Abstract
Most major food-borne outbreaks of listeriosis in Europe and in the United States have been caused by genetically closely related Listeria monocytogenes strains of serotype 4b. In order to assess whether genomic loci exist that could underlie this increased epidemic potential, we subtracted the genome of the virulent prototype L. monocytogenes strain EGD from a prototype epidemic strain. A total of 39 DNA fragments corresponding to 20% of an estimated total of 150 to 190 kb of differential genome material were isolated. For 21 of these fragments, no function on the basis of homology could be predicted. Of the remaining 18 fragments, 15 had homologies to bacterial surface proteins, some of which have been implicated in virulence mechanisms such as cell invasion, adhesion, or immune escape. Southern hybridization of arrays containing the epidemic-clone-specific DNA segments with genomic DNA of different L. monocytogenes strains was consistent with the current lineage division. Surprisingly, however, some of the fragments hybridized in a mosaic-like fashion to genomes of two other Listeria species, the animal pathogen L. ivanovii and the nonpathogen L. innocua. Taken together, our results provide a starting point for the identification of epidemic-trait-associated genes.
The gram-positive, opportunistic bacterium Listeria monocytogenes is responsible for severe food-borne infections with a high case fatality rate in susceptible animal and human hosts (16, 44). Human listeriosis occurs as sporadic disease or in the form of outbreaks (epidemics) which can affect up to several hundred people, in particular elderly, very young, or immunocompromised individuals. Symptoms of primary infection range from barely apparent to distinct flu-like symptoms and self-limiting diarrhea. After an extended incubation period, susceptible individuals may develop listeriosis, which may manifest itself as meningitis, meningoencephalitis, or sepsis, or in the case of pregnancy as abortion, miscarriage, or severe generalized infection of the newborn.
Of the 16 or so L. monocytogenes serotypes known, three serotypes, 4b, 1/2b, and 1/2a, are responsible for most human cases (16, 44). Molecular and other typing methods divide the species L. monocytogenes into distinct phylogenetic lineages separating the serotypes 4b, 3b, and 1/2b from the serotypes 1/2a and 1/2c (6, 38, 42). Serotype 4b strains are responsible for the majority of sporadic disease as well as most major outbreaks of food-borne listeriosis in Europe and North America since 1981. By multilocus enzyme electrophoresis, a specific 4b strain, representing 1 of 82 electrophoretic types of L. monocytogenes, was associated with epidemic disease outbreaks in California in 1985, Switzerland between 1983 and 1987, and France in 1992 (6, 26, 38). A 4b strain with a closely related electrophoretic type was implicated in two outbreaks in Boston in 1979 and 1983 (6, 38).
L. monocytogenes possesses a number of well-characterized virulence factors that play a role in its facultative intracellular lifestyle and its capacity to circumvent the humoral immune system by spreading from cell to cell within tissues (11, 45). Although no correlation between epidemic prevalence and increased virulence in a number of animal models and cell culture tests has been found to date for L. monocytogenes 4b strains (28, 46), one should nevertheless consider the possibility of genetic loci conferring additional pathogenic traits (12) to the strains prevalent in epidemics. These traits may include properties (not measurable by existing virulence tests) that allow the adaptation to specific environmental conditions and that can influence the course or the outcome of an infection.
One way to identify genetic loci encoding such traits is to employ bacterial genome subtraction (1, 9, 36). Despite the apparent genetic distinctness of L. monocytogenes serotype 4b strains, only three genes have been identified to date which are characteristic of serotype 4b strains. These genes were isolated on the basis of their involvement in expression of cell wall teichoic acid-associated, serotype 4b-specific surface antigens (30, 40).
In order to assess whether genetic loci exist that distinguish epidemic L. monocytogenes strains, we subtracted the genome of the virulent, experimentally best-characterized strain (EGD serotype 1/2a, whose completed genome sequence is pending publication) from a prototype epidemic strain (F.4565, serotype 4b) that had caused 142 cases of listeriosis including 48 deaths during an outbreak in Los Angeles in 1985 (31). We found that about 5 to 6% of the genome of strain F.4565 does not hybridize to strain EGD DNA. We have isolated 39 such fragments, 35 of which were absent and 4 of which were strongly divergent from the genome of strain EGD. A large portion of these DNA regions code for bacterial surface determinants and hence are likely to play a role in the interaction with the environment or the host organism. We have devised a screening method to test the occurrence of these fragments in the genomes of other Listeria species or other L. monocytogenes strains, thereby opening the way to search for epidemic-trait-associated genes.
MATERIALS AND METHODS
Bacterial strains.
L. monocytogenes strains F.4565 (bacterial collection no. [BAC] 239), G.3129 (BAC 266), F.7441 (BAC 262), L.4486a (BAC 240), L.2190a (BAC 265), L.2192 (BAC 260), F.1092 (BAC 267), F.8353 (BAC 259), G.0039 (BAC 258), L.5089 (BAC 261), F.7390 (BAC 264), and L.735 (BAC 263) (7; see also Table 2) were provided by J. McLauchlin, Food Hygiene Laboratory, Central Public Laboratory, London, England. L. monocytogenes strain LO28 (BAC 089), L. innocua Rham− (BAC 042) CLIP11262T, L. innocua (BAC 269) CLIP12511 (type strain), L. ivanovii (BAC 085) CLIP12510, L. grayi (BAC 273) ATCC 25401, and B. subtilis 168 trpC2 (BAC 274) were obtained from P. Cossart, Institut Pasteur, Paris, France. L. monocytogenes strain EGD (BAC 146) was obtained from W. Goebel, Theodor-Boveri-Institut, Würzburg, Germany. The Escherichia coli strain used was JM101 (BAC 098). L. monocytogenes L028 ΔactA Δhly (BAC 200) is an isogenic mutant of strain L028 carrying deletions of actA and hly (C. Kocks, unpublished data).
TABLE 2.
Hybridization patterns of 39 L. monocytogenes F.4565-specific gene fragments with DNA from different L. monocytogenes strains
| Original no. | Countryc | Origin | Serotype | Outbreak | Hybridization pattern(s) | Remark | Reference |
|---|---|---|---|---|---|---|---|
| F.4565a | USA | Patient, epidemic | 4b | Los Angeles 1985 | 4b-I | Tester strain | Bille and Rocourt (7) |
| G.3129a | USA | Patient, epidemic | 4b | Los Angeles 1985 | 4b-I | Same outbreak as F.4565 | Bille and Rocourt (7) |
| F.7441 | USA | Patient, sporadic | 4b | 4b-I | Bille and Rocourt (7) | ||
| L.4486a | Switzerland | Patient, epidemic | 4b | Lausanne 1987 | 4b-I | Bille and Rocourt (7) | |
| L.2190ab | England | Food, sporadic | 4b | 4b-I | Same as L.2192 | Bille and Rocourt (7) | |
| L.2192b | England | Patient, sporadic | 4b | 4b-I | Same as L.2190a | Bille and Rocourt (7) | |
| F.1092 | USA | Patient, epidemic | 4b | Boston 1983 | 4b-II | Bille and Rocourt (7) | |
| F.8353 | USA | Food, sporadic | 4b | 4b-II | Bille and Rocourt (7) | ||
| G.0039 | USA | Patient, sporadic | 3b | 3b and 1/2b | Bille and Rocourt (7) | ||
| L.5089 | UK | Patient, sporadic | 1/2b | 3b and 1/2b | Bille and Rocourt (7) | ||
| F.7390 | USA | Food, sporadic | 1/2a | 1/2a-I | Bille and Rocourt (7) | ||
| L.735 | England | Patient, epidemic | 1/2a | Carlisle 1981 | 1/2a-II | Bille and Rocourt (7) | |
| EGD | Rabbit | 1/2a | 1/2a-III and 1/2c | Driver strain | |||
| LO28 | Spain | Fecal isolate | 1/2c | 1/2a-III and 1/2c | Healthy pregnant carrier | Vicente et al. (48) |
Strains with the same electrophoretic type (included for internal control).
Strains have the same electrophoretic type (included for internal control).
USA, United States of America; UK, United Kingdom.
Purification of genomic DNA, Southern blotting, and “reverse” Southern blotting.
Molecular biology methods were carried out using standard procedures (5) unless indicated otherwise. High-molecular-weight DNA was prepared from saturated liquid bacterial culture, taking care to remove all RNA. In order to verify the absence of plasmids, an alkaline lysis type plasmid DNA preparation was carried out with strains F.4565, L.4486a, L. innocua BAC 042, and L. ivanovii BAC 085 and the equivalent of up to 2 ml of satured liquid culture loaded onto an agarose gel. For Southern blotting, 200 ng of RsaI-digested genomic DNA was separated and blotted onto positively charged nylon membranes (Hybond N+; Amersham Pharmacia) by downward capillary transfer (5). Membranes were hybridized in Puregene Hyb-9 DNA hybridization solution (Gentra Systems) to 10 ng of PCR product labeled with [α-32P]ATP (Megaprime kit; Amersham Pharmacia). Washing steps were carried out under low-stringency conditions (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at room temperature), and the membrane was exposed to X-ray film. For reverse Southern blotting, plasmid inserts were amplified by PCR using adapter-specific oligonucleotides. Approximately 20 ng of plasmid DNA or PCR products were spotted onto nylon membranes either by hand or by using a VP408 Multi-Blot replicator (V&P Scientific, Inc.) and hybridized to 25 ng of radioactively labeled, RsaI-digested genomic DNA.
Establishment of genomic subtraction conditions suitable for Listeria genomes.
HaeIII-digested φX174 replicative-form (RF) DNA (5,389 bp) was added to L. monocytogenes EGD DNA (3,150 kb) at a molar ratio of 1:1 and subtracted as described below. Assuming a similar fragment length distribution, the initial ratio of phage-specific fragments to other DNA fragments was 1/589. We cloned and sequenced the PCR-amplified material from this subtraction as described below. Four out of 18 randomly selected clones were identified as tester-specific φX174 RF HaeIII fragments, corresponding to a ratio of 1/4.5. Thus, enrichment for tester-specific fragments was 130-fold. A similar enrichment factor (132-fold) was found in a second control subtraction in which DNA of L. monocytogenes strain L028 was used as tester and DNA of an isogenic mutant, LO28 ΔactA Δhly lacking genes actA and hly, was used as driver. In this case, the subtraction efficiency was assessed for an actA fragment by PCR as described below for plcA, iap, and hly.
Genome subtraction and plasmid library construction.
Genome subtraction was performed with the PCR-Select Bacterial Genome Subtraction kit (Clontech) according to the manufacturer's instructions except for the hybridization temperature. Genomic DNA was digested with RsaI and subtractive hybridization was carried out at 60°C, a temperature adapted to the low GC content of the Listeria genome (17). Tester-specific fragments were amplified by two rounds of suppression PCR that yielded a mixture of fragments corresponding in size to RsaI-restricted genomic DNA. The subtraction efficiency was tested with PCR by comparing the concentration of RsaI fragments of three known genes, plcA (KO79/80 5′-TCGCGTTACCTGGCAAATAGATGG-3′/5′-TCAAATCATCGACGGCAACCTCGG-3′), hly (KO83/84 5′-CCGAACTGCATGCCGAATTTGC-3′/5′-CAATCTCAACATTTACCATGGG-3′), and iap (KO77/78 5′-AAAGCGGTGACACTATTTGGGC-3′/5′-TTGTGTTTGTAGATGGTGCAGG-3′), in equal amounts of PCR-amplified subtracted and unsubtracted material. The PCR products enriched for tester-specific fragments were ligated into plasmid vector pGEM-T Easy (system I; Promega) and transformed into competent E. coli cells. Plasmid DNA of 355 clones from the subtracted library and of 125 clones from the unsubtracted library was prepared with the QIAprep 8 Miniprep kit (Qiagen) and analyzed by restriction with EcoRI to test for the presence of inserts and to normalize plasmid DNA concentration. Insert sizes ranged from approximately 0.2 to 2 kb with an average length of 800 bp (consistent with the size distribution of RsaI fragments of genomic L. monocytogenes DNA).
Sequence analysis and database searches.
Cycle sequencing was carried out using the BigDye Termination kit (PE Applied Biosystems) and an ABI Prism 377 DNA sequencer. Inserts were sequenced by one pass in each direction using vector-specific oligonucleotides. Homology searches at the nucleotide level (all nonredundant GenBank plus EMBL plus DDBJ plus PDB sequences) or at the amino acid level (all nonredundant GenBank CDS translations plus PDB plus SwissProt plus PIR plus PRF sequences) were carried out at the National Center for Biotechnology Information using BLASTN 2.0.12 and BLASTX 2.1.1 (4). Expect values lower than 0.05 were considered to be potentially significant. If necessary, internal fragment-specific oligonucleotides were used to obtain the complete sequence of fragments with homology to known genes. Fragments with homologies to unknown genes or no homology to database entries were sequenced to about 300 to 400 bp from both ends.
Nucleotide sequence accession numbers.
The sequences described in Table 1 have been deposited in the EMBL database and assigned accession numbers AJ410352 to AJ410411. They can also be accessed at http://www.uni-koeln.de/∼aeg14/Kocks/index.html.
TABLE 1.
Gene fragments homologous to virulence determinants, surface proteins, transporters, or other proteinsd
| Fragment no. | Length (bp) | Best matches to homologous proteins (BlastX search) | Amino acid identity (%) (length) | Organism | Reference |
|---|---|---|---|---|---|
| Differential fragmentsb | |||||
| 167 | 378 | Involved in expression of surface antigens | 100 (378 bp) | L. monocytogenes 10527 (serotype 4b) | |
| 157 | 484 | Internalins D, H, A, G, and E (B repeats) | 64 (477 bp) | L. monocytogenes | Dramsi et al. (13) |
| 29B (=U37) | 367 | L. monocytogenes antigen C | 48 (225 bp) | L. monocytogenes | |
| 162 | 396 | Internalins A, E, H, G, and D (leucine-rich repeats) | 45 (393 bp) | L. monocytogenes | Dramsi et al. (13) |
| 104 (=U58B) | 400 | Internalins B, H, E, D, and G (leucine-rich repeats) | 33 (390 bp) | L. monocytogenes | Dramsi et al. (13) |
| 143 | 239 | Internalins B (C repeats) | 33 (222 bp) | L. monocytogenes | Dramsi et al. (13) |
| Amidase (repeats) | 32 (239 bp) | L. monocytogenes | McLaughlan and Foster (33) | ||
| 58 | 326 | Receptor protein kinase-like protein (leucine-rich region) | 35 (240 bp) | Arabidopsis manana | |
| 33B (=54B) | 512 | ORF A (repeats) | 52 (510 bp) | Listeria seeligeri | |
| Alpha C protein (bca) (repeats) | 32 (357 bp) | Streptococcus agalactiae | Michel et al. (34) | ||
| Chitinase B (chiB) (cadherin-like repeat) | 27 (426 bp) | Clostridium paraputrificum | Morimoto et al. (35) | ||
| 97 | 523 | ORF A (repeats) | 47 (516 bp) | L. seeligeri | |
| Immunoreactive 92 kDA antigen PG21 | 27 (384 bp) | Porphyromonas gingivalis | |||
| Chitinase B (chiB) (cadherin-like repeat) | 26 (447 bp) | C. paraputrificum | Morimoto et al. (35) | ||
| 30 | 880 | Chitinases A and B (chiAB) (cadherin-like repeat) | 47 (537 bp) | C. paraputrificum | Morimoto et al. (35) |
| U17B | 560 | Wall-associated protein precursor (wapA) | 34 (560 bp) | B. subtilis | Foster (18) |
| U46 (=65B) | 676 | Wall-associated protein precursor (wapA; repeat) | 40 (676 bp) | B. subtilis | Foster (18) |
| 41 (=98B) | 430 | papE fimbrial protein subunit | 30 (258 bp) | E. coli | |
| 29 | 452 | vgaB pristinamycin resistance (ABC transporter) | 43 (174 bp) | S. aureus | Allignet and El Solh (2) |
| 120 (=108; 27) | 535 | malF homologue cymF (ABC transporter) | 42 (534 bp) | Klebsiella oxytoca | |
| 125 | 1,230 | ABC transporter (bacitracin resistance homologue) | 45 (570 bp) | Bacillus halodurans | |
| Nondifferential fragmentsb | |||||
| 70 | 573 | Membrane transporter homologue yclI | 65 (567 bp) | B. subtilis | |
| 83 | 720 | Internalin B | 100 (507 bp) | L. monocytogenes | |
| Differential fragmentsc | |||||
| 85 | 1,143 | Methyltransferase (R-M system) | 57 (471 bp) | Lactococcus lactis | Twomey et al. (47) |
| 181 | 492 | Exo-α-1,4-glucosidase | 48 (471 bp) | Bacillus halodurans | |
| Nondifferential fragmentsc | |||||
| 48B | 525 | Transketolase | 62 (525 bp) | B. subtilis | |
| 60 | 565 | α-Mannosidase | 61 (565 bp) | B. halodurans | |
| 61 | 929 | Catabolic control protein A | 98 (558 bp) | L. monocytogenes | |
| 62 | 359 | UDP-N-acetylglucosamin-2-epimerase | 64 (360 bp) | B. subtilis | |
| 64 | 675 | Topoisomerase IV subunit | 49 (672 bp) | B. subtilis | |
| 66 | 709 | PduT (1,2-propanediol catabolism) | 32 (501 bp)a | S. enterica | |
| Ethanolamine utilization protein | 47 (255 bp)a | E. coli | |||
| 71 | 456 | Thioredoxin reductase homologue yumC | 62 (456 bp) | B. subtilis | |
| 80 | 483 | Carbamate kinase | 62 (447 bp) | Lactobacillus sakei | |
| 82 | ∼1,000 | β-Glucosidase | 56 (315 bp)a | Streptomyces coelicolor | |
| β-Glucosidase | 41 (302 bp)a | S. coelicolor | |||
| 104B | 533 | comG operon protein 1 (DNA uptake) | 48 (504 bp) | B. subtilis | Chung and Dubnau (10) |
Alignments at different locations.
Gene fragments homologous to virulence determinants, surface proteins, or transporters.
Gene fragments homologous to other proteins.
Gene fragments without homologies or homologous to proteins with unknown function (indicated with #) as of 29 Sept. 2000 included differential fragment numbers 7 (∼1 kb); 15 (787 bp); 16 (192 bp); 17B (∼1.7 kb); 18 (∼0.9 kb); 55 (531 bp); 67# (734 bp); 73 (478 bp); 78B# (∼1.1 kb); 80B (=10B; 294 bp); 110 (∼0.9 kb); 116 (313 bp); 128B (446 bp); 133 (=113; 748 bp); 136B (625 bp); 144# (532 bp); 149 (393 bp); 161 (∼1.1 kb); 166 (597 bp); U5B (625 bp); and U22# (∼0.9 kb) and the nondifferential fragment number 69 (306 bp).
RESULTS
Five to 6% of the L. monocytogenes strain F.4565 genome did not hybridize to the genome of L. monocytogenes strain EGD.
Genomic subtractions were carried out using a PCR-based subtractive hybridization method (1). Effective subtraction conditions for L. monocytogenes were established as detailed above in Materials and Methods. In order to identify DNA fragments specific for an epidemic clone of L. monocytogenes (strain F.4565; serotype 4b) (31) that belongs to a genetically closely related subgroup of strains that has caused major outbreaks of listeriosis in America and Europe (6, 26, 38), we subtracted the virulent prototype L. monocytogenes strain EGD (serotype 1/2a) from this epidemic strain. Subtraction efficiency for the L. monocytogenes genes plcA, hly, and iap (all shared by both strains) ranged from 64-fold for plcA to 8-fold for hly to no detectable subtraction for iap (data not shown).
Plasmids from libraries of PCR-amplified subtracted and unsubtracted material were spotted onto positively charged nylon membranes and hybridized with radioactively labeled whole-genome probes of tester (L. monocytogenes strain F.4565) and driver (L. monocytogenes strain EGD) DNA under low-stringency conditions. A total of 45 of 270 clones from the subtracted library versus 5 of 90 clones from the unsubtracted library did not hybridize with driver DNA, indicating threefold enrichment. Since the differentially hybridizing fragments from the nonsubtracted library were in the average size range (see Materials and Methods), one can conclude from the frequency of these clones (5 out of 90) that in the order of 5 to 6% of the genome of the epidemic L. monocytogenes clone strain F.4565 does not hybridize with the genome of L. monocytogenes strain EGD.
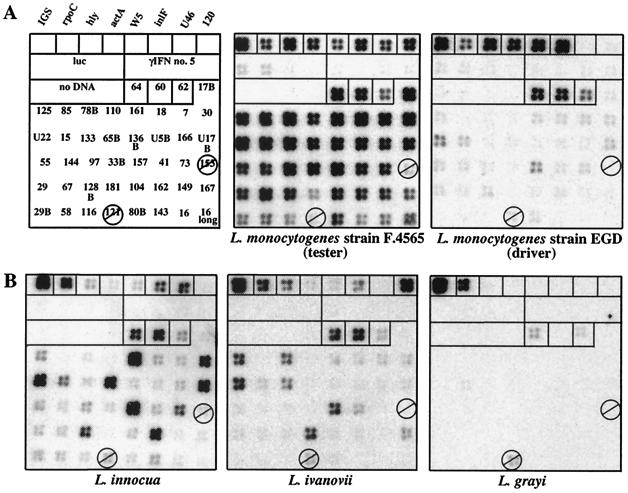
A total of 50 differentially hybridizing clones were isolated and sequenced. Seven of these turned out to be isolated more than once. The inserts of the 41 remaining, unique clones were amplified by PCR, arrayed onto positively charged nylon membrane, and reanalyzed. Figure 1A shows that 39 clones hybridized differentially with the genome of the tester strain L. monocytogenes F.4565. To verify these results, 6 of the 39 fragments were tested by Southern blotting under low-stringency conditions (data not shown). All six fragments hybridized specifically to the tester strain (L. monocytogenes strain F.4565) and to another epidemic strain of serotype 4b (L. monocytogenes strain L.4486a from the Swiss outbreak in 1983), while none of the fragments showed hybridization to the driver strain (L. monocytogenes EGD, serotype 1/2a) or to L. monocytogenes strain L028 (serotype 1/2c). Two of the six fragments cross-hybridized to different extents with L. innocua DNA (U46 and 143; see also Fig. 1 and below).
FIG. 1.
Identification of 39 gene fragments specific for the epidemic L. monocytogenes strain F.4565 and cross-hybridization with other Listeria species, with reverse Southern blots of replica DNA arrays. PCR products were spotted four times each at 20 ng per spot onto positively charged nylon membranes and hybridized to whole-genome probes of Listeria strains. Membranes were washed under low-stringency conditions. The first row of the array contained eight known DNA fragments as hybridization controls. The second row contained two sets of four unrelated DNA sequences at 80, 40, 20, and 10 ng per spot as negative controls. The third row contained four sets with no DNA followed by three fragments common to the tester and driver strains (no. 64, 60, and 62). Epidemic-clone-specific fragments were spotted from left to right, starting with no. 17B and ending with no. 16 containing the 270-bp flanking vector sequence. Fragments 153 and 121 gave no signal and were excluded from the analysis. 65B is the same as U46. (A) A total of 39 fragments hybridized differentially with tester DNA. (B) Nine L. monocytogenes strain F.4565-specific fragments gave strong signals with L. innocua DNA, five gave signals with L. ivanovii DNA, and none gave signals with L. grayi DNA.
The absence of the 39 differentially hybridizing fragments from the EGD genome were confirmed by BLASTN and FASTA nucleotide similarity searches against the completed genome sequence of L. monocytogenes strain EGD-e (which corresponds to our driver strain). Only 4 of the 39 fragments showed significant homologies to EGD-e genome sequences, mainly in coding regions (European Consortium, personal communication). Two of these fragments (125 and 015) had at one end a short sequence region (of about one-sixth of their respective lengths) that had high homology to EGD-e sequences, indicating a junction between conserved and nonconserved F.4565 genome regions. Four differentially hybridizing fragments could be detected with FASTA (162, 60% identity in 394 nucleotides [nt]; 73, 66% identity in 395 nt; 128B, 69% identity in 341 nt; and 157, 71% identity in 416 nt [European Consortium, personal communication]). (Fragment number 157 showed a weak, barely detectable hybridization signal representing our detection limit.) Thus, in these cases, homologous sequences were present in the EGD-e genome but showed a high level of sequence diversity. In contrast to these findings, 13 nondifferential control fragments showed strong homologies to EGD-e sequences with high conservation ranging from 88 to 98% similarity (European Consortium, personal communication).
Many strain F.4565-specific gene fragments had homologies to surface proteins involved in virulence.
The results of amino acid homology searches in translated public nucleotide or protein databases with the 39 translated L. monocytogenes strain F.4565-specific genome fragments are summarized in Table 1. A total of 21 fragments showed no significant homology to database entries (“no hit”) or were homologous to proteins of unknown function. Eleven fragments showed homologies to surface proteins of L. monocytogenes or to other gram-positive bacteria, including one to a leucine-rich protein interaction motif of a plant disease resistance gene, three to ABC transporters (fragments 29, 120, and 125), one to a locus involved in surface antigen expression, one to a component of a type II restriction system, and one to a metabolic enzyme. In contrast to these findings, when we analyzed 13 randomly selected nondifferential clones (i.e., hybridizing to both the tester and driver L. monocytogenes strains) we found that only one was a “no hit” and only two had homology to bacterial surface components, whereas 10 had homology to various enzymes. The distribution of database hits in different protein categories is graphically displayed in Fig. 2. A total of 41% of the differential fragments showed homology to bacterial surface components. Many of these have previously been implicated in virulence mechanisms, such as L. monocytogenes internalins (host cell recognition and invasion) (11, 19), the alpha C protein of group B streptococci (repeats involved in immune escape) (32, 34), papE fimbriae of E. coli (host cell recognition and adhesion) (25), ABC transporters (resistance to bacitracin [39] or pristinamycin [2]).
FIG. 2.
Properties of L. monocytogenes F.4565-specific fragments differ from those common to tester and driver. Functional categories are as assigned by homology (for further details, see Table 1).
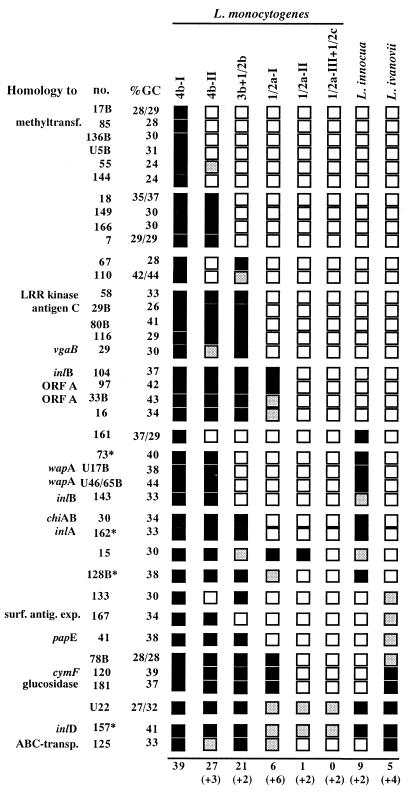
Hybridization patterns of L. monocytogenes F.4565-specific fragments with genomes of different L. monocytogenes strains and species.
In order to obtain information on the occurrence of L. monocytogenes F.4565-specific fragments in the genomes of L. monocytogenes strains of the same or other serotypes, we hybridized the arrays with whole genome probes of 14 L. monocytogenes strains (corresponding to 12 independent isolates; Table 2). According to hybridization patterns, the 4b strains were subdivided into two groups (4b-I and 4b-II), 1/2a strains were divided into three groups (1/2a-I, 1/2a-II, and 1/2a-III), serotype 3b and 1/2b strains grouped together, and the 1/2c strain grouped together with the 1/2a-III strain (Table 2 and Fig. 3). Thus, according to the number of shared hybridization signals, 4b strains appeared more related to 3b and 1/2b strains than to 1/2a and 1/2c strains. These results reflect the established division of L. monocytogenes strains into two groups (one comprising 1/2a and 1/2c and one comprising the 4b, 1/2b, and 3b strains) on the basis of multilocus enzyme electrophoresis, ribotyping, pulse-field gel electrophoresis, and restriction enzyme analysis (6, 8, 21, 38) and, additionally, into subgroups of 1/2a and 4b strains by more resolving methods, such as PCR-restriction enzyme analysis of the in1A-in1B region (14) or sequencing of the in1B gene (15).
FIG. 3.
Summary of hybridization profiles of strain F.4565-specific fragments. Black boxes indicate positive, grey boxes indicate weak, and white boxes indicate negative hybridization signals. The number of hybridizing fragments is indicated below each column, and weakly hybridizing fragments are shown in parentheses. The GC content of the fragments is indicated; in the case of not fully sequenced fragments, the GC contents of the left and right parts of the fragment are given separately. Asterisks indicate fragments present in the EGD genome with high sequence diversity. Abbreviations: methyltransf., methyltransferase; surf. antig. expo.; surface antigen expression; transp., transporter.
Apart from controls, no significant hybridization signal after low-stringency washing was seen with genomic DNA of E. coli, B. subtilis (data not shown), or Listeria grayi, the member of the genus Listeria most distantly related to L. monocytogenes (Fig. 1B). However, DNA from the animal pathogen L. ivanovii and from nonpathogenic L. innocua strongly cross-hybridized to five and nine fragments, respectively, only two of which were common (Fig. 1B and Fig. 3). A second L. innocua strain showed an identical hybridization behavior (data not shown). These latter results raise the possibility that the genomes of at least three different species within the genus Listeria may have a mosaic-like composition with respect to these genes.
DISCUSSION
Five to 6% of DNA segments randomly picked from libraries from the unsubtracted genome of L. monocytogenes strain F.4565 did not hybridize to strain EGD DNA, indicating that about 5% of the F.4565 genome is not present in strain EGD. Physical genomic maps are available for L. monocytogenes strains EGD (serotype 1/2a) (49) and Scott A (serotype 4b) (23) and have yielded genome sizes of 3,000 ± 50 and 3,210 ± 60 kb, respectively. Assuming that strain F.4565 has a genome size similar to that of strain Scott A, which has the same serotype, about 150 to 190 kb of its DNA would be absent or substantially different from the genome of strain EGD. With an average RsaI fragment size of 0.8 kb, we would thus expect about 200 differential fragments to be present in the F.4565 genome. Of these, we have isolated 39 fragments corresponding to about one-fifth of the theoretically expected number. (The latter may explain why we failed to isolate the genes gtcA and gltAB, which are known to be present in serotype 4b but not in serotype 1/2a strains [30, 40].)
Our results are in line with interstrain genome comparison data available for other bacterial species. E. coli strains with different pathogenic potential or different host ranges were found to vary by 300 to more than 1,000 kb (9, 12, 37), and the genomes of different Salmonella sp. serovars can differ by greater than 900 kb (36). Recent pairwise whole-genome comparisons of Helicobacter pylori and Chlamydia isolates showed that strain-specific genes tend to cluster in distinct genomic regions of about 50 to 160 kb termed plasticity zones (3, 43), while E. coli O157:H7 and E. coli K-12 differ by hundreds of islands of DNA that are apparently introgressed into a shared sequence backbone (37). Also, the extent of genome diversification varies: while different Chlamydia isolates showed little strain-specific genome material (43), the two unrelated H. pylori isolates each had 6 to 7% strain-specific genes (3), and the E. coli O157:H7 and K-12 isolates had about 25 and 12% strain-specific genes, respectively (37).
H. pylori isolates seem to differ mainly in components of restriction modification systems, insertion sequences, and DNA repeats (1, 3), while the E. coli isolates do not seem to differ by functionally distinct groups of genes (37). This is in contrast to the situation in L. monocytogenes, where 41% of the strain-specific genes exhibited homology to proteins exposed on the bacterial surface, while for 54% of the genes, no function on the basis of sequence similarity could be predicted (Fig. 2). We found only one DNA segment with homology to a restriction modification system component: a methyltransferase from L. lactis sharing homology with the target recognition domain of M · Sau3A (47). The presence of this DNA fragment in the genomes of the different L. monocytogenes strains used by us (Table 2 and Fig. 3) correlated with resistance of their DNA to Sau3A restriction (data not shown). Resistance to Sau3A restriction has previously been reported as a prevalent trait of epidemic-associated L. monocytogenes serotype 4b strains (50).
Based on the sequences of the large and small intergenic spacer regions between the 16S and 23S rRNA genes, L. monocytogenes strains of different serotypes are more related to each other than to L. innocua (20). The nucleotide diversity of the sigmaB and iap genes accessible in the databases is consistent with this interpretation (data not shown). Within the genus Listeria, the nonpathogen L. innocua is the species most closely related to L. monocytogenes, while the animal pathogen L. ivanovii and nonpathogen L. grayi are more distantly related (20). Surprisingly, we found that two partially overlapping sets of nine and five L. monocytogenes F.4565-specific fragments strongly hybridized with the genomes of L. innocua and L. ivanovii, respectively (Fig. 1 and 3). Also, the other serotype 4b-specific genes isolated to date, gtcA and gltAB, which are loci involved in cell wall teichoic acid-associated surface antigen expression, cross-hybridize to certain strains of L. innocua (29, 30, 40). While some of these fragments may have been lost by some lineages of L. monocytogenes, others may have been acquired by lateral gene transfer. Thus, our data raise the possibility that lateral gene transfer could play a role in the diversification of L. monocytogenes strains. Indeed, this has recently been substantiated for gtcA (29).
Repeat regions are preferential subjects of lateral gene transfer events. Many of the F.4565-specific fragments had homologies to repetitive protein elements (Table 1). Repeats facilitate recombination events, which are thought to occur about 50-fold more frequently than point mutations and therefore to have a much higher impact on bacterial genome diversification, in both gram-negative (E. coli, Neisseria meningitidis) and gram-positive (Streptococcus pneumoniae) bacteria (22). In particular, the set of fragments cross-hybridizing with L. innocua shows homology to multidomain, repeat-containing surface proteins such as internalins and chitinases (Fig. 3). Lateral gene transfer has been implicated in the generation and diversification of chitinases (especially the cadherin-like domain) (35) and proteins containing leucine-rich repeat short sequence motifs thought to be involved in specific protein-protein interactions (27). Moreover, mosaic internalin genes have been described (41). Consistent with this possibility, a large number of transducing Listeria phages have been isolated recently (24). It is also interesting that one of the nondifferential control fragments (no. 104B) shows some homology to a B. subtilis gene involved in DNA uptake (10).
Our hybridization analysis with arrays harboring the 39 F.4565-specific fragments against genomes of different L. monocytogenes strains suggests that L. monocytogenes strains can be subtyped by genomic differences (Fig. 3). Because of its higher resolution, this approach is superior to serotyping for serotype 1/2a and 4b strains. Thus, genomic differences could be exploited for typing L. monocytogenes strains by combinatorial PCRs or by genome hybridizations to premanufactured typing arrays.
Taken together, we have shown that epidemic L. monocytogenes strains differ substantially from the sequenced prototype strain of L. monocytogenes. Many of the isolated gene fragments have homology to bacterial surface components and are therefore likely to confer traits that provide selective advantages in the environment. While this may be a general phenomenon applying to strain diversification of virulent as well as nonvirulent bacterial strains, our results nevertheless provide a good starting point to search for epidemic-trait-associated genes. For example, the complete set of genes specifically present (or absent, since pathogenicity traits may also be due to lack of specific genes [36]) in an epidemic-trait-associated strain can now be identified and screened against a large number of genomes from different L. monocytogenes isolates and Listeria species. Identification of genes consistently present or absent in epidemic-associated L. monocytogenes strains will open the way for mutational and functional analyses to address the question of whether indeed there is a molecular basis for the increased pathogenic potential of epidemic L. monocytogenes strains.
ACKNOWLEDGMENTS
We are grateful to Jim McLauchlin, London, for providing many of the bacterial strains; to Ali Hemmati-Bivanlou, New York, N.Y., and Chris Wilson, Cambridge, England, for technical advice concerning DNA arrays; to our colleagues from the Institute for Genetics, Uli Böhm, Thorsten Klamp, and Gloria Esposito, for advice with subtractive techniques; to Rita Lange for DNA sequencing; and to Olga Vites and Jonathan Howard for stimulating criticism throughout the work. We thank the European consortium involved in sequencing the L. monocytogenes EGD-e genome for the possibility of analyzing the complete genome sequence prior to its release, especially P. Cossart, Paris, France, who made this possible, and P. Glaser, Paris, France, who kindly carried out the analysis and discussed the results with us. Many thanks also to colleagues whose comments have helped to improve the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 274.
REFERENCES
- 1.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet J, El Solh N. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene. 1997;202:133–138. doi: 10.1016/s0378-1119(97)00464-2. [DOI] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 6.Bibb W, Gellin B G, Weaver R, Schwartz B, Plikaytis B D, Reeves M W, Pinner R W, Broome C V. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations Appl. Environ Microbiol. 1990;56:2133–2141. doi: 10.1128/aem.56.7.2133-2141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bille J, Rocourt J. WHO international multicenter Listeria monocytogenes subtyping study—rationale and set-up of the study. Int J Food Microbiol. 1996;32:251–262. doi: 10.1016/s0168-1605(96)01140-3. [DOI] [PubMed] [Google Scholar]
- 8.Brosch R, Chen J, Luchansky J B. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl Environ Microbiol. 1994;60:2584–2592. doi: 10.1128/aem.60.7.2584-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown P K, Curtiss R., 3rd Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 1996;93:11149–11154. doi: 10.1073/pnas.93.20.11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung Y S, Dubnau D. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart P, Lecuit M. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz I, Davies I. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 13.Dramsi S, Dehoux P, Lebrun M, Goossens P L, Cossart P. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect Immun. 1997;65:1615–1625. doi: 10.1128/iai.65.5.1615-1625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ericsson H, Stalhandske P, Danielsson-Tham M L, Bannerman E, Bille J, Jacquet C, Rocourt J, Tham W. Division of Listeria monocytogenes serovar 4b strains into two groups by PCR and restriction enzyme analysis. Appl Environ Microbiol. 1995;61:3872–3874. doi: 10.1128/aem.61.11.3872-3874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericsson H, Unnerstad H, Mattsson J G, Danielsson-Tham M L, Tham W. Molecular grouping of Listeria monocytogenes based on the sequence of the inlB gene. J Med Microbiol. 2000;49:73–80. doi: 10.1099/0022-1317-49-1-73. [DOI] [PubMed] [Google Scholar]
- 16.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feresu S B, Jones D. Taxonomic studies on Brochothrix, Erysipelothrix, Listeria and atypical lactobacilli. J Gen Microbiol. 1988;134:1165–1183. doi: 10.1099/00221287-134-5-1165. [DOI] [PubMed] [Google Scholar]
- 18.Foster S J. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1993;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 19.Galan J E. Alternative strategies for becoming an insider: lessons from the bacterial world. Cell. 2000;103:363–366. doi: 10.1016/s0092-8674(00)00127-6. [DOI] [PubMed] [Google Scholar]
- 20.Graham T A, Golsteyn-Thomas E J, Thomas J E, Gannon V P. Inter- and intraspecies comparison of the 16S–23S rRNA operon intergenic spacer regions of six Listeria spp. Int J Syst Bacteriol. 1997;47:863–869. doi: 10.1099/00207713-47-3-863. [DOI] [PubMed] [Google Scholar]
- 21.Graves L M, Swaminathan B, Reeves M W, Hunter S B, Weaver R E, Plikaytis B D, Schuchat A. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J Clin Microbiol. 1994;32:2936–2943. doi: 10.1128/jcm.32.12.2936-2943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 23.He W, Luchansky J B. Construction of the temperature-sensitive vectors pLUCH80 and pLUCH88 for delivery of Tn917::NotI/SmaI and use of these vectors to derive a circular map of Listeria monocytogenes Scott A, a serotype 4b isolate. Appl Environ Microbiol. 1997;63:3480–3487. doi: 10.1128/aem.63.9.3480-3487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgson D A. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol Microbiol. 2000;35:312–323. doi: 10.1046/j.1365-2958.2000.01643.x. [DOI] [PubMed] [Google Scholar]
- 25.Jacob-Dubuisson F, Kuehn M, Hultgren S J. A novel secretion apparatus for the assembly of adhesive bacterial pili. Trends Microbiol. 1993;1:50–55. doi: 10.1016/0966-842x(93)90032-m. [DOI] [PubMed] [Google Scholar]
- 26.Jacquet C, Catimel B, Brosch R, Buchrieser C, Dehaumont P, Goulet V, Lepoutre A, Veit P, Rocourt J. Investigations related to the epidemic strain involved in the French listeriosis outbreak in 1992. Appl Environ Microbiol. 1995;61:2242–2246. doi: 10.1128/aem.61.6.2242-2246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 28.Lammerding A M, Glass K A, Gendron-Fitzpatrick A, Doyle M P. Determination of virulence of different strains of Listeria monocytogenes and Listeria innocua by oral inoculation of pregnant mice. Appl Environ Microbiol. 1992;58:3991–4000. doi: 10.1128/aem.58.12.3991-4000.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan Z, Fiedler F, Kathariou S. A sheep in wolf's clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J Bacteriol. 2000;182:6161–6168. doi: 10.1128/jb.182.21.6161-6168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei X-H, Fiedler F, Lan Z, Kathariou S. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J Bacteriol. 2001;183:1133–1139. doi: 10.1128/JB.183.4.1133-1139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linnan M J, Mascola L, Lou X D, Goulet V, May S, Salminen C, Hird D W, Yonekura M L, Hayes P, Weaver R, et al. Epidemic listeriosis associated with Mexican-style cheese. N Engl J Med. 1988;319:823–828. doi: 10.1056/NEJM198809293191303. [DOI] [PubMed] [Google Scholar]
- 32.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlan A M, Foster S J. Molecular characterization of an autolytic amidase of Listeria monocytogenes EGD. Microbiology. 1998;144:1359–1367. doi: 10.1099/00221287-144-5-1359. [DOI] [PubMed] [Google Scholar]
- 34.Michel J L, Madoff L C, Olson K, Kling D E, Kasper D L, Ausubel F M. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci USA. 1992;89:10060–10064. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrow B J, Graham J E, Curtiss R., III Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect Immun. 1999;67:5106–5116. doi: 10.1128/iai.67.10.5106-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perna N T, Plunkett III G, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck E J, Davis N W, Lim A, Dimalanta E T, Potamousis K D, Apodaca J, Anantharaman T S, Lin J, Yen G, Schwartz D C, Welch R A, Blattner F R. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 38.Piffaretti J C, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser J M, Selander R K, Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci USA. 1989;86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- 40.Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J Bacteriol. 1999;181:418–425. doi: 10.1128/jb.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raffelsbauer D, Bubert A, Engelbrecht F, Scheinpflug J, Simm A, Hess J, Kaufmann S H, Goebel W. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol Gen Genet. 1998;260:144–158. doi: 10.1007/s004380050880. [DOI] [PubMed] [Google Scholar]
- 42.Rasmussen O F, Skouboe P, Dons L, Rossen L, Olsen J E. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- 43.Read T D, Brunham R C, Shen C, Gill S R, Heidelberg J F, White O, Hickey E K, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg S L, Eisen J, Fraser C M. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 46.Sokolovic Z, Schuller S, Bohne J, Baur A, Rdest U, Dickneite C, Nichterlein T, Goebel W. Differences in virulence and in expression of PrfA and PrfA-regulated virulence genes of Listeria monocytogenes strains belonging to serogroup 4. Infect Immun. 1996;64:4008–4019. doi: 10.1128/iai.64.10.4008-4019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Twomey D P, McKay L L, O'Sullivan D J. Molecular characterization of the Lactococcus lactis LlaKR2I restriction-modification system and effect of an IS982 element positioned between the restriction and modification genes. J Bacteriol. 1998;180:5844–5854. doi: 10.1128/jb.180.22.5844-5854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicente M F, Baquero F, Perez-Diaz J C. Cloning and expression of the Listeria monocytogenes haemolysin in E. coli. FEMS Microbiol Lett. 1985;30:77–79. [Google Scholar]
- 49.von Both U, Otten S, Darbouche A, Domann E, Chakraborty T. Physical and genetic map of the Listeria monocytogenes EGD serotype 1/2a chromosome. FEMS Microbiol Lett. 1999;175:281–289. doi: 10.1111/j.1574-6968.1999.tb13632.x. [DOI] [PubMed] [Google Scholar]
- 50.Zheng W, Kathariou S. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl Environ Microbiol. 1997;63:3085–3089. doi: 10.1128/aem.63.8.3085-3089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]