SUMMARY
The use of an integrated systems biology approach to investigate tissues and organs has been thought to be impracticable in the field of structural biology, where the techniques mainly focus on determining the structure of a particular biomacromolecule of interest. Here, we report the use of cryoelectron microscopy (cryo-EM) to define the composition of a raw bovine retinal pigment epithelium (RPE) lysate. From this sample, we simultaneously identify and solve cryo-EM structures of seven different RPE enzymes whose functions affect neurotransmitter recycling, iron metabolism, gluconeogenesis, glycolysis, axonal development, and energy homeostasis. Interestingly, dysfunction of these important proteins has been directly linked to several neurodegenerative disorders, including Huntington’s disease, amyotrophic lateral sclerosis (ALS), Parkinson’s disease, Alzheimer’s disease, and schizophrenia. Our work underscores the importance of cryo-EM in facilitating tissue and organ proteomics at the atomic level.
In brief
Morgan et al. use cryo-EM to simultaneously identify and solve the structures of seven different enzymes from a raw bovine retinal pigment epithelium lysate. This work highlights the potential of this methodology in facilitating structural-omics of a biological system at the atomic level.
Graphical Abstract
INTRODUCTION
The retinal pigment epithelium (RPE) is a neural epithelial cell layer constituting a single layer of post-mitotic cells that anatomically display as a hexagonal structure. This pigmented cell layer is situated between the retina, which comprises light-sensitive photoreceptors, and the blood-vessel-dense choroid. The RPE plays an important role in the eye by forming a selective barrier and maintaining the function of the photoreceptor layer.1 It participates in several vegetative regulations, including the conversion and storage of retinoid, renewing the outer segment of photoreceptors, protecting the outer retina from excessive light energy and reactive oxygen species, maintaining retinal homeostasis, and governing the transport of nutrients and metabolic waste products in and out of the retina.
The significance of the RPE is particularly underscored by its direct correlation with many retinal disorders. For example, dysfunction of the RPE can cause albinism,2 retinitis pigmentosa,3 fundus albipunctatus,4 gyrate atrophy,5 bestrophinopathy,6 and Stargardt disease.7 In addition, the abnormality of the RPE is involved in the common disease age-related macular degeneration,8,9 a leading cause of blindness.10,11 To study the RPE, bovine eye tissue is commonly used, as the size of the bovine eye is large and the process for tissue extraction is relatively straightforward. The bovine RPE can be easily separated from the choroid layer, facilitating the study of RPE cells.
Recently, the advances of mass spectrometry have provided an opportunity for understanding the details of the eye proteome at the systems level.12 A proteomic landscape study of the human RPE-choroid complex revealed that there are more than 4,000 unique proteins expressed in RPE-choroid tissues.13 This integrated systems strategy for investigating tissue and organ samples has been thought to be inaccessible in the field of structural biology with methods such as X-ray crystallography and cryoelectron microscopy (cryo-EM), where these techniques typically focus on the structural study of a particular target biomacromolecule at the atomic level. Additionally, structural biology methodologies routinely require homogeneous and pure samples. As a result, these structural biology tools have been thought to be inadequate in studying tissue and organ samples, where these biological raw samples contain numerous, diverse proteins and biomacromolecules in a complex, heterogeneous environment.
To develop an approach to employ cryo-EM to study systems proteomics of tissue and organ samples, we recently developed a build-and-retrieve (BaR) cryo-EM methodology.14 This is an iterative methodology capable of performing in silico purification and sorting of images from a large heterogeneous dataset. The BaR methodology is able to deconvolute images and allows us to simultaneously produce near-atomic-resolution cryo-EM maps for several individual proteins originating from a complex, multiprotein sample. BaR is similar to, but also distinct from, cryoID,15 which is a powerful technique to uncover protein identities via high-resolution cryo-EM maps, particularly for relatively large protein complexes of ~750 kDa.
To demonstrate the capability of BaR in elucidating systems structural proteomics of the RPE, we isolated this single layer of cells from bovine eyes and enriched enzymes obtained from this raw lysate using size-exclusion chromatography. Through the use of BaR on the single-particle cryo-EM images collected, we were able to identify and solve cryo-EM structures of seven different RPE enzymes, with functions important for neurotransmitter recycling, iron metabolism, gluconeogenesis, glycolysis, axonal development, and energy homeostasis. Importantly, these essential enzymes are linked to several neurogenerative diseases, suggesting that these enzymes may play critical roles in their pathogenesis.
RESULTS
We enriched the bovine RPE enzymes from our sample using size-exclusion chromatography. From this, we obtained two major peaks with sizes corresponding to 100–250 kDa and 300–650 kDa, respectively. We then performed proteomic analysis of each peak to elucidate its protein composition. The 10 most abundant proteins of these two enriched fractions, based upon the protein identification score, are listed in Table S1A and S1B. We then collected single-particle images (Figure S1) of the two bovine RPE-enriched samples separately using cryo-EM and processed the dataset using BaR.14 Several iterative rounds of 2D classifications allowed us to sort the images into different protein classes (Figures S2 and S3). Together, BaR allowed us to determine cryo-EM structures of seven different enzymes with resolutions ranging between 2.30 and 3.36 Å. These enzymes were identified as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), ferritin (FT), aspartyl aminopeptidase (DNPEP), glutamine synthetase (GS), dihydropyrimidinase-related protein 2 (DPYSL2), fructose-bisphosphate aldolase (FPA), and mitochondrial creatine kinase (MtCK). The presence of these seven enzymes was confirmed by proteomic analysis (Table S1). Cryo-EM maps of the MtCK enzyme before and after the BaR procedures are shown in Figure S4.
To predict if these seven bovine RPE enzymes have disparate or cooperative functions, we constructed an interconnected network based on the STRING database.16 We found that all of the seven identified enzymes participate in a range of interactions within the network (Figure S5).
Glyceraldehyde 3-phosphate dehydrogenase
We collected a total of 374,086 single-particle projections for a class of images that we were able to identify as the bovine GAPDH enzyme (Figure S6; Table S2). Its cryo-EM structure was able to be refined to a resolution of 2.30 Å. Density modification17 allowed us to extend the resolution to 2.16 Å (Figure 1).
Figure 1. Structure of bovine GAPDH.
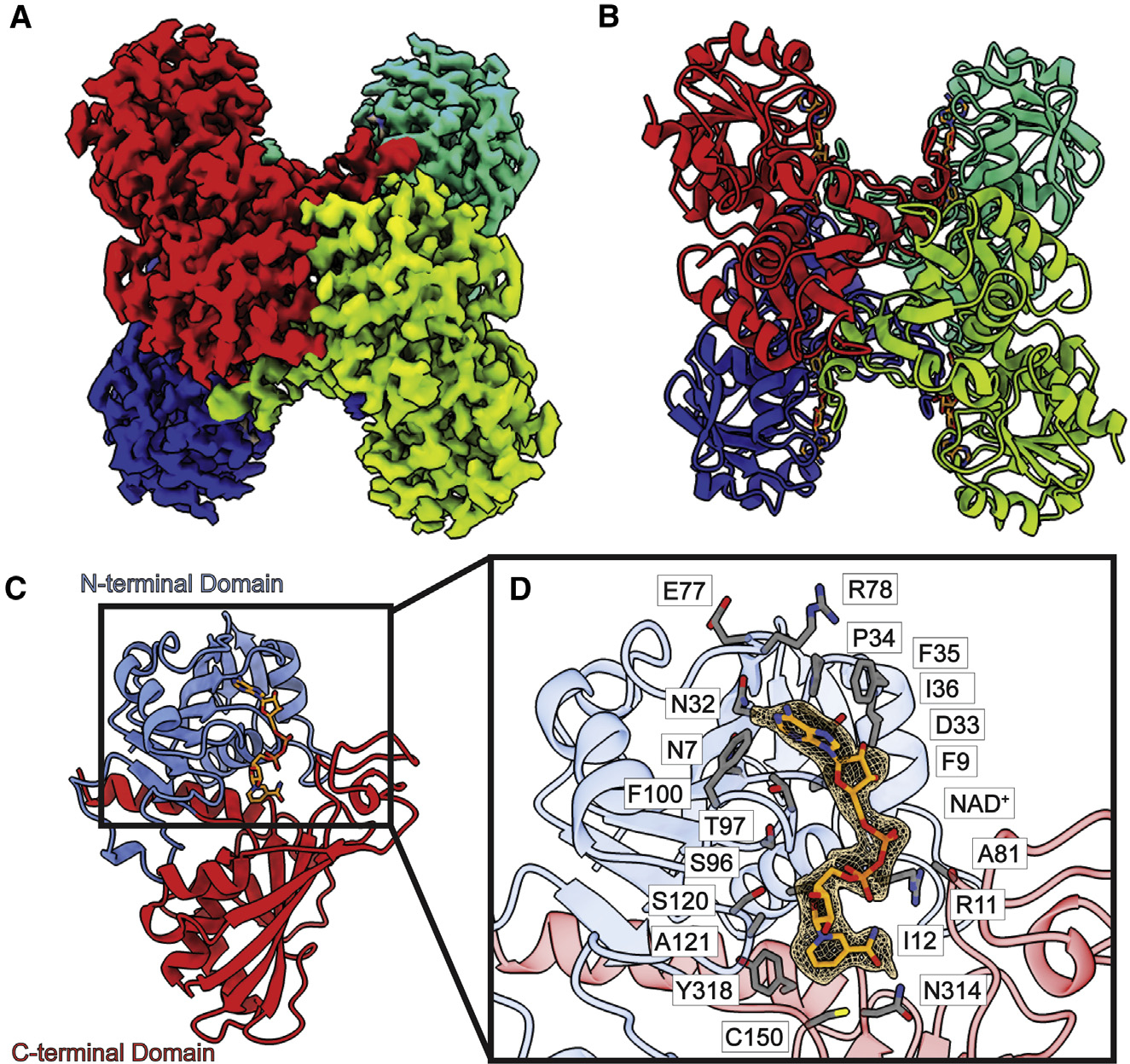
(A) Cryo-EM map of GAPDH.
(B) Structure of GAPDH. GAPDH forms a tetramer with D2 symmetry. In both (A) and (B), protomers are distinguished by individual colors.
(C) Layout of a GAPDH subunit, which can be divided into two domains, N-terminal domain (blue) and C-terminal domain (red). A single NAD+ ligand is identified in each subunit of the GAPDH tetramer (orange sticks).
(D) Zoomed view of the NAD+-binding site. NAD+ is depicted as orange sticks, cryo-EM density of NAD+ (3σ) is shown as orange mesh, and residues within 4 Å of bound NAD+ are shown as gray sticks.
Bovine GAPDH assembles as a tetrameric oligomer (Figures 1A and 1B), which is in good agreement with crystal structures of the human,18 bovine,19 lobster,20 and Bacillus stearothermophilus21 GAPDH enzymes. Superimposition of the cryo-EM structure of bovine GAPDH to its X-ray structure (PDB: 4O59)19 gives rise to a root-mean-square deviation (RMSD) of 0.49 Å (for 328 Cα atoms). Our GAPDH cryo-EM structure represents a substrate-bound conformation. Each subunit is occupied by a nicotinamide adenine dinucleotide (NAD+) endogenous ligand.
GAPDH is a well-known multifunctional protein. Its primary function is to catalyze the oxidative phosphorylation of glyceraldehyde-3-phosphate to mediate the formation of ATP and NADH during glycolysis.22 However, this cytoplasmic protein is also an essential metabolic regulator involved in various cellular processes, including membrane fusion, transport, apoptosis, DNA replication and repair, and regulation of transcription and translation.23 Besides its role in different metabolic activities, GAPDH can serve as a chaperone for labile heme to maintain iron homeostasis.24,25 It also has the ability to bind different macromolecules and interact with the cytoskeleton to influence microtubule and actin polymerization.26,27
Each subunit of bovine GAPDH consists of 332 amino acids that can be divided into two domains. The first 147 residues constitute the N-terminal domain, with residues 148–332 forming the catalytic C-terminal domain (Figure 1C). The N-terminal domain contains five α helices and eight β strands, whereas the C-terminal domain possesses six α helices and seven β strands. An NAD+-binding site, which possesses a strong Ross-mann-folded signature,28 is found in each subunit of this enzyme. The N-terminal domain makes up most of this nucleotide-binding site; however, the C-terminal residues 312–333 are also found to engage in NAD+ binding. The endogenous NAD+ coenzyme is observed to anchor in this NAD+-binding site of each subunit of this tetrameric dehydrogenase, where residues N7, F9, R11, I12, N32, D33, P34, F35, I36, E77, R78, A81 S96, T97, F100, S120, A121, C150, N314, and Y318 specifically participate in NAD+ binding (Figure 1D).
Notably, C150 of GAPDH, located at the NAD+-binding site, is a very critical residue. Mutations of C150 have been associated with Alzheimer’s disease,29 and studies in both human and yeast have shown the catalytic activity of GAPDH is abolished when this amino acid is altered.30,31
Ferritin
We obtained 2,222 single-particle counts for a second protein class in our dataset. These single-particle images originate from the FT enzyme (Figure S7; Table S2). We then solved the cryo-EM structure of this bovine enzyme to a resolution of 2.57 Å. Density modification17 allowed us to extend the structural resolution of the structure to 2.17 Å (Figure 2).
Figure 2. Structure of bovine FT.
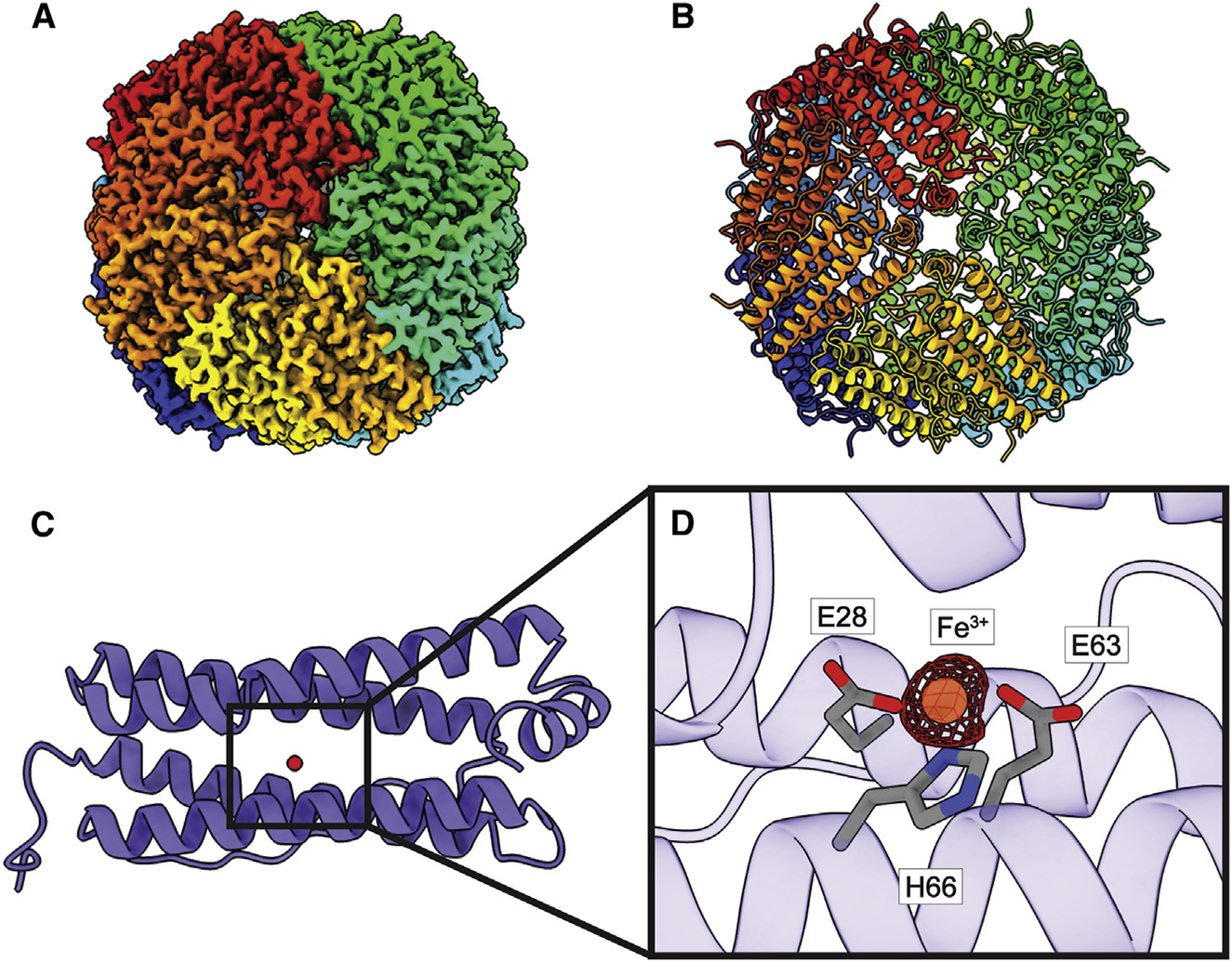
((A) Cryo-EM map of FT.
(B) Structure of FT. FT monomers are individually colored in both (A) and (B).
(C) A single FT subunit is made up of five α helices. A single Fe3+ ion (red sphere) is found in each FT monomer.
(D) Zoomed view of the Fe3+-binding site. Fe3+ is shown as an orange sphere, cryo-EM density of Fe (5σ) is shown as red mesh, and interacting residues are shown as gray sticks.
Similar to the crystal structure of human FT,32 bovine FT is a 181-amino-acid protein, in which 173 residues are included in the final structure. Superimposition of the cryo-EM structure of bovine FT to the X-ray structure of human FT (PDB: 1FHA)32 provides an RMSD of 0.39 Å (for 171 Cα atoms). Bovine FT consists of 24 subunits that display an octahedral symmetry (Figures 2A and 2B). Our cryo-EM data indicate that bovine RPE is composed solely of FT heavy chains without any light chains. Proteomic analysis also confirmed that this enzyme in our sample indeed only contains heavy chains (Table S1). Each FT subunit is composed of five α helices and forms an elongated α-helical bundle. Interestingly, our bovine FT structure is in its ligated form and each subunit is found to bind an Fe3+ ion (Figure 2C).
FT is the major iron storage protein in all kingdoms of life.33 It is widely recognized as a critical protein for iron metabolism, particularly in the brain and the eye.34,35 FT not only participates in providing iron availability for cellular demand but also protects cells against damage from iron-mediated free radicals.36
FT expression and the amount of Fe3+ storage have been reported to be altered strongly in patients with diseases, such as Alzheimer’s, Parkinson’s, and acquired immunodeficiency syndrome (AIDS).37,38 In our bovine FT structure, an Fe3+ ion is found to occupy the Fe-binding site in each subunit (Figure 2C), agreeing with previous X-ray structures.39 This Fe3+-binding site is created by three charged residues, E28, E63, and H66, which stabilize Fe3+ binding via charge-charge interactions (Figure 2D). Previously, mutagenesis studies indicate that the E63 and H66 residues play a major role in iron uptake for both the human and mouse FT enzymes,40,41 highlighting the significance of these charged residues for the function of FT.
Aspartyl aminopeptidase
We obtained 1,029 projections for this class of protein molecules (Figure S8; Table S2). After using BaR, we identified this class as the DNPEP enzyme. We then resolved the structure of bovine DNPEP to a resolution of 3.36 Å. Density modification17 allowed us to improve the resolution to 2.76 Å (Figure 3).
Figure 3. Structure of bovine DNPEP.
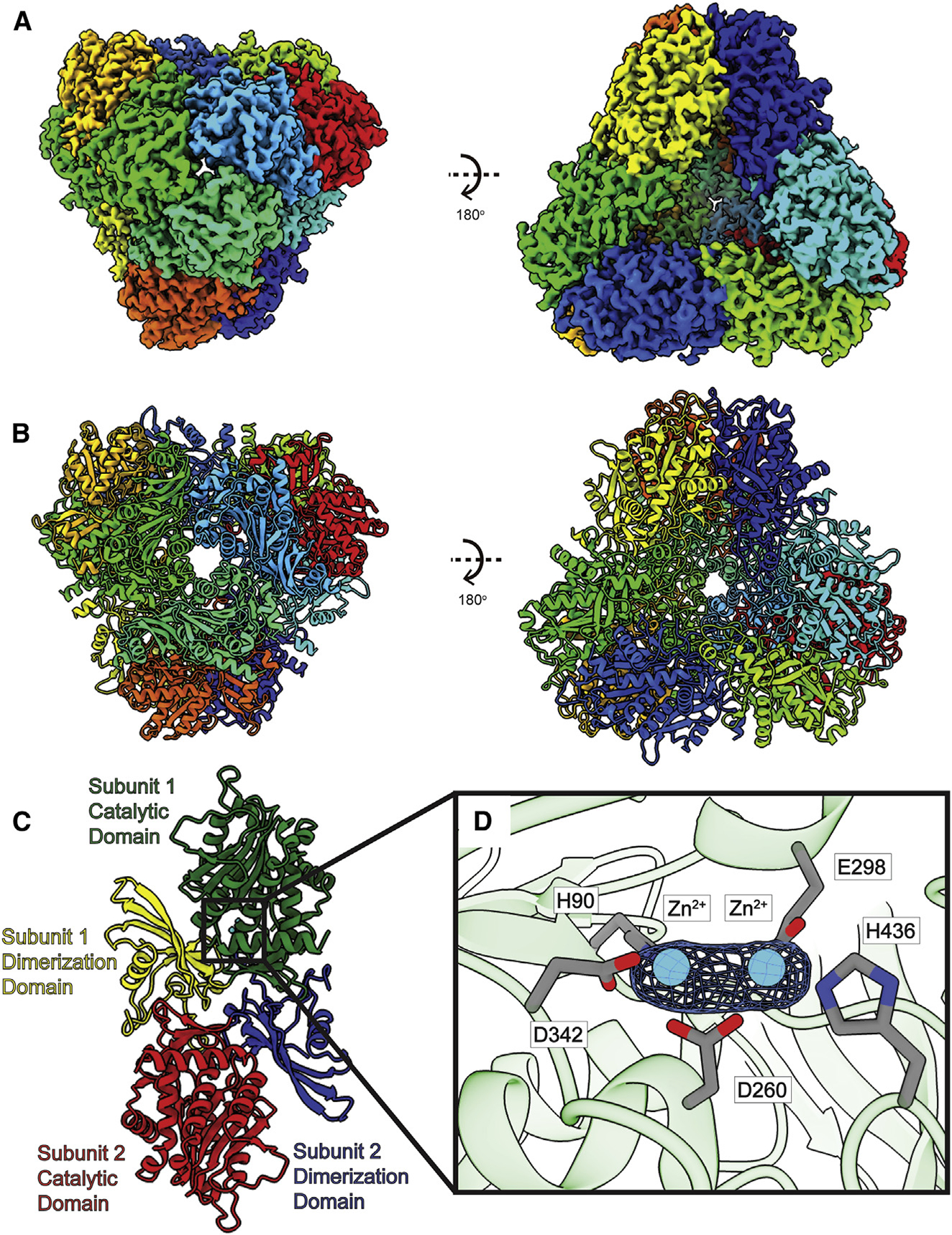
(A) Cryo-EM map of DNPEP.
(B) Structure of DNPEP. Twelve DNPEP monomers form a dodecamer with tetrahedral symmetry. Individual subunits are distinguished with different colors in both (A) and (B).
(C) Layout of DNPEP dimer. A DNPEP subunit can be divided into two units: a catalytic domain and a dimerization domain.
(D) Zoomed view of the Zn2+-binding site in a single DNPEP subunit. Two Zn2+ ions are found in each DNPEP subunit. The Zn2+ ions are colored cyan, cryo-EM densities (4σ) are depicted as blue meshes, and interacting residues are shown as gray spheres.
The cryo-EM structure of bovine DNPEP indicates that this enzyme assembles as a dodecamer (Figures 3A and 3B). This dodecameric oligomerization can be interpreted as a hexamer of dimers. Like crystal structures of DNPEP,42,43 each subunit of the enzyme can be divided into catalytic and dimerization domains (Figure 3C). Superimposition of our cryo-EM structure of bovine DNPEP to its X-ray structure (PDB: 3VAT)43 leads to an RMSD of 0.58 Å (for 419 Cα atoms). The catalytic domain is formed by residues 6–94 and 243–468, which feature a secondary structure consisting of 11 α helices and 12 β strands. The dimerization domain includes residues 95–242, which are folded into two α helices and five β strands. This domain is responsible for securing oligomerization formation. However, a portion of the catalytic domain, including the flexible loops formed by residues 241–247, is also observed to participate in dimerization. Within the catalytic domain, a Zn2+-binding site is identified in each subunit of our bovine DNPEP structure. This site is occupied by two Zn2+ ions (Figure 3D), consistent with the crystal structures of human and bovine DNPEPs.42,43
DNPEP belongs to the M18 metallopeptidase family of enzymes.44,45 These enzymes are particularly important for the regulation of signaling peptide activity, engaging in a variety of significant biological processes, including angiogenesis, blood pressure regulation, memory, reproduction, tumor growth, and metastasis,46 and controlling the function of the renin-angiotensin system.47 In the eye, the ocular renin-angiotensin system plays critical functional roles in regulation of intraocular pressure, angiogenesis, and neuron function,48–51 thereby becoming a potential target for glaucoma and retinopathy diseases.52–54
In the cryo-EM structure of bovine DNPEP, two bound Zn2+ ions are found to house in the Zn2+-binding site within the catalytic domain of each subunit of the enzyme. Particularly, residues H90, D260, E298, D342, and H436 are responsible for creating this binuclear metal-binding site (Figure 3D). Interestingly, it has been observed that mutations of the corresponding histidine residues, H90 and H436, in the Zn2+-binding sites of the human and mouse DNPEPs completely diminished the activity of these enzymes.55 Therefore, these two histidines are deemed important for the function of this enzyme.
Glutamine synthetase
We were able to obtain 35,076 projections for this very abundant protein (Figure S9; Table S2). Using BaR, we generated the final cryo-EM map, which allowed us to validate its identity as the bovine GS enzyme. We then resolved its cryo-EM structure to a resolution of 2.58 Å. Density modification17 permitted us to extend the structural resolution of this enzyme to 2.17 Å (Figure 4).
Figure 4. Structure of bovine GS.
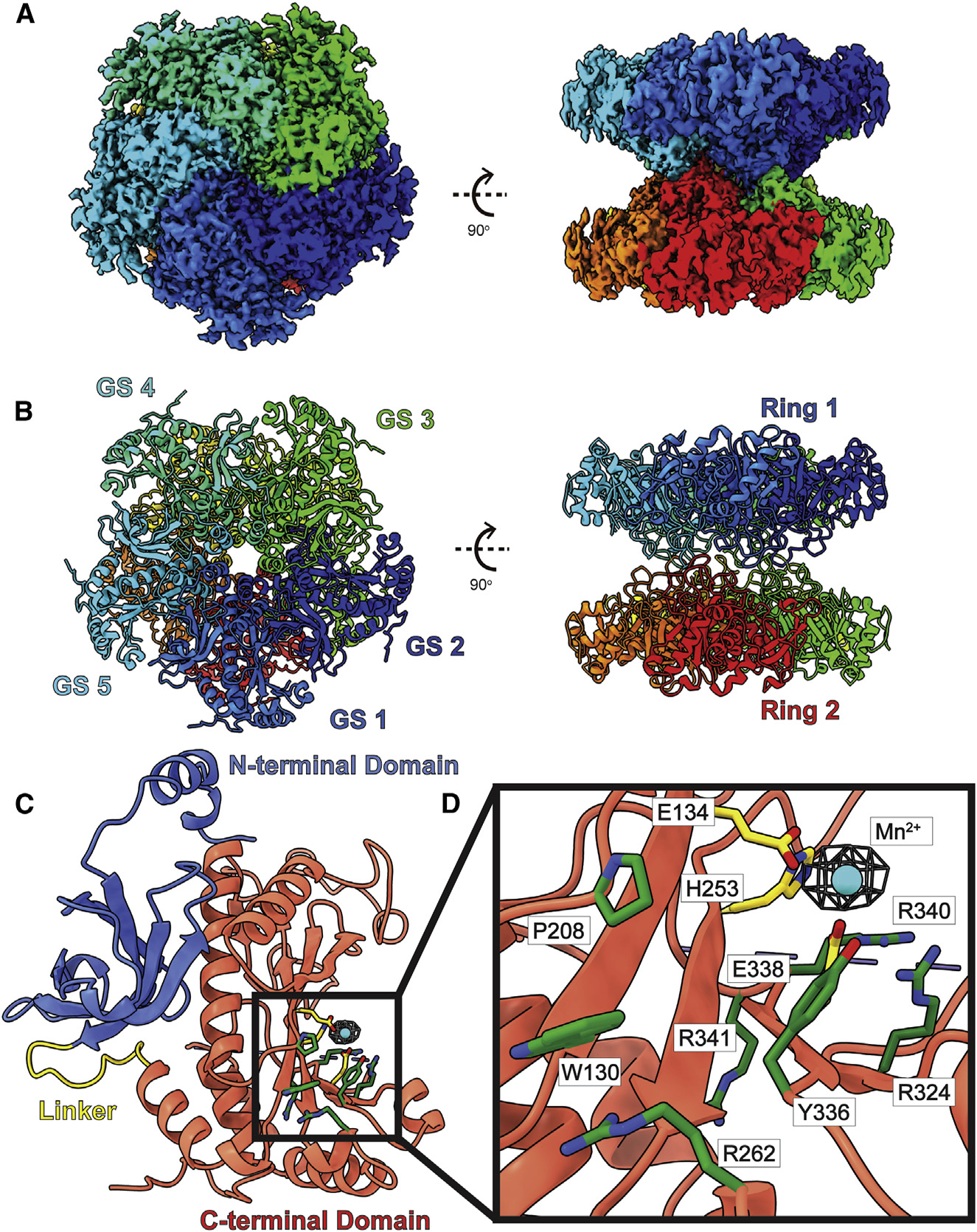
(A) Cryo-EM map of GS.
(B) Structure of GS. GS forms a complex of D5 symmetry with two pentameric rings stacked in a two-ring conformation. In both (A) and (B), subunits are distinguished through individual colors and match accordingly.
(C) A GS protomer is organized into a small N-terminal domain (blue) and a large C-terminal domain (red) connected by a flexible linker (yellow). The N-terminal domain mediates inter-subunit interactions, while the C-terminal domain creates the catalytic site.
(D) Zoomed view of the catalytic site of a GS subunit. Residues responsible for binding Mn2+ (cyan sphere) are in yellow sticks. The remaining residues, which contribute to form the catalytic site, are in green sticks. Cryo-EM density (4σ) of bound Mn2+ is in blue mesh.
GS is a glutamine-synthesizing enzyme involved in the recycling of synaptically released glutamate and γ-aminobutyric acid (GABA) as well as the detoxification of ammonia. In the brain, GS actively participates in the metabolic regulation of glutamate, assimilation of ammonia, recyclization of neurotransmitters, and termination of neurotransmitter signals.56,57 Remarkably, there is a strong link between GS and Alzheimer’s plaque formation, where an increased level of GS was found in the cerebral spinal fluid of Alzheimer’s patients.58 GS is observed to be heavily populated in Müller cells of the neural retina.59 However, it is not present in the RPE of healthy adult mammals.60 Interestingly, GS is highly expressed in the RPE of fetal bovine retinae, indicating that the supply of glutamine by GS to RPE cells is critical for early cell development and proliferation.61 In addition, GS is often detected in pathological diseases of the human retina, where it may be responsible for providing the glutamine needed for proliferation.61
Overall, the structure of bovine GS in the RPE sample has a very similar fold to that of canine GS.62 Bovine GS consists of 373 amino acids, of which 343 residues are ordered and included in the final structure. Superimposition of our cryo-EM structure of bovine GS to the X-ray structure of canine GS (PDB: 2UU7)62 gives an RMSD of 0.46 Å (for 337 Cα atoms). GS assembles as a dimer of pentamers (Figures 4A and 4B). The 10 identical protein chains are arranged as two donut-shaped pentamers that stack to two layers. Each protomer is folded into a small N-terminal domain (residues 2–102) and a large C-terminal domain (residues 114–372) (Figure 4C). These two domains are connected by a flexible loop made up of 11 residues (residues 103–113). The small N-terminal domain forms four α helices and five β strands, whereas the large C-terminal domain possesses seven α helices and seven β strands. It appears that the N-terminal domain is responsible for inter-subunit interactions to secure the pentameric oligomerization within a layer of the pentamer. The two pentameric layers are locked by loops composed of residues 149–157 within the C-terminal domain of each subunit. The five loops from one pentamer contact the respective five loops from the second pentamer to anchor these two layers against each other.
The C-terminal domain of GS forms a catalytic site for substrates. Within the catalytic site, we observed a Mn2+ ion bound by residues E134, H253, and E338. This site is also surrounded with several positively charged arginines, including R262, R324, R340, and R341, which contribute to substrate binding (Figure 4D). Many of these residues are involved in binding ADP and phosphate in the human GS enzyme.62 However, we did not observe any other extra densities corresponding to these ligands in the structure of bovine GS.
Interestingly, two reports of congenital human GS deficiency have been documented. Both cases resulted in severe brain malformations with multiorgan failure and neonatal death.63 Each infant had a homozygous mutation in the GS gene, where residues R324 and R341 of GS were replaced by cysteines.63 These two cases highlighted the significance of these two substrate-binding residues, which are critical for GS activity.
Dihydropyrimidinase-related protein 2
We identified that our bovine RPE sample contains the enzyme DPYSL2. We extracted 12,503 single-particle images for this enzyme and ascertained its identity using the cryo-EM map generated from BaR (Figure S10; Table S2). We then solved its cryo-EM structure to a resolution of 2.66 Å and also extended the resolution of this bovine DPYSL2 structure to a resolution of 2.17 Å by density modification17 (Figure 5).
Figure 5. Structure of bovine DPYSL2.
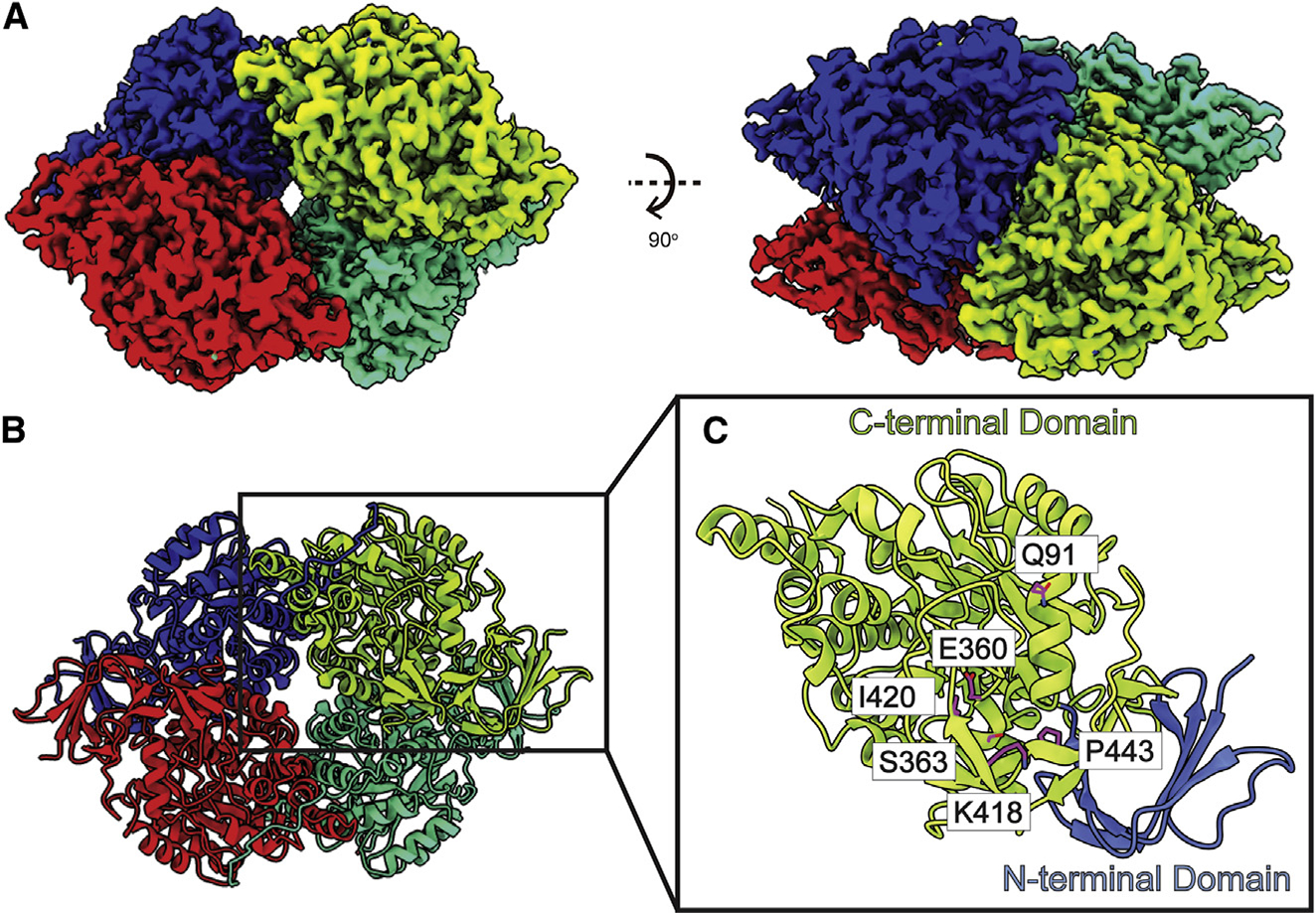
(A) Cryo-EM map of DPYSL2.
(B) Structure of DPYSL2. Individual DPYSL2 subunits are distinguished by different colors.
(C) Organization of a DPYSL2 monomer. This protein can be divided into a small N-terminal β-barrel domain (cyan) followed by a large C-terminal TIM-barrel domain (red). Residues engaged in forming the substrate-binding site are in violet sticks.
DPYSL2, also known as collapsing response mediator protein 2 (CRMP2), has drawn wide attention for its crucial role in the pathogenesis of the serious mental illness schizophrenia.64–66 This important enzyme is able to mediate axonal outgrowth in the developing brain. It regulates neuronal polarity to maintain proper cytoskeletal dynamics and vesical trafficking.67 It has been observed that altering the expression of DPYSL2 in cultured rat hippocampal neurons significantly affects axonal growth.68 The ablation of DPYSL2 leads to neurodevelopmental disorders, including unregulated axon growth and branching.69 Importantly, DPYSL2 is a key regulator for promoting neural stem cell differentiation into neurons, astrocytes, and oligodendrocytes, cells that could then be used to replace necrotic cells caused by brain or spinal cord injuries.69 Therefore, understanding the structural and functional roles of DPYSL2 could provide insight into the design of neural stem cell-based therapeutic treatments for brain and spinal cord injuries.
Similar to crystal structures of murine CRMP170 and human CRMP2,71 our bovine DPYSL2 cryo-EM structure is tetrameric in form (Figures 5A and 5B). Each subunit contains 572 amino acids, and residues 14–506 of each subunit of the DPYSL2 tetramer were included in the final structure. Superimposition of our cryo-EM structure of bovine DPYSL2 to the X-ray structure of murine CRMP1 (PDB: 1KCX)70 allows us to obtain an RMSD of 0.55 Å (for 464 Cα atoms). Each protomer of this enzyme consists of a small N-terminal β sheet domain (residues 14–68) consisting of seven β strands. Following this domain, the majority of the protomer is folded into a mixture of α and β secondary structural elements, resembling the α/β barrel of triosephosphate isomerase (TIM). This C-terminal TIM-barrel domain (residues 69–487) consists of 17 α helices and 14 β strands (Figure 5C). As indicated in the structure of human CRMP2, the active site of bovine DPYSL2 should be located within the α/β TIM barrel. Residues Q91, E360, S363, K418, I420, and P443 are likely engaged in substrate binding.72,73 The C terminus of bovine DPYSL2 forms an extended unstructured random segment (residues 488–506). Interestingly, this C-terminal random tail has no detectable sequence homology to other proteins71 and it is readily cleaved off both in vivo and in vitro.70 Our structure indicates that this C-terminal tail extends to the next subunit of DPYSL2, intimately contacting the TIM-barrel domain of this protomer. As we did not observe any bound ligands, our cryo-EM structure should represent the apo form of bovine DPYSL2.
A missense mutant, Q91R, within the substrate-binding site of human DPYSL2 has been found to cause seizure, where lacosamide, an anti-seizure drug, has been used to effectively reduce seizure frequency in patients with uncontrolled partial-onset seizures.72,73 Residue Q91 appears critical as it is involved in binding this anti-seizure drug.72,73
Fructose-bisphosphate aldolase
We recorded 6,223 images for this class of single particles. BaR allowed us to determine that these particles represent bovine FPA. We then determined the cryo-EM structure of this enzyme to a resolution of 3.12 Å (Figure S11; Table S2). Density modification17 led us to extend the resolution to 2.52 Å (Figure 6).
Figure 6. Structure of bovine FPA.
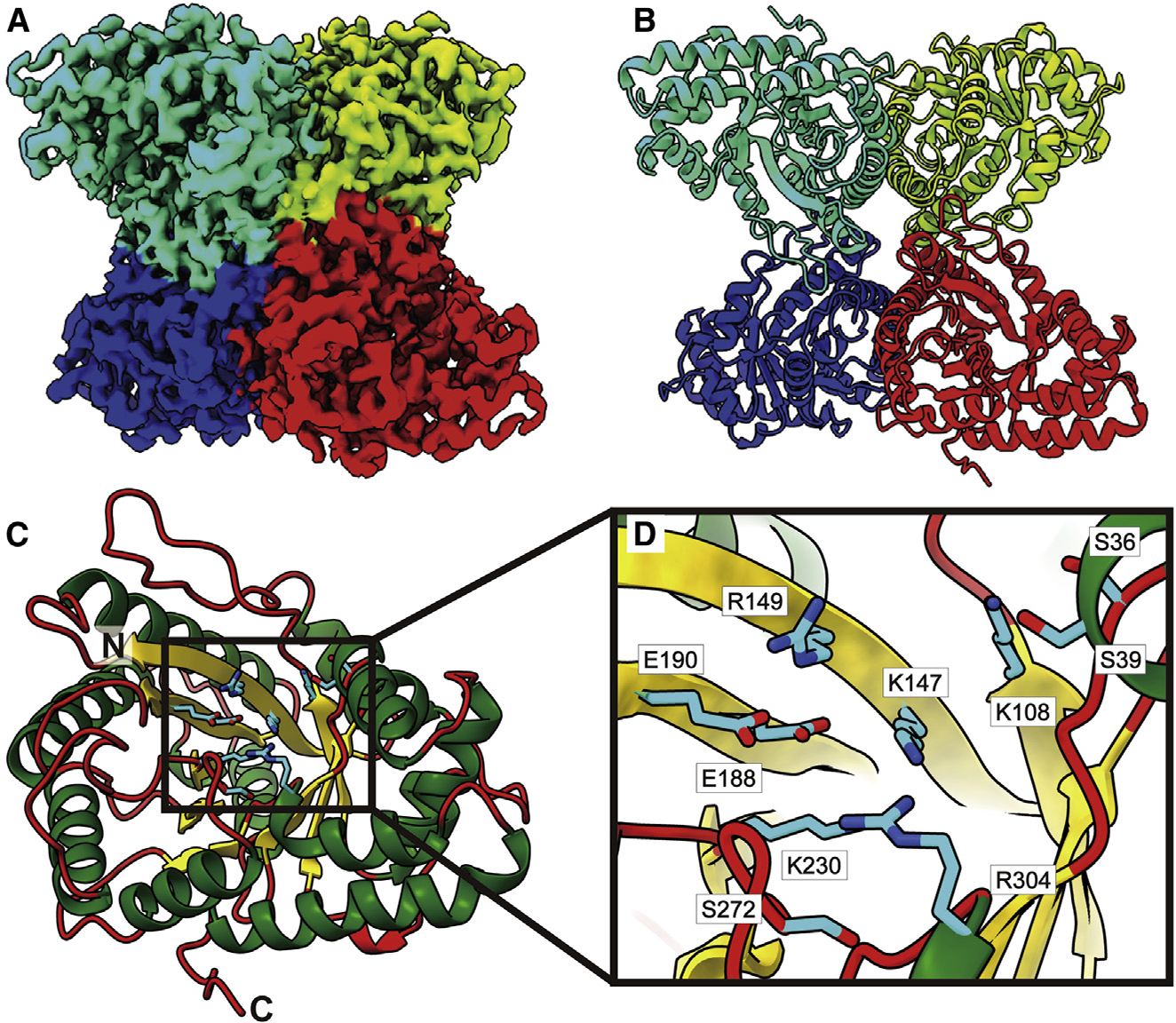
(A) Cryo-EM map of FPA.
(B) Structure of FPA. Individual FPA subunits aredistinguished by different colors.
(C) Structure of an FPA monomer.
(D) Zoomed view of the substrate-binding site. In both (C) and (D), the α helices, β strands, and flexible loops are colored green, yellow, and red, respectively. Residues forming the substrate-binding sites are in cyan sticks.
FPA, often simply called aldolase, is a key enzyme in both gluconeogenesis and glycolysis. It catalyzes a reversible reaction by converting fructose-1,6-bisphosphate into dihydroxyacetone phosphate and glyceraldehyde-3-phosphate.74 These FPA enzymes are multifunctional, involved in other processes in addition to its specific role in carbohydrate metabolism. It has been reported that FPAs participate in a number of non-enzymatic moonlighting functions. They have the ability to bind an array of proteins, interfering with the processes of cellular scaffolding, signaling, transcription, and motility.75–79 Because of the essential and multifunctional nature of FPAs, these enzymes are attractive drug targets for several human diseases, including cancers and pathogenic infections.
Bovine FPA is observed to be tetrameric in oligomerization (Figures 6A and 6B). Each FPA subunit consists of 364 amino acids (345 residues are included in the final structure) and features a β-barrel consisting of nine β strands at its core. This β-barrel is housed by a frame-like structure created by 13 α helices. Therefore, the tetrameric enzyme possesses four β-barrels that are all wrapped by α helices (Figure 6C). Interestingly, each β-barrel is populated with both charged and polar amino acids that likely interact with substrates (Figure 6D). It has been observed that the corresponding charged and polar residues of S36, S39, K108, K147, R149, E188, E190, K230, S272, and R304 in human FPA are engaged in contacting the sugar fructose 1,6-bisphosphate.80 Overall, our cryo-EM structure is in good agreement with crystal structures of the human and rabbit FPA enzymes.80–83 Superimposition of the cryo-EM structure of bovine FPA to the X-ray structure of rabbit FPA (PDB: 1ADO)81 leads to an RMSD of 0.47 Å (for 343 Cα atoms). As we did not observe any bound ligands at the β-barrel substrate-binding site, our cryo-EM structure should depict the ligand-free form of the bovine FPA enzyme.
Mutagenesis and chemical modification studies have shown that the aspartate and lysine residues at the substrate-binding site are critical for the function of the FPA enzymes.84–89 Interestingly, there was a study involving a patient with hereditary fructose intolerance.90 The patient has a six-nucleotide deletion in exon 6, perturbing the position and orientation of the corresponding K147 and E188 residues of human FPA at the active site.90 This missense mutation probably diminishes the binding of substrates leading to the observed disease.
Mitochondrial creatine kinase
We obtained 6,693 single-particle images for this soluble protein. This protein was identified as an isozyme of bovine ubiquitous MtCK (uMtCK) (Figure S12; Table S2). We solved its cryo-EM structure to a resolution of 3.12 Å with density modification17 allowing us to extend the structural resolution of this MtCK enzyme to 2.52 Å (Figure 7).
Figure 7. Structure of bovine uMtCK.
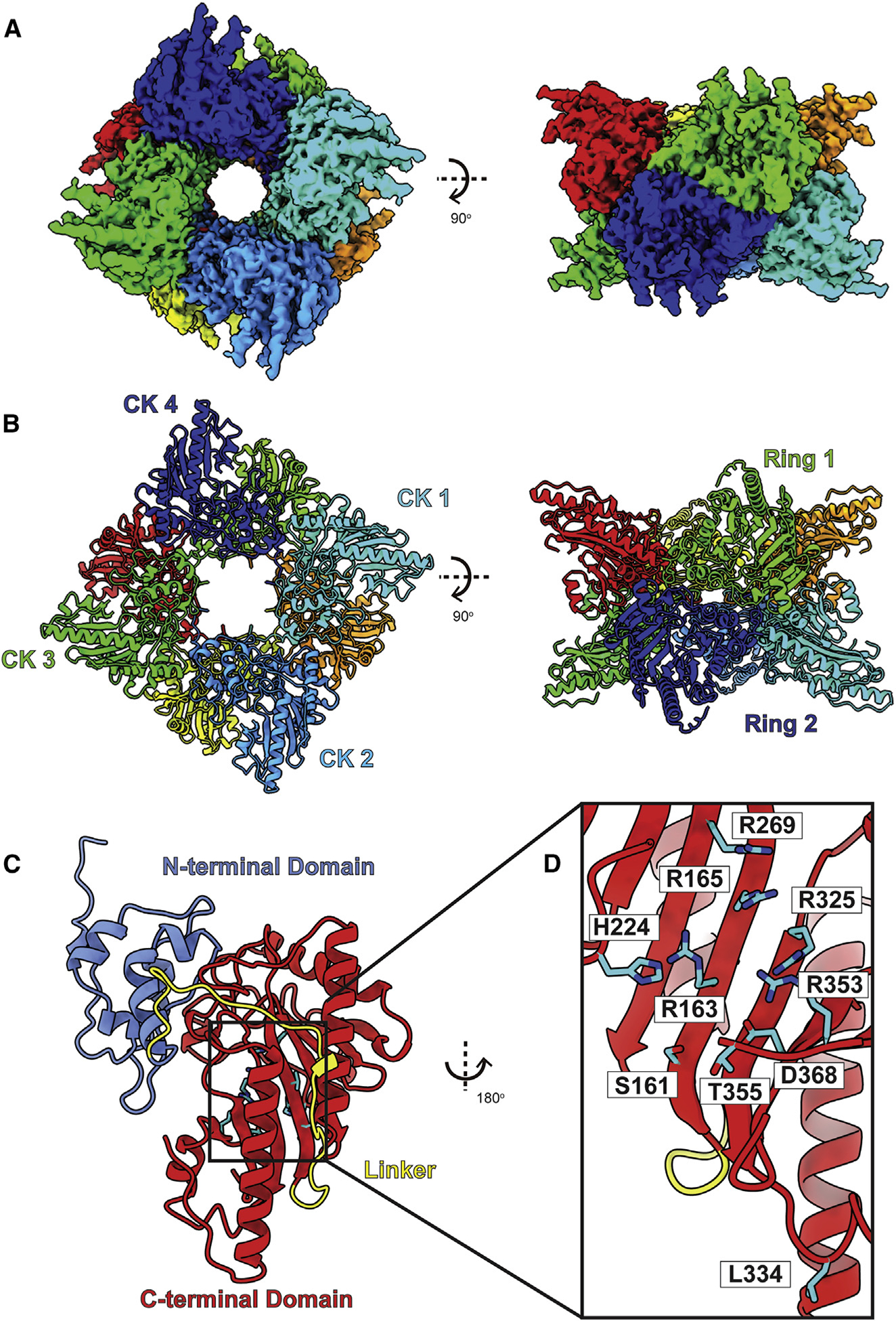
(A) Cryo-EM map of uMtCK.
(B) Structure of uMtCK. Individual uMtCK subunits are distinguished with different colors.
(C) Each bovine uMtCK subunit contains a small N-terminal dimerization domain (cyan) and a large C-terminal catalytic domain (red) connected by a flexible linker (yellow).
(D) Zoomed view of the substrate-binding site. Residues responsible for forming this site are in cyan sticks.
MtCK is a central controller of cellular energy homeostasis.91 It catalyzes the reversible transfer of phosphate between ATP and creatine into ADP and phosphocreatine and provides a large pool of diffusing phosphocreatine to channel energy. This enzyme plays a critical role in energy transduction in tissues with large, fluctuating energy demands, such as the brain, heart, retina, spermatozoa, and skeletal muscle.92–95 In vertebrates, MtCK exists as two isoenzymes, uMtCK and sarcomeric MtCK (sMtCK), which are encoded by the genes CKMT1 and CKMT2, respectively.96–98 uMtCK has been detected in most tissues, including brain, kidney, and sperm, whereas sMtCK has been found to express in heart and skeletal muscles.92,94,95
As an enzyme responsible for controlling cell energetics, MtCK is linked to many human diseases. MtCK is a prime target of oxidative and radical-induced molecular damage.91 Impairment of MtCK has been observed in ischemia, cardiomyopathy, and neurodegenerative disorders, probably due to incompetency in supporting metabolic homeostasis and energy coupling, which leads to further energy depletion.91 Inactivation and aberration of MtCK have often been seen in patients with Alzheimer’s disease and amyotrophic lateral sclerosis (ALS).99,100 On the other hand, overexpression of MtCK has been detected in several solid tumors with poor prognosis such as adenocarcinomas.101 In these cases, MtCK, which has been suggested to be a tumor-associated marker,102 may facilitate energy channeling to maintain the high growth rate of cancer cells.
Bovine uMtCK contains 416 amino acids per subunit (363 residues are included in the final model) and presents an octameric oligomerization (Figures 7A and 7B). Each subunit of uMtCK consists of a small N-terminal domain (residues 42–131) and a large C-terminal domain (residues 159–411) (Figure 7C), similar to those found in crystal structures of the chicken mitochondrial and human mitochondrial enzymes.103,104 These two domains are connected by a flexible loop comprising residues 132–158. The small N-terminal domain contains six α helices, whereas the large C-terminal domain consists of eight α helices and eight β strands. Superimposition of the cryo-EM structure of bovine uMtCK to the X-ray structure of human uMtCK (PDB: 1QK1)103 gives rise to an RMSD of 0.59 Å (for 358 Cα atoms).
The C-terminal domain of uMtCK creates a nucleotide-binding site that anchors ATP and is predominantly cationic in nature. Residues S161, R163, R165, H224, R269, R325, L334, R353, T355, and D368 are responsible for forming this binding site. Our cryo-EM structure indicates that there are no bound ligands located at the substrate-binding site. Therefore, this bovine uMtCK structure represents the apo form of this enzyme.
It has been identified that an alteration of residue L334 in the nucleotide-binding site of human uMtCK to an arginine or a valine leads to a missense mutation that significantly diminishes the function of this creatine kinase.105 Based on our cryo-EM structure, L334 is located at the C-terminal flexible tail of uMtCK and is at least 3.8 Å apart from the rest of the substrate-binding residues. Therefore, it is expected that a major conformational change would occur that shifts the position of this important residue toward the rest of the binding residues to accommodate binding when this enzyme is occupied by substrates.
DISCUSSION
Cellular function is complex and orchestrated by interactions between different proteins, enzymes, and biomolecules that often further coordinate with small molecules and metabolites in order to maintain homeostasis. To unfurl the complexity of living tissues and organs, the approach of systems biology is desirable as it allows a more comprehensive view of the biological process. We previously developed a BaR methodology, a systems structural proteomic approach to identify and obtain near-atomic-resolution structural models of multiple proteins from a raw biological sample. We demonstrated that this methodology is capable of allowing us to solve structures of a number of relatively small and less abundant unidentified proteins within a single, heterogeneous sample. In addition, since this technique uses protein isolated from native tissue, we are able to validate previous structures solved using recombinant proteins and verify their native ligands.
In the current study, we use BaR to investigate the bovine RPE tissue, permitting us to establish a cryo-EM structural biology approach to elucidate the proteome of ocular tissues to high resolution. It appears that the identified seven bovine RPE enzymes are interconnected and participate in a range of interactions based on the predicted interaction network (Figure S2).
Despite these seven enzymes being symmetrical in nature, it is worth noting that BaR has the capability of identifying and solving structures of asymmetric complexes. This has been clearly demonstrated from the previous work, in which BaR was utilized to resolve the cryo-EM structure of cytochrome bo3, which is a 143-kDa asymmetric complex created by four different subunits.14 The work also allowed for a simultaneous determination of a 93-kDa Burkholderia pseudomallei HpnN transporter, a monomeric membrane protein without any symmetrical subunits, from the same sample.14
The BaR methodology can also be used to identify endogenous ligands and/or ions. This is shown by the case of the GAPDH enzyme, where an NAD+ ligand is found to anchor in this protein. In addition, BaR allowed us to observe bound native ions of the FT, DNPEP, and GS in their corresponding binding sites. Additionally, BaR can be used to elucidate structures of posttranslational modifications, glycosylations, and cysteine modifications of enzymes based on the cryo-EM maps (<3.5 Å resolution). These modifications can also be validated by mass spectrometry.
Although we did not observe any previously undefined enzyme structures in this work, BaR is capable of identifying and solving structures of proteins even if the populations of corresponding particles are as low as 5%. This methodology is also able to determine enzyme complexes, allowing us to understand how different enzymes interact within the native tissue sample. Additionally, BaR should permit us to simultaneously elucidate different conformational states of a particular protein in a single cryo-EM grid. This has been demonstrated previously on how a transporter interacts with its respective ligand.106,107 In addition to allowing for the acquisition of additional views and more particles (as seen in the GAPDH, FT, DNPEP, and DYPSL2 proteins), the BaR methodology can substantially improve the cryo-EM map quality by removing contaminating particles. This has been demonstrated in the cases of the GS, FPA, and uMtCK enzymes.
Several of the RPE enzymes identified in our enriched sample are directly linked to neurodevelopment and neurodegenerative disorders. For example, it was observed that GS activity was reduced in the vicinity of amyloid plaques in patients with Alzheimer’s disease.108 Recently, an interesting study indicated that patients with probable Alzheimer’s disease or major depression showed elevated levels of glutamate and glutamine in the cerebrospinal fluid.109 As GS is responsible for converting glutamate to glutamine, removing the toxicity of glutamate at concentrations that may lead to neuronal death, it is appealing to suggest that GS may be a good diagnostic marker for Alzheimer’s disease. In addition to GS, both GAPDH and FT are directly related to several neurodegenerative disorders, such as Huntington’s, Parkinson’s, and Alzheimer’s diseases.110,111 Interestingly, it has been identified that the levels of GAPDH in the hippocampus and FT in the cerebrospinal fluid are significantly higher in Alzheimer’s patients.112,113 Additionally, it is known that DPYSL2 plays a critical role in the pathogenesis of schizophrenia,64–66 and inactivation of MtCK is often found in patients with Alzheimer’s and ALS diseases.99,100
Some of these RPE proteins are interconnected with cancer diseases. For instance, a high level of MtCK has been observed for several solid tumors with poor prognosis, including adenocarcinomas.101 Likewise, GAPDH is found to promote cancer growth and metastasis.110 It is highly expressed in human lung cancers, and its overexpression often correlates with poor prognosis of lung cancer patients.114,115 Further, the FPA enzyme is noted to promote lung cell tumorigenesis and migration.116
Due to the fact that many of these enzymes are tightly connected to neurodegenerative disorders or cancer diseases, our BaR methodology is exciting in that it enables us to study these proteins simultaneously in a single sample. Notably, both MtCK and FPA have been suggested to be promising tumor-associated markers.102,116 Specific to the eye, DNPEP has been proposed to be a drug target for glaucoma and retinopathy diseases,52–54 whereas FT has been suggested to be a strong biomarker for ferritinemia cataract and age-related macular degeneration.117 Our work strongly indicates that BaR can be used to overcome the problem of sample impurity and heterogeneity, enabling us to utilize the cryo-EM structural approach to simultaneously solve structures of a variety of enzymes from a tissue sample at high resolutions.
Limitations of the study
Although BaR allows us to simultaneously solve structures of different proteins from a raw sample, this methodology has its limitations. First, it is not easy, if not impossible, to identify a particular protein if the population of the protein is <5% within the heterogeneous sample. Second, it is difficult to unambiguously identify a protein when the cryo-EM map is not better than 3.5 Å in resolution. In addition, preferential orientation of the protein images can be a problem. In this case, BaR may not be able to offer a good solution for the protein. The problem of preferential orientation of images may also limit the quality of the initial model and create difficulties during initial retrieval, although we may be able to overcome this problem by going through multiple iterations of the BaR protocol. Regardless, coupled with mass spectrometry, we believe that the BaR methodology can be used to illuminate the details of biological pathways, networks, and even mechanisms of diseases at near-atomic resolution.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Edward W. Yu (edward.w.yu@case.edu).
Materials availability
This study did not generate unique reagents.
Data and code availability
Coordinates and EM maps for GS, GAPDH, FT, DPYSL2, FPA, MtCK and DNPEP can be found at PDB accession numbers 7U5N, 7U5M, 7U5L, 7U5K, 7U5J, 7U5I and 7U5H and EMDB accession codes EMD-26356, EMD-26355, EMD-26354, EMD-26353, EMD-26352, EMD-26351, and EMD-26350, respectively. The raw cryo-EM data have been deposited in EMPIAR (https://www.ebi.ac.uk/empiar/). The raw mass spectrometry proteomics data have been deposited in ProteomeXchange via the PRIDE database (https://www.ebi.ac.uk/pride/). Accession numbers, codes, and links to access the structural data, raw cryo-EM images and raw mass spectrometry proteomics data are also listed in the key resources table. All data reported in this paper are available upon requested from the Lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Biological samples | ||
|
| ||
| Fresh bovine eyes | Mahan Packing Company | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Acetonitrile | Millipore Sigma | Cat# 1000291000 |
| Ammonium bicarbonate | Sigma-Aldrich | Cat# 5330050050 |
| Graphene oxide | Sigma-Aldrich | Cat# 763705-100ML |
| Iodoacetamide | GE Healthcare | Cat# RPN6302 |
| Trypsin/Lys-C Mix | Promega | Cat# V5073 |
|
| ||
| Deposited data | ||
|
| ||
| Raw cryo-EM data | This paper | EMPIAR-11251 https://doi.org/10.6019/EMPIAR-11251 |
| Cryo-EM structure and map of GS | This paper | PDB ID: 7U5N EMDB ID: EMD-26356 https://doi.org/10.2210/pdb7U5N/pdb |
| Cryo-EM structure and map of GAPDH | This paper | PDB ID: 7U5M EMDB ID: EMD-26355 https://doi.org/10.2210/pdb7U5M/pdb |
| Cryo-EM structure and map of FT | This paper | PDB ID: 7U5L EMDB ID: EMD-26354 https://doi.org/10.2210/pdb7U5L/pdb |
| Cryo-EM structure and map of DPYSL2 | This paper | PDB ID: 7U5K EMDB ID: EMD-26353 https://doi.org/10.2210/pdb7U5K/pdb |
| Cryo-EM structure and map of FPA | This paper | PDB ID: 7U5J EMDB ID: EMD-26352 https://doi.org/10.2210/pdb7U5J/pdb |
| Cryo-EM structure and map of MtCK | This paper | PDB ID: 7U5I EMDB ID: EMD-26351 https://doi.org/10.2210/pdb7U5I/pdb |
| Cryo-EM structure and map of DNPEP | This paper | PDB ID: 7U5H EMDB ID: EMD-26350 https://doi.org/10.2210/pdb7U5H/pdb |
| Raw mass spectrometry proteomics data | This paper | PXD037186 (Table S3) |
|
| ||
| Software and algorithms | ||
|
| ||
| Coot | Emsley et al.118 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| cryoSPARC v3 | Punjani et al.119 | https://cryosparc.com/ |
| MaxQuant | Cox and Mann | https://www.maxquant.org/ |
| Phenix | Adams et al.120 | https://phenix-online.org/ |
| STRING database | STRING | https://string-db.org/ |
| UCSF-Chimera | Pettersen et al.121 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF-Chimera X | Pettersen et al.122 | https://www.cgl.ucsf.edu/chimerax/ |
|
| ||
| Other | ||
|
| ||
| C18 Microspin column | Nest Group | Cat# SEM SS18V |
| Quantifoil R 2/2 Cu 200 mesh, copper | Quantifoil | Cat# Q210CR2 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fresh bovine eyes (calf or adult) from both male and female animals were obtained from a local slaughterhouse (Mahan Packing Company Inc., Bristolville, OH). Retinal pigmented epithelium (RPE) cells were isolated by dissecting individual bovine eyes. The generated microsome samples were stored at −80°C.
METHOD DETAILS
Isolation of RPE cells from bovine eyes
Fresh bovine eyes (calf or adult) were obtained from a local slaughterhouse (Mahan Packing Company Inc., Bristolville, OH). Individual eyes were dissected with the anterior segment, lens, and vitreous discarded. Exposed retina was gently peeled off with forceps and collated.123 Cold buffered sucrose (1 mL/eye) was added to the eye cup and the retinal pigmented epithelium (RPE) cells were isolated by gently stroking from center to periphery with a small brush. The dark suspension was removed using a Pasteur pipet. After repeating this procedure, cells were homogenized and centrifuged at 20,000 g for 20 min at 4°C. The supernatant was centrifuged at 150,000 g for 1 h at 4°C. The microsomal fraction was then resuspended in 10 mL of 10 mM MOPS, pH 7.0, 2 mM DTT and stored in small aliquots at −80°C.
Proteomic analysis of RPE cell homogenates
The protein of RPE cell homogenate in the two peaks from the size-exclusion chromatography enrichment (200 μL) was digested by trypsin. The resulting peptides were desalted by a C18 Microspin column (Nest Group, Ipswich, MA) per the manufacturer’s instruction and analyzed by LC-MS/MS using a ThermoScientific Fusion Lumos mass spectrometry system.124 Proteins were identified by comparing all of the experimental peptide MS/MS spectra against the UniProt human database using the Andromeda search engine integrated into the MaxQuant version 1.6.3.3.125,126 Carbamidomethylation of cysteine was set as a fixed modification, whereas variable modifications included oxidation of methionine to methionine sulfoxide and acetylation of N-terminal amino groups. For peptide/protein identification, strict trypsin specificity was applied, the minimum peptide length was set to 7, the maximum missed cleavage was set to 2, and the cutoff false discovery rate was set to 0.01. Match between runs (match time window: 0.7 min; alignment time window: 20 min) and label-free quantitation (LFQ) options were enabled. The LFQ minimum ratio count was set to 2. The remaining parameters were kept as default.
Cryo-EM sample preparation
Each lysate sample was collected from the corresponding microsomal fraction. A 100 μL of sample was mixed with 100 μL buffer (10 mM Tris pH 7.5 and 100 mM NaCl) and incubated at 4°C for 1 h. The mixture was spun at 20,000 × g for 30 min to remove the membrane fraction. Sample was then passed through a 0.22 μm centrifugal filter and enriched using size-exclusion chromatography (Superdex 200 increase, GE Healthcare). Protein sizes corresponding to 300–650 kDa and 100–250 kDa were used for cryo-EM analysis. 3.5 μL (0.07–0.08 mg/mL) of each sample was applied to Quantifoil R 2/2 Cu 200 holey grids coated with graphene oxide. Samples were blotted for 8 s and plunge frozen into liquid ethane using a Vitrobot (Thermo Fisher). The resulting grids were stored in liquid nitrogen until data collection.
Data collection
A Titan Krios equipped with a K3 direct electron detector was used to collect data in super-resolution mode at 81,000× magnification (physical pixel size of 1.070 Å/pix, 0.535 Å/pix super-resolution). Micrographs of the two samples were collected in two separate datasets (peak 1, 35 frames, total dose 36 or 37 e−/Å2; peak 2, 37 frames, total dose 36 e−/Å2). Data were collected in correlated-double sampling mode using Serial EM.127
Data processing
Data were binned by 2 and processed using patch motion and patchCTF in cryoSPARC v3.119 All micrographs were picked using Topaz.128 A modified BaR protocol was used to determine the different structures from both samples (Figure S1), closely following its initial application in prokaryotic systems.14 Initially, particles were cleaned using multiple rounds of 2D classification, manually selecting clear classes and repeating classification until no new clear classes were seen. Visually similar classes were manually combined. Initial low-resolution maps for individual proteins were solved using iterative rounds of two-class ab initio 3D reconstruction followed by homogeneous refinement in cryoSPARC. At this stage, most volumes suffered from orientation bias and low particle counts. These low-resolution 3D models were used as targets for four rounds of heterogeneous refinement of the initial particle sets along with noisy junk ab initio classes that served as particle sinks, resulting in more particles for each target protein. These individual particle sets were then cleaned using 2D classification and multi-class 3D ab initio reconstruction and refined using non-uniform refinement in cryoSPARC v3.119 2D classes commonly appeared after a round of BaR, therefore the process of manually solving initial low-resolution classes for inputs to heterogeneous refinement was performed iteratively until no new clear classes were uncovered from the remaining particles by either 2D or 3D classification. A final round of retrieval with all maps was used to separate each dataset and solve the final high-resolution classes using 2D/3D classification followed by non-uniform refinement with applied symmetry in cryoSPARC v3.119
At this stage, protein identities were still unknown. Once sufficient resolution was reached (~3.5 Å), protein identities were determined directly from the BaR maps using Deep Tracer.129 Results were used for sequence alignment in BLASTP,130 using Bos Taurus as the target species, accurately identifying all proteins. The presence of these identified enzymes was also confirmed via mass spectrometry (Table S1).
Interaction network
The interaction network connecting the seven identified enzymes was predicted using the STRING database.16 The enzymes from Bos taurus were input as a multiprotein search and the network was extended to display 50 interactions to show full connectivity. Line thickness was used to depict interaction confidence.
Model building and refinement
Initial models were build using Swiss Model via the Uniprot database and aligned to the cryo-EM maps in Chimera.121,122 Models were refined using phenix.real_space_refine120 and Coot.118 Final structures were evaluated using MolProbity.131 3D FSC was used to quantitatively evaluate each cryo-EM map at distinct viewing directions.132
QUANTIFICATION AND STATISTICAL ANALYSIS
Standard GS-FSC (Gold Standard-Fourier Shell Correlation) curves at a threshold of 0.143 were computed using cryoSPARC v3119 to obtain final resolutions of protein models. The final atomic models were evaluated using MolProbity.131
Supplementary Material
Highlights.
Using cryo-EM to define the composition of a raw bovine RPE lysate
Simultaneously identifying and solving cryo-EM structures of seven RPE enzymes
Noting the importance of cryo-EM in studying structural-omics at the atomic level
ACKNOWLEDGMENTS
We thank Dr. Philip A. Klenotic for proofreading the manuscript. This work was supported by NIH grants R01AI145069 (E.W.Y.) and R01EY023948 (M.G.). The mass spectrometer used was purchased with an NIH Shared Instrument Grant (S10 RR031537). We thank Belinda Willard and Ling Li for the acquisition of mass spectrometry data. We are grateful to the Cryo-Electron Microscopy Core at the CWRU School of Medicine and Dr. Kunpeng Li for access to the sample preparation and cryo-EM instrumentation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111876.
REFERENCES
- 1.Boulton M, and Dayhaw-Barker P (2001). The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye 15, 384–389. 10.1038/eye.2001.141. [DOI] [PubMed] [Google Scholar]
- 2.Figueroa AG, and McKay BS (2019). GPR143 signaling and retinal degeneration. Adv. Exp. Med. Biol. 1185, 15–19. 10.1007/978-3-030-27378-1_3. [DOI] [PubMed] [Google Scholar]
- 3.Talib M, van Schooneveld MJ, van Duuren RJG, Van Cauwenbergh C, Ten Brink JB, De Baere E, Florijn RJ, Schalij-Delfos NE, Leroy BP, Bergen AA, and Boon CJF (2019). Long-term follow-up of retinal degenerations associated with LRAT mutations and their comparability to phenotypes associated with RPE65 mutations. Transl. Vis. Sci. Technol. 8, 24. 10.1167/tvst.8.4.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatz P, Preising M, Lorenz B, Sander B, Larsen M, and Rosenberg T (2011). Fundus albipunctatus associated with compound heterozygous mutations in RPE65. Ophthalmology 118, 888–894. 10.1016/j.ophtha.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Tsang SH, Aycinena ARP, and Sharma T (2018). Inborn errors of metabolism: gyrate atrophy. Adv. Exp. Med. Biol. 1085, 183–185. 10.1007/978-3-319-95046-4_37. [DOI] [PubMed] [Google Scholar]
- 6.Guziewicz KE, Sinha D, Gómez NM, Zorych K, Dutrow EV, Dhingra A, Mullins RF, Stone EM, Gamm DM, Boesze-Battaglia K, and Aguirre GD (2017). Bestrophinopathy: an RPE-photoreceptor interface disease. Prog. Retin. Eye Res. 58, 70–88. 10.1016/j.preteyeres.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain RM, Ciulla TA, Berrocal AM, Gregori NZ, Flynn HW, and Lam BL (2018). Stargardt macular dystrophy and evolving therapies. Expert Opin. Biol. Ther. 18, 1049–1059. 10.1080/14712598.2018.1513486. [DOI] [PubMed] [Google Scholar]
- 8.Ao J, Wood JP, Chidlow G, Gillies MC, and Casson RJ (2018). Retinal pigment epithelium in the pathogenesis of age-related macular degeneration and photobiomodulation as a potential therapy? Clin. Exp. Ophthalmol. 46, 670–686. 10.1111/ceo.13121. [DOI] [PubMed] [Google Scholar]
- 9.De S, Rabin DM, Salero E, Lederman PL, Temple S, and Stern JH (2007). Human retinal pigment epithelium cell changes and expression of αB-crystallin: a biomarker for retinal pigment epithelium cell change in age-related macular degeneration. Arch. Ophthalmol. 125, 641–645. 10.1001/archopht.125.5.641. [DOI] [PubMed] [Google Scholar]
- 10.Ambati J, Ambati BK, Yoo SH, Ianchulev S, and Adamis AP (2003). Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv. Ophthalmol. 48, 257–293. 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 11.Congdon N, O’Colmain B, Klaver CCW, Klein R, Muñoz B, Friedman DS, Kempen J, Taylor HR, and Mitchell P; Eye Diseases Prevalence Research Group (2004). Causes and prevalence of visual impairment among adults in the United States. Arch. Ophthalmol. 122, 477–485. 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad MT, Zhang P, Dufresne C, Ferrucci L, and Semba RD (2018). The human eye proteome project: updates on an emerging proteome. Proteomics 18, 1700394. 10.1002/pmic.201700394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skeie JM, and Mahajan VB (2014). Proteomic landscape of the human choroid–retinal pigment epithelial complex. JAMA Ophthalmol. 132, 1271–1281. 10.1001/jamaophthalmol.2014.2065. [DOI] [PubMed] [Google Scholar]
- 14.Su C-C, Lyu M, Morgan CE, Bolla JR, Robinson CV, and Yu EW (2021). A “Build and Retrieve” methodology to simultaneously solve cryo-EM structures of membrane proteins. Nat. Methods 18, 69–75. 10.1038/s41592-020-01021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho C-M, Li X, Lai M, Terwilliger TC, Beck JR, Wohlschlegel J, Goldberg DE, Fitzpatrick AWP, and Zhou ZH (2020). Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nat. Methods 17, 79–85. 10.1038/s41592-019-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, and Afonine PV (2020). Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ismail SA, and Park HW (2005). Structural analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 61, 1508–1513. 10.1107/S0907444905026740. [DOI] [PubMed] [Google Scholar]
- 19.Baker BY, Shi W, Wang B, and Palczewski K (2014). High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules. Protein Sci. 23, 1629–1639. 10.1002/pro.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moras D, Olsen KW, Sabesan MN, Buehner M, Ford GC, and Rossmann MG (1975). Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 250, 9137–9162. 10.1016/S0021-9258(19)40703-5. [DOI] [PubMed] [Google Scholar]
- 21.Biesecker G (1977). Sequence and Structure of N-Glyceraldehyde 3-phosphate Dehydrogenase from, 266, p. 6. [DOI] [PubMed] [Google Scholar]
- 22.Seidler NW (2013). Basic biology of GAPDH. Adv. Exp. Med. Biol. 985, 1–36. 10.1007/978-94-007-4716-6_1. [DOI] [PubMed] [Google Scholar]
- 23.Sirover MA (1999). New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432, 159–184. 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 24.Boradia VM, Raje M, and Raje CI (2014). Protein moonlighting in iron metabolism: glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Biochem. Soc. Trans. 42, 1796–1801. 10.1042/BST20140220. [DOI] [PubMed] [Google Scholar]
- 25.Sweeny EA, Singh AB, Chakravarti R, Martinez-Guzman O, Saini A, Haque MM, Garee G, Dans PD, Hannibal L, Reddi AR, and Stuehr DJ (2018). Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem. 293, 14557–14568. 10.1074/jbc.RA118.004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huitorel P, and Pantaloni D (1985). Bundling of microtubules by glyceraldehyde-3-phosphate dehydrogenase and its modulation by ATP. Eur. J. Biochem. 150, 265–269. 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 27.Walsh JL, Keith TJ, and Knull HR (1989). Glycolytic enzyme interactions with tubulin and microtubules. Biochim. Biophys. Acta 999, 64–70. 10.1016/0167-4838(89)90031-9. [DOI] [PubMed] [Google Scholar]
- 28.Rao ST, and Rossmann MG (1973). Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256. 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 29.Sen T, Saha P, and Sen N (2018). Nitrosylation of GAPDH augments pathological tau acetylation upon exposure to amyloid-β. Sci. Signal. 11, eaao6765. 10.1126/scisignal.aao6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazarev VF, Guzhova IV, and Margulis BA (2020). Glyceraldehyde-3-phosphate dehydrogenase is a multifaceted therapeutic target. Pharmaceutics 12, 416. 10.3390/pharmaceutics12050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringel AE, Ryznar R, Picariello H, Huang K.l., Lazarus AG, and Holmes SG (2013). Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet. 9, e1003871. 10.1371/journal.pgen.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, and Cesareni G (1991). Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349, 541–544. 10.1038/349541a0. [DOI] [PubMed] [Google Scholar]
- 33.Honarmand Ebrahimi K, Hagedoorn P-L, and Hagen WR (2015). Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 115, 295–326. 10.1021/cr5004908. [DOI] [PubMed] [Google Scholar]
- 34.Fisher J, Devraj K, Ingram J, Slagle-Webb B, Madhankumar AB, Liu X, Klinger M, Simpson IA, and Connor JR (2007). Ferritin: a novel mechanism for delivery of iron to the brain and other organs. Am. J. Physiol. Cell Physiol. 293, C641–C649. 10.1152/ajpcell.00599.2006. [DOI] [PubMed] [Google Scholar]
- 35.Loh A, Hadziahmetovic M, and Dunaief JL (2009). Iron homeostasis and eye disease. Biochim. Biophys. Acta 1790, 637–649. 10.1016/j.bbagen.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison PM, and Arosio P (1996). The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275, 161–203. 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 37.Connor JR, Snyder BS, Arosio P, Loeffler DA, and LeWitt P (1995). A quantitative analysis of isoferritins in select regions of aged, parkinsonian, and Alzheimer’s diseased brains. J. Neurochem. 65, 717–724. 10.1046/j.1471-4159.1995.65020717.x. [DOI] [PubMed] [Google Scholar]
- 38.Drakesmith H, Chen N, Ledermann H, Screaton G, Townsend A, and Xu X-N (2005). HIV-1 Nef down-regulates the hemochromatosis protein HFE, manipulating cellular iron homeostasis. Proc. Natl. Acad. Sci. USA. 102, 11017–11022. 10.1073/pnas.0504823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozzi C, Di Pisa F, Bernacchioni C, Ciambellotti S, Turano P, and Mangani S (2015). Iron binding to human heavy-chain ferritin. Acta Crystallogr. D Biol. Crystallogr. 71, 1909–1920. 10.1107/S1399004715013073. [DOI] [PubMed] [Google Scholar]
- 40.Rucker P, Torti FM, and Torti SV (1996). Role of H and L subunits in mouse ferritin. J. Biol. Chem. 271, 33352–33357. 10.1074/jbc.271.52.33352. [DOI] [PubMed] [Google Scholar]
- 41.Lawson DM, Treffry A, Artymiuk PJ, Harrison PM, Yewdall SJ, Luzzago A, Cesareni G, Levi S, and Arosio P (1989). Identification of the ferroxidase centre in ferritin. FEBS Lett. 254, 207–210. 10.1016/0014-5793(89)81040-3. [DOI] [PubMed] [Google Scholar]
- 42.Chaikuad A, Pilka ES, De Riso A, von Delft F, Kavanagh KL, Vénien-Bryan C, Oppermann U, and Yue WW (2012). Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family. BMC Struct. Biol. 12, 14. 10.1186/1472-6807-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Farquhar ER, Chance MR, Palczewski K, and Kiser PD (2012). Insights into substrate specificity and metal activation of mammalian tetrahedral aspartyl aminopeptidase. J. Biol. Chem. 287, 13356–13370. 10.1074/jbc.M112.347518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowther WT, and Matthews BW (2002). Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem. Rev. 102, 4581–4608. 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- 45.Rawlings ND, Tolle DP, and Barrett AJ (2004). MEROPS: the peptidase database. Nucleic Acids Res. 32, D160–D164. 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hooper NM, and Lendeckel U (2012). Aminopeptidases in Biology and Disease (Springer Science & Business Media; ). [Google Scholar]
- 47.Zini S, Fournie-Zaluski MC, Chauvel E, Roques BP, Corvol P, and Llorens-Cortes C (1996). Identification of metabolic pathways of brain angiotensin II and III using specific aminopeptidase inhibitors: predominant role of angiotensin III in the control of vasopressin release. Proc. Natl. Acad. Sci. USA. 93, 11968–11973. 10.1073/pnas.93.21.11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costagliola C, Di Benedetto R, De Caprio L, Verde R, and Mastropasqua L (1995). Effect of oral captopril (SQ 14225) on intraocular pressure in man. Eur. J. Ophthalmol. 5, 19–25. 10.1177/112067219500500104. [DOI] [PubMed] [Google Scholar]
- 49.Fletcher EL, Phipps JA, Ward MM, Vessey KA, and Wilkinson-Berka JL (2010). The renin-angiotensin system in retinal health and disease: its influence on neurons, glia and the vasculature. Prog. Retin. Eye Res. 29, 284–311. 10.1016/j.preteyeres.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Jacobi PC, Osswald H, Jurklies B, and Zrenner E (1994). Neuromodulatory effects of the renin-angiotensin system on the cat electroretinogram. Invest. Ophthalmol. Vis. Sci. 35, 973–980. [PubMed] [Google Scholar]
- 51.Otani A, Takagi H, Suzuma K, and Honda Y (1998). Angiotensin II potentiates vascular endothelial growth factor-induced angiogenic activity in retinal microcapillary endothelial cells. Circ. Res. 82, 619–628. 10.1161/01.res.82.5.619. [DOI] [PubMed] [Google Scholar]
- 52.Mauer M, Zinman B, Gardiner R, Suissa S, Sinaiko A, Strand T, Drummond K, Donnelly S, Goodyer P, Gubler MC, and Klein R (2009). Renal and retinal effects of enalapril and losartan in type 1 diabetes. N. Engl. J. Med. 361, 40–51. 10.1056/NEJMoa0808400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva KC, Rosales MAB, Biswas SK, Lopes de Faria JB, and Lopes de Faria JM (2009). Diabetic retinal neurodegeneration is associated with mitochondrial oxidative stress and is improved by an angiotensin receptor blocker in a model combining hypertension and diabetes. Diabetes 58, 1382–1390. 10.2337/db09-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaajanen A, and Vapaatalo H (2011). Local ocular renin-angiotensin system - a target for glaucoma therapy? Basic Clin. Pharmacol. Toxicol. 109, 217–224. 10.1111/j.1742-7843.2011.00729.x. [DOI] [PubMed] [Google Scholar]
- 55.Wilk S, Wilk E, and Magnusson RP (2002). Identification of histidine residues important in the catalysis and structure of aspartyl aminopeptidase. Arch. Biochem. Biophys. 407, 176–183. 10.1016/s0003-9861(02)00494-0. [DOI] [PubMed] [Google Scholar]
- 56.Liaw S-H, Kuo I, and Eisenberg D (1995). Discovery of the ammonium substrate site on glutamine synthetase, A third cation binding site. Protein Sci. 4, 2358–2365. 10.1002/pro.5560041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suárez I, Bodega G, and Fernández B (2002). Glutamine synthetase in brain: effect of ammonia. Neurochem. Int. 41, 123–142. 10.1016/S0197-0186(02)00033-5. [DOI] [PubMed] [Google Scholar]
- 58.Gunnersen D, and Haley B (1992). Detection of glutamine synthetase in the cerebrospinal fluid of Alzheimer diseased patients: a potential diagnostic biochemical marker. Proc. Natl. Acad. Sci. USA. 89, 11949–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis GP, Erickson PA, Kaska DD, and Fisher SK (1988). An immunocytochemical comparison of Müller cells and astrocytes in the cat retina. Exp. Eye Res. 47, 839–853. 10.1016/0014-4835(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 60.Riepe RE, and Norenberg MD (1978). Glutamine synthetase in the developing rat retina: an immunohistochemical study. Exp. Eye Res. 27, 435–444. 10.1016/0014-4835(78)90022-2. [DOI] [PubMed] [Google Scholar]
- 61.Fröhlich E, and Klessen C. (2000). Glutamine synthetase and marker enzymes of the blood-retina barrier in fetal bovine retinal pigment epithelial cells. Graefes Arch. Clin. Exp. Ophthalmol. 238, 500–507. 10.1007/pl00007891. [DOI] [PubMed] [Google Scholar]
- 62.Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, and Mowbray SL (2008). Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J. Mol. Biol. 375, 217–228. 10.1016/j.jmb.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 63.Häberle J, Görg B, Rutsch F, Schmidt E, Toutain A, Benoist J-F, Gelot A, Suc A-L, Höhne W, Schliess F, et al. (2005). Congenital glutamine deficiency with glutamine synthetase mutations. N. Engl. J. Med. 353, 1926–1933. 10.1056/NEJMoa050456. [DOI] [PubMed] [Google Scholar]
- 64.Edgar PF, Douglas JE, Cooper GJ, Dean B, Kydd R, and Faull RL (2000). Comparative proteome analysis of the hippocampus implicates chromosome 6q in schizophrenia. Mol. Psychiatr. 5, 85–90. 10.1038/sj.mp.4000580. [DOI] [PubMed] [Google Scholar]
- 65.Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF, and Yolken RH (2000). Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol. Psychiatr. 5, 142–149. 10.1038/sj.mp.4000696. [DOI] [PubMed] [Google Scholar]
- 66.Martins-de-Souza D, Gattaz WF, Schmitt A, Maccarrone G, Hunyadi-Gulyás E, Eberlin MN, Souza GHMF, Marangoni S, Novello JC, Turck CW, and Dias-Neto E (2009). Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J. Psychiatr. Res. 43, 978–986. 10.1016/j.jpsychires.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Hensley K, Venkova K, Christov A, Gunning W, and Park J (2011). Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications. Mol. Neurobiol. 43, 180–191. 10.1007/s12035-011-8166-4. [DOI] [PubMed] [Google Scholar]
- 68.Lee H, Joo J, Nah S-S, Kim JW, Kim H-K, Kwon J-T, Lee H-Y, Kim YO, and Kim H-J (2015). Changes in Dpysl2 expression are associated with prenatally stressed rat offspring and susceptibility to schizophrenia in humans. Int. J. Mol. Med. 35, 1574–1586. 10.3892/ijmm.2015.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiong L-L, Qiu D-L, Xiu G-H, Al-Hawwas M, Jiang Y, Wang Y-C, Hu Y, Chen L, Xia Q-J, and Wang T-H (2020). DPYSL2 is a novel regulator for neural stem cell differentiation in rats: revealed by Panax notoginseng saponin administration. Stem Cell Res. Ther. 11, 155. 10.1186/s13287-020-01652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deo RC, Schmidt EF, Elhabazi A, Togashi H, Burley SK, and Strittmatter SM (2004). Structural bases for CRMP function in plexindependent semaphorin3A signaling. EMBO J. 23, 9–22. 10.1038/sj.emboj.7600021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stenmark P, Ogg D, Flodin S, Flores A, Kotenyova T, Nyman T, Nordlund P, and Kursula P (2007). The structure of human collapsin response mediator protein 2, a regulator of axonal growth. J. Neurochem. 101, 906–917. 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- 72.Ben-Menachem E, Biton V, Jatuzis D, Abou-Khalil B, Doty P, and Rudd GD (2007). Efficacy and safety of oral lacosamide as adjunctive therapy in adults with partial-onset seizures. Epilepsia 48, 1308–1317. 10.1111/j.1528-1167.2007.01188.x. [DOI] [PubMed] [Google Scholar]
- 73.Moutal A, White KA, Chefdeville A, Laufmann RN, Vitiello PF, Feinstein D, Weimer JM, and Khanna R (2019). Dysregulation of CRMP2 post-translational modifications drive its pathological functions. Mol. Neurobiol. 56, 6736–6755. 10.1007/s12035-019-1568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rutter WJ (1964). Evolution of aldolase. Fed. Proc. 23, 1248–1257. [PubMed] [Google Scholar]
- 75.Ahn AH, Dziennis S, Hawkes R, and Herrup K (1994). The cloning of zebrin II reveals its identity with aldolase C. Development 120, 2081–2090. [DOI] [PubMed] [Google Scholar]
- 76.Merkulova M, Hurtado-Lorenzo A, Hosokawa H, Zhuang Z, Brown D, Ausiello DA, and Marshansky V (2011). Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am. J. Physiol. Cell Physiol. 300, C1442–C1455. 10.1152/ajpcell.00076.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rangarajan ES, Park H, Fortin E, Sygusch J, and Izard T (2010). Mechanism of aldolase control of sorting nexin 9 function in endocytosis. J. Biol. Chem. 285, 11983–11990. 10.1074/jbc.M109.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ritterson Lew C, and Tolan DR (2013). Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics. J. Cell. Biochem. 114, 1928–1939. 10.1002/jcb.24538. [DOI] [PubMed] [Google Scholar]
- 79.Wang J, Tolan DR, and Pagliaro L (1997). Metabolic compartmentation in living cells: structural association of aldolase. Exp. Cell Res. 237, 445–451. 10.1006/excr.1997.3811. [DOI] [PubMed] [Google Scholar]
- 80.Dalby A, Dauter Z, and Littlechild JA (1999). Crystal structure of human muscle aldolase complexed with fructose 1, 6-bisphosphate: mechanistic implications. Protein Sci. 8, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blom N, and Sygusch J (1997). Product binding and role of the C-terminal region in class I D-fructose 1, 6-bisphosphate aldolase. Nat. Struct. Biol. 4, 36–39. 10.1038/nsb0197-36. [DOI] [PubMed] [Google Scholar]
- 82.St-Jean M, Lafrance-Vanasse J, Liotard B, and Sygusch J (2005). High resolution reaction intermediates of rabbit muscle fructose-1, 6-bisphosphate aldolase: substrate cleavage and induced fit. J. Biol. Chem. 280, 27262–27270. 10.1074/jbc.M502413200. [DOI] [PubMed] [Google Scholar]
- 83.Sygusch J, Beaudry D, and Allaire M (1987). Molecular architecture of rabbit skeletal muscle aldolase at 2.7-A resolution. Proc. Natl. Acad. Sci. USA. 84, 7846–7850. 10.1073/pnas.84.22.7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta S, Hollenstein R, Kochhar S, and Christen P (1993). Paracatalytic self-inactivation of fructose-1, 6-bisphosphate aldolase. Structure of the crosslink formed at the active site. Eur. J. Biochem. 214, 515–519. 10.1111/j.1432-1033.1993.tb17949.x. [DOI] [PubMed] [Google Scholar]
- 85.Hartman FC, and Brown JP (1976). Affinity labeling of a previously undetected essential lysyl residue in class I fructose bisphosphate aldolase. J. Biol. Chem. 251, 3057–3062. [PubMed] [Google Scholar]
- 86.Lai CY, Nakai N, and Chang D (1974). Amino acid sequence of rabbit muscle aldolase and the structure of the active center. Science 183, 1204–1206. 10.1126/science.183.4130.1204. [DOI] [PubMed] [Google Scholar]
- 87.Lobb RR, Stokes AM, Hill HA, and Riordan JF (1975). Arginine as the C-1 phosphate binding site in rabbit muscle aldolase. FEBS Lett. 54, 70–72. 10.1016/0014-5793(75)81070-2. [DOI] [PubMed] [Google Scholar]
- 88.Morris AJ, and Tolan DR (1993). Site-directed mutagenesis identifies aspartate 33 as a previously unidentified critical residue in the catalytic mechanism of rabbit aldolase A. J. Biol. Chem. 268, 1095–1100. [PubMed] [Google Scholar]
- 89.Morris AJ, and Tolan DR (1994). Lysine-146 of rabbit muscle aldolase is essential for cleavage and condensation of the C3-C4 bond of fructose 1, 6-bis(phosphate). Biochemistry 33, 12291–12297. 10.1021/bi00206a036. [DOI] [PubMed] [Google Scholar]
- 90.Santamaria R, Vitagliano L, Tamasi S, Izzo P, Zancan L, Zagari A, and Salvatore F (1999). Novel six-nucleotide deletion in the hepatic fructose-1, 6-bisphosphate aldolase gene in a patient with hereditary fructose intolerance and enzyme structure-function implications. Eur. J. Hum. Genet. 7, 409–414. 10.1038/sj.ejhg.5200299. [DOI] [PubMed] [Google Scholar]
- 91.Schlattner U, Tokarska-Schlattner M, and Wallimann T (2006). Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta 1762, 164–180. 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Boero J, Qin W, Cheng J, Woolsey TA, Strauss AW, and Khuchua Z (2003). Restricted neuronal expression of ubiquitous mitochondrial creatine kinase: changing patterns in development and with increased activity. Mol. Cell. Biochem. 244, 69–76. [PubMed] [Google Scholar]
- 93.Hornemann T, Stolz M, and Wallimann T (2000). Isoenzyme-specific interaction of muscle-type creatine kinase with the sarcomeric M-line is mediated by nh2-terminal lysine charge-clamps. J. Cell Biol. 149, 1225–1234. 10.1083/jcb.149.6.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaldis P, Stolz M, Wyss M, Zanolla E, Rothen-Rutishauser B, Vorherr T, and Wallimann T (1996). Identification of two distinctly localized mitochondrial creatine kinase isoenzymes in spermatozoa. J. Cell Sci. 109, 2079–2088. [DOI] [PubMed] [Google Scholar]
- 95.Wallimann T, and Hemmer W (1994). Creatine kinase in non-muscle tissues and cells. Mol. Cell. Biochem. 133–134, 193–220. 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- 96.Haas RC, and Strauss AW (1990). Separate nuclear genes encode sarcomere-specific and ubiquitous human mitochondrial creatine kinase isoenzymes. J. Biol. Chem. 265, 6921–6927. [PubMed] [Google Scholar]
- 97.Haas RC, Korenfeld C, Zhang ZF, Perryman B, Roman D, and Strauss AW (1989). Isolation and characterization of the gene and cDNA encoding human mitochondrial creatine kinase. J. Biol. Chem. 264, 2890–2897. [PubMed] [Google Scholar]
- 98.Schlegel J, Wyss M, Schürch U, Schnyder T, Quest A, Wegmann G, Eppenberger HM, and Wallimann T (1988). Mitochondrial creatine kinase from cardiac muscle and brain are two distinct isoenzymes but both form octameric molecules. J. Biol. Chem. 263, 16963–16969. [PubMed] [Google Scholar]
- 99.Bürklen TS, Schlattner U, Homayouni R, Gough K, Rak M, Szeghalmi A, and Wallimann T (2006). The creatine kinase/creatine connection to Alzheimer’s disease: CK inactivation, APP-CK complexes, and focal creatine deposits. J. Biomed. Biotechnol. 2006, 35936. 10.1155/JBB/2006/35936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wendt S, Dedeoglu A, Speer O, Wallimann T, Beal MF, and Andreassen OA (2002). Reduced creatine kinase activity in transgenic amyotrophic lateral sclerosis mice. Free Radic. Biol. Med. 32, 920–926. 10.1016/s0891-5849(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 101.Qian X-L, Li Y-Q, Gu F, Liu F-F, Li W-D, Zhang X-M, and Fu L (2012). Overexpression of ubiquitous mitochondrial creatine kinase (uMtCK) accelerates tumor growth by inhibiting apoptosis of breast cancer cells and is associated with a poor prognosis in breast cancer patients. Biochem. Biophys. Res. Commun. 427, 60–66. 10.1016/j.bbrc.2012.08.147. [DOI] [PubMed] [Google Scholar]
- 102.Kanemitsu F, Kawanishi I, Mizushima J, and Okigaki T (1984). Mitochondrial creatine kinase as a tumor-associated marker. Clin. Chim. Acta 138, 175–183. 10.1016/0009-8981(84)90232-8. [DOI] [PubMed] [Google Scholar]
- 103.Eder M, Fritz-Wolf K, Kabsch W, Wallimann T, and Schlattner U (2000). Crystal structure of human ubiquitous mitochondrial creatine kinase. Proteins 39, 216–225. [DOI] [PubMed] [Google Scholar]
- 104.Fritz-Wolf K, Schnyder T, Wallimann T, and Kabsch W (1996). Structure of mitochondrial creatine kinase. Nature 381, 341–345. 10.1038/381341a0. [DOI] [PubMed] [Google Scholar]
- 105.Yang M, Liu S, Xiong Y, Zhao J, and Deng W (2022). An integrative pan-cancer analysis of molecular characteristics and oncogenic role of mitochondrial creatine kinase 1A (CKMT1A) in human tumors. Sci. Rep. 12, 10025. 10.1038/s41598-022-14346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morgan CE, Glaza P, Leus IV, Trinh A, Su C-C, Cui M, Zgurskaya HI, and Yu EW (2021). Cryoelectron microscopy structures of AdeB illuminate mechanisms of simultaneous binding and exporting of substrates. mBio 12, e036900–20. 10.1128/mBio.03690-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moseng MA, Lyu M, Pipatpolkai T, Glaza P, Emerson CC, Stewart PL, Stansfeld PJ, and Yu EW (2021). Cryo-EM structures of CusA reveal a mechanism of metal-ion export. mBio 12, e004522–21. 10.1128/mBio.00452-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, Floyd RA, and Markesbery WR (1991). Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc. Natl. Acad. Sci. USA. 88, 10540–10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madeira C, Vargas-Lopes C, Brandão CO, Reis T, Laks J, Panizzutti R, and Ferreira ST (2018). Elevated glutamate and glutamine levels in the cerebrospinal fluid of patients with probable Alzheimer’s disease and depression. Front. Psychiatry 9, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Colell A, Green DR, and Ricci J-E (2009). Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 16, 1573–1581. 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- 111.Jurado R, Adamcik J, López-Haro M, González-Vera JA, Ruiz-Arias Á, Sánchez-Ferrer A, Cuesta R, Domínguez-Vera JM, Calvino JJ, Orte A, et al. (2019). Apoferritin protein amyloid fibrils with tunable chirality and polymorphism. J. Am. Chem. Soc. 141, 1606–1613. 10.1021/jacs.8b11418. [DOI] [PubMed] [Google Scholar]
- 112.Ayton S, Faux NG, and Bush AI; Alzheimer’s Disease Neuroimaging Initiative (2015). Ferritin levels in the cerebrospinal fluid predict Alzheimer’s disease outcomes and are regulated by APOE. Nat. Commun. 6, 6760. 10.1038/ncomms7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, Klein JB, Turner DM, and Butterfield DA (2007). An increase in S-glutathionylated proteins in the Alzheimer’s disease inferior parietal lobule, a proteomics approach. J. Neurosci. Res. 85, 1506–1514. 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- 114.Puzone R, Savarino G, Salvi S, Dal Bello MG, Barletta G, Genova C, Rijavec E, Sini C, Esposito AI, Ratto GB, et al. (2013). Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol. Cancer 12, 97. 10.1186/1476-4598-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tokunaga K, Nakamura Y, Sakata K, Fujimori K, Ohkubo M, Sawada K, and Sakiyama S (1987). Enhanced expression of a glyceraldehyde-3-phosphate dehydrogenase gene in human lung cancers. Cancer Res. 47, 5616–5619. [PubMed] [Google Scholar]
- 116.Du S, Guan Z, Hao L, Song Y, Wang L, Gong L, Liu L, Qi X, Hou Z, and Shao S (2014). Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PLoS One 9, e85804. 10.1371/journal.pone.0085804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sergeev YV, Dolinska MB, and Hejtmancik JF (2018). Apoferritin is maintaining the native conformation of citrate synthase in vitro. J. Anal. Pharm. Res. 7, 680–684. [PMC free article] [PubMed] [Google Scholar]
- 118.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 119.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- 120.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 122.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saari JC, and Bredberg DL (1990). Acyl-CoA:Retinol acyltransferase and lecithin:Retinol acyltransferase activities of bovine retinal pigment epithelial microsomes. In In Methods in Enzymology Retinoids Part B: Cell Differentiation and Clinical Applications (Academic Press), pp. 156–163. 10.1016/0076-6879(90)90020-2. [DOI] [PubMed] [Google Scholar]
- 124.Robinson NJ, Miyagi M, Scarborough JA, Scott JG, Taylor DJ, and Schiemann WP (2021). SLX4IP promotes RAP1 SUMOylation by PIAS1 to coordinate telomere maintenance through NF-κB and Notch signaling. Sci. Signal. 14, eabe9613. 10.1126/scisignal.abe9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteomewide protein quantification. Nat. Biotechnol. 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 126.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 127.Mastronarde DN (2003). SerialEM: a program for automated tilt series acquisition on tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183. 10.1017/S1431927603445911. [DOI] [Google Scholar]
- 128.Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, and Berger B (2019). TOPAZ: a positive-unlabeled convolutional neural network CryoEM particle picker that can pick any size and shape particle. Microsc. Microanal. 25, 986–987. 10.1017/S143192761900566X. [DOI] [Google Scholar]
- 129.Pfab J, Phan NM, and Si D (2021). DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl. Acad. Sci. USA. 118, e2017525118. 10.1073/pnas.2017525118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, and Madden TL (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, and Lyumkis D (2017). Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796. 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and EM maps for GS, GAPDH, FT, DPYSL2, FPA, MtCK and DNPEP can be found at PDB accession numbers 7U5N, 7U5M, 7U5L, 7U5K, 7U5J, 7U5I and 7U5H and EMDB accession codes EMD-26356, EMD-26355, EMD-26354, EMD-26353, EMD-26352, EMD-26351, and EMD-26350, respectively. The raw cryo-EM data have been deposited in EMPIAR (https://www.ebi.ac.uk/empiar/). The raw mass spectrometry proteomics data have been deposited in ProteomeXchange via the PRIDE database (https://www.ebi.ac.uk/pride/). Accession numbers, codes, and links to access the structural data, raw cryo-EM images and raw mass spectrometry proteomics data are also listed in the key resources table. All data reported in this paper are available upon requested from the Lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Biological samples | ||
|
| ||
| Fresh bovine eyes | Mahan Packing Company | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Acetonitrile | Millipore Sigma | Cat# 1000291000 |
| Ammonium bicarbonate | Sigma-Aldrich | Cat# 5330050050 |
| Graphene oxide | Sigma-Aldrich | Cat# 763705-100ML |
| Iodoacetamide | GE Healthcare | Cat# RPN6302 |
| Trypsin/Lys-C Mix | Promega | Cat# V5073 |
|
| ||
| Deposited data | ||
|
| ||
| Raw cryo-EM data | This paper | EMPIAR-11251 https://doi.org/10.6019/EMPIAR-11251 |
| Cryo-EM structure and map of GS | This paper | PDB ID: 7U5N EMDB ID: EMD-26356 https://doi.org/10.2210/pdb7U5N/pdb |
| Cryo-EM structure and map of GAPDH | This paper | PDB ID: 7U5M EMDB ID: EMD-26355 https://doi.org/10.2210/pdb7U5M/pdb |
| Cryo-EM structure and map of FT | This paper | PDB ID: 7U5L EMDB ID: EMD-26354 https://doi.org/10.2210/pdb7U5L/pdb |
| Cryo-EM structure and map of DPYSL2 | This paper | PDB ID: 7U5K EMDB ID: EMD-26353 https://doi.org/10.2210/pdb7U5K/pdb |
| Cryo-EM structure and map of FPA | This paper | PDB ID: 7U5J EMDB ID: EMD-26352 https://doi.org/10.2210/pdb7U5J/pdb |
| Cryo-EM structure and map of MtCK | This paper | PDB ID: 7U5I EMDB ID: EMD-26351 https://doi.org/10.2210/pdb7U5I/pdb |
| Cryo-EM structure and map of DNPEP | This paper | PDB ID: 7U5H EMDB ID: EMD-26350 https://doi.org/10.2210/pdb7U5H/pdb |
| Raw mass spectrometry proteomics data | This paper | PXD037186 (Table S3) |
|
| ||
| Software and algorithms | ||
|
| ||
| Coot | Emsley et al.118 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| cryoSPARC v3 | Punjani et al.119 | https://cryosparc.com/ |
| MaxQuant | Cox and Mann | https://www.maxquant.org/ |
| Phenix | Adams et al.120 | https://phenix-online.org/ |
| STRING database | STRING | https://string-db.org/ |
| UCSF-Chimera | Pettersen et al.121 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF-Chimera X | Pettersen et al.122 | https://www.cgl.ucsf.edu/chimerax/ |
|
| ||
| Other | ||
|
| ||
| C18 Microspin column | Nest Group | Cat# SEM SS18V |
| Quantifoil R 2/2 Cu 200 mesh, copper | Quantifoil | Cat# Q210CR2 |


