Abstract
Purpose:
Recently, the Connective Tissue Oncology Society published consensus guidelines for recognizing ultrarare sarcomas (URS), defined as sarcomas with an incidence ≤1 per 1,000,000. We assessed the outcomes of 56 patients with soft tissue, and 21 with bone sarcomas, enrolled in Phase 1 trials.
Experimental Design:
In this Sarcoma-Matched Biomarker Analysis (SAMBA-102 study), we reviewed records from patients on Phase 1 trials at the University of Texas MD Anderson Cancer Center between January 2013 and June 2021.
Results:
Among 587 sarcomas, 106 (18.1%) were classified as URS. Fifty (47%) were male, and the median age was 44.3 years (range, 19–82). The most common subtypes were alveolar soft part sarcoma (ASPS), chordoma, dedifferentiated chondrosarcoma, and sclerosing epithelioid fibrosarcoma. Compared with common sarcomas, median OS was similar 16.1 months [95% confidence interval (CI), 13.6–17.5] versus 16.1 (95% CI, 8.2–24.0) in URS (P = 0.359). Objective response to treatment was higher in URS 13.2% (n = 14/106) compared with common sarcomas 6.9% (n = 33/481; P = 0.029). Median OS for those treated on matched trials was 27.3 months (95% CI, 1.9–52.7) compared with 13.4 months (95% CI, 6.3–20.6) for those not treated on matched trials (P = 0.291). Eight of 33 (24%) molecularly matched treatments resulted in an objective response, whereas 6 of 73 unmatched treatments (8.2%) resulted in an objective response (P = 0.024). Clinical benefit rate was 36.4% (12/33) in matched trials versus 26.0% (19/73) in unmatched trials (P = 0.279).
Conclusions:
The results demonstrate the benefit of genomic selection in Phase 1 trials to help identify molecular subsets likely to benefit from targeted therapy.
Translational Relevance.
Ultrarare sarcomas (URS) are a newly defined category of rare sarcomas for which histology-driven treatments are limited. Individually, these cancer types are scarce, but when combined, they comprise 20% of all bone and soft tissue sarcomas. Genomic sequencing of rare tumors may identify actionable alterations amenable for molecularly targeted therapies. A review of all URS from a large Phase 1 clinic treated on molecularly matched treatments found an improved response rate (24% vs. 8.2% P = 0.024) when URS were treated on matched to unmatched treatments. This translated to a clinically meaningful but not statistically significant improvement in progression-free survival and overall survival and outcomes non-inferior to more common sarcoma subtypes in Phase 1 trials. Therefore, genomic sequencing is essential to unlocking all potential therapeutic avenues for advanced or metastatic URS.
Introduction
Sarcomas are a rare cancer of the tissue or bones of which an estimated 13,460 soft tissue and 3,610 bone sarcomas will be diagnosed in 2021 in the United States (1). Drug development in sarcoma has been challenging given the rarity and heterogeneity. The recent failure of olaratumab, a PDGFRα antagonist that received accelerated approval for advanced soft tissue sarcoma (STS) but was later withdrawn from the market due to disappointing Phase 3 findings in the ANNOUNCE trial, has been a setback as well (2).
The olaratumab/doxorubicin combination missed the trial's primary endpoint of overall survival (OS). The treatment could not confirm a clinical benefit compared with standard doxorubicin in patients with advanced or metastatic STS. However, this was a non–biomarker-driven study in all types of STS. The increasing availability of clinical next-generation sequencing (NGS) has altered the landscape of many common and rare tumors. Biomarker-driven drug approvals in rare indications like BRAF and MEK inhibition in anaplastic thyroid cancer or BRAF inhibitor in Erdheim–Chester disease in a dataset of fewer than 30 patients gives us the enthusiasm to design studies in rare indications (3, 4).
Because more than 150 different sarcoma subtypes exist, the Connective Tissue Oncology Society recently published consensus guidelines for recognizing ultrarare sarcomas (URS; ref. 5). Fifty-six STS and 21 bone sarcoma types were defined as URS based on an incidence ≤1 per 1,000,000. These ultrarare soft tissues and bone sarcoma histologies comprise up to 20% of the total sarcoma population and often lack approved histology-driven treatments. Currently, only three therapies are FDA-approved among URS subtypes. This includes tazemetostat for advanced epithelioid sarcoma with INI/SMARCb1 loss (6) and nab-sirolimus, an mTORC1 inhibitor that has demonstrated activity in metastatic malignant perivascular epithelioid cell tumors that often harbor mTOR-activating TSC1/2 mutations. In this single-arm, multi-center, Phase II trial, 34 patients at 9 centers were treated with nab-sirolimus. The overall response rate (ORR) was 39% (12 of 31), with a clinical benefit of 52% (n = 16/31; ref. 7). Histology-specific or biomarker-driven trials in rare sarcoma subtypes should be considered when testing investigational anticancer therapeutics (8). Most recently, crizotinib was approved for ALK-positive inflammatory myofibroblastic tumors (IMFT) following 7 adult patients with responses [7 complete response (CR) and 1 partial response (PR)] in a Phase 1b trial (9).
Enrollment in histology/subtype-driven trials requires a large and arduous effort given the rarity of these malignancies (10). Given the paucity of evidence-driven guidelines and limited treatment options, patients with URS are often referred for consideration of early-phase clinical trials (11, 12). Incorporation of NGS, and earlier matching of patients to biologically targeted therapies or immunomodulatory agents, has provided an expanded menu of novel treatment options to offer patients battling rare cancer types, including sarcoma (13–16).
In this Sarcoma-Matched Biomarker Analysis 102 (SAMBA 102) study, we evaluated the clinical outcomes of URS versus common sarcomas in early-phase clinical trials and among genomic alterations, which yielded clinical benefits in our cohort.
Materials and Methods
Data collection and eligibility
All patients treated in Phase 1 trials at the Department of Investigational Cancer Therapeutics at the University of Texas MD Anderson Cancer Center between January 2013 and June 2021, were included in analysis. We reviewed clinical and demographic data, including age, sex, cancer type, prior lines of treatment, number of trials treated, and type and target of agent used. In addition, we reviewed the CLIA-certified next NGS data and molecular testing performed on histologic samples during clinical care.
Endpoints and statistical methods
We reviewed each patient's chart for documentation of response via radiologic review. We reviewed the endpoints: specifically, ORR, disease control rate (DCR), progression-free survival (PFS), and OS. ORR was defined as CR plus PR. DCR was defined as ORR plus stable disease (SD). The best response was determined by individual trial protocol data using the RECIST (version 1.1) or immune-related RECIST (irRECIST). PFS and OS for each patient were calculated on the basis of their individual trial participation. Clinical benefit was defined as the sum of objective responses and SD that lasted for ≥6 months on treatment. OS was defined as the time from the first dose of the Phase 1 study drug until death. PFS was defined as the period between the first dose of Phase 1 drug and the date of documented radiologic or clinical progression or death. The Kaplan–Meier method was used to estimate OS and PFS, and a log-rank test was used to compare between groups. Descriptive variables were compared by the χ2 method. Statistical analysis was performed using SPSS version 25 and R software.
AACR GENIE database
For mutational targets associated with clinical benefit, we reviewed the incidence of the mutation in the histologic URS subtype in our Phase 1 cohort and the incidence in the AACR Project GENIE (Genomics Evidence Neoplasia Information Exchange). AACR Project GENIE is an international data-sharing consortium of clinical genomic data (17). Version 11.0, released in January 2022, contains 136,096 samples from 19 cancer institutions. cBioPortal was used to access and analyze data (18, 19).
The MD Anderson Institutional Review Board (IRB) independently reviewed and approved each clinical trial, and the MD Anderson IRB also approved this retrospective review with a waiver of informed consent.
Informed written consent was obtained from patients for each Phase 1 trial they were enrolled to. Each Phase 1 trial was conducted in accordance with the Declaration of Helsinki and separately approved by the IRB at the University of Texas MD Anderson Cancer Center.
Data availability
The data generated in this study are available upon reasonable request from the corresponding author.
Results
Overall characteristics
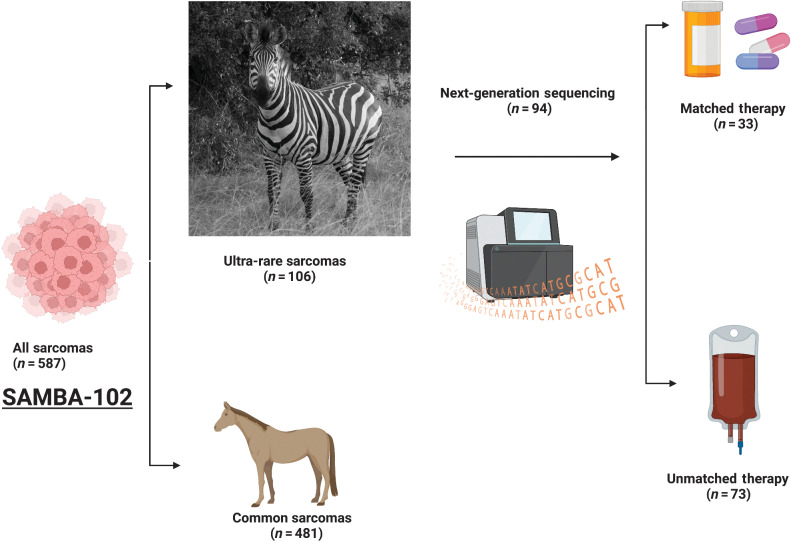
Between January 2013 and June 2021, 587 patients with sarcomas were enrolled in Phase 1 trials. Among them, 481 (81.9%) were common sarcoma histologies, and 106 (18.1%) were classified as URS (Fig. 1). Table 1 summarizes demographic data for URS patients. Supplementary Table S1 summarizes the histologic make-up of common sarcomas. The median age at the first trial was 44 years (SD, 16y; range, 19–82y), mean prior lines of therapy = 2 (range, 0–8), the mean number of Phase 1 trials = 1.5 (range, 1–6), and 47% were male. The most common URS were alveolar soft part sarcoma (ASPS; n = 12), dedifferentiated chondrosarcoma (n = 9), chordoma (n = 9), and sclerosing epithelioid fibrosarcoma (n = 9). Table 2 shows the distribution of URS histologies. Thirty-four different URS types were treated in trials, 7 bone URS and 24 soft tissue URS.
Figure 1.
Outline of SAMBA-102 study and patient allocation from the Phase 1 clinic at MD Anderson Cancer Center. Zebra photograph by Dr. Justin Moyers, Lake Mburu National Park in Uganda (Created with BioRender.com).
Table 1.
Demographic data.
Table 2.
Histology breakdown among URS in Phase 1 population.
| Multiple cases | n | Only one case |
|---|---|---|
| ASPS | 12 | Adamantinoma |
| Chordoma | 9 | BCOR-rearranged endometrial stromal sarcoma |
| De-differentiated chondrosarcoma | 9 | Clear cell chondrosarcoma |
| Sclerosing epithelioid fibrosarcoma | 9 | Giant cell tumor of soft tissue |
| Clear cell sarcoma | 8 | High-grade undifferentiated pleomorphic sarcoma of bone |
| PEComa | 6 | Histiocytic sarcoma |
| Desmoplastic small round cell tumor | 5 | Low-grade fibromyxoid sarcoma |
| Epithelioid sarcoma | 4 | Metastatic phyllodes tumor |
| Alveolar rhabdomyosarcoma | 4 | Parosteal osteosarcoma |
| Embryonal rhabdomyosarcoma | 3 | Periosteal osteosarcoma |
| Epithelioid hemangioendothelioma | 3 | Pleomorphic rhabdomyosarcma |
| Extraskeletal myxoid chondrosarcoma | 3 | Small round cell sarcoma NOS |
| Endometrial stromal sarcoma | 2 | Spindle cell rhabdomyosarcoma |
| Inflammatory myofibroblastic tumor | 2 | |
| Mesenchymal chondrosarcoma | 2 | |
| Malignant giant cell tumor of bone | 2 | |
| NTRK-rearranged sarcoma | 2 | |
| Ossifying fibromyxoid tumor | 2 | |
| Rhabdomyosarcoma | 2 | |
| Round cell sarcoma with EWSR1–non-ETS fusion | 2 | |
| Sarcoma with BCOR genetic alterations | 2 |
URS versus common sarcomas
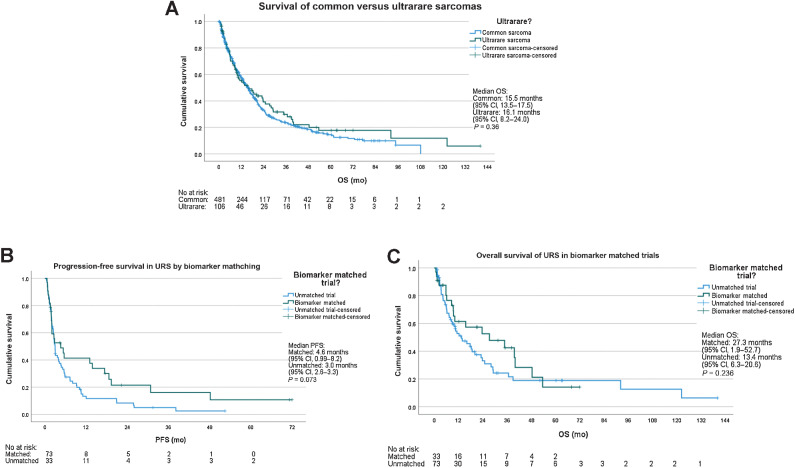
Compared with common sarcomas, median OS from time of Phase 1 enrollment was similar 16.1 months [95% confidence interval (CI), 13.6–17.5] versus 16.1 (95% CI, 8.2–24.0) in URS (P = 0.359) as in Fig. 2A. Median PFS were similar at 3.1 months (95% CI, 2.63–3.51) for common sarcomas and 2.97 months (95% CI, 1.87–4.06) for URS (P = 0.184). Objective response to Phase 1 treatment was significantly higher in URS at 13.2% (n = 14/106) compared with 6.9% (n = 33/481) in common sarcomas (P = 0.029). Tumor genomics with NGS or multi-gene panel was performed for 87% (n = 417/481) of common sarcomas and 89% (n = 94/106) of URS. Thirty-three of 106 (31%) URS were treated on matched trials when compared with 188/481 (39%) of common sarcomas (P = 0.126).
Figure 2.
A, Kaplan–Meier curve for overall survival (OS) between common and ultrarare sarcomas. B and C, Kaplan–Meier curve for OS (B) and progression-free survival (C) for matched versus unmatched trials in URS.
Landscape of genomic alterations in URS
Among the 94 patients who had molecular profiling completed, 69% (n = 73) had mutations present, 16% (n = 17) had no mutations identified, 4% (n = 4) had limited molecular analysis performed, and 2% (n = 2) failed NGS.
The most common alterations were EWSR1 fusions 13.2% (n = 14), TP53 11.3% (n = 12), CDKN2A loss 9.4% (n = 10), KIT 7.5% (n = 8), BRCA1/2 3.4% (n = 7), AR 6.6% (n = 7), CDKN2B loss 5.7% (n = 6), IDH1/2 5.7% (n = 6), mTOR/RAPTOR 6.0% (N = 5), PIK3CA 4% (N = 5), met 4% (n = 4), and NTRK 1/2/3 fusions 1.9% (n = 2). DNA damage repair (DDR) pathway genes (e.g., ATM, ATR, BRCA1/2, RAD51, PALB2) were altered in 13.2% (n = 14). Non-EWS fusions with approved drugs (e.g., ALK, NTRK1/2/3, ROS1) were present in 4% (n = 4) of cases. A complete list of alterations in the dataset is listed in Supplementary Table S2.
Molecularly matching in URS
NGS was performed on 94 of 106 (89%) patients. Thirty-three (31%) patients were treated on molecularly selected trials. Median OS for those treated on matched trials was 27.3 months (95% CI, 1.9–52.7) compared with 13.4 months (95% CI, 6.3–20.6) for those not treated on matched trials (P = 0.29). PFS for those treated on matched trials was 4.6 months (95% CI, 1.0–8.2) versus 3.0 months (95% CI, 2.6–3.3) for unmatched trials (P = 0.073). Figure 2B and C shows Kaplan–Meier curves.
Among 14 responses seen in URS, 13 were PR and one CR, among which targeted biomarker-matched therapies resulted in one CR and seven PR. Eight of 33 (24%) molecularly matched treatments resulted in an objective response, whereas 6 of 73 (8.2%) unmatched treatments resulted in objective responses (P = 0.024). Clinical benefit rate was 36.4% (12/33) in matched trials versus 26.0% (19/73) in unmatched trials (P = 0.279). Table 3 summarizes these results.
Table 3.
Objective response rate (CR + PR) and clinical benefit rate (CR + PR + ≥6 months of SD) between matched and unmatched patients with URS on Phase 1 clinical trials.
Objective responses in matched treatments were seen in clear cell sarcomas with c-met inhibitors (n = 2), NTRK inhibitors in NTRK-rearranged sarcoma (n = 2), EZH2/EED inhibitor in epithelioid sarcomas (n = 1), MDM2 inhibitors in chordomas (n = 1), combination VEGFR inhibitor + mTOR inhibitor in epithelioid hemangioendothelioma (n = 1), and ALK in IMFT (n = 1). In addition, clinical benefit was seen for matched treatments with Sonic Hedgehog (SHH) pathway inhibitor in sclerosing epithelioid fibrosarcoma (n = 1), MET/VEGFR2 inhibitor in clear cell sarcoma (n = 1), EGFR inhibitor in clear cell sarcoma (n = 1), and CDK4/6 inhibitor in low-grade fibromyxoid sarcoma (n = 1). Table 4 summarizes the response and target in the MD Anderson Phase 1 population and the AACR GENIE v11.0 populations. Supplementary Table S3 summarizes non-responders in matched trials.
Table 4.
Sarcomas experiencing clinical benefit from a molecularly matched agent with characteristics of their agent and matching.
| MDACC Phase 1 population | AACR GENIE v11.0 population | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor | Best response | Drug mechanism | Molecular selection | Number with mutation | Total samples | Percentage with mutation | Number with mutation | Total samples | Percentage with mutation |
| Chordoma | PR | MDM2 antagonist | ATM C430S | 1 | 10 | 10% | 2 | 82 | 2.4% |
| Clear cell sarcoma | PR | CMET inhibitor | EWSR1 fusion | 6 | 10 | 60% | 26 | 27 | 96% |
| Clear cell sarcoma | PR | CMET inhibitor | cMET amplification | 2 | 10 | 20% | 0 | 32 | 0% |
| Epithelioid sarcoma | PR | EZH2/EED inhibitor | INI/SMARCB1 loss | 1 | 4 | 25% | 20 | 43 | 47% |
| Epithelioid hemangioendothelioma | PR | VEGF inhibitor + mTOR | TSC1 | 1 | 2 | 50% | 2 | 54 | 4% |
| Inflammatory myofibroblastic tumor | PR | ALK inhibitor | ALK fusion | 2 | 2 | 100% | 17 | 41 | 42% |
| NTRK-rearranged sarcoma | PR, CR | NTRK inhibitor | ETV6-NTRK3,NTRK3-EML4 | 2 | 587 | 0.3% | 27 | 4,036 | 0.66% |
| Clear cell sarcoma | SD > 6 m | MET/VEGFR2 inhibitor | EWSR fusion | 6 | 10 | 60% | 29 | 45 | 64% |
| Clear cell sarcoma | SD > 6 m | EGFR inhbitor | ERBB4 M1017T | 1 | 10 | 10% | 1 | 45 | 2% |
| Low-grade fibromyxoid sarcoma | SD > 6 m | CDK4/6 inhibitor | CDKN2A amplification | 1 | 1 | 100% | 1 | 16 | 6% |
| Sclerosing epithelioid fibrosarcoma | SD > 6 m | SHH inhibitor | PTCH1 | 1 | 9 | 11% | 2 | 19 | 11% |
Immunotherapy
Forty (37.7%) patients received investigational immunotherapy regimens. Four of 40 (10%) experienced objective responses, all PR. Fourteen of 40 (35%) experienced a clinical benefit. Responses are summarized in Supplementary Table S4. A TLR7/8 agonist resulted in a PR in one embryonal rhabdomyosarcoma, as did checkpoint inhibitors in 3 ASPS. Clinical benefit was seen with checkpoint inhibitors alone and in combination in chordoma (n = 2), clear cell sarcoma (n = 1), fibromyxoid sarcoma low grade (n = 1), ossifying fibromyxoid tumor (n = 1), epithelioid hemangioendothelioma (n = 1), sarcoma with BCOR genetic alterations (n = 1), periosteal osteosarcoma (n = 1), and ASPS (n = 2); however, it should be noted that several of these tumor types are traditionally slow growing sarcoma subtypes. Eight of 12 (67%) ASPS in our cohort received checkpoint inhibitors alone or in combination. Five of 8 (63%) ASPS experienced a clinical benefit with 3 PR (38%) when given checkpoint inhibitors.
Discussion
We have reported the largest dataset of patients with URS to specifically characterize the clinical and molecular features and outcomes of patients with URS who are referred to early-phase clinical trials. We found potential benefits to enrolling in an early-phase clinical trial with an ORR of 24.2%, PFS of 4.6 months, and median OS of 27.3 months in this heavily pretreated patient population with limited options before referral for consideration of Phase 1 trials. Patients clinically benefited from a matched therapy, whether in a dose-escalation cohort or a dose-expansion cohort.
Sarcoma treatments are often developed on the basis of clinical trials for other cancers and often involve diverse sarcoma subtypes (20). However, drugs are making their way into later-phase trials for sarcoma, increasingly using targeted mechanisms of action or with immunotherapy (21). Many genomic alterations have been identified in sarcomas, and the number of clinical trials targeting them has expanded significantly (22, 23).
One of the patients in our cohort with epithelioid sarcoma was treated with tazemetostat, the only approved drug for URS. The ezh2/eed inhibitor, approved in advanced epithelioid sarcoma with INI/SMARCb1 loss following an open-label Phase 2 basket study, achieved a response rate of 15% (n = 15/62; ref. 6).
MDM2 is one of the most frequently altered genes in sarcomas and is an attractive target for inhibitors (24). However, MDM2 antagonists have been developed to interact through a different mechanism causing interaction with p53 to activate DDR mechanisms of which sarcoma subtypes have been known to harbor alterations in DDR (25, 26). A chordoma with a DDR alteration in our cohort experienced a PR following treatment with an MDM2 antagonist.
Clear cell sarcomas are characterized by the chimeric transcription factor, EWS–ATF1 translocation. The EWS–ATF1 activates melanocyte-inducing transcription factor, thereby promoting c-MET transcription (27). The EORTC trial 90101 showed a benefit for crizotinib in MET-amplified clear cell sarcoma, though a low ORR of 3.8% (n = 1/28) and DCR of 69.2% (28). STS have also been included in basket trials of c-Met inhibitors with varying responses (29–31).
One patient with an IMFT responded to ALK-targeted therapy, specifically crizotinib in combination with pazopanib (32). Approximately half (47%; 9/22) of myofibroblastic tumors have been reported to harbor ALK gene rearrangements, which we also observed in 50% (n = 15/30) of the AACR GENIE population (33, 34). Indeed, on the basis of the frequency of this gene rearrangement and report of successful use of crizotinib, a non-randomized Phase 2 trial was undertaken in IMFT with and without ALK gene rearrangements for whom no surgical option existed (35). Crizotinib showed an ORR of 50% (n = 6/12) in ALK-rearranged and 14% (n = 1/7) in ALK non-rearranged tumors (36). On the basis of these data, crizotinib is now recently FDA approved for IMFT.
Although NTRK fusions are rare in the broader soft-tissue sarcoma patient population (approximately 0.66% of the AACR GENIE population), those harboring NTRK fusions can exhibit extraordinary responses to the two approved NTRK inhibitors, larotrectinib and entrectinib. Among the 11 patients with STS who received larotrectinib, 10 were shown to have objective responses (37). Likewise, objective responses to entrectinib have also occurred in STS (38). Similarly, RET fusions have also emerged as tissue agnostic targets with sensitivity to selective RET inhibitors with reports of sarcoma-harboring RET fusions benefiting from matched therapies. Selpercatinib recently received FDA approval for all RET-positive cancers, including sarcomas (39, 40).
The clinical benefit of checkpoint inhibition in patients with ASPS has already been noted in the MD Anderson Phase 1 population and is accepted as a potential line of treatment for advanced ASPS (41–43). A single-center, single-arm, Phase 2 study by Wilky and colleagues (44) enrolled 12 patients with ASPS in which they saw a 72.7% 3-month PFS and response rate of 54.5% (n = 6/11) with pembrolizumab with axitinib. Preliminary data exhibiting single-agent activity of atezolizumab have additionally been presented from a Phase 1/2 trial with 28% (n = 5/7) experiencing a confirmed PR (45). Furthermore, dual checkpoint inhibition with durvalumab plus tremilimumab in a single-center Phase 2 study showed ORR of 40% (n = 4/10) for ASPS cohort by irRECIST (46).
In addition, we found that patients with URS responded similarly or better than patients with more common sarcoma diagnoses. This may be in part due to URS subtypes defined by an actionable target (e.g., NTRK-rearranged soft tissue and bone sarcomas). As more agents come to fruition, more URS sarcomas defined by a molecular target, may join the list of current URS diagnoses.
Limitations
Our study is a retrospective single-institution study, so broad characterizations cannot be made. In addition, given the limited numbers of patients with URS in our cohort, analysis of specific sarcoma subtypes survival and response characteristics subtypes cannot be performed. In addition, due to the heterogeneous growth rate patterns in sarcomas, clinical benefit in sarcomas with slow growth rate may not represent clinically beneficial treatments.
Future directions
Drug development for rare sarcoma subtypes will likely continue to rely on histology/subtype-driven treatments and histology-agnostic methods appropriate for sarcomas as a generic class of mesenchymal tumors. Thus far, the sarcoma subtype-specific FDA approvals (e.g., tazemetostat, nab-sirolimus, and crizontinib) relied upon multicenter early-phase trials conducted at large tertiary referral cancer centers with expertise in the care and treatment of patients with sarcoma (6, 7). However, tumor agnostic trials testing checkpoint inhibitors paved the way for regulatory approval in tumors with high tumor mutational burdens and NTRK-rearrangement (37, 47). Basket trials allow investigators to evaluate novel agents in diverse cancer types and can allow for testing of low-incidence alterations, but must ensure that the targeted alteration is an oncogenic driver across multiple cancers (48, 49). Given the success of each strategy, both approaches are likely to be used, taking into account the prevalence of the targeted alteration and subtype prevalence.
When treated on Phase 1 trials, patients with URS experienced similar survival and response rates compared with those diagnosed with more common sarcoma subtypes. In molecularly matched trials, patients with URS experienced a significantly improved ORR, with a trend toward improved OS and PFS. Given the paucity of approved options or data to guide treatment choice, treating in an “n of 1” fashion with molecularly matched agents or enrollment in Phase 1 trials remains an important treatment option for these newly classified “URS.” Clinical NGS, and early referral for consideration of early-phase clinical trials, may offer an additional line of therapy for patients with URS that urgently need better treatment options.
Supplementary Material
Supplement Table 1: Histologic Make-up of common sarcomas Supplement Table 2: List of common alterations among URS population by frequency Supplement Table 3: Sarcomas experiencing without clinical benefit (e.g. SD for <6 months or PD as best response) Supplement Table 4: Immunotherapy Responses and Clinical Benefit
Acknowledgments
V. Subbiah is an Andrew Sabin Family Foundation Fellow at The University of Texas MD Anderson Cancer Center and acknowledges support of The Jacquelyn A. Brady Fund. V. Subbiah is supported by NIH grant R01CA242845. MD Anderson Cancer Center Department of Investigational Cancer Therapeutics is supported by the Cancer Prevention and Research Institute of Texas (RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (1U01 CA180964), NCATS Grant UL1 TR000371 (Center for Clinical and Translational Sciences), and the MD Anderson Cancer Center Support Grant (P30 CA016672). The authors would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the study authors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC Section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
R.C. Pestana reports personal fees from Servier, Bayer, Pfizer, Merck, BMS, Amgen, Manual de Oncologia Brasil (MOC), Oncologia Brasil, and Associao Brasileira de Linfoma e Leucemia (ABRALE) outside the submitted work. D.S. Hong reports grants from AbbVie, Adaptimmune, Aldai-Nortye, Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Deciphera, Endeavor, Erasca, F. Hoffmann-La Roche, Fate Therapeutics, Genentech, Genmab, Immunogenesis, Infinity, Merck, Mirati, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Pyramid Bio, Revolution Medicine, SeaGen, ST-Cube, Takeda, TCR2, Turning Point Therapeutics, and VM Oncology; travel, accommodations, and expenses from Bayer, Genmab, AACR, ASCO, SITC, and Telperian; consulting, speaker, or advisory role at Adaptimmune, Alpha Insights, Acuta, Alkermes, Amgen, Aumbiosciences, Axiom, Baxter, Bayer, Boxer Capital, BridgeBio, COR2ed, COG, Cowen, ECOR1, Erasca, F. Hoffmann-La Roche, Gennao Bio, Genentech, Gilead, GLG, Group H, Guidepoint, HCW Precision, Immunogenesis, Janssen, Liberium, MedaCorp, Medscape, Numab, Oncologia Brasil, Orbi Captial, Pfizer, Pharma Intelligence, POET Congress, Prime Oncology, RAIN, SeaGen, STCube, Takeda, Tavistock, Trieza Therapeutics, Turning Point, Vertical, WebMD, YingLing Pharma, and Ziopharm; and other ownership interests in Molecular Match (advisor), OncoResponse (founder, advisor), and Telperian (founder, advisor). A. Naing reports grants from NCI, EMD Serono, MedImmune, Healios Onc. Nutrition, Atterocor/Millendo, Amplimmune, ARMO BioSciences, Karyopharm Therapeutics, Incyte, Novartis, Regeneron, Merck, Bristol Myers Squibb, Pfizer, CytomX Therapeutics, Neon Therapeutics, Calithera Biosciences, TopAlliance Biosciences, Eli Lilly, Kymab, PsiOxus, Arcus Biosciences, NeoImmuneTech, Immune-Onc Therapeutics, Surface Oncology, Monopteros Therapeutics, BioNTech SE, Seven & Eight Biopharma, and SOTIO Biotech AG; personal fees from Deka Biosciences, NGM Bio, PsiOxus Therapeutics, Immune-Onc Therapeutics, STCube Pharmaceuticals, OncoSec KEYNOTE-695, Pharming, Genome and Company, Horizon Therapeutics, CytomX Therapeutics, Kymab, Nouscom, Merck Sharp & Dohme Corp, OncoNano, Servier, AKH Inc., The Lynx Group, Society for Immunotherapy of Cancer (SITC), Korean Society of Medical Oncology (KSMO), Scripps Cancer Care Symposium, ASCO Direct Oncology Highlights, European Society for Medical Oncology (ESMO), European Society for Immunodeficiencies (ESID), CME Outfitters, and Lynx Health; and non-financial support from ARMO BioSciences and NeoImmuneTech outside the submitted work. In addition, A. Naing reports their spouse received research funding from The Texas Medical Center Digestive Diseases Center, Jeffery Modell Foundation, Immune Deficiency Foundation, Baxalta US Inc., Chao Physician-Scientist Foundation; consultant/advisory board for Takeda, Pharming Healthcare Inc., and Horizon Therapeutics, Inc.; and ad hoc consultancy speaker for Alfaisal University. S. Piha-Paul reports other support from AbbVie, ABM Therapeutics, Acepodia, Alkermes, Aminex Therapeutics, Amphivena Therapeutics, BioMarin Pharmaceutical, Boehringer Ingelheim, Bristol Myers Squibb, Cerulean Pharma, Chugai Pharmaceutical, Curis, Cyclacel Pharmaceuticals, Daiichi Sankyo, Eli Lilly, ENB Therapeutics, Five Prime Therapeutics, F-Star Beta Limited, F-Star Therapeutics, Gene Quantum, Genmab A/S, Gilead Sciences, GlaxoSmithKline, Helix BioPharma, Hengrui Pharmaceuticals, HiberCell, Immunomedics, Incyte, Jacobio Pharmaceuticals, Lytix Biopharma AS, Medimmune, Medivation, Merck Sharp & Dohme, Novartis Pharmaceuticals, Pieris Pharmaceuticals, Pfizer, Phanes Therapeutics, Principia Biopharma, Puma Biotechnology, Purinomia Biotech, Rapt Therapeutics, Replimmune, Seattle Genetics, Silverback Therapeutics, Synlogic Therapeutics, Taiho Oncology, Tesaro, TransThera Bio, and ZielBio, as well as grants from NCI/NIH (P30CA016672) Core Grant (CCSG Shared Resources) outside the submitted work; S. Piha-Paul has also worked as a consultant for CRC Oncology. T.A. Yap reports grants from Acrivon, Artios, AstraZeneca, Bayer, Beigene, BioNTech, Blueprint, BMS, Clovis, Constellation, Cyteir, Forbius, GlaxoSmithKline, Genentech, Haihe, ImmuneSensor, Ionis, Ipsen, Jounce, Karyopharm, KSQ, Kyowa, Mirati, Novartis, Ribon Therapeutics, Regeneron, Rubius, Sanofi, Scholar Rock, Seattle Genetics, Tesaro, Vivace, Zenith, and Eli Lilly; grants and personal fees from EMD Serono, F-Star, Merck, Pfizer, and Repare; personal fees from AbbVie, AstraZeneca, Acrivon, Adagene, Almac, Aduro, Amphista, Artios, Athena, Atrin, Avoro, Axiom, Baptist Health Systems, Bayer, Beigene, Boxer, Bristol Myers Squibb, C4 Therapeutics, Calithera, Cancer Research UK, Clovis, Cybrexa, Diffusion, Genmab, Glenmark, GLG, Guidepoint, Idience, Ignyta, I-Mab, ImmuneSensor, Institut Gustave Roussy, Intellisphere, Janssen, Kyn, MEI pharma, Mereo, Natera, Nexys, Novocure, OHSU, OncoSec, Ono Pharma, Pegascy, PER, Piper-Sandler, Prolynx, resTORbio, Roche, Schrodinger, Theragonostics, Varian, Versant, Vibliome, Xinthera, Zai Labs, and ZielBio; and other support from Seagen and University of Texas MD Anderson Cancer Center outside the submitted work. D. Karp reports grants from NIH; grants and other support from Myriad Drug Companies; personal fees from Black Beret Life Sciences and Affigen; and grants, personal fees, and other support from Phosplatin outside the submitted work. J. Rodon reports personal fees from Peptomyc, Kelun Pharmaceuticals/Klus Pharma, Ellipses Pharma, Molecular Partners, and IONCTURA; grants from Blueprint Medicines, Black Diamond Therapeutics, Merck Sharp & Dohme, Hummingbird, Yingli, Vall d'Hebron Institute of Oncology/Cancer Core Europe, Novartis, Spectrum Pharmaceuticals, Symphogen, BioAtla, Pfizer, GenMab, CytomX, Kelun-Biotech, Takeda-Millenium, GlaxoSmithKline, Taiho, Roche Pharmaceuticals, Bicycle Therapeutics, Merus, Curis, Bayer, Aadi Bioscience, Nuvation, ForeBio, BioMed Valley Discoveries, Loxo Oncology, Hutchinson MediPharma, Cellestia, Deciphera, Ideaya, Amgen, Tango Therapeutics, Mirati, and Linnaeus Therapeutics; and other support from European Society for Medical Oncology, Vall d'Hebron Institute of Oncology/Minestero De Empleo Y Seguridad Social, Chinese University of Hong Kong, Boxer Capital, and Tang Advisors, outside the submitted work. A. Livingston reports grants from REPARE Therapeutics, as well as personal fees from Foghorn Therapeutics outside the submitted work. S. Patel reports personal fees from Deciphera, Daiichi Sankyo, Rain Therapeutics, Adaptimmune, and GSK, as well as grants from Rain Therapeutics and Blueprint Medicines during the conduct of the study. R. Ratan reports personal fees and non-financial support from Springworks Therapeutics; personal fees from Bayer and Epizyme; and non-financial support from Ayala outside the submitted work. N. Somaiah reports personal fees from Epizyme, Boehringer Ingelheim, and Aadi Biosciences, as well as grants from AstraZeneca and Innovent outside the submitted work. A. Conley reports grants from Chordoma Foundation, Eli Lilly, Roche, NCI, Nant Pharma, and EpicentRx; grants and personal fees from Inhibrx; personal fees from Aadi Bioscience, Deciphera Pharmaceuticals, Applied Clinical Intelligence, OncLive, and Guidepoint Global; and other support from Medscape outside the submitted work. F. Meric-Bernstam reports personal fees from AbbVie, Aduro BioTech, Alkermes, AstraZeneca, Daiichi Sankyo, DebioPharm, Ecor1 Capital, eFFECTOR Therapeutics, F. Hoffman-La Roche, GT Apeiron, Genentech, Harbinger Health, IBM Watson, Infinity Pharmaceuticals, The Jackson Laboratory, Kolon Life Science, Lengo Therapeutics, Menarini Group, OrigiMed, PACT Pharma, Parexel International, Pfizer, Protai Bio, Samsung Bioepis, Seattle Genetics, Tallac Therapeutics, Tyra Biosciences, Xencor, Zymeworks, Black Diamond, Biovica, Eisai, FogPharma, Immunomedics, Inflection Biosciences, Karyopharm Therapeutics, Loxo Oncology, Mersana Therapeutics, OnCusp Therapeutics, Puma Biotechnology, Sanofi, Silverback Therapeutics, Spectrum Pharmaceuticals, Chugai Biopharmaceuticals, and Zentalis, as well as grants from Aileron Therapeutics, AstraZeneca, Bayer Healthcare Pharmaceutical, Calithera Biosciences, Curis, CytomX Therapeutics, Daiichi Sankyo, Debiopharm International, eFFECTOR Therapeutics, Genentech, Guardant Health, Klus Pharma, Takeda Pharmaceutical, Novartis, Puma Biotechnology, and Taiho Pharmaceutical outside the submitted work. V. Subbiah reports research funding from Novartis to conduct clinical trials; grant support for clinical trials from AbbVie, Agensys, Alfasigma, Altum, Amgen, Bayer, BERG Health, Blueprint Medicines, Boston Biomedical, Boston Pharmaceuticals, Celgene Corporation, D3 Bio, Dragonfly Therapeutics, Exelixis, Fujifilm, GlaxoSmithKline, Idera Pharmaceuticals, Incyte Corporation, Inhibrx, Loxo Oncology/Eli Lilly, MedImmune, MultiVir, NanoCarrier, National Comprehensive Cancer Network, NCI-CTEP, Novartis, PharmaMar, Pfizer, Relay Therapeutics, Roche/Genentech, Takeda, Turning Point Therapeutics, UT MD Anderson Cancer Center, and Vegenics Pty; travel support from ASCO, ESMO, Helsinn Healthcare, Incyte Corporation, Novartis, and PharmaMar; consultancy or advisory board participation for Helsinn Healthcare, Incyte Corporation, Loxo Oncology/Eli Lilly, MedImmune, Novartis, QED Therapeutics, Relay Therapeutics, Daiichi Sankyo, and R-Pharm US; and other relationship with Medscape. No disclosures were reported by the other authors.
Authors' Contributions
J.T. Moyers: Conceptualization, data curation, formal analysis, writing–original draft, writing–review and editing. R.C. Pestana: Conceptualization, data curation, formal analysis, writing–review and editing. J. Roszik: Writing–review and editing. D.S. Hong: Writing–review and editing. A. Naing: Writing–review and editing. S. Fu: Investigation, writing–review and editing. S. Piha-Paul: Writing–review and editing. T.A. Yap: Writing–review and editing. D. Karp: Writing–review and editing. J. Rodon: Investigation, writing–review and editing. A. Livingston: Writing–review and editing. M.A. Zarzour: Investigation, writing–review and editing. V. Ravi: Writing–review and editing. S. Patel: Writing–review and editing. R.S. Benjamin: Writing–review and editing. J. Ludwig: Investigation, writing–review and editing. C. Herzog: Investigation, writing–review and editing. R. Ratan: Investigation, writing–review and editing. N. Somaiah: Investigation, writing–review and editing. A. Conley: Writing–review and editing. R. Gorlick: Investigation, writing–review and editing. F. Meric-Bernstam: Investigation, writing–review and editing. V. Subbiah: Conceptualization, resources, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Tap WD, Wagner AJ, Schöffski P, Martin-Broto J, Krarup-Hansen A, Ganjoo KN, et al. Effect of doxorubicin plus olaratumab vs. doxorubicin plus placebo on survival in patients with advanced soft tissue sarcomas: the ANNOUNCE randomized clinical trial. JAMA 2020;323:1266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J Clin Oncol 2018;36:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diamond EL, Subbiah V, Lockhart AC, Blay JY, Puzanov I, Chau I, et al. Vemurafenib for BRAF V600-mutant erdheim-chester disease and langerhans cell histiocytosis: analysis of data from the histology-independent, phase 2, open-label VE-BASKET study. JAMA Oncol 2018;4:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stacchiotti S, Frezza AM, Blay JY, Baldini EH, Bonvalot S, Bovée J, et al. Ultra-rare sarcomas: a consensus paper from the Connective Tissue Oncology Society community of experts on the incidence threshold and the list of entities. Cancer 2021;127:2934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gounder M, Schöffski P, Jones RL, Agulnik M, Cote GM, Villalobos VM, et al. Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet Oncol 2020;21:1423–32. [DOI] [PubMed] [Google Scholar]
- 7. Wagner AJ, Ravi V, Riedel RF, Ganjoo K, Tine BAV, Chugh R, et al. nab-Sirolimus for patients with malignant perivascular epithelioid cell tumors. J Clin Oncol 2021;39:3660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pestana RC, Groisberg R, Roszik J, Subbiah V. Precision oncology in sarcomas: divide and conquer. JCO Precis Oncol 2019;3:PO.18.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gambacorti-Passerini C, Orlov S, Zhang L, Braiteh F, Huang H, Esaki T, et al. Long-term effects of crizotinib in ALK-positive tumors (excluding NSCLC): a phase 1b open-label study. Am J Hematol 2018;93:607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Villalobos VM, Hoffner B, Elias AD. We can study ultrarare tumors effectively in this day and age, it just takes a cooperative approach: the role of dasatinib in assorted indolent sarcomas. Cancer 2017;123:20–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subbiah V, Kurzrock R. Phase 1 clinical trials for sarcomas: the cutting edge. Curr Opin Oncol 2011;23:352–60. [DOI] [PubMed] [Google Scholar]
- 12. Kato S, Kurasaki K, Ikeda S, Kurzrock R. Rare tumor clinic: the University of California San Diego Moores Cancer Center experience with a precision therapy approach. Oncologist 2018;23:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groisberg R, Hong DS, Roszik J, Janku F, Tsimberidou AM, Javle M, et al. Clinical next-generation sequencing for precision oncology in rare cancers. Mol Cancer Ther 2018;17:1595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groisberg R, Hong DS, Holla V, Janku F, Piha-Paul S, Ravi V, et al. Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas. Oncotarget 2017;8:39254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subbiah V. Prospects and pitfalls of personalizing therapies for sarcomas: from children, adolescents, and young adults to the elderly. Curr Oncol Rep 2014;16:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dembla V, Groisberg R, Hess K, Fu S, Wheler J, Hong DS, et al. Outcomes of patients with sarcoma enrolled in clinical trials of pazopanib combined with histone deacetylase, mTOR, Her2, or MEK inhibitors. Sci Rep 2017;7:15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Consortium TAPG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee DY, Staddon AP, Shabason JE, Sebro R. Phase I and phase II clinical trials in sarcoma: implications for drug discovery and development. Cancer Med 2019;8:585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Albayrak HC, Gürler F, Sütçüoğlu O, Akyürek N, Özet A. How long does crizotinib work? a rare case of recurrent inflammatory myofibroblastic tumor. Anticancer Drugs 2022;33:109–11. [DOI] [PubMed] [Google Scholar]
- 22. Grünewald TG, Alonso M, Avnet S, Banito A, Burdach S, Cidre-Aranaz F, et al. Sarcoma treatment in the era of molecular medicine. EMBO Mol Med 2020;12:e11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martínez-Trufero J, Jurado JC, Hernández-León CN, Correa R, Asencio JM, Bernabeu D, et al. Uncommon and peculiar soft tissue sarcomas: multidisciplinary review and practical recommendations. Spanish Group for Sarcoma Research (GEIS-GROUP). Part II. Cancer Treat Rev 2021;99:102260. [DOI] [PubMed] [Google Scholar]
- 24. Lucchesi C, Khalifa E, Laizet Y, Soubeyran I, Mathoulin-Pelissier S, Chomienne C, et al. Targetable alterations in adult patients with soft-tissue sarcomas: insights for personalized therapy. JAMA Oncol 2018;4:1398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valentine JM, Kumar S, Moumen A. A p53-independent role for the MDM2 antagonist Nutlin-3 in DNA damage response initiation. BMC Cancer 2011;11:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep 2018;23:239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis IJ, McFadden AW, Zhang Y, Coxon A, Burgess TL, Wagner AJ, et al. Identification of the receptor tyrosine kinase c-Met and its ligand, hepatocyte growth factor, as therapeutic targets in clear cell sarcoma. Cancer Res 2010;70:639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schöffski P, Wozniak A, Stacchiotti S, Rutkowski P, Blay JY, Lindner LH, et al. Activity and safety of crizotinib in patients with advanced clear-cell sarcoma with MET alterations: European Organization for Research and Treatment of Cancer phase II trial 90101 “CREATE.” Annal Oncol 2017;28:3000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong DS, LoRusso P, Hamid O, Janku F, Kittaneh M, Catenacci DVT, et al. Phase I Study of AMG 337, a highly selective small-molecule MET inhibitor, in patients with advanced solid tumors. Clin Cancer Res 2019;25:2403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagner AJ, Goldberg JM, Dubois SG, Choy E, Rosen L, Pappo A, et al. Tivantinib (ARQ 197), a selective inhibitor of MET, in patients with microphthalmia transcription factor-associated tumors: results of a multicenter phase 2 trial. Cancer 2012;118:5894–902. [DOI] [PubMed] [Google Scholar]
- 31. Subbiah V, Holmes O, Gowen K, Spritz D, Amini B, Wang WL, et al. Activity of c-Met/ALK inhibitor crizotinib and multi-kinase VEGF inhibitor pazopanib in metastatic gastrointestinal neuroectodermal tumor harboring EWSR1–CREB1 fusion. Oncology 2016;91:348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subbiah V, McMahon C, Patel S, Zinner R, Silva EG, Elvin JA, et al. STUMP un“stumped”: antitumor response to anaplastic lymphoma kinase (ALK) inhibitor based targeted therapy in uterine inflammatory myofibroblastic tumor with myxoid features harboring DCTN1–ALK fusion. J Hematol Oncol 2015;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov 2014;4:889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coffin CM, Patel A, Perkins S, Elenitoba-Johnson KSJ, Perlman E, Griffin CA. ALK1 and p80 expression and chromosomal rearrangements involving 2p23 in inflammatory myofibroblastic tumor. Mod Pathol 2001;14:569–76. [DOI] [PubMed] [Google Scholar]
- 35. Butrynski JE, D'Adamo DR, Hornick JL, Dal Cin P, Antonescu CR, Jhanwar SC, et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 2010;363:1727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schöffski P, Sufliarsky J, Gelderblom H, Blay JY, Strauss SJ, Stacchiotti S, et al. Crizotinib in patients with advanced, inoperable inflammatory myofibroblastic tumours with and without anaplastic lymphoma kinase gene alterations (European Organisation for Research and Treatment of Cancer 90101 CREATE): a multicentre, single-drug, prospective, non-randomised phase 2 trial. Lancet Respir Med 2018;6:431–41. [DOI] [PubMed] [Google Scholar]
- 37. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Demetri GD, De Braud F, Drilon A, Siena S, Patel MR, Cho BC, et al. Updated integrated analysis of the efficacy and safety of entrectinib in patients with NTRK fusion-positive solid tumors. Clin Cancer Res 2022;28:1302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subbiah V, Wolf J, Konda B, Kang H, Spira A, Weiss J, et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol 2022;3:1261–73. [DOI] [PubMed] [Google Scholar]
- 40. Subbiah V, Cassier PA, Siena S, Garralda E, Paz-Ares L, Garrido P, et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the phase 1/2 ARROW trial. Nat Med 2022;28:1640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Groisberg R, Hong DS, Behrang A, Hess K, Janku F, Piha-Paul S, et al. Characteristics and outcomes of patients with advanced sarcoma enrolled in early-phase immunotherapy trials. J Immunother Cancer 2017;5:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Groisberg R, Roszik J, Conley AP, Lazar AJ, Portal DE, Hong DS, et al. Genomics, morphoproteomics, and treatment patterns of patients with alveolar soft part sarcoma and response to multiple experimental therapies. Mol Cancer Ther 2020;19:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paoluzzi L, Maki RG. Diagnosis, prognosis, and treatment of alveolar soft-part sarcoma: a review. JAMA Oncol 2019;5:254–60. [DOI] [PubMed] [Google Scholar]
- 44. Wilky BA, Trucco MM, Subhawong TK, Florou V, Park W, Kwon D, et al. Axitinib plus pembrolizumab in patients with advanced sarcomas including alveolar soft-part sarcoma: a single-centre, single-arm, phase 2 trial. Lancet Oncol 2019;20:837–48. [DOI] [PubMed] [Google Scholar]
- 45. O'Sullivan Coyne G, Sharon E, Moore N, et al. Phase 2 study of atezolizumab in patients with alveolar soft part sarcoma. Presented at: CTOS 2018 Annual Meeting; Rome, Italy: November 14-17. Paper 021. [Google Scholar]
- 46. Somaiah N, Conley AP, Parra ER, Lin H, Amini B, Soto LS, et al. Durvalumab plus tremelimumab in advanced or metastatic soft tissue and bone sarcomas: a single-centre phase 2 trial. Lancet Oncol 2022;23:1156–66. [DOI] [PubMed] [Google Scholar]
- 47. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 2020;38:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pestana RC, Sen S, Hobbs BP, Hong DS. Histology-agnostic drug development—considering issues beyond the tissue. Nat Rev Clin Oncol 2020;17:555–68. [DOI] [PubMed] [Google Scholar]
- 49. Park JJH, Hsu G, Siden EG, Thorlund K, Mills EJ. An overview of precision oncology basket and umbrella trials for clinicians. CA Cancer J Clin 2020;70:125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table 1: Histologic Make-up of common sarcomas Supplement Table 2: List of common alterations among URS population by frequency Supplement Table 3: Sarcomas experiencing without clinical benefit (e.g. SD for <6 months or PD as best response) Supplement Table 4: Immunotherapy Responses and Clinical Benefit
Data Availability Statement
The data generated in this study are available upon reasonable request from the corresponding author.