Abstract
The development of programmable biomaterials for use in nanofabrication represents a major advance for the future of biomedicine and diagnostics. Recent advances in structural nanotechnology using nucleic acids have resulted in dramatic progress in our understanding of nucleic acid-based nanostructures (NANs) for use in biological applications. As the NANs become more architecturally and functionally diverse to accommodate introduction into living systems, there is a need to understand how critical design features can be controlled to impart desired performance in vivo. In this review, we survey the range of nucleic acid materials utilized as structural building blocks (DNA, RNA, and xenonucleic acids), the diversity of geometries for nanofabrication, and the strategies to functionalize these complexes. We include an assessment of the available and emerging characterization tools used to evaluate the physical, mechanical, physiochemical, and biological properties of NANs in vitro. Finally, the current understanding of the obstacles encountered along the in vivo journey is contextualized to demonstrate how morphological features of NANs influence their biological fates. We envision that this summary will aid researchers in the designing novel NAN morphologies, guide characterization efforts, and design of experiments and spark interdisciplinary collaborations to fuel advancements in programmable platforms for biological applications.
I. INTRODUCTION
A fundamental axiom that holds true across biology and architecture states that “form follows function”; the rational design of systems and choice of materials provides a foundation for structural performance. On the scale of nanomaterials, the ability to construct tailored, self-assembling platforms for use as sensors and drug delivery vehicles in vivo has been catalyzed by advances in the field of nucleic acid (NA) nanotechnology.1 By exploiting the well-defined rules of standard Watson–Crick (WC) base pairing, NA strands can be designed to anneal predictably in solution, forming a near endless array of arbitrary shapes.2,3 These NA nanostructures (NANs) can take on a range of morphologies from simple geometric shapes formed from a few oligonucleotides to more complex nanoparticle platforms by a network of hundreds of annealed strands. Through synthetic chemistry and bioconjugation techniques, versatile component oligos can be modified with moieties that impart a variety of functions, including loading small molecule drugs, to attaching fluorophores, enzymes, antibodies, and nanoparticles.4 While DNA serves as the primary and traditional NA-material for nanofabrication, structural designs have been evolving rapidly with the use of RNA and synthetic NA analogs, broadly referred to as xenonucleic acids (XNA). However, with the increasing molecular complexity of NA-based materials, modification strategies, and nanocomposites, there is a gap in the literature regarding a comprehensive evaluation of the connection between choice of NA materials and morphology with the resulting biological fate of the assembly. A detailed understanding of the fate pathways of NANs within living organisms is critical to advance the technology toward future clinical relevance.5
To narrow our review's scope to provide maximal impact, we direct our focus to discrete nucleic acid nanostructures (NANs), defined herein as planar (2D) or volumetric (3D) shapes, featuring a finite size, formed primarily by annealing NA-based materials with at least four synthetic strands. Although larger megadalton or mesoscale heterogeneous assemblies, such as the products of hybridization chain reactions6 or rolling circle amplification,7–10 tile-based aggregate assemblies,11 spherical NANs built on non-NA nanoparticle platforms,12 and others have significant utility in biological applications, readers are directed to in-depth analysis of these platforms in the prior literature.13,14 We will place further emphasis on nanostructures and nanocomposites that have been demonstrated in cellular or in vivo conditions across applications in biology, sensing, or medicine. As such, this review will be divided into three main sections: (1) an introduction to NA materials and relevant nanostructures, (2) a survey of in vitro and in vivo characterization methods and resulting data, and then (3) an analysis of how this prior research informs our understanding of cross-disciplinary links between morphology and biological fate and what phenomena remain to be elucidated.
A. NAN applications and considerations
NA-based nanomaterials are uniquely different from other materials used in nanofabrication and offer several significant advantages with regard to biological applications.15 Compared to polymers, the well-defined base pairing properties of NAs enable programmable assembly of molecularly defined structures with tunable size, shape, and functional features. The capacity for bottom-up fabrication gives scientists unambiguous control over the position and valency of functional components, including therapeutic agents and ligands for targeting or other activities. This feature is critical for the design of drug delivery vehicles or biosensors and is a unique advantage to NA-based structures in comparison to conventional nanomaterials such as lipid or metal oxide nanoparticles that exhibit significant heterogeneity. Furthermore, synthetic NA strands are generally regarded as biocompatible, biodegradable, and non-cytotoxic16 whereas other systems, such as gold and silica nanoparticles, may have concerns with possible adverse effects of the component materials.17 These attributes highlight the properties of NA materials that are favorable for biological applications as delivery vehicles or other functional structures.
The first example of a NAN used in vivo was in 2011 when a DNA I-switch was demonstrated for pH-responsive fluorescent imaging in C. elegans.18 Since then, functional nanomaterials formed from NAs with molecular addressability in vivo have rapidly expanded through a variety of design strategies.1,19–22 Recent literature is abundant with works emphasizing the range of potential uses of NANs as drug delivery vehicles,23–25 therapeutics,26–30 and technologies for a variety of other biomedical applications.15,31–36 Additionally, NANs have been developed for detection and imaging of a range of analytes in biological systems,37,38 including pH,39,40 ions,40 and small molecules.39,41 For a more in-depth summary of the timeline26 and breadth of applications of NANs in living systems, readers are directed to the following reviews.16,42–46 Applications for NANs as tools in nanofabrication, computing, and other in vitro areas have been thoroughly reviewed elsewhere.47–50 Herein, we will focus on the design, application, and characterization of NA nanostructures that are used for biological applications, including therapeutics and sensing.
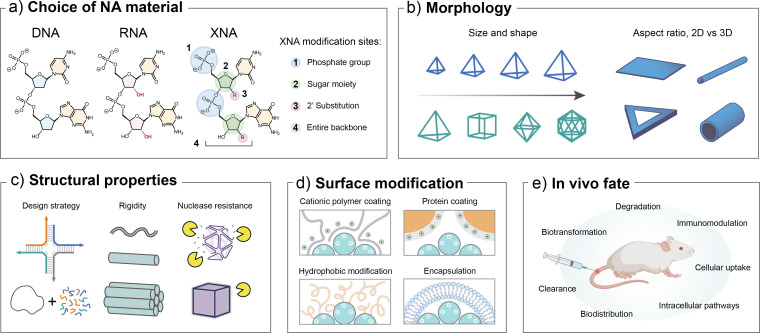
An essential consideration for any material designed for in vivo use is the structural and functional compatibility of the platform with the targeted application. When designing a NAN for performance in a living system, a researcher must broadly reflect on a few key parameters, including choice of material, morphology, surface modification, and structural properties including stability, as highlighted in Fig. 1. Choice of the NA material (e.g., DNA, RNA, or XNA) directly impacts the base mechanics as strand modifications affect the helical orientation, thermal stability, solubility, and geometry of annealed duplexes.51 The sequence of the material must also be carefully screened to control the biological consequences of signaling sequences, such as toxicity and immune stimulation.52 The morphology of a structure as defined by its size and shape is a crucial feature that impacts biodistribution, biotransformation, and cellular interactions.53 These in vivo fate consequences are also directly influenced by surface modifications that impact the charge and hydrophobicity of the structure.54,55 The inherent chemical and biological stability of the NAN is critical to defining the scope of biological applications.56 While NANs are more resistant to degradation compared to their linear counterparts,57 free DNA can be rapidly degraded by endogenous nucleases. Finally, researchers must consider the practicality of their nano-constructs for in vivo performance as defined by a careful balance between the functional efficacy and the structural complexity, mechanisms of actuation, and cost of production. The manufacturing of synthetic nucleic acids as therapeutics is ranked among the costliest platforms and it is anticipated that broader translation of NANs to biological applications may be even more expensive due to the complexity of structures.58 This increased cost could be offset if enhanced targeting and delivery methods enable use of smaller quantities of therapeutics.59 However, design and fabrication efforts to keep nanostructures and methods as simple as practical for their respective applications are likely necessary for broader applicability.
FIG. 1.
An overview of key parameters to consider for the design of nucleic acid nanostructures for biological applications. Critical features that can be tuned include (a) the choice of NA material, (b) morphological features including size and shape, (c) physiochemical properties of the structure, and (d) any modifications to the surface to facilitate delivery or include ligands for functional applications. These features directly impact (e) the in vivo fate of the structures. Part (d) is modified with permission from Jiang et al., Chem 7(5), 1156–1179 (2021). Copyright 2021 Elsevier. Part (e) is created using BioRender.
II. NA MATERIALS FOR NANOFABRICATION
To understand the physical principles underlying the biological fate of NA nanostructures, it is important to first review key aspects of NA nanotechnology assembly. In the three decades since the creation of structural DNA nanotechnology by Seeman et al.,60–62 several design strategies have emerged for bottom-up fabrication of tailored NANs ranging in size, shape, molecular complexity, and functionality. The core commonality among these materials is the use of synthetic biopolymers of nucleobases (i.e., adenine, thymine, guanine, cytosine, and uracil) that form predictable Watson–Crick (WC) base-pairing or other non-canonical interactions. DNA is the predominant form of NA utilized, although there is growing popularity in using materials with variations in backbone architecture, including RNA or XNAs. A comparison of the chemical structures of the different NA materials is highlighted in Fig. 1. Understanding the respective structural and chemical features of the NA building block materials enables engineering synthetic structures with atomic-level control over formation of the complexes.
A. DNA-based nanomaterials
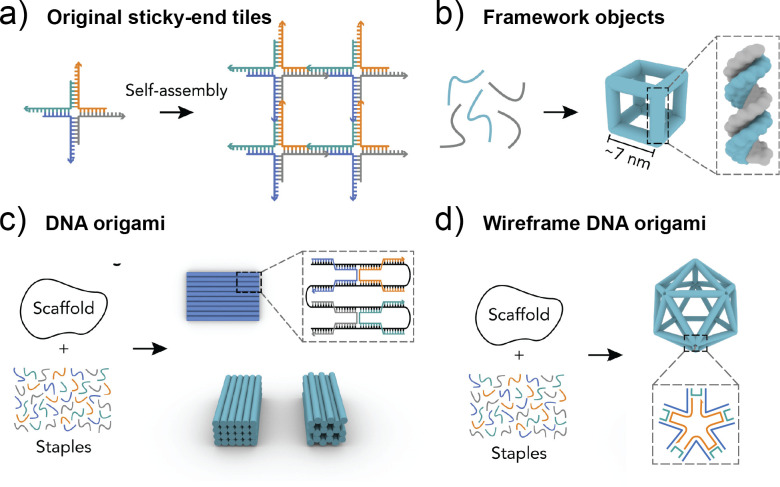
The use of NAs as materials was born out of careful consideration of the well-defined geometry of DNA. Double-stranded DNA (dsDNA) in the B-form features a right-handed helical twist with a rise of approximately 10.5 base pairs per turn, with 0.34 nm spacing between bases and a diameter of about 2.0 nm.8 The observation that DNA can deviate from its standard duplex conformation to form mobile four-way junctions (WJs) during the biological process such as genetic recombination served as the inspiration for building unnatural structural motifs using DNA.62,63 Since its inception, the field of DNA nanotechnology has advanced into a variety of approaches to assemble structures.9,64 Here, we provide a brief overview of the different DNA design motifs to aid later discussion on morphological impact. A summary of the different DNA design motifs for bottom-up nanostructure fabrication is shown in Fig. 2.
FIG. 2.
An overview of different DNA design strategies. (a) DNA tiles are designed with sticky ends to self-assemble into a network structure. (b) Framework DNA objects are assembled using the addition of several component oligonucleotides. (c) The DNA origami method involves the use of a uniform scaffold, that is, folded through addition of partly complementary staple strands. (d) Wireframe DNA origami is a method that combines the DNA origami annealing method with the design of framework objects to form more stable wireframe architectures. Modified with permission from Jiang et al., Chem 7(5), 1156–1179. (2021). Copyright 2021 Elsevier.
The first form of structural DNA nanotechnology came in the form of tile-based assemblies. DNA tiles are generally formed from a series of partly complementary ssDNA that hybridize into three-way or four-way junctions, also termed Holliday junctions. Use of single-stranded DNA (ssDNA) overhangs on the ends of these motifs, referred to as sticky ends, enable hybridization between different junctions to form a network of tiles in a DNA lattice. Variations on the tile-based motifs were further developed to increase structural rigidity though double crossover (DX) motifs between adjacent helices as well as triple crossover (TX), paranemic crossover (PX),65 and other motifs.11,12 Typically, DNA lattices are two-dimensional assemblies that form an expansive periodic crystalline network, although by careful sequence design aided by computational tools,13 finite shapes can be formed from single stranded DNA tiles,14 or through 3D tiled building blocks in the form of DNA bricks.15 While the tile-based design strategy is inherently tailorable, the designs often exhibit low yields dependent on precise stoichiometry and increasing complexity of design upon scaling.9
From the concept of DNA lattice creation through sticky-end ssDNA strands came the DNA framework design strategy. This method uses three-way and four-way junctions with sticky ends to form defined polyhedral shapes, such as a tetrahedron, cube, octahedron, and icosahedron.61,66 Most prominently, fabrications based on the DNA tetrahedron have expanded into a separate subfield of tetrahedral framework NAs (TFNAs).32 Variations on the crossover motifs between component strands have led to more rigid tile-based framework structures. Building from advances in DNA framework structures,67 the method of DNA origami emerged as first described in 2006 by Rothemund.3 This method involves folding a long ssDNA strand, called the scaffold, into a precise shape through the use of shorter ssDNA strands, called staples, which hybridize several nonconsecutive regions of the scaffold in anti-parallel adjacent helices by double crossover motifs. Most commonly, the M13mp18 phage plasmid is used as a scaffold due to its accessible bioproduction, and the series of up to hundreds of staple strands are prepared synthetically.68 The use of a scaffold strand enables higher yields compared to tile-based designs and promotes the formation of more complex shapes through enhanced structural integrity.19,69 Originally, DNA origami enabled a diverse array of flat, 2D-shapes that can be visualized upon surface immobilization. Advances by the Shih and Yan groups brought the technique into the third dimension through developing design rules for multilayered lattices70–73 and complex curvatures.74 The Bathe, Högberg, and Yan groups additionally expanded origami complexity through developing mesh wireframe junctions.75,76 For a more in-depth summary of DNA origami design principles, readers are directed to the following reviews by Dey et al.19 and Castro et al.69 While DNA is the most common NA material for nanofabrication due to its synthetic and biologic accessibility, it may not the most suitable material for all programmable applications. Nanostructure assemblies built from RNA and XNA have emerged as alternatives to DNA to maintain programmability while offering the potential for immune evasion and enhanced biostability.
B. RNA-based nanomaterials
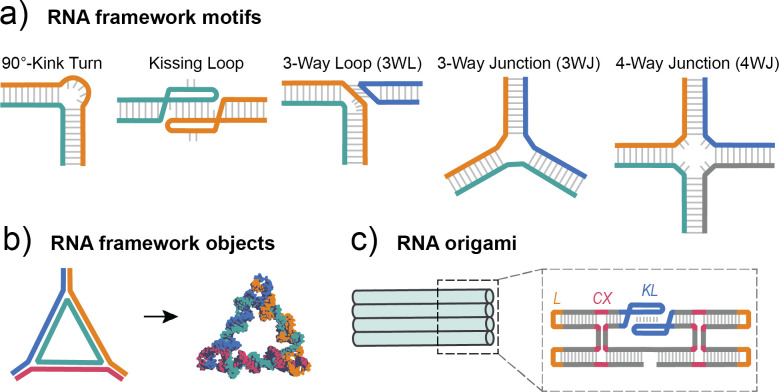
Using similar methodologies as DNA, the field of RNA nanotechnology has emerged for bottom-up assembly of versatile nanoscale materials.77 The presence of a 2′-OH on the sugar-phosphate backbone and the substitution of uracil for thymine distinguishes RNA as chemically different from DNA. Consequently, double-stranded RNA (dsRNA) typically adopts the A-form confirmation with approximately 11 base pairs per turn.78 There are two primary strategies to assemble RNA into nanostructures: (1) folding RNA to form specified structures through standard WC base pairing and (2) the use of naturally occurring RNA motifs as building blocks to construct hierarchal NANs. A brief highlight of these strategies is shown in Fig. 3. As RNA is more chemically labile than DNA, the ability to stabilize RNA duplexes is key to unlocking the potential of R-NANs for nanotechnology applications in biological systems.79 Chemical modifications, such as 2′-modification80 or 2′-4′ linkage,81 improve the stability of RNA without changing folding properties or biological functions.
FIG. 3.
An overview of common RNA design motifs. (a) Commonly utilized fundamental motifs in RNA nanofabrication include the kink tun, kissing loop, three-way loop, three-way junction, and four-way junction. (b) Similar to DNA, framework RNA objects are assembled from the addition of several complementary oligonucleotides. (c) RNA origami structures can be fabricated through methods similar to DNA origami with crossover (CX) regions or they can also be assembled by a single strand and feature loop (L) and kissing loop (KL) regions. Part (a) modified with permission from Li et al., Nature 9, 2196 (2018). Copyright 2018 Springer Nature, licensed under a Creative Commons Attribution (CC-BY) license.
A common strategy for assembly of all RNA nanostructures (R-NANs) on the small scale is to use framework motifs formed from multi-way junctions. One of the most well studied RNA motifs is found in packaging RNA as a three-way junction (pRNA-3WJ), derived from bacteriophage phi29.82 The pRNA-3WJ is a thermodynamically and chemically stable motif83 and can be used to construct a variety of 2D and 3D R-NANs. Additional nanostructured motifs include hairpins, 90°-kinks, open junctions (o3WJ and o4WJ), stacked junctions (s3WJ), and three-way loops (3WL).84 RNAs can also self-assemble into relatively stable complexes using noncanonical base-pairing through tertiary interactions with the loop and bulge regions of the RNA in the form of kissing loops. RNA loops play a critical role in R-NAN stability driven by the underlying sequences.85 Further RNA-based motifs are available in databases and can be additionally tuned by their interchangeable units.86 A method for designing RNA origami was developed by Geary and Andersen using a similar methodology to the original DNA origami technique.87 RNA origami is often formed from a single strand and uses double crossover (DX) motifs much like DNA origami, although additional stabilizing motifs are required due to the structural differences between DNA and RNA. Incorporation of kissing loops in origami enables single RNA strands to be co-transcriptionally folded assembled as building blocks,88 and crossover strands are used to tether and determine the spacing of adjacent helices.89
Research on stability, design, and conformations of R-NANs has greatly facilitated the development of RNA nanotechnology toward in vivo applications. Forming RNA into nanostructures improves the thermal and biological stabilities of RNA,90,91 while the kinetics in the living body change depending on the R-NAN shape and size.92 For more information on RNA's versatility, flexibility, and thermostability in nanostructures and their applications, we direct readers to the following reviews by Haque et al.82 and Binzel et al.93 Notably, Mao and colleagues have developed a programmable strategy for both in vitro and vivo production of R-NANs, a promising method for synthesizing nanostructures on a large scale at a low cost.84 In this review, our focus will be on structures assembled from chemically or enzymatically synthesized oligonucleotides with an emphasis on the structural and functional perspective.
C. XNA-based nanomaterials
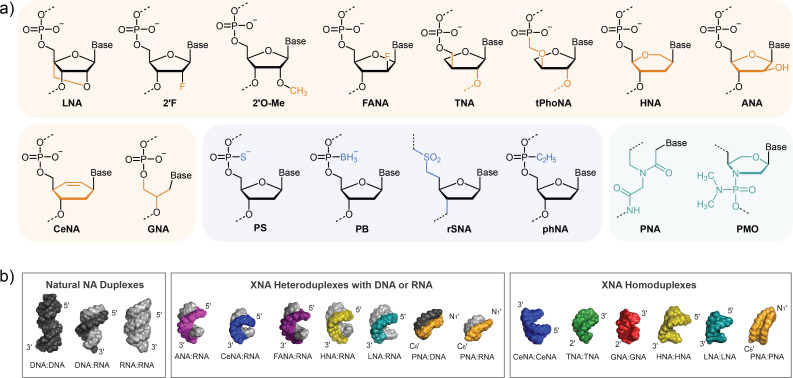
The use of non-natural analogs of NAs as materials for nanostructures has emerged as a strategy to improve upon properties of DNA and RNA based nanostructures94 such as enhancing thermodynamic stability,95 nuclease resistance,56 and preventing adverse biological interactions.96 These analogs, also known as XNAs, feature chemical modifications to the sugar or phosphate or total replacement of the sugar-phosphate backbone with another type of connectivity as highlighted in Fig. 4(a). There is an additional class of XNAs featuring unnatural base pairs (UBPs),97,98 although incorporation of these strands into structures is currently limited. Given the chemical and conformational differences, XNAs retain the ability to anneal into duplex secondary structures similar to DNA and RNA counterparts with geometric differences as shown in Fig. 4(b). Development of XNA-only nanostructures (X-NANs) is not yet widespread due to the limited knowledge of stable XNA:XNA motifs outside of the duplex, so XNAs are typically incorporated into framework NANs as hybrids into established structures with DNA or RNA. Notably, the DNA:XNA, RNA:XNA, and XNA:XNA duplexes present with a wide range of helical conformations,78 so researchers must consider these variations when designing hybrid NANs.
FIG. 4.
Examples of xenonucleic acids (XNAs) used in nanotechnology applications. (a) Molecular structures of XNAs with modifications to either the sugars (orange), phosphates (blue), or total backbone replacement (green). (b) Crystal structures of naturally occurring NA duplexes in comparison to the geometry of homoduplexes and heteroduplexes of XNAs with DNA or RNA. Part (b) is reproduced with permission from Anosova and Kowal, Nucleic Acids Res. 44(3), 1007–1021 (2015). Copyright 2015 Oxford Press.
By modifying the deoxyribose or ribose sugar in the standard NA backbone, researchers can create a wide variety of XNAs with tunability in properties, such as thermodynamic stability, rigidity, and nuclease resistance. Glycerol NA (GNA) was the first XNA assembled into 4WJ X-NAN99 and serves as a promising scaffold for chemical functionalization due to its structural simplicity.100 Locked NA (LNA) features a methylene bridge connection between the 2′O and 4′C, instilling backbone rigidity that has been used to enhance the thermodynamic stability and nuclease resistance of NANs.101,102 Direct modifications to the natural sugar in the form of 2′-fluoro (2′F) and 2′-methoxy (2′O-Me) NAs have been shown to increase thermostability and reduce immunogenicity of component strands.103 The 2′-fluoro-arabinonucleic acid (FANA) analog has also shown promise as a nuclease resistant DNA mimic in several biomedical applications.104–106 Additional XNAs have been utilized in forming stable duplex and stem-loop structures, including arabinonucleic acid (ANA),107 cyclohexene NA (CeNA),108 hexitol NA (HNA),105 α-L-threofuranosyl NA (TNA),109 glycerol NA (GNA),99 and 3′-2′ phosphonomethyl-threosyl NA (tPhoNA).110
Substitutions to the phosphate component of the NA backbone represent another avenue for XNA structural diversity to modify charge and hydrophobicity properties. One of the most common modifications features substitution of a phosphate oxygen for a sulfur in the form of phosphorothioate linkages (PS), which induce bends in the NA backbone and enhances resistance to nuclease degradation.111 Other emerging analogs featuring neutral backbone modification that could prove useful in future NAN designs include P-alkyl phosphonate NA (phNA),112 nonionic sulfone-linked RNA analogs (rSNA),113 and boranephosphonate-linked NA (PB).113 Two of the most common XNAs to feature complete backbone modification include peptide NAs (PNAs) and phosphordiamidite morpholino oligos (PMOs). The PNA technology takes advantage of the solid-phase synthetic accessibility of peptides and the programmable base pairing of NAs to create a self-assembling material.114 PNA nanostructures feature a neutral backbone with PNA:PNA and PNA:DNA hybridization presenting greater stability than the sequence corresponding DNA:DNA strands.115 The PMO structure also features a neutral backbone and is commonly employed in applications of antisense therapy due to its high binding affinity to DNA and RNA.116 Notably, while there have been many advances in expanding the structural diversity of XNAs, there are still several unknowns and cost of production barriers that prevent widespread nanostructure fabrication efforts. While there have been advances in high throughput bioproduction of DNA and RNA scaffolds, preparations of most XNAs are dependent on costly chemical synthesis techniques. Future work in XNA enzyme engineering117,118 and production techniques are necessary for more widespread implementation of XNA-based NANs. Applications using X-NANs are still relatively new to the NA nanotechnology field and as more studies on novel XNAs are emerging, future in vivo applications could prove promising.
III. NA-BASED NANOSTRUCTURES
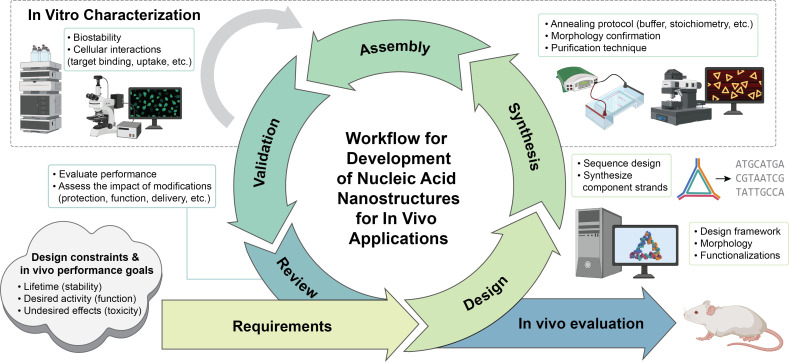
The process by which new NAN platforms move from conceptualization to reality is through many iterations of design, synthesis, and benchtop optimization. This cycle, as shown in Fig. 5, revolves around the critical validation of the intended features of the structure, including morphology, functionalization, and performance.119 The field of structural NA nanotechnology is not at the stage where this process is autonomous—there is still a high degree of trial and error in the assembly process. Advances in computational design tools have rapidly facilitated the design and application of a variety of new NAN morphologies. Furthermore, advances in functionalization strategies have widely expanded the breadth of applications where NANs can be effective. Through more in depth understanding of the design processes and breadth of available structures for NANs, the field can work toward collaborations to bring more functional NANs for biological uses.
FIG. 5.
An overview of the workflow for the design and validation process to develop new NANs for in vivo applications. Researchers first begin with a set of design requirements and constraints that match the needs of their targeted applications. Structures are designed, synthesized, and assembled, with characterization techniques used for validation at each step along the process. Results are reviewed intermittently to determine agreement with predetermined design requirements. The in vitro cycle continues until the requirements are satisfied, then in vivo evaluation is performed. This process is not always straightforward, and often many iterations of design and assembly are needed before validation experiments can be initiated. This figure was created using BioRender.
A. The NAN design and assembly process
As NANs are complex objects formed from self-assembly of numerous component sequences, several software tools have emerged to facilitate the process of design and sequence generation. The conventional approach to convert a desired 2D or 3D shape into a DNA-based structure is done manually by a researcher with the aid of software tools, such as Tiamat120 and CaDNAno70 for framework and DNA origami strategies, respectively. These programs utilize a constrained set of design parameters and enable users to design lattice-based or free-form geometries, then generate a list of corresponding strand sequences. Software solutions for molecular modeling and coarse-grained simulation tools, including CanDo121 and oxDNA,122 can aid in prediction of structural formation, followed by extensive experimental validation to confirm the geometry. Advancements in modeling and simulation software have taken top-down approaches using geometric inputs to automate DNA wireframe design.2,123–125 Further information regarding the breadth and depth of computational D-NAN design approaches has been reviewed elsewhere.126,127
Biological applications of NANs necessitate computational design strategies that can accommodate more than just DNA as RNA, XNA, proteins, and other molecular entities are often incorporated. RNA design algorithms must consider the variety of non-canonical interactions that can occur to facilitate structure assembly.128,129 Design and simulation of 3D RNA motifs has been made possible by the RNAMake algorithm,130 and design of RNA origami structures can be facilitated by a program called ROAD (RNA Origami Automated Design).89 The design capabilities for XNAs have been expanding with the development of the proto-Nucleic Acid Builder,131 the first tool specifically for XNA duplexes. Future integrations of such helical simulation parameters may aid in facilitating XNA hybrid nanostructures in future development. Recent advancements of integrative design, visualization, and simulation platforms such as Adenita,132 oxView,133,134 and the oxDNA.org135 ecosystem are expanding the capabilities of the standard design programs into more user-accessible interfaces with the ability to model DNA and RNA hybrid structures with proteins. Future developments in hybrid NAN design and simulation software will increase the accessibility of these structures to researchers and lower the barrier to assembling new functionalized structures for biomedical applications.
Following the design of a new NAN, structures are assembled from chemically or enzymatically synthesized single strand oligonucleotides through a thermal annealing protocol. Often the intended NAN morphology is thermodynamically stable but not kinetically favorable, so a temperature ramp (e.g., 95–4 °C) and time are adjusted to control the annealing process.136,137 Folding of D-NANs is also sensitive to ionic composition, with critical buffer concentrations of Mg2+ at 5–20 mM or high mM-range NaCl required to mitigate electrostatic repulsion from the folded backbones of adjacent helices.72 For framework NANs assembled primarily from a set of oligos, precise control of stoichiometry is critical to maximize yield of the structure containing all compositional strands. For NANs using origami-based folding methods, a large excess of staple strands to scaffold is required to push the annealing reaction toward thermodynamic completion. Optimization of the annealing conditions is an iterative process, that is, achieved when a high yield of a single thermodynamic product is present as monitored by gel electrophoresis. However, undesired kinetic endpoints can indicate nonideal topology of the assembled strands,138 suggesting that the re-design of the structure could be necessary. In these instances, particularly for highly complex NAN architectures, part-by-part assembly through subsequent additions of staple pools may help to control the mechanical folding pathways.139 Structures comprised of RNA and XNA are annealed with similar variable control as with D-NANs,140,141 with uncharged XNAs such as PNA oligos capable of annealing into simple structural motifs following a rapid isothermal incubation step.142 In most cases of NAN assembly, side products or excess component strands remain in the sample after annealing, thus requiring removal through a purification technique prior to downstream applications. Isolation of the assembled NANs from other constituents in solution can be performed through centrifugal filtration, gel-based extraction, liquid chromatography, affinity-capture methods, or other techniques,143 depending on the yield and resolution required.
Following assembly and purification, NANs encounter a number of challenges in order to remain structurally intact in buffers during storage and especially when introduced to complex biological matrices. The stability of a NAN can be defined in terms of the chemical, biological, and mechanical conditions under which the structure remains fully intact and stability profiles are highly specific to the NAN design. The chemical integrity of a NAN is driven primarily by the ionic composition of the assembly and the secondary media following any purification or application-specific procedures.144 As the formation of a NAN involves the folding and twisting of NA helices through non-natural geometries and structural motifs, cations such as Mg2+ or Na+ are also required to stabilize the intact structure after assembly by mitigating electrostatic repulsions that would unfold the complexes. When stored in an EDTA-free buffer, even with low Mg2+ concentrations NANs, such as DNA origami triangles and helix bundles (HB), have been reported to remain structurally intact at room temperature for over 2 months145 and for extended periods, including at least 32 freeze-thaw cycles with the use of cryoprotectants.146 However, in physiologically relevant conditions, cations are not pervasive at high enough concentrations to sustain NAN integrity over long periods of time.95 Furthermore, a major barrier to the biological stability of a NAN is the presence of nucleases both extra- and intracellularly that can dismantle the structures. Nucleases can latch onto ss toeholds, uncapped 5′- or 3′-ends, as well as exposed dsDNA or dsRNA domains and break down the strands into component bases or oligos through cleavage of phosphodiester bonds.147 The structural integrity of NA duplexes is also linked with applied mechanical stress, such as shearing or unzipping,148 which can occur under flow or through enzymatic activity. To adequately prepare a NAN for biological applications, researchers must find a balance between the chemical, biological, and mechanical stability of the structure and match the stability profile with the desired functional application.
B. NAN morphologies utilized for biological applications
While the molecular addressability of NAs enables near endless design possibilities for NANs, so far there is only a series of morphologies that have been utilized for applications in living systems. This is likely due to the fact that nanomaterials for use in vivo necessitate a strict set of design requirements with regard to stability, toxicity, and practicality.58 Broadly, NAN morphologies can be divided into the following categories: 2D motifs, framework polyhedrons, and 2D or 3D origami objects, with special considerations for complex and dynamic structures. Herein, we will briefly discuss discrete NAN morphologies and considerations for NA nanoplatforms for use in biological applications.
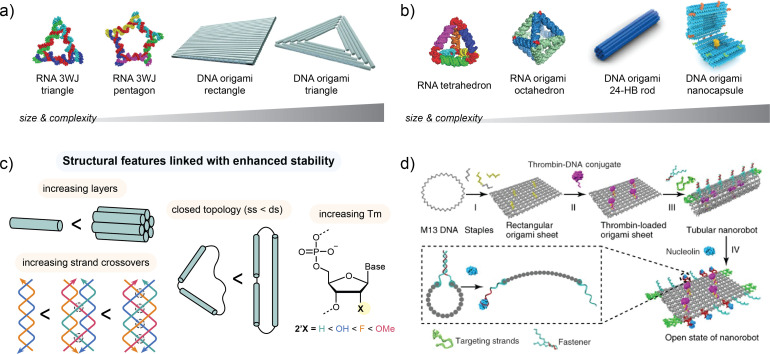
NANs are defined as 2D when the entire composite is formed with a single layer of helices and are typically designed for simplicity or surface-tethered applications. The simplest two-dimensional (2D) framework design motifs are based on the use of unique sequences of oligonucleotides (∼8–60-nt in length) to form multi-strand junctions, such as an immobile Holliday junction,62 RNA three-way junction (3WJ),92 or other motifs formed into planar geometries. Structures that are 2D can be formed over a range of sizes from small framework shapes such as a triangle and pentagon (∼5–10 nm) to large planar DNA origami rectangles and triangles (∼80–120 nm in length) depending on the assembly method and complexity of component strands as shown in Fig. 6(a). Small rigid 2D morphologies, including RNA nanosquares,149,150 are advantageous for certain biological applications, including drug delivery due to their molecular simplicity in assembly and characterization. Larger 2D origami structures can exhibit significant strain and floppiness in solution, making solution size and shape difficult to predict.151 This inherent flexibility of large 2D NANs can be harnessed to facilitate biodistribution in narrow passageways such as those of the kidney.152
FIG. 6.
Overview of the morphological features of NANs used in biological applications. NANs can be assembled in 2D (a) or 3D (b) morphologies depending on the desired goals of stability and size. (c) Structural features linked with improved stability include increasing helix layers, increasing the number of strand crossovers, restricting topology to minimize single-stranded overhangs, and using NA analogs with higher melting temperatures. (d) An example of the assembly of a complex dynamic NAN in the form of a shape-changing DNA origami tubular nanorobot for delivery of a thrombin cargo following nucleolin binding in tumor microvasculature. Part (a) reproduced with permissions from Khisamutdinov et al., Nucleic Acids Res. 42, 15 (2014). Copyright 2014 Oxford University Press, licensed under a Creative Commons Attribution (CC-BY) license and Jiang et al., Nat. Biomed. Eng. 2, 865–877 (2018). Copyright 2018 Springer Nature. Part (b) reproduced with permissions from Li et al., Adv. Mater. 28, 34 (2016). Copyright 2016 John Wiley and Sons; Høiberg et al., Biotechnol. J. 14, 1700634 (2018). Copyright 2018 John Wiley and Sons; Bastings et al., Nano Lett. 18(6), 3557–3564 (2018). Copyright 2018 American Chemical Society; and Ijäs et al., ACS Nano 13(5), 5959–5967 (2019). Copyright 2019 American Chemical Society, licensed under a Creative Commons Attribution (CC-BY) license. Part (d) is modified with permission from Li et al., Nat. Biotechnol. 36, 258–264 (2018). Copyright 2018 Springer Nature.
NANs are considered 3D when multiple layers of helices are utilized, or the structure is designed with a polyhedral framework with a defined length, width, and depth. The motifs utilized in 3D NAN construction are similar to 2D, with consideration for depth to form framework polyhedrons, hollow nanocontainers, or dense 3D objects as shown in Fig. 6(b). A wide range of 3D polyhedral NA nanoparticles have been demonstrated ranging from framework tetrahedrons,66,153 cubes,61,154 and octahedrons155,156 comprised of DNA or RNA through larger origami structures, including the icosahedron.2 Dense 3D origami objects with enhanced rigidity and compactness, such as rods and bricks,157,158 have also been demonstrated in biological applications. Use of 3D nanostructures is advantageous for most biological applications due to nanoparticle-like properties in addition to enhanced transport properties and structural stability. More complex 3D nanodevices, such as a DNA origami pH-controlled nanocapsule159 or nanorobot,160 have not yet been utilized in biomedical applications but have significant potential for future applicability.
The morphological building blocks of the 2D and 3D nanostructures can be controlled to tune expected in vivo fate outcomes. Thus far, there are several structural motifs that have generally been linked with more favorable in vivo outcomes as shown in Fig. 6(c). Stabilizing modifications, including use of multiple helix layers69 or increased numbers of interhelical crossovers,161 are also linked with prolonged in vivo lifetime.56 As such, compact 3D dimensional structures are more likely to achieve higher rates of cellular internalization with improved nuclease resistance over similar 2D or wireframe architectures.158 Minimizing accessible single-stranded loops results in prolonged stability as demonstrated through the closed topology of a DNA nanoswitch.162 Increasing the thermodynamic stability of the component strands through use of RNA instead of DNA or through XNA modifications, including 2′-OMe, L-DNA, or LNA, can further enhance the structural integrity which may lead to more favorable in vivo outcomes.51
In addition to discrete 2D and 3D NANs, dynamic structures that can change shape or topology in response to a stimulus are attractive for biological applications, including biosensors and drug delivery.163 These structures are complex in design and feature multiple modifications for coordinated functions. One prominent example of a dynamic DNA origami nanorobot platform for drug delivery27 is illustrated in Fig. 6(d). The nanorobot is assembled first as a rectangular sheet through DNA origami annealing and loaded with thrombin cargo for cancer therapy. The structure is then folded into a tube through fastener strands in combination with nucleolin-binding aptamers to protect the cargo on the interior. In response to binding with the tumor vessel biomarker nucleolin, the tube opens to expose the interior cargo. This particular opening-based delivery strategy has also been utilized in a number of 3D nanocontainers, including DNA origami boxes164,165 and nanocapsule robots.159,160 Dynamic NANs can be actuated between states in response to analytes, such as pH,18,60,166,167 metal ions,60,166 proteins,168 and target NA sequences through toehold-mediated strand displacement.169,170 Physical stimuli can also induce coordinated motion in NANs, such as through changes in temperature,171,172 photoirradiation,173–175 and magnetic176 or electric177 fields. While many of the dynamic NA structures have been thoroughly evaluated for performance in vitro, limited studies have been conducted looking into how the incorporation of moving or changing parts affects the resulting stability and fate in vivo. One study from the Graugnard group demonstrated that the topology of a dynamic NAN influences the lifetime in human serum due to differences in nuclease accessibility between open and closed states.178 Further research is needed to elucidate the impacts of structural complexity and topology in dynamic NANs in order to predict how such features impact in vivo fate.
Hybrid NANs featuring a mix of DNA or RNA with XNA components have been developed with the goal of use in biological applications for improved physical stability, enzymatic resistance, targeting, or activity in biological systems. For example, a DNA nanosuitcase with LNA insertions, hexaethylene glycol spacers, and loaded siRNA cargo demonstrated enhanced nuclease stability in addition to favorable cargo release profiles in fixed cells.102 In another study, tetrahedral structures comprised of L-DNA or DNA with 2′OMe or 2′F modifications were developed for siRNA delivery and demonstrated in vivo.179 Additional NANs constructed using XNAs have been reported with characterization data, including tetrahedra and octahedra made from 2′F-RNA, FANA, HNA, and CeNA,105 and 4WJ structures made of GNA, TNA, or PNA.142,180 There is an extensive review that discusses the structural diversity of hybrid duplex structures with at least one strand being entirely composed of XNA.78 However, there have been limited studies regarding the biostability and application of all XNA or XNA hybrid nanostructures. As future studies are required to elucidate generalized trends for in vivo fate in these XNA-based nanostructures, our review will focus on the DNA and RNA discrete NANs as discussed above with XNA modifications highlighted as a promising area for future studies.
C. Modification strategies for tailored biological applications
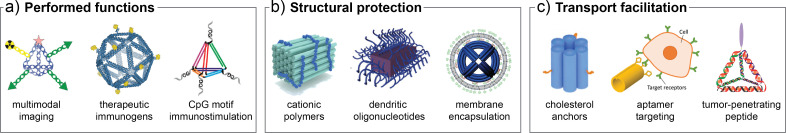
Nanomaterials comprised of solely structural NAs exhibit limited utility for biological applications and can experience issues with stability and toxicity. As such, NANs designed for in vivo use often contain one or more modifications with NA or non-NA materials primarily to enable (1) performed functions, (2) structural protection, or (3) transport facilitation.42 Advances in the diversity of synthetic methods available to modify NAs have enabled a wide range of functionalization strategies. Specific chemical methods utilized to modify NAs for nanotechnology have thoroughly been reviewed by Madsen and Gothelf.4 Furthermore, functional modification strategies and approaches to mitigate nuclease resistance have been detailed in reviews by Kizer et al.181 and Chandrasekaran56 among others.32,42 Herein, we will briefly describe some examples of the modification strategies and components that are utilized to prepare functional NA-based nano-constructs for biological applications.
One of the most common modifications to NA nanoarchitectures is to incorporate components that are designed to perform specific functions ranging from delivery of therapeutic cargo to stimuli response as shown in Fig. 7(a). Functional NAs, such as aptamers,182 siRNA,183 and CpG184 motif oligonucleotides, have been incorporated into NANs to enact targeted binding, gene silencing, and immunostimulatory properties, respectively. Non-NA cargoes have also been loaded onto NANs for delivery of therapeutics ranging from small molecules like doxorubicin185 to larger moieties, including peptides and proteins.186,187 Covalent attachment of optically active materials, such as fluorophores, radiotracers, and gold nanoparticles, is used to enable image-guided tracking of NANs in vivo, as well as to administer photothermal therapy.188–190 Additional functional elements can be incorporated to make structures responsive to stimuli, including photocleavable linkers,175 pH-responsive nanoswitches,18,166,167 aptamers,182 and strand-displacement reactions.191 By incorporating one or more of these functional elements in addition to adding protective or trafficking modifications, researchers can design multi-functional NANs for use as in vivo biosensors or responsive drug delivery platforms.35,192,193
FIG. 7.
Examples of functionalization strategies to improve biological properties of NANs. Modifications to the NAN architecture can be utilized to (a) enable specific functions, such as imaging or stimulation of the immune system, (b) protect the structure from degradation, and (c) facilitate transport to the intended biological location. Part (a) reproduced with permissions by Jiang et al., ACS Appl. Mater. Interfaces 8(7), 4378–4384 (2016). Copyright 2016 American Chemical Society; Veneziano et al., Nat. Nanotechnol. 15, 716–723 (2020). Copyright 2020 Springer Nature; and Li et al., ACS Nano 5(11), 8783–8789 (2011). Copyright 2011 American Chemical Society. Part (b) reproduced with permission by Kiviaho et al., Nanoscale 8, 11674–11680 (2016). Copyright 2016 Royal Society of Chemistry, licensed under a Creative Commons Attribution (CC-BY-NC) license; Kim and Yin, Angew. Chem., Int. Ed., 59, 700–703 (2019). Copyright 2019 John Wiley and Sons; and Perrault and Shih, ACS Nano 8(5), 5132–5140 (2014). Copyright 2014 American Chemical Society. Part (c) reproduced with permission from Whitehouse et al., Bioconjugate Chem. 30(7), 1836–1844 (2019). Copyright 2019 American Chemical Society; Sakai et al., Genes 9, 571 (2018). Copyright 2018 MDPI, licensed under a Creative Commons Attribution (CC-BY) license; and Xia et al., Biochemistry 55(9), 1326–1331 (2016). Copyright 2016 American Chemical Society.
As NANs are prone to degradation through chemical, biological, or mechanical means, strategies for stabilization through structural protection are critical for in vivo applications. Three examples of structural protection strategies are depicted in Fig. 7(b). Surface coating strategies, such as PEGylated oligolysine,194,195 dendritic oligonucleotides,196 cationic polymers,197 peptoids,198 and proteins, aim to protect NANs from nuclease degradation and disassembly. Strand-based modifications including terminal groups, such as hexaethylene glycol162 or use of XNA motifs,51,199 further improve biostability of NANs. Additional approaches such as encapsulation of a D-NAN in a PEGylated lipid bilayer to achieve a “virus-like” morphology200 show promise for preventing nuclease digestion in addition to decreasing immune activation while increasing circulation lifetime. Strategies to mitigate chemical and mechanical instability in addition to nuclease resistance of NANs include cross-linking or ligating component strands as well as silica coatings.201–204 When deploying protection strategies, researchers must find a careful balance stability in one aspect with instability or reduced performance in another area. For example, while lipid encapsulation and PEG-based coatings can effectively protect the NAN from nuclease degradation, there may be undesired physiochemical profiles or reduced accessibility for ligand or antigen interactions.178,205 Furthermore, while increased helical crossovers can greatly stabilize DNA origami to nuclease degradation, this comes at penalty of reduced ionic stability under low-Mg2+ conditions due to lower structural malleability.206
There are several approaches to equip NANs with functional components that facilitate transport to application-specific destinations inside of biological systems with examples shown in Fig. 7(c). Aptamers, signal peptides, and other moieties like folate can be used to traffic the NAN to engage with specific cellular receptors for targeted delivery.186,207,208 Hydrophobic modifications such as dendritic alkyl chains on a DNA nanostructure have been used to bind with human serum albumin for transport and increased serum stability.209 Integrating cholesterol into NANs is a strategy to regulate binding with lipoproteins210 in the blood in addition to enabling tethering or embedding of structures in lipid bilayers for membrane-localized applications.211,212 Furthermore, increased efficiency of cellular uptake has been controlled using cholesterol213 or viral capsid proteins55 among other moieties. When incorporating ligands for targeting or functional applications, it is important to note that the valency, linker length, and receptor accessibility are critical to ensure that the structures are practical and efficient.187,214,215
IV. CHARACTERIZATION TOOLS AND SUMMARY OF NAN ANALYSIS TECHNIQUES
In the development of technology intended for use in a living system, in vitro characterization is used as a predictive tool for in vivo performance. There are a variety of accepted methodologies and analytical techniques that can be used to evaluate the structure and performance of assembled NANs. Herein, we will narrow our focus to some of the most utilized techniques for validating NANs and place particular emphasis on the methods that offer translatable information prior to biological applications.216 Notably, there are many other techniques that can be used for NAN analysis in vitro and more information on those techniques can be found elsewhere.217 As a general guideline provided by Lacroix and Sleiman,5 it is recommended that every new structure should be systematically studied for understanding the: (1) thermal stability by a melting temperature assay, (2) chemical stability by analyzing structural integrity in various buffers, and (3) biological stability in relevant enzyme-containing media, in addition to assessing (4) the morphological features. Furthermore, assays and simulations to understand cellular interactions, immunogenicity, clearance, and biological functions can facilitate greater understanding of the performance potential of a particular NAN.26
A. Analyzing NAN morphology and physical properties
The standard method to validate structure assembly according to designed specifications is through gel electrophoresis, sometimes referred to as an electrophoretic mobility shift assay (EMSA). For NANs built from several oligonucleotides, such as framework polyhedrons, a stepwise assembly experiment is often performed to show cooperative hybridization of each combination of component strands. The complete structure should consist of a single band as the stable thermodynamic product, although bands representing combinatorial intermediates or branched aggregates can form as kinetic endpoints.138 For these oligonucleotide-based structures, precise control of stoichiometry and buffer composition is critical to maximize yield of the full structure. For NANs formed using the origami folding method, gels demonstrate full conversion of the scaffold strand into a uniform product band to optimize assembly conditions. As with oligo-based assemblies, kinetic traps can form intermediate structures as secondary products. However, due to the limited resolving power of gels, high-resolution imaging techniques and other methods are required to identify if any of the annealed products match the designed molecular topology.
There are several morphological features of assembled NANs that are critical to verify prior to application in terms of the nanostructure size, shape, and surface charge. Bulk sample measurement techniques, such as dynamic light scattering (DLS), can be used to assess the hydrodynamic size distributions of larger NA nanoparticles (>10 nm) and detect the presence of aggregates. Although easily accessible, DLS measurements may not always be reliable as they are sensitive to temperature and solvent viscosity and often struggle to distinguish molecules that are similar in size (e.g., monomer from dimer).218 Use of zeta potential measurements can additionally aid in confirmation of the presence of any charge-modifying coatings, such as PEG or dextran.219 Additional methods based on optical, separation, or biophysical characterization can be utilized to confirm the presence of incorporated modifications, such as fluorophores or ligands into the component NAs or structure populations. Advanced molecular imaging techniques are required in order to directly assess the shape and size of individual NANs. Atomic force microscopy (AFM) is utilized most frequently to probe the morphologies of individual structures that are typically dried onto a substrate surface, which affects the conformation, although advances in liquid-mode have made it possible to image hydrated nanoassemblies.220 While AFM can be one of the most powerful morphological assessment methods for NANs, access to instrumentation can be limited and a significant barrier to attaining high-quality images exists for inexperienced researchers. Transmission electron microscopy (TEM) has also been used for analysis of larger structures that have been stained with an electron-dense material, such as uranyl formate and possibly metal nanoparticle labels.221
The physical properties of a nanostructured morphology further offer critical insight into the practical application of a NAN design. Melting curve analysis is a simple and practical approach to assess the thermal stability of a NAN through absorbance222 or fluorescence223 monitoring in a thermal cycler or by thermophoresis.224 This process to determine the melting point of a structure yields insight into the temperature at which the majority of the structure exists as unhybridized domains and is a useful metric for assessing stability and optimizing annealing protocols. Recent advances in data processing methods have demonstrated improved melting curve precision for topology-specific origami analysis.225,226 Additional techniques for probing fundamental NAN mechanical properties often require highly skilled technicians, although these methods can measure force-extension, rigidity, elasticity, and other significant mechanical properties through AFM or optical and magnetic tweezers experiments.227,228 For example, these techniques have been used to investigate the elasticity of DNA- and RNA-based nanosquares229 and demonstrate that RNA and DNA exhibit different elasticity230 and helical twist231 responses. Computational methods have also been applied to a limited subset of DNA motifs to predict the stiffness of domains using coarse-grained and atomistic molecular dynamics simulations.232,233
B. Examining NAN stability and interactions in biological systems
The structural integrity of NANs under application specific in vitro buffer conditions is commonly assessed in bulk using gel electrophoresis as a simple and straightforward approach to track extent of degradation. Validation of specific structural changes upon destabilization can be assessed using a morphology confirmation technique, such as AFM or TEM imaging.234,235 For example, through these techniques, it has been shown that DNA origami triangles and helix bundles (6HB, 24HB) can remain intact in several Mg-free buffers145 while the presence of EDTA, chaotropic agents, and other ionic species can compromise NAN stability by mechanisms that can be difficult to predict based on the topology of the structure.144 When assessing stability of biologically-derived materials in vitro, gel-based analysis can provide an efficient approach to monitor degradation by specific enzyme treatments (e.g., DNaseI) or low concentrations of biofluids such as 10% fetal bovine serum (FBS).236 It is typical to extract NANs from complex or concentrated biofluid matrixes prior to analysis by gel or AFM methods due to the high background signal from the matrix that can interfere with data collection and interpretation.235 If intercalating dyes or conjugated Förster resonance energy transfer (FRET) dye pairs are used to label the structure, fluorescence can be used to track the kinetic profile of NAN degradation by the loss of signal over time during biofluid incubation.178 Recently, a label-free approach was demonstrated by our group using size exclusion chromatography (SEC-HPLC).237 With this method, the degradation profile of a DNA tetrahedron in high concentrations of biological matrix, up to 50% human serum, was achieved through a ratiometric data analysis technique.237 Further advances in separation methods or multimodal analytical techniques will improve the efficiency of in vitro stability analysis and expand the set of conditions that can be screened in the NAN development process.
Probing the structural stability and interactions of NANs in live cells requires a different set of characterization techniques than the bulk biofluid methods due to the relevance of spatial context in the cellular milieu. The standard technique for directly assessing cell membrane interactions and internalization of NANs is fluorescence microscopy (FM) by colocalization of the tagged analyte with the intact stained cells and readout using a minimum of two spectrally distinct fluorophores.238 For example, a method to study lysosomal degradation of NANs within the coelomocytes of C. elegans was demonstrated by FM monitoring.239 Time-lapse live cell imaging by confocal laser scanning microscopy (CLSM) can further aid in real-time monitoring of uptake kinetics or release of cargo over timescales of seconds to minutes.185 While these methods are powerful in probing real-time kinetics of uptake, it is important to consider that labeled degradation products could yield false signals by CLSM,240 and it is analytically challenging to distinguish the uptake of intact from degraded NANs by this method. Recently, an advanced technique using three-color FRET monitoring by CLSM following single-cell microinjections demonstrated stability assessment of NANs in the cytosol, independent of cell uptake dynamics.241 Cellular internalization and intracellular trafficking of DNA origami has also been studied using gold nanoparticle labels to enable visualization by TEM on the single cell level.242 Further method development for direct study of NANs in cells will improve understanding of uptake and cellular trafficking destinies.
Sample processing is often a necessary step before characterization techniques can be used to evaluate the stability, uptake, and interactions of NANs within bulk cell populations and tissues. CLSM can be used in conjunction with flow cytometry to monitor internalization efficiency of different cell types quantitatively.158 Immunohistochemical methods in combination with CLSM enable analysis of fluorescently labeled NA nanostructures in tissues.157 Further techniques probing cell lysates can offer indirect insight into stability and uptake profiles. For example, analysis of uptake of a DNA octahedron nanocage was performed by gel electrophoresis, following extraction of DNA from cell lysates and secondary labeling by biotin–streptavidin dot blot staining.243 Furthermore, quantitative real-time polymerase chain reaction (qPCR) can be used to quantify the number of intracellular DNA origami structures.244 Emerging analytical techniques applied to NA nanotechnology studies, such as proteomic profiling, have begun to pinpoint interactions between NANs and specific proteins in the cellular and biofluid environment.245 Computational approaches using dissipative particle dynamics and molecular docking simulations serve as additional tools to probe interactions between NANs and cell membrane proteins238,246 as well as predict mechanistic interactions between NANs with ligands and cargo, to better inform delivery mechanisms.247,248
C. Contextualizing in vitro characterization results toward in vivo applications
The resulting data from all in vitro characterizations of NA nanostructures must be carefully considered in the context of relevant controls before cross-work comparisons can be made and predictions of in vivo fate can be drawn. This is particularly important when moving to animal studies which require that the in vitro characterizations should be performed under relevant physiological conditions. To highlight this point, Graugnard and colleagues demonstrated that the use of in vitro test media such as FBS for examining nuclease susceptibility of DNA nanostructures grossly underestimates the viability of these structures in human-derived biofluids.178 In addition, there is often an inaccurate assumption that NANs shown to remain intact in serum can remain intact until they reach the targeted site in vivo, considering cell clearance pathways and other mechanisms of biotransformation. Furthermore, despite the wide array of methods and strategies available for characterizing NAN properties, there still remains a lack of standardized protocols249 and validation of these techniques across a wide array of morphologies. Differences in analysis protocols, reported performance metrics, and model systems present as major hurdles toward moving the field forward.5 With this information in mind, validation of new NAN designs should proceed with the end application always in sight, applying careful selection of appropriate controls and metrics for reporting the resulting data.
V. INSIGHT INTO THE IN VIVO FATE OF NUCLEIC ACID NANOSTRUCTURES
The ultimate fates of NA platforms in vivo are contingent on the connection between the biophysical and structural properties of the NANs as they interact with biological components that impact their structure and its intended function. The complete picture of the transport pathways for NA-based materials is emerging and complex, but it is clear that NANs are significantly altered by the conditions of physiological environments. While nuclease resistance is a fundamental consideration for predicting susceptibility to degradation in vivo, there are many additional factors that should be considered. In this section, we aim to outline the current understanding of the in vivo journey of NANs from administration through clearance. We will discuss how factors such as animal model and cell types can set the course for different transport mechanisms and fates. Throughout each section, we will offer insight into how the physiochemical properties of the NANs impact each outcome. We will conclude with a reflection on morphological trends and suggestions for studies to fill the gaps in understanding for future research.
A. In vivo entry: Route of administration and animal model considerations
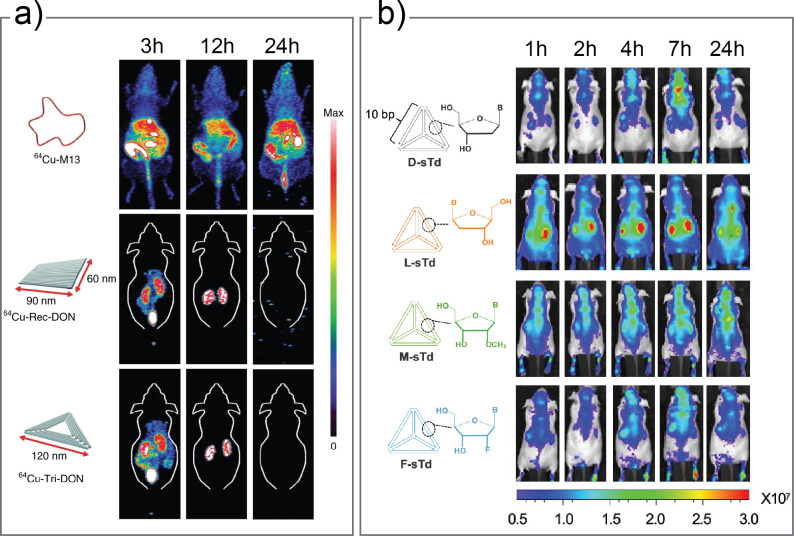
The in vivo journey begins when a NAN is first introduced into an organism. The route of administration sets the course for the initial obstacles that NANs must overcome through different transport pathways.250,251 The most common delivery method used in NAN research is an intravenous (IV) injection into the tail vein of a mouse. Notably, IV introduction leads to NANs first encountering the bloodstream, which can result in rapid clearance due to several biotransformation events which will be discussed in detail in Secs. V B and V C. Alternative routes of administration may be advantageous to consider, depending on the disease model and application being studied. For example, in a study of framework DNA tetrahedrons in a tumor mouse model, transdermal administration resulted in high tumor penetration and carrier efficacy with ∼75% of structures remaining intact compared to IV injection which lead to rapid degradation and clearance.43 Similarly, more localized site delivery of NANs through ophthalmic,252 intraarticular,253 or intrathecal254 routes may be beneficial to bypass the circulation for targeted applications. As the IV injection method is the most common approach to introduce NANs in vivo, it will serve as the basis to guide our discussion of fate. The choice of an animal model system is another consideration that will impact contextualization of the resulting data. The first studies for NANs in vivo centered on I-switch DNA nanostructures in Caenorhabditis elegans, a roundworm.18,255 This model is advantageous as it enables the user to track real-time fluorescence signals as the NANs traverse throughout the transparent organism, although the resulting data may not be directly translatable to mammalian systems. As the most common animal studied is a mouse, our discussion of in vivo fate will be centered around data from mouse models. Other animal models that have been used in NAN performance or toxicity studies include cockroaches,256 rats,254 and miniature pigs.27
B. Degradation: Extracellular nucleases and physiological conditions
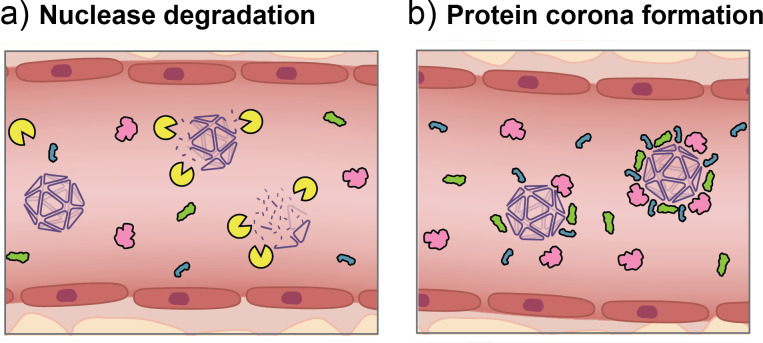
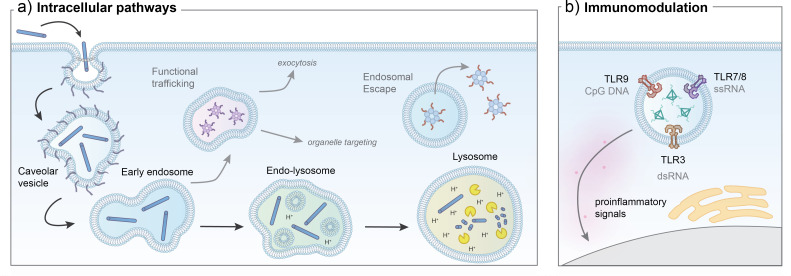
The biochemical composition of biological fluids presents a major barrier to the structural integrity of NANs in vivo. Physiological conditions, such as low salinity, low Mg2+ concentration, and body temperature at 37 °C, can destabilize NANs through unfolding and aggregation processes.95,144,145,257 The use of flexible and high cationic charge density ions such as spermidine or poly-l-lysine (K10) have been shown to improve physiological stability of DNA origami by stabilizing the charge repulsions of closely packed helices.258 Design strategies, such as a high density of helical crossovers,206 dendritic oligonucleotide coatings,196 or use of XNA motifs,106 have further stabilized NANs to unfolding in under low salt increased temperature conditions. Furthermore, the presence of extracellular nucleases in biofluids, such as blood, urine, and saliva, can severely compromise the NA structure as demonstrated in Fig. 8(a). These endogenous nucleases for DNA and RNA are indispensable for life functions such as replication and repair as well as host immune responses, and function through cleavage of the phosphodiester bonds in the NA backbone in sequence-specific or nonspecific mechanisms.147 It is understood that the folding of NA material into non-natural architectures itself confers some degree of nuclease resistance over linear and single-duplex counterparts.57,162 However, the NANs can still be accessible to rapid nuclease digestion, in some cases leading to rapid structure degradation. NA design strategies to improve structural stability as discussed in Sec. III B in this context are typically achievable longer circulation lifetimes. Furthermore, modification strategies to prevent enzymatic degradation of NANs as highlighted in Sec. III C are of critical importance, and have been the subject of numerous studies198,200,239,259 and reviews.56,260,261 As some nucleases utilize specific sequences in the form of restriction sites or strand termini to degrade NA domains, it has been shown that positioning these sites closer to three-way junctions on the corners of a DNA tetrahedron can sterically restrict nuclease access compared to a linear dsDNA domain.57 This implies that stability of a NAN could be tailored to restrict or grant access to nucleases to enhance or promote degradation depending on the application.56,96 Generally, some strategy to mitigate nuclease digestion, such as sequence modification, close-packed helices, cross-linking, XNA strand incorporation, terminal group modification, or coating, will likely be required in order to facilitate IV administration in vivo. Furthermore, as nucleases are abundant in tissues of the pancreas and kidney and present in other organs, including the liver, spleen, heart, and thymus,262 a nuclease resistance strategy is important for maintaining the designed structure throughout the biodistribution process.
FIG. 8.
Two primary challenges faced by NANs in the bloodstream are nuclease degradation and formation of a protein corona. (a) NANs are susceptible to cleavage by endogenous extracellular nucleases in blood and other biofluids. (b) Proteins within the blood interact with and adsorb onto the surface of NANs based on hydrophobicity and charge properties in patterns that are often challenging to predict.
C. Biotransformation: The protein corona
Biofluids contain an abundance of proteins that are known to interact with and adsorb nonspecifically onto foreign materials. This phenomenon, known as the protein corona, represents a complex series of protein interactions with a nanomaterial surface in both reversible and irreversible ways that can be challenging to understand and predict.263,264 The composition of the protein corona is known to directly impact the physiochemical properties of a nanomaterial as well as transport, opsonization, cell interactions, endosomal escape, and clearance mechanisms.265–268 In the broader nanomedicine field, the chemical and biophysical signatures of the protein corona have been studied to elucidate impacts on biological fate.269 It is understood that inherent properties of nanomaterials, such as size, shape, and surface functionalization, are known to have a direct impact on the populations of proteins that bind and interact in the corona.270–272 Protein corona formation has been verified to occur on the surface of NANs, as shown in Fig. 8(b). While characterization studies have expanded over the past decade for a variety of nanomaterials, the impacts of the protein corona on NANs are still emerging. It is known that larger DNA structures show higher comparative abundances of proteins on their surface when incubated in serum, as demonstrated in a proteomics study by Xu et al.273 In that study, the populations of proteins that adsorbed electrostatically were not of a particular charge type and were shown to vary depending on the health status of the serum donor. This suggests that NAN protein corona formation, and thus, in vivo fate of NANs may vary widely when different materials are administered to different groups of subjects.273 Thus, it may be even more challenging to predict in vivo fate than previously thought.
Several studies have explored the use of functionalization strategies to direct NAN interactions with biofluid proteins to achieve modulation of transport properties. For example, modification of the terminal ends of a DNA cube with dendritic alkyl chains resulted in high affinity binding to human serum albumin.209 Similarly, binding to albumin proteins has been achieved with PS-XNA modified oligos in antisense oligonucleotide applications as a result of the increase in strand hydrophobicity.274 These strategies could be used to better understand the impact of the protein corona on NAN transport, as well as serving as a hitchhiking mechanism to prolong circulation half-life. Tuning the NAN surface properties can also serve as a strategy to prevent the corona formation. For example, the use of polythymidine (polyT) ssDNA overhangs on a DNA origami rod was demonstrated to inhibit protein binding.273 Alternative coatings as well as chemical and sequence modifications may offer additional benefits for protein corona modulation, although work in this area is currently limited.26,275 One important consideration regarding the protein corona and its impact on NAN performance has been demonstrated by Smolkova et al.268 In this work, they established that the endosomal escape efficacy of an peptide-functionalized D-NAN can be significantly inhibited if structures were first incubated in a serum-containing medium.268 Thus, careful consideration and analysis of biomolecular interactions with NANs should be utilized in moving platforms forward toward in vivo studies. The formation of a protein corona could be advantageous to increase circulation lifetime by hiding a NAN from degradation or immunosurveillance or it could be detrimental by altering the intended functions of the structure. Future studies in this realm would be beneficial to elucidate trends in formation and improve prediction of the biological fates of NANs.
D. Interactions with cells: Mechanisms of uptake
NANs can interact with cell membranes through both receptor-mediated and non-receptor mediated pathways, leading to cell stimulation and uptake. Understanding the mechanisms of NAN interactions with cells an area of great interest because it informs our understanding of how biological behavior can be tuned using synthetic approaches for targeted applications.31,33,276–278 For an in-depth summary of biophysical phenomena governing DNA nanostructures interacting with cell membranes and subsequent applications, readers are directed to a comprehensive review by the Taylor group.212 Herein, we will summarize these phenomena from the morphological perspective of the uptake of multidimensional NANs designed for in vivo applications.
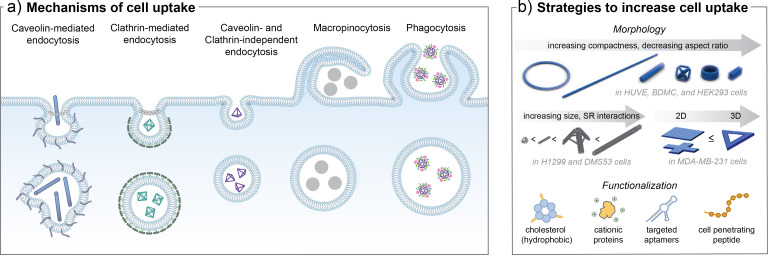
Unmodified single strands of NAs are unlikely to be internalized by cells as a result of electrostatic repulsions with the cell membrane and early macrophage-mediated removal upon administration.279,280 Transfection agents, such as lipofectamine, are typically used to deliver gene-length NAs into cells in vitro; however, NANs are able to enter cells without the need of transfection agents.281 It is hypothesized that the compact 3D geometry of NANs enables cell entry through one of several active internalization pathways as shown in Fig. 9(a). For NANs internalized without targeting ligands, the specific uptake pathway is directed by parameters such as the morphology and surface functionalization of the structure, as well as the physiological conditions, including cell type.278 The biophysical processes that facilitate nonspecific entry of D-NANs into a variety of cell types has been investigated both experimentally and computationally.242,246,282 A group of membrane-bound proteins, known as scavenger receptors (SRs), are known to play a critical role in mediating the endocytotic entry of polyanionic substrates, such as D-NANs.157,283 Higher rates of internalization through SR binding are associated with higher surface polyanionic density and lower levels of serum protein opsonization.284 For geometric 3D structures such as a DNA tetrahedron, “corner attack” engagement with SRs including LOX-1208,243,285 minimizes electrostatic repulsion compared to interactions at one of the faces of the shape, leading to a lipid charge redistribution process that facilitates cell entry.246,282 Similarly, for long, tubular D-NANs, cell entry is permitted by first engaging with surface SRs in the longitudinal orientation, followed by rotation to the narrowest transverse orientation to enter narrow endosomal vesicles with minimal electrostatic repulsion.242 Following surface receptor engagement, NANs undergo one of several mechanisms of uptake through endocytotic and non-endocytotic processes.
FIG. 9.
(a) An overview of mechanisms of cell internalization utilized by NANs. (b) The efficiency and extent of uptake can be enhanced due to modifications to the NAN morphology, such as increasing the compactness, size, and 3D-character. Functional modifications, including cholesterol or other hydrophobic moieties, cationic proteins, aptamers, or cell penetrating peptides, can further increase uptake. Part (b) is modified with permissions by Bastings et al., Nano Lett. 18(6), 3557–3564 (2018). Copyright 2018 American Chemical Society; Wang et al., J. Am. Chem. Soc. 140(7), 2478–2484 (2018), Copyright 2018 American Chemical Society; and Zeng et al., J. Mater. Chem. B 6. 1605–1612 (2018). Copyright 2018 Royal Society of Chemistry.
One of the most commonly reported cellular uptake process for NANs is through caveolin-mediated endocytosis (CVME).278,286 This is an active pathway associated with receptor-dependent, nonspecific entry of negatively charged particles and occurs primarily in epithelial and fibroblast cells.269 Upon recognition of cargo through multivalent binding to LOX-1 or other SRs, “flask-shaped” pockets form in the membrane composed mostly of the caveolin-1 structural protein.287 Studies of the entry mechanisms of viral particles have revealed that caveolar vesicles are flask shaped with a 10–50 nm neck and 60–80 nm base,288 suggesting a potential size cutoff or orientation requirement for cargo entry through this mechanism.238 Once internalized in the cell, caveolar carrier vesicles containing cargo typically fuse with early endosomal compartments and the cargo proceeds down the endo-lysosomal degradation pathway. Deviation from this classical pathway can occur upon recognition of certain ligands to direct caveolar vesicles to fuse with caveosomes for intracellular trafficking to the organelles, such as the endoplasmic reticulum, although the underlying mechanisms are not well understood.289 Another reported uptake mechanism for NANs is through clathrin-mediated endocytosis (CLME). Binding of molecular cargo to proteins in clathrin pockets of mammalian cell membranes leads to formation of acidic clathrin-coated pits that internalize the cargo into the cell in the form of clathrin-coated vesicles.290 These vesicles then disassemble from the clathrin coat and fuse together with early endosomes to follow the endo-lysosomal degradation pathway.278
The specific internalization pathway that NANs proceed through is governed by physiochemical properties of the cargo in addition to cell type, although there are some uncertainties in establishing a central pathway in the NA nanotechnology community. For example, several studies have reported that D-NANs of a variety of shapes and sizes from small framework polyhedral to large DNA origami rods are internalized through primarily CVME mechanisms as tracked in HeLa cells.238,242,291 Alternatively, a recent study validated that CLME is responsible for uptake of several framework DNA polyhedra in a variety of cell types, spheroid models, and an in vivo zebrafish model.292 Other studies suggest CLME is the dominant mechanism, particularly when CVME pathways are inhibited or not,293 or cargo such as siRNA and spermidine are included.294 Variations in experimental conditions including the use of controls and characterization techniques for uptake tracking are likely a significant cause of discrepancies in uptake mechanisms between studies.249 Future cross-study validation of structures and techniques are needed to further assess dominant pathways, which will be critical to inform the intracellular destinies of internalized NANs.
Additional mechanisms for cell entry of NANs that are less commonly studied include caveolin- and clathrin-independent endocytosis (CIE), phagocytosis, and micropinocytosis. Internalization of cargo through CIE processes such as through lipid raft formation may occur in a variety of cell types, although exact mechanisms and applications to NANs are limited.278 Phagocytotic uptake by macrophages is typically observed for particles greater than 250 nm, especially for NANs coated with a protein corona and subsequently opsonized for immune-mediated removal by the reticuloendothelial system (RES).278 Macropinocytosis (MP) is a process driven by actin-rich membrane protrusions that engulfs usually larger particles and facilities transport to the lysosome in cell-type specific mechanisms.278 The role of this process in NAN uptake is not known, although there is evidence that DNA tetrahedra can be internalized through sorting nexin five protein-mediated MP, particularly when caveolin-mediated entry is blocked,245 and the use of an AS1411 aptamer can facilitate MP uptake of tetrahedra in cancer cells.295
Parameters, including cell type and NAN morphology, surface charge, hydrophobicity, and use of targeting ligands, are known to directly impact the efficiency and rate of cellular uptake, as shown in Fig. 9(b). Regarding cell type, immune cells and dendritic cells that are inherently designed to recognize and respond to foreign material will internalize NANs at a higher rate than epithelial cells that are designed to serve as protection barriers.158,181 Concurrently, immune cells have higher expression levels of SRs on their surfaces,283 which are primarily utilized by NANs in cell entry. These SRs are also highly present on the surface of inflamed cells and cancers, which serves as an additional factor to explain why NANs can exhibit preferential tumor uptake in addition to the EPR effect.296 The morphology of a NAN is known to facilitate differential interactions with cells through modulation of cellular internalization biophysical processes. Through screening studies of various DNA origami objects, it has been demonstrated that larger structures with more compact domains such as bricks157 and barrels will exhibit a higher extent of cell uptake compared to less compact rods and rings of similar mass in various epithelial and cancer cells.158 However, in a separate work comparing tetrahedral and rod-like geometries on small (233–240 kDa) and large (3.7–3.9 MDa) scales, the larger structures were more readily internalized in human cancer cell lines with rod-like structures favored presumably due to stronger membrane interactions.242 As seen with the cell uptake mechanism studies, the use of different experimental methods may be a source of this shape trend discrepancy,249 but these results could also represent interesting variations in internalization dependent on cell type. The rigidity of the NAN is also likely to influence the degree of internalization as demonstrated by a 3D DNA origami triangle exhibiting more efficient uptake in breast cancer cells compared to planar 2D rectangle and cross shaped origami structures.185 Regarding smaller framework NANs, DNA tetrahedrons studied in HeLa cells over the range of 13–37 bp edges (10–280 nm3) exhibit nearly size-independent trends in uptake efficiency.286 Similarly in adipose stem cells, TDNs with edges from 7 to 37 bp (∼2–13 nm) showed nearly size-independent uptake, although TDN-21 showed increased internalization compared to the other sizes.297 Across this smaller NAN size range, tetrahedral geometries are considered more favorable to cell entry than prisms or cubes,292 perhaps due to favorable corner-mediated entry processes.282
The efficiency of cellular uptake can be improved through several functionalization strategies,212 including surface charge modulation or use of hydrophobic moieties or targeting ligands. Electrostatic coatings with cationic complexes, such as virus capsid proteins,55 have been shown to increase internalization efficiency of D-NANs. Hydrophobic modifications, such as cholesterol, can be used to anchor NANs into the lipid bilayers of the membrane for applications as nanopores298 or for prolonged imaging applications.299 This anchoring strategy with cholesterol can also improve the uptake efficiency of D-NANs that are not readily internalized.213 Use of cell-penetrating peptide ligands has been used to increase cell entry of DNA and PNA conjugates through receptor-mediated endocytosis.186,300 Further targeting applications using can direct interactions with specific cell types,207,301 such as the use of a MUC1 aptamer to target MCF-7 epithelial cancer cells.302,303 Use of transferrin conjugates has also been demonstrated in planar DNA origami to increase entry into KM carcinoma cells.304 Notably, functionalization strategies may be a requisite for neutral X-NANs to facilitate internalization as there is evidence that they are unlikely to enter cells without a delivery mechanism or carrier, such as incorporation into a DNA tetrahedron.25 However, due to limited studies of NANs comprised of only XNAs, it is unclear if specific morphologies may be developed to facilitate intracellular delivery.
E. Inside the cell: Endo-lysosomal pathways
The specific mechanism of internalization involved in cell entry of NANs sets the course for the fate of NANs inside the cell. Following endocytotic vesicle internalization and fusion, many framework structures can remain stable in the vesicles for several hours.305 If cargo entry was facilitated through CVME, the structures engage with receptors and undergo a sorting process to either the endo-lysosomal degradation pathway, fusion with caveosomes for organelle delivery278 or recycling back to the cell surface (i.e., endosomal escape)306 as shown in Fig. 10(a). Following CLME or other receptor-mediated entry processes, the NANs are processed through the endo-lysosomal pathway. In this process, endocytotic vesicles fuse to form early endosomes which contain low concentrations of available Mg2+ (0.25–1 mM),285 so D-NANs without a stabilizing strategy may rapidly disassemble. As the endosomes mature, fusion with acidic vacuoles containing lysosomal components such as degradative enzymes will transform endo-lysosomes into the final lysosomes. The highly acidic and enzyme-rich environment of the lysosome degrades NANs into component oligos.242 Degraded materials can then be either released into the cytoplasm for reuse or trafficked out of the cell through exosomes. The timescale of this process for NANs relates to the stability of the structure and cell type, although detailed molecular mechanisms are unknown. While some structures are degraded rapidly in the lysosome, others such as a DNA icosahedron formed from framework 5WJ connections239 or crosslinked DNA origami structures285 can sustain longer intracellular lifetimes. For NANs that are ultimately digested in the lysosomes, the degradation process can be utilized as an effective intracellular drug delivery mechanism for cargo that would otherwise not be able to enter cells.307 However, it is important to consider that since NAs are inherently genetic material, DNA nanostructures and their degradation products can be transcribed in cells.308 As such, constituent sequences should be carefully screened to avoid potential toxicities, although more work is needed in this area.309
FIG. 10.
(a) The intracellular fates of NANs can follow one of several pathways, with the primary route following caveolin-mediated endocytosis as the endo-lysosomal route to degradation. Alternative routes (in gray) include functional trafficking and endosomal escape to the cytoplasm, which are modulated by the NAN morphology and functionalization. (b) Immunomodulation in the endosome occurs through toll-like receptors specific to DNA and RNA, leading to the production of proinflammatory signals (e.g., cytokines).
Strategies for NANs to escape from the endo-lysosomal degradation pathway may be critical for sustaining longer bioavailability as well as ensuring the NANs reach their target in vivo destinations. Approaches for endosomal escape of NANs have been through functionalization strategies. In one such example, researchers developed a DNA-based 6HB with a protective poly-lysine (K10) coating and a functional endosomal escape (EE) aurein 1.2 peptide.268 Notably, while this K10-EE coating offered in vitro protection, formation of a protein corona diminished the escape efficiency, suggesting such a coating would not be effective if the structure was administered in serum-containing media.268 Other strategies to escape the fate of lysosomal degradation include using alternative uptake mechanisms entirely, such as dynamin-independent endocytosis through the use of folate ligands,208 which avoids the lysosomal pathway, or localization to specific organelles using signaling peptides.238 Further work is needed to explore additional strategies to increase intracellular lifetimes for NANs, perhaps drawing upon endosomal escape efforts from the broader nanomaterial research community.310,311
F. Immune system interactions: Stimulation and suppression
One of the major challenges in translating NA nanotechnologies into further development for in vivo applications is the limited information on the direct impact of NA materials on the immune system.58 It is well understood that introducing exogeneous materials into a living system leads to varying levels of immunomodulation that ultimately guide the resulting kinetics, dynamics, and toxicity profiles, all of which are important to control.312 For example, immune stimulation by NANs may be advantageous for researchers designing certain therapeutic strategies but detrimental for in vivo biosensor platforms. Thus, it is important to examine the current understanding of NAN-based immunomodulation and strategies to control immune cell interactions.313
Stimulation of the immune system by NA materials commonly occurs intracellularly through endosomal receptor engagements, as shown in Fig. 10(b). For DNA-based nanostructures, the most common pathway for immune system activation following internalization is through recognition by endosomal toll-like receptor 9 (TLR9). Inside the endosomal vesicles, TLR9 detects unmethylated cytosine-phosphate-guanine motifs (“CpG motifs”), which are prominent in the genomes of bacteria and viruses.52 Macrophages can recognize long DNA strands with CpG motifs and endocytose them as a protective strategy against foreign genetic material.280 In response to CpG recognition in macrophages and dendritic cells, the TLR9 pathway induces Th-1 like secretion of cytokines and generation of cytotoxic T lymphocytes.314 This proinflammatory activation has been harnessed by several groups by incorporating single stranded CpG motif cargo on a NAN to act as an immunotherapy or an adjuvant.184,315,316 The extent of TLR9 mediated immune activation is found to be proportional to the valency and accessibility of the CpG-containing strands.184,316 However, concerns for toxic overstimulation of the immune system may arise in cases where CpG stimulation is prolonged, leading to cytokine storms in mice.317 In the absence of CpG motifs or other adjuvants, uncoated DNA origami are known to induce low levels of immune activation in primary mouse splenocytes, leading to production of IL-6 and IL-12 cytokines.200,318 This is hypothesized to be a result of non-TLR9-mediated pathways as evidenced by a TLR9 knockout study,315 perhaps through one of several cytosolic receptor pathways utilized for dsDNA recgonition.96
Notably, TLR9 is the only receptor that recognizes CpG-containing DNA specifically within the endosomes. Other TLRs like TLR3 and TLR7/8 recognize double and single-stranded RNA, respectively, as well as several nucleoside analogs.319 As such, the immune stimulation pathways for RNA and XNA-based NANs should not be assumed to follow all DNA-based TLR9-mediated trends. For example, RNA-based NANs have been shown to induce stronger stimulation of interferons than shape and sequence-identical RNA-DNA hybrids and DNA-based structures in different cell lines, with RNA cubes showing the highest levels.318,320 This suggests that there is a link between the physiochemical NAN properties and immunomodulatory activities, although more research is needed to further elucidate the direct connection. Inside the endosome, double-stranded RNA (dsRNA) recognition is known to trigger RNA interference (RNAi) mechanisms that cleave the dsRNA into smaller fragments as part of innate defense mechanisms against dsRNA viral infections.321 Small interfering RNA strands (siRNAs) can trigger RNAi mechanisms through sequence-specific binding to mRNAs in the cell. If NANs contain siRNA cargo or antisense oligos, they can be functionally degraded and enact changes in expression of proteins and immune signaling.28,102,157,278,322–324
Despite sequence-specific immunostimulatory properties, DNA nanostructures have also been used to suppress inflammatory activity when administered in vivo. Molecularly, DNA is sensitive to reactive oxygen species (ROS) which can form radical species that oxidize bases and induce strand breaks and genetic DNA damage.325 Several researchers have exploited this property to use D-NANs as ROS-scavengers with anti-inflammatory properties. In one example, several DNA origami structures with high renal accumulation demonstrated ROS scavenging behavior to reduce inflammatory activity in a model of acute kidney injury.152 Other structures such as functionalized DNA tetrahedrons have shown similar activities in immunosuppression and reduced apoptosis.326,327 As there have been limited studies exploring ROS-scavenging behavior of NANs, the range of structures for which this trend can extend is uncertain, and the balance between anti- and pro-inflammatory properties of NA-based materials has not been elucidated.
The extent of immune activation can be mitigated through additional modifications outside of morphology and sequence. For example, encapsulation in a PEGylated lipid membrane can prevent inflammatory cytokine production by a DNA NanoOctahedron.200 Approaches for extracellular immune activation through the use of ligand-receptor engagement is a strategy utilized by several groups for drug delivery and immunotherapy. For example, use of eOD-GT8:PNA immunogens arranged on DNA origami structures demonstrated robust activation of B cell responses in vitro.187 Furthermore, the use of aptamers can enable binding to specific immunomodulatory targets, such as the case of a TNA aptamer that can enter the cytoplasm and stimulate immune activity through the PD-1/PD-L1 blockade.328 Notably, the valency, orientation, and spacing of ligands on the surface of NANs has been assessed as critical to control the resulting immunostimulatory activities.187,329,330 Further trends in the link between NAN morphology, sequences, and functionalization on the immunogenicity and subsequent clearance mechanisms have yet to be elucidated and represent a promising subject for future research.96
G. Clearance and biodistribution pathways
In the traditional nanomaterial research community, it is well understood that several major factors impacting patterns of biodistribution and organ accumulation are the size, shape, and surface characteristics of the material.331 Generally, larger particles (>200 nm) have a propensity to be phagocytized or trapped in the lungs, whereas smaller particles (<6 nm) can diffuse through the vasculature and be removed by renal filtration.332 Particles with a size greater than 10 nm are more likely to be removed through the reticuloendothelial system (RES), leading to liver accumulation.333 The biodistribution patterns of NANs are complex and have not been investigated thoroughly, although there are several morphology-centric trends that have been identified. Following intravenous administration, free DNA strands such as the M13mp18 phage ssDNA scaffold can be rapidly sequestered into the liver, likely as a consequence of RES-mediated removal from circulation.152,334 Similarly, as DNA nanostructures without a stabilizing or protection strategy can become quickly degraded in the blood, circulation half-lives are often not much longer than the component oligonucleotides.200 For discrete NA-based objects that withstand immediate degradation in vivo, the biodistribution and clearance destinations are guided through mechanisms of renal filtration, RES-mediated removal, and accumulation in tumors where applicable. While there have been limited studies of in vivo pharmacokinetics of NANs,335 it is understood that several morphology-centric trends have been identified, as shown in Fig. 11.
FIG. 11.
Examples of the biodistribution of NANs in mouse models. (a) PET images of the biodistribution of three 64Cu-labeled objects in healthy mice. The M13 ssDNA scaffold shows RES-mediated removal and accumulation in the liver, while the flexible DNA origami rectangle (Rec-DON) and triangle (Tri-DON) show preferential accumulation in the kidneys. Reproduced with permission from Jiang et al., Nat. Biomed. Eng. 2, 865–877 (2018). Copyright 2018 Springer Nature. (b) Comparison of the biodistribution of sequence-identical tetrahedral NANs made of DNA (D-sTd), L-DNA (L-sTd), 2′-O-Me-RNA (M-sTd), and 2′-F-RNA (F-sTd). The L-DNA structure revealed the longest circulation lifetime of all four structures with preferential kidney accumulation. Reproduced with permission from Thai et al., ACS Central Sci. 6(12), 2250–2258 (2020). Copyright 202 American Chemical Society.
As the primary filtering organ of the bloodstream, the kidneys are responsible for removing the majority of small waste products from the circulation. Intact NA nanoparticles that are much larger than 6 nm are usually size-excluded from renal filtration336 while smaller particles and degradation products are removed and cleared through the urine.200 However, some larger structures such as a DNA origami planar rectangle and 6HB tube have demonstrated preferential accumulation in and removal by the kidneys, thus escaping from hepatic sequestration.152 This phenomenon is hypothesized to be a consequence of the compactness of the structures that can rotate in order to bypass standard glomerular filtration cutoffs.337 For NANs around the 6–10 nm range, there is conflicting evidence regarding kidney accumulation trends particularly with regard to functionalization. For example, one 30-bp DNA tetrahedron with six siRNA-folate motifs (∼8–10 nm) showed preferential kidney uptake in addition to tumor accumulation.322 On the other hand, a similar 20-bp DNA tetrahedron with three dsDNA-folate motifs, a fluorophore, and a technetium-99m-labeled motif showed high levels of accumulation in the liver, followed by gradual clearance through the kidney.189 Both of these studies were performed using intravenous tail vein injection in KB-tumor bearing mice, suggesting that regardless of the specific size, ligand material and valency may play a critical role in determining clearance and biodistribution mechanisms. Furthermore, it is important to make a distinction between structures that are accumulated intact in the kidneys, and cases where kidney uptake and urinary clearance indicate degradation products. In the aforementioned study of preferential uptake of DNA origamis in the kidney,152 researchers collected mouse urine post-injection and verified through FRET analysis that a significant portion of structures remained intact following urinary excretion. In yet another example, a 30-bp DNA tetrahedron with DNA-64Cu labeled motifs (∼8–16 nm) showed such efficient renal uptake that it was demonstrated as a quantitative tool for kidney function assessment in a mouse model, with structures found intact in the urine.188 For other NANs with sizes above the renal filtration cutoffs, rapid kidney accumulation and clearance most likely indicates significant structural degradation upon administration.200
The reticuloendothelial system (RES), also referred to as the mononuclear phagocyte system (MPS), is comprised of a collection of the Kupffer cells of the liver, microglia cells in the brain, and macrophages that aid in removal of foreign material and waste from the bloodstream.54,338 During RES-mediated removal, particles and byproducts are directed to the liver where they are phagocytized and broken down through lysosomal pathways. The RES-mediated removal process is associated with greater hydrophobicity of the material, such as those structures which have been coated with a protein corona and tagged with opsonin labels, including albumin, antibodies, and complement factors, also referred to as the process of opsonization.54,338–340 NANs coated with polymeric protection strategies such as PEGylated lipid bilayer encapsulation also exhibit liver accumulation through RES-mediated processes.200 It is known that partially-folded origami structures and the component M13mp18 ssDNA scaffold can be rapidly sequestered into the liver,152 likely as a consequence of exposing viral-derived DNA motifs that can activate antiviral-like inflammatory responses.96 These findings emphasize that the physiological stability of a NAN is linked to its activation of RES processes, suggesting that instability of the structure in extracellular biofluids will increase accumulation of NAN materials in the liver.
Several studies have begun to compare morphology-based trends in biodistribution and clearance using controlled structural parameters. For example, size-dependent analysis of RNA nanosquares demonstrated that the smallest structure (5 nm) was rapidly filtered by the kidneys while the larger structures (10 and 20 nm) exhibited a slower hepatic clearance mechanism in addition to tumor accumulation.150,341 Differently shaped RNA planar polygons of the same 10-nm size including a triangle, square, and pentagon demonstrated subtle differences in biodistribution profiles. The triangle particles showed the fastest kidney clearance with low RES-mediated activity while the pentagon particles showed slow kidney clearance and high accumulation in the spleen.150 This trend is hypothesized to be a result of more symmetrical nanostructures being more likely to interact with the immune system due to similar morphological patterns to pathogenic microorganisms.342 In an investigation of shape-based mechanical properties, a 20-nm 4WJ-RNA square demonstrated high tumor uptake relative to kidney and liver removal as a function of its enhanced rubbery properties.229 For three dimensional polygons in the 10–20 nm size range, there are limited shape-based biodistribution trends, though the tetrahedral geometry shows a slight improvement in tumor accumulation compared to a trigonal pyramid, cube, and rugby ball-like construct.293
Outside of size- and shape-guided trends in biodistribution, the influence of NA material and functionalization has been investigated. Motifs that enable binding to serum proteins can lead to lower cell uptake by mitigating SR recognition, thus increasing the circulation lifetime and bioavailability of a NAN.209,343 The biodistribution patterns of XNA tetrahedral structures has been demonstrated to be backbone-dependent where an L-DNA structure demonstrated targeted kidney tubular cell uptake with slow urinary clearance.179 The corresponding DNA tetrahedron was rapidly degraded and cleared through the kidney while 2′O-Me-RNA and 2′F-RNA tetrahedra exhibited primarily liver uptake attributed to the increased hydrophobicity of the analog backbones.179 In further XNA polygon screening studies, it has been shown that structures with higher serum stability and RES evasion such as the L-DNA tend to have longer circulation times and thus higher tumor accumulation.199,293 Functional strategies to prevent formation of a protein corona demonstrate reduction in macrophage uptake and thus reduced RES-mediated clearance.273,344 Biodistribution patterns of NANs can also be actively targeted using ligands such as folate to target the folate receptors expressed in high levels on solid tumors.189,322 Tumor accumulation for particles larger than 20 nm can occur through the enhanced permeability and retention (EPR) effect due to leaky vasculature leading to particle uptake. Additionally, SRs are also highly present on the surface of inflamed cells and cancers, which serves as an additional factor to explain why NANs can exhibit preferential tumor uptake in addition to the EPR effect.296 Suppression of RES uptake by small size (under 10 nm), as well as the EPR effect leads to higher accumulation in tumors of cancerous mouse models, as demonstrated by an L-DNA tetrahedron.345 Combinatorial strategies of using the EPR effect with targeting through aptamers or other ligands have been used for a variety of NAN cancer therapeutic applications.28,35,207,346
Accumulation of NANs in vivo through mechanisms outside of renal filtration and the RES system have been investigated, although more validation studies are needed to elucidate trends. Preferential accumulation of NANs into the lymphatic system has been demonstrated for a DNA tetrahedron following subcutaneous injection into a mouse forepaw.347 There is evidence that DNA tetrahedrons are able to enter the brain tissue of tMCAo rats in an ischemic stroke model,348 possibly through CVME into the capillary endothelial cells of the brain.349 Several studies have further shown that use of ligands, including cholesterol, tocopherol, peptides, and aptamers, have been shown to increase delivery of NA cargo across the blood-brain barrier (BBB),254,350–352 although more work is need in this area to validate broad applicability of targeted brain delivery of NANs. Through expanded collaboration and in vitro screening studies, future work may elucidate understanding of mechanisms to control delivery of NAN cargo throughout the organ systems of the body for tailored in vivo applications.
H. Summary and future perspectives
The rapid growth and development in the field of NA nanotechnology over the past two decades has led to greater opportunities for tailored in vivo applications. In this review, we have highlighted the importance of the inherent chemical and biological stability of the NANs based on the choice of NA material, nanostructure morphology, and surface modifications, while considering the practicality for in vivo performance. Methods for characterization of the fundamental properties of NAN platforms were summarized to inform how in vitro tools can expand our understanding of in vivo outcomes. Foundational design strategies and performance analysis then enable assessment of the impact of morphology, physiochemical properties, and functions on biological fate.
From this discussion, we have highlighted the evidence that NAN morphology directly influences the resulting biological fate. Based on current understandings of cellular uptake studies, non-targeted structures that are large (e.g., greater than 20 nm), three-dimensional, and compact are more likely to be internalized into cells than shape-comparable smaller or 2D structures, likely as a result of increased surface receptor interactions. For these larger structures, tetrahedral or rod-like morphologies may result in more internalization into epithelial cells due to corner-mediated entry processes compared to planar, wireframe, or ring-like structures. However, making such generalizations is currently challenging due to the limited scope of studies and factors such as use of different cell types and experimental conditions. For smaller NANs under 20 nm in size, internalization appears to be nearly shape-independent with other physical properties more likely to be the driving forces for setting trends in cell uptake. For example, modifications with RNA, XNA, or other functional motifs represent a promising strategy to promote or prevent internalization of NANs into cells, in addition to directing the intracellular trafficking of internalized NANs to enhance or escape from degradation in lysosomes. The surface charge and degree of hydrophobicity are additional properties that impact biological fates, including stability, cellular interactions, and biodistribution. The use of cationic or hydrophobic surface modifications to several NANs has shown to improve both ionic stability and nuclease resistance in addition to increased rates of cellular uptake.55,213 Without such modifications, negatively charged NANs still retain their ability to be internalized in cells through receptor-mediated endocytotic mechanisms, although rates of clearance through nuclease degradation and immunomodulation may appear to be faster than their neutral or cationic NAN counterparts. Increased surface hydrophobicity is associated with greater occurrence of protein corona formation and, thus, altered biodistribution patterns. Specific trends in NAN biological fate based solely on surface properties, such as charge and hydrophobicity, are challenging to predict, although it is likely that these factors will play a combinatorial role with other morphological features in guiding in vivo fate outcomes.
Further trends have emerged with respect to the influence of the physiochemical properties of NANs on biological interactions. Strategies that increase the stability of NANs by maintaining structural integrity and resistance to nucleases are associated with prolonged in vivo lifetimes. Thus, the degradation of a NAN is a critical aspect that must be attenuated or exploited by a researcher depending on the intended applications. For example, intracellular degradation can be harnessed for effective drug delivery while highly stable architectures would be advantageous for long-term imaging applications.249 The choice of protective strategies is likely to be a balancing act to block nuclease accessibility while ensuring the steric availability of ligands to engage in performed functions. Validation of the impact of these stabilizing modifications presents an additional challenge to distinguish between intact and degraded structures in vivo, such as NANs that are intact and localized in the kidneys from the degraded oligos that are removed through renal filtration.152 Moreover, while intact NA objects generally present with low levels of immune stimulation, unfolded or partially degraded structures can trigger immune-mediated clearance mechanisms.152 As a result, the design of NANs will need to be tuned to prevent or promote immunomodulation based on the intended functions of the construct.96 The final biodistribution and clearance destinies of NANs in vivo are complex and challenging to predict based solely on the morphology and material properties of a NAN. For example, if a researcher was designing a NAN for an application requiring long-term circulation, that structure would need to withstand extracellular nuclease degradation, inhibit cellular uptake or escape from intracellular degradation, bypass renal filtration, and avoid activating the immune system by triggering RES-mediated removal, among overcoming other obstacles, including protein corona formation. Synthetic and analytical strategies to address this multifaceted set of biological challenges and improve understanding of the in vivo fate of NANs represents an exciting and promising area of future work.
There is immense potential in the development of programmable and bioresponsive platforms using NANs, although to date there have been limited validation studies of in vivo applications. To propel the NA nanotechnology field forward toward developing structures for in vivo applications, there are several remaining challenges that need to be addressed, as discussed by others in previous reviews.5,16,44,58,261,275,353 One challenge is in the shortage of standardized protocols and methods for analyzing nanostructure performance.217 To ensure that the data from one analytical method can be accurately assessed against another, it is important to establish a set of standard reporting metrics and controls,5 and perhaps a standardized set of nanostructure morphologies for comparison. In a review by Green, Mathur, and Medintz,249 the authors suggested that a standard set of D-NAN designs to span the range of morphologies including examples such as a framework tetrahedron, 2D origami triangle, 3D origami rod, and wireframe icosahedron may represent sufficient diversity to control for size, shape, and sequence across different methods. Utilizing this set would be particularly impactful for screening the broad applicability of functional modifications, as well as the comparative impacts of incorporating RNA or XNA materials to work toward enhancement of tailored nanostructure performance. Furthermore, with future advances in synthetic and bioengineered methods to reduce costs and scale up production of component NA strands, there is enhanced potential toward more translational studies.58 As a research community, we can also look toward methods developed for therapeutic NAs and other nanomedicines for inspiration on new ways to deliver NANs and characterize their fates.216 We envision that interdisciplinary collaborations across the NA nanotechnology field will help address the existing gaps in knowledge regarding mechanisms of morphology-centric patterns of cellular uptake, immunogenicity, biodistribution, and clearance mechanisms for NANs.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Nicole I. Langlois: Conceptualization (equal); Investigation (equal); Visualization (lead); Writing – original draft (lead); Writing – review & editing (equal). Kristine Y. Ma: Conceptualization (equal); Investigation (equal); Writing – original draft (supporting); Writing – review & editing (equal). Heather A. Clark: Conceptualization (equal); Project administration (equal); Writing – review & editing (equal).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Seeman N. C. and Sleiman H. F., “ DNA nanotechnology,” Nat. Rev. Mater. 3(1), 17068 (2017). 10.1038/natrevmats.2017.68 [DOI] [Google Scholar]
- 2. Veneziano R., Ratanalert S., Zhang K., Zhang F., Yan H., Chiu W., and Bathe M., “ Designer nanoscale DNA assemblies programmed from the top down,” Science 352(6293), 1534–1534 (2016). 10.1126/science.aaf4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothemund P. W. K., “ Folding DNA to create nanoscale shapes and patterns,” Nature 440(7082), 297–302 (2006). 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]
- 4. Madsen M. and Gothelf K. V., “ Chemistries for DNA nanotechnology,” Chem. Rev. 119(10), 6384–6458 (2019). 10.1021/acs.chemrev.8b00570 [DOI] [PubMed] [Google Scholar]
- 5. Lacroix A. and Sleiman H. F., “ DNA nanostructures: Current challenges and opportunities for cellular delivery,” ACS Nano 15(3), 3631–3645 (2021). 10.1021/acsnano.0c06136 [DOI] [PubMed] [Google Scholar]
- 6. Xu G., Lai M., Wilson R., Glidle A., Reboud J., and Cooper J. M., “ Branched hybridization chain reaction—Using highly dimensional DNA nanostructures for label-free, reagent-less, multiplexed molecular diagnostics,” Microsyst. Nanoeng. 5(1), 37 (2019). 10.1038/s41378-019-0076-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu M., Zhang C., Zhang C., Zhao Y., Qi Z., Fan C., Chao J., and Wei B., “ DNA origami nanostructures with scaffolds obtained from rolling circle amplification,” ACS Mater. Lett. 2(10), 1322–1327 (2020). 10.1021/acsmaterialslett.9b00484 [DOI] [Google Scholar]
- 8. Xu Y., Lv Z., Yao C., and Yang D., “ Construction of rolling circle amplification-based DNA nanostructures for biomedical applications,” Biomater. Sci. 10, 3054–3061 (2022). 10.1039/D2BM00445C [DOI] [PubMed] [Google Scholar]
- 9. Paukstelis Paul J. and Ellington Andrew D., “ Rolling out DNA nanostructures in vivo,” Proc. Natl. Acad. Sci. 105(46), 17593–17594 (2008). 10.1073/pnas.0810029105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao W., Ali M. M., Brook M. A., and Li Y., “ Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids,” Angew. Chem. Int. Ed. 47(34), 6330–6337 (2008). 10.1002/anie.200705982 [DOI] [PubMed] [Google Scholar]
- 11. Gu H., Chao J., Xiao S.-J., and Seeman N. C., “ Dynamic patterning programmed by DNA tiles captured on a DNA origami substrate,” Nat. Nanotechnol. 4(4), 245–248 (2009). 10.1038/nnano.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cutler J. I., Auyeung E., and Mirkin C. A., “ Spherical nucleic acids,” J. Am. Chem. Soc. 134(3), 1376–1391 (2012). 10.1021/ja209351u [DOI] [PubMed] [Google Scholar]
- 13. Ebrahimi S. B., Samanta D., and Mirkin C. A., “ DNA-based nanostructures for live-cell analysis,” J. Am. Chem. Soc. 142(26), 11343–11356 (2020). 10.1021/jacs.0c04978 [DOI] [PubMed] [Google Scholar]
- 14. Wilner I. O. and Willner I., “ Functionalized DNA nanostructures,” Chem. Rev. 112(4), 2528–2556 (2012). 10.1021/cr200104q [DOI] [PubMed] [Google Scholar]
- 15. Ma W., Zhan Y., Zhang Y., Mao C., Xie X., and Lin Y., “ The biological applications of DNA nanomaterials: Current challenges and future directions,” Signal Transduction Targeted Ther. 6(1), 351 (2021). 10.1038/s41392-021-00727-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keller A. and Linko V., “ Challenges and perspectives of DNA nanostructures in biomedicine,” Angew. Chem. Int. Ed. 59(37), 15818–15833 (2020). 10.1002/anie.201916390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Santos A., Evdokiou A., and Losic D., “ An overview of nanotoxicity and nanomedicine research: Principles, progress and implications for cancer therapy,” J. Mater. Chem. B 3(36), 7153–7172 (2015). 10.1039/C5TB00956A [DOI] [PubMed] [Google Scholar]
- 18. Surana S., Bhat J. M., Koushika S. P., and Krishnan Y., “ An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism,” Nat. Commun. 2(1), 340 (2011). 10.1038/ncomms1340 [DOI] [PubMed] [Google Scholar]
- 19. Dey S., Fan C., Gothelf K. V., Li J., Lin C., Liu L., Liu N., Nijenhuis M. A. D., Saccà B., Simmel F. C., Yan H., and Zhan P., “ DNA origami,” Nat. Rev. Methods Primers 1(1), 13 (2021). 10.1038/s43586-020-00009-8 [DOI] [Google Scholar]
- 20. Fu D. and Reif J., “ 3D DNA nanostructures: The nanoscale architect,” Appl. Sci. 11(6), 2624 (2021). 10.3390/app11062624 [DOI] [Google Scholar]
- 21. Wang P., Meyer T. A., Pan V., Dutta P. K., and Ke Y., “ The beauty and utility of DNA origami,” Chem 2(3), 359–382 (2017). 10.1016/j.chempr.2017.02.009 [DOI] [Google Scholar]
- 22. Hong F., Zhang F., Liu Y., and Yan H., “ DNA origami: Scaffolds for creating higher order structures,” Chem. Rev. 117(20), 12584–12640 (2017). 10.1021/acs.chemrev.6b00825 [DOI] [PubMed] [Google Scholar]
- 23. Jiang Q., Liu S., Liu J., Wang Z.-G., and Ding B., “ Rationally designed DNA-origami nanomaterials for drug delivery in vivo,” Adv. Mater. 31(45), 1804785 (2019). 10.1002/adma.201804785 [DOI] [PubMed] [Google Scholar]
- 24. Hu Q., Li H., Wang L., Gu H., and Fan C., “ DNA nanotechnology-enabled drug delivery systems,” Chem. Rev. 119(10), 6459–6506 (2019). 10.1021/acs.chemrev.7b00663 [DOI] [PubMed] [Google Scholar]
- 25. Madhanagopal B. R., Zhang S., Demirel E., Wady H., and Chandrasekaran A. R., “ DNA nanocarriers: Programmed to deliver,” Trends Biochem. Sci. 43(12), 997–1013 (2018). 10.1016/j.tibs.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 26. Jiang S., Ge Z., Mou S., Yan H., and Fan C., “ Designer DNA nanostructures for therapeutics,” Chem 7(5), 1156–1179 (2021). 10.1016/j.chempr.2020.10.025 [DOI] [Google Scholar]
- 27. Li S., Jiang Q., Liu S., Zhang Y., Tian Y., Song C., Wang J., Zou Y., Anderson G. J., Han J.-Y., Chang Y., Liu Y., Zhang C., Chen L., Zhou G., Nie G., Yan H., Ding B., and Zhao Y., “ A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo,” Nat. Biotechnol. 36(3), 258–264 (2018). 10.1038/nbt.4071 [DOI] [PubMed] [Google Scholar]
- 28. Wang Z., Song L., Liu Q., Tian R., Shang Y., Liu F., Liu S., Zhao S., Han Z., Sun J., Jiang Q., and Ding B., “ A tubular DNA nanodevice as a siRNA/chemo-drug co-delivery vehicle for combined cancer therapy,” Angew. Chem. Int. Ed. Engl. 60(5), 2594–2598 (2021). 10.1002/anie.202009842 [DOI] [PubMed] [Google Scholar]
- 29. Liu S., Jiang Q., Zhao X., Zhao R., Wang Y., Wang Y., Liu J., Shang Y., Zhao S., Wu T., Zhang Y., Nie G., and Ding B., “ A DNA nanodevice-based vaccine for cancer immunotherapy,” Nat. Mater. 20(3), 421–430 (2021). 10.1038/s41563-020-0793-6 [DOI] [PubMed] [Google Scholar]
- 30. Liu X., Xu Y., Yu T., Clifford C., Liu Y., Yan H., and Chang Y., “ A DNA nanostructure platform for directed assembly of synthetic vaccines,” Nano Lett. 12(8), 4254–4259 (2012). 10.1021/nl301877k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schoenit A., Cavalcanti-Adam E. A., and Göpfrich K., “ Functionalization of cellular membranes with DNA nanotechnology,” Trends Biotechnol. 39(11), 1208–1220 (2021). 10.1016/j.tibtech.2021.02.002 [DOI] [PubMed] [Google Scholar]
- 32. Zhang T., Tian T., and Lin Y., “ Functionalizing framework nucleic-acid-based nanostructures for biomedical application,” Adv. Mater. 34, 2107820 (2022). 10.1002/adma.202107820 [DOI] [PubMed] [Google Scholar]
- 33. Feng L., Li J., Sun J., Wang L., Fan C., and Shen J., “ Recent advances of DNA nanostructure-based cell membrane engineering,” Adv. Healthcare Mater. 10(6), 2001718 (2021). 10.1002/adhm.202001718 [DOI] [PubMed] [Google Scholar]
- 34. He L., Mu J., Gang O., and Chen X., “ Rationally programming nanomaterials with DNA for biomedical applications,” Adv. Sci. 8(8), 2003775 (2021). 10.1002/advs.202003775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen L., Wang P., and Ke Y., “ DNA nanotechnology-based biosensors and therapeutics,” Adv. Healthcare Mater. 10(15), 2002205 (2021). 10.1002/adhm.202002205 [DOI] [PubMed] [Google Scholar]
- 36. Li Y., Yang S., Zheng J., Zou Z., Yang R., and Tan W., “‘ Trojan Horse’ DNA nanostructure for personalized theranostics: Can it knock on the door of preclinical practice?,” Langmuir 34(49), 15028–15044 (2018). 10.1021/acs.langmuir.8b02008 [DOI] [PubMed] [Google Scholar]
- 37. Li F., Li J., Dong B., Wang F., Fan C., and Zuo X., “ DNA nanotechnology-empowered nanoscopic imaging of biomolecules,” Chem. Soc. Rev. 50(9), 5650–5667 (2021). 10.1039/D0CS01281E [DOI] [PubMed] [Google Scholar]
- 38. Xie N., Liu S., Yang X., He X., Huang J., and Wang K., “ DNA tetrahedron nanostructures for biological applications: Biosensors and drug delivery,” Analyst 142(18), 3322–3332 (2017). 10.1039/C7AN01154G [DOI] [PubMed] [Google Scholar]
- 39. Liang H., Zhang X.-B., Lv Y., Gong L., Wang R., Zhu X., Yang R., and Tan W., “ Functional DNA-containing nanomaterials: Cellular applications in biosensing, imaging, and targeted therapy,” Acc. Chem. Res. 47(6), 1891–1901 (2014). 10.1021/ar500078f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saha S., Prakash V., Halder S., Chakraborty K., and Krishnan Y., “ A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells,” Nat. Nanotechnol. 10(7), 645–651 (2015). 10.1038/nnano.2015.130 [DOI] [PubMed] [Google Scholar]
- 41. Xia J., Yang H., Mu M., Micovic N., Poskanzer K. E., Monaghan J. R., and Clark H. A., “ Imaging in vivo acetylcholine release in the peripheral nervous system with a fluorescent nanosensor,” Proc. Natl. Acad. Sci. 118(14), e2023807118 (2021). 10.1073/pnas.2023807118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henry S. J. W. and Stephanopoulos N., “ Functionalizing DNA nanostructures for therapeutic applications,” WIREs Nanomed. Nanobiotechnol. 13(6), e1729 (2021). 10.1002/wnan.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wiraja C., Zhu Y., Lio D. C. S., Yeo D. C., Xie M., Fang W., Li Q., Zheng M., Van Steensel M., Wang L., Fan C., and Xu C., “ Framework nucleic acids as programmable carrier for transdermal drug delivery,” Nat. Commun. 10(1), 1147 (2019). 10.1038/s41467-019-09029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mathur D. and Medintz I. L., “ The growing development of DNA nanostructures for potential healthcare-related applications,” Adv. Healthcare Mater. 8(9), 1801546 (2019). 10.1002/adhm.201801546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang D.-X., Wang J., Wang Y.-X., Du Y.-C., Huang Y., Tang A.-N., Cui Y.-X., and Kong D.-M., “ DNA nanostructure-based nucleic acid probes: Construction and biological applications,” Chem. Sci. 12(22), 7602–7622 (2021). 10.1039/D1SC00587A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu S., Xiang K., Wang C., Zhang Y., Fan G.-C., Wang W., and Han H., “ DNA nanotweezers for biosensing applications: Recent advances and future prospects,” ACS Sens. 7, 3–20 (2022). 10.1021/acssensors.1c01647 [DOI] [PubMed] [Google Scholar]
- 47. Liu W., Duan H., Zhang D., Zhang X., Luo Q., Xie T., Yan H., Peng L., Hu Y., Liang L., Zhao G., Xie Z., and Hu J., “ Concepts and application of DNA origami and DNA self-assembly: A systematic review,” Appl. Bionics Biomech. 2021, 9112407. 10.1155/2021/9112407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xavier P. L. and Chandrasekaran A. R., “ DNA-based construction at the nanoscale: Emerging trends and applications,” Nanotechnology 29(6), 062001 (2018). 10.1088/1361-6528/aaa120 [DOI] [PubMed] [Google Scholar]
- 49. Tapio K. and Bald I., “ The potential of DNA origami to build multifunctional materials,” Multifunct. Mater. 3, 032001 (2020). 10.1088/2399-7532/ab80d5 [DOI] [Google Scholar]
- 50. Wang W., Yu S., Huang S., Bi S., Han H., Zhang J.-R., Lu Y., and Zhu J.-J., “ Bioapplications of DNA nanotechnology at the solid–liquid interface,” Chem. Soc. Rev. 48(18), 4892–4920 (2019). 10.1039/C8CS00402A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pinheiro V. B. and Holliger P., “ Towards XNA nanotechnology: New materials from synthetic genetic polymers,” Trends Biotechnol. 32(6), 321–328 (2014). 10.1016/j.tibtech.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., and Akira S., “ A toll-like receptor recognizes bacterial DNA,” Nature 408(6813), 740–745 (2000). 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 53. Lane L. A., “ Physics in nanomedicine: Phenomena governing the in vivo performance of nanoparticles,” Appl. Phys. Rev. 7(1), 011316 (2020). 10.1063/1.5052455 [DOI] [Google Scholar]
- 54. Li S.-D. and Huang L., “ Nanoparticles evading the reticuloendothelial system: Role of the supported bilayer,” Biochim. Biophys. Acta 1788(10), 2259–2266 (2009). 10.1016/j.bbamem.2009.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mikkilä J., Eskelinen A.-P., Niemelä E. H., Linko V., Frilander M. J., Törmä P., and Kostiainen M. A., “ Virus-encapsulated DNA origami nanostructures for cellular delivery,” Nano Lett. 14(4), 2196–2200 (2014). 10.1021/nl500677j [DOI] [PubMed] [Google Scholar]
- 56. Chandrasekaran A. R., “ Nuclease resistance of DNA nanostructures,” Nat. Rev. Chem. 5(4), 225–239 (2021). 10.1038/s41570-021-00251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Keum J.-W. and Bermudez H., “ Enhanced resistance of DNA nanostructures to enzymatic digestion,” Chem. Commun. 2009(45), 7036–7038. 10.1039/b917661f [DOI] [PubMed] [Google Scholar]
- 58. Afonin K. A., Dobrovolskaia M. A., Ke W., Grodzinski P., and Bathe M., “ Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation,” Adv. Drug Delivery Rev. 181, 114081 (2022). 10.1016/j.addr.2021.114081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Coleridge E. L. and Dunn K. E., “ Assessing the cost-effectiveness of DNA origami nanostructures for targeted delivery of anti-cancer drugs to tumours,” Biomed. Phys. Eng. Express 6(6), 065030 (2020). 10.1088/2057-1976/abbe73 [DOI] [PubMed] [Google Scholar]
- 60. Mao C., Sun W., Shen Z., and Seeman N. C., “ A nanomechanical device based on the B–Z transition of DNA,” Nature 397(6715), 144–146 (1999). 10.1038/16437 [DOI] [PubMed] [Google Scholar]
- 61. Chen J. and Seeman N. C., “ Synthesis from DNA of a molecule with the connectivity of a cube,” Nature 350(6319), 631–633 (1991). 10.1038/350631a0 [DOI] [PubMed] [Google Scholar]
- 62. Kallenbach N. R., Ma R.-I., and Seeman N. C., “ An immobile nucleic acid junction constructed from oligonucleotides,” Nature 305(5937), 829–831 (1983). 10.1038/305829a0 [DOI] [Google Scholar]
- 63. Ortiz-Lombardía M., González A., Eritja R., Aymamí J., Azorín F., and Coll M., “ Crystal structure of a DNA Holliday junction,” Nat. Struct. Biol. 6(10), 913–917 (1999). 10.1038/13277 [DOI] [PubMed] [Google Scholar]
- 64. Li H., Carter J. D., and LaBean T. H., “ Nanofabrication by DNA self-assembly,” Mater. Today 12(5), 24–32 (2009). 10.1016/S1369-7021(09)70157-9 [DOI] [Google Scholar]
- 65. Wang X., Chandrasekaran A. R., Shen Z., Ohayon Y. P., Wang T., Kizer M. E., Sha R., Mao C., Yan H., Zhang X., Liao S., Ding B., Chakraborty B., Jonoska N., Niu D., Gu H., Chao J., Gao X., Li Y., Ciengshin T., and Seeman N. C., “ Paranemic crossover DNA: There and back again,” Chem. Rev. 119(10), 6273–6289 (2019). 10.1021/acs.chemrev.8b00207 [DOI] [PubMed] [Google Scholar]
- 66. Goodman R. P., Berry R. M., and Turberfield A. J., “ The single-step synthesis of a DNA tetrahedron,” Chem. Commun. 2004(12), 1372–1373. 10.1039/b402293a [DOI] [PubMed] [Google Scholar]
- 67. Ge Z., Gu H., Li Q., and Fan C., “ Concept and development of framework nucleic acids,” J. Am. Chem. Soc. 140(51), 17808–17819 (2018). 10.1021/jacs.8b10529 [DOI] [PubMed] [Google Scholar]
- 68. Kick B., Praetorius F., Dietz H., and Weuster-Botz D., “ Efficient production of single-stranded phage DNA as scaffolds for DNA origami,” Nano Lett. 15(7), 4672–4676 (2015). 10.1021/acs.nanolett.5b01461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Castro C. E., Kilchherr F., Kim D.-N., Shiao E. L., Wauer T., Wortmann P., Bathe M., and Dietz H., “ A primer to scaffolded DNA origami,” Nat. Methods 8(3), 221–229 (2011). 10.1038/nmeth.1570 [DOI] [PubMed] [Google Scholar]
- 70. Douglas S. M., Marblestone A. H., Teerapittayanon S., Vazquez A., Church G. M., and Shih W. M., “ Rapid prototyping of 3D DNA-origami shapes with caDNAno,” Nucl. Acids Res. 37(15), 5001–5006 (2009). 10.1093/nar/gkp436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ke Y., Douglas S. M., Liu M., Sharma J., Cheng A., Leung A., Liu Y., Shih W. M., and Yan H., “ Multilayer DNA origami packed on a square lattice,” J. Am. Chem. Soc. 131(43), 15903–15908 (2009). 10.1021/ja906381y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Douglas S. M., Dietz H., Liedl T., Högberg B., Graf F., and Shih W. M., “ Self-assembly of DNA into nanoscale three-dimensional shapes,” Nature 459(7245), 414–418 (2009). 10.1038/nature08016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Han D., Pal S., Yang Y., Jiang S., Nangreave J., Liu Y., and Yan H., “ DNA gridiron nanostructures based on four-arm junctions,” Science 339(6126), 1412–1415 (2013). 10.1126/science.1232252 [DOI] [PubMed] [Google Scholar]
- 74. Dietz H., Douglas S. M., and Shih W. M., “ Folding DNA into twisted and curved nanoscale shapes,” Science 325(5941), 725–730 (2009). 10.1126/science.1174251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang W., Chen S., An B., Huang K., Bai T., Xu M., Bellot G., Ke Y., Xiang Y., and Wei B., “ Complex wireframe DNA nanostructures from simple building blocks,” Nat. Commun. 10(1), 1067 (2019). 10.1038/s41467-019-08647-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang F., Jiang S., Wu S., Li Y., Mao C., Liu Y., and Yan H., “ Complex wireframe DNA origami nanostructures with multi-arm junction vertices,” Nat. Nanotechnol. 10(9), 779–784 (2015). 10.1038/nnano.2015.162 [DOI] [PubMed] [Google Scholar]
- 77. Guo P., “ The emerging field of RNA nanotechnology,” Nat. Nanotechnol. 5(12), 833–842 (2010). 10.1038/nnano.2010.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anosova I., Kowal E. A., Dunn M. R., Chaput J. C., Van Horn W. D., and Egli M., “ The structural diversity of artificial genetic polymers,” Nucl. Acids Res. 44(3), 1007–1021 (2015). 10.1093/nar/gkv1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu J., Guo S., Cinier M., Shlyakhtenko L. S., Shu Y., Chen C., Shen G., and Guo P., “ Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging,” ACS Nano 5(1), 237–246 (2011). 10.1021/nn1024658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kawasaki A. M., Casper M. D., Freier S. M., Lesnik E. A., Zounes M. C., Cummins L. L., Gonzalez C., and Cook P. D., “ Uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets,” J. Med. Chem. 36(7), 831–841 (1993). 10.1021/jm00059a007 [DOI] [PubMed] [Google Scholar]
- 81. Dowler T., Bergeron D., Tedeschi A.-L., Paquet L., Ferrari N., and Damha M. J., “ Improvements in siRNA properties mediated by 2'-deoxy-2'-fluoro-beta-D-arabinonucleic acid (FANA),” Nucl. Acids Res. 34(6), 1669–1675 (2006). 10.1093/nar/gkl033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Haque F., Pi F., Zhao Z., Gu S., Hu H., Yu H., and Guo P., “ RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications,” WIREs RNA 9(1), e1452 (2018). 10.1002/wrna.1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shu D., Shu Y., Haque F., Abdelmawla S., and Guo P., “ Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics,” Nat. Nanotechnol. 6(10), 658–667 (2011). 10.1038/nnano.2011.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li M., Zheng M., Wu S., Tian C., Liu D., Weizmann Y., Jiang W., Wang G., and Mao C., “ In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs,” Nat. Commun. 9(1), 2196 (2018). 10.1038/s41467-018-04652-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shu D., Moll W.-D., Deng Z., Mao C., and Guo P., “ Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology,” Nano Lett. 4(9), 1717–1723 (2004). 10.1021/nl0494497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shapiro B. A., Bindewald E., Kasprzak W., and Yingling Y., “ Protocols for the in silico design of RNA nanostructures,” in Nanostructure Design: Methods and Protocols, edited by Gazit E. and Nussinov R. ( Humana Press, Totowa, NJ, 2008), pp. 93–115. [DOI] [PubMed] [Google Scholar]
- 87. Geary C., Rothemund P. W. K., and Andersen E. S., “ A single-stranded architecture for cotranscriptional folding of RNA nanostructures,” Science 345(6198), 799–804 (2014). 10.1126/science.1253920 [DOI] [PubMed] [Google Scholar]
- 88. Afonin K. A., Viard M., Koyfman A. Y., Martins A. N., Kasprzak W. K., Panigaj M., Desai R., Santhanam A., Grabow W. W., Jaeger L., Heldman E., Reiser J., Chiu W., Freed E. O., and Shapiro B. A., “ Multifunctional RNA nanoparticles,” Nano Lett. 14(10), 5662–5671 (2014). 10.1021/nl502385k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Geary C., Grossi G., McRae E. K. S., Rothemund P. W. K., and Andersen E. S., “ RNA origami design tools enable cotranscriptional folding of kilobase-sized nanoscaffolds,” Nat. Chem. 13(6), 549–558 (2021). 10.1038/s41557-021-00679-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Khisamutdinov E. F., Jasinski D. L., and Guo P., “ RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry,” ACS Nano 8(5), 4771–4781 (2014). 10.1021/nn5006254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Osada E., Suzuki Y., Hidaka K., Ohno H., Sugiyama H., Endo M., and Saito H., “ Engineering RNA–protein complexes with different shapes for imaging and therapeutic applications,” ACS Nano 8(8), 8130–8140 (2014). 10.1021/nn502253c [DOI] [PubMed] [Google Scholar]
- 92. Guo S., Li H., Ma M., Fu J., Dong Y., and Guo P., “ Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles,” Mol. Ther. Nucleic Acids 9, 399–408 (2017). 10.1016/j.omtn.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Binzel D. W., Li X., Burns N., Khan E., Lee W.-J., Chen L.-C., Ellipilli S., Miles W., Ho Y. S., and Guo P., “ Thermostability, tunability, and tenacity of RNA as rubbery anionic polymeric materials in nanotechnology and nanomedicine—Specific cancer targeting with undetectable toxicity,” Chem. Rev. 121(13), 7398–7467 (2021). 10.1021/acs.chemrev.1c00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pinheiro A. V., Han D., Shih W. M., and Yan H., “ Challenges and opportunities for structural DNA nanotechnology,” Nat. Nanotechnol. 6(12), 763–772 (2011). 10.1038/nnano.2011.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hahn J., Wickham S. F. J., Shih W. M., and Perrault S. D., “ Addressing the instability of DNA nanostructures in tissue culture,” ACS Nano 8(9), 8765–8775 (2014). 10.1021/nn503513p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Surana S., Shenoy A. R., and Krishnan Y., “ Designing DNA nanodevices for compatibility with the immune system of higher organisms,” Nat. Nanotechnol. 10(9), 741–747 (2015). 10.1038/nnano.2015.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kimoto M. and Hirao I., “ Genetic alphabet expansion technology by creating unnatural base pairs,” Chem. Soc. Rev. 49(21), 7602–7626 (2020). 10.1039/D0CS00457J [DOI] [PubMed] [Google Scholar]
- 98. Feldman A. W. and Romesberg F. E., “ Expansion of the genetic alphabet: A chemist's approach to synthetic biology,” Acc. Chem. Res. 51(2), 394–403 (2018). 10.1021/acs.accounts.7b00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang R. S., McCullum E. O., and Chaput J. C., “ Synthesis of two mirror image 4-helix junctions derived from glycerol nucleic acid,” J. Am. Chem. Soc. 130(18), 5846–5847 (2008). 10.1021/ja800079j [DOI] [PubMed] [Google Scholar]
- 100. Piotrowicz M., Kowalczyk A., Trzybiński D., Woźniak K., and Kowalski K., “ Redox-active glycol nucleic acid (GNA) components: Synthesis and properties of the ferrocenyl-GNA nucleoside, phosphoramidite, and semicanonical dinucleoside phosphate,” Organometallics 39(6), 813–823 (2020). 10.1021/acs.organomet.9b00851 [DOI] [Google Scholar]
- 101. Astakhova I. K., Santhosh Kumar T., Campbell M. A., Ustinov A. V., Korshun V. A., and Wengel J., “ Branched DNA nanostructures efficiently stabilised and monitored by novel pyrene–perylene 2′-α-l-amino-LNA FRET pairs,” Chem. Commun. 49(5), 511–513 (2013). 10.1039/C2CC37547H [DOI] [PubMed] [Google Scholar]
- 102. Bujold K. E., Hsu J. C. C., and Sleiman H. F., “ Optimized DNA ‘nanosuitcases’ for encapsulation and conditional release of siRNA,” J. Am. Chem. Soc. 138(42), 14030–14038 (2016). 10.1021/jacs.6b08369 [DOI] [PubMed] [Google Scholar]
- 103. Watts J. K., Deleavey G. F., and Damha M. J., “ Chemically modified siRNA: Tools and applications,” Drug Discovery Today 13(19), 842–855 (2008). 10.1016/j.drudis.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 104. Teo R. D., Smithwick E. R., and Migliore A., “ 2′-deoxy-2′-fluoro-arabinonucleic acid: A valid alternative to DNA for biotechnological applications using charge transport,” Phys. Chem. Chem. Phys. 21(41), 22869–22878 (2019). 10.1039/C9CP04805G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Taylor A. I., Beuron F., Peak-Chew S.-Y., Morris E. P., Herdewijn P., and Holliger P., “ Nanostructures from synthetic genetic polymers,” ChemBioChem 17(12), 1107–1110 (2016). 10.1002/cbic.201600136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Q., Chen X., Li X., Song D., Yang J., Yu H., and Li Z., “ 2′-fluoroarabinonucleic acid nanostructures as stable carriers for cellular delivery in the strongly acidic environment,” ACS Appl. Mater. Interfaces 12(48), 53592–53597 (2020). 10.1021/acsami.0c11684 [DOI] [PubMed] [Google Scholar]
- 107. Martín-Pintado N., Yahyaee-Anzahaee M., Campos-Olivas R., Noronha A. M., Wilds C. J., Damha M. J., and González C., “ The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): Effect of arabinoses in duplex-hairpin interconversion,” Nucl. Acids Res. 40(18), 9329–9339 (2012). 10.1093/nar/gks672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang J., Verbeure B., Luyten I., Lescrinier E., Froeyen M., Hendrix C., Rosemeyer H., Seela F., Van Aerschot A., and Herdewijn P., “ Cyclohexene nucleic acids (CeNA): Serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA,” J. Am. Chem. Soc. 122(36), 8595–8602 (2000). 10.1021/ja000018+ [DOI] [Google Scholar]
- 109. Schöning K. U., Scholz P., Guntha S., Wu X., Krishnamurthy R., and Eschenmoser A., “ Chemical etiology of nucleic acid structure: The α-threofuranosyl-(3'→2') oligonucleotide system,” Science 290(5495), 1347–1351 (2000). 10.1126/science.290.5495.1347 [DOI] [PubMed] [Google Scholar]
- 110. Liu C., Cozens C., Jaziri F., Rozenski J., Maréchal A., Dumbre S., Pezo V., Marlière P., Pinheiro V. B., Groaz E., and Herdewijn P., “ Phosphonomethyl oligonucleotides as backbone-modified artificial genetic polymers,” J. Am. Chem. Soc. 140(21), 6690–6699 (2018). 10.1021/jacs.8b03447 [DOI] [PubMed] [Google Scholar]
- 111. Appaiahgari M. B. and Vrati S., “ DNAzyme-mediated inhibition of Japanese encephalitis virus replication in mouse brain,” Mol. Ther. 15(9), 1593–1599 (2007). 10.1038/sj.mt.6300231 [DOI] [PubMed] [Google Scholar]
- 112. Arangundy-Franklin S., Taylor A. I., Porebski B. T., Genna V., Peak-Chew S., Vaisman A., Woodgate R., Orozco M., and Holliger P., “ A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids,” Nat. Chem. 11(6), 533–542 (2019). 10.1038/s41557-019-0255-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhu G., Song P., Wu J., Luo M., Chen Z., and Chen T., “ Application of nucleic acid frameworks in the construction of nanostructures and cascade biocatalysts: Recent progress and perspective,” Front. Bioeng. Biotechnol. 9, 792489 (2022). 10.3389/fbioe.2021.792489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hyrup B. and Nielsen P. E., “ Peptide nucleic acids (PNA): Synthesis, properties and potential applications,” Bioorg. Med. Chem. 4(1), 5–23 (1996). 10.1016/0968-0896(95)00171-9 [DOI] [PubMed] [Google Scholar]
- 115. Demidov V. V., Potaman V. N., Frank-Kamenetskil M. D., Egholm M., Buchard O., Sönnichsen S. H., and Nlelsen P. E., “ Stability of peptide nucleic acids in human serum and cellular extracts,” Biochem. Pharmacol. 48(6), 1310–1313 (1994). 10.1016/0006-2952(94)90171-6 [DOI] [PubMed] [Google Scholar]
- 116. Nan Y. and Zhang Y.-J., “ Antisense phosphorodiamidate morpholino oligomers as novel antiviral compounds,” Front. Microbiol. 9, 750 (2018). 10.3389/fmicb.2018.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Medina E., Yik E. J., Herdewijn P., and Chaput J. C., “ Functional comparison of laboratory-evolved XNA polymerases for synthetic biology,” ACS Synth. Biol. 10(6), 1429–1437 (2021). 10.1021/acssynbio.1c00048 [DOI] [PubMed] [Google Scholar]
- 118. Houlihan G., Arangundy-Franklin S., Porebski B. T., Subramanian N., Taylor A. I., and Holliger P., “ Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity,” Nat. Chem. 12(8), 683–690 (2020). 10.1038/s41557-020-0502-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chou L. Y. T., “ Design verification as foundation for advancing DNA nanotechnology applications,” ACS Nano 15(6), 9222–9228 (2021). 10.1021/acsnano.1c04304 [DOI] [PubMed] [Google Scholar]
- 120. Williams S., Lund K., Lin C., Wonka P., Lindsay S., and Yan H., “ Tiamat: A three-dimensional editing tool for complex DNA structures,” in Proceedings of the International Workshop on DNA-Based Computers ( Springer, 2008), pp. 90–101. [Google Scholar]
- 121. Kim D.-N., Kilchherr F., Dietz H., and Bathe M., “ Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures,” Nucl. Acids Res. 40(7), 2862–2868 (2011). 10.1093/nar/gkr1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ouldridge T. E., Louis A. A., and Doye J. P. K., “ Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model,” J. Chem. Phys. 134(8), 085101 (2011). 10.1063/1.3552946 [DOI] [PubMed] [Google Scholar]
- 123. Benson E., Mohammed A., Gardell J., Masich S., Czeizler E., Orponen P., and Högberg B., “ DNA rendering of polyhedral meshes at the nanoscale,” Nature 523(7561), 441–444 (2015). 10.1038/nature14586 [DOI] [PubMed] [Google Scholar]
- 124. Jun H., Shepherd T. R., Zhang K., Bricker W. P., Li S., Chiu W., and Bathe M., “ Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges,” ACS Nano 13(2), 2083–2093 (2019). 10.1021/acsnano.8b08671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jun H., Zhang F., Shepherd T., Ratanalert S., Qi X., Yan H., and Bathe M., “ Autonomously designed free-form 2D DNA origami,” Sci. Adv. 5(1), eaav0655 (2019). 10.1126/sciadv.aav0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Koh H., Lee J. G., Lee J. Y., Kim R., Tabata O., Kim J.-W., and Kim D.-N., “ Design approaches and computational tools for DNA nanostructures,” IEEE Open J. Nanotechnol. 2, 86–100 (2021). 10.1109/OJNANO.2021.3119913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kekic T. and Barisic I., “ In silico modelling of DNA nanostructures,” Comput. Struct. Biotechnol. J. 18, 1191–1201 (2020). 10.1016/j.csbj.2020.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Laing C. and Schlick T., “ Computational approaches to 3D modeling of RNA,” J. Phys.: Condens. Matter 22(28), 283101 (2010). 10.1088/0953-8984/22/28/283101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Magnus M., Antczak M., Zok T., Wiedemann J., Lukasiak P., Cao Y., Bujnicki J. M., Westhof E., Szachniuk M., and Miao Z., “ RNA-puzzles toolkit: A computational resource of RNA 3D structure benchmark datasets, structure manipulation, and evaluation tools,” Nucl. Acids Res. 48(2), 576–588 (2020). 10.1093/nar/gkz1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yesselman J. D., Eiler D., Carlson E. D., Gotrik M. R., d'Aquino A. E., Ooms A. N., Kladwang W., Carlson P. D., Shi X., Costantino D. A., Herschlag D., Lucks J. B., Jewett M. C., Kieft J. S., and Das R., “ Computational design of three-dimensional RNA structure and function,” Nat. Nanotechnol. 14(9), 866–873 (2019). 10.1038/s41565-019-0517-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Alenaizan A., Barnett J. L., Hud N. V., Sherrill C. D., and Petrov A. S., “ The proto-nucleic acid builder: A software tool for constructing nucleic acid analogs,” Nucl. Acids Res. 49(1), 79–89 (2020). 10.1093/nar/gkaa1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. de Llano E., Miao H., Ahmadi Y., Wilson A. J., Beeby M., Viola I., and Barisic I., “ Adenita: Interactive 3D modelling and visualization of DNA nanostructures,” Nucl. Acids Res. 48(15), 8269–8275 (2020). 10.1093/nar/gkaa593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Poppleton E., Bohlin J., Matthies M., Sharma S., Zhang F., and Šulc P., “ Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation,” Nucl. Acids Res. 48(12), e72 (2020). 10.1093/nar/gkaa417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bohlin J., Matthies M., Poppleton E., Procyk J., Mallya A., Yan H., and Šulc P., “ Design and simulation of DNA, RNA and hybrid protein–nucleic acid nanostructures with oxView,” Nat. Protoc. 17, 1762–1788 (2022). 10.1038/s41596-022-00688-5 [DOI] [PubMed] [Google Scholar]
- 135. Poppleton E., Romero R., Mallya A., Rovigatti L., and Šulc P., “ OxDNA.org: A public webserver for coarse-grained simulations of DNA and RNA nanostructures,” Nucl. Acids Res. 49(W1), W491–W498 (2021). 10.1093/nar/gkab324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Whitelam S. and Jack R. L., “ The statistical mechanics of dynamic pathways to self-assembly,” Annu. Rev. Phys. Chem. 66, 143–163 (2015). 10.1146/annurev-physchem-040214-121215 [DOI] [PubMed] [Google Scholar]
- 137. Wei X., Nangreave J., and Liu Y., “ Uncovering the self-assembly of DNA nanostructures by thermodynamics and kinetics,” Acc. Chem. Res. 47(6), 1861–1870 (2014). 10.1021/ar5000665 [DOI] [PubMed] [Google Scholar]
- 138. Gao L., Liu L., Tian Y., Yang Q., Wu P., Fan C., Zhao Q., and Li F., “ Probing the formation kinetics and thermodynamics with rationally designed analytical tools enables one-pot synthesis and purification of a tetrahedral DNA nanostructure,” Anal. Chem. 93(18), 7045–7053 (2021). 10.1021/acs.analchem.1c00363 [DOI] [PubMed] [Google Scholar]
- 139. Bae W., Kim K., Min D., Ryu J.-K., Hyeon C., and Yoon T.-Y., “ Programmed folding of DNA origami structures through single-molecule force control,” Nat. Commun. 5(1), 5654 (2014). 10.1038/ncomms6654 [DOI] [PubMed] [Google Scholar]
- 140. Taylor A. I. and Holliger P., “ Selecting fully-modified XNA aptamers using synthetic genetics,” Curr. Protoc. Chem. Biol. 10(2), e44 (2018). 10.1002/cpch.44 [DOI] [PubMed] [Google Scholar]
- 141. Duan T., He L., Tokura Y., Liu X., Wu Y., and Shi Z., “ Construction of tunable peptide nucleic acid junctions,” Chem. Commun. 54(23), 2846–2849 (2018). 10.1039/C8CC00108A [DOI] [PubMed] [Google Scholar]
- 142. Kazane S. A., Axup J. Y., Kim C. H., Ciobanu M., Wold E. D., Barluenga S., Hutchins B. A., Schultz P. G., Winssinger N., and Smider V. V., “ Self-assembled antibody multimers through peptide nucleic acid conjugation,” J. Am. Chem. Soc. 135(1), 340–346 (2013). 10.1021/ja309505c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Shaw A., Benson E., and Högberg B., “ Purification of functionalized DNA origami nanostructures,” ACS Nano 9(5), 4968–4975 (2015). 10.1021/nn507035g [DOI] [PubMed] [Google Scholar]
- 144. Ramakrishnan S., Ijäs H., Linko V., and Keller A., “ Structural stability of DNA origami nanostructures under application-specific conditions,” Comput. Struct. Biotechnol. J. 16, 342–349 (2018). 10.1016/j.csbj.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kielar C., Xin Y., Shen B., Kostiainen M. A., Grundmeier G., Linko V., and Keller A., “ On the stability of DNA origami nanostructures in low-magnesium buffers,” Angew. Chem. Int. Ed. 57(30), 9470–9474 (2018). 10.1002/anie.201802890 [DOI] [PubMed] [Google Scholar]
- 146. Xin Y., Kielar C., Zhu S. Q., Sikeler C., Xu X. D., Moser C., Grundmeier G., Liedl T., Heuer-Jungemann A., Smith D. M., and Keller A., “ Cryopreservation of DNA origami nanostructures,” Small 16(13), 7 (2020). 10.1002/smll.201905959 [DOI] [PubMed] [Google Scholar]
- 147. Yang W., “ Nucleases: Diversity of structure, function and mechanism,” Q. Rev. Biophys. 44(1), 1–93 (2011). 10.1017/S0033583510000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mosayebi M., Louis A. A., Doye J. P. K., and Ouldridge T. E., “ Force-induced rupture of a DNA duplex: From fundamentals to force sensors,” ACS Nano 9(12), 11993–12003 (2015). 10.1021/acsnano.5b04726 [DOI] [PubMed] [Google Scholar]
- 149. Severcan I., Geary C., Verzemnieks E., Chworos A., and Jaeger L., “ Square-shaped RNA particles from different RNA folds,” Nano Lett. 9(3), 1270–1277 (2009). 10.1021/nl900261h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jasinski D. L., Li H., and Guo P., “ The effect of size and shape of RNA nanoparticles on biodistribution,” Mol. Ther. 26(3), 784–792 (2018). 10.1016/j.ymthe.2017.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ni H., Fan X., Zhou F., Guo G., Lee J. Y., Seeman N. C., Kim D.-N., Yao N., Chaikin P. M., and Han Y., “ Direct visualization of floppy two-dimensional DNA origami using cryogenic electron microscopy,” iScience 25(6), 104373 (2022). 10.1016/j.isci.2022.104373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Jiang D., Ge Z., Im H.-J., England C. G., Ni D., Hou J., Zhang L., Kutyreff C. J., Yan Y., Liu Y., Cho S. Y., Engle J. W., Shi J., Huang P., Fan C., Yan H., and Cai W., “ DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury,” Nat. Biomed. Eng. 2(11), 865–877 (2018). 10.1038/s41551-018-0317-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Li H., Zhang K., Pi F., Guo S., Shlyakhtenko L., Chiu W., Shu D., and Guo P., “ Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting,” Adv. Mater. 28(34), 7501–7507 (2016). 10.1002/adma.201601976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Afonin K. A., Bindewald E., Yaghoubian A. J., Voss N., Jacovetty E., Shapiro B. A., and Jaeger L., “ In vitro assembly of cubic RNA-based scaffolds designed in silico,” Nat. Nanotechnol. 5(9), 676–682 (2010). 10.1038/nnano.2010.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zhang Y. and Seeman N. C., “ Construction of a DNA-truncated octahedron,” J. Am. Chem. Soc. 116(5), 1661–1669 (1994). 10.1021/ja00084a006 [DOI] [Google Scholar]
- 156. Høiberg H. C., Sparvath S. M., Andersen V. L., Kjems J., and Andersen E. S., “ An RNA origami octahedron with intrinsic siRNAs for potent gene knockdown,” Biotechnol. J. 14(1), 1700634 (2019). 10.1002/biot.201700634 [DOI] [PubMed] [Google Scholar]
- 157. Rahman M. A., Wang P., Zhao Z., Wang D., Nannapaneni S., Zhang C., Chen Z., Griffith C. C., Hurwitz S. J., Chen Z. G., Ke Y., and Shin D. M., “ Systemic delivery of Bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth,” Angew. Chem. Int. Ed. 56(50), 16023–16027 (2017). 10.1002/anie.201709485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Bastings M. M. C., Anastassacos F. M., Ponnuswamy N., Leifer F. G., Cuneo G., Lin C., Ingber D. E., Ryu J. H., and Shih W. M., “ Modulation of the cellular uptake of DNA origami through control over mass and shape,” Nano Lett. 18(6), 3557–3564 (2018). 10.1021/acs.nanolett.8b00660 [DOI] [PubMed] [Google Scholar]
- 159. Ijäs H., Hakaste I., Shen B., Kostiainen M. A., and Linko V., “ Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo,” ACS Nano 13(5), 5959–5967 (2019). 10.1021/acsnano.9b01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Douglas S. M., Bachelet I., and Church G. M., “ A logic-gated nanorobot for targeted transport of molecular payloads,” Science 335(6070), 831–834 (2012). 10.1126/science.1214081 [DOI] [PubMed] [Google Scholar]
- 161. Chandrasekaran A. R., Vilcapoma J., Dey P., Wong-Deyrup S. W., Dey B. K., and Halvorsen K., “ Exceptional nuclease resistance of paranemic crossover (PX) DNA and crossover-dependent biostability of DNA motifs,” J. Am. Chem. Soc. 142(14), 6814–6821 (2020). 10.1021/jacs.0c02211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Conway J. W., McLaughlin C. K., Castor K. J., and Sleiman H., “ DNA nanostructure serum stability: Greater than the sum of its parts,” Chem. Commun. 49(12), 1172–1174 (2013). 10.1039/c2cc37556g [DOI] [PubMed] [Google Scholar]
- 163. DeLuca M., Shi Z., Castro C. E., and Arya G., “ Dynamic DNA nanotechnology: Toward functional nanoscale devices,” Nanoscale Horiz. 5(2), 182–201 (2020). 10.1039/C9NH00529C [DOI] [Google Scholar]
- 164. Kuzuya A. and Komiyama M., “ Design and construction of a box-shaped 3D-DNA origami,” Chem. Commun. 2009(28), 4182–4184. 10.1039/b907800b [DOI] [PubMed] [Google Scholar]
- 165. Andersen E. S., Dong M., Nielsen M. M., Jahn K., Subramani R., Mamdouh W., Golas M. M., Sander B., Stark H., Oliveira C. L. P., Pedersen J. S., Birkedal V., Besenbacher F., Gothelf K. V., and Kjems J., “ Self-assembly of a nanoscale DNA box with a controllable lid,” Nature 459(7243), 73–76 (2009). 10.1038/nature07971 [DOI] [PubMed] [Google Scholar]
- 166. Liu D. and Balasubramanian S., “ A proton-fuelled DNA nanomachine,” Angew. Chem. Int. Ed. 42(46), 5734–5736 (2003). 10.1002/anie.200352402 [DOI] [PubMed] [Google Scholar]
- 167. Idili A., Vallée-Bélisle A., and Ricci F., “ Programmable pH-triggered DNA nanoswitches,” J. Am. Chem. Soc. 136(16), 5836–5839 (2014). 10.1021/ja500619w [DOI] [PubMed] [Google Scholar]
- 168. Morales J. M., Pawle R., Akkilic N., Luo Y., Xavierselvan M., Albokhari R., Calderon I. A. C., Selfridge S., Minns R., Takiff L., and Clark H., “ DNA-based photoacoustic nanosensor for interferon gamma detection,” ACS Sens. 4(5), 1313–1322 (2019). 10.1021/acssensors.9b00209 [DOI] [PubMed] [Google Scholar]
- 169. Yurke B., Turberfield A. J., Mills A. P., Simmel F. C., and Neumann J. L., “ A DNA-fuelled molecular machine made of DNA,” Nature 406(6796), 605–608 (2000). 10.1038/35020524 [DOI] [PubMed] [Google Scholar]
- 170. Zhang D. Y. and Seelig G., “ Dynamic DNA nanotechnology using strand-displacement reactions,” Nat. Chem. 3(2), 103–113 (2011). 10.1038/nchem.957 [DOI] [PubMed] [Google Scholar]
- 171. Viasnoff V., Meller A., and Isambert H., “ DNA nanomechanical switches under folding kinetics control,” Nano Lett. 6(1), 101–104 (2006). 10.1021/nl052161c [DOI] [PubMed] [Google Scholar]
- 172. Turek V. A., Chikkaraddy R., Cormier S., Stockham B., Ding T., Keyser U. F., and Baumberg J. J., “ Thermo‐responsive actuation of a DNA origami flexor,” Adv. Funct. Mater. 28(25), 1706410 (2018). 10.1002/adfm.201706410 [DOI] [Google Scholar]
- 173. You M., Chen Y., Zhang X., Liu H., Wang R., Wang K., Williams K. R., and Tan W., “ An autonomous and controllable light-driven DNA walking device,” Angew. Chem. Int. Ed. Engl. 51(10), 2457–2460 (2012). 10.1002/anie.201107733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Takenaka T., Endo M., Suzuki Y., Yang Y., Emura T., Hidaka K., Kato T., Miyata T., Namba K., and Sugiyama H., “ Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials,” Chem. Eur. J. 20(46), 14951–14954 (2014). 10.1002/chem.201404757 [DOI] [PubMed] [Google Scholar]
- 175. Kohman R. E. and Han X., “ Light sensitization of DNA nanostructures via incorporation of photo-cleavable spacers,” Chem. Commun. 51(26), 5747–5750 (2015). 10.1039/C5CC00082C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lauback S., Mattioli K. R., Marras A. E., Armstrong M., Rudibaugh T. P., Sooryakumar R., and Castro C. E., “ Real-time magnetic actuation of DNA nanodevices via modular integration with stiff micro-levers,” Nat. Commun. 9(1), 1446 (2018). 10.1038/s41467-018-03601-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kopperger E., List J., Madhira S., Rothfischer F., Lamb D. C., and Simmel F. C., “ A self-assembled nanoscale robotic arm controlled by electric fields,” Science 359(6373), 296–301 (2018). 10.1126/science.aao4284 [DOI] [PubMed] [Google Scholar]
- 178. Goltry S., Hallstrom N., Clark T., Kuang W., Lee J., Jorcyk C., Knowlton W. B., Yurke B., Hughes W. L., and Graugnard E., “ DNA topology influences molecular machine lifetime in human serum,” Nanoscale 7(23), 10382–10390 (2015). 10.1039/C5NR02283E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Thai H. B. D., Kim K.-R., Hong K. T., Voitsitskyi T., Lee J.-S., Mao C., and Ahn D.-R., “ Kidney-targeted cytosolic delivery of siRNA using a small-sized mirror DNA tetrahedron for enhanced potency,” ACS Central Sci. 6(12), 2250–2258 (2020). 10.1021/acscentsci.0c00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Iverson D., Serrano C., Brahan A. M., Shams A., Totsingan F., and A. J. Bell, Jr. , “ Supporting data for the characterization of PNA-DNA four-way junctions,” Data Brief 5, 756–760 (2015). 10.1016/j.dib.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kizer M. E., Linhardt R. J., Chandrasekaran A. R., and Wang X., “ A molecular hero suit for in vitro and in vivo DNA nanostructures,” Small 15(26), 1805386 (2019). 10.1002/smll.201805386 [DOI] [PubMed] [Google Scholar]
- 182. Krissanaprasit A., Key C. M., Pontula S., and LaBean T. H., “ Self-assembling nucleic acid nanostructures functionalized with aptamers,” Chem. Rev. 121(22), 13797–13868 (2021). 10.1021/acs.chemrev.0c01332 [DOI] [PubMed] [Google Scholar]
- 183. Zhang H., Demirer G. S., Zhang H., Ye T., Goh N. S., Aditham A. J., Cunningham F. J., Fan C., and Landry M. P., “ DNA nanostructures coordinate gene silencing in mature plants,” PNAS 116(15), 7543–7548 (2019). 10.1073/pnas.1818290116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Li J., Pei H., Zhu B., Liang L., Wei M., He Y., Chen N., Li D., Huang Q., and Fan C., “ Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides,” ACS Nano 5(11), 8783–8789 (2011). 10.1021/nn202774x [DOI] [PubMed] [Google Scholar]
- 185. Zeng Y., Liu J., Yang S., Liu W., Xu L., and Wang R., “ Time-lapse live cell imaging to monitor doxorubicin release from DNA origami nanostructures,” J. Mater. Chem. B 6(11), 1605–1612 (2018). 10.1039/C7TB03223D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Xia Z., Wang P., Liu X., Liu T., Yan Y., Yan J., Zhong J., Sun G., and He D., “ Tumor-penetrating peptide-modified DNA tetrahedron for targeting drug delivery,” Biochemistry 55(9), 1326–1331 (2016). 10.1021/acs.biochem.5b01181 [DOI] [PubMed] [Google Scholar]
- 187. Veneziano R., Moyer T. J., Stone M. B., Wamhoff E.-C., Read B. J., Mukherjee S., Shepherd T. R., Das J., Schief W. R., Irvine D. J., and Bathe M., “ Role of nanoscale antigen organization on B-cell activation probed using DNA origami,” Nat. Nanotechnol. 15(8), 716–723 (2020). 10.1038/s41565-020-0719-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Jiang D., Im H.-J., Boleyn M. E., England C. G., Ni D., Kang L., Engle J. W., Huang P., Lan X., and Cai W., “ Efficient renal clearance of DNA tetrahedron nanoparticles enables quantitative evaluation of kidney function,” Nano Res. 12(3), 637–642 (2019). 10.1007/s12274-019-2271-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Jiang D., Sun Y., Li J., Li Q., Lv M., Zhu B., Tian T., Cheng D., Xia J., Zhang L., Wang L., Huang Q., Shi J., and Fan C., “ Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging,” ACS Appl. Mater. Interfaces 8(7), 4378–4384 (2016). 10.1021/acsami.5b10792 [DOI] [PubMed] [Google Scholar]
- 190. Du Y., Jiang Q., Beziere N., Song L., Zhang Q., Peng D., Chi C., Yang X., Guo H., Diot G., Ntziachristos V., Ding B., and Tian J., “ DNA-nanostructure–gold-nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy,” Adv. Mater. 28(45), 10000–10007 (2016). 10.1002/adma.201601710 [DOI] [PubMed] [Google Scholar]
- 191. Simmel F. C., Yurke B., and Singh H. R., “ Principles and applications of nucleic acid strand displacement reactions,” Chem. Rev. 119(10), 6326–6369 (2019). 10.1021/acs.chemrev.8b00580 [DOI] [PubMed] [Google Scholar]
- 192. Liu Y., Kumar S., and Taylor R. E., “ Mix‐and‐match nanobiosensor design: Logical and spatial programming of biosensors using self‐assembled DNA nanostructures,” Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 10(6), e1518 (2018). 10.1002/wnan.1518 [DOI] [PubMed] [Google Scholar]
- 193. Wang S., Zhou Z., Ma N., Yang S., Li K., Teng C., Ke Y., and Tian Y., “ DNA origami-enabled biosensors,” Sensors 20(23), 6899 (2020). 10.3390/s20236899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Ponnuswamy N., Bastings M. M., Nathwani B., Ryu J. H., Chou L. Y., Vinther M., Li W. A., Anastassacos F. M., Mooney D. J., and Shih W. M., “ Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation,” Nat. Commun. 8, 15654 (2017). 10.1038/ncomms15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Anastassacos F. M., Zhao Z., Zeng Y., and Shih W. M., “ Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation,” J. Am. Chem. Soc. 142(7), 3311–3315 (2020). 10.1021/jacs.9b11698 [DOI] [PubMed] [Google Scholar]
- 196. Kim Y. and Yin P., “ Enhancing biocompatible stability of DNA nanostructures using dendritic oligonucleotides and brick motifs,” Angew. Chem. Int. Ed. Engl. 59(2), 700–703 (2020). 10.1002/anie.201911664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kiviaho J. K., Linko V., Ora A., Tiainen T., Järvihaavisto E., Mikkilä J., Tenhu H., Nonappa, and Kostiainen M. A., “ Cationic polymers for DNA origami coating—Examining their binding efficiency and tuning the enzymatic reaction rates,” Nanoscale 8(22), 11674–11680 (2016). 10.1039/C5NR08355A [DOI] [PubMed] [Google Scholar]
- 198. Wang S.-T., Gray M. A., Xuan S., Lin Y., Byrnes J., Nguyen A. I., Todorova N., Stevens M. M., Bertozzi C. R., Zuckermann R. N., and Gang O., “ DNA origami protection and molecular interfacing through engineered sequence-defined peptoids,” Proc. Natl. Acad. Sci. 117, 6339–6348 (2020). 10.1073/pnas.1919749117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kim K.-R., Kim H. Y., Lee Y.-D., Ha J. S., Kang J. H., Jeong H., Bang D., Ko Y. T., Kim S., Lee H., and Ahn D.-R., “ Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs,” J. Controlled Release 243, 121–131 (2016). 10.1016/j.jconrel.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 200. Perrault S. D. and Shih W. M., “ Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability,” ACS Nano 8(5), 5132–5140 (2014). 10.1021/nn5011914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Nguyen M. K., Nguyen V. H., Natarajan A. K., Huang Y. K., Ryssy J., Shen B. X., and Kuzyk A., “ Ultrathin silica coating of DNA origami nanostructures,” Chem. Mater. 32(15), 6657–6665 (2020). 10.1021/acs.chemmater.0c02111 [DOI] [Google Scholar]
- 202. Rajendran A., Endo M., Katsuda Y., Hidaka K., and Sugiyama H., “ Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly,” J. Am. Chem. Soc. 133(37), 14488–14491 (2011). 10.1021/ja204546h [DOI] [PubMed] [Google Scholar]
- 203. Cassinelli V., Oberleitner B., Sobotta J., Nickels P., Grossi G., Kempter S., Frischmuth T., Liedl T., and Manetto A., “ One-step formation of ‘chain-armor’-stabilized DNA nanostructures,” Angew. Chem. Int. Ed. Engl. 54(27), 7795–7798 (2015). 10.1002/anie.201500561 [DOI] [PubMed] [Google Scholar]
- 204. Gerling T., Kube M., Kick B., and Dietz H., “ Sequence-programmable covalent bonding of designed DNA assemblies,” Sci. Adv. 4(8), eaau1157 (2018). 10.1126/sciadv.aau1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Eklund A. S., Comberlato A., Parish I. A., Jungmann R., and Bastings M. M. C., “ Quantification of strand accessibility in biostable DNA origami with single-staple resolution,” ACS Nano 15(11), 17668–17677 (2021). 10.1021/acsnano.1c05540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Xin Y., Piskunen P., Suma A., Li C., Ijäs H., Ojasalo S., Seitz I., Kostiainen M. A., Grundmeier G., Linko V., and Keller A., “ Environment-dependent stability and mechanical properties of DNA origami six-helix bundles with different crossover spacings,” Small 18(18), 2107393 (2022). 10.1002/smll.202107393 [DOI] [PubMed] [Google Scholar]
- 207. Vindigni G., Raniolo S., Iacovelli F., Unida V., Stolfi C., Desideri A., and Biocca S., “ AS1411 aptamer linked to DNA nanostructures diverts its traffic inside cancer cells and improves its therapeutic efficacy,” Pharmaceutics 13(10), 1671 (2021). 10.3390/pharmaceutics13101671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Raniolo S., Vindigni G., Unida V., Ottaviani A., Romano E., Desideri A., and Biocca S., “ Entry, fate and degradation of DNA nanocages in mammalian cells: A matter of receptors,” Nanoscale 10(25), 12078–12086 (2018). 10.1039/C8NR02411A [DOI] [PubMed] [Google Scholar]
- 209. Lacroix A., Edwardson T. G. W., Hancock M. A., Dore M. D., and Sleiman H. F., “ Development of DNA nanostructures for high-affinity binding to human serum albumin,” J. Am. Chem. Soc. 139(21), 7355–7362 (2017). 10.1021/jacs.7b02917 [DOI] [PubMed] [Google Scholar]
- 210. Kim K.-R., Kim J., Back J. H., Lee J. E., and Ahn D.-R., “ Cholesterol-mediated seeding of protein corona on DNA nanostructures for targeted delivery of oligonucleotide therapeutics to treat liver fibrosis,” ACS Nano 16(5), 7331–7343 (2022). 10.1021/acsnano.1c08508 [DOI] [PubMed] [Google Scholar]
- 211. Birkholz O., Burns J. R., Richter C. P., Psathaki O. E., Howorka S., and Piehler J., “ Multi-functional DNA nanostructures that puncture and remodel lipid membranes into hybrid materials,” Nat. Commun. 9(1), 1521 (2018). 10.1038/s41467-018-02905-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Wang W., Arias D. S., Deserno M., Ren X., and Taylor R. E., “ Emerging applications at the interface of DNA nanotechnology and cellular membranes: Perspectives from biology, engineering, and physics,” APL Bioeng. 4(4), 041507 (2020). 10.1063/5.0027022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Whitehouse W. L., Noble J. E., Ryadnov M. G., and Howorka S., “ Cholesterol anchors enable efficient binding and intracellular uptake of DNA nanostructures,” Bioconjugate Chem. 30(7), 1836–1844 (2019). 10.1021/acs.bioconjchem.9b00036 [DOI] [PubMed] [Google Scholar]
- 214. Cremers G. A. O., Rosier B. J. H. M., Meijs A., Tito N. B., van Duijnhoven S. M. J., van Eenennaam H., Albertazzi L., and de Greef T. F. A., “ Determinants of ligand-functionalized DNA nanostructure–cell interactions,” J. Am. Chem. Soc. 143(27), 10131–10142 (2021). 10.1021/jacs.1c02298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ohmann A., Göpfrich K., Joshi H., Thompson R. F., Sobota D., Ranson N. A., Aksimentiev A., and Keyser U. F., “ Controlling aggregation of cholesterol-modified DNA nanostructures,” Nucl. Acids Res. 47(21), 11441–11451 (2019). 10.1093/nar/gkz914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Mendes B. B., Conniot J., Avital A., Yao D., Jiang X., Zhou X., Sharf-Pauker N., Xiao Y., Adir O., Liang H., Shi J., Schroeder A., and Conde J., “ Nanodelivery of nucleic acids,” Nat. Rev. Methods Primers 2(1), 24 (2022). 10.1038/s43586-022-00104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Mathur D. and Medintz I. L., “ Analyzing DNA nanotechnology: A call to arms for the analytical chemistry community,” Anal. Chem. 89, 2646–2663 (2017). 10.1021/acs.analchem.6b04033 [DOI] [PubMed] [Google Scholar]
- 218. Stetefeld J., McKenna S. A., and Patel T. R., “ Dynamic light scattering: A practical guide and applications in biomedical sciences,” Biophys. Rev. 8(4), 409–427 (2016). 10.1007/s12551-016-0218-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Ahmadi Y., De Llano E., and Barišić I., “ (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures,” Nanoscale 10(16), 7494–7504 (2018). 10.1039/C7NR09461B [DOI] [PubMed] [Google Scholar]
- 220. Chakraborty K., Veetil A. T., Jaffrey S. R., and Krishnan Y., “ Nucleic acid–based nanodevices in biological imaging,” Annu. Rev. Biochem. 85(1), 349–373 (2016). 10.1146/annurev-biochem-060815-014244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Pal S., Deng Z., Wang H., Zou S., Liu Y., and Yan H., “ DNA directed self-assembly of anisotropic plasmonic nanostructures,” J. Am. Chem. Soc. 133(44), 17606–17609 (2011). 10.1021/ja207898r [DOI] [PubMed] [Google Scholar]
- 222. Marmur J. and Doty P., “ Heterogeneity in deoxyribonucleic acids. I. Dependence on composition of the configurational stability of deoxyribonucleic acids,” Nature 183(4673), 1427–1429 (1959). 10.1038/1831427a0 [DOI] [PubMed] [Google Scholar]
- 223. Reed G. H., Kent J. O., and Wittwer C. T., “ High-resolution DNA melting analysis for simple and efficient molecular diagnostics,” Pharmacogenomics 8(6), 597–608 (2007). 10.2217/14622416.8.6.597 [DOI] [PubMed] [Google Scholar]
- 224. Wienken C. J., Baaske P., Duhr S., and Braun D., “ Thermophoretic melting curves quantify the conformation and stability of RNA and DNA,” Nucl. Acids Res. 39(8), e52 (2011). 10.1093/nar/gkr035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Majikes J. M., Patrone P. N., Schiffels D., Zwolak M., Kearsley A. J., Forry S. P., and Liddle J. A., “ Revealing thermodynamics of DNA origami folding via affine transformations,” Nucl. Acids Res. 48(10), 5268–5280 (2020). 10.1093/nar/gkaa283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Majikes J. M., Patrone P. N., Kearsley A. J., Zwolak M., and Liddle J. A., “ Failure mechanisms in DNA self-assembly: Barriers to single-fold yield,” ACS Nano 15(2), 3284–3294 (2021). 10.1021/acsnano.0c10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Li L., Zhang X., Wang H., Lang Q., Chen H., and Liu L. Q., “ Measurement of radial elasticity and original height of DNA duplex using tapping-mode atomic force microscopy,” Nanomaterials 9(4), 561 (2019). 10.3390/nano9040561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Goodman R. P., Schaap I. A. T., Tardin C. F., Erben C. M., Berry R. M., Schmidt C. F., and Turberfield A. J., “ Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication,” Science 310(5754), 1661–1665 (2005). 10.1126/science.1120367 [DOI] [PubMed] [Google Scholar]
- 229. Ghimire C., Wang H., Li H., Vieweger M., Xu C., and Guo P., “ RNA nanoparticles as rubber for compelling vessel extravasation to enhance tumor targeting and for fast renal excretion to reduce toxicity,” ACS Nano 14(10), 13180–13191 (2020). 10.1021/acsnano.0c04863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Herrero-Galán E., Fuentes-Perez M. E., Carrasco C., Valpuesta J. M., Carrascosa J. L., Moreno-Herrero F., and Arias-Gonzalez J. R., “ Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level,” J. Am. Chem. Soc. 135(1), 122–131 (2013). 10.1021/ja3054755 [DOI] [PubMed] [Google Scholar]
- 231. Lipfert J., Skinner G. M., Keegstra J. M., Hensgens T., Jager T., Dulin D., Köber M., Yu Z., Donkers S. P., Chou F.-C., Das R., and Dekker N. H., “ Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA,” Proc. Natl. Acad. Sci. 111(43), 15408 (2014). 10.1073/pnas.1407197111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Naskar S. and Maiti P. K., “ Mechanical properties of DNA and DNA nanostructures: Comparison of atomistic, Martini and oxDNA models,” J. Mater. Chem. B 9(25), 5102–5113 (2021). 10.1039/D0TB02970J [DOI] [PubMed] [Google Scholar]
- 233. Lee C., Lee J. Y., and Kim D.-N., “ Polymorphic design of DNA origami structures through mechanical control of modular components,” Nat. Commun. 8(1), 2067 (2017). 10.1038/s41467-017-02127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Zagorovsky K., Chou L. Y. T., and Chan W. C. W., “ Controlling DNA nanoparticle serum interactions,” PNAS 113(48), 13600–13605 (2016). 10.1073/pnas.1610028113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Mei Q., Wei X., Su F., Liu Y., Youngbull C., Johnson R., Lindsay S., Yan H., and Meldrum D., “ Stability of DNA origami nanoarrays in cell lysate,” Nano Lett. 11(4), 1477–1482 (2011). 10.1021/nl1040836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Chandrasekaran A. R. and Halvorsen K., “ Nuclease degradation analysis of DNA nanostructures using gel electrophoresis,” Curr. Protoc. Nucl. Acid Chem. 82(1), e115 (2020). 10.1002/cpnc.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Langlois N. I. and Clark H. A., “ Characterization of DNA nanostructure stability by size exclusion chromatography,” Anal. Methods 14(10), 1006–1014 (2022). 10.1039/D1AY02146J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Liang L., Li J., Li Q., Huang Q., Shi J., Yan H., and Fan C., “ Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells,” Angew. Chem. Int. Ed. 53(30), 7745–7750 (2014). 10.1002/anie.201403236 [DOI] [PubMed] [Google Scholar]
- 239. Surana S., Bhatia D., and Krishnan Y., “ A method to study in vivo stability of DNA nanostructures,” Methods 64(1), 94–100 (2013). 10.1016/j.ymeth.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Lacroix A., Vengut-Climent E., de Rochambeau D., and Sleiman H. F., “ Uptake and fate of fluorescently labeled DNA nanostructures in cellular environments: A cautionary tale,” ACS Central Sci. 5(5), 882–891 (2019). 10.1021/acscentsci.9b00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Mathur D., Rogers K. E., Díaz S. A., Muroski M. E., Klein W. P., Nag O. K., Lee K., Field L. D., Delehanty J. B., and Medintz I. L., “ Determining the cytosolic stability of small DNA nanostructures in cellula,” Nano Lett. 22(12), 5037–5045 (2022). 10.1021/acs.nanolett.2c00917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Wang P., Rahman M. A., Zhao Z., Weiss K., Zhang C., Chen Z., Hurwitz S. J., Chen Z. G., Shin D. M., and Ke Y., “ Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells,” J. Am. Chem. Soc. 140(7), 2478–2484 (2018). 10.1021/jacs.7b09024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Vindigni G., Raniolo S., Ottaviani A., Falconi M., Franch O., Knudsen B. R., Desideri A., and Biocca S., “ Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells,” ACS Nano 10(6), 5971–5979 (2016). 10.1021/acsnano.6b01402 [DOI] [PubMed] [Google Scholar]
- 244. Okholm A. H., Nielsen J. S., Vinther M., Sørensen R. S., Schaffert D., and Kjems J., “ Quantification of cellular uptake of DNA nanostructures by qPCR,” Methods 67(2), 193–197 (2014). 10.1016/j.ymeth.2014.01.013 [DOI] [PubMed] [Google Scholar]
- 245. Tian T., Zhang C., Li J., Liu Y., Wang Y., Ke X., Fan C., Lei H., Hao P., and Li Q., “ Proteomic exploration of endocytosis of framework nucleic acids,” Small 17(23), 2100837 (2021). 10.1002/smll.202100837 [DOI] [PubMed] [Google Scholar]
- 246. Ding H., Li J., Chen N., Hu X., Yang X., Guo L., Li Q., Zuo X., Wang L., Ma Y., and Fan C., “ DNA nanostructure-programmed like-charge attraction at the cell-membrane interface,” ACS Central Sci. 4(10), 1344–1351 (2018). 10.1021/acscentsci.8b00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Huang H., Belwal T., Li L., Xu Y., Zou L., Lin X., and Luo Z., “ Amphiphilic and biocompatible DNA origami-based emulsion formation and nanopore release for anti-melanogenesis therapy,” Small 17(45), 2104831 (2021). 10.1002/smll.202104831 [DOI] [PubMed] [Google Scholar]
- 248. Vanmeert M., Razzokov J., Mirza M. U., Weeks S. D., Schepers G., Bogaerts A., Rozenski J., Froeyen M., Herdewijn P., Pinheiro V. B., and Lescrinier E., “ Rational design of an XNA ligase through docking of unbound nucleic acids to toroidal proteins,” Nucl. Acids Res. 47(13), 7130–7142 (2019). 10.1093/nar/gkz551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Green C. M., Mathur D., and Medintz I. L., “ Understanding the fate of DNA nanostructures inside the cell,” J. Mater. Chem. B 8(29), 6170–6178 (2020). 10.1039/D0TB00395F [DOI] [PubMed] [Google Scholar]
- 250. Onoue S., Yamada S., and Chan H.-K., “ Nanodrugs: Pharmacokinetics and safety,” Int. J. Nanomed. 9, 1025–1037 (2014). 10.2147/IJN.S38378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Kang H., Mintri S., Menon A. V., Lee H. Y., Choi H. S., and Kim J., “ Pharmacokinetics, pharmacodynamics and toxicology of theranostic nanoparticles,” Nanoscale 7(45), 18848–18862 (2015). 10.1039/C5NR05264E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Liu N., Zhang X., Li N., Zhou M., Zhang T., Li S., Cai X., Ji P., and Lin Y., “ Tetrahedral framework nucleic acids promote corneal epithelial wound healing in vitro and in vivo,” Small 15(31), 1901907 (2019). 10.1002/smll.201901907 [DOI] [PubMed] [Google Scholar]
- 253. Sirong S., Yang C., Taoran T., Songhang L., Shiyu L., Yuxin Z., Xiaoru S., Tao Z., Yunfeng L., and Xiaoxiao C., “ Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis,” Bone Res. 8(1), 6 (2020). 10.1038/s41413-019-0077-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Li S., Jiang D., Rosenkrans Z. T., Barnhart T. E., Ehlerding E. B., Ni D., Engle J. W., and Cai W., “ Aptamer-conjugated framework nucleic acids for the repair of cerebral ischemia-reperfusion injury,” Nano Lett. 19(10), 7334–7341 (2019). 10.1021/acs.nanolett.9b02958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Bhatia D., Surana S., Chakraborty S., Koushika S. P., and Krishnan Y., “ A synthetic icosahedral DNA-based host–cargo complex for functional in vivo imaging,” Nat. Commun. 2(1), 339 (2011). 10.1038/ncomms1337 [DOI] [PubMed] [Google Scholar]
- 256. Amir Y., Ben-Ishay E., Levner D., Ittah S., Abu-Horowitz A., and Bachelet I., “ Universal computing by DNA origami robots in a living animal,” Nat. Nanotechnol. 9(5), 353–357 (2014). 10.1038/nnano.2014.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Myhrvold C., Dai M., Silver P. A., and Yin P., “ Isothermal self-assembly of complex DNA structures under diverse and biocompatible conditions,” Nano Lett. 13(9), 4242–4248 (2013). 10.1021/nl4019512 [DOI] [PubMed] [Google Scholar]
- 258. Roodhuizen J. A. L., Hendrikx P. J. T. M., Hilbers P. A. J., de Greef T. F. A., and Markvoort A. J., “ Counterion-dependent mechanisms of DNA origami nanostructure stabilization revealed by atomistic molecular simulation,” ACS Nano 13(9), 10798–10809 (2019). 10.1021/acsnano.9b05650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Auvinen H., Zhang H., Nonappa, Kopilow A., Niemelä E. H., Nummelin S., Correia A., Santos H. A., Linko V., and Kostiainen M. A., “ Protein coating of DNA nanostructures for enhanced stability and immunocompatibility,” Adv. Healthcare Mater. 6(18), 1700692 (2017). 10.1002/adhm.201700692 [DOI] [PubMed] [Google Scholar]
- 260. Stephanopoulos N., “ Strategies for stabilizing DNA nanostructures to biological conditions,” ChemBioChem 20(17), 2191–2197 (2019). 10.1002/cbic.201900075 [DOI] [PubMed] [Google Scholar]
- 261. Bila H., Kurisinkal E. E., and Bastings M. M. C., “ Engineering a stable future for DNA-origami as a biomaterial,” Biomater. Sci. 7(2), 532–541 (2019). 10.1039/C8BM01249K [DOI] [PubMed] [Google Scholar]
- 262. Nadano D., Yasuda T., and Kishi K., “ Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method,” Clin. Chem. 39(3), 448–452 (1993). 10.1093/clinchem/39.3.448 [DOI] [PubMed] [Google Scholar]
- 263. Ritz S., Schöttler S., Kotman N., Baier G., Musyanovych A., Kuharev J., Landfester K., Schild H., Jahn O., Tenzer S., and Mailänder V., “ Protein corona of nanoparticles: Distinct proteins regulate the cellular uptake,” Biomacromolecules 16(4), 1311–1321 (2015). 10.1021/acs.biomac.5b00108 [DOI] [PubMed] [Google Scholar]
- 264. Latreille P.-L., Le Goas M., Salimi S., Robert J., De Crescenzo G., Boffito D. C., Martinez V. A., Hildgen P., and Banquy X., “ Scratching the surface of the protein corona: Challenging measurements and controversies,” ACS Nano 16(2), 1689–1707 (2022). 10.1021/acsnano.1c05901 [DOI] [PubMed] [Google Scholar]
- 265. Cai R. and Chen C., “ The crown and the scepter: Roles of the protein corona in nanomedicine,” Adv. Mater. 31(45), 1805740 (2019). 10.1002/adma.201805740 [DOI] [PubMed] [Google Scholar]
- 266. Aggarwal P., Hall J. B., McLeland C. B., Dobrovolskaia M. A., and McNeil S. E., “ Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy,” Adv. Drug Delivery Rev. 61(6), 428–437 (2009). 10.1016/j.addr.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Tenzer S., Docter D., Kuharev J., Musyanovych A., Fetz V., Hecht R., Schlenk F., Fischer D., Kiouptsi K., Reinhardt C., Landfester K., Schild H., Maskos M., Knauer S. K., and Stauber R. H., “ Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology,” Nat. Nanotechnol. 8(10), 772–781 (2013). 10.1038/nnano.2013.181 [DOI] [PubMed] [Google Scholar]
- 268. Smolková B., MacCulloch T., Rockwood T. F., Liu M., Henry S. J. W., Frtús A., Uzhytchak M., Lunova M., Hof M., Jurkiewicz P., Dejneka A., Stephanopoulos N., and Lunov O., “ Protein corona inhibits endosomal escape of functionalized DNA nanostructures in living cells,” ACS Appl. Mater. Interfaces 13(39), 46375–46390 (2021). 10.1021/acsami.1c14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Ren J., Andrikopoulos N., Velonia K., Tang H., Cai R., Ding F., Ke P. C., and Chen C., “ Chemical and biophysical signatures of the protein corona in nanomedicine,” J. Am. Chem. Soc. 144(21), 9184–9205 (2022). 10.1021/jacs.2c02277 [DOI] [PubMed] [Google Scholar]
- 270. Tenzer S., Docter D., Rosfa S., Wlodarski A., Kuharev J., Rekik A., Knauer S. K., Bantz C., Nawroth T., Bier C., Sirirattanapan J., Mann W., Treuel L., Zellner R., Maskos M., Schild H., and Stauber R. H., “ Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: A comprehensive quantitative proteomic analysis,” ACS Nano 5(9), 7155–7167 (2011). 10.1021/nn201950e [DOI] [PubMed] [Google Scholar]
- 271. Ferdosi S., Tangeysh B., Brown T. R., Everley P. A., Figa M., McLean M., Elgierari E. M., Zhao X., Garcia V. J., Wang T., Chang M. E. K., Riedesel K., Chu J., Mahoney M., Xia H., O'Brien E. S., Stolarczyk C., Harris D., Platt T. L., Ma P., Goldberg M., Langer R., Flory M. R., Benz R., Tao W., Cuevas J. C., Batzoglou S., Blume J. E., Siddiqui A., Hornburg D., and Farokhzad O. C., “ Engineered nanoparticles enable deep proteomics studies at scale by leveraging tunable nano-bio interactions,” Proc. Natl. Acad. Sci. 119(11), e2106053119 (2022). 10.1073/pnas.2106053119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Ban Z., Yuan P., Yu F., Peng T., Zhou Q., and Hu X., “ Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles,” Proc. Natl. Acad. Sci. 117(19), 10492–10499 (2020). 10.1073/pnas.1919755117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Xu F., Dong B., Li X., Gao F., Yang D., Xue W., and Wang P., “ Profiling and regulating proteins that adsorb to DNA materials in human serum,” Anal. Chem. 93(24), 8671–8679 (2021). 10.1021/acs.analchem.1c02075 [DOI] [PubMed] [Google Scholar]
- 274. Geary R. S., Norris D., Yu R., and Bennett C. F., “ Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides,” Adv. Drug Delivery Rev. 87, 46–51 (2015). 10.1016/j.addr.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 275. Frtús A., Smolková B., Uzhytchak M., Lunova M., Jirsa M., Henry S. J. W., Dejneka A., Stephanopoulos N., and Lunov O., “ The interactions between DNA nanostructures and cells: A critical overview from a cell biology perspective,” Acta Biomater. 146, 10–22 (2022). 10.1016/j.actbio.2022.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Langecker M., Arnaut V., List J., and Simmel F. C., “ DNA nanostructures interacting with lipid bilayer membranes,” Acc. Chem. Res. 47(6), 1807–1815 (2014). 10.1021/ar500051r [DOI] [PubMed] [Google Scholar]
- 277. Huo S., Li H., Boersma A. J., and Herrmann A., “ DNA nanotechnology enters cell membranes,” Adv. Sci. 6(10), 1900043 (2019). 10.1002/advs.201900043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Lee D. S., Qian H., Tay C. Y., and Leong D. T., “ Cellular processing and destinies of artificial DNA nanostructures,” Chem. Soc. Rev. 45(15), 4199–4225 (2016). 10.1039/C5CS00700C [DOI] [PubMed] [Google Scholar]
- 279. Lacroix A., Fakih H. H., and Sleiman H. F., “ Detailed cellular assessment of albumin-bound oligonucleotides: Increased stability and lower non-specific cell uptake,” J. Controlled Release 324, 34–46 (2020). 10.1016/j.jconrel.2020.04.020 [DOI] [PubMed] [Google Scholar]
- 280. Roberts T. L., Dunn J. A., Terry T. D., Jennings M. P., Hume D. A., Sweet M. J., and Stacey K. J., “ Differences in macrophage activation by bacterial DNA and CpG-containing oligonucleotides,” J. Immunol. 175(6), 3569–3576 (2005). 10.4049/jimmunol.175.6.3569 [DOI] [PubMed] [Google Scholar]
- 281. Walsh A. S., Yin H., Erben C. M., Wood M. J. A., and Turberfield A. J., “ DNA cage delivery to mammalian cells,” ACS Nano 5(7), 5427–5432 (2011). 10.1021/nn2005574 [DOI] [PubMed] [Google Scholar]
- 282. Peng X., Fang S., Ji B., Li M., Song J., Qiu L., and Tan W., “ DNA nanostructure-programmed cell entry via corner angle-mediated molecular interaction with membrane receptors,” Nano Lett. 21(16), 6946–6951 (2021). 10.1021/acs.nanolett.1c02191 [DOI] [PubMed] [Google Scholar]
- 283. Canton J., Neculai D., and Grinstein S., “ Scavenger receptors in homeostasis and immunity,” Nat. Rev. Immunol. 13(9), 621–634 (2013). 10.1038/nri3515 [DOI] [PubMed] [Google Scholar]
- 284. Patel P. C., Giljohann D. A., Daniel W. L., Zheng D., Prigodich A. E., and Mirkin C. A., “ Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles,” Bioconjugate Chem. 21(12), 2250–2256 (2010). 10.1021/bc1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Raniolo S., Croce S., Thomsen R. P., Okholm A. H., Unida V., Iacovelli F., Manetto A., Kjems J., Desideri A., and Biocca S., “ Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries,” Nanoscale 11(22), 10808–10818 (2019). 10.1039/C9NR02006C [DOI] [PubMed] [Google Scholar]
- 286. Chen X., Tian F., Li M., Xu H., Cai M., Li Q., Zuo X., Wang H., Shi X., Fan C., Baigude H., and Shan Y., “ Size-independent transmembrane transporting of single tetrahedral DNA nanostructures,” Global Challenges 4(3), 1900075 (2020). 10.1002/gch2.201900075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Sun S.-W., Zu X.-Y., Tuo Q.-H., Chen L.-X., Lei X.-Y., Li K., Tang C.-K., and Liao D.-F., “ Caveolae and caveolin-1 mediate endocytosis and transcytosis of oxidized low density lipoprotein in endothelial cells,” Acta Pharmacol. Sin. 31(10), 1336–1342 (2010). 10.1038/aps.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Predescu S. A., Predescu D. N., and Malik A. B., “ Molecular determinants of endothelial transcytosis and their role in endothelial permeability,” Am. J. Physiol. 293(4), L823–L842 (2007). 10.1152/ajplung.00436.2006 [DOI] [PubMed] [Google Scholar]
- 289. Matthaeus C. and Taraska J. W., “ Energy and dynamics of caveolae trafficking,” Front. Cell Dev. Biol. 8, 614472 (2021). 10.3389/fcell.2020.614472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Kaksonen M. and Roux A., “ Mechanisms of clathrin-mediated endocytosis,” Nat. Rev. Mol. Cell Biol. 19(5), 313–326 (2018). 10.1038/nrm.2017.132 [DOI] [PubMed] [Google Scholar]
- 291. Kim K.-R., Kang S. J., Lee A.-Y., Hwang D., Park M., Park H., Kim S., Hur K., Chung H. S., and Mao C. J. B., “ Highly tumor-specific DNA nanostructures discovered by in vivo screening of a nucleic acid cage library and their applications in tumor-targeted drug delivery,” Biomaterials 195, 1–12 (2019). 10.1016/j.biomaterials.2018.12.026 [DOI] [PubMed] [Google Scholar]
- 292. Rajwar A., Shetty S. R., Vaswani P., Morya V., Barai A., Sen S., Sonawane M., and Bhatia D., “ Geometry of a DNA nanostructure influences its endocytosis: Cellular study on 2D, 3D, and in vivo systems,” ACS Nano 16(7), 10496–10508 (2022). 10.1021/acsnano.2c01382 [DOI] [PubMed] [Google Scholar]
- 293. Hivare P., Rajwar A., Gupta S., and Bhatia D., “ Spatiotemporal dynamics of endocytic pathways adapted by small DNA nanocages in model neuroblastoma cell-derived differentiated neurons,” ACS Appl. Bio Mater. 4(4), 3350–3359 (2021). 10.1021/acsabm.0c01668 [DOI] [PubMed] [Google Scholar]
- 294. Wang D., Liu Q., Wu D., He B., Li J., Mao C., Wang G., and Qian H., “ Isothermal self-assembly of spermidine–DNA nanostructure complex as a functional platform for cancer therapy,” ACS Appl. Mater. Interfaces 10(18), 15504–15516 (2018). 10.1021/acsami.8b03464 [DOI] [PubMed] [Google Scholar]
- 295. Charoenphol P. and Bermudez H., “ Aptamer-targeted DNA nanostructures for therapeutic delivery,” Mol. Pharmaceutics 11(5), 1721–1725 (2014). 10.1021/mp500047b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Zhang Q., Jiang Q., Li N., Dai L., Liu Q., Song L., Wang J., Li Y., Tian J., Ding B., and Du Y., “ DNA origami as an in vivo drug delivery vehicle for cancer therapy,” ACS Nano 8(7), 6633–6643 (2014). 10.1021/nn502058j [DOI] [PubMed] [Google Scholar]
- 297. Shi S., Li Y., Zhang T., Xiao D., Tian T., Chen T., Zhang Y., Li X., and Lin Y., “ Biological effect of differently sized tetrahedral framework nucleic acids: Endocytosis, proliferation, migration, and biodistribution,” ACS Appl. Mater. Interfaces 13(48), 57067–57074 (2021). 10.1021/acsami.1c20657 [DOI] [PubMed] [Google Scholar]
- 298. Burns J. R., Seifert A., Fertig N., and Howorka S., “ A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane,” Nat. Nanotechnol. 11(2), 152–156 (2016). 10.1038/nnano.2015.279 [DOI] [PubMed] [Google Scholar]
- 299. Liu J., Li W., Li R., Yin X., He S., Hu J., and Ruan S., “ Programmable DNA framework sensors for in situ cell-surface pH analysis,” Anal. Chem. 93(36), 12170–12174 (2021). 10.1021/acs.analchem.1c03227 [DOI] [PubMed] [Google Scholar]
- 300. Zhao X.-L., Chen B.-C., Han J.-C., Wei L., and Pan X.-B., “ Delivery of cell-penetrating peptide-peptide nucleic acid conjugates by assembly on an oligonucleotide scaffold,” Sci. Rep. 5(1), 17640 (2015). 10.1038/srep17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Sakai Y., Islam M. S., Adamiak M., Shiu S. C.-C., Tanner J. A., and Heddle J. G., “ DNA aptamers for the functionalisation of DNA origami nanostructures,” Genes 9(12), 571 (2018). 10.3390/genes9120571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Chang M., Yang C.-S., and Huang D.-M., “ Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy,” ACS Nano 5(8), 6156–6163 (2011). 10.1021/nn200693a [DOI] [PubMed] [Google Scholar]
- 303. Liu J., Song L., Liu S., Jiang Q., Liu Q., Li N., Wang Z.-G., and Ding B., “ A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy,” Nano Lett. 18(6), 3328–3334 (2018). 10.1021/acs.nanolett.7b04812 [DOI] [PubMed] [Google Scholar]
- 304. Schaffert D. H., Okholm A. H., Sørensen R. S., Nielsen J. S., Tørring T., Rosen C. B., Kodal A. L. B., Mortensen M. R., Gothelf K. V., and Kjems J., “ Intracellular delivery of a planar DNA origami structure by the transferrin-receptor internalization pathway,” Small 12(19), 2634–2640 (2016). 10.1002/smll.201503934 [DOI] [PubMed] [Google Scholar]
- 305. Du Y., Peng P., and Li T., “ DNA logic operations in living cells utilizing lysosome-recognizing framework nucleic acid nanodevices for subcellular imaging,” ACS Nano 13(5), 5778–5784 (2019). 10.1021/acsnano.9b01324 [DOI] [PubMed] [Google Scholar]
- 306. Huotari J. and Helenius A., “ Endosome maturation,” EMBO J. 30(17), 3481–3500 (2011). 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Halley P. D., Lucas C. R., McWilliams E. M., Webber M. J., Patton R. A., Kural C., Lucas D. M., Byrd J. C., and Castro C. E., “ Daunorubicin-loaded DNA origami nanostructures circumvent drug-resistance mechanisms in a leukemia model,” Small 12(3), 308–320 (2016). 10.1002/smll.201502118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Schaffter S. W., Green L. N., Schneider J., Subramanian H. K. K., Schulman R., and Franco E., “ T7 RNA polymerase non-specifically transcribes and induces disassembly of DNA nanostructures,” Nucl. Acids Res. 46(10), 5332–5343 (2018). 10.1093/nar/gky283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Jiao K., Zhu B., Guo L., Zhou H., Wang F., Zhang X., Shi J., Li Q., Wang L., Li J., and Fan C., “ Programming switchable transcription of topologically constrained DNA,” J. Am. Chem. Soc. 142(24), 10739–10746 (2020). 10.1021/jacs.0c01962 [DOI] [PubMed] [Google Scholar]
- 310. Selby L. I., Cortez-Jugo C. M., Such G. K., and Johnston A. P. R., “ Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles,” WIREs Nanomed. Nanobiotechnol. 9(5), e1452 (2017). 10.1002/wnan.1452 [DOI] [PubMed] [Google Scholar]
- 311. Smith S. A., Selby L. I., Johnston A. P. R., and Such G. K., “ The endosomal escape of nanoparticles: Toward more efficient cellular delivery,” Bioconjugate Chem. 30(2), 263–272 (2019). 10.1021/acs.bioconjchem.8b00732 [DOI] [PubMed] [Google Scholar]
- 312. Wang Y., Wang J., Zhu D., Wang Y., Qing G., Zhang Y., Liu X., and Liang X.-J., “ Effect of physicochemical properties on in vivo fate of nanoparticle-based cancer immunotherapies,” Acta Pharm. Sin. B 11(4), 886–902 (2021). 10.1016/j.apsb.2021.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Tseng C. Y., Wang W. X., Douglas T. R., and Chou L. Y. T., “ Engineering DNA nanostructures to manipulate immune receptor signaling and immune cell fates,” Adv. Healthcare Mater. 11(4), 2101844 (2022). 10.1002/adhm.202101844 [DOI] [PubMed] [Google Scholar]
- 314. Ballas Z. K., Krieg A. M., Warren T., Rasmussen W., Davis H. L., Waldschmidt M., and Weiner G. J., “ Divergent therapeutic and immunologic effects of oligodeoxynucleotides with distinct CpG motifs,” J. Immunol. 167(9), 4878–4886 (2001). 10.4049/jimmunol.167.9.4878 [DOI] [PubMed] [Google Scholar]
- 315. Schüller V. J., Heidegger S., Sandholzer N., Nickels P. C., Suhartha N. A., Endres S., Bourquin C., and Liedl T., “ Cellular immunostimulation by CpG-sequence-coated DNA origami structures,” ACS Nano 5(12), 9696–9702 (2011). 10.1021/nn203161y [DOI] [PubMed] [Google Scholar]
- 316. Mohri K., Nishikawa M., Takahashi N., Shiomi T., Matsuoka N., Ogawa K., Endo M., Hidaka K., Sugiyama H., Takahashi Y., and Takakura Y., “ Design and development of nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory CpG motifs to immune cells,” ACS Nano 6(7), 5931–5940 (2012). 10.1021/nn300727j [DOI] [PubMed] [Google Scholar]
- 317. Behrens E. M., Canna S. W., Slade K., Rao S., Kreiger P. A., Paessler M., Kambayashi T., and Koretzky G. A., “ Repeated TLR9 stimulation results in macrophage activation syndrome–like disease in mice,” J. Clin. Invest. 121(6), 2264–2277 (2011). 10.1172/JCI43157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Johnson M. B., Halman J. R., Satterwhite E., Zakharov A. V., Bui M. N., Benkato K., Goldsworthy V., Kim T., Hong E., Dobrovolskaia M. A., Khisamutdinov E. F., Marriott I., and Afonin K. A., “ Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling,” Small 13(42), 1701255 (2017). 10.1002/smll.201701255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Akira S. and Takeda K., “ Toll-like receptor signalling,” Nat. Rev. Immunol. 4(7), 499–511 (2004). 10.1038/nri1391 [DOI] [PubMed] [Google Scholar]
- 320. Hong E., Halman J. R., Shah A. B., Khisamutdinov E. F., Dobrovolskaia M. A., and Afonin K. A., “ Structure and composition define immunorecognition of nucleic acid nanoparticles,” Nano Lett. 18(7), 4309–4321 (2018). 10.1021/acs.nanolett.8b01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Chattopadhyay S. and Sen G. C., “ dsRNA-activation of TLR3 and RLR signaling: Gene induction-dependent and independent effects,” J. Interferon Cytokine Res. 34(6), 427–436 (2014). 10.1089/jir.2014.0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Lee H., Lytton-Jean A. K. R., Chen Y., Love K. T., Park A. I., Karagiannis E. D., Sehgal A., Querbes W., Zurenko C. S., Jayaraman M., Peng C. G., Charisse K., Borodovsky A., Manoharan M., Donahoe J. S., Truelove J., Nahrendorf M., Langer R., and Anderson D. G., “ Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery,” Nat. Nanotechnol. 7(6), 389–393 (2012). 10.1038/nnano.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Kim K.-R., Jegal H., Kim J., and Ahn D.-R., “ A self-assembled DNA tetrahedron as a carrier for in vivo liver-specific delivery of siRNA,” Biomater. Sci. 8(2), 586–590 (2020). 10.1039/C9BM01769K [DOI] [PubMed] [Google Scholar]
- 324. Zhang H., Zhang H., Demirer G. S., González-Grandío E., Fan C., and Landry M. P., “ Engineering DNA nanostructures for siRNA delivery in plants,” Nat. Protoc. 15(9), 3064–3087 (2020). 10.1038/s41596-020-0370-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Cooke M. S., Evans M. D., Dizdaroglu M., and Lunec J., “ Oxidative DNA damage: Mechanisms, mutation, and disease,” FASEB J. 17(10), 1195–1214 (2003). 10.1096/fj.02-0752rev [DOI] [PubMed] [Google Scholar]
- 326. Zhang Q., Lin S., Shi S., Zhang T., Ma Q., Tian T., Zhou T., Cai X., and Lin Y., “ Anti-inflammatory and antioxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses,” ACS Appl. Mater. Interfaces 10(4), 3421–3430 (2018). 10.1021/acsami.7b17928 [DOI] [PubMed] [Google Scholar]
- 327. Zhang Q., Lin S., Wang L., Peng S., Tian T., Li S., Xiao J., and Lin Y., “ Tetrahedral framework nucleic acids act as antioxidants in acute kidney injury treatment,” Chem. Eng. J. 413, 127426 (2021). 10.1016/j.cej.2020.127426 [DOI] [Google Scholar]
- 328. Li X., Li Z., and Yu H., “ Selection of threose nucleic acid aptamers to block PD-1/PD-L1 interaction for cancer immunotherapy,” Chem. Commun. 56(93), 14653–14656 (2020). 10.1039/D0CC06032A [DOI] [PubMed] [Google Scholar]
- 329. Wang Y., Baars I., Fördös F., and Högberg B., “ Clustering of death receptor for apoptosis using nanoscale patterns of peptides,” ACS Nano 15(6), 9614–9626 (2021). 10.1021/acsnano.0c10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Comberlato A., Koga M. M., Nüssing S., Parish I. A., and Bastings M. M. C., “ Spatially controlled activation of toll-like receptor 9 with DNA-based nanomaterials,” Nano Lett. 22(6), 2506–2513 (2022). 10.1021/acs.nanolett.2c00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Arnida, Janát-Amsbury M. M., Ray A., Peterson C. M., and Ghandehari H., “ Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages,” Eur. J. Pharm. Biopharm. 77(3), 417–423 (2011). 10.1016/j.ejpb.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Schroeder A., Heller D. A., Winslow M. M., Dahlman J. E., Pratt G. W., Langer R., Jacks T., and Anderson D. G., “ Treating metastatic cancer with nanotechnology,” Nat. Rev. Cancer 12(1), 39–50 (2012). 10.1038/nrc3180 [DOI] [PubMed] [Google Scholar]
- 333. Nam J., Won N., Bang J., Jin H., Park J., Jung S., Jung S., Park Y., and Kim S., “ Surface engineering of inorganic nanoparticles for imaging and therapy,” Adv. Drug Delivery Rev. 65(5), 622–648 (2013). 10.1016/j.addr.2012.08.015 [DOI] [PubMed] [Google Scholar]
- 334. Hashida M., Mahato R. I., Kawabata K., Miyao T., Nishikawa M., and Takakura Y., “ Pharmacokinetics and targeted delivery of proteins and genes,” J. Controlled Release 41(1), 91–97 (1996). 10.1016/0168-3659(96)01360-0 [DOI] [Google Scholar]
- 335. Messaoudi S., Greschner A. A., and Gauthier M. A., “ Progress toward absorption, distribution, metabolism, elimination, and toxicity of DNA nanostructures,” Adv. Ther. 2(12), 1900144 (2019). 10.1002/adtp.201900144 [DOI] [Google Scholar]
- 336. Soo Choi H., Liu W., Misra P., Tanaka E., Zimmer J. P., Itty Ipe B., Bawendi M. G., and Frangioni J. V., “ Renal clearance of quantum dots,” Nat. Biotechnol. 25(10), 1165–1170 (2007). 10.1038/nbt1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Ruggiero A., Villa C. H., Bander E., Rey D. A., Bergkvist M., Batt C. A., Manova-Todorova K., Deen W. M., Scheinberg D. A., and McDevitt M. R., “ Paradoxical glomerular filtration of carbon nanotubes,” Proc. Natl. Acad. Sci. 107(27), 12369–12374 (2010). 10.1073/pnas.0913667107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Nie S., “ Understanding and overcoming major barriers in cancer nanomedicine,” Nanomedicine (London) 5(4), 523–528 (2010). 10.2217/nnm.10.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Diebold S. and Brencicova E., “ Nucleic acids and endosomal pattern recognition: How to tell friend from foe?,” Front. Cell. Infect. Microbiol. 3, 37 (2013). 10.3389/fcimb.2013.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Alexis F., Pridgen E., Molnar L. K., and Farokhzad O. C., “ Factors affecting the clearance and biodistribution of polymeric nanoparticles,” Mol. Pharmaceutics 5(4), 505–515 (2008). 10.1021/mp800051m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Gunawan C., Lim M., Marquis C. P., and Amal R., “ Nanoparticle–protein corona complexes govern the biological fates and functions of nanoparticles,” J. Mater. Chem. B 2(15), 2060–2083 (2014). 10.1039/c3tb21526a [DOI] [PubMed] [Google Scholar]
- 342. Fadeel B., “ Clear and present danger? Engineered nanoparticles and the immune system,” Swiss Med. Weekly 142(2526), w13609 (2012). 10.4414/smw.2012.13609 [DOI] [PubMed] [Google Scholar]
- 343. Li X., Xu F., Yang D., and Wang P., “ A DNA-binding, albumin-targeting fusion protein promotes the cellular uptake and bioavailability of framework DNA nanostructures,” Nanoscale 13(12), 6038–6042 (2021). 10.1039/D0NR07967G [DOI] [PubMed] [Google Scholar]
- 344. Sheng Y., Yuan Y., Liu C., Tao X., Shan X., and Xu F., “ In vitro macrophage uptake and in vivo biodistribution of PLA–PEG nanoparticles loaded with hemoglobin as blood substitutes: effect of PEG content,” J. Mater. Sci.: Mater. Med. 20(9), 1881–1891 (2009). 10.1007/s10856-009-3746-9 [DOI] [PubMed] [Google Scholar]
- 345. Kang J. H., Kim K.-R., Lee H., Ahn D.-R., and Ko Y. T., “ In vitro and in vivo behavior of DNA tetrahedrons as tumor-targeting nanocarriers for doxorubicin delivery,” Colloids Surf., B 157, 424–431 (2017). 10.1016/j.colsurfb.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 346. Song L., Wang Z., Liu J., Wang T., Jiang Q., and Ding B., “ Tumor-targeted DNA bipyramid for in vivo dual-modality imaging,” ACS Appl. Bio Mater. 3(5), 2854–2860 (2020). 10.1021/acsabm.9b01096 [DOI] [PubMed] [Google Scholar]
- 347. Kim K.-R., Lee Y.-D., Lee T., Kim B.-S., Kim S., and Ahn D.-R., “ Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron,” Biomaterials 34(21), 5226–5235 (2013). 10.1016/j.biomaterials.2013.03.074 [DOI] [PubMed] [Google Scholar]
- 348. Zhou M., Zhang T., Zhang B., Zhang X., Gao S., Zhang T., Li S., Cai X., and Lin Y., “ A DNA nanostructure-based neuroprotectant against neuronal apoptosis via inhibiting toll-like receptor 2 signaling pathway in acute ischemic stroke,” ACS Nano 16(1), 1456–1470 (2022). 10.1021/acsnano.1c09626 [DOI] [PubMed] [Google Scholar]
- 349. Ke W., Shao K., Huang R., Han L., Liu Y., Li J., Kuang Y., Ye L., Lou J., and Jiang C., “ Gene delivery targeted to the brain using an angiopep-conjugated polyethyleneglycol-modified polyamidoamine dendrimer,” Biomaterials 30(36), 6976–6985 (2009). 10.1016/j.biomaterials.2009.08.049 [DOI] [PubMed] [Google Scholar]
- 350. Nagata T., Dwyer C. A., Yoshida-Tanaka K., Ihara K., Ohyagi M., Kaburagi H., Miyata H., Ebihara S., Yoshioka K., Ishii T., Miyata K., Miyata K., Powers B., Igari T., Yamamoto S., Arimura N., Hirabayashi H., Uchihara T., Hara R. I., Wada T., Bennett C. F., Seth P. P., Rigo F., and Yokota T., “ Cholesterol-functionalized DNA/RNA heteroduplexes cross the blood–brain barrier and knock down genes in the rodent CNS,” Nat. Biotechnol. 39(12), 1529–1536 (2021). 10.1038/s41587-021-00972-x [DOI] [PubMed] [Google Scholar]
- 351. Tam D. Y., Ho J. W.-T., Chan M. S., Lau C. H., Chang T. J. H., Leung H. M., Liu L. S., Wang F., Chan L. L. H., Tin C., and Lo P. K., “ Penetrating the blood–brain barrier by self-assembled 3D DNA nanocages as drug delivery vehicles for brain cancer therapy,” ACS Appl. Mater. Interfaces 12(26), 28928–28940 (2020). 10.1021/acsami.0c02957 [DOI] [PubMed] [Google Scholar]
- 352. Tian T., Li J., Xie C., Sun Y., Lei H., Liu X., Xia J., Shi J., Wang L., Lu W., and Fan C., “ Targeted imaging of brain tumors with a framework nucleic acid probe,” ACS Appl. Mater. Interfaces 10(4), 3414–3420 (2018). 10.1021/acsami.7b17927 [DOI] [PubMed] [Google Scholar]
- 353. Kansara K., Kumar A., and Bhatia D., “ In vivo sojourn of DNA nanodevices: Taking stock of the past and perspective for future challenges and applications,” Appl. Nanomed. 22(2), 337 (2022); available at https://pubs.thesciencein.org/journal/index.php/nanomed/article/view/337 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.