Abstract
While acetylated, RNA binding deficient TDP-43 reversibly phase separates within nuclei into complex droplets (anisosomes) comprised of TDP-43-containing liquid outer shells and liquid centers of HSP70 family chaperones, cytoplasmic aggregates of TDP-43 are hallmarks of multiple neurodegenerative diseases, including ALS. Here we show that transient oxidative stress, proteasome inhibition, or inhibition of HSP70’s ATP-dependent chaperone activity provokes reversible cytoplasmic TDP-43 de-mixing and transition from liquid to gel/solid, independent of RNA binding or stress granules. Isotope labeling mass spectrometry is used to identify that phase separated cytoplasmic TDP-43 is bound by the small heat shock protein HSPB1. Binding is direct, mediated through TDP-43’s RNA binding and low complexity domains. HSPB1 partitions into TDP-43 droplets, inhibits TDP-43 assembly into fibrils, and is essential for disassembly of stress-induced TDP-43 droplets. Decrease of HSPB1 promotes cytoplasmic TDP-43 de-mixing and mislocalization. HSPB1 depletion is identified within ALS-patient spinal motor neurons containing aggregated TDP-43. These findings identify HSPB1 to be a regulator of cytoplasmic TDP-43 phase separation and aggregation.
Introduction
TDP-43, a predominantly nuclear RNA/DNA binding protein with a prion-like domain, is mis-localized to and aggregated within the cytoplasm of motor neurons in almost all (97%) patients with amyotrophic lateral sclerosis (ALS)1. It has been reported to similarly accumulate cytoplasmically in other neurodegenerative diseases, including frontotemporal degeneration (FTD)2, 3, Alzheimer’s disease (AD)4, and a newly recognized dementia found in the oldest population and named limbic-predominant age-related TDP-43 encephalopathy (LATE)5.
ALS/FTD-associated RNA binding proteins (e.g., FUS6, hnRNPA17, and TDP-438-13) have been shown to undergo liquid-liquid phase separation. Work from us8 and others6, 7 has demonstrated that TDP-43-containing, liquid-like, phase separated droplets can convert into gel/solid structures with prolonged stress, or even into amyloid-like fiber structures in vitro. Fibril-induced stress in turn can induce cytoplasmic TDP-43 droplets which provoke inhibition of nucleocytoplasmic transport (accompanied by mislocalization of RanGap1, Ran, and Nup107), clearance of nuclear TDP-43, and cell death8. These findings identify a neuronal cell death mechanism that can be initiated by transient stress-induced cytosolic de-mixing of TDP-43.
In normal cells, phase separation of these RNA binding proteins appears to be tightly controlled, with a proportion of each de-mixed into a liquid droplet within nucleoplasm or cytoplasm. While post-translational acetylation can drive intranuclear phase separation into complex droplets (called anisosomes) comprised of liquid outer shells of TDP-43 and liquid centers of HSP70 family chaperones14, the mechanisms that mediate cytoplasmic phase separation have not yet been determined.
Some RNA binding proteins can phase separate when cells are challenged by stresses, including oxidative stress8 or proteasome inhibition15, 16. Molecular chaperones play a central role in maintaining proteostasis and safeguarding proteins from misfolding and aggregation17-19. In agreement with this, a variety of chaperones (e.g., HSP60, HSPB1, HSPB3, HSPB8, BAG3, and DNAJB2) have been reported to harbor pathogenic mutations in neurodegenerative diseases20, 21, while expression of the HSP70 family has been reported to be transcriptionally downregulated in Alzheimer’s disease22. Moreover, studies using model organisms and human brain samples have proposed an age-dependent decline of proteostasis capacity, including decrease of heat shock proteins during normal aging and in different neurodegenerative diseases22, 23.
Using a proximity labeling approach, our recent work has identified that nuclear RNA-binding deficient TDP-43 interacts with HSP70 family members to enable its phase separation into anisosomes which require the ATP-dependent chaperone activity of the HSP70 family for maintaining liquidity of the shells and cores14. Here we adopt a similar proximity labeling approach to identify that when phase separated in the cytoplasm, TDP-43 binds to the small heat shock protein HSPB1 (also known as HSP27 in human), an ATP-independent chaperone belonging to the small heat shock protein family24, 25. We also determine that HSPB1 de-mixes with TDP-43 in vitro, delays the aging of TDP-43 droplets into gels, inhibits aggregation/assembly of TDP-43’s low complexity domain (LCD) into fibrils, and plays a critical role in disassembly of stressed-induced, gel/solid-like structures formed from phase separated cytoplasmic TDP-43.
Results
Proteotoxic stress-induced liquid droplets/gels of TDP-43
Recognizing that nuclear transport declines during aging26, 27 and that RNA binding of TDP-43 can be modulated by its post-translational acetylation of lysines in each of its RNA binding motifs (RRMs)14, 28, 29, we tested how cytoplasmic TDP-43 (with or without ability to bind RNA) is affected by the proteomic stress accompanying age-dependent reduction in proteasome30, 31 or HSP70 chaperone22, 23 activity. Induction of RNA binding competent cytoplasmic TDP-43 (Figure 1a) produced concentration-dependent phase separation into many rounded droplets of varying diameters (Figure 1b). RNA binding incompetent TDP-43 (generated by mutating five phenylalanines to leucines to block pi-pi interactions with RNA bases32, 33) phase separated into several larger spherical particles (Extended Data Fig. 1a,b) when TDP-43 concentration exceeded a diffuse cytoplasmic concentration (~3 μM) (Extended Data Fig. 1c-e).
Figure 1. Cytoplasmic TDP-43 phase separation and liquid to gel/solid transition are induced by oxidative stress, or reduction in proteasome activity.
(a) Schematic of the design to assess the effects of different stresses on cytoplasmic TDP-43. (b) Representative images of induced expression of cytoplasmic TDP-43 (TDP-43ΔNLS-Clover) for 1 day or 2 days in U2OS cells. (c-e) Representative live cell images of cytoplasmic RNA binding proficient TDP-43 (TDP-43ΔNLS-Clover) or RNA binding incompetent TDP-43 (TDP-43ΔNLS/2KQ-Clover) de-mixing structures induced by reduction of proteasome activity (MG132) (c), inhibition of HSP70 protein folding chaperone activity (VER155008) (d), or arsenite stress (NaAsO2) (e). (f) Quantification of the percentage of cells with TDP-43 de-mixing structures. Cells quantified for each condition from left to right in the bar graph are: 381, 372, 460, 507, 427, 420, respectively, for the top panel; 342, 225, 289, 281, 337, 350, respectively, for the bottom panel. Data are from a live cell imaging experiment.(g-i) Representative examples of FRAP analysis of cytoplasmic TDP-43ΔNLS-Clover droplets under (g) no stress but at higher accumulated level, (h) proteasome inhibition, (i) arsenite stress. (j) FRAP curve of TDP-43ΔNLS-Clover droplets under no stress, proteasome inhibition, or arsenite stress condition. Light color lines were plotted for standard deviation. Number of droplets that were bleached in no stress, proteasome inhibition, HSP70 chaperone inhibition and arsenite stress conditions are: 8, 9, 14 and 8. (k) Representative live images of U2OS cells expressing TDP-43ΔNLS-Clover or TDP-43ΔNLS/2KQ-Clover after treatment with sodium arsenite for increasing times. (l) Relative level of TDP-43ΔNLS-Clover or TDP-43ΔNLS/2KQ-Clover in the diffuse pool of cells before or after exposure to sodium arsenite for 1 hr or 2 hr. Number of cells quantified are 20 and 26, respectively. Cells analyzed are from one live cell image experiment. (m) Level of TDP-43ΔNLS-Clover in the diffuse pool of cells with de-mixed TDP-43 droplets or in cells without de-mixed droplets, or after exposure to sodium arsenite for 1 hr or 2 hr. Number of cells quantified are 20, 71, 21 and 21 in each group from left to right, respectively. (n) Ultrastructural analysis of cytoplasmic TDP-43 de-mixing droplets delineated by correlative light and electron microscopy. U2OS cells expressing TDP-43ΔNLS-Clover were treated with sodium arsenite for 60 min before fixation. (I, m): Medians, 25th and 75th percentiles are shown in the box; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
A lower level of initially diffusely positioned RNA binding competent TDP-43 was induced by transient reduction in proteasome activity (by addition of the inhibitor MG132) to phase separate into many small, rounded cytoplasmic particles (Figure 1c). Proteasome inhibition induced a proportion of RNA binding incompetent TDP-43 (TDP-432KQ with acetylation mimicking conversion of lysines K145 and K192 to glutamine14, 28, 29 [Figure 1c] or TDP-435FL [Extended Data Fig. 1a,f]) to phase separate into nearly spherical, cytoplasmic droplets. Inhibition of HSP70 activity also efficiently induced de-mixing but only of RNA-binding incompetent, cytoplasmic TDP-43 (Figure 1d, Extended Data Fig. 1f), consistent with our prior finding that HSP70 is preferentially bound to RNA-free TDP-4314.
While an even stronger oxidative stress (sodium arsenite) produced robust de-mixing of multiple other RNA binding proteins (including EIF3n and UBAP2L) into polyadenylated mRNA-containing stress granules (Extended Data Fig. 1g-i), arsenite exposure induced phase separation of both RNA binding competent and incompetent TDP-43 (Figure 1e, Extended Data Fig. 1g-i) into cytoplasmic droplets that were clearly distinct from stress granules (Extended Data Fig. 1g-i). A close examination of the droplet size and roundedness revealed that greater than 2/3rds (69%) of TDP-43ΔNLS-Clover droplets had circularity of 0.80 or more, but this decreased to only 49% after 4 hours of arsenite exposure (Extended Data Fig. 1j) accompanied by an increase in droplet size (Extended Data Fig. 1l). A similar trend of changes was seen for the droplets formed by RNA binding deficient TDP-43ΔNLS/2KQ-Clover (Extended Data Fig. 1k, m).
Use of fluorescence recovery after photo-bleaching (FRAP) validated that most molecules of TDP-43 (RNA binding competent or RNA binding incompetent) in cytoplasmic droplets formed by a high level of cytoplasmic TDP-43 had liquid-like character, with ~70% fluorescence recovery with a t1/2 of 12.8 seconds (Figure 1g,j, Extended Data Fig. 2a,d), as we reported previously8. Phase separated, liquid-like TDP-43 droplets displayed viscoelastic properties that included deformation of their shapes as they moved through cytoplasm (Supplementary Movie 1). While at cytoplasmic levels below the saturation concentration TDP-43 was diffusely localized, within 2 to 4 hours after addition of an applied stress [i.e., an inhibitor to reduce proteasome activity (Figure 1h,j, Extended Data Fig. 2b,d) or exposure to sodium arsenite (Figure 1i,j, Extended Data Fig. 2c,d)], the apparent dynamic arrest of initially phase separated TDP-43 produced rounded, cytoplasmic gels/solids, for which there was essentially no fluorescence recovery after photobleaching and no deformation when moved through cytoplasm (Supplementary Movie 2).
Selective permeabilization of the plasma membrane (with digitonin) without disruption of the nuclear envelope lowered the diffuse cytoplasmic pool of TDP-43ΔNLS-Clover and provoked rapid droplet dissolution in unstressed cells, but not in after exposure to arsenite stress (Extended Data Fig. 2e-h). Phase separation into initially liquid TDP-43 droplets8 upon exposure to sodium arsenite was followed by a gradual, time-dependent decrease (by 2-4 fold) in saturation concentration (i.e., diffuse TDP-43) (Figure 1k-m, Extended Data Fig. 2i) and correspondingly increased recruitment of TDP-43 into the rounded droplets (Extended Data Fig. 2j), consistent with stress-induced dynamical arrest after phase separation. Correlative light and electron microscopy confirmed that the cytoplasmic TDP-43 droplets induced by arsenite contained electron dense components closely packed within a membraneless organelle (Figure 1n).
Additionally, in non-stressed cells, accumulation of increasing levels of TDP-43 initially produced small, rounded TDP-43 droplets that, with recruitment of additional TDP-43 coupled with occasional droplet fusion, converted into bigger, brighter, elongated droplets (Extended Data Fig. 2k-m). FRAP analysis revealed that the small, rounded droplets were liquid-like with rapid recovery, while the bigger, elongated ones were gel-like, with less than 10% of TDP-43 molecules within individual droplets rapidly exchanging with the diffuse pool (Extended Data Fig. 2n). Liquid-like and gel-like droplets were present within the same cell (Extended Data Fig. 2i,n). Collectively, this evidence supports phase separation-aided percolation34 as an underlying mechanism for gelation in the absence of an acute proteotoxic stress.
Cytoplasmic TDP-43 droplets deplete nuclear TDP-43
We next tested if continuing cytoplasmic TDP-43 de-mixing was sufficient to sequester nuclear TDP-43 so as to produce a nuclear TDP-43 loss of function (as is seen in surviving neurons in postmortem analyses of ALS tissues2, 35). Cells in which fluorescently tagged wild type TDP-43 replaced endogenous TDP-438 (mediated by the known TDP-43 auto-regulation mechanism36, 37) were cell cycle arrested in G0/G1 (by a combination of serum reduction and addition of the Cdk4/6 inhibitor Palbociclib) and then induced to express cytoplasmic TDP-43 (TDP-43ΔNLS-Clover) (Figure 2a). Increased cytoplasmic TDP-43 drove concentration dependent formation of liquid TDP-43 droplets which sequestered a proportion of nuclear TDP-43 within 24 hours, with almost complete nuclear clearing of TDP-43 within 40 hours (Figure 2b). Continuous addition of a low level of sodium arsenite for a final 4 or 8 hours exacerbated depletion of nuclear TDP-43 and conversion of de-mixed cytoplasmic TDP-43 into larger gel/solid droplets (Figure 2c, d). At all timepoints, TDP-43 phase separation in the cytoplasm was independent of stress granule proteins (e.g., G3BP1) in the absence (Figure 2e,g) or presence (Figure 2f,g) of arsenite.
Figure 2. Slow depletion of nuclear TDP-43 by cytoplasmic TDP-43 phase separation is enhanced by stress-induced liquid to gel/solid transition.
(a) Experimental design to check if cytoplasmic TDP-43 de-mixing could sequester nuclear TDP-43 in cell cycle arrested U2OS cells. (b) Representative images of G1/G0 arrested U2OS cells that stably express wildtype TDP-43mRuby2 being induced to express cytoplasmic TDP-43ΔNLS-Clover for 0, 16, 24, and 40 hours. (c) Representative images of U2OS cells that stably express wildtype TDP-43mRuby2 being induced to express cytoplasmic TDP-43ΔNLS-Clover for 0, 10, 24 hours followed by another 4-hour treatment with 25 μM sodium arsenite. (d) Cytoplasmic to nuclear ratio of TDP-43mRuby2 (total fluorescence intensity) in U2OS cells without cytoplasmic TDP-43ΔNLS-Clover, with 24 hour induced cytoplasmic TDP-43ΔNLS-Clover, or with 24 hour induced cytoplasmic TDP-43ΔNLS-Clover and treated with 25 μM sodium arsenite for 8 hours. Number of cells quantified are 40, 55 and 41, respectively. Data are from two independent live cell imaging experiments. ***P < 0.001 (student t-test, two-tailed). Data are plotted as mean values ± SD. (e-f) Representative fluorescence images of TDP-43mRuby2, TDP-43ΔNLS-Clover and G3BP1 in G1/G0 arrested U2OS cells in the absence (e) or presence (f) of sodium arsenite. (g) Quantification of cells that have stress granule independent TDP-43 droplets in (e) and (f). Cell number for no arsenite group is 77 and for 25 μM arsenite group is 52. Cells are pooled from two independent experiments.
RRM1 is required for gelation of TDP-43 droplets
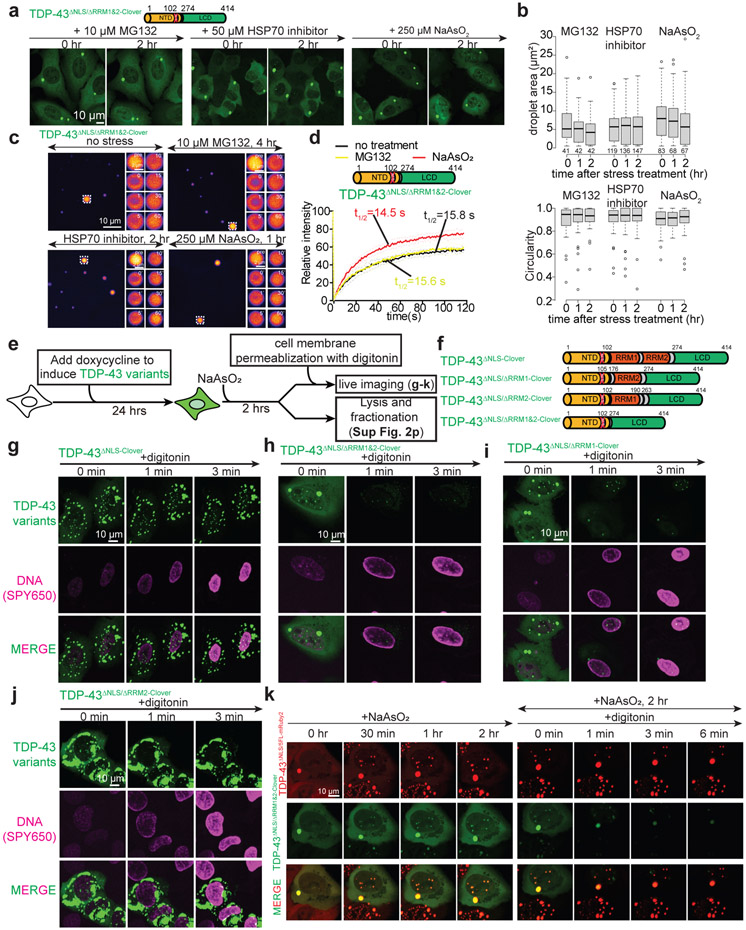
While TDP-43 without both RNA binding domains (TDP-43ΔNLS/ΔRRM1&2-Clover) formed round, liquid-like droplets when expressed in cytoplasm (Extended Data Fig. 2o) (in line with an earlier report38), added stresses (e.g., proteasome inhibition or exposure to arsenite), did not induce additional droplets (Figure 3a-b) or liquid-to-gel conversion of cytoplasmic TDP-43 variants without both RRMs (Figure 3c-d). The reversible liquid character of these RRM-free TDP-43 droplets was further validated by their fast dissolution (within 1 minute) after cell membrane permeabilization with nonionic detergent digitonin. In contrast, arsenite-induced TDP-43ΔNLS-Clover droplets were highly resistant to similar cell permeabilization (Figure 3e-h), yielding gelled droplets that were sedimentable (Extended Data Fig. 2p). While after arsenite exposure full-length RNA binding deficient TDP-43 co-phase separated with RRM-deleted TDP-43, the RRM-deficient TDP-43 remained liquid-like even when the full-length protein had been dynamically arrested into a gelled state (Figure 3k), evidence supporting that the RNA binding domains of TDP-43 are critical for liquid-to-gel/solid transition of initially liquid TDP-43 droplets. Tests of variants missing either RRM revealed RRM1, but not RRM2, is essential for this liquid-to-gel transition (Figure 3i-j, Extended Data Fig. 2p).
Figure 3. RNA binding domains, especially RRM1 is crucial for gelation of TDP-43 droplets.
(a) Representative images of no induction of cytoplasmic RRM-deleted TDP-43ΔNLS/ΔRRM1&2-Clover droplets by proteasome inhibition, HSP70 inhibition or sodium arsenite treatment. (b) Boxplot of area and roundedness of TDPΔNLS/ΔRRM1&2-Clover droplets at 0, 1, 2 hour of 10 μM MG132, 50 μM HSP70 inhibitor or 250 μM sodium arsenite treatment. Number of droplets quantified for each group are 41, 42, 42 for 0, 1, 2 hr of MG132 treatment; 119, 136, 147 for 0, 1, 2 hr of HSP70 inhibitor treatment; 83, 68, 67 for 0, 1, 2 hr of sodium arsenite treatment. Data are from one experiment. (c) Representative images of FRAP analysis of cytoplasmic TDP-43ΔNLS/ΔRRM1&2-Clover droplets under no stress but at higher accumulated level, proteasome inhibition, HSP70 chaperone inhibition and arsenite stress conditions. (d) FRAP curve of TDP-43ΔNLS/ΔRRM1&2-Clover under no stress, proteasome inhibition, HSP70 chaperone inhibition and arsenite stress condition with numbers of droplets of 8, 8, 16 and 9, respectively. Light color lines were plotted for standard deviation. (e) Schematic of experimental design for testing the properties of TDP-43 droplets by cell membrane permeabilization with 50 μg/mL digitonin treatment. (f) Schematic of the TDP-43 variants constructs. (g-j) Representative images of arsenite-stressed U2OS cells containing droplets of TDP-43 variants after cell membrane permeabilization with 50 μg/mL digitonin. Images represent 6 independent frames for each condition. (k) Cells co-expressing full-length RNA binding deficient TDP-43ΔNLS/5FL-mRuby2 and RRM-deleted TDP-43ΔNLS/ΔRRM1&2-Clover were treated with sodium arsenite and their plasma membranes then permeabilized by addition of digitonin. Images represent 6 independent frames. (b) Medians, 25th and 75th percentiles are shown in the box; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
HSPB1 binds and phase separates with cytoplasmic TDP-43
Proximity labeling allows identification of close interactors at ~20 nm spatiotemporal resolution39-42. As we have done for nuclear TDP-43 phase separated droplets14, the components of de-mixed cytoplasmic TDP-43 were identified using proximity labeling and quantitative mass spectrometry. Cell lines were generated that express fluorescent, APEX2-tagged cytoplasmic TDP-43 (TDP-43ΔNLS-Clover-APEX2) or fluorescent, cytoplasmic APEX2 (APEX2Clover-NES) alone. Cytoplasmic RNA binding competent TDP-43 phase separation was induced with addition of sodium arsenite or the proteasome inhibitor MG132 (Extended Data Fig. 3a,b). To enable direct comparisons, proximity labeling was done in six groups: cells with diffuse cytoplasmic TDP-43 (TDP-43ΔNLS-Clover-APEX2) or cytoplasmic APEX2 (APEX2Clover-NES), cells with predominantly de-mixed cytoplasmic TDP-43 (TDP-43ΔNLS-Clover-APEX2) or APEX2 following exposure to sodium arsenite, and cells with partially de-mixed cytoplasmic TDP-43 (TDP-43ΔNLS-Clover-APEX2) or APEX2 after exposure to MG132 (Figure 4a).
Figure 4. The combination of APEX proximity labeling, quantitative mass spectrometry using isotopically labeled tandem mass tags (TMTs), co-expression and immunofluorescence identifies the small heat shock protein HSPB1 to bind cytoplasmic TDP-43 and to de-mix with it into droplets/gels after arsenite stress.
(a) Experiment design of the proximity labeling and TMT quantitative mass spectrometry. Cells inducibly expressing TDP-43ΔNLS-Clover-APEX and APEX2Clover-NES were treated with sodium arsenite for 2 hr or MG132 for four hours before proximity labeling, streptavidin enrichment and TMT labeling for quantitative mass spectrometry analysis. (b) Venn diagram of the proteins that show more than two-fold difference in TDP-43ΔNLS-Clover-APEX labeling comparing to APEX2Clover-NES under arsenite stress or no stress. (c) Volcano plot of statistical significance against fold-change (arsenite v.s. no stress) of each protein labeled by TDP-43ΔNLS-Clover-APEX. Unadjusted P-value is calculated by one-sample t-test, two-sided. (d) Colocalization of TDP-43ΔNLS-Clover and HSPB1mCherry in arsenite-induced de-mixing droplets detected by direct fluorescence signal. Experiments were repeated for three times. (e) Representative fluorescence images of TDP-43 and HSPB1 co-de-mixed droplets and stress granules (indicated by G3BP1 staining). Number of granules for quantification are 370, 631, 449, respectively. Data are plotted as mean values ± SD. Granules are quantified in three independent images from two independent replicates. (f) Representative examples of FRAP analyses of TDP-43ΔNLS-Clover and HSPB1mCherry in co-de-mixed droplets. Dotted circles label the regions that are bleached. (g) Mean relative fluorescence intensity of TDP-43ΔNLS-Clover and HSPB1mCherry over time in FRAP experiments. Number of droplets bleached are 8. Data are from three independent bleaching experiments.
Binding partners of TDP-43 in each condition were identified using six-plex tandem mass tag (TMT) labeling (Supplementary Table 1) and quantitative mass spectrometry (MS3-based). Comparing the combined intensity of all peptides belonging to each protein in cells expressing TDP-43ΔNLS-Clover-APEX2 and APEX2NES-Clover from three biological replicates (each with two technical replicates), 27 proteins were reproducibly labeled by APEX tagged cytoplasmic TDP-43 under no stress conditions, 19 proteins after cell exposure to sodium arsenite, and 53 proteins after proteasome inhibition (Figure 4b, Extended Data Fig. 3c, Source data Fig. 4).
While 15 proteins were both found in no stress and sodium arsenite groups (including RNA binding proteins, translation factors, and β-tubulins - Figure 4b), arsenite-induced TDP-43 phase separation significantly increased proximity labeling more than 3-fold only for one: the small heat shock protein folding chaperone HSPB1 (Figure 4c). Immunostaining for endogenous HSPB1 (in U2OS cells and iPSC-derived cortical and motor neurons - Extended Data Fig. 3d-e) and direct detection of mCherry-tagged HSPB1 (Figure 4d) validated that HSPB1 was recruited to and enriched in cytoplasmic TDP-43 droplets. These HSPB1-positive TDP-43 droplets were independent of stress granules and their constituents (Figure 4e), consistent with decreased labeling of stress granule proteins TIAR and UPF1 by APEX-tagged cytoplasmic TDP-43 after arsenite-induced stress granule assembly (Figure 4c). Use of FRAP revealed that the phase separated droplets of TDP-43 induced by sodium arsenite rapidly converted from liquid to gels in which TDP-43 did not exchange with soluble TDP-43 despite continuing dynamic exchange of half of the de-mixed HSPB1 (45% fluorescence recovery with a t1/2 of 7.5 s) (Figure 4f-g).
HSPB1 de-mixes with TDP-43 and delays droplet aging in vitro
To determine whether, and if so how, HSPB1 affected TDP-43 phase separation, we affinity purified bacterially expressed full-length maltose binding protein (MBP) tagged TDP-43 and HIS tagged HSPB1 (Figure 5a, Extended Data Fig. 4a). Each was then fluorescently-labeled with NHS-Alexa 488 or NHS-Alexa 555, respectively (Figure 5b). MBP-tagged TDP-43 or MBP tag-cleaved TDP-43 spontaneously phase separated into droplets upon addition of a crowding reagent (dextran added to 7.5% final concentration) (Figure 5b, Extended Data Fig. 4b-c). FRAP revealed that freshly prepared TDP-43 droplets exhibited rapid, nearly complete (80%) intensity recovery with a t1/2 of 52 s (Figure 5c). Aging TDP-43 droplets in vitro for as little as 2 h dramatically decreased interior dynamics, with the proportion of recovery after photobleaching diminished (only 20% recovery within 200 s) (Figure 5c,d), consistent with time-dependent gelation after phase separation.
Figure 5. HSPB1 de-mixes in vitro into liquid droplets of full length TDP-43, the TDP-43 LCD alone, or TDP-43 without its LCD and acts to inhibit TDP-43 LCD assembly into amyloid fibrils.
(a) Schematic of TDP-43 variants and HSPB1 that are used for in vitro phase separation assay. ATD, α-crystallin domain; and CTD, C-terminal domain. (b) Fluorescence images of in vitro phase separated TDP-43 (2% NHS-Alexa488 labeled) droplets with/without HSPB1 (2% NHS-Alexa555 labeled). Phase separation of 50 μM TDP-43-MBP was conducted by adding 7.5% dextran with/without 10 μM HSPB1. (c) Representative examples of FRAP analysis of in vitro phase separated TDP-43 droplets with/without HSPB1 at initial timepoint or after 2 hours. (d) FRAP curve of relative TDP-43 intensity of in vitro phase separated TDP-43 droplets with/without HSPB1 at initial timepoint or after 2 hours. Number of droplets that are bleached is 6 for each group. Data are plotted as mean values ± SD. (e) Fluorescence images of in vitro phase separated TDP-43 LCD (50 μM, 2% NHS-Alexa488 labeled) droplets with/without HSPB1 (50 μM, 2% NHS-Alexa555 labeled). (f) Fluorescence images of in vitro phase separated TDP-43 ΔLCD (50 μM, 2% NHS-Alexa647 labeled) droplets with/without HSPB1 (50 μM, 2% NHS-Alexa555 labeled). (g) Thioflavin T aggregation assay to monitor the TDP-43 LCD (10 μM) amyloid assembly over time in the presence or absence of 2 μM or 10 μM HSPB1. Data are plotted as mean values ± SEM. (h) Negative stain electron microscopy images of TDP-43 LCD assemblies at the end point of Thioflavin T aggregation assay. Similar images were taken from three independent experiments.
HSPB1 did not form droplets on its own, but efficiently phase separated into TDP-43 droplets (Figure 5b). Remarkably, HSPB1 co-phase separated with both the amino terminal half of TDP-43 (TDP-43ΔLCD, aa 1-273) and the LCD containing carboxy-terminus of TDP-43 (aa 274-414) (Figure 5e,f). HSPB1 co-de-mixing with TDP-43 altered the properties of TDP-43 droplets, delaying their maturation into gels/solids (Figure 5c,d). Additionally, HSPB1 inhibited phase separation of TDP-43 when present at molar concentrations two-fold or higher above the TDP-43 concentration (Extended Data Fig. 4d-f). After extended incubation, the LCD of TDP-43 spontaneously formed amyloid fibrils (as previously shown43-48) whose presence could be visualized by thioflavin T (ThT) fluorescence (Figure 5g) and negative-stain electron microscopy (Figure 5h). HSPB1 prevented this fibril assembly in a dose-dependent manner (Figure 5g,h).
NMR spectroscopy was then used to determine which region(s) of TDP-43 directly bind HSPB1. Since titration of HSPB1 into 15N-labeled TDP-43 LCD induced co-precipitation of it with the TDP-43 LCD (consistent with the known tendency of HSPB1 to precipitate at high concentration), we mimicked the known stress-induced phosphorylation of HSPB149-51 by converting serines 15, 78, 82 to aspartates (to produce HSPB13D). The resultant phosphorylation-mimicking HSPB1 exhibited behavior similar to wild type HSPB1 in TDP-43 de-mixing and fibrillation assays (Extended Data Fig. 5a-c). Titration of HSPB13D with 15N-labeled TDP-43 LCD enabled acquisition of high-resolution NMR spectra. HSPB13D produced a dose-dependent global decrease in TDP-43 LCD signal intensities (Extended Data Fig. 5d-f) that were most prominently clustered in a highly conserved 20 amino acid region52, 53 (aa 320-340) of TDP-43 (Extended Data Fig. 5e-g) previously reported52 to transiently adopt an α-helical conformation and to be essential in mediating phase separation of TDP-43. Therefore, HSPB1 binding to this conserved sub-domain of the TDP-43 LCD can act to prevent amyloid fibril formation by TDP-43. Beyond the LCD domain, examination by immunostaining and immunoprecipitation of TDP-43 deleted in RRM1 revealed that this domain also plays an important role in binding with HSPB1 (Extended Data Fig. 5h-j).
HSPB1, BAG2, and HSP70 promote TDP-43 droplet disassembly
The small heat shock proteins, like HSPB1, have been shown to hold client substrates in a folding-competent state and to work in concert with the ATP-dependent chaperone HSP70 to facilitate efficient substrate refolding54-57. HSP70, along with the small heat shock protein HSPB8 and the HSP70 nucleotide exchange factor BAG3, have been reported to cooperatively maintain the reversibility of stress granule assembly51. Recognizing this, we tested if members of the HSP70 family, its HSP40 co-chaperones, and the known HSP70 nucleotide exchange factors (NEFs) are recruited to cytoplasmic droplets of TDP-43. For this, we chose: 1) the two abundant members of the HSP40 family (DNAJA1 and DNAJB1); 2) four ubiquitously expressed NEFs (BAG1, BAG2, BAG3 and HSPH1); and 3) HSPB8, a small heat shock protein implicated in stress granule disassembly15 (Extended Data Fig. 6).
HSPB1 phase separated into arsenite-induced TDP-43-containing droplets at early times, with the amount de-mixed increasing in a time-dependent manner (Extended Data Fig. 6a,b). HSP70-HSC70 (Extended Data Fig. 6c) and BAG2 (Extended Data Fig. 6d) were enriched in TDP-43 droplets at early times (with diminishing amounts at later stages). Stress-induced TDP-43 droplets did not recruit HSPB8 (Extended Data Fig. 6e), HSPH1 (Extended Data Fig. 6f), BAG3 (Extended Data Fig. 6g), DNAJA1 (Extended Data Fig. 6h), DNAJB1 (Extended Data Fig. 6i), or BAG1 (Extended Data Fig. 6j).
Stress-induced assemblies of RNA binding proteins, like stress granules, typically dissolve after removal of stress15. Consistent with this, live cell imaging revealed that in approximately half of our cells, arsenite-induced cytoplasmic TDP-43 phase separated droplets/gels dissolved within 8-12 hours after arsenite removal (Figure 6a,b; Supplementary Movie 3). Cells that failed to resolve the TDP-43-containing condensates also failed to recover, instead undergoing cell shrinkage, detachment from the dish, and death.
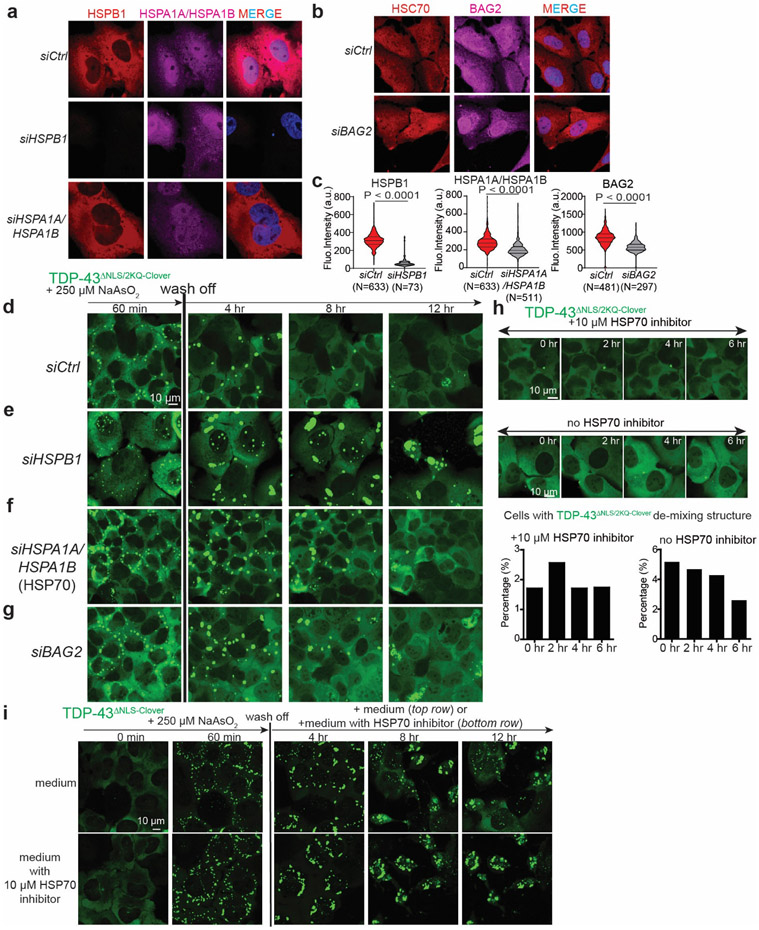
Figure 6. Activities of HSPB1, HSP70, and BAG2 [the nucleotide exchange factor for HSPA1A (HSP70) are essential for disassembly of stress-induced de-mixing and aggregation of TDP-43.
(a) Schematic of experimental design for measuring the disassembly of TDP-43 de-mixing droplets after removal of stress. (b-e) Representative live images of TDP-43ΔNLS-Clover de-mixing droplets induced by arsenite for 1 hour followed by removal of stress. (f) Quantification of cells forming TDP-43 de-mixing droplets in (b-e) after one hour of sodium arsenite treatment. (g) Quantification of cells in which TDP-43 de-mixing droplets disassemble after 12-hr stress removal in (b-e). For f, g: **P < 0.01, ***P < 0.001 (student t-test, two-tailed). For f,g: number of cells quantified in four independent experiments are 346, 425, 309, 354 for siRNA control; 40, 50, 23, 68 for siHSPB1; 283, 248, 100, 144 for siHSPA1A; 72, 145, 68, 86 for siBAG2, respectively. (h-i) Quantification of the relative level of HSPB1 in U2OS cells transduced with HSPB1-IRES-tdTomato lentivirus to stably increase level of HSPB1 compared to un-transduced cells. Medians, 25th and 75th percentiles are shown in the box; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. (j) Representative live images of the disassembly of TDP-43ΔNLS-Clover de-mixing droplets after stress removal in the cells transduced with HSPB1-IRES-tdTomato. (k) Quantification of cells forming TDP-43 de-mixing droplets and cells that disassemble TDP-43 de-mixing droplets after 12-hr stress removal. Data are from three independent replicates; 114, 145 and 109 non-transduced controls, and 123, 161 and 157 HSPB1-IRES-tdTomato-transduced cells were analyzed.
We next tested if depletion of HSPB1, BAG2, or HSPA1A (a major inducible HSP70 family member) affected resolution of stress-induced TDP-43 droplets/gels. For this, we suppressed HSPB1, BAG2, or HSPA1A by transfection of the corresponding siRNAs (Extended Data Fig. 7a-c) and then induced expression of fluorescently tagged cytoplasmic TDP-43 (TDP-43ΔNLS-Clover - Figure 6a). While decrease of HSPB1, HSPA1A, or BAG2 did not affect arsenite induction of cytoplasmic TDP-43 phase separation, reduction in any of the three sharply slowed disassembly after stress removal (Figure 6c-g). Likewise, depletion of HSPB1 strongly inhibited the disassembly of arsenite-induced droplets/gels of RNA binding deficient TDP-43 (Extended Data Fig. 7d,e), while reduction of HSPA1A (Extended Data Fig. 7f) [and to a lesser extent BAG2 (Extended Data Fig. 7g)] delayed their disassembly. Strikingly, partial inhibition of HSP70 family chaperone activity [by addition of the HSP70/HSC70 ATPase inhibitor VER155008 at a level too low to induce TDP-43 de-mixing (Extended Data Fig. 7h)] blocked disassembly of arsenite-induced cytoplasmic TDP-43 droplets (Extended Data Fig. 7i). In addition, increase of HSPB1 in cells by 2-6 fold inhibited arsenite-induced TDP-43 de-mixing and promoted cell recovery (Figure 6h-i).
Although reduction in HSPA1A strongly inhibited disassembly of TDP-43 droplets (Figure 6d), depletion of HSPA8, the constitutive HSP70 family member originally referred to as HSC70, led to induction of HSPA1A (Extended Data Fig. 8a-c) [as previously reported58] and acceleration of disassembly of cytoplasmic TDP-43 droplets/gels (Extended Data Fig. 8d-f). Increased accumulation of HSPA1A (following depletion of HSPA8) also inhibited arsenite induced cytoplasmic droplets of both TDP-43 RNA binding competent (Extended Data Fig. 8d,f,g) and RNA binding deficient TDP-43 (Extended Data Fig. 8h), consistent with a higher chaperone activity of HSPA1A versus HSPA8 for inhibiting TDP-43 phase separation and accelerated resolution of de-mixed droplets/gels (Extended Data Fig. 8i). After stress removal there was enhanced recruitment of HSP70 and stress-inducible co-chaperones of HSP70 (i.e., BAG3 and DNAJB1, but not DNAJA1) to stress-induced TDP-43 droplets (Extended Data Fig. 9a), further supporting the ATP-dependent chaperone activity of HSP70 in the disassembly of TDP-43 droplets. In addition, disruption (by nocodazole addition) of the normal assembled microtubule array did not measurably affect formation and fusion/coalescence of arsenite-induced cytoplasmic TDP-43 droplets, but it did inhibit their disassembly after stress removal (Extended Data Fig. 9b-e).
Taken together, the combined activities of HSPB1, BAG2, and HSPA1A facilitate disassembly of stress-induced, stress granule-independent, phase separated cytoplasmic TDP-43 droplets.
HSPB1 is decreased in motor neurons with TDP-43 pathology
Recognizing that HSPB1 has been implicated to affect neuronal differentiation59, 60, neurite growth61, and axon regeneration62, 63, we determined HSPB1 expression level in motor neurons. Examination of cell type specific translation profiling data from motor neurons64 and single cell transcriptomic data65 identified HSPB1 to be expressed at moderate levels in normal motor neurons (the 509th most expressed mRNA in the translation profiling data64), with ~8 times lower levels in astrocytes and oligodendrocytes (Figure 7a-c), features confirmed by in situ hybridization data of adult mouse spinal cord66.
Figure 7. HSPB1 is highly expressed in normal motor neurons (but not astrocytes or oligodendrocytes) but accumulation of it is sharply decreased in spinal motor neurons with TDP-43 pathology in ALS patients.
(a) Levels of HSPB1 mRNA in motor neurons, astrocytes and oligodendrocytes from spinal cords of BacTrap mice82. Data are plotted as mean values ± SD. (b) mRNA levels of chaperones including all the members of HSP70, HSP90, DNAJ, NEFs and small heat shock proteins in motor neurons from spinal cords of BacTrap mice82. (c) Levels of HSPB1 mRNA in different types of cells in spinal cord from a single nucleus RNA-seq transcriptome83. (d-e) Representative immunofluorescence images of HSPB1 and TDP-43 in control (d) and sporadic ALS patients (e). (f) Representative images of motor neurons with different levels (high, medium or low) of HSPB1, and with different localization of TDP-43 (nuclear, partially cytoplasmic or cytoplasmic). (g) Quantification of TDP-43 localization in motor neurons of control and ALS patients. Data are plotted as mean values ± SD. (h) Quantification of HSPB1 expression levels in motor neurons of control and ALS patients. Data are plotted as mean values ± SD. (i) Quantification of HSPB1 expression levels in motor neurons with nuclear TDP-43 and motor neurons with cytoplasmic TDP-43 in ALS patients. Data are for three control individuals and four patients with ALS. For g-i: the number of motor neurons characterized in control patients are 35, 76, and 59, respectively. The number of motor neurons in ALS patients are 65, 77, 99 and 257, respectively (seen in Source Data Fig. 7).
We tested if HSPB1 amount or subcellular localization changed in motor neurons in ALS patients with TDP-43 pathology (Figure 7d-e). Examination of spinal cord sections from three patients with sporadic ALS and one familial ALS individual with a repeat expansion in the C9orf72 gene (Supplementary Table 2a-b) revealed that a large majority (between 59% (patient #81) and 90% (patient#107)) of the remaining motor neurons had cytoplasmic TDP-43 pathology, with many (44%-78%) displaying obvious loss of nuclear TDP-43 (Figure 7f-g, Source Data Fig. 7). Most remarkably, HSPB1 levels were markedly and reproducibly reduced in the surviving ALS patient motor neurons (Figure 7f-h, Source Data Fig. 7). The fraction of motor neurons accumulating a normal level of HSPB1 decreased to between 13% and 31% in ALS patient motor neurons (Figure 7f-h, Source Data Fig. 7). This was most strongly seen in comparing the level of HSPB1 in ALS patient motor neurons with nuclear TDP-43 to those with TDP-43 pathology (Figure 7i, Source Data Fig. 7). In non-ALS control individuals, no motor neurons had cytoplasmic TDP-43 abnormalities, with almost all (>90%) of them containing both nuclear TDP-43 and high levels of HSPB1.
Reduction in HSPB1 promotes cytoplasmic TDP-43 de-mixing
Transfection of siRNA to HSPB1 (Figure 8a) was used to determine that depletion of HSPB1 promoted phase separation of cytoplasmic, RNA binding incompetent TDP-43 (TDP-43ΔNLS-5FL-Clover) into droplets in both cycling (Figure 8b) and G1/G0 arrested (Extended Data Fig. 10a-d) cells. HSPB1 reduction also induced nuclear TDP-43 (TDP-43Clover-APEX2) mis-localization to the cytoplasm (Figure 8c-e). A similar trend of increased cytoplasmic mis-localization was observed for endogenous TDP-43 (Extended Data Fig. 10e-f) (with the increased size [102 kD for TDP-43Clover-APEX2 versus 43 kD for TDP-43] making it a more sensitive marker for a nuclear import defect).
Figure 8. TDP-43 de-mixing and cytoplasmic mis-localization when HSPB1 level is depleted.
(a) Schematic of the experimental design for assessing the effects of decreasing HSPB1 on cytoplasmic TDP-43 de-mixing. (b) Representative fluorescence images of cells expressing TDP-43ΔNLS/5FL-Clover transfected with control siRNA or siHSPB1. Quantification of the cells with TDP-43ΔNLS/5FL-Clover de-mixing droplets after transfection with control siRNA (N=98) or siHSPB1 (N=88). (c) Representative images of cells expressing TDP-43Clover-APEX2 with and without HSPB1 knockdown detected by immunostaining of HSPB1. (d) Violin plot of the cytoplasmic/nuclear TDP-43Clover-APEX2 intensity ratio of the cells transfected with siHSPB1 or siRNA control. Number of cells quantified are 488 and 1648 for siHSPB1 and siRNA control, respectively. ***P < 0.001 (student t-test, two-tailed). (e) Representative images of FUS and HnRNPU localization in cells transfected with siRNA control or siHSPB1. Images were taken from more than 10 frames with cells showing similar phenotype. (f) Representative images of RanGAP1 localization in U2OS cells transfected with siHSPB1 and control, respectively, and quantification of the cells with RanGAP1 mis-localization. Number of cells quantified in three independent replicate experiments for the siHSPB1 group are 272, 442 and 378, respectively, and 226, 223 and 384 in the siRNA control group. *P=0.0138 (student t-test, two-tailed). Data are plotted as mean values ± SD. (g) Model of HSPB1 regulation of TDP-43 phase separation and the disassembly of TDP-43 de-mixing droplets. HSPB1 maintains the liquid-like state of cytoplasmic TDP-43 and facilitates the ATP-dependent HSP70 complex-mediated disassembly of cytoplasmic TDP-43 de-mixing structures induced by proteomic stress after stress removal. When HSPB1 is depleted like what happens in the motor neurons of patients with ALS, cytoplasmic TDP-43 de-mixing is promoted and disassembly of cytoplasmic TDP-43 de-mixing structures is inhibited, which causes nuclear TDP-43 depletion.
Another nuclear protein with a classical nuclear-localization sequence (e.g., hnRNPU – Figure 8e) was also mis-localized when HSPB1 was reduced, while the PY-NLS-containing FUS was not, suggestive of a potential role for HSPB1 in facilitating import of at least some classical NLS-containing proteins. RanGAP1, which maintains cytoplasmic Ran in a GDP bound form thereby enabling nuclear import by importins67, 68, has been reported to be mis-localized in neurodegenerative diseases69, 70, including ALS71-73. Indeed, beyond decrease in HSPB1 promoting cytoplasmic TDP-43 mis-localization and induction of its cytoplasmic de-mixing, RanGAP1 was mis-localized when HSPB1 level was reduced (Figure 8f), consistent with reduction in Ran-dependent nuclear import.
HSPB1 variants in ALS
HSPB1 variants have previously been reported in ALS patients74-76 (Supplementary Table 3a), including mutation in a heat shock response element sequence in the promoter region and missense mutations in the intrinsically disordered C-terminal domain. Recognizing this, we examined four large human genome projects, including the Project MinE77 (http://databrowser.projectmine.com/), the ALS Variant Server78 (http://als.umassmed.edu/), the ALS Data Browser79 (http://alsdb.org/), and the ALS Knowledge Portal80 (http://alskp.org/), which in total have currently collected complete genome sequences from 8625 ALS patients and 9671 non-ALS individuals. Our search identified a probable enrichment in HSPB1 missense or frameshift variants in ALS patients (p = 0.06877), with 22 variants overall, 14 of which were not found in controls - Supplementary Table 3a-b).
Discussion
Discovery that neurodegenerative disease-causing RNA binding proteins (like TDP-43 and FUS) have intrinsically disordered low complexity domains that can spontaneously undergo de-mixing and subsequently convert into amyloid fibrils in vitro6, 9, 11, 13, 43-48, 53, 81, highlights the unresolved question of how the de-mixed liquid-like state is maintained in normal cells. Prior work has identified reversible acetylation of TDP-43 and the activity of the HSP70 protein folding chaperone family to modulate intranuclear phase separation of TDP-43 into anisosomes comprised of a liquid outer shell of TDP-43 and an inner liquid core of HSP7014. When accumulated into cytoplasm TDP-43 also forms liquid-like phase separated droplets which under stress are dynamically arrested to form gel/solid-like structures. Here we have identified HSPB1, along with HSP70 chaperone activity, to be required for maintaining the liquid state of phase separated TDP-43 droplets in the cytoplasm. Binding of HSPB1 is direct, mediated through TDP-43’s RNA binding and low complexity domains, with HSPB1 partitioning into TDP-43 droplets and inhibiting droplet aging and assembly of TDP-43 into fibrils.
Accumulating evidence points to the importance of the chaperone system for maintaining the neuronal proteome20-23, 82-92. A decreased or impaired heat shock protein system leads to the formation of protein aggregates in models of neurodegenerative diseases, like Alzheimer’s disease22 and Parkinson’s disease83, 84. An increase in HSPB1 has been reported to be beneficial in the early disease stage of a model for SOD1 mutant-mediated ALS90, albeit the benefit was not maintained at later disease stages. Our study suggests that HSPB1 functions together with the HSP70 chaperone machinery to regulate the phase separated state of TDP-43. Recognizing that cytoplasmic TDP-43 inclusion is the most common characteristic feature of ALS and related disorders2, approaches to increase levels of heat shock chaperones and other components of the protein quality control machinery could therefore hold promise for therapy.
HSPB1 is an ATP-independent chaperone predominantly localized within the cytoplasm93, 94 and with a central α-crystalline domain flanked by two intrinsically disordered regions (IDRs) at N- and C-termini24, 51, 95. It normally forms a range of oligomers (12 to 30-mers, ~500-1100 kDa) mediated by its IDRs, with the most common form in cells thought to be the 24 mer93 which is in rapid, reversible equilibrium with monomers, dimers, and other higher order oligomers93, 94. This dynamic behavior of HSPB1 is thought to render it an efficient chaperone when ATP is limited (like after exposure to sodium arsenite96, 97). In line with this, we have identified increased recruitment of HSPB1 to cytoplasmic TDP-43 droplets, along with the recruitment of ATP-dependent HSP70 and its BAG2 co-chaperone, evidence supporting a central role of HSPB1 in maintaining TDP-43 proteostasis under oxidative stress.
HSPB1 has previously been shown (using in vitro phase separation and NMR) to prevent aging of phase separated FUS LCD into fibrils51. To this, we have shown that HSPB1 interaction with TDP-43 is through a region of the LCD which prior NMR efforts had demonstrated to transiently convert into a helical structure81. We also have detected an interaction of HSPB1 with the TDP-43 RRM domain, especially RRM1 (Extended Data Fig. 5h-j). These data are in line with reports of RNA binding inhibiting TDP-43 phase separation98, 99, disease-causing mutations in RRM1 enhancing TDP-43 phase separation100, and TDP-43 RRM1 having two cysteine residues (Cys173, Cys175) that are sensitive to oxidative stress101-103. Oxidation of these cysteines results in disulfide bond formation and a conformational change that facilitates TDP-43 aggregation102, 103, consistent with our demonstration in cells that liquid-to-gel transitions of TDP-43 droplets rely on the RRMs of TDP-43. While HSPB8, another small heat shock protein implicated in neuromuscular disease104, has been recently reported to bind to the unfolded RRM of FUS and delay the aging of FUS condensates105, we did not find HSPB8 recruited to cytoplasmic TDP-43 phase separated droplets.
To prior efforts of how de-mixed RNA/RBP condensates are disassembled in cells15, 106, 107, our demonstration that HSPB1 facilitates disassembly of TDP-43 de-mixed droplets (Figure 6) offers what we believe is strong evidence for the proposal that binding of HSPB1 to TDP-43 in gel/solid structures acts to maintain TDP-43 in a refolding-capable state that facilitates its refolding. After removal of a provoking stress and restoration of ATP levels, HSPB1 enhances droplet resolution (Figure 8g; Extended Data Fig. 9a) acting in conjunction with the HSP70 family chaperones, BAG2, BAG3, and HSP40 (e.g., DNAJB1 but not DNAJA1 [in line with recent report of the unique function DNAJB family proteins108]), whose expression is induced to facilitate ATP-fueled substrate refolding (Figure 8g). Decrease of HSPB1 also leads to mis-localization of RanGAP1, partial inhibition of nuclear import, and a corresponding increase in cytoplasmic TDP-43 accumulation (Figure 8c,f).
HSPB1 mutations have been reported in multiple motor neuron diseases, including Charcot-Marie tooth (CMT) disease109, 110, distal hereditary motor dystrophy111-113, and ALS74, 76, with sequence variants scattered throughout HSPB1 and an enrichment in the α-crystallin domain111, 113. Some CMT mutations (R127W, S135F, and R136W) in this domain have been shown to increase chaperone activity, with mutation in the C-terminus (P182L) enhancing aggregation of HSPB1114. How the HSPB1 sequence variants found in ALS affect chaperone activity and binding to TDP-43 is not established, although some have been reported to have increased binding to tubulin, thereby disturbing microtubule dynamics115, 116. Indeed, we not only determined that retention of an assembled microtubule array was necessary for rapid TDP-43 droplet disassembly, but also in our proximity labeling dataset we identified an increased proximity of cytoplasmic TDP-43 to β-tubulins. These findings raise a future direction to determine whether cytoplasmic TDP-43 affects HSPB1 function in tubulin folding/refolding.
Methods
All experiments performed in this study were approved by the institutional ethical review committee of University of California, San Diego.
Plasmids
Plasmid information is listed in Supplementary table 4. The vectors are built by Gibson assembly or double-restriction digestion cloning method.
Cell culture and stable cell line construction
Cell lines used in this paper are: HEK293T (ATCC: CRL-11268), U2OS (ATCC: HTB-96) and the human inducible Ngn2 iPSC line (ALSTEM, iP11N), and the human inducible NGN2, ISL1, and LHX3 iPSC line117, a gift from Dr. Michael Ward’s group at NIH. Routine maintenance of these model cell lines follows the guideline posted on ATCC. In brief, U2OS and HEK293T cells were cultured in complete DMEM supplemented with 10% fetal bovine serum. iPSC cells are cultured on Matrigel-coated plates in mTeSR™ plus medium.
Lentivirus is produced in HEK293t cells by transfection of lentiviral plasmid and packaging plasmids pMD2.G and psPAX2 using TransIT-VirusGEN® Transfection Reagent (Mirus, MIR6705). After two days of transfection, the culture medium containing the lentivirus was passed through a 0.45 μm filter and was used to infect U2OS cell line. After two days of infection, the medium is exchanged to medium containing 20 μg/μL blasticidin or 2 μg/μL puromycin for selection. Single clones are sorted by SH800S Cell Sorter (Sony).
Neuron differentiation
iPSCs are first induced to differentiate to neural precursor cells117 by culturing into induction medium (DMEM/F12, 1x N2 supplement, 1x GlutaMAX, 1x non-essential amino acids, 0.2 μM compound E (only for iPSC-motor neurons), 2 μg/mL doxycycline, 10 μM ROCK inhibitor Y-27632) for 1 day and then replaced with induction medium (no ROCK inhibitor) for 1 day. Then neural precursor cells are treated with Accutase and plated onto 8-well chamber slide (iBidi, 80827) coated with poly-L-ornithine and laminin with neural culture medium (Neurobasal, 1xN2 supplement, 1xB27 supplement, 1x GlutaMAX, 1x non-essential amino acids, 10 ng/mL BDNF, 10 ng/mL GDNF, 1 μg/mL laminin). Medium is replaced 50% every two days.
Proximity labeling and enrichment of biotinylated protein
Before labeling, U2OS cells were treated with the indicated reagents and biotin phenol (Iris-Biotech, 41994-02-9) containing medium was further added to 250 μM final concentration and treated for 30 min. Then 1 mM hydrogen peroxide was added to the medium to activate APEX labeling reaction for 1 min, followed by immediate quenching of reaction with ice-cold quenching buffer (1xPBS, 10 mM sodium azide, 10 mM sodium ascorbate, 2.5 mM Trolox). After four washes with cold quenching buffer, the cells were collected from plates with scrapers. Cells were lysed in lysis buffer (100 mM NaPO4, PH 8.0, 8 M Urea, 1% SDS, 10 mM sodium azide, 10 mM sodium ascorbate, 5 mM Trolox, 10 mM TCEP) and passed through an insulin syringe for 15 times to break DNA. After sonication at water bath sonicator for 10 mins, protein lysates are cleared by centrifuge. Protein concentration was measured using 2-D quant kit (GE healthcare, Cat# 80648356), by following manufacturer’ instruction. After alkylation with 20 mM iodoacetamide for 15 min, 0.5 mg of protein samples were aliquoted and equilibrated to the same volume with lysis buffer. After dilution with equal volume of ddH2O to reduce the concentration of urea to 4 M and SDS to 0.5%, the samples were incubated with streptavidin magnetic AccuNanobeads (Bioneer, Cat# TA-1015-1) at 4 °C overnight.
Protein digestion and TMT labeling
After three washes with wash buffer 1 (100 mM TEAB, PH 8.0, 4 M Urea, 0.5% SDS) and four washes with wash buffer 2 (100 mM TEAB, PH 8.0, 4 M Urea), the beads were resuspended in 100 mM TEAB, 2 M Urea supplemented with 10 ng/uL Trypsin, 5 ng/uL Lys-C for pre-digestion at 37 °C on a thermomixer shaking at 1,000 rpm. The pre-digested products were collected, and an additional 10 ng/uL Trypsin were added to digest overnight at with 1% 37 °C. Digested peptides from each sample are labeled with TMT six-plex labeling reagents (Thermo, Cat# 90061) following manufacture instruction. Briefly, TMT reagents are solubilized in anhydrous acetonitrile and add to peptides from each sample according to the labeling design in Supplementary Table 1. After 1-hr reaction at RT, 5% hydroxylamine was added and incubated for 15 mins to quench the reaction. Then equal volume of peptides of each sample in the same group are pooled together and speedvac to remove acetonitrile. The samples are acidified with formic acid (1%, final concentration) and desalted using Pierce C18 spin columns (89870).
Liquid chromatography-Mass spectrometry analysis
The TMT labeled samples were analyzed on a Orbitrap Eclipse mass spectrometer (Thermo). Samples were injected directly onto a 25 cm, 100 μm ID column packed with BEH 1.7 μm C18 resin (Waters). Samples were separated at a flow rate of 300 nL/min on a nLC 1200 (Thermo). Buffer A and B were 0.1% formic acid in 5% acetonitrile and 80% acetonitrile, respectively. A gradient of 0–25% B over 75 min, an increase to 40% B over 30 min, an increase to 100% B over another 10 min and held at 100% B for a 5 min was used for a 120 min total run time.
Peptides were eluted directly from the tip of the column and nano-sprayed directly into the mass spectrometer by application of 2.5 kV voltage at the back of the column. The Eclipse was operated in a data dependent mode. Full MS1 scans were collected in the Orbitrap at 120k resolution. The cycle time was set to 3 s, and within this 3 s the most abundant ions per scan were selected for collision-induced dissociation tandem mass spectrometry in the ion trap. MS3 analysis with multi-notch isolation (SPS3) was utilized for detection of TMT reporter ions at 7.5k resolution118. Monoisotopic precursor selection was enabled, and dynamic exclusion was used with exclusion duration of 60 s.
Quantitative mass spectrometry data analysis
The raw data was processed by Rawconverter119 to extract MS2 and MS3 spectra with a correction of each precursor ion peak to its monoisotopic peak when appropriate. MS2 and MS3 mass spectrometry spectra were searched against a complete human protein database downloaded from Uniprot with the addition of APEX2 and Clover protein sequence using the search algorithm ProLuCID120. The searching parameters are: precursor mass tolerance of 50 ppm, fragment ion tolerance of 500 ppm for CID spectra and of 20 ppm for HCD spectra; minimum peptide length of 6 amino acids; static modifications for carbamidomethylation of cysteine and TMT tags on lysine residues and peptide N-termini (+229.162932 Da). The identified peptide-spectrum matches (PSMs) were filtered to an FDR of ≤1% at a PSM level with DTASelect2121. The FDR was calculated based on the number of PSMs that matched to sequences in the reverse decoy database. TMT quantification of reporter ions from MS3 spectra is done by Census2122 with the filter of over 0.6 for isobaric purity. The normalized intensity based on weighted normalization were used to calculate the ratio of reporter ions corresponding to the indicated groups. The ratios of each protein from three forward labeling groups and three reverse labeling groups (Supplementary Table 1) were used to calculate P-value through one sample t-test. The volcano plot was generated with R package.
Human post-mortem tissues
Human tissues were obtained from the UCSD ALS tissue repository that was created following HIPAA-compliant informed consent procedures approved by Institutional Review Boards (either Benaroya Research Institute, Seattle, WA IRB# 10058 or University of California San Diego, San Diego, CA IRB# 120056) and de-identified at time of acquisition. Informed consent was obtained on all subjects and there was no compensation.
Tissue samples were obtained from patients who met the modified El Escorial criteria for definite ALS. Control nervous systems were obtained from non-neurological patients when life support was withdrawn, or from patients on hospice. Tissues were acquired using a short-postmortem interval acquisition protocol usually under 6 hours. Tissues were immediately dissected in the autopsy suite, placed in labelled cassettes and fixed in neutral buffered formalin for at least 2 weeks before being dissected and paraffin embedded for indefinite storage.
Immunofluorescence (IF) in postmortem tissue
On day one, sections were deparaffinized with Citrisolv (FISHER brand #04-355-121) and hydrated through a serial dilution of ethanol. Sections were permeabilized with 1% FBS (Atlanta Biologicals #511150) and 0.2% Triton X-100 (Sigma #65H2616). Following permeabilization, antigen retrieval was performed in a high pH solution (Vector # H- 3301) in a pressure cooker for 20 min at 120 °C. Next, sections were blocked with 2% FBS in 1 × PBS for 60 min and were incubated with primary antibody overnight. Primary antibodies were diluted in 2% FBS in 1X PBS. On day two, slides were incubated with secondary antibodies diluted in 2% FBS in 1X PBS for 60 min at room temperature. We quenched CNS auto-fluorescence with 0.1% Sudan Black in 70% ethanol for 15 seconds. Slides were cover slipped using ProLong Gold Antifade Mountant with or without DAPI. We analyzed two to four 8-μm sections per patient. Motor neurons were identified by their large size, multipolar cytoplasm, presence of lipofuscin, and large nucleus with a less dense DAPI staining in IF studies.
siRNA transfection
ON-TARGETplus SMARTpool siRNAs (Horizon Discovery) targeting HSPB1 (L-005269-00-0005), HSPA1A (L-005168-00-0005), BAG2 (L-011961-00-0005) or HSPA8 (L-017609-00-0005) are transfected to U2OS cells at a concentration of 3 fmol/5,000 cells using Lipofectamine RNAiMAX Transfection Reagent (Thermofisher, 13778075) for three or four days before the experiment.
Cell cycle blocking and FACS analysis
U2OS cells are plated onto SCREENSTAR 96-well microplate (Greiner, #655866) at 10,000 cells/well with normal medium and after 1 day are changed to 1% FBS-supplemented DMEM with 1 μM palbociclib. After three days of treatment, cells are trypsinized, fixed in 70% ethanol, treated with RNase A and stained with propidium iodide (PI) solution. Then DNA contents are analyzed by BD LSRFortessa cell analyzer. The plot was generated with Flowjo; the gating strategies are provided in Supplementary Figure S1.
Live cell imaging
U2OS cells are plated onto the 96-well plate (Greiner, #655866) with DMEM medium, no phenol red, and treated as indicated. Images are taken by CQ1 benchtop high-content analysis with a 40x/1.2 objective under a constant CO2 flow.
Fluorescence recovery after photobleaching (FRAP)
U2OS cells for FRAP experiments were cultured on an 8-well chamber slide (iBidi, 80827) in DMEM supplemented with 10% fetal bovine serum (FBS) and Antibiotic-Antimycotic (Thermofisher, 15240062). Expression of TDP-43 variants was induced 24 hour or 48 hours as indicated by adding 1 μg/mL doxycycline to the culture medium. FRAP analysis was performed on a Zeiss LSM880 Aryscan microscope with 40x/1.2 W objective using Zen software. The intensity of the fluorescent signal is controlled in the detection range through changing the laser power, digital gain and off-set. For green and red fluorescent channels, bleaching was conducted by 488-nm or 561-nm line correspondingly and the laser power and iteration of bleaching are optimized to get an efficient bleaching effect. Fluorescence recovery was monitored at 1 second intervals for 2 minutes. In the focal-bleach experiment, roughly half (partial bleach) or all (full bleach) of de-mixing structures was photobleached to determine the molecular mobility with diffuse pool or inside a condensate.
The FRAP data were quantified using ImageJ. The time series of the fluorescence intensity of condensates were calculated. The intensity of the droplet during the whole experiment was normalized to the one before bleaching and the intensity of the droplet just after bleaching was normalized to zero. At least 6-10 images were analyzed per condition to calculate the mean and standard deviation. The averaged relative intensity and standard error were plotted to calculate dynamics.
Immunofluorescence
For immunofluorescence, U2OS cells were cultured on 8-well chamber slides (iBidi, 80827) in DMEM supplemented with 10% fetal bovine serum (FBS) and Antibiotic-Antimycotic (Thermofisher, 15240062). After treatments of the cells as indicated, cells were fixed with 4% PFA in PBS and permeabilized with 0.2% Triton X-100 for 10 min. After blocking with 2% BSA in PBS, 0.05% Triton X-100 for 2 hours, cells were incubated for 1 hour at room temperature with primary antibody in blocking solution. After three washes with PBS, cells were incubated with Alexa647-labeled, Cy3-labeled or Alexa488-labeled secondary antibody at 1:500 dilution or Alexa647-streptavidin at 1:1000 dilution for APEX labeling experiment in blocking solution for 30 minutes at room temperature. After three washes with PBS and DAPI staining, cells were kept in PBS for imaging. Dilutions for primary antibodies used in this study are 1:300 for anti-HSPB1 (goat polyclonal, Santa Cruz, sc-1048; rabbit polyclonal, Stressmarq, SPC-106B, Lot#1201), 1: 100 for anti-HSP70/HSC70 (mouse monoclonal, Enzo Life Sciences, ADI-SPA-820),anti-HSP70 (mouse monoclonal, Enzo Life Sciences, ADI-SPA-810), anti-HSC70 (rat monoclonal, Enzo Life Sciences, ADI-SPA-815), 1:500 for anti-BAG1 (mouse monoclonal, Novus Biologicals, H00000573-M02, Lot#F8031-2D3), anti-BAG2 (Rabbit polyclonal, Novus Biologicals, NB100-56087, Lot# AR99-091613), anti-BAG3(Rabbit polyclonal, Novus Biologicals, NBP2-27398, Lot#102119), anti-DNAJB1 (rabbit polyclonal, Enzo Life Sciences, ADI-SPA-400, Lot# 07051758), anti-DNAJA1 (rabbit polyclonal, Proteintech, 11713-1-AP), anti-HSP110 (rabbit polyclonal, Proteintech, 13383-1-AP) and anti-HSPB8 (rabbit polyclonal, Novus Biologicals, NBP2-67836). 1: 300 for anti-EIF3η (goat polyclonal, Santa Cruz, sc-16377).
Correlative light microscopy and electron microscopy
U2OS cells were plated on gridded coverslips (Mattek, P35G-1.5-14-CGRD-D), induced by doxycycline to express TDP-43ΔNLS-Clover or Clover alone and pre-incubated with SPY-650 DNA dye for 24 hr. After 1 hour treatment with sodium arsenite, cells were fixed with 4% PFA, 0.25% glutaraldehyde in 0.1 M sodium cacodylate for 20 min at RT and then changed to 0.1 M sodium cacodylate buffer for imaging. High resolution imaging is taken by DeltaVision Elite microscope (Cytiva) with 100x objective at 0.1 μm Z interval for the whole cell volume. Then the cells are fixed by 2% glutaraldehyde in 0.1M sodium cacodylate for 4-5 min at RT and then overnight at 4°C. After fixation, cells were washed five times with 0.1 M sodium cacodylate buffer and stained with 1% OsO4 in 0.1M SC buffer for 45 minutes on ice. Then OsO4-stained cells were washed with 0.1M sodium cacodylate buffer five times and MilliQ water for 2 times. After that, coverslips were stained in 2% uranyl acetate buffer for 45 minutes and then rinsed with Milli-Q water. Samples were dehydrated with 20%, 50%, 70%, 90% ETOH followed by two times with 100% ETOH, 1 min each time, and then dried with acetone for 2x1min. After dehydration, samples were incubated at RT in Ducurpan:Acetone=50/50 for 1 hour, and in fresh 100% Durcupan for two times, 1 hour each time. Coverslips were then embedded in Durcupan resin, and the regions of interest navigated by the grid network were cut into 60nm ultrathin sections by diamond knife. Sections were mounted on 300 mesh grids for TEM imaging on FEI Tecnai Spirit G2 BioTWIN. Fluorescent image and TEM image of the same cell are manually correlated with Image J.
RNA Fluorescence in situ hybridization (FISH)
All hybridization steps were performed following the Stellaris RNA FISH protocol for adherent cells. Briefly, cells were fixed in 4% PFA for 10 minutes and then permeabilized with ethanol 70% overnight at 4°C. Then, cells were washed once with wash buffer A (Biosearch Technologies, SMF-WA1-60) supplemented with 10% deionized formamide (Sigma, F7503) for 5 min, and incubated with Hybridization Buffer (SMF-HB1-10) supplemented with 10% deionized formamide and 1ng/ml Cy5-labeled Cy5-(d)T20 oligonucleotides (gift from Dr. J. Paul Taylor, St. Jude Children Hospital) in the dark at 37°C for 4 hours in a humidified chamber. After cells were washed with wash buffer A in the dark at 37°C for 30 minutes, cells were stained with DAPI in wash buffer A, and then washed once with wash buffer B (Biosearch Technologies, SMF-WB1-20) before imaging.
Cell lysate fractionation
U2Os cells were cultured at 6-well plate and each well of cells were lysed in 300 μL of RIPA buffer (25 mM Tris•HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with 1000 unit/mL Benzonase (Millipore, 70746) and 1x Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermofisher, 78440). The cell lysates were passed through 28G Insulin Syringe (Fisher Scientific, 14-829-1A) for eight times and lysed on ice for 20 min. Then cell lysates were centrifuged at 18,000 g for 20 min and supernatants were collected as the soluble fraction. The pellet fractions were washed once with 500 μL of RIPA buffer and solubilized in 100 μL of 1xLDS (Thermofisher, B0007) running buffer as the insoluble fraction.
Immunoprecipitation
U2OS cells were treated with 250 μM sodium arsenite for 2 hr before collected with cell scraper. Cell pellets from one 10-cm dish were lysed in 1 mL of 1xIP lysis buffer (50 mM Hepes pH 7.4, 300 mM NaCl, 2 mM EDTA, 1% NP-40 and 5% glycerol) supplemented with 1000 unit/mL Benzonase (Millipore, 70746) and 1x Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermofisher, 78440). Cell lysates were passed through 28G Insulin Syringe (Fisher Scientific, 14-829-1A) for eight times and lysed on ice for 20 min. The release of TDP-43 droplets in cell lysates were checked under microscope. Then cell debris were removed by 20 min centrifuge at 1,500 g. The droplets containing supernatant were collected and incubated with GFP-Trap Magnetic Particles M-270 (Chromotek) at 4 °C for 4 hours. After incubation, 5 μL of samples were taken to examine the efficiency of GFP binding on the beads under fluorescence microscope. Then, the beads were collected by magnet stand and washed with 1 mL lysis buffer for 4 times before eluted with 1xLDS (Thermofisher, B0007) running buffer at 80 °C for 30 min on a thermomixer. One percent of input and ten percent of IP sample were used for western analysis.
Western blot
Protein samples were loaded onto 12% Criterion™ TGX™ Precast Midi Protein Gel (Bio-Rad) and transferred onto Immobilon®-FL PVDF Membrane (Millipore). Then membrane was blocked with Intercept Blocking Buffer (LICOR) and incubated with primary antibody (1:1000 dilution for HSPB1 monoclonal antibody, StressMarq, 5D12-A12; 1:2000 dilution for GAPDH polyclonal antibody, Cell Signaling 14C10; 1:1000 dilution for TDP-43 C-terminal antibody, Proteintech, 12892-1-AP) overnight at 4 °C. After four washes with TBST, membrane was incubated with secondary antibody (1:20,000 for IRDye® 800CW Donkey anti-Mouse, LICOR 926-32212, and IRDye® 680RD Donkey anti-Rabbit IgG, LICOR 926-68073 or 1:5,000 for HRP Donkey anti-Mouse, Invitrogen). Then membrane was imaged by LICOR Odyssey® Imager.
Protein expression, purification, and fluorescence labeling
A plasmid encoding MBP (maltose binding protein)-H3C-TDP-TEV-COVER-His6 was transformed into Escherichia coli strain LOBSTR (Kerafast) which were cultured at 37 °C in 2XYT media in the presence of ampicillin and chloramphenicol an optical density value 0.8 at 600 nm, induced with 0.25 mM isopropyl β-d-thiogalactoside (IPTG) and then cultured for a further 16 h at 20 °C. Harvested cells were resuspended in buffer A (25 mM Tris-HCl pH 7.5, 5 mM MgCl2 10% glycerol, 5 mM β-mercaptoethanol, and 1 mM NaN3) plus 1 M NaCl and 5 mM imidazole. For purification, cells were lysed by sonication, then clarified lysates were loaded onto a Ni2+ affinity column (Ni-NTA Superflow; Qiagen), washed in buffer A plus 300 mM NaCl and 20 mM imidazole, and eluted in buffer A plus 300 mM NaCl and 400 mM imidazole. For cleavage of MBP, proteins were buffer exchanged to buffer A plus 300mM NaCl and 40mM imidazole, then incubated 48 hours at 4 °C with GST-tagged Prescission protease (human rhinovirus 3C protease). Cleavage reactions were passed through a Ni2+ affinity column again to remove MBP and GST-tagged Prescission protease. Elution fractions containing TDP-TEV-COVER-His6 were concentrated in centrifugal concentrators (Amicon Ultra, EMD Millipore) and further purified by size-exclusion chromatography (Superdex 200 Increase 10/300 GL; Cytiva) in gel filtration buffer (25 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT). Purified proteins were concentrated and stored at −80 °C for analysis.
TDP-43 LCD and its variant A326P were overexpressed in BL21 (DE3) chemically component cells (Transgene, Beijing). After the addition of 1 mM Isopropyl β-D-thiogalactoside (IPTG), cells were incubated at 37 °C for 12 h to induce the expression of the protein. Cells were harvested and resuspended with buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl), following sonication in 6 M guanidine hydrochloride buffer. The supernatant of cell lysate was loaded onto a Ni2+ column (GE Healthcare, USA) after filtration with a 0.22 μm filter. Proteins were then eluted by the denatured elution buffer consisting of 50 mM Tris-HCl, pH 8.0, 6 M guanidine hydrochloride, and 100 mM imidazole. The eluted fraction was concentrated to a concentration above 30 mg/mL, and proteins were desalted into the buffer (20 mM MES, pH 6.0) for further experiment.
TDP-43 ΔLCD was overexpressed in BL21 (DE3) chemically component cells. Protein expression was induced by adding 0.5 mM IPTG at 25 °C for 12 h. Cells were collected and lysed in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 2 mM β-mercaptoethanol, 0.1 mg/ml RNase A, and 2 mM PMSF. After filtration, the cell lysates were load onto the Ni column and eluted by elution buffer consisting of 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 250 mM imidazole, and 2 mM β-mercaptoethanol. Eluted proteins were stored at −80 °C, and they were desalted into the buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 2 mM DTT) before the experiment. Full-length MBP tagged TDP-43 (TDP-43-MBP) was overexpressed in BL21 (DE3) pLysS Chemically Competent Cells (Transgene, Beijing). Proteins were induced by adding 1mM IPTG, and cells were cultured at 16 °C overnight. Cells were harvested and lysed in 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 2 mM DTT, 10% glycerol, 1 mM EDTA and 2 mM PMSF. The cell lysates were centrifuged and filtered before loading onto the MBP Trap HP column. Proteins were then eluted with 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 2 mM DTT, 10% glycerol, and 10 mM maltose. Eluted fractions were further purified over the size exclusion chromatography (Superdex 200 pg, GE, USA) in buffer containing 50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 2 mM DTT.
HSPB1 wild-type (HSPB1 WT) and its point mutant HSPB13D were overexpressed in BL21 (DE3) chemically component cells and proteins were induced by 0.5 mM IPTG. After cultured at 25 °C for 16 h, cells were collected and lysed in buffer containing 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 5% glycerol. The cell lysates were centrifuged and filtered before loading onto the Ni column. Following elution with the elution buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 2 mM β-mercaptoethanol), the collected fraction was further purified by the size exclusion chromatography (Superdex 75 pg, GE, USA) in buffer containing 50 mM PB, 50 mM NaCl, pH 7.5.
Proteins for NMR spectroscopy were expressed with E. coli, incubated in the M9 minimal medium with 15N-labeled NHCl4 (1 g/L) as the sole nitrogen source. Purification of 15N-labeled proteins followed the same procedures as that for the unlabeled proteins.
For fluorescence labeling, Alexa-488 (A10254, Invitrogen, USA) was for TDP-43-MBP, Alexa-647 (A20347, Invitrogen, USA) was for TDP-43 ΔLCD, OregonGreen488 (Invitrogen, O6149) was for TDP-43 LCD and Alexa-555 (A20346, Invitrogen, USA) was for HSPB1 WT and 3D. All the labeling experiments were performed as described by the manufacturer.
In vitro phase separation of protein and imaging
For in vitro co-liquid-liquid phase separation assay, purified TDP-43 LCD and HSPB1 were diluted into the buffer (150 mM NaCl, 50 mM Tris, pH 7.5) at final concentrations of 50 μM (TDP-43 LCD) and 10 μM (HSPB1), respectively. And the phase separation of TDP-43 ΔLCD (50 μM) and HSPB1 (10 μM) were induced in the buffer containing 60 mM NaCl, 50 mM Tris, pH 7.5. In the co-LLPS system of TDP-43 (MBP-tagged or MBP tag-removed by adding TEV protease) TDP-43 and HSPB1, 50 μM TDP-43 and 10 μM HSPB1 were performed in the buffer with 150 mM NaCl, 50 mM Tris, pH 7.5, and 7.5% Dextran 70. For fluorescence imaging, a 1/50 molar ratio of fluorescence-labeled protein was added to the system. Images were collected by Leica TCS SP8 microscope with a 100 × objective (oil immersion, NA= 1.4) at room temperature.
Saturation concentration measurement
A titration of TDP-43Clover-HIS was imaged by CQ1 confocal microscope to get the standard curve of fluorescence intensity and concentration. The same setting was used to image U2OS cells expressing TDP-43ΔNLS-Clover or TDP-43ΔNLS/5FL-Clover. The concentration of diffuse TDP-43 was calculated from the mean fluorescence intensity of the region of the cytoplasm subtracted of droplets’ region (recognized by auto threshold method) done by Image J.
Nuclear Magnetic Resonance assay
All NMR titration experiments were performed at 298 K on a Bruker 900 MHz spectrometer equipped with a cryogenic probe in an NMR buffer of 50 mM HEPES (pH 7.0), 50 mM NaCl, and 10% D2O. Each NMR sample was made with a volume of 500 μL, containing 10 μM 15N labeled TDP-43 LCD. We performed the titration experiment by addition of 5 μM or 10 μM HSPB1 and its variant respectively. Bruker standard pulse sequence (hsqcetfpf3gpsi) was used to collect the 2D 1H-15N HSQC spectrum with 32 scans, and 2048 × 160 complex points were used for 1H (14 ppm) and 15N (21 ppm) dimension, respectively. Backbone assignment of TDP-43 LCD was accomplished according to the previous publication12. All NMR data were processed by NMRPipe123 and analyzed by Sparky124.
Thioflavin-T (ThT) fluorescence kinetic assay
ThT fluorescence kinetic assay was performed with 10 μM TDP-43 LCDHIS in the buffer containing 20 mM MES, pH 6.0, 50 mM NaCl, 4 mM DTT, 50 mM ThT and 0.05% NaN3. The mixture was applied to a 384-well plate (Thermo Fisher Scientific, USA). The ThT fluorescence was monitored by a FLUO star Omega Rational Microplate Reader (BMG LABTECH) with excitation at 440 nm and emission at 485 nm at 37 °C, and the plate was shaken at 900 rpm with orbital shaking mode.
Transmission Electron Microscopy
After agitation in orbital shaker for 36 hours, TDP-43 LCD and TDP-43 LCD/HSPB1 samples from the ThT assay are loaded on the charged carbon film grids. The uranyl acetate (2%, v/v) is used to stain the samples for 45 s. Then, the grids were washed with ddH2O twice before the experiment. The TEM images are captured by the Tecnai G2 Spirit transmission electron microscope. The accelerating voltage of TEM was set to 120 kV, which could avoid the irradiation damage of samples.
Statistical analysis
Statistical analyses of quantification result were performed using GraphPad Prism. The statistical significance of the differences between two groups was investigated by two-tailed unpaired t test. The number of cells analyzed per experiment is provided in the corresponding figure legends.
Extended Data
Extended Data Fig. 1 ∣. HSP70 inhibition, proteasome inhibition, or arsenite-mediated stress induces cytoplasmic TDP-43 de-mixing independent of stress granule.
(a) Schematic of experimental design to characterize stress induced TDP-43 de-mixing droplets independent of stress granules or RNA binding. (b) Representative images of induced expression of cytoplasmic TDP-43 (TDP-43ΔNLS-Clover) for 1 day or 2 days in U2OS cells. (c) Boxplot of relative mean fluorescence intensity of diffuse TDP-43 in the U2OS cells. Number of cells quantified are 70 and 41, respectively. The cells are from one experiment. (d) Fluorescence intensity curve of recombinant TDP-43Clover at different concentration. (e) Concentration of TDP-43ΔNLS/5FL-Clover in the diffuse pool of cells with de-mixing droplets calculated based on the standard curve. Seven cells are analyzed. (f) Representative images of induction of cytoplasmic RNA binding deficient TDP-43 (TDP-43ΔNLS/2KQ-Clover) droplets by 10 μM proteasome inhibitor (MG132), 50 μM HSP70 inhibitor (VER155008) or 250 μM NaAsO2 treatment. (g) Representative images of stress granules (EIF3η) and cytoplasmic TDP-43ΔNLS-Clover or TDP-43ΔNLS/2KQ-Clover de-mixing droplets under NaAsO2, NaAsO2/cycloheximide, VER155008 or MG132 treatment. Percent of TDP-43 droplets showing no recruitment of EIF3η was labeled on the top of merged images. (h) Representative fluorescence images of TDP-43ΔNLS-Clover de-mixing droplets (green) and Poly-A RNA (oligo-dT FISH; red). Percent of TDP-43 droplets showing no enrichment of Poly-A RNA was labeled on the top of merged images. (i) Representative images of the induction of TDP-43ΔNLS/5FL-Clover (green) de-mixing droplets and stress granules (red) by live cell imaging. (j-k) Circularity of TDP-43 droplets formed after 1 hr, 2 hr, 3 hr and 4 hr of 250 μM NaAsO2 treatment. j: TDP-43ΔNLS-Clover; k: TDP-43ΔNLS/2KQ-Clover (l-m) Area of TDP-43 droplets formed after 1 hr, 2 hr, 3 hr and 4 hr of sodium arsenite treatment. l: TDP-43ΔNLS-Clover; m: TDP-43ΔNLS/2KQ-Clover. Number of droplets quantified are indicated on the figures. Images are from one live cell imaging experiment. (c,e,l,m) Medians, 25th and 75th percentiles are shown in the boxes; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
Extended Data Fig. 2 ∣. Proteasome inhibition, or arsenite-mediated stress rapidly converts liquid droplets of cytoplasmic TDP-43 into gels/solids.
(a-c) Representative images of FRAP analysis of cytoplasmic TDP-43ΔNLS/5FL-Clover droplets under (a) no stress but at higher accumulated level, (b) proteasome inhibition, and (c) arsenite stress. (d) FRAP curves of cytoplasmic TDP-43ΔNLS/5FL-Clover droplets in (a-c). Light color lines, S.D.; Number of droplets analyzed in no stress, proteasome inhibition, HSP70 chaperone inhibition and arsenite stress conditions are 5, 3, 6 and 11, respectively, from three independent experiments. (e) Schematic of experimental design for TDP-43 droplet dissolution assay by mild cell permeabilization. (f-h) Representative images of U2OS cells containing TDP-43ΔNLS-Clover droplets which recruit nuclear TDP-43mRuby2 under no stress (f, h) or 2 hour of 250 μM NaAsO2 treatment (g) after permeabilization with 50 μg/mL digitonin. (i) Relative level of TDP-43 in U2OS cells that did or did not form TDP-43ΔNLS-Clover de-mixing droplets after 2 hour of 250 μM NaAsO2 treatment comparing to the endogenous TDP-43 level in naïve U2OS nucleus. N: 211, 106, 51, respectively, from an experiment. (j) Representative image of dynamical arrest of liquid TDP-43 into droplets after arsenite treatment. (k) Examples of U2OS cells that form large, elongated droplets when cytoplasmic TDP-43ΔNLS/5FL-Clover is accumulated with time. (l) Area of the droplets increased with time. N: 32, 150, 390, 488. Data are from one live cell imaging experiment. (m) Circularity of the droplets in different sizes. N: 244, 158, 28, 15, 13, 11. Data are from an experiment. (n) FRAP of small and big TDP-43 droplets. FRAP curve are from three big droplets formed after three days of expression. (o) Circularity of RRM-del TDP-43 droplets were not changed by size of the droplets. Numbers quantified are indicated in the figure. Data are from an experiment (p) Solubility of TDP-43 variants after arsenite-induced phase separation. Fluorescence images of TDP-43 variants in lysates from U2OS cells treated with 250 μM sodium arsenite and lysed with RIPA buffer. Western blot of the solution and insoluble fractions. Images were taken at 10 frames with similar results in two independent experiments. (l,m,o) Medians, 25th and 75th percentiles are shown in the boxes; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
Extended Data Fig. 3 ∣. Proximity labelling of cytoplasmic TDP-43 de-mixing structures and verification of HSPB1 partition into cytoplasmic TDP-43 de-mixing structures in iPSC-cortical and motor neurons.
(a) Representative images of proximity labeling by cytoplasmic TDP-43ΔNLS-Clover-APEX2 in diffuse (no stress) and de-mixed state (sodium arsenite, MG132). Images represents 10 independent images taken for each condition. (b) Representative images of proximity labeling by Clover-APEX2NES under no stress, sodium arsenite and MG132 treatment conditions. Images represents 10 independent images taken for each condition. (c) Volcano plots of statistical significance against fold-change (TDP-43ΔNLS-Clover-APEX v.s. Clover-APEX2NES) of each protein under no stress, sodium arsenite and MG132 treatment conditions. P-value is calculated by one-sample t-test. (d) Representative immunofluorescence images of HSPB1 enriched in cytoplasmic TDP-43ΔNLS-Clover droplets induced by sodium arsenite. Images represents 10 independent images taken for each condition. (e) Representative immunofluorescence images of HSPB1 enriched in cytoplasmic TDP-43ΔNLS/2KQ-Clover droplets in iPSC-derived cortical neurons and motor neurons. Images represents 5 independent images taken for each condition.
Extended Data Fig. 4 ∣. HSPB1 inhibits TDP-43 phase separation at higher molecular ratio.
(a) SDS-PAGE analysis of all TDP-43 variants and HSPB1 variants used for in vitro phase separation assay and NMR analysis in figure 4 and supplementary figure 4, 5. Images represent analysis from three independent runs. (b) SDS-PAGE analysis of TDP-43 and HSPB1 phase separation samples after adding TEV protease to cleave MBP tag off. Images represent analysis from three independent runs. (c) Fluorescence images of in vitro phase separated droplets of untagged TDP-43 (2% NHS-Alexa488 labeled) and HSPB1 (2% NHS-Alexa555 labeled). (d) DIC images of mixtures of TDP-43 and HSPB1 at different concentrations. (e) Measurement of the size of de-mixed droplets in different conditions. Data of over 20 droplets from three independent experiments are presented as mean values ± SD. *** p<0.001 one-way ANOVA analysis. (f) Phase diagram of TDP-43 in (e). The size of dots represents the size of droplets formed at that condition.
Extended Data Fig. 5 ∣. HSPB1 binds to TDP-43 LCD through the conserved transient α-helix region (320-340 aa) and binds to RRM1 domain.
(a) Fluorescence images of in vitro phase separated TDP-43 (2% NHS-Alexa488 labeled) droplets with or without phosphor-mimetic HSPB13SD (2% NHS-Alexa555 labeled). Phase separation of 50 μM TDP-43-MBP was conducted by adding 7.5% dextran with or without 10 μM HSPB13SD. (b) Fluorescence images of in vitro phase separated TDP-43 LCD (50 μM, 2% NHS-Alexa488 labeled) droplets with or without HSPB13SD (50 μM, 2% NHS-Alexa555 labeled). (c) Thioflavin T aggregation assay to monitor the TDP-43 LCD (10 μM) amyloid assembly over time in the presence or absence of HSPB13SD at different molecular ratios. Data are collected from three biological replicates. Data are presented as mean values ± SD. (d) The 2D 1H15N HSQC spectra of 15N-labeled TDP-43 LCD titrated with increasing concentrations of HSPB1 (left). The representative residues that are markedly attenuated by HSPB1 titration are shown in right panel. (e-f) Profiles of the intensity changes (top) and chemical shift perturbations (bottom) of 20 μM 15N-labeled TDP-43 LCD in the presence of 20 (e) and 10 μM (f) HSPB13D, respectively. (g) Intensity changes of signals in the 2D 1H15N HSQC spectra of 20 μM 15N-labeled TDP-43 LCDA326P with 20 μM or 10 μM HSPB1. Data represents analysis from three independent runs. (h-i) Representative fluorescence images of (h) TDP-43ΔNLS/ΔRRM1-Clover (green) and (i) TDP-43ΔNLS/ΔRRM2-Clover (green) and HSPB1 (red) in U2OS cells. Images represent 10 independent images for each condition. (j) Immunoprecipitation of HSPB1 by full-length TDP-43 and RRM1-containing variant TDP-43ΔNLS/ΔRRM2-Clover but not RRM1-deletion variant TDP-43ΔNLS/ΔRRM1-Clover.
Extended Data Fig. 6 ∣. HSPB1, HSP70/HSC70 and BAG2 are partitioned into the sodium arsenite-induced TDP-43 gels/solids.
(a) Live imaging of TDP-43ΔNLS-Clover and HSPB1mCherry in U2OS cells treated by NaAsO2. (b-j) Immunofluorescence images of TDP-43ΔNLS-Clover and (b) HSPB1, (c) HSP70/HSC70, (d) BAG2, (e) HSPB8, (f) HSPH1, (g) BAG3, (h) DNAJA1, (i) DNAJB1 and (j) BAG1 in U2OS cells treated with sodium arsenite for 80 min or 120 min. Images represent 10 independent images for each condition. (k) Summary of the result in (b-j).
Extended Data Fig. 7 ∣. Reduction in HSPB1, HSPA1A, BAG2 or mild inhibition of HSP70 activity inhibits or delays the disassembly of cytoplasmic TDP-43 de-mixed droplets.
(a) Immunofluorescence images of HSPB1 and HSP70 in U2OS cells transfected with control siRNA, siHSPB1 or siHSPA1A. (b) Immunofluorescence images of BAG2 in U2OS cells transfected with control siRNA, siBAG2. (c) Quantification of fluorescence intensity of HSPB1, HSP70 and BAG2 in cells transfected with control siRNA, siHSPB1, siHSPA1A or siBAG2. Number of cells for quantification are indicated in the figure (633, 73, 633, 511, 481, 297, respectively). Images are from one experiment. P < 0.0001 (student t-test, two-tailed). Medians, 25th and 75th percentiles are labeled as lines in the plots. (d-g) Time lapse images of the disassembly of cytoplasmic TDP-43ΔNLS/2KQ-Clover de-mixing droplets in U2OS cells transfected with (d) control siRNA, (e) siHSPB1, (f) siHSPA1A, (g) siBAG2. (h) Time lapse images of TDP-43ΔNLS/2KQ-Clover in U2OS cells after 10 μM VER155008 treatment. Quantification of U2OS cells containing cytoplasmic TDP-43ΔNLS/2KQ-Clover de-mixing droplets after 10 μM VER155008 treatment. The number of cells for quantification is 117. (i) Time lapse images of the disassembly of cytoplasmic TDP-43ΔNLS-Clover de-mixing droplets in U2OS cells after washing off sodium arsenite with or without 10 μM VER155008 in the medium.
Extended Data Fig. 8 ∣. Reduction in HSC70/HSPA8 induces the expression of the HSP70 family member HSPA1A, inhibits the arsenite-induced de-mixing of cytoplasmic TDP-43, and promotes droplet/gel disassembly.
(a) Immunofluorescence images of HSC70 (HSPA8) and HSP70 (HSPA1A/HSPA1B) in U2OS cells transfected with control siRNA, siHSPA8, siHSPA1A or co-transfected with siHSPA8 and siHSPA1A. (b-c) Quantification of fluorescence intensity of (b) HSP70 and (c) HSC70 in (a). Each dot represents a single cell and orange lines represent mean value and standard error (S.E.M.) of fluorescence intensity. The number of cells quantified are 407, 261, 220 and 149, respectively. Images are taken from one experiment. (d-e) Time lapse imaging of TDP-43ΔNLS-Clover de-mixing droplets induction by arsenite stress and disassembly after removal of sodium arsenite in cells transfected with (d) siHSPA8 or (e) control siRNA. (f) Quantification of U2OS cells containing cytoplasmic TDP-43ΔNLS/2KQ-Clover de-mixing droplets after arsenite treatment (left) and quantification of U2OS cells that have TDP-43 de-mixing droplets disassembled after stress removal (right) in (d-e). Cells are quantified from four independent replicates. N: 346, 425, 309, 354 for siRNA control group; 86,88, 86, 69 for siHSPA8 group. (g) Fluorescence intensity of TDP-43ΔNLS-Clover in U2OS cells transfected with siRNA control or siHSPA8 (up) and percentage of TDP-43ΔNLS-Clover in de-mixing droplets (bottom). ***P < 0.0001, n.s. P=0.2159 (student t-test, two-tailed). Data are presented as mean values ± SEM. Number of cells quantified in siRNA control group are 42 and in siHSPA8 group are 39. Cells are pooled from four independent experiments. (h) Time lapse images of TDP-43ΔNLS/2KQ-Clover expressing U2OS cells transfected with siHSPA8 after sodium arsenite treatment and wash-off. (i) Representative images of U2OS cells expressing TDP-43ΔNLS-Clover together with HSPA1AmRuby2 or HSPA8mRuby2 after 2 hour of sodium arsenite treatment. Images represent 10 independent images.
Extended Data Fig. 9 ∣. enhanced recruitment of HSP70 and its co-chaperones DNAJB1 and BAG3 to phase separated cytoplasmic TDP-43 droplets after stress removal and assembled microtubule array is required for disassembly of TDP-43 droplets.
(a) Fluorescence images of TDP-43ΔNLS-Clover (green) with BAG3 (red), HSP70/HSC70 (red), DNAJB1 (red), and DNAJA1 (red), respectively, in U2OS cells treated with sodium arsenite for 1 hour followed by 4-hour wash-off. Images represent 10 independent images for each condition. (b-c) Microtubule disassembly by nocodazole treatment. Microtubule structures are imaged by sir-Tubulin dye. (d) Schematic design of testing if microtubule disassembly affects TDP-43 droplets fuse/aggregate/coalesce. (e) Microtubule disassembly does not strongly affect TDP-43 droplet fusion/aggregation but affect the resolution of the droplets.
Extended Data Fig. 10 ∣. Reduction in HSPB1 induces cytoplasmic TDP-43 de-mixing and mislocalization.
(a) Experimental design for testing the effect of HSPB1 depletion on cytoplasmic TDP-43 de-mixing in cell cycle arrested U2OS cells. (b) DNA content analysis by FACS. U2OS cells are treated by reduced serum medium and 1 μM G1 cell cycle blocker palbociclib to block cell cycle. Line is drawn to separate 2-N and 4-N cells based on FACS plot of cell population. (c) Fluorescence images of TDP-43ΔNLS/5FL-Clover (upper), TDP-43ΔNLS-Clover (medium) and Clover (bottom) in cell cycle arrested U2OS cells transfected with siRNA control or siHSPB1 after induction with doxycycline for 1 day or 2 days. (d) Quantification of the percentage of cells forming cytoplasmic TDP-43 de-mixing droplets in (c). Numbers of cells quantified are 4108, 3632, 3897 (TDP-43ΔNLS/5FL-Clover, siRNA control, 1 day) and 2901, 2978, 2897 (TDP-43ΔNLS/5FL-Clover, siRNA control, 2 day), 4035, 3788, 3673 (TDP-43ΔNLS/5FL-Clover, siHSPB1, 1 day) and 3663, 3266, 3278 (TDP-43ΔNLS/5FL-Clover, siHSPB1, 2 day), 4203, 4399, 4272 (TDP-43ΔNLS-Clover, siRNA control, 1 day) and 3777, 3957, 3709 (TDP-43ΔNLS-Clover, siRNA control, 2 day), 3913, 3803, 3829 (TDP-43ΔNLS-Clover, siHSPB1, 1 day) and 3469, 3358, 3345 (TDP-43ΔNLS-Clover, siHSPB1, 2 day). Data are presented as mean values ± SD. Each data are from three independent experiments. (e) Fluorescence images of endogenous TDP-43 and HSPB1 in U2OS cells transfected with siRNA control or siHSPB1 and quantification of cytoplasmic/nuclear fluorescence intensity of TDP-43 in cells expressing different levels of HSPB1. The number of cells for plotting are 114, 883, 519 and 292, respectively. <200 group V.S. 200-400 group, n.s. P = 0.2878; <200 group V.S. 400-600 group, **P = 0.0005; <200 group V.S. >600 group, **P=0.027; 200-400 group V.S. 400-600 group, ***P < 0.0001; 200-400 group V.S. >600 group, ***P = 0.0001 (unpaired student t-test, two-tailed). Images are pooled from two independent experiments. (f) Fluorescence images of endogenous TDP-43 and HSPB1 in U2OS cells transfected with control siRNA or siHSPB1 and quantification of cells forming cytoplasmic de-mixing TDP-43 droplets. The number of quantified cells for control siRNA is 586 and the number of cells for siHSPB1 is 175. Data are from an experiment.
Supplementary Material
Acknowledgements
We thank Jennifer Santini at the UCSD Microscopy Core, Eric Griffis at the UCSD Nikon Imaging Center and David Jenkins from SMD group of Ludwig Institute for assistance with imaging and image analysis. We thank Ying Jones at Electron Microscopy Core Facility of UCSD for epoxy resin embedding and sectioning. We are grateful for helpful discussions from Zevik Melamed, Cong Chen, Melinda S. Beccari, Jone Lopez-Erauskin, Dong Hyun Kim and Prasad Trivedi and experimental help from Yifu Jin and Noorhan Monther. DWC acknowledges support from the NIH (R01 NS27036) and the Nomis Foundation, and JRY acknowledges support from NIH (P41 GM103533), JR acknowledges grants from Target ALS (20134792), National Institute of Neurological Diseases and Stroke (NIH R01NS088578 and NS047101), Kraatz Family/Nicholas Martin Jr. Family Foundation and Pam Golden, OAA acknowledges National Science Foundation Graduate Research Fellowship (DGE-1650112), SVS acknowledges ALS association (21-PDF-583). We acknowledge the UCSD School of Medicine Microscopy Core Grant P30 NS047101. We acknowledge staff members of the National Facility for Protein Science in Shanghai, Zhangjiang Laboratory, China for providing technical support and assistance in the NMR data collection.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Quantitative mass spectrometry datasets have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD035001. BacTrap RNA seq data from mouse is available under accession code GSE74724. snRNA seq data of mouse spinal cord is available under accession code GSE161621. The human genome project datasets analyzed are available from Project MinE77 (http://databrowser.projectmine.com/), the ALS Variant Server78 (http://als.umassmed.edu/), the ALS Data Browser79 (http://alsdb.org/), and the ALS Knowledge Portal80 (http://alskp.org/). Further information on the post-mortem samples analyzed are available from the UCSD ALS tissue repository (contact information please find in https://health.ucsd.edu/specialties/neuro/specialty-programs/als-clinic/pages/default.aspx). Source data are provided with this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Ling S-C, Polymenidou M & Cleveland DW Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann M et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Neumann M et al. Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta neuropathologica 113, 543–548 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Josephs KA et al. TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta neuropathologica 127, 811–824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson PT et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 142, 1503–1527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel A et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Molliex A et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasset-Rosa F et al. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357. e337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang A et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. The EMBO journal 37, e97452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann JR et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGurk L et al. Poly (ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Molecular cell 71, 703–717. e709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conicella AE et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proceedings of the National Academy of Sciences 117, 5883–5894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C et al. Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle lncRNA NEAT1-mediated liquid-liquid phase separation. Molecular Cell 79, 443–458. e447 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Yu H et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganassi M et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Molecular cell 63, 796–810 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kedersha N & Anderson P Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochemical Society Transactions 30, 963–969 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Hartl FU & Hayer-Hartl M Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Tyedmers J, Mogk A & Bukau B Cellular strategies for controlling protein aggregation. Nature reviews Molecular cell biology 11, 777–788 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Hartl FU, Bracher A & Hayer-Hartl M Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Macario AJ, Grippo TM & de Macario EC Genetic disorders involving molecular-chaperone genes: a perspective. Genetics in Medicine 7, 3–12 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Sarparanta J, Jonson PH, Kawan S & Udd B.J.I.j.o.m.s. Neuromuscular diseases due to chaperone mutations: a review and some new results. International journal of molecular sciences 21, 1409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brehme M et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell reports 9, 1135–1150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voisine C, Pedersen JS & Morimoto RI Chaperone networks: tipping the balance in protein folding diseases. Neurobiology of disease 40, 12–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haslbeck M, Franzmann T, Weinfurtner D & Buchner J Some like it hot: the structure and function of small heat-shock proteins. Nature structural molecular biology 12, 842–846 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Shashidharamurthy R, Koteiche HA, Dong J & McHaourab HS Mechanism of chaperone function in small heat shock proteins: dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. Journal of Biological Chemistry 280, 5281–5289 (2005). [DOI] [PubMed] [Google Scholar]
- 26.D'Angelo MA, Raices M, Panowski SH & Hetzer MW Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertens J et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell stem cell 17, 705–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen TJ et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nature communications 6, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Wander CM, Yuan C-X, Bereman MS & Cohen TJ Acetylation-induced TDP-43 pathology is suppressed by an HSF1-dependent chaperone program. Nature communications 8, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller JN, Hanni KB & Markesbery WR Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mechanisms of ageing and development 113, 61–70 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Keller JN, Huang F.F.a. & Markesbery WR Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience 98, 149–156 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Buratti E & Baralle FE Characterization and Functional Implications of the RNA Binding Properties of Nuclear Factor TDP-43, a Novel Splicing Regulator ofCFTR Exon 9. Journal of Biological Chemistry 276, 36337–36343 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Elden AC et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon TS, Holehouse AS, Rosen MK & Pappu RV Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai T et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and Biophysical Research Communications 351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Polymenidou M et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayala YM et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 30, 277–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt HB & Rohatgi R In Vivo Formation of Vacuolated Multi-phase Compartments Lacking Membranes. Cell Reports 16, 1228–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee H-W et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam SS et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobingier BT et al. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360. e312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paek J et al. Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell 169, 338–349. e311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson BS et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. Journal of Biological Chemistry 284, 20329–20339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babinchak WM et al. The role of liquid–liquid phase separation in aggregation of the TDP-43 low-complexity domain. Journal of Biological Chemistry 294, 6306–6317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenoy J et al. Structural dissection of amyloid aggregates of TDP-43 and its C-terminal fragments TDP-35 and TDP-16. The FEBS journal 287, 2449–2467 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Cao Q, Boyer DR, Sawaya MR, Ge P & Eisenberg DS Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nature structural & molecular biology 26, 619–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuo X-F et al. Solid-state NMR reveals the structural transformation of the TDP-43 amyloidogenic region upon fibrillation. Journal of the American Chemical Society 142, 3412–3421 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Babinchak WM & Surewicz WK Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nature communications 12, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landry J et al. Human Hsp27 Is Phosphorylated at Serines-78 and Serines-82 by Heat-Shock and Mitogen-Activated Kinases That Recognize the Same Amino-Acid Motif as S6 Kinase-Ii. Journal of Biological Chemistry 267, 794–803 (1992). [PubMed] [Google Scholar]
- 50.Gaestel M et al. Identification of the Phosphorylation Sites of the Murine Small Heat-Shock Protein Hsp25. Journal of Biological Chemistry 266, 14721–14724 (1991). [PubMed] [Google Scholar]
- 51.Liu Z et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nature structural & molecular biology 27, 363–372 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Conicella AE, Zerze GH, Mittal J & Fawzi NL ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt HB, Barreau A & Rohatgi R Phase separation-deficient TDP43 remains functional in splicing. Nature communications 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrnsperger M, Gräber S, Gaestel M & Buchner J Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. Nature structural & molecular biology 16, 221–229 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee GJ, Roseman AM, Saibil HR & Vierling E A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. The EMBO journal 16, 659–671 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng G, Basha E, Wysocki VH & Vierling E Insights into small heat shock protein and substrate structure during chaperone action derived from hydrogen/deuterium exchange and mass spectrometry. Journal of Biological Chemistry 283, 26634–26642 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Żwirowski S et al. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. The EMBO journal 36, 783–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sirtori R, Riva C, Ferrarese C & Sala GJNL HSPA8 knock-down induces the accumulation of neurodegenerative disorder-associated proteins. Neuroscience Letters 736, 135272 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Cheng YC et al. Knocking down of heat-shock protein 27 directs differentiation of functional glutamatergic neurons from placenta-derived multipotent cells. Sci Rep 6, 30314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirbach BB & Golenhofen N Differential expression and induction of small heat shock proteins in rat brain and cultured hippocampal neurons. J Neurosci Res 89, 162–175 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Williams KL, Rahimtula M & Mearow KM Heat shock protein 27 is involved in neurite extension and branching of dorsal root ganglion neurons in vitro. Journal of Neuroscience Research 84, 716–723 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Benn SC et al. Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron 36, 45–56 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Kalmar B, Burnstock G, Vrbova G & Greensmith L The effect of neonatal nerve injury on the expression of heat shock proteins in developing rat motoneurones. J Neurotrauma 19, 667–679 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Sun SY et al. Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. P Natl Acad Sci USA 112, E6993–E7002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blum JA et al. Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat Neurosci 24, 572–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sunkin SM et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41, D996–D1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A & Ponstingl H RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci U S A 91, 2587–2591 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorlich D, Pante N, Kutay U, Aebi U & Bischoff FR Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15, 5584–5594 (1996). [PMC free article] [PubMed] [Google Scholar]
- 69.Grima JC et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron 94, 93–107 e106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gasset-Rosa F et al. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron 94, 48–57 e44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinoshita Y et al. Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 68, 1184–1192 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Zhang K et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shang J et al. Aberrant distributions of nuclear pore complex proteins in ALS mice and ALS patients. Neuroscience 350, 158–168 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Capponi S et al. Molecular chaperones in the pathogenesis of amyotrophic lateral sclerosis: the role of HSPB1. Human mutation 37, 1202–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katz M et al. Mutations in heat shock protein beta-1 (HSPB1) are associated with a range of clinical phenotypes related to different patterns of motor neuron dysfunction: A case series. J Neurol Sci 413, 116809 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Dierick I et al. Genetic variant in the HSPB1 promoter region impairs the HSP27 stress response. Human mutation 28, 830–830 (2007). [DOI] [PubMed] [Google Scholar]
- 77.van der Spek RA et al. The project MinE databrowser: bringing large-scale whole-genome sequencing in ALS to researchers and the public. Amyotrophic Lateral Sclerosis Frontotemporal Degeneration 20, 432–440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicolas A et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97, 1268-+ (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cirulli ET et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farhan SMK et al. Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat Neurosci 22, 1966–1974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conicella AE et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proceedings of the National Academy of Sciences 117, 5883–5894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciechanover A & Kwon YT Protein quality control by molecular chaperones in neurodegeneration. Frontiers in neuroscience 11, 185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roodveldt C et al. Chaperone proteostasis in Parkinson's disease: Stabilization of the Hsp70/α-synuclein complex by Hip. The EMBO journal 28, 3758–3770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Auluck PK, Chan HE, Trojanowski JQ, Lee VM-Y & Bonini NM Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295, 865–868 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Udan-Johns M et al. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Human molecular genetics 23, 157–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hageman J et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Molecular cell 37, 355–369 (2010). [DOI] [PubMed] [Google Scholar]
- 87.Novoselov SS et al. Molecular chaperone mediated late-stage neuroprotection in the SOD1G93A mouse model of amyotrophic lateral sclerosis. PLoS One 8, e73944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wacker JL et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. Journal of Neuroscience 29, 9104–9114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H-J et al. The heat shock response plays an important role in TDP-43 clearance: evidence for dysfunction in amyotrophic lateral sclerosis. Brain 139, 1417–1432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharp PS et al. Protective effects of heat shock protein 27 in a model of ALS occur in the early stages of disease progression. Neurobiology of disease 30, 42–55 (2008). [DOI] [PubMed] [Google Scholar]
- 91.Crippa V et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Human molecular genetics 19, 3440–3456 (2010). [DOI] [PubMed] [Google Scholar]
- 92.Bourdenx M et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 184, 2696–2714 e2625 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hayes D, Napoli V, Mazurkie A, Stafford WF & Graceffa P Phosphorylation dependence of hsp27 multimeric size and molecular chaperone function. Journal of Biological Chemistry 284, 18801–18807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alderson TR et al. Local unfolding of the HSP27 monomer regulates chaperone activity. Nature communications 10, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clouser AF et al. Interplay of disordered and ordered regions of a human small heat shock protein yields an ensemble of ‘quasi-ordered’states. Elife 8, e50259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yih LH, Huang HM, Jan KY & Lee TC Sodium arsenite induces ATP depletion and mitochondrial damage in HeLa cells. Cell Biol Int Rep 15, 253–264 (1991). [DOI] [PubMed] [Google Scholar]
- 97.Chanda D, Kim SJ, Lee IK, Shong M & Choi HS Sodium arsenite induces orphan nuclear receptor SHP gene expression via AMP-activated protein kinase to inhibit gluconeogenic enzyme gene expression. Am J Physiol Endocrinol Metab 295, E368–379 (2008). [DOI] [PubMed] [Google Scholar]
- 98.Mann JR et al. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 102, 321–338 e328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.French RL et al. Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J Biol Chem 294, 6696–6709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen HJ et al. RRM adjacent TARDBP mutations disrupt RNA binding and enhance TDP-43 proteinopathy. Brain 142, 3753–3770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pirie E et al. S-nitrosylated TDP-43 triggers aggregation, cell-to-cell spread, and neurotoxicity in hiPSCs and in vivo models of ALS/FTD. Proceedings of the National Academy of Sciences 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen TJ, Hwang AW, Unger T, Trojanowski JQ & Lee VMY Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. The EMBO journal 31, 1241–1252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang C.-k., Chiang M.-h., Toh EK-W, Chang C-F & Huang T.-h. Molecular mechanism of oxidation-induced TDP-43 RRM1 aggregation and loss of function. FEBS letters 587, 575–582 (2013). [DOI] [PubMed] [Google Scholar]
- 104.Irobi J et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nature genetics 36, 597–601 (2004). [DOI] [PubMed] [Google Scholar]
- 105.Boczek EE et al. HspB8 prevents aberrant phase transitions of FUS by chaperoning its folded RNA binding domain. bioRxiv, 2021.2004.2013.439588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maxwell BA et al. Ubiquitination is essential for recovery of cellular activities after heat shock. 372, eabc3593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gwon Y et al. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. 372, eabf6548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Faust O et al. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 587, 489-+ (2020). [DOI] [PubMed] [Google Scholar]
- 109.Ismailov S et al. A new locus for autosomal dominant Charcot-Marie-Tooth disease type 2 (CMT2F) maps to chromosome 7q11-q21. European Journal of Human Genetics 9, 646–650 (2001). [DOI] [PubMed] [Google Scholar]
- 110.Ylikallio E et al. Truncated HSPB1 causes axonal neuropathy and impairs tolerance to unfolded protein stress. BBA clinical 3, 233–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Evgrafov OV et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nature genetics 36, 602–606 (2004). [DOI] [PubMed] [Google Scholar]
- 112.Benndorf R, Martin JL, Pond SLK & Wertheim JO Neuropathy-and myopathy-associated mutations in human small heat shock proteins: characteristics and evolutionary history of the mutation sites. Mutation Research/Reviews in Mutation 761, 15–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Houlden H et al. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology 71, 1660–1668 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Almeida-Souza L et al. Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot-Marie-Tooth neuropathy. J Biol Chem 285, 12778–12786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Almeida-Souza L et al. Small heat-shock protein HSPB1 mutants stabilize microtubules in Charcot-Marie-Tooth neuropathy. J Neurosci 31, 15320–15328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.d'Ydewalle C et al. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat Med 17, 968–974 (2011). [DOI] [PubMed] [Google Scholar]
Method-only References
- 117.Fernandopulle MS et al. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr Protoc Cell Biol 79, e51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McAlister GC et al. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Analytical Chemistry 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He L, Diedrich J, Chu YY & Yates JR 3rd Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal Chem 87, 11361–11367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu T et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129, 16–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tabb DL, McDonald WH & Yates JR 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1, 21–26 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park SK et al. Census 2: isobaric labeling data analysis. Bioinformatics 30, 2208–2209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 124.Lee W, Tonelli M & Markley JL NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Quantitative mass spectrometry datasets have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD035001. BacTrap RNA seq data from mouse is available under accession code GSE74724. snRNA seq data of mouse spinal cord is available under accession code GSE161621. The human genome project datasets analyzed are available from Project MinE77 (http://databrowser.projectmine.com/), the ALS Variant Server78 (http://als.umassmed.edu/), the ALS Data Browser79 (http://alsdb.org/), and the ALS Knowledge Portal80 (http://alskp.org/). Further information on the post-mortem samples analyzed are available from the UCSD ALS tissue repository (contact information please find in https://health.ucsd.edu/specialties/neuro/specialty-programs/als-clinic/pages/default.aspx). Source data are provided with this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.