Abstract
A confluence of evidence indicates that brain functional connectivity is not static but rather dynamic. Capturing transient network interactions in the individual brain requires a technology that offers sufficient within-subject reliability. Here, we introduce an individualized network-based dynamic analysis technique and demonstrate that it is reliable in detecting subject-specific brain states during both resting state and a cognitively challenging language task. We evaluate the extent to which brain states show hemispheric asymmetries and how various phenotypic factors such as handedness and gender might influence network dynamics, discovering a right-lateralized brain state that occurred more frequently in men than in women and more frequently in right-handed versus left-handed individuals. Longitudinal brain state changes were also shown in 42 patients with subcortical stroke over 6 months. Our approach could quantify subject-specific dynamic brain states and has potential for use in both basic and clinical neuroscience research.
A dynamic analysis technique reliably captures the spontaneous moment-to-moment changes in brain network reconfigurations in individuals.
INTRODUCTION
The coordination of information across multiple spatial and temporal scales is a prerequisite of optimal brain functioning that spans both cognitive and behavioral domains. Synchronization of hierarchically organized electrophysiological oscillations facilitates the dynamic integration of local and distributed brain regions and enables the fluid formation and dissolution of coherent, functional network configurations necessary for fast and efficient cortical processing [see (1, 2) for reviews]. The organizing principles governing the large-scale network dynamics that manifests this time-varying and intrinsically coupled activity over spatially distributed and functionally differentiated cortical regions are only beginning to be understood. Emerging evidence from multiple modalities points toward functional state patterns that are dynamically formed or expressed (3–5). These transient network configurations and their spatiotemporal dynamics that emerge at fine-grained time scales have been observed not only during the so-called “resting-state” (6) but also during various cognitive tasks (7, 8). An emerging extension of standard analytical strategies of resting-state functional magnetic resonance imaging (fMRI) has demonstrated that functional coupling between regions is not static throughout the length of a standard scan but is instead highly dynamic (9, 10). In addition to early evidence establishing possible behavioral (11, 12), physiological (13), consciousness state (14), structural (15), and electrophysiological (16, 17) correlates, the dynamic functional connectivity (FC) approach has been used to reveal slowly fluctuating and reproducible whole-brain configurations that are lost when averaging across time points (6, 18). Recent advances in analytical techniques have allowed for the decomposition of blood oxygenation level–dependent (BOLD) MR images into their underlying time-varying brain coactivated patterns, which sum to form the averaged FC maps of canonical large-scale intrinsic networks, such as the default network (DN) and dorsal attention network (18–22). The examination of single-volume MR images, rather than averaging over a windowed period, enables the detection of transient functional network interactions occurring on a smaller time scale and therefore better evaluation of their temporal dependencies. Analyses based on single volumes are not confounded by the “sampling variability” that affects second-order statistics, such as temporal correlations in a sliding window [see Discussion in (23)]. This approach may therefore be particularly powerful for the investigation of network dynamics that evolve across spatially distinct and functionally differentiated brain regions. However, this approach has been mostly applied to study brain characteristics at a group level (20) and has not demonstrated sufficient reliability at the individual level. More generally, fMRI studies as a whole are facing the challenge of low reliability at the single-subject level (24–27), which has hampered the clinical applications of fMRI and the discovery of meaningful neuroimaging biomarkers (28–31). Although previous studies have suggested that reliability of some fMRI measures may be boosted by increasing scan length (32, 33), it remains unclear whether reliability of dynamic metrics, especially those based on single-volume images, can also benefit from longer scans.
Here, we introduce an “Individualized Network-based Single-frame Coactivation Pattern Estimation” (INSCAPE) approach to begin to explore the spatiotemporal characteristics of dynamic brain state network configurations at the individual level. The goals of the present study are to reliably characterize subject-specific functional properties and capture individual differences in the spatiotemporal dynamics of functional brain states. As a test, we investigated functional lateralization, which is a well-studied and fundamental property of human brain organization and is highly individual-specific (34–36). We explored the extent to which the spatiotemporal dynamics of identified functional brain states using our INSCAPE approach showed hemispheric asymmetry of language processing. Given the link between functional lateralization of language with handedness and gender, we also examined whether these phenotypic traits were related to the occurrence of lateralized brain states. Last, to demonstrate the potential of this technique in future patient-related research, we investigated the longitudinal changes in brain state network dynamics in 42 patients with subcortical stroke to track the time course and spatiotemporal properties associated with poststroke recovery across a 6-month period.
RESULTS
Different brain states have specific coactivation patterns and occurrence rates
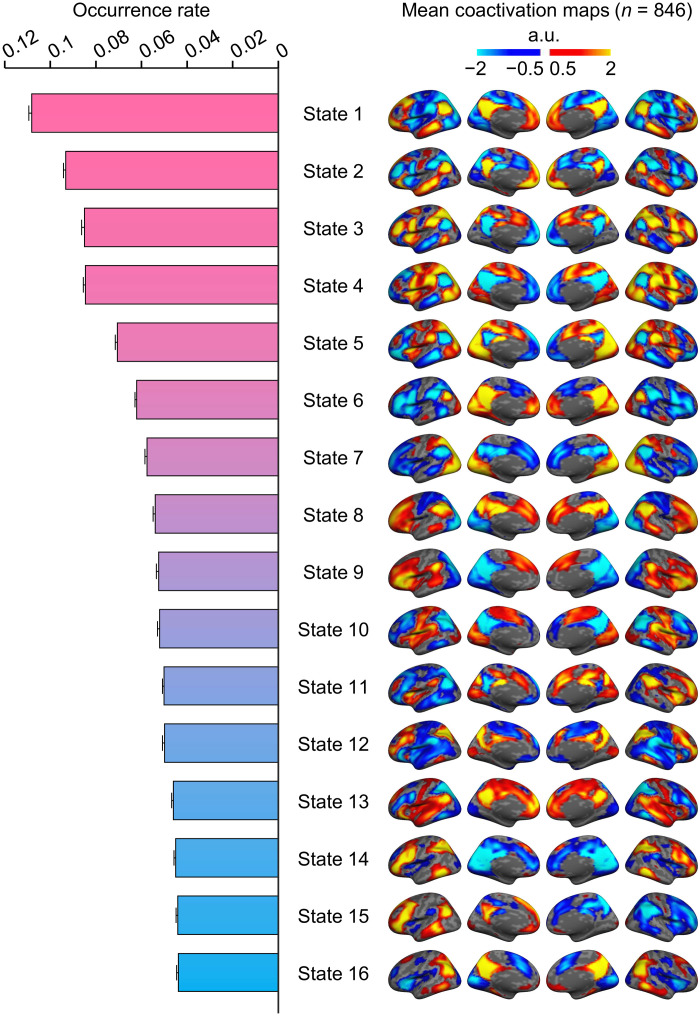
The INSCAPE approach (see Fig. 1 and Materials and Methods) was performed to generate the group templates of the 16 brain states using resting-state fMRI data of 846 healthy individuals from the Brain Genomics Superstruct Project (GSP; Dataset I). The number of brain states was selected according to the test-retest reliability of results (see fig. S1). The occurrence rates of all brain states were then estimated and sorted by rank in decreasing order (Fig. 2). The mean coactivation maps were computed by averaging the fMRI frames assigned to the same brain state and displayed on an inflated cortical surface. The coactivation patterns of all brain states exhibited intrinsic functional network properties. The brain states with DN activation had the highest occurrence rate during resting state, followed by brain states demonstrating coactivations between regions comprising the salience (SN) and fronto-parietal control (FPN) networks. In addition, several different combinations of coactivated brain regions belonging to other networks, such as the ventral attention, visual, and sensorimotor networks, were also identified. Notably, we observed specific brain states showing hemispheric lateralization and discuss these results in detail in the next section.
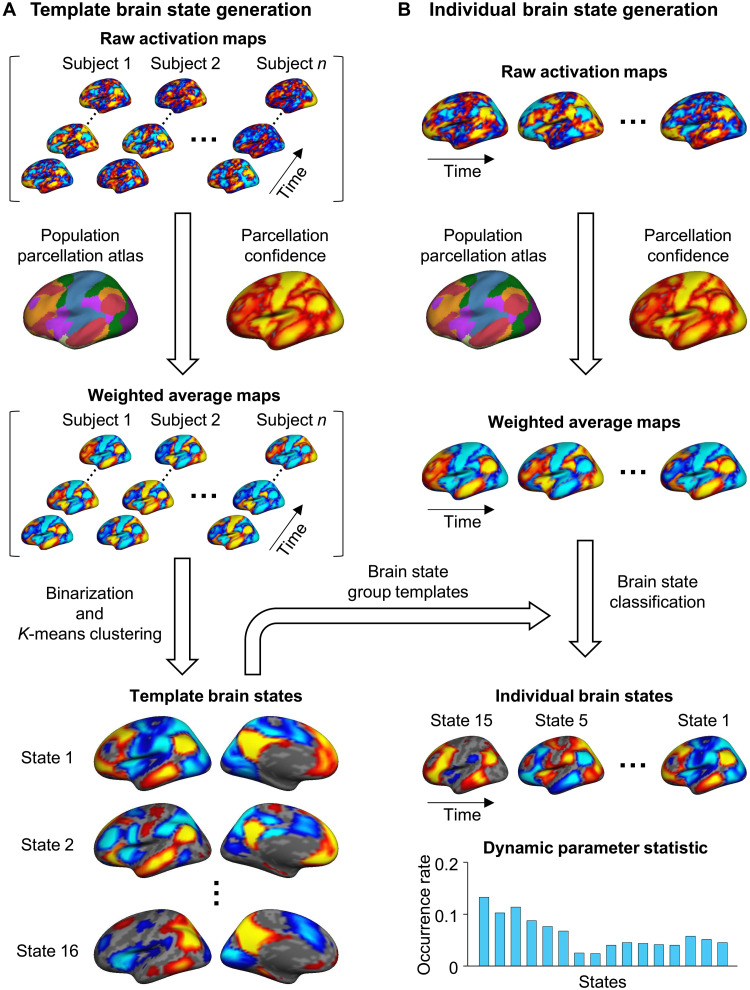
Fig. 1. Schematic of the INSCAPE approach.
(A) The group templates of brain coactivation states were generated using a resting-state fMRI dataset containing 846 healthy subjects from the GSP (Dataset I) (76). Specifically, the preprocessed fMRI data of each subject were projected to the FreeSurfer fsaverage4 surface space, which has 2562 vertices per hemisphere. For each time frame of BOLD data, the raw activation map was weighted by the parcellation confidence derived from a population-based cortical parcellation (82) and averaged within each of the 48 network patches (24 per hemisphere). The mean activations of the 48 patches were then binarized (i.e., values larger than 0 were set to 1 and values smaller than 0 were set to −1) to represent the mean weighted coactivation maps that were then concatenated along with the time series across all subjects. A k-means clustering analysis was then performed to classify the fMRI frames into 16 clusters. The optimal cluster number of 16 was selected on the basis of results from the test-retest reliability analysis (see fig. S1) because it balanced the test-retest reliability with the diversity of the brain states. Last, the maps of fMRI frames assigned to the same cluster were averaged to generate the group templates for the 16 dynamic brain states. (B) At an individual subject level, the maps of the preprocessed fMRI data were first weighted by the parcellation confidence and averaged within the 48 patches using the same procedure described above for group template generation. Each patch map was then assigned to one of the 16 template brain states having the shortest spatial distance to it. The occurrence rate of each brain state was calculated as the percentage of frames assigned to a given brain state out of the total number of frames.
Fig. 2. Occurrence rate and spatial maps of the 16 coactivated brain states.
The group templates of brain coactivation states were computed using the INSCAPE approach across 846 healthy individuals from the GSP dataset. The group-level brain state coactivation maps were generated by averaging the fMRI time frames assigned to the same cluster across subjects and ranking them by their rate of occurrence in descending order. Of the 16 brains states, states 1 through 4 showed canonical DN activation and had the highest occurrence rates across subjects during resting state. a.u., arbitrary units.
To verify the reproducibility of the brain states, we computed the brain state coactivation maps and their occurrence rates in half of the GSP dataset (Dataset I), which was designated as the discovery sample (n = 423), and then replicated this approach independently in the other half of the GSP dataset, designated as the replication sample (n = 423). The discovery and replication occurrence rates for the 16 brain states were strongly correlated, demonstrating high reproducibility at the group level (fig. S2; Pearson correlation, r = 0.995, P < 0.001). The corresponding coactivation maps of the 16 brain states also demonstrated high spatial similarity (mean spatial similarity ± SD, r = 0.992 ± 0.003, P < 0.001).
Although the 16-brain state solution was selected as the optimal cluster number for analysis in the current study because it balanced the test-retest reliability with the diversity of the brain states, we also investigated the 10- and 19-cluster solutions given these two local optimal solutions yielded high permutation test-retest reliability (see fig. S1) in the GSP dataset. Although the 19-cluster solution generated a greater number of brain states than the 10-cluster solution, both solutions yielded brain states that shared common spatiotemporal coactivation profiles. Moreover, the order of occurrence rates for these brain states was highly similar (fig. S3). For example, the mean coactivation maps of the first four brain states with the highest occurrence rates showed similar coactivation profiles in both the 10- and 19-cluster solutions. These findings indicate that additional brain states comprising the 19-cluster solution were likely derived from the decomposition of other brain state configurations identified in the 10-cluster solution.
The occurrence rate of brain states is reliable at the individual level and captures intersubject variability
Next, we estimated the frequency of brain states at the individual level and quantified the reproducibility for each participant. Our INSCAPE approach can apply the group templates generated from the GSP dataset to decode brain states in any new subject even if the subject was scanned using a different protocol. We thus applied the INSCAPE approach to Dataset II, a dataset from the Consortium for Reliability and Reproducibility Project by Hangzhou Normal University (CoRR-HNU) that consisted of a sample of 30 healthy participants scanned on 10 different occasions. This cohort was intentionally different from the GSP dataset, which was used for generating the group template of brain states, in terms of scanner type, scanning protocol, and scan durations.
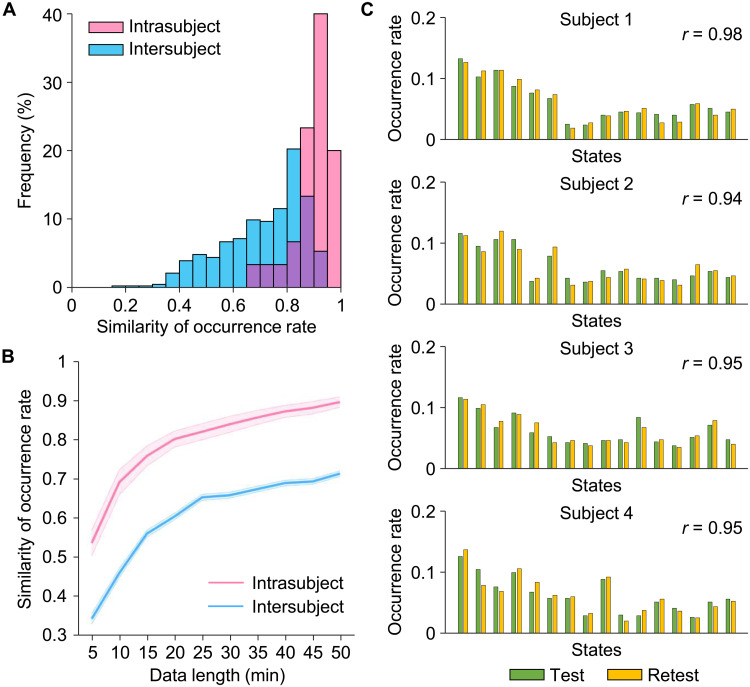
To evaluate the test-retest reliability and sensitivity of our INSCAPE approach for capturing individual differences in the occurrence rate of subject-specific brain states, we equally divided each subject’s resting-state fMRI data from the CORR-HNU dataset (Dataset II) into a test session and a retest session and computed the occurrence rate of brain states in each session. The intra- and intersubject similarities of occurrence rates were tested using Pearson correlation (Fig. 3A). The within-subject occurrence rates of brain states derived from the two sessions showed high consistency with a mean intrasubject similarity of r = 0.90. Between any two individuals, the mean similarity of occurrence was 0.71 (intersubject variability = 0.29). Test-retest results of four participants are displayed in Fig. 3C and illustrate the distribution of brain state occurrence rates for each subject. As expected, the intrasubject similarity of state occurrence rates was significantly higher than the intersubject similarity [two-sample t test, t(463) = 6.621, P < 0.001], indicating that the INSCAPE approach is not only highly reproducible at the single-subject level but can also reliably and robustly capture individual differences across subjects.
Fig. 3. Estimation of the reliability of the INSCAPE analysis.
(A) Test-retest reliability was evaluated using the CoRR-HNU dataset (i.e., Dataset II). We divided each subject’s data equally into two 50-min sessions and calculated the occurrence rates for each session. The intra- and intersubject similarities were quantified by estimating the correlation coefficient of occurrence rates within the same subject and between any two individuals. Correlation analyses yielded a mean intrasubject similarity of r = 0.90 and a mean intersubject similarity of r = 0.71. The frequency distributions of occurrence rates are depicted in a histogram with intrasubject and intersubject similarity denoted by pink bars and blue bars, respectively. (B) The intrasubject (pink line) and intersubject (blue line) similarities in occurrence rates were also examined using the test-retest dataset with 10 different data lengths ranging from 5 to 50 min in duration, in 5-min time increments (mean intrasubject test-retest reliability ± SEM, 5 min: 0.54 ± 0.03, 10 min: 0.69 ± 0.03, 15 min: 0.76 ± 0.03, 20 min: 0.80 ± 0.02, 25 min: 0.82 ± 0.02, 30 min: 0.84 ± 0.02, 35 min: 0.86 ± 0.02, 40 min: 0.87 ± 0.02, 45 min: 0.88 ± 0.02, 50 min: 0.90 ± 0.01). The shaded areas in the figure represent the standard errors. (C) Histograms showing the distributions of test-retest occurrence rates for the 16 brain states extracted from four randomly selected individuals taken from the CoRR-HNU dataset. Green bars represent the occurrence rate of brain states in the test session, while yellow bars represent the occurrence rate of brain states in the retest session.
Because data acquisition length is one of the main factors that affects the reliability of fMRI-based analyses, we estimated the test-retest reliability of occurrence rates using varying lengths of resting-state fMRI data, ranging from 5 to 50 min, in 5-min increments of time. Both intra- and intersubject similarities of occurrence rates increased as data became longer (Fig. 3B). A within-subject test-retest reliability of 0.70 was observed with 15 min of fMRI data, with the mean reliability increasing to 0.90 with 50 min of data.
Brain lateralization is reflected in the network dynamics of coactivated brain states
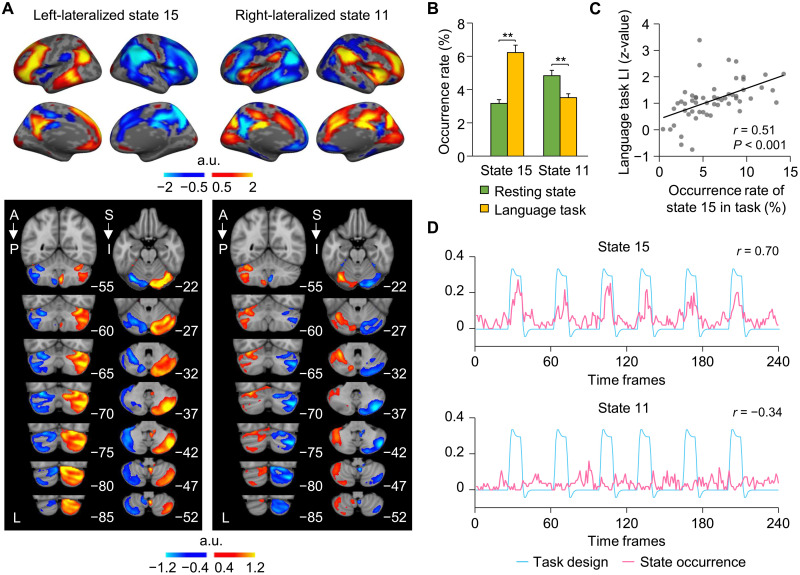
Brain lateralization is an organizing principle of the human brain postulated to contribute to fast and efficient information processing (37). To discern the extent to which the 16 coactivated brain states were functionally lateralized, we calculated the laterality index (LI) by comparing the number of activated vertices between the left and right hemispheres. Among the 16 brain states derived from our INSCAPE analysis of Dataset I, states 15 and 11 displayed the strongest leftward and rightward lateralization, respectively (fig. S4). The coactivated brain regions comprising left-lateralized state 15 included the left lateral prefrontal cortex, temporal parietal junction, and posterior cingulate cortex (Fig. 4A and fig. S5). Right-lateralized state 11 showed strong coactivations in the insula, angular gyrus, and dorsal anterior cingulate cortex. In addition, we examined the activation patterns of these lateralized brain states in the cerebellum by averaging the corresponding raw coactivation maps within a cerebellum mask in MNI152 volumetric space. Both left-lateralized state 15 and right-lateralized state 11 in the cerebrum showed asymmetric functional activations on the contralateral side of the cerebellum (Fig. 4A).
Fig. 4. Cerebral lateralization of coactivated brain states is reflected in the contralateral cerebellum and in language task.
(A) Left-lateralized state 15 showed activation in the DN and FPN, as well as cortical areas known to be important for language processing, while activation of right-lateralized state 11 comprised cortical regions anchored in the SN and ventral attention networks. Both lateralized brain states 15 and 11 showed the strongest coactivations in the contralateral cerebellar hemisphere. (B) The occurrence rates of left-lateralized state 15 and right-lateralized state 11 were calculated during resting state and during a language task. Bar graphs depict mean (±SEM) occurrence rates for both states 15 and 11 during resting state (green bars) and the language task (yellow bars). The occurrence of left-lateralized state 15 was significantly greater during the language task compared to resting state, whereas right-lateralized state 11 showed an opposite pattern. (C) Task-based language LI was significantly correlated with the occurrence rate of left-lateralized state 15 in 55 subjects. (D) The occurrence of left-lateralized state 15 was significantly correlated with language task onsets (r = 0.70, P < 0.001), indicating that coactivated regions comprising left-lateralized brain state 15 may subserve language processing. In contrast, the occurrence of right-lateralized state 11 showed a significant negative correlation with task onsets (r = −0.34, P < 0.001).
Another important finding was that the coactivated regions comprising left-lateralized brain state 15 largely overlapped with traditional language cortical areas. To investigate the extent to which lateralized brain states reflected the degree of language lateralization, we estimated the occurrence rates of brain states 15 and 11 in an independent sample (Dataset III) of 55 healthy participants scanned during both resting state and during a language task (i.e., semantic decision task). Left-lateralized state 15 showed a significantly higher occurrence rate during the language task compared to resting state [Fig. 4B; paired t test, t(54) = 6.633, **P < 0.001], whereas right-lateralized state 11 had a significantly higher occurrence rate during resting state compared to the language task [paired t test, t(54) = 3.875, **P < 0.001]. We then calculated a task-based language LI for each subject based on the asymmetric task-evoked activation in the two hemispheres. The language lateralization was significantly correlated to the occurrence rate of the left-lateralized state 15 during the task (Fig. 4C; r = 0.51, P < 0.001). We also examined the relationship between the occurrence of brain states across all participants at each time point of the language task and the hemodynamic response curves elicited by task processing. Results revealed that the occurrence of left-lateralized state 15 showed a strong, positive correlation with language task onsets (Fig. 4D; r = 0.70, P < 0.001), while the right-lateralized state 11 showed a moderate, negative correlation with language task onsets (r = −0.34, P < 0.001). Brain state 9 also showed a strong, positive correlation with language task onsets (fig. S6; r = 0.78, P < 0.001). On the basis of these findings, we then combined both states 15 and 9 into a single language-related brain state and examined its occurrence in relation to the language task onsets. We found that the occurrence of the combined brain states demonstrated an even greater association with language task onsets than previously observed with either states 15 or 9 alone (r = 0.86, P < 0.001). These results indicate that left-lateralized state 15 and state 9 may be functioning cooperatively to jointly subserve language-related semantic processing.
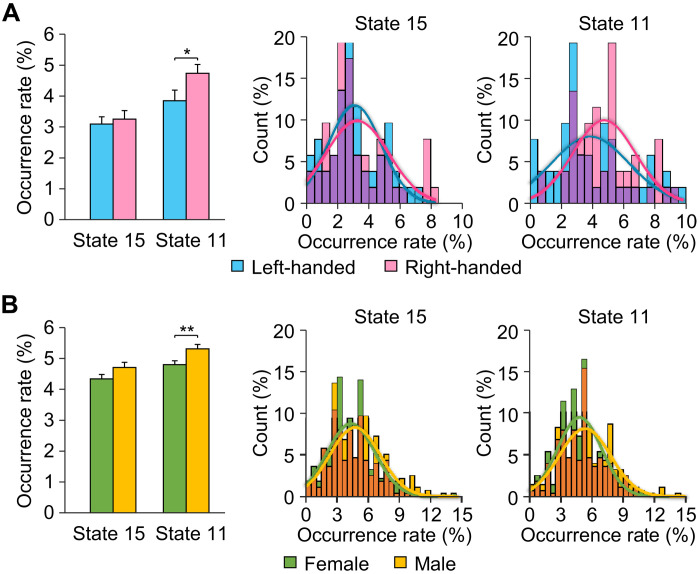
Last, given the known influence of handedness and gender on functional lateralization of language, we investigated the effects of these phenotypic characteristics on lateralization of coactivated brain states in two separate datasets. For the handedness comparison, we computed the occurrence rate of the lateralized brain states in 52 left-handed and 52 demographically matched right-handed subjects from Dataset IV. Compared to left-handers, right-handed subjects showed a significantly higher occurrence rate for right-lateralized state 11 [Fig. 5A; two-sample t test, t(102) = 2.037, *P = 0.044]. The distribution of occurrence rates also showed a higher occurrence of state 11 in right-handed versus left-handed subjects (Kolmogorov-Smirnov test, P = 0.021). We then examined the differences in occurrence rates of lateralized brain states in a sample of 279 males and 279 demographically matched (matched for age, education, and handedness) females from Dataset V. Overall, males compared to females showed a significantly higher occurrence rate for right-lateralized state 11 [two-sample t test, t(556) = 2.674, **P = 0.008] along with a trend toward a significantly higher occurrence rate in left-lateralized state 15 [Fig. 5B; two-sample t test, t(556) = 1.877, P = 0.061]. The distribution of occurrence rate for state 11 in males was significantly different compared to females (Kolmogorov-Smirnov test, P = 0.017), with males demonstrating greater occurrence rates for brain state 11 relative to their female counterparts. In addition, we investigated whether handedness or gender influenced the occurrence rate of state 9; however, no significant differences [paired t test, handedness: t(51) = 0.045, P = 0.965, gender: t(278) = 0.807, P = 0.420] were found for either analysis (fig. S7).
Fig. 5. The influence of handedness and gender on the occurrence rates of lateralized brain states.
(A) The occurrence rate of lateralized brain states was computed in 52 left-handed and 52 demographically matched right-handed subjects. Right-handed subjects showed a significantly higher occurrence rate in right-lateralized state 11 relative to left-handers (two-sample t test, *P = 0.044). The occurrence rate distributions for lateralized states 15 and 11 are displayed in the histograms with left-handed subjects depicted by blue bars and right-handed subjects denoted by pink bars, with the overlap of the two shown in purple. The distributions were fitted using Gaussian curves and demonstrate that right-handed subjects showed a higher occurrence rate in state 11 than left-handed subjects (Kolmogorov-Smirnov test, P = 0.021). (B) Occurrence rate of lateralized brain states 15 and 11 across 279 males and 279 matched females. Bar graph depicts the mean percentage of occurrence (±SEM) of states 15 and 11 in male (yellow bars) and female (green bars) subjects. Males demonstrated a significantly higher occurrence of state 11 (two-sample t test, **P = 0.008) and a trend toward a higher occurrence rate in state 15 that approached significance (paired t test, P = 0.054) compared to females. The occurrence rate distributions for these two states are graphically displayed and demonstrate that males (yellow bars) showed a significantly higher occurrence rate in right-lateralized state 11 than females (green bars) (Kolmogorov-Smirnov test, P = 0.017).
The occurrence rate of dynamic coactivated brain states for assessment of poststroke recovery
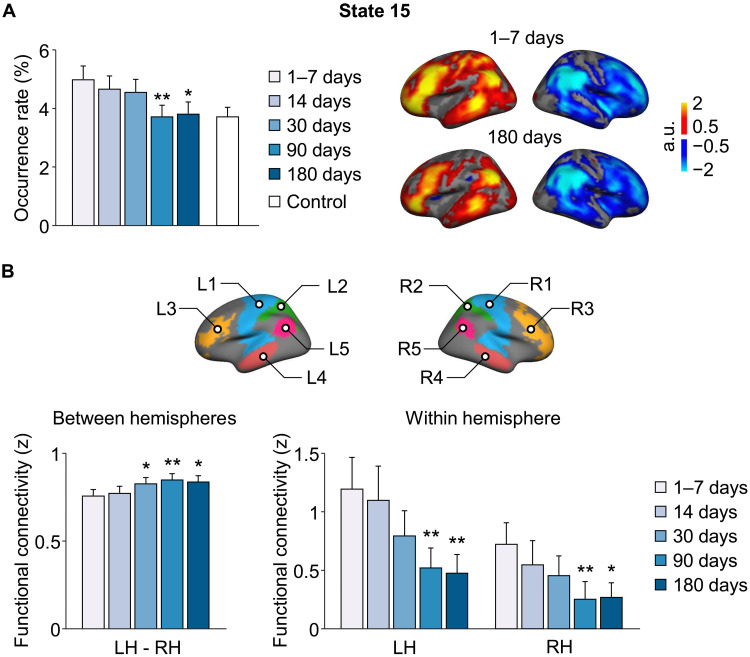
Individualized dynamic brain states may be used to evaluate functional brain status in patients with various brain disorders, especially in monitoring disease progression or recovery after treatment. As a proof of concept, we applied our INSCAPE approach to a dataset (Dataset VI) consisting of 42 subcortical stroke patients and 23 healthy control participants to evaluate the functional changes over a period of 6 months. We chose this stroke dataset because of the clear etiology of the patients and the homogeneity of functional impairment among subjects. The occurrence rates of the 16 brain states were estimated in patients at five time points over a 6-month period and included scans acquired at 1 to 7, 14, 30, 90, and 180 days after stroke. We also estimated the occurrence rates of the 16 brain states in a group of healthy controls at a single time point, which served as a baseline for comparison with the patient group at the 1- to 7-day poststroke time point. Results of the repeated measures analysis of variance (ANOVA) revealed a main effect of time [F(4) = 4.451, P = 0.002, family-wise error rate (FWER) corrected]. Overall, a gradual and sustained diminution in the occurrence rate of the left-lateralized brain state 15 was observed in patients across the 6-month poststroke recovery period. A series of post hoc paired-sample t tests showed that the occurrence rate of brain state 15 was significantly reduced at both 90 [t(41) = 3.662, **P < 0.001] and 180 [t(41) = 2.684, *P = 0.01] days after stroke compared to the baseline (1 to 7 days) poststroke time point (FWER corrected). The initially elevated occurrence rate of state 15 at acute stage normalized over time and became similar to that in healthy controls at 6 months [two-sample t test, 1- to 7-day poststroke: t(63) = 1.883, P = 0.064, 6-month poststroke: t(63) = 0.296, P = 0.768]. Nevertheless, the occurrence rate of state 15 was not directly correlated with motor scores (P > 0.05).
Given brain state 15 is a lateralized brain state, we hypothesized that the continuous decrease in occurrence of this state over the course of the 6-month poststroke period may be indicative of a gradual recovery of function from interhemispheric correspondence deficits sustained by the subcortical stroke. To test this hypothesis, we selected five left-hemispheric patches (L1: sensorimotor, L2: superior parietal lobule, L3: lateral prefrontal cortex, L4: middle temporal gyrus, and L5: angular gyrus) that had the highest activations in brain state 15 at 1 to 7 days after stroke and five symmetric right-hemispheric patches (R1, R2, R3, R4, and R5) (Fig. 6B). FC between each pair of patches (i.e., L1-R1, L2-R2, L3-R3, L4-R4, and L5-R5) was computed and averaged in patients at each of the five time points after stroke to measure the between-hemisphere connectivity changes over time. Results of the repeated measures ANOVA indicated a significant main effect of time [F(4) = 4.504, P = 0.002, FWER-corrected] suggesting a gradual increase in between-hemisphere FC in patients across the 6-month poststroke recovery period (see fig. S8 for FC of each patch-pair in patients at the five time points after stroke). Post hoc paired-sample t tests showed that the FC between hemispheres significantly increased at 30 [t(41) = 2.201, *P = 0.033], 90 [t(41) = 3.540, **P = 0.001], and 180 [t(41) = 2.289, *P = 0.027] days after stroke compared to the baseline (1 to 7 days). Then, FC was estimated between any two patches within each hemisphere and averaged to represent the within-hemisphere connectivity. Both hemispheres showed a gradual decrease in the within-hemispheric FC in patients across the 6-month poststroke recovery period [repeated measures ANOVA, left hemisphere: F(4) = 4.688, P = 0.001, right hemisphere: F(4) = 2.623, P = 0.037]. A series of post hoc paired-sample t tests showed that the FC within both hemispheres was significantly reduced at 90 and 180 days after stroke compared to the acute stage [90 days after stroke versus 7 days after stroke, left hemisphere: t(41) = 2.874, **P = 0.006, right hemisphere: t(41) = 2.987, **P = 0.005; 180 days after stroke versus 7 days after stroke, left hemisphere: t(41) = 2.861, **P = 0.007, right hemisphere: t(41) = 2.456, *P = 0.018].
Fig. 6. Longitudinal changes of brain states in patients with subcortical stroke during the first 6 months of recovery.
(A) The occurrence rates of the 16 brain states were estimated in patients with subcortical stroke at five time points over a 6-month period (i.e., 1 to 7, 14, 30, 90, and 180 days after stroke). The bar graph shows the mean occurrence rate of state 15 (±SEM) in the patient group (n = 42; blue bars) at each successive time point and in the healthy control group (n = 23; white bars). There was a significant reduction in the occurrence of left-lateralized state 15 in stroke patients at the 90 and 180 days poststroke time points relative to baseline (1 to 7 days after stroke) (paired t test, *P < 0.05 and **P < 0.01). The coactivation maps of left-lateralized brain state 15 in patients at 1 to 7 days and 180 days after stroke are displayed in the right. (B) Five left-hemispheric patches (L1: sensorimotor, L2: superior parietal lobule, L3: lateral prefrontal cortex, L4: middle temporal gyrus, and L5: angular gyrus) that had the highest activations in brain state 15 at the 1 to 7 days poststroke time point and five symmetric right-hemispheric patches (R1, R2, R3, R4, and R5) were selected for the estimation of between- and within-hemisphere connectivity. Between-hemisphere FC increased at the 30, 90, and 180 days poststroke time points relative to baseline (paired t test, *P < 0.05 and **P < 0.01). Both hemispheres showed a gradual reduction in the within-hemisphere connectivity over the 6-month period, and the reduction was statistically significant at 90 and 180 days after stroke compared to the baseline (paired t test, *P < 0.05 and **P < 0.01). LH, left hemisphere; RH, right hemisphere.
Control analyses
We compared the performance of our INSCAPE approach with two previously reported dynamic analysis methods—Group CAP (22) and HMM-MAR (38). The comparisons were carried out in two independent datasets—Human Connectome Project (HCP) unrelated 100-subject dataset (Dataset VII) and CoRR-HNU 30-subject dataset (Dataset II). We found that the Group CAP method had lower test-retest reliability in occurrence rate compared to INSCAPE and HMM-MAR (see the Supplementary Materials and fig. S9). Given that HMM-MAR and INSCAPE showed similar test-retest reliability in occurrence rate, these two methods were further compared in terms of spatial characteristics. We found that INSCAPE yielded more stable group-level brain states across two datasets compared to HMM-MAR (see the Supplementary Materials and figs. S10 and S11). Because the HMM-MAR approach does not capture stable group-level brain states across different datasets, the findings from one dataset may not be easily generalized to a different dataset. We also found that INSCAPE is computationally more efficient than the other two methods, making it easier to use in large data samples (see the Supplementary Materials and fig. S12). Last, we performed control analyses to examine how head motion affects our results. We found that when head motion is not excessive, we could still decode the brain states with relatively reliable temporal and spatial characteristics (fig. S13).
DISCUSSION
In the present study, we leveraged our INSCAPE approach for estimating the temporally dynamic functional brain states at the individual level. We demonstrate that the occurrence rate of the 16 brain states identified using our approach showed high intrasubject test-retest reliability while also capturing significant intersubject differences. We applied this approach retrospectively under a number of different experimental contexts and in different subject populations to begin to characterize the spatiotemporal dynamics of these subject-specific brain states. We also showed that a subset of identified transient brain states demonstrating functional lateralization was related to subject-specific phenotypic traits such as handedness and gender. Last, we showed that the INSCAPE approach may be used for investigating longitudinal changes in large-scale network dynamics in patients with neurological or psychiatric disorders. Collectively, these findings indicate that our INSCAPE approach can robustly and reliably detect the dynamic network properties of brain states at the individual level and has potential for use in investigating the neurological and psychiatric sequelae of various brain diseases.
Revealing individual variability in dynamic brain states
A characteristic of complex, self-organizing nonlinear biological systems, such as the human brain, is the capacity to rapidly and adaptively respond to external demands that lead to survival, thereby increasing fitness. The burgeoning new interdisciplinary field of network dynamics, with its origins rooted in information theory and network science, seeks to synthesize data derived from both empirical and computationally based in silico studies to better understand how the spatiotemporal patterning of neural network dynamics unfolds over time and is capable of supporting higher-order cognitive processes during both intrinsic resting-state and task-induced perturbations (39, 40). Previous studies have suggested that dynamic brain network reconfigurations contain a wealth of information underlying critical aspects of human cognition and behavior (9, 41) and can also reflect aberrant changes in various psychiatric (42, 43) and neurological disorders (44). However, the majority of dynamic FC studies are still carried out at the group level. Investigating intersubject variability in human dynamic brain states requires imaging techniques capable of capturing reliable moment-to-moment dynamic network reconfigurations that evolve rapidly and are differentially expressed over time and at the individual level. For over a decade now, the sliding window method has been widely used for analyzing intrinsic temporal fluctuations in FC patterns (6, 10, 45). However, this approach has limited temporal resolution and thus is only able to detect the averaged state over a windowed period, an approach that is potentially confounded by the sampling variability that is known to affect second-order statistics (23, 46). Recently, Janes and colleagues (20) proposed an approach for capturing these transient functional network reconfigurations based on coactivation patterns of resting-state fMRI data and were able to obtain reliable results at the group level; however, intersubject variability was not evaluated. In the present study, we demonstrate that the occurrence rate of brain states derived from our INSCAPE approach is highly reliable within the same subject but also sensitive enough to robustly capture individual differences (Fig. 3). Delineating these moment-to-moment changes in network dynamic brain state reconfigurations using our approach may provide new avenues of research inquiry for discovery of meaningful biomarkers for tracking cognitive abilities or disease states over time by affording greater statistical power for detecting behavioral, physiological, or genetic associations.
Quantitative assessment of brain asymmetry
Hemispheric specialization has been extensively studied given that it is a fundamental organizing principle of human brain function thought to contribute to rapid and efficient information processing. Despite recent progress in our understanding of functional brain lateralization of intrinsic large-scale networks using static FC measures, comparatively little work has been conducted using a dynamic FC framework. An intriguing finding of the present study is that two dynamic states may be related to brain functional lateralization. The strongly left-lateralized state 15 appears to involve traditional language areas, including the inferior frontal gyrus, superior temporal gyrus, inferior parietal lobule, and supplemental motor areas. However, this state also involves some DN regions such as posterior cingulate. In a language task experiment, the occurrence of this state was significantly correlated with the onset of language task. In addition, individual differences in the occurrence rate of this state may predict individual differences in language lateralization observed during task fMRI (Fig. 4). These findings suggest that state 15 may be critical for language processing. In contrast, the strongly right-lateralized state 11 appears to involve traditional attention areas including insular and angular gyrus. In addition, this state also involves some FPN network regions. This state occurred more frequently in men than in women and more frequently in right-handed versus left-handed individuals (Fig. 5). Previous work has indicated that there is a handedness effect and a sex effect on the lateralization of the attention network (34). We speculate that state 11 might be recruited during attention processing.
Hemispheric asymmetry at rest and during various cognitive tasks is not stationary but changes over time in both healthy individuals and in clinical populations diagnosed with various psychiatric (47–49) and neurologic disorders (50, 51). Investigating the moment-to-moment spatiotemporal dynamics in functional asymmetries within and between large-scale networks in healthy individuals may inform our understanding of the pathophysiology underlying a multitude of neurological disorders that alter brain laterality. In the present study, we found that the left-lateralized state 15 showed a high occurrence rate at the early state of stroke, which gradually reduced to the level of healthy controls in later stages (Fig. 6). It has been suggested that lateralization emerges in the human brain as a means to reduce interhemispheric interactions, which can minimize wiring cost and improve the efficiency of information processing (37). Thus, one possible explanation of the increased occurrence of left-lateralized state 15 after stroke onset is that interhemispheric communications were impaired by subcortical stroke, leading to stronger cortical lateralization as a functional compensation. This compensatory effect diminishes as the function recovers.
Together, reliable subject-specific brain states can be used to quantify individual differences in laterality and how they may be related to clinical symptoms or cognitive abilities. This approach may be particularly powerful for monitoring real-time brain changes longitudinally in chronic diseases such as stroke, dementia, and autism.
Intrinsic and task-induced coactivated brain states share common large-scale network configurations
Studies on intrinsic static FC and task-evoked changes in brain networks have both contributed immensely to our current understanding of brain network dynamics. However, little research to date has focused on the network interactions and spatiotemporal dynamic relationships that emerge during spontaneous and task-evoked activity. Previous studies have reported that regions showing intrinsic FC at rest tend to coactivate during a task (52). The network architecture revealed by intrinsic FC is also present across a wide variety of task-induced states (53). In addition, FC network parcellation based on an individual subject’s task-based fMRI data time series largely resembled the networks derived from resting-state data (28), suggesting that spontaneous activity and task-evoked perturbations share commonalities in functional network configurations. An area of debate is a central tendency dominating the functional coupling patterns across markedly different task-specific states, as well as during resting state (54). Examining the spatiotemporal characteristics of dynamic transient coactivation brain states can shed new light on the coordination and possible competition of dynamic brain state reconfiguration. Findings from the present study revealed occurrence rates of lateralized brain states during resting state and during a language task shared common brain coactivation states (see fig. S6). Temporal segregation between left- and right-lateralized states was further supported by the observation that specific brain states were time-locked with the language task onsets, while the opposing right-lateralized brain state showed task-induced deactivations indicative of a decoupling or dissolution of the network brain state configuration (see Fig. 4, B and D). Identifying the common features shared by intrinsic and task-driven dynamic brain states holds promise for elucidating not only how these rapid and fluid transitions through different network configuration and metastable states unfold but also the degree to which dynamic network changes are state dependent and/or trait specific. Moreover, while the shared spatial and temporal properties of brain states may inform us with regard to the commonalities between intrinsic resting and task-driven states, it could also elucidate how these large-scale transient network dynamics evolve over time and reconfigure to specific brain states or energy landscapes to meet increasing task demands. Last, the individual-specific spatiotemporal properties of these transient dynamic brain states derived from our INSCAPE approach could potentially be applied to determine the extent to which a given task can modify a particular brain state at the individual level, especially in patients showing deficits or disruptions in specific network brain state coactivation patterns, and may facilitate the discovery of new personalized targets for various noninvasive therapeutic interventions.
Implications in clinical brain disorder research
Numerous imaging studies have reported disruptions in the coordination of locally segregated and global integrated large-scale network dynamics, which has been implicated in various psychiatric and neurological diseases (55–57). For decades, these abnormal network dynamic spatiotemporal patterns have been routinely investigated by examining the correlations of BOLD signals between distinct brain regions in the resting-state fMRI data time series at coarse-grained time scales, also known as “static” FC (29, 58). However, correlations based on a temporal stationarity assumption cannot detect the rapid changes in functional network reconfiguration that occur at shorter time scales (10, 18). A large body of emerging evidence has shown that psychiatric and mood disorders, in particular, are associated with altered dynamic network spatiotemporal profiles in large-scale brain networks. For example, Kaiser and colleagues (59) reported abnormal patterns in low-frequency spontaneous time-varying dynamic network coactivated patterns correlated with self-reported depression severity in patients diagnosed with major depressive disorder. Braun and coworkers (60) demonstrated that altered dynamic flexibility of large-scale network reconfiguration is related, in part, to the genetic liability for schizophrenia. To enable the detection of network interactions on a smaller time scale, our group recently performed a group-level single-frame coactivation method in patients with schizophrenia and found abnormal changes in the occurrence rate of brain state in patients who was associated with the severity of the psychotic symptoms (19). In the present study, we observed longitudinal changes of dynamic brain states in patients with subcortical stroke. Specifically, these patients showed a strong presence of the leftward lateralized brain state 15 within days following stroke onset, along with a concomitant decrease in the occurrence rate of this lateralized brain state over the course of the 6-month poststroke recovery period to levels observed in healthy cohort (Fig. 6). This finding is consistent with results from a static FC study reported previously using another independent dataset (57). This suggests that our individualized INSCAPE analysis approach is not only capable of detecting changes in the dynamic functional interactions between brain networks during the acute phase of brain disease but can also be used to generate biomarkers to track the development and recovery of various brain diseases over time, as well as clinically inform the guidance of therapeutic treatment options at the individual level.
In addition, another application of the INSCAPE approach is to assist traditional FC analytic methods in the detection of the focal lesion sites or abnormal network configuration in clinical research populations. Data-driven, hypothesis-free FC analysis has been widely adopted in clinical studies to investigate abnormalities in brain function related to various neurological diseases (58, 61). FC-based analysis in these studies is often performed to examine the connectivity across whole-brain voxels or functional segmentations that are selected as seeds. However, with the application of higher-field MRI (62, 63), along with the development of functional imaging acquisition methods (64), and analytical techniques (28, 65), the resolution of functional images is becoming higher, and functional parcellations of the cortex are becoming finer. All of these advances have led to an increasing number of “seeds” that can be explored. On the one hand, this can be advantageous to researchers studying brain diseases in more temporal and spatial detail than ever before imagined but, at the same time, increases the likelihood of tuning into statistical traps—increasing the number of multiple comparisons results in a higher risk of obtaining false-positive results (66). To solve this problem, our INSCAPE approach can be performed as a preanalysis method to identify the abnormal brain coactivation states by comparing the occurrence rate of a given brain state as a biomarker at the individual level, while retaining the sensitivity necessary to detect disease induced variance in patients versus a healthy cohort. Then, traditional static FC analysis can be used to further investigate the functional correspondence among regions demonstrating abnormal brain state dynamics. For example, in this study, we revealed the abnormalities in lateralized brain state in patients with stroke (Fig. 6A) and then further demonstrated the FC changes in the primary motor area and superior parietal lobule during the poststroke recovery period (Fig. 6B and fig. S8). These findings indicate a more optimal strategy for tracking clinical endpoints rather than conducting an aimless search for abnormal focal sites that are related to the disease severity and duration of symptoms.
Limitations and future directions
There are several technical limitations in the present study. First, the number of brain states was chosen somewhat arbitrarily. In the present study, we selected the 16-cluster solution to maximize the spatial similarity of brain states derived from the test and retest data. This procedure can, to some extent, avoid the impact of cluster number selection on the temporal characteristics of the brain states, i.e., the occurrence rate, which is the primary dynamic metric evaluated in the present study. Incorporating other measures, such as the silhouette coefficient, may help to find an optimal cluster number and can be explored in the future (67, 68). Second, in the present study we chose occurrence rate as our primary metric, without evaluating other dynamic metrics such as dwell times and number of state transitions. We found that dwell times and transition probability were highly dependent on the occurrence rate. Nevertheless, given that our approach provides a brain state label for each single time frame at the individual level, one can easily derive a variety of secondary dynamic metrics including dwell time and transition probability. Third, some recent studies suggested that coactivation patterns observed in single-volume MRI data may not be fully attributed to the nonstationarity of resting brain activity (69), and attention should be paid to all time points instead of those time points presumed to contain neural events (70). While we found that brain states derived from the INSCAPE approach could reflect neural events during the language task, the states should be interpreted with care. Fourth, we band-pass–filtered the data as in the conventional resting-state fMRI analyses. Some studies have shown that high-frequency oscillations (>0.1 Hz) of resting-state fMRI signals also encoded meaningful neural information (71–73), although they are more prone to the effects of physiological noise caused by heart beats and respiration (74, 75). Thus, temporal dynamics in high-frequency bands may be of interest to be explored in the future. Last, in our current INSCAPE pipeline, fMRIs were weighted by the parcellation confidence derived from a group-level cortical parcellation. The interindividual variance in the topological organization of functional networks dynamics should be considered. For example, the group parcellation template could be replaced by an individualized cortical parcellation in future investigations.
MATERIALS AND METHODS
Experimental design
Seven fMRI datasets including data from over 1000 subjects and over 2000 scanning sessions were included in the current study. Note that Datasets I, III, IV, and V were all collected from the GSP (76). Datasets III, IV, and V also partially overlapped with Dataset I. The details are outlined below.
Dataset I
The first dataset consisted of 1000 healthy, young adult participants (mean age 21.3 ± 3.1 years; 427 males) from the GSP. All participants provided written informed consent in accordance with guidelines set by the Institutional Review Boards of Harvard University or Partners Healthcare. Each subject performed one or two resting-state fMRI runs (6 min 12 s per run) and a structural run. All data were collected using 3T Tim Trio scanners (Siemens, Erlangen, Germany) equipped with 12-channel head coils. The structural data included a high-resolution multi-echo T1-weighted magnetization-prepared gradient-echo image sequence (MPRAGE; Repetition Time (TR) = 2200 ms, Inversion Time (TI) = 1000 ms, Echo Time (TE) = 1.54 ms for image 1 to 7.01 ms for image 4, Flip Angle (FA) = 7°, 1.2 × 1.2 × 1.2–mm voxels, and Field-of-view (FOV) = 230). The fMRI images were acquired using a gradient-echo echo-planar imaging (EPI) sequence (TR = 3000 ms, TE = 30 ms, flip angle = 85°, 3 × 3 × 3–mm voxels, FOV = 216, and 47 axial slices collected with interleaved acquisition). Participants were instructed to stay awake and keep eyes open during the resting-state scans.
Dataset II
The second dataset included 30 healthy, young adults (mean age 24 ± 2.4 years; 15 males) that were collected as part of the CoRR-HNU. Each participant underwent 10 resting-state fMRI scans (10 min per scan) and a structural MRI scan across a period of 1 month. MRI data were acquired on a GE MR750 3T scanner (GE Healthcare, Milwaukee, USA). Structural images were collected using a T1-weighted Fast Spoiled gradient-echo sequence (TR = 8.1 ms, TI = 450 ms, TE = 3.1 ms, FA = 8°, 1.0 × 1.0 × 1.0–mm voxels, and FOV = 256). Functional data were obtained using an EPI sequence (TR = 2000 ms, TE = 30 ms, FA = 90°, 3.4 × 3.4 × 3.4–mm voxels, FOV = 220). Participants were instructed to keep their eyes open and relax during the resting-state fMRI scans. All participants provided written informed consent in accordance with guidelines set by the Institutional Review Board of Hangzhou Normal University.
Dataset III
The third dataset comprised a subset of subjects from the GSP dataset (i.e., Dataset I) and contained 55 young, healthy adult participants (mean age 21.1 ± 2.7 years; 25 males). Each participant underwent two resting-state fMRI runs (6 min 12 s per run) and two task-based fMRI runs (6 min 12 s per run) wherein a semantic classification language task was performed (77). Following the task runs, a structural MRI was acquired to obtain a high-resolution T1-weighted anatomical image. The MRI data acquisition parameters were identical to the first dataset (GSP dataset; Dataset I) described above. For the language task, each run consisted of four “novel-word” blocks, four “familiar-word” blocks, and four “fixation” blocks; the duration of each block was 30 s. During each novel-word block, five novel concrete words and five novel abstract words were randomly presented. In the familiar-word block, five practiced words were presented repeatedly. All word stimuli were randomly presented for 2 s, with 1-s interstimulus intervals. In each trial, subjects were asked to indicate whether the meaning of the word presented was concrete or abstract. All participants provided written informed consent in accordance with guidelines set by the Institutional Review Boards of Harvard University or Partners Healthcare.
Dataset IV
The fourth dataset consisted of 52 left-handed and 52 right-handed subjects matched by age (left-handed: 19.9 ± 1.9 years, right-handed: 19.9 ± 1.7 years, P = 0.90), gender (24 males per group), ethnicity (five Hispanic subjects per group), education (left-handed: 13.8 ± 1.7 years, right-handed: 13.8 ± 1.5 years, P = 0.67), fMRI data acquisition (matched for scanner, console, and investigator acquiring the data), and data quality (Signal-to-noise ratio (SNR), left-handed: 183.4 ± 36.4, right-handed: 183.4 ± 34.0, P = 0.98; head motion, left-handed: 0.05 ± 0.02 mm, right-handed: 0.05 ± 0.02 mm, P = 0.92) that were acquired as part of the GSP. For each participant, two resting-state fMRI runs (6 min 12 s per run) and a structural MRI run were acquired. MRI data acquisition parameters were identical to those described for the first dataset (Dataset I: GSP dataset). The handedness of each subject was assessed using the Edinburgh handedness inventory (78). All participants provided written informed consent in accordance with guidelines set by the Institutional Review Boards of Harvard University or Partners Healthcare.
Dataset V
The fifth dataset was a subset of the GSP dataset (Dataset I) and consisted of 279 male and 279 female participants matched by age, education, and handedness. For each participant, one or two resting-state fMRI runs (6 min 12 s per run) were acquired along with a structural T1-weighted scan. The MRI data acquisition parameters were identical to those described above for the first dataset (i.e., GSP). All participants provided written informed consent in accordance with guidelines set by the Institutional Review Boards of Harvard University or Partners Healthcare.
Dataset VI
The sixth dataset included 42 patients with first-episode subcortical stroke (mean age 50.7 ± 11.8 years; 39 males) and 23 age-matched healthy control participants (mean age 51.8 ± 6.9 years; 9 males). All participants provided written informed consent in accordance with guidelines of Xuanwu Hospital of Capital Medical University. Eligibility criteria for patients were (i) full admission history (within 7 days after symptom onset) and clinical diagnosis of ischemic stroke, (ii) unilateral infarction involving basal ganglia (we selected the patients with homogeneous lesions to minimize the impact of other factors such as lesion location and lesion size on our results), (iii) absence of other brain lesions or prior infarcts, (iv) absence of MRI contraindications, (v) clear time of symptom onset, and (vi) absence of deafness, blindness, aphasia, or visual field deficits, typical of cortical strokes. Eligibility criteria for healthy control participants included a lack of history of neurologic or psychiatric disease. Each participant underwent five resting-state fMRI scans over a period of 6 months (at 1 to 7, 14, 30, 90, and 180 days after stroke). Each scan session consisted of two to four resting-state fMRI runs (6 min per run) and a structural MRI run. All data were acquired on a 3T Tim Trio scanner (Siemens, Erlangen, Germany) using a 12-channel phased-array head coil. Structural images were collected using a T1-weighted MPRAGE sequence (TR = 1600 ms, TE = 2.15 ms, FA = 9°, 1.0 × 1.0 × 1.0–mm voxels, and FOV = 256). Functional images were acquired using a gradient-echo echo-planar pulse sequence (TR = 3000 ms, TE = 30 ms, FA = 90°, and 3.0 × 3.0 × 3.0–mm voxels). Participants were instructed to stay awake and keep their eyes open during the resting-state scans. All patients underwent rehabilitation therapy according to the Guidelines for Chinese Stroke Rehabilitation. Patients were treated by neurologists at the emergency department during the acute phase until they were stabilized (i.e., with stable vital signs and without neural symptoms progressing within 48 hours). Acute inpatient rehabilitation services were delivered by a multidisciplinary team including neurologists, rehabilitation physicians, physiotherapists, and nurses for 7 to 14 days. Postdischarge rehabilitation was provided by community medical centers. Each patient’s rehabilitation plan was personalized according to symptoms, guided by researchers and professional rehabilitation physicians in our team. These rehabilitation treatments are consistent with the American Heart Association and the American Stroke Association guidelines for adult stroke rehabilitation (79).
Dataset VII
The seventh dataset included 100 young healthy individuals (the “Unrelated 100” group, age between 22 and 35 except one individual from the 36+ group, 54 females) made publicly available by the HCP, supported by the WU-Minn Consortium (80). Written informed consent was obtained from each participant in accordance with relevant guidelines and regulations approved by the local institutional review board at Washington University in St. Louis. Each subject had two resting-state fMRI sessions (each session consisted of one run with left-to-right direction phase encoding and one run with right-to-left direction, 14 min 24 s per run). Subjects were scanned on a customized Siemens 3T “Connectome Skyra” scanner (Siemens, Erlangen, Germany) using a standard 32-channel head coil. Functional images were collected using the gradient-echo EPI sequence (TR = 720 ms, TE = 33.1 ms, FA = 52°, 2 × 2 × 2–mm voxels, FOV = 208 × 180 mm, multiband factor = 8, echo spacing = 0.58 ms, bandwidth = 2290 Hz/Px, time points = 1200, 72 oblique axial slices alternated between phase encoding in a right to left direction in one run and phase encoding in a left to right direction in the other run).
Data preprocessing
The fMRI data (the GSP, CoRR-HNU, and Stroke datasets; the details regarding the preprocessing of HCP dataset—Dataset VII—can be found in the Supplementary Materials) were preprocessed using a previously described analysis pipeline (28), which included the following steps: (i) slice timing correction (Statistical Parametric Mapping, SPM2; www.fil.ion.ucl.ac.uk/spm/software/spm2/), (ii) rigid body correction for head motion (FMRIB Software Library, FSL v5.0.4; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki), (iii) normalization for global mean signal intensity across runs, (iv) band-pass filtering (0.01 to 0.08 Hz), and (v) nuisance signal regression of head-motion parameters and whole-brain, ventricular, and white-matter signals.
Structural data were preprocessed using the FreeSurfer v5.3.0 software package (https://surfer.nmr.mgh.harvard.edu/). For each participant, the surface mesh of the cortical mantle was reconstructed from the structural T1-weighted image and then registered to a common spherical coordinate system (81). The preprocessed functional data were then registered to the FreeSurfer “fsaverage6” cortical surface template, which consisted of 40,962 vertices in each hemisphere. Spatial smoothing was performed in surface space with a 6-mm full width at half maximum Gaussian kernel. To reduce computational costs, the smoothed data were then down-sampled to the “fsaverage4” template, consisting of 2562 vertices per hemisphere.
Network-based single-frame dynamic coactivation analysis
The INSCAPE approach consists of the following two steps, which are described in detail below and illustrated in Fig. 1: (i) generating the group-level templates of brain coactivation states and (ii) estimating individual-specific brain states.
Generating group template of brain states
We used Dataset I to generate the group-level templates of coactivated brain states. Because head motion can confound the BOLD signal, we only included subjects with low head motion in this analysis, defined as having a mean and maximum head motion (framewise displacement) under 0.1 and 0.5 mm, respectively. Of a total of 1000 subjects from the GSP dataset, 154 subjects did not meet these criteria and therefore were excluded from the foregoing analyses, leaving a final sample of 846 subjects. This stringent criterion for head motion control was used only for creating the template brain states, ensuring the template was minimally affected by head motion-related noise. We did not exclude data based on head motion in other datasets involved in the subsequent analyses. The procedures for generating group templates of dynamic coactivated brain states are illustrated in the schematic presented in Fig. 1A. First, a network-based weight dictionary was created to suppress the effect of noise and retain the intrinsic network structure of the raw coactivation data. Specifically, to generate this weight dictionary, the atlas of seven canonical functional networks derived from a population-based cortical parcellation (82) was divided into multiple discontinuous patches. The patches that were represented in both the left and right hemispheres were preserved, resulting in a total of 48 network patches (24 patches per hemisphere). The parcellation confidence of these 48 network patches was then extracted to comprise the network weight dictionary. The parcellation confidence refers to the probability of each vertex to be assigned to a specific functional network during the population-based cortical parcellation (82). Vertices located in the center of a network patch would normally have higher parcellation confidence compared to those near the network boundary. After creating the weight dictionary, maps of preprocessed fMRI data were weighted at each vertex at each time point. The weighted single-frame fMRIs were then averaged within each of the 48 network patches and binarized so that values larger than 0 were set to 1 and values smaller than 0 were set to −1. The binarized fMRIs of all subjects were then concatenated along their time series and classified into 16 clusters, or brain states, using k-means clustering analysis. This cluster number was selected according to the optimal within-subject test-retest reliability of the resulting brain states (see fig. S1). Last, the maps of each cluster were averaged across subjects to comprise the group template of each brain state.
Estimating individual-specific brain states
At the individual level, functional images for each subject were first weighted by the network weight dictionary following the same procedure described above (Fig. 1A). The weighted map of each time frame was then compared with the group template of brain states and assigned to the brain state with the shortest spatial distance to it. Note that at the individual level, images were not binarized. The functional images assigned to the same brain state were averaged to yield an individualized brain state map (Fig. 1B). For each of the 16 brain states, the state occurrence rates were quantified as the number of time frames of a given brain state divided by the total number of fMRI time frames.
Evaluating the reliability of INSCAPE analysis
Test-retest reliability of the INSCAPE approach was evaluated using an independent sample of 30 young, healthy participants from the CoRR-HNU dataset (Dataset II). Each participant underwent 10 separate 10-min resting-state fMRI scans. The first five scans were assigned to the test session, and the last five scans were assigned to the retest session (50 min for each session). Then, the occurrence rates of the 16 brain states were estimated in the test and retest sessions. Intrasubject reliability of the INSCAPE analysis was evaluated using the similarity (Pearson correlation coefficient) of occurrence rates for the 16 brain states within each subject. The intersubject similarity was evaluated using the similarity of occurrence rates estimated between any pair of subjects. Given the known influence of data length on results derived from dynamic analyses, we also quantified the test-retest reliability of the INSCAPE results by computing the reliability of brain state occurrence rates using 10 different test-retest data lengths, ranging from 5 to 50 min, in 5-min increments of time.
Estimating functional lateralization of brain states
To explore the degree of functional lateralization of coactivated brain states, mean maps of each of the 16 brain states (i.e., group template) derived from the GSP dataset were first registered to a FreeSurfer symmetric cortical surface template with 2562 vertices per hemisphere. For each brain state, vertices with positive activity values were counted in the left and right hemispheres. The LI of a given brain state was then calculated as follows
where VL_Pos is the number of positively activated vertices in the left hemisphere, VR_Pos is the number of positively activated vertices in the right hemisphere, and VHemi is the total number of vertices in a single hemisphere (2562 vertices). Note that this LI was evaluated at the group template rather than at the individual level. Positive LI values indicated leftward lateralization, and negative LI values indicated rightward lateralization. Moreover, to verify whether functional laterality of coactivated brain states on the cortex could also be observed in the cerebellum, we computed the mean coactivation maps within the cerebellum mask in MNI152 volumetric space.
Functional lateralization is a fundamental property of the human brain that is also related to handedness and sex (34–36). Here, we investigated whether brain states identified by our INSCAPE approach can reflect functional laterality and whether they show handedness- and sex-related differences. The occurrence rates of lateralized brain states were first estimated in 55 healthy participants scanned during a language task [i.e., semantic decision task (77, 83)] and during resting state (Dataset III). A series of paired sample t tests were used to examine whether the occurrence rates of each of the 16 coactivated brain states differed significantly between task-driven and resting-state conditions. Language LI was then calculated for each individual based on the asymmetric activations in two hemispheres using the approach previously reported (84). Correlation between the language LI and the occurrence rate of the most leftward lateralized brain state during the task was calculated. The occurrence of the most leftward and rightward lateralized brain states was counted across all subjects at each time frame during the language task and then normalized by the number of subjects. To investigate whether the occurrence of lateralized brain states was associated with language-related processing, we compared the normalized occurrence rates of each of the 16 coactivated brain states with the hemodynamic response curves evoked by the language task onsets using Pearson correlation.
In addition, to determine the effects of handedness on functional lateralization of brain states, the occurrence rate was calculated in 52 left-handed and 52 demographically matched right-handed subjects (Dataset IV). Paired sample t tests were performed to examine whether differences in brain state occurrence rates were affected by handedness. We also compared the distributions of the occurrence rates in left- and right-handed subjects using the Kolmogorov-Smirnov test. These same analyses were carried out in a separate dataset of 279 males and 279 demographically matched females (Dataset V; subjects matched by age, education, and handedness) to examine whether gender influenced the occurrence of lateralized brain states.
Tracking longitudinal changes in the occurrence rates of brain states during the poststroke recovery period
We quantified the occurrence rates of brain states in 42 patients with subcortical stroke at five time points following stroke onset (i.e., 1 to 7, 14, 30, 90, and 180 days after stroke) and 23 healthy controls (Dataset VI). The occurrence rates were compared between the first scan session, which served as the baseline scan and was acquired within 1 to 7 days after stroke, and each of the four subsequent scan sessions at 14, 30, 90, and 180 days after stroke. A repeated-measures ANOVA was conducted to investigate whether the occurrence rates of brain states changed over the course of the 6-month poststroke recovery period relative to the baseline time point. We specified time (days 7, 14, 30, 90, and 180) as the within-subject factor and subjects as the fixed factor. A series of post hoc analyses were conducted using paired sample t tests to determine which time points had occurrence rates that differed significantly from the baseline time point. We also compared occurrence rates between the patient group and the group of healthy controls using two-sample t tests to ascertain whether there was a normalization in occurrence rates of dynamic brain states in patients over the course of the 6-month poststroke recovery period to levels seen in control subjects.
Statistical analysis
The reproducibility of the INSCAPE approach was evaluated by calculating the Pearson correlations of occurrence rates for the 16 brain states between the Discovery and Replication datasets. Spatial similarity of brain state maps was quantified by Spearman correlation. Durbin-Watson test was applied to examine the potential impact of spatial dependence between neighboring vertices on correlations. Specifically, we performed a repeated (n = 1000) random sampling of 7% of the vertices and computed the correlation coefficient on the subsets of the vertices. Durbin-Watson test was performed to calculate the spatial dependence for each subset and averaged across the 1000 iterations. In all spatial correlations reported in this study, the values of the Durbin-Watson statistic were close to 2, with P > 0.05, suggesting that there was no significant spatial autocorrelation in the subset of vertices. The difference between intrasubject and intersubject similarity of state occurrence rates was examined using the two-sample t test. Paired t test was calculated to evaluate the state occurrence rate changes during language task compared to resting state. Pearson correlation was used to investigate the relationship between the task-based language LI and the occurrence rate of the left-lateralized state and the relationship between the occurrence of brain states across all participants at each time point of the language task and the hemodynamic response curves elicited by task processing. Paired t test was used to compare the difference in state occurrence rates between left-handed and right-handed, as well as between male and female participants. The distributions of the occurrence rates in handedness and gender were compared using the Kolmogorov-Smirnov test. For the stroke patient data, a repeated-measures ANOVA was conducted to evaluate the changes in the occurrence rate of brain state during poststroke recovery. Paired t test was used to determine which time points had occurrence rates that differed significantly from the baseline time point.
Acknowledgments
Funding: This work was supported by Changping Laboratory, Ministry of Science and Technology of China grant 2021B-01-01, National Natural Science Foundation of China grant 81790652, National Natural Science Foundation of China grant 81790650, National Institutes of Health grant R01DC017991, National Institutes of Health grant P50MH106435, National Institutes of Health grant R21NS120186, National Institutes of Health grant R61MH121640, and National Institutes of Health grant 5K01MH111802.
Author contributions: Conception and design of the study: X.P., Q.L., M.D.F., and HL. Acquisition and analysis of data: X.P., Q.L., D.W., and H.L. Drafting a significant portion of the manuscript or figures: X.P., Q.L., C.S.H., D.W., W.Z., M.D.F., and H.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The GSP dataset is available through the GSP (https://dataverse.harvard.edu/dataverse/GSP). The CoRR-HNU dataset is available through the Consortium for Reliability and Reproducibility Project (http://fcon_1000.projects.nitrc.org/indi/CoRR/html/hnu_1.html). The HCP dataset is available through the Human Connectome Project (https://humanconnectome.org/). The stroke dataset and the codes used in this article are available at https://doi.org/10.5061/dryad.kwh70rz76.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
REFERENCES AND NOTES
- 1.Varela F., Lachaux J.-P., Rodriguez E., Martinerie J., The brainweb: Phase synchronization and large-scale integration. Nat. Rev. Neurosci. 2, 229–239 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Ward L. M., Synchronous neural oscillations and cognitive processes. Trends Cogn. Sci. 7, 553–559 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Bressler S. L., Kelso J. S., Cortical coordination dynamics and cognition. Trends Cogn. Sci. 5, 26–36 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Deco G., Jirsa V. K., McIntosh A. R., Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci. 12, 43–56 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Rabinovich M. I., Varona P., Selverston A. I., Abarbanel H. D., Dynamical principles in neuroscience. Rev. Mod. Phys. 78, 1213V1265 (2006). [Google Scholar]
- 6.Allen E. A., Damaraju E., Plis S. M., Erhardt E. B., Eichele T., Calhoun V. D., Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y., Fu Z., Calhoun V. D., Classification and prediction of brain disorders using functional connectivity: Promising but challenging. Front. Neurosci. 12, 525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Castillo J., Bandettini P. A., Task-based dynamic functional connectivity: Recent findings and open questions. Neuroimage 180, 526–533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchison R. M., Womelsdorf T., Allen E. A., Bandettini P. A., Calhoun V. D., Corbetta M., Della Penna S., Duyn J. H., Glover G. H., Gonzalez-Castillo J., Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage 80, 360–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C., Glover G. H., Time–frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage 50, 81–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Castillo J., Hoy C. W., Handwerker D. A., Robinson M. E., Buchanan L. C., Saad Z. S., Bandettini P. A., Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc. Natl. Acad. Sci. U.S.A. 112, 8762–8767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shine J. M., Koyejo O., Poldrack R. A., Temporal metastates are associated with differential patterns of time-resolved connectivity, network topology, and attention. Proc. Natl. Acad. Sci. U.S.A. 113, 9888–9891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shine J. M., Bissett P. G., Bell P. T., Koyejo O., Balsters J. H., Gorgolewski K. J., Moodie C. A., Poldrack R. A., The dynamics of functional brain networks: Integrated network states during cognitive task performance. Neuron 92, 544–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barttfeld P., Uhrig L., Sitt J. D., Sigman M., Jarraya B., Dehaene S., Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl. Acad. Sci. U.S.A. 112, 887–892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen K., Hutchison R. M., Bezgin G., Everling S., McIntosh A. R., Network structure shapes spontaneous functional connectivity dynamics. J. Neurosci. 35, 5579–5588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C., Liu Z., Chen M. C., Liu X., Duyn J. H., EEG correlates of time-varying BOLD functional connectivity. Neuroimage 72, 227–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laufs H., Krakow K., Sterzer P., Eger E., Beyerle A., Salek-Haddadi A., Kleinschmidt A., Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proc. Natl. Acad. Sci. U.S.A. 100, 11053–11058 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Duyn J. H., Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc. Natl. Acad. Sci. U.S.A. 110, 4392–4397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D., Peng X., Pelletier-Baldelli A., Orlov N., Farabaugh A., Nasr S., Eryilmaz H., Fava M., Holmes A. J., Roffman J. L., Altered temporal, but intact spatial, features of transient network dynamics in psychosis. Mol. Psychiatry 26, 2493–2503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janes A. C., Peechatka A. L., Frederick B. B., Kaiser R. H., Dynamic functioning of transient resting-state coactivation networks in the Human Connectome Project. Hum. Brain Mapp. 41, 373–387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J. E., Chang C., Greicius M. D., Glover G. H., Introducing co-activation pattern metrics to quantify spontaneous brain network dynamics. Neuroimage 111, 476–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Chang C., Duyn J. H., Decomposition of spontaneous brain activity into distinct fMRI co-activation patterns. Front. Syst. Neurosci. 7, 101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laumann T. O., Snyder A. Z., Mitra A., Gordon E. M., Gratton C., Adeyemo B., Gilmore A. W., Nelson S. M., Berg J. J., Greene D. J., On the stability of BOLD fMRI correlations. Cereb. Cortex 27, 4719–4732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott M. L., Knodt A. R., Ireland D., Morris M. L., Poulton R., Ramrakha S., Sison M. L., Moffitt T. E., Caspi A., Hariri A. R., What is the test-retest reliability of common task-functional MRI measures? New empirical evidence and a meta-analysis. Psychol. Sci. 31, 792–806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo X.-N., Biswal B. B., Poldrack R. A., Editorial: Reliability and reproducibility in functional connectomics. Front. Neurosci. 13, 117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Button K. S., Ioannidis J., Mokrysz C., Nosek B. A., Flint J., Robinson E. S., Munafò M. R., Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Dubois J., Adolphs R., Building a science of individual differences from fMRI. Trends Cogn. Sci. 20, 425–443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D., Buckner R. L., Fox M. D., Holt D. J., Holmes A. J., Stoecklein S., Langs G., Pan R., Qian T., Li K., Parcellating cortical functional networks in individuals. Nat. Neurosci. 18, 1853–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., Li M., Wang M., Schoeppe F., Ren J., Chen H., Öngür D., Brady R. O., Baker J. T., Liu H., Individual-specific functional connectivity markers track dimensional and categorical features of psychotic illness. Mol. Psychiatry 25, 2119–2129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gratton C., Laumann T. O., Nielsen A. N., Greene D. J., Gordon E. M., Gilmore A. W., Nelson S. M., Coalson R. S., Snyder A. Z., Schlaggar B. L., Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98, 439–452.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elliott M. L., Knodt A. R., Hariri A. R., Striving toward translation: Strategies for reliable fMRI measurement. Trends Cogn. Sci. 25, 776–787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birn R. M., Molloy E. K., Patriat R., Parker T., Meier T. B., Kirk G. R., Nair V. A., Meyerand M. E., Prabhakaran V., The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage 83, 550–558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller S., Wang D., Fox M. D., Pan R., Lu J., Li K., Sun W., Buckner R. L., Liu H., Reliability correction for functional connectivity: Theory and implementation. Hum. Brain Mapp. 36, 4664–4680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Stufflebeam S. M., Sepulcre J., Hedden T., Buckner R. L., Evidence from intrinsic activity that asymmetry of the human brain is controlled by multiple factors. Proc. Natl. Acad. Sci. U.S.A. 106, 20499–20503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gotts S. J., Jo H. J., Wallace G. L., Saad Z. S., Cox R. W., Martin A., Two distinct forms of functional lateralization in the human brain. Proc. Natl. Acad. Sci. U.S.A. 110, E3435–E3444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomasi D., Volkow N. D., Laterality patterns of brain functional connectivity: Gender effects. Cereb. Cortex 22, 1455–1462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., Buckner R. L., Liu H., Functional specialization in the human brain estimated by intrinsic hemispheric interaction. J. Neurosci. 34, 12341–12352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidaurre D., Smith S. M., Woolrich M. W., Brain network dynamics are hierarchically organized in time. Proc. Natl. Acad. Sci. U.S.A. 114, 12827–12832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badcock P. B., Friston K. J., Ramstead M. J., The hierarchically mechanistic mind: A free-energy formulation of the human psyche. Phys. Life Rev. 31, 104–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassett D. S., Sporns O., Network neuroscience. Nat. Neurosci. 20, 353–364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shine J. M., Breakspear M., Bell P. T., Martens K. A. E., Shine R., Koyejo O., Sporns O., Poldrack R. A., Human cognition involves the dynamic integration of neural activity and neuromodulatory systems. Nat. Neurosci. 22, 289–296 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Yan B., Xu X., Liu M., Zheng K., Liu J., Li J., Wei L., Zhang B., Lu H., Li B., Quantitative identification of major depression based on resting-state dynamic functional connectivity: A machine learning approach. Front. Neurosci. 14, 191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin C., Jia H., Lanka P., Rangaprakash D., Li L., Liu T., Hu X., Deshpande G., Dynamic brain connectivity is a better predictor of PTSD than static connectivity. Hum. Brain Mapp. 38, 4479–4496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Horn H. J., Vergara V. M., Espinoza F. A., Calhoun V. D., Mayer A. R., van der Naalt J., Functional outcome is tied to dynamic brain states after mild to moderate traumatic brain injury. Hum. Brain Mapp. 41, 617–631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutchison R. M., Womelsdorf T., Gati J. S., Everling S., Menon R. S., Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum. Brain Mapp. 34, 2154–2177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindriks R., Adhikari M. H., Murayama Y., Ganzetti M., Mantini D., Logothetis N. K., Deco G., Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage 127, 242–256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribolsi M., Koch G., Magni V., Di Lorenzo G., Rubino I. A., Siracusano A., Centonze D., Abnormal brain lateralization and connectivity in schizophrenia. Rev. Neurosci. 20, 61–70 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Xie W., Peng C.-K., Huang C.-C., Lin C.-P., Tsai S.-J., Yang A. C., Functional brain lateralization in schizophrenia based on the variability of resting-state fMRI signal. Prog. NeuroPsychopharmacol. Biol. Psychiatry 86, 114–121 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Nielsen J. A., Zielinski B. A., Fletcher P. T., Alexander A. L., Lange N., Bigler E. D., Lainhart J. E., Anderson J. S., Abnormal lateralization of functional connectivity between language and default mode regions in autism. Mol. Autism. 5, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen T., Milner B., The role of early left-brain injury in determining lateralization of cerebral speech functions. Ann. N. Y. Acad. Sci. 299, 355–369 (1977). [DOI] [PubMed] [Google Scholar]
- 51.Miller J. W., Jayadev S., Dodrill C. B., Ojemann G. A., Gender differences in handedness and speech lateralization related to early neurologic insults. Neurology 65, 1974–1975 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Smith S. M., Fox P. T., Miller K. L., Glahn D. C., Fox P. M., Mackay C. E., Filippini N., Watkins K. E., Toro R., Laird A. R., Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U.S.A. 106, 13040–13045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole M. W., Bassett D. S., Power J. D., Braver T. S., Petersen S. E., Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krienen F. M., Yeo B. T., Buckner R. L., Reconfigurable task-dependent functional coupling modes cluster around a core functional architecture. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersson-Yeo W., Allen P., Benetti S., McGuire P., Mechelli A., Dysconnectivity in schizophrenia: Where are we now? Neurosci. Biobehav. Rev. 35, 1110–1124 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Menon V., Large-scale brain networks and psychopathology: A unifying triple network model. Trends Cogn. Sci. 15, 483–506 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Park C.-H., Chang W. H., Ohn S. H., Kim S. T., Bang O. Y., Pascual-Leone A., Kim Y.-H., Longitudinal changes of resting-state functional connectivity during motor recovery after stroke. Stroke 42, 1357–1362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker J. T., Holmes A. J., Masters G. A., Yeo B. T., Krienen F., Buckner R. L., Öngür D., Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiat. 71, 109–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaiser R. H., Whitfield-Gabrieli S., Dillon D. G., Goer F., Beltzer M., Minkel J., Smoski M., Dichter G., Pizzagalli D. A., Dynamic resting-state functional connectivity in major depression. Neuropsychopharmacology 41, 1822–1830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun U., Schäfer A., Bassett D. S., Rausch F., Schweiger J. I., Bilek E., Erk S., Romanczuk-Seiferth N., Grimm O., Geiger L. S., Dynamic brain network reconfiguration as a potential schizophrenia genetic risk mechanism modulated by NMDA receptor function. Proc. Natl. Acad. Sci. U.S.A. 113, 12568–12573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lebois L. A., Li M., Baker J. T., Wolff J. D., Wang D., Lambros A. M., Grinspoon E., Winternitz S., Ren J., Gönenç A., Large-scale functional brain network architecture changes associated with trauma-related dissociation. Am. J. Psychiatry 178, 165–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ladd M. E., Bachert P., Meyerspeer M., Moser E., Nagel A. M., Norris D. G., Schmitter S., Speck O., Straub S., Zaiss M., Pros and cons of ultra-high-field MRI/MRS for human application. Prog. Nucl. Magn. Reson. Spectrosc. 109, 1–50 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Shah P., Bassett D. S., Wisse L. E., Detre J. A., Stein J. M., Yushkevich P. A., Shinohara R. T., Pluta J. B., Valenciano E., Daffner M., Mapping the structural and functional network architecture of the medial temporal lobe using 7T MRI. Hum. Brain Mapp. 39, 851–865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feinberg D. A., Yacoub E., The rapid development of high speed, resolution and precision in fMRI. Neuroimage 62, 720–725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glasser M. F., Coalson T. S., Robinson E. C., Hacker C. D., Harwell J., Yacoub E., Ugurbil K., Andersson J., Beckmann C. F., Jenkinson M., A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Head M. L., Holman L., Lanfear R., Kahn A. T., Jennions M. D., The extent and consequences of p-hacking in science. PLOS Biol. 13, e1002106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Charrad M., Ghazzali N., Boiteau V., Niknafs A., NbClust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 61, 1–36 (2014). [Google Scholar]
- 68.Fan F., Liao X., Lei T., Zhao T., Xia M., Men W., Wang Y., Hu M., Liu J., Qin S., Development of the default-mode network during childhood and adolescence: A longitudinal resting-state fMRI study. Neuroimage 226, 117581 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Matsui T., Pham T. Q., Jimura K., Chikazoe J., On co-activation pattern analysis and non-stationarity of resting brain activity. Neuroimage 249, 118904 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Iraji A., Faghiri A., Fu Z., Kochunov P., Adhikari B. M., Belger A., Ford J. M., McEwen S., Mathalon D. H., Pearlson G. D., Moving beyond the ‘CAP’ of the Iceberg: Intrinsic connectivity networks in fMRI are continuously engaging and overlapping. Neuroimage 251, 119013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H.-L., Zahneisen B., Hugger T., LeVan P., Hennig J., Tracking dynamic resting-state networks at higher frequencies using MR-encephalography. Neuroimage 65, 216–222 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Gohel S. R., Biswal B. B., Functional integration between brain regions at rest occurs in multiple-frequency bands. Brain Connect. 5, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baria A. T., Baliki M. N., Parrish T., Apkarian A. V., Anatomical and functional assemblies of brain BOLD oscillations. J. Neurosci. 31, 7910–7919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogawa S., Tank D. W., Menon R., Ellermann J. M., Kim S. G., Merkle H., Ugurbil K., Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci. U.S.A. 89, 5951–5955 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biswal B., Deyoe E. A., Hyde J. S., Reduction of physiological fluctuations in fMRI using digital filters. Magn. Reson. Med. 35, 107–113 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Holmes A. J., Hollinshead M. O., O’keefe T. M., Petrov V. I., Fariello G. R., Wald L. L., Fischl B., Rosen B. R., Mair R. W., Roffman J. L., Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci. Data 2, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Desmond J. E., Sum J. M., Wagner A. D., Demb J. B., Shear P. K., Glover G. H., Gabrieli J., Morrell M., Functional MRI measurement of language lateralization in Wada-tested patients. Brain 118, 1411–1419 (1995). [DOI] [PubMed] [Google Scholar]
- 78.Oldfield R. C., The assessment and analysis of handedness: The Edinburgh inventory. Neuropsychologia 9, 97–113 (1971). [DOI] [PubMed] [Google Scholar]
- 79.Winstein C. J., Stein J., Arena R., Bates B., Cherney L. R., Cramer S. C., Deruyter F., Eng J. J., Fisher B., Harvey R. L., Guidelines for adult stroke rehabilitation and recovery: A guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke 47, e98–e169 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Van Essen D. C., Smith S. M., Barch D. M., Behrens T. E., Yacoub E., Ugurbil K.; WU-Minn HCP Consortium , The WU-Minn Human Connectome Project: An overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischl B., Sereno M. I., Dale A. M., Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 195–207 (1999). [DOI] [PubMed] [Google Scholar]
- 82.Yeo B. T., Krienen F. M., Sepulcre J., Sabuncu M. R., Lashkari D., Hollinshead M., Roffman J. L., Smoller J. W., Zöllei L., Polimeni J. R., The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Demb J. B., Desmond J. E., Wagner A. D., Vaidya C. J., Glover G. H., Gabrieli J., Semantic encoding and retrieval in the left inferior prefrontal cortex: A functional MRI study of task difficulty and process specificity. J. Neurosci. 15, 5870–5878 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Binder J. R., Swanson S. J., Hammeke T. A., Morris G. L., Mueller W. M., Fischer M., Benbadis S., Frost J. A., Rao S. M., Haughton V. M., Determination of language dominance using functional MRI: A comparison with the Wada test. Neurology 46, 978–984 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13