Abstract
The genetic consequences of species-wide declines are rarely quantified because the timing and extent of the decline varies across the species’ range. The sea otter (Enhydra lutris) is a unique model in this regard. Their dramatic decline from thousands to fewer than 100 individuals per population occurred range-wide and nearly simultaneously due to the 18th-19th century fur trade. Consequently, each sea otter population represents an independent natural experiment of recovery after extreme population decline. We designed sequence capture probes for 50 megabases of sea otter exonic and neutral genomic regions. We sequenced 107 sea otters from five populations that span the species range to high coverage (18-76x) and three historic Californian samples from ~1500 and ~200 years ago to low coverage (1.5-3.5X). We observe distinct population structure and find that sea otters in California are the last survivors of a divergent lineage isolated for thousands of years and therefore warrant special conservation concern. We detect signals of extreme population decline in every surviving sea otter population and use this demographic history to design forward-in-time simulations of coding sequence. Our simulations indicate that this decline could lower the fitness of recovering populations for generations. However, the simulations also demonstrate how historically low effective population sizes prior to the fur trade may have mitigated the effects of population decline on genetic health. Our comprehensive approach shows how demographic inference from genomic data, coupled with simulations, allows assessment of extinction risk and different models of recovery.
Keywords: sea otters, conservation genomics, population bottleneck, genetic load, demographic simulations
Introduction
The historical range of the sea otter (Enhydra lutris) spanned the Northern Pacific Rim from the Kuril Islands of Russia to Baja California (Figure 1A; Figure S1A). Across this range, sea otter populations were hunted to extinction or the brink of extinction by the 18th-19th century fur trade (Kenyon, 1969; Riedman & Estes, 1990). The few populations that survived in the Kuril Islands, Kamchatka Peninsula, Commander and Aleutian Islands, central Alaska, and California contained fewer than one hundred survivors apiece, a severe population bottleneck from previous population sizes estimated at 10-20,000 individuals per population (Riedman & Estes, 1990). The loss of sea otters triggered a trophic cascade that led to the disappearance of kelp forests through overgrazing by sea urchins (Estes & Palmisano, 1974). Sea otters were formally protected in 1911 by the International Fur Seal Treaty, and populations have increased and been reintroduced into southeast Alaska, British Columbia, Washington State, and southern California where they had been fully extirpated (Jameson et al., 1982). Sea otters in Baja California, Mexico were declared likely extinct due to the fur trade (Kenyon, 1969), but there have continued to be sporadic sightings in the region (Gallo-Reynoso & Rathbun, 1997; Schramm et al., 2014). The recent history of sea otter populations provides a unique model for species decline, because rarely are bottlenecks so uniform and near-concurrent across an entire species distribution. Each surviving sea otter population represents a natural replicate of an extreme bottleneck event, offering a powerful system to investigate the genomic consequences of near-extinction.
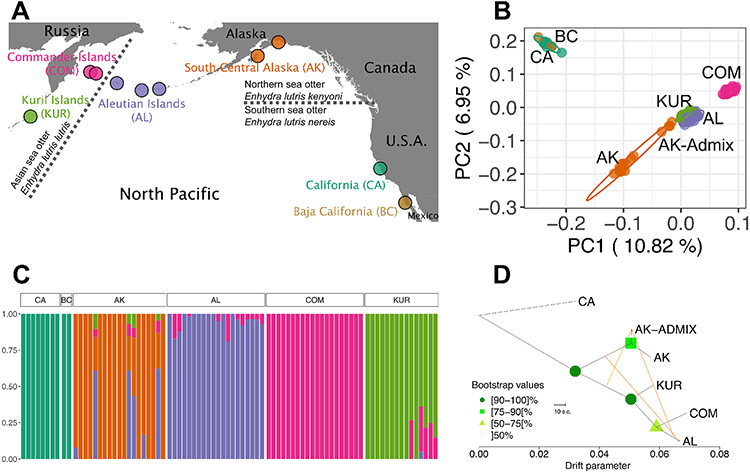
Figure 1. Sea Otter Population Structure Mirrors Historic Distribution.
A) Locations of sea otter samples used in this study. Sea otters were sampled from the Kuril Islands (KUR, n = 13), Commander Islands (COM, n = 45), Aleutian Islands (AL, consisting of Attu, Amchitka and Adak Islands, n = 21), south central Alaska (AK, n = 19), California (CA, n = 7), and Baja California (BC, n = 2). Dashed lines indicate the previously-designated subspecies boundaries between southern, northern and Asian sea otters. See Figure S1 for finer-scale sampling location maps, Supplemental Methods for sample information, and Tables S1 and S3 for sample and coverage details.
B) Principal components analysis of modern sea otter samples. Multivariate t-distribution 95% data ellipses are shown. See Figure S3A-C for PCA analyses of the northern and Asian populations without California which provide greater resolution of their population structure.
C) fastSTRUCTURE analysis of the samples with the number of clusters (K) = 5. See Figure S3F for additional values of K.
D) Bootstrapped Treemix analysis showing drift between populations, with migration events shown as orange lines. California was set as the root. Admixed south central Alaskan individuals are separated in the analysis (AK-Admix) from the non-admixed (AK). Baja California was excluded as it appears to be an expansion of the California population, rather than a distinct population. Treemix residuals are shown in Figure S3G.
Previous genetic studies of the sea otter considered microsatellite loci and mitochondrial control region sequences which showed population structure and low average levels of genetic variation (Bodkin et al., 1999; Gagne et al., 2018; Larson et al., 2012, 2021; Larson, Jameson, Bodkin, et al., 2002; Larson, Jameson, Etnier, et al., 2002). Here, we use a more comprehensive genomic approach to investigate the genomic impacts of the fur trade bottleneck on remnant sea otter populations. Using sequence capture, we assess genic and non-genic regions of the genome in 107 sea otters (Figure 1A; Figure S1A-C; Table S1) sampled from five remnant populations across the species range (Figure 1A), as well as three historical samples from California dating from ~1500-200 years ago (Figure 2A; Table S2). We use these sequence data to analyze sea otter population structure, genetic continuity, and demographic history. These results then serve as the basis for forward-in-time simulations to explore the impacts of the fur trade on genetic load and recovery. We find that although the historical population structure has been preserved through the persistence of survivors in each population, the fur trade bottleneck may have long-lasting effects on population fitness and plans for restoration.
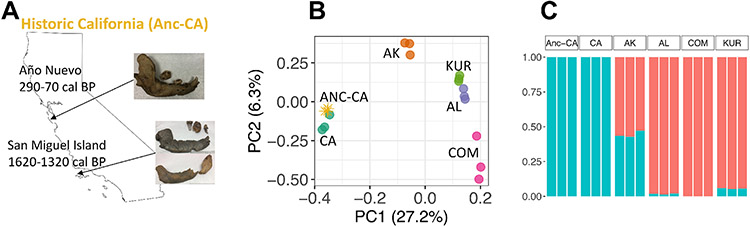
Figure 2. Historical DNA Suggests Genetic Stability in California.
A) Sampling locations and ages of three historic sea otter samples from California shell middens.
B) PCAngsd principal components analysis of three historic California samples (“Anc-CA”, gold stars) and 15 modern sea otter samples based on genotype likelihoods. Each modern population (California (CA), Alaska (AK), Aleutian Islands (AL), and Kuril Islands (KUR), and the Commander Islands (COM)) was downsampled to three individuals to balance the sample size of the three historic samples.
C) PCAngsd admixture analysis, based on genotype likelihoods, including the three historic California samples (“Anc-CA”).
Materials and Methods
Refer to the Supplemental Methods for additional details on all analyses described below.
Sample information.
Sea otters were sampled from the Kuril Islands (KUR, n = 13), Commander Islands (COM, n = 45), Aleutian Islands (AL, consisting of Attu, Amchitka and Adak Islands, n = 21), south central Alaska (AK, n = 19), California (CA, n = 7), and Baja California (BC, n = 2). All samples derive from sea otter research expeditions from 1974 to 1994 (Supplemental Methods; Figure 1A; Figure S1A-C; Table S1), except for the two recent (2008-2011) samples recovered from stranded individuals from Baja California (Schramm et al., 2014) (Table S1; Figure S1D). Three historical Californian samples, excavated from coastal shell middens and dating from ~1500-200 years ago, were also analyzed (Supplemental Methods; Figure 2A; Table S2).
Sequence capture probe design.
We designed a custom sequence capture based on the annotation of the southern sea otter genome from Beichman et al. (2019) (Enhydra lutris nereis, accession GCA_006410715.1). 50Mb of sequence was included in the capture design, including all annotated exonic sequence passing custom filters, 1kb regions upstream of genes, and 10Mb of neutral regions far from genes and therefore unlikely to be under selection. We chose these target regions for three reasons. First, they provided enough information to carry out high resolution analysis of population structure. Second, they enabled us to generate the neutral site frequency spectrum (SFS) from regions far from genes for demographic inference. Third, we could generate the synonymous and missense site frequency spectra from exonic regions, allowing to calibrate our simulations of coding sequence under our inferred demographic models. The custom sequence capture probe pool was built based on the coordinates of these regions by KAPA Biosystems (Wilmington, MA).
Extraction, library preparation, sequence capture, and sequencing.
Blood and tissue samples were extracted using Qiagen’s DNEasy Blood and Tissue kit. Libraries were prepared using the KAPA HyperPlus library preparation kit, and sequence capture was carried out using the KAPA SeqCap EZ Hyperflow workflow kit, with 12-13 uniquely indexed samples per capture pool. Capture success was evaluated using the Agilent Bioanalyzer and Qubit. Captures were sequenced on the Novaseq 5000 (150PE) at Medgenome (Foster City, CA). An average of 6Gb of sequence was generated per sample, with 93% of bases ≥ 30 base quality, and per-individual coverage of 18-76x (Table S3). Four samples from California were excluded from subsequent analysis due to low sequencing yields.
Samples from Baja California were extracted at the Autonomous University of Baja California, Ensenada, Mexico. Library preparation and sequence capture was carried out using the workflow described above and the samples were sequenced on the NextSeq 500 at the National Laboratory of Genomics for Biodiversity in Mexico (LANGEBIO-CINVESTAV, Irapuato, Mexico).
The three historic samples were prepared in UCLA’s ancient DNA lab, a clean room that is separate from all modern DNA labs. DNA was extracted using the Gamba et al. (2014) protocol. The historic samples with the highest concentration were chosen for library preparation at Arbor Biosciences (Madison, WI) using NEB Next Fast DNA library preparation. Sequence capture was carried out and the historic samples were sequenced to 1.5-3.5x coverage of capture regions on a HiSeq3000 at the UCLA Technology Center for Genomics & Bioinformatics. The historic samples were radiocarbon dated at the W.M. Keck Carbon Cycle Accelerator Mass Spectrometer at the University of California, Irvine and dates were calibrated as described in the Supplemental Methods.
Read mapping, genotype calling & filtering (modern samples).
Following the strategy of Beichman et al. (2019), reads were mapped to the domestic ferret reference genome (Mustela putorius furo, accession GCA_000215625.1) in order to use a reference genome that is an outgroup from all sea otter populations with a good annotation and preexisting resources, particularly the Ensembl Variant Effect Predictor database. Reads were mapped using the PALEOMIX pipeline (Schubert et al., 2014) with BWA-MEM (Li & Durbin, 2009) as the alignment algorithm. Genotypes were called using GATK’s HaplotypeCaller (Van der Auwera et al., 2013) and genotypes were hard-filtered as described in the Supplemental Methods. Fifteen individuals with greater than the mean + 1 standard deviation worth of missing genotypes were excluded from downstream analyses.
After genotype filtering, the average coverage per individual of sites was 36x (based on sites with at least 10x coverage). Each individual had an average of 61.4 Mb called genotypes (Table S3). Approximately 60,000 total segregating sites were discovered.
To ensure that our choice of a divergent outgroup reference genome would not bias estimates of heterozygosity or demographic history, we also mapped all individuals to the southern sea otter reference genome (accession GCA_006410715.1) generated in Beichman et al. (2019). Mapping rates were highly similar between the two reference genomes (Table S3). We repeated all analyses described below with reads mapped to each reference genome.
Read mapping, genotype likelihoods & filtering (historic samples).
The historic samples were mapped to both the ferret and sea otter reference genomes using the PALEOMIX pipeline (Schubert et al., 2014) with the BWA backtrack (Li & Durbin, 2009) alignment algorithm. MapDamage (Jónsson et al., 2013) was used to detect DNA-damage profiles and to rescale the quality score of bases that are likely to be misincorporations (Figure S2A). Three modern samples from each remnant population were selected to be analyzed alongside the historic California samples. Genotype likelihoods, genotype posterior probabilities, and per-site depths were determined using ANGSD (Korneliussen et al., 2014). The reads from historic samples showed a mapping preference toward the sea otter reference genome (Table S2), likely due to lower coverage and read quality. However, mapping to an ingroup genome from one sea otter population can cause reference biases. Therefore, analyses are presented mapped to both the domestic ferret genome and to the southern sea otter genome.
Population Structure analyses (modern samples).
Relatedness between individuals was assessed using PLINK’s identity-by-descent method of moments approach (snpgdsIBDMoM) (Purcell et al., 2007) (Table S4). SNPRelate (Zheng et al., 2012) was used to carry out linkage disequilibrium (LD) pruning of SNPs and principal components analysis (PCA) (Figure 1B; Figure S3). Weir & Cockerham’s (1984) FST was calculated for each population-pair using SNPRelate (Zheng et al., 2012) (Table S5; Figure S3). Population structure was assessed using fastSTRUCTURE (Raj et al., 2014) (Figure 1C; Figure S3). Treemix (Pickrell & Pritchard, 2012) was used to assess historical population divergence and gene flow using covariance of allele frequencies (Figure 1D; Figure S3). We constructed a Neighbor-Joining (NJ) phylogenetic tree to explore the relationship between the Baja California sea otters and the California population using the APE package in R (Paradis & Schliep, 2019) (Figure S3).
Population Structure analyses (historic samples).
PCAngsd (Meisner & Albrechtsen, 2018) was used to carry out PCA and structure analyses for the three historic samples and a subsample of three modern individuals from each population. Since the historic data mapped better to the sea otter reference genome than to the ferret reference genome, the analysis was done for each reference genome, with similar qualitative results (Figure 2; Figure S2).
Generating site frequency spectra.
ENSEMBL’s Variant Effect Predictor (VEP) (McLaren et al., 2016) was used to classify SNPs as synonymous, missense or stop-gained based on the domestic ferret genome annotation database. For the neutral SFS, putatively neutral regions >10kb away from ferret exon sequences that did not intersect with CpG islands or repeat regions were chosen. For each population, the neutral, synonymous, and missense folded SFSs were generated using a hypergeometric projection to smooth over missing data (Table S6).
S (total number of segregating sites), π (average pairwise heterozygosity), Watterson’s θ (θW) (Watterson, 1975) and Tajima’s D (Tajima, 1989) were calculated from the projected folded site frequency spectra for each population based on Wakeley (2009).
Demographic inference.
The neutral site frequency spectra were used for demographic inference in ∂a∂i (Gutenkunst et al., 2009) and fastsimcoal2 (Excoffier et al., 2013) for the following single-population models (described forward in time; model diagrams in Figure 3A):
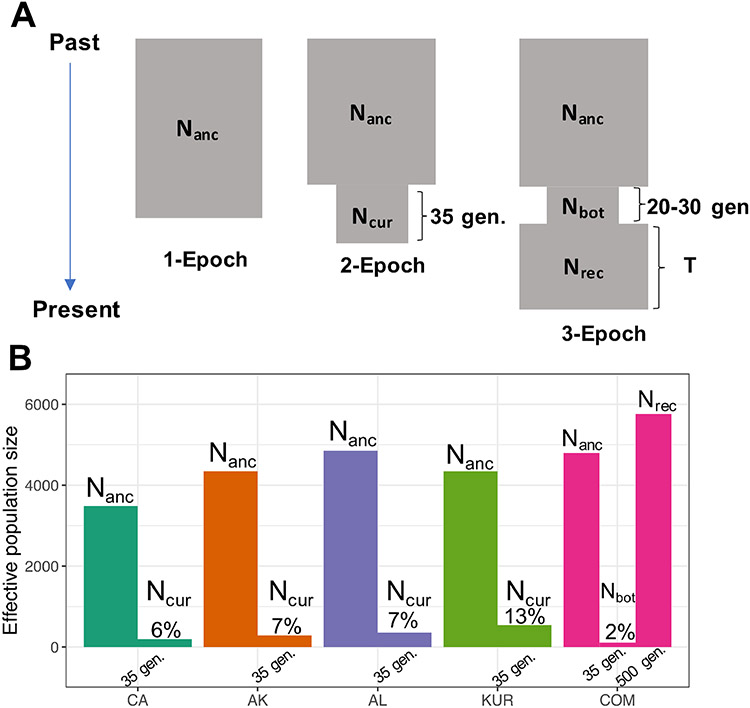
Figure 3. Signals of Extreme Decline Detected in Every Remnant Population Using the Site Frequency Spectrum.
A) The three single-population demographic models that were compared: a 1-Epoch model in which no size change occurs, a 2-Epoch model in which a single contraction occurs, and a 3-Epoch bottleneck model in which a recovery period is modeled. For the 2-Epoch model, a wide variety of contraction durations was explored using a grid search approach (Figure S4B). However, in order to compare the magnitude of decline across populations, results shown have the duration of the contraction fixed to ~35 generations, representing the start of the fur trade ~245 years prior to sampling.
B) For California (CA), Alaska (AK), Aleutian Islands (AL), and Kuril Islands (KUR), a 2-Epoch contraction model with parameters inferred using a grid search in ∂a∂i fit the SFS best. Nanc represents the inferred ancestral size and Ncur is the inferred contraction size if the contraction duration is fixed to ~35 generations to be concordant with the timing of the fur trade. The ratio of the inferred post-contraction effective population size to ancestral effective population size is shown as a percentage for each population. For the Commander Islands (COM), a 3-Epoch model with a bottleneck size (Nbot) and recovery size (Nrec) was a significantly better fit to the SFS, but this signal did not appear when mapped to the southern sea otter genome and therefore may be artefactual (Table S7; Supplemental Methods).
See also Figures S4 and S5 and Tables S6 and S7.
1-Epoch model: no size changes, infers the ancestral size (Nanc) of the population.
2-Epoch model: a single size change from Nanc to the current size (Ncur) occurring T generations ago.
3-Epoch model: two sizes changes from Nanc to a bottleneck (Nbot) followed by recovery (Nrec) that lasts T generations. The duration of the bottleneck is set to 20-30 generations.
For each model, 50 replicates of the inference were carried out to confirm that both parameters and log-likelihoods converged and parameters with the maximum log-likelihood were chosen. Models were compared using the likelihood ratio tests (LRTs) (Table S7).
Since the duration and magnitude of a contraction are difficult to disentangle, we also used a grid search approach in ∂a∂i to explore the range of parameter combinations that fit the data (Figure S4B). To compare the magnitudes of decline across populations, we fixed the duration of contraction at 245 years since the advent of the fur trade in 1741 (Kenyon, 1969) and the time of sampling ~1986 (~35 generations assuming a 7 year generation time (Gagne et al., 2018; Ralls et al., 1983)).
We used the joint site frequency spectrum between California and central Alaska sea otters to infer the shared demographic history and divergence time of the populations using fastsimcoal2 and ∂a∂i, as above (model diagrams in Figure S4D).
Finally, we compared our models inferred here using the SFS to those we previously inferred using MSMC (Schiffels & Durbin, 2014) from a single southern sea otter genome from California and a northern sea otter genome from the translocated southeast Alaska population (Beichman et al., 2019) (Figure S5).
Wright-Fisher simulations with selection.
We designed models based on the inferred demographic parameters for the California population to determine the impacts of population contractions on deleterious variation (model diagram in Figure 4A). We then simulated a period of partial recovery after the contraction to reflect the current recovery status of the population (diagram in Figure 4B).
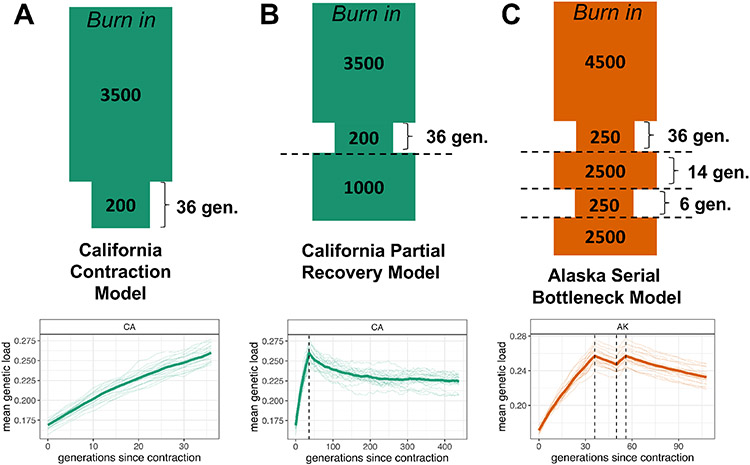
Figure 4. Recessive Genetic Load is Predicted to Increase During Decline and Persist after Recovery.
A) Forward-in-time Wright-Fisher simulations of genetic load (reduction in fitness due to deleterious variation) under the demographic model of recent population decline we inferred for the California sea otter population. The demographic model is shown above, with sizes in diploid individuals and times in generations. The simulation results below the model diagram show an increase in genetic load during the inferred population contraction (contraction occurs at generation 0). The thick line represents the mean of 20 simulation replicates, with individual replicates as faint lines. The contraction duration was increased from 35 to 36 generations to accommodate sampling of load every even-numbered generation in the simulation framework.
B) Simulations of genetic load under a model of partial recovery for the California population in which the contraction modeled in (A) is followed by a partial recovery to an effective size of 1000 individuals. The simulation results below the model are as described in (A). The partial recovery occurs at the dashed line.
C) A model of serial declines in the south central Alaska population, showing the possible impacts of post-fur trade events such as the Exxon Valdez oil spill or orca predation. The simulation results below the model are as described in (A). The first population contraction occurring at generation 0, followed by a brief recovery (starting at the first dashed line), then another rapid contraction and partial recovery (second and third dashed lines).
Results based on alternative distributions of dominance and selection coefficients are shown in Figure S6C-E.
To explore the role of serial bottlenecks, we designed a model loosely based on the history of serial bottlenecks in the Alaskan and Aleutian Island sea otter populations due to the Exxon Valdez oil spill and predation by orcas, respectively (Ballachey et al., 1994; Estes et al., 1998) (diagram in Figure 4C).
Finally, to model the potential benefits of gene flow between populations, we designed a joint simulation between the California and south central Alaska populations in which reciprocal translocations occur at a rate of either 1 individual per generation, 25 individuals per generation, or a single burst of 25 individuals being exchanged for two generations and then ceasing (diagram in Figure 5). Detailed model descriptions are in the Supplemental Methods.
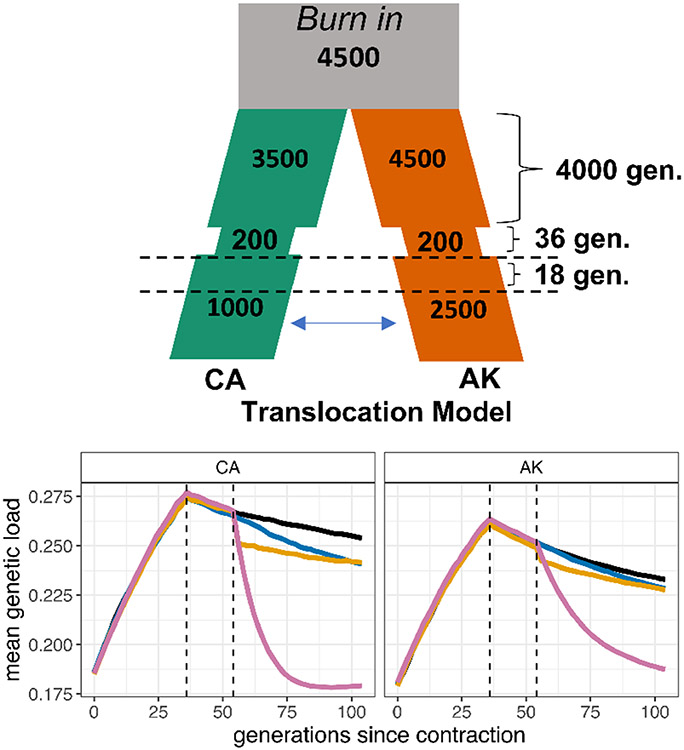
Figure 5. Gene Flow May Mitigate Genetic Load.
An isolation-migration model exploring the theoretical benefits of enabling gene flow between California and south central Alaska, based on the joint population histories of the populations we inferred. The simulated populations split 4000 generations ago, then both experience contractions followed by partial recovery, then hypothetical translocations at varying levels of intensity begin 18 generations after the populations have partially recovered from their respective declines. The forward-in-time Wright-Fisher simulations of genetic load under this model show a decrease in recessive genetic load caused by high levels of gene flow between populations. The partial recovery occurs at the first dashed line, and gene flow begins at the second dashed line. Levels of gene flow range from no gene flow (black), one individual per generation (blue), a burst of 25 individuals exchanged for two generations (yellow), or sustained high levels of gene flow of 25 individuals per generation (pink). Each line represents the mean of 20 simulation replicates. Results based on alternative distributions of dominance and selection coefficients are shown in Figure S9.
Under these different demographic models, we simulated neutral and coding sequence using forward-in-time Wright-Fisher simulations in SLiM 2 (Haller & Messer, 2017). We assumed that the dominance and selection coefficients of the mutations were inversely related, such that the most strongly deleterious mutations are also the most recessive (Henn et al., 2016; Huber et al., 2018) (Figure S6). The selection coefficients of new missense mutations were drawn from the distribution of selection coefficients inferred by Kim et al. (2017). The burden of deleterious variation, known as genetic load (L), was calculated every two generations of the model as described in the Supplemental Methods.
Since the precise distributions of dominance and selection coefficients in sea otters is unknown, we explored a variety of relationships between dominance and selection which strongly affect the genetic load dynamics. We carried out simulations in which deleterious mutations are only partially recessive (Deng & Lynch, 1997), or all mutations are fully additive or fully recessive. We also simulated under a distribution of selection coefficients that contains a higher proportion of strongly deleterious and lethal mutations suggested by Kardos et al. (2021). See Note S1 for additional discussion.
Non-Wright-Fisher simulations with selection.
To explore the extent to which the California sea otter population may be threatened by extinction due an accumulation of deleterious mutations, we conducted simulations with the non-Wright-Fisher (nonWF) models in SLiM 3 (Haller & Messer, 2019), following the approach of Kyriazis et al. (2021) with modifications described in the Supplemental Methods. These models aim to facilitate more ecologically-realistic population genetic simulations by relaxing many of the assumptions of the Wright-Fisher model. The simulated population size is determined by the absolute fitness of individuals in the population and a user-defined carrying capacity. As a result, the simulated population can go extinct when fitness declines. In addition, this model provides a natural framework for incorporating environmental stochasticity and natural catastrophes, which may also impact the persistence of sea otter populations.
Finally, to explore the potential impact of an increased historical population size on levels of inbreeding depression and extinction risk in the California sea otter population, we ran the above simulations with the ancestral carrying capacity increased three-fold, with all other parameters kept the same (diagram in Figure 6; Table S8). See Note S1 for additional discussion.
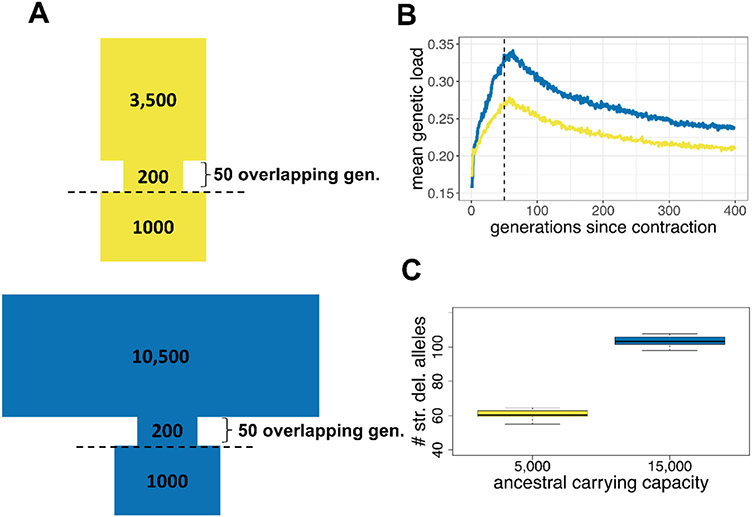
Figure 6. Long-term Low Population Size May Mitigate Recent Bottleneck Effects.
A) More ecologically-realistic non-Wright-Fisher models of the impact of historical population size on genetic load. Values represent effective population sizes, though the model uses carrying capacities (K) (see Table S8 for corresponding K). The yellow (upper) model represents a population with ancestral effective population size based on our demographic inference, and the blue (lower) model is a theoretical population that has a 3x higher ancestral effective size. Each population goes through an extreme decline as in model, then partially recovers. The contraction duration was increased from 35 to 50 generations to account for overlapping generations in the non-Wright-Fisher framework.
B) Non-Wright Fisher simulations of genetic load under the models in (A). The yellow line represents the yellow model in (A) based on our inferred demographic parameters, and the blue line represents the blue model in (A) with the 3x larger ancestral size. Recovery occurs at the dashed line.
C) The average number of strongly deleterious alleles per individual for each ancestral carrying capacity described in (A). Results based on alternative distributions of dominance and selection coefficients are shown in Figure S10.
Results
Genetic structure of sea otter populations
Individuals from each sampling location clustered by genotypes in both PCA (Figure 1B) and fastSTRUCTURE (Figure 1C) analyses, and values of FST increased with distance between populations (Figure S3D; Table S5), agreeing with previous studies (Larson et al., 2012, 2021; Larson, Jameson, Bodkin, et al., 2002). Sea otters in California are the most divergent population and represent the only surviving remnant of the southern sea otter subspecies (Enhydra lutris nereis) (Figure 1B, 1D). All other sea otter populations surveyed, which span the northern (Enhydra lutris kenyoni) and Asian (Enhydra lutris lutris) subspecies range, clustered according to geography once the California individuals were excluded (Figure S3A). Inference of the population split time between California and Alaska using the joint site frequency spectrum (SFS) dates the divergence to ~4000 generations ago (~28kya), with only low amounts of genetic exchange between the populations (Table S7; Figure S4D-E). The best-fitting model was one which also included a recent contraction in both populations (inferred in the single-population models below).
To assess the history of connectivity among the northern and Asian sea otter populations, we inferred historical migration events between these populations using Treemix (Figure 1D) and individual ancestry profiles in fastSTRUCTURE (Figure 1C). We found that northern and Asian sea otter populations have a complex history of connectivity, with evidence for fine-scale spatial structure and a history of admixture events. Twelve individuals showed evidence of admixture, notably four from the Kenai Peninsula and Kodiak Island in south central Alaska which have mixed Alaskan, Aleutian, Kuril, and Commander Island ancestry (multicolored AK columns in Figure 1C; labeled “AK-Admix” in Figure 1B and 1D; Figure S1C), and five in the Kuril Islands that had a portion of Commander Islands ancestry. Interestingly, the highly admixed Alaskan individuals sampled from Kodiak Island all had kinship coefficients ≥0.1 to each other and are unrelated to any other otters sampled in Alaska (Table S4). Historical migration (orange arrows in Figure 1D) from the Aleutian Islands and the common ancestor of the Kuril/Commander/Aleutian populations into the admixed Alaskan group, and in the opposite direction from Alaska into the Aleutian Islands, was inferred using Treemix (Pickrell & Pritchard, 2012) based on the covariance of allele frequencies. Within the Aleutian Islands, samples were obtained from Adak, Amchitka and Attu Islands (Figure S1B). When analyzed alone, we found that the three islands clustered separately in the PCA (Figure S3B) with very low values of FST (0-0.02) (Table S5). We did not observe any divergence between the two Commander Islands (Figure S3C; Table S5).
Historical DNA suggests genetic stability in California
To examine the temporal continuity of the southern sea otter population, we sequenced DNA from three historical sea otter tooth roots sampled from prehistoric and historic shell middens on the California coast. The oldest samples were from San Miguel Island, dating from 1610-1320 calibrated calendar years before present (cal BP), and the third was a historical sample from Ano Nuevo, dating from 290-70 cal BP (Figure 2A; Table S2). The low sequencing coverage (1.5-3.5x) of these samples was not high enough to explore levels of heterozygosity but was sufficient for population structure analyses. We randomly chose three individuals from every modern population, including California, to analyze alongside the historic samples. The three historic samples cluster with modern Californian sea otter samples in the PCAngsd (Figure 2B) and admixture (Figure 2C) analyses based on genotype likelihoods. We found the population structure results were qualitatively similar regardless of the reference genome used for mapping (Figure S2B-C; Table S2).
Expansion of southern sea otters into Baja California
The two modern individuals sampled in Baja California clustered with the California samples in the PCA (Figure 1B) and fastSTRUCTURE (Figure 1C) analyses. They had an FST of ~0 compared to the California sea otters (Table S5) and clustered within the California clade in a neighbor-joining tree (Figure S3E). Although historic levels of genetic differentiation between California and Baja California sea otter populations are unknown, the local population structure we observe in other remnant sea otter populations (e.g. in Figure S3B; Figure S3D) suggests that the two populations would be genetically distinguishable. The lack of differentiation that we observe therefore suggests that the Baja California samples may be migrants or the offspring of migrants from the California population, rather than survivors of a historically differentiated Baja California population. Indeed, we observed moderate levels of kinship (0.08 – 0.15) between them and individuals in the California sample (Table S4), indicating that they may even be relatives of extant California individuals.
Signals of extreme decline detected in every remnant population
Each sea otter population has extremely low neutral genetic diversity, ranging from 1.1-1.6x10−4 heterozygotes/base pair, irrespective of which reference genome (domestic ferret or southern sea otter) was used for read mapping (Table S6). To explore demographic scenarios that could give rise to low diversity, we conducted demographic inference based on the site frequency spectrum (SFS). We detected low ancestral effective population sizes (3500-5000 individuals per population) and a signal of extreme decline in every sea otter population we surveyed (Figure 3A-B; Table S7). Single-size population models were strongly rejected in favor of two-epoch contraction models (likelihood ratio test p-values < 0.005; Figure 3A; Table S7). After fixing the duration of the contraction based on historical records (Methods), we inferred that each population suffered an extreme contraction, with only 2-13% of the ancestral effective population size estimated to have survived the decline (Figure 3B). Qualitatively similar results of a steep decline in every population were obtained using either the domestic ferret or southern sea otter reference genome to map sequence reads (Table S7; Supplemental Methods). Since our inference method averages over the period of decline, sea otter populations in some regions may have reached even lower population sizes than those in our model, as suggested by field observations (Riedman & Estes, 1990).
We used forward-in-time simulations to demonstrate that this extreme bottleneck would only moderately lower neutral heterozygosity (Figure S6A), and therefore long-term small population size, rather than a rapid contraction, is likely responsible for the extremely low genetic diversity we observe. However, a decline of this severity would shift the SFS toward intermediate frequency variants (Figure S4C), with implications for genetic load (discussed below).
Our inference was not able to detect the very recent recovery of sea otter populations using the SFS, except in the case of the Commander Islands. Specifically, a three-epoch model with a recovery period was the best fit to the Commander Islands SFS (Figure 3B; likelihood ratio test p-value = 5x10−7; Table S7). However, the inferred recovery duration is too long (500 generations) to be consistent with the time since the fur trade. Further, the evidence of a recovery in the Commander Islands is diminished because the signal is not found when demographic inference used reads mapped to the southern sea otter reference genome (Table S7; Supplemental Methods).
Finally, we compared our SFS-based demographic models to those previously obtained from single whole genome sequences (Beichman et al., 2019) and found that the fit of the whole-genome models to the empirical SFS was improved by including the recent decline that we detect with the SFS. The combination of these inference methods increases confidence in our findings of an extreme decline in recent time occurring in every sea otter population (Figure S5). For California, we found that a model that included a shallow pre-fur trade decline in population size in addition to the recent extreme bottleneck was more consistent with the whole-genome inference than the simpler two-epoch model inferred using the SFS (Figure S5).
Severe population contraction is predicted to increase genetic load
To explore how changes in population size could affect deleterious variation, we used forward-in-time Wright-Fisher simulations of coding sequence to assess the reduction in fitness due to deleterious mutations (i.e. genetic load) before and after the inferred fur trade bottleneck. Our simulations showed that contractions of the magnitude we infer, while brief, could increase the genetic load of the population up to 54% (Figure 4A). This increase in load is due to deleterious recessive mutations that were shielded from selection in the larger ancestral population being exposed as homozygotes in the contracted population, allowing them to reduce fitness (Figure S7A). This shift in the frequency of missense mutations can be seen in the shift of variants from low to intermediate frequency in the simulated post-contraction and empirical missense SFSs compared to the simulated pre-contraction SFS (Figure S8A). Additional model conditions are presented in Figure S6C, Figure S7B-E and Figure S8B-E and discussed in Note S1.
Simulations indicate genetic load may persist, but not cause extinction
To assess the possible long-term genetic impacts of the fur trade and subsequent bottlenecks, we carried out additional Wright-Fisher simulations of sea otter recovery. We found that even with partial recovery (defined as a return to an effective size of 1000 individuals for the California population), genetic load did not return to pre-fur trade levels even after 400 generations (2,800 years) (Figure 4B, Figure S6D). Subsequent declines in population size further slowed the reduction in genetic load (Figure 4C).
To explore whether the increase in genetic load could cause the extinction of sea otter populations, we made the simulations more ecologically realistic by allowing the simulated population size to depend on population fitness such that the simulated population can go extinct when fitness declines or due to environmental stochasticity, following the approach of Kyriazis et al. (2021). Under this model, the California otter population did not decline to extinction due to genetic factors at any point during its 400 generation-long partial recovery from the decline (Table S8). Additional model conditions are presented in Figures S6D-E and discussed in Note S1.
Gene flow may mitigate impact of genetic load
To explore the potential consequences of restoring gene flow between otter populations, we used Wright-Fisher simulations to model a scenario in which there are varying levels of gene flow between the bottlenecked Alaska and California populations (Figure 5). We found that genetic load decreases more swiftly than in the no-gene flow scenario if at least one migrant is reciprocally exchanged per generation. Equivalently, it is decreased the same amount if a single burst of 25 individuals are reciprocally exchanged for just two generations and then gene flow ceases (a translocation-like scenario) (Figure 5). However, genetic load is still slow to return to pre-fur trade levels in each population except in the case of extremely high gene flow of 25 individuals reciprocally exchanged per generation, which is also highly impractical in reality. Additional model conditions are presented in Figure S9 and discussed in Note S1.
Long-term low population size may mitigate effects of bottlenecks
Finally, we explored the potential role of the relatively small historical effective population sizes that our results suggest on the accumulation of genetic load and extinction risk in the California sea otter population. Specifically, we hypothesized that, if sea otters had a larger historical effective population size, they might have experienced more severe genetic effects from this bottleneck, due to the higher numbers of heterozygous recessive strongly deleterious mutations harbored in large populations (Hedrick, 2002; Hedrick & Garcia-Dorado, 2016; Kyriazis et al., 2021; Robinson et al., 2018). These mutations, when made homozygous by inbreeding following the bottleneck, could lead to a greater increase in genetic load and potentially drive extinction. To explore the consequences of a larger historical population, we ran non-Wright-Fisher simulations with an ancestral population size that was three times larger than our empirically-inferred size (Figure 6A). The historically-larger population had a greater increase in recessive genetic load due to the recent contraction than the historically smaller population (117% vs 61% increase, Figure 6B; Table S8). The higher load is a direct consequence of the higher number of recessive strongly deleterious mutations in the larger ancestral population (Figure 6C). Nevertheless, the historically-larger population did not go extinct in any simulation replicates. Additional model conditions are presented in Figure S10 and discussed in Note S1.
Simulation dynamics under differing dominance and selection distributions
Due to uncertainty in the distribution of selection coefficients and the relationship between dominance and selection in sea otters, we carried out additional simulations under a variety of selection and dominance parameters (Note S1). We found that the population contraction would result in an increase in genetic load if mutations were recessive, but not if they were additive or only partially recessive (Figure S6C). Strongly deleterious additive mutations are not shielded from selection in the heterozygous state and are continuously eliminated by selection. This finding is consistent with theoretical studies suggesting that recent demography may only have a limited impact on additive load (Do et al., 2015; Lohmueller, 2014; Simons et al., 2014) and that recessive load caused by strongly deleterious genotypes in the homozygous state may be more of a threat to population persistence. In addition, if there is a substantial proportion of recessive lethal mutations present in the simulated population (≥ 5%) as proposed by Kardos et al. (2021), we found that overall genetic load would be much higher, but the percent gain in load during the contraction would be moderate (~9%) and rapidly return to baseline after population recovery (Figures S6C-E). Also, translocations and ancestrally-small population sizes would show no benefits (Figures S9-S10). Each of these different model conditions is shown in Figures S6-S10 and discussed fully in Note S1.
Discussion
Sea otters exhibit broad- and fine-scale population structure
Defining management units for sea otters, whether at fine or broader scales, remains a challenge to conservation efforts (Davis et al., 2019). Genetic structure as defined by PCA and admixture analyses can provide insight into natural genetic groupings, though cannot characterize micro-scale structuring of sea otter familial groups (Bodkin, 2015; Davis et al., 2019; Gagne et al., 2018; Larson et al., 2021; Tinker et al., 2019). We found that the geographically-driven population structure of sea otter populations remained after the fur trade bottlenecks, leading to five broad geographic groupings in our sample: the Kuril Islands; Commander Islands; western Aleutian Islands; south central Alaska; and California. This shows that a key element of the successful restoration of sea otters was their survival and numerical increase from small remnant populations distributed throughout their geographic range rather than long-distance dispersal (Larson et al., 2012, 2021; Larson, Jameson, Bodkin, et al., 2002). This fortuitous survival allowed both rapid range-wide restoration of former abundance and preserved historic genetic structure and perhaps even local specializations that enhanced survival (Fujii et al., 2015).
The importance of these remnant populations is exemplified by the southern sea otter, whose former distribution likely spanned from Washington to Mexico (Wilson et al., 1991), but was fully eliminated, except for 30-50 individuals on the California coast (Riedman & Estes, 1990). Our results suggest that southern sea otters, represented by the only surviving population in California, diverged from the four northern and Asian populations before the latter populations differentiated from each other. The ~28kya we infer for this north-south split means it may have occurred during the Last Glacial Maximum (~20-27kya). Sea otters cannot tolerate extensive ice cover that restricts access to foraging habitat (Schneider & Faro, 1975) and much of the Alaskan and British Columbian coast was glaciated during that period (Lesnek et al., 2018; Mann & Hamilton, 1995; Mann & Peteet, 1994). This may have isolated northern and southern populations, with only low levels of gene flow occurring subsequently along the North American Pacific coast after the ice retreated. Since the intermediate populations between Alaska and California are now extinct, we cannot easily differentiate between a demographic model of long-term isolation of northern and southern sea otter populations, or isolation by distance with low dispersal between populations, both of which are consistent with our results. Ancient mitochondrial DNA and morphological characteristics of extinct Oregon sea otters analyzed in Valentine et al. (2008) suggest they were more similar to California sea otters, but recent ancient mitogenome haplotype analyses in Wellman et al. (2020) shows clustering between northern sea otters and ancient Oregon sea otters, possibly indicating that the southern sea otters in California and Mexico may have been genetically distinct from Oregon populations, and/or that Oregon was a location in which northern and southern sea otters mixed (Larson et al., 2012; Lee Lyman, 1988). The genetic legacy of the southern sea otter lineage would have entirely been lost if not for the preservation of the last relict population in California.
Our analysis of population structure in northern and Asian sea otter populations demonstrates a complex and interconnected history shaped by gene flow, recolonization, and fine-scale population structure. Migration appears to have occurred in both directions along the island chains of the north Pacific, likely aided by the high abundance of sea otters in these areas prior to the fur trade. The four interrelated individuals sampled from Kodiak Island, Alaska, that had high levels of Aleutian Island ancestry are particularly intriguing. Conceivably, Kodiak Island was a sink for migration from the Aleutian Islands when otters were abundant. Previously, mitochondrial and microsatellite data identified Kodiak Island otters as a separate stock from those in Prince William Sound and the Kenai Peninsula, with greater similarity to the Aleutian Islands (Gorbics & Bodkin, 2001). Our findings add evidence that the Kodiak Island sea otters are a distinct genetic grouping with mixed south-central Alaskan and Aleutian Island ancestry.
Among the northern populations, the differentiation we observed between the Aleutian Islands indicates that the populations among these islands are highly interconnected, but there may nevertheless exist fine-scale structure at the level of only hundreds of kilometers between the islands. The lack of similar differentiation between the Commander Islands could indicate enduring high levels of gene flow between Bering and Medny Island or is also consistent with the theory that the Bering Island colony was fully extirpated during the fur trade and recolonized by otters from Medny Island (Bodkin et al., 2000).
Historical DNA suggests genetic stability in California
Only 30-50 southern sea otters are thought to have survived the fur trade near Big Sur, CA, a minuscule fraction of their historic range (Riedman & Estes, 1990). We had hypothesized that the fur trade bottleneck may have caused considerable genetic differentiation in extant sea otters which are descended from this relict population compared to historic sea otters sampled from the same region due to founder effects, drift or local extinction and recolonization from genetically differentiated subpopulations (Hastings & Harrison, 1994). Local extinction and recolonization is frequently observed in mammal populations using ancient DNA (Brace et al., 2012; Cooper et al., 2015; Foote et al., 2012; Hofreiter & Barnes, 2010; Hofreiter & Stewart, 2009; Nichols et al., 2007; Pilot et al., 2010) and population replacement has found to be widespread in human populations (Allentoft et al., 2015; Lazaridis et al., 2016; Olalde et al., 2018). However, we found that modern sea otters from the central coast of California cluster closely with a historic sample from the central coast (290-70 cal BP) and two historic samples from southern California (1620-1320 cal BP). Our comparison is global in scale, and therefore cannot detect more subtle signals of differentiation caused by the population bottleneck, but the clustering of historic and extant California sea otter samples from the northern and southern regions of the state does not support extinction after an extreme population bottleneck followed by recolonization from a genetically divergent population.
Southern sea otters are expanding into Baja California
The sea otter population in Baja California, Mexico, was declared ‘probably extinct’ in the late 1960s (Kenyon, 1969) based on aerial surveys, but since the 1970s fishermen and scientists have reported sporadic sightings of sea otters (Gallo-Reynoso & Rathbun, 1997; Rodriguez-Jaramillo & Gendron, 1996) and the population is currently classified as endangered (Norma Oficial Mexicana, 2010). A previous study that analyzed 318-bp of the mitochondrial control region of the two samples used in this study (Schramm et al., 2014) did not have the resolution to determine if these otters were recent migrants from California (Hatfield, 2005; Rathbun et al., 2000) or were survivors of the historic population. We found that the two individuals we sampled are likely migrants or the descendants of migrants from California, representing a southward expansion of the population, rather than survival of the original Baja California sea otter population. Though additional sampling will be necessary, this finding has immediate conservation and management implications for the southern sea otter subspecies. First, binational efforts to protect sea otters in California and Mexico should be increased to assure that the California population remains the source of individuals for Baja California with the objective that in the long term, a permanent self-sustaining Baja California population would be reestablished. Second, the protection of the few individuals in the Mexican Pacific and their habitat is key to increase population numbers, which would facilitate the recolonization of their old geographic range and provide redundancy for regional extinctions should they occur (Eisaguirre et al., 2020). Finally, protecting the southern subspecies could help restore kelp forest ecosystems (Wilmers et al., 2012), which is critical for coastal fisheries in both the US and Mexico.
Low diversity indicates long-term small population size
The extremely low diversity we detect in all sea otter populations likely preceded the fur trade (Beichman et al., 2019). We estimate small ancestral effective sizes (Nanc) from the SFS for every sea otter population prior to the fur trade contraction (3500-5000 individuals per population). The low ancestral effective population sizes we infer are ~20% of the historical census sizes of these populations, which range from 10-20,000 individuals (Riedman & Estes, 1990). This low effective:census ratio is expected (Frankham, 1995; Nunney, 1993, 1996), and may be due to several factors. Resource availability may set biological limits on the amount of successful reproduction for each population (Davis et al., 2019; Tinker et al., 2019), which may explain similar ancestral sizes across all populations. Overlapping generations and high variance in reproductive success between males and females due to a highly polygynous mating system may have limited successful breeding to a smaller fraction of the census population (Nunney, 1993, 1996; Riedman & Estes, 1990). The linear geography of sea otter populations along coastlines or around islands may increase local structure which could further decouple census and effective population sizes (Gagne et al., 2018). Finally, hunting by aboriginal peoples may have kept otter population numbers low or caused local depletions prior to the fur trade (Aguilar et al., 2008; Braje & Rick, 2011; Erlandson et al., 2005; Hildebrandt & Jones, 1992; Larson et al., 2012; Simenstad et al., 1978). Our estimates of effective size provide insight into long-term levels of genetic diversity of these populations, showing that sea otters have maintained low genetic diversity throughout the history of the species.
Signals of extreme decline detected in every remnant population
We detected a strong signal of independent population collapse in every sea otter population surveyed using SFS-based demographic inference methods, consistent with worldwide near-extinction caused by the fur trade. This decline may have had lasting impacts on genetic diversity. As predicted theoretically by Ralls, Ballou & Brownell (1983), this extreme contraction would only moderately lower heterozygosity, and is therefore not responsible for the very low diversity we report above. However, the contraction may have increased genetic load in the surviving populations (discussed below).
For the California population, we found that a model that combines SFS and whole genome-based demographic inference methods supported an extreme recent bottleneck as well as an additional earlier contraction. Several studies have raised the possibility of multiple declines in the California sea otter population, possibly caused by hunting by Native Californians (Aguilar et al., 2008; Beichman et al., 2019; Erlandson et al., 2005; Larson et al., 2012), and we demonstrate that a model incorporating multiple declines could integrate both whole-genome and SFS-based inferences.
Impact of genetic load in sea otter populations
Our simulations show that the extreme fur trade bottleneck, although brief, may have increased the genetic load of surviving sea otter populations due to the exposure of high-frequency deleterious recessive alleles as homozygotes (Balick et al., 2015; Do et al., 2015; Hedrick & Garcia-Dorado, 2016; Kimura et al., 1963; Lynch et al., 1995; Ohta, 1973). Many sea otter populations quickly recovered after their protection in 1911 under the International Fur Seal Treaty (Kenyon, 1969; Riedman & Estes, 1990). The California population, however, has yet to recover beyond one third of its estimated pre-fur trade levels, with a current census size ~3000 individuals (Hatfield et al., 2019), down from an estimated ancestral size of 10,000-20,000 individuals (Riedman & Estes, 1990). Populations in Alaska and the Aleutian Islands have also suffered serial declines due to cataclysms such as the Exxon Valdez oil spill in Prince William Sound in 1989 or due to predation by orca in the Aleutian Islands (Ballachey et al., 1994; Estes et al., 1998). We therefore simulated a variety of recovery models including partial recovery and serial declines to determine their effects on genetic load. We found that under a scenario of partial recovery, recessive genetic load did not return to pre-fur trade levels even after 400 generations (~2,800 years). Serial declines further slowed this process, highlighting the critical importance of protecting sea otter populations from further bottlenecks as robust populations are needed to erode the increase in genetic load caused by the fur trade.
Since selection can act much more strongly on partially recessive and additive mutations, the dynamics of genetic load are strongly affected by the distribution of dominance and selection coefficients (Note S1) (García-Dorado, 2012; Hedrick & Garcia-Dorado, 2016). Additional research on the distribution of dominance and selection coefficients in mammals is therefore needed to gain a more precise understanding of the extent to which the fur trade bottleneck led to an increased genetic load in sea otters.
Despite the persistence of genetic load in our simulations, we found that this accumulation of genetic load is unlikely to cause extinction in more ecologically-realistic non-Wright-Fisher simulations. This result is encouraging, as it indicates that the decline due to the fur trade was not severe enough to cause species extinction due to genetic factors alone. Critically, the environmental threats that sea otters currently confront, particularly predation by sharks and orca, lack of habitat or prey availability, and environmental catastrophes such as oil spills are likely to have a far greater impact on population recovery (Ballachey et al., 1994; Estes et al., 1998; Tinker et al., 2016; U.S. Fish and Wildlife Service, 2003). In particular, our models do not currently incorporate detailed information about local population density or habitat availability, which have been shown to be important for predicting sea otter carrying capacity and range expansion (Tinker et al., 2008, 2021). Incorporating these spatial dynamics into future simulations might provide even greater resolution on the impact of genetic load on sea otter recovery.
Our Wright-Fisher simulations show that translocations between populations could decrease genetic load through heterosis of population-specific recessive deleterious variants. Such heterosis would have the effect of returning sea otter populations to pre-fur trade levels of fitness more quickly (Frankham, 2016; Hedrick & Garcia-Dorado, 2016; Whiteley et al., 2015). Translocated populations of mixed south central Alaskan and Aleutian ancestry in southeast Alaska and British Columbia, which were not analyzed in this study, may have already benefitted in this way from the mixing of populations through translocation (Bodkin et al., 1999; Jameson et al., 1982; Larson et al., 2021). While our simulations suggest a potential benefit of translocation for the southern sea otter, admixture of southern and northern sea otters could also result in the loss of local adaptations and the distinct evolutionary legacy of this long-diverged population (Bell et al., 2019; Edmands, 2007; Waller, 2015). Genetic exchange between southern and northern sea otters may occur naturally in the future if northward migrants from California overlap in range with southward migrants from the translocated northern sea otter population in Washington.
Long-term low ancestral population size may mitigate bottleneck effects
Our simulations indicate that the genetic impacts of the fur trade could have been substantially worse if the pre-bottleneck ancestral populations were larger. Strongly deleterious variants, which tend to be highly recessive, are expected to be present at lower levels in smaller populations, because they are more frequently exposed to selection as homozygotes (García-Dorado, 2012; Hedrick, 2002; Hedrick & Garcia-Dorado, 2016). Recent work by Robinson et al. (2018) and Kyriazis et al. (2021) has suggested that long-term low ancestral population sizes may lead to decreased extinction risk after a population bottleneck due to these populations harboring fewer recessive strongly deleterious variants. We therefore hypothesized that the relatively small (~3500-5000) historical effective population sizes we inferred for all surviving sea otter populations may have contributed to their apparent low risk of extinction due to genetic factors. We found that a simulated historical population three times larger than what we actually inferred had much greater gains in genetic load during the subsequent population decline (117% vs 61% increase), indicating that intermediate-sized populations may gain less genetic load through a population bottleneck if strongly deleterious mutations are recessive (Grossen et al., 2020; Kyriazis et al., 2021; Robinson et al., 2018). However, under models in which deleterious mutations are only partially recessive or additive, there was no such benefit to having had a long-term small ancestral population (Note S1). These results highlight the importance of future work to determine the frequency of recessive deleterious variants in genomes, as they determine the degree to which historical demography impacts inbreeding depression. Interestingly, regardless of the mode of dominance, the larger ancestral population model did not have an elevated rate of extinction, suggesting that the recovery of the population is substantial enough to stave off extinction even when genetic load is significantly increased. Furthermore, there may be an important tradeoff in long-term small populations between the elimination of strongly deleterious variation versus the maintenance of adaptive potential. Large, genetically diverse populations may have greater ability to rapidly adapt to changes in their environment (Hoffmann et al., 2017), though the prediction of adaptive potential based on neutral diversity is not always straightforward (Teixeira & Huber, 2021).
In conclusion, we characterized genome-wide genetic diversity in sea otter populations that survived the fur trade. We found that each northern sea otter remnant population is genetically distinct but interconnected via a complex history of gene flow. The southern sea otter population in California is the last remnant of a long-diverged lineage and is beginning to recolonize Baja California, Mexico. Each of these remnant populations provided a means for rapid recovery of the sea otter and continuity of historical population structure. We detected a strong genetic signal of the fur trade bottleneck in every population, allowing us to estimate its severity and possible impacts on genetic load. We combined these empirical results with genetic simulations to demonstrate an approach for assessing the impacts of extreme population decline and exploring varied recovery scenarios. Several species, such as the Northern elephant seal (Hoelzel et al., 1993), island fox (Robinson et al., 2016, 2018), and North American bison (Hedrick, 2009) have managed to survive extreme population bottlenecks (reviewed in Wiedenfeld et al. (2021)). Our simulations indicate that dramatic declines may not doom a species to eventual extinction due to genetic factors alone, provided the recovery is rapid, but the legacy of increased genetic load caused by a bottleneck may be challenging to erode without gene flow between populations. Our genome- and species-wide analysis indicates the need for varied conservation strategies for each sea otter population that reflect region-specific genomic variation, demographic history, and local environmental challenges.
Supplementary Material
Table S1. Modern Sample Information, Related to Figure 1.
Table S3. Depth, Coverage and Neutral Heterozygosity per Individual, Mapped to Domestic Ferret and Southern Sea Otter, Related to Figure 1. Note that neutral regions are defined slightly differently in sea otter and ferret reference genomes, with 9.3Mb passing neutral region filters in sea otter, and 6.8Mb in ferret due to an intersection with repeat regions in the ferret annotation. See Supplemental Methods for details.)
Table S4. Pairwise Kinship Coefficients (calculated within each population), Related to Figure 1. Only one individual was retained from pairs of individuals with kinship > 0.2.
Acknowledgements
We thank Jacqueline Robinson, Bernard Kim, Clare Marsden, Tanya Phung, Arun Durvasula, Jesse Garcia, Jazlyn Mooney, Jakub Vlček, Eduardo Amorim, Xinjun Zhang, Nicolas Rochette and Kelley Harris for help and advice on analyses. We thank James Estes for insights into study design, sea otter ecology and demographic history and aid in the interpretation of results. We also thank Janet Sinsheimer and Paul Barber for comments on the manuscript, Lilian Carswell for suggesting we examine sea otters in Baja California, and Michelle Staedler, Andy Johnson, Shawn Larson, Tim Tinker, and Michael Murray for advice on study design. We thank Jacob Enk, Alison Devault, Brian Brunelle and Sivakumar Nallasivam for advice on historic DNA experimental design, library preparation, and capture, Seth Newsome and Emma Eliott-Smith for sharing historic sea otter samples, and Brian Holguin for advice on historic sea otter samples. We thank Alexander Burdin, Robert Brownell, Jr. and Glenn VanBlaricom for sample collection. We thank Shana McDevitt for advice on sequencing strategies, John Southon for radiocarbon dating, and Joann Shih for sea otter art. We thank the Ensenada Marine Mammal Research and Conservation for collecting the Ensenada sample.
The work was supported by the National Science Foundation (DEB Small Grant #1556705), Monterey Bay Aquarium, UCLA Academic Senate, and UC Conservation Genomics Consortium. ACB was supported by the National Institutes of Health (NIH) Training Grant in Genomic Analysis and Interpretation (T32 HG-002536), the NSF Graduate Research Fellowship Program, and the Biological Mechanisms of Healthy Aging Training Program (NIH T32AG066574). KPK was partially supported by funding from St. Petersburg State University, Russia (Genome Russia Grant no. 1.52.1647.2016). KEL and CCK were supported by NIH grant R35GM119856 to KEL. SNM was partially funded by the Mexican Council for Science and Technology (Postdoctoral Fellowship 724094).
Footnotes
Declaration of interests
The authors have no conflicts of interest.
DATA AND CODE AVAILABILITY.
All sequencing data are available on the Sequence Read Archive (SRA), BioProject accession PRJNA629776. Data files are available on Dryad (https://doi.org/10.5068/D1ZD4D). All custom scripts are available on Github (https://github.com/LohmuellerLab/SeaOtterSequenceCaptureProject).
BENEFITS SHARED.
The sea otter samples from Baja California, Mexico were collected, sequenced, and analyzed by Mexican researchers, all of whom are co-authors (Gisela Heckel, Yolanda Schramm, Sergio Nigenda-Morales, Andrés Moreno-Estrada), enabling reciprocal exchange of expertise between Mexican and U.S. researchers. The findings will inform sea otter conservation efforts in the United States and Mexico. All collaborators are co-authors, including two undergraduate researchers (Pooneh Kalhori and Amber DeVries). We have disseminated findings via conference talks in the United States and Mexico, and to the public via posts on the Monterey Bay Aquarium blog. Annabel Beichman and co-author Mark Hylkema (Cultural Resources Program Manager & Tribal Liaison/Archaeologist, California State Parks) will coordinate further to disseminate the findings to the public via a virtual webinar. All data and code to generate these analyses are shared publicly (described above).
References
- Aguilar A, Jessup DA, Estes J, & Garza JC (2008). The distribution of nuclear genetic variation and historical demography of sea otters. Animal Conservation, 11(1), 35–45. 10.1111/j.1469-1795.2007.00144.x [DOI] [Google Scholar]
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas A-S, Margaryan A, Higham T, Chivall D, Lynnerup N, Harvig L, Baron J, Casa PD, Dąbrowski P, … Willerslev E (2015). Population genomics of Bronze Age Eurasia. Nature, 522(7555), 167–172. 10.1038/nature14507 [DOI] [PubMed] [Google Scholar]
- Balick DJ, Do R, Cassa CA, Reich D, & Sunyaev SR (2015). Dominance of deleterious alleles controls the response to a population bottleneck. PLOS Genetics, 11(8), e1005436. 10.1371/journal.pgen.1005436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballachey BE, Bodkin JL, & DeGange AR (1994). An overview of sea otter studies. Marine Mammals and the Exxon Valdez, 47–59. [Google Scholar]
- Beichman AC, Koepfli K-P, Li G, Murphy W, Dobrynin P, Kliver S, Tinker MT, Murray MJ, Johnson J, Lindblad-Toh K, Karlsson EK, Lohmueller KE, & Wayne RK (2019). Aquatic adaptation and depleted diversity: A deep dive into the genomes of the sea otter and giant otter. Molecular Biology and Evolution, 36(12), 2631–2655. 10.1093/molbev/msz101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DA, Robinson ZL, Funk WC, Fitzpatrick SW, Allendorf FW, Tallmon DA, & Whiteley AR (2019). The exciting potential and remaining uncertainties of genetic rescue. Trends in Ecology & Evolution, 34(12), 1070–1079. 10.1016/j.tree.2019.06.006 [DOI] [PubMed] [Google Scholar]
- Bodkin JL (2015). Chapter 3—Historic and contemporary status of sea otters in the North Pacific. In Larson S, Bodkin JL, & VanBlaricom GR (Eds.), Sea Otter Conservation (pp. 43–61). Academic Press. 10.1016/B978-0-12-801402-8.00003-2 [DOI] [Google Scholar]
- Bodkin JL, Ballachey BE, Cronin MA, & Scribner KT (1999). Population demographics and genetic diversity in remnant and translocated populations of sea otters. Conservation Biology, 13(6), 1378–1385. [Google Scholar]
- Bodkin JL, Burdin AM, & Ryazanov DA (2000). Age- and sex-specific mortality and population structure in sea otters. Marine Mammal Science, 16(1), 201–219. 10.1111/j.1748-7692.2000.tb00913.x [DOI] [Google Scholar]
- Brace S, Palkopoulou E, Dalén L, Lister AM, Miller R, Otte M, Germonpré M, Blockley SPE, Stewart JR, & Barnes I (2012). Serial population extinctions in a small mammal indicate Late Pleistocene ecosystem instability. Proceedings of the National Academy of Sciences, 109(50), 20532–20536. 10.1073/pnas.1213322109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braje TJ, & Rick TC (2011). Human Impacts on Seals, Sea Lions, and Sea Otters: Integrating Archaeology and Ecology in the Northeast Pacific. University of California Press. [Google Scholar]
- Cooper A, Turney C, Hughen KA, Brook BW, McDonald HG, & Bradshaw CJA (2015). Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science, 349(6248), 602–606. 10.1126/science.aac4315 [DOI] [PubMed] [Google Scholar]
- Davis RW, Bodkin JL, Coletti HA, Monson DH, Larson S, Carswell LP, & Nichol LM (2019). Future directions in sea otter research and management. Frontiers in Marine Science, 5. 10.3389/fmars.2018.00510 [DOI] [Google Scholar]
- Deng H-W, & Lynch M (1997). Inbreeding depression and inferred deleterious-mutation parameters in Daphnia. Genetics, 147(1), 147–155. 10.1093/genetics/147.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Balick D, Li H, Adzhubei I, Sunyaev S, & Reich D (2015). No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nature Genetics, 47(2), 126–131. 10.1038/ng.3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S (2007). Between a rock and a hard place: Evaluating the relative risks of inbreeding and outbreeding for conservation and management. Molecular Ecology, 16(3), 463–475. 10.1111/j.1365-294X.2006.03148.x [DOI] [PubMed] [Google Scholar]
- Eisaguirre JH, Eisaguirre JM, Davis K, Carlson PM, Gaines SD, & Caselle JE (2020). Trophic redundancy and predator size class structure drive differences in kelp forest ecosystem dynamics. Ecology, 101(5), e02993. 10.1002/ecy.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson JM, Rick TC, Estes JA, Graham MH, Braje TJ, & Vellanoweth RL (2005). Sea otters, shellfish, and humans: 10,000 years of ecological interaction on San Miguel Island, California. Proceedings of the Sixth California Islands Symposium, 58–69. [Google Scholar]
- Estes JA, & Palmisano JF (1974). Sea otters: Their role in structuring nearshore communities. Science, 185(4156), 1058–1060. [DOI] [PubMed] [Google Scholar]
- Estes JA, Tinker MT, Williams TM, & Doak DF (1998). Killer whale predation on sea otters linking oceanic and nearshore ecosystems. Science, 282(5388), 473–476. 10.1126/science.282.5388.473 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, & Foll M (2013). Robust demographic inference from genomic and SNP data. PLoS Genetics, 9(10), e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Hofreiter M, & Morin PA (2012). Ancient DNA from marine mammals: Studying long-lived species over ecological and evolutionary timescales. Annals of Anatomy - Anatomischer Anzeiger, 194(1), 112–120. 10.1016/j.aanat.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Frankham R (1995). Effective population size/adult population size ratios in wildlife: A review. Genetics Research, 66(2), 95–107. 10.1017/S0016672300034455 [DOI] [PubMed] [Google Scholar]
- Frankham R (2016). Genetic rescue benefits persist to at least the F3 generation, based on a meta-analysis. Biological Conservation, 195, 33–36. 10.1016/j.biocon.2015.12.038 [DOI] [Google Scholar]
- Fujii JA, Ralls K, & Tinker MT (2015). Ecological drivers of variation in tool-use frequency across sea otter populations. Behavioral Ecology, 26(2), 519–526. 10.1093/beheco/aru220 [DOI] [Google Scholar]
- Gagne RB, Tinker MT, Gustafson KD, Ralls K, Larson S, Tarjan LM, Miller MA, & Ernest HB (2018). Measures of effective population size in sea otters reveal special considerations for wide-ranging species. Evolutionary Applications, 11(10), 1779–1790. 10.1111/eva.12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Reynoso JP, & Rathbun GB (1997). Status of sea otters (Enhydra lutris) in Mexico. Marine Mammal Science, 13(2), 332–340. 10.1111/j.1748-7692.1997.tb00639.x [DOI] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, Whittle A, Dani J, Raczky P, Higham TFG, Hofreiter M, Bradley DG, & Pinhasi R (2014). Genome flux and stasis in a five millennium transect of European prehistory. Nature Communications, 5(1), 1–9. 10.1038/ncomms6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorado A (2012). Understanding and predicting the fitness decline of shrunk populations: Inbreeding, purging, mutation, and standard selection. Genetics, 190(4), 1461–1476. 10.1534/genetics.111.135541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbics CS, & Bodkin JL (2001). Stock structure of sea otters (Enhydra lutris kenyoni) in Alaska. Marine Mammal Science, 17(3), 632–647. 10.1111/j.1748-7692.2001.tb01009.x [DOI] [Google Scholar]
- Grossen C, Guillaume F, Keller LF, & Croll D (2020). Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nature Communications, 11(1), 1–12. 10.1038/s41467-020-14803-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, & Bustamante CD (2009). Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics, 5(10), e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, & Messer PW (2017). SLiM 2: Flexible, interactive forward genetic simulations. Molecular Biology and Evolution, 34(1), 230–240. 10.1093/molbev/msw211 [DOI] [PubMed] [Google Scholar]
- Haller BC, & Messer PW (2019). SLiM 3: Forward genetic simulations beyond the Wright–Fisher model. Molecular Biology and Evolution, 36(3), 632–637. 10.1093/molbev/msy228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings A, & Harrison S (1994). Metapopulation dynamics and genetics. Annual Review of Ecology and Systematics, 25, 167–188. JSTOR. https://www.jstor.org/stable/2097309 [Google Scholar]
- Hatfield BB (2005). The translocation of sea otters to San Nicolas Island: An update. Proceedings of the 6th California Islands Symposium (Garcelon DK and Schwemm CA, Eds.). Institute for Wildlife Studies, Arcata, California National Park Service, Technical Publication CHIS-05-01, 473–475. [Google Scholar]
- Hatfield BB, Yee JL, Kenner MC, & Tomoleoni JA (2019). California sea otter (Enhydra lutris nereis) census results, spring 2019 (USGS Numbered Series No. 1118; Data Series). U.S. Geological Survey. http://pubs.er.usgs.gov/publication/ds1118 [Google Scholar]
- Hedrick PW (2002). Lethals in finite populations. Evolution, 56(3), 654–657. 10.1111/j.0014-3820.2002.tb01374.x [DOI] [PubMed] [Google Scholar]
- Hedrick PW (2009). Conservation genetics and North American bison (Bison bison). Journal of Heredity, 100(4), 411–420. 10.1093/jhered/esp024 [DOI] [PubMed] [Google Scholar]
- Hedrick PW, & Garcia-Dorado A (2016). Understanding inbreeding depression, purging, and genetic rescue. Trends in Ecology & Evolution, 31(12), 940–952. 10.1016/j.tree.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, Excoffier L, Kidd JM, & Bustamante CD (2016). Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proceedings of the National Academy of Sciences, 113(4), E440–E449. 10.1073/pnas.1510805112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt WR, & Jones TL (1992). Evolution of marine mammal hunting: A view from the California and Oregon coasts. Journal of Anthropological Archaeology, 11(4), 360–401. 10.1016/0278-4165(92)90013-2 [DOI] [Google Scholar]
- Hoelzel AR, Halley J, O’Brien SJ, Campagna C, Arnborm T, Le Boeuf B, Ralls K, & Dover GA (1993). Elephant seal genetic variation and the use of simulation models to investigate historical population bottlenecks. Journal of Heredity, 84(6), 443–449. 10.1093/oxfordjournals.jhered.a111370 [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM, & Kristensen TN (2017). Revisiting adaptive potential, population size, and conservation. Trends in Ecology & Evolution, 32(7), 506–517. 10.1016/j.tree.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Hofreiter M, & Barnes I (2010). Diversity lost: Are all Holarctic large mammal species just relict populations? Journal of Biology, 9(3), 24. 10.1186/jbiol228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, & Stewart J (2009). Ecological change, range fluctuations and population dynamics during the Pleistocene. Current Biology, 19(14), R584–R594. 10.1016/j.cub.2009.06.030 [DOI] [PubMed] [Google Scholar]
- Huber CD, Durvasula A, Hancock AM, & Lohmueller KE (2018). Gene expression drives the evolution of dominance. Nature Communications, 9(1), 1–11. 10.1038/s41467-018-05281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson RJ, Kenyon KW, Johnson AM, & Wight HM (1982). History and status of translocated sea otter populations in North America. Wildlife Society Bulletin (1973-2006), 10(2), 100–107. JSTOR. https://www.jstor.org/stable/3781726 [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PLF, & Orlando L (2013). mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics, 29(13), 1682–1684. 10.1093/bioinformatics/btt193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M, Armstrong EE, Fitzpatrick SW, Hauser S, Hedrick PW, Miller JM, Tallmon DA, & Funk WC (2021). The crucial role of genome-wide genetic variation in conservation. Proceedings of the National Academy of Sciences, 118(48). 10.1073/pnas.2104642118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KW (1969). The sea otter in the eastern Pacific Ocean. North American Fauna, 1–352. [Google Scholar]
- Kim BY, Huber CD, & Lohmueller KE (2017). Inference of the Distribution of Selection Coefficients for New Nonsynonymous Mutations Using Large Samples. Genetics, 206, 345–361. 10.1534/genetics.116.197145/-/DC1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Maruyama T, & Crow JF (1963). The mutation load in small populations. Genetics, 48(10), 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, & Nielsen R (2014). ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics, 15(1), 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriazis CC, Wayne RK, & Lohmueller KE (2021). Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evolution Letters, 5(1), 33–47. 10.1002/evl3.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson S, Gagne RB, Bodkin J, Murray MJ, Ralls K, Bowen L, Leblois R, Piry S, Penedo MC, Tinker MT, & Ernest HB (2021). Translocations maintain genetic diversity and increase connectivity in sea otters, Enhydra lutris. Marine Mammal Science, 37(4), 1475–1497. https://doi.org/ 10.1111/mms.12841 [DOI] [Google Scholar]
- Larson S, Jameson R, Bodkin J, Staedler M, & Bentzen P (2002). Microsatellite DNA and mitochondrial DNA variation in remnant and translocated sea otter (Enhydra Lutris) populations. Journal of Mammalogy, 83(3), 893–906. [DOI] [Google Scholar]
- Larson S, Jameson R, Etnier M, Fleming M, & Bentzen P (2002). Loss of genetic diversity in sea otters (Enhydra lutris) associated with the fur trade of the 18th and 19th centuries. Molecular Ecology, 11(10), 1899–1903. [DOI] [PubMed] [Google Scholar]
- Larson S, Jameson R, Etnier M, Jones T, & Hall R (2012). Genetic giversity and population parameters of sea otters, Enhydra lutris, before fur trade extirpation from 1741–1911. PLOS ONE, 7(3), e32205. 10.1371/journal.pone.0032205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, Connell S, Stewardson K, Harney E, Fu Q, Gonzalez-Fortes G, Jones ER, Roodenberg SA, Lengyel G, Bocquentin F, … Reich D (2016). Genomic insights into the origin of farming in the ancient Near East. Nature, 536(7617), 419–424. 10.1038/nature19310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Lyman R (1988). Zoogeography of Oregon coast marine mammals: The last 3,000 years. Marine Mammal Science, 4(3), 247–264. 10.1111/j.1748-7692.1988.tb00205.x [DOI] [Google Scholar]
- Lesnek AJ, Briner JP, Lindqvist C, Baichtal JF, & Heaton TH (2018). Deglaciation of the Pacific coastal corridor directly preceded the human colonization of the Americas. Science Advances, 4(5), eaar5040. 10.1126/sciadv.aar5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE (2014). The distribution of deleterious genetic variation in human populations. Current Opinion in Genetics & Development, 29, 139–146. 10.1016/j.gde.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J, & Burger R (1995). Mutation accumulation and the extinction of small populations. The American Naturalist, 146(4), 489–518. [Google Scholar]
- Mann DH, & Hamilton TD (1995). Late Pleistocene and Holocene paleoenvironments of the North Pacific coast. Quaternary Science Reviews, 14(5), 449–471. 10.1016/0277-3791(95)00016-I [DOI] [Google Scholar]
- Mann DH, & Peteet DM (1994). Extent and timing of the last glacial maximum in southwestern Alaska. Quaternary Research, 42(2), 136–148. 10.1006/qres.1994.1063 [DOI] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, & Cunningham F (2016). The ensembl variant effect predictor. Genome Biology, 17(1), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, & Albrechtsen A (2018). Inferring population structure and admixture proportions in low-depth NGS data. Genetics, 210(2), 719–731. 10.1534/genetics.118.301336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C, Herman J, Gaggiotti OE, Dobney KM, Parsons K, & Hoelzel AR (2007). Genetic isolation of a now extinct population of bottlenose dolphins (Tursiops truncatus). Proceedings of the Royal Society B: Biological Sciences, 274(1618), 1611–1616. 10.1098/rspb.2007.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norma Oficial Mexicana. (2010). 059-SEMARNAT-2010. Protección Ambiental-Especies Nativas de México de Flora y Fauna Silvestres-Categorías de Riesgo y Especificaciones Para Su Inclusión, Exclusión o Cambio-Lista de Especies En Riesgo, 30. [Google Scholar]
- Nunney L (1993). The influence of mating system and overlapping generations on effective population size. Evolution, 47(5), 1329–1341. 10.1111/j.1558-5646.1993.tb02158.x [DOI] [PubMed] [Google Scholar]
- Nunney L (1996). The influence of variation in female fecundity on effective population size. Biological Journal of the Linnean Society, 59(4), 411–425. 10.1111/j.1095-8312.1996.tb01474.x [DOI] [Google Scholar]
- Ohta T (1973). Slightly deleterious mutant substitutions in evolution. Nature, 246(5428), 96–98. 10.1038/246096a0 [DOI] [PubMed] [Google Scholar]
- Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, Altena E, Lipson M, Lazaridis I, Harper TK, Patterson N, Broomandkhoshbacht N, Diekmann Y, Faltyskova Z, Fernandes D, … Reich D (2018). The Beaker phenomenon and the genomic transformation of northwest Europe. Nature, 555(7695), 190–196. 10.1038/nature25738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, & Schliep K (2019). ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics, 35(3), 526–528. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- Pickrell J, & Pritchard J (2012). Inference of population splits and mixtures from genome-wide allele frequency data. Nature Precedings, 1–1. 10.1038/npre.2012.6956.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot M, Branicki W, Jędrzejewski W, Goszczyński J, Jędrzejewska B, Dykyy I, Shkvyrya M, & Tsingarska E (2010). Phylogeographic history of grey wolves in Europe. BMC Evolutionary Biology, 10(1), 104. 10.1186/1471-2148-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, & Sham PC (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, & Pritchard JK (2014). fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics, 197(2), 573–589. 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralls K, Ballou J, & Brownell RL (1983). Genetic diversity in California sea otters: Theoretical considerations and management implications. Biological Conservation, 25(3), 209–232. 10.1016/0006-3207(83)90037-X [DOI] [Google Scholar]
- Rathbun GB, Hatfield BB, & Murphey TG (2000). Status of translocated sea otters at San Nicolas Island, California. The Southwestern Naturalist, 45(3), 322–328. JSTOR. 10.2307/3672835 [DOI] [Google Scholar]
- Riedman M, & Estes JA (1990). The sea otter (Enhydra lutris): Behavior, ecology, and natural history. Biological Report (USA). No. 90 (14). [Google Scholar]
- Robinson JA, Brown C, Kim BY, Lohmueller KE, & Wayne RK (2018). Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Current Biology, 28(21), 3487–3494.e4. 10.1016/j.cub.2018.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Vecchyo DO-D, Fan Z, Kim BY, vonHoldt BM, Marsden CD, Lohmueller KE, & Wayne RK (2016). Genomic flatlining in the endangered island fox. Current Biology, 26(9), 1183–1189. 10.1016/j.cub.2016.02.062 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Jaramillo MDC, & Gendron D (1996). Report of a sea otter, Enhydra Lutris, off the coast of Isla Magdalena, Baja California Sur, Mexico. Marine Mammal Science, 12(1), 153–156. 10.1111/j.1748-7692.1996.tb00314.x [DOI] [Google Scholar]
- Schiffels S, & Durbin R (2014). Inferring human population size and separation history from multiple genome sequences. Nature Genetics, 46(8), 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider KB, & Faro JB (1975). Effects of sea ice on sea otters (Enhydra lutris). Journal of Mammalogy, 56(1), 91–101. 10.2307/1379609 [DOI] [Google Scholar]
- Schramm Y, Heckel G, Sáenz-Arroyo A, López-Reyes E, Baez-Flores A, Gómez-Hernández G, Lazo-de-la-Vega-Trinker A, Lubinsky-Jinich D, & Milanés-Salinas M. de los Á. (2014). New evidence for the existence of southern sea otters (Enhydra lutris nereis) in Baja California, Mexico. Marine Mammal Science, 30(3), 1264–1271. 10.1111/mms.12104 [DOI] [Google Scholar]
- Schubert M, Ermini L, Sarkissian CD, Jónsson H, Ginolhac A, Schaefer R, Martin MD, Fernández R, Kircher M, McCue M, Willerslev E, & Orlando L (2014). Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nature Protocols, 9(5), 1056–1082. 10.1038/nprot.2014.063 [DOI] [PubMed] [Google Scholar]
- Simenstad CA, Estes JA, & Kenyon KW (1978). Aleuts, sea otters, and alternate stable-state communities. Science, 200(4340), 403–411. 10.1126/science.200.4340.403 [DOI] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, & Sella G (2014). The deleterious mutation load is insensitive to recent population history. Nature Genetics, 46(3), 220–224. 10.1038/ng.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3), 585–595. https://www.genetics.org/content/123/3/585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira JC, & Huber CD (2021). The inflated significance of neutral genetic diversity in conservation genetics. Proceedings of the National Academy of Sciences, 118(10). 10.1073/pnas.2015096118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker MT, Doak DF, & Estes JA (2008). Using demography and movement behavior to predict range expansion of the southern sea otter. Ecological Applications, 18(7), 1781–1794. 10.1890/07-0735.1 [DOI] [PubMed] [Google Scholar]
- Tinker MT, Gill VA, Esslinger GG, Bodkin J, Monk M, Mangel M, Monson DH, Raymond WW, & Kissling ML (2019). Trends and carrying capacity of sea otters in southeast Alaska. The Journal of Wildlife Management, 83(5), 1073–1089. 10.1002/jwmg.21685 [DOI] [Google Scholar]
- Tinker MT, Hatfield BB, Harris MD, & Ames JA (2016). Dramatic increase in sea otter mortality from white sharks in California. Marine Mammal Science, 32(1), 309–326. 10.1111/mms.12261 [DOI] [Google Scholar]
- Tinker MT, Yee JL, Laidre KL, Hatfield BB, Harris MD, Tomoleoni JA, Bell TW, Saarman E, Carswell LP, & Miles AK (2021). Habitat features predict carrying capacity of a recovering marine carnivore. The Journal of Wildlife Management, 85(2), 303–323. 10.1002/jwmg.21985 [DOI] [Google Scholar]
- U.S. Fish and Wildlife Service. (2003). Final revised recovery plan for the southern sea otter (Enhydra lutris nereis). US Fish and Wildlife Service, Portland, Oregon USA. https://www.cabdirect.org/cabdirect/abstract/20137204961 [Google Scholar]
- Valentine K, Duffield DA, Patrick LE, Hatch DR, Butler VL, Hall RL, & Lehman N (2008). Ancient DNA reveals genotypic relationships among Oregon populations of the sea otter (Enhydra lutris). Conservation Genetics, 9(4), 933–938. 10.1007/s10592-007-9422-z [DOI] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, & DePristo MA (2013). From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics, 43, 11.10.1–33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J (2009). Coalescent theory: An introduction. Roberts & Co. [Google Scholar]
- Waller DM (2015). Genetic rescue: A safe or risky bet? Molecular Ecology, 24(11), 2595–2597. 10.1111/mec.13220 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975). On the number of segregating sites in genetical models without recombination. Theoretical Population Biology, 7(2), 256–276. 10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Weir BS, & Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38(6), 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Wellman HP, Austin RM, Dagtas ND, Moss ML, Rick TC, & Hofman CA (2020). Archaeological mitogenomes illuminate the historical ecology of sea otters (Enhydra lutris) and the viability of reintroduction. Proceedings of the Royal Society B: Biological Sciences, 287(1940), 20202343. 10.1098/rspb.2020.2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AR, Fitzpatrick SW, Funk WC, & Tallmon DA (2015). Genetic rescue to the rescue. Trends in Ecology & Evolution, 30(1), 42–49. 10.1016/j.tree.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Wiedenfeld DA, Alberts AC, Angulo A, Bennett EL, Byers O, Contreras-MacBeath T, Drummond G, da Fonseca GAB, Gascon C, Harrison I, Heard N, Hochkirch A, Konstant W, Langhammer PF, Langrand O, Launay F, Lebbin DJ, Lieberman S, Long B, … Zhang L (2021). Conservation resource allocation, small population resiliency, and the fallacy of conservation triage. Conservation Biology, 35(5), 1388–1395. 10.1111/cobi.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmers CC, Estes JA, Edwards M, Laidre KL, & Konar B (2012). Do trophic cascades affect the storage and flux of atmospheric carbon? An analysis of sea otters and kelp forests. Frontiers in Ecology and the Environment, 10(8), 409–415. 10.1890/110176 [DOI] [Google Scholar]
- Wilson DE, Bogan MA, Brownell RL, Burdin AM, & Maminov MK (1991). Geographic variation in sea Otters, Enhydra lutris. Journal of Mammalogy, 72(1), 22–36. 10.2307/1381977 [DOI] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, & Weir BS (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics, 28(24), 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Modern Sample Information, Related to Figure 1.
Table S3. Depth, Coverage and Neutral Heterozygosity per Individual, Mapped to Domestic Ferret and Southern Sea Otter, Related to Figure 1. Note that neutral regions are defined slightly differently in sea otter and ferret reference genomes, with 9.3Mb passing neutral region filters in sea otter, and 6.8Mb in ferret due to an intersection with repeat regions in the ferret annotation. See Supplemental Methods for details.)
Table S4. Pairwise Kinship Coefficients (calculated within each population), Related to Figure 1. Only one individual was retained from pairs of individuals with kinship > 0.2.