Abstract
The p53 protein is a transcription factor that preserves the integrity of the genome. The TP53 gene has inactivating mutations in about 50% of all human cancers. Some missense mutations lead to decreased thermal stability in the p53 protein, its unfolding and aggregation under physiological conditions. A general understanding of the impact of point mutations on the stability and conformation of mutant p53 is essential for the design and development of small molecules that target specific p53 mutations. In this work, we determined the thermostability properties of some of the most common mutant forms of the p53 protein—p53(R273H), p53(R248Q), p53(R248W) and p53(Y220C)—that are often considered as attractive therapeutic targets. The results showed that these missense mutations lead to destabilization of the p53 protein and a decrease in its melting temperature.
Keywords: p53, transcription factor, mutant, melting temperature, differential scanning fluorimetry
1. Introduction
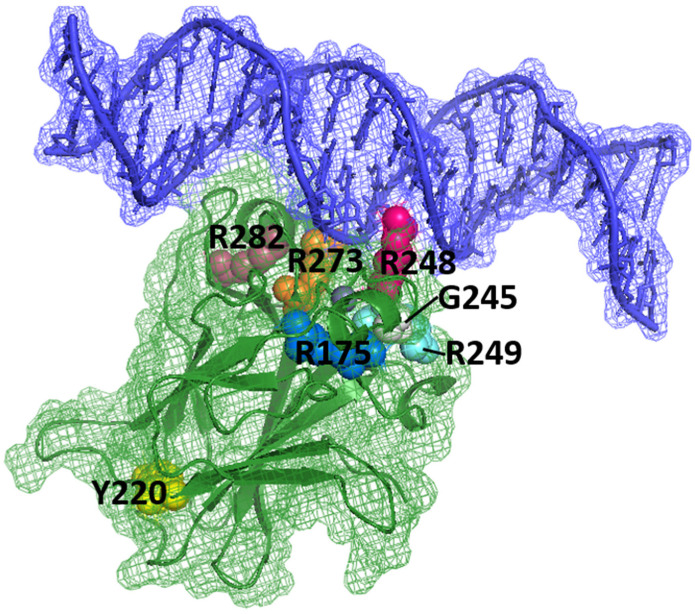
The p53 tumor suppressor plays a key role in multiple signaling pathways that control cell cycle and is responsible for the stability of the human genome. Transcription factor p53 dysfunction occurs in most human malignancies [1]. Two main causes are responsible for this dysfunction: mutations in the TP53 gene and suppression of wild-type p53 mediated by its negative regulators MDM2/MDM4. The p53 protein is inactivated by mutations in about half of all tumors [2]. Most p53 oncogenic mutations are missense mutations in the DNA-binding domain (DBD) of the protein [3]. Despite functional heterogeneity, these mutations can be broadly classified into contact and structural (Figure 1). The former disrupts the binding of p53 to DNA and shows relatively small changes in the overall structure and stability of the protein but contributes to the loss of its transcriptional activity (R248Q, R273H, R248W, R248L, R273C) [4]. The latter reduces the thermal stability of the p53 protein to varying degrees, causing it to rapidly unfold and aggregate under physiological conditions. The most-common structural mutations in p53 are G245S, R175H, R282W and Y220C [5]. Over 40% of missense mutations in the DNA-binding domain of p53 occur at arginine residues, while glycine and cysteine, the next most-common residues to change, mutate up to five-times less often. No p53 missense mutation is found at a frequency greater than 6%, and the 10 most-common and best-studied hotspot mutants together account for approximately 25% of all p53 mutations. Overall, these mutations lead to more than 2.5 million new cases of cancer per year worldwide [6].
Figure 1.
p53 DBD (green) in complex with DNA (blue). The locations of amino acids R175, Y220, G245, R248, R249, R273 and R282, which are sites of key carcinogenic mutations (“hot spots”), are shown (PDB identifier: 1TSR) [4]. DNA is in direct interaction with R273 and R248, and several “hot spots” are present near this interaction (R249, G245, R282). Other “hot spots” are far from the surface involved in DNA binding, for example, Y220.
Most mutant p53 proteins, in addition to losing their transcriptional activity (loss of function, LOF), can also acquire new oncogenic properties (gain of function, GOF). The p53 oncoprotein has a dominant-negative effect on the remaining wild-type p53 proteins. This effect is mediated by hetero-oligomerization of the mutant protein with the wild-type protein, which leads to the appearance of functionally inactive tetrameric complexes [7]. Further, the mutant protein leads to uncontrolled cell proliferation, inhibits apoptosis, confers resistance to specific anticancer compounds, and promotes invasion and metastasis.
According to the data provided in the IARC TP53 database, missense mutations R248Q, R248W and R273H are among the five most-frequent p53 mutations that lead to more than 630,000 newly diagnosed cases of cancer worldwide every year [8]. The R248Q oncogenic mutation is the second-most-common mutation in p53. It is found in many types of cancer, such as colorectal cancer, lymphoid and myeloid leukemia, breast and ovarian cancer. Changing arginine to glutamine at position 248 disrupts binding of p53 to DNA [9]. R273H and R248W are the third- and fourth-most-common hotspot mutations, respectively. They are most often observed in lung, breast and ovarian cancer, as well as gliomas. Mutant proteins p53(R273H) and p53(R248W) not only lose their DNA-binding abilities but can also acquire new oncogenic properties that promote proliferation, chemoresistance and metastasis of tumor cells [10].
The oncogenic Y220C mutation is found in approximately 100,000 cancer cases each year [11]. This mutation changes the tertiary structure of the p53 DNA-binding domain, as a result of which the protein becomes destabilized, partially denatured and loses its cellular activity [8,12,13].
A decrease in the melting temperature (Tm) of the p53 DNA-binding domain is observed in 30% of all missense mutations that inactivate the protein [9,14]. Thus, the purpose of this work was to determine the melting temperature of the most-common mutant forms of p53.
2. Materials and Methods
2.1. Reagents
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), phenylmethylsulfonyl fluoride (PMSF) and fluorescent dye SYPRO Orange were purchased from Sigma-Aldrich (St. Louis, MO, USA). Kanamycin, chloramphenicol and lysozyme were purchased from PanReac AppliChem (Boca Raton, FL, USA). Isopropyl-β-D-thiogalactoside (IPTG) and β-mercaptoethanol were from Anatrace (Maumee, OH, USA). 2-Amino-2-(hydroxymethyl)propane-1,3-diol (Tris) and dithiothreitol (DTT) were from Thermo Fisher Scientific (Waltham, MA, USA).
2.2. Site-Directed Mutagenesis
Plasmids encoding the p53 DBD (residues 94–312) (wild-type and Y220C mutant) were kindly provided by Prof. Matthias Baud from the University of Southampton (UK). Plasmids encoding the p53 core domains with R248Q, R248W and R273H hotspot mutations were prepared by site-directed mutagenesis at Evrogen Ltd. (Moscow, Russia). The primers are listed in Table 1.
Table 1.
Primer sequences for site-directed mutagenesis.
| Mutant | Primer Sequences | Amino Acid Change: wt → Mutant |
|---|---|---|
| R248W | Rev: GTTCATGCCGCCCATGCAGGAACTGTAACACATGTAGTTGTAGTGGATGGT For: CCTGCATGGGCGGCATGAACtGGAGGCCCATCCTCACCATCATC |
CGG (R) → TGG (W) |
| R248Q | Rev: GTTCATGCCGCCCATGCAGGAACTGTAACACATGTAGTTGTAGTGGATGGT For: CTGCATGGGCGGCATGAACCaGAGGCCCATCCTCACCATCATC |
CGG (R) → CAG (Q) |
| R273H | Rev: GCACCTCAAAGCTGTCCCGTCCCAGTAGATTACCACTGGAGTCTTCC For: ACGGGACAGCTTTGAGGTGCaTGTTTGTGCCTGTCCTGGGAGA |
CGT (R) → CAT (H) |
The presence of a mutation in the gene was confirmed by sequencing. Plasmids were transformed into Escherichia coli by the heat-shock method. The pET24A(+) vector containing the p53 DBD gene (wt/mut) was isolated using the Plasmid Midiprep 2.0 Kit (cat. no. BC124).
2.3. Recombinant Production and Purification of p53
Proteins of p53 DBD (residues 94–312) were overexpressed in Escherichia coli BL21 (DE3) pLysS cells in TB medium at 18–20 °C for 16 h. For the primary purification of proteins, Ni Sepharose 6 FastFlow affinity sorbent (GE Healthcare, Chicago, IL, USA) was used. Proteolytic cleavage of hexa-histidine residues of recombinant proteins was performed using TEV protease. Further purification was carried out with HiTrap Heparin HP column (GE Healthcare, Chicago, IL, USA). Finally, size-exclusion chromatography was performed using ENrich SEC 70 column (Bio-Rad, Hercules, CA, USA) using an NGC Discover chromatography system (Bio-Rad, Hercules, CA, USA). The proteins were concentrated using a Vivaspin concentrator with molecular-weight cutoff of 10 kDa. Molecular weight and protein purity were confirmed by SDS gel electrophoresis.
2.4. Differential Scanning Fluorimetry (DSF)
The method is based on measuring the optical density (λ = 570 nm) of a gradually heated solution containing the protein and the hydrophobic fluorescent dye Sypro Orange (Sigma-Aldrich, St. Louis, MO, USA). The unfolding of the protein with increasing temperature and the opening of its hydrophobic regions leads to release of fluorescent signal that is recorded in a characteristic graph [14,15]. The experiment was carried out in 96-well LightCycler 480 Multiwell Plate 96 plates (Roche Diagnostics GmbH, Penzberg, Germany). Protein (final concentration of 10 µM) was mixed with SyproOrange fluorescent dye (up to ×10) (λex = 470 nm, λem = 570 nm) in 25 mM Hepes buffer pH = 7.2, 150 mM NaCl. The fluorescence readings were carried out using Lightcycler 480 thermal cycler (Roche, Penzberg, Germany), in which the plate was smoothly heated from 30 °C to 95 °C at a constant heating rate of 1 °C/min. The excitation and emission filters were set at λ = 460 nm and λ = 510 nm, respectively. All measurements were carried out in triplicate.
3. Results and Discussion
To determine the impact of missense mutations on p53 thermostability, the melting temperatures of the recombinant core domains p53(wt), p53(Y220C), p53(R248Q), p53(R248W) and p53(R273H) were studied via DSF. This technique allows for rapid analysis of protein stability in solutions [16,17]. Thermostable fluorescent dyes can bind nonspecifically to the hydrophobic surface of the protein. At a gradual increase in temperature, the protein thermal denaturation causes unfolding of the hydrophobic core, making it accessible to the dye molecules. When the dye binds to the hydrophobic core, this eliminates fluorescence quenching, and the fluorescence quantum yield increases dramatically. The optical signal is measured at each point and the thermal denaturation curve is generated, which allows for calculation of the melting temperature of a protein globule and, therefore, estimation of the protein thermal stability [18,19].
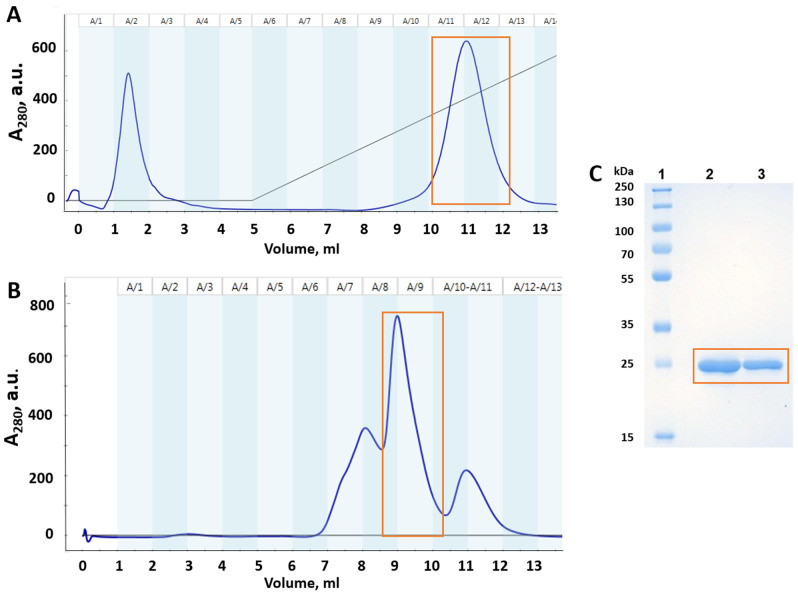
The full-length p53 mutants are known to be thermodynamically unstable, prone to aggregation and have a lot of disordered N- and C-terminal parts [19]. The p53 core domain is key to protein stability, and mutations in this domain directly affect relative stability of the whole protein [20]. Therefore, in this study, we expressed and purified recombinant p53 DBD for wild-type and mutated isoforms. Representative chromatograms and electropherograms of purified p53(Y220C) DBD are shown in Figure 2. The degree of electrophoretic homogeneity of recombinant protein was above 95%, according to the results of densitometric analysis (Image Lab 6.0.1, Bio-Rad, Hercules, CA, USA).
Figure 2.
Purification of mutant p53(Y220C) DBD. Orange square shows the purified mutant p53(Y220C) DBD protein: (A) heparin-sepharose chromatography profile (dark blue line—absorbance at 280 nm); (B) gel filtration chromatography profile; (C) SDS-PAGE monitoring of mutant p53(Y220C) DBD after gel filtration chromatography: Lane 1—Protein molecular-weight marker; Lane 2—p53(Y220C) DBD (10 µg/well); Lane 3—p53(Y220C) DBD (5 µg/well).
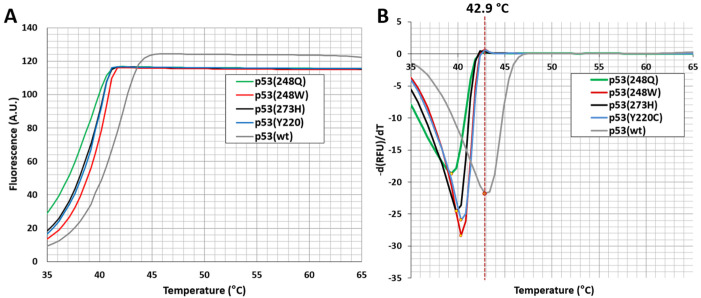
According to the melting curves of the wild-type p53 and its four mutant variants, the wild-type protein exhibits the highest thermal stability among them (Figure 3). The Tm of the wild-type p53 DBD was around 42.9 °C, which is in agreement with the previously reported data (Table 2) [16,20,21,22,23]. The Tm values of DNA contact mutants p53(R248Q), p53(R248W) and p53(R273H) were equal to 38.5 °C, 39.3 °C and 38.8 °C, respectively (Figure 3 and Table 2).
Figure 3.
Analysis of the thermal stability of DNA-binding domains (residues 94–312) of p53 (wild-type) and p53(Y220C), p53(R248Q), p53(R248W) and p53(R273H) mutants by differential scanning fluorimetry. (A) Representative melting curves and (B) derivative curves. Protein (at final concentration of 10 µM) was mixed with SyproOrange fluorescent dye (up to ×10) in 25 mM Hepes buffer pH = 7.2, 150 mM NaCl. Colored curves represent the thermal denaturation patterns of various p53 mutants—p53(Y220C) in light blue, p53(R248Q) in green, p53(R248W) in red and p53(R273H) in black. The red vertical dashed line indicates the Tm of p53 (wild-type) DBD (gray curve). RFU is a relative unit of fluorescence.
Table 2.
Melting temperatures of wild-type p53 and its mutant variants measured by differential scanning fluorimetry.
| Protein | Structural Region | Tm, °C | ΔTm, °C * |
|---|---|---|---|
| p53 (wild-type) | - | 42.9 ± 0.0 | - |
| p53(Y220C) | β-sandwich | 40.3 ± 0.2 | −2.6 |
| p53(R248Q) | DNA contact | 38.5 ± 0.3 | −4.4 |
| p53(R248W) | DNA contact | 39.3 ± 0.0 | −3.6 |
| p53(R273H) | DNA contact | 38.8 ± 0.3 | −4.1 |
* ΔTm = Tm (mutant) − Tm (wild-type). Mean values of triplicate measurements ± SEM are shown.
Structural mutation of p53(Y220C), which is the ninth-most-common p53 mutation, reduces the stability of p53 DBD by 4 kcal/mol [24]. This results in unfolding and transcriptional inactivation of over 80% of the protein under physiological conditions. According to our data, the Tm value of p53(Y220C) mutant was equal to 40.3 °C (Figure 3 and Table 2).
We demonstrate that p53 mutations Y220C, R248Q, R248W and R273W destabilize the protein and the results are consistent with earlier studies [12,25,26,27,28,29]. Most DNA contact mutations are structurally conserved and normally do not have a profound effect on the p53 thermostability [30,31]. However, our data show that the difference in thermostability between DNA contact (R248Q, R248W and R273H) and structural (Y220C) mutations might be less pronounced than previously thought (ΔTm = approx. 1 °C). In support of this, some p53 mutations were indeed described to exhibit characteristics of both contact and structural mutations [10]. Tumor cells can contain a wide range of p53 mutants with reduced thermostability. The development of drugs capable of restoring wild-type function in such a heterogeneous target is, no doubt, a challenging task [32]. Alan Fersht and colleagues proposed several criteria that should be met by compounds considered as mutant p53 reactivators. Compounds should demonstrate binding to mutant p53 and lead to its thermal stabilization, restore the correctly folded state and activate the expression of p53 target genes in cancer cells [33]. Thus, the data on melting temperatures of various mutant forms of p53 allow for estimations of reactivation levels when screening small-molecule stabilizers.
4. Conclusions
Hotspot mutants of the p53 core domain are of considerable interest for the field of cancer research. In this study, we aimed to examine the effect of a series of missense p53 mutations on protein thermostability. For that, we determined the melting temperatures of wild-type, Y220C, R248Q, R248W and R273H variants of the p53 core domain via differential scanning fluorimetry. We confirmed that these missense mutations result in destabilization of the p53 protein and lower its melting temperature. Knowledge of the melting behavior of mutant p53 proteins is important for revealing the mechanisms of p53 inactivation and will further facilitate the development of personalized small-molecule therapeutics targeting p53 mutants.
Abbreviations
| DBD | DNA-binding domain |
| DSF | Differential scanning fluorimetry |
| MDM2 | Mouse double minute 2 homolog |
| MDM4 | Mouse double minute 4 homolog |
| NGC | Next-generation chromatography |
| PCR | Polymerase chain reaction |
| SDS | Sodium dodecyl sulfate |
| TB | Terrific broth |
| Tm | Melting temperature |
| WT | Wild type |
Author Contributions
Conceptualization, R.K. and R.M.; methodology, D.D.; validation, R.M., V.C. and D.D.; formal analysis, R.K.; investigation, E.K.; data curation, R.M.; writing—original draft preparation, R.K.; writing—review and editing, E.B.; visualization, D.D.; supervision, R.M.; project administration, E.B.; funding acquisition, E.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data related to this study can be provided by the corresponding authors on request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by RSF grant 22-24-20034 to RM and performed according to the Russian Government Program of Strategic Academic Leadership (Priority 2030) of Kazan Federal University.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hassin O., Oren M. Drugging P53 in Cancer: One Protein, Many Targets. Nat. Rev. Drug Discov. 2022:1–18. doi: 10.1038/s41573-022-00571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soussi T., Wiman K.G. TP53: An Oncogene in Disguise. Cell Death Differ. 2015;22:1239–1249. doi: 10.1038/cdd.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bykov V.J.N., Eriksson S.E., Bianchi J., Wiman K.G. Targeting Mutant P53 for Efficient Cancer Therapy. Nat. Rev. Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y., Gorina S., Jeffrey P.D., Pavletich N.P. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 5.Joerger A.C., Fersht A.R. The P53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 6.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 7.Chasov V., Mirgayazova R., Zmievskaya E., Khadiullina R., Valiullina A., Clarke J.S., Rizvanov A., Baud M.G.J., Bulatov E. Key Players in the Mutant P53 Team: Small Molecules, Gene Editing, Immunotherapy. Front. Oncol. 2020;10:1460. doi: 10.3389/fonc.2020.01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y., Yao Y., Wei G. Unraveling the Allosteric Mechanism of Four Cancer-Related Mutations in the Disruption of P53-DNA Interaction. J. Phys. Chem. B. 2021;125:10138–10148. doi: 10.1021/acs.jpcb.1c05638. [DOI] [PubMed] [Google Scholar]
- 9.Bullock A.N., Henckel J., Fersht A.R. Quantitative Analysis of Residual Folding and DNA Binding in Mutant P53 Core Domain: Definition of Mutant States for Rescue in Cancer Therapy. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 10.Joerger A.C., Fersht A.R. Structural Biology of the Tumor Suppressor P53 and Cancer-Associated Mutants. Adv. Cancer Res. 2007;97:1–23. doi: 10.1016/s0065-230x(06)97001-8. [DOI] [PubMed] [Google Scholar]
- 11.Joerger A.C., Fersht A.R. Structural Biology of the Tumor Suppressor P53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 12.Bauer M.R., Krämer A., Settanni G., Jones R.N., Ni X., Tareque R.K., Fersht A.R., Spencer J., Joerger A.C. Targeting Cavity-Creating P53 Cancer Mutations with Small-Molecule Stabilizers: The Y220X Paradigm. ACS Chem. Biol. 2020;15:657–668. doi: 10.1021/acschembio.9b00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke J.R.S., Douglas L.R., Duriez P.J., Balourdas D.-I., Joerger A.C., Khadiullina R., Bulatov E., Baud M.G.J. Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic P53 Mutant Y220C. ACS Pharmacol. Transl. Sci. 2022;5:1169–1180. doi: 10.1021/acsptsci.2c00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedler A., Veprintsev D.B., Hansson L.O., Fersht A.R. Kinetic Instability of P53 Core Domain Mutants: Implications for Rescue by Small Molecules. J. Biol. Chem. 2003;278:24108–24112. doi: 10.1074/jbc.M302458200. [DOI] [PubMed] [Google Scholar]
- 15.Vivoli M., Novak H.R., Littlechild J.A., Harmer N.J. Determination of Protein-Ligand Interactions Using Differential Scanning Fluorimetry. J. Vis. Exp. 2014;91:51809. doi: 10.3791/51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S., Wu J.-L., Liang Y., Tang Y.-G., Song H.-X., Wu L.-L., Xing Y.-F., Yan N., Li Y.-T., Wang Z.-Y., et al. Arsenic Trioxide Rescues Structural P53 Mutations through a Cryptic Allosteric Site. Cancer Cell. 2021;39:225–239. doi: 10.1016/j.ccell.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Hamiaux C., Janssen B.J., Snowden K.C. Strigolactones. Humana; New York, NY, USA: 2021. The Use of Differential Scanning Fluorimetry to Assess Strigolactone Receptor Function; pp. 233–243. [DOI] [PubMed] [Google Scholar]
- 18.Niesen F.H., Berglund H., Vedadi M. The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions That Promote Protein Stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 19.Wu J., Song H., Wang Z., Lu M. Three Optimized Assays for the Evaluation of Compounds That Can Rescue P53 Mutants. STAR Protoc. 2021;2:100688. doi: 10.1016/j.xpro.2021.100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ang H.C., Joerger A.C., Mayer S., Fersht A.R. Effects of Common Cancer Mutations on Stability and DNA Binding of Full-Length P53 Compared with Isolated Core Domains. J. Biol. Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 21.Bullock A.N., Henckel J., DeDecker B.S., Johnson C.M., Nikolova P.V., Proctor M.R., Lane D.P., Fersht A.R. Thermodynamic Stability of Wild-Type and Mutant P53 Core Domain. Proc. Natl. Acad. Sci. USA. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemos C., Schulze L., Weiske J., Meyer H., Braeuer N., Barak N., Eberspächer U., Werbeck N., Stresemann C., Lange M., et al. Identification of Small Molecules That Modulate Mutant P53 Condensation. iScience. 2020;23:101517. doi: 10.1016/j.isci.2020.101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luwang J.W., Nair A.R., Natesh R. Stability of P53 Oligomers: Tetramerization of P53 Impinges on Its Stability. Biochimie. 2021;189:99–107. doi: 10.1016/j.biochi.2021.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Babikir H.A., Afjei R., Paulmurugan R., Massoud T.F. Restoring Guardianship of the Genome: Anticancer Drug Strategies to Reverse Oncogenic Mutant P53 Misfolding. Cancer Treat. Rev. 2018;71:19–31. doi: 10.1016/j.ctrv.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Wilcken R., Wang G., Boeckler F.M., Fersht A.R. Kinetic Mechanism of P53 Oncogenic Mutant Aggregation and Its Inhibition. Proc. Natl. Acad. Sci. USA. 2012;109:13584–13589. doi: 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Bykov V.J.N., Wiman K.G., Zawacka-Pankau J. APR-246 Reactivates Mutant P53 by Targeting Cysteines 124 and 277. bioRxiv. 2017:214049. doi: 10.1101/214049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer M.R., Jones R.N., Tareque R.K., Springett B., Dingler F.A., Verduci L., Patel K.J., Fersht A.R., Joerger A.C., Spencer J. A Structure-Guided Molecular Chaperone Approach for Restoring the Transcriptional Activity of the P53 Cancer Mutant Y220C. Future Med. Chem. 2019;11:2491–2504. doi: 10.4155/fmc-2019-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg A., Hazra J.P., Sannigrahi M.K., Rakshit S., Sinha S. Variable Mutations at the P53-R273 Oncogenic Hotspot Position Leads to Altered Properties. Biophys. J. 2020;118:720–728. doi: 10.1016/j.bpj.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palanikumar L., Karpauskaite L., Al-Sayegh M., Chehade I., Alam M., Hassan S., Maity D., Ali L., Kalmouni M., Hunashal Y., et al. Protein Mimetic Amyloid Inhibitor Potently Abrogates Cancer-Associated Mutant P53 Aggregation and Restores Tumor Suppressor Function. Nat. Commun. 2021;12:3962. doi: 10.1038/s41467-021-23985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joerger A.C., Ang H.C., Veprintsev D.B., Blair C.M., Fersht A.R. Structures of P53 Cancer Mutants and Mechanism of Rescue by Second-Site Suppressor Mutations. J. Biol. Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 31.Joerger A.C., Fersht A.R. The Tumor Suppressor P53: From Structures to Drug Discovery. Cold Spring Harb. Perspect. Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiman K.G. Pharmacological Reactivation of Mutant P53: From Protein Structure to the Cancer Patient. Oncogene. 2010;29:4245–4252. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- 33.Liu X., Wilcken R., Joerger A., Chuckowree I., Spencer J., Fersht A. Abstract 2472: Small-Molecule-Induced Reactivation of Mutant P53 in Cancer Cells. Nucleic Acids Res. 2013;73:2472. doi: 10.1158/1538-7445.AM2013-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to this study can be provided by the corresponding authors on request.