Abstract
Ferritin-based, self-assembling protein nanoparticle vaccines are being developed against a range of viral pathogens, including SARS-CoV-2, influenza, HIV-1, and Epstein-Barr virus. However, purification of these nanoparticles is often laborious and requires customization for each potential nanoparticle vaccine. We propose that the simple insertion of a polyhistidine tag into exposed flexible loops on the ferritin surface (His-Fer) can mitigate the need for complex purifications and enable facile metal-chelate-based purification, thereby allowing for optimization of early stage vaccine candidates. Using sequence homology and computational modeling, we identify four sites that can accommodate insertion of a polyhistidine tag and demonstrate purification of both hemagglutinin-modified and SARS-CoV-2 spike-modified ferritins, highlighting the generality of the approach. A site at the 4-fold axis of symmetry enables optimal purification of both protein nanoparticles. We demonstrate improved purification through modulating the polyhistidine length and optimizing both the metal cation and the resin type. Finally, we show that purified His-Fer proteins remain multimeric and elicit robust immune responses similar to those of their wild-type counterparts. Collectively, this work provides a simplified purification scheme for ferritin-based vaccines.
Subunit vaccines, which often utilize only a single protein component of a pathogen, are typically weakly immunogenic.1 Nanoparticle-based multimerization is a common way to increase their immunogenicity.2−4Helicobacter pylori ferritin is frequently used as a self-assembling protein nanoparticle for vaccine development.5−8 The ferritin nanoparticle is an iron-binding protein complex composed of 24 identical protomers.5 For vaccine design, the 3-fold axis of symmetry5 at the N-terminus of ferritin can be functionalized with trimeric viral glycoproteins, such as influenza hemagglutinin. Such fusions display eight copies of the trimeric glycoprotein on the nanoparticle surface. Ferritin nanoparticle vaccines have been applied widely, including for respiratory syncytial virus (RSV),9 HIV-1,6,10,11 influenza,12,13 SARS-CoV-2,15−18 and other viruses.12,19,20 A number of these ferritin nanoparticle vaccines have shown promise and are being introduced into the clinic20,21 (e.g., influenza trials NCT03186781, NCT03814720, and NCT04579250 and Epstein-Barr virus trial NCT04645147).
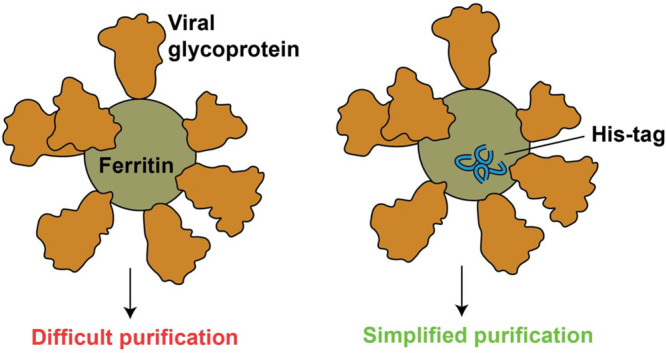
A major limitation, however, to the development of both early and late stage vaccine candidates based on ferritin nanoparticles is the difficulty in purifying them efficiently.5 Purification is often conducted using lectin-affinity chromatography or ion-exchange chromatography10 followed by size-exclusion chromatography and requires optimization for each individual ferritin fusion.5 While effective, these methods are not as simple or generalizable as metal-affinity chromatography. A polyhistidine tag (His tag) has been successfully inserted at the N-terminus of the ferritin protomer14 and at the N-terminus of a functionalized antigen.22 However, this approach is not easily translated to other antigens, as not all N-termini of fusion proteins are amenable to modification with a His tag.23 Moreover, the ferritin C-terminus is not exposed, so it cannot accommodate a His tag for affinity purification.14
Collectively, this demonstrates the need to develop a simple, universal purification scheme for ferritin-based vaccines. Here, we demonstrate that installing His tags into surface-exposed, flexible loops of H. pylori ferritin facilitates metal-chelate-based purification24 of viral-glycoprotein-modified ferritin nanoparticles and that these His tag-installed versions elicit robust immune responses similar to those of the non-installed counterparts. By allowing rapid purification of functionalized ferritin nanoparticles, this method will facilitate development of easily purifiable and scalable vaccine candidates.
Experimental Procedures
Design of His-Ferritin Constructs
Analysis of conservation was performed for H. pylori ferritin (based on accession ID WP_000949190) compared to ferritin derived from other sources, and nonconserved sites were identified as candidate insertion positions. This analysis was done using the consurf server25−27 (HMMER, Uniref-90, 150 sequences with a range of the degree of homology between 95% and 35%). These variable sites were then mapped to the structure to identify which were surface-exposed and located within flexible regions using Rosetta.28 After the H. pylori ferritin monomer had been relaxed in Rosetta [Protein Data Bank (PDB) entry 3BVE], polyhistidine insertions of length 6 (6xHis) were made at each of the sequence-derived loop sites using RosettaRemodel. Also included were three flanking residues on either side of the His residues to facilitate a mobile backbone, and 10 structures were generated for each site. The mutated monomers were then multimerized and relaxed in Rosetta to gauge whether it was possible to resolve clashes at the interfaces. Insertions were considered viable if more than five structures had successful loop closure and all clashes were resolved. Four sites were selected for cloning and further experimental validation. Structures shown in Figure 1 are models of the top-scoring insertion sites.
Figure 1.
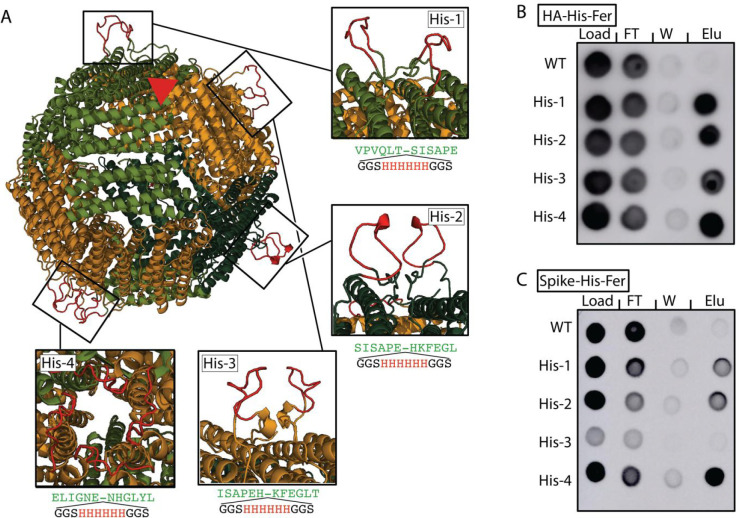
Installation of polyhistidine tags into surface-exposed loops on ferritin. (A) Rendering of the H. pylori ferritin nanoparticle (PDB entry 3EGM) with representative polyhistidine insertion sites in boxes. Two insertions are shown for His-1, His-2, and His-3, which fall along the 2-fold axis of symmetry, and four insertions are shown for His-4, which falls along the 4-fold axis of symmetry. Insertions were modeled using Rosetta. A single 3-fold axis of symmetry is shown (red triangle); the C-termini of HA or spike were installed along the 3-fold axis at residue 5 at the N-terminus of ferritin. Dot blots showing the purification of His-Fer proteins for either (B) HA or (C) spike. Columns show dots from the load, flow-through (FT), wash (W), or elution (Elu). The intensity of the dots on a dot blot, like a Western blot, is a surrogate for concentration. Elution was observed in all His-Fer proteins for HA, but there is a clear preference for His-4 in the case of spike-His-Fer.
Plasmids
All plasmids for mammalian cell expression were cloned into an in-house pADD2 mammalian expression vector using HiFi PCR (Takara) followed by In-Fusion (Takara) cloning with EcoRI/XhoI cut sites.18,39 The pADD2 plasmid contains a CMV promoter and an rBeta-globin intron prior the protein of interest and is flanked by a β-globin poly(A) sequence. Plasmid information can be found in Table S1.
Expression and Purification of His-Tagged Ferritin Constructs
All multimeric antigens for immunization were expressed and purified from Expi293F cells, cultured using 66% FreeStyle 293 expression medium (ThermoFisher) and 33% Expi293 Expression Medium (ThermoFisher), and grown in polycarbonate shaking flasks at 37 °C and 8% CO2. Cells were transfected at a density of approximately 3–4 × 106 cells/mL with 0.6 μg of maxi-prepped DNA (NucleoBond, Takara), and 1.3 μL of FectoPro (Polyplus) per 1 mL of culture medium. After incubation for 10 min, the resulting transfection cocktail was added to 75 mL of cells. Expi293F supernatants were collected 3–5 days post-transfection, harvested by being spun at 7500g for 5 min, and filtered through a 0.22 μm filter.
Expi293F cell culture supernatants were then diluted 1:1 with 10 mM imidazole and 1× PBS (pH 7.4), and 1 mL of vortexed Ni-NTA resin was added to the diluted supernatant and incubated overnight while rotating at 4 °C (HA) or room temperature (spike). All spike purifications for immunization were performed at room temperature with care, as refrigeration is known to alter the spike conformation.40 Nanoparticles were then added to plastic chromatography columns for gravity-flow purification, washed with 30 mL of 20 mM imidazole and 1× PBS, and eluted in 250 mM imidazole and 1× PBS. For immunizations, Ni-NTA elutions were concentrated using Amicon spin concentrators [100 kDa molecular weight cutoff (MWCO)] followed by size-exclusion chromatography on an AKTA Pure FPLC instrument with the SRT SEC-1000 column. Fractions were then pooled on the basis of the A280 signal. Samples for immunizations were supplemented with 10% glycerol, filtered through a 0.22 μm filter, snap frozen, and then stored at −20 °C until immunization.
Expression and Purification of Wild-Type (WT) Ferritin
WT proteins were expressed in the same way as the His-tagged proteins described above. The cell culture supernatant was spun, filtered, and then buffer-exchanged into 20 mM Tris (pH 8.0) overnight at 4 °C in 100 kDa MWCO dialysis tubing. Dialyzed samples were then filtered through a 0.22 μm filter and loaded onto a 5 mL HiTrapQ anion-exchange column (GE) that was equilibrated in 20 mM Tris (pH 8.0) on a GE AKTA Pure system. Antigen-ferritin proteins were then eluted using an NaCl gradient. Fractions that contained the protein of interest were identified by dot blot (below) using MEDI8852 or CR3022 as the primary detection antibody. The relevant fractions were then combined and concentrated using a 100 kDa MWCO Amicon spin filter and then purified on a GE AKTA Pure system using an SRT SEC-1000 SEC column that was equilibrated in PBS. Protein-containing fractions were identified by A280 as well as by dot blot and stored at −20 °C until immunization.
Dot Blot Analysis of Ni-Resin-Purified Antigens
Samples from each step of the Ni-resin gravity-flow purification were collected and analyzed by dot blotting, including the cell supernatant derived after filtration but prior to the addition of resin, the flow-through (FT) after the collection of resin, FT from the wash step with 20 mM imidazole, and the eluate (Elu) upon treatment with 250 mM imidazole. Then, 2 μL of each of these samples was dotted directly onto a nitrocellulose membrane, left to dry for 10–15 min, and then blocked with 5% milk/H2O for at least 1 h or as much as overnight. CR3022 (for SARS-CoV-2 spike) or MEDI8852 (for influenza HA) was added (final concentration of ∼5 μg/mL) and incubated at room temperature for 1 h. Blots were then washed nine times with 10 mL of PBST. Horseradish peroxidase (HRP) rabbit anti-human IgG (Abcam ab6759) was added at a 1:7500 dilution in 5% milk/H2O and incubated for 1 h, before being washed again nine times with 10 mL of PBST. Blots were developed using Pierce ECL Western blotting substrate, imaged using a GE Amersham imager 600, and presented using ImageJ version 1.51.
SEC-MALS of Ferritin-Antigen Nanoparticles
SEC-MALS was performed on an Agilent 1260 Infinity II HPLC instrument with Wyatt detectors for light scattering (miniDAWN) and refractive index (Optilab). The purified antigen (1–10 μg) was loaded onto a SRT SEC-1000 4.6 mm × 300 mm column and equilibrated in 1× PBS (pH 7.4). Columns were used at a rate of 0.35 mL/min, and molecular weights were determined using ASTRA version 7.3.2 (Wyatt Technologies).
Mouse Immunizations
Balb/c female mice (10–12 weeks old) were purchased from Jackson Laboratories. All mice were maintained at Stanford University according to the Public Health Service Policy for “Humane Care and Use of Laboratory Animals”. Mice were immunized with either WT-ferritin-antigen or His-ferritin-antigen via subcutaneous injection with a mixture of 50 μL of antigen (stored at 0.2 mg/mL in 10% glycerol PBS), 10 μg of QuilA (in a 1 μL volume), 10 μg of MPLA (in a 1 μL volume), and 48 μL of PBS. Mice were immunized on days 0 and 21 with the same antigen-adjuvant mixture, and serum was collected on days 0, 21, and 28. Immune responses were analyzed from serum samples collected on day 28.
Enzyme-Linked Immunosorbent Assay (ELISA) Using Mouse Serum
For the HA-ferritin immunizations, antibody titers were analyzed by an ELISA. WT-HA-ferritin was hydrophobically plated onto MaxiSorp 96-well plates (ThermoFisher) at 1 μg/mL in 1× PBS (pH 7.4) and incubated at 37 °C for 2 h. After being coated, plates were washed three times with PBST and blocked overnight with ChonBlock Blocking Buffer (Chondrex). ChonBlock was removed, and then the plates were washed three times with PBST before being plated with serial dilutions of mouse serum in 1× PBS with 1% BSA and 0.1% Tween, starting at 1:10 serum dilution. These were then incubated at room temperature for 90 min and washed three times again with PBST using an ELx 405 Bio-Tex plate washer. HRP goat anti-mouse IgG1 (BioLegend 405306) was added at a 1:7500 dilution in diluent buffer for 90 min at room temperature. After incubation with the secondary antibody, ELISA plates were washed six times with PBST, and plates were developed for 5 min with One-Step Turbo TMB substrate (Pierce) before being quenched with 2 M sulfuric acid. The absorbance at 450 nm (A450) was recorded using a BioTek plate reader, and the background signal was averaged across wells containing preimmunization mouse serum at a 1:100 dilution from all 10 mice and subtracted from all wells on the plate. Background-subtracted values were then imported into GraphPad Prism version 8.4.1 and fitted with three-parameter nonlinear regression to obtain the EC50 values reported.
Off-target activity against the His tag was assessed using an ELISA following the same procedure described above, but with sfGFP-His (as previously described)4 plated at 5 μg/mL, which indicated no significant difference between the control and His-ferritin immunized groups.
SARS-CoV-2 Pseudotyped Lentivirus Production
SARS-CoV-2 spike-pseudotyped lentivirus was produced via calcium phosphate transfection in HEK293T cells. Then, 6 × 106 cells were seeded in 10 cm plates 1 day prior to transfection in DMEM medium with 10% fetal bovine serum, l-glutamate, penicillin, streptomycin, and 10 mM HEPES (D10 medium). As previously reported, the five-plasmid system41 was used for viral production. The five plasmids are the packaging vector (pHAGE_Luc2_IRES_ZsGreen), the SARS-CoV-2 spike, and the helper plasmids (HDM-Hgpm2, HDM-Tat1b, and pRC-CMV_Rev1b). The spike vector contained the full-length wild-type spike sequence from the Wuhan-Hu-1 strain of SARS-CoV-2 (GenBank NC_045512). Ten micrograms of pHAGE_Luc2_IRS_ZsGreen, 3.4 μg of SARS-CoV-2 spike, 2.2 μg of HDM-Hgpm2, 2.2 μg of HDM-Tat1b, and 2.2 μg of pRC-CMV_Rev1b were used in a final volume of 500 μL in doubly distilled H2O. This was followed by dropwise addition of HEPES-buffered saline (2×, pH 7.0) to a final volume of 1 mL. Then, 100 μL of 2.5 M CaCl2 was added dropwise while the solution was gently agitated. These transfection reaction mixtures were incubated at room temperature for 20 min, followed by dropwise addition to plated cells. Medium was replaced ∼24 h post-transfection. Virus-containing culture supernatants were harvested ∼72 h post-transfection via centrifugation at 300g for 5 min and filtered through a 0.45 μm filter. Viral stocks were aliquoted and stored at −80 °C until use.
SARS-CoV-2 Viral Neutralization Assays
For viral neutralization assays, HeLa cells expressing the ACE2 protein42 were plated in 96-well, white-walled, clear-bottom plates at a density of 5000 cells/well 1 day prior to infection. Mouse serum was centrifuged at 2000g for 15 min, heat-inactivated for 30 min at 56 °C, and diluted in DMEM. Dilutions were made starting at an initial dilution of 102, and the final volume was 60 μL. Then, 60 μL of virus, diluted in DMEM and supplemented with Polybrene, was then added to inhibitor dilutions for a final volume of 120 μL. Polybrene was present at a final concentration of 5 μg/mL in diluted samples. Virus/serum dilutions were incubated for 1 h at 37 °C. After incubation, medium was removed from the plate containing the cells, and it was replaced with 100 μL of the incubating inhibitor/virus dilutions. The resultant mixture was incubated at 37 °C for ∼48 h. Infectivity was measured using the BriteLite assay readout solution (PerkinElmer), and luminescence values were measured with a BioTek plate reader; luminescence is inversely correlated with neutralization. Each plate was normalized by averaging RLUs from wells with cells only (0% infectivity) and virus only (100% infectivity). Normalized values were fitted with a three-parameter nonlinear regression inhibitor curve in GraphPad Prism 8.4.1 to obtain NT50 values. Fits for all serum neutralization assays were constrained to have a value of 0% at the bottom of the fit.
Results
To develop a generalizable strategy for purifying functionalized H. pylori ferritin nanoparticles, we aimed to install His tags into flexible loops on its surface. We identified ferritin residues that could accommodate the installation of His tags, using sequence homology25−27 and computational modeling.28 After identifying regions within ferritin with a high degree of sequence variability and backbone flexibility (Figure S1), we selected four positions (Figure S1) to install a six-His tag flanked on each side by a Gly-Gly-Ser flexible linker. In addition, computational modeling of ferritin nanoparticles revealed the solvent accessibility of the installed tag at these positions (Figure 1A).
We installed His tags at these four positions on ferritin displaying either the ectodomains of hemagglutinin (HA) or a C-terminally truncated SARS-CoV-2 spike glycoprotein (spike),18 and we refer to these polyhistidine-tagged ferritin molecules as “His-Fer”. We tested the efficiencies of expressing and purifying the HA (Figure 1B) and spike (Figure 1C) His-Fer complexes using dot blots,18,29 where the sample is spotted directly onto nitrocellulose and probed using antibodies against HA (MEDI8852) or spike (CR3022).
While we observed protein expression for all polyhistidine installation sites, spike-His-Fer tagged at position 3 (His-3) showed a reduced level of expression compared to that of the WT (Figure 1C). All installations facilitated Ni-NTA-based purification of the His-Fer proteins, showing selective enrichment in the eluate compared to the WT sample without a His tag (Figure 1B,C, right column). While the purifications for all installations were similar for the HA His-Fer complexes, installation at site 4 (His-4) at the 4-fold axis of symmetry, farthest from the viral glycoprotein, was ideal for spike His-Fer purification (Figure 1B,C). The probable explanation for the difference in purification efficiency among sites 1–3 compared to site 4 for HA and spike likely results from the differences in molecular weight between HA30 and spike.31 HA is substantially smaller than spike, ∼70 kDa for monomers versus ∼150 kDa, respectively, so the increased steric bulk of the larger spike glycoprotein may occlude the binding of Ni to His tags at the closer installation sites (1–3).
We had two main concerns about using an affinity resin to purify the glycoprotein-modified nanoparticles. First, the numerous copies of the tag on each individual nanoparticle may result in incomplete elution due to avidity. Conversely, there is potential for incomplete binding due to steric occlusion of the tag by the glycoprotein. Alternatively, a combination of both could occur; steric occlusion may mitigate the initial interaction with the resin, but with an increase in effective concentration following initial binding, additional tags may decrease the elution efficiency.
To test these predictions, we modulated the length of the His tag at position His-4 to four, five, six, seven, or eight histidine residues (His-4.4–His-4.8) (Figure 2A). As with the six His, we could efficiently purify HA-His-Fer constructs with four, five, seven, and eight histidines. However, we could not purify spike-His-Fer when the His tag contained either four or five histidine residues, yet His tags of six, seven, and eight residues could facilitate purification, though to varying degrees (Figure 2B,C). Despite the fact that the geometry of the His-4 position would place 16 or 20 histidine residues in the proximity for His-4.4 or His-4.5, respectively, it is clear that steric occlusion on the surface of the nanoparticle, specifically for spike, plays a potentially limiting role in His-Fer purification. The molecular weights of HA-His-Fer and spike-His-Fer were confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis following Ni-NTA purification (Figure 2D). Additionally, we did find that His-4.8 could accommodate more stringent imidazole washing conditions (Figure S2). As such, we used these proteins, termed HA-His-4.8-Fer and spike-His-4.8-Fer, for all future assays.
Figure 2.
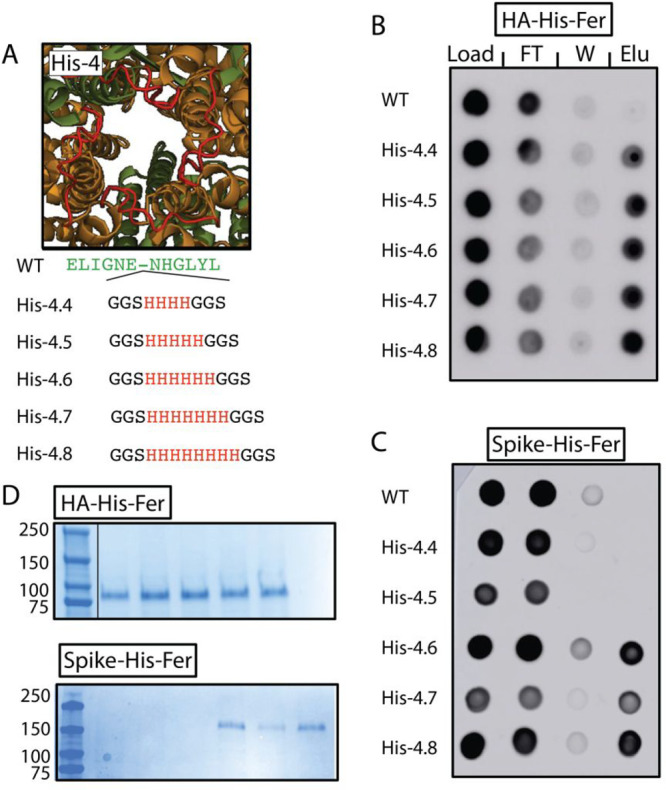
Polyhistidine length impacts purification of spike-His-Fer, but not HA-His-Fer. (A) Site 4, as in Figure 1, highlighting the length of the installed polyhistidine tag, ranging from four to eight histidine residues. The image in panel A shows installation of six histidine residues. Dot blots tracking the purification of His-4.4–His-4.8 proteins for either (B) HA or (C) spike. Columns show dots from the load, flow through (FT), wash (W), or elution (Elu). Expression was detected for all lengths of HA-His-Fer but only for greater than or equal to six histidine residues for spike-His-Fer. (D) SDS–PAGE analysis of the elution from the dot blots shown in panels B and C. Lanes in HA-His-Fer correspond to a molecular weight (MW), in kilodaltons, ladder (lane 1, labels on the left), His-4.4–His-4.8 (lanes 2–6, respectively), and the WT (lane 7). Lanes in spike-His-Fer correspond to the MW ladder (lane 1, labels on the left), the WT (lane 2), and His-4.4–His-4.8 (lanes 3–7, respectively). These results confirm the dot blot analysis and demonstrate that the proteins migrate at the expected MW.
Because of the difficulty in purifying spike-His-Fer with fewer histidines, we tested whether altering the metal ion in the affinity resin would improve the purification. We tested resins containing a range of divalent cations (Co2+, Zn2+, Ni2+, and Cu2+).32,33 In theory, Co2+ and Zn2+ should be weaker than Ni2+ while Cu2+ should bind more strongly. Consistent with these expectations, we found that our spike-His-Fer, both the six- and eight-His tags, purified best in the ascending order of Co2+, Zn2+, and Ni2+ (Figure S3). Surprisingly, however, we did not find any purification off of the Cu2+ resin, which suggested a lack of binding to this resin.
Next, we investigated if the source of the Ni2+ resin played a role in purification efficiency. We tested Ni2+ resins from a variety of vendors with either NTA or IDA coordination ligands. We found ThermoFisher His-Pur to be the optimal purification resin for the HA- and spike-His-4.8-Fer nanoparticles (Figure S4). Resins obtained from some vendors were entirely unable to purify the His-Fer proteins. As such, we utilized ThermoFisher His-Pur for all further purifications.
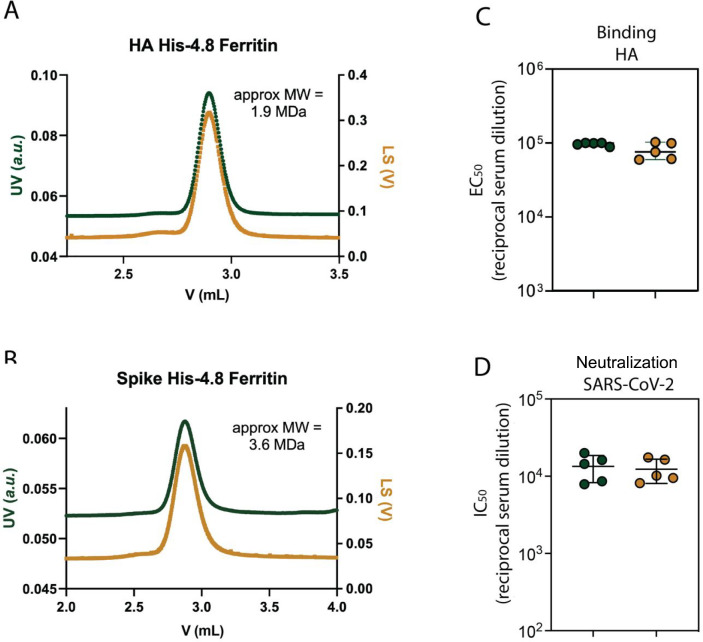
To ensure that installation of the His tag did not alter the multivalency of modified ferritin nanoparticles, we utilized size-exclusion chromatography with multiangle light scattering (SEC-MALS) to confirm nanoparticle size. Both nanoparticles were the expected molecular weight (1.9 MDa for HA-His-4.8-Fer and 3.6 MDa for spike-His-4.8-Fer) (Figure 3A,B), indicating that installation of the polyhistidine tag at the His-4 position did not impact the overall ability of the proteins to self-assemble into nanoparticles.
Figure 3.
His-Fer proteins remain multimeric and elicit immune responses comparable to those of their non-polyhistidine-tagged counterparts. SEC-MALS analysis of (A) HA-His-4.8-Fer and (B) spike-His-4.8-Fer showing a single, monodispersed peak with a good correspondence between UV absorbance (left axis) and light scattering (right axis), suggesting there is minimal aggregation or degradation. Both nanoparticles elute at the expected retention time for their molecular weights. The molecular weight (MW) was calculated using the Wyatt software. (C) ELISA binding EC50 against WT-HA-Fer for mice immunized with WT-HA-Fer (green, left) or HA-His-4.8-Fer (yellow, right). The binding results demonstrate that the two immune responses were similar, suggesting that polyhistidine insertion did not alter the immunogenicity of the nanoparticle. (D) Neutralization NT50 of serum from mice immunized with WT-spike-Fer (green, left) or spike-His-4.8-Fer (yellow, right) against lentivirus pseudotyped with Wuhan-1 SARS-CoV-2 spike showing good agreement between the groups. This supports the conclusion that polyhistidine insertion did not impact the multivalency or immunogenicity.
Finally, given that we intend to apply this method to vaccine development, we aimed to profile the immunogenicity of the functionalized nanoparticles. We immunized mice at days 0 and 21, with either the WT or His-4.8 versions of HA-Fer or spike-Fer. Both the WT and His-Fer proteins elicited a robust immune response (Figure 3C,D). For HA-Fer, the His-4.8 and WT nanoparticles elicited antibodies with similar binding activity by ELISA to WT HA-Fer (Figure 3C) but no binding to an off-target His-tagged protein (Figure S5). For spike-Fer, the His-4.8 and WT nanoparticles showed comparable neutralization of SARS-CoV-2 spike-pseudotyped lentivirus (Figure 3D). These results demonstrate that installation of a polyhistidine tag into the flexible loops on the surface of H. pylori ferritin facilitates purification while not affecting the geometry or short-term immunogenicity of the nanoparticles.
Discussion
The global pandemic caused by SARS-CoV-2 has highlighted the need to quickly design and produce effective vaccines. Converging alongside this still-important need are the promising results achieved with ferritin-based nanoparticle vaccines against other viruses that have moved into the clinic. A primary difficulty in developing ferritin-based vaccines has been their purification. Therefore, there is a significant need for methods that allow for quick and easy purification of potential vaccine nanoparticles. Simplified purification would facilitate rapid testing of many different vaccine candidates, which could help mitigate the impact of emerging diseases. For example, testing of variant vaccines containing individual mutations, like those within the Omicron variant of SARS-CoV-2, would be substantially easier with simplified nanoparticle purification.
While purification of HA-His-Fer and purification of spike-His-Fer were both successful, the latter proved to be more sensitive to the site insertion and length of the His tag, likely given the much greater molecular weight of the spike glycoprotein compared to that of HA. Indeed, because all four chosen installation sites were suitable for purifying HA-His-Fer nanoparticles, we predict that alternative installation sites could be used, depending on the application. For example, given that smaller glycoproteins may utilize a variety of His tag installation sites, it is possible that sequential prime boost vaccine regimes could take advantage of different His tag installation sites for each immunization. Such considerations may help mitigate a major caveat in bringing His-Fer proteins to the clinic by decreasing the risk of eliciting immunity against the neo-epitopes that may result from the His tag installation.
Another consideration for His-Fer is our utilization of H. pylori ferritin. These data demonstrating the flexibility of modifications and installations into ferritin loops may act as the foundation for applications into alternative ferritin sequences to simultaneously help improve purification and also mitigate antiscaffold immunity upon repeated exposure. For example, we similarly applied our analysis to determine sites for installing His tags into a number of other ferritins that have previously been tested for vaccine design, Escherichia coli, Pyrococcus furiosus, Archaeoglobus fulgidus,5 or Trichoplusia ni.34 Such an analysis also suggests His-4 as a potential insertion site (Figure S6).
Additionally, the His tag could be used for screening and removed for future applications. Indeed, these sites could easily be reverted to WT or modified to include other purification tags,35 or T-cell epitopes36 to promote immune stimulation in lead vaccine candidates. The ability to rapidly screen a large number of candidates has facilitated vaccine development in the past,37,38 demonstrating the importance of having early stage tools that enable lead optimization into late stage development. Our work demonstrates the flexibility of the ferritin scaffold to accommodate affinity-purification tags and other modifications, allowing for future design optimization of the ferritin scaffold. We envision that the simplicity of our method will lay the groundwork for novel ferritin-based vaccines.
Acknowledgments
The authors thank J. Bloom and A. Greaney for plasmids and cells related to viral neutralization assays.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00241.
Figures identifying regions of potential His tag installation, data supporting the use of eight histidine resins, Ni resin over other metal resins, ThermoFisher His Pur resin, evidence of the lack of anti-His tag antibodies, identification of potential His tag installation sites in ferritin from other species, and sequences of the proteins utilized in this study (PDF)
Accession Codes
H. pylori ferritin, based on accession ID WP_000949190; H1 hemagglutinin, based on accession ID KF356052; SARS-CoV-2 spike, based on accession ID QJE39038.
Author Contributions
⊥ P.W. and S.M. contributed equally to this work.
This work was supported by the National Institutes of Health (5DPAI15812502), the Virginia & D.K. Ludwig Fund for Cancer Research, the Frank Quattrone and Denise Foderaro Family Research Fund, the Chan Zuckerberg Biohub, Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant K99HD104924 (S.T.), and a Damon Runyon Cancer Research Foundation fellowship DRG-2301-17 (S.T.).
The authors declare no competing financial interest.
Supplementary Material
References
- Vartak A.; Sucheck S. J. Recent advances in subunit vaccine carriers. Vaccines 2016, 4, 12. 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R.; Serruto D. Self-Assembling Nanoparticles Usher in a New Era of Vaccine Design. Cell 2019, 176, 1245–1247. 10.1016/j.cell.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Pati R.; Shevtsov M.; Sonawane A. Nanoparticle Vaccines Against Infectious Diseases. Front. Immunol. 2018, 9, 2224. 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenbacher P. A.-B. Chemically Modified Bacterial Sacculi as a Vaccine Microparticle Scaffold. ACS Chem. Biol. 2022, 17, 1184. 10.1021/acschembio.2c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. Q.; Alves P. M.; Roldão A. Functionalizing Ferritin Nanoparticles for Vaccine Development. Pharmaceutics 2021, 13, 1621. 10.3390/pharmaceutics13101621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Q.; Soistman E.; Carter D. C. Ferritin nanoparticle technology: A new platform for antigen presentation and vaccine development. Ind. Biotechnol 2006, 2, 143–147. 10.1089/ind.2006.2.143. [DOI] [Google Scholar]
- Wang Z.; et al. Functional ferritin nanoparticles for biomedical applications. Front. Chem. Sci. Eng. 2017, 11, 633–646. 10.1007/s11705-017-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I.; Iwahori K.; Kumagai S. Ferritin in the field of nanodevices. Biochim. Biophys. Acta 2010, 1800, 846–857. 10.1016/j.bbagen.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Swanson K. A.; et al. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci. Immunol. 2020, 5, eaba6466. 10.1126/sciimmunol.aba6466. [DOI] [PubMed] [Google Scholar]
- Sliepen K.; et al. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 2015, 12, 82. 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.; et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 2016, 7, 12041. 10.1038/ncomms12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine H. M.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–70. 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Kanekiyo M.; et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. J.; et al. The Crystal Structure of Ferritin from Helicobacter pylori Reveals Unusual Conformational Changes for Iron Uptake. J. Mol. Biol. 2009, 390, 83–98. 10.1016/j.jmb.2009.04.078. [DOI] [PubMed] [Google Scholar]
- Joyce M. G.; et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep 2021, 37, 110143. 10.1016/j.celrep.2021.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen J. M.; et al. A spike-ferritin nanoparticle vaccine induces robust innate immune activity and drives polyfunctional SARS-CoV-2-specific T cells. bioRxiv 2021, 10.1101/2021.04.28.441763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuertz K. M.; et al. A SARS-CoV-2 spike ferritin nanoparticle vaccine protects against heterologous challenge with B.1.1.7 and B.1.351 virus variants in Syrian golden hamsters. bioRxiv 2021, 10.1101/2021.06.16.448525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A. E.; et al. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 2021, 7, 183–199. 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp H. D.; et al. Design of a broadly reactive Lyme disease vaccine. npj Vaccines 2020, 5, 33. 10.1038/s41541-020-0183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K. V.; et al. Safety and immunogenicity of a ferritin nanoparticle H2 influenza vaccine in healthy adults: a phase 1 trial. Nat. Med. 2022, 28, 383. 10.1038/s41591-021-01660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; et al. A milk-based self-assemble rotavirus VP6-ferritin nanoparticle vaccine elicited protection against the viral infection. J. Nanobiotechnol. 2019, 17, 13. 10.1186/s12951-019-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob E.; Unger R. A tale of two tails: why are terminal residues of proteins exposed?. Bioinformatics 2007, 23, e225–e230. 10.1093/bioinformatics/btl318. [DOI] [PubMed] [Google Scholar]
- Porath J.; Carlsson J.; Olsson I.; Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Celniker G.; et al. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013, 53, 199–206. 10.1002/ijch.201200096. [DOI] [Google Scholar]
- Ashkenazy H.; et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules.. Nucleic Acids Res. 2016, 44, W344–W350. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H.; Erez E.; Martz E.; Pupko T.; Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell D. J.; Coutsias E. A.; Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 2009, 6, 551–552. 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M.; et al. Differential glycosylation of envelope gp120 is associated with differential recognition of HIV-1 by virus-specific antibodies and cell infection. AIDS Res. Ther 2014, 11, 23. 10.1186/1742-6405-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriwilaijaroen N.; Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 2012, 88, 226–249. 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst J. A.; Falke J. J. Purification of proteins using polyhistidine affinity tags. Methods Enzymol 2000, 326, 245–254. 10.1016/S0076-6879(00)26058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block H.; et al. Immobilized-metal affinity chromatography (IMAC): a review. Methods Enzymol 2009, 463, 439–473. 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- Georgiev I. S.; et al. Two-Component Ferritin Nanoparticles for Multimerization of Diverse Trimeric Antigens. ACS Infect. Dis 2018, 4, 788–796. 10.1021/acsinfecdis.7b00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple M. E.; Brill A. L.; Pasker R. L. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 2013, 73, 9.9.1–9.9.23. 10.1002/0471140864.ps0909s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemstra H. S.; et al. The identification of CD4+ T cell epitopes with dedicated synthetic peptide libraries. Proc. Natl. Acad. Sci. 1997, 94, 10313–10318. 10.1073/pnas.94.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A.; Rappuoli R. Review Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol 2000, 3, 445–450. 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- Weidenbacher P. A.; Kim P. S. Protect, modify, deprotect (PMD): A strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 9947–9952. 10.1073/pnas.1822062116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S. M.; et al. The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. bioRxiv 2021, 10.1101/2021.07.11.451855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K. H. D.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T. F.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.