Abstract
In addition to the canonical right-handed double helix, other DNA structures, termed ‘non-B DNA’, can form in the genomes across the tree of life. Non-B DNA regulates multiple cellular processes, including replication and transcription, yet its presence is associated with elevated mutagenicity and genome instability. These discordant cellular roles fuel the enormous potential of non-B DNA to drive genomic and phenotypic evolution. Here we discuss recent studies establishing non-B DNA structures as novel functional elements subject to natural selection, affecting evolution of transposable elements, and specifying centromeres. By highlighting the contributions of non-B DNA to repeated evolution and adaptation to changing environments, we conclude that evolutionary analyses should include a perspective of not only DNA sequence, but also its structure.
Keywords: non-canonical DNA structure, G-quadruplexes, Z-DNA, mutations, natural selection
The ubiquity, in vivo formation, and functions of non-B DNA
In addition to the canonical B form described by Watson and Crick, DNA with certain sequence motifs can assume alternative conformations, i.e. ‘non-B DNA’. Non-B DNA includes G-quadruplexes (G4s) formed by guanine-rich sequences, Z-DNA formed by alternating purine and pyrimidine sequences, bent DNA formed by A-phased (adenine-rich) repeats, slipped-strand structures formed by direct repeats, H- or triplex DNA formed by mirror repeats with homopurine:homopyrimidine sequences, and DNA hairpins and cruciforms formed by inverted repeats (Box 1). Recent studies have indicated that motifs capable of forming non-B DNA (‘non-B DNA motifs’) are ubiquitous across the tree of life. They are present in viruses [1], bacteria [2,3], single-cell eukaryotes including human pathogens [2–4], fungi [2,5], plants [5], and animals [2,5]. Approximately 13% of the human genome has the potential to fold into non-B DNA [6].
BOX 1. TYPES OF NON-B DNA.
A variety of different non-B DNA structure types have been described. Six commonly known types are shown in the figure below. In G4 quadruplex structures, four tracts of at least three guanines (‘stems’) interleaved by a variable number of other nucleotides (‘loops’) are the prerequisite for forming a planar structure, in which the guanines belonging to different G-tracts bind to each other via Hoogsteen hydrogen bonds [130]. The average length of G4-forming motifs is 35 bp. Cruciform structures may form via intra-strand pairing of repeat copies in inverted repeats (21 bp average length), while mirror repeats (58 bp average length) with polypurine/polypyrimidine runs may lead to the formation of triple helices, and direct repeats promote hairpins and cruciforms. The left-handed zig-zag helix structure (Z-DNA) is formed at regions containing stretches of purine-pyrimidine repeats such as (CG:CG)n or (CA:TG)n, which are typically around 12 bp long. Finally, A-phased repeats, which are characterized by several iterations of tracts of four to nine adenines with an average length of 26 bp, with centers separated by 11–12 and interleaved by other nucleotides, may form bent DNA structures.
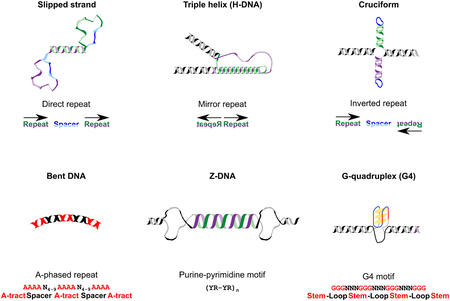
Forms of alternative DNA structures.
For each type of non-B DNA, the respective name, the molecular structure, the underlying sequence name, and the arrangement of the nucleotide sequence are shown.
Non-B DNA structures form transiently, depending on conditions within the cell. For instance, the formation of G4s and H-DNA is stabilized by elevated potassium and magnesium concentrations, respectively [7,8], and the curvature of bent DNA formed by A-tracts in bacteria depends on temperature [9]. Originally, the formation of non-B DNA was shown in vitro, but within the last decade, it has also been unequivocally demonstrated in vivo (Table 1). Presently, there is experimental proof that G4s form in the native chromatin environment in several cancerous and noncancerous human cell lines [10,11], and their formation has been visualized with fluorescence imaging in DNA in human osteosarcoma cells [12] and in RNA in human osteosarcoma and breast cancer cells, as well as in mouse melanoma cells [13]. In vivo formation has also been shown for Z-DNA (e.g., in human HeLa cells [14]), slipped structures, cruciforms, and triplex DNA (reviewed in [15]). According to Permanganate/S1 nuclease footprinting performed in human cancerous Raji cells, 26.4%, 20.0%, and 5.5% of all computationally predicted G4, Z-DNA, and H-DNA motifs form non-B structures respectively, and additional 17.0%, 20.0%, and 5.5% of the respective predicted motifs might also form at sites occupied by RNA polymerase II [16]. Similarly, in activated mouse B cells, 7.0%, 8.9%, and 5.3% of all predicted G4, Z-DNA, and H-DNA motifs form non-B structures respectively, with additional 5.3%, 6.8%, and 1.1% of respective predicted motifs potentially forming at sites occupied by RNA polymerase II [16]. To what extent the formation of non-B DNA structures varies across cell types remains to be explored.
TABLE 1.
Methods for experimental validation of non-B DNA structures.
| Method | Type / Principle | Target non-B structure | Environment | Reference |
|---|---|---|---|---|
| Permanganate/S1 Nuclease Footprinting | Enrichment for single-stranded DNA, chemical stabilization of non-B DNA structures, high-throughput sequencing | All non-B types | in vivo | [16] |
| S1-seq | Enrichment for single-stranded DNA, high-throughput sequencing | predominantly H-DNA, but also other non-B types | in vitro | [120] |
| KAS-seq | N3-kethoxal-assisted labeling of single-stranded DNA, high-throughput sequencing | All non-B DNA types | in vivo | [121] |
| 2D3 monocolonal antibody | bandshift assay and immunoprecipitation | Cruciforms | in vivo | [122–124] |
| G4-seq | Detection based on mismatches between sequence reads, high-throughput sequencing | G4s | in vitro | [125] |
| G4-ChIP seq | ChIP-seq based on G4 antibody | G4s | in vitro | [126] |
| G4P-seq | Small protein probe coupled with ChIP-seq | G4s | in vitro and in vivo | [127] |
| G4 CUT&TAG | Cleavage under targets and tagmentation, high-throughput sequencing | G4s | in vitro | [128] |
| SiR-PyPDS | G4-specific fluorescent probe, microscopy | G4s | in vivo | [12] |
| N-TASQ | G4-specific ligand (NaphthoTASQ), microscopy | G4s | in vivo | [13] |
| PuPy FISH | Polypurine/polypyrimidine-specific probe, fluorescent in situ hybridization | H-DNA | in vitro | e.g., [129] |
| Zaa-ChIP-seq | ChIP-seq based on Z-DNA antibody | Z-DNA | in vivo | [14] |
Non-B DNA is implicated in myriad cellular processes (Figure 1; reviewed in [17]). Lately, the direct evidence supporting genome-wide and locus-specific functions of non-B DNA in vivo has been rapidly accumulating. Several types of non-B DNA are involved in replication initiation. For instance, G4s facilitate firing of origins of replication in the mouse and human genomes [18–20], and a hairpin forming at the light-strand origin (OriL) is required for the replication of the vertebrate mitochondrial DNA [21]. Additionally, non-B DNA has been implicated in telomere end protection [22][23,24] (see also Figure 1), recombination [25–28], and DNA repair [29,30]. A-tract curvature was demonstrated to be a thermosensor of virulence of the human bacterial pathogen Shigella [9]. Furthermore, almost all types of non-B DNA have been shown to be involved in regulation of transcription either by providing specific conformation for transcription-factor binding sites or via other mechanisms [3,4,8,14,31–37]. In fact, Z-DNA is associated with actively transcribed regions of human HeLa cells [14], and G4s were recently called transcription-factor-binding hubs in human chromatin in myelogenous leukemia and hepatocellular carcinoma cells [32]. Relatedly, it has become evident that non-B DNA—particularly G4s, but also some other types—is involved in regulation of local and higher-order chromatin organization in the human genome [11,38–41]. G4s may act as protective cis elements against the methylation of CpG islands, as was shown in human embryonic stem cells [42]. In these cells, G4s may also act as epigenetic markers that determine the transition from the pluripotent to the specialized state [43]. Moreover, at the level of RNA, structured RNA resulting from transcription of non-B DNA motifs can affect translation [3,44,45] and play an active role in the function of non-coding RNA [46]. Interestingly, Z-RNA can regulate interferon I response [47] in humans and mice, or can trigger apoptosis, when detected by mammalian cells infected with some viruses (reviewed in [48]).
FIGURE 1. Non-B DNA structures as a blessing and a curse.
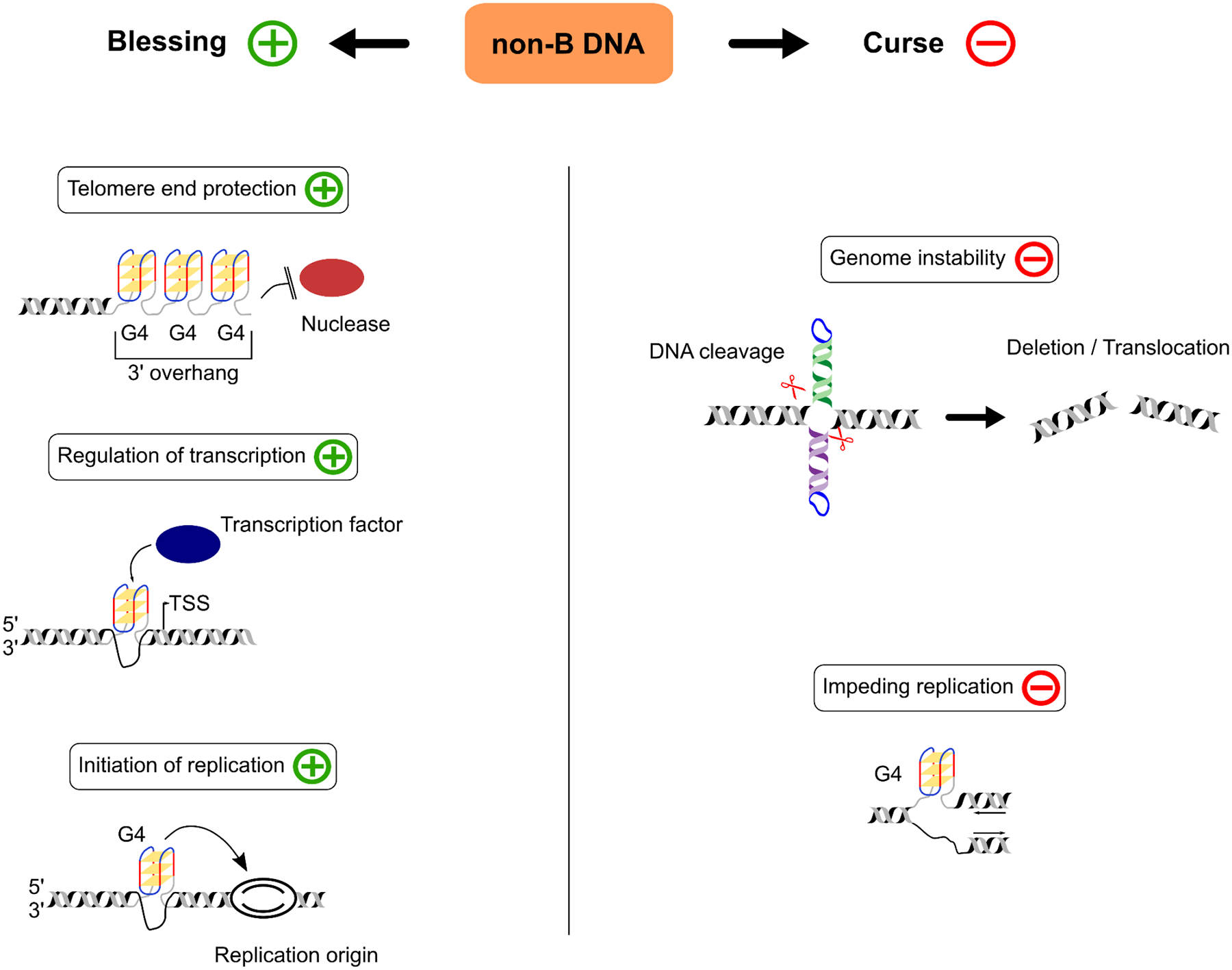
Due to their many functions and molecular effects, non-B DNA structures can be seen as both ‘a blessing’ and ‘a curse’. In this figure, we present schematic examples of vital cellular functions (‘blessing’) as well as of detrimental effects (‘curse’). The former include telomere end protection, where G4 structures may prevent the telomeric 3’ overhang from being degraded by nucleases; the regulation of transcription, in which folded G4s act as transcription factor binding sites in the promoter; and the initiation of replication, where G4s located upstream of replication origins facilitate the firing of the replication machinery. Examples of manifested non-B DNA structures having detrimental effects are cruciform-structure-mediated genome instability leading to deletions and chromosome translocations; and the potential impeding of replication, in which a folded G4 structure on the leading strand stalls the progression of the replication fork. After [18,44,116,118,119].
Despite the growing support for the ubiquity, in vivo formation, and function of non-B DNA (and the corresponding highly structured RNA), its evolution remains understudied. In this opinion piece, we highlight what is currently known about the evolution of non-B DNA evolution and how these structures, in turn, affect the evolution of genomes and phenotypes. We focus on G4s because these structures and their motifs have been most well-studied to date.
Non-B DNA motifs affect mutation rate and facilitate genome instability
Non-B DNA structures can pose obstacles for replicative polymerase progression during replication, increasing pausing and errors [49]. Additionally, replication through such structures is accomplished with the participation of specialized polymerases (e.g., polymerases eta and kappa), which are error-prone [50–53]. Consistent with these mechanisms, non-B DNA motifs at the human CFS-FRA16 common fragile site had increased genetic variation at polymerase pause locations, and a mutation spectrum consistent with the involvement of polymerase eta [50]. Furthermore, non-B DNA structures can be recognized as damaged DNA, which triggers error-generating repair pathways leading to double-strand breaks (DSBs) and genomic instability [15,54,55]. As a result, non-B DNA motifs are sites of elevated mutagenesis and thus are well positioned to be major, yet unrecognized, drivers of genome evolution.
Non-B DNA motifs are emerging as hotspots of single-nucleotide substitutions and small insertions and deletions (‘indels’). An elevated frequency of single-nucleotide polymorphisms (SNPs) and small indels at non-B DNA motifs was observed in the data obtained from 1,000 Genomes Project [56]. This observation remained true even for intergenic, and thus presumably neutrally-evolving regions, arguing for high mutagenicity of non-B DNA. Another recent study also demonstrated elevated rates of nucleotide substitutions (both for SNPs and for fixed differences) at different types of non-B DNA motifs located in the non-coding non-repetitive portion of the human genome [57]. Supporting these findings, the frequency of cancer somatic genetic variants was shown to be elevated at non-B DNA motifs [58]. Albeit potentially affected by selection, an analysis of disease-causing genic mutations confirmed the high mutagenic potential of non-B DNA motifs [59]. A mutation hotspot was recently found at the OriL of macaques, potentially due to the stem-loop structure formation in this region [60].
The pattern of small-scale mutations is non-random along non-B motifs. For direct, inverted, and mirror repeats, the mutation frequency, as proxied by SNP frequency and frequency of fixed differences, is lower in stems than in spacers [57,58,61], potentially reflecting gene conversion acting in stems. Mutation frequencies are elevated towards the edges of Z-DNA motif annotations, likely because of Z-DNA/B-DNA boundaries [57]. Within G4 motifs, the mutation frequency is higher in loops than in stems (guanine stretches), and this is particularly evident for motifs capable of forming thermodynamically stable G4 structures [57]. Similarly, elevation in mutation frequency was observed for G4 motifs (particularly at their loops) capable of forming stable structures in mitochondrial DNA [62]. G4 structure stability is more strongly affected by substitutions in stems (particularly the ones affecting the central guanine in a G-track) than in loops [63]. The overall loop length is inversely proportional to G4 structure stability [64], and G4s with 1-bp loops are particularly stable and have the highest potential to induce genome instability [65].
In addition to their role in small-scale mutation rate variation, non-B DNA motifs are emerging as the preferential sites of large-scale indels and rearrangements, thus contributing to genome instability. For instance, Copy Number Variant (CNV) breakpoints in both Drosophila melanogaster and human are enriched in non-B DNA motifs [66]. Z-DNA in yeast and human cells harbors high frequency of large deletions [54]. G4 motifs in a nematode deficient for the dog-1 helicase were found to be at a site of recurring 50–300 bp deletions [67]. Cruciform-forming inverted repeats have increased chromosomal instability in budding yeast [68]. In cancer genomes, there are elevated frequencies of DSBs at H-DNA motifs [69] and an overrepresentation of non-B DNA motifs in regions of somatic copy number alterations and chromosomal breakage [70][71].
Importantly, caution should be exercised when inferring mutations at non-B DNA motifs from sequencing data [6]. Several major sequencing technologies (e.g., Illumina and Pacific Biosciences) are polymerase-based and thus, if non-B DNA structures form during the sequencing process, their motifs may have elevated sequencing error rates [72,73]. Whereas non-B motifs frequently exhibit increased sequencing error rates, mutations in them can be studied by using a combination of several independent sequencing technologies, increased read depth, and stringent quality filters [72].
Non-B DNA motifs as novel functional genomic elements that evolve under selection
Since non-B DNA contributes to regulating multiple essential cellular processes, it should evolve under purifying selection. Consistent with this prediction, studies of polymorphisms and/or fixed nucleotide differences point towards purifying selection acting on G4 motifs in different parts of the human genome. Single-nucleotide variants in G4s located in promoters can dramatically affect the activity of the host gene, as was shown with luciferase assays in human embryonic kidney cells [33]. A lower SNP frequency at G4-structure-disruptive positions on the template vs. the non-template strand suggests that G4s regulate gene expression at the mRNA level [74]. G4 motifs located in UTRs appear to evolve under purifying selection and are enriched for eQTLs, RNA protein-binding sites, and human pathogenic variants [75]. Moreover, when located in enhancers, replication origins, TAD boundaries, regions upstream of genes, and transcribed strands of exons, G4s evolve under purifying selection, are overrepresented, and thus are likely functional [76]. In the above-mentioned genomic regions, purifying selection is stronger for stable than unstable G4s, whereas in some other regions, including 5’UTRs (Table 1), only stable G4s evolve under purifying selection [76]. Because of purifying selection that acts on them and correlates with their stability, G4s were called ‘novel functional genomic elements’ [76].
To achieve the biochemical specificity needed for their varied cellular functions, G4s likely have sequence constraints related to their required topological structures. Indeed, it was determined that G4s with different functions have distinct but overlapping sequence requirements [77]. In another in vitro study, a G4 motif was found to have different sequence requirements for GTP-binding vs. peroxidase activities [78]. Only a limited number of G4 sequences and biological functions have been so far analyzed to determine biochemical specificity.
Can selection be acting against G4s because of their potentially negative effects on replication and genome stability? Such selection might be operating against the G4s with pyrimidine-containing 1-nt loops, which are absent in yeast and significantly under-represented in the human genome [65]. Moreover, sequences with highly thermodynamically stable G4s are usually present in the genomes outside of repeats, thus preventing an overload of the genome with these elements [79]. Interestingly, because they might interfere with translation, the most stable G4s are selected against (via synonymous codon usage) in protein-coding regions of mRNAs in multiple species [64]. In agreement with this study, the G4 motifs on the nontranscribed strand of human exons are underrepresented, encode predominantly unstable structures, and do not exhibit signatures of purifying selection [76].
Different types of non-B DNA could be utilized by various taxa to fulfill important functions, and thus variability in selective pressure might contribute to the observed differences in the G4 motif repertoire among taxa [2]. It was suggested that the density (BOX 2) and diversity of G4 motifs increases with organismal complexity [80]. The enrichment of G4s in functional regions of the genome also differs among species. For instance, a strong enrichment of G4s in promoters and 5’ UTRs is evident for human, mouse, and Trypanosoma, but not for nematode, zebrafish, and fruit fly [2]. In Archaea, G4s are overrepresented in non-coding RNA [81]. In mammals, G4 motifs with single-adenine loops are overrepresented and thus might have been recruited and selected for their functionality [5]. Other G4s appear to be playing such roles in the non-mammalian genomes [5]. The differences in G4 motif occurrences among viruses may result from selective pressure dictated by the host. In particular, stable G4s are enriched in eukaryotic, but depleted in prokaryotic, viruses because eukaryotic cells can process such G4s with the participation of helicases and other enzymes [82].
BOX 2. VARIATION IN THE DENSITY OF G4S ACROSS THE TREE OF LIFE.
The variability in genomic proportion of non-B DNA-forming sequences is unknown across the tree of life, and the analyses addressing the genomic abundance and distribution of known non-B DNA motifs across species have been lacking. Thus far, studies have focussed on either a specific type of non-B DNA or a certain group of organisms. For example, Marsico et al. [2] investigated the occurrence of G4 motifs sampling representative species across the tree of life and experimentally identified G4s forming in vitro under physiological conditions. In the figure below, their findings on the relationship between G4 motif density nucleotide content in eukaryotic species is summarized. It is notable that there is a strong variation in the G4 motif density among these species even independent of GC content, and this requires further investigation.
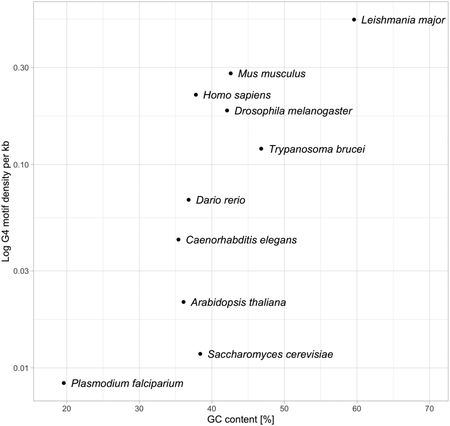
Relationship between GC content and G4 motif density.
On the x-axis, the genomic proportion of guanines plus cytosines is shown, and on the y-axis, the log-scaled density of G4-forming motifs per 1,000 bp (1kb) is shown. Individual dots correspond to the species names.
In general, the taxonomic distribution and selection for and against non-B DNA types other than G4s has been under-investigated. Since the evidence for a variety of non-B DNA structures to be of functional importance and under selective pressure (e.g.,[76]) is increasing, a more thorough investigation of these questions is warranted. Such future analyses are expected to uncover additional roles played by non-B DNA in the cell and other ways by which it affects genome evolution. These potential discoveries should then reinforce the view/notion that non-B DNA ‘epitomizes a non-traditional way of encoding genetic information’ [83].
Non-B DNA and TEs
Another contribution of non-B DNA to genome evolution concerns its multiple relationships with transposable elements (TEs). First, non-B DNA motifs, G4 motifs in particular, are abundant in TEs and may play a role in the TE life cycle. G4 and triplex motifs were found in LTR transposons of 21 plant species [84]. Notably, within these TEs, G4 motifs were overrepresented in the long terminal repeats, which contain promoters [85]. Moreover, long runs of guanines, which form stable G4s, were present mostly in young LTR transposons suggesting the participation of G4s in the life cycle of these TEs [84].
In the human genome, as many as 71% of G4 motifs are located within TEs, with particular abundance in SVAs, L1s, and HERVs [86]. Additionally, Alu elements carry a Z-DNA motif, a non-canonical G4 motif, and a mirror repeat [83]. G4 motifs are more abundant and their predicted structures more stable in evolutionarily younger, as compared with older, SVAs and L1s [86,87]. Thus, G4s may be important for the SVA and L1 TE life cycles (Table 2). Consistent with this prediction, a deletion or an alteration of a G4 motif in human L1 3’UTR decreased L1 transpositional activity in cultured human HeLa cells [87]. The participation of G4s in the life cycle of human SVAs and plant LTR elements is yet to be tested experimentally. Moreover, whether non-B DNA is overrepresented in, and plays a role in the life cycle of, TEs in species outside of primates, plants, and yeast remains to be investigated.
TABLE 2.
| TE class | SVA | L1 | HERV | Alu |
|---|---|---|---|---|
| Abundance(% of elements carrying canonical G4s) | 36.2% | 7.7% | 4.8% | 1.1% |
| Part of an element with particular G4 abundance | VNTR | 3’UTR | LTRs | Left part of left monomer contains a non-canonical G4 |
| Higher abundance in younger elements | Yes | Yes | N/A | No |
| Role in the life cycle | Hypothesize, not tested | Demonstrated | Not tested | Not tested |
Second, non-B DNA is a likely factor affecting integration of TEs in the genome (Table 3). Noncanonical DNA conformations can represent the genomic targets of new TE insertions as they are frequently nucleosome-free and can be recognized by a transposase or an integrase [88–90]. Several types of non-B DNA motifs (e.g. G4 motifs and mirror repeats) were shown to be enriched at integration sites of L1 transposons in the human genome [91]. Additionally, non-B DNA motifs (e.g. mirror repeats and Z-DNA) exhibit an association with the density of human and mouse endogenous retroviruses [92] and of human and bat DNA transposons [93]. Moreover, an enrichment of microsatellites, capable of forming unusual DNA conformations, was found in the vicinity of TEs on the evolutionary young chromosomes of the dioecious plant sorrel (Rumex acetosa) [94]; LTR transposon insertions in 12 plant species showed a weak preference for palindromes [95].
TABLE 3.
Non-B DNA and TE integration preferences with the sign of non-B DNA predictors in regression models and the genomic window size (when relevant) used for the analysis (after [91–93]).
| TE/Non-B DNA type | L1s | DNA transposons* | Human and mouse ERVs** |
|---|---|---|---|
| A-phased repeats | Positive for fixation (±50 kb) | Negative for Charlie distributions (1 Mb and 20 kb) | |
| Direct Repeats | Functional negative predictor for fixation | Positive for Tigger distributions (20 kb) | |
| Positive for Helitron distributions (1 Mb) | |||
| G4s | Positive for integration (±2 kb), negative for fixation (±50 kb) | Negative for Tigger distributions (20 kb) | Functional negative predictor of fixed HERV-K distributions |
| Negative for PiggyBat integrations (20 kb) | |||
| Negative for SB integrations (20 Kb) | |||
| Positive for Helitron distributions (1Mb) | |||
| Inverted Repeats | Negative for Charlie distributions (1 Mb) | ||
| Positive for SB integrations (1 Mb) | |||
| Positive for hAT distributions (20 kb) | |||
| Positive for TcMar distributions (20 kb) | |||
| Positive for Helitron distributions (20 kb) | |||
| Mirror Repeats | Positive for integration (−2 kb - insertion), positive for fixation (insertion - 1 kb) | Negative for Charlie distributions (20 kb) | Functional positive predictor of fixed ETn distributions |
| Negative for Tigger distributions (20 kb) | |||
| Negative for PiggyBat integrations (1 Mb) | |||
| Positive for SB integrations (20 kb) | |||
| Negative for hAT distributions (1 Mb) | |||
| Positive for Helitron distributions (1 Mb) | |||
| Z-DNA | Positive for hAT distributions (1 Mb and 20 kb) | Negative scalar predictor for fixed ETn vs. controls distributions | |
| Negative for Helitron distributions (1 Mb) |
only predictors with RCVE >1% are shown
only predictors with RCDE>1% in multiple functional regression models are shown
SB - Sleeping Beauty
Third, TEs were proposed to serve as vehicles of spreading non-B DNA across the genome [83,88]. Non-B DNA located in TEs may play a role in (a) regulating activity of neighboring genes by affecting the activity of promoters (e.g. plant G4s embedded in TEs are frequently located in the vicinity of promoters [84]), (b) regulating epigenetic state of discrete genomic regions and facilitating genome silencing, including TE silencing, via spreading heterochromatin [85], (c) re-shuffling of genomic DNA via recombination at TEs [84], potentially leading to formation of chimeric TEs [85]. Additionally, G4 motifs within hominid-specific SVA retrotransposons are enriched in cancer genome breakpoints [96].
Non-B DNA motifs located in TEs are also linked to human diseases. For instance, G4s within L1 elements accumulate in Alzheimer’s disease neurons [97]. Additionally, it was shown that Z-RNA located in double-stranded RNA (dsRNA) derived from the adjacent inversely oriented mammalian SINEs (human Alu and mouse B1 and B2 elements) is essential for preventing an autoinflammatory response characterized by chronic type I interferon (IFN-I) production [47]. Upon infection, exogenous (e.g. viral) dsRNA is recognized by a nucleic acid sensor MDA5, which activates the IFN-I response. Without infection, transcribed SINE inverted repeats, which are underrepresented in the genome, undergo adenosine-to-inosine editing by the ADAR1 deaminase [98], which specifically recognizes Z-RNA formed by dsRNA [47]. Such editing masks these molecules from the detection by MDA5, and thus prevents the activation of the IFN-I response and autoinflammation. Mutations in the ADAR1 gene lead to a severe autoinflammatory disease [47].
Together this suggests a strongly intertwined relationship between TEs and non-B DNA. In some cases, non-B DNA stimulates transposition of TEs by being their integral part, in other cases non-B DNA might contribute to specifying TE integration preferences in the genome, and also some non-B motifs in TEs are associated with human diseases. Future studies should determine whether in other instances the abundance of certain non-B DNA motifs in TEs is due to their functional impact, or merely due to hitchhiking a particularly prolific TE group.
Non-B DNA and satellites
With advances in sequencing technologies and assembly algorithms, the prevalence and role of non-B DNA in satellite sequences is being rapidly evaluated. It has long been known that non-B DNA (G4s) forms at telomeres [22,99]. Recently, the role of satellite non-B DNA in specifying centromere identity has been emerging (summarized in [100]). It has been suggested that, depending on the taxon and on the chromosome, the centromere identity is defined either by the recruitment of sequence-specific DNA-binding proteins (e.g. CENP-B binding to CENP-B boxes) or by the recognition of non-B DNA [101]. Consistent with this prediction, the loss of CENP-B boxes (e.g. on the Old World monkeys’ chromosomes as well as on the human Y chromosome) correlates with an increased tendency of centromeric satellites to contain short (<10-bp arm length) inverted repeats (also called ‘dyad symmetries’), which can form cruciforms [101]. In the human genome, most of the centromeres are enriched in CENP-B boxes and not in inverted repeats, however they still form non-B DNA in vivo [101]. In fact, as an alternative to the presence of inverted repeats, CENP-B binding itself was proposed to induce non-B DNA formation [102].
Centromere specification via non-B DNA might represent an ancient mechanism for eukaryotes (Table 4)[101]. Indeed, S. cerevisiae centromeres have high levels of predicted non-B DNA formation [101], and D. melanogaster centromeres are enriched in non-B DNA motifs [103]. Also, the dioecious plant Silene latifolia has accumulated TRAYC satellite sequences with palindromic pattern and capable of forming non-canonical structures, most prominently near the centromere of its Y chromosome [104]. Non-B DNA might define centromeres directly—e.g. via providing cruciform structures recognized by the Holliday Junction Recognition Protein and thus facilitating nucleosome loading with the centromere-specific histone H3 variant—or indirectly—e.g. by initiating transcription (or being part of transcripts) that drives centromere recognition [101,105].
TABLE 4.
| Species/Non-B DNA type | Dyad symmetries forming cruciforms | G4s | Non-B DNA formation by Permanganate-Seq |
|---|---|---|---|
| Human | Y chromosome centromere, neocentromeres | Centromeric alpha-satellite | |
| Great apes | Centromeric alpha-satellite | ||
| Old World monkeys | Centromeric alpha-satellite | ||
| Horse | Centromeres | ||
| Mouse | Centromeric satellite | ||
| Chicken | Neocentromeres and centromeres | ||
| Drosophila | Y chromosome centromere | Centromeres on chromosomes 2, 4, and X | |
| Plants | Centromeres | ||
| Fission yeast | Centromeres |
Telomeres and centromeres apart, the recent telomere-to-telomere assembly of the human genome has unearthed extensive copy number variation of a DNA satellite WaluSat that contains a G4 motif at the junction of individual repeat units [106]. G4s may facilitate frequent non-allelic or ectopic recombination of these satellite arrays, leading to the high variability in their copy number [106]. With multiple telomere-to-telomere assemblies quickly accumulating [107], there is no doubt that the role of non-B DNA in satellite function and evolution will be further uncovered.
Non-B DNA as a driver of phenotypic evolution and disease
Mutations causing phenotypic change provide the substrate for evolution and lead to adaptations to new environments and death. Typically, most mutations are rare, and demographic constraints (e.g., small effective population size) can prevent successful adaptations. Non-B DNA has the potential to challenge this dogma.
The increased mutagenicity of non-B DNA and the genomic instability it promotes have the potential to induce genetic diversity at an unprecedented level [5], and, subsequently, to affect phenotypes and their evolution. Several examples include microsatellites forming slipped strand structures, leading to variation in microsatellite repeat number, which in turn influences phenotypes—e.g., social and sexual behavior in bank voles dependent on microsatellite repeat number in the regulatory regions of the vasopressin 1a receptor and oxytocin receptor genes [108], vocal learning in a transgenic zebrafinch dependent on repeat number in the huntingtin gene (albeit in a transgenic bird [109]), and more globally, variation in gene expression levels dependent on microsatellite repeat number in upstream regions [110]. However, more recently, such instances have included other types of non-B DNA as well.
In a recent study [111], Xie and colleagues demonstrated that elevated mutagenicity at non-B DNA contributes to repeated morphological evolution via adaptation to freshwater environments in stickleback fish. Specifically, the (TG)n repeats capable of forming Z-DNA drastically—by several orders of magnitude—increase the mutability of an enhancer regulating the Pitx1 gene, which encodes a homeodomain transcription factor. Such high mutability facilitates recurrent deletions within the enhancer sequence leading to hindfin loss, which is advantageous for freshwater stickleback populations. Notably, the enhancer sequences, which form non-B DNA in vitro, have an elevated frequency of DSBs and deletions in vivo. Thus, non-B DNA can contribute to repeated evolution, a phenomenon for which mechanistic explanations have been lacking.
Another study [112] suggested (albeit not unequivocally demonstrated) that non-B DNA contributes to explaining the genetic basis of a stable trans-species polymorphism in color morphs of Midas cichlids, as related to their adaptive radiation in Nicaraguan crater lakes. Most Midas cichlids are of the melanistic dark morphs, whereas 1–20% of them transition into gold morphs during development. In the gold morphs, an insertion in an intron of the goldentouch gene is associated with its lower expression and affects the expression of several other genes. The insertion contains two copies of a PiggyBac-like DNA transposon positioned in the inverted orientation and thus capable of forming a cruciform, which might impede gene expression by halting transcription or decreasing the unspliced RNA stability. Therefore, non-B DNA might explain some instances of stable trans-species polymorphisms, whose genetic underpinnings, just as for repeated evolution, have remained elusive [112]. One can envision that non-B DNA may not only cause genetic polymorphisms (as in the Midas cichlid example), but also facilitate their maintenance because of its high mutagenicity, i.e. by constantly providing alternative alleles.
G4s forming in RNA in Arabidopsis thaliana provide an example of non-canonical RNA conformation directly enhancing the adaptive potential of an organism. Yang and colleagues [113] demonstrated that G4s form more readily in cold temperatures (4°C compared to 22°C), leading to increased mRNA stability and decreased root growth. In addition, they found that, globally, G4s are enriched in plant species associated with colder climates, suggesting that G4 frequency in transcriptomes is an indicator of adaptation to such an environment. This study provides conclusive evidence that non-canonical RNA structure formation can serve as a mechanism to respond to environmental changes.
The flipside of the increase in mutation rate and genomic instability, and of the functionality, of non-B DNA structures is their association with diseases. Non-canonical DNA and RNA structures have been linked to neurodegenerative diseases by controlling repeat expansion, gene expression, gene methylation, local translation, and toxic peptide accumulation (reviewed in [114]). An example of non-B DNA directly linked to neurological diseases is the hexanucleotide repeat expansion in C9orf72 in humans, which leads to the formation of a stable G4 quadruplex, causing amyotrophic lateral sclerosis and frontotemporal dementia [115]. Another example is the contribution of a G4 structure to the Fragile X syndrome, caused by the expanded CGG repeat (reviewed in [116]). There are several ways in which non-canonical DNA and RNA structures are implicated in cancer. First, non-B DNA motifs are preferential sites of cancer genomic rearrangements [117]. Second, non-canonical DNA and RNA structures affect the expression of cancer-related genes, and these alterations may influence cancer progression (reviewed in [114]). From a different perspective, there are several human genetic diseases resulting from mutations in genes encoding proteins the cell uses to handle non-B DNA (e.g., helicases)—such as Werner syndrome caused by the mutated WRN helicase (reviewed in [116]).
Together, these examples illustrate that non-B DNA structures indeed can be simultaneously seen as ‘a blessing’ and ‘a curse’ (Figure 1), as their formation can enhance an organism’s ability to adapt and therefore increases its fitness, while in other cases the direct or indirect effects of non-B DNA structures lead to the emergence of diseases or promote the occurrence of deleterious mutations. The effects of mutations in non-B DNA motifs are also highly dependent on the genomic location, as some non-B DNA structures (e.g., the ones located in promoters, enhancers, and origins of replication) are important for function and evolve under purifying selection [76].
Concluding remarks
While studying the relationship between nucleotide sequence and observed phenotype of an organism is non-trivial, adding non-B DNA as another layer of possible interactions is expected to substantially increase the complexity and potential for uncovering the molecular mechanisms of evolutionary adaptations. Similar to other regulatory elements of the genome, the formation of non-B DNA structures has the potential to influence cellular processes depending on whether they are formed and/or are stable. Their formation in turn is dependent on environmental parameters, such as ion concentrations and temperature, enabling an organism to respond to changes in the environment without the need to change the genome, thus considerably increasing its adaptive potential. However, this very feature of transient formation also makes non-B DNA challenging to study, e.g., sophisticated experiments are needed to prove non-B formation in the cell at a given time and its influence on phenotypic traits. Nevertheless, the examples above illustrate the importance of investigating genomes beyond the linear DNA sequence and highlight the enormous potential of non-B DNA to impact the evolutionary trajectory of an organism, population, and species. We expect that many more analogous examples will be discovered in the near future and that non-B DNA will become an important component of the studies of not only genome evolution but also of organismal evolution and phenotypic adaptations. We have summarized the most prominent knowledge gaps in this area in the ‘Outstanding questions’ section.
Outstanding questions.
Are particular types of non-B DNA more common in some but not other taxa? Are different types of non-B DNA, or different non-B DNA motifs of the same type, recruited by various taxa to fulfill important functions? Are there taxa with stronger purifying selection at non-B loci?
Do types of selection other than purifying operate on G4s (e.g., selection against G4 structure formation in exons)? Does selection operate on non-B DNA other than G4s?
What are the sequence and structure requirements for different G4 functions? How is functional specificity achieved by other types of non-B DNA motifs?
Do G4s play a role in the life cycle of primate SVA and plant LTR transposable elements?
Does non-B DNA specify centromeres directly (i.e. via its unusual DNA structure) or via transcription? What other biological roles does non-B DNA play, when it is located in satellite sequences?
How often does non-B DNA affect phenotypic evolution?
FIGURE 2. Evidence suggesting the functionality of G4 motifs in the human genome.
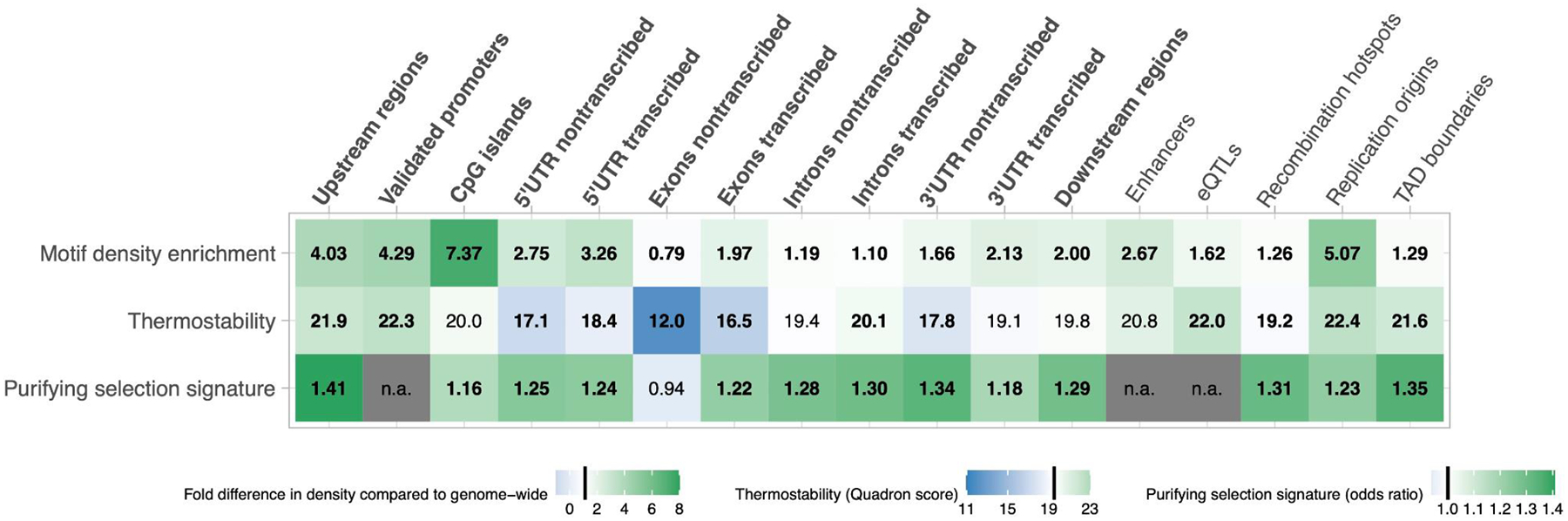
On the x-axis, different genomic regions are shown, with genic regions in bold. The first row depicts the fold-difference between the G-corrected G4 motif density for a particular genomic region as compared to the average genome-wide G4 motif density; a significant increase in representation above 1 indicates overrepresentation and thus potentially functionality. In the second row, median thermostability (as computed by Quadron) is shown; the genome-wide average thermostability is 19.5; a value significantly higher than 19.5 indicates elevated thermostability and thus potentially functionality. The third row depicts the odds ratios of the Hudson-Kreitman-Aquade test used to evaluate purifying selection; odds ratio equal to one is inconsistent with selection; odds ratio significantly higher than one is suggestive of purifying selection. * bold font within tiles denotes a significance level of <0.05 [76]; normal font - not significant; n.a. - not analyzed. Vertical black bars indicate a thermostability value of 19.5, a fold-change of G4 motif density of 1, and an odds ratio of 1, respectively. After [76].
Highlights.
Non-B DNA promotes genomic instability and large-scale rearrangements.
In some genomic regions (e.g. promoters and enhancers), G4 motifs evolve under purifying selection because of their functionality, yet in some other genomic regions (e.g. coding exons) they might be selected against because of their detrimental effects.
Non-B DNA is overrepresented in TEs, participates in the life cycle of some of them, and can affect their integration.
Many eukaryotic centromeres are specified by non-B DNA.
Elevated mutagenicity of non-B DNA may explain some instances of repeated evolution and trans-species polymorphism in fishes, and is likely to contribute to phenotypic evolution in other species.
Acknowledgments
We are grateful to Jean-Christophe Andrau, Kristin Eckert, Wilfried Guiblet, and Eduard Kejnovsky for their critical reading of the manuscript. This work was supported by the NIH grant R01GM136684.
Glossary
- Copy number variation (CNV)
a stretch of sequence in the genome that is repeated a different number of times among individuals
- CpG island
a genomic region that is enriched for cytosine-guanine dinucleotides, which is commonly found associated with mammalian gene promoters
- eQTL
expression quantitative trait locus. A genomic region that explains variation in gene expression levels
- G-quadruplex (G4)
a 3D structure of the DNA molecule with four strands, formed due to Hoogsteen hydrogen bonds within the same or between different DNA molecules
- Repeated (recurrent) evolution
a phenomenon when the same trait, character, or mutation, emerges multiple times across distinct populations of the same species
- SNP
Single Nucleotide Polymorphism, a genetic variant with (typically) two different nucleotides in an organism or a population
- Slipped strand structure
direct repeats interrupted by a spacer leading to pairing between repeat arms and looping out of the spacer
- Trans-species polymorphism
the occurrence of the same allele(s) in different species
- Z-DNA
a left-winding zig-zag double helix structure of the DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Métifiot M et al. (2014) G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 42, 12352–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsico G et al. (2019) Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saranathan N and Vivekanandan P (2019) G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 27, 148–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazanion E et al. (2020) Genome wide distribution of G-quadruplexes and their impact on gene expression in malaria parasites. PLoS Genet. 16, e1008917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puig Lombardi E et al. (2019) Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 47, 6098–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guiblet WM et al. (2018) Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 28, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hänsel-Hertsch R et al. (2017) DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol 18, 279–284 [DOI] [PubMed] [Google Scholar]
- 8.Del Mundo IMA et al. (2017) Alternative DNA structure formation in the mutagenic human c-MYC promoter. Nucleic Acids Res. 45, 4929–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prosseda G et al. (2004) The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol 51, 523–537 [DOI] [PubMed] [Google Scholar]
- 10.Biffi G et al. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem 5, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hänsel-Hertsch R et al. (2016) G-quadruplex structures mark human regulatory chromatin. Nat. Genet 48, 1267–1272 [DOI] [PubMed] [Google Scholar]
- 12.Di Antonio M et al. (2020) Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem 12, 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laguerre A et al. (2015) Visualization of RNA-Quadruplexes in Live Cells. J. Am. Chem. Soc 137, 8521–8525 [DOI] [PubMed] [Google Scholar]
- 14.Shin S-I et al. (2016) Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome. DNA Res. 23, 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poggi L and Richard G-F Alternative DNA Structures In Vivo : Molecular Evidence and Remaining Questions. Microbiology and Molecular Biology Reviews, 85. (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzine F et al. (2017) Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst 4, 344–356.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaushik M et al. (2016) A bouquet of DNA structures: Emerging diversity. Biochem Biophys Rep 5, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prorok P et al. (2019) Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun 10, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai H et al. (2019) Rif1 promotes association of G-quadruplex (G4) by its specific G4 binding and oligomerization activities. Sci. Rep 9, 8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akerman I et al. (2020) A predictable conserved DNA base composition signature defines human core DNA replication origins. Nat. Commun 11, 4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanrooij S et al. (2012) In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep. 13, 1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moye AL et al. (2015) Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun 6, 7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildiz A (2022) Dynamic folding and accessibility of telomeric overhang. Proc. Natl. Acad. Sci. U. S. A 119, e2211219119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiekh S et al. (2022) Emerging accessibility patterns in long telomeric overhangs. Proc. Natl. Acad. Sci. U. S. A 119, e2202317119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boán F and Gómez-Márquez J (2010) In vitro recombination mediated by G-quadruplexes. Chembiochem 11, 331–334 [DOI] [PubMed] [Google Scholar]
- 26.Mani P et al. (2009) Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One 4, e4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kshirsagar R et al. (2017) Probing the Potential Role of Non-B DNA Structures at Yeast Meiosis-Specific DNA Double-Strand Breaks. Biophys. J 112, 2056–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phung HTT et al. (2020) The cruciform DNA-binding protein Crp1 stimulates the endonuclease activity of Mus81-Mms4 in Saccharomyces cerevisiae. FEBS Lett. 594, 4320–4337 [DOI] [PubMed] [Google Scholar]
- 29.Sharma M et al. (2013) DNA bending propensity in the presence of base mismatches: implications for DNA repair. J. Phys. Chem. B 117, 6194–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brázda V et al. (2016) Strong preference of BRCA1 protein to topologically constrained non-B DNA structures. BMC Mol. Biol 17, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haran TE and Mohanty U The unique structure of A-tracts and intrinsic DNA bending. Quarterly Reviews of Biophysics, 42. (2009), 41–81 [DOI] [PubMed] [Google Scholar]
- 32.Spiegel J et al. (2021) G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 22, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong J-Y et al. (2021) G-quadruplex structural variations in human genome associated with single-nucleotide variations and their impact on gene activity. Proc. Natl. Acad. Sci. U. S. A 118, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biswas B et al. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Research, 45. (2017), 11268–11280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulovari A et al. (2019) Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc. Natl. Acad. Sci. U. S. A 116, 23243–23253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts JW Mechanisms of Bacterial Transcription Termination. Journal of Molecular Biology, 431. (2019), 4030–4039 [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto Y et al. Cruciform Formable Sequences within Pou5f1 Enhancer Are Indispensable for Mouse ES Cell Integrity. International Journal of Molecular Sciences, 22. (2021), 3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lago S et al. (2021) Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun 12, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura O et al. A strong structural correlation between short inverted repeat sequences and the polyadenylation signal in yeast and nucleosome exclusion by these inverted repeats. Current Genetics, 65. (2019), 575–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Y et al. (2019) Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics 14, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson J et al. (2021) DNA G-quadruplex structures: more than simple roadblocks to transcription? Nucleic Acids Res. 49, 8419–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jara-Espejo M and Line SR (2020) DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 287, 483–495 [DOI] [PubMed] [Google Scholar]
- 43.Zyner KG et al. (2022) G-quadruplex DNA structures in human stem cells and differentiation. Nat. Commun 13, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bochman ML et al. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet 13, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bugaut A and Balasubramanian S (2012) 5’-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 40, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyu K et al. (2021) RNA G-quadruplexes (rG4s): genomics and biological functions. Nucleic Acids Res. DOI: 10.1093/nar/gkab187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Reuver R et al. (2021) ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep. 36, 109500. [DOI] [PubMed] [Google Scholar]
- 48.Balachandran S and Mocarski ES (2021) Viral Z-RNA triggers ZBP1-dependent cell death. Curr. Opin. Virol 51, 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaushal S and Freudenreich CH (2019) The role of fork stalling and DNA structures in causing chromosome fragility. Genes Chromosomes Cancer 58, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twayana S et al. (2021) Translesion polymerase eta both facilitates DNA replication and promotes increased human genetic variation at common fragile sites. Proc. Natl. Acad. Sci. U. S. A 118, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bournique E et al. Role of specialized DNA polymerases in the limitation of replicative stress and DNA damage transmission. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 808. (2018), 62–73 [DOI] [PubMed] [Google Scholar]
- 52.Tsao W-C and Eckert KA (2018) Detours to Replication: Functions of Specialized DNA Polymerases during Oncogene-induced Replication Stress. Int. J. Mol. Sci 19, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyer A-S et al. (2013) The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. J. Mol. Biol 425, 4767–4781 [DOI] [PubMed] [Google Scholar]
- 54.McKinney JA et al. (2020) Distinct mechanisms of mutagenic processing of alternative DNA structures by repair proteins. Mol Cell Oncol 7, 1743807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G and Vasquez KM (2014) Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 19, 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du X et al. Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation. Nucleic Acids Research, 42. (2014), 12367–12379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guiblet WM et al. (2021) Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome. Nucleic Acids Res. 49, 1497–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgakopoulos-Soares I et al. (2018) Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 28, 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamat MA et al. (2016) A role for non-B DNA forming sequences in mediating microlesions causing human inherited disease. Hum. Mutat 37, 65–73 [DOI] [PubMed] [Google Scholar]
- 60.Arbeithuber B et al. (2022) Advanced age increases frequencies of de novo mitochondrial mutations in macaque oocytes and somatic tissues. Proc. Natl. Acad. Sci. U. S. A 119, e2118740119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou X et al. (2017) Short inverted repeats contribute to localized mutability in human somatic cells. Nucleic Acids Res. 45, 11213–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler TJ et al. (2020) Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro. Hum. Mol. Genet 29, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JY and Kim DS (2009) Dramatic effect of single-base mutation on the conformational dynamics of human telomeric G-quadruplex. Nucleic Acids Res. 37, 3625–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mirihana Arachchilage G et al. (2019) Stable G-quadruplex enabling sequences are selected against by the context-dependent codon bias. Gene 696, 149–161 [DOI] [PubMed] [Google Scholar]
- 65.Piazza A et al. (2015) Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 34, 1718–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cardoso-Moreira M et al. (2012) Mutation spectrum of Drosophila CNVs revealed by breakpoint sequencing. Genome Biol. 13, R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemmens B et al. (2015) Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun 6, 8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ait Saada A et al. (2021) Genetic and Molecular Approaches to Study Chromosomal Breakage at Secondary Structure-Forming Repeats. Methods Mol. Biol 2153, 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao J et al. (2018) Distinct Mechanisms of Nuclease-Directed DNA-Structure-Induced Genetic Instability in Cancer Genomes. Cell Rep. 22, 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bacolla A et al. (2016) Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 44, 5673–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smida J et al. (2017) Genome-wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. Int. J. Cancer 141, 816–828 [DOI] [PubMed] [Google Scholar]
- 72.Weissensteiner MH et al. 16-Jun-(2022), Altered sequencing success at non-B-DNA motifs. bioRxiv, 2022.06.13.495922 [Google Scholar]
- 73.McGinty RJ and Sunyaev SR Mutagenesis at non-B DNA motifs in the human genome: a course correction.. [DOI] [PMC free article] [PubMed]
- 74.Nakken S et al. (2009) The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 37, 5749–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee DSM et al. (2020) Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun 11, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guiblet WM et al. (2021) Selection and thermostability suggest G-quadruplexes are novel functional elements of the human genome. Genome Res. DOI: 10.1101/gr.269589.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volek M et al. (2021) Overlapping but distinct: a new model for G-quadruplex biochemical specificity. Nucleic Acids Res. 49, 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Švehlová K et al. (2016) Altered biochemical specificity of G-quadruplexes with mutated tetrads. Nucleic Acids Res. 44, 10789–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huppert JL and Balasubramanian S (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu F et al. (2021) Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol 4, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brázda V et al. (2020) G-Quadruplexes in the Archaea Domain. Biomolecules 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z et al. (2021) G-quadruplexes in genomes of viruses infecting eukaryotes or prokaryotes are under different selection pressures from hosts. J. Genet. Genomics DOI: 10.1016/j.jgg.2021.08.018 [DOI] [PubMed] [Google Scholar]
- 83.Herbert A (2020) ALU non-B-DNA conformations, flipons, binary codes and evolution. R Soc Open Sci 7, 200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lexa M et al. (2014) Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 42, 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kejnovsky E and Lexa M (2014) Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob. Genet. Elements 4, e28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lexa M et al. (2014) Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics 15, 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sahakyan AB et al. (2017) G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat. Struct. Mol. Biol 24, 243–247 [DOI] [PubMed] [Google Scholar]
- 88.Kejnovsky E et al. (2015) Transposable elements and G-quadruplexes. Chromosome Res. 23, 615–623 [DOI] [PubMed] [Google Scholar]
- 89.Geurts AM et al. (2006) Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 34, 2803–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cree SL et al. (2020) G-quadruplex structures bind to EZ-Tn5 transposase. Biochimie 177, 190–197 [DOI] [PubMed] [Google Scholar]
- 91.Chen D et al. (2020) Human L1 Transposition Dynamics Unraveled with Functional Data Analysis. Mol. Biol. Evol 37, 3576–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Campos-Sánchez R et al. (2016) Integration and Fixation Preferences of Human and Mouse Endogenous Retroviruses Uncovered with Functional Data Analysis. PLoS Comput. Biol 12, e1004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Campos-Sánchez R et al. (2014) Genomic landscape of human, bat, and ex vivo DNA transposon integrations. Mol. Biol. Evol 31, 1816–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kejnovský E et al. Expansion of Microsatellites on Evolutionary Young Y Chromosome. PLoS ONE, 8. (2013), e45519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jedlicka P et al. (2019) Nested plant LTR retrotransposons target specific regions of other elements, while all LTR retrotransposons often target palindromes and nucleosome-occupied regions: in silico study. Mob. DNA 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bacolla A et al. (2019) Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog. Biophys. Mol. Biol 147, 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanna R et al. (2021) G-quadruplexes originating from evolutionary conserved L1 elements interfere with neuronal gene expression in Alzheimer’s disease. Nat. Commun 12, 1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bazak L et al. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paudel BP et al. (2020) A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. Elife 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Talbert PB and Henikoff S (2022) The genetics and epigenetics of satellite centromeres. Genome Res. 32, 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kasinathan S and Henikoff S (2018) Non-B-Form DNA Is Enriched at Centromeres. Mol. Biol. Evol 35, 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Talbert PB and Henikoff S (2020) What makes a centromere? Exp. Cell Res 389, 111895. [DOI] [PubMed] [Google Scholar]
- 103.Patchigolla VSP and Mellone BG (2022) Enrichment of Non-B-Form DNA at D. melanogaster Centromeres. Genome Biol. Evol 14, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hobza R et al. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma, 115. (2006), 376–382 [DOI] [PubMed] [Google Scholar]
- 105.Talbert PB and Henikoff S (2018) Transcribing Centromeres: Noncoding RNAs and Kinetochore Assembly. Trends Genet. 34, 587–599 [DOI] [PubMed] [Google Scholar]
- 106.Hoyt SJ et al. 12-Jul-(2021), From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. bioRxiv, 2021.07.12.451456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rhie A et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Watts PC et al. (2017) Stabilizing selection on microsatellite allele length at arginine vasopressin 1a receptor and oxytocin receptor loci. Proc. Biol. Sci 284, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu W-C et al. (2015) Human mutant huntingtin disrupts vocal learning in transgenic songbirds. Nat. Neurosci 18, 1617–1622 [DOI] [PubMed] [Google Scholar]
- 110.Gymrek M et al. (2016) Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet 48, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xie KT et al. (2019) DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363, 81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kratochwil CF et al. (2022) An intronic transposon insertion associates with a trans-species color polymorphism in Midas cichlid fishes. Nat. Commun 13, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang X et al. 04-Mar-(2022), RNA G-quadruplex structure contributes to cold adaptation in plants. bioRxiv, 2022.03.04.482910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tateishi-Karimata H and Sugimoto N (2021) Roles of non-canonical structures of nucleic acids in cancer and neurodegenerative diseases. Nucleic Acids Res. 49, 7839–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haeusler AR et al. (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maizels N (2015) G4-associated human diseases. EMBO Rep. 16, 910–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheloshkina K and Poptsova M (2021) Comprehensive analysis of cancer breakpoints reveals signatures of genetic and epigenetic contribution to cancer genome rearrangements. PLoS Comput. Biol 17, e1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu S et al. (2015) Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes. Cell Rep. 10, 1674–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Varshney D et al. (2020) The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol 21, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maekawa K et al. (2022) Triple-helix potential of the mouse genome. Proc. Natl. Acad. Sci. U. S. A 119, e2203967119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lyu R et al. (2022) KAS-seq: genome-wide sequencing of single-stranded DNA by N3-kethoxal-assisted labeling. Nat. Protoc 17, 402–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sinden RR et al. (1983) Perfect palindromic lac operator DNA sequence exists as a stable cruciform structure in supercoiled DNA in vitro but not in vivo. Proc. Natl. Acad. Sci. U. S. A 80, 1797–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frappier L et al. (1987) Monoclonal antibodies to cruciform DNA structures. J. Mol. Biol 193, 751–758 [DOI] [PubMed] [Google Scholar]
- 124.Steinmetzer K et al. (1995) Anti-cruciform monoclonal antibody and cruciform DNA interaction. J. Mol. Biol 254, 29–37 [DOI] [PubMed] [Google Scholar]
- 125.Chambers VS et al. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol 33, 877–881 [DOI] [PubMed] [Google Scholar]
- 126.Hänsel-Hertsch R et al. (2018) Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc 13, 551–564 [DOI] [PubMed] [Google Scholar]
- 127.Zheng K-W et al. (2020) Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 48, 11706–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lyu J et al. (2022) Genome-wide mapping of G-quadruplex structures with CUT&Tag. Nucleic Acids Res. 50, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ohno M et al. (2002) Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma 111, 201–213 [DOI] [PubMed] [Google Scholar]
- 130.Davis JT G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angewandte Chemie International Edition, 43. (2004), 668–698 [DOI] [PubMed] [Google Scholar]


