Abstract
Single-cell barcoding technologies enable genome sequencing of thousands of individual cells in parallel, but with extremely low sequencing coverage (<0.05) per cell. While the total copy number of large multi-megabase segments can be derived from such data, important allele-specific mutations – such as copy-neutral loss-of-heterozygosity (LOH) in cancer – are missed. We introduce Copy-number Haplotype Inference in Single-cells using Evolutionary Links (CHISEL), a method to infer allele- and haplotype-specific copy numbers in single cells and subpopulations of cells by aggregating sparse signal across hundreds or thousands of individual cells. We applied CHISEL to 10 single-cell sequencing datasets of cells from two breast cancer patients. We identified extensive allele-specific copy-number aberrations (CNAs) in these samples, including copy-neutral LOHs, whole-genome duplications (WGDs), and mirrored-subclonal CNAs. These allele-specific CNAs affect genomic regions containing well-known breast cancer genes. We also refined the reconstruction of tumor evolution, timing allele-specific CNAs before and after WGDs, identifying low-frequency subpopulations distinguished by unique CNAs, and uncovering evidence of convergent evolution.
Introduction
Single-cell DNA sequencing is a promising technology to quantify tumor heterogeneity and evolution with unprecedented resolution, enabling the identification of rare subpopulations of cells with distinct mutations and the inference of the evolutionary dynamics of cancer1–4. Recently, single-cell barcoding technologies, including the Chromium Single Cell CNV Solution from 10x Genomics 5 and direct library preparation 6,7, have been used to perform low-coverage whole-genome sequencing of thousands of individual cells in parallel, overcoming the limited number of cells and the amplification/coverage biases of previous techniques. Due to technical and financial limitations, these technologies have extremely low sequencing coverage (0.05 per cell) which has thus far limited their application to the detection of large (3–5Mb) CNAs in individual cells. CNAs alter the number of copies of genomic regions, are frequent somatic mutations that drive cancer development 8–11, play a crucial role in cancer treatment and prognosis 12, 13, and provide important markers for reconstruction of cancer evolution 14–18.
Since the human genome is diploid, each CNA affects one allele of a genomic region located on either of the two homologous chromosomes (maternal and paternal), called haplotypes. Many methods have been developed to identify allele-specific copy numbers, which indicate the number of copies of each homolog, from bulk tumor sequencing data 19–25. Moreover, multiple cancer studies have demonstrated the importance of deriving allele-specific copy numbers 9, 26–28. For example, copy-neutral LOH – where one allele is lost and the other duplicated so the total copy number remains 2 – is common in many cancers 26, 29–32. Allele-specific copy numbers have also been shown to be essential for accurate inference of WGDs 20, 33 and timing WGDs relative to other CNAs 9, 20.
Despite the demonstrated importance of allele-specific copy numbers, previous single-cell sequencing studies have assumed that low-coverage data is too shallow to obtain allele-specific information from single cells 6, 7, 34–36. Existing methods for identifying CNAs from single-cell sequencing data 5–7, 35–39 are limited to the inference of total copy number, which indicates only the sum of copy numbers at each locus, by analyzing differences between the observed and expected number of sequencing reads aligned to a locus, or the read-depth ratio (RDR). The signal to detect allele-specific copy numbers is the B-allele frequency (BAF), or relative proportions of reads from the two alleles of a genomic region; however, standard methods to calculate the BAF from individual germline heterozygous single-nucleotide polymorphisms (SNPs) do not work with extremely low coverage sequencing data.
We introduce CHISEL, a method that infers allele-specific and haplotype-specific copy numbers in single cells from low-coverage DNA sequencing data. CHISEL amplifies the extremely weak signal in individual SNPs into a sufficiently strong signal to compute BAFs in genomic regions of modest size (5Mb) by combining reference-based phasing methods with a novel algorithm to phase short haplotype blocks across cells. CHISEL further phases allele-specific copy numbers across cells using an evolutionary model to derive haplotype-specific copy numbers that indicate the number of copies of the alleles located on the same haplotype in individual cells. CHISEL includes several other innovative features, including global clustering of RDRs and BAFs along the whole genome and across all cells, and integrating BAFs in the challenging inference of the genome ploidy of individual cells.
We applied CHISEL to analyze 10 single-cell datasets from 2 breast cancer patients, each dataset containing 2 000 cells. CHISEL identified extensive allele-specific CNAs in these samples, including copy-neutral LOH, WGDs, and mirrored-subclonal CNAs. These latter events are haplotype-specific CNAs that alter the number of copies of the two distinct alleles of the same genomic region in different cells 40. We used the haplotype-specific copy numbers derived by CHISEL to reconstruct a more refined and accurate view of tumor heterogeneity than previous total copy-number analysis. We identified events that alter the copy number of well-known breast cancer genes and characterize key mechanisms in tumor progression, including potential precursors of WGDs and evidence of convergent evolution. Finally, we identified somatic single-nucleotide variants (SNVs) in subpopulations of cells with distinct haplotype-specific copy-number profiles. These SNVs provide orthogonal evidence for the phylogeny inferred by CHISEL compared to the phylogeny inferred by total copy numbers. Additionally, the variant-allele frequencies (VAFs) of SNVs and the spatial distribution of clones further support the CHISEL phylogeny. CHISEL provides a tool to realize the potential of single-cell whole genome sequencing for studies of tumor heterogeneity and evolution.
Results
CHISEL Algorithm
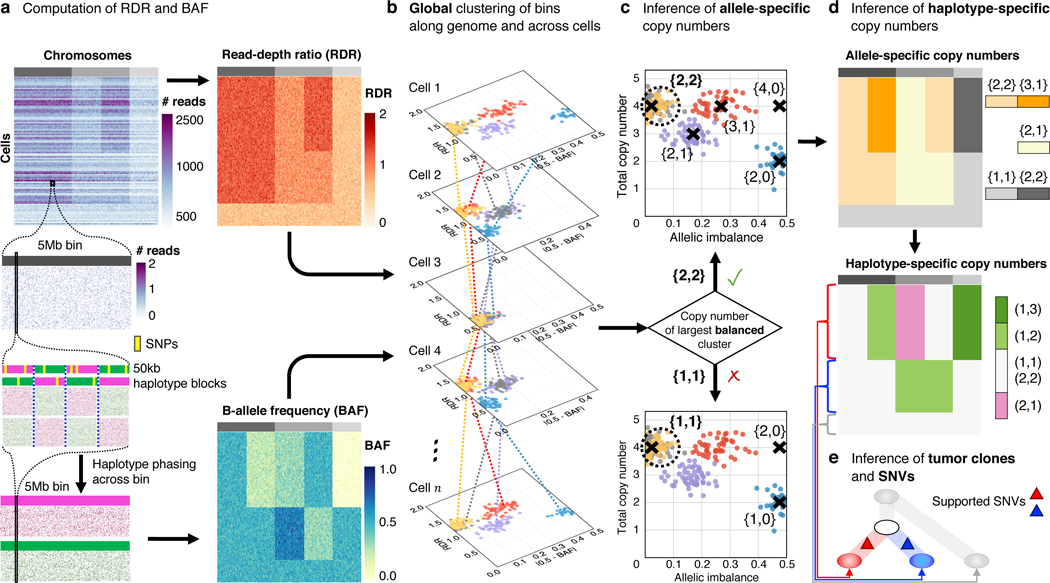
We developed the CHISEL algorithm to identify allele- and haplotype-specific CNAs from low-coverage single-cell DNA sequencing data (Fig. 1). CHISEL leverages information across hundreds to thousands of individual cells to overcome low sequencing coverage (0.05) per cell. As in DNA sequencing of bulk samples 19–25, CHISEL uses two quantities derived from aligned reads to estimate the number of copies, and , of the two alleles of each genomic region . The first quantity, the RDR , is directly proportional to the total copy number . The second quantity, the BAF , measures the imbalance between the number of copies of the two alleles and corresponds to either or . The key steps of CHISEL are: (1) computation of RDR and BAF in low-coverage DNA sequencing data from individual cells; (2) global clustering of RDRs and BAFs into a small number of copy-number states jointly across the entire genome of all cells; (3) inference of the pair of allele-specific copy numbers accounting for varying genome ploidy; (4) inference of haplotype-specific copy numbers by phasing allele-specific copy numbers to their corresponding haplotypes across all cells; (5) inference of tumor clones by clustering of haplotype-specific copy numbers. We briefly describe these steps below and provide additional details in Methods.
Fig. 1: The CHISEL algorithm.
a, CHISEL computes RDRs and BAFs in low-coverage ( per cell) single-cell DNA sequencing data (top left). Read counts from individual cells (rows) in Mb genomic bins (columns) across three chromosomes (grey rectangles in first row) are shown. For each bin in each cell, CHISEL computes the RDR (top) by normalizing the observed read counts. CHISEL computes the BAF in each bin and cell (bottom) by first performing referenced-based phasing of germline SNPs in kb haplotype blocks (magenta and green) and then phasing all these blocks jointly across all cells. b, CHISEL clusters RDRs and BAFs globally along the genome and jointly across all cells resulting here in 5 clusters of genomic bins (red, blue, purple, yellow, and grey) with distinct copy-number states. c, CHISEL infers a pair of allele-specific copy numbers for each cluster by determining whether the allele-specific copy numbers of the largest balanced (BAF ) cluster are equal to (diploid), (tetraploid), or are higher ploidy. d, CHISEL infers haplotype-specific copy numbers by phasing the allele-specific copy numbers consistently across all cells. e, CHISEL clusters tumor cells into clones according to their haplotype-specific copy numbers. Here, a diploid clone (light gray) and two tumor clones (red and blue) are obtained. A phylogenetic tree describes the evolution of these clones. Somatic SNVs are derived from pseudo-bulk samples and placed on the branches of the tree.
In steps (1)-(3), CHISEL infers allele-specific copy numbers in individual cells. First, CHISEL divides the genome into bins of fixed size (here 5Mb). For each bin in every cell, CHISEL computes the RDR using a standard normalization of the number of reads that aligned to . Next, CHISEL computes the BAF by first using reference-based phasing algorithms 41 to aggregate the individual SNPs in bin into haplotype blocks of fixed size (here 50kb) and then phasing these blocks into the two alleles of jointly across all cells (Fig. 1a). To our knowledge, no previous algorithm before CHISEL calculates BAF in low-coverage single-cell DNA sequencing data. In step (2), CHISEL globally clusters RDRs and BAFs into a small number of copy-number states across every bin and cell (Fig. 1b). This global clustering approach extends the one introduced in HATCHet 25 for multi-sample bulk sequencing data. The global clustering leverages information along the entire genome and across all cells; in contrast, current methods 5–7, 35–39 locally cluster RDRs of neighboring bins into segments and, with one recent exception 39, analyze each cell independently. Finally in step (3), CHISEL infers the pair of allele-specific copy numbers for each bin by identifying the largest balanced (BAF ) cluster and using a model-selection criterion to select the copy numbers (Fig. 1c). In contrast, existing approaches rely on the inference of the genome ploidy (or equivalent factors) from only RDRs 5–7, 35–39 and apply restrictive assumptions to select among many equally-plausible solutions without utilizing BAFs; this may result in selecting copy numbers that contradict the underlying allelic balance/imbalance.
In steps (4) and (5), CHISEL infers haplotype-specific copy numbers for each bin in individual cells and clusters cells into clones according to these copy numbers. In contrast to the unordered pair of allele-specific copy numbers, the ordered pair of haplotype-specific copy numbers indicates the number of copies of the alleles on each of the two homologous chromosomes, or haplotypes, and . One cannot directly determine haplotype-specific copy numbers of an individual cell from allele-specific copy numbers as we do not know the phase of each copy number, i.e. whether or . The key insight in deriving haplotype-specific copy numbers is to leverage the shared evolutionary history of the cells in a tumor and thus phase allele-specific copy numbers jointly across cells (Fig. 1d). CHISEL infers the phasing that minimizes the number of CNAs required to explain the haplotype-specific copy numbers using a model of interval events for CNA evolution 16–18. This approach generalizes and extends the method introduced by Jamal-Hanjani et al. 40 to infer haplotype-specific copy numbers in multiple bulk-tumor samples. Finally, in step (5) CHISEL clusters cells into clones according to the inferred haplotype-specific copy numbers (Fig. 1e).
Allele-specific copy-number aberrations
We applied CHISEL to 10x Genomics Chromium single-cell DNA sequencing data from 2 breast cancer patients: patient S0 with 5 publicly available sequenced tumor sections and patient S1 with 5 previously unpublished datasets. Each of the 10 datasets comprises 2 000 cells that were sequenced with coverage ranging from 0.01 to 0.05 per cell. CHISEL identified extensive allele-specific CNAs that were previously uncharacterized by total copy-number analysis. Across all the datasets, we found that allele-specific CNAs alter 25% of the genome on average in at least 100 cells (Supplementary Fig. 1). CHISEL also further improved the inference of total copy numbers (Supplementary Fig. 2‒4 and Supplementary Results 1).
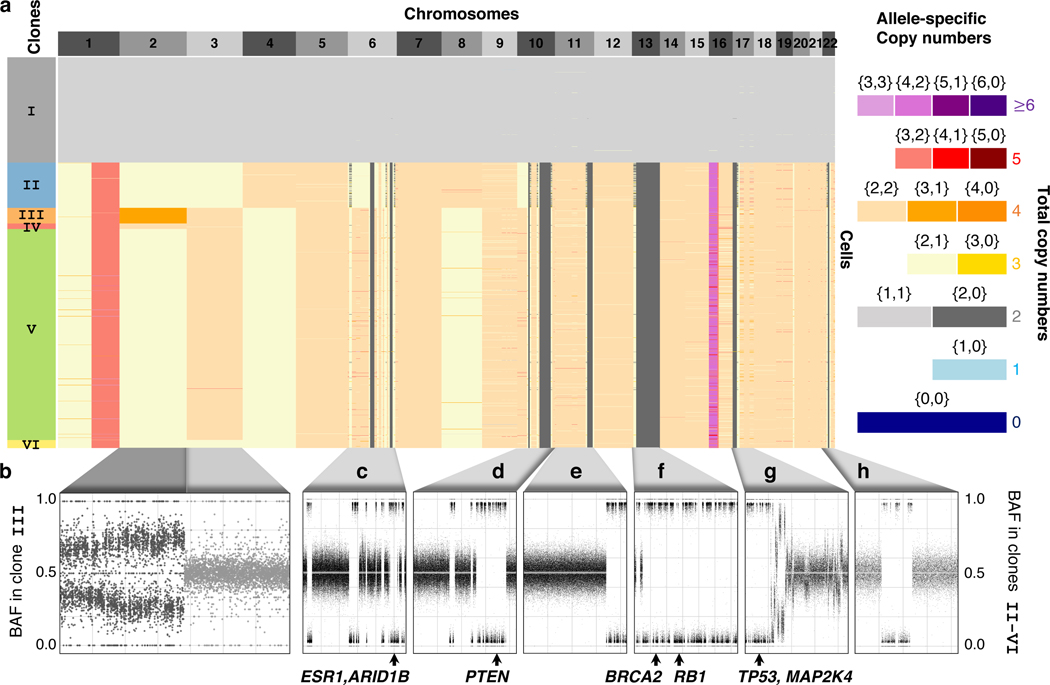
In patient S0, CHISEL identified 5–6 clones in each section that together comprise 70–92% of cells (Supplementary Fig. 5‒9). In patient S1, CHISEL identified 2–3 clones in each section that together comprise 81–93% of cells (Supplementary Fig. 1). For example, in section E of patient S0, CHISEL assigned 1448/2075 of cells to 6 clones, including one diploid clone (labeled I) and 5 aneuploid clones (labeled II - VI) (Fig. 2a). Since a single diploid clone and one or more aneuploid clones were identified in each tumor section, we concluded that the diploid clone comprises mostly normal cells while the aneuploid clones comprise tumor cells, in concordance with previous analysis. The remaining 7–30% of cells are unclassified and the proportions of such cells are consistent with previously reported causes 5, including cells in S-phase of the cell cycle with actively replicating DNA (12–42%), cells with a low number of sequenced reads (8%), and doublets (2%). Interestingly, we found direct evidence of doublets in a small number of cells of patient S1 (Supplementary Fig. 10).
Fig. 2: CHISEL reliably identifies allele-specific copy numbers.
a, Allele-specific copy numbers inferred by CHISEL across 1448 cells from section E of breast cancer patient S0. Clustering of cells according to allele-specific copy numbers reveals 6 clones (colored boxes on left). Clones III and IV are distinguished by an allele-specific CNA affecting the entire chromosome 2; cells in both these clones have 4 total copies of chromosome 2 but distinct allele-specific copy numbers equal to and , respectively. b, The BAF (computed in kb haplotype blocks) of cells from clone III exhibits a clear shift away from BAF=0.5 in chromosome 2, supporting the unequal copy numbers of the two alleles on this chromosome. In contrast, chromosome 3 shows BAF0.5, supporting equal copy numbers of the two alleles. c-h, Copy-neutral LOHs are the most frequent allele-specific CNAs identified by CHISEL with examples shown on six chromosomes. All of these regions have a total copy number equal to 2, the same as the cells in the diploid clone I, but have allele-specific copy numbers equal to . Each of these copy-neutral LOHs is supported by BAFs approximately equal to 0 or 1 indicating the complete loss of one allele across all tumors cells. Known breast-cancer genes in each LOH region are indicated below each plot.
We found that allele-specific CNAs were important for resolving the clonal organization of the tumor. For example, in section E of patient S0, allele-specific copy numbers on chromosome 2 distinguish clones III and IV (Fig. 2a). Chromosome 2 has the same total copy number in the cells of both clones III and IV but different allele-specific copy numbers of and , respectively. Tumor clones III and IV are thus indistinguishable using total copy numbers (Supplementary Fig. 11). We found that BAFs support these allele-specific copy numbers with a clear shift above and below 0.5 observed along the entire chromosome 2 in the cells of clone III (Fig. 2b). The same allele-specific CNA is also observed in other sections from the same patient (Supplementary Fig. 5‒8).
The most common type of allele-specific CNAs identified by CHISEL in both patients S0 and S1 are copy-neutral LOHs (Supplementary Fig. 1), which have allele-specific copy numbers equal to . Copy-neutral LOHs are invisible to total copy-number analysis methods as they are indistinguishable from normal diploid regions of the genome. We found copy-neutral LOHs in patient S0 in regions containing multiple genes implicated in breast cancer 42 including ESR1 and ARID1B on chromosome 6q, PTEN on chromosome 10q, BRCA2 and RB1 on chromosome 13, and MAP2K4 on chromosome 17p (Fig. 2c–h). We also found copy-neutral LOHs in patient S1 in regions containing the genes ESR1 and ARID1B on chromosome 6q. Notably, CHISEL identified most of these copy-neutral LOHs to be clonal as they are present in nearly all tumor cells, suggesting an early acquisition of these mutations during the tumor evolution. These clonal copy-neutral LOHs are strongly supported by BAFs which clearly show the presence of a single allele in these regions across all tumor cells (Fig. 2c–h).
Allele- and haplotype-specific mechanisms of tumor evolution
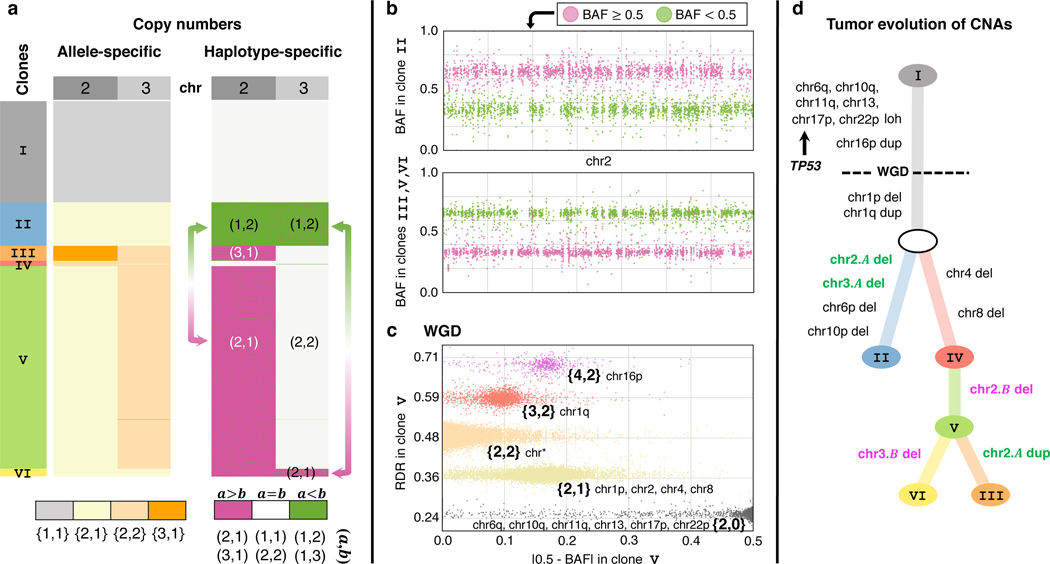
CHISEL derives haplotype-specific copy numbers in individual cells by examining changes in allele-specific copy numbers across cells. A particularly interesting application of haplotype-specific copy numbers is the identification of mirrored-subclonal CNAs: these are haplotype-specific CNAs occurring in different subpopulations of cells and affecting the two distinct alleles of the same genomic region. Such events were previously identified in the TRACERx multi-region sequencing of non-small-cell lung cancer patients and hypothesized to indicate parallel, or convergent, evolution 40. We identified mirrored-subclonal CNAs on chromosomes 2 and 3 in a large number of cells of patient S0 (Fig. 3a). Specifically, in section E we identified 168 cells with haplotype-specific copy numbers on chromosome 2 and 812 cells with haplotype-specific copy numbers . To confirm the presence of this mirrored-subclonal CNA, we pooled sequencing reads from cells with the same haplotype-specific copy numbers and calculated the BAF in these two pseudo-bulk samples. The pooled BAFs across chromosome 2 show a clear switch in frequencies of the two haplotypes in the two subpopulations of cells (Fig. 3b and Supplementary Fig. 12). We observed the same mirrored-subclonal CNA in a large number of cells from the other sections of the same patient S0 (Supplementary Fig. 5‒8). We note that multiple breast cancer tumor suppressor genes 42 are present on these chromosomes, including CASP8, MSH2, and DNMT3A on chromosome 2, and ATR, FOXP1, and SETD2 on chromosome 3. Therefore, mirrored-subclonal CNAs on these chromosomes suggest convergent evolution, as previously seen in non-small cell lung cancer 40. We emphasize that mirrored-subclonal CNAs are not apparent to methods that calculate only total copy numbers or allele-specific copy numbers, and thus CHISEL’s ability to identify haplotype-specific copy numbers provides a refined view of the evolution of this tumor.
Fig. 3: CHISEL reveals haplotype-specific CNAs and WGDs that shape tumor evolution.
a, CHISEL transforms allele-specific copy numbers (left) into haplotype-specific copy numbers (right) in 1448 cells from a breast tumor (section E of patient S0). Haplotype-specific copy numbers reveal mirrored-subclonal CNAs (arrows), or CNAs that alter the two distinct alleles of the same genomic region in different cells. Here, clone II has haplotype-specific copy numbers on chromosome 2, while clones V and VI have haplotype-specific copy numbers . Similarly, clone II has haplotype-specific copy numbers on chromosome 3, while clone VI has . b, BAFs on chromosome 2 support mirrored-subclonal CNAs, with a switch in the haplotype with larger BAF between clone II and clones III, V, and VI; each point in the plot indicates BAF in a 50kb haplotype block. c, RDRs, BAFs, and allele-specific copy numbers inferred by CHISEL along the entire genome and across all cells of clone V support the occurrence of a WGD as the two standard criteria for WGD are met: the larger allele-specific copy number is greater than 2 in of the genome and most of the genome (chr*) has allele-specific copy numbers . d, A phylogenetic tree describes the CNA evolution of the 6 clones identified by CHISEL, with inferred haplotype-specific copy number events indicated on branches. LOH on multiple chromosomes and duplication of chromosome 16p precede the WGD, while deletion of chromosome 1p and duplication of chromosome 1q occur after WGD. Chromosome 17p contains the gene TP53; LOH at this locus supports published reports that TP53 inactivation precedes WGD. Mirrored-subclonal CNAs on chromosomes 2 and 3 separate the 6 clones into two clear evolutionary branches, one containing the deletions of one haplotype of chromosomes 2 and 3 and the other containing the deletions of the other haplotype. These branches are further supported by the presence of specific subclonal CNAs, affecting chromosomes 4, 6p, 8, and 10p.
By integrating allele-specific copy numbers across cells, we inferred that a WGD was a clonal event that doubled the entire genome content of nearly all tumor cells in patient S0. We identified the occurrence of a WGD in every tumor clone of section E using criteria from published studies 20, 33 which demonstrated that allele-specific copy numbers are necessary to accurately infer the occurrence of WGDs. For example, the signal of WGD is clearly shown by pooling reads from all tumor cells of the largest tumor clone V of section E into a pseudo-bulk sample, and observing two imbalanced allele-specific deletion states, and (Fig. 3c). We similarly inferred the presence of a WGD in nearly all tumor clones from the other sections of patient S0 (Supplementary Fig. 5‒8). In contrast, in patient S1 the inferred allele-specific copy numbers do not support the occurrence of a WGD and suggest that this tumor is mostly diploid (Supplementary Fig. 1).
Using the haplotype-specific copy numbers, mirrored-subclonal CNAs, and WGDs identified by CHISEL, we constructed a phylogenetic tree that describes the tumor evolution of all cells in section E of patients S0 (Fig. 3d). The WGD and nine clonal CNAs that are present in all tumor cells are placed on the trunk of the phylogeny. The haplotype-specific copy numbers inferred by CHISEL enable the inference of the temporal order of some of these events 9, 20: the six clonal copy-neutral LOHs as well as the duplication of chromosome 16p are more likely to have occurred before WGD as the allele-specific copy numbers are even integers, while the two CNAs of chromosome 1 occurred after WGD. Interestingly, this inferred temporal order implies that LOH of chromosome 17p, which contains the gene TP53, precedes WGD, an order consistent with previous reports of TP53 inactivation occurring before WGDs 33. The mirrored-subclonal CNAs affecting chromosomes 2 and 3 separate the tumor clones into two clearly distinct branches: one including 168 cells from clone II and the other including 890 cells from the other clones. The two distinct branches are further subdivided by subclonal CNAs that are unique to each branch: CNAs of chromosomes 6p and 10p are unique to clone II, while CNAs of chromosome 4 and chromosome 8 are unique to the other tumor clones. Moreover, since all of the mirrored-subclonal CNAs follow the WGD, the mirrored-subclonal CNAs with allele-specific copy numbers (chromosomes 2 and 3) correspond to losses, while the mirrored-subclonal CNAs with allele-specific copy numbers (chromosome 2 in clone III) correspond to gains.
Clonal evolution across multiple tumor regions and somatic single-nucleotide variants
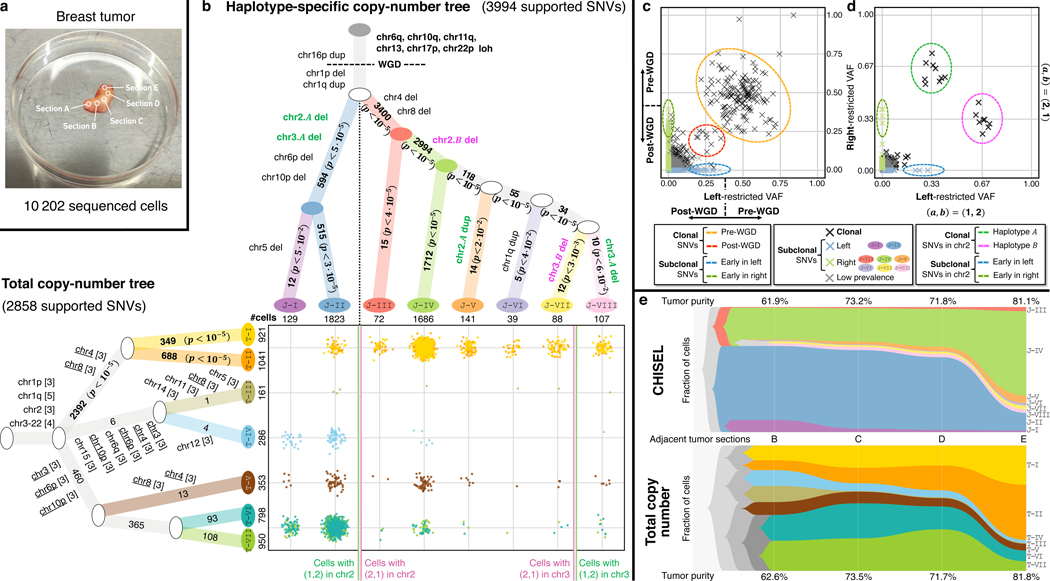
We applied CHISEL to jointly analyze cells from all 5 sequenced sections of breast cancer patient S0 (Fig. 4a). Using the inferred allele- and haplotype-specific copy numbers (Supplementary Fig. 13), we constructed a phylogenetic tree describing the evolution of 8 tumor clones (labeled J-I ‒ J-VIII) that include cells across all 5 sections (Fig. 4b) and one normal diploid clone that includes cells. This tree recapitulates the major features of the tree inferred from only the cells in section E (Fig. 3d), including: clonal CNAs and copy-neutral LOHs that occur before and after a WGD, deletions on chromosomes 4, 6p, 8, and 10p that define the initial split into a left branch containing clones J-I and J-II, and a right branch containing clones J-III ‒ J-VIII, and mirrored-subclonal CNAs on chromosomes 2 and 3 that further subdivide the subclones in these two branches. The larger number of cells in the integration of data from all 5 tumor sections yields a more refined tree with additional clones that contain small numbers of cells and are defined by haplotype-specific CNAs; e.g. clones J-I, J-VI, and J-VIII in Fig. 4b.
Fig. 4: Reconstruction of tumor heterogeneity and evolution across multiple tumor sections.
a, DNA from cells in five adjacent sections of a breast tumor patient S0 was sequenced using the 10x Genomics Chromium Single Cell CNV Solution in previous analysis of total copy numbers. b, Phylogenetic trees describing tumor clones and their CNA evolution as inferred by CHISEL jointly across all cells (top) and as reported in previous analysis using total copy numbers (left). Each branch of the CHISEL tree is labeled by haplotype-specific copy-number events, and each branch of the total copy number tree is labeled by total copy-number changes (in brackets with repeated events underlined). Each non-truncal branch in the trees is also labelled by the number of supported SNVs; -values comparing to the number of such SNVs expected by chance are indicated, with significant values () in boldface. Overall, 3994 SNVs support the haplotype-specific tree inferred by CHISEL, but only 2858 SNVs support the published tree inferred using total copy numbers. (Middle) Comparison of the cellular composition of clones identified in the two analyses, where each point corresponds to a cell colored by its clone assignment. For simplicity, clones T-I and T-II as well as T-VI and T-VII are merged as they are distinguished by only few isolated and small (5Mb) CNAs. Vertical lines separate cells with different haplotype-specific copy numbers in chromosomes 2 and 3 in the CHISEL tree. c, Left-restricted VAF (resp. right-restricted VAF) is computed as the VAF of SNVs using sequencing reads from cells in the left branch (clones J-1 and J-II) (resp. right branch with clones J-III ‒ J-VIII) of the CHISEL tree. Excluding low-prevalence SNVs that are potential false positives, the CHISEL tree partitions SNVs according to their clonal status (clonal or subclonal in each branch) and the restricted VAFs support this classification. Clonal SNVs have restricted VAFs consistent with their occurrence before (0.33) and after (0.33) WGD. All subclonal SNVs have restricted VAF consistent with their occurrence after WGD (0.25). Subclonal SNVs with the highest restricted VAFs (some 0.33 due to CNAs) are inferred to occur early in the evolution of these clones. d, Restricted VAFs of all clonal SNVs on chromosome 2 occurring before WGD are consistent with the distinct haplotype-specific copy numbers and in the left and right branches. SNVs on haplotype A (green circle) have left-restricted VAF 0.33 and right-restricted VAF 0.67, while SNVs on haplotype B (magenta circle) have the opposite restricted VAFs. Early SNVs in both left and right branches have restricted VAFs 0.33, which is consistent with occurrence after WGD. e, The proportions of tumor clones and the corresponding tumor purity identified by CHISEL and total copy-number analysis across the four adjacent tumor sections with highest tumor purity.
We compared the CHISEL tree with a previously described tree derived from the total copy numbers (Supplementary Fig. 14) inferred by Cell Ranger DNA (reported to be consistent 5 with copy numbers obtained with Ginkgo 35) and containing 7 tumor clones (labeled T-I ‒ T-VII). We found that there is good agreement on the initial branch of both trees (Fig. 4b): deletions of chromosomes 4 and 8 occur on the branch containing clones J-III ‒ J-VIII in the joint tree and clones T-I and T-II in the total copy-number tree, while deletions of chromosomes 3, 6p, and 10p occur on the branch containing the remaining clones in both trees. However, CHISEL further subdivided cells into novel clones/subpopulations that are characterized primarily by allele- and haplotype-specific CNAs. Of particular note are the mirrored-subclonal CNAs identified by CHISEL on chromosomes 2 and 3 that distribute cells from clones T-I and T-II in the tree derived from total copy numbers into clones J-III ‒ J-VIII in the tree derived from CHISEL. These mirrored-subclonal CNAs are invisible to the total copy-number analysis and consequently the tree constructed from total copy numbers includes cells with different haplotype-specific copy numbers in the same clone (e.g. T-V) and infers multiple independent occurrences of the same copy-number events on different branches of the tree (e.g. chromosomes 4, 6p, 8, and 10p) (Fig. 4b).
To further quantify the differences between the phylogenetic trees produced by CHISEL and total copy-number analysis, we examined somatic SNVs. Since SNVs were not used in tree construction, they provide an orthogonal signal for subdividing cells into subpopulations. Because of extremely low sequencing coverage, identification of SNVs in individual cells is impossible. Thus, we pooled sequencing reads from all cells into a pseudo-bulk sample and we identified 49k SNVs using standard methods developed for bulk-tumor sequencing data. We assigned each SNV to those cells with a variant read and found that of the SNVs are present only in the tumor clones J-I ‒ J-VIII. This number of SNVs is close to the average of 7 000 (range 500–93 000) somatic SNVs reported in whole-genome sequencing studies of 560 breast tumors 42. Next, for each non-truncal branch in the phylogenetic trees, we computed the number of SNVs that are uniquely assigned to cells in the subtree defined by that branch. We found that 40% more SNVs (3 994 vs. 2 858) are consistent with the tree inferred by CHISEL compared to the tree inferred by total copy numbers. Moreover, we found that all 14 branches in the CHISEL tree are supported by more SNVs than expected by chance (), while only 3/11 branches in the total copy-number tree have significant support (Fig. 4b). Additionally, we found that while clones T-I and T-II in the total copy-number tree are supported by a significant number of SNVs – even though they are not distinguished by any large CNAs (Supplementary Fig. 14c) – these SNVs are the same as those that support the smaller subclones (J-III ‒ J-VIII) identified by CHISEL. In summary, we found that SNVs support nearly all tumor clones inferred by CHISEL in both patients S0 and S1 (Supplementary Fig. 15 and 16).
Next, we examined the relationship between the VAF of each SNV – the proportion of reads covering an SNV locus that contain the variant allele – and the clonal status of the SNV induced by the CHISEL tree. We classified the SNVs according to the partition of cells defined by the initial two branches of the CHISEL tree and, after excluding likely false positive SNVs with low clone prevalence, we obtained: 594 SNVs unique to cells in the left branch (clones J-I and J-II), 1 632 SNVs unique to the right branch (clones J-III ‒ J-VIII), and 2 798 clonal SNVs in both branches. Since each read has a unique cell barcode, we computed the left-restricted VAF (resp. right-restricted VAF) of each SNV using only the sequencing reads from the subpopulation of cells in the left (resp. right) branch of the CHISEL tree. We found that restricted VAFs of SNVs were consistent with the placement of the SNVs on the CHISEL tree (Fig. 4c): clonal SNVs have restricted VAFs consistent with their occurrence before (0.5) or after (0.25) WGD (assuming no other CNAs at the locus) in both branches, while subclonal SNVs have lower restricted VAFs (0.25). In addition, we found that the restricted VAFs of SNVs on chromosome 2 are consistent with the corresponding mirrored-subclonal CNA (Fig. 4d): clonal SNVs that occurred before WGD have restricted VAF equal to either or when they are located on the deleted allele or the other allele, respectively. SNVs with both these values of restricted VAF in the two distinct branches clearly support the deletion of two different haplotypes. We observed similar consistency between the standard VAF computed across all cells and the placement of the SNVs on the CHISEL tree (Supplementary Fig. 17).
We observed an interesting discordance between the number of cells in the left (clones J-I and J-II) and right (clones J-III ‒ J-VIII) branches and the number of SNVs assigned to these branches. While the left and right branches have a very similar number of cells (1952 vs. 2133, respectively), the left branch has fewer SNVs (594 vs. 1 632). This discordance may reflect different rates of growth and/or selection between the clones in these branches. Intriguingly, we found a subclonal CNA affecting the entire HLA gene complex in chromosome 6p that is unique to the left branch and could provide a mechanism for evasion of immune response 43.
Finally, we examined the variation in proportion of cells in each tumor clone across the different sections of patient S0, as these are adjacent sections of the same tumor. We found that the left and right branches in the CHISEL tree are consistent with the spatial distribution of the tumor as all the clones in the same branch consistently expand or contract across the adjacent tumor sections B-E (Fig. 4e): clones J-I and J-II from the left branch contract towards section E, while all the remaining clones from the right branch expand towards section E. In contrast, the clones inferred by total copy-number analysis have more complicated dynamics across the tumor sections (Fig. 4e). While the merge of clones T-VI and T-VII contract towards section E and the merge of clones T-I and T-II expand towards section E, both the proportions of the subclones in these groups and the proportions of the remaining clones fluctuate independently across the sections. This discordance between the spatial and temporal evolution suggests that the clones inferred by the total copy-number analysis are less plausible.
Discussion
New technologies to perform low-coverage whole-genome sequencing from thousands of individual cells provide data to study tumor heterogeneity and evolution at previously unprecedented resolution. However, methods to analyze this data have thus far been limited to the identification of total copy numbers of large genomic regions in individual cells. Here, we introduce CHISEL, an algorithm to infer allele- and haplotype-specific copy numbers in single cells from low-coverage DNA sequencing data. CHISEL integrates the weak allelic signals across thousands of individual cells, leveraging a strength of single-cell sequencing technologies (many cells) to overcome a weakness (low genomic coverage per cell). CHISEL also includes other innovative features, such as global clustering of RDRs and BAFs, and a rigorous model selection procedure for inferring genome ploidy, that improves both the inference of allele-specific and total copy numbers.
We demonstrated the unique features of CHISEL on 10 datasets, each comprising 2 000 cells, from 2 breast cancer patients. CHISEL identified previously uncharacterized CNAs and mutational events that shape the tumor heterogeneity and evolution, including extensive allele-specific CNAs – especially copy-neutral LOHs – that further distinguish novel clones and affect well-known breast cancer genes. In addition, the allele- and haplotype-specific copy numbers inferred by CHISEL reveal mirrored-subclonal CNAs and WGDs that characterize some of the key mechanisms of tumor evolution, including evidence of convergent evolution and potential precursors of WGDs. Many of these events are corroborated by somatic SNVs and the spatial distribution of the inferred clones. To demonstrate CHISEL’s applicability to other sequencing technologies, we analyzed a DOP-PCR 1 single-cell sequencing dataset of a breast tumor 44. This dataset has a much smaller number of cells (i.e. 89 vs. 2 000) but a much higher sequencing coverage per cell (i.e. 0.24 vs. 0.02) than the 10x Genomics datasets. We found that CHISEL identifies allele-specific CNAs that affect multiple breast cancer genes (Supplementary Fig. 18 and Supplementary Results 2). Overall, on all datasets CHISEL provides a more refined and accurate view of tumor evolution than obtained by previous total copy-number analysis.
The single-cell view of allele- and haplotype-specific copy numbers provided by CHISEL offers the opportunity for deeper analysis of tumor evolution. CHISEL enables the identification of allele-specific CNAs, including copy-neutral LOHs and WGDs, and haplotype-specific CNAs, including mirrored-subclonal CNAs, in individual cells without the limitations of bulk tumor-samples where inference of tumor ploidy and purity from admixed signals is extremely challenging 19–25. In addition, previous analysis of haplotype-specific CNAs has been restricted to the special case where these CNAs are present in different samples from the same patient 40. CHISEL may be used to analyze the frequency and function of mirrored-subclonal CNAs and other complex copy-number events across different cancer types, especially for haplotype-specific CNAs which have received scant attention thus far in the analysis of bulk-tumor samples.
While CHISEL enables the accurate inference of allele- and haplotype-specific copy numbers in individual cells, there are a number of areas for future improvements. First, the low coverage of single-cell DNA sequencing data limits the size of the CNAs that can be accurately inferred. One approach to improve the resolution is to iteratively run CHISEL on pseudo-bulk genomes obtained by merging multiple cells with similar haplotype-specific copy-number profiles, as suggested in recent studies 6, 7. Second, BAF estimation could be further improved by using variable-size bins 35, by using the signal from sequencing reads that cover multiple SNPs, or by using larger haplotype blocks that one could infer from the results of CHISEL in regions of allelic imbalance. Third, a more refined model of CNA evolution might be integrated in the inference of haplotype-specific copy numbers, for example reconstructing the full copy-number tree from the model of interval events 18 or integrating the additional signal from breakpoints 24. Fourth, one could improve techniques for classifying cells with highly aberrant copy-number profiles, designing classifiers to distinguish actively replicating regions 5, 7 from cell doublets.
Haplotype-specific copy numbers inferred by CHISEL provide a useful substrate for other analyses of tumor heterogeneity and evolution. In particular, further integration of CNAs and SNVs in single cells would provide higher resolution reconstructions of tumor evolution. While our initial analysis of SNVs using restricted-VAFs showed good consistency between SNVs and the clones inferred from CNAs, a complete and accurate classification of all SNVs remains a challenging problem: distinguishing true SNVs from false positive is difficult for clones with few cells, and also variant read counts are expected to be low in regions of high copy number (e.g. from WGD). The SNV analysis could be further extended to derive the mutant copy number of individual SNVs, a task that is notoriously difficult in bulk tumor sequencing where discordance between VAFs and cancer cell fractions (CCFs) complicates tumor evolution studies 45–48. As an example, we computed CCFs of the SNVs in the pseudo-bulk analysis using methods developed for bulk tumors 48. We found that cells from the same CHISEL-derived tumor clone have different SNVs (Supplementary Fig. 19). Thus, SNVs provide evidence of additional heterogeneity beyond what we detected with CHISEL, possibly motivating the sequencing of cells at higher coverage to better quantify the limitations of detectability for clones with few cells. In addition, integrated analysis of CNA and SNV evolution would help resolve questions about the relative rates of these mutation classes during tumor evolution, including “punctuated evolution" of CNAs in certain tumors 49. Our analysis of tumor patient S0 showed an intriguing discordance between number of SNVs unique to a copy-number clone and the prevalence of the clone in the tumor cell population, and further studies of such phenomenon in additional tumors would be informative. Finally, allele-specific copy numbers provide useful signal for single-cell studies of allele-specific gene expression by combining single-cell DNA sequencing with single-cell RNA sequencing 34, 50.
Methods
CHISEL Algorithm
We introduce the CHISEL algorithm to derive allele- and haplotype-specific copy numbers from low-coverage DNA sequencing of cells by integrating two signals, the RDR and the BAF, jointly across the whole genome of all cells. We divide the reference genome into bins and represent the genome of each cell by two integer vectors, and . Each bin has two alleles, and the entry indicates the number of copies of the allele that is located on haplotype whereas the entry indicates the number of copies of the other allele located on haplotype . We call the haplotype-specific copy numbers of the cell.
CHISEL addresses three major challenges in the derivation of the haplotype-specific copy numbers of each cell from low-coverage single-cell DNA sequencing data. The first is the calculation of the BAF: the standard approach used in bulk sequencing to estimate the BAF from the proportion of alternate reads at individual germline SNPs 19, 20, 22, 23, 25, 53–59 does not work for extremely low-coverage data. The second is the inference of a pair of allele-specific copy numbers for each bin : genome ploidy varies across cells due to CNAs and WGDs and thus the derivation of integer copy numbers from read counts requires care. The third is the inference of an ordered pair of haplotype-specific copy numbers from the unordered pair of allele-specific copy numbers: one does not know a priori which allele is located on haplotype or .
CHISEL has five major steps (Fig. 1) which we detail in the subsections below.
Computation of RDR and BAF
The first step of CHISEL is to compute the RDRs and the BAFs for all bins in every cell (Fig. 1a). The RDR of bin in cell is directly proportional to the total number of reads that align to and is used to estimate the total copy number ; i.e. for some cell-specific scale factor . CHISEL computes by appropriate normalization of the number of reads aligned to sufficiently large bins (Mb in this work, Supplementary Fig. 20) – accounting for GC bias and other biases – similar to other approaches for CNA detection from bulk 19–25, 53–59 or single-cell 5–7, 35–39 sequencing (Supplementary Fig. 21 and Supplementary Methods 1). Additional details on the selection of bin length for different number of cells and sequencing coverage are in Supplementary Methods 2.
The BAF of bin in cell is the fraction of reads belonging to one of the two distinct alleles of and provides an estimate of either or . Previous methods for bulk sequencing data compute the BAF from either individual heterozygous germline SNPs 19, 20, 22, 23, 25, 53–59 or by aggregating SNPs in small haplotype blocks 21, 24. However, methods that compute BAF from individual SNPs are not useful for low-coverage DNA sequencing data because few, if any, reads will cover each SNP; e.g. in the 10x Genomics datasets that we analyzed (0.02 coverage) only 2% of SNPs were covered by at least one read in a single cell and only 0.08% were covered by more than one read. Methods that compute BAF from haplotype blocks are also not directly applicable since the inferred blocks remain too short, containing too few SNPs for accurate calculation of BAF in single cells. Current reference-based phasing methods have switch error rates 60 of 1% meaning that haplotype blocks longer than a few hundred kilobases are likely to contain a phasing error. For example, the Battenberg algorithm 21 for bulk DNA sequencing reported blocks up to 300kb, which is too small to contain enough SNP-covering reads for accurate calculation of BAF in individual cells.
CHISEL computes the BAF of bin in cell using a two-stage procedure. In the first stage, CHISEL uses the reference-based algorithm Eagle2 41 to phase germline heterozygous SNPs in each bin into haplotype blocks. We used blocks of length 50kb in this work (Supplementary Fig. 22) as phasing errors are unlikely at this scale 60; additional details on the selection of the length of haplotype blocks are in Supplementary Methods 2. Each haplotype block is composed of two sequences of nucleotides at consecutive SNPs, called the reference and alternate sequences, with each sequence located on a different allele of . In the second stage, CHISEL computes the BAF by phasing the blocks into the two alleles of bin across all cells. Specifically, we phase the blocks with respect to one of the two alleles of , which we call the minor allele (see below), and we define the phase of every block such that indicates that the alternate sequence of belongs to , and otherwise. Given the phases of the blocks in bin , CHISEL calculates the BAF from the total number of reads covering block in cell and the corresponding number of reads only covering the alternate sequence of in cell as follows
| (1) |
The BAF is typically calculated in bulk sequencing to estimate the proportion of an unknown allele of and corresponds to either or . In contrast, CHISEL calculates to estimate the proportion [1] of the same allele in every cell . As the phases are unknown, CHISEL seeks values of such that in Eq. (1) accurately estimates where is the copy number of in every cell .
CHISEL infers phases for the blocks in bin based on the estimated proportion of the minor allele across all cells, where we assume without loss of generality that is the allele of with the lower proportion so that . We designed an expectation-maximization (EM) algorithm 61 to calculate the maximum likelihood estimate given the observed values of read counts and in every block across every cell , where the phases are unobserved, latent variables (Supplementary Methods 3). One issue that arises in this calculation is that the cases and need to be handled differently. When we compute the maximum-likelihood phases using the idea that the lower (respectively higher) read counts belong to the same allele where there is allelic imbalance, as previously used in bulk sequencing 53, 56, 59. We also showed empirically that this approach accurately identifies both the true phases and the BAF (Supplementary Fig. 23 and 24). When , the allelic origin of each block cannot be determined from read counts 59. Thus, CHISEL selects the haplotype phase of every block uniformly at random. We show that the corresponding is an unbiased estimator of under the assumption that implies in every cell , which is reasonable as violations of this assumption are rare. Further details are in Supplementary Methods 4.
Global clustering of genomic bins into copy-number states
The second step of CHISEL is to cluster bins into a small number of copy-number states according to the RDR and BAF values for each bin in each cell . Clustering helps to overcome measurement errors and variance in the computed RDRs and BAFs. The standard approach used in bulk 19–24, 53, 54, 56–59 and single-cell 5–7, 35–39 copy number analysis is to segment bins locally along the genome grouping neighboring bins with similar values of RDR and/or BAF into segments that are assigned the same copy-number state. This local segmentation leverages the observation that a CNA alters the copy numbers of multiple adjacent bins. Existing methods for single-cell copy numbers 5–7, 35–39 perform this segmentation on RDR values only as these methods do not calculate BAF; moreover, with one recent exception 39, these methods perform this segmentation independently for each cell. Such local and cell-specific clustering is problematic for low-coverage single-cell sequencing data because RDRs (and BAFs) have high variance in individual cells.
For CHISEL, we developed a global clustering approach that simultaneously clusters RDR and BAF values across all bins from all cells (Fig. 1b). Specifically, CHISEL uses a -means algorithm to identify clusters of bins which share the same allele-specific copy numbers in every cell . This global clustering approach leverages two observations: (1) all bins from a genome occupy a small number of copy-number states, regardless of their genomic position; (2) all cells from a tumor share a common evolutionary history. This approach extends the global clustering that we introduced in HATCHet 25 for simultaneous analysis of multiple bulk-tumor samples. We select the number of clusters to minimize the unexplained variance given a certain threshold of tolerance, using standard model selection criteria 62. An important issue that arises in the global clustering is that one cannot directly compare the BAFs across different bins since we do not know whether the BAF of each bin is the proportion of the allele on either haplotype or . To address this issue, we define the mirrored BAF as in previous studies 53, 59 to guarantee that any pair of bins with similar values of RDRs and mirrored BAFs have the same copy-number state (i.e. the same allele-specific copy numbers). We compared the global clustering of CHISEL with the local clustering of a Hidden Markov Model (HMM) 6, 7, 22 on 2 000 subsampled datasets with varying number of cells and bins (Supplementary Results 3). We found that the CHISEL’s global clustering results in substantially lower error rates than HMM local clustering (Supplementary Fig. 25).
Inferring allele-specific copy numbers
The third step of CHISEL is to infer a pair of allele-specific copy numbers for every bin in each cell (Fig. 1c). To do this, one typically needs to know the cell-specific scale factor that transforms the RDR into the corresponding total copy number ; i.e. . Once one calculates , it is then straightforward to separate into the pair of allele-specific copy numbers using the BAF since estimates or . Unfortunately, the scale factor depends on the genome ploidy of the cell, which is generally unknown due to effects of CNAs and WGDs. Existing methods for single-cell sequencing data 5–7, 35–39 infer using only the RDRs and total copy numbers; for example, Ginkgo 35 and Cell Ranger DNA 5 minimize the error between the expected and inferred values of for every bin , i.e. . However, this approach has two main issues. First, there are generally many equally plausible solutions for because depends on which in turn depends on the total copy numbers to be inferred. Current methods 5, 35 use restrictive or biased assumptions on the values of . Second, because current methods do not consider BAFs, the chosen value of may result in total copy numbers that contradict the underlying allelic balance (Supplementary Fig. 3), e.g. a total copy number for a bin with BAF .
CHISEL jointly infers the scale factor and the allele-specific copy numbers of every bin in a two-stage procedure that integrates both RDRs and BAFs. First, we identify candidate values of under the assumption that the genome of every cell contains a reasonable number of balanced bins, i.e. bins with equal copy numbers of both alleles. This assumption follows from the observation that bins unaffected by CNAs in a cell have allele-specific copy numbers without WGD, with one WGD, and so on. CHISEL identifies these bins as the largest cluster among the clusters inferred in the second step whose BAF is approximately equal to 0.5. Second, CHISEL chooses the value among the candidates and the corresponding pair of allele-specific copy numbers for every bin using the Bayesian information criterion (BIC) to select among models of varying complexity. Specifically, given a candidate value and allele-specific copy numbers for all the bins in a cluster , we model the observed RDR and the mirrored BAF of bin as observations from the following two normal distributions
| (2) |
where the sample variances are estimated from the inferred clusters. For every candidate value of we find the maximum likelihood estimates for using an exhaustive search, which is feasible as the number of candidate values of (e.g. 3 when considering the occurrence of at most 2 WGDs) and the number of distinct pairs of allele-specific copy numbers for a cluster are relatively small. Higher values of always have higher likelihood but also higher model complexity, as they induce more combinations of allele-specific copy numbers; thus, we choose the candidate value of with minimum BIC. Further details of this procedure are in Supplementary Methods 5.
Inferring haplotype-specific copy numbers
The fourth step of CHISEL is to infer haplotype-specific copy numbers for every cell (Fig. 1d). The challenge is that given the pair of allele-specific copy numbers for every bin , we do not know whether and , or vice versa. The reason for this unknown phasing is that the BAF indicates whether the copy number of the minor allele is equal to or , but we do not know whether is located on either haplotype or . A naive approach that assigns the of every bin to the same haplotype generally leads to unlikely scenarios as may be determined by different subpopulations of cells in different genomic regions (Supplementary Fig. 26). While it is impossible to determine the correct phasing given only one sample from a tumor, Jamal-Hanjani et al. 40 recently showed how to infer haplotype-specific copy numbers in some cases when given multiple bulk samples from a tumor. However, the approach in Jamal-Hanjani et al. 40 has three main limitations that prevent its applicability on single-cell DNA sequencing data. First, the approach relies on the BAFs computed at individual SNPs, making it unfeasible for low-coverage single-cell DNA sequencing data. Second, the approach only determines the presence of different haplotype-specific copy numbers for a specific genomic region but does not phase neighboring regions on the same chromosome. Third, the approach is only successful when different haplotype-specific CNAs are clearly present in different samples. We overcome these limitations in CHISEL and infer haplotype-specific copy numbers jointly across all cells.
The key idea of CHISEL is to phase the minor allele of every bin to the haplotype that minimizes the number of CNAs required to explain the resulting haplotype-specific copy numbers across all cells. Specifically, we define the phase of a bin such that when is located on haplotype and otherwise. Given the phase of bin , we compute the corresponding haplotype-specific copy numbers in every cell : when and when . Note that we can easily determine from the BAF and the allele-specific copy numbers : assuming without loss of generality that we have that if and otherwise. To count the number of CNAs that explain a phasing, CHISEL uses the model of interval events 16–18 that model CNAs as events that either increase or decrease the copy numbers of neighboring genomic regions on the same haplotype.
We compute the total number of interval events given the phases , as each interval event introduces a difference between or of two neighboring bins and . Thus, we seek the phases which minimize the total number of interval events across all bins, i.e. . We design a dynamic-programming algorithm to solve this problem based on the following recurrence to compute the minimum number of interval events for the first bins given the phase :
| (3) |
Further details and the proof of correctness are in Supplementary Methods 6.
Inferring tumor clones
The fifth step of CHISEL is to infer distinct subpopulations of cells, or clones, with the same complement of CNAs (Fig. 1e). CHISEL uses standard hierarchical clustering to group cells according to their inferred haplotype-specific copy-number profiles. To compute these clusters, we define the distance between two cells as the fraction of the genome with different haplotype-specific copy numbers and set a threshold on the maximum distance between cells in the same cluster to cut the dendrogram. Next, CHISEL selects the groups of cells that correspond to clones using a minimum threshold on the number of included cells, since we expect that groups composed only of few cells are likely due to noise in the data or errors in the measurements. We compute a consensus copy-number profile for each clone; additional details are in Supplementary Methods 7. We investigated the sensitivity and lower limits of detection for a clone by subsampling from the single-cell datasets from patient S0. We found that CHISEL can accurately recover clones containing as few as 10–20 cells (Supplementary Fig. 27). The subsampling approach is available in the CHISEL software and can be used to investigate the lower limits of detection on other datasets; further details are in Supplementary Methods 8.
Analysis of breast cancer datasets
Single-cell DNA sequencing data of breast cancer
We analyzed sequencing data from the 10x Genomics Chromium Single Cell CNV Solution from 10 single-cell datasets of 2 breast cancer patients: (1) 5 adjacent sections from a triple negative ductal carcinoma in situ (patient S0); (2) 3 and 2 technical replicates from two samples of a stage 1 infiltrating ductal carcinoma (patient S1). Each section includes 1 400‒2 300 individual cells, whose genome has been sequenced with a sequencing coverage ranging from 0.01 to 0.05 (0.02 on average) per cell. Sequencing was performed on an Illumina NovaSeq 6000 System using paired sequencing with a 100b (R1), 8b (i7), and 100b (R2) configuration. The details of the sequencing procedure and of the previous total copy-number analysis are available in the Application Note “Assessing Tumor Heterogeneity with Single Cell CNV” at the 10x Genomics website (https://www.10xgenomics.com/solutions/single-cell-cnv). The sequencing reads from each dataset were aligned to the human reference genome (hg38 for S0 and hg19 for S1) using the Cell Ranger DNA pipeline (https://support.10xgenomics.com/single-cell-dna/software/pipelines/latest/what-is-cell-ranger-dna).
Inference of allele- and haplotype-specific CNAs using CHISEL
We applied CHISEL to analyze every single-cell dataset from patients S0 and S1. We selected only those cell barcodes with a sufficient number of sequencing reads using standard approaches for 10x Genomics data (Supplementary Methods 9). In addition to a barcoded BAM file, CHISEL requires two other sources of information: a matched-normal sample and a haplotype phasing for heterozygous germline SNPs. For patient S0, we used section A as a matched-normal sample, as in the previous total copy-number analysis, because this section contains mostly diploid cells (91%), which we assumed are normal (non-cancerous cells). For patient S1, we used the available matched-normal sample. In case of a missing matched-normal sample, CHISEL includes an accurate procedure to identify normal diploid cells (Supplementary Methods 10) and to generate a corresponding pseudo-bulk sample (Supplementary Fig. 28 and Supplementary Results 4). We used BCFtools 63 (v1.9) to identify germline heterozygous SNPs from the matched-normal sample of each patient and we used Eagle2 41 through the Michigan Imputation Server 64 to phase germline SNPs with respect to the HRC reference panel (Version r1.1 2016) comprising haplotypes 65. As the HRC panel currently supports the human reference genome hg19 but not hg38, we used the LiftoverVcf tool from the Picard software package (v2.18, http://broadinstitute.github.io/picard/) to convert the genomic coordinates between the different required builds of the reference genome. For each dataset, we applied CHISEL using the default parameters with haplotype blocks of length kb and genomic bins of length Mb. We also applied CHISEL to jointly analyze all the cells of patient S0.
Reconstruction of copy-number trees
We built copy-number trees that describe the evolution of the clones identified in section E of patient S0 and the clones identified in the joint analysis of all cells of patient S0. A copy-number tree has 3 main features: (1) the root corresponds to the diploid clone, (2) the leaves correspond to the other identified clones, and (3) every branch is labelled by copy-number events, with each event either increasing or decreasing the copy numbers of a genomic segment from the parent to the child. We used the model of interval events for CNAs 16–18 and reconstructed the most parsimonious copy-number tree with the minimum number of events using the consensus haplotype-specific copy numbers inferred by CHISEL. To perform the reconstruction, we separated the two haplotypes of each chromosome and we classified the copy-number events according to the identified WGD as deletions after WGD (i.e. del), deletions before WGD (i.e. loh), and duplications before and after WGD (i.e. dup).
We used the same approach to identify the events labeling the branches of the total copy-number tree for patient S0. Specifically, we fixed the topology of the tree to be the one reported in the total copy-number analysis described above and we represented the events as changes in total copy numbers. In both cases, we ignored small CNAs only affecting few genomic bins.
Analysis of somatic single-nucleotide variants
We pooled the sequencing reads from all cells in each section of patient S0 and ran Varscan 2 (v2.3.9) 66 to identify somatic SNVs. To identify SNVs present in small clones ( cells), we relaxed the default parameters of Varscan 2, selecting the highest confidence somatic SNVs with at least 2 supporting sequencing reads from the cells in the clones of CHISEL (Supplementary Methods 11). We used SAMtools 67 (v1.9) to assign each variant read to the corresponding cell through the related barcode. Among all SNVs, SNVs were present only in the tumor clones of CHISEL, SNVs present only in the diploid clone, and SNVs were in both the diploid clone and the tumor clones. Note that the latter class of mutations likely consist of: germline SNPs that were incorrectly classified by Varscan 2 as somatic, false positive variant calls, and early somatic mutations that preceded tumor aneuploidy.
We examined the correspondence between all the identified SNVs and the copy-number tree in two steps. First, we say that a SNV supports a branch in the tree if all the cells with the SNV are contained in clones of the subtree descended from the branch. Second, we counted the number of SNVs supporting each non-truncal branch and we assessed whether this number is higher than expected by chance using a permutation test with randomly sampled subsets of cells, each subset containing the same number of cells as in the clones of the corresponding subtree.
We examined the relationship between the VAF of each SNV and the clonal status of the SNV induced by the CHISEL tree for the 10 551 SNVs identified in the tumor clones. We calculated the VAF of each SNV using the standard definition as the fraction of variant reads over the total number of reads covering the SNV locus. We also defined a restricted VAF for an SNV with respect to a subpopulation of cells by restricting to sequencing reads with barcodes matching the cells in the subpopulation. In particular, we computed a left-restricted VAF and a right-restricted VAF by restricting to the sequencing reads from cells belonging to the left (clones J-I and J-II) and right (clones J-III ‒ J-VIII) branches of the CHISEL tree. Next, we classified the SNVs according to the CHISEL tree by separating SNVs into clonal SNVs, which are present in all tumor clones, and subclonal SNVs which are unique to either the left or right subtree.
Distinguishing true positive from false positive SNVs is complicated in this dataset due to the low number of variant reads for many SNVs. Thus, we restricted attention to high prevalence SNVs that were present in multiple clones of the same branch, resulting in SNVs unique to the left branch, SNVs unique to the right branch, and clonal SNVs present in both branches. The remaining low-prevalance SNVs have both low VAFs (Supplementary Fig. 17) and low restricted VAFs (Fig. 4c) in both branches, underscoring the low confidence in these mutation calls. Further details of the VAF analysis are in Supplementary Methods 12.
Statistical analysis
For each non-truncal branch of the CHISEL tree and of the total copy-number tree, we computed the probability that the observed number of supporting SNVs is higher than expected by chance using a permutation test. We selected subsets of cells uniformly at random with each subset containing the same number of cells as the clones in the subtree defined by the branch. We counted how many of such subsets contain an equal or larger number of supporting SNVs than observed.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The sequencing data from 10x Genomics Chromium Single Cell CNV Solution for patient S0 are available at https://support.10xgenomics.com/single-cell-dna/datasets. Raw read counts and phased SNP counts for patient S0 are available at https://doi.org/10.5281/zenodo.3817605 and for patient S1 at https://doi.org/10.5281/zenodo.3817536. The DOP-PCR sequencing data of 89 breast tumor cells are available from the NCBI Sequence Read Archive under accession SRA: SRP114962. All the processed data for all datasets of patients S0 and S1 and for the DOP-PCR data, as well as all the results of CHISEL, are available on GitHub at https://github.com/raphael-group/chisel-data.
Code availability
CHISEL is available on GitHub at https://github.com/raphael-group/chisel and on Code Ocean at https://doi.org/10.24433/CO.6796686.v1.
Supplementary Material
Acknowledgments
We thank L. Hepler and K. Ganapathy from 10x Genomics for providing additional data for our study, for providing access to the published data of the total copy-number analysis, and for the useful feedback. This work is supported by a US National Institutes of Health (NIH) grants R01HG007069 and U24CA211000, US National Science Foundation (NSF) CAREER Award (CCF-1053753) and Chan Zuckerberg Initiative DAF grants 2018-182608 (B.J.R.). Additional support was provided by NIH grant (Rutgers) 2P30CA072720-20, the O’Brien Family Fund for Health Research, and the Wilke Family Fund for Innovation (B.J.R.).
Footnotes
Competing interests
B.J.R. is a cofounder of, and consultant to, Medley Genomics.
References
- 1.Navin N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navin NE The first five years of single-cell cancer genomics and beyond. Genome research 25, 1499–1507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gawad C, Koh W. & Quake SR Single-cell genome sequencing: Current state of the science. Nature Reviews Genetics 17, 175 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Andor N. et al. Joint single cell dna-seq and rna-seq of gastric cancer reveals subclonal signatures of genomic instability and gene expression. Preprint at bioRxiv, 10.1101/445932 (2018). [DOI] [Google Scholar]
- 6.Zahn H. et al. Scalable whole-genome single-cell library preparation without preamplification. Nature methods 14, 167 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Laks E. et al. Clonal decomposition and dna replication states defined by scaled single-cell genome sequencing. Cell 179, 1207–1221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zack TI et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics 45, 1134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciriello G. et al. Emerging landscape of oncogenic signatures across human cancers. Nature genetics 45, 1127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor AM et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer cell 33, 676–689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrell RA, McGranahan N, Bartek J. & Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338 (2013). [DOI] [PubMed] [Google Scholar]
- 13.McGranahan N. & Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer cell 27, 15–26 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Desper R. et al. Distance-based reconstruction of tree models for oncogenesis. Journal of Computational Biology 7, 789–803 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury SA et al. Algorithms to model single gene, single chromosome, and whole genome copy number changes jointly in tumor phylogenetics. PLOS Computational Biology 10, e1003740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz RF et al. Phylogenetic quantification of intra-tumour heterogeneity. PLOS Computational Biology 10, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Kebir M. et al. Complexity and algorithms for copy-number evolution problems. Algorithms for Molecular Biology 12, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaccaria S, El-Kebir M, Klau GW & Raphael BJ Phylogenetic copy-number factorization of multiple tumor samples. Journal of Computational Biology 25, 689–708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Loo P. et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences 107, 16910–16915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter SL et al. Absolute quantification of somatic dna alterations in human cancer. Nature biotechnology 30, 413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nik-Zainal S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha G. et al. TITAN: Inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome research 24, 1881–1893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer A, Vázquez-García I, Illingworth CJ & Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell reports 7, 1740–1752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPherson AW et al. ReMixT: Clone-specific genomic structure estimation in cancer. Genome biology 18, 140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaccaria S. & Raphael BJ Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. Preprint at bioRxiv, 10.1101/496174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pleasance ED et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waddell N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dentro SC et al. Portraits of genetic intra-tumour heterogeneity and subclonal selection across cancer types. Preprint at bioRxiv, 10.1101/312041 (2018). [DOI] [Google Scholar]
- 29.Langdon JA et al. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes, Chromosomes and Cancer 45, 47–60 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Kuga D. et al. Prevalence of copy-number neutral loh in glioblastomas revealed by genomewide analysis of laser-microdissected tissues. Neuro-oncology 10, 995–1003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Keefe C, McDevitt MA & Maciejewski JP Copy neutral loss of heterozygosity: A novel chromosomal lesion in myeloid malignancies. Blood 115, 2731–2739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha G. et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome research 22, 1995–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bielski CM et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nature genetics 50, 1189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell KR et al. Clonealign: Statistical integration of independent single-cell rna and dna sequencing data from human cancers. Genome biology 20, 54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garvin T. et al. Interactive analysis and assessment of single-cell copy-number variations. Nature methods 12, 1058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakker B. et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome biology 17, 115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Chen H. & Zhang NR DNA copy number profiling using single-cell sequencing. Briefings in bioinformatics 19, 731–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong X, Zhang L, Hao X, Wang T. & Vijg J. SCCNV: A software tool for identifying copy number variation from single-cell whole-genome sequencing. Preprint at bioRxiv, 10.1101/535807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R, Lin D-Y & Jiang Y. SCOPE: A normalization and copy number estimation method for single-cell dna sequencing. Preprint at bioRxiv, 10.1101/594267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamal-Hanjani M. et al. Tracking the evolution of non–small-cell lung cancer. New England Journal of Medicine 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Loh P-R et al. Reference-based phasing using the haplotype reference consortium panel. Nature genetics 48, 1443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N. et al. Allele-specific hla loss and immune escape in lung cancer evolution. Cell 171, 1259–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim C. et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth A. et al. PyClone: Statistical inference of clonal population structure in cancer. Nature methods 11, 396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deshwar AG et al. PhyloWGS: Reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome biology 16, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Kebir M, Satas G, Oesper L. & Raphael BJ Inferring the mutational history of a tumor using multi-state perfect phylogeny mixtures. Cell systems 3, 43–53 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Dentro SC, Wedge DC & Van Loo P. Principles of reconstructing the subclonal architecture of cancers. Cold Spring Harbor perspectives in medicine 7, a026625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao R. et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature genetics 48, 1119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan J. et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell rna-seq data. Genome research 28, 1217–1227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaccaria S. and Raphael BJ Characterizing allele- and haplotype-specific copy numbers in single cells with CHISEL. Github https://github.com/raphael-group/chisel (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaccaria S. and Raphael BJ Characterizing allele- and haplotype-specific copy numbers in single cells with CHISEL. Code Ocean 10.24433/CO.6796686.v1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staaf J. et al. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome snp arrays. Genome Biology 9, R136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenman CD et al. PICNIC: An algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11, 164–175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popova T. et al. Genome alteration print (gap): A tool to visualize and mine complex cancer genomic profiles obtained by snp arrays. Genome biology 10, R128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter SL, Meyerson M. & Getz G. Accurate estimation of homologue-specific dna concentration-ratios in cancer samples allows long-range haplotyping. Preprint at Nature Precedings, 10.1038/npre.2011.6494.1 (2011). [DOI] [Google Scholar]
- 57.Chen H, Bell JM, Zavala NA, Ji HP & Zhang NR Allele-specific copy number profiling by next-generation dna sequencing. Nucleic acids research 43, e23–e23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen R. & Seshan VE FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput dna sequencing. Nucleic acids research 44, e131–e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y. et al. Quantification of multiple tumor clones using gene array and sequencing data. The annals of applied statistics 11, 967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi Y, Chan AP, Kirkness E, Telenti A. & Schork NJ Comparison of phasing strategies for whole human genomes. PLOS Genetics 14, e1007308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Do CB & Batzoglou S. What is the expectation maximization algorithm? Nature biotechnology 26, 897 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Thorndike RL Who belongs in the family? Psychometrika 18, 267–276 (1953). [Google Scholar]
- 63.Li H. A statistical framework for snp calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das S. et al. Next-generation genotype imputation service and methods. Nature genetics 48, 1284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarthy S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics 48, 1279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koboldt DC et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H. et al. The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data from 10x Genomics Chromium Single Cell CNV Solution for patient S0 are available at https://support.10xgenomics.com/single-cell-dna/datasets. Raw read counts and phased SNP counts for patient S0 are available at https://doi.org/10.5281/zenodo.3817605 and for patient S1 at https://doi.org/10.5281/zenodo.3817536. The DOP-PCR sequencing data of 89 breast tumor cells are available from the NCBI Sequence Read Archive under accession SRA: SRP114962. All the processed data for all datasets of patients S0 and S1 and for the DOP-PCR data, as well as all the results of CHISEL, are available on GitHub at https://github.com/raphael-group/chisel-data.