Abstract
Although gut microbiota has been linked to exercise, whether alterations in the abundance of specific bacteria improve exercise performance remains ambiguous. In a cross-sectional study involving 25 male long-distance runners, we found a correlation between Bacteroides uniformis abundance in feces and the 3000-m race time. In addition, we administered flaxseed lignan or α-cyclodextrin as a test tablet to healthy, active males who regularly exercised in a randomized, double-blind, placebo-controlled study to increase B. uniformis in the gut (UMIN000033748). The results indicated that α-cyclodextrin supplementation improved human endurance exercise performance. Moreover, B. uniformis administration in mice increased swimming time to exhaustion, cecal short-chain fatty acid concentrations, and the gene expression of enzymes associated with gluconeogenesis in the liver while decreasing hepatic glycogen content. These findings indicate that B. uniformis enhances endurance exercise performance, which may be mediated by facilitating hepatic endogenous glucose production.
B. uniformis enhances exercise performance in mice and α-cyclodextrin preferred by the microbe improves the performance in humans.
INTRODUCTION
Although training can improve human exercise performance, other factors can also have an impact. For example, several recent studies have shown that gut microbiota could contribute to exercise performance (1–5). Studies have shown that gut microbes are associated with exercise performance in mice; for example, Hsu et al. (6) reported that germ-free mice have a shorter swimming endurance than specific pathogen–free (SPF) mice and Bacteroides fragilis–monoassociated mice. In addition, Huang et al. (7) showed that inoculation with Eubacterium rectale or Clostridium coccoides improved the swimming time of gnotobiotic mice. Moreover, mice with disrupted gut microbiota due to multiple antibiotic treatments had a shorter treadmill running time than the control (8). Veillonella atypica from the gut microbiome of marathon runners increased treadmill running time in SPF mice by producing propionate from exercise-induced lactate (9). In contrast, other studies have demonstrated that exercise alters the gut microbiome profile in mice (10, 11). Despite the lack of evidence linking gut microbes to human exercise performance, studies have reported that the gut microbiota of athletes are more diverse (12, 13) and have a different composition than nonathletes (12). These findings suggest that the gut microbiota may play a role in the host’s physical performance, and the relationship between the microbiota and exercise appears to be bidirectional. However, there is insufficient evidence to establish a causal relationship between exercise performance and specific gut microbes, and whether other human gut microbes are associated with exercise performance remains unknown.
Therefore, this study aimed to identify additional gut microbes associated with endurance exercise performance and determine whether the identified species can influence exercise performance. To this end, we first conducted a cross-sectional study of Japanese long-distance runners and age- and sex-matched nonathletes and analyzed their gut microbiome, and identified Bacteroides uniformis, a dominant bacterial species in the human gut (14), associated with endurance exercise performance (3000-m run time). We then found that the supplementation with α-cyclodextrin (αCD) in human males increased gut B. uniformis abundance and improved endurance exercise performance and that B. uniformis administration in mice enhanced endurance exercise performance, possibly by facilitating endogenous glucose production in the liver during exercise.
RESULTS
Gut microbiota profile of Japanese long-distance runners was distinct from that of nonathletes
We conducted a cross-sectional study to identify bacterial groups associated with endurance exercise. Fecal samples were collected from 48 male long-distance runners specializing in ekiden (a type of long-distance relay running) who were members of the university ekiden team [athlete group; age, 20.3 ± 1.0 years (mean ± SD); height, 169.9 ± 4.2 cm; body weight, 54.8 ± 3.5 kg; and body mass index (BMI), 19.0 ± 0.9] and 10 males who served as controls (nonathlete group; age, 22.6 ± 0.7 years; height, 169.7 ± 7.6 cm; body weight, 59.5 ± 8.0 kg; and BMI 20.6 ± 2.2).
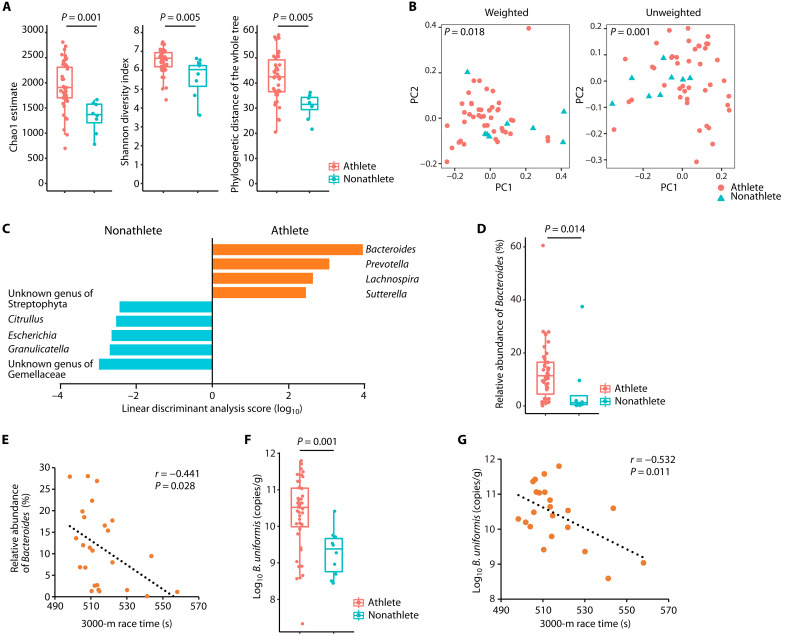
DNA was extracted from the feces, and the V4 region of the bacterial 16S ribosomal RNA (rRNA)–encoding gene was sequenced using the MiSeq platform to generate a gut microbiota profile. Five participants in the athlete group and two in the nonathlete group were excluded from the sequencing process because polymerase chain reaction (PCR) failed to amplify the target region. This failure could be attributed to PCR inhibitors, such as polysaccharides (15). Ultimately, 6,654,772 sequence reads were analyzed. The mean, minimum, and maximum of the reads per sample were 130,485, 46,053, and 294,588, respectively. The Chao1 estimate (P = 0.001), Shannon index (P = 0.005), and phylogenetic distance (PD) of the whole tree (P = 0.005) revealed that the gut microbiota of the athlete group had significantly higher α-diversity than that of the nonathlete group (Fig. 1A). In addition, β-diversity analysis based on weighted and unweighted UniFrac distances showed that the microbiota profiles of the two groups were significantly different (P = 0.018 and P = 0.001, respectively; Fig. 1B). A linear discriminant analysis effect size (LEfSe) analysis demonstrated that the variations in microbiota profiles of the two groups was primarily due to the following nine genera: Bacteroides, Prevotella, Lachnospira, Sutterella, an unknown genus of the family Gemellaceae, Granulicatella, Escherichia, Citrullus, and an unknown genus of the order Streptophyta (Fig. 1C). Among these genera, the relative abundance of the following six genera showed significant differences between the two groups: Bacteroides (P = 0.014; Fig. 1D), Lachnospira, and Sutterella were highly abundant in the athlete group (P = 0.021 and P = 0.021; fig. S1A), whereas Escherichia and two unknown genera of Gemellaceae and Streptophyta were abundant in the nonathlete group (P = 0.002, P = 0.001, and P = 0.008, respectively; fig. S1A).
Fig. 1. Bacteroides abundance was higher in long-distance runners and correlated with the 3000-m race time.
(A) Gut microbiota α-diversity of long-distance male runners (athletes) and nonathlete male controls (nonathletes). The statistical significance of the differences between groups was analyzed using a two-sample t test with Monte Carlo permutations. (B) Fecal microbiota profiles of athletes (red circles) and nonathletes (blue triangles). Principal coordinate analysis plots using weighted or unweighted UniFrac distances are shown. The statistical significance of the differences between groups was analyzed using permutational multivariate analysis of variance (PERMANOVA). (C) LEfSe analysis was performed to isolate genera with | Linear discriminant analysis score | > 2. (D) Relative abundance of the genus Bacteroides in the athlete and nonathlete groups, determined by 16S rRNA-encoding gene amplicon sequencing and compared using the Mann-Whitney U test. (E) Scatterplot of the 3000-m race time and the relative abundance of Bacteroides in the athlete group. Individual participants are represented as circles. (F) B. uniformis abundance in feces collected from the athlete and nonathlete groups, determined by 16S rRNA-encoding gene copy numbers using B. uniformis–specific qPCR. The Mann-Whitney U test was used to compare the groups. (G) Scatterplot of the 3000-m race time and B. uniformis abundance in the athlete group. Individual participants are represented as circles. The distribution of values within each group (A, D, and F) is illustrated by a box-and-whisker plot. (A to D) Athlete group, n = 43; nonathlete group, n = 8. (F) Athlete group, n = 48; nonathlete group, n = 10. (E and G) Athlete group, n = 25. Twenty-three of 48 athletes refused to participate in the trial to measure the race time. All statistical tests were two-tailed. For correlations, the dotted line shows the regression line.
To determine whether the gut microbiota is associated with endurance exercise performance in long-distance runners, we analyzed the correlations of the relative abundances of the nine genera identified in the LEfSe analysis with exercise-related blood parameters and the 3000-m race time, measured during the same period as that in which the fecal sample collection; however, four of the nine genera (Citrullus, unknown genus of the family Gemellaceae, Granulicatella, and unknown genus of the order Streptophyta) were excluded because these were detected only in a few participants in the athlete group (fig. S1A). Although all participants provided blood samples, 23 of 48 athletes refused the measurement of their 3000-m race time. In these analyses, only Bacteroides showed a significant negative correlation with the 3000-m race time [Pearson correlation coefficient (r) = −0.441, P = 0.028; Fig. 1E and fig. S1B]. The overall trend was a negative correlation between Lachnospira and blood parameters; however, the other genera exhibited no patterns (fig. S1C). These findings indicated that the genus Bacteroides could be associated with endurance exercise performance.
Next, we used species-specific quantitative PCR (qPCR) to determine the abundance of 13 commonly detected Bacteroides species in human feces by measuring the copy numbers of their 16S rRNA-encoding gene. Bacteroides coprocola, Bacteroides coprophilus, Bacteroides dorei, Bacteroides plebeius, and Bacteroides vulgatus were given the new genus name Phocaeicola in 2020 (16); however, the Greengenes database used for the gut microbiota analysis in this study does not include the new genus Phocaeicola. Thus, the abundances of these five species are included in the abundance of Bacteroides in Fig. 1 (D and E). The qPCR results showed that the abundance of B. uniformis (Fig. 1F), Bacteroides caccae, Phocaeicola dorei, Bacteroides eggerthii, Bacteroides thetaiotaomicron, and Phocaeicola vulgatus (fig. S2A) was significantly higher in the athlete group than that in the nonathlete group (P = 0.001, P = 0.036, P = 0.009, P =0.005, P = 0.006, and P = 0.037, respectively). Furthermore, among these six bacterial species, only B. uniformis exhibited a significant negative correlation between abundance and 3000-m race time (r = −0.532, P = 0.011; Fig. 1G and fig. S2B). These results suggested that B. uniformis could be associated with endurance exercise performance.
αCD supplementation increased B. uniformis abundance and human male exercise performance
Although B. uniformis administration in mice or rats has no adverse outcomes (17, 18), the safety implications of human ingestion have not been explored. Therefore, to investigate whether increasing B. uniformis abundance in the human gut affects exercise performance, we needed to increase its abundance using other means. Thus, we sought to identify foods containing substances previously shown to specifically favor the proliferation of B. uniformis. Although secoisolariciresinol diglucoside (SDG) is not known to selectively favor the proliferation of B. uniformis in the gut, the microbe expresses a gene encoding β-glucosidase, which releases glucose from SDG via hydrolysis (19, 20). Therefore, we tested whether supplementing with flaxseed lignans (FL), which are particularly rich in SDG (21), would affect B. uniformis abundance in the gut and human endurance exercise performance. To improve the solubility of SDGs in water, the FL used in this experiment contained approximately 40% (w/w) αCD, a cyclic oligosaccharide composed of six glucose subunits. αCD reportedly alters the gut microbiota in mice fed a high-fat diet (22); therefore, we also examined its effects in humans.
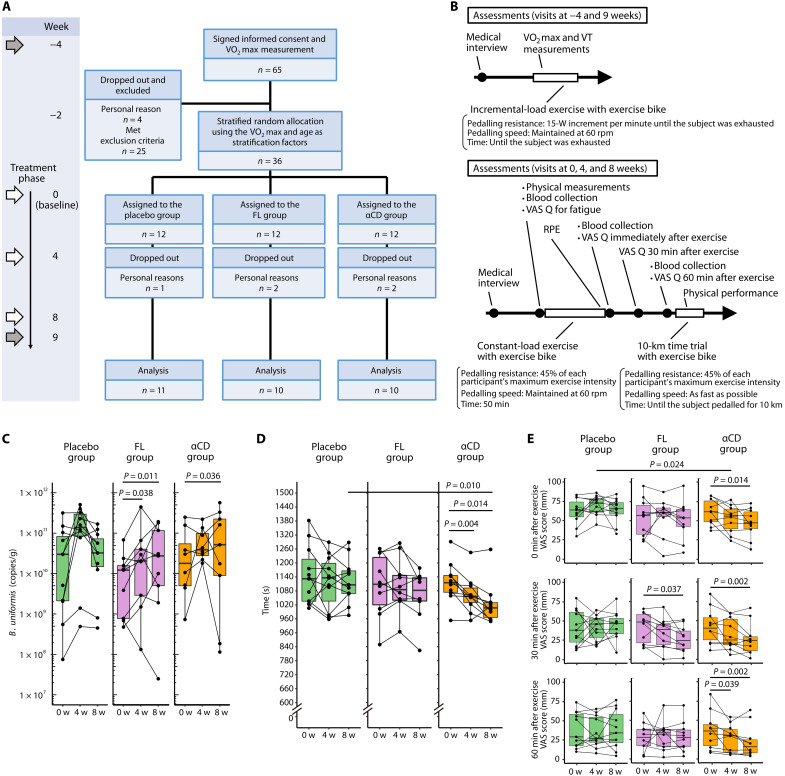
We conducted a randomized, double-blind, placebo-controlled, parallel-group human study to test whether FL or αCD increased intestinal B. uniformis abundance and influenced endurance exercise performance. Thirty-six Japanese males aged 20 to 49 years, who exercised regularly (once or twice per week), were randomly assigned to three different groups using age and maximum oxygen consumption (V̇O2max; the maximum capacity of the body to transport and use oxygen during physical activity), which is one of the factors related to endurance exercise performance (23) and was measured 4 weeks before supplementation (−4 weeks), as stratifying factors (Fig. 2A). At the beginning of the study, no significant differences in age, weight, BMI, heart rate, or V̇O2max were detected (table S1). For 9 weeks, the three groups were administered a placebo (placebo group), FL (200 mg/day, FL group), or αCD (200 mg/day, αCD group) as a supplement. The energy provided by each supplement per daily dose was 3.02, 3.04, and 2.96 kcal for the placebo, FL, and αCD groups, respectively. The assessments and a schedule of the visits at −4, 0, 4, 8, and 9 weeks are shown in Fig. 2B. All exercise tests were completed by all participants who attended each visit. One, two, and two participants in the placebo, FL, and αCD groups, respectively, dropped out of the study for personal reasons. Thus, the analysis ultimately included data from 11, 10, and 10 participants in the placebo, FL, and αCD groups, respectively. The supplement consumption rates in the placebo, FL, and αCD groups were 99.6 ± 1.0, 97.1 ± 4.6, and 97.8 ± 3.0% (mean ± SD), respectively, and the supplements did not cause any adverse events during the experimental period.
Fig. 2. α-Cyclodextrin affected B. uniformis abundance, endurance exercise performance, and postexercise fatigue, as demonstrated in a randomized, double-blind, placebo-controlled, parallel-group study.
(A) The randomized, double-blind, placebo-controlled, parallel-group study protocol. The O2max was evaluated at the weeks indicated by gray arrows (−4 and 9 weeks). Participants who met the inclusion criteria and did not meet the exclusion criteria were randomly allocated to the placebo, FL, or αCD groups. At the weeks indicated with white arrows (0, 4, and 8 weeks), we conducted a VAS questionnaire (VAS Q) related to fatigue before and after a 50-min constant-load exercise session, assessed the RPE, measured the time needed to pedal 10 km on an exercise bike, and collected blood and fecal samples. (B) Assessments performed at each visit in chronological order. (C) qPCR analysis of B. uniformis abundance in feces from each group at baseline (0 weeks) and after 4 and 8 weeks of supplementation. (D) Time required to pedal 10 km on an exercise bike. (E) Results of the VAS Q on fatigue conducted 0, 30, and 60 min after a 50-min constant-load biking exercise session. Fatigue levels measured by the VAS range from 0, indicating “not feel fatigued at all,” to 100, indicating “feel extreme fatigue.” The distribution of values within each group (C to E) is illustrated by a box-and-whisker plot. Consecutive measurements of the same individuals (dots) are connected with lines. The Mann-Whitney U test (placebo group versus FL or αCD group) or Wilcoxon signed-rank test (0 weeks versus 4 or 8 weeks) were used to determine the statistical significance of the different analyses. All statistical tests were two-tailed.
To investigate whether FL or αCD increase the abundance of B. uniformis in the human gut, we extracted DNA from fecal samples and performed B. uniformis–specific qPCR. The qPCR analysis revealed that after 4 and 8 weeks of supplementation, neither the FL nor the αCD groups exhibited a significant increase in B. uniformis abundance compared with the placebo group. However, there was a significant increase in B. uniformis abundance from baseline at 4 and 8 weeks (P = 0.038 and 0.011, respectively) in the FL group and 8 weeks (P = 0.036) in the αCD group. In the placebo group, the increase in B. uniformis abundance at 4 and 8 weeks was not significant (Fig. 2C). Change in B. uniformis abundance, between 0 and 8 weeks (8-week abundance minus baseline abundance), was 3.74 × 108 (−2.56 × 1010 to 3.76 × 1010) [median (interquartile range)], 1.46 × 1010 (1.15 × 109 to 4.84 × 1010), and 3.99 × 1010 (9.73 × 109 to 9.48 × 1010) copies/g in the placebo, FL, and αCD groups, respectively. These alterations did not differ significantly between the placebo and FL or αCD groups. Despite these findings, neither the FL nor the αCD group showed significant differences in the α- or β-diversity of the gut microbiota compared with the placebo group (fig. S3).
We measured the 10-km biking time to verify the effects of FL and αCD on exercise performance. No significant differences were observed in either of the placebo or FL groups at any time points; however, after 4 and 8 weeks of supplementation, the completion times of the αCD group were significantly shorter than those at baseline (P = 0.004 and P = 0.014); at 8 weeks, the completion times were significantly shorter than those of the placebo group (P = 0.010), which suggests that αCD improved endurance exercise performance (Fig. 2D).
Improving endurance exercise performance may have the side benefit of reducing subjective fatigue due to endurance exercise. To confirm this benefit, we measured an individual’s fatigue after a 50-min constant-load bike exercise using a visual analog scale (VAS) questionnaire for fatigue and an individual’s subjective perception of effort during the exercise using Borg’s ratings of perceived exertion (RPE) scale (24). At 0, 30, and 60 min after exercise, the VAS scores for fatigue in the placebo group did not differ significantly from baseline (Fig. 2E). For the FL group, the VAS score at 0 min immediately after the exercise was not significantly affected throughout the study, and no significant differences compared with the placebo group were observed at any of the time points. In contrast, 30 min after the exercise, the VAS score of the FL group after 8 weeks of supplementation was significantly lower than that at baseline (P = 0.037; Fig. 2E). For the αCD group, significantly lower VAS scores at 0, 30, and 60 min after exercise were found after 8 weeks of supplementation than those at baseline (P = 0.014, P = 0.002, and P = 0.002, respectively). In addition, at week 4, the αCD group had a significantly lower VAS score at 0 min than the placebo group (P = 0.024; Fig. 2E). For the αCD group, the RPE score after 50 min of constant road exercise at the 8-week visit was significantly lower than that at baseline (P = 0.004), but the same result was also observed in the placebo group (P = 0.020; fig. S4).
To explore the potential mechanisms underlying the improvements in endurance exercise performance, we measured the V̇O2max and ventilatory threshold (VT) as cardiopulmonary function, muscle mass, blood parameters, and concentrations of 30 amino acids in the blood. However, there were no notable changes in these parameters that led to an understanding of the potential mechanisms in humans (figs. S5 to S8).
To summarize the findings of the randomized, double-blind, placebo-controlled, parallel-group study, we found that supplementing with FL or αCD for 8 weeks increased the B. uniformis abundance compared with its abundance before supplementation and that αCD improved endurance exercise performance.
Administration of αCD increased B. uniformis abundance in the murine intestines and improved endurance exercise performance
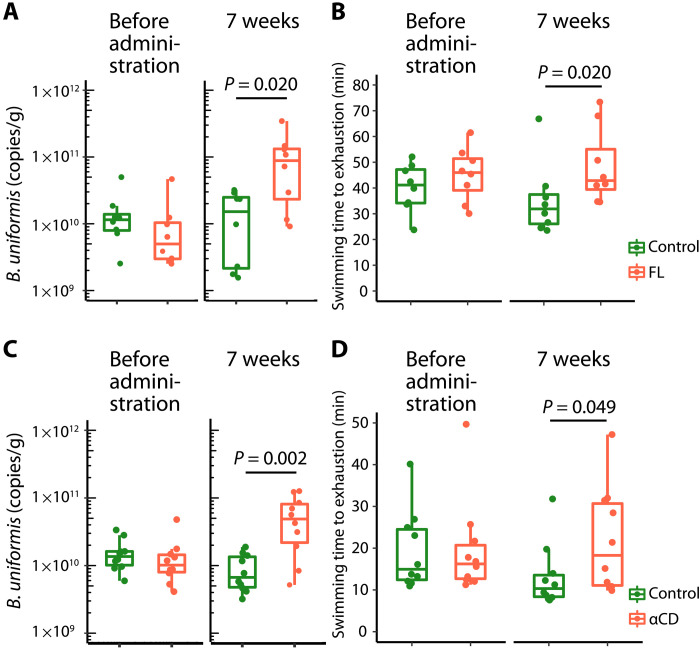
To confirm the effects of FL and αCD on the host, we examined whether supplementation with these substances altered the B. uniformis abundance in the gut and the endurance exercise performance in mice. Male C57BL/6J SPF mice (aged 8 weeks) were acclimated to swimming for 1 week and divided into two groups based on each mouse’s swimming time to exhaustion. The mice were fed either a standard CE-2 diet (control group) or a CE-2 diet containing 5% (w/w) FL (FL group) ad libitum for 7 weeks, and each group underwent a weekly swimming endurance test in a six-lane, flowing water pool (flow rate program: 12 liter/min for 5 min, 13 liter/min for 5 min, and 14 liter/min until exhaustion; water temperature: 34° ± 0.5°C). After 7 weeks, the FL group had a significantly higher fecal B. uniformis abundance as determined by qPCR and higher relative abundance of the genus Bacteroides as determined by 16S rRNA-encoding gene sequencing than the control group (P = 0.020 and P < 0.001; Fig. 3A and fig. S9A). LEfSe analysis results and gut microbiota compositions at the genus level are shown in fig. S9 (B and C). Furthermore, at the end point, the FL group had a significantly longer swimming time to exhaustion than the control group (P = 0.020; Fig. 3B).
Fig. 3. FL and αCD increased the B. uniformis abundance in the murine intestine and prolonged the swimming time to exhaustion.
The mice in the FL or αCD groups were fed a standard diet containing 5% (w/w) FL or αCD, respectively, and subjected to a weekly swimming endurance test (n = 8 for the FL group and associated control group; n = 10 for the αCD group and associated control group). (A and C) Box-and-whisker plots of B. uniformis abundance in feces from each group before administration and at the end point (7 weeks) based on the 16S rRNA-encoding gene copy numbers, calculated using B. uniformis–specific qPCR. (B and D) Box-and-whisker plots of the swimming time to exhaustion for each group before administration and at the end point (7 weeks). The statistical significance of the differences between groups was analyzed using two-tailed Mann-Whitney U tests.
The effects of αCD were also investigated by feeding mice a diet containing 5% (w/w) αCD (αCD group) for 7 weeks. A similar significant increase in the fecal B. uniformis abundance and a longer swimming time to exhaustion was noted at 7 weeks in the αCD group compared with the control group (P = 0.002 and P = 0.049, respectively; Fig. 3, C and D). The relative abundance of Bacteroides in feces, LEfSe analysis results, and the genus-level composition of the gut microbiota are shown in fig. S9 (D to F). The LEfSe analysis revealed that only the relative abundance of Bacteroides was increased in the αCD group compared with the control group.
According to these findings, both FL and αCD increase B. uniformis abundance in the murine intestine and improve mouse endurance exercise ability. In particular, αCD specifically increases the abundance of Bacteroides compared with FL.
B. uniformis administration improved endurance exercise performance in mice
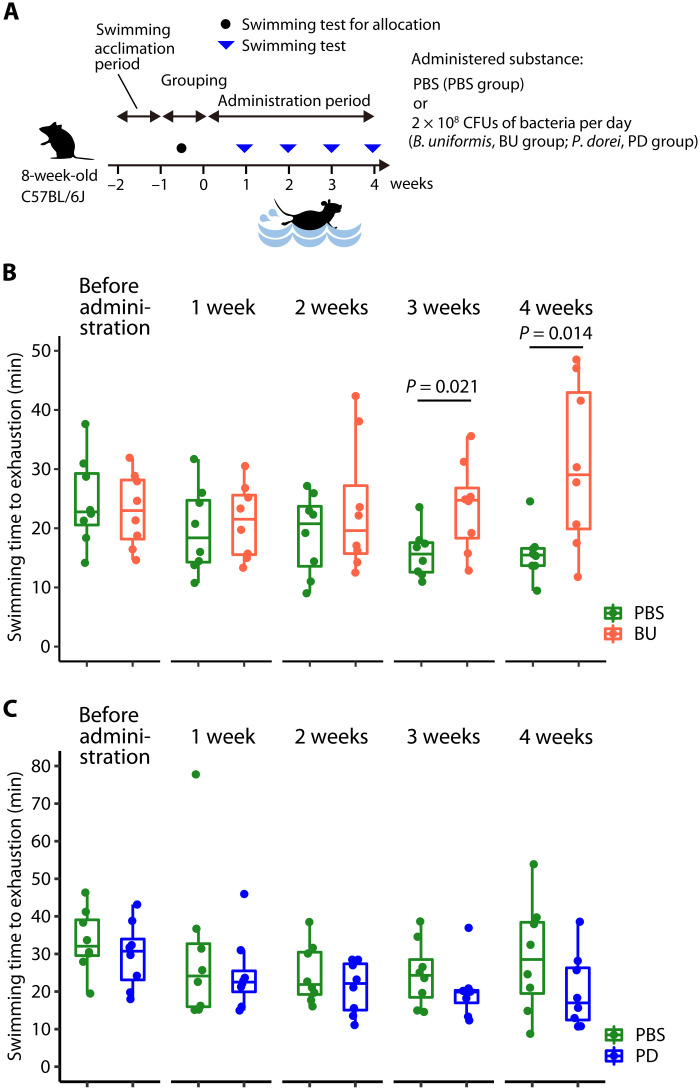
The results from the human and mouse experiments indicated that B. uniformis may improve endurance exercise performance. Thus, we subsequently determined whether B. uniformis administration could improve endurance exercise performance in mice. First, we evaluated the effect of an oral administration of the B. uniformis on the swimming endurance of SPF mice. For this experiment, 8-week-old male C57BL/6J mice were acclimated to swimming and grouped in the same way as in the experiment used to generate the data shown in Fig. 3. For 4 weeks, both groups received either phosphate-buffered saline (PBS) or B. uniformis type strain [2 × 108 colony-forming units (CFU)/day] orally (Fig. 4A). Under the same conditions as the experiment depicted in Fig. 3, a weekly swimming endurance test was performed. At 3 and 4 weeks, the B. uniformis–treated mice swam for a significantly longer duration than the PBS-treated control group (P = 0.021 and P = 0.014, respectively; Fig. 4B). Next, we used P. dorei, which, following B. uniformis, had the second-highest abundance among the Bacteroides genus in the feces of the athlete group, whose abundance was significantly higher than that of the nonathlete group (fig. S2A). However, the swimming time to exhaustion of P. dorei type strain–administered SPF mice (2 × 108 CFU/day) showed no significant differences to PBS-administered mice at any time (Fig. 4C). These results suggest that B. uniformis administration improves endurance exercise performance in mice.
Fig. 4. B. uniformis administration improved endurance exercise performance in mice.
(A) Endurance exercise test schedule. The mice were administered PBS (PBS group), B. uniformis (2 × 108 CFU/day) (BU group), or P. dorei (2 × 108 CFU/day) (PD group) for 4 weeks and tested weekly for swimming endurance. (B and C) Swimming time to exhaustion of PBS-administered (n = 8), B. uniformis–administered (n = 8) (B), and P. dorei–administered (n = 8) (C) mice. The box-and-whisker plot illustrates the distribution of values within each group. The statistical significance of the differences between groups was analyzed using two-tailed Mann-Whitney U tests.
B. uniformis administration and periodic exercise altered the intestinal short-chain fatty acid and hepatic glycogen concentrations in mice
We analyzed the cecal short-chain fatty acid (SCFA) concentrations, hepatic gene expression patterns, hepatic glycogen content, muscle metabolite concentrations, and muscle glycogen content of the mice administered B. uniformis and subjected to exercise to explore the underlying biological phenomena. In this experiment, the mice were forced to swim for 15 min once a week (Fig. 5A) but under milder conditions (flow rate program: 12 liter/min for 5 min and 13 liter/min for 10 min; water temperature: 34° ± 0.5°C) than those used in the swimming endurance test to elicit a consistent response to exercise. Immediately following the swimming exercise in week 10, the mice were euthanized, and their cecal content, liver, and quadriceps femoris were collected.
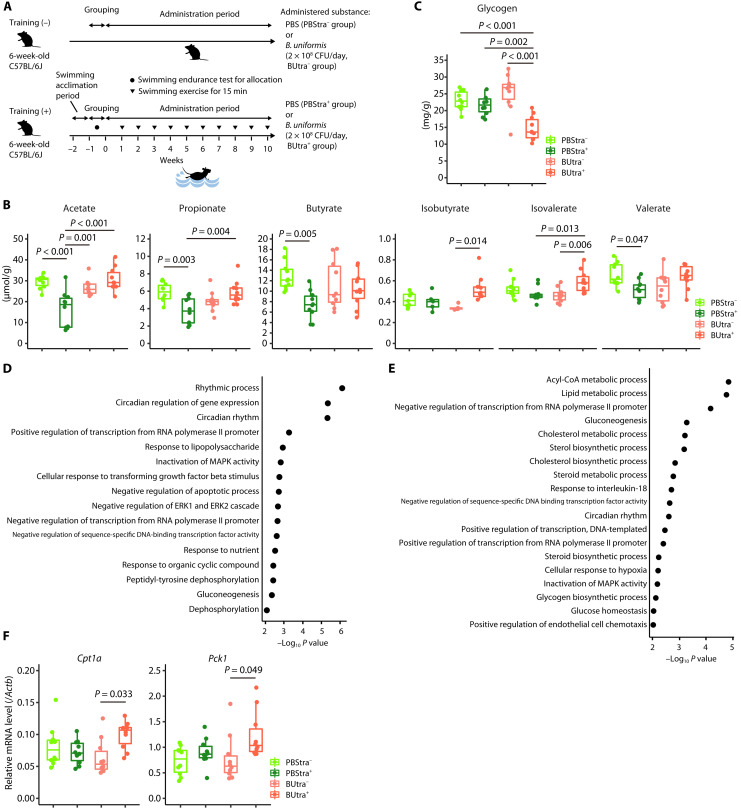
Fig. 5. B. uniformis administration altered intestinal SCFA concentrations and hepatic glycogen levels in SPF mice.
(A) Experimental schedule. The mice were orally administered PBS or B. uniformis (2 × 108 CFU/day) for 10 weeks, and the mice in training (+) groups were subjected to weekly swimming exercises. (B) SCFA concentrations in the cecal content of PBS-administered mice subjected (PBStra+, n = 10) and not subjected to exercise (PBStra−, n = 10) and B. uniformis–administered mice subjected (BUtra+, n = 10) and not subjected to exercise (BUtra−, n = 10). (C) Hepatic glycogen content in PBStra− (n = 10), PBStra+ (n = 10), BUtra− (n = 10), and BUtra+ (n = 10) mice. (D and E) Gene ontology (GO) analysis results of differentially expressed genes in the liver. The enriched GO terms identified using DAVID are listed in (D) [PBStra− (n = 6) versus PBStra+ (n = 6)] and (E) [BUtra− (n = 6) versus BUtra+ (n = 6)]. Statistical significance was analyzed using Fisher’s exact test. P < 0.01 was considered significant. (F) Reverse transcription qPCR analysis of the hepatic Cpt1a and Pck1 expression in PBStra− (n = 10), PBStra+ (n = 10), BUtra− (n = 10), and BUtra+ (n = 10) mice. The box-and-whisker plot illustrates the distribution of values within each group (B, C, and F). In (B), the Tukey-Kramer test was used for the statistical analysis of SCFAs, with the exception of isobutyrate, which was analyzed using the Steel-Dwass test. In (C), the Tukey-Kramer test was used for the analysis. In (F), the Tukey-Kramer test and Steel-Dwass tests were used for the analyses of Cpt1a and Pck1, respectively. These tests were selected on the basis of the outcomes of the normality test. All statistical tests were two-tailed.
Gas chromatography measurements of cecal SCFA concentrations revealed that the concentrations of acetate, propionate, butyrate, and valerate in the PBS-administered mice with exercise (PBStra+) were significantly lower than those in the PBS-administered mice without exercise (PBStra−) (P < 0.001, P = 0.003, P = 0.005, and P = 0.047, respectively; Fig. 5B). However, the concentrations of these four SCFAs were comparable in the B. uniformis–administered mice with (BUtra+) and without exercise (BUtra−), and the acetate and propionate concentrations were significantly higher in the BUtra+ mice than in the PBStra+ mice (P < 0.001 and P = 0.004, respectively; Fig. 5B). The isobutyrate and isovalerate concentrations showed different patterns from the four SCFAs. The two SCFA concentrations were similar in PBStra+ and PBStra− and higher in BUtra+ mice than in BUtra− mice (P = 0.014 and P = 0.006, respectively). These results suggest that exercise reduces the acetate, propionate, butyrate, and valerate concentrations in the cecum, while B. uniformis administration prevents this exercise-induced decrease.
During fasting and exercise, liver glycogen is broken down into glucose and used to restore blood glucose levels. Thus, we assessed the hepatic glycogen content of mice in the four groups. The BUtra+ group, compared with the other three groups, had significantly lower levels of hepatic glycogen (P < 0.001, P = 0.002, and P < 0.001, respectively; Fig. 5C).
Next, we investigated the hepatic gene expression patterns by performing an RNA sequencing (RNA-seq) analysis of mRNAs isolated from the mice livers in four groups and then conducting a gene ontology (GO) analysis of differentially expressed genes with a q value of <0.05. The PBStra− versus PBStra+ and BUtra− versus BUtra+ comparisons identified 91 and 603 differentially expressed genes, respectively. Consistent with prior findings, a GO analysis comparing the PBStra− and PBStra+ groups revealed enrichment in the rhythmic process and circadian rhythm (Fig. 5D) (25, 26). However, the comparison of the BUtra− and BUtra+ groups indicated enrichment of lipid metabolism and gluconeogenesis in addition to GO terms related to the circadian rhythm (Fig. 5E). Moreover, the mRNA levels of Cpt1a (GO term: lipid metabolism) and Pck1 (GO term: gluconeogenesis) measured by reverse transcription qPCR in the livers of the BUtra+ mice were significantly higher than those in the livers of the BUtra− mice (P = 0.033 and P = 0.049, respectively; Fig. 5F). Cpt1a encodes carnitine palmitoyl transferase 1a, which transfers long-chain fatty acids into mitochondria for β-oxidation (27). Pck1 encodes phosphoenolpyruvate carboxykinase, which catalyzes the reversible conversion of oxaloacetate to phosphoenolpyruvate for gluconeogenesis (28).
To determine whether B. uniformis administration and swimming exercise alters muscle metabolism related to energy acquisition, specifically the adenosine triphosphate–creatine phosphate (ATP-CP) system, the tricarboxylic acid (TCA) cycle, glycolysis, and fatty acid β-oxidation, we subsequently performed a capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS)–based metabolome analysis of quadriceps femoris samples from the four groups. However, no significant differences in the muscle metabolites related to energy acquisition were found among the four groups (fig. S10A). Furthermore, there was no significant difference in the transcript levels of Pgc-1α, a regulator involved in energy metabolism, or four genes related to glucose metabolism (Hk1, encoding hexokinase 1; Hk2, encoding hexokinase 2; Pygm, encoding muscle glycogen phosphorylase; and Glut4, encoding solute carrier family 2, member 4) in the quadriceps femoris muscles of the groups (fig. S10B). Moreover, the quadriceps femoris glycogen content showed no significant differences among the four groups (fig. S10C). These results indicate that the B. uniformis administration and weekly exercise performed in this experiment did not affect the ATP-CP system, TCA cycle, glycolysis, fatty acid β-oxidation, or glycogen content in the quadriceps femoris.
Fecal SCFA levels were higher, and hepatic glycogen levels were lower than those of germ-free mice in exercised B. uniformis–monoassociated mice
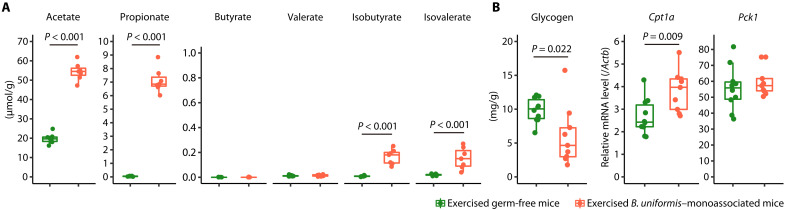
Because the cecal SCFA concentrations and hepatic glycogen levels of the BUtra+ mice differed from those of the PBStra+ mice (Fig. 5, B and C), we used B. uniformis–monoassociated mice to examine whether B. uniformis induces SCFA production in the murine intestines and the reduction of glycogen in the murine liver during exercise. Germ-free grade male C57BL/6N mice (10 weeks old) were injected with a single dose of B. uniformis type strain (2 × 109 CFU) to generate B. uniformis–monoassociated mice; three weeks after dose administration, fecal samples were collected, and the mice and germ-free mice were released from the sterile isolator and allowed to swim in the flowing water pool for 20 min or until exhaustion. Immediately after swimming, the mice were dissected, and livers were collected. We observed that B. uniformis promoted acetate, propionate, isobutyrate, and isovalerate production but not butyrate and valerate in the murine intestine (Fig. 6A). These findings and the results from the aforementioned experiments that B. uniformis administration prevented the exercise-induced reduction of SCFA concentrations in the cecum of SPF mice imply a possibility that the presence of B. uniformis in the gut may induce the production of acetate and propionate at a higher rate than their reduction during exercise. In addition, similar to BUtra+ mice, hepatic glycogen levels in the exercised B. uniformis–monoassociated mice were significantly lower than in exercised germ-free mice (P = 0.022; Fig. 6B). Liver Cpt1a transcription was significantly up-regulated in the exercised B. uniformis–monoassociated mice compared with exercised germ-free mice (P = 0.009), whereas Pck1 transcription in the livers was similar in both groups (Fig. 6B).
Fig. 6. Fecal SCFA and hepatic glycogen levels differed between exercised B. uniformis–monoassociated mice and germ-free mice.
(A) SCFA concentrations in the feces of exercised germ-free mice (n = 8) and B. uniformis–monoassociated mice (n = 7). (B) Hepatic glycogen content and expression of Cpt1a and Pck1 in exercised germ-free mice (n = 8) and B. uniformis–monoassociated mice (n = 7). The box-and-whisker plot illustrates the distribution of values within each group. An unpaired t test was used to statistically analyze the acetate, propionate, isovalerate, glycogen, and Cpt1a levels. The Mann-Whitney U test was used to analyze the valerate, isobutyrate, and Pck1 levels. These tests were selected on the basis of the outcomes of the normality test. All statistical tests were two-tailed.
DISCUSSION
This study focused on B. uniformis because the fecal abundance of B. uniformis was correlated with the 3000-m race time in the athlete group. We showed that αCD, a preferred substrate for B. uniformis, increased the abundance of B. uniformis and improved human and mouse endurance exercise performance. We tested the hypothesis that B. uniformis in the gut may improve endurance exercise performance by administering the bacteria to SPF mice and observing an improvement in their endurance exercise ability. To our knowledge, this is the first study to investigate the effect of the dominant gut microbe, B. uniformis, on endurance exercise performance.
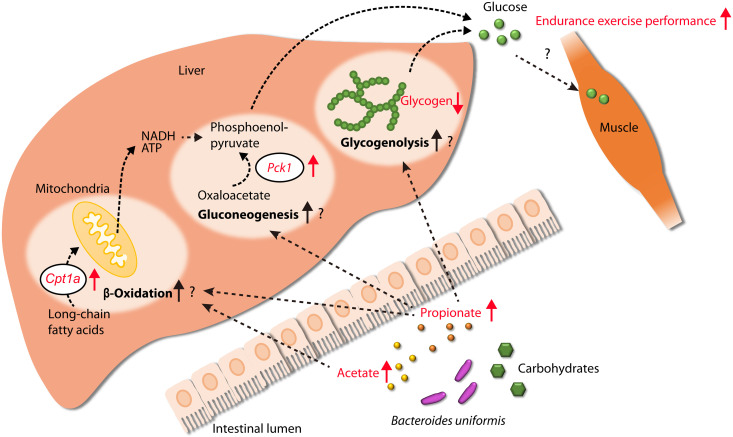
In our animal exercise tests, SPF mice subjected to exercise after PBS administration presented reduced cecal SCFA levels, whereas SPF mice subjected to exercise following the administration of B. uniformis did not exhibit this reduction. Furthermore, B. uniformis administration markedly increased the acetate and propionate levels in the intestines of germ-free mice. These findings suggest that the presence of high levels of B. uniformis in the gut induces increased acetate and propionate production through carbohydrate metabolism at rates that exceed the rates of decrease induced by exercise, which results in the maintenance of SCFA concentrations. Notably, Scheiman et al. (9) showed that the intrarectal administration of propionate improves endurance and exercise performance of mice. In our experiments, although no metabolome changes were observed in the muscle of SPF mice subjected to exercise following B. uniformis administration, their livers showed elevated Cpt1a and Pck1 mRNA levels, which encode rate-limiting enzymes in β-oxidation and gluconeogenesis, respectively, suggesting that β-oxidation and gluconeogenesis may be activated in the murine liver. The expression of these genes reportedly increases after the cecal infusion of propionate in the pig liver (29), and acetate administration also increases Cpt1a expression in the murine liver (30). Furthermore, fatty acid β-oxidation in the livers produces NADH (reduced form of nicotinamide adenine dinucleotide) and ATP, which are used for hepatic gluconeogenesis (31, 32). Another study showed that more than 60% of propionate directly infused in the cecum of mice is used in glucose production, which indicates its role as a gluconeogenic substrate (33). Focusing on hepatic glycogen, B. uniformis did not affect the glycogen levels in mice without exercise, but its presence led to a reduction in glycogen in mice with exercise. Similarly, exercised B. uniformis–monoassociated mice had low hepatic glycogen. These results suggest that B. uniformis administration could not inhibit glycogen synthesis in the liver but induced hepatic glycogenolysis during exercise. In mice, intraperitoneally injected propionate stimulates hepatic glycogenolysis and increases blood glucose by activating the sympathetic nervous system (34). Therefore, the presence of B. uniformis in the gut, likely through stimulation by propionate, may enhance hepatic glycogenolysis during exercise. Together, these findings and previous reports suggest the potential existence of a mechanism through which exercise performance is improved by hepatic endogenous glucose production that is induced by the maintenance of the acetate and propionate concentrations in the gut as a result of bacteria (perhaps B. uniformis)–induced acetate and propionate production. We postulated a potential mechanism by which B. uniformis improves endurance exercise performance (Fig. 7), but further research is needed to determine the actual mechanism.
Fig. 7. Potential mechanism through which B. uniformis enhances host endurance exercise performance.
B. uniformis produces acetate and propionate in the intestine through carbohydrate metabolism. These SCFAs may then migrate to the liver and stimulate Cpt1a and Pck1 gene expression, encoding enzymes involved in fatty acid oxidation and gluconeogenesis (29, 30), respectively. Specifically, propionate is used as a gluconeogenic substrate (33) and stimulates glycogenolysis (34), resulting in glucose production, which can be used as an energy source during exercise in exercise-related organs, such as muscle. Red arrows indicate the phenomena observed in this study.
In a randomized, double-blind, placebo-controlled, parallel-group study, an 8-week αCD supplementation improved the exercise bike time trials compared with baseline and placebo results but did not affect V̇O2max. These phenotypes suggest that factors other than V̇O2max, such as exercise economy, lactate threshold, and power, affect exercise performance (35). Consistent with our results, prior studies have shown that free radical scavengers (36), nitrates (37), and l-citrulline (38) supplementation can improve time trial performance without affecting V̇O2max. Unidentified gut microbes other than B. uniformis affected by αCD may also explain the improved cycling time trial performance in the αCD group. However, energy from the test tablet is unlikely to affect the performance, as the energy of tablets used to supplement placebo, FL, or αCD were approximately equal. The mechanisms behind the beneficial effects of αCD supplementation in humans, such as weight loss in overweight people, are unclear (39–42). Therefore, identifying the mechanism through which αCD improves endurance exercise performance in humans is challenging; however, we postulate that an increase in B. uniformis abundance may be partially responsible. Supplementation with FL did not improve human exercise bike time trials, unlike αCD, but improved mice swimming performance and increased B. uniformis in human feces. The lack of FL effects on endurance exercise performance may be because of differences in individual adaptability to the sport and physical conditions on the trial day. These confounding variables may have hampered the performance of the FL group, rendering the detection of their beneficial effects challenging. In addition, insufficient B. uniformis increase is also a potential reason. The increase of B. uniformis in the FL group may not be sufficient to improve exercise performance noticeably.
This study had a few limitations. First, the cross-sectional study observing long-distance runners had a few biases. The athletes included in the study had a higher B. uniformis abundance in the gut than nonathletes, but the two groups had different diets and living environments, which may affect gut microbiota composition (43, 44). Controlled diets and living environments are needed to determine whether a high abundance of B. uniformis is a characteristic of the gut microbiota of Japanese long-distance runners. In addition, our DNA extraction method using bead-beating, phenol-chloroform-isoamyl, and the glass fiber fleece-based purification columns may not completely remove PCR inhibitors such as polysaccharides (15) from the feces, which may explain PCR failure in five athletes and two nonathletes. Thus, future research focusing on the gut microbiome of Japanese athletes should use a DNA extraction method optimized for Japanese fecal samples, such as the method described by Tourlousse et al. (45). Second, our placebo-controlled study revealed that αCD supplementation improves human exercise performance, but whether the effect was due to B. uniformis in the gut could not be confirmed. After confirming the safety of B. uniformis in humans, further exploration of the effects of B. uniformis on exercise in humans can be conducted using B. uniformis supplementation. Last, we conducted both the mice experiments and human studies involving only males; thus, the findings may not apply to females because of sex differences in athletic ability.
In conclusion, our findings suggest that B. uniformis, a dominant human gut bacterium, improves mice endurance exercise performance and that a substrate that B. uniformis prefers, αCD, may be an effective prebiotic that enhances performance. However, the precise mechanisms through which B. uniformis enhances endurance exercise performance are unclear. We hypothesize that acetate and propionate, associated with increased B. uniformis abundance in the gut, may facilitate glucose supply from the liver during exercise. Future studies are needed to clarify how B. uniformis improves endurance exercise performance, and elucidating the mechanism may pave the way for applications improving endurance exercise performance through gut microbiota modifications.
MATERIALS AND METHODS
Gut microbiome analysis of Japanese long-distance runners
Study design and participants
This cross-sectional study was conducted from July to September 2015. Forty-eight male Japanese long-distance runners (aged 18 to 22 years, defined as the athlete group) from the ekiden team of the Aoyama Gakuin University Track and Field Club participated in the study. The outcomes from the athletes were the fecal microbiota, blood parameters [hemoglobin concentration (Hb), mean corpuscular hemoglobin (MCH), MCH concentration (MCHC), mean corpuscular volume (MCV), hematocrit, red blood cell (RBC) count, creatine phosphokinase (CPK), hematocrit, serum iron (Fe), and ferritin], and their running time in a 3000-m race. No eligibility or exclusion criteria were set; thus, all the team members (i.e., 48 athletes) participated in the study. They resided in team dormitories in Tokyo or Kanagawa Prefecture. Ten Japanese nonathlete healthy males (aged 22 to 24 years, defined as the nonathlete group), who were not related or socially associated with the ekiden team members and lived in individual houses in Tokyo or Kanagawa, participated in this investigation for comparison purposes. The exclusion criteria for the nonathletes were as follows: (i) past or present serious heart, liver, kidney, or gastrointestinal disease; (ii) allergy to any medications or foods; (iii) current disease requiring treatment; (iv) consumption of dietary supplements; (v) current participation in other clinical tests, participation within the previous 4 weeks, or plans to participate; and (vi) subjects whom a physician determined to be unable to follow instructions because of personality or health concerns. All participants gave informed consent to participate in the investigation before enrollment. In addition, if the participant was a minor (age < 20 years in Japan), then his parents provided consent.
Ethics statements
The protocol for investigating athletes was approved by the Ethical Committee for Research in Humans of Shiba Palace Clinic (Tokyo, Japan) in compliance with the Helsinki Declaration (approval no. 2015 AC-001). The protocol for investigating the nonathlete gut microbiota was approved by the Ethical Committee for Research in Humans of Aisei Hospital Ueno Clinic (Tokyo, Japan) in compliance with the Helsinki Declaration (approval no. 27030).
Fecal microbiota analysis
Fecal samples of both athletes and nonathletes were collected using the same collection protocol in July 2015 and September 2015, respectively. Participants collected fecal samples at their convenience in fecal collection containers (Sarstedt, Nümbrecht, Germany) at their dormitories or homes. After collection, the samples were frozen and delivered to our laboratory while still frozen and maintained at −30°C for later use. Subsequently, 0.2 g of each thawed fecal sample was aliquoted into plastic tubes. The samples were washed in 1.0 ml of PBS buffer and centrifuged at 14,000g for 3 min. The pellets were suspended in 500 μl of extraction buffer [166 mM tris-HCl, 66 mM EDTA, and 8.3% (w/v) SDS (pH 9.0)] and tris-EDTA (TE) buffer–saturated phenol. Glass beads (300 mg; diameter, 0.1 mm) were added to the suspension, and the mixture was vigorously vortexed for 60 s using a Multi-beads Shocker (Yasui Kikai, Osaka, Japan). After centrifugation at 14,000g for 5 min, 400 μl of the supernatant was purified using phenol-chloroform-isoamyl alcohol, and DNA was precipitated with isopropanol. The samples were washed in 70% ethanol and dissolved in TE buffer. The High Pure PCR Template Preparation Kit (Roche Diagnostics, Rotkreuz, Switzerland) was used for purification according to the manufacturer’s recommendations. Sequencing of the V4 region of the 16S rRNA-encoding gene was performed according to the manufacturer’s instructions. Briefly, 12.5 ng of purified DNA from each sample was used to amplify the V4 region of the 16S rRNA-encoding genes using PCR (23 cycles) with 515F and 806R primers (46). Amplicons were purified using the AMPure XP system (Beckman Coulter Biomedical GmbH, Munich, Germany), and double barcodes were then added on both the forward and reverse strands via a second round of PCR (eight cycles). The double-barcoded amplicons were purified using the AMPure XP system, pooled with a 1 pM PhiX library, and sequenced using a MiSeq system (Illumina, San Diego, CA, USA). Because of insufficient V4 region amplification, five participants in the athlete group and two in the nonathlete group were excluded from the gut microbiota analysis. QIIME version 1.9.1 was used for sequence filtering and analysis (47). Sequences with a quality score of less than 29 were removed from the fastq files. Chimeric sequences were filtered out using USEARCH, and operational taxonomic units (OTUs) were assigned using an open-reference OTU picking process with a 97% pairwise identity threshold. The α-diversity (Chao1 richness estimates, Shannon diversity index, and PD of the whole tree) within two groups and the distances between participants (unweighted and weighted UniFrac distances as a measurement of the β-diversity) were also estimated using QIIME with data rarefied at 35,000 reads per sample. The β-diversity was visualized using principal coordinates analysis. The LEfSe analysis was performed using the online tool LEfSe (Galaxy version 1.0), available at the following website: http://huttenhower.sph.harvard.edu/galaxy/ (48). The species-specific primers and TaqMan probes shown in table S2, reported previously (49, 50) or designed for this study, were used for qPCR. A LightCycler 480 Instrument II (Roche Diagnostics) was used to amplify and detect specific sequences. The standard curve was generated by amplifying 10-fold serial dilutions of targeted bacterial 16S rRNA-encoding gene sequences synthesized by Thermo Fisher Scientific (Waltham, MA, USA).
Blood analyses
During the same period as that at which fecal sampling was performed, blood samples from athletes were collected by a physician in a clinic. Before blood sample collection, no dietary or behavioral restrictions were given to the participants. Blood samples were collected in ethylenediaminetetraacetic acid disodium salt dihydrate–coated tubes for Hb, MCH, MCHC, MCV, hematocrit, and RBC analyses and in noncoated tubes for CPK, Fe, and ferritin analyses. The collected blood samples were sent directly to a clinical laboratory.
The 3000-m race
The time required by each participant to run 3000 m on the outdoor field track of Aoyama Gakuin University was recorded. Twenty-five of 48 athletes participated in this trial, but the other 23 athletes refused the measurement of their 3000-m race time. All participating athletes started at the same time.
Statistical analysis
To compare the gut microbial α- and β-diversities between groups, we used a two-sample t test with Monte Carlo permutations and permutational multivariate analysis of variance (PERMANOVA), respectively. The number of permutations for both tests was set to 999 to calculate P values. Correlations among the relative abundances of Bacteroides, Prevotella, Escherichia, Lachnospira, and Sutterella and blood data or the 3000-m race time of the athletes were determined using the Pearson correlation coefficient. Correlations among the log10 copy numbers of each Bacteroides and Phocaeicola species and the 3000-m race time were determined using the Pearson correlation coefficient. In these correlation analyses, missing data were deleted by pairwise deletion. Mann-Whitney U tests were used to compare the relative abundances of genera and log10 copy numbers of each Bacteroides and Phocaeicola species between the athlete and nonathlete groups. These statistical analyses were conducted using SPSS Statistics for Windows (version 23; IBM, Armonk, NY, USA) and R (version 4.0.2; R Foundation for Statistical Computing, Vienna, Austria). Differences with P < 0.05 were considered statistically significant. In the box plots, the center line represents the median, and the box boundaries represent the 25th and 75th percentiles. The whiskers correspond to the most extreme values within 1.5 times the interquartile range below the 25th percentile and above the 75th percentile.
Randomized, double-blind, placebo-controlled, parallel-group study with FL or αCD as test supplements
Study design
This study was a randomized, double-blinded, placebo-controlled, parallel-group study conducted at the Chiyoda Paramedical Care Clinic, Tokyo, Japan. Safety and adverse events were monitored by hematology, vital signs, and individual self-reporting at each clinic visit.
Ethics statement
This study was conducted in accordance with the Declaration of Helsinki and Ethical Guidelines for Medical and Health Research Involving Human Subjects (notification from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labour, and Welfare). Before implementation, the Institutional Review Board of Chiyoda Paramedical Care Clinic reviewed and approved the study protocol (approval no.: 18071903; approval date: 20 July 2018). The study was registered in the University Hospital Medical Information Network system (registration ID: UMIN000033748; registration date: 14 August 2018). All the participants provided informed consent before a preliminary assessment (V̇O2max measurement; visit −4 weeks) and were enrolled between 17 and 19 August 2018. After the preliminary test, 36 participants were allocated to one of the three groups on 13 September 2018, and the supplementation period started on 20 to 25 September 2018 and ended on 22 to 27 November 2018.
Participants and sample size
This study was a first clinical trial; therefore, no sample size calculation could be performed. Participants were recruited at the Chiyoda Paramedical Care Clinic. A total of 65 participants who consented to participate after receiving a thorough explanation of the nature of the experiment were enrolled in the study and underwent the preliminary assessment (V̇O2max measurement) approximately 4 weeks before test supplement intake. After the V̇O2max measurement, four participants declined to participate in the study for personal reasons, and 25 met at least one of the exclusion criteria described below. Therefore, 36 Japanese males received a placebo, FL, or αCD as a supplement for 9 weeks. During the study, the participants were asked to maintain their normal dietary and physical activity. One, two, and two participants in the placebo, FL, and αCD groups, respectively, dropped out of the study for personal reasons. The placebo, FL, and αCD groups had 11, 10, and 10 participants in the final analysis, respectively. The inclusion criteria were as follows: (i) healthy males aged 20 to 49 years at the time of providing consent; (ii) a regular exercise habit, that is, one to two sessions (≥30 min per session) per week of exercise of ≥5 metabolic equivalents (METs), excluding resistance (weight) training among the exercises listed in the 2011 Compendium of Physical Activities (51); (iii) continuation of exercise habits during the study period; and (iv) verbal agreement not to consume prohibited foods (listed below) for at least 1 week before the date of providing consent and throughout the study period. The exclusion criteria were as follows: (i) self-report of ≤5 bowel movements per week or proneness to diarrhea; (ii) past or present serious heart, liver, kidney, or gastrointestinal disease; (iii) high alcohol consumption (60 g/day); (iv) extremely irregular eating habits or irregular daily rhythm due to shifting or night work; (v) allergy to any medicines or foods; (vi) current participation in other clinical tests, participation within the past 4 weeks, or plans to participate; (vii) blood donation of more than 200 ml in the past month or over 400 ml in the past 3 months; and (viii) subjects who were determined by a physician to be unable to follow instructions or complete exercise sessions because of their personality or health concerns.
Randomization and blinding
Randomization and allocation were carried out by Okutoeru LLC (Tokyo, Japan). V̇O2max and age were used to stratify 36 participants into three groups (12 participants per group) through stratified randomization (Fig. 2A). The Microsoft Excel randomization function generated the allocation sequence. The study investigators, research coordinators, physicians, clinical staff, and participants were blinded to the allocation of the participants to the treatment groups. The test supplements were provided in an opaque sachet with a numeric code, and each participant’s samples were blinded using a specific code that did not link to the potential treatment.
Supplements
The tested substances were administered to the participants as tablets prepared by Sankyo Co. (Tokyo, Japan). Tablets containing NIPPN flaxseed lignans (Nippon Flour Mills, Tokyo, Japan), a source of dietary SDG, were designated the FL supplements. NIPPN flaxseed lignans contained approximately 40% (w/w) SDG and 40% (w/w) αCD. Tablets containing Dexypearl-α (Ensuiko Sugar Refining, Tokyo, Japan), an almost pure αCD (>98%), were designated the αCD supplements. The placebo tablet contained maltitol instead of NIPPN flaxseed lignans or Dexypearl-α. Table S3 details the compositions of the placebo, FL supplement, and αCD supplement.
Study interventions
All participants who were allocated to one of the three groups ingested the corresponding test supplements (three tablets) daily for 9 weeks beginning on the first day of the experiment. The three tablets were ingested with water at the same time. Participants in the three groups were fed a placebo (placebo group), FL (200 mg/day, FL group), or αCD (200 mg/day, αCD group) as the supplement. Participants received a prescribed dinner on the days before clinic visits (300 g of white rice, a cup of miso soup, a Hamburg steak, and gomoku-ni). On clinic visit days, they received a prescribed breakfast (“cream and brown rice bran,” a nutritional supplement). These meals were delivered to the participants a few days before their consumption.
Prohibited foods
The following foods were prohibited: (i) foods and beverages that improve exercise performance; (ii) pharmaceuticals and quasi-drugs that are used to promote recovery or prevent and improve “fatigue” or “physical strength”; and (iii) dietary supplements (including “Foods with Functional Claims” and “Food for Specified Health Uses”). All participants were prohibited from consuming these foods starting at least 1 week before the start of the supplementation period until the end of the study.
Study outcomes
The primary outcomes were the V̇O2max and questionnaire responses regarding fatigue. The secondary outcomes were physical performance, VT, RPE, blood biochemistry, muscle and body fat mass, and gut microbiota composition. The participants allocated to one of the three groups were subjected to the designated tests (described below) at baseline (0) and after 4, 8, and 9 weeks at the Chiyoda Paramedical Care Clinic. The clinic staff collected the data. A physician monitored the condition of the participants during all tests. The assessments and visit schedules at −4, 0, 4, 8, and 9 weeks are depicted in Fig. 2B. Every participant who attended a visit completed all exercise tests.
O2max, VT, and maximum exercise intensity (performed on the visits at −4 and 9 weeks)
Exercise testing was conducted on an exercise bike (AEROBIKE-75XLIII; Konami Sports Life, Tokyo, Japan) at room temperature (23° ± 2°C) with 45 ± 5% relative humidity. The exercise load was incrementally increased by 15 W every minute until exhaustion. The oxygen and carbon dioxide concentrations in expired gas during exercise were measured using an Aero Monitor expired gas analyzer (AE-310s; Minato Medical Science Co., Osaka, Japan). The heart rate was monitored during the exercise. Exhaustion was determined on the basis of the following criteria: (i) The participant declared himself exhausted or was unable to continue exercising; (ii) the participant’s heart rate reached the estimated maximum for his age, defined as 220 − age; or (iii) a physician declared the participant exhausted. The maximum oxygen consumption and pedal resistance immediately before exhaustion were defined as the V̇O2max and maximum exercise intensity, respectively.
Fatigue questionnaire and RPE (performed on the visits at 0, 4, and 8 weeks)
The exercise testing was conducted with an exercise bike as described above in a room maintained at 23° ± 2°C with 45 ± 5% relative humidity. The pedaling resistance of the exercise bike was set to 45% of each participant’s maximum exercise intensity during testing. The participants maintained a pedal speed as close as possible to 60 rpm for 50 min. The RPE was measured with Borg’s RPE scale at 0, 10, 20, 30, 40, 45, and 50 min during the exercise. In addition, a VAS questionnaire was used to assess fatigue after exercise (0, 30, and 60 min after exercise). The participants completed the questionnaire themselves, and the questionnaire used in this study was created by the Japanese Society of Fatigue Science (www.hirougakkai.com/VAS.pdf).
Exercise performance (performed on the visits at 0, 4, and 8 weeks)
The effects of FL and αCD on exercise performance were evaluated on the basis of the time needed to pedal 10 km on an exercise bike (FBS-101; Fujimori Ltd., Takaoka, Japan). This test was conducted at room temperature (23° ± 2°C) with 45 ± 5% relative humidity. The intensity level (i.e., the resistance of the pedal) was set to 45% of each participant’s maximum exercise intensity. The participants were asked to pedal as fast as possible for 10 km. The clinic staff measured the time.
Blood testing (performed on the visits at 0, 4, and 8 weeks)
Blood was collected immediately before and after exercise to measure lactate, CPK, lactate dehydrogenase, glucose, creatine, free fatty acids, cortisol, amino acids, glucagon, interleukin-6 (IL-6), growth hormone, and diacron-reactive oxygen metabolite (dROM) levels. Blood was collected 60 min after exercise to measure lactate dehydrogenase, CPK, creatine, glucose, and lactate levels. Blood samples were collected in ethylenediaminetetraacetic acid disodium salt dihydrate-coated tubes for amino acid analysis; aprotinin-coated tubes for glucagon analysis; heparin-coated tubes for glucose analysis; perchloric acid–added tubes for lactate analysis; and noncoated tubes for CPK, creatine, lactate dehydrogenase, free fatty acids, cortisol, IL-6, growth hormone, and dROM analysis. All analyses except the dROM analysis were carried out at LSI Medience Co. (Tokyo, Japan), while the dROM analysis was performed with FREE Carrio DUO (Wismerll Co., Tokyo, Japan) at our laboratory.
Muscle and body fat mass (performed on the visits at 0, 4, and 8 weeks)
These parameters were measured using a body composition analyzer (InBody 570; InBody Japan Inc., Japan) before a 50-min constant-load bike exercise.
Gut microbiota (performed on the visits at 0, 4, and 8 weeks)
Fecal samples were collected at 0 and after 4 and 8 weeks of supplementation. Each participant collected the samples in fecal collection containers (Sarstedt) at their houses within 5 days before each visit. After collection, the samples were frozen at the participants’ houses, kept at approximately −30°C, and delivered to the clinic by each participant. As in the cross-sectional study, DNA was extracted from the samples, and gut microbiota was analyzed using the MiSeq platform (Illumina). The total number of analyzed sequence reads was 4,757,361. The mean, minimum, and maximum reads per sample were 52,859, 12,805, and 125,205, respectively; thus, data rarefied at 10,000 reads per sample were used to estimate the α- and β-diversities. B. uniformis abundance was measured using species-specific qPCR, as described above.
Statistical analyses
In this randomized, double-blind, placebo-controlled, parallel-group study, Mann-Whitney U tests were used to compare B. uniformis abundance between the two treatment groups and the placebo group, and Wilcoxon signed-rank tests were used to compare baseline and 4- or 8-week levels within each group. Except for microbial data, normally distributed continuous raw data were assessed using the Shapiro-Wilk test. All subsequent statistical tests were carried out on the basis of the results of normality tests. To determine the significance of the differences in parameters between the two treatment groups and the placebo group, an unpaired t test or the Mann-Whitney U test was used. A paired t test or Wilcoxon signed-rank test was used to compare baseline and 4- or 8-week levels within each group. These statistical analyses were conducted using R (version 4.0.2; R Foundation for Statistical Computing). Differences of P < 0.05 were considered statistically significant. In the box plots, the center line represents the median, and the box boundaries represent the 25th and 75th percentiles. The whiskers correspond to the most extreme values within 1.5 times the interquartile range below the 25th percentile and above the 75th percentile. Pie charts in fig. S3 show the mean relative abundance of each bacterial genus to illustrate gut microbial compositions. The participants’ baseline data are shown in table S1, expressed as the mean and SD values.
Mouse treatment, exercise regimen, and sample collection
B. uniformis and P. dorei cultivation
The B. uniformis JCM 5828 or P. dorei JCM 13471 type strains, provided by RIKEN BioResource Research Center (Ibaraki, Japan), were cultured on glucose blood liver (BL) agar plates (Nissui Pharmaceutical, Tokyo, Japan) or modified EG (Eggerth-Gagnon) agar plates (52) supplemented with 5% (v/v) horse blood for 48 hours at 37°C under anaerobic conditions [10% (v/v) CO2, 4% (v/v) H2, and 86% (v/v) N2], respectively. Colonies from the agar plates were collected and incubated overnight in 25 ml of Gifu anaerobic medium broth (Nissui Pharmaceutical) or modified EG medium broth supplemented with Fildes’ peptic digest of horse blood [5% (v/v)] at 37°C under anaerobic conditions, respectively. The culture was diluted and plated on BL or modified EG with horse blood agar to count viable bacterial cells. After 48 hours of culture, colonies on the plates were manually counted, and the CFU per milliliter was calculated.
Mouse housing
SPF male C57BL/6J mice (Japan SLC, Shizuoka, Japan) were individually housed in open-top cages covered by wire bar lids and maintained on a 12-hour light/12-hour dark cycle at room temperature (22° ± 3°C) with feed and water provided ad libitum.
Endurance exercise test
Physical performance was evaluated using a flowing water pool (length, 92 cm; width, 45 cm; height, 45.5 cm; water depth, 35 cm; Antitec, Otsu, Japan) (53, 54) developed at Kyoto University. The pool had six lanes (length, 22.5 cm; width, 7.0 cm), and the water temperature was maintained at 34° ± 0.5°C. The mice were fed the chow diet CE-2 (CLEA Rodent Diet CE-2; CLEA Japan, Tokyo, Japan). After a 1-week acclimation period, SPF male C57BL/6J mice (aged 7 weeks) were used in the experiment. The mice were then given a 1-week swimming acclimation period. The acclimation swimming sessions were performed every 2 days and consisted of the following: On day 1, the flow rate was maintained at 8 liter/min for 3 min, increased by 1 liter/min every 3 min to 12 liter/min, and maintained at 12 liter/min for 3 min; on day 2, the flow rate was maintained at 10 liter/min for 5 min, increased by 1 liter/min every 5 min to 12 liter/min, and maintained at 12 liter/min for 5 min, and on day 3, the flow rate was maintained at 12 liter/min for 5 min, 13 liter/min for 15 min, and 14 liter/min for 15 min. During these swimming periods, the swimming was stopped if a mouse reached exhaustion (i.e., failure to return to the water surface within 3 s). Three days after swimming acclimatization, the swimming endurance of the mice was tested to divide them into appropriate groups. The swimming test consisted of a flow rate program of 12 liter/min for 5 min, 13 liter/min for 5 min, and 14 liter/min until exhaustion. The time taken by each mouse to reach exhaustion, i.e., failure to return to the water surface within 3 s, was recorded. The mice were tested for their endurance levels and split into two groups with similar swimming times (PBS and B. uniformis or P. dorei treatment groups). The treatment group was orally administered B. uniformis JCM 5828T or P. dorei JCM 13471T (2 × 108 CFU, 200 μl of PBS per mouse) daily for four consecutive weeks, whereas the control group received sterile PBS. During this administration period, mice were subjected to a weekly swimming endurance test under the same conditions as the initial swimming endurance test. These experiments were approved by the Institutional Animal Care and Use Committee of New Drug Research Center Inc. (approval no.: 170428A and 180130A; approval date: May 2017 and January 2018; experimental period: May to July 2017 and February to May 2018, respectively).
B. uniformis– or PBS-administered mice with and without exercise
After the acclimation period, SPF male C57BL/6J mice (aged 5 weeks) were introduced to the CE-2 chow diet and used in the experiment. The mice were allocated into exercising and non-exercising groups based on their body weight. The non-exercising group was further divided into two subgroups (PBStra− and BUtra−). After swimming acclimation, the exercise group mice were divided into two subgroups (PBStra+ and BUtra+) with similar average swimming times. After grouping, mice in the B. uniformis treatment groups (BUtra− and BUtra+) were orally administered B. uniformis JCM 5828T (2 × 108 CFUs, 200 μl of PBS per mouse) daily for 10 weeks, whereas those in the PBS groups (PBStra− and PBStra+) received sterile PBS (Fig. 5A). During this administration period, the PBStra+ and BUtra+ group mice had weekly swimming exercise sessions in the flowing water pool for 10 weeks (Fig. 5A). The weekly exercise consisted of a flow rate program of 12 liter/min for 5 min and 13 liter/min for 10 min. Immediately after the swimming exercise in week 10, mice were euthanized under isoflurane anesthesia, and their liver, cecal content, and quadriceps femoris were collected and frozen in liquid nitrogen. This experiment was approved by the Institutional Animal Care and Use Committee of Sankyo Labo Service Corporation Inc., Tokyo, Japan (approval no.: SKL-A-17095; approval date: February 2018; experimental period: February to May 2018).
Administration of FL
After a 1-week acclimation period, male C57BL/6J SPF mice (aged 7 weeks) were housed in the laboratory. Subsequently, the mice were subjected to a swimming acclimation period, a swimming endurance capacity test for allocation, and grouping using the endurance exercise test protocols. After grouping, mice were fed the chow control diet CE-2 alone or supplemented with 5% (w/w) FL for 7 weeks. NIPPN flaxseed lignans (Nippon Flour Mills) was used as the FL. During the administration period, mice were subjected to a weekly swimming endurance test. Fecal samples were collected at day 0 and week 7 and immediately stored at −80°C. This experiment was performed following approval by the Institutional Animal Care and Use Committee of New Drug Research Center Inc. (approval no.: 171010D; approval date: October 2017; experimental period: October to December 2017).
Administration of αCD
The acclimation of the mice, exercise protocols, and fecal sampling were performed similarly to the administration of FL. For 7 weeks, mice were fed the chow control diet CE-2 alone or supplemented with 5% (w/w) αCD. Dexypearl-α (Ensuiko Sugar Refining) was used in the study. This experiment was performed following approval by the Institutional Animal Care and Use Committee of Asahi Quality and Innovations Ltd. (approval no.: 18-21-01; approval date: May 2018; experimental period: May to August 2018).
B. uniformis–monoassociated mice
Germ-free male C57BL/6N mice (aged 10 weeks; three to four animals per cage; Sankyo Labo Service Co.) were housed and maintained in a vinyl isolator at a constant temperature (23° ± 2°C). After 6 days of acclimation, on day 0, mice in the B. uniformis–monoassociated group were given a single oral dose of B. uniformis JCM 5828T (2 × 109 CFUs, 200 μl of PBS per mouse), while those in the control group (germ-free mouse group) received sterile PBS. On days 0 and 21 (the final day of the experiment), fecal samples were collected and immediately stored at −80°C. On day 21, the B. uniformis–monoassociated and germ-free mice were removed from the isolator and allowed to swim for 20 min or until exhaustion in the flowing water pool with six lanes (water temperature: 34° ± 0.5°C). The flow rate was maintained at 10 liter/min for 5 min, 11 liter/min for 5 min, and 12 liter/min for 10 min. During these exercises, swimming was stopped if a mouse reached exhaustion (i.e., failure to return to the water surface within 3 s). Immediately after the swimming, the mice were euthanized under isoflurane anesthesia, and their livers were collected, frozen, and stored at −80°C. The experiment was conducted following approval from the Institutional Animal Care and Use Committee of Sankyo Labo Service Corporation Inc. (approval no.: SKL-J18004; approval date: April 2018; experimental period: April to May 2018).
Fecal microbiota analysis
Fecal DNA was extracted from 50 mg of mice fecal samples using the same protocol described above for the human fecal samples. The quantification of B. uniformis using qPCR and 16S rRNA-encoding gene sequencing with MiSeq platform (Illumina) was also performed using the same protocol for human fecal samples.
RNA sequencing
Six mice from each group (PBStra−, PBStra+, BUtra−, and BUtra+) were randomly selected. Total liver RNA was extracted using the RNeasy Mini Kit (QIAGEN, Venlo, the Netherlands), and the subsequent steps were performed at Bioengineering Lab. Co. Ltd. (Sagamihara, Japan). Briefly, cDNA libraries were constructed using the KAPA Stranded RNA-Seq Kit (Kapa Biosystems, Wilmington, MA, USA) and FastGene Adapter Kit (Nippon Genetics, Tokyo, Japan), following the manufacturer’s protocol. The fragments were sequenced on the HiSeq X System (Illumina). For data analysis, read sequences (75 base pairs) were aligned to the Mus musculus (GRCm38.p6) reference genome (www.ncbi.nlm.nih.gov/assembly/GCF_000001635.26/) using HISAT2 version 2.1.0 (55). The gene-mapped reads in the reference sequence were counted using featureCounts version 1.6.3 (56). Differentially expressed genes were detected using edgeR version 3.20.9 following iDEGES normalization (57, 58). GO analysis was conducted using the DAVID website (https://david.ncifcrf.gov/) (59).
Reverse transcription qPCR
Total RNA from the liver or quadriceps femoris was extracted using the RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions. cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara Bio, Kusatsu, Japan). Reverse transcription qPCR was performed using the LightCycler 480 System (Roche Diagnostics) and TB Green Premix Ex Taq II (Takara Bio). The primer sequences for Pck1, Cpt1a, Hk1, Hk2, Glut4, and Pgc-1α have been reported previously (60–64). For Pygm, the Pygm Mouse qPCR Primer Pair (OriGene Technologies, Rockville, MD, USA) was used. The relative amounts of these mRNAs were normalized using Actb mRNA.
SCFA analysis
The SCFA measurements were performed at TechnoSuruga Laboratory Co. Ltd. (Shizuoka, Japan). We analyzed 100 mg of the cecal content or fecal samples using a gas chromatography–flame ionization detector system (7890B Gas Chromatograph System, Agilent Technologies, Santa Clara, CA, USA) fitted with a DB-WAXetr column (30 m, 0.25-mm internal diameter, 0.25-μm film thickness; Agilent Technologies) to measure SCFA levels, as previously reported (65).
Glycogen content
The liver and muscle glycogen content was measured using a Glycogen Colorimetric/Fluorometric Assay Kit (BioVision, Abcam, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, 10 mg of liver or quadriceps femoris was homogenized in 200 μl of cooled water and boiled for 10 min. The boiled samples were then centrifuged, and glucoamylase was added to the diluted supernatants to hydrolyze the glycogen into glucose. During a 30-min incubation at room temperature, glucose was oxidized with an OxiRed probe (Abcam) to generate color, and absorbance (optical density at 570 nm) was measured using a Synergy HTX Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT, USA).
Metabolite extraction and CE-TOFMS measurements
A quantitative analysis of charged metabolites in the quadriceps femoris was performed using CE-TOFMS as described previously (66, 67). Quadriceps femoris muscle samples (50 mg) were submerged in methanol (500 μl) containing internal standards [methionine sulfone (Wako, Tokyo, Japan), 2-(N-morpholino)ethane sulfonic acid (Dojindo Laboratories, Tokyo, Japan), and d-camphor-10-sulfonic acid (Wako) each at 20 μM]. The mixture was combined with four 3-mm zirconia beads (BioSpec Products, Bartlesville, OK, USA) and vigorously shaken for 5 min using a Shake Master NEO (Biomedical Science, Tokyo, Japan). Subsequently, deionized water (200 μl) and chloroform (500 μl) were added, and the suspension was vigorously shaken for 5 min and centrifuged at 4600g and 4°C for 30 min. The supernatant was filtered through a 5-kDa-cutoff filter column (Millipore, Burlington, MA, USA). The filtrate was concentrated via centrifugation (3 hours at 40°C) and dissolved in water containing reference compounds [200 μM each of 3-aminopyrrolidine (Sigma-Aldrich, St. Louis, MO, USA) and trimesic acid (Wako)] before CE-TOFMS analysis. As described previously (68), the CE-TOFMS experiments were performed using an Agilent CE-TOFMS system (Agilent Technologies).
Statistical analyses
Mann-Whitney U tests were used to compare B. uniformis or the relative abundance of Bacteroides across the groups. The Benjamin-Hochberg procedure was used to estimate RNA-seq false discovery rates (q values). The raw data from the other mouse experiments were tested for normality using the Shapiro-Wilk test, and subsequent tests were selected on the basis of the outcomes. The swimming endurance across the groups was assessed using the Mann-Whitney U test. The cecal SCFA (excluding isobutyrate); hepatic glycogen; liver Cpt1a transcript; and muscle HK2, Pygm, and Pgc1α levels were assessed using the Tukey-Kramer test. The isobutyrate, liver Pck1 transcript, muscle glycogen, and muscle Hk1 and Glut4 levels were assessed using the Steel-Dwass test. Acetate, propionate, and isovalerate in feces and glycogen and Cpt1a in the livers were compared between germ-free and B. uniformis–monoassociated mice using an unpaired t test; valerate, isobutyrate, and Pck1 levels were compared using the Mann-Whitney U test. These statistical analyses were conducted using SPSS (version 23; IBM) and R (version 4.0.2; R Foundation for Statistical Computing). Differences of P < 0.01 or 0.05 were considered statistically significant in GO analyses or other experiments. In the box plots, the center line represents the median, and the box boundaries represent the 25th and 75th percentiles. The whiskers correspond to the most extreme values within 1.5 times the interquartile range below the 25th percentile and above the 75th percentile.
Acknowledgments
We thank K. Washida and Y. Miyamoto (Asahi Quality & Innovations Ltd.) for the assessment of the αCD content in NIPPN flaxseed lignans. We also thank K. Morimoto (Asahi Quality & Innovations Ltd.) for the outstanding technical assistance. CPCC Company Ltd. served as the Contract Research Organization and Site Management Organization for the placebo-controlled study involving dietary supplements. We thank the company staff members involved in the study, especially the statistical analyst K. Sakano.
Funding: This work was partially supported by JSPS KAKENHI (22H03541 to S.F.), JST ERATO (JPMJER1902 to S.F.), AMED-CREST (JP22gm1010009 to S.F.), the Food Science Institute Foundation (to S.F.), and was funded by Asahi Quality & Innovations Ltd.
Author contributions: H.M., C.K., T.N., and S.F. designed the study. S.F. and T.N. managed the whole project. C.K. and H.M. conducted the animal and clinical studies. S.H. and Y.U. supervised the observational cross-sectional study of athletes. C.I., N.K., T.I., and A.H. conducted the metabolome analysis. H.M., C.K., and S.F. wrote the manuscript, and all the authors reviewed the manuscript.
Competing interests: C.K., H.M., and S.F. have applied for patents (PCT/JP2018/035295 field with the European Patent Office, China, Japan, and the United States) related to the use of antifatigue agents and/or physical strength-improving substances containing B. uniformis or a treated product thereof. H.M., C.K., and T.N. have filed two patent applications (PCT/JP2020/011262 and PCT/JP2020/011263) related to the use of sport performance–improving agents containing FL or αCD. H.M., C.K., and T.N. are current employees of Asahi Quality & Innovations Ltd. S.F. is the founder and CEO of Metagen Inc., a company dedicated to improving human health through gut environmental design. This organization had no control over the interpretation, writing, or publication of this work. Keio University is managing the terms of these arrangements according to its conflict-of-interest policies. All other authors declare that they have no competing interests.
Data and materials availability: The B. uniformis JCM 5828 and P. dorei JCM 13471 can be provided by RIKEN BioResource Research Center pending scientific review and a completed material transfer agreement. Requests for these strains should be submitted to RIKEN BioResource Research Center. The 16S rRNA gene sequence data have been submitted to the DNA Data Bank of Japan (DDBJ) with accession nos. DRA009619, DRA009620, DRA009664, and DRA009669. The accession number for the entire RNA-seq dataset reported in this manuscript is DRA009645. All remaining data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S3
REFERENCES AND NOTES
- 1.O. O’Sullivan, O. Cronin, S. F. Clarke, E. F. Murph, M. G. Molloy, F. Shanahan, P. D. Cotter, Exercise and the microbiota. Gut Microbes 6, 131–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.N. Mach, D. Fuster-Botella, Endurance exercise and gut microbiota: A review. J. Sport Health Sci. 6, 179–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Clark, N. Mach, The crosstalk between the gut microbiota and mitochondria during exercise. Front. Physiol. 8, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V. Cella, V. M. Bimonte, C. Sabato, A. Paoli, C. Baldari, M. Campanella, A. Lenzi, E. Ferretti, S. Migliaccio, Nutrition and physical activity-induced changes in gut microbiota: Possible implications for human health and athletic performance. Foods 10, 3075 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Marttinen, R. Ala-Jaakkola, A. Laitila, M. J. Lehtinen, Gut microbiota, probiotics and physical performance in athletes and physically active individuals. Nutrients 12, 2936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Y. J. Hsu, C. C. Chiu, Y. P. Li, W. C. Huang, Y. T. Huang, C. C. Huang, H. L. Chuang, Effect of intestinal microbiota on exercise performance in mice. J. Strength Cond. Res. 29, 552–558 (2015). [DOI] [PubMed] [Google Scholar]
- 7.W. C. Huang, Y. H. Chen, H. L. Chuang, C. C. Chiu, C. C. Huang, Investigation of the effects of microbiota on exercise physiological adaption, performance, and energy utilization using a gnotobiotic animal model. Front. Microbiol. 10, 1906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T. Okamoto, K. Morino, S. Ugi, F. Nakagawa, M. Lemecha, S. Ida, N. Ohashi, D. Sato, Y. Fujita, H. Maegawa, Microbiome potentiates endurance exercise through intestinal acetate production. Am. J. Physiol. Endocrinol. Metab. 316, E956–E966 (2019). [DOI] [PubMed] [Google Scholar]
- 9.J. Scheiman, J. M. Luber, T. A. Chavkin, T. MacDonald, A. Tung, L. D. Pham, M. C. Wibowo, R. C. Wurth, S. Punthambaker, B. T. Tierney, Z. Yang, M. W. Hattab, J. Avila-Pacheco, C. B. Clish, S. Lessard, G. M. Church, A. D. Kostic, Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 25, 1104–1109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. C. Evans, K. J. LePard, J. W. Kwak, M. C. Stancukas, S. Laskowski, J. Dougherty, L. Moulton, A. Glawe, Y. Wang, V. Leone, D. A. Antonopoulos, D. Smith, E. B. Chang, M. J. Ciancio, Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLOS ONE 9, e92193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. S. Kang, P. R. Jeraldo, A. Kurti, M. E. Miller, M. D. Cook, K. Whitlock, N. Goldenfeld, J. A. Woods, B. A. White, N. Chia, J. D. Fryer, Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol. Neurodegener. 9, 36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. F. Clarke, E. F. Murphy, O. O’Sullivan, A. J. Lucey, M. Humphreys, A. Hogan, P. Hayes, M. O’Reilly, I. B. Jeffery, R. Wood-Martin, D. M. Kerins, E. Quigley, R. P. Ross, P. W. O’Toole, M. G. Molloy, E. Falvey, F. Shanahan, P. D. Cotter, Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920 (2014). [DOI] [PubMed] [Google Scholar]
- 13.S. Mörkl, S. Lackner, W. Müller, G. Gorkiewicz, K. Kashofer, A. Oberascher, A. Painold, A. Holl, P. Holzer, A. Meinitzer, H. Mangge, S. Holasek, Gut microbiota and body composition in anorexia nervosa inpatients in comparison to athletes, overweight, obese, and normal weight controls. Int. J. Eat. Disord. 50, 1421–1431 (2017). [DOI] [PubMed] [Google Scholar]
- 14.J. Qin, R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, S. Li, M. Jian, Y. Zhou, Y. Li, X. Zhang, S. Li, N. Qin, H. Yang, J. Wang, S. Brunak, J. Doré, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach; MetaHIT Consortium, P. Bork, S. D. Ehrlich, J. Wang, A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L. Monteiro, D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnet, R. Vidal, J. Cabrita, F. Mégraud, Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35, 995–998 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Oren, G. M. Garrity, List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 70, 2960–2966 (2020). [DOI] [PubMed] [Google Scholar]
- 17.M. L. Fernández-Murga, Y. Sanz, Safety assessment of Bacteroides uniformis CECT 7771 isolated from stools of healthy breast-fed infants. PLOS ONE 11, e0145503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.E. M. Gómez Del Pulgar, A. Benítez-Páez, Y. Sanz, Safety assessment of Bacteroides uniformis CECT 7771, a symbiont of the gut microbiota in infants. Nutrients 12, 551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. X. Li, H. Y. Zhu, D. H. Yang, X. Q. Ma, C. Z. Wang, S. Q. Cai, G. R. Liu, B. S. Ku, S. L. Liu, Production of secoisolariciresinol from defatted flaxseed by bacterial biotransformation. J. Appl. Microbiol. 113, 1352–1361 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Y. L. Tao, D. H. Yang, Y. T. Zhang, Y. Zhang, Z. Q. Wang, Y. S. Wang, S. Q. Cai, S. L. Liu, Cloning, expression, and characterization of the β-glucosidase hydrolyzing secoisolariciresinol diglucoside to secoisolariciresinol from Bacteroides uniformis ZL1. Appl. Microbiol. Biotechnol. 98, 2519–2531 (2014). [DOI] [PubMed] [Google Scholar]
- 21.P. Johnsson, A. Kamal-Eldin, L. N. Lundgren, P. Aman, HPLC method for analysis of secoisolariciresinol diglucoside in flaxseeds. J. Agric. Food Chem. 48, 5216–5219 (2000). [DOI] [PubMed] [Google Scholar]
- 22.N. Nihei, H. Okamoto, T. Furune, N. Ikuta, K. Sasaki, G. Rimbach, Y. Yoshikawa, K. Terao, Dietary α-cyclodextrin modifies gut microbiota and reduces fat accumulation in high-fat-diet-fed obese mice. Biofactors 44, 336–347 (2018). [DOI] [PubMed] [Google Scholar]
- 23.A. W. Midgley, L. R. McNaughton, A. M. Jones, Training to enhance the physiological determinants of long-distance running performance: Can valid recommendations be given to runners and coaches based on current scientific knowledge? Sports Med. 37, 857–880 (2007). [DOI] [PubMed] [Google Scholar]
- 24.G. Borg, H. Dahlström, A pilot study of perceived exertion and physical working capacity. Acta Soc. Med. Ups. 67, 21–27 (1962). [PubMed] [Google Scholar]
- 25.G. Wolff, K. A. Esser, Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med. Sci. Sports Exerc. 44, 1663–1670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.H. Sasaki, Y. Hattori, Y. Ikeda, M. Kamagata, S. Iwami, S. Yasuda, Y. Tahara, S. Shibata, Forced rather than voluntary exercise entrains peripheral clocks via a corticosterone/noradrenaline increase in PER2::LUC mice. Sci. Rep. 6, 27607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. Eaton, K. Fukumoto, N. Paladio Duran, A. Pierro, L. Spitz, P. A. Quant, K. Bartlett, Carnitine palmitoyl transferase I and the control of myocardial β-oxidation flux. Biochem. Soc. Trans. 29, 245–249 (2001). [DOI] [PubMed] [Google Scholar]
- 28.P. Latorre-Muro, J. Baeza, E. A. Armstrong, R. Hurtado-Guerrero, F. Corzana, L. E. Wu, D. A. Sinclair, P. López-Buesa, J. A. Carrodeguas, J. M. Denu, Dynamic acetylation of phosphoenolpyruvate carboxykinase toggles enzyme activity between gluconeogenic and anaplerotic reactions. Mol. Cell 71, 718–732.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K. Yu, Y. Zhang, H. Chen, W. Zhu, Hepatic metabolomic and transcriptomic responses induced by cecal infusion of sodium propionate in a fistula pig model. J. Agric. Food Chem. 67, 13073–13081 (2019). [DOI] [PubMed] [Google Scholar]
- 30.T. Kondo, M. Kishi, T. Fushimi, T. Kaga, Acetic acid upregulates the expression of genes for fatty acid oxidation enzymes in liver to suppress body fat accumulation. J. Agric. Food Chem. 57, 5982–5986 (2009). [DOI] [PubMed] [Google Scholar]
- 31.L. Rui, Energy metabolism in the liver. Compr. Physiol. 4, 177–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Nocito, A. S. Kleckner, E. J. Yoo, A. R. Jones Iv, M. Liesa, B. E. Corkey, The extracellular redox state modulates mitochondrial function, gluconeogenesis, and glycogen synthesis in murine hepatocytes. PLOS ONE 10, e0122818 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. den Besten, K. Lange, R. Havinga, T. H. van Dijk, A. Gerding, K. van Eunen, M. Müller, A. K. Groen, G. J. Hooiveld, B. M. Bakker, D. J. Reijngoud, Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G900–G910 (2013). [DOI] [PubMed] [Google Scholar]
- 34.A. Tirosh, E. S. Calay, G. Tuncman, K. C. Claiborn, K. E. Inouye, K. Eguchi, M. Alcala, M. Rathaus, K. S. Hollander, I. Ron, R. Livne, Y. Heianza, L. Qi, I. Shai, R. Garg, G. S. Hotamisligil, The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci. Transl. Med. 11, eaav0120 (2019). [DOI] [PubMed] [Google Scholar]
- 35.M. Burnle, A. M. Jones, Oxygen uptake kinetics as a determinant of sports performance. Eur. J. Sport Sci. 7, 63–79 (2007). [Google Scholar]
- 36.H. S. MacRae, K. M. Mefferd, Dietary antioxidant supplementation combined with quercetin improves cycling time trial performance. Int. J. Sport Nutr. Exerc. Metab. 16, 405–419 (2006). [DOI] [PubMed] [Google Scholar]
- 37.K. E. Lansley, P. G. Winyard, S. J. Bailey, A. Vanhatalo, D. P. Wilkerson, J. R. Blackwell, M. Gilchrist, N. Benjamin, A. M. Jones, Acute dietary nitrate supplementation improves cycling time trial performance. Med. Sci. Sports Exerc. 43, 1125–1131 (2011). [DOI] [PubMed] [Google Scholar]
- 38.T. Suzuki, M. Morita, Y. Kobayashi, A. Kamimura, Oral l-citrulline supplementation enhances cycling time trial performance in healthy trained men: Double-blind randomized placebo-controlled 2-way crossover study. J. Int. Soc. Sports Nutr. 13, 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. B. Comerford, J. D. Artiss, K. L. Jen, S. E. Karakas, The beneficial effects α-cyclodextrin on blood lipids and weight loss in healthy humans. Obesity (Silver Spring) 19, 1200–1204 (2011). [DOI] [PubMed] [Google Scholar]
- 40.M. J. Amar, M. Kaler, A. B. Courville, R. Shamburek, M. Sampson, A. T. Remaley, Randomized double blind clinical trial on the effect of oral α-cyclodextrin on serum lipids. Lipids Health Dis. 15, 115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. D. Buckley, A. A. Thorp, K. J. Murphy, P. R. Howe, Dose-dependent inhibition of the post-prandial glycaemic response to a standard carbohydrate meal following incorporation of α-cyclodextrin. Ann. Nutr. Metab. 50, 108–114 (2006). [DOI] [PubMed] [Google Scholar]
- 42.G. Grunberger, K. L. Jen, J. D. Artiss, The benefits of early intervention in obese diabetic patients with FBCx: A new dietary fibre. Diabetes Metab. Res. Rev. 23, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 43.A. A. Kolodziejczyk, D. Zheng, E. Elinav, Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 742–753 (2019). [DOI] [PubMed] [Google Scholar]
- 44.D. Rothschild, O. Weissbrod, E. Barkan, A. Kurilshikov, T. Korem, D. Zeevi, P. I. Costea, A. Godneva, I. N. Kalka, N. Bar, S. Shilo, D. Lador, A. V. Vila, N. Zmora, M. Pevsner-Fischer, D. Israeli, N. Kosower, G. Malka, B. C. Wolf, T. Avnit-Sagi, M. Lotan-Pompan, A. Weinberger, Z. Halpern, S. Carmi, J. Fu, C. Wijmenga, A. Zhernakova, E. Elinav, E. Segal, Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 45.D. M. Tourlousse, K. Narita, T. Miura, M. Sakamoto, A. Ohashi, K. Shiina, M. Matsuda, D. Miura, M. Shimamura, Y. Ohyama, A. Yamazoe, Y. Uchino, K. Kameyama, S. Arioka, J. Kataoka, T. Hisada, K. Fujii, S. Takahashi, M. Kuroiwa, M. Rokushima, M. Nishiyama, Y. Tanaka, T. Fuchikami, H. Aoki, S. Kira, R. Koyanagi, T. Naito, M. Nishiwaki, H. Kumagai, M. Konda, K. Kasahara, M. Ohkuma, H. Kawasaki, Y. Sekiguchi, J. Terauchi, Validation and standardization of DNA extraction and library construction methods for metagenomics-based human fecal microbiome measurements. Microbiome 9, 95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S. M. Owens, J. Betley, L. Fraser, M. Bauer, N. Gormley, J. A. Gilbert, G. Smith, R. Knight, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Peña, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld, R. Knight, QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett, C. Huttenhower, Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. Tong, C. Liu, P. Summanen, H. Xu, S. M. Finegold, Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 17, 64–68 (2011). [DOI] [PubMed] [Google Scholar]
- 50.T. Odamaki, J. Z. Xiao, M. Sakamoto, S. Kondo, T. Yaeshima, K. Iwatsuki, H. Togashi, T. Enomoto, Y. Benno, Distribution of different species of the Bacteroides fragilis group in individuals with Japanese cedar pollinosis. Appl. Environ. Microbiol. 74, 6814–6817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.B. E. Ainsworth, W. L. Haskell, S. D. Herrmann, N. Meckes, D. R. Bassett Jr., C. Tudor-Locke, J. L. Greer, J. Vezina, M. C. Whitt-Glover, A. S. Leon, 2011 Compendium of physical activities: A second update of codes and MET values. Med. Sci. Sports Exerc. 43, 1575–1581 (2011). [DOI] [PubMed] [Google Scholar]
- 52.H. Sugahara, T. Odamaki, S. Fukuda, T. Kato, J. Z. Xiao, F. Abe, J. Kikuchi, H. Ohno, Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci. Rep. 5, 13548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.K. Matsumoto, K. Ishihara, K. Tanaka, K. Inoue, T. Fushiki, An adjustable-current swimming pool for the evaluation of endurance capacity of mice. J. Appl. Physiol. 81, 1843–1849 (1996). [DOI] [PubMed] [Google Scholar]
- 54.K. Ishihara, A. Yamada, Y. Mita, A. Goto, T. Ishimi, H. Mabuchi, K. Inoue, T. Fushiki, K. Yasumoto, Improved swimming pool achieves higher reproducibility and sensitivity to effect of food components as ergogenic AIDS. J. Nutr. Sci. Vitaminol. (Tokyo) 55, 301–308 (2009). [DOI] [PubMed] [Google Scholar]
- 55.D. Kim, J. M. Paggi, C. Park, C. Bennett, S. L. Salzberg, Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Liao, G. K. Smyth, W. Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 57.M. D. Robinson, D. J. McCarthy, G. K. Smyth, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. Sun, T. Nishiyama, K. Shimizu, K. Kadota, TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14, 219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. W. Huang, B. T. Sherman, R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 60.R. W. Hunter, C. C. Hughey, L. Lantier, E. I. Sundelin, M. Peggie, E. Zeqiraj, F. Sicheri, N. Jessen, D. H. Wasserman, K. Sakamoto, Metformin reduces liver glucose production by inhibition of fructose-1-6-bisphosphatase. Nat. Med. 24, 1395–1406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. M. Mihaylova, C. W. Cheng, A. Q. Cao, S. Tripathi, M. D. Mana, K. E. Bauer-Rowe, M. Abu-Remaileh, L. Clavain, A. Erdemir, C. A. Lewis, E. Freinkman, A. S. Dickey, A. R. La Spada, Y. Huang, G. W. Bell, V. Deshpande, P. Carmeliet, P. Katajisto, D. M. Sabatini, Ö. H. Yilmaz, Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R. Dentin, J. P. Pégorier, F. Benhamed, F. Foufelle, P. Ferré, V. Fauveau, M. A. Magnuson, J. Girard, C. Postic, Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J. Biol. Chem. 279, 20314–20326 (2004). [DOI] [PubMed] [Google Scholar]
- 63.J. M. Gurley, B. A. Griesel, A. L. Olson, Increased skeletal muscle GLUT4 expression in obese mice after voluntary wheel running exercise is posttranscriptional. Diabetes 65, 2911–2919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R. Nakao, S. Shimba, K. Oishi, Ketogenic diet induces expression of the muscle circadian gene Slc25a25 via neural pathway that might be involved in muscle thermogenesis. Sci. Rep. 7, 2885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R. García-Villalba, J. A. Giménez-Bastida, M. T. García-Conesa, F. A. Tomás-Barberán, J. Carlos Espín, M. Larrosa, Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 35, 1906–1913 (2012). [DOI] [PubMed] [Google Scholar]
- 66.T. Soga, Y. Ohashi, Y. Ueno, H. Naraoka, M. Tomita, T. Nishioka, Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2, 488–494 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Y. Akiyama, Y. Takeuchi, K. Kikuchi, E. Mishima, Y. Yamamoto, C. Suzuki, T. Toyohara, T. Suzuki, A. Hozawa, S. Ito, T. Soga, T. Abe, A metabolomic approach to clarifying the effect of AST-120 on 5/6 nephrectomized rats by capillary electrophoresis with mass spectrometry (CE-MS). Toxins (Basel) 4, 1309–1322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.E. Mishima, S. Fukuda, H. Shima, A. Hirayama, Y. Akiyama, Y. Takeuchi, N. N. Fukuda, T. Suzuki, C. Suzuki, A. Yuri, K. Kikuchi, Y. Tomioka, S. Ito, T. Soga, T. Abe, Alteration of the intestinal environment by lubiprostone is associated with amelioration of adenine-induced CKD. J. Am. Soc. Nephrol. 26, 1787–1794 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S3