Abstract
The insect cuticle is a key component of their success, being important for protection, communication, locomotion, and support. Conversely, as an exoskeleton, it also limits the size of the insect and must be periodically molted and a new one synthesized, to permit growth. To achieve this, the insect secretes a solution of chitinases, proteases and other proteins, known collectively as molting fluid, during each molting process to break down and recycle components of the old cuticle. Previous research has focused on the degradative enzymes in molting fluid and offered some characterization of their biochemical properties. However, identification of the specific proteins involved remained to be determined. We have used 2D SDS-PAGE and LC/MS-based proteomic analysis to identify proteins in the molting fluid of the tobacco hornworm, Manduca sexta, undergoing the larval to pupal molt. We categorized these proteins based on their proposed functions including chitin metabolism, proteases, peptidases, and immunity. This analysis complements previous reported work on M. sexta molting fluid and identifies candidate genes for enzymes involved in cuticle remodeling. Proteins classified as having an immune function highlight potential for molting fluid to act as an immune barrier to prevent infections during the cuticle degradation and ecdysis processes. Several proteins known to function in melanin synthesis as an immune response in hemolymph were present in molting fluid. We demonstrated that the bacterium Micrococcus luteus and the entomopathogenic fungus Beauveria bassiana can stimulate activation of phenoloxidase in molting fluid, indicating that the recognition proteins, protease cascade, and prophenoloxidase needed for melanin synthesis are present as a defense against infection during cuticle degradation. This analysis offers insights for proteins that may be important not only for molting in M. sexta but for insects in general.
Keywords: cuticle, chitinase, immunity, melanization, molting, peptidase, phenoloxidase, protease, proteomics
Graphical Abstract
1. Introduction
The insect cuticle is an exoskeleton that protects the insect from environmental dangers such as UV radiation, desiccation, physical damage, and assault from pathogens. Internally, it serves as a site for muscle and organ attachment and is important for movements such as walking, jumping and flying. It generally is described of consisting of three layers (Locke, 2001; Moussian, 2010). The outermost layer, the envelope, is composed primarily of proteins, quinones and lipids, and serves mainly as a water barrier, protecting the insect from both desiccation and swelling. The middle layer, the epicuticle, is composed of proteins and lipids that are stabilized by quinones in a process known as sclerotization (Andersen, 2010). The inner layer, the procuticle, comprises the bulk of the cuticle and contains a network of chitin fibers embedded in a matrix of proteins. The procuticle is laid down in layers known as lamellae. The procuticle is further subdivided into two layers referred to as the exo- and endocuticle, with the exocuticle synthesized premolt and the endocuticle post molt; the exocuticle may further undergo various degrees of sclerotizaion, while the endocuticle does not.
As an external skeleton, the cuticle limits the size of an insect and, therefore, in order to grow, the cuticle must be periodically degraded and discarded and a new one synthesized. The degradation of the "old" cuticle is accomplished by the secretion of a solution known as molting fluid. Manuca sexta is ideal for studying molting fluid, as its large size allows for sufficient quantity to be collected for analysis. Early work by Bade and Shoukimas (1974), and Brookhart and Kramer (1990), identified metalloprotease and trypsin-like protease activity in the molting fluid during the larval to pupal molt. Subsequently, Samuels and coworkers (1993a,b) characterized two proteases, which they named molting fluid protease 1 and 2 (MFP-1 and MFP-2), from the molting fluid of pharate adults. In agreement with the previous reports, MFP-1 was classified as trypsin-like based on activity assays with various substrates and inhibitors, while similar analyses with MFP-2 led to its classification as a neutral metalloprotease. MFP-2 by itself showed limited cuticle degrading activity but was greatly enhanced by the addition of MFP-1. Moreover, the peptide fragments produced by the combined MFP-1/MFP-2 digestion were smaller than those produced by MFP-1 alone, leading the authors to hypothesize that the initial cuticle digestion was carried out by MFP-1 and that the generated peptides were then further cleaved by MFP-2. In an analogous manner, a binary enzyme system for the degradation of chitin was proposed: endochitinases first cleave the chitin chains into oligosaccharides, which are in turn hydrolyzed to N-acetylglucosamine monomers by exochitinases (Fukamizo and Kramer, 1985). Three enzymes with endochitinase activity and three enzymes with exochitinase activity having been reported for M. sexta (Dziadik-Turner et al., 1981; Koga et al., 1983a,b).
In spite of the advances made in elucidating the enzymatic properties of M. sexta molting fluid, very few of the specific genes encoding these proteins have been identified, with only two corresponding cDNAs cloned; one each for an endochitinase and an exochitinase (Kramer et al., 1993; Zen et al., 1996). However, with the sequencing and annotation of the M. sexta genome (Kanost et al., 2016), it is now possible to identify many of the proteins present in the molting fluid. Towards this goal, we have used 2D gel analysis and liquid chromatography coupled with tandem mass spectrometry to analyze the proteins present in the molting fluid during the larval to pupal molt. The highly conserved nature of several of the identified proteins suggests they may be important for molting not only in M. sexta but insects in general. In addition, we identified proteins that function in immune responses, suggesting that molting fluid may act as a barrier to microbial infection.
2. Materials and Methods
2.1. Insect rearing and molting fluid collection
Insects were reared on an artificial diet at 26°C with a 16:8 light:dark cycle (Bell and Joachim, 1976). Molting fluid was collected from 5th instar larvae undergoing the larval to pupal molt when pigmentation patterns on the dorsal surface of the third thoracic segment (metathoracic bars) became visible (Jungreis 1978; Reinecke et al., 1980). Individuals, chilled on ice, were pricked with a 25 gauge needle at the base of the horn, or the first thoracic segment, and the fluid droplets that emerged were collected with a glass capillary tube (30 - 40 μl per insect was reliably obtained by this method). Phenylthiourea, prepared in ethanol, was added to a final concentration of 2 mM to inhibit melanization by phenoloxidase, and the samples were stored at −20°C.
2.2. Phenoloxidase activity assay
Molting fluid was collected from individual 5th instar larvae as described above without the addition of phenylthiourea. Phenoloxidase activity assays were performed in a manner similar to that described previously with M. sexta hemolymph (Jiang et al., 2003). Three microliters of molting fluid were mixed with different microorganisms in 20 mM Tris, 5 mM CaCl2, pH 7.5, in a total volume of 20 μl. The microorganisms tested were the bacteria Escherichia coli and Micrococcus luteus (formalin-treated; 2 x 105 cells) and the fungus Beauveria bassiana (6 x 103 conidia). Following an incubation period of 20 minutes at room temperature, 100 μl of 2 mM dopamine in 50 mM sodium phosphate, pH 6.5, was added and the samples incubated for an additional 30 minutes. The oxidation of dopamine by phenoloxidase was monitored by the formation of dopaminochrome at 470 nm. All reactions were set up in individual wells of a 96-well plate, and the spectrophotometer measurements were taken with an Epoch 2 microplate reader (BioTek). Statistical analysis was performed using a paired t-test (GraphPad Prism software, v5.04).
2.3. 2D gel electrophoresis
Molting fluid from three larvae was pooled and diluted with an equal volume of 2X SDS sample buffer (125 mM Tris (pH 7), 20% glycerol, 10% SDS, 10% β-mercaptoethanol). This was sent to Kendrick Laboratories (Madison, WI) for 2D gel electrophoresis. The isoelectric focusing step was carried out in an acrylamide tube gel containing a 2% mix of ampholytes, pH 4-8, (Serva, Heidelberg, Germany) at 700 V for 13.75 h (9600 Vhr). During this step, the SDS is stripped from the proteins (minimizing its effect on protein charge) and forms micelles with the non-ionic detergent NP-40. The micelles migrate to the acidic end of the tube gel where they form a bulb that can be cut off and discarded (Burgess-Cassler et al., 1989). The tube gel was then equilibrated for 10 min in a buffer of 10% glycerol, 50 mM dithiothreitol, 2.3% SDS, 62.5 mM Tris (pH 6.8), and sealed to the top of a stacking gel overlaid on a 0.75 mm thick 10% acrylamide slab gel. Electrophoresis was performed for 4 h at 15 mA. Molecular weight standards used were myosin (220,000), phosphorylase A (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000) and lysozyme (14,000) (Sigma-Aldrich). Following electrophoresis, the gel was stained with Coomassie brilliant blue R250 and sent as a wet gel to the Biotech Core Facility at Kansas State University for proteomic analysis.
2.4. Mass spectrometry analysis and database searching
Protein spot picking, digestion, and tandem mass spectrometry were performed as described previously (Christen et al., 2012). Briefly, excised gel pieces were washed with acetonitrile (ACN), dried under vacuum, then incubated overnight at 30°C with sequencing grade trypsin in 20 mM ammonium bicarbonate (pH 8). Tryptic peptides were extracted in a solution of 50% ACN and 2% trifluoroacetic acid, dried under vacuum, then resuspended in 2% ACN and 0.1 % formic acid (FA). Peptide solutions were loaded onto a C18 reverse phase capillary column (Acclaim PepMap, Thermo Fisher) and eluted with a 6% to 49% linear gradient of ACN in 0.1% FA. Eluted peptides were injected into an HCT Ultra Ion Trap Mass Spectrometer (Bruker Daltronics) in MS/MS mode; peak lists were acquired using DataAnalysis3.4 and Biotools 3.0 softare (Bruker Daltronics). Peptide peak lists were analyzed using Mascot software v2.3.02 (Matrix Science) and compared to a combined M. sexta OGS2 protein and decoy databases (Kanost et al., 2016); the decoy database was generated by randomizing the amino acid sequence of each OGS2 protein. The combined size of the OGS2 and decoy databases was 54,806 sequences and 29,108,670 residues. Search criteria allowed for one missed cleavage, peptide and fragment mass tolerances of ± 1.2 and 0.6 Da, respectively, oxidation of methionine residues, and carbamidomethylation of cysteines. A peptide score ≥ 41 was considered significant (p < 0.05) and a protein was deemed identified with a minimum of two peptide matches. Under these conditions all matches obtained were to the OGS2 database and none to the decoy database; thus, the false discovery rate was considered to be very low. The OGS2 database is available at USDA Ag Data Commons (https://data.nal.usda.gov/dataset/manduca-sexta-official-gene-set-v20) and can be searched at the the i5k Workspace of the National Agricultural Library (https://i5k.nal.usda.gov). All OGS2 hits were next searched against a subset of 2498 manually annotated proteins (Kanost et al., 2016) or the non-redundant protein database at the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) to assign putative identification and function.
3. Results
3.1. Protein classification
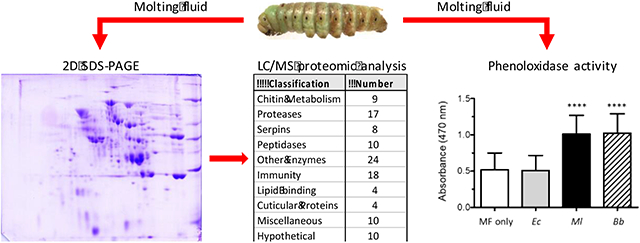
From our analysis of 196 protein spots, 174 yielded identifications representing 121 unique proteins (Figure S1 and Table S1). Forty percent of the proteins (48 of 121) occurred in only one spot, with a majority (74 of 121) in two or fewer (Fig. 1A). Thirty-five percent of the spots (61 of 174) contained only one protein identification, while a majority (105 of 174) contained two or fewer (Fig. 1B). Putative identity or function was inferred by comparison to a list of manually curated OGS2 genes and by searching against the non-redundant protein database at the National Center for Biotechnology Information. The proteins identified were divided into 10 categories (some proteins were placed in more than one category, e.g. "serine protease" and "immunity", when appropriate) (Table S2): chitin metabolism (9), serine proteases and serine protease homologs (17), serine protease inhibitors (serpins) (8), peptidases (10), non-proteolytic enzymes (24), immunity (18), lipid-binding (14), cuticular proteins (4), hypothetical (10), and miscellaneous (10).
Figure 1. Graphical representation of protein abundance and distribution in the 2D gel.
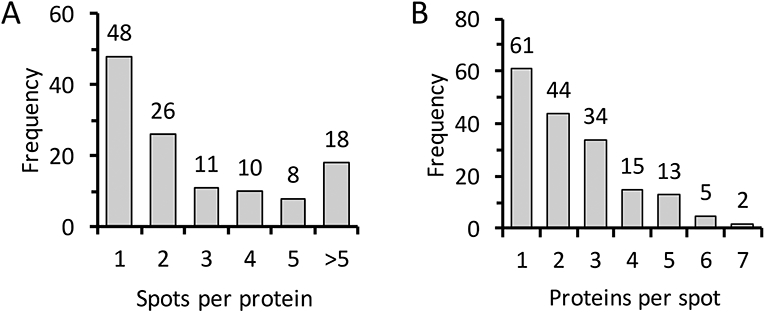
(A) Frequency of the number of proteins identified per spot analyzed. (B) Frequency of the number of spots where a specific protein was found.
3.2. Chitin metabolism
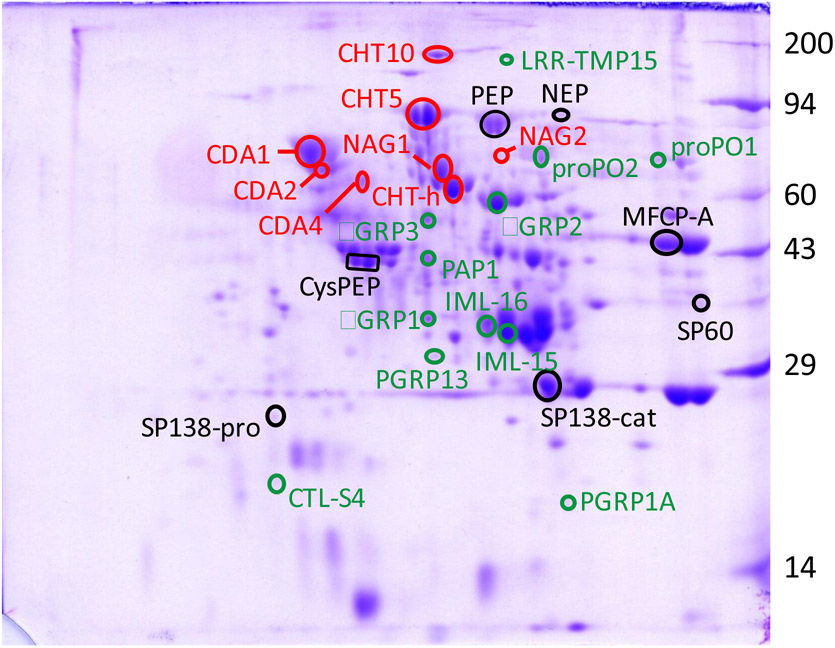
Nine proteins were identified with potential functions in chitin metabolism (Fig. 2, Tables 1 and S2). These were three chitinases (Cht-h, Cht 5 and Cht 10), two N-acetylglucosaminidases (NAG1 and NAG2), three chitin deacetylases (CDA1, CDA2, and CDA4), and a chitinase-like protein, hemocyte aggregation inhibitor protein (HAIP).
Figure 2. Two-dimensional gel electrophoresis of proteins in the molting fluid extracted from pharate pupae.
Proteins were separated by isoelectric focusing (pH 4 to 8), followed by denaturing polyacrylamide gel electrophoresis (10%). Select proteins identified by LC/MS proteomic analysis are circled and annotated in Table 1. Red type: chitin metabolism; black type: proteases and peptidases; green type: immunity. Molecular mass standards, in kDa, are noted on the right side of the gels.
Table 1:
Select list of identified proteins
| Classification | OGS2 ID | GenBank ID | Protein Name |
|---|---|---|---|
| Chitin metabolism | Msex2.02783 | ABB88891.1 | Chitinase-h (Cht-h) |
| Msex2.08301 | AAC04924.1 | Chitinase 5 (Cht 5) | |
| Msex2.04970 | XP_037302083.1 | Chitinase 10 (Cht 10) | |
| Msex2.08131 | AAQ97603.1 | N-acetylglucosaminidase 1 (NAG1) | |
| Msex2.02344 | XP_030019961.1 | N-acetylglucosaminidase 2 (NAG2) | |
| Msex2.03153 | XP_030021435.1 | Chitin deactylase 1 (CDA1) | |
| Msex2.03152 | XP_030021412.1 | Chitin deactylase 2 (CDA2) | |
| Msex2.02671 | XP_030020590.1 | Chitin deactylase 4 (CDA4) | |
| Msex2.03419 | ACW82749.1 | Hemocyte aggregation inhibitor protein (HAIP) | |
| Serine Proteases | Msex2.05571 | AAX18636.1 | Prophenoloxidase activating protease 1 (PAP1) |
| Msex2.11070 | AAB94558.1 | Hemocyte protease 2 (HP2) | |
| Msex2.13600 | AAV91003.1 | Hemolymph proteinase 5 (HP5) | |
| Msex2.10123 | AAZ91694.1 | Hemolymph proteinase 12 (HP12) | |
| Msex2.03587 | AAV91017.1 | Hemolymph proteinase 19 (HP19) | |
| Msex2.14451 | Hemolymph protease 26 (HP26) | ||
| Msex2.15056 | XP_037303322.1 | Hemolymph protease 28 (HP28) | |
| Msex2.14453 | XP_037300168.1 | Gut protease 33 (GP33) | |
| Msex2.12745 | XP_037303327.1 | Serine protease 30 (SP30) | |
| Msex2.14074 | XP_037299567.1 | Serine protease 31 (SP31) | |
| Msex2.09420 | XP_030031627.1 | Serine protease 60 (SP60) | |
| Msex2.14592 | XP_037299464.1 | Serine protease 112 (SP112) | |
| Msex2.03588 | Serine protease 138 (SP138) | ||
| Peptidases | Msex2.05425 | XP_037298002.1 | Serine carboxypeptidase |
| Msex2.03717 | XP_037298718.1 | Prolyl endopeptidase (PEP) | |
| Msex2.13134 | XP_030037348.1 | Cysteine peptidase | |
| Msex2.01424 | AAO21504.1 | Neutral endopeptidase (NEP) | |
| Msex2.01555 | Carboxypeptidase D | ||
| Msex2.03688 | XP_030022076.1 | Dipeptidase | |
| Msex2.14756 | XP_037296180 | Aminopeptidase N | |
| Msex2.07568 | XP_030028755.1 | Similar to angiotensin converting enzyme | |
| Msex2.07569 | XP_030028761.2 | Similar to angiotensin converting enzyme | |
| Msex2.02590 | Molting fluid carboxypeptidase A (MFCP-A) | ||
| Pattern Recognition Proteins | Msex2.13760 | Q9NJ98.1 | Beta-1,3-glucan recognition protein 1 (βGRP1) |
| Msex2.13759 | Q8ISB6.1 | Beta-1,3-glucan recognition protein 2 (βGRP2) | |
| Msex2.06180 | ADK39022.1 | Beta-1,3-glucan recognition protein 3 (βGRP3) | |
| Msex2.10092 | AAO21509.1 | Peptidoglycan recognition protein 1A (PGRP1A) | |
| Msex2.10093 | XP_030032673.1 | Peptidoglycan recognition protein 13 (PGRP13) | |
| Msex2.05487 | XP_030025145.1 | C-type lectin S4 (CTL-S4) | |
| Msex2.01104 | XP_030035841.1 | Immulectin 15 (IML-15) | |
| Msex2.01105 | XP_030035853.1 | Immulectin 16 (IML-16) | |
| Msex2.00830 | XP_037296933.1 | Leucine-rich repeat transmembrane protein 15 (LRR-TMP15) | |
| Melanization | Msex2.11070 | AAB94558.1 | Hemocyte protease 2 (HP2) |
| Msex2.13600 | AAV91003.1 | Hemolymph proteinase 5 (HP5) | |
| Msex2.11800 | AAM69353.1 | Serine protease homolog 2 (SPH2) | |
| Msex2.05571 | AAX18636.1 | Prophenoloxidase activating protease 1 (PAP1) | |
| Msex2.09885 | O44249.3 | Prophenoloxidase subunit 1 (proPO1) | |
| Msex2.11367 | Q25519.3 | Prophenoloxidase subunit 2 (proPO2) |
3.3. Proteases and peptidases
Thirteen proteases and ten peptidases were identified (Fig.2, Tables 1 and S2). All of the proteases were classified as serine type. Seven of these have previously been identified in the hemolymph (PAP1, HP2, HP5, HP12, HP19, HP26 and HP28) (Jiang et al., 1999, 2005; He et al., 2018; Wang et al., 2020); all are predicted to have trypsin-like specificity except HP19, which is predicted to be elastase-like (Cao et al., 2015). Of the remaining six, two each are predicted to have trypsin-like specificity (SP30 and SP60), chymotrypsin-like specificity (SP31 and SP138), and elastase-like specificity (GP33 and SP112) (Cao et al., 2015).
Seven of the 10 peptidases were metallo type, two were serine type, and one cysteine type. Three each were predicted to be carboxypeptidases (serine carboxypeptidase, carboxypeptidase D, molting fluid carboxypeptidase A) or dipeptidases (dipeptidase and two similar to angiotensin converting enzyme), two were predicted to be endopeptidases (prolyl endopeptidase and neutral endopeptidase), and one an aminopeptidase (aminopeptidase N).
3.4. Cuticular proteins
Only 4 cuticular proteins were identified (Table S2); two cuticular proteins analogous to peritrophins, type 3 (CPAP3-A1 and CPAP3-C), knickkopf 2, and a glycine-rich cuticular protein.
3.5. Immunity
Proteins with a putative immune function were grouped into two main categories, pattern recognition proteins (PRPs) and melanization proteins (Fig. 2, Tables 1 and S2). The PRPs included three beta-1,3-glucan recognition proteins (βGRP1, βGRP 2 and βGRP 3), three C-type lectins (C-type lectin CTL-S4 and immulectins IML-15 and IML-16), two peptidoglycan recognition proteins (PGRP1A and PGRP13), a leucine-rich repeat transmembrane protein (LRR-TMP15), hemolin, and histone H4. The proteins involved in melanization were prophenoloxidase subunits 1 and 2 (proPO1 and proPO2), prophenoloxidase activating protease 1 (PAP1), HP2, HP5, and serine protease homolog 2 (SPH2). In addition, transferrin 1 was also included with the immune related proteins as it has bacteriostatic activity and may play a role in immunity by sequestering iron from invading bacteria (Brummett et al., 2017).
3.6. PO activity
We observed that molting fluid gradually darkened in color after collection, suggesting that phenoloxidase in the molting fluid could become active. The appearance of melanin also indicates that a PO substrate such as tyrosine, DOPA, or dopamine is present in molting fluid. PO activity assays were performed to test the competence of molting fluid to respond to microbial infection. Even without the addition of exogenous dopamine, melanization was observed in samples in which the Gram-positive bacterium, M. luteus, or the fungus B. bassiana were added to molting fluid, indicating the activation of proPO and the presence of a PO substrate (data not shown). After the addition of dopamine, samples with M. luteus or B. bassiana had twice the activity compared to samples with no added microorgansim or those incubated with E. coli (Fig. 3); control assays in which the molting fluid was omitted showed no melanization, indicating that the oxidation of dopamine was not due to the microorgansisms (data not shown). These results indicate that the proteins needed to activate proPO are present in molting fluid and respond to microbial elicitors.
Figure 3. Phenoloxidase activity in molting fluid.
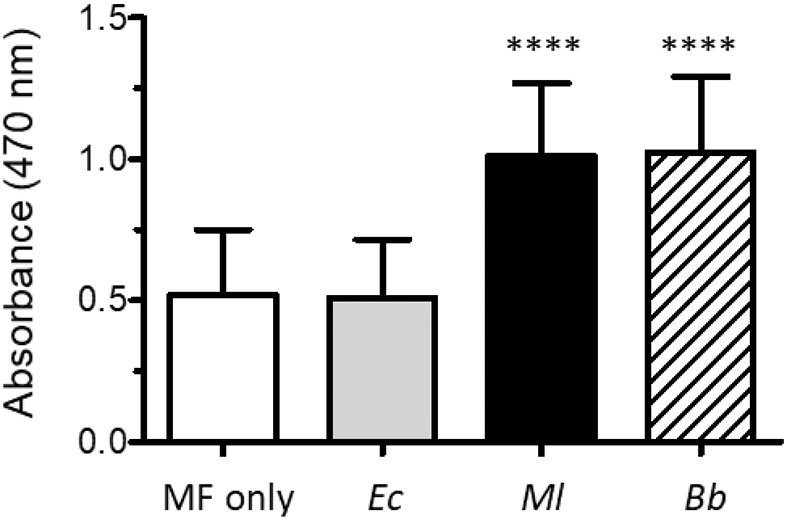
PO activity assays were performed as described in the Materials and Methods. The microorganism added to molting fluid (MF) are indicated below each bar: Ec, E. coli; Ml, M. luteus; Bb, B. bassiana. The values shown are the mean ± SD (n = 15) of the recorded absorbance at 470 nm. Asterisks (****) indicate statistical difference (p < 0.0001).
4. Discussion
4.1. Protein identification and classification
The proteins identified in this analysis were presumed to have been derived from two sources: either as a component of the molting fluid with a role in cuticle degradation and reabsorption, or as a constituent of the cuticle subsequently released upon its breakdown. A third possibility was hemolymph contamination during collection of the molting fluid. However, absent from our analysis were the lipid transport proteins apolipophorin I and II, and the hexamerins arylphorin-α, methionine-rich storage protein (MSRP), and moderately methionine-rich storage protein (MMSRP), which are highly abundant in the hemolymph at this time (Fig. S2A) (Burmester, 2015). Arylphorin-β, which is also highly abundant in the hemolymph, was only detected in two faint spots in the 2D gel (Fig S2B). From this we concluded that hemolymph contamination was minimal or absent. Additionally, nearly all of the detected proteins were predicted to be secreted, with only a few lacking secretion signals (actin, HSPs), indicating lack of contamination due to cell lysis.
One advantage of combining 2D gel electrophoresis with LC/MS-based proteomic analysis was that it allows for identification of proteins which are visibly highly abundant, possibly inferring their importance either to the cuticle proper or its degradation. Conversely, a disadvantage of this method was that proteins of lower abundance, low molecular weight, or small peptides from digested proteins may be missed. In our analysis, it is likely that proteins/peptides less than 20 ng in abundance, or smaller than 14 kDa, were not detected. This may explain why only four cuticular structural proteins were identified as it is likely that following their breakdown, the resulting peptides were of too low abundance to be detected by Coomassie stain, or that their small size resulted in their loss from the gel during electrophoresis. Additionally, we cannot exclude the possibility that the molting fluid was collected late (or at least later) during the molting period. Thus, many of the structural proteins may have already been digested and reabsorbed at the time the molting fluid was collected. Future proteomic analysis may benefit by analyzing molting fluid directly instead of first fractionating it by 2D gel electrophoresis, and by collecting molting fluid at earlier time points; such analysis is likely to identify more cuticular structural proteins, as well as reveal enzymes present earlier in the molting process that our analysis missed.
Given that structural proteins and chitin fibers are the main components of the cuticle, enzymes involved in their breakdown were of keen interest. Proteins that were grouped into chitin metabolism, proteases, and peptidases, represented 29% of those identified. Other enzymes such as glycosidases, oxidoreductases, esterases, and phosphatases represented another 21%. Proteins from two families characterized by binding and transport of small hydrophoboic molecules were also abundant. The Takeout/TULIP family, which includes lepidopteran juvenile hormone binding proteins (Alva and Lupas, 2016), and members of the odorant binding protein family (Vogt et al., 2015; Sun et al., 2018), each represented approximately 12% of the proteins identified. The role of such proteins that may bind hydrophobic ligands in molting fluid is unknown but intriguing. The exchangeable apolipoprotein apoLp-III was also present and may have some function in binding to hydrophobic surfaces. However, for many of the proteins identified, their potential functions are still unknown and in need of further investigation. Of the 121 proteins identified, 90 putative orthologs were found in the silk moth, Bombyx mori, based on reciprocal BLAST analysis (Table S2). There have been three published reports on the proteomic analysis of B. mori molting fluid (Qu et al., 2014; Zhang et al., 2014; Liu et al., 2018). The number of matches between our work and those varied from 42 to 70, with the work of Liu et al. (2018) having the highest number of matches (Table S2). In total, 75 proteins (representing 83% of the identifiable B. mori orthologs) were in common between M. sexta and B. mori, indicating high similarity of molting fluid components between the two species.
4.2. Chitin metabolism
We identified three chitinases (Cht-h, Cht 5 and Cht 10) in our analysis. This is in agreement with a previous report on the purification of three endochitinases from the integument and molting fluid of M. sexta (designated as chitinase I-III; Koga et al., 1983a). It is reasonable to propose that the chitinases we identified correspond to those previously reported, but which lacked sequence information. The observed molecular masses of Cht-h, Cht 5 and Cht 10 in our 2D gel (57, 81 and 178 kDa) differed from the reported masses of chitinase I-III (75, 62 and 50 kDa), likely due to protease degradation during the purification process of the latter. Thus, identification based solely on mass was not possible. Nevertheless, Cht 5 was identical to the deduced amino acid sequence of a cDNA previously identified from an expression library using antisera against chitinase III (Kramer et al, 1993). However, as this antiserum was also immunoreactive against chitinase II, either chitinase III or chitinase II might correspond with chitinase 5.
Both Cht 5 and Cht 10 have orthologs in several other insect species, suggesting that they may have a conserved function (Tetreau et al., 2015a). RNAi experiments with the red flour beetle, Tribolium castaneum, have shown Cht 5 to be necessary for the pupal to adult molt, whereas Cht 10 was critical at all molts (Zhu et al., 2008). Similarly, in the brown planthopper, Nilaparvata lugens, knockdown of Cht 5 and Cht 10 were lethal in 60% and 90% of treated insects (Xi et al., 2015). In Drosophila melanogaster, Cht 5 knockdown resulted in larvae with "double cuticles", an indication of failure to shed the old cuticle during the molt. In pupae, RNAi for Cht 5 was either lethal or resulted in adults with defects in wing extension (Pesch et al., 2016). Knockdown of Cht 10 in the wing buds during metamorphosis resulted in an increase in chitin content and a disruption of procuticle organization (loss of lamellae) of the wing cuticle; such individuals had smaller, crinkled wings and were unable to fly. Additionally, the wings were more permeable to dye penetration test, indicating a breakdown of the "barrier" function of the cuticle (Dong et al., 2020).
To date, Cht-h has only been found in lepidopterans. Its amino acid sequence is similar to that of bacterial and baculoviral chitinases and appears to be the result of horizontal transfer (Daimon et al., 2003). No knockdown of this gene has been reported, and therefore, it's role or importance is unknown. A chitinase-like protein, hemocyte aggregation inhibitor protein (HAIP) was also present in molting fluid. HAIP had previously been identified from M. sexta larval hemolymph (Kanost et al., 1994). It belongs to the imaginal disc growth factor (idgf) group of chitinase-like proteins, being most similar to idgf4. Knockdown of this gene in D. melanogaster larvae produced the double cuticle phenotype, but had no effect on the pupal molt (Pesch et al., 2016).
In addition to the three endochitinases, we identified two N-acetylglucosaminidases (NAG1 and NAG2) that are likely involved in breaking down chitin oligomer fragments into glucosamine monomers. The sequence of NAG1 was identical to the deduced amino acid sequence of a previously cloned cDNA (Zen, at al., 1996). This cDNA was identified by screening an expression library with antibodies raised against the exochitinase E I. Although three enzymes with exochitinase activity (designated E I-III) have been reported for M. sexta, only E I was found in the molting fluid (Dziadik-Turner et al., 1981; Koga et al., 1983b). Thus, we infer that NAG1 is the previously identified E I exochitinase. In support of this, the apparent mass and pI of NAG1 in our 2D gel are approximately 61.7 kDa and 5.8, respectively, similar to the estimated values of 60 kDa and 5.9 reported for E I (Dziadik-Turner et al., 1981). NAG2 was identified in only one faint spot (Fig. 2), and its low abundance in the molting fluid (relative to NAG1) may be the reason why it was not detected in previous activity assays. Orthologs of M. sexta NAG1 and NAG2 have also been detected in the molting fluid of a lepidopteran pest, the Asian corn borer (Ostrinia furnacalis; OfHex1 and OfHex3) (Qu et al., 2013).
Chitin deacetylase activity has not previously been reported from M. sexta, although a family of CDA-like genes has been described following the sequencing of the genome (Tetreau et al., 2015b). The identified CDA1 and CDA2 are orthologs of the D. melanogastger proteins serpentine (serp) and vermiform (verm), which are important for embryonic cuticle formation (Luschnig et al, 2006; Wang et al, 2006). Additionally, knockdown of serp and verm expression during wing development resulted in a reduction of chitosan content (serp), disruption of the laminar organization of the procuticle (verm), and lost/malformed surface nanostructures (serp and verm), leading to increased cuticle permeability (Zhang et al., 2019). Similarly, knockdown of CDA1 and CDA2 affected molting of T. castaneum, N. lugens, and L. migratoria, causing lethality (Arakane et al., 2009; Xi et al., 2014; Yu et al., 2016, 2019). Interestingly, only CDA1 appears to have deacetylase activity (Yu et al., 2019; Zhang et al., 2019). Although CDA2 appears to lack deactylase activity, it has been shown to be important for proper laminar organization in the procuticle (Yu et al., 2016; Noh, et a., 2018; Zhang et al., 2019). Recently, Liu and coworkers (2019) demonstrated that a recombinant form of B. mori CDA1 was not enzymatically active without the addition of accessory factors. The addition of either crude molting fluid or the cuticular protein CPAP3-A1 to BmCDA1 was able to restore deacetylase activity. CPAP3-A1 was also identified in our analysis, suggesting that it may function as a cofactor for M. sexta CDA1.
All nine of the chitin metabolism enzymes we identified were also found in B. mori molting fluid, although some of the naming differed (Qu et al., 2014; Zhang et al., 2014; Liu et al., 2018). Two additional NAG-like proteins were also identified in B. mori (BGIBMGA003990, BGIBMGA014116) that were not found in our analysis. Nevertheless, the proteins found between the two lepidopterans were nearly identical, suggesting that these may be all the enzymes necessary for breaking down chitin in the cuticle.
4.3. Proteases and peptidases
All but one of the proteases and peptidases we identified in molting fluid could be classified as either serine or metallo, consistent with previous published reports for M. sexta (Bade and Shoukimas, 1974; Brookhart and Kramer, 1990; Samuels et al., 1993a, b). All but one of the serine proteases had extended prodomains between 10 - 37 kDa that contain additional modules such as clip (pfam12032), gastrulation defective N -terminus (gd_N; pfam16030), or sushi (pfam00084) domains, that are predicted to remain attached to the protease domain through a disulfide linkage after proteolytic activation of the zymogen. An exception was SP31 (Msex2.14074, XP_037299567.1), which does contain an extended amino terminus of 88 residues upstream of the protease domain, however, this region does not contain any known motifs or cysteines for disulfide bond formation. These additional domains are thought to play a regulatory role, possibly controlling catalytic activity or protein localization. Serine proteases with extended N-terminal domains may be involved in the activation of other proteases rather than the direct digestion of proteins in the cuticle (Jiang and Kanost, 2015; Cao et al., 2015). Additionally, seven of these have been previously reported to be in the hemolymph (PAP1, HP2, HP5, HP12, HP19, HP26 and HP28) and, therefore, may play a role in regulating immunity (e.g. the phenoloxidase cascade) rather than cuticular protein degradation (Jiang et al., 1999, 2005), although these two roles may not be mutually exclusive.
Using gelatin containing SDS-PAGE under non-reducing conditions, Brookhart and Kramer (1990) identified two major zones of protease activity of approximately 50 kDa and 100 kDa. Any of the serine proteases we identified could be responsible for the activity at 50 kDa as they have predicted molecular masses in the range of 40 - 66 kDa. Similarly, Samuels and coworkers (1993a) characterized a trypsin-like serine protease, which they named molting fluid protease 1 (MFP-1), from pharate adults. However, MFP-1 was reported to have the same molecular weight (~41 kDa) under both reducing and non-reducing conditions, which indicates that it is not composed of two peptides held together by a disulfide linkage. Thus, this would seem to preclude the trypsin-like serine proteases we identified. Further investigation is required to determine if any of the serine proteases we identified are the same as those previously reported. Nevertheless, SP60 and SP138 deserve further mention.
SP60 is orthologous to Lm-TSP and BmSP95, proteases found in the molting fluid of the Oriental migratory locust, Locusta migratoria manilensis, and B. mori (Wei et al., 2007; Liu et al., 2017). SP60 shares 77% and 75% identity with Lm-TSP and BmSP95, with clear orthologs present in other insects as well (Liu et al., 2017). (The higher identity to Lm-TSP rather than BmSP95 is likely due to the L. migratoria sequence being only partial and restricted to the more conserved protease domain.) Importantly, both Lm-TSP and BmSP95 caused molting defects when transcript levels were knocked down by RNAi. In L. migratoria manilensis, when dsRNA was injected four days prior to molting, individuals failed to molt and died. Furthermore, a reduction in protease activity and cuticle degradation was observed in whole molting fluid from RNAi treated insects (Wei et al., 2007). In B. mori, two-thirds of the individuals injected with dsRNA for BmSP95 delayed molting from pupa to adult. We propose that SP60, and its orthologs in other species, plays a critical role in cuticle degradation during molting. SP138 was unique in that peptides that mapped to the pro and catalytic domains were found in separate locations in the 2D gel (Fig. 2), consistent with the theoretical mass and pI values for the respective domains. Thus, we postulate that SP138 may have been cleaved at its zymogen activation site and was active at the time the molting fluid was collected, though its role remains unknown.
Another protease of interest was a putative prolyl endopeptidase (PEP). PEPs cleave after proline residues and act on small peptides of no more than 30 amino acid residues. In humans, it has drawn considerable interest for its role in the processing or degradation of peptide hormones affecting memory, depression, and blood pressure (Polgár, 2002). In insects, PEPs have been described from the flesh fly, Sarcophaga peregrina, D. melanogaster, the shield bug, Eurygaster integriceps, and B. mori (Ohtsuki et al., 1997a, b; Amin et al., 1999; Darkoh et al., 2010; Fu et al., 2018). Various roles proposed for insect PEPs include regulation of imaginal disc and nervous system development, inhibition of DNA synthesis, and wheat gluten digestion. We hypothesize that PEP acts to degrade proline-rich cuticular proteins. The CPLCP family (cuticular protein of low complexity, proline rich) contains extensive repeats of Pro-Val or Pro-Tyr (Cornman and Willis, 2009; Willis, 2010). Therefore, we propose that PEP is involved in breaking down oligopeptides resulting from the digestion of CPLCPs by other proteases.
The carboxypeptidase activity reported by Brookhart and Kramer (1990) was resolved into three fractions by gel filtration chromatography between 10 −60 kDa. As the three carboxypeptidases we identified all had an apparent mass of ~44-50 kDa, it seems reasonable that they correspond to only one of these peaks rather than one for each distinct peak. Also described by Brookhart and Kramer (1990) was an aminopeptidase classified as a metalloprotease due to its sensitivity to metal chelators. We propose the aminopeptidase N-like protein, Msex2.14756, (XP_037296180) as a candidate for that enzyme. Knockdown of a putative ortholog in T. castaneum (TC004557, EEZ99336.1) resulted in the inability to shed the old cuticle during molting (Zhang et al., 2014). It is unclear if this peptidase is the same as molting fluid protease 2 (MFP-2) described by Samuels et al. (1993b). MFP-2 was reported to have a mass of approximately 39 kDa as determined by SDS-PAGE, whereas the aminopeptidase we identified had an apparent mass of 65 kDa in our 2D PAGE. Given that both MFP-1 and MFP-2 were purified from the molting fluid of M. sexta pharate adults, it is possible that they may be specific to the pupal to adult molt as the physical properties of the pupal cuticle is much different than that of the larva (hard and rigid versus soft and flexible).
4.4. Immunity
The cuticle serves as a protective physical barrier against pathogens from the environment. Molting represents a vulnerable time in the insect's life cycle as the "old" barrier is being broken down before the "new" barrier is fully constructed. We have found that molting fluid contains many proteins known to function in antimicrobial immunity, suggesting that molting fluid may function as an additional biochemical immune barrier to infection during molting. The first step in the immune response is recognition of pathogens by pattern recognition proteins. Most of the molting fluid proteins we categorized as pattern recognition proteins belonged to one of three groups: βGRPs, PGRPs, or C-type lectins; this diversity allows the insect to recognize a wide range of microorganisms.
βGRPs contain an amino-terminal carbohydrate binding domain and a carboxyl-terminal glucanase-like domain, which lacks key catalytic residues for glucan hydrolysis and is, therefore, enzymatically inactive (Zhang et al., 2015). The amino-terminal domain binds β-1,3-glucan, a component of fungal cell walls (Dai et al., 2013); βGRP3 is unique in that it lacks this N-terminal domain (Rao et al., 2014). All three of the βGRPs we identified have been shown to bind to bacteria and yeast, and to stimulate proPO activation (Ma and Kanost, 2000; Jiang et al., 2004; Rao et al., 2014, Dai et al. 2013). In addition, βGRP3 was also demonstrated to have bacteriostatic activity (Rao et al., 2014). Thus, the involvement of these proteins in the insect immune response is well established.
PGRPs bind to peptidoglycan, a component of bacterial cell walls. Thirteen PGRP genes have been identified for M. sexta, but only PGRP1 has been studied in detail (Sumathipala and Jiang, 2010; Zhang et al., 2015). PGRP1 can bind both the Lys-type and DAP-type of peptidoglycan and stimulate the activation of proPO. A similar protein was also isolated from the molting fluid of another lepidopteran, the Brazilian skipper, Calpodes ethlius (CECP22; Marcu and Locke, 1998). CECP22 was expressed only during the molting periods of larvae and deposited into the cuticle. PGRP13 is unique in that in addition to the peptidoglycan binding domain, it is predicted to also contain a lepidopteran 30 kDa lipoprotein domain (Lp-11; pfam03260) (Zhang et al., 2015). In B. mori, proteins with this domain have been implicated in anti-apoptosis, embryogenesis, and innate immunity (Sun et al., 2007). The protein 6G1 (also known as Bmlp1) was shown to have β-glucan binding activity, to stimulate activation of proPO, and inhibit the hyphal growth of the entomopathogenic fungus Paecilomyces tenuipes (Ujita et al., 2005). It should be noted that in our analysis, all of the peptides mapping to PGRP13 were in the Lp-11 domain and that the apparent molecular mass of the protein in the 2D gel was approximately 30 kDa (Table S1 and Fig. 1). Thus, the architecture of PGRP13 needs further investigation. Nevertheless, work with B. mori indicates that Lp-11 domains can act as pattern recognition proteins.
Immulectins (IML) are C-type lectins that contain two carbohydrate binding domains (Rao et al., 2015, Xia et al., 2018). They have been shown to bind to several types of microorganisms including Gram-positive and Gram-negative bacteria, yeast, and nematodes in a calcium-dependent manner, and to help activate proPO (Xia et al., 2018). Both IML-15 and IML-16 are part of a large expansion in M. sexta and may be unique to this species (Rao et al., 2015, Lu et al., 2020). Although their architecture is similar to that of the well-studied IML-1 through 4 of M. sexta (Yu et al., 1999, 2006; Yu and Kanost, 2004; Shi and Yu, 2012), their involvement in immunity remains to be demonstrated.
CTL-S4 differs from IMLs in that it has only one carbohydrate-binding domain. Its B. mori ortholog, BmCTL7, was also found in molting fluid (Table S2). Orthologs can be found in other insects, including D. melanogaster, T. castaneum, and the mosquitoes Anopheles gambiae and Aedes aegypti (Rao et al., 2015), indicating that it possibly has a conserved function. We have tentatively placed CTL-S4 in the "Immunity" category based on the known ability of lectins to act as microbial pattern recognition proteins. However, its function remains to be determined. In D. melanogaster, a C-type lectin encoded by the gene schlaff (CG3244), an ortholog to M. sexta CTL-S2, serves not as an immune protein but rather as a structural component of the cuticle, necessary for the integrity of soft cuticle (Zuber et al., 2019).
Following detection of microorganisms by the pattern recognition proteins in hemolymph, a serine protease cascade is initiated that leads to immune responses including melanization (via activation of proPO) and antimicrobial peptide synthesis (Jiang et al., 2010; Kanost and Jiang, 2015). The genes encoding HP2, HP5, HP12, and HP19 are up-regulated upon bacterial challenge in M. sexta larvae (Jiang et al., 1999, 2005). Recently, HP5 was reported to be an activator of both the melanization and antimicrobial peptide synthesis pathways (Wang et al., 2020). Additionally, both HP5 and HP19 have orthologs that were detected in the molting fluid of B. mori (Table S2), and we hypothesize they play a conserved role in that insect as well. The presence of PAP-1 and the two subunits of proPO in the molting fluid indicates that the enzymes at the end of the PO cascade are present and can lead to melanin formation if the cuticle is breached. These enzymes have also been previously purified from the cuticle of pharate pupae (Aso et al., 1984; Wang et al., 1998). Proteins from the serpin superfamily regulate the serine proteases that mediate immune responses in hemolymph (Meekins et al. 2017). We identified a number of serpins in molting fluid (Table S1), including serpins 1, 3, 4, 5, 6, and 12, which have been demonstrated to regulate immune responsive proteases in hemolymph (Jiang et al., 2003; Tong et al., 2005; An et al., 2011; Li et al., 2018; Wang et al., 2020), suggesting that these serpins may regulate proteases in the molting fluid; all but serpin 12 were identified in B. mori molting fluid as well.
4.5. Phenoloxidase activity
We further explored the ability of molting fluid to respond to infection by synthesizing melanin. Without the addition of any additional factors, including PO substrate (e.g. tyrosine, DOPA, or dopamine), molting fluid spontaneously melanized, indicating that all of the needed components of the pathway are present. While addition of a Gram-negative bacterium, E. coli, showed no enhancement in proPO activity, inclusion of a Gram-positive bacterium, M. luteus, or of the entomopathogenic fungus, B. bassiana, significantly enhanced PO activity and production of melanin pigment. These results are complimentary to those of Zhang et al. (2014) who observed that B. mori molting fluid could inhibit the growth of the Gram-positive bacterium Bacillus subtilis, and that it could also protect larvae undergoing the molt from infection by B. bassiana. Thus, our results are consistent with a hypothesis that molting fluid can function as an immune barrier to defend against microbial infection during the critical developmental process of molting.
5. Conclusion
Molting is a critical activity during the life cycle of all insects. Understanding the factors involved in this process offers important insights into insect physiology. Our analysis identified several enzymes likely to be involved in the breakdown of chitin and proteins in the cuticle. All of the chitin degrading/modifying enzymes we identified were also found in B. mori molting fluid, and all but one (chitinase-h) have orthologs in other insects outside of Lepidoptera; we hypothesize that these enzymes are critical for molting in other insects as well. All of the serine proteases identified in molting fluid contain extended prodomains that may have a regulatory function, unlike the digestive proteases of the midgut, which characteristically have very short pro-domains. SP60 is likely to be a key protease, given its orthology in other insects as well as prior published studies regarding its role in molting. Nevertheless, whether it acts directly on proteins in the cuticle or functions as an activator of another proenzyme is unresolved. Additionally, as SP60 itself is secreted as a proenzyme, the protease responsible for its activation remains to be determined. Several proteins were identified that likely have an immune function, highlighting the importance of the molting fluid in protection from infection during the molting process. However, a majority of the proteins we identified in molting fluid lack a known function. While some of these are likely unique to M. sexta or lepidopterans, others can be expected to be important to insects in general. The ability to combine proteomics with genomics now allows a more detailed analysis of proteins participating in the molting process. Further studies with M. sexta and other insects should lead to new insights of this critical aspect of insect physiology.
Supplementary Material
Figure S1. Spot numbering of proteins analyzed by LC/MS analysis. 2D SDS-PAGE analysis of molting fluid from pharate adults. Identified peptides and protein annotation are listed in supplementary Tables S1 and S2.
Figure S2. SDS-PAGE analysis of hemolymph and molting fluid. (A) Representative samples of hemolymph (H; 0.25 μl) and molting fluid (MF; 0.5 μl) from a 5th instar larva undergoing the larval to pupal molt were separated on a 4-12% acrylamide gel (NuPAGE, Thermo Fisher) and stained with Coomassie Brilliant Blue. The position of the lipophorins and hexamerins in the hemolymph sample are indicated. The molecular mass standard (SeeBlue, Thermo Fisher) is noted on the left. (B) A portion of the 2D gel shown in Figure 2; protein spots and number containing arylphorin-β are circled in red.
121 proteins were identified from the pharate pupal molting fluid of M. sexta
Chitin metabolism enzymes include exo- and endochitinases, and chitin deacetylases
Proteases and peptidases belonged to the serine and metalloprotease families
Several proteins, including prophenoloxidase, are involved in immunity
Melanization of molting fluid is enhanced by the addition of microorganisms
Acknowledgement
The authors thank Ms. Patil Tawidian and Dr. Kristin Michel for providing a culture of B. bassiana. Research reported in this publication was supported by the National Science Foundation, Award Number IOS 1257961, and the National Institute Of General Medical Sciences of the National Institutes of Health under Award Numbers R37 GM41247 and R35 GM141859. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by the United States Department of Agriculture National Institute of Food and Agriculture, Hatch Project No. 1013197, and the Kansas Agricultural Experiment Station (contribution number 22-237-J).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alva V, Lupas AN, 2016. The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim. Biophys Acta 1861, 913–923. [DOI] [PubMed] [Google Scholar]
- Amin A, Li Y, Finkelstein R, 1999. Identification of a Drosophila prolyl endopeptidase and analysis of its expression. DNA Cell Biol. 18, 605–610. [DOI] [PubMed] [Google Scholar]
- An C, Ragan EJ, Kanost MR, 2011. Serpin-1 splicing isoform J inhibits the proSpätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Dev. Comp. Immunol 35, 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO, 2010. Insect cuticular sclerotization: a review. Insect Biochem Mol. Biol 40, 166–178. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Dixit R, Begum K, Park Y, Specht CA, Merzendorfer H, Kramer KJ, Muthukrishnan S, Beeman RW, 2009. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol 39, 355–365. [DOI] [PubMed] [Google Scholar]
- Aso Y, Kramer KJ, Hopkins TL, Whetzel SZ, 1984. Properties of tyrosinase and DOPA quinone imine conversion factor from pharate pupal cuticle of Manduca sexta L. Insect Biochem. 14, 463–472. [Google Scholar]
- Bade ML, Shoukimas JJ, 1974. Neutral metal chelator-sensitive protease in insect moulting fluid. J. Insect Physiol 20, 281–290. [DOI] [PubMed] [Google Scholar]
- Bell RA, Joachim FG, 1976. Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms. Ann. Entomol. Soc. Am 69, 365–373. [Google Scholar]
- Brookhart GL, Kramer KJ, 1990. Proteinases in molting fluid of the tobacco hornworm, Manduca sexta. Insect Biochem. 20, 467–477. [Google Scholar]
- Brummett LM, Kanost MR, Gorman MJ, 2017. The immune properties of Manduca sexta transferrin. Insect Biochem. Mol. Biol 81, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Cassler A, Johansen JJ, Santek DA, Ide JR, Kendrick NC, 1989. Computerized quantitative analysis of coomassie-blue-stained serum proteins separated by two-dimensional electrophoresis. Clin. Chem 35, 2297–304. [PubMed] [Google Scholar]
- Burmester T, 201. Expression and evolution of hexamerins from the tobacco hornworm, Manduca sexta, and other Lepidoptera. Insect Biochem. Mol. Biol 62, 226–234. [DOI] [PubMed] [Google Scholar]
- Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard GW, Kanost MR, Jiang H, 2015. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem. Mol. Biol 62, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen JM, Hiromasa Y, An C, Kanost MR, 2012. Identification of plasma proteinase complexes with serpin-3 in Manduca sexta. Insect Biochem. Mol. Biol 42, 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH, 2009. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol. Biol 18, 607–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Hiromasa Y, Takahashi D, VanderVelde D, Fabrick JA, Kanost MR, Krishnamoorthi R, 2013. An initial event in the insect innate immune response: structural and biological studies of interactions between β-1,3-glucan and the N-terminal domain of β-1,3-glucan recognition protein. Biochemistry 52,161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon T, Hamada K, Mita K, Okano K, Suzuki MG, Kobayashi M, Shimada T, 2003. A Bombyx mori gene, BmChi-h, encodes a protein homologous to bacterial and baculovirus chitinases. Insect Biochem. Mol. Biol 33, 749–759. [DOI] [PubMed] [Google Scholar]
- Darkoh C, El-Bouhssini M, Baum M, Clack B, 2010. Characterization of a prolyl endoprotease from Eurygaster integriceps Puton (Sunn pest) infested wheat. Arch. Insect Biochem. Physiol 74, 163–178. [DOI] [PubMed] [Google Scholar]
- Dong W, Gao Y-H, Zhang X-B, Moussian B, Zhang J-Z, 2020. Chitinase 10 controls chitin amounts and organization in the wing cuticle of Drosophila. Insect Science 27, 1198–1207. [DOI] [PubMed] [Google Scholar]
- Dziadik-Turner C, Koga D, Mai MS, Kramer KJ, 1981. Purification and characteristics of two beta-N-acetylhexosaminidases from the tobacco hornworm, Manduca sexta (L.) (Lepidoptera: Sphingidae). Arch. Biochem. Biophys 12, 546–560. [DOI] [PubMed] [Google Scholar]
- Fu P, Sun W, Lai J, Shen Y-H, Zhang Z, 2018. Identification of two isoforms of Pop in the domestic silkworm, Bombyx mori: cloning, characterization and expression analysis. Gene 667, 101–111. [DOI] [PubMed] [Google Scholar]
- Fukamizo T, Kramer KJ, 1985. Mechanism of chitin hydrolysis by the binary chitinase system in insect moulting fluid. Insect Biochem. 15, 141–145. [Google Scholar]
- He Y, Wang Y, Hu Y, Jiang H, 2018. Manduca sexta hemolymph protease-2 (HP2) activated by HP14 generates prophenoloxidase-activating protease-2 (PAP2) in wandering larvae and pupae. Insect Biochem. Mol. Biol 101, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR, 1998. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc. Natl. Acad. Sci. USA 95, 12220–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Kanost MR, 1999. Four serine proteinases expressed in Manduca sexta haemocytes. Insect Mol. Biol 8, 39–53. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Kansot MR, 2003. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J. Biol. Chem 278, 3552–3561. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Zhu Y, Kansot MR, 2003. Prophenoloxidase-activating proteinase-3 (PAP-3) from hemolymph of Manduca sexta: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol 33, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR, 2005). Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem. Mol. Biol 35, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR, 2010. Immunity in lepidopteran insects. Adv. Exp. Med. Biol 708, 181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis AM, 1978. The composition of larval-pupal moulting fluid in the tobacco hornworm, Manduca sexta. J. Insect Physiol 24, 65–73. [Google Scholar]
- Kanost MR, Zepp MK, Ladendorff NE, Andersson LA, 1994. Isolation and characterization of a hemocyte aggregation inhibitor from hemolymph of Manduca sexta larvae. Arch. Insect Biochem. Physiol 27, 123–136. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Yu X-Q, 2004. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev 198, 97–105. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, 2015. Clip-domain serine proteases as immune factors in insect hemolymph. Curr. Opin. Insect Sci 11, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Arrese EL, Cao X, Chen Y-R, Chellapilla S, Goldsmith MR, Grosse-Wilde E, Heckel DG, Herndon N, Jiang H, et al. , 2016. Multifaceted biological insights from a draft genome sequence of the tobasso hornworm moth, Manduca sexta. Insect Biochem. Mol. Biol 76, 118–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga D, Jilka J, Kramer KJ, 1983a. Insect endochitinases: glycolproteins from molting fluid, integument and pupal haemolymph of Manduca sexta L. Insect Biochem. 13, 295–305. [Google Scholar]
- Koga D, Mai MS, Kramer KJ, 1983b. Comparative biochemistry of insect exo-β-N-acetylglucosaminidases: characterization of a third enzyme from pupal hemolymph of the tobacco hornworm, Manduca sexta. Comp. Biochem. Physiol 74B, 515–520. [Google Scholar]
- Koga D, Funakoshi T, Mizuki K, Ide A, Kramer KJ, Zen K-C, Choi H, Muthukrishnan S, 1992. Immunoblot analysis of chitinolytic enzymes in integument and molting fluid of the silkworm, Bombyx mori, and the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol 22, 305–311. [Google Scholar]
- Kramer KJ, Corpuz L, Choi HK, Muthukrishnan S, 1993. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manduca sexta. Insect Biochem. Mol. Biol 23, 691–701. [DOI] [PubMed] [Google Scholar]
- Li M, Christen JM, Dittmer NT, Cao X, Zhang X, Jiang H, Kanost MR, 2018. The Manduca sexta serpinome: Analysis of serpin genes and proteins in the tobacco hornworm. Insect Biochem. Mol. Biol 102, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-W, Wang L-L, Meng Z, Tang X, Li Y-S, Xia Q-Y, Zhao P, 2017. A clip domain serine protease involved in moulting in the silkworm, Bombyx mori: cloning, characterization, expression patterns and functional analysis. Insect Mol. Biol, 26, 507–521. [DOI] [PubMed] [Google Scholar]
- Liu H-W, Wang L-L, Tang X, Dong Z-M, Guo P-C, Zhao D-C, Xia Q-Y, Zhao P, 2018. Proteomic analysis of Bombyx mori molting fluid: insights into the molting process. J. of Proteomics 173, 115–125. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhou Y, Qu M, Qiu Y, Guo X, Zhang Y, Liu T, Yang J, Yang Q, 2019. Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family. J. Biol. Chem 294, 5774–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M, 2001. The Wigglesworth lecture: insects for studying fundamental problems in biology. J. Insect Physiol 47, 495–507. [DOI] [PubMed] [Google Scholar]
- Lu Y, Su F, Zhu K, Zhu M, Li Q, Hu Q, Zhang J, Zhang R, Yu X-Q, 2020. Comparative genomic analysis of C-type lectin-domain genes in seven holometabolous insect species. Insect Biochem. Mol. Biol 126, 103451. [DOI] [PubMed] [Google Scholar]
- Luschnig S, Bätz T, Armbruster K, Krasnow MA, 2006. Serpentine and vermiform encode matrix proteins with chitin-binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol 16, 186–194. [DOI] [PubMed] [Google Scholar]
- Meekins DA, Kanost MR, Michel K, 2017. Serpins in arthropod biology. Semin. Cell Dev. Biol 62, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, 2010. Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem. Mol. Biol. 40, 363–375. [DOI] [PubMed] [Google Scholar]
- Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y, 2018. Group 1 chitin deacetylases are essential for higher order organization of chitin fibers in beetle cuticle. J. Biol. Chem 293, 6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Homma K-I, Kurata S, Natori S, 1997a. Molecular cloning of cDNA for Sarcophaga prolyl endopeptidase and characterization of the recombinant enzyme produced by an E. coli expression system. Insect Biochem. Mol. Biol 27, 337–343. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Homma K-I, Kurata S, Natori S, 1997b. Nuclear localization and involvement in DNA synthesis of Sarcophaga prolyl endopeptidase. J. Biochem 121, 1176–1181. [DOI] [PubMed] [Google Scholar]
- Pesch Y-Y, Riedel D, Patil KR, Loch G, Behr M, 2016. Chitinases and imaginal disc growth factors organize the extracellular matrix formation at barrier tissues in insects. Sci. Rep 6, 18340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár L, 2002. The prolyl oligopeptidase family. Cell. Mol. Life Sci 59, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M, Liu T, Chen P, Yang Q, 2013. A sperm-plasma β-N-acetyl-d-hexosaminidase interacting with a chitinolytic β-N-acetyl-d-hexosaminidase in insect molting fluid. PLoS ONE 8(8): e71738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M, Ma L, Chen P, Yang Q, 2014. Proteomic analysis of insect molting fluid with a focus on enzymes involved in chitin degradation. J. Proteome Res 13, 2931–2940. [DOI] [PubMed] [Google Scholar]
- Rao X-J, Cao X, He Y, Hu Y, Zhang X, Chen Y-R, Blissard G, Kanost MR, Yu X-Q, Jiang H, 2015. Structural features, evolutionary relationships, and transcriptional regulation of C-type lectin-domain proteins in Manduca sexta. Insect Biochem. Mol. Biol 62, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke JP, Buckner JS, Grugel SR, 1980. Life cycle of laboratory-reared tobacco hornwoms, Manduca sexta, a study of development and behavior, using time-lapse cinematography. Biol. Bull 158, 129–140. [Google Scholar]
- Samuels RI, Charnley AK, Reynolds SE, 1993a. A cuticle-degrading proteinase from the moulting fluid of the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol 23, 607–614. [DOI] [PubMed] [Google Scholar]
- Samuels RI, Charnley AK, Reynolds SE, 1993b. An Aminopeptidase from the moulting fluid of the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol 23, 615–620. [DOI] [PubMed] [Google Scholar]
- Sun JS, Xiao S, Carlson JR, 2018. The diverse small proteins called odorant-binding proteins. Open Biol. 8:180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhao P, Lin Y, Hou Y, Xia Q-Y, Xiang Z-H, 2007. Analysis of the structure and expression of the 30K protein genes in silkworm, Bombyx mori. Insect Sci. 14, 5–14. [Google Scholar]
- Tetreau G, Cao X, Chen Y-R, Muthukrishnan S, Jiang H, Blissard GW, Kanost MR, Wang P, 2015a. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol 62, 114–126. [DOI] [PubMed] [Google Scholar]
- Tetreau G, Dittmer NT, Cao X, Agrawal S, Chen YR, Muthukrishnan S, Jiang H, Blissard GW, Kanost MR, Wang P, 2015b. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem. Mol. Biol 62, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Jiang H, Kanost MR, 2005. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activaion pathway. J. Biol. Chem 280, 14932–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujita M, Katsuno Y, Kawachi I, Ueno Y, Banno Y, Fujii H, Hara A, 2005. Glucan-binding activity of silkworm 30-kDa apolipoprotein and its involvement in defense against fungal infection. Biosci. Biotechnol. Biochem 69, 1178–1185. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Große-Wilde E, Zhou J-J, 2015. The lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol 62, 142–153. [DOI] [PubMed] [Google Scholar]
- Wang S, Jayaram AS, Hemphala J, Senti KA, Tsarouhas V, Jin H, Samakovlis C, 2006. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol 16, 180–185. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang F, Cao X, Zou Z, Lu Z, Kanost MR, Jiang H, 2020. Hemolymph protease-5 links the melanization and Toll immune pathways in the tobacco hornworm, Manduca sexta. Proc. Natl. Acad. Sci. USA 117, 23581–23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Cao X, Huang R, Paskewitz S, Hartson SD, Kanost MR, Jiang H, 2020. Inhibition of immune pathway-initiating hemolymph protease-14 by Manduca sexta serpin-12, a conserved mechanism for the regulation of melanization and Toll activation in insects. Insect Biochem. Mol. Biol 116, 103261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yin Y, Zhang B, Wang Z, Peng G, Cao Y, Xia Y, 2007. Cloning of a novel protease required for the molting of Locusta migratoria manilensis. Dev. Growth Differ 49, 611–621. [DOI] [PubMed] [Google Scholar]
- Willis JH, 2010. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol 40, 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Pan P-L, Ye Y-X, Yu B, Zhang C-X, 2014. Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol 23, 695–705. [DOI] [PubMed] [Google Scholar]
- Xi Y, Pan P-L, Ye Y-X, Yu B, Xu H-J, Zhang C-X, 2015. Chitinase-like gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol 24, 29–40. [DOI] [PubMed] [Google Scholar]
- Xia X, You M, Rao X-J, Yu X-Q, 2018. Insect C-type lectins in innate immunity. Dev. Comp. Immunol 83, 70–79. [DOI] [PubMed] [Google Scholar]
- Yu R, Liu W, Li D, Zhao X, Ding G, Zhang M, Ma E, Zhu KY, Li S, Moussian B, Zhang J, 2016. Helicoidal organization of chitin in the cuticle of the migratory locust requires the function of the chitin deacetylase2 enzyme (LmCDA2). J. Biol. Chem 291, 24352–24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R-R, Liu W-M, Zhao X-M, Zhang M, Li D-Q, Zuber R, Ma E-B, Zhu KY, Moussian B, Zhang J-Z, 2019. LmCDA1 organizes the cuticle by chitin deacetylation in Locusta migratoria. Insect Mol. Biol 28, 301–312. [DOI] [PubMed] [Google Scholar]
- Zen K-C, Choi HK, Krishnamachary N, Muthukrishnan S, Kramer KJ, 1996. Cloning, expression, and hormonal regulation of an insect β-N-acetylglucosaminidase gene. Insect Biochem. Mol. Biol 26, 435–444. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lu A, Kong L, Zhang Q, Ling E, 2014. Functional analysis of insect molting fluid proteins on the protection and regulation of ecdysis. J. Biol. Chem 289, 35891–35906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Ji Y, Zhang X, Ma P, Wang Y, Moussian B, Zhang J, 2019. The putative chitin deacetylases Serpentine and Vermiform have non-redundant functions during Drosophila wing development. Insect Biochem. Mol. Biol 110, 128–135. [DOI] [PubMed] [Google Scholar]
- Zhang X, He Y, Cao X, Gunaratna RT, Chen Y-R, Blissard G, Kanost MR, Jiang H, 2015. Phylogenetic analysis and expression profiling of the pattern recognition receptors: insights into molecular recognition of invading pathogens in Manduca sexta. Insect Biochem. Mol. Biol 62, 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Arakane Y, Beeman RW, Kramer KJ, Muthukrishnan S, 2008. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc. Natl. Acad. Sci U.S.A 105, 6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber R, Shaik KS, Meyer F, Ho H-N, Speidel A, Gehring N, Bartoszewski S, Sxhwarz H, Moussian B, 2019. The putative C-type lectin Schlaff ensures epidermal barrier compactness in Drosophila. Sci. Rep, 9, 5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Spot numbering of proteins analyzed by LC/MS analysis. 2D SDS-PAGE analysis of molting fluid from pharate adults. Identified peptides and protein annotation are listed in supplementary Tables S1 and S2.
Figure S2. SDS-PAGE analysis of hemolymph and molting fluid. (A) Representative samples of hemolymph (H; 0.25 μl) and molting fluid (MF; 0.5 μl) from a 5th instar larva undergoing the larval to pupal molt were separated on a 4-12% acrylamide gel (NuPAGE, Thermo Fisher) and stained with Coomassie Brilliant Blue. The position of the lipophorins and hexamerins in the hemolymph sample are indicated. The molecular mass standard (SeeBlue, Thermo Fisher) is noted on the left. (B) A portion of the 2D gel shown in Figure 2; protein spots and number containing arylphorin-β are circled in red.