Abstract
Neurodegenerative diseases are characterized by the progressive decline of neuronal function in several brain areas, and are always associated with cognitive, psychiatric, or motor deficits due to the atrophy of certain neuronal populations.
Most neurodegenerative diseases share common pathological mechanisms, such as neurotoxic protein misfolding, oxidative stress, and impairment of autophagy machinery.
Amyotrophic lateral sclerosis (ALS) is one of the most common adult-onset motor neuron disorders worldwide. It is clinically characterized by the selective and progressive loss of motor neurons in the motor cortex, brain stem, and spinal cord, ultimately leading to muscle atrophy and rapidly progressive paralysis.
Multiple recent studies have indicated that the amyloid precursor protein (APP) and its proteolytic fragments are not only drivers of Alzheimer’s disease (AD) but also one of the earliest signatures in ALS, preceding or anticipating neuromuscular junction instability and denervation. Indeed, altered levels of APP peptides have been found in the brain, muscles, skin, and cerebrospinal fluid of ALS patients.
In this short review, we discuss the nature and extent of research evidence on the role of APP peptides in ALS, focusing on the intracellular C-terminal peptide and its regulatory motif 682YENPTY687, with the overall aim of providing new frameworks and perspectives for intervention and identifying key questions for future investigations.
Keywords: Amyloid precursor protein, 682YENPTY687 motif, Amyotrophic lateral sclerosis, Neurodegeneration
1. Introduction
Over the past few years, common pathways involved in neurodegenerative diseases have been highlighted [1]. Indeed, neurodegenerative disorders, such as Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS), show various degrees of overlapping pathology, not only in clinical appearance but also at the single-protein level or in an entire signalling cascade.
One case is that of the amyloid precursor protein (APP), a protein primarily at the center of AD research. An increasing number of studies have proposed APP as an active contributor to certain forms of ALS [2]. In line with this concept, APP is expressed at the neuromuscular junction (NMJ) [3] and is required for the normal development and function of the NMJ [4], [5] suggesting that alterations in the signalling or processing of APP might influence NMJ function and are likely to predispose patients to motor neuron diseases (MND), such as ALS. Accordingly, alterations in the APP pathway have been proposed to represent an ALS signature preceding or anticipating the pathology [1], [6].
ALS and AD are age-associated sporadic disorders with no precisely identified genetic causes but with a large number of susceptibility genes in which selective and progressive dysfunctions of specific neuronal populations occur [1], [7]. Although apparently unrelated, as AD is primarily a central nervous system disease and ALS targets the peripheral nervous system, approximately 30 % of ALS patients show neuritic plaques and neurofibrillary tangles, especially in the amygdala, hippocampus, and entorhinal and insular cortices [6], [8].
In addition, both AD and ALS show accumulation and deposition of a specific misfolded protein, APP in AD and TDP-43 in ALS, conferring vulnerability to specific neuronal populations [9] affecting mitochondrial and autophagy functions [10], [11] and triggering neurotoxic mechanisms [12].
In this short review, we provide evidence for the role of APP peptides in ALS, and underline new frameworks and perspectives for future research.
Findings regarding the pathophysiology of AD and ALS and their similarities are beyond the scope of this review, as many outstanding reports have extensively discussed this area of research [13], [14].
In particular, because APP contains multiple structural and functional domains, we focused our review mainly on the properties of APP intracellular domains and its regulatory motif 682YENPTY687.
2. Lights and shadows of APP in ALS
APP is expressed in both neuronal and non-neuronal cells and is largely distributed in extra-neuronal tissues [15]. APP is present at synaptic sites in both the central and peripheral nervous systems, including the NMJ, and plays an essential role in the development of neuromuscular synapses [3], [4].
APP is post-transcriptionally processed into three major isoforms with differential cellular and tissue expression patterns. The three main isoforms of APP described to date are APP695, APP751 and APP770, depending on the number of amino acids, and are produced through alternative splicing of exons 7 and 8, which encode the Kunitz protease inhibitor and OX-2 domains, respectively [16]. APP695 lacks both domains, whereas the APP751 isoform containes only KPI domain in the extracellular sequence. APP770 in addition to KPI domain, contains an OX-2 domain [17]. APP751 and APP770 are ubiquitously expressed, whereas APP695 is predominantly expressed in neurons [18], [19].
APP belongs to an evolutionarily conserved type I transmembrane glycoprotein family that includes two paralogues, amyloid precursor-like proteins 1 and 2 (APLP1 and APLP2), with similar structures and membrane topologies [20]. Notably, previous studies using knockout mice have emphasized the high functional redundancy of APP, APLP1, and APLP2 [21]. These proteins contain several conserved motifs that are shared between all vertebrates, including E1 and E2 domains in the extracellular region and a short intracellular C-terminal domain (AICD) that contains the highest conserved consensus motif, Y682ENPTY687 [22]. The latter is thought to be crucial for AICD binding to adaptor proteins, and for APP trafficking and localization in cells [23]. Notably, while Aβ originates solely from APP, AICD originates from APP, APLP1, and APLP2 [20].
Aβ peptides, the major components of amyloid fibrils in AD, are the result of sequential cleavage of β- and γ-secretase. Briefly, β-secretase cleavage generates a soluble APP peptide (sAPPβ) secreted in extracellular compartments and an intracellular C-terminal fragment (CTF99). CTF99 is then cleaved by γ-secretase to produce Aβ40 and Aβ42 peptides and AICD, which appears to regulate the transcription of certain genes [24], [25]. AICD is rapidly metabolized by insulin-degrading enzyme [26], [27] and the endosomal/lysosomal system [28]. Alternatively, α-secretase cleaves APP within the Aβ sequence, thus precluding Aβ formation and generating sAPPα with neurotrophic properties and the CTF83 peptide, which is further processed by γ-secretase to yield AICD and p3 fragment [29], [30], [31], [32].
Increased β-secretase activity has been observed in animal models of ALS and nerve injury [33], [34]. Similarly, a lack of α-secretase expression associated with increased β-secretase expression and activation of the amyloid cascade of APP, leading to increases in amyloid-β and AICD peptides, has been reported in the hippocampi of ALS patients [35]. In addition, deficits in lysosomal autophagic pathways have been demonstrated to activate the γ-secretase complex and lead to Aβ42 accumulation in cultured human muscle fibers [36]. Pharmacological inhibition of β-secretase enhances peripheral functional recovery after sciatic nerve ablation and increases axonal sprouting due to partial nerve injury [37]. Treatment with a monoclonal antibody (MAb) that blocks β-secretase cleavage prevents an increase in APP expression, phosphorylation, processing, and inflammatory processes [33], [34].
β-Secretase cleavage to generate Aβ peptides and AICD occurs preferentially in the APP695 isoform, although increased expression of APP751 and APP770 has been detected in the brains of patients with AD and is associated with increased Aβ deposition [38], [39]. Interestingly, prolonged activation of extrasynaptic NMDA receptors, which has been associated to neurodegenerative diseases [40], [41], shifts APP splicing from APP695 to KPI-containing APP isoforms in neurons and triggers APP processing to produce Aβ [40]. This might imply that dysregulated splicing of APP mRNA occurs in pathological conditions and might allow discrimination of different pathologies in which APP has been demonstrated to be involved, including PD and ALS. Indeed, most reports focusing on the role of the APP gene in ALS face difficulties in discriminating between the three isoforms and refer to APP generically [22]. In this regard, a recent study reported the development of a new PCR procedure that can accurately measure and quantify the transcript copy numbers of all three major isoforms, APP695, APP751, and APP770 [42].
It is noteworthy that specific adaptors might bind APP695, APP751, and APP770 because of the differences in their APP sequences, APP/KPI versus APP695, thus affecting APP endocytosis, trafficking, and metabolism in neuronal cells. Accordingly, sequence differences between APP695, APP751, and APP770 may regulate the transport of APP695 along a distinct processing route, leading to β-secretase cleavage, whereas APP/KPI isoforms are excluded from this pathway or located in a distinct subcellular compartment. In this context, the identification of these different adaptor proteins may be useful for designing innovative strategies for the differential diagnosis of neurodegenerative diseases associated with altered APP levels.
Notably, only AICD generated by β- and γ-secretase cleavage translocates to the nucleus, where several potential target genes have been identified Table 1 [25]. Although γ-secretase cuts AICD in several subcellular locations, AICD generated by α-secretase cleavage at the plasma membrane has a lower likelihood of reaching the nucleus because of its short half-life and longer distance from the cell surface [43]. In contrast, AICD produced in the endosomes by β- and γ-secretase cleavage can reach the nuclear vicinity before γ-cleavage releases AICD owing to dynein- and microtubule-mediated transport systems [44].
Table 1.
Putative genes regulated by AICD.
| Protein | References |
|---|---|
| α2-actin and transgelin | [44] |
| Neprilysin (Aβ degradation factor) | [45] |
| GSK3 β (glycogen synthase kinase) | [46], [47], [48], [49], [50] |
| p53 (Tumor suppressor) | [51], [52] |
| AchE1, AchE2 (Acetyl choline esterase) | [53], [54] |
| Thymidylate synthase | [45], [55] |
| (HES) Hairy and Enhancer of split (differentiation factor) | [56] |
| LRP1 (Lipoprotein Receptor) | [57] |
| EGFR (Epidermal growth factor receptor) | [58] |
Source: Adapted from Muller et al. [44].
Interestingly, less AICD is produced in amyloidogenic APP processing than in non-amyloidogenic processing, raising the question of whether a reduction in AICD levels results in the loss of physiological functions or the gain of new functions.
Some of these genes, such as those encoding the Aβ-degrading enzyme, neprilysin (NEP), are implicated in APP metabolism. Although the direct involvement of NEP in ALS has not yet been defined, it is known that NEP not only participates in the regulation of various brain functions but also in movement regulation [59], [60]. Loss of NEP expression results in altered locomotor activity [61].
Other putative AICD target genes are α2-actin and transgelin, which are involved in the regulation of actin cytoskeleton dynamics [44]. Notably, many mutations in ALS-related genes that affect cytoskeletal integrity and dynamics have been identified [62]. For instance, mutations in proteins that regulate actin polymerization, including superoxide dismutase (SOD1), TDP-43, FUS, and Profilin1 (PFN1), have been identified in patients with ALS, causing an increased tendency to aggregate and leading to the formation of cytoplasmic inclusions [63]. Notably, mutations in PFN1 (C71G, M114T, G118V, A20T, T109M, Q139L, and E117G) [64], [65] and other cytoskeletal-related proteins such as Tubulin A4A (TUBA4A) [66] and kinesin family member 5A (KIF5A) [67] have been identified in familial ALS patients. The ability of mutant PFN1 to associate with actin is impaired in ALS, and mutant PFN1 motoneurons exhibit morphological abnormalities characterized by smaller growth cones and shorter axons [68].
Indeed, the disruption of cytoskeletal integrity and/or motor neuron-dependent transport are key features of ALS. This highlights the necessity of potentially differentiating variants of these genes that might act as primary causes of the disease from those that might become risk factors or disease modifiers of the pathology. In addition, the possibility that altered levels of AICD in ALS might influence the expression of some of these genes and activate neurotoxic downstream pathways is an aspect not enough speculated that might deserve attention.
Glycogen synthase kinase 3β (GSK3β) promotes tau hyperphosphorylation and neurofibrillary tangle formation in AD [69]. Dysregulations of GSK3β signalling has also been recognized in ALS [70]. In this regard, increased levels of GSK3β expression and phosphorylation of the Tyr216 residue have been reported in the spinal cord, frontal and temporal cortices, and hippocampus of patients with ALS [71], [72], [73].
The tumor suppressor genes p53 and cyclin B1, and D1 or KAI1 are pro-apoptotic factors and cell cycle reentry, respectively, and are involved in neuronal death processes, also included ALS (Reviewed by Szybińska et al. [74]. Consistently, activation of p53 and an altered Bcl-x/Bax ratio were also observed in the ventral horns of the lumbar spinal cord of SOD1 transgenic mice harboring a single amino acid substitution of glycine to arginine at codon 86 (SOD1 G86R) mice [75]. p53 [76] and other apoptotic markers, such as Rb, Bax, Fas, and caspases [77] are increased in the motor cortex and spinal ventral horns of postmortem brain tissues [74], [78].
APP regulates Cu/Zn SOD1 expression and function, which is one of the major targets of oxidative damage in the brains of AD patients [79], [80] and its mutations have been linked to familial ALS [81]. In ALS neurons, Aβ acts as an early and short-lived change [82] directly interacting with superoxide dismutase 1 (SOD1), decreasing its enzymatic activity [83] and accelerating the onset of motor impairment [84]. Accordingly, increased Aβ immunoreactivity has been reported in the perikaryal region of anterior horn neurons of patients with familial and sporadic forms of ALS, and proximal axonal swelling was detected in mild lesions or in the early stage of the pathology [85] supporting the concept that ALS is a disease not confined to the motor system [86], [87], [88]. Indeed, neurodegeneration in patients with ALS also involves brain areas such as the dorsolateral prefrontal cortex, anterior cingulate, hippocampus, dentate gyrus (DG), parietal lobe, substantia nigra, cerebellum, amygdala, and basal ganglia [86], [89], [90], [91] and amyloid cascade-related biomarkers have been found in the cerebral spinal fluid of patients with ALS and frontotemporal dementia (FTD) [92], [93]. Additionally, an increase in Aβ levels has been observed in the skin and muscles of ALS patients [93], [94].
Similar results were obtained in SOD1 transgenic mice harboring a single amino acid substitution of glycine to alanine at codon 93 (SOD1-G93A), which is commonly used to model ALS, where Aβ peptide accumulation and increased APP levels have been detected in a restricted subpopulation of vulnerable muscle fibers and in the spinal cord [2]. Interestingly, genetic ablation of APP (APP−/−) in SOD1-G93A mice significantly prevents neuromuscular junction loss, reduces disease progression, and promotes motor neuron survival, further supporting the idea that APP and Aβ peptides might contribute to ALS pathology by accelerating muscle denervation [2]. The hypothesis that Aβ can also be neurotoxic in the peripheral nervous system was further supported by evidence from murine models of familial AD overexpressing Aβ, in which the susceptibility of motor neurons to Aβ peptides, progressive degeneration of skeletal muscle, and age-dependent axonal degeneration in the spinal cord have been described [95], [96], [97].
3. The 682YENPTY687-mediated APP processing regulation: possible implications in ALS
As mentioned above, APP processing can result in the production of Aβ peptides, which contribute to AD or the secretion of the sAPPα peptide as well as intracellular AICD.
The production of sAPP(α or β) and AICD metabolites largely depends on the level of Tyr682 phosphorylation of the highly conserved 682YENPTY687 motif on AICD (referred to as neuronal APP695 numbering). The 682YENPTY687 motif represents a docking site for multiple interacting proteins. 682YENPTY687 phosphorylation changes the AICD conformation, which shifts the cis/trans isoforms, resulting in loss of affinity for binding proteins. Notably in both APP695 as well as APP751 and APP771 the 682YENPTY687 motif is preserved.
For instance, Grb2, Shc, Grb7, and Crk interact with APP only when Tyr682 is phosphorylated, whereas Fe65, Fe65L1, and Fe65L2 interact with APP only when this tyrosine is not phosphorylated (reviewed by Matrone et al. [23]) Table 2.
Table 2.
Some of the most common 682YENPTY687 adaptor proteins.
| Protein | References |
|---|---|
| Fe65 (fe65l1, fe65l2) | [98], [99], [100] |
| Mint/X11 (mint1, mint2, mint3) | [101] |
| Numb | [56] |
| JIP (JIP1, JIP3) (Islet-brain1/C-Jun N-terminal kinase interacting protein-1) | [102] |
| PAT (PAT1, PAT1A) (Protein interacting with APP tail 1) | [103] |
| Pin1 (peptidyl-prolyl isomerases. PPIase) | [104] |
| FKBP12 | [105] |
| SHCA/SHCC (Src homology and collagen homology) | [106], [107] |
| GRB2 (growth factor receptor-bound protein 2) | [108] |
| Dab1, dab2 (Disabled Regulator protein) | [109], [110] |
| Crk | [111] |
| cAbl (Tyrosine kinase) | [112] |
| Fyn (Tyrosine kinase) | [113], [114], [115] |
| Clathrin, AP2 (adaptor protein 2) | [116] |
| SorLA (Sortilin-related receptor) | [117] |
In this regard, the 682YENPTY687 binding protein, Fe65 acts as an AICD stabilizer in the nuclear compartment, where it binds to histone acetylase Tip60 to form AFT complexes and prevents APP amyloidogenic processing [118]. Notably, decreased Fe65 expression has been identified in patients with ALS, in which the accumulation of APP and Aβ has also been detected, suggesting that the AICD-Fe65 complex is internalized into the nucleus, as occurs when the APP amyloidogenic signalling pathway is activated [86].
Similarly, the 682YENPTY687 binding proteins Clathrin and AP2 control APP endocytosis, as well as many other transmembrane proteins, and proper trafficking to the early endosome and back to the plasma membrane, thus preventing APP accumulation in the late endosome and lysosome, where because of the acidic environment, APP is preferentially cleaved by β secretase [119] thereby initiating amyloidogenic processing [116], [120]. Although a direct link between ALS and the Clathrin and AP2 adaptors has not yet been demonstrated, alterations in the transport of endosomes or lysosomes have been proposed to be likely causative of the pathology, as in many other neurodegenerative diseases [121]. Accordingly, several genes involved in endosomal maturation, lysosome biogenesis, and vesicle trafficking have been linked to ALS [122], suggesting that these pathways are altered in ALS. In addition, changes in the expression of proteins responsible for endocytic trafficking have been detected in ALS patients [123], [124]. Among others, SorLA, which belongs to the VPS10Ps protein family and interacts with the 682YENPTY687 motif of APP [125], decreases in the anterior horn cells (AHCs) of patients with ALS compared to controls [126]. Notably, abundant SorLA expression has been detected in neurons throughout the central nervous system, including the cortex, hippocampus, cerebellum, and spinal cord, which controls retromer-dependent sorting of APP and prevents APP amyloidogenic processing [125], [127], [128].
Referring to another 682YENPTY687 binding protein, Notch, studies have reported that Notch and APP compete for α- and γ-secretase cleavage. Interestingly, inactivation of the Notch pathway and a reduction in α-secretase expression have been described in the hippocampus of patients with motor neuron deficits. Such alterations are associated with increased β-secretase expression and the activation of the amyloidogenic cascade, leading to Aβ and AICD accumulation [35], [129], [130]. Of note Notch 1 is essential for hippocampal neurogenesis [131], [132] and the Notch receptor is expressed in neural stem cells [131]. Consistently, inactivation of the Notch pathway results in inhibition of neurogenesis, and Notch signalling is repressed in the hippocampi of patients with ALS [133]. Interestingly, some drugs that increase Notch signalling have been found to promote hippocampal neurogenesis [134]. Similarly, a rat model of AD showed that soluble Aβ42 suppresses Notch1 expression [135].
The 682YENPTY687 adaptor protein Numb is involved in stem cell maintenance and differentiation, as well as in neuritogenesis, and antagonizes Notch-1 signalling [136], [137]. Numb is reduced one week after the spinal cord lesion or after motor neuron ablation and then restored at one month [129] in animal models of ALS, in line with other evidence of decreased neurogenesis in patients with ALS [35], [133]. Nevertheless, the role of Numb, as well as the other APP adaptor protein Shh, has also been reported in the regulation of adult neurogenesis [138] and the expression of these proteins has been found to be downregulated in animal models of motor neuron degeneration [129].
Furthermore, c-Abl [49], [139] and Fyn tyrosine kinase (TK) phosphorylate the APP Tyr682 residue of APP under physiological or pathological conditions, although Fyn appears to be primarily responsible for aberrant Tr682 phosphorylation in AD neurons [115]. Interestingly, an increase in the amount of c-Abl mRNA, phosphorylated c-Abl and Fyn TK has been detected in motor neurons of ALS [140], [141], [142]. Consistently, treatment with c-Abl and Fyn inhibitors, such as dasatinib and bosutinib, or the new compound SC75741, has been shown to exert protective effects on motor neuron degeneration in G93A-SOD1 transgenic ALS mice [142] as well as iPSC-derived motor neurons from patients with ALS [141], [143], [144]. In addition, multiple studies have associated mutations in genes encoding different kinases with ALS [145], [146], suggesting that alterations in the function of specific kinases and/or their downstream targets are crucial to neuronal survival, and that protein kinase inhibitors may be a reasonable target for the design of innovative ALS treatment [147], [148].
Multiple lines of evidence indicate that regulation of APP trafficking might prevent Aβ generation. Consistently, increased sAPPα levels appeared to be associated with a reduced risk of developing AD [149], [150], [151], [152].
Interestingly, variations in sAPPα production have also been reported in conditions other than AD such as cerebrovascular and neurodegenerative diseases [153], bipolar disorder [154] and ALS [92], [93].
In particular, sAPPα is upregulated in the muscles of mouse models of familial ALS and in patients [1], [2], [94], whereas low sAPPα concentrations have been found in the CSF of patients with ALS with a rapidly progressive course of the disease [92]. However, whether the increase in sAPPα represents a cell survival response to molecular changes caused by MND [86] or a neurotoxic process to promote neuronal death is a matter of debate.
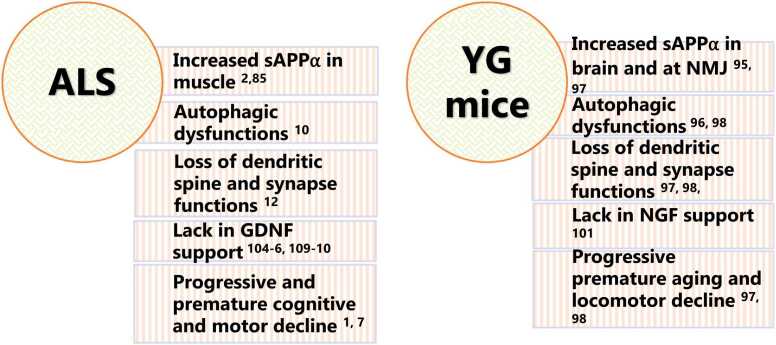
Interestingly, Barbagallo et al. previously demonstrated that sAPPα production largely depends on Tyr682 phosphorylation of the 682YENPTY687 motif of APP in neurons [155]. Accordingly, when Tyr682 is not phosphorylated, APP is largely located in the plasma membrane where it is processed by α-secretase to generate sAPPα. In contrast, when APP is phosphorylated at the Tyr682 residue, APP endocytosis and trafficking inside neurons are affected, resulting in APP accumulation in acidic neuronal compartments, such as late endosomes and lysosomes, where it is preferentially cleaved to generate sAPPβ peptides [114], [116], [125]. Consistently, APP YG knock-in mice, in which Tyr682 is not phosphorylated because it is replaced by glycine (YG), show aberrant sAPPα production in the brain and motor neurons [155], [156]. In addition, YG mice display a progressive reduction in muscular strength, motor functions and abilities, and learning performance [157]. Such deficits are associated with age-dependent cognitive decline, autophagic dysfunction, and progressive dendritic spine loss [125], mirroring some of the crucial features reported in patients [158] (Fig. 1).
Fig. 1.
Similarities between ALS SOD1-G93A and APP YG mice.
Notably, the YG background, when introduced into an APLP2 null background failed to rescue early postnatal lethality or neuromuscular synaptic defects present in APLP2 null mice, supporting the role of the Tyr682 residue and 682YENPTY687 motif in regulating NMJ neurodevelopment and function [156].
In accordance with the importance of Tyr682 phosphorylation on the 682YENPTY687 motif in controlling sAPPα release and preventing aberrant sAPPα secretion, when the APP background lacking the 682YENPTY687 domain was reintroduced into APP-knockout mice, an increased cell surface expression of sAPPα was detected [159].
Interestingly, YG hippocampal neurons fail to differentiate properly in vitro because of deficits in nerve growth factor (NGF) response [160]. In fact, the lack of Tyr682 phosphorylation prevents the association between APP and the NGF receptor TrkA, resulting in TrkA perinuclear accumulation and causing APP redistribution towards the non-amyloidogenic pathway with the accumulation of sAPPα and AICD peptides [160]. This critical role of NGF in APP trafficking, control of neuronal functions, and prevention of dysfunction largely reminds us of the crosstalk between glial cell-derived neurotrophic factor (GDNF) and APP at the neuromuscular junctions [161], [162]. GDNF controls muscle and Schwann cell functions [163]. Deficits in GDNF and APP signalling have been associated with ALS. GDNF is decreased in the serum of patients with ALS, whereas sAPPα levels are increased in the same fluid [164]. APP regulates GDNF gene expression [164], [165]. NGF promotes trophic effects and protects neurons from AD-related processes [166]; when GDNF is administered directly to muscles, it improves muscle-nerve synapse performance and promotes motor neuron activity and survival [167]. In addition, overexpression of GDNF in muscles extends the lifespan of ALS mice [168]. Whether GDNF activity and secretion levels change depending on APP Tyr682 phosphorylation is worth investigating.
4. Conclusions
Considerable knowledge gaps and clinical challenges associated with neurodegenerative diseases remain unaddressed. Perhaps the biggest challenge is to better define and understand the factors that initiate the pathology and drive cellular dysfunction in the disease. Numerous studies suggest that neurodegenerative diseases share not only clinical phenotypes but also molecular mediators (s). Although the findings discussed here portray only part of the broad literature on AD and ALS and their roles in these diseases, it is likely sufficient to delineate some of the critical questions for the next phase of studies.
Herein, we discuss a novel hypothesis that might deserve to be expanded and sustained in the future regarding the potential role of the conserved 682YENPTY687 motif located on the AICD of APP in ALS and speculate that modifications in the 682YENPTY687 peptide might represent an early signature of the disease, as previously described in AD [23], [120].
The 682YENPTY687 peptide has been consistently viewed as an active and critical player in controlling APP function and preventing the switch from the non-amyloidogenic to amyloidogenic pathway through phosphorylation of the Tyr682 residue [23], [120]. However, the idea that this peptide can also regulate APP activity in other pathologies such as ALS has never been speculated. Importantly, evidence regarding the role of 682YENPTY687 peptide in regulating the levels of sAPPα in motor neurons and influencing the correct development of NMJ has been reported previously [33], [34], [82], [164]. Indeed, Tyr682 phosphorylation of the 682YENPTY687 motif controls APP trafficking and prevents amyloidogenic APP processing to generate Aβ [157], [160]. Conversely, the lack of Tyr682 phosphorylation in YG mice causes an increase in sAPPα levels, autophagic deficits, locomotor deficiency, and cognitive deficits, all of which have been observed in ALS patients [155], [157]. Consistently, an aberrant increase in sAPPα levels has been detected in the dysfunctional NMJ of patients with ALS [1], [2], [94].
These findings raise the question of whether a possible malfunction of the 682YENPTY687 pathway might influence Tyr682 phosphorylation and predispose APP to aberrant production of sAPPα in patients with ALS.
Based on these perspectives, this short review provides new and important directions for the investigation of ALS.
Author statement
I declare that this manuscript is original and has not been published before and is not currently, being considered for publication elsewhere., I confirm that the manuscript has only one author and that there are no other persons who, satisfied the criteria for authorship and are not listed., I will be the responsible for communicating with the editor about progress, submissions of revisions and final approval of proofs.
Conflict of Interest
The authors declare that they have no affiliations with or involvement in any organization or entity with any financial interest in the subject matter or materials discussed in this manuscript.
Acknowledgments
The study has been supported by Ministero dell'Istruzione, dell'Università e della Ricerca, Italy (MIUR/PRIN #2017T9JNLT to CM).
References
- 1.Vrillon A., Deramecourt V., Pasquier F., Magnin É., Wallon D., Lozeron P., et al. Association of Amyotrophic Lateral Sclerosis and Alzheimer's Disease: New Entity or Coincidence? A Case Series. J Alzheimers Dis. 2021;84(4):1439–1446. doi: 10.3233/JAD-215226. [DOI] [PubMed] [Google Scholar]
- 2.Bryson J.B., Hobbs C., Parsons M.J., Bosch K.D., Pandraud A., Walsh F.S., et al. Amyloid precursor protein (APP) contributes to pathology in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(17):3871–3882. doi: 10.1093/hmg/dds215. [DOI] [PubMed] [Google Scholar]
- 3.Akaaboune M., Allinquant B., Farza H., Roy K., Magoul R., Fiszman M., et al. Developmental regulation of amyloid precursor protein at the neuromuscular junction in mouse skeletal muscle. Mol Cell Neurosci. 2000;15(4):355–367. doi: 10.1006/mcne.2000.0834. [DOI] [PubMed] [Google Scholar]
- 4.Wang P., Yang G., Mosier D.R., Chang P., Zaidi T., Gong Y.D., et al. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J Neurosci. 2005;25(5):1219–1225. doi: 10.1523/JNEUROSCI.4660-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torroja L., Packard M., Gorczyca M., White K., Budnik V. The Drosophila beta-amyloid precursor protein homolog promotes synapse differentiation at the neuromuscular junction. J Neurosci. 1999;19(18):7793–7803. doi: 10.1523/JNEUROSCI.19-18-07793.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton R.L., Bowser R. Alzheimer disease pathology in amyotrophic lateral sclerosis. Acta Neuropathol. 2004;107(6):515–522. doi: 10.1007/s00401-004-0843-1. [DOI] [PubMed] [Google Scholar]
- 7.Ying W. A new hypothesis of neurodegenerative diseases: the deleterious network hypothesis. Med Hypotheses. 1996;47(4):307–313. doi: 10.1016/s0306-9877(96)90071-7. [DOI] [PubMed] [Google Scholar]
- 8.Coan G., Mitchell C.S. An Assessment of Possible Neuropathology and Clinical Relationships in 46 Sporadic Amyotrophic Lateral Sclerosis Patient Autopsies. Neurodegener Dis. 2015;15(5):301–312. doi: 10.1159/000433581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng C., Trojanowski J.Q., Lee V.M. Protein transmission in neurodegenerative disease. Nat Rev Neurol. 2020;16(4):199–212. doi: 10.1038/s41582-020-0333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicencio E., Beltrán S., Labrador L., Manque P., Nassif M., Woehlbier U. Implications of Selective Autophagy Dysfunction for ALS Pathology. Cells. 2020;9(2) doi: 10.3390/cells9020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briston T., Hicks A.R. Mitochondrial dysfunction and neurodegenerative proteinopathies: mechanisms and prospects for therapeutic intervention. Biochem Soc Trans. 2018;46(4):829–842. doi: 10.1042/BST20180025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyer M.S., Woodhouse A., Blizzard C.A. Cytoplasmic Human TDP-43 Mislocalization Induces Widespread Dendritic Spine Loss in Mouse Upper Motor Neurons. Brain Sci. 2021;11(7) doi: 10.3390/brainsci11070883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muresan V., Ladescu Muresan Z. Shared Molecular Mechanisms in Alzheimer's Disease and Amyotrophic Lateral Sclerosis: Neurofilament-Dependent Transport of sAPP, FUS, TDP-43 and SOD1, with Endoplasmic Reticulum-Like Tubules. Neurodegener Dis. 2016;16(1-2):55–61. doi: 10.1159/000439256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price D.L., Wong P.C., Borchelt D.R., Pardo C.A., Thinakaran G., Doan A.P., et al. Amyotrophic lateral sclerosis and Alzheimer disease. Lessons from model systems. Rev Neurol (Paris) 1997;153(8-9):484–495. [PubMed] [Google Scholar]
- 15.Niederwolfsgruber E., Schmitt T.L., Blasko I., Trieb K., Steger M.M., Maczek C., et al. The production of the Alzheimer amyloid precursor protein (APP) in extraneuronal tissue does not increase in old age. J Gerontol A Biol Sci Med Sci. 1998;53(3):B186–B190. doi: 10.1093/gerona/53a.3.b186. [DOI] [PubMed] [Google Scholar]
- 16.Goate A., Chartier-Harlin M.C., Mullan M., Brown J., Crawford F., Fidani L., et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 17.Sandbrink R., Masters C.L., Beyreuther K. APP gene family. Alternative splicing generates functionally related isoforms. Ann N Y Acad Sci. 1996;777:281–287. doi: 10.1111/j.1749-6632.1996.tb34433.x. [DOI] [PubMed] [Google Scholar]
- 18.Rohan de Silva H.A., Jen A., Wickenden C., Jen L.S., Wilkinson S.L., Patel A.J. Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res Mol Brain Res. 1997;47(1-2):147–156. doi: 10.1016/s0169-328x(97)00045-4. [DOI] [PubMed] [Google Scholar]
- 19.Kang J., Müller-Hill B. Differential splicing of Alzheimer's disease amyloid A4 precursor RNA in rat tissues: PreA4(695) mRNA is predominantly produced in rat and human brain. Biochem Biophys Res Commun. 1990;166(3):1192–1200. doi: 10.1016/0006-291x(90)90992-v. [DOI] [PubMed] [Google Scholar]
- 20.Gralle M., Ferreira S.T. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82(1):11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Heber S., Herms J., Gajic V., Hainfellner J., Aguzzi A., Rülicke T., et al. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J Neurosci. 2000;20(21):7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen K.V. β-Amyloid precursor protein (APP) and the human diseases. AIMS Neurosci. 2019;6(4):273–281. doi: 10.3934/Neuroscience.2019.4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matrone C., Iannuzzi F., Annunziato L. The Y. Ageing Res Rev. 2019;52:120–128. doi: 10.1016/j.arr.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Nalivaeva N.N., Turner A.J. The amyloid precursor protein: a biochemical enigma in brain development, function and disease. FEBS Lett. 2013;587(13):2046–2054. doi: 10.1016/j.febslet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Beckett C., Nalivaeva N.N., Belyaev N.D., Turner A.J. Nuclear signalling by membrane protein intracellular domains: the AICD enigma. Cell Signal. 2012;24(2):402–409. doi: 10.1016/j.cellsig.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Edbauer D., Willem M., Lammich S., Steiner H., Haass C. Insulin-degrading enzyme rapidly removes the beta-amyloid precursor protein intracellular domain (AICD) J Biol Chem. 2002;277(16):13389–13393. doi: 10.1074/jbc.M111571200. [DOI] [PubMed] [Google Scholar]
- 27.Cupers P., Orlans I., Craessaerts K., Annaert W., De, Strooper B. The amyloid precursor protein (APP)-cytoplasmic fragment generated by gamma-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J Neurochem. 2001;78(5):1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 28.Vingtdeux V., Hamdane M., Bégard S., Loyens A., Delacourte A., Beauvillain J.C., et al. Intracellular pH regulates amyloid precursor protein intracellular domain accumulation. Neurobiol Dis. 2007;25(3):686–696. doi: 10.1016/j.nbd.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Chow V.W., Mattson M.P., Wong P.C., Gleichmann M. An overview of APP processing enzymes and products. Neuromolecular Med. 2010;12(1):1–12. doi: 10.1007/s12017-009-8104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh D.M., Selkoe D.J. Amyloid β-protein and beyond: the path forward in Alzheimer's disease. Curr Opin Neurobiol. 2020;61:116–124. doi: 10.1016/j.conb.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Karran E., De, Strooper B. The amyloid cascade hypothesis: are we poised for success or failure? J Neurochem. 2016;139(Suppl 2):237–252. doi: 10.1111/jnc.13632. [DOI] [PubMed] [Google Scholar]
- 32.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 33.Rabinovich-Toidman P., Becker M., Barbiro B., Solomon B. Inhibition of amyloid precursor protein beta-secretase cleavage site affects survival and motor functions of amyotrophic lateral sclerosis transgenic mice. Neurodegener Dis. 2012;10(1-4):30–33. doi: 10.1159/000334774. [DOI] [PubMed] [Google Scholar]
- 34.Rabinovich-Toidman P., Rabinovich-Nikitin I., Ezra A., Barbiro B., Fogel H., Slutsky I., et al. Mutant SOD1 Increases APP Expression and Phosphorylation in Cellular and Animal Models of ALS. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0143420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gómez-Pinedo U., Galán L., Matías-Guiu J.A., Pytel V., Moreno T., Guerrero-Sola A., et al. Notch Signalling in the Hippocampus of Patients With Motor Neuron Disease. Front Neurosci. 2019;13:302. doi: 10.3389/fnins.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogalska A., D'Agostino C., Engel W.K., Askanas V. Activation of the γ-secretase complex and presence of γ-secretase-activating protein may contribute to Aβ42 production in sporadic inclusion-body myositis muscle fibers. Neurobiol Dis. 2012;48(1):141–149. doi: 10.1016/j.nbd.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Tallon C., Marshall K.L., Kennedy M.E., Hyde L.A., Farah M.H. Pharmacological BACE Inhibition Improves Axonal Regeneration in Nerve Injury and Disease Models. Neurotherapeutics. 2020;17(3):973–988. doi: 10.1007/s13311-020-00852-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menéndez-González M., Pérez-Pinera P., Martínez-Rivera M., Calatayud M.T., Blázquez Menes B. APP processing and the APP-KPI domain involvement in the amyloid cascade. Neurodegener Dis. 2005;2(6):277–283. doi: 10.1159/000092315. [DOI] [PubMed] [Google Scholar]
- 39.Devi L., Prabhu B.M., Galati D.F., Avadhani N.G., Anandatheerthavarada H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26(35):9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordji K., Becerril-Ortega J., Nicole O., Buisson A. Activation of extrasynaptic, but not synaptic, NMDA receptors modifies amyloid precursor protein expression pattern and increases amyloid-ß production. J Neurosci. 2010;30(47):15927–15942. doi: 10.1523/JNEUROSCI.3021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bukke V.N., Archana M., Villani R., Romano A.D., Wawrzyniak A., Balawender K., et al. The Dual Role of Glutamatergic Neurotransmission in Alzheimer's Disease: From Pathophysiology to Pharmacotherapy. Int J Mol Sci. 2020;21(20) doi: 10.3390/ijms21207452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tharp W.G., Lee Y.H., Greene S.M., Vincellete E., Beach T.G., Pratley R.E. Measurement of altered AβPP isoform expression in frontal cortex of patients with Alzheimer's disease by absolute quantification real-time PCR. J Alzheimers Dis. 2012;29(2):449–457. doi: 10.3233/JAD-2011-111337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konietzko U. AICD nuclear signaling and its possible contribution to Alzheimer's disease. Curr Alzheimer Res. 2012;9(2):200–216. doi: 10.2174/156720512799361673. [DOI] [PubMed] [Google Scholar]
- 44.Müller T., Meyer H.E., Egensperger R., Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog Neurobiol. 2008;85(4):393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Pardossi-Piquard R., Petit A., Kawarai T., Sunyach C., Alves da Costa C., Vincent B., et al. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46(4):541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Kim H.S., Kim E.M., Lee J.P., Park C.H., Kim S., Seo J.H., et al. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 2003;17(13):1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 47.Chang K.A., Kim H.S., Ha T.Y., Ha J.W., Shin K.Y., Jeong Y.H., et al. Phosphorylation of amyloid precursor protein (APP) at Thr668 regulates the nuclear translocation of the APP intracellular domain and induces neurodegeneration. Mol Cell Biol. 2006;26(11):4327–4338. doi: 10.1128/MCB.02393-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan K.A., Pimplikar S.W. Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol. 2005;171(2):327–335. doi: 10.1083/jcb.200505078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perkinton M.S., Standen C.L., Lau K.F., Kesavapany S., Byers H.L., Ward M., et al. The c-Abl tyrosine kinase phosphorylates the Fe65 adaptor protein to stimulate Fe65/amyloid precursor protein nuclear signaling. J Biol Chem. 2004;279(21):22084–22091. doi: 10.1074/jbc.M311479200. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y., Kim H.S., Joo Y., Choi Y., Chang K.A., Park C.H., et al. Intracellular domains of amyloid precursor-like protein 2 interact with CP2 transcription factor in the nucleus and induce glycogen synthase kinase-3beta expression. Cell Death Differ. 2007;14(1):79–91. doi: 10.1038/sj.cdd.4401928. [DOI] [PubMed] [Google Scholar]
- 51.Alves da Costa C., Sunyach C., Pardossi-Piquard R., Sévalle J., Vincent B., Boyer N., et al. Presenilin-dependent gamma-secretase-mediated control of p53-associated cell death in Alzheimer's disease. J Neurosci. 2006;26(23):6377–6385. doi: 10.1523/JNEUROSCI.0651-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Checler F., Sunyach C., Pardossi-Piquard R., Sévalle J., Vincent B., Kawarai T., et al. The gamma/epsilon-secretase-derived APP intracellular domain fragments regulate p53. Curr Alzheimer Res. 2007;4(4):423–p56. doi: 10.2174/156720507781788945. [DOI] [PubMed] [Google Scholar]
- 53.Zambrano N., Bimonte M., Arbucci S., Gianni D., Russo T., Bazzicalupo P. feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and beta-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J Cell Sci. 2002;115(Pt 7):1411–1422. doi: 10.1242/jcs.115.7.1411. [DOI] [PubMed] [Google Scholar]
- 54.Bimonte M., Gianni D., Allegra D., Russo T., Zambrano N. Mutation of the feh-1 gene, the Caenorhabditis elegans orthologue of mammalian Fe65, decreases the expression of two acetylcholinesterase genes. Eur J Neurosci. 2004;20(6):1483–1488. doi: 10.1111/j.1460-9568.2004.03611.x. [DOI] [PubMed] [Google Scholar]
- 55.Bruni P., Minopoli G., Brancaccio T., Napolitano M., Faraonio R., Zambrano N., et al. Fe65, a ligand of the Alzheimer's beta-amyloid precursor protein, blocks cell cycle progression by down-regulating thymidylate synthase expression. J Biol Chem. 2002;277(38):35481–35488. doi: 10.1074/jbc.M205227200. [DOI] [PubMed] [Google Scholar]
- 56.Roncarati R., Sestan N., Scheinfeld M.H., Berechid B.E., Lopez P.A., Meucci O., et al. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci U S A. 2002;99(10):7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q., Zerbinatti C.V., Zhang J., Hoe H.S., Wang B., Cole S.L., et al. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56(1):66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y.W., Wang R., Liu Q., Zhang H., Liao F.F., Xu H. Presenilin/gamma-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc Natl Acad Sci U S A. 2007;104(25):10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nalivaeva N.N., Zhuravin I.A., Turner A.J. Neprilysin expression and functions in development, ageing and disease. Mech Ageing Dev. 2020;192 doi: 10.1016/j.mad.2020.111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X.Y., Xue Y., Chen H., Chen L. The globus pallidus as a target for neuropeptides and endocannabinoids participating in central activities. Peptides. 2020;124 doi: 10.1016/j.peptides.2019.170210. [DOI] [PubMed] [Google Scholar]
- 61.Fischer H.S., Zernig G., Schuligoi R., Miczek K.A., Hauser K.F., Gerard C., et al. Alterations within the endogenous opioid system in mice with targeted deletion of the neutral endopeptidase ('enkephalinase') gene. Regul Pept. 2000;96(1-2):53–58. doi: 10.1016/s0167-0115(00)00200-7. [DOI] [PubMed] [Google Scholar]
- 62.Mathis S., Goizet C., Soulages A., Vallat J.M., Masson G.L. Genetics of amyotrophic lateral sclerosis: A review. J Neurol Sci. 2019;399:217–226. doi: 10.1016/j.jns.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 63.Taylor J.P., Brown R.H., Cleveland D.W. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu C.H., Fallini C., Ticozzi N., Keagle P.J., Sapp P.C., Piotrowska K., et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488(7412):499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith B.N., Vance C., Scotter E.L., Troakes C., Wong C.H., Topp S., et al. Novel mutations support a role for Profilin 1 in the pathogenesis of ALS. Neurobiol Aging. 2015;36(3):1602. doi: 10.1016/j.neurobiolaging.2014.10.032. e17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith B.N., Ticozzi N., Fallini C., Gkazi A.S., Topp S., Kenna K.P., et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84(2):324–331. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicolas A., Kenna K.P., Renton A.E., Ticozzi N., Faghri F., Chia R., et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 2018;97(6):1268–1283. doi: 10.1016/j.neuron.2018.02.027. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castellanos-Montiel M.J., Chaineau M., Durcan T.M. The Neglected Genes of ALS: Cytoskeletal Dynamics Impact Synaptic Degeneration in ALS. Front Cell Neurosci. 2020;14 doi: 10.3389/fncel.2020.594975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kockeritz L., Doble B., Patel S., Woodgett J.R. Glycogen synthase kinase-3--an overview of an over-achieving protein kinase. Curr Drug Targets. 2006;7(11):1377–1388. doi: 10.2174/1389450110607011377. [DOI] [PubMed] [Google Scholar]
- 70.Choi H.J., Cha S.J., Lee J.W., Kim H.J., Kim K. Recent Advances on the Role of GSK3β in the Pathogenesis of Amyotrophic Lateral Sclerosis. Brain Sci. 2020;10(10) doi: 10.3390/brainsci10100675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W., Leystra-Lantz C., Strong M.J. Upregulation of GSK3beta expression in frontal and temporal cortex in ALS with cognitive impairment (ALSci) Brain Res. 2008;1196:131–139. doi: 10.1016/j.brainres.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 72.Hu J.L., Liu G., Li Y.C., Gao W.J., Huang Y.Q. Dopamine D1 receptor-mediated NMDA receptor insertion depends on Fyn but not Src kinase pathway in prefrontal cortical neurons. Mol Brain. 2010;3:20. doi: 10.1186/1756-6606-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu J.H., Zhang H., Wagey R., Krieger C., Pelech S.L. Protein kinase and protein phosphatase expression in amyotrophic lateral sclerosis spinal cord. J Neurochem. 2003;85(2):432–442. doi: 10.1046/j.1471-4159.2003.01670.x. [DOI] [PubMed] [Google Scholar]
- 74.Szybińska A., Leśniak W. P53 Dysfunction in Neurodegenerative Diseases - The Cause or Effect of Pathological Changes? Aging Dis. 2017;8(4):506–518. doi: 10.14336/AD.2016.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González de Aguilar J.L., Gordon J.W., René F., de Tapia M., Lutz-Bucher B., Gaiddon C., et al. Alteration of the Bcl-x/Bax ratio in a transgenic mouse model of amyotrophic lateral sclerosis: evidence for the implication of the p53 signaling pathway. Neurobiol Dis. 2000;7(4):406–415. doi: 10.1006/nbdi.2000.0295. [DOI] [PubMed] [Google Scholar]
- 76.Martin L.J. p53 is abnormally elevated and active in the CNS of patients with amyotrophic lateral sclerosis. Neurobiol Dis. 2000;7(6 Pt B):613–622. doi: 10.1006/nbdi.2000.0314. [DOI] [PubMed] [Google Scholar]
- 77.Ranganathan S., Bowser R. p53 and Cell Cycle Proteins Participate in Spinal Motor Neuron Cell Death in ALS. Open Pathol J. 2010;4:11–22. doi: 10.2174/1874375701004010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eve D.J., Dennis J.S., Citron B.A. Transcription factor p53 in degenerating spinal cords. Brain Res. 2007;1150:174–181. doi: 10.1016/j.brainres.2007.02.088. [DOI] [PubMed] [Google Scholar]
- 79.Murakami K., Murata N., Noda Y., Tahara S., Kaneko T., Kinoshita N., et al. SOD1 (copper/zinc superoxide dismutase) deficiency drives amyloid β protein oligomerization and memory loss in mouse model of Alzheimer disease. J Biol Chem. 2011;286(52):44557–44568. doi: 10.1074/jbc.M111.279208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spisak K., Klimkowicz-Mrowiec A., Pera J., Dziedzic T., Aleksandra G., Slowik A. rs2070424 of the SOD1 gene is associated with risk of Alzheimer's disease. Neurol Neurochir Pol. 2014;48(5):342–345. doi: 10.1016/j.pjnns.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Arneson D., Zhang Y., Yang X., Narayanan M. Shared mechanisms among neurodegenerative diseases: from genetic factors to gene networks. J Genet. 2018;97(3):795–806. [PMC free article] [PubMed] [Google Scholar]
- 82.Turner B.J., Li Q.X., Laughton K.M., Masters C.L., Lopes E.C., Atkin J.D., et al. Brain beta-amyloid accumulation in transgenic mice expressing mutant superoxide dismutase 1. Neurochem Res. 2004;29(12):2281–2286. doi: 10.1007/s11064-004-7037-z. [DOI] [PubMed] [Google Scholar]
- 83.Yoon E.J., Park H.J., Kim G.Y., Cho H.M., Choi J.H., Park H.Y., et al. Intracellular amyloid beta interacts with SOD1 and impairs the enzymatic activity of SOD1: implications for the pathogenesis of amyotrophic lateral sclerosis. Exp Mol Med. 2009;41(9):611–617. doi: 10.3858/emm.2009.41.9.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Q.X., Mok S.S., Laughton K.M., McLean C.A., Volitakis I., Cherny R.A., et al. Overexpression of Abeta is associated with acceleration of onset of motor impairment and superoxide dismutase 1 aggregation in an amyotrophic lateral sclerosis mouse model. Aging Cell. 2006;5(2):153–165. doi: 10.1111/j.1474-9726.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- 85.Sasaki S., Warita H., Abe K., Iwata M. Impairment of axonal transport in the axon hillock and the initial segment of anterior horn neurons in transgenic mice with a G93A mutant SOD1 gene. Acta Neuropathol. 2005;110(1):48–56. doi: 10.1007/s00401-005-1021-9. [DOI] [PubMed] [Google Scholar]
- 86.Gómez-Pinedo U., Villar-Quiles R.N., Galán L., Matías-Guiu J.A., Benito-Martin M.S., Guerrero-Sola A., et al. Immununochemical Markers of the Amyloid Cascade in the Hippocampus in Motor Neuron Diseases. Front Neurol. 2016;7:195. doi: 10.3389/fneur.2016.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calingasan N.Y., Chen J., Kiaei M., Beal M.F. Beta-amyloid 42 accumulation in the lumbar spinal cord motor neurons of amyotrophic lateral sclerosis patients. Neurobiol Dis. 2005;19(1-2):340–347. doi: 10.1016/j.nbd.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Sasaki S., Iwata M. Immunoreactivity of beta-amyloid precursor protein in amyotrophic lateral sclerosis. Acta Neuropathol. 1999;97(5):463–468. doi: 10.1007/s004010051015. [DOI] [PubMed] [Google Scholar]
- 89.Maekawa S., Al-Sarraj S., Kibble M., Landau S., Parnavelas J., Cotter D., et al. Cortical selective vulnerability in motor neuron disease: a morphometric study. Brain. 2004;127(Pt 6):1237–1251. doi: 10.1093/brain/awh132. [DOI] [PubMed] [Google Scholar]
- 90.Matías-Guiu J.A., Pytel V., Cabrera-Martín M.N., Galán L., Valles-Salgado M., Guerrero A., et al. Amyloid- and FDG-PET imaging in amyotrophic lateral sclerosis. Eur J Nucl Med Mol Imaging. 2016;43(11):2050–2060. doi: 10.1007/s00259-016-3434-1. [DOI] [PubMed] [Google Scholar]
- 91.Kuang G., Murugan N.A., Tu Y., Nordberg A., Ågren H. Investigation of the Binding Profiles of AZD2184 and Thioflavin T with Amyloid-β(1-42) Fibril by Molecular Docking and Molecular Dynamics Methods. J Phys Chem B. 2015;119(35):11560–11567. doi: 10.1021/acs.jpcb.5b05964. [DOI] [PubMed] [Google Scholar]
- 92.Steinacker P., Fang L., Kuhle J., Petzold A., Tumani H., Ludolph A.C., et al. Soluble beta-amyloid precursor protein is related to disease progression in amyotrophic lateral sclerosis. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steinacker P., Hendrich C., Sperfeld A.D., Jesse S., Lehnert S., Pabst A., et al. Concentrations of beta-amyloid precursor protein processing products in cerebrospinal fluid of patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. J Neural Transm (Vienna) 2009;116(9):1169–1178. doi: 10.1007/s00702-009-0271-4. [DOI] [PubMed] [Google Scholar]
- 94.Koistinen H., Prinjha R., Soden P., Harper A., Banner S.J., Pradat P.F., et al. Elevated levels of amyloid precursor protein in muscle of patients with amyotrophic lateral sclerosis and a mouse model of the disease. Muscle Nerve. 2006;34(4):444–450. doi: 10.1002/mus.20612. [DOI] [PubMed] [Google Scholar]
- 95.Wirths O., Weis J., Kayed R., Saido T.C., Bayer T.A. Age-dependent axonal degeneration in an Alzheimer mouse model. Neurobiol Aging. 2007;28(11):1689–1699. doi: 10.1016/j.neurobiolaging.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 96.Seo J.S., Leem Y.H., Lee K.W., Kim S.W., Lee J.K., Han P.L. Severe motor neuron degeneration in the spinal cord of the Tg2576 mouse model of Alzheimer disease. J Alzheimers Dis. 2010;21(1):263–276. doi: 10.3233/JAD-2010-091528. [DOI] [PubMed] [Google Scholar]
- 97.Sugarman M.C., Yamasaki T.R., Oddo S., Echegoyen J.C., Murphy M.P., Golde T.E., et al. Inclusion body myositis-like phenotype induced by transgenic overexpression of beta APP in skeletal muscle. Proc Natl Acad Sci U S A. 2002;99(9):6334–6339. doi: 10.1073/pnas.082545599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bressler S.L., Gray M.D., Sopher B.L., Hu Q., Hearn M.G., Pham D.G., et al. cDNA cloning and chromosome mapping of the human Fe65 gene: interaction of the conserved cytoplasmic domains of the human beta-amyloid precursor protein and its homologues with the mouse Fe65 protein. Hum Mol Genet. 1996;5(10):1589–1598. doi: 10.1093/hmg/5.10.1589. [DOI] [PubMed] [Google Scholar]
- 99.Fiore F., Zambrano N., Minopoli G., Donini V., Duilio A., Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein. J Biol Chem. 1995;270(52):30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 100.McLoughlin D.M., Miller C.C. The intracellular cytoplasmic domain of the Alzheimer's disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;397(2-3):197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 101.Borg J.P., Ooi J., Levy E., Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16(11):6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scheinfeld M.H., Ghersi E., Davies P., D'Adamio L. Amyloid beta protein precursor is phosphorylated by JNK-1 independent of, yet facilitated by, JNK-interacting protein (JIP)-1. J Biol Chem. 2003;278(43):42058–42063. doi: 10.1074/jbc.M304853200. [DOI] [PubMed] [Google Scholar]
- 103.Zheng P., Eastman J., Vande Pol S., Pimplikar S.W.P.A.T.1. a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci U S A. 1998;95(25):14745–14750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pastorino L., Sun A., Lu P.J., Zhou X.Z., Balastik M., Finn G., et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440(7083):528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 105.Liu F.L., Liu P.H., Shao H.W., Kung F.L. The intracellular domain of amyloid precursor protein interacts with FKBP12. Biochem Biophys Res Commun. 2006;350(2):472–477. doi: 10.1016/j.bbrc.2006.09.073. [DOI] [PubMed] [Google Scholar]
- 106.Tarr P.E., Roncarati R., Pelicci G., Pelicci P.G., D'Adamio L. Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem. 2002;277(19):16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- 107.Xie Z., Dong Y., Maeda U., Xia W., Tanzi R.E. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation. J Biol Chem. 2007;282(7):4318–4325. doi: 10.1074/jbc.M609293200. [DOI] [PubMed] [Google Scholar]
- 108.Zhou D., Noviello C., D'Ambrosio C., Scaloni A., D'Adamio L. Growth factor receptor-bound protein 2 interaction with the tyrosine-phosphorylated tail of amyloid beta precursor protein is mediated by its Src homology 2 domain. J Biol Chem. 2004;279(24):25374–25380. doi: 10.1074/jbc.M400488200. [DOI] [PubMed] [Google Scholar]
- 109.Howell B.W., Lanier L.M., Frank R., Gertler F.B., Cooper J.A. The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol Cell Biol. 1999;19(7):5179–5188. doi: 10.1128/mcb.19.7.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97(6):689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 111.Huang Y., Magdaleno S., Hopkins R., Slaughter C., Curran T., Keshvara L. Tyrosine phosphorylated Disabled 1 recruits Crk family adapter proteins. Biochem Biophys Res Commun. 2004;318(1):204–212. doi: 10.1016/j.bbrc.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 112.Zambrano N., Bruni P., Minopoli G., Mosca R., Molino D., Russo C., et al. The beta-amyloid precursor protein APP is tyrosine-phosphorylated in cells expressing a constitutively active form of the Abl protoncogene. J Biol Chem. 2001;276(23):19787–19792. doi: 10.1074/jbc.M100792200. [DOI] [PubMed] [Google Scholar]
- 113.Matrone C., Petrillo F., Nasso R., Ferretti G. Fyn Tyrosine Kinase as Harmonizing Factor in Neuronal Functions and Dysfunctions. Int J Mol Sci. 2020;21(12) doi: 10.3390/ijms21124444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Poulsen E.T., Iannuzzi F., Rasmussen H.F., Maier T.J., Enghild J.J., Jørgensen A.L., et al. An Aberrant Phosphorylation of Amyloid Precursor Protein Tyrosine Regulates Its Trafficking and the Binding to the Clathrin Endocytic Complex in Neural Stem Cells of Alzheimer's Disease Patients. Front Mol Neurosci. 2017;10:59. doi: 10.3389/fnmol.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Iannuzzi F., Sirabella R., Canu N., Maier T.J., Annunziato L., Matrone C. Fyn Tyrosine Kinase Elicits Amyloid Precursor Protein Tyr682 Phosphorylation in Neurons from Alzheimer's Disease Patients. Cells. 2020;9(8) doi: 10.3390/cells9081807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Poulsen E.T., Larsen A., Zollo A., Jørgensen A.L., Sanggaard K.W., Enghild J.J., et al. New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies. Int J Mol Sci. 2015;16(12):29446–29453. doi: 10.3390/ijms161226181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zollo A., Allen Z., Rasmussen H.F., Iannuzzi F., Shi Y., Larsen A., et al. Sortilin-Related Receptor Expression in Human Neural Stem Cells Derived from Alzheimer's Disease Patients Carrying the APOE Epsilon 4 Allele. Neural Plast. 2017;2017:1892612. doi: 10.1155/2017/1892612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.von Rotz R.C., Kohli B.M., Bosset J., Meier M., Suzuki T., Nitsch R.M., et al. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci. 2004;117(Pt 19):4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- 119.Zhang X., Song W. The role of APP and BACE1 trafficking in APP processing and amyloid-β generation. Alzheimers Res Ther. 2013;5(5):46. doi: 10.1186/alzrt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matrone C. A new molecular explanation for age-related neurodegeneration: the Tyr682 residue of amyloid precursor protein. Bioessays. 2013;35(10):847–852. doi: 10.1002/bies.201300041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomez-Navarro N., Miller E. Protein sorting at the ER-Golgi interface. J Cell Biol. 2016;215(6):769–778. doi: 10.1083/jcb.201610031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shi Y., Lin S., Staats K.A., Li Y., Chang W.H., Hung S.T., et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24(3):313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Le Gall L., Anakor E., Connolly O., Vijayakumar U.G., Duddy W.J., Duguez S. Molecular and Cellular Mechanisms Affected in ALS. J Pers Med. 2020;10(3) doi: 10.3390/jpm10030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi B., Conner S.D., Liu J. Dysfunction of endocytic kinase AAK1 in ALS. Int J Mol Sci. 2014;15(12):22918–22932. doi: 10.3390/ijms151222918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.La Rosa L.R., Perrone L., Nielsen M.S., Calissano P., Andersen O.M., Matrone C. Y682G Mutation of Amyloid Precursor Protein Promotes Endo-Lysosomal Dysfunction by Disrupting APP-SorLA Interaction. Front Cell Neurosci. 2015;9:109. doi: 10.3389/fncel.2015.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mori F., Miki Y., Tanji K., Kakita A., Takahashi H., Utsumi J., et al. Sortilin-related receptor CNS expressed 2 (SorCS2) is localized to Bunina bodies in amyotrophic lateral sclerosis. Neurosci Lett. 2015;608:6–11. doi: 10.1016/j.neulet.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 127.Andersen O.M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Andersen O.M., Schmidt V., Spoelgen R., Gliemann J., Behlke J., Galatis D., et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45(8):2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 129.Gulino R., Parenti R., Gulisano M. Novel Mechanisms of Spinal Cord Plasticity in a Mouse Model of Motoneuron Disease. Biomed Res Int. 2015;2015 doi: 10.1155/2015/654637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A.J., Nye J.S., et al. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16(7):846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Traiffort E., Ferent J. [Neural stem cells and Notch signalling] Med Sci (Paris) 2015;31(12):1115–1125. doi: 10.1051/medsci/20153112015. [DOI] [PubMed] [Google Scholar]
- 132.Casares-Crespo L., Calatayud-Baselga I., García-Corzo L., Mira H. On the Role of Basal Autophagy in Adult Neural Stem Cells and Neurogenesis. Front Cell Neurosci. 2018;12:339. doi: 10.3389/fncel.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Galán L., Gómez-Pinedo U., Guerrero A., García-Verdugo J.M., Matías-Guiu J. Amyotrophic lateral sclerosis modifies progenitor neural proliferation in adult classic neurogenic brain niches. BMC Neurol. 2017;17(1):173. doi: 10.1186/s12883-017-0956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xue F., Chen Y.C., Zhou C.H., Wang Y., Cai M., Yan W.J., et al. Risperidone ameliorates cognitive deficits, promotes hippocampal proliferation, and enhances Notch signaling in a murine model of schizophrenia. Pharmacol Biochem Behav. 2017;163:101–109. doi: 10.1016/j.pbb.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 135.Zhang S., Wang P., Ren L., Hu C., Bi J. Protective effect of melatonin on soluble Aβ1-42-induced memory impairment, astrogliosis, and synaptic dysfunction via the Musashi1/Notch1/Hes1 signaling pathway in the rat hippocampus. Alzheimers Res Ther. 2016;8(1):40. doi: 10.1186/s13195-016-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Petersen P.H., Zou K., Hwang J.K., Jan Y.N., Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419(6910):929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- 137.Petersen P.H., Zou K., Krauss S., Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7(8):803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- 138.Nishimura T., Yamaguchi T., Tokunaga A., Hara A., Hamaguchi T., Kato K., et al. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol Biol Cell. 2006;17(3):1273–1285. doi: 10.1091/mbc.E05-07-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yáñez M.J., Belbin O., Estrada L.D., Leal N., Contreras P.S., Lleó A., et al. c-Abl links APP-BACE1 interaction promoting APP amyloidogenic processing in Niemann-Pick type C disease. Biochim Biophys Acta. 2016;1862(11):2158–2167. doi: 10.1016/j.bbadis.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 140.Jiang Y.M., Yamamoto M., Kobayashi Y., Yoshihara T., Liang Y., Terao S., et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57(2):236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 141.Imamura K., Izumi Y., Watanabe A., Tsukita K., Woltjen K., Yamamoto T., et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med. 2017;9(391) doi: 10.1126/scitranslmed.aaf3962. [DOI] [PubMed] [Google Scholar]
- 142.Katsumata R., Ishigaki S., Katsuno M., Kawai K., Sone J., Huang Z., et al. c-Abl inhibition delays motor neuron degeneration in the G93A mouse, an animal model of amyotrophic lateral sclerosis. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0046185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Riancho J., Gil-Bea F.J., Castanedo-Vazquez D., Sedano M.J., Zufiría M., de Eulate G.F.G., et al. Clinical evidences supporting the Src/c-Abl pathway as potential therapeutic target in amyotrophic lateral sclerosis. J Neurol Sci. 2018;393:80–82. doi: 10.1016/j.jns.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 144.Zhou D., Yan H., Yang S., Zhang Y., Xu X., Cen X., et al. SC75741, A Novel c-Abl Inhibitor, Promotes the Clearance of TDP25 Aggregates. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.741219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Higelin J., Catanese A., Semelink-Sedlacek L.L., Oeztuerk S., Lutz A.K., Bausinger J., et al. NEK1 loss-of-function mutation induces DNA damage accumulation in ALS patient-derived motoneurons. Stem Cell Res. 2018;30:150–162. doi: 10.1016/j.scr.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 146.Freischmidt A., Wieland T., Richter B., Ruf W., Schaeffer V., Müller K., et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18(5):631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 147.Guo W., Vandoorne T., Steyaert J., Staats K.A. Van Den Bosch L. The multifaceted role of kinases in amyotrophic lateral sclerosis: genetic, pathological and therapeutic implications. Brain. 2020;143(6):1651–1673. doi: 10.1093/brain/awaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Palomo V., Nozal V., Rojas-Prats E., Gil C., Martinez A. Protein kinase inhibitors for amyotrophic lateral sclerosis therapy. Br J Pharmacol. 2021;178(6):1316–1335. doi: 10.1111/bph.15221. [DOI] [PubMed] [Google Scholar]
- 149.Obregon D., Hou H., Deng J., Giunta B., Tian J., Darlington D., et al. Soluble amyloid precursor protein-α modulates β-secretase activity and amyloid-β generation. Nat Commun. 2012;3:777. doi: 10.1038/ncomms1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Peters-Libeu C., Campagna J., Mitsumori M., Poksay K.S., Spilman P., Sabogal A., et al. sAβPPα is a Potent Endogenous Inhibitor of BACE1. J Alzheimers Dis. 2015;47(3):545–555. doi: 10.3233/JAD-150282. [DOI] [PubMed] [Google Scholar]
- 151.Deng J., Habib A., Obregon D.F., Barger S.W., Giunta B., Wang Y.J., et al. Soluble amyloid precursor protein alpha inhibits tau phosphorylation through modulation of GSK3β signaling pathway. J Neurochem. 2015;135(3):630–637. doi: 10.1111/jnc.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Copanaki E., Chang S., Vlachos A., Tschäpe J.A., Müller U.C., Kögel D., et al. sAPPalpha antagonizes dendritic degeneration and neuron death triggered by proteasomal stress. Mol Cell Neurosci. 2010;44(4):386–393. doi: 10.1016/j.mcn.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 153.Selnes P., Blennow K., Zetterberg H., Grambaite R., Rosengren L., Johnsen L., et al. Effects of cerebrovascular disease on amyloid precursor protein metabolites in cerebrospinal fluid. Cerebrospinal Fluid Res. 2010;7:10. doi: 10.1186/1743-8454-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jakobsson J., Zetterberg H., Blennow K., Johan Ekman C., Johansson A.G., Landén M. Altered concentrations of amyloid precursor protein metabolites in the cerebrospinal fluid of patients with bipolar disorder. Neuropsychopharmacology. 2013;38(4):664–672. doi: 10.1038/npp.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Barbagallo A.P., Weldon R., Tamayev R., Zhou D., Giliberto L., Foreman O., et al. Tyr(682) in the intracellular domain of APP regulates amyloidogenic APP processing in vivo. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barbagallo A.P., Wang Z., Zheng H., D'Adamio L. A single tyrosine residue in the amyloid precursor protein intracellular domain is essential for developmental function. J Biol Chem. 2011;286(11):8717–8721. doi: 10.1074/jbc.C111.219873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Matrone C., Luvisetto S., La Rosa L.R., Tamayev R., Pignataro A., Canu N., et al. Tyr682 in the Abeta-precursor protein intracellular domain regulates synaptic connectivity, cholinergic function, and cognitive performance. Aging Cell. 2012;11(6):1084–1093. doi: 10.1111/acel.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deng Z., Zhou X., Lu J.H., Yue Z. Autophagy deficiency in neurodevelopmental disorders. Cell Biosci. 2021;11(1):214. doi: 10.1186/s13578-021-00726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ring S., Weyer S.W., Kilian S.B., Waldron E., Pietrzik C.U., Filippov M.A., et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci. 2007;27(29):7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Matrone C., Barbagallo A.P., La Rosa L.R., Florenzano F., Ciotti M.T., Mercanti D., et al. APP is phosphorylated by TrkA and regulates NGF/TrkA signaling. J Neurosci. 2011;31(33):11756–11761. doi: 10.1523/JNEUROSCI.1960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tovar-Y-Romo L.B., Ramírez-Jarquín U.N., Lazo-Gómez R., Tapia R. Trophic factors as modulators of motor neuron physiology and survival: implications for ALS therapy. Front Cell Neurosci. 2014;8:61. doi: 10.3389/fncel.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ekestern E. Neurotrophic factors and amyotrophic lateral sclerosis. Neurodegener Dis. 2004;1(2-3):88–100. doi: 10.1159/000080049. [DOI] [PubMed] [Google Scholar]
- 163.Chao M.V. Trophic factors: An evolutionary cul-de-sac or door into higher neuronal function? J Neurosci Res. 2000;59(3):353–355. [PubMed] [Google Scholar]
- 164.Stanga S., Brambilla L., Tasiaux B., Dang A.H., Ivanoiu A., Octave J.N., et al. A Role for GDNF and Soluble APP as Biomarkers of Amyotrophic Lateral Sclerosis Pathophysiology. Front Neurol. 2018;9:384. doi: 10.3389/fneur.2018.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stanga S., Zanou N., Audouard E., Tasiaux B., Contino S., Vandermeulen G., et al. APP-dependent glial cell line-derived neurotrophic factor gene expression drives neuromuscular junction formation. FASEB J. 2016;30(5):1696–1711. doi: 10.1096/fj.15-278739. [DOI] [PubMed] [Google Scholar]
- 166.Calissano P., Amadoro G., Matrone C., Ciafrè S., Marolda R., Corsetti V., et al. Does the term 'trophic' actually mean anti-amyloidogenic? The case of NGF. Cell Death Differ. 2010;17(7):1126–1133. doi: 10.1038/cdd.2010.38. [DOI] [PubMed] [Google Scholar]
- 167.Suzuki M., McHugh J., Tork C., Shelley B., Hayes A., Bellantuono I., et al. Direct muscle delivery of GDNF with human mesenchymal stem cells improves motor neuron survival and function in a rat model of familial ALS. Mol Ther. 2008;16(12):2002–2010. doi: 10.1038/mt.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mohajeri M.H., Figlewicz D.A., Bohn M.C. Intramuscular grafts of myoblasts genetically modified to secrete glial cell line-derived neurotrophic factor prevent motoneuron loss and disease progression in a mouse model of familial amyotrophic lateral sclerosis. Hum Gene Ther. 1999;10(11):1853–1866. doi: 10.1089/10430349950017536. [DOI] [PubMed] [Google Scholar]