Abstract
Cigarette smoking and alcohol use are among the most prevalent substances used worldwide and account for a substantial proportion of preventable morbidity and mortality, underscoring the public health significance of understanding their etiology. Genome-wide association studies (GWAS) have successfully identified genetic variants associated with cigarette smoking and alcohol use traits. However, the vast majority of risk variants reside in non-coding regions of the genome, and their target genes and neurobiological mechanisms are unknown. Chromosomal conformation mappings can address this knowledge gap by charting the interaction profiles of risk-associated regulatory variants with target genes. To investigate the functional impact of common variants associated with cigarette smoking and alcohol use traits, we applied Hi-C coupled MAGMA (H-MAGMA) built upon cortical and newly generated midbrain dopaminergic neuronal Hi-C datasets to GWAS summary statistics of nicotine dependence, cigarettes per day, problematic alcohol use, and drinks per week. The identified risk genes mapped to key pathways associated with cigarette smoking and alcohol use traits, including drug metabolic processes and neuronal apoptosis. Risk genes were highly expressed in cortical glutamatergic, midbrain dopaminergic, GABAergic, and serotonergic neurons, suggesting them as relevant cell types in understanding the mechanisms by which genetic risk factors influence cigarette smoking and alcohol use. Lastly, we identified pleiotropic genes between cigarette smoking and alcohol use traits under the assumption that they may reveal substance-agnostic, shared neurobiological mechanisms of addiction. The number of pleiotropic genes was ~26-fold higher in dopaminergic neurons than in cortical neurons, emphasizing the critical role of ascending dopaminergic pathways in mediating general addiction phenotypes. Collectively, brain region- and neuronal subtype-specific 3D genome architecture helps refine neurobiological hypotheses for smoking, alcohol, and general addiction phenotypes by linking genetic risk factors to their target genes.
Introduction
The National Survey on Drug Use and Health in 2018 estimated that 27.3 million individuals were daily cigarette smokers and 16.6 million individuals were heavy alcohol users [1]. Cigarette smoking and alcohol use are the 1st and 3rd leading causes of mortality and morbidity, accounting for 480,000 and 88,000 deaths per year in the United States, respectively [2, 3]. Despite their public health burden, treatment options for nicotine and alcohol use disorders are limited and often fail. However, existing treatments can be improved and new treatments can be developed with a better understanding of the underlying neurobiology of addiction. Genome-wide association studies (GWAS) on smoking and alcohol use traits have demonstrated that common variation explains a significant proportion of phenotypic variance of substance use [4]. Nearly 400 genomic loci were found to have an impact on smoking and/or alcohol use traits from GWAS sample sizes of up to 1.2 million [5–7]. However, the vast majority of associated variants reside in non-coding DNA, and their target genes and relevant neurobiological mechanisms are poorly understood. Examining higher-order chromatin architecture is crucial to understanding the functional consequences of non-coding variation by linking variants to distal genes based on chromatin interaction profiles [8, 9]. Whereas the three-dimensional (3D) genomic landscape of the human brain has advanced our understanding of neurobiological mechanisms underlying psychiatric disorders [9–12], such approaches have been essentially lacking in explaining the genetic architecture of substance use disorders (SUD).
To understand the functional impact of common variants associated with cigarette smoking and alcohol use, we applied Hi-C coupled MAGMA (H-MAGMA) [11] to GWAS of smoking and alcohol use traits and identified their putative target genes for further characterization [5–7]. Smoking and alcohol use traits likely affect neural circuits that underlie addiction and include the prefrontal cortex (PFC), nucleus accumbens (NAc), amygdala, and midbrain dopaminergic cell groups such as ventral tegmental area (VTA) and substantia nigra (SN) [13, 14]. We reasoned that the characterization of chromatin architecture across the brain reward circuitry is critical to understanding the gene regulatory mechanisms associated with substance use. With neurons being the major drivers of substance use behaviors, we profiled chromatin architecture from cortical neurons (CNs) in the dorsolateral PFC (DLPFC) [15] and dopaminergic neurons (DNs) in the midbrain [16]. We then built H-MAGMA inputs from CNs and DNs, and applied them to GWAS summary statistics of smoking and alcohol use traits. In particular, given the recent work on a potential difference in genetic architecture between substance consumption and clinical diagnosis of use disorder [4], we mapped genetic variants associated with consumption or use (drinks per week [DPW] [5] and cigarettes per week [CPD] [5]) versus use disorder (problematic alcohol use [PAU] [6] and nicotine dependence [ND] [7]) to their associated risk genes. Our analysis of substance use risk genes identified key biological pathways, primary cell types, and brain circuitry that might confer risk for substance use. In addition, we characterized genes and pathways shared between cigarette smoking and alcohol use traits to provide a core neurobiological basis of addiction.
Results
Epigenetic landscape of cortical and midbrain dopaminergic neurons
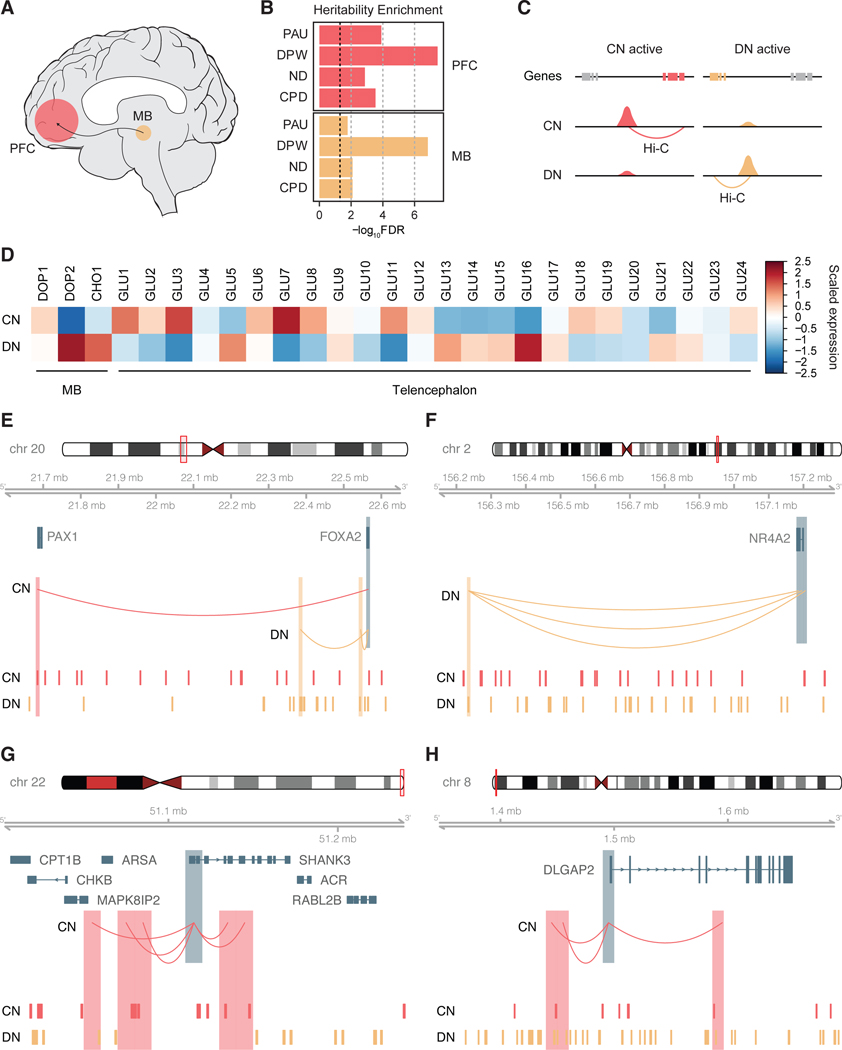
Neural circuitry underlying addiction involves, among others, dopaminergic cell groups in the midbrain, including VTA and SN, as well as neuronal populations in the PFC [13] (Figure 1A). However, the gene regulatory landscape in these two brain regions and its implication in the genetics of cigarette smoking and alcohol use traits have not been studied. To understand the relationship between the reward circuitry and genetic underpinnings of substance use, we evaluated enrichment of genetic risk factors for four traits associated with alcohol use (PAU [6] and DPW [5]) and cigarette smoking (ND [7] and CPD [5]) in cis-regulatory elements (CREs) of the midbrain and PFC [17] using stratified LD score regression (LDSC) [18]. We defined CREs using publicly available data and annotations derived from epigenomic assays and demonstrated that every trait showed significant heritability enrichment for CREs in the midbrain and PFC (Figure 1B, Supplementary Figures 1 and 2).
Figure 1. Gene regulatory landscape in cortical and dopaminergic neurons.
A. Brain reward circuitry encompasses the midbrain (MB) and its projection to the prefrontal cortex (PFC). B. Stratified LDSC analysis determined that cis-regulatory elements (CREs) in the PFC and substantia nigra (SN) are enriched for genetic risk factors for problematic alcohol use (PAU), drinks per week (DPW), nicotine dependence (ND), and cigarettes per day (CPD). The black dotted line represents FDR=0.05. C. Dopaminergic neuronal (DN) differentially accessible regions (DARs) were linked to their target genes using DN Hi-C data, while cortical neuronal (CN) DARs were linked to target genes using CN Hi-C data. D. Genes mapped to DN-DARs were highly expressed in midbrain dopaminergic (DOP2) and cholinergic neurons (CHO1), while genes mapped to CN-DARs were highly expressed in telencephalic glutamatergic neurons (GLU1, 3, 7, 11). E-H. Different enhancer connectivity between CNs and DNs for FOXA2 (E), NR4A2 (F), SHANK3 (G), and DLGAP2 (H) loci. Promoters of genes (FOXA2, NR4A2, SHANK3, DLGAP2) are highlighted in blue, while their interaction targets in CN and DN are highlighted in red and orange, respectively. CN- and DN-DAR are depicted in the bottom tracks.
Midbrain DNs have long been hypothesized to be the major player of the brain reward circuitry [13, 19]. Thus, we investigated whether midbrain DN-CREs explained the heritability enrichment of cigarette smoking and alcohol use traits (Supplementary Methods). Indeed, genetic risk factors for substance use traits were enriched in chromatin accessible regions of DNs derived from human induced pluripotent stem cells [20] (hiPSC, Supplementary Figure 2A).
Given the cellular heterogeneity of the PFC, we also evaluated heritability enrichment of substance use traits in CREs of four major cell types (neurons, astrocytes, microglia, and oligodendrocytes) in the cortex [21]. Neurons showed the strongest heritability enrichment of substance use traits among the four cell types (Supplementary Figure 2B). These results collectively suggest that the gene regulatory relationships in CNs and DNs may provide rich information about genetic underpinnings of substance use traits.
We next sought to compare gene regulatory relationships between CNs and DNs. Substantial differences in chromatin architecture have been observed across different cell types in the human brain [12, 15, 21], but less information is available for the chromatin architecture in different brain regions and/or neuronal subtypes. To interrogate differences in chromatin architecture between CNs and DNs, we identified differential chromatin accessibility peaks between CNs and DNs [20] using DiffBind [22] (Supplementary Table 1). Differentially accessible regions (DARs) in CNs (CN-DARs) were then mapped to their target genes based on CN Hi-C data recently generated by our group [15] (Supplementary Methods, Figure 1C). Since DN Hi-C data with comparable read depths were not available, we generated high-resolution chromosome conformation maps from the midbrain DNs to link DN-DARs to the corresponding genes (Figure 1C, Supplementary Figure 1, sample information provided in Supplementary Methods and Supplementary Table 2). To examine the correlation between neuronal subtype-specific chromatin architecture and gene expression signatures, we then measured cell-type specific expression profiles of the genes linked to CN- and DN-DARs. Genes linked to CN-DARs were highly expressed in cortical pyramidal neurons of the telencephalon (GLU1–3, 6–8), whereas genes linked to DN-DARs were highly expressed in midbrain dopaminergic (DOP2) and cholinergic neurons (CHO1) as well as subcortical-projection glutamatergic neurons in the telencephalon (GLU5, 13–17) [23] (Figure 1D).
We also found evidence of different enhancer wiring between CNs and DNs. For example, FOXA2 and NR4A2, master regulators for dopaminergic neuronal specification and differentiation[24–26], displayed different regulatory connections between CNs and DNs. FOXA2 was linked to two proximal enhancers in DNs as compared to one distal enhancer in CNs (Figure 1E). NR4A2 was linked to multiple distal enhancers only in DNs, but not in CNs (Figure 1F). On the contrary, genes that encode synaptic scaffolding proteins at glutamatergic synapses (e.g. SHANK3 and DLGAP2) displayed enhancer-promoter connectivities only in CNs, but not in DNs (Figure 1G–H).
We next compared topologically associating domains (TADs) between CNs and DNs (Supplementary Methods). Consistent with previous reports [27], TADs were largely conserved between CNs and DNs. However, we noted some differences in TAD boundary strengths (defined by binSignal, see Supplementary Methods for details) between CNs and DNs. For example, EN1 is a critical survival factor for DN differentiation and maintenance [28]. We found that EN1 was located at the TAD boundary whose strength is stronger in DNs than in CNs (Supplementary Figure 3A). FOXA2 also showed strengthened TAD boundaries in DNs (Supplementary Figure 3B), which corresponds to the confinement of loops in proximal space as evidenced in Figure 1E. Importantly, these genes were more highly expressed in DNs than in CNs (Supplementary Figure 3A–B). On the contrary, CREBBP and WNT3A, genes with elevated expression in CNs, were located in TADs that were enlarged in CNs (Supplementary Figure 3C–D). Therefore, these results indicate that different neuronal subtypes involved in substance use traits display distinct chromatin architecture that is coupled with transcriptional regulation.
CN and DN H-MAGMA identifies genes and biological pathways underlying cigarette smoking and alcohol use traits
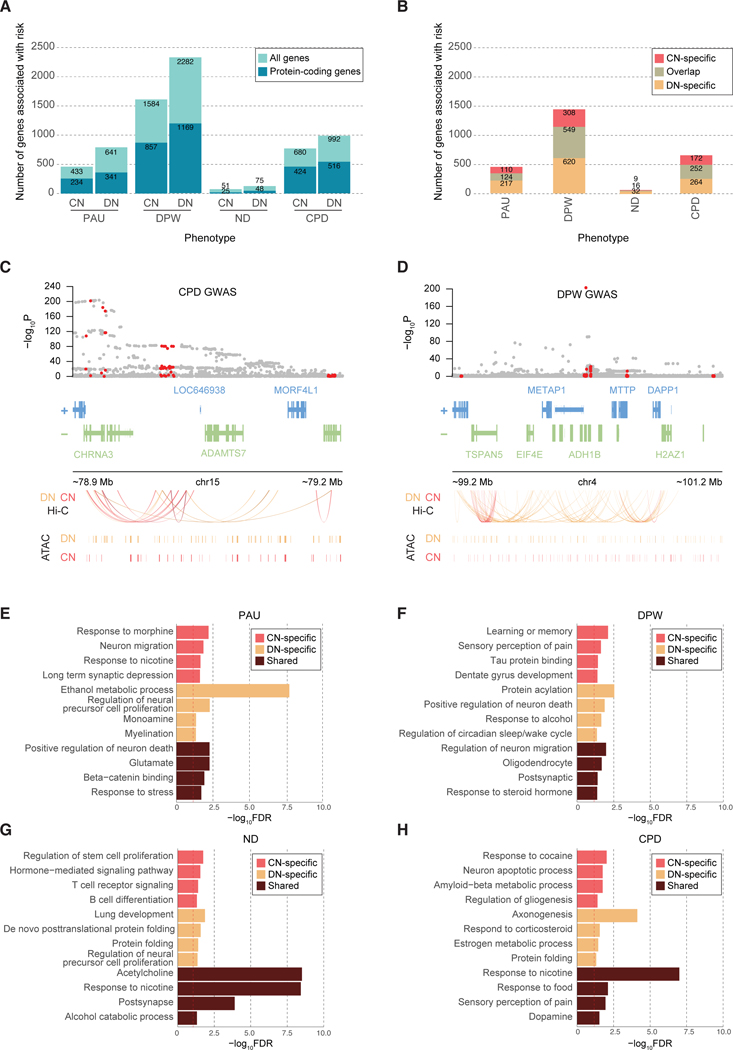
To investigate the functional impact of common variants associated with cigarette smoking and alcohol use traits, we next employed H-MAGMA to assign genetic variants to their target genes based on long-range chromatin interaction [11]. Heritability enrichment results suggested roles for CNs and DNs in cigarette smoking and alcohol use traits (Figure 1B, Supplementary Figure 2). Therefore, we generated H-MAGMA input files from CN Hi-C data previously reported in Hu et al. [15] and newly generated Hi-C libraries from the midbrain DNs (hereafter referred to as CN and DN H-MAGMA, respectively). We applied H-MAGMA to PAU, DPW, ND, and CPD, and identified risk genes for each trait using a false discovery rate (FDR) threshold of 5% (Figure 2A–B, Supplementary Tables 3 and 4). We detected a small number of risk genes for ND in comparison to other GWAS, which can be attributed to the smaller sample size of ND GWAS (see Supplementary Methods for the sample size for each GWAS).
Figure 2. Genes and pathways associated with cigarette smoking and alcohol use traits.
A. The number of risk genes for cigarette smoking and alcohol use traits based on H-MAGMA built from CN and DN Hi-C data (FDR<0.05). For each stacked bar plot, an upper bar plot and number in light blue denote all genes, whereas a lower layer and number in dark blue correspond to protein-coding genes. B. The number of protein-coding genes associated with smoking and alcohol use traits by CN H-MAGMA only (CN-specific), DN H-MAGMA only (DN-specific), and by both CN and DN H-MAGMA (overlap) at an FDR threshold of 0.05. C. H-MAGMA identifies CHRNA3 to be associated with CPD. From top to bottom, we display the GWAS variant association, gene model, Hi-C loops, and CREs. GWAS variant association, gray dots represent all SNPs in the locus, red dots represent SNPs annotated to CHRNA3 via H-MAGMA (either DN or CN). Hi-C loops and CREs, red and orange lines represent the regulatory architecture of CNs and DNs, respectively. D. H-MAGMA identifies ADH1B to be associated with DPW. Red dots in the GWAS association represent SNPs annotated to ADH1B via H-MAGMA (either DN or CN). E-H. Gene ontologies (GO) enriched for PAU (E), DPW (F), ND (G), and CPD (H). CN-specific GO terms represent terms unique to genes identified from H-MAGMA built on CN Hi-C data, while DN-specific GO terms represent terms unique to genes identified from H-MAGMA built from DN Hi-C data. Shared terms denote GO terms detected in both CN and DN H-MAGMA results. The dotted red line denotes FDR=0.05.
Both CN and DN H-MAGMA identified CHRNA3, whose role in increased risk of smoking[7] has been well established, to be associated with CPD (Figure 2C). Similarly, ADH1B, a gene that encodes alcohol metabolizing enzymes, was found to be associated with DPW via both CN and DN H-MAGMA, illustrating H-MAGMA’s ability (Figure 2D). Additionally, some of the risk genes identified by CN and DN H-MAGMA have been directly characterized using rodent models of substance use. For example, DRD2, a gene that encodes a dopaminergic receptor, was identified by CN and DN H-MAGMA to be associated with all traits. Deletion of its ortholog in mice altered alcohol preferences and alcohol-induced ataxia [29]. Moreover, individual variability in DRD2 was found to be associated with response to nicotine replacement therapies[30]. We also identified GABRG1 association with DPW from DN H-MAGMA. In a previous study investigating the role of GABA receptors in alcohol consumption, Gabrg1 knockout mice exhibited decreased alcohol consumption during an operant conditioning testing and home cage drinking assessment [31]. Lastly, BDNF was identified by CN and DN H-MAGMA to be associated with PAU and DPW. Downregulation of this gene in the dorsal striatum has been shown to increase alcohol consumption in rats [32].
While differential gene expression has been detected in the postmortem brain samples of substance abusers compared with controls [33, 34], the extent to which this expression signature is predisposed by genetic risk factors remains unclear. To address this, we compared H-MAGMA gene-level scores to differentially expressed genes from human brain tissue and human iPSC-derived neural cultures after exposure to nicotine and alcohol, respectively. In this comparison, DN H-MAGMA results for DPW showed a significant association with gene expression signatures from hiPSC-derived forebrain neural cells exposed to alcohol (DEG logFC ~ H-MAGMA Z-score + number of SNPs, β=2.31×10−2, p<2.0×10−16) [35]. In parallel, we identified a significant association between CN H-MAGMA results for ND and gene expression signatures from the adult prefrontal cortex of active smokers (DEG logFC ~ H-MAGMA Z-score + number of SNPs, β=3.10×10−3, p<2.0×10−16) [34]. Among the genes that were differentially expressed in ND was CHRNB4, a gene that encodes a nicotinic acetylcholine receptor subunit. Together, these results suggest that genetic risk factors may contribute to the gene expression signatures of cigarette smoking and alcohol use traits.
Next, we mapped risk genes identified from CN and DN H-MAGMA to biological pathways using gene ontology (GO) analysis. Rather than using a specific FDR threshold, we ran ranked-based GO analysis using the Z-score of H-MAGMA output files. Since we used two separate H-MAGMA inputs to assign common variants to their target genes, we obtained two GO results for each trait – one for CN H-MAGMA risk genes and the other for DN H-MAGMA risk genes. We then classified GO terms as CN-specific or DN-specific if they represented biological pathways unique to CN or DN H-MAGMA, respectively.
We validated previous findings that ethanol metabolic processes and response to alcohol were associated with PAU and DPW (Figure 2E–F) [4, 6], and that cholinergic and nicotinic pathways were associated with ND and CPD [5, 7] (Figure 2G–H, see Supplementary Table 5 for full GO outputs). Notably, we also identified alcohol catabolic processes for ND and nicotinic pathways for PAU. Likewise, we further identified GO terms relating to other substances of abuse. For instance, GO terms for PAU included response to morphine (Figure 2E), while GO terms for CPD included response to cocaine (Figure 2H). Taken together, these findings underscore potential genetic overlap and interplay among different substances of abuse.
We identified several similarities across cigarette smoking and alcohol use traits. For example, neuronal processes such as neuronal migration and apoptosis were associated with cigarette smoking and alcohol use, which is in line with studies that have pinpointed the disruption of neuronal migration and neurotransmission in response to substance use [36, 37]. We also observed myelination and gliogenesis to be associated with DPW and CPD, respectively, hinting at the role of neuron-glia interactions in substance use traits. Several immune processes including T and B cell activation were shown to be associated with cigarette smoking and alcohol use, which corroborates the relationship between substance use and suppressed immunity (Figure 2G) [38, 39]. We also identified a potential role of protein folding that has been shown to contribute to the stress response[40]. A potential link between substance use and neurodegeneration emerged, such as amyloid-beta metabolic processes for CPD and tau protein binding for DPW. Lastly, pain perception was associated with DPW and CPD, consistent with prior research linking pain perception and the reward circuitry [41].
We also observed distinct biological processes between cigarette smoking and alcohol use traits. For instance, long term synaptic depression (Figure 2E), as well as learning and memory (Figure 2F), were characteristic of alcohol use traits but not cigarette smoking, highlighting the important role of synaptic plasticity and memory consolidation in the mechanism of alcohol use [42, 43]. We also found GO terms relating to sleep and wake cycle for alcohol use traits, which support a rich body of evidence suggesting that prolonged alcohol use and misuse can cause deleterious effects on sleep quality [44]. Cigarette smoking traits also exhibited distinct associations not observed in alcohol use traits. For instance, we noted lung development to be associated with both ND and CPD which supports epidemiological findings of lung morbidities linked to cigarette smoking [45].
Discrete biological processes were also observed between CN and DN H-MAGMA. Ethanol metabolism and alcohol response were enriched for alcohol use traits in a DN-specific manner (Figure 2E–F). In contrast, the potential link between neurodegeneration and substance use was specific to CNs. These results suggest that the neurobiological basis of cigarette smoking and alcohol use traits may need to be studied in a brain region- and neuronal subtype-specific manner.
Cellular expression profiles of cigarette smoking and alcohol use risk genes convey cell types associated with substance use
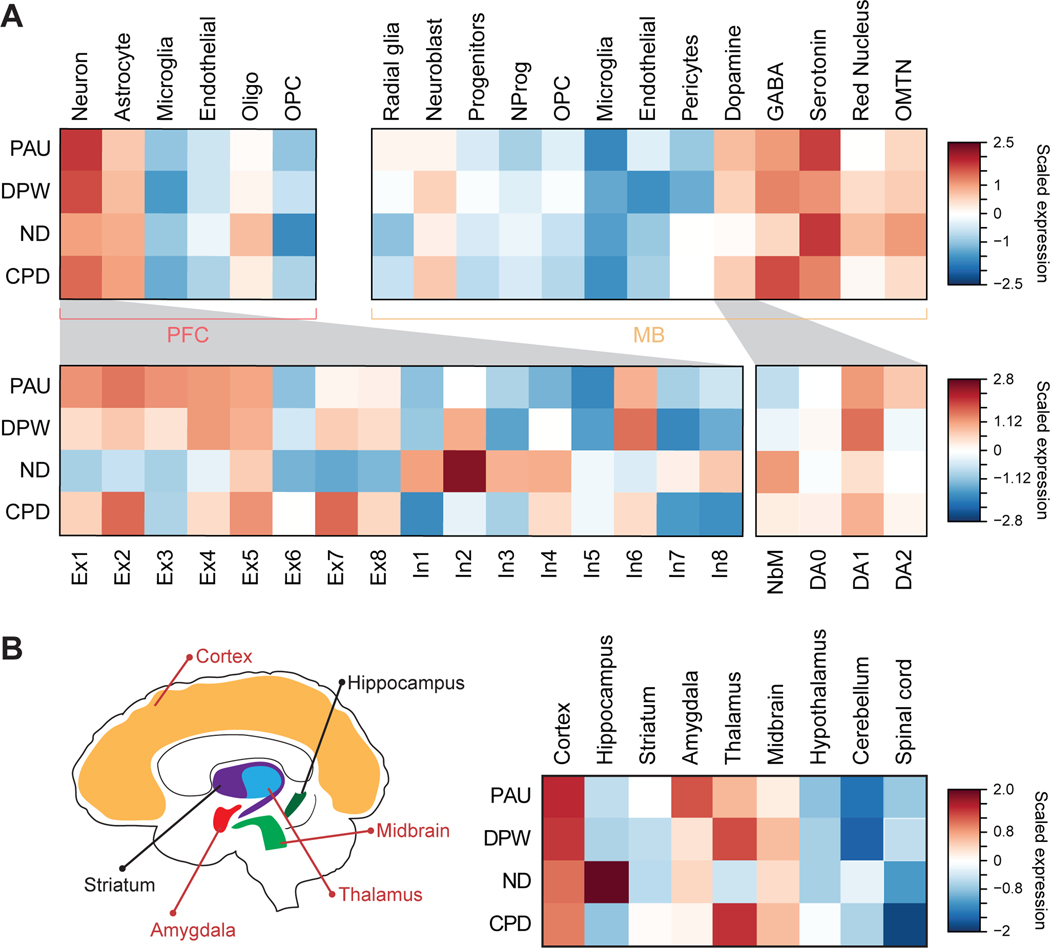
Since CNs and DNs display heterogeneity and act in synchrony with multiple cell types, we leveraged single-cell RNA sequencing (scRNA-seq) datasets to further refine neuronal subtypes that confer risk of substance use. We first evaluated cellular expression profiles of cigarette smoking and alcohol use risk genes identified from CN H-MAGMA using scRNA-seq data from the human cortex[46]. We not only recapitulated our findings that genetic risk variants underlying cigarette smoking and alcohol use are highly expressed in neurons, but also observed that the risk genes were highly expressed in excitatory neurons (Figure 3A). Specifically, we found PAU, DPW, and CPD risk genes to be highly expressed in layer 5 pyramidal neurons (Ex5) that project to both cortical and subcortical areas including the striatum and the midbrain and layer 4 neurons (Ex2) that receive sensory signals from the thalamus, a region that has been shown to be integral to addiction by modulating arousal and motivation [47].
Figure 3. Cellular and brain regional expression profiles of cigarette smoking and alcohol use risk genes.
A. Top left panel represents cellular expression profiles of cigarette smoking and alcohol use risk genes identified from CN H-MAGMA using scRNA-seq data from the adult cortex[46, 70]. Genetic risk factors underlying cigarette smoking and alcohol use influence genes highly expressed in neurons. Bottom left panel represents risk gene expression across neuronal subclusters. OPC, oligodendrocytes progenitor cells; Ex, excitatory neurons; In, inhibitory neurons. Top right panel, cellular expression profiles of cigarette smoking and alcohol use risk genes identified from DN H-MAGMA using scRNA-seq from the ventral midbrain of human embryo. Risk genes are highly expressed in dopaminergic, GABA-ergic, and serotonergic neurons in the midbrain. NProg, neuronal progenitors; OMNT, oculomotor and trochlear nucleus. Bottom right panel, cigarette smoking and alcohol use risk genes were enriched for DA1 across DN development in human embryonic midbrain. NbM, medial neuroblasts and precursors of DNs; DA0, immature DNs; DA1, intermediate DNs; DA2 matured DNs. B. Left, graphic representation of brain regions with elevated expression levels of risk genes for substance use traits. Regions highlighted in red are enriched for at least three of the four traits. Right, brain regional expression profiles of cigarette smoking and alcohol use risk genes using scRNA-seq from the mouse nervous system[23]. Risk gene expression spans multiple brain regions including the cortex, amygdala, and midbrain.
Comparably, we examined expression profiles of cigarette smoking and alcohol use risk genes identified from DN H-MAGMA in midbrain cell types using scRNA-seq data from the human embryonic ventral midbrain[48]. Risk genes for all traits except for ND were highly expressed in DNs, providing additional evidence to support the impact of DNs in modulating substance use via the reward-circuitry [13] (Figure 3A). Within the DN lineage, we found elevated expression of risk genes in intermediate DNs (DA1) for all traits, suggesting that they may be more vulnerable to substance use. Moreover, risk genes showed elevated expression in midbrain GABAergic neurons which have been shown to regulate a diverse set of processes including motor control and inhibition of dopaminergic cells, thereby modulating the reward-circuitry [49]. Similarly, risk genes’ expression in serotonergic neurons is consistent with their reported involvement in substance use vulnerability [50]. We also validated cellular expression profiles with a linear regression model (Supplementary Methods) and identified strong correlations between cellular enrichment and expression patterns (Supplementary Figure 4A).
Furthermore, we leveraged scRNA-seq data from the nucleus accumbens (NAc) of the adult human brain [51] to evaluate the expression profiles of risk genes identified from DN H-MAGMA. We found that cigarette smoking and alcohol use risk genes were highly expressed in medium spiny neurons (MSNs) that express DRD1 (MSN-D1) and DRD2 (MSN-D2) (Supplementary Figure 4B).
To confirm that the cellular expression profiles are not driven by the specific Hi-C data being used, we further evaluated cellular expression profiles of risk genes identified by H-MAGMA built upon cell-type agnostic adult cortical tissue [11, 52]. Overall, we recapitulated similar expression patterns for all traits except for the lack of intermediate DN (DA1) expression of genes associated with cigarette smoking traits (Supplementary Figure 4C).
Next, we extended our approach to a brain-wide fashion by assessing brain regional expression profiles of cigarette smoking and alcohol use risk genes. We leveraged extensive scRNA-seq data from the mouse nervous system to determine brain regions with high expression values of risk genes identified by CN and DN H-MAGMA [23]. Both cigarette smoking and alcohol use risk genes were highly expressed in cortical and midbrain regions as expected (Figure 3B). We also found strong expression in the hippocampus for ND risk genes, highlighting the role of hippocampus-dependent learning in ND [53]. Furthermore, thalamic expression was observed for PAU, DPW, and CPD risk genes, which is consistent with high levels of expression in Ex2 that receives thalamic inputs (Figure 3A) and points to the role of sensory perception in drug-seeking behaviors [54]. Finally, our results highlight the amygdala for elevated expression of risk genes associated with cigarette smoking and alcohol use. The association of risk variants with the amygdala underscores the role of emotional processing in substance use due to its projections to other parts of the reward-circuitry [13].
Shared genetic architecture among substance use
Individuals often become dependent on multiple substances, and these comorbidities may be driven by shared genetic signal [6, 55]. We hypothesized that biological characterization of pleiotropic genes between cigarette smoking and alcohol use traits would identify neurobiological mechanisms underlying the shared genetic architecture of substance use traits.
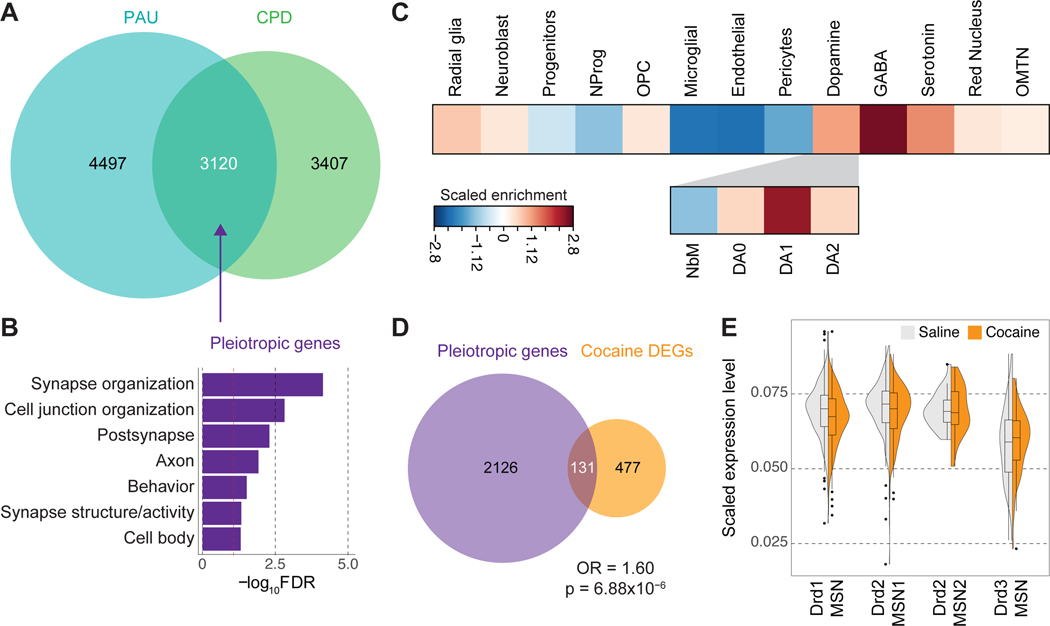
We first calculated genetic correlations and gene-level overlap across cigarette smoking and alcohol use traits using LDSC and rank-rank hypergeometric overlap (RRHO) test, respectively (Supplementary Methods, Supplementary Figure 5). We found that RRHO of DN H-MAGMA outputs gives stronger gene-level overlaps than that of CN H-MAGMA. For example, 119 and 3,120 genes were shared between PAU and CPD using CN and DN H-MAGMA, respectively (Figure 4A, Supplementary Figures 5A and C). These results suggest that DNs may play a central role in explaining comorbidity in substance use. Because the PAU and CPD showed a significant genetic correlation (genetic correlation = 0.19) and gene-level overlap (RRHO Z-score = 12.16), we selected shared genes between PAU and CPD in DN H-MAGMA to serve as pleiotropic genes (Figure 4A, Supplementary Table 6). Pleiotropic genes were enriched for synaptic function and cell junction organization (Figure 4B), suggesting that alterations in synaptic organization may influence core features of substance use. We further evaluated cellular expression profiles of pleiotropic genes in the human embryonic ventral midbrain and the adult NAc. We again found elevated expression of pleiotropic genes in dopaminergic, GABAergic, serotonergic neurons in the midbrain as well as MSN subpopulations in the NAc, indicating their potential function in substance use biology (Figure 4C, Supplementary Figure 5D).
Figure 4. Pleiotropic genes highlight shared neurobiological bases of cigarette smoking and alcohol use.
A. Overlap between PAU and CPD risk genes identified by DN H-MAGMA using a rank rank hypergeometric overlap (RRHO) test. Overlapping genes represent pleiotropic genes. B. Biological processes and molecular functions enriched for pleiotropic genes. Dotted red line denotes FDR=0.05. C. Cellular expression profiles of pleiotropic genes in the midbrain (top plot) and dopaminergic lineage (bottom plot). Pleiotropic genes are highly expressed in GABAergic midbrain neurons and intermediate DNs (DA1). D. Overlap between pleiotropic genes and differentially expressed genes (DEGs) in the rat NAc after cocaine treatment (Fisher’s exact test, p=6.88×10−6; odds ratio [OR]=1.60; 95% confidence interval [CI]=1.34–1.96). E. Cellular expression changes of pleiotropic genes in response to cocaine treatment[56]. The x-axis indicates medium spiny neuronal (MSN) clusters identified in the rat NAc while the y-axis indicates scaled expression values of the pleiotropic genes in each cluster.
Based on our hypothesis that pleiotropic genes between cigarette smoking and alcohol use traits may represent risk genes shared across multiple SUD, we next examined whether they are dysregulated in response to other substances. We overlapped our pleiotropic genes with differentially expressed genes (DEGs) in the rat NAc after cocaine treatment[56]. We found a significant proportion of our pleiotropic genes was dysregulated in response to cocaine (Figure 4D). We also compared the cellular expression profiles of pleiotropic genes in saline versus cocaine treatment conditions [56]. We found that pleiotropic genes were downregulated in response to cocaine in Drd1- and Drd2-expressing MSN (Figure 4E, Supplementary Figure 6). Taken together, these results indicate that pleiotropic genes derived from cigarette smoking and alcohol use traits can provide insights into the core neurobiological mechanism of substance abuse.
Drug repurposing analysis
A fundamental issue facing the treatment of SUD is the limited number of effective medications available. Although medications such as Naltrexone [57] and Nicotine Replacement Therapies (NRT) [58] have been traditionally used to treat alcohol use disorder and nicotine addiction, respectively, their efficacies are lacking or produce severe adverse outcomes, rendering the need for new treatment. To address this challenge, we used the Drug Signature and Drug Matrix databases of EnrichR [59], a comprehensive gene analysis tool to identify potential drug candidates for SUD based on genetic evidence. We identified several significantly enriched drug candidates for cigarette smoking and alcohol use traits (Supplementary Table 7). Among these included mood stabilizers and selective serotonin reuptake inhibitors such as Fluoxetine, Citalopram, and Imipramine, consistent with their potential therapeutic benefits in some patients diagnosed with nicotine or alcohol dependency. We further identified enrichment for antipsychotics such as Chlorpromazine and Clozapine, pointing to some degree of convergence of addiction-relevant risk genes with molecular pathways implicated in other types of psychiatric illnesses. These findings speak to the well-documented epidemiological [60, 61] and genetic [62, 63] evidence supporting the comorbidity between psychiatric illnesses and substance use.
Discussion
We interrogated chromatin interaction profiles of CNs and DNs, two primary neuronal subtypes involved in the neurocircuitry of addiction, to map GWAS risk variants of cigarette smoking and alcohol use traits to their target genes. While we identified cigarette smoking and alcohol use traits to be significantly enriched in CREs in the midbrain and PFC, it is possible for the enrichment pattern to be dependent on the sample size of GWAS used.
We next built enhancer-promoter interaction landscapes in CNs and DNs by combining Hi-C and ATAC-seq, and demonstrated brain region- and neuronal subtype-specific gene regulatory relationships. We then employed these profiles to perform CN and DN H-MAGMA, which was used to identify risk genes and neurobiological pathways underlying PAU, DPW, ND, and CPD. Investigation into the biological pathways underlying cigarette smoking and alcohol use risk genes revealed the important role of drug catabolic process and alcohol metabolic process in substance use. Notably, we found that substance use risk genes were enriched for pathways associated with other neurodegenerative disorders such as tau protein binding for DPW and amyloid-beta metabolic process for CPD. The association between substance use and neurodegenerative disorders has been observed in a mouse model of Alzheimer’s disease where alcohol exposure was shown to heighten neuronal and behavioral deficits related to Alzheimer’s disease [64]. Thus, our results provide additional evidence to support that substance use and neurodegenerative disorders may share underlying genetic risk factors [65, 66] and that risk variants associated with alcohol use may exacerbate neurodegenerative disorders by disrupting protein metabolism. We also identified an association between cigarette smoking and food intake which is in line with the previous reports linking weight gain with smoking cessation [67, 68].
We next surveyed the cellular expression profiles of cigarette smoking and alcohol use risk genes to refine cortical and midbrain neuronal subtypes that confer risk for substance use. Within CNs, we found that cigarette smoking and alcohol use risk genes were highly expressed in glutamatergic neurons, providing an additional level of support for the neuronal basis of addiction [13]. We have previously shown that risk genes of psychiatric disorders were also enriched for glutamatergic neurons [11]. Based on prior epidemiological studies reporting higher substance use among individuals with mental health issues, these results suggest a potential cellular basis of comorbidity between substance use and psychiatric disorders [55]. We also identified potential divergence between ND and CPD such that risk genes associated with ND were enriched in inhibitory neurons, in contrast to the observed excitatory enrichment for CPD risk genes. While this may hint at a distinct biological pattern underlying use (CPD) versus a use disorder (ND), caution should be exercised as this finding could also be influenced by the smaller number of genes associated with ND in comparison to CPD due to the smaller sample size of ND GWAS. Our cellular expression profiles within the DN lineage showed enrichment in intermediate DNs (DA1), suggesting early development as a critical time period linked to heritable risk of substance use that manifests later in life [34, 69]. Finally, we leveraged cigarette smoking and alcohol use risk genes to identify the brain circuitry of addiction based on the hypothesis that defining brain regions most relevant to substance use may help derive better targeted approaches to treating SUD. In addition to the cortical and midbrain enrichment, we found enrichment for the amygdala and thalamus, reinforcing that multiple brain regions are important for understanding substance use and addiction.
To further characterize how cigarette smoking and alcohol use risk genes can expand our understanding of substance use and addiction as a whole, we generated a list of pleiotropic genes between PAU and CPD. In contrast to individual risk genes being more focused on individual substance use traits, we reasoned that pleiotropic genes would provide us with the opportunity to identify principal pathways associated with addiction. Therefore, we generated pleiotropic genes using both CN and DN H-MAGMA output files. DNs, but not CNs, showed strong gene-level overlap between PAU and CPD, conveying that DNs might be the central cell type that mediates pleiotropy. Based on our hypothesis that pleiotropic genes might translate beyond just cigarette smoking and alcohol use traits, we compared them with DEGs in response to cocaine [56]. Indeed, we showed that pleiotropic genes were likely to be dysregulated in response to cocaine in the rat NAc, demonstrating that these genes may be more susceptible to a wide range of substance use.
Lastly, we took advantage of EnrichR to identify potential drug candidates to treat nicotine dependence and alcohol use disorder. We found potential drug candidates including those already on the market to treat various psychiatric illnesses such as depression and schizophrenia, further supporting a shared genetic architecture between psychiatric illnesses and substance use (Supplementary Table 7). These findings could be further prioritized by incorporating pathway analyses and literature review to corroborate the association between potential drug candidates’ mechanism of action and substance use. Additionally, prioritized genes and drug candidates could be validated in model organism experiments. Together, we demonstrate that H-MAGMA built from brain region- and neuronal subtype-specific chromatin architecture can successfully identify risk genes and biologically relevant processes associated with cigarette smoking and alcohol use.
Methods
A full description of the methods is available in the Supplementary Methods. Briefly, we generated Hi-C libraries from the midbrain dopaminergic neuronal nuclei (NeuN+/Nurr1+) and performed integrative genomic analysis. Using chromatin connectivities from the midbrain dopaminergic neurons and cortical neurons, we constructed and applied H-MAGMA to four traits associated with alcohol use (PAU and DPW) and cigarette smoking (ND and CPD).
Data Availability
CN (syn21760712) and DN (syn24184521) Hi-C datasets described in this manuscript are available via the PsychENCODE Knowledge Portal (https://psychencode.synapse.org/). The PsychENCODE Knowledge Portal is a platform for accessing data, analyses, and tools generated through grants funded by the National Institute of Mental Health (NIMH) PsychENCODE program. Data is available for general research use according to the following requirements for data access and data attribution: (https://psychencode.synapse.org/DataAccess). H-MAGMA input and output files are available in the Github repository (https://github.com/thewonlab/H-MAGMA). GWAS summary statistics for DPW and CPD were obtained from https://genome.psych.umn.edu/index.php/GSCAN. GWAS summary statistics for ND and PAU were obtained from dbGaP with the accession numbers and phs001532.v1.p1 and phs001672.v3.p1, respectively. RNA-seq and ATAC-seq data from hiPSC-derived CNs and DNs were obtained from GSE129017.
Code Availability
All custom code used in this work is available in the following Github repository: https://github.com/thewonlab/H-MAGMA.
Supplementary Material
Acknowledgements
We thank members of the Won lab for helpful discussions and comments about this paper, in particular, Nana Matoba, Won Mah, and Jessica McAfee. We also acknowledge helpful advice and discussion from Jonathan Pollock, Amy Lossie, and Susan Wright. We thank Drs. Stefano Marenco and Barbara Lipska from the Human Brain Collection Core (HBCC, Bethesda, MD) for providing postmortem brain specimens; Mette Peters, Kelsey Montgomery, and Juliane Schneider for assisting data deposition into synapse. This research was supported by the National Institute on Drug Abuse (R21DA051921, H.W., D.B.H., E.O.J.; U01DA048279, S.A), National Institute of Mental Health (R00MH113823, DP2MH122403, H.W.), the NARSAD Young Investigator Award from the Brain and Behavior Research Foundation (H.W.). N.Y.A.S. was supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650116 and in part by a grant to the University of North Carolina at Chapel Hill from the Howard Hughes Medical Institute through the James H. Gilliam Fellowship for Advanced Study Program. S.L. was supported by the National Institute of General Medical Sciences (5T32GM067553). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Reference
- 1.Key Substance Use and Mental Health Indicators in the United States: Results from the 2018 National Survey on Drug Use and Health. https://www.samhsa.gov/data/sites/default/files/cbhsq-reports/NSDUHNationalFindingsReport2018/NSDUHNationalFindingsReport2018.pdf. Accessed 17 August 2020.
- 2.Peacock A, Leung J, Larney S, Colledge S, Hickman M, Rehm J, et al. Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction. 2018;113:1905–1926. [DOI] [PubMed] [Google Scholar]
- 3.National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. Atlanta (GA): Centers for Disease Control and Prevention (US); 2014. [PubMed] [Google Scholar]
- 4.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun. 2020;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mah W, Won H. The three-dimensional landscape of the genome in human brain tissue unveils regulatory mechanisms leading to schizophrenia risk. Schizophr Res. 2020;217:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020. 9 March 2020. 10.1038/s41593-020-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, et al. Neuron-specific Signatures in the Chromosomal Connectome Are Associated with Schizophrenia Risk. Science. 2018;Accepted f:eaat4311–eaat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3:760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammel S, Lim BK, Malenka RC. Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology. 2014;76 Pt B:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B, Won H, Mah W, Park RB, Kassim B, Spiess K, et al. Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat Commun. 2021;12:3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espeso-Gil S, Halene T, Bendl J, Kassim B, Ben Hutta G, Iskhakova M, et al. A chromosomal connectome for psychiatric and metabolic risk variants in adult dopaminergic neurons. Genome Med. 2020;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium Roadmap Epigenomics, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Zhang H, Zhou Y, Qiao M, Zhao S, Kozlova A, et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science. 2020;369:561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark R, Brown G, Others. DiffBind: differential binding analysis of ChIP-Seq peak data. R Package Version. 2011;100:4–3. [Google Scholar]
- 23.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, Ericson J, et al. Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development. 2012;139:2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H-S, Bae E-J, Yi S-H, Shim J-W, Jo A-Y, Kang J-S, et al. Foxa2 and Nurr1 synergistically yield A9 nigral dopamine neurons exhibiting improved differentiation, function, and cell survival. Stem Cells. 2010;28:501–512. [DOI] [PubMed] [Google Scholar]
- 26.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon HH, Saueressig H, Wurst W, Goulding MD, O’Leary DD. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci. 2001;21:3126–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer AA, Low MJ, Grandy DK, Phillips TJ. Effects of a Drd2 deletion mutation on ethanol-induced locomotor stimulation and sensitization suggest a role for epistasis. Behav Genet. 2003;33:311–324. [DOI] [PubMed] [Google Scholar]
- 30.Herman AI, DeVito EE, Jensen KP, Sofuoglu M. Pharmacogenetics of nicotine addiction: role of dopamine. Pharmacogenomics. 2014;15:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.June HL, Foster KL, Eiler WJA, Goergen J, Cook JB, Johnson N, et al. Dopamine and Benzodiazepine-Dependent Mechanisms Regulate the EtOH-Enhanced Locomotor Stimulation in the GABAA α1 Subunit Null Mutant Mice. Neuropsychopharmacology. 2007;32:137–152. [DOI] [PubMed] [Google Scholar]
- 32.Jeanblanc J, He D-Y, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, Enoch M-A, Goldman D. Gene expression in the addicted brain. Int Rev Neurobiol. 2014;116:251–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semick SA, Collado-Torres L, Markunas CA, Shin JH, Deep-Soboslay A, Tao R, et al. Developmental effects of maternal smoking during pregnancy on the human frontal cortex transcriptome. Mol Psychiatry. 2018;25:3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen KP, Lieberman R, Kranzler HR, Gelernter J, Clinton K, Covault J. Alcohol-responsive genes identified in human iPSC-derived neural cultures. Transl Psychiatry. 2019;9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skorput AGJ, Gupta VP, Yeh PWL, Yeh HH. Persistent Interneuronopathy in the Prefrontal Cortex of Young Adult Offspring Exposed to Ethanol In Utero. J Neurosci. 2015;35:10977–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazemi T, Huang S, Avci NG, Waits CMK, Akay YM, Akay M. Investigating the influence of perinatal nicotine and alcohol exposure on the genetic profiles of dopaminergic neurons in the VTA using miRNA–mRNA analysis. Scientific Reports. 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox HC, Milivojevic V, Angarita GA, Stowe R, Sinha R. Peripheral immune system suppression in early abstinent alcohol-dependent individuals: Links to stress and cue-related craving. J Psychopharmacol. 2017;31:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasala S, Barr T, Messaoudi I. Impact of alcohol abuse on the adaptive immune system. Alcohol Res. 2015;37:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Díaz-Villanueva JF, Díaz-Molina R, García-González V. Protein Folding and Mechanisms of Proteostasis. Int J Mol Sci. 2015;16:17193–17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elman I, Borsook D. Common Brain Mechanisms of Chronic Pain and Addiction. Neuron. 2016;89:11–36. [DOI] [PubMed] [Google Scholar]
- 42.Goodman J, Packard MG. Memory Systems and the Addicted Brain. Front Psychiatry. 2016;7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morin J-FG, Afzali MH, Bourque J, Stewart SH, Séguin JR, O’Leary-Barrett M, et al. A Population-Based Analysis of the Relationship Between Substance Use and Adolescent Cognitive Development. Am J Psychiatry. 2019;176:98–106. [DOI] [PubMed] [Google Scholar]
- 44.Elmenhorst E-M, Elmenhorst D, Benderoth S, Kroll T, Bauer A, Aeschbach D. Cognitive impairments by alcohol and sleep deprivation indicate trait characteristics and a potential role for adenosine A1 receptors. Proc Natl Acad Sci U S A. 2018;115:8009–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Qi F, Wang Y, Jia X, Lin P, Geng M, et al. Cancer mortality attributable to cigarette smoking in 2005, 2010 and 2015 in Qingdao, China. PLoS One. 2018;13:e0204221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matzeu A, Martin-Fardon R. Drug Seeking and Relapse: New Evidence of a Role for Orexin and Dynorphin Co-transmission in the Paraventricular Nucleus of the Thalamus. Front Neurol. 2018;9:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167:566–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morello F, Partanen J. Diversity and development of local inhibitory and excitatory neurons associated with dopaminergic nuclei. FEBS Lett. 2015;589:3693–3701. [DOI] [PubMed] [Google Scholar]
- 50.Kirby LG, Zeeb FD, Winstanley CA. Contributions of serotonin in addiction vulnerability. Neuropharmacology. 2011;61:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran MN, Maynard KR, Spangler A, Huuki LA, Montgomery KD, Sadashivaiah V, et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron. 2021;109:3088–3103.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gould TJ. Nicotine and hippocampus-dependent learning. Mol Neurobiol. 2006;34:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y, Wienecke CFR, Nachtrab G, Chen X. A thalamic input to the nucleus accumbens mediates opiate dependence. Nature. 2016;530:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abuse S. Mental Health Services Administration.(2018). Key substance use and mental health indicators in the United States: Results from the 2017 National Survey on Drug Use and Health (HHS Publication No. SMA 18–5068, NSDUH Series H-53). Rockville, MD: Center for Behavioral Health Statistics and Quality. Substance Abuse and Mental Health Services Administration; Retrieved from Https://www Samhsa Gov/data.2019.2019. [Google Scholar]
- 56.Savell KE, Tuscher JJ, Zipperly ME, Duke CG, Phillips RA 3rd, Bauman AJ, et al. A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci Adv. 2020;6:eaba4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendershot CS, Wardell JD, Samokhvalov AV, Rehm J. Effects of naltrexone on alcohol self-administration and craving: meta-analysis of human laboratory studies. Addiction Biology. 2017;22:1515–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stead LF, Perera R, Bullen C, Mant D, Hartmann-Boyce J, Cahill K, et al. Nicotine replacement therapy for smoking cessation. Cochrane Database Syst Rev. 2012;11:CD000146. [DOI] [PubMed] [Google Scholar]
- 59.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castillo-Carniglia A, Keyes KM, Hasin DS, Cerdá M. Psychiatric comorbidities in alcohol use disorder. Lancet Psychiatry. 2019;6:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murthy P, Mahadevan J, Chand PK. Treatment of substance use disorders with co-occurring severe mental health disorders. Curr Opin Psychiatry. 2019;32:293–299. [DOI] [PubMed] [Google Scholar]
- 62.Hartz SM, Horton AC, Oehlert M, Carey CE, Agrawal A, Bogdan R, et al. Association Between Substance Use Disorder and Polygenic Liability to Schizophrenia. Biol Psychiatry. 2017;82:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang L-H, Whitfield JB, Liu M, Medland SE, Hickie IB, Martin NG, et al. Associations between polygenic risk for tobacco and alcohol use and liability to tobacco and alcohol use, and psychiatric disorders in an independent sample of 13,999 Australian adults. Drug and Alcohol Dependence. 2019;205:107704. [DOI] [PubMed] [Google Scholar]
- 64.Hoffman JL, Faccidomo S, Kim M, Taylor SM, Agoglia AE, May AM, et al. Alcohol drinking exacerbates neural and behavioral pathology in the 3xTg-AD mouse model of Alzheimer’s disease. Int Rev Neurobiol. 2019;148:169–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicholatos JW, Francisco AB, Bender CA, Yeh T, Lugay FJ, Salazar JE, et al. Nicotine promotes neuron survival and partially protects from Parkinson’s disease by suppressing SIRT6. Acta Neuropathologica Communications. 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piao W-H, Campagnolo D, Dayao C, Lukas RJ, Wu J, Shi F-D. Nicotine and inflammatory neurological disorders. Acta Pharmacologica Sinica. 2009;30:715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bush T, Lovejoy JC, Deprey M, Carpenter KM. The effect of tobacco cessation on weight gain, obesity, and diabetes risk. Obesity. 2016;24:1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Germeroth LJ, Levine MD. Postcessation weight gain concern as a barrier to smoking cessation: Assessment considerations and future directions. Addict Behav. 2018;76:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCrory EJ, Mayes L. Understanding Addiction as a Developmental Disorder: An Argument for a Developmentally Informed Multilevel Approach. Current Addiction Reports. 2015;2:326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CN (syn21760712) and DN (syn24184521) Hi-C datasets described in this manuscript are available via the PsychENCODE Knowledge Portal (https://psychencode.synapse.org/). The PsychENCODE Knowledge Portal is a platform for accessing data, analyses, and tools generated through grants funded by the National Institute of Mental Health (NIMH) PsychENCODE program. Data is available for general research use according to the following requirements for data access and data attribution: (https://psychencode.synapse.org/DataAccess). H-MAGMA input and output files are available in the Github repository (https://github.com/thewonlab/H-MAGMA). GWAS summary statistics for DPW and CPD were obtained from https://genome.psych.umn.edu/index.php/GSCAN. GWAS summary statistics for ND and PAU were obtained from dbGaP with the accession numbers and phs001532.v1.p1 and phs001672.v3.p1, respectively. RNA-seq and ATAC-seq data from hiPSC-derived CNs and DNs were obtained from GSE129017.