ABSTRACT
Background
Periodontitis is caused by a dysbiotic shift in the dental plaque microbiome. Fusobacterium nucleatum is involved in the colonization of Porphyromonas gingivalis, which plays a key role in dysbiosis, via coaggregation and synergy with this microorganism.
Aim
We investigated the effect of diffusible signaling molecules from P. gingivalis ATCC 33277 on F. nucleatum TDC 100 to elucidate the synergistic mechanisms involved in dysbiosis.
Methods
The two species were cocultured separated with an 0.4-µm membrane in tryptic soy broth, and F. nucleatum gene expression profiles in coculture with P. gingivalis were compared with those in monoculture.
Results
RNA sequencing revealed 139 genes differentially expressed between the coculture and monoculture. The expression of 52 genes was upregulated, including the coaggregation ligand-coding gene. Eighty-seven genes were downregulated. Gene Ontology analysis indicated enrichment for the glycogen synthesis pathway and a decrease in de novo synthesis of purine and pyrimidine.
Conclusion
These results indicate that diffusible signaling molecules from P. gingivalis induce metabolic changes in F. nucleatum, including an increase in polysaccharide synthesis and reduction in de novo synthesis of purine and pyrimidine. The metabolic changes may accelerate biofilm formation by F. nucleatum with P. gingivalis. Further, the alterations may represent potential therapeutic targets for preventing dysbiosis.
KEYWORDS: Synergy, biofilm, periodontal disease, Fap2, Porphyromonas gingivalis, Fusobacterium nucleatum
Introduction
Periodontitis is a highly prevalent disease, with a global age-standardized prevalence of approximately 10% [1]. Inflammation caused by this disease disrupts the periodontal ligament, induces resorption of the alveolar bone, and results in tooth loss [2,3]. A major feature of this disease is dysbiosis, which is a shift in the composition and abundance of the subgingival microbiota toward more pathogenic species [4]; however, the details of the dysbiotic shift are yet to be elucidated. Understanding the intricacies of dysbiosis is essential for developing treatment and prevention strategies for this condition. Periodontopathic bacteria implicated in dysbiosis include Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Filifactor alocis, which are abundant in periodontitis lesions [5–7]. Among them, P. gingivalis is a key microorganism for the induction of this dysbiotic shift of the microbiome [8].
Coaggregation, which is adherence between bacteria, is essential for dental plaque formation [9–11]. Fusobacterium nucleatum is one of the most frequently detected bacterial species in both the healthy oral microbiome and periodontal disease lesions. This microorganism is associated with systemic conditions such as preterm birth and colorectal cancer [5,12–14]. In an in vivo study, F. nucleatum was detected between the basal layer and top layer of the dental plaque, and P. gingivalis was detected in the top layer [15]. In another in vivo study, Fusobacterium was detected in the annulus area between the tooth side and peripheral area of the plaque. Porphyromonas was detected in the peripheral area and was found adjacent to Fusobacterium [16]. In vitro studies revealed that F. nucleatum may coaggregate with periodontopathic bacteria such as P. gingivalis, T. denticola, and Prevotella intermedia [17–19] and exert a synergistic effect with periodontopathic bacteria, including P. gingivalis, on pathogenicity or biofilm formation [18,20–22]. F. nucleatum has a coaggregation ligand Fap2 that mediates the adhesion of F. nucleatum to P. gingivalis and red blood cells [17] and promotes the growth of P. gingivalis in aerobic and CO2 depleted conditions [23]. The localization of F. nucleatum and P. gingivalis in the plaque structure and their coaggregation and synergy indicate that F. nucleatum assists the colonization of P. gingivalis. In addition, Feuille et al. found an increase in the virulence of P. gingivalis cocultured with F. nucleatum [24]. Metzger et al. also reported the synergistic pathogenicity of P. gingivalis and F. nucleatum using a murine subcutaneous chamber model [21]. Recent proteomic analyses have provided insights into synergic interactions via cross-feeding among species such as Streptococcus gordonii, F. nucleatum, and P. gingivalis using a chemically defined medium or brain heart infusion broth [25,26]. In these studies, S. gordonii showed synergism with F. nucleatum by providing ArcD-regulated ornithine as an energy source, whereas F. nucleatum facilitated P. gingivalis biofilm formation by providing putrescine. These studies provide insights into how dysbiosis is involved in biofilm formation. However, the effect of P. gingivalis on F. nucleatum was not fully investigated.
Previously, we showed that F. nucleatum TDC 100 isolated from apical periodontitis lesions strongly adheres to type-I collagen and shows enhanced biofilm formation through a synergistic relationship with P. gingivalis [27]. Coinfection of these species also enhanced the invasion of epithelial cells by P. gingivalis [28,29]. Enhanced F. nucleatum biofilm formation by P. gingivalis was observed in a coculture separated with an 0.4-µm membrane, which was significantly higher than that observed for other bacterial species [27], indicating that a factor produced by P. gingivalis plays an important role in the synergy between F. nucleatum and P. gingivalis. Therefore, in this study, we investigated the effect of diffusible signaling molecules of P. gingivalis on the mRNA and protein expression in F. nucleatum to clarify the mechanism of enhanced F. nucleatum biofilm formation by P. gingivalis.
Materials and methods
Bacterial strains and culture conditions
F. nucleatum TDC 100 [27] and P. gingivalis ATCC 33277 [30] were maintained on tryptic soy agar (BD Bioscience, San Jose, CA) containing 10% defibrinated horse blood, 5 µg/mL hemin, and 0.5 µg/mL menadione. All strains were cultivated at 37°C under anaerobic conditions (H2: 10%, CO2: 10%, N2: 80%) in an anaerobic chamber (ANX-4; Hirasawa, Tokyo, Japan).
Induction of biofilm formation using a two-compartment system
The interaction of the diffusible signaling molecules between F. nucleatum and P. gingivalis was evaluated via coculture separated with an 0.4-µm membrane using a two-compartment system as described previously [27]. F. nucleatum and P. gingivalis were inoculated into tryptic soy broth (TSB) containing 5 µg/mL hemin and 0.5 µg/mL menadione (TSBhm) and precultured anaerobically for 1 day at 37°C. Then, each culture was diluted with fresh TSBhm to an OD 660 = 0.1 and cultured anaerobically. After 24 h, each culture was diluted 1:2 with fresh TSBhm. Next, 750 µL of the resulting F. nucleatum culture was added into the wells (lower well) of a 12-well plate, coated with type-I collagen and polystyrene (Iwaki, Tokyo, Japan). Upper wells (Transwell, Corning, NY) were then inserted into the lower well, followed by the addition of 750 µL of the resulting P. gingivalis culture to the upper wells. The organisms were then cocultured while being physically separated by an 0.4-µm pore size membrane. For monoculture, only 750 µL of TSBhm was added to the upper well. After anaerobic incubation at 37°C for 2 days, the upper wells were removed. Biofilm formation was evaluated using crystal violet staining with Spectra MAX M5 (Molecular Device, Sunnyvale, CA) [31].
F. nucleatum biofilm protein profiling
F. nucleatum biofilms were lysed with a sample buffer containing a reducing agent, and the lysate proteins were resolved using 10–20% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE). The protein profiles were determined by staining the gel with Coomassie Blue. In the cocultured F. nucleatum protein profile, the 39-kDa protein showed considerably enhanced staining intensity. To identify the protein band, the electrophoresed bands were transferred from the gel to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore, Billerica, MA) using Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad, Hercules, CA). After staining the membrane with Coomassie Blue, the 39-kDa protein band was excised from the membrane, and the N-terminal amino acids were sequenced using the automatic peptide sequencer Procise 494 cLC (Applied Biosystems, Foster City, CA). The peptide sequences were matched with the genomic sequences of F. nucleatum in the NCBI database (http://blast.ncbi.nlm.nih.gov).
Based on the obtained gene sequence, an antibody against the protein was prepared. The peptide ESEVKNWRWQPTAW showed the highest similarity to residues 352–365 of Fusobacterium periodonticum outer membrane protein A (FomA), which has an additional N-terminal Cys residue. This sequence was identical between F. nucleatum and F. periodonticum except for the sixth residue. The peptide was synthesized using F-moc chemistry and was conjugated to keyhole limpet hemocyanin using maleimidobenzoic acid N-hydroxy succinimide ester, and the antibody was prepared as described previously [32]. To quantify FomA, immunoblotting was performed as described earlier. Ten micrograms of biofilm organized in the lower well, which was monocultured and cocultured with P. gingivalis, was separated using SDS-PAGE and transferred onto a PVDF membrane. FomA was stained with anti-FomA antibody (1:1,000 dilution) and goat anti-rabbit IgG (1:3,000 dilution). The membrane was scanned, and the staining intensity of the developed 39-kDa bands was evaluated using Image-Quant TL software v.8.1 (GE Healthcare, Little Chalfont, UK).
RNA-sequencing
F. nucleatum TDC 100 was grown as a monoculture and cocultured with P. gingivalis ATCC 33277 for 2 days using the two-compartment system. After removing the upper wells, three wells containing F. nucleatum were combined into one sample for RNA extraction, and the total RNA was isolated using TRIzol (ThermoFisher Scientific, Tokyo, Japan). The total RNA was obtained from three independent replicates of monocultured and cocultured F. nucleatum. DNA was removed using the TURBO DNA-free kit (Thermo Fisher Scientific, Waltham, MA), and ribosomal RNA was extracted from the sample using the Ribo-Zero rRNA Removal kit (Bacteria) (Illumina). A library was constructed from the extracted RNA using the Truseq Stranded mRNA Sample Prep kit (Illumina) according to the manufacturer’s instructions. The obtained library was sequenced on HiSeq 2500 (Illumina); 100-bp paired-end reads were trimmed using cutadapt (https://cutadapt.readthedocs.org/en/stable/) and Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic). As the genomic DNA of F. nucleatum TDC 100 has not been sequenced, the genomes of F. nucleatum subsp. nucleatum ATCC 25586, F. nucleatum subsp. vincentii ATCC 49256, F. nucleatum subsp. vincentii 3_1_36A2, and F. nucleatum KCOM 2931 were used as references to map the obtained sequences. Among these subspecies, F. nucleatum KCOM 2931 showed the highest mapping rate (89.7%), and its 16S rRNA sequence showed 99% identity with that of F. nucleatum TDC 100. Thereafter, the obtained sequences were mapped using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) and Bowtie1 (http://bowtie-bio.sourceforge.net/index.shtml) with F. nucleatum KCOM 2931 as the reference strain. From the mapped sequences, the gene expression in monocultured and cocultured F. nucleatum was evaluated using cufflinks and cuffdiff. The difference in expression between monoculture and coculture was compared using TCC-GUI [33]. Normalization was carried out using the TMM method, and differentially expressed genes were identified using edgeR [34], where findings with a p-value of 0.05 were considered significant and the false discovery rate (FDR) was set to 0.05, with differences in gene expression evaluated using log2 fold ≥1.5. Differentially expressed genes were subjected to Gene Ontology (GO) analysis using goatools v. 1.1.6 [35].
Statistical analysis
To investigate the effect of coculture on biofilm formation, the level of biofilm formation and FomA expression were analyzed using Student’s t-test at a 5% level of significance.
Results
Effect of P. gingivalis coculture on F. nucleatum protein profile
Biofilm formation under P. gingivalis coculture was greater than that under F. nucleatum TDC 100 monoculture (Figure 1b, p < 0.01). A comparison of protein profiles revealed a band at approximately 39 kDa with increased staining intensity for F. nucleatum TDC 100 cocultured with P. gingivalis ATCC 33277 (Figure 1b). The N-terminal amino acid sequence of the band at 39 kDa was EVTPAWRPNG. The BLAST analysis revealed that this sequence was identical to that of residues 49–58 of FomA. The predicted molecular mass of amino acids 1–48 was 5179.17, indicating that the 39-kDa band was FomA without these 48 amino acid residues. The antibody reactivity to the 39-kDa band (Figure 2a) also supported this result.
Figure 1.
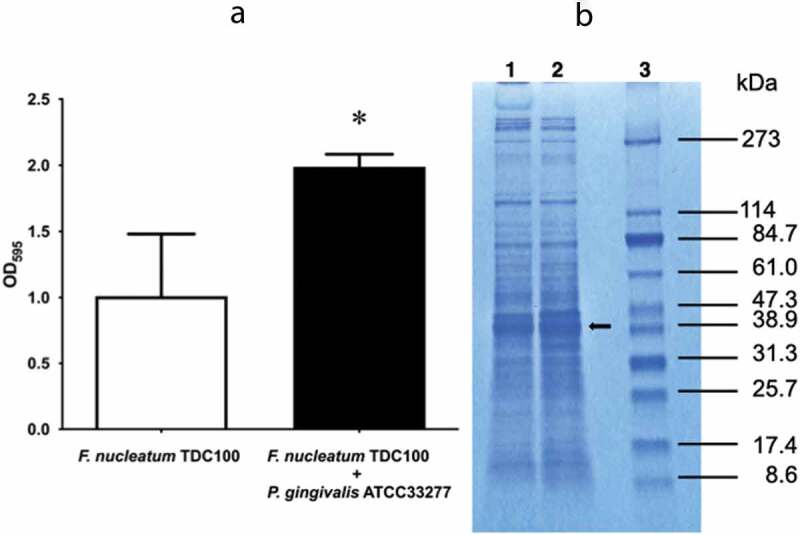
Biofilm formation by Fusobacterium nucleatum with/without Porphyromonas gingivalis in a two-compartment assay system (a) and the protein profile determined using sodium dodecyl sulfate polyacrylamide gel electrophoresis (b). (a) Biofilm mass of cocultured F. nucleatum TDC 100 evaluated using crystal violet staining. Student’s t-test was used for inter-group comparisons. *p < 0.05. Data are shown as mean ± standard deviation. (b) Lanes 1: F. nucleatum TDC 100 alone; 2: F. nucleatum TDC 100 with P. gingivalis ATCC 33277; 3: molecular size marker. Arrow indicates the 39-kDa protein.
Figure 2.
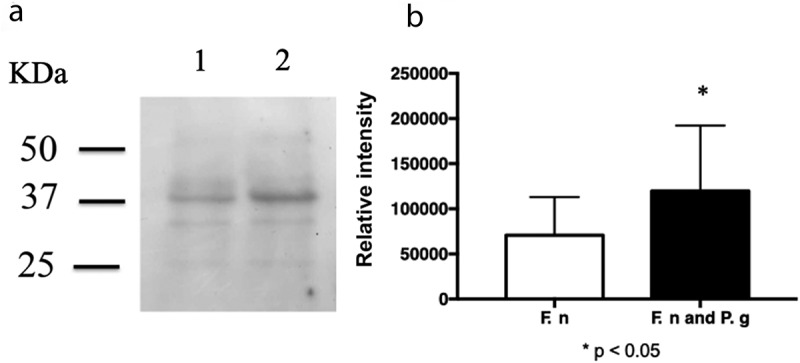
Quantification of FomA using immunoblotting. (a) Immunoblot analysis of Fusobacterium nucleatum TDC 100 cocultured with Porphyromonas gingivalis ATCC 33277. Lanes 1: F. nucleatum TDC 100 alone; 2: F. nucleatum TDC 100 with P. gingivalis ATCC 33277. The blotted membrane was detected using an antibody against F. nucleatum FomA, and the reacted band was detected using horseradish peroxidase conjugated goat anti-rabbit antibody. (b) A comparison of the intensity of the 39-kDa bands. Data are shown as mean ± standard deviation. *p < 0.05 by Student’s t-test.
The level of the major outer membrane protein FomA was investigated using immunoblotting with rabbit anti-FomA antibody. Image analysis of the immunoblots using Imagequant TL showed that staining of the 39-kDa and faint 35-kDa bands detected with the anti-FomA antibody in cocultured F. nucleatum was 1.7 times more intense than that for F. nucleatum monoculture (Figure 2b).
RNA-sequencing of F. nucleatum TDC 100 cocultured with P. gingivalis ATCC 33277
To evaluate the effect of P. gingivalis on F. nucleatum in coculture, RNA-sequencing analysis was performed. At FDR <0.05, 176 differentially expressed genes were obtained (Figure 3a). Among these, the genes from the coculture showing a log2-fold difference of >1.5 compared with their expression in the monoculture of F. nucleatum, p < 0.05, and FDR <0.05 were selected (Figure 3b). In F. nucleatum cocultured with P. gingivalis, 139 genes (4.9%) were differentially expressed compared with those in monocultured F. nucleatum. Among them, 52 genes were upregulated (Table 1), including the gene encoding the galactose inhibitable autotransporter adhesion Fap2, a protein involved in porphyrin metabolism (cobK and Cobart-precorrin 5A hydrolase), chaperone proteins (ClpB, DnaK, and DnaJ), proteins involved in polysaccharide synthesis (glycogen synthase, GlgG, glucose-1-phosphate adenylyltransferase, 1,4-alpha-glucan branching protein GlgB, and GalU), and proteins involved in membrane transport (basic amino acid ABC transporter substrate-binding protein, amino acid ABC transporter ATP-binding protein, DMT family transporter, AzlC family ABC transporter permease, branched-chain amino acid transporter permease, basic amino acid ABC transporter substrate-binding protein, efflux RND transporter permease subunit, ABC transporter ATP-binding protein, energy-coupling factor transporter transmembrane protein EcfT, autotransporter serine protease fusolisin, ABC transporter permease, and ABC transporter ATP-binding protein). Interestingly, the expression of fomA did not show an increase in the RNA-sequencing.
Figure 3.
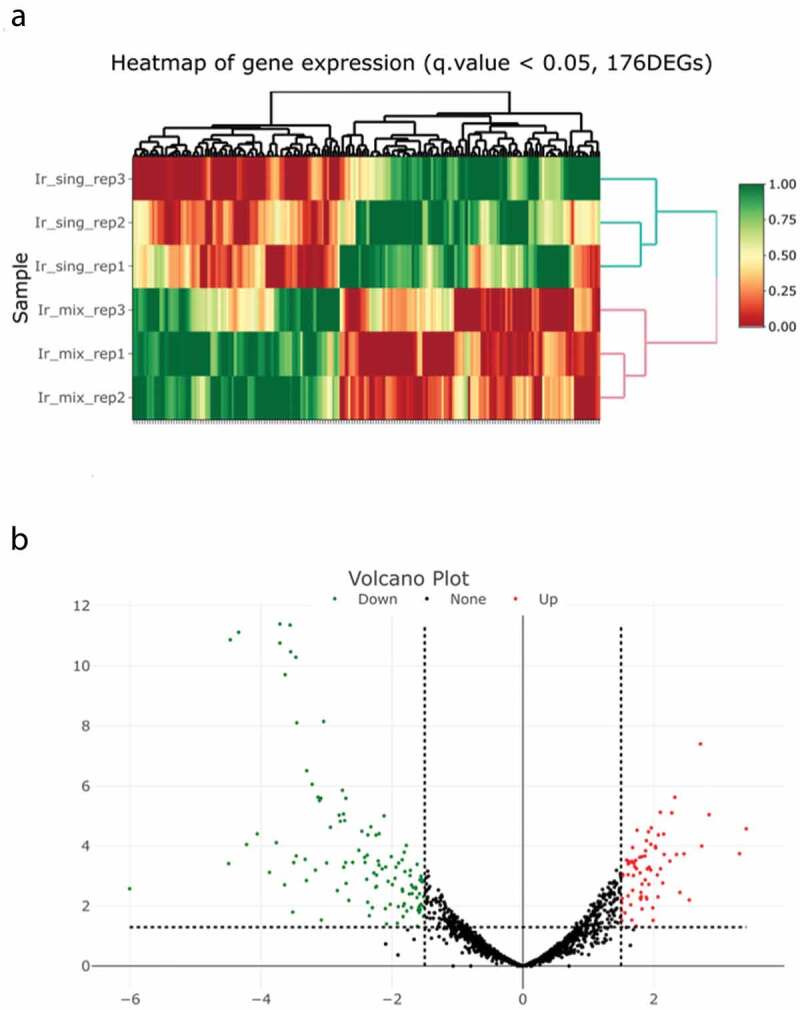
Genes differentially expressed between cocultured and monocultured Fusobacterium nucleatum. (a) Hierarchical clustering of differentially expressed genes at a false discovery rate (FDR) of <0.05 between cocultured and monocultured F. nucleatum. Ir_sing: monocultured, Ir_mix: cocultured, red to green color scale indicates the relative level of gene expression. (b) Volcano plots show differentially expressed genes (p < 0.05, FDR <0.05). Horizontal dotted lines show the p-value cut-off of 0.05, and vertical dotted lines show the expression level log2 (fold change) cut-off of 1.5. Red and green dots indicate upregulated and downregulated, respectively.
Table 1.
Upregulated genes in cocultured Fusobacterium nucleatum.
| Gene no. or ID | Annotation | log2(co/mono*) |
|---|---|---|
| CS401_RS09410 | Hypothetical protein | 3.414 |
| clpB | ATP-dependent chaperone ClpB | 3.312 |
| CS401_RS03075 | Basic amino acid ABC transporter substrate-binding protein | 2.843 |
| DnaK | Molecular chaperone DnaK | 2.733 |
| CS401_RS01585 | Glucose-1-phosphate adenylyltransferase | 2.713 |
| CS401_RS03070 | M20 family metallopeptidase | 2.462 |
| CS401_RS09415 | Hypothetical protein | 2.399 |
| metK | Methionine adenosyltransferase | 2.346 |
| CS401_RS01590 | 1,4-alpha-glucan branching protein GlgB | 2.323 |
| CS401_RS05055 | Galactose inhibitable autotransporter adhesin Fap2 | 2.276 |
| CS401_RS06090 | V-type ATP synthase subunit D | 2.246 |
| CS401_RS06310 | FAD-dependent oxidoreductase | 2.159 |
| CS401_RS07400 | Energy-coupling factor transporter transmembrane protein EcfT | 2.151 |
| CS401_RS06095 | V-type ATP synthase subunit B | 2.136 |
| CS401_RS07100 | DUF1353 domain-containing protein | 2.103 |
| CS401_RS08440 | DUF523 domain-containing protein | 2.101 |
| CS401_RS03615 | Hypothetical protein | 2.07 |
| dnaJ | Molecular chaperone DnaJ | 2.054 |
| CS401_RS07395 | Glutathione peroxidase | 2.029 |
| CS401_RS00690 | MATE family efflux transporter | 2.026 |
| CS401_RS9850 | Multispecies: ABC transporter ATP-binding protein | 1.963 |
| CS401_RS03620 | Cytidylate kinase-like family protein | 1.956 |
| CS401_RS3610 | Efflux RND transporter permease subunit | 1.953 |
| CS401_RS07425 | GDYXXLXY domain-containing protein | 1.93 |
| raiA | Ribosome-associated translation inhibitor RaiA | 1.925 |
| glgG | Glucose-1-phosphate adenylyltransferase subunit GlgD | 1.919 |
| CS401_RS07390 | ABC transporter ATP-binding protein | 1.9 |
| CS401_RS09845 | ABC transporter permease | 1.887 |
| CS401_RS02470 | S1 RNA-binding domain-containing protein | 1.876 |
| CS401_RS02460 | Branched-chain amino acid transporter permease | 1.87 |
| CS401_RS01575 | Glycogen synthase | 1.865 |
| CS401_RS01335 | Basic amino acid ABC transporter substrate-binding protein | 1.821 |
| CS401_RS01340 | Amino acid ABC transporter ATP-binding protein | 1.818 |
| CS401_RS09420 | Hypothetical protein | 1.804 |
| CS401_RS02465 | GNAT family N-acetyltransferase | 1.791 |
| CS401_RS2450 | DMT family transporter | 1.785 |
| CS401_RS08435 | AAA family ATPase | 1.745 |
| CS401_RS03260 | ABC transporter ATP-binding protein/permease | 1.736 |
| cobK | Precorrin-6A reductase | 1.723 |
| CS401_RS07810 | Autotransporter serine protease fusolisin | 1.71 |
| cbiG | Cobalt-precorrin 5A hydrolase | 1.695 |
| CS401_RS02455 | AzlC family ABC transporter permease | 1.689 |
| CS401_RS03215 | AEC family transporter | 1.678 |
| CS401_RS03565 | Hypothetical protein | 1.677 |
| uvrC | Excinuclease ABC subunit | 1.665 |
| galU | UTP-glucose-1-phosphate uridylyltransferase | 1.656 |
| CS401_RS08075 | Txe/YoeB family addiction module toxin | 1.611 |
| nth | Endonuclease III | 1.611 |
| CS401_RS09665 | DNA polymerase III subunit alpha | 1.608 |
| CS401_RS02055 | Putative DNA modification/repair radical SAM protein | 1.585 |
| metG | Methionine – tRNA ligase | 1.531 |
| CS401_RS08695 | CoA transferase subunit A | 1.529 |
*co/mono: coculture/monoculture.
Eighty-seven genes were downregulated (Table 2), including those encoding protein involved in de novo synthesis of purine (phosphoribosyl amine-glucine ligase, purH, class I SAM-dependent methyltransferase, phosphoribosyl glycinamide formyl transferase, purM, amidophosphoribosyltransferase, phosphoribosylaminoimidazole-succinocarboxamide synthase, purE, and phosphoribosylformylglycinamidine synthetase), proteins involved in de novo pyrimidine synthesis (bifunctional pyr operon transcriptional regulator/uracil phosphoribosyltransferase PyrR, aspartate carbamoyltransferase, dihydroorotase, glutamine-hydrolyzing carbamoyl-phosphate synthase small subunit, carbamoyl-phosphate synthase large subunit, dihydroorotate dehydrogenase electron transfer subunit, dihydroorotate dehydrogenase, orotidine 5’-phosphate decarboxylase, and orotate phosphoribosyltransferase), bioA involved in biotin metabolism, and TonB-dependent receptor.
Table 2.
Downregulated genes in cocultured Fusobacterium nucleatum.
| Gene no. or ID | Annotation | log2(co/mono*) |
|---|---|---|
| CS401_RS04350 | DUF2194 domain-containing protein | −6.008 |
| purE | 5-(carboxyamino)imidazole ribonucleotide mutase | −4.496 |
| CS401_RS07440 | Queuosine precursor transporter | −4.47 |
| CS401_RS07445 | Radical SAM protein | −4.342 |
| CS401_RS04345 | Hypothetical protein | −4.221 |
| CS401_RS08470 | CoA-disulfide reductase | −4.058 |
| CS401_RS08465 | Helix-turn-helix transcriptional regulator | −3.871 |
| CS401_RS07475 | Sodium-dependent transporter | −3.767 |
| purF | Amidophosphoribosyltransferase | −3.713 |
| CS401_RS02225 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | −3.712 |
| CS401_RS04965 | Hypothetical protein | −3.641 |
| bioA | Adenosylmethionine—8-amino-7-oxononanoate transaminase | −3.633 |
| purM | Phosphoribosylfomylglycinamidine cyclo-ligase | −3.557 |
| CS401_RS02215 | Hypothetical protein | −3.548 |
| pelF | GT4 family glucosyltransferase PelF | −3.501 |
| CS401_RS02235 | Phosphoribosylformylglycinamidine synthetase | −3.469 |
| CS401_RS04330 | Hypothelical protein | −3.463 |
| CS401_RS02205 | Phosphoribosylglycinamide formyltransferase | −3.454 |
| pelG | Exopolysaccharide Pel transporter PelG | −3.327 |
| CS401_RS04355 | Hypothetical protein | −3.307 |
| CS401_RS10000 | Aspartate carbamoyltransferase catalytic subunit | −3.302 |
| megL | Methionine gamma-lyase | −3.219 |
| CS401_RS06670 | A24 family peptidase | −3.17 |
| CS401_RS10010 | Dihydroorotase | −3.13 |
| CS401_RS09550 | Na+/H+ antiporter NhaC family protein | −3.086 |
| CS401_RS02200 | Class I SAM-dependent methyltransferase | −3.044 |
| CS401_RS04325 | Endo alpha-1,4 polygalactosaminidase | −2.998 |
| CS401_RS10045 | Hypothetical protein | −2.939 |
| CS401_RS07480 | Tyrosine phenol-lyase | −2.834 |
| CS401_RS10015 | Glutamine-hydrolyzing carbamoyl-phosphate synthase small subunit | −2.806 |
| carA | Orotidine-5’-phosphate decarboxylase | −2.788 |
| purH | Bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | −2.757 |
| CS401_RS08865 | ABC transporter ATP-binding protein/permease | −2.741 |
| CS401_RS05375 | 6.7-dimethyl-8-ribityllumazine synthase | −2.736 |
| CS401_RS02575 | Alpha/beta hydrolase | −2.722 |
| CS401_RS08540 | Hypothetical protein | −2.708 |
| CS401_RS00210 | TonB-dependent receptor | −2.704 |
| ribD | Bifunctional diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase | −2.6 |
| CS401_RS10025 | Dihydroorotate dehydrogenase electron transfer subunit | −2.506 |
| CS401_RS10020 | Carbamoyl-phosphate synthase large subunit | −2.46 |
| CS401_RS08600 | 3-aminobutyl-CoA ammonia lyase | −2.419 |
| CS401_RS09220 | Transcription antiterminator | −2.411 |
| CS401_RS10030 | Dihydroorotate dehydrogenase | −2.377 |
| CS401_RS09160 | Cytochrome c biogenesis protein CcdA | −2.368 |
| CS401_RS02190 | Phosphoribosylamine – glucine ligase | −2.322 |
| pflA | Pyrvate formate-lyase-activating protein | −2.276 |
| CS401_RS05390 | Riboflavin synthase | −2.249 |
| cbpF | CEACAM-binding trimeric autotransporter adhesin CbpF | −2.244 |
| pflB | Formate C-acetyltransferase | −2.235 |
| CS401_RS06640 | Hypothelical | −2.234 |
| nifJ | Pyruvate:ferredoxin (flavodoxin) oxidoreductase | −2.21 |
| CS401_RS09995 | Bifunctional pyr operon transcriptional regulator/Uracil phosphoribosyltransferase PyrR | −2.198 |
| CS401_RS02270 | M42 family metallopeptidase | −2.196 |
| pyrE | Orotate phosphoribosyltransferase | −2.15 |
| CS401_RS07595 | HPr family phosphocarrier protein | −2.121 |
| CS401_RS09080 | RNA-binding protein | −2.095 |
| CS401_RS02570 | Hypothetical protein | −2.058 |
| CS401_RS08860 | ABC transporter ATP-binding protein/permease | −2.011 |
| CS401_RS05160 | Hypothetical protein | −1.994 |
| CS401_RS07575 | Hypothetical protein | −1.954 |
| CS401_RS08855 | Flavodoxin | −1.938 |
| CS401_RS06490 | Flavodoxin | −1.898 |
| CS401_RS03225 | Acetyl-CoA hydrolase/transferase family protein | −1.844 |
| CS401_RS08625 | Hypothetical protein | −1.841 |
| CS401_RS06480 | NrdH-redoxin | −1.838 |
| CS401_RS09340 | AAA family ATPase | −1.812 |
| CS401_RS06480 | Ribonucleoside-diphosphate reductase subunit alpha | −1.802 |
| CS401_RS03235 | Mechanosensitive ion channel | −1.781 |
| CS401_RS08655 | 3-keto-5-aminohexanoate cleavage protein | −1.735 |
| CS401_RS01275 | HAD-IB family hydrolase | −1.723 |
| CS401_RS07720 | Hypothetical protein | −1.719 |
| CS401_RS02565 | WGR domain-containing protein | −1.702 |
| CS401_RS04775 | DUF1871 family protein | −1.692 |
| CS401_RS0675 | Ribonucleotide-diphosphate reductase subunit beta | −1.677 |
| CS401_RS07725 | Hypothetical protein | −1.637 |
| CS401_RS08570 | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis protein | −1.635 |
| CS401_RS01055 | DUF1456 family protein | −1.615 |
| CS401_RS09170 | thiol:disulfide interchange protein | −1.579 |
| CS401_RS06365 | Hypothetical protein | −1.573 |
| CS401_RS05800 | ATP-dependent DNA helicase RecG | −1.549 |
| CS401_RS08910 | Osmolarity sensor protein EnvZ | −1.547 |
| CS401_RS00170 | glutamate dehydrogenase | −1.535 |
*co/mono: coculture/monoculture.
The GO enrichment analysis of the 139 differentially expressed genes showed significant changes in de novo synthesis of inosinic acid and uridylic acid and glycogen biosynthesis in biological process (p < 0.05, Figure 4).
Figure 4.
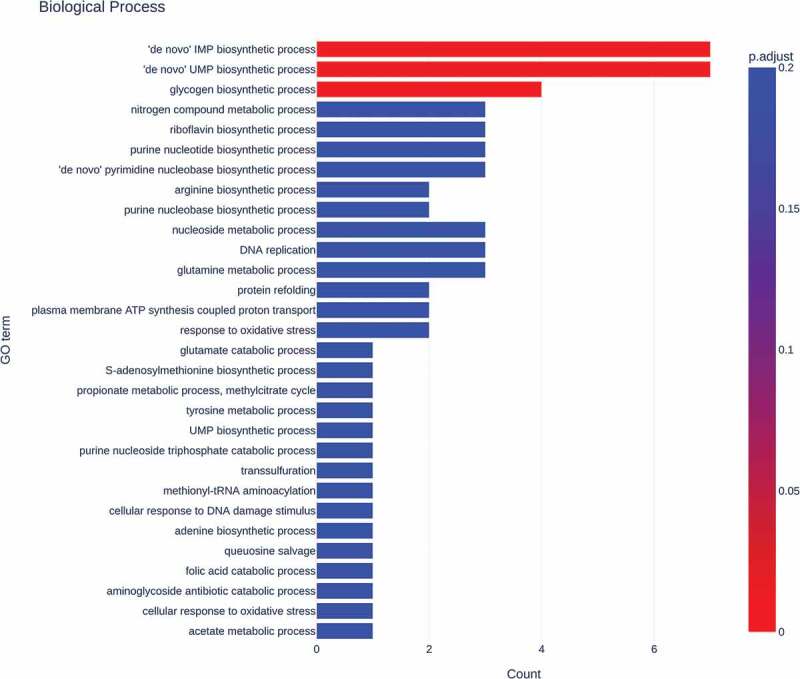
Gene ontology (GO) enrichment of differentially expressed genes. Gene count from GO in biological process. Gradient from red to blue on the bar indicates the adjusted p-value.
Discussion
In the present study, the effect of P. gingivalis diffusible signaling molecules on F. nucleatum was examined to clarify the synergy between these microorganisms involved in a dysbiotic shift of the dental plaque microbiome. F. nucleatum cocultured with P. gingivalis enhanced biofilm formation, and 139 differentially expressed genes were detected compared with those in monocultured F. nucleatum, including the ligand for adherence (fap2) to P. gingivalis. The GO analysis suggested sugar metabolism shift to glycogen biosynthesis, whereas the de novo purine and pyrimidine synthesis were decreased.
The expression of fap2 increased significantly in cocultured F. nucleatum. Fap2 is a galactose inhibitable adherence factor involved in coaggregation between F. nucleatum and P. gingivalis, hemagglutination by F. nucleatum [17], and localization of F. nucleatum to colon tumors [36]. The results indicate that the sensing of diffusible signaling molecules from P. gingivalis by F. nucleatum induces an increase in fap2. The increase in Fap 2 may contribute to binding between the species and enhance these intimate interactions, although further analysis is required to confirm the magnitude of contribution. Fap2 is also involved in hemagglutination by F. nucleatum [17]. P. gingivalis has hemagglutination activity, which is involved in heme acquisition [37]. In the cocultured F. nucleatum, cobK and cbiJ expression increased. These genes are involved in porphyrin metabolism. It is possible that hemagglutination is involved in iron acquisition in F. nucleatum. Therefore, the increase in Fap2 expression may be one of the adaptations for establishing a synergistic community of F. nucleatum and P. gingivalis.
Increased expression of genes involved in the glycogen synthase pathway in cocultured F. nucleatum was detected through the GO analysis, including those encoding GlgG, glucose-1-phosphate adenylyltransferase, GlgB, and GauU. The increase in GlgG and glucose-1-phosphate adenylyltransferase expression in F. nucleatum biofilms formed during coculture with P. gingivalis compared with their expression in a single-species biofilm of F. nucleatum has also been reported in a proteomic study [38]. GalU also plays an important role in glycogenesis and cell wall synthesis. Mutant galU affects biofilm formation in Escherichia coli [39]. In Pseudomonas aeruginosa, galU is required for lipopolysaccharide-core organization [40]. In Xanthomonas citri, galU is involved in exopolysaccharide and capsular polysaccharide synthesis [41]. galU is also essential for biofilm formation in Vibrio cholerae [42]. In F. nucleatum, the major energy source is amino acid catabolism; however, this microorganism can also metabolize fructose [43]. The study showed that fructose metabolism is stopped, and polysaccharides are organized under amino-acid-rich conditions. P. gingivalis has Arg-gingipain; Lys-gingipain [44,45]; dipeptidyl peptidases such as DPP4, DPP5, DPP7, and DPP11; as well as serine exopeptidases [46]. The strong proteolytic activity of P. gingivalis is considered to enhance F. nucleatum growth [12]. Here, the expression of genes involved in amino acid metabolism was not altered, but Hendrickson et al. [38] reported that the coculture of P. gingivalis and F. nucleatum inhibited more numbers of amino acid metabolic pathways compared with F. nucleatum alone. F. nucleatum and P. gingivalis were incubated in PBS for 18 h in their study, whereas in this study, microorganisms were inoculated in TSBhm for 2 days, and therefore, we observed large amounts of amino acids. In addition, the subspecies of F. nucleatum used were different between the studies. These differences in the experimental conditions may contribute to the difference in the results. The increase in the expression of genes involved in glycogen synthesis strongly suggested that the shift of energy source from sugar to amino acid was promoted by the increase in the levels of available amino acids upon coculture with P. gingivalis.
In contrast, the expression of pelF and pelG in F. nucleatum was reduced in the coculture. pelF and pelG are involved in the synthesis and transport of the exopolysaccharides Psl and Pel, and their protein production is regulated via cyclic-di-GMP in P. aeruginosa [47,48]. It is possible that the downregulation of these genes results in an increase in biofilm formation in cocultured F. nucleatum, although further analysis of the role of these genes in F. nucleatum is required.
Fusolisin, which is an autotransporter protease [49], showed increased expression in cocultured F. nucleatum. The expression of branched-chain amino acid transporter permease, basic amino acid ABC transporter substrate-binding protein, and amino acid ABC transporter ATP-binding protein was also increased in cocultured F. nucleatum. As mentioned earlier, P. gingivalis shows a strong proteolytic activity. These genes may be involved in peptide digestion or amino acid import. However, in previous proteomic analyses, the expression levels of these transporters did not change in F. nucleatum cocultured with P. gingivalis [38,50] or S. gordonii. In the previous studies, F. nucleatum was cocultured without a membrane, whereas in this study, F. nucleatum was cocultured with P. gingivalis separated by an 0.4-µm pore membrane. One study used Brucella broth [50] and another used Todd Hewitt broth [38]. In addition, in the former study, the samples of biofilms were harvested after 4 days of culture, and in the latter, samples were obtained after 18 h of incubation in PBS; in this study, we harvested samples after 2 days. This might explain the differences in the results.
Genes for de novo synthesis of inosinic acid and uridylic acid were downregulated in our study. These processes involve the synthesis of pyrimidine and purine using amino acids. Previous proteomic studies have found only minor changes in the proteome of F. nucleatum cocultured with P. gingivalis, primarily a decrease in the production of specific proteins [50]. In the present study, the numbers of decreased and increased proteins were comparable.
Here, the expression of genes involved in riboflavin synthesis from GTP was reduced, such as ribD and the riboflavin synthase gene. In eukaryotic cells, metabolic convergence of glutamine toward nucleotide biosynthesis observed in the process of malignant progression and inhibition of the shift reduced the proliferation of cancer cells [51]. It is possible that these reductions associated with purine and pyrimidine reflect changes in the external conditions caused by P. gingivalis, including the amino-acid-rich condition induced by the proteolytic activity, and the downregulation of genes involved in nucleotide biosynthesis from glutamine supports the glutamine for energy production. Furthermore, P. gingivalis and F. nucleatum produce indole that induces increased biofilm formation [52]. The biofilm mass in cocultured F. nucleatum was almost double that in the monoculture. In the biofilm, gradients of nutrients and oxygen induce a gradient of growth rate that results in fast-growing cells at the surface and slow-growing cells in the deeper layer of the biofilm [53]. It is possible that F. nucleatum growth decreases with increased biofilm formation and, therefore, the expression of the enzymes involved in pyrimidine and purine synthesis is decreased.
The expression of chaperones, including ClpB, DnaK, and DnaJ, was increased in the cocultured F. nucleatum. Although chaperones are generally cytoplasmic, six chaperones, including DnaK and ClpB, were detected in the matrix of F. nucleatum monoculture biofilm [54]. This indicates a relationship between chaperone translocation to the biofilm matrix and increased expression of these genes in F. nucleatum stimulated by P. gingivalis in coculture.
SDS-PAGE and immunoblotting indicated an increase in FomA expression in cocultured F. nucleatum. In the RNA-sequencing analysis, fomA expression did not show a difference between monoculture and coculture. FomA is a major porin protein in the matrix of biofilms formed by F. nucleatum [54]; however, an increase in FomA expression has not been observed in other coculture studies [55]. A recent study reported that the small RNA FoxI acts as a post-transcriptional repressor of FomA induced by oxygen [56]. Therefore, post-transcriptional regulation by FoxI or turnover of the protein in F. nucleatum cells may be responsible for the increase in FomA expression. In this analysis, an internal control was not evaluated, although the quantity of the protein was adjusted and the bands on SDS-PAGE showed almost identical levels. Therefore, further analysis using an internal control is required.
In the present study, F. nucleatum TDC 100 was used because of its strong synergy with P. gingivalis. The result of RNA-sequencing showed that this strain belongs to F. nucleatum subsp. vincentii. Previous studies on such gene or protein expression profiling used F. nucleatum subspecies nucleatum. Subspecies may show differing characteristics, including biofilm formation [57]. Human skeletons from the 18th and 19th centuries show high abundance of F. nucleatum subsp. vincentii with P. gingivalis and Prevotella pleuritidis, and this is maintained in modern samples [58]. F. nucleatum subsp. vincentii and P. gingivalis are useful for diagnosing periodontitis in mass patient screening using saliva samples [59]. High abundance of F. nucleatum subsp. vincentii is associated with periodontal lesions, and it further highlights the importance of this subspecies in the periodontopathic microbiome. To fully understand F. nucleatum-associated dysbiotic shift, further analysis of gene expression using multiple subspecies is required.
In this study, the gene expression was evaluated at a single time point before stationary phase, and therefore a single growth phase, of F. nucleatum. Several factors, other than coculture with P. gingivalis, such as differences in the growth phase, affect gene expression. To elucidate the influence of coculture, the evaluation of gene expression at multiple time points and in different ratios between the bacteria is required. TSBhm was used in the current assay; in other proteomics analyses, Brucella broth [50] or Todd-Hewitt broth [38] containing hemin and menadione were used. In the latter studies, the investigation was conducted in PBS. These media can be used to detect synergistic interactions; however, differences in the medium content may result in different results. In addition, a precise investigation on the synergistic effect requires the conditions that simulate those in the oral cavity. To fully comprehend the synergism between F. nucleatum and P. gingivalis, additional research using media that mimic the physiological condition of the oral cavity is required.
In conclusion, the diffusible signaling molecules from P. gingivalis induced an increase in the ligand for coaggregation with P. gingivalis and led to metabolic changes, including the promotion of polysaccharide synthesis and inhibition of the de novo synthesis of purine and pyrimidine in F. nucleatum. The metabolic changes may play a key role in the acceleration of biofilm formation by F. nucleatum cocultured with P. gingivalis. Therefore, the metabolic pathway in which differentially expressed genes involved in this study could be used as potential therapeutic targets for preventing dysbiosis.
Funding Statement
This work was partially supported by JSPS KAKENHI under Grant numbers 245927783 (KI) and 15K1102 (KI).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The sequence raw data obtained using RNA-Sequencing analysis has been deposited to the Sequence Read Archive in the DNA Data Bank of Japan (DDBJ/DRA) (https://www.ddbj.nig.ac.jp/dra) under submission ID: DRA014805.
References
- [1].Kassebaum NJ, Bernabe E, Dahiya M, et al. Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J Dent Res. 2014;93(11):1045–12. DOI: 10.1177/0022034514552491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–490. [DOI] [PubMed] [Google Scholar]
- [3].Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366(9499):1809–1820. [DOI] [PubMed] [Google Scholar]
- [4].Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7(5):1016–1025. DOI: 10.1038/ismej.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. Isme J. 2012;6(6):1176–1185. DOI: 10.1038/ismej.2011.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–144. DOI: 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- [8].Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bradshaw DJ, Marsh PD, Watson GK, et al. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66(10):4729–4732. DOI: 10.1128/IAI.66.10.4729-4732.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kolenbrander PE, Ganeshkumar N, Cassels FJ, et al. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 1993;7(5):406–413. DOI: 10.1096/fasebj.7.5.8462782 [DOI] [PubMed] [Google Scholar]
- [11].Rickard AH, Gilbert P, High NJ, et al. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100. DOI: 10.1016/S0966-842X(02)00034-3 [DOI] [PubMed] [Google Scholar]
- [12].Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9(1):55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015; 23 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zijnge V, van Leeuwen MB, Degener JE, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5(2):e9321. DOI: 10.1371/journal.pone.0009321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mark Welch JL, Rossetti BJ, Rieken CW, et al. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–800. DOI: 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Coppenhagen-Glazer S, Sol A, Abed J, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83(3):1104–1113. DOI: 10.1128/IAI.02838-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Okuda T, Kokubu E, Kawana T, et al. Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe. 2012;18(1):110–116. DOI: 10.1016/j.anaerobe.2011.09.003 [DOI] [PubMed] [Google Scholar]
- [19].Rosen G, Genzler T, Sela MN. Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol Lett. 2008;289(1):59–66. [DOI] [PubMed] [Google Scholar]
- [20].Horiuchi A, Kokubu E, Warita T, et al. Synergistic biofilm formation by Parvimonas micra and Fusobacterium nucleatum. Anaerobe. 2020;62(102100): DOI: 10.1016/j.anaerobe.2019.102100 [DOI] [PubMed] [Google Scholar]
- [21].Metzger Z, Lin YY, Dimeo F, et al. Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. J Endod. 2009;35(1):86–94. DOI: 10.1016/j.joen.2008.10.015 [DOI] [PubMed] [Google Scholar]
- [22].Okuda T, Okuda K, Kokubu E, et al. Synergistic effect on biofilm formation between Fusobacterium nucleatum and Capnocytophaga ochracea. Anaerobe. 2012;18(1):157–161. DOI: 10.1016/j.anaerobe.2012.01.001 [DOI] [PubMed] [Google Scholar]
- [23].Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–472. [DOI] [PubMed] [Google Scholar]
- [24].Feuille F, Ebersole JL, Kesavalu L, et al. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect Immune. 1996;64(6):2095–2100. DOI: 10.1128/iai.64.6.2094-2100.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sakanaka A, Kuboniwa M, Shimma S, et al. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems. 2022. 30; 7(4): e0017022. 10.1128/msystems.00170-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sakanaka A, Kuboniwa M, Takeuchi H, et al. Arginine-ornithine antiporter ArcD controls arginine metabolism and interspecies biofilm development of Streptococcus gordonii. J Biol Chem. 2015;290(35):21185–21198. DOI: 10.1074/jbc.M115.644401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Saito Y, Fujii R, Nakagawa KI, et al. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23(1):1–6. DOI: 10.1111/j.1399-302X.2007.00380.x [DOI] [PubMed] [Google Scholar]
- [28].Saito A, Inagaki S, Ishihara K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb Pathog. 2009;47(6):329–333. [DOI] [PubMed] [Google Scholar]
- [29].Saito A, Inagaki S, Kimizuka R, et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54(3):349–355. DOI: 10.1111/j.1574-695X.2008.00481.x [DOI] [PubMed] [Google Scholar]
- [30].Shah HN, Collins MD. Proposal for reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new genus. Porphyromonas Int Syst Bacteriol, Int Syst Bacteriol. 1988;38(1):128–131. [Google Scholar]
- [31].Takahashi N, Ishihara K, Kato T, et al. Susceptibility of Actinobacillus actinomycetemcomitans to six antibiotics decreases as biofilm matures. J Antimicrob Chemother. 2007;59(1):59–65. DOI: 10.1093/jac/dkl452 [DOI] [PubMed] [Google Scholar]
- [32].Ishihara K, Wawrzonek K, Shaw LN, et al. Dentipain, a Streptococcus pyogenes IdeS protease homolog, is a novel virulence factor of Treponema denticola. Biol Chem. 2010;391(9):1047–1055. DOI: 10.1515/bc.2010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Su W, Sun J, Shimizu K, et al. TCC-GUI: a shiny-based application for differential expression analysis of RNA-Seq count data. BMC Res Notes. 2019;12(1):133. DOI: 10.1186/s13104-019-4179-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Klopfenstein DV, Zhang L, Pedersen BS, et al. GOATOOLS: a python library for gene ontology analyses. Sci Rep. 2018;8(1):10872. DOI: 10.1038/s41598-018-28948-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Abed J, Emgard JE, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNac. Cell Host Microbe. 2016;20(2):215–225. DOI: 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smalley JW, Olczak T. Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol Oral Microbiol. 2017;32(1):1–23. [DOI] [PubMed] [Google Scholar]
- [38].Hendrickson EL, Wang T, Beck DA, et al. Proteomics of Fusobacterium nucleatum within a model developing oral microbial community. Microbiologyopen. 2014;3(5):729–751. DOI: 10.1002/mbo3.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Genevaux P, Bauda P, DuBow MS, et al. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol. 1999;172(1):1–8. DOI: 10.1007/s002030050732 [DOI] [PubMed] [Google Scholar]
- [40].Dean CR, Goldberg JB. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol Lett. 2002;210(2):277–283. [DOI] [PubMed] [Google Scholar]
- [41].Guo Y, Sagaram US, Kim JS, et al. Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri subsp. citri Appl Environ Microbiol, Appl Environ Microbiol. 2010;76(7):2234–2242. DOI: 10.1128/AEM.02897-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nesper J, Lauriano CM, Klose KE, et al. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 2001;69(1):435–445. DOI: 10.1128/IAI.69.1.435-445.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Robrish SA, Thompson J. Regulation of fructose metabolism and polymer synthesis by Fusobacterium nucleatum ATCC 10953. J Bacteriol. 1990;172(10):5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153–192. [DOI] [PubMed] [Google Scholar]
- [45].Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54(1):15–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nemoto TK, Ohara Nemoto Y. Dipeptidyl-peptidases: key enzymes producing entry forms of extracellular proteins in asaccharolytic periodontopathic bacterium Porphyromonas gingivalis. Mol Oral Microbiol. 2021;36(2):145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Borlee BR, Goldman AD, Murakami K, et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75(4):827–842. DOI: 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Irie Y, Starkey M, Edwards AN, et al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78(1):158–172. DOI: 10.1111/j.1365-2958.2010.07320.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Doron L, Coppenhagen-Glazer S, Ibrahim Y, et al. Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease. PLoS One. 2014;9(10):e111329. DOI: 10.1371/journal.pone.0111329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ali Mohammed MM, Pettersen VK, Nerland AH, et al. Label-free quantitative proteomic analysis of the oral bacteria Fusobacterium nucleatum and Porphyromonas gingivalis to identify protein features relevant in biofilm formation. Anaerobe. 2021;72:102449. [DOI] [PubMed] [Google Scholar]
- [51].Kodama M, Oshikawa K, Shimizu H, et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat Commun. 2020;11(1):1320. DOI: 10.1038/s41467-020-15136-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sasaki-Imamura T, Yano A, Yoshida Y. Production of indole from L-tryptophan and effects of these compounds on biofilm formation by Fusobacterium nucleatum ATCC 25586. Appl Environ Microbiol. 2010;76(13):4260–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ciofu O, Moser C, Jensen PO, et al. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022;20(10):621–635. DOI: 10.1038/s41579-022-00682-4 [DOI] [PubMed] [Google Scholar]
- [54].Mohammed MMA, Pettersen VK, Nerland AH, et al. Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis. Anaerobe. 2017;44:133–142. [DOI] [PubMed] [Google Scholar]
- [55].Pocanschi CL, Apell HJ, Puntervoll P, et al. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J Mol Biol. 2006;355(3):548–561. DOI: 10.1016/j.jmb.2005.10.060 [DOI] [PubMed] [Google Scholar]
- [56].Ponath F, Tawk C, Zhu Y, et al. RNA landscape of the emerging cancer-associated microbe Fusobacterium nucleatum. Nat Microbiol. 2021;6(8):1007–1020. DOI: 10.1038/s41564-021-00927-7 [DOI] [PubMed] [Google Scholar]
- [57].Muchova M, Balacco DL, Grant MM, et al. Fusobacterium nucleatum subspecies differ in biofilm forming ability in vitro. Front Oral Health. 2022;15(3):853618. DOI: 10.3389/froh.2022.853618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shiba T, Komatsu K, Sudo T, et al. Comparison of periodontal bacteria of edo and modern periods using novel diagnostic approach for periodontitis with micro-CT. Front Cell Infect Microbiol. 2021;11:723821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ma J, Kageyama S, Takeshita T, et al. Clinical utility of subgingival plaque-specific bacteria in salivary microbiota for detecting periodontitis. PLoS One. 2021;16(6):e0253502. DOI: 10.1371/journal.pone.0253502 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence raw data obtained using RNA-Sequencing analysis has been deposited to the Sequence Read Archive in the DNA Data Bank of Japan (DDBJ/DRA) (https://www.ddbj.nig.ac.jp/dra) under submission ID: DRA014805.


