Abstract
Radioresistance is the major factor of glioblastoma multiforme (GBM) treatment failure and relapse. Hypoxia and autophagy are linked to radioresistance and poor prognosis in solid tumors, but mechanisms remain unknown. Thus, we hypothesize that hypoxia may activate autophagy through two critical factors, HIF1A and Beclin-1, resulting in radioresistance of GBM in vitro and in vivo. In this study, we first demonstrated that HIF1A was overexpressed in GBM tissues and predicted a poor prognosis via bioinformatics. Secondly, we determined that hypoxia induced high expression of HIF1A and upregulated levels of Beclin-1 and autophagy, while HIF1A knockdown by shRNA reduced the expression of Beclin-1. Then we revealed the crosstalk and mechanisms of HIF1A-associated-Beclin-1 in three aspects: (a) transcriptional regulation, (b) protein interaction, and (c) HIF1A/BNIP3/Beclin-1 signaling pathway. Furthermore, we confirmed that silencing HIF1A enhanced the radiosensitivity of GBM in vitro and in vivo. Additionally, Beclin-1 suppression by 3-MA could reverse radioresistance induced by HIF1A under hypoxia. In conclusion, we demonstrated that hypoxia triggered autophagy via HIF1A-associated Beclin-1, resulting in radioresistance in GBM. HIF1A knockdown improved GBM radiosensitivity, and silencing Beclin-1 could reverse HIF1A-induced radioresistance under hypoxic conditions. These findings may help us comprehend the molecular underpinnings of hypoxia-induced autophagy and provide a novel perspective and prospective treatment for GBM radiosensitization.
Keywords: Autophagy, Beclin-1, Hypoxia, Hypoxia-inducible factor 1 alpha (HIF1A), Radiotherapy
Highlights
-
•
Hypoxia activates autophagy via two pivotal molecules, HIF1A and Beclin-1.
-
•
HIF1A and Beclin-1 crosstalk in transcriptional, proteinic, BNIP3/Bcl-2 signaling.
-
•
Intracerebral human glioblastoma xenograft proves radioresistant.
-
•
HIF1A-associated Beclin-1 elicits glioblastoma radioresistance in vitro and vivo.
-
•
Beclin-1 inhibitor reverses hypoxia-induced radioresistance under oxygen deprival.
1. Introduction
Glioblastoma multiforme (GBM) is a highly aggressive cancer that originates in the brain [1]. Glioblastoma accounts for approximately 15% of brain tumors [2], exhibiting rapid progression and dismal prognosis. The standard treatment consists of surgery followed by adjuvant radiotherapy with or without temozolomide (depending on the MGMT methylation status) [3]. Even with complete resection, the median progression-free survival time is only 6.9 months, and the overall survival time (with or without temozolomide) ranges from 14.6 to 16.6 months [4,5]. However, local recurrence following radiotherapy is the primary sign of tumor progression, and radioresistance indicates a poor prognosis in GBM [[6], [7], [8], [9]]. Therefore, elucidating molecular mechanisms of radioresistance is essential for improving the overall survival of GBM patients.
Recently, much effort has been devoted to the mechanisms underlying GBM radioresistance, including DNA double-strand break repair, autophagy, hypoxia exposure, and more [[10], [11], [12], [13], [14], [15], [16]]. Among the numerous radioresistance-inducing factors, hypoxia-inducible factor 1 alpha (HIF1A) has been extensively reported. HIF1A is the crucial transcriptional regulator responding to hypoxia [17,18] and mediates radioresistance by orchestrating downstream gene activity [19,20]. Consequently, targeting HIF1A is a promising strategy to increase radiosensitivity.
On the other hand, autophagy is a double-edged sword, and Beclin-1 plays various roles depending on the circumstances. Autophagy is considered the adaptive response to stress, for systematic degradation and recycling of cellular components [21]. The underlying role and mechanisms remain to be explored, and some literature suggests that autophagy and Beclin-1 are involved in radioresistance [22–24]. Autophagy demonstrated dual roles in GBM radiotherapy studied, conferring radioresistance and promoting cell survival while also causing cell death; however, the underlying mechanism is controversial [22–24]. Beclin–1 is a critical protein in autophagosome formation, which binds to the class III PI–3 kinase Vps34 and expedites autophagic proceedings [23,25]. We previously observed that targeting Beclin–1 impaired the induction of IR–mediated autophagy and sensitized C6 glioma cells to radiotherapy [26].
Notably, some researchers claim that HIF1A may induce radioresistance via autophagy [[27], [28], [29], [30]]. In Wang’s study on nasopharyngeal carcinoma, a correlation was discovered between the Beclin–1 and HIF1A expression. Moreover, it was reported that HIF1A–associated Beclin–1 high expression might facilitate the survival of NPC cells from chemoradiotherapy, leading to a poor prognosis [31]. On the contrary, for patients with high HIF1A expression but lower Beclin–1, it displayed a favorable advantage for overall survival [31]. Therefore, we hypothesized that autophagy might be the link between hypoxia and radioresistance, with HIF1A and Beclin–1 serving as crucial factors.
Our research aimed to determine the effects of hypoxia–induced autophagy on radiation and to elucidate underlying molecular mechanisms by targeting HIF1A and Beclin–1 in human GBM cells.
2. Materials and Methods
2.1. Cell cultures
GBM cell lines U87, U251, and U373 were kindly provided by Dr. Ren Jinghua, who purchased them from the American Type Culture Collection. U87 and U251 were bred in DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin. And U373 was bred in RPMI–1640 with 10% fetal bovine serum and 1% penicillin/streptomycin. The above cells were cultured at 37 °C with 5% CO2 for normoxic conditions. The hypoxic incubator was applied to construct the hypoxic condition with O2 1%, CO2 5%, and 37 °C.
2.2. RNA interference and construction of lentiviral stably transfected GBM cells
The short hairpin RNAs (shRNAs) lentivirus was constructed and identified by Shanghai GKN Genetics and transfected into U87 and U251. At 48 h post–transfection, lentiviruses harvested were used to infect GBM cells after mixing with polybrene (10 mg/mL; Sigma). Stable cells were screened with media containing 2 μg/ml puromycin and confirmed by qRT–PCR and Western blot.
2.3. Quantitative real–time PCR
Total RNA extraction was accomplished by TRIzol reagent (Invitrogen) and the reverse transcription with Prime RT reagent kit (Vazyme). Quantitative PCR was performed utilizing SYBR Green real–time PCR kit (Vazyme). The relative mRNA levels were computed with the ΔCt method. PCR primer sequences are shown in Table 1.
Table 1.
Primer sequences of the targeted gene
| Primer | Sequence (5’ to 3’) |
|---|---|
| HIF1A_Fw | CACCACAGGACAGTACAGGAT |
| HIF1A_Rv | CGTGCTGAATAATACCACTCACA |
| Beclin–1_Fw | ACCTCAGCCGAAGACTGAAG |
| Beclin–1_Rv | AACAGCGTTTGTAGTTCTGACA |
| ATG5_Fw | TTGACGTTGGTAACTGACAAAGT |
| ATG5_Rv | TGTGATGTTCCAAGGAAGAGC |
| ATG7_Fw | GATCCGGGGATTTCTTTCACG |
| ATG7_Rv | CAGCAATGTAAGACCAGTCAAGT |
| ATG12_Fw | TAGAGCGAACACGAACCATCC |
| ATG12_Rv | CACTGCCAAAACACTCATAGAGA |
| β–Actin_Fw | TCACCCACACTGTGCCCA |
| β–Actin_Rv | ATGTCACGCACGATTTCCC |
2.4. Western blotting
Whole–cell lysates were performed in NETN buffer with 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P–40 separated by SDS–PAGE and transferred to PVDF membranes. The following primary antibodies were used: ß–Actin (Abcam, USA), HIF1A (BD, USA/ Abcam, USA), Beclin–1 (Abcam, USA), LC–3 (Abcam, USA), γ–H2AX (Abcam, USA), HIF1A (ChIP level, Abcam, USA), BNIP3 (Abcam, USA), and Bcl–2 (Abcam, USA).
2.5. Cell growth and colony formation assay
Cells were plated in six–well plates in triplicate. With optimal density and viability, cells were exposed to irradiation 2Gy in a single dose. After being incubated for 14 days, numbers of colonies consisting of above 50 cells were counted.
2.6. EdU assay
EdU (5–ethynyl–2′–deoxyuridine) is a nucleoside analog of thymidine incorporated into DNA during replication and used to test cell proliferation. Cells in the exponential phase were seeded into 96–well plates overnight and treated with 3–MA (0, 4 h) or in hypoxia condition (0, 16 h), subsequently exposed with 2 Gy X–ray irradiation. After 24 h of radiation, cells were incubated with EdU solution (1/1000, RiboBio, Guangzhou, China) for 2 hours, and fixed with 4% paraformaldehyde for 30 min, then permeabilized with 0.5% Triton X–100 for 10 min, followed by stained with Hoechst and detected by fluorescent microscope. EdU–positive cells were counted via ImageJ Software (V1.49, National Institutes of Health).
2.7. Transmission electron microscopy (TEM)
Cells were collected by centrifugation at 1000 rpm, 4 °C for 15 min. Then, samples were added with 2.5% glutaraldehyde solution at 4 °C overnight. After dehydrating, embedding, solidifying, ultrathin slicing, and staining, samples were observed and imaged by transmission electron microscope, on the Sharing Management Platform of Scientific Research (Basic Medical College of Wuhan University).
2.8. Immunofluorescence staining
Cells were seeded on coverslips in 24–well plates and exposed to 2Gy/1F irradiation. Subsequently, cells were fixed with 4% paraformaldehyde at different time points after radiation, then permeabilized with 0.2% Triton X–100 for 5 min, and blocked with 5% BSA for 30 min, next immunostained with mouse monoclonal anti–γ–H2AX (1:500) overnight at 4 °C. Then samples were incubated with Cyanine3–conjugated goat anti–mouse IgG secondary antibody (1:200) for 1 h, finally stained with DAPI and examined by fluorescence microscope.
2.9. Irradiation
Cells were seeded in the 6–cm dishes and exposed to 0, 2, 4, 6, and 8 Gy irradiation using the X–ray irradiator (6MV, Varian) with a dose rate of 600 Mu/min. Next, the number of surviving colonies (over 50 cells) was calculated after 14 days. For animal experiments, the successful intracerebral xenograft models were randomly divided into two groups (un–irradiated/irradiated). The whole brain irradiation with 10 Gy in a single dose was administered to the irradiated group, with the same physical parameters for cells. Then, tumors were measured every week by in vivo fluorescence imaging.
2.10. In vivo xenograft mouse model
The animal experiments were authorized by the Medical Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology, under national standard guidelines for animal welfare (Ethics Approval No. 2016–S034). Female nude mice (5–6 weeks old, BALB/c Nude) were randomly grouped into three groups (KD, MOCK, and WT). Luciferin labeling GBM cells (U87–LUC, U251–LUC, 5 × 105 cells/5 uL suspended in phosphate–buffered saline) were intracerebrally injected into the right caudate nucleus of the brains, in the guidance of The Mouse Brain in Stereotaxic Coordinates Atlas [32]. With Bregma as the origin, the lateral 2 mm, anterior 1 mm, and vertical 3 mm point is appropriate for inoculation. Tumors were monitored by in vivo fluorescence imaging per week. Tumor volume was calculated by the formula: V = (L × W2)×π/6, where L = length (mm), and W = width (mm). Tumor inhibition rate (TIR) was calculated as follows: TIR = [1−VIR(+)/VIR(–)]×100%, where VIR(+) = the average tumor volume of irradiated groups, and VIR(–) = the average tumor volume of un–irradiated groups. There were six nude mice in each group.
2.11. Statistical analyses
The data are presented as the mean ± SD except where otherwise indicated, of at least three replicate independent experiments. And the statistical analysis and graphs were performed with Graph Pad Prism 7.0 (GraphPad Software, CA). The student’s t–test or variance test (ANOVA) was used for statistical analysis, and P < 0.05 was considered statistically significant.
3. Results
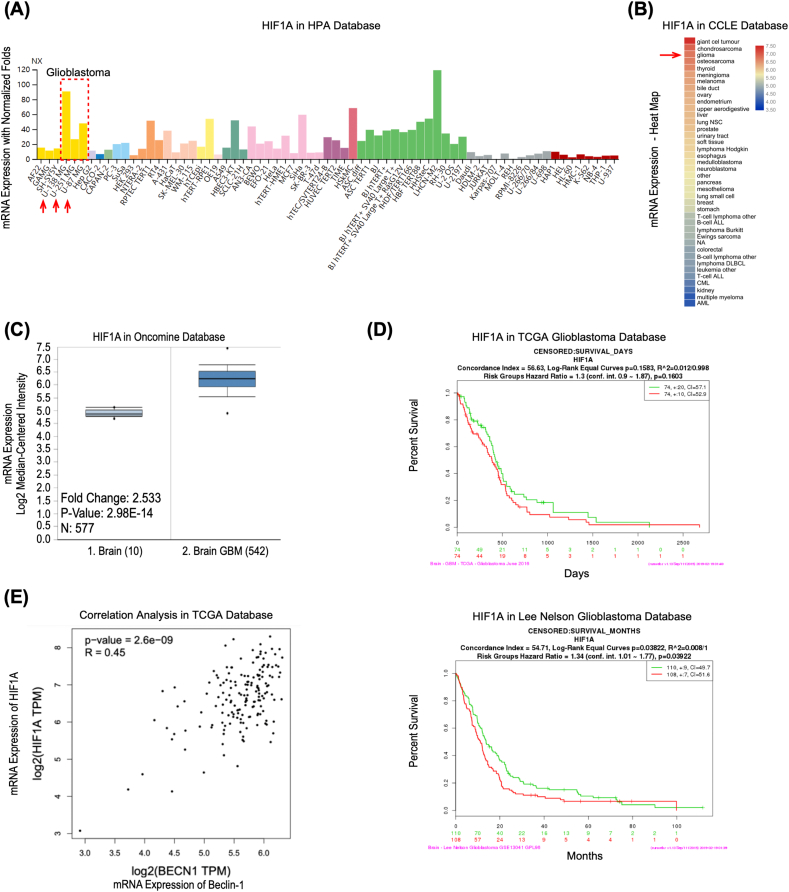
3.1. HIF1A is overexpressed in GBM patients and predicts the adverse prognosis
To investigate the clinical significance of HIF1A in GBM, we first analyzed the HIF1A mRNA level. In human cell lines, we discovered that the levels of HIF1A mRNA were considerably higher in GBM than in other solid tumors (Figure 1 A). We found that glioma tissue HIF1A mRNA levels were significantly greater than other tumor tissues (Figure 1 B). Accordingly, the Oncomine database found that HIF1A mRNA expression was 2.53 times higher in brain glioblastoma than in healthy brain tissue (Figure 1 C). Furthermore, according to the Kaplan–Meier survival curves for overall survival in The Cancer Genome Atlas (TCGA) glioblastoma database, elevated HIF1A gene expression was revealed to mediate a poor prognosis in GBM (HR 1.34, P = 0.039) (Figure 1 D). These findings demonstrated that HIF1A is overexpressed in GBM and predicts an adverse prognosis. Even though there is no significant result to support that Beclin–1 negatively affects overall survival or disease–free survival (Figure S 1). However, we found a strong correlation (R 0.45, P < 0.0001) between HIF1A and Beclin–1 in the transcriptional level of clinical samples in the TCGA database (Figure 1 E).
Figure 1.
HIF1A is overexpressed in GBM and predicts an adverse prognosis. (A) The mRNA expression level of HIF1A in different cell lines based on normalized RNA sequencing data on the Human Protein Atlas (HPA) database. https://www.proteinatlas.org. NX: Normalized expression, the expression level of gene–specific transcripts is given as normalized expression (NX) values, and transcripts with NX values ≥1 are considered as detected. The dotted box and red arrows indicate the Glioblastoma cell lines. (B) Heat map of the HIF1A mRNA expression level in various tissues based on the data in Cancer Cell Line Encyclopedia (CCLE). https://www.broadinstitute.org/ccle/home. GraphPad Prism plotted the heat map. The red arrow indicates the normalized mean value of mRNA expression in glioma tissue datasets. (C) HIF1A mRNA expression was significantly increased in brain glioblastoma tissue compared with normal brain tissue, as revealed by Oncomine data mining analysis (in the reporter 200,989 at probe set in the TCGA brain dataset). https://www.oncomine.org. (D) Multivariate Cox regression analysis of HIF1A gene expression and Kaplan–Meier survival curves for overall survival outcomes in the TCGA glioblastoma and Lee Nelson study datasets (The red and green lines correspondingly indicate a gene expression level above and below the median). https://cancergenome.nih.gov. (E) Correlation Analysis between HIF1A and Beclin–1 in the transcriptional level of clinical samples in TCGA GBM Tumor database, visualized by GEPIA website. https://gepia.cancer-pku.cn; TPM: transcripts per million, a normalized unit to measure the transcript coding gene expression.
3.2. Hypoxia induces expression of HIF1A and upregulates levels of Beclin–1 and autophagy
According to the initial bioinformatics analysis, HIF1A and Beclin–1 have comparable effects in predicting the adverse prognosis of GBM patients. Therefore, based on the background of our study and other reports, we turned to investigate the relationship between hypoxia and autophagy, in terms of two crucial molecules, HIF1A and Beclin–1.
First, we verified the HIF1A high expression induced by hypoxia. According to the results of qRT–PCR and Western blot analysis, we determined that hypoxia exposure elevated the expression of HIF1A at the transcriptional and translational levels (Figure S 2 A–B). Furthermore, the HIF1A protein mainly accumulated in the cytoplasm in GBM cells (Figure S 2C).
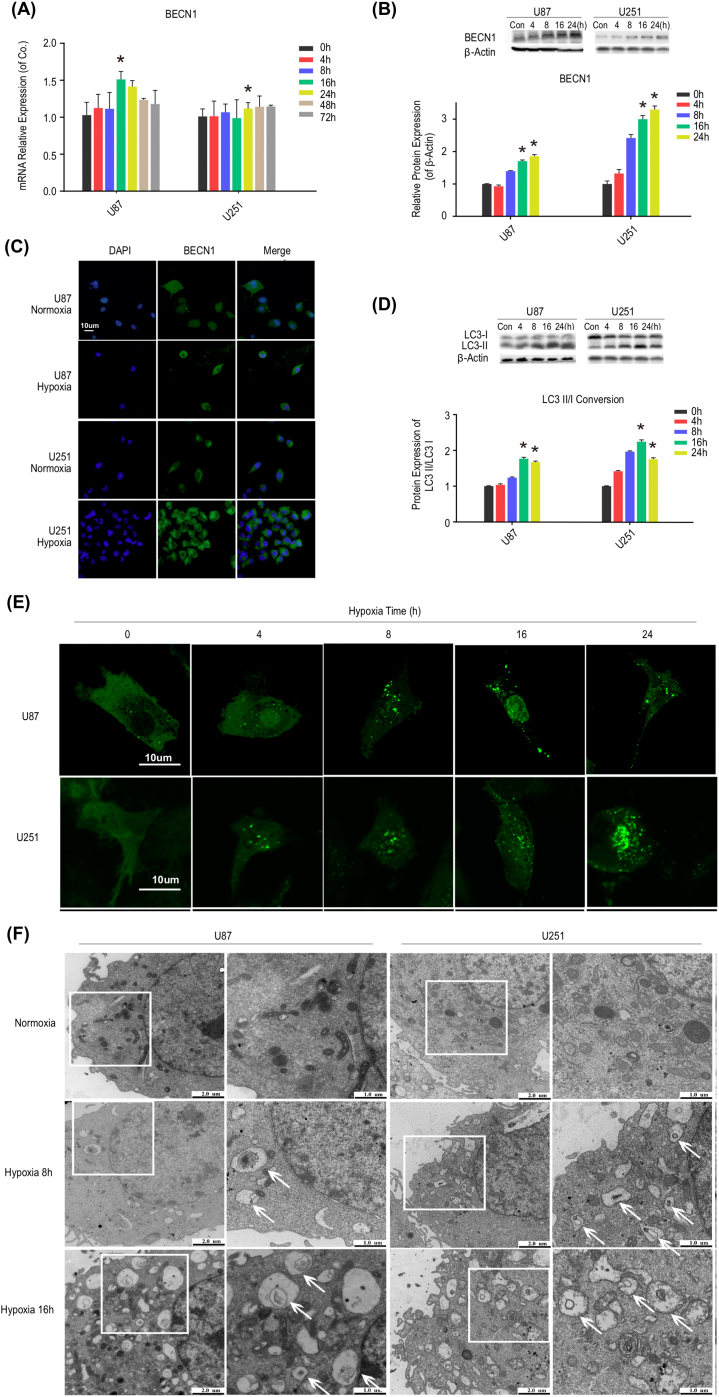
Next, we determined the effects of hypoxia on cellular autophagy and Beclin–1. As depicted in Figure 2 A–C and Figure S 3A, hypoxia increased the mRNA and protein expression levels of autophagy–associated genes, especially Beclin–1. The protein MAPLC3, a well–documented autophagosome marker, associates with the lipid membranes of autophagosomes and initiates autophagic flux by shifting from its cytoplasmic form (LC3–I) to autophagosomal state (LC3–II). The Western blot results showed that hypoxia caused the conversion of LC3–I to LC3–II (Figure 2 D). Consistent with this finding, granular LC3–GFP puncta aggregated under oxygen deprivation (Figure 2 E), with the amount and intensity of puncta increasing in a time–dependent manner, indicating the increased level of autophagy. Additionally, we adopted TEM to confirm the formation of autophagosomes further. Under oxygen deprivation, the results showed the presence of autophagic vesicles with a double membrane (Figure 2 F). Collectively, these results showed that hypoxia elevated HIF1A expression, upregulated Beclin–1 expression, and promoted autophagosome formation.
Figure 2.
Hypoxia upregulates levels of Beclin–1 and autophagy. (A) The relative expression of Beclin–1 mRNA in GBM cells (U87 and U251) after exposure to hypoxia for 0, 4, 8, 16, 24, 48, and 72 h. Hypoxia for 16 or 24 h induced temporary upregulation of Beclin–1 mRNA expression in cells. BECN1: Beclin–1. *P < 0.05 compared with control cells (hypoxia for 0h). (B) The protein expression of Beclin–1 was increased after exposure to hypoxia for both 16 and 24 h. Left panel: Western blot analysis of Beclin–1 protein. Right panel: Quantification of the relative protein expression of Beclin–1. β–Actin was amplified for internal normalization. Con: control. *P < 0.05 compared with control cells. (C) Cells were incubated under normoxia and hypoxia for 16 h, and Beclin–1 staining was then visualized using fluorescence microscopy to detect its protein expression and localization. (D) Hypoxia induced the conversion of LC3–I to LC3–II. Left panel: Western blot analysis of LC3–I and LC3–II protein. Right panel: Quantification of the protein LC3–II/I conversion. *P < 0.05 compared with control cells. (E) Granular LC3–GFP puncta aggregated as the hypoxia time increased. Cells were transfected with GFP–LC3 and incubated under hypoxia for 0, 4, 8, 16, and 24 h. GFP–LC3 staining was visualized using confocal laser scanning fluorescence microscopy. (F) Hypoxia induced the accumulation of autophagosomes. Cells were incubated under hypoxia for 8 and 16 h, and autophagosomes were visualized by transmission electron microscopy.
3.3. HIF1A silencing downregulated Beclin–1 expression with multi–crosstalks behind
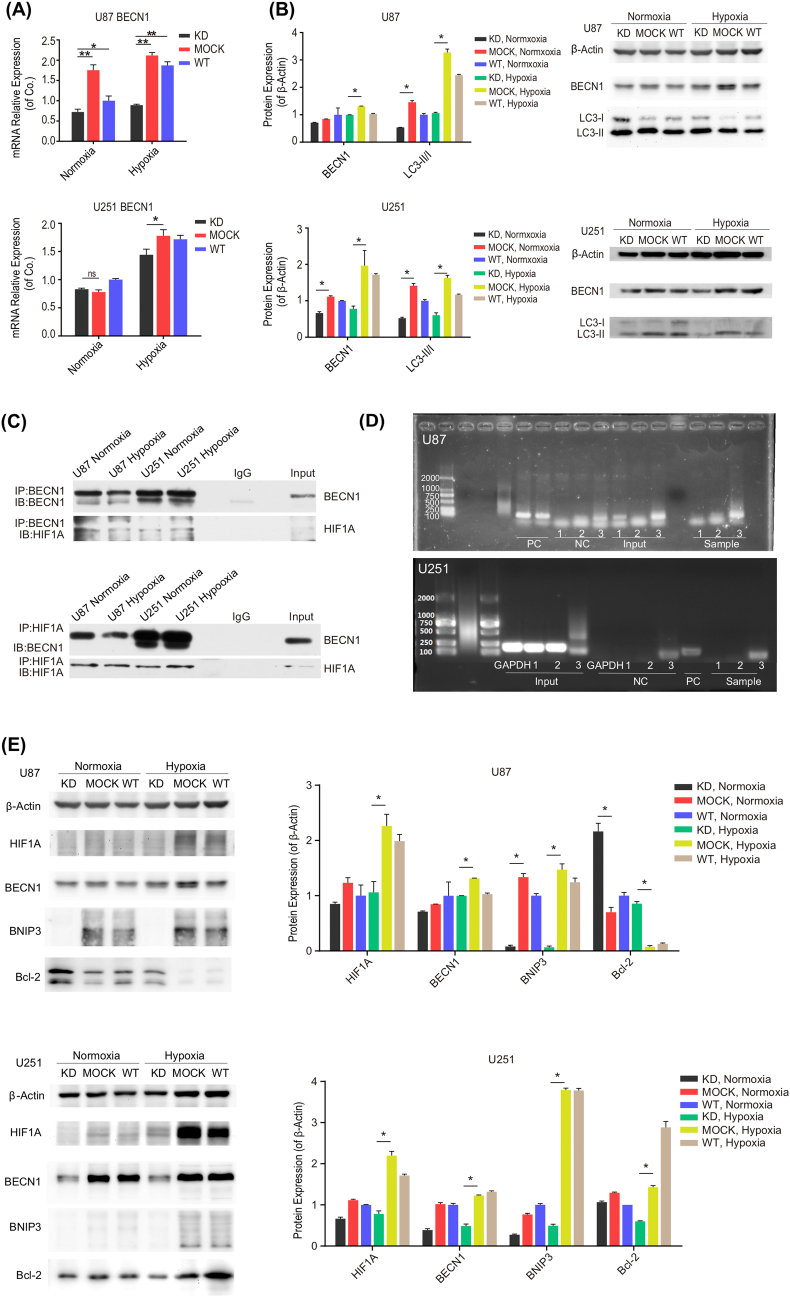
To further identify whether hypoxia exposure induces autophagy via HIF1A, we knocked down HIF1A to test the changes of Beclin–1 and cellular autophagy. First of all, we applied shRNAs to downregulate HIF1A expression in U87 and U251. As shown in Figure S 4 A–D, HIF1A was successfully knocked down both at mRNA and protein levels. Thus, we investigated whether HIF1A knockdown makes changes to cellular autophagy.
As shown in Figure 3, Beclin–1 was downregulated at both the mRNA and protein levels (Figure 3 A and B) in response to HIF1A depletion (KD group) in GBM cells. Similarly, mRNA expression of autophagy–related genes also declined when HIF1A was knocked down (Figure S 3B). Consistent with these observations, we found that the conversion of LC3–I to LC3–II was suppressed in HIF1A deficiency cells (KD) compared with controls (MOCK and WT) (Figure 3 B); taken together, these findings suggested that HIF1A knockdown attenuates Beclin–1 expression and cellular autophagy. To this end, we demonstrated that hypoxia triggered autophagy in HIF1A/Beclin–1 pattern.
Figure 3.
HIF1A silencing downregulated Beclin–1 expression with multi–crosstalks behind. (A) Beclin–1 mRNA expression was downregulated when HIF1A was knocked down in U87 and U251. The relative expression of Beclin–1 mRNA was detected by qRT–PCR after exposure to normoxia and hypoxia for 16 h in KD, MOCK, and WT groups. KD: knock down; WT: wildtype. *P < 0.05, **P < 0.01 compared with MOCK, under normoxia or hypoxia; ns: no statistically significant difference (P > 0.05). (B) Beclin–1 protein expression was attenuated, and LC3–I to LC3–II conversion was suppressed in the HIF1A–KD group after exposure to normoxia and hypoxia for 16 h. Left panel: Quantification of Beclin–1 protein and LC3–II/I conversion. Right panel: Western blot analysis of Beclin–1, LC3–I, and LC3–II protein. *P < 0.05 compared with MOCK, under normoxia or hypoxia. (C) Endogenous HIF1A was coprecipitated in cells ectopically expressing Beclin–1 and vice versa. Coimmunoprecipitation experiments were performed in U87 and U251 cells after exposure to normoxia and hypoxia for 16 h. IP: immunoprecipitation; IB: immunoblotting. (D) Three pairs of primers were designed for potential binding sites in the Beclin–1 promoter. ChIP experiments were performed in U87 and U251 under hypoxia for 16 h. Bands were amplified for primers No. 1 and No. 3, in contrast to the Input and the negative control (NC). Furthermore, primer No. 3 successfully amplified specific bands in the experimental group compared to the Input of U87. However, all primers failed to amplify specific bands in U251. PC: positive control; NC: negative control. (E) BNIP3/Bcl–2 pathway components were evaluated after exposure to normoxia and hypoxia for 16 h. BNIP3 was downregulated when HIF1A was knocked down in U87 and U251, especially under hypoxic conditions. Bcl–2 expression was elevated in U87 HIF1A–KD group but decreased in U251 HIF1A–KD, compared with the corresponding MOCK groups. β–Actin was amplified for internal normalization. *P < 0.05 compared with MOCK cells, under normoxia or hypoxia.
Furthermore, we performed relevant assays to elucidate the underlying mechanisms by which HIF1A cooperates with Beclin–1. First, we employed coimmunoprecipitation to understand how HIF1A controls Beclin–1 at the posttranslational level. Endogenous HIF1A was coprecipitated in cells ectopically expressing Beclin–1 and vice versa (Figure 3 C). Notably, the interaction between endogenous HIF1A and Beclin–1 was observed under both normoxic and hypoxic conditions in U87 and U251. Secondly, to determine whether HIF1A acts as a transcription factor to interact with Beclin–1 mRNA, we performed ChIP–qPCR analysis and revealed that HIF1A protein regulated Beclin–1 promoter activity under hypoxic conditions in U87 (Figure 3 D). Thus, these results confirm that HIF1A physically interacts with Beclin–1 in cells. Moreover, we investigated the potential signaling pathway through which HIF1A regulates Beclin–1. As shown in Figure 3 E, under hypoxic conditions in U87 cells, HIF1A knockdown dramatically attenuated the BNIP3 expression and downregulated Bcl–2 levels. While in HIF1A–depleted U251 cells, an appreciable decline was observed both in BNIP3 and Bcl–2. Thus, we speculate that HIF1A can regulate Beclin–1 via the BNIP3/Bcl–2 signaling pathway.
Taken together, these data demonstrated that the hypoxia could induce autophagy via HIF1A–associated Beclin–1. The crosstalk may occur along the HIF1A/BNIP3/Beclin–1 axis. Additionally, potential crosstalk between HIF1A and Beclin–1 could affect protein stability and transcriptional binding. Nonetheless, the definite mechanisms involved in this pathway need further investigation.
3.4. HIF1A silencing enhances the radiosensitivity of GBM in vitro and in vivo
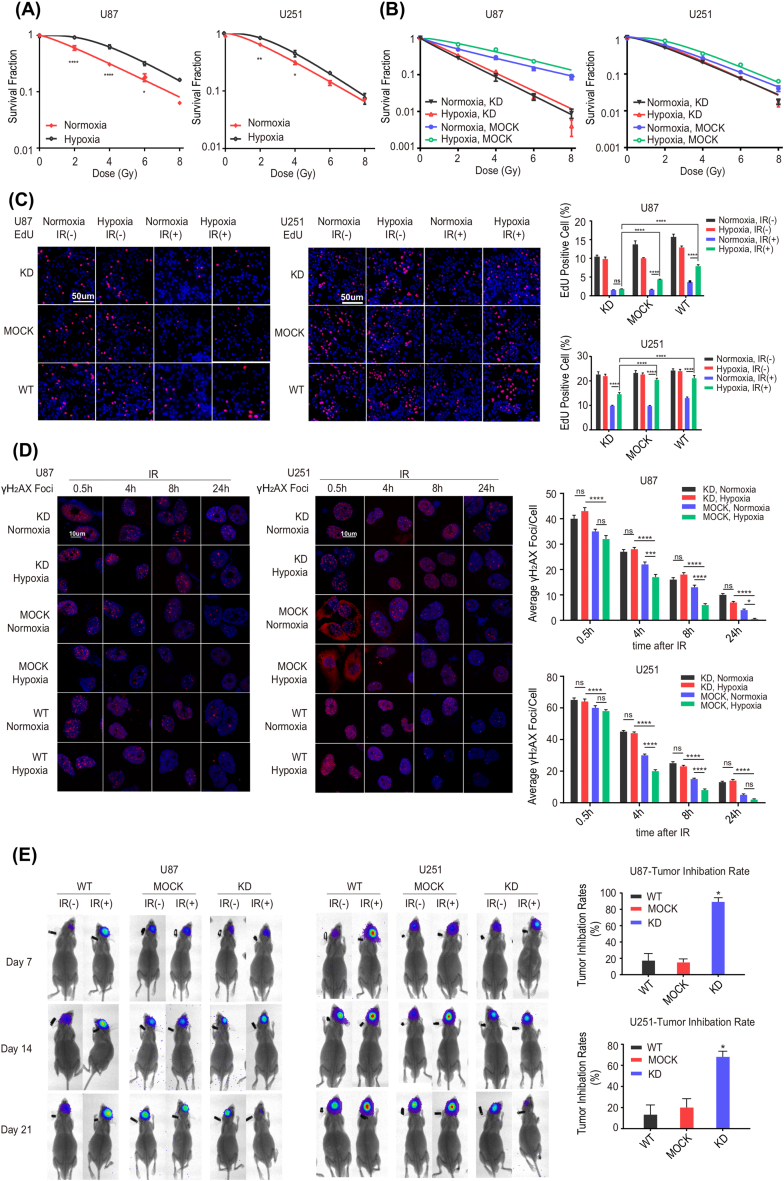
To verify that hypoxia induces radioresistance at the cellular level, we conducted the colony formation assay in U87 and U251 cells. It was demonstrated that the survival rate increased with oxygen deprivation compared to oxygen exposure (Figure 4 A and Table 2), indicating that hypoxia triggered radioresistance in GBM.
Figure 4.
HIF1A silencing enhances the radiosensitivity of GBM cellsin vitroandin vivo. (A) Hypoxia induced radioresistance of GBM cells. Colony formation after irradiation was significantly enhanced in hypoxia–exposed U87 and U251 cells. Colony formation assays were performed to evaluate the radiosensitivity of U87 and U251 after exposure to normoxia and hypoxia for 16h. The survival fraction curves were fitted by the multitarget one–click model equation SF=1–(1–e–D/D0) ^N. *P < 0.05, **P < 0.01, ****P < 0.0001 compared with normoxia cells, under the corresponding radiation doses. (B) Cellular proliferation was impaired, and radiosensitivity was enhanced in the HIF1A–KD group after exposure to normoxia and hypoxia for 16 h. Colony formation assays were performed in U87 and U251 incubated under normoxia or hypoxia for 16 h. (C) The proportion of EdU–positive cells was noticeably decreased in the HIF1A–KD group, especially under hypoxic conditions. Left panel: EdU incorporation was visualized using fluorescence microscopy after 24 h of X–ray irradiation (2Gy/1F). Right panel: quantification of EdU–positive cells. IR: irradiation. **P < 0.01, ****P < 0.0001. (D) The number of γ–H2AX foci was significantly increased in the KD group, especially under hypoxic conditions. Left panel: γ–H2AX foci were visualized using confocal laser scanning fluorescence microscopy after 0.5, 4, 8, and 24 h of X–ray irradiation (2Gy/1F). Right panel: quantification of γ–H2AX foci. * P < 0.05, ***P < 0.001, ****P < 0.0001. (E) In vivo, orthotopic glioblastoma xenografts were generated in nude mice, and brain tumors were measured weekly by in vivo fluorescence imaging. The tumor growth inhibition rates were calculated and were dramatically higher in the KD group than in MOCK and WT. *P < 0.0001 compared with MOCK and WT.
Table 2.
The survival fraction of U87 and U251 cells after X–ray irradiation under normoxia and hypoxia for 16 h.
| SFa (Mean±SEMb) | 2Gy | 4Gy | 6Gy | 8Gy | |
|---|---|---|---|---|---|
| U87 | Normoxia | 0.60 ± 0.06 | 0.31 ± 0.02 | 0.18 ± 0.03 | 0.06 ± 0.00 |
| Hypoxia | 0.92 ± 0.04 | 0.63 ± 0.05 | 0.32 ± 0.02 | 0.16 ± 0.01 | |
| U251 | Normoxia | 0.64 ± 0.01 | 0.32 ± 0.03 | 0.14 ± 0.01 | 0.07 ± 0.00 |
| Hypoxia | 0.83 ± 0.05 | 0.45 ± 0.06 | 0.21 ± 0.01 | 0.07 ± 0.02 |
survival fraction;
standard error of the mean
From above, we identified that the hypoxic condition triggered high expression of HIF1A, resulting in radioresistance. Given that HIF1A is frequently overexpressed in GBM tissues and predicts poor prognosis, we speculated that HIF1A served as the core element to promote radioresistance in GBM. To illustrate the effects of HIF1A on radiosensitivity, we applied HIF1A silencing system in vitro and in vivo to test changes in GBM radiosensitivity. As shown in Figure 4 B and Table 3, under both normoxic and hypoxic conditions, the colony formation viability curves indicated that HIF1A knockdown decreased cell proliferation relative to controls. EdU incorporation is a good metric for detecting cell proliferation. To investigate the effects of hypoxia and irradiation on cell proliferation, EdU incorporation assays were applied. As shown in Figure 4 C, under hypoxic conditions, HIF1A–knockdown cells had lower proportions of EdU–positive cells than WT and MOCK after IR exposure. As predicted, the absence of HIF1A significantly suppressed DNA replication, thereby enhancing cellular sensitivity to radiation, suggesting that HIF1A plays an essential role in promoting radioresistance. Since γ–H2AX is a crucial marker of DNA damage, γ–H2AX foci formation assays were performed to confirm our hypothesis further. As shown in Figure 4 D, after irradiation, the number of γ–H2AX foci was significantly increased in HIF1A–KD cells compared with controls. In addition, the upward tendency was more pronounced under hypoxic than normoxic settings, indicating that HIF1A silencing, especially under hypoxia, hindered double–strand break (DSB) repair. As expected, HIF1A knockdown significantly enhanced cellular sensitivity to radiation. Moreover, the radiosensitizing impact of HIF1A knockdown under hypoxic conditions was accentuated. These data strongly indicate that HIF1A promotes proliferation and radioresistance in vitro, especially under hypoxia.
Table 3.
The survival fraction of U87 and U251 cells after X–ray irradiation whether HIF1A was knocked down under normoxia and hypoxia for 16 h.
| SFa (Mean±SEMb) | 2Gy | 4Gy | 6Gy | 8Gy | |
|---|---|---|---|---|---|
| U87 | Normoxia, KDc | 0.28 ± 0.03 | 0.09 ± 0.02 | 0.03 ± 0.01 | 0.01 ± 0.00 |
| Hypoxia, KD | 0.49 ± 0.04 | 0.30 ± 0.04 | 0.15 ± 0.02 | 0.09 ± 0.01 | |
| Normoxia, MOCK | 0.33 ± 0.02 | 0.12 ± 0.01 | 0.02 ± 0.00 | 0.01 ± 0.00 | |
| Hypoxia, MOCK | 0.67 ± 0.04 | 0.48 ± 0.05 | 0.23 ± 0.01 | 0.09 ± 0.01 | |
| U251 | Normoxia, KD | 0.52 ± 0.05 | 0.21 ± 0.01 | 0.07 ± 0.00 | 0.02 ± 0.00 |
| Hypoxia, KD | 0.60 ± 0.00 | 0.27 ± 0.01 | 0.11 ± 0.01 | 0.04 ± 0.01 | |
| Normoxia, MOCK | 0.57 ± 0.03 | 0.22 ± 0.00 | 0.08 ± 0.01 | 0.02 ± 0.00 | |
| Hypoxia, MOCK | 0.80 ± 0.04 | 0.36 ± 0.00 | 0.18 ± 0.02 | 0.06 ± 0.01 |
a: survival fraction; b: standard error of the mean; c: knockdown
To investigate whether HIF1A promotes radioresistance in vivo, we constructed the orthotopic glioblastoma xenograft model in nude mice. After a single dose of 10 Gy brain irradiation, the brain tumor of mice injected with HIF1A–deficient cells (KD group) was smaller than those injected with control shRNA–transfected cells (MOCK) or wild–type cells (WT) (Figure 4 E). The tumor growth inhibition rate of the HIF1A–KD group was dramatically higher than that of MOCK and WT.
Together, these data imply that HIF1A acts as the core molecule to induce GBM radioresistance, and HIF1A silencing enhances the radiosensitivity of GBM in vitro and in vivo.
3.5. Beclin–1 suppression by 3–MA reverses radioresistance induced by HIF1A under hypoxia
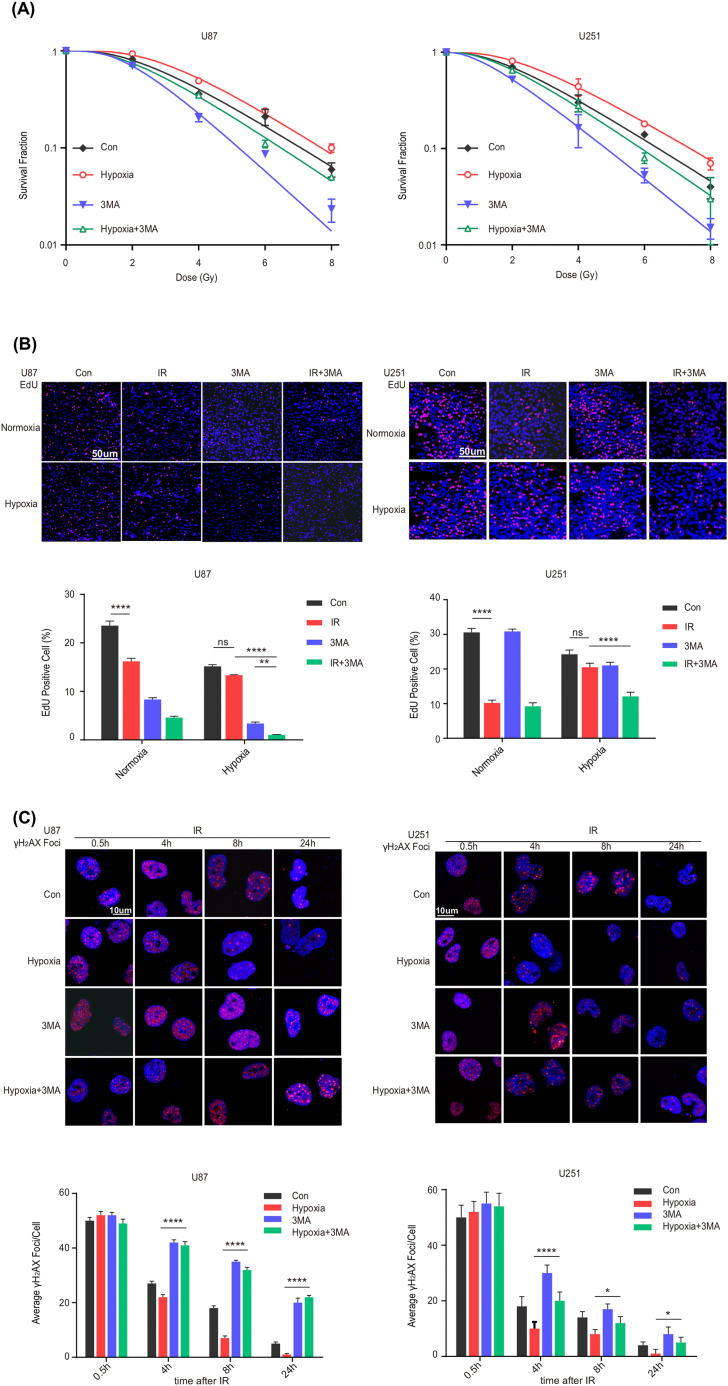
From above, HIF1A resulted in radioresistance of GBM under hypoxia conditions. And according to our previous study, Beclin–1 shared similar effects on radioresistance. Given that hypoxia exposure induces autophagy via HIF1A/Beclin–1 axis, whether downregulating Beclin–1 would reverse the radioresistance generated by HIF1A? Therefore, the following experiments were performed to determine whether Beclin–1 affects the radiosensitivity of GBM cells. Firstly, we suppressed Beclin–1 protein expression in U87 and U251 with 3–MA, a PI3K inhibitor proven to prevent cellular autophagy. As shown in Figure S 5, Beclin–1 was successfully blocked by 3–MA at the transcriptional protein level. Therefore, we investigated the radiosensitizing effects of Beclin–1 suppression on GBM cells. From the data, we found that under hypoxic conditions, Beclin–1 suppression by 3–MA decreased the survival rate of GBM cells (Figure 5 A and Table 4), reduced the proportion of EdU–positive cells (Figure 5 B), and increased the number of γ–H2AX foci (Figure 5 C) in response to IR relative to controls. As suggested, the downregulation of Beclin–1 significantly reduced the colony formation capability, suppressed DNA replication, and increased DSBs while slowing DNA repair. Collectively, these findings indicated that 3–MA reduced the expression of Beclin–1 and enhanced cellular sensitivity to radiation, hence reversing the radioresistance induced by HIF1A under hypoxia; and revealing its critical function in improving radioresistance. In addition, in the normoxia condition with low HIF1A expression, the survival rate was further reduced, and the radiosensitivity can be maximized by blocking Beclin–1, which demonstrated an optimal radiosensitivity when both HIF1A and Beclin–1 were downregulated.
Figure 5.
Beclin–1 suppression by 3–MA reverses radioresistance induced by HIF1A under hypoxia. (A) The survival rate was reduced and radiosensitivity was enhanced when Beclin–1 was suppressed by 3–MA, especially under hypoxic conditions. Colony formation assays were performed in U87 and U251 under normoxia or hypoxia for 16 h. Con: control. (B) The proportion of EdU–stained cells was noticeably decreased when Beclin–1 was suppressed by 3–MA, especially under hypoxic conditions. Upper panel: EdU incorporation was visualized using fluorescence microscopy after 24 h of X–ray irradiation (2Gy/1F). Lower panel: quantification of EdU–positive cells. **P < 0.01, ****P < 0.0001. (C) The number of γ–H2AX foci was significantly increased when Beclin–1 was suppressed by 3–MA, especially under hypoxic conditions. Upper panel: γ–H2AX foci were visualized using confocal laser scanning fluorescence microscopy after 0.5, 4, 8, and 24 h of X–ray irradiation (2Gy/1F). Lower panel: quantification of γ–H2AX foci. * P < 0.05, ****P < 0.0001.
Table 4.
The survival fraction of U87 and U251 cells after X–ray irradiation whether 3–MAa was applied under normoxia and hypoxia for 16 h.
| SFb (Mean±SEMc) | 2Gy | 4Gy | 6Gy | 8Gy | |
|---|---|---|---|---|---|
| U87 | Cond | 0.82 ± 0.04 | 0.36 ± 0.02 | 0.21 ± 0.04 | 0.06 ± 0.01 |
| Hypoxia | 0.93 ± 0.04 | 0.49 ± 0.03 | 0.24 ± 0.01 | 0.10 ± 0.01 | |
| 3–MA | 0.70 ± 0.01 | 0.21 ± 0.02 | 0.09 ± 0.01 | 0.02 ± 0.01 | |
| 3–MA+Hypoxia | 0.74 ± 0.02 | 0.35 ± 0.02 | 0.11 ± 0.01 | 0.05 ± 0.00 | |
| U251 | Con | 0.70 ± 0.03 | 0.30 ± 0.06 | 0.14 ± 0.01 | 0.04 ± 0.01 |
| Hypoxia | 0.81 ± 0.01 | 0.44 ± 0.09 | 0.18 ± 0.01 | 0.07 ± 0.01 | |
| 3–MA | 0.52 ± 0.03 | 0.16 ± 0.06 | 0.05 ± 0.01 | 0.02 ± 0.00 | |
| 3–MA+ Hypoxia | 0.65 ± 0.01 | 0.28 ± 0.04 | 0.08 ± 0.01 | 0.03 ± 0.02 |
3-Methyladenine;
survival fraction;
standard error of the mean;
control;
4. Discussion
Glioblastoma multiforme is one of the most malignant CNS tumors [1,2], and postsurgical radiotherapy is the therapeutic strategy recommended by NCCN Guidelines [3]. However, radioresistance is the major factor of treatment failure [6–9]. Hypoxia is a characteristic of solid tumors, especially glioblastoma. HIF1A is a crucial transcription factor induced by hypoxia and implicated in tumor radiosensitivity [19,20]. Adrian L showed increased expression of HIF1A after irradiation in the hypoxia cells, accompanied by distinct radioresistance mediated by enhanced DSB repair [33,34]. Moreover, it has been reported that the radiation–induced DNA damage was diminished under hypoxia, for the absence of reactive oxygen species (ROS) and free radicals (FRs) [13,35]. The potential mechanisms of HIF1A–mediated radioresistance consist of angiogenesis, DSB repair, cell cycle blockade, apoptosis suppression, and autophagy activation [[36], [37], [38], [39], [40], [41], [42], [43], [44], [45]]. All these results verified that HIF1A induced radioresistance under hypoxia, confirming our findings. The role of autophagy in radioresistance remains a long–standing concern. The autophagy inhibitor 3–MA has been observed to downregulate cellular autophagy and increase radiosensitivity in MDA–231 breast cancer cells [46] and TE–1 esophageal cancer cells [47]. Autophagy was also found to shield GBM cells from irradiation damage by inducing ATP accumulation, inhibiting Beclin–1 and exacerbating DSB injury [48]. Consistent with our previous study, Beclin–1 depletion significantly downregulated autophagy, promoted DNA–PK function, and improved radiosensitivity in C6 rat glioma cells [26]. Some researchers have claimed that HIF1A may induce radioresistance via autophagy [27–30]. In Wang’s study on nasopharyngeal carcinoma, a positive relationship was found between the Beclin–1 and HIF1A expression [31]. Moreover, it reported that HIF1A–associated Beclin–1 high expression might facilitate NPC cells surviving from chemoradiotherapy, leading to a poor prognosis. On the contrary, for patients with high HIF1A expression but lower Beclin–1, it displayed a favorable advantage for overall survival [31]. Therefore, we hypothesized that autophagy might be the link between hypoxia and radioresistance, with HIF1A and Beclin–1 serving as crucial factors.
Given the context above, we hypothesized that hypoxia might induce autophagy through activation of Beclin–1 by HIF1A, which in turn affects the radiosensitivity of GBM; downregulating HIF1A or suppressing Beclin–1 would be expected to improve radiosensitivity. First, by bioinformatics data retrieval and analysis, we confirmed that HIF1A is overexpressed in GBM patients and predicts a poor prognosis. Additionally, HIF1A strongly correlates with Beclin–1 in the transcriptional level of clinical tissue samples. After successfully validating the hypoxic microenvironment, we found that hypoxia triggers the upregulation of Beclin–1 and promotes autophagosome accumulation. To further uncover the role of HIF1A in Beclin–1 and cellular autophagy, we downregulated the expression of HIF1A using shRNAs in GBM cells. The results showed that HIF1A knockdown impaired cellular autophagy, as demonstrated by the decreased Beclin–1 expression and suppressed LC3 II/I conversion. Further investigation was performed to reveal the mechanism and elucidate the crosstalk of the core factors, HIF1A and Beclin–1. The results showed that HIF1A could interact with Beclin–1 in protein levels. Additionally, as a transcription factor, HIF1A interacts with the promoter of Beclin–1 under oxygen–depleted conditions. Moreover, the BNIP3/Bcl–2 pathway functions in signaling between two molecules, thus we conclude hypoxia triggers autophagy via HIF1A–associated–Beclin–1 by three means: a) transcriptional regulation, b) protein interaction, and c) HIF1A/BNIP3/Beclin–1 signaling pathway. However, the underlying mechanisms are yet to be investigated further.
Till now, we shift our focus to radioresistance, which is considered the primary reason for GBM relapse and poor survival. As is widely acknowledged, hypoxia induces radioresistance in many solid tumors. We sought to clarify how hypoxia induces radioresistance, the critical molecules involved, and by which means radiosensitivity could be enhanced. Through the study, we found that the radiosensitivity of GBM was improved after HIF1A was knocked down, in vitro and in vivo. Since prior findings confirmed that autophagy causes radioresistance and can be induced by hypoxia, we looked into how Beclin–1 affected radiosensitivity. We applied the standard autophagy inhibitor 3–MA and verified that it effectively inhibited cellular autophagy and reduced Beclin–1 expression, reversing the radioresistance and achieving radiosensitization under hypoxic conditions. Taken together, we conclude that hypoxia triggers autophagy through the HIF1A–associated Beclin–1, leading to radioresistance in GBM. HIF1A knockdown can improve radiosensitivity. Furthermore, silencing Beclin–1 reverses radioresistance induced by HIF1A under hypoxic conditions.
In the real world, hypoxia is a long–standing but persistent problem in solid tumors. Although HIF1A is extensively focused on attempts to improve hypoxia, some questions remain to be addressed. Since ischemia and necrosis are always accompanied by hypoxia, it is difficult for chemotherapeutic agents or small molecules to reach the core of the tumor via vascular permeability, which poses a long–standing challenge to hypoxia–improving treatment. Therefore, exploring alternative methods to reverse hypoxia–induced radioresistance is vital and merits special consideration. For this reason, we endeavored to find the intermediary molecule between hypoxia and GBM radioresistance; and attempted to reverse the hypoxia–induced radioresistance by modulating this molecule. We proposed this concept, HIF1A–associated Beclin–1, for the fact that we showed that Beclin–1 functions as a gateway molecule for hypoxia–mediated radioresistance in GBM. Even under hypoxic conditions along with strong HIF1A expression, inhibiting Beclin–1 can enhance radiosensitivity and restore the undesirable state of radioresistance. Furthermore, the radiosensitivity can be improved to the fullest extent when both HIF1A and Beclin–1 are downregulated, which is promising to improve patient prognosis and even prolong GBM patient survival. Nevertheless, the mechanisms behind HIF1A–associated Beclin–1 leave much to be discovered. As a transcription factor, HIF1A interacts with the promoter of Beclin–1 under oxygen–depleted conditions. Guillaume Robert reported Bcl–2 and Bcl–X(L) could bind and inhibit Beclin–1–induced autophagy [49]. And Nathalie’s team discovered that in hypoxia, Beclin–1 was displaced from Bcl–X(L) and Bcl–2 by the rapid expression of the atypical BH3–only proteins (BNIP3 and BNIP3L), resulting in autophagy [50]. Consistent with the idea above, the two cell lines U87 and U251 displayed identical decreasing patterns of BNIP3 in hypoxia. But they showed different trends in Bcl–2, demonstrating a distinction. Therefore, we infer that HIF–1A affects the Beclin–1/BNIP3/Bcl–2 interaction to promote autophagy, while the definite mechanisms in this pathway need further investigation.
In conclusion, we provide an approach to breaking the persistent troubled situation of hypoxia, not only by HIF1A but also through HIF1A–associated Beclin–1, to reach the goal of radiotherapy sensitization, and we also clarified the crosstalk between HIF1A and Beclin–1, which is the significance of this article. Given that HIF1A–associated Beclin–1 has significant explicit effects on radiosensitivity, it is expected to be a potential biomarker for the radiotherapy response; nevertheless, further studies and clinical evidence are needed.
Author Contributions
CL and GW designed the study and supervised the experiments; JW, KZ, ZY, YZ, and ZX performed the experiments and analyzed the data; YZ and JR provided advice; JW wrote the manuscript; and CL, GW, and JR revised the manuscript. All authors approved the final manuscript and agreed to be accountable for the content of the work.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81602189]; and CSCO– Genecast Cancer Precision Therapy Research Fund [grant number Y–2019Genecast–039].
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express thanks to co–workers in the lab of the Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, and the animals involved in this study.
Footnotes
Supplementary data related to this article can be found at .https://doi.org/10.1016/j.heliyon.2023.e12820.
Contributor Information
Gang Wu, Email: gangwu_uhcc@163.com.
Cuiwei Liu, Email: cuiweiliu19870620@163.com.
Abbreviations
- 3–MA
3–Methyladenine
- ATG
Autophagy Related Gene
- Bcl–2
B–Cell Lymphoma–2
- BECN1
Beclin–1
- BNIP3
Bcl–2 Nineteen–kilodalton Interacting Protein 3
- BSA
Bovine Serum Albumin
- CCLE
Cancer Cell Line Encyclopedia
- ChIP
Chromatin Immunoprecipitation
- DAPI
4’,6–diamidino–2–phenylindole
- DNA
Deoxyribonucleic Acid
- DNA–PK
DNA–Dependent Protein Kinase
- DSB
Double–Strand Break
- EdU
5–Ethynyl–2–Deoxyuridine
- FR
Free Radical
- GBM
Glioblastoma Multiforme
- GFP
Green Fluorescent Protein
- HIF1A
Hypoxia–Inducible Factor, alpha Subunit
- HPA
Human Protein Atlas
- KD
Knock Down
- LC3
Light Chain–3
- NX
Normalized Expression
- PAGE
Polyacrylamide Gel Electrophoresis
- PCR
Polymerase Chain Reaction
- PVDF
Polyvinylidene Fluoride
- RNAi
RNA Interference
- ROS
Reactive Oxygen Species
- qRT–PCR
Quantitative Real–time PCR
- SDS
Sodium Dodecyl Sulphate
- shRNA
Small Hairpin RNA
- TCGA
The Cancer Genome Atlas
- TPM
Transcripts Per Million
- WT
Wildtype
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Bleeker F.E., Molenaar R.J., Leenstra S. Recent advances in the molecular understanding of glioblastoma. J. Neurooncol. 2012;108(1):11–27. doi: 10.1007/s11060–011–0793–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young R.M., Jamshidi A., Davis G., Sherman J.H. Current trends in the surgical management and treatment of adult glioblastoma. Ann. Transl. Med. 2015;3(9):121. doi: 10.3978/j.issn.2305–5839.2015.05.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nabors L.B., Portnow J., Ahluwalia M., Baehring J., Brem H., Brem S., et al. Central nervous system cancers, version 3.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2020;18(11):1537–1570. doi: 10.6004/jnccn.2020.0052. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Perry J.R., Laperriere N., O’Callaghan C.J., Brandes A.A., Menten J., Phillips C., et al. Short–course radiation plus Temozolomide in elderly patients with glioblastoma. N. Engl. J. Med. 2017;376(11):1027–1037. doi: 10.1056/NEJMoa1611977. [DOI] [PubMed] [Google Scholar]
- 6.Struve N., Riedel M., Schulte A., Rieckmann T., Grob T.J., Gal A., et al. EGFRvIII does not affect radiosensitivity with or without gefitinib treatment in glioblastoma cells. Oncotarget. 2015;6(32) doi: 10.18632/oncotarget.5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn G.P., Rinne M.L., Wykosky J., Genovese G., Quayle S.N., Dunn I.F., et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 9.Giese A., Bjerkvig R., Berens M.E., Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003;21(8):1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 10.Ponnala S., Veeravalli K.K., Chetty C., Dinh D.H., Rao J.S. Regulation of DNA repair mechanism in human glioma xenograft cells both in vitro and in vivo in nude mice. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golding S.E., Rosenberg E., Adams B.R., Wignarajah S., Beckta J.M., O’Connor M.J., et al. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle. 2012;11(6):1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski D.H., Beckett M.A., Jaskowiak N.T., Calvin D.P., Mauceri H.J., Salloum R.M., et al. Blockade of the vascular endothelial growth factor stress response increases the antitumor effects of ionizing radiation. Cancer Res. 1999;59(14):3374–3378. [PubMed] [Google Scholar]
- 13.Moeller B.J., Cao Y., Li C.Y., Dewhirst M.W. Radiation activates HIF–1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5(5):429–441. doi: 10.1016/s1535–6108(04)00115–1. [DOI] [PubMed] [Google Scholar]
- 14.Bajetto A., Barbieri F., Dorcaratto A., Barbero S., Daga A., Porcile C., et al. Expression of CXC chemokine receptors 1–5 and their ligands in human glioma tissues: role of CXCR4 and SDF1 in glioma cell proliferation and migration. Neurochem. Int. 2006;49(5):423–432. doi: 10.1016/j.neuint.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee B., McEllin B., Camacho C.V., Tomimatsu N., Sirasanagandala S., Nannepaga S., et al. EGFRvIII and DNA double–strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69(10):4252–4259. doi: 10.1158/0008–5472.CAN–08–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao G.D., Jiang Z., Fernandes A.M., Gupta A.K., Maity A. Inhibition of phosphatidylinositol–3–OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J. Biol. Chem. 2007;282(29):21206–21212. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Cao P. Clinical and prognostic significance of HIF–1alpha in glioma patients: a meta–analysis. Int. J. Clin. Exp. Med. 2015;8(12):22073–22083. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang E., Zhang C., Polavaram N., Liu F., Wu G., Schroeder M.A., et al. The role of factor inhibiting HIF (FIH–1) in inhibiting HIF–1 transcriptional activity in glioblastoma multiforme. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0086102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hockel M., Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 2001;93(4):266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 20.Jensen R.L. Brain tumor hypoxia: tumorigenesis, angiogenesis, imaging, pseudoprogression, and as a therapeutic target. J. Neuro–Oncol, 2009;92(3):317–335. doi: 10.1007/s11060-009-9827-2. [DOI] [PubMed] [Google Scholar]
- 21.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroemer G., Levine B. Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008;9(12):1004–1010. doi: 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirtoli L., Cevenini G., Tini P., Vannini M., Oliveri G., Marsili S., et al. The prognostic role of Beclin1 protein expression in high–grade gliomas. Autophagy. 2009;5(7):930–936. doi: 10.4161/auto.5.7.9227. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi H., Kondo Y., Fujiwara K., Kanzawa T., Aoki H., Mills G.B., et al. Synergistic augmentation of rapamycin–induced autophagy in malignant glioma cells by phosphatidylinositol 3–kinase/protein kinase B inhibitors. Cancer Res. 2005;65(8):3336–3346. doi: 10.1158/0008–5472.CAN–04–3640. [DOI] [PubMed] [Google Scholar]
- 25.Apel A., Herr I., Schwarz H., Rodemann H.P., Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–1494. doi: 10.1158/0008–5472.CAN–07–0562. [DOI] [PubMed] [Google Scholar]
- 26.Liu C., He W., Jin M., Li H., Xu H., Liu H., et al. Blockage of autophagy in C6 glioma cells enhanced radiosensitivity possibly by attenuating DNA–PK–dependent DSB due to limited Ku nuclear translocation and DNA binding. Curr. Mol. Med. 2015;15(7):663–673. doi: 10.2174/1566524015666150831141112. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y., Xing X., Liu Q., Wang Z., Xin Y., Zhang P., et al. Hypoxia–induced autophagy reduces radiosensitivity by the HIF–1alpha/miR–210/Bcl–2 pathway in colon cancer cells. Int. J. Oncol. 2015;46(2):750–756. doi: 10.3892/ijo.2014.2745. [DOI] [PubMed] [Google Scholar]
- 28.Wu H.M., Jiang Z.F., Ding P.S., Shao L.J., Liu R.Y. Hypoxia–induced autophagy mediates cisplatin resistance in lung cancer cells. Sci. Rep. 2015;5(1) doi: 10.1038/srep12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y.M., Hu G.Y., Zhao X.Q., Lu T., Zhu F., Yu S.Y., et al. Hypoxia–induced autophagy contributes to radioresistance via c–Jun–mediated Beclin1 expression in lung cancer cells. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2014;34(5):761–767. doi: 10.1007/s11596–014–1349–2. [DOI] [PubMed] [Google Scholar]
- 30.Jeong J.K., Gurunathan S., Kang M.H., Han J.W., Das J., Choi Y.J., et al. Hypoxia–mediated autophagic flux inhibits silver nanoparticle–triggered apoptosis in human lung cancer cells. Sci. Rep. 2016;6(1) doi: 10.1038/srep21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan X.-B., Fan X.-J., Chen M.-Y., Xiang J., Huang P.-Y., Guo L., et al. Elevated Beclin1 expression is correlated with HIF–1α in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy. 2010;6(3):395–404. doi: 10.4161/auto.6.3.11303. [DOI] [PubMed] [Google Scholar]
- 32.Paxinos G., Franklin K.B. 2019. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates: Academic Press. [Google Scholar]
- 33.Traub F., Schleicher S., Kirschniak A., Zieker D., Kupka S., Weinmann M., et al. Gene expression analysis in chronic postradiation proctopathy. Int. J. Colorectal. Dis. 2012;27(7):879–884. doi: 10.1007/s00384–011–1387–1. [DOI] [PubMed] [Google Scholar]
- 34.Adrian L., Gorisch H. Microbial transformation of chlorinated benzenes under anaerobic conditions. Res. Microbiol. 2002;153(3):131–137. doi: 10.1016/s0923–2508(02)01298–6. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y., Hilliard G., Ferguson T., Millhorn D.E. Cobalt inhibits the interaction between hypoxia–inducible factor–α and von Hippel–Lindau protein by direct binding to hypoxia–inducible factor–α. J. Biol. Chemistry. 2003;278(18):15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- 36.Unruh A., Ressel A., Mohamed H.G., Johnson R.S., Nadrowitz R., Richter E., et al. The hypoxia–inducible factor–1α is a negative factor for tumor therapy. Oncogene. 2003;22(21):3213–3220. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]
- 37.Nishimoto A., Kugimiya N., Hosoyama T., Enoki T., Li T.S., Hamano K. HIF–1alpha activation under glucose deprivation plays a central role in the acquisition of anti–apoptosis in human colon cancer cells. Int. J. Oncol. 2014;44(6):2077–2084. doi: 10.3892/ijo.2014.2367. [DOI] [PubMed] [Google Scholar]
- 38.Ning S., Laird D., Cherrington J.M., Knox S.J. The antiangiogenic agents SU5416 and SU6668 increase the antitumor effects of fractionated irradiation. Radiat. Res. 2002;157(1):45–51. doi: 10.1667/0033–7587. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths J.R., McSheehy P.M., Robinson S.P., Troy H., Chung Y.L., Leek R.D., et al. Metabolic changes detected by in vivo magnetic resonance studies of HEPA–1 wild–type tumors and tumors deficient in hypoxia–inducible factor–1beta (HIF–1beta): evidence of an anabolic role for the HIF–1 pathway. Cancer Res. 2002;62(3):688–695. [PubMed] [Google Scholar]
- 40.Takasaki C., Kobayashi M., Ishibashi H., Akashi T., Okubo K. Expression of hypoxia–inducible factor–1alpha affects tumor proliferation and antiapoptosis in surgically resected lung cancer. Mol. Clin. Oncol. 2016;5(2):295–300. doi: 10.3892/mco.2016.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee M.-J., Kim J.-Y., Suk K., Park J.-H. Identification of the hypoxia–inducible factor 1α–responsive HGTD–P gene as a mediator in the mitochondrial apoptotic pathway. Mol. Cell Biol. 2004;24(9):3918–3927. doi: 10.1128/MCB.24.9.3918-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erler J.T., Cawthorne C.J., Williams K.J., Koritzinsky M., Wouters B.G., Wilson C., et al. Hypoxia–mediated down–regulation of Bid and Bax in tumors occurs via hypoxia–inducible factor 1–dependent and –independent mechanisms and contributes to drug resistance. Mol. Cell Biol. 2004;24(7):2875–2889. doi: 10.1128/MCB.24.7.2875–2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruick R.K. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Nat. Aca. Sci. 2000;97(16):9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki H., Tomida A., Tsuruo T. Dephosphorylated hypoxia–inducible factor 1α as a mediator of p53–dependent apoptosis during hypoxia. Oncogene. 2001;20(41):5779–5788. doi: 10.1038/sj.onc.1204742. [DOI] [PubMed] [Google Scholar]
- 45.Sohda M., Ishikawa H., Masuda N., Kato H., Miyazaki T., Nakajima M., et al. Pretreatment evaluation of combined HIF‐1α, p53 and p21 expression is a useful and sensitive indicator of response to radiation and chemotherapy in esophageal cancer. Int. J. Cancer. 2004;110(6):838–844. doi: 10.1002/ijc.20215. [DOI] [PubMed] [Google Scholar]
- 46.Chaachouay H., Ohneseit P., Toulany M., Kehlbach R., Multhoff G., Rodemann H.P. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol. 2011;99(3):287–292. doi: 10.1016/j.radonc.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y.S., Song H.X., Lu Y., Li X., Chen T., Zhang Y., et al. Autophagy inhibition contributes to radiation sensitization of esophageal squamous carcinoma cells. Dis. Esophagus. 2011;24(6):437–443. doi: 10.1111/j.1442–2050.2010.01156.x. [DOI] [PubMed] [Google Scholar]
- 48.Katayama M., Kawaguchi T., Berger M.S., Pieper R.O. DNA damaging agent–induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14(3):548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 49.Robert G., Gastaldi C., Puissant A., Hamouda A., Jacquel A., Dufies M., et al. The anti–apoptotic Bcl–B protein inhibits BECN1–dependent autophagic cell death. Autophagy. 2012;8(4):637–649. doi: 10.4161/auto.19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellot G., Garcia–Medina R., Gounon P., Chiche J., Roux D., Pouyssegur J., et al. Hypoxia–induced autophagy is mediated through hypoxia–inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.