Abstract
The precise arrangement of structural subunits is a key factor for the proper shape and function of natural and artificial supramolecular assemblies. In DNA nanotechnology, the geometrically well-defined double-stranded DNA scaffold serves as an element of spatial control for the precise arrangement of functional groups. Here, we describe the supramolecular assembly of chemically modified DNA hybrids into diverse types of architectures. An amphiphilic DNA duplex serves as the sole structural building element of the nanosized supramolecular structures. The morphology of the assemblies is governed by a single subunit of the building block. The chemical nature of this subunit, i.e., polyethylene glycols of different chain length or a carbohydrate moiety, exerts a dramatic influence on the architecture of the assemblies. Cryo-electron microscopy revealed the arrangement of the individual DNA duplexes within the different constructs. Thus, the morphology changes from vesicles to ribbons with increasing length of a linear polyethylene glycol. Astoundingly, attachment of a N-acetylgalactosamine carbohydrate to the DNA duplex moiety produces an unprecedented type of star-shaped architecture. The novel DNA architectures presented herein imply an extension of the current concept of DNA materials and shed new light on the fast-growing field of DNA nanotechnology.
Introduction
Biochemical processes generally rely on the precise arrangement of the molecular subunits within a hierarchically ordered supramolecular assembly held together by noncovalent, directional interactions.1 Illustrative examples are natural light-harvesting complexes, in which the spatial organization of the chromophores is crucial for an efficient transfer of excitation energy within large protein complexes.2,3 In nanotechnology, DNA is widely used as a structural element for the bottom-up assembly of nanostructures because the scaffold of the DNA duplex serves as a versatile, robust, and yet highly reliable tool of spatial control.4−6 The specificity and programmability of nucleic acid folding enables the bottom-up creation of multidimensional structures by the DNA origami approach.7−14 Alternatively, DNA nanostructures can be constructed by a self-assembly approach using DNA tiles with sticky ends.15−19 Despite their elegance, both approaches face some limitations, because they often require a set of many, even up to hundreds, different DNA sequences to assemble a desired nanostructure.20 The integration of unnatural nucleotide surrogates into oligonucleotides introduces additional functionality and extends the scope of application of DNA from the biological context to the field of materials sciences.21−30 Chemically modified DNA conjugates were shown to form supramolecular polymers with potential applications, e.g., in biomedicine for drug delivery systems or in optoelectronic devices.31−40 Recently, we reported the supramolecular assembly of amphiphilic DNA, bearing either phenanthrene or tetraphenylethylene (TPE) hydrophobic ends, into vesicle-shaped objects.41,42 Hydrophobic interactions of DNA sticky ends, as well as spermine-mediated electrostatic interactions,43,44 are among the driving forces that lead to the formation of these DNA-constructed vesicles. Besides serving as sticky ends, hydrophobic TPE overhangs also permit the direct observation of the self-assembly process by fluorescence spectroscopy due to their aggregation-induced emission (AIE) properties.45−47 Potential applications of vesicles consisting of a DNA-constructed membrane comprise, among others, the use as delivery vehicles for biologically active compounds. We therefore explored the effects of modification of the amphiphilic TPE-derived DNA duplex with additional terminal bioconjugate groups that are, e.g., applied in liposome-based drug delivery systems. Here, we describe the profound effects of such bioconjugates, polyethylene glycol (PEG) chains of different length and a carbohydrate-based moiety, on the DNA-based architectures.
Results and Discussion
Supramolecular Assembly of Amphiphilic DNA into Different Types of Architectures
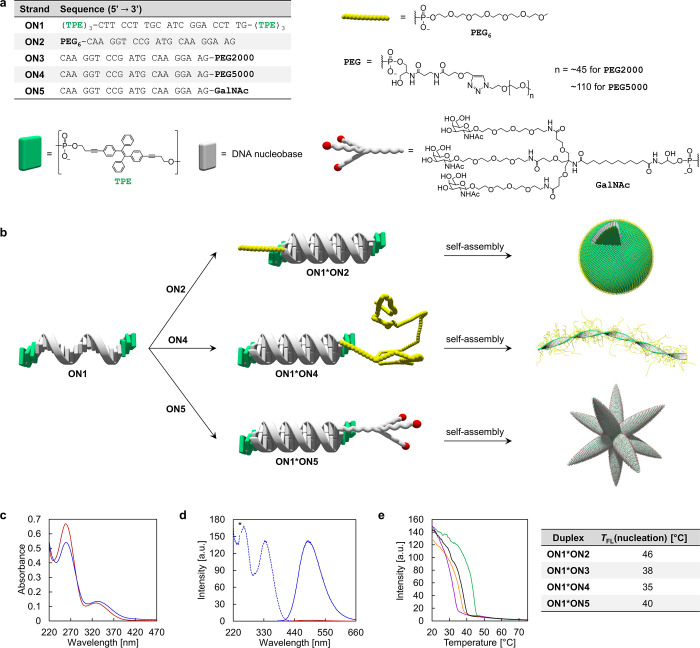
The DNA conjugates studied in this work are displayed in Figure 1a. The TPE-modified oligonucleotide ON1 was prepared via solid-phase synthesis and purified by HPLC following published procedures (see Supporting Information).42 The oligonucleotide contains three phosphodiester-linked TPE moieties at the 3′- and 5′-ends. Oligonucleotides ON2 and ON5 were purchased from commercial suppliers. PEG-DNA conjugates ON3 and ON4 were prepared via copper-catalyzed azide–alkyne cycloaddition from the commercially available 3′-modified alkyne oligonucleotide and the corresponding methoxy-PEG (mPEG) azides.48 mPEG2000 azide (PEG average Mn 2000 Da) and mPEG5000 azide (PEG average Mn 5000 Da) were used for the preparation of ON3 and ON4, respectively, to yield the PEG-DNA conjugates.
Figure 1.
Architectonics of DNA conjugates. (a) Sequences of oligomers and chemical structures of modifications. (b) TPE modified single strand ON1 leads to the formation of diverse DNA architectures after hybridization with different complements. (c,d) Temperature-dependent UV–vis absorption (c), fluorescence emission (d, solid line), and excitation (d, dotted line) spectra of ON1*ON5 at 75 °C (red) and at 20 °C (blue) after thermally controlled assembly (0.5 °C/min; * denotes second-order diffraction). (e) Fluorescence-monitored annealing curves of ON1*ON2 (green), ON1*ON3 (orange), ON1*ON4 (purple), and ON1*ON5 (black) including their respective nucleation temperatures TFL(nucleation) [Nucleation temperature determined by sharp fluorescence onset in cooling curves.] Conditions for (c–e): 1 μM each single strand, 10 mM sodium phosphate buffer pH 7.2, 0.1 mM spermine·4 HCl, 30 vol % ethanol, λex: 335 nm, λem: 490 nm, gradient: 0.5 °C/min.
Hybridization of ON1 with any of the complementary oligonucleotides ON2–ON5 leads to DNA duplexes with TPE overhangs (sticky ends) at both ends (Figure 1b). A further functionality, a PEG or carbohydrate moiety, is attached to the DNA complement, as illustrated in Figure 1b.
Supramolecular assembly of duplex ON1*ON5 is monitored by temperature-dependent UV–vis spectra (Figure 1c) showing a hypochromic effect around 260 nm (DNA and TPE absorption), together with a bathochromic shift of the absorption band from 328 to 333 nm (TPE absorption only) upon cooling (0.5 °C/min). Temperature-dependent fluorescence emission spectra of ON1*ON5 further support the formation of supramolecular assemblies at 20 °C (Figure 1d). At 75 °C, only a marginal emission is detected upon TPE excitation (λex: 335 nm) and in agreement with AIE; this implies that duplex ON1*ON5 is entirely disassembled into separate DNA single strands ON1 and ON5. After cooling to 20 °C, TPE aggregation results in a strong fluorescence signal (centered around 490 nm). The start of the supramolecular assembly process upon cooling of the solution is indicated by a sharp increase in fluorescence. The nucleation temperatures [TFL(nucleation)] of the four different duplexes range between 35 and 46 °C as shown in the corresponding fluorescence-monitored annealing curves (Figure 1e).
Effects of DNA-PEGylation on Supramolecular Architecture
PEGylation of oligonucleotides is a frequently used method for the improvement of the pharmacokinetic properties of therapeutic oligonucleotides.49 The stealth properties of PEG protect oligonucleotides from undesired interactions with enzymes and, thus, from nucleolytic degradation.50
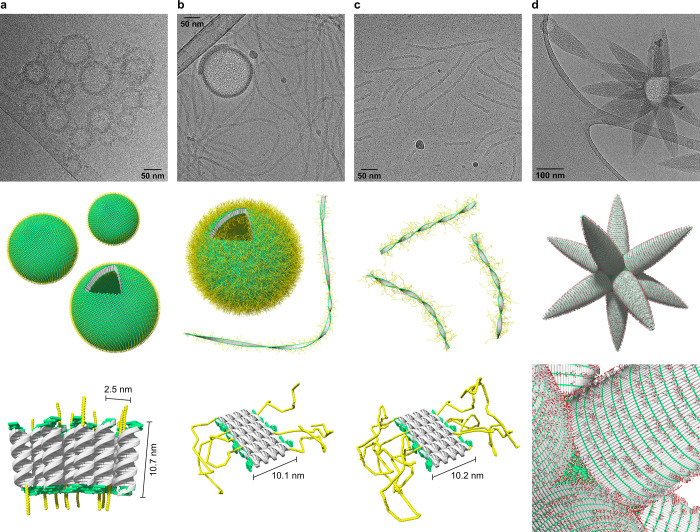
As demonstrated by cryo-electron microscopy (cryo-EM), duplex ON1*ON2, which is functionalized with a short terminal PEG6 chain, self-assembles into vesicular constructs after thermal assembly (Figure 2a). The size of the vesicles ranges between 50 and 100 nm in diameter. The thickness of the vesicular membrane was determined to be 10.7 ± 0.6 nm, which correlates with the length of a 26-mer DNA duplex and is well in agreement with a previously reported, comparable architecture.42 The membrane thickness evidences the presence of unilamellar vesicles formed by a compact, columnar arrangement of ON1*ON2 duplexes. The columnar packing of the DNA is further supported by a regular pattern which is arranged perpendicular to the vesicular membrane. A distance of 2.5 ± 0.3 nm was measured between the parallel aligned rods, which matches the width of a single DNA duplex (Figure 2a, illustration). We assume that the PEG6 chains randomly point toward the inside and outside of the vesicles, thus covering both sides with a thin PEG layer. Cryo-EM imaging shows considerable agglomeration of individual vesicles. Likely, the PEG6 chain is too short to shield individual vesicles entirely from hydrophobic surface interactions.
Figure 2.
Impact of terminal functionalities on the supramolecular DNA architecture revealed by cryo-EM imaging. (a) Formation of unilamellar vesicles by self-assembly of ON1*ON2. (b) Coexistence of ribbons and vesicles after self-assembly of ON1*ON3. (c) Duplex ON1*ON4 assembles into ribbons only. (d) Self-assembly of ON1*ON5 leads to the formation of star-like nanoobjects (asterosomes). Conditions for (a–d): 1 μM each single strand, 10 mM sodium phosphate buffer pH 7.2, 0.1 mM spermine·4 HCl, 30 vol % ethanol; see Supporting Information for additional images.
Duplex ON1*ON3, which contains a PEG2000 chain, substantially changes the predominant morphology of the nanostructures (Figure 2b). While cryo-EM imaging still shows the rare occurrence of vesicles, the main type of supramolecular structure now appears as ribbons. The vesicles of ON1*ON3 exhibit the same kind of DNA packing as those assembled from ON1*ON2 (Figure 2a) and the distance measurements are nearly identical with a membrane thickness of 10.0 ± 0.5 nm and a DNA width of 2.4 ± 0.3 nm. The only observable difference resides in a somewhat larger diameter of 100 to 200 nm, which is about twice the diameter observed for vesicles of ON1*ON3. The ribbons exhibit an overall thickness of 10.1 ± 0.7 nm, which again corresponds very well to the length of the DNA duplex. Therefore, a side-by-side duplex alignment as depicted schematically in (Figure 2b) can be reasonably assigned. The ribbons extend over very long distances, in some cases over a few micrometers.
The coexistence of vesicles and ribbons observed after increasing the size of the PEG chains directed us to test the effect of even longer PEG chains (PEG5000, 110 units). As anticipated, this duplex leads to the formation of ribbons only (Figure 2c). Again, the ribbons possess a thickness of 10.2 ± 0.7 nm, which is similar to the ribbons assembled from ON1*ON3 and, thus, suggests an identical duplex alignment as illustrated in Figure 2c. However, ribbons of ON1*ON4 are considerably shorter than the ones formed by ON1*ON3 (usually less than 350 nm). The long PEG5000 chains are assumed to wrap around individual DNA duplexes and the integration of these wrapped duplexes into the supramolecular ribbons is impeded. This assumption is supported experimentally by the presence of small dark spots (encircled in Figure S21) present in the cryo-EM images, which might be ascribed to individual DNA duplexes.
Conjugation of a Branched Carbohydrate Moiety Leads to Fundamental Morphological Changes
We next turned our attention to the modification of the DNA duplex with a branched tricarbohydrate moiety. In addition to presumably enhance the potential bioapplications of the supramolecular assemblies, the N-acetylgalactosamine (GalNAc) moiety is, due to its branched nature, also sterically more demanding than the linear PEG chains (Figure 1). The aspect of surface crowding is believed to have a strong impact on the supramolecular assembly, as described below.51 The trivalent GalNAc DNA conjugate ON5 was selected due to its well-documented potential for targeted delivery of GalNAc conjugated nucleic acids, in particular, short interfering RNAs (siRNAs), to hepatocytes.52−54 This ligand exhibits an excellent binding affinity toward the asialoglycoprotein receptor (ASGR), which is abundantly expressed in hepatocytes and, thus, responsible for the targeted delivery.55
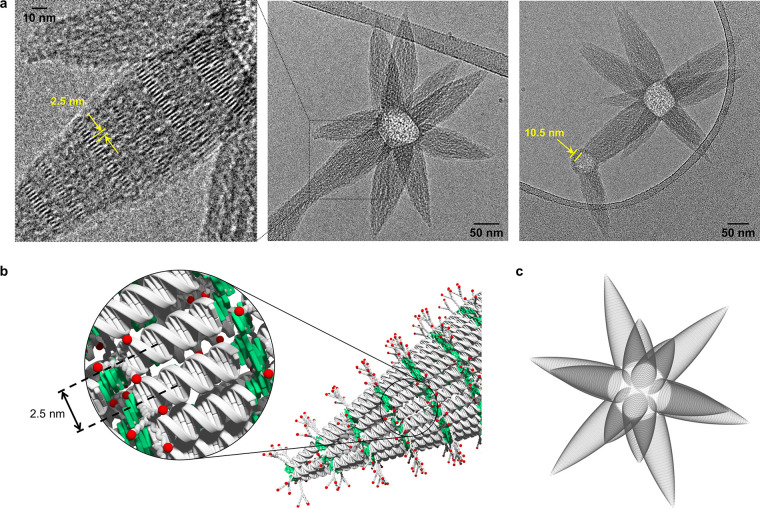
Much to our surprise, the supramolecular assembly of ON1*ON5 leads to an entirely different and novel type of DNA architecture. Cryo-EM imaging (Figure 2d) discloses star-like, or mace-like, supramolecular polymers, in which multiple parabolic cone-shaped extensions are pointing outward from the center of the object. Due to their star-shaped appearance, we will refer to these objects as asterosomes. The number of cones varies among individual asterosomes. The center of the structures appears spherical, and for small spheres, only one or two cones are present. In larger objects, roughly up to 12 protrusions are observed, in which case their arrangement can be approximated by a small stellated dodecahedron.56 Noteworthy, a vesicular membrane is visible around the base of the parabolic cone(s) of structures with only a small number of extensions. The thickness of this membrane amounts to 10.5 ± 0.5 nm, which is consistent with a unilamellar, columnar DNA duplex alignment. The width of the cones at their base ranges between 50 and 75 nm. Regular patterns of discrete bands, which are clearly visible in some areas (Figure 3a), reveal the arrangement of the DNA in the cones. These bands are formed by parallel aligned rods with a distance between these rods of 2.5 ± 0.2 nm (illustration Figure 3b), which corresponds again to the width of the DNA duplex. Hence, the discrete bands are formed by parallel aligned DNA duplexes which, in turn, leads to the conclusion that the cones are characterized by an extended duplex alignment, as illustrated in Figure 3b. Interaction between the DNA segments is ensured by the hydrophobic sticky ends. In the images, the cones of individual asterosomes appear to be of different lengths. Cryo-EM imaging is a projection of three-dimensional (3D) objects onto a two-dimensional (2D) plane, which may lead to a perceived asymmetry of rather uniform-sized cones. To test this possibility, we used a shadow image to simulate the appearance of a regular asterosome at an arbitrary perspective. Regular is defined here as derived from the platonic body (dodecahedron) with all cones pointing toward the center of the surface planes (pentagon). The resulting shadow image of the modeled dodecahedron-type asterosome is depicted in Figure 3c. The obvious similarity between modeled and found pattern suggests that the cones of an individual asterosome are rather uniform in size.
Figure 3.
Asterosomes formed via supramolecular assembly of triantennary GalNAc-functionalized DNA conjugate ON1*ON5. (a) Cryo-EM images of self-assembled ON1*ON5. Conditions: 1 μM ON1*ON5, 10 mM sodium phosphate buffer pH 7.2, 0.1 mM spermine·4 HCl, 30 vol % ethanol. (b) Illustration of the extended DNA duplex alignment in the cones. (c) Shadow image created from a dodecahedral arrangement of cones.
Taking all the morphological features into consideration, we assume that the star-like morphology evolves in a two-step process. Thus, columnar packed vesicular constructs of varying sizes are formed in the first phase of the assembly process. It can be assumed that the trivalent GalNAc moieties lead to a steric crowding on the surface of the vesicles. The resulting strain can be reduced by the formation of the cones. This involves a change of the supramolecular organization from the columnar packing to the extended arrangement of the DNA units. The transition from columnar to extended arrangement results in a significant surface enlargement, hence a redistribution of the bulky GalNAc moieties over a wider area along with a reduction of surface crowding. The growth of the cones can be imagined as a telescopic slide-out process, during which the columnar aligned DNA duplexes slide along each other until reaching the extended arrangement (see SI Video for further illustration). This hypothesis is supported by two additional aspects. First, the number of DNA duplexes assembled in one cone matches roughly the number of duplexes that would be present within the curved surface area of the columnar packed vesicle. Second, fewer cones evolve from small vesicles; the smaller the vesicle, the higher the curvature, and hence, a smaller surface crowding between the branched GalNAc moieties is assumed.
Atomic force microscopy (AFM) was utilized to validate the different morphologies observed by cryo-EM imaging and further support the findings (Figures S24–S26). Vesicles with a size range consistent with the ones detected by cryo-EM were observed for self-assembled ON1*ON2. For the ribbons assembled from ON1*ON3, the average height is around 1 nm, which is less than expected for the width of a single DNA duplex. However, such a flattening has been described in the literature and is probably due to the AFM tip convolution effect.57 Finally, AFM imaging also provided independent evidence of star-shaped objects formed by self-assembly of ON1*ON5 with clearly discernible cones, albeit with lower resolution than cryo-EM.
Conclusion
In conclusion, the supramolecular assembly of amphiphilic DNA conjugates into three distinct, well-defined DNA architectures has been demonstrated. Vesicles, ribbons, and star-shaped objects (asterosomes) are assembled from DNA duplexes formed of a tetraphenylethylene-oligonucleotide and single-stranded complements bearing various types of terminal functionalities. The terminal functionalities dictate the supramolecular assembly process and account for the emergence of the diverse morphologies. Cryo-EM imaging revealed precise information on the exact DNA duplex arrangements within the nanostructures. A morphological change from unilamellar vesicles to ribbons was observed when the length of linear PEG was increased. The use of a trivalent GalNAc moiety in combination with the amphiphilic DNA resulted in a fundamental morphological change. The supramolecular self-assembly of this DNA duplex led to the formation of star-shaped nanostructures (asterosomes), which are characterized by cones emerging radially from the center of the nanoobject. The driving force for the formation of this nanostructure is best explained by a reduction of surface crowding effected by a switch of the DNA duplex alignment from a compact to an extended arrangement during the formation of the cones. A model for the evolution of the cones is presented. So far, comparable star-shaped structures were only reported based on either nanoparticles58,59 or colloidal clusters,60,61 whereas all types of structures presented here represent DNA-built, self-supported architectures. Ongoing research aims at the exploration of the presented vesicular DNA architectures as nanocarriers for drug delivery applications and the investigation of excitation energy transfer along the ribbons to an acceptor dye, creating artificial light-harvesting systems.
Methods
Spectroscopic Measurements
Spectroscopic data were collected from 5 min thermally equilibrated samples at the corresponding temperature using quartz cuvettes with an optical path of 1 cm. UV–vis spectra were measured on an Agilent Cary 100 spectrophotometer, equipped with a Peltier-thermostated multicell holder. Fluorescence spectra were recorded on a Cary Eclipse fluorescence spectrophotometer, equipped with a Peltier-thermostated multicell holder, using an excitation slit of 2.5 nm and an emission slit of 5 nm.
Thermally Controlled Assembly Process
Supramolecular assembly of the DNA architectures was accomplished via thermal disassembly and controlled reassembly. Therefore, the sample solution was heated to 75 °C, before cooling the solution to 20 °C with a gradient of 0.5 °C/min in a Cary Eclipse fluorescence spectrophotometer equipped with a Peltier-thermostated multicell holder. Samples for cryo-EM and AFM were prepared directly after the thermal assembly process.
Cryo-EM Image Acquisition and Analysis
Samples for cryo-EM were plunge-frozen using the FEI Vitrobot Mark 4 at room temperature and 100% humidity. In brief, copper lacey carbon grids were glow-discharged (air −10 mA for 20 s). Three microliters of the sample was pipetted on the grids and blotted for 3 s before plunging into liquid ethane. Sample grids were stored in liquid nitrogen. Images were acquired using a Gatan 626 cryo holder on a Falcon III equipped FEI Tecnai F20 in nanoprobe mode. Due to the nature of the sample, acquisition settings had to be adjusted for a low total electron dose (less than 20 e–/Å2) using EPU software. Distance measurements were done in Fiji62,63 using the multipoint tool to set marks. After the readout of the x- and y-values, the distances between the marks were calculated. The reported distances are mean values with the corresponding standard deviation. A summary of all measured distances as well as representative graphical descriptions where the measurements have been done is provided in the Supporting Information.
Shadow Image
The shadow image was rendered in POV-Ray 3.7 (official POV-Ray Web site: www.povray.org). In brief, the image was created from a dodecahedral 3D assembly, constructed from light-absorbing medium; the triantennary GalNAc linkers were neglected for simplification. Afterward, the 3D illustration was rotated into an arbitrary orientation. The 3D assembly was irradiated with parallel light, orthogonal to the shadow plane, which afforded the shadow image.
AFM Imaging
AFM experiments were conducted on a Nanosurf FlexAFM instrument in tapping mode under ambient conditions. Tap190Al-G cantilevers from BudgetSensors, Innovative Solutions Bulgaria Ltd. were used. AFM samples were prepared on APTES-modified mica sheets (Glimmer “V1”, 20 mm × 20 mm, G250-7, Plano GmbH) according to published procedures, using a sample adsorption time of 7 min.42,64 APTES, (3-aminopropyl)triethoxysilane.
Acknowledgments
Financial support by the Swiss National Foundation (200020_188468 to R.H. and 31003A_179520 to B.Z.) is gratefully acknowledged. Cryo-electron microscopy was performed on equipment supported by the Microscopy Imaging Center (MIC), University of Bern, Switzerland.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.2c00077.
The authors declare no competing financial interest.
Supplementary Material
References
- van der Zwaag D.; Meijer E. W. Fueling Connections between Chemistry and Biology. Science 2015, 349 (6252), 1056–1057. 10.1126/science.aad0194. [DOI] [PubMed] [Google Scholar]
- Lambrev P. H.; Várkonyi Z.; Krumova S.; Kovács L.; Miloslavina Y.; Holzwarth A. R.; Garab G. Importance of Trimer-Trimer Interactions for the Native State of the Plant Light-Harvesting Complex II. Biochim. Biophys. Acta 2007, 1767 (6), 847–853. 10.1016/j.bbabio.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Engel G. S.; Calhoun T. R.; Read E. L.; Ahn T. K.; Mančal T.; Cheng Y.-C.; Blankenship R. E.; Fleming G. R. Evidence for Wavelike Energy Transfer through Quantum Coherence in Photosynthetic Systems. Nature 2007, 446 (7137), 782–786. 10.1038/nature05678. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. DNA in a Material World. Nature 2003, 421, 427–431. 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- Brady R. A.; Brooks N. J.; Cicuta P.; Di Michele L. Crystallization of Amphiphilic DNA C-Stars. Nano Lett. 2017, 17 (5), 3276–3281. 10.1021/acs.nanolett.7b00980. [DOI] [PubMed] [Google Scholar]
- Fabrini G.; Minard A.; Brady R. A.; Di Antonio M.; Di Michele L. Cation-Responsive and Photocleavable Hydrogels from Noncanonical Amphiphilic DNA Nanostructures. Nano Lett. 2022, 22 (2), 602–611. 10.1021/acs.nanolett.1c03314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Kim D.-N.; Kilchherr F.; Dietz H.; Bathe M. Quantitative Prediction of 3D Solution Shape and Flexibility of Nucleic Acid Nanostructures. Nucleic Acids Res. 2012, 40 (7), 2862–2868. 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S.; Fan C.; Gothelf K. V.; Li J.; Lin C.; Liu L.; Liu N.; Nijenhuis M. A. D.; Saccà B.; Simmel F. C.; et al. DNA Origami. Nat. Rev. Methods Prim 2021, 1, 13. 10.1038/s43586-020-00009-8. [DOI] [Google Scholar]
- Schill J.; Rosier B. J. H. M.; Audenis B. G.; Estirado E. M.; de Greef T. F. A.; Brunsveld L. Assembly of Dynamic Supramolecular Polymers on a DNA Origami Platform. Angew. Chem. Int. Ed 2021, 60 (14), 7612–7616. 10.1002/anie.202016244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneziano R.; Moyer T. J.; Stone M. B.; Wamhoff E.-C.; Read B. J.; Mukherjee S.; Shepherd T. R.; Das J.; Schief W. R.; Irvine D. J.; et al. Role of Nanoscale Antigen Organization on B-Cell Activation Probed Using DNA Origami. Nat. Nanotechnol 2020, 15 (8), 716–723. 10.1038/s41565-020-0719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R. P.; Malle M. G.; Okholm A. H.; Krishnan S.; Bohr S. S. R.; Sørensen R. S.; Ries O.; Vogel S.; Simmel F. C.; Hatzakis N. S.; et al. A Large Size-Selective DNA Nanopore with Sensing Applications. Nat. Commun. 2019, 10, 5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum M.; Schwarz R. J.; Schwarz M.; Kramer M.; Richert C. Stabilizing DNA Nanostructures through Reversible Disulfide Crosslinking. Nanoscale 2019, 11 (31), 14921–14928. 10.1039/C9NR05143K. [DOI] [PubMed] [Google Scholar]
- Noteborn W. E. M.; Abendstein L.; Sharp T. H. One-Pot Synthesis of Defined-Length SsDNA for Multiscaffold DNA Origami. Bioconjugate Chem. 2021, 32 (1), 94–98. 10.1021/acs.bioconjchem.0c00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Seeman N. C. Synthesis from DNA of a Molecule with the Connectivity of a Cube. Nature 1991, 350, 631–633. 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- Huang K.; Yang D.; Tan Z.; Chen S.; Xiang Y.; Mi Y.; Mao C.; Wei B. Self-Assembly of Wireframe DNA Nanostructures from Junction Motifs. Angew. Chem. Int. Ed 2019, 58 (35), 12123–12127. 10.1002/anie.201906408. [DOI] [PubMed] [Google Scholar]
- Taylor L. L. K.; Riddell I. A.; Smulders M. M. J. Self-Assembly of Functional Discrete Three-Dimensional Architectures in Water. Angew. Chem. Int. Ed 2019, 58 (5), 1280–1307. 10.1002/anie.201806297. [DOI] [PubMed] [Google Scholar]
- Yao G.; Zhang F.; Wang F.; Peng T.; Liu H.; Poppleton E.; Šulc P.; Jiang S.; Liu L.; Gong C.; et al. Meta-DNA Structures. Nat. Chem. 2020, 12 (11), 1067–1075. 10.1038/s41557-020-0539-8. [DOI] [PubMed] [Google Scholar]
- Gentile S.; Del Grosso E.; Pungchai P. E.; Franco E.; Prins L. J.; Ricci F. Spontaneous Reorganization of DNA-Based Polymers in Higher Ordered Structures Fueled by RNA. J. Am. Chem. Soc. 2021, 143 (48), 20296–20301. 10.1021/jacs.1c09503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA Nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar]
- Simeth N. A.; Kobayashi S.; Kobauri P.; Crespi S.; Szymanski W.; Nakatani K.; Dohno C.; Feringa B. L. Rational Design of a Photoswitchable DNA Glue Enabling High Regulatory Function and Supramolecular Chirality Transfer. Chem. Sci. 2021, 12 (26), 9207–9220. 10.1039/D1SC02194J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürki N.; Grossenbacher E.; Cannizzo A.; Feurer T.; Langenegger S. M.; Häner R. DNA-Organized Artificial LHCs - Testing the Limits of Chromophore Segmentation. Org. Biomol. Chem. 2020, 18 (35), 6818–6822. 10.1039/D0OB01531H. [DOI] [PubMed] [Google Scholar]
- Goodnow R. A.; Dumelin C. E.; Keefe A. D. DNA-Encoded Chemistry: Enabling the Deeper Sampling of Chemical Space. Nat. Rev. Drug Discov 2017, 16 (2), 131–147. 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Burns J. R.; Göpfrich K.; Wood J. W.; Thacker V. V.; Stulz E.; Keyser U. F.; Howorka S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew. Chem. Int. Ed 2013, 52 (46), 12069–12072. 10.1002/anie.201305765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetzger C.; Bäuerlein C.; Höbartner C. Supramolecular Fluorescence Resonance Energy Transfer in Nucleobase-Modified Fluorogenic RNA Aptamers. Angew. Chem. Int. Ed 2020, 59 (17), 6760–6764. 10.1002/anie.201916707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen M.; Gothelf K. V. Chemistries for DNA Nanotechnology. Chem. Rev. 2019, 119 (10), 6384–6458. 10.1021/acs.chemrev.8b00570. [DOI] [PubMed] [Google Scholar]
- Mass O. A.; Wilson C. K.; Roy S. K.; Barclay M. S.; Patten L. K.; Terpetschnig E. A.; Lee J.; Pensack R. D.; Yurke B.; Knowlton W. B. Exciton Delocalization in Indolenine Squaraine Aggregates Templated by DNA Holliday Junction Scaffolds. J. Phys. Chem. B 2020, 124 (43), 9636–9647. 10.1021/acs.jpcb.0c06480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda A. S.; Przypis Ł.; Walczak K.; Jørgensen P. T.; Wengel J. Carbazole Modified Oligonucleotides: Synthesis, Hybridization Studies and Fluorescence Properties. Org. Biomol. Chem. 2020, 18 (35), 6935–6948. 10.1039/D0OB01553A. [DOI] [PubMed] [Google Scholar]
- Kropp H. M.; Diederichs K.; Marx A. The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide. Angew. Chem. Int. Ed 2019, 58 (16), 5457–5461. 10.1002/anie.201900315. [DOI] [PubMed] [Google Scholar]
- Zhu R. Y.; Majumdar C.; Khuu C.; De Rosa M.; Opresko P. L.; David S. S.; Kool E. T. Designer Fluorescent Adenines Enable Real-Time Monitoring of MUTYH Activity. ACS Cent. Sci. 2020, 6 (10), 1735–1742. 10.1021/acscentsci.0c00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H.; Díaz S. A.; Fontana J.; Chiriboga M.; Veneziano R.; Medintz I. L. Utilizing the Organizational Power of DNA Scaffolds for New Nanophotonic Applications. Adv. Opt. Mater. 2019, 7 (18), 1900562. 10.1002/adom.201900562. [DOI] [Google Scholar]
- Mathur D.; Medintz I. L. The Growing Development of DNA Nanostructures for Potential Healthcare-Related Applications. Adv. Healthcare Mater. 2019, 8 (9), 1801546. 10.1002/adhm.201801546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A.; Linko V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed 2020, 59 (37), 15818–15833. 10.1002/anie.201916390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnands S. P. W.; Meijer E. W.; Merkx M. DNA-Functionalized Supramolecular Polymers: Dynamic Multicomponent Assemblies with Emergent Properties. Bioconjugate Chem. 2019, 30 (7), 1905–1914. 10.1021/acs.bioconjchem.9b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensslen P.; Gärtner S.; Glaser K.; Colsmann A.; Wagenknecht H.-A. A DNA-Fullerene Conjugate as a Template for Supramolecular Chromophore Assemblies: Towards DNA-Based Solar Cells. Angew. Chem. Int. Ed 2016, 55 (5), 1904–1908. 10.1002/anie.201509332. [DOI] [PubMed] [Google Scholar]
- Vybornyi M.; Vyborna Y.; Häner R. DNA-Inspired Oligomers: From Oligophosphates to Functional Materials. Chem. Soc. Rev. 2019, 48 (16), 4347–4360. 10.1039/C8CS00662H. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Du T.; Liang F.; Liu S. Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine. Int. J. Mol. Sci. 2018, 19 (8), 2283. 10.3390/ijms19082283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Märcher A.; Nijenhuis M. A. D.; Gothelf K. V. A Wireframe DNA Cube: Antibody Conjugate for Targeted Delivery of Multiple Copies of Monomethyl Auristatin E. Angew. Chem. Int. Ed 2021, 60 (40), 21691–21696. 10.1002/anie.202107221. [DOI] [PubMed] [Google Scholar]
- Appukutti N.; Serpell C. J. High Definition Polyphosphoesters: Between Nucleic Acids and Plastics. Polym. Chem. 2018, 9 (17), 2210–2226. 10.1039/C8PY00251G. [DOI] [Google Scholar]
- Zhang Y.; Peng R.; Xu F.; Ke Y. Hierarchical Self-Assembly of Cholesterol-DNA Nanorods. Bioconjugate Chem. 2019, 30 (7), 1845–1849. 10.1021/acs.bioconjchem.9b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösch C. D.; Jevric J.; Bürki N.; Probst M.; Langenegger S. M.; Häner R. Supramolecular Assembly of DNA-Phenanthrene Conjugates into Vesicles with Light-Harvesting Properties. Bioconjugate Chem. 2018, 29 (5), 1505–1509. 10.1021/acs.bioconjchem.8b00263. [DOI] [PubMed] [Google Scholar]
- Rothenbühler S.; Iacovache I.; Langenegger S. M.; Zuber B.; Häner R. Supramolecular Assembly of DNA-Constructed Vesicles. Nanoscale 2020, 12 (41), 21118–21123. 10.1039/D0NR04103C. [DOI] [PubMed] [Google Scholar]
- Egli M. DNA-Cation Interactions: Quo Vadis?. Chem. Biol. 2002, 9 (3), 277–286. 10.1016/S1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- Li J.; Wijeratne S. S.; Qiu X.; Kiang C.-H. DNA under Force: Mechanics, Electrostatics, and Hydration. Nanomaterials 2015, 5 (1), 246–267. 10.3390/nano5010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J.; Xie Z.; Lam J. W. Y.; Cheng L.; Chen H.; Qiu C.; Kwok H. S.; Zhan X.; Liu Y.; Zhu D.; et al. Aggregation-Induced Emission of 1-Methyl-1,2,3,4,5-Pentaphenylsilole. Chem. Commun. 2001, 1740–1741. 10.1039/b105159h. [DOI] [PubMed] [Google Scholar]
- Li S.; Langenegger S. M.; Häner R. Control of Aggregation-Induced Emission by DNA Hybridization. Chem. Commun. 2013, 49 (52), 5835–5837. 10.1039/c3cc42706d. [DOI] [PubMed] [Google Scholar]
- Liu H.; Xiong L. H.; Kwok R. T. K.; He X.; Lam J. W. Y.; Tang B. Z. AIE Bioconjugates for Biomedical Applications. Adv. Opt. Mater. 2020, 8 (14), 2000162. 10.1002/adom.202000162. [DOI] [Google Scholar]
- Presolski S. I.; Hong V. P.; Finn M. G. Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. 10.1002/9780470559277.ch110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang D.; Jia F.; Miller A.; Tan X.; Chen P.; Zhang L.; Lu H.; Fang Y.; Kang X.; et al. Self-Assembled DNA–PEG Bottlebrushes Enhance Antisense Activity and Pharmacokinetics of Oligonucleotides. ACS Appl. Mater. Interfaces 2020, 12 (41), 45830–45837. 10.1021/acsami.0c13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerantzaki M.; Loth C.; Lutz J.-F. Chemical Conjugation of Nucleic Acid Aptamers and Synthetic Polymers. Polym. Chem. 2021, 12 (24), 3498–3509. 10.1039/D1PY00516B. [DOI] [Google Scholar]
- Stachowiak J. C.; Hayden C. C.; Sasaki D. Y. Steric Confinement of Proteins on Lipid Membranes Can. Drive Curvature and Tubulation. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (17), 7781–7786. 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J. A.; Witzigmann D.; Thomson S. B.; Chen S.; Leavitt B. R.; Cullis P. R.; Van der Meel R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol 2021, 16, 630–643. 10.1038/s41565-021-00898-0. [DOI] [PubMed] [Google Scholar]
- Benizri S.; Gissot A.; Martin A.; Vialet B.; Grinstaff M. W.; Barthélémy P. Bioconjugated Oligonucleotides: Recent Developments and Therapeutic Applications. Bioconjugate Chem. 2019, 30 (2), 366–383. 10.1021/acs.bioconjchem.8b00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A. D.; Dowdy S. F. GalNAc-SiRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther 2018, 28 (3), 109–118. 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.; Prakash T. P.; Donner A. J.; Kinberger G. A.; Gaus H. J.; Low A.; Østergaard M. E.; Bell M.; Swayze E. E.; Seth P. P. Characterizing the Effect of GalNAc and Phosphorothioate Backbone on Binding of Antisense Oligonucleotides to the Asialoglycoprotein Receptor. Nucleic Acids Res. 2017, 45 (5), 2294–2306. 10.1093/nar/gkx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayley A. On Poinsot’s Four New Regular Solids. Philos. Mag 1859, 17 (112), 123–128. 10.1080/14786445908642635. [DOI] [Google Scholar]
- Bustamante C.; Vesenka J.; Tang C. L.; Rees W.; Guthold M.; Keller R. Circular DNA Molecules Imaged in Air by Scanning Force Microscopy. Biochemistry 1992, 31 (1), 22–26. 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Creyer M. N.; Chen A.; Yim W.; Lafleur R. P. M.; He T.; Lin Z.; Xu M.; Abbasi P.; Wu J.; et al. Stereoselective Growth of Small Molecule Patches on Nanoparticles. J. Am. Chem. Soc. 2021, 143 (31), 12138–12144. 10.1021/jacs.1c04272. [DOI] [PubMed] [Google Scholar]
- Lermusiaux L.; Many V.; Barois P.; Ponsinet V.; Ravaine S.; Duguet E.; Tréguer-Delapierre M.; Baron A. Toward Huygens’ Sources with Dodecahedral Plasmonic Clusters. Nano Lett. 2021, 21 (5), 2046–2052. 10.1021/acs.nanolett.0c04666. [DOI] [PubMed] [Google Scholar]
- Kim Y.-J.; Kim J.-H.; Jo I.-S.; Pine D. J.; Sacanna S.; Yi G.-R. Patchy Colloidal Clusters with Broken Symmetry. J. Am. Chem. Soc. 2021, 143 (33), 13175–13183. 10.1021/jacs.1c05123. [DOI] [PubMed] [Google Scholar]
- Liu X.; Moradi M.-A.; Bus T.; Debije M. G.; Bon S. A. F.; Heuts J. P. A.; Schenning A. P. H. J. Flower-like Colloidal Particles through Precipitation Polymerization of Redox-Responsive Liquid Crystals. Angew. Chem. Int. Ed 2021, 60 (52), 27026–27030. 10.1002/anie.202111521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9 (7), 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M.; Rueden C. T.; Allan C.; Burel J.-M.; Moore W.; Patterson A.; Loranger B.; Moore J.; Neves C.; MacDonald D.; et al. Metadata Matters: Access to Image Data in the Real World. J. Cell Biol. 2010, 189 (5), 777–782. 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösch C. D.; Langenegger S. M.; Häner R. Light-Harvesting Nanotubes Formed by Supramolecular Assembly of Aromatic Oligophosphates. Angew. Chem. Int. Ed 2016, 55 (34), 9961–9964. 10.1002/anie.201604508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.