Abstract
Mitochondrial retrograde signaling (MRS) supports photosynthetic function under a variety of conditions. Induction of mitochondrial dysfunction with myxothiazol (a specific inhibitor of the mitochondrial bc1 complex) or antimycin A (an inhibitor of the mitochondrial bc1 complex and cyclic electron transport in the chloroplast under light conditions) in the light and dark revealed diurnal control of MRS. This was evidenced by (1) significantly enhanced binding of ANAC017 to promoters in the light compared with the dark in Arabidopsis plants treated with myxothiazol (but not antimycin A), (2) overlap in the experimentally determined binding sites for ANAC017 and circadian clock regulators in the promoters of ANAC013 and AOX1a, (3) a diurnal expression pattern for ANAC017 and transcription factors it regulates, (4) altered expression of ANAC017-regulated genes in circadian clock mutants with and without myxothiazol treatment, and (5) a decrease in the magnitude of LHY and CCA1 expression in an ANAC017-overexpressing line and protein–protein interaction between ANAC017 and PIF4. This study also shows a large difference in transcriptome responses to antimycin A and myxothiazol in the dark: these responses are ANAC017 independent, observed in shoots and roots, similar to biotic challenge and salicylic acid responses, and involve ERF and ZAT transcription factors. This suggests that antimycin A treatment stimulates a second MRS pathway that is mediated or converges with salicylic acid signaling and provides a merging point with chloroplast retrograde signaling.
Key words: mitochondria, chloroplast, retrograde signaling, diurnal, circadian clock, ANAC017
The mitochondrial stress response is a complex response that involves ANAC017-dependent and -independent pathways. The ANAC017 pathway is co-regulated by circadian clock regulators to coordinate photosynthetic activity with mitochondrial capacity to oxidize excess chloroplast reducing equivalents when required.
Introduction
Organelle signaling has been studied for almost 50 years, with mitochondrial signaling initially studied in Saccharomyces cerevisiae (yeast) and plastid (chloroplast) signaling revealed in Hordeum vulgare (barley) (Börner, 2017). It is now clear that organelle signaling, often referred to as retrograde signaling, occurs in all eukaryotic organisms studied and interacts with other signaling pathways. The distinctive feature of organelle retrograde signaling is that the functional state of the organelle initiates a signal transduction cascade, ultimately resulting in a transcriptional response that effects whole-cell function. By contrast, anterograde control regulates (internal or external) transcriptional and post-transcriptional responses independent of the status of organelle function. In plants, retrograde signaling is essential for optimal growth and development, and key regulators of organelle signaling in Arabidopsis thaliana are essential for flooding, drought, and high light tolerance (Giraud et al., 2008; Crawford et al., 2018; Phua et al., 2018; Meng et al., 2019).
Although definitions of anterograde and retrograde regulatory pathways are useful for classification, stimuli such as light often trigger regulation via a variety of mechanisms to initiate and maintain chloroplast development. By definition, organelle retrograde signaling pathways interact with other signaling pathways to produce transcriptomes that are adjusted to the prevailing conditions. Given the interactions between mitochondria and chloroplasts at a metabolic level (Vanlerberghe et al., 2020), integration of organelle retrograde signaling is likely to be required to coordinate the activities of these organelles. However, studies of chloroplast and mitochondrial retrograde signaling proceed from studies targeted at either organelle, and overlap between pathways is then inferred from overlapping transcriptome results. Although these overlapping transcriptomes are useful for indicating convergence or interactions in different signaling or regulatory pathways, the diversity of experimental conditions may also cause secondary responses reflected in the transcriptomic changes. Also, converging co-expression does not necessarily mean co-regulation (Wang et al., 2020).
Chloroplasts and mitochondria function together in metabolic pathways, with the photorespiratory pathway as a well-known example. Other pathways are also essential to the coordinated function of energy and signaling processes in plant cells (Lim et al., 2020; Selinski and Scheibe, 2019). For example, the malate shuttle forms a communication pathway between chloroplasts and mitochondria via the induction of cell death through mitochondrial oxidation of reducing equivalents exported from the chloroplast (Zhao et al., 2018). Similarly, the alternative oxidase (AOX) in plant mitochondria is induced in a variety of mutants or by treatments that perturb the functioning of chloroplasts, in particular photosynthesis (Vanlerberghe et al., 2020). Consistent with their interlinked functions in metabolism, it is expected that there are also signaling connections between these organelles. It was established early that the SAL1 phosphatase is targeted to both mitochondria and chloroplasts (Estavillo et al., 2011) and thus has the potential to initiate signaling from both organelles, although no definitive studies exist to show that this occurs. A sal1 mutant complemented with the protein exclusively targeted to mitochondria more closely resembles the wild type than a mutant complemented with the protein targeted only to chloroplasts (Ashykhmina et al., 2021). Several dual-targeted proteins, including members of the Whirly transcription factors, are also potential candidates for coordination of organelle signaling (Mackenzie and Kundariya, 2020; Wu et al., 2020). Given the central role of organelle gene expression in retrograde signaling, the dual targeting of proteins associated with organelle transcription, editing, and maturation may also coordinate signaling (Hedtke et al., 2000; Duchene et al., 2005; Colcombet et al., 2013). Indeed, PROLYL-tRNA SYNTHETASE1 (PRORS1) was the first dual-targeted protein shown to have synergistic effects on signaling (Pesaresi et al., 2006).
Although convergence of chloroplast and mitochondrial signaling pathways is likely, identification of regulatory components involved in both is limited at present. ABSCISIC ACID-INSENSITIVE 4 (ABI4) was initially suggested to be involved in both chloroplast and mitochondrial retrograde signaling (Koussevitzky et al., 2007; Giraud et al., 2009), but its role in chloroplast retrograde signaling has been re-evaluated because it does not display the gun gene expression profile as initially reported (Kacprzak et al., 2019). RADICAL CELL DEATH 1 (RCD1) binds to the NAC domain transcription factors ANAC013 and ANAC017 and is a coordination point for chloroplast and mitochondrial reactive oxygen species (ROS) signaling pathways (Shapiguzov et al., 2019). The repression of ANAC017 and ANAC013 by RCD1 raises the question of whether these NAC transcription factors are activators of chloroplast and mitochondrial retrograde signaling. Another convergence point for the integration of chloroplast and mitochondrial retrograde signals is the Mediator complex in the nucleus (He et al., 2021). A subunit of the Mediator complex, CYCLIN-DEPENDENT KINASE E1 (CDKE1), is involved in mitochondrial retrograde signaling through interaction with Sucrose non-fermenting 1 (SNF1)-related protein kinase 1.1 (KIN10) in Arabidopsis thaliana (Ng et al., 2013b). CDKE1 also regulates the expression of LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2.4 (LHCB2.4) in response to changes in redox status in the photosynthetic electron transport chain (Blanco et al., 2014).
Although the light-regulated expression of genes encoding chloroplast proteins was one of the earliest molecular studies in plant molecular biology (Bogorad, 2001), light regulation of genes encoding mitochondrial proteins is less well understood. Some components in mitochondria, such as the subunits of glycine decarboxylase, display classical light responses, linked to their role in photorespiration (Oliver, 1994). Binding of TCP transcription factors to the promoter regions of several genes encoding mitochondrial proteins, and their interaction with components of the circadian clock, results in a diurnal expression pattern for several genes encoding mitochondrial proteins such as TRANSLOCASE OUTER MEMBRANE 20-2 (Tom20-2), Rieske FeS protein, and some mitochondrial carrier proteins (Giraud et al., 2010). As many as 65% of genes that encode mitochondrial proteins display oscillations in transcript abundance, many of which are enriched in the night phase (Cervela-Cardona et al., 2021b). Specific examples of genes with a diurnal expression pattern are FUMARASE 2 (FUM2) (Cervela-Cardona et al., 2021b), NAD-DEPENDENT MALIC ENZYME (NAD-ME) (Tronconi et al., 2008), and ISOCITRATE DEHYDROGENASE (ICDH) (Gibon et al., 2006). Mitochondrial proteins and metabolites also show diurnal rhythms (Lee et al., 2010). Thus, it appears from the above examples that genes encoding mitochondrial proteins show diurnal changes in transcript abundance, and this is also reflected in protein and metabolite signatures. For the latter, it has been proposed that changes in metabolites drive changes in gene expression, not the other way around (Gibon et al., 2006).
Although AOX is an established marker for mitochondrial dysfunction, it is also essential for maintenance of efficient chloroplast (i.e. photosynthetic) function under normal, drought, and high-light conditions (Vanlerberghe et al., 2020). We therefore tested whether mitochondrial retrograde signaling is affected by light (anterograde) regulation. We characterized the transcriptome responses of ANAC017, the master regulator of mitochondrial signaling (Ng et al., 2013b, 2014), after perturbation of mitochondrial function in the light or dark with antimycin A and myxothiazol to determine whether light influences (1) the mitochondrial dysfunction response and (2) the regulatory network of ANAC017. The results revealed significant differences in transcriptome responses to treatment with both inhibitors in the light and dark, as well as differential binding of ANAC017 in the light and dark when mitochondrial dysfunction was imposed by myxothiazol treatment. Analysis of ChIP-seq data sets of circadian clock regulators and the response of marker genes to mitochondrial dysfunction confirmed that the mitochondrial retrograde response (MRR) is coordinately modulated by circadian clock regulators and ANAC017. Also, there is a large ANAC017-independent alteration of gene expression induced by antimycin A in the dark that shares significant overlap with biotic stress responses and salicylic acid signaling, representing an additional convergence point of mitochondrial and chloroplast retrograde signaling.
Results
The transcriptome response to mitochondrial dysfunction differs between antimycin A and myxothiazol treatments in the light and dark
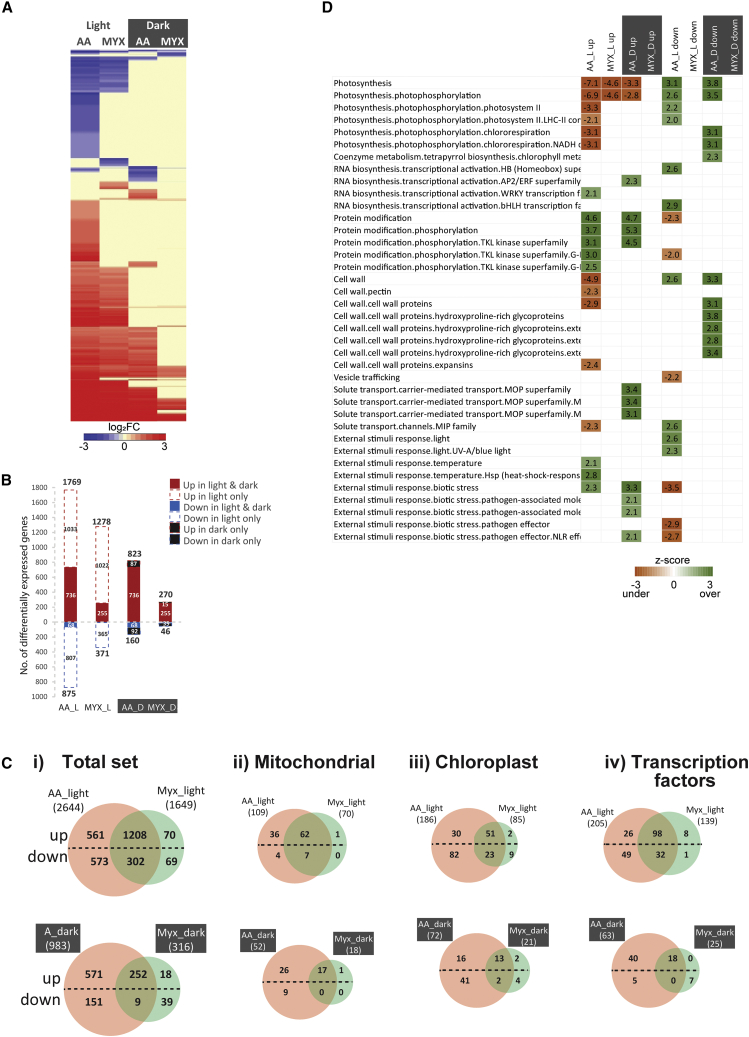
Arabidopsis seedlings treated with antimycin A or myxothiazol that were 1 h into the light or dark phase of a 16 h light:8 h dark growth cycle revealed substantial transcriptomic differences between treatments and between the light and dark phases within each treatment (Figure 1, Supplemental Data 1). Myxothiazol inhibits complex III (cytochrome bc1 complex) of the respiratory chain at the Qo site, and antimycin A inhibits this complex at the Qi site. Antimycin A also inhibits cyclic electron transport at PROTON GRADIENT REGULATION 5 (PGR5), i.e. it inhibits two different functional sites in the light (Labs et al., 2016; Alber and Vanlerberghe, 2019). Thus, an additional effect of antimycin A compared with myxothiazol in the light was likely, although both inhibitors were expected to produce a similar response in the dark. The latter has been shown in Nicotiana tabacum using marker genes (Alber and Vanlerberghe, 2019). However, here, in the dark, antimycin A treatment resulted in 983 differentially expressed genes (DEGs) (defined by a |log2(fold change)| > 1 and FDR < 0.05) compared with 316 DEGs following myxothiazol treatment (Figure 1A, 1B, and 1Ci, Supplemental Data 1). This large difference was unexpected based on known sites of inhibition and previous studies using marker genes (Ng et al., 2013a; Vanlerberghe et al., 2020; Meng et al., 2022). This difference in the dark was also observed when subsets of genes were analyzed (Figure 1Cii–iv). Among genes encoding mitochondrial proteins, chloroplast proteins, and transcription factors, 52, 72, and 63 DEGs were identified under antimycin A treatment, whereas only 18, 21, and 25 DEGs were observed under myxothiazol treatment (Figure 1C, Supplemental Data 1). Although there was a difference in the number of DEGs, myxothiazol treatment still led to the expected induction of marker genes encoding mitochondrial proteins: ALTERNATIVE OXIDASE 1A (AOX1a and AOX1d), NAD(P)H DEHYDROGENASE B2 (NDB2), and cytochrome bc1 synthesis (BCS1)/Outer Membrane 66 kDa (OM66) (Supplemental Data 1). The ANAC013 transcription factor, which acts downstream of ANAC017, was induced, as were WRKY DNA-BINDING PROTEIN 15 (WRKY 15), which is responsible for the mitochondrial stress response under osmotic stress (Vanderauwera et al., 2012), WRKY33, which has been linked to submergence tolerance (Liu et al., 2021), and cytokinin response factors 5 and 6 (Supplemental Data 1), all previously associated with the mitochondrial retrograde response (Selinski et al., 2018). In the dark, the magnitude of increase displayed a pattern of being slightly higher with antimycin A than with myxothiazol. However, the overall difference in the magnitude of response of genes that were commonly induced by antimycin A and myxothiazol in the dark was not significant (Supplemental Data 1).
Figure 1.
Transcriptome responses to antimycin A and myxothiazol treatment in light and dark conditions.
(A) Fold changes in response to treatment compared with mock treatment were hierarchically clustered and visualized in a heatmap.
(B) Numbers of differentially expressed genes (p < 0.05, |log2(fold change)| > 1) are shown, and numbers of genes common and exclusive to light and dark conditions are indicated.
(C) Venn diagram showing the overlap in differentially expressed genes in response to antimycin A and myxothiazol under light and dark conditions for (i) the total gene set and genes encoding (ii) mitochondrial proteins, (iii) chloroplast proteins, and (iv) transcription factors. Upregulated genes are above the dashed lines, whereas downregulated genes are below the dashed lines.
(D) PageMan visualization showing under- and overrepresented functional categories in each treatment. ORA, Fisher’s test, p < 0.05.
Analysis of overrepresented functional categories in response to antimycin A and myxothiazol treatment (PageMan ORA, Fisher’s test, p < 0.05; Usadel et al., 2006) revealed that in the dark, the DEGs upregulated under antimycin A were enriched in external stimuli–biotic response, protein modification, and solute transport, and downregulated genes were enriched in components of photosynthesis and cell wall proteins. By contrast, these were not significantly enriched categories in response to myxothiazol in the dark (Figure 1D). It was notable that many of the DEGs that were responsive to antimycin A (823 + 160 genes) in the dark were also responsive to myxothiazol and antimycin A treatments in the light (Figure 1A and 1B).
Under light conditions, a larger number of DEGs was observed in both treatments, especially compared with the dark for myxothiazol treatment, with 1649 DEGs identified in the light (Figure 1Ci). Nevertheless, no significant enrichment was evidenced in the functional categories for the DEGs in this set (Figure 1D). The greatest number of DEGs (2644) was observed following antimycin A treatment in the light (Figure 1Ci). In addition to photosynthesis being overrepresented in downregulated genes under antimycin A in the light, the downregulation of genes encoding cell wall proteins was also notable (Figure 1D). Genes encoding cell wall proteins were also enriched in the downregulated genes under antimycin A, but not myxothiazol, in the dark (Figure 1D).
Thus, there were significant differences in the transcriptomic responses to inhibition of mitochondrial function. This difference was explored further to determine whether it represented an additional signaling pathway triggered by antimycin A or whether it represented a threshold effect due to the fact that inhibition of mitochondrial function by antimycin A can produce more ROS than inhibition with myxothiazol. These differences in ROS production are related to the different sites of cytochrome bc1 complex inhibition (Moller, 2001; Crofts, 2004), as confirmed experimentally using antimycin A and myxothiazol in tobacco leaves (Alber et al., 2017).
First, we confirmed that myxothiazol could inhibit respiration as effectively as antimycin A. In isolated mitochondria and leaf discs, oxygen consumption decreased by ∼95% under both treatments at a concentration of 50 μM (Supplemental Figure 1). Although antimycin A seemed to be more effective at inhibiting oxygen consumption at low concentrations (i.e., 0.5 μM), inhibition was the same for both inhibitors at the 50-μM concentration used in these experiments. Residual oxygen uptake after addition of antimycin A or myxothiazol is abolished by addition of 10 mM SHAM (Chai et al., 2010). Given that little AOX is present in the plants under these growing conditions (indicated by the low rate of alternative respiration), we could not observe any significant difference between the addition of the cytochrome chain inhibitors (antimycin A and myxothiazol) and the subsequent addition of SHAM (data not shown).
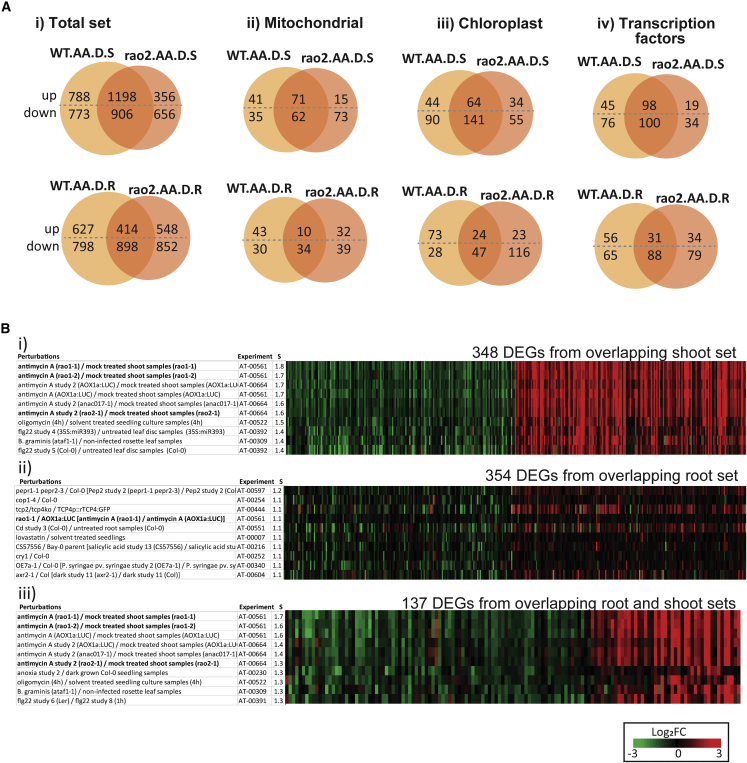
Among the DEGs that were exclusive to antimycin A compared with myxothiazol in the dark, 571 increased and 151 decreased in transcript abundance (Figure 1Ci). Those that had a probeset, i.e. were represented within the Genevestigator database, and encoded mitochondrial proteins (34), chloroplast proteins (53), or transcription factors (41) were analyzed in Genevestigator using the signature tool to identify similar perturbations (Zimmermann et al., 2004). As well as the expected similarity to other antimycin A treatments and hypoxia, the responses also had high similarity to antimycin A treatment responses in rao1 (CDKE1 mutant) and/or rao2 (ANAC017 mutant); both CDKE1 and ANAC017 are previously characterized regulators of the response to antimycin A (Ng et al., 2013a, 2013b) (Supplemental Figure 2, Supplemental Data 2). Similarity to biotic challenges and Flg22, inhibition of ethylene signaling with AgNO3, and ABA/drought treatment, which is well characterized as being antagonistic to ethylene, was also notable (Supplemental Figure 2, Supplemental Data 2). Based on these analyses, we carried out additional RNA-seq experiments to determine (1) whether the genes induced by antimycin A under dark conditions were independent of ANAC017/RAO2 by comparing the wild type with the well-characterized rao2-1 mutant (Ng et al., 2013a; Meng et al., 2019) and (2) whether similar responses took place in roots and shoots by dividing the samples into these organs rather than using whole seedlings.
As expected, a pattern similar to that in Figure 1 was observed, whereby antimycin A produced a substantially larger response in terms of DEGs in all samples compared with myxothiazol (Supplemental Figure 3). In shoots under dark conditions, there was a large antimycin A-specific induction of genes in the wild type (2914 genes, 1288 upregulated and 1626 downregulated), and a similar pattern was observed in rao2 (3039 genes, 1495 upregulated and 1544 downregulated). A similar pattern of antimycin A-specific gene induction was observed in wild-type roots under dark conditions (2232 genes, 773 upregulated and 1459 downregulated), and this was also observed in rao2 roots (2580 genes, 909 upregulated and 1671 downregulated) (Supplemental Figure 3). Thus, we concluded that the antimycin A-specific changes in transcript abundance under dark conditions were independent of ANAC017 regulation, and this was observed in both shoots and roots.
To characterize this antimycin A-specific, ANAC017-independent response, we identified genes that were independent of ANAC017 regulation by comparing wild-type responses to rao2 responses in the dark for shoots and roots (Figure 2A). This resulted in 2104 ANAC017-independent DEG responses in the shoot (1198 upregulated and 906 downregulated) and 1312 ANAC017-independent DEG responses in the root (414 upregulated and 898 downregulated) (Figure 2Ai). When genes encoding mitochondrial proteins, chloroplast proteins, and transcription factors were examined in shoots, the antimycin A-specific, ANAC017-independent gene sets were larger than the ANAC017-dependent gene sets (Figure 2Aii-iv). The ANAC017-independent gene sets in shoots and roots had a small overlap, revealing that antimycin A-specific shoot and root responses were largely distinct (Supplemental Figure 3B). This may reflect differences in metabolic pathways of the two tissues, given the known mitochondrial proteome differences between roots and shoots (Lee et al., 2008, 2011).
Figure 2.
Characterization of the antimycin A-specific response in dark conditions.
(A) Venn diagrams showing numbers of overlapping DEGs between wild-type and ANAC017 mutant (rao2) lines after antimycin A (AA) treatment in the dark (D) in shoots (S) and roots (R).
(B) Fold changes of genes differentially expressed in response to AA in the dark in shoots and roots were analyzed using the signature tool in Genevestigator to identify studies (perturbations) that showed the most similar fold-change responses; similarity scores (S) are indicated. Two hundred randomly selected upregulated genes and 200 randomly selected downregulated genes from the overlapping total sets in (A) were used. (i) Shoots and (ii) roots were viewed (348 and 354 of these matched probesets from the database in shoots and roots, respectively). (iii) The overlapping set of genes responsive in both roots and shoots to antimycin A treatment in the dark from (B) were also examined using the signature tool (137 matched probesets). The top 10 studies in which the most similar fold-change responses were observed for these genes are shown. Each subset contained antimycin A-treated rao mutant studies (indicated in bold).
Because of the large size of the overlapping sets of genes in shoots and roots, 400 genes were randomly selected and examined using the signature tool in Genevestigator (Figure 2Bi and ii). Apart from other antimycin A studies, the response in shoots was dominated by biotic stress-associated studies, such as Flg22 and Pseudomonas syringae (Figure 2B, Supplemental Data 3). Although the antimycin A-specific root set differed from the shoot set and the root and shoot common set, there were still biotic stress-associated studies in the root-specific set, such as eds-1 (enhanced disease susceptibility), fls2-17 (flagellin-sensitive 2), ubc-13 (ubiquitin conjugating), and pad4-1 (phytoalexin deficient). In the shoot, the flu mutant studies revealed overlap with singlet oxygen signaling (1O2) (Wang et al., 2016), which is distinct from the role of ANAC017 for which signaling overlaps with H2O2 signatures (Ng et al., 2013a). This is also consistent with the different sites inhibited by antimycin A and myxothiazol and the production of more superoxide (Brand, 2016). Notably, there was also similarity to ozone treatment, in which constitutive activation of salicylic acid signaling is linked to Arabidopsis accessions that are tolerant to ozone (Xu et al., 2015).
In terms of transcription factors associated with the antimycin A-specific response (Figure 2Aiv), many were associated with biotic defense responses, and some were associated with suppression of ROS responses, including CALMODULIN BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3), which inhibits salicylic acid synthesis (Kim et al., 2013), HBI1, which suppresses ROS by inducing Catalase 2 expression (Chu et al., 2021), WRKY 11, which is a negative regulator of basal resistance to P. syringae (Ali et al., 2018), and ERF11, which balances stress versus growth responses (Dubois et al., 2015). The transcription factors ZAT10 and 12, which are involved in photooxidative and ROS stress responses (Davletova et al., 2005; Rossel et al., 2007; Le et al., 2016), a number of ERF transcription factors involved in thermotolerance (Supplemental Data 3), and WRKY transcription factors associated with salicylic acid and senescence signaling pathways were also observed in this set.
ChIP-seq analysis reveals light-dependent binding of ANAC017 in myxothiazol treatment
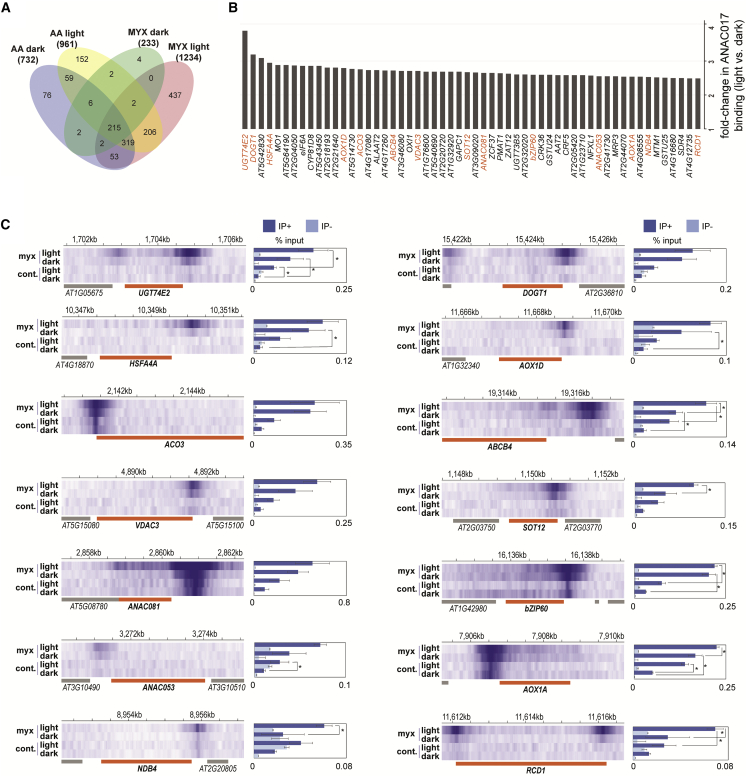
To complement the above transcriptome analyses, we treated transgenic plants expressing a GFP-ANAC017 fusion protein under control of the native promoter (proANAC017:GFP-ANAC017) with myxothiazol or antimycin A after 1 h into the light or dark phase of a 16 h light:8 h dark growth cycle and then performed ChIP-seq experiments to identify ANAC017 binding sites and corresponding target genes (Supplemental Data 4A). After treatment with antimycin A, the number of peaks detected in the dark (732 peaks) was similar to that detected in the light (961 peaks) (Figure 3A, Supplemental Data 4A). By contrast, there was a greater than four-fold difference between treatment in the dark (233 peaks) and in the light (1234 peaks) for myxothiazol (Figure 3A, Supplemental Data 4A).
Figure 3.
Light-dependent quantitative differences in ANAC017 binding to target genes after treatment with myxothiazol.
A ChIP-seq assay was performed using an ANAC017-GFP transgenic line with the ANAC017 native promoter. Plants were treated with myxothiazol (myx) or a solvent control (cont.) for 3 h in the light and dark periods.
(A) Number of peaks detected for the different treatments in the light and dark and their overlapping genes.
(B) Differential binding of ANAC017 to the promoters of target genes in the light versus dark were quantified using the DiffBind algorithm. Genes shown in (C) are highlighted in orange.
(C) ChIP-seq peaks in the promoters of ANAC017 target genes (gene loci indicated with orange bars) were determined after read alignment and are represented by heatmaps. ChIP–qPCR data are shown as percentage of input (% input) for each sample, which consisted of three independent immunoprecipitations. Data are mean ± SE (n = 3, with triplicate qPCR reactions). Asterisks (∗) indicate statistical significance (Student’s t-test with p < 0.05).
To examine the differential binding of ANAC017 to its target sites after myxothiazol treatment, we used a conservative approach of selecting only genes with more than two-fold (FDR < 0.05) stronger binding of ANAC017 to their promoters in the light than in the dark (Figure 3B and 3C; Supplemental Data 4B). For antimycin A, no quantitative light-dependent differences were observed. However, promoters of genes encoding a variety of functions displayed increased binding in the light after myxothiazol treatment (Figure 3B). The gene with the largest difference was URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE 74E2 (UGT74E2), with four-fold enriched binding of ANAC017 in the light (Figure 3B). UGT74E2 is a UDP-glucosyltransferase that transfers glucose to indole-3-butyric acid and is induced upon inhibition of auxin signaling as part of the mitochondrial dysfunction response via the mitochondrial dysfunction motif, to which ANAC017 binds (De Clercq et al., 2013). The second highest was DON-GLUCOSYLTRANSFERASE 1 (DOGT1) (Figure 3B), which glucosylates brassinosteroids (Poppenberger et al., 2005) and plays a central role in growth, development, and stress responses (Nolan et al., 2020). Although it is unknown whether the amount or activity of these enzymes increases with increased transcript abundance, it appears that upon mitochondrial dysfunction in the light, the two growth-promoting hormones auxin and brassinosteroids may be targeted for inactivation by upregulation of conjugating enzymes through ANAC017 (Figure 3). Other stress-responsive transcription factors were also activated, including BASIC REGION/LEUCINE ZIPPER MOTIF 60 (bZIP60) and HEAT SHOCK TRANSCRIPTION FACTOR A4A (HSFA4A). bZIP60 is linked to several chloroplast retrograde signaling pathways, i.e., singlet oxygen (1O2) and β-cyclocitral signaling (Ramel et al., 2012; Beaugelin et al., 2020), heat shock response, and maintenance of chlorophyll synthesis gene expression (Li et al., 2020b), and to methylerythritol cyclodiphosphate signaling (Walley et al., 2015; Benn et al., 2016). HSFA4A mediates salt tolerance and acts downstream of MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and 6 (Pérez-Salamó et al., 2014). ACONITASE 3 (ACO3), which requires ANAC017 signaling for phosphorylation and the negative regulator of endoplasmic reticulum (ER)-bound NAC transcription factors, RCD1, also showed increased binding in the light, as did genes encoding mitochondrial proteins such as AOX1a and NAD(P)H DEHYDROGENASE B4 (NDB4), as well as other NAC transcription factors, including ANAC053 and ANAC081 (Figure 3). To confirm the observed differences in binding, we carried out ChIP–qPCR of three independently grown biological replicates and tested whether a similar pattern was observed. Overall, as with the ChIP-seq, increased binding in light was observed after myxothiazol treatments, although in some cases this difference was not statistically significant (p < 0.05). Thus, the differential binding patterns observed for ANC017 in the light compared with the dark, as well as the number of binding sites (Figure 3A) and the amount of binding (Figure 3B and 3C), suggested light-dependent MRR regulated by ANAC017.
Co-regulation of mitochondrial retrograde signaling by light and the clock via ANAC017
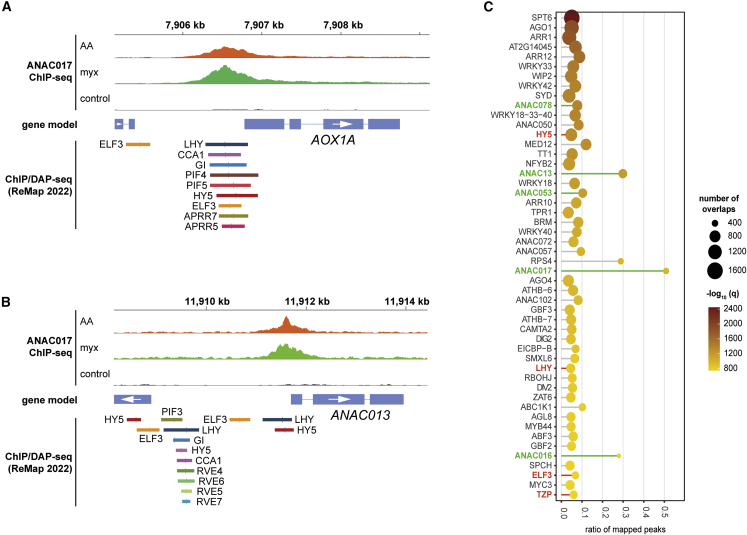
To determine whether MRR was directly regulated by clock or light regulators, we determined binding of ANAC017 to the promoter regions of AOX1a, the pre-eminent MRR marker gene, and ANAC013 after treatment with antimycin A and myxothiazol. The binding site of ANAC017 to the AOX1a promoter has been defined previously (Ng et al., 2013b) and was confirmed by our ChIP-seq analysis (Figure 3C). Analyses of the experimentally determined target sites contained in the ReMap catalog for 423 transcription factors (Hammal et al., 2022) revealed that a variety of clock- or diurnal-related transcription factors bind to the promoter regions of AOX1a and ANAC013 and overlap with ANAC017 binding sites (Figure 4A and 4B, Supplemental Table 1). The binding sites of LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), GIGANTEA (GI), PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), PIF5, ELONGATED HYPOCOTYL 5 (HY5), PSEUDO-RESPONSE REGULATOR 7 (APRR7), and APRR5 were within 100–200 bp of each other and all overlapped with the ANAC017 binding region (Figure 4A). EARLY FLOWERING 3 (ELF3) has two binding sites, one that overlaps with the ANAC017 site and one further upstream (Figure 4A). For ANAC013, a binding site for LHY and HY5 also overlapped with the ANAC017 ChIP-seq peak, whereas several binding sites for LHY, HY5, EARLY FLOWERING 3 (ELF3), CCA1, GIGANTEA (GI), PIF3, and four REVEILLE transcription factors (RVE4,5,6,7) were further upstream in the ANAC013 promoter (Figure 4B).
Figure 4.
Overlap of ANAC017 binding sites with those of light or circadian clock regulators in the promoter regions of AOX1a and ANAC013.
(A and B) Genome browser view showing the ChIP-seq peaks identified in the AOX1a(A) and ANAC013(B) promoters (top panels) and the binding sites of transcription factors involved in circadian clock and light regulation (bottom panels). These binding sites were retrieved from the ReMap2022 database (https://remap.univ-amu.fr/).
(C) Genome-wide analysis of overlap in binding sites of ANAC017 and other transcriptional regulators. Binding sites of the 423 transcriptional regulators contained in the ReMap2022 catalog were compared with ANAC017 ChIP-seq peaks. The x axis represents the ratio of binding sites for the 50 most significant transcription factors included in the catalog and overlapping with ANAC017. Members of the ER-bound ANACs closely related to ANAC017 and with a similar binding motif are highlighted in green. Regulators of light and circadian processes are highlighted in red. The regulators are ordered by dot size, which represents the number of overlapping binding sites across the Arabidopsis genome. The color scale represents the significance of enrichment for these overlapping regions.
For a genome-wide view of the interaction of ANAC017 target genes, the ReMap catalog was further analyzed to determine their overlap with the ANAC017 binding sites identified in our ChIP-seq experiment. As expected, the binding sites of ANAC017 in both datasets overlapped with those of the closely related ER-tethered ANAC013, ANAC053, and ANAC078, as well as ANAC017 itself, all of which share the common mitochondrial dysfunction motif (De Clercq et al., 2013) (Figure 4C, Supplemental Table 2). Also in the list of the 50 most significant regulators that share binding sites with ANAC017 were HY5, LHY, ELF3, and TANDEM ZINC KNUCKLE PROTEIN (TZP), which are important regulators of light-dependent (HY5, TZP) or clock-dependent (LHY, ELF3) transcriptional cascades (Alabadí et al., 2001; Andronis et al., 2008; Loudet et al., 2008; Nusinow et al., 2011), suggesting a role for ANAC017 in the co-regulation of mitochondrial retrograde signaling by light and the clock.
Diurnal control of the mitochondrial stress response
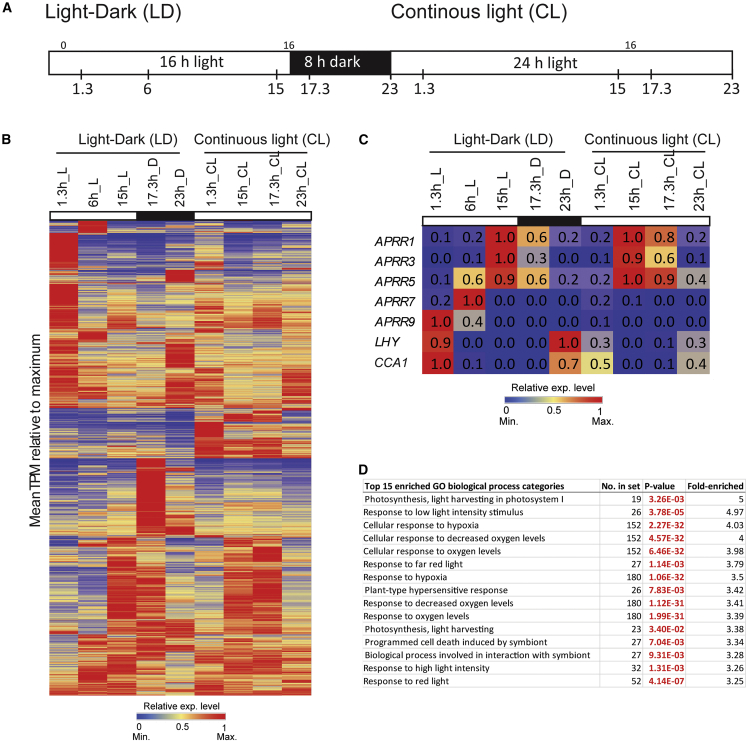
To determine whether the mitochondrial stress response was controlled diurnally, gene expression in wild-type (Col-0) plants was determined over a 48 h time course, first over 24 h in a 16 h light:8 h dark photoperiod with samples harvested at 1.3, 6, and 15 h into the light cycle (L) and then at 17.3 and 23 h, which were 1.3 and 7 h into the dark cycle (Figure 5A). The same sampling times were then used for the next 24 h in continuous light (Figure 5A). After filtering only genes that showed differential expression (|log2(fold change)| > 1, p < 0.05), the transcripts per million values were normalized to the maximum, and the 4446 genes (12.2% of all genes) were visualized in a heatmap, displaying a rhythmic pattern in either the light/dark cycle or continuous light (Figure 5B; Supplemental Data 5). This was consistent with previous microarray studies, which indicated that about 10% of the Arabidopsis transcriptome shows diurnal variation in expression (Harmer et al., 2000; Schaffer et al., 2001; Edwards et al., 2006; Covington and Harmer, 2007). The core circadian clock regulators displayed the expected rhythmic patterns and peak expression times (Figure 5C), as characterized previously (Staiger et al., 2013). The morning genes LHY and CCA1 oscillated with a peak around dawn, followed by sequential expression of the PRR gene family from APRR9 to APRR1/TOC1 (TIMING OF CAB EXPRESSION 1), which peaked around dusk (Figure 5C). As expected, genes encoding proteins involved in photosynthesis were enriched (GO biological process; Fisher’s test, p < 0.05) in the set of 4446 DEGs, but surprisingly, many genes responsive to oxygen and hypoxia were also enriched (Figure 5D). To understand how light responses and specific inhibition of mitochondrial function interact, the 4466 genes that displayed a diurnal pattern were compared with the 1703 genes that showed differential expression upon treatment with myxothiazol. Of the 4446 DEGs over the light-dark and continuous light time series, 746 were also differentially expressed in response to myxothiazol treatments in the light/dark, making up 16.8% of the 4446 genes in the diurnal set. This is a statistically significant overrepresentation (chi-square; p < 0.001) compared with the myxothiazol-responsive genes in the genome (6%). Thus, almost 44% (i.e. 746 genes) of the 1703 genes that were differentially expressed in response to myxothiazol treatment in the light/dark were also differentially expressed in the diurnal set, indicating a possible link between the myxothiazol-responsive genes and light-responsive genes (Supplemental Data 8).
Figure 5.
Diurnal changes in transcript abundance.
(A) Time points at which samples were collected for RNA-seq in the 16 h light (L):8 h dark (D) growth period (LD) and in continuous light (CL).
(B) Average transcripts per million (TPM) were calculated, expressed relative to the maximum, and hierarchically clustered. A total of 4446 genes were significantly differentially expressed (p < 0.05, |log2(fold change)| > 1) at any time point compared with 23h_D or 23h_CL.
(C) The transcript abundance patterns of known diurnally expressed marker genes are shown.
(D) GO overrepresentation analysis (Fisher’s test, Benjamini–Hochberg p < 0.05) was performed, and the top 15 overrepresented categories are indicated.
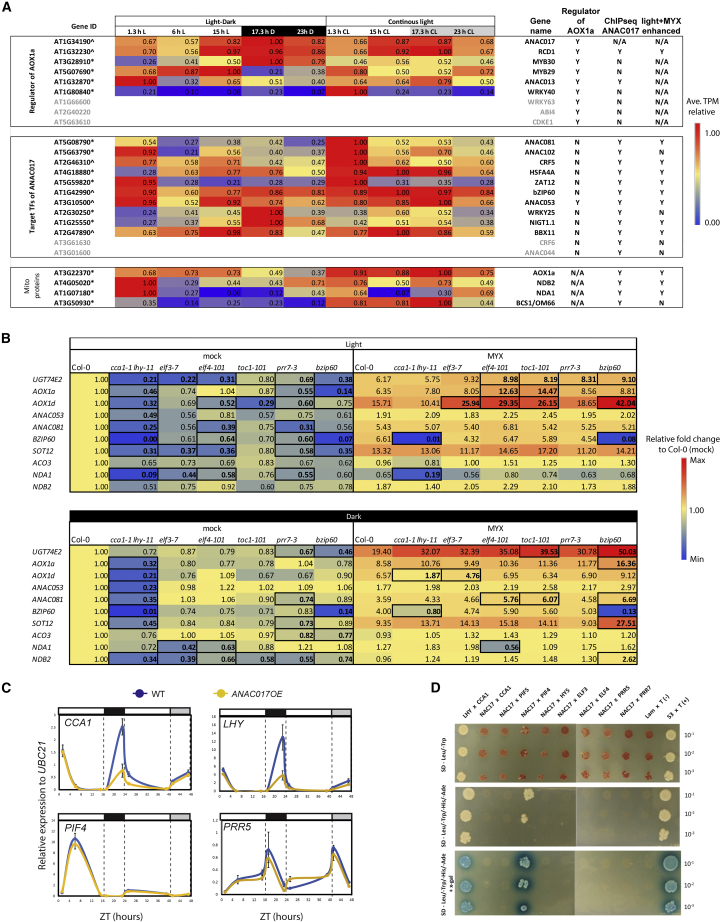
The master regulator ANAC017 displayed variation over the 24 h diurnal period, with a peak in the early night (Figure 6A). ANAC017 transcript abundance is not induced by a variety of stress treatments (Meng et al., 2019), and the slight variation observed over the 24-h diurnal period has been observed only in the current study (|log2(fold-change)| < 1). AOX1a, a marker gene for mitochondrial retrograde signaling, showed a diurnal rhythm during the light–dark cycle, with substantially higher expression in the light than in the dark, and this continued to increase gradually after growth in continuous light (Figure 6A). Two NAD(P)H dehydrogenases, NDA1 and NDB2, and BCS1/OM66 also displayed a significant diurnal rhythm and peaked around dawn (Figure 6A). A list of experimentally characterized regulators of AOX1a and genes encoding transcription factors that are targets of ANAC017 was compiled to investigate whether they displayed diurnal patterns of transcript abundance and whether increased binding of ANAC017 was observed in the light compared with the dark (Figure 6A). Of the nine known regulators of AOX1a, six showed a diurnal rhythm, including ANAC017 and RCD1 with a log2 fold change of less than 1. ANAC017, together with MYB DOMAIN PROTEIN 30 (MYB30) and RCD1, displayed a similar diurnal pattern with a peak of transcript abundance in the dark, and these expression patterns persisted in continuous light (Figure 6A). RCD1 is also a target of ANAC017 and displayed increased binding in the light compared with the dark under myxothiazol treatment (Figure 6A). RCD1 protein was previously shown to interact with and negatively regulate ANAC017 (Shapiguzov et al., 2019). Thus, ANAC017 is a positive regulator of a negative feedback loop that controls its own activity. Although ANAC013 is a target of ANAC017 (Ng et al., 2013b; De Clercq et al., 2013), it was not observed to increase its binding in the light (Figure 3), but it did show a peak in expression at dawn (Figure 6A).
Figure 6.
Expression of genes encoding mitochondrial proteins under light (L)–dark (D) conditions and under continuous light (CL) and their protein interaction with circadian clock components.
(A) Expression levels of regulators of AOX1a and targets of ANAC017. The average TPM values at 23 h D, 1.3 h L, 6 h L, 15 h L, and 17.3 h D and at the same time points under CL were expressed relative to the maximum, and these values are shown. Gene IDs for genes that showed significant differential expression (p < 0.05) are indicated with an ∗ for genes that showed (|log2(fold change)| > 1) or ˆ for genes that showed (|log2(fold change)| < 1) at any time point compared with 23 h dark (D) or 23 h CL.
(B) Transcript abundance of selected ANAC017 target genes measured by qRT–PCR after myxothiazol (MYX) treatment in the light or dark. Transcript abundance is expressed as the fold change relative to wild-type (Col-0) mock after normalization to the reference gene UBC21 (AT5G25760). Bold and boxed cells denote a significant difference versus the wild type for the same treatment using a Student’s t-test (p < 0.05).
(C) Expression of indicated genes was determined relative to the reference gene UBC21 (AT5G25760) by qRT–PCR in the wild type (Col-0) and the ANAC017 overexpression line (ANAC017OE). Plants were harvested at the indicated times under long-day (16 h light:8 h dark) growth conditions or after transfer to continuous light conditions. The mean ± SE of three biological replicates is given.
(D) Coding sequences for ANAC017 and circadian genes were cloned into pGBKT7 and pGADT7, followed by separate transformation into the yeast Y2HGold or Y187 strain. Successful mating was confirmed by growth on SD/-Leu/-Trp medium. Positive interactions were determined by growth on SD/-Leu/-Trp/-His/-Ade (with or without x-Gal) medium with the serial dilutions shown. pGBKT7-53 and pGBKT7-Lam transformed into Y2HGOLD were mated with pGADT7-T in Y187 as positive and negative controls. Self-activation tests are presented in Supplemental Figure 4.
At least 12 genes encoding transcription factors were direct targets of ANAC017, and 8 of these showed increased binding in the light under myxothiazol treatment (Figure 6A, Supplemental Data 4A, 4B, and 6). Many of the 12 transcription factors that are direct targets of ANAC017 displayed more than four-fold diurnal variation in transcript abundance, including ANAC0102, WRKY25, HSFA4A, RESPONSIVE TO HIGH LIGHT 41 (ZAT12), and NITRATE-INDUCIBLE GARP-TYPE TRANSCRIPTIONAL REPRESSOR 1 (NIGT1.1) (Figure 6A). bZIP60 plays a central role in the ER stress response (Howell, 2013), and this has been linked to activation of mitochondrial retrograde signaling (Fuchs et al., 2022). Increased ANAC017 binding at bZIP60 was observed in the light with myxothiazol treatment (Figure 3B and 3C). bZIP60 is a regulator of several chloroplast retrograde signaling pathways involving singlet oxygen (1O2) and β-cyclocitral signaling (Ramel et al., 2012; Beaugelin et al., 2020), heat shock response, maintenance of chlorophyll synthesis gene expression (Li et al., 2020b), and methylerythritol cyclodiphosphate signaling (Walley et al., 2015; Benn et al., 2016). Thus, it appears that mitochondrial retrograde signaling both responds to and is a positive regulator of the unfolded protein response in the ER. The link between bZIP60 and protein homeostasis prompted us to search for any other transcription factors that might be linked to the proteasomal system. Another direct target of ANAC017, ANAC053 (Figures 3B, 6A, and 6B), is a direct regulator of proteotoxic stress (Gladman et al., 2016) and also controls the abundance of Golden2-like transcription factors that promote chloroplast biogenesis (Fitter et al., 2002). Cytosolic protein folding stress is linked to the chloroplast GUN signaling retrograde pathway (Wu et al., 2019). Other proteotoxic-stress-responsive transcription factors that are also direct targets of ANAC017 are ANAC013, ANAC044, ANAC081, and WRKY25 (Supplemental Data 2A) (Gladman et al., 2016). The stress-response roles of CRF6 and the light-associated functions of ZAT12, B-BOX PROTEIN 11 (BBX11), and HSFA4 are all previously documented (Davletova et al., 2005; Pérez-Salamó et al., 2014; Zwack et al., 2016; Job et al., 2022). Thus, these results suggest that most of the regulators of AOX1a and target genes of ANAC017 displayed diurnal rhythms, and many showed increased binding of ANAC017 in the light compared with the dark upon myxothiazol treatment.
Expression of mitochondrial genes is altered in mutants for regulators of the circadian clock
We next examined the effects of mutation of circadian clock regulators on ANAC017 target gene expression following myxothiazol treatment in the light and dark to see whether clock components coordinately regulate MRR. Several circadian clock regulator mutants (toc1-1, cca1-1 lhy-11, elf3-7, elf4-101, and prr7-3) and bzip60 were included (Supplemental Table 3). Target genes that showed increased binding of ANAC017 in the light were selected (Figure 3, Supplemental Data 4B). Interestingly, many of the ANAC017 targets were downregulated or shifted in periodicity of expression in the clock mutants, even before myxothiazol treatment (Figure 6B). This suggests involvement of the clock in regulation of these genes in the absence of perturbed mitochondrial function. In the cca1-1 lhy-11 double mutant, BZIP60 and NDA1 transcript levels were low in the light with and without myxothiazol treatment. In the light with myxothiazol treatment, UGT74E2, AOX1a, and AOX1d showed increased transcript levels in clock mutants compared with the wild type for the same treatment (Figure 6B). In elf4-101 and toc1-101 mutants, there was a two-fold increase in AOX1a and AOX1d abundance compared with the wild type (Col-0) treated with myxothiazol (Figure 6B). In the dark, many target genes were also downregulated or shifted in periodicity of expression, especially in the cca1-1 lhy-11 double mutant (Figure 6B). The bzip60 mutant showed significant differential expression of 6 out of the 10 genes after myxothiazol treatment in the dark. bZIP60 also showed a diurnal pattern that peaked at dusk, similar to that of ANAC017, suggesting the light-dependent co-expression of BZIP60 and ANAC017 in MRR (Figure 6A). We also investigated binding sites of transcription factors that regulate clock- and light-dependent processes by examining the 56 genes (Supplemental Data 4B) that showed enhanced binding of ANAC017 in the light upon myxothiazol treatment, using data retrieved from ReMap2022 (https://remap.univ-amu.fr/) (Supplemental Table 4). Similar binding sites were bound by clock or light components for all 56 genes, further supporting a role for the coordinated regulation of these genes by known clock regulators and ANAC017 (Supplemental Table 4).
The effects of mitochondrial retrograde signaling on circadian gene expression were tested by examining the expression of core clock regulators in an ANAC017 overexpression line (Meng et al., 2019). The peak expression of CCA1 and LHY before dawn was greatly suppressed in the ANAC017 overexpression line (Figure 6C). However, other regulators such as PIF4 and PPR5 were not affected (Figure 6C). To determine whether there was a direct interaction between ANAC017 and regulators of the circadian clock, yeast two-hybrid interaction assays were performed, and interaction of CCA1 and LHY was used as a positive control. Direct interaction was detected only between ANAC017 and PIF4, but not with the other components tested (Figure 6D, Supplemental Figure 4).
Thus, the direct targets of ANAC017 are strongly linked to chloroplast function/dysfunction and retrograde signaling, as well as the ER unfolded protein response. Many genes showed diurnal rhythms and increased binding of ANAC017 in the light compared with the dark upon myxothiazol treatment. Transcript abundance of these target genes was also significantly affected in many clock mutants, with and/or without myxothiazol treatment, and the expression of core clock regulators was also altered in an ANAC017 overexpression line. Collectively, these results suggest coordinated regulation of the mitochondrial retrograde response by circadian clock regulators and ANAC017.
Discussion
Organelle retrograde signaling is initiated by the functional state of organelles to optimize growth in prevailing conditions. Thus, organelle signaling must be integrated with a variety of other signaling pathways to achieve the optimal outcome. Unexpectedly, a large difference in transcriptome response was observed between treatment with antimycin A and myxothiazol in the light and dark (Figure 1). Although both chemicals inhibit mitochondrial electron transport via the cytochrome bc1 complex, they differ in their sites of inhibition. Antimycin A inhibits the formation of an unstable Qo-site semiquinone, whereas inhibition with myxothiazol prevents formation of ubisemiquinone. Thus, inhibition with antimycin A will produce more mitochondrial superoxide than inhibition with myxothiazol (Moller, 2001; Alber and Vanlerberghe, 2019). Also, the topology of ROS production differs. ROS produced by myxothiazol will be almost exclusively on the matrix side of the inner mitochondrial membrane, whereas ROS produced by antimycin A has been demonstrated to be on both the matrix and cytoplasmic sides of the mitochondrial inner membrane (Quinlan et al., 2013; Goncalves et al., 2015; Brand, 2016).
Characteristics of the antimycin A-specific response were:
-
I.
Transcriptomic changes occurred independently of CDKE1 and ANAC017 (Supplemental Figure 2, Supplemental Data 2). This was experimentally confirmed for ANAC017 independence using mutant analysis (Figure 2).
-
II.
Changes occurred in both shoots and roots, and the overlap in transcriptomic response between these organs was small.
-
III.
Overall, the transcriptomic response showed similarities to biotic stresses, particularly Flg22, suggesting the involvement of salicylic acid. Notably, the antimycin A-specific transcription factor set contained many ERF and ZAT transcription factors associated with light and various stress signaling pathways.
There were no significant differences in magnitude in the common genes that were induced by myxothiazol and antimycin A, and it thus appears that the additional DEGs under antimycin A in the dark represent an additional signaling pathway (Figure 7). The presence of a salicylic acid-related mitochondrial signaling pathway has been reported for plant mitochondria (Gleason et al., 2011; Belt et al., 2017). This pathway is associated with the DISRUPTED IN STRESS RESPONSES 1 (DSR1) gene, which encodes subunit 1 of succinate dehydrogenase and whose corresponding mutant has a reduced response to salicylic acid and increased susceptibility to pathogens (Gleason et al., 2011; Belt et al., 2017). It has also been demonstrated that salicylic acid likely interacts with the ubiquinone binding site of succinate dehydrogenase, suggesting that it has a similar effect to antimycin A in increasing the production of the superoxide oxygen radical (Belt et al., 2017). Although superoxide is highly reactive and unlikely to travel beyond mitochondria, it is converted to H2O2, which can leave mitochondria (Bienert et al., 2007). Also, an earlier report noted a salicylic acid-dependent pathway for induction of MRS marker genes that was dependent on PHYTOALEXIN-DEFICIENT 4 (PAD4), which acts upstream of salicylic acid and is essential for salicylic acid defense pathways, and ENHANCED DISEASE SUCEPTIBILITY4 (EDS4), which acts downstream of salicylic acid but not NONEXPRESSER OF PR GENES (NPR1) (Ho et al., 2008). Salicylic acid induction of AOX in the voodoo lily inflorescence was also reported 30 years ago (Rhoads and McIntosh, 1992). In addition, inhibition of QH2 oxidation by antimycin A will cause the QH2/Q ratio to increase, and electrons from the QH2/Q pool may block other centers, such as complex II, linked to salicylic acid signaling as outlined above (Brand, 2016). Thus, the additional DEGs with antimycin A likely result from a combination of more ROS and the topology of ROS production, triggering an additional pathway (Alber et al., 2017; Alber and Vanlerberghe, 2021).
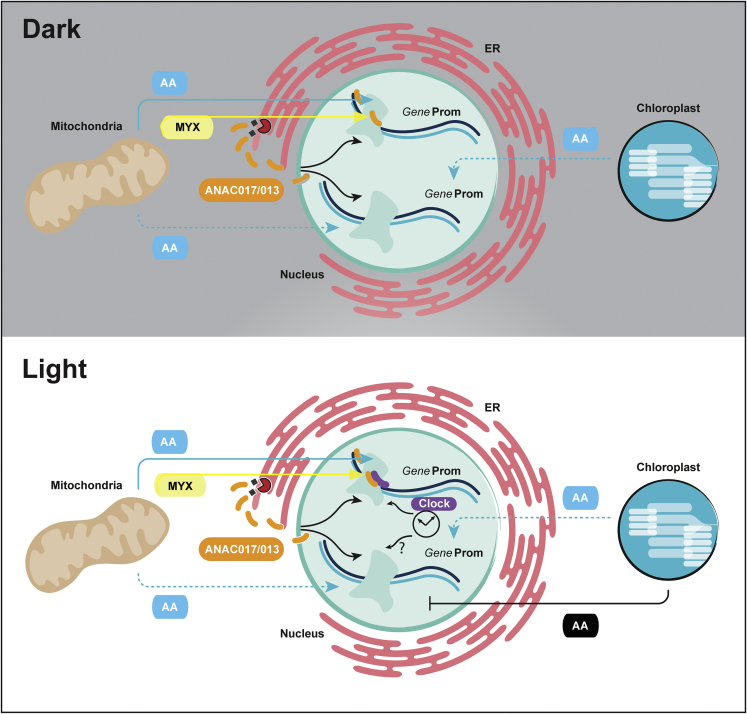
Figure 7.
Model for the mode of action of myxothiazol and antimycin A in plant cells and interaction of mitochondrial signaling with circadian clock regulators.
In the dark, myxothiazol (yellow) and antimycin A (blue) produce a common mitochondrial dysfunction response mediated by ANAC017 and ANAC013 (orange). In the dark, antimycin A also induces an additional mitochondrial response, which displays similarity to salicylic acid signaling responses, indicated by a dotted blue line, and is independent of ANAC017. Antimycin A likely has no inhibitory effect on chloroplasts in the dark, but this cannot be ruled out based on the shoot transcriptome response to antimycin A in the dark that bears similarity to some chloroplast stress responses. In the light, antimycin A inhibits cyclic electron transport that will induce additional responses, indicated by a solid black line. This mitochondrial retrograde response that is regulated by ANAC017 interacts with circadian clock regulators (purple). It is unknown whether the antimycin A ANAC017-independent response interacts with clock regulators, as indicated by the question mark (?). For clarity, the circadian clock is only shown in the bottom panel of the diagram.
Although antimycin A appears to induce at least two distinct mitochondrial signaling pathways, i.e. ANAC017 dependent and independent, there is likely to be overlap between them. AOX1a is induced by salicylic acid (see above), whereas transcripts encoding OUTER MITOCHONDRIAL MEMBRANE PROTEIN OF 66 KDA (OM66), whose induction is sensitive to components of salicylic acid signaling, PAD4, and EDS4 are induced by antimycin A. Likewise, ANAC053, the only NAC transcription factor that is induced after antimycin A treatment in the dark, is also a target of ANAC017. Thus, defining the antimycin A, ANAC017-dependent and -independent gene sets will now provide gene markers to identify components associated with this pathway by forward or reverse genetic approaches, which were used to elucidate the ER-NAC-dependent pathway.
The chloroplast site of inhibition for antimycin A is located in one of the pathways involved in cyclic electron flow and should not be operational in the dark. However, given that the exact mechanistic role of the thylakoid protein PROTON GRADIENT REGULATION 5 (PGR5) is still not fully elucidated (Wu et al., 2021) and that it may have an ancient role in iron delivery (Leister et al., 2022), we cannot exclude the possibility that some of the changes in shoots could be due to binding to PGR5 in the dark, resulting in instability of protein complexes (Rühle et al., 2021). Although the chloroplast is unlikely to be the initiator of the signals that cause these changes under antimycin A in the dark, there are well-defined molecular links between salicylic acid and chloroplast retrograde signaling. Pathogen-associated molecular pattern signals are relayed to chloroplasts, where 1O2 signaling is induced (Nomura et al., 2012; Medina-Puche et al., 2020). This 1O2 signaling pathway is linked to salicylic acid signaling by the dual targeting of SIGMA FACTOR BINDING PROTEIN 1 (SIB1) to the nucleus and chloroplast and antagonistic regulation of Golden2-like transcription factors by LESION-STIMULATING DISEASE 1 (LSD1) and SIB1 (Li et al., 2022). Thus, previous comparisons of transcriptome-level similarity between ANAC017 and chloroplast retrograde signaling need to be re-interpreted, as not all changes caused by antimycin A are mediated by ANAC017 (Van Aken and Pogson, 2017).
When we investigated the effect of light on MRS with the mitochondria-specific inhibitor myxothiazol, there was a clear and significant difference in binding of ANAC017 to promoters of approximately 56 genes in light compared with dark conditions (Figure 3). The functionality of this differential binding was supported by the diurnal expression pattern of transcription factors regulated by ANAC017, the altered response to mitochondrial dysfunction in clock mutants, and the experimentally verified overlap in the binding of clock regulators and ANAC017 to AOX1a and ANAC013 (Figure 3). Overexpression of ANAC017 also caused downregulation of two key clock components, CCA1 and LHY. Protein–protein interaction studies showed that ANAC017 interacts with PIF4. PIF4 was initially characterized as an important element of the phytochrome B signaling pathway and subsequently shown to function in thermomorphogenesis (Huq and Quail, 2002; Han et al., 2019). Studies have also shown that PIF4 transcripts are cooperatively regulated by the evening complex (ELF4-ELF3-LUX) and the PRR gene family of the circadian clock pathway (Nusinow et al., 2011; Li et al., 2020a). Thus, the interaction between ANAC017 and PIF4 may suggest an indirect link by which exogenous signals, such as light and temperature, feed in to control MRS. On the other hand, this may also be presented as an alternative means by which the key MRS regulator ANAC017 coordinates plant growth via PIF4 in response to stress. These results show that circadian clock regulators co-regulate the expression of ANAC017 targets, suggesting coordinated regulation of the circadian clock and ANAC017 in the mitochondrial retrograde response. Some previous studies have shown that the circadian clock is involved in regulating activity in mitochondria and cellular energy production (Lee et al., 2010; Graf et al., 2017; Cervela-Cardona et al., 2021a, 2021b). Metabolite profiling of triple mutants of PRR9, PRR7, and PRR5 showed increased accumulation of tricarboxylic acid cycle intermediates (Fukushima et al., 2009). A study on TOC1 described a mechanism of diurnal regulation of metabolism and mitochondrial activity (Cervela-Cardona et al., 2021b). TOC1 represses expression of FUM2, a tricarboxylic acid-related gene, by binding to its promoter. Diurnal regulation of the abundance of 45 mitochondrial proteins was also reported in a quantitative analysis of the mitochondrial proteome (Lee et al., 2010). These included NDA1, which also showed distinct diurnal changes in transcript abundance in our study (Figure 6A, Supplemental Data 5). Promoter analysis of the TEOSINTE BRANCHED 1, CYCLOIDEA, and PCF (TCP) transcription factors identified site II regulatory elements [TGGGC(C/T)] in the promoter regions of 15 of the mitochondrial proteins, which led to diurnal regulation of their transcripts (Giraud et al., 2010). Several TCP transcription factors have also been shown to interact with key clock components such as CCA1 and PRR5 by yeast two-hybrid assays (Giraud et al., 2010). On the other hand, retrograde signaling also regulates circadian clock gene rhythms. SAL1 was previously shown to localize to both mitochondria and chloroplasts, dephosphorylate 3′-phosphoadenosine 5′-phosphate (PAP), and contribute to retrograde signaling via inhibition of 5′ to 3′ exoribonucleases (XRNs) (Estavillo et al., 2011). The SAL1-PAP-XRN4 retrograde signaling pathway regulates clock genes, LHY, PRR5, and GI expression (Litthauer and Jones, 2018). A previous study also demonstrated that the SAL1 and ANAC017 retrograde signaling pathways may converge to suppress genes involved in programmed cell death (Van Aken and Pogson, 2017). These findings suggest that both mitochondria and chloroplast retrograde signaling interact to regulate circadian rhythms.
The physiological significance of the interaction between mitochondrial retrograde signaling and light anterograde signaling is likely to be linked to the roles of many of the proteins encoded by the genes induced in this pathway. It is well established that the AOX and co-regulated alternative NAD(P)H dehydrogenase B2 (Sweetman et al., 2019) play an important role in optimizing photosynthesis under a variety of conditions (Florez-Sarasa et al., 2016; Dahal et al., 2017; Yamada et al., 2020a; Vanlerberghe et al., 2020; Chadee et al., 2021). Also, other genes that are induced in this pathway, such as the gene encoding the mitochondrial malate carrier (Van Aken et al., 2009; Lee et al., 2021), can play an important role in the malate shuttle, in which excess reducing equivalents from chloroplasts are oxidized by mitochondria to prevent photo-oxidation and maintain photosynthesis (Selinski and Scheibe, 2019). Therefore, integration of the regulation of the mitochondrial stress response pathway with circadian regulators extends the interaction at the metabolic level to the regulatory level. It is noteworthy that both the transcription factor that regulates this response, ANAC017, and AOX1a require post-translational activation (Selinski et al., 2018). Given that the response to high-light stress requires a rapid response to prevent photo-oxidative damage, coordination of the transcription of these components with the light–dark cycle, coupled with post-translational regulation, will provide metabolic plasticity and a fast response when required.
Many of the genes that encode regulatory proteins acting downstream of ANAC017 are associated with both chloroplast retrograde signaling pathways related to light and oxidative stress (e.g. ZAT12) and the reductive stress/unfolded protein response in the ER (e.g. bZIP60). One of the clearly established roles of mitochondrial alternative respiration is the dissipation of excess reducing equivalents produced in chloroplasts under a variety of conditions (Yamada et al., 2020b; Vanlerberghe et al., 2020). ANAC017 is required for the response to reductive stress in the ER (Fuchs et al., 2022), and RCD1 is involved in coordinating ROS responses from chloroplasts and mitochondria by binding to ANAC017 (and ANAC013) (Shapiguzov et al., 2019). Here, we show that ANAC017 links these processes at the regulatory level, in that it acts upstream of several transcription factors that have been shown to participate in both chloroplast and ER stress responses. As previously shown for mitochondrial stress, ANAC017 is not only involved but is also essential: it cannot be compensated for by other factors, as both MRS and the response to reductive ER stress are abolished in the absence of ANAC017. In the case of chloroplast retrograde pathways linked to ANAC017 in this study mediated by bZIP60, ANAC0102, WRKY25, HSFA4A, ZAT12, NIGT1.1, and others, it is unknown whether ANAC017 is directly essential or whether these transcription factors can be activated in separate, albeit converging pathway(s). Previous studies based on analysis of marker genes have shown that ANAC017 is required for the response to methyl viologen, which is considered to be a chloroplast-specific inhibitor at the site of photosystem I, but not for responses to high light (Van Aken et al., 2016). Therefore, although activation of MRS can lead to activation of these pathways, it is also possible that they may be activated independently and converge. The recent demonstration that ANAC017 is required to ameliorate the response to ER stress suggests that the role of ANAC017 in some pathway(s) may previously have been missed, as it is not transcriptionally induced under stress but rather activated by release from the ER.
Overall, these results revealed that the difference in MRS in light and dark is achieved by co-regulation of ANAC017 target genes and clock regulators under mitochondrial dysfunction, as well as differential binding of ANAC013/17 under light and dark conditions (Figures 6 and 7). Antimycin A and myxothiazol induce the mitochondrial dysfunction response in the dark, but there is an additional response to antimycin A treatment that displays the hallmarks of salicylic acid signaling. Our results show a coordinated regulation of the mitochondrial retrograde response induced by the mitochondrial-specific inhibitor myxothiazol via circadian clock regulators and ANAC017.
Methods
Plant material and growth conditions
The full list of T-DNA or EMS mutants and transgenic lines used in this study are listed in Supplemental Table 3. Seeds were sterilized and stratified for 48 h at 4°C in the dark. Seeds were sown on Gamborg (B5) medium with 1% (w/v) sucrose and 0.8% (w/v) agar and grown under a 16 h:8 h day:night photoperiod at 23°C with 100 μmol m−2 s−1 photosynthetic photon flux density. Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild-type control for all experiments unless otherwise stated.
proANAC017:GFP-ANAC017 plants were generated by cloning the open reading frame of ANAC017 into pK7m34GW (Karimi et al., 2002). For this, the 2-kb upstream region of the translational start site of ANAC017 (proANAC017) was amplified by PCR from Arabidopsis Col-0 genomic DNA with primers (Supplemental Table 5) and cloned into the pDONR P4-P1r vector (Invitrogen). GFP-Linker was cloned into pDONR221 (Invitrogen), and the open reading frame of ANAC017 without the start codon was cloned into pDONR P2r-P3 (Invitrogen) (Supplemental Table 5 for cloning primers).
The final proANAC017:GFP-ANAC017 construct was created by recombination of these constructs into the MultiSite destination vector pK7m34GW using Gateway technology according to the manufacturer’s instructions (Invitrogen). Constructs were transformed into Arabidopsis Col-0 by Agrobacterium tumefaciens-mediated floral dipping (Clough and Bent, 1998). Representative transgenic lines were selected from progeny of 30 independent events.
Treatment with antimycin A and myxothiazol
Seeds were sown on Gamborg (B5) medium with 1% (w/v) sucrose and 0.8% (w/v) agar and grown under a 16 h:8 h day:night photoperiod at 23°C with 100 μmol m−2 s−1 photosynthetic photon flux density. Plants grown for 12 days were sprayed with either 50 μM antimycin A or 50 μM myxothiazol in 0.01% Tween 20 solution, or with 0.01% Tween 20 alone for controls, after 1 h into the light or dark phase. Whole seedlings (6–7 seedlings per biological replicate) were harvested 3 h after treatment application for RNA extraction and qRT–PCR. Three biological replicates (10 seedlings each) were harvested for each treatment. For the root and shoot experiment, seeds were sown on Gamborg (B5) medium with 1% (w/v) sucrose and 0.8% (w/v) agar and grown under a 16 h:8 h day:night photoperiod at 23°C with 100 μmol m−2 s−1 photosynthetic photon flux density for 6 days, transferred to vermiculite with ½ B5 medium, and grown for another 6 days. One hour into the light or dark phase, the whole plant was dipped three times in 50 μM antimycin A, 50 μM myxothiazol, or mock medium. The dipped plants were transferred to B5 medium plates, and a small amount of antimycin A/myxothiazol/mock was poured onto the plates. After 3 h of treatment in the light or dark, root and shoot tissues were harvested separately for RNA extraction and RNA-seq. Three biological replicates (10 seedlings each) were harvested for each treatment.
Respiration rate measurement
Oxygen consumption of purified mitochondria or leaf discs was measured by a computer-controlled Clark-type O2 electrode (Hansatech Instruments, Pentney, UK) according to a previously published protocol (Lyu et al., 2018). All reactions were carried out at 25°C using 1 ml of mitochondrial reaction medium (0.3 M sucrose, 10 mM TES, 10 mM NaCl, 4 mM MgSO4, 0.1% [w/v] BSA [pH 7.2]) and mitochondria equivalent to approximately 100 μg of protein. Either antimycin A or myxothiazol (0.5, 5, or 50 μM) was applied to measure inhibition of O2 consumption. Three biological replicates were performed for each treatment.
Yeast two-hybrid interactions
The coding sequences of targeted genes were cloned from Arabidopsis Col-0 cDNA into pGBKT7 and pGADT7, then transformed separately into the Y2HGold strain or Y187 strain (Matchmaker Gold Yeast Two-Hybrid System, Clontech). Cloning primers are listed in Supplemental Table 5. Transformed cells were grown on SD/-Leu or SD/-Trp medium. Mating was carried out in a 96-well flat-bottom plate at 30°C overnight. Positive interactions were identified after 4 days of growth at 30°C on SD/-His/-Leu-/Trp or SD/-Ade/-His/-Leu/-Trp medium.
Diurnal experiment
Col-0 seeds were sown onto B5 medium supplemented with 0.8% (w/v) agar (pH 5.8) and 2% (w/v) sucrose and grown under a 16 h:8 h day:night photoperiod of 100 μmol m−2 s−1 white light at 23°C. After 8 days of growth, 8 to 15 seedlings growing under long-day conditions or continuous light were collected at each time point across 24 h in long-day and 24 h in continuous light. Three biological replicates were collected and frozen in liquid nitrogen for RNA extraction and qRT–PCR.
RNA isolation and qRT–PCR
RNA was isolated using the Spectrum Plant Total RNA Kit with On-Column DNase I Digestion according to the manufacturer’s instructions (Sigma-Aldrich). The cDNA was synthesized using a Tetro cDNA Synthesis Kit (Bioline, UK), and qRT–PCR was performed using SYBR Select Master Mix (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. For quantitative real-time PCR, 1 μg of total RNA was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Real-time PCR was carried out using 1 ng cDNA with SYBR Green PCR Master Mix and a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, Australia) using UBC21 (AT5G25760) as a reference gene. Gene-specific primer pairs were designed using QuantPrime (Arvidsson et al., 2008), and their sequences are given in Supplemental Table 5.
RNA-seq analysis
RNA-seq libraries from three biological replicates were prepared from total RNA according to the manufacturer’s instructions (Illumina) using the TruSeq Stranded mRNA Library Prep Kit and sequenced on a NextSeq 500 system (Illumina) as 70-bp single-end reads. The average read quality score (Q30) was greater than 95%, and there were an average of 13M reads per sample. Quality control was performed using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Transcript abundances as transcripts per million and estimated counts were quantified at the gene level by pseudo-aligning reads against a k-mer index built from representative transcript models of the Araport 11 annotation using the kallisto program (Cheng et al., 2017) with a k-mer length of 31 and 100 bootstraps (Bray et al., 2016). Genes with at least 5 counts in a quarter of all samples per genotype were included in the analysis. The sleuth program with a likelihood ratio test was used to determine differential gene expression (Pimentel et al., 2017). Differentially expressed genes were defined as those with a |log2(fold change)| > 1 and a false discovery rate p < 0.05. Partek Genomics software suite version 6.16 (Partek Incorporated, http://www.partek.com/) was used for hierarchical clustering and generation of heatmaps. For the analysis of genes encoding mitochondrial and plastid proteins, the lists were generated using SUBA4 (Hooper et al., 2017) and CropPAL (Hooper et al., 2016), followed by manual curation (Zhu et al., 2020). For transcription factors, the list was obtained from Plant TFDB (Jin et al., 2017). Panther was used for GO overrepresentation analyses (http://pantherdb.org/), specifically examining biological processes, and significance was defined using Fisher’s test, p < 0.05 (Bonferroni correction).
ChIP-seq
The ChIP experiments were performed as described previously (Bowler et al., 2004; Berckmans et al., 2011) with minor modifications. Approximately 100 mg of proANAC017:GFP-ANAC017 seeds per plate were grown under a 16 h:8 h day:night photoperiod at 23°C with 100 μmol m−2 s−1 photosynthetic photon flux density for 12 days on B5 medium with 1% sucrose and 0.8% (w/v) agar. After 1 h into the light or dark phase, samples were sprayed with 50 μM antimycin A + 0.01% Tween 20, 50 μM myxothiazol + 0.01% Tween 20, or 0.01% Tween 20 alone for the mock control. After 3 h of treatment, the seedlings were scraped off the plates, rinsed twice with 10 mM HEPES-NaOH, and submerged in 20 mL of 10 mM HEPES-NaOH + 1% (v/v) formaldehyde. Three biological replicates were harvested, with pooled seedlings from one plate per replicate. Seedlings in fixation buffer were vacuum infiltrated for 5 min. The vacuum was then released, reapplied for 10 min, and repeated for another 5 min. Formaldehyde was quenched with glycine by adding 1.34 ml of 2 M glycine to 20 ml fixation buffer and vacuum infiltrating again for 2 min. Finally, the samples were washed with 10 mM HEPES-NaOH, dried with a paper towel to remove extra buffer, and snap frozen. Approximately 2 g of 12-day-old proANAC017:GFP-ANAC017 seedling tissue was used. Nuclei were isolated and lysed, and chromatin was fragmented by sonication with a Bioruptor sonicator (Diagenode), resulting in fragments of ∼500 bp. Chromatin samples were pre-cleared with 80 μl Dynal Protein A magnetic beads (10001D, Thermo Fisher Scientific) for at least 2 h at 4°C with gentle agitation. Experiments were conducted with antibodies against GFP (A11122, Thermo Fisher Scientific). Anti-GFP antibody (10 μg) or mock (no antibody) was coupled to 50 μl Dynal Protein A Dynabeads (10001D, Thermo Fisher Scientific) overnight at 4°C and subsequently incubated overnight at 4°C with equal amounts of sonicated chromatin. After overnight incubation, beads were washed twice with low-salt buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100), high-salt buffer (50 mM Tris–HCl [pH 7.4], 500 mM NaCl, 2 mM EDTA, 0.5% Triton X-100), and final wash buffer (50 mM Tris–HCl [pH 7.4], 50 mM NaCl, 2 mM EDTA). After elution, samples were de-crosslinked and digested by proteinase K digestion before DNA purification using the QIAquick PCR Purification Kit (QIAGEN).
ChIP–qPCR was performed as described for qRT–PCR above using the ChIP DNA as a template and primers designed based on the binding region of ChIP-seq analysis (Supplemental Table 5). ChIP–qPCR data were analyzed by the percentage input method using input samples as the positive control (Lin et al., 2012). ChIP-seq libraries were generated with the Accel-NGS 2S Plus DNA Library Kit following the manufacturer’s instructions (Swift Biosciences) and sequenced on the NextSeq 500 platform (Illumina) with an 84-bp read length.
For bioinformatic analysis, reads were mapped to the Arabidopsis reference genome (TAIR10) using Bowtie 2 (Langmead and Salzberg, 2012). ChIP-seq peaks were called with MACS2 software using default parameters (Zhang et al., 2008), and associated genes were identified with the bedtools package (Quinlan 2010). Differential binding of ANAC017 in the light or dark was quantified using the DiffBind package in R (https://bioconductor.org/packages/release/bioc/html/DiffBind.html). For analysis of binding sites shared between ANAC017 and other transcriptional regulators, the complete ReMap2022 catalog of 4.8 million ChIP-seq and DAP-seq peaks was retrieved (Hammal et al., 2022). The ReMap2022 catalog and the ANAC017 datasets were each collapsed by merging redundant overlapping peaks and by the highest FDR using the bedops and bedtools toolkits (Neph 2012, Quinlan 2010). The resulting datasets were filtered for highly significant peaks with a −log10(FDR) > 10 cutoff and subsequently used as input files for enrichment analysis using the ReMapEnrich tool in R (https://github.com/remap-cisreg/ReMapEnrich).
Funding
This work was supported by the facilities of the Australian Research Council Centre of Excellence Program (CE140100008) and Discovery Grant DP210103258. R.N. is supported by an Australian Research Council DECRA fellowship (DE160101536). C.H. is supported by a La Trobe University postgraduate scholarship.
Author contributions
J.W. and L.C.L. conceived the project. Y.Z., C.H., and L.C.L. performed the experiments. L.C.L., R.N., Y.W., O.B., and J.W. analyzed the data. Y.Z., L.C.L., R.N., Y.W., O.B., and J.W. wrote the manuscript with contributions by all authors.
Acknowledgments
We thank Asha Haslem for technical assistance with RNA-seq and ChIP-seq library generation and the La Trobe University Genomics Platform for access to next-generation sequencing instruments. No conflict of interest declared.
Published: December 5, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
ANAC017–At1g34190; ANAC013–At1G32870; ANAC044–At3g01600; ANAC053–At3g10500; ANAC078–At5G04410; ANAC081/ATAF2–At3G10500; ANAC102–At5G63790; AOX1a–At3g22370; AOX1d–At1G32350; UBC–At5g25760; NDB2–At4G05020; BCS1/OM66–At3G50930; UGT74E2–At1G05680; DOGT1–At2G36800; bZIP60–At1g42990; RCD1–At1g32230; NDB4–At2G20800; LHY–At1G01060; CCA1–At2G46830; GI–At1G22770; PIF4–At2G43010; PIF5–At3G59060; HY5–At5G11260; APRR1/TOC1–At5G61380; APRR3–At5G60100; APRR5–At5G24470; APRR7–At5G02810; APRR9–At2G46790; ELF3–At2G25930; MYB30–At3G28910; WRKY25–At2G30250; HSF4A4–At4G18880; ZAT12–At5G59820; NIGT1.1–At1G25550.
Supplemental information
Data availability
RNA-seq and ChIP-seq data have been deposited at the NCBI SRA database under project IDs PRJNA843852, PRJNA843855, PRJNA894307, and PRJNA837635.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Alber N.A., Vanlerberghe G.C. Signaling interactions between mitochondria and chloroplasts in Nicotiana tabacum leaf. Physiol. Plantarum. 2019;167:188–204. doi: 10.1111/ppl.12879. [DOI] [PubMed] [Google Scholar]
- Alber N.A., Vanlerberghe G.C. The flexibility of metabolic interactions between chloroplasts and mitochondria in Nicotiana tabacum leaf. Plant J. 2021;106:1625–1646. doi: 10.1111/tpj.15259. [DOI] [PubMed] [Google Scholar]
- Alber N.A., Sivanesan H., Vanlerberghe G.C. The occurrence and control of nitric oxide generation by the plant mitochondrial electron transport chain. Plant Cell Environ. 2017;40:1074–1085. doi: 10.1111/pce.12884. [DOI] [PubMed] [Google Scholar]
- Ali M.A., Azeem F., Nawaz M.A., Acet T., Abbas A., Imran Q.M., Shah K.H., Rehman H.M., Chung G., Yang S.H., Bohlmann H. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018;226:12–21. doi: 10.1016/j.jplph.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Andronis C., Barak S., Knowles S.M., Sugano S., Tobin E.M. The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant. 2008;1:58–67. doi: 10.1093/mp/ssm005. [DOI] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riaño-Pachón D.M., Mueller-Roeber B. QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashykhmina N., Chan K.X., Frerigmann H., Van Breusegem F., Kopriva S., Flügge U.I., Gigolashvili T. Dissecting the role of SAL1 in metabolizing the stress signaling molecule 3′-phosphoadenosine 5′-phosphate in different cell compartments. Front. Mol. Biosci. 2021;8:763795. doi: 10.3389/fmolb.2021.763795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaugelin I., Chevalier A., D'Alessandro S., Ksas B., Havaux M. Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant J. 2020;102:1266–1280. doi: 10.1111/tpj.14700. [DOI] [PubMed] [Google Scholar]
- Belt K., Huang S., Thatcher L.F., Casarotto H., Singh K.B., Van Aken O., Millar A.H. Salicylic acid-dependent plant stress signaling via mitochondrial succinate dehydrogenase. Plant Physiol. 2017;173:2029–2040. doi: 10.1104/pp.16.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G., Bjornson M., Ke H., De Souza A., Balmond E.I., Shaw J.T., Dehesh K. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. USA. 2016;113:8855–8860. doi: 10.1073/pnas.1602582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B., Vassileva V., Schmid S.P.C., Maes S., Parizot B., Naramoto S., Magyar Z., Alvim Kamei C.L., Koncz C., Bögre L., et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell. 2011;23:3671–3683. doi: 10.1105/tpc.111.088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert G.P., Møller A.L.B., Kristiansen K.A., Schulz A., Møller I.M., Schjoerring J.K., Jahn T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- Blanco N.E., Guinea-Díaz M., Whelan J., Strand Å. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130231. doi: 10.1098/rstb.2013.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L. Emergence of plant molecular biology viewed through the portal of chloroplast research. Curr. Sci. 2001:153–160. [Google Scholar]
- Börner T. The discovery of plastid-to-nucleus retrograde signaling—a personal perspective. Protoplasma. 2017;254:1845–1855. doi: 10.1007/s00709-017-1104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- Brand M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Cervela-Cardona L., Alary B., Mas P. The Arabidopsis circadian clock and metabolic energy: a question of time. Front. Plant Sci. 2021;12:804468. doi: 10.3389/fpls.2021.804468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervela-Cardona L., Yoshida T., Zhang Y., Okada M., Fernie A., Mas P. Circadian control of metabolism by the clock component TOC1. Front. Plant Sci. 2021;12:683516. doi: 10.3389/fpls.2021.683516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee A., Alber N.A., Dahal K., Vanlerberghe G.C. The complementary roles of chloroplast cyclic electron transport and mitochondrial alternative oxidase to ensure photosynthetic performance. Front. Plant Sci. 2021;12:748204. doi: 10.3389/fpls.2021.748204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai T.T., Simmonds D., Day D.A., Colmer T.D., Finnegan P.M. Photosynthetic performance and fertility are repressed in GmAOX2b antisense soybean. Plant Physiol. 2010;152:1638–1649. doi: 10.1104/pp.109.149294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Chu X., Wang J.G., Li M., Zhang S., Gao Y., Fan M., Han C., Xiang F., Li G., Wang Y., et al. HBI transcription factor-mediated ROS homeostasis regulates nitrate signal transduction. Plant Cell. 2021;33:3004–3021. doi: 10.1093/plcell/koab165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., Berthomé R., Lurin C. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 2013;10:1557–1575. doi: 10.4161/rna.26128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Harmer S.L. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford T., Lehotai N., Strand Å. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018;69:2783–2795. doi: 10.1093/jxb/erx481. [DOI] [PubMed] [Google Scholar]
- Crofts A.R. The cytochrome bc1 complex: function in the context of structure. Annu. Rev. Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- Dahal K., Martyn G.D., Alber N.A., Vanlerberghe G.C. Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J. Exp. Bot. 2017;68:657–671. doi: 10.1093/jxb/erw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I., Vermeirssen V., Van Aken O., Vandepoele K., Murcha M.W., Law S.R., Inzé A., Ng S., Ivanova A., Rombaut D., et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013;25:3472–3490. doi: 10.1105/tpc.113.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Van den Broeck L., Claeys H., Van Vlierberghe K., Matsui M., Inzé D. The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 2015;169:166–179. doi: 10.1104/pp.15.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne A.M., Giritch A., Hoffmann B., Cognat V., Lancelin D., Peeters N.M., Zaepfel M., Maréchal-Drouard L., Small I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.D., Anderson P.E., Hall A., Salathia N.S., Locke J.C.W., Lynn J.R., Straume M., Smith J.Q., Millar A.J. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo G.M., Crisp P.A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter D.W., Martin D.J., Copley M.J., Scotland R.W., Langdale J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Florez-Sarasa I., Ribas-Carbo M., Del-Saz N.F., Schwahn K., Nikoloski Z., Fernie A.R., Flexas J. Unravelling the in vivo regulation and metabolic role of the alternative oxidase pathway in C3 species under photoinhibitory conditions. New Phytol. 2016;212:66–79. doi: 10.1111/nph.14030. [DOI] [PubMed] [Google Scholar]
- Fuchs P., Bohle F., Lichtenauer S., Ugalde J.M., Feitosa Araujo E., Mansuroglu B., Ruberti C., Wagner S., Müller-Schüssele S.J., Meyer A.J., Schwarzländer M. Reductive stress triggers ANAC017-mediated retrograde signaling to safeguard the endoplasmic reticulum by boosting mitochondrial respiratory capacity. Plant Cell. 2022;34:1375–1395. doi: 10.1093/plcell/koac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Fukushima A., Kusano M., Sakakibara H., Mizuno T., Saito K., Mizuno T., Saito K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Plant Signal. Behav. 2009;4:660–662. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Usadel B., Blaesing O.E., Kamlage B., Hoehne M., Trethewey R., Stitt M. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 2006;7:R23–R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009;150:1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Ho L.H.M., Clifton R., Carroll A., Estavillo G., Tan Y.-F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., et al. The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008;147:595–610. doi: 10.1104/pp.107.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C.P., Van Aken O., Millar A.H., Murcha M., Whelan J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N.P., Marshall R.S., Lee K.H., Vierstra R.D. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell. 2016;28:1279–1296. doi: 10.1105/tpc.15.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C., Huang S., Thatcher L.F., Foley R.C., Anderson C.R., Carroll A.J., Millar A.H., Singh K.B. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA. 2011;108:10768–10773. doi: 10.1073/pnas.1016060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves R.L.S., Quinlan C.L., Perevoshchikova I.V., Hey-Mogensen M., Brand M.D. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J. Biol. Chem. 2015;290:209–227. doi: 10.1074/jbc.M114.619072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Coman D., Uhrig R.G., Walsh S., Flis A., Stitt M., Gruissem W. Parallel analysis of Arabidopsis circadian clock mutants reveals different scales of transcriptome and proteome regulation. Open Biol. 2017;7:160333. doi: 10.1098/rsob.160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammal F., de Langen P., Bergon A., Lopez F., Ballester B. ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab996. D316-d325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Yu H., Yuan R., Yang Y., An F., Qin G. Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience. 2019;15:611–622. doi: 10.1016/j.isci.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.-S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- He H., Denecker J., Van Der Kelen K., Willems P., Pottie R., Phua S.Y., Hannah M.A., Vertommen D., Van Breusegem F., Mhamdi A. The Arabidopsis mediator complex subunit 8 regulates oxidative stress responses. Plant Cell. 2021;33:2032–2057. doi: 10.1093/plcell/koab079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B., Börner T., Weihe A. One RNA polymerase serving two genomes. EMBO Rep. 2000;1:435–440. doi: 10.1093/embo-reports/kvd086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L.H.M., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 2008;147:1858–1873. doi: 10.1104/pp.108.121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C.M., Castleden I.R., Aryamanesh N., Jacoby R.P., Millar A.H. Finding the subcellular location of barley, wheat, rice and maize proteins: the compendium of crop proteins with annotated locations (cropPAL) Plant Cell Physiol. 2016;57:e9. doi: 10.1093/pcp/pcv170. [DOI] [PubMed] [Google Scholar]
- Hooper C.M., Castleden I.R., Tanz S.K., Aryamanesh N., Millar A.H. SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1041. D1064-d1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw982. D1040-d1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job N., Lingwan M., Masakapalli S.K., Datta S. Transcription factors BBX11 and HY5 interdependently regulate the molecular and metabolic responses to UV-B. Plant Physiol. 2022;189:2467–2480. doi: 10.1093/plphys/kiac195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak S.M., Mochizuki N., Naranjo B., Xu D., Leister D., Kleine T., Okamoto H., Terry M.J. Plastid-to-Nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 2019;179:18–23. doi: 10.1104/pp.18.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S.J., Thomashow M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013;75:364–376. doi: 10.1111/tpj.12205. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Labs M., Rühle T., Leister D. The antimycin A-sensitive pathway of cyclic electron flow: from 1963 to 2015. Photosynth. Res. 2016;129:231–238. doi: 10.1007/s11120-016-0217-2. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le C.T.T., Brumbarova T., Ivanov R., Stoof C., Weber E., Mohrbacher J., Fink-Straube C., Bauer P. ZINC finger of arabidopsis THALIANA12 (ZAT12) interacts with FER-LIKE iron DEFICIENCY-INDUCED transcription factor (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 2016;170:540–557. doi: 10.1104/pp.15.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., Millar A.H. Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol. Cell. Proteomics. 2010;9:2125–2139. doi: 10.1074/mcp.M110.001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., O'Toole N., Millar A.H. Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell. Proteomics. 2008;7:1297–1316. doi: 10.1074/mcp.M700535-MCP200. [DOI] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., O'Toole N., Millar A.H. Combining proteomics of root and shoot mitochondria and transcript analysis to define constitutive and variable components in plant mitochondria. Phytochemistry. 2011;72:1092–1108. doi: 10.1016/j.phytochem.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Lee C.P., Elsässer M., Fuchs P., Fenske R., Schwarzländer M., Millar A.H. The versatility of plant organic acid metabolism in leaves is underpinned by mitochondrial malate-citrate exchange. Plant Cell. 2021;33:3700–3720. doi: 10.1093/plcell/koab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D., Marino G., Minagawa J., Dann M. An ancient function of PGR5 in iron delivery? Trends Plant Sci. 2022;27:971–980. doi: 10.1016/j.tplants.2022.04.006. [DOI] [PubMed] [Google Scholar]
- Li M., Lee K.P., Liu T., Dogra V., Duan J., Li M., Xing W., Kim C. Antagonistic modules regulate photosynthesis-associated nuclear genes via GOLDEN2-LIKE transcription factors. Plant Physiol. 2022;188:2308–2324. doi: 10.1093/plphys/kiab600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang Y., He Y., Wang Y., Wang L. Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiol. 2020;183:686–699. doi: 10.1104/pp.19.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tang J., Srivastava R., Bassham D.C., Howell S.H. The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell. 2020;32:3559–3575. doi: 10.1105/tpc.20.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.L., Voon C.P., Guan X., Yang Y., Gardeström P., Lim B.L. In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD(+) fluorescent protein sensors. Nat. Commun. 2020;11:3238. doi: 10.1038/s41467-020-17056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Tirichine L., Bowler C. Protocol: chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species. Plant Methods. 2012;8:48. doi: 10.1186/1746-4811-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litthauer S., Jones M.A. SAL1-PAP retrograde signalling extends circadian period by reproducing the loss of exoribonuclease (XRN) activity. Plant Signal. Behav. 2018;13:e1500066. doi: 10.1080/15592324.2018.1500066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Jiang Y., Tang H., Tong S., Lou S., Shao C., Zhang J., Song Y., Chen N., Bi H., et al. The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 in Arabidopsis. Plant Cell. 2021;33:1771–1789. doi: 10.1093/plcell/koab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O., Michael T.P., Burger B.T., Le Metté C., Mockler T.C., Weigel D., Chory J. A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008;105:17193–17198. doi: 10.1073/pnas.0807264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu W., Selinski J., Li L., Day D.A., Murcha M.W., Whelan J., Wang Y. Isolation and respiratory measurements of mitochondria from Arabidopsis thaliana. JoVE. 2018 doi: 10.3791/56627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie S.A., Kundariya H. Organellar protein multi-functionality and phenotypic plasticity in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190182. doi: 10.1098/rstb.2019.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Puche L., Tan H., Dogra V., Wu M., Rosas-Diaz T., Wang L., Ding X., Zhang D., Fu X., Kim C., Lozano-Duran R. A defense pathway linking plasma membrane and chloroplasts and Co-opted by pathogens. Cell. 2020;182:1109–1124.e25. doi: 10.1016/j.cell.2020.07.020. [DOI] [PubMed] [Google Scholar]
- Meng X., Li L., De Clercq I., Narsai R., Xu Y., Hartmann A., Claros D.L., Custovic E., Lewsey M.G., Whelan J., et al. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019;180:634–653. doi: 10.1104/pp.18.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Li L., Pascual J., Rahikainen M., Yi C., Jost R., He C., Fournier-Level A., Borevitz J., Kangasjärvi S., et al. GWAS on multiple traits identifies mitochondrial ACONITASE3 as important for acclimation to submergence stress. Plant Physiol. 2022;188:2039–2058. doi: 10.1093/plphys/kiac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller I.M. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- Ng S., De Clercq I., Van Aken O., Law S.R., Ivanova A., Willems P., Giraud E., Van Breusegem F., Whelan J. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant. 2014;7:1075–1093. doi: 10.1093/mp/ssu037. [DOI] [PubMed] [Google Scholar]
- Ng S., Ivanova A., Duncan O., Law S.R., Van Aken O., De Clercq I., Wang Y., Carrie C., Xu L., Kmiec B., et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Giraud E., Duncan O., Law S.R., Wang Y., Xu L., Narsai R., Carrie C., Walker H., Day D.A., et al. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T.M., Vukašinović N., Liu D., Russinova E., Yin Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 2020;32:295–318. doi: 10.1105/tpc.19.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H., Komori T., Uemura S., Kanda Y., Shimotani K., Nakai K., Furuichi T., Takebayashi K., Sugimoto T., Sano S., et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012;3:926. doi: 10.1038/ncomms1926. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D.J. The glycine decarboxylase complex from plant mitochondria. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994;45:323–337. [Google Scholar]
- Pérez-Salamó I., Papdi C., Rigó G., Zsigmond L., Vilela B., Lumbreras V., Nagy I., Horváth B., Domoki M., Darula Z., et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014;165:319–334. doi: 10.1104/pp.114.237891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Masiero S., Eubel H., Braun H.-P., Bhushan S., Glaser E., Salamini F., Leister D. Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell. 2006;18:970–991. doi: 10.1105/tpc.105.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua S.Y., Yan D., Chan K.X., Estavillo G.M., Nambara E., Pogson B.J. The Arabidopsis SAL1-PAP Pathway: a case study for integrating chloroplast retrograde, light and hormonal signaling in modulating plant growth and development? Front. Plant Sci. 2018;9:1171. doi: 10.3389/fpls.2018.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G.L., Vaistij F.E., Hiranuma S., Seto H., Takatsuto S., Adam G., Yoshida S., Bowles D. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA. 2005;102:15253–15258. doi: 10.1073/pnas.0504279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan C.L., Perevoshchikova I.V., Hey-Mogensen M., Orr A.L., Brand M.D. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol. 2013;1:304–312. doi: 10.1016/j.redox.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.M., McIntosh L. Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell. 1992;4:1131–1139. doi: 10.1105/tpc.4.9.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19:4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühle T., Dann M., Reiter B., Schünemann D., Naranjo B., Penzler J.F., Kleine T., Leister D. PGRL2 triggers degradation of PGR5 in the absence of PGRL1. Nat. Commun. 2021;12:3941. doi: 10.1038/s41467-021-24107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Landgraf J., Accerbi M., Simon V., Larson M., Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J., Scheibe R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019;21:21–30. doi: 10.1111/plb.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J., Scheibe R., Day D.A., Whelan J. Alternative oxidase is positive for plant performance. Trends Plant Sci. 2018;23:588–597. doi: 10.1016/j.tplants.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A., Vainonen J.P., Hunter K., Tossavainen H., Tiwari A., Järvi S., Hellman M., Aarabi F., Alseekh S., Wybouw B., et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife. 2019;8:e43284. doi: 10.7554/eLife.43284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Shin J., Johansson M., Davis S.J. The circadian clock goes genomic. Genome Biol. 2013;14:208. doi: 10.1186/gb-2013-14-6-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman C., Waterman C.D., Rainbird B.M., Smith P.M.C., Jenkins C.D., Day D.A., Soole K.L. AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 2019;181:774–788. doi: 10.1104/pp.19.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronconi M.A., Fahnenstich H., Gerrard Weehler M.C., Andreo C.S., Flügge U.I., Drincovich M.F., Maurino V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 2008;146:1540–1552. doi: 10.1104/pp.107.114975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinf. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O., Pogson B.J. Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ. 2017;24:955–960. doi: 10.1038/cdd.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O., Zhang B., Carrie C., Uggalla V., Paynter E., Giraud E., Whelan J. Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant. 2009;2:1310–1324. doi: 10.1093/mp/ssp053. [DOI] [PubMed] [Google Scholar]
- Van Aken O., De Clercq I., Ivanova A., Law S.R., Van Breusegem F., Millar A.H., Whelan J. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 2016;171:2150–2165. doi: 10.1104/pp.16.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S., Vandenbroucke K., Inzé A., van de Cotte B., Mühlenbock P., De Rycke R., Naouar N., Van Gaever T., Van Montagu M.C.E., Van Breusegem F. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:20113–20118. doi: 10.1073/pnas.1217516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G.C., Dahal K., Alber N.A., Chadee A. Photosynthesis, respiration and growth: a carbon and energy balancing act for alternative oxidase. Mitochondrion. 2020;52:197–211. doi: 10.1016/j.mito.2020.04.001. [DOI] [PubMed] [Google Scholar]
- Walley J., Xiao Y., Wang J.Z., Baidoo E.E., Keasling J.D., Shen Z., Briggs S.P., Dehesh K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2015;112:6212–6217. doi: 10.1073/pnas.1504828112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim C., Xu X., Piskurewicz U., Dogra V., Singh S., Mahler H., Apel K. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA. 2016;113:E3792–E3800. doi: 10.1073/pnas.1603562113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Selinski J., Mao C., Zhu Y., Berkowitz O., Whelan J. Linking mitochondrial and chloroplast retrograde signalling in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190410. doi: 10.1098/rstb.2019.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.Z., Meyer E.H., Richter A.S., Schuster M., Ling Q., Schöttler M.A., Walther D., Zoschke R., Grimm B., Jarvis R.P., Bock R. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants. 2019;5:525–538. doi: 10.1038/s41477-019-0415-y. [DOI] [PubMed] [Google Scholar]
- Wu X., Wu J., Wang Y., He M., He M., Liu W., Shu S., Sun J., Guo S. The key cyclic electron flow protein PGR5 associates with cytochrome b6f, and its function is partially influenced by the LHCII state transition. Hortic. Res. 2021;8:55. doi: 10.1038/s41438-021-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Waneka G., Broz A.K., King C.R., Sloan D.B. MSH1 is required for maintenance of the low mutation rates in plant mitochondrial and plastid genomes. Proc. Natl. Acad. Sci. USA. 2020;117:16448–16455. doi: 10.1073/pnas.2001998117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E., Vaahtera L., Hõrak H., Hincha D.K., Heyer A.G., Brosché M. Quantitative trait loci mapping and transcriptome analysis reveal candidate genes regulating the response to ozone in Arabidopsis thaliana. Plant Cell Environ. 2015;38:1418–1433. doi: 10.1111/pce.12499. [DOI] [PubMed] [Google Scholar]
- Yamada S., Ozaki H., Noguchi K. The mitochondrial respiratory chain maintains the photosynthetic electron flow in Arabidopsis thaliana leaves under high-light stress. Plant Cell Physiol. 2020;61:283–295. doi: 10.1093/pcp/pcz193. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Takano Y., Abe J., Hibino M., Harashima H. Therapeutic strategies for regulating mitochondrial oxidative stress. Biomolecules. 2020;10:83. doi: 10.3390/biom10010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Luo L., Xu J., Xin P., Guo H., Wu J., Bai L., Wang G., Chu J., Zuo J., et al. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 2018;28:448–461. doi: 10.1038/s41422-018-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Berkowitz O., Selinski J., Hartmann A., Narsai R., Wang Y., Mao P., Whelan J. Conserved and opposite transcriptome patterns during germination in Hordeum vulgare and Arabidopsis thaliana. Int. J. Mol. Sci. 2020;21:7404. doi: 10.3390/ijms21197404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack P.J., De Clercq I., Howton T.C., Hallmark H.T., Hurny A., Keshishian E.A., Parish A.M., Benkova E., Mukhtar M.S., Van Breusegem F., Rashotte A.M. Cytokinin response factor 6 represses cytokinin-associated genes during oxidative stress. Plant Physiol. 2016;172:1249–1258. doi: 10.1104/pp.16.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited at the NCBI SRA database under project IDs PRJNA843852, PRJNA843855, PRJNA894307, and PRJNA837635.