Abstract
Long-read RNA sequencing (RNA-seq) holds great potential for characterizing transcriptome variation and full-length transcript isoforms, but the relatively high error rate of current long-read sequencing platforms poses a major challenge. We present ESPRESSO, a computational tool for robust discovery and quantification of transcript isoforms from error-prone long reads. ESPRESSO jointly considers alignments of all long reads aligned to a gene and uses error profiles of individual reads to improve the identification of splice junctions and the discovery of their corresponding transcript isoforms. On both a synthetic spike-in RNA sample and human RNA samples, ESPRESSO outperforms multiple contemporary tools in not only transcript isoform discovery but also transcript isoform quantification. In total, we generated and analyzed ~1.1 billion nanopore RNA-seq reads covering 30 human tissue samples and three human cell lines. ESPRESSO and its companion dataset provide a useful resource for studying the RNA repertoire of eukaryotic transcriptomes.
A computational tool, ESPRESSO, robustly discovers and quantifies transcript isoforms from error-prone long-read RNA-seq data.
INTRODUCTION
In higher eukaryotes, a single gene can generate multiple transcript isoforms that diversify the transcriptome and proteome (1). Switches between transcript isoforms and their underlying RNA processing events occur in many biological processes, such as cellular differentiation (2, 3), and are known to be dysregulated in the context of human diseases, including cancer (4, 5). Consequently, it is important to examine the transcriptome diversity of cells not only at the gene level but also at the isoform level.
Tremendous efforts have been made over the past 20 years in developing genomic technologies and computational tools to discover and quantify transcript isoforms (6). In the past decade, short-read RNA sequencing (RNA-seq) has become a widely used approach for profiling eukaryotic transcriptomes, and numerous tools have been developed and optimized to analyze short-read RNA-seq data (7). However, despite having high sequencing quality and throughput, short-read RNA-seq is inherently limited in its ability to discover and quantify transcript isoforms because its limited read lengths often cannot cover more than one splice junction (SJ), let alone full-length transcripts (8). By contrast, rapidly developing single-molecule long-read RNA-seq technologies are capable of generating reads longer than 10 kb (9, 10), which can span the entirety of almost all eukaryotic transcripts, and therefore have emerged as a potentially powerful solution to analyzing transcriptome variation at the isoform level. One major limitation of long-read RNA-seq technologies, however, is that their raw reads have a high sequencing error rate. For example, current long-read sequencing platforms, including Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), are reported to have an average error rate ranging between 5 and 20% (11, 12). To improve sequencing quality, PacBio developed a circular consensus sequencing (CCS) protocol that involves reading circularized complementary DNA (cDNA) molecules multiple times to generate accurate consensus reads (13). Similar strategies are also available in both commercial and customized ONT sequencing methods, such as 1D squared (1D2) and rolling circle amplification to concatemeric consensus (R2C2) (14). While such methods can reduce the sequencing error rate, they lead to a much lower sequencing throughput and are subject to systematic biases due to additional size selection and ligation steps (11). As a result, consensus-free ONT 1D cDNA and direct RNA-seq (15) represent arguably the most cost-friendly long-read RNA-seq protocols.
Alignment-based strategies are commonly adopted for analyzing RNA-seq data (16). Previous studies have demonstrated that aligning long RNA-seq reads against high-quality reference genomes can serve as an initial step in discovering and quantifying transcript isoforms (17, 18). Nevertheless, because of the typically high error rate of long-read sequencing platforms, a nonnegligible proportion of SJs discovered from raw long RNA-seq reads have incorrect positions, based on an evaluation of four state-of-the-art aligners applied to ONT 2D RNA-seq reads (19). The best performer in this evaluation, minimap2, an algorithm specifically designed for mapping error-prone long reads, reported incorrect SJ positions for 6% of detected introns, mainly due to frequent insertion and deletion errors around splice sites. As human protein-coding transcripts have an average of 10 introns (SJs) per transcript (20), the high frequency of incorrect SJ positions poses a major challenge for accurate inference of full-length transcripts and protein products.
In recent years, various computational tools have been developed for transcript isoform analysis using long-read RNA-seq data. However, some of these tools were primarily designed or tested on consensus reads (e.g., PacBio CCS reads or ONT R2C2 reads) (14, 17, 21), which have a higher sequencing accuracy compared to raw long reads. For example, Mandalorion (14, 21) was primarily designed for ONT 2D RNA-seq reads and R2C2 consensus reads obtained from rolling circle amplification (RCA). To mitigate the high sequencing error in raw long RNA-seq reads, some investigators have proposed and adopted a “hybrid sequencing” strategy, which combines long-read and short-read data on the same RNA sample to improve transcript isoform discovery and quantification (22, 23). However, by requiring both long-read and short-read RNA-seq data, this strategy increases the complexity and decreases the flexibility of data acquisition and analysis. For example, Full-Length Alternative Isoform analysis of RNA (FLAIR) uses short-read RNA-seq data to correct putative SJs found in long-read alignments (23), yet in the absence of short-read data, FLAIR cannot detect novel SJs. Two recently published tools, Long-read Isoform Quantification and Analysis (LIQA) (24) and NanoCount (25), quantify the abundances of annotated transcript isoforms using long-read RNA-seq data alone, but neither tool discovers novel transcript isoforms. LIQA focuses on addressing the effects of long-read RNA-seq coverage biases on transcript isoform quantification but does not include any step for improving SJ accuracy or discovering novel transcript isoforms (24). On the other hand, NanoCount aligns long RNA-seq reads against a reference transcriptome followed by transcript abundance estimation but is restricted to annotated transcript isoforms (25). Given the increasingly broad adoption of long-read RNA-seq technologies and the rapid accumulation of error-prone long-read RNA-seq data in public repositories, there is an urgent need to develop robust computational tools for transcript isoform discovery and quantification using error-prone long-read RNA-seq data alone.
Here, we report ESPRESSO (Error Statistics PRomoted Evaluator of Splice Site Options), a new computational tool for transcript isoform analysis using long-read RNA-seq data. Instead of relying on high-accuracy consensus reads or assistance from short-read RNA-seq data, ESPRESSO can robustly discover and quantify transcript isoforms, including novel transcript isoforms, using error-prone long-read RNA-seq data alone. ESPRESSO is motivated by two intuitive observations about long-read RNA-seq data. First, among all putative SJs discovered from long-read RNA-seq data, SJs with perfect alignments around splice sites are much less likely to be incorrect than those with imperfect alignments. Second, by borrowing information from multiple long reads aligned to a gene, it is feasible to improve SJ discovery from each long read. Therefore, ESPRESSO jointly considers alignments of all long reads aligned to a gene and uses the error profiles of individual reads to improve the identification of SJs and quantification of transcript isoforms. We demonstrate the performance and utility of ESPRESSO by generating and analyzing ~1.1 billion nanopore RNA-seq reads on synthetic as well as biological samples across diverse human tissues and cell types.
RESULTS
Computational workflow of ESPRESSO
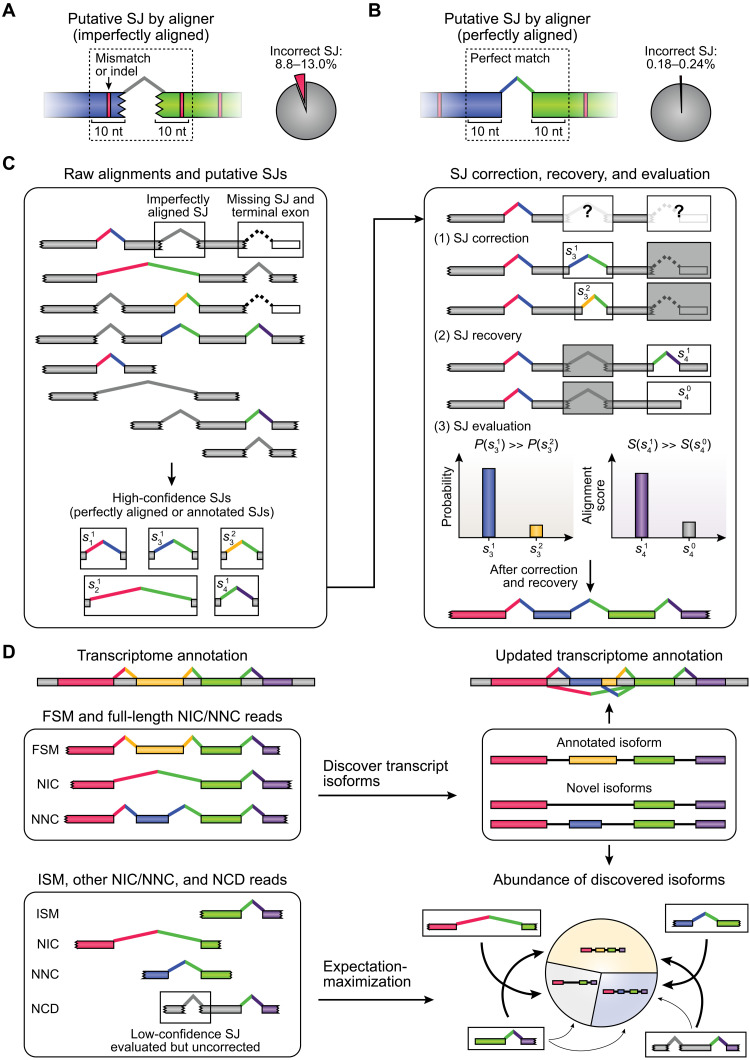
ESPRESSO consists of three major steps (Fig. 1). First, raw long RNA-seq reads are aligned to a reference genome and putative SJs are detected from the long-read-to-genome alignment. A putative SJ is defined as a high-confidence SJ if it is annotated in the existing transcript catalog or if it has the canonical splice site dinucleotide motif (GT/AG, GC/AG, or AT/AC) (26) and is supported by at least two reads (by default) with perfect alignments around splice sites (hereafter referred to as “perfectly aligned reads”). To support the validity of this definition of high-confidence SJs based on alignment features, we generated and analyzed ONT 1D cDNA and direct RNA-seq reads on a sample of Spike-In RNA Variants (SIRVs) (Materials and Methods), which are composed of 68 synthetic transcripts with known transcript structures and concentrations (27). We found that putative SJs supported by perfectly aligned reads were much less likely to be incorrect (0.18 to 0.24%) as compared to putative SJs that were not (8.8 to 13.0%) (Fig. 1, A and B, and table S1).
Fig. 1. Overview of ESPRESSO.
(A and B) Proportion of incorrect splice junctions (SJs) among (A) imperfectly aligned or (B) perfectly aligned putative SJs found in raw long-read-to-genome alignments of ONT 1D cDNA reads (n = 3) and direct RNA reads (n = 3) for Spike-In RNA Variants (SIRVs). Perfectly aligned putative SJs do not have any mismatches or indels within 10 nt of splice sites. (C) High-confidence SJs are identified from raw long-read-to-genome alignments based on whether they are present in the existing transcript catalog, or if they have canonical splice site dinucleotide motifs (GT/AG, GC/AG, or AT/AC) and are supported by at least two (by default) perfectly aligned reads. The resulting set of high-confidence SJs is used to correct, recover, and evaluate SJs found in individual long reads based on each read’s alignment and error profile. (D) First, reads are classified into the following categories on the basis of the annotation statuses of their corresponding SJs in the existing transcript catalog: full splice match (FSM), incomplete splice match (ISM), novel in catalog (NIC), novel not in catalog (NNC), or not completely determined (NCD). Second, FSM and full-length NIC/NNC reads are used to discover annotated and novel transcript isoforms, respectively. Third, all long reads (full-length and non-full-length) are matched to compatible transcript isoforms. Last, abundances of discovered isoforms are quantified using an expectation-maximization (EM) algorithm. Thickness of arrows drawn between reads and compatible transcript isoforms (bottom right) indicates probability of assigning reads to specific transcript isoforms.
Second, for each long RNA-seq read, ESPRESSO considers all high-confidence SJs for the gene to which the read is mapped and determines the read’s most optimal set of SJs. Specifically, each long read is realigned to the sequence of every high-confidence SJ with overlapping coordinates. Matches, mismatches, insertions, and deletions in the long-read-to-high-confidence-SJ realignment are counted. If a long read has a putative SJ with an imperfect alignment around splice sites and multiple options for a high-confidence SJ exist for this putative SJ, then ESPRESSO identifies and selects the most likely high-confidence SJ using a probability calculated from the long read’s alignment and error profile (Fig. 1C). This probability is calculated on the basis of the assumption that a given long read has a similar error profile within and outside its SJ regions. In a hypothetical scenario where a long read has two high-confidence SJ options for a putative SJ (e.g., and ; Fig. 1C), the probability for a particular high-confidence SJ option is calculated from a multivariate hypergeometric distribution, based on the numbers of matches, mismatches, insertions, and deletions in the long-read-to-high-confidence-SJ alignment, within versus outside that SJ region. Among multiple SJ options, the best one with the highest probability is selected for the given long read if its probability is at least 10 times that of the second-best option. Notably, this procedure can also recover missing first or last SJs and their associated terminal exons at alignment ends (Fig. 1C).
Third, after SJ correction and recovery, long reads are classified into multiple categories based on the annotation statuses of their corresponding SJs in the existing transcript catalog. Specifically, we adopted an established classification system for long-read analysis of transcript isoforms: full splice match (FSM), novel in catalog (NIC), novel not in catalog (NNC), and incomplete splice match (ISM) (Materials and Methods) (17). We also introduced a new category, not completely determined (NCD), to classify long reads containing at least one low-confidence SJ that could not be corrected by ESPRESSO. Transcript isoforms are discovered by collapsing full-length long reads into unique chains of high-confidence SJs. To maintain stringency in transcript isoform discovery, novel transcript isoforms that are not annotated in the existing transcript catalog are further required to have at least two perfectly aligned reads supporting each SJ. Once the set of transcript isoforms is determined for a given gene, all long reads (full-length and non-full-length) are matched to compatible transcript isoforms, and the abundances of individual transcript isoforms are quantified using an expectation-maximization (EM) algorithm (Fig. 1D) (28, 29).
ESPRESSO improves SJ identification from error-prone long reads
We evaluated ESPRESSO’s definition of high-confidence SJs based on perfectly aligned reads. For this analysis, we used ONT direct RNA-seq data on SIRV synthetic RNAs. Direct RNA-seq data are known to have a higher error rate but may more accurately preserve transcript structures and abundances as they are free of reverse transcription (RT) and polymerase chain reaction (PCR) amplification biases (30, 31). We first tested the precision and recall of de novo (i.e., annotation-free) SJ identification from raw long-read-to-genome alignments of direct RNA-seq data on SIRVs, using varying read count thresholds based on either the number of perfectly aligned reads or the total number of aligned reads supporting a given SJ. To ensure an unbiased evaluation of SJ identification, we did not use SIRV transcript annotations to guide long-read alignments around SJs.
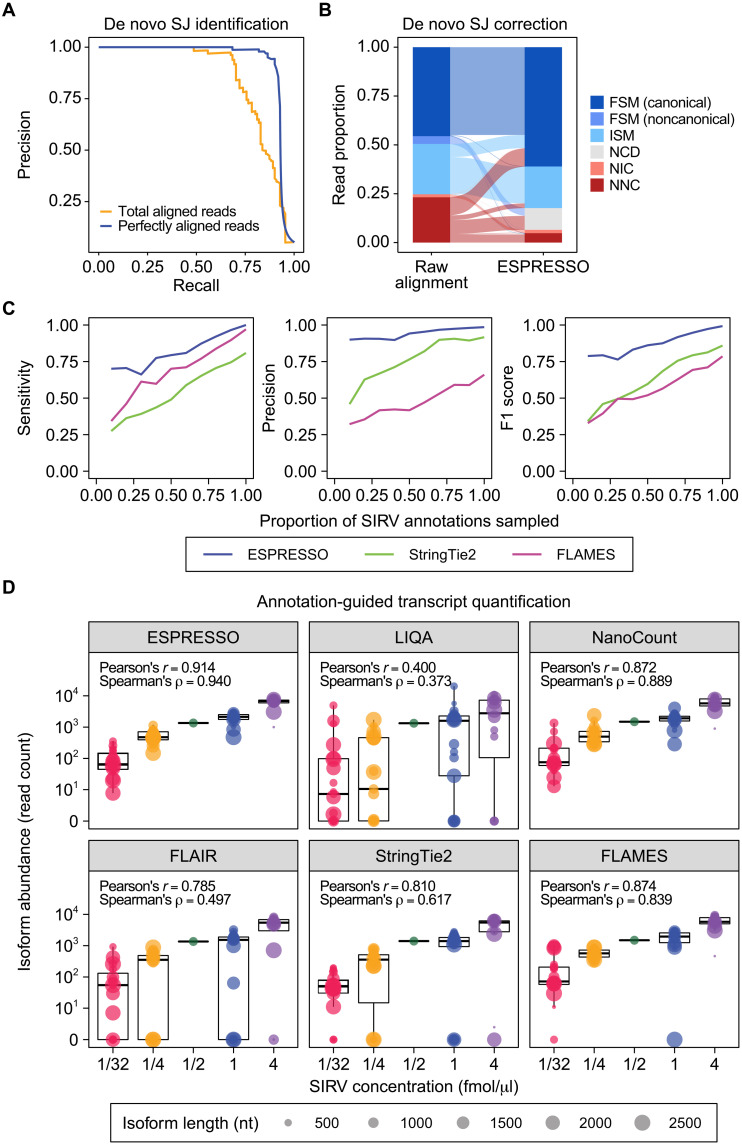
As shown in Fig. 2A, compared to cutoffs based on the total number of aligned reads, cutoffs based on the number of perfectly aligned reads generally yielded a higher precision for SJ identification at the same recall rate. For example, when requiring SJs to be supported by ≥2 perfectly aligned reads, both a high recall (91.9%) and a high precision (85.7%) were achieved. By contrast, at the same recall rate, de novo SJ identification based on total aligned reads achieved a much lower precision of 34.5%. Overall, de novo SJ identification on the SIRV data using perfectly aligned reads had a higher area under the precision-recall curve compared to using total aligned reads (0.930 versus 0.842, respectively). Collectively, these results support ESPRESSO’s definition of high-confidence SJs based on perfectly aligned reads.
Fig. 2. Evaluation of ESPRESSO using ONT direct RNA-seq data of SIRVs.
(A) Precision-recall curves for de novo SJ identification from raw long-read-to-genome alignments, combined over n = 3 direct RNA-seq replicates, using read count thresholds based on total aligned reads or perfectly aligned reads supporting a given SJ. (B) Distribution of transcript isoform categories among aligned reads, combined over n = 3 direct RNA-seq replicates, before and after de novo SJ correction. FSM or ISM indicates that all SJs in a read are consistent with those in an annotated SIRV transcript, with FSM and ISM reads representing full-length and fragmented reads, respectively. FSM reads are further partitioned into two subcategories (canonical and noncanonical) based on whether they contain SJs without the canonical splice site dinucleotide motif. NIC or NNC indicates a novel combination of annotated or novel splice sites, respectively, and reads classified as NIC or NNC have incorrect transcript structures with respect to SIRVs. NCD reads contain at least one putative SJ in the raw alignment that was evaluated as low-confidence but could not be corrected by ESPRESSO. (C) Sensitivity, precision, and F1 score of ESPRESSO and two other tools (StringTie2 and FLAMES) in discovering SIRV transcripts from direct RNA-seq data (n = 3), using random downsamples of different proportions of SIRV annotations as a guide. Each point represents the mean of three random samplings per downsampling level. (D) Box-and-whisker plots (median and interquartile range) and correlation (Pearson’s and Spearman’s) between known concentrations of 68 SIRV transcripts and their estimated abundances from ESPRESSO and five other tools (LIQA, NanoCount, FLAIR, StringTie2, and FLAMES). For each tool, transcript abundance is reported as the sum of assigned read counts over n = 3 direct RNA-seq replicates. Diameters of points in the box-and-whisker plots are scaled according to transcript length.
We next asked whether de novo SJ correction and recovery by ESPRESSO based on high-confidence SJs would improve the accuracy of SJs identified in error-prone long reads. To this end, we compared SJs identified from raw alignments of direct RNA-seq data on SIRVs to SJs called by ESPRESSO after SJ correction and recovery, using SIRV transcript annotations as ground truth. Specifically, we classified individual long RNA-seq reads into distinct categories based on the annotation statuses of their SJs, using a classification system developed by Tardaguila et al. (17). FSM or ISM indicates that all SJs and their combinations in a given read are consistent with those in an annotated SIRV transcript, with FSM and ISM reads representing full-length reads and fragmented reads, respectively. Therefore, reads classified as FSM or ISM are considered as reads with correct transcript structures. On the other hand, NIC or NNC indicates a novel combination of annotated or novel splice sites, respectively. In the context of the SIRV analysis, reads classified as NIC or NNC are considered as reads with incorrect transcript structures.
As shown in Fig. 2B, 49.5% of aligned reads were classified as FSM based on raw alignments. These FSM reads can be further divided into reads containing only SJs with canonical splice site dinucleotide motifs (45.5%) and reads containing at least one SJ without the canonical splice site dinucleotide motif (4.0%). The latter set of “noncanonical FSM” reads exists because some of the artificial SIRV synthetic transcripts contain SJs without the canonical splice site dinucleotide motif. After SJ correction and recovery by ESPRESSO, the proportion of canonical FSM reads increased from 45.5 to 61.0%. The majority of reads reclassified as FSM by ESPRESSO were previously classified as NNC (9.3% of aligned reads) and ISM (6.7% of aligned reads) based on raw alignments, demonstrating the efficacy of ESPRESSO to correct SJ positions and recover missing SJs at alignment ends. Correspondingly, the proportion of NNC and NIC reads decreased substantially from 24.7 to 6.5%, suggesting that ESPRESSO can effectively remove incorrect SJs initially identified from raw alignments. Together, after SJ correction and recovery by ESPRESSO, 18.5% of reads were reclassified from an incorrect type to a correct type (i.e., from NNC/NIC to FSM/ISM) or from an incomplete type to a complete type (i.e., from ISM to FSM). Only 0.96% of reads were reclassified in the opposite direction.
A small proportion of reads contained at least one SJ in the raw alignment that was evaluated by ESPRESSO as low-confidence but could not be corrected by ESPRESSO. These reads were classified as NCD and constituted 11.2% of reads in the SIRV direct RNA-seq data after SJ correction and recovery. The majority of these reads were initially classified as NNC (7.1% of aligned reads) based on raw alignments. In addition, 4.0% of reads initially classified as noncanonical FSM reads were reclassified as NCD by ESPRESSO. This is an expected behavior by ESPRESSO for the SIRV data because ESPRESSO does not consider SJs without the canonical splice site dinucleotide motif as high-confidence SJs in an annotation-free setting. It should be noted that, although NCD reads are not used for transcript isoform discovery, they are subsequently assigned to discovered transcript isoforms and used for quantification (Fig. 1D).
Systematic evaluation of ESPRESSO for transcript isoform discovery and quantification
Next, we evaluated the performance of ESPRESSO in discovering transcript isoforms from ONT direct RNA and 1D cDNA sequencing data on SIRV synthetic RNAs. For this evaluation, we compared ESPRESSO to StringTie2 (32) and FLAMES (33), two recently published tools that can perform transcript isoform discovery and quantification using long-read RNA-seq data alone. In particular, we ran all three tools on the SIRV data using random downsamples of SIRV transcript annotations, which we varied to contain from 10 to 100% of the transcripts. On direct RNA-seq data, ESPRESSO showed the best performance in transcript isoform discovery compared to StringTie2 and FLAMES across all sampling thresholds tested (Fig. 2C). For example, even when only 10% of the SIRV transcript annotations were provided as a guide, ESPRESSO’s sensitivity and precision (0.701 and 0.900, respectively) were far better than those of StringTie2 (0.275 and 0.459, respectively) and FLAMES (0.343 and 0.322, respectively). Similarly, for 1D cDNA sequencing data, ESPRESSO showed the best overall performance in transcript isoform discovery, when sensitivity and precision are jointly considered through the F1 score, compared to StringTie2 and FLAMES for all sampling thresholds tested (fig. S1A). For both direct RNA and 1D cDNA sequencing data of SIRVs, StringTie2 had the lowest sensitivity and FLAMES had the lowest precision. Collectively, these results demonstrate that ESPRESSO shows the best overall performance in discovering transcripts that are not present in annotations.
We also ran ESPRESSO, StringTie2, and FLAMES on ONT direct RNA and 1D cDNA sequencing data on human embryonic kidney (HEK) 293T cells (Materials and Methods) to further evaluate the performance of each tool in discovering transcript isoforms in real human transcriptomes. In the context of RNA-seq data on human samples, we do not know what transcripts are actually expressed. Therefore, to evaluate the performance of each tool in terms of transcript isoform discovery, we adopted an evaluation framework originally used by StringTie2 (32). In this framework, among all discovered transcripts, annotated transcripts are treated as true positives, while novel transcripts are treated as false positives. For the purpose of this evaluation, these definitions are acceptable given that human transcript annotations are of relatively high quality and HEK293T is arguably a well-characterized cell line. We computed for each tool the number of annotated transcripts discovered (proxy for sensitivity) and the proportion of annotated transcripts among all transcripts discovered (proxy for precision). For both ONT direct RNA and 1D cDNA sequencing datasets, ESPRESSO discovered more annotated transcripts compared to StringTie2 and FLAMES (fig. S2A). Moreover, the proportion of annotated transcripts among all transcripts discovered by ESPRESSO was 2 to 3.5 times that of StringTie2 and FLAMES (fig. S2B). These comparisons further indicate that ESPRESSO is a robust tool for transcript isoform discovery.
Besides transcript isoform discovery, another important goal of long-read RNA-seq analysis is to quantify transcript isoforms. To evaluate the accuracy of transcript isoform quantification, we ran ESPRESSO and five other contemporary tools [LIQA (24), NanoCount (25), FLAIR (23), StringTie2 (32), and FLAMES (33)] on ONT direct RNA and 1D cDNA sequencing data of SIRVs. We compared transcript isoform read counts reported by each tool against known SIRV transcript concentrations. As several of the five other tools require existing transcript annotations as their input, all six tools were run in an annotation-guided setting. For direct RNA-seq data of SIRVs, ESPRESSO outperformed all five other tools in terms of quantification accuracy, as evidenced by the correlation between known SIRV transcript concentrations and quantifications by individual tools (Fig. 2D). ESPRESSO yielded SIRV transcript abundance estimates that had a Pearson’s correlation of 0.914 and a Spearman’s correlation of 0.940 with ground-truth spike-in concentrations. By contrast, the other five tools yielded estimates that had Pearson’s correlation values ranging from 0.400 to 0.874 and Spearman’s correlation values ranging from 0.373 to 0.889 and tended to show strong quantification biases for several SIRV transcript isoforms. ESPRESSO also exhibited a lower bias and variability in transcript quantification relative to other tools. For example, the coefficient of variation in estimated abundances of the 19 lowest-concentration SIRV transcript isoforms (spike-in concentration of 1/32 fmol/μl) was 89% for ESPRESSO compared to 151% for NanoCount, which was the second-best performer in this evaluation but was designed to quantify only annotated transcript isoforms. Similarly, we found that ESPRESSO outperformed all five other tools when using 1D cDNA sequencing data for the same SIRV RNAs (fig. S1B). Together, ESPRESSO achieves a higher accuracy in transcript isoform quantification for the two most popular ONT RNA-seq protocols (1D cDNA and direct RNA), as compared to other contemporary tools.
Having evaluated ESPRESSO for long-read RNA-seq-based transcript quantification, we also compared the performance of ESPRESSO to that of short-read RNA-seq-based transcript quantification. Specifically, we analyzed a publicly available short-read RNA-seq dataset on the same set of SIRV RNAs (Materials and Methods) using StringTie2, which can perform transcript isoform analysis using either short or long RNA-seq reads. We found that StringTie2 abundance estimates for SIRV transcripts using short-read RNA-seq data were not as well correlated with ground-truth spike-in concentrations (Pearson’s correlation of 0.687 and Spearman’s correlation of 0.588) compared to StringTie2 abundance estimates using ONT 1D cDNA sequencing data (Pearson’s correlation of 0.755 and Spearman’s correlation of 0.719) (fig. S3). This observation is consistent with the expectation that long-read RNA-seq can quantify transcript isoforms more accurately. Notably, compared to StringTie2, ESPRESSO generated SIRV transcript abundance estimates that had an even higher correlation with ground-truth spike-in concentrations (Pearson’s correlation of 0.777 and Spearman’s correlation of 0.920) (fig. S3).
Last, we performed an additional benchmark evaluation of ESPRESSO and other contemporary tools using simulated ONT RNA-seq datasets, in which we knew the ground-truth set of transcripts that were present and their abundance levels. Specifically, we used NanoSim (34) to simulate ONT direct RNA and 1D cDNA reads of varying sequencing depths (0.5 million, 1 million, 3 million, and 5 million) based on transcript abundance levels, read length distributions, and error profiles observed in our real ONT RNA-seq data on HEK293T cells. Across all simulated ONT RNA-seq datasets, ESPRESSO consistently demonstrated the best performance in transcript isoform discovery compared to StringTie2 and FLAMES, as reflected by the sensitivity, precision, and F1 score (fig. S4). Consistent with our expectation, the sensitivity of transcript isoform discovery improved with increasing sequencing depth for all three tools. Notably, ESPRESSO’s precision in transcript isoform discovery (direct RNA, 0.84 to 0.89; 1D cDNA, 0.93 to 0.95) was much higher than that of StringTie2 (direct RNA, 0.65 to 0.80; 1D cDNA, 0.60 to 0.77) and FLAMES (direct RNA, 0.64 to 0.75; 1D cDNA, 0.69 to 0.78). Furthermore, we evaluated the accuracy of transcript isoform quantification using our simulated ONT direct RNA and 1D cDNA sequencing datasets. On the basis of the correlation of transcript abundance estimates with ground-truth transcript abundance levels, ESPRESSO consistently outperformed LIQA, NanoCount, FLAIR, and FLAMES and was a close second or comparable to StringTie2 (figs. S5 and S6).
Comparison of three nanopore RNA-seq protocols
In addition to direct RNA and 1D cDNA sequencing, the R2C2 protocol is a recently described alternative strategy for long-read RNA-seq on the ONT platform (14). Specifically designed for cDNAs, R2C2 uses RCA of circularized cDNA templates to generate long concatemeric sequences that can be subsequently processed into consensus sequences with improved base accuracy. To investigate how different ONT RNA-seq protocols affect transcript isoform analysis, we generated data on three human cell lines (PC3E, GS689, and HEK293T) using three protocols [1D cDNA, direct RNA, and linear RCA (LRCA), which is an adapted version of the R2C2 protocol; Materials and Methods] and analyzed the resulting data using ESPRESSO.
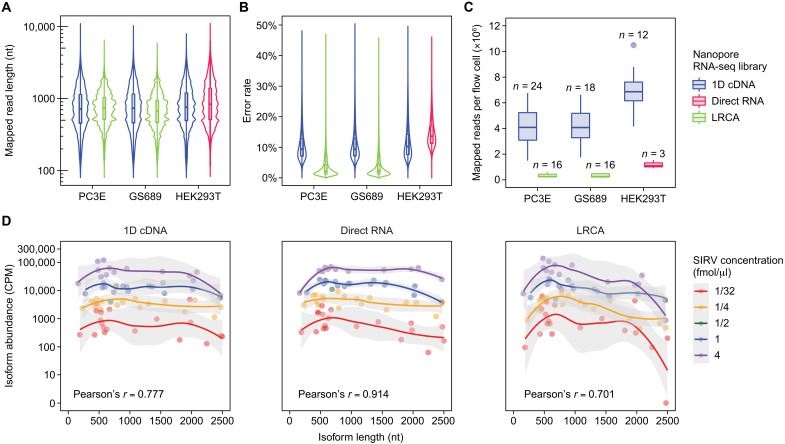
We first assessed basic library statistics such as read length, base error rate, and library yield. Length distributions of mapped raw reads (or consensus reads for LRCA) were comparable across the three protocols (Fig. 3A). As expected, the base error rate was lowest for LRCA libraries (1.3 to 4.4%) and highest for direct RNA-seq libraries (11.3 to 16.9%), while 1D cDNA libraries had an intermediate base error rate (7.3 to 13.2%) (all values shown represent interquartile ranges) (Fig. 3B). However, the improved base accuracy of LRCA relative to 1D cDNA and direct RNA came at the expense of having a significantly lower library yield per flow cell (Fig. 3C). While we obtained an average of 4.7 million mapped reads per flow cell for 1D cDNA libraries, the average number of mapped consensus reads per flow cell was only 0.3 million for LRCA, even fewer than that of direct RNA (1.2 million).
Fig. 3. Comparison of three ONT RNA-seq library types.
(A and B) Violin plots showing distributions of (A) mapped read length and (B) base error rate for ONT 1D cDNA, direct RNA, and linear RCA (LRCA) libraries prepared from PC3E (1D cDNA and LRCA), GS689 (1D cDNA and LRCA), and HEK293T (1D cDNA and direct RNA) cell lines. (C) Box-and-whisker plots (median and interquartile range) showing number of mapped reads per flow cell (×106) for the same ONT 1D cDNA, direct RNA, and LRCA libraries in (A) and (B). The number of flow cells per combination of library type and cell line is shown above each box-and-whisker plot. (D) Scatterplots showing abundance estimates for 68 Spike-In RNA Variant (SIRV) transcripts as a function of transcript length for ONT 1D cDNA, direct RNA, and LRCA libraries. SIRVs were spiked into 1D cDNA and LRCA libraries of PC3E cell lines as well as direct RNA libraries of HEK293T cell lines. Local regression curves were fitted to groups of points corresponding to transcripts with the same SIRV concentration using the geom_smooth function in R (v4.0.3). The gray area represents a 95% confidence interval for predictions from each regression curve. Pearson’s correlations between SIRV concentration and estimated abundance for SIRV transcripts are shown.
Using data on SIRV synthetic RNAs spiked into these libraries, we next investigated how different ONT RNA-seq protocols affect the accuracy of transcript isoform quantification. Transcript abundances estimated by ESPRESSO using direct RNA-seq data had the highest correlation with known SIRV transcript concentrations (0.914), likely attributed to the fact that the direct RNA-seq protocol is free of RT and PCR amplification biases (30, 31). 1D cDNA data yielded an intermediate correlation with known SIRV transcript concentrations (0.777), while the correlation was the lowest for LRCA data (0.701). In addition, transcript abundances estimated from LRCA data exhibited a strong length-dependent bias with respect to transcript length. SIRV transcript isoforms have lengths from 161 nucleotides (nt) to 2498 nt. Leveraging this diverse length distribution, we examined the estimated transcript abundances of all 68 SIRV transcripts as a function of transcript length. Among the three library types, LRCA exhibited the most pronounced length bias for estimated transcript abundances (Fig. 3D), as confirmed by an analysis of variance (ANOVA) test comparing the fits of linear regression models on estimated SIRV transcript abundances with or without transcript length considered (P = 0.016 for LRCA, P = 0.11 for 1D cDNA, and P = 0.36 for direct RNA; table S2). This observed length bias did not appear to be a monotonic function of transcript length, as transcripts shorter than 700 nt or longer than 2000 nt appeared to have the most pronounced biases (Fig. 3D). It is possible that additional circularization and size selection steps during LRCA library preparation may be responsible for the apparent length-dependent bias in transcript abundance estimation.
Transcriptome-wide analysis of transcript isoforms in three human cell lines
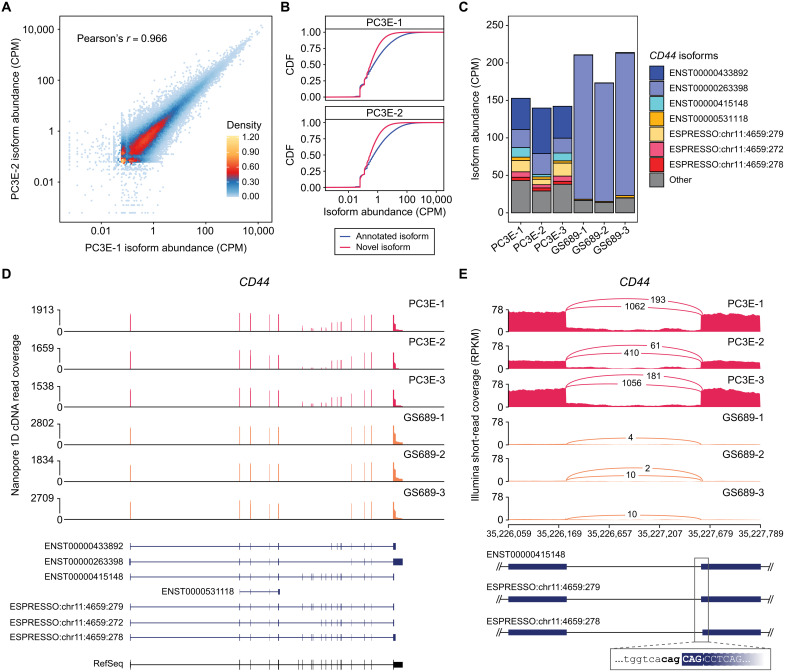
Because the 1D cDNA protocol generates by far the highest library yield per flow cell (Fig. 3C) and is arguably the default ONT RNA-seq protocol, we generated deep 1D cDNA sequencing data on three human cell lines (PC3E, GS689, and HEK293T), with a total of 430 million reads from 54 flow cells (table S3). To assess how sequencing depth affects gene and transcript isoform discovery, we performed a saturation analysis by running ESPRESSO on the full or randomly downsampled data. The numbers of annotated genes and annotated transcript isoforms discovered were trending saturation in all three cell lines, ranging from 24,594 to 25,279 genes and 79,976 to 80,048 transcript isoforms on the full data. By contrast, the number of novel transcript isoforms discovered (82,998 to 90,314 on the full data) was still far from saturation and higher than the number of annotated transcript isoforms discovered (fig. S7 and table S4). These results suggest that a large number of novel transcript isoforms remain to be discovered with even deeper sequencing of these cell lines. The estimated transcript abundances were highly correlated across biological replicates (n = 3) of the same cell line. For example, Pearson’s correlation values ranged from 0.97 to 0.99 for PC3E replicate pairs and GS689 replicate pairs (Fig. 4A and figs. S8 and S9). Similar pairwise correlation values were observed when applied to annotated transcript isoforms (0.97 to 0.99) or novel transcript isoforms (0.95 to 0.99).
Fig. 4. Discovery and quantification of transcript isoforms from deep ONT 1D cDNA data of PC3E and GS689 cell lines.
(A) Density plot comparing estimates of isoform abundance between two PC3E replicates, PC3E-1 and PC3E-2. (B) Cumulative distribution function (CDF) plots showing distributions of isoform abundance for annotated transcript isoforms (blue) or novel transcript isoforms (red) discovered in the PC3E-1 and PC3E-2 replicates. (C) Stacked barplot showing estimated abundances of CD44 transcript isoforms across three PC3E (PC3E-1, PC3E-2, and PC3E-3) and three GS689 (GS689-1, GS689-2, and GS689-3) replicates. Isoforms with CPM ≥ 3 in all three replicates of at least one cell line are represented individually, while the remaining isoforms were grouped together into an “Other” category. (D) ONT 1D cDNA read coverage tracks for three PC3E and three GS689 replicates, showing alignments of long reads that were uniquely assigned to the seven CD44 isoforms shown in (C) and classified as being FSM, NIC, or NNC. Transcript structures for the seven CD44 isoforms are displayed using blue boxes and lines. Displays for read coverage tracks and transcript structures were generated using Integrative Genomics Viewer (v2.8.0). (E) Sashimi plots based on Illumina short-read data for three PC3E and three GS689 replicates, showing an alternative 3′ splice site event involving an NAGNAG alternative splice acceptor site in CD44. This event distinguishes novel isoform ESPRESSO:chr11:4659:278 from annotated isoform ENST00000415148 and novel isoform ESPRESSO:chr11:4659:279. Sashimi plots were generated using rmats2sashimiplot (v2.0.2).
To further characterize novel transcript isoforms discovered from these human cell lines, we next compared the distributions of estimated transcript abundances between annotated and novel transcript isoforms. As expected, annotated transcript isoforms were consistently more abundant than novel transcript isoforms across all three PC3E replicates (Fig. 4B and fig. S8). For example, across the three PC3E replicates, 12.6 to 14.5% of annotated transcript isoforms that were discovered had CPM (counts per million) values > 10. By contrast, 1.7 to 2.4% of novel transcript isoforms that were discovered had CPM values > 10 (Fig. 4B and table S4). While more novel transcript isoforms were discovered compared to annotated transcript isoforms for each PC3E replicate, 83.5 to 86.1% of transcript isoforms with CPM > 10 were annotated. Among the PC3E replicates, a majority (86.6 to 87.0%) of genes had an annotated transcript isoform as the major transcript isoform (fig. S10). However, for genes in which the major transcript isoform’s proportion was <50%, 32.7 to 34.0% had a novel transcript isoform as the major transcript isoform. We observed a similar pattern in GS689 and HEK293T cells.
Using ESPRESSO, we discovered cell type-specific transcript isoforms involving complex alternative splicing patterns. For example, CD44 isoform switching is essential for epithelial-mesenchymal transition and involves changes in inclusion levels of nine variant exons within CD44 (35). Using ESPRESSO, we discovered a total of 52 CD44 transcript isoforms (20 annotated and 32 novel) across replicates of PC3E and GS689 (n = 3 each), two prostate cancer cell lines with contrasting epithelial versus mesenchymal properties (36). Seven CD44 transcript isoforms (four annotated and three novel) were consistently discovered with CPM ≥ 3 in all replicates of either cell line. While the composition of CD44 transcript isoforms in all PC3E replicates appeared largely heterogeneous, CD44 expression in all GS689 replicates shifted almost entirely to a single, annotated transcript isoform, ENST00000263398, which lacks all nine variant exons (Fig. 4C). Using transcript isoform CPM values and raw long-read alignments, we found eight of the nine variant exons to be abundantly expressed across PC3E replicates, but none of the variant exons were detected in any of the GS689 replicates (Fig. 4D). This observation is consistent with previous short-read RNA-seq data of PC3E and GS689 cell lines (37). ESPRESSO was also able to differentiate and quantify CD44 transcript isoforms with highly similar transcript structures at base resolution. For example, three CD44 transcript isoforms (annotated transcript isoform ENST00000415148 and novel transcript isoforms ESPRESSO:chr11:4659:279 and ESPRESSO:chr11:4659:278) that were consistently discovered across PC3E replicates share highly similar transcript structures but differ in combinations of NAGNAG alternative splice acceptor sites for two variant exons. Corresponding short-read RNA-seq data for the same PC3E replicates confirmed alternative splicing at these NAGNAG acceptor splice sites (Fig. 4E and fig. S11).
ESPRESSO enables accurate quantification of intron retention and other types of alternative splicing events
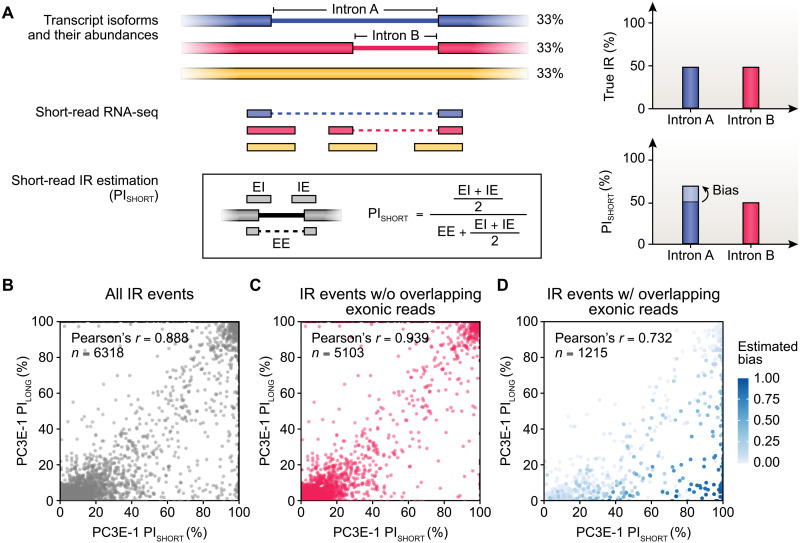
Intron retention (IR) is a form of alternative splicing that fine-tunes gene expression and regulates a variety of cellular processes, such as erythropoiesis and T cell activation (3). However, quantification of IR events using short-read RNA-seq data is challenging, especially in regions with complex alternative splicing patterns (38). An inherent limitation in quantifying IR using short-read RNA-seq data is that some reads counted as deriving from retained introns may originate from overlapping transcripts, such as transcripts with alternative splice donor or acceptor sites within an intron of interest. For example, in the toy example in Fig. 5A, IR estimation for intron A could be inflated by short RNA-seq reads originating from the transcript isoform that uses an alternative splice site to splice out intron B. Long RNA-seq reads can connect multiple alternative splicing events and span entire introns, potentially improving quantification of IR events.
Fig. 5. Long-read RNA-seq improves quantification of intron retention events.
(A) Intron retention (IR) estimation using short-read data may be biased when transcript isoforms with overlapping exonic structures are present. The true IR level should be estimated as the number of reads with the intron fully retained, divided by the number of reads with the intron either fully retained or fully spliced out. For example, consider three equally abundant transcript isoforms. Introns A and B are retained at the same frequency. Intron B retention level is correctly estimated using short reads. However, short-read-based estimate of intron A retention is confounded by the exonic read from the second transcript isoform (red) because this read overlaps with the exon-intron junction of the first transcript isoform (blue). This creates a discrepancy/bias between short-read-based measurement of intron A retention and its true retention level (PI, percent of intron retention; EI/IE, exon-intron/intron-exon junction reads; EE, exon-exon junction reads). (B to D) Comparison of PI values using short versus long reads for (B) all IR events, (C) IR events without overlapping exonic reads, and (D) IR events with overlapping exonic reads detected in a PC3E replicate. Long-read-based PI values are calculated as the number of long reads that retain an intron, divided by the number of long reads with the intron either fully retained or fully spliced out. Points in (D) are colored on the basis of estimated bias, calculated as the difference between observed long-read PI values and biased long-read PI values, which are calculated in a manner that mimics PI value estimation using short-read data (Materials and Methods).
To quantify IR events detected from long-read RNA-seq data on PC3E and GS689 cell lines, we computed the percent of IR (PI), which is the proportion of reads fully retaining an intron among reads in which the intron is either fully spliced or retained (Materials and Methods). For IR events detected in each of the six samples (n = 3 per cell line), PI values calculated from long-read RNA-seq data were overall highly correlated (0.888 to 0.919) with PI values calculated from corresponding short-read RNA-seq data (fig. S12). However, the degree of correlation differed substantially for subsets of IR events, depending on whether the intron of interest overlapped with the exonic region of an overlapping transcript discovered from long-read RNA-seq data. For example, 6318 IR events were detected in a PC3E replicate (PC3E-1), and the PI values calculated from corresponding long-read and short-read data had an overall Pearson’s correlation of 0.888 (Fig. 5B). Of these IR events, 5103 had no overlapping exonic regions of overlapping transcripts, and the PI values calculated from corresponding long-read and short-read data had an even higher Pearson’s correlation of 0.939 (Fig. 5C). By contrast, 1215 IR events detected in this PC3E replicate had overlapping exonic regions of overlapping transcripts. For these IR events, the estimated PI values had a much lower Pearson’s correlation of 0.732 between the long-read and short-read data, with generally higher PI values estimated from short-read data compared to long-read data (Fig. 5D). A consistent pattern was observed for all other PC3E/GS689 replicates (fig. S12). To investigate the potential reason for this lower correlation, for this subset of IR events, we treated long reads supporting overlapping exonic regions of overlapping transcripts as reads supporting the retention of the introns of interest, to calculate a “biased” long-read-based PI value in a manner that mimicked PI value estimation using short-read data (Fig. 5A). Notably, such biased long-read PI values had a much higher correlation (0.943) with short-read PI values (fig. S13). Collectively, these results indicate that long-read RNA-seq data can generate a more reliable quantification of IR events, particularly in regions with overlapping transcripts and complex alternative splicing patterns.
We also examined, more broadly, how long-read RNA-seq-based quantification of other types of alternative splicing events, such as exon skipping and alternative 5′ and 3′ splice site usage, compares to that of short-read RNA-seq. To this end, we estimated the percent spliced in (PSI) values of alternative splicing events detected in PC3E and GS689 cell lines using short-read and long-read RNA-seq data, separately (Materials and Methods). For exon skipping events detected in each of the six samples (n = 3 per cell line), PSI values calculated from long-read data overall agreed well with PSI values calculated from corresponding sample-matched short-read data (Pearson’s correlation between 0.873 and 0.891) (fig. S14, A and B). On the other hand, the correlation between short-read- and long-read-based PSI values was even higher for alternative 5′ splice site usage events (Pearson’s correlation between 0.941 and 0.952) and alternative 3′ splice site usage events (Pearson’s correlation between 0.934 and 0.944).
To further investigate why the correlation between short-read- and long-read-based PSI values was relatively lower for exon skipping events compared to alternative 5′ and 3′ splice site usage events, we noticed that short-read data often yielded higher PSI values for exon skipping events compared to long-read data (fig. S14, A and B). When we partitioned the exon skipping events based on whether they overlap other alternative splicing events (e.g., the cassette exon itself carries alternative 5′ or 3′ splice sites), the degree of correlation differed substantially for the two resulting subsets (fig. S14, C and D). For exon skipping events that do not overlap other alternative splicing events (i.e., “simple” exon skipping events), the Pearson’s correlation between short-read and long-read PSI values ranged between 0.964 and 0.972. In contrast, for exon skipping events that overlap with other alternative splicing events (i.e., “complex” exon skipping events), the correlation was much lower (Pearson’s correlation between 0.825 and 0.847). Notably, for simple exon skipping events, we observed that short-read data did not yield higher PSI values compared to long-read data, yet this pattern still remained for complex exon skipping events.
Last, we sought to investigate how the accuracy of long-read-based quantification of alternative splicing events changes with decreasing long-read RNA-seq depth. Given that short-read RNA-seq is considered the de facto approach for quantifying alternative splicing events (6, 39), we reasoned that the correlation between short-read- and long-read-based measurement of alternative splicing events can be used as a proxy for quantification accuracy. To this end, we randomly sampled between 1 and 100% of long RNA-seq reads generated for PC3E and GS689 cell lines, which were subjected to deep long-read RNA-seq (table S3), and used each set of randomly sampled long reads to quantify transcript isoforms and their associated alternative splicing events. For each of the six samples (n = 3 per cell line), we examined how the number of randomly sampled long reads affected the correlation between short-read and long-read PSI values for simple exon skipping events. We found that, when the number of sampled long reads was greater than 5 million, the correlation between short-read and long-read PSI values remained largely stable. However, when the number of sampled long reads dropped below 5 million, the degree of correlation rapidly declined (fig. S15).
A comprehensive catalog of transcript isoforms across 30 human tissues
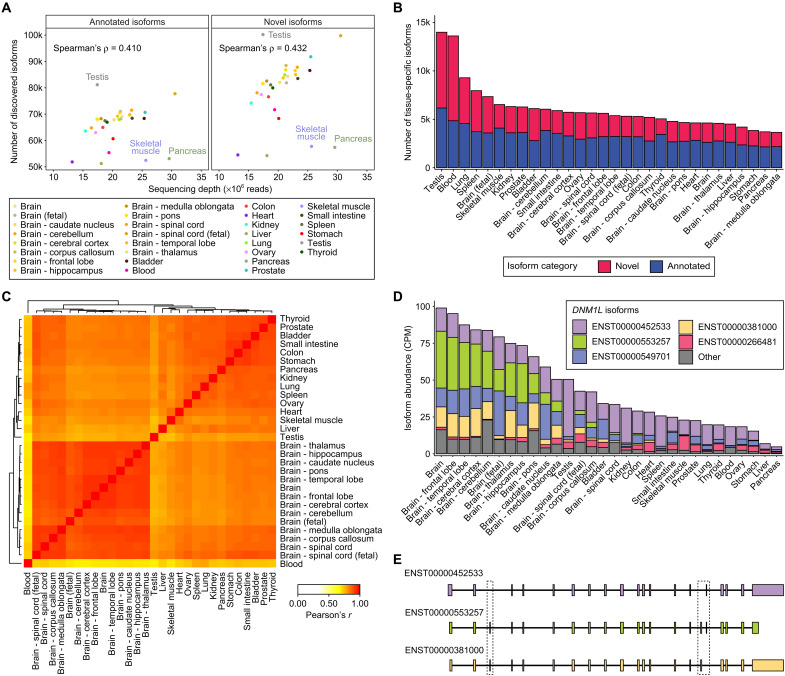
Last, we applied ESPRESSO to discover and quantify transcript isoforms from 30 human tissues representing diverse anatomical sites (table S5). Specifically, we generated a total of 623 million ONT 1D cDNA reads, with the number of reads per tissue ranging from 13.2 million to 30.6 million (table S6). Using ESPRESSO, we discovered a total of 340,149 transcript isoforms from our tissue datasets, including 141,640 annotated transcript isoforms and 198,509 novel transcript isoforms. For 270,932 transcript isoforms discovered from annotated genes, the average transcript length and exon number (1542 nt; 6.2 exons) matched well with those reported for annotated transcript isoforms in GENCODE (1569 nt; 6.0 exons; GENCODE v34) (40). The number of transcript isoforms discovered per tissue ranged from 105,500 in the liver to 181,352 in the testis. In general, tissues with higher RNA-seq depth tended to have more transcript isoforms (both annotated and novel), but a few tissues were outliers (Fig. 6A). For example, the testis had the largest number of transcript isoforms, but also had a relatively low RNA-seq depth (17.4 million reads) compared to other tissues. By contrast, skeletal muscle and pancreas had a high RNA-seq depth but a small number of transcript isoforms. In each tissue, we found more novel than annotated transcript isoforms. However, across all tissues, annotated transcript isoforms had higher RNA-seq read support compared to novel transcript isoforms (fig. S16), consistent with the pattern observed on the cell line data (Fig. 4B and figs. S8 and S9).
Fig. 6. Characterization of transcript isoforms across 30 human tissues.
(A) Scatterplots showing the Spearman’s correlation between ONT 1D cDNA sequencing depth (×106 reads) and the number of discovered transcript isoforms that are annotated (left column) or novel (right column) for 30 human tissues (14 brain tissues and 16 nonbrain tissues). (B) Stacked barplot showing the number of tissue-specific transcript isoforms [false discovery rate (FDR) < 1%, Materials and Methods] that are annotated (blue) or novel (red) for each of the 30 human tissues. (C) Heatmap displaying pairwise Pearson’s correlations in isoform proportions for tissue-specific transcript isoforms across 30 human tissues. Hierarchical clustering was applied to the correlation matrix to group similar tissues. The correlation heatmap with hierarchical clustering was generated using the heatmap.2 function from the gplots package (v3.1.1) on R (v4.1.0). (D) Stacked barplot showing estimated abundances for transcript isoforms of DNM1L discovered across 30 human tissues. Five DNM1L transcript isoforms with the highest average CPM across 30 human tissues are displayed individually, while the remaining transcript isoforms were grouped together into an Other category. (E) Transcript structures of three DNM1L isoforms—ENST00000452533 (purple), ENST00000553257 (green), and ENST00000381000 (yellow)—displayed in a 5′ to 3′ orientation. Boxes are drawn around transcript regions involving alternative splicing events.
A useful feature of our tissue dataset is that it allowed us to examine tissue-specific differences in transcript isoform composition for any gene across 30 human tissues. For genes containing at least two transcript isoforms, we first identified genes with significant shifts in transcript isoform composition in at least one tissue relative to all tissues and subsequently identified specific transcript isoforms responsible for the observed shifts in a given tissue (Materials and Methods). Using this approach, we found a total of 75,111 transcript isoforms exhibiting patterns of tissue-specific inclusion, including 33,519 annotated transcript isoforms and 41,592 novel transcript isoforms. The number of transcript isoforms with tissue-specific inclusion ranged from 3650 in the medulla oblongata to 13,991 in the testis (Fig. 6B). Neither RNA-seq depth nor total number of transcript isoforms was correlated to the number of tissue-specific transcript isoforms identified in each tissue (fig. S17).
We found that the identified tissue-specific transcript isoforms reflect the biological relationships among these 30 human tissues. Specifically, when tissues were clustered using proportions of tissue-specific transcript isoforms, they were largely grouped into clusters reflecting their biological relationships, such as brain tissues and hollow organs (e.g., stomach, intestines, and bladder) (Fig. 6C). Whole blood and testis were both different from the rest of the tissues and from each other, as indicated by the heights of dendrogram branches. A similar clustering pattern was obtained when tissues were clustered using proportions of all transcript isoforms (fig. S18). Functional characterization of tissue-specific transcript isoforms underlying the observed clustering pattern can shed insight into how differential transcript isoform usage is important for tissue function and cellular identity. For example, we found two transcript isoforms of DNM1L (ENST00000553257 and ENST00000381000) that are preferentially used in brain tissues compared to other tissues (Fig. 6D). Relative to the major transcript isoform of DNM1L in nonbrain tissues (ENST00000452533), these brain-specific transcript isoforms involve different combinations of three cassette exons found in two distal regions that are separated by ~1.3 kb in the transcripts (Fig. 6E and fig. S19). More broadly, we found that transcript isoforms involving multiple combinations of alternative splicing events comprised a large category of tissue-specific transcript isoforms discovered across the 30 human tissues (fig. S20). We also observed that the relative proportions of alternative splicing event types associated with tissue-specific transcript isoforms appeared largely consistent across each of the human tissues, with the exception of testis, whose tissue-specific transcript isoforms showed a significant enrichment of alternative first exon usage events (binomial test of overrepresentation, P = 1.16 × 10−146). In total, we found 1910 testis-specific transcript isoforms involving alternative first exon usage, among which 485 transcript isoforms used an alternative first exon that was novel. We randomly selected 12 of these transcript isoforms and validated their novel alternative first exons by RT-PCR and Sanger sequencing (fig. S21).
DISCUSSION
Long-read RNA-seq has emerged as a powerful and increasingly popular technology for transcriptome analysis (9, 10). However, the high sequencing error rate of current long-read sequencing platforms (e.g., PacBio and ONT) can result in alignment artifacts posing as novel splice sites and, consequently, present a major challenge for accurate discovery and quantification of transcript isoforms (11, 12, 19). Existing methods for transcript isoform discovery from error-prone long-read RNA-seq data correct spurious alignments near splice sites by using SJs that are either present in existing transcript annotations or identified in highly accurate short-read RNA-seq data (22–25). However, in the absence of matching short-read RNA-seq data on the same biological samples, it has been difficult to reliably discover novel SJs and their corresponding transcript isoforms using long-read RNA-seq data alone.
Here, we report ESPRESSO, a new computational tool for discovering and quantifying transcript isoforms from error-prone long-read RNA-seq data. ESPRESSO is designed to reliably identify novel SJs and discover novel transcript isoforms using long-read RNA-seq data alone, without relying on short-read RNA-seq data. The core innovation of ESPRESSO lies in its ability to correct putative SJs found in individual long reads by borrowing information from other long reads aligned to the same genomic region. Notably, we observed from direct RNA and 1D cDNA sequencing data of SIRV synthetic RNAs that putative SJs found in long reads with perfect alignments around splice sites are much less likely to be incorrect (0.18 to 0.24%) compared to those with imperfect alignments (8.8 to 13.0%) (Fig. 1, A and B, and table S1). Motivated by this observation, we reasoned that, for any given gene, a set of high-confidence SJs could be reliably defined from a collection of raw long-read-to-genome alignments based on whether there are alignment errors around splice sites. While the proportion of putative SJs with perfect alignments around splice sites could vary among datasets (e.g., 15.4 to 38.6% among all putative SJs identified from the SIRV data; table S1), such SJs have a high reliability and could be used for correcting putative SJs found in individual long reads, circumventing the need to rely on matching short-read data to identify and verify novel SJs.
To this end, ESPRESSO first defines a set of high-confidence SJs from aligned long reads of a given gene. A putative SJ is defined as high-confidence if it is present in existing transcript annotations. If a putative SJ is novel, then ESPRESSO defines it as high-confidence based on several criteria, requiring that the SJ carries the canonical splice site dinucleotide motif (26) and that the number of supporting long reads with perfect alignments around splice sites exceeds a threshold (2 by default). Next, ESPRESSO uses this set of high-confidence SJs to determine the most optimal set of SJs for individual long reads based on each read’s alignment and error profile. After low-confidence SJs are corrected and missing SJs at alignment ends are recovered, ESPRESSO collapses full-length long reads into unique chains of high-confidence SJs representing distinct transcript isoforms, and the abundances of transcript isoforms are estimated using an EM algorithm.
The ESPRESSO computational workflow uses reference transcript annotations for transcript isoform analysis in multiple ways. First, before running ESPRESSO, users are recommended to provide reference transcript annotations when aligning long RNA-seq reads to a reference genome. The long-read RNA-seq aligner used by ESPRESSO is minimap2, arguably the most widely used long-read RNA-seq aligner (19). However, other more recently published long-read RNA-seq aligners [e.g., deSALT (41), 2passtools (42), and uLTRA (43)] exist, and in the future, it would be interesting to investigate whether using these newer alternative aligners could further improve the performance of ESPRESSO. Second, in the ESPRESSO algorithm, reference transcript annotations are used when defining high-confidence SJs from raw long-read-to-genome alignments. If a putative SJ found in a long read is annotated, then ESPRESSO classifies the SJ as high-confidence by default. Third, reference transcript annotations are also used to check whether a putative transcript isoform discovered from long-read RNA-seq data is annotated or novel. Novel transcript isoforms (i.e., not present in reference transcript annotations) are required to have at least two perfectly aligned reads supporting each SJ.
Using real ONT RNA-seq data of SIRV synthetic RNAs, which have known transcript structures and concentrations, as well as a human cell line (HEK293T), we systematically assessed the performance of ESPRESSO for transcript isoform analysis. We demonstrate that, in an annotation-free setting, ESPRESSO improves both the accuracy and completeness of transcript structures discovered from long RNA-seq reads, over the raw long-read-to-genome alignments (Fig. 2B). Furthermore, on ONT direct RNA and 1D cDNA sequencing data, we show that, for transcript isoform discovery, ESPRESSO outperforms two existing state-of-the-art tools that can also discover novel transcript isoforms using long-read RNA-seq data alone (Fig. 2C and figs. S1A and S2). Last, we demonstrate that, in an annotation-guided setting, ESPRESSO achieves the best accuracy in transcript isoform quantification, as compared to five recently published tools for long-read RNA-seq transcript quantification (Fig. 2D and fig. S1B).
By running ESPRESSO on data generated from different ONT RNA-seq protocols, we obtained insights into how differences in library preparation protocols influence transcript isoform analysis (Fig. 3). We observed that, compared to 1D cDNA and direct RNA libraries, LRCA libraries had the lowest base error rate, which is consistent with the expectation that CCS generates consensus reads with a higher base accuracy (13, 14). However, the improved base accuracy of LRCA libraries came at the expense of having a substantially lower library yield, as well as a significant length-dependent bias and the lowest accuracy in transcript quantification. By contrast, both 1D cDNA and direct RNA libraries had a substantially higher base error rate, but they also had a substantially higher library yield and more accurate transcript quantification. Among the three protocols, the 1D cDNA protocol generated the highest library yield while the direct RNA protocol achieved the highest quantification accuracy. The superior performance of the direct RNA protocol for transcript quantification likely reflects the fact that this protocol is free of RT and PCR amplification biases (30, 31). However, the higher error rate of direct RNA libraries is a known issue for transcript isoform discovery (30). Thus, ESPRESSO’s ability to reliably discover and quantify transcript isoforms from data generated by 1D cDNA and direct RNA protocols, which are much more cost-effective (in terms of library yield), but also more error-prone (in terms of base error rate), is expected to benefit future long-read RNA-seq studies of eukaryotic transcriptomes.
We demonstrate that ESPRESSO enables accurate quantification of IR using long-read RNA-seq data (Fig. 5). As a specific mode of alternative splicing, IR has recently emerged as an important mechanism for fine-tuning transcript and protein products in a tissue-specific or developmentally regulated manner (2, 3). While short-read RNA-seq data have been the primary source for IR analysis (38, 44), an inherent limitation is that introns in mammalian genes are large (20) such that individual short RNA-seq reads cannot cover the entirety of most introns. Consequently, some short RNA-seq reads counted as deriving from retained introns may originate from overlapping transcripts, such as transcripts with alternative donor or acceptor splice sites within an intron of interest (38). By contrast, long-read RNA-seq data have an inherent advantage for IR analysis, as they can provide direct evidence of full-length transcripts corresponding to intron-retained or intron-spliced isoforms. By comparing long-read and short-read RNA-seq data generated on the same biological samples, we provide evidence that long-read RNA-seq data can generate more reliable quantifications of IR events, particularly in regions with overlapping transcripts and complex alternative splicing patterns. Notably, the apparent discrepancy in IR quantification between long-read and short-read RNA-seq data for a certain subset of introns can be explained by modeling confounding short-read RNA-seq signals from overlapping transcripts (Fig. 5 and fig. S13).
More broadly, long-read and short-read RNA-seq data overall agree well with each other in quantifying other types of alternative splicing events, such as alternative 5′ and 3′ splice site usage as well as a subset of exon skipping events in which the cassette exon does not overlap other alternative splicing events (i.e., simple exon skipping events) (fig. S14). However, for exon skipping events that overlap other alternative splicing events (i.e., complex exon skipping events), we observed a general tendency for short-read RNA-seq data to yield higher estimates of cassette exon inclusion levels (PSI values) compared to long-read RNA-seq data. It is possible that, for complex exon skipping events, short-read data may yield overestimates of PSI values. Individual short reads are typically not long enough to span both upstream and downstream inclusion junctions associated with an exon skipping event, so short reads covering at least one inclusion junction are typically used for quantifying PSI values. However, for complex exon skipping events, the inclusion junctions may be shared with other overlapping alternative splicing events, which can inflate the number of short reads supporting cassette exon inclusion. By contrast, long reads can provide direct evidence for whether or not a cassette exon is included or not, suggesting that long-read RNA-seq data can provide a more unbiased quantification of complex exon skipping events. This pattern appears to mirror the observation that we made for IR, in which we found that long-read RNA-seq data can yield more reliable quantifications of IR events, particularly in regions with overlapping transcripts and complex alternative splicing patterns.
As low sequencing throughput remains a bottleneck for long-read sequencing technologies (45), an important question is what is the minimum sequencing depth recommended for using long-read RNA-seq to study transcript isoform variation and alternative splicing. Our analysis of short-read and long-read RNA-seq data generated on the same human samples revealed that 5 million long reads represent the threshold below which the correlation between long-read versus short-read quantification of alternative splicing rapidly declines (fig. S15). On the basis of this observation, we would recommend generating at least 5 million long RNA-seq reads for profiling transcript isoform variation and alternative splicing in a human sample. Using read length distributions observed across the long-read RNA-seq datasets generated in this work, we estimate that the “information content” of 5 million long reads corresponds to 22.7 to 25.4 million short-read pairs, assuming a read length of 2 × 101 base pairs for short-read RNA-seq. This sequencing depth is comparable to the short-read RNA-seq depth of the ENCODE phase III project that studied alternative splicing and its regulation by RNA binding proteins (46).
Note that while the SJ correction and recovery procedure in ESPRESSO improves transcript isoform discovery and quantification, it represents a computational bottleneck in the overall workflow, as each individual long read needs to be realigned to high-confidence SJs of the corresponding gene (Materials and Methods). To improve computational efficiency and reduce runtime, we have now implemented ESPRESSO to support multiple threads, such that reads from nonoverlapping genomic regions of the same gene can be processed in parallel during SJ correction and recovery. In addition, individual samples in a large dataset can be processed in parallel. These optimizations led to several times speed-up when running ESPRESSO on an HPC cluster and are integrated in the latest pre-release of ESPRESSO (“version 1.3.0-beta”), which is publicly available at https://github.com/Xinglab/espresso/releases/tag/v1.3.0-beta. Notably, to maintain consistency, all analyses conducted here were done using the original (slower) version of 1.2.2, which remains the latest official release of ESPRESSO and is publicly available at https://github.com/Xinglab/espresso. Moreover, to facilitate easy use of ESPRESSO, we also provide a Snakemake pipeline that covers all of the main steps in ESPRESSO and upstream preprocessing steps, such as basecalling and read alignment.
In summary, we have developed ESPRESSO, a new computational tool for discovering and quantifying transcript isoforms using error-prone long-read RNA-seq data. We assessed the performance and demonstrated the utility of ESPRESSO using extensive data on synthetic and biological samples. We also generated and analyzed ~1.1 billion ONT RNA-seq reads covering 30 diverse human tissues and three human cell lines, providing a useful data resource for studying human transcriptome variation at the level of full-length transcript isoforms. Given the increasingly wide adoption of long-read RNA-seq in biomedical research, we envision that ESPRESSO will be a useful tool for researchers to explore the RNA repertoire of eukaryotic cells in diverse settings.
MATERIALS AND METHODS
Cell lines
All cells were grown at 37°C in a humidified chamber with 5% CO2. Low-passage HEK293T cells [American Type Culture Collection (ATCC), Manassas, VA, #CRL-3216] were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, #11-885-084) supplemented with 10% fetal bovine serum (FBS; Corning, #35-010-CV), 1% GlutaMAX (Gibco, #35050061), and penicillin-streptomycin (100 U/ml; Gibco, #15140122). Human prostate cancer cell lines PC3E and GS689 (gifts of M. D. Henry, University of Iowa) were generated as previously described (36, 47). Briefly, the PC3E cell line was derived from the PC-3 cell line (ATCC, #CRL-1435) by isolating E-cadherin-positive cells by flow cytometry. E-cadherin-positive PC-3 cells (PC3E cells) were confirmed to have an epithelial-like phenotype in subsequent assays (36, 47). The GS689 cell line, a metastatic variant of the PC-3 cell line, was isolated from a secondary metastatic liver tumor by in vivo passaging of PC-3 cells in SCID mice. Low-passage PC3E and GS689 cells were maintained in DMEM/F-12 (Gibco, #11330032) with 10% FBS, 1× nonessential amino acids (Gibco, #11140050), and G418 (400 μg/ml; Gibco, #10131035). The identity of each cell line was validated by short tandem repeat analysis. All cells were examined and confirmed to be negative for mycoplasma.
RNA extraction and preparation
SIRV-Set 1-E2 (Lexogen, lot no. 001418) consists of 68 artificial transcripts from seven model genes with concentrations that cover more than two orders of magnitude. The difference in transcript number for this lot compared to the Lexogen website (68 versus 69 transcripts) was due to the exclusion of SIRV108 during the SIRV production quality assurance process, as per the manufacturer’s communications (“Amendment for SIRV-Set 1 Lot No. 00141N,” Lexogen, Jun 2017). RNA purity and individual concentrations of SIRVs were verified by the manufacturer. Total RNA was extracted from HEK293T, PC3E, and GS689 cell lines using TRIzol reagent (Invitrogen, #15596018), according to the manufacturer’s instructions. RNA concentrations and RNA integrity were measured by NanoDrop 2000 Spectrophotometer and Agilent 4200 TapeStation respectively. Poly(A) + RNA (5 μg) from 30 human tissue samples (table S5) was purchased from Clontech. Most tissue RNA samples were isolated from pooled tissues of multiple donors. Poly(A) + RNA quality (size, 0.2 to 10 kb) was confirmed by denaturing gel electrophoresis, as indicated by the manufacturer.
Direct RNA library construction and nanopore sequencing
A 20-μg aliquot of total RNA extracted from HEK293T cells was subjected to poly(A) + RNA selection using the Dynabeads mRNA DIRECT purification kit (Invitrogen, #61011), in accordance with the manufacturer’s instructions. Approximately 500 ng of the resulting poly(A) + RNA, along with 25 ng of SIRV-Set 1-E2 RNA, was pooled in one tube as input for direct RNA library generation. Libraries were made by following the standard SQK-RNA002 protocol with the optional RT step included. All libraries were loaded onto R9.4.1 flow cells (ONT) and sequenced on MinION/GridION devices.
cDNA synthesis
Templates for cDNA synthesis included the following: (i) 200 ng of total RNA extracted from PC3E cells, together with 100 pg of SIRV-Set 1-E2 RNA; (ii) 500 ng of total RNA extracted from either the HEK293T, PC3E, or GS689 cell line; or (iii) 50 ng of Poly(A) + RNA from one of the 30 human tissues. The cDNA synthesis process followed the SMART-seq2 protocol (48) with some modifications. Briefly, the RT and template-switching reaction was performed with Maxima H minus reverse transcriptase (Thermo Fisher Scientific, #EP0751) under the following conditions: 42°C for 90 min, followed by 85°C for 5 min. PCR amplification of first-strand cDNA using the KAPA HiFi ReadyMix (KAPA Biosystems, #KK2602) was performed by incubating the mixture at 95°C for 3 min, followed by 13 to 15 cycles of (98°C for 20 s, 67°C for 20 s, and 72°C for 4 min), with a final extension at 72°C for 5 min. A 13-cycle PCR was performed for cell line and tissue RNA samples, whereas a 15-cycle PCR was performed for RNA samples with SIRVs. PCR products were treated with Exonuclease I [New England Biolabs (NEB), #M0293] to remove unused primers and then purified using 0.8× volumes of SPRIselect beads (Beckman Coulter, #B23318). Amplified cDNA was measured by using the Qubit double-stranded DNA (dsDNA) High Sensitivity assay and Agilent High Sensitivity D5000 ScreenTape assay on a 4200 TapeStation. Sequences for all oligos/primers are detailed in table S7.
1D library construction and nanopore sequencing
We constructed 1D libraries using 1 μg of amplified cDNA according to the standard SQK-LSK109 protocol. Briefly, cDNA products were end-repaired and dA-tailed using the NEBNext Ultra II End Repair/dA-Tailing Module (NEB, #E7546) by incubating at 20°C for 20 min and 65°C for 20 min. End-repaired cDNA was purified with 1× volume of AMPure XP beads (Beckman Coulter, #A63881) and eluted in 60 μl of nuclease-free water. Adapter ligation was performed using NEBNext Quick T4 DNA ligase (NEB, #E6056) at room temperature for 10 min. After ligation, libraries were purified using 0.45× volumes of AMPure XP beads and short fragment buffer to enrich all fragments equally. The final libraries were loaded onto R9.4.1 or R10.3 flow cells (ONT) and sequenced on MinION/GridION devices for 72 hours. Sequencing statistics, including library information, chemistry, flow cell types, and sequencing output, are detailed in tables S3 and S6.
LRCA library construction and nanopore sequencing
The LRCA protocol is an adapted version of the R2C2 method (14) with the following modifications:
1) First-strand cDNA synthesis. The cDNA was generated in a 20-μl reaction by using the abovementioned cDNA synthesis protocol. Additional reverse-transcribed oligo(dT) primers were digested with 1 μl of Exonuclease I at 37°C for 20 min and 80°C for 10 min. The 3′ end of cDNA was protected from digestion by the hybrid structure of the cDNA and template-switching oligo.
2) Labeling cDNA with unique molecular identifier (UMI) sequences. After RT, a UMI sequence was added to the 3′ end of cDNA by the strand extension reaction using Phusion HiFi DNA polymerase (NEB, #M0530), a template oligo including 60 random bases of C, G, and T (B60), and a 3′ 3C-spacer. The reaction was performed by incubating the mixture at 95°C for 3 min, followed by 5 cycles of (98°C for 15 s, 67°C for 15 s, and 72°C for 15 s), with a final extension at 72°C for 5 min.
3) PCR preamplification of cDNA. UMI-coded cDNA products were aliquoted into multiple tubes and PCR-amplified using the KAPA HiFi ReadyMix by incubating at 95°C for 3 min, followed by 13 cycles of (98°C for 20 s, 67°C for 15 s, and 72°C for 4 min), with a final extension at 72°C for 5 min. PCR products were purified with 1× volume of AMPure XP beads.
4) Circularization of cDNA. Purified cDNA was denatured at 95°C for 5 min and circularized via template-mediated enzymatic ligation by incubating 1 μg of cDNA with 200 ng of template oligos and 2 μl (80 U) of Taq DNA ligase at 50°C for 60 min. The template oligo is composed of sequences that are complementary to the 5′- and 3′-end adapters (table S7). Linear DNA was digested using a mixture of Exonuclease I and III (NEB, #M0206) at 37°C for 4 to 12 hours and 80°C for 15 min.
5) LRCA reaction. To initiate LRCA, a universal oligo that anneals to the PCR adapter sequence of circular dsDNA generated in step 4 was used. The LRCA reaction was performed by incubating circular dsDNA products with 4 μl of Phi29 DNA polymerase (10 U/μl; NEB, #M0269), 8 μl of dNTPs (NEB, #N0447), 8 μl of universal oligo [10 μM; Integrated DNA Technologies (IDT)], 2 μl of bovine serum albumin (20 mg/ml; NEB, #M0269), and 20 μl of 10× Phi29 DNA polymerase buffer (NEB, #M0269) in a 200-μl reaction at 30°C for 16 hours and 65°C for 15 min. LRCA products were purified with 0.7× volumes of SPRIselect beads.
6) Second-strand DNA synthesis and DNA shearing. Second-strand synthesis of LRCA products was performed using Phusion DNA polymerase and Taq DNA ligase (NEB, #M0208) by the gap-filling and ligation reaction at 50°C for 60 min. The dsDNA sequences were then sheared by g-TUBE (Covaris, #520079) to the desired size (around 8 to 10 kb) by following the vendor’s instructions.
7) Library construction and sequencing. Sheared DNA products were purified with 0.5× volumes of SPRIselect beads and subjected to 1D library construction by following the standard SQK-LSK109 protocol. Sequences of all oligos/primers are detailed in table S7. Sequencing statistics, including library information, chemistry, flow cell types, and sequencing output, are detailed in table S3.
RT-PCR and Sanger sequencing validation of novel alternative first exons
cDNA was synthesized from 20 ng of human testis poly(A) + RNA using random hexamer primed RT as described in the Maxima H minus reverse transcriptase protocol. Next, PCR was performed in a 50-μl reaction by using first-strand cDNA synthesized from testis poly(A) + RNA, 25 μl of the KAPA HiFi ReadyMix, and 20 pmol of a primer pair. All primer pairs are listed in table S8. PCR amplification was carried out in a Veriti 96-well Thermal Cycler (Applied Biosystems, catalog no. 43-757-86) by incubating the mixture at 95°C for 2 min, followed by 28 cycles of (98°C for 20 s, 65°C for 20 s, and 72°C for 25 s) with a final extension at 72°C for 2 min. Amplified products were analyzed by 1.5% agarose gel electrophoresis and purified by using the QIAquick Gel Extraction Kit (Qiagen, catalog no. 28706X4). SJ sequences of novel alternative first exons were confirmed by Sanger sequencing of the purified PCR products.
Basecalling of raw nanopore sequencing data
Basecalling was performed in fast mode with Guppy (v3.6.0) for all nanopore sequencing data in this study (https://community.nanoporetech.com/downloads). Basecalling of direct RNA and cDNA libraries was done using config files rna_r9.4.1_70bps_fast.cfg and dna_r9.4.1_450bps_fast.cfg, respectively.
Alignment to the reference genome
Basecalled ONT direct RNA and 1D cDNA sequencing reads generated on SIRV synthetic RNAs (n = 3 per library type) were mapped to the SIRV reference genome using minimap2 (v2.17-r974-dirty), which was run with parameters “-ax splice --splice-flank=no -w 4 -k 14”, as recommended by the developer. When SIRV transcript annotation was used for read mapping, it was converted to BED format using the “paftools.js gff2bed” command (included as part of minimap2) and provided to minimap2 using the “--junc-bed” option. Basecalled ONT RNA-seq reads (i.e., 1D cDNA reads, direct RNA reads, and LRCA consensus reads) generated on human RNA samples were mapped to the GRCh37/hg19 reference genome using minimap2 (v2.17-r974-dirty) with parameters “-ax splice --splice-flank=no -w 4 -k 14” together with transcript annotations from GENCODE v34 (www.gencodegenes.org/human/release_34lift37.html).
A publicly available short-read RNA-seq dataset [Sequence Read Archive (SRA) accession ERR4330671] (49) that contained SIRV synthetic RNAs from SIRV-Set 1 E2 was aligned to the SIRV reference genome using STAR (v2.7.1a) (50) on two-pass mode with parameters “--alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --alignEndsType EndToEnd” together with SIRV transcript annotations. Short-read RNA-seq datasets for PC3E and GS689 cell lines, which were taken from a previous study (37), were mapped to the GRCh37/hg19 reference genome using STAR (v2.7.1a) on two-pass mode with parameters “--alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --alignEndsType EndToEnd” together with transcript annotations from GENCODE v31 (www.gencodegenes.org/human/release_31lift37.html).
ESPRESSO workflow
ESPRESSO was designed to discover and quantify transcript isoforms from long-read RNA-seq data that have been mapped to a reference genome. ESPRESSO is composed of three major steps: high-confidence SJ identification (“S”), SJ correction and recovery (“C”), and transcript isoform discovery and quantification (“Q”).
A cutoff for mapping quality (≥1 by default) is first applied to reads from all input samples. For a read with multiple alignments, only the alignment with the longest mapped length or highest mapping quality (ordered by priority) is used for further analysis. Next, ESPRESSO groups reads into independent clusters based on their mapping positions on the reference genome. Briefly, for each read in an ESPRESSO-defined cluster, the interval between the start and end positions of the read’s alignment must have an overlap of at least 1 nt with the interval of at least one other read in the same cluster. ESPRESSO then determines high-confidence SJs from each cluster using the following criteria. Any putative SJ that is annotated in a user-provided transcript annotation file is considered high-confidence. Novel putative SJs identified from raw long-read-to-genome alignments are considered high-confidence if the following two conditions are met: (i) the SJ has a canonical splice site dinucleotide motif (GT/AG, GC/AG, or AT/AC) with respect to the reference genome, and (ii) the SJ is supported by at least two (by default) long reads with perfect alignments (i.e., no mismatches, insertions, or deletions) within a 10-nt distance of the corresponding splice sites (Fig. 1, A and B).
For each unique high-confidence SJ, ESPRESSO concatenates two 25-nt genomic sequences of putative exons flanking the splice sites. All long reads harboring a putative SJ within a 35-nt distance to the high-confidence SJ with respect to the reference genome are queried against the concatenated sequence using a blastn (51) search with the following settings: blastn -task blastn -word_size 4 -reward 5 -penalty -4 -gapopen 8 -gapextend 6 -evalue 10 -dust no -soft_masking false (Fig. 1C, right). For each putative SJ position on a read, any high-confidence SJ with a blastn hit is listed as a candidate SJ. The probability of using a particular candidate SJ at a given putative SJ position is modeled using a multivariate hypergeometric distribution and is computed as follows. Given a raw long-read-to-genome alignment harboring n putative SJ positions, the probability of using the lth candidate SJ at the ith putative SJ position is calculated using the following formula
where j represents one of the four possible alignment types (mismatch, insertion, deletion, or match) and represents the number of times that the jth alignment type appears within a window of 20 nt (by default) surrounding the candidate SJ (10 nt on each side) within the long-read-to-candidate-SJ alignment generated by blastn. Kj represents the number of times that the jth alignment type appears in any part of the raw long-read-to-genome alignment that is more than 10 nt away from any of the n putative SJ positions. The numerator represents the product of four binomial coefficients, and the denominator represents the binomial coefficient for the sum of all alignment types. For each putative SJ position, all candidate SJs are evaluated based on their calculated probabilities to determine the most likely candidate SJ. Briefly, if a candidate SJ has a probability that is at least 10 times higher than the probability of the second-best candidate SJ, then this candidate SJ is determined to be the most likely candidate SJ at this putative SJ position (Fig. 1C, right).
After evaluating and correcting putative SJs in raw long-read-to-genome alignments, ESPRESSO next recovers missing first or last SJs and their associated terminal exons at alignment ends that the aligner may fail to detect. For each read, a high-confidence SJ that has a blastn hit before the first or after the last putative SJ position on the read is evaluated as a candidate missing first or last SJ if the high-confidence SJ has a probability that is higher than one tenth of the minimum probability across all high-confidence or corrected SJs determined for the read. Next, ESPRESSO uses nhmmer (52) with parameters “-T 3 --max” to query the read’s sequence against the sequences of candidate missing first or last SJs as well as the genomic sequence to which the end of the read was initially aligned. A candidate missing first or last SJ is considered recovered if its corresponding query has an nhmmer score that is at least two points higher than those of all other queries. For reads that have recovered first or last SJs, their mapping start and end positions on the reference genome are updated according to the alignment positions of the nhmmer queries.
Following SJ correction and recovery for individual long reads, ESPRESSO classifies reads into multiple categories by comparing the combination of SJs observed in a read against SJ combinations observed in all annotated transcript isoforms of the corresponding gene. Specifically, we adopted a classification system developed by Tardaguila et al. (17). Full-length or fragmented reads with SJ combinations that are consistent with those observed in annotated transcript isoforms are classified as FSM or ISM, respectively. On the other hand, reads harboring a novel combination of annotated or novel splice sites are classified as NIC or NNC, respectively. Last, reads with at least one low-confidence SJ that could not be corrected by ESPRESSO are classified as NCD (Fig. 1D). An annotated transcript isoform is considered as detected if ESPRESSO identifies at least one supporting read that is classified as FSM. ESPRESSO does not require the supporting read to have the exact 5′ and 3′ ends of the annotated transcript isoform, as annotation of transcript ends, or discovery of them from RNA-seq data, is imprecise (8). To minimize the number of false positive transcript isoforms reported, ESPRESSO adopts more stringent criteria for novel transcript isoforms. Specifically, a SJ combination observed in an NIC or NNC read is defined as a novel transcript isoform only if the following two criteria are met: (i) each SJ is supported by at least two (by default) perfectly aligned reads and (ii) the combination of SJs is not a substring of any other SJ combinations in other novel transcript isoforms discovered. The second criterion means that if one novel transcript isoform is a substring of another novel transcript isoform, ESPRESSO consolidates them and reports the longer transcript isoform as the novel transcript isoform. For each novel transcript isoform discovered using these criteria, ESPRESSO assigns a unique identifier composed of four colon-separated fields, with the string “ESPRESSO” used as the first field. The second and third fields are, respectively, the chromosome and ESPRESSO-defined cluster (represented by a numerical index) associated with the novel transcript isoform. Last, the fourth field is a cluster-specific numerical index representing the SJ combination of the novel transcript isoform.
After transcript isoform discovery, reads of all five categories (FSM, ISM, NIC, NNC, and NCD) are assigned to transcript isoforms based on the compatibility of their SJ combinations. Subsequently, the abundances of individual transcript isoforms are estimated using an EM algorithm (53). The EM algorithm used for transcript isoform quantification was from Xing et al. (28), originally developed for transcript isoform quantification using expressed sequence tag data. This algorithm has been widely used in short-read and long-read RNA-seq models (54). The detailed statistical formulation and algorithmic solution can be found in Xing et al. (28).
Running ESPRESSO
ESPRESSO (v1.2.2) was run on default settings to jointly discover and quantify transcript isoforms from the following sets of ONT RNA-seq alignments: (i) direct RNA data for SIRVs; (ii) 1D cDNA data for SIRVs; (iii) LRCA data for SIRVs; (iv) 1D cDNA data for PC3E, GS689, and HEK293T cell lines; (v) LRCA data for PC3E and GS689 cell lines; (vi) direct RNA data for HEK293T cell line; (vii) 1D cDNA data for HEK293T cell line; and (viii) 1D cDNA data for 30 human tissues. Information on the number of biological samples/replicates and number of ONT RNA-seq libraries prepared per biological sample/replicate can be found in tables S3 and S6. For ONT RNA-seq datasets aligned to the GRCh37/hg19 reference genome, reads that were mapped to the mitochondrial genome or contained large continuous insertions (≥20 nt) in the raw long-read-to-genome alignments were filtered out.
Read count estimates for transcript isoforms discovered by ESPRESSO were normalized into CPM by dividing the read count estimate of a transcript isoform by the sum of read counts assigned across all transcript isoforms discovered in a sample and multiplying this number by 1 million. We also calculated isoform proportions by dividing the CPM value of a transcript isoform by the CPM value for the corresponding gene (i.e., sum of CPM values over all transcript isoforms discovered for the gene). In cases where the gene has a CPM value of 0 in a given sample, we assigned all transcript isoforms of that gene an isoform proportion of 0.
Running LIQA
LIQA (v1.1.22) was run on ONT direct RNA and 1D cDNA sequencing alignment data for SIRVs to estimate the abundances of 68 SIRV transcripts. Before running LIQA, read alignments were filtered using SAMtools with parameters “-F 2308 -q 50” as recommended by the developer. The “quantify” module of LIQA was run on the filtered read alignments with parameters “-max_distance 10 -f_weight 1” as recommended by the developer.
Running NanoCount
Given that NanoCount is designed to work with reads aligned to a reference transcriptome, we first mapped basecalled ONT direct RNA and 1D cDNA sequencing reads for SIRVs to the sequences of 68 SIRV transcripts using minimap2 (v2.17-r974-dirty) with parameters “-ax map-ont -p 0 -N 10” as recommended by the developer of NanoCount. The resulting set of transcriptome alignments was then processed by NanoCount (v1.0.0.post6) using default tool settings.
Running FLAIR
FLAIR (v1.5.1 pre-release) was run directly on basecalled ONT direct RNA and 1D cDNA sequencing reads for SIRVs, as it contains an internal module (“align”) that can align reads to the SIRV reference genome with minimap2 (v2.17-r974-dirty). To ensure that FLAIR was using information from all replicates to discover transcript isoforms, we merged FLAIR corrected read alignments for replicates of a given library type before running the FLAIR “collapse” module, as recommended by the developer. SIRV transcript annotations were provided to all FLAIR modules, which were run with parameter “--nvrna” for direct RNA data and default settings for 1D cDNA data.
Running StringTie2
StringTie2 (v2.2.1) was run with the “-L” option on the following sets of ONT RNA-seq alignments: (i) direct RNA data for SIRVs, (ii) 1D cDNA data for SIRVs, (iii) direct RNA data for HEK293T cell line, and (iv) 1D cDNA data for HEK293T cell line. Before running StringTie2, we merged alignments across all replicates to ensure that StringTie2 uses information from all replicates to discover transcript isoforms. SIRV transcript annotations were provided when processing long-read alignments to the SIRV reference genome, and GENCODE v34 annotations were provided when processing long-read alignments to the GRCh37/hg19 reference genome. We also ran StringTie2 with the “--rf” option and SIRV transcript annotations on a publicly available short-read RNA-seq dataset (SRA accession ERR4330671) (49) that we aligned to the SIRV reference genome.
Running FLAMES
We ran the “bulk_long_pipeline.py” script contained in FLAMES (v0.1, downloaded 11 June 2022) directly on basecalled ONT RNA-seq reads, as FLAMES has an internal module dedicated to aligning reads to a reference genome with minimap2 (v2.17-r974-dirty). Specifically, we ran the Python script on the following sets of basecalled reads: (i) direct RNA data for SIRVs, (ii) 1D cDNA data for SIRVs, (iii) direct RNA data for HEK293T cell line, and (iv) 1D cDNA data for HEK293T cell line. SIRV transcript annotations were provided when processing reads derived from SIRV synthetic RNAs, and GENCODE v34 annotations were provided when processing reads derived from human samples. The bulk_long_pipeline.py script was run with parameters contained in the default config file (“config_sclr_nanopore_default.json”), and we used the following parameters: “has_UMI: false”, “generate_raw_isoform: true”, as well as “strand_specific: 1” for direct RNA data and “strand_specific: 0” for 1D cDNA data.
Benchmarking transcript isoform discovery using downsampled SIRV transcript annotations
We generated downsampled SIRV transcript annotations by randomly sampling a specified proportion (10, 20, 30, 40, 50, 60, 70, 80, or 90%) of SIRV transcripts. This procedure was performed three times per sampling level. Basecalled ONT direct RNA and 1D cDNA sequencing reads for SIRVs were first realigned to the SIRV reference genome using the downsampled annotations as a guide. The resulting long-read-to-genome alignments were subsequently processed by ESPRESSO, StringTie2, and FLAMES as previously described, where for each tool, transcript discovery and quantification were guided by the downsampled annotations. Among transcripts discovered by a given tool, true positives were composed of known SIRV transcripts including those removed from the downsampled annotations. False positives were novel transcripts whose combinations of SJs and/or exons did not match those of known SIRV transcripts. False negatives were known SIRV transcripts that were not discovered.
Evaluating transcript isoform discovery and quantification using simulated ONT RNA-seq data
Simulated ONT direct RNA and 1D cDNA sequencing reads were generated with NanoSim (v3.1.0). We trained NanoSim on real ONT direct RNA and 1D cDNA sequencing data that we generated on HEK293T cells. Specifically, the real data that we used for training were composed of either ONT direct RNA reads or 1D cDNA reads merged across the first technical replicates of three biological replicates of HEK293T cells. We ran the characterization stage of NanoSim (read_analysis.py) on “transcriptome” mode such that reads in the training data were first aligned to the sequences of annotated transcripts from GENCODE v34 using minimap2 (v2.17-r974-dirty). NanoSim uses the resulting set of transcriptome alignments to characterize the length distributions and error profiles of the training reads. We also ran the characterization stage on "quantify" mode to estimate the abundance levels of annotated transcripts from GENCODE v34. These learned features were subsequently used by NanoSim to guide simulation of ONT RNA-seq data. Specifically, using the files generated during the characterization stage as input, we ran the simulation stage of NanoSim (simulator.py) on “transcriptome” mode with parameters “-n 5000000 -k 6 -b guppy -r dRNA --no_model_ir --fastq” to generate 5 million ONT direct RNA reads. The same approach was used to generate 5 million ONT 1D cDNA reads, except that the parameter “-r” was set to “cDNA_1D”. Next, we downsampled each of our 5 million ONT RNA-seq datasets to 3 million, 1 million, and 0.5 million reads. All simulated ONT RNA-seq datasets were subsequently processed by ESPRESSO, LIQA, NanoCount, FLAIR, StringTie2, and FLAMES as previously described, where for each tool, the GRCh37/hg19 reference genome and GENCODE v34 annotations were provided.
To evaluate the performance of a given tool in discovering transcripts from a set of simulated reads, we first defined a “positive” set of transcripts as those whose abundance levels were estimated to be nonzero by NanoSim in the training data. True positives were defined as transcripts in the positive set that were discovered, whereas false negatives were defined as transcripts in the positive set that were not discovered. On the other hand, false positives were defined as discovered transcripts that were not in the positive set. For our assessments of transcript quantification accuracy, we computed the Spearman’s correlation between transcript abundance estimates from a given tool and ground-truth transcript abundance levels used for read simulation, focusing on transcripts whose abundance levels were estimated to be nonzero by NanoSim in the training data.
FASTQ files containing simulated ONT RNA-seq reads, together with the transcript expression profiles [tab-separated values (TSV) format] used to guide read simulation, are available at Zenodo (https://doi.org/10.5281/zenodo.7246437).
SPIRIT pipeline
We also designed the SPIRIT (splint improved repeat identifier for transcripts) pipeline (https://github.com/Xinglab/SPIRIT) to obtain consensus sequences from long concatemeric cDNA reads generated from LRCA libraries. SPIRIT differs from previous methods (14, 55) for generating consensus sequences from RCA concatemers in that it uses known sequences of DNA splints to perform consensus calling. Briefly, nhmmer (52) is used to first identify splint sequences from each concatemeric read using the parameters “-T 8 --max”. Sequences that are separated by pairs of neighboring splint sequences and have a length difference of less than 15% of the length of the longest sequence are considered copies of the same cDNA sequence. If three or more cDNA sequence copies are identified on a concatemeric read, then their consensus sequence is obtained using adaptive banded Partial Order Alignment (abPOA) (56), a fast implementation of the multiple sequence alignment algorithm Partial Order Alignment (POA) (57).
Analysis of transcript length bias on transcript quantification for different ONT RNA-seq library types
CPM values for 68 SIRV transcripts estimated by ESPRESSO (v1.2.2) from 1D cDNA, direct RNA, and LRCA data for SIRV synthetic RNAs were summed across replicates of the same library type. Next, the following two linear regression models were fitted (with intercepts) on summed CPM values of SIRV transcripts for each ONT RNA-seq library type, with spike-in concentration and length of SIRV transcripts used as model covariates
All linear regression analyses were done using R (v4.0.3). Specifically, linear regression models were fitted using the lm function (with default settings), and the fits of both models were compared using the anova function (with default settings).
Detection and quantification of alternative splicing events
We identified IR events from transcript isoforms recorded in the transcript annotation file generated by ESPRESSO for ONT 1D cDNA data of PC3E and GS689 cell line replicates (n = 3 per cell line). For each identified IR event, all long reads in a given cell line replicate with the intron either fully retained (inclusion reads, I) or spliced out (spliced reads, S) were counted. The PI value using long-read data was then calculated using the equation: . For identified IR events in which one or more overlapping exonic reads (I′) were detected, a potentially biased estimate of PI values based on long-read data was calculated using the equation: (fig. S13), in a manner that mimicked PI value estimation using short-read data. We estimated the bias in short-read-based PI values relative to long-read PI values using the equation: bias = PILONG_BIASED − PILONG. Short-read PI values (PISHORT) for the same set of IR events were computed by running SIRI (https://github.com/Xinglab/siri) (38) with default settings on short-read RNA-seq data generated on the same cell line samples (37). To compare short-read and long-read PI values for IR events identified in each of the PC3E and GS689 cell line replicates, we focused on events with at least 20 supporting reads in both long-read and short-read datasets.
We detected and quantified exon skipping as well as alternative 5′ and 3′ splice site usage from short-read RNA-seq data of the same PC3E and GS689 cell line replicates using rMATS (v4.1.2) (58, 59). For each alternative 5′ or 3′ splice site usage event detected by rMATS from short-read data of a given cell line replicate, we used ESPRESSO results generated on long-read data of the same sample to count the number of long reads carrying the inclusion junction (inclusion reads, I) and the number of long reads carrying the skipping junction (skipping reads, S). Similarly, for each exon skipping event detected by rMATS from short-read data of a given cell line replicate, we used ESPRESSO results for long-read data of the same sample to count the number of long reads harboring the cassette exon (inclusion reads, I) and the number of long reads in which the cassette exon is skipped (skipping reads, S). The PSI value of a given event using long-read data was then calculated using the equation: . To compare short-read and long-read PSI values, we focused on events satisfying the following criteria: (i) the number of short reads and the number of long reads supporting the inclusion junction(s) are both nonzero, (ii) the number of short reads and the number of long reads supporting the skipping junction are both nonzero, and (iii) the total number of short reads and the total number of long reads supporting either the inclusion junction(s) or skipping junction are both at least 20. Furthermore, we classified exon skipping events detected in a given cell line replicate as either simple (i.e., the cassette exon does not overlap any other alternative splicing events) or complex (i.e., the cassette exon overlaps other alternative splicing events) based on transcript isoforms discovered by ESPRESSO from long-read data. Specifically, an exon skipping event was considered simple if the number of transcript isoforms harboring both upstream and downstream inclusion junctions was equivalent to the number of transcript isoforms harboring either the upstream or the downstream inclusion junction. If this requirement was not met, then the exon skipping event was considered complex.
Identification of tissue-specific transcript isoforms
We sought to identify transcript isoforms with tissue-specific isoform proportions across ONT 1D cDNA sequencing data for 30 human tissues. For each gene, we generated an m × 30 contingency table composed of read counts (rounded to the nearest integer) for m discovered isoforms across 30 tissues. Using this matrix, we computed gene expression levels in each tissue as the sum of read counts over all transcript isoforms of the gene. We ignored genes that only had one discovered isoform or were expressed in only one tissue. We also omitted tissues from the contingency table if the gene of interest was not expressed in those tissues.
Next, we ran a chi-square test of homogeneity [false discovery rate (FDR) < 1%] on the contingency table to identify genes in which isoform proportions are not homogeneous across the considered tissues. To determine the minimum sample size for running the chi-square test on an R × C matrix, we applied Cohen’s w formula. The threshold on the total matrix read count, N, was chosen to be the smallest value of N for which an effect size of w = 0.5 (commonly interpreted as a large effect size) can be detected from running a chi-square test on an R × C matrix at a significance level of 0.01 (60)
Using the genes identified by the chi-square test with FDR < 1%, we subsequently ran a post hoc test to identify tissue-transcript isoform pairs in which the isoform proportion in the given tissue is significantly higher than the overall isoform proportion across all tissues (i.e., sum of read counts of the transcript isoform over all tissues divided by the sum of read counts of the gene over all tissues) (one-tailed binomial test, FDR < 1%). Our procedure for identifying tissue-specific transcript isoforms is contained in a custom Python script “SampleSpecificIsoforms.py” that can be downloaded from https://github.com/Xinglab/espresso in the folder “tissue_specific_analysis.”
Classifying alternative splicing events underlying tissue-specific transcript isoforms
Using the Ensembl BioMart database (Release 106, April 2022), we obtained canonical transcripts for genes with at least one tissue-specific transcript isoform discovered at FDR < 1% from ONT 1D cDNA sequencing data for 30 human tissues. The genomic coordinates of each canonical transcript were taken from GENCODE v40 transcript annotations (www.gencodegenes.org/human/release_40lift37.html), as Ensembl Release 106 annotations are based on GENCODE v40. We next compared the structure of each tissue-specific transcript isoform with the structure of the canonical transcript isoform for the corresponding gene, and we classified local differences in transcript structure into basic types of alternative splicing events, including exon skipping, alternative 5′ splice site usage, alternative 3′ splice site usage, mutually exclusive exon, intron retention, alternative first exon, and alternative last exon. Any local differences in transcript structure that could not be classified as one of the basic types of alternative splicing events were classified as “complex splicing.” If a tissue-specific transcript isoform harbors multiple transcript regions that differ in transcript structure compared to the canonical transcript isoform, then we classified the tissue-specific transcript isoform as having a combination of events. Notably, in our comparisons of transcript structure, we filtered out tissue-specific transcript isoforms that (i) were also the canonical transcript isoform of the corresponding gene, (ii) only differed in transcript ends relative to the canonical transcript isoform, or (iii) were expressed from a gene whose canonical transcript isoform is not defined in GENCODE v40.
Acknowledgments
Funding: This work was supported by National Institutes of Health grants (R01GM088342, U01CA233074, R01GM121827, and R56HG012310). R.W. is supported by a National Institutes of Health T32 Training Grant in Computational Genomics (T32HG000046).
Author contributions: Y.G. and Y.Xi. conceived the study. Y.G., F.W., L.L., and Y.Xi. designed the research and developed the methodology. Y.G., R.W., E.K., Y.Xu, and S.X. contributed to analytic tools. F.W. generated the data. Y.G., R.W., S.X., Y.W., and Y.Xi. analyzed the data. Y.G., F.W., R.W., K.E.K.-E., and Y.Xi. wrote the paper with input from all other authors.
Competing interests: Y.Xi. is a scientific cofounder of Panorama Medicine. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data (fast5 files) and processed data (abundance estimates and transcript annotation files generated by ESPRESSO) for all ONT RNA-seq data generated on SIRV synthetic RNAs, PC3E, GS689, and HEK293T cell lines, as well as 30 human tissues, were uploaded to GEO (accession number: GSE192955). The ESPRESSO software, together with other scripts used in this study, is available at GitHub (https://github.com/Xinglab/espresso) and archived at Zenodo (version 1.2.2, https://doi.org/10.5281/zenodo.6977552). FASTQ files containing simulated ONT RNA-seq reads, together with the transcript expression profiles (TSV format) used to guide read simulation, are available at Zenodo (https://doi.org/10.5281/zenodo.7246437).
Supplementary Materials
This PDF file includes:
Figs. S1 to S21
Tables S1 to S8
REFERENCES AND NOTES
- 1.S. Djebali, C. A. Davis, A. Merkel, A. Dobin, T. Lassmann, A. Mortazavi, A. Tanzer, J. Lagarde, W. Lin, F. Schlesinger, C. Xue, G. K. Marinov, J. Khatun, B. A. Williams, C. Zaleski, J. Rozowsky, M. Roder, F. Kokocinski, R. F. Abdelhamid, T. Alioto, I. Antoshechkin, M. T. Baer, N. S. Bar, P. Batut, K. Bell, I. Bell, S. Chakrabortty, X. Chen, J. Chrast, J. Curado, T. Derrien, J. Drenkow, E. Dumais, J. Dumais, R. Duttagupta, E. Falconnet, M. Fastuca, K. Fejes-Toth, P. Ferreira, S. Foissac, M. J. Fullwood, H. Gao, D. Gonzalez, A. Gordon, H. Gunawardena, C. Howald, S. Jha, R. Johnson, P. Kapranov, B. King, C. Kingswood, O. J. Luo, E. Park, K. Persaud, J. B. Preall, P. Ribeca, B. Risk, D. Robyr, M. Sammeth, L. Schaffer, L. H. See, A. Shahab, J. Skancke, A. M. Suzuki, H. Takahashi, H. Tilgner, D. Trout, N. Walters, H. Wang, J. Wrobel, Y. Yu, X. Ruan, Y. Hayashizaki, J. Harrow, M. Gerstein, T. Hubbard, A. Reymond, S. E. Antonarakis, G. Hannon, M. C. Giddings, Y. Ruan, B. Wold, P. Carninci, R. Guigo, T. R. Gingeras, Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Kalsotra, T. A. Cooper, Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. E. Baralle, J. Giudice, Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. M. Scotti, M. S. Swanson, RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Cherry, K. W. Lynch, Alternative splicing and cancer: Insights, opportunities, and challenges from an expanding view of the transcriptome. Genes Dev. 34, 1005–1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Park, Z. Pan, Z. Zhang, L. Lin, Y. Xing, The expanding landscape of alternative splicing variation in human populations. Am. J. Hum. Genet. 102, 11–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Conesa, P. Madrigal, S. Tarazona, D. Gomez-Cabrero, A. Cervera, A. McPherson, M. W. Szczesniak, D. J. Gaffney, L. L. Elo, X. Zhang, A. Mortazavi, A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T. Steijger, J. F. Abril, P. G. Engstrom, F. Kokocinski; RGASP Consortium, T. J. Hubbard, R. Guigo, J. Harrow, P. Bertone, Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. O. Pollard, D. Gurdasani, A. J. Mentzer, T. Porter, M. S. Sandhu, Long reads: Their purpose and place. Hum. Mol. Genet. 27, R234–R241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. L. Amarasinghe, S. Su, X. Dong, L. Zappia, M. E. Ritchie, Q. Gouil, Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. L. Weirather, M. de Cesare, Y. Wang, P. Piazza, V. Sebastiano, X. J. Wang, D. Buck, K. F. Au, Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res 6, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.F. J. Rang, W. P. Kloosterman, J. de Ridder, From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. M. Wenger, P. Peluso, W. J. Rowell, P. C. Chang, R. J. Hall, G. T. Concepcion, J. Ebler, A. Fungtammasan, A. Kolesnikov, N. D. Olson, A. Topfer, M. Alonge, M. Mahmoud, Y. Qian, C. S. Chin, A. M. Phillippy, M. C. Schatz, G. Myers, M. A. DePristo, J. Ruan, T. Marschall, F. J. Sedlazeck, J. M. Zook, H. Li, S. Koren, A. Carroll, D. R. Rank, M. W. Hunkapiller, Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Volden, T. Palmer, A. Byrne, C. Cole, R. J. Schmitz, R. E. Green, C. Vollmers, Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc. Natl. Acad. Sci. U.S.A. 115, 9726–9731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Wang, Y. Zhao, A. Bollas, Y. Wang, K. F. Au, Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Srivastava, L. Malik, H. Sarkar, M. Zakeri, F. Almodaresi, C. Soneson, M. I. Love, C. Kingsford, R. Patro, Alignment and mapping methodology influence transcript abundance estimation. Genome Biol. 21, 239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Tardaguila, L. de la Fuente, C. Marti, C. Pereira, F. J. Pardo-Palacios, H. Del Risco, M. Ferrell, M. Mellado, M. Macchietto, K. Verheggen, M. Edelmann, I. Ezkurdia, J. Vazquez, M. Tress, A. Mortazavi, L. Martens, S. Rodriguez-Navarro, V. Moreno-Manzano, A. Conesa, SQANTI: Extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 28, 396–411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D. R. Garalde, E. A. Snell, D. Jachimowicz, B. Sipos, J. H. Lloyd, M. Bruce, N. Pantic, T. Admassu, P. James, A. Warland, M. Jordan, J. Ciccone, S. Serra, J. Keenan, S. Martin, L. McNeill, E. J. Wallace, L. Jayasinghe, C. Wright, J. Blasco, S. Young, D. Brocklebank, S. Juul, J. Clarke, A. J. Heron, D. J. Turner, Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018). [DOI] [PubMed] [Google Scholar]
- 19.H. Li, Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Piovesan, F. Antonaros, L. Vitale, P. Strippoli, M. C. Pelleri, M. Caracausi, Human protein-coding genes and gene feature statistics in 2019. BMC. Res. Notes 12, 315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. Byrne, A. E. Beaudin, H. E. Olsen, M. Jain, C. Cole, T. Palmer, R. M. DuBois, E. C. Forsberg, M. Akeson, C. Vollmers, Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 8, 16027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.K. F. Au, V. Sebastiano, P. T. Afshar, J. D. Durruthy, L. Lee, B. A. Williams, H. van Bakel, E. E. Schadt, R. A. Reijo-Pera, J. G. Underwood, W. H. Wong, Characterization of the human ESC transcriptome by hybrid sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, E4821–E4830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. D. Tang, C. M. Soulette, M. J. van Baren, K. Hart, E. Hrabeta-Robinson, C. J. Wu, A. N. Brooks, Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 11, 1438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Y. Hu, L. Fang, X. Chen, J. F. Zhong, M. Li, K. Wang, LIQA: Long-read isoform quantification and analysis. Genome Biol. 22, 182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Gleeson, A. Leger, Y. D. J. Prawer, T. A. Lane, P. J. Harrison, W. Haerty, M. B. Clark, Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 50, e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N. Sheth, X. Roca, M. L. Hastings, T. Roeder, A. R. Krainer, R. Sachidanandam, Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 34, 3955–3967 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L. Paul, P. Kubala, G. Horner, M. Ante, I. Holländer, S. Alexander, T. Reda, SIRVs: Spike-in RNA variants as external isoform controls in RNA-sequencing. bioRxiv 080747. 2016.
- 28.Y. Xing, T. W. Yu, Y. N. Wu, M. Roy, J. Kim, C. Lee, An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 34, 3150–3160 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. Trapnell, B. A. Williams, G. Pertea, A. Mortazavi, G. Kwan, M. J. van Baren, S. L. Salzberg, B. J. Wold, L. Pachter, Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R. E. Workman, A. D. Tang, P. S. Tang, M. Jain, J. R. Tyson, R. Razaghi, P. C. Zuzarte, T. Gilpatrick, A. Payne, J. Quick, N. Sadowski, N. Holmes, J. G. de Jesus, K. L. Jones, C. M. Soulette, T. P. Snutch, N. Loman, B. Paten, M. Loose, J. T. Simpson, H. E. Olsen, A. N. Brooks, M. Akeson, W. Timp, Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. Soneson, Y. Yao, A. Bratus-Neuenschwander, A. Patrignani, M. D. Robinson, S. Hussain, A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat. Commun. 10, 3359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Kovaka, A. V. Zimin, G. M. Pertea, R. Razaghi, S. L. Salzberg, M. Pertea, Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L. Tian, J. S. Jabbari, R. Thijssen, Q. Gouil, S. L. Amarasinghe, O. Voogd, H. Kariyawasam, M. R. M. Du, J. Schuster, C. Wang, S. Su, X. Dong, C. W. Law, A. Lucattini, Y. D. J. Prawer, C. Collar-Fernandez, J. D. Chung, T. Naim, A. Chan, C. H. Ly, G. S. Lynch, J. G. Ryall, C. J. A. Anttila, H. Peng, M. A. Anderson, C. Flensburg, I. Majewski, A. W. Roberts, D. C. S. Huang, M. B. Clark, M. E. Ritchie, Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 22, 310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Hafezqorani, C. Yang, T. Lo, K. M. Nip, R. L. Warren, I. Birol, Trans-NanoSim characterizes and simulates nanopore RNA-sequencing data. Gigascience 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. L. Brown, L. M. Reinke, M. S. Damerow, D. Perez, L. A. Chodosh, J. Yang, C. Cheng, CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121, 1064–1074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Z. X. Lu, Q. Huang, J. W. Park, S. Shen, L. Lin, C. J. Tokheim, M. D. Henry, Y. Xing, Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res. 13, 305–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Z. Zhang, Z. Pan, Y. Ying, Z. Xie, S. Adhikari, J. Phillips, R. P. Carstens, D. L. Black, Y. Wu, Y. Xing, Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods 16, 307–310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.K. H. Yeom, Z. Pan, C. H. Lin, H. Y. Lim, W. Xiao, Y. Xing, D. L. Black, Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring. Genome Res. 31, 1106–1119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R. Stark, M. Grzelak, J. Hadfield, RNA sequencing: The teenage years. Nat. Rev. Genet. 20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 40.J. Harrow, A. Frankish, J. M. Gonzalez, E. Tapanari, M. Diekhans, F. Kokocinski, B. L. Aken, D. Barrell, A. Zadissa, S. Searle, I. Barnes, A. Bignell, V. Boychenko, T. Hunt, M. Kay, G. Mukherjee, J. Rajan, G. Despacio-Reyes, G. Saunders, C. Steward, R. Harte, M. Lin, C. Howald, A. Tanzer, T. Derrien, J. Chrast, N. Walters, S. Balasubramanian, B. Pei, M. Tress, J. M. Rodriguez, I. Ezkurdia, J. van Baren, M. Brent, D. Haussler, M. Kellis, A. Valencia, A. Reymond, M. Gerstein, R. Guigo, T. J. Hubbard, GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. Liu, Y. Liu, J. Li, H. Guo, T. Zang, Y. Wang, deSALT: Fast and accurate long transcriptomic read alignment with de Bruijn graph-based index. Genome Biol. 20, 274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. T. Parker, K. Knop, G. J. Barton, G. G. Simpson, 2passtools: Two-pass alignment using machine-learning-filtered splice junctions increases the accuracy of intron detection in long-read RNA sequencing. Genome Biol. 22, 72 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. Sahlin, V. Makinen, Accurate spliced alignment of long RNA sequencing reads. Bioinformatics 37, 4643–4651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U. Braunschweig, N. L. Barbosa-Morais, Q. Pan, E. N. Nachman, B. Alipanahi, T. Gonatopoulos-Pournatzis, B. Frey, M. Irimia, B. J. Blencowe, Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24, 1774–1786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. Byrne, C. Cole, R. Volden, C. Vollmers, Realizing the potential of full-length transcriptome sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.E. L. Van Nostrand, P. Freese, G. A. Pratt, X. Wang, X. Wei, R. Xiao, S. M. Blue, J. Y. Chen, N. A. L. Cody, D. Dominguez, S. Olson, B. Sundararaman, L. Zhan, C. Bazile, L. P. B. Bouvrette, J. Bergalet, M. O. Duff, K. E. Garcia, C. Gelboin-Burkhart, M. Hochman, N. J. Lambert, H. Li, M. P. McGurk, T. B. Nguyen, T. Palden, I. Rabano, S. Sathe, R. Stanton, A. Su, R. Wang, B. A. Yee, B. Zhou, A. L. Louie, S. Aigner, X. D. Fu, E. Lecuyer, C. B. Burge, B. R. Graveley, G. W. Yeo, A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. M. Drake, G. Strohbehn, T. B. Bair, J. G. Moreland, M. D. Henry, ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol. Biol. Cell 20, 2207–2217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. Picelli, O. R. Faridani, A. K. Bjorklund, G. Winberg, S. Sagasser, R. Sandberg, Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 49.V. Boehm, S. Kueckelmann, J. V. Gerbracht, S. Kallabis, T. Britto-Borges, J. Altmuller, M. Kruger, C. Dieterich, N. H. Gehring, SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nat. Commun. 12, 3965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. F. Altschul, W. Gish, W. Miller, E. W. Myers, D. J. Lipman, Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 52.T. J. Wheeler, S. R. Eddy, nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. P. Dempster, N. M. Laird, D. B. Rubin, Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B Stat. Methodol. 39, 1–22 (1977). [Google Scholar]
- 54.L. Pachter, Models for transcript quantification from RNA-Seq. arXiv:1104.3889 (2011).
- 55.R. Xin, Y. Gao, Y. Gao, R. Wang, K. E. Kadash-Edmondson, B. Liu, Y. Wang, L. Lin, Y. Xing, isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 12, 266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Gao, Y. Liu, Y. Ma, B. Liu, Y. Wang, Y. Xing, abPOA: An SIMD-based C library for fast partial order alignment using adaptive band. Bioinformatics 37, 2209–2211 (2021). [DOI] [PubMed] [Google Scholar]
- 57.C. Lee, C. Grasso, M. F. Sharlow, Multiple sequence alignment using partial order graphs. Bioinformatics 18, 452–464 (2002). [DOI] [PubMed] [Google Scholar]
- 58.S. Shen, J. W. Park, Z. X. Lu, L. Lin, M. D. Henry, Y. N. Wu, Q. Zhou, Y. Xing, rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. W. Phillips, Y. Pan, B. L. Tsai, Z. Xie, L. Demirdjian, W. Xiao, H. T. Yang, Y. Zhang, C. H. Lin, D. Cheng, Q. Hu, S. Liu, D. L. Black, O. N. Witte, Y. Xing, Pathway-guided analysis identifies Myc-dependent alternative pre-mRNA splicing in aggressive prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 117, 5269–5279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. Cohen, Statistical Power Analysis for the Behavioral Sciences. (L. Erlbaum Associates, ed. 2, 1988), pp. 567. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S21
Tables S1 to S8