Abstract
Advances in genetic code reprogramming have allowed the site-specific incorporation of noncanonical functionalities into polypeptides and proteins, providing access to wide swaths of chemical space via in vitro translation techniques like mRNA display. Prior efforts have established that the translation machinery can tolerate amino acids with modifications to both the peptide backbone and side chains, greatly broadening the chemical space that can be interrogated in ligand discovery efforts. However, existing methods for confirming the translation yield of new amino acid building blocks for these technologies necessitate multistep workups and, more importantly, are not relevant for measuring translation within the context of a combinatorial library consisting of multiple noncanonical amino acids. In this study, we developed a luminescence-based assay to rapidly assess the relative translation yield of any noncanonical amino acid in real time. Among the 59 amino acids tested here, we found that many translate with high efficiency, but translational yield is not necessarily correlated to whether the amino acid is proteinogenic or has high tRNA acylation efficiency. Interestingly, we found that single-template translation data can inform the library-scale translation yield and that shorter peptide libraries are more tolerant of lower-efficiency amino acid monomers. Together our data show that the luminescence-based assay described herein is an essential tool in evaluating new building blocks and codon table designs within mRNA display toward the goal of developing druglike peptide-based libraries for drug discovery campaigns.
Introduction
While natural ribosomal translation is largely limited to the 20 amino acids of the standard genetic code, in vitro translation (IVT) systems such as the flexible in vitro translation (FIT) system1 can polymerize noncanonical amino acids (NCAAs) with exotic and desirable physicochemical properties.1−3 The FIT system consists of cell-free, reconstituted E. coli translation components and utilizes artificial tRNA-aminoacylating ribozymes known as flexizymes4,5 to generate custom aminoacyl-tRNAs, thus allowing the custom incorporation of NCAAs into polypeptides by the ribosome. Many efforts have established that NCAAs such as nonproteinogenic l-α-amino acids, d-amino acids, N-alkylated-amino acids, β- and γ-amino acids, and backbone-modified amino acids6−22 can be site-specifically incorporated into ribosomally translated peptides. Such noncanonical monomers present chemical moieties that are foreign to natural proteins and can impart resistance to proteolysis, stabilize unique secondary structure, and/or improve other druglike23 properties over traditional disulfide-linked peptides.1,9
Unfortunately, the translation yield of polypeptides that contain noncanonical functionalities is often significantly lower than natural peptides,24 limiting our ability to efficiently generate combinatorial libraries with a large number of NCAAs for ligand discovery through methods such as mRNA display. To date, characterizing the translational efficiency of a new NCAA relies on low-throughput methods that measure the translation of a peptide with a predefined sequence using either mass spectrometry or autoradiography methods. Moreover, matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry requires postreaction sample preparation (e.g., desalting and matrix cocrystallization), is relatively insensitive to hydrophobic peptides and macrocycles, and does not provide a quantitative measure of translation efficiency. Autoradiography requires translation reactions to be supplemented with radioisotopes and necessitates a multistep workup to analyze the reaction products via gel electrophoresis. We thus sought to develop assays to directly measure the real-time and combinatorial efficiencies of amino acid translation in situ.
Results and Discussion
Monitoring Translation via Luminescence
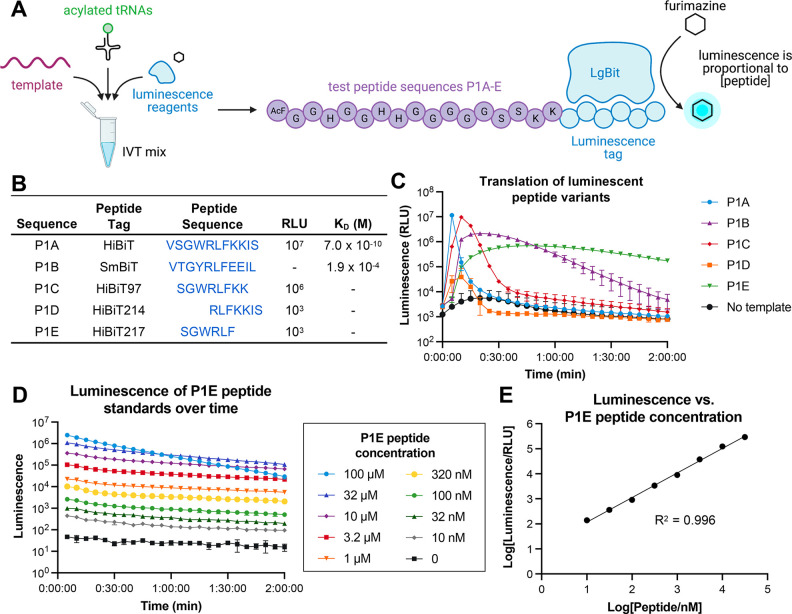
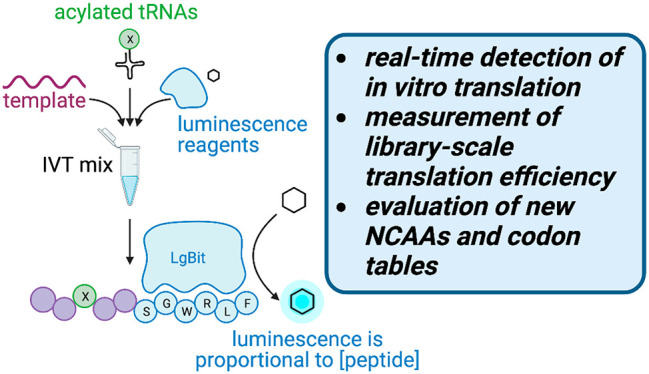
To create a high-throughput, plate-based assay for translation efficiency, we combined our IVT system with the commercially available NanoBiT system,25 a two-component assay consisting of the 18 kDa enzyme subunit LgBiT and its high affinity (KD = 700 pM) 11-residue HiBiT peptide. Complementation between these two partners reconstitutes a functional NanoBiT luciferase enzyme, and when these are incubated in the presence of its substrate, furimazine, a luminescence signal is generated that is proportional to the concentration of HiBiT present over a wide linear dynamic range. We hypothesized that if we used this IVT system to translate HiBiT-containing peptides, luciferase would be reconstituted in situ, and any resulting luminescence signal would act as a proxy for peptide translation in real time (Figure 1A).
Figure 1.
Design of luminescence-based assay for detection of IVT efficiency of noncanonical amino acids. (A) Overall schematic for IVT of luminescent peptides. AcF = N-acetyl-l-phenylalanine. Open blue circles indicate the 5 different luminescent peptide sequences detailed in Figure 1B. (B) Amino acid sequences of luminescent peptide variants tested in the P1A-E scaffolds. RLU and KD values were previously reported in Dixon et al. for complementation with LgBiT; dashed values were not reported.25 (C) Luminescence of IVT reactions for each luminescent peptide variant, monitored over 2 h. (D) Luminescence of a dilution series of a P1E peptide standard, monitored over 2 h. (E) Luminescence of the P1E peptide standard at 1 h (from Figure 1D) has a linear correlation to peptide concentration.
In order to develop this assay, we sought a peptide tag that would have a detectable, stable luminescence signal in our IVT system with a minimal size so it could be encoded in heavily engineered codon tables. We designed five linear peptide scaffolds with different HiBiT-related sequences based off of previously reported work25 that showed varying affinity to LgBiT to determine the correct detection range for our IVT reactions (Figure 1B). Our test IVT scaffolds, P1A-E, included an N-terminal 14 amino acid His/Gly/Ser region that allows for peptide flexibility and ease of translation as well as two Lys residues that aid in peptide solubility (Figure 1A). We appended the HiBiT sequences on the C-terminus such that the luminescent signal would only be detected if the preceding peptide sequence was fully translated. In our IVT mix, we included only the natural amino acids and the required synthetases necessary for translation of these peptide sequences. In all reactions, cyanomethyl ester (CME)-activated N-terminal acetyl-l-phenylalanine (AcF) and l-tryptophan were charged onto tRNAiniCAU and tRNAAsnGCA, respectively, using the eFx flexizyme,4,26 which allows for initiation of translation and Trp-dependent expression of the luminescent peptides. Finally, we supplemented each of the IVT reactions with LgBiT and its substrate, furimazine, and measured the translation of each scaffold independently by luminescence over 2 h at 37 °C (Figure 1C).
Interestingly, we found that translating the full HiBiT sequence (VSGWRLFKKIS) in template P1A led to a significant increase in luminescence followed by a complete loss of signal within 10 min (Figure 1C). This observation is consistent with rapid consumption of the furimazine substrate due to the high concentration of translated peptide (estimated to be ∼10 μM) and its high affinity for LgBiT (Figure 1B). Truncated variants such as P1B or P1C also rapidly dropped in the luminescence signal after an initial spike, while P1D showed a steep drop in the luminescence signal after only 15 min. However, the P1E peptides tagged with the shortest HiBiT217 sequence motif (SGWRLF), which is reported to have ∼104-fold lower luminescence compared to HiBiT,25 could be detected for up to 2 h with minimal loss of signal (Figure 1C).
To confirm that the luminescence signal observed in this assay correlates to peptide concentration, we measured the luminescence of a synthetic peptide standard of P1E at concentrations ranging from 10 nM to 100 μM. We observed a dose-dependent signal on a time scale relevant to the IVT of peptidic combinatorial libraries (Figure 1D-E) for concentrations up to 32 μM. Additionally, luminescence production is dependent on the inclusion of Trp-acylated tRNAAsnGCA but is also attenuated in the absence of AcF-acylated tRNAiniCAU or Phe-acylated tRNAAsnGGC, confirming that the luminescence signal is specifically produced by the NanoBiT complementation reaction and not due to an exogenous source [Figure S1]. Surprisingly, we observed that the “DMSO” negative control for this experiment, in which tRNAiniCAU is present in the IVT reaction but not acylated, had a significantly higher background luminescence signal as compared to the similar controls with the elongation amino acids used in our previous experiments (Figure S1). To investigate the source of this higher background luminescence, we performed MALDI-TOF mass spectrometry and observed many peaks corresponding to the masses of truncated products where the ribosome had initiated translation downstream of the initiator position (Figure S2A). We hypothesize that in the absence of an acylated initiator tRNA, stochastic initiation of translation occurs downstream of the start codon, as was recently reported by Tajima et al.,27 leading to a mixture of peptides with C-terminal HiBiT217 tag. However, when an initiator amino acid is acylated onto tRNAiniCAU, the IVT system is strongly biased toward the correct start position as shown by the correct MALDI-TOF spectrum of the fully translated peptide [Figure S2B]. Other reports have similarly reported translation initiation at downstream codons when using “difficult” initiators in IVT systems.14
Evaluating Relative Yields of Single Amino Acid Incorporations
We next designed a HiBiT217-containing peptide scaffold that could be used to measure the relative translation efficiencies of different NCAAs. We made single codon position variants of the P1E template, replacing the second through ninth codons with the GGC codon to test if the position of NCAA incorporation affected translation efficiency and signal window [Figure S3A]. l-Phenylalanine (Phe), (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic), or no amino acid were acylated onto tRNAAsnGGC as benchmarks for efficient, moderate, or no ribosomal translation, respectively.
IVT reactions were carried out with each of the acylated tRNAs and P2–P9 templates, and luminescence was measured at the 60 min time point to allow for relative translation yield measurements [Figure S3B]. As expected, IVT reactions that did not have an amino acid acylated on the tRNAAsnGGC showed no luminescence signal (Figure S3B, red bars) with the exception of P1E, which lacks the GGC codon. Interestingly, there was an observed variability of the total luminescent signal for each positional scaffold with some templates exhibiting little sensitivity as to which amino acid (Phe or Tic) was acylated onto tRNAAsnGGC (e.g., P2 and P8) and other scaffolds displaying a relative lower luminescence signal (P5, P7). We hypothesize that these differences may be due to a dependence of elongation efficiency on the identity of the immediately preceding amino acids.28−30 However, from this data, we chose to use P3 in subsequent experiments as the Phe-containing translation product P3[Phe] had a relatively high luminescence signal and the Tic-containing peptide P3[Tic] yielded an intermediate signal above the no acylation-containing conditions. We hypothesized that this sensitivity to different amino acids would allow us to rank translation efficiencies of a wide variety of NCAAs. MALDI-TOF mass spectrometry also validated that full-length P3[Phe] and P3[Tic] peptides are the major products in the corresponding luminescent IVT reactions [Figure S4].
We next wanted to benchmark our assay against previously reported autoradiography results,17 which is commonly used to quantitate translational efficiency in our IVT system. We acylated β-2-(S)-homophenylalanine (β2Phe), β-2-(S)-homoleucine (β2Leu), β-2-(S)-homovaline (β2Val), or no amino acid (“DMSO”) onto tRNAAsnGGC, translated each in the P3 sequence context, and measured luminescence for 60 min (Figure 2). As expected, the normalized luminescence signals of our translation assay correlated to the relative yields of peptides in the previously reported autoradiography experiments.17 β2Phe had the highest translation efficiency (64% in luminescence assay compared to l-Phe, 125% in autoradiography normalized to β2Ala), β2Leu had intermediate translation (10% vs 65%), and β2Val exhibited a negligible yield (1% vs 13%). Importantly, our luminescence-based assay can be interpreted in under an hour, as opposed to several days of post-translation reaction workup for autoradiography analysis.
Figure 2.
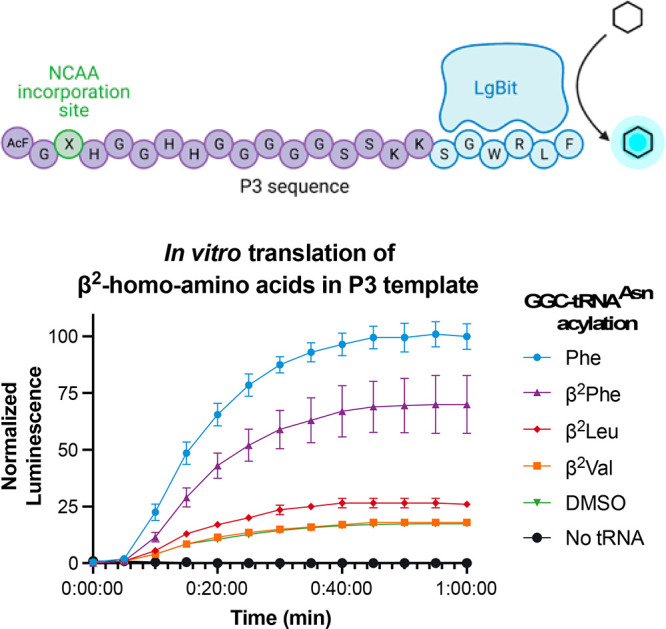
In vitro translation of P3 peptides containing single β2-homo-amino acids was monitored by luminescence. Relative luminescence values reported are normalized to Phe translation at 1 h. The “no tRNA” condition omits AcF-tRNAiniCAU and Trp-tRNAAsnGCA as well.
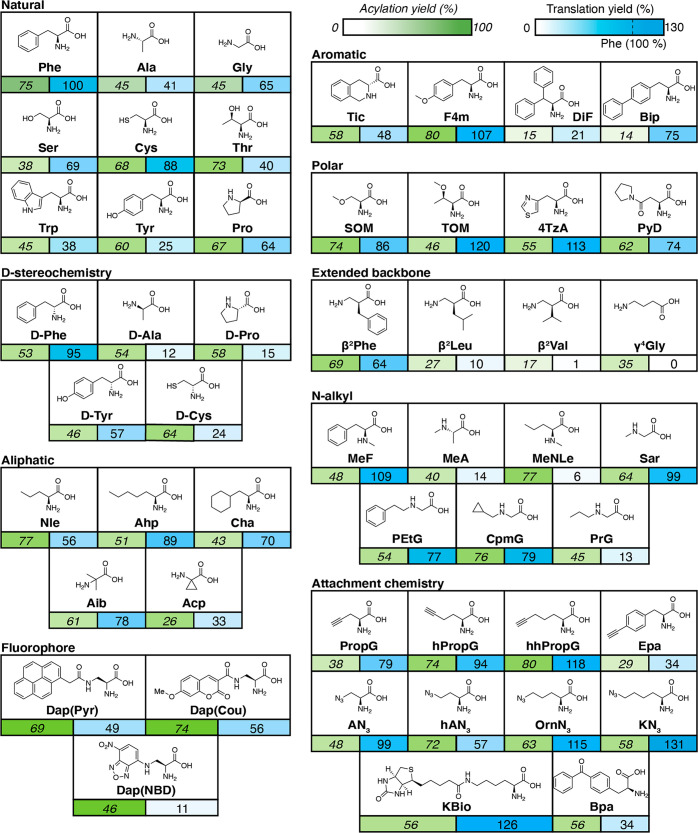
Now that our assay benchmarked against existing methods, we proceeded to test a panel of NCAAs with diverse properties for their translational efficiency in the P3 scaffold. To start, each NCAA was acylated onto the tRNAAsnGGC using the appropriate flexizyme with conditions that were determined using a short microhelix RNA, as previously reported4,17 (Table S1). Acylation percentages were determined using gel electrophoresis and quantifying the gel band shift that results when an amino acid is successfully acylated to the microhelix, with a theoretical maximum of 100% representing all microhelix RNA has been acylated with an amino acid (Figure 3, green). To quantitate IVT efficiency of our NCAAs, we took luminescence values at a 1 h translation, normalized to the luminescence signal of the P3 scaffold with Phe in the same position, and then subtracted the normalized luminescence values of a nonacylated tRNAAsnGGC control reaction (Figure 3, blue). Translation efficiency values that are reported higher or lower than 100 represent NCAAs whose translation efficiency is better or worse than phenylalanine, respectively. We deem translation efficiency values of <30%, 30–60%, and >60% as low, moderate, and high translation efficiency, respectively.
Figure 3.
Acylation and translation efficiencies for all elongation amino acids tested in the P3 sequence context. Reported acylation yields (percentage, green) were calculated based on the relative band intensities of aminoacyl-NCAA-microhelix RNA [MH+aa] and microhelix RNA [MH], presented as [MH+aa]/([MH+aa] + [MH]). Translation yields (percentage, blue) are normalized to translation of Phe in the P3 peptide context with a nonacylated control subtracted. Full names and acylation conditions for all amino acids are listed in Table S1.
Out of the 51 amino acids tested, 12 (24%) had low translation efficiency, 12 (24%) had moderate translation efficiency, and 27 (53%) had high translation efficiency. Surprisingly, some natural amino acids such as Trp or Tyr have a moderate translation efficiency in the P3 context (38% and 25%, respectively). Extended backbone side chains such as β2Val or γ4Gly had negligible peptide yield, which is consistent with prior work.17,18 Some N-methyl or d-amino acids have greatly attenuated translation efficiencies (e.g., d-alanine and N-methyl-l-alanine) compared to their canonical amino acid counterparts, though there are exceptions such as d-tyrosine or sarcosine. As d- and N-methyl amino acids are often extensively utilized in mRNA display efforts,6,7 systematic evaluation of these classes of amino acids may inform future library design.
In addition to directly incorporating NCAAs with moieties that explore druglike property space, many efforts have focused on diversifying peptide libraries through post-translational modification. For example, “click” chemistry has been used to generate glycopeptides from alkyne-containing translated peptides31 as well as redefine peptide topology through side-chain cyclization.21 Among these alkyne- or azide-presenting monomers, hhPropG or KN3 had the highest translation efficiency yields. These alkyne and azide amino acids, along with biotinylated or a benzophenoyl amino acids, could additionally be used for a wide variety of peptide labeling, chemical biology, or pulldown applications. We also report the relative translation efficiencies of three recently reported fluorescent amino acids,19 showing that the yields of translations with the pyrene- or coumarin-derivatized diaminopropanoic acid monomers are markedly greater than with the nitrobenzodioxazole monomer. Thus, our luminescence assay can quickly inform which among functionally equivalent NCAAs may be preferable for post-translational modification applications.
Overall, we did not find strict correlation between the translation yield and factors such as acylation yield (Figure S5) or steric bulk; for example, while the β-substituted diphenylalanine (DiF) has low acylation (15%) and translation yields (21%), the para-phenyl substituted biphenylalanine (Bip) has a high translation yield (75%) despite its similarly modest acylation yield (14%). In our IVT reactions, even inefficiently acylated amino acids are present at sufficient concentrations for detectable peptide yield (for example, 15% acylation yield of 25 μM tRNA would yield 3.75 μM aminoacylated tRNA). Thus, differences in translation efficiency of such closely related structures can be most likely attributed to a poor interaction with the translation machinery such as elongation factors (e.g., EF-Tu/EF-Ts) or the ribosome;30,32 thus, having an empirical, high-throughput method for systematically evaluating incorporation of these NCAAs is highly critical in the design of IVT schemes involving genetic code recoding.
Evaluating Translation Efficiency at the Initiator Codon
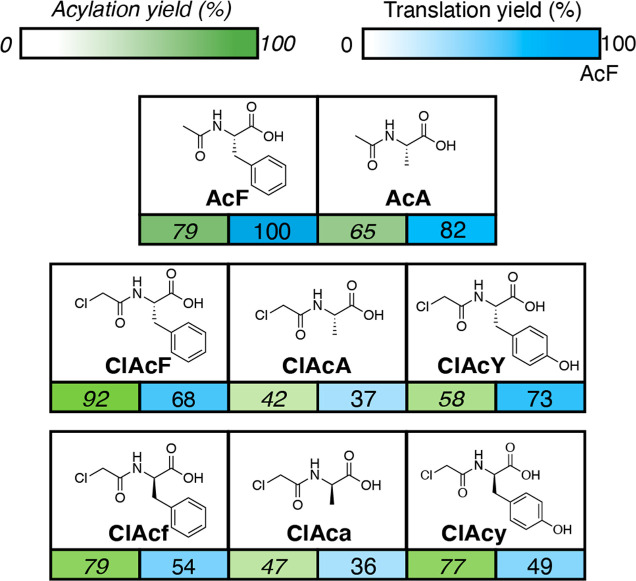
Amino acids used to initiate translation differ from their elongation counterparts in that they must bear an N-terminal acylation, which mimics the native N-formylation of methionine found in vivo.33 Additionally, a unique tRNA, tRNAiniCAU, which has a completely different sequence and structure from the elongation tRNA used previously is required to be recognized by the translation initiation factors that initiate translation.33,34 We thus tested the translation efficiency of several initiator amino acids in the P3 sequence (Figure 4) by acylating a variable initiator amino acid onto tRNAiniCAU, l-Phe onto tRNAAsnGGC, and l-Trp onto tRNAAsnGCA. For this experiment, we chose three natural amino acids (Phe, Ala, Tyr) with varying translation efficiencies in the elongation context (100%, 41%, 25%, respectively) and were curious if similar trends held at the initiator codon. Additionally, we varied their stereochemistry and N-terminal modification in order to see how these modifications affected their translation efficiency; N-acetyl amino acids generate a linear peptide, while N-chloroacetyl amino acids are utilized to generate thioether-linked macrocycles if a cysteine residue is incorporated in the peptide sequence.35 Translation efficiencies of N-chloroacetyl initiators ClAcF (68%) and ClAcA (37%) were lower than their N-acetyl counterparts AcF (100%) and AcA (82%), indicating that cyclized peptides may be synthesized in lower yield than their linear counterparts. In general, alanine-derived initiators displayed a lower translation efficiency as compared to the phenylalanine or tyrosine analogs measured. This suggests that the Phe/Tyr-derivatives may be preferable for usage in combinatorial libraries due to the fact that, as an initiator, these amino acids are incorporated into every single peptide in a library setting.
Figure 4.
Acylation yields and translational efficiency of initiator amino acids. Reported acylation yields (percentage, green) are relative to microhelix RNA, and translation yields (percentage, blue) are relative to the translation of AcF as the initiator in the P3 peptide context. See Table S2 for full names and acylation conditions for all amino acids.
Translation Efficiency of Combinatorial Macrocyclic Peptide Libraries
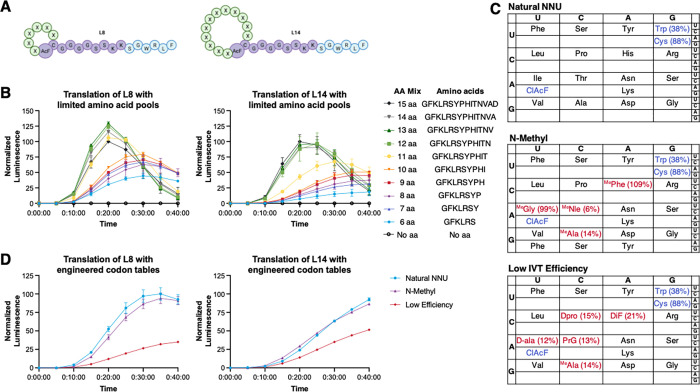
Testing a single defined template such as P3 is a simple assay for measuring the effect of a single amino acid incorporation event on translational yield. However, there is great utility in using a translational efficiency assay to inform the design of combinatorial libraries with multiple exotic NCAAs, as translation efficiency of any one amino acid can be affected by the preceding sequence which could lower the actual diversity of a library well below its theoretical size. To assess this, we designed two combinatorial libraries, L8 and L14, for the translation of thioether-linked 8- or 14-member macrocycles, respectively (Figure 5A). Each library templates either six or 12 variable codons encoded by degenerate NNU codons in the RNA template sequences. The variable amino acids are flanked by a fixed N-terminal N-(chloroacetyl)-l-phenylalanine (ClAcF) and an l-cysteine residue, which allow cyclization to occur spontaneously between the thiol group of the Cys residue and the initiator, forming a thioether linkage. In addition, each template encodes a constant G4S2K2–HiBiT217 C-terminal tail for luminescence detection (Figure 5A). ClAcF, l-Cys, and l-Trp were acylated onto tRNAiniCAU, tRNAAsnCCA, and tRNAAsnGCA, respectively, and only amino acids and aminoacyl-tRNA synthetases necessary for translation of the NNU codon table were included in the IVT reaction. Importantly, with the initiator and the C-terminal cysteine held constant for all library members, L8 has a theoretical library diversity of 1.7 × 107, and L14 has a diversity of 2.8 × 1014. mRNA display typically generates library sizes of 1013–1014 which would give approximately an n = 108 or n = 1 copy number for any one sequence in the L8 and L14 libraries, respectively, allowing us to understand how the translation yield of NCAA building blocks can affect the overall translation yield in libraries of varying diversities.
Figure 5.
Monitoring the translational efficiency of model macrocycle libraries via luminescence. (A) Macrocyclic scaffolds L8 and L14 used for translation and luminescence detection. (B) Translation of L8 and L14 templates using progressively limited amino acid pools, from the 15 monomers encoded by the NNU degenerate codons to only the six natural amino acids required for HiBiT217 scaffold translation. ClAcF, l-Cys, and l-Trp were acylated onto tRNAiniCAU, tRNAAsnCCA, and tRNAAsnGCA for all translations. Translation yields are normalized to translation of the 15 aa libraries at maximum signal. (C) Codon tables used to test translation of L8 and L14 libraries. Variable amino acids are highlighted with translation efficiencies in the P3 sequence context (in red). l-Trp, l-Cys, and ClAcF (blue) were acylated onto tRNAiniCAU, tRNAAsnCCA, and tRNAAsnGCA, respectively, in all tables. (D) Bulk luminescence for translation of L8 and L14 libraries using the three codon tables shown in Figure 5C. Translation yields are normalized to translation of the Natural NNU libraries at maximum signal.
To understand the effects of how poorly translating amino acids affect the overall translation of a combinatorial library, we progressively omitted single amino acids from the IVT mix and monitored luminescence over 40 min. Surprisingly, the bulk luminescence signal for either library is only attenuated when five or four amino acids are removed from the L8 or L14 scaffolds, respectively (Figure 5B). This indicates that several poorly translating amino acids can be included in a codon table without affecting the overall translational yield to a degree; however, it comes with a potential loss of library diversity. For example, in the L8 scaffold with six variable positions, up to four amino acids could be entirely omitted without affecting the overall translation yield. Because the variable region of this combinatorial library is shorter (six vs 12 NNU codons), the higher luminescence signal is consistent with a greater number of competent RNA template molecules remaining in the pool even when subsets of amino acid monomers are unavailable for translation (Table S3). We hypothesize that the relatively high signal observed in the absence of one or more amino acids indicated some level of retranslation of the “competent” mRNA templates. Despite the lack of release factor RF1 in our IVT mix, there may be some previously unaccounted for mechanisms for peptide release and turnover of ribosomes on the limited pool of mRNA templates.
Finally, we translated the same two combinatorial libraries with three different codon tables as models for efficient (“Natural NNU”), moderate (“N-methyl”), or inefficient (“Low IVT Efficiency”) ribosomal translation based on varying the composition of amino acids included in each table (Figure 5C). The bulk luminescence signals observed (Figure 5D) correlated well to our previously tested model libraries (Figure 5B) with the IVT yield decreasing as the number of low-efficiency amino acids increased in the codon table. Whereas the N-methyl libraries, with only two additional low-efficiency amino acids MePhe and MeNle, had minimally attenuated translation efficiencies, the “Low Efficiency” codon table, with five poorly translated amino acids, had a greatly reduced translation yield in both L8 and L14 scaffolds. After the IVT reactions were complete, we measured the final concentrations of these libraries against a HiBit217 standard to be in the range of ∼7–17 μM (Table S4), as the luminescence tag is a simple way to quantitate the yield of IVT reactions. Notably, these peptide concentrations are higher than the input mRNA concentration (2 μM), which would be consistent with multiple translation events and stochastic peptide release from the mRNA templates despite the previously mentioned lack of RF1 in our IVT reactions. Overall, these results indicate that our luminescence assay can measure the IVT efficiency of engineered codon tables in the library translation context and help inform the design of more druglike libraries with exotic NCAAs.
Conclusions
Peptide macrocycles and other “beyond-rule-of-five” molecules can potentially target proteins that cannot be engaged by traditional small molecule modalities.36 However, there are significant hurdles to their deployment as in vivo probes or orally bioavailable drugs, due to their properties such as their size or lipophilicity. To maximize the probability of discovering new leads, peptidic libraries used in drug discovery efforts should maximally sample such favorable property space.
While the chemical space of unnatural amino acids is vast, the number of validated monomers for ribosomal incorporation is comparatively limited. In order to rapidly triage NCAAs with desirable properties for inclusion in combinatorial libraries, we developed a luminescence-based assay for quantitating translation by the ribosomal machinery. Our assay is consistent with prior efforts7,17 to compare the translational efficiencies of NCAAs, in a format that is much faster than MALDI-MS or autoradiography. Unlike the parental HiBiT peptide, the HiBit217 tag is compatible with endpoint quantitation or kinetic monitoring of the relatively high (∼micromolar) concentrations of peptides produced in IVT reactions. Because we are able to quantitate IVT peptides over greater than 4 orders of magnitude, this also could enable downstream assays such as KD measurements. Additionally, the small size of the HiBiT217 hexapeptide minimally perturbs polypeptides onto which it is appended thus making it compatible with recoding large portions of the natural codon table.
Among our exploratory set of 59 amino acids, many with noncanonical side chains translated as well or better than proteinogenic amino acids, and we did not find strong correlations between tRNA acylation and translation efficiency. This may be due to factors such as accommodation of the aminoacyl-tRNA by the ribosome,32 interactions with various translation factors, or varying rates of peptidyl transfer.30 To this point, recent efforts have shown that these hurdles can be overcome in part by engineered tRNAs that favorably interact with translation factors, EF-Tu or EF-P.32,37 Other sizable efforts have been focused on engineering ribosomes to better tolerate and elongate NCAAs.38,39 Additionally, because translation efficiency of any given monomer is dependent on its sequence context, translation efficiency should be tested in the specific sequence contexts researchers are interested in. We anticipate that our assay could empower further optimization of any in vitro or cellular model of translation.
In addition to comparing the effects of single amino acid differences on peptide translation, we also monitored the combinatorial translation efficiency of peptide libraries with various scaffolds or codon tables. We found that the L8 scaffolds with lower combinatorial diversity were more tolerant of monomers with lower translation efficiency than the larger L14 scaffolds. It is important to note that in the context of larger libraries like L14 with lower copy numbers (on the order of n = 1), any loss of overall library concentration necessarily represents a loss of unique library members. Thus, we would not recommend using higher numbers (>4) of difficult-to-translate amino acids when making combinatorial libraries of larger sizes such as L14. Conversely, libraries of a comparable or smaller size than L8 may be more tolerant of low efficiency codon tables with even higher numbers of low translation efficiency NCAAs. Many ligand discovery efforts limit the number of NCAAs in a single library to six or fewer13,20 (though some have reported larger codon tables16), but our data suggest that more NCAAs could generally be included if shorter macrocycle scaffolds are used or the NCAAs all have high translation efficiency. This may also allow for strategic usage of NCAAs that translate poorly but have physicochemical properties desirable in biologically active ligands. Altogether, these results highlight that when incorporating a high number of NCAAs, it is imperative to measure library-scale translation efficiency before embarking on in vitro affinity selection efforts.
It is an ongoing challenge to identify “beyond-rule-of-five” macrocyclic peptides that demonstrate the optimal balance of potency, cell permeability, protease stability, and other favorable druglike properties. In order to bias our discovery efforts toward finding more druglike starting points, mRNA display-generated peptide libraries will require extensive reprogramming to include monomers that contribute favorably both to target binding and balance physicochemical properties upfront. Thus, we anticipate that our luminescence assay, which can not only assess translation of novel monomers on an individual level but also measure their effects on combinatorial library expression, will be an invaluable tool in future drug discovery efforts.
Methods
All oligonucleotides (DNA, tRNAs, flexizymes, and microhelix) were synthesized and purified by Integrated DNA Technologies. LgBiT protein and the furimazine substrate were purchased from Promega.
Synthesis of Active Esters and Initiators of Amino Acids
Aliphatic amino acids were activated with 3,5-dinitrobenzyl ester (DNB), and aromatic amino acids were activated with cyanomethyl ester (CME), with the exceptions of N-(2-phenylethyl)glycine (PEtG) and (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic), which were activated with (2-aminoethyl)amidocarboxybenzyl thioester (ABT). All active esters and initiators of amino acids were synthesized using the previously published procedures.4,17−19,35,40−42 The characterization data of new compounds are provided in the Supporting Information.
Peptide Synthesis
Peptides were synthesized using standard solid-phase Fmoc chemistry.43 The obtained crude peptides were purified by reversed-phase (RP) high-performance liquid chromatography (HPLC) (C18 column, 21 mm × 250 mm, 5 μm, 100 Å, 10 mL/min, 60 min gradient from 30 to 60% aqueous acetonitrile containing 0.1% TFA) and characterized by analytical HPLC (C18 column, 4.6 mm × 150 mm, 5 μm, 100 Å, 1.0 mL/min, 20 min gradient from 25 to 65% aqueous acetonitrile containing 0.1% TFA, 214 nm) and electrospray ionization mass spectrometry (ESI-MS).
Preparation of dsDNA Templates
For PCR of individual templates, 800 μL of amplification was performed using 1X Hot Start Taq 2X Master Mix (New England Biolabs), 0.5 μM each forward and reverse primer, and 2.5 nM template (see Table S5). The mixture was incubated at 95 °C for 5 min and then subjected to 12 cycles of 95 °C for 40 s, 50 °C for 40 s, and 68 °C for 40 s.
For preparation of library pools (L8, L14), a 20 μL extension reaction was first performed using 1 μM each template primer and reverse primer, 1X KOD Plus buffer (Toyobo), 0.02 U/μL KOD plus DNA polymerase, 0.2 mM dNTPs, and 1.5 mM MgSO4. The mixture was incubated at 95 °C for 2 min and then subjected to six cycles of 95 °C for 40 s, 60 °C for 40 s, and 68 °C for 60 s. The entire extension mix was then amplified in an 800 μL reaction containing 1X Hot Start Taq 2X Master Mix (New England Biolabs), 250 nM FW primer, and 250 nM reverse primer.
All PCR products were confirmed by gel electrophoresis (E-Gel, ThermoFisher Scientific), purified using the MinElute PCR Purification Kit (Qiagen), and quantified by UV absorbance (NanoDrop).
Preparation of RNA Templates for Library IVT Reactions
One mL transcription reactions were prepared with 200 μL of DNA template (250 ng/μL), 40 mM Tris-HCl pH 8, 1 mM spermidine, 0.01% Triton X-100, 10 mM DTT, 20 mM MgCl2, 25 nM KOH, 3.75 μM NTPs, and 240 nM T7 RNA polymerase. The reaction was incubated at 37 °C overnight. DNase I enzyme (0.025 U/μL) and buffer (40 mM Tris-HCL pH 8.0, 10 mM MgSO4, and 1 mM CaCl2) were added, and the reaction was incubated at 37 °C for an hour. RNA was purified using RNeasy spin columns (Qiagen) according to the manufacturer’s instructions and quantitated using UV absorbance.
Reconstituted in Vitro Translation System
The in vitro translation system of recombinant E. coli was reconstituted in 50 mM HEPES-KOH (pH 7.6), 10 mM Mg(OAc)2, 100 mM KOAc, 1 mM DTT, 2 mM spermidine, 20 mM creatine phosphate, 2 mM ATP, 2 mM GTP, 1 mM CTP, 1 mM UTP, and 1.5 mg mL–1E. coli total tRNA with final protein concentrations of 0.03 μM ArgRS, 0.09 μM GlyRS, 0.02 μM HisRS, 0.4 μM IleRS, 0.02 μM LeuRS, 0.11 μM LysRS, 0.68 μM PheRS, 0.04 μM SerRS, 0.02 μM ValRS, 0.6 μM MTF, 2.7 μM IF1, 3 μM IF2, 1.5 μM IF3, 0.1 μM EF-G, 20 μM EF-Tu/Ts, 0.25 μM RF2, 0.17 μM RF3, 0.5 μM RRF, 1 μM T7 RNA polymerase, 4 μg/mL creatine kinase, and 1.2 μM ribosome. All necessary amino acids were included at 0.2 mM. Reagents and protocols for this in vitro translation system were performed as described in the literature.17,26,37,44,45
Aminoacylation of Microhelix RNA with Amino Acids
Aminoacylation reactions (10 μL) of microhelix RNA with amino acids were carried out with 20 μM microhelix and 20 μM dFx or eFx in 0.1 M HEPES, pH 7.2 or 0.1 M Bicine, pH 9.0 (see Table S5). Flexizymes were preincubated with microhelix RNA, heated at 95 °C for 3 min, and cooled to RT over 5 min. 600 mM (for eFx reactions) or 20 mM MgCl2 (for dFx) was added, and the mixture was stored at 25 °C for 1 min. The reaction was initiated by addition of 25 mM amino acid substrates in DMSO and incubated on ice for the listed times in Tables S1–S2. The aminoacylation reaction was stopped by adding 1 μL of 3 M sodium acetate buffer. The resulting product was mixed with 11 μL of formamide loading buffer and analyzed by 20% denaturing acid PAGE. RNA was stained by 0.01% SYBR Green II for 15 min, and the bands were visualized by a UV transilluminator. Reported acylation yields were calculated based on the relative band intensities of aminoacyl-NCAA-microhelix RNA [MH+aa] and microhelix RNA [MH], presented as [MH+aa]/([MH+aa] + [MH]).
Synthesis of Aminoacyl-Amino Acids-tRNA
Aminoacylation reactions (20 μL) were carried out as follows: 20 μM tRNA and 20 μM flexizyme were combined in 0.1 M buffer (see Tables S1–S2), heated at 95 °C for 3 min, and cooled to RT over 5 min. 600 mM MgCl2 (for eFx reactions) or 20 mM MgCl2 (for dFx reactions) was added, and the mixture was chilled on ice for 5 min. The reaction was initiated by addition of 25 mM substrate amino acid in DMSO and incubated on ice for the listed times. After the acylation reaction, 0.3 M NaOAc (pH 5.2) and 100% EtOH were added and centrifuged for 15 min at 10,000g to pellet the aminoacyl-aa-tRNA. The pellet was rinsed with a 70% EtOH aqueous solution containing 0.1 M NaOAc (pH 5.2) and then dried.
IVT Reactions for Ribosomal Synthesis of Peptides
In vitro translation reactions were carried out in 20 μL of the in vitro translation system described above, additionally containing either the 0.04 μM DNA template (P3) or the 2 μM RNA template (L8 or L14), 0.2 mM each of the amino acids necessary for translation, and 250 μM each aminoacyl-aa-tRNA and incubated at 37 °C for up to 2 h.
For luminescence monitoring, the reaction was supplemented with LgBiT protein and the furimazine substrate diluted 100-fold from stock (Promega). The translation reaction mixture was divided into two 9 μL replicate wells, incubated at 37 °C for up to 2 h, and luminescence monitored via a plate reader. To normalize between experiments and quantitate IVT efficiency, raw luminescence values at 1 h were normalized to translation of an S3[Phe] peptide, with a nonacylated tRNA control (“DMSO”) subtracted.
MALDI-TOF Mass Spectrometry of in Vitro Translated Peptides
After in vitro translation, the reaction volume (10–20 μL) was acidified to 0.1% trifluoroacetic acid and desalted on a ZipTipC18 (Millipore) into 70% acetonitrile/water saturated with α-cyano-4-hydroxycinnamic acid as the MALDI carrier. MALDI-TOF-MS analysis was performed using a RAPID-FLEX (Bruker) in reflector/positive mode.
For FLAG-tagged peptides, the reaction mixture (20 μL) was first diluted to 100 μL with PBS buffer and incubated for 1 h with anti-FLAG M2 magnetic beads (Sigma). The beads were washed three times with 100 μL of PBS buffer before the bound peptides were eluted in 20 μL of Pierce IgG Elution Buffer. Purified peptides were subjected to the same desalting and analysis protocol as above for MALDI.
Acknowledgments
The figures were created in part using BioRender. HiBiT variant sequences HiBit97, HiBit214, and HiBiT217 were used under license from Promega Corporation. We thank E. Adaligil, K. Hallenbeck, and L. Zhou for assistance with the acylation of NCAAs.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.2c00712.
Experimental protocols; synthesis of amino acid esters and peptides; Figures S1–S4, validation and optimization of luminescence-based detection of in vitro translation; Tables S1–S2, aminoacylation conditions for amino acids; Figure S5, comparison of translation efficiency vs acylation efficiency; Tables S3–S4, library-scale diversity and translation yields; and Table S5, sequences of primers (PDF)
Author Present Address
§ Department of Antibody and Protein Engineering, 23andMe, Inc., South San Francisco, California 94080, United States
The authors declare the following competing financial interest(s): A.I.C., A.S., J.T., D.J.B., and C.N.C. are employees of Genentech, Inc. and shareholders of Roche.
Supplementary Material
References
- Goto Y.; Suga H. The RaPID Platform for the Discovery of Pseudo-Natural Macrocyclic Peptides. Acc. Chem. Res. 2021, 54 (18), 3604–3617. 10.1021/acs.accounts.1c00391. [DOI] [PubMed] [Google Scholar]
- Chin J. W. Expanding and Reprogramming the Genetic Code. Nature 2017, 550 (7674), 53–60. 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- Obexer R.; Walport L. J.; Suga H. Exploring Sequence Space: Harnessing Chemical and Biological Diversity towards New Peptide Leads. Curr. Opin Chem. Biol. 2017, 38, 52–61. 10.1016/j.cbpa.2017.02.020. [DOI] [PubMed] [Google Scholar]
- Murakami H.; Ohta A.; Ashigai H.; Suga H. A Highly Flexible TRNA Acylation Method for Non-Natural Polypeptide Synthesis. Nat. Methods 2006, 3 (5), 357–359. 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- Niwa N.; Yamagishi Y.; Murakami H.; Suga H. A Flexizyme That Selectively Charges Amino Acids Activated by a Water-Friendly Leaving Group. Bioorg. Med. Chem. Lett. 2009, 19 (14), 3892–3894. 10.1016/j.bmcl.2009.03.114. [DOI] [PubMed] [Google Scholar]
- Kawakami T.; Murakami H.; Suga H. Messenger RNA-Programmed Incorporation of Multiple N-Methyl-Amino Acids into Linear and Cyclic Peptides. Chem. Biol. 2008, 15 (1), 32–42. 10.1016/j.chembiol.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Fujino T.; Goto Y.; Suga H.; Murakami H. Reevaluation of the D-Amino Acid Compatibility with the Elongation Event in Translation. J. Am. Chem. Soc. 2013, 135 (5), 1830–1837. 10.1021/ja309570x. [DOI] [PubMed] [Google Scholar]
- Torikai K.; Suga H. Ribosomal Synthesis of an Amphotericin-B Inspired Macrocycle. J. Am. Chem. Soc. 2014, 136, 17359. 10.1021/ja508648s. [DOI] [PubMed] [Google Scholar]
- Josephson K.; Ricardo A.; Szostak J. W. MRNA Display: From Basic Principles to Macrocycle Drug Discovery. Drug Discov Today 2014, 19 (4), 388–399. 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Rogers J. M.; Passioura T.; Suga H. Nonproteinogenic Deep Mutational Scanning of Linear and Cyclic Peptides. Proc. National Acad. Sci. 2018, 115 (43), 10959–10964. 10.1073/pnas.1809901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. M.; Kwon S.; Dawson S. J.; Mandal P. K.; Suga H.; Huc I. Ribosomal Synthesis and Folding of Peptide-Helical Aromatic Foldamer Hybrids. Nat. Chem. 2018, 10, 405. 10.1038/s41557-018-0007-x. [DOI] [PubMed] [Google Scholar]
- Tsiamantas C.; Rogers J. M.; Suga H. Initiating Ribosomal Peptide Synthesis with Exotic Building Blocks. Chem. Commun. 2020, 56 (31), 4265–4272. 10.1039/D0CC01291B. [DOI] [PubMed] [Google Scholar]
- Nitsche C.; Passioura T.; Varava P.; Mahawaththa M. C.; Leuthold M. M.; Klein C. D.; Suga H.; Otting G. De Novo Discovery of Nonstandard Macrocyclic Peptides as Noncompetitive Inhibitors of the Zika Virus NS2B-NS3 Protease. ACS Med. Chem. Lett. 2019, 10 (2), 168–174. 10.1021/acsmedchemlett.8b00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Schwieter K. E.; Watkins A. M.; Kim D. S.; Yu H.; Schwarz K. J.; Lim J.; Coronado J.; Byrom M.; Anslyn E. V.; Ellington A. D.; Moore J. S.; Jewett M. C. Expanding the Limits of the Second Genetic Code with Ribozymes. Nat. Commun. 2019, 10 (1), 5097. 10.1038/s41467-019-12916-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Schwarz K. J.; Kim D. S.; Moore J. S.; Jewett M. C. Ribosome-Mediated Polymerization of Long Chain Carbon and Cyclic Amino Acids into Peptides in Vitro. Nat. Commun. 2020, 11 (1), 4304. 10.1038/s41467-020-18001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T.; Liu W.; Dunkelmann D.; Higuchi T.; Suga H. Display Selection of Exotic Macrocyclic Peptides Expressed under a Radically Reprogrammed 23 Amino Acid Genetic Code. J. Am. Chem. Soc. 2018, 140 (37), 11551–11555. 10.1021/jacs.8b03367. [DOI] [PubMed] [Google Scholar]
- Adaligil E.; Song A.; Hallenbeck K. K.; Cunningham C. N.; Fairbrother W. J. Ribosomal Synthesis of Macrocyclic Peptides with Β2- and Β2,3-Homo-Amino Acids for the Development of Natural Product-Like Combinatorial Libraries. ACS Chem. Biol. 2021, 16 (6), 1011–1018. 10.1021/acschembio.1c00062. [DOI] [PubMed] [Google Scholar]
- Adaligil E.; Song A.; Cunningham C. N.; Fairbrother W. J. Ribosomal Synthesis of Macrocyclic Peptides with Linear Γ4- and Β-Hydroxy-γ4-amino Acids. ACS Chem. Biol. 2021, 16 (8), 1325–1331. 10.1021/acschembio.1c00292. [DOI] [PubMed] [Google Scholar]
- Lee J.; Schwarz K. J.; Yu H.; Krüger A.; Anslyn E. V.; Ellington A. D.; Moore J. S.; Jewett M. C. Ribosome-Mediated Incorporation of Fluorescent Amino Acids into Peptides in Vitro. Chem. Commun. 2021, 57 (21), 2661–2664. 10.1039/D0CC07740B. [DOI] [PubMed] [Google Scholar]
- Diao J.; Komura R.; Sano T.; Pantua H.; Storek K. M.; Inaba H.; Ogawa H.; Noland C. L.; Peng Y.; Gloor S. L.; Yan D.; Kang J.; Katakam A. K.; Volny M.; Liu P.; Nickerson N. N.; Sandoval W.; Austin C. D.; Murray J.; Rutherford S. T.; Reichelt M.; Xu Y.; Xu M.; Yanagida H.; Nishikawa J.; Reid P. C.; Cunningham C. N.; Kapadia S. B. Inhibition of Escherichia Coli Lipoprotein Diacylglyceryl Transferase Is Insensitive to Resistance Caused by Deletion of Braun’s Lipoprotein. J. Bacteriol. 2021, 203 (13), e00149–21. 10.1128/JB.00149-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y.; Morimoto J.; Murakami H.; Suga H. Ribosomal Synthesis of Bicyclic Peptides via Two Orthogonal Inter-Side-Chain Reactions. J. Am. Chem. Soc. 2008, 130 (23), 7232–7234. 10.1021/ja800953c. [DOI] [PubMed] [Google Scholar]
- Katoh T.; Suga H. Consecutive Ribosomal Incorporation of Α-Aminoxy/α-Hydrazino Acids with l/D-Configurations into Nascent Peptide Chains. J. Am. Chem. Soc. 2021, 143, 18844. 10.1021/jacs.1c09270. [DOI] [PubMed] [Google Scholar]
- Muttenthaler M.; King G. F.; Adams D. J.; Alewood P. F. Trends in Peptide Drug Discovery. Nat. Rev. Drug Discov 2021, 20 (4), 309–325. 10.1038/s41573-020-00135-8. [DOI] [PubMed] [Google Scholar]
- Katoh T.; Suga H. Engineering Translation Components Improve Incorporation of Exotic Amino Acids. Int. J. Mol. Sci. 2019, 20 (3), 522. 10.3390/ijms20030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A. S.; Schwinn M. K.; Hall M. P.; Zimmerman K.; Otto P.; Lubben T. H.; Butler B. L.; Binkowski B. F.; MacHleidt T.; Kirkland T. A.; Wood M. G.; Eggers C. T.; Encell L. P.; Wood K. V. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11 (2), 400–408. 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Goto Y.; Katoh T.; Suga H. Flexizymes for Genetic Code Reprogramming. Nat. Protoc 2011, 6 (6), 779–790. 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- Tajima K.; Katoh T.; Suga H. Drop-off-Reinitiation Triggered by EF-G-Driven Mistranslocation and Its Alleviation by EF-P. Nucleic Acids Res. 2022, 50, 2736. 10.1093/nar/gkac068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Duc K.; Song Y. S. The Impact of Ribosomal Interference, Codon Usage, and Exit Tunnel Interactions on Translation Elongation Rate Variation. Plos Genet 2018, 14 (1), e1007166 10.1371/journal.pgen.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov A. A.; Sumbatyan N. V.; Shishkina A. V.; Karpenko V. V.; Korshunova G. A. Ribosomal Tunnel and Translation Regulation. Biochem Mosc 2010, 75 (13), 1501–1516. 10.1134/S0006297910130018. [DOI] [PubMed] [Google Scholar]
- Verma M.; Choi J.; Cottrell K. A.; Lavagnino Z.; Thomas E. N.; Pavlovic-Djuranovic S.; Szczesny P.; Piston D. W.; Zaher H. S.; Puglisi J. D.; Djuranovic S. A Short Translational Ramp Determines the Efficiency of Protein Synthesis. Nat. Commun. 2019, 10 (1), 5774. 10.1038/s41467-019-13810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S.; Bailey J. K.; Temme J. S.; Schlippe Y. V. G.; Krauss I. J. Directed Evolution of Multivalent Glycopeptides Tightly Recognized by HIV Antibody 2G12. J. Am. Chem. Soc. 2014, 136 (14), 5407–5415. 10.1021/ja500678v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwane Y.; Kimura H.; Katoh T.; Suga H. Uniform Affinity-Tuning of N -Methyl-Aminoacyl-TRNAs to EF-Tu Enhances Their Multiple Incorporation. Nucleic Acids Res. 2021, 49, 10807. 10.1093/nar/gkab288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A.; Pavlov M. Y.; Lovmar M.; Ehrenberg M. How Initiation Factors Tune the Rate of Initiation of Protein Synthesis in Bacteria. EMBO J. 2006, 25 (11), 2539–2550. 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Thijssen V.; Jongkees S. A. K. Suppression of Formylation Provides an Alternative Approach to Vacant Codon Creation in Bacterial In Vitro Translation. Angew. Chem., Int. Ed. Engl. 2020, 59 (49), 21870–21874. 10.1002/anie.202003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y.; Ohta A.; Sako Y.; Yamagishi Y.; Murakami H.; Suga H. Reprogramming the Translation Initiation for the Synthesis of Physiologically Stable Cyclic Peptides. ACS Chem. Biol. 2008, 3 (2), 120–129. 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- Doak B. C.; Kihlberg J. Drug Discovery beyond the Rule of 5 - Opportunities and Challenges. Expert Opin Drug Dis 2017, 12 (2), 115–119. 10.1080/17460441.2017.1264385. [DOI] [PubMed] [Google Scholar]
- Katoh T.; Iwane Y.; Suga H. Logical Engineering of D-Arm and T-Stem of TRNA That Enhances d-Amino Acid Incorporation. Nucleic Acids Res. 2017, 45 (22), 12601 10.1093/nar/gkx1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Aquino A. E.; Kim D. S.; Jewett M. C. Engineered Ribosomes for Basic Science and Synthetic Biology. Annu. Rev. Chem. Biomol 2018, 9 (1), 311. 10.1146/annurev-chembioeng-060817-084129. [DOI] [PubMed] [Google Scholar]
- Maini R.; Dedkova L. M.; Paul R.; Madathil M. M.; Chowdhury S. R.; Chen S.; Hecht S. M. Ribosome-Mediated Incorporation of Dipeptides and Dipeptide Analogues into Proteins in Vitro. J. Am. Chem. Soc. 2015, 137 (35), 11206–11209. 10.1021/jacs.5b03135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H.; Saito H.; Suga H. A Versatile TRNA Aminoacylation Catalyst Based on RNA. Chem. Biol. 2003, 10 (7), 655–662. 10.1016/S1074-5521(03)00145-5. [DOI] [PubMed] [Google Scholar]
- Murakami H.; Kourouklis D.; Suga H. Using a Solid-Phase Ribozyme Aminoacylation System to Reprogram the Genetic Code. Chem. Biol. 2003, 10 (11), 1077–1084. 10.1016/j.chembiol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Goto Y.; Murakami H.; Suga H. Initiating Translation with D-Amino Acids. Rna 2008, 14 (7), 1390–1398. 10.1261/rna.1020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.; White P.. Fmoc Solid Phase Peptide Synthesis: A Practical Approach; Oxford University Press: 2000; 10.1093/oso/9780199637256.001.0001. [DOI] [Google Scholar]
- Shimizu Y.; Inoue A.; Tomari Y.; Suzuki T.; Yokogawa T.; Nishikawa K.; Ueda T. Cell-Free Translation Reconstituted with Purified Components. Nat. Biotechnol. 2001, 19 (8), 751–755. 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K.; Reid P.. Rapid Display Method in Translational Synthesis of Peptide, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.