Abstract
Immune checkpoint inhibitors (ICIs) have revolutionized cancer therapeutics, triggering studies to understand the molecular and cellular wiring of response and resistance. Our increased understanding of the underlying biology of response to ICI has enabled the investigation of tumor-intrinsic and -extrinsic features that may predict therapeutic outcomes. In parallel, liquid biopsy measurements of circulating tumor DNA (ctDNA) can be used to assess real-time molecular responses and guide clinical decisions during ICI. The combination of these approaches provides a deeper understanding of cancer biology, immunoediting, and evolution during ICI and promise to extend the utility of immunotherapies for patients with cancer.
CLINICAL LANDSCAPE OF IMMUNE CHECKPOINT INHIBITORS
Immune checkpoint inhibitors (ICIs) block regulatory pathways that dampen T cell activity and immune responses, mechanisms that are in place to achieve immunological tolerance (1). Cancer cells can co-opt immune checkpoints to evade immune surveillance, which has opened a therapeutic window of opportunity for ICI over the past decade. Since the 2011 approval of the prototypical ICI, the anti–CTLA-4 antibody ipilimumab, for the treatment of advanced melanoma (2), immune checkpoint inhibition has become a standard treatment option across solid tumors. As of early 2022, ICIs have been approved by the U.S. Food and Drug Administration (FDA) for the treatment of 17 distinct solid tumor histologies in addition to two tumor-agnostic indications for microsatellite instability (MSI)–high tumors (3).
Focusing on non–small cell lung cancer (NSCLC) as a notable example, the therapeutic landscape for advanced disease has radically changed. In 2012, treatment for most patients diagnosed with advanced NSCLC consisted of first-line platinum doublet chemotherapy followed by docetaxel single-agent chemotherapy at the time of progression, with no other standard treatment options available. After the demonstration in 2015 that the anti-programmed cell death protein-1 (PD-1) ICI, nivolumab, improved survival for patients with pretreated NSCLC, there has been a succession of new approvals to the extent that there are now multiple first-line combination and single-agent ICI treatment options for patients with newly diagnosed advanced NSCLC (4). Currently, with the exception of oncogene-addicted NSCLC, the vast majority of patients with NSCLC receive PD-1 or programmed cell death ligand 1 (PD-L1) containing therapy as a first-line treatment (5, 6). Clinical decision-making with respect to treatment selection for NSCLC is based on PD-L1 expression and genomic testing for tumor-specific (somatic) mutations; as such, PD-L1 immunohistochemistry is recommended for all newly diagnosed advanced NSCLC alongside with tumor next-generation sequencing (7).
Translation of ICI therapies from advanced to earlier stage disease has been slow but is lastly coming to fruition with improved outcomes associated with consolidation anti–PD-L1, durvalumab, for unresectable NSCLC after definitive chemoradiation and eradication of disease and both neoadjuvant and adjuvant ICI for resectable disease (8-10). Recent findings from the CheckMate 816 trial, comparing neoadjuvant chemo-immunotherapy to standard chemotherapy, have highlighted the potential for in vivo assessment of pathological complete response (pCR) to therapy as a potential early indicator of benefit from therapy (10).
In contrast to the clear clinical benefits reported with PD-1 pathway blockade, trial results from single-agent ICIs targeting nonredundant coinhibitory checkpoints have been largely disappointing thus far (11-13). Combined PD-1 and CTLA-4 blockade has been shown to be effective (14, 15); however, patients receiving anti–CTLA-4 agents have a lower incidence of immune-related adverse events, with ICI combinations increasing the incidence, severity, and onset of toxicities (16). In 2022, we have seen the first approval of an ICI-targeting LAG-3, relatlimab, in combination with anti–PD-1, nivolumab, for advanced melanoma, and thus it appears that the paradigm of adding new agents to a PD-1–blocking antibody backbone will continue to be implemented in clinical practice (17).
CLINICALLY INTEGRATED BIOMARKERS OF ICI RESPONSE
PD-L1 expression
To date, the most commonly used predictive biomarker for ICI response is PD-L1 expression as determined by immunohistochemistry; however, the clinical utility of PD-L1 testing varies based on the cancer type evaluated and the ICI therapy considered (18, 19). PD-L1 is expressed on tumor cells, and through its interaction with PD-1, the PD-1/PD-L1 axis regulates adaptive immune responses, ultimately promoting tumor escape from immune surveillance (20). Inhibition of PD-1/PD-L1 by anti–PD-1 or anti–PD-L1 monoclonal antibodies has been shown to be the most effective in tumors with high PD-L1 expression in cancer cells or tumor-infiltrating immune cells (21, 22), establishing the use of PD-L1 assays as a companion diagnostics test for ICI. PD-L1 status can be used to select patients most likely to attain longer survival with pembrolizumab across cancers—atezolizumab for urothelial cancer, NSCLC, and triple-negative breast cancer; ipilimumab and nivolumab for NSCLC; and cemiplimab for NSCLC (23). However, several phase 3 trials failed to reproduce the association between PD-L1 expression and outcomes with ICI treatment in these scenarios (4, 24). Variability with PD-L1 scoring and reporting, interassay heterogeneity especially with respect to immune cell staining, assay sensitivity, and analytical characteristics call for further standardization and harmonization of PD-L1 immunohistochemistry assays (25). PD-L1 expression can be evaluated together with genomic and tumor microenvironment features to strengthen its predictive value within multimodal models of ICI response.
Tumor mutation burden and MSI
Tumor mutation burden (TMB) is the prototypic measure of tumor foreignness where a higher tissue-based TMB has been associated with benefit from ICI in multiple studies (26-28) including randomized (14, 29) and nonrandomized clinical trials (30). TMB has been shown to predict response to ICI in a dose-dependent fashion, as patients with the highest TMB tumors attained longer survival after immunotherapy (31). This phenomenon is markedly exemplified in hypermutated tumors in the context of patients with mismatch repair deficiency that typically have the highest fraction of responses to ICI (32). TMB is defined as the number of nonsynonymous mutations per megabase of coding sequence. The premise of TMB as a predictive biomarker for ICI relies on the potential of these mutations to generate antigens, known as mutation-associated neoantigens (MANAs), that are foreign to the immune system and can therefore elicit an antitumor immune response (33). Together, these findings led to a tissue-agnostic FDA approval for TMB as a companion diagnostic biomarker for the ICI, pembrolizumab for tumors with TMB >10 mutations per megabase. Nevertheless, although the value of TMB in predicting ICI response is well documented for cancer types such as NSCLC and melanoma, its predictive value is not well supported for patients with glioma, prostate cancer, and breast cancer (34).
Moreover, several technical and biological limitations preclude the broad use of TMB as a universal biomarker of response for ICI (35). TMB is an imperfect biomarker of ICI response with inherent challenges related to lack of standardization and a universal threshold that defines TMB-high tumors (31, 36). Conceptually, the generalizability of the threshold of 10 mutations per megabase included in the FDA approval for pembrolizumab is limited by the cancer lineage–dependent dynamic range of TMB that renders a pan-cancer TMB threshold challenging. To add to the biological complexities, tissue TMB estimates are subject to sampling bias and are affected by low tumor purity, as the power of detection of mutations markedly decreases with decreasing tumor purity especially in the context of clonally heterogeneous tumors harboring a higher fraction of subclonal mutations (37). Although the dilution effect of tumor purity may be compensated bioinformatically (37) or through deeper targeted next-generation sequencing and improved machine learning–based TMB determination (38), TMB remains part of the constellation of features that determines an effective antitumor immune response.
Endogenous mutagenic processes, such as mismatch repair deficiency, induce a higher TMB in the context of DNA repair errors, with about 4% of human cancers harboring an MSI footprint that confers sensitivity to ICI (39, 40). MSI tumors have a high number of alterations throughout the genome, including in microsatellite regions that result from a deficiency in mismatched DNA repair machinery. MSI-high tumors harbor a higher somatic mutation burden, which generates a higher immunogenic neoantigen burden (41), thus predisposing to tumor clearance in the context of ICI (39). A number of clinical trials have shown the clinical utility of MSI as a biomarker of response to ICI and patient selection criterion (42, 43), exemplified in the tumor-agnostic FDA approval for pembrolizumab for patients with tumors harboring MSI and for nivolumab for patients with MSI-high colorectal cancer.
GENOMIC LANDSCAPES OF THERAPEUTIC RESPONSE AND RESISTANCE REVEAL A CONSTELLATION OF EMERGING HOST AND TUMOR FEATURES
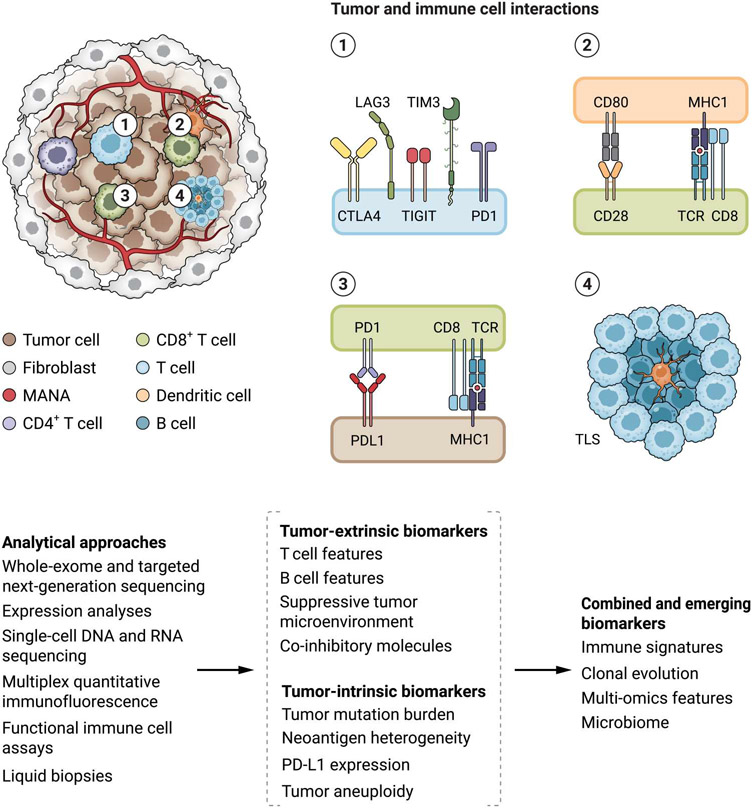
Emergence of primary and acquired resistance to ICI constitutes the key barrier to further improving patient outcomes. Clinical outcomes with ICI depend on the complex cancer-immune system interactions and are driven by several tumor-intrinsic and tumor-extrinsic features that mediate immunoediting mechanisms (Fig. 1). This dynamic and multifaceted process mandates the development of combined predictive models by means of integrative molecular and cellular analyses that capture the interplay between cancer and immune cells during therapy (Table 1).
Fig. 1. Tumor and tumor microenvironment features driving tumor immunoediting and contributing to ICI clinical outcomes.
A number of tumor intrinsic and tumor extrinsic features, such as the genomic landscape of the cancer cells and composition of the immune cell infiltrate, orchestrate the antitumor immune response in the context of immune checkpoint blockade. Tumor foreignness is determined by tumor mutation burden (TMB) and mutation-associated neoantigen (MANA) density. The phenotype and functional state of T cells (shown in 1), together with the antigen presentation potential (shown in 2 and 3) determine in part a tumor’s visibility by the immune system. The composition and phenotype of the T and B cell infiltrates and their interaction in tertiary lymphoid structures (TLSs; shown in 4) in the tumor microenvironment are key components differentiating immunologically “hot” tumors that eventually regress with ICI. These nuanced genomic and tumor microenvironment features can be captured by different analytical approaches such as bulk and single-cell multi-omic approaches, functional assays, and liquid biopsies, ultimately converging in multimodal biomarkers of therapeutic response.
Table 1. Established and emerging biomarkers for cancer immunotherapy.
| Biomarker | Context | References |
|---|---|---|
| Clinically integrated | ||
| PD-L1 expression by IHC | Pan-cancer, NSCLC, urothelial carcinoma, triple-negative breast cancer | (18, 21-23) |
| MSI | Pan-cancer, colorectal cancer | (39, 42, 43) |
| TMB | Pan-cancer, melanoma, NSCLC | (14, 26-30) |
| Emerging and context-specific | ||
| Clonal TMB | Pan-cancer | (36, 44, 45) |
| MANA quality | Pan-cancer | (53, 54, 59, 61, 62) |
| HLA diversity | Pan-cancer | (37, 64, 65) |
| Oncogenic drivers of immune suppression (PTEN, Wnt, JAK1/2, STK11, MYC, and EGFR) | Pan-cancer | (37, 73-76) |
| Neoantigen loss | NSCLC | (85) |
| Gene expression profiles | Pan-cancer | (87) |
| Tertiary lymphoid structures | Melanoma, renal cell carcinoma, sarcoma | (88-90) |
| T cell repertoire | Pan-cancer | (96, 97) |
| ctDNA | Pan-cancer | (99, 102-104, 108, 109, 113, 114) |
Nuanced TMB subsets and global genomic features
Emerging studies support differential weights of somatic mutations within TMB in shaping antitumor immune responses and ultimately therapeutic outcomes (36, 44). Low intratumoral clonal heterogeneity and a higher clonal mutation load have been associated with favorable clinical outcomes with ICI, especially in NSCLC and melanoma tumors (44, 45). Mutations in haploid regions of the genome of ICI-treated mesotheliomas have also been reported to predict response to ICI, as these are less likely to be lost during tumor evolution and may drive sustained tumor elimination in the tumor microenvironment (46). Tied into TMB, mutational spectra signatures have also been linked with therapeutic response to ICI, especially in relation to tobacco and ultraviolet exposures for patients with NSCLC and melanoma, respectively (37, 47). These environmental carcinogen exposures induce a higher mutation accumulation rate that, in turn, predisposes to therapeutic response in the context of high TMB (37). Subclonal mutagenesis may be reflected in a higher contribution of the APOBEC3 signature in the tumor’s mutational spectra and, as such, may be linked with inferior outcomes with ICI in NSCLC and breast cancer (48).
In addition to TMB, genome-wide copy number analyses of cancers have revealed that tumor aneuploidy may be associated with poor outcomes to ICI. This may be, in part, attributed to deletions of key immune-regulating genes and reduced cytotoxic immune cell infiltration resulting in tumor immune evasion (49). Nevertheless, the contribution of tumor aneuploidy to ICI resistance, especially in the setting of homologous recombination deficiency and an increased loss-of-heterozygosity (LOH) genomic content, may be context dependent and merits further evaluation (50). Together, TMB can be refined by considering both sequence and structural genomic alterations in cancer genomes, and additional studies are needed to support this notion and clinical translation.
Tumor foreignness and neoantigen repertoire as a predictor of ICI response
MANAs stem from somatic mutations and are presented by major histocompatibility complex (MHC) class I or MHC class II molecules to T cell receptors (TCRs) of CD8+ or CD4+ T cells, respectively (33). Because neoantigens are specific to cancer cells, T cells are able to recognize them as nonself antigens and elicit an antitumor immune response (33). Although several machine learning approaches have been developed to computationally derive MHC class I– and II–restricted neopeptides from next-generation sequencing data, these efforts have largely yielded similar prediction accuracy for computationally identified MANA load compared to TMB (27, 37, 51, 52). Clonal neoantigens, shared among all tumor cells, may confer sensitivity to ICI, whereas subclonal neoantigens, only present in a subset of tumor cells, may not be sufficient to elicit effective and robust immune responses (45). Neoantigen MHC binding and presentation are key features that determine tumor foreignness, and an increased neoantigen binding affinity for MHC class I molecules has been reported to be linked with ICI response (53, 54). Neoantigen fitness, primarily determined by differential presentation, “nonselfness,” and microbial antigen mimicry, has been shown to drive sustained tumor rejection and clinical responses in patients with pancreatic cancer (55, 56). Neoantigens derived from oncogenic hotspots, such as KRAS and PIK3CA, have also been shown to elicit T cell responses (57, 58). In addition to MANAs encoded by single-nucleotide variants, neoantigens derived from frameshift mutations have been shown to attain high-affinity MHC binding and contribute to ICI sensitivity (59). In tumors with a low burden of single-nucleotide variants, MANAs derived from gene fusions have been shown to drive cytotoxic T cell responses (60). Furthermore, as only mutations in expressed genes would yield MANAs that could potentially be presented and induce the priming and activation of neoantigen-specific T cells, the expression of transcripts containing singlebase substitutions may be more informative than TMB (61).
Historically, antitumor immune responses have been considered to be driven by human leukocyte antigen (HLA) class I–restricted neoantigen-driven cytotoxic CD8+ T cell responses. Nevertheless, tumor rejection in the context of immunotherapy has been shown to require the activity of both antigen-specific CD8+ and CD4+ T cells, suggesting the nonoverlapping but complementary role of HLA class I– and class II–restricted neoantigens (62). In addition to MANAs, cancer cells reexpress cancer germline antigens as a mechanism of immune evasion; these antigens, although less studied, may determine clinical outcomes with ICI (63). Collectively, these studies support the notion that, similar to somatic mutation burden, neoantigen quality rather than quantity is a key feature driving tumor elimination in the context of ICI, and understanding neoantigen features that predominantly contribute to ICI response can enhance their translation to more robust predictors of ICI response.
HLA genetic variation and antigen presentation potential
The metastatic potential of the tumor and the eventual clinical outcome of the host does not merely depend on tumor cells intrinsically endowed with the ability to proliferate, invade, and metastasize but rather on a dynamic equilibrium between the tumor and the host’s immune system. Central to this notion is the role of antigen presentation capacity in potentiating antitumor immune responses. Germline HLA class I evolutionary divergence may determine response to immune checkpoint blockade (64, 65) but requires further study. In addition, disruptive neoantigen presentation due to HLA down-regulation or loss through LOH events, ß-2-microglobulin loss, or dysregulation of other proteins involved in protein cleavage and neopeptide transport have been described as potential mediators of primary and acquired resistance to immune checkpoint blockade, but these mechanisms are rare and potentially occur in a context-specific manner (66).
Nevertheless, the impact of germline HLA class I and II zygosity in clinical responses with ICI has not been universally documented, as germline HLA genotypes and diversity alone may not be independent biomarkers of anti–PD-1 clinical efficacy (37, 67, 68). Integration of HLA germline variation with somatic status in a TMB-stratified manner has been shown to be informative in identifying tumors most likely to respond to ICI (37). Furthermore, germline HLA variation can shape the evolutionary landscape of cancer by exerting a negative selective pressure on mutations related to highly presented peptides (69, 70). In considering features that affect tumor visibility by the immune system and ultimately lead to tumor rejection in the context of ICI treatment, one has to consider the constellation of tumor and immune-related processes that modulate functional antigen presentation capacity in the tumor microenvironment. Germline HLA diversity and promiscuity has to be combined with somatic LOH events in the tumor cells or epigenetic silencing of the HLA loci and considered in the context of a given host and an evolving tumor.
Oncogenic drivers associated with ICI therapeutic resistance
Specific genomic features are linked with therapeutic response to ICI in a pan-cancer manner, whereas others are cancer type specific. Mutations in oncogenic drivers that have immune regulatory functions have been linked with resistance to ICI (71). PTEN loss has been linked to ICI resistance through induction of an immunosuppressive tumor microenvironment, a decrease in T cell infiltration and inhibition of T cell mediated tumor killing, and cytotoxic activity from loss of immunosuppressive cytokines, such as vascular endothelial growth factor (72, 73). Similarly, activation of the Wnt/β-catenin pathway has been linked with exclusion of tumor-infiltrating T cells, suppressed recruitment of dendritic cells impairing T cell priming, and ultimately an ineffective antitumor immune response (74). Inactivating mutations in the JAK1/2 genes have been linked with defective interferon-γ signaling resulting in ICI resistance in both the primary and acquired resistance settings (66, 75, 76). STK11/KEAP1 comutations have been shown to drive primary ICI resistance through PD-L1 regulation and subsequent T cell exclusion (77). Up-regulation of MYC contributes to an immunosuppressive environment by up-regulating PD-L1 and CD47 that both inhibit T cell activity (78), whereas suppression of MYC signaling through epigenetic therapy may reverse immune suppression (79). More broadly, oncogene-addicted tumors, such as those tumors harboring epidermal growth factor receptor (EGFR)–activating mutations, are thought to be less responsive to ICI, which is mainly attributed to the lower TMB of these tumors and tumor immune exclusion (80). Intriguingly, in EGFR mutant NSCLC, response varies among different EGFR mutant alleles, again highlighting the importance of considering the specific genomic context (81). In addition to considering oncogenic drivers independently, evaluation of co-occurring mutations in a context-dependent manner is key in understanding the genomically heterogeneous landscape of response and resistance to ICI (82). Patients with NSCLC harboring KEAP1 mutations have a shorter survival compared to patients with single KEAP1 mutation or wild-type tumors (83). Collectively, these findings support the notion that understanding the nuances of the genetic landscape of tumors in terms of oncogenic drivers and comutation patterns may collectively provide important predictors of response to ICI. Nevertheless, the significance of each alteration’s contribution to sculpting the tumor microenvironment and drive tumor immunoediting has yet to be uncovered.
Evolving cancer genomes and neoantigen loss
Patient selection strategies tailored to the molecular footprint of cancer genomes may nevertheless fail when based on analyses of a single time point due to the challenges of using static biomarkers to interpret dynamic processes of the tumor-immune system cross-talk. The evolutionary trajectories of cancer cells as they go through bottlenecks imposed by immunotherapy represent an avenue of biomarker identification for response to ICI, as understanding how cancer cells gain a fitness advantage and escape immune surveillance allows for timely translation to clinical practice and the rational design of ICI treatment strategies. Cancer lineages have been extensively studied in the context of natural clonal evolution or during targeted therapies (84), with fewer studies in the context of immunotherapy. Acquired resistance to immune checkpoint blockade has been shown to emerge in the setting of clonal neoantigen loss by chromosomal deletions and LOH, followed by selection and expansion of the resistant clone (85). These observations highlight the importance of evolving changes in cancer genomes as a mechanism of secondary resistance to ICI and exemplify the importance of considering the evolving cancer genome during ICI rather than solely relying on snapshot analyses of tumors before ICI initiation.
HALLMARKS OF ICI RESISTANCE POINT TOWARD A SUPPRESSIVE TUMOR MICROENVIRONMENT
The phenotype of the tumor microenvironment, assessed through gene expression analyses, has been linked with response to ICI, either alone (86) or in combination with TMB (87). B and T cell interactions in tertiary lymphoid structures have also been linked with response to immunotherapy (88-90). Several transcriptomic signature-based models, predominantly related to adaptive immunity, have been proposed to predict therapeutic response for patients treated with immune checkpoint blockade (91-94); however, generalizability of these approaches has been limited by lack of validation in independent cohorts (95). In addition to bulk gene expression approaches, single-cell RNA sequencing approaches are gaining momentum for tumor microenvironment phenotyping and deconvolution of the heterogeneity of immune cell populations. Single-cell RNA profiling together with TCR sequencing have been used to determine the phenotype of CD8+ tumor–specific tumor-infiltrating lymphocytes and their respective properties, allowing for the recognition of specific cells in the tumor microenvironment that elicit an antitumor immune response (96, 97). Although in-depth RNA or TCR sequencing–based single-cell resolution analyses have shed light into the tumor microenvironment complexity and heterogeneity in the context of immunotherapy, these approaches have not yet been translated into biomarker selection strategies (98).
INTEGRATIVE APPROACHES TO CAPTURE ICI RESPONSE
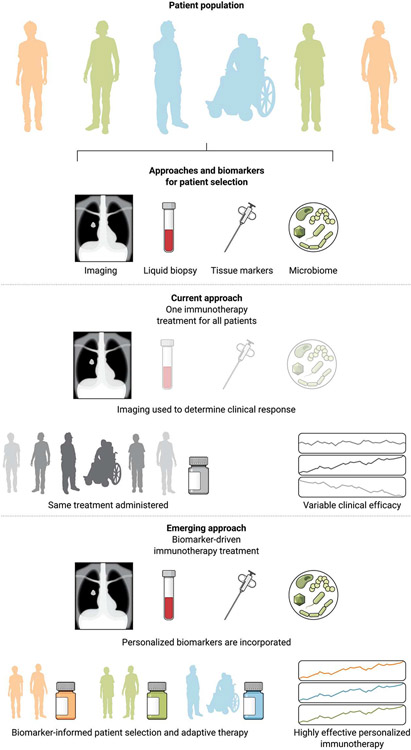
Despite the growing body of molecular studies using single time-point “snapshot” analyses, there is a paucity of studies that investigate the evolutionary trajectory of cancer cells in conjunction to the evolving characteristics of the host. These challenges are particularly pronounced in the context of therapies that induce global host effects, such as immunotherapy. Tumor-host interactions in the context of immunotherapy extend beyond the tumor microenvironment; however, our understanding of molecular mechanisms of response and resistance to these therapies primarily comes from modeling local rather than systemic interactions. Multi-omic features can be integrated by machine learning approaches to more accurately classify patients at risk of disease progression on ICI. This approach is exemplified in integrative genomic meta-analyses where smaller studies are pooled and sequence data are reanalyzed to study tumor-intrinsic and tumor microenvironment features of response and resistance to ICI in a variety of human cancers (44). Clonal TMB, total TMB, and frameshift nonsense-mediated decay-escaping mutation load together with CXCL9 expression, indicative of priming and recruitment of cytotoxic T cells, have been shown to significantly contribute to ensemble models that capture tumor and tumor microenvironment features (44). The overarching clinical question remains as to how to incorporate this biologic complexity in clinical trial design and navigate away from one-immunotherapy-fits-all approaches toward precision immuno-oncology (Fig. 2).
Fig. 2. Paradigm shift toward a precision immuno-oncology approach.
Current treatment strategies for cancer immunotherapy remain unselected for biomarkers, with the exception of PD-L1 expression and TMB-selected ICI. The current standard of care for assessment of therapeutic response is determined on the basis of radiographic imaging, which does not always capture the nature and timing of response. There are several other biomarkers such as liquid biopsy, tissue markers, and the microbiome that could be used to better monitor and predict patient outcome that are not used. This one-treatment-fits-all approach results in variable clinical efficacy. Patient selection and stratification based on the genomic and molecular make up of tumors and their microenvironment may enhance the clinical efficacy of immunotherapy approaches and improve clinical outcomes. To this end, biomarker-driven clinical trials have the potential to further improve the therapeutic efficacy and long-term outcomes with cancer immunotherapy.
LIQUID BIOPSIES ARE EMERGING DYNAMIC APPROACHES FOR DETERMINING AND TRACKING ICI RESPONSE
Circulating tumor burden as a real-time cancer biomarker
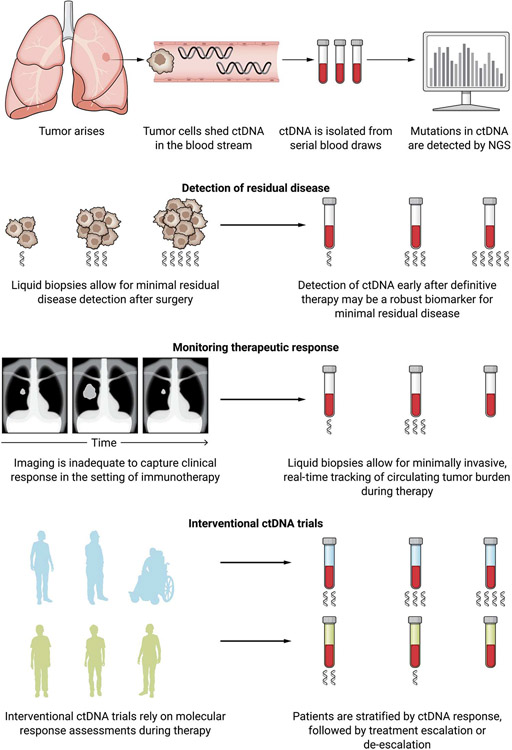
Despite important progress in understanding the mechanisms of ICI resistance, the underlying cause of therapeutic resistance for many patients may not be determined, and our ability to predict clinical responses is currently limited. In this setting, there may be value in the use of noninvasive cell-free DNA (cfDNA) approaches to capture dynamic changes in tumor burden during therapy. Liquid biopsies have emerged as powerful noninvasive means of detecting and measuring the presence of tumor-derived DNA in the circulation and tracking tumor evolution during therapy. In patients with cancer, a portion of cfDNA originates from the tumor and is called ctDNA. ctDNA is shed into the blood stream through cellular apoptosis and necrosis and can be noninvasively sampled and analyzed. There is an ever-increasing number of studies that demonstrate the clinical utility of liquid biopsies at almost every stage of management of patients with cancer, including screening and early detection, detection of minimal residual disease, treatment selection, therapeutic response monitoring, and identification of resistance (Fig. 3). The development of digital-based polymerase chain reaction and next-generation sequencing technologies combined with bioinformatic analyses has provided sensitive and specific quantitative approaches to detect mutant sequences from ctDNA typically in the background of vast amounts of wild-type DNA in the circulation. The development of ultrasensitive platforms that use deep and redundant sequencing together with sequencing error suppression algorithms allow for sensitive and specific detection of low-frequency sequence and structural alterations in ctDNA (99-106). Because mutations in cfDNA can be related to clonal hematopoiesis, parallel deep sequencing of white blood cells can allow for accurate classification of variants by origin (107), which further enables tracking of bona fide tumor-specific alterations in the circulation. Liquid biopsy approaches are gaining momentum in immuno-oncology as they can be used to rapidly and accurately determine clinical responses. In looking at the landscape of ICI clinical trials, molecular response–driven approaches are urgently needed to interpret outcomes and guide therapy. Liquid biopsies not only can provide an independent measure of therapeutic response but may also improve on current radiographic response criteria that may underestimate the benefit from ICI and the unique patterns and timing of response (99, 102).
Fig. 3. Integration of liquid biopsies approaches with cancer immunotherapy.
During carcinogenesis and cancer evolution, tumor cells release their DNA into the bloodstream, providing the opportunity to sample tumor DNA through noninvasive analyses of blood. Liquid biopsies enable the detection and analysis of mutations in circulating tumor DNA (ctDNA) using ultrasensitive next-generation sequencing (NGS) technologies. For minimal residual disease, liquid biopsies can detect the recrudescence of tumor cells after surgery or definitive therapy in ways that may be difficult to capture through imaging or other available cancer biomarkers. Minimally invasive ctDNA detection methods can detect real-time changes in circulating tumor burden during therapy that would be otherwise missed with imaging alone. Longitudinal liquid biopsies can be informative for detecting minimal residual disease for patients with early-stage cancer receiving cancer immunotherapy in the neoadjuvant setting, monitoring response for patients with metastatic disease, and patient stratification for ICI clinical trials. Given the challenges of conventional imaging for capturing responses to immunotherapies, liquid biopsies provide an alternative strategy to detect early signs of disease progression and therapeutic resistance as well as disease clearance that would otherwise not be identified on the basis of imaging alone. To this end, collection of serial blood draws over time introduces opportunities for disease monitoring that can guide clinical decisions. The recent emergence of interventional trials that assess ctDNA dynamics and use ctDNA molecular response to guide clinical decision-making can further extend the premise of precision immunotherapy to differentiate patients most likely to attain long-term clinical outcomes (in green) from the ones that experience disease progression (in blue).
Blood-derived TMB
Blood-derived determination of MSI and TMB (bTMB) has been evaluated as a surrogate for tissue-derived MSI and TMB (108-112). The blood-based MSI status and bTMB provides several advantages over tissue analyses including the accessibility and noninvasive nature of liquid biopsy collection as well as capturing tumor heterogeneity that may otherwise be lost due to tumor biopsy sampling and tumor purity. Blood-based MSI has been shown to correlate well with tissue-based analyses and may be used to identify patients who have a high likelihood to attain a response with ICI (110). In principle, bTMB evaluates the same components as tissue TMB, mainly the number of somatic mutations per megabase, with bTMB requiring the removal of clonal hematopoietic mutations that may confound analyses. A higher bTMB has been shown to correlate with longer survival after immunotherapy (108, 109). bTMB predicted response to atezolizumab in the POPLAR (NCT01903993) and OAK (NCT02008227) NSCLC clinical trials using a threshold of 16 mutations per megabase as the optimal cutpoint. Similarly, bTMB analyses from the MYSTIC trial that assessed first-line durvalumab ± tremelimumab for metastatic NSCLC (NCT02453282) supported the predictive value of bTMB, however, using a different threshold. In addition, the B-F1RST trial of atezolizumab in NSCLC (NCT02848651) failed to validate the association between progression-free survival and bTMB ≥ 16 mutations per megabase (113). High bTMB has been shown to correlate with longer overall survival in the Impower110 (NCT02409342) and B-F1RST (NCT02848651) clinical trials assessing first-line atezolizumab in NSCLC (113, 114). Although these results are promising, further validation and standardization of bMTB as a predictor of ICI response is warranted.
Promise of ctDNA as a dynamic real-time biomarker of response to ICI
Tumor-derived variant mutation allele fractions (MAFs) measured at serial time points during ICI provide insights into tumor burden kinetics and enable the monitoring of changes in MAFs over time that are reflective of therapeutic response (99, 102-104). Collectively, these analyses have revealed distinct patterns of ctDNA-based molecular responses that are reflective of patients’ clinical outcome, such that patients with a ctDNA molecular response have a reduction in ctDNA, which is reflective of long-term clinical benefit. In contrast, for patients with primary resistance to immune checkpoint blockade, ctDNA has limited fluctuations or displays a rise after therapy initiation (99, 102). Although the quantitative variation in ctDNA has been clearly linked with therapeutic outcome in the context of ICI, a unified consensus of the definition for molecular response is lacking, which may be explained by differences in overall design among reported studies, the tumors and therapies that were evaluated, the timing of liquid biopsy assessments, and the specific assay characteristics (99, 103). Similarly, the optimal timing to measure ctDNA response and the durability of ctDNA molecular responses are not well documented. Further standardization and prospective trials will ultimately be needed to develop and validate molecular response criteria that will be useful clinically.
As ICI is now incorporated in the therapeutic armamentarium for patients with early stage cancer, there is an unmet need for noninvasive approaches to inform outcomes. ctDNA clearance has been associated with tumor regression at the time of resection for patients with NSCLC receiving neoadjuvant ICI (99), and analyses from the CheckMate 816 trial of neoadjuvant nivolumab with platinum-doublet chemotherapy showed that ctDNA clearance was reflective of longer event-free survival (10). Implementation of liquid biopsies in the setting of adjuvant immunotherapy for assessing minimal residual disease is already in progress. Representative examples include the LUN0115 (NCT04585477), CtDNA lung RCT (NCT04966663), and AAAT0800 (NCT04625699) phase 2 trials that select patients with NSCLC and detectable ctDNA after surgery for adjuvant immunotherapy (Table 2). The IMvigor011 phase 3 trial evaluates adjuvant atezolizumab for patients with muscle-invasive bladder cancer who have detectable ctDNA after cystectomy (NCT04660344). The PERSEVERE trial stratifies patients with triple-negative breast cancer by ctDNA positivity to optimize outcomes (NCT04849364). Additional trials include the c-TRAK TN trial, where patients with triple-negative breast cancer and detectable ctDNA are randomized to pembrolizumab versus observation (NCT03145961), and a study of pembrolizumab after surgery in patients with MSI-high solid tumors for individuals with detectable ctDNA (NCT03832569; Table 2). These clinical trials emphasize the potential utility of liquid biopsies for detecting minimal residual disease and molecular responses during adjuvant immunotherapy.
Table 2. Selected clinical trials that incorporate ICI biomarkers in the trial eligibility criteria or in the trial intervention.
This table summarizes tissue and blood biomarker–based immuno-oncology clinical trials. TMB, tumor mutation burden; MSI, microsatellite instability; dMMR, mismatch repair deficient; HRD, homologous recombination deficiency; NSCLC, non–small cell lung cancer.
| NCT | Phase | Biomarker | Intervention | Condition | Status |
|---|---|---|---|---|---|
| NCT05197322 | II | TMB, MSI | Pembrolizumab | Colorectal cancer | Not yet recruiting |
| NCT03638297 | II | MSI-H/dMMR | BAT 1306, pembrolizumab | Colorectal cancer | Recruiting |
| NCT04940637 | II | PD-L1, HRD | Dostarlimab | NSCLC | Recruiting |
| NCT03911557 | II | TMB | Durvalumab, tremelimumab | Somatically hypermutated tumors | Recruiting |
| NCT04589845 | II | TMB | Atezolizumab | Solid tumors | Recruiting |
| NCT02693535 | II | TMB | Pembrolizumab | Solid tumors | Recruiting |
| NCT04006262 | II | MSI | Nivolumab, ipilimumab | Localized esophagogastric adenocarcinoma | Recruiting |
| NCT04730544 | II | MSI | Nivolumab, ipilimumab | Colorectal cancer | Recruiting |
| NCT05468138 | II | MSI | Anti–PD-1 | Operable gastric cancer | Recruiting |
| NCT04008030 | III | MSI | Nivolumab, ipilimumab | Colorectal cancer | Recruiting |
| NCT05118724 | II | MSI | Atezolizumab | Colorectal cancer | Recruiting |
| NCT02912572 | II | MSI, POLE mutation | Avelumab | Endometrial cancer | Recruiting |
| NCT02997228 | III | dMMR | Atezolizumab | Colorectal cancer | Recruiting |
| NCT05201612 | II | HRD | Pembrolizumab | Solid tumors | Not yet recruiting |
| NCT04317105 | I/II | PTEN, PIK3CA mutations | Copanlisib, nivolumab, ipilimumab | Solid tumors | Recruiting |
| NCT05472623 | II | KRAS mutation | Adagrasib, nivolumab | NSCLC | Not yet recruiting |
| NCT04059887 | IV | blood TMB | Atezolizumab | Lung cancer | Recruiting |
| NCT04940286 | II | ctDNA | Durvalumab | Pancreatic adenocarcinoma | Recruiting |
| NCT04638582 | II | ctDNA | Pembrolizumab | NSCLC | Not yet recruiting |
| NCT04644289 | II | ctDNA | Durvalumab | Epithelial ovarian cancer | Recruiting |
| NCT04849364 | II | ctDNA | Atezolizumab | Triple-negative breast cancer | Recruiting |
| NCT04434040 | II | ctDNA | Atezolizumab | Triple-negative breast cancer | Recruiting |
| NCT04660344 | III | ctDNA | Atezolizumab | High-risk muscle-invasive bladder cancer | Recruiting |
| NCT04585477 | II | ctDNA | Durvalumab | Early-stage NSCLC | Recruiting |
| NCT04966663 | II | ctDNA | Nivolumab | Early-stage NSCLC | Recruiting |
| NCT03832569 | I | ctDNA | Pembrolizumab | Solid tumors | Recruiting |
| NCT04625699 | II | ctDNA | Durvalumab, tremelimumab | NSCLC | Not yet recruiting |
| NCT04966676 | II | ctDNA | Nivolumab and ipilimumab | NSCLC | Not yet recruiting |
| NCT04166487 | II | ctDNA | Pembrolizumab | NSCLC | Recruiting |
| NCT03808441 | II | ctDNA | Nivolumab and ipilimumab | Melanoma | Recruiting |
Liquid biopsy–informed ICI clinical trials
The expanding clinical utility of ctDNA approaches has set the foundation for a paradigm shift toward interventional trials that rely on liquid biopsy–informed molecular responses to guide therapy (Table 2). The integration of serial liquid biopsies to actively guide clinical decisions represents a new approach whereby patients may benefit from the detection of response earlier and more accurately than routine computed tomography scans and modify treatment modality should resistance emerge. Interventional trials that coordinate clinical decisions with liquid biopsy detected molecular responses are currently underway. Examples include the BR36 trial that investigates first-line pembrolizumab in metastatic NSCLC where early ctDNA dynamics are used to identify patients with molecular response who continue single-agent immunotherapy and patients with molecular progression who are randomized to receive pembrolizumab or pembrolizumab and platinum doublet therapy (NCT04093167). Similarly, the plasma-adapted first-line pembrolizumab clinical trial (NCT04166487) in patients with metastatic NSCLC assesses serial liquid biopsies from patients treated with pembrolizumab to determine molecular responses with non-responders changing treatment to pembrolizumab and chemotherapy. The ATLAS interventional trial evaluates metastatic NSCLC response to nivolumab and ipilimumab using ctDNA with addition of chemotherapy for individuals who do not attain a molecular response (NCT04966676). The phase 3 MERMAID trial uses a tumor-informed ctDNA panel to direct postoperative therapy for patients with resected NSCLC (NCT04385368). The ctDNA-guided (CAcTUS) interventional trial for patients with melanoma uses ctDNA to guide clinical decisions on when to switch from targeted therapy to immunotherapy (NCT03808441; Table 1). The multitude of emerging trials in this space reflects the enthusiasm in using ctDNA analyses for earlier and more efficient determination of response to ICI and modifying patient intervention accordingly.
FUTURE DIRECTIONS
Most therapeutic strategies aiming to overcome ICI resistance are not biomarker-based (115); therefore, despite their conceptual relevance, personalized immuno-oncology approaches using tumor analyses and ctDNA measurements are most likely to be successful in predicting, preventing, and overcoming resistance. Given the strong association reported in the CheckMate 816 trial between pCR and event-free survival together with a subset analysis from the same study suggesting an association between ctDNA clearance and clinical outcomes, one could envisage the use of dynamic biomarkers in future neoadjuvant and adjuvant trials. ctDNA approaches have the potential to enrich the trial population with patients most likely to benefit from adjuvant therapy while minimizing exposure to toxicity for those already cured by neoadjuvant therapy and surgery.
Given the incremental integration of liquid biopsies in clinical cancer care, it is important to note that we may be reaching a plateau in sensitivity of mutation-based liquid biopsy approaches and their relatively high cost may limit their widespread use in clinical trials. To this end, a new generation of liquid biopsy approaches may offer new avenues to further use cfDNA analyses in clinical practice. For example, blood cfDNA fragmentome analyses in healthy individuals have revealed DNA fragments derived from hematopoietic cells, whereas cfDNA in patients with cancer contains an admixture of hematopoietic and tumor preserved DNA fragments (116, 117). The unique chromatin landscape reflected in these fragmentation profiles can be identified by means of whole-genome sequencing and leveraged using machine learning approaches to distinguish between healthy and cancer state with high performance (116, 117). These approaches may greatly reduce the design complexity of targeted next-generation sequencing panel assays and negate the need of matched tumor tissue for removal of hematopoietic artifacts, allowing for more sensitive and lower cost analyses. As initial studies have demonstrated the use of fragmentome approaches in response monitoring for targeted therapies, the evaluation of these methods in ICI response monitoring will be of great interest in the future.
Acknowledgments
Funding: This work was supported, in part, by the U.S. National Institutes of Health grants CA121113 (to V.A. and V.E.V.) and CA006793 (to V.E.V.), the Bloomberg-Kimmel Institute for Cancer Immunotherapy (to V.A and P.M.F.), the Department of Defense Congressionally Directed Medical Research Programs grant CA190755 (to V.A. and P.M.F.), the ECOG-ACRIN Thoracic Malignancies Integrated Translational Science Center grant UG1CA233259 (to V.A. and V.E.V.), the V Foundation (to V.A. and V.E.V.), the LUNGevity Foundation (to V.A. and V.E.V.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (V.E.V.), the Cole Foundation (V.E.V.), and a research grant from Delfi Diagnostics (V.E.V.).
Footnotes
Competing interests: V.A. receives research funding to Johns Hopkins University from AstraZeneca, Personal Genome Diagnostics, and Delfi Diagnostics and has received research funding to Johns Hopkins University from Bristol-Myers Squibb in the past 5 years. V.A. is an inventor on patent applications (63/276,525; 17/779,936; 16/312,152, 16/341,862; 17/047,006; and 17/598,690) submitted by Johns Hopkins University related to cancer genomic analyses, ctDNA therapeutic response monitoring, and immunogenomic features of response to immunotherapy that have been licensed to one or more entities. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. P.M.F. has received research funding to Johns Hopkins University from AstraZeneca, Bristol-Myers Squibb, Novartis, Corvus, and Kyowa. He has also served as a consultant for Amgen, AstraZeneca, Bristol-Myers Squibb, Daiichi Sankyo, Genentech, G1 Therapeutics, Iteos, Janssen, Merck, Surface Oncology, Mirati, Novartis, and Sanofi and as a DSMB member for Polaris and Flame Therapeutics. V.E.V. is a founder of Delfi Diagnostics, serves on the board of directors and as a consultant for this organization and owns Delfi Diagnostics stock, which are subject to certain restrictions under university policy. In addition, Johns Hopkins University owns equity in Delfi Diagnostics. V.E.V. divested his equity in Personal Genome Diagnostics (PGDx) to LabCorp in February 2022. V.E.V. is an inventor on patents and patent applications submitted by Johns Hopkins University related to cancer genomic analyses and cell-free DNA for cancer detection that have been licensed to one or more entities, including Delfi Diagnostics, LabCorp, Qiagen, Sysmex, Agios, Genzyme, Esoterix, Ventana and ManaT Bio. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. V.E.V. is an advisor to Danaher, Takeda Pharmaceuticals, and Viron Therapeutics. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. All other authors declare that they have no competing interest.
REFERENCES AND NOTES
- 1.Topalian SL, Drake CG, Pardoll DM, Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJM, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ, Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullard A, 2021 FDA approvals. Nat. Rev. Drug Discov 21, 83 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Frontera OA, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR, Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Remon J, Hellmann MD, First-line immunotherapy for non-small-cell lung cancer. J. Clin. Oncol 40, 586–597 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D’Amico TA, De Camp M, Dilling TJ, Dowell J, Gettinger S, Grotz TE, Gubens MA, Hegde A, Lackner RP, Lanuti M, Lin J, Loo BW, Lovly CM, Maldonado F, Massarelli E, Morgensztern D, Ng T, Otterson GA, Pacheco JM, Patel SP, Riely GJ, Riess J, Schild SE, Shapiro TA, Singh AP, Stevenson J, Tam A, Tanvetyanon T, Yanagawa J, Yang SC, Yau E, Gregory K, Hughes M, Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 20, 497 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Chakravarty D, Johnson A, Sklar J, Lindeman NI, Moore K, Ganesan S, Lovly CM, Perlmutter J, Gray SW, Hwang J, Lieu C, André F, Azad N, Borad M, Tafe L, Messersmith H, Robson M, Meric-Bernstam F, Somatic genomic testing in patients with metastatic or advanced cancer: ASCO provisional clinical opinion. J. Clin. Oncol 40, 1231–1258 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Spigel DR, Faivre-Finn C, Gray JE, Vicente D, Planchard D, Paz-Ares L, Vansteenkiste JF, Garassino MC, Hui R, Quantin X, Rimner A, Wu Y-L, Özgüiroğlu M, Lee KH, Kato T, de Wit M, Kurata T, Reck M, Cho BC, Senan S, Naidoo J, Mann H, Newton M, Thiyagarajah P, Antonia SJ, Five-year survival outcomes from the PACIFIC trial: Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. J. Clin. Oncol 40, 1301–1311 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felip E, Altorki N, Zhou C, Csőszi T, Vynnychenko I, Goloborodko O, Luft A, Akopov A, Martinez-Marti A, Kenmotsu H, Chen YM, Chella A, Sugawara S, Voong D, Wu F, Yi J, Deng Y, McCleland M, Bennett E, Gitlitz B, Wakelee H; IMpower010 Investigators, Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): A randomised, multicentre, open-label, phase 3 trial. Lancet 398, 1344–1357 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Forde PM, Spicer J, Lu S, Provencio M, Mitsudomi T, Awad MM, Felip E, Broderick SR, Brahmer JR, Swanson SJ, Kerr K, Wang C, Ciuleanu T-E, Saylors GB, Tanaka F, Ito H, Chen K-N, Liberman M, Vokes EE, Taube JM, Dorange C, Cai J, Fiore J, Jarkowski A, Balli D, Sausen M, Pandya D, Calvet CY, Girard N; CheckMate 816 Investigators, Neoadjuvant nivolumab plus chemotherapy in resectable lung cancer. N. Engl. J. Med 386, 1973–1985 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD, Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A, Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med 372, 2521–2532 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Robert C, Ribas A, Schachter J, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil CM, Lotem M, Larkin JMG, Lorigan P, Neyns B, Blank CU, Petrella TM, Hamid O, Su SC, Krepler C, Ibrahim N, Long GV, Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 20, 1239–1251 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Hellmann MD, Paz-Ares L, Caro RB, Zurawski B, Kim S-W, Costa EC, Park K, Alexandru A, Lupinacci L, de la Mora Jimenez E, Sakai H, Albert I, Vergnenegre A, Peters S, Syrigos K, Barlesi F, Reck M, Borghaei H, Brahmer JR, O’Byrne KJ, Geese WJ, Bhagavatheeswaran P, Rabindran SK, Kasinathan RS, Nathan FE, Ramalingam SS, Nivolumab plus Ipilimumab in advanced non-small-cell lung cancer. N. Engl. J. Med 381, 2020–2031 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Paz-Ares L, Ciuleanu T-E, Cobo M, Schenker M, Zurawski B, Menezes J, Richardet E, Bennouna J, Felip E, Juan-Vidal O, Alexandru A, Sakai H, Lingua A, Salman P, Souquet P-J, De Marchi P, Martin C, Pérol M, Scherpereel A, Lu S, John T, Carbone DP, Meadows-Shropshire S, Agrawal S, Oukessou A, Yan J, Reck M, First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): An international, randomised, open-label, phase 3 trial. Lancet Oncol. 22, 198–211 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Schneider BJ, Naidoo J, Santomasso BD, Lacchetti C, Adkins S, Anadkat M, Atkins MB, Brassil KJ, Caterino JM, Chau I, Davies MJ, Ernstoff MS, Fecher L, Ghosh M, Jaiyesimi I, Mammen JS, Naing A, Nastoupil LJ, Phillips T, Porter LD, Reichner CA, Seigel C, Song J-M, Spira A, Suarez-Almazor M, Swami U, Thompson JA, Vikas P, Wang Y, Weber JS, Funchain P, Bollin K, Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: ASCO guideline update. J. Clin. Oncol 39, 4073–4126 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, Rutkowski P, Gogas HJ, Lao CD, de Menezes JJ, Dalle S, Arance A, Grob JJ, Srivastava S, Abaskharoun M, Hamilton M, Keidel S, Simonsen KL, Sobiesk AM, Li B, Hodi FS, Long GV; RELATIVITY-047 Investigators, Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N. Engl. J. Med 386, 24–34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS, Hirsch FR, PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol 18, 345 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Davis AA, Patel VG, The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 7, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G Jr., Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB, Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 387, 1540–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR; KEYNOTE-024 Investigators, Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 23.US Food and Drug Administration, List of cleared or approved companion diagnostic devices (in vitro and imaging tools) (2022); www.fda.gov/medical-devices/vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-vitro-and-imaging-tools.
- 24.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, Cortinovis DL, Leach J, Polikoff J, Barrios C, Kabbinavar F, Frontera OA, De Marinis F, Turna H, Lee J-S, Ballinger M, Kowanetz M, He P, Chen DS, Sandler A, Gandara DR; OAK Study Group, Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 389, 255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, Homer R, West WW, Wu H, Roden AC, Fujimoto J, Yu H, Anders R, Kowalewski A, Rivard C, Rehman J, Batenchuk C, Burns V, Hirsch FR, Wistuba II, A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol. 3, 1051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA, Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA, Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarchoan M, Hopkins A, Jaffee EM, Tumor mutational burden and response rate to pd-1 inhibition. N. Engl. J. Med 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, Borghaei H, Ramalingam SS, Brahmer J, Reck M, O’Byrne KJ, Geese WJ, Green G, Chang H, Szustakowski J, Bhagavatheeswaran P, Healey D, Fu Y, Nathan F, Paz-Ares L, Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med 378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, Chung HC, Kindler HL, Lopez-Martin JA, Miller WH Jr., Italiano A, Kao S, Piha-Paul SA, Delord J-P, McWilliams RR, Fabrizio DA, Aurora-Garg D, Xu L, Jin F, Norwood K, Bang Y-J, Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21, 1353 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, AI-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, De Angelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, De Matteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT, Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousseau B, Foote MB, Maron SB, Diplas BH, Lu S, Argilés G, Cercek A, Diaz LA Jr., The spectrum of benefit from checkpoint blockade in hypermutated tumors. N. Engl. J. Med 384, 1168–1170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher TN, Schreiber RD, Neoantigens in cancer immunotherapy. Science 348, 69 (2015). [DOI] [PubMed] [Google Scholar]
- 34.McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, Jonasch E, Khasraw M, Heimberger AB, Lim B, Ueno NT, Litton JK, Ferrarotto R, Chang JT, Moulder SL, Lin S-Y, High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol 32, 661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R, The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 39, 154–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anagnostou V, Bardelli A, Chan TA, Turajlic S, The status of tumor mutational burden and immunotherapy. Nat. Cancer 3, 652 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Anagnostou V, Niknafs N, Marrone K, Bruhm DC, White JR, Naidoo J, Hummelink K, Monkhorst K, Lalezari F, Lanis M, Rosner S, Reuss JE, Smith KN, Adleff V, Rodgers K, Belcaid Z, Rhymee L, Levy B, Feliciano J, Hann CL, Ettinger DS, Georgiades C, Verde F, Illei P, Li QK, Baras AS, Gabrielson E, Brock MV, Karchin R, Pardoll DM, Baylin SB, Brahmer JR, Scharpf RB, Forde PM, Velculescu VE, Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat. Cancer 1, 99–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keefer LA, White JR, Wood DE, Gerding KMR, Valkenburg KC, Riley D, Gault C, Papp E, Vollmer CM, Greer A, Hernandez J, McGregor PM III, Zingone A, Ryan BM, Deak K, McCall SJ, Datto MB, Prescott JL, Thompson JF, Cerqueira GC, Jones S, Simmons JK, Elhinny AM, Dickey J, Angiuoli SV, Diaz LA Jr., Velculescu VE, Sausen M, Automated next-generation profiling of genomic alterations in human cancers. Nat. Commun 13, 2830 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr., Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA Jr.; KEYNOTE-177 Investigators, Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med 383, 2207–2218 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Kloor M, von Knebel Doeberitz M, The immune biology of microsatellite-unstable cancer. Trends Cancer 2, 121–133 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, El Dika IH, Segal N, Shcherba M, Sugarman R, Stadler Z, Yaeger R, Smith JJ, Rousseau B, Argiles G, Patel M, Desai A, Saltz LB, Widmar M, Iyer K, Zhang J, Gianino N, Crane C, Romesser PB, Pappou EP, Paty P, Garcia-Aguilar J, Gonen M, Gollub M, Weiser MR, Schalper KA, Diaz LA Jr., PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N. Engl. J. Med 386, 2363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maio M, Ascierto PA, Manzyuk L, Motola-Kuba D, Penel N, Cassier PA, Bariani GM, Acosta ADJ, Doi T, Longo F, Miller WH, Oh D-Y, Gottfried M, Xu L, Jin F, Norwood K, Marabelle A, Pembrolizumab in microsatellite instability high or mismatch repair deficient cancers: Updated analysis from the phase II KEYNOTE-158 study. Ann. Oncol 33, 929–938 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, Watkins TBK, Rosenthal R, Biswas D, Rowan A, Lim E, al Bakir M, Turati V, Guerra-Assunção JA, Conde L, Furness AJS, Saini SK, Hadrup SR, Herrero J, Lee SH, van Loo P, Enver T, Larkin J, Hellmann MD, Turajlic S, Quezada SA, McGranahan N, Swanton C, Metaanalysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TBK, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C, Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forde PM, Anagnostou V, Sun Z, Dahlberg SE, Kindler HL, Niknafs N, Purcell T, Santana-Davila R, Dudek AZ, Borghaei H, Lanis M, Belcaid Z, Smith KN, Balan A, White JR, Cherry C, Sivakumar IKA, Shao XM, Chan HY, Singh D, Thapa S, Illei PB, Pardoll DM, Karchin R, Velculescu VE, Brahmer JR, Ramalingam SS, Durvalumab with platinum-pemetrexed for unresectable pleural mesothelioma: Survival, genomic and immunologic analyses from the phase 2 PrE0505 trial. Nat. Med 27, 1910–1920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anagnostou V, Bruhm DC, Niknafs N, White JR, Shao XM, Sidhom JW, Stein J, Tsai H-L, Wang H, Belcaid Z, Murray J, Balan A, Ferreira L, Ross-Macdonald P, Wind-Rotolo M, Baras AS, Taube J, Karchin R, Scharpf RB, Grasso C, Ribas A, Pardoll DM, Topalian SL, Velculescu VE, Integrative tumor and immune cell mutli-omic analyses to predict melanoma response to immune checkpoint blockade. Cell Rep. Med 1, 100139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesan S, Angelova M, Puttick C, Zhai H, Caswell DR, Lu WT, Dietzen M, Galanos P, Evangelou K, Bellelli R, Lim EL, Watkins TBK, Rowan A, Teixeira VH, Zhao Y, Chen H, Ngo B, Zalmas LP, al Bakir M, Hobor S, Grönroos E, Pennycuick A, Nigro E, Campbell BB, Brown WL, Akarca AU, Marafioti T, Wu MY, Howell M, Boulton SJ, Bertoli C, Fenton TR, de Bruin RAM, Maya-Mendoza A, Santoni-Rugiu E, Hynds RE, Gorgoulis VG, Jamal-Hanjani M, McGranahan N, Harris RS, Janes SM, Bartkova J, Bakhoum SF, Bartek J, Kanu N, Swanton C; TRACERx Consortium, Induction of APOBEC3 exacerbates DNA replication stress and chromosomal instability in early breast and lung cancer evolution. Cancer Discov. 11, 2456–2473 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davoli T, Uno H, Wooten EC, Elledge SJ, Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-David U, Amon A, Context is everything: Aneuploidy in cancer. Nat. Rev. Genet 21, 44–62 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, Liu C, Sauter JL, Rekhtman N, Chang E, Callahan MK, Chaft JE, Voss MH, Tenet M, Li XM, Covello K, Renninger A, Vitazka P, Geese WJ, Borghaei H, Rudin CM, Antonia SJ, Swanton C, Hammerbacher J, Merghoub T, McGranahan N, Snyder A, Wolchok JD, Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33, 843–852.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA, Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I, Loquai C, Grabbe S, Vokes N, Margolis CA, Conway J, He MX, Elmarakeby H, Dietlein F, Miao D, Tracy A, Gogas H, Goldinger SM, Utikal J, Blank CU, Rauschenberg R, von Bubnoff D, Krackhardt A, Weide B, Haferkamp S, Kiecker F, Izar B, Garraway L, Regev A, Flaherty K, Paschen A, van Allen EM, Schadendorf D, Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghorani E, Ghorani E, Rosenthal R, McGranahan N, Reading JL, Lynch M, Peggs KS, Swanton C, Quezada SA, Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann. Oncol 29, 271–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Riaz N, Smoragiewicz M, Kelley ZL, Basturk O; Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer, Gönen M, Levine AJ, Allen PJ, Fearon DT, Merad M, Gnjatic S, Iacobuzio-Donahue CA, Wolchok JD, De Matteo RP, Chan TA, Greenbaum BD, Merghoub T, Leach SD, Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luksza M, Sethna ZM, Rojas LA, Lihm J, Bravi B, Elhanati Y, Soares K, Amisaki M, Dobrin A, Hoyos D, Guasp P, Zebboudj A, Yu R, Chandra AK, Waters T, Odgerel Z, Leung J, Kappagantula R, Makohon-Moore A, Johns A, Gill A, Gigoux M, Wolchok J, Merghoub T, Sadelain M, Patterson E, Monasson R, Mora T, Walczak AM, Cocco S, Iacobuzio-Donahue C, Greenbaum BD, Balachandran VP, Neoantigen quality predicts immunoediting in survivors of pancreatic cancer. Nature 606, 389–395 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran E, Robbins PF, Lu Y-C, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, Kriley IR, Rosenberg SA, T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med 375, 2255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandran SS, Ma J, Klatt MG, Dündar F, Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, Banks LB, Yi F, Vercher E, Etxeberria I, Bestman WD, da Cruz Paula A, Aricescu IS, Drilon A, Betel D, Scheinberg DA, Baker BM, Klebanoff CA, Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med 28, 946–957 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, al Bakir M, Chambers T, Salgado R, Savas P, Loi S, Birkbak NJ, Sansregret L, Gore M, Larkin J, Quezada SA, Swanton C, Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D, Makarov V, Hoen D, Dalin MG, Wexler L, Ghossein R, Katabi N, Nadeem Z, Cohen MA, Tian SK, Robine N, Arora K, Geiger H, Agius P, Bouvier N, Huberman K, Vanness K, Havel JJ, Sims JS, Samstein RM, Mandal R, Tepe J, Ganly I, Ho AL, Riaz N, Wong RJ, Shukla N, Chan TA, Morris LGT, Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med 25, 767–775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anagnostou V, Bruhm DC, Niknafs N, White JR, Shao XM, Sidhom JW, Stein J, Tsai HL, Wang H, Belcaid Z, Murray J, Balan A, Ferreira L, Ross-Macdonald P, Wind-Rotolo M, Baras AS, Taube J, Karchin R, Scharpf RB, Grasso C, Ribas A, Pardoll DM, Topalian SL, Velculescu VE, Integrative tumor and immune cell multi-omic analyses predict response to immune checkpoint blockade in melanoma. Cell Rep Med 1, 100139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao XM, Huang J, Niknafs N, Balan A, Cherry C, White J, Velculescu VE, Anagnostou V, Karchin R, HLA class II immunogenic mutation burden predicts response to immune checkpoint blockade. Ann. Oncol 33, 728–738 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kortleve D, Coelho RML, Hammerl D, Debets R, Cancer germline antigens and tumor-agnostic CD8(+) T cell evasion. Trends Immunol. 43, 391 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, Kuo F, Kendall SM, Requena D, Riaz N, Greenbaum B, Carroll J, Garon E, Hyman DM, Zehir A, Solit D, Berger M, Zhou R, Rizvi NA, Chan TA, Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chowell D, Krishna C, Pierini F, Makarov V, Rizvi NA, Kuo F, Morris LGT, Riaz N, Lenz TL, Chan TA, Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med 25, 1715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Moreno BH, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TNM, Lo RS, Ribas A, Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chhibber A, Huang L, Zhang H, Xu J, Cristescu R, Liu X, Mehrotra DV, Shen J, Shaw PM, Hellmann MD, Snyder A, Germline HLA landscape does not predict efficacy of pembrolizumab monotherapy across solid tumor types. Immunity 55, 56–64.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Sivapalan L, Anagnostou V, Genetic variation in antigen presentation and cancer immunotherapy. Immunity 55, 3–6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marty R, Kaabinejadian S, Rossell D, Slifker MJ, van de Haar J, Engin HB, de Prisco N, Ideker T, Hildebrand WH, Font-Burgada J, Carter H, MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, Swanton C, Allele-specific HLA Loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spranger S, Gajewski TF, Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 18, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.George S, Miao D, Demetri GD, Adeegbe D, Rodig SJ, Shukla S, Lipschitz M, Amin-Mansour A, Raut CP, Carter SL, Hammerman P, Freeman GJ, Wu CJ, Ott PA, Wong K-K, Van Allen EM, Loss of PTEN Is associated with resistance to anti-PD-1 check-point blockade therapy in metastatic uterine leiomyosarcoma. Immunity 46, 197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL Jr., Bosenberg MW, Woodman SE, Overwijk WW, Lizée G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P, Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 6, 202–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spranger S, Bao R, Gajewski TF, Melanoma-intrinsic β-catenin signalling prevents antitumour immunity. Nature 523, 231 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, le DT, Pardoll DM, Diaz LA Jr., Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A, Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 7, 188–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, Chen P-L, Hwu P, Allison JP, Futreal A, Wargo JA, Sharma P, Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, Schrock AB, Hartmaier RJ, Trabucco SE, Gay L, Ali SM, Elvin JA, Singal G, Ross JS, Fabrizio D, Szabo PM, Chang H, Sasson A, Srinivasan S, Kirov S, Szustakowski J, Vitazka P, Edwards R, Bufill JA, Sharma N, Ou S-HI, Peled N, Spigel DR, Rizvi H, Aguilar EJ, Carter BW, Erasmus J, Halpenny DF, Plodkowski AJ, Long NM, Nishino M, Denning WL, Galan-Cobo A, Hamdi H, Hirz T, Tong P, Wang J, Rodriguez-Canales J, Villalobos PA, Parra ER, Kalhor N, Sholl LM, Sauter JL, Jungbluth AA, Mino-Kenudson M, Azimi R, Elamin YY, Zhang J, Leonardi GC, Jiang F, Wong K-K, Lee JJ, Papadimitrakopoulou VA, Wistuba II, Miller VA, Frampton GM, Wolchok JD, Shaw AT, Jänne PA, Stephens PJ, Rudin CM, Geese WJ, Albacker LA, Heymach JV, STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casey SC, Tong L, Li Y, do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW, MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Topper MJ, Vaz M, Chiappinelli KB, Shields CEDS, Niknafs N, Yen R-WC, Wenzel A, Hicks J, Ballew M, Stone M, Tran PT, Zahnow CA, Hellmann MD, Anagnostou V, Strissel PL, Strick R, Velculescu VE, Baylin SB, Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gavralidis A, Gainor JF, Immunotherapy in EGFR-mutant and ALK-positive lung cancer: Implications for oncogene-driven lung cancer. Cancer J. 26, 517–524 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Hastings K, Yu HA, Wei W, Sanchez-Vega F, DeVeaux M, Choi J, Rizvi H, Lisberg A, Truini A, Lydon CA, Liu Z, Henick BS, Wurtz A, Cai G, Plodkowski AJ, Long NM, Halpenny DF, Killam J, Oliva I, Schultz N, Riely GJ, Arcila ME, Ladanyi M, Zelterman D, Herbst RS, Goldberg SB, Awad MM, Garon EB, Gettinger S, Hellmann MD, Politi K, EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann. Oncol 30, 1311–1320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skoulidis F, Heymach JV, Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19, 495–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marinelli D, Mazzotta M, Scalera S, Terrenato I, Sperati F, D’Ambrosio L, Pallocca M, Corleone G, Krasniqi E, Pizzuti L, Barba M, Carpano S, Vici P, Filetti M, Giusti R, Vecchione A, Occhipinti M, Gelibter A, Botticelli A, De Nicola F, Ciuffreda L, Goeman F, Gallo E, Visca P, Pescarmona E, Fanciulli M, De Maria R, Marchetti P, Ciliberto G, Maugeri-Saccà M, KEAP1-driven co-mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann. Oncol 31, 1746–1754 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, Lund T, Tanić M, Reading JL, Joshi K, Henry JY, Ghorani E, Wilson GA, Birkbak NJ, Jamal-Hanjani M, Veeriah S, Szallasi Z, Loi S, Hellmann MD, Feber A, Chain B, Herrero J, Quezada SA, Demeulemeester J, Van Loo P, Beck S, Granahan NM, Swanton C; The TRACERx consortium, Neoantigen-directed immune escape in lung cancer evolution. Nature 567, 479–485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE, Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK, IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, Zhang C, Lunceford JK, Joe A, Cheng J, Webber AL, Ibrahim N, Plimack ER, Ott PA, Seiwert TY, Ribas A, McClanahan TK, Tomassini JE, Loboda A, Kaufman D, Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cabrita R, Lauss M, Sanna A, Donia M, Larsen MS, Mitra S, Johansson I, Phung B, Harbst K, Vallon-Christersson J, van Schoiack A, Lövgren K, Warren S, Jirström K, Olsson H, Pietras K, Ingvar C, Isaksson K, Schadendorf D, Schmidt H, Bastholt L, Carneiro A, Wargo JA, Svane IM, Jönsson G, Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, Gopalakrishnan V, Xi Y, Zhao H, Amaria RN, Tawbi HA, Cogdill AP, Liu W, Le Bleu VS, Kugeratski FG, Patel S, Davies MA, Hwu P, Lee JE, Gershenwald JE, Lucci A, Arora R, Woodman S, Keung EZ, Gaudreau P-O, Reuben A, Spencer CN, Burton EM, Haydu LE, Lazar AJ, Zapassodi R, Hudgens CW, Ledesma DA, Ong SF, Bailey M, Warren S, Rao D, Krijgsman O, Rozeman EA, Peeper D, Blank CU, Schumacher TN, Butterfield LH, Zelazowska MA, McBride KM, Kalluri R, Allison J, Petitprez F, Fridman WH, Sautès-Fridman C, Hacohen N, Rezvani K, Sharma P, Tetzlaff MT, Wang L, Wargo JA, B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petitprez F, de Reyniès A, Keung EZ, Chen TWW, Sun CM, Calderaro J, Jeng YM, Hsiao LP, Lacroix L, Bougoüin A, Moreira M, Lacroix G, Natario I, Adam J, Lucchesi C, Laizet Y′, Toulmonde M, Burgess MA, Bolejack V, Reinke D, Wani KM, Wang WL, Lazar AJ, Roland CL, Wargo JA, Italiano A, Sautès-Fridman C, Tawbi HA, Fridman WH, B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020). [DOI] [PubMed] [Google Scholar]
- 91.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS, Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med 24, 1550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS, Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, de Macedo MP, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba II, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA, Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, Tian T, Wei Z, Madan S, Sullivan RJ, Boland G, Flaherty K, Herlyn M, Ruppin E, Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med 24, 1545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JH, Shklovskaya E, Lim SY, Carlino MS, Menzies AM, Stewart A, Pedersen B, Irvine M, Alavi S, Yang JYH, Strbenac D, Saw RPM, Thompson JF, Wilmott JS, Scolyer RA, Long GV, Kefford RF, Rizos H, Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat. Commun 11, 1897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, le PM, Forman J, Huang T, Li S, Zhang W, Xu Q, Cieri N, Clauser KR, Shukla SA, Neuberg D, Justesen S, MacBeath G, Carr SA, Fritsch EF, Hacohen N, Sade-Feldman M, Livak KJ, Boland GM, Ott PA, Keskin DB, Wu CJ, Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caushi JX, Zhang J, Ji Z, Vaghasia A, Zhang B, Hsiue EH-C, Mog BJ, Hou W, Justesen S, Blosser R, Tam A, Anagnostou V, Cottrell TR, Guo H, Chan HY, Singh D, Thapa S, Dykema AG, Burman P, Choudhury B, Aparicio L, Cheung LS, Lanis M, Belcaid Z, Asmar ME, Illei PB, Wang R, Meyers J, Schuebel K, Gupta A, Skaist A, Wheelan S, Naidoo J, Marrone KA, Brock M, Ha J, Bush EL, Park BJ, Bott M, Jones DR, Reuss JE, Velculescu VE, Chaft JE, Kinzler KW, Zhou S, Vogelstein B, Taube JM, Hellmann MD, Brahmer JR, Merghoub T, Forde PM, Yegnasubramanian S, Ji H, Pardoll DM, Smith KN, Transcriptional programs of neo-antigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lahnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, Pinello L, Skums P, Stamatakis A, Attolini CS-O, Aparicio S, Baaijens J, Balvert M, de Barbanson B, Cappuccio A, Corleone G, Dutilh BE, Florescu M, Guryev V, Holmer R, Jahn K, Lobo TJ, Keizer EM, Khatri I, Kielbasa SM, Korbel JO, Kozlov AM, Kuo T-H, Lelieveldt BPF, Mandoiu II, Marioni JC, Marschall T, Mölder F, Niknejad A, Raczkowski L, Reinders M, de Ridder J, Saliba A-E, Somarakis A, Stegle O, Theis FJ, Yang H, Zelikovsky A, McHardy AC, Raphael BJ, Shah SP, Schönhuth A, Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, Marrone K, Sivakumar IKA, Bruhm DC, Rosner S, Phallen J, Leal A, Adleff V, Smith KN, Cottrell TR, Rhymee L, Palsgrove DN, Hann CL, Levy B, Feliciano J, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Isbell JM, Sauter JL, Taube J, Scharpf RB, Karchin R, Pardoll DM, Chaft JE, Hellmann MD, Brahmer JR, Velculescu VE, Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res. 79, 1214–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, Speir S, Reinert T, Orntoft M-BW, Woodward BD, Murphy D, Parpart-Li S, Riley D, Nesselbush M, Sengamalay N, Georgiadis A, Li QK, Madsen MR, Mortensen FV, Huiskens J, Punt C, van Grieken N, Fijneman R, Meijer G, Husain H, Scharpf RB, Diaz LA Jr., Jones S, Angiuoli S, Ørntoft T, Nielsen HJ, Andersen CL, Velculescu VE, Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med 9, eaan2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Phallen J, Leal A, Woodward BD, Forde PM, Naidoo J, Marrone KA, Brahmer JR, Fiksel J, Medina JE, Cristiano S, Palsgrove DN, Gocke CD, Bruhm DC, Keshavarzian P, Adleff V, Weihe E, Anagnostou V, Scharpf RB, Velculescu VE, Husain H, Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res. 79, 1204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, Lheureux S, Spreafico A, Razak AA, Shchegrova S, Louie M, Billings P, Zimmermann B, Sethi H, Aleshin A, Torti D, Marsh K, Eagles J, Cirlan I, Hanna Y, Clouthier DL, Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 1, 873 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Nabet BY, Esfahani MS, Moding EJ, Hamilton EG, Chabon JJ, Rizvi H, Steen CB, Chaudhuri AA, Liu CL, Hui AB, Almanza D, Stehr H, Gojenola L, Bonilla RF, Jin MC, Jeon Y-J, Tseng D, Liu C, Merghoub T, Neal JW, Wakelee HA, Padda SK, Ramchandran KJ, Das M, Plodkowski AJ, Yoo C, Chen EL, Ko RB, Newman AM, Hellmann MD, Alizadeh AA, Diehn M, Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 183, 363–376.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, Abdullah SE, Higgs BW, Dennis PA, van der Heijden MS, Segal NH, Chaft JE, Hembrough T, Barrett JC, Hellmann MD, Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 10, 1842–1853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Al Zoughbi W, Fox J, Beg S, Papp E, Hissong E, Ohara K, Keefer L, Sigouros M, Kane T, Bockelman D, Nichol D, Patchell E, Bareja R, Karandikar A, Alnajar H, Cerqueira G, Guthrie VB, Verner E, Manohar J, Greco N, Wilkes D, Tagawa S, Malbari MS, Holcomb K, Eng KW, Shah M, Altorki NK, Sboner A, Nanus D, Faltas B, Sternberg CN, Simmons J, Houvras Y, Molina AM, Angiuoli S, Elemento O, Mosquera JM, Validation of a circulating tumor DNA-based next-generation sequencing assay in a cohort of patients with solid tumors: A proposed solution for decentralized plasma testing. Oncologist 26, e1971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, Meng W, Dewal N, Milbury C, Clark T, Donahue A, Stover D, Kennedy M, Dacpano-Komansky J, Burns C, Vietz C, Alexander B, Hegde P, Dennis L, Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLOS ONE 15, e0237802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, Broeckaert MAM, Anagnostou V, Adleff V, Bruhm DC, Canzoniero JV, Fiksel J, Nordsmark M, Warmerdam FARM, Verheul HMW, van Spronsen DJ, Beerepoot LV, Geenen MM, Portielje JEA, Jansen EPM, van Sandick J, Kranenbarg EM-K, van Laarhoven HWM, van der Peet DL, van de Velde CJH, Verheij M, Fijneman R, Scharpf RB, Meijer GA, Cats A, Velculescu VE, White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat. Commun 11, 525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G, Malboeuf C, Lieber DS, Lipson D, Silterra J, Amler L, Riehl T, Cummings CA, Hegde PS, Sandler A, Ballinger M, Fabrizio D, Mok T, Shames DS, Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med 24, 1441 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Si H, Kuziora M, Quinn KJ, Helman E, Ye J, Liu F, Scheuring U, Peters S, Rizvi NA, Brohawn PZ, Ranade K, Higgs BW, Banks KC, Chand VK, Raja R, A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC Treatment: Results from the MYSTIC study. Clin. Cancer Res 27, 1631–1640 (2021). [DOI] [PubMed] [Google Scholar]
- 110.Georgiadis A, Durham JN, Keefer LA, Bartlett BR, Zielonka M, Murphy D, White JR, Lu S, Verner EL, Ruan F, Riley D, Anders RA, Gedvilaite E, Angiuoli S, Jones S, Velculescu VE, le DT, Diaz LA Jr., Sausen M, Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin. Cancer Res 25, 7024–7034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, Catenacci DVT, Fakih M, Barbacioru C, Zhao J, Sikora M, Fairclough SR, Lee H, Kim K-M, Kim ST, Kim J, Gavino D, Benavides M, Peled N, Nguyen T, Cusnir M, Eskander RN, Azzi G, Yoshino T, Banks KC, Raymond VM, Lanman RB, Chudova DI, Talasaz AA, Kopetz S, Lee J, Odegaard JI, Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin. Cancer Res 25, 7035 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Silveira AB, Bidard F-C, Kasperek A, Melaabi S, Tanguy M-L, Rodrigues M, Bataillon G, Cabel L, Buecher B, Pierga J-Y, Proudhon C, Stern M-H, High-accuracy determination of microsatellite instability compatible with liquid biopsies. Clin. Chem 66, 606–613 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Kim ES, Velcheti V, Mekhail T, Yun C, Shagan SM, Hu S, Chae YK, Leal TA, Dowell JE, Tsai ML, Dakhil CSR, Stella P, Jin Y, Shames DS, Schleifman E, Fabrizio DA, Phan S, Socinski MA, Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: The phase 2 B-F1RST trial. Nat. Med 28, 939 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herbst RS, Giaccone G, de Marinis F, Reinmuth N, Vergnenegre A, Barrios CH, Morise M, Felip E, Andric Z, Geater S, Özgüroğlu M, Zou W, Sandler A, Enquist I, Komatsubara K, Deng Y, Kuriki H, Wen X, Cleland MM, Mocci S, Jassem J, Spigel DR, Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N. Engl. J. Med 383, 1328 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Passaro A, Brahmer J, Antonia S, Mok T, Peters S, Managing resistance to immune checkpoint inhibitors in lung cancer: Treatment and novel strategies. J. Clin. Oncol 40, 598 (2022). [DOI] [PubMed] [Google Scholar]
- 116.Cristiano S, Leal A, Phallen J, Adleff JFV, Bruhm DC, Jensen SØ, Medina JE, Hruban C, White JR, Palsgrove DN, Niknafs N, Anagnostou V, Forde P, Naidoo J, Marrone K, Brahmer J, Woodward BD, Husain H, van Rooijen KL, Ørntoft M-BW, Madsen AH, van de Velde CJH, Verheij M, Cats A, Punt CJA, Vink GR, van Grieken NCT, Koopman M, Fijneman RJA, Johansen JS, Nielsen HJ, Meijer GA, Andersen CL, Scharpf RB, Velculescu VE, Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mathios D, Johansen JS, Cristiano S, Medina JE, Phallen J, Larsen KR, Bruhm DC, Niknafs N, Ferreira L, Adleff V, Chiao JY, Leal A, Noe M, White JR, Arun AS, Hruban C, Annapragada AV, Jensen SØ, Ørntoft MBW, Madsen AH, Carvalho B, de Wit M, Carey J, Dracopoli NC, Maddala T, Fang KC, Hartman AR, Forde PM, Anagnostou V, Brahmer JR, Fijneman RJA, Nielsen HJ, Meijer GA, Andersen CL, Mellemgaard A, Bojesen SE, Scharpf RB, Velculescu VE, Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun 12, 5060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]