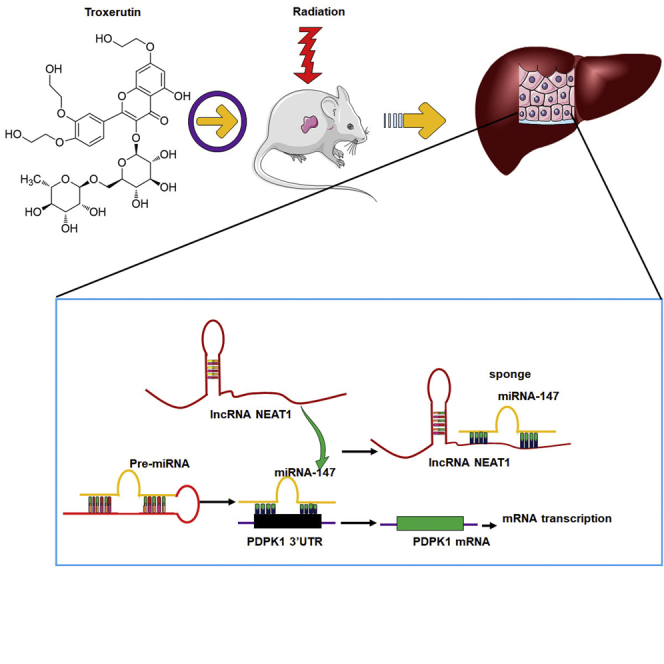
Summary
A better understanding of the molecular mechanism involving the lncRNA-miRNA-mRNA network underlying radiation damage can be beneficial for radioprotection. This study was designed to investigate the potential role of lncRNA NEAT1, miR-147 and Phosphoinositide Dependent Protein Kinase 1 (PDPK1) interaction in radioprotection by troxerutin (TRT). We first demonstrated that NEAT1 sponged miR-147, and PDPK1 mRNA was the primary target of miR-147. In the cells, the NEAT1 and PDPK1 levels were downregulated after the radiation but increased after the treatment with TRT. The miR-147 level was significantly induced by radiation and inhibited by TRT. NEAT1 negatively regulated the expression of miR-147, whereas miR-47 targeted PDPK1 to downregulate its expression. In radioprotection, TRT effectively upregulated NEAT1 to inhibit miR-147 and to upregulate PDPK1. We concluded that TRT could promote radioprotection by stimulating NEAT1 to upregulate PDPK1 expression by suppressing miR-147. NEAT1 could be a critical therapeutic target of radiation damage.
Subject areas: Biological sciences, Cell biology, Molecular biology, Molecular mechanism of gene regulation
Graphical abstract
Highlights
-
•
NEAT1 and PDPK1 levels were downregulated after the radiation
-
•
The miR-147 level was significantly induced by radiation
-
•
NEAT1 targeted miR-147, whereas miR-147 targeted PDPK1
Biological sciences; Cell biology; Molecular biology; Molecular mechanism of gene regulation
Introduction
The liver is often incidentally irradiated during radiation therapy (RT) for treating the tumors in the upper abdomen, right lower lung, distal esophagus, or whole abdomen or whole-body RT. RT-induced liver disease (RILD) involves anicteric hepatomegaly and ascites, typically occurring between 2 weeks and 3 months after therapy.1 Treatment options for RILD are limited, leading to liver failure and death. A previous study has indicated that abnormal expression of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), as well as their potential interactions in radiation, could serve as molecular targets in radioprotection.2 Thus, a better understanding of molecular mechanisms underlying radiation might be beneficial to improve its therapeutic applications.
Long non-coding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with limited protein-coding potential. Recent studies have revealed that lncRNAs can play multiple roles in various biological processes, such as controlling cell division, apoptosis, migration, carcinogenesis, healing, and tissue repair. It also shown that lncRNAs are frequently dysregulated in several diseases.3 Moreover, published studies of whole genome expression analysis have identified that lncRNA transcripts can change in response to either ultraviolet (UV) or ionizing radiation in the cultured peripheral blood mononuclear cells (PBMCs),4 thymocytes,5 melanocytes6 and human bronchial epithelial cells.7 High- and low-dose X-rays also induced different subsets of lncRNAs in the primary breast epithelial cells.8 It has been established that the nuclear paraspeckle assembly transcript 1 (NEAT1) was highly expressed in cancer tissues, non-sensitive tissues and radio-resistant cancer cells. NEAT1 was found to enhance the radio-resistance of cervical cancer by promoting cell proliferation, causing cell-cycle arrest and triggering cell apoptosis.9 Of interest, NEAT1 depletion enhanced the radio-sensitivity of gastric cancer by negatively regulating miR-27b-3p under both in vitro and in vivo studies.10 In brief, lncRNA NEAT1 was identified as an important molecule in radiation-induced cancer cells. However, the change in NEAT1 level in the normal cells has not been reported.
miRNAs are highly conserved small single-stranded non-coding RNAs, which could directly regulate gene expression by inhibiting the translation or promoting the degradation of mRNA.11 Several reports have suggested that miRNAs are responsible for regulating more than 50% of all protein-coding genes and can control multiple biological processes, such as cell proliferation, differentiation, apoptosis and stress resistance.12 For instance, miR-147 can inhibit the PI3K/AKT/mTOR signaling pathway to prevent the proliferation and migration of breast cancer cells, but its direct target has not been elucidated.13,14miR-147 was also upregulated in mouse thymus responses to ionizing radiation by deep sequencing analysis.15
3-phosphoinositide-dependent protein kinase-l (PDPK1) plays an important role in controlling cell proliferation and survival.16,17 Gene knockout PDPK1 −/− mouse embryos died at 9.5 days. The PDPK1−/+ mice were 40–50% smaller than the normal mice, and the inhibition of cell proliferation was associated with low expression of PDPK1.18 In PDPK1 gene knockout cells or mice, AKT was inactivated, thus indicating that PDPK1 could act as an important upstream kinase of AKT, and the loss of AKT activity was related to its reduced phosphorylation.18,19 Elevated PDPK1 expression-driven PI3K/AKT/MTOR signaling cascade has been reported to promote radiation-resistant and dedifferentiate phenotype of hepatocellular carcinoma.20
NEAT1 contains few specific sequences that can sponge miR-147, and the 3′UTR region of PDPK1 has a targeting sequence of miR-147. Therefore, it is important to study the interaction mechanism of NEAT1, miR-147 and PDPK1 during radiation damage. Troxerutin is a flavonoid with anti-radiation and antioxidant properties, and it has aroused substantial research interest owing to its extensive pharmacological activities.21,22 Our previous results have also indicated that TRT enhanced radioprotection at least partially by activating AKT to inhibit the activation of JNK.23 PDPK1 was an important upstream kinase of AKT. Therefore, we investigated the potential interaction between lncRNA NEAT1, miR-147 and PDPK1 in TRT radioprotection and identified their possible roles in liver radioprotection. Design of the study was shown in Table 1. These findings can provide new insights for understanding the mechanisms involved in radiation treatment.
Table 1.
Design of the study
| Relationship | Cells |
|---|---|
| NEAT1-miR-147 (Luciferase) | 293T, BNL CL2, L02 |
| miR-147-PDPK1(Luciferase) | 293T, BNL CL2, L02 |
| NEAT1-miR-147 (Radiation) | BNL CL2 |
| miR-147-PDPK1(AKT) (Radiation) | L02 |
| PDPK1-AKT (Radiation) | L02 |
| NEAT1-miR-147-PDPK1 (Radiation) | BNL CL2, L02, mice |
Results
NEAT1 targeted miR-147, whereas miR-147 targeted PDPK1
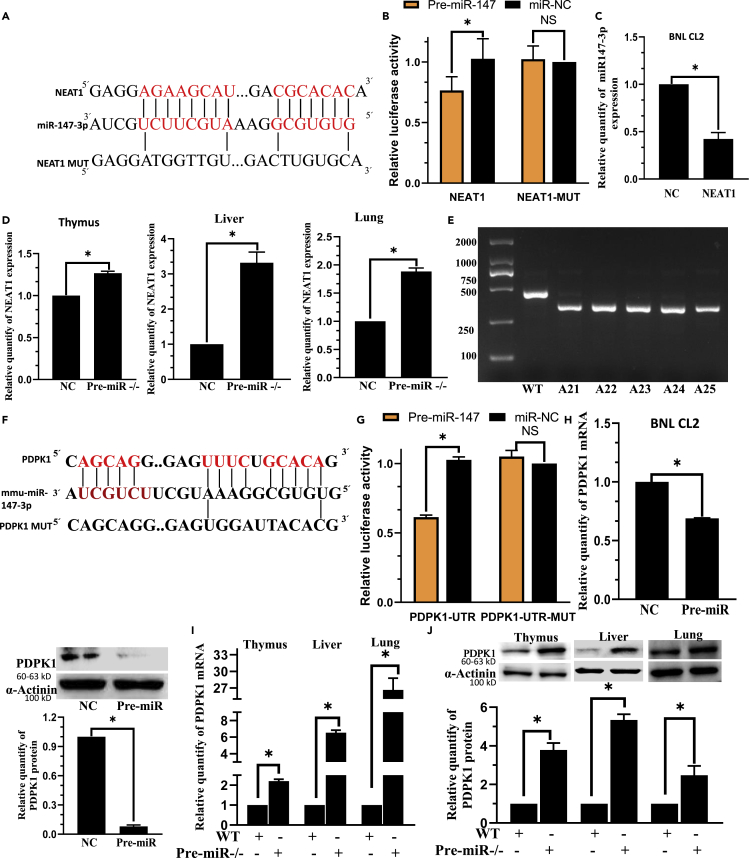
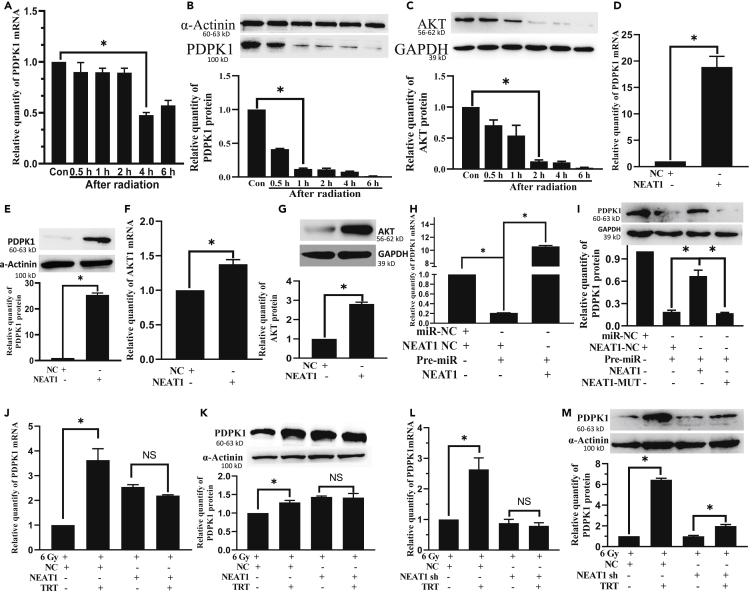
We searched RNAhybrid 2.2: https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html to acquire the predicted potential target lncRNAs of miR-147. We observed that NEAT1 was expected to be a target of miR-147, but the relationship between NEAT1 and miR-147 has not been functionally validated (Figures 1A, S1A, S1B, and S2A). HEK293T cells co-transfected with pre-miR-147 plasmid and NEAT1-WT exhibited a significant reduction in luciferase activity, confirming the target relationship between NEAT1 and miR-147. On the contrary, the co-transfection of miR-147 and NEAT1-MUT showed little influence on the luciferase activity (Figures 1B andS2B), thus establishing that miR-147 was the target miRNA of NEAT1. After overexpression of NEAT1 in BNL CL2 and L02 cells, miR-147 expression was significantly decreased (Figures 1C andS2C), thus further verifying the regulatory relationships between NEAT1 and miR-147. NEAT1 expression was considerably higher in the thymus, liver, and lung of pre-miR−/− mice compared to WT mice (Figure 1D) (pre-miR−/− mice, amplicon size = 324 bp; WT mice, amplicon size = 435 bp; Figures 1E andS1C).
Figure 1.
NEAT1 targets miR-147 and miR-147 can target PDPK1
(A) miR-147 is predicted to be a target miRNA of NEAT1.
(B) The relative luciferase activity in 293T cells co-transfected with miR-147 and NEAT1-WT plasmid decreased whereas that in the cells co-transfected with miR-47 and NEAT1-MUT did not exhibit any significant change.
(C) The relative expression of miR-147 in BNL CL2 cells was significantly decreased in NEAT1 group. U6 is the internal control.
(D) The relative expression of NEAT1 in the thymus, liver and lung was significantly increased in Pre-miR−/− group.
(E) Pre-miR−/− mice genotyping. Pre-miR−/− mice had a band at 324 bp, whereas WT mice displayed a band at 435 bp.
(F) PDPK1 is predicted to be a target miRNA of miR-147.
(G) The relative luciferase activity in 293 T cells co-transfected with miR-147 and PDPK1-WT plasmid decreased significantly whereas that in cells co-transfected with miR-47 and PDPK1-MUT showed no significant change.
(H) The relative expression of PDPK1 mRNA and protein in BNL CL2 cells significantly decreased in Pre-miR group.
(I) The relative expression of PDPK1 mRNA in the thymus, liver and lung significantly increased in Pre-miR−/− group.
(J) The relative expression of PDPK1 protein in the thymus, liver and lung significantly increased in Pre-miR−/− group. C, H, n = 3; B, G, n = 5; D, I, n = 8. ∗p < 0.01; NS: not significant. Data were shown as mean ± SEM.
Similarly, through TargetScan:https://www.targetscan.org/vert_80/ and RNAhybrid 2.2: https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html, we predicted PDPK1 as a target gene of miR-147 (Figures 1F andS2D). The luciferase activity depicted the potential interaction between miR-147 and PDPK 3′UTR. HEK293 T cells co-transfected with pre-miR-147 plasmid and PDPK1-WT exhibited a significant reduction in the luciferase activity. On the contrary, the co-transfection of miR-147 and PDPK1-MUT displayed minimal influence on the luciferase activity, thus proving that PDPK1 was the target of miR-147 (Figures 1G andS2E).
After overexpression of miR-147 in BNL CL2 and L02 cells, PDPK1 mRNA and protein levels were substantially decreased (Figures 1H, S2F, and S2G), thus further verifying the regulatory relationships between PDPK1 and miR-147. After 293 T cells were transfected with miR-147 overexpression plasmid for 48 hours (h), Act D (actinomycin D) 10 μg/mL was added to each group to inhibit the synthesis of mRNA. It was found that compared with the miR-NC group, the miR-147a overexpression plasmid group significantly inhibited PDPK1 mRNA expression (Figure S2G). In pre-miR−/− mice, PDPK1 mRNA and protein increased considerably in the thymus, liver and lung compared to WT mice (Figures 1I and 1J).
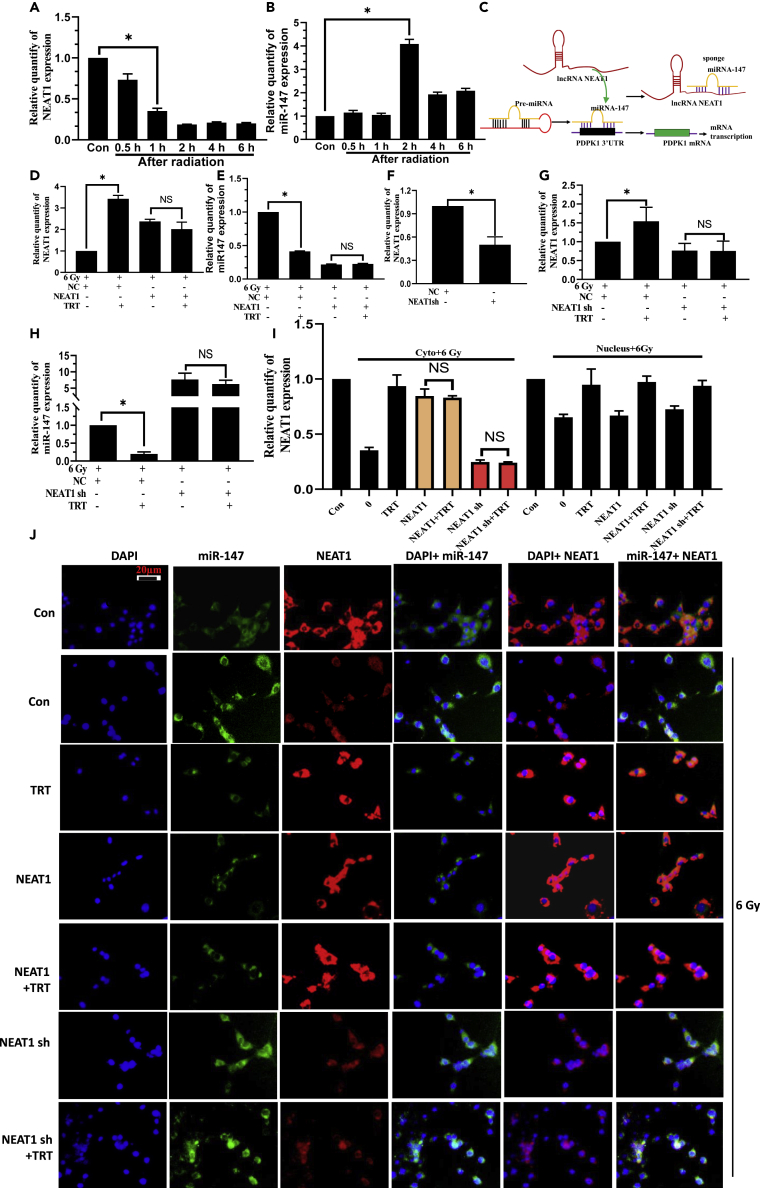
TRT downregulated miR-147 by targeting NEAT1 in BNL CL2 cells to facilitate radioprotection
The relative expression of NEAT1 was observed to be downregulated in BNL CL2 cells after the radiation (Figure 2A). The relative expression of miR-147was confirmed to be upregulated in BNL CL2 cells after the radiation (Figure 2B). Based on the result of Figure 1, we have proposed a schematic diagram of potential interaction mechanism between NEAT1 and miR-147 (Figure 2C). The expression of NEAT1 was significantly increased, and miR-147 was significantly decreased by treatment with TRT 10 μg/mL as compared with the radiation alone group. There was no significant difference observed in the expression of NEAT1 and miR-147 in NEAT1 +TRT groups, as compared with the NEAT1 group after radiation (Figures 2D and 2E). We used optimized NEAT1 shRNA to knockdown NEAT1 (Figure 2F). No significant difference was found in the expression of NEAT1 and miR-147 in NEAT1 shRNA + TRT groups, compared with the NEAT1 shRNA group after the radiation (Figures 2G and 2H). No significant difference was found in the expression of NEAT1 in the cytoplasm of BNL CL2 cells when NEAT1 shRNA or NEAT1 was transfected. But for the nucleus, there’s a difference (Figure 2I).
Figure 2.
TRT downregulated miR-147 by targeting NEAT1 in BNL CL2 cells after exposure to the radiation
(A) RT-qPCR was used to analyze NEAT1 expression at the different time points after the radiation.
(B) RT-qPCR analyzed miR-147 expression at the different time points after the radiation.
(C) A schematic diagram depicting the interaction mechanism between NEAT1 and miR-147.
(D) The effect of TRT on NEAT1 in the experimental group of radiation and impact of overexpression of NEAT1.
(E) The effect of TRT on miR-147 in the experimental group of radiation and overexpression of NEAT1.
(F) NEAT1 shRNA was transfected to facilitate the knockdown of NEAT1.
(G) The effect of TRT on NEAT1 in the experimental group of radiation and knockdown of NEAT1.
(H) The effect of TRT on miR-147 in the experimental group of radiation and knockdown of NEAT1.
(I) The effect of TRT on NEAT1 in the cytoplasm and nucleus in NEAT1 and NEAT1 shRNA groups co-treated with TRT and 6Gy.
(J) Fluorescence in situ hybridization analyze was used to detect NAET1 and miR-147 in irradiated BNL CL2 cells pretreated with TRT. Scale bar, 20 μm n = 3. ∗p < 0.01; NS: not significant. Data were shown as mean ± SEM.
Finally, to determine whether TRT prevented radiation in a NEAT1-dependent manner, we treated cells with TRT or NEAT1. As shown in Figure 2J, TRT reduced the radiation-induced elevation of miR-147 and elevated the radiation-induced reduction of NEAT1, compared to that in the 6 Gy group. As expected, co-treatment with NAET1 completely abolished the beneficial effects of TRT on miR-147 in radiation cells, suggesting that overexpression of NAET1 downregulated miR-147 expression and TRT downregulated miR-147 in a NAET1-dependent manner after radiation. Thus, it was concluded that TRT downregulated miR-147 by directly targeting NEAT1 for radioprotection.
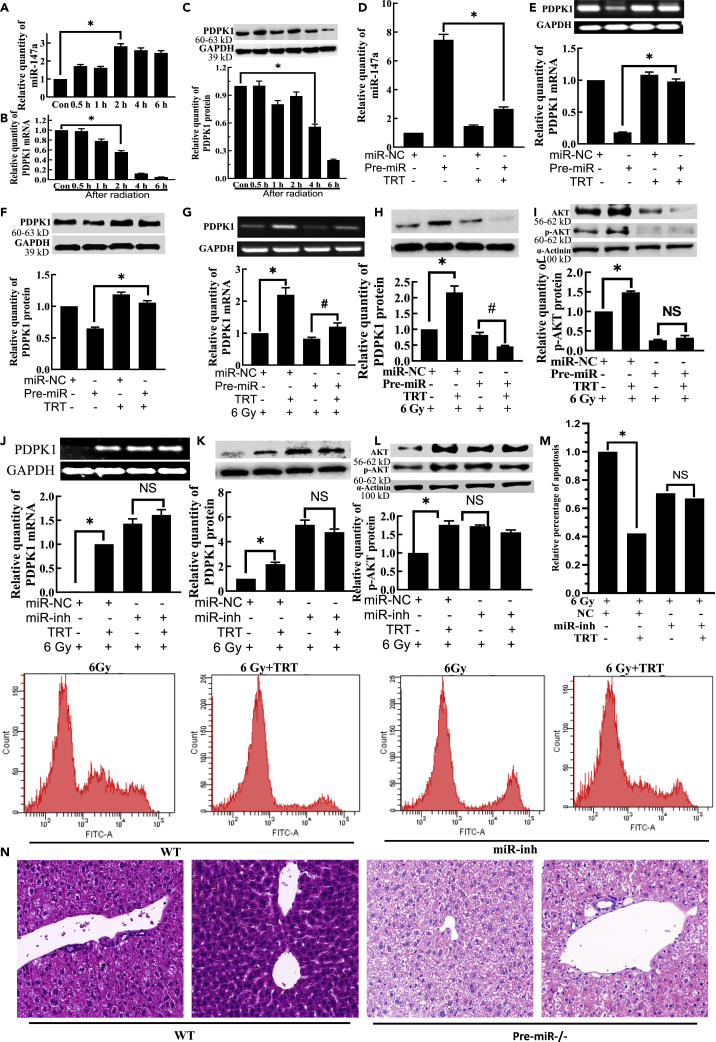
TRT upregulated PDPK1 by targeting miR-147 to activate AKT during the radioprotection
The relative expression of miR-147 was also found to be upregulated in cells after the radiation (Figures 3A andS3A). The relative expression of PDPK1 was confirmed to be downregulated in L02 cells after the radiation (Figures 3B, 3C, S3B, and S3C). In non-irradiated cells, pre-miR was transfected with TRT treatment, followed by 48 h incubation. It was found that TRT significantly reduced the expression of miR-147 (Figures 3D andS3D) and increased the expression of PDPK1 (Figures 3E, 3F, S3E, and S3F). However, in irradiated cells, the expressions of PDPK1 and p-AKT were significantly increased, whereas that of miR-147 was significantly decreased by TRT 10 μg/mL compared to the radiation alone group.
Figure 3.
TRT upregulated PDPK1 by targeting miR-147 to activate AKT during the radioprotection in L02 cells
(A) The expression of miR-147a in L02 cells at the different time points after radiation.
(B) The expression of PDPK1 mRNA in L02 cells at different time points after radiation.
(C) The expression of PDPK1 protein in L02 cells at different time points after radiation.
(D) The effect of TRT on miR-147a after miR-147a overexpression. Pre-miR was transfected with TRT treatment, and after 48 h, total RNA was extracted. TRT significantly reduced the expression of miR-147a.
(E) The effect of TRT on PDPK1 mRNA after miR-147a overexpression.
(F) The impact of TRT on PDPK1 protein after miR-147a overexpression.
(G) Effect of TRT on the expression of PDPK1 mRNA after transfection of pre-miR and radiation injury.
(H) Effect of TRT on the expression of PDPK1 protein after the transfection of pre-miR and radiation injury.
(I) Effect of TRT on the expression of p-AKT protein after transfection of pre-miR and radiation injury.
(J) Effect of TRT on the expression of PDPK1 mRNA after the transfection of miR-inh and radiation injury.
(K) The impact of TRT on the expression of PDPK1 protein after transfection of miR-inh and radiation injury.
(L) Effect of TRT on the expression of p-AKT protein after the transfection of miR-inh and radiation injury.
(M) Effect of TRT on the apoptosis of L02 cells after radiation injury by flow cytometry.
(N) Effect of TRT on the liver histopathological changes after radiation in pre-miR−/− mice treated with TRT. Scale bar, 50 μm. A-M, n = 3; N, n = 8. ∗p < 0.01, # <0.05; NS: not significant. Data were shown as mean ± SEM.
There were minimal or no significant differences in the expression of miR-147 (Figures S4A–S4C), PDPK1 (Figures 3G, 3H, S5A, and S5B) and p-AKT (Figures 3I andS5C) in pre-miR + TRT group, compared with the pre-miR group. There were no significant differences in the expression of miR-147 (Figures S4D–S4F), PDPK1 (Figures 3J, 3K, S5D, and S5E) and p-AKT (Figures 3L andS5F) in miR-inh + TRT group, compared with the miR-inh group, and only the difference in the expression of PDPK1 mRNA was found to be significant in V79 cells. From the analysis of PDPK1 mRNA (Figure S5D), the degree of PDPK1 mRNA was enhanced by TRT decreased significantly, compared with that of the non-transfected miR-inh group. As shown in Figure 3M, apoptosis was significantly reduced by exposure to TRT 10 μg/mL compared with the radiation alone group. There were no significant differences in apoptosis in the group co-treated with miR-147 inh, compared with the miR-147 inh group after the radiation.
These findings indicated that TRT reduced the apoptosis of liver tissue cells by decreasing the expression of miR-147 after the radiation. The same observation was true for the liver, thymus and lungs (Figure S6). The administration of TRT decreased the damage caused by radiation, with the mice’s tissues preserving their normal shape and less pathological variations. In Pre-miR−/− mice, the changes of TRT on tissue damage were not as obvious as those in WT mice (Figure 3N). Thus, it was concluded that TRT upregulated PDPK1 by targeting miR-147 to activate AKT in radioprotection.
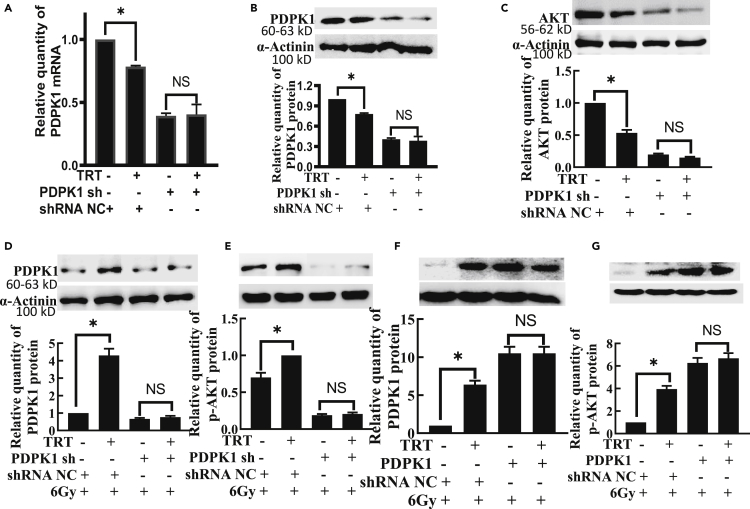
TRT upregulated p-AKT by targeting PDPK in radioprotection
In non-irradiated cells, pre-miR was transfected, and after 46 h incubation, TRT was added and incubated for an additional 2 h. It was observed that TRT significantly reduced the expression of PDPK1 (Figures 4A, 4B, S7A, and S7B) and AKT (Figures 4C andS7C), but after PDPK1 silencing, there were no significant differences in expression of PDPK1 and AKT by TRT. However, in irradiated cells, the expression of both PDPK1 and p-AKT was significantly increased by treatment with TRT 10 μg/mL compared with the radiation alone group. There were no significant differences in the expression of PDPK1 (Figures 4D andS7D) and p-AKT (Figures 4E andS7E) in the PDPK1 sh + TRT group, compared with the PDPK1 sh group. There were no significant differences in the expression of PDPK1 (Figures 4F andS7F) and p-AKT (Figures 4G,S7G, and S7H) in the PDPK1 + TRT group, compared with the PDPK1 group. The effect of TRT on p-AKT was found to disappear completely after co-administration of PDPK1 sh or PDPK1, thereby suggesting that TRT activated AKT through PADPK1 in radioprotection.
Figure 4.
TRT caused AKT activation by targeting PDPK1 in radioprotection in L02 cells
(A) The effect of TRT on PDPK1 mRNA after PDPK1 knockdown.
(B) The effect of TRT on PDPK1 protein after PDPK1 knockdown.
(C) The effect of TRT on AKT protein after PDPK1 knockdown.
(D) Effect of TRT on the expression of PDPK1 protein after PDPK1 knockdown and radiation injury.
(E) Influence of TRT on the expression of p-AKT protein after PDPK1 knockdown and radiation injury.
(F) Effect of TRT on the expression of PDPK1 protein after PDPK1 overexpression and radiation injury.
(G) Effect of TRT on the expression of p-AKT protein after PDPK1 overexpression and radiation injury. n = 3. ∗p < 0.01; NS: not significant. Data were shown as mean ± SEM.
TRT downregulated p-JNK expression by targeting AKT in radioprotection
After 46 h of AKT1-sh transfection, the cells were treated with TRT. After 2 h of incubation, the total protein was extracted. The results showed that TRT could significantly reduce the expression of p-JNK protein after the radiation. In addition, after AKT1 gene silencing, AKT1-sh +TRT group displayed no significant difference in the effect of p-JNK protein compared with the AKT1-sh group alone (Figures S8A–S8C). After overexpression of the AKT1 gene, AKT1 +TRT group showed no significant difference in the effect of p-JNK protein compared with the AKT1 overexpression group alone (Figures S8D–S8F). In addition, as expected, co-treatment with AKT1-sh or AKT1, TRT completely abolished the beneficial effects on the radiation, thereby suggesting that TRT downregulated p-JNK by targeting AKT1 in radioprotection.
NEAT1 elevated PDPK1 expression through targeting miR-147 during radioprotection conferred by TRT
The optimal time to identify PDPK1 mRNA was 4 h after radiation (Figure 5A). PDPK1 protein detection was carried out at 1h after radiation (Figure 5B). The AKT proteins were detected 2 h after radiation (Figure 5C). The relative expression of PDPK1 mRNA and protein significantly increased in NEAT1 group, as compared with the NC group (Figures 5D and 5E). Moreover, transfection with NEAT1, AKT1 mRNA and AKT protein was dramatically increased (Figures 5F and 5G). The inhibition of miR-147 on PDPK1 was significantly neutralized by NEAT1. In the group of NEAT1 MUT, the effect was canceled (Figures 5H, 5I, S9A, and S9B). On the contrary, in irradiated cells, the expression of PDPK1 was significantly increased by treatment with TRT 10 μg/mL compared with the radiation alone group. There were no significant differences in the expression of PDPK1 (Figures 5J, 5K, S9C, and S9D) NEAT1 + TRT group, compared with the NEAT1 group. The same observation was true for the experimental group transfected with NEAT1 shRNA (Figures 5L and 5M). These results suggested that TRT upregulated PDPK1 expression by causing down-regulation of miR-147 affected by up-regulating NEAT1.
Figure 5.
NEAT1 elevated PDPK1 expression through targeting miR-147 after radiation in BNL CL2 cells
(A) The expression of PDPK1 mRNA at the different time points after radiation.
(B) The expression of PDPK1 protein at the different time points after radiation.
(C) The expression of AKT protein at the different time points after radiation.
(D) The relative expression of PDPK1 mRNA significantly increased in the NEAT1 group.
(E) The relative expression of PDPK1 protein significantly increased in the NEAT1 group.
(F) The relative expression of AKT1 mRNA significantly increased in the NEAT1 group.
(G) The relative expression of AKT1 protein significantly increased in the NEAT1 group.
(H) The relative expression of PDPK1 mRNA decreased in the miR-147 group, compared with NC, and increased in the miR-147 + NEAT1 group, compared with miR-147 + NC.
(I) The inhibition of miR-147 on PDPK1 was significantly neutralized by NEAT1. In the group of NEAT1 MUT, the effect was canceled.
(J) Effect of TRT on the expression of PDPK1 mRNA after transfection of NEAT1 and radiation injury.
(K) Effect of TRT on the expression of PDPK1 protein after transfection of NEAT1 and radiation.
(L) Impact of TRT on the expression of PDPK1 mRNA after transfection of NEAT1 shRNA and radiation injury.
(M) Effect of TRT on the expression of PDPK1 protein after the transfection of NEAT1 shRNA and radiation injury. n = 3. ∗p < 0.01; NS: not significant. Data were shown as mean ± SEM.
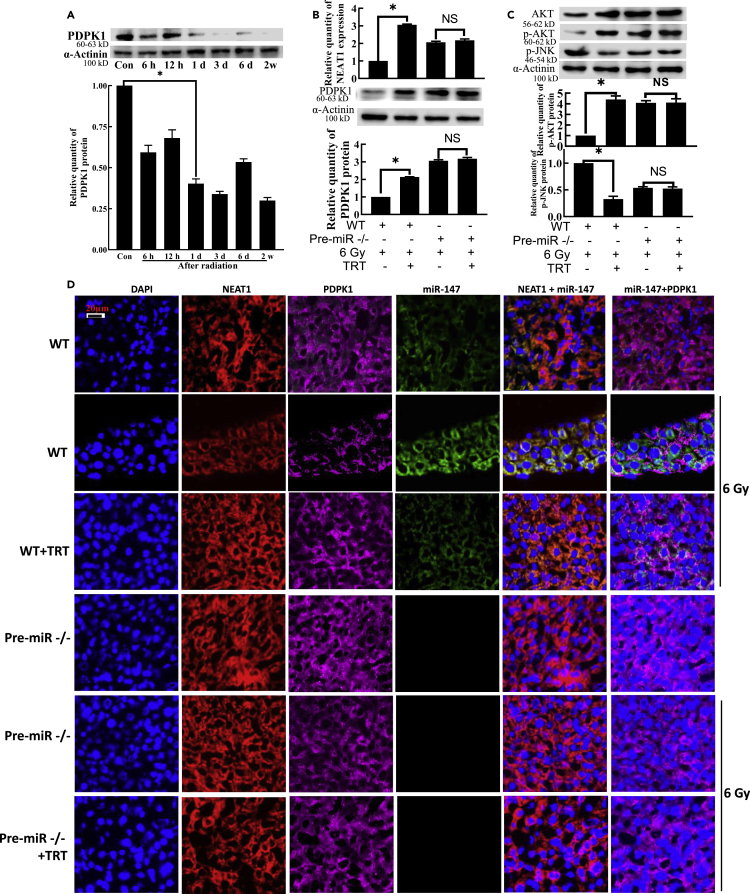
TRT interfered with the radiation damage in vivo by modulating the NEAT1-miR-147-PDPK1 pathway
After 6 Gy irradiation, PDPK1 was found to be significantly downregulated at 6 h. Still, the downregulated level was relatively stable at 1 day in the liver (Figure 6A) and 6 h in the thymus (Figure S10A). After C57BL/6N mice were irradiated, the expression of NEAT1 and PDPK1 protein in TRT 10 mg/kg group was found to be significantly higher than that in radiation alone group. For pre-miR−/− mice, the expressions of NEAT1 and PDPK1 had no significant difference between the TRT-irradiated and irradiated groups, except that PDPK1 in the thymus was significantly increased (Figures 6B, S10B, and S10C). The same observation was true for p-AKT and p-JNK expression (Figures 6C andS10C).
Figure 6.
TRT upregulated PDPK1 expression by targeting miR-147 during the radioprotection in the mice liver
(A) The expression of PDPK1 protein at different time points in the liver after radiation.
(B) TRT’s effect on the NEAT1 expression and PDPK1 protein expression after the radiation in pre-miR−/− mice.
(C) Effect of TRT on the expression of p-AKT and p-JNK proteins after the radiation in pre-miR−/− mice.
(D) Immunofluorescence and in situ hybridization analyses were used to detect NAET1, miR-147 and PDPK1 in irradiated liver tissues of pre-miR−/− mice pretreated with TRT. Scale bar, 20 μm n = 8. ∗p < 0.01; NS: not significant. Data were shown as mean ± SEM.
For immunofluorescence and in situ hybridization, in C57BL/6N mice, up-regulation of NEAT1 and down-regulation of miR-147 by TRT treatment at 10 mg/kg significantly increased PDPK1 expression compared with the irradiated group. For pre-miR−/− mice, there was no significant difference in PDPK1 protein expression between the TRT-irradiated group and the irradiated group in the liver (Figure 6D). The integrity of the thymus tissue was found to be severely damaged. The number of cells obviously decreased, and the boundary between the medulla and cortex was unclear. Administration of TRT was observed to reduce the number of thymocytes in mice caused by radiation, maintain the normal morphology of thymocytes in mice, decrease the diffusion of cortex and medulla, maintain clear boundaries, and improve the pathological changes of the thymus in the mice. For pre-mir−/− mice, the performance of TRT was not obvious (Figure S10D). No significant difference was found in the expression of NEAT1 in the cytoplasm and nucleus of pre-miR−/− mice (Figure S10E).
Discussion
The major finding of this study was that lncRNA NEAT1 can sponge mir-147, and PDPK1 mRNA is a potential target of miR-147. TRT can also activate NEAT1 sponging microRNA-147 to inhibit radiation damage by targeting PDPK1.
FISH assays further demonstrated the transposition of NEAT1 from the nucleus to the cytoplasm during BMSCs senescence and differentiation.24 Neat1, which normally resides in the paraspeckles, disassociates from these nuclear bodies and translocates to the cytoplasm to modulate inflammasome activation.25 The radiation treatment induced significant cell apoptosis and inflammation.26 So NEAT1 sponges miR-147 mainly in the cytoplasm in radiation. But it is unclear if FRT affects miRNA synthesis by interfering with Drosha and Dicer in our experiment. It will be considered in future studies. There is also recent data that radiation induces p53 in BNL CL2 cells27 and that NEAT1 is a direct target of p53.28,29,30 However, we detected a decrease in NEAT1 in response to radiation. After radiation, the change of molecules is a dynamic process, changing with time, and the change rules are also closely related to the radiation dose. Within 6 h after 6 Gy radiation, NEAT1 decreases with time, and there may be a rebound later, which is an adaptive response to radiation damage.
After radiation exposure, NEAT1 was found to be upregulated in cancer cells, thus resulting in radiation resistance. Loss of NEAT1 or upregulation of miR-27b-3p substantially increased the effect of radiation on cell survival inhibition and apoptosis promotion.10 NEAT1 has been previously reported to facilitate the radio-resistance of cervical cancer via competitively binding miR-193b-3p to upregulate the expression of cyclin D1.9 NEAT1 upregulation can promote proliferation but block cell apoptosis by regulating the miR-34a-5p/Bcl2 axis.31 Overall, these studies have suggested that NEAT1 may serve as a promising biomarker for prognosis and the target of treatment for cancers. However, the change of NEAT1 expression in the normal irradiated cells and its possible relationship with radiation injury has not been reported previously. The expression of NEAT1 in BNL CL2 cells decreased, and miR-147 increased after the radiation exposure (Figures 2A and 2B).
Moreover, NEAT1 contains several target sequences of miR-147, so we further confirmed that miR-147 was a putative target of NEAT1. This new relationship in radiation injury was verified, and TRT upregulated NEAT1 to effectively downregulate miR-147 and thus interfered with the radiation injury. This study was the first to ascertain that NEAT1 could primarily determine miR-147 stability. This discovery uncovered a new mechanism of miR-147 gene regulation and provided important insights into the various biological functions of NEAT1.
miR-147 was upregulated in the mouse thymus in response to ionizing radiation by deep sequencing analysis.15 It was also found that up-regulation of miR-147 inhibited the expression of pro-inflammatory cytokines (such as TNF-α and IL-6), indicating that miR-147 exhibited potent anti-inflammatory properties and functions as a negative regulator of TLR/NF-κB-mediated pro-inflammatory cytokines.32 In the radiation damage model, miR-147 and PI3K/PDPK1/AKT expression trends are exactly the opposite. It was observed that the level of miR-147 was significantly upregulated,15 whereas activation of the PI3K/AKT signaling pathway was inhibited.33,34 No study has analyzed miR-147 expression in response to radiation exposure. In this study, RT-qPCR analysis indicated that the expression of miR-147 in L02, TC, and V79 cells was increased after the radiation. RT-qPCR and western blot analysis revealed that the expression of PDPK1 was markedly inhibited. There is a target area of miR-147 in the 3′UTR region of PDPK1, so we speculated that PDPK1 is one target of miR-147. In addition, we have previously reported that TRT can enhance radioprotection at least partially by modulating PI3K/AKT pathway.23 We found that TRT could downregulate miR-147 and upregulate PDPK1 after radiation. Based on the above findings, we speculated that TRT could activate PDPK1 by down-regulating miR-147 to reduce radiation damage. In the present study, we established a negative regulatory relationship between miR-147 and PDPK1, and miR-147-mediated action of TRT to improve the effect of radioprotection.
Elevated PDPK1 expression can activate PI3K/AKT/MTOR signaling pathway to promote radiation resistance.20 In the last decade, there has been an increased focus on the possible role of phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB, AKT)/mammalian target of Rapamycin (mTOR) signaling in the acquisition of an IR-resistant phenotype by the different cancerous cells of the different histological origin.35,36,37,38 Accruing evidence suggests activated AKT’s role in predicting sensitivity to anticancer chemo- or radio-therapy. It has been established that the activation of AKT through phosphorylation of the threonine residue (Thr)-308 in the AKT activation loop by 3-phosphoinositide-dependent protein kinase-1 (PDK1/PDPK1) can effectively enhance AKT activity by over 100-fold. When followed by phosphorylation of Serine (Ser)-473 at the C terminus, which is the AKT hydrophobic motif, it can induce AKT activity by an additional 7- to 10-fold and thus stabilize AKT in its active conformation.39 Consistent with these findings, PDPK1 has been shown to be dysregulated in various malignancies, and accumulating evidence also suggests that the underexplored PDK1 may serve as a therapeutic target and is a probable modulator of sensitivity in response to cancer therapy.40 TRT can effectively reverse the cognitive dysfunction caused by high cholesterol levels by activating the PI3K/AKT signaling pathway and inhibiting the activation of the mouse hippocampal endoplasmic reticulum stress pathway JNK1. It can be recommended as a candidate drug for both the prevention and treatment of cognitive dysfunction in patients with type 2 diabetes and Alzheimer’s disease.41 TRT was found to activate the nerve growth factor receptor TrkA, which can activate AKT to improve the memory ability of the galactose-treated mouse brain and significantly reduce oxidative stress.42 We have reported that pre-treatment of thymocytes with PI3K/AKT inhibitor before the radiation could substantially increase the apoptosis rate of cells after exposure to the radiation. In this study, the regulation of PDPK1 by TRT was further verified.
Conclusions
This study uncovered a common mechanism by which radiation damage can promote lncRNA-miRNA-mRNA interaction. NEAT1 could indirectly regulate PDPK1 expression by directly targeting miR-147. TRT upregulated NEAT1 level, resulting in the downregulation of miR-147 and consequent PDPK1 upregulation after radiation exposure. Thus, TRT controlled the interaction of lncRNA NEAT1-miRNA-147-PDPK1 mRNA after the radiation and reduced the occurrence of radiation damage (Figure S11). TRT activated NEAT1 sponging miRNA-147 to inhibit the radiation damage by targeting PDPK1 in radioprotection. In conclusion, targeting NEAT1 or miR-147 could form the basis of a strategy to improve radioprotection clinically.
Limitations of the study
First, we acknowledge that in the case of mir-147 gene knockout mice, there could potentially be other targets that may have a sequence or functional relationship with PDPK1, AKT and NEAT1. Second, further investigations are required to elucidate the underlying mechanisms of lncRNA-mRNA. In addition to the interaction between lncRNA-miRNA, lncRNA can also interact with mRNA. We did not study the interaction between lncRNA-mRNA, in fact, these effects may affect the final mRNA expression. Future experiments can be designed to study the mechanism of lncRNA-mRNA and jointly explore the effects of lncRNA and miRNA on mRNA.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-AKT | Proteintech Group Inc. | Cat#10176-2-AP; RRID: AB_2224574 |
| Anti-p-AKT | Proteintech Group Inc. | Cat#66444-1-1g; RRID: AB_2919327 |
| Anti-p-JNK | Wan-Lei Biotechnology Co., Ltd. | Cat#WL01813; RRID: AB_2910628 |
| Anti-α-Actintin | Proteintech Group Inc. | Cat#11313-2-AP; RRID: AB_2223815 |
| Anti-GAPDH | Proteintech Group Inc. | Cat#10494-1-AP; RRID: AB_2263076 |
| Anti-PDPK1 | Proteintech Group Inc. | Cat#17086-1-AP; RRID: AB_2161289 |
| Goat anti-rabbit IgG-Cy3 | Bi-Yun-Tian Biotechnology Co., Ltd. | Cat#A0516; RRID: AB_2893015 |
| Horseradish peroxidase (HRP)-labeled antibody | Cell Signaling Technology | Cat#7074S; RRID: AB_2099233 |
| Chemicals, peptides, and recombinant proteins | ||
| Troxerutin | MedChemExpress | Lot#27664 |
| TRIzol | Life Technologies | Lot#350302 |
| Critical commercial assays | ||
| ReverTra Ace qPCR RT Master Mix kit | Toyobo, Tonga,Japan | FSQ-201 |
| RT First Strand cDNA Synthesis Kit | Wuhan Servicebio Technology Co., Ltd. Wuhan, China | G3330-50 |
| SYBR1 Green Real-time PCR Master Mix kit | Toyobo, Tonga,Japan | QPK-201 |
| A protease inhibitor cocktail was obtained from Roche Diagnostics GmbH | Mannheim, Germany | 11836145001 |
| TurboFect Transfection Reagente | Thermo Fisher Scientific, America | R0531 |
| PMSF | Bi-Yun-Tian Biotechnology Co., Ltd. | ST506 |
| BCA protein concentration determination kit BCA protein concentration determination kit | Bi-Yun-Tian Biotechnology Co., Ltd. | P0010 |
| Beyo ECL Plus | Bi-Yun-Tian Biotechnology Co., Ltd. | P0018m |
| Experimental models: Cell lines | ||
| BNL CL2 | Beina Chuanglian Biotechnology Co., Ltd. | No.: BNCC 337873 |
| L02 | Beina Chuanglian Biotechnology Co., Ltd. | No.: BNCC 351907 |
| MTEC-1 | Shanghai Hongshun Biotechnology Co., Ltd. | No.: HSC9942 |
| V79 | Beijing Dingguo Changsheng Biotechnology Co., Ltd. | No.: CS0199 |
| Experimental models: Organisms/strains | ||
| C57BL/6N mouse | Beijing Wei-tong-li-Hua Experimental Animal Technology Co. | http://www.lascn.com/Category_1785/Index.aspx |
| Pre-mir-147−/− mouse | Chinese-French Immunoregulatory Genes Laboratory of Xinxiang Medical University | MGI number: 2676832 |
| Oligonucleotides | ||
| H-NEAT1- sequencing: Forward:5′-GGGGTGGTCTGAGGAGTGATG-3′ Reverse:5′-CATTACCAACAATACCGACTCCAA-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| M-Neat1 sequencing: Forward:5′-TAGGTTCCGTGCTTCCTCTTCT-3′ Reverse:5′-CCCTCTGTGAATCAACACCATC-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| mmu-miR-147-3p-RT sequencing:5′-CTCAACTGGTGTCGTGGAGTCGGCA ATTCAGTTGAGTAGCAGAAGCATTT-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| mmu-miR-147-3p-Ssequencing:5′-GTGT GCGGAAATGCTTCTGCTA-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| hsa-miR-147a-RT sequencing:5′-CTCAACTGGTGTCGTGGAGTCGGCA ATTCAGTTGAGGCAGAAGC-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| hsa-miR-147a-S sequencing:5′-ACACTCCAG CTGGGGTGTGTGGAAATGC-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| universal primer-A sequencing:5′-TGGTGTCGTGGAGTCG-3′ | Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| M-PDPK1 sequencing: Forward:5′-AGGAGGACGCTGAGGAGG-3′ Reverse:5′-AATGGGCACAGCGTCATACA-3′ |
Wuhan Servicebio Technology Co., Ltd.’ Wuhan, China | N/A |
| H-PDPK1 sequencing: Forward:5′-CAGAGGTCAGGCAGCAACATA-3′ Reverse:5′-CTGTTAGGCAAGGGTTTCCG-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| H-GAPDH sequencing: Forward:5′-GGAAGCTTGTCATCAATGGAAATC-3′ Reverse:5′-TGATGACCCTTTTGGCTCCC-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| M-GAPDH sequencing: Forward:5′-CCTCGTCCCGTAGACAAAATG-3′ Reverse:5′-TGAGGTCAATGAAGGGGTCGT-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| U6 sequencing: Forward:5′-CTCGCTTCGGCAGCACA-3′ Reverse:5′-AACGCTTCACGAATTTGCGT-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| SgRNA-1(matching forward strand of the gene)sequencing:5′-AGAACACTCTATGAATCTAG TGG-3′ | Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| SgRNA-2(matching reverse strand of the gene)sequencing:5′-GCAGAAGCATTTCCGCACAC TGG-3′ | Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| SgRNA-3(matching forward strand of the gene)sequencing:5′-GATCCTGCATTAGCAAGTGA AGG-3′ | Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| (miR-147, genotyping) Forward primer sequencing:5′-TGAGGAGCTGCAAAAAGTCC-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| (miR-147, genotyping) Reverse primer sequencing:5′-CAAAATGTGCTTGTTCCCATAA-3′ |
Wuhan Servicebio Technology Co., Ltd. Wuhan, China | N/A |
| Recombinant DNA | ||
| mmu-miR-147-3p-inh-IMAT0004857(GV249) | Shanghai Genechem Co., Ltd. | GMDE0168564 |
| mmu-mir-147-MI0005482(GV268) | Shanghai Genechem Co., Ltd. | GMUE0284900 |
| hsa-miR-147a-inh-MIMAT0000251(GV249) | Shanghai Genechem Co., Ltd. | GMDE0189784 |
| has-mir-147a-MI0000262(GV268) | Shanghai Genechem Co., Ltd. | GMUE0103344 |
| AKT1-NM_009652(GV146) | Shanghai Genechem Co., Ltd. | GOSE0217154 |
| AKT1 sh-NM_009652(GV102) | Shanghai Genechem Co., Ltd. | GIEE0217153 |
| Pdpk1 sh-NM_011062(GV102) | Shanghai Genechem Co., Ltd. | GIEE0197550 |
| Pdpk1-NM_011062(GV146) | Shanghai Genechem Co., Ltd. | GOSE0168969 |
| NEAT1-NR_131012(miR-147a) (GV272) | Shanghai Genechem Co., Ltd. | GOSE0284892 |
| NEAT1-NR_131012(miR-147a)-mut(GV272) | Shanghai Genechem Co., Ltd. | GOSE0284897 |
| Neat1-NR_131012(miR-147-3p) (GV272) | Shanghai Genechem Co., Ltd. | GOSE0284902 |
| Neat1-NR_131212(miR147-3p)mut (GV272) | Shanghai Genechem Co., Ltd. | GOSE0284903 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ping Xu (13273730271@163.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cells
TC, MTEC1, V79 and L-02 cells were cultured in RPMI-1640 medium containing 100 U/mL penicillin, 100 U/mL streptomycin and 10% fetal bovine serum, at 37°C in a humidified atmosphere of 5% CO2. BNL CL2 were cultured in DMEM medium in the same condition with other cells.
Animals
Pre-mir-147−/− mice were obtained from the Chinese-French Immunoregulatory Genes Laboratory of Xinxiang Medical University. WT mice were purchased from Beijing Wei-tong-li-Hua Experimental Animal Technology Co., Ltd, and all animals were housed in temperature-controlled cages with a 12-hour light-dark cycle. Male mice (4–6 weeks old, 20 ± 2 g) were used in in vivo experiments, which were conducted following the Guide for the Care and Use of Laboratory Animals. The study was approved by the Animal Center of Xinxiang Medical University (Henan, China).
Irradiation
The cells were irradiated in vitro at room temperature at a dose rate of 100 cGy/min for a total dose of 6 Gy. The cells in the treatment group were exposed to 10 μg/mL of TRT for 2–24 h. For the in vivo studies, animals were placed in bespoke boxes before exposure to 6 Gy of total body radiation at a dose rate of 100 cGy/min. TRT was administered orally for 4 days at a 10 mg/kg dose before irradiation.
Method details
Plasmid transfection into HEK293 and reporter assays
The plasmid constructs (NEAT1-UTR or MUT-NEAT1-UTR; PDPK1-UTR or MUT-PDPK1-UTR) were co-transfected in HEK293 cells with the GV272 (SV40-Luc-MCS) plasmid and miR-147 (or negative control) cloned to GV268 (CMV-MCS-SV40-Neomycin) by using transfection reagent turbofect (DNA 2.5 μg, turbofect 5 μL). After 48-hour transfection, the cells’ luciferase activity of fireflies and sea kidneys was measured. The sequence of pre-mmu-mir-147 used was TATGAATCTAGTGGAAACATTTCTGCACAAACTA.
GATGTTGATGCCAGTGTGCGGAAATGCTTCTGCTACATTTGTAGG.
The sequence of pre-hsa-mir-147a used was AATCTAAAGACAACATTTCTGCACACAC.
ACCAGACTATGGAAGCCAGTGTGTGGAAATGCTTCTGCTAGATT.
RNA quantifications
Total RNA was isolated using a TRIzol-based (Invitrogen) RNA isolation protocol. RNA was analyzed by Nanodrop 2000C (Thermo Scientific). After that, 2 μg RNA was reversely transcribed using the ReverTra Ace qPCR RT Master Mix Kit according to the manufacturer’s instructions. Reverse transcription of miRNA was performed by cervical ring method, using RT First Strand cDNA Synthesis Kit. PCR cycling conditions for NEAT1 and PDPK1 were 95°C for 1 min, 40 cycles of 95°C for 15 sec and 60°C for 30 sec, and 72°C for 45 sec. The sequences of the various primers used (all the primers were designed and synthesized by Wuhan Servicebio Biotechnology Co., Ltd (Wuhan, China) have been listed in key resources table.
Western blotting
The cells were harvested and lysed in lysis buffer containing NP40 and cocktail protease inhibitor at 50 mL:1 tablet (20 mM Tris-HCl, pH 7.4, 137 mM NaCl, 2 mM Na2EDTA, 1% NP-40, 10% Glycerol; approximately 100 μL of lysis buffer per 106 cells were used. Before use, PMSF (Phenylmethanesulfonyl fluoride) was added to NP40 buffer, and the final concentration was set at 1 mM. The cell was lysed with NP40 cell-lysis buffer. The protein content was assayed by BCA (Bicinchoninic acid) protein assay reagent. 100 μg proteins were loaded to SDS-PAGE and then transferred to the PVDF membrane. The membrane was incubated with a dilution of the primary antibody, followed by a 1:5000 dilution of horseradish peroxidase-conjugated secondary antibody. The protein bands were then visualized by ECL (Enhanced Chemiluminescence, Shanghai, China). The intensity (area X density) of the individual band on the western blots was measured by ImageJ software. The background was subtracted from the calculated area.
Fluorescence in situ hybridization (FISH) and immunofluorescence (IF)
FISH of NEAT1 and miR-147and co-immunofluorescence with apoptosis inhibitory molecular marker PDPK1 was performed on 4-μm paraffin-embedded liver sections. Briefly, the sections of paraffin-embedded tissue were cut using a slice, mounted on the polylysine microscope slides and stored at 4°C for FISH and IF analyses. Tissue repair fluid was used to repair the various antigens, and protease K digestion facilitated the entry of miRNA hybridization probes. The paraffin was removed in xylene, and the slices were rehydrated in a series of progressively decreasing ethanol solutions, washed with DEPC water, and then fixed in 4% paraformaldehyde.
The pre-hybridization solution was incubated at 37°C for 1 hour. The pre-hybridization solution was then removed, and the hybridization solution containing miR-147 probe (5-GUGUGCGGAAAUGCUUCUGCUA-3-DIG labelled) and NEAT1 probe(cy3-5-GCCCCCACUAAGUGCAGUCCUGUCAUCC-3-cy3) was dropped (1 μM), and the hybridization was conducted overnight at 37°C. The slides were washed with gradient-decreasing SSC (Saline Sodium Citrate Buffer) at 37°C. After blocking, the slides were incubated with anti-DIG-488 and shielded from light for 50 min, followed by washing with PBS three times. PDPK1 primary antibody diluent was then added and incubated at 4°C overnight. Then the slides were washed with PBS for 3 × 5min. The corresponding secondary antibody-cy5 dilution was added and incubated at room temperature for 50 min. After that, the slides were washed again with PBS for 3 × 10 min. The slices were added with DAPI dye and incubated for 8 min in darkness. After rinsing, anti-fluorescence quenching agents were added to the slides. The sections and captured images were observed under the MIDI Panoramic scanner (3DHISTECH, Budapest, Hungary). The two ends of the NEAT1 probe are labeled with cy3, and red light is excited. miR-147 was labeled with digoxin, digoxin antibody was labeled with 488, and the green light is excited. PDPK1 antibody was labeled with cy5, and purple light is excited.
Analysis of apoptosis of the tissues by Hoechst and HE staining
On day 6 after irradiation, the mice’ liver and thymus tissues were removed and immediately rinsed and fixed. Hoechst 33342 and HE staining were performed after the paraffin embedding and sectioning.
Flow cytometry assays (FCM)
An Annexin V-FITC flow cytometry assay was used to detect cellular apoptosis. Cells were incubated with treatment drugs for 2 h and then irradiated with 6 Gy radiation. After 24 h, cells were treated according to the Annexin V-FITC apoptosis kit’s instructions. Apoptosis was identified by flow cytometry (FCM) as green fluorescence, cell death was represented by red fluorescence while living cells emitted no fluorescence.
Identification of miR-147 in the mouse tail
Preparation and identification of pre-miR-147−/− mice
C57BL/6N-miR-147 mice, referred to as pre-miR-147−/− mice, were prepared by the Chinese-French Immunoregulatory Genes Laboratory of Xinxiang Medical University, with MGI number: 2676832. CRISPR/Cas9 technology was used to knock out pre-miR-147 of the miR-147 gene by sgRNAs. The final 0.2–1 cm of the tails of experimental mice were placed in 60 μL Alkaline lysis reagent at 100°C for 30min, cooled down to 4°C. Neutralizing reagent 80 μL was added, centrifuged at 4°C 10,000 RPM for 5min, and 2–4 μL supernatant was taken for PCR amplification (94°C, 2min; 35 cycles of 94°C for 30 S, 60°C for 30 S and 72°C for 12 S; 72°C, 2min; 4°C, hold).
The final 0.2–1 cm of the tails of experimental mice were placed in 60 μL alkaline lysis reagent at 100°C for 30 min and then cooled down to 4°C. After that, 80 μL of the neutralizing agent was added and centrifuged at 4°C 10,000 rpm for 5min, and 2–4 μL of supernatant was used for PCR amplification (94°C, 2min; 35 cycles of 94°C for 30 S, 60°C for 30 S and 72°C for 12 S; 72°C, 2min). The sequence and sites of sgRNAs and primers were listed in key resources table.
QUANTIFICATION AND STATISTICAL ANALYSIS
Randomization was used to assign the various samples to the experimental groups and treatment conditions for all in vivo studies. All quantitative results were expressed as mean ± Standard Error of Mean (SEM). The data was analyzed using Unpaired Student’s t-test or analysis of variance (ANOVA) on GraphPad Prism 8.0 and ImageJ software. A value of p < 0.05 was considered to be statistically significant.
Acknowledgments
We thank Dr Shuangxi Wang’s insightful assistance in initiating this study. This work was supported by the Natural Science Foundation of China (NSFC) (Nos. 81773358, 11705158, U1504824) and the Young Teachers Plan of Higher Schools in Henan Province (2014GGJS-098).
Author contributions
P.X. designed and performed experiments, analyzed data and wrote the paper. Y-J.H., G-Y.S. and F.Z. performed experiments and analyzed the data. They contribute equally. N.Z., F.W., J-L.W., X.W., T-Y.W., Y-F.L., F.Z., Y-D.Y., W-T.D., and C-Y.C. performed some experiments. All authors read and approved the final manuscript.
Declaration of interests
The authors of this study declare that there is no conflict of interest.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105932.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Lawrence T.S., Robertson J.M., Anscher M.S., Jirtle R.L., Ensminger W.D., Fajardo L.F. Hepatic toxicity resulting from cancer treatment. Int. J. Radiat. Oncol. Biol. Phys. 1995;31:1237–1248. doi: 10.1016/0360-3016(94)00418-K. [DOI] [PubMed] [Google Scholar]
- 2.Tian B., Fu H., Liu B., Zhu J., Zheng X., Ge C. Effects of amifostine pre-treatment on MIRNA, LNCRNA, and MRNA profiles in the hypothalamus of mice exposed to 60Co gamma radiation. Health Phys. 2020;119:297–305. doi: 10.1097/HP.0000000000001233. [DOI] [PubMed] [Google Scholar]
- 3.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer L., Nemec L., Wagner T., Ristl R., Altenburger L.M., Ankersmit H.J., Mildner M. Ionizing radiation regulates long non-coding RNAs in human peripheral blood mononuclear cells. J. Radiat. Res. 2017;58:201–209. doi: 10.1093/jrr/rrw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao H., Dong Z., Wei W., Shao L., Jin L., Lv Y., Zhao G., Jin S. Integrative analysis for the role of long non-coding RNAs in radiation-induced mouse thymocytes responses. Acta Biochim. Biophys. Sin. 2017;49:51–61. doi: 10.1093/abbs/gmw114. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Q., Wang Q., Chen X., Xia K., Tang J., Zhou X., Cheng Y., Chen Y., Huang L., Xiang H., et al. Analysis of lncRNAs expression in UVB-induced stress responses of melanocytes. J. Dermatol. Sci. 2016;81:53–60. doi: 10.1016/j.jdermsci.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Nie J., Peng C., Pei W., Zhu W., Zhang S., Cao H., Qi X., Tong J., Jiao Y. A novel role of long non-coding RNAs in response to X-ray irradiation. Toxicol. Vitro. 2015;30:536–544. doi: 10.1016/j.tiv.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Terradas M., Martín M., Repullès J., Huarte M., Genescà A. Distinct sets of lncRNAs are differentially modulated after exposure to high and low doses of X rays. Radiat. Res. 2016;186:549–558. doi: 10.1667/RR14377.1. [DOI] [PubMed] [Google Scholar]
- 9.Han D., Wang J., Cheng G. LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget. 2018;9:2395–2409. doi: 10.18632/oncotarget.23416. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Jiang Y., Jin S., Tan S., Xue Y., Cao X. Long noncoding RNA NEAT1 regulates radio-sensitivity via microRNA-27b-3p in gastric cancer. Cancer Cell Int. 2020;20:581. doi: 10.1186/s12935-020-01655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahimian P., He J.J. HIV-1 Tat-shortened neurite outgrowth through regulation of microRNA-132 and its target gene expression. J. Neuroinflammation. 2016;13:247. doi: 10.1186/s12974-016-0716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams B.D., Kasinski A.L., Slack F.J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlmann S., Mannsperger H., Zhang J.D., Horvat E.Á., Schmidt C., Küblbeck M., Henjes F., Ward A., Tschulena U., Zweig K., et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang H.E., Liu Z. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol. Lett. 2016;11:405–410. doi: 10.3892/ol.2015.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C., Lu J., Hao L., Zheng Z., Zhang N., Wang Z. Discovery and characterization of miRNAs in mouse thymus responses to ionizing radiation by deep sequencing. Int. J. Radiat. Biol. 2016;92:548–557. doi: 10.1080/09553002.2016.1207821. [DOI] [PubMed] [Google Scholar]
- 16.Lu Z., Cox-Hipkin M.A., Windsor W.T., Boyapati A. 3-phosphoinositide-dependent protein kinase-1 regulates proliferation and survival of cancer cells with an activated mitogen-activated protein kinase pathway. Mol. Cancer Res. 2010;8:421–432. doi: 10.1158/1541-7786.MCR-09-0179. [DOI] [PubMed] [Google Scholar]
- 17.Qian X.J., Li X.L., Xu X., Wang X., Feng Q.T., Yang C.J. α-SMA-Cre-mediated excision of PDK1 reveals an essential role of PDK1 in regulating morphology of cardiomyocyte and tumor progression in tissue microenvironment. Pathol. Biol. 2015;63:91–100. doi: 10.1016/j.patbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Lawlor M.A., Mora A., Ashby P.R., Williams M.R., Murray-Tait V., Malone L., Prescott A.R., Lucocq J.M., Alessi D.R. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mora A., Komander D., van Aalten D.M.F., Alessi D.R. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. Cell Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Bamodu O.A., Chang H.L., Ong J.R., Lee W.H., Yeh C.T., Tsai J.T. Elevated PDK1 expression drives PI3K/AKT/MTOR signaling promotes radiation-resistant and dedifferentiated phenotype of hepatocellular carcinoma. Cells. 2020;9:746. doi: 10.3390/cells9030746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ping X., Junqing J., Junfeng J., Enjin J. Radioprotective effects of troxerutin against gamma irradiation in V79 cells and mice. Asian Pac. J. Cancer Prev. 2011;12:2593–2596. [PubMed] [Google Scholar]
- 22.Ping X., Junqing J., Junfeng J., Enjin J. Radioprotective effects of troxerutin against gamma irradiation in mice liver. Int. J. Radiat. Biol. 2012;88:607–612. doi: 10.3109/09553002.2012.692494. [DOI] [PubMed] [Google Scholar]
- 23.Xu P., Zhang W.B., Cai X.H., Qiu P.Y., Hao M.H., Lu D.D. Activating AKT to inhibit JNK by troxerutin antagonizes radiation-induced PTEN activation. Eur. J. Pharmacol. 2017;795:66–74. doi: 10.1016/j.ejphar.2016.11.052. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Xu R., Li B., Xin Z., Ling Z., Zhu W., Li X., Zhang P., Fu Y., Chen J., et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 2022;29:351–365. doi: 10.1038/s41418-021-00858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., Cao L., Zhou R., Yang X., Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 2019;10:1495. doi: 10.1038/s41467-019-09482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu S.J., Zhang F., Wang L.J., Hao M.H., Yang X.J., Li N.N., Ji H.L., Xu P. Flavonoids of Rosa Roxburghii Tratt offers protection against radiation induced apoptosis and inflammation in mouse thymus. Apoptosis. 2018;23:470–483. doi: 10.1007/s10495-018-1466-7. [DOI] [PubMed] [Google Scholar]
- 27.Urabe M., Hikita H., Saito Y., Kudo S., Fukumoto K., Mizutani N., Myojin Y., Doi A., Sato K., Sakane S., et al. Activation of p53 after irradiation impairs the regenerative capacity of the mouse liver. Hepatol. Commun. 2022;6:411–422. doi: 10.1002/hep4.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W.G., Radaelli E., Vermi W., et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 29.Mello S.S., Sinow C., Raj N., Mazur P.K., Bieging-Rolett K., Broz D.K., Imam J.F.C., Vogel H., Wood L.D., Sage J., et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan J., Zhu Q., Zhang X., Wen Z., Zhang G., Li N., Pei Y., Wang Y., Pei S., Xu J., et al. Ezh2 competes with p53 to license lncRNA Neat1 transcription for inflammasome activation. Cell Death Differ. 2022;29:2009–2023. doi: 10.1038/s41418-022-00992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding N., Wu H., Tao T., Peng E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. OncoTargets Ther. 2017;10:4905–4915. doi: 10.2147/OTT.S142446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu G., Friggeri A., Yang Y., Park Y.J., Tsuruta Y., Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. USA. 2009;106:15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L., Wang R., Gao Y., Xu X., Fu K., Wang S., Li Y., Peng R. The protective role of interleukin-11 against neutron radiation injury in mouse intestines via MEK/ERK and PI3K/Akt dependent pathways. Dig. Dis. Sci. 2014;59:1406–1414. doi: 10.1007/s10620-013-3015-0. [DOI] [PubMed] [Google Scholar]
- 34.Li K.R., Yang S.Q., Gong Y.Q., Yang H., Li X.M., Zhao Y.X., Yao J., Jiang Q., Cao C. 3H-1, 2-dithiole-3-thione protects retinal pigment epithelium cells against Ultra-violet radiation via activation of Akt-mTORC1-dependent Nrf2-HO-1 signaling. Sci. Rep. 2016;6:25525. doi: 10.1038/srep25525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu S., Li Y., Lu Y., Huang J., Ren J., Zhang S., Yin Z., Huang K., Wu G., Yang K. LZTS2 inhibits PI3K/AKT activation and radioresistance in nasopharyngeal carcinoma by interacting with p85. Cancer Lett. 2018;420:38–48. doi: 10.1016/j.canlet.2018.01.067. [DOI] [PubMed] [Google Scholar]
- 36.Horn D., Hess J., Freier K., Hoffmann J., Freudlsperger C. Targeting EGFR-PI3K-AKT-mTOR signaling enhances radiosensitivity in head and neck squamous cell carcinoma. Expert Opin. Ther. Targets. 2015;19:795–805. doi: 10.1517/14728222.2015.1012157. [DOI] [PubMed] [Google Scholar]
- 37.Chang L., Graham P.H., Hao J., Ni J., Bucci J., Cozzi P.J., Kearsley J.H., Li Y. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875. doi: 10.1038/cddis.2013.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szymonowicz K., Oeck S., Malewicz N.M., Jendrossek V. New insights into protein kinase B/Akt signaling: role of localized Akt activation and compartment-specific target proteins for the cellular radiation response. Cancers. 2018;10:78. doi: 10.3390/cancers10030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J., Feng J., Li Y., Hammarsten O., Brazil D.P., Hemmings B.A. DNA-dependent protein kinase-mediated phosphorylation of protein kinase B requires a specific recognition sequence in the C-terminal hydrophobic motif. J. Biol. Chem. 2009;284:6169–6174. doi: 10.1074/jbc.C800210200. [DOI] [PubMed] [Google Scholar]
- 40.Gagliardi P.A., Puliafito A., Primo L. PDK1: at the crossroad of cancer signaling pathways. Semin. Cancer Biol. 2018;48:27–35. doi: 10.1016/j.semcancer.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Lu J., Wu D.M., Zheng Z.H., Zheng Y.L., Hu B., Zhang Z.F. Troxerutin protects against high cholesterol-induced cognitive deficits in mice. Brain. 2011;134:783–797. doi: 10.1093/brain/awq376. [DOI] [PubMed] [Google Scholar]
- 42.Lu J., Wu D.M., Hu B., Zheng Y.L., Zhang Z.F., Wang Y.J. NGF-Dependent activation of TrkA pathway: a mechanism for the neuroprotective effect of troxerutin in D-galactose-treated mice. Brain Pathol. 2010;20:952–965. doi: 10.1111/j.1750-3639.2010.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.