Abstract
Genetic drugs based on nucleic acid biomolecules are a rapidly emerging class of medicines that directly reprogramme the central dogma of biology to prevent and treat disease. However, multiple biological barriers normally impede the intracellular delivery of nucleic acids, necessitating the use of a delivery system. Lipid and polymer nanoparticles represent leading approaches for the clinical translation of genetic drugs. These systems circumnavigate biological barriers and facilitate the intracellular delivery of nucleic acids in the correct cells of the target organ using passive, active and endogenous targeting mechanisms. In this Review, we highlight the constituent materials of these advanced nanoparticles, their nucleic acid cargoes and how they journey through the body. We discuss targeting principles for liver delivery, as it is the organ most successfully targeted by intravenously administered nanoparticles to date, followed by the expansion of these concepts to extrahepatic (non-liver) delivery. Ultimately, this Review connects emerging materials and biological insights playing key roles in targeting specific organs and cells in vivo.
Subject terms: Drug delivery, Biomedical engineering, Drug delivery
Nucleic acids for gene silencing, expression and editing can precisely target disease at the molecular level but require effective delivery systems. This Review discusses the material and biological principles used to design delivery systems to target specific organs in the body.
Introduction
The targeted delivery of therapeutic molecules to specific sites in the body represents an active area of research in the biomedical sciences involving scientists, clinicians and engineers alike. To be successful, drug delivery systems must overcome limitations that normally render a pharmaceutical agent ineffective, such as biological barriers and poor biodistribution to the desired site of action. At present, several delivery platforms have been incorporated into clinically used products, including viral vectors, molecular conjugates, antibody–drug conjugates and nanoparticles. Of those, nanoparticles are a particularly promising delivery platform because of their capacity to load different types of drugs separately or in combination, their minimal immunogenicity and their easily modulated properties through controlled chemical synthesis1.
One application that has received substantial attention recently is the delivery of nucleic acids for therapeutics and vaccines. Most notably, the rapid and successful deployment of the messenger RNA (mRNA) vaccines against SARS-CoV-2 by Pfizer–BioNTech and Moderna represented a turning point for drug delivery2,3. These vaccines rely on lipid nanoparticles (LNPs) as a delivery system to shuttle mRNA past multiple biological barriers that would otherwise prevent efficacy. With over a billion doses of mRNA–LNP vaccines administered globally, LNPs for nucleic acid delivery represent one of the most widely used drug products in history. Additionally, LNPs as delivery platforms have been leveraged in multiple clinical trials for applications such as vaccination, protein replacement therapy, cancer immunotherapy, RNA interference and gene editing4. LNPs feature high potency, biocompatibility and the ability to be administered repeatedly, all of which are properties that favour clinical translation4. Therefore, it is timely to reflect on the lessons learnt from the successful development of LNPs to help devise design criteria for new clinically translatable nanoparticles based on LNPs and related materials. Polymers represent another class of materials with emerging utility for nucleic acid delivery. Multiple biomedical products, such as controlled release depots and absorbable sutures, already incorporate polymers, laying a foundation for successful clinical translation, and their properties can be controlled using chemical synthesis to produce efficacious delivery systems5,6. Collectively, lipid and polymer nanoparticles represent the largest share of non-viral systems studied for nucleic acid delivery.
In this Review, we discuss recent advances in the engineering of lipid and polymer nanoparticles for the delivery of genetic drugs to specific organs and cell types in the body. Other approaches and materials have been excellently summarized elsewhere7–10. Genetic drugs based on nucleic acid biomolecules must reach the cytosol of target cells while avoiding bystander cells, which necessitates the design of tissue-targeted delivery systems. As cell-based assays can be a poor predictor of in vivo performance, we limit our scope to delivery systems whose organ-targeting properties have been validated in animal models11. Although we emphasize targeting specific organs and cell types via intravenous (i.v.) administration, alternative modes of administration can also be used to control the biological fate of a delivery system12. Three mechanisms, passive, active and endogenous targeting (Box 1), are leveraged to engineer nanoparticles that reach specific organs through the vascular network. Delivery using passive and active targeting has been extensively investigated for decades. However, endogenous targeting represents an emerging paradigm based on a growing understanding of how the protein corona impacts delivery. The examples we provide highlight mechanisms for organ-specific delivery of genetic drugs, but we anticipate that these concepts can be extended to other therapeutic modalities such as small-molecule and protein drugs.
Box 1 Mechanisms of nanoparticle targeting.
Multiple mechanisms can be utilized separately or in combination to control where in the body nanoparticles accumulate (see the figure). The final nanoparticle design will reflect how these mechanisms are used to achieve a desired organ-targeting outcome.
Passive targeting
Passive targeting relies on tuning the physical properties of a nanoparticle, such as size, shape, stiffness and surface charge, to favourably interface with anatomy and physiology221. For example, adjusting nanoparticle size can determine the tendency of a nanoparticle to extravasate from discontinuous blood vessels, such as those in the liver and the spleen (see the figure, part a).
Active targeting
Active targeting involves modifying the surface of a nanoparticle with a chemical or biological moiety that specifically binds to receptors or other cellular features highly expressed by cells of a target organ222 (see the figure, part b). For example, nanoparticles have been modified with monoclonal antibodies to enable nucleic acid delivery into difficult-to-transfect immune cells.
Endogenous targeting
Endogenous targeting involves engineering the composition of a nanoparticle such that it binds to a distinct subset of plasma proteins on injection, which direct it to a target organ and promote uptake by specific cells97 (see the figure, part c). For example, proteins involved in cholesterol transport in the body have been demonstrated as essential for efficacious liver hepatocyte delivery of lipid nanoparticles.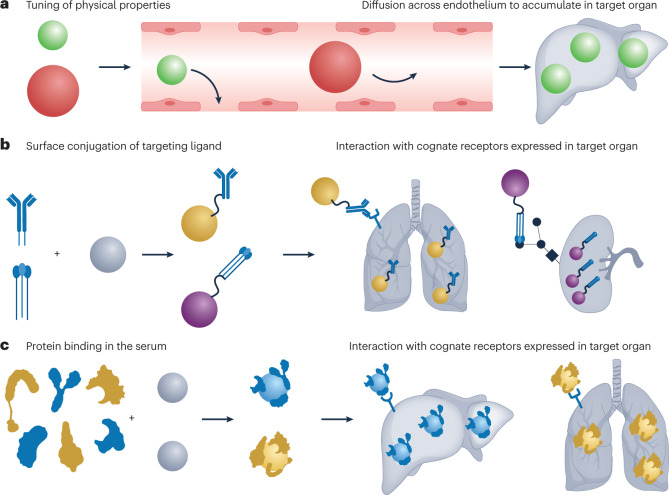
Molecular building blocks
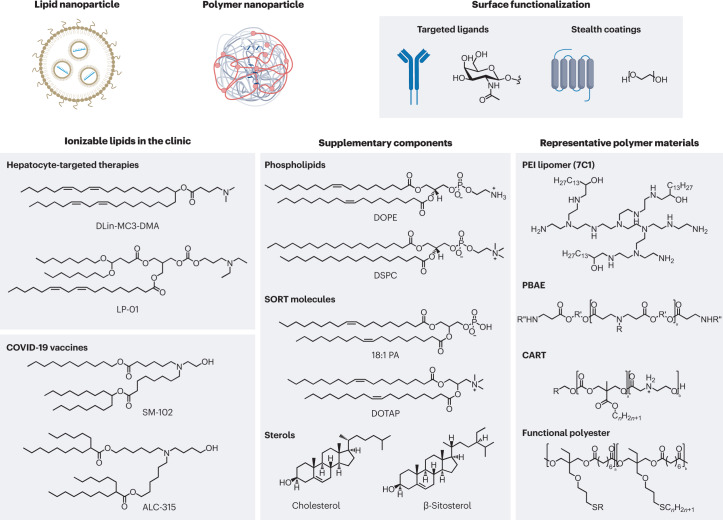
Selecting suitable materials is a critical first step in the design of organ-targeted delivery systems. Optimal nanoparticles must integrate multiple functions to allow for efficacious delivery to the target organ: they must be able to encapsulate the cargo (here, the nucleic acids) with high efficiency and remain stable in the serum during circulation. They must have a mechanism to enter cells and escape into the cytosol and should also degrade into biocompatible metabolites following cargo release. Finally, they should be amenable to large-scale manufacturing when envisioning clinical utility1,4. Presently, lipids and polymers (Fig. 1) represent the most advanced materials that satisfy these criteria, leading to their evaluation in multiple clinical trials13 (Table 1). The synthesis of novel molecular components with diverse structures and functionalities provides a large chemical space for engineering materials with desired performance. Understanding the key features of these materials is an important first step in designing nanoparticles for organ-targeted delivery.
Fig. 1. Self-assembled nanoparticles based on lipid and polymer materials are the state of the art for the delivery of genetic drugs.
Currently, lipid nanoparticles that incorporate an ionizable lipid are the most advanced delivery system for genetic drugs in the clinic. These materials feature a tertiary amine that can acquire charge at acidic pH to facilitate nucleic acid loading during formulation and promote endosomal escape following cellular uptake. Dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA) is a component of the US Food and Drug Administration-approved drug Onpattro. LP-01 is a component of Intellia Therapeutics’ clinical candidates NTLA-2001 and NTLA-2002 for gene editing in the liver, and SM-102 and ALC-315 are the ionizable lipid components of the Moderna and Pfizer–BioNTech vaccines, respectively. Alternatively, certain polymers that incorporate ionizable amine groups can also be used to formulate nanoparticles, and the choice of monomers will impact nanoparticle delivery efficiency and tissue selectivity. For both ionizable lipids and polymers, supplementary components can be included to improve the stability, fusogenicity (the ability to facilitate fusion with cellular membranes) and selectivity of the formulated nanoparticle. Additionally, the surfaces of these nanoparticles can be further modified using synthetic or biological targeting ligands and stealth coatings to alter nanoparticle circulation time, biodistribution and cellular uptake. Nucleic acid biomolecules can be loaded into nanoparticles to reprogramme the central dogma of biology through gene silencing, expression and editing to correct the course of disease. 18:1 PA, 1,2-dioleoyl-sn-glycero-3-phosphatidic acid; CART, charge-altering releasable transporter; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; PBAE, poly(beta-amino ester); PEI, polyethyleneimine; SORT, selective organ targeting.
Table 1.
Injected nanoparticles for organ-targeted drug delivery studied in clinical trials
| Drug name | Cargo | Delivery target | Application | Clinical trial |
|---|---|---|---|---|
| Patisiran (Onpattro) | TTR siRNA | Liver hepatocytes | Silencing of the TTR gene to treat transthyretin-mediated amyloidosis | NCT04201418 |
| NCT03997383 | ||||
| NCT03862807 | ||||
| NCT03759379 | ||||
| NCT02939820 | ||||
| NCT02510261 | ||||
| NCT02053454 | ||||
| NCT01961921 | ||||
| NCT01960348 | ||||
| NCT01617967 | ||||
| NCT01559077 | ||||
| BMS-986263 | HSP47 siRNA | Hepatic stellate cells | Silencing of HSP47 expression to inhibit collagen secretion in hepatic fibrosis | NCT01858935 |
| NCT02227459 | ||||
| NCT03420768 | ||||
| NCT03241264 | ||||
| NCT03142165 | ||||
| NCT04267393 | ||||
| NCT04225936 | ||||
| mRNA-3927 | PCC subunit-α mRNA, PCC subunit-β mRNA | Liver hepatocytes | Replacement of the PCC enzyme to treat propionic acidaemia | NCT04159103 |
| mRNA-1944 | CHKV-24 light chain mRNA, CHKV-24 heavy chain mRNA | Liver hepatocytes | Systemic secretion of an anti-chikungunya virus antibody for passive immunization | NCT03829384 |
| ARCT-810 | OTC mRNA | Liver hepatocytes | Replacement of the OTC enzyme to treat ornithine transcarbamylase deficiency | NCT04442347 |
| NCT04416126 | ||||
| NTLA-2001 | Cas9 mRNA, TTR sgRNA | Liver hepatocytes | Gene editing of the TTR locus to treat transthyretin-mediated amyloidosis | NCT04601051 |
| NTLA-2002 | Cas9 mRNA, KLKB1 sgRNA | Liver hepatocytes | Gene editing of the KLKB1 locus to treat hereditary angioedema | NCT05120830 |
| VERVE-101 | ABE mRNA, PCSK9 sgRNA | Liver hepatocytes | Gene editing of the PCSK9 locus to lower LDL cholesterol levels and prevent cardiovascular disease | NCT05398029 |
| mRNA-3705 | MCM mRNA | Liver hepatocytes | Replacement of the MCM enzyme to treat methylmalonic acidemia | NCT04899310 |
| Lipo-MERIT | Melanoma-associated antigens mRNA | Dendritic cells | Expression of a cocktail of tumour-associated antigens to stimulate an immune response against advanced melanoma | NCT02410733 |
| CLDN6 RNA-LPX | CLDN6 mRNA | Dendritic cells | Expression of CLDN6 to amplify the activity of CAR-T cells directed against solid tumours | NCT04503278 |
| Autogene cevumeran | Individualized neoantigen mRNA | Dendritic cells | Expression of personalized tumour neoantigens as an individualized cancer vaccine | NCT03289962 |
ABE, adenine base editor; CAR-T, chimeric antigen receptor T; Cas9, CRISPR-associated 9; CHKV, Chikungunya virus; CLDN, Claudin; HSP47, heat shock protein 47; LDL, low-density lipoprotein; MCM, methylmalonyl-CoA mutase; mRNA, messenger RNA; OTC, ornithine transcarbamylase; PCC, propionyl-CoA carboxylase; sgRNA, single guide RNA; siRNA, small interfering RNA.
Lipids
Lipids are small amphiphilic or hydrophobic molecules that have structural, signalling and energy storage functions in cellular physiology. There are several classes of natural lipid molecules with distinct chemical structures and biological functions, including phospholipids, sterols, fatty acids, glycolipids and sphingolipids14. Among the natural lipids, phospholipids and cholesterol are most commonly used for delivery systems in the clinic15. Additionally, synthetic lipid molecules that have functions tailored to delivery applications, such as ionizable cationic lipids or polyethylene glycol (PEG) lipids, have also been prepared using various chemical techniques15. In an aqueous solvent, lipid molecules will self-assemble into various nanoscale structures to sequester their hydrophobic domains from the surrounding aqueous environment while minimizing unfavourable interactions (electrostatic repulsion and steric crowding) between individual molecules16. The structural features of lipids, such as head group, alkyl chain length and unsaturation, along with the molar ratio of lipid components in a mixture, will impact the morphology of the fully assembled nanostructure16.
The development of LNPs as a delivery system for nucleic acids builds on early work involving the use of liposomes to improve the therapeutic index (that is, the ratio between the effective dosage of a drug and toxic dosage) of cytotoxic small-molecule drugs for cancer chemotherapy. Liposomes are bilayer vesicles of variable lamellarity that enclose an aqueous phase, entrapping drugs within the aqueous compartment or the lipid bilayer based on the hydrophobicity of drugs. Liposomal doxorubicin (Doxil) was the first nanoparticle for drug delivery to enter the clinic, and, as of December 2022, seven different liposomal nanomedicines have been approved by either the US Food and Drug Administration (FDA) or the European Medicines Agency (EMA)13,17. Liposomes are not discussed, as their clinical use has largely applied to small-molecule drug cargoes, but several reviews have examined their use for the delivery of chemotherapies, anti-fungals and other drugs17–22. In addition to the zwitterionic phospholipid, cholesterol and PEG lipid components typically constituting liposomes, LNPs also include an ionizable cationic lipid to promote electrostatic loading of anionic nucleic acid molecules4. This yields a more complex internal structure compared with the bilayer morphology of liposomes that appears to depend on the choice of nucleic acid cargo, the lipid composition of the LNP, formulation method and processing23.
The structure and properties of LNPs depend on both the chemistry of the constituent lipids and their molar proportions. Thus, it is important to understand the function of each lipid in an LNP, identify or synthesize appropriate lipid molecules to execute those functions and test the efficacy of different LNP chemical compositions and combinations in vivo24. Optimization of the chemistry of the ionizable cationic lipid component of LNPs has been the focus of extensive efforts15. A common feature of these lipid materials is an ionizable amine (amines) with varying charge: it acquires a positive charge at low pH to enable complexation with anionic cargoes during the formulation process, is neutral at physiological pH to reduce toxicity in the circulation and is positively charged again in the acidifying endosome following cellular uptake, enabling cargo release. By tailoring the chemistry of the amine component (components), the hydrophobic lipidic tails and the linkers between these two moieties, novel lipid materials can be obtained. Additional motifs, such as biodegradable linkages, branching chains and other functional groups, also impact the potency, targeting and biocompatibility of lipid-based carriers. Selected examples of this chemical diversity include: ionizable amino lipids25–27, lipidoids28–31, lipopeptides32,33, modular degradable dendrimer-based ionizable amino lipids34–36, bioreducible lipids37–40, TT lipids41, zwitterionic amino lipids42, cationic lipid aminoglycosides43,44 and ionizable phospholipids45. Some of the lead lipid materials that have entered the clinic are: dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA; Onpattro), SM-102 (a component of the Moderna COVID-19 vaccine)26,46, ALC-0315 (a component of the Pfizer–BioNTech vaccine)47 and LP-01 (Intellia’s clinical candidates NTLA-2001 and NTLA-2002)48,49 (Fig. 1).
Although the large design space created by the many different lipid molecules and possible molar compositions of those molecules is advantageous, it is impractical to sample in totality. Design-of-experiments approaches, such as fractional factorial design or Box–Behnken design, allow robust sampling of the LNP design space with minimal experiments to yield an optimal formulation for a desired application50. Another solution to minimize the total number of experiments performed while facilitating the high-throughput screening of chemically distinct LNPs in vivo involves the encapsulation of DNA or mRNA barcodes that are extracted from the tissues of interest and quantified using deep sequencing to measure LNP biodistribution to target organs in vivo51–53. Although initial iterations of the barcoding approach were unable to resolve inactive versus functional LNP delivery52, the integration of a functional readout into the assay allows quantification of LNP activity alongside biodistribution54. Because of the extensive clinical experience of LNPs, future iterations of this technology are well poised for translation to patients.
Polymers
Polymers are macromolecules consisting of multiple repeating units known as monomers. Homopolymers are formed from identical monomer units and copolymers are composed of two or more different monomer units often arranged in defined sequences, configurations and architectures. Polymers are inherently a chemically flexible molecular platform, as their size, structure and functionality can be varied to tune their physicochemical properties. The chemical structure of polymers as well as the selection of supplemental components (including lipids and polymers) added to a formulation can influence nanoparticle potency, stability and organ-targeting properties. Similar to LNPs, this inherent intramolecular and intermolecular flexibility generates a large chemical space that researchers can explore8.
Polymeric nanoparticles can spontaneously assemble into various different structures. The thermodynamic principles that govern the self-assembly of LNPs also apply to polymeric nanoparticles. However, polymeric nanoparticles can be made of diverse chemical groups and use covalent and non-covalent interactions, which provide additional opportunities for organization. Examples of structures include solid matrix systems, micelles and polyplex nanoparticles. Solid matrix systems are condensed particles held together by hydrophobic interactions between individual polymer macromolecules. Meanwhile, micelles are produced from amphiphilic block polymers, forming a hydrophobic core with a hydrophilic shell to minimize unfavourable interactions with the surrounding aqueous environment55. Both solid matrix systems and micelles have proven useful for the loading and delivery of small-molecule drugs with poor water solubility56–58. Finally, polyplex nanoparticles assemble as a result of electrostatic interactions between the polymer material itself and oppositely charged macromolecular cargoes (such as nucleic acids) and can be further stabilized and/or enhanced through the addition of other molecular components59. Although polymers have less clinical experience for the delivery of genetic drugs compared with LNPs, their wide possible chemical space allows the discovery of materials with new properties, such as cell and tissue tropism60 and capacity for endosomal escape61, beneficial for the organ-specific delivery of genetic drugs.
The toolbox of genetic drugs
The central dogma of molecular biology specifies the direction of the flow of information in the cell; genes are encoded within DNA, stored in the nucleus and transcribed into mRNA to facilitate the synthesis of functional proteins in the cytosol by ribosomes. Conventional drugs, such as small molecules and monoclonal antibodies, typically act directly on proteins at the tail end of this process to have a pharmacological effect62. By contrast, genetic drugs use nucleic acid molecules that directly affect protein expression in cells. By acting on the processes of information flow in the cell, rather than the final product, genetic drugs are not hampered by druggability concerns (that is, the ability to bind a protein target to exert a biological effect) faced by conventional drug modalities63. Nucleic acids can be used to inactivate genes driving a disease64–66, produce a protein that is missing or non-functional67,68 or express an exogenous protein not typically made by the target cells69,70 (Box 2). However, nucleic acids have very poor pharmacokinetic properties. On their own, nucleic acids are readily degraded by enzymes in the serum and do not easily diffuse across the lipid membrane of cells because of their high molecular weight, repeated number of negatively charged phosphate groups and hydrophilic nature1. Their incorporation in organ-targeted nanoparticles can overcome these limitations by protecting them from enzymatic degradation and enabling their uptake and release into the cytosol of target cells1. Innovations in both the chemical modification of nucleic acids71,72 and the synthesis of materials with novel properties have been instrumental in translating small interfering RNA (siRNA) and mRNA therapies into the clinic2,3,73 (Table 1). Among these promising advances, we underline in particular the development of ionizable cationic lipids and polymers to load anionic nucleic acids into nanoparticles and promote their endosomal escape following cellular uptake27,28. Despite these successes, limitations in intracellular delivery74–77, owing to poor endosomal escape and exocytosis, unwanted immune responses78 and difficulty targeting extrahepatic tissue hamper the full potential of nucleic acid therapies.
Because all nucleic acid biomolecules have similar physicochemical characteristics, both the delivery system and the manufacturing process can be easily adapted by simply changing the genetic sequence and type of modification79–81. Thus, nucleic acids are an inherently flexible therapeutic modality. Unlike small-molecule and protein drugs, they do not require the synthesis of an entirely new molecule to execute different functions, and the associated delivery system can then be repurposed for the delivery of alternative nucleic acid sequences. Using this modular approach could expedite the drug approval process, as was seen for the bivalent mRNA vaccines that were developed for emerging variants of SARS-CoV-2. This approach could also be used to develop personalized medicines for small cohorts of patients. It is important to note that some variations to the molecular composition of the nanoparticle may be required depending on the length of the nucleic acid35,50.
Box 2 Genetic drug mechanisms of action.
Nucleic acid biomolecules can be loaded into nanoparticles to reprogramme the central dogma of biology through gene silencing, expression and editing (see the figure) to correct the course of disease.
Gene silencing
Gene silencing is the process of inactivating protein expression by degrading messenger RNA (mRNA) before it undergoes translation into a functional protein. Both small interfering RNAs (siRNAs) and microRNAs (miRNAs) leverage common enzymatic machinery to degrade a target mRNA transcript using complementary Watson–Crick base pairing. siRNA provides highly potent, durable and specific silencing of virtually any gene in the body, including proteins that cannot be drugged by small molecules. The first RNA drug approved by the US Food and Drug Administration was an siRNA delivered to liver hepatocytes by lipid nanoparticles and used to treat polyneuropathy of hereditary transthyretin-mediated amyloidosis216. In the short time since then, four more siRNA drugs against liver targets have entered the clinic, with many more in the pipeline across diverse settings including oncology, neurology, ophthalmology, cardiology and rare diseases. However, current delivery technologies in the clinic largely target the liver, and extrahepatic delivery remains a bottleneck for realizing the full potential of siRNA.
Unlike siRNA, no genetic drugs utilizing miRNA as the active agent have entered the clinic as a commercial product yet. However, although siRNA requires full complementarity with the target mRNA, miRNA needs only to be partially complementary223. As a result, a single miRNA sequence can repress multiple genetic targets to produce complex effects on gene expression. miRNAs have the potential to target complex diseases driven by multiple genes, such as cancer, neurodegenerative disorders and cardiovascular conditions223.
Gene expression
Both DNA and mRNA can encode proteins for therapeutic gene expression. Over 75,000 pathogenic mutations are known to disrupt the function of native proteins to cause disease and could possibly be treated by the exogenous expression of the healthy protein using DNA or mRNA68. Additionally, non-native proteins such as antigens (for vaccination) or monoclonal antibodies (to bind a target protein) can be expressed for therapeutic benefit. The DNA or mRNA sequence can be engineered to maximize therapeutic efficacy by improving mRNA translation rates, transcript half-life and function and properties of the resultant protein. As such, sequence engineering can play a key role in defining the efficacy of a genetic drug, while also minimizing dose frequency to improve patient compliance.
Both gene expression modalities feature inherent advantages and disadvantages. DNA can produce multiple copies of mRNA, hypothetically amplifying protein production, and is more stable in the cell, which promotes prolonged gene expression. However, DNA must enter the nucleus to be functionally active, which constitutes an additional delivery barrier that mRNA does not face as it only has to reach the cytosol. In cases in which a therapeutic protein is only required for a short time frame, such as vaccination or gene editing, the shorter half-life of mRNA serves as an advantage2,3,48. Ultimately, the specific application and the desired outcome must be considered before selecting DNA or mRNA as a modality.
Gene editing
Compared with gene silencing and gene expression, which are transient by their very nature, direct editing of the genomic DNA enables durable changes in the genetic programme of a cell85. Thus, gene editing can yield functional cures to diseases for which a molecular defect is known and is amenable to correction. However, gene editing may require the delivery of multiple components, including an expression system for the gene editor, a guide molecule to target the correct genomic locus and a template specifying the genetic repair224. Additionally, gene-editing systems must edit the target genomic locus with high fidelity in a predictive manner as there is great risk associated with multiple off-target edits in the DNA of healthy cells. As a result, nanoparticles must be designed to accommodate multiple cargoes that each have distinct physicochemical properties while minimizing delivery to bystander cells224.
Currently, clustered regularly interspaced short palindromic repeat (CRISPR) gene-editing systems represent the most advanced and widely adopted strategy for genome engineering85. CRISPR-associated (Cas) proteins can associate with a short strand of RNA that guides the protein to the target genomic locus using Watson–Crick base pairing, enabling gene editing to be performed in a facile and a programmable manner by engineering the guide RNA. New gene-editing tools based on CRISPR–Cas systems, such as base editing and prime editing, have been developed to increase the precision and efficiency of gene editing by either directly altering the chemical structure of single nucleotides225 or precisely inserting nucleotide sequences into genomic DNA from an RNA template226. In vivo gene editing has entered the clinic at a remarkable pace. The use of Cas9 for programmable gene editing was initially reported in 2012 (ref. 227), and in less than 10 years, the first preliminary data on Cas9-mediated gene editing in human patients were reported48. Advances in delivery technologies and in the understanding of the fundamental mechanisms whereby they operate played an instrumental role in accelerating the clinical entry of in vivo gene editing and lay the foundation for additional clinical trials against liver targets48,216.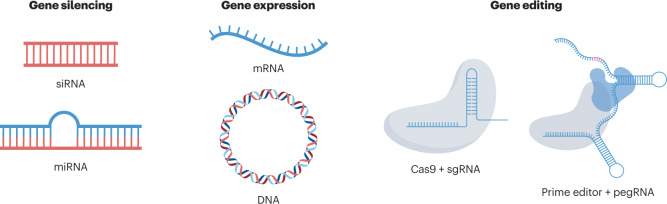
pegRNA, prime editing guide RNA; sgRNA, single guide RNA.
The nanoparticle journey
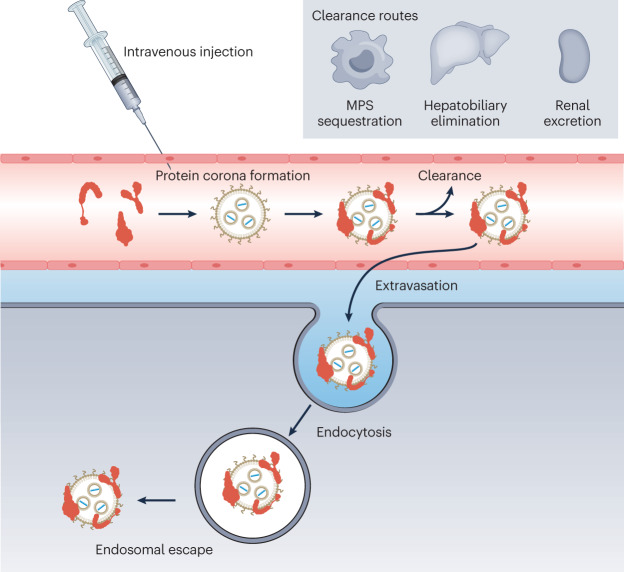
Following i.v. injection, nanoparticles are subject to biological processes at the organ, cellular and molecular levels that ultimately dictate their fate in the body82. Because the physical and chemical properties of a nanoparticle govern how they are affected by these processes, there is a continued need to mechanistically study the relationship between the properties of nanoparticles and their biological fate. Knowledge of these processes and their mechanics is essential for the design of nanoparticles for organ-specific drug delivery. Major processes include protein corona formation, clearance from the circulation, extravasation, cellular uptake and intracellular trafficking (Fig. 2).
Fig. 2. The journey of a nanoparticle through the human body following intravenous injection.
Upon contact with the blood, plasma proteins often adsorb to the nanoparticle surface to form an interfacial layer known as the protein corona. The composition of the protein corona is influenced by the surface properties and composition of the nanoparticle. To reach the target organ, nanoparticles must exit the vasculature (extravasation) by passing through gaps in the endothelium, a size-limited process, or by active transcytosis, involving interaction with specific receptors expressed on the endothelium. Following extravasation, nanoparticles must interact with and be internalized by target cells. They must then escape the endosome into the cytosol and release their genetic payload. Throughout this journey, nanoparticles can be cleared from the systemic circulation by the mononuclear phagocytic system (MPS), hepatobiliary elimination (by the faeces) or renal excretion (by urine). These processes limit how much of the injected nanoparticle dose reaches the desired target site; thus, steps must be taken to minimize their action.
Ultimately, the amount of nanoparticles delivered to the target site should be enough to induce a therapeutic benefit against the disease being treated83. Organ-targeted drug delivery is less about getting all of an administered dosage to a target organ, but rather about delivering a sufficient amount for a desired biological effect while limiting toxicity from off-target accumulation. This is a key pharmacological concept; even if the majority of the injected dose does not reach the target cells, it should still be sufficient to induce a physiological effect and provide benefits to patients84. Keeping this distinction in mind is key for ensuring the clinical feasibility of any nanoparticle design for drug delivery. However, there are some cases in which off-target delivery must be minimized, for example, in gene editing, as permanent changes to the DNA in undesired cell types may pose notable risk85.
Protein corona formation
Upon contact with the blood, the surface of a nanoparticle rapidly adsorbs plasma proteins and other biomolecules to form an entity known as the biomolecular or protein corona86. Although the term biomolecular corona covers a broader array of molecular classes that may bind a nanoparticle, we refer to protein corona here as the majority of studies on nanoparticle organ targeting have focused on how plasma proteins impact delivery outcomes. The composition of the protein corona endows the nanoparticle with a ‘biological identity’ that has properties distinct from its original, pristine ‘synthetic identity’ and defines the subsequent fate of a nanoparticle11,87–89. The protein corona can even shield active targeting ligands, reducing their efficacy90,91. The hydrophobicity, surface chemistry and charge and molecular composition of a nanoparticle all impact the set of plasma proteins that bind to it92–94.
Historically, the protein corona has been associated with detrimental delivery outcomes because of opsonization. Opsonins are proteins that bind to the surface of foreign particulates (including synthetic nanomaterials) in the blood, tagging them for sequestration and degradation by cells of the mononuclear phagocyte system (MPS; also known as the reticuloendothelial system (RES))95. Coating the nanoparticle surface with PEG or other non-fouling materials is a well-established strategy to limit protein corona formation and subsequent phagocytosis. For example, PEG-modified lipids enabled the prolonged plasma circulation of Doxil by reducing opsonization, resulting in one of the first FDA-approved nanoparticle therapeutics17. However, growing evidence suggests that proteins distinct from opsonins, such as apolipoproteins, can be incorporated into the protein corona and may play an active role in targeting nanoparticles to specific cells and organs in the body, forming the bedrock of the emerging concept of endogenous targeting96,97. As PEGylation can impair protein corona formation, strategies for the controlled removal of PEG to promote adsorption of specific proteins to the nanoparticle surface, such as PEG shedding97,98, may be necessary to fully leverage endogenous targeting.
The conformation of a protein can change following adsorption, resulting in the masking or revealing of epitopes responsible for functional receptor interactions. This conformational change suggests that the structure of the protein corona, beyond its composition, is important for cultivating a desired biological identity99–101. Understanding the link among the synthetic identity of a nanoparticle, the protein corona and the subsequent fate of a nanoparticle in the body remains an active and necessary area of study101,102.
Clearance from the circulation
The circulation behaviour of a nanoparticle in the blood plays a major role in organ targeting103. Following systemic injection, a dynamic competition ensues between the accumulation of the nanoparticle within the target or off-target tissues, sequestration by the MPS or excretion from the body by either the renal or hepatobiliary systems104. The kinetics of each process determine the total amount of nanoparticles reaching the desired site of action. Optimizing nanoparticle circulation involves reducing the rate of elimination by the MPS, the renal system and the hepatobiliary system. Improving the serum half-life of a nanoparticle thereby increases the probability of reaching the target organ.
Macrophages, monocytes and dendritic cells constitute the major cell types of the MPS105. These cells play a crucial role in innate immunity by binding opsonized particulates and removing them from the blood106. The MPS is a substantial barrier to organ-targeted drug delivery; the presence of opsonins in the nanoparticle protein corona results in the notable accumulation of nanoparticles in the liver or spleen (two major organs of the MPS)95. Thus, it is critical to implement strategies to circumnavigate the MPS to maximize the targeting efficacy of nanoparticle drug delivery systems.
As mentioned previously, PEGylation of the nanoparticle surface can reduce opsonization, thus minimizing interactions with the MPS and enhancing nanoparticle circulation time. The PEG chain length, surface density and the stability of the linker anchoring PEG to the nanoparticle surface all impact the circulation time of a nanoparticle, serving as inputs to tune clearance behaviour98,107. However, repeated administration of PEGylated nanoparticles has been shown in some cases to induce an immunogenic response known as accelerated blood clearance (ABC). The initial dose of PEGylated nanoparticles can stimulate marginal zone B cells in the spleen108, resulting in the potential formation of anti-PEG IgM antibodies that can bind subsequently administered PEGylated nanoparticles and flag them for MPS sequestration109. The magnitude and extent of ABC can be affected by several parameters including dosage time interval, total number of doses, PEG surface density and nanoparticle composition and structure110. The comprehensive characterization of PEGylated nanoparticle pharmacokinetics is necessary to estimate the impact of ABC on organ-targeting efficacy.
The adverse impact of ABC on nanoparticle delivery has prompted the development of alternative strategies to overcome MPS clearance. For example, novel materials such as zwitterionic polymers including poly(carboxybetaine)s and poly(sulfobetaine)s, polysarcosines, poly(phosphoester)s and poly(oxazoline)s have been created and shown to be capable of resisting protein adsorption while minimizing immunogenicity111–115. Additionally, biomimetic surface coatings have been devised to ‘camouflage’ nanoparticles from the MPS. Grafting of the CD47 molecule, a known ‘don’t eat me’ signal to avoid phagocytosis, onto the nanoparticle surface has been proposed as an alternative strategy for engineering stealth materials116. By contrast, coating nanoparticles with membranes derived from endogenous blood cells, such as platelets117, red blood cells118,119 and leukocytes120–122, is thought to enable multivalent presentation of markers of self to immune cells, resulting in evasion of the MPS.
Alternatively, overwhelming the phagocytic rates of MPS cells, by either pre-administering or co-administering a material that can occupy cellular receptors in lieu of the delivery system, can reduce MPS elimination123–126. Notably, a one trillion nanoparticle dose threshold has been discovered in mice: nanoparticle dosages below this threshold result in high Kupffer cell accumulation but dosages above the threshold saturate Kupffer cell uptake, resulting in prolonged nanoparticle circulation time and improved biodistribution to the target site126. It is important to consider particle dose and other factors in the design and evaluation of future nanomaterials.
Nanoparticles that are not phagocytosed will eventually undergo clearance from the body by either the renal or hepatobiliary system in a size-dependent manner pending material degradation. Renal excretion involves filtration of nanoparticles through the narrow sieves of the kidney glomerulus, with only nanoparticles of a small size (typically less than 6 nm) capable of passing into the urinary track for elimination127. Additionally, degradation products of nanoparticles, such as polymer chains or other small molecules that are not metabolized intracellularly, can be cleared by the renal system because of their small size128. Larger particles are eliminated by the liver to the faeces by the hepatobiliary system, and the kinetics of this process depend on nanoparticle physical properties such as size, shape and surface charge104. Accumulation of nanoparticles in off-target tissues is also possible through other mechanisms, and some plasma proteins have been shown to affect the clearance behaviour of injected nanoparticles in vivo107. To maximize the probability of reaching the target organ, promoting tissue accumulation by enhancing circulation time and reducing the mechanisms leading to off-target clearance are crucial.
Extravasation
While in systemic circulation, injected nanoparticles must reach and cross the endothelium of the vasculature perfusing a target organ to accumulate at the site of therapeutic action, a process known as extravasation. Endothelial cells lining blood vessels form a barrier that regulates the flux of substances into and out of an organ, and the endothelium of different organs displays great heterogeneity depending on the physiological function of the organ in question129–131. These differences include the size and density of fenestrae132,133, the capacity for endocytosis and transcytosis134,135 and the expression of cellular receptors involved in active molecular transport136–138. Understanding the unique characteristics of the endothelial beds of target organs can help guide nanoparticle design, for example, by tuning size and shape to enhance extravasation via fenestrations or by altering surface chemistry to engage specific endothelial transporters for organ-specific uptake. Even further, disease can disrupt endothelium integrity to generate distinct features compared with physiological conditions139, providing new guideposts for optimizing nanoparticle design to reach the target site.
Cellular uptake and trafficking
Once nanoparticles exit the systemic circulation and arrive at the target organ, the therapeutic cargo must be liberated in the cytosol of the proper cell type140. Because cellular uptake typically occurs by receptor-mediated endocytosis, nanoparticles often have a mechanism by which they bind cellular receptors expressed on the plasma membrane. This can be accomplished by both endogenous ligands (such as plasma proteins adsorbed to the nanoparticle surface) and exogenous ligands that have been conjugated to the nanoparticle surface before administration96.
Following receptor-mediated endocytosis, nanoparticles and their cargo are sequestered in the endosome. Over time, the endosomes mature, acidify and eventually fuse with lysosomes, a degradative organelle141. Escape from the endosome is necessary for genetic drugs to avoid breakdown in the lysosome and be functionally active142. Poor endosomal escape is a key barrier for efficacious nucleic acid delivery that has been overcome through the design of ionizable materials that can acquire charge within maturing endosomes to promote endosome destabilization and cargo release143,144. Thus, designing nanoparticles that can enter cells and escape from the endosome is crucial for engineering successful organ-targeted drug delivery systems.
Delivery to the liver
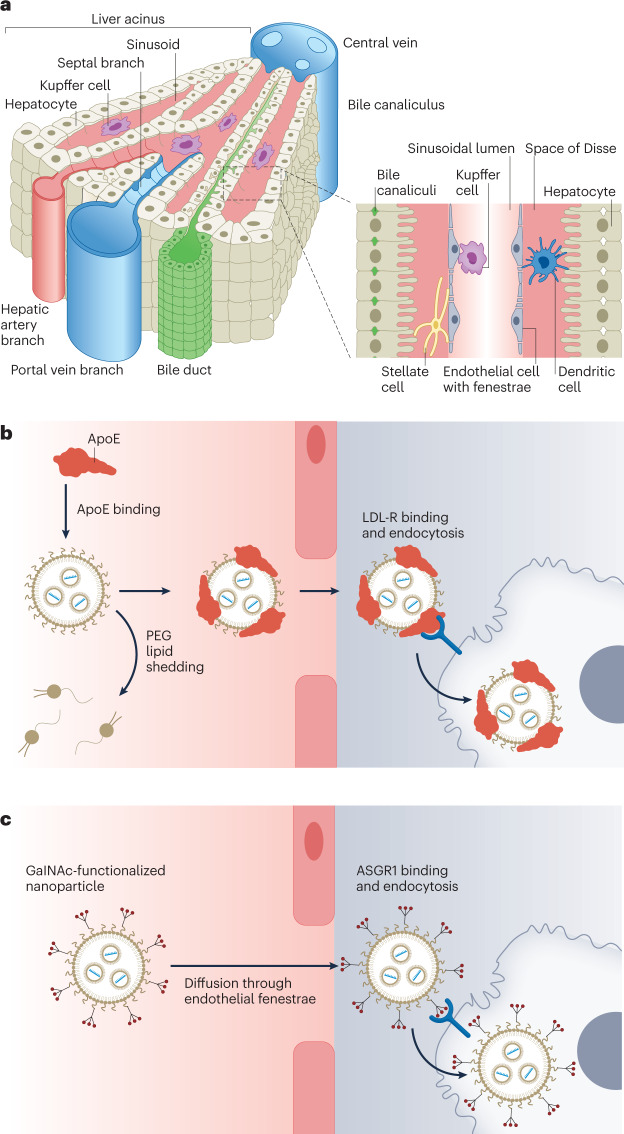
The liver plays a role in many physiological processes such as nutrient break down and storage, blood detoxification, immune surveillance and the synthesis of plasma proteins145. Several anatomical features of the liver promote interactions with injected nanoparticles — the liver is highly perfused with fenestrated endothelium, promoting widespread extravasation and organ accumulation, and multiple cell types are involved in the clearance of both foreign and endogenous particulates from the bloodstream128 (Fig. 3a). Despite extensive liver accumulation, it is important to ensure that injected nanoparticles transfect the proper cell type for therapeutic intervention and avoid off-target cells to prevent unwanted toxicity and immune responses. Four key cell types coordinate the functions of the liver: hepatocytes, Kupffer cells, liver sinusoidal endothelial cells (LSECs) and hepatic stellate cells (HSCs)145. These cells form the functional subunits of the liver known as hepatic lobules (Fig. 3a). Presently, LNPs represent the most advanced system for delivery of genetic drugs to the liver and, as such, are the focus of this section, but relevant examples of polymer nanoparticles are highlighted for specific applications.
Fig. 3. Lipid nanoparticles are a clinically mature technology for genetic drug delivery to the liver.
a, The liver microanatomy is composed of four distinct cell types. Nanoparticles in the blood can either be sequestered by Kupffer cells, taken up by liver sinusoidal endothelial cells, or extravasate through the wide fenestrations in liver endothelium into the Space of Disse. There, the nanoparticles can target hepatic stellate cells or hepatocytes. The hepatobiliary system can eliminate nanoparticles from the body via the bile duct220. b, Endogenous targeting of liver cells is a clinically validated mechanism for small interfering RNA delivery to hepatocytes. For example, delivery of Onpattro lipid nanoparticles occurs by exchange of polyethylene glycol (PEG) lipid at the nanoparticle surface with apolipoprotein E (ApoE) in the blood. Adsorption of ApoE to the nanoparticle surface results in binding of the nanoparticle by low-density lipoprotein receptor (LDL-R), highly expressed by hepatocytes, and subsequent endocytosis. c, Active targeting of hepatocytes can also be achieved by functionalizing the nanoparticle surface with an N-acetylgalactosamine (GalNAc) ligand and reducing non-specific protein binding through extensive PEGylation. GalNAc binds asialoglycoprotein receptor 1 (ASGR1) to facilitate nanoparticle uptake by hepatocytes220. Part a reprinted from ref. 220, Springer Nature Limited.
Targeting hepatocytes
Liver hepatocytes are an attractive cellular target for delivery owing to their metabolic and secretory functions. A great many genetic disorders affect hepatocytes, most of which cannot be treated using conventional small-molecule drugs. Thus, the use of nanoparticles to deliver nucleic acid therapeutics and gene-editing systems has emerged to meet this clinical need146. Alternatively, researchers have sought to leverage the physiological role of the liver in secretion of molecules into the systemic circulation, treating hepatocytes as an endogenous bioreactor for producing therapeutic biomolecules.
LNPs are the most advanced delivery system for targeting hepatocytes. Most notably, the first siRNA drug approved by the FDA, Onpattro, utilizes a four-component LNP carrier composed of an ionizable amino lipid (DLin-MC3-DMA), 1,2-distearoyl-sn-glycero-3-phosphocholine, cholesterol and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000, to treat hereditary transthyretin-mediated amyloidosis73. The successful clinical translation of Onpattro and of the two LNP-based SARS-CoV-2 vaccines2,3 has established that LNPs are a safe, efficacious and scalable technology platform, laying a firm foundation that can ease the translation of other LNP-based therapeutics. Because of the extensive experience and varied applications of LNPs as delivery systems to liver hepatocytes, the remainder of this section focuses on LNPs.
LNPs are now being utilized as delivery systems in a diverse array of preclinical and clinical studies of nucleic acid therapeutics to hepatocytes. Many of these studies focus on mRNA delivery in several clinical contexts including protein replacement therapy, such as in clotting disorders147–149 or in-born errors of metabolism35,150–156, in situ gene editing49,157 and systemic secretion of monoclonal antibodies against infectious diseases or cancer69,70,158.
There are multiple ways to target liver hepatocytes. A major mechanism proposed for targeting liver hepatocytes involves the formation of a protein corona enriched in apolipoprotein E (ApoE), a protein involved in endogenous cholesterol transport (Fig. 3b). First, solvent-exposed PEG lipid desorbs from the LNP surface, unshielding the LNP for interactions with plasma proteins98. ApoE is among the subset of proteins that most abundantly adsorb to the LNP surface, and it can bind the low-density lipoprotein receptor (LDL-R)96 that is highly expressed by hepatocytes. LNPs enter the cell by receptor-mediated endocytosis, localizing to the endosome. As the endosome acidifies, ionizable lipids in the LNP are protonated, resulting in electrostatic interactions with negatively charged endosome lipids27 and, eventually, disrupting the endosomal bilayer143. An LNP pKa of around 6.4 has been determined as optimal for endosomal escape, leading to cargo release into the hepatocyte cytosol to exert its therapeutic effect159. Knowing this mechanism has allowed researchers to improve the efficacy of LNP delivery to the liver159, highlighting the importance of mechanistically studying nanoparticle targeting.
Other mechanisms for targeting hepatocytes that are ApoE-independent have also been developed. For example, incorporating ‘synergistic’ alkyne lipids into a cKK-E12 lipopeptide-based LNP promotes the binding of serum albumin to enhance mRNA delivery to the liver160, demonstrating that endogenous targeting of hepatocytes is possible using multiple proteins.
Instead of altering the molecular composition of an LNP to yield a specific protein corona for hepatocyte delivery (endogenous targeting), conjugating ligands that interact with receptors expressed by hepatocytes to the nanoparticle surface serves as an alternative mechanism for delivery (active targeting). Targeting of the asialoglycoprotein receptor has been achieved using N-acetylgalactosamine (GalNAc) as an active targeting ligand (Fig. 3c). Appropriate chemical design of GalNAc ligands is needed to maximize their targeting efficacy, with features such as valency, spatial arrangement and linker length playing crucial roles. Additionally, the rapid endocytic rate and the high recycling frequency of asialoglycoprotein receptor promote sustained liver accumulation of GalNAc-based delivery systems161. To date, five GalNAc-siRNA molecular conjugates have entered the clinic for therapeutic gene silencing in hepatocytes, demonstrating the feasibility of this targeting mechanism. However, larger nucleic acid cargoes cannot be readily incorporated into the molecular conjugate architecture. Thus, using nanoparticles functionalized with GalNAc could extend this well-validated active targeting mechanism for other classes of genetic drugs.
Using a GalNAc ligand to target hepatocytes, instead of ApoE, can help overcome pathophysiological features that impair ApoE-mediated targeting162 or dose-limiting toxicities163. However, protein corona formation and subsequent endogenous targeting of the liver appear to be a competitive process. ‘Deactivation’ of endogenous targeting through extensive PEGylation has been shown to prevent this competition, ensuring that GalNAc-mediated active targeting is the dominant mechanism of delivery96,161. Deactivating endogenous targeting using PEGylation probably applies to other active targeting systems that target either the liver or extrahepatic organs.
Delivery to cells beyond hepatocytes
Other major cell types of the liver can also benefit from genetic drugs. For example, LSECs have been proposed as a target for immunomodulation owing to their physiological role in antigen presentation and innate immunity164. Because LSECs highly express the mannose receptor, which recognizes and binds endogenous glycoproteins and infectious microorganisms, functionalizing nanoparticles with mannose moieties promotes targeting of LSECs. In one example, conjugating mannose to the end of the PEG lipid component of an LNP shifted mRNA delivery from hepatocytes to LSECs for Cre recombinase gene editing165. Further cell specificity was achieved by increasing the PEG density on the LNP surface to reduce ApoE binding and subsequent hepatocyte targeting.
Manipulating the molecular composition of nanoparticles also leads to LSEC delivery. For example, altering the pKa of LNPs away from 6.4, which is optimal for hepatocyte delivery, to approximately 7.1 corresponded to a shift in the intrahepatic biodistribution and gene-silencing efficacy of LNPs from hepatocytes to LSECs. This altered ionization behaviour was obtained through either the synthesis of new ionizable cationic lipids166 or the mixing of lipids with different pKa values167. Alternatively, modifying the hydrocarbon side chain of cholesterol with a hydroxyl group at the 20α position also resulted in the retargeting of LNPs from hepatocytes to LSECs168. Although these studies demonstrate how changes in LNP chemistry affect intrahepatic cell-targeting properties, further elucidation of the biological interactions involved, namely, the formation of the protein corona, is necessary for the rational design of nanoparticles for LSEC delivery.
HSCs are another cell type that could benefit from genetic drugs. On detecting liver injury, HSCs transdifferentiate to a myofibroblast-like phenotype, a process known as activation, and secrete collagen and matrix metalloproteinases to create scar tissue. Sustained sensing of injury leads to liver fibrosis, which progresses to cirrhosis and, ultimately, to the development of hepatocellular carcinoma and organ failure as scar tissue replaces healthy liver tissue169. Given the lack of effective therapies to attenuate liver fibrosis, and its growing prominence around the globe, there is a substantial clinical need for HSC-targeted genetic drugs169. Normally, HSCs serve as reservoirs for 50–80% of the vitamin A in the body170. Because of this physiological hallmark, researchers have incorporated vitamin A into nanoparticles for the endogenous targeting of anti-fibrotic nucleic acids to HSCs. Of note, LNPs that incorporate vitamin A as a helper lipid have delivered siRNA against heat shock protein 47, a molecular chaperone involved in collagen secretion by activated HSCs, to resolve liver fibrosis in rats171. This type of LNP has been the subject of multiple phase II clinical trials in therapeutic indications related to liver fibrosis (Table 1).
Vitamin A also retargets polymer nanoparticles to HSCs. For example, the end modification of polyethyleneimine (PEI) with vitamin A significantly increased the enrichment of retinol-binding protein 4 in the protein corona compared with unmodified PEI, enabling the targeting of HSCs and the treatment of liver fibrosis172. Identifying a suitable molecular structure with high affinity for a distinct protein in the blood can lead to the controlled formation of a protein corona that promotes targeting of a specific cell type in vivo. Identifying other combinations of material compositions, plasma proteins and target cells is essential for the rational design of novel delivery systems that leverage endogenous targeting.
Extrahepatic delivery
Despite the extraordinary progress in engineering nanoparticles for targeted delivery to the liver, developing new systems that access extrahepatic (non-liver) tissues is essential to fully realize the promise and potential of genetic drugs. Elucidating key design rules to reduce liver uptake of nanoparticles and/or target extrahepatic organs using both active and endogenous mechanisms remains an active area of research. To date, the lungs and the lymphoid tissues are the two extrahepatic organs that have been most successfully targeted by lipid and polymer nanoparticles (Table 2).
Table 2.
Nanoparticle compositions for extrahepatic delivery
| Organ | Material | Refs. |
|---|---|---|
| Lungs | LNPs incorporating quaternary ammonium lipids | 45,97,173,174 |
| Lungs | LNPs based on ionizable lipids with amide linkages | 176 |
| Lungs | Low-molecular-weight PEI oligomers | 52,54,178 |
| Lungs | PBAE nanoparticles | 181–183 |
| Lungs | Functional polyester nanoparticles | 184,185 |
| Lungs | Ionizable amino polyester nanoparticles (IDD-3 building block) | 60 |
| Lungs | GALA peptide-modified LNPs | 186,187 |
| Lungs | Anti-PECAM1-modified LNPs | 188 |
| Lungs | Anti-PV1-modified LNPs | 189 |
| Spleen | LNPs incorporating permanently anionic lipids | 97,173 |
| Spleen | DOPE–DOTMA lipoplex | 197–200 |
| Spleen | PACE polymer nanoparticles | 201–203 |
| Spleen | Charge-altering releasable transporters (CARTs) | 204–206 |
| Spleen, lymph nodes | Zwitterionic phospholipidated polymers | 61 |
| Spleen (T cells) | Anti-CD3-modified or anti-CD8-modified PBAE nanoparticles | 208,209 |
| Spleen (T cells) | Anti-CD4-modified or anti-CD5-modified LNPs | 210,211 |
DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DOTMA, 1,2-di-O-octadecenyl-3-trimethylammonium propane; GALA, glutamic acid-alanine-leucine-alanine repeat peptide; LNP, lipid nanoparticle; PACE, poly(amine-co-ester); PBAE, poly(beta-amino ester); PECAM1, platelet endothelial cell adhesion molecule; PEI, polyethyleneimine; PV1, plasmalemmal vesicle-associated protein 1.
Delivery to the lungs
The lungs are at the centre of the respiratory system and enable gas exchange between air in the respiratory tract and blood in the circulation. The two major functional regions of the lung are the bronchi and the alveoli. External air travels from the bronchi to the alveoli, which are sac-like structures with high surface area enshrouded in a capillary network so as to promote efficient transfer of oxygen to the blood. The entirety of the cardiac output must pass through the lungs to oxygenate red blood cells, rendering the lungs readily accessible for intravenously administered nanoparticles82. Some diseases that interfere with proper lung function that are amenable to targeting with genetic drugs are cystic fibrosis, α1 anti-trypsin deficiency and lung cancer.
Re-targeting LNPs from the liver to the lungs has been achieved through the incorporation of atypical chemical motifs that alter protein corona composition. Notably, doping LNPs with a lipid containing a quaternary ammonium headgroup, a component termed a selective organ targeting (SORT) molecule, enables the lung-selective delivery of either mRNA or Cas9–single guide RNA (sgRNA) ribonucleoproteins for gene editing in endothelial, epithelial and immune cells45,173,174. In the case of Cas9–sgRNA ribonucleoproteins, the positively charged quaternary ammonium lipids permit their formulation in a neutral buffer, which preserves their function and integrity. LNPs loaded with Cas9 were used to generate complex murine lung cancer models through multiplexed gene editing174. The lung-selective delivery of LNPs that contain quaternary ammonium lipids occurs by endogenous targeting, wherein desorption of PEG lipid from the LNP surface enables the binding of distinct proteins, such as vitronectin, at the expense of ApoE and other proteins implicated in liver targeting97. These large-scale changes in the functional composition of the protein corona are driven by the chemical structure of the SORT molecule and the ionization behaviour of the LNP97. The distinct protein corona composition promotes interactions with specific cellular receptors highly expressed within the lungs, such as receptor αVβ3 integrin by vitronectin, to yield tissue-specific mRNA delivery97. Notably, these mechanistic studies demonstrate that the protein corona can provide a link between the molecular composition of nanoparticles and their organ-targeting properties.
Similarly, zwitterionic amino lipids42 and cationic quaternary sulfonamide amino lipids175, both of which feature a permanently cationic quaternary ammonium functional group, also biodistribute and deliver either mRNA or siRNA to the lungs, suggesting that lung-targeting is a key property of quaternary ammonium-containing materials. Alternatively, LNPs incorporating lipidoids that feature amide linkages could impact protein corona composition and re-direct mRNA delivery from the liver to lungs172. Interestingly, these LNPs have a different protein corona compared with lung-targeting SORT LNPs and transfect lung cells in different proportions (mainly endothelium), suggesting the need to fully characterize how the protein corona impacts both organ-level and cell-level tropism97,176.
Multiple polymer nanoparticles have been identified for delivering nucleic acids to the lungs. One of the first cationic homopolymers used to package genetic material for intracellular delivery was PEI. However, a trade-off exists between the potency and toxicity of PEI polyplexes, in which higher molecular weight materials are potent but toxic because they have a high cationic charge density and are not biodegradable177.To minimize these off-target effects of high-molecular-weight PEI polyplexes while maintaining potency, a library of low-molecular-weight lipidated PEI600 materials were prepared to self-assemble into nanoparticles alongside C14-PEG2000 lipid and cholesterol. A specific oligomer, called 7C1, was identified as most efficacious for gene silencing in lung endothelial cells of both mice178 and non-human primates179. Gene silencing in kidney and heart endothelial cells178 was also achieved to a lesser extent. In addition, nanoparticles incorporating 7C1 have been leveraged to deliver mRNA to pulmonary endothelial cells54. Adjusting the chemistry and the amount of PEG lipid in 7C1-based formulations enabled the re-targeting of the nanoparticles from the pulmonary endothelium to the bone marrow endothelium54,180. Further studies that reveal the mechanism responsible for shifts in organ-targeting properties following adjustments to nanoparticle PEGylation remain necessary.
Similarly, potent lung-specific mRNA delivery was achieved using poly(beta-amino ester) (PBAE) terpolymers that were designed for enhanced serum stability when formulated with C14-PEG2000 lipid181. The inclusion of additional lipid components (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and cholesterol) further enhanced mRNA delivery potency in lung endothelial and immune cells182. The same PBAE terpolymers were used successfully for plasmid DNA (pDNA) delivery, but the molecular composition of the most potent formulation was distinct from that which was best for mRNA delivery183. This result suggests that the composition of a delivery system may need to change to account for physicochemical differences in specific nucleic acid biomolecules.
Because of their biocompatibility and their extensive use in FDA-approved products, polyester materials with tunable molecular weight and chemical functionality were also prepared for extrahepatic nucleic acid delivery184,185. For example, low-molecular-weight (4.2 kDa) polyester materials with cysteamine side chains were proven effective for lung-specific delivery of mRNA185. Crucially, the chemical features of polymers are important for determining cellular selectivity and delivery potency of a nanoparticle184,185. Similarly, the link between nanoparticle composition and organ-targeting properties was demonstrated using ionizable amino-polyester materials synthesized from the ring-opening polymerization of lactones with tertiary amino-alcohols60. The selection of amino-alcohol building block allows to vary the number of ionizable amines, charge density and polymer branching and molecular features that have been previously shown to impact delivery efficacy. The polyester molecular weight and lipophilicity are, meanwhile, controlled through the degree of polymerization and structure of the lactone monomer. One compound, known as IDD-3, was formulated with three additional lipid components to enable lung-selective mRNA delivery. Interestingly, compounds that share the same degree of polymerization and lactone monomer as IDD-3, differing in only the choice of the amino-alcohol building block, no longer target the lung.
Less progress has been made using active targeting strategies to the lungs. Although both peptide and monoclonal antibody ligands have been tested in vivo, they have only demonstrated the capacity for transfecting lung endothelial cells, necessitating the development of targeting ligands for other cell types of the lungs. The utility of the GALA peptide, designed to mimic the haemagglutinin protein of influenza virus, has been studied as a lung-targeting ligand for the delivery of siRNA and pDNA in vivo by LNPs186,187. Alternatively, monoclonal antibodies directed against platelet endothelial cell adhesion molecule (PECAM1)188 or plasmalemmal vesicle-associated protein 1 (PV1)189 can be conjugated to the PEG lipid component of LNPs to deliver mRNA to the pulmonary endothelium. Interestingly, the functional delivery of luciferase mRNA to the lungs by anti-PECAM-1-modified LNPs is not impacted by the genetic knockout of ApoE in mice, reinforcing that ApoE-independent mechanisms are involved in extrahepatic targeting188. Despite the acquired capacity to target the lungs through the inclusion of these active targeting ligands, off-target delivery still occurs to the liver and the spleen; as is the case with GalNAc ligands, preventing protein corona formation through denser and more stable PEG shells may help to reduce this off-target delivery.
Delivery to lymphoid tissue and cells
Lymphoid organs, such as the spleen and the lymph nodes, are a staging ground for coordinating the adaptive immune response, facilitating communication between immune cell subpopulations involved in both collecting immunological information and acting on it190. Because immune cells have highly specialized functions in identifying, seeking and destroying unwanted invaders, the delivery of genetic payloads to lymphoid organs provides unique opportunities to treat disease190. Some applications of targeting the spleen and lymph nodes with genetic drugs include vaccination against infectious diseases or cancer, in situ immunoengineering of B cells or T cells or the abrogation of pro-inflammatory signalling involved in driving disease.
Multiple chemical structures, when formulated into LNPs without targeting ligands, enable the spleen-targeting of nucleic acids. Different classes of lipid materials, including diketopiperazine lipids with degradable ester linkages191,192, diethylamino lipids with adamantane tails193, lipidoids with an imidazole headgroup194 and piperazine lipids with either hydrazine or ethanolamine linkers195, can deliver nucleic acids to various cell types in the spleen. Interestingly, the chemical structure of the lipid material affects the cell types in which functional delivery occurs within the spleen. As such, characterizing the specific immune cell types transfected remains important because of their specialized nature and the possible functions they can be engineered to execute.
Tuning the internal ratio of lipid components also promotes the targeting of lymphoid tissues, with mechanistic experiments demonstrating the importance of endogenous targeting using plasma proteins. In one example, the enhanced binding of LNPs by the complement component C3, a plasma protein involved in innate immunity, led to increased interactions with the complement receptors expressed by splenocytes and their subsequent transfection with pDNA196. SORT LNPs that incorporate an anionic phospholipid, such as phosphatidic acid, have a protein corona that is enriched with β2-glycoprotein I, an anionic phospholipid binding protein. The subsequent spleen-specific delivery of mRNA by these SORT LNPs results in the gene editing of B cells and T cells for possible immunoengineering applications173. However, splenic targeting is antagonized by the presence of ApoE in the protein corona, cementing the importance of reducing ApoE binding to de-target the liver97. These studies demonstrate that endogenous targeting of the spleen is feasible through rational manipulation of the molecular composition of LNPs.
Combining the phospholipid DOPE and the permanently cationic lipid 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) in a lipoplex enables mRNA delivery to antigen-presenting cells in the spleen, with potential applications in cancer immunotherapy197–200. Reducing the ratio of DOTMA to DOPE in the formulation, which leads to a negative net charge, allows functional mRNA delivery to the spleen without active targeting ligands197. When loaded with mRNA encoding four tumour-associated antigens, the spleen-specific delivery of this nanoparticle resulted in objective responses in patients with melanoma, demonstrating the potential of mRNA as a modality for personalized cancer vaccines198. In another trial, an mRNA vaccine was used to boost the activity of chimeric antigen receptor T (CAR-T) cells targeting claudin 6 in solid tumours, in which CAR-T cells typically lack efficacy, leading to partial responses in patients with testicular or ovarian cancer199,200. The promising human trials data generated for these genetic vaccines suggest that organ-targeted delivery to lymphoid tissues by LNPs can unlock previously unattainable immunotherapy strategies in the clinic (Table 1).
Certain copolymers have also demonstrated the capacity to transfect cells in the spleen. Poly(amine-co-ester) (PACE) polymers are synthesized by enzyme-catalysed polymerization and have a lower charge density than conventional cationic polymers used for nucleic acid delivery. By relying on hydrophobic interactions, rather than electrostatic forces, to load nucleic acids201,202, acute toxicity events in vivo are limited. PACE polymers with different end groups all delivered mRNA to the spleen in mice, suggesting that the polymer backbone determines the organ-targeting properties of PACE-based nanoparticles203. Similarly, charge-altering releasable transporters (CARTs), polycations capable of packaging long negatively charged molecules such as mRNA, can be used for splenic delivery, indicating utility for immunotherapy applications204. CARTs degrade to non-toxic small molecules by a controlled ester-to-amide isomerization on cellular entry, liberating the mRNA cargo204–206. The use of alternative CARTs, which incorporate mixtures of lipid side chains or an oligo(serine ester) backbone, improved the delivery to lymphocytes204,206. Collectively, these studies demonstrate that the composition of the polymer nanoparticles can be directly engineered to target various immune cell populations in the spleen without the need for active targeting ligands.
Alternatively, nanoparticles formulated from modified cationic polymers with phospholipid side chains resulted in the systemic delivery of mRNA to the spleen and lymph nodes61. These nanoparticles possess increased serum stability, through the formation of a non-fouling sphere of hydration, and enhanced membrane fusion, thereby transforming ineffective polymers into potent mRNA delivery systems in vivo61. The organ-targeting properties of these polymers appear to generalize to the entire material class, with individual variations in the polymer side chains affecting mRNA delivery potency. Within the lymph nodes, both CD4+ and CD8+ T cells are transfected, suggesting potential applications for the in situ programming of immune cells61. The development of this class of polymer materials demonstrates the feasibility of delivering genetic drugs to the lymph nodes for immunotherapy using systemic injection.
Meanwhile, the targeting of immune cells, particularly in the spleen, has been achieved through active targeting. One area of substantial medical interest is the in situ engineering of CAR-T cells. Even though CAR-T cells have had an important medical impact in multiple liquid cancers, their lengthy and bespoke manufacturing process poses practical challenges that limit the availability of these cell therapies. These challenges could be overcome through direct reprogramming of endogenous T cells in the body of a patient207, using both polymers and LNPs208–211. For example, the surface deposition of antibody-functionalized polyglutamic acid onto nucleic acid–PBAE polyplexes enabled the delivery of DNA and mRNA to T cells by targeting either CD3 (ref. 208) or CD8 (ref. 209). Alternatively, modifying the PEG lipid component of LNPs with antibodies directed against either CD4 (ref. 210) or CD5 (ref. 211) achieved mRNA delivery to splenic T cells. Transient expression of a CAR against fibroblast activation protein using CD5-targeting LNPs led to reduced fibrosis and improved function of the heart following cardiac injury211. Despite these advances, off-target delivery to other immune cells and to other organs, particularly the liver, suggests the need for greater specificity.
Given the molecular diversity of receptors expressed on the surface of leukocytes, it is difficult to develop a universal platform to target specific subsets of immune cells involved in distinct physiological and disease processes. To address this challenge, a modular platform for the engineering of active targeting LNPs for nucleic acid delivery was invented by incorporating Anchored Secondary scFv Enabling Targeting (ASSET), a lipidated protein that non-covalently binds targeting antibodies, into the outer membrane of an LNP212. By incubating ASSET-LNPs with an antibody targeting a desired cell type, the intracellular delivery of siRNA or mRNA to different subsets of hard-to-transfect leukocytes was achieved in vivo, leading to the therapeutic modulation of inflammatory signalling212,213. Other cell types have been targeted using the ASSET-LNP platform by swapping out which antibodies are mixed with the LNPs, expanding the applicability of this technology214,215. To date, active targeting has enabled the functional delivery of genetic cargoes into difficult-to-transfect immune cells, but improving the organ-targeting specificity of these delivery systems will maximize the success of clinical translation.
Outlook
The rapid and successful development of mRNA vaccines for the COVID-19 pandemic has promoted research and development efforts to commercialize additional genetic drugs80. Lipid and polymer materials are promising platforms for supporting the clinical development of genetic drugs because they circumnavigate biological barriers, such as cellular membranes and physiological clearance systems, that would otherwise render nucleic acids ineffective. Advances in materials synthesis have driven improvements in nanoparticle potency and organ selectivity15. Meanwhile, the identification of the biological processes involved in nanoparticle transport has provided mechanistic rationales for refining the material compositions of nanoparticles96,97,104,107. Moving forward, continued innovations in genetic drug delivery will play a crucial role in expanding the clinical applications of this therapeutic approach. The ability to innovate is greatly aided by fundamental knowledge linking nanoparticle material properties with organ-targeting outcomes.
The widespread application of genetic drugs has largely been limited by the preferential accumulation of nanoparticles in the liver following i.v. administration. As such, genetic drugs directed against liver targets, such as Onpattro, were among the first to be approved by regulatory authorities and still constitute the majority of the clinical pipeline of nanoparticle therapeutics. These systems have leveraged endogenous lipid transport proteins to target hepatocytes, highlighting the utility of endogenous targeting as a delivery mechanism48,216. Recent discoveries have demonstrated that extrahepatic delivery, particularly to the lungs and lymphoid tissues, is feasible, but it often requires some means of de-targeting the liver by disrupting ApoE binding97. Although some nanoparticles used for extrahepatic delivery have been evaluated in preclinical trials that examine their therapeutic efficacy, the organ-targeting outcomes for many systems have only been measured using reporter models, such as fluorescent proteins or luciferase, and further validation of their therapeutic utility is required. Additionally, some compounds used for extrahepatic nanoparticles can exhibit toxicity, necessitating medicinal chemistry efforts to find biocompatible alternatives that maintain targeting and potency.
Current technologies largely target at the organ level. Advancements can be made to refine delivery to specific cell types within a target organ165. This is a key consideration for when a disease only affects a subpopulation of cells or the function of a specific cell type needs to be modulated to yield a therapeutic benefit. Delivery to cells beyond hepatocytes within the liver has been achieved through the incorporation of active targeting ligands or altering the protein corona. The factors influencing cell-level targeting in non-liver organs still have not been thoroughly studied, but can likely be affected by passive, active and endogenous mechanisms. Additionally, identifying nanoparticles that target as-of-yet inaccessible tissues, such as the muscles, the heart, the central nervous system and the gastrointestinal tract, remains an active area of investigation. The current generation of extrahepatic-targeting nanoparticles provides some mechanistic insights on the criteria necessary to overcome the delivery barrier of liver accumulation that can aid in the design of new carriers to other organs.
Continued identification of the mechanistic factors undergirding delivery will pave the way for the rational engineering of nanoparticles. For passive targeting, linking nanoparticle physical properties to biological behaviour, such as circulation time and organ accumulation, will impact the general design of nanoparticle carriers. For active targeting, the identification of suitable ligands to avidly target a specific cell type, mechanisms to avoid protein corona and off-target accumulation and the ideal ligand valency all remain to be addressed. Interestingly, nanoparticles that use active targeting have not advanced beyond clinical trials; this may be due to various reasons such as added complexity of the formulations, limited targeting efficacy and possible off-target delivery217. For endogenous targeting, identifying which protein corona composition yields a desired organ-targeting outcome and relating it to the nanoparticle composition will be essential to fully capitalize on this targeting mechanism. Only recently has the importance of the protein corona been demonstrated for nanoparticles with distinct organ-targeting properties, suggesting the need for more mechanistic studies that clarify how specific corona proteins influence delivery in vivo97.
The inability to accurately predict the in vivo targeting and efficacy of nanoparticles from in vitro models necessitates the use of animal models for the discovery of new materials11. As such, nanoparticle discovery is a largely empirical process that requires a non-trivial number of animals to yield meaningful advances. Barcoding strategies represent one approach to increase the throughput of animal-based screening of nanoparticles52. Alternatively, the refinement of organ-on-a-chip and organoid models, which better recapitulate the complexity of physiology, could help identify promising nanoparticle candidates before animal testing218. Furthermore, it is possible that certain delivery outcomes demonstrated in murine models, which have been the emphasis of most studies, may not translate in higher order species. Given the high cost of non-human primate models and human clinical trials, developing models that accurately predict species-to-species translation of nanoparticle organ-targeting properties can increase the probability of clinical success219. Ultimately, combining both insights in the materials chemistry of nanoparticle systems and the biological processes of organ-targeted drug delivery, engineers and scientists can provide solutions to unmet clinical needs that deliver the full promises of nanoparticle technologies to patients.
Acknowledgements
D.J.S. acknowledges support from the National Institutes of Health (NIH) (R01 EB025192-01A1, R01 CA269787-01), the Cystic Fibrosis Foundation (SIEGWA21XX0) and the Cancer Prevention and Research Institute of Texas (RP190251). S.A.D. acknowledges financial support from the NIH Pharmacological Sciences Training Grant (GM007062) and the NIH Molecular Medicine Training Grant (GM109776).
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Materials thanks Michael Mitchell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
D.J.S.: ReCode Therapeutics, co-founder, consultant and Scientific Advisory Board; Tome Biosciences, Scientific Advisory Board. D.J.S., S.A.D. and the Regents of the University of Texas System have filed patent applications related to delivery technologies.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yin H, et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 2.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anselmo AC, Mitragotri S. An overview of clinical and commercial impact of drug delivery systems. J. Control. Rel. 2014;190:15–28. doi: 10.1016/j.jconrel.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenton OS, Olafson KN, Pillai PS, Mitchell MJ, Langer R. Advances in biomaterials for drug delivery. Adv. Mater. 2018 doi: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Large DE, Abdelmessih RG, Fink EA, Auguste DT. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021;176:113851. doi: 10.1016/j.addr.2021.113851. [DOI] [PubMed] [Google Scholar]
- 8.Cheng CJ, Tietjen GT, Saucier-Sawyer JK, Saltzman WM. A holistic approach to targeting disease with polymeric nanoparticles. Nat. Rev. Drug Discov. 2015;14:239–247. doi: 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croissant JG, Fatieiev Y, Almalik A, Khashab NM. Mesoporous silica and organosilica nanoparticles: physical chemistry, biosafety, delivery strategies, and biomedical applications. Adv. Healthc. Mater. 2018;7:1700831. doi: 10.1002/adhm.201700831. [DOI] [PubMed] [Google Scholar]
- 10.Jeong H-H, Choi E, Ellis E, Lee T-C. Recent advances in gold nanoparticles for biomedical applications: from hybrid structures to multi-functionality. J. Mater. Chem. B. 2019;7:3480–3496. doi: 10.1039/C9TB00557A. [DOI] [Google Scholar]
- 11.Paunovska K, et al. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 2018;18:2148–2157. doi: 10.1021/acs.nanolett.8b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardi N, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Rel. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anselmo AC, Mitragotri S. Nanoparticles in the clinic: an update. Bioeng. Transl. Med. 2019;4:e10143. doi: 10.1002/btm2.10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim. Biophys. Acta. 2011;1811:637–647. doi: 10.1016/j.bbalip.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Sun C, Wang C, Jankovic KE, Dong Y. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 2021;121:12181–12277. doi: 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilcock CP. Lipid polymorphism. Chem. Phys. Lipids. 1986;40:109–125. doi: 10.1016/0009-3084(86)90066-6. [DOI] [PubMed] [Google Scholar]
- 17.Barenholz Y. Doxil® — the first FDA-approved nano-drug: lessons learned. J. Control. Rel. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 19.Crommelin DJA, van Hoogevest P, Storm G. The role of liposomes in clinical nanomedicine development. What now? Now what? J. Control. Rel. 2020;318:256–263. doi: 10.1016/j.jconrel.2019.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Venditto VJ, Szoka FC. Cancer nanomedicines: so many papers and so few drugs! Adv. Drug Deliv. Rev. 2013;65:80–88. doi: 10.1016/j.addr.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda H. Toward a full understanding of the EPR effect in primary and metastatic tumors as well as issues related to its heterogeneity. Adv. Drug Deliv. Rev. 2015;91:3–6. doi: 10.1016/j.addr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1:16014. doi: 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- 23.Eygeris Y, Patel S, Jozic A, Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samaridou E, Heyes J, Lutwyche P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv. Drug Deliv. Rev. 2020;154-155:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Conway A, et al. Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabnis S, et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 28.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajj KA, et al. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15:e1805097. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 30.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci. USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead KA, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl Acad. Sci. USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenton OS, et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou K, et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl Acad. Sci. USA. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Q, et al. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 2018;30:e1805308. doi: 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- 36.Zhou K, et al. Hydrophobic domain structure of linear-dendritic poly(ethylene glycol) lipids affects RNA delivery of lipid nanoparticles. Mol. Pharm. 2020;17:1575–1585. doi: 10.1021/acs.molpharmaceut.9b01288. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019;31:e1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, et al. Efficient delivery of antisense oligonucleotides using bioreducible lipid nanoparticles in vitro and in vivo. Mol. Ther. Nucleic Acids. 2020;19:1357–1367. doi: 10.1016/j.omtn.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu M, et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl Acad. Sci. USA. 2021;118:e2020401118. doi: 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JB, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, et al. Lipid-modified aminoglycosides for mRNA delivery to the liver. Adv. Healthc. Mater. 2020;9:e1901487. doi: 10.1002/adhm.201901487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Lipid-modified aminoglycoside derivatives for in vivo siRNA delivery. Adv. Mater. 2013;25:4641–4645. doi: 10.1002/adma.201301917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hassett KJ, et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buschmann MD, et al. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillmore JD, et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 49.Finn JD, et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Kauffman KJ, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 51.Yaari Z, et al. Theranostic barcoded nanoparticles for personalized cancer medicine. Nat. Commun. 2016;7:13325. doi: 10.1038/ncomms13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dahlman JE, et al. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl Acad. Sci. USA. 2017;114:2060–2065. doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guimaraes PPG, et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Rel. 2019;316:404–417. doi: 10.1016/j.jconrel.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sago CD, et al. Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J. Am. Chem. Soc. 2018;140:17095–17105. doi: 10.1021/jacs.8b08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang D, Ramsey JD, Kabanov AV. Polymeric micelles for the delivery of poorly soluble drugs: from nanoformulation to clinical approval. Adv. Drug Deliv. Rev. 2020;156:80–118. doi: 10.1016/j.addr.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hrkach J, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci. Transl. Med. 2012;4:128ra139. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 57.Ashton S, et al. Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci. Transl. Med. 2016;8:325ra317. doi: 10.1126/scitranslmed.aad2355. [DOI] [PubMed] [Google Scholar]
- 58.Kalra AV, et al. Preclinical activity of nanoliposomal irinotecan is governed by tumor deposition and intratumor prodrug conversion. Cancer Res. 2014;74:7003. doi: 10.1158/0008-5472.CAN-14-0572. [DOI] [PubMed] [Google Scholar]
- 59.Hall A, Lächelt U, Bartek J, Wagner E, Moghimi SM. Polyplex evolution: understanding biology, optimizing performance. Mol. Ther. 2017;25:1476–1490. doi: 10.1016/j.ymthe.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowalski PS, et al. Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv. Mater. 2018 doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, et al. Zwitterionic phospholipidation of cationic polymers facilitates systemic mRNA delivery to spleen and lymph nodes. J. Am. Chem. Soc. 2021;143:21321–21330. doi: 10.1021/jacs.1c09822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santos R, et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blanco M-J, Gardinier KM. New chemical modalities and strategic thinking in early drug discovery. ACS Med. Chem. Lett. 2020;11:228–231. doi: 10.1021/acsmedchemlett.9b00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 65.Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 67.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 68.Dunbar CE, et al. Gene therapy comes of age. Science. 2018;359:eaan4672. doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 69.Rybakova Y, et al. mRNA delivery for therapeutic anti-HER2 antibody expression in vivo. Mol. Ther. 2019;27:1415–1423. doi: 10.1016/j.ymthe.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stadler CR, et al. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 71.Karikó K, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong Y, Siegwart DJ, Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019;144:133–147. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams D, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 74.Sahay G, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wittrup A, et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilleron J, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 77.Patel S, et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karikó K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Dev. 2007;10:523–532. [PubMed] [Google Scholar]
- 79.Leung AKK, Tam YYC, Chen S, Hafez IM, Cullis PR. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 80.Damase TR, et al. The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kulkarni JA, Witzigmann D, Chen S, Cullis PR, van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 82.Bertrand N, Leroux J-C. The journey of a drug-carrier in the body: an anatomo-physiological perspective. J. Control. Rel. 2012;161:152–163. doi: 10.1016/j.jconrel.2011.09.098. [DOI] [PubMed] [Google Scholar]
- 83.McNeil SE. Evaluation of nanomedicines: stick to the basics. Nat. Rev. Mater. 2016;1:16073. doi: 10.1038/natrevmats.2016.73. [DOI] [Google Scholar]
- 84.Lammers T, et al. Cancer nanomedicine: is targeting our target? Nat. Rev. Mater. 2016;1:16069. doi: 10.1038/natrevmats.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doudna JA. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tenzer S, et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 87.Walkey CD, et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 88.Jain P, et al. In-vitro in-vivo correlation (IVIVC) in nanomedicine: is protein corona the missing link? Biotechnol. Adv. 2017;35:889–904. doi: 10.1016/j.biotechadv.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 89.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 90.Mirshafiee V, Mahmoudi M, Lou K, Cheng J, Kraft ML. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013;49:2557–2559. doi: 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salvati A, et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 92.Cedervall T, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl Acad. Sci. USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen D, Ganesh S, Wang W, Amiji M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale. 2019;11:8760–8775. doi: 10.1039/C8NR09855G. [DOI] [PubMed] [Google Scholar]
- 94.Ban Z, et al. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc. Natl Acad. Sci. USA. 2020;117:10492–10499. doi: 10.1073/pnas.1919755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 96.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dilliard SA, Cheng Q, Siegwart DJ. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl Acad. Sci. USA. 2021;118:e2109256118. doi: 10.1073/pnas.2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mui BL, et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fleischer CC, Payne CK. Nanoparticle–cell interactions: molecular structure of the protein corona and cellular outcomes. Acc. Chem. Res. 2014;47:2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kelly PM, et al. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat. Nanotechnol. 2015;10:472–479. doi: 10.1038/nnano.2015.47. [DOI] [PubMed] [Google Scholar]
- 101.Li Z, et al. Emerging well-tailored nanoparticulate delivery system based on in situ regulation of the protein corona. J. Control. Rel. 2020;320:1–18. doi: 10.1016/j.jconrel.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 102.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/C1CS15233E. [DOI] [PubMed] [Google Scholar]
- 103.Tietjen GT, Bracaglia LG, Saltzman WM, Pober JS. Focus on fundamentals: achieving effective nanoparticle targeting. Trends Mol. Med. 2018;24:598–606. doi: 10.1016/j.molmed.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Poon W, et al. Elimination pathways of nanoparticles. ACS Nano. 2019;13:5785–5798. doi: 10.1021/acsnano.9b01383. [DOI] [PubMed] [Google Scholar]
- 105.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat. Rev. Immunol. 2011;11:788–798. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 106.Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bertrand N, et al. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 2017;8:777. doi: 10.1038/s41467-017-00600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ishida T, Wang X, Shimizu T, Nawata K, Kiwada H. PEGylated liposomes elicit an anti-PEG IgM response in a T cell-independent manner. J. Control. Rel. 2007;122:349–355. doi: 10.1016/j.jconrel.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 109.Ishida T, et al. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control. Rel. 2006;112:15–25. doi: 10.1016/j.jconrel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 110.Abu Lila AS, Kiwada H, Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J. Control. Rel. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 111.Jiang SY, Cao ZQ. Ultralow-fouling, functionalizable, and hydrolyzable zwitterionic materials and their derivatives for biological applications. Adv. Mater. 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- 112.Nogueira SS, et al. Polysarcosine-functionalized lipid nanoparticles for therapeutic mRNA delivery. ACS Appl. Nano Mater. 2020;3:10634–10645. doi: 10.1021/acsanm.0c01834. [DOI] [Google Scholar]
- 113.Bauer KN, Simon J, Mailänder V, Landfester K, Wurm FR. Polyphosphoester surfactants as general stealth coatings for polymeric nanocarriers. Acta Biomater. 2020;116:318–328. doi: 10.1016/j.actbio.2020.09.016. [DOI] [PubMed] [Google Scholar]
- 114.Viegas TX, et al. Polyoxazoline: chemistry, properties, and applications in drug delivery. Bioconjug. Chem. 2011;22:976–986. doi: 10.1021/bc200049d. [DOI] [PubMed] [Google Scholar]
- 115.Barz M, Luxenhofer R, Zentel R, Vicent MJ. Overcoming the PEG-addiction: well-defined alternatives to PEG, from structure–property relationships to better defined therapeutics. Polym. Chem. 2011;2:1900–1918. doi: 10.1039/c0py00406e. [DOI] [Google Scholar]
- 116.Gheibi Hayat SM, Bianconi V, Pirro M, Sahebkar A. Stealth functionalization of biomaterials and nanoparticles by CD47 mimicry. Int. J. Pharm. 2019;569:118628. doi: 10.1016/j.ijpharm.2019.118628. [DOI] [PubMed] [Google Scholar]
- 117.Hu C-MJ, et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature. 2015;526:118–121. doi: 10.1038/nature15373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu C-MJ, et al. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl Acad. Sci. USA. 2011;108:10980. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rao L, et al. Red blood cell membrane as a biomimetic nanocoating for prolonged circulation time and reduced accelerated blood clearance. Small. 2015;11:6225–6236. doi: 10.1002/smll.201502388. [DOI] [PubMed] [Google Scholar]
- 120.Parodi A, et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 2013;8:61–68. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Molinaro R, et al. Biomimetic proteolipid vesicles for targeting inflamed tissues. Nat. Mater. 2016;15:1037–1046. doi: 10.1038/nmat4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corbo C, et al. Unveiling the in vivo protein corona of circulating leukocyte-like carriers. ACS Nano. 2017;11:3262–3273. doi: 10.1021/acsnano.7b00376. [DOI] [PubMed] [Google Scholar]
- 123.Proffitt Richard T, et al. Liposomal blockade of the reticuloendothelial system: improved tumor imaging with small unilamellar vesicles. Science. 1983;220:502–505. doi: 10.1126/science.6836294. [DOI] [PubMed] [Google Scholar]
- 124.Germain M, et al. Priming the body to receive the therapeutic agent to redefine treatment benefit/risk profile. Sci. Rep. 2018;8:4797. doi: 10.1038/s41598-018-23140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saunders NRM, et al. A nanoprimer to improve the systemic delivery of siRNA and mRNA. Nano Lett. 2020;20:4264–4269. doi: 10.1021/acs.nanolett.0c00752. [DOI] [PubMed] [Google Scholar]
- 126.Ouyang B, et al. The dose threshold for nanoparticle tumour delivery. Nat. Mater. 2020;19:1362–1371. doi: 10.1038/s41563-020-0755-z. [DOI] [PubMed] [Google Scholar]
- 127.Choi HS, et al. Renal clearance of quantum dots. Nat. Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang YN, Poon W, Tavares AJ, McGilvray ID, Chan WCW. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J. Control. Rel. 2016;240:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 129.Aird WC. Endothelial cell heterogeneity. CSH Perspect. Med. 2012;2:a006429. doi: 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jambusaria A, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Life. 2020;9:e51413. doi: 10.7554/eLife.51413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sukriti S, Tauseef M, Yazbeck P, Mehta D. Mechanisms regulating endothelial permeability. Pulm. Circ. 2014;4:535–551. doi: 10.1086/677356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sarin H. Physiologic upper limits of pore size of different blood capillary types and another perspective on the dual pore theory of microvascular permeability. J. Angiogenes. Res. 2010;2:14. doi: 10.1186/2040-2384-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wisse E, Jacobs F, Topal B, Frederik P, De Geest B. The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther. 2008;15:1193–1199. doi: 10.1038/gt.2008.60. [DOI] [PubMed] [Google Scholar]
- 134.Sindhwani S, et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020;19:566–575. doi: 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- 135.Bendayan M. Morphological and cytochemical aspects of capillary permeability. Microsc. Res. Tech. 2002;57:327–349. doi: 10.1002/jemt.10088. [DOI] [PubMed] [Google Scholar]
- 136.Bien-Ly N, et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J. Exp. Med. 2014;211:233–244. doi: 10.1084/jem.20131660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mommaas-Kienhuis AM, Krijbolder LH, Van Hinsbergh VW, Daems WT, Vermeer BJ. Visualization of binding and receptor-mediated uptake of low density lipoproteins by human endothelial cells. Eur. J. Cell Biol. 1985;36:201–208. [PubMed] [Google Scholar]
- 138.Vogel SM, Minshall RD, Pilipovic M, Tiruppathi C, Malik AB. Albumin uptake and transcytosis in endothelial cells in vivo induced by albumin-binding protein. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281:L1512–L1522. doi: 10.1152/ajplung.2001.281.6.L1512. [DOI] [PubMed] [Google Scholar]
- 139.Rajendran P, et al. The vascular endothelium and human diseases. Int. J. Biol. Sci. 2013;9:1057–1069. doi: 10.7150/ijbs.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Behzadi S, et al. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 2017;46:4218–4244. doi: 10.1039/C6CS00636A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Smith SA, Selby LI, Johnston APR, Such GK. The endosomal escape of nanoparticles: toward more efficient cellular delivery. Bioconjug. Chem. 2019;30:263–272. doi: 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- 143.Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 144.Bus T, Traeger A, Schubert US. The great escape: how cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B. 2018;6:6904–6918. doi: 10.1039/C8TB00967H. [DOI] [PubMed] [Google Scholar]
- 145.Trefts E, Gannon M, Wasserman DH. The liver. Curr. Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ruiz de Galarreta M, Lujambio A. Therapeutic editing of hepatocyte genome in vivo. J. Hepatol. 2017;67:818–828. doi: 10.1016/j.jhep.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 147.DeRosa F, et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ramaswamy S, et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen CY, et al. Treatment of hemophilia A using factor VIII messenger RNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2020;20:534–544. doi: 10.1016/j.omtn.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.An D, et al. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 2017;21:3548–3558. doi: 10.1016/j.celrep.2017.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jiang L, et al. Systemic messenger RNA as an etiological treatment for acute intermittent porphyria. Nat. Med. 2018;24:1899–1909. doi: 10.1038/s41591-018-0199-z. [DOI] [PubMed] [Google Scholar]
- 152.Prieve MG, et al. Targeted mRNA therapy for ornithine transcarbamylase deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cao J, et al. mRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol. Ther. 2019;27:1242–1251. doi: 10.1016/j.ymthe.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Truong B, et al. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl Acad. Sci. USA. 2019;116:21150–21159. doi: 10.1073/pnas.1906182116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Balakrishnan B, et al. Novel mRNA-based therapy reduces toxic galactose metabolites and overcomes galactose sensitivity in a mouse model of classic galactosemia. Mol. Ther. 2020;28:304–312. doi: 10.1016/j.ymthe.2019.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang L, et al. Dual mRNA therapy restores metabolic function in long-term studies in mice with propionic acidemia. Nat. Commun. 2020;11:5339. doi: 10.1038/s41467-020-19156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schuh RS, et al. In vivo genome editing of mucopolysaccharidosis I mice using the CRISPR/Cas9 system. J. Control. Rel. 2018;288:23–33. doi: 10.1016/j.jconrel.2018.08.031. [DOI] [PubMed] [Google Scholar]
- 158.Kose N, et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019;4:eaaw6647. doi: 10.1126/sciimmunol.aaw6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miao L, et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sato Y, Kinami Y, Hashiba K, Harashima H. Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J. Control. Rel. 2020;322:217–226. doi: 10.1016/j.jconrel.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 162.Kasiewicz LN, et al. Lipid nanoparticles incorporating a GalNAc ligand enable in vivo liver ANGPTL3 editing in wild-type and somatic LDLR knockout non-human primates. bioRxiv. 2021 doi: 10.1101/2021.11.08.467731. [DOI] [Google Scholar]
- 163.Sato Y, et al. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J. Control. Rel. 2017;266:216–225. doi: 10.1016/j.jconrel.2017.09.044. [DOI] [PubMed] [Google Scholar]
- 164.Wohlleber D, Knolle PA. The role of liver sinusoidal cells in local hepatic immune surveillance. Clin. Transl. Immunol. 2016;5:e117. doi: 10.1038/cti.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim M, et al. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv. 2021;7:eabf4398. doi: 10.1126/sciadv.abf4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sato Y, Hatakeyama H, Hyodo M, Harashima H. Relationship between the physicochemical properties of lipid nanoparticles and the quality of siRNA delivery to liver cells. Mol. Ther. 2016;24:788–795. doi: 10.1038/mt.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shobaki N, Sato Y, Harashima H. Mixing lipids to manipulate the ionization status of lipid nanoparticles for specific tissue targeting. Int. J. Nanomed. 2018;13:8395–8410. doi: 10.2147/IJN.S188016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Paunovska K, et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater. 2019;31:1807748. doi: 10.1002/adma.201807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang CY, Yuan WG, He P, Lei JH, Wang CX. Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J. Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Senoo H, et al. Hepatic stellate cell (vitamin A-storing cell) and its relative — past, present and future. Cell Biol. Int. 2010;34:1247–1272. doi: 10.1042/CBI20100321. [DOI] [PubMed] [Google Scholar]
- 171.Sato Y, et al. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat. Biotechnol. 2008;26:431–442. doi: 10.1038/nbt1396. [DOI] [PubMed] [Google Scholar]
- 172.Zhang Z, et al. Corona-directed nucleic acid delivery into hepatic stellate cells for liver fibrosis therapy. ACS Nano. 2015;9:2405–2419. doi: 10.1021/nn505166x. [DOI] [PubMed] [Google Scholar]
- 173.Cheng Q, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ. Systemic nanoparticle delivery of CRISPR–Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020;11:3232. doi: 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miller JB, Kos P, Tieu V, Zhou K, Siegwart DJ. Development of cationic quaternary ammonium sulfonamide amino lipids for nucleic acid delivery. ACS Appl. Mater. Interfaces. 2018;10:2302–2311. doi: 10.1021/acsami.7b15982. [DOI] [PubMed] [Google Scholar]
- 176.Qiu M, et al. Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl Acad. Sci. USA. 2022;119:e2116271119. doi: 10.1073/pnas.2116271119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc. Natl Acad. Sci. USA. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xue W, et al. Small RNA combination therapy for lung cancer. Proc. Natl Acad. Sci. USA. 2014;111:E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Khan OF, et al. Endothelial siRNA delivery in nonhuman primates using ionizable low-molecular weight polymeric nanoparticles. Sci. Adv. 2018;4:eaar8409. doi: 10.1126/sciadv.aar8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Krohn-Grimberghe M, et al. Nanoparticle-encapsulated siRNAs for gene silencing in the haematopoietic stem-cell niche. Nat. Biomed. Eng. 2020;4:1076–1089. doi: 10.1038/s41551-020-00623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kaczmarek JC, et al. Polymer-lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew. Chem. Int. Ed. 2016;55:13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaczmarek JC, et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 2018;18:6449–6454. doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kaczmarek JC, et al. Systemic delivery of mRNA and DNA to the lung using polymer-lipid nanoparticles. Biomaterials. 2021;275:120966. doi: 10.1016/j.biomaterials.2021.120966. [DOI] [PubMed] [Google Scholar]
- 184.Yan Y, et al. Functional polyesters enable selective siRNA delivery to lung cancer over matched normal cells. Proc. Natl Acad. Sci. USA. 2016;113:E5702. doi: 10.1073/pnas.1606886113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yan Y, Xiong H, Zhang X, Cheng Q, Siegwart DJ. Systemic mRNA delivery to the lungs by functional polyester-based carriers. Biomacromolecules. 2017;18:4307–4315. doi: 10.1021/acs.biomac.7b01356. [DOI] [PubMed] [Google Scholar]
- 186.Kusumoto K, et al. Lipid envelope-type nanoparticle incorporating a multifunctional peptide for systemic siRNA delivery to the pulmonary endothelium. ACS Nano. 2013;7:7534–7541. doi: 10.1021/nn401317t. [DOI] [PubMed] [Google Scholar]
- 187.Hagino Y, Khalil IA, Kimura S, Kusumoto K, Harashima H. GALA-modified lipid nanoparticles for the targeted delivery of plasmid DNA to the lungs. Mol. Pharm. 2021;18:878–888. doi: 10.1021/acs.molpharmaceut.0c00854. [DOI] [PubMed] [Google Scholar]
- 188.Parhiz H, et al. PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake. J. Control. Rel. 2018;291:106–115. doi: 10.1016/j.jconrel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li Q, et al. Engineering caveolae-targeted lipid nanoparticles to deliver mRNA to the lungs. ACS Chem. Biol. 2020;15:830–836. doi: 10.1021/acschembio.0c00003. [DOI] [PubMed] [Google Scholar]
- 190.Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J. Immunol. 2009;183:2205. doi: 10.4049/jimmunol.0804324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fenton OS, et al. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 2017 doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 192.Fenton OS, et al. Customizable lipid nanoparticle materials for the delivery of siRNAs and mRNAs. Angew. Chem. Int. Ed. 2018;57:13582–13586. doi: 10.1002/anie.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lokugamage MP, Sago CD, Gan Z, Krupczak BR, Dahlman JE. Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv. Mater. 2019;31:e1902251. doi: 10.1002/adma.201902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao X, et al. Imidazole-based synthetic lipidoids for in vivo mRNA delivery into primary T lymphocytes. Angew. Chem. Int. Ed. 2020;59:20083–20089. doi: 10.1002/anie.202008082. [DOI] [PubMed] [Google Scholar]
- 195.Ramishetti S, et al. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv. Mater. 2020;32:e1906128. doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 196.Kimura S, Khalil IA, Elewa YHA, Harashima H. Novel lipid combination for delivery of plasmid DNA to immune cells in the spleen. J. Control. Release. 2021;330:753–764. doi: 10.1016/j.jconrel.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 197.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 198.Sahin U, et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 199.Reinhard K, et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science. 2020;367:446–453. doi: 10.1126/science.aay5967. [DOI] [PubMed] [Google Scholar]
- 200.Haanen JBAG, et al. BNT211: a phase I trial to evaluate safety and efficacy of CLDN6 CAR-T cells and CARVac-mediated in vivo expansion in patients with CLDN6-positive advanced solid tumors. Cancer Res. 2022;82:CT002. doi: 10.1158/1538-7445.AM2022-CT002. [DOI] [Google Scholar]
- 201.Jiang Y, et al. A ‘top-down’ approach to actuate poly(amine-co-ester) terpolymers for potent and safe mRNA delivery. Biomaterials. 2018;176:122–130. doi: 10.1016/j.biomaterials.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Grun MK, et al. PEGylation of poly(amine-co-ester) polyplexes for tunable gene delivery. Biomaterials. 2021;272:120780. doi: 10.1016/j.biomaterials.2021.120780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jiang Y, et al. Quantitating endosomal escape of a library of polymers for mRNA delivery. Nano Lett. 2020;20:1117–1123. doi: 10.1021/acs.nanolett.9b04426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.McKinlay CJ, Benner NL, Haabeth OA, Waymouth RM, Wender PA. Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters. Proc. Natl Acad. Sci. USA. 2018;115:E5859–E5866. doi: 10.1073/pnas.1805358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.McKinlay CJ, et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc. Natl Acad. Sci. USA. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Benner NL, et al. Oligo(serine ester) charge-altering releasable transporters: organocatalytic ring-opening polymerization and their use for in vitro and in vivo mRNA delivery. J. Am. Chem. Soc. 2019;141:8416–8421. doi: 10.1021/jacs.9b03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Parayath NN, Stephan MT. In situ programming of CAR T cells. Annu. Rev. Biomed. Eng. 2021;23:385–405. doi: 10.1146/annurev-bioeng-070620-033348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Smith TT, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 2017;12:813–820. doi: 10.1038/nnano.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Parayath NN, Stephan SB, Koehne AL, Nelson PS, Stephan MT. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 2020;11:6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tombácz I, et al. Highly efficient CD4+ T cell targeting and genetic recombination using engineered CD4+ cell-homing mRNA-LNPs. Mol. Ther. 2021;29:3293–3304. doi: 10.1016/j.ymthe.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rurik Joel G, et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375:91–96. doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Veiga N, et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Veiga N, et al. Leukocyte-specific siRNA delivery revealing IRF8 as a potential anti-inflammatory target. J. Control. Rel. 2019;313:33–41. doi: 10.1016/j.jconrel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 214.Kampel L, et al. Therapeutic inhibitory RNA in head and neck cancer via functional targeted lipid nanoparticles. J. Control. Rel. 2021;337:378–389. doi: 10.1016/j.jconrel.2021.07.034. [DOI] [PubMed] [Google Scholar]
- 215.Singh MS, et al. Therapeutic gene silencing using targeted lipid nanoparticles in metastatic ovarian cancer. Small. 2021;17:2100287. doi: 10.1002/smll.202100287. [DOI] [PubMed] [Google Scholar]
- 216.Akinc A, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 217.Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bhise NS, et al. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Rel. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hatit MZC, et al. Species-dependent in vivo mRNA delivery and cellular responses to nanoparticles. Nat. Nanotechnol. 2022;17:310–318. doi: 10.1038/s41565-021-01030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Adams DH, Eksteen B. Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat. Rev. Immunol. 2006;6:244–251. doi: 10.1038/nri1784. [DOI] [PubMed] [Google Scholar]
- 221.Sun W, Hu Q, Ji W, Wright G, Gu Z. Leveraging physiology for precision drug delivery. Physiol. Rev. 2016;97:189–225. doi: 10.1152/physrev.00015.2016. [DOI] [Google Scholar]
- 222.Li J, Kataoka K. Chemo-physical strategies to advance the in vivo functionality of targeted nanomedicine: the next generation. J. Am. Chem. Soc. 2020;143:538–559. doi: 10.1021/jacs.0c09029. [DOI] [PubMed] [Google Scholar]
- 223.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Farbiak L, et al. All-in-one dendrimer-based lipid nanoparticles enable precise HDR-mediated gene editing in vivo. Adv. Mater. 2021;33:2006619. doi: 10.1002/adma.202006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Musunuru K, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 226.Anzalone AV, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]