Abstract
Background
Helicobacter pylori is one of the most common bacterial infections and is widespread globally. It causes a variety of gastrointestinal disorders, though a great proportion of infections are asymptomatic. A total of 143 fresh stool samples were collected from apparently healthy farm and pet animals (43 cattle, 50 buffaloes, 50 sheep, 50 dogs, and 50 cats), in addition to 768 human stool samples. The samples were examined using stool antigen and rapid antibody tests, and further confirmation of glmM “human antigen-positive samples and animal milk samples” was conducted by polymerase chain reaction (PCR).
Results
The prevalence rates of H. pylori infection in animals were 22.2% and 16% in antibody and stool antigen tests, respectively. The detection rates were 28%, 24%, 12%, 10%, and 4.7% in cats, dogs, buffaloes, sheep, and cattle, respectively. On the other hand, the prevalence rate of H. pylori infection in human stool samples was 74.8%, and a statistically significant association was observed between prevalence and several factors, such as sex, age, and locality. PCR was performed to detect the glmM gene of H. pylori, and this gene was found in 21 of 27 human antigen-positive samples and 5 of 13 animal milk samples.
Conclusions
H. pylori was detected in both human and animal samples. Furthermore, glmM was found in milk and human samples. Our findings suggest that pet and farm animals could transmit H. pylori infection to humans.
Keywords: Animal, glmM gene, H. pylori, Human, Milk, Zoonosis
Background
H. pylori infection is a major public health concern in both developed and developing countries [1]. H. pylori is a microaerophilic gram-negative helix bacterium that specifically infects the stomach lining. The clinical manifestations are associated with the development of chronic gastritis as well as gastric and duodenal ulcers and carcinoma. However, a large proportion of infections are asymptomatic [2, 3]. The severity of clinical manifestations varies depending on several factors, such as host genetic susceptibility, the immune system, the bacterial load, and virulence [4].
The prevalence of H. pylori differs among societies and geographical areas and is controlled by many factors, e.g., ecological factors, sociodemographic features, population lifestyle, and hygienic practices [5, 6]. The prevalence rate of H. pylori is estimated to be approximately 20%–50% in developed countries, reaching 90% in Africa [7], whereas in Egypt, it is reported to be 60%–80% in the adult population [2, 8]. This global increase in prevalence may be attributed to efforts to increase the productivity of food derived from animals to meet the steadily growing population, disregarding hygienic requirements, either for the herd or personnel [9, 10].
H. pylori colonizes the gut of both humans and animals [11, 12]. In former studies, H. pylori was isolated from the milk of different farming animals, such as cows, camels, ewes, and sows [13, 14]. Previous studies have also documented isolation of H. pylori from domestic cats [15]. Professionals handling animals and food of animal origin (e.g., veterinarians, butchers, and slaughterhouse workers) exhibit high titers of H. pylori antibodies (Ab). This suggests that ruminants may be the source of infection in humans [9, 16, 17].
Despite the availability of several diagnostic techniques, only tests with specificity and sensitivity beyond 90% are recommended for clinical diagnosis. H. pylori stool antigen tests are rapid and accurate noninvasive techniques with high sensitivity and specificity for diagnosis of active and recent H. pylori infection [18, 19]. Moreover, anti-H. pylori antibody test detection has a specificity and sensitivity of 79–90% and 76–84%, respectively [20].
Molecular techniques represent highly sensitive methods for H. pylori detection; indeed, they can directly detect the organism in clinical specimens. The targets of polymerase chain reaction (PCR) include glmM, which is the most significant indicator of H. pylori infection in milk and stool [9, 21, 22]. The protein encoded by H. pylori glmM directly participates in cell wall synthesis and plays an important and unique role in the growth and survival of the organism [23, 24].
This study aimed to investigate the prevalence of H. pylori infection in humans and factors associated with the infection, such as sex, age, and locality, as well as the possibility of different animal species transmitting H. pylori to humans.
Results
Detection of H. pylori antigen in different animal species
In this study, 39 (16%) of 243 animal stool samples tested positive when using the H. pylori stool antigen test. Among the 39 samples, 2, 6, 5, 12, and 14 were from cattle, buffaloes, sheep, dogs, and cats, respectively (Table 1). Statistically significant differences were observed in the prevalence of H. pylori infection among these animal species (P < 0.0001).
Table 1.
Occurrence of H. pylori antigen in stool samples from apparently healthy farm and pet animals
| Species | No. of examined samples | Positive samples | |
|---|---|---|---|
| No | % | ||
| Cattle | 43 | 2 | 4.7 |
| Buffaloes | 50 | 6 | 12.0 |
| Sheep | 50 | 5 | 10.0 |
| Dogs | 50 | 12 | 24.0 |
| Cats | 50 | 14 | 28.0 |
| Total | 243 | 39 | 16.0 |
| Chi2 value | 28.87* | P < 0.0001 *Statistically significant | |
*Indicates significant differences with a P value < 0.0001 using the chi-squared test
Detection of anti-H. pylori antibodies in different animal species
H. pylori antibodies were detected in 54 (22.2%) of 243 animal serum samples tested for infection using the H. pylori rapid antibody test. The antibodies were found to be more prevalent in pet animals (cats, 28%; dogs, 24%) than in farm animals (buffaloes, 12%; sheep, 10%; cattle, 4.7%) (Table 2). We observed a higher prevalence for the antibody test (22.2%) than the antigen test (16%) in animals, with the antibody test detecting 15 samples that were negative for H. pylori using the antigen test.
Table 2.
Prevalence of H. pylori antibodies in different animals using the H. pylori rapid antibody test
| Species | No. of serum samples | Positive serum samples | |
|---|---|---|---|
| No | % | ||
| Cattle | 43 | 8 | 18.6 |
| Buffaloes | 50 | 10 | 20.0 |
| Sheep | 50 | 8 | 16.0 |
| Dogs | 50 | 15 | 30.0 |
| Cats | 50 | 13 | 26.0 |
| Total | 243 | 54 | 22.2 |
| Chi2 value | 8.27* | P < 0.01 | |
*Indicates significant differences with a P value < 0.01 using the chi-squared test
Prevalence of H. pylori in human stool samples
In this study, 575 (74.8%) of 768 human stool samples were positive for the H. pylori antigen. Furthermore, a statistically significant association was observed between the prevalence of H. pylori infection and several risk factors, such as sex, age, and locality (P < 0.0001) (Table 3). With respect to sex, females had a higher detection rate (79.7%) than males (65. 5%). Additionally, the occurrence of H. pylori increased with age: the age group > 50 years exhibited the highest prevalence (87.3%), followed by the age group 15–50 years (76.8%) and finally the age group < 15 years (16.1%). H. pylori was also more frequently detected in individuals living in rural areas (88.5%) than in those living in urban areas (51.7%). Therefore, H. pylori infection was considered a dependent variable.
Table 3.
Prevalence of H. pylori infection in humans in relation to sex, age, and locality
| Variables | No. of examined samples | No. of positive samples (%) | Chi2 value | P value |
|---|---|---|---|---|
| Sex | ||||
| Male | 264 | 173 (65.5) | 18.65* | P < 0.0001 |
| Female | 504 | 402 (79.7) | ||
| Age group (years) | ||||
| < 15 | 93 | 15 (16.1) | 200.4* | P < 0.0001 |
| 15 and ≤ 50 | 302 | 232 (76.8) | ||
| > 50 | 372 | 325 (87.3) | ||
| Locality | ||||
| Urban | 288 | 149 (51.7) | 129.15* | P < 0.0001 |
| Rural | 480 | 425 (88.5) | ||
*Indicates significant differences with a P value < 0.01 using the chi-squared test
Molecular detection of the glmM gene
A total of 13 milk samples from farm animals with a positive H. pylori stool test and 27 positive human stool samples were subjected to H. pylori glmM detection via PCR. glmM was detected in 5 and 21 milk and stool samples, respectively (Table 4 and Fig. 1).
Table 4.
Detection frequency of glmM specific for H. pylori in milk samples and human stool samples
| Species | No. of examined samples | Positive samples | |
|---|---|---|---|
| No | % | ||
| Cattle | 2 | 1 | 50.0 |
| Buffaloes | 6 | 2 | 33.3 |
| Sheep | 5 | 2 | 40.0 |
| Total | 13 | 5 | 38.4 |
| Human | 27 | 21 | 77.8 |
Fig. 1.
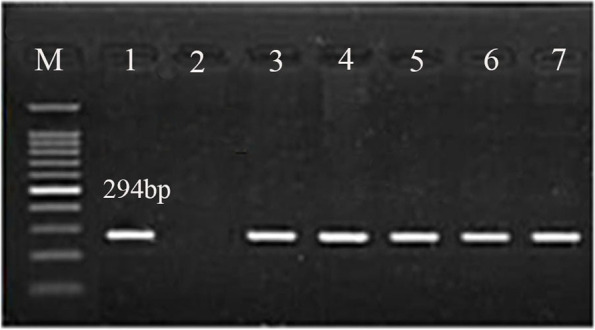
Agarose gel electrophoresis of PCR products of glmM (294 bp) for characterization of H. pylori in animal milk and human stool samples. Lane M: 100-bp ladder, Lane 1: control positive, Lane 2: control negative, Lanes 3 and 4: positive milk samples, Lanes 5–7 positive human stool samples
Discussion
H. pylori infects nearly half of the population at an early age [25]. Food, particularly milk, is one of the most important potential sources of infection in humans [9, 14].
Regarding animal stool samples, the H. pylori antigen (Table 1) was most frequently detected in cats (28%) in our study, followed by dogs (24%), buffaloes (12%), sheep (10%), and cattle (4.7%). In Iran, detection frequencies were reportedly 3% and 16% in cows and sheep, respectively [17]. In Italy, a higher prevalence was recorded in dog and sheep stool samples, at 100% and 82%, respectively [26].
In the sera of diverse animals (Table 2), H. pylori antibodies were most frequently detected in dogs (30%), followed by cats (26%), buffaloes (20%), cattle (18.6%), and sheep (16%), with a statistically significant association between these frequency rates. These rates were in agreement with those in cow sera (27%) in Iran [27]; detection rate in Algerian local-breed cows is 12% [9]. In contrast, higher detection rates have been detected in sheep (68%), cattle (70.3%), and goats (96.4%) in Sudan [28]. Additionally, H. pylori was found in 100% of dogs and 95% of sheep in northern Sardinia [26]. Overall, the antibody test results in our study were higher than those of the rapid Ag test, which could be explained by the fact that Ab seropositivity persists for a longer period of time, possibly up to 3 years [29].
Regarding human stool samples, the overall detection rate of H. pylori was 74.8% using the rapid antigen test. Nearly similar results were recorded among Egyptian patients with dyspepsia (73.7%) [2]. Furthermore, in west Iran, the H. pylori antigen positivity was 64.2% [30]. However, these results were higher than those reported in Japan (56.4%) [31], Ethiopia (49.7%) [32], and Khuzestan province, Iran (53.5%) [33]. In addition, our results were much higher than those reported in Turkey (36.6%) [34] and India (42.8%) [35], though the results in Pakistan (25%) [36] and Ethiopia (30.4%) were lower [37]. This significant variation in frequency might be due to the use of various techniques for H. pylori detection, socioeconomic status of the studied population, geographical variations, and environmental changes due to poor control measures, leading to exposure to raw sewage particles.
The association between the prevalence of H. pylori infection in humans and the sex of participants is illustrated in Table 3, with the infection rate in females (79.7%) and males (65.5%) showing a statistically significant association (P < 0.0001). The same finding was obtained in studies conducted in Oman [38] and Egypt [2]. This could be explained by the fact that women are more caring about their health than are men and often seek treatment for health problems. However, in a study on Australian patients, H. pylori infection rates were higher in males than in females [39]. This could be attributed to the fact that males tend to work outside of the home for longer periods of time than do women; thus, they are more exposed to environmental factors. In Ethiopia [37] and Sudan [40], no relationship between H. pylori infection and patient sex was observed.
Statistical analysis of the data presented in Table 3 revealed a significant difference (P < 0.0001) between various age groups and H. pylori prevalence. The highest prevalence was observed in the age group > 50 years (87.3%), followed by the age group 15–50 years (76.8%) and the age group < 15 years (16.1%). However, our study disagrees with previous studies in Egypt [2] and Sudan [40]. In addition, no statistically significant association was observed between the prevalence of H. pylori infection and age in Ethiopia [37]. These contradictory findings may be due to differences in the studied population and sample size.
The effect of participants' habitat on the prevalence of H. pylori infection revealed a statistically significant association (P < 0.0001). The infection was prevalent in those living in rural areas, where "there is an increased likelihood of contact with animals" (88.5%), and in urban areas (51.7%) (Table 3). Our findings were in agreement with those of a study conducted in Egypt reporting that farmers are more prone to H. pylori infection (82.1%) than non-farmers (64.9%) [2]. The higher prevalence of infection among rural inhabitants supports the theory that such an infection is a zoonosis. This was proposed by some previous studies in Egypt that isolated the studied organism from stool and milk samples of cattle [13]. Furthermore, a study in Iran confirmed isolation of H. pylori from gastric samples of several animals, such as sheep and cows, and considered them potential sources of H. pylori infection in humans [17]. In addition, the higher prevalence of H. pylori among rural inhabitants in our study may be attributed to the conditions that promote transmission, such as lack of income, poor hygiene, and low educational level. These results indicate that farm animals may be sources of H. pylori infection among humans, especially those who are handling them or consuming their products, such as milk.
glmM is important for cell wall development and microorganism growth. It has also been used extensively for molecular confirmation of H. pylori owing to its high specificity [41].
Milk is often consumed by humans, especially children. Thus, consuming milk represents the most proposed theory for H. pylori transmission from animals to humans. H. pylori has been detected in both pasteurized milk and raw milk in addition to the milk products of goats, sheep, and cows using conventional methods, molecular techniques, or both [42, 43]. Regarding milk, 13 samples were examined for the presence of H. pylori glmM in our study, with detection in five (38.4%) samples (one from cattle, two from buffaloes, and two from sheep). This finding was supported by former studies in Italy [43] and Sudan, which detected the presence of glmM in 22% of milk samples examined [44]. On the other hand, lower rates were recorded in Iran, including 8.7% in goat, 12.2% in sheep, and 14.1% in cow milk samples [45]. Furthermore, 13% of the Algerian local-breed cows were glmM positive, representing a possible zoonosis [9]. The presence of H. pylori in milk indicates the increasing hazard of human infection via consumption of contaminated raw milk [39]. Therefore, food safety and quality standards as well as good dairy farming practices are required for the production of milk that is safe for human consumption.
In this study, the prevalence of glmM in human stool samples was 77.8%. This finding is consistent with a study conducted in Iran that confirmed the existence of glmM in antigen-positive stool samples [27] and a study in Egypt that confirmed the presence of glmM in 64.7% of H. pylori-infected stool samples from sewage workers [46]. A slightly higher prevalence (93.7%) was observed in stool samples obtained from patients with gastritis [47]. In Iran, lower levels of glmM (39%) were found in stool samples from pediatric patients [48].
Conclusion
In El-Beheira Governorate, Egypt, H. pylori is frequently detected in both humans and animals. Factors such as sex, age, and locality were found to be associated with H. pylori infection in humans. H. pylori was more frequently detected in pet animals than in farm animals, and milk is a possible source of transmission to humans. However, further studies are warranted to provide more information regarding the exact sources of infection and the role of animals in disease transmission. H. pylori poses a great public hazard; therefore, we recommend that public health authorities prioritize the development of preventive measures against H. pylori infection.
Methods
Ethical declaration
This research was conducted according to guidelines of the Institutional Animal Care and Use Committee of Alexandria University (ALEXU-IACUC, 3312020) in Egypt. Informed consent was obtained from the human participants and/or their legal guardian after receiving detailed information about the aims of the study.
Study area and design
The present study was conducted from January to December 2021 at El-Beheira Governorate, Egypt, to determine the prevalence of H. pylori in different animal species and humans.
Sampling
Animal samples
A total of 143 samples were obtained from apparently healthy farm animals, namely, cattle (n = 43), buffaloes (n = 50), and sheep (n = 50). The animals were Egyptian native breeds and reared in a semi-intensive system. Samples, such as stool, serum, and milk, were collected from each animal investigated. Stool samples were collected in sterile dry, disposable, leak-proof, wide-mouthed, plastic containers, and 5–7-mL blood samples were collected in plain Vacutainer tubes and kept at room temperature for 1–2 h before storage; raw milk samples were placed in sterile Falcon tubes. Furthermore, 100 stool samples were randomly collected from pet animals in veterinary clinics, including dogs and cats (50 samples each). All the collected samples were labeled and immediately transferred to the laboratory in an icebox for examination.
Human samples
A total of 768 fresh stool samples were collected from patients with dyspepsia (264 males and 504 females) aged 5–80 years who visited the Gastroenterology Department in hospitals in El-Beheira Governorate, Egypt. Personal and sociodemographic data relevant to risk factors for infection were collected, such as sex, age, and locality. The patients were divided into three age groups (< 15, 15–50, and > 50 years). The samples were collected in sterile airtight cups, labeled, and then immediately transferred to the laboratory in an icebox.
H. pylori antigen detection
Stool samples (from animals and humans) were examined for the presence of H. pylori-specific antigen using H. pylori Ag Rapid Test-Cassette (OnSite®, CTK Biotech, San Diego, USA), a lateral-flow chromatographic immunoassay for the qualitative detection of H. pylori antigen in stool samples [49, 50]. The procedures and reading of the results were performed according to the manufacturer’s protocols.
H. pylori rapid antibody test (serological examination)
An antibody rapid test device (serum/plasma) (Atlas Medical, Cambridge, England, and Model No. 8.04.21) was used as a qualitative membrane-based immunoassay to investigate the presence of IgM and IgG antibodies specific for H. pylori in serum samples [51]. The procedures and interpretation of the results were performed according to the manufacturer’s protocols.
Detection of H. pylori glmM via PCR
DNA was extracted from milk samples using a DNA isolation kit (Cat. No. ABIN412492, Roche, Mannhein, Germany) according to the manufacturer’s protocols. Furthermore, a QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) was used to extract DNA from stool samples according to the manufacturer’s protocols. Specific oligonucleotide primers (Table 5) were used to amplify a 294-bp region of the H. pylori glmM gene [45]. PCR for glmM was performed under the following conditions: initial denaturation at 94 °C for 10 min, 35 cycles of denaturation at 94 °C for 60 s, annealing at 55 °C for 60 s, extension at 72 °C for 60 s, and a final extension at 72 °C for 10 min. The PCR products were subjected to 1.5% agarose gel electrophoresis (Bio-Rad, France) with ethidium bromide staining.
Table 5.
Oligonucleotide primer sequences used for the detection of glmM
| Target gene | Oligonucleotide sequence 5′–3′ | Size (bp) |
|---|---|---|
| glmM |
GAATAAGCTTTTAGGGGTGTTAGGGG GCTTACTTTCTAACACTAACGCGC |
294 |
Statistical analysis
Statistical analysis was conducted using the chi-squared (Chi2) test to examine significant differences between the studied groups according to SAS (2014). P values < 0.05 were considered to indicate significant differences.
Acknowledgements
The authors would like to thank the staff members of the faculty of veterinary medicine at Alexandria University for their valuable cooperation in this study.
Authors’ contributions
All authors collaborated in the planning of the work, experimental design, measurement of parameters, and writing of the manuscript. SIS, MAN, and MSD conceived of and designed the experiments. DT and MAA measured the parameters. DT and SAK statistically analyzed the data. SIS, MAN, RME and MSD wrote the manuscript. All authors read, reviewed, and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). No funding has been provided.
Availability of data and materials
The datasets used and/or analyzed in the current study were not publicly published to preserve the privacy of the participants but are available upon reasonable request from the corresponding author.
Declarations
Ethics approval and consent to participate
This research was conducted according to the guidelines of the Institutional Animal Care and Use Committee of Alexandria University (ALEXU-IACUC, 3312020), Egypt. All farm owners included in the study were informed of all the study procedures and aims, and permission to collect animal samples was obtained from them verbally. All methods were performed in accordance with the ARRIVE guidelines for the reporting of animal experiments (https://arriveguidelines.org). In addition, a comprehensive discussion was conducted with each worker, and they received detailed information about the aims of the study. Informed consent was obtained from the human participants and/or their legal guardian after receiving detailed information about the aims of the study. The study was conducted in accordance with the Declaration of Helsinki for medical research involving human subjects. The animal and human experiments were approved by the ethics committee of Alexandria University, Egypt.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sabah I. Shaaban, Email: sabah_ibrahim@vetmed.dmu.edu.eg
Dalia Talat, Email: dalia_talat@vetmed.dmu.edu.eg.
Shymaa A. Khatab, Email: shymaa.genetics@gmail.com
Mohamed A. Nossair, Email: mohammadnossair@alexu.edu.eg
Mousa A. Ayoub, Email: mousaayoub2007@yahoo.com
Rania M. Ewida, Email: r_ewida@vet.nvu.edu.eg
Mohamed Said Diab, Email: mohameddiab333@gmail.com.
References
- 1.Hunt R, Xiao S, Megraud F, Leon-Barua R, Bazzoli F, Van der Merwe S, Coelho LV, Fock M, Fedail S, Cohen H. Helicobacter pylori in developing countries. World gastroenterology organisation global guideline. J Gastrointestin Liver Dis. 2011;20(3):299–304. [PubMed] [Google Scholar]
- 2.Salem E, Sakr A, Younis FaMA. Prevalence of helicobacter pylori infection among farmers and non-farmers with dyspepsia. Egypt J Occup Med. 2019;43(2):229–244. doi: 10.21608/ejom.2019.31419. [DOI] [Google Scholar]
- 3.Fagoonee S, Pellicano R. Helicobacter pylori: molecular basis for colonization and survival in gastric environment and resistance to antibiotics. A short review. Infect Dis. 2019;51(6):399–408. doi: 10.1080/23744235.2019.1588472. [DOI] [PubMed] [Google Scholar]
- 4.Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39(1):14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olokoba A, Gashau W, Bwala S, Adamu A, Salawu F. Helicabacter pylori Infection in Nigerians with Dyspepsia. Ghana Med J. 2013;47(2):79–81. [PMC free article] [PubMed] [Google Scholar]
- 6.Alsulaimany FA, Awan ZA, Almohamady AM, Koumu MI, Yaghmoor BE, Elhady SS, Elfaky MA. Prevalence of Helicobacter pylori infection and diagnostic methods in the Middle East and North Africa Region. Medicina. 2020;56(4):169. doi: 10.3390/medicina56040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idowu A, Mzukwa A, Harrison U, Palamides P, Haas R, Mbao M, Mamdoo R, Bolon J, Jolaiya T, Smith S. Detection of Helicobacter pylori and its virulence genes (cagA, dupA, and vacA) among patients with gastroduodenal diseases in Chris Hani Baragwanath Academic Hospital, South Africa. BMC Gastroenterol. 2019;19(1):1–10. doi: 10.1186/s12876-019-0986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emara MH, Salama RI, Salem AA. Demographic, endoscopic and histopathologic features among stool H. pylori positive and stool H. pylori negative patients with dyspepsia. Gastroenterology Res. 2017;10(5):305. doi: 10.14740/gr886w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guessoum M, Guechi Z, Adnane M. First-time serological and molecular detection of Helicobacter pylori in milk from Algerian local-breed cows. Vet World. 2018;11(9):1326. doi: 10.14202/vetworld.2018.1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payao SL, Rasmussen LT. Helicobacter pylori and its reservoirs: a correlation with the gastric infection. World J Gastrointest Pharmacol Ther. 2016;7(1):126–132. doi: 10.4292/wjgpt.v7.i1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mladenova-Hristova I, Grekova O, Patel A. Zoonotic potential of Helicobacter spp. J Microbiol Immunol Infect. 2017;50(3):265–269. doi: 10.1016/j.jmii.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Ranjbar R, Chehelgerdi M. Genotyping and antibiotic resistance properties of Helicobacter pylori strains isolated from human and animal gastric biopsies. Infect Drug Resist. 2018;11:2545–2554. doi: 10.2147/IDR.S187885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Gohary AH, Yousef MA, Mohamed AA, Abou El-Amaiem WE, Abdel-Kareem LM. Epidemiological study on H. pylori in cattle and its milk with special reference to its zoonotic importance. Biol Med. 2015;7(5):1. doi: 10.4172/0974-8369.1000251. [DOI] [Google Scholar]
- 14.Talaat Al Sherief LM, Thabet SS. Isolation of Helicobacter Pylori from Raw Milk and Study on Its Survival in Fermented Milk Products. J Appl Vet Sci. 2022;7(2):73–81. [Google Scholar]
- 15.Canejo-Teixeira R, Oliveira M, Pissarra H, Niza MM, Vilela CL. A mixed population of Helicobacter pylori, Helicobacter bizzozeronii and "Helicobacter heilmannii" in the gastric mucosa of a domestic cat. Ir Vet J. 2014;67(1):25. doi: 10.1186/2046-0481-67-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeidi E, Sheikhshahrokh A. VacA genotype status of Helicobacter pylori isolated from foods with animal origin. BioMed Res Int. 2016;2016:8701067. doi: 10.1155/2016/8701067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14(1):1–7. doi: 10.1186/1471-230X-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirak MY, Akyön Y, Mégraud F. Diagnosis of Helicobacter pylori. Helicobacter. 2007;12:4–9. doi: 10.1111/j.1523-5378.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 19.Dore MP, Pes GM. What is new in Helicobacter pylori diagnosis. An overview. J Clin Med. 2021;10(10):2091. doi: 10.3390/jcm10102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaker Y, Moon A, Afzali A. Helicobacter pylori: a review of epidemiology, treatment, and management. J Clin Gastroenterol Treat. 2016;2(2):1–5. doi: 10.23937/2469-584X/1510019. [DOI] [Google Scholar]
- 21.Malfertheiner P, Megraud F, O’morain C, Gisbert J, Kuipers E, Axon A, Bazzoli F, Gasbarrini A, Atherton J, Graham DY. Management of Helicobacter pylori infection—the Maastricht V/Florence consensus report. Gut. 2017;66(1):6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 22.Leonardi M, La Marca G, Pajola B, Perandin F, Ligozzi M, Pomari E. Assessment of real-time PCR for Helicobacter pylori DNA detection in stool with co-infection of intestinal parasites: a comparative study of DNA extraction methods. BMC Microbiol. 2020;20(1):1–8. doi: 10.1186/s12866-020-01824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer B, Meyer TF. The human gastric pathogen Helicobacter pylori and its association with gastric cancer and ulcer disease. Ulcers. 2011;2011:Article ID 340157, 23 pages. 10.1155/2011/340157.
- 24.Patel SK, Mishra GN, Pratap CB, Jain AK, Nath G. Helicobacter pylori is not eradicated after triple therapy: a nested PCR based study. BioMed Res Int. 2014;2014:483136. doi: 10.1155/2014/483136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guillermo I, Rothenbacher D, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9(2):1–6. doi: 10.1111/j.1083-4389.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- 26.Dore M, Pes G, Sanna G, Sepulveda A, Graham D. Helicobacter pylori infection in shepherds, sheep and sheep-dogs. Microb Health Dis. 2020;2:e306. [Google Scholar]
- 27.Safaei HG, Rahimi E, Zandi A, Rashidipour A. Helicobacter pylori as a zoonotic infection: the detection of H. pylori antigens in the milk and faeces of cows. J Res Med Sci. 2011;16(2):184. [PMC free article] [PubMed] [Google Scholar]
- 28.Hussien MO, Hassan AD, Ibrahim AT, Musa AB, Abukunna FE, Mohammed NM, Seidahmed SI, Hassan HM, Taha KMaEH, A.M Seroprevalence of Helicobacter pylori infection in cattle, sheep, goats andchickens in Marawi area, Northern State, Sudan. Sudan J Vet Res 2014, 29:47‐48.
- 29.Ahmet A, Mehmet A, Güneş T, Özkan S, Dündar N. Comparison of antigen and antibody detection tests used for diagnosing the Helicobacter pylori infection in symptomatic patients. Basic Clin Sci. 2010;1(4):61–70. [Google Scholar]
- 30.Soltani J, Amirzadeh J, Nahedi S, Shahsavari S. Prevalence of helicobacter pylori infection in children, a population-based cross-sectional study in west iran. Iran J Pediatr. 2013;23(1):13. [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoyama T, Oyama T, Matsuzaka M, Danjo K, Nakaji S, Fukuda S. Comparison of a stool antigen test and serology for the diagnosis of Helicobacter pylori infection in mass survey. Helicobacter. 2009;14(2):87–90. doi: 10.1111/j.1523-5378.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- 32.Mathewos B, Moges B, Dagnew M. Seroprevalence and trend of Helicobacter pylori infection in Gondar University Hospital among dyspeptic patients, Gondar, North West Ethiopia. BMC Research Notes. 2013;6(1):1–4. doi: 10.1186/1756-0500-6-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talaiezadeh A, Borhani M, Moosavian M, Rafiei A, Neisi A, Hajiani E, Alavi SSM, Nik KA. Prevalence of Helicobacter pylori Infection evaluated by Stool antigen test in Khuzestan Province since September to October 2009, south-west of Iran: a population based study. 2013. [Google Scholar]
- 34.Büyükbaba-Boral Ö, Küçüker-Anğ M, Aktaş G, Işsever H, Anğ Ö. HpSA fecoprevalence in patients suspected to have Helicobacter pylori infection in Istanbul, Turkey. Int J Infect Dis. 2005;9(1):21–26. doi: 10.1016/j.ijid.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Mishra S, Singh V, Rao G, Dixit V, Gulati A, Nath G. Prevalence of Helicobacter pylori in asymptomatic subjects—A nested PCR based study. Infect Genet Evol. 2008;8(6):815–819. doi: 10.1016/j.meegid.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Hassan MK, Haq M, Saifullah Z, Khan AG, Khattak AK, Khan AG. Frequency of Helicobacter pylori infection by testing stool antigen in patients with functional dyspepsia. Gomal J Med Sci. 2013;11:183–187. [Google Scholar]
- 37.Seid A, Demsiss W. Feco-prevalence and risk factors of Helicobacter pylori infection among symptomatic patients at Dessie Referral Hospital, Ethiopia. BMC Infect Dis. 2018;18(1):1–9. doi: 10.1186/s12879-018-3179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joob B, Wiwanitkit V. Helicobacter pylori seropositivity among the patients with acute dyspepsia. Asian Pac J Trop Dis. 2014;4:S435–S436. doi: 10.1016/S2222-1808(14)60484-9. [DOI] [Google Scholar]
- 39.Windsor HM, Morrow SD, Marshall BJ, Abioye-Kuteyi EA, Leber JM, Bulsara MK. Prevalence of Helicobacter pylori in Indigenous Western Australians: comparison between urban and remote rural populations. Med J Aust. 2005;182(5):210–213. doi: 10.5694/j.1326-5377.2005.tb06668.x. [DOI] [PubMed] [Google Scholar]
- 40.Abdallah TM, Mohammed HB, Mohammed MH, Ali AAA. Sero-prevalence and factors associated with Helicobacter pylori infection in Eastern Sudan. Asian Pac J Trop Dis. 2014;4(2):115–119. doi: 10.1016/S2222-1808(14)60326-1. [DOI] [Google Scholar]
- 41.Córdova Espinoza MG, González Vazquez R, Morales Mendez I, Ruelas Vargas C, Giono Cerezo S. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J Clin Microbiol. 2011;49(4):1650–1652. doi: 10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimura S, Kawamura T, Kato S, Tateno H, Watanabe A. Detection of Helicobacter pylori in cow's milk. Lett Appl Microbiol. 2002;35(6):504–507. doi: 10.1046/j.1472-765X.2002.01229.x. [DOI] [PubMed] [Google Scholar]
- 43.Quaglia N, Dambrosio A, Normanno G, Parisi A, Patrono R, Ranieri G, Rella A, Celano G. High occurrence of Helicobacter pylori in raw goat, sheep and cow milk inferred by glmM gene: a risk of food-borne infection? Int J Food Microbiol. 2008;124(1):43–47. doi: 10.1016/j.ijfoodmicro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Osman EY, El-Eragi A, Musa AM, El-Magboul SB. Detection of Helicobacter pylori glmM gene in bovine milk using nested polymerase chain reaction. Vet World. 2015;8(7):913. doi: 10.14202/vetworld.2015.913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahimi E, Kheirabadi EK. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog Dis. 2012;9(5):453–456. doi: 10.1089/fpd.2011.1060. [DOI] [PubMed] [Google Scholar]
- 46.Agha S, Foad MF, Awadalla NJ, Saudy N. Helicobacter pylori cagA gene in Egyptian sewage workers. Afr J Pathol Microbiol. 2013;2:1–6. doi: 10.4303/ajpm/235847. [DOI] [Google Scholar]
- 47.Makristathis A, Hirschl AM, Mégraud F, Bessède E. Diagnosis of Helicobacter pylori infection. Helicobacter. 2019;24:e12641. doi: 10.1111/hel.12641. [DOI] [PubMed] [Google Scholar]
- 48.Falsafi T, Favaedi R, Mahjoub F, Najafi M. Application of stool-PCR test for diagnosis of Helicobacter pylori infection in children. World J Gastroenterol: WJG. 2009;15(4):484. doi: 10.3748/wjg.15.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelmalek S, Hamed W, Nagy N, Shokry K, Abdelrahman H. Evaluation of the diagnostic performance and the utility of Helicobacter pylori stool antigen lateral immunochromatography assay. Heliyon. 2022;8(3):e09189. doi: 10.1016/j.heliyon.2022.e09189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang YJ, Chen MJ, Chen CC, Lee JY, Yang TH, Yu CC, Chiu MC, Kuo CC, Weng YJ, Bair MJ, et al. Accuracy of rapid Helicobacter pylori antigen tests for the surveillance of the updated prevalence of H. pylori in Taiwan. J Formos Med Assoc. 2020;119(11):1626–1633. doi: 10.1016/j.jfma.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Naji EN. Comparative Study for the Accuracy of Helicobacter pylori Diagnostic Methods Associated with Some Inflammatory Factors. Al-Mustansiriyah J Sci. 2017;28(2):16–28. doi: 10.23851/mjs.v28i2.495. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the current study were not publicly published to preserve the privacy of the participants but are available upon reasonable request from the corresponding author.


