Abstract
Background
While major advances have been made in improving the quality of life and survival of children with most forms of medulloblastoma (MB), those with MYC-driven tumors (Grp3-MB) still suffer significant morbidity and mortality. There is an urgent need to explore multimodal therapeutic regimens which are effective and safe for children. Large-scale studies have revealed abnormal cancer epigenomes caused by mutations and structural alterations of chromatin modifiers, aberrant DNA methylation, and histone modification signatures. Therefore, targeting epigenetic modifiers for cancer treatment has gained increasing interest, and inhibitors for various epigenetic modulators have been intensively studied in clinical trials. Here, we report a cross-entity, epigenetic drug screen to evaluate therapeutic vulnerabilities in MYC amplified MB, which sensitizes them to macrophage-mediated phagocytosis by targeting the CD47-signal regulatory protein α (SIRPα) innate checkpoint pathway.
Methods
We performed a primary screen including 78 epigenetic inhibitors and a secondary screen including 20 histone deacetylase inhibitors (HDACi) to compare response profiles in atypical teratoid/rhabdoid tumor (AT/RT, n=11), MB (n=14), and glioblastoma (n=14). This unbiased approach revealed the preferential activity of HDACi in MYC-driven MB. Importantly, the class I selective HDACi, CI-994, showed significant cell viability reduction mediated by induction of apoptosis in MYC-driven MB, with little-to-no activity in non-MYC-driven MB, AT/RT, and glioblastoma in vitro. We tested the combinatorial effect of targeting class I HDACs and the CD47-SIRPa phagocytosis checkpoint pathway using in vitro phagocytosis assays and in vivo orthotopic xenograft models.
Results
CI-994 displayed antitumoral effects at the primary site and the metastatic compartment in two orthotopic mouse models of MYC-driven MB. Furthermore, RNA sequencing revealed nuclear factor-kB (NF-κB) pathway induction as a response to CI-994 treatment, followed by transglutaminase 2 (TGM2) expression, which enhanced inflammatory cytokine secretion. We further show interferon-γ release and cell surface expression of engulfment (‘eat-me’) signals (such as calreticulin). Finally, combining CI-994 treatment with an anti-CD47 mAb targeting the CD47-SIRPα phagocytosis checkpoint enhanced in vitro phagocytosis and survival in tumor-bearing mice.
Conclusion
Together, these findings suggest a dynamic relationship between MYC amplification and innate immune suppression in MYC amplified MB and support further investigation of phagocytosis modulation as a strategy to enhance cancer immunotherapy responses.
Keywords: Macrophages; Phagocytosis; Brain Neoplasms; Cytotoxicity, Immunologic; Immunotherapy
WHAT IS ALREADY KNOWN ON THIS TOPIC
Despite detailed molecular subclassification of group 3 (MYC amplified), medulloblastoma development of molecularly targeted therapy has proved challenging. Low mutational load, tumor immune suppression, and immune evasion have all contributed to poor response to the current generation of immune checkpoint inhibitors in brain tumors.
WHAT THIS STUDY ADDS
Using a comparative high throughput epigenetic drug screen, we identified tacedinaline to have specific antineoplastic activity against MYC-amplified medulloblastoma. We further establish epigenetic inhibitors as a viable mechanism to induce tumor inflammation to enhance macrophage checkpoint immunotherapy against pediatric brain tumors.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
We have identified a potential mechanism by which epigenetically driven pediatric tumors with poor host antitumor response can be immunologically activated and made susceptible to phagocytosis checkpoint immunotherapy. This opens a new avenue of immune-epigenetic therapy paradigms in children with therapy-resistant malignant tumors.
Introduction
Epigenetics is defined as the heritable changes in gene expression that occur without changes in the DNA and has gained increasing attention in the context of tumor initiation and progression. Traditionally cancer has been defined as a genetic disease. However, disruption of epigenetic mechanisms can influence tumorigenesis likewise by inducing malignant cellular transformation and driving tumor progression.1 Epigenetic mechanisms, including DNA methylation, histone modifications, nucleosome positioning, and post-transcriptional gene regulation by non-coding RNAs, directly influence gene expression patterns and cell identity. The best-studied epigenetic modifications are the methylation of the DNA base cytosine within so-called CpG island of gene promoter regions and the acetylation of lysine side chains of histone tails. The methylation of cytosine is mediated by DNA methyltransferases (DNMTs), and the hypermethylation of CpG island in promoter regions is associated with transcriptional silencing of genes.2 DNA methylation profiling has been widely applied for classifying brain tumors and helped to identify further layers of heterogeneity within entities.3 Furthermore, repression of gene transcription is also mediated by histone deacetylases (HDACs), which catalyze the removal of acetyl groups from the lysine side chains of histone proteins. Exploiting the reversible nature of epigenetic modifications has emerged as a promising therapeutic strategy for cancer treatment. The Food and Drug Administration (FDA) has already approved DNAMT inhibitors (azacytidine and decitabine) for acute myeloid leukemia and myelodysplastic syndrome and histone deacetylase inhibitors (HDACi) romidepsin, Zolinza (vorinostat), and belinostat (PXD101) for T-cell lymphomas such as cutaneous T-cell lymphoma and peripheral T-cell lymphoma. In addition, dysregulation of the epigenetic landscape has been reported in several pediatric and adult brain tumor entities and constitutes a promising therapeutic approach.
Medulloblastoma (MB) is the most common malignant pediatric brain tumor and is composed of four distinct molecular subgroups, with additional layers of intertumoral heterogeneity. The four consensus subgroups, wingless, sonic hedgehog, group 3, and group 4, are characterized by distinct DNA copy number aberrations, mutational alterations, DNA methylation patterns, and gene expression profiles.4 5 Relative to all disease subgroups, group 3 MB is associated with a comparatively poor prognosis in current multimodal therapeutic regimens and aberrant activation of the proto-oncogene MYC in distinct group 3 subtypes, particularly confers dismal outcomes5–7 with metastasis incidence of 39% and a less than 45% 5-year overall survival rate in tumor with MYC amplification. Metastatic dissemination at diagnosis is a well-established clinical predictor of poor prognosis in patients with MB.8 Previous studies have demonstrated consistently that group 3 MBs display the highest propensity for metastatic dissemination at diagnosis.7 9 Furthermore, most group 3 MBs recur metastatically10 and maintain or even acquire MYC amplification at recurrence11–13 while curative second-line treatment options are lacking and desperately needed for these patients.
In the current study, we performed drug screening to compare the antitumoral activity of epigenetic inhibitors (n=78) in atypical teratoid/rhabdoid tumor (AT/RT), MB, and glioblastoma. With our multientity study design, we aimed to provide an unbiased evaluation of selective activity emerging from the biological differences of the entities rather than identifying inhibitors with broad, unspecific cytotoxicity across entities. In our comparative analysis, we demonstrate a preferential therapeutic activity of HDACi in MYC-driven MB compared with the other brain tumors tested. We performed a targeted rescreen of commercially available preclinical and clinically approved HDACi (n=20) and identified CI-994 (tacedinaline), a class I specific HDACi, as the most selectively active compound for MYC-driven MB. CI-994 demonstrated in vivo efficacy against the primary tumor and, importantly, metastatic lesions in orthotopic xenograft models of MYC-driven MB. Lastly, we uncovered that CI-994 overcomes immune-evasion mechanisms in MYC-driven MB potentially by restoring the nuclear factor-kB (NF-kB) pathway and inducing the expression of immunogenic damage-associated molecular patterns (DAMPS) on the tumor cell surface and secretion of pro-inflammatory cytokines. This, in combination with immune checkpoint blockade (CD47-signal regulatory protein α (SIRPα) phagocytosis blockers), provides a promising approach to improving the therapeutic efficacy against high-risk MB.
Material and methods
Cell lines and culture conditions
A detailed description of all cell models and culture conditions is provided in online supplemental tables 1,2. The MYC status of all MB models was annotated according to the initial model descriptions (online supplemental table 1). Cell line authentication was conducted by short tandem report profiling, and mycoplasma contaminations were ruled out by PCR-based evaluation. Briefly MB primary cells were grown in Neurobasal (-A) supplemented with epidermal growth factor (EGF, 20 ng/mL), basic fibroblast growth factor (bFGF, 20 ng/mL) and leukemia inhibitory factor (LIF, 20 ng/mL). MED8A was cultured in Rosewell Park Memorial Institute (RPMI) supplemented with 10% fetal bovine serum (FBS). Cells were expanded every 10–14 days. The cell lines D425 MED and MED8A used for in vivo experiments were infected with EF1-GFP-T2A-Luc2 lentivirus and double-sorted for green flourescent protein (GFP) signal.
jitc-2022-005871supp001.pdf (211.5KB, pdf)
jitc-2022-005871supp002.pdf (198.2KB, pdf)
Inhibitor libraries and drug screening
A complete overview of the libraries is provided in online supplemental tables 3-5.
jitc-2022-005871supp003.xlsx (22.5KB, xlsx)
jitc-2022-005871supp004.pdf (152.7KB, pdf)
jitc-2022-005871supp005.xlsx (738.6KB, xlsx)
Inhibitors were dispensed using the D300(e) Digital Dispenser (Tecan, Crailsheim, Germany) in 8–11 dilution steps (0.0043–25 µM). The dimethyl sulfoxide (DMSO) concentration was normalized to 0.25% in each well. The inhibitors were dispensed in a randomized manner in white 384-well plates (#3570, Corning, Wiesbaden, Germany). Library plates were stored at −80°C and thawed 1 hour before seeding of cell lines using the Multidrop Combi Reagent Dispenser (Thermo Scientific, Schwerte, Germany). For each cell line, the optimal seeding number was determined prior to the individual screen (online supplemental table 2) to ensure exponential growth during exposure to the inhibitors for 72 hours. The final assay volume was 30 µL per well.
CellTiter-Glo luminescent cell viability assay
The CellTiter-Glo reagent (#G7573, Promega, Mannheim, Germany) was prepared as per the manufacturer’s instructions and used diluted with phosphate-buffered saline (PBS) (1:2 to 1:4) for the library screens and undiluted for validation. 30 µL/well of the readout reagent was dispensed using the Multidrop Combi Reagent Dispenser (Thermo Scientific), and luminescence signals were measured using a Spark 10M microplate reader (Tecan).
ONE-Glo luciferase assay
For evaluation of NF-κB activation following inhibitor or tumor necrosis factor (TNF)-α treatment, the NF-κB reporter cells were plated into 384-well plates and treated for 48 hours. The ONE-Glo (#E6110, Promega) reagent was prepared as per the manufacturer’s instructions, added to the wells, and after an incubation time of 10 min, luminescence signals were measured. Fold change was calculated relative to DMSO control wells.
RNA extraction, complementary DNA synthesis, and quantitative real-time PCR
Extraction of RNA from treated or untreated cells was conducted using Trizol (#15 596–018, Thermo Scientific) or the Maxwell RSC Instrument (RSC simplyRNA Tissue, #AS1340, Promega), and complementary DNA (cDNA) was synthesized from 0.5 µg RNA using M-MLV Reverse Transcriptase (#M3683, Promega) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad, Munich, Germany) with TaqMan probes for MYC (Hs.PT.58.26770695, IDT, Leuven, Belgium) and TGM2 (Hs.PT.58.23141755, IDT). Samples were amplified in triplicate, and relative quantification to housekeeping genes PPIA (Hs.PT.39a.22214851, IDT) and PGK1 (Hs.PT.58.606641, IDT) was assessed using the ΔΔCT method.
Cell lysis, protein extraction, and protein quantification
Cell lysates were generated after treatment of D425 MED and MED8A cells with CI-994 (5 µM and 7.5 µM) and the corresponding DMSO control (0.1%) for 24 hours and 48 hours. Cells were lysed, and protein was extracted using RIPA lysis buffer (#20–188, Merck Millipore, Darmstadt, Germany) supplemented with protease and phosphatase inhibitor cocktail from Roche (#04693132001 and #04906837001, Sigma-Aldrich, Taufkirchen, Germany). Protein was quantified with the Bradford method using the Protein Assay Dye Reagent (#500–0006, Bio-Rad). Samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane (#10600002, Sigma-Aldrich) by wet blot using the Mini Gel Tank and Blot Module (#A25977 and #B1000, Thermo Scientific). The membrane was incubated with mouse anti-MYC (#MA1-980, 9E10, 1:1000, Thermo Scientific) or rabbit anti-TGM2 (#3557S, D11A6, 1:1000, Cell Signaling, Leiden, The Netherlands), and mouse anti-Actin (#MAB1501, clone 4, 1:5000, EMD Millipore, Darmstadt, Germany) primary antibodies overnight at 4°C. Following incubation with species-specific, peroxidase-coupled secondary antibodies (anti-rabbit-HRP, #7074S, 1:5000, Cell Signaling or anti-mouse-HRP, #H2014, 1:5000, Santa Cruz Biotech, Heidelberg, Germany) for 1 hour at room temperature, proteins were visualized using the SuperSignal West Femto Maximum Sensitivity Substrate (#34095, Thermo Scientific) and detected using the LAS-3000 Imaging System from Fujifilm (Düsseldorf, Germany).
Cell apoptosis assay
MED8A and D425 MED cells were plated into 6-well plates (#657160, Greiner Bio-One, Frickenhausen, Germany) and treated with CI-994 or DMSO. After 48 hours, cells were harvested, stained with propidium iodide (PI) (#P4864, Sigma-Aldrich) and fluorescein isothiocyanate (FITC)-labeled annexin V (#556 419 and #51-66121E, BD Pharmingen, Heidelberg, Germany) as per manufacturer’s instructions and analyzed by flow cytometry using a CytoFLEX flow cytometer (Beckmann Coulter, Krefeld, Germany).
Recombinant lentiviral vector construction for stable overexpression of MYC
A lentiviral vector was used for gene delivery to induce stable overexpression of MYC. The plasmid LeGO-iG2 (a kind gift from Boris Fehse, derived from Addgene, Cambridge, Massachusetts, USA) was used to construct the recombinant lentiviral vector. The cDNA sequence of MYC was amplified from the pcDNA3.3_c_MYC plasmid (a kind gift from Derrick Rossi, derived from Addgene) using PCR. PCR primers were designed (online supplemental table 6) to include the BamHI and EcoRI restriction endonuclease sites. The PCR product and plasmid LeGO-iG2 were digested with BamHI and EcoRI (NEB, Frankfurt am Main, Germany). The digested PCR product and plasmid were purified using Wizard SV Gel and PCR Clean-Up System (Promega) and were then ligated. The sequence of all expression vectors was confirmed by DNA sequencing and restriction enzyme analysis (data not shown).
jitc-2022-005871supp006.pdf (152.7KB, pdf)
Stable transduction
Stable overexpression of MYC in UW228-3 cells and stable expression of NF-κB reporter construct pHAGE NF-κB-TA-LUCUBC-GFP-W (a kind gift from Darrell Kotton, derived from Addgene) in D425 MED and MED8A cells were achieved by lentiviral transduction. The NF-κB reporter plasmid contains four copies of a consensus NF-κB binding sequence (gggaatttcc) in addition to a minimal promoter (minimal TATA-box promoter with low basal activity). Pure populations of each stable cell line were sorted by flow cytometry for stable GFP expression using the Moflo XDP (Beckman Coulter).
RNA sequencing and gene set analysis
RNA sequencing data were generated and analyzed as described.14 Statistical gene set analysis was performed using the non-parametric Kruskal-Wallis test to determine differential expression at the gene level (p<0.05, fold change ±2). Partek flow default settings were used in all analyses. The data have been deposited in the gene expression omnibus (GEO)data repository (accession number GSE123760).
Pathway analysis
Ingenuity pathway analysis (IPA, Qiagen) was conducted using genes with significant differential expression (p≤0.05 and fold change ±2). The significance cut-off for IPA was set to p≤0.05 and z score of ±2 for identification of canonical pathways and p≤0.05 and z score of ±1.5 for upstream regulators. For upstream regulators of biological drugs, all chemical and miRNA entries were filtered out. Heatmap visualization and unsupervised hierarchical clustering were performed after normalizing mean expression to 0 with a SD of 1 and using Pearson’s dissimilarity algorithm and average linkage in Partek Genomics Suite (Partek Incorporated).
In vitro phagocytosis
In vitro phagocytosis assay. In vitro phagocytosis assay was performed as described before15 with human macrophages analyzed by flow cytometry. To obtain human monocytes, peripheral blood mononuclear cells (PBMCs) collected from venous blood of healthy volunteers were separated on a Ficoll density gradient (GE Healthcare). CD14+ monocytes were positively selected to >95% purity by magnetic-activated cell sorting (MACS) using anti-CD14 microbeads (Miltenyi), then plated at 1×106/ml in 150×25 mm tissue culture plates in RPMI 1640 with 10% FBS, penicillin/streptomycin, glutamine, and HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid). To generate monocyte-derived macrophages, isolated monocytes were treated for 7 days with human recombinant macrophage colony-stimulating factor (M-CSF) (25 ng/mL). For phagocytosis assay, carboxyfluorescein succinimidyl ester or CFSE-labeled tumor cells were incubated with or without CI-994 and anti-CD47 Mab (10 µg/mL) for 30 min at 37°C before co-incubation with macrophages. Adherent macrophages were collected using TrypLE Express (Life Technologies) and incubated in serum-free medium. 5×104 macrophages were added to 1×105 CFSE-labeled live tumor cells per well for 4 hours and returned to the incubator. Analysis was carried out by flow cytometry. Human macrophages were identified using anti-human CD11b-Alexa647 and anti-human CD14- APC/Cy7 (BioLegend). Phagocytosis assays for each tumor type were performed in triplicates and repeated at least two times using PBMCs from three different donors.
Orthotopic xenograft models for brain tumors
All mice were housed in specific pathogen-free conditions at a barrier facility at the Lokey Stem Cell Building (SIM1) at Stanford University School of Medicine (Stanford, California, USA) and the University of Colorado, School of Medicine Research Tower 1 north Animal facility. All animal handling, surveillance, and experimentation were performed in accordance with and approval from the Stanford University Administrative Panel on Laboratory Animal Care (APLAC No. 26548)) and University of Colorado AMC administrative panel on laboratory animal care (APLAC No. 000777). Male mice were used in all experiments D425 MED-GFP-Luc2 or MED8A-GFP-Luc2 cells were orthotopically injected into 6–10 weeks old NOD SCID gamma (NSG) mice. Mice were anesthetized with 3% isoflurane (Minrad International, Buffalo, New York, USA) and maintained at 2% on the stereotactic frame (David Kopf Instruments, Tujunga, California, USA) delivered through a nose adaptor. A burr hole was placed 2 mm posterior to lambda on the midline. A blunt-ended needle (75N, 26 s/2”/2.5 µL; Hamilton, Reno, Nevada, USA) was lowered into the burr hole to a depth of 3 mm below the dura surface and retracted 0.5 mm. Using a microinjection pump (UMP-3; World Precision Instruments, Sarasota, Florida, USA), 3×104 D425 MED-GFP-Luc or MED8A-GFP-Luc cells were injected in a volume of 3 µL at 30 nL/s. After leaving the needle in place for 1 min, it was retracted at 3 mm/min.
Tumor formation was followed by bioluminescence imaging on an IVIS spectrum instrument (Caliper Life Science, Hopkinton, Massachusetts, USA) and quantified with Live Image V.4.0 software (Living Image, PerkinElmer, Waltham, Massachusetts, USA). MB-engrafted mice were given a daily treatment of CI-994 (30 mg/kg) or vehicle control per oral gavage until they reached morbidity.
The drug solution of CI-994 was prepared according to the manufacturer’s instructions. Briefly, CI-994 powder was first dissolved in DMSO and stored at −20°C. On the day of administration, a final solution was prepared using diH20 and administered at a concentration of 30 mg/kg per oral gavage. Control mice received DMSO alone. For combination studies with anti-CD47 mAb, anti-CD47 (B6H12) was administered as before at 10 mg/kg every 48 hours.
Data analysis
Dose-response curves were generated using non-linear regression (log(inhibitor) versus normalized response) with mean luminescence signals from DMSO control wells (≥3) as 100% cell viability. Inhibitor response was calculated relative to the control (GraphPad Prism software, V.5.03). All data are presented as mean±SD unless stated otherwise. Comparisons between different groups were made using employing Student’s t-test or analysis of variance as appropriate. Statistical significance between different dose-response curves was assessed with regards to the fitted midpoints (log IC50) using the sum-of-squares F test. The statistical significance of Kaplan-Meier survival curves was evaluated using the log-rank (Mantel-Cox) test. P values≤0.05 were considered significant. Heatmaps were generated using the Morpheus online tool (https://software.broadinstitute.org/morpheus/).
Results
Comparative cross-entity drug screening of epigenetic modulators reveals preferential antitumoral activity of HDACi in MB
To comprehensively evaluate epigenetic modifiers as therapeutic targets for the most common malignant brain tumor entities across all age groups, we performed a drug screen using our in-house semi-automated platform. Specifically, a unique collection of AT/RT (n=11), MB (n=14), and glioblastoma (n=11) in vitro models were systematically exposed to a library of 78 inhibitors that target epigenetic writers, readers, erasers, and transcriptional regulators (online supplemental table 3).
To identify inhibitors that are selectively active in one entity or subgroup, we compared the median response (IC50 value) in one entity against the median of the other tested entities together. Out of the 78 inhibitors screened, 48 drugs displayed a median IC50 greater than 25 µM (online supplemental table 7), revealing that the majority of inhibitors showed negligible activity. However, 17 inhibitors showed significantly lower IC50 values in MB compared with glioblastoma and AT/RT in vitro. Remarkably, 11 out of the 17 (65%) selective compounds were HDACi, demonstrating a clear over-representation of this inhibitor class (online supplemental table 7). The preferential therapeutic activity of HDACi was even more notable when MB models were subdivided according to MYC status. We found significantly decreased IC50 values for these 11 inhibitors when comparing MYC-driven MB with non-MYC-driven MB (p=0.0019, online supplemental figure 1). Thus, we next focused on epigenetic inhibitors that exhibit preferential sensitivity in MYC-driven MB (figure 1A, B). We identified 32 inhibitors with higher median activity in MYC-driven MB (figure 1A, group 1), 40 inhibitors with no preferential activity (group 2), and 4 inhibitors that were less active in MYC-driven MB when compared with the median response in the other tested entities (group 3). Among the group of inhibitors with higher median activity in MYC MB, we identified 13 epigenetic inhibitors that were significantly more active when compared with the other tested entities. Again, the majority of these significantly active inhibitors were HDACi (n=9, 69%) (figure 1A, B). Only six inhibitors demonstrated selective activity when in glioblastoma or AT/RT models were compared against the other cell lines together. Three inhibitors of the histone methyltransferase G9a were preferentially active in glioblastoma cell lines, validating our screening approach since G9a has already been shown to be a promising therapeutic target in glioblastoma (online supplemental figure 2 and online supplemental table 7).16 In addition, we identified two DNMTs inhibitors (5-azacytidine and fisetin) and the Jumonji inhibitor GSKJ4 as selectively active in AT/RT models (online supplemental figure 3 and online supplemental table 7). In conclusion, as in previous corroborating studies,17–19 our cross-entity screen emphasizes the remarkable in vitro antitumoral potential of HDACi, in particular for MYC-driven MB.
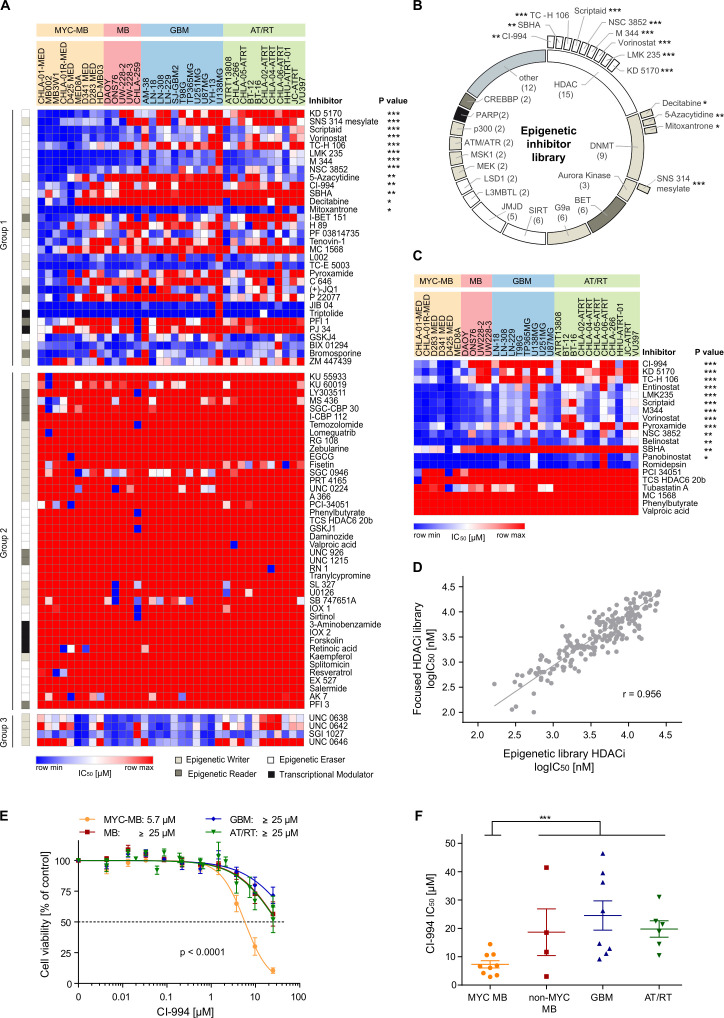
Figure 1.
Drug screening of distinct malignant brain tumor entities reveals the significant vulnerability of MYC-driven medulloblastoma towards HDAC inhibition in vitro. (A) Heatmap representing the activity of 78 epigenetic modulators across 36 brain tumor cell lines derived from medulloblastoma (MB, n=14), glioblastoma (GBM, n=11), and atypical teratoid/rhabdoid tumors (AT/RT, n=11). Inhibitor response was subdivided into three groups. Group 1 consists of inhibitors that showed a lower median IC50 for MYC-driven MB compared with the remaining entities. Group 2 inhibitors were largely inactive across the entities, and group 3 inhibitors were less active in the MYC-driven MB cell lines compared with the other entities. Inhibitors are sorted according to their respective p value when comparing the IC50 values of MYC-driven MB against the IC50 values of all other cell lines (unpaired t-test). (B) The screened epigenetic library is composed of various inhibitor classes targeting different epigenetic modulators (the inner circle). Out of the 78 inhibitors screened, 13 drugs were significantly more active in MYC-driven MB (outer layer). Nine of those selective inhibitors are HDAC inhibitors (HDACi), three compounds inhibit DNA methyltransferases, and one drug is an aurora kinase inhibitor. (C) A focused re-screen of selected HDACi in brain tumor cell lines identified CI-994 as a selective inhibitor for MYC-driven MB. The heatmap represents the IC50 values (µM) of 20 HDACi in 29 cell lines. Inhibitors are sorted according to the respective p value derived from comparing IC50 values of the MYC-driven MB against the IC50 values of all other tested cell lines (unpaired t-test). (D) The response of the HDACi was highly correlated between the primary screen (Epigenetic library, x-axis) and the secondary screen (HDACi library, y-axis). The graph displays the log IC50 (nM) values of the 15 overlapping HDACi in the 29 cell lines included on both screens. (E) Mean dose-response profile and mean IC50 (µM) of CI-994 for each entity/subgroup. (F) The validation screen of CI-994 confirmed the selective response in MYC-driven MB. The values shown represent mean±SEM. *, p<0.05; **, p<0.01; ***, p<0.001. HDAC, histone deacetylases.
jitc-2022-005871supp007.xlsx (738.6KB, xlsx)
jitc-2022-005871supp008.pdf (56.6KB, pdf)
jitc-2022-005871supp009.pdf (50.6KB, pdf)
jitc-2022-005871supp010.pdf (55.1KB, pdf)
Targeted HDACi library screen reveals CI-994 as a selective inhibitor for MYC-driven MB
Given the selective antitumoral activity of HDACi in MYC-driven MB, we performed a second screen, exclusively evaluating HDACi, using our cross-entity panel of brain tumor cell lines (figure 1C). The focused HDACi library included the previously evaluated HDACi (n=15) and was extended with the FDA-approved drugs panobinostat, romidepsin, and belinostat, as well as the well-characterized HDACi entinostat and tubastatin A. IC50 values derived from both screens, are highly correlated (Pearson’s correlation r=0.956, figure 1D). The FDA-approved drugs vorinostat, panobinostat, romidepsin, and belinostat showed substantial inhibition of cell viability across the three tumor entities, with IC50 values in the low nanomolar range for panobinostat and romidepsin (online supplemental table 8). In contrast, valproic acid, phenylbutyrate, and MC 1568 failed to inhibit cell viability at the maximal concentration of 25 µM in any of the screened cell lines (online supplemental table 8). Notably, again none of the HDACi were selectively active against glioblastoma or AT/RT cell lines in vitro (online supplemental table 8). In contrast, 13 of the total 20 HDACi were identified as preferentially active in MYC-driven MB, compared with the other brain tumor models, namely the inhibitors CI-994, KD5170, TC-H 106, entinostat, LMK235, scriptaid, M344, vorinostat, pyroxamide, NSC 3852, belinostat, SBHA and panobinostat (figure 1C, online supplemental table 8). Among these preferentially active HDACi, the class I HDAC selective inhibitor CI-994 demonstrated the most selective reduction of cell viability in MYC-driven MB (p=1.34×10–8, fold-change 3.6; figure 1C, online supplemental table 8). While CI-994 exhibited a mean IC50 of 5.97 µM in the MYC-driven MB, the maximal HDACi concentration of 25 µM induced, on average less than 50% cell viability reduction in any of the other brain tumor entity models (p<0.0001, figure 1E). Notably, three class I selective HDACi (CI-994, TC-H106, and entinostat) were among the top five most preferentially active inhibitors in MYC-driven MB. In a validation screen of CI-994 with an extended concentration range of up to 50 µM, we verified the preferential activity of CI-994 in MYC-driven MB versus other brain tumor entities as demonstrated by significantly lower IC50 values(figure 1F). We also observed minimal cell killing of non-malignant cells (online supplemental figure 4) with human fetal fibroblasts exhibiting a mean IC50 of 4800 µM. Greater than 75% viability was seen at 5 µM treatment of normal human astrocytes and neonatal mouse neuronal cultures compared with <20% viability seen in MB002 cells. In conclusion, our data indicate that CI-994 selectively inhibits cell viability of MYC-driven MB in vitro.
jitc-2022-005871supp011.pdf (47.6KB, pdf)
CI-994 induces apoptosis and decreases MYC expression in MYC-driven MB cell lines
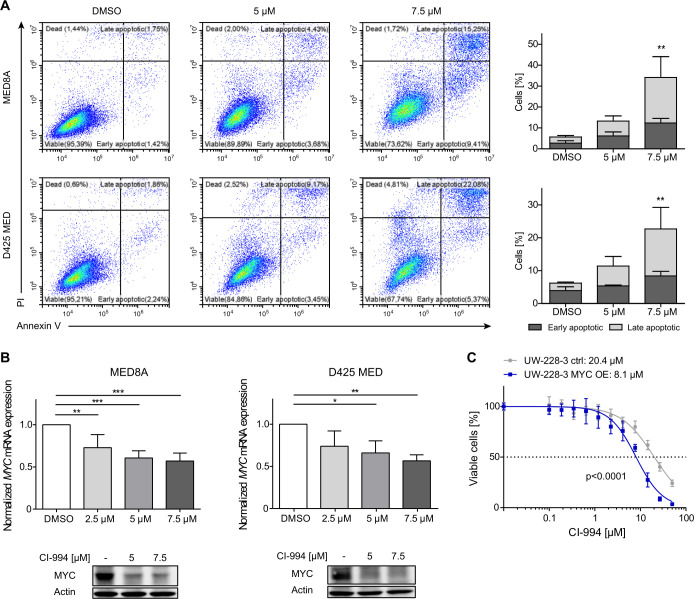
To elucidate the antitumoral effect of CI-994 on MYC-driven MB, we determined the induction of apoptosis following HDACi treatment in two well-characterized, MYC-amplified MB models, namely D425 MED and MED8A (online supplemental table 1). These cells were treated with 5 or 7.5 µM of CI-994 for 48 hours, stained with annexin V and PI, and subsequently analyzed by flow cytometry. We observed a significant increase in annexin V-positive and PI-positive cells in both cell lines on inhibitor treatment at 7.5 µM (p<0.01, figure 2A), indicating that induction of apoptosis contributes to the antitumoral activity of CI-994. Since CI-994 exerts selective activity against MYC-driven MB cells, we examined the impact of CI-994 treatment on MYC expression levels in D425 MED and MED8A. Notably, treatment with CI-994 significantly decreased MYC messenger RNA (mRNA) and protein levels in these two models (p<0.05 for each comparison, figure 2B), suggesting that the antitumoral effect of class I specific HDACi CI-994 is partly mediated by transcriptional repression of MYC. Consistent with the reduced MYC expression, on lentiviral-based MYC overexpression in UW228-3, an MB cell line model with low endogenous MYC level, we observed a twofold decrease in IC50 value compared with the isogenic control (p<0.0001, figure 2C). Thus, our results indicate that the antitumoral activity of CI-994 in MB is in part MYC-dependent, suggesting that aberrant MYC activation in group 3 MB may comprise a predictive biomarker for the response to CI-994.
Figure 2.
CI-994 induces apoptosis and downregulates MYC expression in MYC-driven medulloblastoma cell lines. (A) Representative FACS analysis of MED8A and D425 MED cells treated either with DMSO (control), 5 or 7.5 µM of CI-994 for 48 hours, and bar graphs displaying the mean of n=4 replicates. CI-994 treatment resulted in a dose-dependent increase in mean early apoptotic and late apoptotic cells on treatment in both cell lines. (B) For MYC expression analyses MED8A and D425 MED cells were treated with 2.5, 5, or 7.5 µM of CI-994 for 48 hours. MYC mRNA expression values were normalized to housekeeping controls, and expression was calculated relative to DMSO control. Representative Western blots for MYC and ACTIN loading controls for MED8A and D425 MED cells following treatment with 5 or 7.5 µM CI-994 treatment for 48 hours. (C) MYC overexpressing UW-228–3 cells (UW-228–3 MYC OE) were more sensitive toward CI-994 treatment than the respective isogenic control cells with low endogenous MYC expression (UW-228–3 ctrl). Values shown represent mean±SD of 3–4 replicates per condition. *, p<0.05; **, p<0.01; ***, p<0.001. DMSO, dimethyl sulfoxide; FACS. flow cytometry; mRNA, messenger RNA.
CI-994 inhibits growth and reduces leptomeningeal dissemination of MYC-driven MB in vivo
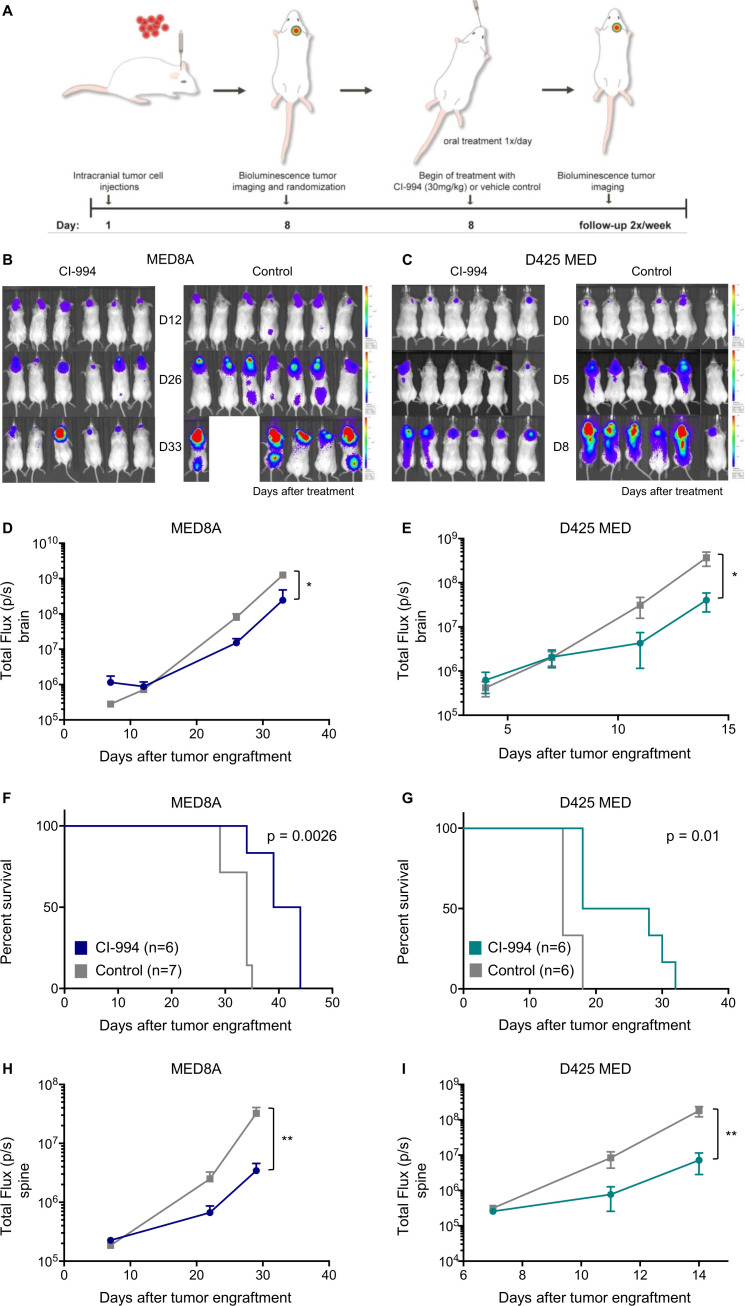
Considering the selective inhibitory effect of CI-994 in vitro, we next tested its efficacy in two orthotopic xenograft mouse models of MYC-driven MB in vivo. D425 MED and MED8A cells expressing GFP and luciferase were orthotopically injected into the cerebella of NSG mice (figure 3A). Tumor engraftment was detected by bioluminescence imaging (BLI) after 7 days. Subsequently, tumor-bearing mice were randomized to daily treatment with either vehicle control or CI-994 (30 mg/kg, by mouth). Treatment with CI-994 significantly decreased tumor growth in both mouse models (figure 3B, C). Control mice exhibited a median survival of 34 and 15 days for MED8A and D425 MED after tumor engraftment, respectively. CI-994 treatment resulted in a significantly prolonged median survival of 41 days for MED8A (p=0.0026, figure 3D) and 23 days for D425 MED (p=0.01, figure 3D). In addition to the reduced tumor growth at the primary site, we detected a significant reduction of spinal dissemination in the CI-994 treatment group in both MYC-driven models (both models p<0.01, figure 3E). Thus, our data demonstrate the anti-tumoral activity of CI-994 against the primary site and, importantly, against the metastatic compartment of MYC-driven MB in vivo. As metastatic recurrences are predominantly observed in group 3 MB, and effective therapeutic strategies are lacking,10 identification of a novel therapeutic agent targeting the metastatic compartment is particularly relevant.
Figure 3.
CI-994 inhibits tumor growth and reduces leptomeningeal dissemination of MYC-driven medulloblastoma in vivo. (A) Treatment schedule for the evaluation of in vivo efficacy of CI-994 in MYC-amplified medulloblastoma. (B) Bioluminescence imaging and luminescence signal quantification of MED8A or D425 MED (C) xenograft from CI-994 treated and control mice. CI-994 treatment reduced tumor growth significantly compared with control mice (MED8A p=0.0015; D425 MED p=0.0344). (D) CI-994 significantly increased survival in both models of MYC-amplified medulloblastoma (MED8A p=0.0026; D425 MED p=0.01). (E) The formation of spinal metastases was significantly reduced in CI-994 treated mice compared with control mice, as determined by quantification of bioluminescence intensity along the spine (MED8A p=0.005; D425 MED p=0.009).
CI-994 induces NF-κB pathway activation in MYC-driven MB
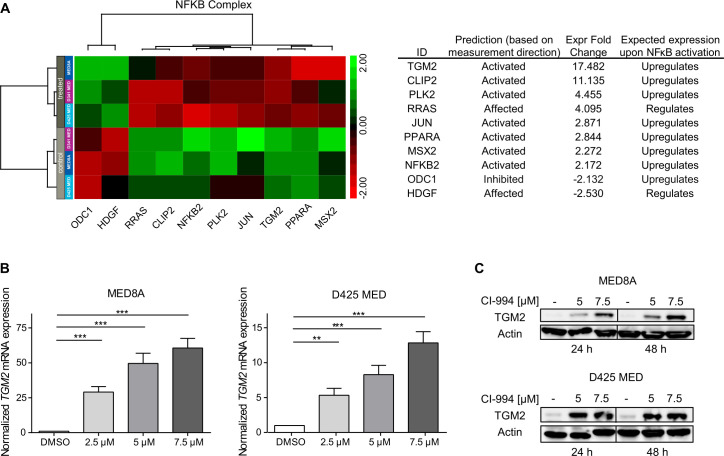
To assess the molecular effects of CI-994 on transcription, we performed RNA sequencing on cells treated with CI-994 and vehicle control. The MYC-driven MB cell lines MED8A, D425 MED, and D341 MED were treated with the respective IC50 concentrations of CI-994 for 48 hours. Treatment with CI-994 resulted in global gene expression changes as detected by RNA sequencing. We identified 173 genes as differentially expressed in DMSO versus CI-994-treated cells using a minimal fold-change ±2 and p≤0.05 as a cut-off (online supplemental table 9). To elucidate canonical pathways and upstream regulators controlled by CI-994 treatment, we performed IPA on the differentially expressed gene sets. Among the canonical pathways and upstream regulators significantly dysregulated following CI-994 treatment, NF-κB pathway activation was consistently identified (online supplemental tables 10,11). Moreover, 26 of 45 canonical pathways (58%) are functionally linked to NF-κB pathway activity (online supplemental table 10). Among the significantly dysregulated NF-κB pathway genes, transglutaminase 2 (TGM2) was the most upregulated target gene in CI-994-treated versus DMSO-treated cells (fold-change 17.5, p<0.05, figure 4A).20 Furthermore, differentially expressed NF-κB pathway genes were sufficient to subdivide all models into treatment groups using unsupervised hierarchical clustering (figure 4A). We next confirmed significant CI-994-mediated upregulation of TGM2 mRNA and protein levels in D425 MED and MED8A cells (figure 4B, C). Hence, our differential expression analysis, in combination with pathway enrichment analyses, points towards a CI-994-mediated NF-κB pathway activation as a potential resistance mechanism in MYC-driven MB. Increase in TGM2 and decrease in cMYC and Ki67 was observed in tumor cells in vivo (online supplemental figure 5).
Figure 4.
CI-994 treatment induces the expression of NF-κB pathway genes in MYC-driven medulloblastoma. (A–C) MED8A, D425 MED, and D341 MED treated with CI-994 or vehicle control were analyzed by RNA sequencing. The non-parametric supervised analysis identified 130 upregulated and 43 downregulated genes with a fold change ±2 and p≤0.05, which were analyzed by ingenuity pathway analysis (IPA). (A) Table showing top 20 out of 44 canonical pathways with activation z-score ≥2 and ranked by p value. (B) Table of top 10 upstream regulators identified by IPA with an activation score ≥1.8 and p≤0.05. (C/D) Unsupervised hierarchical clustering of genes regulated by NF-κB complex. (E) MED8A and D425 MED cells were treated with 2.5, 5, or 7.5 µM of CI-994 for 48 hours. TGM2 mRNA expression values are normalized to housekeeping controls, and expression is calculated relative to DMSO control. The values shown represent the mean±SD of three replicates per condition. (F) Western blots for TGM2 and Actin loading controls following CI-994 treatment for 24 and 48 hours. *, p<0.05; **, p<0.01; ***, p<0.001. DMSO, dimethyl sulfoxide; mRNA, messenger RNA; NF-kB, nuclear factor-kB; TGM2, transglutaminase 2.
jitc-2022-005871supp012.pdf (1.1MB, pdf)
CI-994 induces tumor inflammation in MYC-MB cell lines and increases infiltration of inflammatory macrophages in vivo
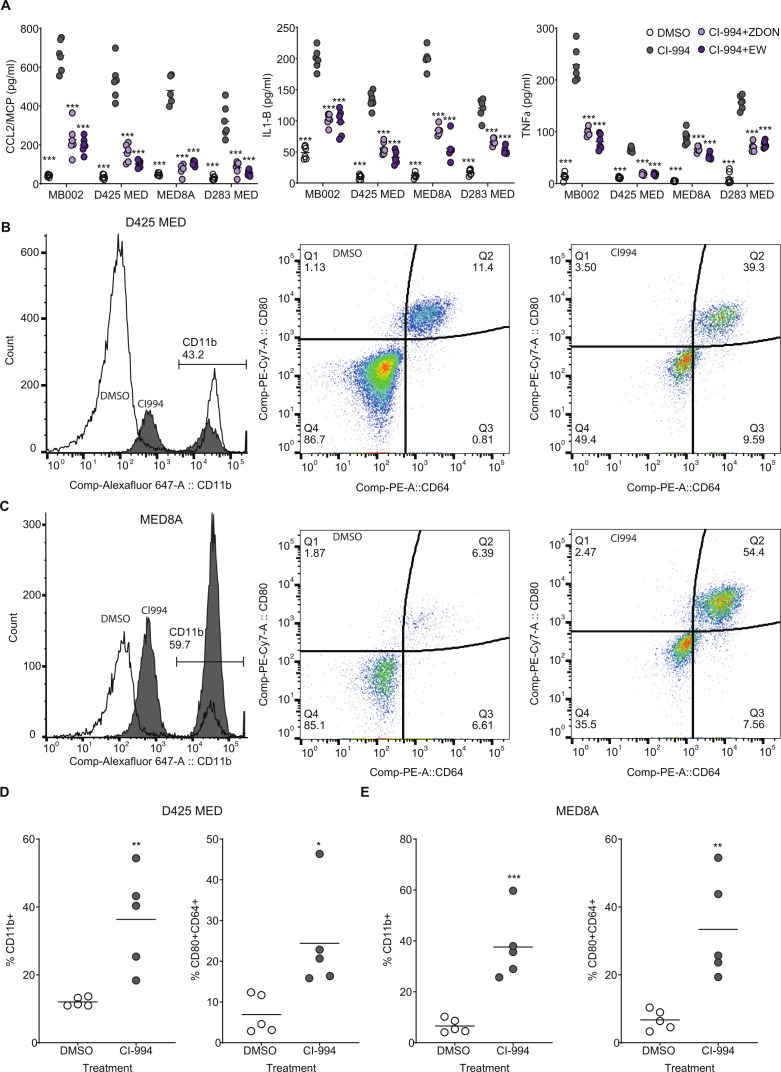
Previously it has been shown that amplification of TGM2 correlated with increased inflammation in gastric cancers.21 We next investigated whether TGM2 upregulation by CI-994 played a role in tumor inflammation. MED8A, D425MED, D283MED and MB002 cells were treated with either DMSO, CI-994 (5 μM), CI-994+ZDON (TGM2 inhibitor: 1 μM) or CI-994+ERW1041E (TGM2 inhibitor: 10 μM) for 48 hours. ZDON is a cell-permeable, peptide-based irreversible inhibitor that reacts with the active site cysteine of TGM2 and ERW1041E is a cell-permeable, active-site directed, acyl-donor substrate competitive inhibitor of TGM2. Cell culture supernatant was collected and analyzed for the secretion of pro-inflammatory/T helper 1 (TH-1)-promoting cytokines monocyte chemoattractant protein-1 (CCL2/MCP-1), interleukin (IL)-1β, and ΤΝF-α. Treatment with CI-994 (5 μM) alone significantly increased secretion of CCL2/MCP-1, IL-1βand TNF-α (figure 5A). Secretion of all three cytokines induced by CI-994 was suppressed by two (functionally independent) TGM2 inhibitors ZDON and ERW1041E. No significant change in inhibitory cytokines such as IL-10 was observed. We next investigated whether treatment of MYC-MB xenograft tumor-bearing mice by CI-994 induced macrophage infiltration (figure 5B–D: CD11b) and a macrophage polarization profile toward a favorable antitumorigenic microenvironment. Mice (n=5 each) that had been orthotopically xenografted with MYC-MB cell lines D425 MED or MED 8a and confirmed by BLI were subsequently treated with either CI-994 or control for 3 weeks after confirmation of tumor engraftment. The resulting tumor-bearing brain tissue was analyzed by flow cytometry for the polarization profile of infiltrating mouse macrophages using cell surface marker expression of CD11b (total macrophages), CD80, and CD64, as shown previously.22 Mice treated with CI-994 were found to have an increased infiltration of macrophages. Furthermore, an increased number of macrophages with M1 phenotype markers (CD80+CD64+) relative to untreated mice were observed in xenografts from D425 MED (figure 5D) and MED8A (figure 5E). Increase in TGM2 and decrease in cMYC and Ki67 was observed in tumor cells in vivo (online supplemental figure 4).
Figure 5.
CI-994 induces a TGM2-dependent pro-inflammatory cytokine response from tumor cells and an increase in infiltration of pro-inflammatory macrophages within tumors: (A) Essay for secretion of CCL1, interleukin-1B, and TNF-α was carried out by ELISA. Significant increase (***p<0.001: ordinary one-way analysis of variance) in the secretion of all three cytokines was observed in MB002, D425MED, MED8A, and D283MED cell lines on treatment with 5 µM CI-994. We also observed a significant drop in secretion of all three cytokines on inhibition of TGM2 using two different inhibitors, ZDON and ERW1041E. (B–C) Representative data from In vivo analysis of macrophage infiltration of macrophages in NSG mice with D425MED (B) or MED8A (C) cells xenografted into the cerebellum. Tumors were harvested and analyzed for total macrophage infiltration as identified being GFP– (tumors cells) and CD11b+. Increase in pro-inflammatory macrophages (CD45+CD11b+CD80+CD64+) is seen in CI-994 treated animals as compared with DMSO control. (D) Quantitative presentation of flow cytometry analysis from B and C. significant increase in CD11b+ macrophages (**p<0.01; n=5) and CD80+CD64+ (*p<0.05; n=5) was observed in D425MED cells whereas higher infiltration was observed in CD11b+ (***p<0.001; n=5) and CD80+CD64+ (**p<0.01; n=5 each). Each data point is an individual tumor-bearing mouse. *p<0.05; **p<0.01; ***p<0.001. DMSO, dimethyl sulfoxide; TGM2, transglutaminase 2; TNF, tumor necrosis factor.
CI-994 enhances anti-CD47 mediated phagocytosis and survival of MYC-MB tumor bearing mice
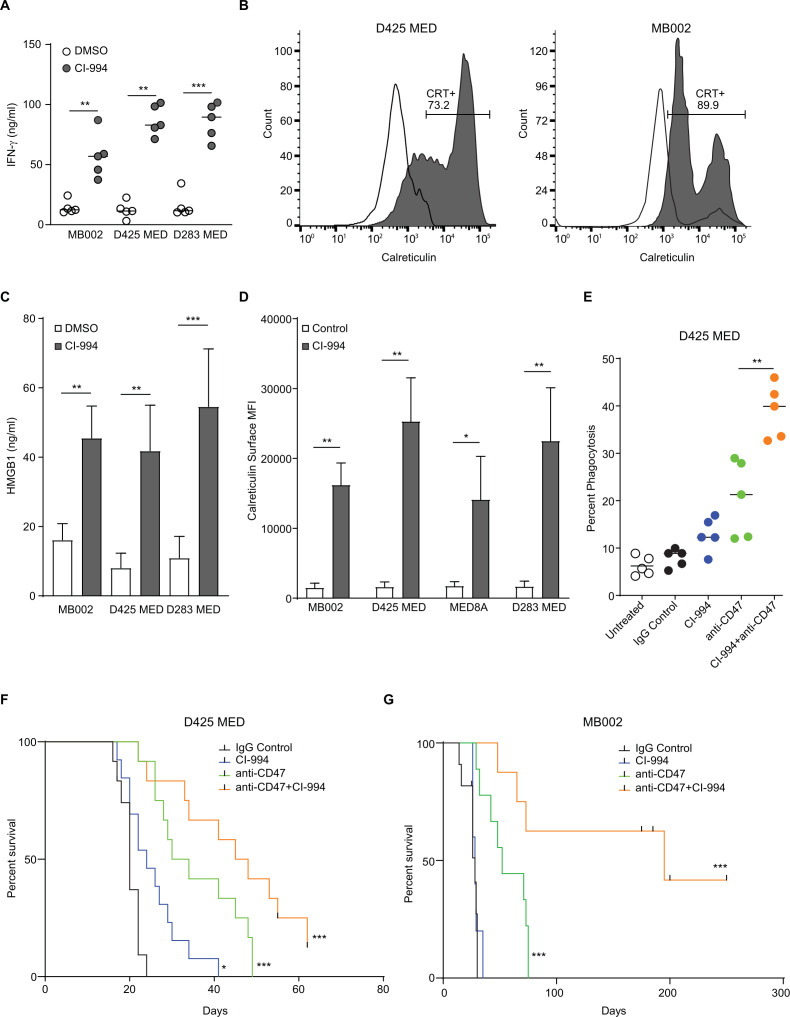
To identify the effect of CI-994 mediated NF-kB activation on macrophage checkpoint immunotherapy, supernatants were harvested from MYC amplified MB cell lines (MB002, D425, and D283) that had been treated with CI-994 for 24 hours, and levels of tumor secreted interferon (IFN)-γ was analyzed. Significantly higher levels of IFN-γ were detected in all three cell lines (figure 6A), with the highest response in D283 cells. The release of damage-associated molecular patterns is crucial for the success of the CD47-SIRPα phagocytosis checkpoint immunotherapy. We assessed if CI-994 treatment induces phagocytosis of tumor cells and causes the release of DAMPs. We found that calreticulin was expressed on the surface of MYC-driven MB cells, and its cell-surface exposure was upregulated by CI-994 treatment (p<0.05, figure 5B, C). Furthermore, a significant increase in the release of another DAMP protein, high mobility group box 1 protein, was noticed in CI-994 treated groups (p<0.05, figure 5D). The combination of CI-994 and anti-CD47 monoclonal antibody (mAb) strongly enhanced phagocytosis of D425 cells by human PBMC-derived macrophages, whereas the treatment of anti-CD47 showed only a slight increase in phagocytosis (figure 5E). In vivo experiments in two different MYC-MB orthotopic xenograft models corroborated these results and showed that the combination treatment of CI-994 and anti-CD47 significantly enhanced survival as compared with CD47 or CI-994 alone (figure 5F, G).
Figure 6.
CI-994 sensitizes MYC-driven medulloblastoma cells to anti-CD47 phagocytosis checkpoint immunotherapy: (A) Tumor inflammation on CI-994 treatment was evaluated by an ELISA-based assay for IFN-γ secretion. A significant increase (**p<0.05: non-parametric t-test) is seen in MB002, D425, and D283 cell lines on treatment at 2.5 μM CI-994. (B) An increase in expression of surface calreticulin is seen in MB002 (left panel) and D425 (right panel) as assayed by flow cytometry. (C) An increase in the relative surface calreticulin expression per cell was assayed by measuring the MFI for four different cell lines. (D) Levels of HMGB1 were assayed by a standard colorimetric ELISA. (E) D425 cells were incubated with human peripheral blood mononuclear cells-derived macrophages and tumor cells pretreated with either IgG control, CI-994, anti-CD47 mAb (B6H12), or a combination of CI-994+ anti-CD47 mAb. Phagocytosis was assayed using flow cytometry. (F) Survival analysis of D425 tumor-bearing mice treated with either control, CI-994, anti-CD47, or a combination of anti-CD47 and CI-994.(p=0.0002 using the log-rank (Mantel-Cox) test with a HR of 3.669). DMSO, dimethyl sulfoxide; IFN, interferon; mAb, monoclonal antibody; MFI, mean fluorescence intensity.
Discussion
Amplification of MYC constitutes a widely accepted biomarker indicating poor prognosis and driver of aggressive tumor biology in patients with MB, as demonstrated in several clinical trial cohorts and retrospective analyses.7 23 24 Hence, patients with MYC-driven MB are commonly treated with intensified high-dose chemotherapy and radiotherapy protocols.4 However, 5-year overall survival does not exceed 40% in this patient cohort, and the aggressive treatment strategies often result in severe long-term side effects and secondary malignancies in survivors.25–28
Our understanding of the underlying intertumoral heterogeneity is constantly expanding, with the recognition of molecularly defined MB subgroups in the current WHO classification,29 and currently emerging subgroup-specific subtypes.9 24 30 A refined, molecular-based classification acknowledging this biological heterogeneity allows the prioritization of therapeutic targets and improves current risk stratification with prognostically-relevant strata in the context of current treatment approaches.31 32 Clinical translation of disease subgroups or subtypes to allocate specific treatments is now emerging to be integrated into clinical trials (eg, NCT02212574, NCT01878617, and SJMB12 trial).33
In order to identify novel therapeutics for the most common malignant brain tumors of all age groups, we performed a targeted drug screen to evaluate epigenetic-based therapies in AT/RT, MB, and glioblastoma. Our approach significantly enhances recent HDACi-related insights for MYC-driven MB19 as we identify drugs that are specifically active in entities or even molecularly-defined subsets of tumors. In our primary screen, we demonstrated that HDACi are specifically active in MYC-driven MB compared with non-MYC MB, AT/RT, and glioblastoma models. Although epigenetic dysregulation caused by, for example, aberrant methylation patterns or alterations of chromatin modulators is also commonly identified in AT/RT and glioblastoma, cell lines from these entities were, on average, less susceptible to our epigenetic inhibitor library. Our cross-entity comparison, therefore, indicates that targeting epigenetic modulators might be particularly promising for MB. In a targeted re-screen with an extended HDACi library, we identified CI-994 as the most promising and selective HDACi in MYC-driven MB. CI-994 is an orally bioavailable class I specific HDACi, which crosses the blood-brain barrier.34 Previous studies reported that CI-994 inhibits proliferation and induces apoptosis in vitro and in vivo in other tumor entities,35 36 but it was not examined in MB up to now.
Based on the selective activity of CI-994 in MYC-driven MB and the attractive pharmacological features as well as the known side effect profiles of CI-994 for treating children with brain tumors, we prioritized this drug for in vivo testing. Our in vivo data not only demonstrate significantly extended survival in two orthotopic MYC-driven MB mouse models (figure 3D) but, importantly, reveal significant antitumoral activity against metastatic dissemination (figure 3E). This addresses a pressing clinical challenge, as these tumors are commonly metastatic at diagnosis and at recurrence, which is associated with dismal prognosis, and therapeutic strategies are urgently required for these high-risk patients.
Downstream effects of class I specific HDACi and potential resistance mechanisms are currently unknown in MB. Using a transcriptomic approach, our data highlighted a significant induction of the NF-κB pathway on CI-994 treatment, which was detected in multiple curated gene sets, and significant transcriptional activation was demonstrated using IPA (online supplemental table 10). The NF-kB pathway plays a crucial role in the regulation of tumor inflammation. As a controller of the transcription of cytokines and antimicrobial effectors, NF-kB is involved in promoting differentiation, development, and response in cells of innate and adaptive immune systems. MYC-driven neoplastic cells can bypass immune detection by regulating immune checkpoints through CD47 and programmed death-ligand 1.37 38 Therefore, activation of NF-kB in MYC-driven MB cells may restore the immune response and sensitize these cells to immunotherapy. Here we observed an upregulation of tissue TGM2, a direct transcriptional target of NF-kB that is frequently upregulated during inflammation and wounding. Treatment with CI-994 increased pro-inflammatory cytokine secretion by Grp3-MB tumor cells, which was inhibited by the blocking of TGM2 function. The CCL2/MCP-1, IL-1β, ΤΝF-α and IFN-γ TH-1 responses explain the infiltration of pro-inflammatory macrophage response when Grp-3 MB xenograft bearing mice were treated with CI-994.
Recently, the important roles of macrophages in the surveillance and elimination of tumor cells, which was termed ‘programmed cell removal’ (PrCR), have been revealed by us and other laboratory studies.39–42 The upregulation of anti-phagocytic ‘don’t eat me’ signals on cancer cells protects them from PrCR, while blockade of such pathways enables PrCR and leads to the elimination of tumor cells by macrophages. The CD47/SIRPα axis serves a broader role as a myeloid immune checkpoint frequently co-opted by tumor cells to avoid immune surveillance.43 Previously we demonstrated that anti-CD47 monoclonal antibody has potent antitumor activity against a wide range of solid tumors, including adult glioblastoma44 and five etiologically distinct human pediatric brain tumors, including Grp3-MB (6). More recently, a number of studies have shown that inducing immunogenic cell death is an attractive approach to activating the immune microenvironment of ‘cold’ tumors. Inducing immunogenic cell death and expression of DAMPs, such as calreticulin and phosphatidyl-serine on the cell surface, increase the immunogenicity of tumor cells to trigger immune responses engaging both the innate and adaptive immune responses. Here we demonstrate that sublethal doses of CI-994 caused tumor inflammation presumably through the activation of the NF-kB pathway leading to secretion of the pro-inflammatory cytokine IFN-γ. We further observe cell surface expression of calreticulin as well secretion of HMGB1. These, combined with the blockade of the CD47-SIRPα phagocytosis checkpoint pathway, enhance macrophage-mediated tumor cell phagocytosis in vitro and in vivo, thereby enhancing the survival of mice with MYC-driven MB xenografts. While the blood-brain barrier is a major concern for the delivery of drugs from the systemic circulation to the brain prior, our previous studies have shown accessibility of the anti-CD47 Mab to the tumor bed. Furthermore, previous studies suggest that the central nervous system (CNS) is immune-accessible and interacts dynamically with the systemic immune system.45 Inflammation also induces immune cells to migrate into the parenchyma by traveling a chemotactic response through (among others) IFN-γ inducible cytokine gradients, a key pro-inflammatory myeloid activator.46 47 Additionally, FcRn, a ubiquitous immunoglobulin receptor highly expressed in blood vessels in the brain, can facilitate IgG transport into the CNS.48 Currently there are multiple clinical investigations targeting the CD47/SIRPα axis alone or in combination with reagents that enhance the do not eat me signal.49 50
Our study demonstrates the efficacy of combining macrophage-checkpoint inhibition with HDACi for cancer immunochemotherapy against MYC-driven MB. The combination of anti-CD47 and HDACi eliminated MYC-driven cancer cells with increased efficacy due to the enhancement of the NF-kB mediated ‘eat-me’ signals and antibody-dependent block of ‘don’t eat-me’ signals. This may be an effective approach for treating high-risk MYC-driven MB.
jitc-2022-005871supp013.pdf (97.9KB, pdf)
Acknowledgments
Anna Kaufhold and Sarah Göbbels are acknowledged for excellent technical assistance. We thank Tim Doyle, Stanford Center for Innovation in In-Vivo Imaging (SCI3), for technical support during bioluminescence imaging. We thank Michael Alvarez and the Stanford University veterinary services staff for their help with animal husbandry.
Footnotes
Twitter: @mitralabcuamc
Contributors: VM, JT, DPa, DPi, NQ, MR and SSM designed, executed, analyzed data, and wrote the manuscript. LB, MM, JB, UA, ML, F-DM, AC, JC-C and CMG provided technical assistance and support. MW, TM, OW, AE-E, GL, UK, AS, KS, STK, DDB, JH, TB, CBK-T, UF, JF, FKH, RV, SV, SHC, GR, AB and TK provided material support. MR and SM were responsible for the overall conduct of research and reproducibility and act as guarantor.
Funding: This research project was supported by the German Cancer Aid awarded to GR and MR (grant no. 111537). JT was supported by the German Cancer Aid (grant no. P-91650709). The work of the UK is supported by the BMBF (03VP0731), Wegener Foundation, and Hempel Foundation. CBK-T was supported by the Max Eder Program of the German Cancer Aid (grant no. 110663) as well as by a grant from the BMBF (01ZX1401B). AB and JH are supported by the German Cancer Aid (Translational Oncology Program 70112951) and the DKTK German Cancer Consortium Joint funding program ‘Targeting MYC’ L*10. CMG was supported by the German Academic Scholarship Foundation. VM received a PhD scholarship from the Düsseldorf School of Oncology (funded by the Comprehensive Cancer Center Düsseldorf/German Cancer Aid and the Medical Faculty HHU Düsseldorf). SSM was supported by grants from the Morgan Adams Foundation, the Cancer League of Colorado, The B+ Foundation, The American Cancer Society-IRG, The Broncos Foundation, the Plachy-Rubin Foundation, and the V Foundation for Cancer Research. The UCD Cancer Center core facilities are supported by NCI Cancer Center Support grant no. P30-CA046934.
Competing interests: SHC and SSM hold a Patent entitled ‘Treatment of pediatric brain tumors with targeting of CD47 pathway’. Other authors hold no potential conflict of interest.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Studies were carried out under the following: Stanford University IACUC # 26548, IRB protocol ID 18672; IRB Number 350: Panel 3. University of Colorado, AMC IACUC # 00777 and Institutional Biosafety Committee No. IBC-1324.
References
- 1.Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol 2009;4:229–49. 10.1146/annurev.pathol.3.121806.151442 [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Zhang Y. Role of mammalian DNA methyltransferases in development. Annu Rev Biochem 2020;89:135–58. 10.1146/annurev-biochem-103019-102815 [DOI] [PubMed] [Google Scholar]
- 3.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramaswamy V, Remke M, Bouffet E, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol 2016;131:821–31. 10.1007/s00401-016-1569-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012;123:465–72. 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y-J, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 2011;29:1424–30. 10.1200/JCO.2010.28.5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of Wnt, Shh, group 3, and group 4 medulloblastomas. Acta Neuropathol 2012;123:473–84. 10.1007/s00401-012-0958-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeltzer PM, Boyett JM, Finlay JL, et al. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the children's cancer group 921 randomized phase III study. J Clin Oncol 1999;17:832–45. 10.1200/JCO.1999.17.3.832 [DOI] [PubMed] [Google Scholar]
- 9.Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 2017;31:737–54. 10.1016/j.ccell.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramaswamy V, Remke M, Bouffet E, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol 2013;14:1200–7. 10.1016/S1470-2045(13)70449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill RM, Kuijper S, Lindsey JC, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015;27:72–84. 10.1016/j.ccell.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korshunov A, Remke M, Kool M, et al. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathol 2012;123:515–27. 10.1007/s00401-011-0918-8 [DOI] [PubMed] [Google Scholar]
- 13.Morrissy AS, Garzia L, Shih DJH, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016;529:351–7. 10.1038/nature16478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forget A, Martignetti L, Puget S, et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell 2018;34:379–95. 10.1016/j.ccell.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 15.Gholamin S, Mitra SS, Feroze AH, et al. Disrupting the CD47-SIRPalpha anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci Transl Med 2017;9. doi: 10.1126/scitranslmed.aaf2968. [Epub ahead of print: 15 Mar 2017]. [DOI] [PubMed] [Google Scholar]
- 16.Ciechomska IA, Przanowski P, Jackl J, et al. BIX01294, an inhibitor of histone methyltransferase, induces autophagy-dependent differentiation of glioma stem-like cells. Sci Rep 2016;6:38723. 10.1038/srep38723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milde T, Lodrini M, Savelyeva L, et al. HD-MB03 is a novel group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. J Neurooncol 2012;110:335–48. 10.1007/s11060-012-0978-1 [DOI] [PubMed] [Google Scholar]
- 18.Ecker J, Oehme I, Mazitschek R, et al. Targeting class I histone deacetylase 2 in MYC amplified group 3 medulloblastoma. Acta Neuropathol Commun 2015;3:22. 10.1186/s40478-015-0201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei Y, Liu K-W, Wang J, et al. HDAC and PI3K antagonists cooperate to inhibit growth of Myc-driven medulloblastoma. Cancer Cell 2016;29:311–23. 10.1016/j.ccell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Chen Z, Miranda RN, et al. TG2 and NF-κB signaling coordinates the survival of mantle cell lymphoma cells via IL6-Mediated autophagy. Cancer Res 2016;76:6410–23. 10.1158/0008-5472.CAN-16-0595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S-Y, Oh Y, Jeong EM, et al. Amplification of transglutaminase 2 enhances tumor-promoting inflammation in gastric cancers. Exp Mol Med 2020;52:854–64. 10.1038/s12276-020-0444-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Hutter G, Kahn SA, et al. Anti-CD47 treatment stimulates phagocytosis of glioblastoma by M1 and M2 polarized macrophages and promotes M1 polarized macrophages in vivo. PLoS One 2016;11:e0153550. 10.1371/journal.pone.0153550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih DJH, Northcott PA, Remke M, et al. Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol 2014;32:886–96. 10.1200/JCO.2013.50.9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwalbe EC, Lindsey JC, Nakjang S, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 2017;18:958–71. 10.1016/S1470-2045(17)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulhern RK, Palmer SL, Merchant TE, et al. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol 2005;23:5511–9. 10.1200/JCO.2005.00.703 [DOI] [PubMed] [Google Scholar]
- 26.Eaton BR, Esiashvili N, Kim S, et al. Endocrine outcomes with proton and photon radiotherapy for standard risk medulloblastoma. Neuro Oncol 2016;18:881–7. 10.1093/neuonc/nov302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer SL, Armstrong C, Onar-Thomas A, et al. Processing speed, attention, and working memory after treatment for medulloblastoma: an international, prospective, and longitudinal study. J Clin Oncol 2013;31:3494–500. 10.1200/JCO.2012.47.4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Packer RJ, Zhou T, Holmes E, et al. Survival and secondary tumors in children with medulloblastoma receiving radiotherapy and adjuvant chemotherapy: results of children's Oncology Group trial A9961. Neuro Oncol 2013;15:97–103. 10.1093/neuonc/nos267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131:803–20. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 30.Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7. 10.1038/nature22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goschzik T, Schwalbe EC, Hicks D, et al. Prognostic effect of whole chromosomal aberration signatures in standard-risk, non-WNT/non-SHH medulloblastoma: a retrospective, molecular analysis of the HIT-SIOP PNET 4 trial. Lancet Oncol 2018;19:1602–16. 10.1016/S1470-2045(18)30532-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remke M, Hielscher T, Korshunov A, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol 2011;29:3852–61. 10.1200/JCO.2011.36.2798 [DOI] [PubMed] [Google Scholar]
- 33.Morfouace M, Shelat A, Jacus M, et al. Pemetrexed and gemcitabine as combination therapy for the treatment of Group3 medulloblastoma. Cancer Cell 2014;25:516–29. 10.1016/j.ccr.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riva L, Blaney SM, Dauser R, et al. Pharmacokinetics and cerebrospinal fluid penetration of CI-994 (N-acetyldinaline) in the nonhuman primate. Clin Cancer Res 2000;6:994–7. [PubMed] [Google Scholar]
- 35.Hubeek I, Comijn EM, Van der Wilt CL, et al. CI-994 (N-acetyl-dinaline) in combination with conventional anti-cancer agents is effective against acute myeloid leukemia in vitro and in vivo. Oncol Rep 2008;19:1517–23. 10.3892/or.19.6.1517 [DOI] [PubMed] [Google Scholar]
- 36.Gediya LK, Belosay A, Khandelwal A, et al. Improved synthesis of histone deacetylase inhibitors (HDIs) (MS-275 and CI-994) and inhibitory effects of HDIs alone or in combination with RAMBAs or retinoids on growth of human LNCaP prostate cancer cells and tumor xenografts. Bioorg Med Chem 2008;16:3352–60. 10.1016/j.bmc.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casey SC, Baylot V, Felsher DW. The MYC oncogene is a global regulator of the immune response. Blood 2018;131:2007–15. 10.1182/blood-2017-11-742577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227–31. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng M, Chen JY, Weissman-Tsukamoto R, et al. Macrophages eat cancer cells using their own calreticulin as a guide: roles of TLR and Btk. Proc Natl Acad Sci U S A 2015;112:2145–50. 10.1073/pnas.1424907112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao MP, Weissman IL, Majeti R. The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol 2012;24:225–32. 10.1016/j.coi.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer 2012;12:58–67. 10.1038/nrc3171 [DOI] [PubMed] [Google Scholar]
- 42.Chao MP, Alizadeh AA, Tang C, et al. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res 2011;71:1374–84. 10.1158/0008-5472.CAN-10-2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng M, Jiang W, Kim BYS, et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer 2019;19:568–86. 10.1038/s41568-019-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willingham SB, Volkmer J-P, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 2012;109:6662–7. 10.1073/pnas.1121623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carson MJ, Doose JM, Melchior B, et al. CNS immune privilege: hiding in plain sight. Immunol Rev 2006;213:48–65. 10.1111/j.1600-065X.2006.00441.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein RS. Regulation of neuroinflammation: the role of CXCL10 in lymphocyte infiltration during autoimmune encephalomyelitis. J Cell Biochem 2004;92:213–22. 10.1002/jcb.20052 [DOI] [PubMed] [Google Scholar]
- 47.Klein RS, Izikson L, Means T, et al. IFN-inducible protein 10/CXC chemokine ligand 10-independent induction of experimental autoimmune encephalomyelitis. J Immunol 2004;172:550–9. 10.4049/jimmunol.172.1.550 [DOI] [PubMed] [Google Scholar]
- 48.Lampson LA. Monoclonal antibodies in neuro-oncology: getting past the blood-brain barrier. MAbs 2011;3:153–60. 10.4161/mabs.3.2.14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lentz RW, Colton MD, Mitra SS, et al. Innate immune checkpoint inhibitors: the next breakthrough in medical oncology? Mol Cancer Ther 2021;20:961–74. 10.1158/1535-7163.MCT-21-0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Duijn A, Van der Burg SH, Scheeren FA. CD47/SIRPalpha axis: bridging innate and adaptive immunity. J Immunother Cancer 2022;10:e004589. 10.1136/jitc-2022-004589 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005871supp001.pdf (211.5KB, pdf)
jitc-2022-005871supp002.pdf (198.2KB, pdf)
jitc-2022-005871supp003.xlsx (22.5KB, xlsx)
jitc-2022-005871supp004.pdf (152.7KB, pdf)
jitc-2022-005871supp005.xlsx (738.6KB, xlsx)
jitc-2022-005871supp006.pdf (152.7KB, pdf)
jitc-2022-005871supp007.xlsx (738.6KB, xlsx)
jitc-2022-005871supp008.pdf (56.6KB, pdf)
jitc-2022-005871supp009.pdf (50.6KB, pdf)
jitc-2022-005871supp010.pdf (55.1KB, pdf)
jitc-2022-005871supp011.pdf (47.6KB, pdf)
jitc-2022-005871supp012.pdf (1.1MB, pdf)
jitc-2022-005871supp013.pdf (97.9KB, pdf)
Data Availability Statement
Data are available upon reasonable request.