Abstract
The alphaviruses are a widely distributed group of positive-sense, single stranded, RNA viruses. These viruses are largely arthropod-borne and can be found on all populated continents. These viruses cause significant human disease, and recently have begun to spread into new populations, such as the expansion of Chikungunya virus into southern Europe and the Caribbean, where it has established itself as endemic. The study of alphaviruses is an active and expanding field, due to their impacts on human health, their effects on agriculture, and the threat that some pose as potential agents of biological warfare and terrorism. In this systematic review we will summarize both historic knowledge in the field as well as recently published data that has potential to shift current theories in how alphaviruses are able to function. This review is comprehensive, covering all parts of the alphaviral life cycle as well as a brief overview of their pathology and the current state of research in regards to vaccines and therapeutics for alphaviral disease.
Keywords: alphaviruses, viral replication, positive-sense RNA viruses, viral life cycle
1. Introduction
Alphaviruses are positive sense, single stranded, RNA viruses in the family Togaviridae, which are classified as members of the domain Riboviria1. The alphaviruses currently encompass more than thirty members that infect a wide range of host and vector species, both terrestrial and aquatic. These viruses are widely dispersed geographically as well, with at least one alphavirus being present on every populated continent2–6. Alphaviruses are continually emerging into naïve populations, and there are currently no licensed treatments or vaccines for alphaviral disease, making these viruses important research targets.
The alphaviruses have historically been divided into two clades based upon the location of their isolation. The Old-World viruses, which were first isolated in the eastern hemisphere, and the New-World viruses which were first discovered in the Americas. The Old-World viruses generally cause arthralgia and fever, with some also causing a rash7. There is some recent evidence that Old-World members may be able to cause encephalitis as well, with the adaptation of neurologically invasive Sindbis virus (SINV) for use in mice, as well as its association with rare cases of viral encephalitis in Europe8–10. The New-World viruses are known to cause encephalitis; the three most prominent members of this clade, Eastern (EEEV), Western (WEEV), and Venezuelan Equine Encephalitis viruses (VEEV) demonstrate high levels of neurological pathogenicity7. Recently it has been proposed that certain South American clades of EEEV be split into the new species Madariaga virus, as they are genetically distinct and are less pathogenic11
Continuing efforts of alphavirus discovery, characterization, and sequencing have now indicated that the division between the Old and New-World viruses may be losing usefulness, as there are now several known New-World alphaviruses that don’t cause any disease12, as well as the recent discovery of alphaviruses that are native to the New-World but have disease phenotypes that are similar to the Old-World clade, such as Mayaro virus2,13.
2. Alphaviral Disease
The Old-World virus of most concern is Chikungunya virus (CHIKV), which has recently expanded into naive populations across Asia, southern Europe, and most dramatically, in the Caribbean3–6. This has resulted in CHIKV becoming endemic in several of these regions. The primary risk of CHIKV is a sustained arthralgia that can last for months, with one study in Mexico indicating that over a third of confirmed CHIKV cases have arthralgia twelve months after acute disease14. Similar pathologies have also been reported after infection with Ross River virus15.
The New-World alphaviruses generally cause more severe disease than the Old-World viruses; however, the three most common (EEEV, WEEV, VEEV) are noted for a high rate of asymptomatic infection12. This asymptomatic infection rate varies between the three viruses and in the two primary populations of interest, equids and humans, with equids having significantly higher rates of symptomatic disease12. Disease is also generally more severe in equid hosts than in humans, with most equid cases being lethal7. EEEV has the highest reported rate of neurological involvement and lethality, VEEV has the least, and WEEV falls between the two12. However, VEEV is the New-World virus of most concern as it has historically caused the largest and most frequent outbreaks affecting both human and equid populations, and resulting in many thousands of human cases and equid deaths12,16. EEEV remains rare in the human population, but there was a significant increase in the number of cases reported in 2019. This increase retreated in 2020, the most recent year with data available from the CDC, with that year having an average number of reports17. WEEV has virtually disappeared from the human population, and also become much rarer in its enzootic hosts in North America18,19
VEEV is also a high risk for accidental release and exposure due to it propensity to infect via aerosols20. This led to VEEV being developed as a bioweapon by both the former USSR and the USA, making VEEV a select agent, a classification it shares with EEEV21. Select agents are those toxins and organisms that “could threaten public health and safety”22 and have additional security regulations in addition to those controls indicated by biosafety. VEEV generally causes a mild febrile illness that occasionally results in encephalitic infection, resulting in death in approximately 10% of cases when encephalitis is present7, however the total case fatality rate is only around 1%23. Those patients that do survive neurological symptoms are likely to suffer from long term sequelae24.
3. Natural Transmission of the Alphaviruses
Alphaviruses are vector-borne viruses that generally require the use of an intermediate species to transmit to a naïve vertebrate host7 (Figure 1). Due to this cycle, the viruses must efficiently infect and replicate in multiple species. Alphaviruses infect a wide variety of both vector and host species. Single species of alphavirus can often infect multiple different species of vector, and different vector species are typically responsible for endemic maintenance and epidemic/epizootic outbreaks of disease23.
Figure 1.
The transmission cycle of encephalitic alphaviruses. A) Alphaviruses are maintained in nature by cycling between a host species, typically a bird or small mammal, and a mosquito vector species. B) Spillover events often occur into livestock, which reach high viral titers and readily transmit the virus to additional vectors. In the case of the New-World viruses this infection almost always leads to death27. C) Typically, after infection of livestock, humans that work in close association with these animals can also be infected by vector species. Humans are regarded as dead-end hosts for most alphaviruses. In humans these infections may lead to disease, and, in severe cases, death.
The virus first enters the mosquito or other vector through a blood meal that is taken from an infected host. The virus then encounters the cells of the mosquito midgut, before passing into the haemocoel, the circulatory system of the mosquito. Eventually virus arrives in the salivary glands where it replicates to high levels and is transmitted to the next vertebrate host during a blood meal29,30. Not only does vector transmission complicate control of these viruses, but infection of the mosquito is an important selection process, and different strains of these viruses can behave differently in the vector. In particular, epidemic VEEV strains behave very differently in the mosquito than those are isolated from enzootic infection29–31.
Upon blood meal from an infected mosquito, the alphavirus is injected into the skin of a naïve host. As these viruses have various cellular tropisms that will result in differing pathologies; we will here outline the general series of events that occur during infection of a susceptible and permissive host cell.
4. Alphaviral replication
The steps outlined here are common to all alphaviruses unless otherwise indicated. After inoculation into the vertebrate host alphaviruses enter permissive and susceptible host cells to manufacture new virions. The alphaviruses are noted to have highly efficient infection. While in infection there is an excess of genomes produced compared to plaque forming units, as time goes on these numbers reach near parity32. The alphaviral replication process is here described in detail, and a summary can be found in Figure 2.
Figure 2.
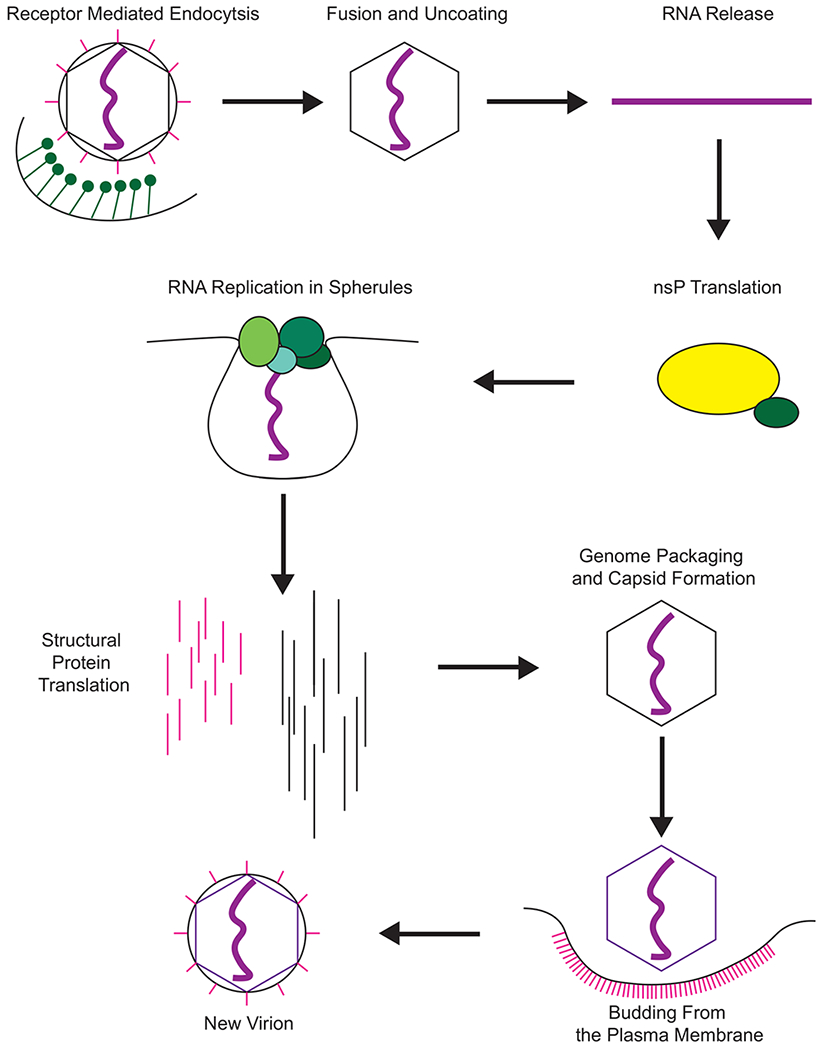
The replication cycle of alphaviruses. The virion enters a susceptible cell via receptor mediated endocytosis, primarily mediated by clathrin (green) and due to pH changes of the endosome releases its RNA (purple) into the cytoplasm of the host cell. The positive sense genomic RNA is first used by ribosomes to translate the viral nsPs as a polyprotein (yellow = nsP123, green = nsP4). The polyprotein will undergo cleavage events that control the synthesis of the viral RNA species (individual nsPs represented as single green circles). This RNA synthesis occurs in membrane invaginations that are termed spherules. These spherules protect the viral RNA and nsPs from detection by the host cell. Late in infection the structural genes are synthesized (pink = the E proteins, black = capsid). The capsid will form into nucleocapsid cores as it packages the viral RNA, and the glycoproteins are transported to the cell membrane. The nucleocapsid cores translocate to the cellular membrane where they bud off, collecting their envelope and glycoproteins and forming new infectious virions.
4.1. Receptor-mediated endocytosis
The primary mechanism by which alphaviruses enter naïve host cells is via receptor mediated endocytosis, and the viruses are highly promiscuous33. Of particular interest are DC-SIGN and L-SIGN, which may influence myeloid cell infection34. Additional receptors continue to be discovered35,36. Heparan sulfate is an important binding partner in cell culture, and this appears to be a specific adaptation that occurs in response to passaging virus repeatedly in cells33,37–39. The exception to this is EEEV, which has been found to have affinity for heparan sulfate in naturally circulating strains40
After receptor binding, the alphaviruses are then transported into the cell via clathrin mediated endocytosis41,42, resulting in a virus-containing endosome passing through the stages of acidification and maturation. The New-World viruses remain in the vacuoles until they reach the endosome stage, whereas the Old-World viruses escape from the early endosomal compartment43.
There is also now evidence that some alphaviruses may be able to utilize alternative entry strategies, such as direct entry at the host cell plasma membrane44,45. This has been well characterized in CHIKV, with research indicating that even though an acidification step is required, it can occur in a manner that is independent of the activity of clathrin46. However, the importance of this entry method remains unclear, but it shows one way these viruses may be able to display such wide cell tropisms. This does indicate that these viruses may have the potential to develop resistance to entry inhibitors rapidly.
4.2. Viral entry as a target for antiviral drug development.
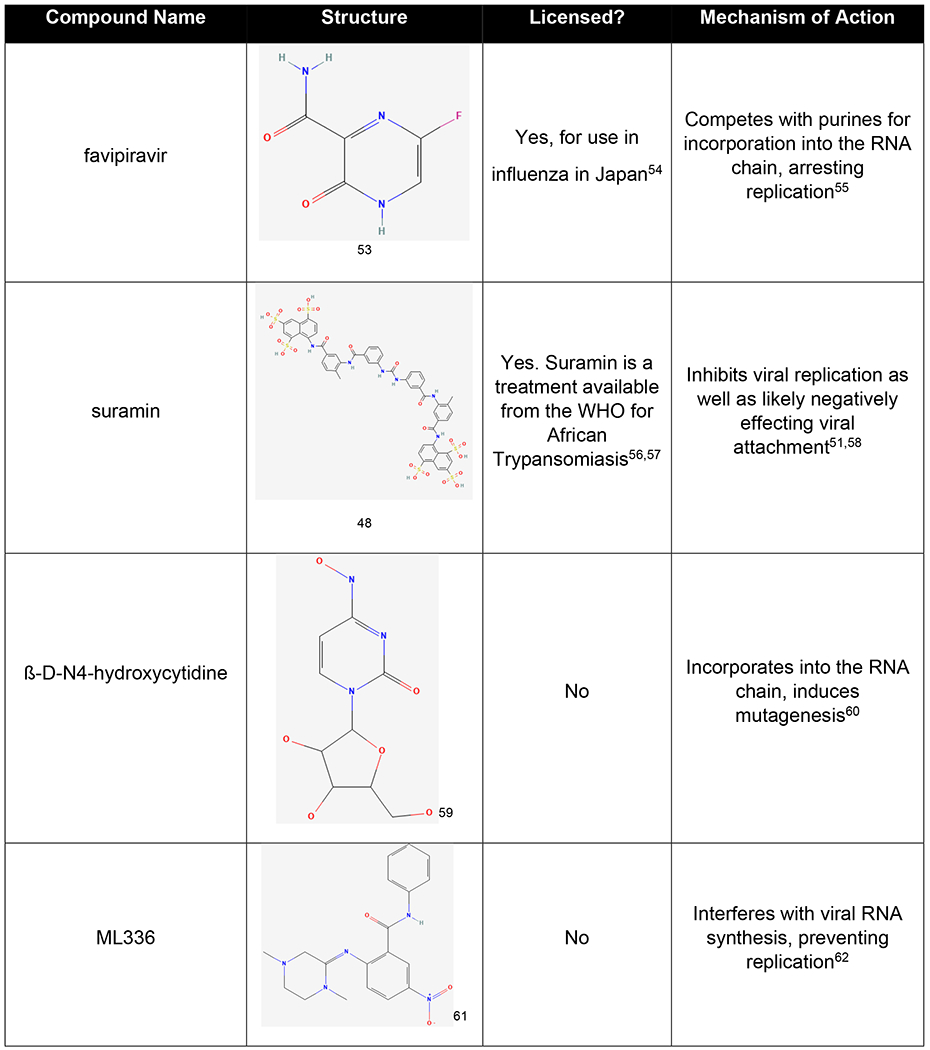
Receptor attachment and entry are common drug targets, and an example of a drug that targets this activity in the alphaviruses is suramin. Suramin was initially discovered as a treatment for trypanosomal diseases in Africa, and it is also known by the names naganol, suramine, forneau, and germanin47,48. Suramin has a long history of being tested for potential therapeutic effects in many different diseases.
Before it was tested in the treatment of alphaviral disease, suramin was already known to function to inhibit trypanosomal disease, to potentially act as an inhibitor of the HIV reverse transcriptase, and to have anticancer effects47,49,50. Suramin has recently been studied in the treatment of CHIKV infection and was found to inhibit multiple stages of the replication cycle, including viral entry. Suramin was also mildly efficacious in the treatment of SINV and SFV. Treatment with suramin was also found to reduce viral load in a mouse model of infection, and reduced infection relation swelling in the foot51,52. As it has a known safety profile, suramin is a promising lead compound for further refinement via modeling and medicinal chemistry. The structure of suramin can be found in table 2 below.
Table 2:
A summary of selected anti-alphaviral compounds and their mechanisms of action. All structures are published in PubChem.
|
4.3. Fusion/Uncoating and RNA release
Fusion of the viral and host cell membranes is achieved by the activity of the E1 protein, and expression of E1 without the other glycoproteins is enough to mediate viral membrane fusion63,64. This fusogenic activity is initially prevented by the interaction of E1 with E2, but this interaction is disrupted at low pH42,65.
After escape from the endosome, the nucleocapsid interacts with ribosomes, which disassemble the capsid in a non-catalytic manner, which is dependent on conserved capsid sequences66,67. The disassembly of the nucleocapsid is enhanced by low pH, and the pore forming activities of the E proteins are implicated to induce these pH changes68–70.
4.4. Translation and processing of the nsPs
Upon release the alphavirus genome is available as an mRNA for cellular ribosomes and recruits all the factors required for protein synthesis in a similar manner as a cellular RNA7. First to be translated is the nonstructural polyprotein, which contains the proteins that are responsible for the replication of the viral RNA. The viral nsPs are numbered in the order that they occur in the genome from 5’ to 3’, 1-4. The genomic organization of alphaviruses can be found in Figure 3 A. The initial polyprotein is translated as either nsP123 or nsP1234, depending on read through of a stop codon that may or may not be present in the genome depending on the alphavirus in question71–73,.
Figure 3.
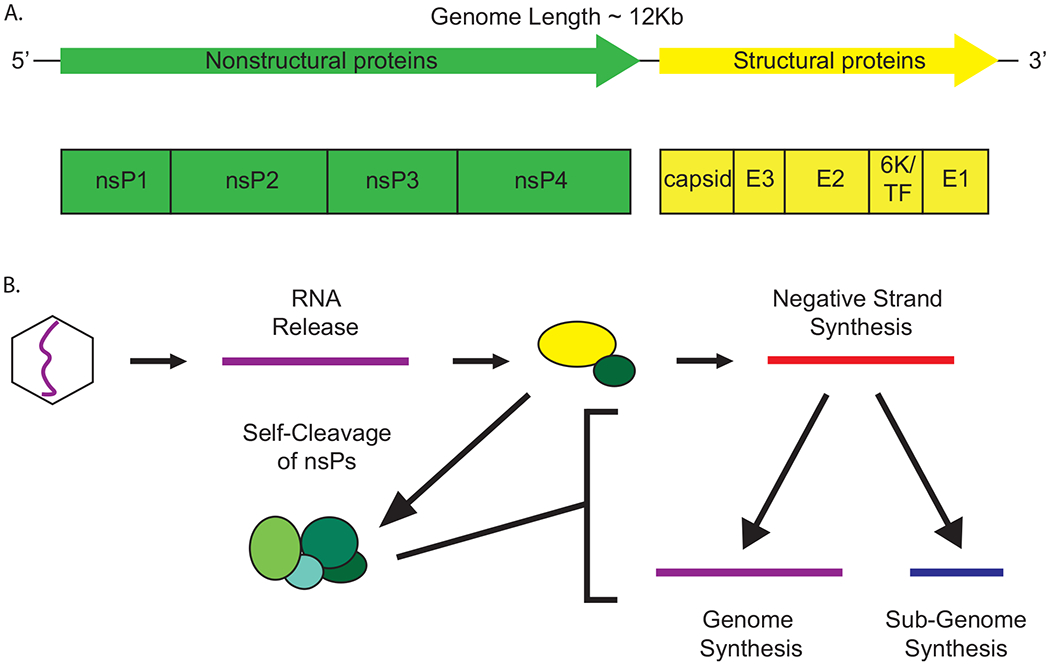
A) The genetic structure of the alphaviruses. The alphaviruses have an approximately 12kb, linear, positive-sense genome. The genome has two open reading frames, the nonstructural and the structural. The nonstructural open reading frame is here displayed in green, and encodes the four nonstructural proteins, which are responsible for replication of the viral RNA. The structural open reading frame is displayed here in yellow and encodes for the E proteins and capsid as well as the 6K and TF proteins. The capsid and E proteins form the structure of the viral particle. B) RNA synthesis of alphaviruses. This RNA synthesis activity is carried out in spherules on the membranes of cellular organelles. After release into the cytoplasm the genomic RNA (purple) is used to synthesize the initial nonstructural polyprotein. nsP2 initially cleaves between nsP3 and 4 leading to nsP123/4 (nsP123 = yellow, nsP4 = green), which synthesizes primarily negative-sense template RNA (red). The protein undergoes rapid cleavage through intermediate states to reach the final replicase complex nsP1/2/3/4 (represented as individual green circles). This complex synthesizes new positive-sense genomic (purple) and subgenomic (blue) RNA and can no longer synthesize negative-sense RNA. The genomic RNA is used to synthesize additional genomes and is packaged into progeny virions. The subgenomic RNA is used to synthesize the structural genes that form the new virions.
After the initial translation of these proteins, they undergo tightly controlled autologous cleavage events which are independent from cellular processes and result in the formation of multiple intermediates as well as the final mature replicase complex nsP1/2/3/474,75. This fully cleaved, mature complex is highly stable. Control of this cleavage process is important as it controls the levels of viral RNA species that are present at different times during infection76,77; this regulation is discussed in more detail in the following section. This cleavage process appears to have unique regulatory features such as having morphological cleavage recognition instead of sequence specificity78. This regulatory process is highly important to viral biology as altering it leads to attenuation79. Proper cleavage is also important to immune evasion, as viruses with incomplete cleavage result in alterations of the viral RNA species prevalence, increasing type I interferon induction as well as the sensitivity of the viruses to interferon80.
4.5. Viral RNA Replication
The process of viral RNA synthesis is outlined in Figure 3 B. To make additional molecules of RNA genome, the virus is required to first transcribe the positive-sense genome into negative-sense template strand. This activity is performed by the partially cleaved polyprotein nsp123/477. However, the protein cleavage activity of nsP2 rapidly degrades the polyprotein into its constitutive parts. This initially produces intermediate forms of the replicase complex that are short lived and produce both negative and positive-sense RNA76,77. The final cleavage between nsP2 and nsP3 leads to the formation of the mature replicase complex nsP1/2/3/4 which produces only positive-sense RNA74,75,81. This self-proteolytic behavior creates a distinct expression profile of the viral RNA. Initially the immature forms of the complex produce higher levels of negative-sense RNA. As the complex is processed the synthesis of negative-sense RNA is reduced and eventually eliminated. This causes most negative-sense RNA to be produced early in infection, as well as less negative-sense RNA being produced overall62. Following cleavage and assembly of the mature replicase complex, RNA synthesis converts to the synthesis of positive-sense genomic and subgenomic RNA82.
The positive-sense genomic RNA functions primarily as the genetic material of the next generation of virus, as well as being translationally active in the cell to produce additional nonstructural proteins. However, recent work has indicated that the genomic RNA may have biological functions that are not dependent on its function as a viral mRNA, as increasing the amount of capped RNA decreased viral fitness, indicating that there is some important role played by the noncapped RNA which isn’t replicatively competent83.
Late in infection an additional positive sense RNA is produced, the small subgenomic RNA, which is used to produce the structural proteins71. Additionally, when viral RNA is examined on agarose gels, there is a third RNA species that appears, the so called replication intermediate RNA 62,84. This an RNA species that runs at very large size, however its significance is unknown, and there is no research indicating its role in the viral life cycle.
4.6. Viral RNA replication as a target of antiviral drug development.
One of the most common targets of antiviral drug development has been reducing replication of the viral genetic material, as this limits viral spread in the host and thus disease. A common drug class that has been found to have these effects is the nucleoside analogs. This class includes the drug ribavirin, used in the treatment of many viruses, but which is largely ineffective as an anti-alphaviral therapy85. Multiple nucleoside analogues have been tested as treatments for the alphaviruses and two will be described here, favipiravir, and β-D-N4-hydroxycytidine.
Favipiravir is a nucleoside analogue that is approved for use by the Pharmaceuticals and Medical Devices Agency, the Japanese drug regulatory body, for the treatment of influenza under the tradename Avigan54. Favipiravir has also been tested against a wide variety of other viruses, including the alphaviruses, both in vitro and in vivo. Favipiravir inhibits the activity of viral polymerases by competing with purines for incorporation in to the viral RNA. This activity locks the strand and prevents its use in further viral replication55,86.
When tested in models of alphaviral infection, favipiravir has been found to be mildly efficacious against WEEV, and treatment with this drug resulted in the clearance of CHIKV from infected mice when given in the acute phase of infection, but had no effect in the chronic phase86–88. However, favipiravir treatment for alphaviruses has also only been tested via intraperitoneal injection, which is not a preferred method of delivery in humans87,89. There have been no tests with the oral formulation currently approved for use in humans for influenza. Sensitivity to favipiravir in the alphaviruses also appears to be strain dependent. With the strains of CHIKV that are of most concern being less sensitive to treatment than strains that are less involved in human outbreaks90. While favipiravir is currently not clinically approved for use in the alphaviruses, the knowledge about its efficacy could lead to future compounds with enhanced anti-alphaviral properties.
Not all nucleoside analogues function by the same mechanism to inhibit viral replication. Another nucleoside analogue that has been examined in alphaviral infection is β-D-N4-hydroxycytidine or NHC60. NHC has been tested for therapeutic efficacy against many viruses including influenza and respiratory syncytial virus, and is structurally highly similar to molnupiravir, one of the currently approved antiviral treatments for SARS-CoV-291,92. NHC functions by multiple mechanisms, one of which is the inducement of hypermutation during replication of the viral genome60. This mutagenesis as well as the secondary effect of reduced viral infectivity results in inducing minimal resistance to treatment in the viral population60 which indicates that NHC and its derivatives/related compounds show promise as potential treatments of the alphaviruses. The structures of both favipiravir and NHC can be found above in table 2.
4.7. Importance of the untranslated regions of alphaviruses
The genomes of alphaviruses have large 5’and 3’ untranslated regions (UTRs) which are biologically active. The 3’ UTR is important in avoiding the immune bottlenecks that exist in the arthropod phase of the life cycle93, and the sequence variability that occurs in this region can have significant effects on transmission in mosquito vectors and on vector specificity93. The main variation in this region is due to size, which directly relates to the number of repeated regions that occur in the sequence94,95. This variation in the 3’ UTR does not only occur between different viruses but can also vary significantly within viral species and has been well documented to differ in the differently pathogenic strains of CHIKV96. The 3’ UTR is also involved in pathogenicity of alphaviruses, such as in EEEV where it binds to micro RNA and promotes neurological disease97, and is involved in the synthesis of the negative sense RNA98.
The 5’ UTR functions in many ways to promote translation of the viral RNA both via its structure and sequence99,100. These structures can vary significantly between species and aren’t necessarily interchangeable101. The structure of the 5’ UTR is also important to evasion of the interferon response in alphaviruses, with the stability of the structure playing an important role in preventing recognition of the cap structure of the viral RNA100,102. The importance of this region to virulence is seen in the attenuation of VEEV strain TC-83, a mutation in this region alters the ratios of viral RNA types and results in a significant increase in sensitivity to interferon compared to wild-type viruses103,104
4.8. Localization of genome replication
Alphaviruses demonstrate a sequestration of their replication to intracellular membranes, which is similar to other RNA viruses which also largely replicate in and on membranous structures105–107. The alphaviruses utilize microinvaginations called spherules105. These are sites where the viral RNA has been found to localize in infected cells108,109. It has been confirmed in vitro that these structures contain viral RNA synthetic activity through the use of purified spherules to produce viral RNA110. It has been recently determined that the initial formation of the spherules is dependent solely on the activity of the nsPs with no requirement for viral RNA being present111. However, the size of the individual spherules is dependent on the length of the RNA that is transcribed within, which appears to be a feature unique to alphaviruses112. These spherules have been suggested to play a role in viral immune evasion by isolating viral double-stranded RNA away from cytoplasmic pattern recognition receptors113,114. There is also evidence from the flaviviruses that in general, membranous association of viral RNA replication can protect RNA from enzymatic digestion115. Many of these phenotypes have also now been confirmed in CHIKV using cutting edge microscopy and structural biology methods116
Spherules were initially identified on large, endosomal-like compartments in infected cells. In several of the alphaviruses these spherules form at the plasma membrane and later traffic to intracellular compartments117. In vertebrate cells, recent work has indicated the movement of the spherules away from the plasma membrane is dependent on the activation of PI3K-Akt-mTOR, and reduction of this activation is associated with an increased proportion of the spherules remaining at the cellular membrane118. However, inhibition of this activity has no effect on viral titer, indicating that localization of replication may not be important to replicative success119
4.9. Translation of the structural genes
The structural genes of the alphaviruses are produced via translation of the subgenomic RNA. The synthesis of this RNA initiates independently from the full-length genomic RNA, but how this secondary initiation happens remains unknown. The initial gene product is a polyprotein that contains the capsid, E proteins, 6K, and TF proteins7,71. This translation is carried out in the cytosol and the structural proteins are synthesized at very high levels and are found throughout the cell120,121. Production of the structural genes occurs primarily later in infection due to the increased expression of the subgenomic RNA7. The capsid protein contains a serine protease domain and uses this to rapidly cleave itself from the other structural genes after it is translated71.
After cleavage of the capsid protein, the glycoproteins pass into the endoplasmic reticulum, and move through the Golgi apparatus before being embedded into the plasma membrane of the cell71. The glycoproteins are also highly post-translationally modified via glycosylation and palmitoyaltion71.
4.10. Packaging of the viral RNA and release of the virion
Unlike some other viruses, the alphaviruses don’t readily produce empty particles122. However, they are known to produce defective interfering (DI) particles when passaged at high concentrations repeatedly, although this occurs less frequently than in other viruses such as vesicular stomatitis virus and influenza123. These particles have a range of sizes and are often smaller than normal virions, and they usually incorporate deletions in the viral RNA. The deletions in these DI particles get larger with increased passages123. The alphaviruses are noted for generally producing highly uniform, icosahedral particles124 though this can be altered by mutating the structural proteins125. Current data indicate that while multicore, and other irregular virion shapes do occur with these viruses, these are highly selected against, and particles largely only contain a single unit of genomic RNA7 and capsid cores select for carrying only single cargo units122.
After translation of the structural proteins, the viral RNA and capsid undergo interactions based on molecule size and charge, resulting in nucleocapsid like structures occurring in the cytoplasm71,126,127. Alphaviruses bud directly from the plasma membrane of the infected cell7. However, it is unclear how this budding process is initiated127. It has been found that both the preformed nucleocapsid like structures and the glycoproteins are able to drive budding127,128. However, when either of these functions occurs independently of the other, there is a marked reduction in efficiency, indicating that it is likely that these two mechanisms interact to allow for the maximal budding of virions128.
Transport of the structural proteins to the plasma membrane requires the host secretory system128. However the exact proteins that are used remain unknown128. Release of virions can also be inhibited by host proteins. In particular tetherin has been shown to prevent the release of virions from infected cells128. The general replication scheme of alphaviruses is outlined in Figure 2.
Reducing viral release and decreasing the infectivity of viral particles is an activity of some anti-alphaviral compounds, in particular ß-D-N4-hydroxycytidine, a compound discussed earlier in section 4.6.
5. Functions of the alphaviral nonstructural proteins
The alphaviruses make four nonstructural proteins. These proteins are responsible for viral RNA replication as well as many other enzymatic functions. The nonstructural proteins are also intimately involved in the pathogenesis of the alphaviruses. The functions of these proteins will now be described in greater detail. While the functions of the nsPs are highly conserved, differences between the Old and New-World viruses will be indicated when necessary.
5.1. Nonstructural protein 1
NsP1 is the capping enzyme for the viral genomic RNA, and this activity occurs independent of the activities of the other nsPs129. The activity of this protein has recently been examined in VEEV, having previously been studied only in Old-World viruses. This was also the first time that each individual step, including the final guanyl transfer, has been described130. The steps occur as follows. 1) The transfer of a methyl group from S-adenosylmethionine to position N7 of a molecule of GTP is catalyzed; 2) nsP1 receives the methyl-GTP becoming guanylated, releasing pyrophosphate in the process; 3) the 7 methyl-GMP is transferred to the 5’ end of the target RNA130,131. For this reaction to occur properly, the RNA being capped must have had its 5’ terminal phosphate removed by nsP2132.
NsP1 is also responsible for the anchoring of the viral replicase complex to cellular membranes which are the site of RNA replication, and this activity is required for capping to be carried out as well133–135. Very recently a cryo-em structure was published that showed how nsP1 influences the structure of the membrane spherules and potentially controls entry and exit of materials136. NsP1 was found to form a ring-like structure that appears to act as a gate and controls movement to and from the compartment136. This detailed structure has now also been used to examine the capping activity of CHIVK in depth as well, as can be seen in recent publication from the Law lab137
5.2. Nonstructural protein 2
NsP2 is a multifunctional protein with several distinct domains with discreet enzymatic activities. First, nsP2 is responsible for host cell transcriptional shutoff in the Old-World viruses, and loss of this phenotype reduces viral cytotoxicity138,139. In the New-World viruses this activity is instead carried out by the capsid protein, and nsP2 is responsible for shutoff of host cell protein synthesis and may have a role in packaging of viral RNA138,140–142. In VEEV this translational shutdown has been shown to mediate resistance to a pre-existing antiviral state141. Interestingly, a recent publication has indicated that both expression of a heterologous nsP2 as well overexpression of an nsP2 in the presence of infection is inhibitory for the alphaviruses in mosquito cells, and is likely one way that the infected cells resist superinfection143
There are three recognized domains in nsP2. The N-terminal region contains a helicase domain and NTPase activity that serves to provide energy for the helicase144,145. This same region also has RNA 5’-triphosphatase activity which prepares RNA for capping, allowing for translation and packaging in virions132. The previous 2 years have seen significant advances in the understanding of the structure of alphaviral nsP2 protein. First, the N terminal region of nsP2 from CHIKV, including the helicase domain, has recently been crystalized146, and this portion of the protein was used to characterize RNA binding activity147. Following this, there is now a full length crystallization of nsP2 available for CHIKV released by the same research group148. Here it was revealed that the N and C termini of this protein are connected by a flexible linker, and that this linker is highly important for normal protein function and viral pathogenicity148. Structures from this group have already proven useful for prediction of the structures of the nsP2 proteins of other alphaviruses62. These structures are also already being used to further research into potential therapeutics, in particular using computer modeling for drug selection and binding prediction149,150.
The most interesting feature of the nsP2 N-terminal crystal structure was the large number of accessory domains that were present, as these domains had not previously been predicted by structural modeling. Of particular interest is the so called stalk domain, which based upon recent research appears to have an important function in viral RNA synthesis as shown by the activity of the recently characterized antiviral compound ML33662. This compound is discussed later in section 5.5
Large portions of the N-terminal region of nsP2 remain poorly characterized. Studies have implicated that in VEEV this region may be important to packaging of the viral genome142. However, in SINV a transposon insertion approach using the sequence for GFP found that this region was involved in the cleavage between nsP2 and nsP3, controlling the ratio of genomic and subgenomic RNA, and regulation of RNA synthesis151. This range of phenotypes indicates that this region is highly important to these viruses, but further characterization and research are needed.
NsP2 also contains a cysteine protease domain that is responsible for the cleavage of the nsPs from the polyprotein into its constitutive members152–154. As described earlier, this cleavage is responsible for the transition from the synthesis of negative-sense viral RNA to positive-sense viral RNA77,155. The protease has also been shown to target cellular proteins, a common feature of viral proteases, and this is related to resistance to innate immune responses156.
Lastly, nsP2 contains a putative methyltransferase domain that was predicted using homology modeling154. However, it is currently thought to be inactive as it lacks a key active site residue129. Recently there has been work that has shown a potential alternative activity for this region. There is data that indicates this domain may play a role in interferon shutoff due to its interaction with signal transducer and activator of transcription proteins 1 (STAT1)157. This activity appears to be mediated by enhancing the nuclear export of STAT1, which prevents the magnification of downstream immune signaling, including the interferon response157. Work with CHIKV nsP2 has also continued with groups now using the complete crystal structure to predict potential interactions with innate immune proteins158, and this structure can now be utilized for studies altering the structures and components of the nsP2 protein to further elucidate its functions159
5.3. Nonstructural protein 3
NsP3 is poorly understood, but mutations within this protein have resulted in defects in both negative-sense and subgenomic RNA synthesis160.
NsP3 contains a macrodomain with both adenosine diphosphate ribose (ADPr) binding and hydrolase activity129,161,162, and these activities have begun to be characterized in the context of infection models. The ADP ribosylase activity is necessary for infection of neural cells and the hydrolase activity results in an increase in replicase complexes163. In a SINV model, reductions in hydrolase activity led to reduced neurovirulence while increases in ADP ribosylase activity increased neurovirulence164.
NsP3 also contains the highly conserved alphavirus unique domain, or AUD129. This domain is maintained across all alphaviruses129. Recent work using CHIKV has indicated that it potentially has many functions, particularly in subgenomic RNA replication165. Disruption of the AUD resulted in decreased infectivity, potentially due to decreased interaction with the viral RNA and the subgenomic promoter165.
The last feature of note in nsP3 is the hypervariable domain or HVD. This domain is so varied that it can be distinct between strains of a single viral species, such as in VEEV166. This region is tolerant of significant mutation and even deletion, which is unique compared to the rest of the nsPs and their domains167. Natural duplications and insertions in this region can even have positive effects on viral fitness168. The HVD is also involved in interaction with host cell proteins, resulting in the formation of distinct protein complexes in Old and New-World viruses169. These interactions include the cellular machinery responsible for the formation of stress granules, which alphaviruses utilize to their own replicative benefit170–173. These interactions are highly specific to viral species as well, and may partially drive the differences in pathogenesis seen between species173–175.
5.4. Nonstructural protein 4
NsP4 is produced in small amounts by most alphaviruses due to the inclusion of a stop codon between nsP3 and nsP471,176. Altering expression levels of nsP4 decreases viral fitness, indicating that tight control of expression is highly important177. The tight limit on expression of nsP4 is also promoted by it being targeted by N-end rule degradation178. Recent work performed using trans replication systems has elegantly shown how nsP4 is involved in RNA selectivity, yet this occurs in tandem with the other nsPs179. Interestingly these authors have shown that in trans and in a transfection system, increasing nsP4 levels does not have these same detrimental effects179
NsP4 is the RNA dependent RNA polymerase (RDRP) of the alphaviruses and is active in both positive and negative-sense RNA synthesis, with the specificity being determined by the cleavage state of the other nsPs77,129,180. NsP4 can display RNA synthesis activity alone, but its activity is enhanced by the presence of the other nsPs180–182. There has been recent publication of crystal structures of the RDRP domains of both RRV and SINV183. This work indicated that the RDRP pocket of these viruses was highly dynamic and flexible, as well as being well conserved between the two viruses183.
NsP4 also has a large N terminal region that lacks predicted structure or function. Recent work has predicted that this region is somehow involved in viral RNA synthesis, as mutations in this region result in resistance to the effects of a drug that inhibits the production of new viral RNA62,184. However, the function of this region remains unclear. Work by others has also shown that mutations in this region has a broad range of effects on viral RNA synthesis185. This work, as well as the antiviral resistant VEEV isolates that have been recovered184, indicate that this region plays an important role in RNA synthesis of these viruses, potentially in tandem with nsP2. This indicates that these proteins have additional, complex interactions and roles in viral biology that remain to be understood. The functions of the nsPs are summarized in Table 3.
Table 3:
A summary of the structures and functions of the alphaviral nsPs
| Protein | Structures | Functions |
|---|---|---|
| nsP1 | methyl transferase domain, guanyl transferase domain, membrane association domains | caps viral RNA making it usable by ribosomes, anchors the replication machinery to cellular membranes111,129–131,133–136 |
| nsP2 | helicase domain, ADP binding region, cysteine protease, methyl transferase like domain | unwinds viral RNA for replication, cleaves the polyprotein into its constitutive parts, digests host cell proteins77,132,138–142,144–146,152–157 |
| nsP3 | macrodomain, alphavirus unique domain, hypervariable domain | poorly described, necessary for replication, highly involved in host cell interactions129,160–168,170–172,174,175 |
| nsP4 | RNA dependent RNA polymerase domain | synthesizes new viral RNA71,77,129,176–178,180–182,185 |
5.5. The nsPs as potential antiviral drug targets
In recent years there has been increasing interest in viral proteins as targets for antiviral drug development. This is primarily due to their being largely distinct from cellular proteins, as well as being required for viral replication. Additionally, many nucleoside and non-nucleoside analogs broadly target replication of RNA and DNA resulting in significant side effects and toxicity, making novel targets attractive. Targeting viral proteins directly theoretically allows for increased specificity and reduced side effect potential. The approval of sofosbuvir/ledipasvir combination treatment for hepatitis C infection and its incredible success in the clinic has increased interest in these targets, as it has proved that viral protein targets are therapeutically viable. Of particular note is that sofosbuvir/ledipasvir combination treatment has fewer side effects and higher cure rates than the previous standard of care using ribavirin/interferon combination treatment186.
With regards to the alphaviruses, researchers have so far reported a novel class of anti-VEEV drugs based around a highly aromatized core184. These compounds were then further developed into the benzamidine drug ML336, which is highly effective at inhibiting RNA synthesis, likely due to interactions with nsP2 and nsP462. This compound is highly specific as well, having only minimal effects on cellular RNA synthesis62, and this combined with its low EC50 value indicate that it is unlikely to have significant side effects. While ML336 is highly specific to VEEV, further manipulation of these drugs has potential to lead to treatments for additional alpahviruses62,184. The structure of ML336 can be found in table 2 above.
There is also work looking at the other nsPs for potential antiviral targets. These include nsP3 in CHIKV, which has been used for modeling studies examining small molecule libraries187. The other nsPs of CHIKV have been examined as well, and a review summarizing these investigations has been published by Sundar, Piramnanyagam, and Natarajan, and it also compares these efforts to dengue and zika188. Further review of CHIKV drug discovery can be found in Kovacikova and Hemert189. A review of promising pre-clinical antivirals against the encephalitic alphaviruses can be found in Kehn-Hall and Bradfute190.
6. Functions of the alphavirus structural proteins
As outlined above, the alphaviruses manufacture their structural proteins through a second open reading frame derived from the genomic RNA termed the subgenomic RNA7. The subgenome encodes six proteins: capsid, E3/2, 6K/TF, and E1. These proteins are produced as a polyprotein similar to the nonstructural proteins, and the capsid has self-cleavage functionality to release itself from the polyprotein71. The function and processing of these proteins will be further described below.
6.1. Capsid protein
The capsid is the first structural protein that is translated after subgenomic RNA production. This protein is responsible for forming the primary structure of the virion and selects for packaging of the viral genomic RNA by recognition of a conserved packaging signal191, but the formation of nucleocapsid cores is not dependent on this signal192–194. Encapsidation selects specifically for only single units of cargo to be packaged122, and in infected cells, this packaging activity is highly specific for viral RNA. After packaging the RNA, the nucleocapsid cores translocate to the plasma membrane where they will bud into the extracellular environment while collecting a membrane as well as the glycoprotein spikes7. The initiation of budding remains poorly defined and there are contributions from both the nucleocapsid core and the glycoproteins. While either can initiate budding independently, they likely function in a synergistic manner127,128.
The capsid protein is one of the most produced proteins during alphaviral infection, and it has several pathogenic roles in addition to its structural function. In the New-World alphaviruses the capsid protein is able to block the nuclear pores and thus prevent the translation of new cellular proteins, enhancing viral pathogenesis, cytopathic effect, and assisting in immune evasion138,140,195. This protein synthesis inhibition functions in tandem with the nsPs which actively inhibit the synthesis of cellular proteins141,196,197. While several mechanisms have been proposed for this activity there is currently no definitive evidence to support one hypothesis over another and it is likely multifactorial. The capsid of the New-World viruses also contains sequences which result in its importation to the nucleus itself, which inhibits the transcription of cellular RNA195. Exactly how this activity is caried out remains unknown.
While the capsid of the Old-World viruses contains a nuclear transport signal and is able to enter the nucleus, why it does this remains unclear as it is not involved in transcriptional shutoff in infection198. This transcriptional shutoff in the Old-World viruses is carried out by nsP2, which ubiquinates subunits of the cellular RNA polymerase, targeting them for degradation195,199.
These non-packaging activities of the capsid protein have been targeted by certain preclinical anti-alphaviral compounds, such as mifepristone and compounds derived from it, by inhibiting the normal trafficking of the capsid protein200.
6.2. The glycoproteins
The alphaviruses manufacture three glycoproteins during infection, E1, E2, and E3. E3 and 2 are made as a single fused peptide that undergoes later processing after translation in the endoplasmic reticulum. E1 is encoded singly7. These proteins are translated in the ER and then traffic through the Golgi apparatus and undergo various glycosylation and palmytoilation events201–207. E2 is cleaved from E3 although the two remain in close association. E2 and E3 also may not always fully separate and this appears to be pH dependent124,208. The proteins are then arrayed at the plasma membrane and form heterotrimers of E1 and E2209 where they are picked up by budding nucleocapsid cores and integrated into the mature virions7.
As mentioned above, the glycoproteins are involved in the fusion of the viral and host cell membranes in the endosomal pathway, which mediates release of the viral genome into the cytoplasm. E1 alone is sufficient to induce this fusogenic activity63,64. E1 is also able to induce membrane pores, and this pore forming activity is likely to contribute to pH dependent viral particle disassembly as well as induce various physiological changes in the cells due to these membrane disruptions68–70,210.
E3 is known to function as a signal sequence that guides the structural polyprotein into the endoplasmic reticulum after capsid cleavage211. E3 remains associated to E2 until furin cleavage separates the two proteins in the Golgi apparatus211,212. However, E3 must have additional functions as replacing it with an endoplasmic reticulum signal sequence results in the other glycoproteins being trapped in the ER213. Swapping the E3 sequences between alphaviral clades also results in attenuation while swapping within a clade does not have the same effect, and this phenotype is dependent on interactions that occur between E2 and E3214. Finally E3 has an important role in protecting E1 from the acidic pH of the secretory pathway and allowing for its secretion to the cell surface215.
6.3. 6K/TF protein
The 6K gene splits the E2 and E1 genes and forms two different protein products due to a frameshift that occurs in response to read through of a so called slippery sequence in the middle of the protein7,216. This frameshift is actively enhanced by the folding of the polyprotein that occurs simultaneously with translation of the viral RNA217.
The more common form of the protein is simply termed 6K as it has a size of roughly six kilodaltons216. 6K is known to be important to budding of the mature virions and is hypothesized to act as a spacer for the glycoproteins216,218. There is also strong evidence that the 6K protein directly associates with the E2 glycoprotein, and a loss of this association reduced budding efficiency216,219–222. However, this relationship and the resulting reduction in budding has yet to be fully described.
The second protein is called TF for “trans frame,” as it is generated from a one nucleotide frame shift216,223. This protein was relatively recently identified as a separate entity from 6K216. TF is found incorporated into the virion structure after release from the cell216,224. Recent work has also indicated that there is a role for TF protein in the evasion of the interferon response and that this is due to specific palmitoylation patterns225, with mutant viruses causing no disease in interferon intact animals, but disease pathology being restored in ifnar knockout mice225. Additionally palmitoylation of this protein is self-regulated, with specific domains of TF protein affecting its own palmitoylation levels226
One or both of these proteins also function as a viroporin ,and when either or both of these proteins is over expressed in bacteria they are cytotoxic to the cultured cells216.
It is currently understood that many of the phenotypes attributed to 6K are likely mediated instead by the TF protein; however, the exact differences in their activities remain poorly defined. Both are important for efficient release of progeny virus, both are known to be highly cytotoxic when expressed ectopically, and both are important to viral virulence, with viruses that lack these proteins causing less severe disease phenotypes216.
Perhaps the most interesting finding about these two proteins is that they are the only alphaviral proteins that are not necessary to produce viable viruses in cell culture216. A deletion of both proteins does result in reduced viral titers in the supernatant of infected cells, but virus is produced216,227. If any of the other proteins coded for by the alphaviruses are deleted, however, it results in nonviability. The functions of the structural proteins are summarized in table 4.
Table 4:
Summary of the functions of the structural proteins of alphaviruses
| Protein | Function |
|---|---|
| Capsid | Forms the virion structure, selects for packaging of viral RNA, involved in initiating budding from the cell, in the New-World viruses inhibits cellular translation and transcription127,128,140,191–194,198,228 |
| E1 | Forms heterotrimers with E2 to form the glycoprotein spikes, fuses the viral and host cell membranes, forms pores in the endosome to induce pH changes63,68–70,210,229 |
| E2 | Forms heterotrimers with E1 to from the glycoprotein spikes209 |
| E3 | Functions as the signal sequence for the structural polypeptide, protects E1 from acidic pH, additional functions involved in trafficking and virulence213–215 |
| 6K | Involved in budding, may help space the glycoproteins on the virion, may function as a viroporin216,218 |
| TF | Involved in evading interferon due to specific palmitoylation, may be a viroporin, incorporated into the virion216,224,225 |
7. Screening assays for antiviral drug development
As highlighted in the above sections, there is extensive ongoing work on discovery and testing of anti-alphaviral therapeutics. As with all antiviral development, this work has seen an explosion in productivity thanks to the development of high-throughput drug discovery assays and technology. The introduction of these techniques has allowed for an exponential increase in the number of compounds that can be screened, and has played roles in most of the already mentioned antiviral compounds. A summary of selected assays can be found in table 5 below. For a thorough review of the state of antiviral development studies more information can be found in Andersen et al.230.
Table 5:
Selected antiviral screening assays
| antiviral assay | readout type | Use | references |
|---|---|---|---|
| CelTiter-Glo | luminescence | cell viability | 62,231,232 |
| DNA staining and TUNEL | fluorescence | cell viability | 233–235 |
| MTT | colorimetric | cell viability | 236,237 |
| Crystal violet | cell staining | cell viability | 238 |
| viral reporter systems | fluorescence or luminescence | tracking viral protein production tracking viral replication determining stages of antiviral interference | 233,237,239–243 |
8. The study of alphaviruses in animal models
A critical part of studying infectious agents and the development of drug treatments is the availability of animal models that recapitulate disease as seen in human patients. The alphaviruses have been extensively studied since their initial discovery in mid 20th century. Due to this there have been a large number of animal models investigated for potential study of these viruses. VEEV in particular was widely studied in a variety of animal models in the mid-20th century in an attempt to determine a model that would closely mimic human infection244. Currently mice and non-human primates are the most common alphavirus models. A summary of commonly used and well described animal models for alphaviral infection can be found in table 6 below.
Table 6:
Animal models of alphavirus infection
| virus | model | Pathology | references |
|---|---|---|---|
| VEEV | guinea pig | high lethality, no encephalitis symptoms | 244–246 |
| hamster | high lethality, no encephalitis symptoms | 244–246 | |
| mice (various immune competent strains) | similar to human infection, route dependent pathology | 247–255 | |
| cynomolgus macaques | Similar to human infection lymphatic pathology, myocarditis develop CNS lesions regardless of encephalitis symptoms | 244,246,254,256,257 | |
| various equids | high lethality but distinct from human disease | 244 | |
| EEEV | mice (various immune competent strains) | develop seizures, no vasculitis | 247,254,258,259 |
| hamsters | high lethality, no encephalitis symptoms | 254 | |
| guinea pigs | lethal with no obvious encephalitis | 254 | |
| macaques | similar to human infection | 254,260–263 | |
| owl monkeys | similar to human infection | 262 | |
| marmosets | similar to human infection | 261 | |
| NHP models | additional readouts such as brainwaves under development | 264 | |
| WEEV | mice (Swiss, Balb/c, others) | Similar to human infection Also develop myocarditis | 254,265–268 |
| hamsters | high mortality when infected in the periphery, rescuable with interferon | 258,269–271 | |
| macaques | Similar to human infection, Also develop myocarditis, lethality is strain and experiment dependent | 254,257,272,273 | |
| CHIKV | neonatal mice | high lethality | 274–277 |
| interferon deficient mice | high lethality | 275 | |
| mice (various immune competent strains) | similar to human arthralgia | 275,278–280 | |
| Rag knockout mice and similar | Similar to human arthralgia increased viral persistence | 275,281–283 | |
| adult macaques | similar to mild human disease, high dose mimics severe disease | 275,284–288 | |
| aged macaques | Similar to human disease, developed to study disease in specific human populations | 275,289 | |
| pregnant macaques | Similar to human disease, developed to study disease in specific human populations | 287 | |
| SINV | mice (various immune competent strains) | specialized viral strains develop encephalitis can also be used for arthralgia, which is more similar to human pathology | 8,290 |
| neonate and weanling mice | high lethality, some encephalitis | 291–294 | |
| RRV | mice (various immune competent strains) | similar to human disease | 295–297 |
| BFV | mice (various immune competent strains) | similar to human disease | 298 |
| ONNV | mice (various immune competent strains) | similar to human disease | 299 |
| MAYV | mice (various immune competent strains) | similar to human disease | 300 |
| cynomolgus macaques | similar to human disease | 284 | |
| SFV | mice (various immune competent strains) | similar to human disease | 301 |
9. The Development of alphaviral vaccines
While there are no approved vaccines against alphaviruses for human use, there are veterinary vaccines available for livestock. These vaccines are available as a trivalent dose for the three Equine Encephalitis viruses and are widely used in North, Central, and South America302. While this vaccine is highly effective at disease prevention it does require annual boosting302. The livestock vaccine is based on inactivated virus and is not replicatively competent303;this is due to previous vaccination with the live TC-83 vaccine strain of VEEV resulting in detectable virus occurring in mosquitos in the area of testing, indicating that use of live attenuated virus could potentially result in outbreaks of disease if there were reversion events304.
There have been many attempts at creating vaccines for the alphaviruses for human use. The most advanced vaccine for VEEV is the strain TC-83, a cell culture attenuated VEEV that was developed by the United States Army305. However, this vaccine is poorly immunogenic and has less than an 85% seroconversion rate when given as a single dose306. It also has a very high side effect rate with more than 20%, and in some studies more than 80%, of treated individuals reporting a side effect upon use306,307. TC-83 has also been noted to have the potential to revert to wild-type, epidemic strain VEEV308,309. There is also a specific booster for T-83, C-84, a formalin inactivated vaccine for those who have received TC-83 but did not seroconvert306.
Despite its shortcomings, TC-83 remains in use for certain high-risk individuals such as those who frequently work with wild-type VEEV in high-risk research applications310. A rationally designed attenuated strain of VEEV termed V3526 was also tested as a vaccine, but it was found to have some remaining neurovirulence in a non-human primate model and was later abandoned early in clinical trials due to a high rate of side effects311,312. There are also candidate vaccines available for human use under special circumstances for both EEEV and WEEV, inactivated PE-6 and inactivated CM-4884, respectively. Though like TC-83 these vaccines have low response rates and these two inactivated vaccines often fail to produce durable immunity313.
There has also been recent work examining the safety profile and immunogenicity of a trivalent virus like particle vaccine against VEEV, WEEV, and EEEV314. While this work is in the early stages of clinical development, the vaccine was extremely safe with only minimal and primarily localized side effects, and also demonstrated the production of a durable immune response as measure by neutralizing antibody titers against the three viruses. One caveat is that as time went on subjects became less likely to respond to all three of the viruses and instead responded preferentially to only one or two of the viruses314. A more detailed review of the state of vaccine development for the encephalitic alphaviruses can be found in Stromberg et al.315.
With the spread of CHIKV across the tropics, it has also become a target of intense vaccine development research. The first candidate was the 181/25 strain of CHIKV which was abandoned due to a high rate of side effects316,317. Recently there has been significant promise shown by a variety of CHIKV vaccines based on live attenuated, vector launched, and subunit platforms318–321. Some of these have shown great promise for clinical approval321.
10. Remaining questions.
While the alphaviruses have been well studied for many years, they remain significant public health threats due to challenges in the development of either vaccines or antiviral treatments. While vaccines are available for veterinary use322,323, none of the available vaccine candidates have been found to meet the more stringent standards for humans due to significant side effects and relatively poor immunogenicity324–326. This lack of treatment means that these viruses require significant ongoing study.
A major challenge that remains in the field is the characterization of the remaining regions of the nsPs that have no predicted function. These proteins remain challenging to study, however recent advances in protein expression have ameliorated this somewhat146,154,327. There is evidence that these uncharacterized regions are important to viral replication62,231 , which makes them promising targets for antiviral drug development.
In addition to being potential drug targets, further characterization of these uncharacterized protein regions has the potential to further our understanding of positive-sense RNA viruses generally. While these viruses are highly variable in their biology and pathogenesis, they retain highly similar replication strategies, as exemplified by the similarities of viral RNA dependent RNA polymerase proteins328 and the consistent use of host cell membranes as scaffolding to develop their replication centers329,330.
Lastly, there has been some recent work to show that the alphaviral RNAs themselves are likely to have biological activities in addition to their use for translation of the viral proteins. This can be seen in the necessarily tight control of capping83, as well as in the highly complex structures that form in the RNA and affect viral replication331,332. This all indicates that there remain many outstanding questions about these viruses, and that further research will be needed as they remain significant threats to public health and for emergence into naïve populations.
Table 1:
A comparison of representative alphaviruses.
| Virus | Old or New World | Arthritic | Neurologic | Mortality | Location |
|---|---|---|---|---|---|
| Chikungunya | OLD | + | − | Reports vary, as low as 0.03%. Some claim this is underreport ed.25 | Tropics world wide |
| Semliki Forest | OLD | + | rarely | extremely rare with neurologic involvement26 | Africa |
| Sindbis | OLD | + | rarely | extremely rare with neurological involvement8–10 | Africa and Europe |
| Ross River | OLD | + | − | none27 | Oceania |
| O’nyong-nyong | OLD | + | − | none28 | Sub-Saharan Africa |
| Venezuelan Equine Encephalitis | NEW | − | + | ≤1%23 | South and Central America |
| Western Equine Encephalitis | NEW | − | + | 3-7%23 | Western North America and parts of South America |
| Eastern Equine Encephalitis | NEW | − | + | 50-75%23 | Eastern North and South America |
Highlights:
A broad review of all aspects of the alphaviral life cycle
Particularly focused on the functions of the structural and nonstructural proteins
Highlights recent discoveries in alphaviral replication
Summarizes the current state of vaccine and antiviral development targeting the alphaviruses
Points out current knowledge gaps in relation to alphaviral replication
Poses additional areas of research focus to further the field
Funding:
This work was supported by the following grants: Department of Defense grant HDTRA1-0-1-0015 (SB) and National Institutes of Health grant K12 GM088021 (AS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.International Committee on Taxonomy of Viruses. Taxonomy. (2020). [Google Scholar]
- 2.Powers AM et al. Evolutionary Relationships and Systematics of the Alphaviruses. J. Virol 75, 10118–10131 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosby L. et al. Severe manifestations of chikungunya virus in critically ill patients during the 2013–2014 Caribbean outbreak. Int. J. Infect. Dis 48, 78–80 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Rezza G. et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. The Lancet 370, 1840–1846 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Grandadam M. et al. Chikungunya Virus, Southeastern France. Emerg. Infect. Dis 17, 910–913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staples JE & Fischer M Chikungunya Virus in the Americas — What a Vectorborne Pathogen Can Do. N. Engl. J. Med 371, 887–889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston Robert E., C. J. P Fields Virology, vol. 1 (Lippincott-Raven, 2018). [Google Scholar]
- 8.Griffin DE & Johnson RT Role of the Immune Response in Recovery from Sindbis Virus Encephalitis in Mice. 7 (1977). [PubMed] [Google Scholar]
- 9.Laine M. et al. Prolonged arthritis associated with Sindbis-related (Pogosta) virus infection. Rheumatology 39, 1272–1274 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Adouchief S, Smura T, Sane J, Vapalahti O & Kurkela S Sindbis virus as a human pathogen—epidemiology, clinical picture and pathogenesis. Rev. Med. Virol 26, 221–241 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Arrigo NC, Adams AP & Weaver SC Evolutionary Patterns of Eastern Equine Encephalitis Virus in North versus South America Suggest Ecological Differences and Taxonomic Revision. J. Virol 84, 1014–1025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Go YY, Balasuriya UBR & Lee C Zoonotic encephalitides caused by arboviruses: transmission and epidemiology of alphaviruses and flaviviruses. Clin. Exp. Vaccine Res 3, 58–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta-Ampudia Y. et al. Mayaro: an emerging viral threat? Emerg. Microbes Infect. 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murillo-Zamora E. et al. Persistent Arthralgia and Related Risks Factors: A Cohort Study at 12 Months from Laboratory-Confirmed Chikungunya Infection. Arch. Med. Res 49, 65–73 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Barber B, Denholm JT & Spelman D RACGP - Ross River virus. (2009). [PubMed] [Google Scholar]
- 16.Aguilar PV et al. Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol. 6, 721–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CDC. Statistics & Maps | Eastern Equine Encephalitis | CDC. https://www.cdc.gov/easternequineencephalitis/statistics-maps/index.html (2021). [Google Scholar]
- 18.Bergren NA et al. “Submergence” of Western equine encephalitis virus: Evidence of positive selection argues against genetic drift and fitness reductions. PLOS Pathog. 16, e1008102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brault AC, Fang Y & Reisen WK Multiplex qRT-PCR for the Detection of Western Equine Encephalomyelitis, St. Louis Encephalitis, and West Nile Viral RNA in Mosquito Pools (Diptera: Culicidae). J. Med. Entomol 52, 491–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusnak JM, Dupuy LC, Niemuth NA, Glenn AM & Ward LA Comparison of Aerosol- and Percutaneous-acquired Venezuelan Equine Encephalitis in Humans and Nonhuman Primates for Suitability in Predicting Clinical Efficacy under the Animal Rule. Comp. Med 68, 380–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CDC & USDA. Federal Select Agent Program - Select Agents and Toxins List. https://www.selectagents.gov/SelectAgentsandToxinsList.html (2020).
- 22.CDC & USDA. About Us | Federal Select Agent Program. https://www.selectagents.gov/overview/index.htm (2022).
- 23.Zacks MA & Paessler S ENCEPHALITIC ALPHAVIRUSES. Vet. Microbiol 140, 281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steele KE & Twenhafel NA REVIEW PAPER: Pathology of Animal Models of Alphavirus Encephalitis. Vet. Pathol 47, 790–805 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Lima Neto AS, Sousa GS, Nascimento OJ & Castro MC Chikungunya-attributable deaths: A neglected outcome of a neglected disease. PLoS Negl. Trop. Dis 13, e0007575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willems WR et al. Semliki Forest Virus: Cause of a Fatal Case of Human Encephalitis. Science 203, 1127–1129 (1979). [DOI] [PubMed] [Google Scholar]
- 27.Harley D, Sleigh A & Ritchie S Ross River Virus Transmission, Infection, and Disease: a Cross-Disciplinary Review. Clin. Microbiol. Rev 14, 909–932 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbet PS, Williams MC & Gillett JD O’nyong-nyong Fever: An Epidemic Virus Disease in East Africa. Trans. R. Soc. Trop. Med. Hyg 55, 463–480 (1961). [DOI] [PubMed] [Google Scholar]
- 29.Smith DR, Adams AP, Kenney JL, Wang E & Weaver SC Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: Infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology 372, 176–186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrester NL, Coffey LL & Weaver SC Arboviral Bottlenecks and Challenges to Maintaining Diversity and Fitness during Mosquito Transmission. Viruses 6, 3991–4004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrigo NC, Smith DR, Weaver SC, Muehlberger LE & Leal G Infection and Dissemination of Venezuelan Equine Encephalitis Virus in the Epidemic Mosquito Vector, Aedes taeniorhynchus. Am. J. Trop. Med. Hyg 77, 176–187 (2007). [PubMed] [Google Scholar]
- 32.Jose J, Taylor AB & Kuhn RJ Spatial and Temporal Analysis of Alphavirus Replication and Assembly in Mammalian and Mosquito Cells. mBio 8, e02294–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung JY-S, Ng MM-L & Chu JJH Replication of Alphaviruses: A Review on the Entry Process of Alphaviruses into Cells. Adv. Virol 2011, 1–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDonald GH & Johnston RE Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol 74, 914–922 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R. et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma H. et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernard KA, Klimstra WB & Johnston RE Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276, 93–103 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Heil ML, Albee A, Strauss JH & Kuhn RJ An Amino Acid Substitution in the Coding Region of the E2 Glycoprotein Adapts Ross River Virus To Utilize Heparan Sulfate as an Attachment Moiety. J. Virol 75, 6303–6309 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimstra WB, Ryman KD & Johnston RE Adaptation of Sindbis Virus to BHK Cells Selects for Use of Heparan Sulfate as an Attachment Receptor. J. Virol 72, 7357–7366 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner CL, Ebel GD, Ryman KD & Klimstra WB Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci 108, 16026–16031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helenius A, Kartenbeck J, Simons K & Fries E On the entry of semliki forest virus into BHK-21 cells. J. Cell Biol 84, 404–420 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung JY-S, Ng MM-L & Chu JJH Replication of Alphaviruses: A Review on the Entry Process of Alphaviruses into Cells. Adv. Virol 2011, 1–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolokoltsov AA, Fleming EH & Davey RA Venezuelan equine encephalitis virus entry mechanism requires late endosome formation and resists cell membrane cholesterol depletion. Virology 347, 333–342 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Hernandez R, Luo T & Brown DT Exposure to low pH is not required for penetration of mosquito cells by Sindbis virus. J. Virol 75, 2010–2013 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paredes AM et al. Conformational changes in Sindbis virions resulting from exposure to low pH and interactions with cells suggest that cell penetration may occur at the cell surface in the absence of membrane fusion. Virology 324, 373–386 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Bernard E. et al. Endocytosis of Chikungunya Virus into Mammalian Cells: Role of Clathrin and Early Endosomal Compartments. PLOS ONE 5, e11479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nok AJ Arsenicals (melarsoprol), pentamidine and suramin in the treatment of human African trypanosomiasis. Parasitol. Res 90, 71–79 (2003). [DOI] [PubMed] [Google Scholar]
- 48.PubChem. Suramin. https://pubchem.ncbi.nlm.nih.gov/compound/5361.
- 49.De Clercq E Suramin: A potent inhibitor of the reverse transcriptase of RNA tumor viruses. Cancer Lett. 8, 9–22 (1979). [DOI] [PubMed] [Google Scholar]
- 50.Stein CA, LaRocca RV, Thomas R, McAtee N & Myers CE Suramin: and anticancer drug with a unique mechanism of action. J. Clin. Oncol 7, 499–508 (1989). [DOI] [PubMed] [Google Scholar]
- 51.Albulescu IC et al. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antiviral Res. 121, 39–46 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Kuo S-C et al. Suramin treatment reduces chikungunya pathogenesis in mice. Antiviral Res. 134, 89–96 (2016). [DOI] [PubMed] [Google Scholar]
- 53.PubChem. Favipiravir. https://pubchem.ncbi.nlm.nih.gov/compound/492405.
- 54.PMDA. AVIGAN Tablets 200mg. 1–6 https://www.sukl.cz/file/92989_1_1/download/ (2019).
- 55.FURUTA Y, KOMENO T & NAKAMURA T Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci 93, 449–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.CDC - African Trypanosomiasis - Resources for Health Professionals. https://www.cdc.gov/parasites/sleepingsickness/health_professionals/index.html (2022).
- 57.Büscher P, Cecchi G, Jamonneau V & Priotto G Human African trypanosomiasis. The Lancet 390, 2397–2409 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Kovacikova K & van Hemert MJ Small-Molecule Inhibitors of Chikungunya Virus: Mechanisms of Action and Antiviral Drug Resistance. Antimicrob. Agents Chemother 64, e01788–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.PubChem. N(4)-Hydroxycytidine. https://pubchem.ncbi.nlm.nih.gov/compound/197020. [Google Scholar]
- 60.Urakova N. et al. β-d-N4-Hydroxycytidine Is a Potent Anti-alphavirus Compound That Induces a High Level of Mutations in the Viral Genome. J. Virol 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.PubChem. (E)-2-(( 1,4-dimethylpiperazin-2-ylidene)-amino)-5-nitro-n-phenylbenzamide. https://pubchem.ncbi.nlm.nih.gov/compound/71301451. [DOI] [PMC free article] [PubMed]
- 62.Skidmore AM, Adcock RS, Jonsson CB, Golden JE & Chung D-H Benzamidine ML336 inhibits plus and minus strand RNA synthesis of Venezuelan equine encephalitis virus without affecting host RNA production. Antiviral Res. 174, 104674 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omar A & Koblet H Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology 166, 17–23 (1988). [DOI] [PubMed] [Google Scholar]
- 64.Sanz MA, Rejas MT & Carrasco L Individual Expression of Sindbis Virus Glycoproteins. E1 Alone Promotes Cell Fusion. Virology 305, 463–472 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Wahlberg JM, Boere WA & Garoff H The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol 63, 4991–4997 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh I & Helenius A Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol 66, 7049–7058 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wengler G, Würkner D & Wengler G Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology 191, 880–888 (1992). [DOI] [PubMed] [Google Scholar]
- 68.Wengler G & Wengler G In vitro analysis of factors involved in the disassembly of Sindbis virus cores by 60S ribosomal subunits identifies a possible role of low pH. J. Gen. Virol 83, 2417–2426 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Schlegel A, Omar A, Jentsch P, Morell A & Kempf C Semliki Forest virus envelope proteins function as proton channels. Biosci. Rep 11, 243–255 (1991). [DOI] [PubMed] [Google Scholar]
- 70.Spyr CA, Käsermann F & Kempf C Identification of the pore forming element of Semliki Forest virus spikes. FEES Lett. 375, 134–136 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Strauss JH & Strauss EG The Alphaviruses: Gene Expression, Replication, and Evolution. MICROBIOL REV 72 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strauss EG, Rice CM & Strauss JH Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci 80, 5271–5275 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Myles KM, Kelly CLH, Ledermann JP & Powers AM Effects of an Opal Termination Codon Preceding the nsP4 Gene Sequence in the O’Nyong-Nyong Virus Genome on Anopheles gambiae Infectivity. J. Virol 80, 4992–4997 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vasiljeva L, Valmu L, Kääriäinen L & Merits A Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem 276, 30786–30793 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Vasiljeva L. et al. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J. Biol. Chem 278, 41636–41645 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Lemm JA & Rice CM Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol 67, 1916–1926 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirako Y & Strauss JH Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol 68, 1874–1885 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lulla A, Lulla V & Merits A Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J. Virol 86, 553–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lulla V. et al. Timeliness of Proteolytic Events Is Prerequisite for Efficient Functioning of the Alphaviral Replicase. J. Virol 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X. et al. Decreased Virulence of Ross River Virus Harboring a Mutation in the First Cleavage Site of Nonstructural Polyprotein Is Caused by a Novel Mechanism Leading to Increased Production of Interferon-Inducing RNAs. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shin G. et al. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc. Natl. Acad. Sci 109, 16534–16539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pietilä MK, Hellström K & Ahola T Alphavirus polymerase and RNA replication. Virus Res. 234, 44–57 (2017). [DOI] [PubMed] [Google Scholar]
- 83.LaPointe AT, Moreno-Contreras J & Sokoloski KJ Increasing the Capping Efficiency of the Sindbis Virus nsP1 Protein Negatively Affects Viral Infection. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barton DJ, Sawicki SG & Sawicki DL Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol 65, 1496–1506(1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Markland W, McQuaid TJ, Jain J & Kwong AD Broad-Spectrum Antiviral Activity of the IMP Dehydrogenase Inhibitor VX-497: a Comparison with Ribavirin and Demonstration of Antiviral Additivity with Alpha Interferon. Antimicrob. Agents Chemother 44, 859–866 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Furuta Y. et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 100, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Julander JG, Smee DF, Morrey JD & Furuta Y Effect of T-705 treatment on western equine encephalitis in a mouse model. Antiviral Res. 82, 169–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdelnabi R, Jochmans D, Verbeken E, Neyts J & Delang L Antiviral treatment efficiently inhibits chikungunya virus infection in the joints of mice during the acute but not during the chronic phase of the infection. Antiviral Res. 149, 113–117 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Delang L, Abdelnabi R & Neyts J Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res 153, 85–94 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Julander JG et al. Strain-dependent disease and response to favipiravir treatment in mice infected with Chikungunya virus. Antiviral Res. 182, 104904 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cavazzoni P. Emergency Use Authorization 108. [Google Scholar]
- 92.Yoon J-J et al. Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob. Agents Chemother 62, e00766–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Filomatori CV, Merwaiss F, Bardossy ES & Alvarez DE Impact of alphavirus 3’UTR plasticity on mosquito transmission. Semin. Cell Dev. Biol 111, 148–155 (2021). [DOI] [PubMed] [Google Scholar]
- 94.Ou J-H, Trent DW & Strauss JH The 3’-non-coding regions of alpharivus RNAs contain repeating sequences. J. Mol. Biol 156, 719–730 (1982). [DOI] [PubMed] [Google Scholar]
- 95.Pfeffer M, Kinney RM & Kaaden O-R The Alphavirus 3’-Nontranslated Region: Size Heterogeneity and Arrangement of Repeated Sequence Elements. Virology 240, 100–108 (1998). [DOI] [PubMed] [Google Scholar]
- 96.Chen R, Wang E, Tsetsarkin KA & Weaver SC Chikungunya Virus 3’ Untranslated Region: Adaptation to Mosquitoes and a Population Bottleneck as Major Evolutionary Forces. PLOS Pathog. 9, e1003591 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trobaugh DW et al. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506, 245–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hardy RW & Rice CM Requirements at the 3’ End of the Sindbis Virus Genome for Efficient Synthesis of Minus-Strand RNA. J. Virol 79,4630–4639(2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frolov I, Hardy R & Rice CM Cis-acting RNA elements at the 5’ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7, 1638–1651 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hyde JL et al. The 5’ and 3’ ends of alphavirus RNAs – Non-coding is not non-functional. Virus Res. 206, 99–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gorchakov R, Hardy R, Rice CM & Frolov I Selection of Functional 5′ cis-Acting Elements Promoting Efficient Sindbis Virus Genome Replication. J. Virol 78, 61–75 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hyde JL et al. A viral RNA stmctural element alters host recognition of non-self RNA. Science 343, 783–787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kulasegaran-Shylini R, Thiviyanathan V, Gorenstein DG & Frolov I The 5′UTR-specific mutation in VEEV TC-83 genome has a strong effect on RNA replication and subgenomic RNA synthesis, but not on translation of the encoded proteins. Virology 387, 211–221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.White LJ, Wang J-G, Davis NL & Johnston RE Role of Alpha/Beta Interferon in Venezuelan Equine Encephalitis Virus Pathogenesis: Effect of an Attenuating Mutation in the 5′ Untranslated Region. J. Virol (2001) doi: 10.1128/JVI.75.8.3706-3718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paul D & Bartenschlager R Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol 2, 32–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salonen A, Ahola T & Kääriäinen L Viral RNA Replication in Association with Cellular Membranes. Membr. Traffick. Viral Replication 285, 139–173 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johan A den Boon & Ahlquist P Organelle-Like Membrane Compartmentalization of Positive-Strand RNA Virus Replication Factories. Annu. Rev. Microbiol 64, 241–256 (2010). [DOI] [PubMed] [Google Scholar]
- 108.Kopek BG, Perkins G, Miller DJ, Ellisman MH & Ahlquist P Three-Dimensional Analysis of a Viral RNA Replication Complex Reveals a Virus-Induced Mini-Organelle. PLoS Biol. 5, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ertel KJ et al. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife 6, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pietilä MK, van Hemert MJ & Ahola T Purification of Highly Active Alphavirus Replication Complexes Demonstrates Altered Fractionation of Multiple Cellular Membranes. J. Virol 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hellström K et al. Partially Uncleaved Alphavirus Replicase Forms Spherule Structures in the Presence and Absence of RNA Template. J. Virol 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kallio K et al. Template RNA Length Determines the Size of Replication Complex Spherules for Semliki Forest Virus. J. Virol 87, 9125–9134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deshmukh D et al. New insights into culture negative endophthalmitis by unbiased next generation sequencing. Sci Rep 9, 844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scutigliani EM & Kikkert M Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Factor Rev. 37, 17–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uchil PD & Satchidanandam V Architecture of the Flaviviral Replication Complex: PROTEASE, NUCLEASE, AND DETERGENTS REVEAL ENCASEMENT WITHIN DOUBLE-LAYERED MEMBRANE COMPARTMENTS *. J. Biol. Chem 278, 24388–24398 (2003). [DOI] [PubMed] [Google Scholar]
- 116.Laurent T et al. Architecture of the chikungunya virus replication organelle. eLife 11, e83042 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S & Frolov I Functional Sindbis Virus Replicative Complexes Are Formed at the Plasma Membrane. J. Virol 84, 11679–11695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thaa B et al. Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization. J. Virol 89, 11420–11437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spuul P, Balistreri G, Kääriäinen L & Ahola T Phosphatidylinositol 3-Kinase-, Actin-, and Microtubule-Dependent Transport of Semliki Forest Virus Replication Complexes from the Plasma Membrane to Modified Lysosomes. J. Virol 84, 7543–7557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martinez MG & Kielian M Intercellular Extensions Are Induced by the Alphavirus Structural Proteins and Mediate Virus Transmission. PLOS Pathog. 12, e1006061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng Y & Kielian M Imaging of the Alphavirus Capsid Protein during Virus Replication. J. Virol 87, 9579–9589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Button JM & Mukhopadhyay S Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core. Viruses 12, 846 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stollar V Defective Interfering Alphaviruses. in The Togaviruses Biology, Structure, Replication 427–457 (Academic Press, 1980). [Google Scholar]
- 124.Zhang R et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J 30, 3854–3863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Snyder AJ, Sokoloski KJ & Mukhopadhyay S Mutating Conserved Cysteines in the Alphavirus E2 Glycoprotein Causes Virus-Specific Assembly Defects. J. Virol 86, 3100–3111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cheng F et al. The Packaging of Different Cargo into Enveloped Viral Nanoparticles. Mol. Pharm 10, 51–58 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Garoff H, Sjöberg M & Cheng RH Budding of alphaviruses. Virus Res. 106, 103–116 (2004). [DOI] [PubMed] [Google Scholar]
- 128.Brown RS, Wan JJ & Kielian M The Alphavirus Exit Pathway: What We Know and What We Wish We Knew. Viruses 10, 89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rupp JC, Sokoloski KJ, Gebhart NN & Hardy RW Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol 96, 2483–2500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li C et al. mRNA Capping by Venezuelan Equine Encephalitis Virus nsP1: Functional Characterization and Implications for Antiviral Research. J. Virol 89, 8292–8303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Decroly E, Ferron F, Lescar J & Canard B Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol 10, 51–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vasiljeva L, Merits A, Auvinen P & Kääriäinen L Identification of a novel function of the alphavirus capping apparatus. RNA 5’-triphosphatase activity of Nsp2. J. Biol. Chem 275, 17281–17287 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Ahola T, Lampio A, Auvinen P & Kääriäinen L Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18, 3164–3172 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spuul P et al. Role of the Amphipathic Peptide of Semliki Forest Virus Replicase Protein nsP1 in Membrane Association and Virus Replication. J. Virol 81, 872–883 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kujala P et al. Biogenesis of the Semliki Forest Virus RNA Replication Complex. J. Virol 75, 3873–3884 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jones R, Bragagnolo G, Arranz R & Reguera J Capping pores of alphavirus nsP1 gate membranous viral replication factories. Nature 1–5 (2020) doi: 10.1038/s41586-020-3036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang K et al. Molecular basis of specific viral RNA recognition and 5′-end capping by the Chikungunya virus nsP1. Cell Rep. 40, 111133 (2022). [DOI] [PubMed] [Google Scholar]
- 138.Garmashova N et al. Analysis of Venezuelan Equine Encephalitis Virus Capsid Protein Function in the Inhibition of Cellular Transcription. J. Virol 81, 13552–13565 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Akhrymuk I, Lukash T, Frolov I & Frolova EI Novel Mutations in nsP2 Abolish Chikungunya Virus-Induced Transcriptional Shutoff and Make the Virus Fess Cytopathic without Affecting Its Replication Rates. J. Virol 93, e02062–18, /jvi/93/4/JVI.02062-18.atom (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Atasheva S, Fish A, Fornerod M & Frolova EI Venezuelan Equine Encephalitis Virus Capsid Protein Forms a Tetrameric Complex with CRM1 and Importin α/β That Obstmcts Nuclear Pore Complex Function. J. Virol 84,4158–4171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bhalla N et al. Host translation shutoff mediated by non-structural protein 2 is a critical factor in the antiviral state resistance of Venezuelan equine encephalitis virus. Virology 496, 147–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim DY, Atasheva S, Frolova EI & Frolov I Venezuelan Equine Encephalitis Virus nsP2 Protein Regulates Packaging of the Viral Genome into Infectious Virions. J. Virol 87, 4202–4213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cherkashchenko L, Rausalu K, Basu S, Alphey L & Merits A Expression of Alphavirus Nonstructural Protein 2 (nsP2) in Mosquito Cells Inhibits Viral RNA Replication in Both a Protease Activity-Dependent and -Independent Manner. Viruses 14, 1327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gomez de Cedrón M, Ehsani N, Mikkola ML, García JA & Kääriäinen L RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448, 19–22 (1999). [DOI] [PubMed] [Google Scholar]
- 145.Das PK, Merits A & Lulla A Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J. Biol. Chem 289, 5635–5653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Law Y-S et al. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc. Natl. Acad. Sci 116, 9558–9567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Law Y-S et al. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc. Natl. Acad. Sci 116, 9558–9567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Law Y-S et al. Interdomain Flexibility of Chikungunya Virus nsP2 Helicase-Protease Differentially Influences Viral RNA Replication and Infectivity. J. Virol 95, e01470–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ivanova L et al. Novel Analogues of the Chikungunya Virus Protease Inhibitor: Molecular Design, Synthesis, and Biological Evaluation. ACS Omega 6, 10884–10896 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Silva Muniz L & Pita S. S. da R. In silico studies revealed interaction mechanisms of benzylidene-acrylohydrazide derivatives and nsP2 CHIKV. New J. Chem 46, 6414–6423 (2022). [Google Scholar]
- 151.Atasheva S, Gorchakov R, English R, Frolov I & Frolova E Development of Sindbis Viruses Encoding nsP2/GFP Chimeric Proteins and Their Application for Studying nsP2 Functioning. J. Virol 81, 5046–5057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hahn YS, Strauss EG & Strauss JH Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol 63, 3142–3150 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hardy WR & Strauss JH Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol 63, 4653–4664 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Russo AT, White MA & Watowich SJ The Crystal Structure of the Venezuelan Equine Encephalitis Alphavirus nsP2 Protease. Structure 14, 1449–1458 (2006). [DOI] [PubMed] [Google Scholar]
- 155.Kim KH, Rümenapf T, Strauss EG & Strauss JH Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323, 153–163 (2004). [DOI] [PubMed] [Google Scholar]
- 156.Morazzani EM et al. Proteolytic cleavage of host proteins by the Group IV viral proteases of Venezuelan equine encephalitis virus and Zika virus. Antiviral Res. 164, 106–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Göertz GP et al. The Methyltransferase-Like Domain of Chikungunya Virus nsP2 Inhibits the Interferon Response by Promoting the Nuclear Export of STAT1. J. Virol 92, e01008–18, /jvi/92/17/e01008-18.atom (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Modeling interaction between non-structural protein 2 of Chikungunya Virus and various protein factors of innate pathway. Biomed. Lett 8, 162–169 (2022). [Google Scholar]
- 159.Wang S & Merits A G3BP/Rin-Binding Motifs Inserted into Flexible Regions of nsP2 Support RNA Replication of Chikungunya Virus. J. Virol 96, e01278–22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.LaStarza MW, Lemm JA & Rice CM Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol 68, 5781–5791 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Malet H et al. The Crystal Structures of Chikungunya and Venezuelan Equine Encephalitis Virus nsP3 Macro Domains Define a Conserved Adenosine Binding Pocket. J. Virol 83, 6534–6545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Eckei L et al. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep 7, 1–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Abraham R et al. ADP-ribosyl–binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci 115, E10457–E10466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Abraham R et al. Both ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus nsP3 Macrodomain Affect Neurovirulence in Mice. mBio 11, e03253–19, /mbio/11/1/mBio.03253-19.atom (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gao Y, Goonawardane N, Ward J, Tuplin A & Harris M Multiple roles of the non-structural protein 3 (nsP3) alphavirus unique domain (AUD) during Chikungunya virus genome replication and transcription. PLOS Pathog 15, e1007239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Oberste MS, Parker MD & Smith JF Complete Sequence of Venezuelan Equine Encephalitis Virus Subtype IE Reveals Conserved and Hypervariable Domains within the C Terminus of nsP3. Virology 219, 314–320 (1996). [DOI] [PubMed] [Google Scholar]
- 167.Foy NJ, Akhrymuk M, Shustov AV, Frolova EI & Frolov I Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J. Virol 87, 7569–7584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Aaskov J, Jones A, Choi W, Lowry K & Stewart E Lineage replacement accompanying duplication and rapid fixation of an RNA element in the nsP3 gene in a species of alphavirus. Virology 410, 353–359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Foy NJ et al. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J. Virol 87, 1997–2010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim DY et al. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog. 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Montero H & Trujillo-Alonso V Stress Granules in the Viral Replication Cycle. Viruses 3, 2328–2338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Valiente-Echeverria F, Melnychuk L & Mouland AJ Viral modulation of stress granules. Virus Res. 169, 430–437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Frolov I, Kim DY, Akhrymuk M, Mobley JA & Frolova EI Hypervariable Domain of Eastern Equine Encephalitis Virus nsP3 Redundantly Utilizes Multiple Cellular Proteins for Replication Complex Assembly. J. Virol 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Götte B, Utt A, Fragkoudis R, Merits A & McInerney GM Sensitivity of Alphaviruses to G3BP Deletion Correlates with Efficiency of Replicase Polyprotein Processing. J. Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Meshram CD et al. Mutations in Hypervariable Domain of Venezuelan Equine Encephalitis Virus nsP3 Protein Differentially Affect Viral Replication. J. Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Myles KM, Kelly CLH, Ledermann JP & Powers AM Effects of an Opal Termination Codon Preceding the nsP4 Gene Sequence in the O’Nyong-Nyong Virus Genome on Anopheles gambiae Infectivity. J. Virol 80, 4992–4997 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li GP & Rice CM Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol 63, 1326–1337 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.de Groot RJ, Rümenapf T, Kuhn RJ, Strauss EG & Strauss JH Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. U. S. A 88, 8967–8971 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lello LS et al. nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity. J. Virol 95,e00355–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rubach JK et al. Characterization of purified Sindbis Virus nsP4 RNA-dependent RNA Polymerase activity in vitro. Virology 384, 201–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tomar S, Hardy RW, Smith JL & Kuhn RJ Catalytic Core of Alphavirus Nonstructural Protein nsP4 Possesses Terminal Adenylyltransferase Activity. J. Virol 80, 9962–9969 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sreejith R et al. Mapping interactions of Chikungunya virus nonstructural proteins. Virus Res. 169, 231–236 (2012). [DOI] [PubMed] [Google Scholar]
- 183.Tan YB et al. Crystal structures of alphavirus nonstructural protein 4 (nsP4) reveal an intrinsically dynamic RNA-dependent RNA polymerase fold. Nucleic Acids Res. 50, 1000–1016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chung D-H et al. Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstmctural Protein 2. PLOS Pathog. 10, e1004213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rupp JC, Jundt N & Hardy RW Requirement for the amino-terminal domain of sindbis virus nsP4 during virus infection. J Virol 85, 3449–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gritsenko D & Hughes G Ledipasvir/Sofosbuvir (Harvoni): Improving Options for Hepatitis C Virus Infection. Pharm. Ther 40, 256–276 (2015). [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang S et al. Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design. PLOS ONE 16, e0245013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sundar S, Piramanayagam S & Natarajan J A review on stmctural genomics approach applied for drug discovery against three vector-borne viral diseases: Dengue, Chikungunya and Zika. Virus Genes 58, 151–171 (2022). [DOI] [PubMed] [Google Scholar]
- 189.Kovacikova K & van Hemert MJ Small-Molecule Inhibitors of Chikungunya Virus: Mechanisms of Action and Antiviral Drug Resistance. Antimicrob. Agents Chemother 64, e01788–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kehn-Hall K & Bradfute SB Understanding host responses to equine encephalitis virus infection: implications for therapeutic development. Expert Rev. Anti Infect. Ther 0, 1–16 (2022). [DOI] [PubMed] [Google Scholar]
- 191.Kim DY, Firth AE, Atasheva S, Frolova EI & Frolov I Conservation of a Packaging Signal and the Viral Genome RNA Packaging Mechanism in Alphavirus Evolution. J. Virol 85, 8022–8036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Frolova E, Frolov I & Schlesinger S Packaging signals in alphaviruses. J. Virol 71, 248–258 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Volkova E, Gorchakov R & Frolov I The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1–3 synthesis. Virology 344, 315–327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mendes A & Kuhn RJ Alphavirus Nucleocapsid Packaging and Assembly. Viruses 10, 138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Garmashova N et al. The Old World and New World Alphaviruses Use Different Virus-Specific Proteins for Induction of Transcriptional Shutoff. J. Virol 81, 2472–2484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Carrasco L, Sanz MA & González-Almela E The Regulation of Translation in Alphavirus-Infected Cells. Viruses 10, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Akhrymuk I, Frolov I & Frolova EI Sindbis Virus Infection Causes Cell Death by nsP2-Induced Transcriptional Shutoff or by nsP3-Dependent Translational Shutoff. J. Virol 92, e01388–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rao S & Taylor A Arthritogenic Alphavirus Capsid Protein. Life 11, 230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Akhrymuk I, Kulemzin SV & Frolova EI Evasion of the Innate Immune Response: the Old World Alphavirus nsP2 Protein Induces Rapid Degradation of Rpb1, a Catalytic Subunit of RNA Polymerase II. J. Virol 86, 7180–7191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.DeBono A et al. Novel RU486 (mifepristone) analogues with increased activity against Venezuelan Equine Encephalitis Virus but reduced progesterone receptor antagonistic activity. Sci. Rep 9, 1–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Curtis I. de & Simons K Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci 85, 8052–8056 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ivanova L & Schlesinger MJ Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol (1993) doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mulvey M & Brown DT Formation and rearrangement of disulfide bonds during maturation of the Sindbis virus E1 glycoprotein. J. Virol (1994) doi: 10.1128/jvi.68.2.805-812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pletnev SV et al. Locations of Carbohydrate Sites on Alphavirus Glycoproteins Show that E1 Forms an Icosahedral Scaffold. Cell 105, 127–136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang X, Fugère M, Day R & Kielian M Furin Processing and Proteolytic Activation of Semliki Forest Virus. J. Virol (2003) doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang X & Kielian M Mutations that promote furin-independent growth of Semliki Forest virus affect p62–E1 interactions and membrane fusion. Virology 327, 287–296 (2004). [DOI] [PubMed] [Google Scholar]
- 207.Soonsawad P et al. Stmctural Evidence of Glycoprotein Assembly in Cellular Membrane Compartments prior to Alphavirus Budding. J. Virol 84, 11145–11151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Holmes AC, Basore K, Fremont DH & Diamond MS A molecular understanding of alphavirus entry. PLOS Pathog. 16, e1008876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mukhopadhyay S et al. Mapping the Structure and Function of the E1 and E2 Glycoproteins in Alphaviruses. Structure 14, 63–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wengler G, Koschinski A, Wengler G & Dreyer F Entry of alphaviruses at the plasma membrane converts the viral surface proteins into an ion-permeable pore that can be detected by electrophysiological analyses of whole-cell membrane currents. J. Gen. Virol 84, 173–181 (2003). [DOI] [PubMed] [Google Scholar]
- 211.Bonatti S & Blobel G Absence of a cleavable signal sequence in Sindbis virus glycoprotein PE2. J. Biol. Chem 254, 12261–12264 (1979). [PubMed] [Google Scholar]
- 212.Jain SK, DeCandido S & Kielian M Processing of the p62 envelope precursor protein of Semliki Forest virus. J. Biol. Chem 266, 5756–5761 (1991). [PubMed] [Google Scholar]
- 213.Lobigs M, Zhao HX & Garoff H Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol 64, 4346–4355 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Snyder AJ & Mukhopadhyay S The Alphavirus E3 Glycoprotein Functions in a Clade-Specific Manner. J. Virol (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Uchime O, Fields W & Kielian M The Role of E3 in pH Protection during Alphavirus Assembly and Exit. J. Virol (2013) doi: 10.1128/JVI.01507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ramsey J & Mukhopadhyay S Disentangling the Frames, the State of Research on the Alphavirus 6K and TF Proteins. Viruses 9, 228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Harrington HR et al. Cotranslational folding stimulates programmed ribosomal frameshifting in the alphavirus structural polyprotein. J. Biol. Chem 295, 6798–6808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gaedigk-Nitschko K & Schlesinger MJ Site-directed mutations in sindbis virus E2 glycoprotein’s cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology 183, 206–214 (1991). [DOI] [PubMed] [Google Scholar]
- 219.Ivanova L, Le L & Schlesinger MJ Characterization of revertants of a Sindbis virus 6K gene mutant that affects proteolytic processing and virus assembly. Virus Res. 39, 165–179 (1995). [DOI] [PubMed] [Google Scholar]
- 220.Ivanova L, Lustig S & Schlesinger MJ A Pseudo-Revertant of a Sindbis Virus 6K Protein Mutant, Which Corrects for Aberrant Particle Formation, Contains Two New Mutations That Map to the Ectodomain of the E2 Glycoprotein. Virology 206, 1027–1034 (1995). [DOI] [PubMed] [Google Scholar]
- 221.Schlesinger MJ, London SD & Ryan C An In-Frame Insertion into the Sindbis Virus 6K Gene Leads to Defective Proteolytic Processing of the Virus Glycoproteins, a Trans-Dominant Negative Inhibition of Normal Virus Formation, and Interference in Virus Shut off of Host-Cell Protein Synthesis. Virology 193, 424–432 (1993). [DOI] [PubMed] [Google Scholar]
- 222.Yao JS, Strauss EG & Strauss JH Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol (1996) doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Firth AE, Chung BY, Fleeton MN & Atkins JF Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J 5, 108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ramsey J, Renzi EC, Arnold RJ, Trinidad JC & Mukhopadhyay S Palmitoylation of Sindbis Virus TF Protein Regulates Its Plasma Membrane Localization and Subsequent Incorporation into Virions. J. Virol 91, e02000–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rogers KJ, Jones-Burrage S, Maury W & Mukhopadhyay S TF protein of Sindbis virus antagonizes host type I interferon responses in a palmitoylation-dependent manner. Virology 542, 63–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ramsey J, Chavez M & Mukhopadhyay S Domains of the TF protein important in regulating its own palmitoylation. Virology 531, 31–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liljeström P, Lusa S, Huylebroeck D & Garoff H In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol (1991) doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Garmashova N et al. Analysis of Venezuelan Equine Encephalitis Virus Capsid Protein Function in the Inhibition of Cellular Transcription. J. Virol 81, 13552–13565 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jose J, Taylor AB & Kuhn RJ Spatial and Temporal Analysis of Alphavirus Replication and Assembly in Mammalian and Mosquito Cells. mBio 8, e02294–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Andersen PI et al. Discovery and development of safe-in-man broad-spectrum antiviral agents.. Int. J. Infect. Dis 93, 268–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chung D-H et al. Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2. PLOS Pathog. 10, e1004213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Promega. CellTiter-Glo® 2.0 Cell Viability Assay | ATP Assay | Promega. https://www.promega.com/products/cell-health-assays/cell-viability-and-cytotoxicity-assays/celltiter_glo-2_0-assay/ (2022).
- 233.Andersen PI et al. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis 93, 268–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bernatchez JA et al. Development and Validation of a Phenotypic High-Content Imaging Assay for Assessing the Antiviral Activity of Small-Molecule Inhibitors Targeting Zika Virus. Antimicrob. Agents Chemother 62, e00725–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bulanova D et al. Antiviral Properties of Chemical Inhibitors of Cellular Anti-Apoptotic Bcl-2 Proteins. Viruses 9, 271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Aggarwal M et al. Evaluation of antiviral activity of piperazine against Chikungunya virus targeting hydrophobic pocket of alphavirus capsid protein. Antiviral Res. 146, 102–111 (2017). [DOI] [PubMed] [Google Scholar]
- 237.Shen L et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol 93, e00023–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Saotome K, Morita H & Umeda M Cytotoxicity test with simplified crystal violet staining method using microtitre plates and its application to injection drugs. Toxicol. In Vitro 3, 317–321 (1989). [DOI] [PubMed] [Google Scholar]
- 239.Li J-Q et al. Development of a replicon cell line-based high throughput antiviral assay for screening inhibitors of Zika virus. Antiviral Res. 150, 148–154 (2018). [DOI] [PubMed] [Google Scholar]
- 240.Cruz DJM et al. Identification of Novel Compounds Inhibiting Chikungunya Virus-Induced Cell Death by High Throughput Screening of a Kinase Inhibitor Library. PLoS Negl. Trop. Dis 7, e2471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Delekta PC et al. The Combined Use of Alphavirus Replicons and Psuedoinfectious Particles for the Discovery of Antivirals Derived from Natural Products. J. Biomol. Screen. 20, 673–680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pohjala L et al. Inhibitors of Alphavirus Entry and Replication Identified with a Stable Chikungunya Replicon Cell Line and Virus-Based Assays. PLOS ONE 6, e28923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Varghese FS et al. The Antiviral Alkaloid Berberine Reduces Chikungunya Virus-Induced Mitogen-Activated Protein Kinase Signaling. J. Virol 90, 9743–9757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gleiser CA, Gouchenour WS Jr, Berge TO & Tigertt WD The Comparative Pathology of Experimental Venezuelan Equine Encephalomyelitis Infection in Different Animal Hosts. J. Infect. Dis 110, 80–97 (1962). [DOI] [PubMed] [Google Scholar]
- 245.Jackson AC, Sengupta SK & Smith JF Pathogenesis of Venezulean Equine Encephalitis Virus Infection in Mice and Hamsters. Vet. Pathol 410–418 (1991). [DOI] [PubMed] [Google Scholar]
- 246.Victor J, Smith DG & Pollack AD The Comparative Pathology of Venezuelan Equine Encephalomyelitis. J. Infect. Dis 98, 55–66 (1956). [DOI] [PubMed] [Google Scholar]
- 247.Biodefense: Research Methodology and Animal Models. (CRC Press, 2005). doi: 10.1201/9781420038118. [DOI] [Google Scholar]
- 248.Vogel P et al. Venezuelan equine encephalitis in BALB/c mice: kinetic analysis of central nervous system infection following aerosol or subcutaneous inoculation. Arch. Pathol. Lab. Med 120, 164–172 (1996). [PubMed] [Google Scholar]
- 249.Charles PC, Walters E, Margolis F & Johnston RE Mechanism of Neuroinvasion of Venezuelan Equine Encephalitis Virus in the Mouse. Virology 208, 662–671 (1995). [DOI] [PubMed] [Google Scholar]
- 250.Davis NL et al. A molecular genetic approach to the study of Venezuelan equine encephalitis virus pathogenesis, in Positive-Strand RNA Viruses (eds. Brinton MA, Calisher CH & Rueckert R) 99–109 (Springer, 1994). doi: 10.1007/978-3-7091-9326-6_l1. [DOI] [PubMed] [Google Scholar]
- 251.Grieder FB et al. Specific Restrictions in the Progression of Venezuelan Equine Encephalitis Virus-Induced Disease Resulting from Single Amino Acid Changes in the Glycoproteins. Virology 206, 994–1006 (1995). [DOI] [PubMed] [Google Scholar]
- 252.MacDonald GH & Johnston RE Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol 74, 914–922 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Steele KE et al. Comparative Neurovirulence and Tissue Tropism of Wild-type and Attenuated Strains of Venezuelan Equine Encephalitis Virus Administered by Aerosol in C3H/HeN and BALB/c Mice. Vet. Pathol 35, 386–397 (1998). [DOI] [PubMed] [Google Scholar]
- 254.Steele KE & Twenhafel NA REVIEW PAPER: Pathology of Animal Models of Alphavirus Encephalitis. Vet. Pathol 47, 790–805 (2010). [DOI] [PubMed] [Google Scholar]
- 255.Ryzhikov AB, Tkacheva NV, Sergeev AN & Ryabchikova EI Venezuelan equine encephalitis virus propagation in the olfactory tract of normal and immunized mice. Biomed. Sci 2, 607–614 (1991). [PubMed] [Google Scholar]
- 256.Danes L, Rychterová V, Kufner J & Hrusková V The role of the olfactory route on infection of the respiratory tract with Venezuelan equine encephalomyelitis virus in normal and operated Macaca rhesus monkeys. II. Results of histological examination. Acta Virol. 17, 57–60 (1973). [PubMed] [Google Scholar]
- 257.Smith DR et al. Comparative pathology study of Venezuelan, eastern, and western equine encephalitis viruses in non-human primates. Antiviral Res. 182, 104875 (2020). [DOI] [PubMed] [Google Scholar]
- 258.Liu C, Voth DW, Rodina P, Shauf LR & Gonzalez G A Comparative Study of the Pathogenesis of Western Equine and Eastern Equine Encephalomyelitis Viral Infections in Mice by Intracerebral and Subcutaneous Inoculations. J. Infect. Dis 122, 53–63 (1970). [DOI] [PubMed] [Google Scholar]
- 259.Gardner CL et al. Eastern and Venezuelan Equine Encephalitis Viruses Differ in Their Ability To Infect Dendritic Cells and Macrophages: Impact of Altered Cell Tropism on Pathogenesis. J. Virol 82, 10634–10646 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Reed DS et al. Severe Encephalitis in Cynomolgus Macaques Exposed to Aerosolized Eastern Equine Encephalitis Virus. J. Infect. Dis 196, 441–450 (2007). [DOI] [PubMed] [Google Scholar]
- 261.Adams AP et al. Common Marmosets (Callithrix jacchus) as a Nonhuman Primate Model To Assess the Virulence of Eastern Equine Encephalitis Virus Strains. J. Virol 82, 9035–9042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Espinosa BJ et al. Susceptibility of the Aotus nancymaae owl monkey to eastern equine encephalitis. Vaccine 27, 1729–1734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Nathanson N, Stolley PD & Boolukos PJ Eastern equine encephalitis: Distribution of central nervous system lesions in man and rhesus monkey. J. Comp. Pathol 79, 109–115 (1969). [DOI] [PubMed] [Google Scholar]
- 264.Trefry JC et al. The utilization of advance telemetry to investigate critical physiological parameters including electroencephalography in cynomolgus macaques following aerosol challenge with eastern equine encephalitis virus. PLoS Negl. Trop. Dis 15, e0009424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Aguilar MJ Pathological Changes in Brain and Other Target Organs of Infant and Weanling Mice After Infection with Non-Neuroadapted Western Equine Encephalitis Virus. Infect. Immun 2, 533–542 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Logue CH et al. Virulence variation among isolates of western equine encephalitis virus in an outbred mouse model. J. Gen. Virol 90, 1848–1858 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Monath TP & Kemp GE Necrotizing Myocarditis in Mice Infected with Western Equine Encephalitis Virus: Clinical, Electrocardiographic, and Histopathologic Correlations. J. Infect. Dis 158, 59–66 (1978). [DOI] [PubMed] [Google Scholar]
- 268.Nagata LP et al. Infectivity variation and genetic diversity among strains of Western equine encephalitis virus. J. Gen. Virol 87, 2353–2361 (2022). [DOI] [PubMed] [Google Scholar]
- 269.Hardy JL, Presser SB, Chiles RE & Reeves WC Mouse and Baby Chicken Virulence of Enzootic Strains of Western Equine Encephalomyelitis Virus from California. Am. J. Trop. Med. Hyg 57, 240–244 (1997). [DOI] [PubMed] [Google Scholar]
- 270.Julander JG et al. Effect of exogenous interferon and an interferon inducer on western equine encephalitis virus disease in a hamster model. Virology 360, 454–460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zlotnik I, Peacock S, Grant DP & Batter-Hatton D The Pathogenesis of Westem Equine Encephalitis Virus (W.E.E.) in Adult Hamsters with Special Reference to the Long and Short Term Effects on the C.N.S. of the Attenuated Clone 15 Variant. Br. J. Exp. Pathol 53, 59–77 (1972). [PMC free article] [PubMed] [Google Scholar]
- 272.Reed DS et al. Aerosol exposure to western equine encephalitis virus causes fever and encephalitis in cynomolgus macaques. J. Infect. Dis 192, 1173–1182 (2005). [DOI] [PubMed] [Google Scholar]
- 273.Wyckoff RWG & Tesar WC Equine Encephalomyelitis in Monkeys. J. Immunol 37, 329–343 (1939). [Google Scholar]
- 274.Couderc T et al. A Mouse Model for Chikungunya: Young Age and Inefficient Type-I Interferon Signaling Are Risk Factors for Severe Disease. PLOS Pathog. 4, e29 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Haese NN et al. Animal Models of Chikungunya Virus Infection and Disease. J. Infect. Dis 214, S482–S487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Levitt NH et al. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4, 157–162 (1986). [DOI] [PubMed] [Google Scholar]
- 277.Werneke SW et al. ISG15 Is Critical in the Control of Chikungunya Virus Infection Independent of UbE1L Mediated Conjugation. PLOS Pathog. 7, e1002322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Gardner J et al. Chikungunya Virus Arthritis in Adult Wild-Type Mice. J. Virol 84, 8021–8032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hallengärd D et al. Novel Attenuated Chikungunya Vaccine Candidates Elicit Protective Immunity in C57BL/6 mice. J. Virol 88, 2858–2866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Morrison TE et al. A Mouse Model of Chikungunya Virus–Induced Musculoskeletal Inflammatory Disease: Evidence of Arthritis, Tenosynovitis, Myositis, and Persistence. Am. J. Pathol 178, 32–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hawman DW et al. Chronic Joint Disease Caused by Persistent Chikungunya Virus Infection Is Controlled by the Adaptive Immune Response. J. Virol 87, 13878–13888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Poo YS et al. Multiple Immune Factors Are Involved in Controlling Acute and Chronic Chikungunya Virus Infection. PLoS Negl. Trop. Dis 8, e3354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Seymour RL, Adams AP, Leal G, Alcorn MDH & Weaver SC A Rodent Model of Chikungunya Virus Infection in RAG1 −/− Mice, with Features of Persistence, for Vaccine Safety Evaluation. PLoS Negl. Trop. Dis 9, e0003800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Binn LN, Harrison VR & Randall R Patterns of viremia and antibody observed in rhesus monkeys inoculated with Chikungunya and other serologically related group A arboviruses. Am. J. Trop. Med. Hyg 16, 782–5 (1967). [PubMed] [Google Scholar]
- 285.Kam Y-W et al. Unique Epitopes Recognized by Antibodies Induced in Chikungunya Virus-Infected Non-Human Primates: Implications for the Study of Immunopathology and Vaccine Development. PLOS ONE 9, e95647 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Labadie K et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. https://www.jci.org/articles/view/40104/table/1 (2010) doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed]
- 287.Messaoudi I et al. Chikungunya Virus Infection Results in Higher and Persistent Viral Replication in Aged Rhesus Macaques Due to Defects in Anti-Viral Immunity. PLoS Negl. Trop. Dis 7, e2343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Paul SD & Singh KRP Experimental infection of Macaca radiala with chikungunya virus and transmission of virus by mosquitoes. Indian J. Med. Res 56, 802–11 (1968). [PubMed] [Google Scholar]
- 289.Chen C-I et al. Comparative Pathogenesis of Epidemic and Enzootic Chikungunya Viruses in a Pregnant Rhesus Macaque Model. Am. J. Trop. Med. Hyg 83, 1249–1258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Heise MT, Simpson DA & Johnston RE Sindbis-Group Alphavirus Replication in Periosteum and Endosteum of Long Bones in Adult Mice. J. Virol 74, 9294–9299 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Jackson AC, Morench TR, Griffen DE & Johnson RT Tha Pathogenesis of Spinal Cord Involvement in the Encephalomyelitis of Mice Caused by Neuroadapted Sindbis Virus Infection. Lab. Invest 56, 418–423 (1987). [PubMed] [Google Scholar]
- 292.Klimstra WB et al. Infection of Neonatal Mice with Sindbis Virus Results in a Systemic Inflammatory Response Syndrome. J. Virol 73, 10387–10398 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Orvedahl A et al. Autophagy Protects against Sindbis Virus Infection of the Central Nervous System. Cell Host Microbe 7, 115–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Trgovcich J, Aronson JF & Johnston RE Fatal Sindbis Infection of Neonatal Mice in the Absence of Encephalitis. Virology 73–83 (1996). [DOI] [PubMed] [Google Scholar]
- 295.Jupille HJ et al. Mutations in nsP1 and PE2 are critical determinants of Ross River virus-induced musculoskeletal inflammatory disease in a mouse model. Virology 410, 216–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Morrison TE et al. Characterization of Ross River Virus Tropism and Virus-Induced Inflammation in a Mouse Model of Viral Arthritis and Myositis. J. Virol 80, 737–749 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Morrison TE, Fraser RJ, Smith PN, Mahalingam S & Heise MT Complement Contributes to Inflammatory Tissue Destruction in a Mouse Model of Ross River Virus-Induced Disease. J. Virol 81, 5132–5143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Herrero LJ et al. Characterization of Barmah Forest virus pathogenesis in a mouse model. J. Gen. Virol 95, 2146–2154 (2022). [DOI] [PubMed] [Google Scholar]
- 299.Seymour RL, Rossi SL, Bergren NA, Plante KS & Weaver SC The Role of Innate versus Adaptive Immune Responses in a Mouse Model of O’Nyong-Nyong Virus Infection. Am. J. Trop. Med. Hyg 88, 1170–1179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Santos FM et al. Animal model of arthritis and myositis induced by the Mayaro virus. PLoS Negl. Trop. Dis 13, e0007375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Fazakerley JK Semliki Forest virus infection of laboratory mice: a model to study the pathogenesis of viral encephalitis | SpringerLink. 10.1007/978-3-7091-0572-6_16 (2022). [DOI] [PubMed]
- 302.USDA. USDA APHIS | Equine Encephalitis (EEE/WEE/VEE). https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/equine/eee-wee-vee/equine-encephalitis (2021).
- 303.MERCK. Compendium of Veterinary Products - Encevac® TC-4+VEE (MERCK ANIMAL HEALTH). https://merckusa.cvpservice.com/product/basic/view/1047320 (2021).
- 304.PEDERSEN CE JR, ROBINSON DM & COLE FE JR. ISOLATION OF THE VACCINE STRAIN OF VENEZUELAN EQUINE ENCEPHALOMYELITIS VIRUS FROM MOSQUITOES IN LOUISIANA. Am. J. Epidemiol 95, 490–496 (1972). [DOI] [PubMed] [Google Scholar]
- 305.BERGE TO, BANKS IS & TIGERTT WD ATTENUATION OF VENEZUELAN EQUINE ENCEPHALOMYELITIS VIRUS BY IN VITRO CULTIVATION IN GUINEA-PIG HEART CELLS 1. Am. J. Epidemiol 73, 209–218 (1961). [Google Scholar]
- 306.Pittman PR et al. Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 14, 337–343 (1996). [DOI] [PubMed] [Google Scholar]
- 307.Mckinney RW, Berge TO, Sawyer WD, Tigertt WD & Crozier D Use of an Attenuated Strain of Venezuelan Equine Encephalo-Myelitis Virus for Immunization in Man. Am. J. Trop. Med Hyg 12, 597–603 (1963). [DOI] [PubMed] [Google Scholar]
- 308.Kautz TF et al. Low-fidelity Venezuelan equine encephalitis virus polymerase mutants to improve live-attenuated vaccine safety and efficacy. Virus Evol. 4, vey004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mckinney RW, Berge TO, Sawyer WD, Tigertt WD & Crazier D Use of an Attenuated Strain of Venezuelan Equine Encephalo-Myelitis Virus for Immunization in Man. Am. J. Trop. Med. Hyg 12, 597–603 (1963). [DOI] [PubMed] [Google Scholar]
- 310.Pittman PR & Plotkin SA Biodefense and Special Pathogen Vaccines. Plotkins Vaccines 149–160.e7 (2018) doi: 10.1016/B978-0-323-35761-6.00012-2. [DOI] [Google Scholar]
- 311.Fine DL et al. Neurovirulence evaluation of Venezuelan equine encephalitis (VEE) vaccine candidate V3526 in nonhuman primates. Vaccine 26, 3497–3506 (2008). [DOI] [PubMed] [Google Scholar]
- 312.Holley H Safety of an Attenuated Venezuelan Equine Encephalitis Virus (VEEV) Vaccine in Humans. in (Idsa, 2008). [Google Scholar]
- 313.Steele KE et al. Alphavirus Encephalitides. in Medical Aspects of Biological Warfare 241–270 (Office of the Surgeon General, 2007). [Google Scholar]
- 314.Coates EE et al. Safety and immunogenicity of a trivalent virus-like particle vaccine against western, eastern, and Venezuelan equine encephalitis viruses: a phase 1, open-label, dose-escalation, randomised clinical trial. Lancet Infect. Dis 22, 1210–1220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Stromberg ZR, Fischer W, Bradfute SB, Kubicek-Sutherland JZ & Hraber P Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines 8, 273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Perry JG et al. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med Hyg 62, 681–685 (2000). [DOI] [PubMed] [Google Scholar]
- 317.Stromberg ZR, Fischer W, Bradfute SB, Kubicek-Sutherland JZ & Hraber P Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines 8, 273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Plante K et al. Novel Chikungunya Vaccine Candidate with an IRES-Based Attenuation and Host Range Alteration Mechanism. PLOS Pathog. 7, e1002142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Reyes-Sandoval A 51 years in of Chikungunya clinical vaccine development: A historical perspective. Hum. Vaccines Immunother 15, 2351–2358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hallengärd D et al. Novel Attenuated Chikungunya Vaccine Candidates Elicit Protective Immunity in C57BL/6 mice. J. Virol 88, 2858–2866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Folegatti PM et al. A single dose of ChAdOxl Chik vaccine induces neutralizing antibodies against four chikungunya virus lineages in a phase 1 clinical trial. Nat. Commun 12, 4636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Barber TL, Walton TE & Lewis KJ Efficacy oftrivalent inactivated encephalomyelitis virus vaccine in horses. Am. J. Vet. Res 39, 621–625 (1978). [PubMed] [Google Scholar]
- 323.AAEP. Eastern & Western Equine Encephalomyelitis | AAEP. https://aaep.org/guidelines/vaccination-guidelines/core-vaccination-guidelines/easternwestern-equine-encephalomyelitis (2021).
- 324.Alevizatos AC, McKinney RW & Feigin RD Live, Attenuated Venezuelan Equine Encephalomyelitis Virus Vaccine: I. Clinical Effects in Man. Am. J. Trop. Med. Hyg 16, 762–768 (1967). [PubMed] [Google Scholar]
- 325.Cole FE, May SW & Eddy GA Inactivated Venezuelan Equine Encephalomyelitis Vaccine Prepared from Attenuated (TC-83 Strain) Virus 27, 4 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Martin SS et al. Comparison of the immunological responses and efficacy of gamma-irradiated V3526 vaccine formulations against subcutaneous and aerosol challenge with Venezuelan equine encephalitis virus subtype IAB. Vaccine 28, 1031–1040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Law Y-S et al. Interdomain Flexibility of Chikungunya Virus nsP2 Helicase-Protease Differentially Influences Viral RNA Replication and Infectivity. J. Virol 95, e01470–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Koonin EV, Dolja VV & Morris TJ Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences. Crit. Rev. Biochem. Mol. Biol 28, 375–430 (1993). [DOI] [PubMed] [Google Scholar]
- 329.Mackenzie JM, Jones MK & Westaway EG Markers for trans-Golgi Membranes and the Intermediate Compartment Localize to Induced Membranes with Distinct Replication Functions in Flavivirus-Infected Cells. J. Virol 73, 9555–9567 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lin W, Feng Z, Prasanth KR, Liu Y & Nagy PD Dynamic interplay between the co-opted Fis1 mitochondrial fission protein and membrane contact site proteins in supporting tombusvirus replication. PLOS Pathog 17, e1009423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kutchko KM et al. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res 46, 3657–3670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Madden EA et al. Using SHAPE-MaP To Model RNA Secondary Structure and Identify 3′UTR Variation in Chikungunya Virus. J. Virol 94, e00701–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


