Abstract
In some lower eukaryotes, d-erythroascorbic acid, a five-carbon analog of l-ascorbic acid, is present instead of l-ascorbic acid. We have cloned ALO1, the gene encoding d-arabinono-1,4-lactone oxidase, which catalyzes the final step of d-erythroascorbic acid biosynthesis in Candida albicans. The ALO1 gene contained a continuous open reading frame of 1,671 bp that encodes a polypeptide consisting of 557 amino acids with a calculated molecular mass of 63,428 Da. To investigate the functional roles of d-erythroascorbic acid in C. albicans, we disrupted or overexpressed the ALO1 gene. In the alo1/alo1 null mutants, the activity of d-arabinono-1,4-lactone oxidase was completely lost and d-erythroascorbic acid could not be detected. When ALO1 on a multicopy plasmid was transformed in C. albicans, the enzyme activity and the intracellular d-erythroascorbic acid level were increased up to 3.4-fold and 4.0-fold, respectively. The alo1/alo1 null mutants of C. albicans showed increased sensitivity towards oxidative stress. Overexpression of ALO1 made the cells more resistant to the same stress. The alo1/alo1 mutants showed defective hyphal growth and attenuated virulence. Taken together, our results suggest that d-erythroascorbic acid functions as an important antioxidant and can be considered one of the virulence factors enhancing the pathogenicity of C. albicans.
l-Ascorbic acid (ASC) is produced in all higher plants and in nearly all higher animals except human, other primates, guinea pig, some birds, and fish (1, 3). In animals, a microsomal l-gulono-1,4-lactone oxidase catalyzes the final step of ASC biosynthesis (15, 29). Koshizaka et al. (17) isolated and characterized a cDNA encoding l-gulono-1,4-lactone oxidase from rat liver. Recently, a biosynthetic pathway for ASC involving l-galactose and l-galactono-1,4-lactone in plants has been proposed (39). It is believed in plants that the final step of ASC biosynthesis is catalyzed by l-galactono-1,4-lactone dehydrogenase (25, 30). The cDNAs encoding l-galactono-1,4-lactone dehydrogenase in cauliflower (31) and sweet potato (11) have been isolated and analyzed. In some eukaryotic microorganisms, ASC is rare or absent but d-erythroascorbic acid (EASC), a five-carbon analog of ASC, is present (5, 24, 27, 28). In Candida albicans and Saccharomyces cerevisiae, the biosynthetic pathway of EASC from d-arabinose by d-arabinose dehydrogenase and d-arabinono-1,4-lactone oxidase (ALO) has been established (9, 10, 13, 14). ALO can also catalyze the production of ASC when l-galactono-1,4-lactone is supplied as a substrate (20).
ASC is known to carry out a number of biochemical functions that are a consequence of its ability to donate one or two electrons. Some known or proposed functions of ASC include its utilization as a free radical scavenger, a cofactor for a number of enzymes, and a controlling factor in plant cell development (26). However, many other functions of the ASC system as well as the precise mechanisms of its functions are still elusive. According to Shao et al. (33), EASC is almost as readily oxidized as ASC in an aqueous system and has reducing power similar to that of ASC. In a bioassay using tobacco hornworm (Manduca sexta) to determine the vitamin C activity of EASC, EASC supported the larval growth of the hornworm almost as well as ASC. This report suggests that EASC has biological properties similar to those of ASC. Considering that some eukaryotic microorganisms produce EASC instead of ASC, it is presumed that EASC may take the place of ASC in these microorganisms. In our previous study, EASC has been proved an important antioxidant molecule in S. cerevisiae (10), like ASC in animals and plants.
C. albicans is a well-known opportunistic fungal pathogen of humans that does not usually cause disease in immunocompetent hosts but causes serious diseases in immunocompromised patients. A number of factors have been implicated to be associated with the virulence properties of C. albicans, such as adhesion to the host tissues, secretion of proteases, and reversible morphological transitions between yeasts, pseudohyphae, and hyphae (4). Recent studies have led to the identification of several genes involved in the transition from yeast-like growth to hyphal growth in C. albicans. Deletion of the Candida genes in a MAPK pathway, such as CST20, HST7, and CPHl, results in impairment of the ability to make hyphae under some conditions, albeit not in response to serum (16, 19, 22), suggesting that there is more than one pathway controlling hyphal growth. Another gene, EFG1, a homolog of S. cerevisiae PHD1, has been found in C. albicans, and its reduced expression causes loss of hyphal growth (36). The cphl/cphl efgl/efgl double mutants of C. albicans are unable to form hyphae under almost all laboratory conditions tested and are avirulent in a mouse model (23). These studies demonstrate the importance of the transition from yeast-like to hyphal growth in the virulence of C. albicans. The ability to adhere to the host tissues has been also proved important in the pathogenicity of C. albicans. Recently, Int1p, a surface protein with limited similarity to vertebrate integrins, has been found in C. albicans. Disruption of INT1 in C. albicans suppresses hyphal growth, adhesion to epithelial cells, and virulence in mice (8). Another hypha-specific surface protein, Hwplp, with similarities to small mammalian proline-rich proteins, has been found in C. albicans and shown to serve as a substrate for mammalian transglutaminases. The hwpl/hwpl mutants of C. albicans are unable to form stable attachments to human buccal epithelial cells and have a reduced capacity to cause systemic candidiasis in mice (35).
To fully understand the pathogenicity of C. albicans, survival traits should also be taken into consideration, in addition to virulence traits. Survival indicates the ability of C. albicans to defend itself against the host immune system and grow in the host successfully. In the present study, we describe the isolation and characterization of the gene encoding ALO (ALO1), which catalyzes the final step of EASC biosynthesis in C. albicans. We show that EASC serves as an important antioxidant, contributes to hyphal growth, and is essential for C. albicans to exhibit full virulence, presumably by enhancing survival of the organism in the host.
MATERIALS AND METHODS
Yeast strains and culture conditions.
C. albicans strains used in this study are listed in Table 1. The strains were routinely cultured on YPD medium (1% yeast extract, 2% peptone, and 2% glucose) at 28°C. Cells containing plasmids or disrupted genes were cultured in minimal defined medium containing 0.67% yeast nitrogen base without amino acids (Difco), 2% glucose, and appropriate supplements (34). Solid media were prepared by adding 1.8% agar to liquid broth. To assess filamentation on solid media, 104 cells in 2 μl of water were spotted onto the plates and incubated for 3 to 4 days.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| ATCC 10231 | Wild-type isolate | ATCCa |
| SC5314 | Wild-type isolate | 6 |
| CAI4 | Δura3::imm434/Δura3::imm434 | 6 |
| JKC19 | Δura3::imm434/Δura3::imm434 cphl::hisGlcph1::hisG-URA3-hisG | 22 |
| HLC52 | Δura3::imm434/Δura3::imm434 efg1::hisGlefgl::hisG-URA3-hisG | 23 |
| WH201 | Δura3::imm434/Δura3::imm434 Δalo1::hisG-URA3-hisG/ALO1 | This study |
| WH202 | Δura3::imm434/Δura3::imm434 Δalo1::hisG/ALO1 | This study |
| WH203 | Δura3::imm434/Δura3::imm434 Δalo1::hisG/Δalo1::hisG-URA3-hisG | This study |
| WH204 | Δura3::imm434/Δura3::imm434 Δalo1::hisG/Δalo1::hisG | This study |
| WH205 | Δura3::imm434/Δura3::imm434 (pRC2312) | This study |
| WH206 | Δura3::imm434/Δura3::imm434 (pWK203) | This study |
| WH207 | Δura3::imm434/Δura3::imm434 Δalo1::hisG/Δalo1::hisG::ALO1::URA3 | This study |
American Type Culture Collection.
Isolation, subcloning, and sequencing of ALO1 from C. albicans.
To construct a C. albicans genomic library, the genomic DNA from C. albicans ATCC 10231 was partially digested with Sau3AI and DNA fragments of 10 to 23 kb were ligated into dephosphorylated λEMBL3 vector (Stratagene) generated by BamHI cleavage. The ligated DNA was packaged using Gigapack II packaging extracts (Stratagene) and replicated according to the manufacturer's instructions. Then, degenerate oligonucleotide primers corresponding to residues 52 to 58 (VGSGHSP) and 444 to 450 (GGKPHWA) of S. cerevisiae ALO1 (10) were synthesized: 5′-GTTGGTTCYGGCCAYTCYCC-3′ and 5′-GGCCCARTGTGGCTTACCTCC-3′, respectively, where Y represents C or T and R represents A or G. PCR amplification was carried out using the genomic DNA from C. albicans ATCC 10231 as a template under the following conditions: denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min. Among the amplified DNA products, a DNA fragment of 1,359 bp was cloned into pGEM-T vector (Promega). The insert DNA fragment was labeled with digoxigenin (Roche Molecular Biochemicals) and used as a probe to screen the λEMBL3 genomic library. Four positive clones were selected, and the common 3.8-kb HindIII fragment giving a positive signal was isolated and cloned into pGEM-7Zf(+) (Promega) at the HindIII site, yielding pCALO. Both stands of the cloned DNA were sequenced by dideoxy chain termination method with an automatic sequencer (ALFexpress; Amersham Pharmacia Biotech).
Disruption, overexpression, and reintegration of C. albicans ALO1.
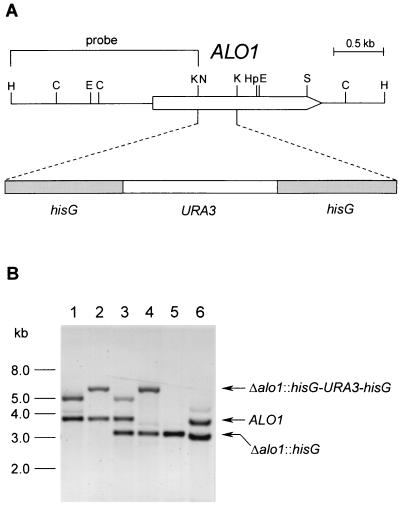
Both alleles of ALO1 were disrupted by using the URA blaster technique (6). A 4.1-kb fragment containing the hisG-URA3-hisG gene disruption cassette from p5921 (6) was inserted in place of a portion of ALO1 within the genomic clone (see Fig. 2A). The resulting plasmid, pWK202, was cut with ApaI and SacI to remove the vector and transformed into the ura3/ura3 C. albicans strain CAI4 (6). Ura+ transformants were selected on uracil-deficient medium, and the integration of the cassette into the ALO1 locus was verified by either PCR or Southern blot analysis. Spontaneous Ura− derivatives of the heterozygous disruptants were selected on minimal defined medium supplemented with 625 mg of 5-fluoroorotic acid and 30 mg of uridine per liter. This procedure was repeated to delete the remaining functional allele of ALO1.
FIG. 2.
Disruption of the ALO1 gene in C. albicans. (A) Restriction map of the ALO1 locus and insertion of the hisG-URA3-hisG cassette at the KpnI sites in ALO1 coding sequence. Endonuclease restriction sites: C, ClaI; E, EcoRI; H, HindIII; Hp, HpaI; K, KpnI; N, NcoI; S, SpeI. (B) Southern blot analysis with the sequence bracketed in panel A used as a probe. The DNA digested with HindIII was from the following strains: CAI4 ALO1/ALO1 (lane 1), WH201 Δalo1::hisG-URA3-hisG/ALO1 (lane 2), WH202 Δalo1::hisG/ALO1 (lane 3), WH203 Δalo1::hisG/Δalo1::hisG-URA3-hisG (lane 4), WH204 Δalo1::hisG/Δalo1::hisG (lane 5), and WH207 Δalo1::hisG/Δalo1::hisG::ALO1::URA3 (lane 6).
In order to overexpress ALO1, a 3.8-kb HindIII fragment containing the entire coding sequence of ALO1 and its 5′ and 3′ flanking regions was isolated from pCALO and inserted into the HindIII site of pRC2312, which contains LEU2 and URA3 from C. albicans as selectable markers and an autonomously replicating sequence from C. albicans for replication in C. albicans and S. cerevisiae (2). The resulting plasmid pWK203 was transformed into CA14 strain and selected for the Ura+ phenotype.
For reintegration of the ALO1 gene into the genome, a 3.8-kb NsiI/XhoI fragment with the ALO1 gene from pCALO was inserted into pURA3, a plasmid containing the C. albicans URA3 gene, at the NsiI/SalI sites to yield pWK204. pWK204 was linearized at the unique HpaI site within the ALO1 coding region and used to integrate into the ALO1 locus in the Ura− alo1/alo1 strain. The occurrence of the desired integration events in all the transformants was verified by Southern blot analysis.
Measurement of ALO activity and intracellular EASC level.
The activity of ALO was measured spectrophotometrically in 0.2 M potassium phosphate (pH 6.1), 1 mM EDTA, 50 mM d-arabinono-1,4-lactone, and an aliquot of enzyme. The production of EASC (ɛ265 = 13,150 M−1 · cm−1) was monitored by the increase in the absorbance at 265 nm during the first 1 min of the reaction at 36°C. One unit of the enzyme was defined as the amount of enzyme that produced 1 μmol of EASC per min. The lower limit for assay of ALO activity was 0.1 mU · mg of protein−1.
The amount of EASC was measured as described previously (10). C. albicans cells (1 g [wet mass]) grown in liquid minimal defined medium were recovered by centrifugation at 6,000 × g for 5 min at 4°C, washed twice with distilled water, and resuspended in 2 ml of 10% trichloroacetic acid. This suspension was stored for 20 min at 4°C. The insoluble residue was removed by centrifugation at 10,000 × g for 10 min at 4°C, and the soluble extracts were subjected to analytical high-performance liquid chromatography using a Waters Associates liquid chromatography system linked to a Waters 460 electrochemical detector. The potential of the detector was set at +0.70 V versus the Ag/AgCl reference electrode. Ten microliters of trichloroacetic acid-soluble extracts was passed through two tandemly linked Hewlett Packard octyldecylsilane Hypersil columns (10 cm by 4.6 mm) and eluted with 0.1% trifluoroacetic acid at a flow rate of 0.7 ml · min−1. The lower limit for detection of EASC was 0.1 nmol · g (wet wt) of cells−1.
Assay of resistance to oxidative stress.
The susceptibility of the cells to H2O2 and menadione was measured as described previously (10), with some modifications. Cells were grown in minimal defined medium to mid-logarithmic phase (5 × 106 cells · ml−1), harvested, and resuspended in 0.1 M potassium phosphate buffer, pH 7.0, to obtain an initial optical density at 600 nm of 0.1. To observe the sensitivity of the cells to oxidants, various concentrations of H2O2 or menadione were added to the cell suspensions. After incubation for 1 h at 30°C, aliquots were taken from the cell suspensions, diluted appropriately in the same buffer, and plated onto solid minimal defined medium. Colonies were counted after incubation for 3 days at 28°C.
Assay of C. albicans virulence.
Inbred female BALB/c mice (Seoul National University Laboratory Animal Center) weighing between 18 and 20 g were used for testing the virulence of C. albicans strains according to the method described previously (23). Statistical analyses of the differences in survival between paired groups were performed with the Kaplan-Meier log-rank test. A P value of 0.05 was taken to indicate statistical significance.
Nucleotide sequence accession number.
The nucleotide sequence data of the ALO1 gene have been deposited in the GenBank/EMBL/DDBJ database under accession no. AF031228.
RESULTS
Isolation and characterization of ALO1, which encodes ALO in C. albicans.
From the comparison of the predicted amino acid sequence of rat l-gulono-1,4-lactone oxidase (17) and that of S. cerevisiae ALO (10), two highly conserved regions were identified. PCR using the oligonucleotide primer pair corresponding to residues 52 to 58 (VGSGHSP) and 444 to 450 (GGKPHWA) of S. cerevisiae ALO could amplify a DNA fragment of 1,359 bp from the chromosomal DNA of C. albicans ATCC 10231. When cloned and sequenced, the fragment showed a high degree of amino acid sequence similarity to S. cerevisiae ALO upon BLAST searches of the GenBank database. The cloned PCR product was used as a probe to screen the λEMBL3 genomic library of C. albicans. From positive clones, the common 3.8-kb HindIII fragment was subcloned in pGEM-7Zf(+) and sequenced.
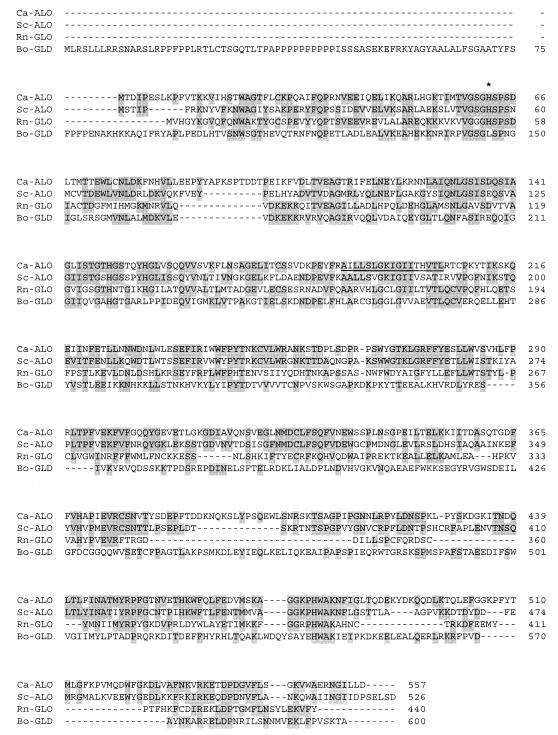
The cloned ALO1 gene contains a continuous open reading frame of 1,671 bp that encodes a polypeptide consisting of 557 amino acids with a calculated molecular mass of 63,428 Da. When the nucleotide sequence of the cloned ALO1 gene was compared with that of the same gene obtained from the Candida genome project (http://www-sequence.stanford.edu/group/candida), five mismatches were found within the coding region of the gene. However, the five mismatches of nucleotide sequence did not alter the amino acid sequence of the gene. C. albicans ALO1 contained no CUG codon, which encodes serine in C. albicans but encodes leucine in S. cerevisiae and elsewhere (32). The nucleotide sequence of ALO1 did not have the consensus sequence for splicing. The fact that the gene contains no intron was confirmed by reverse transcription-PCR (data not shown). The predicted amino acid sequence of C. albicans ALO shared 53, 32, and 24% identity with those of S. cerevisiae ALO (10), rat l-gulono-1,4-lactone oxidase (17), and cauliflower l-galactono-1,4-lactone dehydrogenase (31), respectively (Fig. 1). The hydropathy plot of ALO using the method of Kyte and Doolittle (18) predicted that the enzyme should be an integral membrane protein with a transmembrane segment corresponding to amino acid residues 188 to 204. This prediction agreed well with a previous report (9) in which ALO was suggested to be a mitochondrial membrane protein. Like S. cerevisiae ALO and rat l-gulono-1,4-lactone oxidase, C. albicans ALO also had a putative binding site for covalently bound flavin adenine dinucleotide (FAD) of oxygen-dependent oxidoreductases (7), corresponding to amino acid residues 29 to 62. The site has been found in some oxygen-dependent oxidoreductases, all of which contain a covalently bound FAD group that is attached to a histidine via an 8α-[N(1)-histidyl]FAD or 8α-[N(3)-histidyl]FAD linkage. Kenney et al. (12) reported that S. cerevisiae ALO contains a covalently bound FAD linked to the N(1) position of histidine. In case of C. albicans ALO, however, FAD proved to be covalently linked to the N(3) position of histidine (S.-T. Kim, W.-K. Huh, and S.-O. Kang, unpublished data). Based on these facts, we suggest that the region of amino acid residues 29 to 62 is the covalent FAD-binding site and that the histidine at position 62 is the amino acid covalently linked to FAD in C. albicans ALO.
FIG. 1.
Alignment of the deduced amino acid sequence of ALO from C. albicans (Ca-ALO) with those of other enzymes with similar activity: ALO from S. cerevisiae (10) (Sc-ALO), l-gulono-1,4-lactone oxidase from rat (17) (Rn-GLO), and l-galactono-1,4-lactone dehydrogenase from cauliflower (31) (Bo-GLD). Numbers on the right are amino acid positions. The regions where the sequences have been extended to allow optimal sequence alignment are indicated with dashes. Identical residues are shaded. The asterisk indicates the histidine residue believed to be responsible for covalent attachment of FAD. A 17-residue putative transmembrane segment predicted according to the method by Kyte and Doolittle (18) is underlined.
On genomic Southern blot analysis, only one band was detected when the genomic DNA was digested with BglII, HindIII, NdeI, or XbaI, and two bands were detected when DNA was digested with EcoRI (data not shown). Since the nucleotide sequence of the ALO1 open reading frame contained one EcoRI and no BglII, HindIII, NdeI, or XbaI site, the hybridization pattern indicates that there is a single copy of ALO1 per C. albicans genome. On Northern blot analysis, a single band corresponding to a size of 2.0 kb was detected (data not shown). As in S. cerevisiae (10), the activity of ALO and the ALO1 transcript level remained essentially unchanged whether C. albicans cells were treated with oxidants such as H2O2 and menadione (data not shown), suggesting that ALO1 is constitutively expressed and not regulated in response to oxidative stress.
Disruption and overexpression of C. albicans ALO1.
For the gene disruption, a disruption construct was prepared by replacing a portion of the coding region of ALO1 with the hisG-URA3-hisG sequence (Fig. 2A) and used to transform the ura3/ura3 C. albicans strain CAI4. The resulting Ura+ transformants were screened by PCR or Southern blot analysis, and the spontaneous Ura− “pop-out” revertants from them were selected on minimal defined medium containing 5-fluoroorotic acid. A homozygous disruption of ALO1 was generated by repeating the above procedure and confirmed by Southern blot analysis (Fig. 2B). The alo1/alo1 mutants did not show any auxotrophy and grew normally in minimal defined medium as well as in complex medium. They also showed normal growth patterns when grown in the media with nonfermentable carbon sources such as ethanol and glycerol. In order to overexpress ALO in C. albicans, we constructed the plasmid pWK203 by inserting the entire ALO1 gene and its flanking sequences into the plasmid pRC2312, as described in Materials and Methods. C. albicans cells were transformed with the parental plasmid pRC2312 or pWK203, and transformants containing either plasmid were selected by plating on uracil-deficient medium. As originally reported by Cannon et al. (2), transformation with either pRC2312 or pWK203 resulted in small, slow-growing colonies at high frequency and larger, fast-growing colonies at a lower frequency. According to Cannon et al. (2), the small colonies are replicative transformants with a plasmid copy number of 2 or 3 per genome, and the larger colonies are integrative transformants, with the copy number of the integrated sequence being estimated to be 7 to 12 per diploid genome. For further experiments, we selected the larger, fast-growing colonies.
We measured the activity of ALO and the intracellular level of EASC in the strains in which the ALO1 gene has been disrupted or overexpressed. As expected, the alo1/alo1 mutant strain WH203 did not show any ALO activity. The ALO activity of WH206 carrying the plasmid pWK203 increased up to 3.4-fold compared with that of WH205 carrying the parental vector pRC2312 (Table 2). When the intracellular content of EASC was measured with electrochemical detector, it was impossible to detect EASC in WH203. On the other hand, WH206 showed a marked increase of EASC compared with the control strain WH205. The content of EASC in WH206 was estimated to be 4.0-fold higher than that of WH205 (Table 2). The ALO1 reintegrant strain WH207 showed intermediate levels of ALO activity and EASC content.
TABLE 2.
Activity of ALO and amount of EASC in C. albicans strains of different genetic backgrounda
| Strain | Activity of ALO (mU · mg of protein−1) | Amt of EASC (nmol · g [wet wt] of cells−1) |
|---|---|---|
| SC5314 (ALO1/ALO1) | 37.3 ± 5.1 | 427 ± 35 |
| WH203 (alo1/alo1) | <0.1b | <0.1c |
| WH205 (CAI4 carrying pRC2312) | 34.8 ± 4.2 | 336 ± 15 |
| WH206 (CAI4 carrying pWK203) | 119.0 ± 7.3 | 1,330 ± 52 |
| WH207 (ALO1 reintegrant) | 19.4 ± 3.8 | 237 ± 26 |
Activity of ALO and amount of EASC in C. albicans were measured from exponentially growing cells in minimal defined medium. Values are means ± standard errors (n = 3).
Lower limit for assay of ALO activity.
Lower limit of detection for EASC.
Effect of EASC on resistance to oxidative stress.
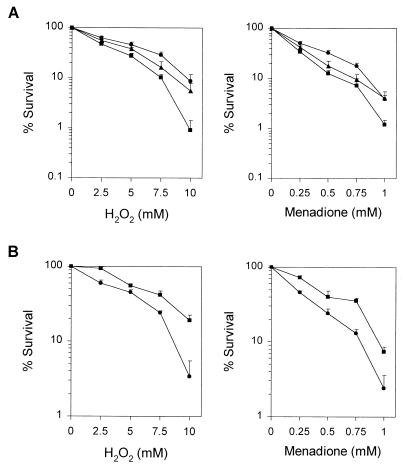
We tested whether disruption or overexpression of ALO1 affects the survival of the cells under oxidative stress conditions. For this purpose, exponentially growing cells were treated with various concentrations of H2O2 or menadione, a redox-cycling agent, and the viable cells were counted. As shown in Fig. 3A, the alo1/alo1 mutant strain WH203, which is devoid of EASC, was more sensitive to H2O2 and menadione than the parental wild-type strain SC5314, and the susceptibility of WH207 to oxidative stress was intermediate. When WH206 with a high EASC content was challenged with the same oxidants, it showed increased resistance to oxidative stress compared with the control strain WH205 (Fig. 3B). These results indicate that EASC functions as an important antioxidant in C. albicans. However, disruption or overexpression of ALO1 did not affect the cell survival under other stress conditions, e.g., heat shock (40°C for 30 min) or osmotic shock (1 M NaCl).
FIG. 3.
Effect of disruption or overexpression of ALO1 on the cell survival against oxidative stress. (A) Sensitivities of the ALO1/ALO1 SC5314 strain (●), the alo1/alo1 WH203 strain (■), and the ALO1 reintegrant WH207 strain (▴) to H2O2 and menadione. (B) Sensitivities of the WH205 strain carrying parental vector pRC2312 (●) and the WH206 strain carrying pWK203 (■) to H2O2 and menadione. Exponentially growing cells were treated with each oxidant at various concentrations for 1 h at 30°C. Data are means plus standard errors of three independent experiments.
Effect of EASC on hyphal growth of C. albicans.
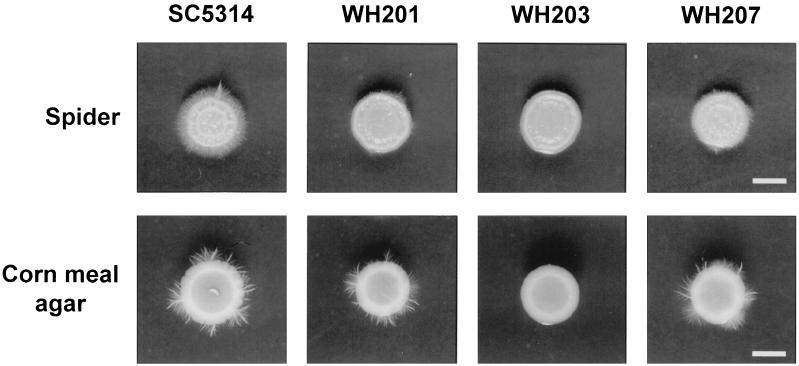
To investigate the effect of the alo1 mutation on hyphal growth of C. albicans, isogenic Ura+ prototrophs were grown on liquid and solid media that induce hyphal growth, e.g., 20% serum, Lee's medium (21), Spider medium (22), corn meal agar (Difco), and RPMI 1640 (Gibco BRL). When grown on solid Spider medium, the parental wild-type strain SC5314 formed extensive agar-invasive hyphae after 3 days (Fig. 4). The Ura+ alo1/ALO1 heterozygote strain WH201 showed a slight reduction in the extent of hyphal formation. The Ura+ alo1/alo1 strain WH203 showed little hyphal formation compared with SC5314. The hyphal growth of the ALO1 reintegrant strain WH207 was similar to that of WH201, regaining the ability to form extensive hyphae. Growth on corn meal agar gave similar results (Fig. 4). These results indicate that EASC contributes to the hyphal growth of C. albicans. However, in spite of the defective hyphal growth of WH203 on solid Spider medium and corn meal agar, the mutant strain exhibited hyphal growth patterns little different from SC5314 in other liquid and solid media, suggesting that EASC is not needed for hyphal growth of C. albicans under all inducing conditions.
FIG. 4.
Effect of ALO1 mutation on hyphal growth of C. albicans. The indicated strains were spotted and incubated on Spider medium at 37°C for 3 days (bar = 3 mm) and on corn meal agar at 28°C for 4 days (bar = 5 mm).
It is known that two major signaling pathways, dependent on CPH1 (22) and EFG1 (36), activate hyphal growth of C. albicans. We compared hyphal growth of the alo1/alo1 strain WH203 with that of the strain carrying a mutation on CPH1 or EFG1. On solid Spider medium, the cph1/cph1 mutant strain showed reduced hyphal formation comparable to that of the alo1/alo1 strain WH203 (data not shown). As reported previously (23), the efg1/efg1 mutant strain did not form hyphae at all. Taken together with the report that the cph1/cph1 strain shows defective hyphal formation on solid Spider medium but exhibits normal growth patterns in other liquid and solid media (22), these observations suggest that, as far as hyphal growth is concerned, the alo1/alo1 strain is similar to the cph1/cph1 strain.
To test a possibility that the expression of ALO1 may be under the control of CPH1 or EFG1, we measured the ALO activity and EASC content of the corresponding null mutants. However, there was no difference in the ALO activity and EASC content between the wild-type strain and the cph1/cph1 or the efg1/efg1 strain (data not shown), indicating that the expression of ALO1 is not influenced by the signaling pathways dependent on CPH1 and EFG1.
Virulence studies in a mouse model.
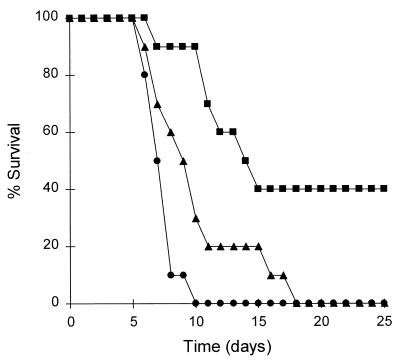
To test the effect of EASC deficiency on the virulence of C. albicans in a mouse model, the wild-type strain SC5314, the alo1/alo1 mutant strain WH203, and the ALO1 reintegrant strain WH207 were intravenously injected into immunocompetent mice. Since the ura3/ura3 mutants show decreased virulence, isogenic Ura+ prototrophs were used to infect mice. As illustrated in Fig. 5, all the mice injected with SC5314 died within 10 days after infection. In contrast, 40% of the mice injected with the EASC-deficient strain WH203 survived to the end of the experiment. The survival difference between SC5314 and WH203 was significant (P < 0.001 by the Kaplan-Meier log-rank test). The ALO1 reintegrant strain WH207 was more virulent than WH203 (P < 0.05 by the Kaplan-Meier log-rank test). These results indicate that EASC contributes to the virulence of C. albicans in a mouse model of intravenous infection.
FIG. 5.
Virulence assay of C. albicans in a mouse model. BALB/c mice were inoculated with 106 cells of SC5314 (●), WH203 (■), and WH207 (▴) in a final volume of 100 μl through the lateral tail vein. Curves are the compiled results of two replicate experiments (five mice for each strain in each experiment).
DISCUSSION
In the present study, the ALO1 gene, which encodes the enzyme (ALO) that catalyzes the final reaction of EASC biosynthesis, was identified and cloned in C. albicans. The ALO1 gene is 1,671 bp in size and encodes 557 amino acids with a calculated molecular mass of 63,428 Da, which is comparable to the molecular mass of the enzyme purified from the mitochondrial fraction of C. albicans (66.7 kDa) (9). The results of sequence comparisons show that C. albicans ALO is more similar to l-gulono-1,4-lactone oxidase from animals than to l-galactono-1,4-lactone dehydrogenase from plants, which has also been suggested by investigating the substrate specificity of C. albicans ALO (9). Through disruption of ALO1, we could obtain C. albicans strains devoid of EASC. Also, we could make C. albicans strains with high intracellular levels of EASC by overexpression of ALO1. The alo1/alo1 mutant strain was more sensitive to oxidative stress, and the strain carrying ALO1 on a multicopy plasmid showed a significant increase in survival under oxidative stress compared with the control strain. In S. cerevisiae, EASC has been reported to function as an important antioxidant (10), like ASC in higher animals and plants. The present study shows that it still holds true for C. albicans.
The alo1/alo1 mutants show defective hyphal growth on solid Spider medium and corn meal agar, although not under all inducing conditions. Conditional defects in hyphal growth have been already observed in some other mutants, e.g., the cph1/cph1 (22) and the int1/int1 mutants (8). It is rather interesting that EASC affects the hyphal growth of C. albicans under some conditions. Some possibilities can be suggested: EASC may be required for proper operation of the components in a signal transduction pathway involved in the transition from yeast-like growth to hyphal growth, or a strong reductant activity of EASC may be needed in constituting normal cell wall structure in hyphal growth. It remains to be determined how EASC deficiency causes defective hyphal growth of C. albicans.
C. albicans is a member of the normal microbial flora and does not usually cause disease in immunocompetent hosts. However, C. albicans causes serious diseases in immunocompromised hosts such as leukemic, diabetic, organ transplant, and human immunodeficiency virus-infected patients. Elimination of C. albicans from an infected host requires the cooperation of many immune cells and several candidacidal mechanisms, among which oxygen-dependent killing mechanisms, mediated by a superoxide anion radical myeloperoxidase-H2O2-halide system, and reactive nitrogen intermediates, are crucial (37). Therefore, antioxidant defense systems are assumed to be essential for C. albicans to resist the host immune response and exhibit full virulence. In accordance with this view, exogenous antioxidants impair killing of C. albicans by neutrophils (38) and a catalase-deficient C. albicans strain is far less virulent for mice than the parental wild-type strain (40). The present study shows that the EASC-deficient alo1/alo1 mutant strain exhibits attenuated virulence. These results, taken together with the proved function of EASC as an important antioxidant molecule in C. albicans, suggest that EASC may be essential for C. albicans to stand against the oxidant-mediated killing actions of the host immune system.
Nevertheless, there is a possibility that attenuated virulence of the alo1/alo1 strain may be attributed to its defective hyphal growth, considering the well-established fact that the transition from yeast-like to hyphal growth is important to C. albicans virulence (23). However, this possibility does not seem to be acceptable for the following reasons. (i) Although the alo1/alo1 strain exhibits suppressed hyphal growth on solid Spider medium and corn meal agar, it shows no difference from the wild-type strain when cultured on other media, including the one containing serum. This result strongly suggests that, when inoculated into the vein of a mouse, the alo1/alo1 strain will show a normal transition from yeast-like to hyphal growth. (ii) The cph1/cph1 strain shows defective hyphal formation similar to that of the alo1/alo1 strain but does not suffer any damage in its virulence for mice (23). Therefore, it is not likely that attenuated virulence of the alo1/alo1 strain is attributed to its defective hyphal growth.
The overall virulence of C. albicans can be defined as the sum of survivability and virulence. The former indicates the ability of C. albicans to defend itself against the host immune system and to grow in the host successfully. The latter allows C. albicans to adhere to and penetrate the host tissues and cause the symptoms of disease. Up to now, most studies on C. albicans have been focused on its virulence traits, including adhesion to the host tissues, secretion of proteases, and reversible morphological transitions between yeasts, pseudohyphae, and hyphae. The present study shows that EASC functions as an important antioxidant and is essential for C. albicans to exhibit full virulence, presumably by enhancing survival of the organism in the host. Therefore, we suggest that EASC can be regarded as an important virulence factor and that closer investigation of the defense mechanisms against the host immune system will broaden our understanding of the pathogenicity of C. albicans.
ACKNOWLEDGMENTS
We thank William A. Fonzi for providing strain CAI4 and plasmid p5921, Richard D. Cannon for plasmid pRC2312, and Gerald R. Fink for strains JKC19 and HLC52.
This work was supported by a grant of the Korea Health 21 R & D Project, Ministry of Health & Welfare, Republic of Korea (HMP-00-B-20200-0010), and by Research Fellowship of the BK21 project.
REFERENCES
- 1.Burns J J. Missing step in man, monkey and guinea pig required for the biosynthesis of l-ascorbic acid. Nature. 1957;180:533. doi: 10.1038/180553a0. [DOI] [PubMed] [Google Scholar]
- 2.Cannon R D, Jenkinson H F, Shepherd M G. Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol Gen Genet. 1992;235:453–457. doi: 10.1007/BF00279393. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee I B. Evolution and the biosynthesis of ascorbic acid. Science. 1973;182:1271–1272. doi: 10.1126/science.182.4118.1271. [DOI] [PubMed] [Google Scholar]
- 4.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 5.Dumbrava V-A, Pall M L. Control of nucleotide and erythroascorbic acid pools by cyclic AMP in Neurospora crassa. Biochim Biophys Acta. 1987;926:331–338. doi: 10.1016/0304-4165(87)90219-4. [DOI] [PubMed] [Google Scholar]
- 6.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraaije M W, van Berkel W J, Benen J A, Visser J, Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- 8.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 9.Huh W-K, Kim S-T, Yang K-S, Seok Y-J, Hah Y C, Kang S-O. Characterisation of d-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur J Biochem. 1994;225:1073–1079. doi: 10.1111/j.1432-1033.1994.1073b.x. [DOI] [PubMed] [Google Scholar]
- 10.Huh W-K, Lee B-H, Kim S-T, Kim Y-R, Rhie G-E, Baek Y-W, Hwang C-S, Lee J-S, Kang S-O. d-Erythroascorbic acid is an important antioxidant molecule in Saccharomyces cerevisiae. Mol Microbiol. 1998;30:895–903. doi: 10.1046/j.1365-2958.1998.01133.x. [DOI] [PubMed] [Google Scholar]
- 11.Imai T, Karita S, Shiratori G, Hattori M, Nunome T, Ôba K, Hirai M. l-Galactono-γ-lactone dehydrogenase from sweet potato: purification and cDNA sequence analysis. Plant Cell Physiol. 1998;39:1350–1358. doi: 10.1093/oxfordjournals.pcp.a029341. [DOI] [PubMed] [Google Scholar]
- 12.Kenney W C, Edmondson D E, Singer T P, Nishikimi M, Noguchi E, Yagi K. Identification of the covalently-bound flavin of l-galactonolactone oxidase from yeast. FEBS Lett. 1979;97:40–42. doi: 10.1016/0014-5793(79)80047-2. [DOI] [PubMed] [Google Scholar]
- 13.Kim S-T, Huh W-K, Kim J-Y, Hwang S-W, Kang S-O. d-Arabinose dehydrogenase and biosynthesis of erythroascorbic acid in Candida albicans. Biochim Biophys Acta. 1996;1297:1–8. doi: 10.1016/0167-4838(96)00077-5. [DOI] [PubMed] [Google Scholar]
- 14.Kim S-T, Huh W-K, Lee B-H, Kang S-O. d-Arabinose dehydrogenase and its gene from Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1429:29–39. doi: 10.1016/s0167-4838(98)00217-9. [DOI] [PubMed] [Google Scholar]
- 15.Kiuchi K, Nishikimi M, Yagi K. Purification and characterization of l-gulonolactone oxidase from chicken kidney microsomes. Biochemistry. 1982;21:5076–5082. doi: 10.1021/bi00263a035. [DOI] [PubMed] [Google Scholar]
- 16.Köhler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshizaka T, Nishikimi M, Ozawa T, Yagi K. Isolation and sequence analysis of a complementary DNA encoding rat liver l-gulono-γ-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis. J Biol Chem. 1988;263:1619–1621. [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B-H, Huh W-K, Kim S-T, Lee J-S, Kang S-O. Bacterial production of d-erythroascorbic acid and l-ascorbic acid through functional expression of Saccharomyces cerevisiaed-arabinono-1,4-lactone oxidase in Escherichia coli. Appl Environ Microbiol. 1999;65:4685–4687. doi: 10.1128/aem.65.10.4685-4687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 23.Lo H-J, Köhler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 24.Loewus F A, Saito K, Suto R K, Maring E. Conversion of d-arabinose to d-erythroascorbic acid and oxalic acid in Sclerotinia sclerotiorum. Biochem Biophys Res Commun. 1995;212:196–203. doi: 10.1006/bbrc.1995.1956. [DOI] [PubMed] [Google Scholar]
- 25.Mapson L W, Breslow E. Biological synthesis of ascorbic acid; l-galactono-γ-lactone dehydrogenase. Biochem J. 1958;68:395–406. doi: 10.1042/bj0680395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moser U, Bendich A. Vitamin C. In: Machlin L J, editor. Handbook of vitamins. New York, N.Y: Marcel Dekker; 1991. pp. 195–232. [Google Scholar]
- 27.Murakawa S, Sano S, Yamashita H, Takahashi T. Biosynthesis of d-erythroascorbic acid by Candida. Agric Biol Chem. 1977;41:1799–1800. [Google Scholar]
- 28.Nick J A, Leung C T, Loewus F A. Isolation and identification of erythroascorbic acid in Saccharomyces cerevisiae and Lypomyces starkeyi. Plant Sci. 1986;46:181–187. [Google Scholar]
- 29.Nishikimi M, Tolbert B, Udenfriend S. Purification and characterization of l-gulono-γ-lactone oxidase from rat and goat liver. Arch Biochem Biophys. 1976;175:427–435. doi: 10.1016/0003-9861(76)90530-0. [DOI] [PubMed] [Google Scholar]
- 30.Ôba K, Ishikawa S, Nishikawa M, Mizuno H, Yamamoto T. Purification and properties of l-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J Biochem (Tokyo) 1995;117:120–124. doi: 10.1093/oxfordjournals.jbchem.a124697. [DOI] [PubMed] [Google Scholar]
- 31.O/stergaard J, Persian G, Davey M W, Bauw G, van Montagu M. Isolation of a cDNA coding for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J Biol Chem. 1997;272:30009–30016. doi: 10.1074/jbc.272.48.30009. [DOI] [PubMed] [Google Scholar]
- 32.Santos M A, Tuite M F. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Y-Y, Seib P A, Kramer K J, van Galen D A. Synthesis and properties of d-erythroascorbic acid and its vitamin C activity in the tobacco hornworm (Manduca sexta) J Agric Food Chem. 1993;41:1391–1396. [Google Scholar]
- 34.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 35.Staab J F, Bradway S D, Fidel P L, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwpl. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 36.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efglp, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vazquez-Torres A, Balish E. Macrophages in resistance to candidiasis. Microbiol Mol Biol Rev. 1997;61:170–192. doi: 10.1128/mmbr.61.2.170-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner D K, Collins-Lech C, Sohnle P G. Inhibition of neutrophil killing of Candida albicans pseudohyphae by substances which quench hypochlorous acid and chloramines. Infect Immun. 1986;51:731–735. doi: 10.1128/iai.51.3.731-735.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler G L, Jones M A, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 40.Wysong D R, Christin L, Sugar A M, Robbins P W, Diamond R D. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]