Abstract
Radiations are especially important for generating species biodiversity in mountainous ecosystems. The contribution of hybridization to such radiations has rarely been examined. Here, we use extensive genomic data to test whether hybridization was involved in evolutionary radiation within Rhododendron subgenus Hymenanthes, whose members show strong geographic isolation in the mountains of southwest China. We sequenced genomes for 143 species of this subgenus and 93 species of four other subgenera, and found that Hymenanthes was monophyletic and radiated during the late Oligocene to middle Miocene. Widespread hybridization events were inferred within and between the identified clades and subclades. This suggests that hybridization occurred both early and late during diversification of subgenus Hymenanthes, although the extent to which hybridization, speciation through mixing-isolation-mixing or hybrid speciation, accelerated the diversification needs further exploration. Cycles of isolation and contact in such and other montane ecosystems may have together promoted species radiation through hybridization between diverging populations and species. Similar radiation processes may apply to other montane floras in this region and elsewhere.
Keywords: evolutionary radiation, hybridization, rhododendron, subgenus Hymenanthes, montane flora
INTRODUCTION
A central question in evolution concerns the mechanisms underlying rapid radiation, i.e. the generation of large numbers of morphologically diverse species from a single lineage over a short evolutionary time span, and often within a relatively narrow geographical range [1,2]. Such radiations are especially common in mountainous regions, oceanic islands and rift lakes [3–8]. Genomic admixtures caused by hybridization have repeatedly been found to drive radiations in the latter two ecosystems [9–11]. However, although hybridizaion has been reported in many continental plant groups [12–14], this possibility has not been fully explored based on genomic data in mountainous systems, and even less so in alpine plant radiations. Ecological and geographical isolation are assumed to drive species radiations in mountainous regions [15,16] because tectonic uplift and accompanying environmental changes generate high mountains alternating with deep valleys that provide more available niches [17]. Such mountains can form so-called ‘sky islands’ in which climatic oscillations can cause cycles of connection and isolation between populations or species occurring there, which may result in hybridization [18,19]. Moreover, recent modelling of speciation shows that such mixing-isolation-mixing (MIM) cycles could greatly accelerate species diversification [20]. Hence, the role of hybridization as a contributor to other major plant radiations requires investigation, especially in mountainous ecosystems, where there are strong effects from ecological and geographic isolation, plus frequent hybridization events [21].
Testing the role of hybridization in plant radiations remains challenging because genomic admixture can result from any of four evolutionary outcomes after hybridization, depending on timescales. First, hybrid zones may form containing many hybrid derivatives, but without necessarily altering the parent species or leading to speciation [22]. Second, backcrossing might lead to germplasm transfer through introgression, altering the recipient species either locally or, ultimately, throughout its range [22], but without causing speciation. Third, hybridization may occur between diverging populations through natural selection and geographic isolation and such divergence with gene flow can promote speciation [20]. Finally, hybrid species may arise between two already well-differentiated species [23] and this new lineage can then generate more species through subsequent speciation events [24]. Only in the latter two scenarios, where it triggered or promoted the origin of extant species or clades, can hybridization be said to have contributed significantly to evolutionary radiation. However, the signatures of such events can be hard to distinguish from recent hybridization events that did not generate new species, such as recent introgression, especially if only a small number of individuals are sampled. This is a particular issue in groups where hybridization and introgression are very common, even between morphologically and phylogenetically distant species [25].
Rhododendron subgenus Hymenanthes is an iconic example of plant radiation, containing around 302 species, most of them diploid and highly interfertile, with the great majority occurring in the mountains of southwest China, where many are endemic to single mountains [26–29]. All species of this subgenus are outcrossing, with extreme diversity in flowers, pollinators and ecological preference [30,31]. Small and light rhododendron seeds are mainly dispersed by wind and animals [32] giving the potential to reach sites relatively distant from the parent plant [33]. As found for other groups with evolutionary radiation [34], species delimitation in this subgenus is challenging due to morphological variation within species, and even populations [35]. All previous phylogenetic studies indicate that Hymenanthes species comprise a monophyletic group [36–38], but within the group, discordance between cpDNA, nuclear DNA and morphology appear to indicate ancient hybridization events [36–39]. In addition, numerous natural hybrids have also been reported [40,41], so there is great potential for recent interspecific gene flow as well. In this study, we aimed to examine the contribution of historical hybridization to the evolutionary radiation of Hymenanthes. We assembled a high-quality genome for one Hymenanthes species (Rhododendron prattii). We then performed whole-genome sequencing for 164 individuals of 143 species through selecting typical samples after genotyping available populations for the widespread species of this subgenus. Next, we constructed a dated phylogeny of the subgenus, and used this to seek signals of hybridization within and between clades, subclades and species. We uncovered extensive hybridization, both early and late during the diversification of subgenus Hymenanthes.
RESULTS
Reference genome assembly and annotation
We generated a chromosome level genome assembly for one member of subgenus Hymenanthes, Rhododendron prattii, by integrating single-molecule real-time (SMRT) sequencing, Illumina sequencing, 10x Genomics and high-throughput chromosome conformation capture (Hi-C mapping) techniques (Table S1). This de novo assembly was 673 Mb in size with 631 Mb sequences anchored onto 13 chromosomes and scaffolds N50 of 47.1 Mb (Figs S1 and S2, and Tables S2 and S3). Benchmarking universal single-copy orthologs (BUSCO) analysis indicated that the assembled genome obtained 97.3% of the embryophyta universal single-copy orthologs, confirming the high completeness of the assembly; 57.26% of the genome sequences were identified as repetitive elements. Among them, long terminal repeat (LTR) retrotransposons were the most abundant, accounting for 32.14% of the genome (Table S4). A total of 37 092 protein-coding genes were predicted for the assembly and 36 523 (96.1%) could be annotated by at least one public database (Tables S5 and S6). The repeat content and predicted gene number are similar to those of the other five reported species in genus Rhododendon (Table S7). All annotated genes in these six high-quality Rhododendron genomes could be assigned to 26 372 gene families, which comprise 5415 single-copy orthologous genes across them in the total genus.
Calibrated-phylogeny and recent radiation of subgenus hymenanthes
To ensure a sufficient species sampling for the radiation analyses, we examined around 12 000 individuals comprising ca. 800 populations of 292 species of subgenus Hymenanthes distributed in southwest China and other regions [28]. If a species had clear morphological gaps from other species, it was selected for further investigation. To rule out recent cryptic introgression, we genotyped widespread species that had more than four examined populations using 15 pairs of nuclear simple sequence repeat (SSR) markers [25] and chose one to three individuals that represent a relatively pure genetic composition of each species for final genome sequencing (see Supplementary Materials and Methods; Fig. S3). In total, we sequenced genomes of 164 individuals of 143 species (including subspecies and variety) for subgenus Hymenanthes, with a mean sequencing depth of 38× per individual (Table S8). In addition, 113 individuals of 93 species from four other Rhododendron subgenera were also selected for genome sequencing by examining wild populations in the same way (Table S8). By aligning all sequencing data to the R. prattii reference genome, we inferred the ploidy for each individual and found that all genome sequenced samples within Hymenanthes are diploid. The polyploid species detected in other subgenera were excluded from subsequent analyses (Table S9). After variants calling and stringent quality filtering, 14.8 million high-quality biallelic SNPs were obtained for the whole genus. The average sequence divergences (dXY) between subgenus Hymenanthes and other subgenera ranged from 0.035 to 0.048 (Fig. S4) and that between species within subgenus Hymenanthes was 0.018 (Fig. S5). We found that sequence diversities within individuals (heterozygosity) of R. agastum (0.022), R. catawbiense (0.026) and R. adenopodum (0.026) were obviously higher than those of the other sampled species—from 0.005 to 0.018 per base pair. Therefore, these three ‘species’ may comprise F1s as suggested before [41] and were further excluded in the following analyses (see Supplementary Materials and Methods).
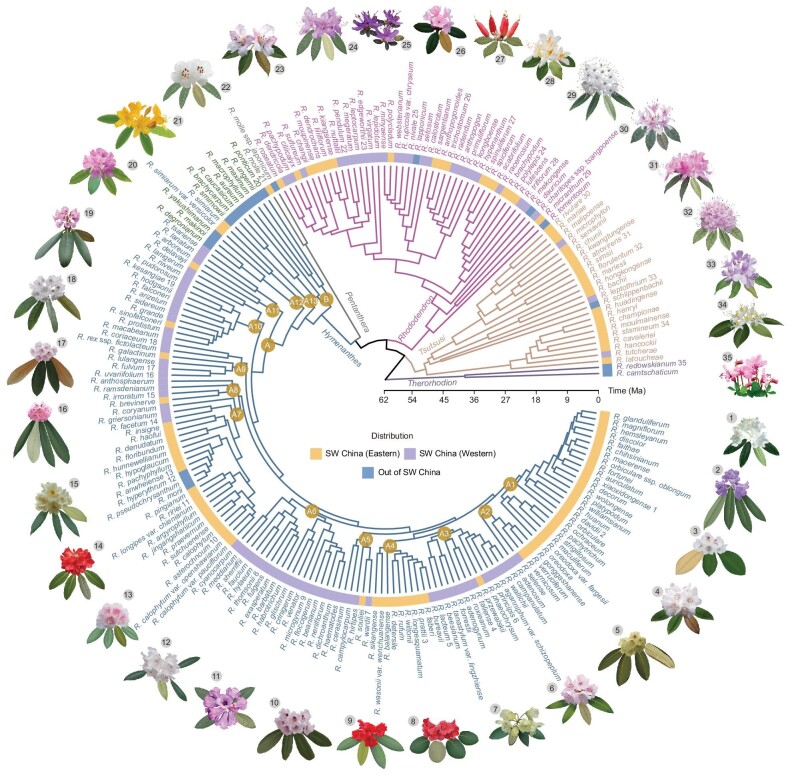
Principal component analysis (PCA) of genome-wide polymorphisms distinctly separated subgenus Hymenanthes from the four other subgenera by the first two PCs (Fig. S6). We inferred phylogenetic relationships among all sampled species based on three datasets: all nuclear genome SNPs (6 448 516), SNPs from protein-coding regions of the 4 468 single-copy nuclear genes only (483 944), and all SNPs from the chloroplast genome (12 273). Both concatenation and coalescent analyses supported the monophyly of subgenus Hymenanthes with a sister relationship to subgenus Pentanthera (Fig. 1 and Figs S7–S15). We used the concatenated nuclear genome tree to date the crown divergence and diversification dynamics within subgenus Hymenanthes, using the ∼56 Ma (million years ago) old fossil of Rhododendron newburyanum for calibration [42]. We found that the basal clade within Hymenanthes diverged from others at about 35.4 Ma (95% confidence interval: 31.5 to 41.3 Ma; Fig. 1). Since then, subgenus Hymenanthes showed increased diversification rates from the late Oligocene to middle Miocene (Fig. S16). This acceleration of diversification occurred during a time of increased geological and climatic oscillations in southwest China [21].
Figure 1.
Time-calibrated species tree of subgenus Hymenanthes. The species tree was calibrated based on a maximum-likelihood topology inferred from whole-genome SNPs. Branches are coloured according to different subgenera and those marked in blue belong to subgenus Hymenanthes. The outer circle represents the distribution of each species. Names of each clade and subclade are indicated in the ocher circles. The outer illustrations are 35 representative species from the tree for which the illustration IDs are marked behind species names. Names of Hymenanthes species from Tertiary relict regions are marked in green. SW China, southwest China and adjacent regions.
Phylogenetic discordance and shared polymorphisms
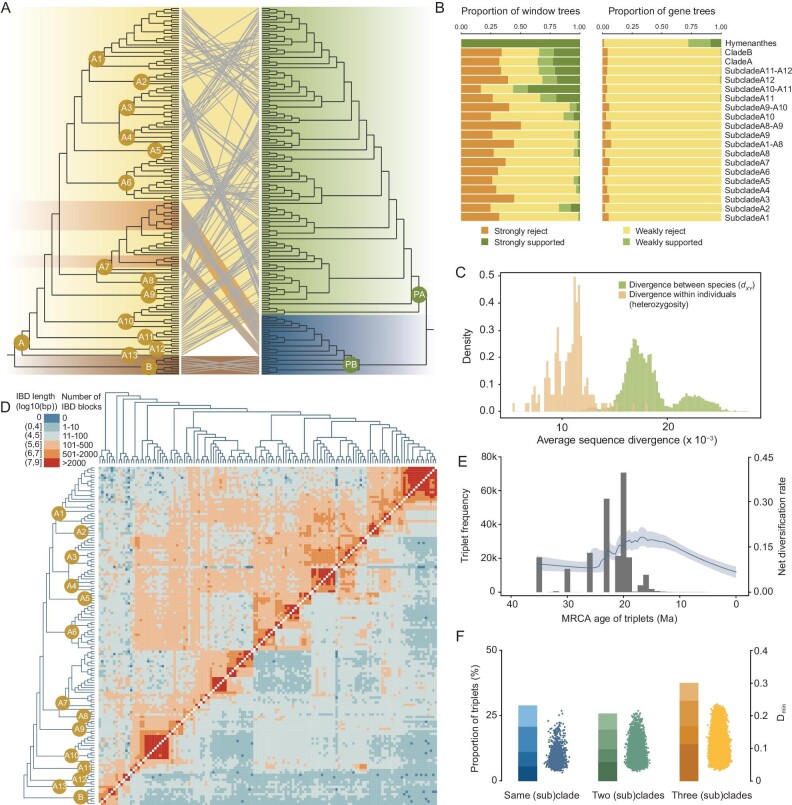
Relationships among species within subgenus Hymenanthes were largely incongruent between phylogenies inferred from different methods or datasets, especially for species from southwest China (Figs 1 and 2A, and Figs S7–S15). Based on the nuclear genome ML tree, two clades were identified within subgenus Hymenanthes. Clade A comprised all sampled species from southwest China and surrounding regions along with four species from Japan and southwestern Eurasia which occupied basal positions, while clade B was formed by seven species distributed in North America, southwestern Eurasia and northeastern Asia. Thirteen subclades were further recovered within clade A (Fig. 1). The plastome topology, however, differs markedly from the phylogenetic trees obtained with nuclear genome, although all sampled species are again divided into two clades (PA and PB; Fig. 2A). The clade PA contained the same four species from southwestern Eurasia and northeastern Asia in basal positions as did clade A, plus a huge monophyletic subclade comprising all but 11 of the sampled species from southwest China and adjacent regions. The remaining 11 species from southwest China and neighboring areas formed a monophyletic subclade within clade PB, nested among the seven members of nuclear clade B, from other parts of the Northern Hemisphere (Figs 1 and 2A and Fig. S15). In addition, topological conflicts among local SNP window trees and gene trees were also widely observed among species within the subgenus (Fig. S17). In the nuclear genome ML tree, we found high levels of discordance for window trees as well as gene trees in most focal splits among major clades/subclades, except for the ancestral node of the subgenus (Fig. 2B and Fig. S18). However, for all of the focal splits, the majority of the gene/window trees are compatible with the species tree after contracting those branches with low support (below 75%).
Figure 2.
Topological conflicts and extensive hybridization during evolutionary diversification of subgenus Hymenanthes. (A) Comparison of a maximum-likelihood phylogeny inferred from whole-genome SNPs (left) and a ML phylogeny based on the plastome SNPs (right). Eleven species within subclade A7 descended from an ancient hybridization event between the two initial diverging lineages of Hymenanthes are highlighted with orange background. (B) Local 10k-SNP window tree and gene tree compatibility revealed by the portion of trees for which clades (y-axis) are highly (weakly) supported or rejected. Weakly rejected clades are those not in the tree but being compatible if low support branches (<75%) are contracted. (C) Distributions of genomic divergence between species (dXY) and sequence diversity within individuals (heterozygosity) for subgenus Hymenanthes. (D) Estimated haplotype sharing in all sampled species within subgenus Hymenanthes. Heatmap colors represent the total length (above the diagonal) and the total number (below the diagonal) of identity-by-descent blocks for each species pairwise comparison. (E) Timescale distribution of the most recent common ancestor (MRCA) ages for the triplets with significantly elevated D values resulting from hybridization against the dynamic of net diversification rate of Hymenanthes. Ma, million years ago. (F) Excess allele sharing suggests non-bifurcating relationships among triplets of species within subgenus Hymenanthes. The bars denote the proportion of significantly elevated Dmin values with the shading representing family-wise error rate (FWER, see Supplementary Materials and Methods) of 5 × 10−2, 5 × 10−4, 5 × 10−8 and 5 × 10−14 (from light to dark). The scatterplots represent the Dmin scores that were significantly different from zero with FWER < 5 × 10−2. Results are shown separately for cases where the three members of a triplet come from the same clade (or subclade), two clades (or subclades), or three separate clades (or subclades).
Consistent with the expectations of reticulate evolution, we observed extensive shared polymorphisms among species. The average sequence divergences between species (dXY) within this subgenus ranged from 0.009 to 0.028, and partially overlapped with the distribution of sequence diversity within individuals (heterozygosity; Fig. 2C). In certain cases, sequence divergence within an individual is higher than divergence between species (Fig. 2C), suggesting that some of the species produced during radiation are genetically very closely related. The dXY values follow a bimodal distribution. The first peak mainly indicates the sequence differences between species distributed in southwest China, i.e. all of clade A except for the two basal subclades (A12 and A13). The second peak is composed of dXY values between species from southwest China and those with a Tertiary relict distribution (species from subclades A12 and A13, and clade B), and between species within the Tertiary relict group (Fig. S5). The nucleotide diversities of each clade and subclade ranged from 0.007 to 0.020 and no correlation was found between nucleotide diversity and the number of species in each clade/subclade. We then explored the identical-by-descent (IBD) haplotypes, of which 2 480 269 were detected within subgenus Hymenanthes (Fig. 2D). As expected, extensive sharing of haplotypes between species was detected, showing evidence of recent interspecific gene flow, for example between R. delavayi and R. irroratum (number of IBD = 140, total IBD length = 1.05 Mb, maximum IBD length = 135.75 kb), and between R. decorum and R. vernicosum (number of IBD = 299, total IBD length = 2.08 Mb, maximum IBD length = 121.02 kb). Furthermore, excess haplotype sharing was widely recovered between species from distinct subclades with overlapping distributions. For example, there was considerable haplotype sharing between subclades A1 and A7 in the eastern part of southwest China, and strong signatures of genetic admixture between subclades A6 and A10 in the Hengduan Mountains and the Himalayan region (Fig. 2D).
Tests of historical hybridization events during evolutionary radiation
To further determine the historical role of hybridization in causing these shared polymorphisms and gene tree discordance, we calculated Patterson's D statistics (the ABBA-BABA test) for every possible triplet among the sampled species. The lowest absolute value of the D-statistic (Dmin) across all possible tree topologies was determined for each triplet. In 153 186 of 447 581 triplets (34.2%) within subgenus Hymenanthes, Dmin values differed significantly from zero, indicating hybridization among the triplets. Furthermore, most triplets with an elevated D score had common ancestors dated mainly from the late Oligocene to middle Miocene, when diversification within Hymenanthes was starting to accelerate (Fig. 2E and Fig. S16). Based on these results, we also calculated the Reticulation Index for each node in the inferred species tree, to quantify the intensity of gene flow at each node. We found that the reticulation indices were high throughout the whole phylogeny, especially for lineages distributed in southwest China, suggesting ancient and persistent hybridization contributed substantially to the radiation of this subgenus (Fig. S19). Furthermore, widespread excess allele sharing between species was detected, both within and across major lineages, clades and subclades, revealing reticulate evolution at multiple levels (Fig. 2F).
We then examined whether such multi-level hybridization events occurred between different branches of the species tree using the ‘f-branch’ metric. This method detects excess allele sharing between a species C and a branch b when compared to the branch that is sister to b on a given species tree. In total, 22.7% (8 042 out of 35 316) of species-branch pairs had significantly elevated fb(C) scores (a summary of f4 admixture ratios; see Supplementary Materials and Methods; Fig. 3), and a total of 238 of 278 branches in the nuclear species tree showed significant excess allele sharing with at least one other species. Among these, 109 ancestral branches of multiple species showed significant elevated fb(C) scores, again suggesting that hybridization events frequently occurred among ancestral lineages of clades or subclades, and moreover that in some cases one single ancestral hybridization event may have been involved in the origins of multiple daughter species. Many signals of excess allele sharing for a specific branch are hierarchical, suggesting repeated hybridization events between different clades/subclades during radiation.
Figure 3.
Quantification of gene flow between branches in the species tree of subgenus Hymenanthes. The branch-specific statistic fb(C) value shows excess sharing of derived alleles between a given branch (b) on the y-axis, relative to its sister branch, and the species C on the x-axis. The ML tree based on whole-genome SNPs was used as a basic topology for the branch statistic. Branches marked in red on the y-axis represent the ancestral branch for each clade or subclade. Blue rectangles indicate four cases of significant signals of excess allele sharing, between the common ancestors of (i) subclade A10 and species from subclade A11, (ii) subclade A6 and species from subclades A10 and A11, (iii) subclade A5 and species from subclades A10 and A11, and (iv) subclade A5 and species from subclade A6.
DISCUSSION
Hybridization has long been assumed to promote species radiation, and this has been well studied in animals in the oceanic islands and rift lakes [9–11], but rarely tested within the species-rich clades that occur in mountainous ecosystems. In this study, we used whole-genome resequencing data to examine historical hybridization events underlying evolutionary radiation within Rhododendron subgenus Hymenanthes, which has high species diversity, especially in the mountains of southwest China. We found extensive hybridization and genomic admixture during the historical radiation of this subgenus, and for the first time revealed that repeated isolation and hybridization may have promoted rapid species diversification in mountainous regions. Hybridization can lead to various outcomes, including introgression of material between species and possible homogenization, but also promotion of species diversification through gene flow, and the origin of new lineages via homoploid or polyploid hybrid speciation [20,22–24]. The signature of ancient hybridization promoting such species radiations can be masked by more recent hybridization that leads to hybrid zones, and introgression within and between extant species [24]. Therefore, we genotyped multiple natural populations of the widespread species, in order to exclude possible recent introgression and hybrids, and from this we selected individuals for whole-genome sequencing and final analyses. This minimized the confounding effects of recent introgression, aiding the detection of more ancient hybridization that contributed to species diversification.
Although variant calling bias cannot be completely avoided by mapping the genome-sequencing data from other subgenera to one reference genome from subgenus Hymenanthes, our phylogenetic analyses of three different genome-scale datasets obtained the same relationships among different subgenera as previous studies, and supported the monophyly of subgenus Hymenanthes as found previously [36–38]. In particular, with genome-wide sampling across the nuclear and plastid genomes and a greater taxon coverage than any previous study of the subgenus, we obtained the most detailed phylogenetic relationships between lineages and species within the subgenus so far (Fig. 1 and Figs S7–S15). Our results strongly rejected most taxonomic treatments that were based on morphological traits [27] and likewise most subsections were not monophyletic. For example, subsections Fortunea and Argyrophylla, were highly polyphyletic with the sampled species of each placed in different clades/subclades. We also recovered extensive topological conflicts between phylogenies based on different datasets, especially between nuclear genome and plastome trees (Fig. 2A and B), which indicated that multiple hybridization events might have occurred during the diversification of this subgenus. For example, our nuclear phylogeny revealed a clade of seven Tertiary relict species (clade B in Fig. 1; distributed in North America, southwestern Eurasia, and northeastern Asia), which was not present in previous nuclear phylogenies based on single nuclear regions [36,37]. Similar to a previous cpDNA phylogeny [39], a clade containing the same seven species was also basal in the plastome phylogeny, but here it included a subclade of 11 sino-Himalayan species nested within it, indicating that these 11 species descended from a hybridization event between the two initial diverging lineages of Hymenanthes, probably around central Sichuan where most of these species occur. These 11 species occupy a derived position in the nuclear phylogeny, so the event occurred relatively late during the diversification of Hymenanthes. Furthermore, another 11 species with clade PA plastome types are nested among them in the nuclear phylogeny, indicating several other hybridization events in this clade later on. Any such event might have been hybrid speciation or introgression of nuclear germplasm and/or plastids; however, the initial event certainly represents contact between lineages with markedly different geographical ranges. In addition, extensive IBD haplotype sharing across species and significant signals of excess allele sharing between species or lineages indicated by ABBA-BABA tests further confirmed that hybridization events had occurred between clades, subclades and species, and hence to have occurred at different times throughout the diversification of Hymenanthes (Figs 2 and 3). However, it should be noted that as instances of historical hybridization are so pervasive, the signal of the oldest such events might be obscured by slightly younger events.
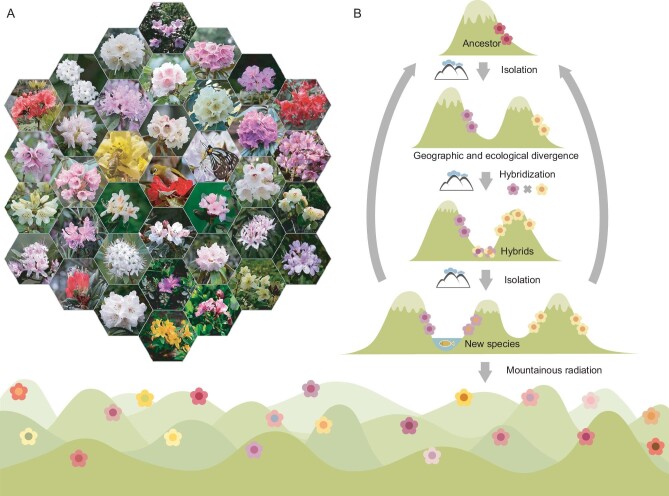
In addition to extensive hybridization and genomic admixture at deep and shallow timescales, our results also reveal that this subgenus radiated mainly from the late Oligocene to middle Miocene (Fig. S16), a period of significant geological and climatological change in southwest China [43]. These events might have promoted the accumulation of specific mutations by applying novel selection pressures, and new ecological niches that are regularly generated in such montane ecosystems [21]. However, climatic oscillations might have caused range expansions of species that had been previously isolated, leading to hybridization events, and hence the potential for alteration of lineages by introgression, and the formation of hybrid lineages [21]. Additionally, dispersing seeds across geographical barriers such as mountain chains or deep valleys [6] might have promoted hybridization and possible speciation. Rhododendrons are outcrossing but employ a diverse range of pollinators for sexual reproduction and seed set (Fig. 4A) [44–46]. Different birds and insects were observed to visit the same species, but birds seem to be more effective pollinators than insects for large-flowered species at high-altitudes where the temperature is low [44]. If birds travel long distances and are not faithful to particular species, these traits might promote gene flow via pollination between populations or species occurring on the isolated mountains. Equally, ecological and geographic isolation between populations, and variation between their niches, might have promoted further divergence among isolated lineages, including those generated or altered by hybridization. Such cycles of isolation and hybridization (Fig. 4B) might thus have created a positive feedback loop that generated many morphologically similar species.
Figure 4.
Simplified schematic of radiative diversification of subgenus Hymenanthes in the mountains of southwest China. (A) Diverse flowers and pollinators of Hymenanthes and other subgenera. (B) Cycles of isolation and hybridization that drive radiative diversification and produce many morphologically similar species.
Speciation with gene flow, i.e. hybridization between diverging populations before the formation of reproductive isolation, can accelerate speciation based on MIM modeling results [20], and hybrid speciation between already genetically diverged species without complete reproductive isolation can likewise contribute to the accumulation of species diversity [22–24]. A major question that remains is the extent to which hybridization accelerated the radiative diversification of Hymenanthes, and how many reticulation events could be discerned among all current lineages. Homoploid hybrid speciation is favored by the geographic and ecological isolation that is readily attained in montane ecosystems like the Himalaya and Hengduan Mountains of southwest China [47,48], but to prove it occurred within Hymenanthes, it must be shown that hybridization events contributed to reproductive isolation in addition to the genomic admixture revealed here [23,24]. It should be noted that cycles of isolation and hybridization may have been common throughout the world in mountainous regions that experienced similar geological dynamics and climatic oscillations [5]. Indeed, hybridization was reported to have frequently occurred in many more plant and animal species-rich genera in southwest China [21], and also the European Alps [49], the Northern American Rockies and Sierras [50], the Southern American Andes [51] and other montane ecosystems [17]. Therefore, such a model of evolutionary radiation involving isolation and hybridization is likely to apply not just to Hymenanthes in southwest China, but to other mountainous floras and even faunas worldwide.
METHODS AND MATERIALS
Detailed descriptions of methods are available as Supplementary data.
DATA AVAILABILITY
Whole-genome sequencing data, and transcriptome data have been deposited the Genome Sequence Archive in the National Genomics Data Center (https://bigd.big.ac.cn/) with accession number CRA005762. Genome assembly and annotation have been deposited in the Genome Warehouse under accession number GWHBHLU00000000.
Supplementary Material
Contributor Information
Yazhen Ma, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China; State Key Laboratory of Grassland Agro-ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Xingxing Mao, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Ji Wang, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Lei Zhang, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Yuanzhong Jiang, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Yuying Geng, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Tao Ma, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Liming Cai, Department of Organismic and Evolutionary Biology and Harvard University Herbaria, Harvard University, Cambridge, MA 02138, USA.
Shuangquan Huang, Institute of Evolution and Ecology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Pete Hollingsworth, Royal Botanic Garden Edinburgh, Edinburgh EH3 5LR, UK.
Kangshan Mao, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Minghui Kang, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Yiling Li, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Wenlu Yang, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Haolin Wu, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Yang Chen, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Charles C Davis, Department of Organismic and Evolutionary Biology and Harvard University Herbaria, Harvard University, Cambridge, MA 02138, USA.
Nawal Shrestha, State Key Laboratory of Grassland Agro-ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Richard H Ree, Negaunee Integrative Research Center, Field Museum, Chicago, IL 60605, USA.
Zhenxiang Xi, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Quanjun Hu, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China.
Richard I Milne, Royal Botanic Garden Edinburgh, Edinburgh EH3 5LR, UK; Institute of Molecular Plant Sciences, The University of Edinburgh, Edinburgh EH9 3JH, UK.
Jianquan Liu, Key Laboratory for Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, China; State Key Laboratory of Grassland Agro-ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
FUNDING
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0502), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000), the National Natural Science Foundation of China (31590821 and 91731301), the National Key Research and Development Program of China (2017YFC0505203), and further by the Fundamental Research Funds for the Central Universities (SCU2022D003 and 2020SCUNL207), and the National High-Level Talents Special Support Plan.
AUTHOR CONTRIBUTIONS
J.L. led the project. J.L., R.M. and Q.H. designed the research. X.M., L.Z., J.W., Y.M., P.H. and K.M. collected the materials. Y.G. and S.H. tested the monophyly of studied samples. Y.M., J.W and Y.J. collected sequencing data. Y.M., J.W, Q.H., T.M., L.C., M.K., Y.L., W.Y., H.W., Y.C. and N.S. performed data analyses. Y.M. and J.L. wrote the manuscript. R.M., C.C.D., R.H.R., Q.H. and Z.X. revised the manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rundell RJ, Price TD. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol Evol 2009; 24: 394–9. 10.1016/j.tree.2009.02.007 [DOI] [PubMed] [Google Scholar]
- 2. Givnish TJ. Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol 2015; 207: 297–303. 10.1111/nph.13482 [DOI] [PubMed] [Google Scholar]
- 3. Rahbek C, Borregaard MK, Antonelli Aet al. Building mountain biodiversity: geological and evolutionary processes. Science 2019; 365: 1114–9. 10.1126/science.aax0151 [DOI] [PubMed] [Google Scholar]
- 4. Antonelli A, Kissling WD, Flantua SGAet al. Geological and climatic influences on mountain biodiversity. Nat Geosci 2018; 11: 718–25. 10.1038/s41561-018-0236-z [DOI] [Google Scholar]
- 5. Igea J, Tanentzap AJ. Global topographic uplift has elevated speciation in mammals and birds over the last 3 million years. Nat Ecol Evol 2021; 5: 1530–5. 10.1038/s41559-021-01545-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flantua S, Hooghiemstra H. Historical connectivity and mountain biodiversity. In: Hoorn C, Perrigo A, Antonelli A (eds.). Mountains, Climate and Biodiversity. Chichester: Wiley-Blackwell, 2018, 171–85. [Google Scholar]
- 7. Brawand D, Wagner CE, Li YIet al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014; 513: 375–81. 10.1038/nature13726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lamichhaney S, Berglund J, Almén MSet al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature 2015; 518: 371–5. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- 9. Stryjewski KF, Sorenson MD. Mosaic genome evolution in a recent and rapid avian radiation. Nat Ecol Evol 2017; 1: 1912–22. 10.1038/s41559-017-0364-7 [DOI] [PubMed] [Google Scholar]
- 10. Malinsky M, Svardal H, Tyers AMet al. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat Ecol Evol 2018; 2: 1940–55. 10.1038/s41559-018-0717-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ronco F, Matschiner M, Böhne Aet al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature 2021; 589: 76–81. 10.1038/s41586-020-2930-4 [DOI] [PubMed] [Google Scholar]
- 12. Feng S, Ru D, Sun Yet al. Trans-lineage polymorphism and nonbifurcating diversification of the genus Picea. New Phytol 2019; 222: 576–87. 10.1111/nph.15590 [DOI] [PubMed] [Google Scholar]
- 13. Wang M, Zhang L, Zhang Zet al. Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol 2020; 225: 1370–82. 10.1111/nph.16215 [DOI] [PubMed] [Google Scholar]
- 14. Zhou B, Yuan S, Crowl AAet al. Phylogenomic analyses highlight innovation and introgression in the continental radiations of Fagaceae across the Northern Hemisphere. Nat Commun 2022; 13: 1320. 10.1038/s41467-022-28917-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nürk NM, Michling F, Linder HP. Are the radiations of temperate lineages in tropical alpine ecosystems pre-adapted? Glob Ecol Biogeogr 2018; 27: 334–45. 10.1111/geb.12699 [DOI] [Google Scholar]
- 16. Xing Y, Ree RH. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc Natl Acad Sci USA 2017; 114: E3444–51. 10.1073/pnas.1616063114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes CE, Atchison GW. The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytol 2015; 207: 275–82. 10.1111/nph.13230 [DOI] [PubMed] [Google Scholar]
- 18. Zhang D, Hao G, Guo Xet al. Genomic insight into “sky island” species diversification in a mountainous biodiversity hotspot. J Syst Evol 2019; 57: 633–45. 10.1111/jse.12543 [DOI] [Google Scholar]
- 19. Chen J, Huang Y, Brachi Bet al. Genome-wide analysis of cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat Commun 2019; 10: 5230. 10.1038/s41467-019-13128-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He Z, Li X, Yang Met al. Speciation with gene flow via cycles of isolation and migration: insights from multiple mangrove taxa. Natl Sci Rev 2019; 6: 275–88. 10.1093/nsr/nwy078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu S, Wang Y, Wang Zet al. Species divergence with gene flow and hybrid speciation on the Qinghai–Tibet Plateau. New Phytol 2022; 234: 392–404. [DOI] [PubMed] [Google Scholar]
- 22. Arnold ML. Natural Hybridization and Evolution. Oxford: Oxford University Press, 1997. [Google Scholar]
- 23. Wang Z, Jiang Y, Bi Het al. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol Plant 2021; 14: 208–22. 10.1016/j.molp.2020.11.008 [DOI] [PubMed] [Google Scholar]
- 24. Wang Z, Kang M, Li Jet al. Genomic evidence for homoploid hybrid speciation between ancestors of two different genera. Nat Commun 2022; 13: 1987. 10.1038/s41467-022-29643-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J, Luo J, Ma Yet al. Nuclear simple sequence repeat markers are superior to DNA barcodes for identification of closely related Rhododendron species on the same mountain. J Syst Evol 2019; 57: 278–86. 10.1111/jse.12460 [DOI] [Google Scholar]
- 26. De Riek J, De Keyser E, Calsyn Eet al. Azalea. In: Van Huylenbroeck J (ed.). Ornamental Crops. Cham: Springer, 2018, 237–72. [Google Scholar]
- 27. Chamberlain D, Hyam R, Argent G. et al. The Genus Rhododendron: Its Classification and Synonymy. Edinburgh: Royal Botanic Garden Edinburgh, 1996. [Google Scholar]
- 28. Geng Y. The Plants of Chinese Rhododendron. Shanghai: Science Press, 2016. [Google Scholar]
- 29. Khan G, Nolzen J, Schepker Het al. Incongruent phylogenies and their implications for the study of diversification, taxonomy, and genome size evolution of Rhododendron. Am J Bot 2021; 108: 1957–81. 10.1002/ajb2.1747 [DOI] [PubMed] [Google Scholar]
- 30. Shrestha N, Su X, Xu Xet al. The drivers of high Rhododendron diversity in south-west China: does seasonality matter? J Biogeogr 2018; 45: 438–47. 10.1111/jbi.13136 [DOI] [Google Scholar]
- 31. Song Y, Huang Z, Huang S. Pollen aggregation by viscin threads in Rhododendron varies with pollinator. New Phytol 2019; 221: 1150–9. 10.1111/nph.15391 [DOI] [PubMed] [Google Scholar]
- 32. Wang Y, Wang J, Lai Let al. Geographic variation in seed traits within and among forty-two species of Rhododendron (Ericaceae) on the Tibetan plateau: relationships with altitude, habitat, plant height, and phylogeny. Ecol Evol 2014; 4: 1913–23. 10.1002/ece3.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stephenson C, Kohnb D, Park Ket al. Testing mechanistic models of seed dispersal for the invasive Rhododendron ponticum (L.). Perspect Plant Ecol Evol Syst 2007; 9: 15–28. 10.1016/j.ppees.2007.07.004 [DOI] [Google Scholar]
- 34. Wang A, Yang M, Liu J. Molecular phylogeny, recent radiation and evolution of gross morphology of the rhubarb genus Rheum (Polygonaceae) inferred from chloroplast DNA trnL–F sequences. Ann Bot 2005; 96: 489–98. 10.1093/aob/mci201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao X, Wang J, Shrestha Net al. Species identification in the Rhododendron vernicosum–R. decorum species complex (Ericaceae). Front Plant Sci 2021; 12: 608964. 10.3389/fpls.2021.608964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goetsch L, Eckert AJ, Hall BD. The molecular systematics of Rhododendron (Ericaceae): a phylogeny based upon RPB2 gene sequences. Syst Bot 2005; 30: 616–26. 10.1600/0363644054782170 [DOI] [Google Scholar]
- 37. Shrestha N, Wang Z, Su Xet al. Global patterns of Rhododendron diversity: the role of evolutionary time and diversification rates. Glob Ecol Biogeogr 2018; 27: 913–24. 10.1111/geb.12750 [DOI] [Google Scholar]
- 38. Xia X, Yang M, Li Cet al. Spatiotemporal evolution of the global species diversity of Rhododendron. Mol Biol Evol 2021; 2021: msab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Milne RI, Davies C, Prickett Ret al. Phylogeny of Rhododendron subgenus Hymenanthes based on chloroplast DNA markers: between-lineage hybridisation during adaptive radiation? Plant Syst Evol 2010; 285: 233–44. 10.1007/s00606-010-0269-2 [DOI] [Google Scholar]
- 40. Zha H, Milne RI, Sun H. Morphological and molecular evidence of natural hybridization between two distantly related Rhododendron species from the Sino-Himalaya. Bot J Linn Soc 2008; 156: 119–29. 10.1111/j.1095-8339.2007.00752.x [DOI] [Google Scholar]
- 41. Zha H, Milne RI, Sun H. Asymmetric hybridization in Rhododendron agastum: a hybrid taxon comprising mainly F1s in Yunnan, China. Ann Bot 2010; 105: 89–100. 10.1093/aob/mcp267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Collinson ME, Crane PR. Rhododendron seeds from the palaeocene of southern England. Bot J Linn Soc 1978; 76: 195–205. 10.1111/j.1095-8339.1978.tb02361.x [DOI] [Google Scholar]
- 43. Mao K, Wang Y, Liu J. Evolutionary origin of species diversity on the Qinghai–Tibet Plateau. J Syst Evol 2021; 59: 1142–58. 10.1111/jse.12809 [DOI] [Google Scholar]
- 44. Huang Z, Song Y, Huang S. Evidence for passerine bird pollination in Rhododendron species. AoB Plants 2017; 9, DOI: 10.1093/aobpla/plx062 10.1093/aobpla/plx062. 10.1093/aobpla/plx062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stevens PF. The altitudinal and geographical distributions of flower types in Rhododendron section Vireya, especially in the Papuasian species, and their significance. Bot J Linn Soc 1976; 72: 1–33. 10.1111/j.1095-8339.1976.tb01392.x [DOI] [Google Scholar]
- 46. Stevens PF, Luteyn J, Oliver EGHet al. Ericaceae. In: Kubitzki K (ed.). Flowering Plants · Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales. Berlin, Heidelberg: Springer, 2004, 145–94. [Google Scholar]
- 47. Kadereit JW. The geography of hybrid speciation in plants. Taxon 2015; 64: 673–87. 10.12705/644.1 [DOI] [Google Scholar]
- 48. Wang D, Xu X, Zhang Het al. Abiotic niche divergence of hybrid species from their progenitors. Am Nat 2022; 200: 635–45. 10.1086/721372 [DOI] [PubMed] [Google Scholar]
- 49. Xu J, Hausdorf B. Repeated hybridization increased diversity in the door snail complex Charpentieria itala in the Southern Alps. Mol Phylogenet Evol 2021; 155: 106982. 10.1016/j.ympev.2020.106982 [DOI] [PubMed] [Google Scholar]
- 50. Drummond CS, Eastwood RJ, Miotto STSet al. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovations with incomplete taxon sampling. Sys Biol 2012; 61:443–60. 10.1093/sysbio/syr126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nevado B, Contreras-Ortiz N, Hughes Cet al. Pleistocene glacial cycles drive isolation, gene flow and speciation in the high-elevation Andes. New Phytol 2018; 219: 779–93. 10.1111/nph.15243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data, and transcriptome data have been deposited the Genome Sequence Archive in the National Genomics Data Center (https://bigd.big.ac.cn/) with accession number CRA005762. Genome assembly and annotation have been deposited in the Genome Warehouse under accession number GWHBHLU00000000.