Abstract
Objective
Accurate and rapid phenotyping is a prerequisite to leveraging electronic health records for biomedical research. While early phenotyping relied on rule-based algorithms curated by experts, machine learning (ML) approaches have emerged as an alternative to improve scalability across phenotypes and healthcare settings. This study evaluates ML-based phenotyping with respect to (1) the data sources used, (2) the phenotypes considered, (3) the methods applied, and (4) the reporting and evaluation methods used.
Materials and methods
We searched PubMed and Web of Science for articles published between 2018 and 2022. After screening 850 articles, we recorded 37 variables on 100 studies.
Results
Most studies utilized data from a single institution and included information in clinical notes. Although chronic conditions were most commonly considered, ML also enabled the characterization of nuanced phenotypes such as social determinants of health. Supervised deep learning was the most popular ML paradigm, while semi-supervised and weakly supervised learning were applied to expedite algorithm development and unsupervised learning to facilitate phenotype discovery. ML approaches did not uniformly outperform rule-based algorithms, but deep learning offered a marginal improvement over traditional ML for many conditions.
Discussion
Despite the progress in ML-based phenotyping, most articles focused on binary phenotypes and few articles evaluated external validity or used multi-institution data. Study settings were infrequently reported and analytic code was rarely released.
Conclusion
Continued research in ML-based phenotyping is warranted, with emphasis on characterizing nuanced phenotypes, establishing reporting and evaluation standards, and developing methods to accommodate misclassified phenotypes due to algorithm errors in downstream applications.
Keywords: electronic health records, phenotyping, cohort identification, machine learning
BACKGROUND AND SIGNIFICANCE
Electronic health records (EHRs) are a central data source for biomedical research.1 In recent years, EHR data have been used to support discovery in disease genomics, to enable rapid and more inclusive clinical trial recruitment, and to facilitate epidemiological studies of understudied and emerging diseases.2–6 EHRs are also positioned to be a key source of data for the development of personalized treatment strategies and the generation of real-world evidence.7,8 A critical aspect of any secondary use of EHR data is phenotyping, the process of identifying patients with a specific phenotype (eg, the presence or onset time of a clinical condition or characteristic) based on information in their EHR.9–11 Phenotyping is one of the first steps of an EHR-based application as it is used to both identify and characterize the population of interest.
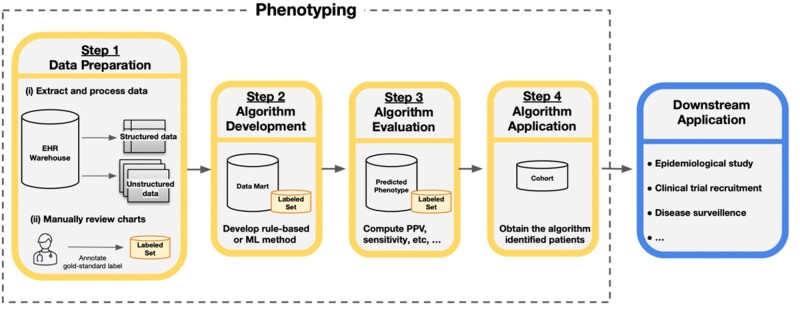
Generally, the phenotyping process consists of 4 steps: (1) data preparation, (2) algorithm development, (3) algorithm evaluation, and (4) algorithm application (Figure 1). The focus of our article is on the use of machine learning (ML) for algorithm development. Traditionally, phenotypes have been inferred from rule-based algorithms consisting of inclusion and exclusion criteria handcrafted by clinical and informatics experts.12 However, given the complexity and variation in documentation across phenotypes, providers, and institutions, developing a sufficient set of rules is prohibitively resource-intensive and difficult to scale across conditions and healthcare settings.13,14 For example, the Electronic Medical Records and Genomics (eMERGE) Network was an early leader in phenotyping in creating a public phenotype library called PheKB. A key finding from this effort was the time intensiveness of rule-based phenotyping, sometimes requiring up to 6–10 months of manual effort depending on the complexity of the condition.14 Similar findings have been reported by other large research networks such as OHDSI (Observational Health Data Science and Informatics).10
Figure 1.
Overview of the phenotyping process. Step 1 involves data preparation which includes (i) extraction and processing of relevant data from records of candidate patients from the data warehouse and (ii) manual review of a subset of charts to obtain gold-standard phenotype labels. Step 2 is the algorithm development phase in which researchers use the data from Step 1, often referred to as the data mart, to develop the phenotyping algorithm with a rule-based or machine learning (ML) method. Step 3 evaluates the accuracy of an algorithm by comparing the assigned phenotype from the algorithm to the gold-standard label, often with estimates of the positive predictive value (PPV), sensitivity, and other accuracy metrics. Step 4 applies the algorithm from Step 2 to obtain the cohort of patients with the phenotype for downstream analysis. The identified cohort can then be used in a variety of downstream applications.
To address this barrier to EHR-based research, there has been increasing interest in phenotyping algorithms derived from ML models.15,16 In contrast to rule-based approaches, ML methods aggregate multiple sources of information available in patient records in a more automated and generalizable fashion to improve phenotype characterization.17 While there has been substantial progress in ML approaches designed to make phenotyping more efficient, accurate, and portable in recent years, these advances have yet to be formally synthesized.18 To the best of our knowledge, 5 articles surveyed EHR-based phenotyping methods through 2018.11,15–17,19 These articles provide conceptual summaries of rule-based methods and early ML approaches and do not capture advances in semi-supervised, weakly supervised, and deep learning that were popularized after publication (Supplementary Table S1). Moreover, in light of the wave of EHR-based studies prompted by the COVID-19 pandemic and the increased complexity of ML approaches relative to their rule-based counterparts, there is a pressing need to survey the landscape of phenotyping given its ubiquity in EHR-based applications.20,21
OBJECTIVE
Our work fills this gap in current literature through a methodical review of ML-based phenotyping with respect to (1) the data sources used, (2) the phenotypes considered, (3) the methods applied, and (4) the reporting and evaluation methods used. Based on our analysis of 37 items recorded across 100 selected articles, we also identify areas of future research.
MATERIALS AND METHODS
Working definitions
To situate our discussion, key terminology related to EHR data and ML is provided in Table 1. We broadly classified an ML method as either (1) supervised, (2) semi-supervised, (3) weakly supervised, or (4) unsupervised according to the model used and the data available for training.22,23 We further classified each method as deep learning if it is neural network-based and as a traditional ML approach otherwise. Consistent with recent literature, we used an inclusive definition of phenotyping as a procedure that uses EHR data to “assert characterizations about patients.”18 Our study therefore includes binary phenotypes such as the presence of disease and nuanced phenotypes such as disease severity, disease progression, and social determinants of health (SDOHs). We focused solely on literature using EHRs, defined as longitudinal records of a patient’s interactions with a healthcare institution or system primarily authored by health professionals. We regard our work as a “methodical review” as it does not qualify as a Cochrane-style review, but closely adheres to the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines.24
Table 1.
Descriptions of (a) terms used to describe EHR data and (b) ML methods in the context of phenotyping
| (a) | |
|---|---|
| Term | Description |
| Structured data | Data that utilize a controlled vocabulary. Structured data are readily available and searchable in an EHR research database, but often have variable accuracy in characterizing phenotypes. Examples include diagnosis codes, procedure codes, demographics, prescriptions, and laboratory values. |
| Unstructured data | Data that are not organized in a specific manner and require substantial processing prior to analysis. In the context of phenotyping, the most common form of unstructured data is free-text, such as progress notes, admission and discharge summaries, and radiology reports. Medical images are another form of unstructured data, but were not used in the selected articles. |
| Gold-standard label | The best classification available for phenotype status, most often derived from manual review of patient records by a clinical expert. |
| Silver-standard label | Proxy for the gold-standard phenotype label that is less accurate in characterizing the phenotype, but that can be obtained without time-consuming chart review. Examples include billing codes specific to the phenotype and laboratory values. |
| Feature | Data elements that are potentially predictive of the phenotype and used for algorithm development. Examples include structured data elements such as diagnosis codes and prescriptions as well as information derived from unstructured free-text such as the number of times a phenotype is positively mentioned in a patient’s record. |
| Labeled data | Data that contain information on both the gold-standard phenotype labels and the features. |
| Weakly labeled data | Data that contain information on silver-standard labels and the features. |
| Unlabeled data | Data that contain information on only the features. |
| (b) | ||
|---|---|---|
| ML category | Description | Motivation for use in phenotyping |
| Supervised learning | Includes methods used to characterize a phenotype with algorithms trained with labeled data. | More automated and potentially more accurate than rule-based methods. |
| Semi-supervised learning | Includes methods used to characterize a phenotype with algorithms trained with both labeled and unlabeled data. | Reduces the amount of labeled data for model training. |
| Weakly supervised learning | Includes methods used to characterize a phenotype with algorithms trained with weakly labeled data. | Eliminates the need for labeled data for model training. |
| Unsupervised learning | Includes methods used to identify structure relevant to a phenotype, such as subtypes or clusters of disease progression trajectories, using unlabeled data. | Enables phenotype discovery. |
| Deep learning | A type of ML method that includes methods based on multilayer neural networks. Can be either supervised, semi-supervised, weakly supervised, or unsupervised. | Alleviates the need for feature engineering and can yield high accuracy on phenotyping tasks. |
| Traditional machine learning | ML methods that are not constructed based on multilayer neural networks. | Simpler to implement and interpret than deep learning methods. |
Search strategy
Due to the broad and evolving definition of phenotyping, early systematic reviews employed a manual review of all full-text articles published in a small number of informatics venues.12,17 This manual approach was later expanded to a PubMed query using an overly inclusive search designed to capture all articles that (1) used EHR as the primary data source and (2) utilized ML or natural language processing (NLP) or considered phenotyping.15 The PubMed query was similarly restricted to a subset of informatics venues in order to target articles focused on phenotyping rather than clinical applications. We followed an analogous strategy, but increased the scope of our search by including Web of Science as we found articles were missed by PubMed. We also added additional strings related to ML.25
Specifically, our search of PubMed and Web of Science identified full-text articles that employed ML or NLP or considered phenotyping with EHR data published between January 1, 2018, and April 14, 2022. The range of publication year was specified to not overlap with existing reviews and focused on the same major informatics venues: (1) Journal of American Medical Informatics Association (JAMIA), (2) Journal of Biomedical Informatics (JBI), (3) PLoS One, (4) Proceedings of the American Medical Informatics Association’s Annual Symposium (AMIA), and (5) JAMIA Open.12,15,16,26,27 The complete search queries are provided in Supplementary Table S2.
Study selection
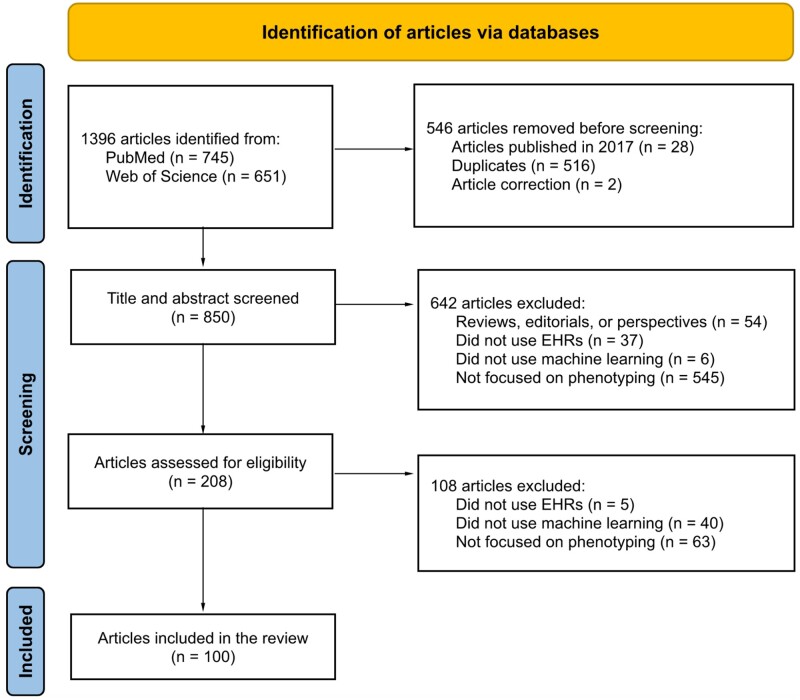
Our overall search strategy is depicted in a PRISMA diagram (Figure 2).
Figure 2.
PRISMA diagram for article selection. Only 1 exclusion reason was chosen for each record during the screening process, although the reasons are not mutually exclusive.
Title and abstract screening
After removing duplicates, articles were retrieved and underwent title and abstract screening by 2 authors (SY and JG). A third author (PV) resolved disagreements. Articles were excluded if they (1) were reviews, perspectives, or editorials, (2) did not use EHRs as a primary data source, (3) did not use ML methods, or (4) did not consider phenotyping. Supplementary Table S3 provides a list of article exclusions.
Full-text review
One author (SY) reviewed the full-text articles and another author (JG) verified the information from the full-text review when necessary. After excluding papers that did not focus on ML approaches for EHR phenotyping, 100 papers were selected (Supplementary Table S4). During the full-text review, we extracted information on (1) the data sources used, (2) the phenotypes considered, (3) the methods applied, and (4) the reporting and evaluation methods used. A list of the 37 recorded variables is included in Supplementary Table S5.
RESULTS
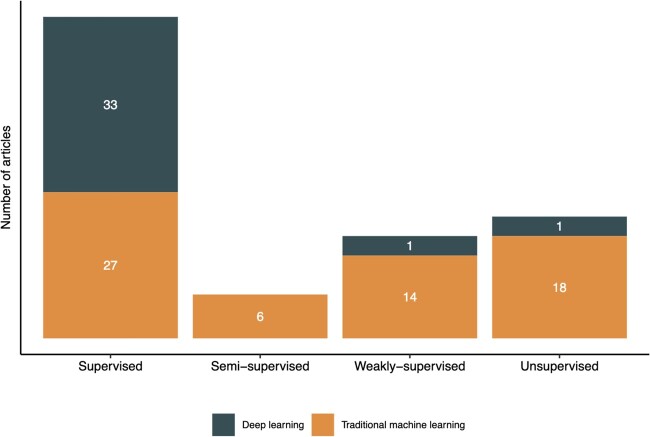
In reviewing the literature, we found that all but 2 deep-learning approaches were supervised (Figure 3). We therefore summarize contributions in traditional supervised, deep supervised, semi-supervised, weakly supervised, and unsupervised learning in the subsequent sections.
Figure 3.
Number of articles that used the various machine learning paradigms.
Data sources
Sixty-three of the 100 articles relied on EHR data from a single institution, while 8 articles used data from multiple institutions, including research networks such as the OHDSI28 and eMERGE.29 The remaining articles leveraged publicly available data from the Medical Information Mart for Intensive Care (MIMIC-III) database and NLP competitions (Supplementary Table S6). A small number of studies utilized additional data sources, including administrative claims30–36 and registry databases.37–40 Ninety-four studies were conducted in the United States.
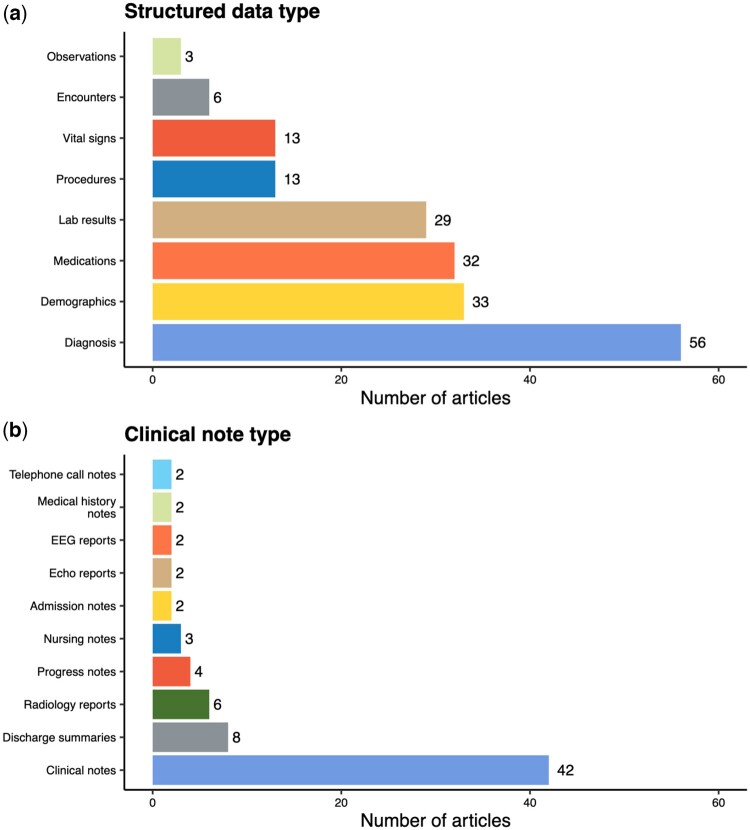
With respect to the data types used for developing phenotyping algorithms, 70 of the 100 articles utilized unstructured free-text data, and half of these articles also incorporated information from structured data. Unsurprisingly, diagnoses were the most common structured data element and were typically derived from the International Classification of Diseases, 9th or 10th Revision (ICD-9/10) billing codes (Figure 4(a)). Clinical note types (eg, progress notes and discharge summaries) used for algorithm development were rarely specified (Figure 4(b)). However, most articles reported on the NLP software that was used to process free-text. The clinical Text Analysis and Knowledge Extraction System (cTAKEs) was the most popular. Frequently used terminologies and NLP software are detailed in Supplementary Tables S7 and S8, respectively.
Figure 4.
Types of structured data and clinical notes used to develop phenotyping algorithms in the selected articles (excluding articles using competition data). A data type is presented if it is used in more than 1 article. Encounters include encounter metadata, while medical history notes include both social history and cardiac surgical history.
Phenotypes
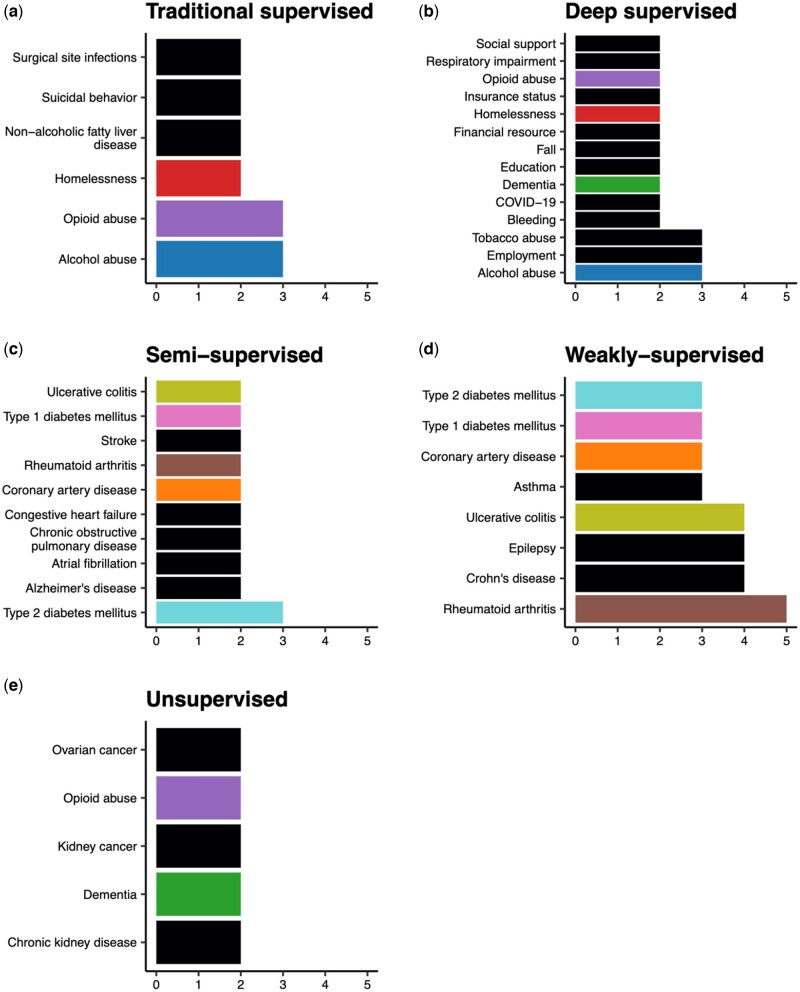
The articles in our study considered 157 phenotypes, with 40 articles focusing on more than 1 phenotype. Studies using data from NLP competitions focused on adverse drug events41 and clinical trial eligibility,42 while studies using MIMIC-III characterized phenotypes seen in the intensive care unit.43 Outside of the articles using publicly available data, chronic conditions with a large burden on the healthcare system, such as heart diseases and type II diabetes mellitus, were most frequently considered overall. Sixty-nine of the 100 articles aimed to identify binary phenotypes (eg, case/control disease status), while few focused on the severity or temporal phenotypes (4 and 11 articles, respectively). Although this finding coincides with previous reviews, there were considerable differences in the top phenotypes across the 5 ML paradigms (Figure 5). The phenotypes considered in articles utilizing traditional supervised learning were not identified in previous reviews.12,15 These include phenotypes primarily documented in free-text such as suicidal behavior44,45 and SDOHs.30,46–49 Deep supervised learning papers similarly considered SDOHs50–57 as well as episodic conditions58–61 and COVID-19.62,63 The phenotypes considered by articles using semi- or weakly supervised methods aiming to expedite algorithm development included common, chronic conditions64–66 that had been previously identified with a rule-based or traditional supervised learning method.13,67 Most unsupervised methods considered progressive conditions associated with multiple comorbidities or phenotypic heterogeneity such as dementia and chronic kidney disease.68,69
Figure 5.
Top phenotypes considered within each machine learning category and the number of articles of each phenotype (excluding articles using competition data sources). Phenotypes are colored if they appear in more than 1 ML paradigm.
ML methods
Traditional supervised learning
Sixty articles employed supervised learning methods, with 27 articles using traditional models. In contrast to rule-based algorithms, phenotyping algorithms derived from supervised learning are less burdensome to develop as they are learned from the data.15 Traditional supervised learning is also more amenable to incorporating a greater number of features predictive of the phenotype into the algorithm, such as information in clinical notes.17,154–160 Among the articles using traditional supervised learning, half of them mapped terms in free-text to clinical concepts in the Unified Medical Language System (UMLS)70 for use in algorithm development. Similar to features derived from structured data elements, the extracted concepts were typically engineered into patient-level features (eg, total number of positive mentions of a concept in a patient’s record) based on the consensus of domain experts.71 Gold-standard labels for model training were predominantly annotated through a manual review of patient records.72 In some instances, labels were also derived from registry data,37 laboratory results,35,36,73 or rule-based algorithms.47
The most commonly used methods were random forest, logistic regression, and support vector machine (SVM) (Table 2). A common trend among selected articles was the use of a selective sampling method, such as undersampling or the Synthetic Minority Oversampling Technique (SMOTE), to address class imbalance for rare phenotypes such as surgical site infections and rhabdomyolysis.31,33,35,37,48,76,77 Several models, including SVM, single-layer perceptron, and logistic regression, were also extended to accommodate federated analysis of distributed EHR data held locally at multiple institutions to identify adverse drug reactions.33
Table 2.
Common methods in each machine learning category
| Machine learning type | Methods | Number of articles |
|---|---|---|
| Traditional supervised learning | Random forest | 14 |
| Logistic regression | 11 | |
| Support vector machine (SVM) | 11 | |
| L1-penalized logistic regression | 8 | |
| Decision trees | 4 | |
| Extreme gradient boosting (XGBoost) | 4 | |
| Naive Bayes | 3 | |
| Deep supervised learning | Recurrent neural networks (RNNs) and variants | 19 |
| Convolutional neural networks (CNNs) and variants | 11 | |
| BERT and variants | 7 | |
| Feed-forward neural networks (FFNNs) | 3 | |
| Weakly supervised learning | PheNorm74 | 3 |
| MAP75 | 2 | |
| Random forest (with silver-standard labels) | 2 | |
| Unsupervised learning | Latent Dirichlet Allocation (LDA) | 5 |
| K-means | 4 | |
| UPGMA (Unweighted Pair Group Method with Arithmetic mean) hierarchical clustering | 2 |
Note: A method is presented if it appeared in more than 1 article. Several papers used more than 1 method. The table does not include any semi-supervised methods as each article used a distinct method. Semi-supervised methods are presented in Table 3.
Deep supervised learning
While traditional supervised learning methods can streamline algorithm development, they are limited by their inability to handle raw input data. Deep learning models consist of many processing layers that discover intrinsic patterns within data to alleviate the burden of feature engineering.78,79 This is particularly valuable in the context of EHR data as models can learn rich representations of the clinical language in free-text.80 All but 2 articles employing deep supervised learning articles leveraged clinical notes. The articles utilized word embeddings to represent words or clinical concepts as real-valued vectors based on their context.81 Word embeddings are typically learned from a large corpus in an unsupervised fashion and used as the input layer to a neural network. Common corpora within the selected articles included clinical notes53,57,63,82–86 as well as external sources such as biomedical publications56,61,62,87,88 and Wikipedia articles51,58,89–92 (Supplementary Table S9). Word2vec,93,161,163 Global Vectors (GloVE),94,162 and Bidirectional Encoder Representations from Transformers (BERT)95–98 were the most frequently used methods for training embeddings (Supplementary Table S10).
Among neural network architectures, feed-forward networks were only used in 3 studies (Supplementary Table S11)99 while BERT and variants were frequently used for phenotypes documented in clinical notes such as SDOHs (eg, education50,57) and symptoms (eg, chest pain92 and bleeding58) Recurrent neural networks (RNNs), convolutional neural networks (CNNs), and their variants were the most prevalent architectures as they accommodate sequential data in longitudinal patient records and clinical text.24,78 For instance, the bidirectional long-short term memory (Bi-LSTM), an RNN variant that captures previous and future information in a sequence, was used to characterize phenotypes evolving over time such as dementia34 and substance abuse.54 In terms of text-based phenotyping, the Bi-LSTM with a conditional random field layer (Bi-LSTM-CRF) was used to improve the identification of adverse drug events.82,83,90 Similarly, Gehrmann et al improved text-based phenotyping with a CNN designed to identify phrases relevant to substance abuse, depression, and other chronic conditions with the MIMIC-III phenotype data set.55
Semi-supervised learning
Despite its widespread use, supervised learning is difficult to scale due to the time and resources required to obtain gold-standard labeled data.100 Semi-supervised methods are trained with a large amount of unlabeled data (ie, unreviewed medical records) and a small amount of labeled data to minimize the burden of chart review.101 Three types of semi-supervised learning methods were used in 6 articles (Table 3). The first type performed feature selection using “silver-standard labels” that can be automatically extracted from patient records, such as the frequency of phenotype-specific diagnostic codes, prior to supervised training.102,103 For instance, PheCAP processed openly available knowledge sources such as Wikipedia articles to generate a candidate list of related UMLS concepts. An ensemble sparse regression approach using silver-standard labels was then used to identify relevant concepts for supervised learning. PheCAP was used to phenotype over 20 conditions using EHR data from 4 institutions.102,105 The second type of semi-supervised learning applied self-learning to train a generative model with labeled data to create pseudolabels for the unlabeled data set in order to train a traditional supervised model. Self-learning performed on par with supervised learning for 18 phenotypes.64,65 In contrast, the third type of semi-supervised learning directly incorporated unlabeled data into the algorithm through modification of the loss function.66,104 For example, a semi-supervised tensor factorization (PSST) approach used the information in unlabeled data to incorporate cannot link constraints into tensor factorization for the classification of hypertension and type-2 diabetes.66 PSST performed similarly to supervised tensor factorization, but with fewer labeled examples.
Table 3.
Semi-supervised methods used in the selected articles as well as the phenotypes considered and the size of the labeled and unlabeled data sets
| Method | Paper | Phenotype(s) | Unlabeled data set size | Labeled data set size |
|---|---|---|---|---|
| Silver-standard based feature selection | Cade et al102 | Sleep apnea | 15 741 | 300 |
| Cohen et al103 | Acute hepatic porphyria | 22 372 | 200 | |
| Self-learning | Estiri et al64 | Alzheimer's disease; atrial fibrillation; asthma; bipolar disorder; breast cancer; coronary artery disease; Crohn's disease; congestive heart failure; chronic obstructive pulmonary disease; epilepsy; gout; hypertension; rheumatoid arthritis; schizophrenia; stroke; type 1 diabetes mellitus; type 2 diabetes mellitus; ulcerative colitis | 5732 (Average) | 360 (Average) |
| Estiri et al65 | Alzheimer's disease; atrial fibrillation; coronary artery disease; congestive heart failure; chronic obstructive pulmonary disease; rheumatoid arthritis; stroke; type 1 diabetes mellitus; type 2 diabetes mellitus; ulcerative colitis | 6000 (Average) | 351 (Average) | |
| Modified loss function | Zhang et al104 | Aldosteronism | 6391 | 185 |
| Henderson et al66 | Resistant hypertension; type 2 diabetes mellitus | 1622 | 400 |
Weakly supervised learning
Analogous to semi-supervised learning, the goal of weakly supervised learning is to expedite the phenotyping process by eliminating the need for gold-standard labeled data. Weakly supervised methods rely on a silver-standard label that can be easily extracted from patients records in place of a gold-standard label.106 The silver-standard label is selected based on clinical expertise as a proxy for the phenotype.106–109 Common silver-standard labels included phenotype-specific diagnosis codes, lab results, and free-text mentions of the phenotype.74,75,110
Two types of weakly supervised learning approaches were used in 15 articles (Table 4). The first type assumed the silver-standard label follows a mixture model representing phenotype cases and controls.74,75,110–114 For example, PheNorm utilized Gaussian mixture models with denoising self-regression for phenotyping 4 chronic conditions.74 MAP later improved upon PheNorm with an ensemble of mixture models and was validated across 16 phenotypes and 2 phenome-wide association studies.40,75 PheVis extended the resolution of PheNorm from patient-level to visit-level by incorporating past medical history information into estimation.112 The second type of weakly supervised methods used silver standards to directly train supervised models.51,107,108,115–119 For instance, APHRODITE employs noisy label learning with an anchor feature with a near-perfect positive predictive value (PPV), but potentially imperfect sensitivity to train L1-penalized logistic regression models.115 APHRODITE is available in openly available R software for users of the OMOP common data model. Similar approaches have been used to identify phenotypes poorly documented in structured data such as systemic lupus erythematosus.51,116 In general, weakly supervised models exhibit similar or improved performance to their rule-based and supervised counterparts (Supplementary Figures S1 and S2).
Table 4.
Weakly supervised methods used in the selected articles, as well as the phenotypes considered and the silver-standard label used
| Method | Paper | Phenotype(s) | Silver-standard label(s) |
|||
|---|---|---|---|---|---|---|
| ICD code | SNOMED code | Relevant concept or word in free-text | Other | |||
| Mixture modeling | PheNorm74 | Rheumatoid arthritis; Crohn's disease; ulcerative colitis; coronary artery disease | ✓ | ✓ | ||
| PheProb111 | Rheumatoid arthritis | ✓ | ||||
| Multimodel Automated Phenotyping (MAP)75 | Asthma; Crohn's disease; ulcerative colitis; cardiomyopathy; congestive heart failure; epilepsy; juvenile rheumatoid arthritis; chronic pulmonary heart disease; type 1 diabetes mellitus; cardiovascular disease; inflammatory bowel disease | ✓ | ✓ | |||
| Geva et al40 | Asthma; bipolar disorder; Schizophrenia; breast cancer; chronic obstructive pulmonary disease; congestive heart failure; coronary artery disease; hypertension; depression; epilepsy; multiple sclerosis; rheumatoid arthritis; type 1 diabetes mellitus; type 2 diabetes mellitus; Crohn's disease; ulcerative colitis | ✓ | ✓ | |||
| PheMAP110 | Type 2 diabetes mellitus; dementia; hypothyroidism | ✓ | ||||
| PheVis112 | Rheumatoid arthritis; tuberculosis | ✓ | ✓ | |||
| Surrogate-guided ensemble latent Dirichlet allocation (sureLDA)113 | Asthma; breast cancer; chronic obstructive pulmonary disease; depression; epilepsy; hypertension; schizophrenia; stroke; type 1 diabetes mellitus; obesity | Phenotype probabilities derived from PheNorm | ||||
| Ning et al114 | Coronary artery disease; rheumatoid arthritis; Crohn's disease; ulcerative colitis; pulmonary hypertension | ✓ | ✓ | |||
| Noisy labeling | Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation (APHRODITE)115 | Appendicitis; type 2 diabetes mellitus; cataracts; heart failure; abdominal aortic aneurysm; epilepsy; peripheral arterial disease; obesity; glaucoma; venous thromboembolism | ✓ | |||
| Murray et al116 | Systemic lupus erythematosus | ✓ | ||||
| Ling et al38 | Metastatic breast cancer | ✓ | ||||
| Banerjee et al117 | Urinary incontinence; Bowel dysfunction | ✓ | ||||
| NimbleMiner118 | Fall | ✓ | ||||
| Annapragada et al51 | Child physical abuse | ✓ | ||||
| Sanyal et al119 | Insulin pump failure | ✓ | ||||
Unsupervised learning
In contrast to the previously discussed ML approaches, unsupervised learning is used for phenotype discovery, including identification of subphenotypes,39,76,120–128 co-occurring conditions,69,129 and disease progression patterns.68,130–134 Among the 19 articles utilizing unsupervised learning, Latent Dirichlet Allocation (LDA)69,124,125,127,133 and K-means were the most frequently used methods.120,121,123,125 LDA was applied to identify the co-occurrence of allergic rhinitis and osteoporosis among patients with kidney disease69 as well as to capture trends in mental health and end-of-life care among dementia patients.133 K-means was used to discover subphenotypes such as patients with different symptoms of acute kidney injury.120 Model-derived subpopulations were commonly used in downstream prediction tasks.39,68,121,122,125,131 For example, a SVM was used to identify sepsis using features of subpopulations with distinct dysfunction patterns discovered from a self-organizing map.128 Only 1 article utilized a deep learning approach, specifically a deep autoencoder to discover subtypes of depression.132
Reporting and evaluation methods
As the articles primarily focused on identifying disease cases (excluding unsupervised learning articles), most evaluated algorithm performance with PPV, sensitivity, and/or F-score (70/81 articles reported at least 1 of these metrics; Supplementary Table S12). The area under the ROC curve (AUROC) was also reported as an overall summary of discriminative performance (42/81 articles), while calibration was rarely assessed (7/81 articles). Additionally, several studies linked EHR data to administrative claims30–36 or registry databases37–40 to validate algorithm accuracy. Biorepositories were also used to demonstrate the validity of a derived phenotype in replicating a genetic association study.75,110,111,135 Only 5 studies performed external validation or evaluated algorithmic fairness.36,40,52,61,136 We also found limited reporting of the data descriptors necessary to assess the feasibility of implementing an algorithm in a new setting. Patient demographics were only reported in 38 of 71 papers using private data sources and only 20 articles released their analytic code. A majority of these articles used complex deep-learning models (9 articles) and/or free-text data (9 articles).
With respect to performance comparisons, 21 articles compared an ML approach to a rule-based method (Supplementary Table S13). Traditional ML was used in 10 of these articles and outperformed rule-based algorithms in 8 articles with respect to PPV, sensitivity, or both (Supplementary Figure S3). Two supervised deep learning models were compared to rules, with a Bi-LSTM performing similarly to a rule-based approach for substance abuse54 and a bidirectional gated recurrent unit model significantly decreasing performance in identifying insulin rejection.137 Twenty articles also provided comparisons between deep learning and traditional baselines (Supplementary Table S14). Deep learning outperformed traditional ML across all reported accuracy metrics for 18 of 33 phenotypes considered (Supplementary Figure S4(a)). Deep learning improved sensitivity with a corresponding decrease in PPV or vice-versa (Supplementary Figure S4(b, c)) for the remaining phenotypes, with the exception of 1 study demonstrating that elastic net logistic regression outperformed an RNN for phenotyping fall risk (Supplementary Figure S4(d)).61 It is important to note that a meaningful gain in accuracy must be interpreted in the context of the use case of the algorithm and the target metric of performance. Moreover, improvements in accuracy must be weighed against additional challenges brought on by deep learning, including data demands, decreased interpretability, and limited generalizability over time and across healthcare settings.72,138–140
DISCUSSION
This review highlights the substantial ongoing work in ML-based phenotyping. A broad range of phenotypes have been considered and the use of unstructured information in clinical notes is widespread. While ML approaches did not uniformly outperform rule-based methods, deep learning provided marginal improvement over traditional baselines. Moreover, semi-supervised and weakly supervised learning have expedited the phenotyping process while unsupervised learning has been effective for phenotype discovery. Progress withstanding, most articles focused on binary phenotypes and few studies evaluated external validity or used multi-institution data. Study settings were infrequently reported and analytic code was rarely released. Future work is warranted in “deep phenotyping,” reporting and evaluation standards, and methods to accommodate misclassified phenotypes due to algorithm errors in downstream applications.
Deep phenotyping
“Deep phenotyping” moves beyond binary identification to the characterization of nuanced phenotypes, such as the timing or severity of a condition, using advanced methods leveraging interoperable and multimodal data types.20,122,141,142 From a methodological viewpoint, studies of nuanced phenotypes will face similar, but more substantial challenges in obtaining gold-standard labeled data. Further work in semi- and weakly supervised deep learning methods is necessary.143,144 Moreover, given the privacy constraints associated with EHRs and other health data sources, leveraging interoperable and multimodal data calls for advancements in federated learning methods that can accommodate distributed data sources stored locally across institutions.145
Reporting and evaluation standards
Research networks, such as eMERGE, have long advocated for transparent and reusable phenotype definitions. Most recently, in response to the wave of COVID-19 studies, Kohane et al146 proposed a checklist for evaluating the quality of EHR-based studies, emphasizing phenotypic transparency as a key concern. However, we found most articles did not release the necessary details for a complete evaluation of an approach or implementation in other settings. As a step towards reporting standards that increase transparency and reproducibility, OHDSI proposed Findable, Accessible, Interoperable, and Reusable (FAIR) phenotype definitions based on APHRODITE. All of the necessary tooling, data models, software and vocabularies are publicly available and released with open-source licenses.147 As noted in Kashyap et al in their evaluation of the APHRODITE framework, effective reporting of phenotyping models should include a detailed recipe for data preparation and model training, rather than the pretrained models themselves, given substantial differences in EHR data across institutions.115
Additionally, we observed a lack of rigorous evaluation of phenotyping algorithms, with most studies using standard metrics to evaluate internal validity. We stress further model interrogation for phenotyping, including external validation as well as evaluation of fairness. However, reliable performance evaluation requires a substantial amount of gold-standard labeled data. There is very little work on semi-supervised and weakly supervised methods for evaluating model performance and further research is warranted.148–150
Accounting for misclassified phenotypes due to algorithm errors
As ML phenotyping expands the scope of EHR research, care must be taken when using derived phenotypes for downstream tasks as they are inevitably misclassified due to algorithm errors. In the context of association studies, it is well known in the statistical community that misclassification can lead to diminished statistical power and biased estimation.151–153 However, statistical methods are often siloed from the informatics community. We advocate for the dissemination of existing methods and for methodological developments in “post-phenotyping” inferential and predictive modeling studies.
Limitations
As the definition of phenotyping is variable within the literature,12 we used a broad search capturing articles focusing on ML or NLP or phenotyping using EHRs. Following prior work, we limited our scope to select informatics venues.12,15 Although we have missed articles outside of these journals, our aim is to rigorously characterize the general landscape of ML-based phenotyping, which we believe is captured in the venues considered and in our detailed analyses.
CONCLUSION
This review summarizes the landscape of ML-based phenotyping between 2018 and 2022. Current literature has laid the groundwork for “deep phenotyping,” but developing standards and methodology for the reliable use of a diverse range of phenotypes derived from ML models is necessary for continued EHR-based research.
FUNDING
The project described was supported by an NSERC Discovery Grant (RGPIN-2021-03734) and a Connaught New Researcher Award.
AUTHOR CONTRIBUTIONS
JG conceived and designed the study. SY performed the full-text review. JG and SY analyzed and interpreted the data. JG, PV, and SY drafted and revised the manuscript. JG, PV, SY, ES, and KT approved the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Prof. Lei Sun for her useful comments.
CONFLICT OF INTEREST STATEMENT
JG received scientific consulting fees from Alphabet’s Verily Life Sciences.
Contributor Information
Siyue Yang, Department of Statistical Sciences, University of Toronto, Toronto, Ontario, Canada.
Paul Varghese, Verily Life Sciences, Cambridge, Massachusetts, USA.
Ellen Stephenson, Department of Family & Community Medicine, University of Toronto, Toronto, Ontario, Canada.
Karen Tu, Department of Family & Community Medicine, University of Toronto, Toronto, Ontario, Canada.
Jessica Gronsbell, Department of Statistical Sciences, University of Toronto, Toronto, Ontario, Canada; Department of Family & Community Medicine, University of Toronto, Toronto, Ontario, Canada; Department of Computer Science, University of Toronto, Toronto, Ontario, Canada.
DATA AND CODE AVAILABILITY
The underlying data and R code to replicate our analyses can be found at: https://github.com/jlgrons/ML-EHR-Phenotyping-Review.
REFERENCES
- 1. Institute of Medicine, Roundtable on Value and Science-Driven Health Care. Clinical Data as the Basic Staple of Health Learning: Creating and Protecting a Public Good: Workshop Summary. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 2. Mc Cord KA, Hemkens LG.. Using electronic health records for clinical trials: where do we stand and where can we go? CMAJ 2019; 191 (5): E128–E133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li R, Chen Y, Ritchie MD, et al. Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet 2020; 21 (8): 493–502. [DOI] [PubMed] [Google Scholar]
- 4. Beesley LJ, Salvatore M, Fritsche LG, et al. The emerging landscape of health research based on biobanks linked to electronic health records: existing resources, statistical challenges, and potential opportunities. Stat Med 2020; 39 (6): 773–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu R, Rizzo S, Whipple S, et al. Evaluating eligibility criteria of oncology trials using real-world data and AI. Nature 2021; 592 (7855): 629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geva A, Abman SH, Manzi SF, et al. Adverse drug event rates in pediatric pulmonary hypertension: a comparison of real-world data sources. J Am Med Inform Assoc 2020; 27 (2): 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogers JR, Lee J, Zhou Z, et al. Contemporary use of real-world data for clinical trial conduct in the United States: a scoping review. J Am Med Inform Assoc 2021; 28 (1): 144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boland MR, Hripcsak G, Shen Y, et al. Defining a comprehensive verotype using electronic health records for personalized medicine. J Am Med Inform Assoc 2013; 20 (e2): e232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015; 350: h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei W-Q, Denny JC.. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med 2015; 7 (1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pendergrass SA, Crawford DC.. Using electronic health records to generate phenotypes for research. Curr Protoc Hum Genet 2019; 100 (1): e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014; 21 (2): 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denaxas S, Gonzalez-Izquierdo A, Direk K, et al. UK phenomics platform for developing and validating EHR phenotypes: CALIBER. J Am Med Inform Assoc 2019; 26 (12): 1545–59. [DOI] [PMC free article] [PubMed]
- 14. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013; 20 (e1): e147–54–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banda JM, Seneviratne M, Hernandez-Boussard T, et al. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alzoubi H, Alzubi R, Ramzan N, et al. A review of automatic phenotyping approaches using electronic health records. Electronics 2019; 8 (11): 1235. [Google Scholar]
- 17. Robinson JR, Wei W-Q, Roden DM, et al. Defining phenotypes from clinical data to drive genomic research. Annu Rev Biomed Data Sci 2018; 1: 69–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hripcsak G, Albers DJ.. High-fidelity phenotyping: richness and freedom from bias. J Am Med Inform Assoc 2018; 25 (3): 289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng Z, Deng Y, Li X, et al. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans Comput Biol Bioinform 2019; 16 (1): 139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weng C, Shah NH, Hripcsak G.. Deep phenotyping: embracing complexity and temporality-towards scalability, portability, and interoperability. J Biomed Inform 2020; 105: 103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leslie D, Mazumder A, Peppin A, et al. Does ‘AI’ stand for augmenting inequality in the era of covid-19 healthcare? BMJ 2021; 372: n304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bishop CM, Nasrabadi NM. Pattern Recognition and Machine Learning. Vol. 4. No. 4. New York: springer; 2006. [Google Scholar]
- 23. Zhou Z-H. A brief introduction to weakly supervised learning. Natl Sci Rev 2018; 5 (1): 44–53. [Google Scholar]
- 24. Wu S, Roberts K, Datta S, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc 2020; 27 (3): 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irwin AN, Rackham D.. Comparison of the time-to-indexing in PubMed between biomedical journals according to impact factor, discipline, and focus. Res Social Adm Pharm 2017; 13 (2): 389–93. [DOI] [PubMed] [Google Scholar]
- 26. McBrien KA, Souri S, Symonds NE, et al. Identification of validated case definitions for medical conditions used in primary care electronic medical record databases: a systematic review. J Am Med Inform Assoc 2018; 25 (11): 1567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ford E, Carroll JA, Smith HE, et al. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 2016; 23 (5): 1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 29. McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erickson J, Abbott K, Susienka L.. Automatic address validation and health record review to identify homeless Social Security disability applicants. J Biomed Inform 2018; 82: 41–6. [DOI] [PubMed] [Google Scholar]
- 31. Fialoke S, Malarstig A, Miller MR, et al. Application of machine learning methods to predict non-alcoholic steatohepatitis (NASH) in non-alcoholic fatty liver (NAFL) patients. AMIA Annu Symp Proc 2018; 2018: 430–9. [PMC free article] [PubMed] [Google Scholar]
- 32. Prenovost KM, Fihn SD, Maciejewski ML, et al. Using item response theory with health system data to identify latent groups of patients with multiple health conditions. PLoS One 2018; 13 (11): e0206915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choudhury O, Park Y, Salonidis T, et al. Predicting adverse drug reactions on distributed health data using federated learning. AMIA Annu Symp Proc 2019; 2019: 313–22. [PMC free article] [PubMed] [Google Scholar]
- 34. Nori VS, Hane CA, Sun Y, et al. Deep neural network models for identifying incident dementia using claims and EHR datasets. PLoS One 2020; 15 (9): e0236400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gibson TB, Nguyen MD, Burrell T, et al. Electronic phenotyping of health outcomes of interest using a linked claims-electronic health record database: findings from a machine learning pilot project. J Am Med Inform Assoc 2021; 28 (7): 1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mahesri M, Chin K, Kumar A, et al. External validation of a claims-based model to predict left ventricular ejection fraction class in patients with heart failure. PLoS One 2021; 16 (6): e0252903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seneviratne MG, Banda JM, Brooks JD, et al. Identifying cases of metastatic prostate cancer using machine learning on electronic health records. AMIA Annu Symp Proc 2018; 2018: 1498–504. [PMC free article] [PubMed] [Google Scholar]
- 38. Ling AY, Kurian AW, Caswell-Jin JL, et al. Using natural language processing to construct a metastatic breast cancer cohort from linked cancer registry and electronic medical records data. JAMIA Open 2019; 2 (4): 528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lyudovyk O, Shen Y, Tatonetti NP, et al. Pathway analysis of genomic pathology tests for prognostic cancer subtyping. J Biomed Inform 2019; 98: 103286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Geva A, Liu M, Panickan VA, et al. A high-throughput phenotyping algorithm is portable from adult to pediatric populations. J Am Med Inform Assoc 2021; 28 (6): 1265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henry S, Buchan K, Filannino M, et al. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc 2020; 27 (1): 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stubbs A, Filannino M, Soysal E, et al. Cohort selection for clinical trials: n2c2 2018 shared task track 1. J Am Med Inform Assoc 2019; 26 (11): 1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harutyunyan H, Khachatrian H, Kale DC, et al. Multitask learning and benchmarking with clinical time series data. Sci Data 2019; 6 (1): 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buckland RS, Hogan JW, Chen ES.. Selection of clinical text features for classifying suicide attempts. AMIA Annu Symp Proc 2020; 2020: 273–82. [PMC free article] [PubMed] [Google Scholar]
- 45. Carson NJ, Mullin B, Sanchez MJ, et al. Identification of suicidal behavior among psychiatrically hospitalized adolescents using natural language processing and machine learning of electronic health records. PLoS One 2019; 14 (2): e0211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Afshar M, Phillips A, Karnik N, et al. Natural language processing and machine learning to identify alcohol misuse from the electronic health record in trauma patients: development and internal validation. J Am Med Inform Assoc 2019; 26 (3): 254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. To D, Joyce C, Kulshrestha S, et al. The addition of United States census-tract data does not improve the prediction of substance misuse. AMIA Annu Symp Proc 2021; 2021: 1149–58. [PMC free article] [PubMed] [Google Scholar]
- 48. Badger J, LaRose E, Mayer J, et al. Machine learning for phenotyping opioid overdose events. J Biomed Inform 2019; 94: 103185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feller DJ, Zucker J, Don't Walk OB, et al. Towards the inference of social and behavioral determinants of sexual health: development of a gold-standard corpus with semi-supervised learning. AMIA Annu Symp Proc 2018; 2018: 422–9. [PMC free article] [PubMed] [Google Scholar]
- 50. Han S, Zhang RF, Shi L, et al. Classifying social determinants of health from unstructured electronic health records using deep learning-based natural language processing. J Biomed Inform 2022; 127: 103984. [DOI] [PubMed] [Google Scholar]
- 51. Annapragada AV, Donaruma-Kwoh MM, Annapragada AV, et al. A natural language processing and deep learning approach to identify child abuse from pediatric electronic medical records. PLoS One 2021; 16 (2): e0247404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thompson HM, Sharma B, Bhalla S, et al. Bias and fairness assessment of a natural language processing opioid misuse classifier: detection and mitigation of electronic health record data disadvantages across racial subgroups. J Am Med Inform Assoc 2021; 28 (11): 2393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lybarger K, Yetisgen M, Ostendorf M.. Using neural multi-task learning to extract substance abuse information from clinical notes. AMIA Annu Symp Proc 2018; 2018: 1395–404. [PMC free article] [PubMed] [Google Scholar]
- 54. Ni Y, Bachtel A, Nause K, et al. Automated detection of substance use information from electronic health records for a pediatric population. J Am Med Inform Assoc 2021; 28 (10): 2116–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gehrmann S, Dernoncourt F, Li Y, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One 2018; 13 (2): e0192360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stemerman R, Arguello J, Brice J, et al. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 2021; 4 (3): ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu Z, Yang X, Dang C, et al. A study of social and behavioral determinants of health in lung cancer patients using transformers-based natural language processing models. AMIA Annu Symp Proc 2021; 2021: 1225–33. [PMC free article] [PubMed] [Google Scholar]
- 58. Mitra A, Rawat BPS, McManus D, et al. Bleeding entity recognition in electronic health records: a comprehensive analysis of end-to-end systems. AMIA Annu Symp Proc 2020; 2020: 860–9. [PMC free article] [PubMed] [Google Scholar]
- 59. Chen T, Dredze M, Weiner JP, et al. Identifying vulnerable older adult populations by contextualizing geriatric syndrome information in clinical notes of electronic health records. J Am Med Inform Assoc 2019; 26 (8-9): 787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gao J, Xiao C, Glass LM, et al. Dr. Agent: clinical predictive model via mimicked second opinions. J Am Med Inform Assoc 2020; 27 (7): 1084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin JA, Crane-Droesch A, Lapite FC, et al. Development and validation of a prediction model for actionable aspects of frailty in the text of clinicians’ encounter notes. J Am Med Inform Assoc 2021; 29 (1): 109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Obeid JS, Davis M, Turner M, et al. An artificial intelligence approach to COVID-19 infection risk assessment in virtual visits: a case report. J Am Med Inform Assoc 2020; 27 (8): 1321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lybarger K, Ostendorf M, Thompson M, et al. Extracting COVID-19 diagnoses and symptoms from clinical text: a new annotated corpus and neural event extraction framework. J Biomed Inform 2021; 117: 103761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Estiri H, Vasey S, Murphy SN.. Generative transfer learning for measuring plausibility of EHR diagnosis records. J Am Med Inform Assoc 2021; 28 (3): 559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Estiri H, Strasser ZH, Murphy SN.. High-throughput phenotyping with temporal sequences. J Am Med Inform Assoc 2021; 28 (4): 772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Henderson J, He H, Malin BA, et al. Phenotyping through semi-supervised tensor factorization (PSST). AMIA Annu Symp Proc 2018; 2018: 564–73. [PMC free article] [PubMed] [Google Scholar]
- 67. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou F, Gillespie A, Gligorijevic D, et al. Use of disease embedding technique to predict the risk of progression to end-stage renal disease. J Biomed Inform 2020; 105: 103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bhattacharya M, Jurkovitz C, Shatkay H.. Co-occurrence of medical conditions: Exposing patterns through probabilistic topic modeling of snomed codes. J Biomed Inform 2018; 82: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 22 (5): 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ghassemi M, Naumann T, Schulam P, et al. A review of challenges and opportunities in machine learning for health. AMIA Jt Summits Transl Sci Proc 2020; 2020: 191–200. [PMC free article] [PubMed] [Google Scholar]
- 73. Lu S, Chen R, Wei W, et al. Understanding heart failure patients EHR clinical features via SHAP interpretation of tree-based machine learning model predictions. AMIA Annu Symp Proc 2021; 2021: 813–22. [PMC free article] [PubMed] [Google Scholar]
- 74. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc 2018; 25 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26 (11): 1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ni Y, Alwell K, Moomaw CJ, et al. Towards phenotyping stroke: leveraging data from a large-scale epidemiological study to detect stroke diagnosis. PLoS One 2018; 13 (2): e0192586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shi J, Liu S, Pruitt LCC, et al. Using natural language processing to improve EHR structured data-based surgical site infection surveillance. AMIA Annu Symp Proc 2019; 2019: 794–803. [PMC free article] [PubMed] [Google Scholar]
- 78. Yan LC, Yoshua B, Geoffrey H.. Deep learning. Nature 2015; 521: 436–44. [DOI] [PubMed] [Google Scholar]
- 79. Khalid S, Khalil T, Nasreen S. A survey of feature selection and feature extraction techniques in machine learning. In: 2014 Science and information conference. 2014; Doha, Qatar. doi: 10.1109/sai.2014.6918213 [DOI]
- 80. Khattak FK, Jeblee S, Pou-Prom C, et al. A survey of word embeddings for clinical text. J Biomed Inform X 2019; 100: 100057. [DOI] [PubMed] [Google Scholar]
- 81. Teller V. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. 2000. https://direct.mit.edu/coli/article-abstract/26/4/638/1680. Accessed April 14, 2022.
- 82. Wei Q, Ji Z, Li Z, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc 2020; 27 (1): 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ju M, Nguyen NTH, Miwa M, et al. An ensemble of neural models for nested adverse drug events and medication extraction with subwords. J Am Med Inform Assoc 2020; 27 (1): 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xiong Y, Shi X, Chen S, et al. Cohort selection for clinical trials using hierarchical neural network. J Am Med Inform Assoc 2019; 26 (11): 1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen L, Gu Y, Ji X, et al. Extracting medications and associated adverse drug events using a natural language processing system combining knowledge base and deep learning. J Am Med Inform Assoc 2020; 27 (1): 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang X, Bian J, Fang R, et al. Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. J Am Med Inform Assoc 2020; 27 (1): 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xie K, Gallagher RS, Conrad EC, et al. Extracting seizure frequency from epilepsy clinic notes: a machine reading approach to natural language processing. J Am Med Inform Assoc 2022; 29 (5): 873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Soni S, Roberts K.. Patient cohort retrieval using transformer language models. AMIA Annu Symp Proc 2020; 2020: 1150–9. [PMC free article] [PubMed] [Google Scholar]
- 89. Kim Y, Meystre SM.. Ensemble method-based extraction of medication and related information from clinical texts. J Am Med Inform Assoc 2020; 27 (1): 31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dai H-J, Su C-H, Wu C-S.. Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. J Am Med Inform Assoc 2020; 27 (1): 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou S, Wang N, Wang L, Liu H, Zhang R. CancerBERT: a cancer domain-specific language model for extracting breast cancer phenotypes from electronic health records. J Am Med Inform Assoc 2022; 29 (7): 1208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eisman AS, Shah NR, Eickhoff C, et al. Extracting angina symptoms from clinical notes using pre-trained transformer architectures. AMIA Annu Symp Proc 2020; 2020: 412–21. [PMC free article] [PubMed] [Google Scholar]
- 93. Mikolov T, Sutskever I, Chen K, et al. Distributed representations of words and phrases and their compositionality. In: Burges CJC, Bottou L, Welling M, et al. , eds. Advances in Neural Information Processing Systems. Curran Associates, Inc.; 2013. https://proceedings.neurips.cc/paper/2013/file/9aa42b31882ec039965f3c4923ce901b-Paper.pdf [Google Scholar]
- 94. Pennington J, Socher R, Manning CD. Glove: global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP). 2014: 1532–43; Doha, Qatar.
- 95. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of NAACL-HLT; June 2018: 4171–86; Minneapolis, MN.
- 96. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 36 (4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. arXiv [cs.CL]. 2019. http://arxiv.org/abs/1904.03323.
- 98. Liu Z, Lin W, Shi Y, Zhao J. A robustly optimized BERT pre-training approach with post-training. In: China National Conference on Chinese Computational Linguistics; Cham: Springer; August 2021: 471–84. doi: 10.1007/978-3-030-84186-7_31. [DOI] [Google Scholar]
- 99. Ogunyemi OI, Gandhi M, Lee M, et al. Detecting diabetic retinopathy through machine learning on electronic health record data from an urban, safety net healthcare system. JAMIA Open 2021; 4 (3): ooab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cai T, Cai F, Dahal KP, et al. Improving the efficiency of clinical trial recruitment using an ensemble machine learning to assist with eligibility screening. ACR Open Rheumatol 2021; 3 (9): 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhu X. Semi-supervised learning literature survey. Published Online First: 2008. https://minds.wisconsin.edu/handle/1793/60444. Accessed April 19, 2022.
- 102. Cade BE, Hassan SM, Dashti HS, et al. Sleep apnea phenotyping and relationship to disease in a large clinical biobank. JAMIA Open 2022; 5 (1): ooab117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cohen AM, Chamberlin S, Deloughery T, et al. Detecting rare diseases in electronic health records using machine learning and knowledge engineering: case study of acute hepatic porphyria. PLoS One 2020; 15 (7): e0235574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang L, Ding X, Ma Y, et al. A maximum likelihood approach to electronic health record phenotyping using positive and unlabeled patients. J Am Med Inform Assoc 2020; 27 (1): 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017; 24 (e1): e143–9–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Halpern Y, Horng S, Choi Y, et al. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 23 (4): 731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 23 (6): 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Banda JM, Halpern Y, Sontag D, et al. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 110. Zheng NS, Feng Q, Kerchberger VE, et al. PheMap: a multi-resource knowledge base for high-throughput phenotyping within electronic health records. J Am Med Inform Assoc 2020; 27 (11): 1675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sinnott JA, Cai F, Yu S, et al. PheProb: probabilistic phenotyping using diagnosis codes to improve power for genetic association studies. J Am Med Inform Assoc 2018; 25 (10): 1359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ferté T, Cossin S, Schaeverbeke T, et al. Automatic phenotyping of electronical health record: PheVis algorithm. J Biomed Inform 2021; 117: 103746. [DOI] [PubMed] [Google Scholar]
- 113. Ahuja Y, Zhou D, He Z, et al. sureLDA: a multidisease automated phenotyping method for the electronic health record. J Am Med Inform Assoc 2020; 27 (8): 1235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ning W, Chan S, Beam A, et al. Feature extraction for phenotyping from semantic and knowledge resources. J Biomed Inform 2019; 91: 103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kashyap M, Seneviratne M, Banda JM, et al. Development and validation of phenotype classifiers across multiple sites in the observational health data sciences and informatics network. J Am Med Inform Assoc 2020; 27 (6): 877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Murray SG, Avati A, Schmajuk G, et al. Automated and flexible identification of complex disease: building a model for systemic lupus erythematosus using noisy labeling. J Am Med Inform Assoc 2019; 26 (1): 61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Banerjee I, Li K, Seneviratne M, et al. Weakly supervised natural language processing for assessing patient-centered outcome following prostate cancer treatment. JAMIA Open 2019; 2 (1): 150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Topaz M, Murga L, Gaddis KM, et al. Mining fall-related information in clinical notes: comparison of rule-based and novel word embedding-based machine learning approaches. J Biomed Inform 2019; 90: 103103. [DOI] [PubMed] [Google Scholar]
- 119. Sanyal J, Rubin D, Banerjee I.. A weakly supervised model for the automated detection of adverse events using clinical notes. J Biomed Inform 2022; 126: 103969. [DOI] [PubMed] [Google Scholar]
- 120. Xu Z, Chou J, Zhang XS, et al. Identifying sub-phenotypes of acute kidney injury using structured and unstructured electronic health record data with memory networks. J Biomed Inform 2020; 102: 103361. [DOI] [PubMed] [Google Scholar]
- 121. Apostolova E, Uppal A, Galarraga JE, et al. Towards reliable ARDS clinical decision support: ARDS patient analytics with free-text and structured EMR data. AMIA Annu Symp Proc 2019; 2019: 228–37. [PMC free article] [PubMed] [Google Scholar]
- 122. Zhao J, Zhang Y, Schlueter DJ, et al. Detecting time-evolving phenotypic topics via tensor factorization on electronic health records: cardiovascular disease case study. J Biomed Inform 2019; 98: 103270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mullin S, Zola J, Lee R, et al. Longitudinal K-means approaches to clustering and analyzing EHR opioid use trajectories for clinical subtypes. J Biomed Inform 2021; 122: 103889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Afshar M, Joyce C, Dligach D, et al. Subtypes in patients with opioid misuse: a prognostic enrichment strategy using electronic health record data in hospitalized patients. PLoS One 2019; 14 (7): e0219717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang Y, Zhao Y, Therneau TM, et al. Unsupervised machine learning for the discovery of latent disease clusters and patient subgroups using electronic health records. J Biomed Inform 2020; 102: 103364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Maurits MP, Korsunsky I, Raychaudhuri S, et al. A framework for employing longitudinally collected multicenter electronic health records to stratify heterogeneous patient populations on disease history. J Am Med Inform Assoc 2022; 29 (5): 761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Liu Q, Woo M, Zou X, et al. Symptom-based patient stratification in mental illness using clinical notes. J Biomed Inform 2019; 98: 103274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ibrahim ZM, Wu H, Hamoud A, et al. On classifying sepsis heterogeneity in the ICU: insight using machine learning. J Am Med Inform Assoc 2020; 27 (3): 437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shen F, Peng S, Fan Y, et al. HPO2Vec+: leveraging heterogeneous knowledge resources to enrich node embeddings for the human phenotype ontology. J Biomed Inform 2019; 96: 103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hubbard RA, Xu J, Siegel R, et al. Studying pediatric health outcomes with electronic health records using Bayesian clustering and trajectory analysis. J Biomed Inform 2021; 113: 103654. [DOI] [PubMed] [Google Scholar]
- 131. Ben-Assuli O, Jacobi A, Goldman O, et al. Stratifying individuals into non-alcoholic fatty liver disease risk levels using time series machine learning models. J Biomed Inform 2022; 126: 103986. [DOI] [PubMed] [Google Scholar]
- 132. Gong J, Simon GE, Liu S.. Machine learning discovery of longitudinal patterns of depression and suicidal ideation. PLoS One 2019; 14 (9): e0222665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wang L, Lakin J, Riley C, et al. Disease trajectories and end-of-life care for dementias: latent topic modeling and trend analysis using clinical notes. AMIA Annu Symp Proc 2018; 2018: 1056–65. [PMC free article] [PubMed] [Google Scholar]
- 134. Meaney C, Escobar M, Moineddin R, et al. Non-negative matrix factorization temporal topic models and clinical text data identify COVID-19 pandemic effects on primary healthcare and community health in Toronto, Canada. J Biomed Inform 2022; 128: 104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li R, Chen Y, Moore JH.. Integration of genetic and clinical information to improve imputation of data missing from electronic health records. J Am Med Inform Assoc 2019; 26 (10): 1056–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Klann JG, Estiri H, Weber GM, et al. ; Consortium for Clinical Characterization of COVID-19 by EHR (4CE) (CONSORTIA AUTHOR). Validation of an internationally derived patient severity phenotype to support COVID-19 analytics from electronic health record data. J Am Med Inform Assoc 2021; 28 (7): 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Malmasi S, Ge W, Hosomura N, et al. Comparing information extraction techniques for low-prevalence concepts: the case of insulin rejection by patients. J Biomed Inform 2019; 99: 103306. [DOI] [PubMed] [Google Scholar]
- 138. Ghassemi M, Oakden-Rayner L, Beam AL.. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health 2021; 3 (11): e745–50–e750. [DOI] [PubMed] [Google Scholar]
- 139. Rajpurkar P, Chen E, Banerjee O, et al. AI in health and medicine. Nat Med 2022; 28 (1): 31–8. [DOI] [PubMed] [Google Scholar]
- 140. Nestor B, McDermott MBA, Boag W, et al. Feature robustness in non-stationary health records: caveats to deployable model performance in common clinical machine learning tasks. In: Doshi-Velez F, Fackler J, Jung K, et al. , eds. Proceedings of the 4th machine learning for healthcare conference, PMLR 09–10. 2019: 381–405. [Google Scholar]
- 141. Mate S, Bürkle T, Kapsner LA, et al. A method for the graphical modeling of relative temporal constraints. J Biomed Inform 2019; 100: 103314. [DOI] [PubMed] [Google Scholar]
- 142. Meng W, Ou W, Chandwani S, et al. Temporal phenotyping by mining healthcare data to derive lines of therapy for cancer. J Biomed Inform 2019; 100: 103335. [DOI] [PubMed] [Google Scholar]
- 143. Liang L, Hou J, Uno H, Cho K, Ma Y, Cai T. Semi-supervised approach to event time annotation using longitudinal electronic health records. Lifetime Data Anal 2022; 28 (3): 428–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ahuja Y, Wen J, Hong C, et al. SAMGEP: a novel method for prediction of phenotype event times using the electronic health record. Research Square. 2021. https://www.researchsquare.com/article/rs-1119858/latest.pdf. Accessed April 14, 2022. [DOI] [PMC free article] [PubMed]
- 145. Tong J, Luo C, Islam MN, et al. Distributed learning for heterogeneous clinical data with application to integrating COVID-19 data across 230 sites. NPJ Digit Med 2022; 5 (1): 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kohane IS, Aronow BJ, Avillach P, et al. ; Consortium For Clinical Characterization Of COVID-19 By EHR (4CE). What every reader should know about studies using electronic health record data but may be afraid to ask. J Med Internet Res 2021; 23 (3): e22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Weaver J, Potvien A, Swerdel J, et al. Best practices for creating the standardized content of an entry in the OHDSI Phenotype Library. In: 5th OHDSI annual symposium. 2019. https://www.ohdsi.org/wp-content/uploads/2019/09/james-weaver_a_book_in_the_phenotype_library_2019symposium.pdf. Accessed April 14, 2022.
- 148. Swerdel JN, Hripcsak G, Ryan PB.. PheValuator: development and evaluation of a phenotype algorithm evaluator. J Biomed Inform 2019; 97: 103258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Gronsbell JL, Cai T.. Semi-supervised approaches to efficient evaluation of model prediction performance. J R Stat Soc B 2018; 80 (3): 579–94. [Google Scholar]
- 150. Gronsbell J, Liu M, Tian L, et al. Efficient evaluation of prediction rules in semi‐supervised settings under stratified sampling. J R Stat Soc B 2022; 84 (4): 1353–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Manuel DG, Rosella LC, Stukel TA.. Importance of accurately identifying disease in studies using electronic health records. BMJ 2010; 341: c4226. [DOI] [PubMed] [Google Scholar]
- 152. Sinnott JA, Dai W, Liao KP, et al. Improving the power of genetic association tests with imperfect phenotype derived from electronic medical records. Hum Genet 2014; 133 (11): 1369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hubbard RA, Tong J, Duan R, et al. Reducing bias due to outcome misclassification for epidemiologic studies using EHR-derived probabilistic phenotypes. Epidemiology 2020; 31 (4): 542–50. [DOI] [PubMed] [Google Scholar]
- 154. Koola JD, Davis SE, Al-Nimri O, et al. Development of an automated phenotyping algorithm for hepatorenal syndrome. J Biomed Inform 2018; 80: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Afshar M, Joyce C, Oakey A, et al. A computable phenotype for acute respiratory distress syndrome using natural language processing and machine learning. AMIA Annu Symp Proc 2018; 2018: 157–65. [PMC free article] [PubMed] [Google Scholar]
- 156. Hong N, Wen A, Stone DJ, et al. Developing a FHIR-based EHR phenotyping framework: a case study for identification of patients with obesity and multiple comorbidities from discharge summaries. J Biomed Inform 2019; 99: 103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bucher BT, Shi J, Pettit RJ, et al. Determination of marital status of patients from structured and unstructured electronic healthcare data. AMIA Annu Symp Proc 2019; 2019: 267–74. [PMC free article] [PubMed] [Google Scholar]
- 158. Dai H-J, Wang F-D, Chen C-W, et al. Cohort selection for clinical trials using multiple instance learning. J Biomed Inform 2020; 107: 103438. [DOI] [PubMed] [Google Scholar]
- 159. Hassanzadeh H, Karimi S, Nguyen A.. Matching patients to clinical trials using semantically enriched document representation. J Biomed Inform 2020; 105: 103406. [DOI] [PubMed] [Google Scholar]
- 160. Kulshrestha S, Dligach D, Joyce C, et al. Comparison and interpretability of machine learning models to predict severity of chest injury. JAMIA Open 2021; 4 (1): ooab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chu J, Dong W, He K, et al. Using neural attention networks to detect adverse medical events from electronic health records. J Biomed Inform 2018; 87: 118–30. [DOI] [PubMed] [Google Scholar]
- 162. Chen C-J, Warikoo N, Chang Y-C, et al. Medical knowledge infused convolutional neural networks for cohort selection in clinical trials. J Am Med Inform Assoc 2019; 26 (11): 1227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Segura-Bedmar I, Colón-Ruíz C, Tejedor-Alonso MÁ, et al. Predicting of anaphylaxis in big data EMR by exploring machine learning approaches. J Biomed Inform 2018; 87: 50–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The underlying data and R code to replicate our analyses can be found at: https://github.com/jlgrons/ML-EHR-Phenotyping-Review.