Abstract
Lanthipeptides are ribosomally synthesized and posttranslationally modified peptide natural products characterized by the presence of lanthionine and methyllanthionine crosslinked amino acids formed by dehydration of Ser/Thr residues followed by conjugate addition of Cys onto the resulting dehydroamino acids. Class I lanthipeptide dehydratases utilize glutamyl-tRNAGlu as a cosubstrate to glutamylate Ser/Thr followed by glutamate elimination. The vast majority of lanthipeptides identified from class I synthase systems have been from Gram-positive bacteria. Herein, we report the heterologous expression and modification in Escherichia coli of two lanthipeptides from the Gram-negative Bacteroidetes Pedobacter lusitanus NL19. These peptides are representative of a group of compounds frequently encoded in Pedobacter genomes. Structural characterization of the lanthipeptides revealed a novel ring pattern as well as an unusual LL-lanthionine stereochemical configuration and a cyclase that lacks the canonical zinc ligands found in most LanC enzymes.
Graphical Abstract
Introduction
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) encompass a rapidly expanding class of natural products.1 RiPPs are biosynthesized from a genetically encoded precursor peptide, which contains an N-terminal leader peptide and a C-terminal core peptide. This precursor peptide is acted upon by an array of enzymes, typically encoded within the same biosynthetic gene cluster (BGC), that often bind to the leader peptide region and chemically modify the core peptide.1, 2 The recent expansion of characterized RiPPs is fueled by the ever-increasing availability of genomic data and by synthetic biology methods to access compounds by heterologous production.1, 3, 4
One of the most extensively studied classes of RiPPs is the lanthipeptides, which are characterized by the presence of lanthionine (Lan) and methyllanthionine (MeLan) residues that result in complex polycyclic peptide structures that may be further tailored with additional posttranslational modifications.5, 6 The (Me)Lan cross-links can be installed by one of five classes of lanthipeptide synthase systems (class I-V), which first dehydrate target Ser and Thr residues in the core peptide to generate dehydroalanine (Dha) and dehydrobutyrine (Dhb).5, 7–9 Cysteine residues located in the core peptide then react with these dehydrated residues via intramolecular Michael-type addition to form the thioether rings.
Class I lanthipeptide biosynthetic machinery is generally composed of a LanB dehydratase, which is responsible for activation and elimination of the side chain hydroxyl groups of Ser and Thr residues,10 and a LanC cyclase that is responsible for thioether formation.11 LanB dehydratases utilize glutamyl-tRNAGlu to first glutamylate the side chains of target Ser/Thr prior to glutamate elimination (Figure 1A).12, 13 Understanding of this unusual transformation has been enhanced through biochemical, crystallographic, microscopy, and bioinformatics studies,14–21 which in turn have facilitated exploration of the chemical space of lanthipeptides produced from class I BGCs by heterologous expression strategies. The overwhelming majority of class I lanthipeptides isolated to date are derived from BGCs from Gram-positive bacteria (predominantly Firmicutes and Actinobacteria).22–26 However, surveys of genomic data have revealed that Gram-negative phyla (Bacteroidetes, in particular) contain a wealth of under-explored LanB-containing BGCs.26, 27 As such, these BGCs represent a potential source of lanthipeptides containing novel chemical structures and bioactivities. This potential is highlighted by the characterization of the pinensins from the Bacteroidetes Chitinophaga pinensis, which are the first reported lanthipeptides that exhibit antifungal activity.28
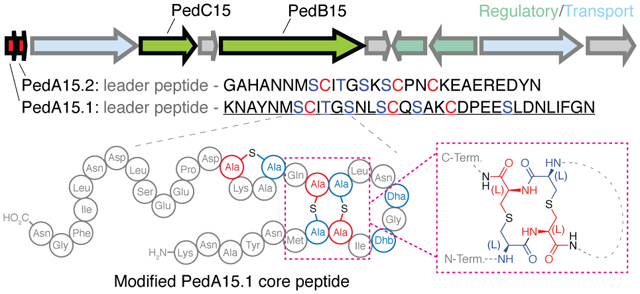
Figure 1: Class I lanthipeptide biosynthesis in Pedobacter lusitanus NL19.
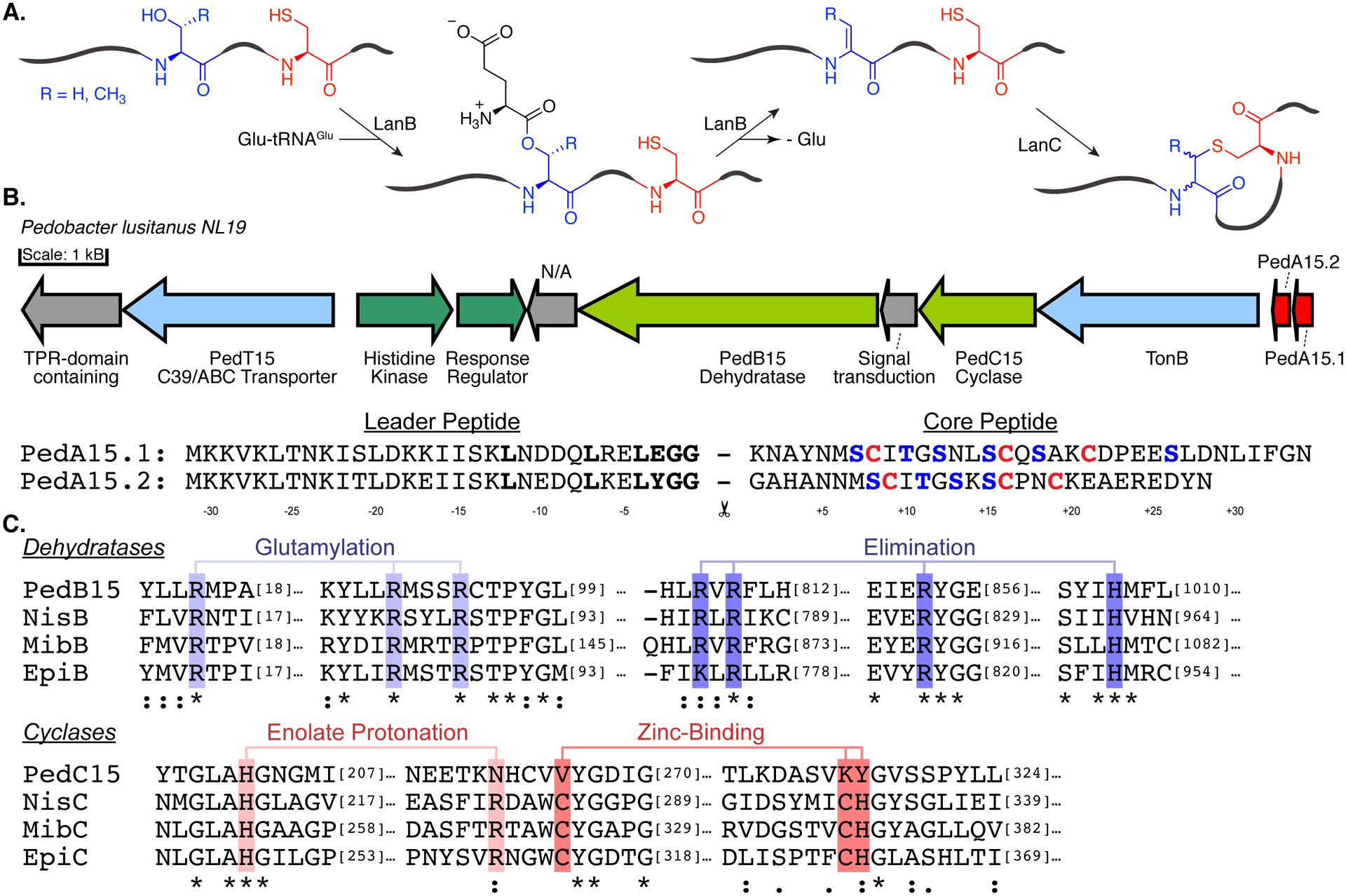
(A) Schematic overview of (methyl)lanthionine formation by class I lanthipeptide synthases. (B) Ped15 biosynthetic gene cluster and the precursor peptides examined in this study. Red, precursor peptides; Blue, transport; Olive, core biosynthetic machinery; Green, regulatory proteins. N/A no annotation. BGC components visualized using the Gene Graphics web tool.85 Previously identified recognition motif for LahT150 is shown in bold. (C) Sequence conservation of catalytic residues in the core biosynthetic enzymes of the Ped15 biosynthetic gene cluster. Key catalytic residues are highlighted in blue (LanBs) and red (LanC’s) and are labeled according to their proposed mechanistic role.
An example of the untapped abundance and diversity of class I lanthipeptides is illustrated by genomic analysis of Pedobacter lusitanus NL19, a Bacteroidetes originally isolated from depleted uranium mine runoff.29 This strain was shown to encode at least five class I lanthipeptide BGCs with potentially novel lanthionine topologies based on the putative precursor peptide sequences.27 Herein we describe the heterologous expression and structural characterization of two novel lanthipeptides from this strain. The precursor peptide-encoding genes pedA15.1 and pedA15.2 were identified previously within a single BGC (Figure 1B) that also encodes a putative class I dehydratase (PedB15) and cyclase (PedC15). They were chosen for study as representative members of a group of lanthipeptides encoded in Bacteroidetes that have no sequence similarity with known compounds. In the current study, coexpression of these components in E. coli resulted in full modification of the precursor peptides only if glutamyl-tRNA synthetase (GluRS) and tRNAGlu from P. lusitanus were co-expressed. Structural analysis of the product revealed a hitherto uncharacterized Lan pattern as determined by mass spectrometry and NMR spectroscopy. This ring pattern appears common in Pedobacter. Interestingly, while the dehydratase contained all of the requisite catalytic residues, a sequence alignment of the PedC15 cyclase and its orthologs in Pedobacter with previously known lanthipeptide cyclases demonstrates the absence of a canonical zinc-binding site.30–32 Despite this absence, the PedA15 peptides were fully cyclized whereas cyclization was incomplete in the absence of the PedC15 cyclase. Most structurally characterized lanthipeptides have been shown to contain (methyl)lanthionine in the DL-configuration (D at the α-carbon that used to be Ser/Thr, and L on the α-carbon that used to be Cys).33 For some lanthipeptides, the stereochemical outcome of the cyclization reaction is influenced by the substrate sequence itself and leads to (methyl)lanthionines in the LL-configuration.34 Given the previous reports of substrate control over the stereochemical outcomes of lanthionine formation,35 the stereochemistry of the lanthionine products from the Pedobacter BGC was analyzed demonstrating that both PedA15 peptides contain lanthionine residues with the uncommon LL-stereochemical configuration.
Results and Discussion
Heterologous Expression of PedA15.1 and PedA15.2.
Previous studies did not succeed in eliciting production of the lanthipeptides encoded in the BGCs in Pedobacter lusitanus NL19.27, 36 Therefore in this work we focused on heterologous expression in E. coli (Figure S1).37–40 We chose the Ped15 BGC because of the unusual features mentioned above. Comparison of related BGCs from other Pedobacter strains (Figure S2) suggests that the cluster shown in Figure 1B contains the minimal genes required for biosynthesis. The two precursor peptides PedA15.1 and PedA15.2 both contain leader peptides ending in a characteristic double Gly motif (Figure 1B) consistent with the presence of a gene encoding an ABC transporter with a C-terminal C39 protease domain that typically removes this family of leader peptides.41 The sequences of the core peptides of PedA15.1 and PedA15.2 are quite different although they do share similar potential ring-forming motifs (Figure 1B). The discovery that LanB dehydratases utilize tRNAGlu to carry out dehydration of LanA precursor peptides has greatly facilitated heterologous expression strategies. Inclusion of the aminoacyl tRNA synthetase gene and an appropriate tRNA isoacceptor from the species of origin often considerably improves production of the fully modified natural product.14, 15 The improved activity of the LanB dehydratases is likely due to the high degree of conservation in tRNA sequences within phyla and the likelihood that LanB enzymes have coevolved to recognize specific tRNAs based on nucleotide sequence in the species of origin. In keeping with these observations, our initial attempts to obtain modified His6-tagged PedA15.1 and PedA15.2 through coexpression with PedB15 and PedC15 in E. coli were unsuccessful, resulting in minimally dehydrated peptides (Figure 2A). This poor activity of PedB15 with the glutamyl-tRNA of E. coli is likely the result of differences in the tRNAGlu discriminator-base sequence between E. coli and P. lusitanus NL19 (Figure 2B),15 and not because of non-functionality of the enzyme, which contains all previously identified catalytic residues (Figure 1C). Indeed, the degree of dehydration of both precursor peptides improved greatly when P. lusitanus GluRS and tRNAGlu were coexpressed with the lanthionine synthase machinery (Figure 2C). Whereas no dehydration was observed in the presence of endogenous E. coli tRNAGlu, four to five dehydrations were observed with PedA15.1 when using GluRS/tRNAGlu from P. lusitanus NL19. Since the core peptide of PedA15.1 contains six Ser/Thr residues, some Ser/Thr residues escape dehydration. With PedA15.2, up to four dehydrations were observed indicating all four Ser/Thr in the core peptide were enzymatically modified.
Figure 2. Heterologous expression of modified PedA15 peptides in E. coli and analysis by MALDI-TOF MS.
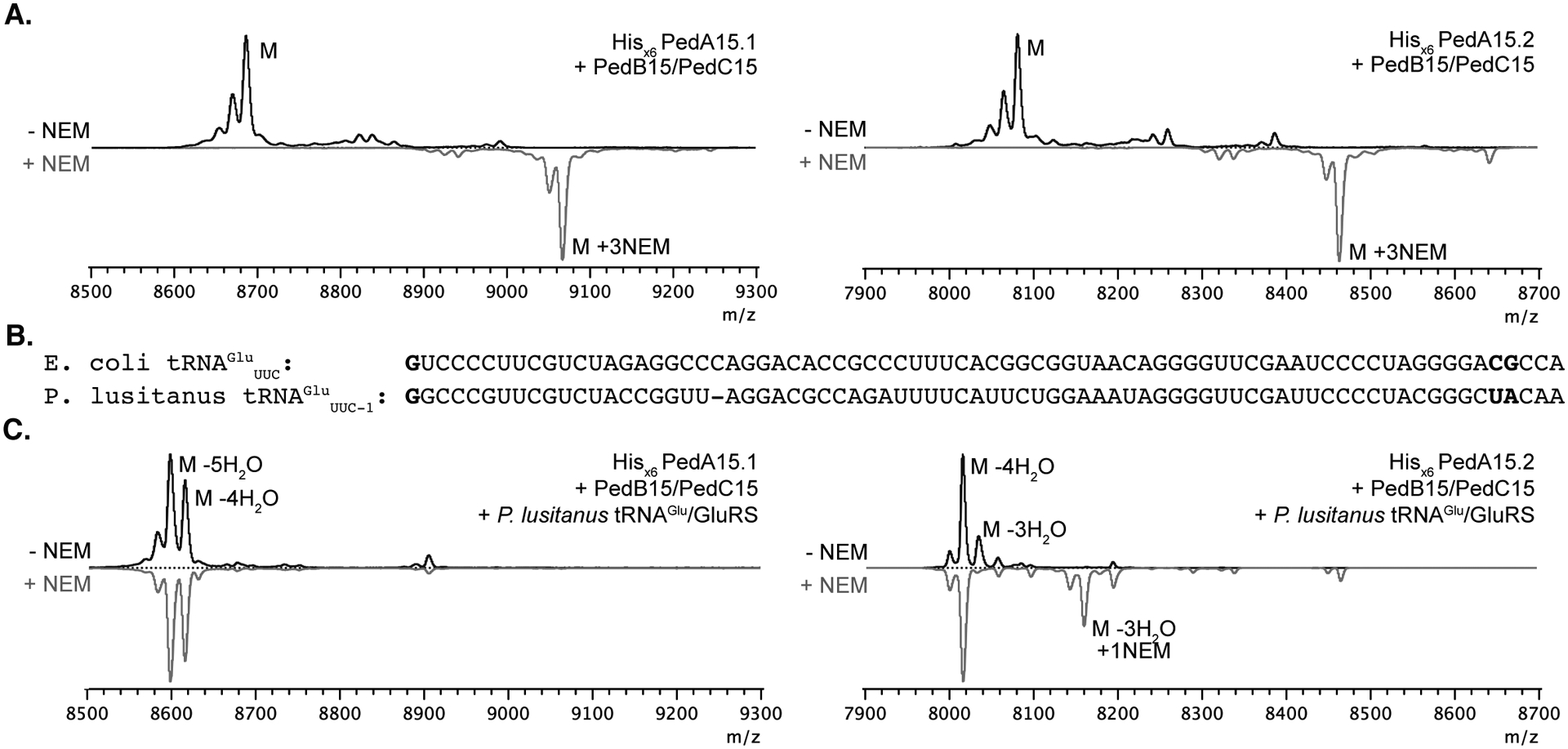
(A) Coexpression in E. coli BL21 (DE3) of PedA15 precursor peptides, PedB15 and PedC15 in the absence of P. lusitanus tRNAGlu/GluRS. Both PedA15.1 (left; avg. m/z 8687.7 calc.; 8686.6 obs.) and PedA15.2 (right; avg. m/z 8085.0 calc.; 8085.4 obs.) were unmodified in the absence of aminoacyl-tRNA from the native producer. Consequently, NEM treatment of these samples resulted in full alkylation of all Cys residues (grey spectra). (B) Comparison of tRNAGlu sequences in E. coli (Proteobacterium) and P. lusitanus NL19 (Bacteroidetes). The anticodon is denoted in subscript. Nucleotide positions previously shown to be important for LanB compatibility are in bold.15 (C) PedA15 precursor peptides co-expressed with PedB15, PedC15 and P. lusitanus tRNAGlu/GluRS. Isolated product for PedA15.1 (left) contained a mixture of 4x (avg. m/z 8615.7 calc.; 8615.8 obs.) and 5x (avg. m/z 8597.6 calc.; 8597.0 obs.) dehydrated peptide and was fully cyclized as evidenced by a lack of NEM alkylation. PedA15.2 (right) was purified as a mixture of 3x (avg. m/z 8030.9 calc.; 8030.5 obs.) and 4x dehydrated (avg. m/z 8012.9 calc.; 8012.7 obs.) peptide, with a minor amount of partially cyclized peptide product observed by NEM alkylation. M, parental mass (avg. m/z, unmodified peptide); NEM, N-ethylmaleimide (grey traces); “-n” H2O, indicates the number (n) of dehydrations consistent with the m/z of the labeled peak.
The degree of cyclization in the purified peptides was examined using an N-ethylmaleimide (NEM) alkylation assay that probes for free cysteine within the core peptide region.42 Modified PedA15.1 that had been coexpressed with PedB15, PedC15, and P. lusitanus GluRS/tRNAGlu did not react with NEM suggesting it was completely cyclized (Figure 2C). In contrast, metal affinity and HPLC-purified full-length PedA15.2 obtained from co-expression showed partial reactivity towards NEM leading to a minor product with one NEM adduct, suggesting the partially cyclized minor peptide contained two of three possible lanthionine rings (Figure 2C). This mixture of products could not be separated by HPLC of the full-length peptide, but removal of the leader peptide through protease digestion enabled removal of the partially modified PedA15.2 product as discussed below.
Proteolytic removal of the leader peptide.
To access the core peptides of PedA15.1 and PedA15.2 for structural analysis, leader peptide removal was optimized. Initial attempts made use of the N-terminal C39-protease domain of the bifunctional transporter LahT (LahT150), which was previously demonstrated to be highly substrate tolerant toward a variety of lanthipeptide precursor peptides containing the conserved GG motif of Nif11-family leader peptides.43–45 Leader peptide removal with LahT150 was only observed in the case of PedA15.2 (Figure 3B, Figure S3). No proteolysis was observed in the case of PedA15.1, even though it contains the conserved amino acid sequence within the leader region (L-12, L-7, L-4, G-2, G-1) that has been previously shown to constitute the recognition motif for LahT150 and related proteases (Figure S3).45–47 The residue at the P1’ position of PedA15.1 is Lys and it is possible that the positively charged side chain interferes with LahT150 recognition since none of the nine cognate LahA substrates has a positive charge at position P1’.45 A second clear difference between PedA15.1 and 15.2 near the leader peptide cleavage site is the Glu at position P2 in the former, which is a Tyr in the latter (Figure 1B). The Glu may prevent cleavage as this residue is an Ala in all LahA substrates, but a previous mutagenesis study on LahA peptides showed that mutation to Glu was tolerated by LahT150. Regardless, an alternative approach was adopted and endoproteinase LysN was effective at removing the leader peptide of PedA15.1 (Figure 3A). Antimicrobial activity assays against a panel of bacteria and fungi did not result in any detectable activity for either modified core peptide or their combination (Table S3).
Figure 3.
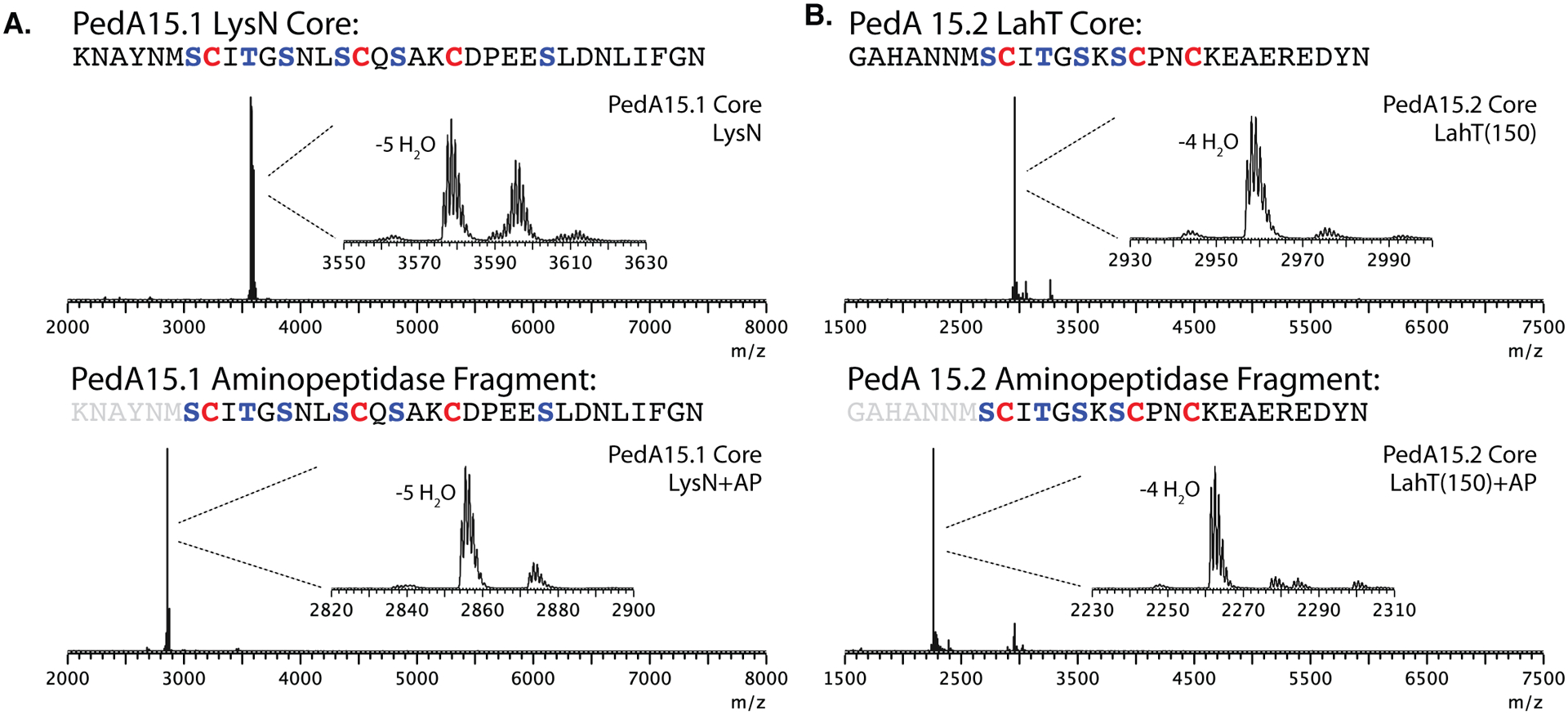
Isolation and analysis of modified PedA15 core peptides by mass spectrometry. (A) Proteolytic digestion and MALDI-TOF MS analysis of PedA15.1 core peptide fragments. Modified PedA15.1 peptide was treated with LysN and analyzed (top; 5x dehydrated, [M+H] mono. m/z 3578.9 calc.; 3578.5 obs.) followed by aminopeptidase treatment and analysis by MALDI-TOF MS (bottom; 5x dehydrated, [M+H] mono. m/z 2855.2 calc.; 2855.5 obs.). (B) Analysis of PedA15.2 core peptide fragments generated through proteolysis. Modified PedA15.2 was treated with LahT150 and analyzed by MALDI-TOF MS (top; 4x dehydrated, [M] avg., m/z 2958.2 calc.; 2958.5 obs.) followed by aminopeptidase treatment (bottom; 4x dehydrated, [M+H] mono., m/z 2261.9 calc.; 2262.5 obs.). AP, aminopeptidase.
For most lanthipeptides containing a Gly-Gly motif leader peptide, cleavage with the C39 protease domain of the transporter provides the mature natural product. However, for a subset of compounds, such as the enterococcal cytolysin, haloduracin, lichenicidin, pinensin, carnolysin and bicereucin a short N-terminal segment of the core peptide-region is removed as part of a second proteolytic event by a protease that is not always encoded in the BGC.28, 40, 48–55 Since the products of the ped15 BGC have not been isolated from the native organism,36 it is not known whether such proteolysis occurs during PedA15 precursor maturation. To explore this possibility and access the potential products of such a modification, PedBC-modified PedA15.1 and PedA15.2 were treated with aminopeptidase following initial leader peptide removal at the Gly-Gly motif using LahT150 or LysN as described above (Figure 3A, 3B). Such treatment resulted in the removal of the N-terminal 6–7 amino acids following the GlyGly sequence, with aminopeptidase activity terminating at the first lanthionine rings in both peptides. Unfortunately, no antimicrobial activity for these peptides, individually or in combination, could be observed against any of the bacterial or fungal strains tested (Table S3).
MS/MS and analysis of lanthionine pattern by mutational analysis.
The PedA peptides do not have any clear sequence homology with characterized lanthipeptides.26 However, analogous peptides are encoded in other Bacteroidetes (Figure 4D). Given the potential for novel ring patterns with the PedA15 peptides, we initially tried to determine their structures using a combination of tandem mass spectrometry (MS/MS) and mutational analysis. Modified wild-type PedA15.1 and PedA15.2 core peptides were first purified by HPLC. For wild-type PedA15.1, upon leader peptide removal five- and fourfold dehydrated core peptide was isolated (Figure 3A). NEM assay of the purified core peptide confirmed the previous results on full-length peptide that PedA15.1 was fully cyclized, regardless of the dehydration state (Figure S4A). In the case of wild-type PedA15.2, the mixture of fully and partially modified core peptides were separable by HPLC. This procedure provided access to a four-fold dehydrated and fully-cyclized core peptide and a three-fold dehydrated core peptide lacking one of its three possible lanthionine rings, as determined by NEM assay (Figure S4B). Determination of the residues involved in Lan/MeLan formation can sometimes be achieved by site-directed mutagenesis. However, alanine scanning of potential ring forming residues in PedA15.1 still resulted in fully cyclized peptide as well as buildup of undesired glutathionylated peptide adducts formed in E. coli, suggestive of alternate topologies for alanine variants and making structural assessment by this approach impossible (Figure S5).
Figure 4. NMR analysis of cyclized PedA15.1 core peptide.
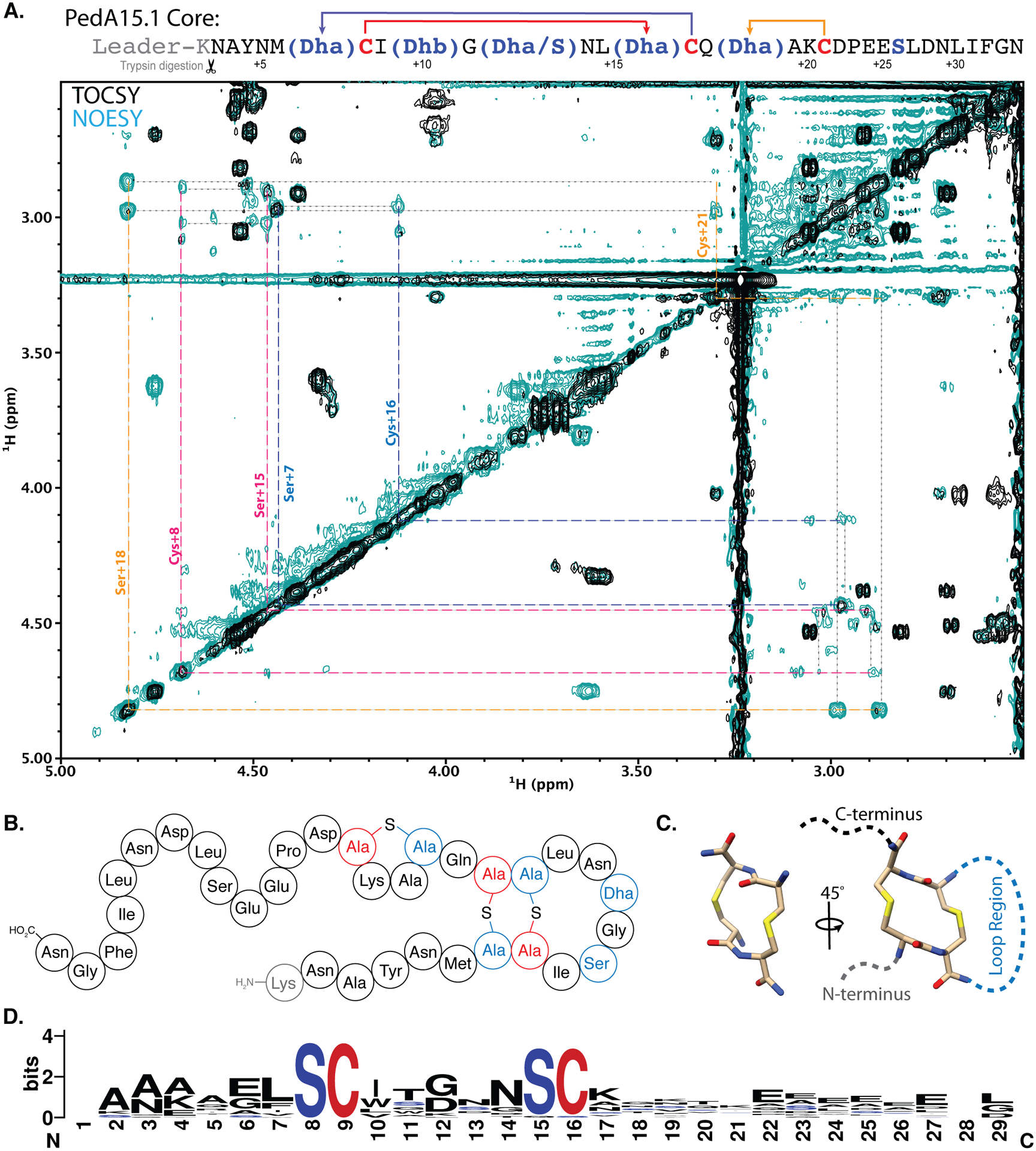
(A) 2D-NMR overlay (TOCSY, black; NOESY, teal) of the aliphatic region highlighting CH2-S-CH2 intra-bridge correlations used to infer lanthionine ring pattern. Intra-bridge NOEs are denoted with black dotted lines. Residues and lanthionine ring systems are color coded according to the sequence diagram at the top. The NMR data also showed that the difference between 4 and 5-fold dehydrated peptide is at Ser12, and that this residue is not involved in ring formation. For full structural assignment data, see Supporting Information. (B) Diagram representing the pattern of cyclized PedA15.1 that is dehydrated five times, as determined by NMR and MS/MS analysis (C) Representation of the unusual 14-membered bis-lanthionine heterocycle identified in the PedA15 peptides. 3D model generated in Avogadro86 representing the LL-Lan/LL-Lan stereochemical configuration. (D) Sequence logo for the core peptide of a family of class I lanthipeptide precursor peptides encoded in Bacteroidetes. See Figure S13 for individual peptides. The SCXnSC motif (n = 3–6, Fig. S13) is highlighted in red and blue.
Analysis of the isolated core peptide of PedA15.1 by MS/MS provided mass fragments (Figures S6, S8) consistent with a lack of dehydration at Ser26 (counting from the Gly-Gly leader peptide-removal site). Having ruled out this residue as being involved in ring formation, further analysis indicated no fragmentation between Ser7 and Cys21. As fragmentation is generally not observed within (Me)Lan rings, these data suggest that the three possible rings formed in this peptide must be confined within the Ser7 to Cys21 region. Likewise, fully modified PedA15.2 showed a lack of fragmentation within the putative lanthionine formation region (Figures S7, S9). However, MS/MS analysis of a three-fold-dehydrated and partially cyclized PedA15.2 core peptide (Figure S4B) isolated after HPLC purification revealed a characteristic y12 ion, suggesting that the missing ring in this intermediate involves Cys19 (Figure S10).
Given these observations and the similarities in the precursor sequences, we surmised that maturation likely yields similar overlapping or nested ring topologies confined between Ser7 and Cys16 in PedA15.1 and Ser8 and Cys16 in PedA15.2. Given the potential for multiple different ring topologies within this sequence window, we could not determine the exact structures of the final product through MS/MS analysis of the WT peptides alone. Therefore, we elucidated the structure of the PedA15.1 peptide by 2D NMR spectroscopy and stereochemical analysis.
NMR analysis of cyclized PedA15.1.
NMR analysis was carried out on a core fragment obtained by trypsin digestion of PedBC-modified PedA15.1 and subsequent HPLC purification. The fragment was predominantly composed of four-fold dehydrated core peptide with also some 5-fold dehydrated peptide and the spectra were acquired in DMSO-d6 at 37 °C. Analysis of TOCSY data identified 32 spins systems attributed to the 33-mer core peptide (Figure S11; Table S4). Subsequent NOESY analysis allowed assignment confirmation by cross-peak correlation with TOCSY α-proton signals (Figure S12). Lanthionine cross-links were then identified by nuclear Overhauser effects (NOEs) observed between methylene protons across the thioether linkage (-CH2-S-CH2-), allowing for correlation of the otherwise distinct TOCSY spin systems of lanthionine. By this approach three lanthionine linkages were identified, with the first between Dha7 (formerly Ser7) and Cys16. The second ring was nested within the first ring, with lanthionine formation between Cys8 and Dha15 (formerly Ser15). The final ring was located between Dha18 (formerly Ser18) and Cys21 (Figure 4). The lanthionines between Ser7/Cys16 and Cys8/Ser15 result in formation of an unusual lanthipeptide ring pattern that has not been observed in previously characterized family members (Figure 4B). This motif results in the formation of a 14-membered bis-lanthionine ring moiety composed of two directly adjacent lanthionine residues (Figure 4C). Importantly, this motif is highly conserved in a family of lanthipeptides encoded predominantly in the genus Pedobacter (Figure 4D and S12). The NMR data also clearly showed that Thr10 had been converted into Dhb, and that Ser26 escapes dehydration, consistent with the MS/MS fragmentation data (Figure S8). The signals of the 4-fold dehydrated peptide in the sample were too weak to assign its structure, but the data did reveal that Ser12 escaped dehydration whereas the rings were still formed in this minor product, consistent with the NEM assays.
Stereochemical analysis of the modified PedA15 peptides.
Gas chromatography-mass spectrometry (GC-MS) analysis with a chiral stationary phase56 was performed next on derivatized peptide hydrolysate to ascertain the stereochemical configuration of the cross-linked residues within cyclized PedA15 peptides and to support the conclusions of the NMR assignments and MS/MS fragmentation data. This analysis revealed that modified PedA15.1 and PedA15.2 both contained lanthionines, with no evidence of methyllanthionine present in either peptide. This finding is in agreement with the NMR data indicating that Thr10 in PedA15.1 had been dehydrated but not cyclized (Figures 4, 5; Table S4). Interestingly, co-injection with synthetic standards revealed the presence of lanthionine in both the LL- and DL-configuration. Integration of the relative peak areas of lanthionine isomers revealed that the relative stoichiometry of LL-lanthionine to DL-lanthionine was approximately 2:1 in both modified PedA15.1 and PedA15.2 peptides (Figure 5A). The LL-stereochemistry for (methyl)lanthionine in lanthipeptides is usually confined to ring formation from DhxDhxXxxXxxCys sequences (Dhx = Dha or Dhb) in class II lanthipeptides.34, 35, 54, 57 The only example thus far of LL-stereochemistry that is not derived from such a sequence has been reported for a class I lanthipeptide from Streptomyces olivaceus NRRL B-3009.58 This latter peptide has no sequence similarity with the PedA15 peptides.
Figure 5. Chiral GC-MS analysis of modified PedA15 peptides.
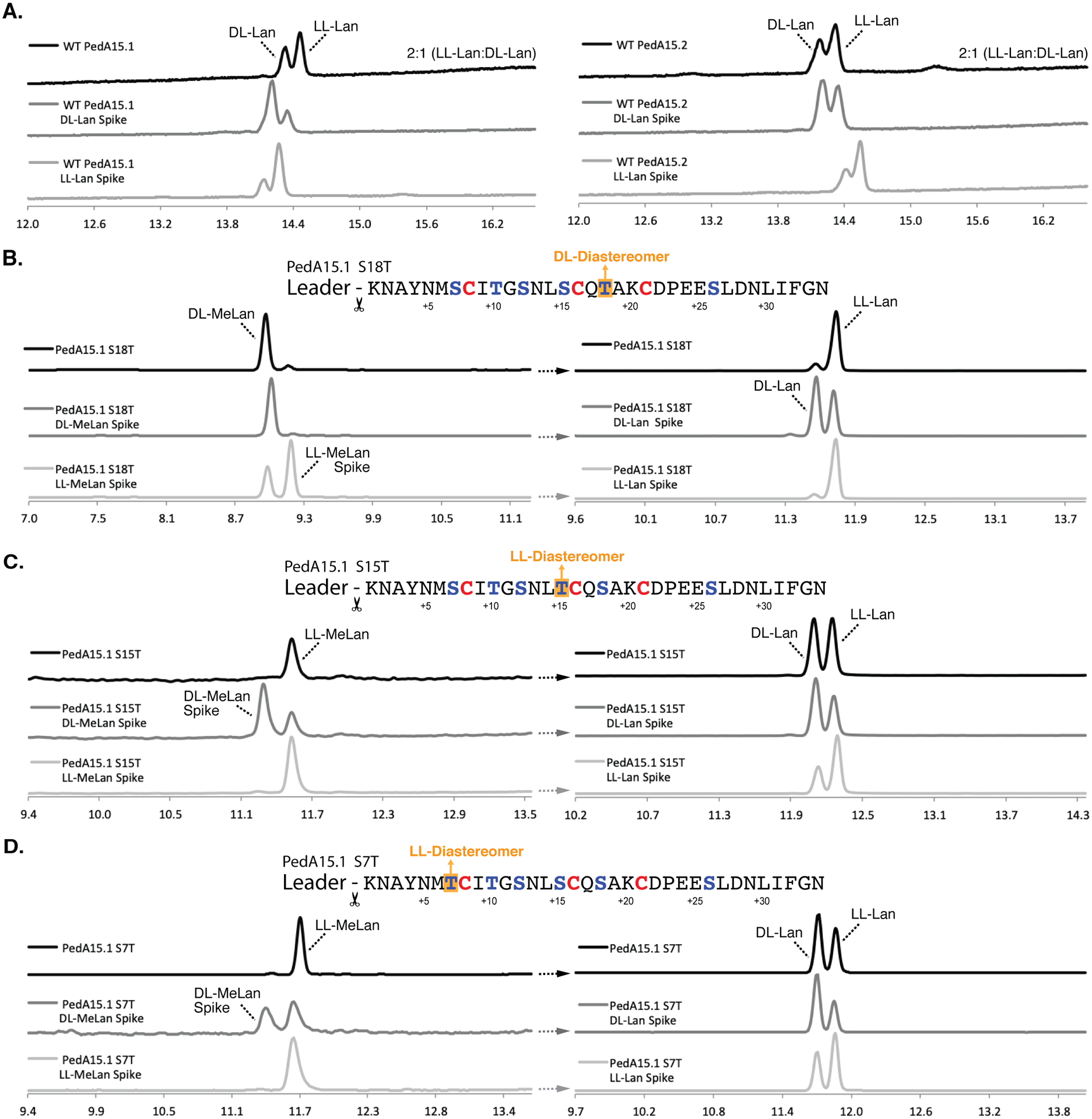
(A) Chiral GC-MS analysis of Ped15B/C-modified PedA15.1 (left) and PedA15.2 (right) core peptide fragments. See Materials and Methods for derivatization of the amino acids. Relative peak integration ratios for LL- and DL-Lan isomers are shown for each trace. (B) Ser-to-Thr mutation at position +18 from the GlyGly site resulted in the production of the DL-MeLan diastereomer (left). Lan in this mutant displayed solely the LL-Lan configuration (right). (C) Ser-to-Thr mutation at position +15 resulted in the production of the LL-MeLan diastereomer (left). Analysis of Lan in this sample revealed an approximately 1:1 ratio of DL- to LL-Lan (right). (D) Ser-to-Thr mutation at position +7 also resulted in the production of the LL-MeLan diastereomer (left). Analysis of Lan revealed an approximately 1:1 ratio of DL- to LL-Lan (right). For each set of traces, derivatized samples are shown in black and samples spiked with synthetic standards (DL- or LL-(Me)Lan, as indicated) are shown below in dark grey and light grey, respectively. Traces are selected-ion monitoring chromatograms for m/z = 365 (Lan) or 379 (MeLan), which correspond to the characteristic mass fragment for each derivatized compound. Retention times drift slightly and therefore coinjections with authentic standards were used to verify assignments.
We next determined the location of the LL-lanthionine linkages in the modified PedA15.1 peptide by analysis of Ser-to-Thr variants in the core-peptide. As neither wild-type PedA15 peptide contains methyllanthionine, Ser-to-Thr mutation of ring-forming residues should introduce a single MeLan into the molecule, the stereochemistry of which can then be identified by GC-MS analysis. As this substitution would represent a minimal structural alteration of the overall peptide (one additional methyl group), we hypothesized that it would not alter the overall ring pattern and thus allow us to infer the stereochemistry of individual crosslinks by accounting for the previously observed DL-/LL-Lan stoichiometry in the wild-type peptide (Figure 5A). Following this approach, complete cyclization of individual Ser-to-Thr variants after co-expression was first confirmed by NEM assay and MALDI-TOF MS analysis (Figure S14). Subsequent hydrolysis, derivatization of the amino acids, and analysis by GC-MS indicated the presence of DL-MeLan upon introduction of the Ser18Thr mutation (Figure 5B). Complementary analysis of lanthionine in this sample showed only LL-lanthionine. Thus, we infer that the DL-Lan observed in wild-type PedA15.1 must be associated with the ring formed between Cys21 and Dha18. Using this same strategy, we were able to show that Ser-to-Thr mutation at positions 7 and 15 resulted in the formation of solely LL-MeLan (Figure 5C, 5D). Analysis of lanthionine from both the Ser7Thr and Ser15Thr variants indicated an approximately 1:1 ratio of LL- to DL-Lan (down from 2:1 LL- to DL-Lan observed in wild-type peptide). Thus, the Lan linkages formed between Cys8 and Dha15 and between Cys16 and Dha7 are formed with LL-Lan stereochemistry in the wild-type PedA15.1 peptide.
Collectively, the data indicate that the most C-terminal ring (between Dha18 and Cys21) formed in PedA15.1 is in the DL-Lan conformation, whereas the two remaining lanthionine residues that form the unusual 14-membered intertwined ring-system are both in the LL-Lan configuration (Figure 4C). These observations suggest that the LL-configuration, which until recently was thought to be accessed only in very special cases for class II lanthipeptides where even non-enzymatic cyclization gives the LL-stereochemistry,59 may be more common than anticipated and demonstrates the importance of stereochemical investigations of new lanthipeptides. Furthermore, the number of systems in which a single cyclase makes rings of both LL and DL stereochemistry is increasing and provides additional support for a model in which the substrate sequence determines the stereochemical outcome of the cyclization process.60
PedC is required for efficient cyclization
NEM alkylation assays show that both modified PedA15 peptides are only partially cyclized when the cyclase PedC15 is not coexpressed (Figures S15–S16). In addition, glutathione adducts are observed in these peptides now that a subset of dehydroamino acids have not reacted with Cys residues. This finding suggests that despite the absence of the conserved zinc-binding residues in PedC15 that are essential for cyclization in other class I lanthipeptide cyclases (Figure 1C),32, 61, 62 PedC15 is important for cyclization. How this enzymatic control over cyclization is achieved without the canonical Zn2+ ligands will be an interesting topic for future mechanistic studies. We note that nearly all the LanC cyclases associated with PedA15-like precursors lack the canonical zinc binding ligands (Figure S17), but they all contain the His that is believed to protonate the enolate.61, 63 The absence of a Zn2+ site in lanthipeptide cyclases is increasingly reported for other classes of lanthipeptides, but no information on the molecular mechanisms used by these cyclases is currently available.7, 64
Conclusion
Herein, we report the heterologous expression and structural characterization of two lanthipeptides from Pedobacter lusitanus NL19. Our data indicate that expression of the ped15 biosynthetic machinery in E. coli produces modified peptides only if the tRNAGlu and GluRS from the native producer are coexpressed. Such recognition of only the cognate glutamyl-tRNA from the producing organisms by the LanB dehydratase had previously been shown for the Actinobacteria Microbispora sp. 107891 and Thermobispora bispora,14, 15 whereas LanB enzymes from the Firmicutes Lactococcus lactis and Geobacillus thermodenitrificans NG80–2 accepted glutamyl-tRNA from E. coli.13, 65 Thus, like the investigated enzymes from Actinobateria, the LanB of the Bacteroidetes P. lusitanus is also highly selective for its cognate tRNA. This observation is important for future synthetic biology studies to uncover the diverse lanthipeptides encoded in the BGCs from this phylum.26
The PedA15.1 product contains a novel ring pattern consisting of a 14-membered bis-lanthionine moiety, as indicated by MS/MS and NMR structure determination. Based on MS/MS data, GC-MS analysis, and sequence similarity, the PedA15.2 peptide is very likely to contain this same structure, and this pattern appears well-conserved in lanthipeptides encoded in Pedobacter and related Bacteroidetes. In addition, chiral GC-MS analysis demonstrated that both Lan components of this 14-membered bis-lanthionine ring system possess the uncommon LL-Lan configuration.
At present we have not detected bioactivity in our preliminary screening for antimicrobial and antifungal activity of the modified core peptides. It is possible that we screened organisms that are not targeted by the compounds, or that the N-termini of the natural products do not correspond to the two sites of leader peptide removal we explored herein (after the GlyGly motif or right before the first lanthionine). The conservation of the ring pattern in multiple organisms strongly suggests that these peptides have a beneficial function for their hosts. We therefore cannot rule out that these compounds have other, non-antimicrobial activities, such as reported for the morphogenic lanthipeptides SapB and SapT,66, 67 or the anti-allodynic labyrinthopeptins.68 The potential need for other post-translational modifications to impart activity is not supported by the collective BGCs, which do not show any other conserved genes (Figure S2), but since modification enzymes are sometimes encoded remotely,1, 43, 69–71 we cannot completely rule out this possibility. These observations highlight both the challenges of a genome mining strategy that prioritizes lanthipeptide precursor peptides that are as different as possible from previously characterized family members, and the opportunities for discovering new functions of natural products in microbial ecology. Indeed, only a subset of lanthipeptides identified thus far by genome mining have identifiable bioactivities and this subset has often consisted of structural analogs of known lantibiotics,51, 52, 65, 72–76 whereas many others with novel ring patterns are still awaiting identification of their functions.58, 65, 69, 70, 77–80 Historically natural products with antimicrobial activity have been prioritized for their potential application as therapeutics, but the RiPPs now discovered by genome mining provide the opportunity to learn more about chemical ecology in the microbial world.81–84
Materials and Methods:
A detailed description of all materials and methods used in this work is provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM 058822 to W. van der Donk and F32 GM117765 to I. Bothwell). We thank A. Ulanov and Z. Li from the Roy J. Carver Biotechnology Center (Metabolomics Facility) of the University of Illinois Urbana-Champaign (UIUC) for assistance with chiral GC-MS analysis. We also thank L. Zhu (UIUC School of Chemical Sciences NMR Laboratory) and J. Acedo (Mount Royal University, Canada) for assistance with the collection and interpretation of NMR data. T. Caetano was funded by national funds (OE), through FCT in the scope of the framework contract foreseen in the numbers 4, 5 and 6 of article 23, of the Decree-Law 57/2016, of August 29, changed by Law 57/2017, of July (CEECIND/01463/2017) and a postdoctoral scholarship funded by FCT, EU and POPH (SFRH/BPD/77900/2011). CESAM is a research unit financially supported by FCT/MCTES (UIDP/50017/2020+UIDB/50017/2020), through national funds.
Footnotes
Associated Content:
The Supporting Information, which includes materials, methods, supplemental tables and figures, is available free of charge at https://pubs.acs.org
References
- (1).Montalbán-López M, Scott TA, Ramesh S, Rahman IR, van Heel AJ, Viel JH, Bandarian V, Dittmann E, Genilloud O, Goto Y, Grande Burgos MJ, Hill C, Kim S, Koehnke J, Latham JA, Link AJ, Martínez B, Nair SK, Nicolet Y, Rebuffat S, Sahl H-G, Sareen D, Schmidt EW, Schmitt L, Severinov K, Süssmuth RD, Truman AW, Wang H, Weng J-K, van Wezel GP, Zhang Q, Zhong J, Piel J, Mitchell DA, Kuipers OP, and van der Donk WA (2021) New developments in RiPP discovery, enzymology and engineering, Nat. Prod. Rep 138, 130 – 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Oman TJ, and van der Donk WA (2010) Follow the leader: the use of leader peptides to guide natural product biosynthesis, Nat. Chem. Biol 6, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hetrick KJ, and van der Donk WA (2017) Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era, Curr. Opin. Chem. Biol 38, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hudson GA, and Mitchell DA (2018) RiPP antibiotics: biosynthesis and engineering potential, Curr. Opin. Microbiol 45, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Repka LM, Chekan JR, Nair SK, and van der Donk WA (2017) Mechanistic understanding of lanthipeptide biosynthetic enzymes, Chem. Rev 117, 5457–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Funk MA, and van der Donk WA (2017) Ribosomal natural products, tailored to fit, Acc. Chem. Res 50, 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ortiz-López FJ, Carretero-Molina D, Sánchez-Hidalgo M, Martín J, González I, Román-Hurtado F, de la Cruz M, García-Fernández S, Reyes F, Deisinger JP, Müller A, Schneider T, and Genilloud O (2020) Cacaoidin, first member of the new lanthidin RiPP family, Angew. Chem. Int. Ed 59, 12654–12658. [DOI] [PubMed] [Google Scholar]
- (8).Xu M, Zhang F, Cheng Z, Bashiri G, Wang J, Hong J, Wang Y, Xu L, Chen X, Huang SX, Lin S, Deng Z, and Tao M (2020) Functional genome mining reveals a class V lanthipeptide containing a D-amino acid introduced by an F420H2-dependent reductase, Angew. Chem. Int. Ed 59, 18029–18035. [DOI] [PubMed] [Google Scholar]
- (9).Kloosterman AM, Cimermancic P, Elsayed SS, Du C, Hadjithomas M, Donia MS, Fischbach MA, van Wezel GP, and Medema MH (2020) Expansion of RiPP biosynthetic space through integration of pan-genomics and machine learning uncovers a novel class of lanthipeptides, PLoS Biol. 18, e3001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sen AK, Narbad A, Horn N, Dodd HM, Parr AJ, Colquhoun I, and Gasson MJ (1999) Post-translational modification of nisin. The involvement of NisB in the dehydration process, Eur. J. Biochem 261, 524–532. [DOI] [PubMed] [Google Scholar]
- (11).Koponen O, Tolonen M, Qiao M, Wahlstrom G, Helin J, and Saris PEJ (2002) NisB is required for the dehydration and NisC for the lanthionine formation in the post-translational modification of nisin, Microbiology 148, 3561–3568. [DOI] [PubMed] [Google Scholar]
- (12).Garg N, Salazar-Ocampo LM, and van der Donk WA (2013) In vitro activity of the nisin dehydratase NisB, Proc. Natl. Acad. Sci. U. S. A 110, 7258–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, and Nair SK (2015) Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB, Nature 517, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hudson GA, Zhang Z, Tietz JI, Mitchell DA, and van der Donk WA (2015) In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin, J. Am. Chem. Soc 137, 16012–16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ortega MA, Hao Y, Walker MC, Donadio S, Sosio M, Nair SK, and van der Donk WA (2016) Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis, Cell Chem. Biol 23, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bothwell IR, Cogan DP, Kim T, Reinhardt CJ, van der Donk WA, and Nair SK (2019) Characterization of glutamyl-tRNA-dependent dehydratases using nonreactive substrate mimics, Proc. Natl. Acad. Sci. U. S. A 116, 17245–17250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mavaro A, Abts A, Bakkes PJ, Moll GN, Driessen AJ, Smits SH, and Schmitt L (2011) Substrate recognition and specificity of NisB, the lantibiotic dehydratase involved in nisin biosynthesis, J. Biol. Chem 286, 30552–30560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Khusainov R, van Heel AJ, Lubelski J, Moll GN, and Kuipers OP (2015) Identification of essential amino acid residues in the nisin dehydratase NisB, Front Microbiol. 6, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Reiners J, Abts A, Clemens R, Smits SH, and Schmitt L (2017) Stoichiometry and structure of a lantibiotic maturation complex, Sci. Rep 7, 42163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Repka LM, Hetrick KJ, Chee SH, and van der Donk WA (2018) Characterization of leader peptide binding during catalysis by the nisin dehydratase NisB, J. Am. Chem. Soc 140, 4200–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chen J, van Heel AJ, and Kuipers OP (2020) Subcellular Localization and Assembly Process of the Nisin Biosynthesis Machinery in Lactococcus lactis, mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Marsh AJ, O’Sullivan O, Ross RP, Cotter PD, and Hill C (2010) In silico analysis highlights the frequency and diversity of type 1 lantibiotic gene clusters in genome sequenced bacteria, BMC Genomics 11, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang Q, Doroghazi JR, Zhao X, Walker MC, and van der Donk WA (2015) Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria, Appl. Environ. Microbiol 81, 4339–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Singh M, and Sareen D (2014) Novel LanT associated lantibiotic clusters identified by genome database mining, PLoS One 9, e91352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang Q, Yang X, Wang H, and van der Donk WA (2014) High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis, ACS Chem. Biol 9, 2686–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Walker MC, Eslami SM, Hetrick KJ, Ackenhusen SE, Mitchell DA, and van der Donk WA (2020) Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family, BMC Genomics 21, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Caetano T, van der Donk W, and Mendo S (2020) Bacteroidetes can be a rich source of novel lanthipeptides: The case study of Pedobacter lusitanus, Microbiol. Res 235, 126441. [DOI] [PubMed] [Google Scholar]
- (28).Mohr KI, Volz C, Jansen R, Wray V, Hoffmann J, Bernecker S, Wink J, Gerth K, Stadler M, and Müller R (2015) Pinensins: the first antifungal lantibiotics, Angew. Chem. Int. Ed 54, 11254–11258. [DOI] [PubMed] [Google Scholar]
- (29).Viana AT, Caetano T, Covas C, Santos T, and Mendo S (2018) Environmental superbugs: The case study of Pedobacter spp, Environ. Pollut 241, 1048–1055. [DOI] [PubMed] [Google Scholar]
- (30).Okeley NM, Paul M, Stasser JP, Blackburn N, and van der Donk WA (2003) SpaC and NisC, the cyclases involved in subtilin and nisin biosynthesis, are zinc proteins, Biochemistry 42, 13613–13624. [DOI] [PubMed] [Google Scholar]
- (31).Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, and Nair SK (2006) Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis, Science 311, 1464–1467. [DOI] [PubMed] [Google Scholar]
- (32).Paul M, Patton GC, and van der Donk WA (2007) Mutants of the zinc ligands of lacticin 481 synthetase retain dehydration activity but have impaired cyclization activity, Biochemistry 46, 6268–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chatterjee C, Paul M, Xie L, and van der Donk WA (2005) Biosynthesis and mode of action of lantibiotics, Chem. Rev 105, 633–684. [DOI] [PubMed] [Google Scholar]
- (34).Tang W, and van der Donk WA (2013) The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry, Nat. Chem. Biol 9, 157–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tang W, Jiménez-Osés G, Houk KN, and van der Donk WA (2015) Substrate control in stereoselective lanthionine biosynthesis, Nat. Chem 7, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Covas C, Almeida B, Esteves AC, Lourenço J, Domingues P, Caetano T, and Mendo S (2021) Peptone from casein, an antagonist of nonribosomal peptide synthesis: a case study of pedopeptins produced by Pedobacter lusitanus NL19, N. Biotechnol 60, 62–71. [DOI] [PubMed] [Google Scholar]
- (37).Nagao J, Harada Y, Shioya K, Aso Y, Zendo T, Nakayama J, and Sonomoto K (2005) Lanthionine introduction into nukacin ISK-1 prepeptide by co-expression with modification enzyme NukM in Escherichia coli, Biochem. Biophys. Res. Commun 336, 507–513. [DOI] [PubMed] [Google Scholar]
- (38).Shi Y, Yang X, Garg N, and van der Donk WA (2011) Production of lantipeptides in Escherichia coli, J. Am. Chem. Soc 133, 2338–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lin Y, Teng K, Huan L, and Zhong J (2011) Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge, Microbiol. Res 166, 146–154. [DOI] [PubMed] [Google Scholar]
- (40).Caetano T, Krawczyk JM, Mosker E, Süssmuth RD, and Mendo S (2011) Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli, Chem. Biol 18, 90–100. [DOI] [PubMed] [Google Scholar]
- (41).Håvarstein LS, Diep DB, and Nes IF (1995) A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export, Mol. Microbiol 16, 229–240. [DOI] [PubMed] [Google Scholar]
- (42).Li B, Cooper LE, and van der Donk WA (2009) In vitro studies of lantibiotic biosynthesis, Methods Enzymol. 458, 533–558. [DOI] [PubMed] [Google Scholar]
- (43).Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, and van der Donk WA (2010) Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria, Proc. Natl. Acad. Sci. U.S.A 107, 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Haft DH, Basu MK, and Mitchell DA (2010) Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family, BMC Biol. 8, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Bobeica SC, Dong SH, Huo L, Mazo N, McLaughlin MI, Jimenéz-Osés G, Nair SK, and van der Donk WA (2019) Insights into AMS/PCAT transporters from biochemical and structural characterization of a double glycine motif protease, eLife 8, e42305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kieuvongngam V, Olinares PDB, Palillo A, Oldham ML, Chait BT, and Chen J (2020) Structural basis of substrate recognition by a polypeptide processing and secretion transporter, eLife 9, e51492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Furgerson Ihnken LA, Chatterjee C, and van der Donk WA (2008) In vitro reconstitution and substrate specificity of a lantibiotic protease, Biochemistry 47, 7352–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Segarra RA, Booth MC, Morales DA, Huycke MM, and Gilmore MS (1991) Molecular characterization of the Enterococcus faecalis cytolysin activator, Infect. Immun 59, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Tang W, Dong S-H, Repka LM, He C, Nair SK, and van der Donk WA (2015) Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage, Chem. Sci 6, 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tang W, Bobeica SC, Wang L, and van der Donk WA (2019) CylA is a sequence-specific protease involved in toxin biosynthesis, J. Ind. Microbiol. Biotechnol 46, 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, and van der Donk WA (2006) Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic, Proc. Natl. Acad. Sci. U. S. A 103, 17243–17248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Lawton EM, Cotter PD, Hill C, and Ross RP (2007) Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125, FEMS Microbiol. Lett 267, 64–71. [DOI] [PubMed] [Google Scholar]
- (53).Dischinger J, Josten M, Szekat C, Sahl HG, and Bierbaum G (2009) Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13, PLoS One 4, e6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lohans CT, Li JL, and Vederas JC (2014) Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with D-amino acids, J. Am. Chem. Soc 136, 13150–13153. [DOI] [PubMed] [Google Scholar]
- (55).Huo L, and van der Donk WA (2016) Discovery and characterization of bicereucin, an unusual D-amino acid-containing mixed two-component lantibiotic, J. Am. Chem. Soc 138, 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Liu W, Chan ASH, Liu H, Cochrane SA, and Vederas JC (2011) Solid supported chemical syntheses of both components of the lantibiotic lacticin 3147, J. Am. Chem. Soc 133, 14216–14219. [DOI] [PubMed] [Google Scholar]
- (57).Garg N, Goto Y, Chen T, and van der Donk WA (2016) Characterization of the stereochemical configuration of lanthionines formed by the lanthipeptide synthetase GeoM, Biopolymers 106, 834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Acedo JZ, Bothwell IR, An L, Trouth A, Frazier C, and van der Donk WA (2019) O-methyltransferase-mediated incorporation of a β-amino acid in lanthipeptides, J. Am. Chem. Soc 141, 16790–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Tang W, Thibodeaux GN, and van der Donk WA (2016) The enterococcal cytolysin synthetase coevolves with substrate for stereoselective lanthionine synthesis, ACS Chem. Biol 11, 2438–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Yu Y, Zhang Q, and van der Donk WA (2013) Insights into the evolution of lanthipeptide biosynthesis, Protein Sci. 22, 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Li B, and van der Donk WA (2007) Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin, J. Biol. Chem 282, 21169–21175. [DOI] [PubMed] [Google Scholar]
- (62).Helfrich M, Entian KD, and Stein T (2007) Structure-function relationships of the lanthionine cyclase SpaC involved in biosynthesis of the Bacillus subtilis peptide antibiotic subtilin, Biochemistry 46, 3224–3233. [DOI] [PubMed] [Google Scholar]
- (63).Yang X, and van der Donk WA (2015) Michael-type cyclizations in lantibiotic biosynthesis are reversible, ACS Chem. Biol 10, 1234–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Müller WM, Schmiederer T, Ensle P, and Süssmuth RD (2010) In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification, Angew. Chem., Int. Ed 49, 2436–2440. [DOI] [PubMed] [Google Scholar]
- (65).Garg N, Tang W, Goto Y, Nair SK, and van der Donk WA (2012) Lantibiotics from Geobacillus thermodenitrificans, Proc. Natl. Acad. Sci. U.S.A 109, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, and Willey JM (2004) The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor, Proc. Natl. Acad. Sci. U.S.A 101, 11448–11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kodani S, Lodato MA, Durrant MC, Picart F, and Willey JM (2005) SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the Streptomycetes, Mol. Microbiol 58, 1368–1380. [DOI] [PubMed] [Google Scholar]
- (68).Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Guhring H, Vertesy L, Wink J, Hoffmann H, Bronstrup M, Sheldrick GM, and Süssmuth RD (2010) Labyrinthopeptins: a new class of carbacyclic lantibiotics, Angew. Chem. Int. Ed 49, 1151–1154. [DOI] [PubMed] [Google Scholar]
- (69).Cubillos-Ruiz A, Berta-Thompson JW, Becker JW, van der Donk WA, and Chisholm SW (2017) Evolutionary radiation of lanthipeptides in marine cyanobacteria, Proc. Natl. Acad. Sci. USA 114, E5424–E5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Merwin NJ, Mousa WK, Dejong CA, Skinnider MA, Cannon MJ, Li H, Dial K, Gunabalasingam M, Johnston C, and Magarvey NA (2020) DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products, Proc. Natl. Acad. Sci. USA 117, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Lu J, Wu Y, Li Y, and Wang H (2021) The utilization of lanthipeptide synthetases Is a general strategy for the biosynthesis of 2-aminovinyl-cysteine motifs in thioamitides, Angew. Chem. Int. Ed 60, 1951–1958. [DOI] [PubMed] [Google Scholar]
- (72).Singh M, Chaudhary S, and Sareen D (2020) Roseocin, a novel two-component lantibiotic from an actinomycete, Mol. Microbiol 113, 326–337. [DOI] [PubMed] [Google Scholar]
- (73).Xu B, Aitken EJ, Baker BP, Turner CA, Harvey JE, Stott MB, Power JF, Harris PWR, Keyzers RA, and Brimble MA (2018) Genome mining, isolation, chemical synthesis and biological evaluation of a novel lanthipeptide, tikitericin, from the extremophilic microorganism Thermogemmatispora strain T81, Chem. Sci 9, 7311–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Vikeli E, Widdick DA, Batey SFD, Heine D, Holmes NA, Bibb MJ, Martins DJ, Pierce NE, Hutchings MI, and Wilkinson B (2020) In situ activation and heterologous production of a cryptic lantibiotic from an African plant ant-derived Saccharopolyspora species, Appl. Environ. Microbiol 86, e01876–01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Wang J, Zhang L, Teng K, Sun S, Sun Z, and Zhong J (2014) Cerecidins, novel lantibiotics from Bacillus cereus with potent antimicrobial activity, Appl. Environ. Microbiol 80, 2633–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).van Heel AJ, Kloosterman TG, Montalbán-López M, Deng J, Plat A, Baudu B, Hendriks D, Moll GN, and Kuipers OP (2016) Discovery, production and modification of five novel lantibiotics using the promiscuous nisin modification machinery, ACS Synth. Biol 5, 1146–1154. [DOI] [PubMed] [Google Scholar]
- (77).Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, and van der Donk WA (2010) Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights, PLoS Biol. 8, e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Völler GH, Krawczyk JM, Pesic A, Krawczyk B, Nachtigall J, and Süssmuth RD (2012) Characterization of new class III lantibiotics-erythreapeptin, avermipeptin and griseopeptin from Saccharopolyspora erythraea, Streptomyces avermitilis and Streptomyces griseus demonstrates stepwise N-terminal leader processing, ChemBioChem 13, 1174–1183. [DOI] [PubMed] [Google Scholar]
- (79).Krawczyk B, Völler GH, Völler J, Ensle P, and Süssmuth RD (2012) Curvopeptin: a new lanthionine-containing class III lantibiotic and its co-substrate promiscuous synthetase, ChemBioChem 13, 2065–2071. [DOI] [PubMed] [Google Scholar]
- (80).Iftime D, Jasyk M, Kulik A, Imhoff JF, Stegmann E, Wohlleben W, Süssmuth RD, and Weber T (2015) Streptocollin, a type IV lanthipeptide produced by Streptomyces collinus Tü 365, ChemBioChem 16, 2615–2623. [DOI] [PubMed] [Google Scholar]
- (81).van der Meij A, Worsley SF, Hutchings MI, and van Wezel GP (2017) Chemical ecology of antibiotic production by actinomycetes, FEMS Microbiol. Rev 41, 392–416. [DOI] [PubMed] [Google Scholar]
- (82).Scherlach K, and Hertweck C (2018) Mediators of mutualistic microbe-microbe interactions, Nat. Prod. Rep 35, 303–308. [DOI] [PubMed] [Google Scholar]
- (83).Li Y, and Rebuffat S (2020) The manifold roles of microbial ribosomal peptide-based natural products in physiology and ecology, J. Biol. Chem 295, 34–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Cao L, Do T, and Link AJ (2021) Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs), J. Ind. Microbiol. Biotechnol, 10.1093/jimb/kuab1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Harrison KJ, Crécy-Lagard V, and Zallot R (2018) Gene Graphics: a genomic neighborhood data visualization web application, Bioinformatics 34, 1406–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, and Hutchison GR (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform, J. Cheminform 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


