Abstract
In recent years, the use of artificial intelligence (AI) in health care has risen steadily, including a wide range of applications in the field of pharmacology. AI is now used throughout the entire continuum of pharmacology research and clinical practice and from early drug discovery to real‐world datamining. The types of AI models used range from unsupervised clustering of drugs or patients aimed at identifying potential drug compounds or suitable patient populations, to supervised machine learning approaches to improve therapeutic drug monitoring. Additionally, natural language processing is increasingly used to mine electronic health records to obtain real‐world data. In this mini‐review, we discuss the basics of AI followed by an outline of its application in pharmacology research and clinical practice.
In recent decades, artificial intelligence (AI), and in particular machine learning (ML), has rapidly gained traction in healthcare applications. This includes the field of pharmacology, where AI and ML approaches are particularly useful to analyze data from different sources, ranging from the chemical structure of a drug to clinical patient characteristics and from genomic data to disease characteristics. The rise of AI applications in pharmacology is also evident in the number of studies published on the topic. A PubMed search for “artificial intelligence” AND “pharmacology” shows that whereas in 2017 only 49 papers on the subject were published, by 2021 this had increased 10‐fold to 502 publications (https://pubmed.ncbi.nlm.nih.gov).
AI is already successfully applied in drug discovery and target identification for several years. More recently AI models which help to characterize patient populations and predict an individual's drug response are emerging, thereby covering the entire pipeline from drug discovery to personalized medicine (Figure 1). In 2020, the use of AI in clinical pharmacology was extensively discussed. 1 Many of these applications are still used and relevant. However, there have been many new developments since. In this mini‐review, we will discuss the current application of different AI and ML approaches in the field of pharmacology.
FIGURE 1.
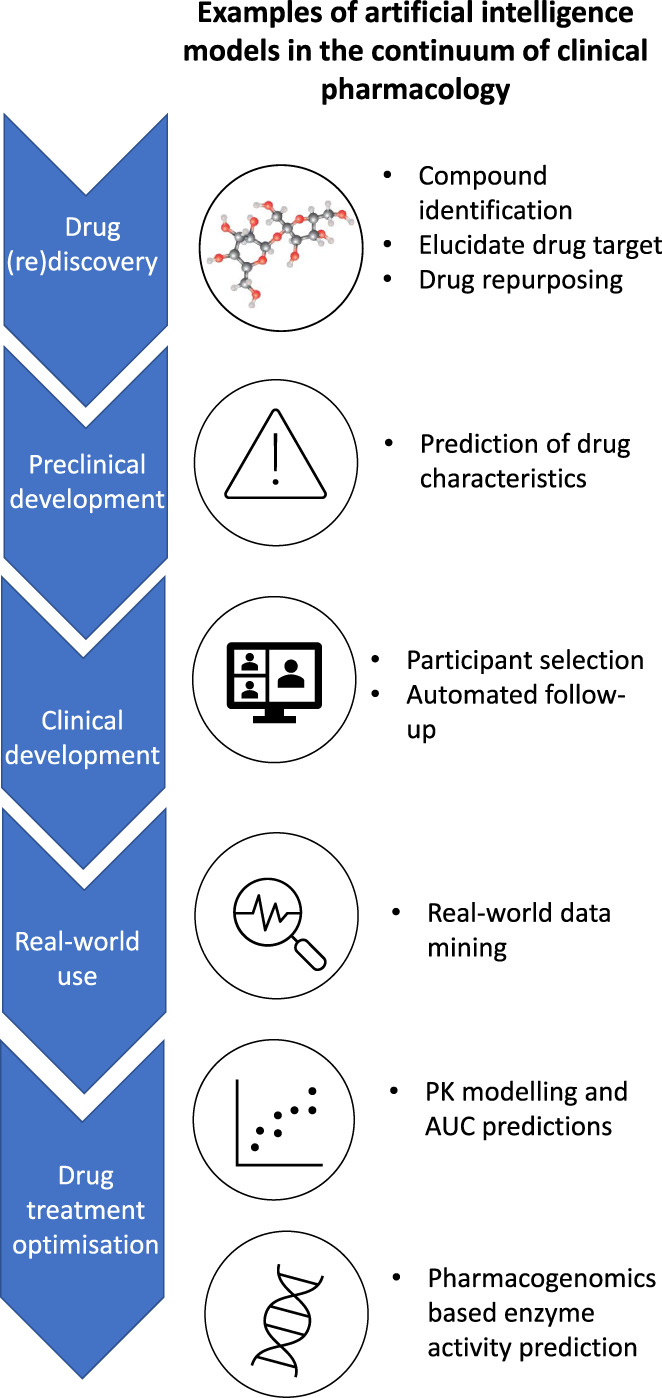
Applications of artificial intelligence in the continuum of clinical pharmacology. AUC, area under the curve; PK, pharmacokinetic.
THE BASICS OF AI
AI describes all forms of intelligence in machines. This ranges from reasoning and language processing to learning based applications. ML is one of the most commonly used forms of AI in clinical applications. ML is a type of AI which is heavily based on statistical methods and relies on the capability of computers to infer relations and make predictions rather than human efforts. ML approaches can be further subdivided in unsupervised learning to explore and cluster data, and supervised learning, aimed at predicting outcomes (Figure 2). Unsupervised learning is often used to develop hypotheses, for example, if there are subgroups of patients with good response that can be identified in a larger cohort. Supervised learning, on the other hand, is most commonly applied to develop models that can accurately predict one or more clinical outcomes, such as predictions of drug response for an individual patient. One commonly used example of supervised learning is a neural network. An artificial neural network is structured to mimic biological neural networks. A neural network consists of input‐ and output layer and hidden layers in between, the nodes connecting the different layers get different weights which are optimized during the training. Neural networks are particularly good at recognizing patterns in the data they are trained on and can process fast amounts of data. This makes them useful and commonly applied in a wide range of applications. Each ML model is based on a theoretical understanding of the problem at hand, and on an experimental part that the model learns upon training. The larger the experimental part, the more difficult a model is to understand, the so‐called black‐box models. Although it is still understood what goes into the model and what comes out, the internal reasoning and weighing of factors is not transparent unless measures are taken to make these factors transparent.
FIGURE 2.
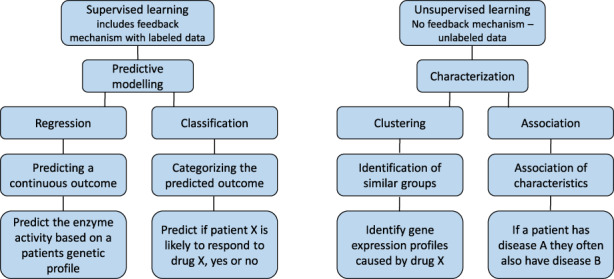
Comparison of supervised and unsupervised learning approaches in clinical pharmacology.
Besides ML, natural language processing (NLP) is also increasingly used. NLP is the domain within AI that focusses on processing and interpreting natural human language and can, for example, help to “read” and process the huge amount of (un)structured text. In pharmacology, NLP can be used to mine databases and electronic health records (EHRs) to obtain data related to pharmacology, such as clinical outcomes. 2 , 3 Advanced NLP approaches can also help to interpret the data and filter the most important items and structure the data to allow for interpretation and further studies.
AI IN DRUG (RE)DISCOVERY
Drug discovery is a long process that involves an extensive pipeline with multiple selection steps. Although the process starts with many lead compounds, at each step of the development pipeline, a stringent selection is applied to select the most favorable compounds to finally deliver one compound receiving market approval. The more compounds that need to be tested, the higher the costs of drug development. AI can be applied to help select the compounds that are most likely to be successful by analyzing the chemical structure and properties. The way AI has been applied in drug discovery is multifaceted. First, if the target is known, AI can be used to predict what type of chemical structures might bind the target in the desired way. Second, the chemical structure of known and efficient drugs or endogenic factors can be used to identify the targets, thereby elucidating the structure of a potential drug target. Last, the in vivo characteristics of a novel compound can be predicted by using knowledge of known drug compounds, pharmacokinetics (PKs), and pharmacodynamics. If in the early stages of drug discovery, it can be predicted if a compound is likely to fail, either by not binding to the desired target or by potentially undesirable absorption, distribution, metabolism, and excretion (ADME) characteristics, the investment in the development can be stopped before expensive trials have started. These applications of AI are already standard practice in many pharmaceutical companies 4 , 5 , 6 to the point that companies (e.g., Cytoreason) have been founded to specifically develop and offer models to pharmaceutical companies to help them study disease and drug pathways.
In a later phase of the development pipeline, the toxicity profile of a novel compound needs to be assessed. This requires substantial datasets with in vivo data obtained in clinical studies. However, as chemical structure plays a large role in the occurrence of toxicity, the same approaches as in drug discovery could be applied. Two important toxicities that are assessed during drug development are cardiotoxicity and hepatotoxicity. Therefore, being able to predict if a compound causes these toxicities in an early stage of the development pipeline would greatly reduce the risk of compound failure, and several groups applied AI approaches to predict them. 7 , 8 , 9 Mamoshina et al. have tested the feasibility of an AI‐based model to predict cardiotoxicity of different compounds. They developed a model to predict cardiotoxicity based on drug characteristics obtained from publicly available data (e.g., Drugbank and medDRA). The model was able to predict cardiotoxicity with high accuracy (area under the curve [AUC] 79% for validation data and AUC 66% for unseen data) by classifying drugs as safe or risky. 10 Similar approaches have been used to predict drug‐induced liver injury from drug characteristics reaching even higher accuracies with 89% correct classifications. 11
The application of AI cannot be limited to selection of the most optimal compound and predict its' in vivo activity. It can also be used to find new applications for existing drugs with an established risk/benefit ratio. This drug repositioning is particularly valuable for small patient populations for which clinical trials are not feasible and the costs for drug development are often considered too high. There are different forms of drug repositioning, all of which require large datasets and intensive computational analyses. First, with a drug‐focused approach, existing drugs are compared to novel compounds to identify compounds with similar properties that may be successfully used to treat the therapeutic indications of the comparator drug. Second, a disease‐focused approach aims to compare disease characteristics and pathways to identify drugs that work for diseases with similar characteristics. Most often these two strategies are combined, for example, by matching gene expression profiles of a disease with the profiles caused by different drugs to find a match. Al‐taie et al. used this combinatorial approach to identify new therapies for colorectal cancer (CRC). 12 They combined clinical data and RNA‐sequencing data with publicly available information regarding drug expression profiles (Hetionet – neo4j.het.io). 13 An AI model was trained to identify subpopulations in the clinical data. Next, the unique gene profiles of each subgroup were compared with the drug database to identify potential candidates for repurposing. From the top 16 drugs identified this way, 12 are cancer‐related drugs of which eight are registered for CRC treatment illustrating the potential of this approach. 12 Similar applications of AI models have been used for other disease areas as well. For example, a study focusing on Alzheimer's disease found 103 hits of which three were supported with population‐based validation studies. 14 For diabetes, Zhang et al. used an approach to identify potential drug targets followed by the identification of drugs which are known to impact these targets. This led to 58 drugs identified of which nine were found to be relevant based on connectivity in gene expression profiles. 15
CLINICAL TRIALS AND REAL‐WORLD EVIDENCE
After the preclinical development of a new drug, the clinical trial phase starts, which brings even higher costs. Some of the biggest challenges in clinical trials are related to patient selection and recruitment. If insufficient patients can be recruited, the trial might need to be stopped. AI applications can help to improve the patient recruitment, for example, by processing large amounts of EHR data to select the patients that meet the eligibility criteria that are most likely to participate. This ensures that less time and money are lost in patient recruitment. Second, AI can help to monitor patients during the trial. This can be done by datamining the EHR but also by processing real‐time data from participants that are collected by trackers, such as smartwatches and other wearables. Finally, AI approaches can help to collect real‐world evidence by mining EHRs for relevant data (as described below), as well as by processing these data into clinically meaningful outcomes. 16 , 17 An example are the abundance of AI‐based approaches available for automated processing of imaging data which are shown to have similar accuracy rates as a radiologist.
Similar to the use in clinical trials, AI can also be used for the analysis of real‐world clinical data. EHRs are a rich source of information regarding disease progress and drug response, making them an ideal source for postmarket studies. However, the sheer amount of (unstructured) data collected in the EHRs makes it very challenging and time intensive to conduct large and thorough studies. Recently, programs that use NLP have been developed to process the data in an EHR to make it useful for analysis. This does, however, come with challenges. An EHR contains many different sources of data which all have different levels of reliability and robustness. Laboratory values, for example, are numeric and objective, making them relatively easy to extract. Unstructured text on the other hand is subject to many different variables, not in the least the use of abbreviations and typos. Van Laar et al. evaluated the accuracy of datamining in EHRs by comparing automated extractions with manual extractions. They focused on treatment outcomes of renal cell carcinoma treatment. In general, they found high precision and recall for structured data with and F1 score of 100% for variables as sex, death, and greater than 90% for variables, such as laboratory measurements. The accuracy was significantly lower for unstructured data, such as adverse drug events and comorbidities (F1 score 53%–90%) which are often part of the free text in an EHR and therefore more challenging to extract accurately. 2
Besides the use of AI in conventional clinical trials and datamining, AI can also be used to predict drug response for a specific patient by using digital twins. A digital twin is a digital profile that matches the real‐world patient and can be used, after training, to model the impact of different treatments on disease outcome for that one specific patient. 18
DRUG TREATMENT OPTIMIZATION
For many marketed drugs, it is useful and important to individualize the treatment. For example, for drugs with a small therapeutic window, therapeutic drug monitoring (TDM) is applied to individualize the dose. Methods to extrapolate TDM data to calculate drug exposure and optimal treatment strategies are often based on statistical prediction models. The application of AI in this field is less developed compared to drug discovery, mainly due to the fact that large clinical datasets are needed to train the models which are not readily available. 9 , 19 For example, Labriffe et al. have trained several XGBoost models to simulate patient PK profiles of everolimus. An XGBoost model is a form of supervised learning which works by combining multiple (weak) decision trees to obtain a strong overall model. They trained the models with different combinations of simulated and real patient PK profiles and TDM measurements of everolimus at three timepoints (predose, 1, and 2 h). The best performing model was trained using 5016 simulated PK profiles and resulted in a model that was able to accurately predict the everolimus AUC for an external validation set (n = 114, R 2 = 0.956, root mean squared error [RMSE] = 10.3%). 20 Similar approaches have been developed for other drugs. 21 , 22 , 23 However, by design, these models will always have the same error margin as the TDM measurements that were used to train the model. Meaning that an AI model can never be more accurate than the outcome on which it was trained.
Besides TDM‐based treatment optimization there is also a shift ongoing in the field of pharmacogenomics (PGx). The field of PGx mainly focusses on explaining PKs by studying genetic variants in the genes encoding for drug metabolizing enzymes. Current clinical PGx practice still relies on prediction models using a small subset of genetic variants to predict an individual's enzyme activity in a categorical model. The sheer abundance of genetic variants and the complexity of enzyme activity, which is a continuous variable and not categorical, makes it impossible to develop basic prediction models without the help of AI. A good example of a pharmacogene suffering from these limitations is the CYP2D6 gene, of which the resulting enzyme is responsible for the metabolism of 25%–30% of commonly prescribed drugs. Due to CYP2D6's complexity and the high amount of variants in this gene it is still challenging to accurately group individuals into predicted enzyme activity groups. Recently, two different approaches to use AI for the prediction of CYP2D6 activity have been published. First, McInnes et al. aimed to develop a model which can predict the activity of novel haplotypes which are not yet curated in the publicly available PGx variant database PharmVar (https://www.pharmvar.org/). A network model was trained on curated alleles from the PharmVar database, followed by the prediction of uncurated haplotypes. The model was able to predict the activity of the validation set with 88% accuracy, indicating that this model can help to assign activities to uncurated alleles. 24 Second, in our own study, we developed a neural network model to predict enzyme activity of CYP2D6 based on long‐read sequencing data. The model was trained using full length CYP2D6 sequencing as its input in the form of two alleles of 77 variants each, followed by a combiner model that combined the result of the two alleles. As output parameter, the metabolic ratio between endoxifen and desmethyltamoxifen was used as a proxy for CYP2D6 activity. The final model was able to explain 79% of the variability in CYP2D6 mediated metabolism, whereas the conventional categorical model was able to explain only 54%. 25 The advantage of neural network‐based approaches in pharmacogenetics is that these models are capable of identifying patterns in the input data which goes beyond human capability. Traditionally, the field of pharmacogenetics relies on curated patterns (haplotypes) to assign activities to individual combinations of variants. By applying AI to perform pattern recognition and activity assignment, higher accuracies can be achieved, as is clearly illustrated by the two examples above.
Finally, it would be possible to combine models to improve treatment optimization, as described above, with real‐world data mined from EHRs using NLP. This would make it possible to not only predict the drug metabolism and optimal dosing based on PKs but to also link these models to real‐world outcomes, as reported in the EHRs.
CONCLUSION
In this mini‐review, we have shown that AI approaches are widely used in all aspects of pharmacology, from drug discovery to real‐world evidence and personalized medicine. In recent years, AI is rapidly becoming a standard analytical tool in drug development. For pharmacology, this comes with many new advancements and improvements in current knowledge. However, care should be taken when applying AI models, and one should be aware of the pitfalls. Specifically, any AI model can only perform as good as the data that is used to train it. If there are innate inaccuracies in the data to train a model (e.g., ethnic, gender, or disease bias, measurement inaccuracies), then those will also be present in the model, making the model less general and more difficult to apply. Therefore, care should always be taken when selecting the data used to train an AI model. Another clear limitation for the application of AI in healthcare is the “accuracy‐interpretability” trade‐off. As a rule of thumb, the more accurate an AI model is, the more difficult it is to interpret. This is less of a problem in the preclinical pharmacology field as patients are not yet involved. In the clinical field, however, transparency and interpretability become of high importance. This trade‐off will force healthcare providers to choose between a highly accurate model of which it is difficult to understand what exactly it does, or a simplified model which resembles conventional statistics more closely which is easy to interpret but less accurate. Nonetheless, when one is aware of the strengths and limitations of AI approaches, the applications for which of these types of models can be used are endless. In the near future, we can expect that AI approaches slowly take over the more conventional models currently in use. Moreover, AI will start to make its way into clinical pharmacology in the form of in silico clinical trials and AI‐based decision support tools that can be regarded as medical devices. For the latter application, the US Food and Drug Administration (FDA) is currently working on a guideline for the credibility of computational models used in medical devices and regulatory applications in anticipation of more widely applied AI in healthcare in general. This will ultimately result in more effective drug discovery pathways and allows us to better optimize drug treatment of the individual patient.
AUTHOR CONTRIBUTIONS
M.vdL. and J.S. wrote the manuscript.
FUNDING INFORMATION
M.vdL. and J.S. are supported by ZonMw (Horizon 2020 INSPIRATION grant 9003035202) under the frame of ERACoSysMed.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
van der Lee M, Swen JJ. Artificial intelligence in pharmacology research and practice. Clin Transl Sci. 2023;16:31‐36. doi: 10.1111/cts.13431
REFERENCES
- 1. Zhavoronkov A, Vanhaelen Q, Oprea TI. Will artificial intelligence for drug discovery impact clinical pharmacology? Clin Pharmacol Ther. 2020;107:780‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Laar SA, Gombert‐Handoko KB, Guchelaar HJ, Zwaveling J. An electronic health record text mining tool to collect real‐world drug treatment outcomes: a validation study in patients with metastatic renal cell carcinoma. Clin Pharmacol Ther. 2020;108:644‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noorbakhsh‐Sabet N, Zand R, Zhang Y, Abedi V. Artificial intelligence transforms the future of health care. Am J Med. 2019;132:795‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim H, Kim E, Lee I, Bae B, Park M, Nam H. Artificial intelligence in drug discovery: a comprehensive review of data‐driven and machine learning approaches. Biotechnol Bioprocess Eng. 2020;25:895‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mak KK, Balijepalli MK, Pichika MR. Success stories of AI in drug discovery—where do things stand? Expert Opin Drug Discov. 2021;17(1):79‐92. [DOI] [PubMed] [Google Scholar]
- 6. Miljković F, Rodríguez‐Pérez R, Bajorath J. Impact of artificial intelligence on compound discovery, design, and synthesis. ACS Omega. 2021;6:33293‐33299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams DP, Lazic SE, Foster AJ, Semenova E, Morgan P. Predicting drug‐induced liver injury with Bayesian machine learning. Chem Res Toxicol. 2020;33:239‐248. [DOI] [PubMed] [Google Scholar]
- 8. Semenova E, Williams DP, Afzal AM, Lazic SE. A Bayesian neural network for toxicity prediction. Comput Toxicol. 2020;16:100133. [Google Scholar]
- 9. Smith GF. Artificial intelligence in drug safety and metabolism. Methods Mol Biol. 2022;2390:483‐501. [DOI] [PubMed] [Google Scholar]
- 10. Mamoshina P, Bueno‐Orovio A, Rodriguez B. Dual transcriptomic and molecular machine learning predicts all major clinical forms of drug cardiotoxicity. Front Pharmacol. 2020;11:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammann F, Schöning V, Drewe J. Prediction of clinically relevant drug‐induced liver injury from structure using machine learning. J Appl Toxicol. 2019;39:412‐419. [DOI] [PubMed] [Google Scholar]
- 12. Al‐Taie Z, Liu D, Mitchem JB, et al. Explainable artificial intelligence in high‐throughput drug repositioning for subgroup stratifications with interventionable potential. J Biomed Inform. 2021;118:103792. [DOI] [PubMed] [Google Scholar]
- 13. Himmelstein DS, Lizee A, Hessler C, et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6. doi: 10.7554/eLife.26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang J, Zhang P, Wang Q, et al. Artificial intelligence framework identifies candidate targets for drug repurposing in Alzheimer's disease. Alzheimers Res Ther. 2022;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang M, Luo H, Xi Z, Rogaeva E. Drug repositioning for diabetes based on 'omics' data mining. PLoS One. 2015;10:e0126082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrer S, Shah P, Antony B, Hu J. Artificial intelligence for clinical trial design. Trends Pharmacol Sci. 2019;40:577‐591. [DOI] [PubMed] [Google Scholar]
- 17. Kolla L, Gruber FK, Khalid O, Hill C, Parikh RB. The case for AI‐driven cancer clinical trials—the efficacy arm in silico. Biochim Biophys Acta Rev Cancer. 2021;1876:188572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venkatapurapu SP, Iwakiri R, Udagawa E, et al. A computational platform integrating a mechanistic model of Crohn's disease for predicting temporal progression of mucosal damage and healing. Adv Ther. 2022;39:3225‐3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Gelder T, Vinks AA. Machine learning as a novel method to support therapeutic drug management and precision dosing. Clin Pharmacol Ther. 2021;110:273‐276. [DOI] [PubMed] [Google Scholar]
- 20. Labriffe M, Woillard JB, Debord J, Marquet P. Machine learning algorithms to estimate everolimus exposure trained on simulated and patient pharmacokinetic profiles. CPT Pharmacometrics Syst Pharmacol. 2022;11(8):1018‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bououda M, Uster DW, Sidorov E, et al. A machine learning approach to predict Interdose vancomycin exposure. Pharm Res. 2022;39:721‐731. [DOI] [PubMed] [Google Scholar]
- 22. Woillard JB, Labriffe M, Debord J, Marquet P. Mycophenolic acid exposure prediction using machine learning. Clin Pharmacol Ther. 2021;110:370‐379. [DOI] [PubMed] [Google Scholar]
- 23. Woillard JB, Labriffe M, Prémaud A, Marquet P. Estimation of drug exposure by machine learning based on simulations from published pharmacokinetic models: the example of tacrolimus. Pharmacol Res. 2021;167:105578. doi: 10.1016/j.phrs.2021.105578 [DOI] [PubMed] [Google Scholar]
- 24. McInnes G, Dalton R, Sangkuhl K, et al. Transfer learning enables prediction of CYP2D6 haplotype function. PLoS Comput Biol. 2020;16:e1008399. doi: 10.1371/journal.pcbi.1008399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Lee M, Allard WG, Vossen RHAM, et al. Toward predicting CYP2D6‐mediated variable drug response from CYP2D6 gene sequencing data. Sci Transl Med. 2021;13. doi: 10.1126/scitranslmed.abf3637 [DOI] [PubMed] [Google Scholar]


