Abstract
BACKGROUND & AIMS:
Next-generation sequencing (NGS) of pancreatic cyst fluid is a useful adjunct in the assessment of patients with pancreatic cyst. However, previous studies have been retrospective or single institutional experiences. The aim of this study was to prospectively evaluate NGS on a multi-institutional cohort of patients with pancreatic cyst in real time.
METHODS:
The performance of a 22-gene NGS panel (PancreaSeq) was first retrospectively confirmed and then within a 2-year timeframe, PancreaSeq testing was prospectively used to evaluate endoscopic ultrasound–guided fine-needle aspiration pancreatic cyst fluid from 31 institutions. PancreaSeq results were correlated with endoscopic ultrasound findings, ancillary studies, current pancreatic cyst guidelines, follow-up, and expanded testing (Oncomine) of postoperative specimens.
RESULTS:
Among 1933 PCs prospectively tested, 1887 (98%) specimens from 1832 patients were satisfactory for PancreaSeq testing. Follow-up was available for 1216 (66%) patients (median, 23 months). Based on 251 (21%) patients with surgical pathology, mitogen-activated protein kinase/GNAS mutations had 90% sensitivity and 100% specificity for a mucinous cyst (positive predictive value [PPV], 100%; negative predictive value [NPV], 77%). On exclusion of low-level variants, the combination of mitogen-activated protein kinase/GNAS and TP53/SMAD4/CTNNB1/mammalian target of rapamycin alterations had 88% sensitivity and 98% specificity for advanced neoplasia (PPV, 97%; NPV, 93%). Inclusion of cytopathologic evaluation to PancreaSeq testing improved the sensitivity to 93% and maintained a high specificity of 95% (PPV, 92%; NPV, 95%). In comparison, other modalities and current pancreatic cyst guidelines, such as the American Gastroenterology Association and International Association of Pancreatology/Fukuoka guidelines, show inferior diagnostic performance. The sensitivities and specificities of VHL and MEN1/loss of heterozygosity alterations were 71% and 100% for serous cystadenomas (PPV, 100%; NPV, 98%), and 68% and 98% for pancreatic neuroendocrine tumors (PPV, 85%; NPV, 95%), respectively. On follow-up, serous cystadenomas with TP53/TERT mutations exhibited interval growth, whereas pancreatic neuroendocrine tumors with loss of heterozygosity of ≥3 genes tended to have distant metastasis. None of the 965 patients who did not undergo surgery developed malignancy. Postoperative Oncomine testing identified mucinous cysts with BRAF fusions and ERBB2 amplification, and advanced neoplasia with CDKN2A alterations.
CONCLUSIONS:
PancreaSeq was not only sensitive and specific for various pancreatic cyst types and advanced neoplasia arising from mucinous cysts, but also reveals the diversity of genomic alterations seen in pancreatic cysts and their clinical significance.
Keywords: Pancreas, Early Detection, Pancreatic Neoplasm, Diagnosis, Pancreatic Cancer
Graphical Abstract
The detection of pancreatic cysts by cross-sectional imaging has become increasingly frequent and represents a significant public health challenge. In the United States, it is estimated that up to 2.5% of the general population harbors a pancreatic cyst.1,2 The prevalence of pancreatic cysts increases with age and up to 40% of patients who are 70 years and older have a pancreatic cyst.3 In addition, approximately half of all pancreatic cysts are mucinous cysts, such as intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs). IPMNs and MCNs are noninvasive precursor neoplasms to pancreatic ductal adenocarcinoma (PDAC).4 Consequently, the identification of mucinous cysts is a source of psychological stress for both the patient and the physician, but most mucinous cysts are indolent in nature and only a minority will transform into PDAC.1,5
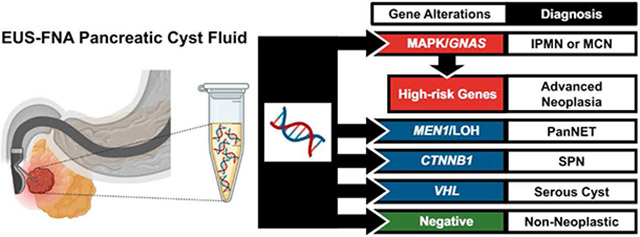
A multidisciplinary approach is currently advocated for the diagnosis and management of pancreatic cysts6-9; however, the evaluation of pancreatic cyst fluid is critical to the classification of pancreatic cysts and early detection of PDAC. Among ancillary studies, targeted DNA-based next-generation sequencing (NGS) is a useful tool in the assessment of pancreatic cysts.10-13 Mutations in the mitogen-activated protein kinase (MAPK) genes and/or GNAS are specific for mucinous cysts, whereas alterations in TP53, SMAD4, and the mammalian target of rapamycin (mTOR) genes are associated with advanced neoplasia (high-grade dysplasia and PDAC arising from a mucinous cyst).14-17 Targeted NGS can also be used to identify other pancreatic cyst types, such as serous cystadenomas (SCAs), solid-pseudopapillary neoplasms, and cystic pancreatic neuroendocrine tumors (PanNETs) that are characterized by mutations in VHL, CTNNB1, and MEN1, respectively.10,12,13,18
To date, several studies have evaluated targeted DNA-based NGS of pancreatic cysts, but published reports have largely been limited to retrospective analyses or single institutional experiences.10,11,13,19 In addition, most NGS studies have been focused on the assessment of IPMNs and IPMN-associated PDACs. The aims of this study were to (1) develop an expanded, targeted NGS panel (PancreaSeq) that can improve not only the assessment of IPMNs and IPMN-associated PDACs, but also other cyst types; (2) on confirmation of PancreaSeq performance using a retrospective cohort, to prospectively evaluate a multi-institutional cohort of pancreatic cyst patients in real time to determine the diagnostic performance of PancreaSeq testing; and (3) perform repeat PancreaSeq testing and expanded targeted DNA/RNA-based NGS (Oncomine) of paired postoperative specimens to establish concordance rates and identify additional genomic alterations that may further improve the assessment of pancreatic cysts.
Methods
Study Population
Study approval was obtained from the authors’ respective institutional review boards and the study design is outlined in Figure 1. For retrospective PancreaSeq testing (Supplementary Material and expected results are summarized in Supplementary Table 1), pancreatic cyst fluid specimens with corresponding clinical, imaging, and diagnostic surgical pathology follow-up were obtained through searching the molecular archives of the Molecular and Genomic Pathology (MGP) laboratory at the University of Pittsburgh Medical Center (UPMC) and cross-referencing the surgical pathology archives of UPMC Department of Pathology. These retrospective molecular specimens were previously reported in 2 large patient cohort studies.10,15 Prospective PancreaSeq testing was performed between January 2018 and February 2020 and consisted of 1933 pancreatic cyst fluid specimens obtained by endoscopic ultrasound (EUS)–fine-needle aspiration (FNA) that were submitted to the UPMC MGP laboratory from 31 medical institutions. In all cases, the indication for PancreaSeq testing was a clinical concern for a pancreatic cyst. Corresponding patient data were collected to include demographics, clinical presentation, EUS findings, fluid viscosity (as noted by the endoscopist using the string sign), carcinoembryonic antigen (CEA) analysis and cytopathological diagnoses. Endoscopic criterion of main duct dilatation was defined by a diameter ≥5 mm. In addition, the presence of a mural nodule was defined as a uniform echogenic nodule of any size without a lucent center or hyperechoic rim. A value >192 ng/mL was used as a cutoff for an elevated pancreatic cyst fluid CEA; however, CEA analysis was not centralized and performed at the submitting institution or reference laboratory. Cytopathologic findings were recorded from the respective submitting institutions and malignant cytopathology was defined as at least suspicious for adenocarcinoma. Diagnostic surgical pathology diagnoses were also obtained from each participating institution and were based on the 2019 World Health Organization (WHO) Classification of Tumors of the Digestive System.20 Cases diagnostic for a mucinous pancreatic cyst (IPMN and MCN) with high-grade dysplasia and/or an associated invasive adenocarcinoma were interpreted as “advanced neoplasia.” In comparison with PancreaSeq testing, absolute surgical resection criteria for the American Gastroenterology Association (AGA) guidelines (cytopathologic evaluation of at least suspicious for adenocarcinoma and/or 2 of the following features: dilated main pancreatic duct, >3.0 cm cyst size, and a solid component) and 2017 revised International Consensus Fukuoka (IAP/Fukuoka) guidelines (high-risk stigmata: jaundice in a patient with a cystic lesion of the pancreatic head, the presence of a mural nodule, main duct dilation suspicious for involvement, and/or cytopathologic evaluation of at least suspicious for adenocarcinoma) were retrospectively applied to the prospectively collected surgical resection study cohort.7,21
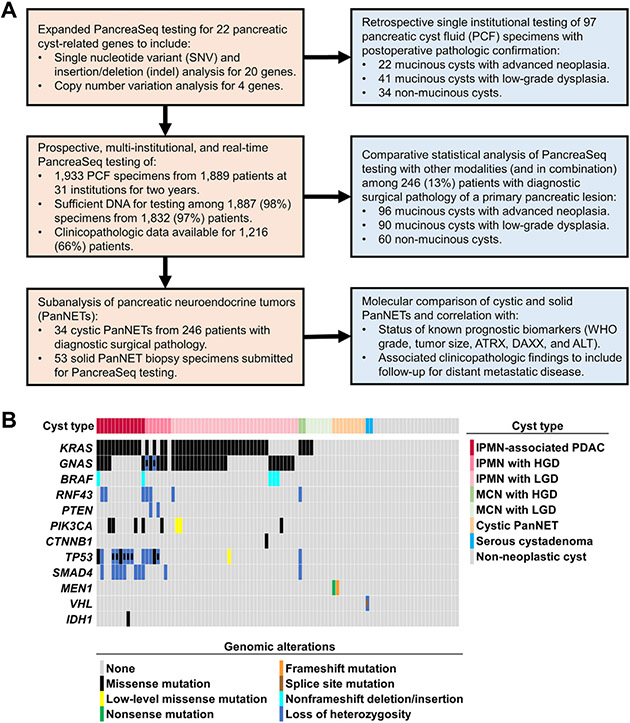
Figure 1.
(A) A summary of the study design to include details of individual patient cohorts used for PancreaSeq testing (tan) and individual analyses performed (blue). (B) Correlative genomic findings based on retrospective PancreaSeq testing of 97 preoperative pancreatic cyst fluid specimens from 63 mucinous cysts and 34 nonmucinous cysts. Among the 63 mucinous cysts, 22 cysts also harbored high-grade dysplasia and/or invasive adenocarcinoma (advanced neoplasia). Genomic alterations in KRAS, GNAS, and/or BRAF were 100% specific for mucinous cysts, whereas alterations in TP53, SMAD4, and/or the mTOR genes were preferentially seen in mucinous cysts with advanced neoplasia. Similarly, genomic alterations in MEN1 and VHL were highly specific for cystic PanNETs and SCAs, respectively. The mTOR genes include PIK3CA and PTEN. HGD, high-grade dysplasia; LGD, low-grade dysplasia.
Nucleic Acid Extraction
Nucleic acid extraction, as well as subsequent DNA- and RNA-based targeted NGS, was performed within the Clinical Laboratory Improvement Amendments- and College of American Pathologists–accredited MGP laboratory at UPMC. Genomic DNA and mRNA were isolated from either pancreatic cyst fluid obtained by EUS-FNA (preoperative specimens) or formalin-fixed paraffin-embedded tissue (surgical resection specimens) using the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche, Indianapolis, IN) on the Compact MagNA Pure (Roche) or the DNeasy Blood and Tissue kit on the automated QIAcube instrument (QIAGEN, Germantown, MD). Extracted DNA and RNA were quantitated on the Glomax Discover using the QuantiFluor ONE dsDNA System and the QuantiFluor RNA system, respectively (Promega, Madison, WI).
PancreaSeq Testing
Amplification-based targeted DNA-based NGS for PancreaSeq was performed with custom AmpliSeq primers for genomic regions of interest within AKT1, APC, BRAF, CTNNB1, GNAS, HRAS, IDH1, IDH2, KRAS, MEN1, MET, NF2, NRAS, PIK3CA, PTEN, STK11, TERT, TP53, TSC2, and VHL with primer sequences and performance characteristics as previously described to include single nucleotide variants, insertions, deletions, and loss of heterozygosity (LOH)/copy number alteration.10,12,13,22 Amplicons were barcoded, ligated with specific adapters, and purified. DNA library quantity and quality checks were performed using the 4200 TapeStation (Agilent Technologies, Santa Clara, CA). The Ion Chef was used to prepare and enrich templates and enable testing via Ion Sphere Particles on a semiconductor chip. Massive parallel sequencing was carried out on an Ion GeneStudio S5 Prime System according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA) and data were analyzed with an in-house bioinformatics program. Variant Explorer (UPMC). Each variant was prioritized according to the 2017 AMP/ASCO/CAP joint consensus guidelines for interpretation of sequence variants in cancer using a tier-based system.23 Tier I, Tier II, and Tier III variants were identified; however, only Tier I and Tier II variants were used for subsequent analysis. The limit of detection of the assay was at 1% mutant allele frequency (AF). The minimum depth of coverage for testing was 1000×. For each mutation identified, an AF was calculated based on the number of reads of the mutant allele versus the wild-type allele and reported as a percentage.10 A low-level variant was classified based on a 10-fold lower AF as compared with the AF for a MAPK/GNAS mutation.10 LOH analysis was performed as previously described.24,25
Oncomine Testing
Expanded targeted NGS-based testing from DNA and mRNA was also performed within the MGP lab at UPMC using the Oncomine Comprehensive Assay v3 (Oncomine) DNA and RNA primer sets (Thermo Fisher Scientific) according to the manufacturer’s protocol. The Oncomine panel evaluates 161 cancer-relevant driver genes to include 760 fusion genes. Briefly, total DNA and mRNA that is reverse transcribed into complementary DNA are subjected to multiplex polymerase chain reaction to amplify the regions of interest. Amplicons were barcoded, ligated with specific adapters, and purified. RNA library quantity and quality check were performed using the 4200 TapeStation (Agilent Technologies, Santa Clara, CA). The Ion Chef was used to prepare and enrich templates and enable testing via Ion Sphere Particles on a semiconductor chip. Massive parallel sequencing was carried out on an Ion GeneStudio S5 Prime System according to the manufacturer’s instructions (Thermo Fisher Scientific) and data were analyzed with Variant Explorer (UPMC) for single nucleotide variant, insertions, deletions, copy number alterations, and RNA fusion genes. The limit of detection of this DNA/RNA assay was 1% to 5% neoplastic cells.
Statistical Analysis
χ2 analysis or Fisher’s exact tests were used to compare categorical data, and Mann-Whitney U test was used to compare continuous variables. Sensitivity and specificity were calculated using standard 2×2 contingency tables for cases with confirmed diagnostic pathology. All statistical analyses were performed using the SPSS Statistical software, V.26 (IBM, Armonk, NY) and statistical significance was defined as a P value of <.05.
Results
Retrospective PancreaSeq Testing of 97 Patients With Diagnostic Surgical Pathology
A retrospective diagnostic performance confirmation cohort of 97 patients who underwent EUS-FNA for a pancreatic cyst and had follow-up diagnostic surgical pathology was evaluated using an expanded NGS panel (PancreaSeq) of 22 pancreatic cyst-associated genes (Supplementary Material and expected results are summarized in Supplementary Table 1). The results of retrospective PancreaSeq testing are summarized in Figure 1 (and Supplementary Table 2). Genomic alterations in KRAS, GNAS, and/or BRAF were detected in 56 of 63 (89%) mucinous cysts. Among mucinous cysts with advanced neoplasia, alterations in TP53, SMAD4, and the mTOR genes were identified in 19 of 22 (86%) cases. Further, 3 of 31 (10%) IPMNs with low-grade dysplasia harbored PIK3CA (n = 2) and TP53 (n = 1) mutations; but, in comparison with KRAS missense mutations, alterations in PIK3CA and TP53 were at a lower AF (low-level). Mutations in VHL and MEN1 were also seen, but specific to SCAs (1 of 2, 50%) and cystic PanNETs (2 of 9, 22%), respectively. Twenty-three non-neoplastic cysts were negative for genomic alterations. The sensitivity and specificity of MAPK/GNAS alterations for a mucinous cyst was 89% and 100%, respectively. In addition, mutations in GNAS and/or BRAF were 100% specific for IPMNs. In conjunction with MAPK/GNAS mutations, alterations in TP53, SMAD4, and the mTOR genes had 86% sensitivity and 96% specificity for a mucinous cyst with advanced neoplasia. However, on exclusion of low-level TP53 and PIK3CA mutations, the sensitivity and specificity for advanced neoplasia was 86% and 100%, respectively.
Prospective, Real-Time, Multi-institutional PancreaSeq Testing of 1832 Patients
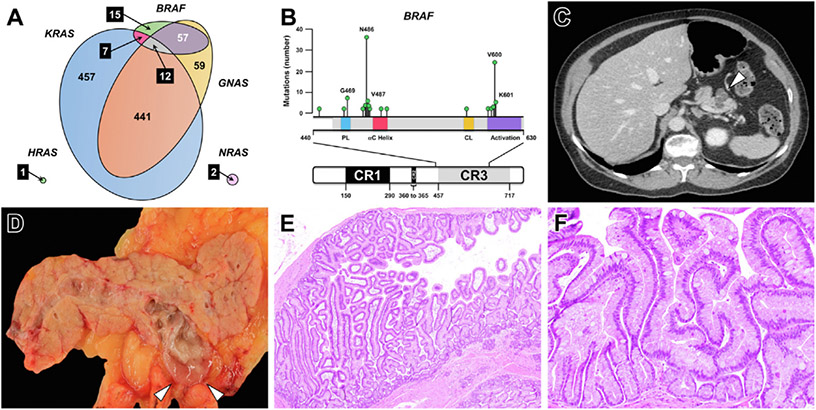
Prospective PancreaSeq testing was attempted for 1933 EUS-FNA obtained pancreatic cyst fluid specimens from 1889 patients and collected from 31 institutions over a 2-year time frame. Sufficient DNA for PancreaSeq testing was identified in 1887 (98%) specimens from 1832 patients (Supplementary Table 3). Two pancreatic cysts were sampled for 55 (3%) patients at the same EUS-FNA procedure with the clinical indication that the 2 cysts were identified in a different region of the pancreas (head/uncinate/neck versus body/tail). Overall, genomic alterations were detected in 1220 (65%) specimens. Genomic alterations in KRAS, BRAF, NRAS, and HRAS were seen in 917 (49%), 91 (5%), 2 (<1%), and 1 (<1%) cysts, respectively (Figure 2 and Supplementary Material). In contrast to other gastrointestinal neoplasms, a minority of BRAF alterations were V600E/L/M/R mutations (class I mutations), and instead were predominantly class II and class III BRAF mutations (n = 83, 91%) (Supplementary Table 4). The most prevalent BRAF alteration was an in-frame deletion involving codon 486. Activating GNAS mutations were seen in 569 (30%) cyst fluid specimens, and co-occurred with either KRAS, BRAF, or both genes in 441 (of 569, 78%), 57 (10%), and 12 (2%) cases. Among GNAS-mutant cysts, 510 (90%) harbored a genomic alteration in at least 1 gene involved within the MAPK pathway. In total, mutations in the MAPK genes and GNAS were detected in 1050 (56%) cases (Supplementary Table 5). Multiple mutations in KRAS and GNAS were found in 138 (7%) and 26 (1%) cysts, respectively. In addition, a concurrent LOH in KRAS and GNAS was seen in 4 and 1 case, respectively.
Figure 2.
(A) An area-proportional Venn diagram demonstrates the distribution of KRAS, GNAS, BRAF, NRAS, and HRAS mutations identified through prospective PancreaSeq testing of 1887 pancreatic cysts. In addition to KRAS and GNAS, BRAF alterations were often identified in EUS-FNA obtained pancreatic cyst fluid specimens and frequently co-occurred with GNAS mutations. (B) Most BRAF alterations found in pancreatic cysts were non-V600E mutations and were predominantly categorized as class II and class III BRAF mutations (n = 83, 91 %). (C) Based on correlative imaging and pathologic studies, BRAF-mutant pancreatic cysts (white arrowhead) were commonly found to communicate with the main pancreatic duct, and (D) on gross pathology, exhibited abundant, thick mucin (white arrowheads). (E and F) Microscopically, BRAF-mutant cysts corresponded to an intraductal papillary mucinous neoplasm with prominent papillary fronds and often lined by both gastric and intestinal epithelium. (E) Hematoxylin and eosin stain, magnification 40×. (F) Hematoxylin and eosin stain, magnification 200×.
Among 1050 MAPK/GNAS-mutant cysts, 158 (15%) were found to have TP53, SMAD4, and/or mTOR gene alterations (Supplementary Table 6). With respect to MAPK/GNAS AF, low-level point mutations in TP53 and PIK3CA were seen in 18 (of 158, 11%) and 8 (5%) cases, respectively. In addition to TP53, SMAD4, and the mTOR genes, 11 MAPK/GNAS-mutant cysts had CTNNB1 mutations. Five of 11 MAPK/GNAS/CTNNB1-mutant cysts had low-level CTNNB1 missense mutations as compared with the AF for the MAPK/GNAS gene(s). Further, none of the MAPK/GNAS/CTNNB1-mutant cysts had co-occurring TP53, SMAD4, and/or mTOR gene alterations (Supplementary Table 7).
In the absence of a MAPK/GNAS mutation (n = 837), alterations in VHL, MEN1, or both genes were seen in 125 (15%), 19 (2%), and 11 (1%) cysts, respectively. Co-occurring alterations were identified in 37 of 125 (30%) VHL-mutant/MEN1 wild-type cysts and included point mutations in TP53 (n = 5), the TERT promoter (n = 5), and PTEN (n = 1) as well as LOH for PTEN (n = 19), TP53 (n = 18), SMAD4 (n = 18), and RNF43 (n = 15). Six of 19 (32%) MEN1-mutant/VHL wild-type cysts also harbored co-occurring alterations that included a TP53 missense mutation (n = 1) and LOH in SMAD4 (n = 6). Interestingly, the VHL alterations in all 11 VHL/MEN1-mutant cysts consisted of LOH alterations. Further, 9 of 11 (82%) VHL/MEN1-mutant cysts had co-occurring LOH in TP53 (n = 6), SMAD4 (n = 5), RNF43 (n = 5), and/or PTEN (n = 9). In the absence of VHL and/or MEN1 alterations, LOH in TP53 (n = 5), SMAD4 (n = 13), RNF43 (n = 5), and/or PTEN (n = 4) was identified in 21 cysts. Point mutations in TP53 as the sole genomic alteration were seen in 7 cases. Finally, IDH1 and IDH2 missense mutations were detected in 1 cyst each without co-occurring alterations.
Clinicopathologic Correlation and Follow-up Information for 1216 Patients
Associated clinicopathologic data were available for 1216 of 1832 (66%) patients (Supplementary Material and Supplementary Table 3) that includes 1253 EUS-FNA obtained pancreatic cyst fluid specimens with genomic alterations detected in 851 specimens, whereas the remaining 402 specimens were negative for detectable mutations. In addition, follow-up information ranged between 2 and 35 months (mean, 20 months; median, 21 months). Diagnostic surgical pathology was available for 251 of 1216 (21%) patients who underwent surgery within 2 to 34 months (mean, 9 months; median, 4 months) from initial EUS-FNA and PancreaSeq testing. This cohort of surgical resected lesions consisted of 246 cysts arising within the pancreas (Figure 3) and 5 metastatic carcinomas involving the pancreas. Alterations in KRAS, BRAF, and/or GNAS were preoperatively detected in 159 of 167 (95%) IPMNs and KRAS missense mutations were seen in 9 of 19 (47%) MCNs. In addition to MAPK/GNAS mutations, alterations in TP53, SMAD4, and/or the mTOR genes were identified in 77 of 90 (86%) IPMNs with advanced neoplasia, 6 of 6 (100%) MCNs with advanced neoplasia, and 5 of 77 (6%) IPMNs with low-grade dysplasia (Figure 4 and Supplementary Figure 1). CTNNB1 missense mutations were also detected in 2 IPMNs with high-grade dysplasia and 1 IPMN with low-grade dysplasia. Both IPMNs with high-grade dysplasia were negative for alterations in TP53, SMAD4, and the mTOR genes. Low-level point mutations in TP53, P1K3CA, PTEN, and CTNNB1 corresponded to either an IPMN with low-grade dysplasia or an MCN with low-grade dysplasia. LOH in KRAS or GNAS was also observed in 4 IPMNs with an associated adenocarcinoma; however, 1 of 4 IPMNs was preoperatively negative for alterations in TP53, SMAD4, CTNNB1, and the mTOR genes.
Figure 3.
A summary of clinical presentation, imaging findings, pathologic features, preoperative PancreaSeq testing, and postoperative PancreaSeq/Oncomine results for 251 patients with pancreatic cyst with diagnostic surgical pathology. Preoperative genomic alterations involving KRAS, GNAS, and/or BRAF corresponded to the presence of a mucinous cyst, whereas additional alterations in TP53, SMAD4, CTNNB1, and/or the mTOR genes were preferentially found in mucinous cysts with advanced neoplasia. Other key findings were the preoperative detection of LOH for multiple genes that correlated with the presence of a cystic PanNET, and the identification of TP53 and TERT promoter mutations in large SCAs. Postoperative PancreaSeq/Oncomine testing revealed the presence of novel BRAF fusion genes and ERBB2 amplification in RAS wild-type IPMNs (Supplementary Figure 3). Moreover, CDKN2A alterations were preferentially found in IPMNs with advanced neoplasia. MAPK genes include KRAS, BRAF, FIRAS, ERBB2, and MAPK1, and mTOR genes include PTEN, PIK3CA, and AKT1.
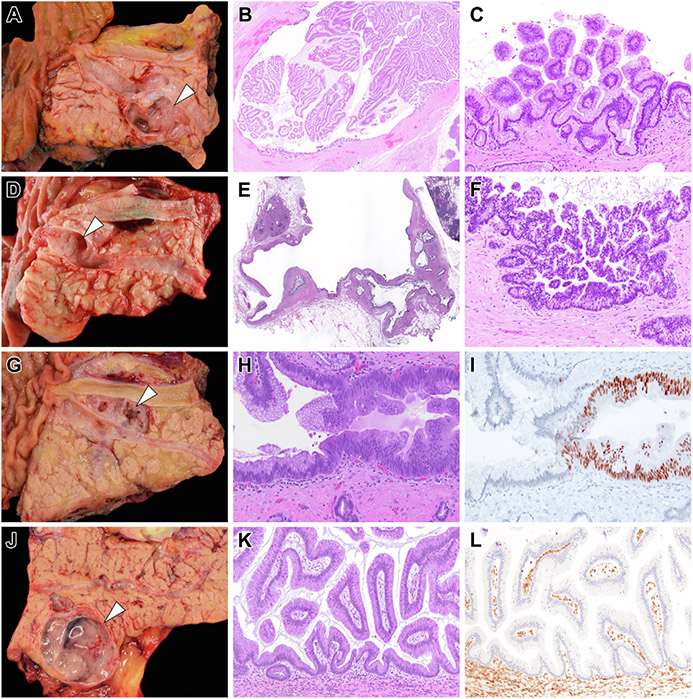
Figure 4.
Representative examples of diagnostic surgical pathology for IPMNs that had preoperative PancreaSeq testing. (A) A branch-duct IPMN that was resected because of the presence of a mural nodule (white arrowhead) detected on preoperative imaging. (B) The mural nodule corresponded to collapsed papillary fronds and (C) microscopically, correlated with low-grade dysplasia. Preoperative PancreaSeq testing detected the presence of KRAS and GNAS mutations, but an absence of TP53, SMAD4, CTNNB1, with mTOR gene alterations. (D) A branch-duct IPMN (white arrowhead) with focal ductal dilation and otherwise no concerning preoperative clinical, imaging, or preoperative pathologic findings. Preoperative PancreaSeq testing identified mutations in KRAS and GNAS, and LOH for PTEN and TP53. (E and F) Diagnostic surgical pathology revealed the presence of high-grade dysplasia. (G) A branch-duct IPMN (white arrowhead) with focal ductal dilatation and otherwise no concerning preoperative clinical, imaging, or preoperative pathologic findings. PancreaSeq testing detected a KRAS mutation and a low-level TP53 mutation. Although the submitting surgical pathology report documented the presence of an IPMN with low-grade dysplasia, a (H) focal area of cytologic atypia was identified and (I) corresponded to aberrant nuclear p53 expression. (J) A 3.0-cm branch-duct IPMN (white arrowhead) with otherwise no concerning preoperative clinical, imaging, or preoperative pathologic findings; however, PancreaSeq testing identified a KRAS mutation and SMAD4 LOH. (K) Although histologically consistent with an IPMN with low-grade dysplasia, (L) diffuse loss of Smad4 expression was seen throughout the IPMN. The mTOR genes include PIK3CA and PTEN. (B) Hematoxylin and eosin stain, magnification 20×. (C) Hematoxylin and eosin stain, magnification 200×. (E) Hematoxylin and eosin stain, magnification 20×. (F) Hematoxylin and eosin stain, magnification 200×. (H) Hematoxylin and eosin stain, magnification 200×. (I) p53 immunolabeling, magnification 200×. (K) Hematoxylin and eosin stain, magnification 200×. (L) SMAD4 immunolabeling, magnification 200×.
All 13 (100%) SCAs harbored VHL alterations. In addition to VHL, 4 SCAs harbored point mutations in either TP53 (n = 2) or the TERT promoter (n = 2). Before surgical resection, all 4 SCAs with a TP53 or TERT promoter mutation demonstrated an interval increase in cyst size (Supplementary Figure 2). Further, 1 TP53-mutant SCA exhibited progressive stricturing of the main pancreatic duct and both acute and chronic pancreatitis. Thirty-four patients who underwent surgery were found to have a cystic PanNET. Genomic alterations found in preoperative cyst fluid specimens from these 34 cystic PanNETs included 7 with MEN1 mutations and 16, 14, 13, 12, and 11 cases with LOH for SMAD4, VHL, TP53, PTEN, and RNF43, respectively. Collectively, the presence of an MEN1 mutation and/or LOH were seen in 24 of 34 (71%) cases.
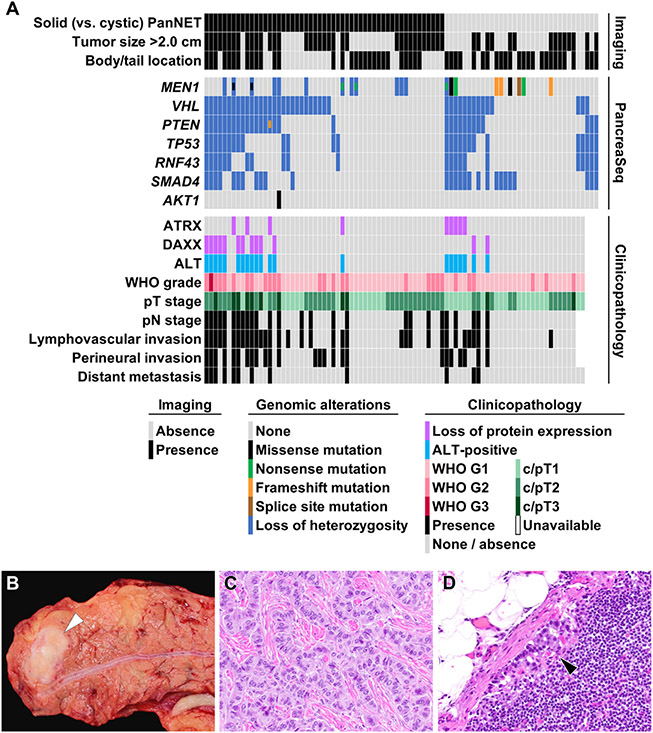
To further analyze the clinicopathologic features of PanNETs harboring LOH for SMAD4, VHL, TP53, PTEN, and/or RNF43, 53 preoperative biopsies from patients with a solid PanNET encountered during the study period were tested using PancreaSeq and correlated with surgical outcome and associated follow-up (Supplementary Material and Supplementary Table 8). Based on a total of 87 PanNETs (34 cyst fluid specimens and 53 biopsies), MEN1 alterations were identified in 21 (42%) cases, whereas LOH of SMAD4, VHL, TP53, PTEN, and/or RNF43 was seen in 51 (59%) cases (Figure 5). The presence of LOH for ≥1 gene correlated with perineural invasion, lymphovascular invasion, regional lymph node metastases, and distant organ metastasis (P < .012). LOH for ≥1 gene was also associated with loss of protein expression for ATRX and DAXX, and the presence of alternative lengthening of telomeres (ALT) by telomere-specific fluorescence in situ hybridization (P < .001). Of note, within this solid and cystic PanNET study cohort, 21 of 51 (41%) PanNETs with LOH of ≥1 gene were 1.0 to 2.0 cm in greatest dimension.
Figure 5.
(A) A summary of imaging findings, preoperative PancreaSeq testing, and postoperative clinicopathologic features of 87 PanNET patients. Both solid and cystic PanNETs exhibited similar genomic alterations; however, LOH for multiple genes correlated with several adverse clinicopathologic features, such as lymphovascular invasion, perineural invasion, higher T- and N-stage, distant metastases, loss of ATRX/DAXX expression, and the presence of ALT. (B) A representative example of a 1.5-cm PanNET (white arrowhead) in the pancreatic body that preoperative PancreaSeq testing revealed LOH for 4 genes. (C) Microscopically and immunohistochemically, the PanNET was classified as WHO grade 1. (D) However, within a single regional lymph node, a metastasis was identified (black arrowhead). In addition, the PanNET exhibited loss of ATRX expression and the presence of ALT. (C) Hematoxylin and eosin stain, magnification 400×. (D) Hematoxylin and eosin stain, magnification 200×.
The remaining 965 patients had clinical follow-up data, but no diagnostic surgical pathology. Of the 965 patients, 2 pancreatic cysts were sampled from 37 patients, and 495 (51%) patients had a pancreatic cyst with a MAPK/GNAS alteration. For the 37 patients with 2 pancreatic cyst specimens, both specimens harbored mutations in the MAPK and/or GNAS genes. Twelve of the 495 (2%) patients also had mutations in TP53 (n = 6) or PIK3CA (n = 6), but all except 1 case with a PIK3CA mutation were low-level point mutations. Co-occurring CTNNB1 missense mutations were seen in 6 cases, and 4 of 6 cases were low-level alterations. For the 470 patients with a MAPK/GNAS wild-type cyst, alterations in VHL, MEN1, or both genes were seen in 79 (17%), 8 (2%), and 8 (2%) cysts, respectively. Six VHL-mutant/MEN1 wild-type cysts also harbored point mutations in TP53 (n = 3) and the TERT promoter (n = 3). During follow-up, 4 of these 6 VHL-mutant/MEN1 wild-type cysts exhibited an increase in cyst size.
Comparison and Combination of PancreaSeq Testing With Other Diagnostic Modalities
Excluding 5 metastatic carcinomas, preoperative PancreaSeq detection of MAPK/GNAS mutations had 90% sensitivity and 100% specificity for a mucinous cyst (Table 1). Increased fluid viscosity and an elevated CEA of >192 ng/mL had lower sensitivities (77% and 73%, respectively) and lower specificities (92% and 94%, respectively). In conjunction with MAPK /GNAS mutations, alterations in TP53, SMAD4, and/or the mTOR genes had 85% sensitivity and 96% specificity for a mucinous cyst with advanced neoplasia. The sensitivity and specificity for advanced neoplasia increased to 87% and 99%, respectively, on inclusion of MAPK/GNAS LOH or TP53, SMAD4, and/or mTOR gene alterations with equivalent allele frequencies to MAPK/GNAS. Further, the inclusion of CTNNB1 with equivalent allele frequencies to MAPK/GNAS achieved a sensitivity of 89% and a specificity of 98% for advanced neoplasia. In comparison, the presence of associated clinical symptoms, jaundice for pancreatic head cysts, cyst size of >3.0 cm, main pancreatic duct dilatation, a mural nodule on EUS, increasing cyst size, and a cytopathologic diagnosis of at least suspicious for adenocarcinoma were all associated with lower sensitivities and lower specificities. Moreover, combining PancreaSeq testing with the aforementioned parameters improved the overall sensitivity of detecting advanced neoplasia (Supplementary Table 9). The highest sensitivity of 93% while maintaining a high specificity of 95% was attained using both PancreaSeq testing and cytopathologic examination (Supplementary Table 10).
Table 1.
Diagnostic Performance of PancreaSeq Testing and Other Modalities Based on 246 Confirmed Pancreatic Cysts
| Parameter | Sensitivity, % (95% CI) |
Specificity, % (95% CI) |
PPV, % (95% CI) |
NPV, % (95% CI) |
|---|---|---|---|---|
| IPMN | ||||
| MAPK/GNAS mutations | 95 (0.91–0.98) | 89 (0.78–0.94) | 95 (0.90–0.97) | 90 (0.42–0.66) |
| Presence of multiple cysts (n = 245)a | 54 (0.46–0.62) | 80 (0.69–0.88) | 85 (0.76–0.91) | 45 (0.37–0.54) |
| Increased fluid viscosity (n = 238)a | 79 (0.72–0.85) | 81 (0.70–0.89) | 89 (0.83–0.94) | 66 (0.55–0.75) |
| Elevated CEA (n = 173)a | 74 (0.65–0.81) | 73 (0.59–0.84) | 86 (0.78–0.92) | 54 (0.42–0.66) |
| IPMN with advanced neoplasia | ||||
| TP53, SMAD4, and/or mTOR gene alterations | 87 (0.78–0.93) | 76 (0.69–0.83) | 68 (0.58–0.76) | 91 (0.84–0.95) |
| TP53, SMAD4, CTNNB1, and/or mTOR gene alterations | 89 (0.80–0.94) | 74 (0.67–0.81) | 67 (0.57–0.75) | 92 (0.86–0.96) |
| MAPK/GNAS mutations with TP53, SMAD4, and/or mTOR gene alterations | 84 (0.75–0.91) | 92 (0.87–0.96) | 86 (0.77–0.93) | 91 (0.85–0.95) |
| MAPK/GNAS mutations with TP53, SMAD4, CTNNB1, and/or mTOR gene alterations | 87 (0.78–0.93) | 91 (0.85–0.95) | 85 (0.75–0.91) | 92 (0.87–0.96) |
| MAPK/GNAS LOH or TP53, SMAD4 and/or mTOR gene AFs = MAPK/GNAS AFs | 86 (0.76–0.92) | 95 (0.90–0.98) | 91 (0.82–0.96) | 92 (0.86–0.96) |
| MAPK/GNAS LOH or TP53, SMAD4, CTNNB1, and/or mTOR gene AFs = MAPK/GNAS AFs | 88 (0.79–0.94) | 94 (0.89–0.97) | 90 (0.81–0.95) | 93 (0.88–0.96) |
| Associated clinical symptoms (n = 244)a | 38 (0.28–0.49) | 71 (0.64–0.78) | 44 (0.33–0.55) | 66 (0.59–0.73) |
| Jaundice (n = 131)b | 31 (0.20–0.44) | 89 (0.78–0.95) | 70 (0.50 – 0.86) | 60 (0.50–0.69) |
| Index cyst size >3.0 cm (n = 242)a | 56 (0.45–0.66) | 55 (0.46–0.63) | 41 (0.32–0.51) | 68 (0.59–0.76) |
| Main pancreatic duct dilatation (n = 244)a | 71 (0.60–0.80) | 65 (0.57–0.73) | 54 (0.44–0.63) | 80 (0.71–0.86) |
| Presence of a mural nodule (n = 245)a | 44 (0.34–0.55) | 80 (0.72–0.85) | 55 (0.43–0.67) | 71 (0.64–0.78) |
| Increasing index cyst size (n = 125)a | 50 (0.34–0.66) | 54 (0.43–0.65) | 36 (0.24–0.49) | 68 (0.55–0.79) |
| Malignant cytopathologyc | 46 (0.35–0.56) | 95 (0.90–0.98) | 84 (0.70–0.92) | 75 (0.68–0.81) |
| IPMN and MCN | ||||
| MAPK/GNAS mutations | 90 (0.85–0.94) | 100 (0.93–1.00) | 100 (0.97–1.00) | 77 (0.66–0.85) |
| Increased fluid viscosity (n = 238)a | 77 (0.70–0.83) | 92 (0.81–0.97) | 97 (0.92–0.99) | 57 (0.47–0.67) |
| Elevated CEA (n = 173)a | 73 (0.64–0.80) | 94 (0.79–0.99) | 98 (0.93–1.00) | 46 (0.34–0.58) |
| IPMN and MCN with advanced neoplasia | ||||
| TP53, SMAD4, and/or mTOR gene alterations | 88 (0.79–0.93) | 79 (0.72–0.85) | 73 (0.74–0.81) | 91 (0.84–0.95) |
| TP53, SMAD4, CTNNB1, and/or mTOR gene alterations | 90 (0.81–0.95) | 77 (0.70–0.84) | 72 (0.63–0.79) | 92 (0.86–0.96) |
| MAPK/GNAS mutations with TP53, SMAD4, and/or mTOR gene alterations | 85 (0.76–0.92) | 96 (0.91–0.98) | 93 (0.85–0.97) | 91 (0.85–0.95) |
| MAPK/GNAS mutations with TP53, SMAD4, CTNNB1, and/or mTOR gene alterations | 88 (0.79–0.93) | 95 (0.89–0.98) | 91 (0.83–0.96) | 92 (0.87–0.96) |
| MAPK/GNAS LOH or TP53, SMAD4, and/or mTOR gene AFs = MAPK/GNAS AFs | 87 (0.78–0.92) | 99 (0.95–1.00) | 98 (0.91–1.00) | 92 (0.86–0.96) |
| MAPK/GNAS LOH or TP53, SMAD4, CTNNB1, and/or mTOR gene AFs = MAPK/GNAS AFs | 89 (0.80–0.94) | 98 (0.94–1.00) | 97 (0.90–0.99) | 93 (0.88–0.96) |
| Associated clinical symptoms (n = 244)a | 38 (0.28–0.48) | 72 (0.64–0.79) | 46 (0.35–0.58) | 64 (0.56–0.71) |
| Jaundice (n = 131)b | 31 (0.20–0.44) | 89 (0.78–0.95) | 70 (0.50–0.86) | 60 (0.50–0.69) |
| Index cyst size >3.0 cm (n = 242)a | 59 (0.48–0.68) | 57 (0.48–0.65) | 46 (0.37–0.56) | 68 (0.59–0.76) |
| Main pancreatic duct dilatation (n = 244)a | 68 (0.58–0.77) | 65 (0.57 – 0.73) | 56 (0.46–0.65) | 76 (0.68–0.83) |
| Presence of a mural nodule (n = 245)a | 45 (0.35–0.56) | 81 (0.74–0.87) | 61 (0.48–0.72) | 70 (0.63–0.77) |
| Increasing index cyst size (n = 125)a | 52 (0.37–0.67) | 56 (0.44–0.67) | 39 (0.27–0.53) | 68 (0.55–0.79) |
| Malignant cytopathologyc | 46 (0.36–0.56) | 97 (0.92–0.99) | 90 (0.77–0.96) | 74 (0.67–0.80) |
NOTE. MAPK genes include KRAS, BRAF, and NRAS; while mTOR genes include PIK3CA, PTEN, and AKT1.
n designates the number of patients with data available for analysis.
Jaundice was evaluated for patients with a cyst in the pancreatic head, uncinate and/or neck.
Malignant cytopathology was defined as at least suspicious for adenocarcinoma.
Considering current pancreatic cyst guidelines have primarily focused on detecting advanced neoplasia in IPMNs, a subanalysis of combined PancreaSeq testing and cytopathologic evaluation among the 167 resected IPMNs revealed a sensitivity and a specificity of 88% and 96%, respectively (Supplementary Table 11). A comparison of the absolute criteria for surgical management from the AGA guidelines and the IAP/Fukuoka guidelines showed lower sensitivities (72% and 86%) and lower specificities (66% and 36%) than PancreaSeq and cytopathologic evaluation. Incorporating PancreaSeq testing as another criterion to the AGA guidelines did increase the sensitivity of each alone to 96%, but the specificity was 62%. Similarly, combining PancreaSeq testing to the IAP/Fukuoka guidelines improved the sensitivity to 98%, but at a specificity of 34%. However, in the prospective clinical setting, distinguishing between IPMNs with advanced neoplasia and for that matter mucinous cysts with advanced neoplasia from other neoplastic and non-neoplastic pancreatic cysts can be challenging. Therefore, we evaluated the AGA guidelines, the IAP/Fukuoka guidelines, and PancreaSeq testing in their ability to identify IPMNs and MCNs with advanced neoplasia among the 246 pancreatic cysts with diagnostic pathology. As per the AGA guidelines, the sensitivity and specificity for advanced neoplasia within a mucinous cyst was 72% and 75%, respectively, while the IAP/Fukuoka guidelines yielded a sensitivity of 84% and a specificity of 52%. The addition of PancreaSeq testing to the AGA guidelines and the IAP/Fukuoka guidelines increased the sensitivities of both guidelines to 96% and 98%, respectively, but the specificities remained essentially the same at 73% and 51%, respectively.
Although the number of resected serous neoplasms was limited, the preoperative identification of VHL alterations in the absence of other genomic alterations had a sensitivity and specificity of 71% and 100%, respectively. Further, the inclusion of point mutations in TP53 or the TERT promoter increased the sensitivity to 100% and the specificity remained at 100%. In comparison, cytopathology was consistent with a serous neoplasm for only 1 patient, whereas the mixed serous-neuroendocrine neoplasm was misdiagnosed as a PDAC in another patient.
For cystic PanNETs, MEN1 alterations in preoperative pancreatic cyst fluid were associated with a sensitivity and specificity of 27% and 100%, respectively. However, the inclusion of LOH for TP53, SMAD4, PTEN, and/or RNF43 improved the sensitivity to 68%, while the specificity remained high at 98%. A preoperative cytopathologic diagnosis of a neuroendocrine tumor had an 85% sensitivity and 100% specificity, and combination of PancreaSeq testing and cytopathology yielded a sensitivity of 97% and a specificity of 98%. Further, the association with metastatic progression increased with the number of genes exhibiting LOH. An LOH of ≥3 genes had a sensitivity and specificity of 83% and 76%, respectively, for distant metastasis (Table 2). Comparatively, preoperative tumor size of >2.0 cm and preoperative histologic grade of at least G2 had sensitivities of 92% and 75%, respectively, and specificities of 50% and 74%, respectively, for distant metastasis. Interestingly, among 31 patients with cystic PanNET, 19 patients had tumors of 1.0 to 2.0 cm and only 1 of the 19 patients developed metastatic progression. This WHO grade 1, cystic PanNET harbored LOH for VHL, TP53, SMAD4, PTEN, and RNF43. Overall, the key genomic alterations detected by PancreaSeq and clinical significance are summarized in Supplementary Figure 3.
Table 2.
Diagnostic Performance of PancreaSeq Testing and Other Modalities for Serous Neoplasms and PanNETs
| Parameter | Sensitivity, % (95% CI) |
Specificity, % (95% CI) |
PPV, % (95% CI) |
NPV, % (95% CI) |
|---|---|---|---|---|
| Serous cystadenoma/neoplasma | ||||
| VHL alteration in the absence of other alterations | 71 (0.42–0.90) | 100 (0.97–1.00) | 100 (0.66–1.00) | 98 (0.95–1.00) |
| VHL alteration w/ or w/o point mutations in TP53 and TERT promoter | 100 (0.73–1.00) | 100 (0.97–1.00) | 100 (0.73–1.00 | 100 (0.97–1.00) |
| PanNETb | ||||
| MEN1 alteration in the absence of other alterations | 27 (0.14–0.45) | 100 (0.98–1.00) | 100 (0.63–1.00) | 90 (0.85–0.93) |
| LOHc in the absence of other alterations | 59 (0.41–0.75) | 98 (0.95–0.99) | 83 (0.62–0.95) | 94 (0.89–0.96) |
| MEN1 alteration w/ or w/o LOHc in the absence of other alterations | 68 (0.49–0.82) | 98 (0.95–0.99) | 85 (0.65–0.95) | 95 (0.91–0.97) |
| Cytopathology positive for neuroendocrine tumor | 85 (0.68–0.95) | 100 (0.97–1.00) | 97 (0.81–1.00) | 98 (0.94–0.99) |
| MEN1 alteration w/ or w/o LOHc and cytopathology | 97 (0.83–1.00) | 98 (0.95–0.99) | 89 (0.74–0.97) | 100 (0.97–1.00) |
| Metastatic PanNETd | ||||
| LOH of at least 1 genee | 92 (0.60–1.00) | 49 (0.37–0.61) | 23 (0.13–0.38) | 97 (0.84–1.00) |
| LOH of at least 2 genese | 92 (0.60–1.00) | 68 (0.56–0.78) | 32 (0.18–0.51) | 98 (0.88–1.00) |
| LOH of at least 3 genese | 83 (0.51–0.97) | 76 (0.65–0.85) | 37 (0.20–0.57) | 97 (0.87–0.99) |
| LOH of at least 4 genese | 58 (0.29–0.84) | 88 (0.77–0.94) | 44 (0.21–0.70) | 93 (0.83–0.97) |
| LOH of at least 5 genese | 33 (0.11–0.64) | 93 (0.84–0.97) | 44 (0.15–0.77) | 89 (0.80–0.95) |
| Preoperative tumor size >2.0 cm | 92 (0.60–1.00) | 50 (0.38–0.62) | 23 (0.13–0.38) | 97 (0.84–1.00) |
| Preoperative cytopathology WHO grades 2 and 3 | 75 (0.43–0.93) | 74 (0.62–0.83) | 32 (0.17–0.52) | 95 (0.84–0.99) |
Based on 246 diagnostically confirmed pancreatic cysts that includes 13 serous cystadenomas and 1 mixed serous cystadenoma-neuroendocrine neoplasm.
Based on 246 diagnostically confirmed pancreatic cysts that includes 34 cystic PanNETs.
LOH of TP53, SMAD4, PTEN, and/or RNF43.
Based on 87 preoperative specimens (34 cystic PanNETs and 53 solid PanNETs) with patient follow-up.
LOH of VHL, TP53, SMAD4, PTEN, and/or RNF43.
Comparative PancreaSeq/Oncomine Testing of Paired Pancreatic Cyst Fluid and Diagnostic Surgical Pathology Specimens
Repeat PancreaSeq and expanded targeted DNA/RNA-based (Oncomine) NGS testing were performed for 192 of 251 (77%) diagnostic surgical pathology specimens (Supplementary Table 12). Discordances between preoperative and postoperative testing were identified in 25 cases and exclusively seen in IPMNs (Figure 3). Of interest, 9 discrepant cases were due to the lack of detectable MAPK/GNAS mutations in preoperative pancreatic cyst fluid specimens. For the remaining 16 cases, discrepancies were seen in RNF43 (n = 8), TP53 (n = 7), SMAD4 (n = 2), CTNNB1 (n = 1), and the mTOR genes (n = 3), but did not affect the overall sensitivity and specificity of PancreaSeq testing. In addition, Oncomine testing found 4 MAPK-negative IPMNs harboring BRAF fusions (n = 3) and ERBB2 amplification (n = 1) (Supplementary Figure 4). To further characterize BRAF-mutant IPMNs, whole transcriptome sequencing revealed a similar gene expression profile as KRAS-mutant IPMNs (Supplementary Material and Supplementary Figure 5). Additional genomic alterations found among IPMNs included those involving CDKN2A (18 of 131 IPMNs, 14%) and ARID1A (n = 6, 4%). CDKN2A alterations were only detected in IPMNs with advanced neoplasia (18 of 75 cases). Two IPMNs with advanced neoplasia that harbored CDKN2A alterations also lacked alterations in TP53, SMAD4, CTNNB1, and the mTOR genes.
Discussion
Despite retrospective studies and single institutional experiences, questions remain as to whether DNA-based targeted NGS can improve pancreatic cyst classification and the detection of advanced neoplasia arising in a mucinous cyst.10-13,19 Based on a multi-institutional, prospectively collected cohort of patients with pancreatic cyst who were evaluated using a centralized molecular laboratory, mutations in the MAPK genes and/or GNAS achieved a sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for mucinous cysts of 90%, 100%, 100%, and 77%, respectively. Both fluid viscosity and elevated CEA levels demonstrated lower sensitivities and lower specificities. Combining PancreaSeq testing with CEA analysis increased the sensitivity to 99%, but at a loss in specificity of 73%. Similarly, MAPK/GNAS LOH or TP53, SMAD4, and/or mTOR gene alterations with equivalent allele frequencies to MAPK/GNAS mutations attained 87% sensitivity, 99% specificity, 98% PPV, and 92% NPV for advanced neoplasia. The identification of advanced neoplasia was further improved with the inclusion of CTNNB1 mutations and yielded a sensitivity, specificity, PPV, and NPV of 89%, 98%, 97%, and 93%, respectively. Moreover, the combination of PancreaSeq testing and cytopathologic evaluation achieved a 93% sensitivity, a 95% specificity, a 92% PPV, and a 95% NPV for advanced neoplasia.
More importantly, the incorporation of PancreaSeq testing to current IPMN-specific guidelines, such as those by the AGA guidelines and the IAP/Fukuoka guidelines, increased the sensitivities of detecting advanced neoplasia from 72% to 96% and 86% to 98%, respectively, whereas the specificities of both guidelines remained essentially the same. Considering the challenges of classifying pancreatic cysts within the preoperative setting, a separate analysis of mucinous cysts (IPMNs and MCNs) with advanced neoplasia also revealed an improvement in the sensitivities of the AGA guidelines (72% to 96%) and the IAP/Fukuoka guidelines (84% to 98%) when applying PancreaSeq testing data, while the specificities of both guidelines once again remained essentially the same. The advantage of PancreaSeq testing is its high specificity for advanced neoplasia. In contrast, the AGA guidelines and the IAP/Fukuoka Guidelines exhibit low-to-moderate specificity, but moderate-to-high sensitivity. The low-to-moderate specificity of both guidelines is not surprising, as they rely on subjective and indirect features of advanced neoplasia, such as large (>3.0 cm) pancreatic cyst size, main pancreatic duct dilatation, and the presence of a mural nodule on EUS. As reported in the AGA technical review, cyst size of >3.0 cm has a pooled sensitivity of 74% for malignancy, but a poor pooled specificity of 49%.8 Main pancreatic duct dilatation and the presence of a mural nodule have pooled specificities of 80% and 91%, respectively, but poor pooled sensitivities of 32% and 48%, respectively.16 The sensitivity and specificity of a mural nodule can be highly variable and largely attributable to the challenges in differentiating a mural nodule from adherent mucus within the pancreatic cyst by EUS.26 The issues with EUS are compounded when factoring interobserver variability and operator dependence.27 However, the utility of EUS is enhanced when coupled with FNA and cytopathologic evaluation of pancreatic cyst fluid. Cytopathologic evaluation for advanced neoplasia closely approaches 100% specificity, but it is often hampered by the low cellular content of pancreatic cyst fluid.28 Nevertheless, in the absence of overt malignancy, differentiating high-grade from low-grade dysplasia can be problematic. In addition, distinguishing neoplastic cells from gastrointestinal tract contamination is often problematic, but imperative to establishing a diagnosis. Thus, the reported sensitivity of cytopathology for malignancy can vary widely from 25% to 88%.8,10,11,19,29,30
Although this study confirms the diagnostic utility of DNA-based targeted NGS, it also expands the compendium of MAPK alterations among pancreatic cysts. For instance, BRAF alterations were found in 5% of all pancreatic cysts and only 8% of BRAF-mutant cysts had co-occurring KRAS mutations. Most BRAF alterations were categorized as class II and class III and included in-frame deletions of codon 486. Previous studies have found class II and class III BRAF alterations, especially in-frame deletions, are often mutually exclusive of KRAS mutations and activate the MAPK signaling pathway.31,32 Based on diagnostic surgical pathology, BRAF alterations detected within this study correlated with the presence of an IPMN. Comparative RNA sequencing revealed BRAF-mutant IPMNs had similar gene expression profiles as KRAS-mutant IPMNs. In addition, through expanded targeted DNA/RNA-based NGS testing of MAPK-negative IPMNs, the spectrum of BRAF alterations was expanded to include fusion genes. The relationship between BRAF alterations and IPMNs is also interesting. For the entire prospectively collected pancreatic cyst cohort, 77% of BRAF-mutant pancreatic cysts harbored GNAS mutations, which are known to be specific for IPMNs. Although diagnostic surgical pathology was unavailable, Ren et al33 reported the association between BRAF and GNAS mutations for 6 pancreatic cysts that were clinically consistent with IPMNs. Hence, BRAF alterations are likely to substitute for KRAS mutations as a driver of the MAPK pathway in the pathogenesis of IPMNs.
An unexpected finding from this study was the identification of pancreatic cysts harboring VHL alterations and either TP53 or TERT promoter mutations. Consistent with prior studies, alterations in VHL alone were specific to serous cystic neoplasms.12,13,18 In addition, the combination of VHL alterations and mutations in TP53 or the TERT promoter correlated with an SCA. However, based on surveillance imaging, SCAs with these additional alterations demonstrated interval growth in size. In fact, the growth of one VHL/TP53-mutant SCA resulted in progressive stricturing of the main pancreatic duct, and, consequently, the patient developed acute and chronic pancreatitis. Although SCAs are benign and the overwhelming majority are asymptomatic, and slow growing, a subset can demonstrate increased growth and associated symptomatology.34 Tseng et al35 reported that patients with SCAs demonstrating a high growth rate (1.98 cm/y) and presented with abdominal pain, fullness and/or jaundice. Similarly, El-Hayek et al36 found symptomatic patients often exhibited rapid growth of their SCA. In both studies, correlative molecular testing was not performed and, therefore, it is intriguing to surmise that clinically significant growth of an SCA and, consequently, symptomatology due to an SCA, may be associated with the development of a mutation in TP53 or the TERT promoter.
Finally, MEN1 alterations were highly specific for cystic PanNETs, but the sensitivity was only 27%. The sensitivity for cystic PanNETs improved to 68% on inclusion of LOH at the TP53, SMAD4, PTEN, and/or RNF43 genomic loci. In comparison, cytopathologic evaluation achieved a sensitivity and specificity of 85% and 100%, respectively. However, the combination of cytopathologic evaluation and PancreaSeq testing yielded a 97% sensitivity and a 98% specificity for a cystic PanNET. To date, available sequencing data for cystic PanNETs are limited, but solid PanNETs are reported to harbor recurrent LOH at multiple genomic loci with a prevalence greater than MEN1 alterations.37-39 As described herein, LOH was similarly present in cystic PanNETs and more frequently seen than alterations in MEN1. Moreover, within a combined cohort of solid and cystic PanNETs, LOH for at least 1 gene was associated with several adverse prognostic features. Both Pea et al38 and Lawrence et al40 published related findings with LOH of multiple genomic loci correlating with an increased risk of distant metastasis. LOH of ≥3 genes within the PanNET study cohort had a sensitivity and specificity of 83% and 76%, respectively, for metastatic spread.
Analogous to mucinous cysts of the pancreas, both solid and cystic PanNETs are increasing in prevalence and often incidentally identified by radiographic imaging. While many patients with PanNET develop rapid and widely metastatic disease, other patients may present with indolent and slow-growing disease.41,42 In fact, the overtreatment of PanNETs has been a subject of debate and an observational approach may be warranted for a subset of patients.43-46 Despite the development of PanNET prognostic classification systems, such as WHO histologic grading, and tumor staging systems, such as those based on tumor size of >2.0 cm, these parameters do not necessarily reflect the pathobiology of these tumors.47,48 LOH of at least 3 genes was associated with a higher specificity (76%) for distant metastasis than >2.0 cm tumor size (50%) and advanced WHO grade (grades 2 and 3, 74%). Moreover, LOH was superior in sensitivity (83%) than advanced WHO grade (75%). Interestingly, LOH was also associated with loss of expression of ATRX/DAXX and the presence of ALT. Although the exact mechanism has not been fully elucidated, ATRX and DAXX play an integral role in telomere maintenance, and loss of protein expression coincides with the presence of ALT, a telomerase-independent telomere maintenance mechanism.49,50 Interestingly, ALT results in broad chromosomal abnormalities, and, therefore, it is plausible that the LOH found at multiple genomic loci in PanNETs is the sequelae of ALT and may reflect a common genomic pathway in the metastatic progression of PanNETs.
We acknowledge that there are several limitations to this study. Although a large number of pancreatic cysts were analyzed, diagnostic surgical pathology was available for only 14% of patients and represents a surgical selection bias. However, clinical follow-up was also obtained for an additional 52% of patients. Our study also suffers from a testing selection bias, as pancreatic cyst fluid specimens satisfactory for targeted NGS were used for analysis. Considering a 2% failure rate of PancreaSeq testing, the effect of this selection bias is likely to be minimal. Nonetheless, molecularly discordant results were identified when comparing preoperative and postoperative specimens. For instance, MAPK/GNAS alterations were not detected in 9 surgically resected IPMNs, but present within the corresponding surgical specimen, which underscores a potential issue of sensitivity for PancreaSeq testing. Alternative explanations for this discordance are the absence of exfoliated neoplastic cells within the pancreatic cyst fluid, degraded mutant DNA within the cyst, and adequate sampling of the pancreatic cyst by the gastroenterologist. In addition, the follow-up period of this study is relatively short to assess the clinical impact of detecting specific genomic alterations, such as TP53, SMAD4, CTNNB1, and the mTOR genes. Although we plan to continue monitoring patients with these genomic alterations, the median duration of follow-up was 23 months or close to 2 years, which by many pancreatic cyst guidelines is sufficient as the initial time interval for imaging surveillance.6,7,9,21 Another limitation is the relative paucity of certain genomic alterations to determine their true clinical significance. For example, the inclusion of CTNNB1 to the assessment of MAPK/GNAS-mutant pancreatic cysts improved the identification of advanced neoplasia, but this was based on only 4 diagnostically confirmed IPMNs harboring CTNNB1 alterations. Moreover, despite PancreaSeq consisting of 22 pancreatic cyst-related genes, it did not include other potentially important genes, such as CDKN2A. Several studies have reported recurrent genomic alterations in CDKN2A in a subset of mucinous cysts and preferentially those with advanced neoplasia.12 Similarly, we found CDKN2A alterations were detected in only IPMNs and those IPMNs with advanced neoplasia at a prevalence of 24%. In addition, 2 IPMNs with advanced neoplasia that were negative for alterations in TP53, SMAD4, CTNNB1, and the mTOR genes harbored CDKN2A alterations. Hence, further studies are required to determine the clinical significance of CDKN2A alterations among pancreatic cysts. Moreover, as the identification of BRAF alterations to include fusion genes highlights, the full breadth of genomic alterations that characterize pancreatic cysts has yet to be determined. A complicated issue with this study is the incorporation of allele frequencies to improve the performance of PancreaSeq testing. As we reported previously, low-level genomic alterations in TP53 and PIK3CA with respect to MAPK/GNAS mutations can be seen in the setting of IPMNs with low-grade dysplasia and it is plausible that these IPMNs are at an increased risk of progression to advanced neoplasia. Admittingly, the current study does not address the malignant potential of this patient population but highlights the increasing complexity of genomic alterations that characterize pancreatic cystic neoplasms. To simplify reporting of key alterations to include allele frequencies, our group is in the process of developing a pancreatic cyst molecular classifier to aid in the interpretation of genomic variants and provide surveillance/treatment guidance to both gastroenterologists and surgeons (Nikiforova and Singhi, unpublished results, 2022). Last, this study does not address the optimal approach of integrating targeted NGS testing to current pancreatic cyst surveillance protocols. As an example, the European evidence-based guidelines could not be applied to this study cohort due to the lack of sufficient data to determine “relative indications” for surgical management. None of the guidelines, however, have sufficient accuracy to dictate appropriate surveillance and management of pancreatic cysts, are admittingly based on “very low quality of evidence,” and, not surprisingly, the institutions participating within this study followed different pancreatic cyst guidelines and, in many cases, utilized a personalized approach for their patients.6,7,9,21,51-53 A major step forward in delineating an optimal pancreatic cyst protocol is the ECOG-ACRIN pancreatic cyst surveillance clinical trial of >4000 patients that will compare the effectiveness between the AGA guidelines and the IAP/Fukuoka guidelines.54 As a secondary aim of this study, biospecimens will be collected from enrolled patients to assess the utility of promising pancreatic cyst biomarkers.
In summary, we report the results of a large, multi-institutional, prospective, and real-time study that clinically applies targeted NGS testing of EUS-FNA-obtained preoperative pancreatic cyst fluid to the evaluation of pancreatic cysts. Overall, our results underscore the clinical utility of targeted NGS given its high sensitivity and high specificity in the diagnosis of mucinous cysts and the identification of advanced neoplasia within a mucinous cyst. This study also broadens the number of genomic alterations that characterize not only mucinous cysts, but SCAs and cystic PanNETs. Although we recognize that additional studies are required, the data reported herein combined with previous studies support the integration of targeted NGS into the establishment of evidence-based pancreatic cyst guidelines.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
While previous studies have shown targeted next-generation sequencing is a useful adjunct to the preoperative evaluation of pancreatic cysts, these studies have largely been retrospective analyses, single institutional experiences, and focused on intraductal papillary mucinous neoplasms.
NEW FINDINGS
Through prospective, real-time, multi-institutional next-generation sequencing (PancreaSeq) of a large patient cohort, a diverse number of genomic alterations were identified in intraductal papillary mucinous neoplasms (eg, BRAF), serous cystadenomas (eg, TP53 and TERT), and pancreatic neuroendocrine tumors (eg, loss of heterozygosity of multiple genes) and are of associated clinical significance.
LIMITATIONS
Considering most pancreatic cysts follow a benign clinical course, diagnostic surgical pathology was available for 14% of tested patients. However, clinical follow-up with a median of 23 months was available for an additional 52% of patients.
IMPACT
The results of this study support the clinical utility of targeted next-generation sequencing in the evaluation of not only pancreatic mucinous cysts, but other cyst types. This study also broadens the number of genomic alterations that characterize pancreatic cysts.
Acknowledgments
The authors thank the clinical coordinating staff (Tracy Hoteck, Laura A. Osman, Nicole R. Habel, Leslie Minteer, and Kara Kirkpatrick) at the UPMC Digestive Disorders Center. In addition, special thanks to Mrs. Lynn Wolkenstein for outstanding administrative assistance.
Funding
This study was supported in part by the National Cancer Institute (1R37CA263622 and 3U01CA200466), Department of Defense (W81XWH-20-PCARP-TRPA), Pancreatic Cancer Action Network, the National Pancreas Foundation, Western PA Chapter, the Sky Foundation, and the Pittsburgh Liver Research Center at the University of Pittsburgh (P30DK120531).
Abbreviations used in this paper:
- AF
allele frequency
- ALT
alternative lengthening of telomeres
- CEA
carcinoembryonic antigen
- EUS
endoscopic ultrasound
- FNA
fine-needle aspiration
- IPMN
intraductal papillary mucinous neoplasm
- LOH
loss of heterozygosity
- MAPK
mitogen-activated protein kinase
- MCN
mucinous cystic neoplasm
- MGP
Molecular and Genomic Pathology
- mTOR
mammalian target of rapamycin
- NGS
next-generation sequencing
- NPV
negative predictive value
- PanNET
pancreatic neuroendocrine tumor
- PDAC
pancreatic ductal adenocarcinoma
- PPV
positive predictive value
- SCA
serous cystadenoma
- UPMC
University of Pittsburgh Medical Center
- WHO
World Health Organization
Footnotes
CRediT Authorship Contributions
Order of Authors (with Contributor Roles):
Alessandro Paniccia, MD (Conceptualization: Equal; Data curation: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Patricio M. Polanco, MD (Conceptualization: Equal; Data curation: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Brian A. Boone, MD (Conceptualization: Equal; Data curation: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Abigail I. Wald, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Equal; Writing – review & editing: Equal).
Kevin McGrath, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Randall E. Brand, MD (Conceptualization: Supporting; Data curation: Supporting; Funding acquisition: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Asif Khalid, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Nisa Kubiliun, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Anne Marie O’Broin-Lennon, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Walter G. Park, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Jason Klapman, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Benjamin Tharian, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Sumant Inamdar, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Kenneth Fasanella, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
John Nasr, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Jennifer Chennat, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Rohit Das, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
John DeWitt, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Jeffrey J. Easier, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Benjamin Bick, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Harkirat Singh, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Kimberly J. Fairley, DO (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Savreet Sarkaria, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Tarek Sawas, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Wasseem Skef, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Adam Slivka, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Anna Tavakkoli, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Shyam Thakkar, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Victoria Kim, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Hendrikus Dutch Vanderveldt, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Allyson Richardson, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Michael B. Wallace, MD, MPH (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Bhaumik Brahmbhatt, MBBS (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Megan Engels, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Charles Gabbert, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Mohannad Dugum, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Samer El-Dika, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Yasser Bhat, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Sanjay Ramrakhiani, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Gennadiy Bakis, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Daniil Rolshud, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Gordon Millspaugh, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Thomas Tielleman, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Carl Schmidt, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
John Mansour, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Wallis Marsh, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Melanie Ongchin, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Barbara Centeno, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Sara E. Monaco, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
N. Paul Ohori, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Sigfred Lajara, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Elizabeth D. Thompson, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Ralph H. Hruban, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Phoenix D. Bell, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Katelyn Smith, BA (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Jennifer Permuth, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Christopher Vandenbussche, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Wayne Ernst, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Maria Grupillo, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Cihan Kaya, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Melissa Hogg, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Jin He, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Christopher L. Wolfgang, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Kenneth K. Lee, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Herbert Zeh, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Amer Zureikat, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Marina N. Nikiforova, MD (Conceptualization: Supporting; Data curation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Aatur Singhi, MD PhD (Conceptualization: Lead; Data curation: Lead; Funding acquisition: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Conflicts of interest
These authors disclose the following: Aatur D. Singhi has received an honorarium from Foundation Medicine, Inc. Ralph H. Hruban has the potential to receive royalty payments from Thrive Earlier Detection for the GNAS invention in an arrangement reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. The remaining authors disclose no conflicts.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2022.09.028.
Data Availability
Study data not present within this manuscript to include but not limited to genomic data and other associated clinical and imaging metadata are available on request.
References
- 1.Gardner TB, Glass LM, Smith KD, et al. Pancreatic cyst prevalence and the risk of mucin-producing adenocarcinoma in US adults. Am J Gastroenterol 2013;108:1546–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laffan TA, Horton KM, Klein AP, et al. Prevalence of unsuspected pancreatic cysts on MDCT. AJR Am J Roentgenol 2008;191:802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee KS, Sekhar A, Rofsky NM, et al. Prevalence of incidental pancreatic cysts in the adult population on MR imaging. Am J Gastroenterol 2010;105:2079–2084. [DOI] [PubMed] [Google Scholar]
- 4.Singhi AD, Koay EJ, Chari ST, et al. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology 2019;156:2024–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marinelli V, Secchettin E, Andrianello S, et al. Psychological distress in patients under surveillance for intraductal papillary mucinous neoplasms of the pancreas: The "Sword of Damocles" effect calls for an integrated medical and psychological approach a prospective analysis. Pancreatology 2020;20:505–510. [DOI] [PubMed] [Google Scholar]
- 6.Elta GH, Enestvedt BK, Sauer BG, et al. ACG clinical guideline: diagnosis and management of pancreatic cysts. Am J Gastroenterol 2018;113:464–479. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Fernandez-Del Castillo C, Kamisawa T, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017;17:738–753. [DOI] [PubMed] [Google Scholar]
- 8.Scheiman JM, Hwang JH, Moayyedi P. American gastroenterological association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015;148:824–848.e22. [DOI] [PubMed] [Google Scholar]
- 9.European Study Group on Cystic Tumours of the Pancreas. European evidence-based guidelines on pancreatic cystic neoplasms. Gut 2018;67:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singhi AD, McGrath K, Brand RE, et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018;67:2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones M, Zheng Z, Wang J, et al. Impact of next-generation sequencing on the clinical diagnosis of pancreatic cysts. Gastrointest Endosc 2016;83:140–148. [DOI] [PubMed] [Google Scholar]
- 12.Springer S, Masica DL, Dal Molin M, et al. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med 2019;11:eaav4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer S, Wang Y, Dal Molin M, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015;149:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011;3:92ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikiforova MN, Khalid A, Fasanella KE, et al. Integration of KRAS testing in the diagnosis of pancreatic cystic lesions: a clinical experience of 618 pancreatic cysts. Mod Pathol 2013;26:1478–1487. [DOI] [PubMed] [Google Scholar]
- 16.Singhi AD, Nikiforova MN, Fasanella KE, et al. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin Cancer Res 2014;20:4381–4389. [DOI] [PubMed] [Google Scholar]
- 17.Amato E, Molin MD, Mafficini A, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol 2014;233:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Jiao Y, Dal Molin M, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A 2011;108:21188–21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbaum MW, Jones M, Dudley JC, et al. Next-generation sequencing adds value to the preoperative diagnosis of pancreatic cysts. Cancer 2017;125:41–47. [DOI] [PubMed] [Google Scholar]
- 20.Lokuhetty D, White V, Watanabe R, et al. Digestive system tumours: WHO classification of tumours. Lyon: International Agency for Research on Cancer, 2019. [Google Scholar]
- 21.Vege SS, Ziring B, Jain R, et al. American Gastroenterological Association institute guideline on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015;148:819–822; quiz 12–3. [DOI] [PubMed] [Google Scholar]
- 22.Singhi AD, Wood LD, Parks E, et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology 2020;158:573–582.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasso C, Butler T, Rhodes K, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn 2015;17:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikiforova MN, Wald AI, Melan MA, et al. Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol 2016;18:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong N, Zhang L, Takahashi N, et al. Histologic and imaging features of mural nodules in mucinous pancreatic cysts. Clin Gastroenterol Hepatol 2012;10:192–198; 198.e1–2. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad NA, Kochman ML, Brensinger C, et al. Interobserver agreement among endosonographers for the diagnosis of neoplastic versus non-neoplastic pancreatic cystic lesions. Gastrointest Endosc 2003;58:59–64. [DOI] [PubMed] [Google Scholar]
- 28.Pitman MB, Lewandrowski K, Shen J, et al. Pancreatic cysts: preoperative diagnosis and clinical management. Cancer Cytopathol 2010;118:1–13. [DOI] [PubMed] [Google Scholar]
- 29.Maker AV, Lee LS, Raut CP, et al. Cytology from pancreatic cysts has marginal utility in surgical decision-making. Ann Surg Oncol 2008;15:3187–3192. [DOI] [PubMed] [Google Scholar]
- 30.Khalid A, Brugge W. ACG practice guidelines for the diagnosis and management of neoplastic pancreatic cysts. Am J Gastroenterol 2007;102:2339–2349. [DOI] [PubMed] [Google Scholar]
- 31.Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singhi AD, George B, Greenbowe JR, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology 2019;156:2242–2253.e4. [DOI] [PubMed] [Google Scholar]
- 33.Ren R, Krishna SG, Chen W, et al. Activation of the RAS pathway through uncommon BRAF mutations in mucinous pancreatic cysts without KRAS mutation. Mod Pathol 2021;34:438–444. [DOI] [PubMed] [Google Scholar]
- 34.Fukasawa M, Maguchi H, Takahashi K, et al. Clinical features and natural history of serous cystic neoplasm of the pancreas. Pancreatology 2010;10:695–701. [DOI] [PubMed] [Google Scholar]
- 35.Tseng JF, Warshaw AL, Sahani DV, et al. Serous cystadenoma of the pancreas: tumor growth rates and recommendations for treatment. Ann Surg 2005;242:413–419; discussion 419–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Hayek KM, Brown N, O’Rourke C, et al. Rate of growth of pancreatic serous cystadenoma as an indication for resection. Surgery 2013;154:794–800; discussion 800–802. [DOI] [PubMed] [Google Scholar]
- 37.Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017;543:65–71. [DOI] [PubMed] [Google Scholar]
- 38.Pea A, Yu J, Marchionni L, et al. Genetic analysis of small well-differentiated pancreatic neuroendocrine tumors identifies subgroups with differing risks of liver metastases. Ann Surg 2020;271:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy S, LaFramboise WA, Liu TC, et al. Loss of chromatin-remodeling proteins and/or CDKN2A associates with metastasis of pancreatic neuroendocrine tumors and reduced patient survival times. Gastroenterology 2018;154:2060–2063.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence B, Blenkiron C, Parker K, et al. Recurrent loss of heterozygosity correlates with clinical outcome in pancreatic neuroendocrine cancer. NPJ Genom Med 2018;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dasari A, Shen C, Halperin D, et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol 2017;3:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heaphy CM, Singhi AD. The diagnostic and prognostic utility of incorporating DAXX, ATRX, and alternative lengthening of telomeres (ALT) to the evaluation of pancreatic neuroendocrine tumors (PanNETs). Hum Pathol 2022;129:11–20. [DOI] [PubMed] [Google Scholar]
- 43.Zhang IY, Zhao J, Fernandez-Del Castillo C, et al. Operative versus nonoperative management of nonfunctioning pancreatic neuroendocrine tumors. J Gastrointest Surg 2016;20:277–283. [DOI] [PubMed] [Google Scholar]
- 44.Sadot E, Reidy-Lagunes DL, Tang LH, et al. Observation versus resection for small asymptomatic pancreatic neuroendocrine tumors: a matched case-control study. Ann Surg Oncol 2016;23:1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz H, Howe JR, Pawlik TM. Surgery vs observation for patients with small pancreatic neuroendocrine tumors. JAMA Surg 2021;156:412–413. [DOI] [PubMed] [Google Scholar]
- 46.Gaujoux S, Partelli S, Maire F, et al. Observational study of natural history of small sporadic nonfunctioning pancreatic neuroendocrine tumors. J Clin Endocrinol Metab 2013;98:4784–4789. [DOI] [PubMed] [Google Scholar]
- 47.Assi HA, Mukherjee S, Kunz PL, et al. Surgery versus surveillance for well-differentiated, nonfunctional pancreatic neuroendocrine tumors: an 11-year analysis of the National Cancer Database. Oncologist 2020;25:e276–e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scarpa A, Mantovani W, Capelli P, et al. Pancreatic endocrine tumors: improved TNM staging and histopathological grading permit a clinically efficient prognostic stratification of patients. Mod Pathol 2010;23:824–833. [DOI] [PubMed] [Google Scholar]
- 49.Marinoni I, Kurrer AS, Vassella E, et al. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology 2014;146:453–460.e5. [DOI] [PubMed] [Google Scholar]
- 50.Hackeng W, Brosens LA, Kim JY, et al. Non-functional pancreatic neuroendocrine tumors: ATRX/DAXX and alternative lengthening of telomeres (ALT) assess prognosis independently from islet-cell subtype and tumor size. Gut 2022;71:961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singhi AD, Zeh HJ, Brand RE, et al. American Gastroenterological Association guidelines are inaccurate in detecting pancreatic cysts with advanced neoplasia: a clinicopathologic study of 225 patients with supporting molecular data. Gastrointest Endosc 2016;83:1107–1117.e2. [DOI] [PubMed] [Google Scholar]
- 52.Lee A, Kadiyala V, Lee LS. Evaluation of AGA and Fukuoka Guidelines for EUS and surgical resection of incidental pancreatic cysts. Endosc Int Open 2017;5:E116–E122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Wang Y, Li Z, et al. Accuracy of Fukuoka and American Gastroenterological association guidelines for predicting advanced neoplasia in pancreatic cyst neoplasm: a meta-analysis. Ann Surg Oncol 2019;26:4522–4536. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg DS, Gatsonis C, Zeh HJ, et al. Comparing the clinical impact of pancreatic cyst surveillance programs: A trial of the ECOG-ACRIN cancer research group (EA2185). Contemp Clin Trials 2020;97:106144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study data not present within this manuscript to include but not limited to genomic data and other associated clinical and imaging metadata are available on request.