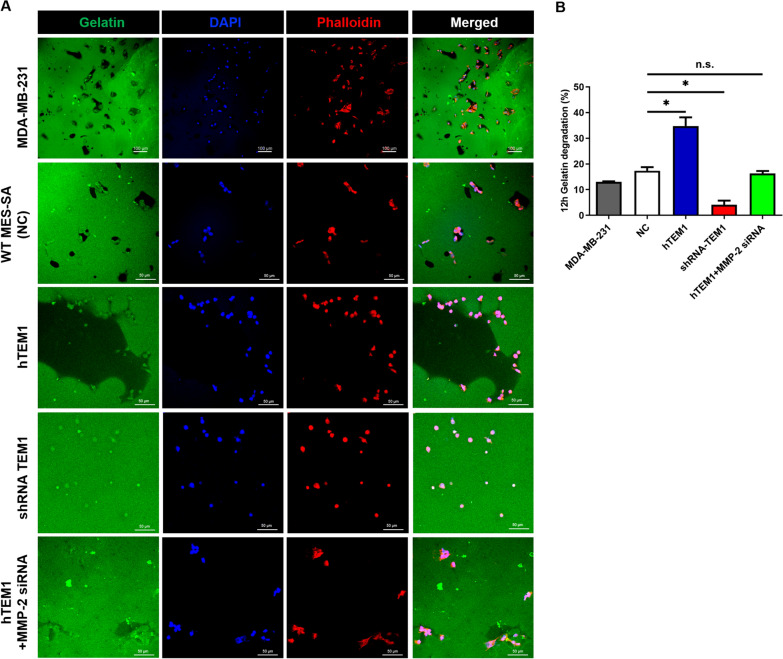
To study the ability of MES-SA cells to degrade the matrix, cells were cultured on coverslips coated with FITC-conjugated gelatin matrix for 12 h and evaluated with confocal microscopy (Fig. 5A). Breast cancer cell line MDA-MB-231 was included as a positive control (PC) because its stable capability to degrade ECM has been confirmed in literature [30, 38–40]. After 12 h of incubation, black areas of degradation were shown underneath the cell body, and cells were found within the degraded areas. Similar degradation pattern was seen in WT MES-SA (NC) cells as black holes were observed in gelatin matrix where the cells were located. Interestingly, in hTEM1 MES-SA cells, we observed pool-like areas of degradation in gelatin matrix, while when TEM1 was knocked-down (shRNA-TEM1 group), gelatin degradation decreased significantly compared with WT MES-SA and hTEM1 MES-SA cells. To clarify the role of MMP-2 in TEM1-mediated ECM degradation, we knocked down MMP-2 expression using MMP-2 siRNA in hTEM1 MES-SA cells and found that gelatin degradation was decreased compared with hTEM1 MES-SA group. To quantify the degraded area in different cells, we calculated the percentage of degradation in each field as a ratio of degraded regions to the total region using Image J software. According to the results showed in Fig. 5B, hTEM1 MES-SA cells displayed the highest percentage of degradation (P < 0.05, Fig. 5B) followed by MDA-MB-231 cells. While shRNA-TEM1 MES-SA cells had the lowest degradation compared with MES-SA NC cells (P < 0.05, Fig. 5B). No significant difference was observed between NC MES-SA cells and hTEM1 + MMP-2 siRNA cells (P > 0.05, Fig. 5B).
Fig. 5.
(A) Gelatin degradation assays of MDA-MB-231, WT MES-SA (NC), hTEM1 MES-SA, shTEM1 MES-SA and hTEM1 MES-SA knocked down MMP-2 expression by MMP-2 siRNA (hTEM1 + MMP-2 siRNA) after 12 h. MDA-MB-231 served as positive control. Pool-like areas of degradation in gelatin matrix formed around hTEM-MES-SA cells. Immunofluorescence staining of FITC- conjugated gelatin (green), Phalloidin (red), nuclei (blue), and merged images. (B) Quantification of gelatin degraded area (n = 10) by Image J. Data are presented as mean ± standard deviation (SD). *, P < 0.05 in Student t-test; n.s., none-sense, P > 0.05; Scale bar, 100 μm or 50 μm
Notably, in our degradation assays, sometimes we only observed shadows or lower intensity compared with the rest of the green gelatin in gelatin matrix instead of black holes, which is consistent with previous studies [39, 41, 42]. Mazurkiewicz E et al. [41] pointed out that this phenomenon could be an incomplete degradation of gelatin in the three dimension, leading to degraded areas not shown as black hole but only seem to be less intense than the rest of the green FITC-gelatin layer. What is more, when aggressive degradation was seen in some certain cell types including MES-SA, pool-like clear area might be seen in gelatin matrix [40, 43]. This indicates that sometimes two-dimensional images might not efficiently reveal cell behaviors in gelatin matrix as it is impossible to determine whether there are cells inside the pool on a different gelatin layer. In order to better elucidate TEM1-mediated ECM degradation, we reconstructed three-dimensional images by collecting sequential Z-sections based on each field of TEM1-hTEM1 cells and positive control MDA-MB-231 cells (Supplementary video 1 and 2). Compared with two-dimensional (2D) image of a pool-like black degradation area with cells close to the matrix boundaries, the three-dimensional (3D) video (Supplementary video 2) depicted the depth of degraded gelatin inside a pool, with cells presented on different gelatin layers, confirming that hTEM1 cells degraded ECM not only horizontally but also vertically. Moreover, the three-dimensional image clearly outlined the track of aggressive cell migration on different layers of gelatin matrix. We also captured three-dimensional images of gelatin degradation by MES-SA cells, which clearly outlined the cells inside of degraded areas and different layers of degradation (Supplementary images).
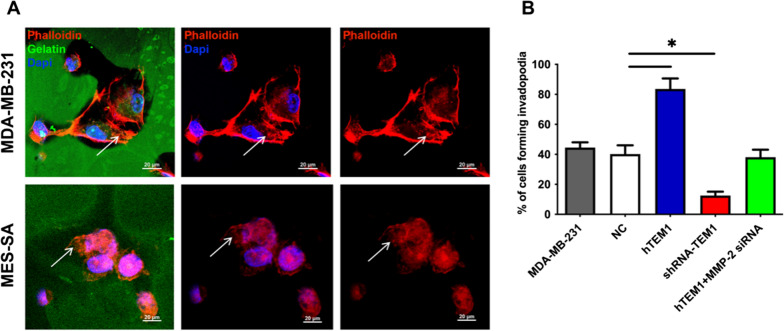
To find out whether TEM1 promoted invadopodia formation ability, representative images of MDA-MB-231 and hTEM1 MES-SA cells were shown in Fig. 6A. F-actin-enriched invadopodia (big rosettes) were observed at the cell edge in both cells (Fig. 6A, white arrows). To quantify invadopodia formation, cells forming more than five F-actin positive invadopodia were counted and normalized to the total cell number of each field. The results (Fig. 6B) showed that compared with WT MES-SA (NC) group, the highest percentage of invadopodia was formed by hTEM1 MES-SA cells (P < 0.05, Fig. 6B) and the lowest proportion was found in shRNA-TEM1 cells (P < 0.05, Fig. 6B). When MMP-2 was knocked-down, the invadopodia formation ability of hTEM1 MES-SA cells was impaired. There was no significant difference in invapodia formation ability between hTEM1 + MMP-2 siRNA cells and NC cells (P > 0.05, Fig. 6B). These results demonstrated that invadopodia formation was involved in TEM1-mediated degradation and could be inhibited by MMP-2 knockdown.
Fig. 6.
Immunofluorescence of invadopodia. A The merged images of F-actin (red), and nuclei (blue) are shown. Arrows indicate invadopodia locations. B Quantification of invadopodia in MDA-MB-231 and MES-SA cells. Data are presented as mean ± standard deviation (SD). *, P < 0.05. Scale bar, 20 μm
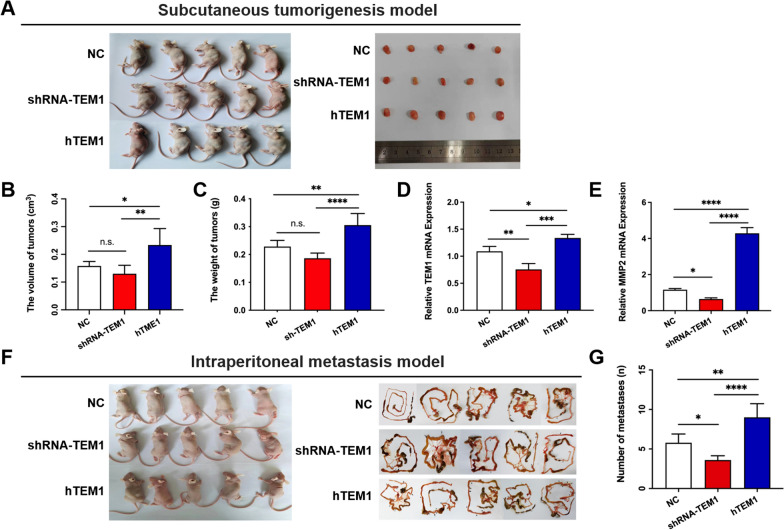
TEM1 overexpression promotes the metastasis of uterine sarcoma in the animal model
To validate the role of TEM1 in uterine sarcoma proliferation and metastasis, we conducted in vivo study using BALB/C nude female mice. MES-SA NC, shRNA-TEM1 and hTEM1 cells were injected subcutaneously in three groups of mice respectively (n = 5 in each group) (Fig. 7A). Although the volume and weight of tumors significantly increased in hTEM1 group compared to group (Fig. 7B and C, both p < 0.05), the tumor size and weight were not significantly affected in shRNA-TEM1 group (Fig. 7B and C, p > 0.05). RT-qPCR analysis was used to confirm the overexpression of TEM1 mRNA levels in tumors of hTEM1 group and deletion effect of TEM1 mRNA in those of shRNA-TEM1 group (Fig. 7D). MMP-2 mRNA expression was up-regulated in tumors of hTEM1 group (Fig. 7E, p < 0.0001) and down-regulated in tumors of shRNA-TEM1 group (Fig. 7E, p < 0.05), confirming the TEM1 regulation effect on MMP-2.
Fig. 7.
TEM1 promotes the metastasis of uterine sarcoma cells rather than tumor growth. A MES-SA NC, shTEM1 and hTEM1 cells were injected subcutaneously in three groups of mice respectively (n = 5 in each group). Tumor weight B and volume C were measured and calculated. TEM1 mRNA D and MMP-2 mRNA E expression levels of subcutaneous tumors from each group were determined by RT-qPCR. F MES-SA NC, shRNA-TEM1 and hTEM1 cells were injected intraperitoneally in three groups of mice respectively (n = 5 in each group). Metastasis sites were displayed in black arrows and the number of metastases were calculated G. n.s., none-sense; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001
Furthermore, MES-SA NC, shRNA-TEM1 and hTEM1 cells were injected intraperitoneally in three groups of mice respectively (n = 5 in each group) (Fig. 7F). Tumor cells spread to several peritoneal organs 28 days after injection (Fig. 7F, right). The number of metastasis sites significantly increased in hTEM1 group (Fig. 7G, p < 0.01) and decreased in shRNA-TEM1 group (Fig. 7E, right, p < 0.05).
Collectively, the in vivo data suggested that TEM1 promotes the metastasis of uterine sarcoma cells without affecting tumor proliferation.
Taken together, these assays demonstrated the TEM1-mediated invasion and migration at two-dimensional and three-dimensional levels on gelatin matrix in MES-SA cells and in vivo xenografts. They also demonstrated the critical role of MMP-2 in TEM1-mediated ECM degradation.
Discussion
MMP-2, not MMP-9, might participate in the TEM1-mediated progression of uterine leiomyosarcoma
TEM1 has been considered a potential therapeutic target for sarcoma in the last decade [7, 44, 45]. Ontuxizumab (MORAb-004), has recently been applied as an effective humanized anti-TEM-1 monoclonal antibody in preclinical and clinical studies [11, 12]. However, to date, the role of TEM1 in tumor progression has not been clarified sufficiently. Yumiko Kondo et al. [46] suggested that TEM1 might be associated with tumor progression and metastasis in osteosarcoma and MORAb-004 could inhibit tumor cell migration in vitro. Consistent with our previous work [15] in which we found that TEM1 was overexpressed in uterine sarcoma specimens and promoted the migration and invasion of uterine sarcoma MES-SA cells, our present data showed that TEM1 expression level was correlated with tumor stage and MMP-2 expression. Leiomyoma specimens were included to allow comparison with its stromal origin as a benign tumor. TEM1 was highly expressed in leiomyosarcoma cells and correlated with tumor stage but not or weakly expressed in leiomyoma cells, implying that TEM1 might be a reliable and specific marker for tumor malignancy.
TEM1 and MMPs were reported to be co-expressed in tumor models. Margarida Maia et al. [19] reported a decrease in activated MMP-9 release in mice lacking the TEM1 cytoplasmic domain. Brian Tomkowicz [6] confirmed that MMP-9 activity was enhanced in generated CHO-TEM1 cells stably expressing TEM1 compared with wide-type CHO-K1 cells. However, our IHC results showed no correlation between TEM1 expression and MMP-9, but a positive correlation between TEM1 and MMP-2 in human uterine leiomyosarcoma specimens. Furthermore, we found that TEM1 and MMP-2 expression was positively correlated with tumor stage, implying that MMP-2, rather than MMP-9, might participate in TEM1-mediated tumorigenesis of uterine leiomyosarcoma.
MMP-2 has been reported to correlate with tumor metastasis in many cancer types. Yang Hyun Kee et al. [47] found significantly higher MMP-2 expression and slightly higher MMP-9 expression in metastatic malignant fibrous histiocytoma (MFH) compared with non-metastatic MFH (P < 0.05). Zhe Song [48] found a positive association of lymph nodes metastasis and the degree of lymphatic metastasis with the expression intensity of MMP-2 in hypopharyngeal carcinoma. Activated MMP-2 were highly expressed in breast cancers and were associated with invasive potential [49–51].
In our study, to explore the mechanism underlying the metastasis and invasion of uterine sarcoma, a poorly differentiated uterine sarcoma cell line MES-SA expressing moderate levels of TEM1 was used. We utilized these cells to establish TEM1 overexpression and knock-down in vitro models by lentivirus transfection. Our gelatinase zymography data displayed two bands corresponding to pro-MMP-2 and active-MMP-2 in MES-SA cells. However, MMP-9 bands were absent, which was consistent with previous findings by Roomi et al. [52] and our IHC staining results for MMP-2 and MMP-9 in uterine sarcoma specimens. The results confirmed the role of MMP-2 in promoting the uterine sarcoma progress.
TEM1 promotes migration and invasion of MES-SA cells by upregulating MMP-2
MMP-2 regulation and mechanism of action is complex. Tissue inhibitor of metalloproteases 2 (TIMP-2) and MMP-14 are common modulators of MMP-2 [53–55]. Recent studies have shown that other molecules, including heparan sulfate [56], cytokines [57, 58], and estradiol [59] also modulate MMP-2 expression or activity.
MMP-2 can degrade ECM components. In an ovarian metastasis model, ECM components such as fibronectin, vitronectin and collagen I were cleaved into small fragments by activated MMP-2 and facilitate cancer cell adhesion and invasion by binding to integrin receptors [60]. TEM1, MMP-2 and collagen IV have been reported to co-exist in early angiogenesis, implying that TEM1 might interact with MMPs during angiogenesis and cell migration [6].
Our gain-of-function and loss-of-function experiments showed that high expression of TEM1 was accompanied by elevated secretion of activated MMP-2 followed by increased migration and invasion. Conversely, low expression of TEM1 is followed by lower expression level of MMP-2 and decreased aggressiveness of MES-SA cells. Downregulation of MMP-2 inhibited the tumorigenesis-promoting effect of TEM1. These results confirmed that MMP-2 up-regulated TEM1, further suggesting that TEM1 might promote the migration and invasion of uterine sarcoma through an MMP-2-dependent pathway.
TEM1 promotes ECM remodeling by modulating MMP-2
Recent studies of TEM1 have been largely confined to promoting angiogenesis rather than ECM transformation [29, 39]. Robust TEM1-ECM interactions have been demonstrated in vivo by Brian Tomkowicz et al.’s [6] study [54]. They demonstrated that the extracellular region of TEM1 (Fc-TEM1) could directly bind to collagen I, IV, and fibronectin, but not to laminin and vitronectin. In contrast, we found that TEM1 promoted MES-SA cell adhesion to fibronectin, laminin and vitronectin. Moreover, we observed aggressive degradation activity of TEM1-overexpressing MES-SA cells on gelatin. A possible explanation for this observation is that malignant cells behave differently from non-malignant cells. Besides, other molecules, such as integrin, might also participate in modulating TEM1-ECM interactions in uterine sarcoma cells.
ECM degradation and remodeling caused by bioactive molecules such as MMPs released from cancer cells are crucial not only to normal development but also to tumor invasion and metastasis [42, 61, 62]. Invadopodia are podosome-like membrane protrusions formed by invasive malignant cells with matrix degradation ability [63]. Invadopodia were first described in breast cancer cell lines seeded on fibronectin-coated gelatin [64]. It has been demonstrated that MMPs, specially MMP-2, MMP-9 and MMP-13 are localized in invadopodia and play an important role in ECM proteolysis [65].
Mazurkiewicz E [41] described in her protocol that, dark spots or areas on a thin layer of fluorescent-labeled gelatin can be observed under fluorescence microscopy after cells are seeded in a given time, which is a result of active gelatin digesting by invadopodia. Invadopodia and focal degradation of gelatin matrix have been observed in PC-3 prostate cancer cells [66, 67], breast cancer MDA-MB-231 cells [40, 68, 69], and other malignant cell [70–72]. In the present study, gelatin degradation of MDA-MB-231 cells were found within the cell region, which was confirmed by literatures [3, 30, 39, 43]. However, aggressive gelatin degradation caused by hTEM1-MES-SA cells can cause pool-like degradation areas similar to studies of Albrechtsen R [40], possibly due to degradation on different layers and active cell migration. TEM1-overexpressed (hTEM1) MES-SA cell line displayed the highest percentage of degradation and invadopodia formation. Pool-like degradation areas were commonly seen on gelatin matrix in hTEM1 MES-SA cells, reflecting a tree-like migrating track or/and pool-like vertical invasion of cell behaviors. From three-dimensional videos, clear migrating routes of a single cell and different layers of gelatin-matrix degradation were revealed. Treatment with MMP-2 siRNA significantly impaired the ability of matrix degradation and invadopodia formation of the cells. Our results of migration and invasion transwell assay on matrix also confirmed that TEM1 promoted the migration and invasion ability of MES-SA cells, which was inhibited by MMP-2 knockdown.
TEM1 promotes the metastasis of uterine sarcoma cells rather than tumor growth in vivo
It is clear that inhibition of TEM1 in cancer-associated fibroblasts significantly impaired the tumor growth of hepatocellular carcinoma [5] and lung cancer [73] in xenograft models. To evaluate the role of TEM1 in promoting uterine sarcoma proliferation and metastasis in vivo, we established MES-SA cells-bearing subcutaneous and intraperitoneal mice models. Our results showed that TEM1 promoted the metastasis of uterine sarcoma cells, while tumor proliferation was not affected. The MMP-2 mRNA expression was increased in hTEM1 subcutaneous tumors, which was in agreement of the MMP-2 up-regulation effect of TEM1 in vitro. The in vivo data validated the role of TEM1 in regulating MMP-2 and promoting metastasis in uterine sarcoma.
These findings suggested that TEM1-overexpressing MES-SA cells displayed an enhanced capacity to degrade ECM. The enhanced cell migration and invasion by TEM1 overexpression might be the result of ECM remodeling upregulated by MMP-2.
Conclusion
Our analysis revealed a high expression pattern of TEM1 and MMP-2 in uterine leiomyosarcoma with percentage of 92% (23/25) and 88% (22/24). TEM1 and MMP-2 were highly expressed in all 17 of high stage (III-IV) uterine sarcoma specimens. TEM1 promoted uterine sarcoma migration and invasion via increasing MMP-2 activity and ECM remodeling/degradation. The above provides valuable insights into a potential TEM1-targeted therapy for uterine sarcoma.
Supplementary Information
Acknowledgements
We would like to thank Home for Researcher editorial team (www.home-for-researchers.com) for partial English language editing service.
Author contributions
YG conceived, designed the study and revised the manuscript. YG, CW, WS, DS performed the experiments and analyzed the data. HL and XT collected clinical specimens and data. YG and CW prepared all the figures and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (81802583, 82071630, 81771560) and Construction of Key Disciplines of Tongji Hospital in the Fifth Cycle (ZD16-FC-1). The funding body did not contribute to any of the following: design of the study, data collection, data analysis, interpretation of data, writing the manuscript or decision to publish.
Data availability
The original contributions presented in the study are included in the article/supplementary file. Further inquiries can be directed to the corresponding author.
Declarations
Ethics approval and consent for participation
The study protocol was approved by the Ethics Committee of Tongji Hospital, Tongji University School of Medicine. Clinical specimens were obtained with patients’ written informed consents and all experiments regarding patients’ tissue samples were conducted in accordance with the Declaration of Helsinki.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenghao Wu, Wenhuizi Sun and Dongsheng Shen have contributed equally to this work and share the first authorship
References
- 1.Mallmann P. Uterine sarcoma—difficult to diagnose, hard to treat. Oncol Res Treat. 2018;41(11):674. doi: 10.1159/000494393. [DOI] [PubMed] [Google Scholar]
- 2.Hosh M, Antar S, Nazzal A, Warda M, Gibreel A, Refky B. Uterine sarcoma: analysis of 13,089 cases based on surveillance, epidemiology, and end results database. Int J Gynecol Cancer. 2016;26(6):1098–1104. doi: 10.1097/IGC.0000000000000720. [DOI] [PubMed] [Google Scholar]
- 3.Abeler VM, Royne O, Thoresen S, Danielsen HE, Nesland JM, Kristensen GB. Uterine sarcomas in Norway. a histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology. 2009;54(3):355–64. doi: 10.1111/j.1365-2559.2009.03231.x. [DOI] [PubMed] [Google Scholar]
- 4.Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S, et al. Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvasc Res. 2008;76(3):180–188. doi: 10.1016/j.mvr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Yang F, Wei Y, Han D, Li Y, Shi S, Jiao D, et al. Interaction with CD68 and regulation of GAS6 expression by Endosialin in fibroblasts drives recruitment and polarization of macrophages in hepatocellular carcinoma. Cancer Res. 2020;80(18):3892–3905. doi: 10.1158/0008-5472.CAN-19-2691. [DOI] [PubMed] [Google Scholar]
- 6.Tomkowicz B, Rybinski K, Foley B, Ebel W, Kline B, Routhier E, et al. Interaction of endosialin/TEM1 with extracellular matrix proteins mediates cell adhesion and migration. Proc Natl Acad Sci U S A. 2007;104(46):17965–17970. doi: 10.1073/pnas.0705647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouleau C, Gianolio DA, Smale R, Roth SD, Krumbholz R, Harper J, et al. Anti-endosialin antibody-drug conjugate: potential in sarcoma and other malignancies. Mol Cancer Ther. 2015;14(9):2081–2089. doi: 10.1158/1535-7163.MCT-15-0312. [DOI] [PubMed] [Google Scholar]
- 8.Carson-Walter EB, Winans BN, Whiteman MC, Liu Y, Jarvela S, Haapasalo H, et al. Characterization of TEM1/endosialin in human and murine brain tumors. BMC Cancer. 2009;9:417. doi: 10.1186/1471-2407-9-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiyohara E, Donovan N, Takeshima L, Huang S, Wilmott JS, Scolyer RA, et al. Endosialin expression in metastatic melanoma tumor microenvironment vasculature: potential therapeutic implications. Cancer Microenviron. 2015;8(2):111–118. doi: 10.1007/s12307-015-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthaiou EI, Guo Y, Barar J, Sandaltzopoulos R, Kandalaft LE, Li C, et al. TEM1-targeting PEGylated PLGA shikonin nanoformulation for immunomodulation and eradication of ovarian cancer. Bioimpacts. 2022;12(1):65–86. doi: 10.34172/bi.2021.23511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris RE, Fox E, Reid JM, Ralya A, Liu XW, Minard C, et al. Phase 1 trial of Ontuxizumab (MORAb-004) in children with relapsed or refractory solid tumors: a report from the children’s oncology group phase 1 pilot consortium (ADVL1213) Pediatr Blood Cancer. 2018;65(5):e26944. doi: 10.1002/pbc.26944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones RL, Chawla SP, Attia S, Schoffski P, Gelderblom H, Chmielowski B, et al. A phase 1 and randomized controlled phase 2 trial of the safety and efficacy of the combination of gemcitabine and docetaxel with ontuxizumab (MORAb-004) in metastatic soft-tissue sarcomas. Cancer. 2019;125(14):2445–2454. doi: 10.1002/cncr.32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz LA, Jr, Coughlin CM, Weil SC, Fishel J, Gounder MM, Lawrence S, et al. A first-in-human phase I study of MORAb-004, a monoclonal antibody to endosialin in patients with advanced solid tumors. Clin Cancer Res. 2015;21(6):1281–1288. doi: 10.1158/1078-0432.CCR-14-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grothey A, Strosberg JR, Renfro LA, Hurwitz HI, Marshall JL, Safran H, et al. A randomized, double-blind, placebo-controlled phase II study of the efficacy and safety of monotherapy Ontuxizumab (MORAb-004) plus best supportive care in patients with chemorefractory metastatic colorectal cancer. Clin Cancer Res. 2018;24(2):316–325. doi: 10.1158/1078-0432.CCR-17-1558. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Hu J, Wang Y, Peng X, Min J, Wang J, et al. Tumour endothelial marker 1/endosialin-mediated targeting of human sarcoma. Eur J Cancer. 2018;90:111–121. doi: 10.1016/j.ejca.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 16.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11(1):5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kontsekova S, Polcicova K, Takacova M, Pastorekova S. Endosialin: molecular and functional links to tumor angiogenesis. Neoplasma. 2016;63(2):183–192. doi: 10.4149/202_15090N474. [DOI] [PubMed] [Google Scholar]
- 19.Maia M, DeVriese A, Janssens T, Moons M, Lories RJ, Tavernier J, et al. CD248 facilitates tumor growth via its cytoplasmic domain. BMC Cancer. 2011;11:162. doi: 10.1186/1471-2407-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Xu C, Zhang K, Han D, Yang F, Li Y, et al. CD248 as a bridge between angiogenesis and immunosuppression: a promising prognostic and therapeutic target for renal cell carcinoma. Ann Transl Med. 2021;9(23):1741. doi: 10.21037/atm-21-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naylor AJ, McGettrick HM, Maynard WD, May P, Barone F, Croft AP, et al. A differential role for CD248 (Endosialin) in PDGF-mediated skeletal muscle angiogenesis. PLoS ONE. 2014;9(9):e107146. doi: 10.1371/journal.pone.0107146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Benedetto P, Ruscitti P, Liakouli V, Del Galdo F, Giacomelli R, Cipriani P. Linking myofibroblast generation and microvascular alteration: the role of CD248 from pathogenesis to therapeutic target (Review) Mol Med Rep. 2019;20(2):1488–1498. doi: 10.3892/mmr.2019.10429. [DOI] [PubMed] [Google Scholar]
- 24.Prat J. FIGO staging for uterine sarcomas. Int J Gynaecol Obstet. 2009;104(3):177–178. doi: 10.1016/j.ijgo.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Jin Y, Liang ZY, Zhou WX, Zhou L. High MMP14 expression is predictive of poor prognosis in resectable hepatocellular carcinoma. Pathology. 2020;52(3):359–365. doi: 10.1016/j.pathol.2020.01.436. [DOI] [PubMed] [Google Scholar]
- 26.Fielder T, Butler J, Tierney G, Holmes M, Lam KY, Satgunaseelan L, et al. ROS1 rearrangements in lung adenocarcinomas are defined by diffuse strong immunohistochemical expression of ROS1. Pathology. 2021 doi: 10.1016/j.pathol.2020.01.190. [DOI] [PubMed] [Google Scholar]
- 27.De Michele S, Remotti HE, Del Portillo A, Lagana SM, Szabolcs M, Saqi A. SATB2 in neoplasms of lung, pancreatobiliary, and gastrointestinal origins. Am J Clin Pathol. 2021;155(1):124–132. doi: 10.1093/ajcp/aqaa118. [DOI] [PubMed] [Google Scholar]
- 28.Cheung CC, Smith AC, Albadine R, Bigras G, Bojarski A, Couture C, et al. Canadian ROS proto-oncogene 1 study (CROS) for multi-institutional implementation of ROS1 testing in non-small cell lung cancer. Lung Cancer. 2021;160:127–135. doi: 10.1016/j.lungcan.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Davidson B, Reich R, Berner A, Givant-Horwitz V, Goldberg I, Risberg B, et al. Ovarian carcinoma cells in serous effusions show altered MMP-2 and TIMP-2 mRNA levels. Eur J Cancer. 2001;37(16):2040–2049. doi: 10.1016/S0959-8049(01)00235-0. [DOI] [PubMed] [Google Scholar]
- 30.Artym VV, Yamada KM, Mueller SC. ECM degradation assays for analyzing local cell invasion. Methods Mol Biol. 2009;522:211–219. doi: 10.1007/978-1-59745-413-1_15. [DOI] [PubMed] [Google Scholar]
- 31.Rouleau C, Smale R, Fu YS, Hui G, Wang F, Hutto E, et al. Endosialin is expressed in high grade and advanced sarcomas: evidence from clinical specimens and preclinical modeling. Int J Oncol. 2011;39(1):73–89. doi: 10.3892/ijo.2011.1020. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, Lin JZ, Wang AQ, Xu WY, Long JY, Luo YF, et al. Threonine and tyrosine kinase may serve as a prognostic biomarker for gallbladder cancer. World J Gastroenterol. 2017;23(31):5787–5797. doi: 10.3748/wjg.v23.i31.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson B. Ovarian carcinoma and serous effusions Changing views regarding tumor progression and review of current literature. Anal Cell Pathol. 2001;23(3–4):107–28. doi: 10.1155/2001/418547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y, Chen KW, Zhang Y, Guo S, Wang JJ, Chu L, et al. TEM1 knock-down inhibits the proliferation and metastasis of MES-SA uterine sarcoma cells. Int J Clin Exp Med. 2017;10(9):13133–13142. [Google Scholar]
- 35.Humphries MJ. Cell adhesion assays. Methods Mol Biol. 2009;522:203–210. doi: 10.1007/978-1-59745-413-1_14. [DOI] [PubMed] [Google Scholar]
- 36.Aktary Z, Alaee M, Pasdar M. Beyond cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis and metastasis. Oncotarget. 2017;8(19):32270–32291. doi: 10.18632/oncotarget.15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18(9):533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontana R, Yang J. Matrix degradation assay to measure the ability of tumor cells to degrade extracellular matrix. Methods Mol Biol. 2021;2294:151–163. doi: 10.1007/978-1-0716-1350-4_11. [DOI] [PubMed] [Google Scholar]
- 39.Machado RAC, Stojevski D, De Landtsheer S, Lucarelli P, Baron A, Sauter T, et al. L-plastin Ser5 phosphorylation is modulated by the PI3K/SGK pathway and promotes breast cancer cell invasiveness. Cell Commun Signal. 2021;19(1):22. doi: 10.1186/s12964-021-00710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrechtsen R, Kveiborg M, Stautz D, Vikeså J, Noer JB, Kotzsh A, et al. ADAM12 redistributes and activates MMP-14, resulting in gelatin degradation, reduced apoptosis and increased tumor growth. J Cell Sci. 2013;126(Pt 20):4707–4720. doi: 10.1242/jcs.129510. [DOI] [PubMed] [Google Scholar]
- 41.Mazurkiewicz E, Mrówczyńska E, Simiczyjew A, Nowak D, Mazur AJ. A fluorescent gelatin degradation assay to study melanoma breakdown of extracellular matrix. Methods Mol Biol. 2021;2265:47–63. doi: 10.1007/978-1-0716-1205-7_3. [DOI] [PubMed] [Google Scholar]
- 42.Desai B, Ma T, Chellaiah MA. Invadopodia and matrix degradation, a new property of prostate cancer cells during migration and invasion. J Biol Chem. 2008;283(20):13856–13866. doi: 10.1074/jbc.M709401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacheco-Fernandez N, Pakdel M, Blank B, Sanchez-Gonzalez I, Weber K, Tran ML, et al. Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J Cell Biol. 2020 doi: 10.1083/jcb.201907058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouleau C, Smale R, Sancho J, Fu YS, Kurtzberg L, Weber W, et al. Endosialin: a novel malignant cell therapeutic target for neuroblastoma. Int J Oncol. 2011;39(4):841–851. doi: 10.3892/ijo.2011.1091. [DOI] [PubMed] [Google Scholar]
- 45.Capone E, Piccolo E, Fichera I, Ciufici P, Barcaroli D, Sala A, et al. Generation of a novel antibody-drug conjugate targeting endosialin: potent and durable antitumor response in sarcoma. Oncotarget. 2017;8(36):60368–60377. doi: 10.18632/oncotarget.19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo Y, Honoki K, Kishi S, Mori S, Fujiwara-Tani R, Tsukamoto S, et al. Endosialin/CD248 may be a potential therapeutic target to prevent the invasion and metastasis in osteosarcoma. Oncol Lett. 2022;23(2):42. doi: 10.3892/ol.2021.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang HK, Jeong KC, Kim YK, Jung ST. Role of matrix metalloproteinase (MMP) 2 and MMP-9 in soft tissue sarcoma. Clin Orthop Surg. 2014;6(4):443–454. doi: 10.4055/cios.2014.6.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Z, Wang J, Su Q, Luan M, Chen X, Xu X. The role of MMP-2 and MMP-9 in the metastasis and development of hypopharyngeal carcinoma. Braz J Otorhinolaryngol. 2021;87(5):521–528. doi: 10.1016/j.bjorl.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alrehaili AA, Gharib AF, Karam RA, Alhakami RA, El Sawy WH, Abd Elrahman TM. Clinical significance of plasma MMP-2 and MMP-9 levels as biomarkers for tumor expression in breast cancer patients in Egypt. Mol Biol Rep. 2020;47(2):1153–1160. doi: 10.1007/s11033-019-05216-5. [DOI] [PubMed] [Google Scholar]
- 50.Dofara SG, Chang SL, Diorio C. Gene polymorphisms and circulating levels of MMP-2 and MMP-9: a review of their role in breast cancer risk. Anticancer Res. 2020;40(7):3619–3631. doi: 10.21873/anticanres.14351. [DOI] [PubMed] [Google Scholar]
- 51.Middleton JD, Sivakumar S, Hai T. Chemotherapy-induced changes in the lung microenvironment the role of MMP-2 in facilitating intravascular arrest of breast cancer cells. Int J Mol Sci. 2021 doi: 10.3390/ijms221910280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roomi MW, Kalinovsky T, Roomi NW, Rath M, Niedzwiecki A. Anticancer effects of a specific mixture of nutrients in the multidrug-resistant human uterine sarcoma MES-SA/Dx5 and the drug-sensitive MES-SA cell lines. Oncol Rep. 2012;27(1):17–27. doi: 10.3892/or.2011.1471. [DOI] [PubMed] [Google Scholar]
- 53.Yu M, Sato H, Seiki M, Thompson EW. Complex regulation of membrane-type matrix metalloproteinase expression and matrix metalloproteinase-2 activation by concanavalin A in MDA-MB-231 human breast cancer cells. Can Res. 1995;55(15):3272–3277. [PubMed] [Google Scholar]
- 54.Sato H, Takino T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010;101(4):843–847. doi: 10.1111/j.1349-7006.2010.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Figueira RC, Gomes LR, Neto JS, Silva FC, Silva ID, Sogayar MC. Correlation between MMPs and their inhibitors in breast cancer tumor tissue specimens and in cell lines with different metastatic potential. BMC Cancer. 2009;9:20. doi: 10.1186/1471-2407-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koo BH, Han JH, Yeom YI, Kim DS. Thrombin-dependent MMP-2 activity is regulated by heparan sulfate. J Biol Chem. 2010;285(53):41270–41279. doi: 10.1074/jbc.M110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roomi MW, Monterrey JC, Kalinovsky T, Niedzwiecki A, Rath M. Modulation of MMP-2 and MMP-9 by cytokines, mitogens and inhibitors in lung cancer and malignant mesothelioma cell lines. Oncol Rep. 2009;22(6):1283–1291. doi: 10.3892/or_00000566. [DOI] [PubMed] [Google Scholar]
- 58.Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol Rep. 2010;23(3):605–614. doi: 10.3892/or_00000675. [DOI] [PubMed] [Google Scholar]
- 59.Nilsson UW, Garvin S, Dabrosin C. MMP-2 and MMP-9 activity is regulated by estradiol and tamoxifen in cultured human breast cancer cells. Breast Cancer Res Treat. 2007;102(3):253–261. doi: 10.1007/s10549-006-9335-4. [DOI] [PubMed] [Google Scholar]
- 60.Kenny HA, Lengyel E. MMP-2 functions as an early response protein in ovarian cancer metastasis. Cell Cycle. 2009;8(5):683–688. doi: 10.4161/cc.8.5.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui J, Dean D, Hornicek FJ, Chen Z, Duan Z. The role of extracelluar matrix in osteosarcoma progression and metastasis. J Exp Clin Cancer Res. 2020;39(1):178. doi: 10.1186/s13046-020-01685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vishnubhotla R, Bharadwaj S, Sun S, Metlushko V, Glover SC. Treatment with Y-27632, a ROCK inhibitor, increases the proinvasive nature of SW620 cells on 3D collagen type 1 matrix. Int J Cell Biol. 2012;2012:259142. doi: 10.1155/2012/259142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Revach OY, Grosheva I, Geiger B. Biomechanical regulation of focal adhesion and invadopodia formation. J Cell Sci. 2020 doi: 10.1242/jcs.244848. [DOI] [PubMed] [Google Scholar]
- 64.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Can Res. 2006;66(6):3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 65.Vishnubhotla R, Sun S, Huq J, Bulic M, Ramesh A, Guzman G, et al. ROCK-II mediates colon cancer invasion via regulation of MMP-2 and MMP-13 at the site of invadopodia as revealed by multiphoton imaging. Lab Invest. 2007;87(11):1149–58. doi: 10.1038/labinvest.3700674. [DOI] [PubMed] [Google Scholar]
- 66.Manuelli V, Cahill F, Wylie H, Gillett C, Correa I, Heck S, et al. Invadopodia play a role in prostate cancer progression. BMC Cancer. 2022;22(1):386. doi: 10.1186/s12885-022-09424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burger KL, Learman BS, Boucherle AK, Sirintrapun SJ, Isom S, Diaz B, et al. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate. 2014;74(2):134–148. doi: 10.1002/pros.22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karamanou K, Franchi M, Vynios D, Brezillon S. Epithelial-to-mesenchymal transition and invadopodia markers in breast cancer: Lumican a key regulator. Semin Cancer Biol. 2020;62:125–133. doi: 10.1016/j.semcancer.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Choi JA, Lim IK. TIS21/BTG2 inhibits invadopodia formation by downregulating reactive oxygen species level in MDA-MB-231 cells. J Cancer Res Clin Oncol. 2013;139(10):1657–1665. doi: 10.1007/s00432-013-1484-3. [DOI] [PubMed] [Google Scholar]
- 70.Huang SS, Liao WY, Hsu CC, Chan TS, Liao TY, Yang PM, et al. A novel invadopodia-specific marker for invasive and pro-metastatic cancer stem cells. Front Oncol. 2021;11:638311. doi: 10.3389/fonc.2021.638311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan X, Cao N, Chen Y, Lan HY, Cha JH, Yang WH, et al. MT4-MMP promotes invadopodia formation and cell motility in FaDu head and neck cancer cells. Biochem Biophys Res Commun. 2020;522(4):1009–1014. doi: 10.1016/j.bbrc.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 72.Benzing C, Lam H, Tsang CM, Rimmer A, Arroyo-Berdugo Y, Calle Y, et al. TIMP-2 secreted by monocyte-like cells is a potent suppressor of invadopodia formation in pancreatic cancer cells. BMC Cancer. 2019;19(1):1214. doi: 10.1186/s12885-019-6429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J, Liu X, Wu J, Lou C, Zhang Q, Chen H, et al. CXCL12 derived from CD248-expressing cancer-associated fibroblasts mediates M2-polarized macrophages to promote nonsmall cell lung cancer progression. Biochim Biophys Acta Mol Basis Dis. 2022;1868(11):166521. doi: 10.1016/j.bbadis.2022.166521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary file. Further inquiries can be directed to the corresponding author.