Abstract
Epigenetic processes allow plasticity in gene regulation in response to significant environmental events. Accumulating evidence suggests that effective psychotherapy is accompanied by epigenetic changes, rendering DNA methylation a potential biomarker of therapy success. Due to the central role of glucocorticoid dynamics in stress regulation and the alteration of aversive memories, glucocorticoid receptors are likely involved in the molecular processes that are required to successfully treat Posttraumatic Stress Disorder (PTSD). This study aimed to investigate the relationship between methylation at the glucocorticoid receptor gene (NR3C1) and PTSD treatment success of evidence-based psychotherapy. A sample of N = 153 conflict survivors from Northern Uganda (98 females and 55 males) with PTSD were treated with Narrative Exposure Therapy (NET). Diagnostic interviews and saliva sampling took place at pretreatment and 4 and 10 months after treatment completion. We investigated potential associations between PTSD symptom development and methylation changes at 38 CpG sites spanning NR3C1 over the three times of measurement using the repeated measures correlation. After accounting for multiple comparisons, DNA methylation at CpG site cg25535999 remained negatively associated with PTSD symptoms. These results were followed up by mixed models as well as structural equation modelling. These analyses revealed that treatment responders had a significant cg25535999 methylation increase after treatment with NET. Furthermore, lower methylation at cg25535999 pretreatment predicted a higher symptom improvement. Our results suggest different epigenetic profile dynamics at NR3C1 cg25535999 in therapy responders compared to non-responders and underscore the central role of glucocorticoid signaling in trauma-focused therapy.
Subject terms: Predictive markers, Genetics, Human behaviour
Introduction
The repeated experience of extremely stressful events can result in the development of Posttraumatic Stress Disorder (PTSD). Intrusive, emotionally intense memories, which torment trauma survivors in the form of thoughts, pictures, nightmares, flashbacks, intense feelings or body sensations represent a hallmark of PTSD [1]. The modification of such aversive memory traces by means of exposure-based psychotherapy, such as Prolonged Exposure, Cognitive Therapy for PTSD, Eye Movement Desensitization and Reprocessing and Narrative Exposure Therapy (NET), has the highest evidence-base from controlled trials for the treatment of PTSD [2–4]. Of the aforementioned treatments, NET has particularly been developed to meet the needs of survivors of multiple and complex trauma, and can be effectively delivered by trained local health workers in low resource settings affected by humanitarian crises [5], such as Northern Uganda [6, 7].
Successful exposure-based therapy requires the reorganization of associative and contextual memories, thereby reducing the conditioned fear response to a stimulus that was previously associated with danger. During this process, an extinction memory is built up and competes with the original fear memory [1]. In exposure-based psychotherapy for PTSD, the trauma memory and the associated emotions, sensations, and physiological responses (i.e., the stress reaction) need to be activated by imaginary exposure to allow for their modification [8, 9]. The hypothalamus-pituitary-adrenal (HPA) axis is centrally implied in the stress reaction as well as in memory formation, retrieval and extinction. In more detail, glucocorticoids prepare the body for an adaptive response in life-threatening situations [10]. At the same time, binding of glucocorticoids to glucocorticoid receptors (GRs) mediate the termination of the stress reaction via a negative feedback loop [10]. Further, glucocorticoids facilitate the consolidation of new emotional memories and extinction learning, but inhibit memory retrieval [1, 11]. Due to the central role of glucocorticoids in the stress reaction as well as the development and extinction of aversive memories, they are likely to also be crucially involved in PTSD etiology and its treatment by means of exposure-based psychotherapy [12, 13].
While evidence regarding changes in basal cortisol levels and their relation to PTSD is mixed and seems to depend on many influencing factors including the timing of sample collection, the tissue studied and the time of trauma exposure [14], the majority of studies indicates an increased glucocorticoid receptor sensitivity in PTSD, particular in the central nervous system and the immune system [15–17]. In this line, genetic variations in the gene encoding the GR (NR3C1) were found to be robustly associated with PTSD risk in a recent meta-analysis [18].
In recent years, research increasingly focused on epigenetic modifications of NR3C1 in response to life stress and PTSD psychopathology, which could result in altered GR expression. The majority of studies found evidence for an association between reduced cytosine methylation of the NR3C1 promoter region and PTSD risk [19–22], that was in turn associated with increased expression of the GR [23]. However, it has to be noted that in a first longitudinal study, development of PTSD after deployment was not significantly associated with epigenetic changes at the NR3C1 promoter after correction for multiple comparisons of the 52 sites investigated [24]. Further, to the best of our knowledge, so far only one pilot study investigated whether epigenetic alterations of NR3C1 impact the outcome of exposure-based psychotherapy for PTSD and/or are reversible by effective treatment [25]. Therapy responders (N = 8) showed higher NR3C1 promoter methylation at pretreatment compared to non-responders (N = 8). However, treatment response was not associated with significant methylation changes at the NR3C1 site in this small sample. Due to the central role of glucocorticoid signaling in extinction learning and stress response regulation, the association of NR3C1 methylation and response to exposure-based psychotherapy warrants further investigation.
In general, research regarding methylation changes in response to psychotherapy is scarce, but the first results are promising. Kumsta [26] summarized first evidence regarding epigenetic alterations in response to psychotherapy across different disorders. While the research methods, therapy approaches and genes under investigation differed between the six studies included in the review, a commonality of all studies was that responders and non-responders differed in the methylation changes in response to psychotherapy. This renders methylation changes a potential biomarker of therapy response [26]. The present study therefore aimed to investigate whether PTSD symptom improvement following exposure-based treatment by means of Narrative Exposure Therapy (NET) is accompanied by NR3C1 methylation changes.
Patients and methods
Sample
The sample for this study was derived from a large treatment study with survivors of the war between the rebel group Lord’s Resistance Army (LRA) and the governmental forces in Northern Uganda cf [6]. We analyzed all study participants who were included into the treatment study (inclusion criteria: diagnosis of PTSD according to DSM-IV, no signs of severe current alcohol or drug abuse, no clinical signs of an acute psychosis, no psychotropic medication) with valid epigenetic data at the pretest and at least one follow-up assessment. In total, the resulting sample consisted of N = 153 individuals (98 females and 55 males), with a mean age of 32.45 (SD = 8.77). Of this sample, N = 2 individuals had missing epigenetic data for the first follow-up assessment, while N = 4 had missing epigenetic data for the second follow-up assessment. Following the recommendations for power calculations by Bakdash and Marusich [27], our sample was adequately powered to detect associations between PTSD symptoms and NR3C1 methylation for an effect size of at least |ρ | = 0.2.
Diagnostic interviews
Diagnostic interviews were conducted by intensely trained local interviewers. The interview training included general quantitative data collection methods as well as the concept of PTSD and relevant differential diagnoses. An event-list adapted for the local context was employed in order to assess the number of traumatic event types experienced and in order to identify the worst traumatic experiences [28]. We further employed the Posttraumatic Diagnostic Scale (PDS; [29]) as an interview in order to assess the diagnosis and severity of PTSD according to DSM-IV. Depressive symptoms were assessed by the respective section of the Hopkins Symptom Checklist (HSCL; [30]). All study instruments were translated into the local language (Luo), followed by blind back translations and group discussions. The interviews took place prior to the beginning of NET treatment (t1), and four (t2) and ten months (t3) after the end of treatment.
NET treatment
In brief, NET is an exposure-based psychotherapy for PTSD that aims at chronologically reconstructing the autobiographical memory of trauma survivors and was particularly developed for survivors of multiple and complex trauma [9, 31]. After obtaining an overview of the client’s life story by means of the lifeline exercise, the following sessions entail exposure therapy to the most stressful traumatic experiences of the client. In these sessions, the therapist guides the client to connect the fragmented emotional, cognitive, sensory and interoceptive memories of the traumatic events with the corresponding context information. In the process of NET, a detailed chronological narration of the survivor’s life story is developed, which focusses on recounting the most arousing experiences across the entire life-span. For more details regarding the treatment, the reader is referred to Schauer et al. [9].
NET treatments were provided by local lay mental health workers under supervision of expert psychologists according to a field version of the treatment manual [9]. Treatment quality was assured by an in-depth training of all study therapists which included the theoretical concepts of NET as well as the practical implementation that was trained intensely in role-plays. Furthermore, treatment fidelity was monitored in weekly supervision meetings with expert psychologists, as well as by a detailed review of the treatment documentation. On average, participants received 12 sessions of NET.
The study procedures were approved by the Institutional Review Board of Gulu University, the Lacor Hospital Institutional Research Committee, the Ugandan National Council for Science and Technology, Uganda, the ethics committee of the German Psychological Society (Deutsche Gesellschaft fur Psychologie), and the ethics committee of the University of Konstanz. All participants provided written informed consent prior to participation.
Epigenetic analyses
DNA isolation
Saliva DNA was collected using an Oragene DNA Kit (DNA Genotek, Ottawa, ONT) and initially extracted using the precipitation protocol recommended by the manufacturer. High-purity DNA was obtained by additional re-purification. For this purpose, 2 µg of DNA isolated via the Oragene procedure was incubated overnight at 50 °C with proteinase K (lysis buffer: 30 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, 150 ng/l proteinase K), agitated by gentle orbital shaking. Next, the DNA was purified using a Genomic DNA Clean & Concentrator Kit (Zymo Research, Irvine, CA). The DNA quality and concentration were assessed using spectrophotometry (Nanodrop 2000; ThermoScientific, Waltham, MA) and fluorometry (Qubit dsDNA BR Assay Kit, Invitrogen, Carlsbad, CA), respectively.
Infinium EPIC 850 K BeadChip methylation analyses
DNA isolated from the saliva samples was investigated with the Infinium Human Methylation EPIC 850 K array (Illumina, Inc., San Diego, CA). All subjects were processed in a single batch, with a single bisulfite conversion, and with balanced randomized plate assignment.
During pre-processing, data were extracted and analysed from the generated idat files using the R package RnBeads version 0.99.9 [32]. CpG annotation was based on the manufacturer’s annotation file (Infinium MethylationEPIC v1.0 B5 Manifest File). During pre-processing, the background was subtracted using the “noob” method in the methylumi package version 2.42.0; [33] and the signal was further normalized using the SWAN algorithm [34]. The following probe categories were excluded from the final data sets, based on the annotation provided within the RnBeads package: non-CpG context probes due to underrepresentation, [35], functional differences when compared to the CpG context as well as very low abundance of non-CpG methylation in somatic tissues [36], probes with a SNP mapping directly to the target CpG site; gonosomal probes; non-specific probes. Using the Greedycut algorithm, we iteratively removed the probes and data sets of the highest impurity (rows and columns in the detection p-value table that contain the largest fraction of unreliable measurements; p < 0.05; for each sample [32].
The B-values were further post-processed step-by-step in order to correct for further influential and putative confounding factors: 1) using logit-transformation, M-value [37] done with the R-package car [38] 2) z-transformation per plate (correcting for plate and batch effects); 3) regressing out the first 11 axes of a principal component analysis (PCA, done with the R-package pcaMethods [39]). The PCA was based on CpGs with no missing values (> 95% of the included CpGs). This approach additionally corrected for technical biases as well as for part of the variability induced by heterogenous cell composition. The given number of axes to regress out was based on a larger cross-sectional sample from the same population that included 47 technical replicate pairs [40] using a previously described quantitative trait loci driven approach [41] 4) regressing out the effects of sex and age. The accepted missing rate per CpG was set to < 1%. Only samples and CpGs surviving all filtering steps were used for the downstream analyses.
Finally, a re-measurement quality assessment was made based on the 47 technical replicate pairs as described above. For the downstream analysis we applied a relatively lower re-test threshold of r = 0.2, given that the NR3C1 includes many sites with methylation values from the extreme regions of the bimodal distribution [41]. This resulted in the final dataset of N = 38 CpGs annotated to the NR3C1 gene that were included in the downstream analysis.
Statistics
All statistical analyses were conducted using the statistical environment R version 4.1.0 [42]. Model assumptions were checked by visual inspection of scatter plots and residual plots. We initially evaluated the PTSD symptom change in the whole therapy sample using mixed models with the PDS score as the outcome variable and time as a fixed effect. In order to further investigate the PTSD symptom changes between the three points of measurement (t1: before treatment; t2: 4 months after treatment and t3: 10 months after treatment) we calculated planned contrasts with corrections for multiple comparisons using the R package multcomp version 1.4-17 [43]. The analyses were repeated with depressive symptoms (HSCL depression score) as the outcome variable.
Identification of CpG sites associated with treatment response
In order to identify associations between the change of PTSD symptoms in response to NET on the one hand, and potential corresponding epigenetic alterations on the other hand, we calculated the Repeated Measures Correlation (rmcorr; [27]; R Package rmcorr version 0.4.3). rmcorr determines the common intra-individual relationship between pairs of repeated measurement. Thereby, the best linear relationship between the two repeated measurements is calculated assuming varying intercepts but the same regression slope for each study participant. In the case of our data, this implies that rmcorr computes the association between the paired repeated measures, i.e., PTSD symptoms and methylations, and reveals if there are similar correlations for each individuum. Like the Pearson correlation coefficient, the absolute value of rmcorr can vary between 0 (no relationship) and 1 (perfect relationship). rmcorr was calculated for each investigated CpG site, and the resulting p-values were corrected for multiple comparisons using the FDR correction. CpGs with an FDR of < 0.05 were further investigated in subsequent analyses.
Linear mixed models
The identified CpG site was further investigated using linear mixed models with methylation level as the outcome variable, time as a fixed factor and study participants as a random effect using the R package nlme version 3.1-152; [44]. Following a recent review regarding epigenetic alterations in response to psychotherapy [26] who noted that a strikingly similarity in epigenetic studies on therapy outcome were different epigenetic alterations in responders and non-responders, we analyzed the data for the whole sample, as well as for responders and non-responders separately.
Responders were defined as individuals with a symptom decline of > 5.37 from pretest (t1) to the last follow-up (t3). This cutoff was identified as a clinically significant change according to the reliable change index in the same population in a previous study by Schneider et al. [6]. Accordingly, only individuals with complete data points at the last follow-up (N = 149) were classified as responders or non-responders. The sample of responders included N = 116 individuals, whereas N = 33 individuals were classified as non-responders.
Structural equation modelling
Finally, in order to get first hints regarding any potential directional relationship between PTSD symptom change and methylation change over time, we conducted cross-lagged analyses using structural equation modelling (SEM) implied in the package lavaan version 0.6-8 [45]. In cross-lagged designs [46], a variable measured at a later time can be predicted by a variable assessed at an earlier time. The comparison of these lagged associations can provide first hints on potential predictive relationships [47, 48].
The SEM included PTSD symptom severity and NR3C1 methylation at each time point (t1, t2, and t3). Auto-regressive paths were added from each previous assessment of a given variable (e.g. PTSD symptoms assessed at t1) to the consecutive time point (e.g., PTSD symptoms assessed at t2). Further, we allowed correlations between PTSD symptoms and NR3C1 methylation levels at the same time point. Finally, the cross-lags were included, which modeled the longitudinal relationship between PTSD symptoms and NR3C1 methylation. The parameter estimates were calculated by means of maximum likelihood estimation. Since the assumption of multivariate normality was not met, robust standard errors were reported and model test statistics were Satorra-Bentler scaled as recommended by Rosseel [45].
Results
Before treatment, the mean PDS score was 16.67 (SD = 4.66). PTSD symptoms declined after the treatment with NET (mean PDS score four months after treatment = 7.87 [SD = 5.51]; mean PDS score ten months after treatment = 6.83 [SD = 5.31]). In linear mixed models, we found a significant effect of time (F = 175.79, p < 0.0001). Treatment response was not influenced by time and sex (i.e., we did not find evidence for time × age or time × sex interaction effects). Thus, for reasons of parsimony, these covariates were not included in the downstream statistical analyses. Planned contrasts revealed a significant symptom decline between t1 and t2, t2 and t3, as well as between t1 and t3. According to the conventions of Cohen, the symptom decline from pretest to the last follow-up can be considered as large (d = 1.97). A significant symptom decline was also observed for depressive symptoms (F = 29.20, p < 0.0001) with a medium effect size (d = 0.70).
In order to investigate whether this symptom improvement was associated with NR3C1 methylation changes, we calculated the repeated measurement correlation between the variables PDS Score and methylation at each CpG site. After applying FDR correction for multiple comparisons, the methylation at CpG site cg25535999 was significantly negatively correlated with the PDS Score across the three repeated measurements (rmcorr = −0.195, p = 0.0007, FDR = 0.025, cf. Supplemental Table 1 and Supplemental Fig. 1). Similarly, a negative association between cg25535999 and depressive symptoms was found across the three repeated measurements (rmcorr = −0.14, p = 0.014, FDR = 0.51). However, this association did not survive correction for multiple comparison and was therefore not followed up by subsequent analyses.
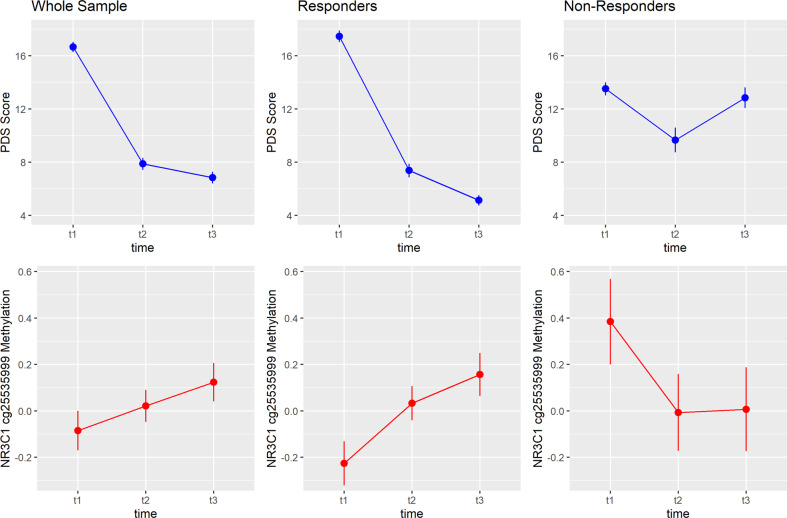
We next calculated linear mixed models with cg25535999 methylation as the outcome and time as a fixed effect to further elaborate the relationship between symptom improvement and methylation changes. In the sample of therapy responders, cg25535999 methylation increased after NET (Fig. 1, F = 5.03, p = 0.007). Planned contrasts indicated a significant increase from t1 to t2 (z = 2.16, p = .04) as well as from t1 to t3 (z = 3.08, p = 0.003). The methylation increase from t1 to t3 corresponds to a small effect (d = 0.20).
Fig. 1. Mean values and standard errors of measurement for PTSD symptoms (upper panel) and NR3C1 cg25535999 methylation (lower panel) over the three points of measurement.
The plot separately displays the results for the whole sample (N = 153), responders (N = 116), and non-responders (N = 33). PDS, Posttraumatic Diagnostic Scale; t1, pretreatment; t2, 4 months posttreatment, t3, 10 months posttreatment.
In non-responders, a non-significant decrease of methylation was observed. Accordingly, the methylation increase in the whole sample was less pronounced than in the sample of responders and did not reach statistical significance. Thus, as illustrated in Fig. 1, the negative relationship between symptom development and methylation identified by the repeated measurement correlation is driven by the sample of therapy responders: the strong reduction of PTSD symptoms over time was accompanied by an increase of NR3C1 cg25535999 methylation.
We did not find any differences between responders and non-responders regarding the number of traumatic events reported before therapy (t = 1.4284, p = 0.16), regarding depressive symptom severity before therapy (t = −0.94, p = 0.3534), or in the occurrence of new events after treatment (for t2: Χ² = 0.35, p = 0.56; for t3: Χ² = 1.51, p = 0.22).
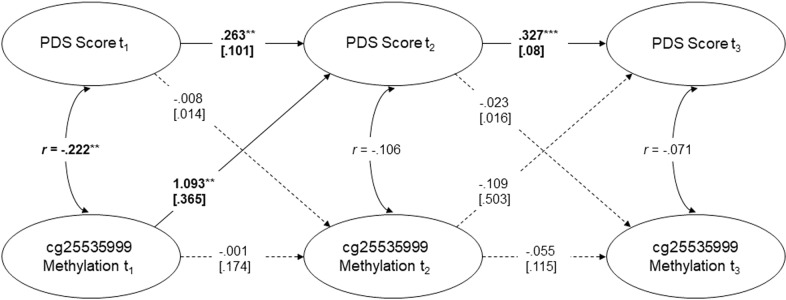
Finally, we employed a cross-lagged analysis in order to examine whether a model assuming predictive relationships between PTSD symptoms and methylation shows a good fit to the data. Furthermore, this model may contribute to answer the questions whether NR3C1 cg25535999 methylation is predictive for PTSD symptoms and vice versa. The constructed SEM, visible in Fig. 2, had an excellent fit to the data with a root mean square error of approximation (RMSEA) of < 0.001 (90% - CI = [0, 0.105]), a comparative fit index (CFI) of approximately 1, and a non-significant deviation from the empirical covariance structure (χ2(4) = 2.915, p = 0.572) (cf. cut-offs proposed by Hu & Bentler who consider CFI > 0.95 and RMSEA < 0.06 as relatively good). We identified significant auto-regressive relationships for the PTSD symptoms assessed over time, but not for NR3C1 cg25535999 methylation. Regarding the cross-lagged relationships, the only significant association was found between methylation status before therapy (t1) and PTSD symptoms four months after the end of treatment (t2) (β = 1.093, SE(β) = 0.365, z = 2.991, p = 0.003). Indeed, responders and non-responders differed significantly regarding cg25535999 methylation before therapy (t = 2.95, p = 0.005, cf. Fig. 1). No predictive influences were identified between t2 and t3.
Fig. 2. Displayed is the constructed Structural Equation Model (SEM) together with the regression or correlation coefficients.
Significant associations are marked in bold. *p < 0.05, **p < 0.01, ***p < 0.001.
NR3C1 cg255535999 methylation and GR expression
cg25535999 is located in the intron 7 of the gene body of NR3C1 (see Supplementary Figure 2). In order to investigate the potential impact of cg25535999 methylation on NR3C1 expression, we investigated publicly available data generated by The Cancer Genome Atlas (TGCA) project (https://www.cancer.gov/tcga), by using the SMART web-interface [49]. These analyses revealed that cg25535999 methylation is significantly correlated with NR3C1 gene expression across 17 TCGA datasets. Further, the majority of the identified correlations was positive (for the distribution of Pearsons’ r across 17 TCGA datasets: μ = 0.41, x˜ = 0.43, σ = 0.22, see Supplementary Fig. 3). This finding agrees with the position of the given CpG site in the NR3C1 gene body, and further corroborates the possible functional relevance of this epigenetic modification for NR3C1 gene expression, and thus the regulation of glucocorticoid signaling in PTSD.
Discussion
In line with a recent meta-analysis [5], we found evidence for a strong PTSD symptom reduction after treatment with NET. According to a cut-off of clinically significant change, the majority (78%) of our sample were classified as therapy responders. This sub-group showed a distinct epigenetic pattern: On the one hand, responders showed a significant NR3C1 cg25535999 methylation increase after treatment with NET, which was not observed in non-responders. On the other hand, this group also presented with significantly lower methylation levels pre-treatment. This is in line with the evidence summarized by Kumsta [26], suggesting that therapy responders and non-responders show distinct epigenetic profiles that can be measured in the periphery. Our SEM indicated that pre-treatment DNA methylation of the cg25535999 site in the gene body of NR3C1 was predictive of PTSD symptoms after NET treatment, with higher values in initial methylation being associated with higher PTSD scores after NET. One earlier pilot study also identified NR3C1 methylation as a predictor of psychotherapy response [25], yet this studies focused on promoter methylation.
What might be the biomolecular mechanisms underlying our observations? First and foremost, it is important to note that the methylation differences observed in this study were measured in saliva samples. While we can assume that the consolidation of extinction memory is accompanied by epigenetic changes linked to synaptic plasticity in the brain [50], central changes are not necessarily reflected by similar changes occurring in the periphery [26]. Yet, one plausible mechanism how interventions are likely to impact methylation in peripheral cells are top-down effects on brain-body communication systems, i.e., the stress axes, which will stimulate physiological changes in the periphery [26, 50]. Accordingly, it is plausible to assume that the glucocorticoid receptor gene NR3C1, which holds a central function in the sensitivity of the HPA axis, could be subject to epigenetic changes in response to psychotherapy that can be measured in the periphery. Nevertheless, caution is generally required when interpreting epigenetic data in the periphery and premature conclusions about underlying processes in the brain should be avoided. Of course, this also has to be considered when interpreting the finding of a decreased NR3C1 cg25535999 methylation status of treatment responders at pretreatment. Bearing that in mind, we found evidence that cg25535999 methylation correlated positively with NR3C1 expression across different TGCA datasets. Accordingly, therapy responders, who had lower cg25535999 methylation before therapy, might also present with a lower expression of the GR before therapy, which could in turn be associated with higher levels of circulating cortisol in contrast to non-responders cf [23, 51]. These higher levels of cortisol might facilitate extinction learning processes during treatment [1, 52]. Therapy success was further accompanied by an increase in cg25535999 methylation, potentially associated with a higher expression of GRs and reduced cortisol levels. Follow-up studies should thus also include a direct measurement of NR3C1 expression and cortisol related to the treatment timeline, in order to further substantiate the current findings.
Nevertheless, the results of this study highlight the central role of glucocorticoids in exposure-based treatments [52]. Further, they correspond with accumulating evidence indicating that the effectiveness of exposure-based therapy for varying anxiety disorders, including spider phobia [53, 54], social phobia [55] and fear of heights [56] is enhanced by glucocorticoid administration. Similar effects were observed in a pilot trial on exposure therapy for PTSD, however it has to be noted that the effect was likely mediated via a higher treatment retention rate in the glucocorticoid group as opposed to the placebo group [57].
Limitations and future directions
The findings of this study have to be interpreted in light of its limitations. Similar to previous studies investigating epigenetic changes in response to psychotherapy, the study was of observational nature and did not investigate an additional or non-treatment patient control group. Accordingly, PTSD symptom changes as well as methylation changes cannot be causally attributed to the treatment.
Future randomized controlled trials are needed to substantiate findings regarding the relationship between treatment and methylation changes which can be adequately powered using the effect sizes reported in this manuscript. In addition, a higher number and density of assessments post therapy would be helpful to identify the chronological order of symptom and methylation changes.
Further, the epigenetic analyses were conducted on saliva samples. Therefore, the assessed epigenetic alterations may vary with a changed life-style, e.g., more sleep, altered physical activity etc. In particular conclusions regarding brain processes as well as overall HPA-functioning remain speculative. In this line, data on NR3C1 expression and cortisol levels would have been helpful to interpret our findings. However, these biological samples are difficult to collect, process and store in a field study.
Finally, it has to be noted that glucocorticoid signaling is regulated by complex pathways. For instance, FKBP5, a co-chaperone of the GR, influences the negative feedback mechanisms of the HPA axis [58], and FKBP5 methylation has been shown to be associated with PTSD symptom development [59, 60]. Future studies with even larger sample sizes might be adequately powered to investigate more complex epigenetic pathways including the interplay of several candidate genes.
Conclusions
In sum, this study identified a distinct epigenetic profile of therapy responders in contrast to non-responders. Therapy response was predicted by lower NR3C1 cg25535999 methylation at baseline, and correlated with a methylation increase. Our findings highlight the central role of glucocorticoids in relation to the effectiveness of trauma-focused therapy.
Supplementary information
Acknowledgements
This research was supported by the German Research Foundation (DFG grant KO3895-4) and funds of the University of Basel. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We would like to thank our team of local mental health workers for conduction the diagnostic interviews and treatments with outstanding scientific quality and therapeutic competence and empathy. We acknowledge support for the publication costs by the Open Access Publication Fund of Bielefeld University and the Deutsche Forschungsgemeinschaft (DFG).
Author contributions
SW, I-TK, TE, VV, DdQ, and AP developed the study concept. SW, AS and APf conducted the data collection in Northern Uganda. VV, DdQ, and AP conducted the epigenetic analyses. SW and SI conducted the statistical analyses, with critical input by MP, I-TK, and VV. SW drafted the manuscript. All authors contributed to the interpretation of the results, critically revised the manuscript, and approved the final version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Due to the highly sensitive nature of this research, participants of this study did not agree for their data to be made available in a data repository. However, participants gave their informed consent that excerpts of the data that guarantee anonymity can be shared with other researchers upon reasonable request. Please address requests to sarah.wilker@uni-bielefeld.de.
Code availability
The R code can be requested from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02316-6.
References
- 1.de Quervain DJ-F, Schwabe L, Roozendaal B. Stress, glucocorticoids and memory: Implications for treating fear-related disorders. Nat Rev Neurosci. 2017;18:7–19. [DOI] [PubMed]
- 2.Bradley R, Greene J, Russ E, Dutra L, Westen D. A multidimensional meta-analysis of psychotherapy for PTSD. AJP. 2005;162:214–27. doi: 10.1176/appi.ajp.162.2.214. [DOI] [PubMed] [Google Scholar]
- 3.Lee DJ, Schnitzlein CW, Wolf JP, Vythilingam M, Rasmusson AM, Hoge CW. Psychotherapy versus pharmacotherapy for posttraumatic stress disorder: Systemic review and meta-analyses to determine first-line treatments. Depress Anxiety. 2016;33:792–806. doi: 10.1002/da.22511. [DOI] [PubMed] [Google Scholar]
- 4.Cusack K, Jonas DE, Forneris CA, Wines C, Sonis J, Middleton JC, et al. Psychological treatments for adults with posttraumatic stress disorder: A systematic review and meta-analysis. Clin Psychol Rev. 2016;43:128–41. doi: 10.1016/j.cpr.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Siehl S, Robjant K, Crombach A. Systematic review and meta-analyses of the long-term efficacy of narrative exposure therapy for adults, children and perpetrators. Psychother Res. 2021;31:695–710. doi: 10.1080/10503307.2020.1847345. [DOI] [PubMed] [Google Scholar]
- 6.Schneider A, Pfeiffer A, Conrad D, Elbert T, Kolassa I-T, Wilker S. Does cumulative exposure to traumatic stressors predict treatment outcome of community-implemented exposure-based therapy for PTSD? Eur J Psychotraumatol. 2020;11:1789323. doi: 10.1080/20008198.2020.1789323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ertl V, Pfeiffer A, Schauer E, Elbert T, Neuner F. Community-implemented trauma therapy for former child soldiers in Northern Uganda: A randomized controlled trial. JAMA. 2011;306:503–12. doi: 10.1001/jama.2011.1060. [DOI] [PubMed] [Google Scholar]
- 8.Schnyder U, Ehlers A, Elbert T, Foa EB, Gersons BPR, Resick PA, et al. Psychotherapies for PTSD: What do they have in common? Eur J Psychotraumatol. 2015;6:28186. doi: 10.3402/ejpt.v6.28186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schauer M, Neuner F, Elbert T. Narrative exposure therapy: A short-term treatment for traumatic stress disorders. Hogrefe Publishing, 2011.
- 10.Herman JP, McKlveen JM, Ghosal S, Kopp B, Wulsin A, Makinson R, et al. Regulation of the Hypothalamic‐Pituitary‐Adrenocortical Stress Response. In Terjung R (ed). Compr Physiol, 2011 Wiley, pp 603–21. [DOI] [PMC free article] [PubMed]
- 11.Quaedflieg CWEM, Schwabe L. Memory dynamics under stress. Memory. 2018;26:364–76. doi: 10.1080/09658211.2017.1338299. [DOI] [PubMed] [Google Scholar]
- 12.Rasmusson AM, Marx CE, Pineles SL, Locci A, Scioli-Salter ER, Nillni YI, et al. Neuroactive steroids and PTSD treatment. Neurosci Lett. 2017;649:156–63. doi: 10.1016/j.neulet.2017.01.054. [DOI] [PubMed] [Google Scholar]
- 13.Singewald N, Schmuckermair C, Whittle N, Holmes A, Ressler KJ. Pharmacology of cognitive enhancers for exposure-based therapy of fear, anxiety and trauma-related disorders. Pharm Ther. 2015;149:150–90. doi: 10.1016/j.pharmthera.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlop BW, Wong A. The hypothalamic-pituitary-adrenal axis in PTSD: Pathophysiology and treatment interventions. Prog Neuropsychopharmacol Biol Psychiatry. 2019;89:361–79. doi: 10.1016/j.pnpbp.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Rohleder N, Wolf JM, Wolf OT. Glucocorticoid sensitivity of cognitive and inflammatory processes in depression and posttraumatic stress disorder. Neurosci Biobehav Rev. 2010;35:104–14. doi: 10.1016/j.neubiorev.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, et al. Biological studies of post-traumatic stress disorder. Nat Rev Neurosci. 2012;13:769–87. doi: 10.1038/nrn3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro-Vale I, van Rossum EFC, Machado JC, Mota-Cardoso R, Carvalho D. Genetics of glucocorticoid regulation and posttraumatic stress disorder—What do we know? Neurosci Biobehav Rev. 2016;63:143–57. doi: 10.1016/j.neubiorev.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Sheerin CM, Lind MJ, Bountress KE, Marraccini ME, Amstadter AB, Bacanu S-A, et al. Meta-analysis of associations between hypothalamic-pituitary-adrenal axis genes and risk of posttraumatic stress disorder. J Traum Stress. 2020;33:688–98. doi: 10.1002/jts.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labonté B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl Psychiatry. 2014;4:e368–e368. doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, et al. Lower Methylation of Glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77:356–64. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Vukojevic V, Kolassa I-T, Fastenrath M, Gschwind L, Spalek K, Milnik A, et al. Epigenetic modification of the Glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J Neurosci. 2014;34:10274–84. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schechter DS, Moser DA, Paoloni-Giacobino A, Stenz L, Gex-Fabry M, Aue T, et al. Methylation of NR3C1 is related to maternal PTSD, parenting stress and maternal medial prefrontal cortical activity in response to child separation among mothers with histories of violence exposure. Front Psychol. 2015;6:690. [DOI] [PMC free article] [PubMed]
- 23.Watkeys OJ, Kremerskothen K, Quidé Y, Fullerton JM, Green MJ. Glucocorticoid receptor gene (NR3C1) DNA methylation in association with trauma, psychopathology, transcript expression, or genotypic variation: A systematic review. Neurosci Biobehav Rev. 2018;95:85–122. doi: 10.1016/j.neubiorev.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Schür RR, Boks MP, Rutten BPF, Daskalakis NP, Nijs L, de, van Zuiden M, et al. Longitudinal changes in glucocorticoid receptor exon 1F methylation and psychopathology after military deployment. Transl Psychiatry. 2017;7:e1181. doi: 10.1038/tp.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, et al. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Front Psychiatry 2013;4:118. [DOI] [PMC free article] [PubMed]
- 26.Kumsta R. The role of epigenetics for understanding mental health difficulties and its implications for psychotherapy research. Psychol Psychother. 2019;92:190–207. doi: 10.1111/papt.12227. [DOI] [PubMed] [Google Scholar]
- 27.Bakdash JZ, Marusich LR. Repeated measures correlation. Front Psychol. 2017;8:456. doi: 10.3389/fpsyg.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilker S, Pfeiffer A, Kolassa S, Koslowski D, Elbert T, Kolassa I-T. How to quantify exposure to traumatic stress? Reliability and predictive validity of measures for cumulative trauma exposure in a post-conflict population. Eur J Psychotraumatol. 2015;6:28306. doi: 10.3402/ejpt.v6.28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foa EB, Cashman L, Jaycox L, Perry K. The validation of a self-report measure of posttraumatic stress disorder: The posttraumatic diagnostic scale. Psychol Assess. 1997;9:445–51. doi: 10.1037/1040-3590.9.4.445. [DOI] [Google Scholar]
- 30.Derogatis LR, Lipman RS, Rickels K, Uhlenhuth EH, Covi L. The Hopkins Symptom Checklist (HSCL): a self-report symptom inventory. Behav Sci. 1974;19:1–15. doi: 10.1002/bs.3830190102. [DOI] [PubMed] [Google Scholar]
- 31.Robjant K, Fazel M. The emerging evidence for narrative exposure therapy: A review. Clin Psychol Rev. 2010;30:1030–39. doi: 10.1016/j.cpr.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–40. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis S, Du P, Bilke S, Triche, Jr T, Bootwalla M. methylumi: Handle Illumina methylation data, 2022.
- 34.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–95. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT-Y, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinforma. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox J, Weisberg S. An R companion to applied regression. Sage: Los Angeles, London, New Delhi, Singapore, Washington, DC, Melbourne, 2019.
- 39.Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–67. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 40.Vukojevic V, Mastrandreas P, Arnold A, Peter F, Kolassa I-T, Wilker S, et al. Evolutionary conserved role of neural cell adhesion molecule-1 in memory. Transl Psychiatry 2020;10:217. [DOI] [PMC free article] [PubMed]
- 41.Milnik A, Vogler C, Demougin P, Egli T, Freytag V, Hartmann F, et al. Common epigenetic variation in a European population of mentally healthy young adults. J Psychiatr Res. 2016;83:260–68. doi: 10.1016/j.jpsychires.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 42.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2021.
- 43.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–63. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 44.Pinheiro J, Bates D, DebRoy S, Sarkar D., R Core Team. nlme: Linear and Nonlinear Mixed Effect Models. R package version 3.1-152, 2021.
- 45.Rosseel Y. Lavaan: An R Package for Structural Equation Modeling. J Stat Soft. 2012;48:1–36.
- 46.Kenny DA. Cross‐Lagged Panel Design. In Balakrishnan N, et al. (eds). Wiley StatsRef: Statistics Reference Online. Wiley, 2014.
- 47.Newsom JT. Cross-lagged Panel Analysis. In Whitbourne SK (ed). The encyclopedia of adulthood and aging. John Wiley & Sons Inc: Hoboken, 2015, pp 1–6.
- 48.Selig J, Little T. Autoregressive and cross-lagged panel analysis for longitudinal data. In Laursen, B, Little, TD & Card, NA (eds). Handbook of developmental research methods. The Guilford Press, 2012, pp 265–78
- 49.Li Y, Di GE, Lu C. The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin. 2019;12:71. doi: 10.1186/s13072-019-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiele MA, Gottschalk MG, Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - A review and synthesis on psychosocial stress, psychotherapy, and prevention. Clin Psychol Rev. 2020;77:101830. doi: 10.1016/j.cpr.2020.101830. [DOI] [PubMed] [Google Scholar]
- 51.Liu PZ, Nusslock R. How stress gets under the skin: Early life adversity and glucocorticoid receptor epigenetic regulation. Curr Genomics. 2018;19:653–64. doi: 10.2174/1389202919666171228164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Quervain DJ-F, Wolf OT, Roozendaal B. Glucocorticoid-induced enhancement of extinction-from animal models to clinical trials. Psychopharmacol (Berl) 2019;236:183–99. doi: 10.1007/s00213-018-5116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soravia LM, Heinrichs M, Winzeler L, Fisler M, Schmitt W, Horn H, et al. Glucocorticoids enhance in vivo exposure-based therapy of spider phobia. Depress Anxiety. 2014;31:429–35. doi: 10.1002/da.22219. [DOI] [PubMed] [Google Scholar]
- 54.Steudte-Schmiedgen S, Fay E, Capitao L, Kirschbaum C, Reinecke A. Hydrocortisone as an adjunct to brief cognitive-behavioural therapy for specific fear: Endocrine and cognitive biomarkers as predictors of symptom improvement. J Psychopharmacol. 2021;35:641–51. doi: 10.1177/02698811211001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soravia LM, Heinrichs M, Aerni A, Maroni C, Schelling G, Ehlert U, et al. Glucocorticoids reduce phobic fear in humans. PNAS. 2006;103:5585–90. doi: 10.1073/pnas.0509184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Quervain DJ-F, Bentz D, Michael T, Bolt OC, Wiederhold BK, Margraf J, et al. Glucocorticoids enhance extinction-based psychotherapy. PNAS. 2011;108:6621–25. [DOI] [PMC free article] [PubMed]
- 57.Yehuda R, Bierer LM, Pratchett LC, Lehrner A, Koch EC, van Manen JA, et al. Cortisol augmentation of a psychological treatment for warfighters with posttraumatic stress disorder: Randomized trial showing improved treatment retention and outcome. Psychoneuroendocrinology. 2015;51:589–97. doi: 10.1016/j.psyneuen.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34:S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Miller O, Shakespeare-Finch J, Bruenig D, Mehta D. DNA methylation of NR3C1 and FKBP5 is associated with posttraumatic stress disorder, posttraumatic growth, and resilience. Psychol Trauma. 2020;12:750–55. doi: 10.1037/tra0000574. [DOI] [PubMed] [Google Scholar]
- 60.Mehta D, Miller O, Bruenig D, David G, Shakespeare-Finch J. A Systematic Review of DNA Methylation and Gene Expression Studies in Posttraumatic Stress Disorder, Posttraumatic Growth, and Resilience. J Traum Stress. 2020;33:171–80. doi: 10.1002/jts.22472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the highly sensitive nature of this research, participants of this study did not agree for their data to be made available in a data repository. However, participants gave their informed consent that excerpts of the data that guarantee anonymity can be shared with other researchers upon reasonable request. Please address requests to sarah.wilker@uni-bielefeld.de.
The R code can be requested from the corresponding author.