In this study, Pekarek et al. demonstrate a novel oxytocin-dependent mechanism of adult-born neuron synaptic maturation and circuit integration. They demonstrate that oxytocin receptor signaling promotes synaptic maturation of newly integrating adult-born neurons by regulating their morphological development and expression of mature synaptic AMPARs and other structural proteins.
Keywords: development, neurogenesis, neuropeptides, olfactory, oxytocin, synaptogenesis
Abstract
Neural circuit plasticity and sensory response dynamics depend on forming new synaptic connections. Despite recent advances toward understanding the consequences of circuit plasticity, the mechanisms driving circuit plasticity are unknown. Adult-born neurons within the olfactory bulb have proven to be a powerful model for studying circuit plasticity, providing a broad and accessible avenue into neuron development, migration, and circuit integration. We and others have shown that efficient adult-born neuron circuit integration hinges on presynaptic activity in the form of diverse signaling peptides. Here, we demonstrate a novel oxytocin-dependent mechanism of adult-born neuron synaptic maturation and circuit integration. We reveal spatial and temporal enrichment of oxytocin receptor expression within adult-born neurons in the murine olfactory bulb, with oxytocin receptor expression peaking during activity-dependent integration. Using viral labeling, confocal microscopy, and cell type-specific RNA-seq, we demonstrate that oxytocin receptor signaling promotes synaptic maturation of newly integrating adult-born neurons by regulating their morphological development and expression of mature synaptic AMPARs and other structural proteins.
Synapse and circuit plasticity in the central nervous system underlies the ability to adapt to ever-changing environmental stimuli. The olfactory bulb (OB) is a highly plastic sensory domain of the brain and has a remarkable capacity to maintain plasticity into adulthood via continuous integration of adult-born neurons (Lois and Alvarez-Buylla 1994; Alvarez-Buylla and Temple 1998). In rodents, adult-born neurons migrate from the subventricular zone (SVZ) of the lateral ventricles through the rostral migratory stream (RMS), differentiate, and integrate into OB circuitry, where they give rise to two classes of interneurons: granule cells and periglomerular cells (Luskin 1993; Lois and Alvarez-Buylla 1994; Petreanu and Alvarez-Buylla 2002). During maturation, adult-born granule cells (abGCs) undergo a sequence of morphological and transcriptional changes that drive synapse formation with OB principle mitral and tufted cells (Whitman and Greer 2007; Panzanelli et al. 2009; Tepe et al. 2018). A key developmental epoch for the survival and integration of abGCs occurs between 2 and 4 wk after migrating from the SVZ. During this period, activity-dependent alterations serve as a foundation for downstream developmental programs (Yamaguchi and Mori 2005; Kelsch et al. 2009) that promote the integration of abGCs into existing circuitry and drive the functional reorganization of the OB (Arenkiel et al. 2011; Huang et al. 2016). As such, this ever-changing circuit architecture makes the OB a good model system for studying neuronal development and synaptogenesis (Abrous et al. 2005; Ming and Song 2005) and for analyzing the effects of direct stimulation and manipulation of these circuits via sensory cues.
Historically, much focus has been applied to fast neurotransmitters and how they shape circuit function and plasticity within the OB (Lepousez et al. 2014; Pallotto and Deprez 2014; Andreae and Burrone 2018; Hanson et al. 2020). However, neuropeptidergic contribution to OB plasticity and development remains poorly understood. Neuropeptides are chemical messengers capable of conveying physiological stimuli and brain state information to neural circuits. The biological functionality of neuropeptides is broad and their half-life is long, making them potent regulators of circuit activity (Neumann 2008; van den Pol 2012). Additionally, neuropeptides are found throughout the brain and are enriched in areas involved in sensory processing, likely serving to modulate and maintain long-term plasticity (Smith et al. 2019, 2020). Previous work has defined a synaptogenic role for local corticotropin-releasing hormone (CRH) signaling in driving integration of adult-born neurons in the OB (Garcia et al. 2014, 2016). These results and others reveal CRH as a potent modulator of circuit plasticity, while also implicating neuropeptides as effector molecules within this process.
The neuropeptide oxytocin plays an important neuromodulatory role in sensory circuit plasticity (Zheng et al. 2014; Marlin et al. 2015; Mitre et al. 2016; Carcea et al. 2021) and is a regulator of hippocampal adult neurogenesis (Sánchez-Vidaña et al. 2016; Lin et al. 2018). Oxytocin has diverse roles in the mammalian brain ranging from modulating social behavior, aggression, and parturition to synaptic development and circuit plasticity (Nelson and Trainor 2007; Marlin et al. 2015; Oettl et al. 2016; Lin et al. 2017; Ripamonti et al. 2017; Carcea et al. 2021). Additionally, oxytocin signaling dynamically increases the representation of social olfactory inputs in the OB, thereby generating an optimized state for processing social information (Oettl et al. 2016; Oettl and Kelsch 2018). Here we show that oxytocin receptors are selectively expressed by abGCs throughout their activity-dependent critical period, and provide evidence that oxytocinergic fibers directly innervate the RMS. Using conditional loss-of-function analyses, viral genetics, imaging, electrophysiology, and transcriptomic analysis, we further show a functional role for oxytocin signaling within abGCs to drive morphological and synaptic development. Collectively, these data provide insight into how oxytocin drives adaptation of the mammalian olfactory bulb by promoting abGC developmental maturation and circuit integration.
Results
Oxytocin receptors are dynamically expressed in developing adult-born neurons throughout the critical period
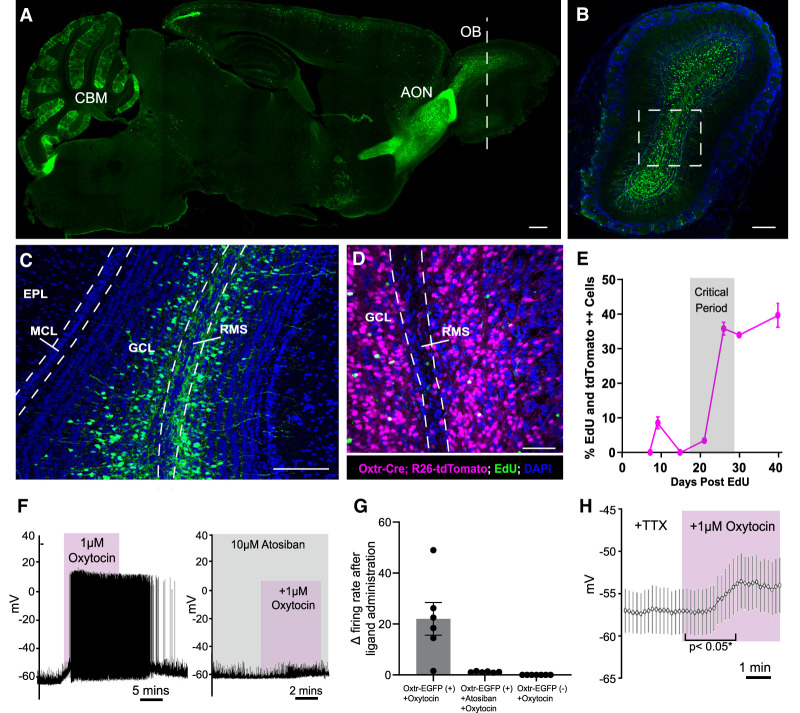
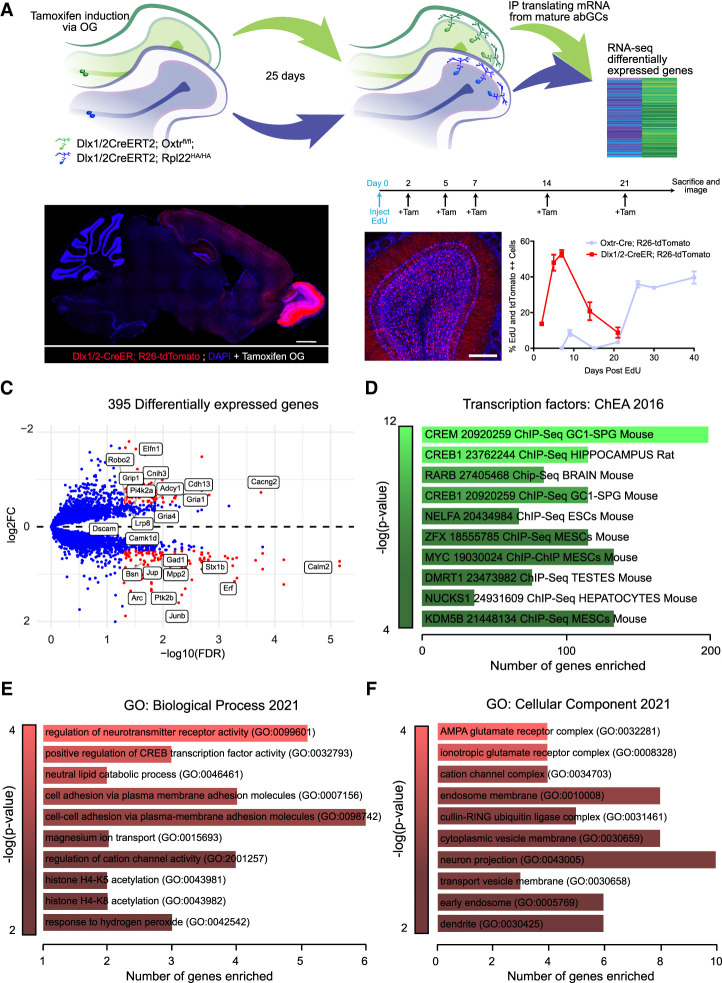
Oxytocin receptor reporter alleles have previously revealed robust expression within the mouse olfactory system (Gong et al. 2003). However, the cast of cell types that express oxytocin and its receptor, as well as the developmental time course of their expression, have not been thoroughly documented. To determine the distribution of oxytocin receptor (Oxtr) expression in the mouse olfactory system, we imaged the mouse brain using a previously described Oxtr-EGFP reporter allele (Fig. 1A; Gong et al. 2003). We noted dense expression of Oxtr-EGFP in previously reported areas such as the anterior olfactory nucleus (AON), the cerebellum, and, interestingly, the OB. Within the OB, we found that Oxtr expression was enriched within abGCs and periglomerular cells (Fig. 1B). Additionally, Oxtr-EGFP-expressing cells were localized deep within the granule cell layer, a domain that is populated by immature abGCs undergoing radial migration to the outer layers of the OB (Fig. 1C). To determine the developmental stage of Oxtr-positive abGCs, we stained against known markers of both mature (NeuN) and immature (doublecortin [Dcx]) abGCs as well as other subpopulation markers of abGCs (Supplemental Fig. S1). We found maximal coexpression of Oxtr-EGFP with NEUN, minimal coexpression of Oxtr-EGFP and DCX, and zero coexpression of Oxtr-EGFP with CALB2 or CALB1, suggesting that Oxtr marks later stage maturing subtypes of abGCs (Supplemental Fig. S1B). To more precisely assess the timing of Oxtr expression in abGC development, we next performed a birth dating experiment in Oxtr-Cre transgenic mice (Gong et al. 2003). Using a Rosa26-tdTomato conditional reporter allele (Madisen et al. 2010) crossed into the Oxtr-Cre background, we labeled all abGC lineages that have ever expressed Oxtr and found that Oxtr was expressed in a large proportion of granule cells within the bulb (Fig. 1D). We then birthdated newborn abGCs using pulses of EdU to delineate the developmental time course of Oxtr expression. Toward this end, we collected bulb tissue from animals at 7, 9, 15, 21, 26, 30, and 40 d after EdU injection to span the developmental progression of abGCs from migration and early synaptogenesis and through circuit integration (Carleton et al. 2003). Notably, we found that Oxtr expression was minimal through 9 d (8.55% ± 1.72%) (Fig. 1E), sharply increased between 21 and 26 d (35.83% ± 1.84%), and finally plateaued between 30 and 40 d (39% ± 3.48%). Moreover, Oxtr reached maximal expression levels throughout the activity-dependent critical period.
Figure 1.
Oxytocin receptor is dynamically expressed in developing adult-born neurons, and oxytocin selectively depolarizes Oxtr+ abGCs. (A) Sagittal section of an entire mouse brain, revealing Oxtr expression using the Oxtr-EGFP reporter allele. (OB) Oflactory bulb, (AON) anterior olfactory nucleus, (CBM) cerebellum. Scale bar, 200 µm. (B) Coronal OB section of the Oxtr-EGFP reporter allele in the plane denoted by the white dashed line in A. Scale bar, 200 µm. (C) Magnified view of the white box in B. (GCL) Granule cell layer, (EPL) external plexiform layer, (MCL) mitral cell layer, (RMS) rostral migratory stream. Scale bar, 200 µm. (D) Coronal view of the Oxtr-Cre;Rosa26-tdTomato reporter allele injected with EdU. (GCL) Granule cell layer, (RMS) rostral migratory stream. Scale bar, 200 µm. (E) Quantification of EdU and Oxtr-Cre;Rosa26-tdTomato-double-positive abGCs over the course of developmental time. n = 4 animals per time point. (F, left) Representative current clamp recording from Oxtr-EGFP+ abGCs. Oxytocin (1 µM) was washed into the bath. (Right) Representative current clamp recording from Oxtr-EGFP+ abGCs. Atosiban (10 µM) was washed into the bath 5 min before oxytocin application. (G) Quantification of the change in frequency after administration of oxytocin or oxytocin + atosiban in Oxtr-EGFP+ and Oxtr-EGFP− abGCs. n = 6–8 cells from three animals. (H) Representative current clamp recording from Oxtr-EGFP+ abGCs. Oxytocin (1 µM) and TTX were washed into the bath. (*) P < 0.05, Student's t test. n = 20 EGFP+ cells from four animals. See also Supplemental Figure S1.
Given the selective expression of Oxtr in abGCs, we next asked whether Oxtr-positive abGCs are responsive to oxytocin. To address this, we prepared acute brain slices from Oxtr-EGFP reporter mice and made visually guided whole-cell recordings from OB granule cells expressing EGFP with application of oxytocin while monitoring for voltage and action potential firing changes in a current clamp mode. Notably, we found that oxytocin selectively depolarized Oxtr-EGFP neurons, eliciting sustained trains of action potentials (Fig. 1F, left). This effect was completely blocked by infusing the chamber with the Oxtr-specific antagonist atosiban (Manning et al. 2008) prior to oxytocin delivery (Fig. 1F, right). All cells expressing Oxtr-EGFP responded to oxytocin with a mean ± SEM change in firing frequency of 21.5 Hz ± 8.6 Hz as compared with baseline (Fig. 1G). Importantly, no oxytocin-induced firing was observed in cells lacking the Oxtr-EGFP reporter (Fig. 1G). To further prove the direct, cell-autonomous effect of oxytocin, we performed additional recordings in the presence of sodium channel blocker TTX, thus excluding indirect, network-dependent changes in firing. Under such conditions, oxytocin application elicited a reproducible depolarization of Oxtr-expressing abGCs (Fig. 1H), likely contributing to their increased activity as highlighted in Figure 1F.
Oxytocinergic fibers directly innervate the RMS
Given that Oxtr is expressed in abGCs of the OB and selectively activated by oxytocin, we next sought to identify the potential source of oxytocin in these cells. Neurophysin is a carrier protein coexpressed with oxytocin to facilitate its loading into dense core vesicles and serves as a surrogate marker for oxytocinergic cell types (Castel and Morris 1988). Immunohistochemical analysis revealed no neurophysin immunoreactive cells in the main olfactory bulb, which suggested a lack of local oxytocin-releasing cells (data not shown). The primary oxytocinergic system resides within the paraventricular (PVN) and supraoptic nuclei (SONs) of the hypothalamus. PVN oxytocin cells project throughout central brain structures, and SON oxytocin cells project primarily to the pituitary gland (Rhodes et al. 1981). A previous study demonstrated functional oxytocinergic input from the PVN into the AON but did not detail any input directly into the OB (Oettl et al. 2016). Because of the OB's long distance from the PVN, we next looked for either direct or indirect points of oxytocin delivery to the OB (Fig. 2A). The PVN contains a dense population of oxytocinergic cells (Fig. 2B) that extend long-range projections throughout many domains of the brain (Liao et al. 2020). Thus, we asked whether projections from oxytocinergic hypothalamic neurons deliver oxytocin to the olfactory system. Since we did not observe direct oxytocinergic projections to the OB, we queried whether the oxytocinergic neurons innervated the RMS. Toward this end, we labeled the RMS using an antidoublecortin (DCX) antibody to visualize migrating adult-born neurons and an antineurophysin antibody to identify oxytocinergic fibers. Indeed, we noted oxytocinergic 10+ fibers per section innervated the AON proximal to the OB in five out of five animals profiled, with on average one to three fibers per section directly innervating the RMS (Fig. 2C–E).
Figure 2.
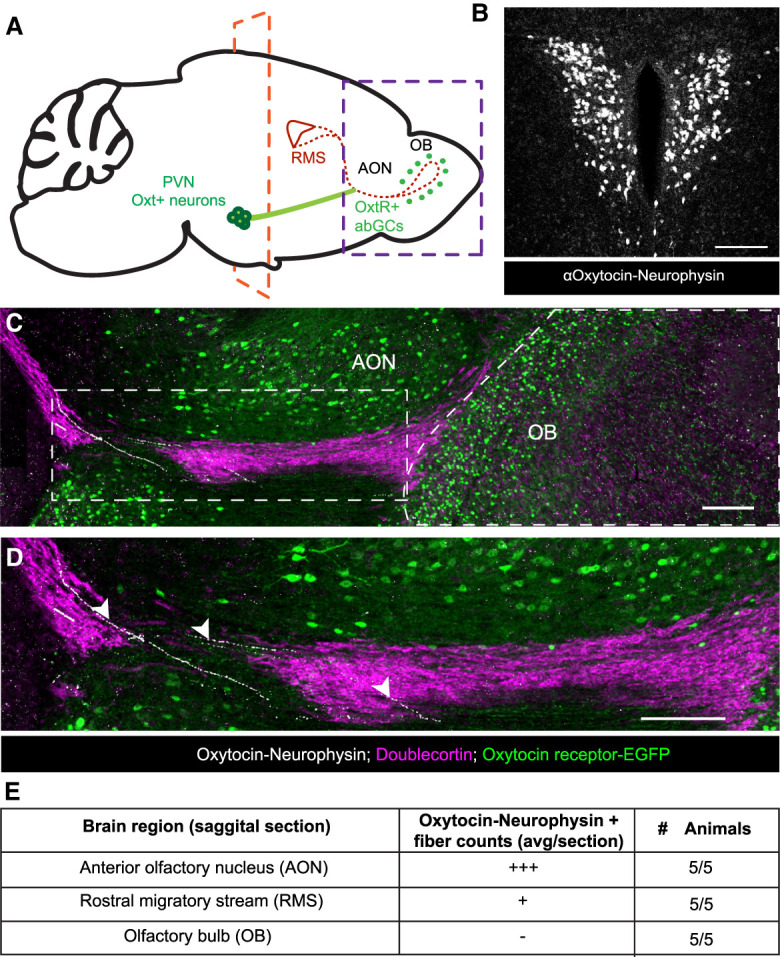
Oxytocin+ fibers innervate the rostral migratory stream. (A) Schematic diagram of a tracing experiment. (PVN) Paraventricular nucleus of the hypothalamus, (RMS) rostral migratory stream, (AON) anterior olfactory nucleus, (OB) olfactory bulb. (B) Coronal slice of the PVN. The plane is denoted by the orange dashed line in F, showing Oxt-immunopositive neurons in the PVN. Scale bar, 100 µm. (C) Sagittal slice. The plane is denoted by the purple dashed line in A, showing Oxt-immunopositive fibers in white, Oxtr-EGFP-immunopositive cells in green, and doublecortin-immunopositive neurons in purple to label neurons within the RMS and OB. Scale bar, 200 µm. (D) Magnified view of the white box in C. White arrowheads show oxytocinergic fibers directly innervating the RMS. Scale bar, 200 µm. (E) Semiquantitative count of oxytocin−neurophysin+ fibers in the AON, RMS, and OB. (+++) 10+ fibers per section, (+) one to three fibers per section,(-) zero fibers per section.
Thus far, these data suggest functional innervation of oxytocinergic fibers into the olfactory system via the RMS, validate functional and cell type-specific expression of Oxtr in abGCs, and support an activating property of oxytocin in developing adult-born neurons.
Oxtr KO does not affect survival of abGCs but promotes excessive growth and branching of dendritic arbors and spines
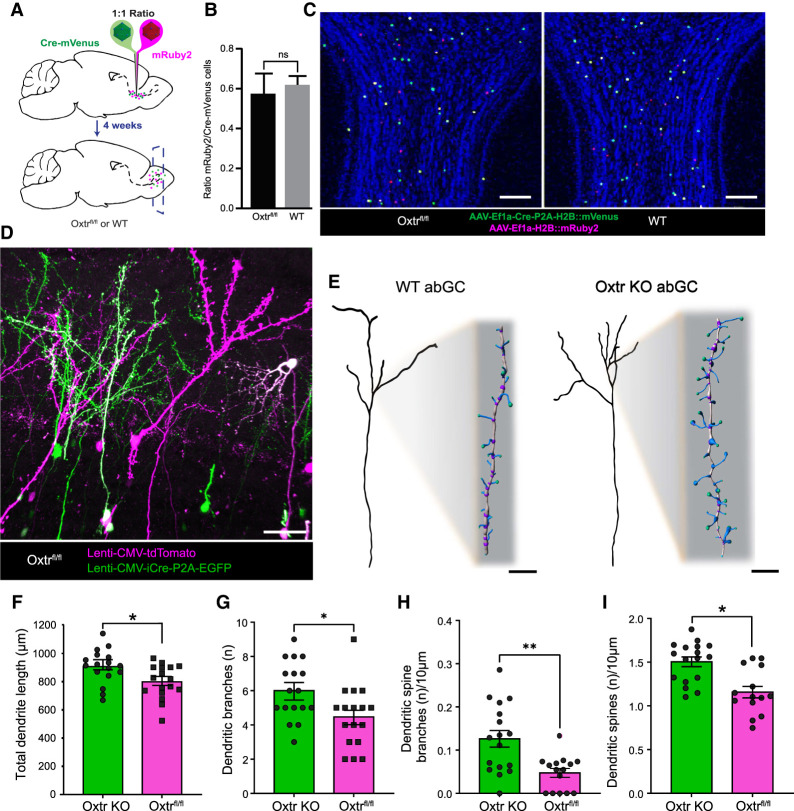
After revealing that Oxtr-positive abGCs of the OB are directly stimulated by oxytocin, we next sought to investigate its developmental function. Considering that Oxtr is expressed throughout the critical period of abGC development and robustly activates abGCs, we queried whether Oxtr may play a role in the survival of abGCs. To address this, we targeted abGCs for Oxtr deletion early in abGC development by injecting a 1:1 mixture of AAVs expressing Cre-mVenus and mRuby2 into the RMS of both Oxtrfl/fl and wild-type animals (Fig. 3A). By targeting the RMS in this way, we selectively targeted Oxtr deletion from young ∼5- to 10-d-old abGCs, well before Oxtr expression peaks in abGCs (Fig. 1E). After allowing 30 d for complete abGC migration and integration, we found no difference in the ratios of mRuby-labeled control neurons to conditional Oxtr KO (mVenus+ and mVenus mRuby++) neurons between the Oxtrfl/fl and WT animals (Oxtrfl/fl mean ± SEM = 0.582 ± 0.100; WT mean ± SEM = 0.627 ± 0.044; P > 0.05) (Fig. 3B), supporting that loss of Oxtr did not affect the survival of abGCs (Fig. 3C). Interestingly, however, we found that these two populations of labeled neurons appeared morphologically distinct, indicating that oxytocin influences abGC development independent of survival. To further investigate this, we first evaluated differences in the dendritic morphologies between conditional Oxtr KO and WT neurons. Toward this end, we injected a mixture of EGFP-Cre and tdTomato lentiviruses into the RMS of Oxtrfl/fl mice and evaluated morphological differences between wild-type and knockout cells, which were identified via differential reporter expression (Fig. 3D). Analyses of differentially labeled neurons showed that abGCs lacking Oxtr had longer dendrites (P < 0.05; Oxtr KO mean ± SEM = 907.235 µm ± 29.27 µm; Oxtrfl/fl mean ± SEM = 798.824 µm ± 28.884 µm) (Fig. 3F), increased numbers of branch points (P < 0.05; Oxtr KO mean ± SEM = 6 ± 0.411; Oxtrfl/fl mean ± SEM = 4.471 ± 0.438), and more complex dendritic trees (Fig. 3G). Complementing the increase in dendritic length and complexity, Oxtr-ablated abGCs also exhibited more branched dendritic spines (P < 0.05; Oxtr KO mean ± SEM = 0.126 ± 0.019 branches/10 µm; Oxtrfl/fl mean ± SEM = 0.047 ± 0.01 branches/10 µm) (Fig. 3H) and greater density of dendritic spines (P < 0.05; Oxtr KO mean ± SEM = 1.504 ± 0.056 spines/10 µm; Oxtrfl/fl mean ± SEM = 1.157 ± 0.066 spines/10 µm) (Fig. 3I). Taken together, these morphological data suggest that Oxtr-ablated abGCs develop more synapses, suggesting that Oxtr regulates synaptic development and circuit plasticity (Fig. 3E).
Figure 3.
Selective Oxtr knockout in developing abGCs yields larger and more complex dendritic arbors with increased spine densities. (A) Schematic diagram of a knockout experiment. (Cre-mVenus) AAV DJ8-Ef1a-Cre-P2A-H2B::mVenus, (mRuby2) AAV DJ8-Ef1a-H2B::mRuby2. (B) Quantification of data derived from C. Student's t-test, P > 0.05. n = 4/group. (C) Coronal view of OB 4 wk after RMS lentivirus injection outlined in A. Scale bar, 200 μm. (D) Representative confocal image for 3D reconstruction. Lenti-CMV-tdTomato and Lenti-CMV-GFP-Cre-P2A-EGFP. Scale bar, 30 μm. (E) Flattened representation of WT and Oxtr KO abGCs. Scale bar, 10 μm for inset magnified spines. (F–I) Quantification of total dendritic morphology. Student's t-test, (*) P < 0.05, (**) P > 0.005. nKO = 17 cells, nKO = 4 animals, nFlox = 17cells, nFlox = 5 animals.
Oxtr KO results in less synaptic input onto developing abGCs
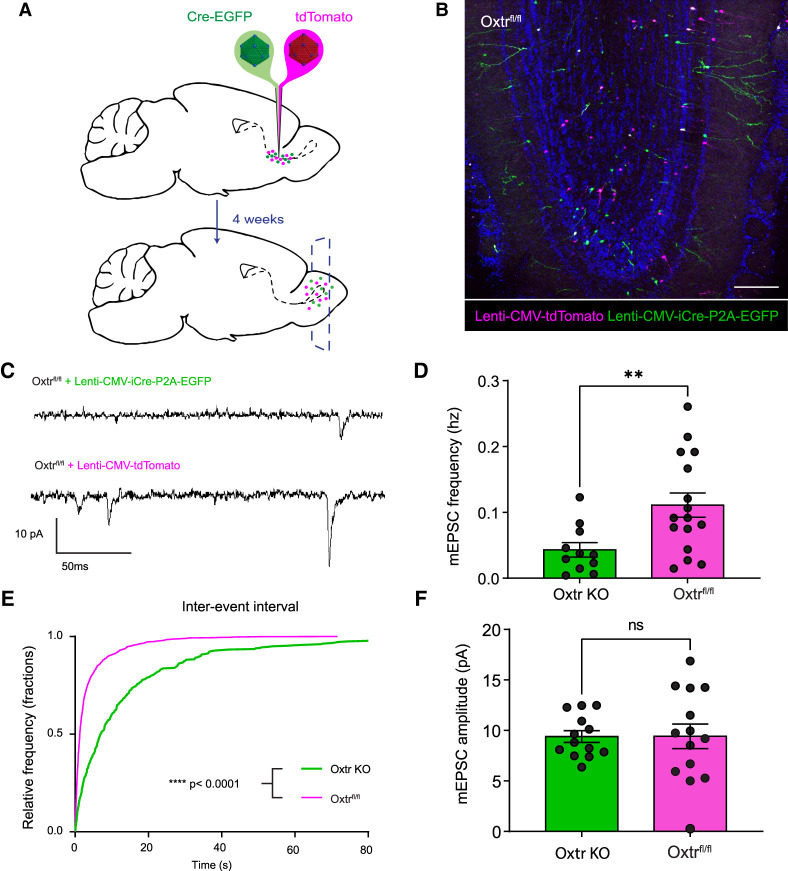
To further investigate the functional synaptic consequence of knocking out Oxtr in abGCs, we recorded frequencies of postsynaptic currents in the Oxtr-ablated abGCs compared with WT cells. Given that the primary local inputs onto abGC spines in the bulb come from excitatory mitral and tufted cells (Yokoi et al. 1995; Doetsch and Hen 2005), we selectively recorded miniature excitatory postsynaptic currents (mEPSCs) in the presence of TTX to suppress an action potential-dependent activity and GABAergic blocker bicuculine to eliminate inhibitory postsynaptic currents. For this, we made visually guided whole-cell recordings from either Oxtr-ablated or WT abGCs by injecting the RMS of Oxtrfl/fl animals with tdTomato and EGFP-Cre lentiviruses as described previously (Fig. 4A,B). Holding targeted cells at −70 mV, we found that Oxtr-ablated abGCs showed a lower frequency of mEPSCs (Oxtr KO mean ± SEM = 0.04 Hz ± 0.01 Hz) relative to their developmental age-matched controls (Oxtrfl/fl mean ± SEM = 0.11 Hz ± 0.02 Hz) (Fig. 4C–E). While mEPSC frequency was markedly lower in Oxtr-ablated abGCs, average amplitudes remained unchanged (Oxtr KO mean ± SEM = 9.40 pA ± 0.58 pA; Oxtrfl/fl mean ± SEM = 9.42 pA ± 1.22 pA) (Fig. 4F), suggesting that oxytocin may be important for maintaining the number of active synaptic sites formed during abGC circuit integration. With the decrease in functional synapses in Oxtr KO abGCs, it seemed plausible that abGC lateral inhibition would be compromised, resulting in impaired odorant discrimination and sensitivity. To test this, we fed tamoxifen to Dlx1/2-CreER;Oxtrfl/fl and Oxtrfl/fl control animals for 4 wk (Supplemental Fig. S2A,B) and subjected them to a go/“no go” paradigm to test their olfactory discrimination abilities as previously described (Supplemental Fig. S2C; Hanson et al. 2021). After testing discrimination of five different odor pairs, we saw no significant differences between groups (Supplemental Fig. S2D–F). Additionally, we subjected Dlx5/6-Cre;Oxtrfl/fl and Oxtrfl/fl control animals to a three-chamber test for sociability, as oxytocin is commonly involved in social behavior processing, but saw no significant differences between groups (Supplemental Fig. S3). Taken together, these assays suggest that Oxtr expression in abGCs does not regulate broad olfactory discrimination or general mouse social behavior. While these experiments provide evidence that Oxtr-ablated abGCs form fewer functional connections with existing cells, these results contradicted morphological data, leading us to consider potential mechanisms that might account for increased spine density but decreased mEPSC frequencies in Oxtr-ablated abGCs.
Figure 4.
Oxtr KO in abGCs results in lower frequency of mEPSCs. (A) Schematic diagram of lentiviral injections into Oxtrfl/fl animals. (B) Representative coronal slice 4 wk after lentiviral RMS injection. Scale bar, 150 μm. (C) Representative miniature excitatory postsynaptic current trace of Oxtr KO and Oxtr Flox/WT cells. (D) Frequency of miniature excitatory postsynaptic current. Students t-test P < 0.05. NOxtr KO = 6 animals, NOxtr KO = 12 cells, NWT = 7 animals, NWT = 20 cells. (E) Interevent interval graph. Kolmogorov-Smirnov test, P < 0.0001. NOxtr KO = 6 animals, NOxtr KO = 11 cells, NWT = 7 animals, NWT = 10 cells. (F) Graph of miniature excitatory postsynaptic current amplitude. Students t-test, P > 0.05. NOxtr KO = 6 animals, NOxtr KO = 14 cells, NWT = 7 animals, NWT = 14 cells.
Single-cell sequencing reveals that abGC Oxtr regulates synaptogenic gene expression
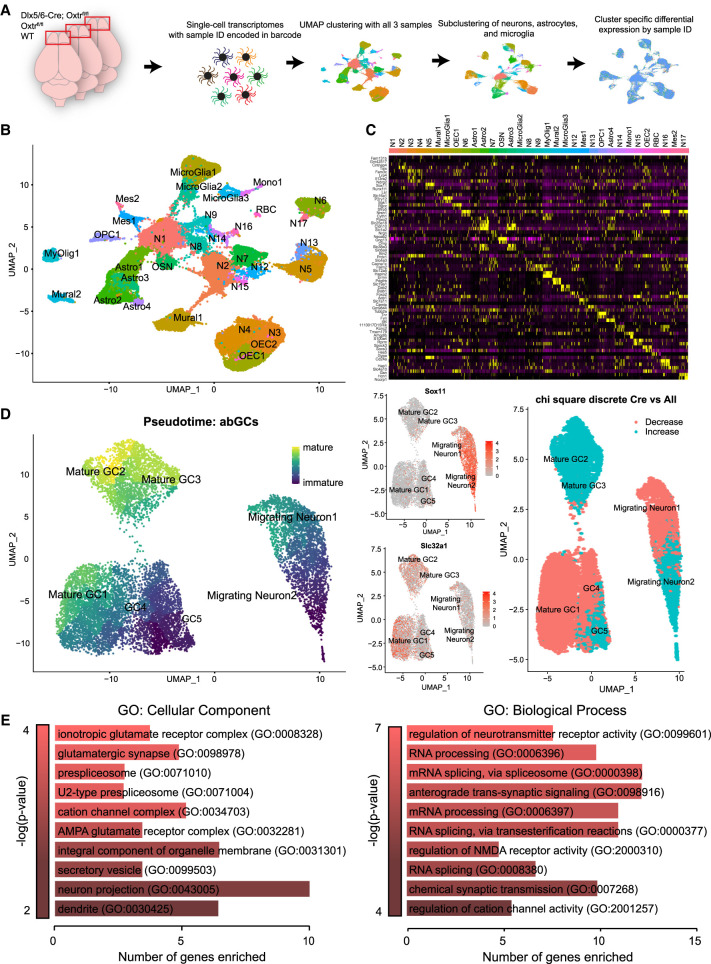
To reveal candidate developmental mechanisms whereby oxytocin signaling regulates morphological and synaptic development of abGCs, we turned to single-cell RNA sequencing. Toward this end, we targeted Oxtr knockout from interneuron populations by generating mice harboring the forebrain-specific interneuron Dlx5/6-Cre driver crossed with Oxtrflox/flox mice, Oxtrflox/flox alone, and wild-type animals, which served as controls (Fig. 5A; Monory et al. 2006). We processed single-cell transcriptomic data and clustered sorted cells using UMAP reduction in the Seurat single-cell informatics package version 4.1.0 (Hao et al. 2021). We clustered all sorted samples into known cell types, identifying each cluster using expression of known canonical markers (Fig. 5B,C). Next, we subclustered adult-born neurons and performed a pseudo-time line analysis using Monocle3 to determine their developmental age (Fig. 5D; Supplemental Table S1; Cao et al. 2019). We observed two unique immature migrating neuron clusters, three mature granule cell clusters, and two clusters of intermediate developmental age (Fig. 5D, left). To verify the developmental ages assigned to these clusters, we plotted expression of the immature marker Sox11 and mature granule cell marker Slc32a1 and noted robust expression in immature and mature clusters, respectively (Fig. 5D, middle). Using a χ2 test, we observed statistically different differential representation of the Dlx5/6-Cre;Oxtrflox/flox sample in multiple clusters within our data set (Fig. 5D, right; Supplemental Table S2). Specifically, we noted an increase in some cell clusters with an immature identity, as well as a decrease in other clusters with a mature cell cluster identity. These data revealed a shift in cluster composition dependent on Oxtr expression, suggesting that the distribution of cellular identities was not equivalent between Oxtr KO and other samples. Together, these data indicated a potential developmental identity shift after knockout of Oxtr. To support this, we next extracted a list of differentially expressed genes (DEGs) from Oxtr KO samples versus WT and Oxtrflox/flox controls within all cell clusters (Supplemental Table S3). The down-regulated DEGs from just abGC clusters were then subjected to gene ontology enrichment (all DEGs, including up-regulated genes, are shown in Supplemental Table S3) using the online gene set enrichment analysis and GO term analysis tool ErichR (Chen et al. 2013; Xie et al. 2021). Interestingly, many of the GO terms resulting from this analysis revealed selective changes in both neurodevelopmental and synaptogenic programs. Moreover, a prominent cohort of differentially expressed genes comprised those that give rise to proteins normally localized to synaptic and glutamate receptor compartments (Fig. 5E). Overall, these data suggested a shift in cellular identity upon Oxtr knockout and revealed down-regulation of synaptic genes as a candidate for the observed altered abGC integration and function.
Figure 5.
Single-cell sequencing of Oxtr KO and Oxtr WT OBs demonstrates transcriptional changes associated with synapse morphology and formation within abGCs. (A) Schematic diagram for 10X Chromium single-cell sequencing workflow. (B) Uniform manifold projection of all OB cells. Dlx5/6-Cre;Oxtrfl/fl, Oxtrfl/fl, and WT samples clustered concurrently. (C) Heat map of the top differentially expressed genes for each cluster in B. (D, left) Uniform manifold projection of subclustered abGCs colored in pseudotime. (Middle) Expression of Sox11 and Slc32a1 maturation markers mapped onto abGC clusters. (Right) χ2 analysis of Dlx5/6-Cre;Oxtrfl/fl cluster composition versus WT and Oxtrfl/fl samples. (E) Differential expression GO analysis from just abGCs adapted from EnrichR.
Cell type-specific and developmental-specific mRNA enrichment in abGCs reveals disrupted postsynaptic compartments and AMPA receptors after KO of Oxtr
Given that we identified selective expression changes in genes associated with synaptic development and function, we next sought to use a more targeted approach to evaluate gene expression in a cell type-specific manner in abGCs. As single-cell sequencing requires tissue dissociation, the synaptic compartments and transcripts are lost in this preparation (Armand et al. 2021). Indeed, many transcripts are transported, stored, and then translated locally at the synapse in response to activity, enabling rapid synaptic protein expression, which drives neuronal adaptation and development (Holt and Schuman 2013; Ifrim et al. 2015; Shigeoka et al. 2016; Sambandan et al. 2017; Holt et al. 2019). To generate known abGC-specific expression profiles at defined developmental stages and circumvent some of the limitations associated with typical dissociation and sorting methods, we turned to cell type-specific RNA enrichment using a Ribotag approach (Sanz et al. 2009, 2019). The Ribotag approach uses a Cre-dependent allele to tag a large ribosomal subunit with the HA epitope. When this allele is combined with a cell type-specific Cre driver, translating ribosomal polysome complexes along with their associated mRNAs can be selectively immunoprecipitated from cells of interest within a whole-tissue lysate.
To selectively enrich for mRNA from abGCs, we introduced a Dlx1/2-CreER allele (Batista-Brito et al. 2008) into mice harboring the Rpl22-HA allele (Sanz et al. 2009) to create Dlx1/2-CreER;Rpl22-HA animals (Fig. 6A). Dlx1/2-CreER is largely expressed in forebrain interneurons and enriched in adult-born interneurons within the OB (Batista-Brito et al. 2008). To first determine the timing of Dlx1/2 expression relative to Oxtr, we crossed Dlx1/2-CreER with a Rosa26-tdTomato conditional reporter (Madisen et al. 2010) and injected these animals with EdU. Importantly, using EdU birth dating and tamoxifen oral gavage, we confirmed that Dlx1/2-CreER is expressed between 5 and 7 d of abGC development, well before the peak expression of Oxtr in abGCs (Fig. 6B). These animals were then crossed with Oxtrfl/fl mice to generate the final experimental models. By administering tamoxifen, we both knocked out Oxtr in abGCs of a known developmental age and selectively labeled polysomes for immunoprecipitation (IP) of actively translating mRNA. The Dlx1/2-CreER driver facilitated inducible and abGC-specific recombination of both Rpl22-HA and Oxtrfl/fl alleles. This approach enabled bulk RNA sequencing of enriched translating mRNA samples from both Oxtr KO and control wild-type 30-d-old abGCs.
Figure 6.
Actively translating enriched mRNA from abGCs reveals alterations in genes necessary for synaptic morphology development, postsynaptic density organization, and AMPA receptor function upon Oxtr KO. (A) Schematic diagram of the Ribotag immunoprecipitation experiment. (B, left) Representative confocal image of Dlx1/2-CreER;Rosa26-tdTomato olfactory bulbs after tamoxifen oral gavage. Saggital view: scale bar, 1000 μm. Coronal image: scale bar, 100 μm. (Right) Representative image and EdU time course of abGCs expressing Dlx1/2-CreER. (Tam) Tamoxifen, administered at 200 mg/kg via oral gavage. N = 4 mice/time point. (C) Volcano plot for the top differentially expressed genes. Red dots represent genes with an abs[log2(FC)] >0.25 and a −log10(FDR) >1. (D–F) Gene ontology enrichment adapted from EnrichR using differentially expressed genes between Dlx1/2-CreER;Oxtrfl/fl;Rpl22-HA/HA and Dlx1/2-CreER;Rpl22-HA/HA immunoprecipitated mRNA. NExp = 11 animals, NControl = 8 animals. See also Supplemental Figure S4.
To ensure specificity of our analysis, we first performed background gene expression removal by comparing paired input versus immunoprecipitated samples, including only genes that were significantly enriched in immunoprecipitated and depleted in input for further downstream analysis as previously described (Sanz et al. 2019). To validate the selective and cell type-specific enrichment of RNA, we first used the DESeq2 R package to perform a differential expression analysis between immunoprecipitated and input samples. Importantly, we showed that our immunoprecipitated samples, regardless of experimental condition, clustered separately from input samples (Supplemental Fig. S4A). Leveraging our previous single-cell sequencing results from the mouse OB (Tepe et al. 2018), we identified distinct cluster marker genes to reliably annotate OB cell types. We found a significant enrichment of abGC-specific genes, as well as a significant depletion of all other cell type markers within our immunoprecipitated group as compared with input (Supplemental Fig. S4B).
With mature 30-d-old abGC-specific mRNA from both Oxtr KO and WT backgrounds, we compared gene expression changes and identified DEGs associated with glutamatergic synapse function and abGC development, including Ntm, Cnih3, Gria1, and Gria4 (Horger et al. 1998; Flavell et al. 2006; Kelsch et al. 2008; Herring et al. 2013; Chater and Goda 2014) (Fig. 6C). Additionally, we observed distinct up-regulation of genes known to induce neuronal differentiation, along with activity-dependent proteins and transcription factors that have been reported to cause changes in cell morphology. We analyzed all DEGs regardless of differential expression direction using EnrichR gene ontology enrichment and observed that transcription factors Creb1, Crem, and Rarb, among others, were significantly dysregulated (Fig. 6D). Intrigued by the enrichment of cAMP binding transcription factors and their known association with abGC development, we next analyzed the down-regulated DEGs separately and observed significant differences in gene expression in categories important for glutamate receptor function, including regulation of the AMPA receptor complex and cell–cell adhesion molecules known to regulate morphological development (Fig. 6E,F; Supplemental Table S4; Chen et al. 2013; Xie et al. 2021). Additionally, many DEGs clustered into synaptic compartments important for the structural integrity of postsynaptic densities, such as AMPA receptors, dendrites, and vesicular compartments (Fig. 6F). Together, these data reveal that Oxtr KO induces changes in gene translation patterns resulting in dysregulation of transcriptional programs and key genetic pathways associated with synaptic and morphological development, including AMPA receptor structure and function.
Oxtr knockout in developing abGCs alters AMPA/NMDA current ratios from mitral/tufted cells
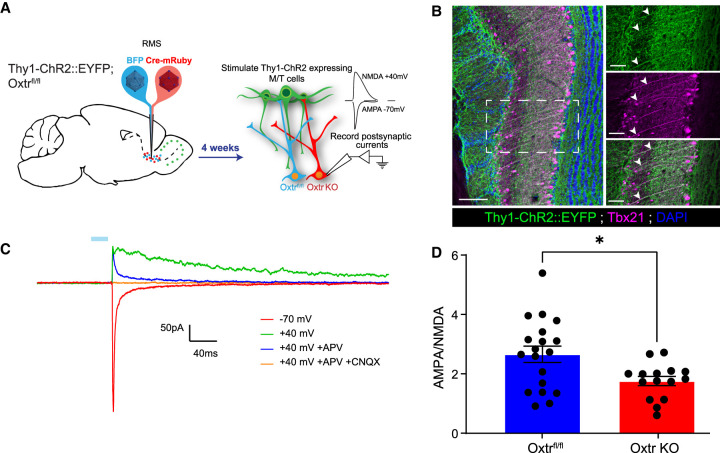
Because our whole-cell recording data show that Oxtr KO abGCs receive decreased postsynaptic current and gene expression analyses revealed down-regulation of transcripts necessary for AMPA receptor function, we next sought to evaluate whether Oxtr KO cells may have impaired transduction of glutamatergic input. abGCs undergo dramatic changes in gene expression that govern their morphological and synaptic properties throughout development (Tepe et al. 2018). A noted example is the switch in synaptic protein expression from mostly NMDA to both AMPA and NMDA ionotropic glutamate receptors (Katagiri et al. 2011; Kovalchuk et al. 2015) in the maturation of abGC synapses. To evaluate potential roles for oxytocin in such developmental processes, we measured elicited AMPA/NMDA currents in both wild-type and Oxtr KO abGCs. To facilitate this, we introduced channelrhodopsin2 (ChR2) into M/T cells (Balu et al. 2007; Panzanelli et al. 2009; Pallotto et al. 2012). For this, we crossed Thy1-ChR2::EYFP into the Oxtrfl/fl background to yield Thy1-ChR2::EYFP;Oxtrfl/fl mice (Fig. 7A). These animals express ChR2::EYFP in mitral/tufted cells of the OB (Arenkiel et al. 2007). Concurrently, lentiviruses expressing either Ef1a-BFP or Ef1a-Cre-P2A-mRuby2 were injected into the RMS either to label wild-type abGCs or to both label and knock out Oxtr, respectively (Fig. 7A). After allowing 4 wk to complete abGC development and integration, fixed cryosections were obtained and stained with the M/T cell marker Tbx21 (Fig. 7B). Slices obtained from Thy1-ChR2::EYFP;Oxtrfl/fl transgenics had robust colocalization of ChR2::EYFP and Tbx21, suggesting that ChR2+ cells from Thy1-ChR2::EYFP;Oxtrfl/fl transgenics were M/T cells (Fig. 7B). To validate that ChR2-elicited current was indeed glutamatergic, we measured evoked synaptic responses via whole-cell recordings from visually targeted abGCs in acute brain slices from both experiments in the presence of AMPA and NMDA blockers. Evoked responses from BFP+ control cells held at +40 mV were partially abolished with bath application of the selective NMDAR blocker APV and completely abolished with both APV and the AMPAR blocker CNQX (Fig. 7C). We then measured postsynaptic responses at either +40 mV or −70 mV to isolate NMDA and AMPA currents, respectively. When recording from Ef1a-Cre-P2A-mRuby2-expressing Oxtr knockout abGCs, we found a significant decrease in the AMPA:NMDA ratio when compared with Ef1a-BFP-expressing controls in the Thy1-ChR2::EYFP;Oxtrfl/fl experiment (Oxtr KO mean ± SEM = 1.782 ± 0.186; Oxtrfl/fl mean ± SEM = 2.657 ± 0.274) (Fig. 7D). These findings reflect an altered AMPA/NMDA component to the postsynaptic current at specifically the M/T and abGC synapse, supporting a model whereby Oxtr signaling promotes maturation of M/T synapses in abGCs.
Figure 7.
Oxtr knockout in developing abGCs reduces the functional AMPA:NMDA ratio from mitral/tufted cells. (A) Schematic diagram of experimental setup using the Thy1-ChR2:EYFP transgenic allele. (B, left) Coronal slice of the OB demonstrating expression of Thy1-ChR2::EYFP. Scale bar, 100 µm. (Top right) Magnified image derived from the white box. Scale bar, 50 µm. (Middle right) Coronal slice of the OB demonstrating expression of Tbx21. Scale bar, 50 µm. (Bottom right) Magnified image derived from the white box. Note the white arrowheads denoting colocalization. Scale bar, 70 µm. (C) Representative trace from the visually guided whole-cell patch clamp experiment. The blue bar represents 470-nm LED stimulation. (APV) Selective NMDAR antagonist, (CNQX) selective AMPAR antagonist. (D) Graph of the AMPA:NMDA ratio. Students t-test, P = 0.0272. nFlox = 5 mice, nFlox = 19 cells, nKO = 4 mice, nKO = 15 cells.
Discussion
Here, we present a novel oxytocin-dependent mechanism that promotes the development and synaptic integration of abGCs within the murine OB. Collectively, our data reveal that the oxytocin receptor coordinates abGC circuit integration by regulating morphological, synaptic, and transcriptional development. Selectively ablating Oxtr within abGCs disrupts gene expression programs essential for dendrite growth, spine development, and AMPA/NMDA receptor balance. The resultant cells have underdeveloped synapses and impaired function. While others have shown that adult-born neurons develop, migrate, and integrate in an activity-dependent manner, much remains unknown about the underlying molecular mechanisms. Neuropeptides such as oxytocin are uniquely poised to adaptively influence activity-dependent circuit integration.
Selective expression of Oxtr sensitizes developing abGCs to oxytocin
Previous studies have shown that oxytocin drives programs necessary for circuit integration in the hippocampus; however, little is known about its role in olfactory adult-born neuron development (Leuner et al. 2012; Lin et al. 2017, 2018; Ripamonti et al. 2017; Tirko et al. 2018). Our data indicate that oxytocin receptor expression within the mammalian OB both is specific to adult-born neurons and coincides with their activity-dependent critical period. Additionally, when oxytocin is presented to these cells, they exhibit enhanced firing and prolonged trains of action potentials. We have shown that oxytocinergic fibers innervate the RMS, providing a source of oxytocin to the OB. Collectively, these data support a mechanism of extrasynaptic information transfer whereby expression of the oxytocin receptor sets the specificity of signal reception, effectively sensitizing abGCs to existing oxytocinergic fiber release during the critical period of their development. Indeed, this method of extrasynaptic transmission is typical for neuropeptides throughout the mammalian brain (Ludwig 1998; Ludwig and Leng 2006; Smith et al. 2019). Such tightly regulated expression of Oxtr is elegantly poised to allow the oxytocinergic system dynamic access to influence adult-born neuron integration, thus facilitating an adaptive sensory dimension.
Oxytocin drives dendritic remodeling and synaptic integration by regulating development of immature synapses
Our data show that selective ablation of Oxtr in immature abGCs yields neurons with overgrown dendritic trees and a greater density of spines. Despite this increased arborization, Oxtr-ablated abGCs do not integrate into the existing circuitry as effectively as their wild-type counterparts, as indicated by electrophysiological recordings. Initially, we speculated that the contradictory phenotypes of Oxtr-ablated abGCs (e.g., having more complex and spine-dense dendritic arbors with lower synaptic frequencies) might be explained by a developmental stalling of abGCs. Indeed, it is well known that abGCs form an abundance of dendritic spines that are then selectively pruned at inactive synapses as they mature (Reshef et al. 2017; Wallace et al. 2020). Given the critical period of Oxtr expression and potent excitatory effect of oxytocin, it is conceivable that Oxtr may regulate or relay activity-dependent signals to developing abGCs to facilitate and/or promote synaptic and morphological development. Supporting this, our single-cell data indicate that Oxtr expression regulates abGC developmental trajectories. For example, knocking out Oxtr alters the ratios of immature to mature cells as characterized by pseudotime analysis. Of note, we noticed that some immature and mature clusters were increased in representation within the Oxtr KO group compared with controls, while others experienced the opposite. Given that not all abGCs express Oxtr, this discrepancy highlights differences in subpopulations of abGCs that may be uniquely susceptible based on expression of Oxtr. These developmental identity changes are accompanied by altered gene expression in programs that govern glutamatergic synapse formation, maintenance, and stability. Specifically, we observed the genes Gria2, Gria3, Gria4, and Dlgap4, among others, as down-regulated in knockout versus WT and floxed controls, indicating an AMPAR-specific impairment. Synaptic development and integration of abGCs are considered stereotyped programs in which new immature synapses express mostly NMDA receptors (Grubb et al. 2008). As abGCs mature, immature synapses begin to express AMPA and NMDA receptors in response to local activity while concurrently mobilizing cytoskeletal binding proteins to promote neurite extension and postsynaptic density formation (Grubb et al. 2008). To support and further interrogate gene expression changes from our single-cell data, we immunoprecipitated actively translating mRNA from 30-d-old mature abGCs from Oxtr KO and WT controls. Differential expression of these two samples revealed that knocking out Oxtr disrupts genes essential for postsynaptic cytoskeletal structures and synaptic function. Supporting our single-cell data, we also observed genes integral for AMPA receptor function such as Gria1, Gria4, Cacng2, and Cnih3 as significantly down-regulated in the KO groups. Conversely, genes important for neuron growth and postsynaptic density formation such as Egr2, Egr1, Pi4k2a, and Dlgap2 (Lindholm et al. 1994; Thakker-Varia et al. 2007; Ao et al. 2014) were significantly up-regulated. Interestingly, we demonstrate a significant difference in AMPA/NMDA ratio upon Oxtr KO. Additionally, we found that the average coefficient of variation for AMPA responses was indeed higher than for NMDA responses, suggesting that adult-born neurons in both the control and Oxtr KO groups do form a certain number of silent immature synapses. While these results are based on recordings of multiquantal synaptic responses and do not rule out silent synapses as a possible explanation, they do suggest some level of AMPA receptor plasticity dysregulation is occurring. After observing dysregulation of AMPAR genes in response to Oxtr KO in both single-cell and Ribotag-enriched RNA-seq data sets, we hypothesized that this drives a paucity of functional glutamatergic synapses, while the increased expression of cytoskeletal binding proteins and growth factors may account for Oxtr KO abGCs forming larger dendritic arbors and supernumerary spines.
Oxytocin influences developmental programs in neuronal subsets
Oxytocin is known to have organizational effects on developing neurocircuits, with loss-of-function mutations giving rise to early developmental abnormalities in animal models and humans (Keebaugh and Young 2011; Tyzio et al. 2014; Guastella and Hickie 2016; Nunes et al. 2020). Given our electrophysiological evidence suggesting poor integration of Oxtr KO abGCs combined with critical period-specific expression of Oxtr, it is likely that oxytocin is an important regulatory signal to integrating abGCs. However, the molecular cascade of events translating Oxtr activation to abGC integration remains elusive.
Mechanistically, OXTR signaling is coupled to Gαq or Gαi, where, upon receptor binding, oxytocin depolarizes neurons via inhibition of KIR7.1 and subsequent activation of voltage-dependent calcium channels through a PLC/PKC-dependent mechanism (Gimpl and Fahrenholz 2001; Stoop 2012; Bakos et al. 2018). Interestingly, studies show that oxytocin bound to OXTR phosphorylates CREB in neurons to drive downstream activity-responsive genes (Tomizawa et al. 2003; Jurek et al. 2015). Our data show that Creb1 and Crem were the two most enriched transcription factor pathways derived from differential expression analysis. Co-opting Creb-responsive activity regulators, such as Arc, Junb, and Fosl2, were also found to be dysregulated in Oxtr KO abGCs, Oxtr signaling dynamically regulates expression of synaptic remodeling genes. It is conceivable that abGCs expressing Oxtr are a specialized developmental subset responsible for transducing specific environmental stimuli by sensitizing abGCs to oxytocin during their critical period. However, it must be considered that because only a subset of abGCs expresses Oxtr, manipulating only this subset may not result in broad effects on general olfactory behavior, which may explain the lack of an olfactory discrimination/sensitivity phenotype. Perhaps supporting a specific environmental stimulus underlying oxytocin's function in abGC integration, a previous study found that motherhood increases circuit integration of abGCs (Kopel et al. 2012). Large amounts of oxytocin are released during pregnancy and parturition, which may act directly on abGCs. In such a scenario, oxytocinergic projections into the RMS may function to convey the physiological state of parturition, parental care, stress, or social status to the olfactory system.
The release of oxytocin into the OB may indeed serve as an extrasynaptic signaling substrate to reshape OB circuitry over time by modulating abGC activity-dependent development through the Creb-related pathway, AMPAR plasticity, and synaptic integration. In this way, oxytocin would serve as a stimulus-specific cue, initiating a plasticity window in the OB by co-opting continually integrating abGCs to remodel circuitry. This type of circuit remodeling could happen rapidly and have wide-ranging effects on specific odor representations. In any case, our current findings reveal a role for oxytocin as a key signaling molecule that supports sensory network development and plasticity at the cellular level. Classically, neuropeptides convey brain state and physiology by signaling to receptive brain circuits. Given the synaptogenic and morphological programs we have uncovered, it is likely that oxytocin contributes to abGC plasticity to drive OB circuit integration and adaptation. Future work will determine whether this adaptation is in response to social, parental, aggressive, or other physiological states.
Materials and methods
Oxtr-Cre EdU birth dating experiments
All birth dating experiments were performed on male and female adult mice 6–10 wk old. Oxtr-EGFP (MGI 4847297) and Oxtr-Cre (MGI 5311702) were used for the initial pharmacological and spatial expression characterizations. The EdU birth dating experiments made use of Oxtr-Cre crossed to a conditional Rosa26-tdTomato reporter allele. We then injected 50 mg/kg EdU (Thermo Fisher C10340) intraperitoneally five times over the course of 1 d.
Antibodies
The antibodies used in this study were rabbit antioxytocin–neurophysin (1:4000; Abcam ab2078), chicken antidoublecortin (1:500; Synaptic Systems 326-006), chicken anti-EGFP (1:1000; Abcam ab13970), mouse anti-NeuN (1:500; Millipore MAB377), rabbit anticalretinin (1:1000; Millipore AB5054), and mouse anticalbinidin (1:500; Abcam ab9481).
Electrophysiology
For acute olfactory bulb slice preparation, animals were anesthetized with isoflurane and perfused with cold artificial cerebrospinal fluid (ACSF) solution containing 125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 25 mM NaHCO3. Brains were rapidly removed and transferred into sucrose-based cutting solution (pH 7.35) containing 87 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, 7 mM MgCl2–6H2O, 13 mM ascorbic acid, 75 mM sucrose, 10 mM glucose, and 25 mM NaHCO3. Both solutions were pH-equilibrated with a 5% CO2/95% O2 gas mixture for at least 10 min prior to experiment. Three-hundred-micrometer-thick coronal olfactory bulb slices were prepared using a Leica VT1200 vibratome and placed for at least 15 min at 37°C in 5% CO2/95% O2 bubbled ACSF solution. They were then gradually lowered to room temperature (25°C) and allowed to acclimate for at least 15 min before recording. For recording, slices were transferred into a recording chamber continuously perfused with ACSF at 1–2 mL/min at 25°C. Neurons were identified by optiMOS camera (QImaging) controlled by Micro-Manager version 1.4.22 software (University of California at San Francisco) using transmitted light DIC (Olympus BX50WI) and fluorescence reporter using Thorlabs DC4100 470-nm or 565-nm LED illumination with 49002-ET-EGFP (FITC/Cy2) or HcRed1 emission filters (Chroma Technology), respectively. Recordings were acquired using an Axon MultiClamp 700B amplifier digitized at 10 kHz (Axon Digidata 1440A). Recording electrodes (3–5 MΩ) were fabricated from borosilicate glass microcapillaries (outer diameter 1.5 mm) with a micropipette puller (Sutter Instruments). The internal solution contained 120 mM Cs-MeSO4, 10 mM CsCl, 2 mM MgCl2, 4 mM ATP-Mg, 0.5 mM GTP-Na, 10 mM phosphocreatine-di-Na, 1 mM EGTA, and 20 mM HEPES; pH was adjusted to 7.3 with CsOH, and the osmolarity was adjusted to 305 mOsm/L with CsMeSO4. The access resistance was monitored throughout the experiment and was typically in a range of 10–20 MΩ and considered to be acceptable up to 30 MΩ. Obtained recordings were processed offline using Clampfit 10.7.0.3 (Molecular Devices) and Synaptosoft Mini Analysis 6.0.7. For pharmacological experiments, 1 µM oxytocin (Tocris 1910) was dissolved into ACSF and perfused into the bath solution. Atosiban (Sigma A3480) was perfused at 10 µM into the bath solution to antagonize Oxtr receptors. For miniature or evoked EPSC analysis, the detection threshold detection was set at 4 pA. All recorded traces were observed by eye, and artifacts were corrected as necessary.
To assess the AMPA:NMDA ratio, we used “high” (4 mM) Mg bath solution to ensure a block of NMDA receptors at negative membrane voltage. As with miniEPSC recordings, bicuculine was added to inhibit GABAergic current. To assess the AMPA receptor component of synaptic response, cells were held at −70 mV, and EPSCs were repeatedly evoked by 470-nm LED and 1-msec light pulses every 30 sec for 5 min. The resulting current was analyzed for maximal amplitude, decay time, and success:failure ratio. The holding voltage was then changed to +40 mV, and repeated 470-nm light pulses of the same intensity were administered at the same rate to evoke outward current, representing a mixed AMPA and NMDA postsynaptic response. In a series of preliminary recordings using specific NMDA and AMPA receptor blockers, we confirmed that (1) no currents other than AMPA and NMDA were evoked under the described stimulation protocol, and (2) pharmacologically isolated AMPA current had a similar inactivation time at −70 mV and +40 mV. Consequently, we measured the NMDA current at the time interval when the AMPA current (judging by corresponding traces acquired at −70 mV) decayed to at least 20% of maximum.
AAVs, lentiviruses, and stereotactic injections
Male and female 6- to 10-wk-old Oxtr-flox (Jax 008471) mice were used for all loss-of-function morphology studies. All AAVs and lentiviruses were cloned and packaged in-house unless otherwise noted. In vivo viral transduction was performed via bilateral stereotactic injections with all listed coordinates relative to bregma. For the loss-of-function survival experiment, we injected 50 nL of AAV-Ef1a-Cre-H2B::mRuby2 and AAV-Ef1a-H2B::mVenus three times into the RMS at coordinates relative to bregma (ML: ± −0.80, AP: 2.58, DV: −3.62). For morphology, we injected 12 nL of Lenti-Ef1a-EGFP-IRES-Cre and Lenti-Ef1a-tdTomato into the RMS 10 times. For AMPA/NMDA recordings, we injected 12 nL of Lenti-Ef1a-tdTomato-P2A-Cre and Lenti-Ef1a-BFP into the RMS 10 times to label abGCs. All analyses and experimentation after viral injections were performed 28 d after injections to ensure full abGC integration. To ensure that differential and uncontrollable lentiviral integration did not affect survival of abGCs, we turned to AAVs for cell survival experiments. We used both platforms and saw no significant differences between the two but chose the AAV platform for survival and lentiviruses for reconstruction, as lentiviruses more faithfully reconstructed 3D volumes in our hands.
Cryosectioning and immunohistochemistry
All tissue used for IHC or morphology analyses were harvested from mice sacrificed by deep anesthesia using isoflurane. Mice were first transcardially perfused with 10 mL of phosphate-buffered saline (PBS) followed by 10 mL of 4% paraformaldehyde (PFA; Electron Microscopy Sciences 15710). Brains were then fixed for an additional 3 h at 4°C in 4% PFA before being transferred to a 30% sucrose solution for cryoprotection. After cryoprotecting overnight, OBs were frozen in OCT and sliced into sections ≤40-µm on a cryostat (Leica CM1860). Tissue to be used in IHC was prepared by blocking in 10% NGS, 0.1% Triton X-100, and 0.3 M glycine for 2 h at room temperature. The tissue was then stained with primary antibody using the above-mentioned dilutions overnight at 4°C, diluted in blocking solution. Primary antibody was washed three times with PBST. The tissue was then incubated in secondary antibodies (Thermo Fisher A32731, A32740, and A32733) at 1:500 for 2 h at room temperature. Finally, the tissue was washed three times with PBST and mounted onto slides suitable for confocal microscopy.
Single-cell sequencing and data analysis
All animals were deeply anesthetized with isoflurane and perfused intracardially with PBS. Olfactory bulbs were dissected, and tissue was dissociated using the 10X Genomics Chromium dissociation protocol. Dissociated cells were counted using a hemocytometer and stained with trypan blue to visualize debris and dead cells. Cells were loaded onto a 10X Genomics Chromium instrument for preparation of cell-barcoded bead complexes. Libraries were then generated using the 10X Chromium single cell 30 v2 reagent kit according to the manufacturer's instructions and sequenced on an Illumina NextSeq 500. After sequencing, reads were aligned using 10X Genomics Cell Ranger software and exported as a digital gene expression matrix. The expression matrix was then loaded into the single-cell sequencing analysis package Seurat v4.1.0 (Hao et al. 2021). Briefly, quality control was first performed by removing doublets, cells with >20% mitochondrial gene expression, and cells with <200 genes expressed. Samples from each experimental group were first normalized using regularized negative binomial regressing and integrated based on common anchors to normalize data from multiple samples into a single reference as previously described (Hafemeister and Satija 2019; Stuart et al. 2019). Principle component analysis (PCA) was then performed, followed by uniform manifold projection for graph-based clustering and two-dimensional visualization.
Pseudo-time line analysis was performed by first inputting the normalized single-cell data set into the Monocole3 environment using the SeuratWrapper R package (Cao et al. 2019). Cells were ordered with starting clusters based on expression of immature abGC markers. Monocle3 pseudotime information was saved in the original Seurat object metadata and used for further visualization.
χ2 analysis was performed using R via Pearson's χ2 test to first calculate continuous χ2 values, with a P-value cutoff of 0.05. We compared Cre groups versus Flox and WT and then assigned discrete designations to create a discrete scaling of continuous data for easier visualization, with resulting residuals binned by sign (i.e., residual >0 became “increased,” and residual <0 became “decreased”). Binned residuals were then visualized using the existing UMAP reduction.
Differentially expressed genes were determined using the Seurat in-package function FindMarkers. Arguments for FindMarkers function that are different from default were as follows: FindMarkers(logfc.threshold = 0.25, test.use = “DESeq2,” min.pct = “0.1”). Subsequent differentially expressed genes were subjected to GO enrichment using EnrichR (Xie et al. 2021).
Ribotag RNA immunoprecipitation and RNA sequencing analysis
RNA immunoprecipitation
Dlx1/2-Cre;Rpl22-HA/+ and Dlx1/2-Cre;Rpl22-HA/+;Oxtrfl/fl animals were gavaged orally with 200 mg/kg tamoxifen (Sigma T5648-1G) dissolved in corn oil. Four weeks after oral gavage, animals were deeply anesthetized using isoflurane. Olfactory bulbs were quickly dissected and immediately transferred to ice-cold HB-S buffer for subsequent immunoprecipitation and preservation of RNA as previously described (Sanz et al. 2019). Samples were mixed gently on a rotator for no more than 14 h after time of dissection. Subsequent immunoprecipitated RNA was sent for sequencing and quality control.
RNA sequencing and analysis
The following critical commercial assays were used: Trio RNA-seq system (NuGEN 0507-96), standard sensitivity NGS fragment analysis kit (Agilent DNF-473-0500), Quant-it dsDNA assay kit (Thermo Fisher Q33120), and High-Output v2 kit (Illumina FC-404-2002).
For RNA-seq library preparation and sequencing, input of ∼30 ng of total RNA was used for cDNA synthesis and construction of Illumina sequencing libraries using the Trio RNA-seq system (NuGEN 0507-96) according to the manufacturer's instructions. The resulting libraries were validated based on size using the standard sensitivity NGS fragment analysis kit (Agilent DNF-473-0500 [formerly AATI]) on a 12-capillary fragment analyzer and quantified using Quant-it dsDNA assay kit (Q33120). Equimolar concentrations (2 nM) of each library were pooled, denatured, and diluted to a final 1.3 pM concentration. Library pools were subjected to paired-end (2 × 75) sequencing of ∼30 million reads per sample using the High-Output v2 kit (Illumina FC-404-2002) on a NextSeq 500 according to the manufacturer's instructions.
Sequencing files from each flow cell lane were downloaded, and the resulting FastQ files were merged and quality-controlled using FastQC (v0.10.1). Paired-end reads were aligned to the mouse genome (mm10) using STAR (v2.5.0a) (Dobin et al. 2013). The resulting BAM files were used to build count matrices in R (v3.5.2) with Bioconductor packages GenomicAlignments (v1.16.0) and GenomicFeatures (v1.32.2) (Lawrence et al. 2013).
Gene counts obtained from STAR aligner were used for unsupervised clustering and differential gene expression (DGE) analyses. Genes with a total read count of <10 across all samples (by sex) were excluded, resulting in count data for 17,020 genes in females and 16,325 genes in males. Raw counts were normalized for library depth and composition using the DESeq2 median to ratio method (Love et al. 2014). DGE analyses were performed using default parameters in DESeq2, and Benjamini–Hochberg (BH)-adjusted P-values were used to define DGE at P < 0.05. DGE comparisons were performed within respective sexes and batch-adjusted by linear regression. Gene enrichment in Ribotag immunoprecipitation (IP) were determined by DGE analysis between control IP and whole-sample input. Genes depleted in control IP when compared with input (log2 fold change < 0, P < 0.05) were considered background and removed, with 1794 genes depleted in females and 1685 genes depleted in males. DGE analysis between control and oxytocin KO were performed without background genes. To evaluate sample heterogeneity, normalized gene counts were rlog transformed (DESeq2), and samples were embedded in 2D space via uniform manifold approximation and projection (UMAP) (Becht et al. 2019). Hierarchical clustering was performed to organize samples in heat maps.
Behavior and analyses
Tamoxifen administration for go/‘no go’
All experimental animals for behavior were 8- to 10-wk-old male and female adult mice. Animals were fed tamoxifen chow formulated for a 250 mg/kg per d tamoxifen diet, assuming a 20- to 25-g body weight and 3- to 4-g intake (Teklad Global TD.130855) for 4 wk mixed with standard chow.
Go/‘no go’ training
Freely moving mice were trained on an olfactory-cued go/“no go” discrimination task using olfactory conditioning boxes (Med Associates) controlled by MedPC software as previously described (Liu et al. 2018). Conditioning boxes included one odor delivery port and one water reward port, both of which were equipped with IR beams to record port entries. The odor delivery port received a constant stream of clean room air, cleared by a vacuum. Odors were injected into the steady air stream by opening valves to one of two odor reservoirs, allowing the air stream to pass through the head space of the reservoir carrying the volatized odorant. Reservoirs were 20 mL in volume and included 2.5 mL of odorant diluted to 1% by volume in mineral oil. Water rewards were dispensed at 5 µL in volume in the separate reward port upon reward port entry only on correctly decided hit trials. Conditioning boxes were housed within larger behavior isolation boxes (Med Associates). Box fans in the isolation boxes continuously circulated ambient air away from the conditioning boxes and out of the isolation boxes to reduce the accumulation of ambient odor.
Over the course of 10–14 d, mice underwent five stages of behavior shaping. Mice were water-restricted to no less than 85% of their body weight during the 48 h prior to the start of training. Mice were first trained to seek water rewards and then to initiate trials by poking their noses into the odor port. Mice were then trained to respond to an S+ odor cue (1% eugenol in mineral oil) to receive a water reward. Upon forming this association, the S− odor cue (1% methylsalicylate in mineral oil) was introduced, in which mice were trained to refrain from seeking a reward and instead initiate another trial. Trials required mice to sample the odor cues for ≥100 msec before responding to the cue and were categorized based on the S+/− odor cue and the mouse's response. When mice were presented with the S+ odor cue, trials were considered “hits” when mice sought a water reward within 5 sec and “misses” if mice did not enter the reward port. Trials in which the S− odor cue was delivered were considered “false alarms” when mice incorrectly attempted to retrieve water and “correct rejects” if mice did not seek water. False alarms resulted in a 4-sec time-out punishment. Accuracy was calculated as percent correct trials (hit + correct reject trials) for blocks of 20 trials. For each training session, mice completed 10–20 blocks. Training was considered complete after mice completed a session with the training odors (eugenol [S+] and methysalicylate [S−]) in which they achieved >85% accuracy in at least two consecutive blocks. Testing sessions followed the same pattern as the last stage of training, but new odor pairs were introduced, requiring mice to learn new S+ and S− odor associations.
Go/‘no go’ testing
Novel odor pairs were presented on consecutive days until mice achieved at least two consecutive blocks of 85% accuracy (when the odor pair was considered learned) or until the mice failed to learn for three consecutive days. Sessions of the same odor pair were then concatenated into a single session spanning up to 3 d. Concatenated sessions were excluded from analysis if mice failed to complete at least 100 trials. Behavior was quantified using the signal detection theory measures d′ [d′ = Z(hit rate) − Z(false alarm rate)] and criterion {C = −1[Z(hit rate) + Z(false alarm rate)]} (Stanislaw and Todorov 1999), where d′ measures the sensitivity of discrimination and reflects overall task performance, and C measures discrimination bias. More negative C values reflect a bias toward reward seeking, C values close to zero reflect optimal decision bias, and positive C values reflect bias away from reward seeking in favor of trial reinitiation. To show change in sensitivity over time within a session, d′ was calculated over a rolling window of 40 trials. For experimental and control mice, d′ and C were calculated for each odor pair, and values were averaged for each mouse.
Three-chamber test for sociability
All behavioral testing was performed on 7- to 12-wk-old male and female mice. Testing was performed as previously described (Buffington et al. 2016; Sgritta et al. 2019). Mice were initially habituated for 10 min in an empty 60 × 40 × 23-cm Plexiglass arena separated into three connected chambers referred to here as “left,” “center,” and “right.” Baseline sociability was analyzed with a 10-min period in which the subject could interact either with an empty cup or a cup containing a genotype-, age-, and sex-matched stranger conspecific. The interaction time was quantified by measuring the time the subject mouse spent sniffing or climbing on the empty cup or the cup with the stranger conspecific, counterbalancing the position of the empty cup/stranger mouse between trials. Preference for social novelty was determined using a 10-min interval by introducing a second stranger mouse into the previously empty cup. Finally, the reciprocal interaction time was measured in the same way for a 10-min period, but this time the interactions between mice of the same genotype were measured. The time spent interacting with either stranger mice, conspecific mice, or the empty cups was recorded and measured using AnyMaze software. The independent human observers were blind to genotype groups.
Software
The following software was used in this study: Seurat (v4.1.0; https://github.com/satijalab/seurat) (Hao et al. 2021), SeuratWrapper (https://github.com/satijalab/seurat-wrappers), Monocle3 (1.0.0; https://cole-trapnell-lab.github.io/monocle3) (Cao et al. 2019), fastQC (v0.10.1; https://www.bioinformatics.babraham.ac.uk/projects/fastqc), STAR (v2.5.0a; https://github.com/alexdobin/STAR) (Dobin et al. 2013), GenomicAlignments (v1.16.0; https://bioconductor.org/packages/release/bioc/html/GenomicAlignments.html) (Lawrence et al. 2013), and GenomicFeatures (v1.32.2; https://bioconductor.org/packages/release/bioc/html/GenomicFeatures.html) (Lawrence et al. 2013).
All scripts are available on request
Microscopy and imaging
Confocal imaging was performed on a Leica confocal SP8X equipped with either a 20×/0.75 NA dry objective or a 63×/1.4 NA oil objective. Briefly, images are acquired using Leica LAS X software with 3-µm step sizes between optical sections. For morphology, tissue was imaged with a fully closed pinhole and 0.5-µm step sizes to ensure optimal z-capture and faithful 3D reconstruction of dendritic spines. For IHC, cell counting, and other tracing experiments, images were acquired at 20× magnification with 5-µm step sizes.
Image processing and analysis
Images were processed using Imaris version 9.7.2 (Bitplane Software). Leica image files (LIF) were first preprocessed using the background subtraction filter within Imaris image processing. For dendritic morphology and spine reconstruction, we used the filament tracer extension to first trace the dendritic arbors of abGCs with the autopath function. Next, dendrite diameters and center points were recalculated via local contrast thresholding. Finally, dendritic spines were manually assigned to the new filament by using a combination of the autopath and autodepth features. After manual assignment of spines, the spine diameters were recomputed using local contrast thresholding. After complete reconstruction of dendritic trees, data were exported in CSV format directly from Imaris and imported into GraphPad Prism 9 software for graphing and statistical tests.
Supplementary Material
Acknowledgments
This work was supported by the McNair Medical Institute, a National Institute of Neurological Disorders and Stroke grant (R01NS078294) to B.R.A., and an American Heart Association grant to P.J.H. We thank the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (National Institute of Child Health grant U54HD083092) for viral packaging. We thank Dr. Matthew McGinley, Dr. Burak Tepe, Dr. Kevin Ung, and Dr. Juan Romero for critical review of the manuscript.
Author contributions: B.T.P. performed the vector design and cloning, stereotactic injections, confocal imaging, CLARITY brain clearing, single-cell sequencing analysis, Ribotag RNA immunoprecipitation and analysis, Imaris image quantification, immunohistochemistry, and analysis; designed the experiments; and wrote the manuscript with help from the other authors. M.K. performed electrophysiological recordings. B.L. and T.W. performed RNA sequencing and data analysis and helped with figure design. M.C.H., B.T.P., and B.T. performed the single-cell RNA sequencing and data analysis. P.J.H. helped with Ribotag immunoprecipitation, figure design, and critical manuscript edits. B.T. helped with vector design, cloning, and conceptualization of experiments. S.W.D., E.H.M., and E.K.T. performed behavior experiments and helped with analysis. J.L.S. helped with vector design, viral injections, and critical manuscript edits. M.P. assisted with experimental design and with conducting monosynaptic tracing and viral targeting experiments. J.M.R. and B.D.W.B. helped with viral tracing and oxytocin receptor expression quantification experiments. S.B. helped with viral olfactory bulb injections and quantification. B.T.P. and B.R.A. conceived the project, helped with experimental design, and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349930.122.
Freely available online through the Genes & Development Open Access option.
Competing interests statement
The authors declare no competing interests.
References
- Abrous DN, Koehl M, Le Moal M. 2005. Adult neurogenesis: from precursors to network and physiology. Physiol Rev 85: 523–569. 10.1152/physrev.00055.2003 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Temple S. 1998. Stem cells in the developing and adult nervous system. J Neurobiol 36: 105–110. [DOI] [PubMed] [Google Scholar]
- Andreae LC, Burrone J. 2018. The role of spontaneous neurotransmission in synapse and circuit development. J Neurosci Res 96: 354–359. 10.1002/jnr.24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X, Liu Y, Qin M, Li C, Chen X, Xiao L, Liu J. 2014. Expression of Dbn1 during mouse brain development and neural stem cell differentiation. Biochem Biophys Res Commun 449: 81–87. 10.1016/j.bbrc.2014.04.152 [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. 2007. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54: 205–218. 10.1016/j.neuron.2007.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Hasegawa H, Yi JJ, Larsen RS, Wallace ML, Philpot BD, Wang F, Ehlers MD. 2011. Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PLoS One 6: e29423. 10.1371/journal.pone.0029423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand EJ, Li J, Xie F, Luo C, Mukamel EA. 2021. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron 109: 11–26. 10.1016/j.neuron.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakos J, Srancikova A, Havranek T, Bacova Z. 2018. Molecular mechanisms of oxytocin signaling at the synaptic connection. Neural Plast 2018: 4864107. 10.1155/2018/4864107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu R, Pressler RT, Strowbridge BW. 2007. Multiple modes of synaptic excitation of olfactory bulb granule cells. J Neurosci 27: 5621–5632. 10.1523/JNEUROSCI.4630-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Close J, Machold R, Fishell G. 2008. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci 28: 3966–3975. 10.1523/JNEUROSCI.5625-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. 2016. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165: 1762–1775. 10.1016/j.cell.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566: 496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcea I, Caraballo NL, Marlin BJ, Ooyama R, Riceberg JS, Mendoza Navarro JM, Opendak M, Diaz VE, Schuster L, Alvarado Torres MI, et al. 2021. Oxytocin neurons enable social transmission of maternal behaviour. Nature 596: 553–557. 10.1038/s41586-021-03814-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton A, Petreanu LT, Lansford R, Alvarez-Buylla A, Lledo P-M. 2003. Becoming a new neuron in the adult olfactory bulb. Nat Neurosci 6: 507–518. 10.1038/nn1048 [DOI] [PubMed] [Google Scholar]
- Castel M, Morris JF. 1988. The neurophysin-containing innervation of the forebrain of the mouse. Neuroscience 24: 937–966. 10.1016/0306-4522(88)90078-4 [DOI] [PubMed] [Google Scholar]
- Chater TE, Goda Y. 2014. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front Cell Neurosci 8: 401. 10.3389/fncel.2014.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. EnrichR: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Hen R. 2005. Young and excitable: the function of new neurons in the adult mammalian brain. Curr Opin Neurobiol 15: 121–128. 10.1016/j.conb.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim T-K, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. 2006. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311: 1008–1012. 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Garcia I, Quast KB, Huang L, Herman AM, Selever J, Deussing JM, Justice NJ, Arenkiel BR. 2014. Local CRH signaling promotes synaptogenesis and circuit integration of adult-born neurons. Dev Cell 30: 645–659. 10.1016/j.devcel.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Bhullar PK, Tepe B, Ortiz-Guzman J, Huang L, Herman AM, Chaboub L, Deneen B, Justice NJ, Arenkiel BR. 2016. Local corticotropin releasing hormone (CRH) signals to its receptor CRHR1 during postnatal development of the mouse olfactory bulb. Brain Struct Funct 221: 1–20. 10.1007/s00429-014-0888-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpl G, Fahrenholz F. 2001. The oxytocin receptor system: structure, function, and regulation. Physiol Rev 81: 629–683. 10.1152/physrev.2001.81.2.629 [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925. 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- Grubb MS, Nissant A, Murray K, Lledo P-M. 2008. Functional maturation of the first synapse in olfaction: development and adult neurogenesis. J Neurosci 28: 2919–2932. 10.1523/JNEUROSCI.5550-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastella AJ, Hickie IB. 2016. Oxytocin treatment, circuitry, and autism: a critical review of the literature placing oxytocin into the autism context. Biol Psychiatry 79: 234–242. 10.1016/j.biopsych.2015.06.028 [DOI] [PubMed] [Google Scholar]
- Hafemeister C, Satija R. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20: 296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson E, Swanson J, Arenkiel BR. 2020. GABAergic input from the basal forebrain promotes the survival of adult-born neurons in the mouse olfactory bulb. Front Neural Circuits 14: 17. 10.3389/fncir.2020.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson E, Brandel-Ankrapp KL, Arenkiel BR. 2021. Dynamic cholinergic tone in the basal forebrain reflects reward-seeking and reinforcement during olfactory behavior. Front Cell Neurosci 15: 635837. 10.3389/fncel.2021.635837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell 184: 3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng C-Y, Blankenship SM, Roche KW, Nicoll RA. 2013. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77: 1083–1096. 10.1016/j.neuron.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. 2013. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80: 648–657. 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Martin KC, Schuman EM. 2019. Local translation in neurons: visualization and function. Nat Struct Mol Biol 26: 557–566. 10.1038/s41594-019-0263-5 [DOI] [PubMed] [Google Scholar]
- Horger BA, Nishimura MC, Armanini MP, Wang LC, Poulsen KT, Rosenblad C, Kirik D, Moffat B, Simmons L, Johnson E, et al. 1998. Neurturin exerts potent actions on survival and function of midbrain dopaminergic neurons. J Neurosci 18: 4929–4937. 10.1523/JNEUROSCI.18-13-04929.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Ung K, Garcia I, Quast KB, Cordiner K, Saggau P, Arenkiel BR. 2016. Task learning promotes plasticity of interneuron connectivity maps in the olfactory bulb. J Neurosci 36: 8856–8871. 10.1523/JNEUROSCI.0794-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim MF, Williams KR, Bassell GJ. 2015. Single-molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci 35: 7116–7130. 10.1523/JNEUROSCI.2802-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurek B, Slattery DA, Hiraoka Y, Liu Y, Nishimori K, Aguilera G, Neumann ID, van den Burg EH. 2015. Oxytocin regulates stress-induced Crf gene transcription through CREB-regulated transcription coactivator 3. J Neurosci 35: 12248–12260. 10.1523/JNEUROSCI.1345-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri H, Pallotto M, Nissant A, Murray K, Sassoè-Pognetto M, Lledo P-M. 2011. Dynamic development of the first synapse impinging on adult-born neurons in the olfactory bulb circuit. Neural Syst Circuits 1: 6. 10.1186/2042-1001-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh AC, Young LJ. 2011. Increasing oxytocin receptor expression in the nucleus accumbens of pre-pubertal female prairie voles enhances alloparental responsiveness and partner preference formation as adults. Horm Behav 60: 498–504. 10.1016/j.yhbeh.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Lin C-W, Lois C. 2008. Sequential development of synapses in dendritic domains during adult neurogenesis. Proc Natl Acad Sci 105: 16803–16808. 10.1073/pnas.0807970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Lin C-W, Mosley CP, Lois C. 2009. A critical period for activity-dependent synaptic development during olfactory bulb adult neurogenesis. J Neurosci 29: 11852–11858. 10.1523/JNEUROSCI.2406-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel H, Schechtman E, Groysman M, Mizrahi A. 2012. Enhanced synaptic integration of adult-born neurons in the olfactory bulb of lactating mothers. J Neurosci 32: 7519–7527. 10.1523/JNEUROSCI.6354-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk Y, Homma R, Liang Y, Maslyukov A, Hermes M, Thestrup T, Griesbeck O, Ninkovic J, Cohen LB, Garaschuk O. 2015. In vivo odourant response properties of migrating adult-born neurons in the mouse olfactory bulb. Nat Commun 6: 6349. 10.1038/ncomms7349 [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepousez G, Nissant A, Bryant AK, Gheusi G, Greer CA, Lledo P-M. 2014. Olfactory learning promotes input-specific synaptic plasticity in adult-born neurons. Proc Natl Acad Sci 111: 13984–13989. 10.1073/pnas.1404991111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner B, Caponiti JM, Gould E. 2012. Oxytocin stimulates adult neurogenesis even under conditions of stress and elevated glucocorticoids. Hippocampus 22: 861–868. 10.1002/hipo.20947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P-Y, Chiu Y-M, Yu J-H, Chen S-K. 2020. Mapping central projection of oxytocin neurons in unmated mice using Cre and alkaline phosphatase reporter. Front Neuroanat 14: 559402. 10.3389/fnana.2020.559402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-T, Chen C-C, Huang C-C, Nishimori K, Hsu K-S. 2017. Oxytocin stimulates hippocampal neurogenesis via oxytocin receptor expressed in CA3 pyramidal neurons. Nat Commun 8: 537. 10.1038/s41467-017-00675-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-T, Hsieh T-Y, Tsai T-C, Chen C-C, Huang C-C, Hsu K-S. 2018. Conditional deletion of hippocampal CA2/CA3a oxytocin receptors impairs the persistence of long-term social recognition memory in mice. J Neurosci 38: 1218–1231. 10.1523/JNEUROSCI.1896-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Harikka J, da Penha Berzaghi M, Castrén E, Tzimagiorgis G, Hughes RA, Thoenen H. 1994. Fibroblast growth factor-5 promotes differentiation of cultured rat septal cholinergic and raphe serotonergic neurons: comparison with the effects of neurotrophins. Eur J Neurosci 6: 244–252. 10.1111/j.1460-9568.1994.tb00267.x [DOI] [PubMed] [Google Scholar]
- Liu G, Patel JM, Tepe B, McClard CK, Swanson J, Quast KB, Arenkiel BR. 2018. An objective and reproducible test of olfactory learning and discrimination in mice. J Vis Exp 2018: e57142. 10.3791/57142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. 1994. Long-distance neuronal migration in the adult mammalian brain. Science 264: 1145–1148. 10.1126/science.8178174 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. 1998. Dendritic release of vasopressin and oxytocin. J Neuroendocrinol 10: 881–895. 10.1046/j.1365-2826.1998.00279.x [DOI] [PubMed] [Google Scholar]
- Ludwig M, Leng G. 2006. Dendritic peptide release and peptide-dependent behaviours. Nat Rev Neurosci 7: 126–136. 10.1038/nrn1845 [DOI] [PubMed] [Google Scholar]
- Luskin MB. 1993. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron 11: 173–189. 10.1016/0896-6273(93)90281-U [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning M, Stoev S, Chini B, Durroux T, Mouillac B, Guillon G. 2008. Peptide and non-peptide agonists and antagonists for the vasopressin and oxytocin V1a, V1b, V2 and OT receptors: research tools and potential therapeutic agents. Prog Brain Res 170: 473–512. 10.1016/S0079-6123(08)00437-8 [DOI] [PubMed] [Google Scholar]
- Marlin BJ, Mitre M, D'amour JA, Chao MV, Froemke RC. 2015. Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature 520: 499–504. 10.1038/nature14402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G, Song H. 2005. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci 28: 223–250. 10.1146/annurev.neuro.28.051804.101459 [DOI] [PubMed] [Google Scholar]
- Mitre M, Marlin BJ, Schiavo JK, Morina E, Norden SE, Hackett TA, Aoki CJ, Chao MV, Froemke RC. 2016. A distributed network for social cognition enriched for oxytocin receptors. J Neurosci 36: 2517–2535. 10.1523/JNEUROSCI.2409-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monory K, Massa F, Egertová M, Eder M, Blaudzun H, Westenbroek R, Kelsch W, Jacob W, Marsch R, Ekker M, et al. 2006. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron 51: 455–466. 10.1016/j.neuron.2006.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Trainor BC. 2007. Neural mechanisms of aggression. Nat Rev Neurosci 8: 536–546. 10.1038/nrn2174 [DOI] [PubMed] [Google Scholar]
- Neumann ID. 2008. Brain oxytocin: a key regulator of emotional and social behaviours in both females and males. J Neuroendocrinol 20: 858–865. 10.1111/j.1365-2826.2008.01726.x [DOI] [PubMed] [Google Scholar]
- Nunes AR, Carreira L, Anbalagan S, Blechman J, Levkowitz G, Oliveira RF. 2020. Perceptual mechanisms of social affiliation in zebrafish. Sci Rep 10: 3642. 10.1038/s41598-020-60154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettl L-L, Kelsch W. 2018. Oxytocin and olfaction. Curr Top Behav Neurosci 35: 55–75. 10.1007/7854_2017_8 [DOI] [PubMed] [Google Scholar]
- Oettl L-L, Ravi N, Schneider M, Scheller MF, Schneider P, Mitre M, da Silva Gouveia M, Froemke RC, Chao MV, Young WS, et al. 2016. Oxytocin enhances social recognition by modulating cortical control of early olfactory processing. Neuron 90: 609–621. 10.1016/j.neuron.2016.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotto M, Deprez F. 2014. Regulation of adult neurogenesis by GABAergic transmission: signaling beyond GABAA-receptors. Front Cell Neurosci 8: 166. 10.3389/fncel.2014.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotto M, Nissant A, Fritschy J-M, Rudolph U, Sassoè-Pognetto M, Panzanelli P, Lledo P-M. 2012. Early formation of GABAergic synapses governs the development of adult-born neurons in the olfactory bulb. J Neurosci 32: 9103–9115. 10.1523/JNEUROSCI.0214-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzanelli P, Bardy C, Nissant A, Pallotto M, Sassoè-Pognetto M, Lledo P-M, Fritschy J-M. 2009. Early synapse formation in developing interneurons of the adult olfactory bulb. J Neurosci 29: 15039–15052. 10.1523/JNEUROSCI.3034-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Alvarez-Buylla A. 2002. Maturation and death of adult-born olfactory bulb granule neurons: role of olfaction. J Neurosci 22: 6106–6113. 10.1523/JNEUROSCI.22-14-06106.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef R, Kudryavitskaya E, Shani-Narkiss H, Isaacson B, Rimmerman N, Mizrahi A, Yirmiya R. 2017. The role of microglia and their CX3CR1 signaling in adult neurogenesis in the olfactory bulb. Elife 6: e30809. 10.7554/eLife.30809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes CH, Morrell JI, Pfaff DW. 1981. Immunohistochemical analysis of magnocellular elements in rat hypothalamus: distribution and numbers of cells containing neurophysin, oxytocin, and vasopressin. J Comp Neurol 198: 45–64. 10.1002/cne.901980106 [DOI] [PubMed] [Google Scholar]
- Ripamonti S, Ambrozkiewicz MC, Guzzi F, Gravati M, Biella G, Bormuth I, Hammer M, Tuffy LP, Sigler A, Kawabe H, et al. 2017. Transient oxytocin signaling primes the development and function of excitatory hippocampal neurons. Elife 6: e22466. 10.7554/eLife.22466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez-Castelao B, Heckel A, Schuman EM. 2017. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 355: 634–637. 10.1126/science.aaf8995 [DOI] [PubMed] [Google Scholar]
- Sánchez-Vidaña DI, Chan N-MJ, Chan AHL, Hui KKY, Lee S, Chan H-Y, Law YS, Sze MY, Tsui W-CS, Fung TKH, et al. 2016. Repeated treatment with oxytocin promotes hippocampal cell proliferation, dendritic maturation and affects socio–emotional behavior. Neuroscience 333: 65–77. 10.1016/j.neuroscience.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. 2009. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci 106: 13939–13944. 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Bean JC, Carey DP, Quintana A, McKnight GS. 2019. RiboTag: ribosomal tagging strategy to analyze cell-type-specific mRNA expression in vivo. Curr Protoc Neurosci 88: e77. 10.1002/cpns.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgritta M, Dooling SW, Buffington SA, Momin EN, Francis MB, Britton RA, Costa-Mattioli M. 2019. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101: 246–259.e6. 10.1016/j.neuron.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE. 2016. Dynamic axonal translation in developing and mature visual circuits. Cell 166: 181–192. 10.1016/j.cell.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Sümbül U, Graybuck LT, Collman F, Seshamani S, Gala R, Gliko O, Elabbady L, Miller JA, Bakken TE, et al. 2019. Single-cell transcriptomic evidence for dense intracortical neuropeptide networks. Elife 8: e47889. 10.7554/eLife.47889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Hawrylycz M, Rossier J, Sümbül U. 2020. New light on cortical neuropeptides and synaptic network plasticity. Curr Opin Neurobiol 63: 176–188. 10.1016/j.conb.2020.04.002 [DOI] [PubMed] [Google Scholar]
- Stanislaw H, Todorov N. 1999. Calculation of signal detection theory measures. Behav Res Methods Instrum Comput 31: 137–149. 10.3758/BF03207704 [DOI] [PubMed] [Google Scholar]
- Stoop R. 2012. Neuromodulation by oxytocin and vasopressin. Neuron 76: 142–159. 10.1016/j.neuron.2012.09.025 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepe B, Hill MC, Pekarek BT, Hunt PJ, Martin TJ, Martin JF, Arenkiel BR. 2018. Single-cell RNA-seq of mouse olfactory bulb reveals cellular heterogeneity and activity-dependent molecular census of adult-born neurons. Cell Rep 25: 2689–2703.e3. 10.1016/j.celrep.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker-Varia S, Krol JJ, Nettleton J, Bilimoria PM, Bangasser DA, Shors TJ, Black IB, Alder J. 2007. The neuropeptide VGF produces antidepressant-like behavioral effects and enhances proliferation in the hippocampus. J Neurosci 27: 12156–12167. 10.1523/JNEUROSCI.1898-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirko NN, Eyring KW, Carcea I, Mitre M, Chao MV, Froemke RC, Tsien RW. 2018. Oxytocin transforms firing mode of CA2 hippocampal neurons. Neuron 100: 593–608.e3. 10.1016/j.neuron.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa K, Iga N, Lu Y-F, Moriwaki A, Matsushita M, Li S-T, Miyamoto O, Itano T, Matsui H. 2003. Oxytocin improves long-lasting spatial memory during motherhood through MAP kinase cascade. Nat Neurosci 6: 384–390. 10.1038/nn1023 [DOI] [PubMed] [Google Scholar]
- Tyzio R, Nardou R, Ferrari DC, Tsintsadze T, Shahrokhi A, Eftekhari S, Khalilov I, Tsintsadze V, Brouchoud C, Chazal G, et al. 2014. Oxytocin-mediated GABA inhibition during delivery attenuates autism pathogenesis in rodent offspring. Science 343: 675–679. 10.1126/science.1247190 [DOI] [PubMed] [Google Scholar]
- van den Pol AN. 2012. Neuropeptide transmission in brain circuits. Neuron 76: 98–115. 10.1016/j.neuron.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J, Lord J, Dissing-Olesen L, Stevens B, Murthy VN. 2020. Microglial depletion disrupts normal functional development of adult-born neurons in the olfactory bulb. Elife 9: e50531. 10.7554/eLife.50531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman MC, Greer CA. 2007. Synaptic integration of adult-generated olfactory bulb granule cells: basal axodendritic centrifugal input precedes apical dendrodendritic local circuits. J Neurosci 27: 9951–9961. 10.1523/JNEUROSCI.1633-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. 2021. Gene set knowledge discovery with EnrichR. Curr Protoc 1: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mori K. 2005. Critical period for sensory experience-dependent survival of newly generated granule cells in the adult mouse olfactory bulb. Proc Natl Acad Sci 102: 9697–9702. 10.1073/pnas.0406082102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Mori K, Nakanishi S. 1995. Refinement of odor molecule tuning by dendrodendritic synaptic inhibition in the olfactory bulb. Proc Natl Acad Sci 92: 3371–3375. 10.1073/pnas.92.8.3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J-J, Li S-J, Zhang X-D, Miao W-Y, Zhang D, Yao H, Yu X. 2014. Oxytocin mediates early experience-dependent cross-modal plasticity in the sensory cortices. Nat Neurosci 17: 391–399. 10.1038/nn.3634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.