Djannatian et al. show by ultrastructural analysis of mouse optic nerves and in vivo imaging of zebrafish spinal cords that developmental myelination is an error-prone process and microglia contribute to the removal of aberrant structures. This may be an important mechanism to generate properly functioning myelin.
Abstract
To enable rapid propagation of action potentials, axons are ensheathed by myelin, a multilayered insulating membrane formed by oligodendrocytes. Most of the myelin is generated early in development, resulting in the generation of long-lasting stable membrane structures. Here, we explored structural and dynamic changes in central nervous system myelin during development. To achieve this, we performed an ultrastructural analysis of mouse optic nerves by serial block face scanning electron microscopy (SBF-SEM) and confocal time-lapse imaging in the zebrafish spinal cord. We found that myelin undergoes extensive ultrastructural changes during early postnatal development. Myelin degeneration profiles were engulfed and phagocytosed by microglia using exposed phosphatidylserine as one “eat me” signal. In contrast, retractions of entire myelin sheaths occurred independently of microglia and involved uptake of myelin by the oligodendrocyte itself. Our findings show that the generation of myelin early in development is an inaccurate process associated with aberrant ultrastructural features that require substantial refinement.
Introduction
Central nervous system (CNS) myelination, a critical step in nervous system development, has enabled vertebrates to expand higher brain functions in a cost-efficient way (Zalc, 2016; Stadelmann et al., 2019). Myelin, produced by oligodendrocytes in the CNS, electrically insulates axons to facilitate saltatory conduction (Rushton, 1951; Cohen et al., 2020; Hartline and Colman, 2007) and also provides metabolic support (Fünfschilling et al., 2012; Lee et al., 2012; Saab et al., 2016). Most of myelin is generated during early postnatal development according to a pre-defined genetic program. Once formed, myelin is a highly stable multilayered membrane, reflected by the unusually long lifetime of myelin-specific proteins (Toyama et al., 2013) and the low turnover of the sheaths. However, myelin remodeling can occur during learning and neuronal circuit refinement, thereby contributing to brain plasticity (Gibson et al., 2014; Etxeberria et al., 2016; McKenzie et al., 2014; Xiao et al., 2016; Scholz et al., 2009; Hill et al., 2018; Steele et al., 2013; Makinodan et al., 2012; Hughes et al., 2018; Lakhani et al., 2016; Monje, 2018; Wake et al., 2015; Fields, 2008). In addition, a recent study in zebrafish demonstrated that developmental myelin is generated in excess and modified by microglial pruning (Hughes and Appel., 2020). We have previously provided a model of myelin wrapping, in which the leading edge moves around the axon underneath the previously deposited membrane, coupled with the lateral extension of myelin membrane layers along the axon (Snaidero et al., 2014). However, little is known about the error-rate and potential correction mechanisms that ensure proper myelin formation.
Here, we investigated the ultrastructural changes of CNS myelin during development using 3D serial block face scanning electron microscopy (SBF-SEM), focusing on abnormal myelination profiles. We show that myelin biogenesis is associated with the formation of abundant aberrant ultrastructure. Live imaging in zebrafish showed that microglia phagocytose myelin, and phosphatidylserine (PS) presents one “eat me” signal that promotes myelin phagocytosis. Myelin remodeling can also occur independently of microglia, in a process in which oligodendrocytes retract entire myelin sheaths into their cell body, followed by trafficking into lysosomes.
Results
Aberrant myelin ultrastructure occurs during development
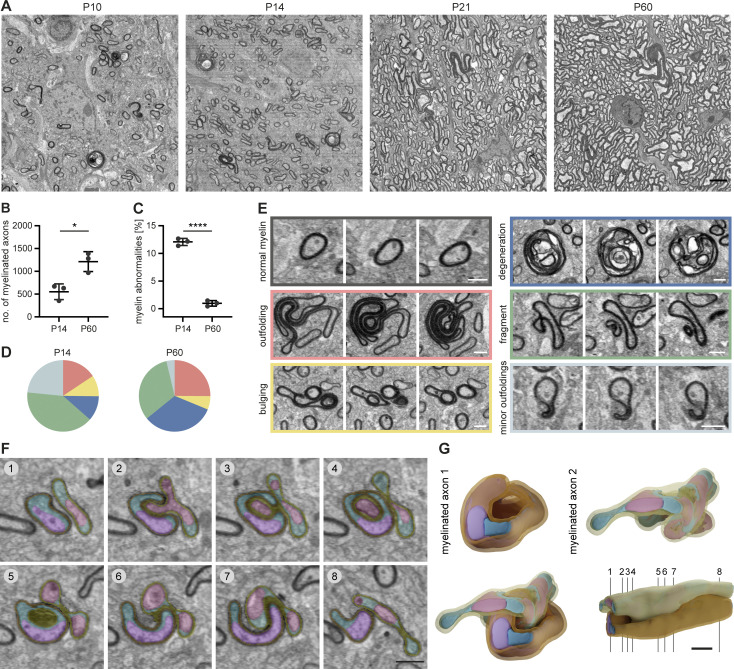
To analyze myelin 3D ultrastructure in the developing CNS, we acquired volume stacks of 80 × 80 × 40–210 µm size of postnatal day 10 (P10), P14, P21, and P60 mouse optic nerves by SBF-SEM (Fig. 1). As expected, myelination was sparse at P10 and sharply increased during the following 2 wk, while all axons were myelinated by P60 (Fig. 1, A and B; and Fig. S1 A). Notably, we found aberrant myelin structures in 24% of the myelinated axons in SEM sub-volumes at P10, which declined to 1.2% by P60 (Fig. 1, A and C; Fig. S1 B; and Video 1). Among these abnormal myelin profiles, we found myelin outfoldings (large sheets of excess membrane that face away from the axon) and whorls of degenerated myelin, as previously described (Edgar et al., 2009; Snaidero et al., 2014; Erwig et al., 2019). The 3D nature of SBF-SEM allowed us to identify three additional structural abnormalities: first, 0.2–0.5 µm fragments that pinch off from a myelin sheath or are found in the extracellular space, second, “myelin bulging,” and third, minor outfolding, that may represent an early form of outfolding and bulging (Fig. 1, D and E; and Video 1). Myelin bulging is characterized by an enlargement of the inner tongue and eccentric dilations of the myelin membrane, which partially folds back on itself, generating a myelin sheath that enwraps non-axonal structures (e.g., small processes of glial cells or even parts of another bulging myelin sheath; Fig. 1, F and G; and Video 2). In a volume reconstruction, it is clearly distinguishable from double myelin, which occurs when two myelin sheaths grow into each other (Djannatian et al., 2019). Bulging is typically found adjacent to normal-appearing myelin on the same sheath. While the frequency of all types of myelin abnormalities decreased between P10 and P60, the fraction of degenerated myelin significantly increased from 11.3 ± 5.2% at P14 to 32.9 ± 6.7% at P60 (Fig. 1 D). We found that large whorl-like structures of degenerated myelin as well as small myelin sheath dystrophies and even tiny, detached fragments were engulfed by microglia (Fig. 2, A−D; and Fig. S1, E and F). Microglia were identified in SEM volumes by their unique features, mainly the electron-dense cytoplasm, bean-shaped nuclei, characteristic pattern of heterochromatin, and presence of phagocytic inclusions (Savage et al., 2018). We confirmed this by reconstructing a whole microglial cell in 3D (Fig. 2 A). Microglia often wrapped around several of these profiles at the same time (Fig. 2 A and Video 3) and contained electron-dense fragments that likely represent products of myelin phagocytosis (Fig. 2 A, section 7, and Fig. 2 E). When we quantified which myelin profiles were associated with microglia at P14, we found that the vast majority exhibited a pathological ultrastructure. Only 9.6 ± 1.9% of myelin segments in contact with microglia exhibited normal myelin ultrastructure. All of the above-described types of myelin aberrations were associated with microglia, but only the category “myelin fragments” was associated significantly more compared to normal myelin (Fig. 2 F). Microglia numbers increased in a similar manner as the number of myelinated axons (Fig. S1 D), except from a temporarily increased microglia abundance at P10 (Fig. 1 G).
Figure 1.
Myelin abnormalities occur temporarily during development in the mouse CNS. SBF-SEM of P10, P14, P21, and P60 wt mouse optic nerves (80 × 80 × 40–210 μm volumes with a 10 × 10 × 80 nm resolution). (A) Representative cross-sections. (B) Mean number of myelinated axons within a 40 × 40 μm area at P14 and P60 (quantified on 8–10 evenly dispersed cross-sections within the SBF-SEM volumes, n = 3 optic nerves). Two-sided Student’s t test: *, P = 0.0145. In total, we quantified 4,655.3 ± 785.4 myelinated axons at P14 and 10,560 ± 2,099.8 at P60. (C) Mean percentage of myelin abnormalities at P14 and P60 (within 8–10 evenly dispersed volumes of 40 × 40 × 8 μm, normalized by the number of myelinated axons in reference sections, n = 3 optic nerves). Two-sided Student’s t test: ****, P < 0.0001. (D) Percentage of error subtypes among the myelin errors quantified in C: outfoldings (red), bulgings (yellow), degenerations (dark blue), fragments pinching off from a sheath or lying in the vicinity of a sheath (green), and minor outfoldings (light blue). Means of n = 3 optic nerves. Color code refers to example images in E. (E) Cross-sections at different z levels show examples of myelin abnormalities: outfolding, bulging, degeneration, fragment detaching from an intact sheath, and minor outfoldings that likely represent an early stage of outfolding and bulging. (F and G) 3D reconstruction of a myelinated axon bulging into another myelinated axon. (F) Pseudo-colored cross-sections at different z levels (violet/pink: axons, petrol/cyan: glial cytoplasm, orange/yellow: myelin). Note the excess glial cytoplasm at the location of bulging. (G) 3D reconstruction. Numbering refers to cross-sections in F. Images in A and F are 4 × 4 binned. Scale bars: 3 µm (A), 2 µm (G), 1 µm (E, F). See also Videos 1 and 2.
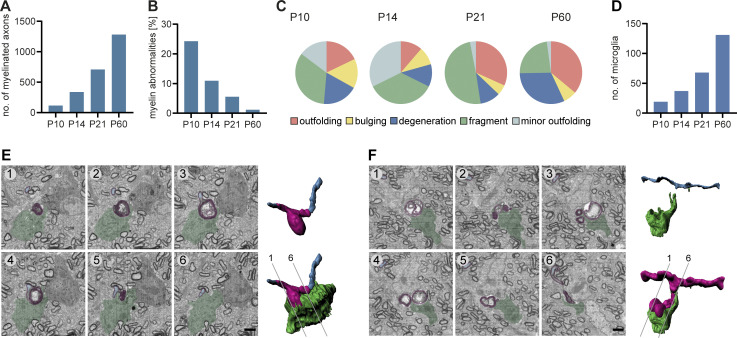
Figure S1.
Temporal dynamics and microglial engulfment of myelin abnormalities during CNS development in the mouse. (A−C) SBF-SEM of P10, P14, P21, and P60 wt mouse optic nerves (80 × 80 × 40–210 μm volumes with a 10 × 10 × 80 nm resolution). Quantifications include all time points for n = 1 optic nerve. (A) Mean number of myelinated axons within a 40 × 40 μm area (quantified on 8–10 evenly dispersed cross-sections within the SBF-SEM volumes). In total, we quantified 1,150 (P10), 3,559 (P14), 7,075 (P21), and 12,808 (P60) myelinated axons. (B) Mean percentage of myelin abnormalities (within 8–10 evenly dispersed volumes of 40 × 40 × 8 μm, normalized by the number of myelinated axons in reference sections). (C) Percentage of error subtypes among the myelin errors quantified in B: outfoldings (red), bulgings (yellow), degenerations (dark blue), fragments pinching off from a sheath or lying in the vicinity of a sheath (green), and minor outfoldings (light blue). (D) Quantification shows the number of microglia in a volume of 80 × 80 × 160 µm at P10, P14, P21, and P60 (n = 1). (E and F) SBF-SEM of a P14 wt mouse optic nerve shows two examples of a microglia engulfing degenerated myelin. Left: Pseudocolored cross-sections (blue: axon, magenta: myelin, green: microglia). Right: 3D reconstruction. Numbering refers to cross-sections. Scale bars: 2 µm.
Video 1.
Aberrant myelin ultrastructure is abundant during development and is resolved until young adulthood. SBF-SEM of mouse optic nerves at P10, P14, P21, and P60 shows abundant myelin aberrations at P10 that are mostly resolved until young adulthood. The number of myelinated axons is steadily increasing with age. Subtypes of myelin aberrations comprise outfoldings, bulgings, degenerations, fragments, and minor outfoldings. Related to Fig. 1.
Video 2.
3D reconstruction of a myelin bulging. SBF-SEM of a mouse optic nerve at P10. Myelin bulging is characterized by an enlargement of the inner tongue and eccentric dilations of the myelin membrane, which partially folds back on itself, generating a myelin sheath that enwraps another myelinated sheath. Video shows raw data with super-imposed pseudo-coloring and 3D reconstructions. Pseudo-coloring of myelinated axons 1 and 2 from Fig. 1 shows axons in violet/pink, glial cytoplasm in petrol/cyan, and myelin in orange/yellow.
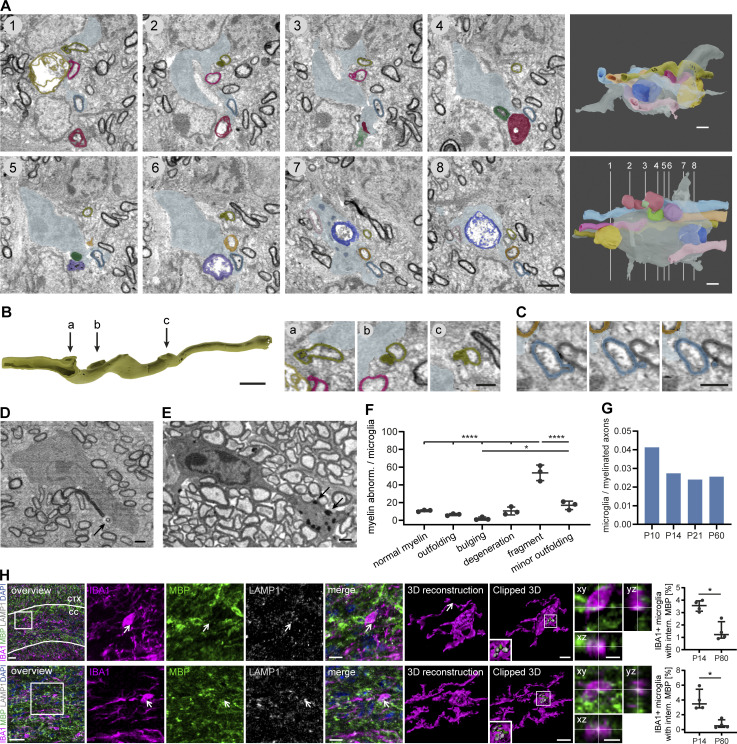
Figure 2.
Microglia engulf and phagocytose developmental myelin abnormalities in the mouse CNS. (A−D) SBF-SEM of a P14 wt mouse optic nerve shows a microglia contacting several myelinated axons with aberrant myelin ultrastructure. (A) Pseudocolored cross-sections show examples of myelin abnormalities engulfed or contacted by a microglia. The microglia is displayed in gray blue, while the other colors designate individual myelin sheaths. 3D reconstructions (right) show different orientations and numbering refers to cross-sections. (B) Focal myelin dystrophies on a reconstructed myelin sheath. The dystrophy in “a” is sliced to display the 3D structure. Arrows refer to cross-sections on the right side. Cross-sections show details of the cross-sections 1, 2, and 6 in A. (C) Myelin fragment pinching off a myelin sheath on three consecutive sections. (D) Microglia engulfing a detached myelin fragment (arrow). (E) Microglia with myelin inclusions at P60 (arrows). (F) Quantification shows the percentage of myelin aberration subtypes associated with microglia in SBF-SEM of P14 wt mouse optic nerves (n = 3 mice). One-way ANOVA with post-hoc Tukey’s test: normal myelin, outfoldings, bulgings, degeneration, and minor outfolding vs. fragment: P < 0.0001, bulging vs. minor outfolding: P = 0.0165, all other comparisons were non-significant. (G) Quantification shows the total number of microglia within a volume of 80 × 80 × 160 µm normalized by the number of myelinated axons at P10, P14, P21, and P60 (n = 1 optic nerve). (H) Sections of P14 wt corpus callosum (CC; top), cortex (CTX), and optic nerve (bottom) co-immunostained for MBP and LAMP1 together with IBA1 for microglia. Clipped 3D view shows MBP-positive staining inside microglia and in close association with lysosomes (arrows). Orthogonal views display co-localization of IBA1 and MBP staining. Quantifications show the percentage of microglia with internalized MBP in P14 vs. P80 brains (n = 4 mice). Mann–Whitney U-test: P = 0.0286 (H, corpus callosum and optic nerve). Data represent median and IQR. *, P < 0.05, ****, P < 0.0001. Images in A–E are 4 × 4 binned. Scale bars: 30 μm (H, overview), 10 μm (H, merge), 5 μm (H, clipped 3Ds and orthogonal views), 2 μm (A, B, 3D reconstruction), and 1 μm (B, cross-sections, C–E). See also Video 3.
Video 3.
3D reconstruction of a microglia that is associated with aberrant myelin ultrastructure in mouse optic nerve. SBF-SEM of a P14 wild-type mouse optic nerve shows a microglia contacting several myelinated axons with aberrant myelin ultrastructure. Video shows raw data with super-imposed pseudo-coloring and 3D reconstructions. Pseudo-coloring shows the microglia in gray blue, while the other colors designate individual myelin sheaths. Related to Fig. 2.
To confirm myelin membrane uptake by microglia, we co-immunostained tissue sections from mouse P14 and P80 corpus callosum (Fig. 2 D, top row) and optic nerve (Fig. 2 D, bottom row) with myelin-basic protein (MBP), IBA1, and LAMP1 to label microglia and late endosomes/lysosomes. About 3.5% of the microglia contained MBP+ fragments within LAMP1+ organelles at P10, while around 1.2% or less did at P80 (corpus callosum: P14: median 3.56% [interquartile range (IQR) 3.10–3.94%], P80: median 1.22% [IQR 0.93–2.27%], P = 0.0286, optic nerve: P14: median 3.48% [IQR 2.98–5.44%], P80: median 0.49% [IQR 0.31–1.32%], P = 0.0286, Mann–Whitney U-test, n = 4 mice). Together, these data show aberrant myelin ultrastructure during development, and myelin debris inside of phagocytes.
Microglia surveil and phagocytose myelin in zebrafish
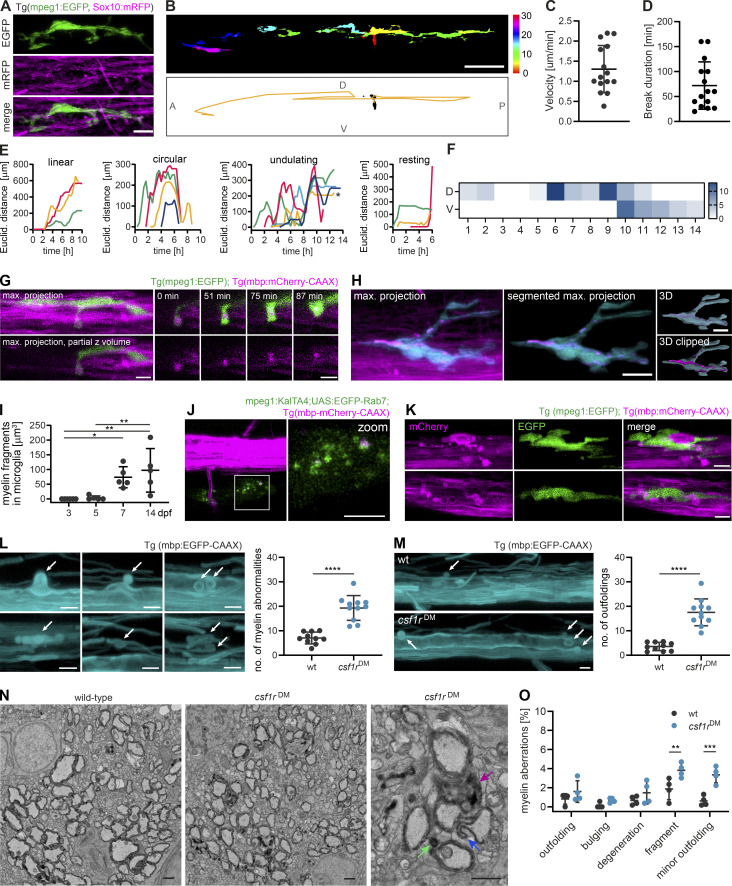
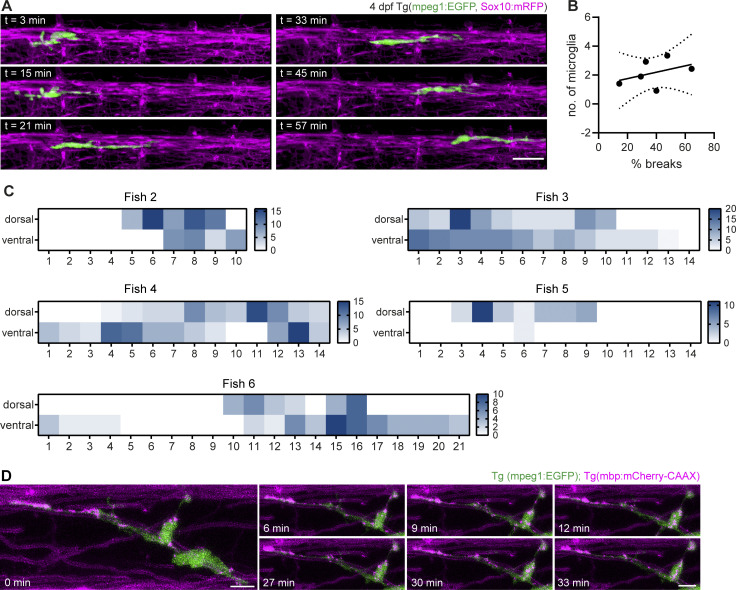
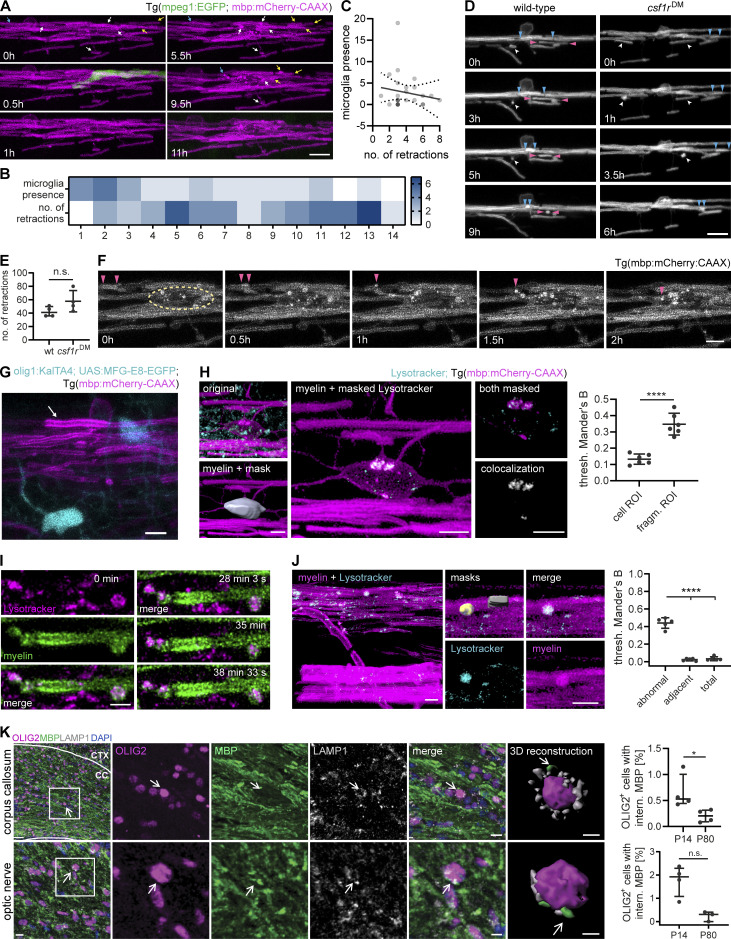
In order to gain insight into how microglia interact with myelin in vivo, we conducted further experiments in the zebrafish spinal cord. This enabled us to follow myelination in a relatively short developmental window by expressing fluorescent reporters and performing time-lapse imaging. When we imaged transgenic zebrafish larvae expressing mpeg1:EGFP (labeling microglia) and sox10:mRFP (labeling myelin) sheaths starting from the onset of myelination at 3 d post-fertilization (dpf), we found that microglia were often elongated and in contact with myelin sheaths (Fig. 3 A). During time-lapse imaging at 3-min intervals, microglia extended and retracted their processes along myelin sheaths and, in parallel, moved their cell body forward (Fig. S2 A and Video 4). In order to monitor microglia movement along the spinal cord, individual microglia at 3–4 dpf were followed in 20–30 min intervals over 10–14 h (Video 4). Movements of individual microglia were tracked by their soma displacement in consecutive time frames (Fig. 3 B). Microglia moved with a mean velocity of 1.30 ± 0.59 µm/min (Fig. 3 C) and occasionally took breaks with negligible soma displacement over one or several consecutive time frames (average break duration: 72.00 ± 47.44 min; Fig. 3 D). We were able to identify distinct migrating patterns (Fig. 3 E): some microglia migrated from anterior to posterior (or vice versa) in a “linear” fashion, while others performed a “circular” movement, returning to approximately their original position within hours. We also observed combinations of these two patterns (“undulating”) and microglia that remained at the same position during most of the acquisition (“static”). The observation that some microglia returned to their previous position or remained static over longer periods prompted the question whether myelin sheaths in some oligodendrocyte territories are more intensely screened than others. We therefore analyzed microglial presence in quadrants of the dorsal and ventral spinal cord. Strikingly, we saw that microglial presence strongly deviated from a random distribution, with some quadrants being highly covered and others not at all during the entire acquisition (Fig. 3 F, Fig. S2 C, and Video 4). Along the same line, high amounts of static phases were not compensated by a higher number of microglia (Fig. S2 B). Thus, microglia travel over considerably large distances in the spinal cord while screening myelin sheaths and they show preference for some myelin sheaths over others.
Figure 3.
Microglia screen myelin sheaths and phagocytose myelin in the zebrafish spinal cord. (A) Maximum projection of a microglia (green) making contact with myelin sheaths (magenta) in a 4 dpf Tg (mpeg1:EGFP; sox10:mRFP) zebrafish spinal cord. (B−F) Microglia scanning behavior along myelin sheaths in the spinal cord was analyzed by confocal time-lapse imaging of 3–4 dpf Tg (mpeg1:EGFP; mbp:mCherry-CAAX) or Tg (mpeg1:EGFP; sox10:mRFP) larvae over 14.5–15 h. Tile scans were acquired every 20 min. Individual microglia were followed through all time frames in which they were contacting myelin sheaths of one hemi-spinal cord (15 microglia from n = 6 larvae). (B) Example of the movement of a single microglia followed over 10 h. Top: Individual time frames are color-coded to highlight microglia movement over time. Bottom: Binary image of the microglia in time frame 1. For the analysis of the microglia motility along the spinal cord, tracks were manually drawn in the Manual Tracking plugin in Fiji by marking soma position in each time frame (overlaid as a yellow line in this image). A: anterior, P: posterior, D: dorsal, V: ventral. (C) Mean velocity of microglia movement along the spinal cord. (D) Mean duration of breaks in motility taken by individual microglia. (E) Tracking profiles (as shown in B) were plotted as the Euclidean distance of the microglia soma in each frame from the position of the soma in the first frame. The shape of the resulting curves was used to classify microglia movements into patterns. Colors identify individual microglia. Asterisk marks sheath displayed in B. (F) Microglia presence per time frame was added up in quadrants of the spinal cord (1 to 14, anterior to posterior) and displayed as heatmap to visualize preferential screening behavior. D: dorsal, V: ventral. (G) Confocal time-lapse imaging of a microglia taking up a myelin fragment over 90 min. Tilescans were acquired every 3 min. Images show maximum projections. For better visualization, only a part of the hemi-spinal cord was projected. Details show the myelin fragment inside the microglial process at distinct time points (top row: merged images, bottom row: 552 nm channel). (H and I) Myelin fragments accumulate inside microglia. (H) Example image from a 10 dpf Tg (mpeg1:EGFP; sox10:mRFP) spinal cord. To quantify myelin fragments within microglia, microglia were manually segmented in Imaris and the resulting surfaces were used to mask the 552 nm channel. (I) Quantification shows the sum of myelin fragment volumes within microglia in Tg (mpeg1:EGFP; mbp:mCherry-CAAX) spinal cords (n = 5 larvae per time point). One-way ANOVA with post-hoc Tukey’s test: 3 vs. 7 dpf: P = 0.0345, 3 vs. 14 dpf: P = 0.0046, 5 vs. 14 dpf: P = 0.0089, all other comparisons were non-significant. (J) Colocalization of mbp:mCherry-CAAX+ fragments with microglial lysosomes, labeled by transiently expressed mpeg1:KalTA4;UAS:EGFP-Rab7. (K) Microglia engulfing an enlarged, seemingly unwrapping myelin sheath (top row) and surrounding round mbp-mCherry-CAAX–positive structures budding off from the ventral Mauthner axon (bottom row). (L) Examples of myelin abnormalities (marked by arrows) observed in wt and csf1rDM Tg (mbp:EGFP-CAAX) larvae (top row: myelinosomes budding off from the Mauthner axon, bottom row left: sheath degeneration, bottom row middle + left: “myelin flaps,” which may represent locally unwrapped myelin). Quantification of the number of myelin abnormalities in 10 dpf dorsal and ventral spinal cord of wt (black) and csf1rDM (blue) larvae, normalized by the myelinated area (wt: n = 10 larvae, csf1rDM: n = 11 larvae). Two-sided Student’s t test: P < 0.0001. (M) Maximum projections of wt and csf1rDM Tg (mbp:EGFP-CAAX) ventral spinal cords showing myelin outfoldings budding off the Mauthner axon (arrows). Quantification of the number of myelin outfoldings in 10 dpf ventral spinal cord of wt and csf1rDM larvae, normalized by the myelinated area (wt: n = 10 larvae, csf1rDM: n = 11 larvae). Two-sided Student’s t test: P < 0.0001. (N) Examples of myelin ultrastructure in SEM cross-sections of the wt and csf1rDM zebrafish spinal cord at P18. Arrows indicate an outfolding (purple), a myelin fragment (green), and a minor outfolding (blue). (O) Quantification shows the percentage of myelin aberrations, categorized by subtypes, in single SEM cross-sections of the wt and csf1rDM zebrafish spinal cord at P18 (n = 4 larvae). One-way ANOVA with post-hoc Tukey’s test: fragments, wt vs. csf1rDM: P = 0.0068, minor outfoldings, wt vs. csf1rDM: P = 0.0001, all other comparisons were non-significant. Data represent means ± SD. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Scale bars: 5 µm (G, J–M), 10 µm (A, H), 50 µm (B). See also Video 4.
Figure S2.
Screening behavior of microglia along myelin sheaths. (A) Microglia motility along myelin sheaths was monitored in 3-min acquisitions over 1 h by confocal time-lapse imaging of a Tg (mpeg1:EGFP; sox10:mRFP) zebrafish spinal cord. (B) XY plot correlates the mean number of microglia present along the spinal cord with the mean percentage of frames in which microglia remained static. Data points represent individual larvae. Linear regression analysis: R2: 0.1669, P = 0.4213. Dotted lines represent the standard error of the linear regression. (C) Heatmaps of microglial presence in quadrants of the dorsal and ventral spinal cord from five other fish (see Fig. 3 F for Fish 1). (D) Confocal time-lapse imaging of motile mbp:mCherry-CAAX fragments within a microglia in a 7 dpf Tg (mpeg1:EGFP; mbp:mCherry-CAAX) spinal cord. Z-stacks were acquired every 3 min. Images show maximum projections. Scale bars: 5 µm (D), 20 µm (A). See also Video 4.
Video 4.
Microglia screen myelin along the spinal cord and phagocytose myelin debris. Confocal time-lapse imaging of zebrafish spinal cords shows microglial processes extending and retracting along myelin sheaths (in 3-min intervals), few microglia migrating along the spinal cord to scan myelin in non-random patterns (in 20-min intervals), microglial uptake of a myelin fragment (indicated by arrow, acquired in 3-min intervals), and trafficking of myelin fragments within microglia (in 3-min intervals). Pseudo-coloring shows microglia in green and myelin in magenta. Frame rate is 2.5/s. Related to Figs. 3 and S2.
We next asked whether microglia phagocytose myelin during their surveillance. Because the sox10:mRFP reporter transiently labels sensory neurons in addition to oligodendrocytes, we analyzed myelin phagocytosis in Tg (mpeg1:EGFP; mbp:mCherry-CAAX) animals. With time-lapse imaging, we observed the intake of a dense mbp:mCherry-CAAX+ myelin piece by a microglial process toward the microglial soma (Fig. 3 G and Video 4). While no microglia contained myelin fragments at 3 dpf, all of them did by 7 dpf (n = 5 fish). Microglia showed increasing volumes of internalized mbp:mCherry-CAAX+ fragments between 5 and 7, or 14 d (Fig. 3, H and I). Microglia did not change their morphology toward an activated phenotype during myelin phagocytosis. Myelin fragments were highly motile inside microglia (Fig. S2 D and Video 4) and co-localized with microglial lysosomes, as shown by the transient expression of the macrophage-specific reporter mpeg1:KalTA4; UAS-EGFP-Rab7 (Fig. 3 J). Thus, microglia in zebrafish phagocytose myelin under physiological conditions and accumulate it in lysosomes over time. These findings are consistent with a recent study by Hughes et al., which showed that myelin phagocytosis by microglia is dependent on neuronal activity (Hughes and Appel, 2020).
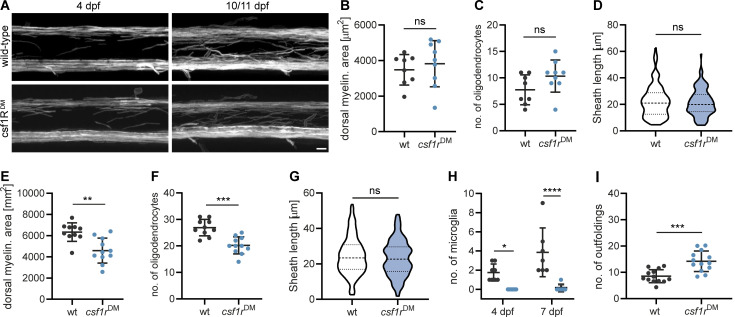
We occasionally found microglia engulfing abnormal myelin sheaths that had either lost their uniform 3D structure or showed signs of fragmentation or local outfoldings (Fig. 3 K). However, in our time-lapse imaging experiments, we did not observe microglia that actively strip off such myelin fragments. Because these events might be too rare to be captured by time-lapse imaging, we asked whether structural myelin abnormalities increase in the absence of microglia. For this, we analyzed a double mutant of the csf1 receptor, a duplicated gene in zebrafish, which is essential for microglia proliferation and development. These mutants maintain only around 5% of microglia and macrophages in the CNS compared to wild-type (wt) animals (Oosterhof et al., 2018). Our characterization of the myelin in csf1rDM; Tg (mbp:EGFP-CAAX) larvae revealed a normal extent of myelination (Fig. S3, A and B), normal oligodendrocyte numbers (Fig. S3 C), and normal sheath length at 4 dpf (Fig. S3 D). However, at 10 dpf, csf1rDM mutants showed a mild spinal cord hypomyelination (Fig. S3, A and E) and reduced oligodendrocyte numbers (Fig. S3 F). Myelin sheath length remained unaffected (Fig. S3 G). The number of microglia associated with myelin in the spinal cord was reduced as expected (wt: 3.9 ± 2.5%, csf1rDM: 0.14 ± 0.38% at 10 dpf, P < 0.0001, n = 7 fish; Fig. S3 H). Interestingly, csf1rDM double mutant fish showed significantly more myelin abnormalities in the 10 dpf ventral spinal cord compared to wt animals (wt: 5.5 ± 1.72, csf1rDM: 11.27 ± 2.76, P < 0.0001, n = 10 [wt] and n = 11 [csf1rDM] fish; Fig. 3 L). These aberrant structures included fully degenerating sheaths and myelin segments that partially lost their 3D structure. The increased myelin abnormalities in csf1rDM mutants were primarily due to the presence of bulb-like, focal myelin outfoldings at the Mauthner axon (Fig. 3 M) and persisted as far as we could follow them up by live imaging, at 18 dpf (Fig. S3 I). We further assessed myelin ultrastructure in single cross-sections of 18 dpf wt and csf1rDM zebrafish spinal cord. Here, we found more than twice as many myelin aberrations in csf1rDM mutants as in wt larvae (wt: 4.1 ± 2.3%, csf1rDM: 10.9 ± 2.1%, P = 0.0048, n = 4 fish; Fig. 3, N and O). Interestingly, we could identify all profiles of myelin aberrations that we had previously described for mouse optic nerves. The increased number of myelin aberrations in csf1rDM mutants mainly represented small dystrophies of the myelin sheath or fragments pinching off (designated as “fragments”; Fig. 3 O). Thus, ultrastructural myelin aberrations can be found in developing zebrafish larvae as well and microglia contribute to their resolution.
Figure S3.
Characterization of csf1rDM mutant. (A) Representative images of myelin in wt and csf1rDM Tg (mbp:EGFP-CAAX) spinal cords. (B−D) Quantifications of myelinated area (B), number of oligodendrocytes (C), and sheath lengths (60 individual myelin sheaths from n = 3 fish; D) in the dorsal spinal cord of 4 dpf wt and csf1rDM larvae (wt: n = 8, csf1rDM: n = 9 larvae in B and C). Two-sided Student’s t test: P = 0.5359 (B), P = 0.0905 (C), Mann–Whitney U-test: P = 0.8445 (D). (E−G) Quantifications of myelinated area (E), number of oligodendrocytes (F), and sheath lengths (60 individual myelin sheaths from n = 3 fish; G) in the dorsal spinal cord of 10/11 dpf wt and csf1rDM larvae (wt: n = 10, csf1rDM: n = 11 larvae in E and F). Two-sided Student’s t test: P = 0.0012 (E), P = 0.0001 (F), Mann–Whitney U-test: P = 0.3419 (G). (H) Number of mpeg1:EGFP-positive microglia per spinal cord in 4 vs. 7 dpf wt and csf1rDM larvae (4 dpf: n = 8, 7 dpf: n = 7 larvae). Two-way ANOVA with post-hoc Sidak’s test: 4 dpf, wt vs. csf1rDM: P = 0.0266, 7 dpf, wt vs. csf1rDM: P < 0.0001. (I) Quantification of the number of myelin abnormalities in 18 dpf ventral spinal cord of wt and csf1rDM larvae, normalized by the myelinated area (n = 13 larvae). Two-sided Student’s t test: P = 0.0002. Data represent means ± SD. ns, not significant, *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Scale bar: 10 µm (A).
Myelin degeneration profiles are phagocytosed by microglia using phosphatidylserine as an “eat me” signal
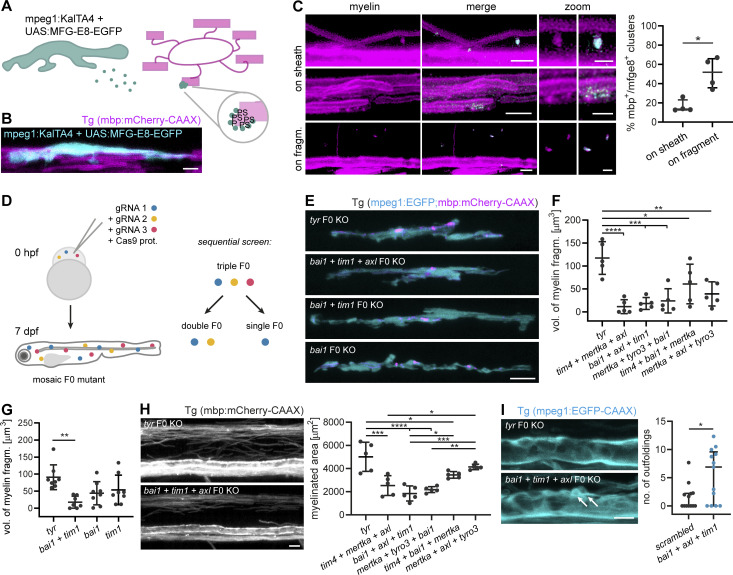
Next, we asked which molecular cues on myelin sheaths are recognized by microglia and promote myelin phagocytosis. Myelin is rich in the phospholipid PS. When PS is exposed on the outer membrane leaflet, it may serve as an “eat me” signal for microglia, which express multiple PS receptors on their surface (Prinz et al., 2019). To determine whether PS during development is exposed on degenerating myelin sheaths, we used a recently described PS reporter that can be applied in vivo. We took advantage of secreted glycoprotein MFG-E8, which recognizes PS and, when fused to EGFP, can be delivered as a recombinant protein for PS detection (Kranich et al., 2020). In order to specifically assess PS recognition, we used the C1 C2 domain of MFG-E8 that contains the PS-binding site but lacks an integrin receptor-binding site. Because injections of recombinant protein into the spinal cord may induce damage, we modified this approach by expressing the genetically encoded secreted MFG-E8-EGFP in microglia (Fig. 4, A and B). When we expressed mpeg1:KalTA4;UAS:MFG-E8-EGFP in transgenic mbp:mCherry-CAAX zebrafish larvae and masked MFG-E8-EGFP fluorescence by the mbp:mCherry-CAAX channel, we found that MFG-E8-EGFP occasionally localized to myelin sheaths (16.34 ± 6.58%) and to small, extracellular fragments positive for the myelin reporter (51.06 ± 16.15%; Fig. 4 C). MFG-E8-EGFP–positive spots on myelin sheaths were focal and their inspection in 3D revealed that they marked small protrusions of the myelin sheath.
Figure 4.
PS recognition contributes to myelin phagocytosis. (A) Scheme depicting the in vivo detection of PS, as performed in B and C. Microglia secrete MFG-E8-EGFP, which binds to PS exposed on the extracellular leaflet of the membrane. In compacted myelin membranes, extracellular exposure of PS is sterically hindered, but may occur due to structural abnormalities of the myelin sheath. (B) A microglial cell expressing MFG-E8-EGFP contacts mbp:mCherry-CAAX–positive myelin sheaths. (C) Images show binding of MFG-E8-EGFP (cyan) to myelin sheaths or fragments (magenta) in Tg (mbp:mCherry-CAAX) larvae. In the top row “on sheath” images, MFG-E8-EGFP labeled several protuberances on two myelin sheaths; the relative amounts of labeled structures in these images are not representative of the quantification. Images are masked by the 552 nm channel. Zoomed images show details of myelin (left) and merged (right) images. Quantifications show the percentage of MFG-E8-EGFP–labeled structures that exhibited mbp:mCherry-CAAX co-staining, categorized by their localization on myelin sheaths or mbp:mCherry-CAAX–positive fragments (n = 4 larvae). Data represent median with IQR. Mann–Whitney U-test: P = 0.0286. (D) Schematic of the CRISPR/Cas9 F0 mutant screen for PS receptors. Fertilized Tg (mpeg1:EGFP; mbp:mCherry-CAAX) zebrafish eggs were injected with Cas9-gRNA RNP complexes at one-cell stage. F0 larvae were assessed at 7 dpf for myelin phagocytosis by microglia. We performed a sequential screen, starting with triple mutants and further dissecting one of them into double and single mutants. (E) Representative images of single, double, and triple mutants of bai1, tim1, and axl. Tyr F0 mutant was used as a control. (F) Quantification shows the total volume of myelin fragments within all microglia in different triple mutants (n = 5 larvae). One-way ANOVA with post-hoc Tukey’s test: tyr vs. tim4+bai1+mertka: P = 0.0462, tyr vs. mertka + axl + tyro3: P = 0.0028, tyr vs. mertka + tyro3+bai1: P = 0.0004, tyr vs. bai1+axl + tim1: P = 0.0002, tyr vs. tim4+mertka + axl: P < 0.0001, all other comparisons were non-significant. (G) Quantification shows the total volume of myelin fragments within all microglia in double and single mutants of bai1 and tim1 (n = 7 larvae for bai1+tim1, n = 8 larvae for tyr, bai1, and tim1). One-way ANOVA with post-hoc Tukey’s test: tyr vs. bai1+tim1: P = 0.0019, all other comparisons were non-significant. (H) Example images and quantification show the myelinated area of the dorsal spinal cord of 7 dpf triple mutants (n = 5 larvae). One-way ANOVA with post-hoc Tukey’s test: tyr vs. tim4+bai1+mertka: P = 0.0187, tim4+mertka + axl vs. mertka + axl + tyro3: P = 0.0145, bai1+axl + tim1 vs. tim4+bai1+mertka: P = 0.0151, mertka + tyro3+bai1 vs. mertka + axl + tyro3: P = 0.0022, tyr vs. tim4+mertka + axl: P = 0.0001, bai1+axl + tim1 vs. mertka + axl + tyro3: P = 0.0003, tyr vs. bai1+axl + tim1 and tyr vs. mertka + tyro3+bai1: P < 0.0001, all other comparisons were non-significant. (I) Maximum projections of wt and bai1;axl;tim1 mutant Tg (mbp:EGFP-CAAX) ventral spinal cords showing myelin outfoldings budding off the Mauthner axon (arrows). Quantification of the number of myelin abnormalities in 10 dpf ventral spinal cord of wt and bai1;axl;tim1 larvae, normalized by the myelinated area (n = 13 larvae). Mann-Whitney U-test: P = 0.0362. Data represent median and IQR (C and I) and means ± SD (F–H). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Scale bars: 10 µm (E and H), 5 µm (B and C, merge, I), 2 µm (C, zoom).
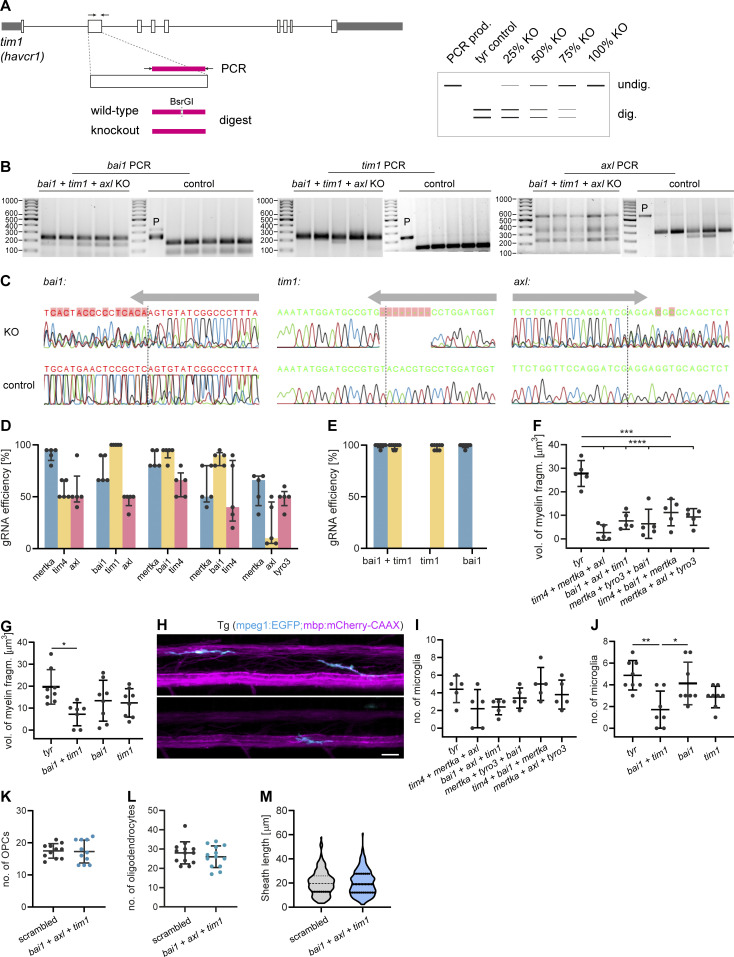
To determine whether PS exposure is critical for developmental myelin phagocytosis by microglia, we performed a targeted candidate CRISPR/Cas9 screen against receptors that use PS as direct or indirect ligand (through bridging molecules) in zebrafish. Targeted mutagenesis by CRISPR/Cas9 induces biallelic mutations, which allows for phenotypic analysis directly in the filial generation zero (F0) generation (Jao et al., 2013; Shah et al., 2015; Hoshijima et al., 2019; Kroll et al., 2021; Keatinge et al., 2021). We generated global knockouts for the different PS receptors and analyzed myelin phagocytosis phenotypes by confocal imaging in F0 larvae (Fig. 4 D). To maximize phenotype penetrance in these mosaic animals (Naert et al., 2020), we selected gRNAs based on their predicted on-target efficiency and frameshift frequency, using the CHOPCHOP and InDelphi prediction tools (Labun et al., 2016; Shen et al., 2018). A gRNA against the tyrosinase (tyr) gene, which causes hypopigmentation, was used as a negative control. Mutants were identified by genotyping after confocal imaging (Fig. S4, A and B) and the ability of gRNAs to induce on-target frameshift mutations was confirmed by Sanger sequencing (Fig. S4 C). Because microglia express several PS receptors with redundant functions (Lemke, 2019), we started by investigating triple F0 mutants of the PS receptor genes mertka, axl, tyro3, tim1, tim4, and bai1 in different combinations. All five triple mutants for different PS receptor combinations that we analyzed resulted in a reduction of myelin fragments in microglia at 7 dpf. This was quantified by the total volume of phagocytosed myelin, summed up over all microglia in a spinal cord (Fig. 4, F and G), or determined as the average volume of phagocytosed myelin per microglia (Fig. S4, F and G). We observed the most marked reduction of the total volume of internalized myelin in microglia of tim4;mertka;axl (90.4%, P < 0.0001) and bai1;axl;tim1 (84.5%, P < 0.0001) mutant fish (n = 5 fish; Fig. 4 F). We further explored bai1;axl;tim1 mutant fish because of the superior gRNA efficiencies (Fig. S4 D). We compared double bai1;tim1 and single bai1 and tim1 mutant fish, and found, as expected, that gRNA efficiencies improved compared to the triple mutants, ranging above 90% in all mutants (Fig. S4 E). Notably, myelin phagocytosis was diminished by 80.6% in the bai1;tim1 double mutants (n = 7 fish, P = 0.0019), but did not significantly change compared to tyr controls in the bai1 (P = 0.0528) and tim1 (P = 0.0163) single mutant fish (n = 8 fish; Fig. 4 G). We also analyzed the number of microglia in the spinal cord, and only found a reduction in bai1;tim1 but not in the other mutants (tyr vs. bai1;tim1: P = 0.0026; Fig. S4, H−J). Similar to the csf1rDM mutants, oligodendrocyte differentiation and myelin sheath growth were unaffected by the ablation of PS receptors (Fig. S4, K−M).
Figure S4.
Details of the CRISPR/Cas9 F0 mutant screen for PS receptors in zebrafish. (A) Workflow of F0 mutant genotyping. Target regions were amplified from genomic DNA. Gene knockout in F0 mutants differed from controls by loss of a unique restriction site. gRNA efficiency was estimated from the ratio of undigested to digested bands on the agarose gel. (B) Agarose gels show genotyping results of bai1, tim1, and axl in the bai1+tim1+axl mutants and tyr controls. P, PCR product. (C) Sanger sequencing of bai1, tim1, and axl target sites in mutant and control animals, showing frameshift mutations in the mutants (KO). Arrows indicate gRNA position. Dashed lines mark Cas9 cleavage sites. (D) gRNA efficiencies in the triple F0 mutant screen (n = 5 larvae). (E) gRNA efficiencies in the double and single F0 mutant screen (n = 8 larvae). (F and G) Average volume of myelin fragments within individual microglia in triple F0 mutants (F), and double and single mutants of bai1 and tim1 (G). n = 5 larvae in F, n = 7 larvae in bai1+tim1, n = 8 larvae for tyr, bai1, and tim1 in G. One-way ANOVA with post-hoc Tukey’s test: tyr vs. tim4+bai1+mertka: P = 0.0002, tyr vs. tim4+mertka + axl, tyr vs. bai1+axl + tim1, tyr vs. mertka + tyro3+bai1 and tyr vs. mertka + axl + tyro3: P < 0.0001, tyr vs. bai1+tim1: P = 0.0160, all other comparisons in F and G were non-significant. (H−J) Microglia in the spinal cord of control and F0 mutant zebrafish larvae in the PS receptor KO screen. Example images (H) and quantification of the number of microglia in triple mutants (I) and double and single mutants of bai1 and tim1 (J). n = 5 larvae in I, n = 7 larvae for bai1+tim1, n = 8 larvae for tyr, bai1, and tim1 in J. One-way ANOVA with post-hoc Tukey’s test: tyr vs. bai1+tim1: P = 0.0026, bai1+tim1 vs. tim1: P = 0.0262, all other comparisons in I and J were non-significant. (K−M) Quantifications of number of OPCs in Tg (olig1:memEYFP) (K; n = 11 larvae), number of oligodendrocytes (L; n = 13 larvae), and sheath lengths in Tg (mbp:EGFP-CAAX) (60–65 individual myelin sheaths from n = 3 fish; M) in the dorsal spinal cord of 10 dpf control larvae injected with a three scrambled gRNAs and bai1;axl;tim1 F0 larvae. Two-sided Student’s t test: P = 0.8874 (K), P = 0.3769 (L), P = 0.8051 (M). Data represent means ± SD. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Scale bar: 20 µm (H). Source data are available for this figure: SourceData FS4.
Interestingly, triple PS receptor mutants were found to be hypomyelinated at 7 dpf, which was most pronounced in tim4;mertka;axl (49.6%, P = 0.0001), bai1;axl;tim1 (63.2%, P < 0.0001), and mertka;tyro3;bai1 (56.5%, P < 0.0001) mutants (n = 5 fish; Fig. 4 H). Despite the hypomyelination, bai1;axl;tim1 triple mutants exhibited significantly more outfoldings at 10 dpf as the control larvae that were injected with a combination of three different scrambled gRNAs (control: 0.0 [0.0–2.2, median and IQR], bai1;axl;tim: 6.9 [0.0–9.6, median and IQR], P = 0.0362, n = 13; Fig. 4 I).
Oligodendrocytes retract myelin sheaths independently of microglia and degrade myelin membrane
Next, we asked whether microglia play a role in myelin sheath retractions. Such structural changes of existing myelin have also been implicated in mediating nervous system plasticity (Czopka et al., 2013; Hines et al., 2015; Liu et al., 2013; Auer et al., 2018; Yang et al., 2020; Orthmann-Murphy et al., 2020). We therefore analyzed retractions in 3–4 dpf Tg (mpeg1:EGFP; mbp:mCherry-CAAX) dorsal spinal cords in time-lapses of 30-min intervals over 10–14 h. The fine microglia processes that extended and retracted along myelin sheaths did not seem to be directly involved in any of the myelin retractions (Fig. 5 A and Video 5). Microglial presence and the number of retractions along the spinal cord were entirely disconnected (Fig. 5 B), which we confirmed by a correlation analysis in 25 distinct territories of 1–2 oligodendrocytes, summed up over the entire acquisition (R2 = 0.0256, P = 0.4453, n = 5 fish; Fig. 5 C). Furthermore, retractions did not significantly differ between microglia-depleted csf1rDM and wt larvae (wt: 42.25 ± 7.50, csf1rDM: 57.75 ± 15.90, P = 0.1429; Fig. 5, D and E; and Video 5). Inspecting retractions more closely revealed that myelin sheaths were entirely withdrawn into oligodendrocyte cell bodies and no visible fragments splitting off to the exterior (Fig. 5 F and Video 5). Among 17 retracting myelin sheaths in 7 larvae, none exhibited MFG-E8 labeling (Fig. 5 G). However, bright fragments appeared and shuttled within the oligodendrocyte soma in the course of retractions (Fig. 5 F and Video 5). We stained Tg (mbp:mCherry-CAAX) larvae with LysoTracker and analyzed the colocalization of these bright membrane fragments with lysosomes. We found that major portions of the fragments, but not the rest of the oligodendrocyte membrane, colocalized with lysosomes (Fig. 5 H). Intriguingly, we also found LysoTracker staining of aberrant structures on myelin sheaths, but not of normal-appearing adjacent sheaths or total myelin in the same image (Fig. 5, I and J), raising the possibility that some of the aberrations are resolved by the oligodendrocyte itself. We next assessed whether myelin membrane fragments occur inside of the oligodendrocyte soma beyond the zebrafish model, and co-immunostained mouse tissue sections of the optic nerve and the brain for MBP, the oligodendrocyte marker OLIG2, and LAMP1. In the corpus callosum, we found lysosome-associated MBP+ fragments in 0.53% (0.44–1.00%, median and IQR) of the OLIG2+ oligodendrocytes at P14, but only 0.20% (0.09–0.31%, median and IQR) at P80 (P = 0.0286; Fig. 5 K). Similarly, 1.92% (1.08–2.29%, median and IQR) of the oligodendrocytes in the optic nerve contained MBP+ fragments at P14, but only 0.30% (0.00–0.40%, median and IQR) did at P80 (P = 0.0571; Fig. 5 K). Because MBP is a cytosolic protein interacting with the inner leaflet of the myelin membrane bilayer, MBP+ fragments within lysosomes are likely a result of myelin degradation.
Figure 5.
Oligodendrocytes retract myelin sheaths independent of microglia and degrade membrane fragments. (A−C) Time-lapse confocal imaging of 3 dpf wt Tg (mpeg1:EGFP; mbp:mCherry-CAAX) zebrafish spinal cord is used to analyze retractions of dorsal myelin sheaths with respect to microglial presence. Images were acquired every 30 min during 14 h. (A) Representative images. Arrows identify individual retractions by color and the position of the arrowheads. A microglia contacting myelin sheaths was only observed during a single frame (at 0.5 h). (B) Heatmap shows microglial presence and the number of retractions in 5 × 5 mm quadrants along the dorsal spinal cord (1 to 12, anterior to posterior). (C) XY plot correlates the number of time frames, in which a microglia was present, with the number of myelin sheath retractions in 26 oligodendrocyte territories (n = 5 larvae). Linear regression analysis: R2: 0.02556, P = 0.4453. Dotted lines represent the standard error of the linear regression. (D and E) Time-lapse confocal imaging of 3 dpf wt and csf1rDM;Tg (mbp:EGFP-CAAX) larvae. Images (D) were acquired every 30 min over 8 h. Arrowheads identify individual retractions by different colors. Quantification in E shows the number of retractions during 8 h. Mann-Whitney U-test: P = 0.1429. (F) Time-lapse confocal imaging from A shows how retractions result in bright fragments inside the oligodendrocyte cell body. Arrowheads mark a retracting sheath and its fragments. Dashed circle indicates the oligodendrocyte soma. (G) Example of a retracting sheath (arrow) exhibiting normal ultrastructure and lacking MFG-E8-EGFP labeling in a 4 dpf Tg (mbp:mCherry-CAAX) spinal cord with transient expression of olig1:KalTA; UAS:MFG-E8-EGFP. (H) Colocalization analysis of mbp:mCherry-CAAX–positive fragments inside oligodendrocyte cell bodies with LysoTracker Green. Oligodendrocyte cell bodies were manually segmented from the 552 nm channel. Channels were masked by the whole cell body (cell ROI) or by the bright mbp:mCherry-CAAX fragments inside (fragment ROI), followed by colocalization analysis. Thresholded Mander’s B coefficient reflects the amount of colocalization within the ROI of the thresholded 552 nm channel (43 cell bodies from n = 6 larvae). Student’s t test: P < 0.0001. (I) Time-lapse imaging of a single myelin sheath stained in vivo with LysoTracker Red (maximum projections). Z-stacks were acquired every 3 s. (J) Quantification of LysoTracker colocalization with structural myelin abnormalities compared to a normal-appearing adjacent myelin sheath and total myelin in the same image (n = 5 larvae). One-way ANOVA with post-hoc Tukey’s test: abnormal vs. adjacent, abnormal vs. total: P < 0.0001, adjacent vs. total: P = 0.9074. (K) Free-floating sections of P14 wt mouse brains co-immunostained for MBP and LAMP1 together with OLIG2 for oligodendrocytes. Top row: corpus callosum, bottom row: optic nerve. 3D reconstruction shows MBP-positive staining in close association with lysosomes (marked by arrow) and the oligodendrocyte nucleus. Quantifications show the percentage of oligodendrocytes with internalized MBP in P14 vs. P80 brains (n = 4 mice). Mann–Whitney U-test: P = 0.0286 (corpus callosum), P = 0.0571 (optic nerve). Data represent means ± SD (H and J) and median and IQR (E and K). n.s., not significant, *, P < 0.05, ****, P < 0.0001. Scale bars: 30 µm (K, corpus callosum overview), 10 µm (A, D, K, optic nerve overview and corpus callosum merge), 5 µm (F, G, H, J, K, optic nerve merge), 3 µm (H, clipped 3Ds), 2 µm (I). See also Video 5.
Video 5.
Myelin retractions occur independently from microglia and are broken down into fragments in the oligodendrocyte cell body.
Confocal time-lapse imaging of zebrafish spinal cords shows that myelin retractions occur independently from microglia (in 3-min intervals), microglia ablation does not affect the number of myelin retractions (in 30-min intervals), and retracted myelin fragments are transported to the oligodendrocyte cell body (indicated by arrow, acquired in 3-min intervals). Pseudo-coloring shows microglia in green and myelin in magenta. Frame rate is 2.5/s. Related to Fig. 5.
Discussion
Myelin biogenesis is a sequential process that involves membrane wrapping around and lateral growth along the axon, resulting in the formation of myelin sheaths that are placed in regularly spaced segments on axons in the nervous system. Here, we provide evidence that this process is error prone and subject to substantial refinements. By performing volume electron microscopy in mouse optic nerves and in vivo confocal light microscopy in zebrafish spinal cord, we observed ultrastructural myelin abnormalities at early stages of development (found in up to 25% of the myelin sheaths at P10) that almost disappeared by young adulthood. Our analyses revealed shedding of abnormal myelin profiles and clearance by microglia. We found that exposed PS on myelin presents a phagocytic signal for microglia, which express several PS-mediated receptors with redundant functions in myelin uptake. In addition, we observed that oligodendrocytes remove entire myelin sheaths by retractions into their cell body and lysosomal degradation therein, in a process that does not involve microglia.
One possible explanation for the high rate of myelin remodeling could be the adaptation of myelination to neural circuit function. Previous studies have shown that myelin remodeling can occur during learning and neuronal circuit refinement (Gibson et al., 2014; Etxeberria et al., 2016; McKenzie et al., 2014; Xiao et al., 2016; Scholz et al., 2009; Hill et al., 2018; Steele et al., 2013; Makinodan et al., 2012; Hughes et al., 2018; Lakhani et al., 2016; Monje, 2018). These adaptations are associated with modifications of myelin sheath length and thickness, the addition and the retraction of myelin segments along an axon (Etxeberria et al., 2016; Ford et al., 2015; Sinclair et al., 2017; Auer et al., 2018; Kaller et al., 2017). Such mechanisms may operate in areas of the brain that are incompletely myelinated, such as the cortex, where the addition and/or removal of myelin segments could provide a mechanism to tune network function. In brain regions that are continuously myelinated, e.g., optic nerve or spinal cord, myelin refinement is more likely to be required for the synchronization of conduction velocity across all axons within the same fiber bundle. However, there are also alternative explanations. The relatively large fraction of myelin abnormalities at very early stages of optic nerve and spinal cord development, when large parts of the axons are still unmyelinated, raises the question whether such ultrastructural abnormalities are intrinsic to the process of myelin biogenesis. Features such as myelin outfoldings may represent a membrane reservoir to allow the completion of the lateral growth and secure unperturbed radial growth of the myelinated axon itself (Snaidero et al., 2014). It is also possible that myelination is an error-prone process that occasionally requires several attempts. Such adjustments could result in myelin retractions when myelin formation is still at an early stage of its development. Once myelin formation has advanced, and the multilayered membrane structure is compacted and anchored at the paranodes, it is not easy to envision how coordinated retractions could occur. At this stage, abnormal myelin structure may be subject to degeneration and clearance by microglia. The large whorls of degenerated myelin are a likely example of such a destructive process. Using SBF-SEM, we found microglia that associate with and internalize degenerated myelin whorls while the myelin sheath is still on the axon. It is tempting to speculate that microglia actively strip myelin off the axons. Evidence for this is provided by a recent study of Hughes et al. in which microglia were found to phagocytose entire myelin sheaths (Hughes and Appel, 2020). Our time-lapse imaging experiments indicate that such events could be rare, as microglia actively stripping off entire myelin sheaths from axons were not easy to find. Occasionally, we observed condensed myelin debris in distal processes of the microglia that suggest it might previously have been a myelin sheath, but were unable to visualize the uptake in our time-lapse imaging experiments.
Our data indicate that some of the altered myelin is pinched off into the extracellular space where it is phagocytosed by microglia. Indeed, our SBF-SEM reconstructions show a large number of small protuberances on normal-appearing sheaths as well as detached fragments that lie in the direct vicinity of a myelin sheath. The extracellular fragments, which are abundantly found and labeled by both the PS and oligodendrocytes/myelin reporter in zebrafish, could represent degenerated and detached myelin. These fragments may resemble “shedosomes” that occur during dendritic pruning (Han et al., 2011) or “axosomes” which form during axon branch removal (Bishop et al., 2004). Shedding of myelin fragments could explain why only relatively few myelin sheath abnormalities were observed after microglia ablation. The uptake of aberrant myelin into microglia is reminiscent of trogocytosis, a process described as one cell physically taking and ingesting little pieces (“bites”) from another cell (Dance, 2019). While the exposure of PS on myelin appears to represent one “eat me” signal for microglia, it will be interesting to know whether “find me” signals are released from degenerating myelin sheaths. “Find me” signals establish chemotactic gradients that attract phagocytes. Potential candidates known from apoptosis research include fractalkine, the nucleotides ATP and UTP, lysophosphatidylcholine, or sphingosine-1-phosphate (Hochreiter-Hufford and Ravichandran, 2013). The migration pattern of microglia, showing a non-random scanning behavior along myelinated axons, could point to such long-range acting signals.
Microglia do not appear to be critical to initiate myelination, as the extent of myelination in microglia-ablated csf1rDM fish appears unaltered at an early developmental time point (4 dpf). Hughes et al. even reported an increased number of myelin sheaths at 4 dpf, which they attributed to the lack of myelin pruning in the absence of microglia (Hughes and Appel, 2020). However, when examining later time points in development (10 dpf), we found that microglial ablation leads to hypomyelination (and reduced oligodendrocyte numbers). How can these apparent discrepancies be explained? Whereas myelin stripping may represent the dominant form of myelin remodeling early in development, shedding of degenerated myelin fragments into the extracellular space may become more frequent with time. In the absence of microglia, an increasing fraction of myelin debris may start to accumulate and progressively inhibit the differentiation of oligodendrocyte precursor cells (OPCs) and myelination, as observed in models of remyelination (Kotter et al., 2001, 2006; Lampron et al., 2015). In addition, microglia are known to interact with OPCs and secrete growth factors that enhance OPC proliferation or differentiation (Hagemeyer et al., 2017; Wlodarczyk et al., 2017; Giera et al., 2018; Sherafat et al., 2021).
Oligodendrocytes have an intrinsic capacity to modify their own myelin sheaths by retracting entire sheaths into their cell bodies, a process that mainly occurs during early development and is related to neuronal activity (Czopka et al., 2013; Hines et al., 2015; Mensch et al., 2015; Baraban et al., 2018; Hughes et al., 2018; Krasnow et al., 2018). This process occurs independently of microglia and retracting sheaths do not display any obvious structural abnormalities detectable by light microscopy prior to retractions. However, we cannot exclude that retracting myelin sheaths bear ultrastructural aberrations that cannot be resolved by light microscopy.
Taking the results together, myelin sheaths appear to remodel or be removed in multiple ways during development. In addition to the function of microglia in clearing degenerated myelin during development, it remains unclear to what extent astrocytes participate in this process. The use of global knockouts in our studies so far does not allow us to clearly dissect this. In the zebrafish spinal cord, astrocytes are regularly tiled and, early on, largely outnumber microglia (Chen et al., 2020). They could therefore aid microglia in myelin debris phagocytosis, as they do in synapse pruning (Lee and Chung, 2019). Cell-type-specific knockouts will be needed to address this in the future. It will also be important to determine whether structural changes to existing myelin operate in adult and contribute to myelin plasticity in response to environmental cues. In addition, future studies are necessary to understand the molecular details of these myelin remodeling pathways.
Materials and methods
Zebrafish husbandry
We used the existing transgenic zebrafish lines Tg (mpeg1:EGFP), Tg (sox10:mRFP), Tg (mbp:mCherry-CAAX), and Tg (olig1:memEYFP) as well as the mutant line csf1rDM (containing mutant alleles csf1ra and csf1rb), which was previously outcrossed to Tg (mbp:EGFP-CAAX; Ellett et al., 2011; Kucenas et al., 2008; Mensch et al., 2015; Oosterhof et al., 2018; Almeida et al., 2011; Marisca et al., 2020). All zebrafish procedures were carried out with approval and according to the regulations of the District Government of Upper Bavaria. Zebrafish were housed at the fish facility of the German Center for Neurodegenerative Diseases in Munich according to local animal welfare regulations. Embryos were obtained by natural spawning and raised at 28.5°C in E3 medium.
Mouse husbandry
Male and female C57BL/6 mice were group-housed in a 12-h dark/light cycle with ad libitum access to food and water in the animal facility of the Max Planck Institute of Experimental Medicine in Göttingen. All animal procedures were handled with approval and in accordance with the regulations of the state government of Lower Saxony.
Generation of transgenic constructs
To generate pTol2-mpeg1:KalTA4, we first cloned the middle entry vector pME-KalTA4GI by a BP reaction of pTol2-sox10:KalTA4GI (kind gift of Tim Czopka [Almeida and Lyons, 2015]) with pDONR 221, using Gateway BP Clonase II Enzyme mix (Thermo Fisher Scientific). pME-KalTA4GI was then recombined with p5E-mpeg1 and p3E-polyA into pDestTol2CG2 (Kawakami, 2007), using LR Clonase II Plus enzyme (Thermo Fisher Scientific). pTol2-UAS:EGFP-Rab7 was generated by recombining p5E-UAS, pME-EGFP no stop and p3E-Rab7 (kind gift of Brian Link [Clark et al., 2011]) into pDestTol2-CG2 in an LR reaction, as described above. To obtain pTol2-UAS:MFG-E88-C1C2-EGFP, we first cloned pME-MFG-E8-C1C2-EGFP by PCR amplification of the mouse MFG-E8 C1 and C2 domain (insert) and of the middle entry vector backbone (vector), using Q5 polymerase (New England Biolabs Inc). Fragments were generated with the following primers (template plasmids indicated in parentheses): insert fragment (pD2523-mMFG-E8_C1C2-EGFP, kind gift of Jan Kranich [Kranich et al., 2020]): 5′- ATGCAAGTCTCTAGGGTAC-3′ (fwd) and 5′-CTTATAAAGTTCATCCATGCCA-3′ (rev), vector fragment (pME-KalTA4GI): 5′-GCATGGATGAACTTTATAAGTAAACCCAGCTTTCTTG-3′ (fwd), and 5′- AGTACCCTAGAGACTTGCATGGTGGCGGCAGCCT-3′ (rev). This was followed by a 2-fragment Gibson assembly, using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs, Inc.) according to the manufacturer’s protocol. pTol2-UAS:MFG-E8-C1C2-EGFP was then generated by recombining p5E-UAS, pME-MFG-E8-C1C2-EGFP and p3E-polyA into pDestTol2-CG2 in a Gateway LR reaction. All sequences were verified by Sanger sequencing.
gRNA design and assembly of Cas9-gRNA RNP complexes
gRNAs were designed to optimize gRNA efficiency and frameshift frequency, combining predictions from the CHOPCHOP and InDelphi online tools (Labun et al., 2016; Shen et al., 2018). Target-specific 20 nt crispr RNA (Alt-R CRISPR-Cas9 crRNA, IDT, see Table S1 for sequences) were annealed with trans-activating crispr RNA (Alt-R CRISPR-Cas9 tracrRNA, IDT) according to the manufacturer’s instructions and stored at −80°C. These gRNAs were mixed 1:1 with 1.5 mg/ml Cas9 protein (PNA Bio, Cat. CP02) and incubated at room temperature for 5 min to assemble RNP complexes.
Microinjection of zebrafish embryos
Transgenic reporter constructs were transiently expressed in zebrafish by injecting a mixture of plasmid DNA (25 ng/μl each), transposase mRNA (25 ng/μl), and 0.2 M KCl into fertilized eggs at the one-cell stage. In CRISPR/Cas9 experiments, pre-assembled RNP complexes were injected into fertilized eggs at the one-cell stage. Tyrosinase gRNA, targeting a pigmentation gene unrelated to myelination (Jao et al., 2013), or a mixture of three different scrambled gRNAs (Parvez et al., 2021), were used as a control. All embryos designated for imaging were treated with phenylthiourea (PTU) from 8 to 24 h post-fertilization onward to prevent pigmentation.
Genotyping of CRISPR/Cas9 F0 mutant larvae
Genotyping was performed as described in (Hruscha and Schmid, 2015). Briefly, after imaging at 7 dpf, larvae were lysed in 1x Tris-EDTA buffer + 10% proteinase K. We amplified target regions by PCR using the primers specified in Table S1. PCR products were digested with unique restriction enzymes that cut close to the Cas9 cleavage site (see Table S1). CRISPR/Cas9 mutations resulted in a loss of the restriction site. gRNA efficiencies were determined by the ratio of undigested/digested bands on an agarose gel. The dataset for triple F0 mutants was reduced to exclude samples with very poor gRNA efficiency based on the genotyping results.
LysoTracker staining in zebrafish
Zebrafish larvae were incubated for 2 h with 1:100 LysoTracker Green DND-26 or LysoTracker Red DND-99 (Thermo Fisher Scientific) in E3 medium (final concentration of 10 µM). Thereafter, larvae were washed three times with E3 medium, mounted, and immediately imaged.
Immunohistochemistry of mouse tissue
30-μm free-floating brain sections and 14-μm mounted optic nerve sections were rinsed with 1x PBS containing 0.2% Tween-20 and permeabilized in 0.5% Triton X-100 for 30 min. Sections were blocked for 1 h at room temperature in a solution containing 2.5% FCS, 2.5% BSA, and 2.5% fish gelatin in PBS. Primary antibodies, diluted in 10% blocking solution, were added and incubated overnight at 4°C. On the following day, sections were incubated with secondary antibodies, diluted in 10% blocking solution, for 1 h at room temperature. After washing with PBS, the free-floating sections were mounted on superfrost plus slides using fluorescence mounting medium (Dako). Primary antibodies: rabbit IBA1 (Cat. 019-19741, 1:1000; Wako), chicken MBP (Cat. PA1-10008, 1:1000; Thermo Fisher Scientific), rabbit Olig2 (Cat. AB9610, 1:250; Millipore), and rat LAMP1 (1D4B; sc-19992, 1:100; Santa Cruz). Secondary antibodies: Alexa Fluor 488–, 555–, and 647–conjugated antibodies (Thermo Fisher Scientific, Abcam, Jackson ImmunoResearch Labs, 1:1,000).
Confocal image acquisition
Zebrafish larvae at 3 to 18 dpf were anesthetized with tricaine and mounted laterally in 1% low melting point agarose (Thermo Fisher Scientific) on a glass bottom dish (#1.5 cover glass, IBL). Fish were imaged at a Leica TCS SP8 confocal laser scanning microscope with automated moving stage and climate chamber (28.5°C), using a resonant scanner, 1.1 NA 40 × water immersion objective with distilled water or Immersol W 2010 immersion medium (Zeiss) and 488 (for EGFP), 514 (for EYFP), and 552 nm (for mCherry and mRFP) lasers. Images were acquired using the Leica Application Suite X 3.1.5.16308 software. Single images or tilescans (1248 × 1248 pixels, pixel size 40–78 nm, z step size 250–320 nm) were acquired using a hybrid detector in counting mode, line accumulation of 4–8, and the pinhole at 0.8 airy units. Overnight time-lapse imaging (800–1248 × 800–1248 pixels, pixel size 78–120 nm, z step size 0.5–1 μm) was done with a hybrid detector in photon counting mode, line accumulation of 4–8, and a pinhole at 1.0–1.2 airy units. Z-stack tiles (z-step = 1 µm) along the spinal cord, starting at the neck, were taken every 15–30 min. Time-lapses were registered with the Correct 3D drift plugin in Fiji. Images were stitched using the LAS X (v3.5) software. For better representation and to improve colocalization analysis, images showing fine details or colocalizations (Fig. 3, A, G, H, K−M; Fig. 4, A, E−H; Fig. S2 D; and Fig. S4) were deconvolved using the default settings of the Huygens Essential software (Scientific Volume Imaging). All images of zebrafish show lateral views of the spinal cord with anterior to the left and dorsal to the top. Images of mouse immunostainings were acquired through a Leica TCS SP5 confocal microscope and were processed and analyzed with Imaris (64x version 9.2.0). To estimate the number of Iba1 positive cells with internalized myelin particles, confocal stacks (step size: 0.8 μm) were captured in the z-direction from the region of interest (corpus callosum and optic nerve) with 20× objective of the confocal microscope. A 3D view was created from stacks of each image using the surpass option in Imaris.
Tissue preparation for SBF-SEM and image acquisition
Animals were sacrificed by cervical dislocation, and the optic nerves were dissociated from the eyeballs and the chiasm. They were immersion fixed for 24 h in Karlsson Schultz solution (4% PFA, 2.5% glutaraldehyde and 0.5% NaCl in 0.1 M PBS [pH 7.3]; Karlsson and Schultz, 1965) at 4°C. Afterwards, they were processed with a modified reduced osmium-thiocarbohydrazide-osmium (rOTO) protocol (Mikula et al., 2012) to increase the contrast. At first, they were washed three times with 0.1 M phosphate buffer followed by 3 h of 2% OsO4 and 0.25% K4 [Fe(CN)6] incubation at 4°C to reduce OsO4 to OsO2. After a wash step in ddH20, samples were incubated with 0.1% thiocarbohydrazide (in double-distilled water [ddH2O]) for 1 h at room temperature. Contrast was achieved by incubation with 2% OsO4 for 90 min followed by (after washing with ddH2O) 2.5% uranyl acetate incubation over night at 4°C. After washing again in ddH2O, samples were dehydrated using increasing concentrations of acetone (always 15 min at 30, 50, 75, 90, 3 × 100%). Samples were embedded using an increasing concentration of Durcupan resin (without component D) in acetone (1.5 h at 25, 50, 75%, and overnight at 100% Durcupan), followed by incubation with 100% Durcupan + component D for 5 h and final embedding in silicone molds with fresh resin from the same composition. They were polymerized for 48 h at 60°C until the blocks hardened. The sample in the resin was exposed over a block length of ∼400 μm on four sides by trimming away the excess resin using a trimming diamond knife (Ultratrimm 90°, Diatome). The trimmed block was mounted on an aluminum Gatan pin with CW2400 conductive epoxy resin and baked for additional 3 d at 60°C. Finally, the block was sputter coated with 20 nm of platinum (ACE600 Leica Microsystems, Vienna) before imaging at the Zeiss Merlin VP Compact SEM equipped with a Gatan 3View2XP system. SEM images of the block face were acquired at the variable pressure mode by an alternating procedure of cutting 80 nm and imaging with a pixel time of 0.5–0.7 μs and an image size of 8000 × 8000 pixels with 10-nm pixel size. About 500–2,700 sections per block were acquired.
3D reconstruction of SEM images
Volumes were 4 × 4 binned for 3D image reconstruction. Cropped volumes containing the regions of interest were aligned with the TrakEM plugin (Cardona et al., 2012) of Fiji. Aligned stacks were manually segmented in VAST (Berger et al., 2018) and renderings were done using the Blender software.
SEM of zebrafish spinal cords
18 dpf wild-type;Tg (mbp:mCherry-CAAX) and csf1rDM;Tg (mbp:mCherry-CAAX) fish larvae were anesthetized and fixed (4% PFA and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4; Science Services) in a laboratory microwave (PELCO BioWave). The tail and head were dissected and the remaining larvae immersion fixed for 6–8 d. We applied a standard rOTO microwave staining protocol (Kislinger et al., 2020; Hua et al., 2015) including postfixation in 2% osmium tetroxide (EMS) in 0.1 M sodium cacodylate (Science Services) buffer (pH 7.4) and a separate incubation step in 2.5% potassium ferricyanide (Sigma-Aldrich) in the same buffer (1.5 h each). Staining was enhanced by reaction with 1% thiocarbohydrazide (Sigma-Aldrich) for 45 min at 40°C. The tissue was washed in water and incubated in 2% aqueous osmium tetroxide (1.5 h), washed and further contrasted by overnight incubation in 1% aqueous uranyl acetate at 4°C and 2 h at 50°C. Samples were dehydrated in an ascending ethanol series and infiltrated with LX112 (LADD), with two overnight steps at 50 and 100% resin. At the level of the proximal spinal cord, 80-nm-thick cross-sections were cut on a Leica UC7 and deposited onto carbon nanotube tape (Science Services). Carbon nanotube tape stripes were assembled onto adhesive carbon tape (Science Services) attached to 4-inch silicon wafers (Siegert Wafer) and grounded by adhesive carbon tape strips (Science Services). SEM images of the entire cross-section were acquired on a Crossbeam 340 (Zeiss) with a four-quadrant backscatter detector at 8 kV using the Atlas 5 software (Fibics). A wafer overview map was generated (1.5 µm/pixel) and regions of interest acquired at 100 × 100 nm and 4 × 4 nm lateral resolution.
Quantifications
Analysis of mouse optic nerve SBF-SEM
The total number of myelinated axons was counted on 10 40 × 40 µm sections (reference sections). Structural defects were assessed within small volumes of in total 100 sections around the reference section and normalized by the total number of myelinated axons. Microglia numbers were counted in a volume of 80 × 80 × 160 µm and normalized of myelinated axons. Microglia-associated myelin aberrations were quantified for five microglia per optic nerve. Counting was done with the Cell Counter tool in Fiji.
Analysis of myelin fragments in mouse immunohistochemistry
The number of cells with internalized myelin particles, labeled by anti-MBP and colocalizing with anti-LAMP1, was quantified and normalized by the total numbers of cells in an area of 0.6 mm2.
Tracking of microglia motility in zebrafish
Tracking of microglia soma displacement was done with the Manual Tracking plugin in Fiji (F. Cordelières, Institut Curie, Orsay, France). Velocity between two consecutive time frames was an output of the Manual Tracking plugin, and mean velocity represents the mean velocity of an individual microglia over all time frames. A time frame was defined as a break when no soma displacement had taken place compared to the previous frame. Break duration was calculated for individual microglia by adding consecutive time frames in which breaks occurred, multiplied by the acquisition interval. Euclidean distances were calculated from x,y coordinates recorded by the plugin. For heatmaps, a 5 × 5 mm grid was placed over the spinal cord, separating the dorsal and ventral spinal cord. Presence of a microglia (or parts of it) in each quadrant was added up in all time frames.
Analysis of csf1rDM mutants and PS receptor triple mutants
Analysis was performed blinded for genotypes. Myelinated area in the ventral or dorsal spinal cord was determined in Fiji as the total area of mbp:EGFP-CAAX–positive pixels from regions of interest (ROIs) of manually thresholded maximum intensity projections. Number of OPCs, oligodendrocytes, sheath lengths, and myelin abnormalities were quantified using the 3D surpass view of IMARIS (Bitplane). For sheath lengths, distances between sheath ends were measured in the dorsal spinal cord, including sheaths of commissural neurons that could be followed along their entire length. Aberrant myelin structure was assessed in the 3D view of Imaris. The number of myelin abnormalities and myelinosomes was normalized by the myelinated area in the ventral spinal cord.
Ultrastructural myelin aberrations were quantified on single SEM cross-sections comprising the whole spinal cord, using the Cell Counter tool in Fiji.
Quantification of myelin sheath retractions
Myelin sheaths in the dorsal spinal cord showing negative growth over consecutive time frames were quantified as retractions. These were assessed on maximum intensity projections and verified on z-stacks in Fiji. For the heatmap, the spinal cord was divided into 5 × 5 mm quadrants. Per quadrant, we quantified the sum of time frames in which a microglia was present and the number of retractions over the whole acquisition. We did a similar quantification to assess the correlation between microglial presence and retractions, but on the basis of oligodendrocyte territories (containing the myelin sheaths made by 1–2 oligodendrocytes) instead of quadrants.
Volume of myelin fragments inside microglia
Microglial volumes were manually or automatically segmented in Imaris, and the resulting surfaces were used to mask both channels (488 nm channel for microglia and 552 nm channel for myelin). The binary masked 488 nm channel was then despeckled in Fiji to remove random noise. Microglia and myelin volumes were calculated with the Voxel Counter plugin in Fiji. Data are represented as means per individual larvae.
Colocalization of LysoTracker staining with myelin fragments inside oligodendrocytes
Oligodendrocyte cell bodies were manually segmented in Imaris, and the resulting surfaces were used to mask both channels as “cell ROI” (488 nm channel for LysoTracker and 552 nm channel for myelin fragments). A second surface encompassing the bright myelin fragments within oligodendrocyte cell bodies was created by thresholding the first surface by intensity. Masking of both channels by this surface resulted in “fragment ROIs.” Masked channels were manually thresholded for colocalization analysis in ImarisColoc. Thresholded Mander’s B coefficients represent the colocalized fraction within the myelin channel after thresholding. Data are represented as means per individual larvae.
Colocalization of LysoTracker staining with structural myelin abnormalities
Myelin abnormalities and parts of adjacent myelin sheaths were manually segmented in Imaris. Additionally, an automatic surface was created for the 552 nm channel, representing total myelin sheaths (and membranes of oligodendrocytes somas). The resulting surfaces from these three segmentations were used to mask both channels (488 nm channel for LysoTracker and the 552 nm channel). Masked channels were manually thresholded in ImarisColoc using the same thresholding parameters for myelin abnormalities, adjacent sheaths, and total myelin. Colocalization analysis for these three categories was carried out in ImarisColoc. Thresholded Mander’s B coefficients represent the colocalized fraction within myelin. Data are represented as means per individual larvae.
Analysis of mbp:mCherry-CAAX+/MFG-E8-EGFP+ clusters
MFG-E8-EGFP–labeled structures and mbp:mCherry-CAAX+/MFG-E8-EGFP+ clusters were assessed on images masked by the 552 nm channel by a surface automatically created in Imaris. Clusters were counted in the 3D view in Imaris. Quantifications show the percentage of MFG-E8-EGFP–labeled structures that exhibited mbp:mCherry-CAAX co-staining, categorized by their localization on myelin sheaths or mbp:mCherry-CAAX–positive fragments. Data are represented as means per individual larvae.
Statistical analysis
Statistical analysis was performed in GraphPad Prism. All samples were tested for normality using the Shapiro–Wilk test. Comparison of two independent groups was carried out using two-sided Student’s t test (for normally distributed data) and Mann–Whitney U-test (for non-normally distributed data). Groups with small sample sizes that were visibly non-normally distributed were tested by Mann–Whitney U-test. Comparisons between three or more groups were done by one-way ANOVA, followed by Tukey’s post-hoc test for multiple comparisons. Data in the text are presented as mean ± SD or median and interquartile range (25th percentile–75th percentile) as indicated in the legends. Correlations were tested by linear regression, and the coefficient of determination (R2) and P values are reported in the legends. P values are indicated throughout the figures as follows: n.s.: P ≥ 0.05, *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001.
Online supplemental material
Fig. S1 shows the quantification of myelin abnormalities at developmental time points P10, P14, P21, and P60 (n = 1) and two examples of microglia engulfing myelin abnormalities (3D reconstructions from SBF-SEM stacks). Fig. S2 shows the screening behavior of microglia along myelin sheaths. Fig. S3 shows the phenotypic characterization of the csf1rDM mutants. Fig. S4 shows details of the CRISPR/Cas9 F0 mutant screen for PS receptors and a characterization of the hypomyelination phenotype in zebrafish. Table S1 lists gRNA sequences and genotyping details for F0 mutants from the CRISPR/Cas9 screen. Video 1 shows that aberrant myelin ultrastructure is abundant during development and is resolved until young adulthood. Video 2 shows the 3D reconstruction of a myelin bulging. Video 3 shows the 3D reconstruction of a microglia that is associated with aberrant myelin ultrastructure in mouse optic nerve. Video 4 shows how microglia screen myelin along the spinal cord and phagocytose myelin debris. Video 5 shows that myelin retractions occur independently from microglia and are broken down into fragments in the oligodendrocyte cell body.
Supplementary Material
shows gRNA sequences and genotyping specifications of the CRISPR/Cas9 screen for PS receptors.
contains original blots for Fig. S4.
Acknowledgments
We thank Tim Czopka (University of Edinburgh, Scotland), David Lyons (University of Edinburgh, Scotland), Brian Link (Medical College of Wisconsin, Milwaukee, WI, USA), Jan Kranich (Ludwig Maximilian University of Munich, Germany), Benjamin Odermatt (University of Bonn, Germany), Randall Peterson (University of Utah, Salt Lake City, UT, USA), Francesca Peri (Universtity of Zürich, Switzerland), and Michel Reimer (Technical University of Dresden, Germany) for providing reagents. We thank members of the Simons lab for their support and critical discussions.
The work was supported by grants from the German Research Foundation (SPP2191, TRR128-2, TRR 274, SyNergy Excellence Cluster, EXC2145, Projekt ID390857198), the Human Frontier Science Program, the European Research Council Advanced Grant (to M. Simons), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. D.S. Campbell was funded by Research Grant CA (1530/1-1) from the German Research Foundation.
Author contributions: M. Simons and M. Djannatian designed the project. M. Djannatian, S. Radha, U. Weikert, S. Safaiyan, C. Wrede, C. Deichsel, G. Kislinger, A. Rhomberg, T. Ruhwedel, and D.S. Campbell performed experiments and/or analyzed the data. T. van Ham, M. Schifferer, W. Möbius, J. Hegermann, B. Schmid, and M. Simons analyzed the data or supervised data acquisition. M. Djannatian wrote the first draft. M. Simons and M. Djannatian edited the manuscript with input from all authors.
References
- Almeida, R.G., Czopka T., Ffrench-Constant C., and Lyons D.A.. 2011. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development. 138:4443–4450. 10.1242/dev.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, R.G., and Lyons D.A.. 2015. Intersectional gene expression in zebrafish using the split KalTA4 system. Zebrafish. 12:377–386. 10.1089/zeb.2015.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer, F., Vagionitis S., and Czopka T.. 2018. Evidence for myelin sheath remodeling in the CNS revealed by In Vivo imaging. Curr. Biol. 28:549–559.e3. 10.1016/j.cub.2018.01.017 [DOI] [PubMed] [Google Scholar]
- Baraban, M., Koudelka S., and Lyons D.A.. 2018. Ca 2+ activity signatures of myelin sheath formation and growth in vivo. Nat. Neurosci. 21:19–23. 10.1038/s41593-017-0040-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, D.R., Seung H.S., and Lichtman J.W.. 2018. VAST (volume annotation and segmentation tool): Efficient manual and semi-automatic labeling of large 3D image stacks. Front. Neural Circuits. 12:88. 10.3389/fncir.2018.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D.L., Misgeld T., Walsh M.K., Gan W.B., and Lichtman J.W.. 2004. Axon branch removal at developing synapses by axosome shedding. Neuron. 44:651–661. 10.1016/j.neuron.2004.10.026 [DOI] [PubMed] [Google Scholar]
- Cardona, A., Saalfeld S., Schindelin J., Arganda-Carreras I., Preibisch S., Longair M., Tomancak P., Hartenstein V., and Douglas R.J.. 2012. TrakEM2 software for neural circuit reconstruction. PLoS One. 7:e38011. 10.1371/journal.pone.0038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Poskanzer K.E., Freeman M.R., and Monk K.R.. 2020. Live-imaging of astrocyte morphogenesis and function in zebrafish neural circuits. Nat. Neurosci. 23:1297–1306. 10.1038/s41593-020-0703-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, B.S., Winter M., Cohen A.R., and Link B.A.. 2011. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240:2452–2465. 10.1002/dvdy.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, C.C.H., Popovic M.A., Klooster J., Weil M.T., Möbius W., Nave K.A., and Kole M.H.P.. 2020. Saltatory conduction along myelinated axons involves a periaxonal nanocircuit. Cell. 180:311–322.e15. 10.1016/j.cell.2019.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka, T., Ffrench-Constant C., and Lyons D.A.. 2013. Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths in vivo. Dev. Cell. 25:599–609. 10.1016/j.devcel.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance, A. 2019. Core Concept: Cells nibble one another via the under-appreciated process of trogocytosis. Proc. Natl. Acad. Sci. USA. 116:17608–17610. 10.1073/pnas.1912252116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djannatian, M., Timmler S., Arends M., Luckner M., Weil M.T., Alexopoulos I., Snaidero N., Schmid B., Misgeld T., Möbius W., et al. 2019. Two adhesive systems cooperatively regulate axon ensheathment and myelin growth in the CNS. Nat. Commun. 10:4794. 10.1038/s41467-019-12789-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, J.M., McLaughlin M., Werner H.B., McCulloch M.C., Barrie J.A., Brown A., Faichney A.B., Snaidero N., Nave K.-A., and Griffiths I.R.. 2009. Early ultrastructural defects of axons and axon-glia junctions in mice lacking expression of Cnp1. Glia. 57:1815–1824. 10.1002/glia.20893 [DOI] [PubMed] [Google Scholar]
- Ellett, F., Pase L., Hayman J.W., Andrianopoulos A., and Lieschke G.J.. 2011. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 117:e49–e56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig, M.S., Patzig J., Steyer A.M., Dibaj P., Heilmann M., Heilmann I., Jung R.B., Kusch K., Möbius W., Jahn O., et al. 2019. Anillin facilitates septin assembly to prevent pathological outfoldings of central nervous system myelin. Elife. 8:e43888. 10.7554/eLife.43888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria, A., Hokanson K.C., Dao D.Q., Mayoral S.R., Mei F., Redmond S.A., Ullian E.M., and Chan J.R.. 2016. Dynamic modulation of myelination in response to visual stimuli alters optic nerve conduction velocity. J. Neurosci. 36:6937–6948. 10.1523/JNEUROSCI.0908-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, R.D. 2008. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 31:361–370. 10.1016/j.tins.2008.04.00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, M.C., Alexandrova O., Cossell L., Stange-Marten A., Sinclair J., Kopp-Scheinpflug C., Pecka M., Attwell D., and Grothe B.. 2015. Tuning of Ranvier node and internode properties in myelinated axons to adjust action potential timing. Nat. Commun. 6:8073. 10.1038/ncomms9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fünfschilling, U., Supplie L.M., Mahad D., Boretius S., Saab A.S., Edgar J., Brinkmann B.G., Kassmann C.M., Tzvetanova I.D., Möbius W., et al. 2012. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 485:517–521. 10.1038/nature11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, E.M., Purger D., Mount C.W., Goldstein A.K., Lin G.L., Wood L.S., Inema I., Miller S.E., Bieri G., Zuchero J.B., et al. 2014. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 344:1252304. 10.1126/science.1252304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giera, S., Luo R., Ying Y., Ackerman S.D., Jeong S.J., Stoveken H.M., Folts C.J., Welsh C.A., Tall G.G., Stevens B., et al. 2018. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. Elife. 7:e33385. 10.7554/eLife.33385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer, N., Hanft K.-M., Akriditou M.-A., Unger N., Park E.S., Stanley E.R., Staszewski O., Dimou L., and Prinz M.. 2017. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134:441–458. 10.1007/s00401-017-1747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, C., Jan L.Y., and Jan Y.N.. 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. USA. 108:9673–9678. 10.1073/pnas.1106386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline, D.K., and Colman D.R.. 2007. Rapid conduction and the evolution of giant axons and myelinated fibers. Curr. Biol. 17:R29–R35. 10.1016/j.cub.2006.11.042 [DOI] [PubMed] [Google Scholar]
- Hill, R.A., Li A.M., and Grutzendler J.. 2018. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat. Neurosci. 21:683–695. 10.1038/s41593-018-0120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines, J.H., Ravanelli A.M., Schwindt R., Scott E.K., and Appel B.. 2015. Neuronal activity biases axon selection for myelination in vivo. Nat. Neurosci. 18:683–689. 10.1038/nn.3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter-Hufford, A., and Ravichandran K.S.. 2013. Clearing the dead: Apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 5:a008748. 10.1101/cshperspect.a008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima, K., Jurynec M.J., Klatt Shaw D., Jacobi A.M., Behlke M.A., and Grunwald D.J.. 2019. Highly efficient CRISPR-cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev. Cell. 51:645–657.e4. 10.1016/j.devcel.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha, A., and Schmid B.. 2015. Generation of zebrafish models by CRISPR /Cas9 genome editing. Neuronal Cell Death. 1254:341–350. 10.1007/978-1-4939-2152-2_24 [DOI] [PubMed] [Google Scholar]
- Hua, Y., Laserstein P., and Helmstaedter M.. 2015. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat. Commun. 6:7923. 10.1038/ncomms8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.N., and Appel B.. 2020. Microglia phagocytose myelin sheaths to modify developmental myelination. Nat. Neurosci. 23:1055–1066. 10.1038/s41593-020-0654-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, E.G., Orthmann-Murphy J.L., Langseth A.J., and Bergles D.E.. 2018. Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat. Neurosci. 21:696–706. 10.1038/s41593-018-0121-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao, L.-E., Wente S.R., and Chen W.. 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 110:13904–13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller, M.S., Lazari A., Blanco-Duque C., Sampaio-Baptista C., and Johansen-Berg H.. 2017. Myelin plasticity and behaviour-connecting the dots. Curr. Opin. Neurobiol. 47:86–92. 10.1016/j.conb.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, U., and Schultz R.L.. 1965. Fixation of the central nervous system for electron microscopy by aldehyde perfusion. I. Preservation with aldehyde perfusates versus direct perfusion with osmium tetroxide with special reference to membranes and the extracellular space. J. Ultrastruct. Res. 12:160–186. 10.1016/S0022-5320(65)80014-4 [DOI] [PubMed] [Google Scholar]
- Kawakami, K. 2007. Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. 8:S7. 10.1186/gb-2007-8-s1-s7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge, M., Tsarouchas T.M., Munir T., Porter N.J., Larraz J., Gianni D., Tsai H.-H., Becker C.G., Lyons D.A., and Becker T.. 2021. CRISPR gRNA phenotypic screening in zebrafish reveals pro-regenerative genes in spinal cord injury. PLoS Genet. 17:e1009515. 10.1371/journal.pgen.1009515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger, G., Gnägi H., Kerschensteiner M., Simons M., Misgeld T., and Schifferer M.. 2020. ATUM-FIB microscopy for targeting and multiscale imaging of rare events in mouse cortex. STAR Protoc. 1:100232. 10.1016/j.xpro.2020.100232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotter, M.R., Li W.-W., Zhao C., and Franklin R.J.M.. 2006. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J. Neurosci. 26:328–332. 10.1523/JNEUROSCI.2615-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotter, M.R., Setzu A., Sim F.J., Van Rooijen N., and Franklin R.J.M.. 2001. Macrophage depletion impairs oligodendrocyte remyelination following lysolecithin-induced demyelination. Glia. 35:204–212. 10.1002/glia.1085 [DOI] [PubMed] [Google Scholar]
- Kranich, J., Chlis N.K., Rausch L., Latha A., Schifferer M., Kurz T., Foltyn-Arfa Kia A., Simons M., Theis F.J., and Brocker T.. 2020. In vivo identification of apoptotic and extracellular vesicle-bound live cells using image-based deep learning. J. Extracell. Vesicles. 9:1792683. 10.1080/20013078.2020.1792683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow, A.M., Ford M.C., Valdivia L.E., Wilson S.W., and Attwell D.. 2018. Regulation of developing myelin sheath elongation by oligodendrocyte calcium transients in vivo. Nat. Neurosci. 21:24–28. 10.1038/s41593-017-0031-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, F., Powell G.T., Ghosh M., Gestri G., Antinucci P., Hearn T.J., Tunbak H., Lim S., Dennis H.W., Fernandez J.M., et al. 2021. A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. Elife. 10:1–34. 10.7554/eLife.59683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas, S., Takada N., Park H.-C., Woodruff E., Broadie K., and Appel B.. 2008. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat. Neurosci. 11:143–151. 10.1038/nn2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun, K., Montague T.G., Gagnon J.A., Thyme S.B., and Valen E.. 2016. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44:W272-6. 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani, B., Borich M.R., Jackson J.N., Wadden K.P., Peters S., Villamayor A., MacKay A.L., Vavasour I.M., Rauscher A., and Boyd L.A.. 2016. Motor skill acquisition promotes human brain myelin plasticity. Neural Plast. 2016:7526135. 10.1155/2016/7526135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampron, A., Larochelle A., Laflamme N., Préfontaine P., Plante M.-M., Sánchez M.G., Yong V.W., Stys P.K., Tremblay M.-È., and Rivest S.. 2015. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 212:481–495. 10.1084/jem.20141656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., and Chung W.S.. 2019. Glial control of synapse number in healthy and diseased brain. Front. Cell. Neurosci. 13:42. 10.3389/fncel.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Morrison B.M., Li Y., Lengacher S., Farah M.H., Hoffman P.N., Liu Y., Tsingalia A., Jin L., Zhang P.W., et al. 2012. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 487:443–448. 10.1038/nature11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, G. 2019. How macrophages deal with death. Nat. Rev. Immunol. 19:539–549. 10.1038/s41577-019-0167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Du J.-L., and He C.. 2013. Developmental pruning of early-stage myelin segments during CNS myelination in vivo. Cell Res. 23:962–964. 10.1038/cr.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan, M., Rosen K.M., Ito S., Corfas G., Plant S.R., Kaing S., Raine C.S., Rowitch D.H., Franklin R.J., Stiles C.D., et al. 2012. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science. 337:1357–1360. 10.1126/science.1220845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisca, R., Hoche T., Agirre E., Hoodless L.J., Barkey W., Auer F., Castelo-Branco G., and Czopka T.. 2020. Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 23:363–374. 10.1038/s41593-019-0581-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, I.A., Ohayon D., Li H., de Faria J.P., Emery B., Tohyama K., and Richardson W.D.. 2014. Motor skill learning requires active central myelination. Science. 346:318–322. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch, S., Baraban M., Almeida R., Czopka T., Ausborn J., El Manira A., and Lyons D.A.. 2015. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat. Neurosci. 18:628–630. 10.1038/nn.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikula, S., Binding J., and Denk W.. 2012. Staining and embedding the whole mouse brain for electron microscopy. Nat. Methods. 9:1198–1201. 10.1038/nmeth.2213 [DOI] [PubMed] [Google Scholar]
- Monje, M. 2018. Myelin plasticity and nervous system function. Annu. Rev. Neurosci. 41:61–76. 10.1146/annurev-neuro-080317-061853 [DOI] [PubMed] [Google Scholar]
- Naert, T., Tulkens D., Edwards N.A., Carron M., Shaidani N.-I., Wlizla M., Boel A., Demuynck S., Horb M.E., Coucke P., et al. 2020. Maximizing CRISPR/Cas9 phenotype penetrance applying predictive modeling of editing outcomes in Xenopus and zebrafish embryos. Sci. Rep. 10:14662. 10.1038/s41598-020-71412-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof, N., Kuil L.E., van der Linde H.C., Burm S.M., Berdowski W., van Ijcken W.F.J., van Swieten J.C., Hol E.M., Verheijen M.H.G., and van Ham T.J.. 2018. Colony-stimulating factor 1 receptor (CSF1R) regulates microglia density and distribution, but not microglia differentiation In Vivo. Cell Rep. 24:1203–1217.e6. 10.1016/j.celrep.2018.06.113 [DOI] [PubMed] [Google Scholar]
- Orthmann-Murphy, J., Call C.L., Molina-Castro G.C., Hsieh Y.C., Rasband M.N., Calabresi P.A., and Bergles D.E.. 2020. Remyelination alters the pattern of myelin in the cerebral cortex. Elife. 9:1–61. 10.7554/eLife.56621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez, S., Herdman C., Beerens M., Chakraborti K., Harmer Z.P., Yeh J.J., MacRae C.A., Yost H.J., and Peterson R.T.. 2021. MIC-drop: A platform for large-scale in vivo CRISPR screens. Science. 373:1146–1151. 10.1126/science.abi8870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, M., Jung S., and Priller J.. 2019. Microglia biology: One century of evolving concepts. Cell. 179:292–311. 10.1016/j.cell.2019.08.053 [DOI] [PubMed] [Google Scholar]
- Rushton, W.A.H. 1951. A theory of the effects of fibre size in medullated nerve. J. Physiol. 115:101–122. 10.1113/jphysiol.1951.sp004655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab, A.S., Tzvetavona I.D., Trevisiol A., Baltan S., Dibaj P., Kusch K., Möbius W., Goetze B., Jahn H.M., Huang W., et al. 2016. Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron. 91:119–132. 10.1016/j.neuron.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J.C., Picard K., González-Ibáñez F., and Tremblay M.-È.. 2018. A brief history of microglial ultrastructure: Distinctive features, phenotypes, and functions discovered over the past 60 Years by electron microscopy. Front. Immunol. 9:803. 10.3389/fimmu.2018.00803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz, J., Klein M.C., Behrens T.E.J., and Johansen-Berg H.. 2009. Training induces changes in white-matter architecture. Nat. Neurosci. 12:1370–1371. 10.1038/nn.2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A.N., Davey C.F., Whitebirch A.C., Miller A.C., and Moens C.B.. 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods. 12:535–540. 10.1038/nmeth.3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., and Sherwood R.I.. 2018. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 563:646–651. 10.1038/s41586-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherafat, A., Pfeiffer F., Reiss A.M., Wood W.M., and Nishiyama A.. 2021. Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor. Nat. Commun. 12:2265. 10.1038/s41467-021-22532-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, J.L., Fischl M.J., Alexandrova O., Heβ M., Grothe B., Leibold C., and Kopp-Scheinpflug C.. 2017. Sound-evoked activity influences myelination of brainstem axons in the trapezoid body. J. Neurosci. 37:8239–8255. 10.1523/JNEUROSCI.3728-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaidero, N., Möbius W., Czopka T., Hekking L.H.P., Mathisen C., Verkleij D., Goebbels S., Edgar J., Merkler D., Lyons D.A., et al. 2014. Myelin membrane wrapping of CNS axons by PI (3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 156:277–290. 10.1016/j.cell.2013.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmann, C., Timmler S., Barrantes-Freer A., and Simons M.. 2019. Myelin in the central nervous system: Structure, function, and pathology. Physiol. Rev. 99:1381–1431. 10.1152/physrev.00031.2018 [DOI] [PubMed] [Google Scholar]
- Steele, C.J., Bailey J.A., Zatorre R.J., and Penhune V.B.. 2013. Early musical training and white-matter plasticity in the corpus callosum: Evidence for a sensitive period. J. Neurosci. 33:1282–1290. 10.1523/JNEUROSCI.3578-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama, B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R. III, and Hetzer M.W.. 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154:971–982. 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake, H., Ortiz F.C., Woo D.H., Lee P.R., Angulo M.C., and Fields R.D.. 2015. Nonsynaptic junctions on myelinating glia promote preferential myelination of electrically active axons. Nat. Commun. 6:7844. 10.1038/ncomms8844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarczyk, A., Holtman I.R., Krueger M., Yogev N., Bruttger J., Khorooshi R., Benmamar-Badel A., de Boer-Bergsma J.J., Martin N.A., Karram K., et al. 2017. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36:3292–3308. 10.15252/embj.201696056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L., Ohayon D., McKenzie I.A., Sinclair-Wilson A., Wright J.L., Fudge A.D., Emery B., Li H., and Richardson W.D.. 2016. Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat. Neurosci. 19:1210–1217. 10.1038/nn.4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.M., Michel K., Jokhi V., Nedivi E., and Arlotta P.. 2020. Neuron class-specific responses govern adaptive myelin remodeling in the neocortex. Science. 370:eabd2109. 10.1126/science.abd2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc, B. 2016. The acquisition of myelin: An evolutionary perspective. Brain Res. 1641:4–10. 10.1016/j.brainres.2015.09.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows gRNA sequences and genotyping specifications of the CRISPR/Cas9 screen for PS receptors.
contains original blots for Fig. S4.