Abstract
Objective
Trisomy 21 is one of the most complex genetic perturbations compatible with postnatal survival. Dosage imbalance arising from the triplication of genes on human chromosome 21 (Hsa21) affects multiple organ systems. Much of Down syndrome (DS) research, however, has focused on addressing how aneuploidy dysregulates CNS function leading to cognitive deficit. Although obesity, diabetes, and associated sequelae such as fatty liver and dyslipidemia are well documented in the DS population, only limited studies have been conducted to determine how gene dosage imbalance affects whole-body metabolism. Here, we conduct a comprehensive and systematic analysis of key metabolic parameters across different physiological states in the Ts65Dn trisomic mouse model of DS.
Methods
Ts65Dn mice and euploid littermates were subjected to comprehensive metabolic phenotyping under basal (chow-fed) state and the pathophysiological state of obesity induced by a high-fat diet (HFD). RNA sequencing of liver, skeletal muscle, and two major fat depots were conducted to determine the impact of aneuploidy on tissue transcriptome. Pathway enrichments, gene-centrality, and key driver estimates were performed to provide insights into tissue autonomous and non-autonomous mechanisms contributing to the dysregulation of systemic metabolism.
Results
Under the basal state, chow-fed Ts65Dn mice of both sexes had elevated locomotor activity and energy expenditure, reduced fasting serum cholesterol levels, and mild glucose intolerance. Sexually dimorphic deterioration in metabolic homeostasis became apparent when mice were challenged with a high-fat diet. While obese Ts65Dn mice of both sexes exhibited dyslipidemia, male mice also showed impaired systemic insulin sensitivity, reduced mitochondrial activity, and elevated fibrotic and inflammatory gene signatures in the liver and adipose tissue. Systems-level analysis highlighted conserved pathways and potential endocrine drivers of adipose-liver crosstalk that contribute to dysregulated glucose and lipid metabolism.
Conclusions
A combined alteration in the expression of trisomic and disomic genes in peripheral tissues contribute to metabolic dysregulations in Ts65Dn mice. These data lay the groundwork for understanding the impact of aneuploidy on in vivo metabolism.
Keywords: Aneuploidy, Trisomy, Down syndrome, Insulin resistance, Diabetes, Obesity
Highlights
-
•
Ts65Dn Down syndrome mice exhibit dysregulated systemic metabolism.
-
•
Obese Ts65Dn mice develop insulin resistance and mild dyslipidemia.
-
•
Obese Ts65Dn mice show reduced mitochondrial activity in liver and fat.
-
•
Ts65Dn male mice have more pronounced phenotypes than female mice.
1. Introduction
The presence of a supernumerary human chromosome 21 (Hsa21) occurs in ∼1/700 live births resulting in Down syndrome (DS) and is the leading cause of intellectual disability [1]. Aneuploidy, due to trisomy 21, is among the most complex genetic perturbations compatible with postnatal survival [2]. Elevated expression of ∼500–600 triplicated genes on Hsa21, combined with their cascading effects on disomic gene expression in other parts of the genome, profoundly influence cellular physiology, leading to functional deficits across multi-organ systems [[3], [4], [5], [6], [7]]. While cognitive deficits, craniofacial dysmorphology, hypotonia, and the development of Alzheimer (AD)-like pathology in mid-life are common core features of DS, additional abnormalities with variable degree of penetrance and expressivity include congenital heart defects, hearing and vision loss, leukemia, reduced bone mass, and gastrointestinal disease [2,8].
Given that intellectual disability is the most prominent feature of DS, much of DS research has centered on understanding the impact of trisomy 21 in the brain [[9], [10], [11], [12], [13], [14], [15]]. As individuals with DS live longer lives [16], other health-related issues also become prominent, including obesity, diabetes, and dyslipidemia [[17], [18], [19], [20]]. While the higher incidence of obesity, insulin resistance, and diabetes in adolescent and adult DS populations is extensively documented [[21], [22], [23], [24], [25]], the underlying cause of impaired metabolic homeostasis in DS remains largely unknown and understudied.
At the cellular level, trisomy 21 has been shown to alter mitochondrial respiration, and this is generally attributed to dosage imbalance arising from Hsa21 genes (e.g., CBS, RCAN1, DSCAM, NRIP1) whose protein products are thought to impair mitochondrial fusion/fission dynamics, and OXPHOS complex expression and/or function [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]]. While these links remain compelling mechanistically, the studies have been performed exclusively in cell culture. It is unclear whether a deficit in mitochondrial activity seen in these cultured cells also holds true in tissues and in metabolically active cell types such as hepatocytes, myocytes, and adipocytes.
Information concerning the impact of trisomy 21 on whole-body metabolism is also limited, with metabolic assessments largely focused on adiposity, food intake, physical activity level, and energy expenditure in adolescent or adult Individuals with DS [[37], [38], [39], [40], [41], [42], [43], [44], [45]]. One study, however, has used phosphorus magnetic resonance spectroscopy to demonstrate a functional deficit in the skeletal muscle mitochondria of adults with DS [46]. It remains unclear whether alterations in these metabolic parameters are the result of specific genetic contexts or lifestyles, or whether altered gene expression due to trisomy 21 in peripheral tissues is causally linked to whole-body metabolic changes seen in DS. Despite the availability of DS mouse models in the past two decades, only a few recent studies reported on how aneuploidy alters baseline (chow-fed) metabolic parameters, and the in vivo response to obesogenic high-fat diet (HFD) [[47], [48], [49], [50]]. A systematic and comprehensive assessment of glucose and lipid metabolism, and other key metabolic parameters, across different physiological states is currently lacking.
Since the generation of the first segmental trisomic Ts65Dn mouse model [[51], [52], [53]], more than twenty additional DS models trisomic for varying amount of orthologous Hsa21 gene contents have been generated [[54], [55], [56], [57], [58], [59], [60], [61]]. Each mouse model has its limitation in recapitulating the full spectrum of human DS. Additionally, two transchromosomic mouse models (Tc1 and TcMA21) carrying Hsa21 have also been generated [62,63]. The Tc1 mouse model appears to have limited utility as the Hsa21 is missing >50 protein-coding genes (PCGs) due to deletions, and it also contains large numbers of structural rearrangements and duplications [64]. In addition, Tc1 mice show extensive mosaicism across tissues (i.e., the human chromosome is randomly lost from cells during development), complicating the results obtained from this mouse model [62]. The TcMAC21 mouse model overcomes the drawbacks of Tc1 in that it is not mosaic and contains a near complete (93%) Hsa21 [63]. Of the DS mouse models generated to date, however, only Ts65Dn, Tc1, and TcMAC21 are true aneuploid models with a freely segregating supernumerary chromosome [65], whereas other DS models have gene dosage imbalance due to intrachromosomal segmental duplications of either mouse chromosome (Mmu)16, Mmu17, or Mmu10 region orthologous to Hsa21 [54].
Behavioral and phenotypic profiling of various DS mouse models have highlighted a variable spectrum of features seen in DS, most notably those related to deficits in learning and memory, motor function, developmental heart defects, reduced bone mass, and altered gastrointestinal and immune function [50,54,57,60,[66], [67], [68], [69], [70], [71], [72], [73], [74], [75]]. In contrast, the metabolic phenotypes of DS mouse models remain largely unexplored. Here, we undertake a systematic analysis across a wide range of metabolic parameters and tissues in Ts65Dn mice under baseline (chow-fed) condition and in the pathophysiological state of obesity induced by chronic high-fat feeding. Notwithstanding the limitations of the Ts65Dn mouse model [[76], [77], [78]], we chose to perform our comprehensive metabolic phenotyping on this DS model for three reasons: 1) Ts65Dn is a true aneuploid model with a freely segregating marker chromosome [53]. The presence of this supernumenary marker chromosome may be important for some of the DS features not found in other DS mouse models with an intrachromosomal segmental duplication [79]; 2) The Ts65Dn model recapitulates many of the features found in DS, in a manner equal to or better than other trisomic mouse models [66,68]; and 3) Ts65Dn is the most widely studied DS mouse model for testing therapeutic interventions [66,[80], [81], [82]]. Although the recently generated TcMAC21 mouse model contains a near complete Hsa21, there is uncertainty regarding the interactions of proteins encoded by human and mouse orthologs, and the presence of a significant number of non-coding human genes (>400) with uncertain effects on the mouse transcriptome. However, this potential issue is obviated in the Ts65Dn mouse model since all the cells express only mouse genes.
Our study shows that obese Ts65Dn mice fed a high-fat diet showed impaired insulin sensitivity, low-grade adipose inflammation, pro-fibrotic gene signature in liver, reduced mitochondrial activity in liver and adipose tissue, and alterations in multiple stress-related pathways. These data provide valuable mechanistic insights concerning the impact of aneuploidy on systemic metabolism in a DS mouse model. This critical baseline information lays the groundwork for future studies addressing which combination of orthologous gene(s) on Hsa21, when triplicated, contribute to dysregulated metabolism at the tissue and organismal level in DS, as well as across different DS mouse models.
2. Materials and methods
2.1. Mouse model
B6EiC3Sn a/A-Ts (1716)65Dn mice (Ts65Dn) were obtained from the Jackson Laboratory (Strain # 001924) and maintained as a C57BL/6J Ei x C3H/HeSnJ (B6EiC3H) advanced intercross. Euploid littermates were used throughout for controls. Mice were fed a standard chow (Envigo; 2018SX) or high-fat diet (HFD; 60% kcal derived from fat, #D12492, Research Diets, New Brunswick, NJ). Mice were housed in polycarbonate cages on a 12h:12h light–dark photocycle with ad libitum access to water and food. Mice were fed either a standard chow or a HFD. For the HFD-fed group, HFD was provided for 16 weeks, beginning at 5 months of age. At termination of the study, all mice were fasted for 2 h and euthanized. Tissues were collected, snap-frozen in liquid nitrogen, and kept at -80 °C until analysis.
All mouse protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine (animal protocol # MO19M481). All animal experiments were conducted in accordance with the National Institute of Health guidelines and followed the standards established by the Animal Welfare Acts.
2.2. Body composition analysis
Body composition analyses for total fat mass, lean mass, and water content were determined using a quantitative magnetic resonance instrument (Echo-MRI-100, Echo Medical Systems, Waco, TX) at the Mouse Phenotyping Core facility at Johns Hopkins University School of Medicine.
2.3. Indirect calorimetry
Chow or HFD-fed Ts65Dn male and female mice and euploid littermates were used for simultaneous assessments of daily body weight change, food intake (corrected for spillage), physical activity, and whole-body metabolic profile in an open flow indirect calorimeter (Comprehensive Laboratory Animal Monitoring System, CLAMS; Columbus Instruments, Columbus, OH) as previously described [83]. In brief, data were collected for three days to confirm mice were acclimatized to the calorimetry chambers (indicated by stable body weights, food intakes, and diurnal metabolic patterns), then data were analyzed for the subsequent three days. Mice were observed with ad libitum access to food, throughout the fasting process, and in response to refeeding. Rates of oxygen consumption (O2; mL·kg−1·h−1) and carbon dioxide production (CO2; mL·kg−1·h−1) in each chamber were measured every 24 min. Respiratory exchange ratio (RER = CO2/O2) was calculated by CLAMS software (version 4.93) to estimate relative oxidation of carbohydrates (RER = 1.0) versus fats (RER = 0.7), not accounting for protein oxidation. Energy expenditure (EE) was calculated as EE = O2 × [3.815 + (1.232 × RER)] and normalized to lean mass. Physical activities (total and ambulatory) were measured by infrared beam breaks in the metabolic chamber. Average metabolic values were calculated per subject and averaged across subjects for statistical analysis by Student's t-test. Meal pattern data were analyzed for average meal frequency and meal size; a meal was defined as being at least 0.04 g and having a post-meal intermeal interval of at least 10 min, as previously described [84,85]. A food intake event was considered a meal only when both criteria were met. Intermeal interval was defined as time between consecutive meals. Satiety ratio was defined as intermeal interval divided by meal size (min/g).
2.4. Body temperature
Deep colonic temperature was measured by inserting a lubricated (Medline, water soluble lubricating jelly, MDS032280) probe (Physitemp, BAT-12 Microprobe Thermometer) into the anus of mice at a depth of 2 cm. Stable numbers were recorded on three separate days in both the dark and light cycle for each mouse.
2.5. Bone length measurements
Tibias were dissected and excess tissue was cleared away. A Mitutoyo Corp. digital caliper (500-196-30) was used to measure end-to-end bone length.
2.6. Glucose, insulin, pyruvate, and lipid tolerance tests
All tolerance tests were conducted as previously described [[86], [87], [88]]. For glucose tolerance tests (GTTs), mice were fasted for 6 h before glucose injection. Glucose (Sigma, St. Louis, MO) was reconstituted in saline (0.9 g NaCl/L) to a final concentration of 1 g/10 mL (for the chow-fed mice) or 2 g/10 mL (for the HFD-fed mice), sterile-filtered, and injected intraperitoneally (i.p.) at 1 mg/g body weight (i.e., 10 μL/g body weight for chow-fed mice or 5 μL/g body weight of HFD-fed mice). Blood glucose was measured at 0, 15, 30, 60, and 120 min after glucose injection using a glucometer (NovaMax Plus, Billerica, MA). Blood was collected at 0, 15, and 30 min time points for serum isolation followed by Insulin ELISA. For insulin tolerance tests (ITTs), food was removed 2 h before insulin injection. 8.62 μL of insulin stock (4 mg/mL; Gibco) was diluted in 10 mL of saline, sterile-filtered, and injected i. p. at 1.0 U/kg body weight (i.e., 10 uL/g body weight). Blood glucose was measured at 0, 15, 30, 60, and 90 min after insulin injection using a glucometer (NovaMax Plus). For pyruvate tolerance tests (PTTs), mice were fasted overnight for 16 h before sodium pyruvate injection. Sodium pyruvate (Sigma) was reconstituted in saline to a final concentration of 1 g/10 mL and injected i. p. into mice at a dose of 1 g/kg body weight (i.e., 10 μL/g body weight). Blood glucose was measured at the indicated time points using a glucometer. For lipid tolerance tests (LTTs), mice were fasted for 12 h and then injected i. p. with 20% emulsified Intralipid (soybean oil; Sigma; 10 μL/g of body weight). Sera were collected via tail bleed using a Microvette® CB 300 (Sarstedt) at 0, 1, 2, 3, and 4 h post-injection. Serum triglyceride levels were quantified using kits from Infinity Triglycerides (Thermo Scientific).
2.7. Fasting-refeeding insulin tests
Mice were fasted overnight (∼16 h) then reintroduced to food as described [85]. Blood glucose was monitored at the 16 h fast time point (time = 0 h refed) and at 1, 2, and 3 h into the refeeding process. Serum was collected at the 16 h fast and 2 h refed time points for insulin ELISA, as well as for the quantification of triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate concentrations.
2.8. Blood and tissue chemistry analysis
Tail vein blood samples were allowed to clot on ice and then centrifuged for 10 min at 10,000×g. Serum samples were stored at −80 °C until analyzed. Serum triglycerides (TG) and cholesterol were measured according to manufacturer's instructions using an Infinity kit (Thermo Fisher Scientific, Middletown, VA). Non-esterified free fatty acids (NEFA) were measured using a Wako kit (Wako Chemicals, Richmond, VA). Serum β-hydroxybutyrate (ketone) concentrations were measured with a StanBio Liquicolor kit (StanBio Laboratory, Boerne, TX). Serum insulin (Crystal Chem, 90,080), T3 (Calbiotech, T3043T-100), and T4 (Calbiotech, T4044T-100) levels were measured by ELISA according to manufacturer's instructions. Serum alanine transaminase (ALT) was quantified using an ALT assay (Abcam, ab105134) according to the manufacturer's instructions. Serum and tissue levels of IL-1β, IL-6, TNF-α, and MCP-1 were quantified using ELISA kits from R&D Systems (Cat # DY401-05, DY406-05, DY410-05, DY479-05) according to the manufacturer's instructions. Pancreatic insulin was isolated from pancreas samples using acid-ethanol extraction. Briefly, pancreatic samples were in a solution of acid-ethanol (1.5% HCl in 70% EtOH) and incubated overnight at −20 °C. They were then homogenized and incubated for an additional night at −20 °C. The following day, samples were centrifuged at 4 °C to pellet debris. The aqueous protein-rich solution was transferred to a new tube and neutralized with 1M Tris pH7.5. Prior to insulin quantification by ELISA, samples were diluted 1:1000 with ELISA sample diluent. Following ELISA quantification of pancreatic insulin (Mercodia, 10-1247-01), protein concentrations of pancreatic samples were quantified using a Bradford assay (Sigma-Aldrich, B6916). Insulin values were then normalized to protein concentration, averaged, and compared across groups. Hydroxyproline assay (Sigma Aldrich, MAK008) was used to quantify total collagen content in liver tissues from Ts65Dn and euploid mice according to the manufacturer's instructions. Lipid peroxidation levels in the liver of Ts65Dn and euploid mice were assessed by the quantification of malondialdehyde (MDA) via Thiobarbituric Acid Reactive Substances (TBARS) assay (Cayman Chemical, 700,870) according to the manufacturer's instructions.
2.9. Serum triglyceride and cholesterol analysis by FPLC
Food was removed for 2–4 h (in the light cycle) prior to blood collection. Sera collected from mice were pooled (n = 8–10/group) and sent to the Mouse Metabolism Core at Baylor College of Medicine for fast protein liquid chromatography (FPLC) separation. A total of 45 fractions were collected, and TG and cholesterol in each fraction were quantified.
2.10. Histology and quantification
Inguinal (subcutaneous) white adipose tissue (iWAT), gonadal (visceral) white adipose tissue (gWAT), liver, and gastrocnemius muscle were dissected and fixed in formalin. Paraffin embedding, tissue sectioning, and staining with hematoxylin and eosin were performed at the Pathology Core facility at Johns Hopkins University School of Medicine. Images were captured with a Keyence BZ-X700 All-in-One fluorescence microscope (Keyence Corp., Itasca, IL). Adipocyte (gWAT and iWAT) and gastrocnemius muscle fiber cross-sectional area (CSA), as well as the total area covered by lipid droplets in hepatocytes were measured on hematoxylin and eosin-stained slides using ImageJ software [89]. For cross sectional area (CSA) measurements, all cells in one field of view at 100× magnification per tissue section per mouse were analyzed. For iWAT and gWAT adipocyte CSA, at least 150 cells were quantified per mouse. For lipid droplet quantification, 3 randomly selected euploid and Ts65n liver samples were imaged at 200× magnification. Cross-sectional area of individual lipid droplets (at least 500 lipid droplets per image) was quantified using custom ImageJ software. For collagen staining of liver fibrosis, tissue sections were stained with Sirius Red and imaged at 100×. Total fibrotic area was quantified using custom ImageJ software from large stitched 100× images of mouse livers. Each stitched image was comprised of at least 50 separate regions of the same liver. All histological sectioning, imaging, and quantification was conducted blinded to genotypes.
2.11. Immunohistochemistry
Ts65Dn and euploid liver sections were deparaffinized with SafeClear (Fisher HealthCare, 23–044,192), rehydrated using graded concentrations of ethanol in water (100% EtOH, 95%, 75%, 50%, then dH2O), then subjected to antigen retrieval with a pressure cooker for 5 min in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH6.0). Samples were stained with a rabbit monoclonal anti-collagen type I primary antibody (Abcam, ab260043) at a concentration of 1:500, followed by a goat anti-rabbit secondary antibody (Invitrogen, Alexa Fluor 488, A11008) at a concentration of 1:1000, and mounted with a coverslip using ProLong Gold antifade reagent with DAPI (Invitrogen, P36935). Images were captured with a Keyence BZ-X700 All-in-One fluorescence microscope (Keyence Corp., Itasca, IL). Total fluorescent area as a percent of total liver area per focal plane was quantified with custom ImageJ software.
2.12. RNA-sequencing analysis
A total of 48 samples were sequenced with paired-end 50-bp reads (2 × 50bp) across two batches that were balanced for genotype (n = 48 across liver, iWAT, gWAT, and skeletal muscle). Raw sequencing reads from all samples were aligned to a custom concatenated Gencode hg38 + mm10 reference genome using HISAT2 2.0.4 [90]. Genes were quantified with featureCounts v1.5.0-p3 [91] to a custom concatenated hg38 + mm10 gtf file within each sample. We retained 20,680 genes with mean RPKM >0.1 across the mouse genome above this cut-off. High-throughput sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA899581.
We interrogated the effects of Ts65Dn genotype within and across tissue, further adjusting for confounders of gene assignment rate and mitochondrial mapping rate, using the limma voom approach [92]. For our across-tissue model, we assumed the same effect of genotype within each tissue (even if the expression of each tissue could vary) using linear mixed effect modeling with the animal ID as a random intercept. We accounted for multiple testing by controlling for the Benjamini-Hochberg false discovery rate across all expressed genes. As a secondary analysis, we modeled the effect of Ts65Dn genotype within each tissue (also adjusting for the same two sequencing-derived confounders as the full model) using linear regression (since there were no repeated measures within a tissue). Gene set enrichment analyses were performed using clusterProfiler [93] using input genes that were marginally significant (at p < 0.05) stratified by directionality – subsequent gene set enrichment results adjusted for multiple testing using the false discovery rate.
2.13. Respirometry of frozen tissue samples
Respirometry was conducted on frozen tissue samples to assay for mitochondrial activity as described previously [94]. Briefly, all tissues were dissected, snap frozen in liquid nitrogen, and stored at −80 °C for later analysis. Samples were thawed in MAS buffer (70 mM sucrose, 220 mM mannitol, 5 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA, 2 mM HEPES pH 7.4), finely minced with scissors, and then homogenized with a glass Dounce homogenizer. The resulting homogenate was spun at 1000 g for 10 min at 4 °C. The supernatant was collected and immediately used for protein quantification by BCA assay (Thermo Scientific, 23,225). Each well of the Seahorse microplate was loaded with 8 μg of homogenate protein for all tissue types. Each biological replicate is comprised of three technical replicates. Samples from all tissues were treated separately with NADH (1 mM) as a complex I substrate or Succinate (a complex II substrate, 5 mM) in the presence of rotenone (a complex I inhibitor, 2 μM), then with the inhibitors rotenone (2 μM) and Antimycin A (4 μM), followed by TMPD (0.45 mM) and Ascorbate (1 mM) to activate complex IV, and finally treated with Azide (40 mM) to assess non-mitochondrial respiration.
Mitochondrial content of homogenates used for respirometry was quantified using MitoTracker Deep Red (MTDR, ThermoFisher, M22426) as described [94]. Briefly, lysates were incubated with MTDR (1 μM) for 10 min at 37 °C, then centrifuged at 2000 g for 5 min at 4 °C. The supernatant was carefully removed and replaced with 1× MAS solution and fluorescence was read with excitation and emission wavelengths of 625 nm and 670 nm, respectively. To minimize non-specific background signal contribution, control wells were loaded with MTDR and 1× MAS and subtracted from all sample values.
2.14. Pan-tissue RNA-seq integration with human data
A detailed walk-through of all scripts and datasets used are available at: https://github.com/Leandromvelez/ts65Dn-gene-integration. Briefly, human orthologues were identified for mouse tissue-specific differentially expressed genes (logistic regression p-value <0.01) [95]. To visualize co-clustering of these genes in GTEx [96], all orthologues were correlated using the biweight midcorrelation (bicor) coefficient to account for outlier inflation [97] and hierarchically clustered using R packages ‘pheatmap’ and ‘hclust’. Next, gene and pathway centrality estimates were calculated by correlating all genes in the 4 tissues with Ts65Dn DEGs (also using bicor coefficient and associated regression p-value) and adjusting these p-value for FDR <0.01 using the R package ‘qvalue’. The top-ranked 500 genes correlated in each tissue were identified based on average of -log10 (qvalue) with all Ts65Dn DEGs, then used for pathway enrichments among Gene Ontology, KEGG and Reactome databases via overrepresentation tests on WebGestalt [98]. Given that these data consist of pan-tissue RNA-seq, the genome was used as a background set. Top-ranked secreted proteins enriched for these processes were identified by intersection of the top-ranked correlated gene set based on average of -log10 (qvalue) with all Ts65Dn DEGs with known secreted proteins annotated from UniProt [99]. Based on these, undirected networks were constructed based on the distribution of bicor coefficients between central secreted proteins and tissue-specific pathways identified via GO Terms using the R package ‘qgraph’.
2.15. Statistical analyses
All results are expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed with Prism 9 software (GraphPad Software, San Diego, CA). Data were analyzed with two-tailed Student's t-tests or by repeated measures ANOVA. For two-way ANOVA, we performed Sidek or Bonferroni post hoc tests. P < 0.05 was considered statistically significant.
3. Results
3.1. Chow-fed Ts65Dn mice have elevated physical activity and energy expenditure
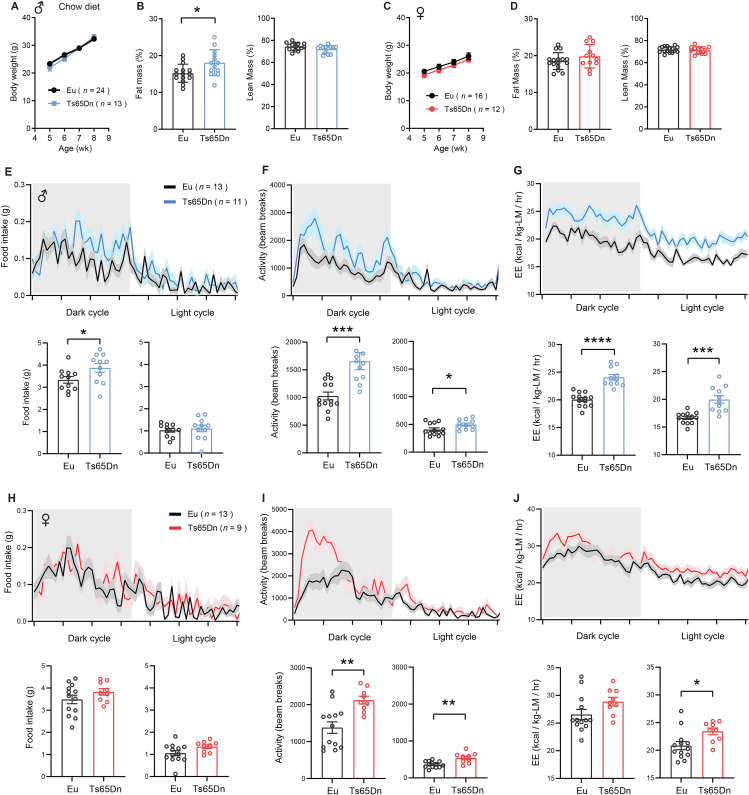
From 5 weeks of age onward, chow-fed Ts65Dn mice of both sexes had body weights indistinguishable from euploid littermates (Figure 1A,C). Body composition analysis by NMR method revealed greater adiposity in male Ts65Dn mice, but no difference in lean mass (Figure 1B). Unlike male mice, in female mice neither fat mass nor lean mass differed between Ts65Dn and euploid littermates (Figure 1D). Indirect calorimetry was conducted to assess food intake, physical activity level, metabolic rate (VO2), and energy expenditure across the circadian cycle (light and dark phase) and metabolic states (ad libitum fed, fasted, and refed). In the dark (active) phase of the circadian cycle, ad libitum fed Ts65Dn male mice consumed a greater amount of food (Figure 1E), and had significantly elevated physical activity level and energy expenditure in both the dark and light cycle (Figure 1F,G). Ts65Dn male mice also showed increased physical activity relative to euploid controls during the 24 h periods of fasting and refeeding (Figures S1 and S2). Interestingly, despite higher physical activity and energy expenditure, chow-fed male mice had lower body temperature during the dark (active) cycle (Figure S3).
Figure 1.
Chow-fed Ts65Dn mice have elevated physical activity and energy expenditure. A) Body weight of chow-fed male Ts65Dn and euploid (Eu) mice over time. B) Relative (%) fat and lean mass after normalization to body weight in male mice. C) Body weight of chow-fed female Ts65Dn and euploid (Eu) mice over time. D) Relative (%) fat and lean mass after normalization to body weight in female mice. E-G) Food intake (E), total physical activity level (F), and energy expenditure (G) of male Ts65Dn and euploid mice across the circadian cycle. Bar graphs indicate cumulative food intake, physical activity, and energy expenditure in the dark and light photocycle. H-J) Food intake (H), total physical activity level (I), and energy expenditure (J) of female Ts65Dn and euploid mice across the circadian cycle. Bar graphs indicate cumulative food intake, physical activity, and energy expenditure in the dark and light photocycle. All data are presented as mean ± SEM. Male mice (Eu, n = 24; Ts65Dn, n = 13). Female mice (Eu, n = 16; Ts65Dn, n = 12). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Food intake in ad libitum chow-fed Ts65Dn female mice, however, was not different from euploid controls (Figure 1H). Like the male mice, Ts65Dn female mice also had elevated physical activity level and energy expenditure relative to euploid controls (Figure 1I–J). Ts65Dn female mice also showed increased physical activity relative to euploid controls during the 24 h periods of fasting and refeeding (Figures S1 and S2). Although energy expenditure continued to be elevated in Ts65Dn female mice during the fasting period, it was not different from euploid controls during the refeeding period (Figures S1 and S2). Body temperatures were not different between genotypes in female mice (Figure S3). These data indicate that, at baseline, chow-fed Ts65Dn mice of either sex are physically more active and expend a greater amount of energy.
3.2. Reduced fasting cholesterol level and mild glucose intolerance in chow-fed Ts65Dn mice
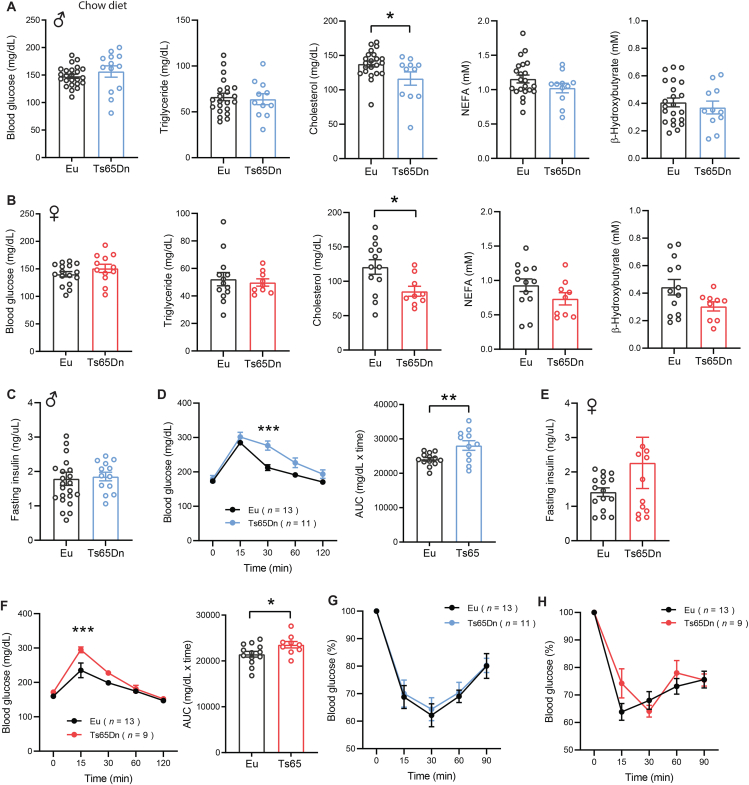
To assess baseline glucose and lipid profile, we measured serum glucose, triglyceride (TG), cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate (BHB) in overnight fasted (16 h) mice. While fasting serum glucose, TG, NEFA, and BHB levels were not different between genotypes of either sex, fasting serum cholesterol levels were significantly lower in both male and female Ts65Dn mice relative to euploid controls (Figure 2A–B). Fasting insulin levels were also not different between genotypes of either sex (Figure 2C,E). We performed glucose tolerance tests to determine whether there is a deficit in glucose clearance in response to an acute glucose loading. Both male and female Ts65Dn mice exhibited mild glucose intolerance (Figure 2D,F). The modest impairments in the kinetics of glucose disposal could be due to mild insulin resistance. We therefore conducted insulin tolerance tests to directly assess insulin sensitivity in these animals. Insulin-stimulated glucose disposal in peripheral tissues, however, was not different between genotypes of either sex (Figure 2G,H). These data indicate that, at baseline, Ts65Dn mice of either sex have altered cholesterol metabolism and a modest impairment in glucose homeostasis.
Figure 2.
Reduced fasting cholesterol level and mild glucose intolerance in chow-fed Ts65Dn mice. A-B) Fasting blood glucose, serum triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate (BHB) in male (A) and female (B) Ts65Dn mice and euploid (Eu) littermate controls. Sample size for male mice (Eu, n = 22; Ts65Dn, n = 11) and female mice (Eu, n = 13; Ts65Dn, n = 9). C) Fasting serum insulin levels in male Ts65Dn (n = 13) and euploid (n = 24) mice. D) Glucose tolerance tests showing glucose clearance over time in male mice after intraperitoneal injection of glucose. The bar graph indicates area-under-curve (AUC), reflecting cumulative glucose disposal. E) Fasting serum insulin levels in female Ts65Dn (n = 12) and euploid (n = 16) mice. F) Glucose tolerance tests showing glucose clearance over time in female mice after intraperitoneal injection of glucose. The bar graph indicates area-under-curve (AUC), reflecting cumulative glucose disposal. G-H) Insulin tolerance tests showing glucose disposal over time in response to intraperitoneal injection of insulin in male (G) and female (H) Ts65Dn and euploid mice. All data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. For glucose tolerance tests, data were analyzed by 2-way ANOVA with Sidek post hoc tests.
3.3. Sexually dimorphic response to high-fat diet in Ts65Dn mice
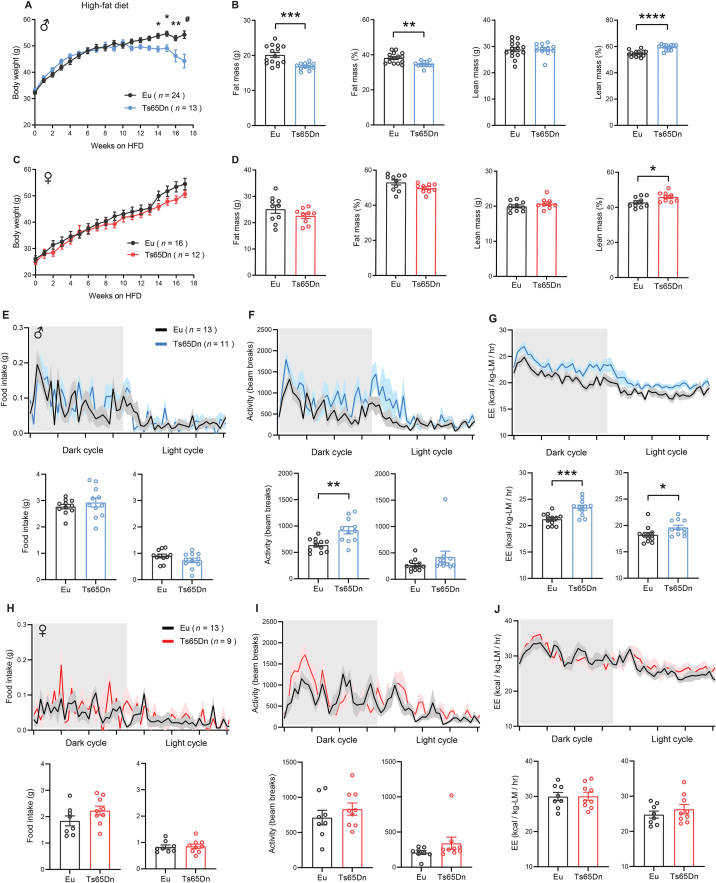
Since obesity is well documented in the DS population [22], we challenged the mice with a high-fat diet (HFD) to induce obesity and assessed how Ts65Dn mice respond to a high caloric diet. For the first eleven weeks of high-fat feeding, the body weight gain of Ts65Dn male mice was not different from euploid controls (Figure 3A). However, beginning at week 12 on HFD, the average body weight of Ts65Dn male mice started to plateau and by week 14 onward, we observed a reduction in body weight. In contrast, euploid male mice continued to gain weight over the entire 16-week period of high-fat feeding (Figure 3A). Body composition analysis at week 12 on HFD showed that Ts65Dn male mice have lower adiposity and a modest increase in lean mass (after normalization to body weight) (Figure 3B). Unlike the male mice, body weight gain in Ts65Dn female mice over the entire 16-week period on HFD was not different from euploid controls (Figure 3C). Body composition analysis of Ts65Dn female mice at week 12 on HFD revealed no difference in adiposity but a modest increase in lean mass (after normalization to body weight) (Figure 3D).
Figure 3.
Sexually dimorphic response to high-fat die in Ts65Dn mice. A) Body weight of male Ts65Dn and euploid (Eu) mice over time fed a high-fat diet. B) Body composition analysis of absolute and relative (%) fat and lean mass in male Ts65Dn (n = 15) and euploid (n = 12) mice. C) Body weight of female Ts65Dn and euploid (Eu) mice over time fed a high-fat diet. D) Body composition analysis of absolute and relative (%) fat and lean mass in female Ts65Dn (n = 10) and euploid (n = 9) mice. E-G) Food intake (E), total physical activity level (F), and energy expenditure (G) of male Ts65Dn and euploid mice across the circadian cycle. Bar graphs indicate cumulative food intake, physical activity, and energy expenditure in the dark and light photocycle. H-J) Food intake (H), total physical activity level (I), and energy expenditure (J) of female Ts65Dn and euploid mice across the circadian cycle. Bar graphs indicate cumulative food intake, physical activity, and energy expenditure in the dark and light photocycle. All data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001. For body weight over time, data were analyzed by 2-way ANOVA with Sidek post hoc tests.
We performed indirect calorimetry analysis on mice after they had been fed HFD for 12 weeks. While food intake in ad libitum fed Ts65Dn male mice was not different from euploid controls (Figure 3E), physical activity and energy expenditure were significantly higher (Figure 3F–G). Elevated physical activity in Ts65Dn male mice persisted during the fasting and refeeding period, whereas energy expenditure remained higher during the refeeding, but not fasting, period (Figures S4 and S5). Body temperatures and serum triiodothyronine (T3; active form) and thyroxine (T4; precursor of T3) levels that could affect activity and energy expenditure were not different between genotypes (Figure S6). Although the amount of food consumed was not different between genotypes, a recent study showed that Ts65Dn mice have altered swallowing function [100]. Therefore, we performed detailed meal pattern analyses to assess if there is any difference in the microstructure of food intake behavior. None of the parameters assessed (meal number, meal size, meal duration, intermeal interval, ingestion rate, satiety ratio) were significantly different between genotypes in male mice (Figure S7). In contrast to the male mice, we did not observe a significant difference in food intake, physical activity, and energy expenditure across different metabolic states (ad libitum fed, fasted, refed) between genotypes in female mice (Figure 3H–J and Figure S4 and S5). Meal pattern analysis also indicated no difference between genotypes in female mice (Figure S8). Taken together, these data indicate that male and female Ts65Dn mice respond differently to chronic high-fat feeding in terms of weight gain, physical activity, and energy expenditure.
3.4. Ts65Dn male mice develop insulin resistance in response to a high-fat diet
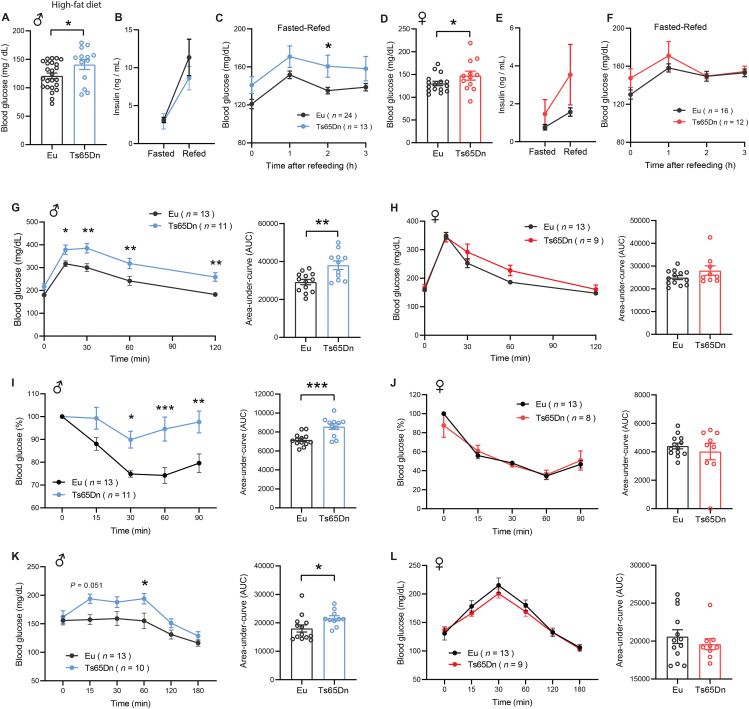
Individuals with DS have higher prevalence of type 2 diabetes [21,23,24]. We therefore addressed whether diet-induced obesity differentially affects glucose homeostasis in Ts65Dn mice fed an HFD. In overnight fasted (16 h) male mice, blood glucose was significantly higher in Ts65Dn relative to euploid controls (Figure 4A). Fasting insulin was not different between genotypes (Figure 4B). Total pancreatic insulin content was also not different between genotypes (Figure S9), suggesting that no overt developmental alteration in the pancreatic islets of Ts65Dn mice that could affect insulin production. While insulin secretion in response to fasting-refeeding was not different between genotypes in male mice (Figure 4B), blood glucose during the first 3 h of refeeding following a fast was consistently higher (Figure 4C), suggesting impaired insulin action. Although female Ts65Dn mice had higher fasting blood glucose levels (Figure 4D), their fasting serum insulin levels, total pancreatic insulin content, insulin secretion in response to fasting-refeeding, and blood glucose over the first 3 h of refeeding were not different from euploid controls (Figure 4E–F and Figure S9).
Figure 4.
Ts65Dn male mice develop insulin resistance in response to a high-fat diet. A) Fasting blood glucose in male Ts65Dn (n = 13) and euploid (n = 24) mice. B) Fasting serum insulin levels in male Ts65Dn (n = 11) and euploid (n = 10) mice. C) Serum glucose levels in overnight fasted and refed (3 h) male Ts65Dn (n = 13) and euploid (n = 24) mice. D) Blood glucose levels in overnight fasted and refed male Ts65Dn (n = 13) and euploid mice (n = 24). E) Fasting serum insulin levels in female Ts65Dn (n = 9) and euploid (n = 11) mice. F) Serum glucose levels in overnight fasted and refed (3 h) female Ts65Dn (n = 12) and euploid (n = 16) mice. G-H) Glucose tolerance tests showing glucose clearance over time in male (G) and female (H) mice after intraperitoneal injection of glucose. The bar graphs indicate area-under-curve (AUC), reflecting cumulative glucose disposal. I-J) Insulin tolerance tests showing glucose disposal over time in male (I) and female (J) Ts65Dn and euploid mice in response to intraperitoneal injection of insulin. The bar graphs indicate area-under-curve (AUC), reflecting cumulative glucose disposal. K-L) Pyruvate tolerance tests showing the rise of blood glucose over time in overnight fasted male (K) and female (L) Ts65Dn and euploid mice injected with pyruvate. The bar graphs indicate area-under-curve (AUC). All data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. For all tolerance tests, data were analyzed by 2-way ANOVA with Sidek post hoc tests.
Next, we subjected HFD-fed Ts65Dn mice to glucose tolerance tests. Ts65Dn male, but not female, mice clearly exhibited glucose intolerance when injected with a bolus of glucose (Figure 4G–H). Impaired glucose clearance in male mice suggests reduced insulin sensitivity in the peripheral tissues. We performed insulin tolerance tests to directly assess insulin sensitivity in Ts65Dn mice. Relative to euploid controls, Ts65Dn male, but not female, mice clearly showed impaired insulin action, confirming the development of insulin resistance (Figure 4I–J). Further, a pyruvate tolerance test was conducted to determine insulin sensitivity in the liver. In the fasted state, pyruvate is used as a major substrate for the de novo synthesis of glucose in liver to maintain euglycemia. Insulin signaling suppresses hepatic gluconeogenesis, and in the insulin resistant state, hepatic glucose output is significantly elevated due to impaired insulin action [101,102]. In overnight fasted mice, pyruvate administration resulted in significantly higher blood glucose levels in Ts65Dn male, but not female, mice relative to euploid controls (Figure 4K–L), suggesting reduced hepatic insulin sensitivity. Taken together, these data highlight a sexually dimorphic metabolic response to HFD, with male Ts65Dn mice showing exacerbated insulin resistance relative to euploid controls.
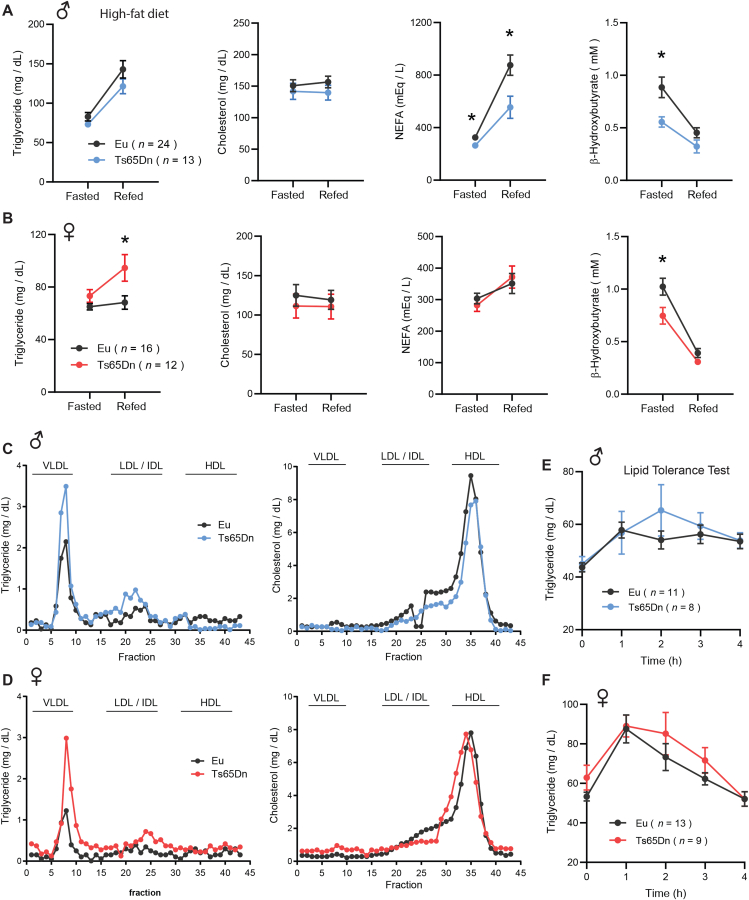
3.5. Dyslipidemia in Ts65Dn mice fed a high-fat diet
Altered lipid profile has been documented in Individuals with DS [18,19]. Thus, we investigated whether aneuploidy impacts lipid homeostasis in Ts65Dn mice during fasting and refeeding, two opposing physiological states. While fasting and refeeding serum TG and cholesterol levels were not different between genotypes in male mice fed a HFD, serum free fatty acids (NEFA) levels in the fasted state were lower in Ts65Dn male mice and continued to remain significantly lower in the refeeding period (Figure 5A). In contrast to males, female Ts65Dn mice had significantly higher serum TG levels during the refeeding period following a fast (Figure 5B). Synthesis of ketones (e.g., β-hydroxybutyrate; BHB) is markedly increased during fasting and suppressed by refeeding [103]. Ketones are made in the liver and used by the brain and peripheral tissues as an additional energy source during starvation. In Ts65Dn mice of both sexes, we observed significantly lower levels of serum BHB in the fasted state (Figure 5A–B). Since ketogenesis is biochemically coupled to hepatic mitochondrial fat oxidation [103], our data suggests reduced β-oxidation in Ts65Dn mouse liver during fasting.
Figure 5.
Dyslipidemia in Ts65Dn mice fed a high-fat diet. A-B) Overnight fasted and refed (2 h) serum triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate (BHB) in male (A) and female (B) Ts65Dn mice. Sample size for male (Eu, n = 24; Ts65Dn, n = 13) and female (Eu, n = 16; Ts65Dn, n = 12) mice. C-D) Pooled mouse sera from male (C) and female (D) Ts65Dn and euploid mice were fractionated by fast protein liquid chromatography (FPLC), and the triglyceride and cholesterol content of each fraction was quantified. Fractions corresponding to very-low density lipoprotein (VLDL), low-density lipoprotein (LDL), intermediate-density lipoprotein (IDL), and high-density lipoprotein (HDL) are indicated. E-F) Lipid tolerance tests showing triglyceride disposal in male (E) and female (F) Ts65Dn and euploid mice over time in response to peritoneal injection of emulsified intralipid. All data are presented as mean ± SEM. ∗P < 0.05 (2-way ANOVA with Sidek post hoc tests).
Large fractions of TG and cholesterol are transported in plasma in lipoprotein particle form. To determine the TG and cholesterol content of lipoprotein particles, pooled mouse sera were subjected to size exclusion chromatographic (FPLC) separation. TG content in very-low density lipoprotein (VLDL) particle fractions was much higher in Ts65Dn mice of either sex relative to euploid controls (Figure 5C–D). TG content in low-density lipoprotein (LDL) and intermediate-density lipoprotein (IDL) particle fractions also appeared to be higher in Ts65Dn mice, although to a lesser extent compared to that of VLDL-TG. In contrast, cholesterol content in high-density lipoprotein (HDL) particle fractions was not different between genotypes of either sex (Figure 5C–D).
Because steady-state and fasting-refeeding lipid profiles were altered in Ts65Dn mice, we performed lipid tolerance tests to determine whether Ts65Dn mice have altered capacity to handle an acute lipid load. Overnight fasted mice were injected with a bolus of emulsified intralipid, and plasma lipid clearance was monitored over time. The kinetics of lipid disposal in peripheral tissues appeared to be similar between genotypes of either sex (Figure 5E–F). All together, these data indicate similarities and sex-dependent differences in lipid profiles between HFD-fed male and female Ts65Dn mice relative to their euploid controls.
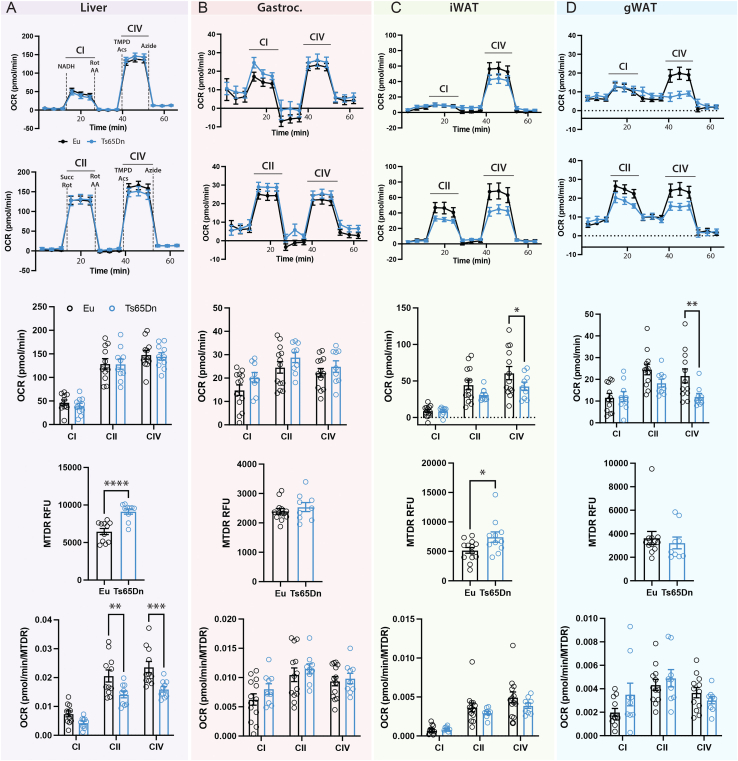
3.6. Reduced mitochondrial activity in Ts65Dn male mice fed a high-fat diet
In vitro studies have suggested mitochondrial dysfunction in DS-derived cells [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]]. To address if dyslipidemia and impaired glucose metabolism seen in Ts65Dn mice are associated with changes in mitochondrial activity in vivo, we assessed mitochondrial respiration in liver, skeletal muscle, subcutaneous and visceral white adipose tissues. We focused this analysis on Ts65Dn male mice because they have more pronounced metabolic phenotypes compared to female mice. The absolute activities (per 8 μg protein input) of liver mitochondrial complex I, II, and IV were not different between genotypes (Figure 6A, top three panels). Mitochondrial content as determined by mitotracker dyes (MTDR), however, was significantly higher in Ts65Dn mouse liver tissue (Figure 6A, fourth panel). Thus, when normalized to mitochondrial content, the relative mitochondrial activities of complex II and IV were significantly lower in the liver of Ts65Dn male mice relative to euploid controls (Figure 6A, bottom panel). This indicates that higher mitochondrial content in Ts65Dn mouse liver is needed to achieve the same amount of mitochondrial respiration as seen in the euploid controls.
Figure 6.
Reduced mitochondrial activity in Ts65Dn male mice fed a high-fat diet. A-D) Mitochondrial respiration in the liver (A), gastrocnemius skeletal muscle (B), inguinal white adipose tissue (iWAT; C), and gonadal white adipose tissue (gWAT; D) of male Ts65Dn (n = 11–13) and euploid (n = 9–10) mice. For each tissue column, the following panels (from top to bottom) are shown: First and top panel: oxygen consumption rate of mitochondrial complex CI and CIV. Second panel: oxygen consumption rate of mitochondrial complex CII and CIV. Third panel: Quantification of oxygen consumption rate of mitochondrial complex CI, CII, and CIV. Fourth panel: Quantification of mitochondrial content by MitoTracker. Fifth and bottom panel: Oxygen consumption rate of mitochondrial complex CI, CII, and CIV after normalization to mitochondrial content. All data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
For skeletal muscle (gastrocnemius), the mitochondrial content, as well as the absolute and relative mitochondrial activities of complex I, II, and IV were not significantly different between genotypes (Figure 6B). For subcutaneous (inguinal) white adipose tissue (iWAT), the absolute mitochondrial activity (per μg protein) of complex IV was significantly lower in Ts65Dn male mice relative to euploid controls (Figure 6C, top three panels). Mitochondrial content, however, was significantly higher in Ts65Dn mouse iWAT (Figure 6C, fourth panel). After normalization to mitochondrial content, the relative mitochondrial activity of complex of IV was not different between genotypes (Figure 6C, bottom panel). For visceral (gonadal) white adipose tissue (gWAT), the absolute mitochondrial activity (per μg protein) of complex IV was significantly lower in Ts65Dn male mice relative to euploid controls (Figure 6D, top three panels). Mitochondrial content was modestly lower in gWAT (Figure 6D, fourth panel). When normalized to mitochondrial content, the relative mitochondrial activity of complex IV was not significantly between genotypes (Figure 6D, bottom panel). Taken together, these data indicate an absolute and relative reduction in mitochondrial function in visceral fat depot and liver, respectively, of Ts65Dn male mice.
3.7. Altered tissue weight in Ts65Dn mice fed a high-fat diet
At termination of study, we dissected and quantified tissue weights to assess possible gross changes. Body weights of Ts65Dn male, but not female, mice were significantly lower compared to euploid controls (Table S1). Subcutaneous (inguinal) fat mass was significantly smaller in Ts65Dn mice of either sex. The spleen in Ts65Dn male mice was significantly larger relative to euploid controls. When normalized to body weight, the relative (%) weight of liver, kidney, and spleen of Ts65Dn male mice were significantly higher compared to euploid controls. The average tibia length of Ts65Dn male mice was shorter than that of the euploid littermates. In female Ts65Dn mice, the size of the pancreas and spleen was larger relative to euploid controls (Table S1). When normalized to body weight, the relative (%) weight of heart, kidney, pancreas, and spleen were significantly higher in Ts65Dn female mice relative to euploid controls. Tibia length, however, was not different between genotypes in female mice. These data indicate a relative enlargement of multiple organs and tissues in HFD-fed Ts65Dn mice.
3.8. Selective expression of trisomic genes in the adipose tissue, liver, and skeletal muscle of Ts65Dn mice
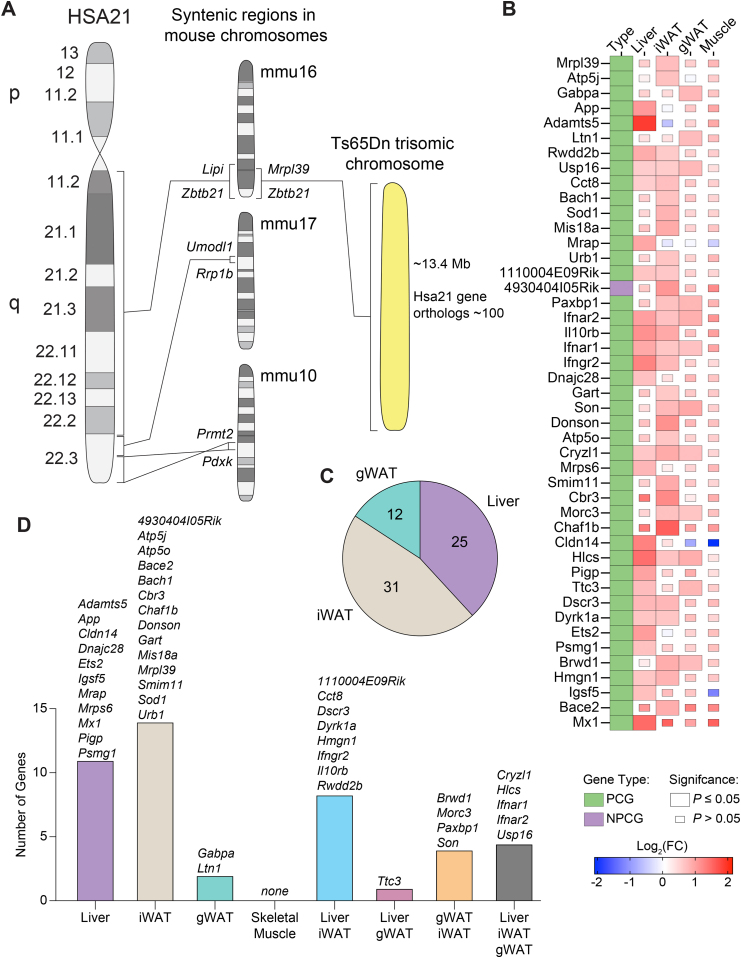
Having established the impact of segmental trisomy on metabolic and physiological function at the tissue and organismal level, we next addressed the impact of gene triplications on the transcriptomes of adipose tissue, liver, and skeletal muscle. First, we performed RNA sequencing to determine which Mmu16 genes triplicated in Ts65Dn are expressed and at what level in peripheral tissues. The entire Hsa21 maps to three conserved segments on mouse chromosomes Mmu16 (22.5 Mb, 119 genes), Mmu10 (2.1 Mb, 39 genes), and Mmu17 (1.1 Mb, 19 genes), with the bulk of it on Mmu16 (Figure 7A). Ts65Dn mice are trisomic for ∼104 orthologous genes [76].
Figure 7.
Selective expression of trisomic genes in the adipose tissue, liver, and skeletal muscle of Ts65Dn mice. A) The orthologous genes on human chromosome 21 (Hsa21) are distributed across three syntenic segments located on mouse chromosomes Mmu10, Mmu16, and Mmu17. Ts65Dn mice are trisomic for ∼104 genes (from Mrpl39 to Zbtb21) out of ∼140 (Lipi to Zbtb21) on Mmu16 that are syntenic to Hsa21. B) Heat map of all trisomic genes in Ts65Dn mice fed a high-fat diet that are differentially expressed relative to euploid littermate controls by at least one tissue. Green box denotes protein coding gene (PCG) and purple box denotes non-protein coding gene (NPCG). The size of the box indicates statistical significance in trisomic gene expression between male Ts65Dn mice relative to euploid controls. Full-size box denotes significantly different expression (P ≤ 0.05), and small box size denotes non-significant expression between genotypes (P > 0.05). The color gradient indicates Log2 fold-change of each gene in Ts65Dn relative to euploid controls, with upregulated genes in the red and downregulated genes in the blue color spectrum. Tissue sample size for all RNA sequencing (Euploid, n = 6; Ts65Dn, n = 6). iWAT, inguinal white adipose tissue; gWAT, gonadal white adipose tissue. C) A pie chart indicating the number of trisomic genes significantly (P ≤ 0.05) and differentially (Log2(FC) ≤ 0.5 or ≥ 0.5) expressed in Ts65Dn mice relative to euploid controls by each tissue. None of the trisomic genes were differentially expressed in the skeletal muscle of Ts65Dn mice. D) Overlap analysis of trisomic genes significantly and differentially expressed by each tissue with lists of all genes contained in each group.
RNA sequencing revealed that 86% (77 out of 89) of the trisomic genes (excluding the Krtap gene cluster) in Ts65Dn were expressed in the liver, skeletal muscle, and visceral and subcutaneous white adipose tissue (Table S2). The expression of 12 genes (Girk1, Cldn17, Cldn8, Olig2, Kcne2, Kcne1, Clic6, Kcnj6, B3galt5, Dscam, Fam3b, zfp295) and the Krtap cluster genes (encoding keratin associated proteins of hair fibers) were not detected by RNA-seq. As expected, a majority of the trisomic genes had an average expression level increased by about 1.5-fold across tissues due to increased gene dosage (Figure S10 and Table S2). However, only 45 genes were significantly upregulated (adjusted p-value <0.05) in at least one tissue (Table S2). Relative to euploid controls, there were 31 orthologs of Hsa21 genes expressed at significantly higher levels by inguinal white adipose tissue (iWAT), 25 by liver, 12 by gonadal white adipose tissue (gWAT), and none by skeletal muscle (Figure 7C). Overlap analysis highlighted tissue-specific and shared gene expression across the four tissues. Of the 45 orthologous trisomic genes differentially expressed in Ts65Dn mice relative to euploids controls, 11 were uniquely expressed in the liver, 14 in the iWAT, 2 in the gWAT, and none in the skeletal muscle (Figure 7D). The liver and iWAT shared expression of 8 genes, whereas liver and gWAT shared the expression of 1 gene, and gWAT and iWAT share the expression of 4 genes (Figure 7D). The liver, iWAT, and gWAT shared the expression of five common genes (Cryzl1, Hlcs, Ifnar1, Ifnar2, and Usp16). Overall, only a subset (∼43%) of the trisomic genes in Ts65Dn were differentially expressed relative to the euploid controls in three out of the four peripheral tissues, and none were differentially expressed in the skeletal muscle. It is known that Ts65Dn mice also contain 46 extra trisomic genes from Mmu17 [77,78], and some of these genes were also significantly upregulated in adipose tissue and liver (Table S3). These data indicate that most of the significant differentially expressed genes (DEGs) in Ts65Dn are expressed in a tissue-specific manner, and that dosage compensation may account for some of the trisomic genes with expression levels close to or below that of euploid controls [104].
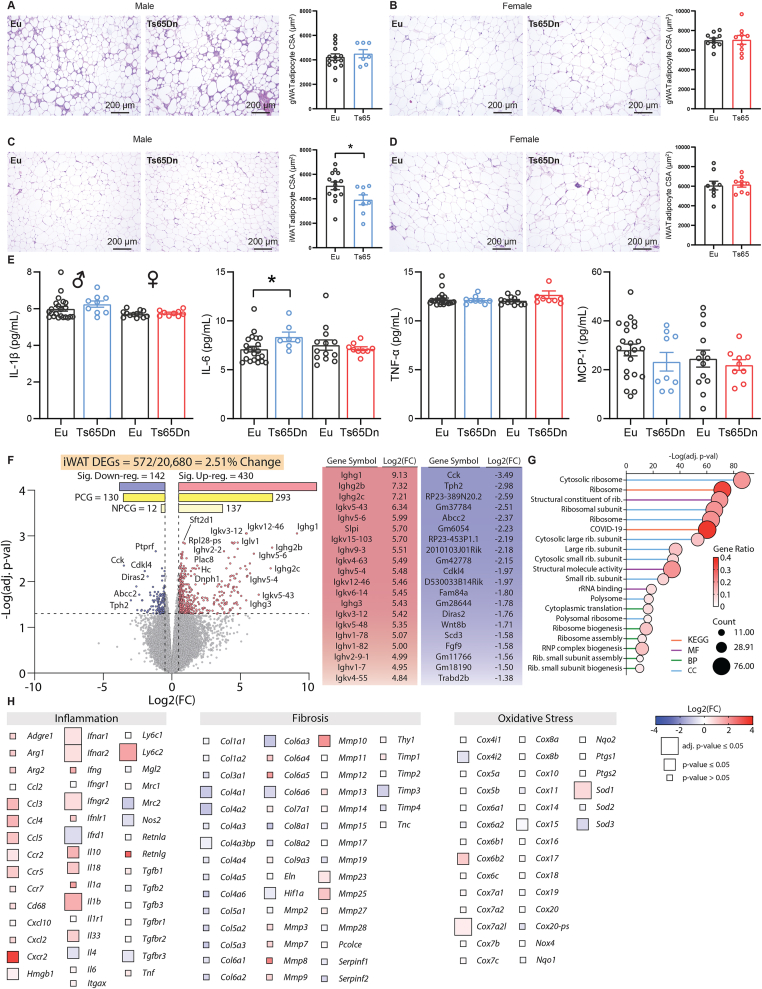
3.9. Adipose tissue inflammation in Ts65Dn male mice fed a high-fat diet
Since HFD-fed Ts65Dn male mice had lower body weight and adiposity (Figure 3A–B and Table S1), we performed histological analysis on two of the major fat depots to determine possible changes in cellularity and cell size. In visceral white adipose tissue (gWAT), adipose size and gross histology were not different between genotypes of either sex (Figure 8A–B). In contrast, adipocyte size was significantly smaller in subcutaneous white adipose tissue (iWAT) of Ts65Dn male, but not female, mice relative to euploid controls (Figure 8C–D). Chronic low-grade inflammation is a notable feature of obesity [105]. Despite lower body weight, we observed a significant, though modest, increase in serum IL-6 in Ts65Dn male mice relative to euploid controls (Figure 8E). Serum levels of other classic pro-inflammatory cytokines (IL-1β, TNF-α, and MCP-1) were not different between genotypes. At the tissue level, MCP-1 protein levels trended higher (P = 0.051) in iWAT, but IL-1β, IL-6, and TNF-α levels in gWAT and iWAT were not different between genotypes (Figure S11).
Figure 8.
Adipose tissue inflammation in Ts65Dn male mice fed a high-fat diet. A-B) Representative H&E stained histological sections of visceral (gonadal) white adipose tissue (gWAT) of male (A) and female (B) Ts65Dn and euploid mice. The quantifications of adipocyte cell size (CSA, cross sectional area) in male (Euploid, n = 14; Ts65Dn, n = 7) and female (Euploid, n = 10; Ts65Dn, n = 9) mice are indicated by bar graphs. C-D) Representative H&E stained histological sections of subcutaneous (inguinal) white adipose tissue (gWAT) of male (C) and female (D) Ts65Dn and euploid mice. The quantification of adipocyte CSA in male (Euploid, n = 14; Ts65Dn, n = 8) and female (Euploid, n = 9; Ts65Dn, n = 9) mice are indicated by bar graphs. E) Serum concentrations of IL-1β, IL-6, TNF-α, and macrophage chemotactic protein 1 (MCP-1) in male (Euploid, n = 22; Ts65Dn, n = 9) and female (Euploid, n = 13; Ts65Dn, n = 9) mice. F) Volcano plot of the iWAT RNA-seq data. The dotted y-axis line shows the adjusted p-value cut-off for significance (adj. p-value = 0.05). The two dotted x-axis lines represent a Log2(FC) of −0.5 or 0.5. A total of 572 out of 20,680 genes (2.51% of the iWAT transcriptome) are differentially expressed in male Ts65Dn mice relative to euploid controls, with 430 genes up-regulated and 142 genes down-regulated. PCG, protein coding genes; NPCG, non-protein coding genes. The adjacent table shows the top 20 up- and down-regulated genes. G) The top 20 most significant Gene Ontology (GO) and KEGG categories based on differentially expressed genes (DEGs) seen in the iWAT of Ts65Dn. The stalk color represents the category the title belongs to (KEGG pathway, MF = molecular function, BP = biological process, CC = cell component). The size of the circle represents how many genes (or counts) are altered in the group. The color of the circle represents the direction of change, and the magnitude of the gene ratio. A high gene ratio means more genes in that gene family are affected in this group. H) Heat maps of genes associated with inflammation, fibrosis, and oxidative stress. The size of the box indicates statistical significance in trisomic gene expression between male Ts65Dn mice relative to euploid controls. Full-size box denotes significantly different expression (Adjusted p-value ≤0.05), medium size box denotes significantly different expression (non-Adjusted p-value ≤0.05), and small box size denotes non-significant expression between genotypes (P value > 0.05). The color gradient indicates Log2(FC) of each gene in Ts65Dn relative to euploid controls, with up-regulated genes in the red and down-regulated genes in the blue color spectrum. Tissue sample size for iWAT RNA sequencing (Euploid, n = 6; Ts65Dn, n = 6).
The histological and serum cytokine data led us to assess potential transcriptomic changes in the two major fat depots. Overall, there were only a very small number (15 upregulated and 1 downregulated) of differentially expressed genes in the gWAT (Table S4). In striking contrast, 2.51% (572) of the transcriptome in iWAT was significantly altered in Ts65Dn mice relative to euploid controls. Of this, 293 protein coding genes (PCGs) and 137 non-protein coding genes (NPCGs) were significantly upregulated, and 130 PCGs and 12 NPCGs were significantly downregulated, in Ts65Dn mice (Figure 8F and Table S5). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses highlighted translational machinery (ribosome and ribosome-related processes) to be the top pathway altered in iWAT of Ts65Dn mice (Figure 8G). Of the other relevant pathways that would affect adipose tissue function, inflammation was notably elevated in the iWAT of Ts65Dn, as indicated by the upregulated expression of multiple cytokines, cytokine receptors, and interferon pathway genes (Figure 8H). Expression of other genes related to lipid metabolism (synthesis, lipolysis, and oxidation), mitochondrial function, fibrosis, oxidative and ER stress were largely not different between genotypes (Figure 8H and Figure S12). These data indicate that chronic high-fat feeding promotes the expression of inflammatory genes in the subcutaneous adipose tissue of Ts65Dn mice. Unlike the liver and iWAT, only a total of 16 genes were differentially expressed in the visceral (gonadal) white adipose tissue (gWAT) (Table S4) and only 1 was differentially expressed in the skeletal muscle of Ts65Dn mice relative to euploid controls.
3.10. Hepatic inflammation and fibrosis in Ts65Dn male mice fed a high-fat diet
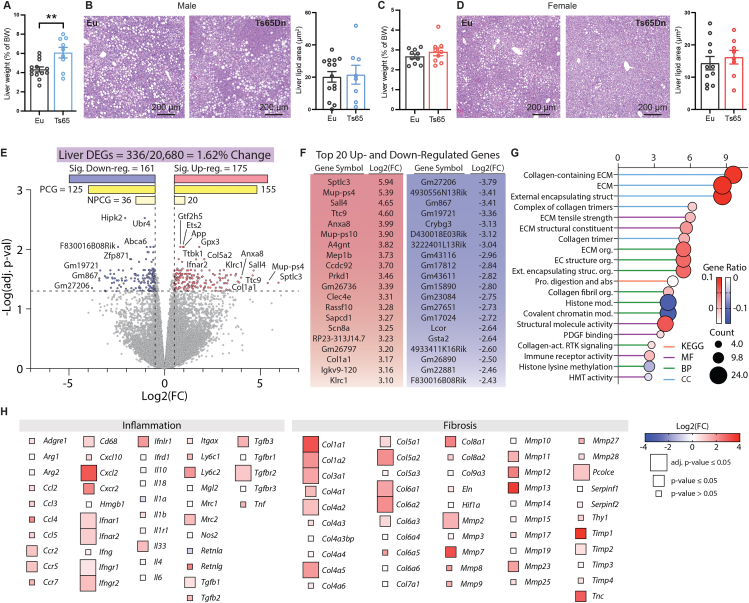
Ts65Dn male, but not female, mice fed a high-fat diet had a relative enlargement of liver when compared to euploid controls (Figure 9A,C). At the tissue level, quantification of lipid content (% lipid area) and lipid droplet size in liver tissue sections revealed no significant differences between genotypes of either sex (Figure 9B,D and Figure S13). However, we observed significant changes in the liver transcriptome of Ts65Dn mice. Of the 20,680 genes detected by RNA sequencing in liver, 336 (1.62%) were differentially expressed between Ts65Dn and euploid controls (Figure 9E). Of the differentially expressed genes (DEGs), 155 protein coding genes (PCGs) and 20 non-protein coding genes (NPCGs) were significantly upregulated, and 125 PCGs and 36 NPCGs were significantly downregulated, in Ts65Dn mice relative to euploid controls (Figure 9F and Table S6).
Figure 9.
Hepatic inflammation, fibrosis, and oxidative stress in Ts65Dn male mice fed a high-fat diet. A) Liver weight of male mice (% of body weight). B) Representative H&E stained liver sections of male Ts65Dn and euploid mice. The quantifications of lipid area in male (Euploid, n = 14; Ts65Dn, n = 8) are indicated by the bar graph. C) Liver weight of female mice (% of body weight). D) Representative H&E stained liver sections of female Ts65Dn and euploid mice. The quantifications of lipid area in female (Euploid, n = 11; Ts65Dn, n = 8) mice are indicated by the bar graph. E) Volcano plot of the liver RNA-seq data. The dotted y-axis line shows the adjusted p-value cut-off for significance (adj. p-value = 0.05). The two dotted x-axis lines represent a Log2(FC) of −0.5 or 0.5. A total of 336 out of 20,680 genes (1.62% of the liver transcriptome) are differentially expressed in male Ts65Dn mice relative to euploid controls, with 175 genes upregulated and 161 genes downregulated. PCG, protein coding genes; NPCG, non-protein coding genes. F) The table shows the top 20 up- and down-regulated genes. G) The top 20 most significant Gene Ontology (GO) and KEGG categories based on differentially expressed genes (DEGs) seen in the liver of Ts65Dn. The stalk color represents the category the title belongs to (KEGG pathway, MF = molecular function, BP = biological process, CC = cell component). The size of the circle represents how many genes (or counts) are altered in the group. The color of the circle represents the direction of change, and the magnitude of the gene ratio. A high gene ratio means more genes in that gene family are affected in this group. H) Heat maps of genes associated with inflammation and fibrosis. The size of the box indicates statistical significance in trisomic gene expression between male Ts65Dn mice relative to euploid controls. Full-size box denotes significantly different expression (adjusted p-value ≤0.05), medium size box denotes significantly different expression (non-adjusted p-value ≤0.05), and small box size denotes non-significant expression between genotypes (p-value >0.05). The color gradient indicates Log2(FC) of each gene in Ts65Dn relative to euploid controls, with upregulated genes in the red and downregulated genes in the blue color spectrum. Tissue sample size for liver RNA sequencing (Euploid, n = 6; Ts65Dn, n = 6).
GO and KEGG analyses highlighted prominent changes in collagen and extracellular matrix (ECM) related processes in the liver of Ts65Dn mice, suggesting elevated signature of fibrosis (Figure 9G). Indeed, many of the collagen genes (Col1, Col3, Col4, Col5, Col6), and their regulators (Tgfbr2, Mmp2, Pcolce) were significantly upregulated in Ts65Dn mouse liver relative to euploid controls (Figure 9H). In addition, we observed an inflammatory gene signature in Ts65Dn mouse liver, as indicated by the upregulation of multiple inflammatory cytokines and their receptors. Hepatic oxidative stress also appeared to be elevated in Ts65Dn mouse liver, as multiple oxidative stress genes (Cox4i2, Cox6b2, Cox20-ps, Sod3) were upregulated, with the concomitant downregulation of multiple antioxidant genes (Gsta1, Gsta2, Gsta3, Gsta4, Gstm1, Gstm2-ps1, Gstm4, Gsto2) (Figure S14). In contrast to fibrotic, inflammatory, and oxidative stress genes, expression of other genes related to lipid metabolism (lipid uptake, synthesis, lipolysis, oxidation), glucose metabolism (gluconeogenesis), ER stress, and mitochondrial function were largely not different between genotypes (Figure S14). Together, these data indicate that chronic high-fat feeding elevates the expression of genes involved in inflammation and fibrosis, and to a lesser extent oxidative stress, in the liver of Ts65Dn mice. Interestingly, and opposite to iWAT and liver, only a single gene (Rai14) was differentially expressed (upregulated) in the skeletal muscle of Ts65Dn mice relative to euploids. Thus, RNA sequencing across four peripheral tissues revealed a tissue-specific impact of trisomy on the expression of disomic genes. In contrast to the gene signature data, protein markers of fibrosis (Sirius red staining of collagen, hydroxyproline content, and collagen type I immunostaining), oxidative stress (malondialdehyde level), and liver damage caused by inflammation (serum ALT level) were not significantly different between genotypes, despite the values being slightly higher in Ts65Dn mice (Figure S15).
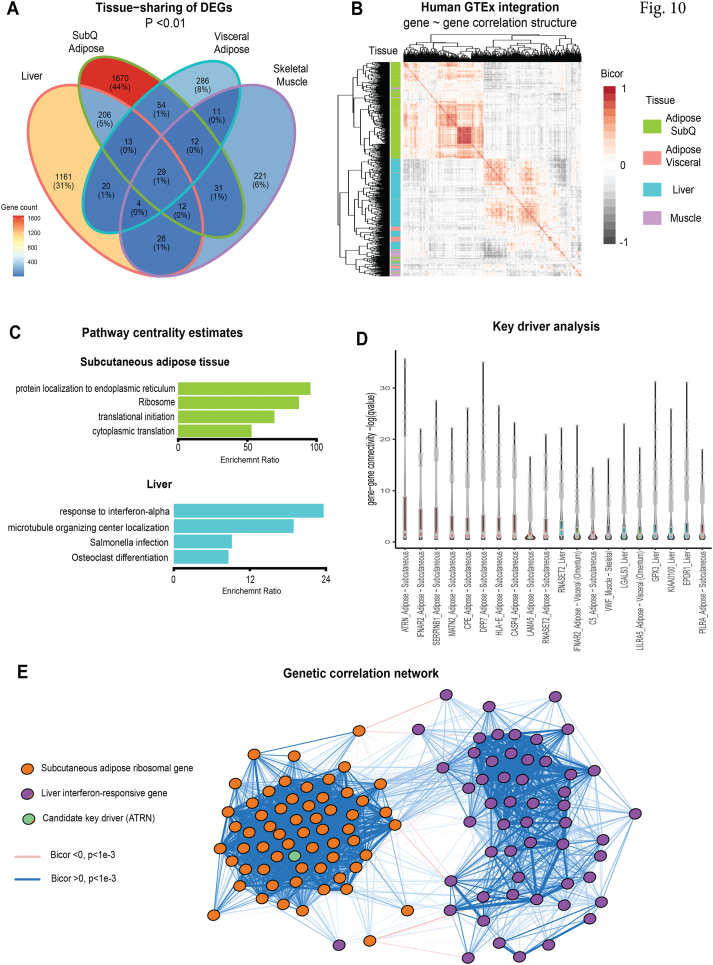
3.11. Coregulation of pan-tissue differentially expressed genes (DEGs) are conserved in humans
Finally, we evaluated whether DEGs from RNA-seq were altered similarly across tissues. Binning genes based on p-value (non-adjusted) cutoffs demonstrated that of the ∼3,000 genes altered in Ts65Dn vs euploid mice, nearly none changed across the 4 tissues measured (Figure 10A). Most genes, however, showed tissue-selective patterns of differential expression, primarily in subcutaneous (inguinal) white adipose tissue (iWAT) and liver (Figure 10A). Given the significant changes in systemic metabolism and the differential patterns of gene expressions across tissues observed in Ts65Dn mice, we sought to elucidate metabolic mechanisms conserved in humans and prioritize putative central driver genes. To accomplish this, we leveraged publicly available datasets of human cross-tissue RNA-sequencing [96] where correlated patterns of specific gene sets could be used to inform metabolic signaling between tissues [[106], [107], [108], [109]]. Analysis of genetic correlation structure of Ts65Dn DEGs showed clustering patterns consistent with gene regulation specific to individual tissues (Figure 10B). This was most notable by unbiased clustering based on correlation coefficients assigning iWAT and liver genes to discrete blocks of co-expression (Figure 10B). To identify pathways and genes specific for iWAT and liver that might affect insulin sensitivity and glucose homeostasis, all DEGs were ranked by gene centrality estimates [97], and this analysis highlighted ribosomal function in adipose tissue and inflammation in liver as core pathways contributing to the metabolic phenotypes (Figure 10C). Gene-based centrality and key driver estimates allowed us to prioritize several candidate endocrine signaling proteins as potential mediators of tissue crosstalk between iWAT and liver (Figure 10D). These included IFNAR2, SERPIN1B and ATRN in adipose tissue as top-ranked genes encoding secreted proteins which showed the strongest levels of significant correlations across all pan-tissue DEGs (Figure 10D.). For example, adipose ATRN (encoding attractin protein) was observed to sit centrally within a subset of adipose ribosomal genes (GO:0005840), which in turn, were strongly enriched with liver genes annotated to be involved in interferon responses (GO:0034,340). Collectively, these analyses indicate that metabolic pathways perturbed in the Ts65Dn mice show genetic conservation to human populations, where adipose-to-liver crosstalk may contribute to the underlying metabolic effects.
Figure 10.
Integration of mouse differentially expressed genes (DEGs) with human genetic correlation structure. A) Venn diagram showing number of differentially expressed (logistic regression p-value <0.01) genes per tissue analyzed. B) Heatmap showing co-correlation structure of all human orthologues from the Genotype-Tissue Expression (GTEx) encoding mouse DEGs. Filled color indicates biweight midcorrelation (bicor) coefficient and row color along y-axis indicates tissue of origin for each gene. Rows and columns of genes are clustered hierarchically to order by similarities (mclust). C) All genes among 4 tissues indicated were correlated with DEGs and ranked by significance (mean q-value of regression), the top 500 genes were then assayed for pathway enrichments using overrepresentation analyses in Gene Ontology (Biological Processes), KEGG and Reactome. The 4 strongest pathways enriched are shown for either subcutaneous (inguinal) white adipose tissue (top) or liver (bottom). D) Key driver analysis showing top 20 genes encoding secreted proteins ranked by -log10 (p-value of regression coefficient) across all DEG orthologues in GTEx. E) Undirected network showing gene correlation structure of adipose tissue ribosomal and liver interferon-responsive pathway genes, where a key driver (Attractin, ATRN) is shown as a central component. Logistic regression p-values calculated using DESeq2, n = 6 mice per group. Bicor and associated p-values calculated using WGCNA package and q-values using q-value package. GTEx = 310 individuals.
4. Discussion
One major aspect of DS research that is understudied and largely overlooked concerns the impact of aneuploidy on metabolic processes occurring in peripheral tissues. Our systematic analyses here show that the triplication of a large fraction (∼44%) of Hsa21 orthologous genes in the segmental trisomic mouse model, Ts65Dn, perturbs systemic glucose and lipid metabolism in a sex- and diet-dependent manner. Under the basal state when mice were fed a standard chow, Ts65Dn male and female mice showed largely similar phenotypes, except for Ts65Dn male mice having higher food intake relative to euploid controls. Despite no differences in body weight, chow-fed male and female Ts65Dn mice had higher physical activity level and energy expenditure relative to their euploid littermates. Interestingly, elevated physical activity level and energy expenditure in Ts65Dn mice did not contribute to a healthier metabolic profile as would be expected. To the contrary, chow-fed Ts65Dn mice of either sex showed a modest, but significant, impairment in glucose tolerance, a finding consistent with a recent study [110]. While the overall impact of aneuploidy on systemic metabolism in the basal state is relatively mild, it is worth noting that all the chow-fed data were collected from Ts65Dn mice that are relatively young (between 2 and 4 months old). Metabolic homeostasis is known to decline with age [111]. Whether the impairment in glucose metabolism would become progressively worse as chow-fed Ts65Dn mice get older (e.g., 1 or 1.5 years of age) remains to be determined.
The hyperactivity we observed at baseline was noted in previous studies of Ts65Dn mice [6,53,81,[112], [113], [114]], and this feature of hyperactivity accompanied by attentional deficit is documented in children with DS [115,116]. The increase in energy expenditure seen in chow-fed Ts65Dn mice of either sex has also been recently reported for chow-fed male Ts65Dn mice [47] and Dp1Tyb mice (a DS mouse model containing a segmental duplication of Mmu16) [50]. Elevated physical activity level likely contributes, at least in part, to the increase in energy expenditure seen in Ts65Dn mice. However, whether similar changes in energy expenditure are observed in individuals with DS remains equivocal. Resting energy expenditure appears to be lower in children with DS [37], and in adults with DS, resting metabolic rate is either lower [41], unchanged [39], or higher (when normalized to lean mass) [39]. Because metabolic rate and energy expenditure are associated with general physical activity level, and individuals with DS tend to be more sedentary [117], the conflicting reports on energy expenditure may be related to differences in physical activity level of the cohorts being studied.
The interactions of genes, genetic sex, and environments have a major influence on phenotypes, and diet is an important environmental factor that dictates metabolic outcomes. The negative impact of aneuploidy becomes much more apparent and accentuated in Ts65Dn male mice when obesity is induced by a high-fat diet. There was a clear sexually dimorphic response to high-fat feeding in Ts65Dn mice. Female Ts65Dn mice fed an HFD had relatively mild phenotypes; these include a modest increase in fasting blood glucose level, a significantly higher serum triglyceride level in the refed state following a fast, and also higher TG level in the VLDL particle fraction. In contrast to females, male Ts65Dn mice had a much more pronounced deterioration in metabolic homeostasis in response to high-fat feeding. Male Ts65Dn mice on HFD exhibited fasting and refeeding hyperglycemia, glucose and pyruvate intolerance, insulin resistance, and higher TG level in the VLDL particle fractions. These metabolic phenotypes—insulin resistance and higher circulating TG—have been previously documented in children and/or adults with DS [19,25,118,119]. Given the complex interplay of multiple factors governing metabolic homeostasis, and the difficulty of establishing causal effects due to various confounding factors in human DS studies, our controlled mouse studies thus provide experimental support for a direct contributive role of aneuploidy in dysregulating some, but not all, aspects of systemic metabolism seen in individuals with DSs.
Given that sex is an important biological variable affecting metabolic outcomes [120], with female sex hormones generally having a protective role [121], it is not surprising that female Ts65Dn mice exhibit a much better metabolic profile than male Ts65Dn mice relative to euploid controls. It appears that only a few studies of Ts65Dn mice have included females in their phenotypic analysis [68,[122], [123], [124]]. With regard to the DS population, information is often lacking on whether sex affects severity, penetrance, and expressivity of various DS phenotypes. Thus, the inclusion of females in our metabolic studies are valuable in informing future studies in individuals with DS.
The insulin resistance phenotype we observed in HFD-fed Ts65Dn male mice is due to impaired insulin action and not the result of reduced insulin production and secretion. Insulin secretion dynamics in response to physiological fasting and refeeding were not different between genotypes. Quantification of pancreatic insulin content also revealed no significant difference between Ts65Dn and euploid mice. In a recent study, Peiris et al. have suggested that one of the Hsa21 genes (Regulator of calcineurin 1, RCAN1, formerly known as DSCR1) links hyperglycemia to functional changes in pancreatic β-cells in type 2 diabetes [110]. The authors showed that RCAN1 overexpression reduces in vivo glucose-stimulated insulin secretion and impairs mitochondrial function in β-cells [110]. These results were obtained outside of the context of trisomy (i.e., not in trisomic cells or tissues) and the level of RCAN1 overexpression (∼2.5-fold higher) is significantly higher than the 50% increase seen for most of the trisomic genes due to dosage imbalance. While Rcan1 clearly plays an important role in β-cells, as seen in overexpression mouse and cell models [110,125,126], its contribution to the metabolic phenotypes seen in the context of trisomy should be interpreted with caution. We concur with the authors that the DS screening approach used serves as a valuable tool to help narrow down and identify genes relevant for β-cell dysfunction in type 2 diabetes [110]. Likewise, transgenic overexpression of Rcan1 has also been shown to enhance hepatic glucose production and impairs glucose homeostasis in chow-fed mice [127]. Overexpression of a single Hsa21 gene ortholog provides valuable insights into its potential function. However, the contribution to phenotypes of a modest (∼50%) increase in a large number of trisomic genes due to dosage imbalance and their complex interactions and epistasis often results in unexpected outcomes [6,55,61,128]. This highlights the importance of dissecting Hsa21 gene(s) functions in the context of trisomy and the appropriate disomic control.
The higher prevalence of obesity is well documented in adults with DS [22]. It is also known that elderly adults with DS experience significant weight loss, due to changes in food intake behaviors (e.g., loss of appetite, difficulty in feeding), and weight loss is markedly more pronounced in individuals with DS who have dementia [129]. Food intake and energy expenditure are two of the major components that fine tune body weight, and these are regulated by central and peripheral mechanisms [130,131]. Increased body mass index (BMI) and adiposity in adults with DS likely reflects a combination of gene–environment interactions, diet, and the extent of sedentariness. Interestingly, chronic high-fat feeding did not result in exacerbated weight gain in Ts65Dn mice. Instead, we observed a divergence in body weight between male Ts65Dn and euploid mice, beginning at 14 weeks on HFD, due to an unexpected decrease in body weight. This reduction in body weight is not due to reduced caloric intake, as food intake was not different between genotypes, thus ruling out unexplained sickness that suppresses appetite. Rather, Ts65Dn male mice on HFD maintained a significantly higher physical activity level and energy expenditure relative to euploid controls. These parameters were measured at 12 weeks on HFD, prior to body weight divergence between male Ts65Dn and euploid mice. Thus, differences in physical activity level and energy expenditure could account, at least in part, for the eventual divergence in body weight between male Ts65Dn and euploid controls. To our knowledge, there is only one other study reported to date where Ts65Dn mice were subjected to an HFD, and none in other DS mouse models. Intriguingly, Fructuoso et al. observed an increase in food intake, energy expenditure, adiposity, and body weight in Ts65Dn male mice fed an HFD for 8 weeks [47]. The magnitude of body weight difference between Ts65Dn and euploid male mice is very small (∼0.15 g), and the difference in adiposity between the two groups is less than 0.05% [47]. The physiological significance of this small change is uncertain; moreover, these results are based on limited sample size (5–6 mice per genotype) and therefore likely underpowered.
Mitochondrial dysfunction has been postulated to be one of the underlying causes for some of the DS phenotypes, including cognitive deficits [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]]. All these prior studies are based on in vitro cultured cells derived mostly from DS fetal fibroblasts and iPSCs. Our present study provides the first evidence documenting changes in mitochondrial content and function in trisomic tissues derived from Ts65Dn mice. We confirmed that mitochondrial activity is indeed reduced in subcutaneous (inguinal) white adipose tissue (iWAT), visceral (gonadal) white adipose tissue (gWAT), and liver, but not skeletal muscle. The causes for reduced mitochondrial respiration in iWAT, gWAT, and liver appear to be different. The absolute, but not relative (when normalized to mitochondrial content), mitochondrial activity at complex IV was reduced in the iWAT and gWAT of Ts65Dn mice. In the liver of Ts65Dn mice, mitochondrial content is significantly increased relative to euploid controls; thus, the oxygen consumption rate at complex II and IV per unit of liver mitochondria is significantly reduced. Increased mitochondrial content in Ts65Dn mouse liver presumably reflects compensatory response to a decrease in mitochondrial activity or efficiency; such a compensatory response, however, is also seen in iWAT of Ts65Dn mice. RNA sequencing of liver, iWAT, and gWAT transcriptomes did not reveal any significant changes in genes related to mitochondrial OXPHOS and function. Thus, a posttranscriptional mechanism is likely responsible for the changes in mitochondrial content and activity in iWAT, gWAT and liver of Ts65Dn mice. In support of this, a systematic proteome profiling of DS fibroblasts showed significant downregulation of the mitochondrial proteome—many are constituents of the OXPHOS complexes I to IV—without the concomitant downregulation of corresponding genes [132]. Our data, along with published studies on cultured cells [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]], support a functional deficit in mitochondria as one of the potential contributive factors affecting DS phenotypes.
Chronic low-grade inflammation, oxidative stress, and fibrosis in adipose tissue are mechanistically linked to impaired adipocyte function in obesity [105,133,134]. The inflammatory milieu within fat depots caused by the infiltrating immune cells (e.g., macrophages), and the inflammatory cytokines they secrete, disrupts the normal fat storage and endocrine function of adipocytes [135]. Because adipocytes secrete large numbers of adipokines with local and systemic effects, altered secretion of adipokines in obesity adversely affects inter-organ communications and whole-body metabolic homeostasis [136,137]. In Ts65Dn male mice, we observed a clear gene signature of inflammation within the subcutaneous fat depot, with signatures of fibrosis and oxidative stress also present, but to a much lesser degree. These changes likely contribute to impaired insulin sensitivity and glucose tolerance seen in Ts65Dn mice, as adipose tissue and skeletal muscle are the two major organs that take up glucose in response to insulin.
Among the macrophage subtypes infiltrated into the subcutaneous fat depot, we observed a significant enrichment of marker genes (e.g., Il1b, Ccl3, Ccl4, Ccr2) for pro-inflammatory M1-type macrophages [[138], [139], [140]]. It has been documented that Individuals with DSs have elevated plasma levels of pro-inflammatory cytokines, such as MCP-1, IL-22, IL-6, and TNF-α [141], indicative of systemic low-grade inflammation. Interestingly, four of the interferon receptor subunit genes (Ifnar1, Ifnar2, Ifngr2, Il10rb) are among the pro-inflammatory genes upregulated in iWAT. Three of the genes (IFNAR1, IFNAR2, IFNGR2) are encoded on chromosome 21, and the corresponding mouse genes are trisomic in Ts65Dn mice. Recent studies have revealed a hyperactive interferon response in DS-derived fibroblasts, lymphoblastoid cell lines, and circulating monocytes and T-cells, and these are thought to be due to triplicated interferon receptor subunit genes in trisomy 21 [[142], [143], [144]]. Presumably, the elevated transcript levels of IFN receptor subunit genes in iWAT of Ts65Dn mice are induced by the infiltrating myeloid cells frequently seen in obesity [145,146]. Increased expression of IFN receptor subunit genes appears to be fat depot specific, as this is only seen in subcutaneous, but not the visceral, fat depot. Interestingly, increased expression of IFN receptor subunit genes in monocytes and T-cells derived from individuals with DS is associated with a global downregulation of ribosomal protein genes of the translational machinery [143]. In our pathway analysis, ribosome biogenesis, assembly, and function are the top pathways being altered in iWAT of Ts65Dn mice, suggesting that a heightened interferon response within the fat depot may potentially contribute to this effect. Whether obesity promotes a hyperactive interferon response in the adipose tissue of individuals with DS and mouse models remains to be determined. Nevertheless, outside of the context of trisomy, interferon signaling axis promoted by immune-adipose crosstalk in obesity apparently have both positive [147,148] and negative [[149], [150], [151]] impacts on systemic metabolism. Thus, the biological consequence of triplicated IFN receptor subunit genes is likely to be tissue-specific and context-dependent.
Liver plays a critical role in whole-body glucose and lipid metabolism, and increased inflammation and fibrosis compromises liver function [[152], [153], [154]]. In Ts65Dn mice, we observed clear inflammatory and fibrotic gene signatures, as indicated by the marked increase in expression of many pro-inflammatory and extracellular matrix-related genes. Despite the robust gene signatures, protein markers of fibrosis, inflammation, and oxidative stress were only marginally higher in Ts65Dn mice. Therefore, future studies are needed to uncover the underlying mechanism that contributes to diminished insulin action in liver with concomitant increase in hepatic glucose output, and elevated fasting blood glucose levels seen in Ts65Dn male mice. As with the iWAT, we also observed a significant increase in the expression of interferon receptor subunit genes (Ifnar1, Ifnar2, Ifngr1, Ifngr2) in Ts65Dn mouse liver. A heightened interferon response could potentially contribute to impaired liver metabolic function and the fibrotic gene signature we observed in Ts65Dn mouse liver. While this remains to be confirmed, recent studies have shown that interferon treatment promotes liver fibrosis in rat [155] and its deficiency attenuates hepatic inflammation, fibrosis, and insulin resistance in mice lacking interferon or its receptor [156,157]. Although the prevalence of non-alcoholic fatty liver disease (NAFLD) is significantly higher in people with DS [17], it is not known what percentage of those with NAFLD develop non-alcoholic steatohepatitis (NASH) characterized by liver steatosis, inflammation, and fibrosis. Future studies are warranted to assess whether Ts65Dn mice or other DS mouse models develop a more severe liver pathology with an accelerated time course when given a NASH-provoking diet.
The integrative and dynamic control of systemic metabolism involves inter-organ communication [158]. Our analysis showed that both tissue autonomous and non-autonomous mechanisms contribute to the metabolic phenotypes of Ts65Dn mice. The genetic correlation structure of Ts65Dn DEGs showed discrete blocks of co-expression specific to liver and adipose tissue. Our gene-centrality and key driver estimate analyses, however, also identified potential endocrine drivers (e.g., ATRN) that mediate an adipose-to-liver crosstalk, connecting the top pathways dysregulated in adipose tissue (ribosome and translational machinery) and liver (inflammation). Thus, the triplicated genes and their effects on other disomic genes in peripheral tissues likely affect whole-body metabolism via a combination of tissue-specific and cross-tissue mechanisms.
The freely segregating marker chromosome in Ts65Dn mouse arose from translocation of the Mmu16 telomeric region to the centromeric region of Mmu17 [51]. Despite being the major workhorse for the DS research community for the past 25 years, limitations of the Ts65Dn mouse model have been recognized with the mapping of the translocation breakpoint [77,78]. The presence of 46 trisomic protein-coding genes in the centromeric region of Mmu17 that are unrelated to Hsa21 may complicate the genotype–phenotype relationship. To circumvent this major limitation, CRISPR/Cas9-based genome editing approach has recently been used to remove the 46 genes from the Mmu17 centromeric region of the marker chromosome [159]. Behavioral and neurocognitive characterization of the refined Ts66Yah mice (trisomic for only Mmu16 genes) showed the conservation of key DS features, but with an overall milder phenotype compared to Ts65Dn mice [159], [160]. It should be noted that a modified Ts65Dn strain, 5252, which shows attenuated severity of phenotypes compared to its original version and to the original Ts65Dn strain, was used to create Ts66Yah. The studies performed here were carried out in the original 1924 strain of Ts65Dn. While we cannot rule out the potential contribution of the trisomic genes on Mmu17 to the metabolic phenotypes we observed in Ts65Dn, the new Ts66Yah mouse model could be useful in resolving this issue. However, no single DS mouse model fully recapitulates the karyotype, genotype, and phenotypes of DS. The continued generation and availability of improved DS mouse models, combined with omics tools and technologies, will catalyze further discoveries of the molecular, biochemical, and physiological underpinnings of DS phenotypes in trisomy 21.
In summary, we have provided the most in-depth and comprehensive analysis to date of the metabolic phenotypes of Ts65Dn mice across sex and diet. We show that dosage imbalance arising from the triplication of genes on the extra marker chromosome contributes to dysregulated systemic glucose and, to a much lesser degree, lipid metabolism in Ts65Dn mice, with more pronounced effects seen in HFD-fed male mice. The complex interactions of trisomic and disomic genes in major metabolic tissues such as adipose and liver likely contribute, at least in part, to the metabolic phenotypes of Ts65Dn mice. These data provide a valuable context to inform future metabolic studies of individuals with DS and other DS mouse models.
Author contributions
DCS, RHR, GWW: Conceptualization; GWW, DCS, SA, AJ, LMV, MMS: Formal analysis; GWW, RHR, MMS: Funding acquisition; DCS, CX, SA, GWW: Investigation; DCS, AJ, LMV, MMS: Methodology; GWW: Project administration; GWW: Supervision; DCS, GWW, MMS, LMV: Visualization; GWW, DCS, MMS: Roles/Writing - original draft; DCS, RHR, GWW, CX, SA, AJ, LMV, MMS: Writing - review & editing.
Acknowledgements
The work was funded, in part, by grants from the National Institutes of Health (DK084171 to GWW, HD098540 and HD038384 to RHR, HL138193 and DK130640 to MMS). The FPLC/serum analyses were conducted by the Mouse metabolic Phenotyping Center (MMPC) at Baylor College of medicine, funded by NIH grants DK114356 and UM1HG006348.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101666.
Conflict of interest
We declared that none of the authors has conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sherman S.L., Allen E.G., Bean L.H., Freeman S.B. Epidemiology of down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13(3):221–227. doi: 10.1002/mrdd.20157. [DOI] [PubMed] [Google Scholar]
- 2.Antonarakis S.E., Skotko B.G., Rafii M.S., Strydom A., Pape S.E., Bianchi D.W., et al. Down syndrome. Nat Rev Dis Prim. 2020;6(1):9. doi: 10.1038/s41572-019-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J., Tsai H.J., Gordon M.R., Li R. Cellular stress associated with aneuploidy. Dev Cell. 2018;44(4):420–431. doi: 10.1016/j.devcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korenberg J.R., Chen X.N., Schipper R., Sun Z., Gonsky R., Gerwehr S., et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A. 1994;91(11):4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonarakis S.E. Down syndrome and the complexity of genome dosage imbalance. Nat Rev Genet. 2017;18(3):147–163. doi: 10.1038/nrg.2016.154. [DOI] [PubMed] [Google Scholar]
- 6.Duchon A., Del Mar Muniz Moreno M., Martin Lorenzo S., Silva de Souza M.P., Chevalier C., Nalesso V., et al. Multi-influential genetic interactions alter behaviour and cognition through six main biological cascades in Down syndrome mouse models. Hum Mol Genet. 2021;30(9):771–788. doi: 10.1093/hmg/ddab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korbel J.O., Tirosh-Wagner T., Urban A.E., Chen X.N., Kasowski M., Dai L., et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A. 2009;106(29):12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaCombe J.M., Roper R.J. Skeletal dynamics of Down syndrome: a developing perspective. Bone. 2020;133 doi: 10.1016/j.bone.2019.115215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potier M.C., Reeves R.H. Editorial: intellectual disabilities in Down syndrome from birth and throughout life: assessment and treatment. Front Behav Neurosci. 2016;10:120. doi: 10.3389/fnbeh.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardiner K., Herault Y., Lott I.T., Antonarakis S.E., Reeves R.H., Dierssen M. Down syndrome: from understanding the neurobiology to therapy. J Neurosci. 2010;30(45):14943–14945. doi: 10.1523/JNEUROSCI.3728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olmos-Serrano J.L., Kang H.J., Tyler W.A., Silbereis J.C., Cheng F., Zhu Y., et al. Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron. 2016;89(6):1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti L., Galdzicki Z., Haydar T.F. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27(43):11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang P., Bush D., Schorge S., Good M., Canonica T., Shing N., et al. Altered Hippocampal-prefrontal neural dynamics in mouse models of Down syndrome. Cell Rep. 2020;30(4):1152–1163. doi: 10.1016/j.celrep.2019.12.065. e1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belichenko P.V., Kleschevnikov A.M., Becker A., Wagner G.E., Lysenko L.V., Yu Y.E., et al. Down syndrome cognitive phenotypes modeled in mice trisomic for all HSA 21 homologues. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0134861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das I., Park J.M., Shin J.H., Jeon S.K., Lorenzi H., Linden D.J., et al. Hedgehog agonist therapy corrects structural and cognitive deficits in a Down syndrome mouse model. Sci Transl Med. 2013;5(201) doi: 10.1126/scitranslmed.3005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q., Rasmussen S.A., Friedman J.M. Mortality associated with Down's syndrome in the USA from 1983 to 1997: a population-based study. Lancet. 2002;359(9311):1019–1025. doi: 10.1016/s0140-6736(02)08092-3. [DOI] [PubMed] [Google Scholar]
- 17.Valentini D., Alisi A., di Camillo C., Sartorelli M.R., Crudele A., Bartuli A., et al. Nonalcoholic fatty liver disease in Italian children with Down syndrome: prevalence and correlation with obesity-related features. J Pediatr. 2017;189:92–97. doi: 10.1016/j.jpeds.2017.05.077. e91. [DOI] [PubMed] [Google Scholar]
- 18.Buonuomo P.S., Bartuli A., Mastrogiorgio G., Vittucci A., Di Camillo C., Bianchi S., et al. Lipid profiles in a large cohort of Italian children with Down syndrome. Eur J Med Genet. 2016;59(8):392–395. doi: 10.1016/j.ejmg.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Adelekan T., Magge S., Shults J., Stallings V., Stettler N. Lipid profiles of children with Down syndrome compared with their siblings. Pediatrics. 2012;129(6):e1382–e1387. doi: 10.1542/peds.2011-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross T.J., Doran E., Cheema A.K., Head E., Lott I.T., Mapstone M. Plasma metabolites related to cellular energy metabolism are altered in adults with Down syndrome and Alzheimer's disease. Dev Neurobiol. 2019;79(7):622–638. doi: 10.1002/dneu.22716. [DOI] [PubMed] [Google Scholar]
- 21.Van Goor J.C., Massa G.G., Hirasing R. Increased incidence and prevalence of diabetes mellitus in Down's syndrome. Arch Dis Child. 1997;77(2):186. doi: 10.1136/adc.77.2.183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertapelli F., Pitetti K., Agiovlasitis S., Guerra-Junior G. Overweight and obesity in children and adolescents with Down syndrome-prevalence, determinants, consequences, and interventions: a literature review. Res Dev Disabil. 2016;57:181–192. doi: 10.1016/j.ridd.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Fonseca C.T., Amaral D.M., Ribeiro M.G., Beserra I.C., Guimaraes M.M. Insulin resistance in adolescents with Down syndrome: a cross-sectional study. BMC Endocr Disord. 2005;5:6. doi: 10.1186/1472-6823-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milunsky A., Neurath P.W. Diabetes mellitus in Down's syndrome. Arch Environ Health. 1968;17(3):372–376. doi: 10.1080/00039896.1968.10665244. [DOI] [PubMed] [Google Scholar]
- 25.Real de Asua D., Parra P., Costa R., Moldenhauer F., Suarez C. Evaluation of the impact of abdominal obesity on glucose and lipid metabolism disorders in adults with Down syndrome. Res Dev Disabil. 2014;35(11):2942–2949. doi: 10.1016/j.ridd.2014.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Helguera P., Seiglie J., Rodriguez J., Hanna M., Helguera G., Busciglio J. Adaptive downregulation of mitochondrial function in down syndrome. Cell Metabol. 2013;17(1):132–140. doi: 10.1016/j.cmet.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valenti D., Tullo A., Caratozzolo M.F., Merafina R.S., Scartezzini P., Marra E., et al. Impairment of F1F0-ATPase, adenine nucleotide translocator and adenylate kinase causes mitochondrial energy deficit in human skin fibroblasts with chromosome 21 trisomy. Biochem J. 2010;431(2):299–310. doi: 10.1042/BJ20100581. [DOI] [PubMed] [Google Scholar]
- 28.Valenti D., Manente G.A., Moro L., Marra E., Vacca R.A. Deficit of complex I activity in human skin fibroblasts with chromosome 21 trisomy and overproduction of reactive oxygen species by mitochondria: involvement of the cAMP/PKA signalling pathway. Biochem J. 2011;435(3):679–688. doi: 10.1042/BJ20101908. [DOI] [PubMed] [Google Scholar]
- 29.Panagaki T., Randi E.B., Augsburger F., Szabo C. Overproduction of H2S, generated by CBS, inhibits mitochondrial Complex IV and suppresses oxidative phosphorylation in Down syndrome. Proc Natl Acad Sci U S A. 2019;116(38):18769–18771. doi: 10.1073/pnas.1911895116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra V., Altamirano F., Hernandez-Fuentes C.P., Tong D., Kyrychenko V., Rotter D., et al. Down syndrome critical region 1 gene, Rcan1, helps maintain a more fused mitochondrial network. Circ Res. 2018;122(6):e20–e33. doi: 10.1161/CIRCRESAHA.117.311522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L., Huo H.Q., Lu K.Q., Tang X.Y., Hong Y., Han X., et al. Abnormal mitochondria in Down syndrome iPSC-derived GABAergic interneurons and organoids. Biochim Biophys Acta, Mol Basis Dis. 2022;1868(6) doi: 10.1016/j.bbadis.2022.166388. [DOI] [PubMed] [Google Scholar]
- 32.Mollo N., Esposito M., Aurilia M., Scognamiglio R., Accarino R., Bonfiglio F., et al. Human trisomic iPSCs from Down syndrome fibroblasts manifest mitochondrial alterations early during neuronal differentiation. Biology. 2021;10(7) doi: 10.3390/biology10070609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson C.C., Marentette J.O., Prutton K.M., Rauniyar A.K., Reisz J.A., D'Alessandro A., et al. Trisomy 21 results in modest impacts on mitochondrial function and central carbon metabolism. Free Radic Biol Med. 2021;172:201–212. doi: 10.1016/j.freeradbiomed.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piccoli C., Izzo A., Scrima R., Bonfiglio F., Manco R., Negri R., et al. Chronic pro-oxidative state and mitochondrial dysfunctions are more pronounced in fibroblasts from Down syndrome foeti with congenital heart defects. Hum Mol Genet. 2013;22(6):1218–1232. doi: 10.1093/hmg/dds529. [DOI] [PubMed] [Google Scholar]
- 35.Izzo A., Manco R., Bonfiglio F., Cali G., De Cristofaro T., Patergnani S., et al. NRIP1/RIP140 siRNA-mediated attenuation counteracts mitochondrial dysfunction in Down syndrome. Hum Mol Genet. 2014;23(16):4406–4419. doi: 10.1093/hmg/ddu157. [DOI] [PubMed] [Google Scholar]
- 36.Izzo A., Nitti M., Mollo N., Paladino S., Procaccini C., Faicchia D., et al. Metformin restores the mitochondrial network and reverses mitochondrial dysfunction in Down syndrome cells. Hum Mol Genet. 2017;26(6):1056–1069. doi: 10.1093/hmg/ddx016. [DOI] [PubMed] [Google Scholar]
- 37.Hill D.L., Parks E.P., Zemel B.S., Shults J., Stallings V.A., Stettler N. Resting energy expenditure and adiposity accretion among children with Down syndrome: a 3-year prospective study. Eur J Clin Nutr. 2013;67(10):1087–1091. doi: 10.1038/ejcn.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Aguero A., Ara I., Moreno L.A., Vicente-Rodriguez G., Casajus J.A. Fat and lean masses in youths with Down syndrome: gender differences. Res Dev Disabil. 2011;32(5):1685–1693. doi: 10.1016/j.ridd.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 39.Fernhall B., Figueroa A., Collier S., Goulopoulou S., Giannopoulou I., Baynard T. Resting metabolic rate is not reduced in obese adults with Down syndrome. Ment Retard. 2005;43(6):391–400. doi: 10.1352/0047-6765(2005)43[391:RMRINR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 40.Ptomey L.T., Willis E.A., Sherman J.R., White D.A., Donnelly J.E. Exploring the effectiveness of an 18-month weight management intervention in adults with Down syndrome using propensity score matching. J Intellect Disabil Res. 2020;64(3):221–233. doi: 10.1111/jir.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allison D.B., Gomez J.E., Heshka S., Babbitt R.L., Geliebter A., Kreibich K., et al. Decreased resting metabolic rate among persons with Down Syndrome. Int J Obes Relat Metab Disord. 1995;19(12):858–861. [PubMed] [Google Scholar]
- 42.Gutierrez-Hervas A., Gomez-Martinez S., Izquierdo-Gomez R., Veiga O.L., Perez-Bey A., Castro-Pinero J., et al. Inflammation and fatness in adolescents with and without Down syndrome: UP & DOWN study. J Intellect Disabil Res. 2020;64(2):170–179. doi: 10.1111/jir.12697. [DOI] [PubMed] [Google Scholar]
- 43.Magenis M.L., Machado A.G., Bongiolo A.M., Silva M.A.D., Castro K., Perry I.D.S. Dietary practices of children and adolescents with Down syndrome. J Intellect Disabil. 2018;22(2):125–134. doi: 10.1177/1744629516686571. [DOI] [PubMed] [Google Scholar]
- 44.Fox B., Moffett G.E., Kinnison C., Brooks G., Case L.E. Physical activity levels of children with Down syndrome. Pediatr Phys Ther. 2019;31(1):33–41. doi: 10.1097/PEP.0000000000000556. [DOI] [PubMed] [Google Scholar]
- 45.Luke A., Sutton M., Schoeller D.A., Roizen N.J. Nutrient intake and obesity in prepubescent children with Down syndrome. J Am Diet Assoc. 1996;96(12):1262–1267. doi: 10.1016/S0002-8223(96)00330-6. [DOI] [PubMed] [Google Scholar]
- 46.Phillips A.C., Sleigh A., McAllister C.J., Brage S., Carpenter T.A., Kemp G.J., et al. Defective mitochondrial function in vivo in skeletal muscle in adults with Down's syndrome: a 31P-MRS study. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0084031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fructuoso M., Rachdi L., Philippe E., Denis R.G., Magnan C., Le Stunff H., et al. Increased levels of inflammatory plasma markers and obesity risk in a mouse model of Down syndrome. Free Radic Biol Med. 2018;114:122–130. doi: 10.1016/j.freeradbiomed.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 48.Lanzillotta C., Tramutola A., Di Giacomo G., Marini F., Butterfield D.A., Di Domenico F., et al. Insulin resistance, oxidative stress and mitochondrial defects in Ts65dn mice brain: a harmful synergistic path in down syndrome. Free Radic Biol Med. 2021;165:152–170. doi: 10.1016/j.freeradbiomed.2021.01.042. [DOI] [PubMed] [Google Scholar]
- 49.Menzies C., Naz S., Patten D., Alquier T., Bennett B.M., Lacoste B. Distinct basal metabolism in three mouse models of neurodevelopmental disorders. eNeuro. 2021;8(2) doi: 10.1523/ENEURO.0292-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lana-Elola E., Cater H., Watson-Scales S., Greenaway S., Muller-Winkler J., Gibbins D., et al. Comprehensive phenotypic analysis of the Dp1Tyb mouse strain reveals a broad range of Down syndrome-related phenotypes. Dis Model Mech. 2021;14(10) doi: 10.1242/dmm.049157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davisson M.T., Schmidt C., Akeson E.C. Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Prog Clin Biol Res. 1990;360:263–280. [PubMed] [Google Scholar]
- 52.Davisson M.T., Schmidt C., Reeves R.H., Irving N.G., Akeson E.C., Harris B.S., et al. Segmental trisomy as a mouse model for Down syndrome. Prog Clin Biol Res. 1993;384:117–133. [PubMed] [Google Scholar]
- 53.Reeves R.H., Irving N.G., Moran T.H., Wohn A., Kitt C., Sisodia S.S., et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11(2):177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 54.Herault Y., Delabar J.M., Fisher E.M.C., Tybulewicz V.L.J., Yu E., Brault V. Rodent models in Down syndrome research: impact and future opportunities. Dis Model Mech. 2017;10(10):1165–1186. doi: 10.1242/dmm.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson L.E., Richtsmeier J.T., Leszl J., Reeves R.H. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306(5696):687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sago H., Carlson E.J., Smith D.J., Kilbridge J., Rubin E.M., Mobley W.C., et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci U S A. 1998;95(11):6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Yu T., Morishima M., Pao A., LaDuca J., Conroy J., et al. Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum Mol Genet. 2007;16(11):1359–1366. doi: 10.1093/hmg/ddm086. [DOI] [PubMed] [Google Scholar]
- 58.Yu T., Li Z., Jia Z., Clapcote S.J., Liu C., Li S., et al. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19(14):2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu T., Liu C., Belichenko P., Clapcote S.J., Li S., Pao A., et al. Effects of individual segmental trisomies of human chromosome 21 syntenic regions on hippocampal long-term potentiation and cognitive behaviors in mice. Brain Res. 2010;1366:162–171. doi: 10.1016/j.brainres.2010.09.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lana-Elola E., Watson-Scales S., Slender A., Gibbins D., Martineau A., Douglas C., et al. Genetic dissection of Down syndrome-associated congenital heart defects using a new mouse mapping panel. Elife. 2016;5 doi: 10.7554/eLife.11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereira P.L., Magnol L., Sahun I., Brault V., Duchon A., Prandini P., et al. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum Mol Genet. 2009;18(24):4756–4769. doi: 10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M.L., Cooke S., et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309(5743):2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazuki Y., Gao F.J., Li Y., Moyer A.J., Devenney B., Hiramatsu K., et al. A non-mosaic transchromosomic mouse model of down syndrome carrying the long arm of human chromosome 21. Elife. 2020;9 doi: 10.7554/eLife.56223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gribble S.M., Wiseman F.K., Clayton S., Prigmore E., Langley E., Yang F., et al. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tosh J., Tybulewicz V., Fisher E.M.C. Mouse models of aneuploidy to understand chromosome disorders. Mamm Genome. 2022;33(1):157–168. doi: 10.1007/s00335-021-09930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das I., Reeves R.H. The use of mouse models to understand and improve cognitive deficits in Down syndrome. Dis Model Mech. 2011;4(5):596–606. doi: 10.1242/dmm.007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa A.C., Walsh K., Davisson M.T. Motor dysfunction in a mouse model for Down syndrome. Physiol Behav. 1999;68(1–2):211–220. doi: 10.1016/s0031-9384(99)00178-x. [DOI] [PubMed] [Google Scholar]
- 68.Aziz N.M., Guedj F., Pennings J.L.A., Olmos-Serrano J.L., Siegel A., Haydar T.F., et al. Lifespan analysis of brain development, gene expression and behavioral phenotypes in the Ts1Cje, Ts65Dn and Dp(16)1/Yey mouse models of Down syndrome. Dis Model Mech. 2018;11(6) doi: 10.1242/dmm.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watson-Scales S., Kalmar B., Lana-Elola E., Gibbins D., La Russa F., Wiseman F., et al. Analysis of motor dysfunction in Down Syndrome reveals motor neuron degeneration. PLoS Genet. 2018;14(5) doi: 10.1371/journal.pgen.1007383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brault V., Duchon A., Romestaing C., Sahun I., Pothion S., Karout M., et al. Opposite phenotypes of muscle strength and locomotor function in mouse models of partial trisomy and monosomy 21 for the proximal Hspa13-App region. PLoS Genet. 2015;11(3) doi: 10.1371/journal.pgen.1005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schill E.M., Wright C.M., Jamil A., LaCombe J.M., Roper R.J., Heuckeroth R.O. Down syndrome mouse models have an abnormal enteric nervous system. JCI Insight. 2019;5 doi: 10.1172/jci.insight.124510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas J.R., LaCombe J., Long R., Lana-Elola E., Watson-Scales S., Wallace J.M., et al. Interaction of sexual dimorphism and gene dosage imbalance in skeletal deficits associated with Down syndrome. Bone. 2020;136 doi: 10.1016/j.bone.2020.115367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore C.S. Postnatal lethality and cardiac anomalies in the Ts65Dn Down syndrome mouse model. Mamm Genome. 2006;17(10):1005–1012. doi: 10.1007/s00335-006-0032-8. [DOI] [PubMed] [Google Scholar]
- 74.Chakrabarti L., Best T.K., Cramer N.P., Carney R.S., Isaac J.T., Galdzicki Z., et al. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13(8):927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Edie S., Klinedinst D., Jeong J.S., Blackshaw S., Maslen C.L., et al. Penetrance of congenital heart disease in a mouse model of Down syndrome depends on a trisomic potentiator of a disomic modifier. Genetics. 2016;203(2):763–770. doi: 10.1534/genetics.116.188045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta M., Dhanasekaran A.R., Gardiner K.J. Mouse models of Down syndrome: gene content and consequences. Mamm Genome. 2016;27(11–12):538–555. doi: 10.1007/s00335-016-9661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duchon A., Raveau M., Chevalier C., Nalesso V., Sharp A.J., Herault Y. Identification of the translocation breakpoints in the Ts65Dn and Ts1Cje mouse lines: relevance for modeling Down syndrome. Mamm Genome. 2011;22(11–12):674–684. doi: 10.1007/s00335-011-9356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reinholdt L.G., Ding Y., Gilbert G.J., Czechanski A., Solzak J.P., Roper R.J., et al. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm Genome. 2011;22(11–12):685–691. doi: 10.1007/s00335-011-9357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodliffe J.W., Olmos-Serrano J.L., Aziz N.M., Pennings J.L., Guedj F., Bianchi D.W., et al. Absence of prenatal forebrain defects in the Dp(16)1Yey/+ mouse model of Down syndrome. J Neurosci. 2016;36(10):2926–2944. doi: 10.1523/JNEUROSCI.2513-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gardiner K.J. Pharmacological approaches to improving cognitive function in Down syndrome: current status and considerations. Drug Des Dev Ther. 2015;9:103–125. doi: 10.2147/DDDT.S51476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Faizi M., Bader P.L., Tun C., Encarnacion A., Kleschevnikov A., Belichenko P., et al. Comprehensive behavioral phenotyping of Ts65Dn mouse model of Down syndrome: activation of beta1-adrenergic receptor by xamoterol as a potential cognitive enhancer. Neurobiol Dis. 2011;43(2):397–413. doi: 10.1016/j.nbd.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez F., Morishita W., Zuniga E., Nguyen J., Blank M., Malenka R.C., et al. Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat Neurosci. 2007;10(4):411–413. doi: 10.1038/nn1860. [DOI] [PubMed] [Google Scholar]
- 83.Sarver D.C., Stewart A.N., Rodriguez S., Little H.C., Aja S., Wong G.W. Loss of CTRP4 alters adiposity and food intake behaviors in obese mice. Am J Physiol Endocrinol Metab. 2020;319(6):E1084–E1100. doi: 10.1152/ajpendo.00448.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei Z., Lei X., Petersen P.S., Aja S., Wong G.W. Targeted deletion of C1q/TNF-related protein 9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice. Am J Physiol Endocrinol Metab. 2014;306(7):E779–E790. doi: 10.1152/ajpendo.00593.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarver D.C., Xu C., Aja S., Wong G.W. CTRP14 inactivation alters physical activity and food intake response to fasting and refeeding. Am J Physiol Endocrinol Metab. 2022;322(6):E480–E493. doi: 10.1152/ajpendo.00002.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodriguez S., Lei X., Petersen P.S., Tan S.Y., Little H.C., Wong G.W. Loss of CTRP1 disrupts glucose and lipid homeostasis. Am J Physiol Endocrinol Metab. 2016;311:E678–E697. doi: 10.1152/ajpendo.00087.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lei X., Wong G.W. C1q/TNF-related protein 2 (CTRP2) deletion promotes adipose tissue lipolysis and hepatic triglyceride secretion. J Biol Chem. 2019;294(43):15638–15649. doi: 10.1074/jbc.RA119.009230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan S.Y., Little H.C., Lei X., Li S., Rodriguez S., Wong G.W. Partial deficiency of CTRP12 alters hepatic lipid metabolism. Physiol Genom. 2016;48(12):936–949. doi: 10.1152/physiolgenomics.00111.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 92.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Acin-Perez R., Benador I.Y., Petcherski A., Veliova M., Benavides G.A., Lagarrigue S., et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 2020;39(13) doi: 10.15252/embj.2019104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Mouse Genome Database G. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42(Database issue):D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Consortium G.T. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glass T.J., Valmadrid L.C.V., Connor N.P. The adult Ts65Dn mouse model of Down syndrome shows altered swallow function. Front Neurosci. 2019;13:906. doi: 10.3389/fnins.2019.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Magnusson I., Rothman D.L., Katz L.D., Shulman R.G., Shulman G.I. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest. 1992;90(4):1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pilkis S.J., el-Maghrabi M.R., Claus T.H. Hormonal regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Biochem. 1988;57:755–783. doi: 10.1146/annurev.bi.57.070188.003543. [DOI] [PubMed] [Google Scholar]
- 103.McGarry J.D., Foster D.W. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 104.Sultan M., Piccini I., Balzereit D., Herwig R., Saran N.G., Lehrach H., et al. Gene expression variation in Down's syndrome mice allows prioritization of candidate genes. Genome Biol. 2007;8(5):R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 106.Velez L.M., Van C., Moore T., Zhou Z., Johnson C., Hevener A.L., et al. Genetic variation of putative myokine signaling is dominated by biological sex and sex hormones. Elife. 2022;11 doi: 10.7554/eLife.76887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seldin M.M., Lusis A.J. Systems-based approaches for investigation of inter-tissue communication. J Lipid Res. 2019;60(3):450–455. doi: 10.1194/jlr.S090316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Petrus P., Cervantes M., Samad M., Sato T., Chao A., Sato S., et al. Tryptophan metabolism is a physiological integrator regulating circadian rhythms. Mol Metabol. 2022;64 doi: 10.1016/j.molmet.2022.101556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao Y., Wang Y., Zhou Z., Pan C., Jiang L., Zhou Z., et al. Liver-heart cross-talk mediated by coagulation factor XI protects against heart failure. Science. 2022;377(6613):1399–1406. doi: 10.1126/science.abn0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peiris H., Duffield M.D., Fadista J., Jessup C.F., Kashmir V., Genders A.J., et al. A syntenic cross species aneuploidy genetic screen links RCAN1 expression to beta-Cell mitochondrial dysfunction in type 2 diabetes. PLoS Genet. 2016;12(5) doi: 10.1371/journal.pgen.1006033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coussons-Read M.E., Crnic L.S. Behavioral assessment of the Ts65Dn mouse, a model for Down syndrome: altered behavior in the elevated plus maze and open field. Behav Genet. 1996;26(1):7–13. doi: 10.1007/BF02361154. [DOI] [PubMed] [Google Scholar]
- 113.Escorihuela R.M., Fernandez-Teruel A., Vallina I.F., Baamonde C., Lumbreras M.A., Dierssen M., et al. A behavioral assessment of Ts65Dn mice: a putative Down syndrome model. Neurosci Lett. 1995;199(2):143–146. doi: 10.1016/0304-3940(95)12052-6. [DOI] [PubMed] [Google Scholar]
- 114.Moran T.H., Capone G.T., Knipp S., Davisson M.T., Reeves R.H., Gearhart J.D. The effects of piracetam on cognitive performance in a mouse model of Down's syndrome. Physiol Behav. 2002;77(2–3):403–409. doi: 10.1016/s0031-9384(02)00873-9. [DOI] [PubMed] [Google Scholar]
- 115.Pueschel S.M., Bernier J.C., Pezzullo J.C. Behavioural observations in children with Down's syndrome. J Ment Defic Res. 1991;35(Pt 6):502–511. doi: 10.1111/j.1365-2788.1991.tb00447.x. [DOI] [PubMed] [Google Scholar]
- 116.Ekstein S., Glick B., Weill M., Kay B., Berger I. Down syndrome and attention-deficit/hyperactivity disorder (ADHD) J Child Neurol. 2011;26(10):1290–1295. doi: 10.1177/0883073811405201. [DOI] [PubMed] [Google Scholar]
- 117.Izquierdo-Gomez R., Martinez-Gomez D., Acha A., Veiga O.L., Villagra A., Diaz-Cueto M., et al. Objective assessment of sedentary time and physical activity throughout the week in adolescents with Down syndrome. The UP&DOWN study. Res Dev Disabil. 2014;35(2):482–489. doi: 10.1016/j.ridd.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 118.Pueschel S.M., Craig W.Y., Haddow J.E. Lipids and lipoproteins in persons with Down's syndrome. J Intellect Disabil Res. 1992;36(Pt 4):365–369. doi: 10.1111/j.1365-2788.1992.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 119.Zamorano A., Guzman M., Aspillaga M., Avendano A., Gatica M. [Concentrations of serum lipids in children with Down's syndrome] Arch Biol Med Exp. 1991;24(1):49–55. [PubMed] [Google Scholar]
- 120.Mauvais-Jarvis F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol Sex Differ. 2015;6:14. doi: 10.1186/s13293-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barros R.P., Gustafsson J.A. Estrogen receptors and the metabolic network. Cell Metabol. 2011;14(3):289–299. doi: 10.1016/j.cmet.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 122.Hawley L.E., Prochaska F., Stringer M., Goodlett C.R., Roper R.J. Sexually dimorphic DYRK1A overexpression on postnatal day 15 in the Ts65Dn mouse model of Down syndrome: effects of pharmacological targeting on behavioral phenotypes. Pharmacol Biochem Behav. 2022;217 doi: 10.1016/j.pbb.2022.173404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kelley C.M., Powers B.E., Velazquez R., Ash J.A., Ginsberg S.D., Strupp B.J., et al. Sex differences in the cholinergic basal forebrain in the Ts65Dn mouse model of Down syndrome and Alzheimer's disease. Brain Pathol. 2014;24(1):33–44. doi: 10.1111/bpa.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Block A., Ahmed M.M., Dhanasekaran A.R., Tong S., Gardiner K.J. Sex differences in protein expression in the mouse brain and their perturbations in a model of Down syndrome. Biol Sex Differ. 2015;6:24. doi: 10.1186/s13293-015-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peiris H., Raghupathi R., Jessup C.F., Zanin M.P., Mohanasundaram D., Mackenzie K.D., et al. Increased expression of the glucose-responsive gene, RCAN1, causes hypoinsulinemia, beta-cell dysfunction, and diabetes. Endocrinology. 2012;153(11):5212–5221. doi: 10.1210/en.2011-2149. [DOI] [PubMed] [Google Scholar]
- 126.Keating D.J., Dubach D., Zanin M.P., Yu Y., Martin K., Zhao Y.F., et al. DSCR1/RCAN1 regulates vesicle exocytosis and fusion pore kinetics: implications for Down syndrome and Alzheimer's disease. Hum Mol Genet. 2008;17(7):1020–1030. doi: 10.1093/hmg/ddm374. [DOI] [PubMed] [Google Scholar]
- 127.Seo D.S., Chau G.C., Baek K.H., Um S.H. A single extra copy of Down syndrome critical region 1-4 results in impaired hepatic glucose homeostasis. Mol Metabol. 2019;21:82–89. doi: 10.1016/j.molmet.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reeves R.H., Baxter L.L., Richtsmeier J.T. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet. 2001;17(2):83–88. doi: 10.1016/s0168-9525(00)02172-7. [DOI] [PubMed] [Google Scholar]
- 129.Prasher V.P., Metseagharun T., Haque S. Weight loss in adults with Down syndrome and with dementia in Alzheimer's disease. Res Dev Disabil. 2004;25(1):1–7. doi: 10.1016/j.ridd.2003.04.005. [DOI] [PubMed] [Google Scholar]
- 130.Morton G.J., Cummings D.E., Baskin D.G., Barsh G.S., Schwartz M.W. Central nervous system control of food intake and body weight. Nature. 2006;443(7109):289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 131.Coll A.P., Farooqi I.S., O'Rahilly S. The hormonal control of food intake. Cell. 2007;129(2):251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y., Borel C., Li L., Muller T., Williams E.G., Germain P.L., et al. Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat Commun. 2017;8(1):1212. doi: 10.1038/s41467-017-01422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Houstis N., Rosen E.D., Lander E.S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440(7086):944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 134.Sun K., Tordjman J., Clement K., Scherer P.E. Fibrosis and adipose tissue dysfunction. Cell Metabol. 2013;18(4):470–477. doi: 10.1016/j.cmet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 136.Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hill D.A., Lim H.W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L., et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci U S A. 2018;115(22):E5096–E5105. doi: 10.1073/pnas.1802611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lumeng C.N., Bodzin J.L., Saltiel A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sullivan K.D., Evans D., Pandey A., Hraha T.H., Smith K.P., Markham N., et al. Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-13858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Araya P., Waugh K.A., Sullivan K.D., Nunez N.G., Roselli E., Smith K.P., et al. Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity. Proc Natl Acad Sci U S A. 2019;116(48):24231–24241. doi: 10.1073/pnas.1908129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sullivan K.D., Lewis H.C., Hill A.A., Pandey A., Jackson L.P., Cabral J.M., et al. Trisomy 21 consistently activates the interferon response. Elife. 2016;5 doi: 10.7554/eLife.16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Waugh K.A., Araya P., Pandey A., Jordan K.R., Smith K.P., Granrath R.E., et al. Mass cytometry reveals global immune remodeling with multi-lineage hypersensitivity to type I interferon in Down syndrome. Cell Rep. 2019;29(7):1893–1908. doi: 10.1016/j.celrep.2019.10.038. e1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mathis D. Immunological goings-on in visceral adipose tissue. Cell Metabol. 2013;17(6):851–859. doi: 10.1016/j.cmet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wieser V., Adolph T.E., Grander C., Grabherr F., Enrich B., Moser P., et al. Adipose type I interferon signalling protects against metabolic dysfunction. Gut. 2018;67(1):157–165. doi: 10.1136/gutjnl-2016-313155. [DOI] [PubMed] [Google Scholar]
- 148.Ying W., Kanameni S., Chang C.A., Nair V., Safe S., Bazer F.W., et al. Interferon tau alleviates obesity-induced adipose tissue inflammation and insulin resistance by regulating macrophage polarization. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0098835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chan C.C., Damen M., Moreno-Fernandez M.E., Stankiewicz T.E., Cappelletti M., Alarcon P.C., et al. Type I interferon sensing unlocks dormant adipocyte inflammatory potential. Nat Commun. 2020;11(1):2745. doi: 10.1038/s41467-020-16571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bradley D., Smith A.J., Blaszczak A., Shantaram D., Bergin S.M., Jalilvand A., et al. Interferon gamma mediates the reduction of adipose tissue regulatory T cells in human obesity. Nat Commun. 2022;13(1):5606. doi: 10.1038/s41467-022-33067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li C., Wang G., Sivasami P., Ramirez R.N., Zhang Y., Benoist C., et al. Interferon-alpha-producing plasmacytoid dendritic cells drive the loss of adipose tissue regulatory T cells during obesity. Cell Metabol. 2021;33(8):1610–1623. doi: 10.1016/j.cmet.2021.06.007. e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chang M.L., Yang S.S. Metabolic signature of hepatic fibrosis: from individual pathways to systems biology. Cells. 2019;8(11) doi: 10.3390/cells8111423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wree A., Broderick L., Canbay A., Hoffman H.M., Feldstein A.E. From NAFLD to NASH to cirrhosis-new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol. 2013;10(11):627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 154.Koyama Y., Brenner D.A. Liver inflammation and fibrosis. J Clin Invest. 2017;127(1):55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mohlenberg M., Eriksen P.L., Laursen T.L., Nielsen M.B., Hamilton Dutoit S.J., Gronbaek H., et al. The presence of interferon affects the progression of non-alcoholic fatty liver disease. Gene Immun. 2022;23(5):157–165. doi: 10.1038/s41435-022-00176-6. [DOI] [PubMed] [Google Scholar]
- 156.Luo X.Y., Takahara T., Kawai K., Fujino M., Sugiyama T., Tsuneyama K., et al. IFN-gamma deficiency attenuates hepatic inflammation and fibrosis in a steatohepatitis model induced by a methionine- and choline-deficient high-fat diet. Am J Physiol Gastrointest Liver Physiol. 2013;305(12):G891–G899. doi: 10.1152/ajpgi.00193.2013. [DOI] [PubMed] [Google Scholar]
- 157.Ghazarian M., Revelo X.S., Nohr M.K., Luck H., Zeng K., Lei H., et al. Type I interferon responses drive intrahepatic T cells to promote metabolic syndrome. Sci Immunol. 2017;2(10) doi: 10.1126/sciimmunol.aai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Priest C., Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nat Metab. 2019;1(12):1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 159.Duchon A., Muniz Moreno M.D.M., Chevalier C., Nalesso V., Andre P., Fructuoso-Castellar M., et al. Ts66Yah, an upgraded Ts65Dn mouse model for down syndrome, for only the region homologous to human chromosome 21. Dis Model Mech. 2022 doi: 10.1242/dmm.049721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guedj F., Kane E., Bishop L.A., Pennings J.L., Herault Y., Bianchi D.W. The impact of Mmu17 non-Hsa21 orthologous genes in the Ts65Dn mouse model of Down syndrome: the “gold standard” revisited. bioRxiv. 2022 doi: 10.1101/2022.09.23.509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.