Summary
The concept of cell fate relates to the future identity of a cell, and its daughters, which is obtained via cell differentiation and division. Understanding, predicting, and manipulating cell fate has been a long-sought goal of developmental and regenerative biology. Recent insights obtained from single-cell genomic and integrative lineage-tracing approaches have further aided to identify molecular features predictive of cell fate. In this perspective, we discuss these approaches with a focus on theoretical concepts and future directions of the field to dissect molecular mechanisms underlying cell fate.
Keywords: cell fate decision, mathematical and computational models, multi-omics, clonal tracking, haematopoiesis, embryogenesis
In this perspective, Laleh Haghverdi and Leif Ludwig discuss recent advances in cell fate research with a focus on the roles of single-cell multi-omics and lineage tracing. They further discuss mathematical and theoretical concepts and future perspectives of this field to dissect cellular mechanisms underlying cell fate.
Introduction
Cell fate decision-making describes the process by which cells develop into a particular fate while rejecting possible alternative fates over the course of cellular differentiation and division (Soldatov et al., 2019). As such, cell fate is a fundamental aspect of multi-cellular organismal development and its homeostatic maintenance, also in response to environmental perturbations, which may trigger adaptive or regenerative cellular processes. The blueprint that underlies the development of a human body, consisting of over 30 trillion cells and organized into a myriad of organ systems each composed of diverse cell types and states, is orchestrated by information encoded in our genomes (Zeng, 2022). A complete reconstruction of the molecular chain of cell-intrinsic and -extrinsic events that underlie cell fate decision-making would thereby outline the required steps to effectively engineer and guide cells toward desired properties, in particular for cell-based (regenerative) therapies from diverse sources of (adult) stem cells or the reprogramming of differentiated cells (Beumer and Clevers, 2020; Liu et al., 2021; Ng et al., 2020; Yu et al., 2021). As such, substantial advances have been made in this field, with new experimental technologies, mathematical concepts, and analytical approaches emerging to identify the molecular determinants and modulators of cell fate (Camp et al., 2019; Packer et al., 2019; Wagner and Klein, 2020; Weinreb et al., 2020).
Here, we highlight recent advances in single-cell omics and lineage tracing and discuss notions related to cell fate in development and homeostasis, as exemplified in mammalian embryogenesis and blood production (hematopoiesis), respectively. We discuss theoretical concepts and possible future directions in light of emerging multi-omics technologies and their integration with lineage-tracing approaches that provide powerful means to experimentally validate predictive features as well as extend the time length over which course such predictions may be amenable.
Principles of cell fate
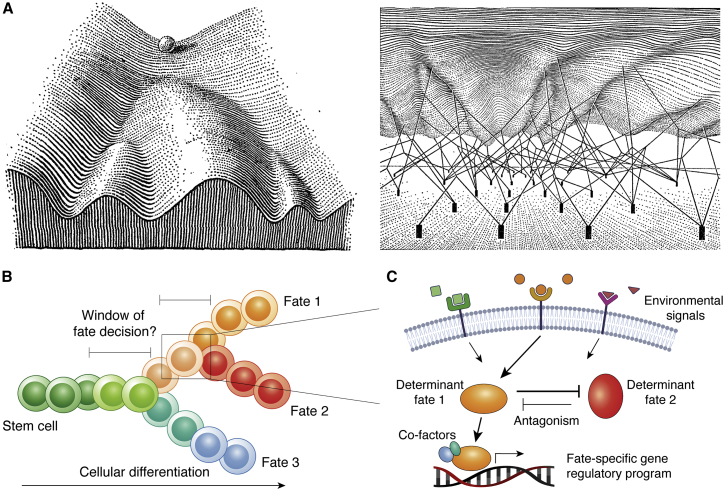
The acquisition of a particular terminal cell fate is the result of the integration of a cell’s intrinsic molecular properties and its interaction with its surroundings, from which the cell derives signals that direct and modulate a cell’s inner workings (Beumer and Clevers, 2020). In biology, the acquisition of such fates has thereby been depicted as marbles rolling down a potential hill shaped by regulatory forces beneath, with the marbles eventually coming to rest at different lowest points of the hill (basins of attraction), representing the various terminally differentiated fates that cells may acquire (Buenrostro et al., 2018; Verd et al., 2014; Waddington, 1957) (Figure 1A). Along these different paths of the so-called Waddington landscape, cells may reach genomic barriers that separate two or multiple distinct cellular fate directions. Understanding the molecular mechanisms underlying how and when cells decide on which path to travel has thereby been a long-sought goal of biology (Figure 1B). Simplistically, a particular differentiation outcome may be driven by individual key factors or determinants of cell fate (e.g., transcription factors [TFs]) that become activated upon cell signaling events triggered by environmental signals (Figure 1C). Antagonists may counteract a determinant and/or drive a cell toward an alternative fate, whereas a consolidator may reinforce the action of a determinant (e.g., transcriptional co-factors). TFs are recognized as key determinants of cell fate, as their loss of expression can completely ablate the presence of a specific cell type. Likewise, their over- or ectopic expression may skew or even reprogram a cell’s identity to an alternative terminal or back to a progenitor or stem(-like) fate (Ng et al., 2020; Orkin and Zon, 2008; Spitz and Furlong, 2012; Stadhouders et al., 2019). TFs further shape the genomic landscape with the aid of co-regulatory factors (i.e., consolidators such as additional TFs or chromatin remodelers) to reconfigure chromatin and rewire the cellular circuits toward a specific fate (Figure 1C). Here, we will exemplarily discuss the role of cell fate decisions in two differentiating systems, the mammalian developing embryo, and adult hematopoiesis.
Figure 1.
Waddington’s landscape and cell fates during cellular differentiation
(A) Waddington’s landscape with the marble representing a cell to take on a set of alternative developmental paths, with the three basins at the base of the hill, representing the alternative differentiated cell fates (left). The shape of the landscape is thereby determined by interacting gene products that pave the path toward the cell attaining a particular differentiated cell fate (right). Figure from Waddington (1957).
(B) Schematic depiction of stem cells undergoing cellular differentiation to obtain one of three cell fates, upon which multiple cell fate decisions may have to be made. The window in which the fate decision is being made and during which the process may be altered or antagonized may thereby currently not be well defined.
(C) Cell fate determination involves the integration of environmental signals and the current cellular state, upon which fate determinants may act or also antagonize each other, before executing a particular fate-specific gene regulatory program.
Cell fate in embryogenesis
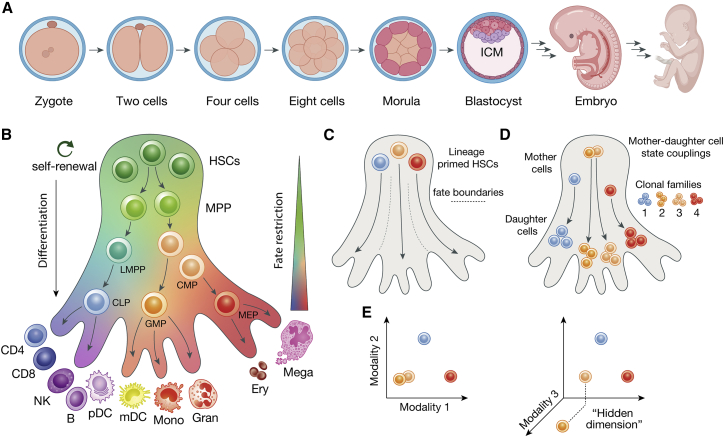
Embryonic development (embryogenesis) starts with the zygote, which undergoes a remarkably organized sequence of cell divisions and initiation of differentiation processes through developmental stages that ultimately form all the organ systems and cells within the human body (Gerri et al., 2020; Shahbazi, 2020) (Figure 2A). As such, the developing cells appear to undergo a largely pre-determined sequence of consecutive cell fate decisions within a self-contained ecosystem fueled by the placenta. If not perturbed (e.g., due to toxins or genetic aberrations), the cells collectively create a self-organizing interdependent environment for every (new) cell to make appropriate decisions to enable full maturation of the embryo. The first cell fate decision takes place at the 16-cell stage and is a function of the cells’ (radial) polarity and geometrical position. Cells on the outside of the so-called morula are thereby fated to develop the extraembryonic trophectoderm, while the inner cells are fated to constitute the inner cell mass (ICM) and, being pluripotent, represent the precursor of the embryo. The ICM cells subsequently localize to one pole of the forming blastocyst, engendering a topological asymmetry that orients the further formation of the embryo (Figure 2A). A multitude of signaling factors, including morphogens forming concentration gradients for patterning and transcriptional regulators, have been identified to orchestrate subsequent oriented cell divisions (Dewey et al., 2015) and cell fate choices (Gerri et al., 2020; Shahbazi, 2020) with an increasing appreciation of the mechanical forces at play (Valet et al., 2021). The system then rapidly grows in complexity as the number of cells and extrinsic and intrinsic cellular features interacting with each other (to further constitute the function and fate of each cell) increase exponentially. Within this complexity, cell death (apoptosis) events are also programmed (Ellis and Horvitz, 1986) to regulate the right balance of cell types to properly initiate specific developmental events. The intricate interplay of signaling, TF activity, and chromatin organization is thereby well described during developmental limb (re)generation (Gerber et al., 2018; Petit et al., 2017). Ultimately, these complex cell fate interaction networks are tightly regulated, leading to a growing organism that becomes increasingly refocused on homeostatically maintaining the developed organ systems.
Figure 2.
Cell fate in embryogenesis and hematopoiesis
(A) Simplified schematic of early developmental steps, including the formation of the morula and blastocyst with the inner cell mass (ICM), which subsequently gives rise to the embryo proper and the fully developed organism.
(B) Simplified schematic of hematopoietic differentiation with lineage commitment occurring across a continuum of states (color gradient) with arrows intimating possible transition paths of differentiation, upon which cells become increasingly fate-restricted. Adapted from Bao et al. (2019). HSC, hematopoietic stem cell; MPP, multi-potent progenitor; LMPP, lymphoid-primed multi-potent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythroid progenitor; CD4, CD4+ T cell; CD8, CD8+ T cell; B, B cell; NK, natural killer cell; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; mono, monocyte; gran, granulocyte; ery, erythroid; mega, megakaryocyte.
(C) Schematic depiction of lineage-primed HSCs suggesting that fate boundaries may already be present early during differentiation, for example also within the HSC pool.
(D) Schematic depiction of how clonal- and lineage-tracing methodologies enable identification of clonal families, including mother-daughter cell state couplings. The capture of early stem and progenitor cell states may thereby aid to identify relevant features of future cell fate. However, in practice, the capture of such states may be impeded by the rather low frequency of, in particular, the HSC state.
(E) Cells may show high similarity in a lower-dimensional space (left), with the measurement of an additional dimension revealing “hidden” features that were previously not accounted for. Such hidden features are expected to enhance the prediction of cell fates (right).
Cell fate in hematopoiesis
Hematopoiesis has long served as a paradigm for our understanding of cellular processes ranging from stem cell maintenance to multi-lineage differentiation and its dysregulation in disease (Orkin and Zon, 2008). A pool of self-renewing hematopoietic stem cells (HSCs) that forms during development sustains life-long blood production of a diverse repertoire of cells with distinct functions, including platelets, erythrocytes, monocytes, granulocytes, natural killer cells, and adaptive T and B lymphocytes (Liggett and Sankaran, 2020) (Figure 2B). Analogous to the Waddington landscape, the acquisition of these cellular fates was traditionally thought to be the result of a stepwise decision-making process through defined cellular stages with increasingly restricted lineage potential as hematopoietic differentiation progressed. In part, this model was supported by the results of cultures and transplantation experiments of bulk populations of cells with defined surface marker expression profiles, which were considered to be characteristic of a homogeneous cellular stage. Ultimately, the fate of individual cells remained masked, but the increasing application of single-cell-based functional and genomic approaches subsequently revealed an unappreciated genomic and functional heterogeneity within the hematopoietic stem and progenitor cell pool (Amann-Zalcenstein et al., 2020; Liggett and Sankaran, 2020; Loeffler and Schroeder, 2019). Today, we recognize multiple distinct lineage hierarchies, with many early hematopoietic progenitor cells displaying advanced priming or restriction to an individual or a few lineage fates (Carrelha et al., 2018; Rodriguez-Fraticelli et al., 2018) (Figure 2C). As such, cell fate may at least in part already be pre-determined within the stem cell pool “long” before the respective cellular fate is attained. However, a detailed molecular understanding of such “clonal memory” (Fennell et al., 2022; Yu et al., 2016), and instructive signals by the bone marrow microenvironmental niche (Haas et al., 2018), including in disease contexts, is yet to be attained.
TFs are the major orchestrators of cell fate decisions in hematopoiesis (Orkin and Zon, 2008), and individual TFs have been recognized to drive the development of a particular hematopoietic lineage. For example, the TF GATA1 is essential to regulate red blood cell development, as alterations of protein levels and function lead to severe forms of congenital anemia (Khajuria et al., 2018; Ludwig et al., 2014; Sankaran et al., 2012). How GATA1 drives lineage choice and at what stage of hematopoiesis, for example, cross-antagonizing alternative lineage determinants such as PU.1, have been the subjects of investigation and debate (Hoppe et al., 2016; Strasser et al., 2018; Wheat et al., 2020). Its role as a major fate determinant is, however, reinforced by its ability to convert fibroblasts into an erythroid progenitor cell fate, together with important co-regulatory TFs such as TAL1 (Capellera-Garcia et al., 2016). Moving forward, cell-state to -fate couplings will be essential to further enhance our understanding of gene regulatory mechanisms that govern hematopoietic cell fate decisions, as we discuss below (Figures 2D and 2E).
Current approaches in single-cell genomics and cell fate
Several omic modalities and analytical approaches have been developed to predict cellular fates. However, we still lack a detailed appreciation of the molecular events underlying cell fate, and current approaches may suffer from inherent limitations such as “missing data” that prevent more reliable predictions (Tritschler et al., 2019; Weinreb et al., 2018). Here, we briefly review core concepts in the field.
Molecular state trajectories of cellular differentiation
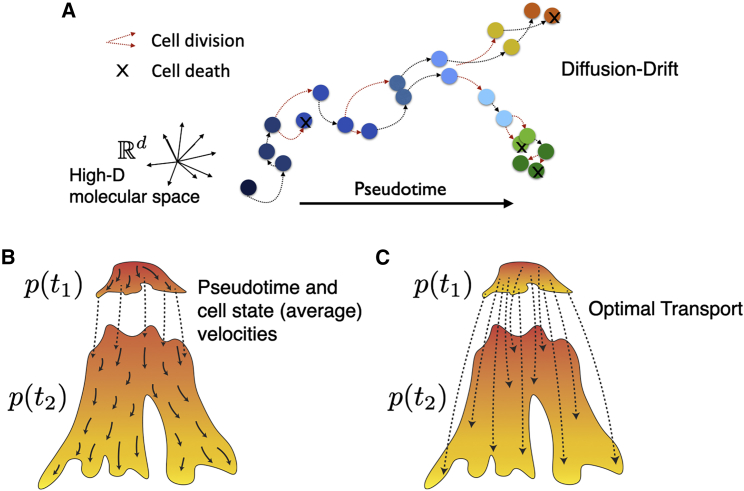
Transcriptomics is the most widely used data modality for defining cell states, as cellular gene expression profiles are closely associated with their identity and function. A snapshot of differentiating cells often includes a range of cells from different maturation stages. Leveraging this asynchrony of the captured cells’ progression stage, pseudotime, and trajectory analysis allow defining comprehensive maps of accessible transcriptional states and the cell state transitions across the cellular differentiation landscape (Saelens et al., 2019). These methods are based on cell state adjacencies, with the general assumption that some form of distance (e.g., Euclidean or diffusion distance) is related to the biological “travel” time between two cell states. Several studies have aimed for a more complete dynamical description of cell differentiation processes using the Fokker-Planck, or diffusion-drift, equation, which integrates the drift (i.e., the directed energy-consuming transition of cells toward more differentiated states) as well as estimations of cell birth and death rates with the merely geometrical (i.e., neighborhoods) information being considered by pseudotime methods (Cho et al., 2018; Farrell et al., 2018; Fischer et al., 2019; Lange et al., 2022) (Note S1). Taking advantage of simultaneous unspliced and spliced mRNA measurements to infer differentiation dynamics via RNA velocity (La Manno et al., 2018) has been another important step toward describing cell differentiation and fate dynamics as a directed diffusion process in which a Langevin equation (Gillespie, 2000) for velocities’ relation with noise and directed force applies (Note S2). However, several technological and computational challenges remain toward reliable quantification of cell state velocities that are actively being refined (Gorin et al., 2022.; Gu et al., 2022; Marot-Lassauzaie et al., 2022; Qiu et al., 2022). Among existing challenges, we highlight (1) the unreliable assignment and thus quantification of mRNA reads as unspliced versus spliced in input data; (2) extensions of cell velocity inference models for more accurately measurable data modalities such as simultaneous mRNA and protein measurements; (3) accounting for gene stochasticity in the inference model as well as the identifiability of its parameters; and (4) inferring the cells’ (average) velocities over defined time intervals. Once reliable single-cell velocities are at hand, they can be used to infer every cell state’s propensity toward different cell fates, for example by chaining different velocities together along multiple possible differentiation paths as in CellRank (Lange et al., 2022). A full diffusion-drift model of cell differentiation including all three components of diffusion, drift, cells’ birth, and death rates would enable a probabilistic description and simulation of cell differentiation as a function of any given initial state and quantify cell state plasticities (captured by the noise or diffusion term) or differentiation bias (captured by the drift term) toward specific fates (Figures 3A and 3B). An alternative model for cell differentiation has been suggested using optimal transport (OT) (Mittnenzweig et al., 2021; Schiebinger et al., 2019), which tries to infer a correspondence between the distribution of cells in the phase space at two different time points, hence inferring probabilities for each cell in the later time point to be a descendant of each cell in the first time point (Figure 3C). There is a mathematical correspondence between the diffusion-drift description of a dynamical process and OT, as the latter can be interpreted as a maximum likelihood solution of the former, integrating transition probabilities among cell states over the interval between two observation time points (Notes S1 and S3). Consequently, unlike the diffusion-drift descriptions, OT does not consider cell state transitions and pseudotime relations among the cell within each measurement time point but rather has a conceptual affinity to uncover ancestral relationships between cells from two time points, similar to clonal tracking.
Figure 3.
Dynamical models of cellular differentiation
(A) The dynamics of cell differentiation in the high-dimensional molecular features space include diffusion (stochastic), drift (directed), and cells’ birth/death events. Cell colors indicate different cell states.
(B and C) Given cell samples from two distributions, p(t1) and p(t2) from time points t1 and t2, respectively, (B) pseudotime and velocity analysis methods infer transition probabilities among all cell states (color gradient indicates the overall pseudotime) , whereas (C) OT infers transition probabilities from cell states in the first time point to the cell states in the second time point. For visual guidance of where arrows (indicating high-probability transitions) start and end, the color gradient indicates pseudotime for each time point separately.
Note that several computational concepts and methods initially developed for application to transcriptomics data (e.g., pseudotime, OT, etc.) have been (or can be) adapted for other high-throughput data modalities or combinations thereof. Consequently, researchers tend to incorporate recent technologies for the multi-modal profiling of cells into the development of respective computational methods. For example, Co-Spar combines multi-modal cell state information (i.e., transcriptome, epigenome) with clonal information to predict future cell states (Wang et al., 2022). In Co-Spar, transition probabilities between cells collected at two time points are calculated based on a constrained OT scheme on the multi-modal space of cell states. Additionally, when clonal data is available, Co-Spar uses it to restrict the transitions to cell pairs that belong to the same clone across two differentiation time points, thereby improving transition predictions.
Transcriptional profiles reflect a cell’s gene regulatory machinery and chromatin/epigenetic state that regulate the activity of TFs on cis-regulatory elements of DNA, including promoters and enhancers (Spitz and Furlong, 2012). In single-cell genomics, the assay for transposase accessible chromatin by sequencing (ATAC-seq) has emerged as a popular tool to survey the heterogeneity of accessible chromatin landscapes across differentiating cells (Lareau et al., 2019; Satpathy et al., 2019). Recently, several methodologies have enabled the simultaneous measurement of the transcriptome and the epigenome (Cao et al., 2018; Ma et al., 2020; Zhu et al., 2019) and opened up new avenues for inferring the regulatory and consecutive feature relations between the two modalities. For example, the notion of “chromatin potential” to infer cell fate choices alludes to the observation of chromatin becoming accessible before the expression of its corresponding gene, as illustrated in hair follicle differentiation (Ma et al., 2020). Combining chromatin accessibility and gene expression information, chromatin potential thus closely relates to the concept of cell state velocity as a fluctuating quantity that depends on the time interval considered in its inference. Whereas RNA velocity reports cell state velocities at the relatively short timescale of splicing events (Marot-Lassauzaie et al., 2022), “chromatin potential” may refer to a cell’s more long-term future plans.
Analogously, changes in chromatin organization, DNA methylation, histone modifications, and the activity of TFs create poised or primed chromatin states that may bias or instruct downstream lineage choice (Bernstein et al., 2006; Parry et al., 2020; Stadhouders et al., 2019). While TF activity can be inferred from single-cell ATAC-seq data, including across pseudotime differentiation trajectories (Satpathy et al., 2019), recent advances now enable us to interrogate the heterogeneity of deposition of TFs and histone modifications on the chromatin of single cells (Bartosovic et al., 2021; Grosselin et al., 2019; Rotem et al., 2015; Wu et al., 2021). As such, a detailed understanding of the events surrounding chromatin regulation and preceding the expression of a cell fate determinant may enable predictions before the determinant is in fact detectable via transcriptomics.
Integrative single-cell omics-based lineage tracing
Lineage- and clonal-tracing approaches aim to track the progeny of single cells, for example to reconstruct the developmental path or the direct ancestral relationships among cells (Baron and van Oudenaarden, 2019). As such, they enable the quantitative assessment of which stem cells give rise to which organ and to what extent over the course of embryogenesis or enable dissection of the clonal population structure in cancers or of physiologic blood production sustained by adult HSCs. Aside from the direct visual tracking of developmental and differentiation processes (Loeffler and Schroeder, 2019; Sulston and Horvitz, 1977), the genetic introduction of heritable tags (e.g., reporter genes, DNA barcodes, transposons, evolving CRISPR arrays) have enabled the prospective lineage tracing of a labeled population of cells of interest (Busch et al., 2015; Chow et al., 2021; Sun et al., 2014). As these genetic engineering-based strategies are prohibitive in humans, with the rare exception of gene therapy (Scala et al., 2018), naturally occurring somatic mutations have been leveraged to retrospectively infer the phylogeny or clonal relationships among cells (retrospective lineage tracing) (Lee-Six et al., 2018; Ludwig et al., 2019). Notably, numerous approaches have now integrated barcoding strategies with classical single-cell transcriptome profiling (Fennell et al., 2022; He et al., 2022; Quinn et al., 2021). These have enabled concomitant cell typing with (developmental) lineage inference (Spanjaard et al., 2018) or the direct coupling of the cellular fate of mother-daughter cell pairs and/or of sister cells with their genomic state represented via their transcriptome and/or chromatin profile (Lareau et al., 2021; Tian et al., 2021; Weinreb et al., 2020). In self-renewing cellular systems such as hematopoiesis, the concomitant characterization of the stem cell pool (mother cells) and its cellular output (daughter cells), these techniques enable us to relate molecular features of a stem cell to the downstream fate of its descending daughter cells (Figure 2D). For example, such strategies identified the TF TCF15 as an important regulator of HSC quiescence and long-term self-renewal (Rodriguez-Fraticelli et al., 2020), determined Bcor as a negative regulator of emergency dendritic cell development (Tian et al., 2021), and revealed two different pathways of monocyte differentiation with distinct clonal relationships and gene expression dynamics (Weinreb et al., 2020). The latter study further revealed sister cells to be more similar in their fate choice than pairs of cells with similar transcriptional profiles (Weinreb et al., 2020). These results strongly suggest that single-cell transcriptomics alone is limited to reliably predicting progenitor cells with distinct fate bias and that computational approaches may misidentify fate decision boundaries in the absence of lineage information. Conceivably, decisive features may also lie substantially “upstream” of determinants that we currently associate with cell fate decisions, such as the detectable expression of a particular TF, which may only emerge as differentiation progress. While in particular lowly expressed but otherwise important transcripts modulating cell choice may not be readily detected by current single-cell RNA sequencing (scRNA-seq) profiling methods, the co-detection of additional cellular properties may provide a more complete means to decode cell fate decision-making (Figure 2E).
Future directions
Our molecular understanding of cellular fate decisions remains incomplete. While key determinants such as TFs or gene signatures predictive of cell fate have been recognized, we remain relatively limited in terms of the distances in the differentiation landscapes over which such predictions may be reliably made. As such, we discuss the challenge of predicting cell fate decisions over longer distances or even consecutive cell fate decisions, before the activity of key determinants may be even recognizable and what molecular features we may have to account for, that are currently not readily amenable to single-cell measurements.
Mathematical models and philosophical frameworks in cell fate decision-making
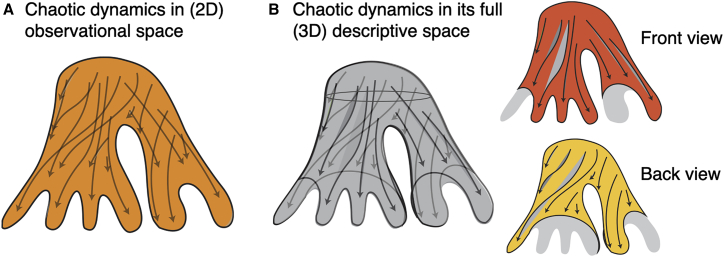
In some deterministic dynamical systems, a small change in the initial state can lead to a vast divergence from the state the system acquires later in time, thus making long-term predictions impossible. Chaos theory (Box 1) (Grebogi et al., 1987; Shinbrot et al., 1992; Thiétart and Forgues, 1995) links unpredictability to inaccuracy and incompleteness of measurements. Conceivably, with awareness of the full dimensionality of features governing the system’s behavior, the dynamics would be fully predictable. It basically suggests that intermingling and crossing dynamical paths in high-dimensional chaotic systems are an artifact of reducing the dimensions of the systems from their (inaccessible) full governing space to the smaller observational space (Figure 4).
Box 1. Chaos theory.
“Chaos” explains deterministic dynamical systems in which a small change in the initial state can lead to a vast divergence from the state the system acquires later in time. This implies that in a real world where our measurements inevitably contain errors (e.g., position measurements up to the precision of millimeters in a particular experiment), long-term prediction of a chaotic system’s behavior is not possible, although its dynamical rules of progression are fully deterministic. Had we been able to exactly characterize the current state, predictions of infinite time would have been possible. In a non-chaotic system, an error in measurement of the initial state would bear a (e.g., linear or quadratic) relation with the error in the prediction of the future state. In a chaotic system, however, no such relation holds, as close-by paths do not stay correlated for long and diverge exponentially over time, producing complex intermingling dynamical paths in the observational space (“topological mixing”).
Chaotic systems can exist both in low and high dimensions. The motion of a double-rod pendulum is an example of deterministic, but chaotic, dynamics in low dimensions (Shinbrot et al., 1992; Thiétart and Forgues, 1995). As the number of dimensions defining the state of a system becomes larger, full characterization of the system’s state becomes more challenging in the real world, leading to chaotic behavior when only a subset of the full features is measured. A well-known example of such a high-dimensional chaotic system is the “butterfly effect” (Shen et al., 2018), which implies that a small perturbation such as (metaphorically) a butterfly flapping its wing can affect the weather at a later time in another place. Consequently, given our lack of awareness of the full feature space, weather predictions remain valid only for a finite time interval such as a few days.
Figure 4.
Incomplete measurements in high-dimensional systems introduce chaos
(A and B) (A) Chaotic (seemingly crossing and intermingling) dynamical trajectories in an observational space (here, the two-dimensional data manifold in orange) are resolved (B) when the same system is measured in its full descriptive space (here, the three-dimensional data manifold in gray), which disentangles the front (red) and back (yellow) views of the three-dimensional data manifold on which the dynamics take place.
Hematopoiesis is a great example where our understanding of the system has been reshaped from a less predictable (formerly associated with stochasticity in fate decisions) toward a more deterministically resolved view via moving from bulk measurements to a high-resolution single-cell description of the feature space and associated cell state-fate couplings (Rodriguez-Fraticelli et al., 2020; Tian et al., 2021; Weinreb et al., 2020). As such, the to-date unavailability of full predictive models for most given biological contexts has been at least partly due to the incompleteness of our measurements. Recent research on sister cells’ fate choice as described in the previous section as well as identical twins’ phenotypic divergence as they age (Martin, 2005) are exemplary phenomena that imply chaos to be a favorable model for cell fate.
Relying on advancements of measurements, it sounds increasingly relevant to design and apply deterministic models such as the aforementioned Co-Spar cell fate prediction method, the assumptions (i.e., coherence and sparsity of state transitions) of which also correspond to non-chaotic settings (i.e., accessibility of a fully descriptive feature space). Nevertheless, it is important to validate how well the assumption of non-chaotic behavior holds for a particular data type for the application of such models. In a chaotic system, state prediction accuracy drops exponentially with time, whereas in non-chaotic systems, neighboring dynamical paths (thus the possibility of future state prediction) remain long-term correlated. This provides an avenue for quantifying the level of chaos for each data type and the time length over which non-chaotic behavior can be assumed. Moreover, new computational approaches and modeling algorithms may be designed to account for chaotic characteristics of the system, for example by considering the exponential decay of prediction accuracy over time.
Whereas chaos theory, diffusion-drift, and OT each describe aspects of cell differentiation dynamics, we further require comprehensive mathematical formulations and theories around the highly deterministically programmed, self-contained unfolding of cell differentiation, for example of how a single cell (zygote) gives rise to a complex organism consisting of multiple tissues and cell types. Furthermore, one may confer multi-scale descriptions to relate molecular observations, which constitute a large stochastic component, to larger-scale observations that appear to behave more deterministically such as homeostasis of cell types at the level of tissues or organisms via adaptation of frameworks from for example statistical physics (Rau, 2017).
Navigating the complex high-dimensional features space
The complexity of cell differentiation poses the challenge of efficiently navigating the high-dimensional space in which the system is embedded. Several computational approaches such as non-linear dimension reduction techniques, pseudotime, and OT are based on cell-to-cell distances. Signals from early determinants of cell fate may, however, fade when calculating cell-cell distances in a space where noise from multiple dimensions adds up. Thus, refining the feature space via prior knowledge of the potential relevance of critical features as well as the application of supervised methods may be more powerful to identify weak, but predictive, signals. As such, the rationale would be to eliminate the redundancy of multiple correlated features as opposed to accounting for all measurable features at once and in an unsupervised manner. The application of supervised-learning methods to high-dimensional data nonetheless requires large data collection efforts.
Further, we note that the most predictive feature set may (and does) vary piecewise in time. For example, geometry and morphogens play an important role in early embryogenesis, whereas their role may be rather marginal in fully structured organ systems, which, however, continue to integrate environmental signals (Dewey et al., 2015). Therefore, building local prediction models is necessary and compatible with short-term predictability if one assumes a chaotic model of cell differentiation and fate decisions.
Imagine a cell was a simple non-chaotic system consisting of only a dozen of regulatory elements (e.g., TFs) with correlated and antagonistic regulatory relations among them. Such a system would have a few attractor basins corresponding to stable fully differentiated cell types. Any imaginary cell at a specific position of this dozen-dimensional phase space would eventually converge to one of those basins. It would then be rather easy to experimentally position (artificial) cells in different regions of this space of all possible cell states (considering the bounds of maximum gene expression levels) and test how and toward which attractor basin they move. This would imply that by sampling a representative amount of initial cell states and observing their future states, we may attain a satisfactory understanding of the dynamics anywhere within this space. We could then arguably more confidently infer the regulatory rules underlying the observed dynamics and simulate the dynamical path from any initial cell state. To do so, we would assume a mathematical model (e.g., a model considering up to 3rd-order feature interactions). If the model is set up in such a way that the number of features, and thus the number of parameters to be inferred, is much smaller than the number of sampled cells (and the transition trajectories between them as additional “velocity” observations), we have a good chance to have an “identifiable” mathematical model (Gábor et al., 2017) for inferring the regulatory mechanisms that have generated the observed data. However, difficulties arise because a real-world cell is complex and chaotic when observed at lower dimensions. This is, in fact, forcing us to talk about “context-specific” and “cell-type-specific” gene regulatory networks, acknowledging our inability to sample the huge governing feature space and the “universal” laws of regulatory interactions and cells’ dynamics in the observation space. Sampling and investigating only small neighborhoods around the known attractors (cell types) in the huge phase space sounds like a more viable strategy but at the cost of disregarding the other dimensions and passages that relate the region to other more distant cell states. In this view, healthy differentiation paths and natural determinants of cell fate constitute only a small part of many other (not sampled) potential dynamical paths that could lead to the same cell type (basin of attraction).
One approach to reducing the complexity of a system is thus to both (1) restrict it to a group of its most relevant players (e.g., TFs as core regulators of cell fate at a particular differentiation stage) as well as (2) considering the regulatory interactions only in a certain context or neighborhood, for example specific cell types, as exemplified by two recent methods for inference of gene regulatory networks, CellOracle (Kamimoto et al., 2020) and spliceJAC (Bocci et al., 2022). Considering multiple sets of key players act throughout differentiation, multiple local or time-resolved consecutive regulatory networks would emerge. However, in most instances, our understanding of the events leading up to the expression of a TF, for instance, and its pleiotropic downstream consequences and interactions, is limited. Meaning that even identifying a set of key players in the neighborhood of a chosen cell state, which is rather isolated from the rest of the players in the universal regulatory network, is not straightforward. As such, we may want to ideally consider the signaling events a stem cell receives, how it integrates them, understand how they prime the chromatin for TF transcription, and be aware of all post-transcriptional and post-translational modulators that ultimately regulate TF activity off (e.g., before translocation to the nucleus) and on chromatin to shape cellular identity. While reduced context-specific regulatory networks are conceptually intriguing, current methodologies are not able to adequately capture the complexity of features, leaving us often with chaos.
Advancing the feature set via multi-omics
Single-cell multi-omics have been evolving rapidly, and multiple methodologies now capture information from up to four data modalities (Mimitou et al., 2021; Swanson et al., 2021). Nevertheless, many features implicated in cell fate decisions remain not accounted for. Single-cell proteomics may provide more robust means to infer a determinant’s activity but currently are less scalable, and TFs that tend to be present in low copy numbers will remain challenging to quantify (Derks et al., 2022). Proteogenomics-based approaches enable signal amplification, but require affinity reagents (e.g., nanobodies, antibodies) to reliably quantify the protein of interest or its activity-modulating post-translational modification (Fiskin et al., 2022; Mimitou et al., 2021), and are still relatively scarce for intracellular signaling or TF molecules. Increasingly sensitive experimental spatial and analytical approaches are rapidly enhancing conventional single-cell omic workflows and will more readily enable inferences about cell-cell communication and interactions with, for example, a stem cell’s niche that may modulate its fate (Moffitt et al., 2022; Nitzan et al., 2019). Geometric position, cell shape, polarity, size/volume, mechanical forces (Chan et al., 2019; Valet et al., 2021; Yang et al., 2021), and metabolic activity are further regulatory factors that determine cell fate (Chakrabarty and Chandel, 2021), with their relative contribution likely being context dependent. Moving forward, identifying and accounting for all relevant features appears heartening toward a non-chaotic understanding of cell fate decisions.
In this realm, lineage- and clonal-tracing approaches have demonstrated their promise to identify predictive features of cell fates by linking clonal cells together over different intervals over the differentiation trajectory. However, current approaches may not enable inferences of detailed phylogenetic trees but rather divide cells into a few clonal subgroups, hence providing limited power in locating the initial state where each current cell originated from and where different subclones diverged from each other in the past. For example, in human hematopoiesis, we are currently able to readily capture the full diversity of cell types and states for select data modalities. However, accounting for the high polyclonality of the system, with tens to hundreds of thousands of stem cells actively contributing to blood production (Lee-Six et al., 2018; Mitchell et al., 2022), we will require more scalable measurements to account for this clonal complexity and do so in conjunction with readouts compatible with the different feature sets implicated in cell fate (Lareau et al., 2021). Moreover, we must consider that cell fate decisions are also orchestrated on a systems level, as perturbations (e.g., infection, blood loss) may require adaptations of the stem cell pool to meet a specific short-term demand. More generally we shall account for the interplay among different cells in a tissue to maintain homeostasis (Jerby-Arnon and Regev, 2022).
Yet, we do acknowledge the intuition that due to the complexity of the system (e.g., high-dimensional feature space, cell-cell communication, cell-environmental interactions, cell migration, etc.) and potentially difficult to overcome technical limitations, one will not be able to fully characterize the full phase space (i.e., the complete governing feature set for an entire range of possible and dynamically accessible cell states) of cell fate dynamical paths. If we accept this paradigm, the question should therefore be which combination of measurable features provides the best predictive power on the future state of the cell in finite time (rather than seeking a predictive power into arbitrary long time intervals, which is as overly ambitious as predicting whether a child will become a mechanic or a musician at birth), and to quantify for a specific set of features, over which time length their predictions remain reliable.
Summary and conclusions
Cell fate decisions are central to development, homeostasis, and adaptive responses to sustain the cellular integrity of multi-cellular organisms. A deeper understanding of the molecular circuits underlying these decisions is thereby of substantial value for cell and regenerative medicine, where specific types of cells are required in large amounts and/or engineered with desired properties. The ability of single-cell genomic approaches to resolve cellular heterogeneity and associate molecular features with differentiation outcomes has already showcased their potential to enhance our understanding of the molecular underpinnings of cell fate.
In this perspective, we, therefore, discussed select methodologies and current practices in cell fate research. We put popular mathematical models of cell differentiation dynamics (pseudotime, diffusion-drift, cell state velocities, and OT) into perspective and clarify their relations with each other. We propose future investigations of “chaos” models for cell differentiation, given the unmeasurably high complexity of the system, analogous to other high-dimensional chaotic problems like weather forecasting. Adapting such higher-level general frameworks could help us refine our expectations and problem sets in the studies of cell fate. We finally highlight the need to reduce the complexity of the problem to tractable approximations and models and discuss related contemporary approaches.
Moving forward, we await multi-omics-based integration of additional molecular features with lineage/clonal tracing and mathematical modeling with concomitant experimental validations including in vitro systems of human developmental processes (He et al., 2022; Liu et al., 2021; Yu et al., 2021) to further catalyze discoveries within this realm.
Author contributions
L.H. and L.S.L. contributed equally to this manuscript.
Acknowledgments
We would like to thank Nikolaus Rajewsky for the useful feedback and discussion as well as the reviewers of this manuscript who helped us improve the quality of this work. L.H. and L.S.L. are supported by the Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association (MDC), Berlin Institute for Medical Systems Biology (BIMSB). L.S.L. is supported by the Berlin Institute of Health at Charité Universitätsmedizin Berlin, an Emmy Noether fellowship by the German Research Foundation (Deutsche Forschungsgemeinschaft, LU 2336/2-1); a Longevity Impetus grant; the National Institutes of Health (1UM1HG012076-01); and a Hector Research Career Development Award by the Hector Fellow Academy. L.H. is supported by the Bundesministerium für Bildung und Forschung (BMBF) consortium grant “LeukoSyStem.” Select figure panels were created with BioRender.com. We apologize to all our colleagues whose work could not be specifically mentioned due to space restrictions.
Conflict of interests
L.S.L. is a consultant to Cartography Biosciences, with no competing interests related to this manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.12.003.
Contributor Information
Laleh Haghverdi, Email: laleh.haghverdi@mdc-berlin.de.
Leif S. Ludwig, Email: leif.ludwig@bih-charite.de.
Supplemental information
References
- Amann-Zalcenstein D., Tian L., Schreuder J., Tomei S., Lin D.S., Fairfax K.A., Bolden J.E., McKenzie M.D., Jarratt A., Hilton A., et al. A new lymphoid-primed progenitor marked by Dach1 downregulation identified with single cell multi-omics. Nat. Immunol. 2020;21:1574–1584. doi: 10.1038/s41590-020-0799-x. [DOI] [PubMed] [Google Scholar]
- Bao E.L., Cheng A.N., Sankaran V.G. The genetics of human hematopoiesis and its disruption in disease. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201910316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C.S., van Oudenaarden A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 2019;20:753–765. doi: 10.1038/s41580-019-0186-3. [DOI] [PubMed] [Google Scholar]
- Bartosovic M., Kabbe M., Castelo-Branco G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 2021;39:825–835. doi: 10.1038/s41587-021-00869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Beumer J., Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2021;22:39–53. doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- Bocci F., Zhou P., Nie Q. spliceJAC: transition genes and state-specific gene regulation from single-cell transcriptome data. Mol. Syst. Biol. 2022;18 doi: 10.15252/msb.202211176. e11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Corces M.R., Lareau C.A., Wu B., Schep A.N., Aryee M.J., Majeti R., Chang H.Y., Greenleaf W.J. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173:1535–1548.e16. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.-R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Camp J.G., Platt R., Treutlein B. Mapping human cell phenotypes to genotypes with single-cell genomics. Science. 2019;365:1401–1405. doi: 10.1126/science.aax6648. [DOI] [PubMed] [Google Scholar]
- Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J., Daza R.M., McFaline-Figueroa J.L., Packer J.S., Christiansen L., et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellera-Garcia S., Pulecio J., Dhulipala K., Siva K., Rayon-Estrada V., Singbrant S., Sommarin M.N.E., Walkley C.R., Soneji S., Karlsson G., et al. Defining the minimal factors required for erythropoiesis through direct lineage conversion. Cell Rep. 2016;15:2550–2562. doi: 10.1016/j.celrep.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrelha J., Meng Y., Kettyle L.M., Luis T.C., Norfo R., Alcolea V., Boukarabila H., Grasso F., Gambardella A., Grover A., et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554:106–111. doi: 10.1038/nature25455. [DOI] [PubMed] [Google Scholar]
- Chakrabarty R.P., Chandel N.S. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28:394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.J., Costanzo M., Ruiz-Herrero T., Mönke G., Petrie R.J., Bergert M., Diz-Muñoz A., Mahadevan L., Hiiragi T. Hydraulic control of mammalian embryo size and cell fate. Nature. 2019;571:112–116. doi: 10.1038/s41586-019-1309-x. [DOI] [PubMed] [Google Scholar]
- Cho H., Ayers K., de Pills L., Kuo Y.-H., Park J., Radunskaya A., Rockne R. Modelling acute myeloid leukaemia in a continuum of differentiation states. Lett. Biomath. 2018;5:S69–S98. doi: 10.1080/23737867.2018.1472532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.-H.K., Budde M.W., Granados A.A., Cabrera M., Yoon S., Cho S., Huang T.-H., Koulena N., Frieda K.L., Cai L., et al. Imaging cell lineage with a synthetic digital recording system. Science. 2021;372 doi: 10.1126/science.abb3099. eabb3099. [DOI] [PubMed] [Google Scholar]
- Derks J., Leduc A., Wallmann G., Huffman R.G., Willetts M., Khan S., Specht H., Ralser M., Demichev V., Slavov N. Increasing the throughput of sensitive proteomics by plexDIA. Nat. Biotechnol. 2022:1–10. doi: 10.1038/s41587-022-01389-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey E.B., Taylor D.T., Johnston C.A. Cell fate decision making through oriented cell division. J. Dev. Biol. 2015;3:129–157. doi: 10.3390/jdb3040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H.M., Horvitz H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Farrell J.A., Wang Y., Riesenfeld S.J., Shekhar K., Regev A., Schier A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360 doi: 10.1126/science.aar3131. eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell K.A., Vassiliadis D., Lam E.Y.N., Martelotto L.G., Balic J.J., Hollizeck S., Weber T.S., Semple T., Wang Q., Miles D.C., et al. Non-genetic determinants of malignant clonal fitness at single-cell resolution. Nature. 2022;601:125–131. doi: 10.1038/s41586-021-04206-7. [DOI] [PubMed] [Google Scholar]
- Fischer D.S., Fiedler A.K., Kernfeld E.M., Genga R.M.J., Bastidas-Ponce A., Bakhti M., Lickert H., Hasenauer J., Maehr R., Theis F.J. Inferring population dynamics from single-cell RNA-sequencing time series data. Nat. Biotechnol. 2019;37:461–468. doi: 10.1038/s41587-019-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin E., Lareau C.A., Ludwig L.S., Eraslan G., Liu F., Ring A.M., Xavier R.J., Regev A. Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC. Nat. Biotechnol. 2022;40:374–381. doi: 10.1038/s41587-021-01065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gábor A., Villaverde A.F., Banga J.R. Parameter identifiability analysis and visualization in large-scale kinetic models of biosystems. BMC Syst. Biol. 2017;11:54. doi: 10.1186/s12918-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362 doi: 10.1126/science.aaq0681. eaaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerri C., Menchero S., Mahadevaiah S.K., Turner J.M.A., Niakan K.K. Human embryogenesis: a comparative perspective. Annu. Rev. Cell Dev. Biol. 2020;36:411–440. doi: 10.1146/annurev-cellbio-022020-024900. [DOI] [PubMed] [Google Scholar]
- Gillespie D.T. The chemical Langevin equation. J. Chem. Phys. 2000;113:297–306. [Google Scholar]
- Gorin G., Fang M., Chari T., Pachter L. RNA velocity unraveled. bioRxiv. 2022 doi: 10.1101/2022.02.12.480214. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebogi C., Ott E., Yorke J.A. Chaos, strange attractors, and fractal basin boundaries in nonlinear dynamics. Science. 1987;238:632–638. doi: 10.1126/science.238.4827.632. [DOI] [PubMed] [Google Scholar]
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., Dahmani A., Lameiras S., Reyal F., Frenoy O., et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019;51:1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- Gu Y., Blaauw D., Welch J. Variational mixtures of ODEs for inferring cellular gene expression dynamics. arXiv. 2022 doi: 10.48550/arXiv.2207.04166. Preprint at. [DOI] [Google Scholar]
- Haas S., Trumpp A., Milsom M.D. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22:627–638. doi: 10.1016/j.stem.2018.04.003. [DOI] [PubMed] [Google Scholar]
- He Z., Maynard A., Jain A., Gerber T., Petri R., Lin H.-C., Santel M., Ly K., Dupré J.S., Sidow L., et al. Lineage recording in human cerebral organoids. Nat. Methods. 2022;19:90–99. doi: 10.1038/s41592-021-01344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N., et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535:299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- Jerby-Arnon L., Regev A. DIALOGUE maps multicellular programs in tissue from single-cell or spatial transcriptomics data. Nat. Biotechnol. 2022;40:1467–1477. doi: 10.1038/s41587-022-01288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto K., Hoffmann C.M., Morris S.A. CellOracle: dissecting cell identity via network inference and in silico gene perturbation. bioRxiv. 2020 doi: 10.1101/2020.02.17.947416. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria R.K., Munschauer M., Ulirsch J.C., Fiorini C., Ludwig L.S., McFarland S.K., Abdulhay N.J., Specht H., Keshishian H., Mani D.R., et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173:90–103.e19. doi: 10.1016/j.cell.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M., Bergen V., Klein M., Setty M., Reuter B., Bakhti M., Lickert H., Ansari M., Schniering J., Schiller H.B., et al. CellRank for directed single-cell fate mapping. Nat. Methods. 2022;19:159–170. doi: 10.1038/s41592-021-01346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau C.A., Duarte F.M., Chew J.G., Kartha V.K., Burkett Z.D., Kohlway A.S., Pokholok D., Aryee M.J., Steemers F.J., Lebofsky R., Buenrostro J.D. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019;37:916–924. doi: 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau C.A., Ludwig L.S., Muus C., Gohil S.H., Zhao T., Chiang Z., Pelka K., Verboon J.M., Luo W., Christian E., et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 2021;39:451–461. doi: 10.1038/s41587-020-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Six H., Øbro N.F., Shepherd M.S., Grossmann S., Dawson K., Belmonte M., Osborne R.J., Huntly B.J.P., Martincorena I., Anderson E., et al. Population dynamics of normal human blood inferred from somatic mutations. Nature. 2018;561:473–478. doi: 10.1038/s41586-018-0497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett L.A., Sankaran V.G. Unraveling hematopoiesis through the lens of genomics. Cell. 2020;182:1384–1400. doi: 10.1016/j.cell.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Tan J.P., Schröder J., Aberkane A., Ouyang J.F., Mohenska M., Lim S.M., Sun Y.B.Y., Chen J., Sun G., et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature. 2021;591:627–632. doi: 10.1038/s41586-021-03372-y. [DOI] [PubMed] [Google Scholar]
- Loeffler D., Schroeder T. Understanding cell fate control by continuous single-cell quantification. Blood. 2019;133:1406–1414. doi: 10.1182/blood-2018-09-835397. [DOI] [PubMed] [Google Scholar]
- Ludwig L.S., Gazda H.T., Eng J.C., Eichhorn S.W., Thiru P., Ghazvinian R., George T.I., Gotlib J.R., Beggs A.H., Sieff C.A., et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig L.S., Lareau C.A., Ulirsch J.C., Christian E., Muus C., Li L.H., Pelka K., Ge W., Oren Y., Brack A., et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell. 2019;176:1325–1339.e22. doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Zhang B., LaFave L.M., Earl A.S., Chiang Z., Hu Y., Ding J., Brack A., Kartha V.K., Tay T., et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103–1116.e20. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marot-Lassauzaie V., Bouman B.J., Donaghy F.D., Demerdash Y., Essers M.A.G., Haghverdi L. Towards reliable quantification of cell state velocities. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1010031. e1010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.M. Epigenetic drift in aging identical twins. Proc. Natl. Acad. Sci. USA. 2005;102:10413–10414. doi: 10.1073/pnas.0504743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Lareau C.A., Chen K.Y., Zorzetto-Fernandes A.L., Hao Y., Takeshima Y., Luo W., Huang T.-S., Yeung B.Z., Papalexi E., et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 2021;39:1246–1258. doi: 10.1038/s41587-021-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell E., Spencer Chapman M., Williams N., Dawson K.J., Mende N., Calderbank E.F., Jung H., Mitchell T., Coorens T.H.H., Spencer D.H., et al. Clonal dynamics of haematopoiesis across the human lifespan. Nature. 2022;606:343–350. doi: 10.1038/s41586-022-04786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnenzweig M., Mayshar Y., Cheng S., Ben-Yair R., Hadas R., Rais Y., Chomsky E., Reines N., Uzonyi A., Lumerman L., et al. A single-embryo, single-cell time-resolved model for mouse gastrulation. Cell. 2021;184:2825–2842.e22. doi: 10.1016/j.cell.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J.R., Lundberg E., Heyn H. The emerging landscape of spatial profiling technologies. Nat. Rev. Genet. 2022;23:741–759. doi: 10.1038/s41576-022-00515-3. [DOI] [PubMed] [Google Scholar]
- Ng A.H.M., Khoshakhlagh P., Rojo Arias J.E., Pasquini G., Wang K., Swiersy A., Shipman S.L., Appleton E., Kiaee K., Kohman R.E., et al. A comprehensive library of human transcription factors for cell fate engineering. Nat. Biotechnol. 2021;39:510–519. doi: 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzan M., Karaiskos N., Friedman N., Rajewsky N. Gene expression cartography. Nature. 2019;576:132–137. doi: 10.1038/s41586-019-1773-3. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J., et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365 doi: 10.1126/science.aax1971. eaax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry A., Rulands S., Reik W. Active turnover of DNA methylation during cell fate decisions. Nat. Rev. Genet. 2021;22:59–66. doi: 10.1038/s41576-020-00287-8. [DOI] [PubMed] [Google Scholar]
- Petit F., Sears K.E., Ahituv N. Limb development: a paradigm of gene regulation. Nat. Rev. Genet. 2017;18:245–258. doi: 10.1038/nrg.2016.167. [DOI] [PubMed] [Google Scholar]
- Qiu X., Zhang Y., Martin-Rufino J.D., Weng C., Hosseinzadeh S., Yang D., Pogson A.N., Hein M.Y., Hoi Joseph Min K., Wang L., et al. Mapping transcriptomic vector fields of single cells. Cell. 2022;185:690–711.e45. doi: 10.1016/j.cell.2021.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.J., Jones M.G., Okimoto R.A., Nanjo S., Chan M.M., Yosef N., Bivona T.G., Weissman J.S. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science. 2021;371 doi: 10.1126/science.abc1944. eabc1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau J. Oxford University Press; 2017. Statistical Physics and Thermodynamics. [Google Scholar]
- Rodriguez-Fraticelli A.E., Wolock S.L., Weinreb C.S., Panero R., Patel S.H., Jankovic M., Sun J., Calogero R.A., Klein A.M., Camargo F.D. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553:212–216. doi: 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A.E., Weinreb C., Wang S.-W., Migueles R.P., Jankovic M., Usart M., Klein A.M., Lowell S., Camargo F.D. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature. 2020;583:585–589. doi: 10.1038/s41586-020-2503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A., Ram O., Shoresh N., Sperling R.A., Goren A., Weitz D.A., Bernstein B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Sankaran V.G., Ghazvinian R., Do R., Thiru P., Vergilio J.-A., Beggs A.H., Sieff C.A., Orkin S.H., Nathan D.G., Lander E.S., et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Invest. 2012;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Granja J.M., Yost K.E., Qi Y., Meschi F., McDermott G.P., Olsen B.N., Mumbach M.R., Pierce S.E., Corces M.R., et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S., Basso-Ricci L., Dionisio F., Pellin D., Giannelli S., Salerio F.A., Leonardelli L., Cicalese M.P., Ferrua F., Aiuti A., et al. Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat. Med. 2018;24:1683–1690. doi: 10.1038/s41591-018-0195-3. [DOI] [PubMed] [Google Scholar]
- Schiebinger G., Shu J., Tabaka M., Cleary B., Subramanian V., Solomon A., Gould J., Liu S., Lin S., Berube P., et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell. 2019;176:1517. doi: 10.1016/j.cell.2019.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N. Mechanisms of human embryo development: from cell fate to tissue shape and back. Development. 2020;147 doi: 10.1242/dev.190629. dev190629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.W., Pielke R.A., Sr., Zeng X., Faghih-Naini S., Shie C.L., Atlas R., Baik J.J., Reyes T.A.L. 11th Chaotic Modeling and Simulation International Conference. 2018. Butterfly effects of the first and second kinds: new insights revealed by high-dimensional lorenz models.https://www.researchgate.net/profile/Bo-Wen-Shen/publication/326274429_Butterfly_Effects_of_the_First_and_Second_Kinds_New_Insights_Revealed_by_High-dimensional_Lorenz_Models/links/5c04336c92851c63cab5cd7d/Butterfly-Effects-of-the-First-and-Second-Kinds-New-Insights-Revealed-by-High-dimensional-Lorenz-Models.pdf [Google Scholar]
- Shinbrot T., Grebogi C., Wisdom J., Yorke J.A. Chaos in a double pendulum. Am. J. Phys. 1992;60:491–499. doi: 10.1119/1.16860. [DOI] [Google Scholar]
- Soldatov R., Kaucka M., Kastriti M.E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Häring M., Dyachuk V., et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science. 2019;364 doi: 10.1126/science.aas9536. eaas9536. [DOI] [PubMed] [Google Scholar]
- Spanjaard B., Hu B., Mitic N., Olivares-Chauvet P., Janjuha S., Ninov N., Junker J.P. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol. 2018;36:469–473. doi: 10.1038/nbt.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stadhouders R., Filion G.J., Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- Strasser M.K., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Schroeder T., Theis F.J., Marr C. Lineage marker synchrony in hematopoietic genealogies refutes the PU.1/GATA1 toggle switch paradigm. Nat. Commun. 2018;9:2697. doi: 10.1038/s41467-018-05037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.-J., Klein A., Hofmann O., Camargo F.D. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson E., Lord C., Reading J., Heubeck A.T., Genge P.C., Thomson Z., Weiss M.D., Li X.-J., Savage A.K., Green R.R., et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. Elife. 2021;10 doi: 10.7554/eLife.63632. e63632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiétart R.A., Forgues B. Chaos theory and organization. Organ. Sci. 1995;6:19–31. doi: 10.1287/orsc.6.1.19. [DOI] [Google Scholar]
- Tian L., Tomei S., Schreuder J., Weber T.S., Amann-Zalcenstein D., Lin D.S., Tran J., Audiger C., Chu M., Jarratt A., et al. Clonal multi-omics reveals Bcor as a negative regulator of emergency dendritic cell development. Immunity. 2021;54:1338–1351.e9. doi: 10.1016/j.immuni.2021.03.012. [DOI] [PubMed] [Google Scholar]
- Tritschler S., Büttner M., Fischer D.S., Lange M., Bergen V., Lickert H., Theis F.J., Klein A., Treutlein B. Concepts and limitations for learning developmental trajectories from single cell genomics. Development. 2019;146 doi: 10.1242/dev.170506. dev170506. [DOI] [PubMed] [Google Scholar]
- Valet M., Siggia E.D., Brivanlou A.H. Mechanical regulation of early vertebrate embryogenesis. Nat. Rev. Mol. Cell Biol. 2021;23:169–184. doi: 10.1038/s41580-021-00424-z. [DOI] [PubMed] [Google Scholar]
- Verd B., Crombach A., Jaeger J. Classification of transient behaviours in a time-dependent toggle switch model. BMC Syst. Biol. 2014;8:43. doi: 10.1186/1752-0509-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C.H. Gorge Allen and Unwin (London); 1957. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. [Google Scholar]
- Wagner D.E., Klein A.M. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet. 2020;21:410–427. doi: 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-W., Herriges M.J., Hurley K., Kotton D.N., Klein A.M. CoSpar identifies early cell fate biases from single-cell transcriptomic and lineage information. Nat. Biotechnol. 2022;40:1066–1074. doi: 10.1038/s41587-022-01209-1. [DOI] [PubMed] [Google Scholar]
- Weinreb C., Wolock S., Tusi B.K., Socolovsky M., Klein A.M. Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl. Acad. Sci. USA. 2018;115:E2467–E2476. doi: 10.1073/pnas.1714723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C., Rodriguez-Fraticelli A., Camargo F.D., Klein A.M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 2020;367 doi: 10.1126/science.aaw3381. eaaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat J.C., Sella Y., Willcockson M., Skoultchi A.I., Bergman A., Singer R.H., Steidl U. Single-molecule imaging of transcription dynamics in somatic stem cells. Nature. 2020;583:431–436. doi: 10.1038/s41586-020-2432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.J., Furlan S.N., Mihalas A.B., Kaya-Okur H.S., Feroze A.H., Emerson S.N., Zheng Y., Carson K., Cimino P.J., Keene C.D., et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 2021;39:819–824. doi: 10.1038/s41587-021-00865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Xue S.L., Chan C.J., Rempfler M., Vischi D., Maurer-Gutierrez F., Hiiragi T., Hannezo E., Liberali P. Cell fate coordinates mechano-osmotic forces in intestinal crypt formation. Nat. Cell Biol. 2021;23:733–744. doi: 10.1038/s41556-021-00700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wei Y., Duan J., Schmitz D.A., Sakurai M., Wang L., Wang K., Zhao S., Hon G.C., Wu J. Blastocyst-like structures generated from human pluripotent stem cells. Nature. 2021;591:620–626. doi: 10.1038/s41586-021-03356-y. [DOI] [PubMed] [Google Scholar]
- Yu V.W.C., Yusuf R.Z., Oki T., Wu J., Saez B., Wang X., Cook C., Baryawno N., Ziller M.J., Lee E., et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell. 2016;167:1310–1322.e17. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
- Zeng H. What is a cell type and how to define it? Cell. 2022;185:2739–2755. doi: 10.1016/j.cell.2022.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Yu M., Huang H., Juric I., Abnousi A., Hu R., Lucero J., Behrens M.M., Hu M., Ren B. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 2019;26:1063–1070. doi: 10.1038/s41594-019-0323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.