Abstract
The DNA-origami technique has enabled the engineering of transmembrane nanopores with programmable size and functionality, showing promise in building biosensors and synthetic cells. However, it remains challenging to build large (>10 nm), functionalizable nanopores that spontaneously perforate lipid membranes. Here, we take advantage of pneumolysin (PLY), a bacterial toxin that potently forms wide ring-like channels on cell membranes, to construct hybrid DNA-protein nanopores. This PLY-DNA-origami complex, in which a DNA-origami ring corrals up to 48 copies of PLY, targets the cholesterol-rich membranes of liposomes and red blood cells, readily forming uniformly-sized pores with an average inner diameter of ~22 nm. Such hybrid nanopores facilitate the exchange of macromolecules between perforated liposomes and their environment, with the exchange rate negatively correlating with the macromolecule size (diameters of gyration: 8–22 nm). Additionally, the DNA ring can be decorated with intrinsically disordered nucleoporins to further restrict the diffusion of traversing molecules, highlighting the programmability of the hybrid nanopores. PLY-DNA pores provide an enabling biophysical tool for studying the cross-membrane translocation of ultra-large molecules and open new opportunities for analytical chemistry, synthetic biology, and nanomedicine.
Introduction
Over the past decade, nanopores have enabled real-time, single-molecule analysis of small molecules, proteins, or nucleic acids.1–5 For example, accurate, ultra-long genome reads provided by the MinION (Oxford Nanopore Technologies) sequencer have become commercially available.6 Nearly all of today’s nanopore implementations can be categorized into two types: synthetic and biological.7, 8 The bulk of synthetic nanopores are solid-state nanopores, typically made by beam milling on silicon chips to achieve pores with controllable geometry.9 Biological pores10,11, exemplified by transmembrane protein channels fabricated through molecular biology methods, allow for site-specific and atomically-precise modifications.9, 12 In addition to enabling nucleic acid and peptide sequencing and label-free biomolecule detection, nanopore technology provides a platform to build devices with sustained directional rotary motions, forming essential building blocks for more advanced nanomachines or synthetic cells.13, 14
Realizing the full potential of nanopore technology relies on the ability to precisely control the nanopore’s physical and biochemical characteristics, including channel diameter and depth, mechanical stiffness, and surface chemistry.7, 8 As an information-rich molecule for building self-assembled nanostructures with programmable geometry, mechanics, and chemical modifications, DNA has emerged as a promising material for nanopore engineering. DNA nanochannels with well-controlled width (0.5–30 nm) can be coupled to solid-state nanopores by an electric current or embedded into the lipid bilayer through hydrophobic interactions.15–23 With the site-specific modification of receptor-specific ligands or mechanical switches, these DNA nanopores have shown molecular gating effects with user-defined selectivity.16, 17, 20, 21, 24
We set out to address a long-standing challenge in nanopore engineering: to readily incorporate wide (>10 nm) DNA channels with customizable functionality into lipid membranes. To date, embedding a highly charged, hydrophilic DNA channel into the hydrophobic core of lipid bilayers was achieved with a high energy input provided by amphipathic membrane anchors (e.g., cholesterol, porphyrin, alkyl chains) displayed on the exterior of the DNA channel.25, 26 The energy requirement grows quadratically with increasing channel width27, necessitating a large number of amphipathic labels on the DNA channel, which brings additional design and experimental challenges to conceal the membrane anchors in order to mitigate hydrophobicity-mediated channel aggregation.16, 17, 22, 23, 28–30 Moreover, these DNA channels often require external stimuli (e.g., electric voltage, solvent, or detergent) to drive membrane insertion or suffer slow pore-forming kinetics. Inspired by previous work that uses DNA scaffold to guide the assembly of protein pores31–33, here we built a DNA-toxin hybrid pore with a rigid, functionalizable DNA-origami channel carrying membrane-targeting pneumolysins (PLYs).34, 35 Powered by the potent PLY toxins, the PLY-DNA complex spontaneously formed large pores (lumen width >20 nm) on cholesterol-rich membranes. Furthermore, the DNA-origami channel promoted the uniform PLY pore size and accommodated additional diffusion-restricting molecules (e.g., Nsp1), thus forming size-selective gatekeepers to control macromolecule flux in and out of a synthetic membrane compartment. By integrating bacterial toxins into DNA nanostructures, our work not only adds a robust ultrawide pore to the DNA nanopore toolkit, but also expands its design space and possible applications.
Results
In our modular hybrid nanopore design, PLY is responsible for perforating the lipid membrane, while the DNA-origami channel displays handles (ssDNA extensions) to corral a defined number of PLYs to limit pore size and host additional modifications. PLY is a cholesterol-dependent cytolysin (CDC),36 a pore-forming protein derived from Streptococcus pneumoniae.34, 35 Structural studies revealed that approximately 42 PLY monomers form a transmembrane channel with an inner diameter of about 25 nm.37 We utilized PLY because its monomers are reasonably stable in solution and remain monodispersed after covalently conjugated to a DNA oligonucleotide (Figure S1). PLY monomers targets cholesterol-rich membranes and form a circular assembly upon membrane binding, which undergoes further conformational changes to form a β-barrel that electrostatically repels lipids to open a pore.37, 38 Additionally, PLY necessitates a lower cholesterol level (30%) than other common CDCs (approximately 50%)39–41, allowing the hybrid nanostructures to function on a broader range of lipid compositions.
To facilitate PLY-DNA conjugation, we recombinantly expressed PLY containing a single cysteine (Cys) residue at the N-terminus within the soluble domain (Figure 1A, sequence in Table S1). Previous studies showed that removing the only internal Cys could reduce its cytotoxicity, but the Cys-free PLY retained the oligomerization and membrane insertion behaviors of the wild-type protein.42, 43 We then chemically crosslinked the N-terminal Cys of PLY to a 5’-amino-modified DNA oligonucleotide (termed “anti-handle” for its sequence complementarity to the DNA handles) and purified the anti-handle-conjugated PLY by size exclusion chromatography (SEC) to remove unreacted oligonucleotides (Figure S1).
Figure 1: The construction and membrane-insertion activities of PLY-DNA rings.
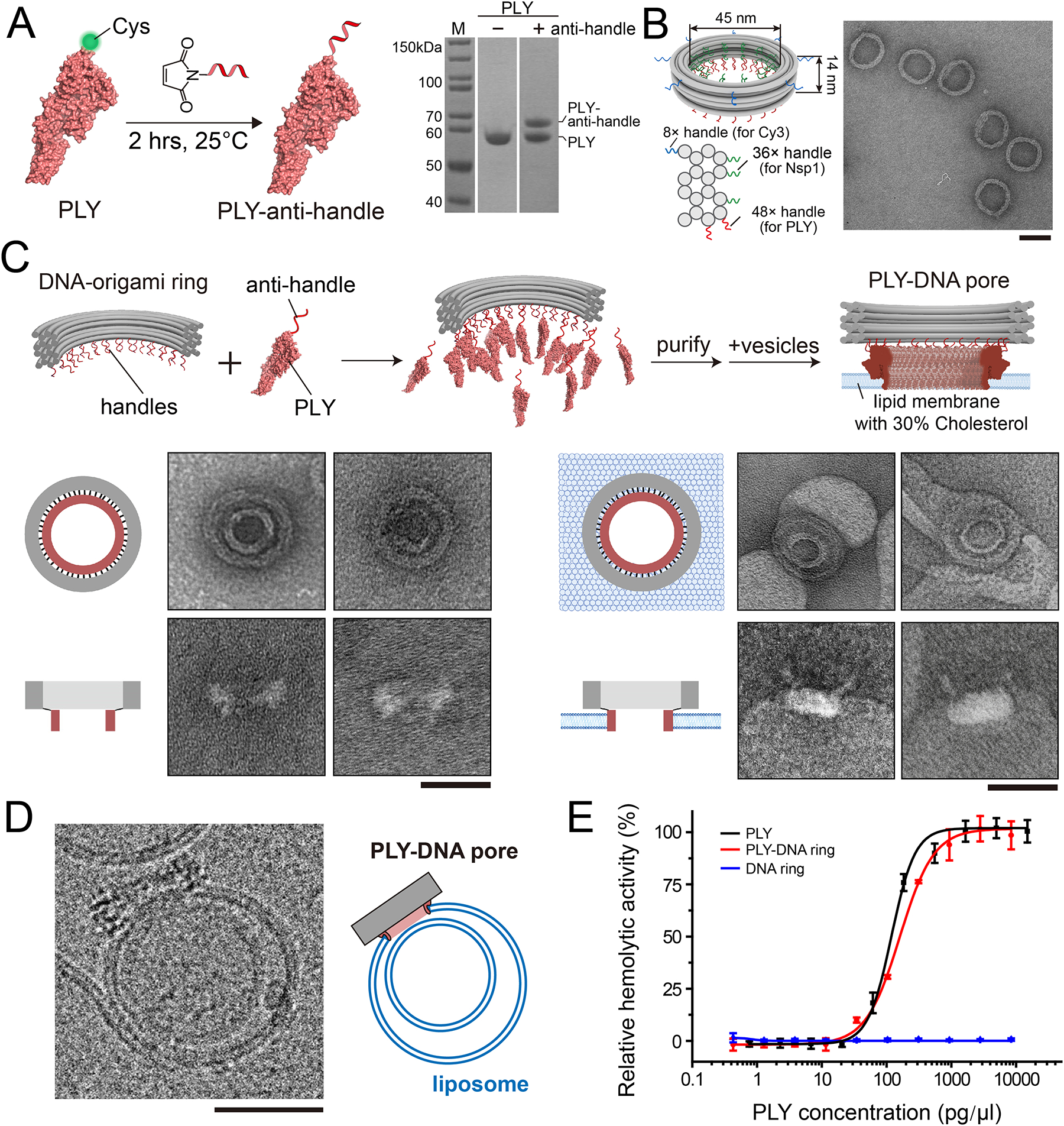
(A) Schematical illustration of PLY-anti-handle conjugation (left) and SDS-PAGE assay (right) showing the gel mobility shift of PLY before and after ssDNA anti-handle conjugation.
(B) Cartoon models showing the dimensions and handle positions of the DNA-origami ring (left) and a negative-stain TEM micrograph of the purified DNA rings. Three sets of handles with orthogonal sequences (blue, green, and red curls) were strategically placed for attaching fluorophores and proteins via handle/anti-handle hybridization. Scale bar: 50 nm.
(C) Schematics and negative-stain EM micrographs showing the assembly of PLY-modified DNA-origami rings and their membrane insertion. Scale bars: 50 nm.
(D) A cryo-EM image showing a liposome perforated by a DNA-origami-corralled PLY pore. Scale bar: 50 nm.
(E) Sheep erythrocytes lysis assay comparing membrane puncturing by unconjugated PLY and PLY-DNA rings. DNA-origami rings without PLY modification serve as a negative control. Data points are shown with mean and standard deviation from three independent trials.
We built a ring-shaped DNA channel (adapted from our previous studies44) with an inner diameter of 45 nm and a height of 14 nm. We extended 48 handles from two helices at the bottom of the DNA channel (red curls in Figure 1B, also see Figure S2 for the caDNAno45 diagram), to which the PLY-anti-handles bound after 2-hour incubation at room temperature or 37 °C (Figure 1C). Importantly, we purified the DNA-origami-PLY complex by density gradient centrifugation to enrich the correctly assembled products and remove any free PLY (Figure S3). The purified DNA-corralled PLY structure migrated as a retarded band compared to the empty DNA channel during agarose gel electrophoresis (Figure S3). Negative-stain transmission electron microscopy (TEM, Figure 1C and Figure S4) of the same sample revealed additional density at the designed locations of the DNA channel, confirming the DNA-guided arrangement of PLYs as expected.
To verify that our assemblies can insert into membranes, we incubated purified PLY-DNA rings (5 nM) with large unilamellar vesicles (LUVs) containing 70% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 30% cholesterol (total lipid concentration = 1 mg/mL, homogenized by extrusion through 100 nm filters) at 37°C for 10 minutes and imaged the mixture under TEM. From the EM micrographs, we observed that nearly all PLY-DNA rings had attached to the LUV membranes. In most cases, the DNA rings and their interior PLY rings were clearly discernable (Figure 1C and Figure S5). These findings were corroborated by cryo-EM studies of LUV-bound PLY-DNA rings, which further captured a DNA-encircled PLY pore penetrating the LUV membrane (Figure 1D and Figure S6).
Next, we compared the pore-formation behaviors of our PLY-DNA rings to that of unconjugated PLYs. We expected minimal differences between the two as the 48 copies of PLY monomers in our PLY-DNA ring design closely match those quantified from the cryo-EM structures of PLY complexes (38, 42, or 44 copies) in bilayers.37, 46 While we designed the DNA-origami channel to carry a few more PLY monomers than the wild-type PLY complexes, variations of several monomers are typical of CDCs and have only mild effects on the subunit packing within the pore complex.46 Indeed, our negative-stain TEM and cryo-EM measurements determined the inner diameter of DNA-corralled PLY complexes to be 21.2±2.39 nm (n=50) and 22.3±1.96 nm (n=24) (Figure S5), respectively. These numbers were slightly larger than the 20.2±0.97 nm (n=48) we measured for PLY-only pores on the same membranes (Figure S5) and close to the inner diameter of wild-type 42-mer PLY pores previously reported (~25 nm).37 The small deviations may be attributed to variations in imaging conditions and measurement techniques, relocation of the single Cys in PLY, imperfect DNA-origami labeling efficiency, and steric constraints imposed by the DNA ring. We further performed a classical cell-lysis assay to confirm the PLY’s membrane-puncturing function, where the rupture of the cell membrane was reported by the absorbance of heme in the released hemoglobin (Figure 1E). By mixing purified PLY-DNA rings, free PLY (positive control), or unmodified DNA rings (negative control) with sheep erythrocytes, we verified that the PLY-DNA ring was almost as effective as free PLYs at rupturing red blood cells (EC50=160.6±3.63 and 119.2±7.35 pg/μL, respectively), showing that DNA-origami attachment did not substantially hinder the pore-forming activities of PLY.
To further verify that PLY-DNA rings punctured holes on lipid bilayers that permit macromolecules to pass through, we mixed purified Cy3-labeled PLY-DNA rings with giant unilamellar vesicles (GUVs, containing 69.2% DOPC, 30% cholesterol, and 0.8% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-Cy5) and simultaneously added FITC-labeled dextran with a nominal molecular weight of 20 kDa to the mixture. After 20 minutes, we imaged the GUVs under a confocal fluorescence microscope to measure the adhesion of PLY-DNA rings and the influx of dextrans (Figure 2). Integrating the Cy3 fluorescence accumulated on GUV membranes and comparing that to the Cy3 signals in solution obtained from a DNA-only sample, we estimated the surface density of PLY-DNA rings to be 40.3±12.8/μm2 or a surface coverage of 12.2±3.9% (n = 11, Figure S7). Moreover, most GUVs co-incubated with PLY-DNA rings were permeable to the 20 kDa dextran, as indicated by the equal fluorescence intensities inside and outside of the GUVs (Figure 2, GUV + PLY-DNA ring). Compared to free PLY proteins (50 nM; Figure 2, GUV + PLY), the PLY-DNA ring (1 nM) showed comparable pore-forming efficiency, consistent with results from our cell lysis assay (Figure 1E). In contrast, most GUVs remained impermeable (with unfilled interiors) in the absence of PLY (Figure 2, GUV only and GUV+DNA ring), confirming the function of PLY in mediating the DNA-membrane binding and pore formation. We note that the PLY-DNA rings produced transmembrane nanopores in the absence of detergent or electric field within minutes, suggesting the robustness and efficacy of PLY-mediated formation of hybrid nanopores.
Figure 2: PLY-DNA-origami produces transmembrane nanopores that facilitate the diffusion of macromolecules into GUVs.
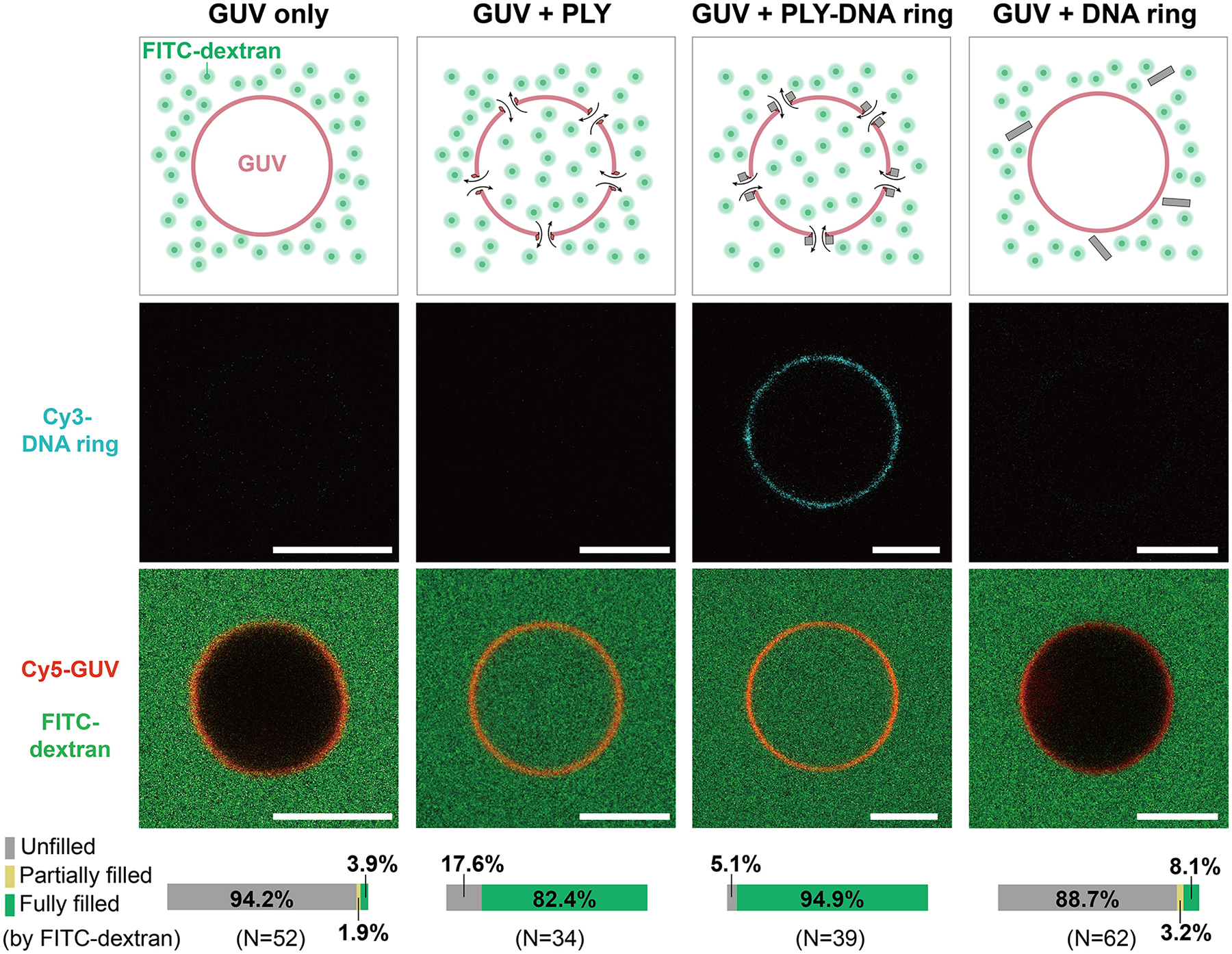
Shown from left to right are the schematics and representative images of GUVs 20 minutes after adding buffer only, anti-handle-conjugated PLY, PLY-DNA rings, and DNA-origami rings without PLY. Pseudo-colors: Cyan = Cy3 (DNA ring), red = Cy5 (GUV), and green = FITC (dextran). Scale bars: 5 μm. Bar graphs at the bottom denote the percentages of GUVs with observable dextran influx.
Subsequently, we studied the diffusion of macromolecules through the PLY-DNA pores. Dextrans of various diameters of gyration—8.4 nm (20 kDa), 11.6 nm (40 kDa), 15.2 nm (70 kDa), and 22.0 nm (150 kDa)17, 47—were separately purified by SEC (Figure S8) and mixed with GUVs and PLY-DNA rings. The cross-nanopore diffusion of dextrans met greater impedance as their sizes increased, showing a considerable variation in the influx rate (Figure S9). For example, the 20 kDa dextran could completely fill a perforated GUV around 10 minutes, while the 150 kDa variant did not reach equilibrium after 90 min (Figure S9). Given the average size of the 150 kDa dextrans (~22.0 nm), we consider these observations consistent with the inner diameters of DNA-corralled PLY pores measured by negative-stain TEM and cryo-EM (21–22 nm, Figure S5).
To quantify the size-dependent cross-nanopore diffusion kinetics, we performed fluorescence recovery after photobleaching (FRAP) assays using the three FITC-dextran species (20, 40, and 70 kDa) that can enter GUVs through the PLY-DNA pores within 1 hour (Figure 3A). After fluorescence intensities reached equilibrium in and outside GUVs, we bleached the GUVs’ interiors and measured the FITC fluorescence recovery. From representative FRAP traces (Figure 3B), it is evident that larger dextran molecules took longer to diffuse through PLY-DNA pores. We normalized the initial linear slope of the recovery traces by multiplying the GUV radii and used this value as a proxy of diffusion rate, under the assumption that GUVs have the same nanopore densities (Figure 3C and Figure S10). By this measurement, the 20 kDa dextran diffuses 4-times and 14-times more rapidly than the 40 kDa and 70 kDa counterparts, respectively. Variations in the apparent diffusion rate within each group could be attributed to the heterogeneous nanopore density on GUVs. To estimate the number of pores on each GUV, we used an established method17 to find the total porous area of the membrane by fitting FRAP traces (n=7) with an exponential function derived from Fick’s first law48. By this method, we estimated 29–184 nanopores per GUV (diameter: 5–9 μm) or a pore density of 0.56±0.34/μm2, assuming a total PLY-DNA channel length of 25 nm (see Supporting Information for details). Compared to the estimated 40.3±12.8/μm2 PLY-DNA rings attached to GUVs, this suggests an estimated pore-formation efficiency of 1.39%. While this reduction in pore formation efficiency compared to the 50–60% of wild-type PLY38 might be related to the C428A mutation42, 43 (necessary for the site-specific DNA conjugation to the N-terminal Cys), we attribute this discrepancy chiefly to the methods we used to quantify the pores. Here the pore-forming efficiency was derived from two indirect measurements based on bulk fluorescence and diffusion kinetics instead of direct single-pore counting, and therefore should be treated as a very rough estimation. Nonetheless, the PLY-DNA complexes robustly penetrated membranes without the help of electric field, detergent or solvent, supporting its potential as an adaptable platform for building functionalized nanopores.
Figure 3: Diffusion of macromolecules across PLY-DNA-origami pores is size-dependent.
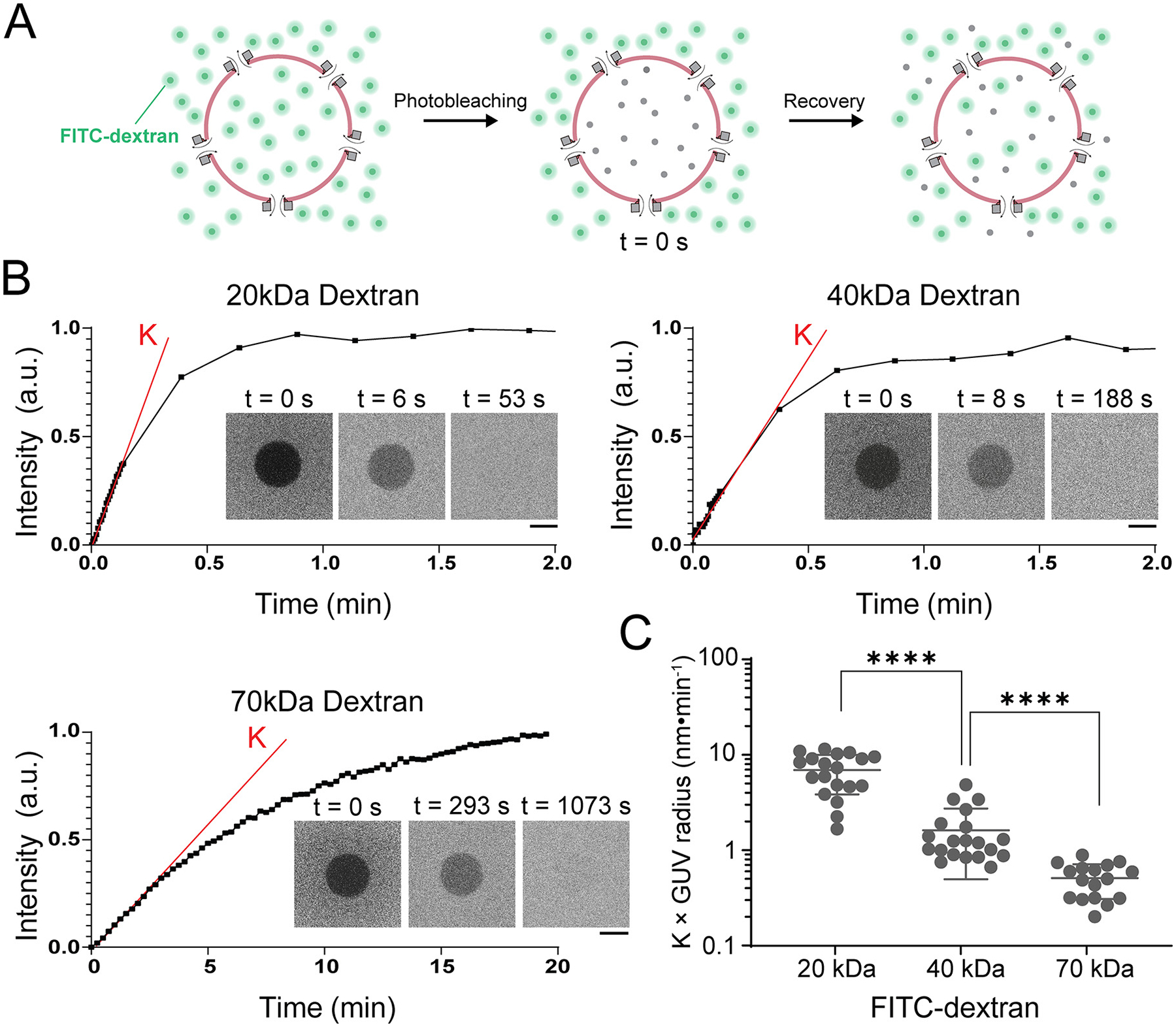
(A) Schematics of the FRAP assay on a GUV with PLY-DNA nanopores. After the interior of the GUV is equilibrated with the exterior containing FITC-dextran, the interior is photobleached and monitored for the fluorescence signal recovery caused by the exchange of FITC-dextran through the nanopores.
(B) Representative FRAP traces of perforated GUVs mixed with FITC-dextran of various sizes. The slope K (red line) denotes the initial rate of FITC fluorescence recovery after photobleaching (time 0). Insets: Confocal micrographs of GUVs at selected time points during the FRAP experiment. Scale bars: 5 μm.
(C) The initial recovery rates (K) normalized by the radii of GUVs are used to compare the diffusion rates of dextrans of various sizes through PLY-DNA pores. Data points are shown with mean and standard deviation. ****: P < 0.0001, two-tailed Mann–Whitney tests, U = 17 (left) and 9 (right). See Supporting Information for details of statistical analyses.
The programmability of DNA origami provides the opportunity to incorporate putative gating molecules into the lumen of the PLY-attached DNA channel. In eukaryotic cells, massive protein channels called nuclear pore complexes (NPCs) control entry and exit from the nucleus through the function of intrinsically disordered phenylalanine-glycine (FG) rich nucleoporin/nup proteins (FG-nups) that fill the central channel. The FG-nups are therefore ideal candidates to form a size-selective diffusion barrier that might be capable of gating the 20 nm wide PLY-DNA origami channels. Indeed, in prior work, we successfully built reductionist NPCs by grafting size-defined DNA-origami channels with FG-nups. Building on this NuPOD (NucleoPorins Organized on DNA) platform,44, 49 we hybridized an anti-handle-conjugated Nsp1, a model FG-nup,50–54 to the 36 copies of complementary handles on the inner face of the purified PLY-DNA origami (Figure 4A). As in previous studies18, 44, 49, 55, we confirmed the Nsp1 incorporation into the PLY-DNA ring by agarose gel electrophoresis and TEM (Figure 4B and Figure 4C). We then used such Nsp1-modified PLY-DNA rings to perforate GUVs and tested their permeability against dextrans of various sizes. While unmodified PLY-DNA rings allowed a sufficient influx of 20 and 40 kDa dextrans into GUVs to reach equilibrium within 15 minutes, Nsp1 severely delayed this process, resulting in substantially reduced entry after the same duration (Figure 4D). Within the timeframe of our measurement (~60 min), the Nsp1-modified pores were effectively impermeable to the 70 kDa dextran (Figure S11). These data suggest that Nsp1 altered the size selectivity of the nanopore, reducing the effective pore size to further hinder cross-pore diffusion.
Figure 4: Incorporating Nsp1 into PLY-DNA rings alters the nanopore size selectivity.
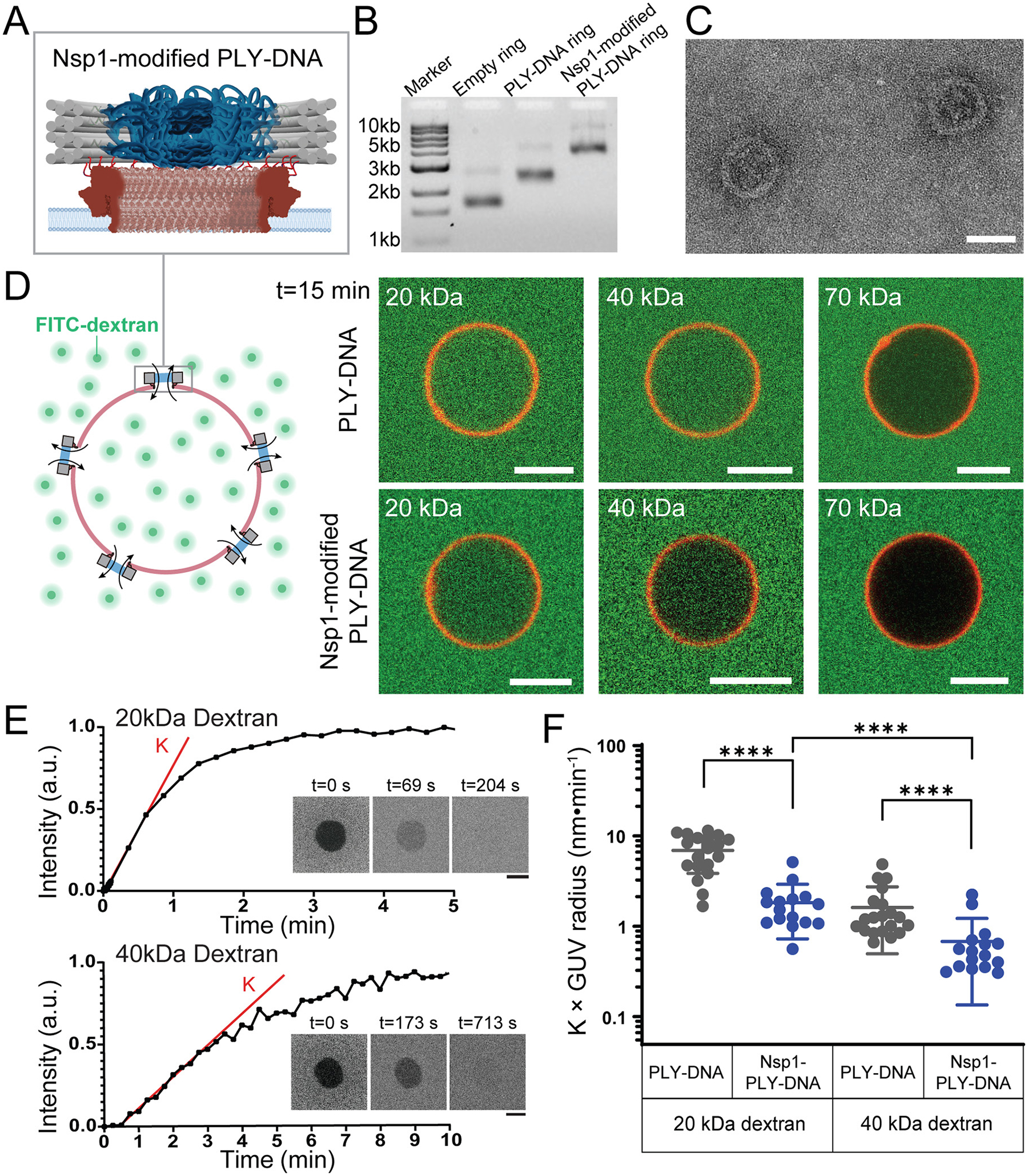
(A) A schematic illustrating a PLY-DNA nanopore modified by 36 copies of Nsp1 grafted to the DNA central channel.
(B) An SDS-agarose gel showing the empty DNA ring, PLY-DNA ring, and Nsp1-modified PLY-DNA ring structures.
(C) Negative-stain EM micrographs of PLY-DNA rings modified by Nsp1. Scale bar: 50 nm.
(D) Schematics (left) and representative fluorescence confocal images showing the influx of FITC-dextran (green) through PLY-DNA pores (top) and Nsp1-modified PLY-DNA pores (bottom) into Cy5-labeled GUVs (red) after 15 min co-incubation. Scale bars: 5 μm.
(E) Representative FRAP traces of GUVs containing Nsp1-modified PLY-DNA nanopores mixed with 20kDa (top) and 40kDa (bottom) FITC-dextran. K (red line) denotes the initial rate of FITC fluorescence recovery after photobleaching (time 0). Insets: Confocal micrographs of GUVs at selected time points during the FRAP experiment. Scale bars: 5 μm.
(F) The initial rates of fluorescence recovery (K) normalized by the radii of GUVs are used to compare the diffusion rates of 20 kDa and 40 kDa dextrans through empty (grey, same as those shown in Figure 3C) and Nsp1-modified (blue) PLY-DNA pores. Data points are shown with mean and standard deviation. ****: P < 0.0001, two-tailed Mann–Whitney test, U = 17 (left), 27 (middle), and 34 (right). See Supporting Information for details of statistical analyses.
We then subjected the GUVs with Nsp1-PLY-DNA pores to a FRAP assay to quantitatively measure the diffusion kinetics of 20 and 40 kDa dextrans through the Nsp1-modified pores (Figure 4E). The 20 kDa dextran passed through the Nsp1-modified pores about 2.7 times faster than the 40 kDa dextran. Compared to empty PLY-DNA pores, the presence of Nsp1 slowed the diffusion of 20 and 40 kDa dextrans by approximately 74% and 58%, respectively (Figure 4F), suggesting that Nsp1 reduced the effective pore size in a way that more heavily impacted diffusing molecules of smaller sizes. These measurements are in qualitative agreement with the size selectivity of Nsp1-NuPOD we previously reported: Nsp1 forms a “soft” diffusion barrier with lower permeability to larger molecules.49 The reduction of diffusion introduced by Nsp1 also matched expectations from established models that describe the diffusion coefficient of a solute through a porous membrane as a function of the solute-to-pore-size ratio.56, 57 Together, these results highlight the programmability of the hybrid nanopore design that enables the customization of the nanopore’s selectivity.
Conclusion
Membrane-spanning nanopores with increasing size and complexity are needed for basic research, such as experimentally modeling nuclear transport, and advanced translational applications, including nanopore detection of macromolecules and their assemblages (e.g., viruses). Our modular toxin-DNA hybrid pores provide a practical method to build well-behaved transmembrane pores over 20 nm in width, featuring the design flexibility and precision to functionalize the nanopore with gating or recognition molecules. The PLY-DNA rings readily penetrate bilayer membranes using the specialized pore-forming toxins and, with the densely grafted Nsp1 in the DNA channel, exhibit altered size selectivity against macromolecules. Thus, the robust pore-forming capability of PLY and the tunable permeability afforded by the DNA channel bode well for future applications that require a stable nanopore with controllable dimensions and gating functions. For example, other nucleoporins can be incorporated into the DNA-PLY pores to mimic the nuclear pore more closely and to ultimately reconstitute a nucleus-like synthetic compartment with controlled molecular uptake and export. It is also conceivable to extend the toxin pore size beyond 20 nm using DNA-origami channels carrying a larger number of PLY or other CDCs (wild-type or engineered). The spacings between the DNA ring, toxins, and gating molecules can be further reduced (e.g., by reducing DNA handle length or coating DNA with block co-polymers58, 59) to mitigate possible leakage of small molecules. Finally, cell targeting molecules and on-off switches similar to those previously described16, 20, 21, 24, 60 may be added to the DNA-origami channel. Activation of nanopores by biomarkers will enable the perforation of diseased cells61 or control the release of liposome-confined therapeutics.62
Supplementary Material
Acknowledgments
We thank members of the Chenxiang Lin and Yong Xiong labs for discussions. We are also grateful to Frederic Pincet, Jean Goder, and members of the James E Rothman lab for their guidance on GUV preparation, as well as to Joerg Nikolaus and the Yale West Campus Imaging Core for the assistance with fluorescence microscopy. This work was supported by National Institutes of Health grants R01 GM132114, and P50 AI150481 (via a collaboration development program of Pittsburgh Center for HIV Protein Interactions) to C.L., R01 AI162260 to C.L. and Y.X., R01 GM100930 to T.J.M., R01 GM105672 to C.P.L., and R21 GM109466 to C.L., C.P.L., and T.J.M., as well as a Singapore Agency for Science, Technology and Research Graduate Scholarship to Q.X.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Supporting Information Available: Materials and methods, DNA origami design, gel images, confocal and TEM micrographs, details of statistical tests, and the diffusion model used in this study.
Data availability
All original data associated with this study are available from the corresponding authors upon reasonable request.
References
- (1).Wang YH; Zhao Y; Bollas A; Wang YR; Au KF Nanopore sequencing technology, bioinformatics and applications. Nature Biotechnology 2021, 39 (11), 1348–1365. DOI: 10.1038/s41587-021-01108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Howorka S Building membrane nanopores. Nat Nanotechnol 2017, 12 (7), 619–630. DOI: 10.1038/nnano.2017.99 [DOI] [PubMed] [Google Scholar]
- (3).Deamer D; Akeson M; Branton D Three decades of nanopore sequencing. Nat Biotechnol 2016, 34 (5), 518–524. DOI: 10.1038/nbt.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schmid S; Dekker C Nanopores: a versatile tool to study protein dynamics. Essays Biochem 2021, 65 (1), 93–107. DOI: 10.1042/EBC20200020 [DOI] [PubMed] [Google Scholar]
- (5).Ying YL; Hu ZL; Zhang S; Qing Y; Fragasso A; Maglia G; Meller A; Bayley H; Dekker C; Long YT Nanopore-based technologies beyond DNA sequencing. Nat Nanotechnol 2022, 17 (11), 1136–1146. DOI: 10.1038/s41565-022-01193-2 [DOI] [PubMed] [Google Scholar]
- (6).Jain M; Koren S; Miga KH; Quick J; Rand AC; Sasani TA; Tyson JR; Beggs AD; Dilthey AT; Fiddes IT; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology 2018, 36 (4), 338–345. DOI: 10.1038/nbt.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Varongchayakul N; Song J; Meller A; Grinstaff MW Single-molecule protein sensing in a nanopore: a tutorial. Chem Soc Rev 2018, 47 (23), 8512–8524. DOI: 10.1039/c8cs00106e [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fragasso A; Schmid S; Dekker C Comparing Current Noise in Biological and Solid-State Nanopores. ACS Nano 2020, 14 (2), 1338–1349. DOI: 10.1021/acsnano.9b09353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Xue L; Yamazaki H; Ren R; Wanunu M; Ivanov AP; Edel JB Solid-state nanopore sensors. Nat Rev Mater 2020, 5 (12), 931–951. DOI: 10.1038/s41578-020-0229-6 [DOI] [Google Scholar]
- (10).Kasianowicz JJ; Brandin E; Branton D; Deamer DW Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A 1996, 93 (24), 13770–13773. DOI: 10.1073/pnas.93.24.13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yan S; Zhang J; Wang Y; Guo W; Zhang S; Liu Y; Cao J; Wang Y; Wang L; Ma F; et al. Single Molecule Ratcheting Motion of Peptides in a Mycobacterium smegmatis Porin A (MspA) Nanopore. Nano Lett 2021, 21 (15), 6703–6710. DOI: 10.1021/acs.nanolett.1c02371 [DOI] [PubMed] [Google Scholar]
- (12).Venkatesan BM; Bashir R Nanopore sensors for nucleic acid analysis. Nat Nanotechnol 2011, 6 (10), 615–624. DOI: 10.1038/Nnano.2011.129 [DOI] [PubMed] [Google Scholar]
- (13).Shi X; Pumm AK; Isensee J; Zhao WX; Verschueren D; Martin-Gonzalez A; Golestanian R; Dietz H; Dekker C Sustained unidirectional rotation of a self-organized DNA rotor on a nanopore. Nat Phys 2022, 18 (9), 1105–1111. DOI: 10.1038/s41567-022-01683-z [DOI] [Google Scholar]
- (14).Shi X; Pumm A-K; Maffeo C; Kohler F; Zhao W; Verschueren D; Aksimentiev A; Dietz H; Dekker C A nanopore-powered DNA turbine. Submission Date: 2022-06-14. arXiv (Biological Physics). 10.48550/arXiv.2206.06612 (accessed 2022-12-13). [DOI] [Google Scholar]
- (15).Langecker M; Arnaut V; Martin TG; List J; Renner S; Mayer M; Dietz H; Simmel FC Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338 (6109), 932–936. DOI: 10.1126/science.1225624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xing YZ; Dorey A; Jayasinghe L; Howorka S Highly shape- and size-tunable membrane nanopores made with DNA. Nat Nanotechnol 2022, 17 (7), 708–713. DOI: 10.1038/s41565-022-01116-1 [DOI] [PubMed] [Google Scholar]
- (17).Fragasso A; De Franceschi N; Stommer P; van der Sluis EO; Dietz H; Dekker C Reconstitution of Ultrawide DNA Origami Pores in Liposomes for Transmembrane Transport of Macromolecules. ACS Nano 2021, 15 (8), 12768–12779. DOI: 10.1021/acsnano.1c01669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ketterer P; Ananth AN; Trip DSL; Mishra A; Bertosin E; Ganji M; van der Torre J; Onck P; Dietz H; Dekker C DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nature Communications 2018, 9, 902. DOI: 10.1038/s41467-018-03313-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wei R; Martin TG; Rant U; Dietz H DNA origami gatekeepers for solid-state nanopores. Angew Chem Int Ed Engl 2012, 51 (20), 4864–4867. DOI: 10.1002/anie.201200688 [DOI] [PubMed] [Google Scholar]
- (20).Dey S; Dorey A; Abraham L; Xing YZ; Zhang I; Zhang F; Howorka S; Yan H A reversibly gated protein-transporting membrane channel made of DNA. Nature Communications 2022, 13 (1), 2271. DOI: 10.1038/s41467-022-28522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Iwabuchi S; Kawamata I; Murata S; Nomura SIM A large, square-shaped, DNA origami nanopore with sealing function on a giant vesicle membrane. Chem Commun 2021, 57 (24), 2990–2993. DOI: 10.1039/d0cc07412h [DOI] [PubMed] [Google Scholar]
- (22).Thomsen RP; Malle MG; Okholm AH; Krishnan S; Bohr SSR; Sorensen RS; Ries O; Vogel S; Simmel FC; Hatzakis NS; et al. A large size-selective DNA nanopore with sensing applications. Nature Communications 2019, 10, 5655. DOI: 10.1038/s41467-019-13284-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gopfrich K; Li CY; Ricci M; Bhamidimarri SP; Yoo J; Gyenes B; Ohmann A; Winterhalter M; Aksimentiev A; Keyser UF Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10 (9), 8207–8214. DOI: 10.1021/acsnano.6b03759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Burns JR; Seifert A; Fertig N; Howorka S A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat Nanotechnol 2016, 11 (2), 152–156. DOI: 10.1038/nnano.2015.279 [DOI] [PubMed] [Google Scholar]
- (25).Diederichs T; Ahmad K; Burns JR; Nguyen QH; Siwy ZS; Tornow M; Coveney PV; Tampe R; Howorka S Principles of Small-Molecule Transport through Synthetic Nanopores. ACS Nano 2021, 15 (10), 16194–16206. DOI: 10.1021/acsnano.1c05139 [DOI] [PubMed] [Google Scholar]
- (26).Shen Q; Grome MW; Yang Y; Lin C Engineering Lipid Membranes with Programmable DNA Nanostructures. Adv Biosyst 2020, 4 (1), 1900215. DOI: 10.1002/adbi.201900215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Akimov SA; Volynsky PE; Galimzyanov TR; Kuzmin PI; Pavlov KV; Batishchev OV Pore formation in lipid membrane I: Continuous reversible trajectory from intact bilayer through hydrophobic defect to transversal pore. Sci Rep-Uk 2017, 7, 12152. DOI: 10.1038/s41598-017-12127-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chidchob P; Offenbartl-Stiegert D; McCarthy D; Luo X; Li JN; Howorka S; Sleiman HF Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions. Journal of the American Chemical Society 2019, 141 (2), 1100–1108. DOI: 10.1021/jacs.8b11898 [DOI] [PubMed] [Google Scholar]
- (29).Diederichs T; Pugh G; Dorey A; Xing YZ; Burns JR; Nguyen QH; Tornow M; Tampe R; Howorka S Synthetic protein-conductive membrane nanopores built with DNA. Nature Communications 2019, 10, 5018. DOI: 10.1038/s41467-019-12639-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Krishnan S; Ziegler D; Arnaut V; Martin TG; Kapsner K; Henneberg K; Bausch AR; Dietz H; Simmel FC Molecular transport through large-diameter DNA nanopores. Nature Communications 2016, 7, 12787. DOI: 10.1038/ncomms12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Henning-Knechtel A; Mazin JK; Magzoub M DNA-assisted oligomerization of pore-forming toxin monomers into precisely-controlled protein channels. Nucleic Acids Research 2017, 45 (21), 12057–12068. DOI: 10.1093/nar/gkx990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Spruijt E; Tusk SE; Bayley H DNA scaffolds support stable and uniform peptide nanopores. Nat Nanotechnol 2018, 13 (8), 739–745. DOI: 10.1038/s41565-018-0139-6 [DOI] [PubMed] [Google Scholar]
- (33).Fennouri A; List J; Ducrey J; Dupasquier J; Sukyte V; Mayer SF; Vargas RD; Fernandez LP; Bertani F; Gonzalo SR; et al. Tuning the Diameter, Stability, and Membrane Affinity of Peptide Pores by DNA-Programmed Self-Assembly. ACS Nano 2021, 15 (7), 11263–11275. DOI: 10.1021/acsnano.0c10311 [DOI] [PubMed] [Google Scholar]
- (34).Rossjohn J; Gilbert RJ; Crane D; Morgan PJ; Mitchell TJ; Rowe AJ; Andrew PW; Paton JC; Tweten RK; Parker MW The molecular mechanism of pneumolysin, a virulence factor from Streptococcus pneumoniae. J Mol Biol 1998, 284 (2), 449–461. DOI: 10.1006/jmbi.1998.2167 [DOI] [PubMed] [Google Scholar]
- (35).Marriott HM; Mitchell TJ; Dockrell DH Pneumolysin: a double-edged sword during the host-pathogen interaction. Curr Mol Med 2008, 8 (6), 497–509. DOI: 10.2174/156652408785747924 [DOI] [PubMed] [Google Scholar]
- (36).Gilbert RJC Inactivation and activity of cholesterol-dependent cytolysins: What structural studies tell us. Structure 2005, 13 (8), 1097–1106. DOI: 10.1016/j.str.2005.04.019 [DOI] [PubMed] [Google Scholar]
- (37).van Pee K; Neuhaus A; D’Imprima E; Mills DJ; Kuhlbrandt W; Yildiz O CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. eLife 2017, 6, e23644. DOI: 10.7554/eLife.23644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).van Pee K; Mulvihill E; Muller DJ; Yildiz O Unraveling the Pore-Forming Steps of Pneumolysin from Streptococcus pneumoniae. Nano Lett 2016, 16 (12), 7915–7924. DOI: 10.1021/acs.nanolett.6b04219 [DOI] [PubMed] [Google Scholar]
- (39).Heuck AP; Hotze EM; Tweten RK; Johnson AE Mechanism of membrane insertion of a multimeric beta-barrel protein: Perfringolysin O creates a pore using ordered and coupled conformational changes. Mol Cell 2000, 6 (5), 1233–1242. DOI: 10.1016/S1097-2765(00)00119-2 [DOI] [PubMed] [Google Scholar]
- (40).Alving CR; Habig WH; Urban KA; Hardegree MC Cholesterol-dependent tetanolysin damage to liposomes. Biochim Biophys Acta 1979, 551 (1), 224–228. DOI: 10.1016/0005-2736(79)90368-7 [DOI] [PubMed] [Google Scholar]
- (41).Rottem S; Cole RM; Habig WH; Barile MF; Hardegree MC Structural characteristics of tetanolysin and its binding to lipid vesicles. J Bacteriol 1982, 152 (2), 888–892. DOI: 10.1128/jb.152.2.888-892.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Saunders FK; Mitchell TJ; Walker JA; Andrew PW; Boulnois GJ Pneumolysin, the Thiol-Activated Toxin of Streptococcus-Pneumoniae, Does Not Require a Thiol-Group for Invitro Activity. Infection and Immunity 1989, 57 (8), 2547–2552. DOI: 10.1128/Iai.57.8.2547-2552.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Berry AM; Alexander JE; Mitchell TJ; Andrew PW; Hansman D; Paton JC Effect of defined point mutations in the pneumolysin gene on the virulence of Streptococcus pneumoniae. Infect Immun 1995, 63 (5), 1969–1974. DOI: 10.1128/iai.63.5.1969-1974.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Fisher PDE; Shen Q; Akpinar B; Davis LK; Chung KKH; Baddeley D; Saric A; Melia TJ; Hoogenboom BW; Lin C; et al. A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano 2018, 12 (2), 1508–1518. DOI: 10.1021/acsnano.7b08044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Douglas SM; Marblestone AH; Teerapittayanon S; Vazquez A; Church GM; Shih WM Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 2009, 37 (15), 5001–5006. DOI: 10.1093/nar/gkp436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tilley SJ; Orlova EV; Gilbert RJ; Andrew PW; Saibil HR Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 2005, 121 (2), 247–256. DOI: 10.1016/j.cell.2005.02.033 [DOI] [PubMed] [Google Scholar]
- (47).Hanselmann R; Burchard W; Lemmes R; Schwengers D Characterization of Deae-Dextran by Means of Light-Scattering and Combined Size-Exclusion Chromatography Low-Angle Laser-Light Scattering Viscometry. Macromol Chem Physic 1995, 196 (7), 2259–2275. DOI: 10.1002/macp.1995.021960715 [DOI] [Google Scholar]
- (48).Dechadilok P; Deen WM Hindrance factors for diffusion and convection in pores. Ind Eng Chem Res 2006, 45 (21), 6953–6959. DOI: 10.1021/ie051387n [DOI] [Google Scholar]
- (49).Shen Q; Tian T; Xiong Q; Ellis Fisher PD; Xiong Y; Melia TJ; Lusk CP; Lin C DNA-Origami NanoTrap for Studying the Selective Barriers Formed by Phenylalanine-Glycine-Rich Nucleoporins. J Am Chem Soc 2021, 143 (31), 12294–12303. DOI: 10.1021/jacs.1c05550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Akey CW; Singh D; Ouch C; Echeverria I; Nudelman I; Varberg JM; Yu Z; Fang F; Shi Y; Wang J; et al. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 2022, 185 (2), 361–378 e325. DOI: 10.1016/j.cell.2021.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Strawn LA; Shen T; Shulga N; Goldfarb DS; Wente SR Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 2004, 6 (3), 197–206. DOI: 10.1038/ncb1097 [DOI] [PubMed] [Google Scholar]
- (52).Patel SS; Belmont BJ; Sante JM; Rexach MF Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 2007, 129 (1), 83–96. DOI: 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- (53).Frey S; Gorlich D FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. Embo J 2009, 28 (17), 2554–2567. DOI: 10.1038/emboj.2009.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Frey S; Richter RP; Gorlich D FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 2006, 314 (5800), 815–817. DOI: 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- (55).Shen Q; Xu C; Jang S; Xiong Q; Devarkar SC; Tian T; Bedwell GJ; Tripler TN; Hu Y; Yuan S; et al. A DNA-origami nuclear pore mimic reveals nuclear entry mechanisms of HIV-1 capsid. Submission Date: 2020-08-11. bioRxiv. 10.1101/2020.08.10.245522 (accessed 2022-12-13). [DOI] [Google Scholar]
- (56).Renkin EM Filtration, Diffusion, and Molecular Sieving through Porous Cellulose Membranes. J Gen Physiol 1954, 38 (2), 225–243. [PMC free article] [PubMed] [Google Scholar]
- (57).Davidson MG; Deen WM Hindered Diffusion of Water-Soluble Macromolecules in Membranes. Macromolecules 1988, 21 (12), 3474–3481. DOI: 10.1021/ma00190a022 [DOI] [Google Scholar]
- (58).Agarwal NP; Matthies M; Gur FN; Osada K; Schmidt TL Block Copolymer Micellization as a Protection Strategy for DNA Origami. Angew Chem Int Edit 2017, 56 (20), 5460–5464. DOI: 10.1002/anie.201608873 [DOI] [PubMed] [Google Scholar]
- (59).Ponnuswamy N; Bastings MMC; Nathwani B; Ryu JH; Chou LYT; Vinther M; Li WA; Anastassacos FM; Mooney DJ; Shih WM Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nature Communications 2017, 8, 15654. DOI: 10.1038/ncomms15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Li S; Jiang Q; Liu S; Zhang Y; Tian Y; Song C; Wang J; Zou Y; Anderson GJ; Han JY; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat Biotechnol 2018, 36 (3), 258–264. DOI: 10.1038/nbt.4071 [DOI] [PubMed] [Google Scholar]
- (61).Ohmann A; Li CY; Maffeo C; Al Nahas K; Baumann KN; Gopfrich K; Yoo J; Keyser UF; Aksimentiev A A synthetic enzyme built from DNA flips 10(7) lipids per second in biological membranes. Nat Commun 2018, 9 (1), 2426. DOI: 10.1038/s41467-018-04821-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Baumann KN; Schroder T; Ciryam PS; Morzy D; Tinnefeld P; Knowles TPJ; Hernandez-Ainsa S DNA-Liposome Hybrid Carriers for Triggered Cargo Release. ACS Appl Bio Mater 2022, 5 (8), 3713–3721. DOI: 10.1021/acsabm.2c00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data associated with this study are available from the corresponding authors upon reasonable request.


