Abstract
Eukaryotic genomics frequently revealed historical spontaneous endogenization events of external invading nucleic acids, such as viral elements. In plants, an extensive occurrence of endogenous plant pararetroviruses (EPRVs) is usually believed to endow hosts with an additional layer of internal suppressive weaponry. However, an actual demonstration of this activity remains speculative. We analyzed the EPRV component and accompanying silencing effectors of Solanum lycopersicum, documenting that intronic/intergenic pararetroviral integrations bearing inverted-repeats fuel the plant's RNA-based immune system with suitable transcripts capable of evoking a silencing response. A surprisingly small set of rearrangements explained a substantial fraction of pararetroviral-derived endogenous small-interfering (si)RNAs, enriched in 22-nt forms typically associated with anti-viral post-transcriptional gene silencing. We provide preliminary evidence that such genetic and immunological signals may be found in other species outside the genus Solanum. Based on molecular dating, bioinformatics, and empirical explorations, we propose that homology-dependent silencing emerging from particular immuno-competent rearranged chromosomal areas that constitute an adaptive heritable trans-acting record of past infections, with potential impact against the unlocking of plant latent EPRVs and cognate-free pararetroviruses.
Keywords: Caulimoviridae, epigenetics, epigenetic silencing, EPRVs, endogenized pararetrovirus, introns, long non-coding RNA, pararetrovirus, post-transcriptional gene silencing, PTGS, siRNA, RNA interference, Solanum, tomato
Significance.
In the plant kingdom, the serendipitous genomic integration of plant pararetroviruses is usually assumed to provide an immunological arsenal against cognate aggressors. However, direct evidence supporting such supposition is scarce. We recognized in tomato a strong accumulation of endogenous small-interfering (si)RNAs mapping to a surprisingly small set of transcriptionally active pararetroviral integrations arranged as inverted-repeats. These were particularly enriched in 22-nt small-interfering RNAs, previously associated with anti-viral post-transcriptional gene silencing. Based on molecular dating, bioinformatics, and empirical explorations, we propose that effective broad homology-dependent silencing emerges from these expressed pararetroviral-related repurposed chromosomal areas, presumably constituting an adaptive heritable record of past experiences.
Introduction
RNA-based silencing is essential to the survival and evolution of eukaryotes. In plants, a convoluted biomachinery drives immunity emerging from small-interfering (si)RNAs, which display sequence complementarity against invading nucleic acids such as viruses (Ding and Voinnet 2007; Ghoshal and Sanfaçon 2015). The anti-viral RNA-based response proceeds after transcription/replication of viruses and further biosynthesis of double-stranded RNA (dsRNA), subsequently diced by distinct plant dicer-like proteins (DCLs) (Deleris, et al. 2006; Gasciolli, et al. 2005). DCL processing is a crucial step because their characteristic siRNA products often mediate separate functionalities mostly depending on size (Deleris, et al. 2006; Ding and Voinnet 2007; Gasciolli, et al. 2005; Ghoshal and Sanfaçon 2015; Xie, et al. 2004). Thus, 24-nt sized siRNAs derived from DCL3 activity mediate transcriptional gene silencing (TGS) against viral mini-chromosomes (for DNA viruses like geminiviruses or pararetroviruses), through suppressive epigenetic chromatin marks such as DNA 5-methylcytosine and H3K9me2; while 21–22-nt forms produced by DCL4 and DCL2, respectively, trigger defensive post-transcriptional gene silencing (PTGS) effected through viral mRNA cleavage or translation inhibition (for both DNA and RNA viruses) (Deleris, et al. 2006; Ding and Voinnet 2007; Ghoshal and Sanfaçon 2015; Raja, et al. 2008). The biogenesis and roles of such virus-derived siRNAs (viRNAs) generated upon infection have been extensively studied in plants (Deleris, et al. 2006; Ding and Voinnet 2007; Ghoshal and Sanfaçon 2015; Sanan-Mishra, et al. 2021; Xie, et al. 2004).
Beyond viRNAs, other naturally occurring plant siRNAs accumulate endogenously under normal or stress conditions. Among them, we highlight 21–22-nt micro (mi)RNAs exerting sequence-specific mRNA regulations impacting development and stress–response (Li, et al. 2017), 22-nt siRNAs translationally repressing functional coding genes (Wu, et al. 2020), and heterochromatic 24-nt forms affecting TGS against potentially mutagenic parasitic intra-genomic transposable elements (Matzke, et al. 2015). Still, the roles of many endogenous siRNAs remain to be uncovered, particularly in crops and non-models.
Ubiquitous transposable elements are not the only alien inhabitants of plant genomes. There exists a wide-spread incidence of endogenous plant pararetroviruses (EPRVs), which presumably originate from sporadic integrations of cognate-free counterparts (Chen and Kishima 2016; Diop, et al. 2018; Geering, et al. 2014; Harper, et al. 2002; Jakowitsch, et al. 1999; Richert-Pöggeler, et al. 2021; Staginnus and Richert-Pöggeler 2006). They belong to the Caulimoviridae family, currently composed of eleven taxonomic genera (https://talk.ictvonline.org/); although considerably more may exist as inferred from extensive chromosome integrations, numerous with yet unrecovered free viruses (Diop, et al. 2018; Gong and Han 2018). Pararetroviruses encapsidate a double-stranded DNA genome and display an episomal replication cycle that does not require an integrative phase. Therefore, chromosomal integrations are thought to proceed through illegitimate recombination (Jakowitsch, et al. 1999; Richert-Pöggeler, et al. 2021; Staginnus and Richert-Pöggeler 2006). Once integrated, EPRVs presumably become inactivated by the host's silencing and are assumed to contribute to immunity (Chen and Kishima 2016; Harper, et al. 2002; Mette, et al. 2002; Richert-Pöggeler, et al. 2021; Staginnus and Richert-Pöggeler 2006). Eventually, they decay turning into relic sequences after partial deletion/truncation and/or acquisition of mutations (Diop, et al. 2018; Jakowitsch, et al. 1999). However, fascinating instances of EPRV reactivation have been described. Pararetroviral infections compatible with vertically transmitted endogenized latent proviruses were reported for interspecific hybrids of the genera Musa, Nicotiana, and Petunia (Kuriyama, et al. 2020; Lockhart, et al. 2000; Ndowora, et al. 1999; Richert-Pöggeler, et al. 2003). In hybrid plants from Solanum such unlocking has so far not been detected (Staginnus, et al. 2007), suggesting that EPRV-containing genomes do not necessarily suffer infections from latent EPRVs. Although the establishment of stable transgenerational silencing or the general absence of endogenous competent reactive copies could imaginably underlie these observations (Staginnus, et al. 2007), their causative molecular basis has long remained understudied.
Here, we explored the endogenized pararetroviral component of exemplary genomes within Solanum, featuring tomato (Solanum lycopersicum), potato (Solanum tuberosum), and eggplant (Solanum melongena) sequenced crops (Barchi, et al. 2019; Bolger, et al. 2014a; Hardigan, et al. 2016; Tomato Genome Sequencing, et al. 2014). Our analysis supports the idea that an adaptive heritable strategy evolved to aid plants in the contest against pararetroviral threats, leveraging on spontaneous integrations with transcriptionally competent rearrangements. These appear to fuel the plant's immune system with endogenous hairpin transcripts, enabling the profuse generation of siRNAs—particularly 22-nt species—expressing complementarities to EPRVs and potentially cognate episomal forms. From comparative molecular dating, bioinformatics, and empirical explorations, we shed light on a rather underappreciated genetic interaction between plant hosts and accompanying immunologically relevant inhabitants.
Results
Exploration of Non-truncated EPRVs Enable Comparative Clade-level Molecular Dating
During de novo calling of long-terminal-repeat (LTR) retrotransposons within Solanum (Sanchez, et al. 2019), we appreciated that some resulting elements harbouring LTR-like motifs encoded viral movement proteins (MPs), naturally unrelated to retrotransposons. Such terminal-repeat bearing elements were predicted as plant EPRVs belonging to Caulimoviridae (Harper, et al. 2002; Staginnus and Richert-Pöggeler 2006). To genome-wide capture integrated pararetroviral sequences through refined in silico searches within four representative Solanum genomes, we used as query these firstly called elements as well as various known members of Caulimoviridae, including some from the endogenous Florendovirus genus (Geering, et al. 2014). Final recognized sequences spanned sizes between 139–23,156 bp, certainly comprising non-truncated elements as well as aged fragmented historical remnants (supplementary table S1, Supplementary Material online). We estimated that these represented around ∼0.79%, ∼0.49%, ∼0.69%, and ∼1.00% of the total genome for S. lycopersicum, Solanum pennellii, S. tuberosum, and S. melongena current genomes, respectively.
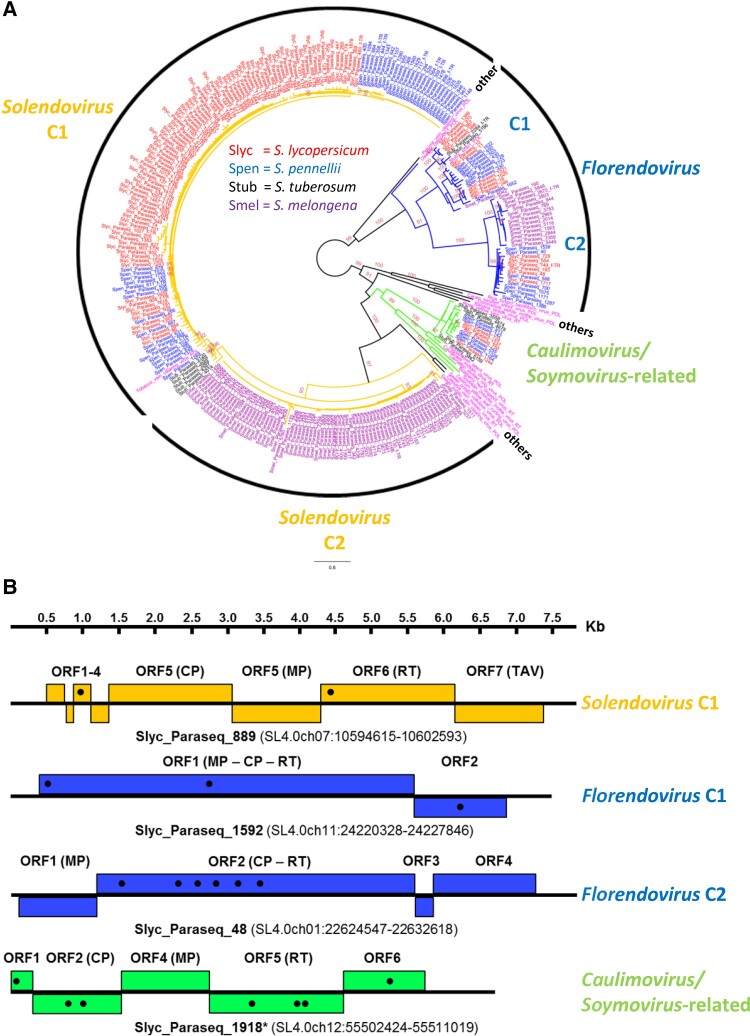
To provide a summarized robust snapshot of endogenized Caulimoviridae variants in Solanum, we retrieved a restricted sub-set of pararetroviral coding sequences containing reverse-transcriptase (RT) open reading frames (ORFs) with accompanying MPs motifs. Inferred RTs and MPs phylogenies had overall comparable topology (supplementary fig. S1A, Supplementary Material online), and suggested that most commonly endogenized pararetrovirus belonged to Solendovirus (orange, fig. 1A). Significant bootstrap support (≥90) pointed at two mayor Solendovirus sub-clades: sub-clade C1 often sub-clustered according to host species (and included Tobacco vein clearing virus [Lockhart, et al. 2000]) while sub-clade C2 comprised only S. melongena sequences; indicating that most of these insertions did not take place in a common host ancestor. Other sequences belonged to Florendovirus and presented two supported mayor sub-clades (blue, fig. 1A), but only sub-clade C1 encompassed previously described Solanum Florendo elements (Geering, et al. 2014). Fewer seemed related to the Caulimovirus and Soymovirus genera (including Cauliflower mosaic virus and Soybean chlorotic mottle virus [Hasegawa, et al. 1989]) (green, fig. 1A); to the best of our knowledge, this Caulimovirus/Soymovirus-related family was not described before (Diop, et al. 2018), and we failed to find it in S. melongena. Finally, note that a RT phylogeny constructed with the addition of representative Solanum LTR retrotransposon sequences demonstrated significant support for the separation of Pseudoviridae and Metaviridae from the Caulimoviridae phyletic lines, highlighting the specificity/selectivity of our detection pipeline (supplementary fig. S1B, Supplementary Material online).
Fig. 1.
Summarized overview of endogenized pararetroviral sequences in exemplary Solanum genomes. (A) Unrooted phylogenetic relationship of recognized pararetroviral-related reverse-transcriptases (RTs) protein sequences with >300 amino-acids (as a conservative threshold to ensure robust confidence in alignment) called within current Solanum lycopersicum (red), Solanum pennellii (blue), Solanum tuberosum (black), and Solanum melongena (violet) genomes. Poly-protein (POL) or RT sequences from 16 different pararetroviruses, along with two complete reported endogenized elements from Florendovirus (StubV_scSt1 and OsatBV_compAsc1 from S. tuberosum and Oryza sativa, respectively), were added in pink to expose phylo-group relatedness. Shown ≥ 90 bootstrap support was considered significant. Phyletic relationships are highlighted for Solendovirus (orange; sub-clades C1 and C2, but note that Sweet potato vein clearing virus would represent an additional sub-clade), Florendovirus (blue; sub-clades C1 and C2) and Caulimovirus/Soymovirus-related group (green). Black phyletic lines represent other/s (Badnavirus, Tungrovirus, Cavemovirus, Petuvirus and Rosadnavirus). f_TR denotes sequences bearing recognized flanking tandem-repeats. (B) Schematic representation of exemplary S. lycopersicum non-truncated EPRVs assembled from distinct phylo-groups. Boxes represent ORFs, with black dots indicating positions with premature stop codons, manually curated using as template other EPRV relatives of the corresponding phylogenetic clade. ORFs encoding conserved domains are indicated between brackets (CP, capsid protein; MP, movement protein; RT, reverse-transcriptase; TAV, transactivator/viroplasmin; ORFX, unknown). Chromosomal coordinates of each non-truncated EPRV are indicated. Note that Slyc_Paraseq_1918* is a portion of an original parasequence which presented a rearrangement at the 5′ end, additionally extending the otherwise complete non-truncated element here presented.
From the universe of registered pararetroviral sequences, we aimed at recovering substantially intact “non-truncated” EPRVs (i.e., elements encoding a complete set of typical pararetroviral ORFs, independently of mutations/indels generating premature stop codons). Sequences were size-filtered (6,500–9,600 bp) and restricted to those showing identity (≥70%) and alignment coverage (≥70%) to three categories of useful archetypal templates (known pararetroviruses, a set of prior manually assembled exemplary non-truncated EPRVs from different phylo-groups, and published complete Solanum Florendovirus). This recovered 135, 51, 4, and 29 non-truncated EPRVs from S. lycopersicum, S. pennellii, S. tuberosum, and S. melongena, respectively (supplementary table S2, Supplementary Material online). Such figures are certainly underestimations, because genome assemblies are not yet complete and the stringent size/identity/coverage thresholds undoubtedly omit potential whole tandemizations and more degenerated elements (i.e., older integrations). Nevertheless, they permit the robust recognition of fairly recent integrations enabling a structural overview. Careful comparative architectural examination allowed the assembly of some marginally mutated non-truncated EPRVs (fig. 1B, supplementary fig. S2 and supplementary dataset S1, Supplementary Material online). Although non-truncated EPRVs within Solendovirus seemed to share the same structural blueprint, novelty manifested in the Florendovirus group, represented by two phylogenetically informative coding organizations: sub-clade C2 with four ORFs instead of the reported two ORFs configuration characteristic of sub-clade C1 (Geering, et al. 2014) (fig. 1B and supplementary fig. S2, Supplementary Material online). Additionally, novel EPRVs closer to Caulimovirus and Soymovirus—yet separated by significant bootstraps (fig. 1A)—presented a core coding organization more comparable to Solendovirus, supporting the notion that they belong to an independent genus (supplementary fig. S3, Supplementary Material online). A comprehensive manual analysis of recovered non-truncated EPRVs showed that most were defective with premature stop codons, save few exceptions (e.g., Spen_Paraseq_947 and Smel_Paraseq_1563; supplementary fig. S2, Supplementary Material online). Modern and older integration events could be deduced from the relative degree of accumulated disruptive mutations, where the occurrence of seemingly non-degenerated elements implies evolutionary recent integrations.
Terminal-repeated sequences have been found in EPRVs, presumably arising from head-to-tail concatemers or from integration of circular life-cycle intermediaries with terminal redundancy in pregenomic RNA (Jakowitsch, et al. 1999; Richert-Pöggeler, et al. 2003). Indeed, in our non-truncated EPRV hits, we recognized the occasional occurrence of variable-sized flanking terminal-repeats (i.e., at beginning/end; supplementary table S2, Supplementary Material online). Given that no reported free pararetrovirus harbours sizable alike structures, said rearrangements must have taken place during and/or after genomic integration. Assuming that flanking terminal-repeats originated from the same template—at integration or posterior chromosomal rearrangement—affords the opportunity to track the elapsed time since repeat expansion. We ventured an approach analogous to that applied for LTR retrotransposons, where the initial transposition presents identical repeats that independently degenerate at a rate proportional to the element's age (Drost and Sanchez 2019; Sanchez, et al. 2017). Such molecular dating hinted at non-truncated EPRVs steady gain of flanking terminal-repeats, evidenced by differential sequence similarity spanning ∼89–100% identity (supplementary table S2, Supplementary Material online); highly similar terminal-repeats being diagnostic of more recent repeat expansion.
Taking all observations together, we suggest that the integration and rearrangement of endogenous pararetroviruses (EPRVs) within Solanum represents not only a pervasive historical property contributing to non-trivial genome enlargement, but possibly also an ongoing evolutionary phenomenon.
Integrated Pararetrovirus-Related Features Deliver Distinct siRNA Patterns
We focused on S. lycopersicum as a representative model to study properties and significance of Solanum EPRVs. It has been shown that EPRVs can be transcriptionally active (Noreen, et al. 2007; Staginnus, et al. 2007), so we first mined available high quality RNA-seq libraries (Sanchez, et al. 2019). We found that only a very minor proportion (∼2%) of pararetroviral sequences could be called present, but most showing very low or near negligible expression (supplementary table S3, Supplementary Material online). Given that EPRV-derived endogenous siRNAs have also been recovered from various plants including S. lycopersicum (Becher, et al. 2014; Noreen, et al. 2007; Richert-Pöggeler, et al. 2021; Staginnus, et al. 2007), we then mined a series of available S. lycopersicum small-RNA libraries comprising vegetative and gametic tissues (Lunardon, et al. 2020). The total pararetroviral siRNA component represented ∼1.84% of mapped 18–25-nt forms from pooled libraries. Using exemplary assembled S. lycopersicum non-truncated EPRVs from different phylo-groups as mapping targets, we detected a non-random siRNAs distribution with remarkable enrichment over distinctive areas depending on template (supplementary fig. S4, Supplementary Material online). We interpreted this as evidence of unbalanced representation or sourcing, suggesting that certain sequence fragments may generate most endogenous probes. To investigate this, we preliminarily explored siRNA expression at listed S. lycopersicum pararetroviral sequences, and appreciated that a substantial proportion mapped across tissues to a surprisingly small set of chromosomal spaces, later detected transcriptionally (see below). Such rare “transcriptionally competent siRNA areas”, henceforth referred to as TSAs (currently from TSA1 to TSA22 in S. lycopersicum; supplementary dataset S2, Supplementary Material online), represented only 3.6% of the pararetroviral-related sequence space and did not necessarily overlap with non-truncated EPRVs (see below). Remarkably, said TSAs exhibited in most cases inverted-repeat conformations usually with matching siRNA peaks, suggesting they behave as bona fide self-complementary hairpins upon expression.
Aiming at assessing the involvement of TSAs in evoking silencing, we counted across libraries the number of “private” siRNAs—restricted to primary alignments with high likelihood of correct mapping—thus increasing the chances to chart their real chromosomal source. TSAs accounted for a major proportion of the endogenous pararetroviral private siRNAs space, although bearing variability across tissues (fig. 2AandB). Distinct developmental patterns of private siRNAs expression between individual TSAs were noticeable, seemingly unrelated to some degree of disparity among private libraries’ sizes (fig. 2B and supplementary fig. S5, Supplementary Material online). TSAs explained almost ∼80% of the pooled libraries’ private pararetroviral siRNA space typically associated with PTGS (21–22-nt forms; supplementary fig. S6, Supplementary Material online); size profiles showed TSAs conspicuously enriched in 22-nt over 21-nt siRNAs (fig. 2C and supplementary fig. S7A, Supplementary Material online). 22-nt forms in S. lycopersicum represent hallmarks of anti-viral PTGS critically biosynthesized by SlDCL2a and SlDCL2b (Wang, et al. 2018). Our analysis of available sldcl2ab double mutant siRNA libraries confirmed that private TSA-mapping 22-nt probes remarkably decreased in this background (supplementary fig. S7B, Supplementary Material online). Furthermore, genome-browsers showed that probes generally charted TSAs’ inverted-repeated segments (fig. 3A and supplementary fig. S8, Supplementary Material online).
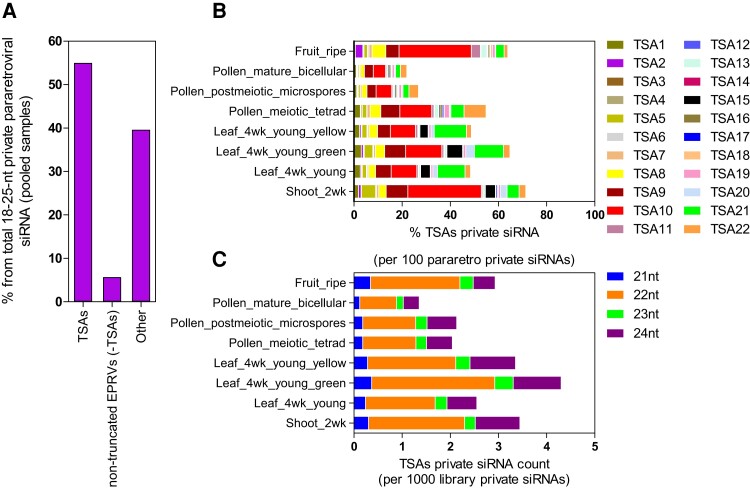
Fig. 2.
Solanum lycopersicum TSA silencing effectors data. (A) Percentage of S. lycopersicum private 18–25-nt siRNA from pooled tissue samples mapping to annotated pararetrovial-related sequences. TSAs, transcriptionally competent siRNA areas; non-truncated EPRVs (-TSAs), recognized non-truncated EPRVs not belonging to any TSA; Other, remaining pararetroviral-related sequences not included in the previous groups, comprising historical remnants. (B) Private 18–25-nt siRNA mapping to each S. lycopersicum TSA across eight tissues, expressed as adding percentage of all pararetroviral-related private siRNAs. (C) Private siRNA counts distinguished by size (21-nt = blue, 22-nt = orange, and 24-nt = indigo) mapping to all S. lycopersicum TSAs across eight tissues.
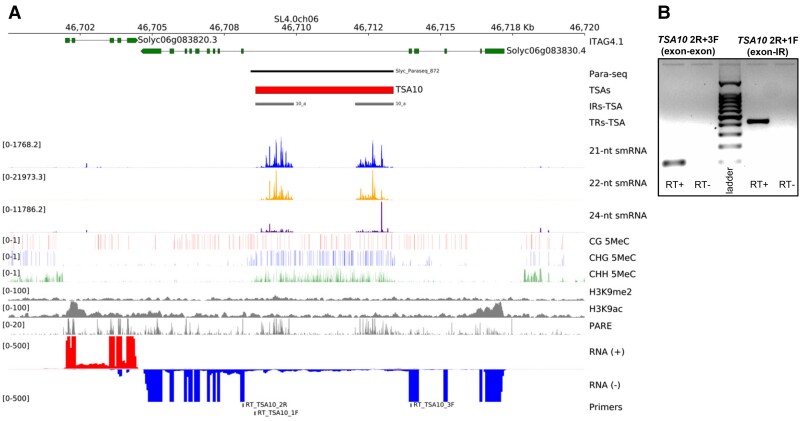
Fig. 3.
Genomics of an exemplary Solanum lycopersicum TSA. (A) Genome-browser data around TSA10 coordinates. Data visualized includes ITAG4.1 gene models (green), recognized pararetroviral sequences (Para-seq, black), proposed positions for TSAs (red), inverted- and tandem-repeats in TSAs (IRs-TSA and TRs-TSA, grey and white), mapping siRNA signals (21-nt = blue, 22-nt = orange, and 24-nt = indigo), DNA methylation in CG (red), CHG (blue), and CHH (green) sequence contexts (H denoting any base but G), leaves’ histone modifications signals for H3K9me2 and H3K9ac (grey), degradome PARE-seq signals (grey), positive (red), and negative (blue) strands RNA-seq signals from pooled tissues (leaves, flowers and meristems), and primers positions for RT-PCR assays. (B) TSA10 RT-PCR transcript signals. Exon–exon, splicing product; exon-IR (inverted-repeat), non-splicing product; with expected size of 102 and 434 bp, respectively. Ladder = 1,000 bp ladder; RT+, reaction with reverse-transcriptase; RT–, negative control reaction without addition of reverse-transcriptase.
Noteworthy, TSAs mapped also 24-nt siRNAs (fig. 3A, and supplementary figs. S6 and S7, Supplementary Material online). Since these represent hallmarks of TGS mediated by RNA-directed-DNA methylation (RdDM) (Matzke, et al. 2015), we explored suppressing epigenetic marks such as DNA 5-methylcitosine and H3K9me2 which usually oppose active chromatin marks like H3K9ac (Wang and Baulcombe 2020). TSAs typically presented DNA methylation in all sequence contexts (CG, CHG and CHH) much like transposable elements (Sanchez, et al. 2019), enriched in CHG and CHH methylation as compared to coding areas that frequently showed gene-body GC methylation, particularly evident in those exons surrounding intronic TSAs (fig. 3A and supplementary fig. S8, Supplementary Material online). This implied the activity of TGS pathways, although we recognized no correlated extraordinary enrichment in H3K9me2, suggesting that these ranges may be marginally heretocromatic.
On the other hand, pararetroviral “non-TSA” sequences included non-truncated EPRVs (but excluding four which were structural parts of TSAs, see below) and the remaining historical integration remnants, comprising, respectively, 16.6% and 79.8% of the endogenous pararetrovirus sequence space. Private size profiles matching non-TSA sequences across tissues were in most cases enriched in heterochromatic 24-nt siRNAs (supplementary fig. S9AandB, Supplementary Material online). Usually, non-TSA features lacked RNA-seq signals, and although sometimes presented inverted-repeats, these did not conspicuously manifest strong enrichment of correlated 22-nt forms; exemplary cases showed more noticeable heterochromatic siRNAs in terminal-repeats (supplementary fig. S9EandF, Supplementary Material online). A particular increase of private 24-nt siRNAs in certain pollen samples was perceptible, presumably paralleling the epigenetic reprogramming that characterize this tissue (Joseph, et al. 2012) (supplementary fig. S9CandD, Supplementary Material online). As expected, non-TSAs normally presented DNA methylation in all sequence contexts (Staginnus, et al. 2007) (supplementary fig. S9EandF, Supplementary Material online). Note that the recognized non-truncated EPRVs accounted for only 5.6% of the total pararetroviral private siRNA universe (fig. 2A; considering 131 out of the 135 because four belonged to TSAs, see below), suggesting that endogenized whole elements may not evoke the strongest silencing response.
Taken together, we conclude that a substantial fraction of pararetroviral S. lycopersicum endogenous silencing effectors derive from chromosomal sections that we baptized TSAs, representing siRNAs clusters particularly enriched in 22-nt species arising through a known anti-viral pathway. In turn, the majority of recognized recent non-truncated EPRVs, and the bulk of fragmented historical remnants, more characteristically present enrichment of heterochromatic 24-nt probes.
Most TSAs Belong to Expressed Intronic/Intergenic Micro-scale Rearrangements
We then aimed at further exploring S. lycopersicum TSA structural properties. We inferred from genome-browsers that TSA4, TSA5, TSA7, TSA8, TSA9, TSA10, TSA13, TSA14, TSA17, TSA19, TSA21, and TSA22 were likely intronic (fig. 3A and supplementary fig. S8, Supplementary Material online). Since intron retention is not uncommon in plants (Jia, et al. 2020b), it may be expected that these are expressed. RNA-seq signals and additional RT-PCR experiments corroborated that exemplary TSA-bearing introns occurred in expressed transcripts, also confirming splicing (fig. 3B, supplementary fig. S10 and table S3, Supplementary Material online). Even though typically weak, RNA-seq established that intronic TSAs transcribed in the same strand as the corresponding exonic portions, while active H3K9ac chromatin marks were frequently associated with initial exonic zones rather than those introns bearing TSAs (fig. 3A and supplementary fig. S8, Supplementary Material online). Therefore, we envision that gene promoters drive whole RNA production, while TSA areas yield silencing probes directly from splice-competent ranges. On the other hand, TSA1, TSA2, TSA3, TSA6, TSA11, TSA12, TSA15, TSA16, TSA18, and TSA20 seemed blocks constituting intergenic regions with no major associated ORF (supplementary fig. S8, Supplementary Material online). As before, RNA-seq and RT-PCR of exemplary cases validated their occurrence in expressed transcripts, also confirming splicing (supplementary fig. S10 and table S3, Supplementary Material online). Hence, they most likely comprise bona fide spliced long non-coding RNAs (lncRNAs).
As mentioned, almost all TSAs presented inverted-repeats (exception TSA14), with conspicuous dispersed siRNA matching peaks and sometimes corresponding RNA degradome signals as mapped from available PARE-seq data (Seo, et al. 2018) (fig. 3A and supplementary fig. S8, Supplementary Material online). Such features are compatible with the idea that these are processed as precursor stem-loops primarily sourcing silencing probes (Axtell 2013). Some TSAs were highly rearranged and also presented tandem-repeats (TSA12, TSA15, and TSA20; supplementary fig. S8, Supplementary Material online). We further analyzed TSA's inverted-repeats in silico, which presumably sourced siRNAs regardless of variable sizes—from few hundred to several thousand basepairs. Constituent inverted-repeats showed different degrees of identity across TSAs pointing to distinct temporal origins; cases bearing more pronounced divergent repeats implied the evolutionary conservation of older structures (see TSA3, TSA15, and TSA20; supplementary fig. S11, Supplementary Material online). The alignment to exemplary assembled non-truncated EPRVs enabled the recognition of viral functional sequences that constituted inverted-repeats, but any coding or non-coding area appeared represented so no general pattern emerged (supplementary fig. S11, Supplementary Material online).
We conclude that central pararetroviral siRNA-associated clusters comprise spliced intronic or intergenic features. With one exception, these presented inverted-repeats assembled from varied portions of integrated elements, which would then presumably generate stem-loop transcripts. Worth noting, only TSA2, TSA8, TSA9, and TSA21 from both intronic and intergenic types corresponded to listed non-truncated EPRVs, indicating that some endogenizations may rapidly became especially devoted regions, acquiring inverted-repeats at integration or chiefly afterwards.
TSA-derived siRNAs Display Broad Specificity for EPRVs and Promote Interference on a Viral Replicon Reporter
We then asked whether TSAs might provide combined silencing coverage across tissues. Aiming at revealing putative targets in silico, we initially analyzed from siRNA pooled libraries the TSA-mapping universe matching reverse-complement sequences of known pararetroviruses or exemplary assembled non-truncated EPRVs. Given that effective RNA interference and RdDM silencing may accommodate siRNAs with mismatches and proceed not only through perfect but also partial complementarity (Fei, et al. 2021; Liu, et al. 2014), we explored 21–22–24-nt probes with perfect-matching complementarity but also allowed for a single SNP (only possible in the last 10 bp of siRNAs) as a conservative lower-bound specificity threshold (Liu, et al. 2014).
This analysis confirmed that TSAs exhibit endogenous probes potentially targeting specific pararetroviral phylo-groups. TSA1, TSA3, TSA4, TSA11, TSA12, TSA14, TSA15, TSA17, and TSA19 presented variable titres of mapping siRNAs with perfect/1-mismatch complementarity to S. lycopersicum assembled Solendovirus EPRVs (supplementary fig. S12, Supplementary Material online). A smaller but still relevant number of hits matched other assembled Solendovirus C1/C2 from related species and even Tobacco vein clearing virus (Lockhart, et al. 2000), although this precise viral sequence is not integrated in Solanum. TSA2, TSA6, TSA9, TSA10, and TSA22 targeted Florendovirus; again, Florendo-EPRVs occurring in related species sometimes stood as possible targets (supplementary fig. S12, Supplementary Material online). In turn, TSA5, TSA8, TSA13, TSA20, and TSA21 displayed specificities for exemplary assembled Caulimovirus/Soymovirus-related EPRVs (supplementary fig. S12, Supplementary Material online). Furthermore, we tested the complete list of S. lycopersicum non-truncated EPRVs (-TSAs), also finding evidence of complementary matching in many cases (supplementary fig. S12, Supplementary Material online). However, TSA7 remained without a recognized target. Additional manual surveys revealed that this was due to its inverted-repeats being derived from a three ORFs Florendovirus (Geering, et al. 2014), not initially recovered given its highly degraded sequences (a complete non-truncated element with this configuration was eventually found in S. pennellii; supplementary fig. S13, Supplementary Material online). 17.4% of TSA7-mapping 21–22–24-nt siRNAs were perfect/1-mismatch complementary to this target. These observations suggest that the host could preserve the production of siRNAs matching an elderly pararetrovirus. We envisage an even broader TSA end target spectrum since not all TSA-mapping probes matched the tested templates, while secondary siRNAs biogenesis may be elicited after primary targeting—particularly in primary 22-nt siRNAs (Axtell 2013; Chen, et al. 2010; Sanan-Mishra, et al. 2021). Nevertheless, our examination already emphasized trans-targeting potentialities with various degrees of specificities and cross coverage against occurring EPRVs.
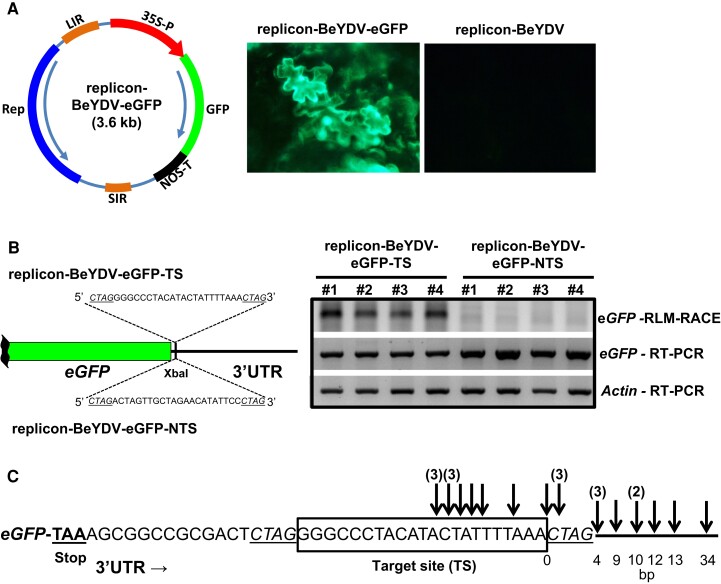
Aiming at empirically evaluate TSAs’ trans-acting capabilities upon operative mRNAs, we assayed a self-replicating artificially modified variant of Bean yellow dwarf virus (BeYDV, genus Mastrevirus from the Geminiviridae family). We used a pRIC3.0-eGFP plasmid carrying a MP-less version of BeYDY with the addition of a reporter cassette (Lamprecht, et al. 2016); this could be readily infiltrated with Agrobacterium tumefaciens into S. lycopersicum cotyledons generating an active cell-autonomous viral replicon that efficiently expresses eGFP reporter (fig. 4A). The construct was manipulated in a way that the viral replicon would express a short 22 bp target-site (TS) just downstream of eGFP (replicon-BeYDV-eGFP-TS, fig. 4B). Such target-site was chosen because its unique sequence occurred only within TSA10, and appeared abundantly as perfect-matching complementary private siRNAs in pooled libraries (one 22-nt and two 21-nt forms). A negative control, as non-target-site (NTS), was also generated with the same nucleotides randomly placed (replicon-BeYDV-eGFP-NTS, fig. 4B). Independent S. lycopersicum plants infiltrated with these two constructs substantially expressed the reporter's transcript, as estimated by RT-PCR (fig. 4B, eGFP RT-PCR), and thus showed tissue GFP signals (supplementary fig. S14A, Supplementary Material online). However, precise GFP transcript quantification using RT-qPCR evidenced decreased levels for replicon-BeYDV-eGFP-TS, as would be expected from a certain degree of interference (supplementary fig. S14B, Supplementary Material online). Accordingly, only samples bearing replicon-BeYDV-eGFP-TS revealed a defined PCR band after RLM-RACE, designed to specifically amplify cleaved mRNA 5′ termini around the target area (Llave, et al. 2011) (fig. 4B, eGFP RLM-RACE). This indicates that only the composite eGFP-target-site transcript was specifically sliced. RLM-RACE products were further cloned and Sanger-sequenced to establish the exact slicing position across mRNA molecules (fig. 4C); as expected for bona fide RNA interference, cleavage appeared in many instances processed at the center region of the target-site (predicted between 9–11 bp in 3′–5′ orientation (Liu, et al. 2014)). In addition, we also found molecules processed downstream, but not upstream, of the relevant area (fig. 4C), implicating PTGS transitivity which directs further slicing at sequences 3′ adjacent to the targeted point (Moissiard, et al. 2007).
Fig. 4.
Exemplary target-site for private siRNAs mapping TSA10 promotes cleavage of a viral replicon-derived reporter transcript. (A) Schematic representation of the self-replicating artificial BeYD-based viral replicon generated after infiltration of Solanum lycopersucum cotyledons with Agrobacterium tumefaciens strain carrying a pRIC3.0-eGFP-derived plasmid. Microscope pictures of cotyledons under UV light were taken three days post-infiltration and demonstrated eGFP expression in S. lycopersicum cells only when the plasmid carried the reporter (left); an eGFP-less pRIC3.0-derived plasmid was assayed as negative control (right). (B) Left: schematic representation of the manipulated segment of the viral replicon in which a short target-site (TS) for TSA10-derived siRNAs and a corresponding non-target-site (NTS, negative control) were introduced. Right: agarose gels are shown for RLM-RACE and RT-PCR from cotyledon total RNA sampled three days post-infiltration in independent S. lycopersicum plants. Upper gel, eGFP mRNA cleaved products detected by RLM-RACE with expected size ∼240 bp; middle gel, eGFP RT-PCR; lower gel, Actin RT-PCR (housekeeping gene). (C) Sanger sequencing analysis of 23 independent RLM-RACE products after cloning DNA fragments from the upper gel. Arrows represent exact cleavage position and above numbers indicate clones’ count if beyond one. Distance to the 3′ end of target-site in basepairs.
Taken together, we interpret the above results as evidence that TSAs represent a rich source of effective endogenous siRNAs targeting all families of endogenized pararetroviruses, potentially able to direct trans-cleavage of viral-originated mRNAs bearing complementary sequences.
Genomics Highlight Evolutionary TSA Dynamics and Trends
In order to explore the prevalence of identified TSAs, we first attempted to estimate their occurrence in a set of S. lycopersicum accessions using available high deep-coverage short-read DNA sequencing (Gao, et al. 2019; Lin, et al. 2014; Tomato Genome Sequencing, et al. 2014). Mapping and counting private DNA reads, we inferred that most accessions presented signals from all TSAs (supplementary fig. S15, Supplementary Material online). However, from lower end outliers, it became evident that certain TSA areas were missing in few instances, implying presence/absence variability at species level. We therefore examined recently generated assemblies from 12 S. lycopersicum accessions (comprising nine var. lycopersicum and three early domesticated forms var. cerasiforme) (Alonge, et al. 2020). In these, we failed to detect some syntenic TSAs, while others presented various degrees of completeness (supplementary table S4, Supplementary Material online). Although current alternative assemblies may still be incomplete and perhaps imperfections could hinder certain deductions, our observations suggest a degree of TSA pan-genome fluidity but with some more commonly fixed. Additionally, from the short-read re-sequencing we estimated the classic neutral summary statistic Tajima's D, which serves as a molecular-level proxy of non-random events arising from natural selection and/or population growth/shrinkage (Korneliussen, et al. 2014; Korneliussen, et al. 2013). Care must be taken to interpret this test though, given the confounding effects from bottlenecks and expansions during domestication, and the potential population structure in S. lycopersicum (Alonge, et al. 2020; Razifard, et al. 2020). Nonetheless, when compared to neighbouring ranges, we noted that some TSAs areas were associated to local Tajima's D score relative drops or peaks (within TSAs, or occasionally in closely adjacent expressed sequences not necessarily linked to evident genes) (supplementary fig. S16, Supplementary Material online). This observation warrants future research on putative selective/population pressures impacting such endogenous siRNAs clusters.
We then searched for endogenous pararetroviral sequences similar to S. lycopersicum TSAs, probing recently available assemblies from three accessions of the closest wild relative Solanum pimpinellifolium, which represents an estimated ∼57–97 KY divergence time (Alonge, et al. 2020; Razifard, et al. 2020; Wang, et al. 2020b). The simplest evolutionary model assuming a molecular clock predicts their homologous sequences in the range of ∼99.75–99.85% identity. We detected all S. lycopersicum (ITAG4.0) TSAs syntenic in S. pimpinellifolium, although in some or all accessions few were absent or incomplete, yet on occasion still showing at least some of the recognizable inverted-repeats distinctive of their S. lycopersicum counterparts (supplementary table S5, Supplementary Material online). Shared complete or incomplete TSA sequences presented identities in the ∼93.50–100% range (supplementary table S5, Supplementary Material online). Aligning homologous/syntenic TSAs—recognized across all explored 13 S. lycopersicum and three S. pimpinellifolium assemblies (supplementary dataset S3, Supplementary Material online)—enabled the estimation of Fay and Wu's H parameter (Fay and Wu 2000); designed to assess non-random molecular derivation from an ancestral allele probing an out-group species (in our case, S. pimpinellifolium). Although a consistent bias might be expected from demography, domestication and population changes that characterized S. lycopersicum history (Alonge, et al. 2020; Razifard, et al. 2020), we noted that Fay and Wu's H scores across verified TSAs showed at times medium or large positive/negative values, compatible with exemplary deficit/excess of high-frequency non-ancestral polymorphisms (supplementary table S6, Supplementary Material online). Broad in-group dissimilarities in nucleotide diversity also manifested. These observations support the idea of contrasting selective or population growth/shrinkage pressures experienced by individual TSA sequences.
A comparable exploration against the more distant relative S. pennelli failed to retrieve any syntenic TSA hit. We interpret this and previous points as an indication that S. lycopersicum TSAs—or endogenizations which would later support inverted-repeats—were already present in the common ancestor shared with S. pimpinellifolium, all presumably gained after the separation from S. pennelli phyletic line that diverged ∼2–3 Ma (Bolger, et al. 2014a; Sarkinen, et al. 2013; Tomato Genome Sequencing, et al. 2014). Also, it can be suggested that the TSA acquisition dynamics parallel speciation trends, and each Solanum may therefore present own species-specific TSAs implying overall evolutionary fluidity with mechanisms in place for gaining and loosing such areas.
Finally, we considered a recent study that identified two Caulimoviridae-related inverted-repeat regions bearing siRNAs in soybean (Glycine max) (Jia, et al. 2020a), so we asked whether TSA-like structures may be present in other dicot genomes outside Solanum. We explored the EPRV component of G. max and tobacco (Nicotiana tabacum), both with reliable genome assemblies and available siRNA libraries (Edwards, et al. 2017; Lunardon, et al. 2020; Shen, et al. 2019). This examination indicated that occasional pararetroviral TSA-like features bearing 22-nt siRNA enrichment indeed occur (supplementary fig. S17, Supplementary Material online). For G. max, we documented some over-sized highly rearranged areas, whereas N. tabacum exhibited several relatively short-sized and devoid of inverted-repeats. Despite such distinctive characteristics, the observations support the idea that plant species in other genera present similar TSA-related functionalities comparable to those of S. lycopersicum.
Discussion
It has been long presumed that EPRVs provide a beneficial role in the contest against pararetroviruses, although evidence is lacking beyond the detection of endogenous siRNAs (Chen and Kishima 2016; Harper, et al. 2002; Mette, et al. 2002; Richert-Pöggeler, et al. 2021; Staginnus and Richert-Pöggeler 2006). Our work offers novel clues supporting the view that EPRVs may contribute to host adaptive structural variation selected as a record of past experiences. Inverted duplications in partially deleted versions or composite insertions of “cut-and-paste” DNA transposable elements may mediate silencing through hairpin transcripts processed into siRNAs (Slotkin, et al. 2005; Wang, et al. 2020a). In a related line of evidence, our observations point to the importance of integrated pararetroviruses that serendipitously acquired micro-scale inverted conformations, not unlike the first steps of the target-gene inverted duplication model explaining de novo emergence of some miRNA genes (Allen, et al. 2004; Baldrich, et al. 2018). We propose that these naturally occurring chromosomal reorganizations—frequently constituents of areas that we baptized TSAs—fuel the plant's immune system with fitting transcripts functionally akin to classical artificial hairpins, from which efficient PTGS and TGS activities typically emerge and are known to counteract viral infections and even silence protein-coding genes (Pooggin, et al. 2003; Smith, et al. 2000; Waterhouse, et al. 1998). Since S. lycopersicum TSA14 presented no repeats, we venture to predict an additional genetic memory strategy at play; this idea is supported by the preliminary observation that some N. tabacum features also lack inverted-repeats. It is reasonable to assume that any alternative to the hairpins may still be based on emerging RNA duplexes processed by the defensive machinery (Axtell 2013). The evolutionary persistence of these anticipated immunologically relevant schemes is presumably contingent on plant–pararetrovirus selective pressures and the occurrence likelihood of relevant genetic rearrangements; micro-scale reorganizations may achieve higher representation upon greater incidence of pararetroviral infections and subsequent germ-line integration events, increasing the chances of future functional exaptation.
We believe that said features, with or without inverted-repeats, are efficiently expressed systemically thanks to their intronic/lncRNA nature inherently connected to transcription and splicing, thus providing a putative primed state of heritable trans-acting PTGS against potential pararetrovirus/EPRV threats. In S. lycopersicum, this is realized mostly by a reduced number of reactive TSAs bearing a characteristic 22-nt siRNA signature as effected by anti-viral DCL2 (Taochy, et al. 2017; Wang, et al. 2018); probably explaining the 22-nt enrichment on EPRV sequences recently documented by siRNA surveys in Glycine and Solanum hybrids confronting genomic-shock (Jia, et al. 2020a; Lopez-Gomollon, et al. 2022). Still, recognized TSAs did not account for the whole universe of S. lycopersicum pararetroviral-derived siRNAs, since other regions—mostly comprising historical remnants individually less conspicuous regarding siRNA pools—proved a significant source of probes when considered in toto. Of course, at present we cannot rule out that few here considered merely remnants might actually represent TSA areas, which passed below our radar of detection if bearing discreet siRNA and/or RNA-seq signals. The relative extent to which S. lycopersicum TSAs and non-TSAs may collectively contribute to effective global PTGS remains to be empirically explored; however, counted interference-associated 21–22-nt forms suggest that non-TSA contribution may be minor but not negligible. On the other hand, both TSA and non-TSA features may conceivably deliver TGS trans-recognition against episomal pararetroviral forms (Richert-Pöggeler, et al. 2003), although the bulk of heterochromatic 24-nt probes were predominantly associated with non-TSA remnant sequences. Accordingly, cis-activity can be inferred from conspicuous all-context DNA methylation signals present in both types of features.
The properties of the anticipated genetic memory not only encompass micro-scale chromosomal reorganizations and/or the occurrence of appropriate messengers eventually resulting in correlated endogenous siRNA enrichment, but sometimes also the parallel progression toward controlled expression. Although intronic features may take immediate advantage of already-in-place regulation from their hosting genes, S. lycopersicum intergenic TSAs were not usually surrounded by nearby same strand gene-coding sequences that may enable spill-over transcriptional activity. Given the seeming lack of significant global EPRV expression, we interpret this as evidence for the fast evolution of coherent transcriptional competence. Non-genic pararetroviral inverted duplications may emerge and then rapidly advance transcriptional capabilities, or alternatively occur in already lowly transcribed sequences with later gain of activity. Regardless, the progression to developmentally harmonized expression may not be surprising, as it has been documented for endogenous miRNAs and inverted-repeat loci selected to control plant development (Voinnet 2009).
Conceivably, pararetroviruses and endogenizing hosts engage in a gradual reciprocal Red Queen conflict, where the former evolve new genetic variants selected to evade immunological pressures while the latter progressively develop improved counter-adapted defences (Daugherty and Malik 2012). However, it stands to reason that the remarkable sequence space occupied by EPRVs and their historical remnants, as estimated in Solanum genomes, would not occur if these antagonists had evolved absolute lethality or absolute resistance, respectively. Therefore, bearing in mind that integration must imply past effective viral infections, and that the recovery from infections may entail the persistence of competent viruses in non-symptomatic tissues expressing a tolerance state (Ghoshal and Sanfaçon 2015; Kørner, et al. 2018), we entertain the idea that such proposed primed immunity possibly often brings about only a certain degree of tolerance just permitting host survival, rather than full complete resistance. Within the conceptual context of conditional mutualistic/symbiogenic viral partners (Roossinck 2011), it is tempting to speculate that endogenization might convey a non-zero-sum scenario serving both contestants: perhaps contributing for pararetoviruses the access to a potential road of “hidden” vertical transmission enabling reactivation in appropriate moments (Lockhart, et al. 2000; Richert-Pöggeler, et al. 2003), while for hosts the means to evolve new genetic variants expressing transgenerational tolerance to relatively recent threats. Unfortunately, current opportunities for empirically testing some particulars arising from our previous suggestions are constrained by the lack of an established pararetovirus-S. lycopersicum pathosystem (Rivarez, et al. 2021). Nonetheless, some lines of evidence may collectively converge on the notion of operative TSA activity in S. lycopersicum; most remarkably, the structural and transcriptional properties compatible with other known functional genomic features under adaptive evolution—such as miRNAs and lncRNAs—the inferred broad homology-dependent silencing potential and experimental slicing activity, and the apparent close cross-species conservation. The observed idiosyncratic allele (site) frequency spectrum summary statistics across TSAs may point to differential selective, demographic or population dynamic pressures. The additional recognition of TSA7 matching extremely derived EPRVs, which we did not routinely document, may be perhaps interpreted as an infrequent historical event of host success in diminishing the integration occurrence likelihood for those cognate episomal elements.
It is becoming increasingly evident that the endogenization of external invading nucleic acids repurposed as immunological weaponry may commonly characterize the tree of life; consider the integration of small viral sequences that constitute the molecular bases of prokaryotic CRISPR-Cas systems (Marraffini 2015), or the anti-viral activity derived from endogenized elements demonstrated in an insect model (Suzuki, et al. 2020). Given the global occurrence of EPRVs (Diop, et al. 2018; Gong and Han 2018), the pervasiveness of inverted-repeat rearrangements within plant genomes (Huang and Rieseberg 2020), and the shared properties of silencing (Svoboda 2020), we believe that potential TSA-associated pararetroviral counter activity may represent a wider phenomenon rather than a peculiarity from S. lycopersicum. This idea is supported by our preliminary results in exemplary Glycine and Nicotiana genomes. However, note that EPRV-containing Petunia hybrida presumably lacks constitutive pararetroviral siRNAs in non-symptomatic tissues (Noreen, et al. 2007). Such observation provides not only the initial evidence that some species may not fully support the proposed priming, but also a prospective underlying causation for the unlocking of latent Petunia EPRVs.
To conclude, we used Solanum species as models for the analysis of the endogenous pararetrovirus component and accompanying silencing effectors, and suggest that homology-dependent silencing emerging from precise repurposed chromosomal areas constitute an adaptive heritable record of past experiences potentially targeting all forms of plant pararetroviruses. The observations indicate that the natural occurrence of inverted-repeats may constitute a first step for de novo emergence not only of regulatory miRNA genes, but also of immunologically relevant genetic memory, perhaps pointing to an evolutionary advantage for the random generation of micro-scale chromosomal rearrangements.
Materials and Methods
Data Sources
We retrieved from public repositories viral accessions representing current genera of Caulimoviridae (https://talk.ictvonline.org/) (Teycheney, et al. 2020). These were Banana streak virus (NC 008018.1), Blueberry red ringspot virus (NC 003138.2), Cacao swollen shoot virus (NC 001574.1) Carnation etched ring virus (NC 003498.1), Cassava vein mosaic virus (NC 001648.1), Cauliflower mosaic virus (NC 001497.2), Cestrum yellow leaf curling virus (NC 004324.3), Commelina yellow mottle virus (NC 001343.1), Horseradish latent virus (NC 018858.1), Petunia vein clearing virus (NC 001839.2), Rice tungro bacilliform virus (NC 001914.1), Rose yellow vein virus (NC 020999.1), Soybean chlorotic mottle virus (NC 001739.2), Sweet potato collusive virus (NC 015328.1), Sweet potato vein clearing virus (MH 188860.1) and Tobacco vein clearing virus (NC 003378.1). Described Florendovirus were recovered from a specific report (Geering, et al. 2014). Scrutinized plant genomes were S. lycopersicum (ITAG4.0 (Hosmani, et al. 2019)), S. pimpinellifolium (LA2093 v1.5 (Wang, et al. 2020b), and PAS014479 and BGV006775 (Alonge, et al. 2020)), S. pennellii (Spenn v2.0 [Bolger, et al. 2014a]), S. tuberosum (DM v4.04 [Hardigan, et al. 2016], S. melongena (Eggplant v3 [Barchi, et al. 2019]), N. tabacum (Nitab v4.5 [Edwards, et al. 2017]), N. benthamiana (Niben v1.01 [Bombarely, et al. 2012]), N. attenuata (Niatt r2 [Xu, et al. 2017]), G. max (Gmax_508 v4.0 and ZH13 a1 [Schmutz, et al. 2010; Shen, et al. 2019]) and G. soja (PI483463 a1 and W05 a1 [Valliyodan, et al. 2019; Xie, et al. 2019]). S. lycoperscum genic features were recovered from ITAG4.1 annotation, whereas assemblies from other various accessions were recently reported (Brandywine, M82, Floradade, EA00371, LYC1410, EA00990, PI303721, PI169588, Fla.8924, BGV006865, BGV007989, and BGV007931; the last three var. cerasiforme) (Alonge, et al. 2020); all available at Solgenomics (www.solgenomics.net).
We obtained small-RNA libraries from publically available resources comprising data from different laboratories (Lunardon, et al. 2020) while sldcl2ab mutant libraries were from (Wang, et al. 2018), and further filtered for 18–25-nt sizes. S. lycopersicum RNA-seq tissue-specific libraries were reported previously (Sanchez, et al. 2019). PARE-seq raw data (Seo, et al. 2018) were analyzed only for trimmed reads ≥14 bp. H3K9me2 and H3K9ac ChIP-seq and BS-seq raw data were publicly available (Wang and Baulcombe 2020). Short-read DNA sequencing of different S. lycopersicum accessions were recovered from pan-genome and breeding history reports (Gao, et al. 2019; Lin, et al. 2014; Tomato Genome Sequencing, et al. 2014). In all cases, independent biological samples were merged to increase sequencing depth. supplementary table S7, Supplementary Material online lists the different NGS datasets analyzed.
Bioinformatics Analyses
De novo annotation of plant EPRVs was initiated with the search of apparent LTR retrotransposon sequences, taking advantage of EPRVs bearing terminal-repeats. The tool LTRharvest (Ellinghaus, et al. 2008), was used with paramenters -v -mintsd 3 -maxtsd 6 -seed 30 -xdrop 5 -mat 2 -mis −2 -ins −3 -del −3 -minlenltr 100 -maxlenltr 7000 -mindistltr 1000 -maxdistltr 30000 -similar 97 -overlaps best -vic 60 -longoutput. The output was mined for sequences presenting an ORF >100 amino-acids with significant similarity to Pfam PF01107.18 representing viral MPs (El-Gebali, et al. 2019); recognized applying HMMER3 (Eddy 2011) hmmscan function with parameter -T 40. Hits also presenting homologies to Pfam entries related to GAG and integrases from transposable elements were filtered out. The outcome was used as input for the initial pararetroviral sequences search within Solanum clade, performed with BLASTN (Altschul, et al. 1990) at -evalue 1e-3, and then collapsing results from all genomes together with the sequences of 16 viral species from Caulimoviridae (www.ictvonline.org/) and different reported elements from the Florendovirus genus (Geering, et al. 2014). Then, two rounds of such consecutive Pfam + BLASTN searches were repeated. Subsequently, using a preliminary phylogenetic analysis of RT proteins of >300 amino-acids (selected as a conservative threshold to ensure robust confidence in alignment) inferred from listed pararetroviral sequences, we recovered exemplary whole EPRVs from distinct phylogenetic groups in each species. Such elements were assembled from marginally mutated sequences after careful manual structural examination. Finally, another two consecutive rounds of Pfam + BLASTN searches were conducted using as input the collapsed results from all Solanum genomes, but now with a closing filtering step accepting only those sequences with at least 70% identity and ≥150 bp total alignment length to the above pararetrovirus species, exemplary assembled EPRVs, or previously recognized complete Florendovirus elements from Solanum (Geering, et al. 2014). The identity threshold was placed conservatively above the Caulimoviridae genera call (40–65% nucleotide identity [Sukal, et al. 2018]) but below the species call within genera (80% nucleotide identity; https://talk.ictvonline.org/), making it very restrictive to this viral family. On the other hand, the size filter avoided output inflation with small fragmented sequences that might code for protein motifs shared with retrotransposons. A similar workflow was applied to available Glycine and Nicotiana genomes, collapsing results from more than one species to finally document the endogenized pararetroviral sequence space in G. max and N. tabacum.
Putative whole non-truncated EPRV candidates were recovered by aligning sequences against pararetroviruses, some initially exemplary assembled EPRVs, or complete Florendovirus elements from Solanum. First, the final pararetroviral sequence list was size-filtered in the range between the smallest assembled EPRVs and the largest virus explored (between 6,500–9,600 bp) ruling out those with ambiguous “N”. Then, those presenting at least 70% identity in 70% of their length using BLASTN were selected, subsequently performing a global alignment with EMBOSS (Rice, et al. 2000) package needle function with parameters -gapopen 10 -gapextend 0.5, accepting only hits above score 25,000 with at least 70% similarity. As a mean to assess the specificity/selectivity of our detection pipeline, the resulting 135 S. lycopersicum whole non-truncated EPRVs were manually confirmed, and then compared with BLASTN against very recent complete non-truncated insertions of S. lycopersicum LTR retrotransposons representing both Copia (Pseudoviridae) and Gypsy (Metaviridae) superfamilies. These LTR retrotransposons were called elements with extremely high LTR similarities (>99.5%) as recognized by LTRharvest, and were annotated by the significant best blast hit (>80% identity and alignment length of at least 500 bp) against Copia/Gypsy reported sequences from Repbase (Bao, et al. 2015); finally accepting only those with translated in-between LTRs (with at least 100 amino-acids) showing compatibility to retrotransposon domains, as evidence by Pfam recognition (but ruling out those with recognized MP domain). Importantly, the vast majority of non-truncated EPRVs showed no significant similarities to any LTR retrotransposon listed at -evalue 1e-3, save few irrelevant matches against fragments of extreme short length (29–31 bp); however with one exception. This exception was revealing, presumably representing a particular element called within coordinates SL4.0ch03:18539939–18553584, where the alignments suspiciously matched only its extreme portions. It was later recognized that it was in fact a composite chromosomal area, comprising pararetroviral-related sequences (recognized by our pipeline: Slyc_Paraseq_325 to Slyc_Paraseq_328) genetically rearranged as similar terminal-repeats around an historical remnant derived from a Gypsy element. We conclude that such curious case defeated once our LTR retrotransposon discovery pipeline but did not defeat our EPRV discovery pipeline, which showed correct detection of pararetroviral-related portions while avoiding those originated in a LTR retrotransposon. As an exception that confirmed the rule, this single hit highlighted that our recognized EPRVs presented no truly relevant similarities to other potentially confounding intra-genomic retroelements. In addition, S. lycopersicum whole non-truncated EPRVs were cross-compared to the REPET pipeline report on repeated tomato sequences (Amselem, et al. 2019) (ITAG4.0_REPET_repeats_aggressive file, available at www.solgenomics.net); where they were represented by 521 REPET fragments, but only 60% of them were correctly distinguished as EPRVs. The rest presented non-declared origins (16.5%), or were incorrectly annotated as retrotransposons (14.6% Gypsy, 2.1% Copia, and 0.6% LINE) or as different types of DNA transposable elements or simple sequence repeats. The analysis extrapolated to all our listed S. lycopersicum pararetroviral-related sequences resulted in a very similar figure, with only 55.9% of REPET fragments being called EPRV-related, suggesting a substantial number of database global misannotations. Taken together, this demonstrates that automatic annotations may represent a major constrain to independent call validation; underscoring the need of a dedicated analysis, such as the one we described above, in order to appropriately recognize EPRVs.
Flanking tandem-repeats were recognized with BLASTN through custom-made python scripts splitting elements in halves (considering only cases of no less than 100 bp aligning tandem-repeats above 85% identity, and occurring no further away than 20 bp from element's edge), whereas for inverted-repeats we initially used EMBOSS einverted with -gap 12 -threshold 200 -match 3 -mismatch -4 or -2 and -maxrepeat 30,000; 5,000; or 1,000, with additional BLASTN and manual assessments.
Protein sequences were aligned with MAFFT (Katoh, et al. 2005) applying parameters -localpair -maxiterate 1000. Maximum likelihood phylogenetic analyses were performed in RAxML-NG (Kozlov, et al. 2019) with -model LG + G + F -tree pars(50), rand(50); no less than 1,000 bootstraps were calculated till convergence at—bs-cutoff 0.02, which were mapped to the best reported tree and visualized in FigTree (https://github.com/rambaut/figtree).
Estimated boundaries of informative chromosomal areas were manually projected from siRNA signals visualized in genome-browsers, conservatively keeping as relevant those coordinates limited by inverted-repeats and/or pararetroviral sequences (depending on chromosomal context). Searches for S. lycopersicum TSAs in assemblies other than Heinz 1706 genome (ITAG4.0)—comprising nine var. lycopersicum, three var. cerasiforme, and the three S. pimpinellifolium LA2093, PAS014479 and BGV006775—were carried out by BLASTN, filtering for those with at least 90% similarity and 50% alignment length and further manually assessing syntheny and uniqueness. The expected similarity between close homologous sequences was estimated from the presumed divergence time following the basic equation: time = p genetic distance/(2×substitution rate), using 1.3 × 10−8 mutations per site per year as inferred previously (Ma and Bennetzen 2004).
Estimates of Tajima's D summary statistic (Tajima 1989) were calculated genome-wide in overlapping sliding windows of 10,000 bp with 1,000 bp steps, from 124 S. lycopersicum accesions’ private DNA-seq mapped data (supplementary table S7, Supplementary Material online), using ANGSD software (Korneliussen, et al. 2014; Korneliussen, et al. 2013). The allele (site) frequency spectrum was estimated from allele frequency likelihoods, obtained with parameters -uniqueOnly 1 -remove_bads 1 -only_proper_pairs 1 -trim 0 -C 50 -baq 1 -minMapQ 20 -minQ 20 -GL 2 -doMajorMinor 1 -doCounts 0 -doSaf 1. The ancestral state was inferred from the analyzed population's most common bases (i.e., majority-frequency allele for each SNP), with -doFasta 2 and parameters -uniqueOnly 1 -remove_bads 1 -only_proper_pairs 1 -trim 0 -C 50 -baq 1 -minMapQ 20 -minQ 20 -basesPerLine 100 -explode 1 -seed 0 -doCounts 1. For this, mappings were randomly subsampled beforehand toward the lowest available sequencing depth, to equalize mapping depth differences across samples. Fay and Wu's H summary test (Fay and Wu 2000) was calculated using Variscan 2.0 (Hutter, et al. 2006), with pameters UseMuts = 1, UseLDSinglets = 1, CompleteDeletion = 1 and the maximum number of NumNuc; aligning with MAFFT—globalpair—maxiterate 1000 those synthenic TSAs’ nucleotide sequences from S. lycopersicum and S. pimpinellifolium accessions when reasonably complete.
Custom-made workflows for data explorations including Python scripts are available at https://github.com/diegohernansanchez/.
Next-Generation Sequencing and Expression Analyses
Next-generation sequencing reads were trimmed using Trimmomatic (Bolger, et al. 2014b) ILLUMINACLIP parameters:2:10:5:1, and further processed with open-source software such as BEDtools (Quinlan and Hall 2010), SAMtools (Li, et al. 2009) and Picard (http://picard.sourceforge.net). Small-RNA-seq, DNA-seq and ChIP-seq data were mapped with Bowtie2 (Langmead and Salzberg 2012), using parameters—very-sensitive—non-deterministic. For counting and size profiling of “private” reads, these were filtered for only primary alignments with high MAPQ likelihood (SAMtools view parameters -q 5 -F 256), further applying BEDtools intersect with -c parameter. Counts per feature were then adjusted to the sum of total filtered counts per library (as counts-per-million, cpm) or per counts of EPRV-related sequences (as fraction or percentage). For DNA-seq, only libraries presenting at least 40 million mapped private pair-end reads were explored (representing a minimum estimated primary alignment of ∼5× fold coverage). For PARE-seq and ChIP-seq, mapped data were collapsed with the bamCoverage function from deepTools2 suit (Ramirez, et al. 2016). BS-seq libraries were mapped, de-duplicated and methylation-called using Bismark (Krueger and Andrews 2011) with mapping parameters—bowtie2 -N 1 -L 20 -X 1000 -score_min L,0, -0.8 -R 3, while bismark_methylation_extractor—were run as comprehensive.
RNA-seq mapping was performed using STAR (Dobin, et al. 2013), with parameters -alignEndsType EndToEnd -twopassMode Basic -outReadsUnmapped None -outFilterMultimapNmax 10 -outMultimapperOrder Random. Reads were counted applying HT-seq count (Anders, et al. 2015) and normalized to the sum of HT-seq total counted library; present-call threshold for robust expression was set to >1 cpm in at least two independent samples under edgeR environment (Robinson, et al. 2010).
The expression and splicing of intronic/intergenic TSAs were validated by RT-PCR, performed with specific primers on cDNA template prepared from total DNA-free RNA from 3-week-old S. lycopersicum leaves. Primers are available from supplementary table S8, Supplementary Material online.
RNA Interference
To build pRIC3.0-eGFP-TS and pRIC3.0-eGFP-NTS, a 22-bp potential target-site for TSA10-derived siRNAs or its randomized sequence were introduced in the available unique XbaI restriction site within pRIC3.0-eGFP (Lamprecht, et al. 2016). Inserts were generated as small double-stranded DNAs obtained by in vitro annealing of specific oligos (TS/TSrevcomp and NTS/NTSrevcomp pairs, respectively; supplementary table S8, Supplementary Material online).
S. lycopersicum cv M82 plants were grown in greenhouse with 16 h/8 h light/dark cycles at 20–24 °C, with supplemental light. A. tumefaciens GV3101-pMP90RK (DSMZ) carrying pRIC3.0 (no reporter) or pRIC3.0-eGFP derivatives were infiltrated in 3-week-old plant cotyledons, following classical reported protocols for agroinfiltration of Nicotiana benthamiana (Sparkes, et al. 2006). Portions of treated cotyledons were examined with a Leica DMR epifluorescence microscope, using excitation and barrier filters at 450/490 and 500/550 nm, respectively, and photographed with an Olympus DP70 digital camera.
RLM-RACE was conducted as previously described (Llave, et al. 2011). Briefly, 1 μg of total DNA-free RNAs extracted with FavorPrep kit (Favorgene) from agroinfiltrated tissues were ligated to a 5′ RACE adapter with T4 RNA ligase (NEB), and then reverse transcribed to cDNA with random primers using M-MuLV (NEB). Naturally cleaved products were amplified by two consecutive PCRs using 5′ RACE forward/eGFP-3′ UTR reverse and 5′ RACE forward-nested/eGFP-3′ UTR reverse-nested primers. RLM-RACE products were gel-purified, cloned into pCRII-TOPO (ThermoFisher) and Sanger-sequenced (Macrogen Europe). The expression of eGFP reporter and Actin housekeeping gene were confirmed by RT-PCR and RT-qPCR from cDNA samples. Primers and oligos are available in supplementary table S8, Supplementary Material online.
Supplementary Material
Acknowledgements
We thank Ed Rybicki and Ann Meyers for providing pRIC3.0 and pRIC3.0-eGFP, Albert Vilella and Julio Rozas for assistance on neutral summary tests, and Juan Antonio García for general support, discussions, and critical reading of the manuscript. Gratitude is extended to Gustavo A. Marcello and Sebastian Nullo, for IT support. We also greatly appreciate the constructive feedback from David Baulcombe and two anonymous reviewers. This work was funded by PID2019-110979RB-I00/AEI/10.13039/501100011033 and RYC2018-025523-I grants from the Spanish Ministerio de Ciencia e Innovación (to A.A.V.), and PICT-2018-02401 and PICT-2019-01736 grants from the Argentinian Agencia Nacional de Promoción Científica y Tecnológica (to D.H.S.).
Contributor Information
Adrian A Valli, Centro Nacional de Biotecnología (CNB-CSIC), Calle Darwin 3, 28049 Madrid, Spain.
Irene Gonzalo-Magro, Centro Nacional de Biotecnología (CNB-CSIC), Calle Darwin 3, 28049 Madrid, Spain.
Diego H Sanchez, IFEVA (CONICET-UBA), Facultad de Agronomía, Universidad de Buenos Aires, Av San Martín 4453, C1417DSE Buenos Aires, Argentina.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author contributions
A.A.V. and D.H.S. designed the project. A.A.V. and I-G.M. performed wet experiments, while D.H.S. executed bioinformatics. A.A.V. and D.H.S. analyzed the data. D.H.S. wrote the paper with contribution from A.A.V. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplemental information files), or are available upon request.
References
- 100 Tomato Genome Sequencing Consortium, Aflitos S, Schijlen E, de Jong H, de Ridder D, Smit S, Finkers R, Wang J, Zhang G, Li N, et al. 2014. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80:136–148. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung G-H, Spatafora JW, Carrington JC. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 36:1282–1290. [DOI] [PubMed] [Google Scholar]
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 182:145–161.e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Amselem J, Cornut G, Choisne N, Alaux M, Alfama-Depauw F, Jamilloux V, Maumus F, Letellier T, Luyten I, Pommier C, et al. 2019. RepetDB: a unified resource for transposable element references. Mob DNA. 10:6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 64:137–159. [DOI] [PubMed] [Google Scholar]
- Baldrich P, Beric A, Meyers BC. 2018. Despacito: the slow evolutionary changes in plant microRNAs. Curr Opin Plant Biol. 42:16–22. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi L, Pietrella M, Venturini L, Minio A, Toppino L, Acquadro A, Andolfo G, Aprea G, Avanzato C, Bassolino L, et al. 2019. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci Rep. 9:11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher H, Ma L, Kelly LJ, Kovarik A, Leitch IJ, Leitch AR. 2014. Endogenous pararetrovirus sequences associated with 24 nt small RNAs at the centromeres of Fritillaria imperialis L. (Liliaceae), a species with a giant genome. Plant J. 80:823–833. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014b. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A, Scossa F, Bolger ME, Lanz C, Maumus F, Tohge T, Quesneville H, Alseekh S, Sørensen I, Lichtenstein G, et al. 2014a. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet. 46:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact. 25:1523–1530. [DOI] [PubMed] [Google Scholar]
- Chen H-M, Chen L-T, Patel K, Li Y-H, Baulcombe DC, Wu S-H. 2010. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 107:15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Kishima Y. 2016. Endogenous pararetroviruses in rice genomes as a fossil record useful for the emerging field of palaeovirology. Mol Plant Pathol. 17:1317–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 46:677–700. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. 2006. Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science. 313:68. [DOI] [PubMed] [Google Scholar]
- Ding S-W, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell. 130:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop SI, Geering ADW, Alfama-Depauw F, Loaec M, Teycheney P-Y, Maumus F. 2018. Tracheophyte genomes keep track of the deep evolution of the caulimoviridae. Sci Rep. 8:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H-G, Sanchez DH. 2019. Becoming a selfish clan: recombination associated to reverse-transcription in LTR retrotransposons. Genome Biol Evol. 11:3382–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, et al. 2017. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 18:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47:D427–d432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. 2000. Hitchhiking under positive darwinian selection. Genetics. 155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Y, Nyikó T, Molnar A. 2021. Non-perfectly matching small RNAs can induce stable and heritable epigenetic modifications and can be used as molecular markers to trace the origin and fate of silencing RNAs. Nucleic Acids Res. 49:1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, Burzynski-Chang EA, Fish TL, Stromberg KA, Sacks GL, et al. 2019. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet. 51:1044–1051. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 15:1494–1500. [DOI] [PubMed] [Google Scholar]
- Geering ADW, Maumus F, Copetti D, Choisne N, Zwickl DJ, Zytnicki M, McTaggart AR, Scalabrin S, Vezzulli S, Wing RA, et al. 2014. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat Commun. 5:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal B, Sanfaçon H. 2015. Symptom recovery in virus-infected plants: revisiting the role of RNA silencing mechanisms. Virology. 479–480:167–179. [DOI] [PubMed] [Google Scholar]
- Gong Z, Han GZ. 2018. Euphyllophyte paleoviruses illuminate hidden diversity and macroevolutionary mode of caulimoviridae. J Virol. 92:e02043–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan MA, Crisovan E, Hamilton JP, Kim J, Laimbeer P, Leisner CP, Manrique-Carpintero NC, Newton L, Pham GM, Vaillancourt B, et al. 2016. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell. 28:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper G, Hull R, Lockhart B, Olszewski N. 2002. Viral sequences integrated into plant genomes. Annu Rev Phytopathol. 40:119–136. [DOI] [PubMed] [Google Scholar]
- Hasegawa A, Verver J, Shimada A, Saito M, Goldbach R, Van Kammen A, Miki K, Kameya-Iwaki M, Hibi T. 1989. The complete sequence of soybean chlorotic mottle virus DNA and the identification of a novel promoter. Nucleic Acids Res. 17:9993–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, Flores-Gonzalez M, van de Geest H, Maumus F, Bakker LV, Schijlen E, van Haarst J, Cordewener J, Sanchez-Perez G, Peters S, et al. 2019. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv. 10.1101/767764 [DOI] [Google Scholar]
- Huang K, Rieseberg LH. 2020. Frequency, origins, and evolutionary role of chromosomal inversions in plants. Front Plant Sci. 11:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Vilella AJ, Rozas J. 2006. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics. 7:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakowitsch J, Mette MF, van der Winden J, Matzke MA, Matzke AJM. 1999. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc Natl Acad Sci USA. 96:13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Ji R, Li Z, Yu Y, Nakano M, Long Y, Feng L, Qin C, Lu D, Zhan J, et al. 2020a. Soybean DICER-LIKE2 regulates seed coat color via production of primary 22-nucleotide small interfering RNAs from long inverted repeats. Plant Cell. 32:3662–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Long Y, Zhang H, Li Z, Liu Z, Zhao Y, Lu D, Jin X, Deng X, Xia R, et al. 2020b. Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat Plants. 6:780–788. [DOI] [PubMed] [Google Scholar]
- Joseph PC, Borges F, Donoghue Mark TA, Van Ex F, Jullien Pauline E, Lopes T, Gardner R, Berger F, Feijó José A, Becker Jörg D, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 151:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT Version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Moltke I, Albrechtsen A, Nielsen R. 2013. Calculation of Tajima's D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics. 14:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner CJ, Pitzalis N, Peña EJ, Erhardt M, Vazquez F, Heinlein M. 2018. Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nat Plants. 4:157–164. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35:4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama K, Tabara M, Moriyama H, Kanazawa A, Koiwa H, Takahashi H, Fukuhara T. 2020. Disturbance of floral colour pattern by activation of an endogenous pararetrovirus, petunia vein clearing virus, in aged petunia plants. Plant J. 103:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht RL, Kennedy P, Huddy SM, Bethke S, Hendrikse M, Hitzeroth II, Rybicki EP. 2016. Production of human papillomavirus pseudovirions in plants and their use in pseudovirion-based neutralisation assays in mammalian cells. Sci Rep. 6:20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X. 2017. The functions of plant small RNAs in development and in stress responses. Plant J. 90:654–670. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. 2014. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 46:1220–1226. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang F, Axtell MJ. 2014. Analysis of complementarity requirements for plant MicroRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell. 26:741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Franco-Zorrilla JM, Solano R, Barajas D. 2011. Target validation of plant microRNAs. Methods Mol Biol. 732:187–208. [DOI] [PubMed] [Google Scholar]
- Lockhart BE, Menke J, Dahal G, Olszewski NE. 2000. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J Gen Virol. 81:1579–1585. [DOI] [PubMed] [Google Scholar]
- Lopez-Gomollon S, Müller SY, Baulcombe DC. 2022. Interspecific hybridization in tomato influences endogenous viral sRNAs and alters gene expression. Genome Biol. 23:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardon A, Johnson NR, Hagerott E, Phifer T, Polydore S, Coruh C, Axtell MJ. 2020. Integrated annotations and analyses of small RNA-producing loci from 47 diverse plants. Genome Res. 30:497–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 101:12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature. 526:55–61. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. 2015. RNA-Directed DNA methylation: the evolution of a Complex epigenetic pathway in flowering plants. Annu Rev Plant Biol. 66:243–267. [DOI] [PubMed] [Google Scholar]
- Mette MF, Kanno T, Aufsatz W, Jakowitsch J, van der Winden J, Matzke MA, Matzke AJM. 2002. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 21:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Parizotto EA, Himber C, Voinnet O. 2007. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 13:1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndowora T, Dahal G, LaFleur D, Harper G, Hull R, Olszewski NE, Lockhart B. 1999. Evidence that badnavirus infection inMusaCan originate from integrated pararetroviral sequences. Virology. 255:214–220. [DOI] [PubMed] [Google Scholar]
- Noreen F, Akbergenov R, Hohn T, Richert-Pöggeler KR. 2007. Distinct expression of endogenous Petunia vein clearing virus and the DNA transposon dTph1 in two Petunia hybrida lines is correlated with differences in histone modification and siRNA production. Plant J. 50:219–229. [DOI] [PubMed] [Google Scholar]
- Pooggin M, Shivaprasad PV, Veluthambi K, Hohn T. 2003. RNAi targeting of DNA virus in plants. Nat Biotechnol. 21:131–132. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P, Sanville BC, Buchmann RC, Bisaro DM. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol. 82:8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razifard H, Ramos A, Della Valle AL, Bodary C, Goetz E, Manser EJ, Li X, Zhang L, Visa S, Tieman D, et al. 2020. Genomic evidence for Complex domestication history of the cultivated tomato in Latin America. Mol Biol Evol. 37:1118–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Richert-Pöggeler KR, Noreen F, Schwarzacher T, Harper G, Hohn T. 2003. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 22:4836–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Pöggeler KR, Vijverberg K, Alisawi O, Chofong GN, Heslop-Harrison JS, Schwarzacher T. 2021. Participation of multifunctional RNA in replication, recombination and regulation of endogenous plant pararetroviruses (EPRVs). Front Plant Sci. 12:689307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarez MPS, Vučurović A, Mehle N, Ravnikar M, Kutnjak D. 2021. Global advances in tomato virome research: current Status and the impact of high-throughput sequencing. Front Microbiol. 12:1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ. 2011. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol. 9:99–108. [DOI] [PubMed] [Google Scholar]
- Sanan-Mishra N, Abdul Kader Jailani A, Mandal B, Mukherjee SK. 2021. Secondary siRNAs in plants: biosynthesis, Various functions, and applications in virology. Front Plant Sci. 12:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Gaubert H, Drost HG, Zabet NR, Paszkowski J. 2017. High-frequency recombination between members of an LTR retrotransposon family during transposition bursts. Nat Commun. 8:1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Gaubert H, Yang W. 2019. Evidence of developmental escape from transcriptional gene silencing in MESSI retrotransposons. New Phytol. 223:950–964. [DOI] [PubMed] [Google Scholar]
- Sarkinen T, Bohs L, Olmstead RG, Knapp S. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol Biol. 13:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature. 463:178–183. [DOI] [PubMed] [Google Scholar]
- Seo E, Kim T, Park JH, Yeom S-I, Kim S, Seo M-K, Shin C, Choi D. 2018. Genome-wide comparative analysis in Solanaceous species reveals evolution of microRNAs targeting defense genes in Capsicum spp. DNA Res. 25:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Du H, Liu Y, Ni L, Wang Z, Liang C, Tian Z. 2019. Update soybean Zhonghuang 13 genome to a golden reference. Sci China Life Sci. 62:1257–1260. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D. 2005. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 37:641–644. [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM. 2000. Total silencing by intron-spliced hairpin RNAs. Nature. 407:319–320. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 1:2019–2025. [DOI] [PubMed] [Google Scholar]
- Staginnus C, Gregor W, Mette MF, Teo CH, Borroto-Fernández EG, Machado ML, Matzke M, Schwarzacher T. 2007. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staginnus C, Richert-Pöggeler KR. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci. 11:485–491. [DOI] [PubMed] [Google Scholar]
- Sukal AC, Kidanemariam DB, Dale JL, Harding RM, James AP. 2018. Characterization of a novel member of the family caulimoviridae infecting Dioscorea nummularia in the pacific, which may represent a new genus of dsDNA plant viruses. PLoS One. 13:e0203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Baidaliuk A, Miesen P, Frangeul L, Crist AB, Merkling SH, Fontaine A, Lequime S, Moltini-Conclois I, Blanc H, et al. 2020. Non-retroviral endogenous viral element limits cognate virus replication in aedes aegypti ovaries. Curr Biol. 30:3495–3506.e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P. 2020. Key mechanistic principles and considerations concerning RNA interference. Front Plant Sci. 11:1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taochy C, Gursanscky NR, Cao J, Fletcher SJ, Dressel U, Mitter N, Tucker MR, Koltunow AMG, Bowman JL, Vaucheret H, et al. 2017. A genetic screen for impaired systemic RNAi highlights the crucial role of DICER-LIKE 2. Plant Physiol. 175:1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teycheney P-Y, Geering ADW, Dasgupta I, Hull R, Kreuze JF, Lockhart B, Muller E, Olszewski N, Pappu H, Pooggin MM, et al. 2020. ICTV Virus taxonomy profile: caulimoviridae. J Gen Virol. 101:1025–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Cannon SB, Bayer PE, Shu S, Brown AV, Ren L, Jenkins J, Chung CYL, Chan T-F, Daum CG, et al. 2019. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 100:1066–1082. [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell. 136:669–687. [DOI] [PubMed] [Google Scholar]
- Wang Z, Baulcombe DC. 2020. Transposon age and non-CG methylation. Nat Commun. 11:1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gao L, Jiao C, Stravoravdis S, Hosmani PS, Saha S, Zhang J, Mainiero S, Strickler SR, Catala C, et al. 2020b. Genome of Solanum pimpinellifolium provides insights into structural variants during tomato breeding. Nat Commun. 11:5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hardcastle TJ, Canto Pastor A, Yip WH, Tang S, Baulcombe DC. 2018. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 32:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang J, Zuo T, Zhao M, Lisch D, Peterson T. 2020a. Small RNA-mediated De Novo silencing of Ac/Ds transposons is initiated by alternative transposition in maize. Genetics. 215:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci U S A. 95:13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Li B, Iwakawa HO, Pan Y, Tang X, Ling-Hu Q, Liu Y, Sheng S, Feng L, Zhang H, et al. 2020. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature. 581:89–93. [DOI] [PubMed] [Google Scholar]
- Xie M, Chung CY-L, Li M-W, Wong F-L, Wang X, Liu A, Wang Z, Leung AK-Y, Wong T-H, Tong S-W, et al. 2019. A reference-grade wild soybean genome. Nat Commun. 10:1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104-E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Brockmöller T, Navarro-Quezada A, Kuhl H, Gase K, Ling Z, Zhou W, Kreitzer C, Stanke M, Tang H, et al. 2017. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc Natl Acad Sci U S A. 114:6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplemental information files), or are available upon request.