Summary
Post-translational modifications (PTMs) on histones play essential roles in cell fate decisions during development. However, how these PTMs are recognized and coordinated remains to be fully illuminated. Here, we show that BRPF1, a multi-histone binding module protein, is essential for pluripotency in human embryonic stem cells (ESCs). BRPF1, H3K4me3, and H3K23ac substantially co-occupy the open chromatin and stemness genes in hESCs. BRPF1 deletion impairs H3K23ac in hESCs and leads to closed chromatin accessibility on stemness genes and hESC differentiation as well. Deletion of the N terminal or PHD-zinc knuckle-PHD (PZP) module in BRPF1 completely impairs its functions in hESCs while PWWP module deletion partially impacts the function. In sum, we reveal BRPF1, the multi-histone binding module protein that bridges the crosstalk between different histone modifications in hESCs to maintain pluripotency.
Subject areas: Genetics, Molecular genetics, Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
BRPF1 maintains pluripotency in human embryonic stem cells (hESCs)
-
•
BRPF1 co-localizes with H3K4me3 and H3K23ac on open-chromatin in hESCs
-
•
BRPF1 deletion impairs H3K23ac and reduces chromatin accessibility in stemness genes
Genetics; Molecular genetics; Cell biology; Stem cells research
Introduction
Epigenetics is critical to regulate cell fate decisions and cell identities during development. At the early embryonic stage , the inner cell mass (ICM) cells in the blastocyst give rise to three primary germ cell layers (i.e. endoderm, mesoderm, and ectoderm) that further contribute to the whole embryo.1 Derived from ICM, embryonic stem cells (ESCs) retain the pluripotency to differentiate into various functionally distinct cell types while possessing the unique capability of indefinite self-renewal in vitro.2,3,4,5 The developmental pluripotency of cultured mouse ESCs (mESCs) has been proven by generating an entire mouse.6 Human ESCs are also capable of generating three germ layers in teratoma formation in vivo or differentiation in vitro.2,3 Accumulated evidence demonstrates that ESC pluripotency is maintained by a set of pluripotent stemness genes, such as Oct4, Nanog, and Sox2.7,8,9,10 Forced expression of these stemness factors successfully induced pluripotency in somatic cells (iPSCs).11,12,13
The stemness gene network in ESCs involves a complex interplay between transcription factors (TFs) and epigenetic mechanisms such as post-translational modifications (PTMs) on histones.10,14,15 For instance, ESCs exhibit unique chromatin modifications that the active promoters of stemness genes are hallmarked by histone H3K4 tri-methylation (me3).14,15 A subset of H3K4me3 associated genes are also occupied by an inactive histone modification, H3K27 tri-methylation (me3).14,15,16 These so-called “bivalent” genes contain critical developmental genes to be maintained at a “low” but “ready” transcription level in ESCs.14,15,16,17 Epigenetic regulators that mediate H3K4me3 including Trithorax group (TrxG) components, or H3K27me3 including Polycomb group (PcG) proteins play critical roles in regulating cell differentiation during developmental processes.18 Deletion of these factors impaired pluripotency and down-regulated stemness genes in mouse or human ESCs,19,20,21,22,23 demonstrating their essential roles in maintaining pluripotency in PSCs.
Histone modifications are recognized by specific proteins (readers) to carry out their intracellular biological processes.24,25,26 Readers that recognize histone lysine methylation have been characterized containing one or more specific domains, such as PHD (plant homeodomain), PWWP (Pro-Trp-Trp-Pro), BAH (bromo-adjacent homology), MBT (malignant brain tumor), WD40, and so forth.25,26,27 Malfunction of these histone readers has been linked to various developmental abnormalities and human diseases including cancer.28,29 For example, EED, a WD40 containing protein that recognizes H3K27me3, plays an essential role in ensuring the repressive function of H3K27me3 during normal embryonic development.30,31,32 The well-characterized reading domain for H3K4me3 is the PHD finger module.33,34,35,36,37 For example, TAF3, a member of the basal transcription complex (TFIID) containing a PHD finger, recognizes H3K4me3 and promotes gene transcription.37 Other PHD finger proteins, such as ING2, ING4, and ING5, were reported to regulate H3K4me3 functions in different cell models.35,38,39 To date, as a hallmark for stemness genes in ESCs, how H3K4me3 is recognized and coordinated with other histone PTMs remains largely unknown.
BRPF1 is a large multi-histone binding module protein and conserved from C elegans to humans.40,41 It is known as a core subunit of the native monocytic leukemic zinc-finger (MOZ) and related factor (MORF) acetyltransferase complexes that catalyze histone acetylation (ac), particularly H3K23ac.41,42,43 Interestingly, in addition to a MOZ/MORF binding domain, it contains three potential histone binding modules, i.e. PHD-zinc finger knuckle-PHD (PZP) module, bromodomain (BD) and PWWP (Pro-Trp-Trp-Pro) module (Figure 1A). Brpf1−/− mouse embryos were lethal at E9.5, and the phenotype was much more severe than those of Moz and Morf deficient mice,40,44,45,46 indicating its functions independent of MOZ/MORF. Human BRPF1 gene was shown to be recurrently mutated in childhood leukemia28 and adult medulloblastoma.47 Mutations in Human BRPF1 are also linked to intellectual disabilities and congenital abnormalities.48,49,50 Conditional genetic studies based on mouse and fish models revealed that BRPF1 regulates various adult tissues such as brain development, axial skeleton, and the hematopoietic system.40,46,51,52,53,54
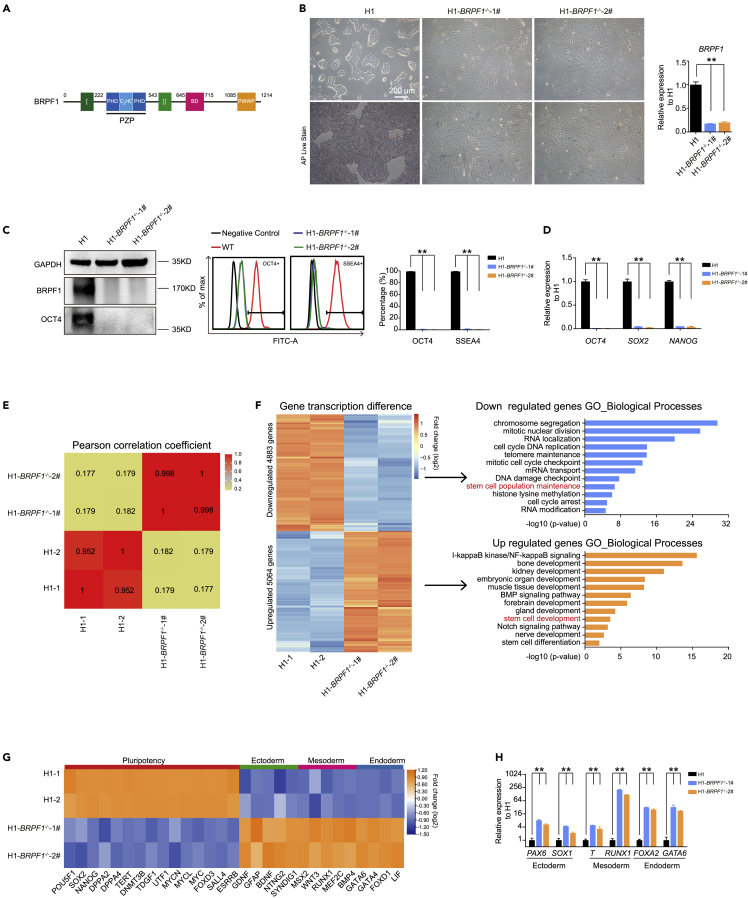
Figure 1.
BRPF1 is essential in human ESC pluripotency and self-renewal
(A) Overview of the BRPF1 domain architecture. I, enhancer of polycomb-I; II, enhancer of polycomb-II; PZP , PHD-zinc knuckle-PHD; BD, bromodomain; PWWP, Pro-Trp-Trp-Pro containing domain.
(B) Morphology of WT and BRPF1−/− H1 hESCs. Alkaline phosphatase (ALP) staining and expression of BRPF1 were examined in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t-tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3). Scale bar, 200 μm.
(C). OCT4 and BRPF1 proteins were examined in WT and two BRPF1−/− H1 hESC clones by western blot or FACS analysis. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(D) Examination of these genes’ expression of OCT4, NANOG, and SOX2 in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(E) Pearson’s correlation coefficient analysis of transcriptomes in WT and two BRPF1−/− H1 hESC clones.
(F) Heatmap and gene ontology (GO) analysis of up- or down-regulated genes in two BRPF1−/− H1 hESC clones compared with WT H1 hESCs.
(G) RNA-seq data of selected pluripotent or three germ layer genes in WT and two BRPF1−/− H1 hESC clones.
(H) Examination of the expression of lineage genes in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3). All error bars throughout the figure represent the SD from three independent replicates (n = 3).
In this study, we reveal that BRPF1 occupies H3K4me3 and coordinates the crosstalk between H3K4me3 and H3K23 acetylation (ac) in human ESCs, which is essential for pluripotency.
Results
BRPF1 is required to maintain pluripotency and self-renewal in human embryonic stem cells
To examine the role of BRPF1 in human ESCs, we generated BRPF1 knock-out in hESCs (Figures 1B and S1A–S1C). BRPF1−/− hESCs displayed a typical differentiated morphology and lost alkaline phosphatase (ALP) staining (Figure 1B). OCT4, the critical pluripotent factor, was significantly down-regulated or lost in BRPF1−/− hESCs based on western blot or FACS assays (Figure 1C). Other PSC genes such as NANOG and SOX2 also lost expression in BRPF1−/− hESCs (Figure 1D). RNA-seq analysis revealed a distinct transcriptome profile between wild-type (WT) and BRPF1−/− hESCs (Figure 1E). The down-regulated genes in BRPF1−/− hESCs contained biological functions related to stem cell self-renewal as well as fundamental cellular processes, while the developmental genes for somatic tissues were up-regulated (Figures 1F and S1F). Selected genes related to PSC pluripotency were reduced while those for three early germ layers were up-regulated in BRPF1−/− hESCs (Figure 1G). The up-regulation of selected differentiation genes was further confirmed by qRT-PCR and Immunofluorescence (Figures 1H and S1D). Self-renewal was also impaired in BRPF1−/− hESCs (Figure S1E). Together, we demonstrate that the multi-histone binding module protein, BRPF1 is essential for maintaining hESC pluripotency and self-renewal.
Histone H3 lysine 23 acetylation is impaired in BRPF1−/− human embryonic stem cells
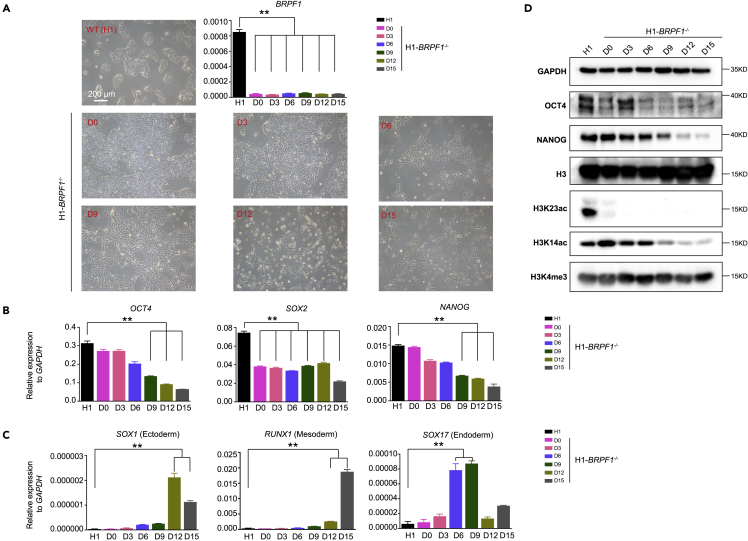
BRPF1 is known as a core subunit of MOZ/MORF or HBO1 acetyltransferase complexes that mainly catalyzes H3K23ac.42,48,53,55 We then examined the H3K23ac level in BRPF1−/− hESCs as well as other histone modifications such as H3K4me3 and H3K14ac. We noticed that the pluripotency loss in BRPF1−/− hESCs was a gradual process (Figure 2A). The pluripotent genes such as OCT4, NANOG, and SOX2 showed a gradual down-regulation in BRPF1−/− hESCs upon continued culture (Figure 2B). Three germ layer genes were gradually up-regulated in this process (Figure 2C). Consistently, the OCT4 and NANOG proteins exhibited a gradual decrease during the culture of BRPF1−/− hESCs (Figure 2D). In terms of histone PTMs, H3K4me3 was well maintained while in contrast, H3K23ac was completely lost in BRPF1−/− hESCs in BRPF1−/− hESCs (Figure 2D). H3K14ac was well maintained at the initial culture stage but lost later when pluripotency exited (Figure 2D). These data indicate that BRPF1 is essential for hESC pluripotency by regulating H3K23ac. To further confirm the role of BRPF1 in hESCs, we performed a rescue experiment in BRPF1−/− hESCs. We introduced an inducible system to over-express (OE) BRPF1 in H1 hESCs (Figure S2A). We performed gene targeting to knockout the endogenous BRPF1 in H1-BRPF1-OE cells (KO-OE-BRPF1) (Figure S2B). The expression of BRPF1 was maintained by DOX treatment. Upon DOX treatment, BRPF1−/− hESCs maintained a typical pluripotent state, while withdrawal of DOX led to complete loss of pluripotency (Figures S2C and S2D). Furthermore, H3K14ac and H3K23ac were restored upon DOX treatment but lost again in the absence of DOX (Figure S2E). Together, these data demonstrate that BRPF1 maintains hESC pluripotency by regulating H3K23ac.
Figure 2.
Loss of histone H3 lysine 23 acetylation in BRPF1−/− hESCs
(A) Morphology of WT and BRPF1−/− H1 hESCs cultured at different time points. Scale bar, 200 μm. qRT-PCR analysis of the expression level of BRPF1 in WT and BRPF1−/− H1 hESCs cultured at different time points. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(B and C) Examination of the expression of the selected (B) pluripotent and (C) lineage genes in BRPF1−/− H1 hESCs cultured at different time points. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(D) Examination of H3K23ac, H3K14ac, H3K4me3 or OCT4, NANOG in BRPF1−/− H1 hESCs cultured at different time points by western blot. All error bars throughout the figure represent the SD (standard deviation) from three independent replicates (n = 3).
Reduced chromatin accessibility of stemness genes in BRPF1−/− human embryonic stem cells
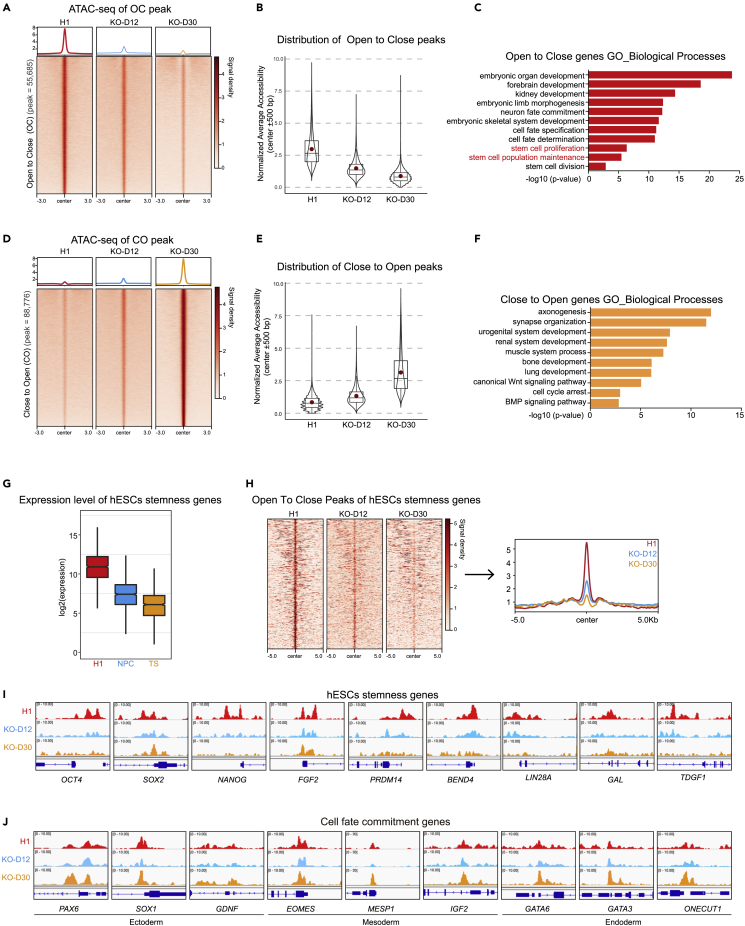
To understand the underlying molecular mechanism of pluripotency loss in BRPF1−/− hESCs, we first examined the global chromatin accessibility through ATAC-seq. We performed the ATAC-seq assay in WT and BRPF1−/− hESCs at two culture time points (Day 12 and Day 30), as BRPF1−/− hESCs completely exited pluripotency at day 12 in culture (Figure 2). Based on ATAC-seq, a panel of genes showed reduced chromatin accessibility in BRPF1−/− hESCs at day 12 and day 30 of culture (Figures 3A and 3B). These open to close (OC) genes were enriched in functions related to stem cell maintenance as well as development (Figure 3C). Another panel of genes showed increased chromatin accessibility in BRPF1−/− hESCs (close to open, CO) (Figures 3D and 3E). These CO genes contained adult tissue developmental genes (Figure 3F). We selected a panel of hPSC stemness genes that were highly expressed in hESCs56,57 but not neural progenitor cells (NPCs)58 or the hESC-derived trophoblast stem cells (TSCs)59 (Figure 3G). These selected hPSC stemness genes exhibited dramatically reduced chromatin accessibility in BRPF1−/− hESCs cultured at day 12 and day 30 (Figure 3H). For example, pluripotent genes such as OCT4, NANOG as well as other stemness genes showed dramatically reduced chromatin accessibility in BRPF1−/− hESCs (Figure 3I). In contrast, the developmental lineage genes showed more opened chromatin in BRPF1−/− hESCs (Figure 3J).60 Together, these data demonstrate that BRPF1 promotes the chromatin accessibility of stemness genes in hESCs.
Figure 3.
Decreased chromatin accessibility of stemness genes in BRPF1−/− H1 hESCs
(A) Heatmap and signal densities of ATAC-seq data for OC accessible chromatin regions in WT and BRPF1−/− H1 hESCs (D12, D30). The signal around a ±3 kb window centered on OC peaks center in the indicated cells. OC, open to closed, indicates regions with reduced accessibility in BRPF1−/− H1 hESCs.
(B) Box-plot of signal densities of the ATAC-seq data for OC accessible chromatin regions in the indicated cells.
(C) GO analysis of genes associated with OC regions is shown.
(D) Heatmap and signal densities of ATAC-seq data for CO accessible chromatin regions in WT and BRPF1−/− H1 hESCs (D12, D30). The signal around a ±3 kb window centered on CO peaks center in the indicated cells. CO, close to open, indicates regions with increased accessibility in BRPF1−/− H1 hESCs.
(E) Box-plot of signal densities of the ATAC-seq data for CO accessible chromatin regions in the indicated cells.
(F) GO analysis of genes associated with CO regions is shown.
(G) Box-plot of transcription levels of selected stemness genes in H1 hESCs, H1 hESC-derived NPCs, and trophoblast stem (TS) cells.
(H) Heatmap and signal densities of ATAC data in regions associated with selected stemness genes. ATAC-seq signal around a ±5 kb window centered on OC peaks in the indicated cells.
(I and J) Genomic view of ATAC-seq data on selected genes with different functions in H1 and BRPF1−/− hESCs. These selected genes include (I) hESCs stemness genes and (J) cell fate commitment genes.
BRPF1, H3K4me3, and H3K23ac co-occupy fundamental and stemness genes in human embryonic stem cells
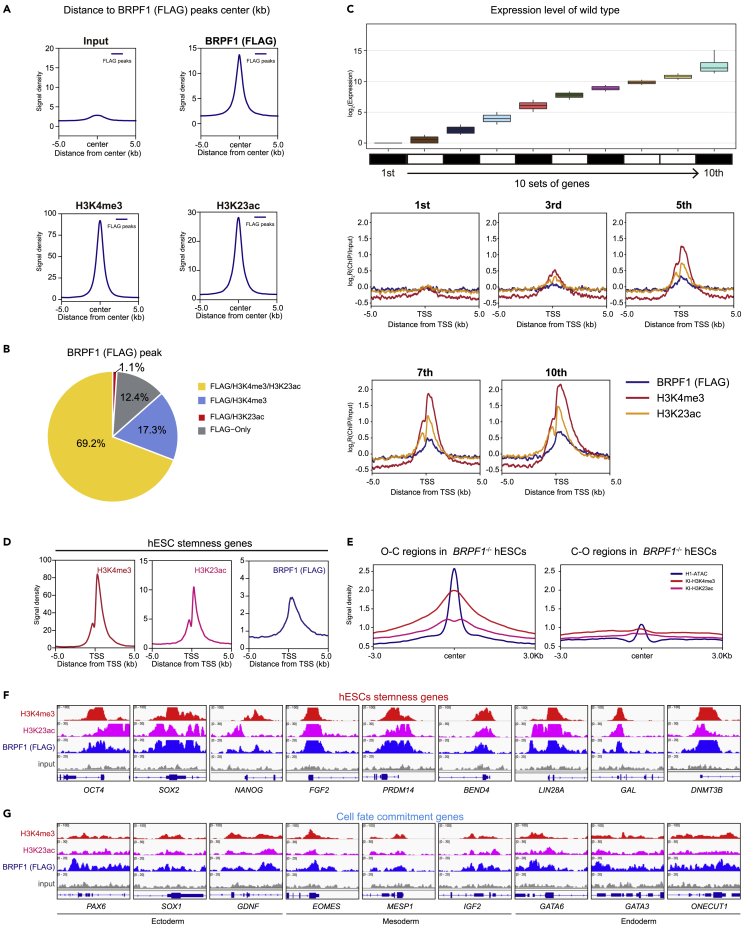
To examine the whole genome binding of BRPF1 in hESCs, we generated a triple-FLAG knock-in (KI) into the BRPF1 locus in hESCs (KI_H1-hESCs) through gene targeting (Figures S3A–S3F). The expression of Flag-tagged BRPF1 was verified by western blotting (Figure S3D). Through ChIP-seq, we revealed a substantial co-localization between BRPF1, H3K4me3, and H3K23ac across the whole genome in hESCs (Figure 4A). Specifically, over 80% of BRPF1 peaks were co-occupied by H3K4me3 and/or H3K23ac (Figure 4B). We then examined the co-relation between gene activity and BRPF1 and/or H3K23ac, H3K4me3 occupancy. We broke down the whole genes by decile based on their expression level detected by RNA-seq (Figure 4C, upper panel). As shown in Figure 4C, the expression activities were highly co-related with the binding intensities of triple factors on the promoters, as the lowest expressors showed lowest binding intensities of triple factors, while as the expression increased, triple-factor intensities increased (Figure 4C, lower panel). These data indicate that BRPF1 binds and promotes the expression of those self-renewal and stemness genes in hESCs. Indeed, the selected stemness genes (Figure 3C) were highly enriched for triple-factors (Figure 4D), and H3K4me3 and H3K23ac were co-enriched in the open chromatin region in WT KI_hESCs (Figure 4E). For example, many well-known pluripotent genes, such as OCT4, SOX2, NANOG, and so forth, were highly enriched for triple factors (Figure 4F). Notably, the developmental genes that were associated with H3K4me3 and H3K27me3 (bivalent)14,15,16,17 showed medium binding of BRPF1 and H3K23ac, which is consistent with their low but steady transcription levels in hESCs (Figure 4G).
Figure 4.
Co-localization of BRPF1, H3K4me3, and H3K23ac on fundamental and stemness genes in human ESCs
(A) Signal densities for input, H3K4me3, BRPF1 (FLAG), and H3K23ac ChIP-seq data around ±5 kb centered on BRPF1 (FLAG) associated peaks in KI hESCs.
(B) BRPF1 (FLAG) associated peaks classification, H3K4me3/H3K23ac/FLAG co-localization peaks (yellow); H3K4me3/FLAG co-localization peaks (indigo); H3K23ac/FLAG co-localization peaks (red); and only FLAG localization peaks (none, gray).
(C) Upper panel, all genes from the H1 RNA-sep in Figure 1 broken down by decile of expression (1st being lowest 10% and 10th being highest 10%). lower panel, The average signal intensity of H3K4me3, BRPF1 (FLAG), and H3K23ac ± 5 kb around the TSS of 1st, 3st, 5st, 7st, and 10st.
(D) Signal densities for H3K4me3, BRPF1 (FLAG), and H3K23ac ChIP-seq data ±5 kb around the TSS of selected stemness genes in KI hESCs.
(E) Signal densities of H3K4me3, H3K23ac, and ATAC-seq data ±3 kb around the peak center of open to close (O-C) and close to open (C-O) in WT hESCs.
(F and G) Genomic views of signal densities of ChIP-seq data of H3K4me3 (red), BRPF1 (FLAG) (blue), and H3K23ac (purple) on selected genes in KI hESCs. These selected genes include (F) hESCs stemness genes and (G) cell fate commitment genes.
We then examined the genomic association of H3K4me3 in BRPF1−/− hESCs since BRPF1−/− hESCs showed steady levels of H3K4me3 (Figure 2D). We examined the localization of H3K4me3 in BRPF1−/− hESCs cultured at day 12 by ChIP-seq. H3K4me3-associated genes in BRPF1−/− hESCs were highly overlapped with those in WT cells (Figures S4A and S4B). However, H3K4me3 density was slightly reduced in BRPF1−/− hESCs, consistent with the western blot data. The open to closed (OC) genes based on ATAC-Seq showed a reduced but substantial level of H3K4me3 densities (Figure S4C). Particularly, even though chromatin accessibly was closed in pluripotent genes such as OCT4, SOX2, NANOG, and TERT, there were also substantial H3K4me3 modifications on these genes in BRPF1−/− hESCs (Figure S4D). These data suggest that BRPF1 is critical for reading H3K4me3 and medicating H3K23ac to maintain the open chromatin state in stemness genes.
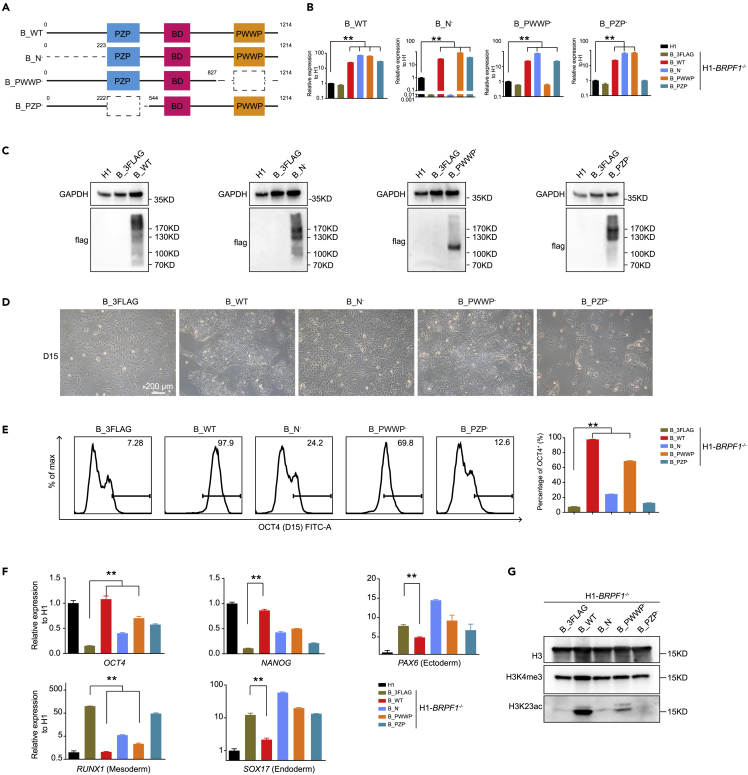
N terminal and plant homeodomain-zinc knuckle- plant homeodomain module are essential for BRPF1 to maintain human embryonic stem cell pluripotency
BRPF1 contains multiple histone binding modules (Figure 1A). How these modules interact with H3K4me3 and maintain chromatin accessibility of stemness genes in hESCs remains unknown. We then generated a panel of BRPF1 mutants with the deletion of each aforementioned individual module (Figure 5A). We then introduced lentiviral-based expression of WT or each mutant BRPF1 into the BRPF1−/− hESCs. The expression of lentiviral-based WT or each mutant BRPF1 was verified by qRT-PCR and western blotting (Figures 5B and 5C). The cultured BRPF1−/− hESCs exhibited distinct phenotypes upon expressing different BRPF1 mutants (Figure 5D). BRPF1−/− hESCs expressing WT BRPF1 fully restored pluripotency, while the control vector showed complete loss of pluripotency, consistent with the data shown in Figure S2 (Figure 5D). BRPF1−/− hESCs expressing N module or PZP module deleted BRPF1 mutants showed a completely differentiated phenotype, while PWWP deleted BRPF1 largely rescued pluripotency (Figure 5D). FACS data for OCT4 staining confirmed that the majority of BRPF1−/− hESCs expressing N module or PZP module deleted BRPF1 mutants lost pluripotency (Figure 5E), while the majority of BRPF1−/− hESCs expressing PWWP deleted BRPF1 mutants maintained pluripotency state (Figure 5E). These phenotypes were further confirmed by qRT-PCR data of the selected pluripotent genes as well as lineage genes (Figure 5F). Lastly, we revealed that WT BPRF1 restored the H3K23ac in BRPF1−/− hESCs, while the N or PZP module deleted mutant showed little effect on H3K23ac rescue (Figure 5G). In contrast, PWWP deleted BRPF1 largely rescued H3K23ac in BRPF1−/− hESCs (Figure 5G). Together, these data demonstrate that the N terminal and PZP module are most essential to the biological functions of BRPF1 in hESCs.
Figure 5.
Functional analysis of BRPF1 with the deletion of different histone binding modules
(A) Overview of wild-type BRPF1 and the BRPF1 mutants with domain architecture. Wild-type BRPF1 (B_WT); BRPF1 with N terminal deletion (B_N−); BRPF1 with PWWP module deletion (B_PWWP−); BRPF1 with PZP module deletion (B_PZP−). The dotted line indicates the deleted regions.
(B) qRT-PCR analysis of the expression with domain-specific primers. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(C) Western blot analysis of FLAG-tagged of overexpression of wild-type BRPF1 and each BRPF1 mutant in BRPF1−/− H1 hESCs. Overexpression of the negative control in BRPF1−/− H1 hESCs (B_3FLAG).
(D) Morphology of BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. Scale bar, 200 μm.
(E) FACS analysis of OCT4 in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(F) qRT-PCR analysis of the expression of the pluripotent genes OCT4, NANOG, and cell fate commitment genes PAX6, RUNX1, and SOX17 in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(G) Western blot analysis of H3K4me3 and H3K23ac in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. All error bars throughout the figure represent the SD (standard deviation) from three independent replicates (n = 3).
Discussion
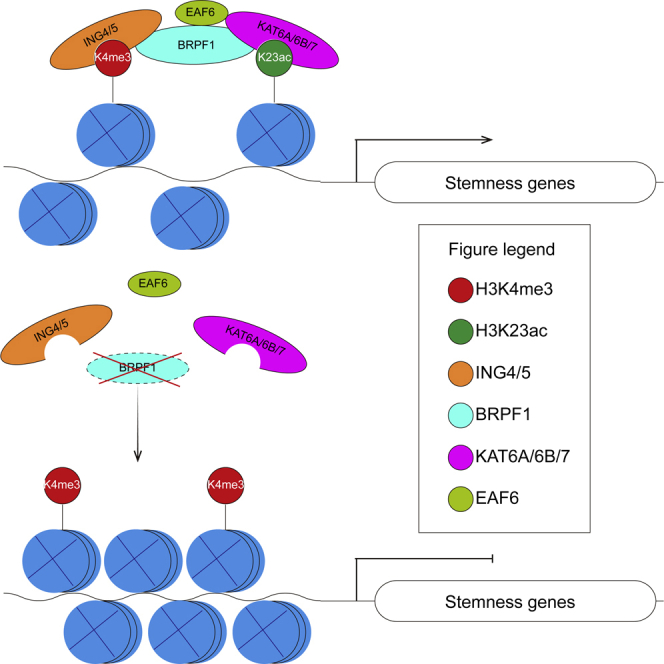
Histone recognition is critical for gene regulation and cell fate decisions through epigenetic mechanisms. The pluripotent stem cells represent a unique cell fate with both pluripotency and indefinite self-renewal. Over the past two decades, remarkable progresses have been made in the characterization of histone modifications in both mouse and human PSCs.14,16,61,62,63,64 H3K4me3, catalyzed by TrxG complex, was mapped to the stemness and fundamental gene promoters in PSCs.14,15 However, less has been known regarding how H3K4me3 is recognized in PSCs. In this study, we reveal that BRPF1, a multiple-histone module protein, co-occupies with H3K4me3 and H3K23ac on stemness and fundamental gene promoters in human ESCs. Deletion of BRPF1 impairs H3K23ac and leads to a closed chromatin state and down-regulation of pluripotent genes and then pluripotency loss in human ESCs. Our studies provide evidence that the cross-talk between different histone modifications mediated by the multiple histone reader, such as BRPF1, is essential for their biological readout to regulate cell fate decisions.
BRPF1 is an interesting large protein containing multi-histone reorganization modules, including an MOZ/MORF interaction domain and three potential histone-binding modules targeting different post-translational modifications (PTMs) (Figure 1A). Mouse embryos with Brpf1−/− died at E9.5, indicating an essential role of Brpf1 in embryogenesis.46 Our data that human ESCs with BRPF1−/− could not be maintained indicate that BRPF1 might play an essential role at the very early stage of human development. In addition, mutations in human BRPF1 have been linked to various cancers, such as leukemia and medulloblastoma.28,47 With its multiple-histone binding modules, BRPF1 might act as an adaptor to bridge the interaction of different epigenetic regulators. A short motif in its N terminal module is implicated in the interaction of MORF subunits.65 Consistently, BRPF1 with N terminal module deletion lost full function in hESCs (Figure 5). The PZP domain was shown to be responsible for interacting with the H3 tail as well as DNA.55,66,67 BRPF1PZP was proposed to stabilize MOZ/MORF complexes at chromatin with accessible DNA.67 In hESCs, BRPF1 promotes chromatin accessibility and BRPF1PZP is essential for its functions (Figure 5). BRPF1 also contains a PWWP module that is known to interact with H3K36me3,68 a PTM that is usually localized across the gene body and indicates transcription elongation.69,70 However, PWWP deletion did not severely impair BRPF1 function in hPSCs (Figure 5), indicating that BRPF1 mainly functions to regulate promoter activities rather than elongations, consistent with its major localization on H3K4me3 associated promoters (Figure 4). In summary, we reveal a critical role of the multi-histone binding modular protein BRPF1 to co-occupy with H3K4me3 and H3K23ac in human ESCs to support self-renewal. It will be interesting in future studies to explore its functions in fate decisions in adult stem cells or lineages such as neural lineages and hematopoietic lineages in the human background.
Limitations of the study
In this study, we show that BRPF1 co-localizes with H3K4me3 and H3K23ac on open-chromatin in human ESCs. Due to technical challenge, we lack a detailed mechanism how BRPF1 reads H3K4me3 and acts as a bridge for MYST acetyltransferase complexes to catalyze H3K23ac at the same site. Additionally, how the loss of H3K23ac reduces chromatin accessibility is also less clear based on current studies. Future studies, particularly biochemical experiments are needed to uncover the detailed molecular mechanisms that how BRPFs reads and co-ordinates different histone PTMs to realize their biological functions in cell fate decisions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-Conjugated GAPDH Monoclonal Antibody | Proteintech | Cat#HRP-60004; RRID: AB_2737588 |
| Rabbit Peregrin antibody [N1N2], N-term | GeneTex | Cat#GTX102778; RRID: AB_1949759 |
| mouse anti-OCT-3/4 | Santa Cruz Biotechnology | Cat#sc-5279; RRID: AB_628051 |
| Rabbit anti-HIST3H3 (H3) | Abcam | Cat#ab1791; RRID: AB_302613 |
| Rabbit anti-Histone H3 (acetyl K23) | Abcam | Cat#ab177275; RRID: AB_2927706 |
| Rabbit anti-Histone H3 (tri methyl K4) | Abcam | Cat#ab8580; RRID: AB_306649 |
| Rabbit anti-NANOG | Cell Signaling Technology | Cat#3580; RRID: AB_2150399 |
| mouse Monoclonal ANTI-FLAG | Sigma | Cat#F1804; RRID: AB_262044 |
| mouse anti-SSEA4 | Santa Cruz Biotechnology | Cat#sc-21704; RRID: AB_628289 |
| mouse anti-Glial Fibrillary Acidic Protein | Millipore | Cat#MAB360; RRID: AB_11212597 |
| Rabbit anti-CALPONIN | Abcam | Cat#AB46794; RRID: AB_2291941 |
| Rabbit anti-HNF3β/FOXA2 | Millipore | Cat#07–633; RRID: AB_390153 |
| Rabbit anti-Histone H3 (trimethyl K4) | Abcam | Cat#ab8580; RRID: AB_306649 |
| Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A-11004; RRID: AB_2534072 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A-11011; RRID: AB_143157 |
| Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11001; RRID: AB_2534069 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | Cat#354277 |
| DMEM/F12 basic | Gibco | Cat#C11330500BT |
| mTeSR1 | STEMCELL Technology | Cat#85850 |
| Accutase | Sigma | Cat#A6964 |
| PBS | GENOM | Cat#GNM20012-2 |
| EDTA | Genstar | Cat#VA17876-500g |
| Penicillin-Streptomycin (100×) | Hyclone | Cat#SV30010 |
| Y27632 | Selleck | Cat#S6390 |
| DMSO | Sigma | Cat#D1435 |
| Thiazovivin | Selleck | Cat#S1459 |
| DAPI | Sigma | Cat#D9542 |
| NcTarget | Nuwacell | Cat#RP01020 |
| Puromycin | Gibco | Cat#A1113803 |
| TRI Reagent | Molecular Research Center | Cat#TR118 |
| Critical commercial assays | ||
| FastPure Gel DNA Extraction Mini Kit | Vazyme | Cat#DC301 |
| pEASY-Uni Seamless Cloning and Assembly Kit | TransGen Biotech | Cat#CU101 |
| E.Z.N.A. Endo-Free Plasmid DNA Midi Kit | OMEGA | Cat#D6915 |
| DNA Ligation Kit Ver.2.1 | TaKaRa | Cat#6022 |
| Equalbit dsDNA HS Assay Kit | Vazyme | Cat#EQ111 |
| AMAXA Basic Nucleofector Kit for Prim. Mammalian Epithelial cells | Lonza | Cat#VPI-1005 |
| mRNA-seq V3 Library Prep Kit for Illumina | Vazyme | Cat#NR611 |
| RNA Adapters set3 - set6 for Illumina | Vazyme | Cat#N809 |
| EdU Flow Cytometry Assay Kits | Invitrogen | Cat#C10425 |
| Universal DNA Library Prep Kit for Illumina | Vazyme | Cat#ND607 |
| TruePrep DNA Library Prep Kit V2 for Illumina | Vazyme | Cat#TD501 |
| Deposited data | ||
| RNA-Seq, ATAC-seq, and ChIP-seq data | This paper | GEO: GSE213695 |
| RNA-Seq, ATAC-seq, and ChIP-seq data | This paper | HRA001928 |
| Experimental models: Cell lines | ||
| Human embryonic stem cell line H1 (WA01) | WiCell | RRID: CVCL_9771 |
| Oligonucleotides | ||
| KO-BRPF1-sgRNA: ATGGTGGAGGTGGACTTGCA |
This paper | N/A |
| KO-BRPF1-LA forward primer: GAGCATGCCTAGATTGTCGCG |
This paper | N/A |
| KO-BRPF1-LA reverse primer: CTTTTTCTTGTGCTTGCGGAGTG |
This paper | N/A |
| KO-BRPF1-RA forward primer: GGAGAACACTGAGACACCAGCTGC |
This paper | N/A |
| KO-BRPF1-RA reverse primer: CATCAGAATGCCTTTTTTGCCTTG |
This paper | N/A |
| KO-BRPF1 forward primer: CCACAACTTGCGGGCGACTA |
This paper | N/A |
| KO-BRPF1 reverse primer: GGTGGGGCATCAGGGGTGT |
This paper | N/A |
| KI-BRPF1-sgRNA: TGCGGTGCTGCAGAGCCCTG |
This paper | N/A |
| KI-BRPF1-LA forward primer: CTGGATGCTCTGGACCTCGTGT |
This paper | N/A |
| KI-BRPF1-LA reverse primer: ATCACTATCGCTGGTCTCACTGCTC |
This paper | N/A |
| KI-BRPF1-RA forward primer: TACTGCTCAACACAGCCCAACCTATA |
This paper | N/A |
| KI-BRPF1-RA reverse primer: GCAGCGGATTGACCCTTGTGT |
This paper | N/A |
| KI-BRPF1 forward primer: CTTGTTCTCCCTGAGATGATTTATTTGAT |
This paper | N/A |
| KI-BRPF1 reverse primer: TTACAGTATTTACAACAGCAGGGGAGG |
This paper | N/A |
| OE-BRPF1-WT forward primer: ATGGGGGTGGACTTTGATGTGAAGA |
This paper | N/A |
| OE-BRPF1-WT reverse primer: TCAATCACTATCGCTGGTCTCACTG |
This paper | N/A |
| OE-BRPF1-N- forward primer: ATCTGGCTGGATATCATGAATGAGC |
This paper | N/A |
| OE-BRPF1-N- reverse primer: TCAATCACTATCGCTGGTCTCACTG |
This paper | N/A |
| OE-BRPF1-PWWP- forward primer: ATGGGGGTGGACTTTGATGTGAAGA |
This paper | N/A |
| OE-BRPF1-PWWP- reverse primer: TCGCTGATGGGCAAGCTTCC |
This paper | N/A |
| OE-BRPF1-PZP- −1 forward primer: ATGGGGGTGGACTTTGATGTGAAGA |
This paper | N/A |
| OE-BRPF1-PZP- −1 reverse primer: GTAGTCCTCCTCGTCCATGTCATACTCT |
This paper | N/A |
| OE-BRPF1-PZP- −2 forward primer: AGAGTATGACATGGACGAGGAGGACT ACTGGACACTGAAGCGGCAGTC |
This paper | N/A |
| OE-BRPF1-PZP- −2 reverse primer: TCAATCACTATCGCTGGTCTCACTG |
This paper | N/A |
| FUW-BRPF1 forward primer: ATGGGGGTGGACTTTGATGTGAA |
This paper | N/A |
| FUW-BRPF1 reverse primer: ATCACTATCGCTGGTCTCACTGCTCT |
This paper | N/A |
| GAPDH forward primer: GGAGCGAGATCCCTCCAAAAT |
This paper | N/A |
| GAPDH reverse primer: GGCTGTTGTCATACTTCTCATGG |
This paper | N/A |
| OCT4 forward primer: CCTCACTTCACTGCACTGTA |
This paper | N/A |
| OCT4 reverse primer: CAGGTTTTCTTTCCCTAGCT |
This paper | N/A |
| SOX2 forward primer: CCCAGCAGACTTCACATGT |
This paper | N/A |
| SOX2 reverse primer: CCTCCCATTTCCCTCGTTTT |
This paper | N/A |
| NANOG forward primer: TGAACCTCAGCTACAAACAG |
This paper | N/A |
| NANOG reverse primer: TGGTGGTAGGAAGAGTAAAG |
This paper | N/A |
| PAX6 forward primer: ATGTGTGAGTAAAATTCTGGGCA |
This paper | N/A |
| PAX6 reverse primer: GCTTACAACTTCTGGAGTCGCTA |
This paper | N/A |
| SOX1 forward primer: AATTTTATTTTCGGCGTTGC |
This paper | N/A |
| SOX1 reverse primer: TGGGCTCTGTCTCTTAAATTTGT |
This paper | N/A |
| T forward primer: TATGAGCCTCGAATCCACATAGT |
This paper | N/A |
| T reverse primer: CCTCGTTCTGATAAGCAGTCAC |
This paper | N/A |
| RUNX1 forward primer: CTGCCCATCGCTTTCAAGGT |
This paper | N/A |
| RUNX1 reverse primer: GCCGAGTAGTTTTCATCATTGCC |
This paper | N/A |
| FOXA2 forward primer: ACTACCCCGGCTACGGTTC |
This paper | N/A |
| FOXA2 reverse primer: AGGCCCGTTTTGTTCGTGA |
This paper | N/A |
| GATA6 forward primer: CTCAGTTCCTACGCTTCGCAT |
This paper | N/A |
| GATA6 reverse primer: GTCGAGGTCAGTGAACAGCA |
This paper | N/A |
| SOX17 forward primer: CGCACGGAATTTGAACAGTA |
This paper | N/A |
| SOX17 reverse primer: GGATCAGGGACCTGTCACAC |
This paper | N/A |
| BRPF1 forward primer: CCCCTGAGAATGGCAGCAACA |
This paper | N/A |
| BRPF1 reverse primer: TGATATCCAGCCAGATGTAGTCCTCCT |
This paper | N/A |
| BRPF1 (WT) forward primer: GACACAGGCAACATCTTCAGCGA |
This paper | N/A |
| BRPF1 (WT) reverse primer: GCGGTAAGCCTCCAAGTTCTGC |
This paper | N/A |
| B_N− forward primer: GCTACTCCCAAGTCAGGCAAACATA |
This paper | N/A |
| B_N− reverse primer: ATGTCATACTCTACTTCCTCGTCCAGC |
This paper | N/A |
| B_PWWP− forward primer: GTGGAAACCAACCAGTGAAGAAGAGT |
This paper | N/A |
| B_PWWP− reverse primer: GGTATCCTCACTGCTGTCCTCTGG |
This paper | N/A |
| B_PZP− forward primer: ACGCCTCCAGGTTCAGCACG |
This paper | N/A |
| B_PZP− reverse primer: GGTCAGGCGGTTGGTGATTTTACT |
This paper | N/A |
| Software and algorithms | ||
| Integrative Genomics Viewer (IGV) | Robinson et al., 201160 | http://software.broadinstitute.org/software/igv/ |
| R version 4.1.0 | R Core Team, 202271 | https://www.r-project.org/ |
| Samtools | Li et al., 200972 | https://samtools.sourceforge.net/ |
| DeepTools | Ramirez et al., 201473 | https://deeptools.readthedocs.io/en/develop/ |
| Other | ||
| Culture plates (6 well) | Greiner Bio-one | Cat#657160 |
| Culture plates (12 well) | Greiner Bio-one | Cat#665180 |
| Culture plates (24 well) | Greiner Bio-one | Cat#662160 |
| Counting chambers | Marienfeld | Cat#0650030 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guangjin Pan (pan_guangjin@gibh.ac.cn).
Materials availability
The plasmids and supplementary information files generated in this study are available from the lead contact on reasonable request.
Experimental model and subject details
Cell culture of hESCs
We used mTeSR1 (STEMCELL Technologies) or ncTarget (nuwacell) to culture the human embryonic stem cell line WA01 (H1-male WiCell) on Matrigel (Corning)-coated plates at 37 °C and 5% CO2. The cell line used in the experiment were passaged every 3 days and the medium was changed every day.
Method details
Gene knock-out and knock-in in human ESCs
Guide RNAs (gRNAs) for the knock-out and knock-in of BRPF1 were designed on the website (CCTop, http://crispr.cos.uni-heidelberg.de).74 The gRNAs for CRISPR genome editing were separately inserted into the vector pX330, which can express the Cas9 protein. The Left and right homology arms of BRPF1 and a LoxP-flanked PGK-puromycin cassette were cloned into the donor plasmid, which was used for targeting. For 3xFLAG knock-in, the left homologous arm contained a 3 × FLAG tag. For the knock-out and knock-in of BRPF1, 4 μg of pX330 plasmid containing corresponding gRNAs and 4 μg of donor plasmid were co-electroporated into 1 million H1 hES cells by a Nucleofector 2b Device (Lonza). Then, these electroporated cells were cultured in mTeSR1 medium with 0.5 μM thiazovivin (Selleck) on Matrigel-coated six-well plates for 1 day. After 3–4 days, puromycin (1 μg mL−1, Gibco) was used to screen positive clones. These individual colonies were picked and cultured for further validation.
Genomic DNA from the knock-out and knock-in of BRPF1 cell clones extracted by the TIANamp Genomic DNA Kit (Tiangen) was used in all PCR experiments. For gene knock-out, the primers KO-F/R were used to amplify an ∼ 1.7 kb product of the targeted integration. For gene knock-in, the primer KI-F/R was used to amplify an ∼2 kb product of the targeted integration. All gRNA sequences and primer sequences are listed in the key resources table.
Inducible system for gene knockout in hESCs
We used an inducible over-expression (OE) system to over-expressing BRPF1 in H1 hESCs. These cells were selected with 2 μg/mL doxycycline (DOX) and puromycin (H1-BRPF1-OE). Then, endogenous BRPF1 was deleted in H1-BRPF1-OE cells (KO-OE-BRPF1). For the knock-out of BRPF1, 4 μg pX330 plasmid containing corresponding gRNA and 4 μg of donor plasmid were co-electroporated into 1 million H1 hES cells by Nucleofector 2b Device (Lonza). Then, these electroporated cells were cultured in mTeSR1 medium with 0.5 μM thiazovivin (Selleck) on Matrigel-coated six-well plates for 1 day. After 3–4 days, puromycin (1 μg mL−1, Gibco) was used to screen positive clones. These individual colonies were picked and cultured in mTeSR1 with 2 μg/mL DOX. All primer sequences are listed in the key resources table.
Overexpression of BRPF1 mutants in the BRPF1−/− hESCs
Wild type and all mutant cDNAs of human BRPF1 were obtained by PCR using high fidelity polymerase (Vazyme). These PCR products were inserted into the psin-3xFLAG vector using homologous recombination technology, respectively. Then, these plasmids were packaged into lentiviruses in 293T cells. 5×105 BRPF1−/− hESCs were cultured in mTeSR1 medium on Matrigel-coated 6-well plates for 1 day and infected with these lentivirus containing these above cDNAs for 1 day. Then, we used puromycin (1 μg mL−1, Gibco) to screen positive cells. These primer sequences are listed in the key resources table.
Quantitative real-time PCR
Total RNA of these cells was extracted by TRIzol (Invitrogen). Two micrograms of total RNA was reverse-transcribed into cDNA with oligo dT (Takara) and RT ACE (Toyobo), and then cDNA was amplified by qRT-PCR using ChamQ SYBR qPCR Master Mix (Vazyme) and a CFX96 machine (Bio-Rad). The qRT-PCR results were normalized to GAPDH. All data were analyzed with three replicates. These primer sequences are listed in the key resources table.
Alkaline phosphatase staining
To detect alkaline phosphatase (ALP) activity, The H1 and BRPF1 knock-out cells were cultured in ncTarget medium on Matrigel-coated 6-well plates. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. After washing three times with TBST, the ALP activity of cells were detected according to the instructions of the alkaline phosphatase assay kit (Beyotime).
EdU assay
A total of 5 × 105 H1 and BRPF1 knock-out cells were seeded in Matrigel-coated 6-well plates and cultured in ncTarget medium with 10 μM EdU or without EdU as a negative control for 1.5 h at 37 °C. The EdU assay was performed according to the specifications of the Click-iT™ EdU Pacific Blue™ flow cytometry assay kit (Invitrogen). Then, the samples were analyzed with a CytoFlox-S flow cytometer.
Western blot analysis
To detect the expression levels of related proteins, Knock-out and knock-in of BRPF1 cell clones were harvested. Cells were lysed on ice with RIPA buffer (Beyotime) for 10 minutes. After boiling with a third volume of 4×SDS loading buffer (Novex), whole-cell extracts were electrophoresed through 12% SDS-PAGE (Beyotime) and then transferred to PVDF membranes (Millipore). These PVDF membranes were blocked in 5% nonfat milk for approximately 2 h and incubated with primary antibodies overnight at 4 °C. After washing three times with TBST for 10 minutes each time, the membranes were incubated with HRP-conjugated secondary antibodies. After washing three times with TBST for 10 minutes each time, ECL (Beyotime) was used as a substrate for detection. The membranes were visualized with a GelView 6000Plus (BLT). The information for antibodies is listed in key resources table.
Flow cytometry analysis
These samples of cells were digested into single cells by Accutase (Sigma). After washing with PBS, the fixation buffer (BD Biosciences) was used to fix these single cells at room temperature for 20 min. Then, the cells were washed with PBS, and the single-cell suspension was permeabilized in perm/wash buffer (BD Biosciences) for 15 min at 4 °C. After washing, these single cells were incubated with primary antibodies and isotype control antibodies for 30 min at 37 °C. After washing, the cells were incubated with secondary antibodies in the dark for 30 min at 37 °C. The single-cell suspension was washed twice. Subsequently, the supernatant was discarded by centrifugation at 500 rpm for 5 min at room temperature. The cells were resuspended in 200 μL PBS, and then analyzed by CytoFlox-S (BD Biosciences). The information for antibodies was listed in key resources table.
Immunostaining assay
The H1 and BRPF1 knock-out cells were seeded in Matrigel-coated 24-well plates and cultured in ncTarget medium. Using 4% paraformaldehyde fixed these samples of cells were fixed for 20 min at room temperature. After washing three times with PBS, these samples were permeabilized and blocked with 0.3% Triton X-100 (Sigma) and 10% goat serum in PBS and incubated with primary antibodies overnight at 4 °C. After washing three times with PBS, these samples were incubated with secondary antibodies and DAPI for 1.5 h at room temperature. After washing three times with PBS, images of these samples were captured with an LSM 800 microscope (Zeiss). These antibodies are listed in key resources table.
RNA-seq and heatmap analyses
We used TRIzol (Invitrogen) to lyse wild-type and BRPF1 knock-out cells and extracted total RNA. Then, sequencing libraries were established according to the manufacturer’s recommendations for the VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme #NR605). These libraries of samples were run on a NextSeq system with a NextSeq 500 Mid Output kit (Illumina). Subsequently, we analyzed these RNA-seq data. The number of raw reads were filtered by Trimmomatic (v0.35) and then aligned to the human reference genome (hg38) using Hisat2 (v2.0.4) with default parameters and using SAMtools (v1.3.1),72 and htseq-count (v0.6.0) were used to calculate gene expression. EDASeq (v2.24.0) filtered by a threshold of at least 10 average raw read counts among samples and normalized. DESeq2 (v 1.30.0) was used to determain differential expression, and PCA plots were prepared. Differences in gene expression were considered significant by a p value < 0.01 and a fold-change > 1.5.71
ATAC-seq
Briefly, 50,000 cells from each sample were processed and then used to generate DNA libraries with a Nextera DNA library preparation kit (Illumina) according to the manufacturer’s specifications. Subsequently, these DNA library samples were used for sequencing on a NextSeq 500 platform. Adapter sequences were removed from the raw data using cutadapt (v1.13) and aligned to the human genome (UCSC hg38) using Bowtie2 (v2.4.1). Duplicates were removed using SAMtools (v1.3.1) and Picard tools (v2.2.4). Signals were compiled using MACS2 callpeak and bdgcmp, and differential open peaks were called using MACS2 bdgdiff. A signal density heatmap and profile were plotted using deepTools (v3.5.0),73 and motifs were found using homer. Peaks were annotated with gene annotation using ChIPpeakAnno (v3.24.2), and gene ontology analysis was performed using clusterProfiler (v3.18.0).
ChIP-seq
We performed ChIP-seq to analyze whole-genome binding and enrichment of histone modifications such as H3K4me3, H3K23ac and BRPF1 and its mutations. In brief, 1 × 107 cells from each sample were crosslinked in 1% formaldehyde with rotation for 10 min at room temperature, and crosslinking reactions were stopped with 0.125 M glycine and rotation at room temperature for 5 min. After washing twice with cooled PBS, each sample was sonicated in 1% SDS lysis buffer plus 1 mM PMSF with protease inhibitor cocktail using a picoruptor to obtain 200–500 bp chromatin fragments. Then, these sonicated samples were dialyzed with ChIP dilution buffer. The sonicated fragments from these samples were coincubated with magnetic beads (Dynabeads protein A and G (1:1)) (Invitrogen) and 5 μg corresponding specific antibody with rotation overnight at 4 °C. These antibody-bound complexes were washed with rotation at 4 °C for 5 min with low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and TE buffer in turn. Then, these samples were eluted with elution buffer (1% SDS, 0.1 M NaHCO3). These eluted samples were reverse-crosslinked with 5 M NaCl overnight at 65 °C. RNase A (20 mg/mL) was added at 37 °C for 1–2 h. Subsequently, 1 M Tris–HCl (PH 6.5), 0.5 M EDTA and Proteinase K were added at 45 °C for 2 h. Then, we purified the ChIPed DNA from these samples for ChIP-Seq. ChIPed DNA and corresponding input DNA were measured by a Qubit fluorometer (Invitrogen), and DNA libraries were generated with this DNA using a VAHTS Universal DNA Library Prep Kit for Illumina (Vazyme #ND607). These DNA libraries were measured with a Qubit fluorometer and sequenced on a NextSeq 500 platform. The following is a detailed description of the reagent composition. low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 150 mM NaCl); high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 500 mM NaCl); LiCl wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris-HCl (pH 8.0)) and TE buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA). These antibodies are listed in the key resources table.
The raw data were cut into adaptors using cutadapt (v1.13) and then aligned to the human reference genome (hg38) using Bowtie2 (v2.2.5). Uniquely mapped reads were retained using SAMtools (v1.3.1) and Picard tools MarkDuplicates (v2.2.4). We performed peak calling of transcription factor ChIP-seq data using the MACS2 (v2.1.0) callpeak module and then only peaks with a -log (p-value) > 4 were retained. SICER2 was used for peak calling of histone modifications.75 The bigWig signal files were visualized using the computeMatrix, plotHeatmap and plotProfile modules in DeepTools (v3.5.0). ChIPpeakAnno (v3.24.2) was used to identify nearby genes from the peaks obtained from MACS. Gene Ontology (GO) analysis was performed using clusterProfiler (v3.18.0).
Ethics approval and consent to participate
The WT human ES cell line used in the study was reviewed and approved by the Human Subject Research Ethics Committee at GIBH.
Quantification and statistical analysis
Statistical analysis
In general, data are shown as the mean ± SD (standard deviation) calculated using Microsoft Excel and GraphPad Prism from at least three biological repeats. Two-tailed Student’s t tests were used to calculate statistical significance. Differences with a p value <0.05 were considered statistically significant. No samples were excluded for any analysis.
Acknowledgments
We thank the lab members in GIBH for their kind help. This work was supported by Research Funds from the Health@InnoHK Program launched by the Innovation Technology Commission of the Hong Kong SAR, P. R. China; the Youth Innovation Promotion Association of the Chinese Academy of Sciences (to Y.S., 2022360); the National Natural Science Foundation of China (32270624, 31971374); Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine (2020B1212060052); Guangzhou Science and Technology Program General project (20180304001, 201904020045); Innovative Team Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR110104005); and the Guangdong Province Special Program for Outstanding Talents (to G.P., 2019JC05Y463).
Author contributions
G.P. , Y.S., and C.Z. initiated and designed the project, and wrote the article. C.Z. performed most experiments and analyzed the resulting data. Y.S. and C.Z. performed the gene knockout of the BRPF1 in hESCs. C.Z., Y.L., and Q.X. Performed overexpression BRPF1 WT and mutants of BRPF1 in BRPF1−/− H1 hESCs. J.Z., J.W., D.Z., and C.Z. performed the FACS and RT-qPCR. T.Z. performed karyotype analysis. C.Z., Q.C., and Y.Z. performed the RNA sequencing, ATAC sequencing. and ChIP sequencing. H.L and C.Z. analyzed the RNA-seq, ATAC-seq, and ChIP-seq data. All authors read and approved the final article.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105939.
Contributor Information
Yongli Shan, Email: shan_yongli@gibh.ac.cn.
Guangjin Pan, Email: pan_guangjin@gibh.ac.cn.
Supplemental information
Data and code availability
The RNA-Seq, ATAC-seq, and ChIP-seq data have been deposited in the Genome Sequence Archive for Human database under the accession code HRA001928. We also deposited these data in the Gene Expression Omnibus database under accession code GSE213695. All the datasets are publicly accessible.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Lu C.C., Brennan J., Robertson E.J. From fertilization to gastrulation: axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 2001;11:384–392. doi: 10.1016/s0959-437x(00)00208-2. [DOI] [PubMed] [Google Scholar]
- 2.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Reubinoff B.E., Pera M.F., Fong C.Y., Trounson A., Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 4.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 5.Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L.R., Hadjantonakis A.K., Reinholdt L.G. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy A., Gócza E., Diaz E.M., Prideaux V.R., Iványi E., Markkula M., Rossant J. Embryonic stem-cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 7.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 8.Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 9.Pan G.J., Chang Z.Y., Schöler H.R., Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 10.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Giulitti S., Pellegrini M., Zorzan I., Martini P., Gagliano O., Mutarelli M., Ziller M.J., Cacchiarelli D., Romualdi C., Elvassore N., Martello G. Direct generation of human naive induced pluripotent stem cells from somatic cells in microfluidics. Nat. Cell Biol. 2019;21:275–286. doi: 10.1038/s41556-018-0254-5. [DOI] [PubMed] [Google Scholar]
- 14.Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A., et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Gifford C.A., Ziller M.J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A.K., Kelley D.R., Shishkin A.A., Issner R., et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and Trithorax: 70 Years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Shan Y., Liang Z., Xing Q., Zhang T., Wang B., Tian S., Huang W., Zhang Y., Yao J., Zhu Y., et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morey L., Santanach A., Di Croce L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol. Cell Biol. 2015;35:2716–2728. doi: 10.1128/Mcb.00266-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., Di Croce L. Nonoverlapping functions of the polycomb group cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Ang Y.S., Tsai S.Y., Lee D.F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan M., Liang J., Xiong Y., Shi F., Zhang Y., Lu W., He Q., Yang D., Chen R., Liu D., et al. The Trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J. Biol. Chem. 2013;288:5039–5048. doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews F.H., Strahl B.D., Kutateladze T.G. Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol. 2016;12:662–668. doi: 10.1038/Nchembio.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musselman C.A., Lalonde M.E., Côté J., Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huether R., Dong L., Chen X., Wu G., Parker M., Wei L., Ma J., Edmonson M.N., Hedlund E.K., Rusch M.C., et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi P., Allis C.D., Wang G.G. Covalent histone modifications - miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faust C., Lawson K.A., Schork N.J., Thiel B., Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- 31.Morin-Kensicki E.M., Faust C., LaMantia C., Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- 32.Margueron R., Justin N., Ohno K., Sharpe M.L., Son J., Drury W.J., Voigt P., Martin S.R., Taylor W.R., De Marco V., et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merson T.D., Dixon M.P., Collin C., Rietze R.L., Bartlett P.F., Thomas T., Voss A.K. The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 2006;26:11359–11370. doi: 10.1523/Jneurosci.2247-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peña P.V., Davrazou F., Shi X., Walter K.L., Verkhusha V.V., Gozani O., Zhao R., Kutateladze T.G. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M.R., et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 37.Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W.M.P., van Schaik F.M.A., Varier R.A., Baltissen M.P.A., Stunnenberg H.G., Mann M., Timmers H.T.M. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Doyon Y., Cayrou C., Ullah M., Landry A.J., Côté V., Selleck W., Lane W.S., Tan S., Yang X.J., Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Champagne K.S., Saksouk N., Peña P.V., Johnson K., Ullah M., Yang X.J., Côté J., Kutateladze T.G. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins. 2008;72:1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You L., Yan K., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. The chromatin regulator Brpf1 regulates embryo development and cell proliferation. J. Biol. Chem. 2015;290:11349–11364. doi: 10.1074/jbc.M115.643189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X.J. MOZ and MORF acetyltransferases: molecular interaction, animal development and human disease. Biochim. Biophys. Acta. 2015;1853:1818–1826. doi: 10.1016/j.bbamcr.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Klein B.J., Jang S.M., Lachance C., Mi W., Lyu J., Sakuraba S., Krajewski K., Wang W.W., Sidoli S., Liu J., et al. Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat. Commun. 2019;10:4724. doi: 10.1038/s41467-019-12551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang F., Abmayr S.M., Workman J.L. Regulation of KAT6 acetyltransferases and their roles in cell cycle progression, stem cell maintenance, and human disease. Mol. Cell Biol. 2016;36:1900–1907. doi: 10.1128/Mcb.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas T., Voss A.K., Chowdhury K., Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 45.Katsumoto T., Aikawa Y., Iwama A., Ueda S., Ichikawa H., Ochiya T., Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You L., Chen L., Penney J., Miao D., Yang X.J. Expression atlas of the multivalent epigenetic regulator Brpf1 and its requirement for survival of mouse embryos. Epigenetics. 2014;9:860–872. doi: 10.4161/epi.28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kool M., Jones D.T.W., Jäger N., Northcott P.A., Pugh T.J., Hovestadt V., Piro R.M., Esparza L.A., Markant S.L., Remke M., et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan K., Rousseau J., Littlejohn R.O., Kiss C., Lehman A., Rosenfeld J.A., Stumpel C.T.R., Stegmann A.P.A., Robak L., Scaglia F., et al. Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am. J. Hum. Genet. 2017;100:91–104. doi: 10.1016/j.ajhg.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naseer M.I., Abdulkareem A.A., Guzmán-Vega F.J., Arold S.T., Pushparaj P.N., Chaudhary A.G., AlQahtani M.H. Novel missense variant in heterozygous state in the BRPF1 gene leading to intellectual developmental disorder with dysmorphic facies and ptosis. Front. Genet. 2020;11:368. doi: 10.3389/fgene.2020.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattioli F., Schaefer E., Magee A., Mark P., Mancini G.M., Dieterich K., Von Allmen G., Alders M., Coutton C., van Slegtenhorst M., et al. Mutations in histone acetylase modifier BRPF1 cause an autosomal-dominant form of intellectual disability with associated ptosis. Am. J. Hum. Genet. 2017;100:105–116. doi: 10.1016/j.ajhg.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You L., Yan K., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. The lysine acetyltransferase activator Brpf1 governs dentate gyrus development through neural stem cells and progenitors. PLoS Genet. 2015;11:e100503410. doi: 10.1371/journal.pgen.1005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You L., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. Deficiency of the chromatin regulator Brpf1 causes abnormal brain development. J. Biol. Chem. 2015;290:7114–7129. doi: 10.1074/jbc.M114.635250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You L., Li L., Zou J., Yan K., Belle J., Nijnik A., Wang E., Yang X.J. BRPF1 is essential for development of fetal hematopoietic stem cells. J. Clin. Invest. 2016;126:3247–3262. doi: 10.1172/Jci80711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hibiya K., Katsumoto T., Kondo T., Kitabayashi I., Kudo A. Brpf1, a subunit of the MOZ histone acetyl transferase complex, maintains expression of anterior and posterior Hox genes for proper patterning of craniofacial and caudal skeletons. Dev. Biol. 2009;329:176–190. doi: 10.1016/j.ydbio.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 55.Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B.J., et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assou S., Le Carrour T., Tondeur S., Ström S., Gabelle A., Marty S., Nadal L., Pantesco V., Réme T., Hugnot J.P., et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cell. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh A., Som A. RNA-Seq analysis reveals pluripotency-associated genes and their interaction networks in human embryonic stem cells. Comput. Biol. Chem. 2020;85:107239. doi: 10.1016/j.compbiolchem.2020.107239. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y., Wang T., Zhang Y., Shi L., Zhang C., Zhang J., Yao J., Chen Q., Zhong X., Wei Y., et al. Coordination of EZH2 and SOX2 specifies human neural fate decision. Cell Regen. 2021;10:30. doi: 10.1186/s13619-021-00092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y., Wang T., Ma L., Zhang Y., Zhao Y., Lye K., Xiao L., Chen C., Wang Z., Ma Y., et al. Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., Mckay R.D.G., Buetow K.H., et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mas G., Blanco E., Ballaré C., Sansó M., Spill Y.G., Hu D., Aoi Y., Le Dily F., Shilatifard A., Marti-Renom M.A., Di Croce L. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 2018;50:1452–1462. doi: 10.1038/s41588-018-0218-5. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Z.H., Sam T.W., Zeng Y., Chu J.J.H., Loh Y.H. Chromatin regulation in development: current understanding and approaches. Stem Cell. Int. 2021;2021:8817581. doi: 10.1155/2021/8817581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ullah M., Pelletier N., Xiao L., Zhao S.P., Wang K., Degerny C., Tahmasebi S., Cayrou C., Doyon Y., Goh S.L., et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell Biol. 2008;28:6828–6843. doi: 10.1128/Mcb.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin S., Jin L., Zhang J., Liu L., Ji P., Wu M., Wu J., Shi Y. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF) J. Biol. Chem. 2011;286:36944–36955. doi: 10.1074/jbc.M111.244400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein B.J., Cox K.L., Jang S.M., Côté J., Poirier M.G., Kutateladze T.G. Molecular basis for the PZP domain of BRPF1 association with chromatin. Structure. 2020;28:105–110.e3. doi: 10.1016/j.str.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vezzoli A., Bonadies N., Allen M.D., Freund S.M.V., Santiveri C.M., Kvinlaug B.T., Huntly B.J.P., Göttgens B., Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 69.Bannister A.J., Schneider R., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 70.Li J., Moazed D., Gygi S.P. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 71.R Core Team R: A Language and Environment for Statistical Computing. 2022. https://www.r-project.org/contributors.html
- 72.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq, ATAC-seq, and ChIP-seq data have been deposited in the Genome Sequence Archive for Human database under the accession code HRA001928. We also deposited these data in the Gene Expression Omnibus database under accession code GSE213695. All the datasets are publicly accessible.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.