Abstract
Helicobacter pylori vacuolating cytotoxin (VacA) is a secreted protein that induces vacuolation of epithelial cells. To study VacA structure and function, we immunized mice with purified type s1-m1 VacA from H. pylori strain 60190 and generated a panel of 10 immunoglobulin G1κ anti-VacA monoclonal antibodies. All of the antibodies reacted with purified native VacA but not with denatured VacA, suggesting that these antibodies react with conformational epitopes. Seven of the antibodies reacted with both native and acid-treated VacA, which suggests that epitopes present on both oligomeric and monomeric forms of the toxin were recognized. Two monoclonal antibodies, both reactive with epitopes formed by amino acids in the carboxy-terminal portion of VacA (amino acids 685 to 821), neutralized the cytotoxic activity of type s1-m1 VacA when toxin and antibody were mixed prior to cell contact but failed to neutralize the cytotoxic activity of type s1-m2 VacA. Only 3 of the 10 antibodies consistently recognized type s1-m1 VacA toxins from multiple H. pylori strains, and none of the antibodies recognized type s2-m2 VacA toxins. These results indicate that there is considerable antigenic diversity among VacA toxins produced by different H. pylori strains.
Helicobacter pylori is a gram-negative bacterium that colonizes the mucosal layer of the human stomach and induces chronic superficial gastritis (10, 20). Colonization with this bacterium is a risk factor for the development of peptic ulcer disease and gastric cancer (20). One virulence factor produced by H. pylori is a secreted protein toxin (VacA) that induces the formation of large cytoplasmic vacuoles in epithelial cells (9, 40). At neutral pH, VacA assembles into large, water-soluble oligomeric complexes composed predominantly of 12 or 14 identical monomers (14, 32). When exposed to acidic or alkaline pH, these oligomeric complexes disassemble into component monomers (14, 38, 58). Acid-activated VacA can insert into lipid bilayers and the plasma membrane of eukaryotic cells to form anion-selective membrane channels (17, 29, 38, 51, 52).
The mature secreted VacA toxin has a molecular mass of 88 kDa and consists of about 821 amino acids (11, 41). In HeLa cells transiently transfected with vacA-containing plasmids, expression of the amino-terminal 422 amino acids of VacA is sufficient to induce vacuole formation (18, 59). Rabbit antiserum generated against a recombinant peptide containing amino acids 476 to 803 of VacA inhibits the binding of VacA to cells and neutralizes toxin activity (23). When taken together, these data suggest that a carboxy-terminal VacA domain mediates binding of the toxin to cells and an amino-terminal domain mediates the intracellular activity of VacA.
H. pylori strains isolated from different human stomachs are genetically very heterogeneous (2, 3, 36). For essentially any gene selected for analysis, the sequences from different strains exhibit 95 to 98% nucleotide identity (1, 4, 22, 31). Suerbaum et al. analyzed a 450-nucleotide segment of vacA (nucleotides 802 to 1245; GenBank accession no. Z26883) in 69 H. pylori strains isolated from two different geographic locations and found that very few sequences were identical (50). Within this region of vacA, 25.4% of the nucleotide positions were polymorphic among the different strains, but almost all of these polymorphisms represented synonymous substitutions. Another study of 153 isolates by Göttke et al. revealed similar levels of sequence diversity in this region of vacA (25). Both studies concluded that genetic recombination has occurred more frequently in H. pylori than in most other bacteria analyzed thus far.
Certain regions in vacA exhibit greater sequence diversity than the segments analyzed by Suerbaum et al. (50) and Göttke et al. (25). Within a 0.7-kb region of vacA known as the midregion, the sequences of vacA alleles from different H. pylori strains can exhibit <70% nucleotide identity (5, 7, 42, 49). Diversity is also prominent in the 5′ portion of vacA that encodes the amino-terminal signal sequence and the amino terminus of the mature toxin (5, 7, 54, 55). Based on analysis of vacA alleles from large numbers of H. pylori strains, two families of midregions (m1 and m2) and two families of signal sequence regions (s1 and s2) are currently recognized (5–7, 53, 55).
Classification of vacA alleles into families (s1, s2, m1, and m2) has proven useful as a method for predicting levels of cytotoxin activity in vitro. Broth culture supernatants from H. pylori strains containing type s1-m1 vacA alleles typically exhibit a high level of cytotoxic activity for multiple cell types, whereas supernatants from strains containing type s2-m2 vacA alleles lack cytotoxic activity (5, 21). Some type s1-m2 vacA toxins exhibit cytotoxic activity toward selected cell types, including RK-13 and Vero, but relatively little activity for HeLa cells (references 30 and 42 and our unpublished data). The basis for these differences in cytotoxic activity among H. pylori strains is probably multifactorial and may reflect differences in vacA transcription, expression, or secretion (21) or may be directly related to polymorphisms in VacA amino acid sequences (5).
Heterogeneity among vacA alleles may be an important factor in understanding variations in clinical manifestations among H. pylori-infected persons. Several studies have demonstrated that gastric infection with H. pylori strains containing type s1 vacA alleles is associated with a higher risk for development of peptic ulcer disease than is infection with strains containing type s2 vacA alleles (5, 24, 47, 53). This association seems to be less apparent in many Asian countries than in Europe and the Americas (27, 43).
Thus far, nearly all studies of VacA diversity have been based on analysis of vacA nucleotide sequences, rather than on analysis of VacA proteins. In this study we sought to analyze VacA structure, function, and diversity by using a panel of anti-VacA monoclonal antibodies. We report here that VacA activity can be neutralized by monoclonal antibodies reactive with the carboxy-terminal portion of the toxin and demonstrate that VacA proteins from different H. pylori strains exhibit considerable antigenic diversity, even within the same vacA family.
MATERIALS AND METHODS
Growth of H. pylori and purification of VacA.
H. pylori strains were grown routinely on Trypticase soy agar plates containing 5% sheep blood in room air containing 5% CO2 at 37°C. H. pylori broth culture supernatants were prepared by growing the bacteria in sulfite-free brucella broth containing either 5% fetal bovine serum (FBS) or 0.5% charcoal for 48 h and removing the bacteria by centrifugation (11). VacA was purified from broth culture supernatant of H. pylori strain 60190 (ATCC 49503), as described previously (14). Acid activation of purified VacA was accomplished by dropwise addition of 250 mM HCl until the pH was reduced to 2.0 (14, 19).
Production and purification of monoclonal antibodies.
Female BALB/c mice (6 to 8 weeks old) were injected intraperitoneally with 15 μg of purified VacA from H. pylori strain 60190 (ATCC 49503) in 0.5 ml of complete Freund's adjuvant. Mice were boosted 4 and 8 weeks after the initial immunization with VacA in phosphate-buffered saline (PBS). Twelve weeks after the initial immunization, spleens were harvested from immunized mice, and the cells were fused with SP2/0 cells. Hybridoma cells were selected by growth in medium containing hypoxanthine-azaserine-thymidine and screened for production of anti-VacA antibodies by enzyme-linked immunosorbent assay (ELISA), as described below. Single clonal populations of anti-VacA-producing hybridoma cells were isolated by the limiting-dilution method. Ten different clonal populations of anti-VacA-producing hybridoma cells were injected intraperitoneally into pristane-primed BALB/c mice in order to generate ascites fluid. Antibodies in ascites fluid were isotyped by ELISA as described below, using a panel of different secondary antibody-horseradish peroxidase (HRP) conjugates (Southern Biotechnology Associates, Inc.). To purify monoclonal antibodies from ascites fluid, 1.5% caprylic acid in 45 mM sodium acetate (pH 4.8) was mixed with ascites fluid to precipitate contaminating proteins, and anti-VacA IgG antibodies were then precipitated from the supernatant with a 50% saturated solution of ammonium sulfate (26). Antibody concentrations were determined by measuring absorbance at 280 nm and confirmed by Micro BCA assay (Pierce). Purified monoclonal antibodies (1 mg/ml) were stored at −20°C in 0.15 M NaCl and 50% glycerol.
Construction of H. pylori strains expressing mutant VacA proteins.
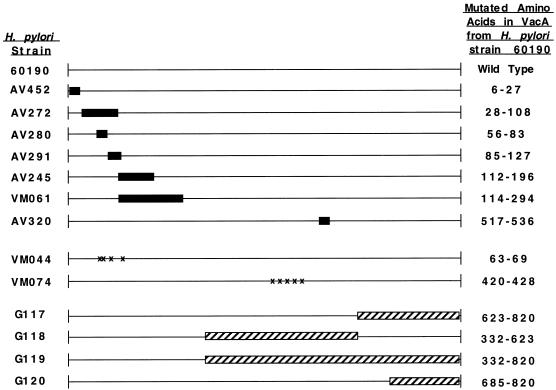
A panel of H. pylori strains containing in-frame deletion mutations in vacA has been described previously (56). In addition, a panel of H. pylori strains expressing mutant VacA proteins in which various clusters of amino acids were changed to alanine residues has been constructed (M. S. McClain, P. Cao, D. Choate, and T. L. Cover, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. D/B-192, 1999). To construct these strains, vacA-containing plasmids were mutated by using the method of Perrin and Gilliland (45), and these plasmids were then transformed into H. pylori strains containing sacB-kan insertions in vacA (8, 56). The desired transformants, in which sacB-kan cassettes were replaced by vacA sequences via a double-crossover event, were selected by growth on Columbia blood agar plates containing 6% sucrose (8, 56). H. pylori strain VM044 expressed a mutant VacA protein in which the underlined residues (63 to 69) of the mature toxin were changed to alanine (KEYDLYK), and H. pylori strain VM074 expressed a mutant toxin in which the underlined residues (420 to 428) of the mature VacA protein were changed to alanine (RVNNQVGGY) (Fig. 1). Based on secondary structure predictions (MacVector), the latter region is predicted to form an amphipathic β sheet.
FIG. 1.
H. pylori strains expressing mutant forms of VacA used in this study. The dark boxes represent regions deleted in the translated vacA product from H. pylori strain 60190 (56), striped boxes represent VacA sequences from H. pylori strain 60190 replaced with corresponding sequences from H. pylori Tx30a in VacA chimeras, and the X's represent sites where selected amino acids were replaced with alanine. The amino acid numbering system is based on designation of the N-terminal alanine of the mature toxin from H. pylori strain 60190 as amino acid 1.
To construct chimeric vacA genes containing portions of m1 vacA and portions of m2 vacA, vacA fragments from H. pylori Tx30a (ATCC 51932) (containing a type s2-m2 vacA allele) (5) were PCR amplified using the following primer pairs, which were designed to contain the indicated restriction enzyme sites (underlined): primer C7386 (5′CCTCCCGAAGGCGGTTATGAG) (EcoNI) and primer C6908 (5′GCTAGCGAAACGCGCGTTATTAG) (NheI), primer C7386 (EcoNI) and primer C6906 (5′GACTATAGTCCATGCTTGCGTTG) (PshAI), primer C6904 (5′GACTATAGTCAAGATTTGGATTTAACC) (PshAI) and primer C6908 (NheI), and primer C6905 (5′AGATCTCACTAAAAATAAAGAACATG) (BglII) and primer C6908 (NheI). The resulting PCR products encode amino acids 337 to 876, 337 to 674, 671 to 876, and 735 to 876, respectively, of the VacA protoxin from Tx30a (5). The PCR products were subcloned into pGEM-T Easy (Promega). Following digestion with the restriction enzymes listed above, the vacA fragments were cloned into pCTB6, containing a vacA fragment from H. pylori strain 60190 (16), using the same sites. The resulting plasmids were termed pMM446, pMM443, pMM439, and pMM441, respectively. The chimeric sequences were introduced into H. pylori strain VM002 or VM018, containing sacB-km cassettes in the vacA gene of H. pylori 60190, using allelic exchange as described previously (56). The resulting H. pylori strains contained type s1 vacA alleles with portions of the m1 regions replaced by m2 vacA sequences and were designated as follows: G117 contains amino acids 671 to 876 from Tx30a, G118 contains amino acids 337 to 674 from Tx30a, G119 contains amino acids 337 to 876 from Tx30a, and G120 contains amino acids 735 to 876 from Tx30a (Fig. 1).
ELISAs.
Purified VacA or various VacA-containing preparations were diluted in PBS and adsorbed to microtiter wells in 96-well ELISA plates (Dynex) by incubation at 4°C for 18 h. Purified oligomeric VacA from H. pylori strain 60190 was adsorbed to wells at a final concentration of 2 μg/ml, and in experiments using unconcentrated H. pylori broth culture supernatants as antigens, undiluted supernatants were adsorbed directly to microtiter plates. Nonspecific protein binding sites were blocked by incubating plates with PBS (pH 7) containing 0.05% (vol/vol) Tween 20 (PBS-Tween) at room temperature for 5 min. Purified anti-VacA monoclonal antibodies, standardized by protein concentration, were diluted in PBS, added to the wells, and incubated for 1 h at room temperature. After being washed with PBS-Tween, bound monoclonal antibodies were detected with goat anti-mouse immunoglobulin G1 (IgG1) conjugated to HRP (Southern Biotechnology Associates, Inc.). Hydrogen peroxide (Sigma) and 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) (Sigma) were added, and color development was quantified at 410 nm with an MRX microplate reader (Dynatech).
Several variations in ELISA methodology were also utilized. (i) To screen hybridoma cells for production of anti-VacA antibodies, purified VacA was adsorbed to an ELISA plate as described above. Aliquots of undiluted growth media from hybridoma cells were then added to the wells for 1 h, followed by goat anti-mouse Ig antibodies conjugated to HRP (Boehringer Mannheim). (ii) To analyze the reactivity of the monoclonal antibodies to acidified VacA, purified VacA from H. pylori strain 60190 was acid activated by the addition of 250 mM HCl to pH 2, diluted to 2 μg/ml in 5% glacial acetic acid, and adsorbed to the microtiter plates, and the liquid was allowed to evaporate. Subsequent steps were performed as described in the previous paragraph. (iii) The ELISA reactivity of antibodies with denatured VacA was examined by two different approaches. As a first approach, purified VacA from H. pylori strain 60190 (final concentration, 2 μg/ml) was boiled in 6 M urea for 15 min and added to microtiter plates, and the liquid was allowed to evaporate. As a second approach, purified VacA (final concentration, 2 μg/ml) was heated at 95°C for 5 min in 1.5% Tris (pH 8.5) containing 2% sodium dodecyl sulfate and 10% glycerol, added to microtiter plates, and incubated at 4°C for 18 h. All subsequent steps were performed as described above. (iv) In the antigen capture ELISA, goat anti-mouse IgG (Southern Biotechnology Associates, Inc.) (5 μg/ml in PBS) was first adsorbed to a microtiter plate. Then, anti-VacA monoclonal antibodies (10 μg/ml), purified VacA (1 μg/ml), anti-VacA rabbit sera (958), and goat anti-rabbit Ig conjugated to HRP (Boehringer Mannheim) (5 μg/ml) were sequentially added to the ELISA plate and incubated for 1 h each. Plates were washed with PBS-Tween between each component.
Immunoprecipitations of VacA–anti-VacA monoclonal antibody complexes.
Purified VacA was diluted in PBS-Tween containing 2% (wt/vol) ammonium sulfate (pH 7), to yield a final concentration of 0.5 μg/ml. Anti-VacA monoclonal antibody (1 μg) was added to 1 ml of the VacA preparation and incubated at 4°C for 1 h. Protein G-Sepharose beads (Zymed) (25 μl), washed twice with PBS-Tween, were then added to the toxin-antibody mixtures and incubated for an additional hour at 4°C. Beads were then washed three times in PBS-Tween containing 2% ammonium sulfate. Immunoprecipitated proteins were separated from the beads by being boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and analyzed by immunoblot analysis with rabbit anti-VacA serum, as described previously (11).
Toxin neutralizing activity of monoclonal antibodies.
Purified VacA was acid activated (14, 19) and reneutralized by diluting the acid-activated VacA in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS and 10 mM ammonium chloride to yield a final VacA concentration of 6 μg/ml. Alternatively, proteins in H. pylori broth culture supernatants from cultures grown in sulfite-free brucella broth containing 5% FBS were precipitated with a 50% saturated ammonium sulfate solution and diluted in DMEM containing 10% FBS. Purified monoclonal antibodies (standardized by protein concentration) were added to the toxin preparations and incubated at 37°C for 1 h prior to addition to HeLa or RK-13 cells. To test the capacity of antibodies to neutralize VacA that was already bound to cells, acid-activated VacA (final concentration, 18 μg/ml) was incubated with HeLa cells for 4 h at 4°C, cells were washed with DMEM containing 10% FBS, and then monoclonal antibodies (10 μg/ml) were added to tissue culture medium overlying the cells. The medium overlying cells was routinely supplemented with 10 mM ammonium chloride for these assays (11). Following incubation at 37°C and 5% CO2 for 18 h, vacuolation was quantified by a neutral red uptake assay, as described previously (15).
RESULTS
Production of anti-VacA monoclonal antibodies.
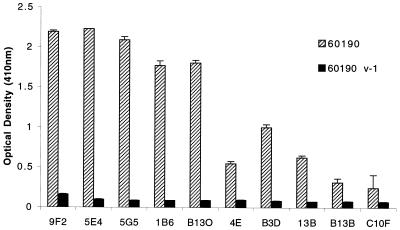
BALB/c mice were immunized with purified, oligomeric VacA from H. pylori strain 60190 as described in Materials and Methods. Thirteen hybridoma cell lines that secreted anti-VacA antibodies were isolated and characterized. A total of 10 antibodies were of the IgG1 isotype with a κ light chain, and 3 were of the IgM isotype. Compared with the IgG antibodies, the three IgM antibodies reacted weakly with VacA and therefore were not studied further. Ascites fluid was generated from each of the 10 IgG-producing hybridomas, and monoclonal antibodies were purified as described in Materials and Methods. Each of the 10 IgG1κ antibodies reacted significantly more strongly in an ELISA with broth culture supernatant from wild-type H. pylori strain 60190 than with broth culture supernatant from H. pylori strain 60190v-1, an isogenic vacA-null mutant strain (16) (Fig. 2), which indicated that the antibodies reacted specifically to VacA. None of the antibodies recognized denatured VacA in Western blot assays, nor did they react with two different preparations of denatured VacA in ELISAs (data not shown). These results suggest that the 10 IgG monoclonal antibodies all recognized conformational epitopes of VacA.
FIG. 2.
Specificity of monoclonal antibodies for VacA. H. pylori strain 60190 (wild type) and strain 60190 v-1 (an isogenic vacA-null mutant) (16) were grown in sulfite-free brucella broth containing 5% FBS. ELISA wells were coated with unconcentrated broth culture supernatants that were standardized by protein concentration. Reactivity of the purified monoclonal antibodies (standardized by protein concentration) to the broth culture supernatants was determined by an ELISA. Values represent the means ± standard deviation from triplicate determinations.
As an initial approach to characterize the panel of monoclonal antibodies, we first compared the reactivity of the purified antibodies (standardized by protein concentration) to VacA in three different types of ELISAs: (i) purified VacA from H. pylori strain 60190 adsorbed directly to ELISA plates in an oligomeric form (at pH 7), (ii) acidified VacA adsorbed to ELISA plates (at pH 2), and (iii) oligomeric VacA in an antigen capture ELISA (Table 1). Seven of the antibodies reacted strongly with VacA in all three ELISAs. The titers of reactivity for four of these antibodies (5G5, B13O, 4E, and B3D) were slightly higher in ELISAs using native (oligomeric) VacA than in ELISAs using acid-treated VacA. In contrast, antibodies 13B, B13B, and C10F reacted weakly with oligomeric VacA in the pH 7 ELISA but had no detectable reactivity with either acid-treated (monomeric) VacA or VacA in an antigen capture ELISA. All of the antibodies with the exception of the latter three (13B, B13B, and C10F) were able to immunoprecipitate VacA (data not shown). Thus, at least two different classes of monoclonal antibodies could be distinguished based on their reactivity to different forms of VacA.
TABLE 1.
Titers of monoclonal antibody reactivity by different ELISAs
| Antibody | Reciprocal titer of antibody by ELISA:a
|
||
|---|---|---|---|
| pH 7b | pH 2c | Antigen captured | |
| 9F2 | >106 | >106 | >106 |
| 5E4 | >106 | >106 | 105 |
| 5G5 | >106 | 105 | >106 |
| 1B6 | 104 | 104 | 104 |
| B130 | 105 | 104 | 104 |
| 4E | 105 | 103 | 105 |
| B3D | >106 | 104 | >106 |
| 13B | 102 | <10 | <10 |
| B13B | 103 | <10 | <10 |
| C10F | 102 | <10 | <10 |
Numbers refer to the maximum dilution of antibody (titer 1:10 = 0.1 mg/ml) that yielded a positive signal, designated as >0.2 at an optical density of 410 nm, which is >4-fold above background. Results are the median of three independent experiments.
VacA (2 μg/ml) from H. pylori strain 60190 in its native, oligomeric state was adsorbed directly to an ELISA plate at pH 7.
VacA from H. pylori strain 60190 was acid activated and adsorbed to an ELISA plate in 5% glacial acetic acid (final concentration of 2 μg/ml at pH 2). Subsequent steps were performed at pH 7.
In the antigen capture ELISA, serial dilutions of each monoclonal antibody were bound to the ELISA plate via a goat anti-mouse IgG antibody. VacA bound to the monoclonal antibodies was detected with rabbit polyclonal anti-VacA antiserum and goat anti-rabbit Ig-HRP.
Neutralization of VacA cytotoxic activity by monoclonal antibodies.
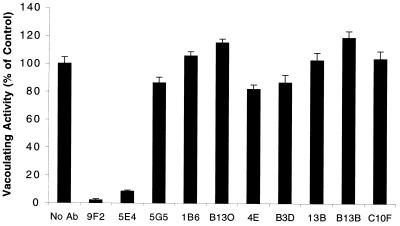
Rabbits immunized with VacA produce serum anti-VacA antibodies with toxin-neutralizing activity (11, 23, 33), and toxin-neutralizing antibodies are also present in serum from some H. pylori-infected humans (13). To determine if any of the monoclonal antibodies could neutralize VacA activity, each antibody was mixed with VacA from H. pylori strain 60190, and the toxin-antibody mixtures were then added to HeLa cells (Fig. 3). Only antibodies 9F2 and 5E4 exhibited detectable toxin-neutralizing activity under these conditions. In contrast, when toxin was first bound to HeLa cells at 4°C, a procedure that prevents toxin internalization (23, 37, 56), and the cells were then incubated with antibodies 9F2 or 5E4, no neutralization of toxin activity was detectable (data not shown).
FIG. 3.
Neutralization of VacA activity by monoclonal antibodies. Monoclonal antibodies were mixed with acid-activated, purified VacA from H. pylori strain 60190 that was diluted and adjusted to neutral pH by the addition of DMEM containing 10% FBS and incubated for 1 h at 37°C before being added to HeLa cells. Final concentrations of monoclonal antibodies and VacA were 10 and 6 μg/ml, respectively. After incubation for 18 h at 37°C, the extent of vacuolation was determined by a neutral red uptake assay (15). Values represent the means ± standard deviation of triplicate determinations. No Ab, no antibody.
Mapping monoclonal antibody epitopes.
To map the regions of VacA to which the monoclonal antibodies bound, we first examined the reactivity of the antibodies with a panel of VacA mutant proteins containing internal deletions (Fig. 1). Antibodies 9F2 and 5E4 reacted to all of the mutant proteins tested. Five antibodies reacted strongly to all but one mutant protein (Table 2); antibodies 1B6, B13O, 4E, and B3D did not react with VacA Δ517-536 and antibody 5G5 did not recognize VacA Δ114-294, which suggests that the relevant epitopes for these antibodies may be located in these regions. Antibodies 13B, B13B, and C10F reacted with VacA Δ6-27 and VacA Δ517-536 but not with the other five VacA deletion mutants (all of which contained deletions in the region comprising amino acids 28 to 294). Notably, a previous study demonstrated that the latter five mutant toxins (i.e., those not recognized by antibodies 13B, B13B, or C10F) were unable to form oligomers similar to wild-type toxin (56).
TABLE 2.
Reactivity of monoclonal antibodies to VacA deletion mutantsa
| Antibody | Reactivity to VacA antigen:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | Δ6-27 | Δ28-108 | Δ56-83 | Δ85-127 | Δ112-196 | Δ114-294 | Δ517-536 | |
| 9F2 | + | + | + | + | + | + | + | + |
| 5E4 | + | + | + | + | + | + | + | + |
| 5G5 | + | + | + | + | + | + | − | + |
| 1B6 | + | + | + | + | + | + | + | − |
| B13O | + | + | + | + | + | + | + | − |
| 4E | + | + | + | + | + | + | + | − |
| B3D | + | + | + | + | + | + | + | − |
| 13B | + | + | − | − | − | − | − | + |
| B13B | + | + | − | − | − | − | − | + |
| C10F | + | + | − | − | − | − | − | + |
H. pylori strains were grown in sulfite-free brucella broth containing 5% FBS, and the supernatant proteins were precipitated with a 50% saturated solution of ammonium sulfate. The VacA content of each sample was standardized so that each preparation yielded a similar optical density (OD) when reacted with polyclonal anti-VacA antiserum. +, OD410 of > 0.2, which is >4-fold above the background; −, OD410 of <0.2. Experiments were repeated three times with similar results.
To further define the epitopes to which the antibodies bound, we analyzed the reactivity of the antibodies with a panel of 12 mutant VacA proteins in which various clusters of amino acids throughout the toxin were changed to alanine residues. All of the monoclonal antibodies reacted with all of the VacA alanine substitution mutants tested (data not shown), with two exceptions. Antibodies 5G5, 1B6, B13O, 4E, and B3D did not react with VacA from H. pylori strain VM074, in which five residues between amino acids 420 and 428 were changed to alanine, as described in Materials and Methods (data not shown). Antibodies 13B, B13B, and C10F did not react with VacA from strain VM044, in which polar or charged residues between amino acids 63 and 69 were changed to alanine, as described in Materials and Methods (data not shown). Thus, it appears that the relevant epitopes for these antibodies are localized in these two regions. Overall, these results are consistent with the patterns of reactivity described above with the VacA deletion mutations (Table 2). Any apparent discrepancies in localization of reactive epitopes are probably attributable to the reactivity of the antibodies with conformational epitopes.
Antigenic diversity of VacA.
Nucleotide sequence analyses of vacA alleles from different H. pylori strains have revealed considerable genetic diversity (5, 16, 28, 42, 53). To experimentally determine whether there is antigenic diversity among VacA proteins, we analyzed the reactivity of the anti-VacA monoclonal antibodies with VacA proteins from a panel of 12 H. pylori strains, which were each isolated from different human stomachs (Table 3) (5, 21). To verify that VacA was present in the culture supernatants of each of these strains, the supernatants were tested for reactivity in an ELISA with pooled polyclonal antisera from rabbits immunized with purified VacA from H. pylori strains 60190 (s1-m1), 95-54 (s1-m2), and 86-338 (s2-m2) (Table 3). The pooled polyclonal antisera yielded a positive signal with each of the 12 wild-type strains tested but not with supernatant from strain 60190-v1, a vacA-null mutant strain (Table 3) (16). All 10 of the monoclonal antibodies recognized type s1-m1 VacA proteins from strains 60190, 87-81, 92-29, and 87-29 (Table 3). However, only 3 of the 10 antibodies (9F2, 5E4, and 5G5) recognized s1-m1 VacA toxins from all six of the s1-m1 strains tested. None of the monoclonal antibodies recognized type s2-m2 VacA proteins, and only four antibodies (5G5, 13B, B13B, and C10F) reacted with any of the s1-m2 proteins. In accordance with these results, the two antibodies (9F2 and 5E4) with neutralizing activity for s1-m1 toxins lacked any detectable neutralizing activity for an s1-m2 toxin from H. pylori strain 95-54 (data not shown). These results imply that there is considerable antigenic diversity in VacA proteins, even within the same vacA family.
TABLE 3.
Reactivity of monoclonal antibodies to VacA proteins from different wild-type H. pylori strainsa
| Antibody | Reactivity to VacA from H. pylori type and strain:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1-m1
|
s1-m2
|
s2-m2
|
60190-vl (vacA-null mutant) | ||||||||||
| 60190 | 84-183 | 87-81 | 92-29 | 87-199 | 87-29 | 95-54 | 95-55 | 87-91 | Tx30a | 86-313 | 86-338 | ||
| 9F2 | + | + | + | + | + | + | − | − | − | − | − | − | − |
| 5E4 | + | + | + | + | + | + | − | − | − | − | − | − | − |
| 5G5 | + | + | + | + | + | + | + | − | + | − | − | − | − |
| 1B6 | + | + | + | + | − | + | − | − | − | − | − | − | − |
| B13O | + | + | + | + | − | + | − | − | − | − | − | − | − |
| 4E | + | + | + | + | − | + | − | − | − | − | − | − | − |
| B3D | + | + | + | + | − | + | − | − | − | − | − | − | − |
| 13B | + | − | + | + | + | + | − | + | − | − | − | − | − |
| B13B | + | − | + | + | + | + | − | + | − | − | − | − | − |
| C10F | + | − | + | + | + | + | − | + | − | − | − | − | − |
| Polyclonal anti-VacA | + | + | + | + | + | + | + | + | + | + | + | + | − |
H. pylori strains were grown in sulfite-free brucella broth containing 5% FBS to an optical density at 600 nm (OD600) of 0.4, and ELISA wells were coated with unconcentrated supernatants. The VacA content of each sample was confirmed by ELISA with pooled rabbit polyclonal anti-VacA antisera 958 (to 60190 VacA), 977 (to 95-54 VacA), and 957 (to 86-338 VacA). +, OD410 of >0.2, which is >4-fold above the background; −, OD410 of <0.2. Experiments were repeated three times with similar results.
Epitope mapping using chimeric VacA proteins.
As noted above, none of the antibodies reacted with the type s2-m2 VacA from H. pylori strain Tx30a (Table 3). Therefore, in an effort to further map the regions to which the antibodies bound, we constructed four chimeric vacA genes in H. pylori 60190, such that segments of the original vacA sequence were replaced with vacA sequences from H. pylori Tx30a (Fig. 1). Each of the four chimeric strains expressed a VacA product, and each of these chimeric proteins oligomerized similarly to wild-type VacA, as determined by elution in the same high-molecular-mass fractions from a size exclusion column (data not shown). Antibodies 9F2 and 5E4 (which exhibited toxin-neutralizing activity for s1-m1 VacA) did not react with chimeric proteins from strains G117, G119, and G120, which all had residues 685 to 820 replaced with type m2 sequences (Table 4 and Fig. 1). Antibodies 5G5, 1B6, B13O, 4E, and B3D did not react with chimeric proteins from strains G118 and G119, which all had amino acid residues 332 to 623 replaced with type m2 sequences (Table 4 and Fig. 1). These results are consistent with previous mapping data using the deletion mutations and alanine substitution mutations (Table 2). Thus, the mapping data for the two neutralizing antibodies, 9F2 and 5E4, suggest that relevant epitopes for neutralization of VacA activity contain amino acids located between amino acids 685 and 820.
TABLE 4.
Reactivity of monoclonal antibodies to VacA chimerasa
| Antibody | Reactivity to VacA from H. pylori strain (type):
|
|||||
|---|---|---|---|---|---|---|
| 60190 (s1-m1) | Tx30a (s2-m2) | G117 (s1-m2) | G118 (s1-m2) | G119 (s1-m2) | G120 (s1-m2) | |
| 9F2 | + | − | − | + | − | − |
| 5E4 | + | − | − | + | − | − |
| 5G5 | + | − | + | − | − | + |
| 1B6 | + | − | + | − | − | + |
| B13O | + | − | + | − | − | + |
| 4E | + | − | + | − | − | + |
| B3D | + | − | + | − | − | + |
| 13B | + | − | + | + | + | + |
| B13B | + | − | + | + | + | + |
| C10F | + | − | + | + | + | + |
H. pylori strains were grown in sulfite-free brucella broth containing 5% FBS, and the supernatant proteins were precipitated with a 50% saturated solution of ammonium sulfate. The VacA content of each sample was standardized so that each preparation yielded a similar optical density (OD) when reacted with pooled polyclonal anti-VacA antiserum. +, OD410 of >0.2, which is >4-fold above the background; OD410 of <0.2. Experiments were repeated three times with similar results.
DISCUSSION
In this study, we generated a panel of 10 monoclonal anti-VacA antibodies, all of which were of the IgG1κ isotype. An intriguing finding was that all of these antibodies reacted only with conformational epitopes of VacA and thus failed to react with denatured VacA in immunoblot assays. The failure to identify any antibodies recognizing linear VacA epitopes potentially reflects a relative resistance of the VacA oligomer to proteolytic degradation during antigen processing or may result from the ability of VacA to alter antigen presentation in mice (39, 48). Alternatively, a bias may have been introduced by our screening methodology, in which oligomeric VacA (hypothesized to contain surface-exposed epitopes comprising predominantly highly folded rather than linear peptide structures) was used as the antigen to identify reactive hybridoma cells. The characteristics of the monoclonal antibodies described in this study are consistent with those of a previously described anti-VacA monoclonal antibody, C1G9, which was also an IgG1κ isotype and reacted with a conformational VacA epitope (46).
Several antibodies described in this study recognized both oligomeric and disassembled (acidified) forms of VacA, whereas other antibodies reacted detectably only with the oligomeric form of VacA (Table 1). One possible explanation for the differential reactivity of the latter antibodies (13B, B13B, and C10F) is that certain conformational epitopes might be displayed only when VacA is in an oligomeric state. This model is consistent with the failure of these antibodies to react with nonoligomerizing mutant forms of VacA (Table 2). However, we cannot exclude the possibility that antibodies 13B, B13B, and C10F have a relatively low affinity for VacA in general and that reactivity with disassembled or nonoligomerizing forms of VacA was simply below the threshold of detection.
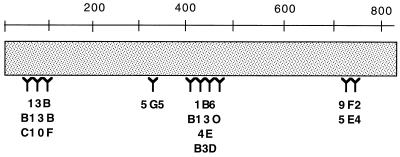
By analyzing the reactivity of the 10 monoclonal antibodies with various mutant VacA proteins, we were able to recognize at least four distinct patterns of reactivity. Based on these patterns, we are able to tentatively map the regions of VacA to which these antibodies bind (Fig. 4). The patterns of reactivity of the monoclonal antibodies to the VacA deletion mutations (Table 2), alanine substitution mutations (data not shown), and VacA chimeras (Table 4) were generally in agreement. Reactivity of the antibodies with conformational epitopes probably accounts for occasional instances when the mapping data were not completely concordant with these different types of mutant VacA proteins. At present, it is not known whether the antibodies with similar patterns of reactivity recognize identical epitopes or different epitopes within the same region of VacA.
FIG. 4.
Schematic representation of the regions in VacA from H. pylori strain 60190 to which the monoclonal antibodies bind, based upon data shown in Tables 2 and 4 as well as reactivity of antibodies with mutant VacA proteins containing alanine substitution mutations (see text). The numbers represent amino acids in the mature VacA toxin from H. pylori strain 60190.
In a previous study, polyclonal antiserum reactive with a recombinant peptide of the C-terminal region of VacA (amino acids 476 to 803) was reported to neutralize VacA cytotoxic activity (23). In the present study, two of the monoclonal antibodies (9F2 and 5E4) that exhibited toxin-neutralizing activity are predicted to react with a portion of VacA located between amino acids 685 and 820. Therefore, it seems likely that amino acid sequences in this portion of VacA mediate toxin binding to cells and that these antibodies, 9F2 and 5E4, block toxin activity by inhibiting toxin binding to cells. This hypothesis is supported by a recent study which suggested that the C-terminal 100 amino acids of the mature VacA protein may be important for the binding of recombinant VacA fusion proteins to HeLa cells (57).
H. pylori vacA is known to exhibit considerable allelic diversity (5, 53, 55). However, the extent of antigenic diversity among VacA toxins has not previously been investigated in detail. In this study, we demonstrate that there is considerable variation among VacA toxins in reactivity with our panel of monoclonal antibodies. The most striking finding was that all of the monoclonal antibodies produced against type s1-m1 VacA failed to recognize type s2-m2 VacA toxins (Table 3). This result is not completely unexpected because mature type s1-m1 and type s2-m2 VacA toxins are about 75% identical in amino acid sequences overall and exhibit <70% amino acid identity in the VacA midregion (5). Another notable result was that various monoclonal antibodies were unable to consistently recognize type s1-m1 toxins from multiple different strains (Table 3). Type s1-m1 toxins produced by different H. pylori strains are typically >90% identical in amino acid sequence (5,7). To account for the lack of uniform antibody reactivity, we speculate that amino acid diversity among VacA proteins from different H. pylori strains occurs disproportionately in surface-exposed epitopes.
Anti-VacA antibodies are present in serum and gastric juice from the majority of H. pylori-infected humans (12, 44), and neutralizing IgG anti-VacA antibodies are sometimes detectable in human serum (13). At present, it is not known whether these antibodies play an important role in the biology of H. pylori-host interactions. Clearly the presence of anti-VacA antibodies in physiologic concentrations does not result in the eradication of established H. pylori infection in humans. However, it seems plausible that such antibodies might modulate the course of H. pylori infection or might inhibit colonization by newly ingested H. pylori strains. In the latter case, antigenic variation among VacA toxins from different strains might render preexisting antibodies relatively ineffective.
In an effort to prevent serious complications of H. pylori infection, such as peptic ulcer disease and distal gastric adenocarcinoma, there has been an interest in development of an H. pylori vaccine (34). Immunization with VacA confers protective immunity in an experimental mouse model of H. pylori infection, and therefore VacA is considered a candidate antigen for inclusion in an H. pylori vaccine (34, 35). In one study, immunization with purified type s1-m1 VacA conferred protection against subsequent challenge by two H. pylori strains that produced vacuolating toxin activity in vitro but failed to protect against challenge with a wild-type Tox− strain that lacked detectable toxic activity for HeLa cells (34, 35). It is now known that many H. pylori strains that lack toxic activity (Tox−) for HeLa cells contain type s1-m2 or s2-m2 vacA alleles and express detectable VacA products (5, 21, 42). Based on the results of the present study, we speculate that the failure of an s1-m1 VacA antigen to induce protective immunity against a Tox− strain may be due to antigenic diversity among different VacA proteins. In particular, it seems clear that many antibodies that are reactive with type s1-m1 VacA fail to recognize type s2-m2 VacA. Therefore, if VacA is to be included as a vaccine component, it might be appropriate to immunize with pooled VacA antigens derived from several different VacA families.
ACKNOWLEDGMENTS
We thank Donna Choate, Phillip Budge, and Beverly Hosse for technical support.
This work was supported by the National Institutes of Health (RO1 AI-39657 and DK-53623), the Medical Research Service of the Department of Veterans Affairs, and a grant from the Vanderbilt-Ingram Cancer Center (2P30 CA69485-04) (R.L.M.).
REFERENCES
- 1.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz N, Bukanov N O, Westblom T U, Berg D E. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20:6221–6225. doi: 10.1093/nar/20.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 5.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 6.Atherton J C, Cover T L, Twells R J, Morales M R, Hawkey C J, Blaser M J. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J Clin Microbiol. 1999;37:2979–2982. doi: 10.1128/jcm.37.9.2979-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atherton J C, Sharp P M, Cover T L, Gonzalez-Valencia G, Peek R M, Jr, Thompson S A, Hawkey C J, Blaser M J. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr Microbiol. 1999;39:211–218. doi: 10.1007/s002849900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 10.Cover T L, Berg D E, Blaser M J, Mobley H L T. Helicobacter pylori pathogenesis. In: Groisman E, editor. Principles of bacterial pathogenesis. New York, N.Y: Academic Press; 2001. pp. 509–558. [Google Scholar]
- 11.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 12.Cover T L, Cao P, Lind C D, Tham K T, Blaser M J. Correlation between vacuolating cytotoxin production by Helicobacter pylori isolates in vitro and in vivo. Infect Immun. 1993;61:5008–5012. doi: 10.1128/iai.61.12.5008-5012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cover T L, Cao P, Murthy U K, Sipple M S, Blaser M J. Serum neutralizing antibody response to the vacuolating cytotoxin of Helicobacter pylori. J Clin Investig. 1992;90:913–918. doi: 10.1172/JCI115967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cover T L, Puryear W, Pérez-Pérez G I, Blaser M J. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect Immun. 1991;59:1264–1270. doi: 10.1128/iai.59.4.1264-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 17.Czajkowsky D M, Iwamoto H, Cover T L, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc Natl Acad Sci USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bernard M, Burroni D, Papini E, Rappuoli R, Telford J, Montecucco C. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect Immun. 1998;66:6014–6016. doi: 10.1128/iai.66.12.6014-6016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, Fontana A, Rappuoli R, Montecucco C. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J Biol Chem. 1995;270:23937–23940. doi: 10.1074/jbc.270.41.23937. [DOI] [PubMed] [Google Scholar]
- 20.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsyth M H, Atherton J C, Blaser M J, Cover T L. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–3094. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garner J A, Cover T L. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J Infect Dis. 1995;172:290–293. doi: 10.1093/infdis/172.1.290. [DOI] [PubMed] [Google Scholar]
- 23.Garner J A, Cover T L. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect Immun. 1996;64:4197–4203. doi: 10.1128/iai.64.10.4197-4203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhard M, Lehn N, Neumayer N, Boren T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Göttke M U, Fallone C A, Barkun A N, Vogt K, Loo V, Trautmann M, Tong J Z, Nguyen T N, Fainsilber T, Hahn H H, Korber J, Lowe A, Beech R N. Genetic variability determinants of Helicobacter pylori: influence of clinical background and geographic origin of isolates. J Infect Dis. 2000;181:1674–1681. doi: 10.1086/315425. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 27.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazaki Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- 29.Iwamoto H, Czajkowsky D M, Cover T L, Szabo G, Shao Z. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS Lett. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 30.Ji X, Fernandez T, Burroni D, Pagliaccia C, Atherton J C, Reyrat J M, Rappuoli R, Telford J L. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect Immun. 2000;68:3754–3757. doi: 10.1128/iai.68.6.3754-3757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kersulyte D, Mukhopadhyay A K, Velapatino B, Su W, Pan Z, Garcia C, Hernandez V, Valdez Y, Mistry R S, Gilman R H, Yuan Y, Gao H, Alarcon T, Lopez-Brea M, Balakrish Nair G, Chowdhury A, Datta S, Shirai M, Nakazawa T, Ally R, Segal I, Wong B C, Lam S K, Olfat F O, Boren T, Engstrand L, Torres O, Schneider R, Thomas J E, Czinn S, Berg D E. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupetti P, Heuser J E, Manetti R, Massari P, Lanzavecchia S, Bellon P L, Dallai R, Rappuoli R, Telford J L. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manetti R, Massari P, Burroni D, de Bernard M, Marchini A, Olivieri R, Papini E, Montecucco C, Rappuoli R, Telford J L. Helicobacter pylori cytotoxin: importance of native conformation for induction of neutralizing antibodies. Infect Immun. 1995;63:4476–4780. doi: 10.1128/iai.63.11.4476-4480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetti M, Aricò B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 35.Marchetti M, Rossi M, Giannelli V, Giuliani M M, Pizza M, Censini S, Covacci A, Massari P, Pagliaccia C, Manetti R, Telford J L, Douce G, Dougan G, Rappuoli R, Ghiara P. Protection against Helicobacter pylori infection in mice by intragastric vaccination with Helicobacter pylori antigens is achieved using a non-toxic mutant of Escherichia coli heat-labile enterotoxin (LT) as adjuvant. Vaccine. 1998;16:33–37. doi: 10.1016/s0264-410x(97)00153-9. [DOI] [PubMed] [Google Scholar]
- 36.Marshall D G, Dundon W G, Beesley S M, Smyth C J. Helicobacter pylori—a conundrum of genetic diversity. Microbiology. 1998;144:2925–2939. doi: 10.1099/00221287-144-11-2925. [DOI] [PubMed] [Google Scholar]
- 37.McClain M S, Schraw W, Ricci V, Boquet P, Cover T L. Acid-activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol Microbiol. 2000;37:433–442. doi: 10.1046/j.1365-2958.2000.02013.x. [DOI] [PubMed] [Google Scholar]
- 38.Molinari M, Galli C, de Bernard M, Norais N, Ruysschaert J M, Rappuoli R, Montecucco C. The acid activation of Helicobacter pylori toxin VacA: structural and membrane binding studies. Biochem Biophys Res Commun. 1998;248:334–340. doi: 10.1006/bbrc.1998.8808. [DOI] [PubMed] [Google Scholar]
- 39.Molinari M, Salio M, Galli C, Norais N, Rappuoli R, Lanzavecchia A, Montecucco C. Selective inhibition of Ii-dependent antigen presentation by Helicobacter pylori toxin VacA. J Exp Med. 1998;187:135–140. doi: 10.1084/jem.187.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montecucco C, Papini E, de Bernard M, Telford J L, Rappuoli R. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors. In: Alouf J E, Freer J H, editors. The comprehensive sourcebook of bacterial protein toxins. London, United Kingdom: Academic Press; 1999. pp. 264–286. [Google Scholar]
- 41.Nguyen V Q, Caprioli R M, Cover T L. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun. 2001;69:543–546. doi: 10.1128/IAI.69.1.543-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover T L, Papini E, Rappuoli R, Telford J L, Reyrat J M. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc Natl Acad Sci USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Z J, Berg D E, van der Hulst R W, Su W W, Raudonikiene A, Xiao S D, Dankert J, Tytgat G N, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Perez G I, Peek R M, Jr, Atherton J C, Blaser M J, Cover T L. Detection of anti-VacA antibody responses in serum and gastric juice samples using type s1/m1 and s2/m2 Helicobacter pylori VacA antigens. Clin Diagn Lab Immunol. 1999;6:489–493. doi: 10.1128/cdli.6.4.489-493.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrin S, Gilliland G. Site-specific mutagenesis using asymmetric polymerase chain reaction and a single mutant primer. Nucleic Acids Res. 1990;18:7433–7438. doi: 10.1093/nar/18.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyrat J M, Charrel M, Pagliaccia C, Burroni D, Lupetti P, de Bernard M, Ji X, Norais N, Papini E, Dallai R, Rappuoli R, Telford J L. Characterisation of a monoclonal antibody and its use to purify the cytotoxin of Helicobacter pylori. FEMS Microbiol Lett. 1998;165:79–84. doi: 10.1111/j.1574-6968.1998.tb13130.x. [DOI] [PubMed] [Google Scholar]
- 47.Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle P R, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satin B, Norais N, Telford J, Rappuoli R, Murgia M, Montecucco C, Papini E. Effect of Helicobacter pylori vacuolating toxin on maturation and extracellular release of procathepsin D and on epidermal growth factor degradation. J Biol Chem. 1997;272:25022–25028. doi: 10.1074/jbc.272.40.25022. [DOI] [PubMed] [Google Scholar]
- 49.Strobel S, Bereswill S, Balig P, Allgaier P, Sonntag H G, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–1289. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford J L, Rappuoli R, Montecucco C, Papini E, Zoratti M. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tombola F, Carlesso C, Szabo I, de Bernard M, Reyrat J M, Telford J L, Rappuoli R, Montecucco C, Papini E, Zoratti M. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: possible implications for the mechanism of cellular vacuolation. Biophys J. 1999;76:1401–1409. doi: 10.1016/S0006-3495(99)77301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Doorn L J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M, Carneiro F, Vanderborght B, Pegado M D, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K, Atherton J, Blaser M J, Quint W G. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 54.van Doorn L J, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa J C, Carneiro F, Quint W G. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Doorn L J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K, Atherton J C, Blaser M J, Quint W G. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinion-Dubiel A D, McClain M S, Czajkowsky D M, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke S R, Shao Z, Cover T L. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J Biol Chem. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 57.Wang H J, Wang W C. Expression and binding analysis of GST-VacA fusions reveals that the C-terminal approximately 100-residue segment of exotoxin is crucial for binding in HeLa cells. Biochem Biophys Res Commun. 2000;278:449–454. doi: 10.1006/bbrc.2000.3820. [DOI] [PubMed] [Google Scholar]
- 58.Yahiro K, Niidome T, Kimura M, Hatakeyama T, Aoyagi H, Kurazono H, Imagawa K, Wada A, Moss J, Hirayama T. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J Biol Chem. 1999;274:36693–36699. doi: 10.1074/jbc.274.51.36693. [DOI] [PubMed] [Google Scholar]
- 59.Ye D, Willhite D C, Blanke S R. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J Biol Chem. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]