Abstract
Background and Objectives:
Uropathogenic Escherichia coli (UPEC) is divided into different phylogenetic groups that differ in their antibiotic resistance patterns, serogroups and pathogenicity. This study aimed to investigate the prevalence of phylogenetic groups of UPEC isolates and their relationship with serogroups and virulence factors in patients with UTIs.
Materials and Methods:
Of the 412 urine samples tested a total of 150 UPEC were isolated and confirmed with PCR using 16S rRNA gene. Antibiotic resistance of the isolates was tested using disk diffusion method and the isolates were divided into phylogenetic groups by the quadruplex PCR method. The prevalence of serogroups and virulence genes were investigated using multiplex PCR.
Results:
87 (58%) of the isolates belonged to phylogroup B2. Virulence genes fimH (95.3%), aer (49.3%) and serogroups O8 (22.3%), O25 (21.5%) showed the highest prevalence. The lowest drug resistance was observed against imipenem (4.6%) and meropenem (3.3%). The prevalence of multidrug-resistant and extended-spectrum beta-lactamases isolates were 60% and 61.3%, respectively. We also found a significant relationship between phylogenetic groups, serogroups and virulence factors among our isolates.
Conclusion:
The high abundance of phylogenetic group B2, serogroups O8 and O25, and virulence genes fimH and aer indicate their importance in the pathogenesis of UPEC in this country.
Keywords: Urinary tract infections, Uropathogenic Escherichia coli, Phylogenetic groups, Serogroup, Drug resistance, Virulence factors
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is the main causative agent (75–90%) of urinary tract infections (UTIs) (1). UPECs are diverting from their commensal status in gastrointestinal (GI) tract, grow and persist in the urinary tract and may carry diverse virulence factors and pathogenic strategies, which allow them to infect and cause infection in the urinary tract (2). The high incidence of UTI highlights the importance of investigating the virulence properties of UPEC.
Today, UTIs have become very difficult to treat due to their high antimicrobial resistance of UPEC against commonly used antibacterial drugs and causing a serious challenge in management of the infection (3). Extended-spectrum β-lactamases (ESBLs) are plasmid-encoded enzymes that are resistant to penicillins, broad-spectrum cephalosporins, and monobactams. In addition, ESBL-producing isolates are resistant to aminoglycosides, quinolones, tetracyclines, nitrofurantoin, and trimethoprim-sulfamethoxazole. The multidrug-resistant (MDR) phenotype is due to the presence of large plasmids that usually carry resistance genes for β-lactams, quinolones, aminoglycosides, and cotrimoxazole (4). Broad-spectrum antibiotics are commonly used in the treatment of UTIs; however, the inappropriate and excessive use of these antibiotics has led to a widespread increase in antimicrobial resistance in recent years. The emergence of resistant strains leads to treatment failure of UTI and causes serious clinical complications (5).
Clermont et al. In 2000, developed a triple PCR method using chuA, yjaA genes, and DNA fragment TspE4.C2, which divided E. coli into four main phylogenies groups A, B1, B2, and D (6). In 2013, these researchers developed an improved phylogenetic group by adding a new gene called arpA to their previous three markers, resulting in a quadruple PCR by which E. coli strains were divided into eight phylogenetic groups A, B1, B2, C, D, E, F, and clade (7).
E. coli contains surface antigens, O (lipopolysaccharide), H (flagellum), and in some cases K (capsule), contribute to serological typing (8). Variation in the structure of the O antigen makes O-serotyping an important component in the typing of E. coli strains in taxonomic and epidemiology fields and has made it an essential tool in prevalence and surveillance research. In the 1940s, Fritz Kaufmann and colleagues developed the E. coli serotyping scheme, and so far, 187 O antigens have been identified and numbered 1 to 187. Serogroups O31, O47, O67, O72, O94, and O122 are no longer known, so there are currently 181 O antigens in E. coli (9). In previous studies, it has been reported that UPEC strains most belong to serogroups O1, O2, O4, O6, O7, O8, O15, O16, O18, O21, O22, O25, O75, and O83 (10, 11).
E. coli genome consists of the core nucleus genome and the mobile gene pool, which are involved in determining whether a bacterium is a pathotype or ecotype. UPEC isolates require the help of virulence genes to colonize or invade host cells, escape or disrupt the host immune system, damage host tissue, and/or stimulate inflammatory responses (9). UPEC strains have significant genetic diversity that gives bacteria the ability to colonize and persist in the patient's urinary tract, especially in highly immunocompromised individuals. These virulence factors include both structural (Fimbriae, pili, curli and flagella) and secretory (toxins and iron sequestering systems), which are related to the colonization and persistence of bacteria in the urinary system. In addition, E. coli strains that cause UTIs have been shown to have a higher prevalence of virulence genes than commensal E. coli strains (1, 12).
To the best of our knowledge, this study is the first complete effort that aims to determine the phylogenetic groups, the prevalence of serogroups, important virulence factors and antibiotic resistance of UPEC strains isolated from outpatients with UTIs in Baghdad, Iraq. Efforts was also made to identify any relationship that may exists between these factors.
MATERIALS AND METHODS
Bacterial culture and identification.
In this study, midstream urine samples were taken from 420 patients with community-acquired (CA) UTI referred to Ibn Balady hospital, Baghdad during two years 2018 and 2019. One hundred and fifty of these samples yielded UPEC. Among patients, 90 were females aged between 4 months and 78 years (median, 42 years) and 60 patients were males aged between 1 year and 70 years (median, 51 years). The microscopic observation of urine (presence of more than three pus cells in each microscopic field) and Gram staining was considered as an early sign of UTI infection. Urine samples were cultured separately on nutrient agar, blood agar, and eosin methylene blue (EMB) (Merck, German) medium and plates with a growth of more than 105 colony forming units (CFU) per ml were considered as positive samples for UTI. An uninoculated culture medium was used as a negative control in all sample processing. Presumptive identification of UPEC strains were done Analytical Profile Index (API 20E) kit (bioMerieux, France) and confirmed using PCR (16S ribosomal RNA gene). The identified isolates were cultured in Lysogeny broth (LB) (Merck, German) and after incubation for 18–24 hours, kept in 20% glycerol at −70°C until they were tested.
Antimicrobial resistance.
Antibiotic resistance was determined by the disk diffusion method according to the criteria of the European Committee for Antimicrobial Suspension Test (EUCAST) version 6.0. According to the size of the growth inhibition of each antibiotic disk on Muller-Hinton agar (Merck, German) and based on the CLSI (2020) standard, the results were recorded as resistant or sensitive. The standard strain of E. coli ATCC25922 was used as quality control. Strains with resistance to three or more classes of antibiotics were defined as MDR. The production of ESBL using the double-disk synergy test was phenotypically confirmed according to EUCAST guidelines (8). An augmentin (amoxicillin/clavulanate) disc with third-generation cephalosporin discs (cefotaxime, ceftazidime) was placed 20 mm from center to center on the Muller-Hinton agar plate on opposite sides of the center plate. The plates were incubated for 18–24 hours at 37°C. All strains with a zone inhibition diameter of ≥ 5 mm from the edge of the third-generation cephalosporin inhibition zone toward the augmentin disc were positively interpreted for ESBL production. According to CLSI instructions, Klebsiella pneumoniae ATCC 700603 was used as positive control and E. coli ATCC 25922 as a negative control for the ESBL test (13). In all, 11 classes of antibiotics including 19 antibiotics (Bioanalyse, Turkey) were used. These included macrolides (erythromycin, ER; 10 μg), quinolones (nalidixic acid, NA; 30 μg), penicillins (ticarcillin, TI; 75 μg, ampicillin, AM; 25 μg, piperacillin, PI; 30 μg, amoxicillin / clavulanate (augmentin, AMC); 20 μg/10 μg), cephalosporins (cefepime, CPM; 30 μg, cefotaxime, CTX; 30 μg, ceftazidime, CAZ; 30 μg), carbapenems (imipenem, IPM; 10 μg, meropenem, MEM; 10 μg), fluoroquinolones (norfloxacin, NOR; 10 μg, ciprofloxacin, CIP; 5 μg), nitrofurans (nitrofurantoin, NIT; 300 μg), aminoglycoside (amikacin, AK; 30 μg, gentamicin, GEN; 10 μg), chloramphenicol (chloramphenicol, C; 30 μg), sulfonamides (co-trimoxazole, COT; 30 μg) and monobactam (aztreonam, ATM; 30 μg).
DNA extraction and confirmation of E. coli.
DNA extraction was performed according to the instructions of the Genomic DNA Minikit (Invitrogen, USA), and the purity of the DNA was measured by spectrophotometer (Eppendorf, German) at a wavelength of 260 nm. All primers (Merck, German) used in this study were blasted at the NCBI site. The 16S ribosomal RNA gene was used to confirm the identity of the isolates. Classification of UPEC strains into phylogenetic groups was performed by the quadruplex PCR method presented by Clermont et al. in 2013. Since the annealing temperatures of the PCR program for serogroups and virulence primers were somewhat similar, serogroups and virulence primers were divided into four groups and multiplex PCR (Bio-Red, German) was used to detect their prevalence. The PCR products were analyzed in 1%–1.5% agarose gel of electrophoresis. The primers sequence and annealing temperature of each set are described in Table 1.
Table 1.
The primer sequences used for phylogenetic grouping, serogrouping and detection of virulence factors, and their annealing temperature.
| Set | Primer name | Primer sequence (5’-3’) | Size of product (bp) | Ref. | Annealing temperature (°C) |
|---|---|---|---|---|---|
| A | ChuA | F: GAC GAA CCA ACG GTC AGG AT | 279 | (6) | 54 |
| R: TGC CGC CAG TAC CAA AGA CA | |||||
| YjaA | F: TGAAGTGTCAGGAGACGCTG | 211 | (14) | ||
| R: ATG GAG AAT GCG TTC CTC AAC | |||||
| TspE4.C2 | F: GAG TAA TGT CGG GGC ATT CA | 152 | (6) | ||
| R: CGC GCC AAC AAA GTA TTA CG | |||||
| AceK | F: AACGCTATTCGCCAGCTTGC | 400 | (7) | ||
| R: TCTCCCCATACCGTACGCTA | |||||
| B | ArpAgpE | F: GATTCCATCTTGTCAAAATATGCC | 301 | (7) | 50 |
| R: GAAAAGAAAAAGAATTCCCAAGAG | |||||
| trpAgpC | F: AGTTTTATGCCCAGTGCGAG | 219 | (7) | ||
| R: TCTGCGCCGGTCACGCCC | |||||
| trpBA | F: CGGCGATAAAGACATCTTCAC | 489 | (7) | ||
| R: GCAACGCGGCCTGGCGGAAG | |||||
| C | fimH | F: TGCAGAACGGATAAGCCGTGG | 508 | (15) | 62 |
| R: GCAGTCACCTGCCCTCCGGTA | |||||
| hly | F: AACAAGGATAAGCACTGTTCTGGCT | 1177 | (15) | ||
| R: ACCATATAAGCGGTCATTCCCGTCA | |||||
| afa | F: GCTGGGCAGCAAACTGATAACTCTC | 750 | (15) | ||
| R: CATCAAGCTGTTTGTTCGTCCGCCG | |||||
| D | papC | F: GTGGCAGTATGAGTAATGACCGTTA | 200 | (15) | 60 |
| R: ATATCCTTTCTGCAGGGATGCAATA | |||||
| aer | F: TACCGGATTGTCATATGCAGACCGT | 602 | (15) | ||
| R: AATATCTTCCTCCAGTCCGGAGAAG | |||||
| cnf1 | F: AAGATGGAGTTTCCTATGCAGGAG | 498 | (15) | ||
| R: CATTCAGAGTCCTGCCCTCATTATT | |||||
| E | O1 | F: GTGAGCAAAAGTGAAATAAGGAACG | 1098 | (16) | 55 |
| R: CGCTGATACGAATACCATCCTAC | |||||
| O2 | F: AGTGAGTTACTTTTTAGCGATGGAC | 770 | (16) | ||
| R: AGTTTAGTATGCCCCTGACTTTGAA | |||||
| O6 | F: GGATGACGATGTGATTTTGGCTAAC | 783 | (16) | ||
| R: TCTGGGTTTGCTGTGTATGAGGC | |||||
| O7 | F: CTATCAAAATACCTCTGCTGGAATC | 610 | (16) | ||
| R: TGGCTTCGAGATTAAACCTATTCCT | |||||
| O8 | F: CCAGAGGCATAATCAGAAATAACAG | 448 | (16) | ||
| R: GCAGAGTTAGTCAACAAAAGGTCAG | |||||
| O16 | F: GGTTTCAATCTCACAGCAACTCAG | 302 | (16) | ||
| R: GTTAGAGGGATAATAGCCAAGCGG | |||||
| O75 | F: GAGATATACATGGGGAGGTAGGCT | 511 | (16) | ||
| R: ACCCGATAATCATATTCTTCCCAAC | |||||
| F | O21 | F: CTGCTGATGTCGCTATTATTGCTG | 209 | (16) | 56 |
| R: TGAAAAAAAGGGAAACAGAAGAGCC | |||||
| O4 | F: TTGTTGCGATAATGTGCATGTTCC | 664 | (16) | ||
| R: AATAATTTGCTATACCCACACCCTC | |||||
| O15 | F: TCTTGTTAGAGTCATTGGTGTATCG | 183 | (16) | ||
| R: ATAAAACGAGCAAGCACCACACC | |||||
| O18 | F: GTTCGGTGGTTGGATTACAGTTAG | 551 | (16) | ||
| R: CTACTATCATCCTCACTGACCACG | |||||
| O22 | F: TTCATTGTCGCCACTACTTTCCG | 468 | (16) | ||
| R: GAAACAGCCCATGACATTACTACG | |||||
| O25 | F: AGAGATCCGTCTTTTATTTGTTCGC | 230 | (16) | ||
| R: GTTCTGGATACCTAACGCAATACCC | |||||
| O83 | F: GTACACCAGGCAAACCTCGAAAG | 362 | (16) | ||
| R: TTCTGTAAGCTAATGAATAGGCA | |||||
| 16S rRNA | F: AGAGTTTGATCMTGGCTCAG | 919 | (17) | 62 | |
| R: CCGTCAATTCATTTGAGTTT |
Statistical analyses.
Data were analyzed using SPSS22 software (IBM, Armonk, NY, USA). The relationship between phylogenetic factors, serogroups, virulence genes, and antibiotic resistance was examined by Chi-squares or exact Fisher test. Statistically, P values less than 0.05 were considered significant.
RESULTS
Study population.
Distribution of identified UPEC isolates; in women and men was 90 (60%) and 60 (40%), respectively. A significant relationship was observed between the prevalence of UPEC and the patient's gender (P <0.001). The prevalence of UPEC isolates based on patient gender and age showed that UPEC isolates were higher in women than men in all age groups, except in age group 40–50 years. UTI showed its highest incidence in age groups 0–10 years (42%) and 20–30 years (28%) old.
Antibiotic resistance.
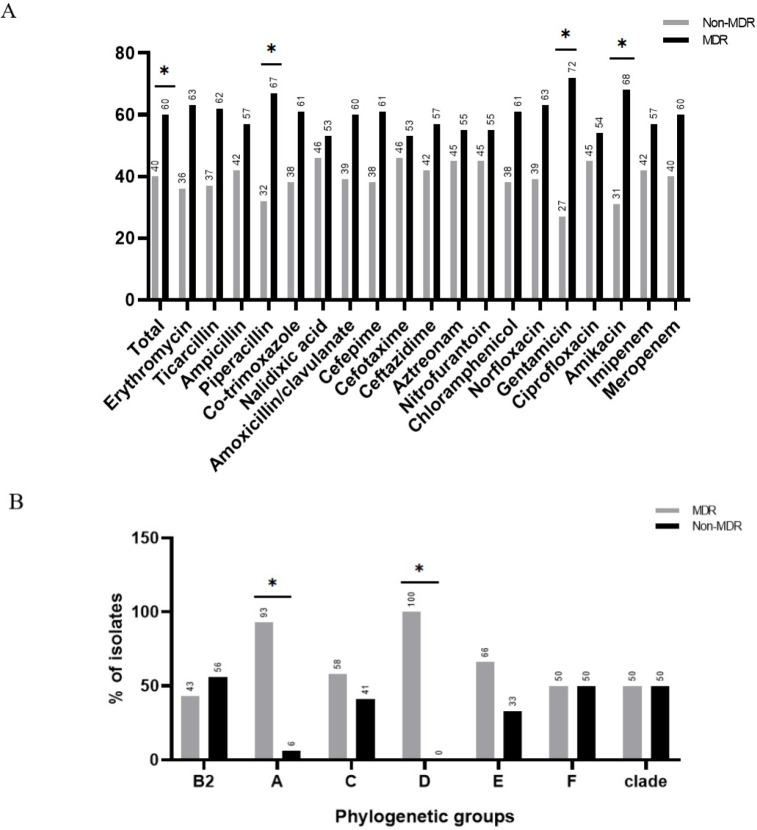
The results of antibiotic resistance showed resistance to different antibiotics ranging from meropenem (3.3%) to erythromycin (92.6%). Among the nineteen antibiotics studied, eleven included: erythromycin, ticarcillin, ampicillin, piperacillin, co-trimoxazole, nalidixic acid, amoxicillin-clavulanate, cefepime, cefotaxime, ceftazidime, and aztreonam were showed above 50% resistance. Ninety strains (60%) were resistant to three classes or more of antibiotics and were regarded as multi-drug resistant (MDR). The level of resistance to all classes of antibiotics was significantly higher among MDR strains than non-MDRs (Fig. 1A). Ninety-two (61.3%) of strains were ESBL positive. Strains resistant to co-trimoxazole, ceftazidime, and aztreonam were found to carry significantly more ESBL resistance genes (Table 2).
Fig. 1.
Prevalence of multidrug resistance (MDR) between strains resistant to different antibiotics (A) and phylogenetic groups (B).
Table 2.
Prevalence of antibiotic resistance among ESBL-positive and ESBL-negative UPEC strains. Only statistically significant values (p <0.05) are shown.
| Antibiotics | Total resistance R (%) | ESBL R (%) | Non-ESBL R (%) | P-Value |
|---|---|---|---|---|
| Erythromycin | 139 (92.6) | 81 (58.2) | 58 (41.7) | |
| Ticarcillin | 133 (88.6) | 92 (69.1) | 41 (30.8) | |
| Ampicillin | 132 (88) | 88 (66.6) | 44 (33.3) | |
| Piperacillin | 132 (88) | 85 (64.3) | 47 (35.6) | |
| Co-trimoxazole | 130 (86.6) | 90 (69.2) | 40 (30.7) | 0.021 |
| Nalidixic acid | 129 (85) | 76 (58.9) | 53 (41) | |
| Amoxicillin / clavulanate | 127 (84.6) | 71 (55.9) | 56 (44) | |
| Cefepime | 95 (63.3) | 54 (56.8) | 41 (43.1) | |
| Cefotaxime | 88 (58.6) | 40 (45.4) | 48 (54.5) | |
| Ceftazidime | 83 (55.3) | 64 (77.1) | 19 (22.8) | 0.011 |
| Aztreonam | 80 (53.3) | 55 (68.7) | 25 (31.2) | 0.017 |
| Nitrofurantoin | 68 (45.3) | 33 (48.5) | 35 (51.4) | |
| Chloramphenicol | 63 (42) | 40 (63.4) | 23 (36.5) | |
| Norfloxacin | 38 (25.3) | 21 (55.2) | 17 (44.7) | |
| Gentamicin | 36 (24) | 18 (50) | 18 (50) | |
| Ciprofloxacin | 35 (23.3) | 20 (57.1) | 15 (42.8) | |
| Amikacin | 16 (10.6) | 9 (56.2) | 7 (43.7) | |
| Imipenem | 7 (4.6) | 5 (71.4) | 2 (28.5) | |
| Meropenem | 5 (3.3) | 4 (80) | 1 (20) |
Prevalence of phylogenetic groups, serogroups, and virulence factors.
87 (58%) strains belonged to phylogenetic group B2 followed by phylogroups A (33 strains; 22%), C (12 strains; 8%), D (8 strains; 5.3%), E (6 strains; 4%), F (2 strains; 1.3%) and clade (2 strains; 1.3%). Phylogenetic group B1 was not found among the UPEC strains in this study.
More than 92% of the strains belonged to 14 serogroups. Serogroup O8 (31 strains; 22.3%) accounted for the most prevalent serogroup among the strains. Serogroups O6, O18, and O75 were not present among the strains.
All virulence genes were found among the strains. fimH (143 strains; 95.3%) and afa (21 strains; 14%) showing the highest and lowest frequencies among the strains.
Antibiotic resistance and phylogenetic groups.
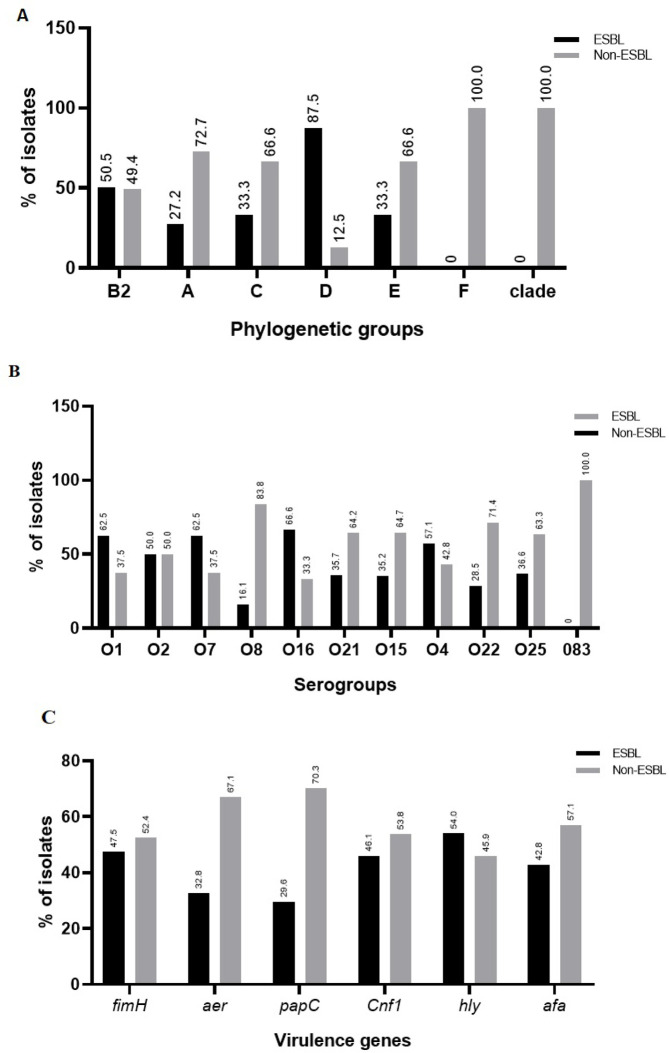
Phylogenetic groups B2 and A showed at least one resistant strain to all antibiotics studied (Table 3). The highest prevalence of ESBL strains in phylogenetic groups is shown in B2 and D. Distribution of the phylogenetic groups among the ESBL-positive and ESBL-negative strains are given in Fig. 2A.
Table 3.
Comparison of antibiotic resistance patterns between different phylogenetic groups. The statistically significant values (p <0.05) are shown in bold numbers.
| Antibiotics | Phylogenetic groups | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| B2 87 (58%) | A 33 (22%) | C 12 (8%) | D 8 (5.3%) | E 6 (4%) | F 2 (1.3%) | Clade 2 (1.3%) | |
| Erythromycin | 81 (93.1) | 29 (87.8) | 11 (91.6) | 7 (87.5) | 5 (83.3) | 2 (100) | 2 (100) |
| Ticarcillin | 75 (86.2) | 31 (93.9) | 10 (83.3) | 8 (100) | 5 (83.3) | 2 (100) | 1 (50) |
| Ampicillin | 79 (90.8) | 27 (81.8) | 10 (83.3) | 6 (75) | 5 (83.3) | 2 (100) | 2 (100) |
| Piperacillin | 75 (86.2) | 30 (90.9) | 11 (91.6) | 7 (87.5) | 5 (83.3) | 2 (100) | 1 (50) |
| Co-trimoxazole | 77 (88.5) | 27 (81.8) | 12 (100) | 7 (87.5) | 2 (33.3) | 2 (100) | 2 (100) |
| Nalidixic acid | 75 (86.2) | 25 (75.7) | 11 (91.6) | 8 (100) | 6 (100) | 1 (50) | 2 (100) |
| Amoxicillin / clavulanate | 74 (85) | 29 (87.8) | 9 (75) | 5 (62.5) | 6 (100) | 2 (100) | 1 (50) |
| Cefepime | 56 (64.3) | 17 (51.5) | 7 (58.3) | 6 (75) | 4 (66.6) | 2 (100) | 2 (100) |
| Cefotaxime | 51 (58.6) | 18 (54.5) | 6 (50) | 5 (62.5) | 4 (66.6) | 1 (50) | 2 (100) |
| Ceftazidime | 48 (55.1) | 15 (45.4) | 7 (58.3) | 4 (50) | 4 (66.6) | 2 (100) | 2 (100) |
| Aztreonam | 47 (54) | 21 (63.6) | 5 (41.6) | 3 (37.5) | 3 (50) | 1 (50) | - |
| Nitrofurantoin | 42 (48.2) | 16 (48.4) | 4 (33.3) | 4 (50) | 2 (33.3) | - | - |
| Chlorampheni-col | 34 (39) | 12 (36.3) | 5 (41.6) | 3 (37.5) | 4 (66.6) | 1 (50) | 2 (100) |
| Norfloxacin | 22 (25.2) | 8 (24.2) | 5 (41.6) | 2 (25) | 1 (16.6) | - | - |
| Gentamicin | 19 (21.8) | 9 (27.2) | 3 (25) | 2 (25) | 3 (50) | 1 (50) | - |
| Ciprofloxacin | 20 (22.9) | 8 (24.2) | 4 (33.3) | 2 (25) | 1 (16.6) | - | - |
| Amikacin | 10 (11.4) | 3 (9) | - | 1 (12.5) | 2 (33.3) | - | - |
| Imipenem | 4 (4.5) | 2 (6) | 1 (8.3) | - | - | - | - |
| Meropenem | 1 (1.1) | 4 (12.1) | - | - | - | - | - |
Fig. 2.
Distribution of ESBL-positive strains among phylogenetic groups (A), serogroups (B), and virulence genes (C).
The distribution of MDR strains were the highest in phylogenetic groups D, A, E and C. The prevalence of MDR strains was less prevalent in the phylogenetic group of B2. The rate of MDR / non-MDR strains was equal in F and clade groups Fig. 1B.
Antibiotic resistance and serogroups.
Strains belonging to serogroup O15 showed the highest level of resistance to almost all antibiotics tested. Similar pattern was found among serogroups O8 and O25 that were resistant to all antibiotics, except imipenem and serogroup O21 which was also resistant to all antibiotics except meropenem (Table 4). ESBL-positive strains had the highest prevalence among serogroups O8, O15, O21, O22 and O25. ESBL-negative strains on the other hand had the highest prevalence among serogroups O1, O4, O7 and O16 (Fig. 2B).
Table 4.
Prevalence of antibiotic resistance strains among different serogroups. The statistically significant values (p <0.05) are shown in bold numbers.
| Antibiotics | Serogroups | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| O1 8 (5.7%) | O2 12 (8.6%) | O7 8 (5.7%) | O8 31 (22.3%) | O16 3 (3.1%) | O21 14 (10%) | O15 17 (12.2%) | O4 7 (5%) | O22 7 (5%) | O25 30 (21.5%) | O83 2 (1.4%) | |
| Erythromycin | 8 (100) | 11 (91.6) | 7 (87.5) | 28 (90.3) | 3 (100) | 13 (92.8) | 17 (100) | 7 (100) | 3 (42.8) | 27 (90) | 1 (50) |
| Ticarcillin | 8 (100) | 9 (75) | 6 (75) | 30 (96.7) | 3 (100) | 12 (85.7) | 12 (70.5) | 6 (85.7) | 7 (100) | 27 (90) | 2 (100) |
| Ampicillin | 8 (100) | 11 (91.6) | 5 (62.5) | 28 (90.3) | 3 (100) | 12 (85.7) | 17 (100) | 7 (100) | 3 (42.8) | 26 (86.6) | 1 (50) |
| Piperacillin | 8 (100) | 9 (75) | 8 (100) | 30 (96.7) | 3 (100) | 13 (92.8) | 12 (70.5) | 6 (85.7) | 7 (100) | 27 (90) | 1 (50) |
| Co-trimoxazole | 6 (75) | 8 (66.6) | 8 (100) | 26 (83.8) | 3 (100) | 13 (92.8) | 15 (88.2) | 5 (71.4) | 7 (100) | 26 (86.6) | 2 (100) |
| Nalidixic acid | 6 (75) | 7 (58.3) | 6 (75) | 29 (93.5) | 2 (66.6) | 14 (100) | 14 (82.3) | 7 (100) | 2 (28.5) | 29 (96.6) | 2 (100) |
| Amoxicillin/clavulanate | 7 (87.5) | 11 (91.6) | 6 (75) | 27 (87) | 2 (66.6) | 13 (92.8) | 16 (94.1) | 4 (57.1) | 3 (42.8) | 28 (93.3) | - |
| Cefepime | 6 (75) | 6 (50) | 5 (62.5) | 16 (51.6) | 1 (33.3) | 11 (78.5) | 7 (41.1) | 4 (57.1) | 5 (71.4) | 21 (70) | 1 (50) |
| Cefotaxime | 6 (75) | 9 (75) | 5 (62.5) | 14 (45.1) | 2 (66.6) | 8 (57.1) | 8 (47) | 3 (42.8) | 3 (42.8) | 20(73.3) | - |
| Ceftazidime | 7 (87.5) | 6 (50) | 5 (62.5) | 15 (48.3) | 1 (33.3) | 7 (50) | 7 (41.1) | 2 (28.5) | 5 (71.4) | 19 (63.3) | - |
| Aztreonam | 5 (62.5) | 8 (66.6) | 4 (50) | 15 (48.3) | 1 (33.3) | 5 (35.7) | 10 (58.8) | 1 (14.2) | 1 (14.2) | 21 (70) | 1 (50) |
| Nitrofurantoin | 3 (37.5) | 6 (50) | 2 (25) | 11 (35.4) | 1 (33.3) | 3 (21.4) | 10 (58.8) | 2 (28.5) | 3 (42.8) | 13 (43.3) | 1 (50) |
| Chlorampheni-col | 5 (62.5) | 7 (58.3) | 4 (50) | 12 (38.7) | 1 (33.3) | 3 (21.4) | 6 (35.2) | 2 (28.5) | 3 (42.8) | 14 (46.6) | - |
| Norfloxacin | - | - | 4 (50) | 14 (45.1) | - | 1 (7.1) | 5 (29.4) | - | - | 12 (40) | - |
| Gentamicin | 3 (37.5) | 5 (41.6) | 3 (37.5) | 7 (22.5) | - | 2 (14.2) | 5 (29.4) | - | - | 8 (26.6) | - |
| Ciprofloxacin | - | - | 4 (50) | 11 (35.4) | - | 2 (14.2) | 5 (29.4) | - | - | 10 (33.3) | - |
| Amikacin | 3 (37.5) | 3 (25) | - | 3 (9.6) | - | 1 (7.1) | 1 (5.8) | - | - | 4 (13.3) | - |
| Imipenem | - | - | 1 (12.5) | - | - | 1 (7.1) | 1 (5.8) | - | 2 (28.5) | - | - |
| Meropenem | - | 1 (8.3) | - | 1 (3.2) | - | - | 1 (1.8) | - | 1 (14.2) | 1 (3.3) | - |
Antibiotic resistance and virulence genes.
All strains carrying afa and aer genes were sensitive to meropenem. Statistically, a significant relationship was observed between genes fimH, papC, hly and afa with antibiotic resistance (Table 5A). Most of the virulence genes were the highest prevalence among ESBL-positive strains Fig. 2C.
Table 5.
Association of strains carrying different antibiotic-resistant and virulence genes (A) and prevalence of strains carrying different virulence genes within each phylogenetic group (B).
| Virulence genes | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| fimH 143 (95.3%) | PapC 64 (42.6%) | cnf1 27 (18%) | hly 26 (17.3) | aer 74 (49.3%) | afa 21 (14%) | ||
| (A) Antibiotics | Erythromycin | 133 (95.6) | 58 (41.7) | 27 (19.4) | 24 (17.2) | 68 (48.9) | 20 (14.3) |
| Ticarcillin | 126 (94.7) | 51 (38.3) | 22 (16.5) | 24 (18) | 63 (47.3) | 20 (15) | |
| Ampicillin | 126 (95.4) | 54 (40.9) | 24 (18.1) | 24 (18.1) | 67 (50.7) | 19 (14.3) | |
| Piperacillin | 126 (95.4) | 53 (40.1) | 23 (17.4) | 26 (19.6) | 64 (48.4) | 18 (13.6) | |
| Co-trimoxazole | 127 (97.6) | 57 (43.8) | 23 (17.6) | 23 (17.6) | 68 (52.3) | 16 (12.3) | |
| Nalidixic acid | 123 (95.3) | 55 (42.6) | 22 (17) | 22 (17) | 66 (51.1) | 19 (14.7) | |
| Amoxicillin / clavula-nate | 119 (93.7) | 50 (39.3) | 23 (18.1) | 23 (18.1) | 60 (47.2) | 17 (13.3) | |
| Cefepime | 91 (95.7) | 39 (41) | 20 (21) | 16 (16.8) | 47 (49.4) | 14 (14.7) | |
| Cefotaxime | 83 (94.1) | 34 (38.6) | 18 (20.4) | 17 (19.3) | 41 (46.5) | 13 (14.7) | |
| Ceftazidime | 77 (92.7) | 35 (42.1) | 17 (20.4) | 15 (18) | 38 (45.7) | 10 (12) | |
| Aztreonam | 75 (93.7) | 36 (45) | 16 (20) | 17 (21.2) | 36 (45) | 6 (7.5) | |
| Nitrofurantoin | 65 (95.5) | 27 (39.7) | 9 (13.2) | 9 (13.2) | 31 (45.5) | 11 (16,1) | |
| Chloramphenicol | 57 (90.4) | 26 (41.2) | 14 (22.2) | 11 (17.4) | 27 (42.8) | 7 (11.1) | |
| Norfloxacin | 37 (97.3) | 18 (47.3) | 8 (21) | 9 (23.6) | 18 (47.3) | 3 (7.8) | |
| Gentamicin | 31 (86.1) | 17 (47.2) | 9 (25) | 9 (25) | 13 (36.1) | 4 (11.1) | |
| Ciprofloxacin | 34 (97.1) | 17 (48.5) | 8 (22.8) | 7 (20) | 16 (45.7) | 2 (5.7) | |
| Amikacin | 13 (81.2) | 8 (50) | 4 (25) | 6 (37.5) | 7 (43.7) | 2 (12.5) | |
| Imipenem | 7 (100) | 4 (57.1) | 2 (28.5) | 1 (14.2) | 2 (28.5) | 1 (14.2) | |
| Meropenem | 5 (100) | 1 (20) | 1 (20) | 1 (20) | - | - | |
| (B) Phylogenetic groups | B2 | ||||||
| A | 85 (97.7) | 57 (65.5) | 23 (26.4) | 21 (24.1) | 66 (75.8) | 18 (20.6) | |
| C | 32 (96.9) | 1 (3) | - | - | - | - | |
| D | 12 (100) | 4 (33.3) | 2 (16.6) | 2 (16.6) | 3 (25) | - | |
| E | 8 (100) | 1 (12.5) | 2 (25) | 3 (37.5) | 1 (12.5) | 2 (25) | |
| F | 3 (50) | 1 (16.6) | - | - | 1 (16.6) | 1 (16.6) | |
| Clade | 1 (50) | - | - | - | 1 (50) | - | |
| 2 (100) | - | - | - | 1 (50) | - | ||
Phylogenetic and virulence genes.
The distribution of virulence genes among phylogenetic groups showed that strains belonging to phylogenetic groups B2 and D carried all virulence genes tested in this study. On the other hand, strains belonging to phylogenetic group A, that was the second most prevalent group. F and clade had the lowest prevalence of virulence genes (Table 5B).
Phylogenetic / serogroups and virulence genes.
Phylogenetic groups B2 and A were found among all serogroups except O83 (Table 6A). Among virulence genes fimH gene was present in all strains belonging to serogroups O2, O16, O21, O15, O4, O22, and O83. The highest frequency of remaining virulence genes was PapC (O22), cnf1 (O4), hly (O15), aer (O21, O22), afa (O4) Table 6B.
Table 6.
Distribution of phylogroup (A) and virulence genes (B) among different serogroups.
| Serogroups | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| O1 8 (5.7%) | O2 12 (8.6%) | O7 8 (5.7%) | O8 31 (22.3%) | O16 3 (3.1%) | O21 14 (10%) | O15 17 (12.2%) | O4 7 (5%) | O22 7 (5%) | O25 30 (21.5%) | O83 2 (1.4%) | ||
| (A) Phylogenetic Groups | B2 | 3 (37.5) | 7 (58.3) | 3 (37.5) | 18 (58) | 2 (66.6) | 12 (85.7) | 14 (82.3) | 4 (57.1) | 5 (71.4) | 12 (40) | - |
| A | 1 (12.5) | 4 (33.3) | 1 (12.5) | 9 (29) | 1 (33.3) | 1 (7.1) | 2 (11.7) | 1 (14.2) | 2 (28.5) | 9 (30) | - | |
| C | 1 (12.5) | 1 (8.3) | 3 (37.5) | 1 (3.2) | - | - | - | - | - | 6 (20) | - | |
| D | - | - | 1 (12.5) | 1 (3.2) | - | - | 1 (5.8) | 2 (28.5) | - | 1 (3.3) | 2 (100) | |
| E | 1 (12.5) | - | - | 1 (3.2) | - | 1 (7.1) | - | - | - | 1 (3.3) | - | |
| F | 1 (12.5) | - | - | 1 (3.2) | - | - | - | - | - | - | - | |
| Clade | 1 (12.5) | - | - | - | - | - | - | - | - | 1 (3.3) | - | |
| (B) virulence genes | fimH | 7 (87.5) | 12 (100) | 8 (87.5) | 28 (90.3) | 3 (100) | 14 (100) | 17 (100) | 7 (100) | 7 (100) | 29 (96.6) | 2 (100) |
| papC | 2 (25) | 6 (50) | 5 (62.5) | 7 (22.5) | 1 (33.3) | 9 (64.2) | 9 (52.9) | 2 (28.5) | 5 (71.4) | 13 (43.3) | - | |
| cnf1 | 3 (37.5) | 3 (25) | 2 (25) | 3 (9.6) | 1 (33.3) | 4 (28.5) | 5 (29.4) | 3 (42.8) | - | 3 (10) | - | |
| Hly | 2 (25) | 3 (25) | - | 2 (6.4) | 1 (33.3) | 4 (28.5) | 7 (41.1) | 2 (28.5) | - | 4 (13.3) | - | |
| Aer | 3 (37.5) | 5 (41.6) | 4 (50) | 18 (58) | 1 (33.3) | 10 (71.4) | 8 (47) | 2 (28.5) | 5 (71.4) | 11 (36.6) | - | |
| Afa | - | 1 (8.33) | - | 7 (22.5) | - | 3 (21.4) | 3 (17.6) | 3 (42.8) | - | 1 (3.3) | - | |
DISCUSSION
Here we investigated the prevalence of antibiotic resistance among the UPEC strains isolated from patients with CA-UTI. The highest sensitivity of the strains was found against amikacin, meropenem, and imipenem which is consistent with other findings in Ethiopia (14), Iraq (15), and Turkey (12, 18). In contrast, high resistance was found against erythromycin, ticarcillin, ampicillin, and piperacillin, a finding which is commonly reported for UPEC strains in Iraq (15, 19), Romania (4), Turkey (12), Thailand (20), and India (21) and it could be partly due to the common use of these antibiotics for treatment of UTI in these countries. The high level of ESBL-positive UPEC in our study is of great concern and we found similar results in less developed countries (22–24). The results of multidrug resistance showed that 60% of the strains were MDR. The prevalence of MDR-UPEC strains in some countries such as Mexico 92.7% (25), Thailand 62% (23), China 77.2% (26) and southern Iraq 94% (15) was higher than our result, while Nepal 51% (2) and Turkey 34.6% (12) reported lower results than ours. In recent years, there has been an increase in drug resistance of E. coli strains around the world. Numerous factors are involved which include improper and repeated use of antibiotics, horizontal transfer of resistance genes, and mutations. The difference in ESBL levels between our study and other studies may also be due to differences in local antibiotic prescribing policy, abuse of broad-spectrum antibiotics, especially penicillin and third-generation cephalosporins, geographical differences and sample type (outpatient/inpatient) (18, 27, 28).
The prevalence of MDR-UPEC rate in Iraq was considered almost an intermediate compared to other countries, however, it is reported that Asian and African countries recorded the highest MDR-UPEC rates while the United States and European countries have the lowest rates (29). The use of prophylactic antibiotics in UTIs caused by ESBL-producing UPEC is not recommended because they contribute to the emergence and spread of antibiotic-resistant strains. Excessive use of antimicrobials in general medicine, agriculture, or veterinary medicine is among the main factors associated with the increasing in the prevalence of bacterial resistance to antibiotics in a society (30). On the other hand, some factors may be related to the host, including risk factors associated with urinary catheterization, previous hospitalization, previous antibiotic treatment, age and previous urinary tract infection (31). In addition, according to the guidelines of the World Health Organization, the inappropriate use of antibiotics has been shown to play an important role in the emergence of MDR bacteria. For instance, in the United States alone during 2014, more than 266 million antimicrobial prescriptions were given, about 30 percent of which were later described as unnecessary. Similarly, in United Kingdom, 69% of all prescriptions contained antimicrobials, and more than 20% were described as inappropriate. In Iraq, about 70% of all medical prescriptions contain antibiotics, but there is insufficient data considering the real need to use them and their health consequences (19).
In our study, phylogenetic group B2 had the highest prevalence among the UPEC strains with strains belonging to group A being the second most prevalent among the strains. These results are consistent with other studies in Iran (32) and Iraq (15). Commensal strains may become virulent by random functional point mutations adaptive for pathogenic environments and a genomic deletion that enhances pathogenicity. In furtherance to that, it is unclear whether E. coli strains should be defined as commensals or pathogens based wholly on the source of the specimen and/or phylogenetic group they belong to since phylogroup A can cause extraintestinal infections in immunocompromised hosts (33, 34). In our study, phylogenetic groups F and clade had the lowest frequency compared to other phylogenetic groups. Cryptic clades are primarily associated with peripheral E. coli; therefore, the observed results may be related to the lack of practicing good hygienic measures. In addition, the lack of phylogenetic group B1 is probably because strains of this group are considered as commensal strains (4). As stated by Clermont et al. (2013), unallocated strains are likely to be the result of large-scale recombination events from two different groups or genome flexibility due to gene loss and amplification (7). The different distribution of phylogenetic groups in different studies may depend on differences in the geographical area, patient health status, use of antibiotics and differences in research design and sample size of studies (20, 35).
In the present study, fourteen serogroups associated with UTI constituted more than 92% of the strains with serogroups O8, O25, and O15 having the highest prevalence suggesting the spread of three dominant clonal groups of UPEC in Baghdad. In other studies serogroups O25 and O15 (Iran) (8, 17), O8, O25 and O21 (Iraq) (16) and O6 (India) (21) showed the highest prevalence. Momtaz et al. (17) in Iran reported that serogroups O25 and O15 had the highest prevalence whereas serogroups O75, O18 with one strain each having the lowest prevalence. In our study, O83 had the lowest prevalence and strains of this serogroup had only fimH gene, indicating that this serogroup may be less important in developing UTI in Baghdad. The presence of different serogroups in different countries could be related to autochthonous bacteria that are part of the intestinal biota which may incidentally cause UTI (36).
Identification of virulence factors encoded by UPEC is of particular importance in the pathogenesis and the severity of UTIs in the host. Virulence factors can also be used as targets for the development of vaccines and drugs against infection (37). UPEC strains have different abilities to cause UTI depending on the type of virulence genes carried by plasmids, pathogenicity islands, transposons, and bacteriophages (9). The distribution of virulence genes among our UPEC strains showed that fimH and aer genes were the most common genes similar to those reported in Thailand (20, 30), Iraq (15) and Ethiopia (37). A comparison of the prevalence of virulence genes among UPEC in our student and those isolated in southern Iraq showed that aer, papC, cnf1 and hly genes were more prevalent in Baghdad (15). The presence of fimH gene among a high percentage of UPEC strains in our study may be due to the key importance of this gene in the early infection stage. fimH is responsible for identifying the surface receptors on bladder cells. This gene helps colonization of bacteria in the urinary tract and protects it against rinsing by the urine stream. The amount of iron in the urinary tract is limited, and the acquisition of sufficient iron is a vital requirement for the survival of UPEC (38). The UPEC supplies the iron it needs by encoding the aer gene, a gene that is strongly associated with pyelonephritis, cystitis, and bacteremia and is involved in bacterial survival and colonization (38, 39). Differences in the prevalence of virulence factors, including fimH and aer in UPEC in our study and other studies such as Iran (40) and Turkey (18) could be due to geographical differences, climate, public health, diet, sampling techniques, etiquette, and traditions. However, negative PCR does not always mean the absence of virulence gene because mutation of the gene may lead to negative PCR (37). Due to the expression of the fimH gene in most UPEC strains, it can be considered as a potential candidate for the development of a vaccine against UTI. In fact, some studies suggest that anti-fimH antibodies can act to prevent UPEC colonization in the urinary system (9).
The findings of this study show that UPEC strains with high pathogenic potential are common in patients with UTIs in Baghdad and measures should be taken to combat these pathogens through the design and implementation of prevention and control strategies. If possible, it is better to study samples from wider community in different parts of the city and different provinces. Also, it is desirable to study more virulence factor genes. These findings may help to understand the pathogenicity of clonal groups of UPEC in this country to rationalize a proper management of UTI patients, thus reducing the misuse of antibiotics.
CONCLUSION
Our results showed that the prevalence of different phylogenetic groups within serogroups of UPEC. The high prevalence of phylogenetic group A among our strains not carrying many virulence genes suggest that host factors can be more important a factor for development of UTI by E. coli in this country. The high prevalence of antibiotic resistance among UPEC is also pointing as a serious challenge of treatment of UTI in this country.
REFERENCES
- 1.Nascimento JAS, Santos FF, Valiatti TB, Santos-Neto JF, M Santos AC, Cayô R, et al. Frequency and diversity of hybrid Escherichia coli strains isolated from urinary tract infections. Microorganisms 2021; 9: 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah C, Baral R, Bartaula B, Shrestha LB. Virulence factors of uropathogenic Escherichia coli (UPEC) and correlation with antimicrobial resistance. BMC Microbiol 2019; 19: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambite I, Butler D, Wan MLY, Rosenblad T, Tran TH, Chao SM, et al. Molecular determinants of disease severity in urinary tract infection. Nat Rev Urol 2021; 18: 468–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristea VC, Gheorghe I, Czobor Barbu I, Popa LI, Ispas B, Grigore GA, et al. Snapshot of phylogenetic groups, virulence, and resistance markers in Escherichia coli uropathogenic strains isolated from outpatients with urinary tract infections in Bucharest, Romania. Biomed Res Int 2019; 2019: 5712371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serretiello E, Folliero V, Santella B, Giordano G, Santoro E, De Caro F, et al. Trend of bacterial uropathogens and their susceptibility pattern: study of single academic high-volume center in Italy (2015–2019). Int J Microbiol 2021; 2021: 5541706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000; 66: 4555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013; 5: 58–65. [DOI] [PubMed] [Google Scholar]
- 8.Noie Oskouie A, Hasani A, Ahangarzadeh Rezaee M, Soroush Bar Haghi MH, Hasani A, Soltani E. A relationship between O-serotype, antibiotic susceptibility and biofilm formation in Uropathogenic Escherichia coli. Microb Drug Resist 2019; 25: 951–958. [DOI] [PubMed] [Google Scholar]
- 9.Rezatofighi SE, Mirzarazi M, Salehi M. Virulence genes and phylogenetic groups of uropathogenic Escherichia coli isolates from patients with urinary tract infection and uninfected control subjects: a case-control study. BMC Infect Dis 2021; 21: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe CM, Salvador FA, Falsetti IN, Vieira MAM, Blanco J, Blanco JE, et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol 2008; 52: 397–406. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto S. Molecular epidemiology of uropathogenic Escherichia coli. J Infect Chemother 2007; 13: 68–73. [DOI] [PubMed] [Google Scholar]
- 12.Yılmaz EŞ, Aslantaş Ö. Phylogenetic group/subgroups distributions, virulence factors, and antimicrobial susceptibility of Escherichia coli strains from urinary tract infections in hatay. Rev Soc Bras Med Trop 2020; 53:e20190429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Furevi A, Perepelov AV, Guo X, Cao H, Wang Q, et al. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev 2020; 44: 655–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa, Ethiopia. BMC Infect Dis 2020; 20: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allami M, Bahreini M, Sharifmoghadam MR. Antibiotic resistance, phylogenetic typing, and virulence genes profile analysis of uropathogenic Escherichia coli isolated from patients in southern Iraq. J Appl Genet 2022; 63: 401–412. [DOI] [PubMed] [Google Scholar]
- 16.Mohammed EJ, Allami M, Sharifmoghadam MR, Bahreini M. Relationship between antibiotic resistance patterns and O-serogroups in uropathogenic Escherichia coli strains isolated from Iraqi patients. Jundishapur J Microbiol 2021; 14(8):e118833. [Google Scholar]
- 17.Momtaz H, Karimian A, Madani M, Dehkordi FS, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob 2013; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Düzgün AÖ, Okumuş F, Saral A, Çiçek A Ç, Cinemre S. Determination of antibiotic resistance genes and virulence factors in Escherichia coli isolated from Turkish patients with urinary tract infection. Rev Soc Bras Med Trop 2019; 52:e20180499. [DOI] [PubMed] [Google Scholar]
- 19.Al-Naqshbandi AA, Chawsheen MA, Abdulqader HH. Prevalence and antimicrobial susceptibility of bacterial pathogens isolated from urine specimens received in rizgary hospital-Erbil. J Infect Public Health 2019; 12: 330–336. [DOI] [PubMed] [Google Scholar]
- 20.Tewawong N, Kowaboot S, Pimainog Y, Watanagul N, Thongmee T, Poovorawan Y. Distribution of phylogenetic groups, adhesin genes, biofilm formation, and antimicrobial resistance of uropathogenic Escherichia coli isolated from hospitalized patients in Thailand. PeerJ 2020; 8: e10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Kaur N, Malhotra S, Madan P, Ahmad W, Hans C. Serotyping and antimicrobial susceptibility pattern of Escherichia coli isolates from urinary tract infections in pediatric population in a tertiary care hospital. J Pathog 2016; 2016: 2548517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrestha R, Khanal S, Poudel P, Khadayat K, Ghaju S, Bhandari A, et al. Extended spectrum β-lactamase producing uropathogenic Escherichia coli and the correlation of biofilm with antibiotics resistance in Nepal. Ann Clin Microbiol Antimicrob 2019; 18: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halabi MK, Lahlou FA, Diawara I, El Adouzi Y, Marnaoui R, Benmessaoud R, et al. Antibiotic resistance pattern of extended spectrum Beta Lactamase producing Escherichia coli isolated from patients with urinary tract infection in Morocco. Front Cell Infect Microbiol 2021; 11: 720701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh N, Pattnaik D, Neogi DK, Jena J, Mallick B. Prevalence of ESBL in Escherichia coli isolates among ICU patients in a tertiary care hospital. J Clin Diagn Res 2016; 10: DC19–DC22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballesteros-Monrreal MG, Arenas-Hernández MM, Enciso-Martínez Y, Martínez-de la Peña C F, Del C Rocha-Gracia R, Lozano-Zarain P, et al. Virulence and resistance determinants of uropathogenic Escherichia coli strains isolated from pregnant and non-pregnant women from two states in Mexico. Infect Drug Resist 2020; 13: 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhao S, Han L, Guo X, Chen M, Ni Y, et al. Drug resistance and virulence of uropathogenic Escherichia coli from Shanghai, China. J Antibiot (Tokyo) 2014; 67: 799–805. [DOI] [PubMed] [Google Scholar]
- 27.Lee DS, Lee SJ, Choe HS. Community-acquired urinary tract infection by Escherichia coli in the era of antibiotic resistance. Biomed Res Int 2018; 2018: 7656752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ndzime YM, Onanga R, Kassa RFK, Bignoumba M, Nguema PPM, Gafou A, et al. Epidemiology of community origin Escherichia coli and uropathogenic strains resistant to antibiotics in Franceville, Gabon. Infect Drug Resist 2021; 14: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T 2015; 40: 277–283. [PMC free article] [PubMed] [Google Scholar]
- 30.Petca RC, Negoiță S, Mareș C, Petca A, Popescu RI, Chibelean CB. Heterogeneity of antibiotics multidrug-resistance profile of uropathogens in Romanian population. Antibiotics (Basel) 2021; 10: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenney J, Hudson N, Alnifaidy H, Li JTC, Fung KH. Risk factors for aquiring multidrug-resistant organisms in urinary tract infections: a systematic literature review. Saudi Pharm J 2018; 26: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boroumand M, Naghmachi M, Ghatee MA. Detection of phylogenetic groups and drug resistance genes of Escherichia coli causing urinary tract infection in southwest Iran. Jundishapur J Microbiol 2021; 14(2):e112547. [Google Scholar]
- 33.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A 1998; 95: 8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabaté M, Moreno E, Pérez T, Andreu A, Prats G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect 2006; 12: 880–886. [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty A, Saralaya V, Adhikari P, Shenoy S, Baliga S, Hegde A. Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann Med Health Sci Res 2015; 5: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández-Chiñas U, Pérez-Ramos A, Belmont-Monroy L, Chávez-Berrocal ME, González-Villalobos E, Navarro-Ocaña A, et al. Characterization of auto-agglutinating and non-typeable uropathogenic Escherichia coli strains. J Infect Dev Ctries 2019; 13: 465–472. [DOI] [PubMed] [Google Scholar]
- 37.Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Drug resistance and plasmid profile of uropathogenic Escherichia coli among urinary tract infection patients in Addis Abeba. J Infect Dev Ctries 2018; 12: 608–615. [DOI] [PubMed] [Google Scholar]
- 38.Terlizzi ME, Gribaudo G, Maffei ME. UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol 2017; 8: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabasi M, Karam M RA, Habibi M, Mostafavi E, Bouzari S. Genotypic characterization of virulence factors in Escherichia coli isolated from patients with acute cystitis, pyelonephritis and asymptomatic bacteriuria. J Clin Diagn Res 2016; 10: DC01–DC07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghavidel M, Gholamhosseini-Moghadam T, Nourian K, Ghazvini K. Virulence factors analysis and antibiotic resistance of uropathogenic Escherichia coli isolated from patients in northeast of Iran. Iran J Microbiol 2020; 12: 223–230. [PMC free article] [PubMed] [Google Scholar]