Abstract
Advances in cryogenic electron microscopy (cryo-EM) enabled routine near-atomic structure determination of membrane proteins, while nanodisc technology has provided a way to provide membrane proteins with a native or native-like lipid environment. After giving a brief history of membrane mimetics, we present example structures of membrane proteins in nanodiscs that revealed information not provided by structures obtained in detergent. We describe how the lipid environment surrounding the membrane protein can be custom designed during nanodisc assembly and how it can be modified after assembly to test functional hypotheses. Because nanodiscs most closely replicate the physiologic environment of membrane proteins and often afford novel mechanistic insights, we propose that nanodiscs ought to become the standard for structural studies on membrane proteins.
Keywords: Single-particle cryo-electron microscopy, membrane scaffold proteins, amphiphilic copolymers, native nanodiscs
The Case for Nanodiscs When Studying the Structure of Membrane Proteins
Structural information on membrane proteins is a high priority because of their importance in diverse biological processes and as pharmaceutical drug targets. However, the number of structures for membrane proteins has long lagged far behind that of soluble proteins, the result of numerous complications associated with membrane proteins: from production of sufficient quantity for structural studies, to stabilization in solution, to the growth of crystals for diffraction analysis.
Recently, the number of new membrane protein structures has skyrocketed due to advances in cryogenic electron microscopy (cryo-EM). Specifically, direct detectors so drastically improved the quality of cryo-EM data that it finally became possible to determine structures at near-atomic resolution from images of individual particles. This “resolution revolution” [1] transformed single-particle cryo-EM from a fringe technique to the method of choice for membrane protein structure determination. However, the native lipid bilayer environment usually remained neglected in structural studies. This, too, is rapidly changing as biochemical progress in mimicking native membranes has paired with the advances in cryo-EM to herald a new era in the structural biology of membrane proteins. Here, we provide a historical perspective on membrane protein structural biology with a focus on approaches previously used to mimic membrane environments, contrasted with current techniques to recreate bona fide native or native-like lipid environments, collectively referred to as “nanodiscs.” We argue that, because nanodiscs more accurately replicate the physiologic milieu of membrane proteins, they afford novel mechanistic insights. Moreover, manipulation of the nanodisc lipid environment allows testing of biophysical hypotheses that had previously been impossible. We conclude that nanodiscs ought to become the field standard for membrane protein structure determination and identify areas for future growth.
A Brief History of Membrane Mimetics in Structural Biology
Lipid bilayers create complex physiochemical environments for their constituent proteins. Lipids are chemically diverse and non-homogenously distributed within bilayers, resulting in lateral subdomains and asymmetric leaflets. Lipid bilayers thus have varied architectures, nuanced mechanical properties, and anisotropic electrostatics. Faced with the challenges inherent to the study of proteins inhabiting this environment, membrane protein structural biologists have pursued strategies to stabilize targets in ways that made them amenable to structure-determination approaches developed for soluble proteins. For membrane proteins with a single transmembrane span, such as many cell-surface receptors, the transmembrane helix has typically been cut off. While facilitating separate structure determination of the cytoplasmic and extracellular domains, information on their coupling is lost. For membrane proteins with more extensive transmembrane domains, such as channels and transporters, the architecture of their membrane-integral regions is central to their structure and function, and thus their presence is requisite for these analyses. Most commonly, detergents have been used to extract integral proteins from the membrane and stabilize them in solution, so that their structures could be studied in the same way as those of soluble proteins.
However, detergents can destabilize proteins and distort their structure. The development of mild detergents with very low critical micellar concentrations (e.g., lauryl maltose neopentyl glycol and glyco-diosgenin) did advance X-ray crystallography of labile membrane proteins, including G protein-coupled receptors (GPCRs) [2,3]. Efforts to find alternatives to detergents also led to the introduction of other membrane mimetics, such as amphipols [4], Salipro [5] and most recently the peptidisc [6]; however, none perfectly recapitulates the physicochemical characteristics of lipid bilayers.
To recapitulate elements of membrane chemistry, lipids were occasionally supplemented during membrane protein purification and crystallization, ensuring that lipids critical for protein stability were retained. Sometimes added lipids revealed regulatory processes, such as the opening of the classical inward rectifier potassium channel Kir2.2 by phosphatidylinositol 4,5-bisphosphate (PIP2) [7]. Despite capturing certain protein–lipid interactions, this approach still fails to mimic most characteristics of lipid bilayers.
More sophisticated approaches used lipid bilayer-like matrices, such as lipidic cubic phases for 3D crystallization [8] and lipid/detergent bicelles for NMR studies [9]. The most native-like environment was provided by the reconstitution of membrane proteins with lipids into two-dimensional (2D) crystals that could then be analyzed by electron crystallography to yield near-atomic structures [10]. Unlike detergents and membrane mimetics, the lipids in 2D crystals form a true bilayer and recapitulate many of the physicochemical characteristics of a native membrane, such as lateral pressure and anisotropic electrostatic profile across the bilayer. In addition, in the case of the water channel aquaporin-0, 2D crystals could be grown with different lipids, thus allowing for systematic studies on lipid–protein interactions [11,12]. However, growing high-quality 2D crystals is exceptionally challenging and collecting the data to determine a high-resolution structure requires so much microscope time that electron crystallography is no longer viable.
Reconstitution of membrane proteins into lipid vesicles for cryo-EM analysis has provided another means to structure determination in a native-like environment (Figure 1A). Notably, vesicles also impose spherical constraints on incorporated membrane proteins, which were exploited to determine their orientation parameters for single-particle reconstruction [13]. Indeed, this approach yielded the first single-particle cryo-EM structure of a membrane protein in a lipid bilayer, that of the BK potassium channel [14] (Figure 1B). A much higher-resolution map was later obtained using data recorded with a direct detector [15] (Figure 1C). This method also has the unique advantage that it provides a way to expose the two extramembranous surfaces of a membrane protein to different environments, for example, to generate a transmembrane potential and visualize its effect on voltage-sensing membrane proteins [16]. Unfortunately, this technique is challenging in practice, as variance in vesicle diameter complicates spherical reconstruction, and preparing good specimens of vitrified vesicles is non-trivial.
Figure 1. Cryo-EM studies of membrane proteins in vesicles.

(A) Detergent removal from a mixture of detergent-solubilized membrane proteins and lipids induces the reconstitution of the integral proteins into lipid vesicles. Note that the two sides of the membrane protein are exposed to different environments. (B) The structure of the BK potassium channel was the first structure determined for a membrane protein in a lipid bilayer. The quality of cryo-EM images at the time was poor (left panel; white boxes indicate BK channels; inset shows a simulation of the vesicle below containing three BK channels). Indeed, after computationally subtracting the density for the lipid membrane, the channels were barely visible (middle panel, white boxes), explaining the limited resolution of the resulting map of ~2 nm (right panel). (C) Data collected with a direct detector yielded images with much better contrast (left panel) and individual BK channels were clearly visible after computational membrane subtraction (middle panel, white boxes), leading to a density map of the BK channel at 3.5 Å resolution (right panel). The images in panel (B) have been reproduced by permission from Springer Nature Customer Service Centre GmbH: Nature, Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy, Wang and Sigworth, 2009 [14]. Images in panel (C) have been reproduced from [15] with kind permission from Liguo Wang.
The Advent of Nanodiscs
Nowadays, the more commonly used approach to visualize membrane proteins in the context of a native lipid bilayer is to reconstitute it into a membrane-scaffold protein (MSP)-based nanodisc. MSPs are modified apolipoprotein A-1 variants that surround and stabilize discoidal membrane patches containing target proteins [17] (Figure 2A). Before direct detectors, nanodiscs were not beneficial for single-particle cryo-EM, because they increased structural heterogeneity and added substantial amorphous density that obscured the transmembrane domain, both complicating particle alignment. For many years, the only cryo-EM structure of a membrane protein in a nanodisc was that of the SecYEG protein translocon in complex with a ribosome, which provided the signal needed for particle alignment [18].
Figure 2. Cryo-EM studies of membrane proteins in membrane-scaffold protein (MSP)-based nanodiscs.
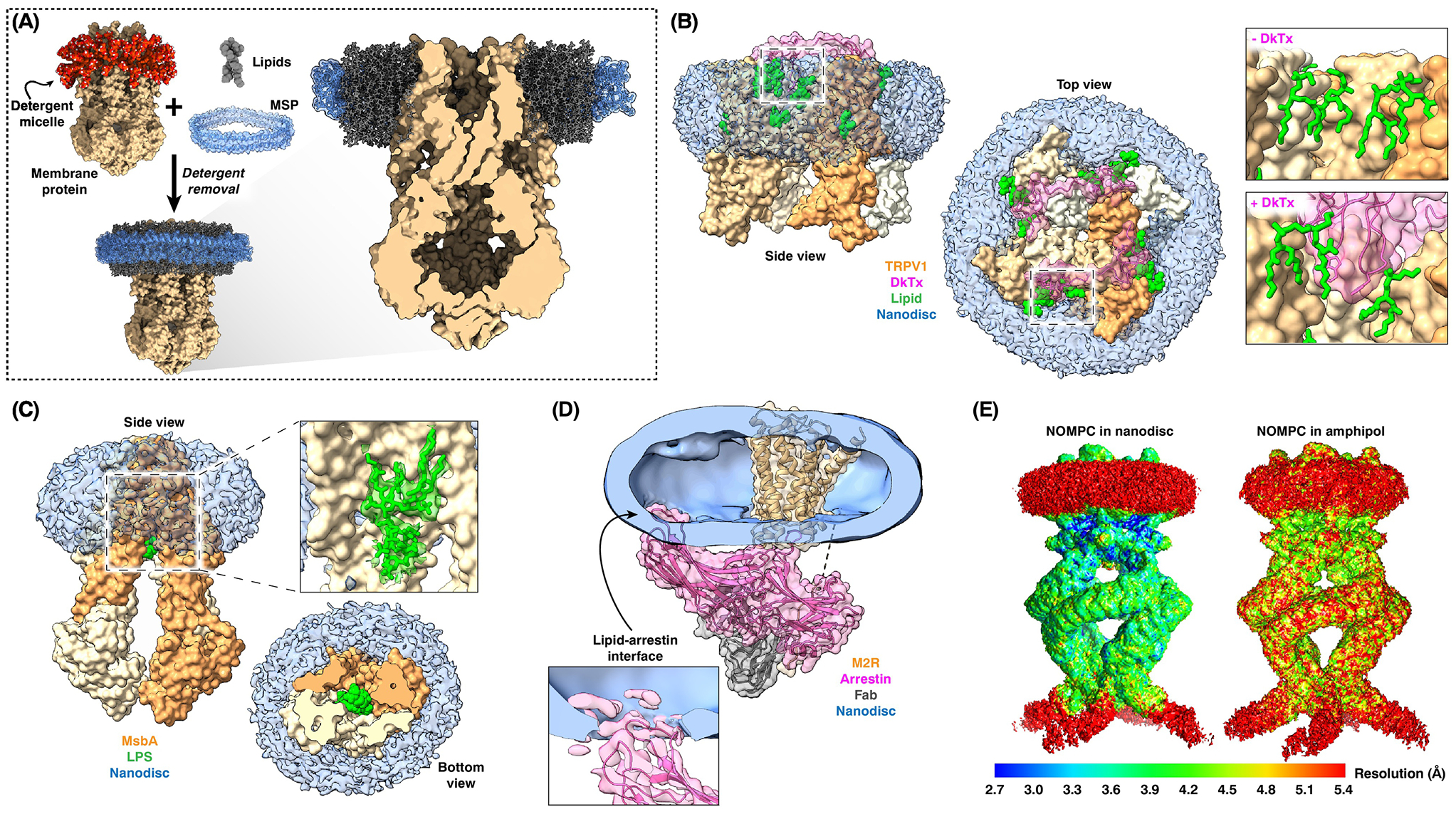
(A) Detergent removal from a mixture of detergent-solubilized membrane proteins and lipids in the presence of MSPs results in the reconstitution of the integral proteins into MSP-bounded discoidal membrane patches. (B) Nanodisc-embedded TRPV1 ion channel in complex with double-knot toxin (DkTx), showing that the toxin (pink) forms a tripartite complex with the TRPV1 channel (brown) and lipids (green) (left and middle panels). Toxin binding results in a rearrangement of the lipids (right panels). (C) The structure of E. coli MsbA determined in the context of a nanodisc revealed bound lipopolysaccharide substrate (green). (D) The structure of β-arrestin bound to the M2 muscarinic GPCR in a nanodisc revealed that β-arrestin also interacts with the membrane. (E) Local resolution maps of the NOMPC ion channel in a nanodisc (left panel) and in amphipol show that the nanodisc has a stabilizing effect on the extramembranous ankyrin-repeat domains. The maps in panel (E) have been reproduced by permission from Springer Nature Customer Service Centre GmbH: Nature, Electron cryo-microscopy structure of the mechanotransduction channel NOMPC, Jin et al., 2017 [24].
However, with the improved data provided by direct detectors, nanodiscs became increasingly used in cryo-EM studies of membrane proteins and have since yielded numerous high-resolution structures, often revealing features that would not have been apparent with detergent-solubilized protein. For example, the structure of the TRPV1 ion channel in a nanodisc revealed that annular lipids occupy defined locations, that specific lipids enhance the binding of a spider toxin to the channel by forming a ternary complex (Figure 2B), and that phosphatidylinositol lipids occupy the binding site for capsaicin and thus need to be released for channel activation [19]. The structure of nanodisc-embedded E. coli ATP-binding cassette transporter MsbA revealed bound lipopolysaccharide substrate not observed in X-ray structures (Figure 2C) and showed the MsbA conformational changes that underlie lipopolysaccharide flipping [20]. Two cryo-EM structures of the bacterial mechanosensitive channel MscS in nanodiscs revealed functionally important lipids [21,22]. A recent cryo-EM structure of β-arrestin bound to the M2 muscarinic GPCR in a nanodisc revealed that β-arrestin not only interacts with the receptor but also with the membrane (Figure 2D), which was shown to be functionally important [23]. Intriguingly, the lipid environment can affect protein structure beyond membrane-integral or -associated regions. In the case of the mechanotransduction channel NOMPC, the extramembranous ankyrin-repeat domains were better resolved in the context of a nanodisc than in amphipols (Figure 2E), even though these domains make no lipid contacts, underscoring the potential impact of the membrane environment on protein structure globally [24].
A related exciting development is the use of amphiphilic copolymers to solubilize membrane proteins into stable 10–30 nm membrane discs. The first copolymers used for this purpose were styrene maleic acid (SMA) copolymers and the resulting lipid-containing nanoparticles became known as SMALPs (for SMA lipid particles) [25] (Figure 3A), but several other copolymers have since been developed. The beauty of using amphiphilic copolymers to generate “native nanodiscs” is that it maintains the membrane proteins in their native lipid environment without any exposure to the potentially distorting effects of detergents. However, as for membrane proteins solubilized in detergents or reconstituted into nanodiscs, only functional assays can establish whether membrane proteins in native nanodiscs retained their biological function.
Figure 3. Cryo-EM studies of membrane proteins in native nanodiscs.

(A) Incubation of biological membranes with amphiphilic copolymers, such as styrene maleic acid (SMA) copolymers, generates SMA lipid particles (SMALPs) that contain membrane proteins in their native lipid environments. (B) The structure of E. coli AcrB (brown) isolated by using amphiphilic copolymers (pink) revealed a complete lipid bilayer patch (green) in the center of the trimer. (C) Reconstitution of purified glycine receptor (yellow and brown) into MSP-based nanodiscs (blue) only allowed visualization of the receptor in the desensitized conformation (left panels). However, isolation of the glycine receptor using SMA resulted in SMALPs (pink) that allowed visualization of the glycine receptor both in the desensitized and open conformations (middle and right panels). Note the different surface areas covered by the nanodisc and the two SMALPs.
Membrane proteins in native nanodiscs have been successfully used for high-resolution structure determination by single-particle cryo-EM (reviewed in [26,27]). For example, the structure of the multidrug exporter AcrB obtained by this approach illustrated their remarkable capacity to retain the native lipid environment by visualizing a membrane patch containing 24 lipids [28] (Figure 3B). This quality can be critical for stabilizing physiologically relevant conformations: whereas glycine receptors extracted in detergent and reconstituted into MSP-based nanodiscs were found to be exclusively in desensitized-like conformations, the same receptors extracted with SMA into native nanodiscs revealed both open and closed conformations [29] (Figure 3C).
Engineering the Nanodisc Lipid Environment
Nanodiscs provide not only a native or native-like membrane environment, but they also permit custom design of the membrane. Nearly all parameters of the MSP-stabilized membrane can be defined during nanodisc assembly (Figure 4, Section I). The size of the nanodisc is defined by the surrounding scaffold proteins and can be varied almost at will (e.g., [30–32]) (Figure 4A). Likewise, lipid composition can be freely varied (Figure 4B). For example, the inclusion of signaling lipids, such as phosphoinositides, allows visualization of how such lipids regulate membrane proteins, as was recently done to show how PIP2 regulates mammalian G protein-gated inward rectifier potassium channels [33]. By including cholesterol and sphingolipids, it should be possible to generate a raft-like membrane environment and to visualize its effect on incorporated proteins. Membrane thickness can be altered by using lipids with longer or shorter acyl chains to visualize the effect of hydrophobic mismatch on a protein (Figure 4C).
Figure 4. Engineering custom membranes.

I: During nanodisc assembly, many lipid bilayer characteristics can be custom designed. (A) The MSP will define the nanodisc diameter. (B) The lipid composition can be freely chosen, allowing for incorporation of special lipids. (C) The length of the lipid acyl chain will define the hydrophobic thickness of the membrane. Nanodisc reconstitution with a short-chain lipid stabilized MscS in the shown subconducting state. In contrast, reconstitution with a long-chain lipid preserved MscS in the closed state, shown in the nanodisc at the center. II: The membrane can be modified after nanodisc assembly, which will affect both leaflets. (D) Incubation with empty cyclodextrins (CDs) will extract lipids from the membrane, thus increasing membrane tension. This approach stabilized MscS in the shown desensitized conformation. (E) Incubation of nanodiscs with lipid-loaded CDs will exchange lipids in the nanodiscs. Coincubation with lipid-loaded CDs and lipid vesicles would drive lipid addition to the nanodiscs. III: To modify just one leaflet, the protein is reconstituted into vesicles, the outer leaflet is modified, and amphiphilic copolymers are used to form native nanodiscs. (F-H) YnaI reconstituted into vesicles has a closed conformation (F). Incubation with lysophospholipids induced channel opening (G). Extraction with amphiphilic copolymers generated native nanodiscs preserving YnaI in the shown open conformation (H). (I) CDs catalyze the exchange of the outer leaflet lipids between acceptor proteoliposomes and donor vesicles, generating an asymmetric membrane for the embedded protein. (J) Subsequent solubilization with amphiphilic copolymers should yield native nanodiscs of defined leaflet asymmetry.
Such ideas were recently employed in a study of the bacterial mechanosensitive channel MscS, which opens in response to an increase in membrane tension [34]. Cryo-EM analysis of MscS in nanodiscs formed with lipids of different acyl-chain lengths confirmed that the channel does not respond to changes in the hydrophobic thickness of the membrane, which had previously been shown by electrophysiology [35]. However, the membrane formed using a lipid with just 10-carbon atom acyl chains serendipitously stabilized the channel in a sub-conducting state, revealing the structure of MscS in a new functional state [36] (Figure 4C).
For some applications, the lipid environment must be modified after nanodisc assembly. Here, modifications will affect both bilayer leaflets (Figure 4, Section II). For example, in the aforementioned study, β-cyclodextrin (βCD) was used to remove lipids from MscS-containing nanodiscs to mimic membrane tension (Figure 4D). The sustained tension drove the channel through the open into the desensitized state, which could then be visualized by cryo-EM [36]. Importantly, in a follow-up experiment designed to show that βCD creates membrane tension by removing lipids from the membrane rather than by simply interdigitating in between the lipids, excised membrane patches were incubated with βCD that was preloaded with cholesterol. Electrophysiological recordings showed that cholesterol-loaded βCD did not activate MscS, but rather the channels became more resistant to activation [37]. Because cholesterol is known to decrease the mechanosensitivity of MscS, this result suggests that βCD deposited cholesterol into the nanodiscs (presumably in equilibrium with extracting lipids from the nanodiscs). More importantly, this experiment shows that βCD can be used not only to remove lipids from nanodiscs, but also to exchange and add lipids to preformed nanodiscs (Figure 4E).
In some cases, it may be desired to modify only one of the two leaflets. This can be achieved by modifying vesicles, which only allow access to their outer leaflet, followed by extraction with amphiphilic copolymers to generate native nanodiscs containing asymmetric membranes (Figure 4, Section III). This idea was beautifully demonstrated for the bacterial mechanosensitive channel YnaI. YnaI solubilized from E. coli plasma membranes with diisobutylene/maleic acid (DIBMA) copolymer yielded a cryo-EM structure of the channel in the closed resting conformation [38]. To visualize its open state, YnaI was reconstituted into vesicles (Figure 4F), which were then incubated with lysophosphatidylcholine (Figure 4G), known to activate mechanosensitive channels (e.g., [35,39]). After solubilizing the proteoliposomes with DIBMA (Figure 4H), cryo-EM structures of the resulting native nanodiscs showed YnaI not only in the closed state, but also in open-like and intermediate states. This study thus visualized how a change in the surrounding membrane environment directly affects the structure of the embedded membrane protein [38]. Importantly, in view of studies that explored characteristics of membrane solubilization by amphiphilic copolymers (e.g., [40]) and the kinetics of lipid exchange, which established much faster lipid exchange between native nanodiscs than between MSP-based nanodiscs and vesicles [41], this study also demonstrated that the effect of membrane perturbation on a vesicle-embedded protein can persist through its solubilization into native nanodiscs.
Concluding Remarks
The confluence of advances in cryo-EM and nanodisc technology promise to improve our mechanistic understanding of membrane proteins in the context of physiologic lipid bilayers. Initial results have already provided insights into the effects of the membrane environment on protein structure unattainable with detergents or membrane mimetics and have facilitated direct testing of novel structural hypotheses through membrane engineering. We therefore argue that nanodiscs should become the field standard for the stabilization of membrane proteins for structural studies.
Despite their promise, there are still other membrane characteristics that have not yet been reproduced in nanodiscs. For example, there is currently no method that would make it possible to recreate membrane potential in nanodiscs, for which the best hope lies in the use of small vesicles [16,42]. Small vesicles may also be needed to study the effect of membrane curvature on the embedded membrane proteins, but it may well be that curvature-inducing proteins could do so in sufficiently large nanodiscs. Perhaps the final frontier in mimicking biological membranes may be the generation of asymmetric leaflets. In mammalian cells, charged lipids are almost exclusively localized to the cytoplasmic leaflet, whereas the extracellular leaflet contains neutral lipids, including sphingolipids, in a tight packing [43]. One approach to engineer lipid asymmetry in vitro employs shuttling lipids with CDs. CDs are well known as a tool to remove cholesterol from the plasma membrane, but they have also been used to introduce lipids into cells [44] and to prepare small unilamellar vesicles with stable asymmetric lipid compositions [45,46]. The principle is to mix unilamellar acceptor vesicles with the lipid composition desired for the inner leaflet with an excess of typically multilamellar donor vesicles with the composition desired for the outer leaflet and to use CD to exchange the lipids between the outer leaflets of the two vesicle populations. After removal of the multilamellar donor vesicles, the remaining unilamellar vesicles will feature an asymmetric membrane. By incorporating a membrane protein into the acceptor vesicles, exchanging the outer leaflet lipids (Figure 4I) and solubilizing them with amphiphilic copolymers, this approach may make it possible to produce native nanodiscs in which the membrane protein is embedded in a defined, asymmetric lipid bilayer (Figure 4J). While open questions remain, such as whether the membrane asymmetry is retained throughout this procedure and persists long enough for structural studies, this example illustrates the almost unlimited opportunities to study the structure of membrane proteins in custom-designed lipid bilayers.
Outstanding Questions.
How do membrane environments affect the embedded proteins, and how might we best use the tools at our disposal to replicate these diverse states in vitro for structural studies?
Could the techniques used to produce and vitrify proteoliposomes be further improved to facilitate their use in structural studies, particularly those requiring membrane curvature, leaflet asymmetry, or transmembrane gradients?
Are curvature-inducing proteins a good approach to curve lipid bilayers in large nanodiscs? Are there other ways to introduce curvature into a nanodisc membrane?
Can we better understand the mechanism by which copolymers extract lipid islands from membranes? Can we further define the preferences with which amphiphilic copolymers extract certain lipids over others? When amphiphilic copolymers (e.g., SMA) are used to generate native protein-containing nanodiscs, for how long is leaflet asymmetry maintained? How are these properties affected by the chemical composition of the copolymer?
Are there lipid environments that are not chemically compatible with MSP-based or native nanodiscs? For example, can they be used to recreate the unique physiochemical properties of lipid rafts?
Why have native nanodiscs allowed visualization of protein conformations not observed in MSP-based nanodiscs? What membrane characteristic(s) do native nanodiscs preserve that are not reproduced in chemically defined MSP nanodiscs?
Is the innate heterogeneity of hydrophilic copolymers and their derived native nanodiscs a hurdle to be overcome, or a technical benefit in the era of high-throughput data collection and advanced image processing?
What effect do bilayer mechanics have on protein dynamics and stability, and how do membrane mechanics affect protein domains not directly in contact with the membrane?
Highlights.
Single-particle cryo-electron microscopy (cryo-EM) studies of membrane proteins have become routine and often yield structures at near-atomic resolution.
Cryo-EM structures of membrane proteins in lipid nanodiscs often yield information not seen in structures obtained with detergent-solubilized proteins.
Nanodiscs not only provide a native-like lipid environment for membrane proteins, but also make it possible to custom design and modify most aspects of the lipid bilayer.
Because of the facility of generating and imaging nanodiscs by cryo-EM and because of the novel mechanistic insights into membrane protein function they can provide, nanodiscs ought to become the standard for membrane protein structure determination by single-particle cryo-EM.
Acknowledgments
RQN acknowledges support from the T32 Investigational Cancer Therapeutics Training Program at Memorial Sloan-Kettering Cancer Center (NIH T32CA009207).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kühlbrandt W (2014) The resolution revolution. Science 343, 1443–1444 [DOI] [PubMed] [Google Scholar]
- 2.Chae PS et al. (2010) Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods 7, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chae PS et al. (2012) A new class of amphiphiles bearing rigid hydrophobic groups for solubilization and stabilization of membrane proteins. Chemistry 18, 9485–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tribet C et al. (1998) Scanning transmission electron microscopy study of the molecular mass of amphipol/cytochrome b6f complexes. Biochimie 80, 475–482 [DOI] [PubMed] [Google Scholar]
- 5.Frauenfeld J et al. (2016) A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 13, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson ML et al. (2018) The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife 7, e34085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen SB et al. (2011) Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landau EM and Rosenbusch JP (1996) Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A 93, 14532–14535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders CR and Landis GC (1995) Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry 34, 4030–4040 [DOI] [PubMed] [Google Scholar]
- 10.Raunser S and Walz T (2009) Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys 38, 89–105 [DOI] [PubMed] [Google Scholar]
- 11.Hite RK et al. (2010) Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J 29, 1652–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hite RK et al. (2015) Effect of lipid head groups on double-layered two-dimensional crystals formed by aquaporin-0. PLoS One 10, e0117371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang QX et al. (2001) Spherical reconstruction: a method for structure determination of membrane proteins from cryo-EM images. J Struct Biol 133, 119–131 [DOI] [PubMed] [Google Scholar]
- 14.Wang L and Sigworth FJ (2009) Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature 461, 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonggu L and Wang L (2018) Broken symmetry in the human BK channel. SSRN Journal DOI: 10.2139/ssrn.3310822 [DOI] [Google Scholar]
- 16.Wang L (2018) Random spherically constrained single-particle (RSC) method to study voltage-gated ion channels. Methods Mol Biol 1684, 265–277 [DOI] [PubMed] [Google Scholar]
- 17.Civjan NR et al. (2003) Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques 35, 556–560, 562–563 [DOI] [PubMed] [Google Scholar]
- 18.Frauenfeld J et al. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y et al. (2016) TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi W et al. (2017) Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen T et al. (2019) Structure of the Mechanosensitive Channel MscS Embedded in the Membrane Bilayer. J Mol Biol 431, 3081–3090 [DOI] [PubMed] [Google Scholar]
- 22.Reddy B et al. (2019) Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS. Elife 8, e50486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staus DP et al. (2020) Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin P et al. (2017) Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles TJ et al. (2009) Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc 131, 7484–7485 [DOI] [PubMed] [Google Scholar]
- 26.Brown CJ et al. (2021) Structural biology of endogenous membrane protein assemblies in native nanodiscs. Curr Opin Struct Biol 69, 70–77 [DOI] [PubMed] [Google Scholar]
- 27.Guo Y (2021) Detergent-free systems for structural studies of membrane proteins. Biochem Soc Trans 49, 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu W et al. (2018) Structure and activity of lipid bilayer within a membrane-protein transporter. Proc Natl Acad Sci U S A 115, 12985–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J et al. (2021) Mechanism of gating and partial agonist action in the glycine receptor. Cell 184, 957–968.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denisov IG et al. (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 31.Hagn F et al. (2018) Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat Protoc 13, 79–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padmanabha Das KM et al. (2020) Large Nanodiscs: A Potential Game Changer in Structural Biology of Membrane Protein Complexes and Virus Entry. Front Bioeng Biotechnol 8, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu Y et al. (2020) Cryo-EM analysis of PIP2 regulation in mammalian GIRK channels. Elife 9, e60552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukharev SI et al. (1993) Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys J 65, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nomura T et al. (2012) Differential effects of lipids and lyso-lipids on the mechanosensitivity of the mechanosensitive channels MscL and MscS. Proc Natl Acad Sci U S A 109, 8770–8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y et al. (2021) Visualization of the mechanosensitive ion channel MscS under membrane tension. Nature 590, 509–514 [DOI] [PubMed] [Google Scholar]
- 37.Cox CD et al. (2021) Cyclodextrins increase membrane tension and are universal activators of mechanosensitive channels. Proc Natl Acad Sci U S A 118, e2104820118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flegler VJ et al. (2020) The MscS-like channel YnaI has a gating mechanism based on flexible pore helices. Proc Natl Acad Sci U S A 117, 28754–28762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perozo E et al. (2002) Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol 9, 696–703 [DOI] [PubMed] [Google Scholar]
- 40.Cuevas Arenas R et al. (2016) Influence of lipid bilayer properties on nanodisc formation mediated by styrene/maleic acid copolymers. Nanoscale 8, 15016–15026 [DOI] [PubMed] [Google Scholar]
- 41.Cuevas Arenas R et al. (2017) Fast Collisional Lipid Transfer Among Polymer-Bounded Nanodiscs. Sci Rep 7, 45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao X et al. (2020) Cryo-EM analysis of a membrane protein embedded in the liposome. Proc Natl Acad Sci U S A 117, 18497–18503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doktorova M et al. (2020) Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat Chem Biol 16, 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kainu V et al. (2010) Introduction of phospholipids to cultured cells with cyclodextrin. J Lipid Res 51, 3533–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng H-T et al. (2009) Preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. J Biol Chem 284, 6079–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doktorova M et al. (2018) Preparation of asymmetric phospholipid vesicles for use as cell membrane models. Nat Protoc 13, 2086–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]


