Abstract
[Aims] Flavonoid apigenin (API) has a wide range of biological functions, particularly anti-inflammation. Indoleamine 2,3-dioxygenase (IDO) and 2-Amino-3-carboxymuconate-6-semialdehyde decarboxylase (ACMSD) are important tryptophan metabolic enzymes that play pivotal roles in the production of toxic metabolite quinolinic acid. However, the relationship between inflammation and ACMSD remains unclear. The present study investigated the relationship between inflammation and tryptophan metabolic key enzymes. Similarly, the anti-inflammatory effect of API on important tryptophan metabolic enzymes was examined in lipopolysaccharide (LPS)-treated microglial cells.
[Main methods] MG6 cells were exposed to LPS with or without API treatment for 24–48 h. IDO and ACMSD mRNA expression and production of inflammatory mediators were analyzed. Activation of inflammatory signaling pathways, such as mitogen-activated protein kinase (MAPK) and nuclear factor-κB (NF-κB), was also examined to study the mechanism of API in the inflammatory state.
[Key findings] LPS suppressed ACMSD expression and enhanced IDO expression. However, API elevated ACMSD mRNA expression and suppressed IDO mRNA expression in LPS-treated MG6 cells. Furthermore, API suppressed interleukin-6 and nitric oxide production, whereas overproduction of inflammatory mediators enhanced IDO expression and assisted tryptophan degradation. API also inhibited activation of extracellular signal-regulated kinase (Erk) and jun N-terminal kinase (JNK) MAPK, and degradation of IκBα.
[Significance] These results indicate alteration of ACMSD expression under inflammatory conditions. Moreover, API recovers expression of tryptophan metabolic key enzymes, which may be mediated by inhibition of proinflammatory mediator production via inactivation of Erk, JNK MAPK, and NF-κB pathways in LPS-stimulated microglial cells.
Keywords: Apigenin, ACMSD, IDO, Microglial cells, Neuroinflammation
Abbreviations: ACMSD, 2-Amino-3-carboxymuconate-6-semialdehyde decarboxylase; IDO, Indoleamine 2,3-dioxygenase
1. Introduction
Apigenin (API) is a natural flavonoid that has received immense attention because of its biological activities, which include anti-inflammation [1], anti-cancer [2], anti-diabetes [3], its roles in amnesia and Alzheimer's disease [4], and its effects on depression and insomnia [5]. API is abundantly present in various vegetables and fruits. Parsley is a major source of this flavone, which contains 215.46 mg API/100 g [6]. Other rich sources of API are kumquats, celery, peppermint, oregano, and perilla that contain 21.87, 19.1, 5.39, 2.57, and 0.07 mg API/100 g, respectively [6]. Previous studies have reported that API protects neurons and glial cell from Aβ oligomers and inflammatory stimuli [7] and ameliorates cognitive dysfunction and neuronal damage in AD mouse model via apoptosis, amyloidogenesis, and BDNF/TrkB pathways [8]. Nuclear factor-κB (NF-κB) and mitogen-activated protein kinases (MAPK) pathways have been suggested to be involved in API-mediated anti-inflammatory effects [9,10].
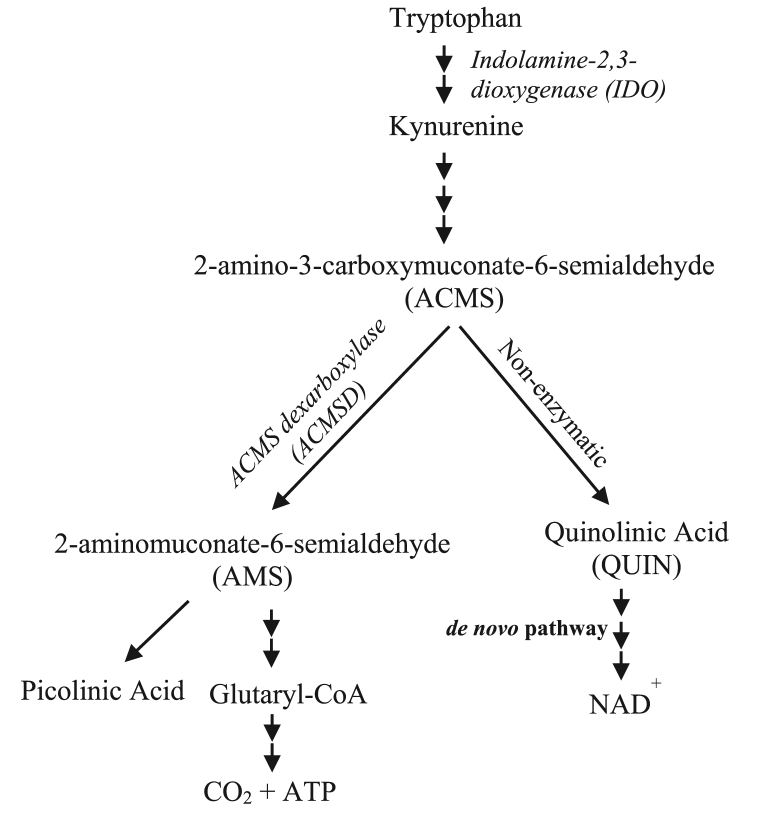
The kynurenine pathway (KP) is the main pathway in the catabolism of tryptophan and responsible for ≥90% of tryptophan degradation (Fig. 1) [11]. KP participates in the inflammatory mechanisms of neurogenic and autoimmune diseases through the action of its critical metabolic enzymes. Alterations in KP metabolites and enzyme activities in diseases are accompanied by changes in immune functions that include the neuroimmune system [12].
Fig. 1.
Simplified diagram of the Tryptophan-kynurenine and -NAD pathway.
Indole amine 2,3 dioxygenase (IDO), a rate-limiting enzyme in tryptophan degradation, is expressed in various cell types including macrophages, which is induced by proinflammatory cytokines [13] and bacterial lipopolysaccharide (LPS) [14]. A high level of IDO in the brain is associated with neuronal disorders, such as depression, Alzheimer's disease, Huntington's disease, and Parkinson's diseases, which are partially caused by upregulation of toxic quinolinic acid (QUIN) production in the kynurenine pathway [15–16]. QUIN, a potent excitotoxin, is an agonist of the N-methyl-d-aspartate receptor [17] and is often implicated in the pathogeneses of neuronal diseases [15–18].
2-Amino-3-carboxymuconate-6-semialdehyde decarboxylase (ACMSD) is a critical enzyme in the NAD+ pathway (Fig. 1), which regulates and limits the formation of QUIN by catalyzing the production of another metabolite. An increased level of QUIN due to reduced ACMSD activity may act as a major trigger of excitotoxic neuronal death in Parkinson's disease and other neurodegenerative diseases [19–20]. ACMSD is expressed in the liver, kidney, and central nervous system (CNS) [21]. The expression and activity of ACMSD are influenced by dietary and metabolic conditions [22–23], and have recently begun to be investigated in neurological disorders. However, the relationship between this enzyme and inflammation remains unclear. In the present study, we clarified the direct relationship between ACMSD and inflammation.
Because of the pivotal role of IDO and ACMSD enzymes in the immune system, suppression of IDO and elevation of ACMSD appear to be effective to suppress the pathogeneses of neurodegenerative diseases. Therefore, we investigated the alterations of ACMSD and IDO enzymes in LPS-induced neuroinflammation of microglial cells. Similarly, the potential molecular mechanism that underlies the anti-inflammatory effect of API on tryptophan metabolism and inflammatory processes was examined because the anti-inflammatory mechanism of API in LPS-stimulated microglial cells has not been clarified.
2. Materials and methods
2.1. Reagents
API and LPS were purchased from Sigma-Aldrich (St Louis, MO, USA). API and LPS were dissolved in dimethyl sulfoxide (DMSO) and phosphate-buffered saline (PBS), respectively. Antibodies against IκBα (sc-371) was obtained from Santa Cruz Biotechnology (Dallas, Texas, USA). Antibodies against p-p38 (#9215), p38 (#9212S), p-Erk (#9101S), Erk (#4695), p-JNK (#9251), JNK (#9252), and β-actin (#4967) were obtained from Cell Signaling Technology (Danvers, MA, USA). Dulbecco's modified Eagle's medium (DMEM) was purchased from Nissui Pharmaceutical (Tokyo, Japan). Fetal bovine serum (FBS) was purchased from Biological Industries (Kibbutz Beit Haemek, Israel). CellTiter 96® AQueous One Solution was purchased from Promega (Tokyo, Japan). ELISA MAX™ was purchased from BioLegend (San Diego, CA, USA). RNAiso plus reagent was purchased from Takara Biotechnology (Shiga, Japan). ReverTraAce® qPCR RT Master Mix with gDNA Remover and THUNDERBIRD® SYBR qPCR MIX were from TOYOBO CO., Ltd. (Tokyo, Japan). The remaining chemicals and reagents were analytical grade and commercially available.
2.2. Cell culture and treatment
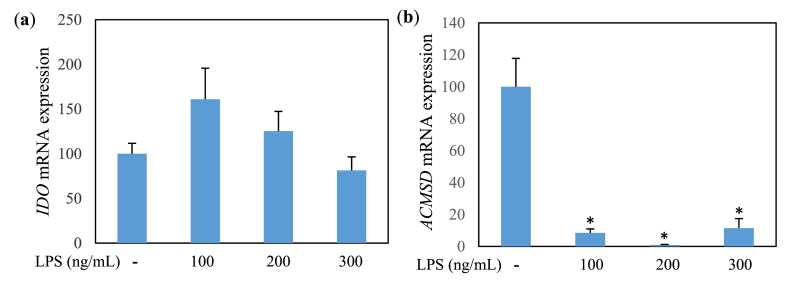
Mouse microglial cell line MG6 cells [24,25] were obtained from RIKEN Cell Bank (Tokyo, Japan). The cells were cultured in DMEM supplemented with 10% FBS and incubated at 37 °C with 5% CO2 for 2–3 days. Dishes with 80%–90% confluent cells were subcultured and cells from passages 6–7 were used for the treatments. MG6 cells were incubated with 100 ng/mL of LPS concomitant by API at various concentrations for either 30 min, 24 h, 30 h, or 48 h, depending on the experiment. We chose the above-mentioned dose of LPS that was shown remarkable alteration of tryptophan metabolic key enzymes (Fig. 4) and effectively induced the release of proinflammatory mediators in previously reported studies [26,27]. Untreated cells and LPS-treated cells without API co-treatment were cultured in parallel as negative and positive controls.
Fig. 4.
IDO (a) and ACMSD (b) mRNA expression (relative value) in MG6 cells treated with LPS for 24 h. Values indicate mean ± SE (n = 5–6). *p < 0.05 vs LPS (−).
2.3. Cell viability assay
Cell viability was assessed by a Cell Titer 96® AQueous One Solution Cell Proliferation Assay in accordance with the manufacturer's protocol. Briefly, MG6 cells (0.4 × 105/well) were seeded on 96-well plates and incubated overnight at 37 °C with 5% CO2. Cells were then treated with or without API (5, 10, 30, and 50 μM) and/or LPS (100 ng/mL). After 24 h of treatment, the cells were incubated with 20 μL Cell Titer reagent for 15 min at 37 °C. Absorbance was then measured at 450 nm using a microplate reader (Infinite® F200 PRO, Tecan Group Ltd., Männedorf, Switzerland).
2.4. Interleukin-6 (IL-6) detection
MG6 cells (0.5 × 106/well) were seeded on 12-well plates and incubated at 37 °C with 5% CO2 for 24 h. The medium was then replaced with LPS (100 ng/mL)- and/or API (5, 10, and 20 μM)-containing media and the cells were incubated for another 24 h. The culture media were collected at the end of treatments and used to detect IL-6 secretion. IL-6 concentrations in the culture media were measured using an ELISA MAX™ in accordance with the manufacturer's protocol. Absorbance was measured at 450 nm using the microplate reader.
2.5. Nitric oxide (NO) assay
After incubation of MG6 cells for 24 h, NO concentration in the medium was measured by the Griess reaction method [28]. In brief, 100 μL of culture medium was applied to each well of a 96-well plate. Then, 100 μL of an equal mixture of Griess reagent:0.2% N-1-naphthyl ethylenediamine (Wako, Osaka, Japan) and 2% sulfanilamide (Sigma-Aldrich) in 10% H3PO4 (Wako) was added to each well. After incubation for 10 min at room temperature, the absorbance was measured at 550 nm using the microplate reader.
2.6. RNA extraction and quantitative real-time PCR analysis
MG6 cells (0.5 × 106/well) were seeded on 12-well plates and incubated at 37 °C with 5% CO2 for 24 h. The medium was then replaced with LPS (100 ng/mL)- and/or API (5, 10, and 20 μM)-containing media and the cells were incubated for another 30 or 48 h. Subsequently, RNAiso plus reagent was used to extract total RNA from the cells in accordance with the manufacturer's protocol. The total RNA was reverse transcribed to cDNA using ReverTra Ace® qPCR RT Master Mix with gDNA Remover in accordance with the manufacturer's protocol. cDNA samples were subjected to PCR using Applied Biosystem StepOnePlus™ Real-time PCR System (Life Technologies, Waltham, MA, USA) and THUNDERBIRD® SYBR qPCR MIX. The following primers were used: IDO forward, 5ʹ-AAGGGCTTCTTCCTCGTCTC-3ʹ and reverse, 5ʹ-AAAAACGTGTCTGGGTCCAC-3ʹ; ACMSD forward 5ʹ-GCCTCCCACAGTTGGATAGA-3ʹ and reverse 5ʹ-GCCTCAAACACAGACCCATT-3ʹ; GAPDH forward 5ʹ-ACCCAGAAGACTGTGGATGG-3ʹ and reverse 5ʹ-CACATTGGGGGTAGGAACAC-3ʹ and 18S rRNA forward 5ʹ-CGCGGTTCTATTTTGTTGGT-3ʹ and reverse 5ʹ-AGTCGGCATCGTTTATGGTC-3ʹ as an internal standard for normalization of IDO and ACMDS mRNA expression levels, respectively. The amplification conditions were 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 60 °C for 31 s, and 95 °C for 15 s, followed by 60 °C for 1 min.
2.7. Protein extraction
MG6 cells (4 × 106/dish) were seeded on 60-mm dishes and incubated at 37 °C with 5% CO2 for 24 h. Then, the medium was replaced with LPS (100 ng/mL)- and/or API (5, 10, and 20 μM)-containing media. Protein collection of MAPK and NF-κB signaling was performed after 30 min of LPS stimulation when activation of these proteins was clearly observed according to the results of preliminary experiments (data not shown). Subsequently, cells were washed with PBS and collected in protein extraction buffer that contained protease (Calbiochem, San Diego, CA, USA) and phosphatase (Wako) inhibitor cocktails. Aggregates were lysed by an ultrasonicator and centrifuged at 13,000×g at 4 °C for 5 min. The supernatants were collected as whole protein samples.
2.8. Western blot analysis
Protein samples were applied to gels at 20–50 μg/well and then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After transferring to a polyvinylidene difluoride membrane (Millipore, Darmstadt, Germany), the blot was blocked with 5% dry skim milk for 1 h and then washed three times with PBS with 0.05% Tween 20 (PBST). Subsequently, membranes were treated overnight with antibodies against p-p38 (1:300), p38 (1:1000), p-Erk (1:2000), Erk (1:1000), p-JNK (1:2000), JNK (1:1000), IκBα (1:300), or β-actin (1:1000). After washing with PBST, the membranes were treated with anti-rabbit IgG horseradish peroxidase conjugate (1:3000) (W4018) (Promega) as secondary antibody. Blots were developed using Immobilon Western Chemiluminescent HRP Substrate (Millipore) and detected by a Light Capture III (ATTO Co., Ltd., Tokyo, Japan). Luminescence intensity was quantified using CS Analyzer 3.0 (ATTO Co., Ltd.).
2.9. Statistical analysis
Data are represented as the mean ± standard error (SE). Statistics were performed using the Bell Curve for Excel ver. 2.10 (Social Survey Research Information Co., Ltd., Tokyo, Japan). Data were analyzed by one-way ANOVA with Dunnett test or Steel's test. Data of Fig. 5 were analyzed by Student's t-test. *p < 0.05 and **p < 0.01 were considered statistically significant.
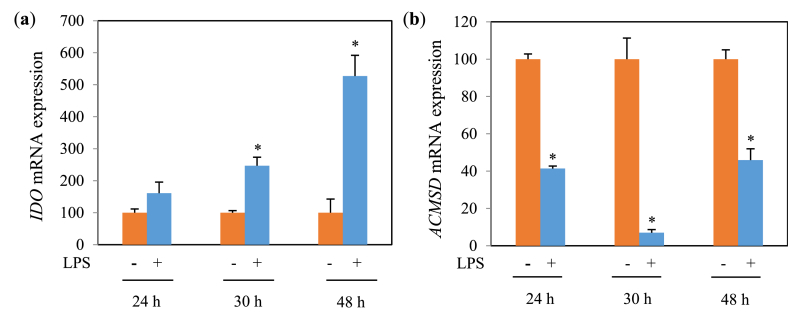
Fig. 5.
IDO (a) and ACMSD (b) mRNA expression (relative value) in MG6 cells treated with 100 ng/mL of LPS for various stimulation times. Values indicate mean ± SE (n = 5–6). *p < 0.05 vs LPS (−).
3. Results
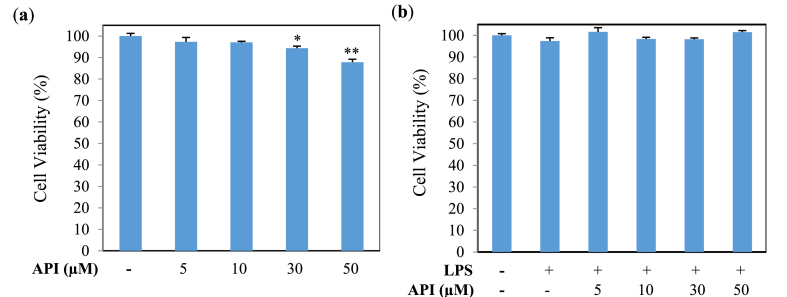
3.1. Effect of API and LPS on cell viability
The cytotoxicity of API and LPS on the viability of MG6 cells was investigated using an MTT assay. Cells were treated with API (0, 5, 10, 30, and 50 μM) and/or LPS (100 ng/mL) for 24 h. The results revealed that 30 and 50 μM API treatments significantly decreased the viability of MG6 cells compared with the control group (Fig. 2a). However, API treatments up to 50 μM in the presence of LPS had no significant effect on cell viability (Fig. 2b). Therefore, API concentrations of lower than 30 μM were selected for the next stage of study, which are also supported by earlier experiments [1,27].
Fig. 2.
Viability of MG6 cells treated for 24 h (a) with or without API and (b) with or without LPS (100 ng/mL). Values indicate the mean ± SE (n = 6). *p < 0.05 and **p < 0.01 vs API (−).
3.2. Production of inflammatory mediators
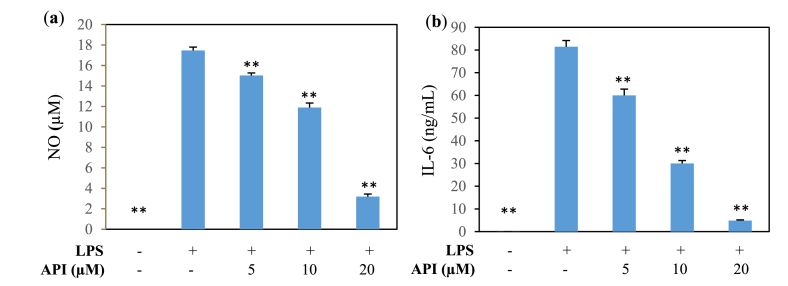
The levels of inflammatory mediators, such as NO, IL-6, and TNF, were measured. Many in vitro studies reported that the production of inflammatory mediators was sufficiently induced at 24 h of LPS exposure [1,27,29]. Stimulation of MG6 cells with LPS resulted in significant increases in the concentrations of NO (Fig. 3a), IL-6 (Fig. 3b), and TNF (data not shown) in the culture media. API treatment led to significant inhibition of NO and IL-6 releases induced by LPS in a dose-dependent manner. However, API did not affect LPS-induced TNF production (data not shown).
Fig. 3.
Effect of API on proinflammatory mediator secretion from MG6 cells after 24 h of LPS treatment (100 ng/mL). (a) NO and (b) IL-6 secretion. Values indicate the mean ± SE (n = 6). **p < 0.01 vs LPS (+).
3.3. IDO and ACMSD mRNA expression
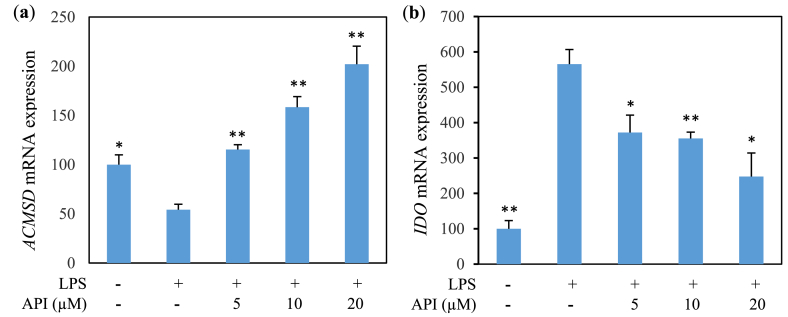
Since this study mainly aims to investigate the relationship of inflammation with tryptophan metabolic enzymes, particularly ACMSD, we carried out qPCR analyses of mRNAs isolated from MG6 cells treated with LPS. To obtain the optimum experimental conditions, the preliminary experiment of LPS in several doses and stimulation time on the mRNA expression level of IDO and ACMSD was performed. LPS treatment with a dose of 100 ng/mL resulted in the optimum induction effect to increase IDO expression level (p < 0.0857) (Fig. 4a) followed by reduction of ACMSD level (Fig. 4b) in MG6 cells at 48 h (5a) and 30 h (Fig. 5b) of LPS stimulation, respectively. LPS treatment suppressed ACMSD mRNA expression (Figs. 5a and 6a) and induced IDO mRNA expression (Figs. 5b and 6b) compared with the LPS untreated group. Similarly, we examined the anti-inflammatory effect of API on tryptophan metabolism in LPS-induced microglial cells. API treatment results in a significant increment in ACMSD mRNA expression in a dose-dependent manner (Fig. 6a) and inhibition of IDO mRNA expression (Fig. 6b) in LPS-treated MG6 cells.
Fig. 6.
Effect of API on mRNA expression in MG6 cells treated with LPS (100 ng/mL). (a) ACMSD (30 h) and (b) IDO (48 h) expression (relative value). Values indicate the mean ± SE (n = 6). *p < 0.05 and **p < 0.01 vs LPS (+).
3.4. Expression of NF-κB and MAPK pathway proteins
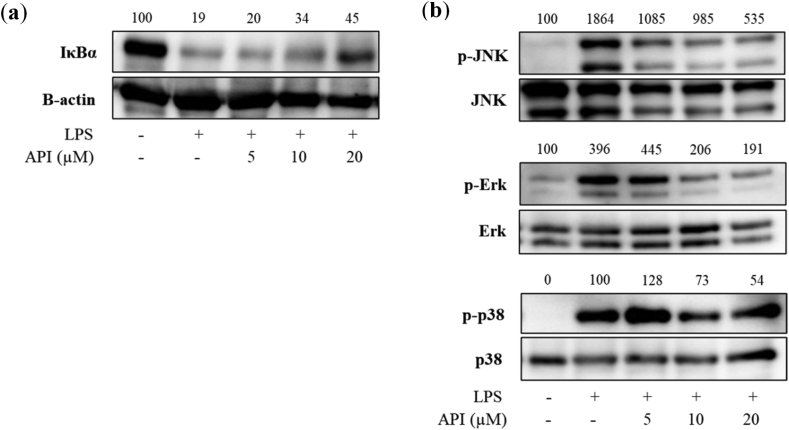
NF-κB and MAPK pathways control the production of inflammatory mediators that include inflammatory cytokines and chemokines [9,30]. The cytoplasmic level of IκBα protein was measured by western blotting to determine the inhibitory effect of API on NF-κB pathways. After stimulation of MG6 cells with LPS for 30 min, cytosolic IκBα protein was degraded. The LPS-induced IκBα degradation was lightly inhibited by co-treatment of LPS and API at high doses (Fig. 7a). LPS treatment for 30 min also facilitated the phosphorylation of MAPK proteins, which included JNK, Erk and p38 in MG6 cells (Fig. 7b). API attenuated the levels of phosphorylated JNK and ERK with the highest decrease in phosphorylation intensity at 72% and 52%, respectively, in 20 μM API-treated group in comparison with those in LPS-treated group. However, LPS-induced phosphorylation of p38 was not remarkable affected by API with a 46% reduction in phosphorylation level of p38 at 20 μM.
Fig. 7.
Effect of API on expression of NF-κB and MAPK pathway proteins in MG6 cells treated with LPS (100 ng/mL). (a) IκBα and (b) phospho-JNK, phospho-Erk, and phospho-p38 protein expression (Appendix_Supplementary Data).
4. Discussion
Flavonoids are abundant in vegetables, fruits, and herbs. A high intake of diets rich in flavonoids is reported to prevent numerous chronic diseases that include metabolic disorders, cardiovascular diseases, neurodegenerative diseases, cancer, and allergies [1,[31], [32], [33], [34]]. API is a natural flavonoid that has received immense attention because of its low intrinsic toxicity and various biological activities [1–5]. Previous studies have reported that API remarkably inhibits cytokine production and astrocyte activation stimulated by LPS via NF-κB, MAPK and STAT3 pathways [9]. It has also been shown that API provides protection against neuroinflammation associated with AD in vitro and in vivo [7,8]. API is proposed to be an important dietary flavonoid with potent neuroprotective, chemopreventive, and anti-inflammatory abilities [1,2,4,9,10].
LPS is produced by gram-negative bacteria, which binds to a surface receptor called Toll-like receptor 4 (TLR4) and triggers the secretion of cytokines [35]. Activating of microglial cells by LPS leads to increased secretion of inflammatory mediators that include NO, IL-6, and TNF [36,37]. In the present study, microglial cells were treated with LPS to mimic the inflammatory state in neurodegenerative diseases. API treatment led to significant reductions of LPS-induced NO and IL-6 levels in MG6 cells. However, API did not affect TNF production. LPS induced massive NO production in an immune cell that participates in pathological acute and chronic inflammatory disorders [38]. During inflammation, high production of NO causes cytotoxicity, tissue and DNA damage, and mutations [39]. Moreover, IL-6 via a specific receptor stimulates various signaling pathways activating gene [40]. In CNS disorders, various molecules elevate the IL-6 level, including LPS and TNF, which increases the central production of inflammatory cytokines [37,40]. Therefore, our results suggest that API attenuates LPS-induced activation of glial cells and the consequential production of inflammatory metabolites, whereby reducing of chronic inflammation is an effective strategy to prevent the pathological progression of chronic diseases.
Several studies have demonstrated the significance of kynurenine metabolism in aging and neurodegenerative diseases through the action of its metabolic enzymes [12]. In this study, the increased IDO mRNA expression induced by LPS was significantly reduced by API treatment of microglial cells. IDO is generally low or even undetectable under normal conditions [41]. Induction of IDO expression by LPS and inflammatory mediators, such as TNF and IL-6, promotes the tryptophan conversion to kynurenine leading to upregulation of neurotoxic metabolites, including 3-hydroxykynurenine (3-HK) and QUIN concentration in the kynurenine pathway [42–43]. High levels of QUIN and 3-HK lead to neuronal damage via oxidative stress [44] and hyperactivation of N-methyl-ᴅ-aspartate receptors [44,45]. Thus, repression of IDO expression appears to improve the condition of neuroinflammation.
Conversely, proinflammatory IFN-γ downregulates ACMSD expression in macrophages and dendritic cells [46]. A depleted amount or activity of ACMSD within the CNS can affect the development of a chronic inflammatory environment by promoting higher non-enzymatic conversion of amino carboxy-muconate-semialdehyde to neurotoxic QUIN [47]. To our best knowledge, the present study provides the first evidence of a reduced level of ACMSD mRNA expression induced by LPS in microglial cells. Furthermore, API treatment upregulated ACMSD expression level suppressed by LPS. The upregulation of ACMSD activity may be protective by contributing to the production of neuroprotective picolinic acid away from neurotoxic QUIN, which would prevent the generation of inflammatory metabolites [47,48].
To examine the regulatory mechanism of IDO and ACMSD expression by API in LPS-stimulated microglia, we investigated the protein expression of NF-κB and MAPKs pathways. Inflammatory signals in the brain, such as NF-κB and MAPKs, are activated by substances such as LPS and proinflammatory cytokines in neuroglial cells [49,50].
NF-κB inactivates in the cytoplasm through an association with the inhibitor protein IκBα. In stimulated cells, activation of the canonical NF-κB is promoted by cytokines and endotoxins such as LPS via proteasomal degradation of IκBα triggered through phosphorylation of its specific site by a multi-subunit IκB kinase (IKK) complex [51–52]. Upon activation, this kinase complex phosphorylates IκBα at two N-terminal serines resulting in cytosolic IκBα degradation in the proteasome. This state allows the release and translocation of NF-κB members, mainly p50/RelA and p50/c-Rel dimers, into the nucleus. Translocated NF-κB bind to selective target gene promoters and enhances genes transcription, including inflammatory mediators [53]. We found that API reduced LPS-induced degradation of IκBα, which inhibited NF-κB nuclear translocation. In the MAPK pathway, API treatment attenuated the LPS-induced phosphorylation of Erk and JNK. However, phosphorylation of p38 was unaffected by API. Activation of TLR4 by LPS triggers the MyD88-dependent pathway through MAPK. MAPK signaling is composed of three major groups that regulate gene expression: Erk activated by MKK1 and MKK2, JNK by MKK4 and MKK7, and p38 kinase by MKK3, MKK4, and MKK6. The prominent example of MAPK modulation in target genes expression is phosphorylation and activation of the transcription factor in the nucleus by JNK MAPK, such as c-Jun, activating transcription factor-2 (ATF-2), and activator protein-1 (AP-1), which are required to target promoters of many genes including various cytokines and chemokines [54,55]. Therefore, inhibition of LPS-induced NF-κB nuclear translocation and MAPK activation by API subsequently suppressed the central production of inflammatory mediators in microglia.
Moreover, these inflammatory signaling pathways are involved in the regulation of IDO expression. For example, induction of IDO is mediated by JNK/p38 MAPK and NF-κB protein signals via activation of the transcription factor c-Jun or NF-κB, respectively, located in the IDO promoter region [56–57]. Conversely, no direct relationship has been found between ACMSD and inflammatory pathways. However, MAPK and NF-κB signaling proteins regulate transcriptional factors of the ACMSD gene promoter [58–59]. Constitutive expression of ACMSD is positively regulated by activation of hepatocyte nuclear factor-4α (HNF4α) [60]. The transcriptional activity of HNF4α is inhibited by the activation of Erk [58,59] and JNK MAPK [59] as well as the NF-κB pathway [61], which might affect the expression of ACMSD. Furthermore, suppression of NF-κB activation enhances PGC1α expression [62], which is positively implicated in regulating ACMSD expression in cooperation with HNF4α [63]. Thus, controlling the transcriptional regulatory region of IDO and ACMSD genes may modulate the expression level of these enzymes.
Taken together, our results indicated that API recovered the LPS-induced decrease in ACMSD and increased IDO mRNA expression. These actions may be mediated by attenuation of IL-6 and NO production via inhibition of Erk and JNK MAPK phosphorylation as well as nuclear translocation of NF-κB.
5. Conclusion
ACMSD and IDO tryptophan metabolic key enzyme's gene expressions are altered under the inflammatory condition induced by LPS. Additionally, API recovered the gene expression of tryptophan metabolic enzymes and suppressed the production of IL-6 and NO. The anti-inflammatory effect of API may be mediated by inhibiting pro-inflammatory mediators via inactivation of Erk, JNK MAPK, and NF-κB pathways in LPS-stimulated microglial cells, which prevents a vicious cycle of generating inflammatory metabolites. Ultimately, API is a promising therapeutic target for inflammatory diseases related to neurodegeneration.
Credit author statement
Dian Kurniati: Performed the experiments; Analyzed and interpreted the data; Wrote the paper; Shizuka Hirai: Contributed reagents, materials, analysis tools or data; Yukari Egashira: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Professor Yukari Egashira was supported by JSPS KAKENHI Grant Number JP21K05420, Japan.
Data availability statement
Data included in article/supp. Material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank Mitchell Arico for editing a draft of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2022.e12743.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Chen P., Huo X., Liu W., Li K., Sun Z., Tian J. Apigenin exhibits anti-inflammatory effects in LPS-stimulated BV2 microglia through activating GSK3β/Nrf2 signaling pathway. Immunopharmacol. Immunotoxicol. 2020;42:9–16. doi: 10.1080/08923973.2019.1688345. [DOI] [PubMed] [Google Scholar]
- 2.Madunić J., Madunić I.V., Gajski G., Popić J., Garaj-Vrhovac V. Apigenin: a dietary flavonoid with diverse anticancer properties. Cancer Lett. 2018;413:11–22. doi: 10.1016/j.canlet.2017.10.041. [DOI] [PubMed] [Google Scholar]
- 3.Malik S., Suchal K., Irfan Khan S., Bhatia J., Kishore K., Kumar Dinda A., Singh Arya D. Apigenin ameliorates streptozotocin-induced diabetic nephropathy in rats via MAPK-NF-KB-TNF-alpha and TGF-beta1-MAPK-fibronectin pathways. Am. J. Physiol. Ren. Physiol. 2017;313:414–422. doi: 10.1152/ajprenal.00393.2016.-Diabetic. [DOI] [PubMed] [Google Scholar]
- 4.Nabavi S.F., Khan H., D’onofrio G., Šamec D., Shirooie S., Dehpour A.R., Argüelles S., Habtemariam S., Sobarzo-Sanchez E. Apigenin as neuroprotective agent: of mice and men. Pharmacol. Res. 2018;128:359–365. doi: 10.1016/j.phrs.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Bijani S., Dizaji R., · Ali Sharafi. · Mir-Jamal Hosseini. Sharafi A., Hosseini M.-J. Neuroprotective effect of apigenin on depressive-like behavior: mechanistic approach. Neurochem. Res. 2022;47:644–655. doi: 10.1007/s11064-021-03473-0. [DOI] [PubMed] [Google Scholar]
- 6.U. Us Department of Agriculture, USDA database for the flavonoid content of selected foods. Release 3.2 (November 2015) | Ag Data Commons, (n.d.). https://data.nal.usda.gov/dataset/usda-database-flavonoid-content-selected-foods-release-32-november-2015 (accessed July 25, 2022).
- 7.Dourado N.S., Souza C. dos S., de Almeida M.M.A., Bispo da Silva A., dos Santos B.L., Silva V.D.A., De Assis A.M., da Silva J.S., Souza D.O., Costa F.D., Butt A.M., Costa S.L. Neuroimmunomodulatory and neuroprotective effects of the flavonoid apigenin in in vitro models of neuroinflammation associated with Alzheimer's disease. Front. Aging Neurosci. 2020;12:1–14. doi: 10.3389/fnagi.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y.J., Kim J.H., He M.T., Lee A.Y., Cho E.J. Apigenin ameliorates scopolamine-induced cognitive dysfunction and neuronal damage in mice. Mol. 2021;26:5192. doi: 10.3390/MOLECULES26175192. 26 (2021) 5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Che D.N., Cho B.O., Kim J.S., Shin J.Y., Kang H.J., Il Jang S. Luteolin and apigenin attenuate LPS-induced astrocyte activation and cytokine production by targeting MAPK, STAT3, and NF-κB signaling pathways. Inflammation. 2020;43:1716–1728. doi: 10.1007/s10753-020-01245-6. [DOI] [PubMed] [Google Scholar]
- 10.Che D.N., Cho B.O., Kim J.S., Shin J.Y., Kang H.J., Il Jang S. Multidisciplinary Digital Publishing Institute (MDPI); 2020. Effect of Luteolin and Apigenin on the Production of IL-31 and IL-33 in Lipopolysaccharides-Activated Microglia Cells and Their Mechanism of Action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badawy A.A.B. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res. 2017;10:1–20. doi: 10.1177/1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Oliveira F.R., Fantucci M.Z., Adriano L., Valim V., Cunha T.M., Louzada-Junior P., Rocha E.M. Neurological and inflammatory manifestations in sjögren’s syndrome: the role of the kynurenine metabolic pathway. Int. J. Mol. Sci. 2018;19:60–80. doi: 10.3390/ijms19123953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banzola I., Mengus C., Wyler S., Hudolin T., Manzella G., Chiarugi A., Boldorini R., Sais G., Schmidli T.S., Chiffi G., Bachmann A., Sulser T., Spagnoli G.C., Provenzano M. Expression of indoleamine 2,3-dioxygenase induced by IFN-γ and TNF-α as potential biomarker of prostate cancer progression. Front. Immunol. 2018;9:1051. doi: 10.3389/FIMMU.2018.01051/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar F., Awuah D., Negm O.H., Shakib F., Ghaemmaghami A.M. The role of indoleamine 2,3-dioxygenase-aryl hydrocarbon receptor pathway in the TLR4-induced tolerogenic phenotype in human DCs. Sci. Rep. 2017;71:1–11. doi: 10.1038/srep43337. 7 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemin G.J., Brew B.J., Noonan C.E., Takikawa O., Cullen K.M. Indoleamine 2,3 dioxygenase and quinolinic acid immunoreactivity in Alzheimer's disease hippocampus. Neuropathol. Appl. Neurobiol. 2005;31:395–404. doi: 10.1111/J.1365-2990.2005.00655.X. [DOI] [PubMed] [Google Scholar]
- 16.Fertan E., Stover K.R.J., Brant M.G., Stafford P.M., Kelly B., Diez-Cecilia E., Wong A.A., Weaver D.F., Brown R.E. Effects of the novel Ido inhibitor DWG-1036 on the behavior of male and female 3xTg-AD mice. Front. Pharmacol. 2019;10:1044. doi: 10.3389/FPHAR.2019.01044/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganong A.H., Cotman C.W. Kynurenic acid and quinolinic acid act at N-methyl-D-aspartate receptors in the rat hippocampus. J. Pharmacol. Exp. Therapeut. 1986;236:293–299. [PubMed] [Google Scholar]
- 18.Heilman P.L., Wang E.W., Lewis M.M., Krzyzanowski S., Capan C.D., Burmeister A.R., Du G., Escobar Galvis M.L., Brundin P., Huang X., Brundin L. Tryptophan metabolites are associated with symptoms and nigral pathology in Parkinson's disease. Mov. Disord. 2020;35:2028–2037. doi: 10.1002/MDS.28202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thirtamara-Rajamani K., Li P., Escobar Galvis M.L., Labrie V., Brundin P., Brundin L. Is the enzyme ACMSD a novel therapeutic target in Parkinson's disease? J. Parkinsons Dis. 2017;7:577–587. doi: 10.3233/JPD-171240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrosi G., Cerri S., Blandini F. A further update on the role of excitotoxicity in the pathogenesis of Parkinson's disease. J. Neural. Transm. 2014;121:849–859. doi: 10.1007/s00702-013-1149-z. [DOI] [PubMed] [Google Scholar]
- 21.Pucci L., Perozzi S., Cimadamore F., Orsomando G., Raffaelli N. Tissue expression and biochemical characterization of human 2-amino 3-carboxymuconate 6-semialdehyde decarboxylase, a key enzyme in tryptophan catabolism. FEBS J. 2007;274:827–840. doi: 10.1111/j.1742-4658.2007.05635.x. [DOI] [PubMed] [Google Scholar]
- 22.Tanabe A., Egashira Y., Fukuoka S.I., Shibata K., Sanada H. Expression of rat hepatic 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase is affected by a high protein diet and by streptozotocin-induced diabetes. J. Nutr. 2002;132:1153–1159. doi: 10.1093/jn/132.6.1153. [DOI] [PubMed] [Google Scholar]
- 23.Egashira Y., Murotani G., Tanabe A., Saito K., Uehara K., Morise A., Sato M., Sanada H. Differential effects of dietary fatty acids on rat liver α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase activity and gene expression. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2004;1686:118–124. doi: 10.1016/j.bbalip.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Nakamichi K., Saiki M., Kitani H., Kuboyama Y., Morimoto K., Takayama-Ito M., Kurane I. Suppressive effect of simvastatin on interferon-β-induced expression of CC chemokine ligand 5 in microglia. Neurosci. Lett. 2006;407:205–210. doi: 10.1016/j.neulet.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 25.Takenouchi T., Ogihara K., Sato M., Kitani H. Inhibitory effects of U73122 and U73343 on Ca2+ influx and pore formation induced by the activation of P2X7 nucleotide receptors in mouse microglial cell line. Biochim. Biophys. Acta Gen. Subj. 2005;1726:177–186. doi: 10.1016/j.bbagen.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Minogue A.M., Barrett J.P., Lynch M.A., Am M., Jp B., Ma L. LPS-induced release of IL-6 from glia modulates production of IL-1β in a JAK2-dependent manner.pdf. J. Neuroinflammation. 2012;9 doi: 10.1186/1742-2094-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Wang G., Gurley E.C., Zhou H. Flavonoid apigenin inhibits lipopolysaccharide-induced inflammatory response through multiple mechanisms in Macrophages. PLoS One. 2014;9:1–18. doi: 10.1371/journal.pone.0107072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guevara I., Iwanejko J., Dembińska-Kieć A., Pankiewicz J., Wanat A., Anna P., Gobek I., Bartuś S., Malczewska-Malec M., Szczudlik A. Determination of nitrite/nitrate in human biological material by the simple Griess reaction. Clin. Chim. Acta. 1998;274:177–188. doi: 10.1016/S0009-8981(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 29.Hirai S., Horii S., Matsuzaki Y., Ono S., Shimmura Y., Sato K., Egashira Y. Anti-inflammatory effect of pyroglutamyl-leucine on lipopolysaccharide-stimulated RAW 264.7 macrophages. Life Sci. 2014;117:1–6. doi: 10.1016/J.LFS.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Cheng S.C., Huang W.C., Pang J.H.S., Wu Y.H., Cheng C.Y. Quercetin inhibits the production of IL-1β-induced inflammatory cytokines and chemokines in ARPE-19 cells via the MAPK and NF-κB signaling pathways. Int. J. Mol. Sci. 2019;20:2957. doi: 10.3390/IJMS20122957. 20 (2019) 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang K.L., Yu S.J., Huang W.C., Yen H.R. Luteolin attenuates allergic nasal inflammation via inhibition of interleukin-4 in an allergic rhinitis mouse model and peripheral blood from human subjects with allergic rhinitis. Front. Pharmacol. 2020;11:291. doi: 10.3389/FPHAR.2020.00291/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A.U., Dagur H.S., Khan M., Malik N., Alam M., Mushtaque M. Therapeutic role of flavonoids and flavones in cancer prevention: current trends and future perspectives. Eur. J. Med. Chem. Reports. 2021;3 doi: 10.1016/J.EJMCR.2021.100010. [DOI] [Google Scholar]
- 33.Ciumărnean L., Milaciu M.V., Runcan O., Vesa S.C., Răchisan A.L., Negrean V., Perné M.G., Donca V.I., Alexescu T.G., Para I., Dogaru G., L C., Mv M., O R., Șc V., AL R., V N., Mg P., VI D., Tg A., I P., G D. The effects of flavonoids in cardiovascular diseases. Molecules. 2020;25 doi: 10.3390/MOLECULES25184320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher P. The potential of flavonoids for the treatment of neurodegenerative diseases. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20123056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park B.S., Lee J.O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013;45:e66–e69. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J., Bi W., Xiao S., Lan X., Cheng X., Zhang J., Lu D., Wei W., Wang Y., Li H., Fu Y., Zhu L. Neuroinflammation induced by lipopolysaccharide causes cognitive impairment in mice. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-42286-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y., Taylor N., Yao X., Bhattacharya A. Mouse primary microglia respond differently to LPS and poly(I:C) in vitro. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-89777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iizumi T., Takahashi S., Mashima K., Minami K., Izawa Y., Abe T., Hishiki T., Suematsu M., Kajimura M., Suzuki N. A possible role of microglia-derived nitric oxide by lipopolysaccharide in activation of astroglial pentose-phosphate pathway via the Keap1/Nrf2 system. J. Neuroinflammation. 2016;13:1–20. doi: 10.1186/S12974-016-0564-0/FIGURES/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M.Y. Intracellular and extracellular factors influencing the genotoxicity of nitric oxide and reactive oxygen species. Oncol. Lett. 2017;13:1417–1424. doi: 10.3892/OL.2017.5584/HTML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y., Zheng S.G. Hall of fame among pro-inflammatory cytokines: interleukin-6 gene and its transcriptional regulation mechanisms. Front. Immunol. 2016;7:604. doi: 10.3389/FIMMU.2016.00604/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meireson A., Devos M., Brochez L. Ido expression in cancer: different compartment, different functionality. Front. Immunol. 2020;11:2340. doi: 10.3389/FIMMU.2020.531491/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y.S., Ogbechi J., Clanchy F.I., Williams R.O., Stone T.W. Ido and kynurenine metabolites in peripheral and CNS disorders. Front. Immunol. 2020;11:388. doi: 10.3389/FIMMU.2020.00388/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parrott J.M., Redus L., Santana-Coelho D., Morales J., Gao X., O'Connor J.C. Neurotoxic kynurenine metabolism is increased in the dorsal hippocampus and drives distinct depressive behaviors during inflammation. Transl. Psychiatry. 2016;610:e918. doi: 10.1038/tp.2016.200. 6 (2016) e918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castellano-Gonzalez G., Jacobs K.R., Don E., Cole N.J., Adams S., Lim C.K., Lovejoy D.B., Guillemin G.J. Kynurenine 3-monooxygenase activity in human primary neurons and effect on cellular bioenergetics identifies new neurotoxic mechanisms. Neurotox. Res. 2019;35:530–541. doi: 10.1007/S12640-019-9997-4/FIGURES/7. [DOI] [PubMed] [Google Scholar]
- 45.Behl T., Kaur I., Sehgal A., Singh S., Bhatia S., Al-Harrasi A., Zengin G., Bumbu A.G., Andronie-Cioara F.L., Nechifor A.C., Gitea D., Bungau A.F., Toma M.M., Bungau S.G. The footprint of kynurenine pathway in neurodegeneration: janus-faced role in Parkinson's disorder and therapeutic implications. Int. J. Mol. Sci. 2021;22:6737. doi: 10.3390/IJMS22136737. 22 (2021) 6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braidy N., Rossez H., Lim C.K., Jugder B.E., Brew B.J., Guillemin G.J. Characterization of the kynurenine pathway in CD8+ human primary monocyte-derived dendritic cells. Neurotox. Res. 2016;30:620–632. doi: 10.1007/s12640-016-9657-x. [DOI] [PubMed] [Google Scholar]
- 47.Sundaram G., Sundaram G., Lim C.K., Brew B.J., Brew B.J., Brew B.J., Guillemin G.J., Guillemin G.J. Kynurenine pathway modulation reverses the experimental autoimmune encephalomyelitis mouse disease progression. J. Neuroinflammation. 2020;17:1–14. doi: 10.1186/s12974-020-01844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brundin L., Sellgren C.M., Lim C.K., Grit J., Pålsson E., Landén M., Samuelsson M., Lundgren K., Brundin P., Fuchs D., Postolache T.T., Traskman-Bendz L., Guillemin G.J., Erhardt S. An enzyme in the kynurenine pathway that governs vulnerability to suicidal behavior by regulating excitotoxicity and neuroinflammation. Transl. Psychiatry. 2016;6 doi: 10.1038/tp.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Zhang T., Wang D., Jiang Y., Guo T., Zhang Y., Zhu F., Han K., Mu L., Wang G. IFN-γ regulates the transformation of microglia into dendritic-like cells via the ERK/c-myc signaling pathway during cerebral ischemia/reperfusion in mice. Neurochem. Int. 2020;141 doi: 10.1016/J.NEUINT.2020.104860. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y.L., Liu M., Cheng X., Li W.H., Zhang S.S., Wang Y.H., Du G.H. Myricitrin blocks activation of NF-κB and MAPK signaling pathways to protect nigrostriatum neuron in LPS-stimulated mice. J. Neuroimmunol. 2019;337 doi: 10.1016/J.JNEUROIM.2019.577049. [DOI] [PubMed] [Google Scholar]
- 51.Didonato J.A., Hayakawa M., Rothwarf D.M., Zandi E., Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 52.Karin M., Delhase M. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin. Immunol. 2000;12:85–98. doi: 10.1006/SMIM.2000.0210. [DOI] [PubMed] [Google Scholar]
- 53.Dorrington M.G., Fraser I.D.C. NF-κB signaling in macrophages: dynamics, crosstalk, and signal integration. Front. Immunol. 2019;10:705. doi: 10.3389/FIMMU.2019.00705/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaminska B., Gozdz A., Zawadzka M., Ellert-Miklaszewska A., Lipko M. MAPK signal transduction underlying brain inflammation and gliosis as therapeutic target. Anat. Rec. 2009;292:1902–1913. doi: 10.1002/ar.21047. [DOI] [PubMed] [Google Scholar]
- 55.Pua L.J.W., Mai C.W., Chung F.F.L., Khoo A.S.B., Leong C.O., Lim W.M., Hii L.W. Functional roles of JNK and p38 MAPK signaling in nasopharyngeal carcinoma. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornyák L., Dobos N., Koncz G., Karányi Z., Páll D., Szabó Z., Halmos G., Székvölgyi L. The role of indoleamine-2,3-dioxygenase in cancer development, diagnostics, and therapy. Front. Immunol. 2018;9:151. doi: 10.3389/FIMMU.2018.00151/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Opitz C.A., Litzenburger U.M., Opitz U.A., Sahm F., Ochs K., Lutz C., Wick W., Platten M. The Indoleamine-2,3-dioxygenase (Ido) inhibitor 1-methyl-D-tryptophan upregulates Ido1 in human cancer cells. PLoS One. 2011;6 doi: 10.1371/JOURNAL.PONE.0019823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vetö B., Bojcsuk D., Bacquet C., Kiss J., Sipeki S., Martin L., Buday L., Bálint B.L., Arányi T. The transcriptional activity of hepatocyte nuclear factor 4 alpha is inhibited via phosphorylation by ERK1/2. PLoS One. 2017;12:1–19. doi: 10.1371/journal.pone.0172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Won K.J., Park J.S., Jeong H. Repression of hepatocyte nuclear factor 4 alpha by AP-1 underlies dyslipidemia associated with retinoic acid. J. Lipid Res. 2019;60:794. doi: 10.1194/JLR.M088880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin M., Kim I., Inoue Y., Kimura S., Gonzalez F.J. Regulation of mouse hepatic α-amino-β-carboxymuconate-ε- semialdehyde decarboxylase, a key enzyme in the tryptophan-nicotinamide adenine dinucleotide pathway, by hepatocyte nuclear factor 4α and peroxisome proliferator-activated receptor α. Mol. Pharmacol. 2006;70:1281–1290. doi: 10.1124/mol.106.026294. [DOI] [PubMed] [Google Scholar]
- 61.Lv D.D., Zhou L.Y., Tang H. Hepatocyte nuclear factor 4α and cancer-related cell signaling pathways: a promising insight into cancer treatment. Exp. Mol. Med. 2021;531:8–18. doi: 10.1038/s12276-020-00551-1. 53 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barroso W.A., Victorino V.J., Jeremias I.C., Petroni R.C., Ariga S.K.K., Salles T.A., Barbeiro D.F., de Lima T.M., de Souza H.P. High-fat diet inhibits PGC-1α suppressive effect on NFκB signaling in hepatocytes. Eur. J. Nutr. 2018;57:1891–1900. doi: 10.1007/S00394-017-1472-5. [DOI] [PubMed] [Google Scholar]
- 63.Koshiguchi M., Hirai S., Egashira Y. PGC1α regulates ACMSD expression through cooperation with HNF4α. Amino Acids. 2018;50:1769–1773. doi: 10.1007/s00726-018-2652-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. Material/referenced in article.