Abstract
During mosquito transmission, malaria ookinetes must cross a chitin-containing structure known as the peritrophic matrix (PM), which surrounds the infected blood meal in the mosquito midgut. In turn, ookinetes produce multiple chitinase activities presumably aimed at disrupting this physical barrier to allow ookinete invasion of the midgut epithelium. Plasmodium chitinase activities are demonstrated targets for human and avian malaria transmission blockade with the chitinase inhibitor allosamidin. Here, we identify and characterize the first chitinase gene of a rodent malaria parasite, Plasmodium berghei. We show that the gene, named PbCHT1, is a structural ortholog of PgCHT1 of the avian malaria parasite Plasmodium gallinaceum and a paralog of PfCHT1 of the human malaria parasite Plasmodium falciparum. Targeted disruption of PbCHT1 reduced parasite infectivity in Anopheles stephensi mosquitoes by up to 90%. Reductions in infectivity were also observed in ookinete feeds—an artificial situation where midgut invasion occurs before PM formation—suggesting that PbCHT1 plays a role other than PM disruption. PbCHT1 null mutants had no residual ookinete-derived chitinase activity in vitro, suggesting that P. berghei ookinetes express only one chitinase gene. Moreover, PbCHT1 activity appeared insensitive to allosamidin inhibition, an observation that raises questions about the use of allosamidin and components like it as potential malaria transmission-blocking drugs. Taken together, these findings suggest a fundamental divergence among rodent, avian, and human malaria parasite chitinases, with implications for the evolution of Plasmodium-mosquito interactions.
After ingestion of infectious Plasmodium gametocytes by the mosquito, motile ookinetes develop in the midgut lumen and traverse the chitin-containing peritrophic matrix (PM), the microvillus-associated network, and the midgut epithelium to form sporozoite-producing oocysts on the hemocoel side of the midgut (11, 18). After the demonstration that ookinetes secrete multiple chitinase activities (6), two distinct Plasmodium chitinase genes were isolated. The first was isolated from the human malaria parasite Plasmodium falciparum (PfCHT1) (14), while the second was found in the avian malaria parasite Plasmodium gallinaceum (PgCHT1) (15). The primary structures of these two chitinase genes are markedly different: PgCHT1 encodes putative amino-terminal proenzyme and carboxy-terminal chitin-binding domains, which are both absent in PfCHT1. P. gallinaceum secretes a second chitinase activity provisionally named PgCHT2, believed to be orthologous to that encoded by PfCHT1 based on its molecular mass and physiological properties (pH optimum and sensitivity to the chitinase inhibitor allosamidin), and it may have additional chitinase activities (15).
The Streptomyces-produced molecule allosamidin is a 622-dalton pseudo-oligosaccharide that inhibits Plasmodium chitinase activities in vitro (10, 14, 15). Moreover, the presence of allosamidin in an infected blood meal inhibited oocyst formation of P. gallinaceum in Aedes aegypti and of P. falciparum in Anopheles freeborni, a process that was reversed when the PM was prevented from forming by the addition of exogenous chitinase to the blood meal (10). Although these inhibitor studies identified Plasmodium chitinases as potential malaria transmission-blocking targets, dissection of the roles of the individual chitinase activities in mosquito infection remains a prerequisite for a rational chitinase-based transmission-blocking vaccine or drug design. For this purpose, we have isolated a chitinase gene of the rodent malaria parasite Plasmodium berghei, a Plasmodium species amenable to such experiments because of its suitability for obtaining stable transgenic gene knockout parasites and its ability to form large numbers of ookinetes in vitro for study. We show that the isolated P. berghei chitinase gene, named PbCHT1, contains putative proenzyme and chitin-binding domains and is a structural ortholog of PgCHT1. The construction of transgenic PbCHT1 null mutants has allowed us to establish that P. berghei ookinetes have only one apparent chitinase activity. We show that these parasites have significantly reduced, but not abolished, infectivity in mosquitoes. Our data further suggest that PbCHT1 may play a role other than PM penetration and is insensitive to allosamidin. The biological significance of these findings is discussed.
MATERIALS AND METHODS
Parasite maintenance, culturing, and purification; differential screening; RNA extraction and purification; Southern, Northern, and Western blotting; and mosquito infections were performed as described previously (3).
Gene isolation and sequence analysis.
From the partial cDNA, the complete PbCHT1 sequence was obtained with the gene-specific primer F02-RACE (CGATACCAGGTGCCCGTGTTGAATAG) using SMART rapid amplification of cDNA ends (RACE) (Clontech Laboratories) according to the manufacturer's instructions. Sequence analyses were carried out with the MacVector package (Oxford Molecular).
Construction of transgenic parasites.
A 600-bp fragment corresponding to the 5′ portion of the PbCHT1 mRNA was amplified with primers CHIT-BAM (GGATCCATTTTTTTGGAGACTTTATAACA) and CHIT-ERI (GAATTCTAAAATTTCCCTTGGAGA), digested with BamHI and EcoRI, and ligated into BamHI/EcoRI-digested pBS-DHFR (3) to give pCHT1-BE. A 530-bp fragment corresponding to the 3′ portion of the PbCHT1 mRNA was amplified with primers CHIT-KPN (GGTACCAAATATATGCAATGTAACTAAAA) and CHIT-HIND (AAGCTTAAACAATGGCATGGAGG), digested with KpnI and HindIII, and ligated into KpnI/HindIII-digested pCHT1-BE to give the transfection plasmid pPbCHT1-KO. Fifty micrograms of pCHT1-KO was digested with KpnI and BamHI to excise the plasmid backbone and transfected into purified schizonts as described previously (16). Pyrimethamine selection of transformed parasites and limiting dilution cloning were performed as described previously (16).
RT-PCR.
One microgram of ookinete total RNA was reverse transcribed with Superscript II (Life Technologies) in the presence of d(T25) according to the manufacturer's instructions and then diluted to 100 μl with Tris-glycine buffer (pH 8.0). One microliter was subjected to PCR amplification. The PbCHT1-specific primers CHT1-PF (GCCAAGGAGCTAGCGGG) and CHT1-PR (CGATACCAGGTGCCCG) were used to amplify the PbCHT1 probe used in Southern and Northern blotting and for semiquantitative reverse transcription (RT)-PCR. Gene Pbs25-specific primers were described previously (9). Degenerate primers CHT2-Forward [GGTAT(A/T/C)AT(A/T/C)(G/C)(G/C)IGGITA(C/T) TA(C/T)(G/C)(G/C)ITCITGG] (where I is inosine) and CHT2-Reverse [GG(C/T)TCI(C/T)A(A/G)TCIA(C/T)(A/G)TCIA(C/T)CC(A/G)TC] were used to amplify Plasmodium chitinase genes.
In vitro chitinase activity assay.
Ten million ookinetes were homogenized in phosphate-buffered saline (PBS) (pH 7.5) containing 1% Nonidet P-40 (Sigma) and centrifuged at 20,000 × g for 2 min; the supernatant was loaded into 1% agarose gels in PBS containing 0.01% ethylene glycol chitin (Seikagaku). After overnight incubation at 37°C, the gels were stained for 5 min in PBS containing 0.01% Fluorescent Brightener 28 (Sigma) and destained in distilled water. Chitin hydrolysis was visualized under UV light. Relative intensities of hydrolysis were measured by pixel density scanning with NIH Image software. Serratia marcescens chitinase A (SmChiA; 10 mU; Sigma) was used as a positive control for allosamidin activity.
Nucleotide sequence accession number.
Sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession number AJ305256.
RESULTS
Identification and sequence analysis of PbCHT1.
A partial cDNA corresponding to PbCHT1 was obtained by differential screening of a subtracted P. berghei cDNA library enriched for ookinete-specific sequences (4). Subsequently, the remainder of the PbCHT1 sequence was obtained by rapid amplification of cDNA ends. PbCHT1 is encoded by a single large open reading frame of 1,947 nucleotides encoding a 649-amino-acid protein with a calculated Mr of 72,127. It contains a predicted amino-terminal signal peptide of 18 amino acids resulting in a mature protein with an Mr of 70,072. Homology searches revealed high levels of sequence homology with the other Plasmodium chitinases, PgCHT1 and PfCHT1. Amino acid sequence identity of PbCHT1 is substantially higher with PgCHT1 (58%) than with PfCHT1 (19%), although the differences at sequence similarity levels are less pronounced (81 and 76%, respectively).
In other respects also, PbCHT1 is more similar to PgCHT1 than to PfCHT1 (Fig. 1A). First, it contains a putative carboxy-terminal chitin-binding domain (residues 495 to 648; PbCHT1 numbering) very similar to that present in PgCHT1. This domain is absent in PfCHT1. Second, PbCHT1 contains a region of low complexity and low sequence conservation between residues 37 and 152; this region contains nine imperfect repeats of the amino acid sequence E(NG)NGNGA/V (one-letter amino acid code, with parentheses enclosing acids not always present and a shill between two acids indicating the presence of one or the other acid). It is likely that this amino-terminal region downstream of the signal peptide constitutes a proenzyme domain like that described for PgCHT1 (15). Given the overall structural features and sequence homologies, it is clear that PbCHT1 and PgCHT1 are orthologs, whereas PfCHT1 is a paralog.
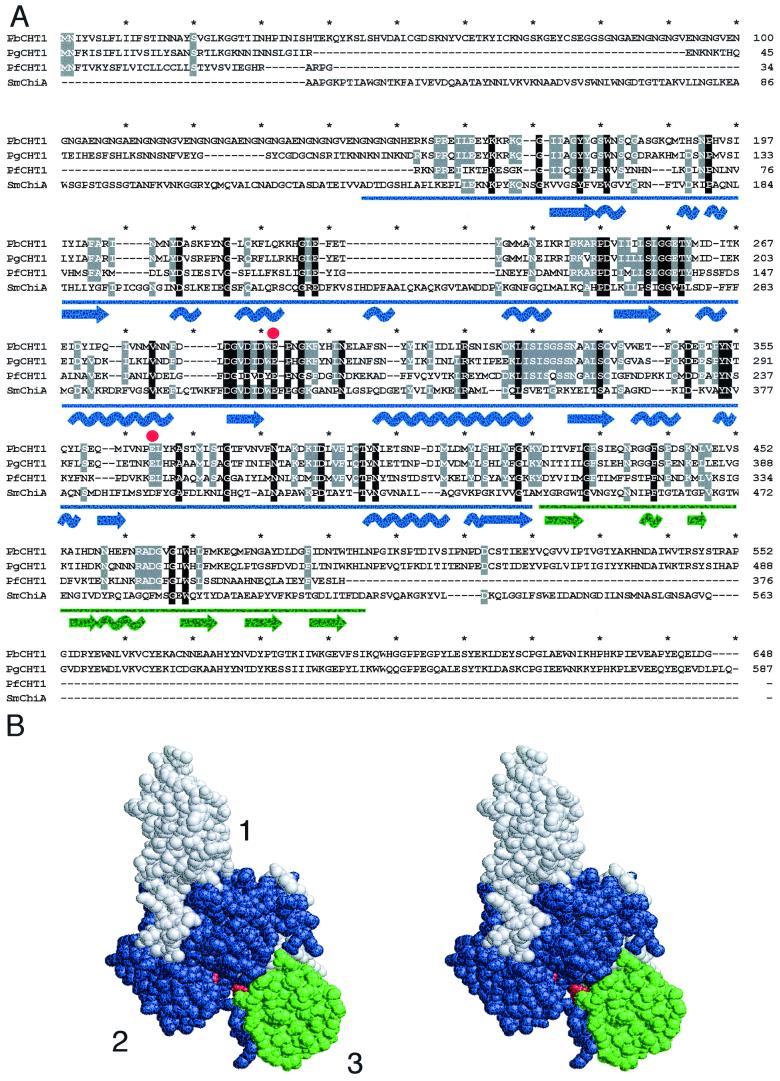
FIG. 1.
Comparison of PbCHT1, PgCHT1, PfCHT1, and SmChiA, a bacterial family 18 chitinase from S. marcescens. (A) Multiple amino acid alignment (Clustal W). Residue identities are indicated by shading (grey, 75%; black, 100%), and secondary structure features (coils represent helices; arrows represent sheets) are shown below the sequences. The predicted catalytic dyad (SmChiA residues 315 and 391) is marked with red dots. Plasmodium chitinases show a high structural conservation with subdomains 2 (blue) and 3 (green) of SmChiA. (B) Stereo-space-filled image of the atomic structure of SmChiA showing subdomains 2 (blue) and 3 (green) and the catalytic dyad (red). These domains correspond to the areas of strong conservation in Plasmodium chitinases and are similarly colored in panel A. The numbers refer to the three subdomains.
A comparison of the three Plasmodium chitinases with SmChiA, a closely related family 18 glycosylhydrolase for which the crystal structure has been determined (8), reveals extended sequence homology for all three parasite chitinases with α/β barrel subdomain 2 and α + β subdomain 3 of SmChiA (Fig. 1). Based on similarities with lysozyme, SmChiA subdomain 2 contains two catalytic dyad residues that are juxtaposed in the substrate-binding groove: glutamate 315 and aspartate 391 (Fig. 1). These residues are conserved in the Plasmodium chitinases, although aspartate 391 has been substituted with glutamate. β-Sheet subdomain 1 of SmChiA is structurally related to the fibronectin III domain; in the parasite chitinases this domain is absent. Notably, sequence homology starts just downstream of the putative proenzyme sequences and extends to include the entire SmChiA subdomains 2 and 3. Thus, all Plasmodium chitinases contain a hydrolytic domain equivalent to SmChiA subdomains 2 and 3, while PbCHT1 and PgCHT1 contain additional amino- and carboxy-terminal subdomains corresponding to the putative proenzyme and chitin-binding domains, respectively.
Expression of PbCHT1.
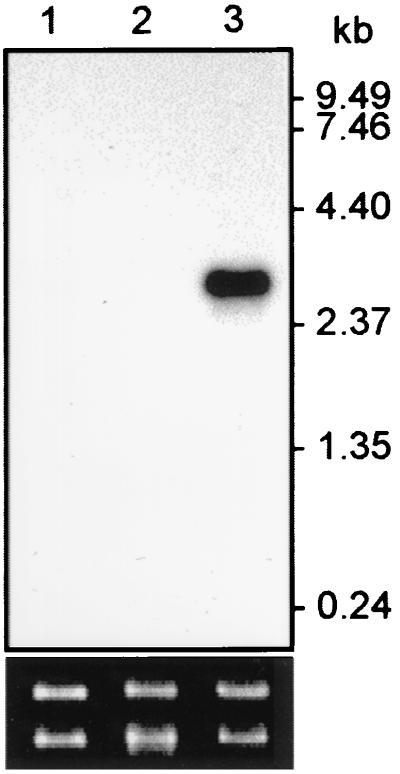
To determine the pattern of expression of PbCHT1, we carried out a Northern blot analysis of RNA samples purified from asexual blood-stage parasites, gametocytes, or in vitro-cultured ookinetes. This analysis identified an abundant mRNA of approximately 2.5 kb in the ookinete sample, while no signal was obtained in either asexual blood-stage parasites or gametocytes (Fig. 2A). These results strongly indicate the expression of PbCHT1 in ookinetes, which is also the case for the other Plasmodium chitinases (14, 15). In fact, PgCHT1 was recently shown to be transported via micronemes to the electron-dense area of the apical complex for extracellular secretion (7).
FIG. 2.
Differential expression of PbCHT1. Total RNA from asexual blood-stage parasites (lane 1), gametocytes (lane 2), and in vitro-cultured ookinetes (lane 3) was subjected to Northern blot analysis using a probe corresponding to PbCHT1. RNA amounts were normalized using large- and small-subunit rRNAs (ethidium bromide stained), as shown at the bottom of the figure.
Targeted disruption of PbCHT1.
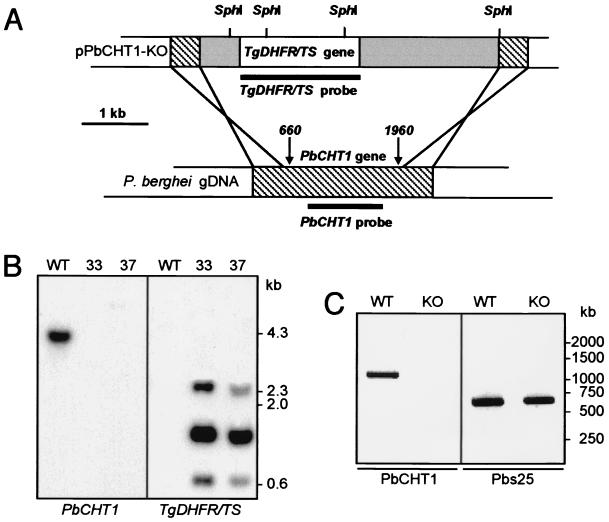
To investigate the function of PbCHT1, we generated transgenic PbCHT1-disrupted parasites by insertion of a modified Toxoplasma gondii dihydrofolate reductase-thymidylate synthase gene cassette (DHFR/TS) (13, 16) that confers resistance to the antimalarial drug pyrimethamine into the PbCHT1 gene by double homologous recombination (Fig. 3A). The DHFR/TS cassette was inserted between nucleotide positions 660 and 1960 of PbCHT1 (Fig. 3A), thereby removing 1.3 kb of the PbCHT1 central coding sequence, including the sequences encoding the putative binding pocket and the catalytic site.
FIG. 3.
Targeted disruption of PbCHT1 and molecular analyses. (A) Schematic diagram of the targeting strategy. Indicated is the transfection vector pPbCHT1-KO containing the T. gondii DHFR/TS gene cassette (white box), P. berghei DHFR flanking sequences (gray boxes), and PbCHT1-specific sequences (hatched boxes). The double homologous recombination crossover sites (crossed lines), the integration sites (arrows with nucleotide positions), the SphI restriction sites, and the probes used in Southern blot analysis (thick lines) are shown. gDNA, genomic DNA. (B) Southern blot analysis of SphI-digested genomic DNA from WT and PbCHT1-KO parasites using probes corresponding to PbCHT1 (left panel) and to the DHFR/TS cassette (right panel). (C) RT-PCR analysis of total RNA derived from ookinete-enriched midgut stages of WT (left lanes) and PbCHT1-KO (right lanes) parasites. Amplicons corresponding to PbCHT1(∼1,100 bp) and Pbs25 (∼600 bp) are shown.
Subsequently, two independent clonal transgenic parasite populations (termed PbCHT1-KO clones 33 and 37) were assessed for their integrity by Southern blot analysis of SphI-digested genomic DNA. A probe corresponding to nucleotide positions 710 to 1840 of PbCHT1 (no internal SphI sites present) gave rise to a single band in the parental (wild-type [WT]) parasites but no bands in the PbCHT1-KO parasites (Fig. 3B), demonstrating the successful removal of the PbCHT1 central sequence by the insertion of the DHFR/TS cassette. Cross-hybridization with other putative chitinase genes was not observed under the conditions used. Conversely, a probe corresponding to the DHFR/TS cassette (two SphI sites present) gave rise to three DHFR/TS-specific bands in the PbCHT1-KO parasites but no signal in the WT sample (Fig. 3B). Together, these results confirmed correct integration of the DHFR/TS cassette into the target gene.
PbCHT1-KO parasites developed gametocytes and formed ookinetes in vivo and in vitro, in numbers similar to and indistinguishable from those of WT parasites in Giemsa-stained blood films (data not shown). To confirm that PbCHT1 expression was abolished in the transgenic parasites, total RNA was extracted from in vitro-cultured ookinetes and subjected to RT-PCR using PbCHT1-specific primers. In the WT parasites, a band of approximately 1,100 bp corresponding to PbCHT1 was amplified, while no band was amplified in the PbCHT1-KO parasites. In contrast, a 600-bp band corresponding to the reference ookinete gene Pbs25 (9) was amplified in both parasite samples (Fig. 3C). Clearly, the absence of PbCHT1 mRNA is in full agreement with the genotype of the PbCHT1-KO parasites (Fig. 3B) and supports the successful knockout of PbCHT1 expression.
Are there other chitinases of P. berghei?
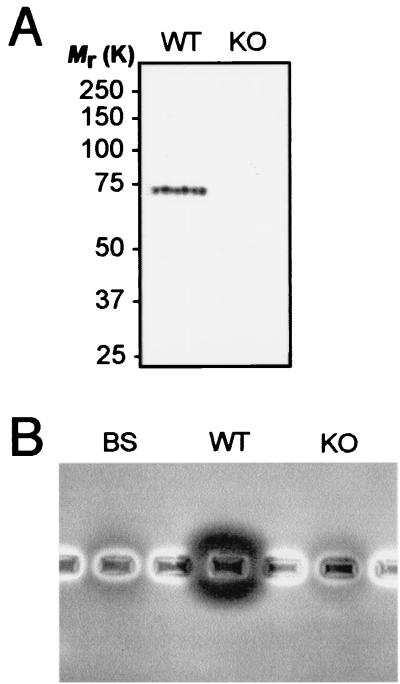
We performed several experiments to investigate whether P. berghei had any additional chitinase genes. First, we performed Western blotting with ookinete homogenates and antiserum raised against a PgCHT1 active-site peptide. This antiserum detects at least two distinct chitinases in P. gallinaceum and cross-reacts with PfCHT1 (7, 15). Thus, it is likely that this antiserum would cross-react with PbCHT1 and other chitinases of P. berghei. However, only a single band with an approximate Mr of 70,000 was detected in the WT parasites; as expected, this band was absent in the PbCHT1-KO parasites (Fig. 4A). Based on its apparent size and its absence in the PbCHT1-KO parasites, this 70-kDa band likely corresponds to PbCHT1 and confirms that the transgenic parasites are PbCHT1 null mutants. Prolonged development of the blot revealed a weak band with an approximate Mr of 60,000 in the WT sample (data not shown), indicating the processing of PbCHT1 and possibly reflecting the cleavage of the putative proenzyme domain. No other proteins were recognized, arguing against the presence of additional chitinases in P. berghei.
FIG. 4.
Analysis of P. berghei WT and PbCHT1-KO ookinete homogenates for additional chitinase activity. (A) Western blot analysis with PgCHT1 active-site antiserum. (B) In vitro chitinase activity assay with glycol chitin-containing agarose. Also included is a homogenate from similarly purified blood stages (BS).
Second, we designed degenerate primers for amino acid sequences conserved among the three Plasmodium chitinases and performed RT-PCR with RNA purified from WT and PbCHT1-KO ookinetes. While we were able to amplify a PbCHT1-specific sequence from the WT parasites, no product could be amplified from the PbCHT1-KO parasites (data not shown), again arguing against the expression of other chitinase genes in this stage of the life cycle.
Third, we performed an in vitro chitinase activity assay with ookinete homogenates and glycol chitin-containing agarose. In this assay, chitinase activity is demonstrated by a dark area around the well containing the homogenate, resulting from hydrolysis of the chitin substrate by the diffusing chitinase. An approximately 80% reduction in activity was observed in the PbCHT1-KO ookinetes (Fig. 4B), again confirming that the PbCHT1-KO parasites are PbCHT1 null mutants. This residual chitinase activity corresponds to background activity, as it was also observed in homogenates from similarly purified blood-stage parasites. This chitinase activity likely is derived from contaminating mouse leukocytes or serum proteins, as has been reported for human leukocytes and serum (5). Thus, the apparent absence of residual ookinete-derived chitinase activity in the PbCHT1-KO sample, combined with the results from the Western and RT-PCR analyses, indicates that PbCHT1 is the sole chitinase gene expressed in P. berghei ookinetes.
Infectivity of PbCHT1-KO parasites to Anopheles stephensi mosquitoes.
To assess the effects of PbCHT1 disruption on mosquito infection, PbCHT1-KO and WT parasites were fed to A. stephensi mosquitoes and compared for their ability to form oocysts, a measure of parasite infectivity. In seven experiments, significant (P < 0.01) reductions in oocyst numbers of between 30 and 90% were obtained with the PbCHT1-KO parasites (Table 1). The two independent clonal populations of transgenic parasites (clones 33 and 37) had very similar transmission phenotypes, indicating that the reduction in infectivity is unlikely to be a result of clonal phenotypic variation.
TABLE 1.
Effect of PbCHT1 knockout on P. berghei infectivity to A. stephensi
| Expt | Type of feed (clone)a | Mean ± SEM no. of oocysts (no. of midguts dissected)b in mosquitoes infected with:
|
% of WT value | |
|---|---|---|---|---|
| WT | PbCHT1-KO | |||
| 1 | gct (33) | 95.2 ± 5.8 (99) | 63.0 ± 3.9c (150) | 66 |
| 2 | gct (37) | 96.0 ± 6.3 (125) | 68.2 ± 4.0c (147) | 71 |
| 3 | gct (33) | 54.9 ± 4.5 (130) | 28.2 ± 2.5c (133) | 51 |
| 4 | gct (37) | 49.6 ± 3.0 (142) | 25.1 ± 2.2c (150) | 50 |
| 5 | ook (33) | 49.2 ± 2.4 (107) | 14.1 ± 0.9c (109) | 29 |
| 6 | ook (33) | 33.8 ± 1.7 (137) | 7.3 ± 0.6c (115) | 22 |
| 7 | ook (33) | 2.41 ± 0.2 (90) | 0.34 ± 0.06c (85) | 14 |
gct, gametocyte; ook, ookinete.
Each experiment included pooled data from three mice (gct) and three membrane feeders (ook).
Significantly different (P < 0.01) from the value for the WT-infected control group, as calculated by Student's t test.
Surprisingly, we observed significant levels of reduction in the infectivity of the PbCHT1-KO parasites in both gametocyte and ookinete feeds (Table 1). In A. stephensi, PM formation is first detectable by electron microscopy at 12 h and continues up to 48 h after blood feeding (1). In ookinete feeds, we observed ookinetes in the midgut epithelium as early as 3 h postfeeding, and by 12 h the majority had reached the midgut epithelium (data not shown). Thus, in ookinete feeds most ookinetes invade the midgut epithelium in the absence of a developed PM. In contrast, in gametocyte feeds 20 to 30 h is required for ookinete development in the midgut lumen (11); consequently, the majority of ookinetes invade the midgut epithelium in the presence of a developed PM. Clearly, if PbCHT1 played a role in PM disruption, then we would anticipate infection levels more comparable to those of the WT parasites in ookinete feeds. As this was clearly not the case, the results suggest that the PM is not a target of PbCHT1 activity in A. stephensi.
PbCHT1-KO oocysts formed normal numbers of sporozoites, which were infectious to mice upon mosquito bite. Moreover, parasites from sporozoite-induced infections retained their phenotype in subsequent mosquito transmissions (data not shown). This result demonstrates that PbCHT1 functions predominantly in the ookinete stage, an observation consistent with its expression profile (Fig. 2). Indeed, chitinase activity is unlikely to be required for downstream sporozoite invasion of the salivary gland ducts of A. stephensi, as these have been reported not to contain chitin (17).
Allosamidin does not inhibit PbCHT1.
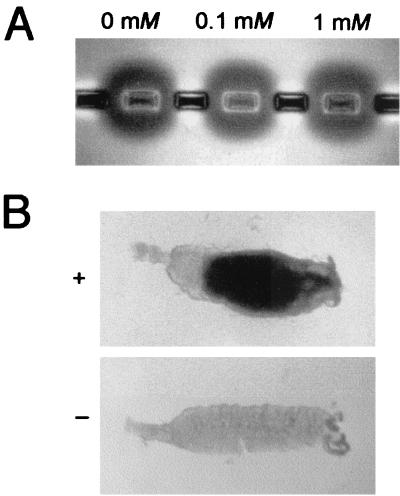
When we assessed the effects of the chitinase inhibitor allosamidin on P. berghei infectivity in A. stephensi, we found no decrease in oocyst numbers (data not shown). Interestingly, we used an allosamidin concentration (0.1 mM) that effectively abolished oocyst development of P. falciparum and P. gallinaceum in A. freeborni and A. aegypti, respectively (10). This result suggested that allosamidin does not inhibit PbCHT1. To test this suggestion, we added allosamidin to ookinete homogenates at concentrations of up to 1 mM in our in vitro chitinase activity assay and observed no inhibition of chitinase activity (Fig. 5A), while a control chitinase activity (SmChiA) was inhibited (data not shown). The concentration of 1 mM is far in excess (200-fold) of that found to reduce P. gallinaceum chitinase activity in vitro by more than 90% (10). The addition of allosamidin to blood feeds did, however, have a clear effect on the ability of the mosquitoes to digest the blood meal. At an allosamidin concentration of 0.1 mM, PMs and partially digested blood meals were still present at 9 days postinfection in one-third of the mosquitoes examined, while none of the control mosquitoes contained blood meal remnants (Fig. 5B). A similar observation was made for A. freeborni and A. aegypti mosquitoes fed allosamidin (10) and is indicative of the allosamidin inhibition of mosquito-derived chitinase(s).
FIG. 5.
Effects of allosamidin. (A) In vitro chitinase activity assay with P. berghei WT ookinete homogenates in the presence of 0, 0.1, and 1 mM allosamidin. (B) Effect of allosamidin on blood meal digestion. Shown are dissected guts of mosquitoes at 9 days after blood feeding in the presence (+) or absence (−) of 0.1 mM allosamidin.
DISCUSSION
In this paper, we describe and characterize PbCHT1, the first chitinase gene isolated from a rodent malaria species. We show that the gene product, PbCHT1, contains putative proenzyme and chitin-binding domains and is orthologous to a previously described endochitinase, PgCHT1, from P. gallinaceum. Targeted disruption of the PbCHT1 gene by double homologous recombination has allowed us to study the existence of other putative chitinase activities in P. berghei ookinetes as well as the role of PbCHT1 in mosquito infection in the absence and presence of a PM.
Our findings indicate that P. berghei ookinetes have a single chitinase activity because (i) no additional bands were recognized in Western blottings by a PgCHT1 active-site antibody, (ii) no specific products could be amplified from PbCHT1-KO ookinetes with Plasmodium chitinase gene-specific degenerate primers, and (iii) most importantly, no residual ookinete-derived chitinase activity was observed in PbCHT1-KO parasites (Fig. 4). In fact, the same may be true for P. falciparum because only a single chitinase gene has thus far been described in the Malaria Genome Project databases, which now contain about 95% of the genome. Intuitively, if this assumption is correct and P. gallinaceum does indeed possess both types of chitinase genes, then it can be suggested that both P. falciparum and P. berghei share an avian Plasmodium ancestor and that each has retained a different one of the two chitinases.
Transmission experiments with A. stephensi have shown that PbCHT1 null mutants are significantly impaired in oocyst formation but that PbCHT1 is not essential for mosquito infection. The residual infectivity of the PbCHT1-KO ookinetes may, at least to some extent, be the result of mouse-derived chitinase activity; however, it is just as conceivable that A. stephensi midgut chitin simply does not provide a foolproof barrier for P. berghei infection. It should be noted that A. stephensi is highly susceptible to P. berghei. In contrast, P. falciparum infection of its anopheline vectors gives substantially lower oocyst numbers, which could have implications for the role of PfCHT1 in mosquito infection.
Reductions in infectivity were observed both in the presence of a PM (gametocyte feeds) and in its absence (ookinete feeds). Although in the absence of complementation we cannot rule out the possibility that the reduced infectivity is the result of pleiotropic effects, the findings suggests that PbCHT1 plays a role other than PM disruption. We cannot rule out the possibilities that chitin is present in the microvillus-associated network or the epithelial cells themselves and that chitinase activity is required to allow ookinete egress from these tissues. In this respect, it should be noted that chitin precursors are synthesized by epithelial cells and must traverse the microvillus-associated network to form the mature PM. Experiments are in progress to investigate these hypotheses.
Our data appear to conflict with previous chitinase inhibitor studies conducted with P. gallinaceum and P. falciparum in A. aegypti and A. freeborni mosquitoes, respectively, which did not indicate a role for Plasmodium chitinases downstream of PM disruption (10). In those experiments, however, transmission in the absence of a PM was achieved not by conducting ookinete feeds but instead by adding exogenous Streptomyces griseus chitinase to the blood meal, thereby preventing PM formation. Clearly, it is conceivable that the S. griseus chitinase may also have affected potential chitin integrity in other midgut tissues. Moreover, there may be substantial differences in chitin composition or deposition between these mosquito species and A. stephensi. Thus, we cannot truly compare those experiments with ours.
Allosamidin is a chitin-like metabolite that inhibits numerous chitinase enzymes with different efficacies by binding to the active site (12). Although it is known that allosamidin does not universally inhibit chitinase enzymes (for example, S. griseus chitinase is not inhibited [10]), it was surprising to discover that PbCHT1, the ortholog of the efficiently inhibited PgCHT1, was insensitive in our assay (Fig. 5A). It is known that single amino acid replacements can alter substrate specificity and enable catalytic turnover of compounds that previously strongly inhibited enzyme activity (2). Modeling the homologous Plasmodium chitinases on the atomic structure of the related bacterial chitinase SmChiA (Fig. 1) highlights a number of candidate residue replacements that may elicit such an effect. Notable among these is a unique Lys366Pro replacement, which occurs adjacent to the predicted catalytic Glu367 residue in PbCHT1. This substitution would significantly alter the orientation of the catalytic residue in the active site, which could change the specificity for allosamidin; future site-directed mutagenesis studies are needed to verify this hypothesis. In any event, these findings may have implications for the long-term use of allosamidin or compounds like it as malaria transmission-blocking drugs. The natural allelic variation of P. falciparum chitinase enzymes is unknown, and under selective pressure by allosamidin, alleles containing enabling mutations could rise to a high frequency in human malaria. Moreover, simple mutations that render the enzymes insensitive to the inhibitor could arise and be selected for.
ACKNOWLEDGMENTS
We thank Meiji Arai for help with the photography.
This work was supported by a grant from the European Union TMR program. C.C. was supported by a grant from the NHMRC, Canberra, Australian Capital Territory, Australia. E. K. was supported by a grant from the Islamic Development Bank Merit Scholarship Program in High Technology, Jeddah, Saudi Arabia.
ADDENDUM IN PROOF
As this paper was going to press, a highly conserved sequence orthologous to PbCHT1 appeared in the P. yoelii database accessible through The Institute for Genomic Research website (www.tigr.org). This sequencing program is carried on in collaboration with the Naval Medical Research Center and is supported by the U.S. Department of Defense. At this time, no other chitinase genes in this genome have been identified.
REFERENCES
- 1.Berner R, Rudin W, Hecker H. Peritrophic membranes and protease activity in the midgut of the malaria mosquito, Anopheles stephensi (Liston) (Insecta: Diptera), under normal and experimental conditions. J Ultrastruct Res. 1983;83:195–204. doi: 10.1016/s0022-5320(83)90077-1. [DOI] [PubMed] [Google Scholar]
- 2.Claudianos C, Russell R J, Oakeshott J G. The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochem Mol Biol. 1999;29:675–686. doi: 10.1016/s0965-1748(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 3.Dessens J T, Beetsma A L, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos F C, Sinden R E. CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 1999;18:6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dessens J T, Margos G, Rodriguez M C, Sinden R E. Identification of differentially regulated genes of Plasmodium by suppression subtractive hybridization. Parasitol Today. 2000;16:354–356. doi: 10.1016/s0169-4758(00)01710-5. [DOI] [PubMed] [Google Scholar]
- 5.Escott G M, Adams D J. Chitinase activity in human serum and leukocytes. Infect Immun. 1995;63:4770–4773. doi: 10.1128/iai.63.12.4770-4773.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber M, Cabib E, Miller L H. Malaria parasite chitinase and penetration of the mosquito peritrophic membrane. Proc Natl Acad Sci USA. 1991;88:2807–2810. doi: 10.1073/pnas.88.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langer R C, Hayward R E, Tsuboi T, Tachibana M, Torii M, Vinetz J M. Micronemal transport of Plasmodium ookinete chitinases to the electron-dense area of the apical complex for extracellular secretion. Infect Immun. 2000;68:6461–6465. doi: 10.1128/iai.68.11.6461-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M C, Gerold P, Dessens J, Kurtenbach K, Schwartz R T, Sinden R E, Margos G. Characterisation and expression of Pbs25, a sexual and sporogonic stage specific protein of Plasmodium berghei. Mol Biochem Parasitol. 2000;110:147–159. doi: 10.1016/s0166-6851(00)00265-6. [DOI] [PubMed] [Google Scholar]
- 10.Shahabuddin M, Toyoshima T, Aikawa M, Kaslow D C. Transmission-blocking activity of a chitinase inhibitor and activation of malarial parasite chitinase by mosquito protease. Proc Natl Acad Sci USA. 1993;90:4266–4270. doi: 10.1073/pnas.90.9.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinden R E. Infection of mosquitoes with rodent malaria. In: Crampton J M, Beard C B, Louis C, editors. The molecular biology of insect disease vectors. London, United Kingdom: Chapman & Hall; 1997. pp. 261–267. [Google Scholar]
- 12.Spindler K D, Spindler-Barth M. Inhibitors of chitinases. EXS. 1999;87:201–209. doi: 10.1007/978-3-0348-8757-1_14. [DOI] [PubMed] [Google Scholar]
- 13.Van Dijk M R, Waters A P, Janse C J. Stable transfection of malaria parasite blood stages. Science. 1995;268:1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 14.Vinetz J M, Dave S K, Specht C A, Brameld K A, Xu B, Hayward R, Fidock D A. The chitinase PfCHT1 from the human malaria parasite Plasmodium falciparum lacks proenzyme and chitin-binding domains and displays unique substrate preferences. Proc Natl Acad Sci USA. 1999;96:14061–14066. doi: 10.1073/pnas.96.24.14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinetz J M, Valenzuela J G, Specht C A, Aravind L, Langer R C, Ribeiro J M, Kaslow D C. Chitinases of the avian malaria parasite Plasmodium gallinaceum, a class of enzymes necessary for parasite invasion of the mosquito midgut. J Biol Chem. 2000;14:10331–10341. doi: 10.1074/jbc.275.14.10331. [DOI] [PubMed] [Google Scholar]
- 16.Waters A P, Thomas A W, van Dijk M R, Janse C J. Transfection of malaria parasites. Methods. 1997;13:134–147. doi: 10.1006/meth.1997.0506. [DOI] [PubMed] [Google Scholar]
- 17.Wright K A. The anatomy of salivary glands of Anopheles stephensi Liston. Can J Zool. 1969;47:579–587. [Google Scholar]
- 18.Zieler H, Garon C F, Fischer E R, Shahabuddin M. A. tubular network associated with the brush-border surface of the Aedes aegypti midgut: implications for pathogen transmission by mosquitoes. J Exp Biol. 2000;203:1599–1611. doi: 10.1242/jeb.203.10.1599. [DOI] [PubMed] [Google Scholar]